ST93P 10.1186/1471 2180 14 31
User Manual: ST93P
Open the PDF directly: View PDF ![]() .
.
Page Count: 13
- Abstract
- Background
- Results and discussion
- Exotoxin expression in ST93 CA-MRSA strains
- PVL
- PSMα3
- Hla
- Comparative virulence of ST93 isolates with differential exotoxin expression
- Impact of exotoxin expression on virulence of ST93 CA-MRSA in the murine skin infection model
- Hla
- α-type PSMs
- PVL
- Genome sequencing of three additional ST93 isolates
- Comparative genomics of ST93 and the importance of agr in the virulence of ST93 CA-MRSA
- The AraC/XylS regulator (AryK) enhanced Hla expression and virulence in ST93 CA-MRSA
- RNAseq demonstrates global regulatory impact of AryK
- Conclusions
- Methods
- Ethics statement
- Bacterial strains and culture
- Detection of LukF-PV and Hla by western blotting
- Detection of PSMα3 expression
- DNA methods, molecular techniques and construction of mutants
- Mouse skin infection assay
- Genome sequencing
- Comparative genomics
- Quantitative RT-PCR for RNAIII expression
- RNA sequencing
- Statistical analysis
- Availability of supporting data
- Additional files
- Competing interest
- Authors’ contributions
- Authors’ information
- Acknowledgements
- Author details
- References
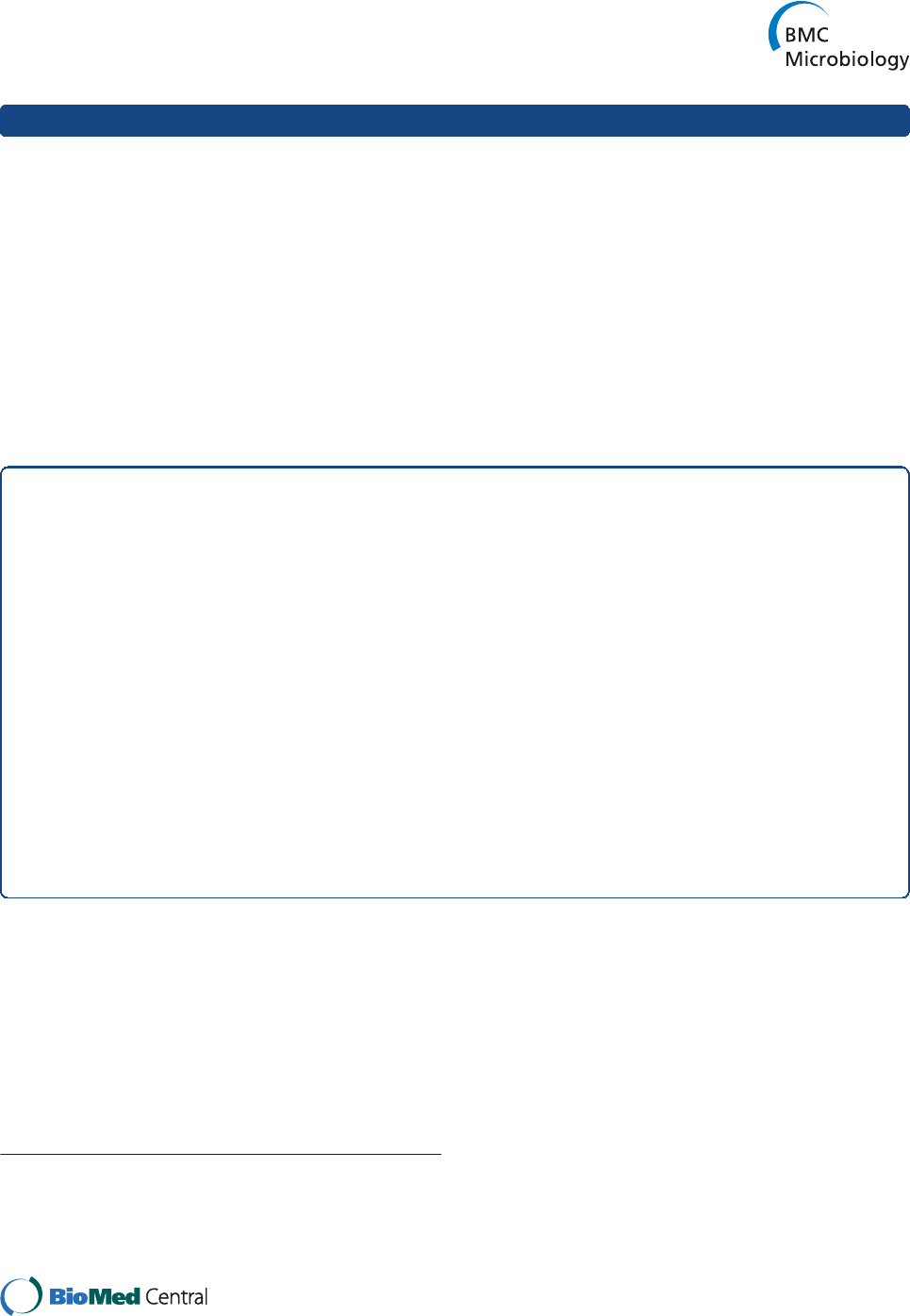
R E S E A R C H A R T I C L E Open Access
Hyperexpression of α-hemolysin explains
enhanced virulence of sequence type 93
community-associated methicillin-resistant
Staphylococcus aureus
Kyra YL Chua
1,2,3,4
, Ian R Monk
1
, Ya-Hsun Lin
1
, Torsten Seemann
5
, Kellie L Tuck
6
, Jessica L Porter
1
, Justin Stepnell
1
,
Geoffrey W Coombs
7,8
, John K Davies
2
, Timothy P Stinear
1,2
and Benjamin P Howden
1,2,3,4*
Abstract
Background: The community-associated methicillin-resistant S. aureus (CA-MRSA) ST93 clone is becoming dominant
in Australia and is clinically highly virulent. In addition, sepsis and skin infection models demonstrate that ST93
CA-MRSA is the most virulent global clone of S. aureus tested to date. While the determinants of virulence have
been studied in other clones of CA-MRSA, the basis for hypervirulence in ST93 CA-MRSA has not been defined.
Results: Here, using a geographically and temporally dispersed collection of ST93 isolates we demonstrate that
the ST93 population hyperexpresses key CA-MRSA exotoxins, in particular α-hemolysin,incomparisontoother
global clones. Gene deletion and complementation studies, and virulence comparisons in a murine skin infection
model, showed unequivocally that increased expression of α-hemolysin is the key staphylococcal virulence
determinant for this clone. Genome sequencing and comparative genomics of strains with divergent exotoxin
profiles demonstrated that, like other S. aureus clones, the quorum sensing agr system is the master regulator of
toxin expression and virulence in ST93 CA-MRSA. However, we also identified a previously uncharacterized
AraC/XylS family regulator (AryK) that potentiates toxin expression and virulence in S. aureus.
Conclusions: These data demonstrate that hyperexpression of α-hemolysin mediates enhanced virulence in
ST93 CA-MRSA, and additional control of exotoxin production, in particular α-hemolysin, mediated by regulatory
systems other than agr have the potential to fine-tune virulence in CA-MRSA.
Keywords: Staphylococcus aureus, CA-MRSA, Pathogenesis, Alpha-hemolysin
Background
Community-associated methicillin-resistant Staphylococcus
aureus (CA-MRSA) is an emerging global problem with
very similar clinical presentations across different clones,
despite significant genetic diversity[1].ManyCA-MRSA
strains carry lukSF-PV in the accessory genome, which en-
codes the Panton-Valentine leukocidin (PVL), an exotoxin
that causes neutrophil lysis [1]. Although there has been
considerable controversy as to the role of this toxin in CA-
MRSA pathogenesis, some of this may be explained by a
variable, species dependent susceptibility to PVL –human
and rabbit neutrophils are lysed by PVL at very low con-
centrations whilst mouse and monkey neutrophils are less
susceptible, making the interpretation of animal model data
difficult in some cases [2]. Additionally, the importance of
PVLisalsolikelytobedependentonthesiteofinfection.
In the rabbit pneumonia model, PVL has been demon-
strated to have a clear role in mediating severe lung necro-
sis and inflammation [3]. In contrast, in skin infection, even
in the rabbit model, its role remains less clear [4,5].
Notwithstanding PVL, the increased expression of other
core genome virulence determinants also contributes sig-
nificantly to the increased virulence of CA-MRSA strains
* Correspondence: benjamin.howden@austin.org.au
1
Department of Microbiology and Immunology, University of Melbourne,
Melbourne, Victoria 3052, Australia
2
Department of Microbiology, Monash University, Clayton, Victoria 3800,
Australia
Full list of author information is available at the end of the article
© 2014 Chua et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative
Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and
reproduction in any medium, provided the original work is properly credited. The Creative Commons Public Domain
Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article,
unless otherwise stated.
Chua et al. BMC Microbiology 2014, 14:31
http://www.biomedcentral.com/1471-2180/14/31
[6,7]. These include α-hemolysin (Hla) and α-type phenol
soluble modulins (PSMs). Hla is a pore-forming exotoxin
that lyses many cells including red cells, platelets, mono-
cytes and endothelial cells [8]. Hla has been demonstrated
to be an important mediator of virulence in skin infection
and pneumonia [9,10]. The α-type PSMs have been re-
cently characterized and they lyse neutrophils and red
cells [11,12]. The α-type PSMs also mediate virulence in
skin infection and septicemia and of these, PSMα3isthe
most potent [11].
The study of unique, distantly related CA-MRSA clones
that also demonstrate enhanced virulence, may provide
insights into the emergence of the global CA-MRSA
phenomenon, and also help define the genomic determi-
nants of enhanced virulence. In Australia, the singleton
ST93 CA-MRSA clone ST93 (“Queensland clone”) has be-
come dominant in the community [13], and we and others
have demonstrated that our reference ST93 strain JKD6159
was highly virulent and caused severe skin infection in a
mouse model compared to other CA-MRSA strains includ-
ing USA300 [14,15].
In this study we used exotoxin analysis, functional geno-
mics and a murine infection model to investigate the rela-
tive contribution of α-hemolysin, α-type phenol soluble
modulins and Panton-Valentine leukocidin to the enhanced
virulence of ST93 CA-MRSA. We show that increased
virulence in the BALB/c mouse skin infection model is less
dependent on α-type phenol soluble modulin or Panton-
Valentine leukocidin production but is instead due to high-
level expression of α-hemolysin in this clone, controlled
predominantly by the agr system.
Results and discussion
The emergence of CA-MRSA is a major public health
issue, and there is a clear need to understand the basis for
both virulence and transmission of global clones of CA-
MRSA. The genetically distinct CA-MRSA clone ST93-IV
[2B] has rapidly become the dominant clone in Australia
and its rise accounts for the increase in incidence of CA-
MRSA as a whole in this country [13]. We, and others
have previously shown that ST93 strain JKD6159 is the
most virulent global clone of S. aureus in murine models
[14,15]. To determine the mediators of virulence in this
clone we initially studied exotoxin expression in a large
collection of ST93 S. aureus from around Australia, and
compared representative high and low expressing strains
to an international selection of clones.
Exotoxin expression in ST93 CA-MRSA strains
Staphylococcus aureus expresses a wide range of exotoxins
that may contribute to virulence. Because Hla, PVL and α-
type PSMs have been found by others to be important viru-
lence factors in CA-MRSA strains [9,11,16], we measured
in vitro expression of these exotoxins by the wildtype ST93
strains and non-ST93 comparator strains. The main
isolates used in this study are described in Table 1,
while the collection of ST93 isolates from around
Australia used for comparative exotoxin expression is
from a study by Coombs et al. [17] and summarized in
Additional file 1. The comparison of expression of
international clones to the ST93 reference strain
JKD6159 and three additional ST93 strains selected for
genome sequencing (see below) are shown in Figure 1,
while the results for all 59 ST93 isolates compared to
USA300 are shown in Additional file 2 (α-type PSMs)
and Additional file 3 (Hla). The results of PVL analysis
for the ST93 collection has been previously reported
[17]. Because PVL is a 2-component exotoxin and both
LukS-PV and LukF-PV are required for activity, we
chose to measure LukF-PV expression by quantitative
Western blot. LukF-PV was chosen over LukS-PV to
obtain anti-LukF-PV antibody with increased specifi-
city of binding as there was more sequence divergence
between lukF-PV and the orthologous 2-component S.
aureus exotoxins compared to lukS-PV. Although there
are four α-type PSMs, PSMα3 causes the most sig-
nificant neutrophil lysis [11] and we measured de-
formylated and N-formylated PSMα3expressionby
high performance liquid chromatography (HPLC). The
proportion of deformylated and formylated forms of
PSMs depends on the growth conditions, the activity
of staphylococcal peptide deformylase and strain back-
ground [7,18].
PVL
As previously reported [17], PVL expression was consist-
ent across most ST93 strains. We found that there was
no significant difference in the LukF-PV expression in
the PVL positive strains JKD6159, TPS3104, USA300
and JKD6177. Although USA300 appeared to produce
less LukF-PV than JKD6159, the difference was not sta-
tistically significant (p = 0.0943, Figure 1A).
PSMα3
We found that the deformylated form of PSMα3 was al-
most always more abundant than the N-formylated form
(Figure 1B and Additional file 2). The ST30 CA-MRSA
strain JKD6177 did not produce any PSMα3. There was
no significant difference in PSMα3 expression between
JKD6159 compared to USA300, however JKD6159 pro-
duced more PSMα3 compared to JKD6272 (p = 0.0003)
and JKD6009 (p = 0.0003). Compared to the other ST93
MRSA strains, JKD6159 produced more PSMα3 com-
pared to TPS3105 (p < 0.0001), and TPS3106 (p = 0.01)
but less than TPS3104 (p = 0.0029) (Figure 1B). Expres-
sion levels across the whole ST93 collection were vari-
able, although many isolates produced levels at least
equivalent to USA300 (Additional file 2).
Chua et al. BMC Microbiology 2014, 14:31 Page 2 of 13
http://www.biomedcentral.com/1471-2180/14/31

Table 1 Bacterial strains used in this study
S. aureus strain Molecular type Date of isolation Place of isolation Site of isolation Relevant characteristics lukSF-PV reference
Clinical isolates
JKD6159 ST93-IV [2B] 2004 Victoria, Australia Blood Dominant Australian CA-MRSA clone + [14]
TPS3104 ST93-IV [2B] 2009 Western Australia, Australia Nasal cavity Dominant Australian CA-MRSA clone + This study
TPS3105 ST93-IV [2B] 2005 New South Wales, Australia Blood Australian CA-MRSA clone - This study
TPS3106 ST93-V [5C2&5] 2008 Western Australia, Australia Nasal cavity Australian CA-MRSA clone - This study
JKD6272 ST1-IV [2B] 2002 Victoria, Australia Blood Australian CA-MRSA clone - [14]
JKD6260 ST1-IV [2B] 2008 Western Australia, Australia Skin Australian CA-MRSA clone + [14]
JKD6177 ST30-IV [2B] 2003 Melbourne, Australia Blood Australian CA-MRSA clone + [14]
FPR3757 USA300 ST8-IV [2B] NA San Francisco, USA Wrist abscess Dominant North American CA-MRSA clone + [19]
JKD6009 ST239-III [3A] 2002 New Zealand Wound Dominant Australian hospital-associated MRSA clone, AUS2/3 - [20]
Mutant strains
JKD6159ΔlukSF-PV ST93-IV [2B] Isogenic unmarked lukSF-PV KO of JKD6159 - This study
JKD6159Δhla ST93-IV [2B] Isogenic unmarked hla KO of JKD6159. Deletion encompassed
genome coordinates 1121291–1120441.
+ This study
JKD6159Δhla r ST93-IV [2B] Isogenic unmarked hla KO repaired in JKD6159Δhla. Introduction
of a novel PstI site within hla.
+ This study
JKD6159ΔpsmαST93-IV [2B] Isogenic unmarked psm-αKO in JKD6159. Deletion encompassed
genome coordinates 453364–45378.
+ This study
JKD6159ΔpsmαrST93-IV [2B] Isogenic unmarked psm-αKO repaired in JKD6159Δpsm-α. Introduction
of a novel SalI site within psm-α
+ This study
JKD6159Δ00043 ST93-IV [2B] Isogenic unmarked SAA6159_00043 KO of JKD6159. Deletion encompassed
genome coordinates 53156 - 54561
+ This study
JKD6159_AraCrST93-IV [2B] Isogenic AraC/XylS regulator repaired in JKD6159 + This study
TPS3105rST93-IV [2B] Isogenic agrA repair of TPS3105 - This study
KO: knockout, NA: not available.
Chua et al. BMC Microbiology 2014, 14:31 Page 3 of 13
http://www.biomedcentral.com/1471-2180/14/31
Hla
Hla expression appeared high for the majority of ST93
isolates, with the exception of four strains where expres-
sion was low (Additional file 3). JKD6159 produced
greater levels of Hla than all the wildtype strains, inclu-
ding USA300 (p < 0.0001 for all strains except JKD6177,
p = 0.0107, Figure 1C). There was no difference in Hla
expression between JKD6159 and TPS3104.
Here we have demonstrated that the majority of ST93
strains consistently produce higher levels of Hla com-
pared to other clones, including USA300, while produc-
tion of PVL and α-type PSM is similar, suggesting that
enhanced expression of Hla may be responsible for in-
creased virulence of ST93 CA-MRSA.
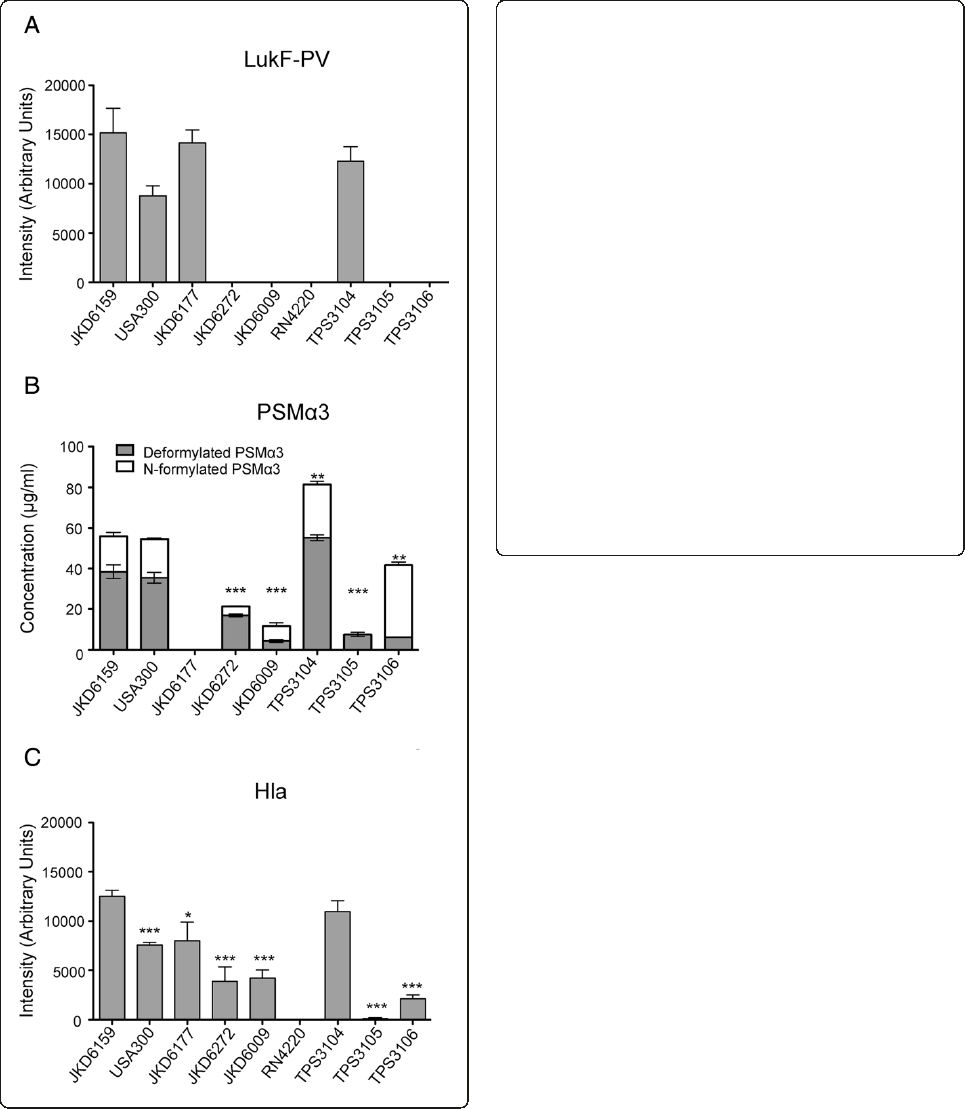
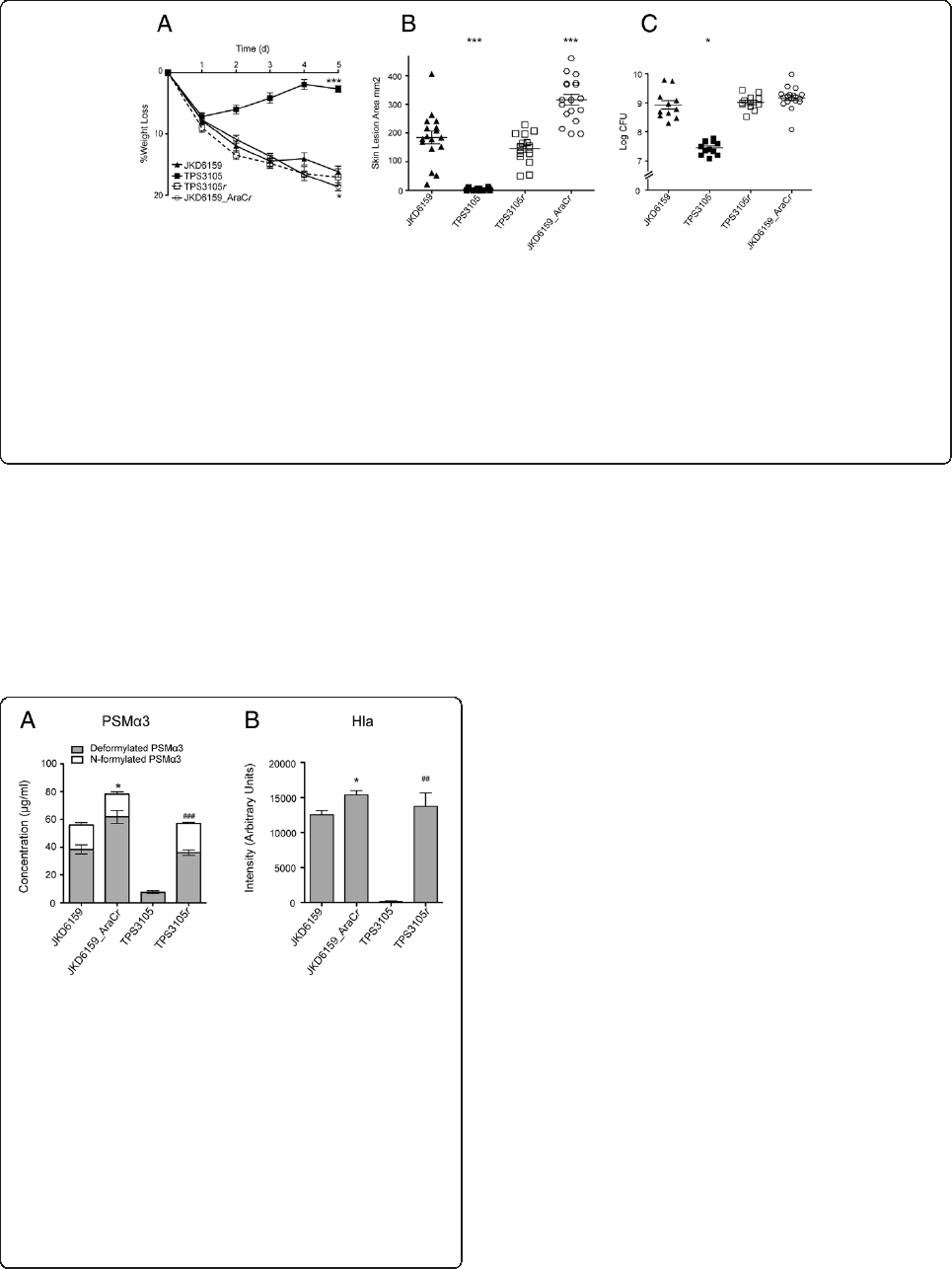
Comparative virulence of ST93 isolates with differential
exotoxin expression
Tofurtherexaminetheroleoftheseselectedexotoxinsin
our mouse skin infection assay, we compared the virulence
of four ST93 isolates selected based on their exotoxin
expression profiles (high exotoxin expression, JKD6159
and TPS3104; low exotoxin expression, TPS3105 and
TPS3106). TPS3104 was as virulent as JKD6159 in the
mouse model in all outcome measures (Figure 2). In
contrast, the strains with reduced exotoxin expression
TPS3105 and TPS3106 were significantly less virulent
compared to JKD6159, with less weight loss at day 5 of in-
fection (p < 0.0001), smaller lesion size (p < 0.0001) and
Figure 1 In vitro exotoxin expression of wildtype S. aureus
isolates. JKD6159 (ST93-IV [2B]) compared with non-ST93 CA-MRSA
strains FPR 3757 USA300 (ST8-IV [2B]), JKD6177 (ST30-IV [2B]), and
JKD6272 (ST1-IV [2B]); Hospital-associated MRSA strain JKD6009
(ST239-III [3A]), wildtype ST93 strains TPS3104 (ST93-IV [2B]), TPS3105
(ST93-IV [2B]), and TPS3106 (ST93-V [5C2&5]). (A) LukF-PV expression
measured by quantitative Western blot. RN4220 was included as a
negative control because it does not contain lukF-PV. All PVL negative
strains did not express LukF-PV.There was no significant difference in
the amount of LukF-PV expressed by the S. aureus strains containing
lukSF-PV. Data shown are mean intensity of bands in arbitrary units
and SEM. (B) PSMα3 expression measured by HPLC. JKD6177 did not
produce PSMα3. JKD6272 (p = 0.0003), JKD6009 (p = 0.0003), TPS3105
(p < 0.0001) and TPS3106 (p = 0.0100) produced less deformylated and
N-formylated PSMα3 compared to JKD6159. There was no difference
between PSMα3 production by JKD6159 and USA300. TPS3104
expressed more PSMα3 than JKD6159 (p = 0.0029). Data shown are
mean concentration (μg/ml), presented as vertical stacked bars and
SEM. Deformylated PSMα3 is shown in grey bars. N-formylated PSMα3
is shown in white bars. (C) Hla expression measured by quantitative
Western blot. RN4220 was included as a negative control because it
does not express Hla. JKD6159 expressed more Hla compared to all
non-ST93 wildtype strains (p < 0.0001 for all strains except JKD6177
p = 0.0107). TPS3105 and TPS3106 produced significantly less Hla
(p < 0.0001). There was no difference in Hla production between
JKD6159 and TPS3104. Data shown are mean intensity of bands in
arbitrary units and SEM. Note, ***p < 0.001, **p < 0.01, *p < 0.05.
Chua et al. BMC Microbiology 2014, 14:31 Page 4 of 13
http://www.biomedcentral.com/1471-2180/14/31
less CFU recovery from lesions (TPS3105, p = 0.0177;
TPS3106, p = 0.0328) in the model (Figure 2).
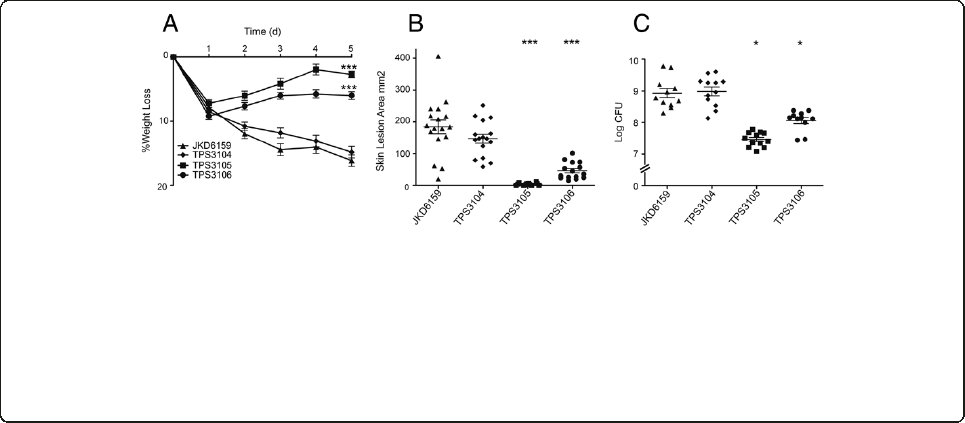
Impact of exotoxin expression on virulence of ST93
CA-MRSA in the murine skin infection model
To further characterize the contribution of each of the
exotoxins to disease in the murine model, genetic deletion
and complementation experiments were performed for
each of the selected toxins.
Hla
Given the increased in vitro expression of Hla by JKD6159
and TPS3104 and the apparent correlation of this increased
expression with increased virulence in the mouse skin in-
fection model, we generated JKD6159Δhla and assessed
this mutant in the mouse skin infection assay (Figure 3).
There was a marked attenuation in virulence in all out-
come measures with significantly decreased weight loss
(p < 0.0001), lesion size (p < 0.0001) and CFU recovery
(p = 0.0177). To confirm that an unintentional mutation
introduced during the procedure to knock-out hla was
not responsible for the reduced virulence in this strain,
complete genome sequencing of the strain using Ion
Torrent sequencing was performed. Mapping of se-
quence reads from JKD6159Δhla against JKD6159 (40×
genome coverage) demonstrated no additional differences
between JKD6159 and JKD6159Δhla. We also restored Hla
function in this strain by using allelic replacement to repair
the deletion in hla and create strain JKD6159Δhla r.Asex-
pected, Hla expression was absent in JKD6159Δhla and ex-
pression was restored in JKD6159Δhla r when tested by
Western Blot (Additional file 4A).JKD6159Δhla r also
reverted to high virulence in the mouse skin infection assay
(Figure 3). The apparent slight reduction in virulence of
this hla repaired strain compared to wild type JKD6159 is
explained by incomplete penetration of the restored hla
allele in JKD6159Δhla r, resulting in mixed bacterial popu-
lations and reversion to JKD6159Δhla for some of the
mice (Additional file 4B and C).
α-type PSMs
In order to determine the contribution of α-type PSMs to
virulence of JKD6159, we generated JKD6159Δpsmα
(deletion of the whole α-type PSM locus) and assessed
this mutant in the mouse skin infection assay (Figure 3).
There was no significant difference in virulence in all
outcome measures; weight loss (p = 0.06), lesion size
(p = 0.8174) and CFU recovery (p = 0.1925). The absence
of PSMα3 production was verified by HPLC and the in-
tegrity of the JKD6159Δpsmαwas confirmed by com-
plementation (Additional file 5). It is noteworthy that
the level of expression of PSMα3byJKD6159wassimi-
lar to USA300 (Figure 1), a strain that produces high
levels of PSMs and where a contribution to virulence
has been demonstrated [7,11]. Despite this, the deletion
mutant (JKD6159Δpsmα) demonstrated no attenuation
of virulence compared to JKD6159 (Figure 3). The sig-
nificantly divergent genetic background of ST93 com-
pared with USA300 may account for this difference
in the importance of α-type PSMs to the virulence of
JKD6159 [6].
PVL
We constructed an isogenic PVL negative mutant in
JKD6159 by deleting lukSF-PV. Western Blot analysis con-
firmed the absence of LukF-PV in the mutant (Additional
file 6). Assessment of the JKD6159ΔlukSF-PV mutant
in the mouse skin infection model showed no decrease
in virulence (Figure 3). Therefore PVL was not contrib-
uting to the increased virulence in JKD6159 in this
Figure 2 Virulence characteristics of wildtype ST93 CA-MRSA isolates. S. aureus JKD6159 compared with three other wildtype ST93
CA-MRSA isolates, TPS3104, TPS3105 and TPS3106 in a BALB/c mouse skin infection assay. At least 10 mice were used for each bacterial strain.
(A) Weight loss induced by intradermal infection with S. aureus strains is demonstrated as percentage loss of weight over 5 days. The difference
in percentage weight loss between JKD6159 and TPS3105 and TPS3106 was significant (p < 0.0001). There was no difference in weight loss
between JKD6159 and TPS3104. Data shown are mean weight loss and SEM. (B) Skin lesion area (mm
2
) at 5 days after infection was significantly
greater with JKD6159 infected mice compared to TPS3105 and TPS3106 (p < 0.0001). There was no difference in lesion area between JKD6159
and TPS3104. Data shown are mean area and SEM. (C) Recovery of S. aureus (log CFU) from infected tissues at 5 days after infection from
JKD6159 infected mice was greater than with TPS3105 (p = 0.0177) and TPS3106 infected mice (p = 0.0328). There was no difference between
JD6159 and TPS3104 infected mice. Data shown are mean CFU and SEM. Note, ***p < 0.001, *p < 0.05.
Chua et al. BMC Microbiology 2014, 14:31 Page 5 of 13
http://www.biomedcentral.com/1471-2180/14/31
murine model. Murine neutrophils, unlike rabbit and
human neutrophils are relatively resistant to the effects
of PVL so it is difficult to draw firm conclusions as to
the human importance of this result [2]. However, the
aim of this study was to uncover the mechanisms for
the observed increased virulence of ST93 previously
demonstrated using this mouse model [14]. Our results
reinforce the results of others who have examined dif-
ferent S. aureus clones which indicate that Hla, rather
than PVL is the main mediator of virulence in CA-MRSA
in a mouse skin infection model [9,10,21,22]. It should be
noted that other authors have concluded that the rabbit
skin infection model gave very similar results to the mouse
model for infection at the same site [4]. Nonetheless, testing
of our PVL deletion mutant in a rabbit model may be
warranted in future.
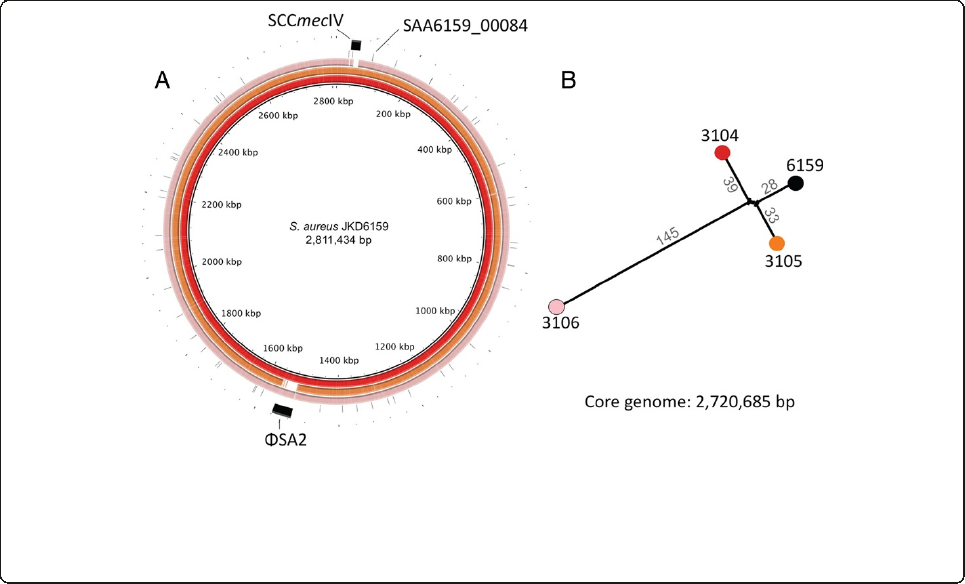
Genome sequencing of three additional ST93 isolates
We have previously fully sequenced and annotated the
genome of ST93 strain JKD6159 [14,23]. The differential
virulence and exotoxin expression of some ST93 isolates
compared to JKD6159 was then exploited by using whole
genome sequencing and comparative genomics to deter-
mine the genetic basis for exotoxin expression in this
clone. We selected the high expression strain TPS3104
and the low virulence and expression strains TPS3105 and
TPS3106 to compare to JKD6159. De novo assembly of
each of these strains resulted in ~700 contigs per isolate,
with a genome length of 2.8 Mbp. The de novo assembly
metrics are summarized in Additional file 7. The contigs
were aligned to JKD6159 using BLASTN, with some im-
portant differences demonstrated between the strains
(Figure 4A). TPS3104 contained SCCmecIV and ϕSA2
with lukSF-PV; TPS3105 contained SCCmecIV but lacked
ϕSA2 and lukSF-PV; TPS3106 contained SCCmecV, and
ϕSA2 without lukSF-PV. In addition, read-mapping
against the complete JKD6159 genome (chromosome and
plasmid: 2,832,164 bp) was then employed to define a
2,720,685 bp core genome among these four isolates (96%
of the JKD6159 genome), revealing 253 polymorphic nu-
cleotide positions, some of which were common to two
strains. A phylogeny was inferred that confirmed the close
relationship among all isolates, with TPS3106 more dis-
tantly related to the others (Figure 4B). The unmapped
reads from each isolate were also subjected to de novo as-
sembly to identify DNA not present in JKD6159. TPS3104
and TPS3105 contained no new sequences, while TPS3106
contained 34 kb of additional DNA, predominantly span-
ning the SCCmecVregion.
Comparative genomics of ST93 and the importance of agr
in the virulence of ST93 CA-MRSA
We next explored the contribution of specific mutations to
the differential virulence of the ST93 strains. Using our
read mapping approach described above, we compared the
genome sequences of TPS3104, TPS3105 and TPS3106
Figure 3 Virulence characteristics of S. aureus JKD6159 and isogenic exotoxin mutants derived from JKD6159. JKD6159 compared
to isogenic PVL knockout (JKD6159ΔlukSF-PV), isogenic Hla knockout (JKD6159Δhla), isogenic Hla complemented strain (JKD6159Δhla r)and
isogenic PSM-αknockout (JKD6159Δpsmα)inaBALB/cmouseskininfectionassay.(A) Weight loss induced by intradermal infection with
S. aureus strains is demonstrated as percentage loss of weight over 5 days. There was no significant difference between JKD6159,
JKD6159ΔlukSF-PV and JKD6159Δpsmαinfected mice. There was significantly less weight loss in mice infected with JKD6159Δhla compared
to JKD6159 (p < 0.0001). There was also less weight loss in mice infected with JKD6159Δhla compared to JKD6159Δhla r (p = 0.0063). Mice
infected with JKD6159Δhla r had less weight loss compared to JKD6159 (p = 0.0004). Data shown are mean weight loss and SEM. (B)
There was no difference in skin lesion area (mm
2
) at 5 days after infection in mice infected with JKD6159 and JKD6159ΔlukSF-PV and
JKD6159Δpsmα.Mice infected with JKD6159Δhla had significantly smaller lesions (p < 0.0001). In some mice, there was no cutaneous lesion
seen. There were significantly smaller lesions in mice infected with JKD6159Δhla compared to JKD6159Δhla r (p < 0.0001). Mice infected with
JKD6159Δhla r had smaller lesions compared to JKD6159 (p = 0.024). Data shown are mean area and SEM. (C) Recovery of S. aureus (log CFU)
from infected tissues at 5 days after infection from JKD6159 infected mice was no different to that from JKD6159ΔlukSF-PV, JKD6159Δpsmαand
JKD6159Δhla r. There was significantly less S. aureus recovered from JKD6159Δhla infected mice (p = 0.0177). There was also significantly less S. aureus
recovered from JKD6159Δhla infected mice compared to JKD6159Δhla r (p = 0.0018). Data shown are mean CFU and SEM. Note, ***p < 0.001, *p < 0.05,
compared to JKD6159.
Chua et al. BMC Microbiology 2014, 14:31 Page 6 of 13
http://www.biomedcentral.com/1471-2180/14/31
with each other and with JKD6159. There were a number
of single nucleotide polymorphisms (SNPs) and insertions
and deletions (indels) differentiating the strains from
JKD6159 (Additional files 8, 9, 10). We searched for muta-
tions in regulatory genes that could potentially explain the
different virulence phenotypes of the strains. Notably, both
avirulent ST93 strains, TPS3105 and TPS3106 contained
mutations within the agr locus. We have since completed
whole genome sequencing of TPS3151 and TPS3161 and
found they contain predicted amino acid substitutions in
AgrC that might disrupt agr function (Stinear et al., sub-
mitted). These isolates demonstrated low expression of
Hla (Additional file 3).
Additionally, TPS3106 also contained a mutation in
a gene encoding a previously uncharacterized AraC/
XylS family regulatory protein. This was also of par-
ticular interest as members of this class have been
shown to contribute to the regulation of exotoxin ex-
pression [24,25].
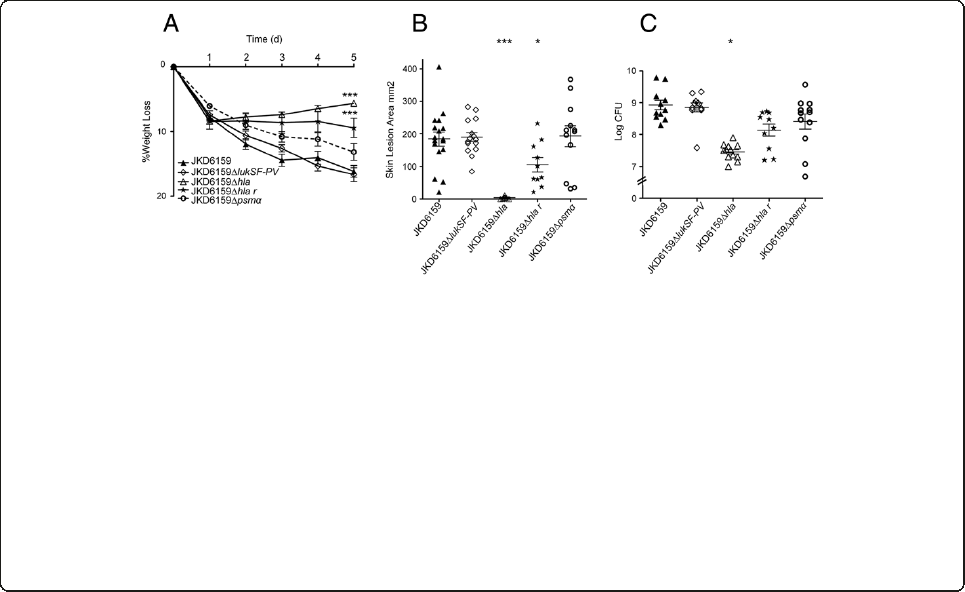
TPS3105 contained a frame-shift mutation within agrA
(Sa_JKD6159 nucleotide 2096502) and a further substi-
tution (G to A) within agrA at nucleotide 2096569),
while TPS3106 contained an ~356 bp deletion spanning
the agr effector molecule, RNAIII (deletion spanning nu-
cleotides 2093372 to 2093728). These mutations sug-
gested these isolates were agr deficient. To demonstrate
that these changes explained the reduced exotoxin ex-
pression and loss of virulence in TPS3105 we repaired
agrA using allelic exchange to create TPS3105r(the par-
ental strain TPS3105 now containing an intact agrA).
Quantitative real time RT-PCR for RNAIII demonstrated
that TPS3105rproduced 325-fold more RNAIII than
TPS3105. Virulence was also restored and TPS3105r
caused greater weight loss, skin lesion area and CFU re-
covery from lesions compared to the parental strain
TPS3105 (p < 0.0001, Figure 5). There was no significant
difference between JKD6159 and TPS3105rin all outcome
measures in the mouse skin infection model (Figure 5).
These experiments show that intact agr is essential for
the virulence of ST93 CA-MRSA. The agrA repaired
mutant of TPS3105, TPS3105rexpressed significantly
greater amounts of PSMα3 (p < 0.0001) and Hla (p =
0.0019), consistent with agr control of these virulence
determinants (Figure 6). Thus, despite the genetic diver-
gence of ST93 from other S. aureus [14], the molecular
foundation of virulence for this CA-MRSA clone is
similar in this respect to USA300 [9,26,27] and other S.
aureus strains [28,29], where the importance of agr has
been very well established.
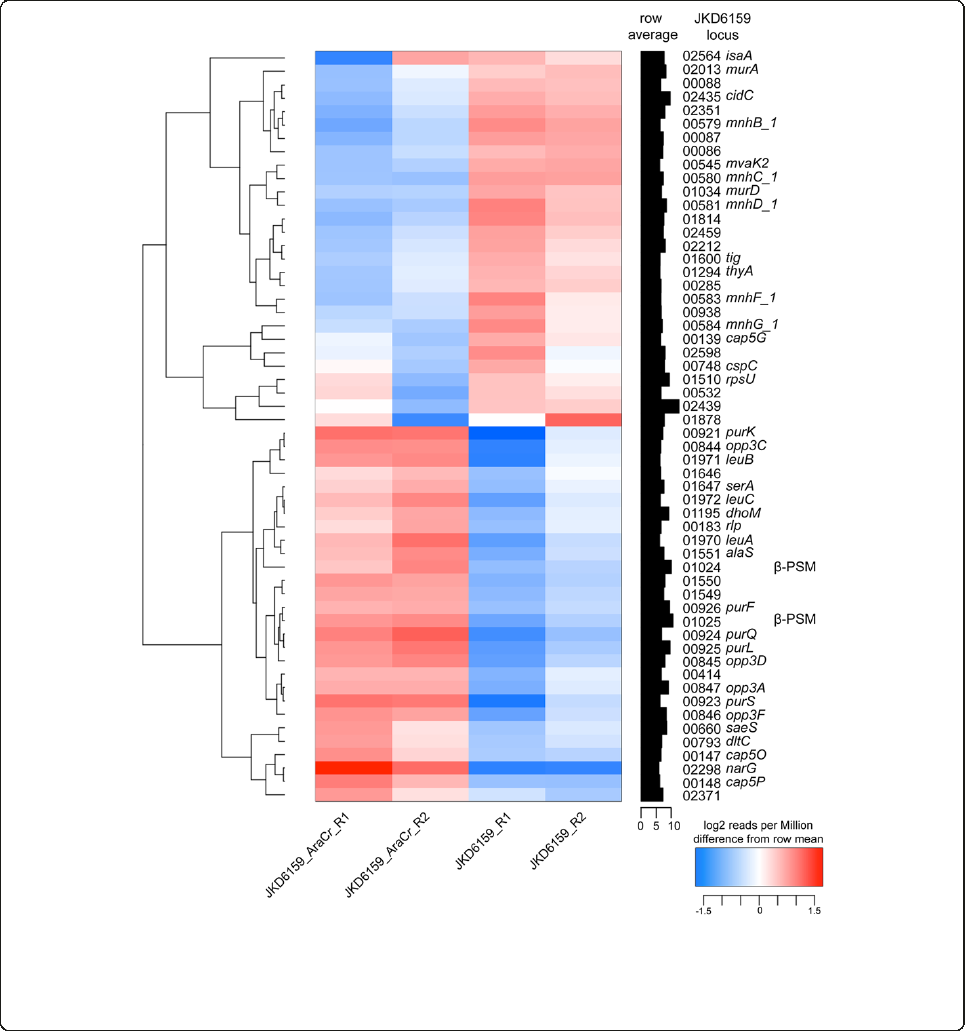
The AraC/XylS regulator (AryK) enhanced Hla expression
and virulence in ST93 CA-MRSA
The SNP at position 92551 in SAA6159_00084 introduced
a premature stop codon and created a pseudogene within
SAA6159_00084 in JDK6159, however the gene was intact
in TPS3106. The intact version of this gene, which was also
Figure 4 Whole genome sequence analysis and comparison of JKD6159 with other ST93 CA-MRSA isolates. (A) Circular diagram of the
JKD6159, TPS3104, TPS3105 and TPS3106 chromosomes (from inner to outer circles). TPS3104, TPS3105 and TPS3106 contigs were mapped by
BLASTN to JKD6159. TPS3104 contained SCCmecIV and ϕSA2 with lukSF-PV; TPS3105 contained SCCmecIV but lacked ϕSA2 and lukSF-PV; TPS3106
contained SCCmecV, and ϕSA2 without lukSF-PV.(B) ST93 S. aureus phylogeny inferred by split decomposition analysis from pairwise comparisons
of the 253 variable nucleotide positions identified from the ST93 core chromosome of 2,720,685 bp. Figures indicate the number of nucleotide
substitutions per branch. All nodes have 100% bootstrap support.
Chua et al. BMC Microbiology 2014, 14:31 Page 7 of 13
http://www.biomedcentral.com/1471-2180/14/31
intact in 19 other publically available S. aureus genome
sequences we examined, encodes a previously uncharac-
terized AraC/XylS family regulatory protein. While the
virulence attenuation in TPS3106 was likely a direct re-
sult of the agr deficiency, we also wanted to determine
if the novel regulator mutation in SAA6159_00084 im-
pacted the virulence in ST93 S. aureus.
To test the hypothesis that SAA6159_00084 encoded a
regulator of virulence, we repaired the premature stop
codon in SAA6159_00084 in JKD6159 using allelic ex-
change to generate strain JKD6159_AraCr. To confirm we
had not introduced additional DNA changes during al-
lelic exchange we sequenced the whole genome of
JKD6159_AraCrand found no additional mutations (35×
coverage). JKD6159_AraCrencoding an intact copy of
SAA6159_00084 demonstrated a modest, but significant
increase in virulence as indicated by lesion size (p <
0.0001) and weight loss in the mouse skin infection
assay (p = 0.0311, Figure 5), suggesting that this pro-
tein is a positive regulator of virulence in CA-MRSA
strains. JKD6159_AraCrexpressed more PSMα3(p=
0.0325) and Hla (p = 0.0473) than its parental strain
JKD6159 that was consistent with an increase mouse
skin lesion size (Figure 6). We propose the name aryK
for SAA6159_00084 (AraC family-like gene).
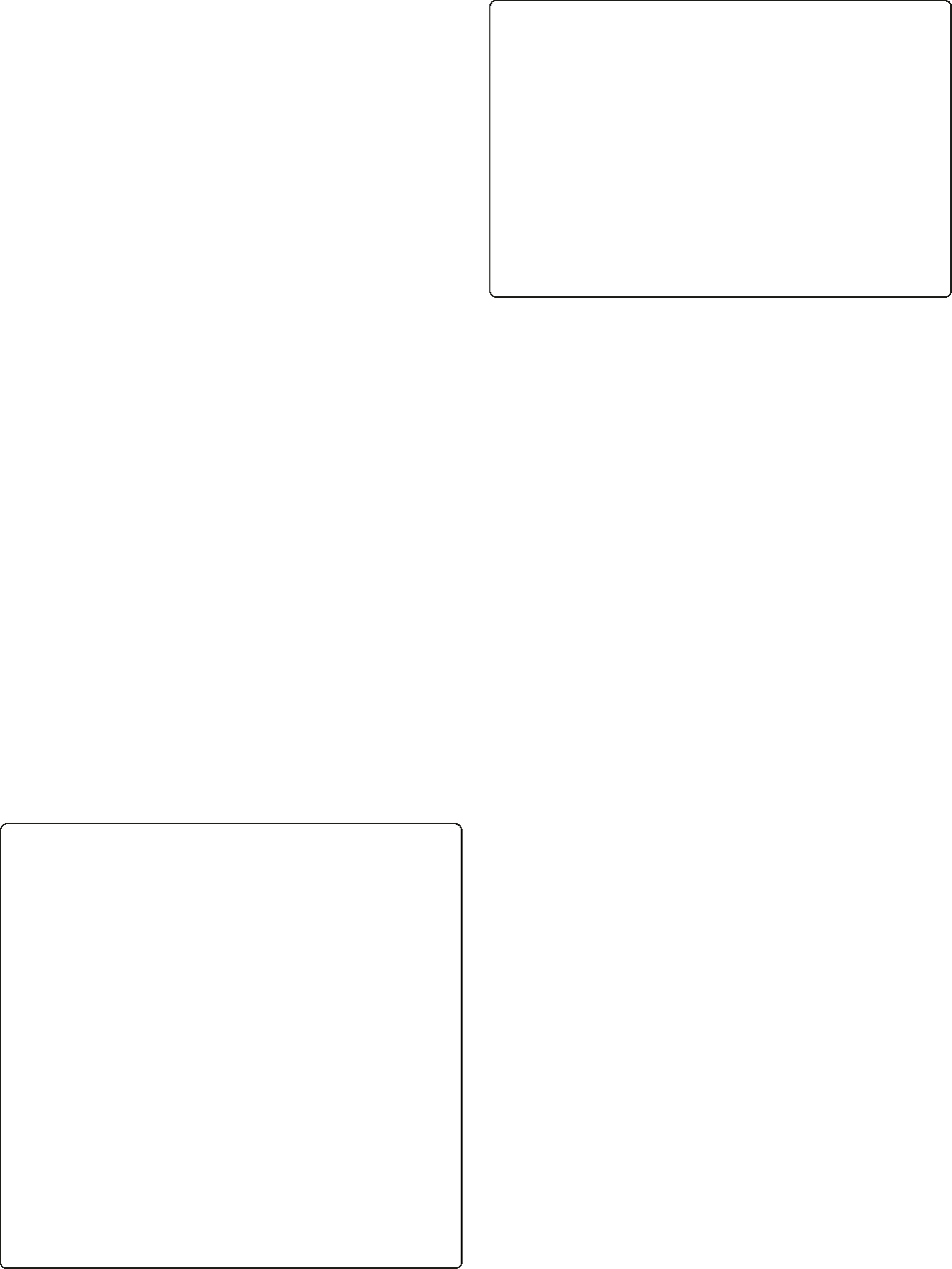
RNAseq demonstrates global regulatory impact of AryK
To investigate the regulatory impact of AryK, RNAseq
was performed using RNA extracted from stationary
phase cultures (Figure 7). This growth phase was se-
lected as we reasoned that AryK might be interacting
with agr and thus any impacts on Hla expression would
be greatest at this time. A small number of virulence-
associated loci were down regulated in the aryK mutant
(JKD6159), including beta-type phenol soluble modulins
(SAA6159_01024 and SAA6159_01025), and the viru-
lence regulator saeS. However, the most dramatic and
significant transcriptional changes were found in genes
involved in central metabolic functions. Using the Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway
Figure 6 In vitro PSMα3 and Hla expression of mutant S.
aureus isolates. JKD6159 compared with JKD6159_AraCr. TPS3105
compared with TPS3105r.(A) PSMα3 expression measured by HPLC.
JKD6159_AraCrexpressed more PSMα3 than JKD6159 (p = 0.0325).
TPS3105rexpressed more PSMα3 than TPS3105 (p < 0.0001). Data
shown are mean concentration (μg/ml), presented as vertical
stacked bars and SEM. Deformylated PSMα3isshowningreybars.
N-formylated PSMα3 is shown in white bars. (B) Hla expression
measured by quantitative Western blot. There was a small but
statistically significant increase in Hla production by JKD6159_AraCr
(p = 0.0473). TPS3105rexpressed more Hla than TPS3105 (p = 0.0019)
Data shown are mean intensity of bands in arbitrary units and SEM.
Note, *p < 0.05, compared to JKD6159. Note also,
###
p < 0.001 and
##
p < 0.01, compared to TPS3105.
Figure 5 The importance of agr and aryK in the virulence of ST93 CA-MRSA. Isogenic repaired agr mutant TPS3105rcompared to TPS3105
and JKD6159, and JKD6159 compared with isogenic repaired AraC/XylS family regulator mutant (JKD6159_AraCr) in a BALB/c mouse skin
infection assay. At least 10 mice were used for each bacterial strain. (A) Weight loss induced by intradermal infection with S. aureus strains is
demonstrated as percentage loss of weight over 5 days. There was no difference between JKD6159 and TPS3105rin all outcome measures.
TPS3105rinfected mice had significantly increased weight loss compared to TPS3105 (p < 0.0001). There was a small increase in weight loss in mice
infected with JKD6159_AraCrcompared to JKD6159 (p = 0.0311). Data shown are mean weight loss and SEM. (B) Skin lesion area (mm
2
) at 5 days after
infection in TPS3105rinfected mice was significantly increased compared to TPS3105 (p < 0.0001). Mice infected with JKD6159_AraCrhad increased
lesion area compared with JKD6159 (p < 0.0001). Data shown are mean area and SEM. (C) Recovery of S. aureus (log CFU) from infected tissues at
5 days after infection from TPS3105r was significantly greater than from TPS 3105 infected mice (p < 0.0001). There was no difference in S. aureus
recovered from mice infected with JKD6159 and JKD6159_AraCr. Data shown are mean CFU and SEM. Note, *** p < 0.001, * p < 0.05.
Chua et al. BMC Microbiology 2014, 14:31 Page 8 of 13
http://www.biomedcentral.com/1471-2180/14/31
analysis (www.genome.jp) [30] the major pathways where
differential transcription occurred were down regulation
of genes linked to purine metabolism (purK, purS, purQ,
purL, purF), valine, leucine and isoleucine biosynthesis
(leuA,leuB,leuC), and oligopeptide transport (opp3C,
opp3D, opp3F, opp3A). In contrast up regulation of genes
encoding cation transport systems (mnhB_1, mnhC_1,
mnhD_1, mnhF_1, mnhG_1) was found.
Here, we have clearly demonstrated that agr is the major
“on-off”switch for virulence in ST93 CA-MRSA, but we
also found that other genetic changes are impacting
virulence gene regulation in a clone-specific manner.
We speculate that the inactivation of aryK may have
been an evolutionary response by ST93 CA-MRSA to
modulate or fine-tune the amount of Hla and other fac-
tors required for host persistence. There are six AraC/
XylS family regulators in S. aureus (SA0097, SA0215,
SA0622, SA1337, SA2092, SA2169; S. aureus strain N315
locus tags). Two of these, Rbf (SA0622) and Rsp (SA2169)
have been studied and demonstrated in other S. aureus
Figure 7 Heatmap of RNA Sequencing comparing JKD6159 (aryK inactive) to JKD6159_AraCr(aryK intact). RNA seq was performed in
duplicate from stationary phase cultures. This heatmap, clustered on expression profiles, was created based on log
2
transformed counts to
identify consistent changes in expression profiles between strains. To be included in the heat map, genes were required to have at least 1000
counts (reads), totaled over all samples, where the standard deviation of log
2
expression differences had to exceed two. The heatmap highlights
significant aryK-dependent changes, in particular genes involved in the regulation of central metabolic functions.
Chua et al. BMC Microbiology 2014, 14:31 Page 9 of 13
http://www.biomedcentral.com/1471-2180/14/31
strains to regulate biofilm formation and modulate expres-
sion of surface-associated proteins [24,25,31]. In contrast,
we found that aryK increases Hla expression and virulence,
acting as a positive regulator of virulence by directly or in-
directly upregulating exotoxin expression, without an ap-
parent effect on agr expression in stationary phase.
Conclusions
In this study, we have obtained insights into the genetic
basis for the increased virulence of ST93 by using a
combination of comparative and functional genomics.
We have demonstrated the key role of Hla and agr and
shown how an additional novel regulatory gene, aryK
by a loss-of-function point mutation, is modulating
virulence in this clone. Quantification of exotoxin ex-
pression in a larger collection of ST93 strains demon-
strated that the findings in strain JKD6159 are relevant
to the majority of the ST93 population isolated from
around Australia as exotoxin expression in JKD6159 is
representative of most of the ST93 population. Our
study highlights the power of comparative genomics to
uncover new regulators of virulence but it also shows
the complex nature of these changes even in closely re-
lated bacterial populations. Careful strain selection, de-
tailed comparative genomics analyses, and functional
genomic studies by creating multiple genetic changes in
one strain will be required to gain a full insight into the
genetic basis for the emergence and hypervirulence of
ST93 CA-MRSA.
Methods
Ethics statement
This study was performed in accordance with the Australian
Prevention of Cruelty to Animals Act 1986 and the
Australian code of practice for the care and use of animals
for scientific purposes. The protocol was approved by the
Animal Ethics Committee of the University of Melbourne
(Permit Number: 0911248.2).
Bacterial strains and culture
Bacterial strains used in this study are summarized in
Table 1 (international clone collection) and Additional file
1 (ST93 strain collection), and include the ST93 reference
strains JKD6159, USA300 strain FPR3757 [19], ST30 strain
JKD6177, and the HA-MRSA ST239 clone JDK6009 [20],
as well as 58 additional ST93 collected from around
Australia and previously reported [17]. For all experi-
ments except exotoxin expression bacteria were grown
in brain heart infusion broth (BHI, Oxoid). For the
mouse skin infection assay, S. aureus were harvested at
the stationary phase of growth after 18 hours incuba-
tion (OD
600
~ 2.0), washed, diluted and resuspended in
PBS. The bacterial inoculum (CFU) and viable counts
were determined by plating onto BHI agar and colony
enumeration.
For LukF-PV expression experiments, bacteria were
growninCCYmedia(3%yeastextract(Oxoid),2%
BactoCasaminoAcids(Difco),2.3%sodiumpyruvate
(Sigma-Aldrich), 0.63% Na
2
HPO
4
, 0.041% KH
2
PO
4
,pH6.7).
For α-hemolysin (Hla) and PSMα3 expression experi-
ments, bacteria were grown in tryptone soy broth (TSB,
Oxoid). Overnight cultures were diluted 1:100 into fresh
media and then incubated at 37°C with shaking (180 rpm)
until stationary phase was achieved. For LukF-PV detection,
isolates were cultured for 8 hours (OD
600
~1.8);for
Hla detection, isolates were cultured for approximately
3 hours (OD
600
~ 1.8); and for PSMα3 detection, isolates
were cultured for 24 hours (OD
600
~2.0).Culturesuperna-
tants were harvested by centrifugation and filter sterilized.
These were performed in at least triplicate for each S.
aureus strain.
Detection of LukF-PV and Hla by western blotting
Trichloroacetic acid was added to culture supernatants
and incubated at 4°C overnight. Precipitates were then
harvested by centrifugation, washed with acetone, air-
dried and solubilized in a SDS and 2-mercaptoethanol
containing sample buffer. The proteins were separated on
12% SDS-PAGE. A peptide sequence specific to LukF-PV,
HWIGNNYKDENRATHT was synthesized and HRP
conjugated polyclonal chicken IgY was raised against
this peptide (Genscript). This antibody was used to de-
tect LukF-PV with enhanced chemiluminescence. Im-
ages generated from the western blots were quantitated
using GS800 Calibrated Densitometer (BioRad) and
Image J [32].
Hla was detected using a polyclonal rabbit anti-Hla
(Sigma-Aldrich), in buffer containing 20 mM DEPC to
inhibit non-specific protein A binding and HRP conju-
gated sheep anti-rabbit secondary antibody (Millipore)
with enhanced chemiluminescence detection [33]. For
comparison of JKD6159 versus international clone col-
lection (Table 1), images generated from the Hla western
blots were quantitated using GS800 Calibrated Densito-
meter (BioRad) and Image J [32].
Subsequently, for comparison of JKD6159 and other
ST93 strains (Table 1), detection of chemiluminescence
was performed using the MF-ChemiBIS 3.2 platform
(DNR Bioimaging systems). Quantitation was performed
using Image J [32].
Detection of PSMα3 expression
HPLC chromatography was performed on an Agilent
Technology 1200 Series system with an analytical Agilent
Eclipse XDB-C18 (4.6 mm × 150 mm) column. A water/
acetonitrile gradient (0.1% trifluoroacetic acid) from 0 –
100% acetonitrile over 28 min at a flow rate of 1 mL/min
Chua et al. BMC Microbiology 2014, 14:31 Page 10 of 13
http://www.biomedcentral.com/1471-2180/14/31
was used. The total run time was 32 min, and peaks were
quantified at a wavelength of 214 nm. The deformylated
and formylated form of PSMα3 MEFVAKLFKFFKDLL
GKFLGNN was identified in the S. aureus TSB culture su-
pernatants by comparison of their retention times to a
commercially synthesized PSMα3 standard (GenScript)
and by spiking the samples with the synthesized standards.
The identity of the deformylated peptide present in the
samples was confirmed by analysing collected fractions by
ESI-MS. There was only one peptide present in this frac-
tion; the deformylated form of PSMα3. In contrast, other
peptides were observed in the fractions of USA300,
JKD6272, TPS3104, TPS3105r,andJKD6159_AraCrcon-
taining the N-formylated form of PSMα3. In these cases,
the percentage of N-formylated PSMα3 peptide was deter-
mined using the total ion count of the major peaks in the
ESI-MS and the peak area of the HPLC chromatogram
was adjusted accordingly. The concentrations of the defor-
mylated and formylated forms of PSMα3 were determined
by comparison of their peak areas to those of the synthe-
sized standards. The standard curves were constructed in
the concentration range of 6.2 –100 μg/ mL and were lin-
ear over this range.
DNA methods, molecular techniques and construction
of mutants
DNA was extracted using the GenElute kit according
to the manufacturer’s instructions (Sigma-Aldrich). A
lukSF-PV knockout, hla knockout and a repaired agrA
of TPS3105 were generated according to the published
method [34]. For the knockouts, flanking sequences
were amplified and ligated prior to cloning with pKOR1.
For allelic replacement to generate TPS3105r, aPCR
product of agrA from JKD6159 was cloned with pKOR1.
For allelic replacement JKD6159_AraCr, a PCR product
of this AraC regulator from TPS3106 was cloned with
pKOR1. The deletion of the whole psmαlocus in JKD6159,
chromosomal restoration of psmαin JKD6159Δpsmαand
the restoration of Hla expression in JKD6159Δhla were
conducted using the pIMAY protocol described by
Monk et al. [35]. Knockout and restoration amplimers
were cloned into pIMAY by SLIC [36]. The primers
used are listed in Additional file 11. The knockout and
restoration clones were confirmed by PCR and Sanger
sequencing of the mutated locus. Mutations were fur-
ther validated with functional assays of activity, which
included sheep blood hemolysis, western blot, and/or
HPLC.
Mouse skin infection assay
Mice were infected with S. aureus as previously de-
scribed [14]. Briefly, six-week-old female BALB/c mice
were infected by intradermal injection with 10
8
CFU of
S. aureus. Mice were assessed and weighed daily for five
days. Mice were culled on the 5th day and lesion size
measured and CFU recovered from infected tissues by
homogenization and colony enumeration on BHI. For
each S. aureus strain, at least 10 mice were assessed.
Genome sequencing
Genome sequences for three ST93 strains (TPS3104,
TPS3105, TPS3106) were obtained from an Illumina
GAIIx analyzer using 100 bp paired-end chemistry with a
mean fold coverage of 331×. Genome sequencing of the
two laboratory-induced mutants JKD6159Δhla (TPS3265)
and JKD6159_AraCr(TPS3268) was performed using Ion
Torrent sequencing technology.
Comparative genomics
A read mapping approach was used to compare the se-
quences from all isolates used in this study, as previously
described [14,37]. Briefly, the reads from all genomes
were aligned to the JKD6159 reference using SHRiMP
2.0 [38]. SNPs were identified using Nesoni v0.60 [www.
bioinformatics.net.au]. Using the whole genome sequence
of JKD6159 as a reference, a global SNP analysis was
performed, and allelic variability at any nucleotide position
was tallied to generate a global SNP analysis for every gen-
ome compared to JKD6159.
Quantitative RT-PCR for RNAIII expression
To investigate activity of the agr locus (RNAIII) qRT-PCR
was performed for RNAIII as previously described [37].
Briefly, RNA was prepared as previously described with two
on-column DNase I digestion steps and cDNA synthesis
using SuperScript II reverse transcriptase (Invitrogen). Rela-
tive expression was determined as previously described and
was normalised against gyrB. Results were obtained from 3
biological replicates each performed in triplicate.
RNA sequencing
Staphylococcus aureus strains JKD6159 and JKD6159_Ar-
aCrwere grown to early stationary culture as described
above. For RNA protection, 0.5 volumes of RNAlater®
RNA stabilization reagent (Qiagen) was added immediately
to the liquid culture and allowed to incubate with the bac-
terial suspension for 15 minutes at room temperature. Cells
were pelleted at 5,000 × g for 5 minutes followed by RNA
extraction using RNeasy mini kit (Qiagen) and two rounds
of DNase I digestion (Qiagen) according to the manu-
facturer’s instruction. RNA concentration was quantified
using Qubit® 2.0 Fluorometer and RNA quality assessed
using Agilent 2100 Bioanalyzer. Ten μgoftotalRNA
from the stationary growth phase with RNA intergrity
number (RIN) greater than 7 was used in RNA-seq.
Ribosomal depletion, cDNA library preparation and
pair ended sequencing using HiSeq2000 sequencing
platform was performed by Beijing Genome Institute
Chua et al. BMC Microbiology 2014, 14:31 Page 11 of 13
http://www.biomedcentral.com/1471-2180/14/31
(Hong Kong, China). RNAseq was performed on two bio-
logical samples for each strain.
RNAseq reads were mapped onto the JKD6159 refe-
rence genome [23], using SHRiMP 2.2.2 [39]. Alignment
to CDS features from each biological replicate of each
strain provided counts that were a measure of mRNA
levels. Counts were normalized using the trimmed-mean
normalization function in edgeR, part of the BioConduc-
tor package [40]. A heat map was created based on log
2
transformed counts to identify consistent changes in ex-
pression profiles between strains. To be included in the
heat map, genes were required to have at least 1000
counts, totaled over all samples, where and the standard
deviation of the log
2
expression levels had to exceed two.
Statistical analysis
Percentage mouse weight change at day 5, viable counts
of S. aureus in mouse tissues and skin lesion area of
each isolate, Hla, LukF-PV and PSMα3 expression versus
JKD6159 were analyzed using an unpaired t test. A simi-
lar analysis was used to analyze virulence outcome mea-
sures and exotoxin expression between TPS3105 and
TPS3105r. (There was no difference in results when
Bonferonni analysis was performed). All analyses were
performed using Prism 5 for Macintosh v5.0b (GraphPad
Software Inc.).
Availability of supporting data
The data sets supporting the results of this article are in
the NCBI Sequence Read Archive under study accession
SRP004474.2 and the NCBI BioProject Archive under
study accession PRJNA217697.
Additional files
Additional file 1: Staphylococcus aureus ST93 strains used in
this study.
Additional file 2: Expression of PSMα3 by ST93 strains and USA300.
(A) Expression of deformylated PSMα3. (B) Expression of N-formylated
PSMα3. Data shown are mean concentration (μg/ml) and SEM.
Additional file 3: Expression of Hla by ST93 strains and USA300.
Hla expression measured by quantitative Western blot. Data shown are
mean intensity of bands in arbitrary units and SEM.
Additional file 4: Hla Western Blot of JKD6159, JKD6159Δhla and
JKD6159Δhla r (A) Western Blot demonstrating that JKD6159Δhla
does not express Hla by Western Blot and that complementation of
this mutant (JKD6159Δhla r) results in restoration of Hla expression.
(B) Arrangement of PCR primers used PCR screen of JKD6159Δhla and
JKD6159Δhla r. (C) PCR screen of 25 randomly selected S. aureus colonies
obtained from two mice (mouse 4 and mouse 7) post skin infection with
JKD6159Δhla r. The PCR primers used flank the region deleted in hla for
the mutant and show incomplete penetration of the bacterial population
with the repaired version of hla (17/25 with an intact allele for mouse 4
and 21/25 for mouse 7), thereby explaining the inability of the repaired
mutant to fully restore the virulence phenotype in this infection model.
Additional file 5: Detection of formylated PSMα3 in JKD6159,
JKD6159Δpsmαand JKD6159Δpsmαrby HPLC of culture filtrates.
JKD6159Δpsmαdid not produce formylated PSMα3. Complementation of
this strain resulted in restoration of formylated PSMα3 expression. In all
strains δ-toxin expression was maintained.
Additional file 6: LukF-PV Western Blot of JKD6159 and
JKD6159ΔlukSF-PV. Western Blot demonstrating that JKD6159ΔlukSF-PV
does not express LukF-PV.
Additional file 7: Table of de novo assembly characteristics for S.
aureus strains TPS3104, TPS3105 and TPS3106.
Additional file 8: Table of single nucleotide differences between
JKD6159 and TPS3104.
Additional file 9: Table of single nucleotide differences between
JKD6159 and TPS3105.
Additional file 10: Table of single nucleotide differences between
JKD6159 and TPS3106.
Additional file 11: Table of primers used in this study.
Competing interest
No author has any competing interests to declare.
Authors’contributions
Conceived the project, TPS, BPH, KYLC, JKD; performed the experiments,
KYLC, IRM, YHL, JLP, GWC, JS, KLT; analysed the data, KYLC, YHL, TPS, BPH, TS,
KLT; wrote the manuscript, KYLC, BPH, TPS. All authors read and approved
the final manuscript.
Authors’information
Timothy P. Stinear and Benjamin P. Howden are the Joint Senior Authors.
Acknowledgements
We thank Kirstie Mangas and Brian Howden for expert technical assistance.
Author details
1
Department of Microbiology and Immunology, University of Melbourne,
Melbourne, Victoria 3052, Australia.
2
Department of Microbiology, Monash
University, Clayton, Victoria 3800, Australia.
3
Austin Centre for Infection
Research (ACIR), Infectious Diseases Department, Austin Health, PO Box 5555,
Heidelberg, Victoria 3084, Australia.
4
Microbiology Department, Austin Health,
Heidelberg, Victoria 3084, Australia.
5
Victorian Bioinformatics Consortium,
Monash University, Clayton, Victoria 3800, Australia.
6
School of Chemistry,
Monash University, Clayton, Victoria 3800, Australia.
7
Australian Collaborating
Centre for Enterococcus and Staphylococcus Species (ACCESS) Typing and
Research, PathWest Laboratory Medicine-WA, Royal Perth Hospital, Perth,
Western Australia 6000, Australia.
8
School of Biomedical Sciences, Curtin
University, Bentley, Western Australia 6102, Australia.
Received: 6 December 2013 Accepted: 5 February 2014
Published: 10 February 2014
References
1. David MZ, Daum RS: Community-associated methicillin-resistant
Staphylococcus aureus: epidemiology and clinical consequences of an
emerging epidemic. Clin Microbiol Rev 2010, 23(3):616–687.
2. Loffler B, Hussain M, Grundmeier M, Bruck M, Holzinger D, Varga G, Roth J,
Kahl BC, Proctor RA, Peters G: Staphylococcus aureus Panton-Valentine
leukocidin is a very potent cytotoxic factor for human neutrophils.
PLoS Pathog 2010, 6(1):e1000715.
3. Diep BA, Chan L, Tattevin P, Kajikawa O, Martin TR, Basuino L, Mai TT, Marbach H,
Braughton KR, Whitney AR, et al:Polymorphonuclear leukocytes mediate
Staphylococcus aureus Panton-Valentine leukocidin-induced lung
inflammation and injury. Proc Natl Acad Sci U S A 2010, 107(12):5587–5592.
4. Kobayashi SD, Malachowa N, Whitney AR, Braughton KR, Gardner DJ, Long
D, Bubeck Wardenburg J, Schneewind O, Otto M, Deleo FR: Comparative
analysis of USA300 virulence determinants in a rabbit model of skin and
soft tissue infection. J Infect Dis 2011, 204(6):937–941.
5. Lipinska U, Hermans K, Meulemans L, Dumitrescu O, Badiou C, Duchateau L,
Haesebrouck F, Etienne J, Lina G: Panton-Valentine leukocidin does play a
role in the early stage of Staphylococcus aureus skin infections: a rabbit
model. PLoS One 2011, 6(8):e22864.
Chua et al. BMC Microbiology 2014, 14:31 Page 12 of 13
http://www.biomedcentral.com/1471-2180/14/31

6. Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF,
Lu Y, Otto M: Evolution of virulence in epidemic community-associated
methicillin-resistant Staphylococcus aureus.Proc Natl Acad Sci U S A 2009,
106(14):5883–5888.
7. Li M, Cheung GY, Hu J, Wang D, Joo HS, Deleo FR, Otto M: Comparative
analysis of virulence and toxin expression of global community-associated
methicillin-resistant Staphylococcus aureus strains. JInfectDis2010,
202(12):1866–1876.
8. Bhakdi S, Tranum-Jensen J: Alpha-toxin of Staphylococcus aureus.Microbiol
Rev 1991, 55(4):733–751.
9. Bubeck Wardenburg J, Bae T, Otto M, Deleo FR, Schneewind O: Poring over
pores: alpha-hemolysin and Panton-Valentine leukocidin in
Staphylococcus aureus pneumonia. Nat Med 2007, 13(12):1405–1406.
10. Kennedy AD, Bubeck Wardenburg J, Gardner DJ, Long D, Whitney AR,
Braughton KR, Schneewind O, DeLeo FR: Targeting of alpha-hemolysin by
active or passive immunization decreases severity of USA300 skin infection
in a mouse model. JInfectDis2010, 202(7):1050–1058.
11. Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD,
Dorward DW, Klebanoff SJ, Peschel A, et al:Identification of novel cytolytic
peptides as key virulence determinants for community-associated MRSA.
Nat Med 2007, 13(12):1510–1514.
12. Cheung GY, Duong AC, Otto M: Direct and synergistic hemolysis caused
by Staphylococcus phenol-soluble modulins: implications for diagnosis
and pathogenesis. Microbes Infect 2012, 14(4):380–386.
13. Coombs GW, Nimmo GR, Pearson JC, Christiansen KJ, Bell JM, Collignon PJ,
McLaws ML, Resistance AGfA: Prevalence of MRSA strains among
Staphylococcus aureus isolated from outpatients, 2006. Commun Dis Intell
2009, 33(1):10–20.
14. Chua KY, Seemann T, Harrison PF, Monagle S, Korman TM, Johnson PD,
Coombs GW, Howden BO, Davies JK, Howden BP, et al:The dominant
Australian community-acquired methicillin-resistant Staphylococcus
aureus clone ST93-IV [2B] is highly virulent and genetically distinct.
PLoS One 2011, 6(10):e25887.
15. Tong SY, Sharma-Kuinkel BK, Thaden JT, Whitney AR, Yang SJ, Mishra NN,
Rude T, Lilliebridge RA, Selim MA, Ahn SH, et al:Virulence of endemic
nonpigmented northern Australian Staphylococcus aureus clone (clonal
complex 75, S. argenteus) is not augmented by staphyloxanthin. JInfectDis
2013, 208(3):520–527.
16. Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM,
VazquezV,HookM,EtienneJ,et al:Staphylococcus aureus Panton-Valentine
leukocidin causes necrotizing pneumonia. Science 2007, 315(5815):1130–1133.
17. Coombs GW, Goering RV, Chua KY, Monecke S, Howden BP, Stinear TP,
Ehricht R, O’Brien FG, Christiansen KJ: The molecular epidemiology of the
highly virulent ST93 Australian community Staphylococcus aureus strain.
PLoS One 2012, 7(8):e43037.
18. Somerville GA, Cockayne A, Durr M, Peschel A, Otto M, Musser JM: Synthesis
and deformylation of Staphylococcus aureus delta-toxin are linked to
tricarboxylic acid cycle activity. J Bacteriol 2003, 185(22):6686–6694.
19. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J,
Carleton HA, Mongodin EF, et al:Complete genome sequence of USA300,
an epidemic clone of community-acquired meticillin-resistant
Staphylococcus aureus.Lancet 2006, 367(9512):731–739.
20. Howden BP, Stinear TP, Allen DL, Johnson PD, Ward PB, Davies JK: Genomic
analysis reveals a point mutation in the two-component sensor gene graS
that leads to intermediate vancomycin resistance in clinical Staphylococcus
aureus.Antimicrob Agents Chemother 2008, 52(10):3755–3762.
21. Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD,
Dorward DW, Gardner DJ, Lina G, et al:Is Panton-Valentine leukocidin the
major virulence determinant in community-associated methicillin-resistant
Staphylococcus aureus disease? JInfectDis2006, 194(12):1761–1770.
22. Bubeck Wardenburg J, Palazzolo-Ballance AM, Otto M, Schneewind O, DeLeo
FR: Panton-Valentine leukocidin is not a virulence determinant in murine
models of community-associated methicillin-resistant Staphylococcus
aureus disease. JInfectDis2008, 198(8):1166–1170.
23. Chua K, Seemann T, Harrison PF, Davies JK, Coutts SJ, Chen H, Haring V,
Moore R, Howden BP, Stinear TP: Complete genome sequence of
Staphylococcus aureus strain JKD6159, a unique Australian clone of
ST93-IV community methicillin-resistant Staphylococcus aureus.
JBacteriol2010, 192(20):5556–5557.
24. Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O’Donnell S,
Rowe S, O’Gara JP, Lee CY: Rbf promotes biofilm formation by
Staphylococcus aureus via repression of icaR, a negative regulator of
icaADBC.J Bacteriol 2009, 191(20):6363–6373.
25. Lei MG, Cue D, Roux CM, Dunman PM, Lee CY: Rsp inhibits attachment
and biofilm formation by repressing fnbA in Staphylococcus aureus MW2.
J Bacteriol 2011, 193(19):5231–5241.
26. Montgomery CP, Boyle-Vavra S, Daum RS: Importance of the global
regulators agr and saeRS in the pathogenesis of CA-MRSA USA300
infection. PLoS One 2010, 5(12):e15177.
27. Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M: Role of the accessory
gene regulator agr in community-associated methicillin-resistant
Staphylococcus aureus pathogenesis. Infect Immun 2011, 79(5):1927–1935.
28. Cheung AL, Eberhardt KJ, Chung E, Yeaman MR, Sullam PM, Ramos M, Bayer
AS: Diminished virulence of a sar-/agr- mutant of Staphylococcus aureus in
the rabbit model of endocarditis. JClinInvest1994, 94(5):1815–1822.
29. Wright JS 3rd, Jin R, Novick RP: Transient interference with staphylococcal
quorum sensing blocks abscess formation. Proc Natl Acad Sci U S A 2005,
102(5):1691–1696.
30. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M: KEGG for integration
and interpretation of large-scale molecular data sets. Nucleic Acids Res
2012, 40(Database issue):D109–D114.
31. Lim Y, Jana M, Luong TT, Lee CY: Control of glucose- and NaCl-induced biofilm
formation by rbf in Staphylococcus aureus.J Bacteriol 2004, 186(3):722–729.
32. Rasband WS: ImageJ. Bethesda, Maryland, USA: U S National Institutes of Health.
available at http:/imagej.nih.gov/ij/, accessed 9 December 2009 1997–2011.
33. Nguyen HM, Rocha MA, Chintalacharuvu KR, Beenhouwer DO: Detection and
quantification of Panton-Valentine leukocidin in Staphylococcus aureus
cultures by ELISA and Western blotting: diethylpyrocarbonate inhibits
binding of protein A to IgG. J Immunol Methods 2010, 356(1–2):1–5.
34. Bae T, Schneewind O: Allelic replacement in Staphylococcus aureus with
inducible counter-selection. Plasmid 2006, 55(1):58–63.
35. Monk IR, Shah IM, Xu M, Tan MW, Foster TJ: Transforming the
untransformable: application of direct transformation to manipulate
genetically Staphylococcus aureus and Staphylococcus epidermidis.
MBio 2012, 3(2):e00277–00211.
36. Li MZ, Elledge SJ: Harnessing homologous recombination in vitro to
generate recombinant DNA via SLIC. Nat Methods 2007, 4(3):251–256.
37. Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G,
Bennett-Wood V, Porter JL, et al:Evolution of multidrug resistance during
Staphylococcus aureus infection involves mutation of the essential two
component regulator WalKR. PLoS Pathog 2011, 7(11):e1002359.
38. RumbleSM,LacrouteP,DalcaAV,FiumeM,SidowA,BrudnoM:SHRiMP:
accurate mapping of short color-space reads. PLoS Comput Biol 2009,
5(5):e1000386.
39. David M, Dzamba M, Lister D, Ilie L, Brudno M: SHRiMP2: sensitive yet
practical SHort Read Mapping. Bioinformatics 2011, 27(7):1011–1012.
40. Robinson MD, McCarthy DJ, Smyth GK: edgeR: a Bioconductor package for
differential expression analysis of digital gene expression data.
Bioinformatics 2010, 26(1):139–140.
doi:10.1186/1471-2180-14-31
Cite this article as: Chua et al.:Hyperexpression of α-hemolysin explains
enhanced virulence of sequence type 93 community-associated
methicillin-resistant Staphylococcus aureus.BMC Microbiology 2014 14:31.
Submit your next manuscript to BioMed Central
and take full advantage of:
• Convenient online submission
• Thorough peer review
• No space constraints or color figure charges
• Immediate publication on acceptance
• Inclusion in PubMed, CAS, Scopus and Google Scholar
• Research which is freely available for redistribution
Submit your manuscript at
www.biomedcentral.com/submit
Chua et al. BMC Microbiology 2014, 14:31 Page 13 of 13
http://www.biomedcentral.com/1471-2180/14/31