UCLA Physics 4AL Lab Manual V37
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 126 [warning: Documents this large are best viewed by clicking the View PDF Link!]
- Introduction to the Data Acquisition (DAQ) system
- Determining and Reporting Measurement Uncertainties
- Experiment 0: Sensor Calibration and Linear Regression
- Experiment 1: Uniform Acceleration
- Experiment 2: Measurement of g
- Experiment 3: Conservation of Mechanical Energy
- Experiment 4: Momentum and Impulse
- Experiment 5: Harmonic Oscillator Part I. Spring Oscillator.
- Experiment 6: Harmonic Oscillator Part II. Physical Pendulum.
- Experiment 7: Waves on a Vibrating String
- Appendix A
- Appendix B
- Appendix C
Physics 4AL: Mechanics Lab Manual1
UCLA Department of Physics & Astronomy
May 12, 2017
1This manual is an adaptation of the work of William Slater by W. C. Campbell, Priscilla Yitong
Zhao, Julio S. Rodriguez, Jr., Chandler Schlupf, and Anthony Ransford
Abstract
Physics 4AL is designed to give students an introduction to laboratory experiments on me-
chanics. This course is intended to be taken after completion of Physics 1A or 1AH and
concurrently to the student taking Physics 1B or 1BH. We will be experimentally investigat-
ing some of the most foundational concepts in physics, including gravity, energy, momentum,
harmonic oscillation and resonance. These concepts are the basis for the classical motion of
everything from electrons to galaxies. There are a lot of “science skeptics” in the world; I
invite you to come to class with a critical eye and to experimentally test whether we, your
instructors, have been lying to you all these years!

i. Introduction to the Data
Acquisition (DAQ) system
A crucial part of modern experimental physics is the use of computers to aide in data
acquisition and experimental control. Computers can allow us to take data faster, more
accurately, and with far less tedium than was possible before their integration into the
laboratory. It is therefore crucial that an introductory physics laboratory course include the
use of computer-aided data acquisition (DAQ), and we will be using the DAQ throughout
the course to allow us to focus on the physics. This chapter will introduce the system we
will use and can be regarded as a user manual and general reference for later experiments.
i.1 PASCO DAQ System: Capstone and the 850 Uni-
versal Interface
Figure i.1: The PASCO 850 Universal Interface
The data acquisition (DAQ) system we will use in this course is produced by PASCO
Scientific for instructional lab courses. The software is called Capstone, and provides the
1
Figure i.2: The default startup screen for PASCO Capstone version 1.0.1. You may be using
a slightly newer version that looks a little different, but the basics should be the same.
communication with the hardware interface. This guide is based on PASCO Capstone 1.0.1
and the PASCO 850 Universal Interface, which is the piece of hardware that connects the
computer to your instruments, shown in Figure i.1.
i.2 Startup
To begin, ensure that the 850 is connected to the computer (via USB) and
power it on by pressing the power button on the front if the 850 is not already
on. If the unit is powered up and connected to the computer, the power button
will be backlit in blue and the connection indicator light just below the power
button will be illuminated in green. If you encounter problems with either of
these, check that cables have not come loose or ask for help from your TA.
Next, start the Capstone program, which has a logo that looks like, well, a capstone. Once
running, the program should look something like Figure i.2.
2
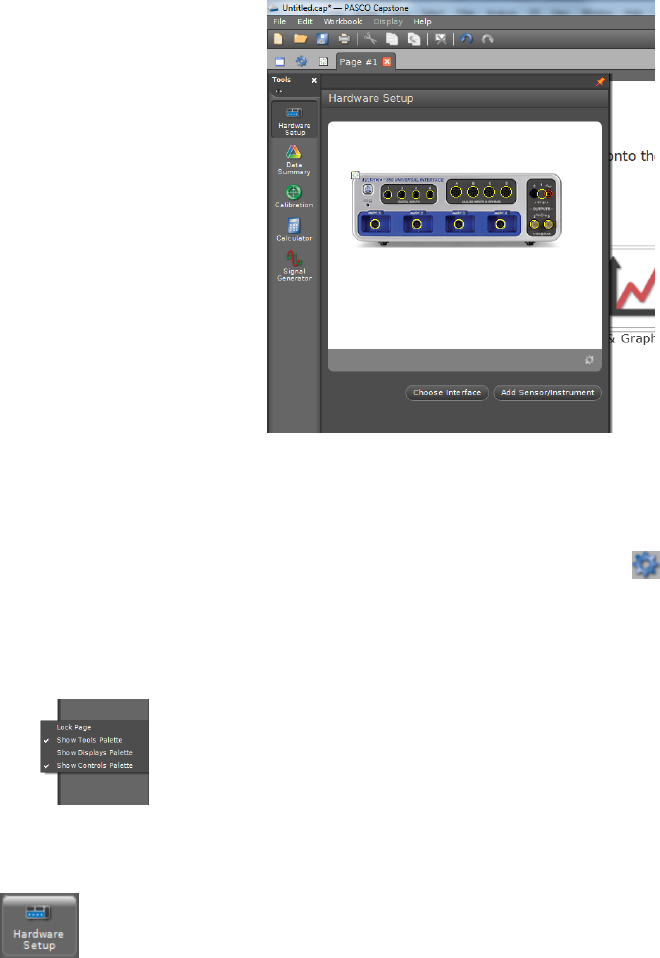
Figure i.3: Clickable Hardware Setup screen shot
If the Tools palette on the left is missing, click the gear icon in the upper left (“Change
properties of current page and Tools Palette”) and check Show Tools Palette under Page
Options. You can also right-click on the gray Capstone desktop and check Show Tools
Palette. The same thing goes for the Controls and Displays palettes.
If your palettes are not shown on the screen, right-click the
gray desktop and select the appropriate palette.
i.3 Configuring Hardware
To set up the hardware for a specific experiment, click on the Hardware Setup
icon in the Tools palette. If the 850 interface is turned on and is communicating
with the computer, this will bring up a clickable picture of the front panel of the
850 interface, as shown in Fig. i.3.
The yellow circles are clickable and will enable you to tell the computer what hardware
is attached to each connector on the 850 interface. Clicking on the yellow circle will bring
up a drop-down menu showing the possible types of hardware connected to each port. Once
you select the appropriate type of hardware from the list, icons will appear indicating how
the software thinks the hardware is connected. For example, to configure Capstone for the
vibrating string lab, you could choose Light Sensor from the drop down menu of the analog
input channel used for the photodiode and Output Voltage Current Sensor for output
channel 1.
3
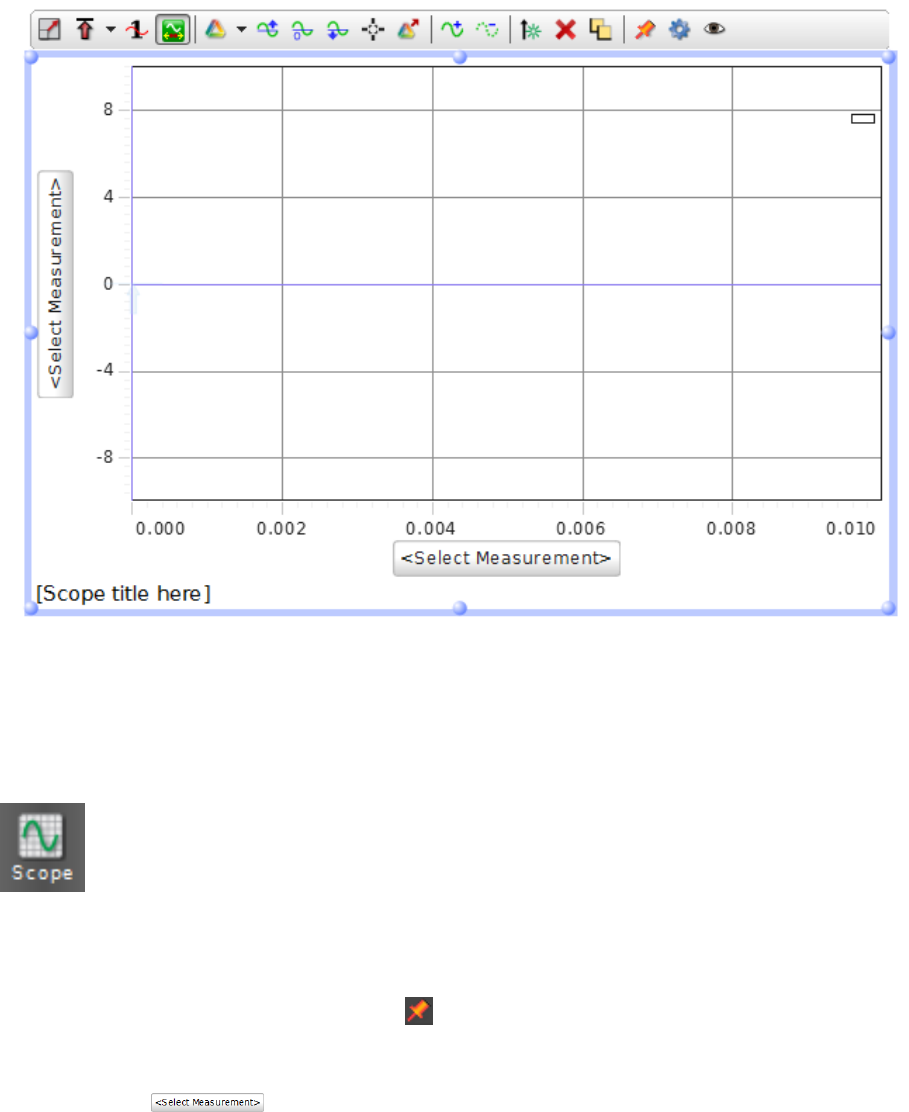
Figure i.4: Clickable Scope screen shot
i.4 Scope Display
The Scope feature is useful for running diagnostics of your apparatus and setting
things up with real-time readout of the results. When all else fails, you can try
using the scope to see if your DAQ is receiving input from its transducers. To
start a full-screen Scope session, double click the Scope icon from the Displays
palette (typically upper right, as shown in Fig. i.2). This will produce a large set of axes on
the screen to display the measurement, such as shown in Fig. i.4. If you have the Hardware
Setup or Signal Generator screens open and the left side of the plot is not visible, either
close those screens or click the push pin at the upper right side of those windows to force
the scope plot to shrink to fit the available space.
To configure what is being plotted on each axis, click the axis label, which is the but-
ton that says to bring up the drop down menu. This menu will have the
names of devices that can be used for plotting, but the only selectable items are the ac-
tual measurements listed below each device name. Selecting the proper measurement for
each axis will display the name of that measurement as the axis label. There is a menu
bar for the scope trace that will appear when it is clicked or moused over in the display.
4
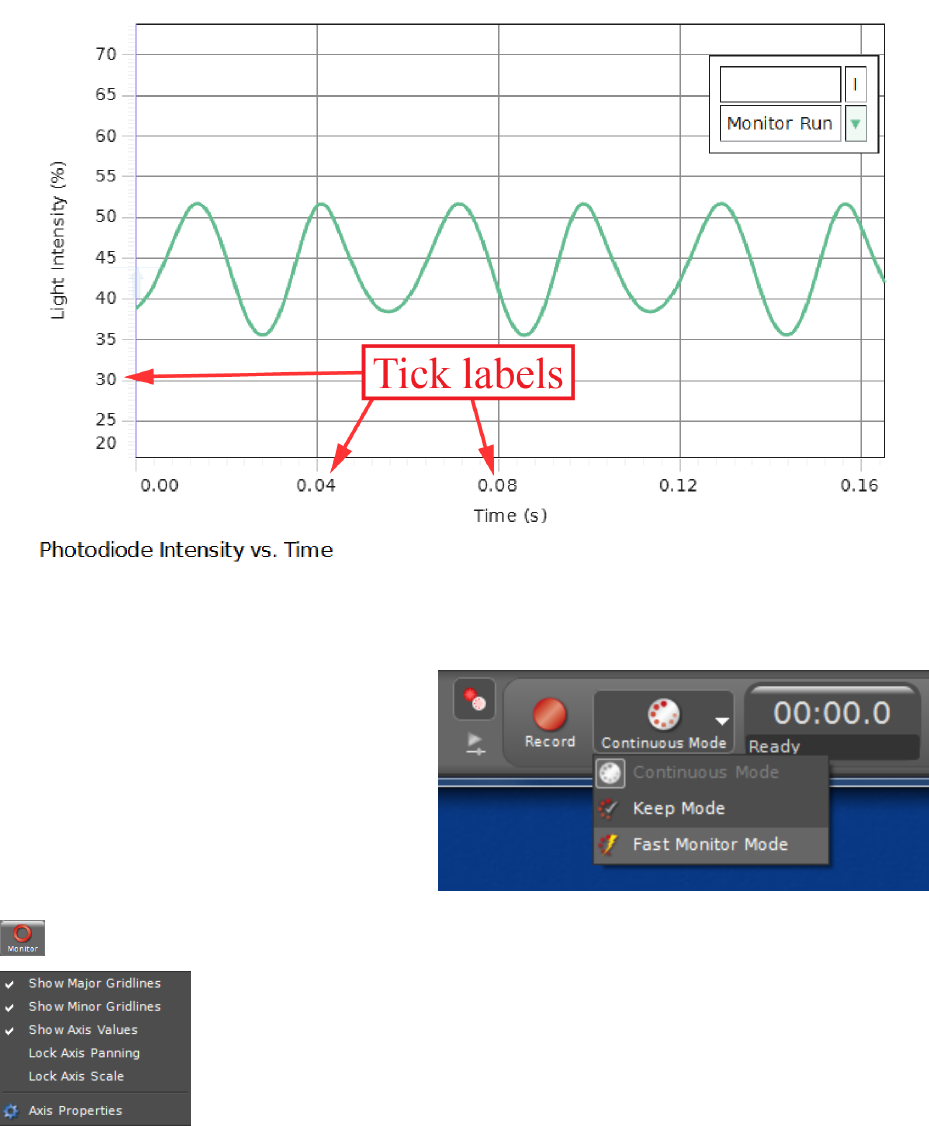
Figure i.5: The scope tool in action, with tick labels pointed out in red.
The x-axis will almost always be time
for our purposes, so to begin plotting
whatever is on the y-axis vs. time in
real-time, you may need to change from
Keep Mode or Continuous Mode to
Fast Monitor Mode, which can be se-
lected in the Controls palette, usually
found at the bottom of the screen. This
will allow you to start seeing real-time traces on the scope by clicking the Monitor button:
.
Changing the scale and offset of the axes is a little tricky. To change
the offset (i.e. to move the location of the origin), you can grab the plot
region itself and drag it around. If this doesn’t work, make sure that
axis panning is not locked by right-clicking on a tick label and making
sure that Lock Axis Panning is not checked. To change the scale of
an axis, either mouse over the tick labels and use the scroll wheel, or
grab an axis tick label and drag it toward or away from the origin to change the scale.
If nothing appears on the scope trace, make sure the expected signal would be shown in
the range that is being plotted. If there is still no trace shown, mouse over the plot area to
5

bring up the pop-up menu at the top and check to make sure that the trigger button
is not depressed. If it is, click it to unselect “normal trigger” mode.1
To plot multiple traces on the same set of axes, click the Add new y-axis to scope
display button on the pop-up menu for the scope trace. (If you don’t see the pop-up
menu at the top of the scope trace such as is shown in Fig. i.4, mouse over the plot region.)
The new y-axis will appear on the right side. To change the color of the plot, click on the
sensor data summary, right click the run, and the option “color picker” will come up.
1If you understand how to use a trigger on an oscilloscope, feel free to use it; it can be quite handy. The
trigger level is indicated by a very faint vertical arrow on the y-axis (at x= 0, shown in Fig. i.5 at t= 0.00 s,
y= 44%)) that can be dragged around to change the trigger level. The arrow starts at (0,0) on the plot, so
if you click the trigger button and can’t find the arrow, make sure you have x= 0 and y= 0 visible in your
plot region.
6
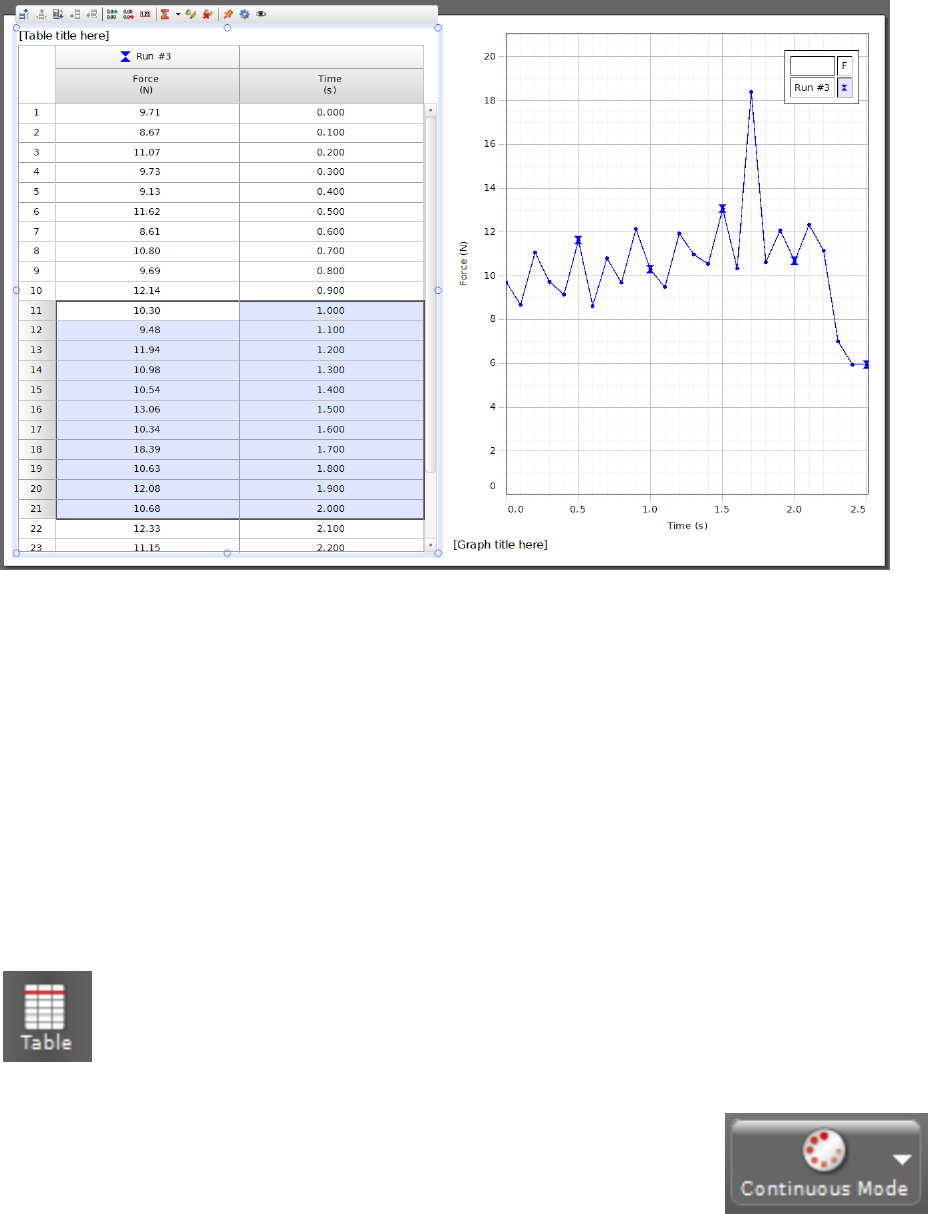
Figure i.6: A Table and Graph showing the same data. If you only want to save the data
between, say, t= 1 s and t= 2 s, you can highlight just the relevant portion in the Table
on the left and copy-and-paste it into Excel.
i.5 Exporting your Data
PASCO has not yet provided a way to save your data in a format that is useful. Ideally,
you should be able to import your data into an analysis program, such as Matlab, Excel,
Mathematica, Open Office Calc, IGOR Pro, Google Spreadsheets, or whatever else is your
preferred program. The way we will get around this software limitation is by displaying the
data in a table, from which you can later copy and paste the data directly into Excel on the
lab computer. Then you can save your Excel file in whatever format you prefer (.xls, .csv,
etc.) and, for instance, email it to yourself for analysis at home.
To display the acquired data in a table, first start a new table by click-
ing the Table icon in the Displays palette. This will open a new, blank,
two-column table on the workspace. The heading of each column will be a
clickable drop-down menu just like the Scope axes, and you can choose the
data you want to save for each column.
For example, Figure i.6 shows the results of a short measurement of
force vs. time. Both the Table column headings and the Graph axis
labels have been configured to display force (in Newtons) and time (in
seconds). The data were then recorded using, for instance, Continuous
7
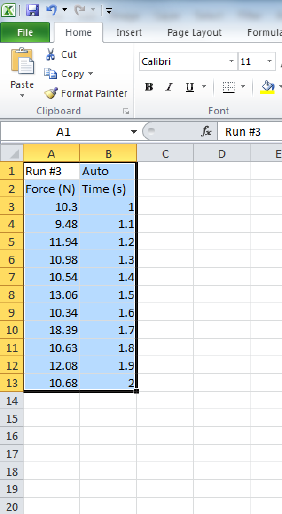
Figure i.7: The same data highlighted in Fig. i.6 right after being pasted into a new workbook
in Excel. Note that the column headers from the table in PASCO were automatically pasted
with the data. This spreadsheet can now be used for plotting, analysis, and saving for further
analysis and inclusion in a lab report.
Mode from the Controls palette, though it may also be acquired using Fast Monitor
Mode and Keep Mode if those are better suited to the particular experiment.
If we only want to save the section of data with the large peak in it for analysis and
plotting in a lab report, we can do this by highlighting the relevant portion of the Table
and copying it to Excel using copy (ctrl-c) and paste (ctrl-v). Figure i.7 shows the data
that was highlighted in Fig. i.6 as it appears in Excel directly after pasting it into a new
workbook. The data are now available for saving, plotting, curve fitting, and inclusion in a
lab report.
8

ii. Determining and Reporting
Measurement Uncertainties
Throughout this course, we will be making and reporting quantitative measurements of
experimental parameters. In order to interpret the results of a measurement or experiment,
it is crucial to specify the uncertainty (often called the “error” or “error bars”) with which
the measurement claims to be a report of the “true value” of the quantity being measured.
This chapter is designed to be a quick reference for the assignment and propagation of errors
for your lab reports. For a more detailed treatment, I recommend the excellent books by
Taylor [9] or Bevington and Robinson [1].
ii.1 Statement of measured values in this course
Every measurement is subject to constraints that limit the precision and accuracy with which
the measured “best value” corresponds to the “true value” of the quantity being measured.
It is fairly standard in physics to use the following notation to specify both the measured
best value and the uncertainty with which this value is known:
q=qbest ±δq. (ii.1)
Here, qis the quantity for which we are reporting a measurement, qbest is the measured
best value (often an average, but not infrequently generated in other ways) and δq is the
uncertainty in the best value, which is defined to be positive and always has the same units
as qbest. For our purposes in this course, the uncertainty will always be symmetric about the
measured best value, so the notation of Eq. ii.1 will be used throughout.
For example, the most accurately measured quantity in the world is currently the ratio
of the energy splittings between pairs of special states in two atomic ions [7], which is given
by νAl+
νHg+
= 1.052 871 833 148 990 44 ±0.000 000 000 000 000 06.(ii.2)
Here, the measured best value is 1.052 871 833 148 990 44 and the uncertainty is 6 ×10−17.
This level of precision and accuracy is far beyond anything we will be measuring in this
9
course, but the notation in nonetheless understandable because it is given in the form of
Eq. ii.1.
In this course, you will almost exclusively be measuring quantities with units. Measured
values for quantities that have units must be stated with their units. Failing to do so results
in complete nonsense, the loss of $300 million space probes [6], dogs and cats living together,
mass hysteria [5], not to mention the loss of points on your lab report grade. It is fine for the
units to appear as abbreviations, words, in column headers, after the numerical values, or in
some combination of these just so long as it is clear what the units are. A good example of
how to report a measured value is given by the 2010 CODATA recommended value for the
proton mass:
mp= (1.672 621 78 ±0.000 000 07) ×10−27 kg.(ii.3)
Note that the parentheses are used around the measured best value and the uncertainty
because they have the same units (which is always true) and are written with the same
exponential factor (which can help to make it easy to read).
There are two last points to make here about reporting measured values. First, in these
two examples, the uncertainties are written with the same precision (in scientific notation,
the number of digits where we include any leading zeros) as the measured best value. This
should always be the case, and when you report a measured value with an uncertainty, you
must make sure the uncertainty is reported with the same precision as the measured best
value.
Second, if we disregard all of the leading zeros, the uncertainty is presented with one
nonzero digit. There are times when it is appropriate to use up to two digits for this
(particularly when the first digit of the uncertainty is small, say, a 1 or 2), but uncertainties
should never have three significant digits. This is because the size of the uncertainty itself sets
the scale of where the measurement can no longer claim to be providing useful information.
ii.1.1 Other notation you will encounter
A quick glance at Eq. ii.2 and ii.3 reveals that it is difficult to read off the absolute value of
the uncertainty, since one has to count a lot of zeros. For this course, you will be expected to
report your measurements in the form of Eq. ii.1, but you should be aware of other methods
that are used so that you can interpret their meaning.
Concise notation is useful when the fractional uncertainty becomes very small (as in the
examples above). In concise notation, only the significant digit or digits of the uncertainty are
written, and they are written in parentheses directly after the best value, which is written to
the same precision. For example, the following shows two ways to express the same measured
value for the frequency of a laser
ν= (3.842 30 ±0.000 02) ×1014 Hz (ii.4)
ν= 3.842 30(2) ×1014 Hz.(ii.5)
The second line uses concise notation, where since the number in parentheses has one digit,
we are being told that this is the uncertainty in the last digit of the best value. This gets
10
more complicated when uncertainties are quoted with two digits, which seems to be getting
more common in the literature. As a concrete example, the mass of the electron reported
by CODATA is actually reported in the following form:
me= 9.109 382 91(40) ×10−31 kg (ii.6)
Here, we are being told that the last two digits have an uncertainty given by the two digits
in parentheses. This notation can seem somewhat confusing for a number of reasons, but if
we always think of writing the digits in parentheses below the best value so that they end
at the same place, it is easier to keep from getting confused, as in
me= 9.109 382 91 ×10−31 kg
±0.000 000 40 ×10−31 kg (ii.7)
Asymmetric uncertainties are also sometimes encountered in the scientific literature,
where the upper uncertainty may have a different size than the lower one. In this case, the
following three examples show how one may see this written, using an example of a measured
radiative decay lifetime:
τ= (37.0 +2.0/−0.8) ms (ii.8)
τ= (37.0+2.0
−0.8) ms (ii.9)
τ= 3.70(+20
−08)×10−2s (ii.10)
ii.1.2 “Sig Figs”
There is a system for implicitly including the order of magnitude of the uncertainty of a
measured quantity by simply stating the value with a certain number of digits, which is
often called the “sig figs” method for reporting uncertainty. Since our uncertainties will be
determined with higher precision than their order of magnitude, we will not be using this
shorthand method in this course, and you will be expected to explicitly write the uncertainty
of your measured quantities, either in the form of Eq. ii.1 or as a separate column entry in
a table. However, you should be aware of this shorthand since it does get used, though
typically outside of formal scientific literature.
Aside from its lack of specificity, an additional drawback of the sig figs method for report-
ing uncertainties is that all too often it leads to laziness and people reporting numbers with
implicit uncertainties far better than are actually merited. Your author starts to become
suspicious that this might be happening when measurements of continuous quantities are
reported to 4 or more digits, particularly by non-scientists. At or after the 4th digit, one
has to think seriously about things such as the calibration and resolution of measurement
equipment, thermal expansion of tape measures and rulers, finite response time and jitter of
timing systems, the linearly and stability of spring constants, surface roughness and cleanli-
ness, not to mention whether it makes sense at all to define the quantity you’re measuring
to that scale.
11
Example ii.1 Your author just looked at espn.com and noticed that they report the
speeds of NASCAR racers to 6 digits. They claim that, for instance, Kyle Busch just
completed a lap with an average speed of 126.648 mph. Can you think of reasons
to suspect that some (perhaps half) of these digits should not be believed? Hint:
was the length of the track even known to 6 digits at that particular time on that
particular day [10]?
ii.1.3 Accuracy and Precision
In physics and other sciences, the words accuracy and precision mean different things, and
it is important to distinguish between them. The accuracy of a measurement is how close
the measured value is to the true value of the quantity being measured. If the true value is
within the uncertainty that is reported, we find that the measurement was accurate.
Estimating the accuracy of a measurement is notoriously difficult without knowing the
true value. The standards that must be met to produce quantitative accuracy estimates vary
from field to field, but they generally consist of trying to estimate the size of the impact of
every possible source of error.
One cheap way to make a measurement that won’t be inaccurate is to construct the
measurement poorly on purpose so that the uncertainty is gigantic and is thereby highly
likely to overlap with the true value. However, this illustrates the point that accuracy is not
very useful without another key ingredient of a good measurement: precision.
The precision of a measurement is how small the range is that a statistical spread of
repeated measurements will fall into. The precision tells us nothing about the accuracy of
a measurement, and is determined entirely without knowing the true value of the quantity
being measured. Measurement precision is often determined by the statistical spread of
repeated measurements, and it will be fairly standard in this course to use the standard
deviation of a collection of repeated measurements to set the precision of the reported mea-
sured value (see §ii.1.6 for a more sophisticated treatment). However, the resolution of the
measurement tool itself could be larger than this spread, in which case the quoted precision
will be dominated by the instrument resolution.
Exercise ii.1 A researcher needs to determine the diameter of a 10 cm long quartz
rod to make sure it will fit the mirror mounts for an optical cavity. The calipers
used have a digital display whose most-precise digit is in the 0.01 mm place. 10
measurements along the length of the rod all yield the same reading on the display,
5.98 mm. It is clear that the measured best value is dbest = 5.98 mm, but how can
the uncertainty δd of the measurement be determined?
ii.1.4 Computer use and too many digits
A common mistake that students make in reporting measured values is to have too many
(way too many) digits on their numbers. After all, the computer will report a measured value
often to 16-bit precision, which gives a relative precision of 10−5. However, just because a
12
computer gives you a number with 5 digits of precision does not mean that the measurement
is accurate at that level. If you have a computer multiply a raw measured value of something
by π, for instance, the computer will happily tell you the answer to 50 digits. This may look
impressive at first, but if the original number is only measured to 2 digits, all of that extra
stuff is nonsense.
For example, if you see that repeated measurements of the same quantity fluctuate at
about the 3rd decimal place, there is not going to be much useful information in the 8th
decimal place and it should probably not find its way into your report in any form. In this
case, it would make sense to hang on to maybe 4 or 5 digits during the calculations and for
making tables of your raw data. However, the uncertainty is likely to be in the 3rd digit in
the end, in which case you will be throwing out everything after the 3rd digit in your final
reported number and there is no reason to keep hauling those 8th digits around, cluttering
up your spreadsheets and implying an unrealistic uncertainty in your numbers. If you keep
things simple, you will find that the physics is easier to see!
ii.1.5 Sources of uncertainty
There are many factors that can contribute to uncertainty in measured quantities. These
sources are often separated into two types, systematic uncertainty and statistical uncertainty.
This is a slight oversimplification, but for our purposes in this course, sources of statistical
uncertainty tend to produce a random distribution of data points about the mean upon
repeated measurements of the same quantity. As such, statistical uncertainty affects the
precision of measured values, but not the accuracy.
Systematic uncertainty, on the other hand, affects all of the measured data points in the
same way and therefore does not contribute to the statistical spread. Systematic uncertainty
limits the accuracy of a measurement, but not the precision of the measured value. If you
need to compare your measured value to a known true value (for instance, perhaps in a
measurement of g), you will need to consider systematic effects, particularly if you find that
your measurement was inaccurate (the true value does not fall within your uncertainty of
your measured best value).
However, we will primarily be concerned with quantitative assessments of statistical un-
certainty in this course. To reiterate an earlier point, you are likely to encounter two main
types of statistical uncertainty in this course, those due to finite instrument resolution and
those due to “noise” sources that cause repeated measurements of the same quantity to fluc-
tuate. The quantitative method for combining multiple sources of uncorrelated uncertainties
is covered in section ii.2.
ii.1.6 Estimation of Statistical Uncertainty in a Mean
One way to estimate the statistical uncertainty in a measurement is to repeat the mea-
surement many times and to look at the spread in measured points. This method is only
applicable in cases where the precision is limited by statistics (instead of, for instance, in-
strument resolution), but will be commonly encountered and so we provide a summary of
13
the procedure here.
Let us assume we have a set of Ndata points xithat all measure some quantity x. If
we do not have a good estimate for the uncertainty in each xi, we can still combine them to
come up with a best value and uncertainty in xby looking at their statistics.
For our purposes in this course, we may assume xbest = ¯x, which is to say that the best
value for our measurement of xis the mean of our measured points
¯x=1
N
N
X
i=1
xi.(ii.11)
One commonly used method for estimating the uncertainty in our knowledge of xis to
use the standard deviation (σx) of the collection of points xi. The standard deviation is a
measure of the spread of points around a mean value. In our case, we should use the so-called
sample standard deviation1, given by
σx≡v
u
u
t
1
N−1
N
X
i=1
(xi−¯x)2.(ii.12)
The reason we divide by N−1 instead of Nis that ¯xwas determined from the data
points themselves and not independently. In Microsoft Excel, the functions STDEV() and
STDEV.S() both calculate the sample standard deviation using the formula above and you
may use either one if you don’t want to enter that formula manually.
While it is probably okay in most cases in this course to use the sample standard deviation
as an estimate of the statistical uncertainty in the mean, there is something about this that
is missing. Specifically, the problem with using δx =σxis that it does not really get smaller
as you collect more data. However, if we take 100 times as many data points as we had and
they have the same spread as our original data set, we should be able to use this to come
up with a significantly better measurement of xsince we get to average over 100 times as
many individual data points. Specifically, we should get a statistical uncertainty that is 10
times smaller if we take 100 times as many data points, which fact is evident when we use
the following formula to calculate our statistical uncertainty:
δx =σx
√N=1
√Nv
u
u
t
1
N−1
N
X
i=1
(xi−¯x)2.(ii.13)
You may apply Eq. ii.13 anytime you are trying to estimate the statistical uncertainty in the
mean of a collection of data points based only on the distribution of those points.
ii.1.7 Summary
When writing your lab report, you may at times choose to include raw data in your report.
This will likely take the form of a table or plot of some kind. Raw data does not need to have
1There is also something called a population standard deviation, which differs from Eq. ii.12 in that the
factor of 1/(N−1) is replaced by 1/N. The population standard deviation is appropriate in cases where the
mean of the parent distribution is determined independently from the data.
14

explicit uncertainties for each entry unless you are specifically asked to provide it. However,
even your raw data must have units labeled. This can be done in column headers or plot
axis labels, but there should not be any room for ambiguity on this point.
•Every single number in your report that describes a quantity that has units must have
its units clearly labeled in your report.
When reporting the results of measurements in your report (as opposed to just showing
examples of raw data), you will be expected to report the measured best value and the
uncertainty. When you quote the final measured value of a quantity in your report, be sure
that
•You report the measured best value and the uncertainty: q=qbest ±δq
•You include proper units
•The measured best value and the uncertainty are written with the same absolute
precision
•The uncertainty has no more than two significant digits
ii.2 Propagation of uncertainties
Fairly often in the laboratory we will be measuring many different quantities and combining
those measurements together in some mathematical way to come up with a measured value
for a composite quantity. For instance, let’s say we wanted to measure the average velocity
of a glider on an air track (Fig. 1.1) by using a stopwatch to measure the time tit takes
the glider to travel some distance x. Using the guidelines above, we will have assigned an
uncertainty to each of these quantities, so we want to know how to turn our measured values
for the parameters xbest ±δx and tbest ±δt into a measured value for the composite quantity,
the average velocity v=vbest ±δv.
The way we do this is by using the functional form of how the composite quantity
depends upon the input parameters to mathematically determine the resulting uncertainty
in the composite quantity. It is important to use the following methods only in the cases
where the uncertainties are uncorrelated, so be sure that the uncertainties are generated by
physically independent mechanisms (statistical uncertainties typically fall firmly into this
category).
We will write the expression relating the uncertainty δf of some composite quantity fto
the uncertainties (δx, ···, δz) of the parameters used to compute f=f(x, ···, z) without
proof or derivation (see, e.g., [1]):
δf =v
u
u
t ∂f
∂x δx!2
+··· + ∂f
∂z δz!2xbest,···,zbest
(ii.14)
15
where the vertical line on the right side instructs us to evaluate the resulting expression at
x=xbest,···, z =zbest.2
If you are not familiar with the notation of those derivatives (∂f/∂x), they are called
partial derivatives and simply instruct you to treat everything except the variable of differ-
entiation as a constant when taking the derivative. If we come back to our example of the
measurement of the average velocity of a glider, we first identify f=v=x/t. We need
to determine δv in terms of xbest,δx,tbest, and δt. We can begin by taking the partial
derivatives: ∂v
∂x =∂
∂x x
t=1
t(ii.15)
and ∂v
∂t =∂
∂t x
t=−x
t2.(ii.16)
This gives us
δv =s1
tδx2
+−x
t2δt2xbest ,tbest
=1
√t2s(δx)2+x2
t2(δt)2xbest ,tbest
=sx2
t2v
u
u
t δx
x!2
+ δt
t!2xbest ,tbest
.(ii.17)
Normally, we would say that qx2/t2=±x/t to reflect the two solutions of that equation.
However, we have defined all uncertainties (δq in Eq. ii.1) to be positive, so here we take the
absolute value: sx2
t2
=
x
t.(ii.18)
We can now evaluate our expression at x=xbest and t=tbest to give us
δv =
xbest
tbest v
u
u
t δx
xbest !2
+ δt
tbest !2
.(ii.19)
We have identified vbest =xbest/tbest, so dividing both sides by the absolute value of this
quantity and noting that we can add an absolute value sign to both terms under the radical
since they are squared anyway gives us the following symmetric-looking form:
δv
|vbest|=v
u
u
t δx
|xbest|!2
+ δt
|tbest|!2
.(ii.20)
2The assumption we are going to invoke here is that fbest =f(xbest,···, zbest). This is sometimes not
strictly correct, but will serve for the purposes of this course.
16
The quantity δq/|qbest|is called the fractional uncertainty or relative uncertainty of some
measurement of q, so we can restate Eq. ii.20 in words by saying that the fractional uncer-
tainty in a ratio of two quantities is given by the quadrature sum of the fractional uncer-
tainties of the two quantities.
ii.2.1 Specific formulas
While form given in Eq. ii.14 is sufficiently general to allow it to be applied directly for every
case encountered in this course, we can summarize some of the most common results that
can be derived from it as follows.
•Measured quantity times an exact number: f=Ax
δf =|A|δx (ii.21)
•Sums and differences (they follow the same rule): f=x+y−z+···
δf =q(δx)2+ (δy)2+ (δz)2+··· (ii.22)
•Products and ratios (they follow the same rule): f=x×···×z
u×···×w
δf
|fbest|=v
u
u
t δx
|xbest|!2
+··· + δz
|zbest|!2
+ δu
|ubest|!2
+··· + δw
|wbest|!2
(ii.23)
•Measured quantity raised to an exact number power: f=Axn
δf
|fbest|=|n|δx
|x|(ii.24)
•Exponential with measured quantity in the exponent f=Aeax
δf
|fbest|=|a|δx (ii.25)
You may recognize that Eq. ii.20 is a special case of Eq. ii.23, as is Eq. ii.21. And, of
course, all of these are special cases of Eq. ii.14. These cases are not the only possibilities
for functional forms of the relationships between measured values, but they are the most
common for us in this course. You should be sure to keep in mind that these formulas are
valid only for uncertainties that are uncorrelated.
17
Experiment 0: Sensor Calibration and
Linear Regression
Your first experiment is intended to familiarize you with some of the basics of the DAQ
system. There will not be a formal report due next week. Instead, there is a short set of
homework problems (“Homework 0”) at the end of this chapter that you will complete and
turn in online. Be sure to read through the homework problems to make sure you have saved
all of the data you will need to complete Homework 0.
For many experiments in this course, you will be using a force sensor similar to the one
shown in Fig. 0.1. This device is designed to output a voltage that is proportional to the
force applied between the hook and the body of the sensor. The DIN cable allows your
computer to read this voltage via the 850 interface. If Force Sensor is selected as the type
of input, the 850 will assume it knows how to convert voltage to force and will simply read
in units of force (N). However, to illustrate how the interface really works, we will not trust
this conversion implicitly and will instead be looking at the raw voltage from the sensor.
The process of determining how voltage corresponds to force is an example of a process
called calibration, and Experiment 0 walks you through a manual calibration procedure for
this force sensor. This procedure is not the same as the Calibration feature in Capstone,
and the manual calibration described here will be far more accurate than that supplied
by the manufacturer. Plastic deformations of the sensor components, changes in ambient
conditions, and electrical offset sources may all change between experiments, so to increase
the accuracy of your force measurements, you may consider checking the sensor calibration
each week that it is used. Your TA may or may not ask you to do this explicitly.
0.1 Hardware setup
To set up the force sensor to read its raw voltage, start your DAQ
system using the instructions in §i.2 and connect the DIN cable to one
of the Analog Inputs of the 850. Set up the 850 to read voltage from
the appropriate analog input (see §i.3) by choosing User Defined Sensor
from the drop-down menu of the appropriate input (Do not choose
Force Sensor), at which point your Hardware Setup pane will show a
little electricity-looking icon with a line to the channel you chose.
To test that you have set up the connection correctly and that there are no software or
hardware errors happening to keep you from reading the sensor correctly, use the Scope
18
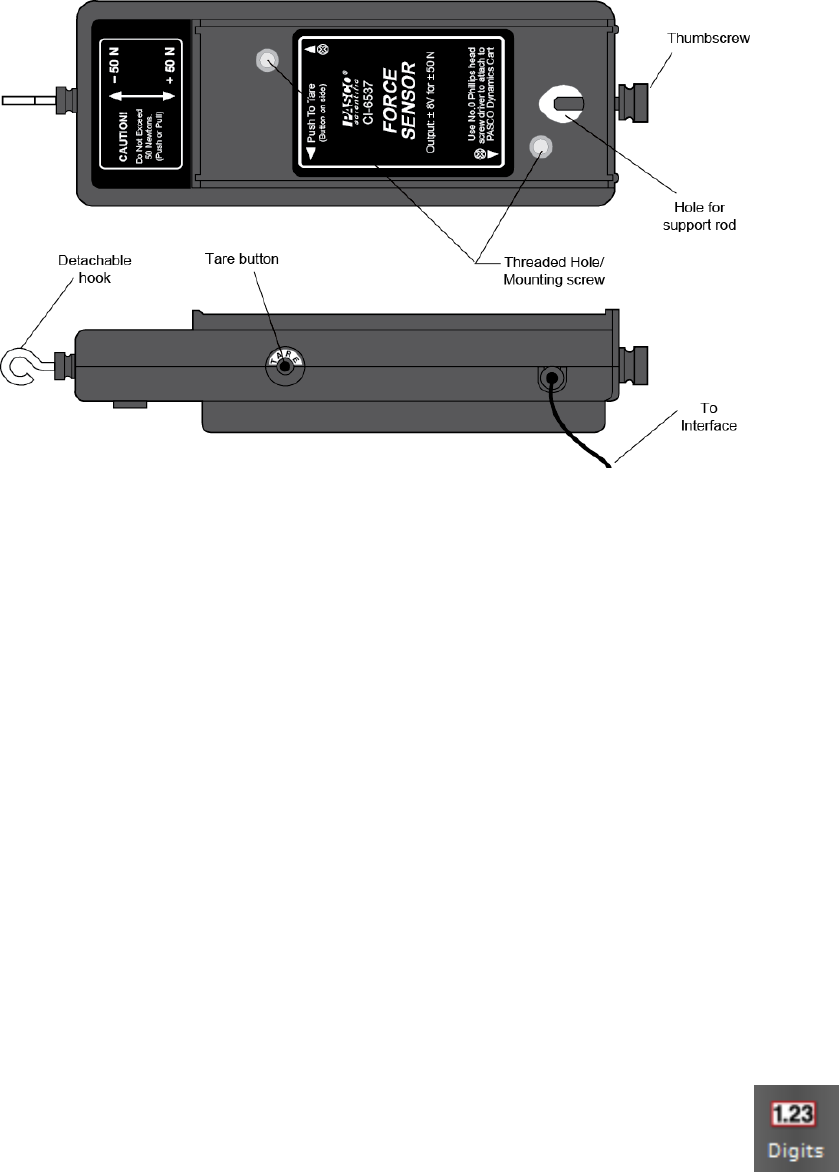
Figure 0.1: PASCO CI-6537 force sensor. This transducer can sense both tension and
compression applied between the hook and the body of the unit, up to 50 N in either mode.
feature in Fast Monitor Mode to see that the voltage changes when you push or pull a
little on the hook. An overview of how to use the Scope feature can be found in section
i.4. If you cannot see any signal from your sensor on the screen, go back through these steps
carefully to see if you missed something; if not, ask your TA for help.
0.2 Manual calibration procedure
Once your DAQ is able to read voltage from the force sensor you are ready to begin the
calibration. The procedure will be to attempt to zero, or tare the sensor, and then to hang
known weights from the hook while monitoring the voltage. You will record several readings,
then use Excel or whatever other program you like to fit a line to the data. The slope and
offset of this line is the calibration curve for your sensor. The offset will almost certainly not
be zero, which will tell you something about how well the tare procedure worked.
0.2.1 Measurement Apparatus
First, attach the force sensor to a horizontal post so that the hook hangs vertically
downward, appropriate for hanging weights from it. The best software tool for
measuring the voltage is probably the Digits feature in the Displays palette. If
the Displays palette is missing, see section i.2 for instructions on how to get it
back onto the screen. You can configure your Digits display to show the voltage produced
19

by the sensor by choosing User Defined (V) from the drop-down menu of the
button.
Your calibration of the force sensor will be capable of accounting for an
offset in the reading caused by a nonzero voltage reading with no applied force.
However, it is useful to know how to tare the sensor in hardware for calibration
and making differential measurements. Using Fast Monitor Mode with your
Digits display, you should be able to read the offset voltage in real-time by clicking Monitor
in the controls palette.
The voltage you see displayed will fluctuate around some value that is not necessarily
zero. You can define this offset to be 0 V by pressing the tare button on the side of the sensor,
shown in Fig. 0.1. The voltage reading will now be centered near zero. Your calibration will
tell you how well this procedure worked when we produce a fit line.
0.2.2 Changing the Display Precision
You may notice that your sensor reading fluctuates around as a function of time, even
after taring the sensor. This gives you some idea of the precision of the sensor, which
determines the minimum size of the steps between distinct values. You will need to observe
and record the magnitude of fluctuations like this in various sensor readings throughout the
course, because they are an important part of your uncertainty determination for measured
quantities.
Aside from the precision of the sensor, it is possible that Capstone is only displaying a
subset of the digits it actually gets from the sensor. This is called the display precision of
your DAQ. It will be crucial in later labs to be able to change the display precision to avoid
becoming limited by this (which is known as quantization error ).
To change the display precision, mouse-over the Digits display to pop up
the menu bar at the top. On the far left side of the menu bar, there will be two
buttons to control the display precision, one to increase it and one to reduce
it. Click each one a few times while watching your sensor reading to get a feel for how the
system works. Determine the maximum number of digits that Capstone is willing to display
by increasing the precision until the button no longer changes the display. Typically, the
ideal display precision is just at the point where the reading fluctuations show up in the last
digit.
Next, we will record the voltage reading for a series of known weights hung from the
hook. To get a steady reading, it will be helpful to turn down the sample rate to something
on the order of a couple of times per second. This can be done in the Controls palette at
the bottom of the screen.
20
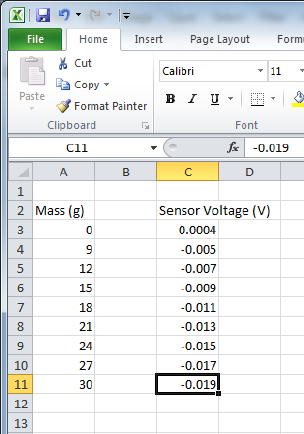
Figure 0.2: Some fake data entered into Excel in two columns with an empty column next
to the mass column. Note the unit labels on the column header.
0.2.3 Entering and Plotting Data in Excel
Next, we will be collecting our data in a table, so start a new workbook in Excel and record
the voltage reading and mass of a series of weights hung from the force sensor hook. Now is
probably a good time to get in the habit of labeling the columns of tables with descriptive
labels and appropriate units, as shown in Fig. 0.2. Since you will be converting mass to force
using F=mg, it is a good idea to leave a blank column next to the column where masses
are recorded so that we can make Excel do the work of filling in the force column.
To have excel fill in the force column with the calculated force in N, highlight the top
cell of the new column (right next to the cell with your first mass value entered) and enter
the formula you want Excel to execute. Formulas in Excel begin with an =sign, followed
by the mathematical expression you want, where variables can be entered either manually,
or through reference to the cell address in which the variable value has been entered.
For instance, for the data shown in Fig. 0.2, to make cell B3 display the gravitational
force (in N) acting on the mass entered in cell A3, one could type in “=A3*0.001*9.80” and
hit enter. In this case, the factor of 0.001 comes from converting g into kg and the factor of
9.80 is for g= 9.80 m/s2. The formula can now be copied down the column by highlighting
the cell with the formula, grabbing the bottom right corner of the cell, and dragging it down
the column. Excel will recognize that “A3” should be changed to “A4” in the formula for
cell B4 and so forth.
Once you have two columns recording the applied force and the output voltage, you can
plot your data. Excel calls this a “chart” (because Excel is made for businesspeople) and
you can create one by highlighting the two columns with your data and choosing Scatter
21
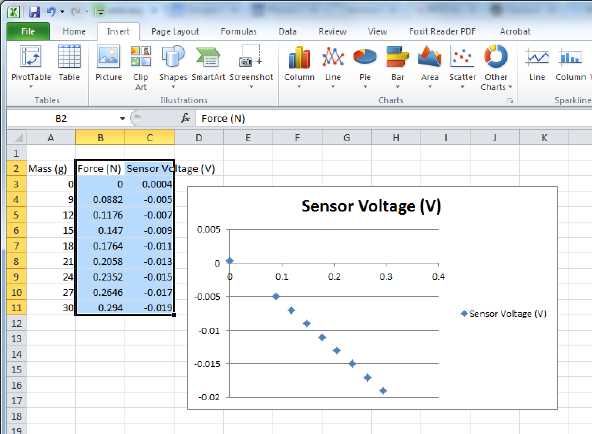
Figure 0.3: Excel has created a terrible-looking plot. In the opinion of your author, what
Excel lacks in the appearance of its plots it makes up for in ease-of-use for students, which
is why it is recommended here. If you have other programs you prefer to do your own data
analysis, feel free to use them.
from the Insert →Charts menu or something similar in whatever version of Excel you are
using.
If you had Excel make a scatter chart for you in this way, your workbook probably looks
similar to that in Fig. 0.3 This plot has no axis labels, so it takes a little looking to even figure
out which column was used for the y-axis and which for the x-axis. I would recommend that
the first thing you do when creating a plot in Excel is to label the axes and units for those
axes. How this is done will depend upon your version of Excel, but you’re probably looking
for something like Chart Tools →Layout. Using the features you have at your disposal
and a bit of web searching, you can hopefully figure out how to make a reasonably clear plot
of your data, such as that shown in Fig. 0.4. All plots that end up in your report must have
axis labels and appropriate units, as well as a figure caption with a title, description of what
is displayed, and possibly a fit line equation. Instructions for how to incorporate plots into
your reports can be found in Appendix B.
0.2.4 Linear Fit to Numerical Data
Next, we would like to have Excel calculate the equation of the best fit line to our data. To
do this, you will have Excel “add a trendline” to your data. Since we expect the voltage to
be linear in the applied force, we are looking for an expression in the form
V=aF +b(0.1)
22
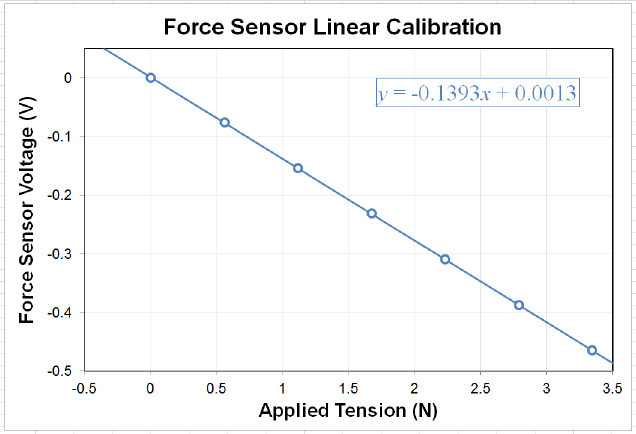
Figure 0.4: A fairly clear plot of some data shown with a linear fit line. Note the title, axis
labels, units, and fit line equation are all clearly visible, and none of the tick labels or text
is overlapped with anything that makes it difficult to read.
to relate the measured voltage Vto the applied tension Fin terms of the slope aand offset
bof our sensor. Note that aand bwill have units, that they constitute the results of a
measurement, and that they need to have a quantitative uncertainty associated with them.
We will use Excel to calculate aand bfor us by having it add a linear trendline (a.k.a. a
linear best fit) to the data and display its equation on the chart, as shown in Fig. 0.4. The
procedure for adding a trendline will vary from version to version of Excel, but right-clicking
on one of the data points or going to Chart Tools →Layout are likely places for this
option to be hiding. In the trendline options, remember that we want a linear trendline,
we do not want the offset set to a particular value (so it will remain a fitting parameter;
your sensor may still have an offset despite the tare procedure), and we want the equation
displayed on the chart.
A guide to making figures and captioning them using Word and Excel by Julio S. Ro-
driguez, Jr. and Anthony Ransford can be found in Appendix B.
0.2.5 Calculating Uncertainties in a Linear Fit with Excel
For the example data of Fig. 0.4, we find that the linear fit line is described by the equa-
tion y=−0.1393x+ 0.0013. Comparing this to Eq. 0.1, we are tempted to identify
a=−0.1393 V/N and b= 0.0013 V and declare victory. However, we still need the un-
certainty in the measured values (δa and δb) used to generate this fit before we can report
the results of a measurement. As is always the case, we cannot claim we measured something
(in this case, the sensor calibration) without also describing the uncertainty associated with
23

this measurement.
One question that we can ask ourselves is “why is there an uncertainty at all? After
all, there is a single, unique fit line that is the very best fit to these data. Why would that
have an uncertainty?” This is a very good question. The answer is that the scatter in the
data will effectively permit a series of different lines that all essentially model the data to
the same degree. For data with a lot of scatter, the “goodness of fit” of the best fit line is
only a very tiny amount “more good” than a whole collection of lines with similar slopes
and offsets, so the uncertainty will be large. For data with very little scatter, a small change
in the slope or offset will substantially degrade the “goodness of fit,” and the uncertainty is
correspondingly small. Excel provides you with the quantitative tools to evaluate statements
like this in a scientific manner, and we will use this throughout the term.
The trendline feature in Excel was a quick and easy way to get the best-fit slope and
intercept from a least-squares fit, but it gives us no quantitative way to determine the
uncertainty in the slope and intercept that it found. There are two ways your author knows
of to get the uncertainties in linear fits in Excel. The first is to use an Excel function called
LINEST that returns an array and is powerful, but not incredibly user-friendly. The other
way is to use the Regression tool.
The regression analysis tool will take as its input your two columns of data and give you
lots of information about how well your data can be described by a linear relationship. For
instance, it will calculate the same equation for the best fit line that we got from the Add
Trendline feature, but it will provide more digits, and (most importantly) it will provide
the uncertainties in the fit parameters. These uncertainties are based entirely on the scatter
in your data points around the fit line and therefore do not contain any information about
systematic uncertainties. If your data span a range much larger than the resolution of your
measurement tool (probably true in this case), the scatter in your data points already reflects
this resolution limit implicitly.1
To perform a linear regression analysis in Excel, look for the regression tool somewhere
like Data →Data Analysis or Tools →Data Analysis. One of the data analysis tools
should be Regression. You will enter the location of your xand ydata, and I would
recommend having the output option set to new worksheet, such as shown in Fig. 0.5.
Figure 0.6 shows the results of the linear regression analysis, along with some high-
lighting of the parameters we were interested in calculating. The regression tool gives the
uncertainties in the slope and intercept in the column called Standard Error .
At this point, it would again be tempting to think we are done and write a= (−0.139320505±
0.0000776577) V/N and b= (0.001334872±0.000181541) V and call it a day. After all, we’ve
got the uncertainties and the units, right? The problem with this is that the precision of
these reported numbers is much, much better than the uncertainty indicates it should be.
The statistical uncertainty given by, for instance, δb = 0.000181541 V tells us immediately
that the last four or five digits of this number are meaningless. The data itself fluctuates at
1If your data do not span a range larger than your instrument resolution, all of your data points are
probably identical, and the instrument resolution is likely the largest source of statistical uncertainty in this
case.
24
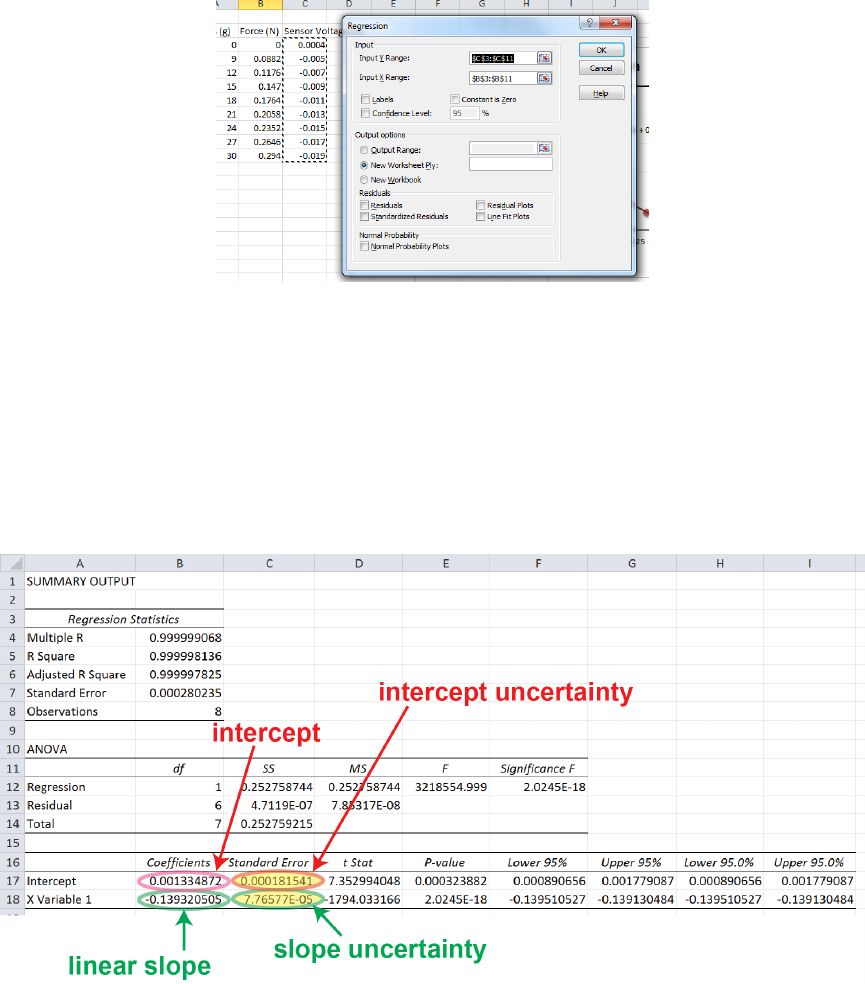
Figure 0.5: The linear regression tool in some version of Excel. Your version may look a
little different, but the basics will be the same.
Figure 0.6: The results of using the linear regression tool in Excel. The best-fit line slope
and intercept are highlighted, along with their uncertainties.
25
a level that is way too large for us to say anything about bon the 10−9V scale. You can
always keep just one nonzero digit on an uncertainty and not get yourself into any trouble
in this course, so that would give us δb = 0.0002 V and δa = 0.00008 V/N. The best values
should then be written with the same absolute precision, and we have our final result for the
measured calibration of this force sensor:
V=aF +b(0.2)
a= (−139.32 ±0.08) mV/N
b= (1.3±0.2) mV (0.3)
As a quick sanity check on these measured results, we note that the last digit of abest is in
the 0.01 mV/N place, which is the same position as the last digit in δa, as it should be. The
same is true for b: the last digit we quote for bis in the 0.1 mV place, as is the last digit of
δb.
0.3 Homework 0
Read through these problems before you leave the lab to ensure that you have
saved all of the data you will need. This short exercise is designed to prepare
you for your first real experiment, Experiment 1. Your completed homework
must be a short document that is typed and formatted using a computer.
During your lab section for Experiment 0, your TA will give you a due date
and instructions for how to turn in your completed homework. You may find
that the information in Appendix B is useful for completing your homework,
so be sure to take a look at Appendix B.
1. You will need a cover page for your reports in this course, so now is a good time to
make a template for yourself you can reuse later. Make a cover sheet that includes the
following information:
•Experiment number and title. This week, this will be something like “Experiment
0: Sensor Calibration and Linear Regression”
•your name and UID
•the date the lab was performed
•your lab section (for instance, “Wednesday 9am.”)
•your TA’s name
•your lab partners’ names
2. A student is trying to measure the constant acceleration of some object in the lab by
measuring the change in velocity of the object over some fixed time interval. Repetition
of the experiment many times and careful error analysis has resulted in a measurement
26
of the velocity change (∆v= ∆vbest ±δ∆v) and the time duration (∆t= ∆tbest ±δ∆t)
with uncertainties. Knowing that the measured acceleration is given by a= ∆v/∆t,
what is the proper expression for the uncertainty in the measured acceleration, δa?
3. What is the maximum number of digits Capstone will display (recall §0.2.2)? If the
sensor precision is exactly 4 digits and you take data with 10 displayed digits, describe
what your actual measurement precision is and what you would expect to see in digits
5 through 10. If the display precision is turned down far enough, it may be possible
to completely eliminate sensor fluctuations. Describe why doing this might not be the
best idea for taking good data.
4. Present your final plotted results of the calibration procedure as a figure (include the
data, the fit line, and the fit line equation on the plot). You should take the time to
make the plot look clear and have all of the axes labeled. Present a brief line or two
of text that could be used as a figure caption for this plot.
5. State the results of your calibration (with uncertainty from your fit). This may require
a sentence or two of explanation to clearly define your parameters, but you do not need
to describe the whole procedure of the experiment. Your fit should have included a
possible nonzero y-intercept. What does your value for this intercept tell you about
the effectiveness of the taring procedure (§0.2.1)?
6. Equations 0.1 and 0.3 tell us how to convert a known tension Finto a predicted output
voltage of the sensor V(including the uncertainty in this conversion), but to use this
as a “force sensor,” we really want to know how to convert Vinto F, right? Use the
measured values of abest ±δa and bbest ±δb that you presented in your response to the
previous question to rewrite your calibration curve in the form F=cV +d, including
the appropriate uncertainties in cand d. Hint: look at Eq. ii.23.
7. You have been provided with information on how to access the syllabus for this course.
Read the syllabus, paying special attention to the text in red. Consider a case where two
students, Frankie Fivefingers and Avril Armstrong are in different sections of Physics
4AL this quarter. At the end of the quarter, they both have an average numerical score
of 84, but Frankie gets a B+ in the course while Avril gets a C+. How is this possible?
What can you conclude about the mean numerical scores of the other students in their
sections?
27

Experiment 1: Uniform Acceleration
Experiment 1 is the first experiment for which you will be expected to submit a full-credit
report. Your report is due about a week from now, and your TA or instructor will give you
specific instructions on how to submit your report as well as when the due date is.
In Experiment 1, you will be measuring the acceleration of a composite system composed
of some masses connected by a string passing over a pulley. The position of the accelerating
masses and string will be monitored as a function of time by the DAQ system, and these
data will be recorded for a series of 5 different masses. A bit of data analysis is required to
produce a plot that can be fit to a straight line to obtain a measurement of the acceleration.
You should already have experience working with the DAQ (chapter i), producing linear
fits with uncertainties (§0.2.4 & §0.2.5) and doing a proper uncertainty analysis of your
complete measurement (chapter ii), so you may find those sections to be helpful references
for Experiment 1.
In order to measure constant acceleration, the friction between the accelerating object
and everything that is stationary must be kept to a minimum. While there will still be some
residual friction (as well as drag forces), the air track setup depicted in Fig. 1.1 does a good
job of minimizing its effect. A “glider” placed on this track will float along on a cushion
of air when the pump is pressurizing the track, exhibiting nearly-ideal linear motion. The
glider and air track setup will be used for many experiments in this course.
Fig. 1.2 shows the air track set up for Experiment 1. A glider with total mass Mis
supported on a cushion of air on a level air track and is attached to a hanging weight (mass
m) by a string that passes over a pulley.
The equation of motion for this system is given by:
a=gm
m+M(1.1)
where ais the acceleration of the glider-string-weight system and gis the earth’s gravitational
acceleration for a free-falling body.
In this lab, you will be measuring the acceleration for 5 different values of m(do not
exceed m= 50 g). Feel free to vary Mas well for any of these runs. The acceleration
measurement will require that you convert position vs. time data into a measured value for
acceleration (with uncertainty, of course!). These position vs. time data will be recorded by
the DAQ using a “smart pulley.”
28
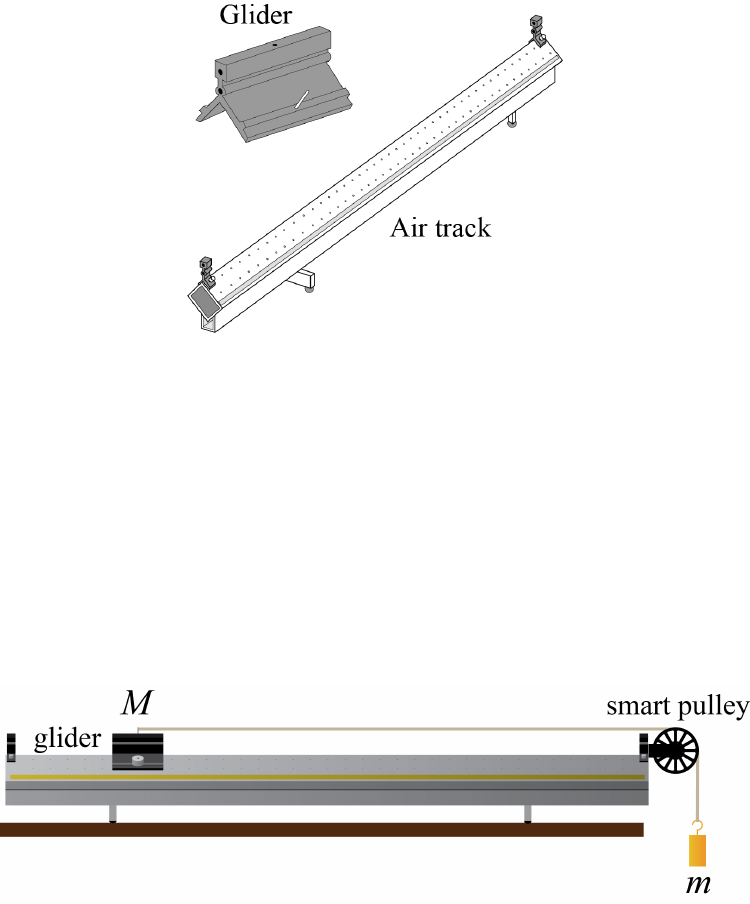
Figure 1.1: Apparatus for linear motion with very low friction. The air track is pressurized
by a continuously-flowing pump, causing air to flow out through the array of holes on the
surface. When a glider is placed on this track, it moves freely in 1D with very little friction,
just like an air-hockey puck.
Figure 1.2: Experiment 1 setup. The glider mass can be changed by putting different
amounts of weight on its sides, and the hanging mass can be changed by adding more
weights with hooks. The smart pulley is attached to the DAQ and records the timing as the
spokes pass by a sensor.
29
1.1 Procedure
In order for Eq. 1.1 to be valid, we have assumed that the air track is level. This is generally
not the case, and there will be multiple experiments in this course that use the air track.
Your first task is to level the air track. You can accomplish this by placing a glider on the
track and turning on the blower. The track itself is not completely flat, so there is really no
such thing as “perfectly level” over the whole length of the track, so do the best you can.
You will find that the two feet under the cross-bar of the air track (one is visible sticking
out from the bottom of Fig. 1.1) can be turned to change their height. Your goal is to get
to the point where a free glider won’t move either direction with the blower on.
You will also need to set up your DAQ to work with the smart pulley, so plug the smart
pulley cord into a digital channel on the 850, turn it on, and start Capstone. Configure the
appropriate channel for Photogate with Pulley.
The smart pulley really only has the ability to measure whether or not a wheel spoke is
blocking a little infrared LED from shining into a photodiode. The LED and photodiode
are set up with a gap between them, which is called a photogate. Every time the photogate
goes from being unblocked by a spoke to being blocked by it, the 850 tells Capstone the time
stamp for that event, so the raw data in this case is really the time stamp for each time when
the photodiode detects a “falling edge.” Therefore, no data is collected if the pulley isn’t
moving, or during the time when the spoke is moving but hasn’t yet blocked the photogate.
Furthermore, the photogate has no idea which direction the pulley is moving, so you should
be careful when interpreting results. You may assume that the linear distance the perimeter
of the pulley moves between successive times when the photogate goes from being unblocked
to blocked is given by
κSP = (1.50 ±0.05) cm/block.(1.2)
You may notice that Equation 1.2 is given in the from κSP =κSP,best ±δκSP, but you are not
told whether the uncertainty δκSP should be treated as systematic or statistical in nature.
You may take this uncertainty to be statistical for the purposes of your analysis, in which
case it will already be taken into account when you do your regression analysis, because this
effect will show up as scatter of your data points.
Capstone has the ability to perform some computational tasks on the data and to use its
own algorithms and calibration of the hardware to display position, velocity, acceleration,
etc. This will be useful for future tasks, but for Experiment 1, you should save the raw
data (Block Count () vs. Time (s)) and use your own data analysis to convert this into 5
acceleration measurements (with uncertainties, of course!). This is designed to help you
understand how to turn raw experimental data into a measured value with uncertainties,
where you have full knowledge and control over the algorithms being applied to the raw
data.
Fig. 1.3 shows an example with some fake data. It is always helpful to have a visual
representation of the data you are collecting in real-time, because this can give you a quick
feel for whether something is not set up properly. For this, it might be a good idea to open
aScope or Graph display to show, for instance, Linear Speed (m/s) vs. Time (s).Since
30
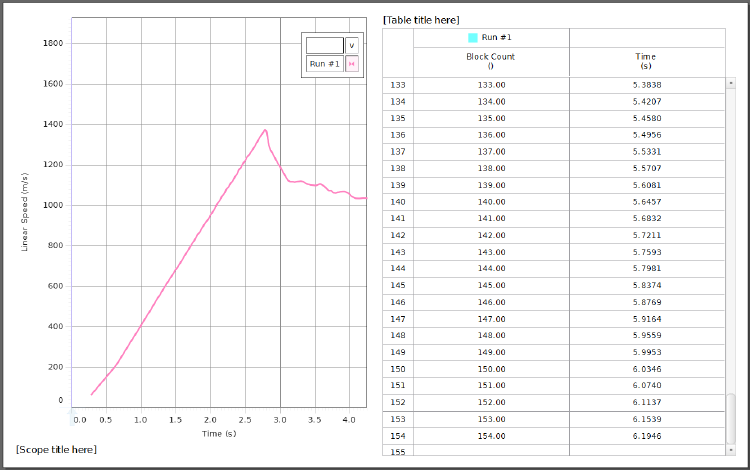
Figure 1.3: Example data from Capstone showing the simultaneous generation of a Scope
plot qualitatively showing the portion of the data that will be useful and the Table that can
be used to cut-and-paste your data into Excel for saving and analysis. Note that the Scope
plot shown here is showing a glider moving at about Mach 4, so don’t trust it! It is only a
qualitative tool to help you view your data in real-time.
31

you do not know how Capstone calculates linear speed, you will not use this. The only data
you should use will be Block Count () vs. Time (s). The display of speed is only to give you
a visual cue of your data. Since you will also be saving your data to analyze it , you will
also need to open (simultaneously) a Table for collecting your data. Remember, the two
columns you will need to generate are Block Count () and Time (s). Do not save a column for
Position,Linear Speed,Linear Acceleration, or any other pre-processed quantity. Continuous
Mode in the Controls palette should work for gathering your data. The sample rate seems
to have no effect for this hardware.
With your lab partner, you will be saving 5 data sets (each with a different set of masses).
Keep in mind that you will need to measure the mass of the glider, so don’t forget! Once the
DAQ is set up, you should be able to generate a plot something like that shown in Fig. 1.3.
The Scope plot in that figure shows the point where the velocity stops increasing linearly,
which is when the weight hits the floor or the glider hits the pulley, whichever comes first.
You may use this plot to decide which data to save (you are only interested in the linear
portion). Be sure to increase the number of displayed digits in the table until you are no
longer precision limited before exporting your data (look at the Time (s) column in Fig. 1.3).
Highlight the data you want to save in the Table and copy and paste it into Excel. You
should then save it somewhere safe. Each person should get 5 data sets with different masses
for mand/or Mthat are unique. You may use the same masses as your lab partners, but
the raw data should be unique for each person.
1.2 Analysis
The data you saved is in the form of something called “Block Number” vs. time. Using
Eq. 1.2, you can create a new column for position by simply multiplying block number by
κSP,best. Remember, you have been instructed to ignore δκSP for this analysis since your
regression analysis will automatically take this effect into account, so you will really only be
using κSP,best. Next, you will need to numerically differentiate the position data. To do this,
you may calculate a single average velocity for every two position vs. time points using
¯vi=∆x
∆t=xi+1 −xi
ti+1 −ti
.(1.3)
Note that since each velocity measurement requires two position data points, you will have
one fewer data points for velocity than position. The column of velocities you generate now
needs a column of times associated with them. Since the formula in Eq. 1.3 gives you the
average velocity in the time interval between tiand ti+1, the best time to associate with
velocity ¯viis the average of tiand ti+1,
¯
ti=ti+ti+1
2.(1.4)
Armed with your two new columns, velocity and time, you can now generate a scatter
plot of your data. Be sure to truncate your data to only show the linear portion of your plot.
32
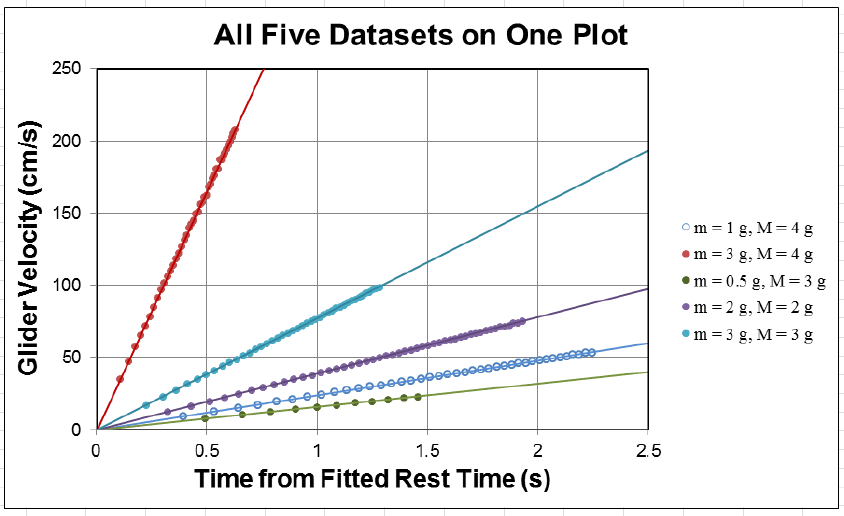
Figure 1.4: An example of five different data sets on one Excel scatter chart. Making the fit
lines the same color as the data points lends clarity to what would otherwise be an incredibly
cluttered plot. This is fake data with fake units, so please do not try to make your plot look
just like this one! The important thing is to make your results as clear as possible.
33
Using the techniques that were discussed in chapter 0, fit a line to the velocity vs. time for
each of your 5 data sets to produce 5 measurements of the acceleration (with uncertainties
obtained from the fits). Keep in mind that the reason we are obtaining uncertainties from the
fits here is that we expect the uncertainty to be dominated by the scatter of the individual
data points, not the systematic uncertainty of each data points. The fit line uncertainty
captures this effect beautifully. Note that since you are only interested in the slope, it is
probably best to leave the intercept as a free fitting parameter and then to ignore it since
the initial velocity may not have been zero when the spoke crossed the photogate. Compare
these measured accelerations to your predicted values.
1.3 Report guidelines
For experiments 1 through 5, your reports will be a single document but will have two sec-
tions. The first section will be called the worksheet, the second will be called the presentation
mini-report. The worksheet will contain your main results presented as numbered items. The
worksheet section is intended to be a basic report of what you did in the lab and how you
analyzed your data to draw conclusions, but does not need to be a self-contained, full lab
report (you will be writing full lab reports for experiments 5, 6, and 7 in later weeks). Most
of your grade for the worksheet section of your report will be based on your data taking,
analysis, and conclusions.
Each week until experiment 5 we will focus on one aspect of a formal written lab report
(such as the abstract, or figures, or citations), and you will be asked to produce some example
of this aspect for the presentation mini-report section of your assignment. The details of this
section will change from week to week and may or may not involve your data. Most of your
grade for this section will be based on the degree to which your presentation mini-report is
on par with high-quality published scientific presentations in physics journal articles.
This week, the worksheet will be worth 40% of your grade and the presentation mini-
report will be worth 60%.
1.3.1 Worksheet guidelines
Your worksheet should contain the following sections
1. Cover Sheet
•experiment number and title
•your name and UID
•the date the lab was performed
•your lab section (for instance, “Monday 3pm”)
•your TA’s name
•your lab partners’ names
34
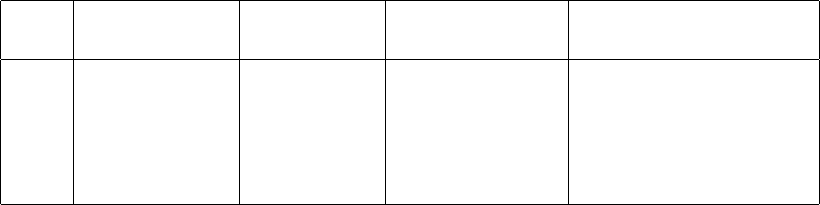
2. Plots
Include plots of all five of your data runs showing velocity vs. time and the fit lines.
This could be a single plot with five data sets on it or five plots with one data set on
each. Report the slope of the fit line for each (with its uncertainty, units, etc.) in a
figure caption.
3. Data Table
Report the main results from your experiment in a table such as the one below, where
your entires will be from your measurements and analysis. Your table does not need
to be exactly like this one, but should contain essentially the same information.
Hanging mass Glider mass Fit acceleration Predicted acceleration
Trial mbest (g) Mbest (g) afit (m/s2)apredict (m/s2)
1 5.0 15.0 0.23 ±0.01 0.25 ±0.08
2 4.0 20.0 0.19 ±0.01 0.20 ±0.07
3 3.0 25.0 0.100 ±0.005 0.08 ±0.07
4 2.0 35.0 0.300 ±0.006 0.29 ±0.05
5 1.0 45.0 0.200 ±0.002 0.30 ±0.06
4. Derivations
Derive equation 1.1 in this manual as well as your propagation of uncertainties used in
the table above.
5. Conclusions
Write a brief discussion of your results. Compare the accelerations you measure from
fitting your velocity vs. time data sets to what you would predict from Eq. 1.1 with
your measurements for the masses. Do they agree? If not, try to convince your reader
that the reason for the disagreement is understood and suggest a way to improve the
agreement in a future experiment.
6. Extra Credit (always optional)
Instead of obtaining a measurement of the acceleration by fitting a slope to the cal-
culated velocity vs. time, which only works in the case of constant acceleration, we
could have differentiated the data once more to obtain a direct measure of acceleration
vs. time. Try this with just one of your data runs and present a plot of your results.
Discuss your results and explain why the noise looks the way it does. Obtain a mea-
surement of the acceleration by using the mean of your calculated accelerations and
an uncertainty based on the procedure outlined in §ii.1.6. Compare the results of the
two methods and use this to draw a conclusion of which measure you believe is better.
1.3.2 Presentation Mini-Report Guidelines
Credit where credit is due
35
Formal scientific writing contains proper citation of the works whose ideas and results
are used. In a highly interconnected world, students (and professors alike, I might add) have
a vast wealth of material available to them that can occasionally find its way into their work
without appropriate citation of the person who deserves credit. Any time you use material
that has been generated by somebody else, you will be expected to either give appropriate
credit to that person’s work or to be able to successfully argue that it is common knowledge
that does not require citation.
For instance, if a student were to take a figure from the lab manual and to use it in their
report without giving credit to the author(s), a person reading the report will see the figure
and assume the student made this themselves. The student would be implicitly claiming
credit for something somebody else did and passing it off as their own. Claiming credit for
another person’s work is probably the second biggest “no-no” in science (behind fabricating
results). Furthermore, whether on North campus or South, a student’s turning in another
person’s work as their own is a clear violation of the UCLA standard for academic honesty,
and can carry with it fairly severe consequences. We will therefore be particularly diligent
about giving credit where credit is due in Physics 4AL, which is the main focus of this week’s
presentation mini-report.
A word of caution
It is easy to find many lab reports for Physics 4AL written by former and current students.
As discussed in the syllabus, your lab report must represent your own work. This includes
the logical presentation of the report, which is often the most difficult part of writing. If a
student uses another person’s work as source material for their own (even if they re-word
everything), they are using another person’s presentation and passing it off as their own,
which is a violation of academic and scientific ethics.
The same thing is true of figures (and their captions). You must generate all of the figures
in your reports on your own (the only exception to this is pictures from the lab manual that
depict the apparatus, as discussed below in §1.3.5). It is not okay to have, for instance, the
same figures as your lab partners, even if you cite them in an effort to give them credit; the
entirety of your reports must be generated by you.
verbatim text
It will almost certainly not be appropriate for you to include verbatim text from any
source in your work for this course (please see the syllabus if you are confused as to why
this is). It is important to understand that one cannot pull verbatim text from a source and
just drop a reference number and the end of the sentence or paragraph with a bibliography
citation – the reader deserves to know where the quotation started, as well as whether or not
you are taking someone else’s words. If you need to use verbatim text in a scientific paper, it
should probably have quotation marks around it and the full author’s name (in addition to a
reference designator and bibliography entry), or maybe even be its own paragraph, probably
with different indentation and font (italics are standard) than the rest of the paper. The
point is that the reader needs to be able to clearly identify when another person’s words are
36
being used.
Presentation mini-report on a current topic in physics research
For this week’s presentation mini-report you are tasked with writing a short review (no
more than 700 words) about a topic of your choice in contemporary physics research. This
will be a review-style paper where you can discuss the basics of some result in physics in the
past decade or so and what the likely next steps may be in the field. Your target readers are
scientifically-literate non-specialists; your goal is to inform them of the main points driving
the field and the likely directions it will go in the future. This should be written as if it were
to be published in a scientific journal, which means you will need to properly cite research
and review articles in scientific and engineering journals, with full citations in a bibliography
(the bibliography does not count toward the word count). Please do not cite any source that
is not either a journal article or a book. Websites, patents, overpass graffiti, and the like are
not appropriate as they typically raise questions about source permanence and whether the
name of the author is the one who really deserves credit (highly questionable for patents).
You are not expected to be responsible for understanding the technical details of the
physics topic you choose, which is beyond the scope of this course. Your grade will primarily
be based on how well you are able to produce a short review that covers the main topics
and cites relevant and important sources properly. Your writing will be graded, and should
be concise, objective, scientific prose. Please state the word count of your review paragraph
below the last sentence.
1.3.3 How to write a bibliography
Scientific bibliographies and citation styles differ from journal to journal. In physics 4AL,
for your reports you will use the format from the journal Nature. The full format guide for
Nature citations can be found at http://www.nature.com, but since we only care about a
subset of those instructions, the important points are as follows.
•References are each numbered in the order in which they are first cited in the text,
tables, and figure legends of the document that references them.
•When cited in the text, reference numbers are superscript with no space between
the superscript numeral and the word, not in brackets unless they are likely to be
confused with a superscript number. Multiple citations are separated by commas in
the superscript, unless they are part of three or more consecutive bibliography items,
in which case the range of citation numbers may be indicated by a dash between the
first and last citation numbers.
•The bibliography will follow the text of your review and is a list of the articles cited
in order of the citation numbers.
•Each entry starts with its number on a new line, followed by the authors’ names, the
article title, journal name, volume, starting page (or full page range) of the article, and
then the year of publication.
37
•All authors should be included in reference lists unless there are more than five, in
which case only the first author should be given, followed by “et al.”. Authors should
be listed surname (typically their last name) first, followed by a comma and initials
of given names (typically their first name or first and middle names), in the order in
which the names appeared on the paper.
•Titles of articles should be in upright (not italic) text. The first word of the title is
capitalized, followed by the rest of the title written exactly as it appears in the work
cited (aside from capitalization), ending with a full stop. If there are words or symbols
in the title whose capitalization is necessary to properly convey their meaning, they
may be capitalized as needed.
•Journal titles are italic and either abbreviated according to common usage or written
in full. Volume numbers are bold, upright font, followed by a bold comma and then the
page numbers (or at least the number of the first page) in regular (not bold), upright
font. This is followed by a space and then the year of publication in parentheses in
regular, upright font. The entire entry is terminated by a full stop.
•Citations of whole books begin with the authors’ or editors’ names (subject to the
same formatting as journal article authors). The book title follows in italic font with
all main words capitalized. Next, the publisher, city of publication, and publication
year appear parentheses and upright font. The entire entry is terminated by a full
stop.
1.3.4 Resources
Nature Format Guidelines
http://www.nature.com/nature/authors/gta/#a5.4
Scientific typesetting guidelines from NIST
http://physics.nist.gov/cuu/pdf/typefaces.pdf
http://physics.nist.gov/cuu/pdf/sp811.pdf
Appendix C shows an example bibliography (a very long one!) from a Nature paper [2].
You are expected to use the formatting found in this bibliography. This bibliography is a
good example of how to format entries, but is also a good example because of its makeup.
Almost all (>99%) of the entires are published journal articles or published books, which is
what is expected in scientific writing. Citation of another student’s lab report, for instance,
would be entirely inappropriate. Cited material needs to be static, available to the readers
of your paper, have clear authorship, and should ideally be peer-reviewed (such as is true
for scientific journals, and is generally close to being true for published books). Sources that
are published only online are in almost no cases appropriate. This is because online content
changes with time and often has unknown authorship, which explains why citing Wikipedia
is not appropriate. An author of a scientific paper needs to make sure that somebody reading
38
their writing 70 years from now will be able to read the exact source they are citing in their
writing.
1.3.5 Exception: Citing this lab manual for reproduced figures
Students in 4AL are welcome to use the images from this lab manual that depict the Physics
4AL equipment in their reports if they properly cite the lab manual as the source of these
figures. This is an exception to the rules for citations given above (the lab manual is not
actually appropriate, strictly speaking, but I have decided to allow students to use the figures
of the apparatus unless they are specifically requested not to). The rest of the content of the
lab report should probably not appear in your lab reports and therefore should probably not
be cited. Copying text from this lab manual (which includes figure captions, even for figures
that are borrowed and properly cited) is not appropriate for a report in physics 4AL, since
the syllabus clearly indicates that each student should be writing their reports in their own
words.
If you use an image of a piece of scientific apparatus from this lab manual in your report,
that is considered a sufficiently direct re-use of material as to warrant a mention of the
authors’ names in the figure caption, followed by a citation number and of course a bibli-
ography item. For instance, if Figure 1.1 is reproduced in a student’s lab report (whether
they modify it or not), the figure caption should say something like “Figure reproduced
(with permission) from Fig. 1.1 by Campbell, W. C. et al.4” and then there would be a
bibliography item following the format below:
4. Campbell, W. C. et al. Physics 4AL: Mechanics Lab Manual (ver. May 12, 2017).
(Univ. California Los Angeles, Los Angeles, California).
1.4 Epilogue
Experiment 2 will be a measurement of the free-fall acceleration due to earth’s gravity, g.
However, it should be possible for you to use your results from Experiment 1 (along with
a little bit of theory) to produce a measurement of g. A measurement of this type would
be considered somewhat more “indirect” than what you will do in Experiment 2 to measure
the acceleration due to gravity, but it is worthwhile thinking about this and considering the
advantages of each method as you perform Experiment 2 next week.
39
Experiment 2: Measurement of g
In the previous experiment, you measured the uniform acceleration of a mechanical system
subjected to a constant force. Since the force in Experiment 1 was produced by gravity
acting on a measured mass m, you should be able to turn your results into a measurement
of the gravitational acceleration g. However, the actual acceleration (a) that was measured
in Experiment 1 was much less than gbecause the system was not in free-fall. This week,
you will be directly measuring a free-falling body to determine (a) if the acceleration due to
gravity is a constant during free fall and, (b) what is the value of the acceleration g.
Experiment 2 will involve making a few choices about how you set up the experiment.
Consider, for instance, a single photogate, similar to that used on the smart pulley in Exper-
iment 1. If the gap between the LED and the photodiode is large enough to permit a falling
object to occult the LED, this device can be used with the 850 to record the arrival time of
the object at the photogate. One way to look at this is that a single photogate can be used
to determine the position of the object at that arrival time, which we can write as (x1, t1).
This does not even give us enough information to measure the average velocity, much less
the acceleration of the ball. Just like your data analysis from Experiment 1, we need at least
two points, (x1, t1) and (x2, t2) to measure a velocity, and at least two velocities to measure
an acceleration, etc.
We therefore imagine that we now have two such photogates, vertically separated by some
distance d. If we drop, say, a plastic ball from above so that it goes through one and then the
other photogate as it accelerates in free-fall, the difference between the arrival times at each
photogate (recorded by the DAQ) should permit us to measure the average velocity between
the gates, essentially given by Eq. 1.3. But we still only have a single velocity measurement,
which is not enough information for us to measure the acceleration.
One could imagine measuring a series of velocities as a function of the height above the
top photogate from which the ball is dropped, but this would not necessarily allow you to
determine if the acceleration is independent of the height from which the ball is dropped.
Another approach would be to take many measurements from a given height while changing
the spacing between the photogates, then repeating this process for a series of different
heights to see if the acceleration is dependent upon the initial height.
We would, however, prefer a method that can provide a measured acceleration in a single
trial, and that does not require us to drop a ball from a well-calibrated height and particular
initial velocity, which would be difficult to perform. In this experiment, you will be using
40
Figure 2.1: Apparatus for measuring acceleration of a ball in free-fall. A plastic ball passes
through two photogates before striking an impact sensor mat. The DAQ should be able to
measure the arrival time of the ball at all three transducers. Measured values for x2−x1
and x3−x2can then be used to determine the acceleration. By changing the height of the
pair of photogates above the impact sensor, one can test if the acceleration depends upon
initial height.
two different arrangements to measure g: multiple sensors that record the arrival of a simple
object (a ball), and a single senor that records the arrival of various different parts of a
complex object (a photogate comb).
First, you will be using a set of three transducers to measure the acceleration of a falling
ball (as shown in Fig. 2.1). The first two transducers are photogates, with the last transducer
being an impact sensor mat. You will show through derivation in your report that if the
gravitational acceleration is constant (independent of initial height or velocity), your mea-
surement method does not require knowledge of the initial height or velocity, which makes
it robust against these difficult to control systematics. You will also vary the height of the
pair of photogates above the impact sensor to test this claim.
Second, you will be using a single photogate to record the arrival time of each slot edge
in a slotted bar called a photogate comb (Fig. 2.3). By measuring the distance between
successive slot edges, the arrival time of each at the photogate can be used to track the
overall position of the comb vs. time. A polynomial fit to the data will have a quadratic
term that is proportional to g.
41
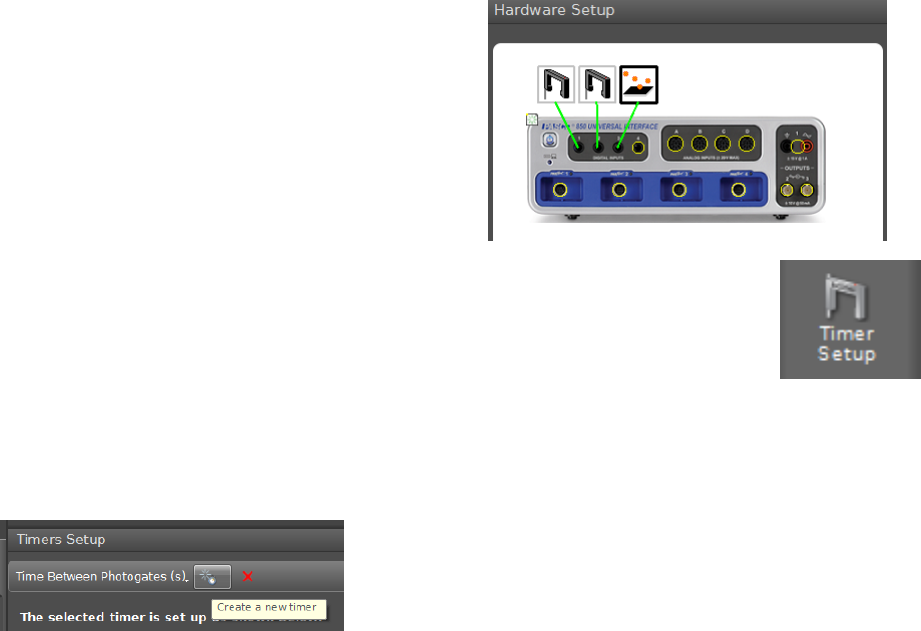
2.1 Procedure
2.1.1 Multi-sensor Method
An example apparatus configuration for the first method of determining gis shown in Fig. 2.1.
Each photogate is fitted with a slotted mask that the light must pass through to hit the
photodiode. When the bottom of the ball blocks this slit, the DAQ will record the time of
this event, so the relevant height that corresponds to, say, x1in Fig. 2.1 is the height of this
slit. You will need to measure the distances x2−x1≡dand x3−x2≡Dand record your
measurements, along with your best estimate of the systematic error in these measurements
(note that the slit has some width).
To setup the DAQ, we will actually be
recording the time between photogate trig-
gers (T1) and the time between the second
photogate trigger and the impact sensor trig-
ger (T2). Plug the three transducers into
three digital channels on the 850. Choose
Photogate and Time Of Flight Accessory for
the appropriate channels in Capstone Hard-
ware Setup.
The two measurements we want will be prepared in Timer Setup, which
can be found in the Tools palette. Your first measurement to set up is to have
the DAQ record the time between when the ball blocks the first photogate to
when it blocks the second photogate. Set up a <New Timer>, choose Build
your own timer in step 1, and select your two photogates in step 2. By clicking
on the little drop-down menu next to each photogate in step 3, you can select Blocked for
each photogate (as shown in Fig. 2.2a.). You can then give your timer a descriptive name
(this will appear as a column header) and click Finish. Capstone will automatically identify
the units it uses for this display (seconds, normally).
To configure your second timing measurement (the
time between when the ball reaches the second pho-
togate and when it hits the impact sensor), click the
Create a new timer button at the top of the Timer
Setup area. Your second timer will measure the time between when the second photogate is
blocked to when the ball hits the impact sensor (which Capstone calls Blocked for the Time
Of Flight Accessory), as shown in Fig. 2.2b.
Once your timers are prepared, you can start a Table from the Displays palette with
your two timer measurements in the table. You may now record data in Continuous Mode.
Capstone will put the two measured times in the table every time the ball is dropped through
the sensor setup. You should set the number of displayed digits on the text in the columns
to one or two more digits than you think you need so that you do not limit your precision.
One thing to keep in mind is that the impact sensors tend to work best and have the
fastest response when the ball lands right above the position of the sensor on the bottom of
42
Figure 2.2: Setting up the two timers for Experiment 2. a) Example parameters to have the
DAQ measure the time between when the ball reaches the upper and lower photogates. b)
Same idea, but for the DAQ to measure the time between when the ball reaches the second
photogate and hits the impact sensor. Note that the software refers to this as producing a
Blocked state for the impact sensor.
43
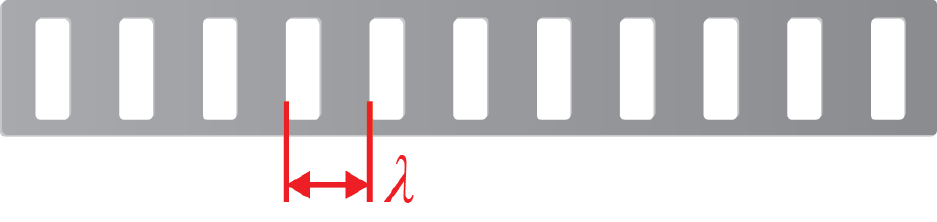
Figure 2.3: A photogate comb with uniformly-spaced through-slots. By dropping this verti-
cally through a photogate, the position of the comb can be tracked as it accelerates in free
fall by monitoring the slots passing through the photogate as a function of time. The spacing
between successive leading (or trailing) slot edges is shown as λ.
the pad. The pads tend to be marked with a little circle on the top where this is, but you
may want to aim for that spot to reduce your systematic uncertainty associated with the
transit time of the acoustic wave between where the ball hits and the sensor location.
Each person should record at least 10 drops for each of 5 different heights of the pair
of photogates above the impact sensor Dfor a total of at least 50 data points. Do not
share data points, but feel free to use the same values of Das your lab partners. Try to
spread these heights out over a wide range to really provide a stringent test of whether the
acceleration is independent of height. Be sure to record your measured values for Dwith
uncertainties.
2.1.2 Photogate Comb Method
Now we will attempt the same measurement with a different apparatus. Remove the impact
sensor and one of the photogates from the ring stand and the 850 so that you have just one
photogate. You will be dropping a fairly long photogate comb through the gap, so be sure
to set the height of the remaining photogate high enough that the comb can fall all the way
through it before hitting the table. Please feel free to move the whole setup to the floor if
you like since it takes a good bit of vertical space to measure gwith this method.
An example photogate comb is shown in Fig. 2.3. First, measure the spacing between
successive edges of the slots in your comb (shown as λin Fig. 2.3). You may assume that
the placement of the slot edges is uniform across the whole length of the comb to 0.1 mm,
so the best way to determine λis probably not by measuring λ, but rather by measuring
nλ (for some integer n) and dividing by n. If you do this, the uncertainty in your measured
value for nλ will clearly be a systematic uncertainty, since it will apply to all of your data
points. You should take this into account when you think about assigning an uncertainty to
your measurement.
Next, set up the DAQ to record the arrival time of each slot edge. In Timer Setup,
make a new Pre-Configured Timer. For timer type, select Picket Fence. You will record Block
44

Event Times in a table. This means you may need to throw out your first data point (since
the bottom edge of the comb is not a slot), so it is worth thinking about this a little bit. The
flag spacing field does not matter for your data since you will be recording times. You will
use your measurement of λwhen you perform your analysis, which will allow you to account
for your uncertainty in that measurement.
One person can hold the photogate comb above the photogate and the other can use a
Table to record the Block Event Times by clicking Fast Monitor Mode a little before
their partner lets go of the photogate comb. Each person should use unique data set for
their report (do not share these data with your partners). Take at least 5 data sets each.
You can experiment with the starting height above the photogate to maximize your chances
that you get at least one data set with as small an uncertainty as possible.
2.2 Analysis
2.2.1 Multi-sensor Method
For the ball drop experiments, you will have sets of data for 5 different values of D. The
data the DAQ has recorded for each trial is T1and T2, the time between the photogates and
the time between the second photogate and the impact sensor. These data points can be
combined with your measured values for the distances Dand dto measure the acceleration
g=2
T1+T2 D
T2−d
T1!.(2.1)
You will need to derive this expression in your worksheet, but you will find that it corresponds
to the difference between the average velocities in both regions divided by the travel time
between the middle of the upper region to the middle of the lower one.
You will have two contributions (statistical and systematic) to the uncertainty in your
measurement of gfor each distance D, which you may add together to get your total un-
certainty. Since we have to this point not spent a lot of time dealing with systematic
uncertainties, we will start by discussing this rather dicey topic.
Unlike statistical uncertainties, for which there are provably reliable methods for estima-
tion and combination, there are no generally-applicable rules for systematic uncertainties.
Recall from §ii.1.5 that systematic uncertainties can generally not be quantified by repeating
measurements, and are therefore typically estimated by using logical arguments about the
influence of various effects on the measurement. If there is more than one source of system-
atic uncertainty in a measurement, you, the scientist, must decide how to combine them. An
aggressive method that is applicable in the case that the uncertainties are uncorrelated and
normally distributed, is to add the uncertainties in quadrature, which produces a relatively
small uncertainty in the measured value and a danger that the true value does not fall within
this uncertainty about a third of the time. A more conservative method that is often used is
to add the uncertainties directly, which produces a larger uncertainty in the final measure-
ment, but a stronger guarantee that the true value falls within this uncertainty. We will use
45
a very conservative method in this case by estimating the total influence of the systematic
uncertainties and stating a large uncertainty that is almost guaranteed to contain the true
value.
You may estimate the systematic uncertainty in your measured value of gfor a particular
value of Ddue to uncertainty in Dand dby using their upper and lower limits (for instance,
using d=dbest +δd and d=dbest −δd) in your calculation of g. Plugging the upper and lower
limits for dand Dinto Excel with your best values for T1and T2for each height is a good
way to see how these affect your measured value for g. You can then take (gmax −gmin)/2 as
your systematic contribution to δg for each D.
There is also a statistical contribution to δg for each Ddue to the spread in your 10
measured values. Use the method of §ii.1.6 to calculate your statistical contribution to
δg. Compare the size of these two contributions. If they are of comparable size, add them
together to make your final estimate of δg. If either the statistical or systematic contribution
to δg is more than 10 times smaller than the other, you may ignore it, in which case your
value for δg is just equal to the larger one. If you think about this, there is no difference
between these two methods since you will almost certainly be stating your uncertainty with
only one significant digit. You will be asked to discuss the process by which you determined
δg in your worksheet.
After determining a measured value for g(with uncertainty) for each value of D, plot
your measured best values gbest vs. D. Is there a trend? One way to determine if there
is a trend in a collection of data is to try to fit your data to a line and see if the slope is
consistent with zero. For our purposes in this course, “consistent with zero” would mean
that the uncertainty in the slope overlaps with zero.
2.2.2 Photogate Comb Method
For each data set, you will now have a table of time stamps for when the photogate was
blocked by the edge of a slot. Using your measured value of λ, construct a table of position
vs. time. It is not necessarily clear what object’s position is being tracked (it’s probably the
top edge of the first slot, though this depends upon your definition of x= 0), but that’s not
important for your measurement; what you care about is the quadratic part of the curve,
not the offset in x(or t).
For each of your data sets, make a plot of position vs. time. Does this look like (part
of) a parabola? If not, you may have something amiss with your data. Remember to think
about the first data point and whether or not to include it.
Excel provides an easy way to fit your data to a quadratic through the Add Trendline
option. You can choose Polynomial and make it of order 2, and even display the resulting
equation on the chart to get gbest. The problem with this is that it doesn’t provide you with
an uncertainty in g! In order to do that, we will need to do a full regression analysis for a
2nd order polynomial, which is not as easy to implement as the Add Trendline option.
46
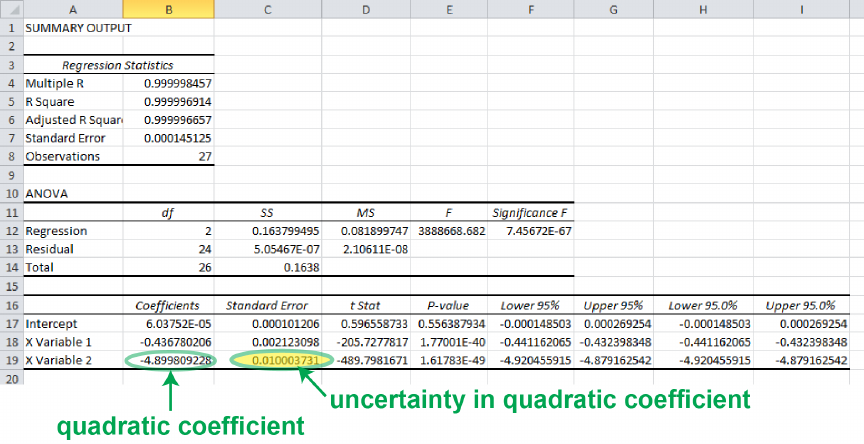
Figure 2.4: The results of a quadratic regression in Excel. The third row of results shows
the best value and uncertainty of the quadratic term. See Fig. 0.6 for a description of the
constant and linear terms.
2.2.3 Quadratic Regression Analysis
When we introduced the linear regression analysis in Excel in §0.2.5, the LINEST function
was mentioned as an alternative way to calculate linear fit parameters, and the same is true
for a quadratic fit. However, in the interest of ease-of-use, we will again choose to describe
how to do this using the Regression tool, but feel free to research how to do this with
LINEST if you like.
The trick to making a quadratic regression is to make a third column of data that has
the square of the original horizontal-axis data in it. In our case, if our time data is in cells
A1:A100, we can make another column right next to this one whose first entry is =A1ˆ2
and copy that down the rest of the column by, e.g dragging its corner down to row 100.
Assuming that A1:A100 has our measured Block Times,B1:B100 has the squares of these
block times, and C1:C100 has the distance data, we start a quadratic regression analysis the
same way as was explained for a linear fit in §0.2.5, but enter the range $A$1:$B$100 for the
Input X Range. This can be easily accomplished by highlighting both columns.
An example quadratic regression result is shown in Fig. 2.4. Comparing this to Fig. 0.6,
we can immediately see the constant and linear terms and their uncertainties in the same
places, but there is a third row beneath them now. This row contains the information about
the quadratic term, with the best value and uncertainty shown in Fig. 2.4.
Using quadratic regression on your data, extract a measured value for gfrom your data.
Remember, the uncertainty given by the regression is only the statistical uncertainty. If you
have a systematic uncertainty in, for instance, the distance column due to uncertainty in
47
λ, you may want to perform separate regressions for different values of nλ at the top and
bottom of your range of uncertainty in nλ and compare the resulting range in measurements
of gbest to the statistical uncertainty in ggiven by the regression.
2.3 Report Guidelines
Last week, we covered citations. From this point forward in 4AL, if you need to reference
prior work by somebody, you will be expected to do so using the rules from last week
(even in your worksheet). If you cite somebody’s work, you will of course need to include a
bibliography at the end of your report in Nature format, and this bibliography will never be
counted toward a word count if there is a limit for that section of that week’s report.
This week’s worksheet will be worth 30% of your grade and the presentation mini-report
will be worth 70%.
2.3.1 Worksheet Guidelines
Your worksheet should contain the following sections
1. Cover Sheet
•experiment number and title
•your name and UID
•the date the lab was performed
•your lab section (for instance, “Wednesday 12pm.”)
•your TA’s name
•your lab partners’ names
2. Derivation Derive Eq. 2.1. Please include just enough detail to demonstrate how this
equation describes what you care about, no more than that.
3. Plots The focus of this week’s presentation mini-report is plots, but the plots you are
asked to include in your worksheet this week do not have to meet the same standard
as the one you will make for your presentation mini-report.
Include a plot of gvs. Dfor the ball drop method. Do you expect that gshould
depend upon D? Briefly discuss this and what your data tell you. Can you find a way
to quantitatively confirm or rule out a linear dependence on D?
You will include a plot of your best dataset of photogate comb results in your presen-
tation mini-report, so decide which set is your best and then include your four other
data sets as a plot (or plots) in your worksheet. Include the fit curves on your plots.
48
4. Data Tables
Include a table of your ball drop results from different heights. An example is shown
below. If an entire column shares the same systematic uncertainty, this should be
stated in the table caption. If the uncertainty differs row-by-row, it is best to state the
uncertainty explicitly in each entry.
Photogate spacing Gap to impact sensor Measured acceleration
Trial d(cm) D(cm) g(m/s2)
1 1.1 3.0 9.6±0.7
2 1.1 5.0 9.8±0.3
3 1.1 15.1 10.1±1.3
4 1.1 20.3 8.6±1.8
5 1.1 30.7 9.843 ±0.004
Include a table of your five photogate-method measured values of g. Briefly describe
your uncertainty analysis and whether the uncertainty is dominated by statistical or
systematic uncertainties.
5. Conclusion
Write a brief discussion of your uncertainty analysis and results. Compare the ac-
celerations you measured with the expected value of g. Which method (ball drop or
photogate comb) do you this is the most accurate? Which do you think is the most
precise? Did you choose to use a combination of systematic and statistical error or did
you just use one? Explain why.
6. Extra Credit (optional)
One of the main challenges associated with getting an accurate measurement of gwith
the photogate comb method is keeping the comb vertical as it drops. You will find
some string and other things around the lab that you can use to experiment with ways
to make sure the comb doesn’t swing or tip once it has been released. For instance,
some students have experimented with using paper strips, tape, string, and other tools
they have constructed to try to get the best measurement.
Design and perform your own experimental test comparing a couple of methods, and
analyze the data to draw conclusions about whether there are methods that perform
better than others.
2.3.2 Presentation Mini-Report Guidelines
Many people reading a journal article will immediately jump to the figures before deciding
to read the article. The figures will often give a quick visual representation of the content of
the paper. It is important for an author to use this knowledge to guide the reader through
what they are presenting by laying out the figures in a logical and possibly chronological
way.
49
Make a figure showing your best photogate comb measurement (the distance vs. time)
to the standards of a scientific journal (see below) and write a paragraph with fewer than
400 words of body text introducing the figure and discussing what it shows. This does not
need to be a completely self-explanatory paragraph, but should instead resemble a paragraph
taken verbatim from a (non-existent) longer paper that you would have written about this
experiment. Your figure should be referenced in your paragraph and will need a caption.
The quadratic fit should be shown, and its results should be described. The caption does
not count toward the word count, and should probably not exceed 200 words or so. Please
write the number of words used in your figure caption and the number used in your body
text on a separate line at the end of your presentation mini-report.
2.3.3 Plots in scientific papers
All figures in well-written scientific journal articles are both clear and necessary. Along with
the guidelines below, an important part of clarity is that the text in the figure must be as
large or larger than the font of the body text. This usually means that the figure itself is
formatted with very large font sizes for its labels since the figure is typically reduced in size
when inserted into the text. A guide for how to insert Microsoft Excel figures into Microsoft
word documents with captions and figure labels can be found in Appendix B.
In Physics 4AL, it should be possible for students to complete their assignments with only
two different types of figures: linear-axis scatter or line plots and apparatus diagrams. We
will focus on the former, though many of the points discussed below regarding the caption
will apply to diagrams as well.
1. Plots should include the following items unless there is a good reason to omit any of
them:
•axes and a bounding box with tick marks pointing inward
•axis labels (see below)
•tick labels (these are the numbers)
•data points and possibly lines joining subsequent points (when needed for clarity)
•a fit or theory curve may be included
•If multiple related data sets can be plotted on the same axes without creating
confusion, they should share the same axes. If multiple plots or figures are closely
related, they should be combined into one figure if doing so does not come at the
expense of clarity.
2. Every figure (and table) needs a caption directly below the item it describes, and plot
captions should include the following items:
•“Figure XX.” or some other appropriate designator with a unique number that is
referenced in the body text, followed by
50
•a descriptive title, followed by
•a description of all lines and symbols on the plot and what they mean
•a brief discussion of what the data (and curves) show
Typically, about five labeled tick marks will be sufficient for fully-specifying an axis.
Tick labels should have an appropriate number of digits for the range they describe.
Though you are unlikely to encounter this in physics 4AL, if the plot range covers a
small fraction of some large absolute scale, you can subtract an overall offset from the
tick labels and explain what this is in the axis label. For instance, if frequency is being
plotted from fmin = 12.642 808 GHz to fmax = 12.642 816 GHz, the tick labels should
probably be every 2 kHz, so the tick labels would be the numbers 8, 10, 12, 14, and
16 and then the axis label would say something like “Oscillator frequency, f(kHz)
−12 642 800 kHz.”
3. Common mistakes to avoid:
•Do not include any lines or curves that are not well-defined (such as splines that
are not fully described, etc.). All lines and curves must be completely specified.
•Do not put a title at the top, as the title is the first part of the caption. Note
that this is one of many differences between a figure in a scientific journal and the
figures in this lab manual, which is not a scientific journal article.
•If possible, avoid the use of yellow or light green (or any color that will be hard
to see).
4. Plot Axis
The format for an axis label in physics 4AL will be “Quantity name, Symbolic Des-
ignation (Unit).” The quantity name should be plain English words (e.g. “Distance
to stop,” “Delay between pulses,” “Glider speed,” “Number of events,” etc.). The
symbolic designation is optional, and if used is the mathematical symbol you be use to
represent this quantity in the body text and its equations (D,t,v,N, etc.). Last (and
not optional), the units symbol for the tick mark labels should be written in parenthe-
ses. Though you are unlikely to encounter this in Physics 4AL, if the units of an axis
(almost always the vertical axis) are suppressed because they are truly irrelevant and
would be distracting, it is conventional to write “(arb.)” for the units.
Axis limits should be chosen to highlight the important points of the plotted material
as well as possible. This may involve cutting off some of your data (typically on the far
left or far right), so you will have to think about whether you can omit parts of your
data set without changing what the data show. Omitting data to change conclusions
is not okay – data may only be omitted if doing so improves clarity of presentation
without altering the interpretation of what the data support.
5. Adding the caption
51

A good figure caption tells the reader not only what is on the plot, but why it is
important and what conclusions can be drawn from the results. The first thing after
the designation (e.g. “Figure 1.”) should be a short, descriptive title. The title should
not simply be “(y-axis label) vs. (x-axis label)” – your reader will already know that
from looking at your figure. Your goal is to give the reader a good idea of what it
is you are presenting and why you are presenting it. For instance, for a figure from
Experiment 0, instead of “Voltage vs. Weight,” a much more descriptive figure title
would be “Calibration of the force sensor.”
Next, describe what is on the plot. The reader should be able to tell what every data
point and curve on the plot is. If there is a fit curve on the plot, it needs to be discussed
in the figure caption. In contrast to many of the figures you see in this lab manual
(which is not a scientific paper), please do not put your fit line equation on the body
of the plot; the fit line equation and the fit parameters need to either appear in the
caption or the caption should clearly indicate to the reader where they can be found.
One good way to add a fitted curve’s equation to the caption is to write the equation
using symbols, then to present the values for these symbols (and their associated
units and uncertainties). For instance, “The solid red curve is a fit of the data to
an exponential decay, J(t) = Ae−t/τ with fit parameters A= (43 ±4) km and τ=
(22.60 ±0.06) fs.” Alternatively, this can be accomplished by of referencing specific
equations in the body text instead (see, e.g. Eqs. 0.2 and 0.3 of this manual).
6. Final notes
A guide to graphing in Excel and adding captions in Word by Julio S. Rodriquez, Jr.
and Anthony Ransford can be found in Appendix B of this lab manual.
2.4 Epilogue
2.4.1 Specialized defined units for gravitational acceleration
The “law of falling bodies” is usually attributed to Galileo, by which we mean that he
is often described as the first to discover that gravitational acceleration is constant and
independent of mass. In recognition for this achievement, he has been given the highest
honor in physics (even higher than the Nobel prize!): a unit named after him. Naturally, it
is a unit of acceleration, the “Galileo.” 1 Gal ≡1 cm/s2. Gravimetric maps often display
their gravity anomaly data in units of mGal, or “milli-Galileos.” There is also a defined unit
called Standard Gravity [8] that is given by go≡9.806 65 m/s2.
2.4.2 gin Knudsen 1-2381
1This analysis and section were contributed by Chandler Schlupf (2015).
52
Standard Gravity (go) is some sort of average of the apparent gravitational acceleration over
the surface of the earth, but the gravitational acceleration at any given point can change
by up to about a percent depending where the point is due to several factors. The first is
that the Earth is not perfectly round, but rather an oblate spheroid (shaped like an onion),
so its radius is not constant and the surface is not equidistant from the geometric center.
Second, since the Earth is spinning, a centrifugal pseudo-potential that varies with latitude
is produced, decreasing the apparent gravitational acceleration near the equator compared
to the poles.
Furthermore, local variations in the makeup of the Earth’s crust can influence the value
of gby changing the average density of the ground below, as well as the distance from
the center of the earth. For example, a pocket of low-density material such as a liquid or
gas underneath the Earth’s surface can cause a tiny decrease in the measured gravitational
acceleration right above it. Gravimetric measurements are therefore sometimes used to locate
petroleum deposits.
All of this means that the value of gin a given location is not, in general, equal to
Standard Gravity go. In fact, these effects (and others) essentially guarantee that gois
actually the gravitational acceleration almost nowhere on the surface of the earth (look how
many digits there are on go)! Luckily, the only important thing for Experiment 2 is knowing
the value in our lab room.
Measured data of variations in the Earth’s gravitational field are collected on gravimetric
maps. One such map for our location created by the Southern California Areal Mapping
Project can be found at http://pubs.usgs.gov/of/2003/0269/. Unfortunately, there is
not a data point on UCLA’s campus (to obtain accurate measurements, they only take
data points on solid bedrock). However, we can use surrounding points to get a good
estimate. Averaging the three closest points to UCLA and taking their standard deviation
as an estimate of the systematic uncertainty gives us a value for gravity in Knudsen 1-238
of g= (9.7955 ±0.0003) m/s2.
53
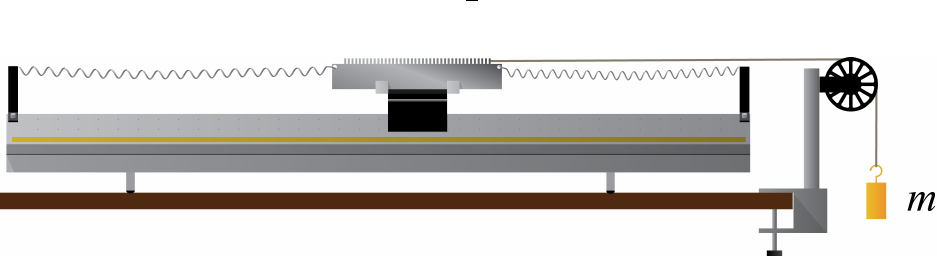
Experiment 3: Conservation of
Mechanical Energy
Conservation of energy is a powerful conceptual and computational tool that is useful in
essentially every field of science and engineering. Dissipation of energy into random thermal
motion still obeys conservation of energy, but it is sometimes difficult to keep track of the
dissipated energy, so we will be focusing here on the conservation of mechanical energy. In
this experiment, you will measure the kinetic and potential energy of a harmonic oscillator
during half of an oscillation cycle, where you will try to determine to what degree the
initial potential energy is converted entirely into kinetic energy and then back into potential
energy. The experiment has been designed in a way to try to minimize dissipation, but you
will be able to analyze your data to quantify how much energy is being dissipated from the
mechanical energies.
A glider is provided that has a comb of photogate flags mounted to the top. You will be
using a photogate to measure the passing of each flag, which will enable you to determine
the position and velocity of the glider. Two springs are also provided to produce a restoring
force on the glider to some point on the air track. By measuring the spring constant of this
restoring system, you will be able calculate potential energy as a function of position
U=1
2kx2.(3.1)
Figure 3.1: Apparatus for measuring the effective spring constant providing the restoring
force for your glider. Once the spring constant has been determined, the hanging mass,
string, and pulley should be removed.
54

You will also measure the mass of the glider (with photogate comb attached!), which will
enable you to determine the kinetic energy as a function of velocity:
K=1
2Mv2.(3.2)
The tricky part of Experiment 3 will be the data analysis, where you will need to figure
out how to interpret your raw data (a series of time stamps for when the photogate goes from
being unblocked to blocked) to calculate these quantities. You will need to determine how
to find the kinetic energy as a function of position so that for every data point where you
calculate the potential energy, you can also calculate the kinetic energy. By plotting these
together, you can also plot their sum, which should be constant in the absence of dissipation.
You will find that despite the fact that the amount of energy of each type changes drastically
over the course of half an oscillation, their sum is remarkably flat. The slight decrease in total
energy over your measurement will enable you (through fitting) to determine the coefficient
of friction of your nearly-lossless air track setup.
3.1 Procedure
The first experimental task to complete is to determine the mass of the glider with its
photogate comb and record this for your analysis. You will then need to determine the
spring constant associated with the restoring force of your spring setup, as shown in Fig.
3.1. The spring constant measurement does not utilize the DAQ at all, and will require you
to write down a series of measurements for your analysis and report.
Figures 3.1 and 3.3 show the basic idea. By measuring the position of the glider for a series
of different masses m, a plot of applied force (mg) vs. displacement can be fit to determine
the slope of this trend, which has units of force per displacement and is the effective spring
constant of your system. Be sure to get data for at least 5 different masses. Once you have
gathered enough data for your spring constant measurement, you may disconnect the pulley
system and hanging mass from the glider and move it out of the way. You may share data
for the spring constant measurement with your lab partners, but will be expected to use
your own data sets for the rest of Experiment 3.
To measure the position and velocity of the glider for your analysis and report, we will
be using a photogate to tell us when each tooth first blocks the photogate. This is a tricky
business, and you will want to be very careful that you describe your procedure in your own
notes so that you can use them to interpret your data later.
The teeth on the photogate comb are 2 mm wide, as are the spaces between teeth, both
to an uncertainty of 30 µm. You will only be using half of a single oscillation of the glider
for Experiment 3, so you may pick a direction for the glider to be moving when it passes the
photogate. Remember, the photogate cannot tell the difference between the glider moving
left or right, so this will simplify your analysis by confining the movement to one direction
only. You will pull the glider back just beyond the photogate, start the DAQ recording, then
release the glider. You may stop recording as soon as the glider stops to reverse its direction.
55
Figure 3.2: An example photogate comb shown with an example for a position of the pho-
togate when the glider is in its equilibrium position. In this case, it is probably easiest to
analyze the data if one chooses to pull the glider back to the right instead of the left.
First, it is important to set and record the equilibrium position of the glider relative to
the photogate beam. A good choice is to put the photogate so that a tooth near the center
of the comb is just on the verge of blocking it (the light on the top of the photogate turns on
when it is blocked). You will thank yourself later if you are very careful about recording (a)
which tooth that is and (b) which direction the glider needs to move to block the beam when
it is at equilibrium. Beware that the teeth may have marks on them that are meaningless;
be sure to do your own counting to determine which tooth number you are using.
Figure 3.2 shows an example configuration where the photogate is set up to trigger on
the 27th tooth if the glider is displaced to the left a tiny amount from equilibrium. In this
example, pulling the glider all the way to the right so that tooth number 1 is just past
the photogate position and then letting go and having the DAQ record the time stamps
every time the photogate goes from being unblocked to blocked means that the 27th time
stamp happens at exactly the equilibrium position, which one could define as being x≡0.
This would mean that the first time stamp happened at a displacement from equilibrium
of x=−104 mm. Likewise, if for the same configuration the glider had been pulled off to
the left and then let go for recording data, the equilibrium position would be reached about
halfway between time stamp 35 and 36, because the photogate triggers when it goes from
being unblocked to being blocked. In this case, the first time stamp indicates the glider is
at a position of x=−138 mm from equilibrium.
To set up the DAQ for recording these time stamps, connect your photogate to a digital
56
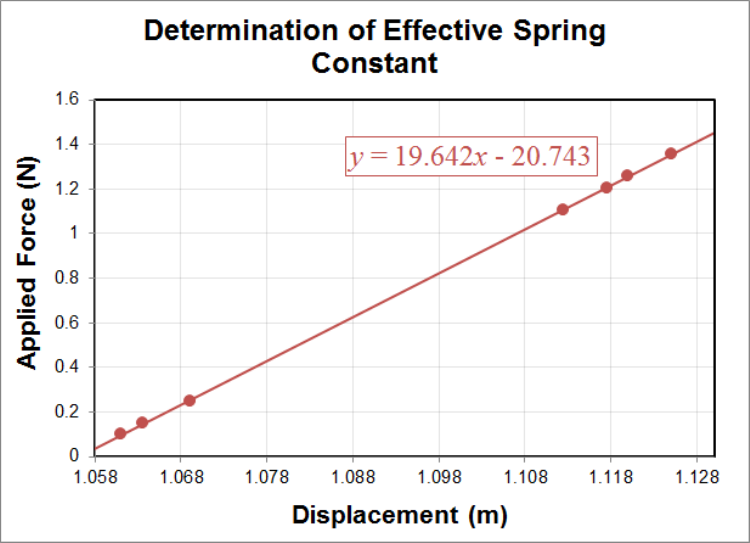
Figure 3.3: Determination of a spring constant by fitting a line to measured values of force vs.
displacement. In this case, the spring constant was determined to be k= (19.64±0.04) N/m.
channel and select Photogate in Hardware Setup. In Timer Setup, you should make a
new Pre-Configured Timer based on your photogate channel that is of the Picket Fence type.
You will record Block Event Times in a table. The flag spacing field does not matter for
your data since you will be recording time stamps. Once this timer has been created, you
may create a Table and use Fast Monitor Mode. One person can pull the glider back in
the chosen direction past the photogate, then their lab partner can click fast monitor mode.
The glider can then be released. Once the glider reverses its direction, stop the DAQ and
you may copy and paste your data into Excel to save for your analysis. Each person needs
their own data set, but all three may be recorded in a row using the same apparatus setup.
3.2 Analysis
For your measurement of the spring constant of the restoring force, plot your force vs.
displacement data. You will need to make sure your plot has the appropriate data as xand
y, because it is easy to get these mixed up, especially in Excel. Figure 3.3 shows an example
with some fake data. The y-intercept is of no interest here as we are only concerned with
determining the slope of the fit line. Remember, the SI units for spring constant are N/m, so
be sure to convert your masses into applied force correctly (remember the difference between
57
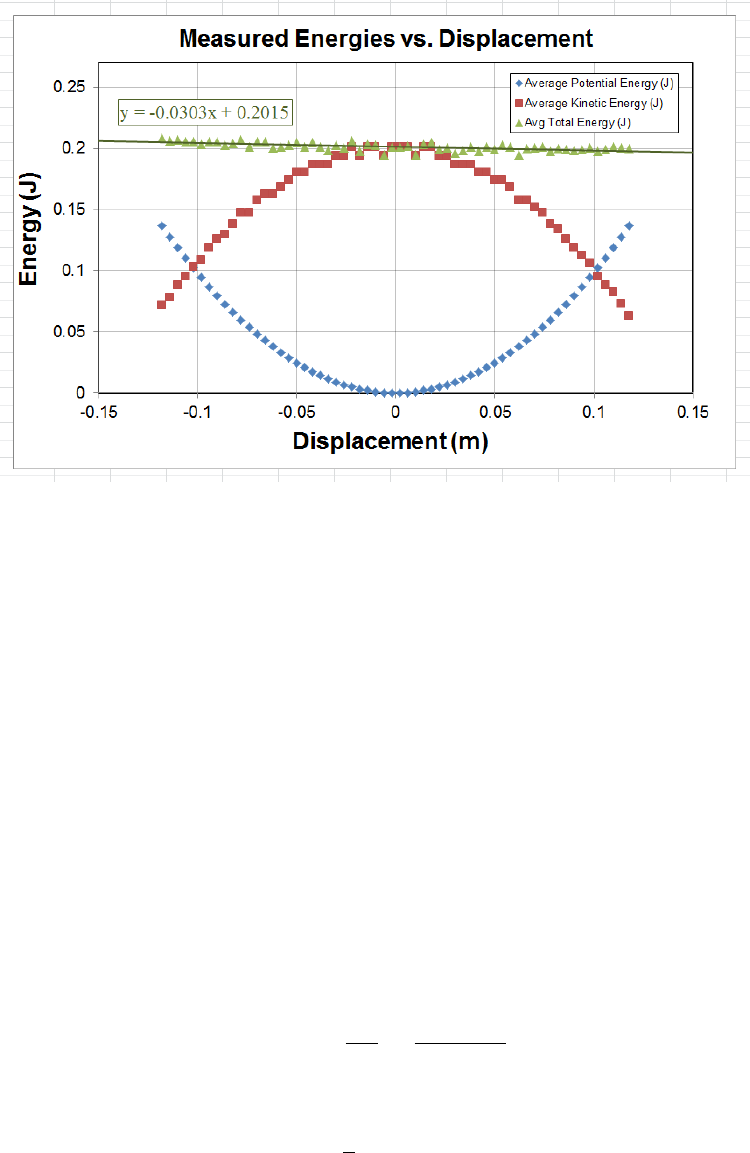
Figure 3.4: Mechanical energy of a glider vs. position. The potential energy shows the
harmonic well for this oscillator. The kinetic energy is based on differentiating the same
data that was used to calculate the potential energy, so it is more noisy, as expected when
differentiating data.
grams and kilograms), and you hopefully found in Experiment 2 that g= 9.8 m/s2(you may
of course use this value even if you got a different answer in Expt. 2).
Your data for the moving glider should consist of a column of time stamps. Using
your knowledge of where the photogate was relative to the comb when the glider was at
equilibrium, you should be able to convert these time stamps into positions, where you
should define the equilibrium position to be x= 0. This is important because you will be
calculating the potential energy using Eq. 3.1, so x= 0 should correspond to U= 0.
It is certainly true that we could calculate the potential energy as a function of distance
based on these data (since you now have a value for the spring constant k). However, we
will be calculating velocity by numerically differentiating your data, which is the same as
saying we will be using the following expression (as in previous experiments)
v(¯x(i)) = ∆x
∆t=xi+1 −xi
ti+1 −ti
(3.3)
where ¯x(i) is the average position in this interval,
¯x(i)≡1
2(xi+1 +xi).(3.4)
So we will have a value of Uat every xi, but a value for Konly halfway between every xi
and xi+1 (which we call ¯x(i)). Since we want to calculate the potential, kinetic, and total
58

energy at the same place, we can, for instance, determine the potential energy for each value
of ¯x(i). To do this, create a new column for the average positions ¯x(i) that is the average of
each successive pair of points in the xicolumn. You can then create a column that calculates
the potential energy U(¯x(i)) = 1
2k¯x(i)2at each value of ¯x(i) using Eq. 3.1. Create another
column and fill it with calculated values of the kinetic energy
K(¯x(i)) = 1
2M xi+1 −xi
ti+1 −ti!2
(3.5)
using your measured value for the mass of the glider with its comb on top. Last, create a
third column that calculates the total mechanical energy for each position ¯x(i).
Create a plot that shows kinetic energy, potential energy, and the total energy as a
function of displacement ¯x(i). An example is shown in Fig. 3.4. Your potential energy
should look like a parabola, characteristic of a harmonic oscillator. The kinetic energy will
be more noisy than the potential energy since it is based on numerical differentiation of data,
which magnifies noise in the data. The total energy should be very linear. If your calculated
total energy does not look linear, it is likely that you have set your zero point in the wrong
place when trying to convert between time stamps and displacement from equilibrium. Fit
a line to the total energy vs. distance and determine its slope (with uncertainty). The slope
of this line has units of J/m = N, and represents a constant force that dissipates energy
from your oscillator, probably due to friction. Using your known value for the glider mass,
calculate the coefficient of friction needed to produce this slope.
3.3 Report Guidelines
For both the worksheet and the presentation mini-report for Experiment 3, you will be
expected to apply what you learned in the previous experiments about citations and plots.
For instance, your worksheet for Experiment 3 will contain two plots, and those plots should
conform to all of the guidelines from §2.3.3 (aside from the body text, which will often not
be necessary on a worksheet).
This experiment’s worksheet will be worth 30% of your grade, with the presentation
mini-report worth 70%.
3.3.1 Worksheet Guidelines
Your worksheet should contain the following sections
1. Cover Sheet
•experiment number and title
•your name and UID
•the date the lab was performed
59
•your lab section (for instance, “Monday 9am.”)
•your TA’s name
•your lab partners’ names
2. Discussion
•Include a description of which direction your glider moved and where the photo-
gate was compared to which tooth.
•Describe how you calculated the potential energy and kinetic energy at the same
positions in space.
3. Plots and Tables
•State your measured value for the mass of the glider with its photogate comb.
•Make a table of your data used to calculate the spring constant.
•Include your plot showing the fit line for determining the spring constant. State
your measured value for the spring constant (as always, with its uncertainty and
units).
•Include your plot of the three energies vs. position, along with a linear fit to the
total energy vs. position.
•Present your calculated value for the coefficient of friction of the glider on the air
track (with its uncertainty, of course!).
4. Extra Credit
Since the photogate does not distinguish between movement of the glider in the +x
and −xdirections, we confined our analysis to half of an oscillation. However, by
appropriately converting the measured times stamps into displacement (warning: this
step is very tricky!), it is possible to generate a plot with multiple oscillations. Save
data for at least two full oscillations of your glider and use this to plot the two types
of energy vs. time (as opposed to plotting them vs. distance). Estimate how long it
would take your glider to decrease its oscillation amplitude by a factor of e(one digit
of precision is okay for this). Fig. 3.5 shows the results of doing something like this,
where the decay of the energy in the mechanical degrees of freedom is clearly visible.
3.3.2 Presentation Mini-Report Guidelines
The first page of a scientific paper will almost always contain the title, author list, and an
abstract for the paper. The title and abstract are where you will tell your story to the largest
audience. The title and abstract must stand alone and the main points should be able to be
understood without the bulk of the report. An emphasis on clear, specific, and minimalist
60
writing will be important not only to communicate what you have done and why it is im-
portant but also to convince the reader that the rest of your paper is worth their time. In
Physics 4AL, we will follow the guideline that no abstract is to contain more than 200 words.
Please always write the number of words contained in your abstracts on a separate line below
the last sentence. For this week’s presentation mini-report, please write a title, author list,
and abstract for your work on experiment 3. Please put this mini-report on a single, new
page, with the title at the top, followed by the author list (centered), followed by the abstract.
Abstract
An abstract is a recap of all the information of a scientific article, conference, presentation
or research update. The abstract should be a standalone document where the reader could
broadly understand all major points (including the conclusions) of the research without
reading further. You are tasked with writing an abstract for Experiment 3 (the one you have
just performed) with fewer than 200 words. In this abstract you will describe what was done
and why. Were you testing some model or another for the behavior of some physical system?
Were you testing a fundamental principle of physics, or measuring a property of something?
Were you investigating the degree to which a system exhibits some desired effect?
In this course, we will depart somewhat from the Nature style in that your abstracts
should not include citations or background. An abstract should be succinct with no un-
needed words. Scrutinize every word and use the minimum number of words required to
express your ideas. Remove words that are ambiguous or imprecise. Even though an ab-
stract is short, it is worth your while as a scientist to make sure you put the time in to make
something you are proud of, as many times it is the only published evidence of conference
talks, presentations or research updates.
Title
Above your abstract you will include a title. The title is sometimes the single most
important thing in a paper. By the pigeonhole principle it is at least the most read part
of a paper. The title should be explicit and understood by the most general of your target
audiences. You need to tie yourself to the similar research out there while distinguishing
what you have done as something different. Like the abstract it is of utmost importance to
scrutinize every word with a focus on clarity. The title should not be an English sentence.
Author List
We will follow the standard U.S. author list convention of initials followed by surname.
You will be the only author on your Physics 4AL papers. At the end of your name include a
superscript that indicates a footnote with your affiliation. For example “F. D. Roosevelt1”.
On the bottom of your abstract you would note the affiliation, for example “1Department of
Physics Port Chester University”. Your lab partners’ names will be on your cover sheet, so
there is no need to worry about giving them credit for helping you to take data. Since all of
the analysis, figures, text, and so forth for your report have to be entirely generated by you,
we will let the cover page suffice for crediting lab partners.
61
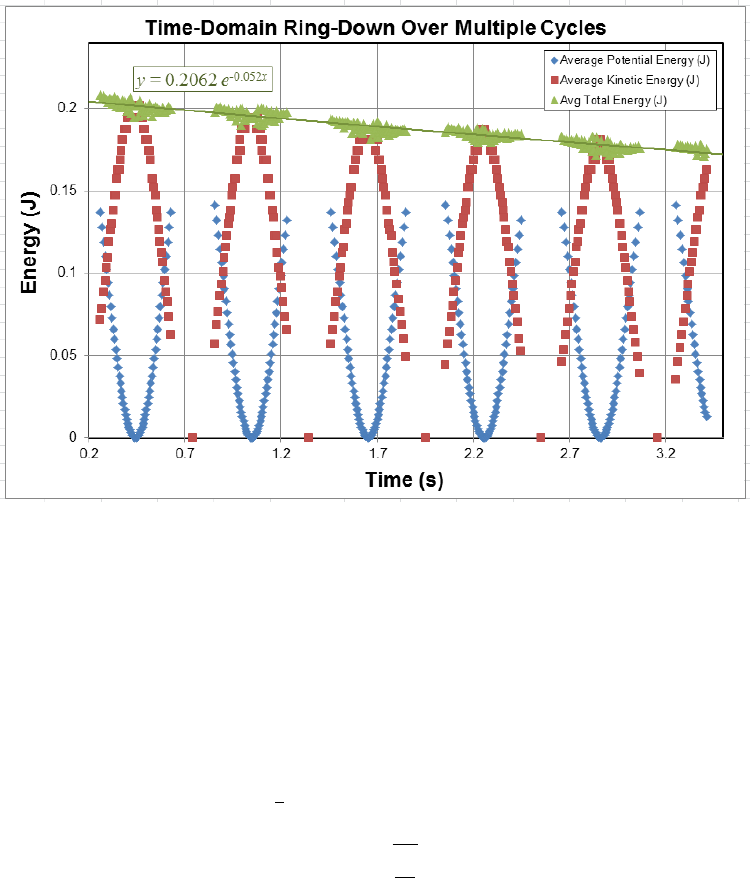
Figure 3.5: The total mechanical energy of a glider slowly decreases due to dissipation (see
Extra Credit). In Experiments 5 & 6, you will be measuring properties of harmonic oscillators
that have similarities to this one.
3.4 Epilogue
A plot of the potential energy vs. position (such as shown in Fig. 3.4) gives a visual repre-
sentation of the “potential well” in which the glider oscillates. The frequency of a mass m
oscillating in a parabolic well (U=1
2kx2) is given by
ω=sk
m.(3.6)
Using the measured value for kassociated with the data in Fig. 3.3 and the measured mass
gives a prediction for the oscillation frequency that matches the frequency shown in Fig. 3.5
at the 10−2level, demonstrating that this system is well-modeled as a simple harmonic
oscillator.
62
Experiment 4: Momentum and
Impulse
In the previous experiment, you investigated conservation of energy by monitoring the trade-
off between kinetic and potential energy in a harmonic oscillator. However, as you were able
to quantitatively verify in Experiment 3, mechanical energy can be dissipated into heat in
a number of ways. It is often fairly difficult to keep track of these dissipation mechanisms,
and so the conservation of energy balance sheet can end up with a kind of catch-all entry for
“dissipation” that limits its usefulness as a predictive tool in all but the most well-controlled
environments.
Compared to conservation of energy, conservation of momentum is often said to be more
“fundamental.” There are a few reasons why you may hear this claim, but for our purposes
it should suffice that it’s often easier to keep track of and predict the flow of momentum
in real-world situations than energy. For instance, both elastic and inelastic collisions are
required to conserve linear momentum with respect to the movement of the colliding objects,
while conservation of energy allows kinetic energy to be converted into, for example, heat
during the collision.
In Experiment 4, you will be investigating the relationship between two different ways of
measuring an impulse, which is a momentum change (and therefore has units of momentum).
You will measure the initial and final velocities of a glider on an air track that impacts a
rigidly-mounted force sensor, which will give you a first method for measuring impulse:
∆P=Pf−Pi.(4.1)
You will also record the reading on the force sensor during the collision as a function of time,
allowing you to integrate the force to provide you with your second method for measuring
the impulse:
∆P=Zdt F (t).(4.2)
It is interesting to note that we expect the two measurements, Eq. 4.1 and Eq. 4.2 to be
equal to each other no matter how inelastic the collision with the force sensor is. It should
not matter if this collision conserves the kinetic energy of the glider, increases it, or decreases
it; we should always expect to be able to use Eq. 4.2 to predict Pfbased on Pi, regardless of
63
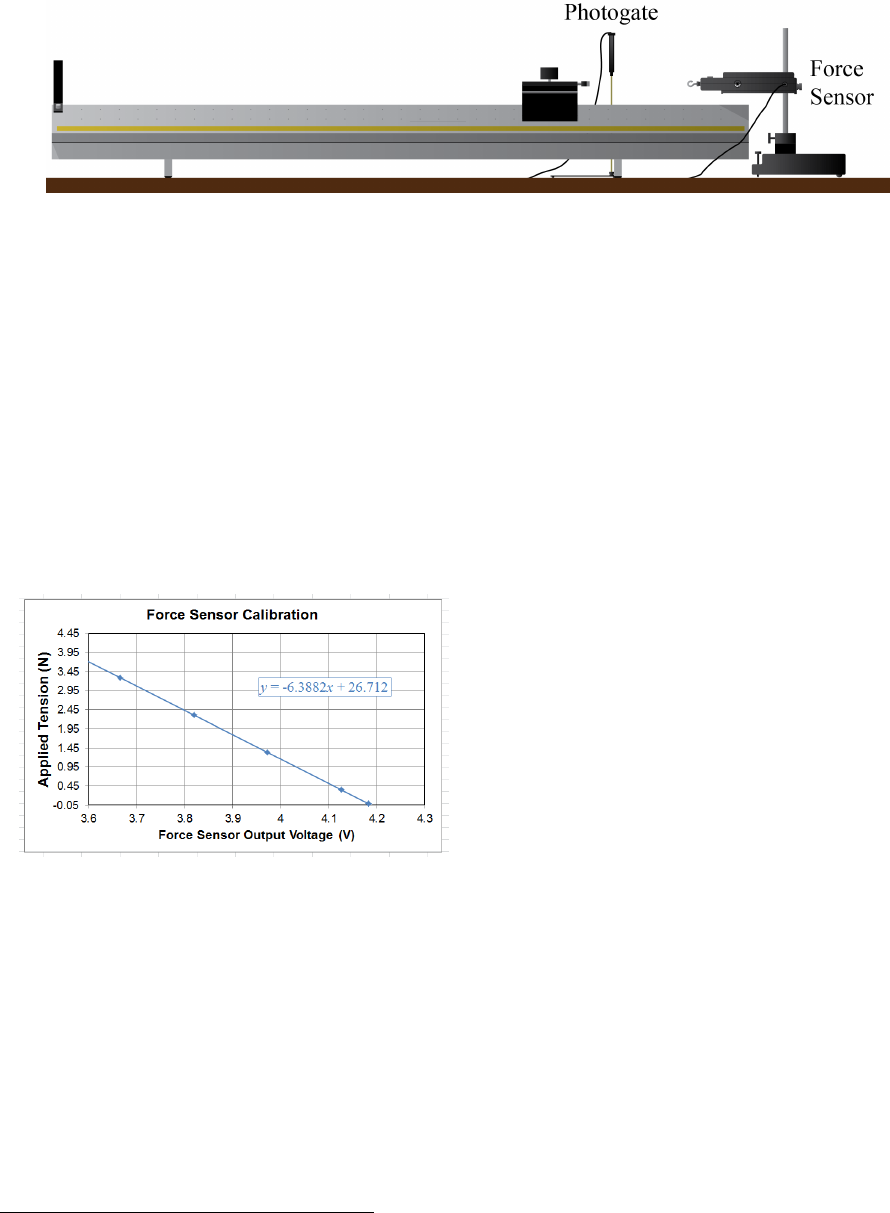
Figure 4.1: Apparatus for measuring the impulse of a glider colliding with a force sensor.
The flag on the glider passes through the photogate both before and after the collision,
providing a means to measure the initial and final momentum. The force sensor allows you
to record F(t), which can be integrated numerically to obtain an independent measure of
the impulse.
whether there is a piece of bubble gum or a diamond on the end of the glider that hits the
force sensor1.
4.1 Procedure
Your first step will be to calibrate your
force sensor, whose internal calibration is
not trustworthy on the level needed for this
experiment. You will essentially follow the
same procedure as that used in Chapter 0.
You will set up your force sensor as a User
Defined Sensor whose output will be in volts.
You should record the voltage reading for at
least 5 different weights hanging from the
force sensor hook spanning the range from
0 to not more than 1 kg. You may assume that the calibration slope found by measuring
tension is also appropriate for compression. You should be sufficiently familiar with the
hardware at this point to be able to do this step without detailed instructions, but you may
want to take a look back at Chapter 0 if you have questions. You may share your sensor
calibration data with your lab partners, but you should perform the analysis and fitting on
your own.
Figure 4.1 shows the experimental setup for producing your two, independent measure-
ments of the impulse of the collision between the glider and the hook of the force sensor.
The photogate flag on top of the glider allows you to use the photogate to measure the speed
of the glider both before and after the collision. For this, you will need to know the mass of
the glider (with all of its attached flags and bumpers) and the length of the photogate flag.
1Please do not put gum on the lab equipment. However, please do feel free to leave diamonds all over it.
64
Figure 4.2: The DAQ will rapidly fill in the first two columns of your table once you tell it to
record. By scrolling through the table while watching the speed column, you should be able
to find your measured initial and final speeds, which will be the two entries in this column.
You will use the Speed function of the photogate timer, so you will need to enter the length
of the flag into Capstone during setup.
To configure the photogate, select Photogate for the appropriate channel and then move
on to Timer Setup. Choose a Pre-Configured Timer for one photogate with a single flag.
In Step 4, you will choose Speed as the measurement that will be visible, at which point
you will need to type in the length of the flag (in meters) to complete your timer setup.
The Speed measurement that Capstone uses is simply the entered flag length divided by
Time in Gate, so feel free to use either, keeping in mind that the important thing is that
the fractional uncertainty in flag length will be the same as the fractional uncertainty in the
speed measurement (assuming the timing has a much smaller fractional uncertainty).
For the force sensor, you should keep the same settings you had for the calibration, which
is to call it a User Defined Sensor on an analog channel.
Your data will consist of three columns in a Table, which will be filled in Continuous
Mode once you hit Record. You will want a column for Time (s),User Defined (V), and
Speed (m/s). The first two columns will start getting filled in as soon as you hit Record,
where the Speed column will only get two entries, which will show up only at the time
the glider has passed through the photogate. In the Controls palette, be sure the sample
rate for the User Defined Sensor is set fast enough to generate enough samples to give you
good results without creating an excess of data. Capstone has been known to have sporadic
problems on this experiment for high sample rates (≥10 kHz), so you may want to stick to
sample rates of a few kHz and fairly gentle collisions to get good data for this experiment.
To take your data, start the glider moving toward to photogate and start recording just
65
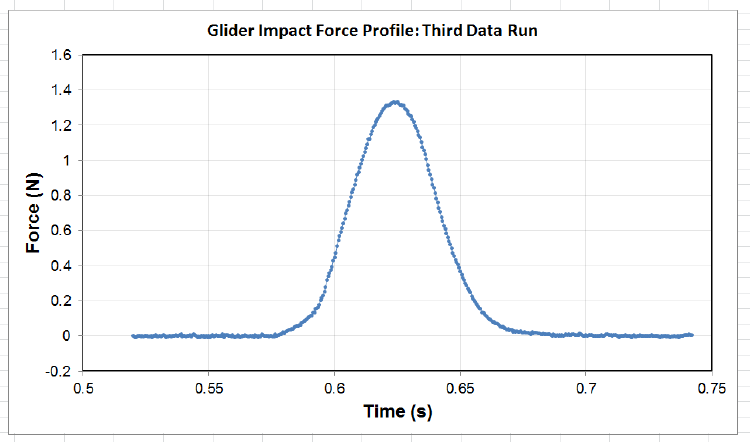
Figure 4.3: Force sensor reading before, during, and after the collision. The first 100 ms
or so of data was averaged to provide a measurement of the force sensor baseline, which
was subtracted from the data before creating this plot. The area under this curve is a
measurement of the impulse of the collision.
before it reaches it. As soon as the glider has passed through the photogate on its way back
the second time, you may stop recording. You will now have three columns of data, two
of which are very full with the third one almost completely empty. Be sure to increase the
number of displayed digits on your table to high enough precision so that you are not limited
by this. Figure 4.2 shows an example of what part of your data might look like. You should
be able to find two entries in the Speed (m/s) column. Write these down somewhere for your
analysis, and be sure to think about whether or not to enter a minus sign on one of them
(remember, the photogate does not know which direction your glider is moving).
Your two remaining columns of data may be quite long. If there are more than about
5000 rows, you should probably try re-taking your data with a faster trigger-finger on the
record button so that you only get the part of the data you really need. You may find that
you need to move the photogate closer to the force sensor to minimize the dead time, but
be sure your photogate measures the glider speed when it is freely moving, not while it is
impacting the sensor. By scrolling through your data, you should be able to find a section
that shows lots of changes in the User Defined (V) section. That section is what you will
need for your report, so copy and paste it (both columns) into Excel for saving. Be sure to
get enough data before and after the collision to give you a good feel for your baseline (see
Fig. 4.3). Each person will need two data sets of their own for their analysis. Try
to get two different speeds to compare them to one another.
66
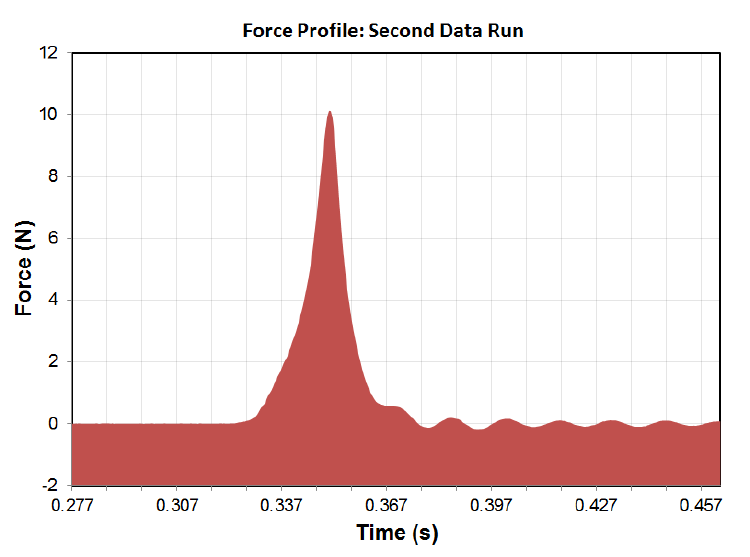
Figure 4.4: Force sensor reading before, during, and after a fast collision. The reading shows
oscillations, or “ringing” after the collision that can be ignored for your integration.
4.2 Analysis
For your determination of the calibration of the force sensor, plot your data and run a
regression fit to determine the slope (with uncertainty). You will probably want your slope to
have units of N/V so that you can just multiply it by your column of recorded voltages to get
force. Remember to flip the sign appropriately to convert between tension and compression.
Your first measurement of the impulse comes from your two speed measurements given
by the photogate. Multiply your measured velocities by the mass of the glider to create a
measured value of the initial and final momentum, then use Eq. 4.1 to convert this into a
measurement of the impulse.
Using your calibration of the force sensor, you can create a new column in Excel that
converts your measured voltages into force. Create a scatter plot of your measured force
vs. time. Even if you tared the force sensor before taking your data, you will want to choose
a big group of data points at the beginning to average to define your baseline, so use your
scatter plot to choose an appropriate range to average for this and create a new column of
forces that have the background subtracted. You can now create a new scatter plot of your
results, as shown in Fig. 4.3.
You may notice that your plot shows some “ringing” in the reading after the collision,
67
such as is shown in Fig. 4.4. For your numerical integration of your data, this can be safely
ignored as long as the amplitude of the oscillations is much smaller than your impact (such
as is shown in Fig. 4.4), and your integration really only needs to include the big peak from
when it starts to rise to about the point where it crosses zero again.
To complete your analysis, you will want to integrate your data to find the area under
the big peak of your force vs. time plot (Eq. 4.2). To do this integral numerically, simply
find the sum of all of the force measurements during the big peak and multiply this by the
duration of the small time step ∆tbetween successive points. This method of numerical
integration is called a Riemann sum, and we have assumed that the force measurements are
all equally spaced in time by ∆t. Mathematically, we are using
∆P=Ztn
t1
dt F (t)≈∆t
n
X
i=1
F(ti) (4.3)
where ∆t≡ti+1 −ti. You should compare your impulse measured with both methods. If they
are off by a large factor, make sure you got all your units correct and that you subtracted
the background force reading appropriately.
4.3 Report Guidelines
As usual, you will be expected to use what you learned from the presentation mini-reports
of Experiments 1 and 2 to format your report (citations and plot guidelines). However, you
will not be called upon to produce an abstract, etc. (covered in Experiment 3’s presentation
mini-report) again until you begin writing full lab reports in Experiment 5.
This week’s worksheet will be worth 40% of your grade, with the presentation mini-report
representing the remaining 60%.
4.3.1 Worksheet Guidelines
Your worksheet should contain the following sections
1. Cover Sheet
•experiment number and title
•your name and UID
•the date the lab was performed
•your lab section (for instance, “Wednesday 9am”)
•your TA’s name
•your lab partners’ names
2. Discussion
•Present your measured values for the mass of the glider (with all its accessories
on it) and the width of the photogate flag, with uncertainties.
68
•Present a plot of your calibration of the force sensor. You do not need to make a
table, but be sure to include the fit line on your plot.
•Be sure to state your calibration constant for converting the force sensor voltage
into newtons with the uncertainty you found for this constant. This can either
be in the plot caption or can be written as body text and referenced in the plot
caption.
•Present your two calculated impulses that were based on the photogate speed
measurements, one for each data run.
•Include a plot of each of your two measured force vs. time curves with the back-
ground subtracted from the force sensor.
•Describe how you used the force vs. time data to determine the impulse, i.e. how
you performed the numerical integration.
•Present your two measured values for the impulses based on numerical integration
of the force profiles. To assign an uncertainty, you may estimate this by assuming
the fractional uncertainty of your integral is the same as the fractional uncertainty
of your force sensor calibration coefficient.
•State the results of the two different methods for both runs in a table (with
uncertainties).
3. Extra Credit (optional, as always)
The inelasticity of a collision can be quantified through a unitless number called the
coefficient of restitution,CR. When two objects collide, CRis defined as the ratio of
the final relative speed of the colliding objects to the initial relative speed. As such,
it tells you something about how elastic the collision was, where CR= 1 for perfectly
elastic collisions and CR= 0 for completely inelastic collisions. Team up with another
table and use two gliders and two photogates on the same setup to measure CRfor
two gliders colliding with different bumpers (including one collision with no bumpers).
Compare the degree to which energy and momentum have been conserved for each
collision.
4.3.2 Presentation Mini-Report Guidelines
For this week’s presentation mini-report, please write an introduction and a methods section
for your work on Experiment 4. Please write the word count for your introduction on a new
line below its last sentence.
Introduction
The introduction section explains, in your own words, the purpose of your experiment
and how you will demonstrate this purpose. Try to be as brief as possible while getting
your point across. The main idea is to leave the reader with a good idea of what you were
trying to measure and how you went about it without going into painfully extreme detail.
69
According to Nature letters this should be less than 300 words so we will stick to that.
The introduction should give historical background, explain what problem you are trying to
solve, motivate how this work solves that problem and briefly explain what you did.
Methods
The methods section should outline in full technical detail how you set up the experiment,
what equipment you used and how you avoided systematic errors. How you analyze your
data is normally included in the analysis section that is not required this week. This means
you may reference analysis techniques in your methods section even if though the analysis
section does not exist! The methods section should have enough detail that a student with
the same equipment should be able to reproduce your results, after all science demands
reproducibility. Be sure that any images of equipment, charts or graphs are properly cited
unless explicitly created by you.
4.4 Epilogue
Smart phones these days are amazing. Your author could go on about this for days, but one
of the neat features available is the vector accelerometer. By attaching a smart phone to the
glider and performing the same experiment you did this week, your author was able to take
data at the same time with the smart phone and the PASCO force sensor. By multiplying
the measured acceleration by the mass of the smart phone and glider system, very good
agreement between these two methods was achieved (see Fig. 4.5).
70
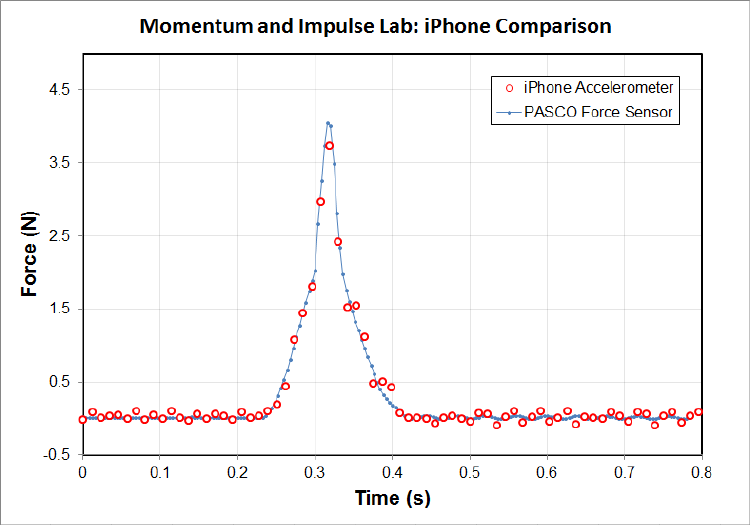
Figure 4.5: Comparison between the PASCO force sensor reading and the accelerometer from
an iPhone 4 that was riding along with the glider. The only free parameter for simultaneously
plotting the two data sets was the time offset.
71

Experiment 5: Harmonic Oscillator
Part I. Spring Oscillator.
The career of a young theoretical physicist consists of treating the harmonic
oscillator in ever-increasing levels of abstraction. -Sidney Coleman
You have already briefly encountered a harmonic oscillator in Experiment 3, where a
glider was held between two springs on an air track. The next three experiments in this
course are spent going into greater detail in the study of oscillating systems. You will study
the effects of damping, resonance, driving, superposition, normal modes, and phase shifts
in these systems. Here, in Experiment 5, you will be measuring the resonant frequency of
an undamped and damped harmonic oscillator. You will measure the damping term and
compare your results to predicted values based on a simple model. You will want to keep
your calculations from this experiment handy because you will need to apply some of these
techniques in your analysis of Experiment 6.
The oscillator for Experiment 5 is a mass hanging from a spring, shown in Fig. 5.1. You
will measure the mass and the spring constant, which will allow you to make a prediction for
the resonant frequency (fo, the cyclic frequency in Hz) of this undamped harmonic oscillator:
fo=1
2πsk
m.(5.1)
The spring will hang from a force sensor, providing you with a means to monitor the
oscillations of the hanging mass as a function of time using the DAQ. In your analysis, you
will be able to produce a measurement of foand compare it to your predicted value based
on Eq. 5.1.
We will then add a damping force F=−bv that is proportional to the velocity vof the
oscillating mass. We can model this system by an equation of motion of the form
m¨x=−kx −b˙x(5.2)
where kis the spring constant and xis the position of the mass in the vertical direction1.
The damping term, b, has units of mass/time, and tells you something about how much
1If you are not familiar with the dotted notation, each dot is a shorthand for a derivative with respect to
time. So ˙x≡dx/dtand ¨x≡d2x/dt2, etc.
72
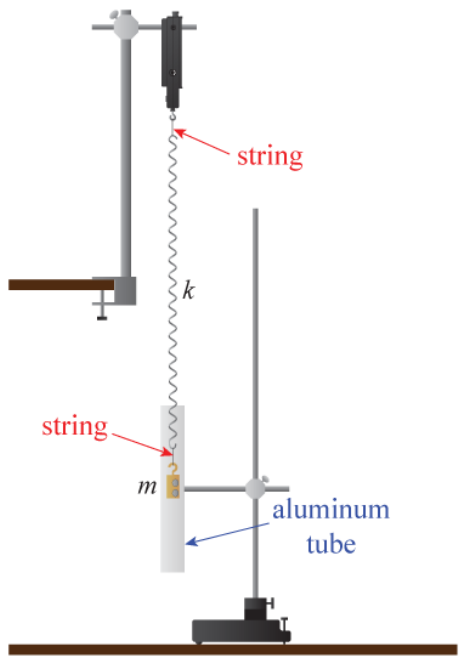
Figure 5.1: A weight hanging from a spring can exhibit oscillatory motion that is well-
described as being a simple harmonic oscillator. The force sensor from which the weight
hangs provides readout of the oscillatory motion. The weight has strong magnets attached
so that when the motion occurs inside a hollow aluminum tube, eddy-current heating provides
a dissipative force that is proportional to velocity. It is important to decouple the rotation
of the spring from the mass by using thin strings to attach the spring at both ends.
73
time it takes to damp out oscillations of some given mass. We will typically be concerned
not with bitself, but with combinations of b,m,fo, and kthat are more straightforward to
interpret.
Experimentally, it is a little bit challenging to come up with a method for producing
a pure damping term of this form without, for instance, adding a friction term (which
would not depend upon velocity). Your hanging mass for this experiment has some strong
magnets embedded in it, which we will use to produce the necessary damping force. The
hanging, oscillating mass will be surrounded by a vertical conducting tube that is made from
aluminum, a “nonmagnetic” material.
The effect of this tube on a magnet that is not moving will be very small and we will
neglect it here. However, when the magnetic field is changing in time due to the motion
of your oscillating mass, it causes eddy currents in the aluminum to be created due to the
tube’s inductance. The induced current is proportional to the velocity of the hanging mass.
Since aluminum has some finite resistivity, these currents are dissipated into heat. You will
not be required to derive these effects, but it is useful for you to have some idea why a
non-magnetic material might have a profound effect on the motion of a magnet.
By monitoring the amplitude of oscillations as a function of time, you will be able to test
whether the damping really is well-described by Eq. 5.2, and to provide a measured value
for 2m/b, which is the characteristic “damping time” of your damped oscillator. This will
also permit you to measure the Q-factor (or, simply the Q) of your resonance, which allows
you to complete your simple model of this system.
5.1 Theoretical Background
Your TA will probably cover this in more detail before you begin your experiment, but this
section is a brief discussion of how to derive the relationships needed for your analysis. Given
Equation 5.2:
m¨x=−kx −b˙x,
we want to find the form of the solutions for x(t). As is often the case when dealing with
differential equations, we can resort to a method of guess-and-check to find solutions. We
will use complex notation for our oscillator to make the algebra easier, choosing to try the
following form for our solution
x(t) = Aeiωt.(5.3)
We are in this case only interested in finding the (possibly complex) frequency ω, which
characterizes the time-dependence of the oscillator’s behavior. To find ω, we can simply
plug this into 5.2 to get
−ω2mAeiωt =−kAeiωt −iωbAeiωt
mω2=k+iωb. (5.4)
74

We can solve this quadratic equation for ωusing the quadratic formula to get
ω=ib
2m±sk
m−b2
4m2.(5.5)
Exercise 5.1 Sanity check: what does Eq. 5.5 give us if we remove the damping?
This should make some sense given what you know about oscillating spring-and-mass
systems. Also, how should we interpret the ±in Eq. 5.5? As a hint, consider the fact
that we chose a solution of the form x(t) = Aeiωt instead of x(t) = Aeiωt +Be−iωt.
We will take the plus sign of the ±for concreteness, but all of this analysis works just as well
if we chose the minus sign. Since this frequency has a nonzero imaginary part, we would like
to interpret what this means. Consider what happens if we plug Eq. 5.5 back into Eq. 5.3:
x(t) = Ae
iib
2m+qk
m−b2
4m2t
=Aeiqk
m−b2
4m2t×e−bt/2m.(5.6)
The second term in Eq. 5.6 is a pure exponential decay and does not contain any oscillating
terms. This comes from the imaginary part of Eq. 5.5, and demonstrates that imaginary
frequencies are usually used to express dissipative terms. We can therefore introduce the
damping time τ, defined as
τ≡2m
b.(5.7)
The damping time is the amount of time it takes this undriven, damped oscillator to have
its amplitude decrease by a factor of 1/e.
The first term in Eq. 5.6 is oscillatory and suggests that we adopt the following definition:
fdamped =ωdamped
2π≡1
2πsk
m−b2
4m2=fos1−b2
4km (5.8)
where fois given by Eq. 5.1. It is helpful to look back at Eq. 5.6 with these new definitions,
giving us
x(t) = Aeiωdamped t×e−t/τ .(5.9)
The real part of this expression is clearly a sinusoidal oscillation at frequency ωdamped that
has an exponentially-decaying amplitude with time constant τ.
Last, we will define something called the Q-factor (also known as the “quality factor”
or simply as “the Q”) of this resonance, which is 2πtimes the ratio of the total mechanical
energy to the amount of energy that is dissipated in a single cycle. Without proof, we will
define Qto satisfy
fdamped ≡fos1−1
4Q2,(5.10)
75
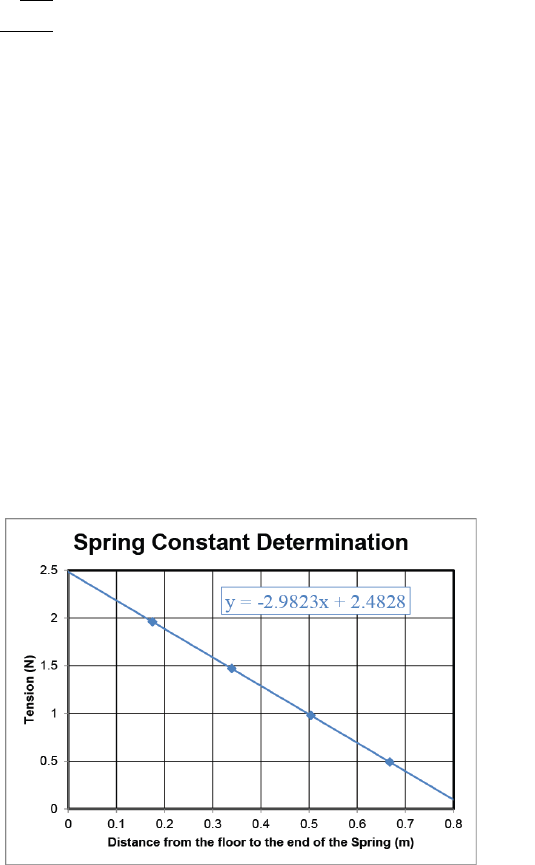
which gives us
Q=√km
b.(5.11)
In the limit of Q1, the Qof a resonance is also the ratio of the resonant frequency
(fdamped) to ∆f, the full-width-at-half-maximum (FWHM) of the (power) resonance curve.
The power resonance curve would be a plot of, for instance, energy dissipated per cycle
vs. frequency. You will explore this in more detail in Experiment 6.
5.2 Procedure
Since you will be measuring the frequency of a weight hanging from a spring, you will need
to measure the mass of the weight and the spring constant to provide something against
which to compare your direct time-domain measurement. While it may not be important
for your measurement of the spring constant, make sure that the spring is attached to the
hook of the force sensor via a string instead of being attached directly (and likewise for the
weight on the bottom of the spring), as shown in Fig. 5.1. Using a string to decouple the
rotation of the spring as it stretches from the mass and hook is important for making sure
your system is well-modeled by the simple equations above.
Measure the spring constant by hanging
at least 5 different masses from the spring
and recording its length. You may find it is
easiest to record the distance from the end
of the spring to the floor, because this re-
moves the need to hold the meter stick up
in the air. Since this distance is linearly re-
lated to the length of the spring, the spring
constant is still the slope, but there will be
an overall minus sign (the spring constant
is positive for these springs). You will per-
form the usual fitting in your analysis to turn
these measurements into a measurement of
the spring constant.
Next, you will set up the DAQ to measure the tension applied to the hook on the force
sensor as a function of time. For Experiment 5, you do not need to know the value of the
force, so feel free to set up the force sensor to display voltage or (uncalibrated) force units
as you see fit. For this discussion, we will assume the output is being recorded in volts.
In order to measure the frequency of oscillations, you will start the mass oscillating freely
and record the force sensor reading vs. time, as shown in Fig. 5.2. Be sure to keep the
oscillation amplitude small enough that the spring stays linear, which means the coils should
not touch one another and the spring should not be anywhere close to being plastically
deformed, etc. For this measurement, it is good to have both a Table and a Scope display
running simultaneously. You will want a sample rate that is high enough to really capture
76
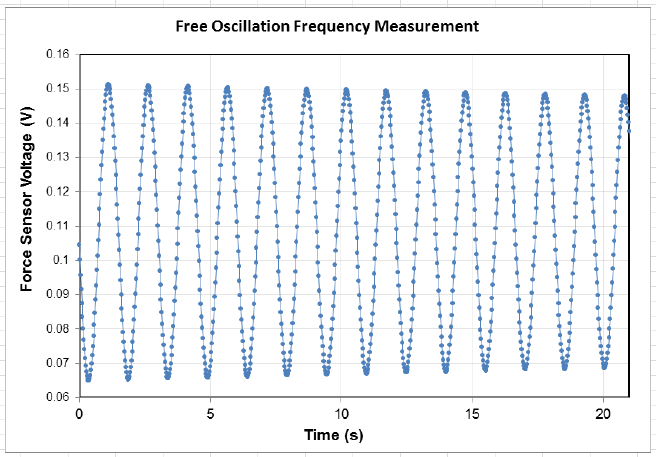
Figure 5.2: Free oscillation of a mass hanging from a spring. The oscillation frequency can be
obtained by, for instance, zooming in on the first and 14th oscillation minima and dividing 13
by the difference between these two times. Note the subtle decay of the oscillation amplitude
due to dissipation; this oscillator has a Qof more than 500.
77
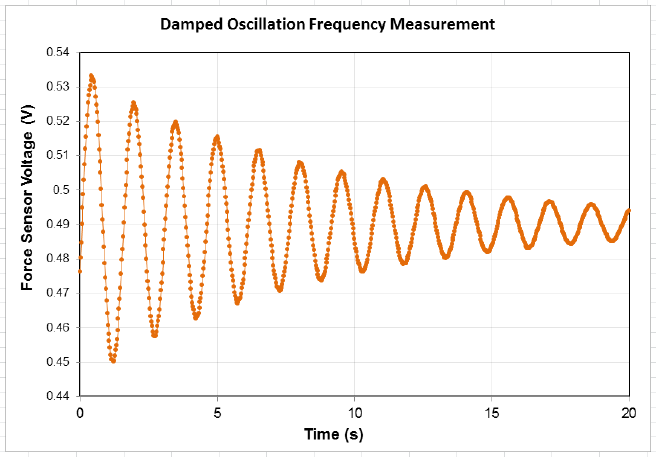
Figure 5.3: Damped harmonic motion. The characteristic damping time in this case is about
9.5 seconds, which corresponds to a Qof about 20.
the force maxima and minima (the “extrema”), so be sure your sample rate is at least 20
Hz in the Controls palette. If you exceed 50 Hz sample rate, it is likely that the amount
of data you generate will be large enough to be a headache for you and you will not get
a better measurement, so try something in the middle. Record oscillations for at least 20
seconds and up to 1 minute and then copy your voltage vs. time data into Excel for your
records. Sometimes the force sensors show a significant drift in their mean value over the
course of 20 seconds, so you may want to write down an estimate for this drift or to “warm
up” the sensor by letting your weight oscillate on the force sensor for a few minutes. This
effect is more important for the damped motion, so be sure to keep an eye on it and to either
quantify the drift or find a way to remove it for the next part.
Next, you will record the same time-domain data with the damping term added. Arrange
the aluminum tube to surround the oscillating weight without touching it. The magnets in
the weight will produce a damping force when the tube surrounds the moving weight. Record
the force sensor reading vs. time again and save your data. You will notice a significant decay
in the oscillation amplitude as a function of time, as shown in Fig. 5.3.
5.3 Analysis
Plot your displacement vs. weight data and fit a line to determine the spring constant. Use
a regression analysis to get an uncertainty on the slope, as usual. Combine this with your
measured mass of the weight with embedded magnets to come up with a prediction for the
78
free oscillation frequency fousing Eq. 5.1.
Next, you will measure the frequency of free oscillations directly from your time-domain
voltage vs. time data. Plot your data and use the positions of the extrema to determine the
oscillation frequency. You can do this by, for instance, zooming in on the first maximum
to determine the time when this occurred, doing the same for the nth maximum, then
dividing n−1 by the time difference, which will yield fo. You should estimate uncertainty
by using some reasonable method, such as repeated measurements with different extrema or
estimating your uncertainty in identifying exactly when each extremum occurred.
Do the same for your damped motion data. You will find that it is more difficult to
determine the damped motion frequency (fdamped) than the free oscillation frequency since
the signal-to-noise ratio gets worse as the signal amplitude decays, so your uncertainty is
likely to be quite a bit larger for the damped motion than the free oscillations. Nonetheless, it
should be possible to obtain a pretty good measurement of the damped oscillation frequency.
Next, you will see if the damping is well described by a velocity-dependent force and
estimate the damping time τ(see Eq. 5.7). τis the 1/e decay time of the amplitude of
the oscillations, and the amplitude will only decay exponentially in time if the damping is
indeed caused by a velocity-dependent damping force (as opposed to, say, a friction force).
To do this, your first step will be to find the voltage that corresponds to the midpoint in the
oscillations, as there is almost certainly a significant offset in your measurement. You need
to subtract this offset from the peak voltages to measure the amplitude.
For instance, consider the data shown in Fig. 5.3. These oscillations are clearly not
centered around 0 volts, and it looks like there is an offset of about 0.49 V. Try to find the
proper offset voltage to subtract from your data to give you oscillations that are centered
around zero. You may find that the force sensor reading drifted during your data run, in
which case you may need to choose a sort of mean value or try subtracting a linear drift
from the data. If your data includes points after essentially all of the oscillations died out,
that may provide you will a good measure of the offset voltage. Subtract your offset from
your data to produce oscillations that are centered around zero.
One of the things about an exponential decay is that the ratio of the heights of all pairs
of successive maxima should be the same since the maxima are separated by the same time
interval (T= 1/f). Record the heights of at least 6 successive maxima (more if you have
good data). Calculate the ratio of the height of peak 1 to peak 2, then peak 2 to peak 3,
and so forth for your data set. If the damping is indeed proportional to the velocity, these
ratios should all give the same result.
Plot your ratios as a function of which extrema were being used as shown in Fig. 5.4.
If the damping force is proportional to velocity, there should not be a discernible nonzero
slope to the data in the plot and the scatter will be centered around some mean value. If
you do detect a systematic trend upward or downward, be sure that you subtracted your
offset voltage correctly, as doing this improperly will also result in a drift of the peak hight
ratios with extremum number.
If you recorded amplitudes once per oscillation period T, the ratio of successive amplitude
79
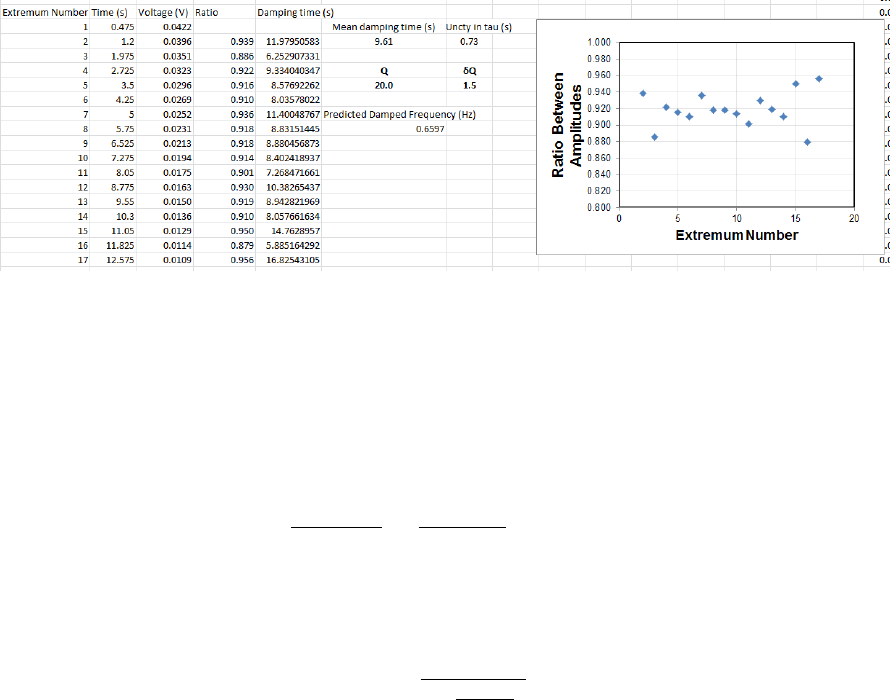
Figure 5.4: Analysis of the damped motion shown in Fig. 5.3. The peak height ratios are all
centered around 0.92. Notice that the scatter seems to increase with extremum number, as
expected since the peak height uncertainty increases.
measurements in an exponentially-decaying oscillation will be given by
V(t+T)
V(t)=e−(t+T)/τ
e−t/τ =e−T/τ .(5.12)
Since you already have a measurement of T= 1/fdamped, you can convert each ratio mea-
surement into a measurement of τby solving for the amplitude damping time
τ=−T
ln hV(t+T)
V(t)i.(5.13)
If your individual data points for the amplitude ratio did not show a trend (such as shown
in Fig. 5.4), you can use your first and last measured amplitude to determine the damping
time. Alternatively, you may combine all of your data points into a measurement of the
damping time with uncertainty, τ=τbest ±δτ. For this, you may use the procedure outlined
in §ii.1.6 to help you come up with an uncertainty in your mean value of τ.
Next, determine Qfrom your measurements. There are a couple of ways to do this, so
find a method that makes sense to you. Last, using your measured Q, predict the frequency
of damped oscillations based on your measured frequency of free oscillations fo. Is your
measurement of fdamped consistent with this? Is the difference between your best value for
foand fdamped statistically significant, or is the difference smaller than their uncertainties?
5.4 Report guidelines
This week, you will turn in your first full lab report. There will not be a
worksheet this week. If you do the extra credit, include it in your lab report
as if it were a normal part of your experiment. Your lab report should contain
the following items.
80
1. Cover Sheet
•experiment number and title
•your name and UID
•the date the lab was performed
•your lab section (for instance, “Tuesday 3pm”)
•your TA’s name
•your lab partners’ names
•the word counts for your abstract and for the body text of your whole report
2. Full Lab Report
This week you will write a full lab report including abstract, introduction, methods,
analysis and conclusions sections. Your completed lab report should look like an ar-
ticle submitted to a scientific journal. All guidelines from all of the previous weeks’
presentation mini-reports apply to this report, with the exception of reporting word
counts, which are to be reported in the cover page as described above. Since you will
be adding the analysis and conclusions sections, we will discuss some of the important
points of these parts of a journal article.
Analysis
This section should contain an explanation of how the data were analyzed to test
the experimental results against theoretical models. The methods section should have
explained how the data were obtained, but the analysis section presents the data itself
to communicate what the data show about the relevant physics. This section will
also typically include the discussion of uncertainty analysis, including any relevant
derivations of error propagation equations that were used.
The analysis section is typically where you would put your “main plot” for the paper.
The methods section can have plots, but the point of those is to describe how the data
were taken, whereas the goal of a plot in the analysis section is to show how the data
illustrate some point about physics.
Conclusions
Briefly restate the goals of the experiment and describe how your results demonstrate
(or fail to demonstrate) these objectives. Briefly describe at least one possible source
of systematic uncertainty that could affect your measurement (and which direction
the effect would tend to systematically shift your observations) and suggest a future
measurement to eliminate it.
Suggested content
81
Your report should be a fully self-contained description of your work this week. If
you performed a derivation or a measurement as part of this, it should probably find
its way into your report. While this is probably all the instructions you need at this
point to write a high-quality report, the following is a list of some of the specific things
from this experiment that you should probably include in your report in one form or
another. That said, anything that is needed to communicate to the reader what was
done and what it showed should be included.
•A derivation of two or three equations that show how the quantities fo,Q,τ, and
bare related to one another. You may take Eq. 5.10 to be a definition of Q.
•A description of how you determined the frequency of oscillations and how you
measured the decay time in the damped case.
•Your measured mass of the hanging weight.
•A plot of the spring constant measurement and a statement of the result, with
uncertainty.
•Your predicted value for the free oscillation frequency.
•A plot of the free oscillatory motion vs. time. You do not need to worry about
calibrating the y-axis and may simply plot it as being the force sensor reading in
volts.
•How did you determine the free oscillation frequency from the data in this plot?
What was your result?
•Compare your predicted free oscillation period and your measured value. Do they
agree? If not, try to explain what is the cause of the mismatch.
•A plot of the damped oscillatory motion in the same manner as your previous
plot. Use these data to measure the damped oscillation frequency.
•A plot of the measured ratios of successive extrema in your damped oscillation
measurement. Is there a trend? If so, what does this mean? If not, what does
this tell you?
•Present your measured value for the damping time with uncertainty.
•State your measured value for Q(with uncertainty) and use this to make a pre-
diction of the damped oscillation frequency.
•Compare your measured damped oscillation frequency to this prediction. Do they
agree? Are the two oscillation frequencies distinguishable given their uncertain-
ties?
3. Extra Credit
It is possible to determine the frequency response of this system to any periodic driving
force using the data you obtained. One way that this frequency response may be
obtained is by taking the Fourier transform of the time-domain signal, which is called
82
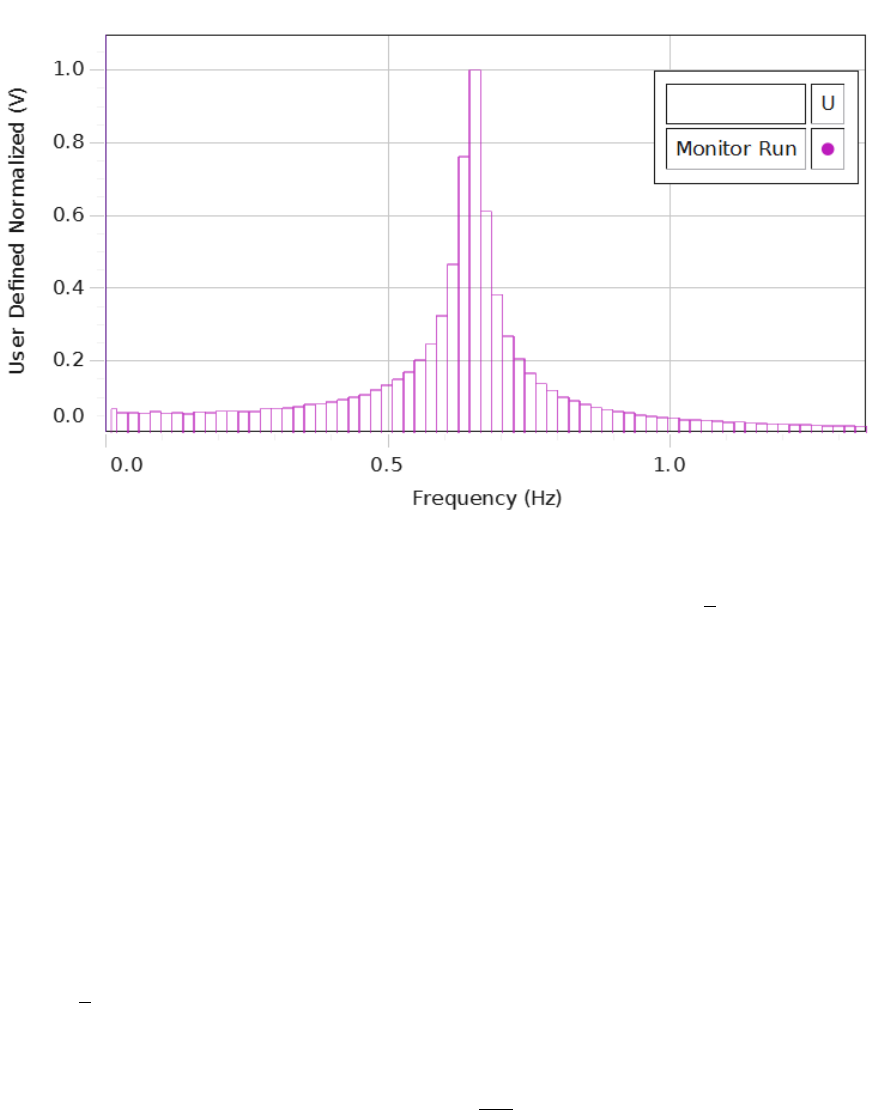
Figure 5.5: Fast Fourier Transform (FFT) of the data from Fig. 5.3. Based on the analysis,
we would predict this amplitude spectrum to have a full width at 1/√2 of its maximum of
about fo/Q ≈33 Hz, which would be about two frequency bins wide on this plot, consistent
with the spectrum shown.
the step response of your system. In Experiment 6, you will be looking at the amplitude
response of another oscillator in the frequency domain, but it may be instructive to
see what this would look like for the oscillator in Experiment 5. Capstone provides
an algorithm for showing the frequency content in a time-domain signal called a Fast
Fourier Transform (or FFT). Figure 5.5 shows the results of using this tool, which
shows a very nice resonance curve.
Use the Capstone FFT tool in the Displays palette to see if you can generate a
frequency-domain representation of your data such as that shown in Fig. 5.5. Estimate
the width of the resonance (∆f), defined as the full width of this peak at a height of
1/√2 of its maximum value. If Qis large, one way to calculate Qis as the ratio of the
resonance frequency to the resonance width:
Q=fo
∆f.(5.14)
Use this to calculate Qand compare your answer to that obtained from your time-
domain analysis.
83
5.5 Epilogue
Q-factors for resonators span a large range of values. The highest Qof which your author
is aware for a man-made oscillator is an optical cavity that is made out of a single crystal
of silicon [3]. This oscillator has a center frequency of fo= 200 THz and a frequency width
of ∆f < 40 mHz, which gives us Q > 5×1015. It is by using cavities such as these that
researchers are able to obtain the passive stability required to make measurements such as
those reported in [7] (Eq. ii.2).
84

Experiment 6: Harmonic Oscillator
Part II. Physical Pendulum.1
The harmonic motion you have studied in previous experiments in this course was typically
analyzed in the time domain, meaning you acquired and studied data as a function of time.
It is also possible to analyze the properties of a harmonic oscillator in the frequency domain,
which you will begin to explore in Experiment 6. You will also study the effects of damping in
more detail, allowing you to demonstrate underdamped, overdamped, and critically-damped
motion by experimentally controlling the damping coefficient. These concepts are important
in control systems for physics and engineering applications, where we are often concerned
with trying to find the fastest way to bring a system to a desired state.
The spring-and-mass oscillator you used in Experiment 5 was very well modeled by
Eq. 5.2. However, you were not able to adjust the damping coefficient in a well-controlled
manner using the aluminum tube, so we will use a more flexible setup in Experiment 6, shown
in Fig. 6.1. An aluminum pendulum shaped like an anchor will serve as your oscillator. The
oscillation frequency of this pendulum will be approximately independent of amplitude so
long as the amplitude is small, where we will say that “small” means the angular motion does
not move the bottom further than the width of the curved bottom portion of the pendulum.
A “wave driver” will be coupled to the pendulum through a vee-shaped torsion spring.
This will allow us to not only drive the oscillator with a periodic signal, but also to read
out the instantaneous position of the oscillator via the rotation sensor. Even though the
wave driver will not be used for your study of damping regimes, the spring that couples the
wave driver to the pendulum will be a factor in determining the resonant frequency of the
pendulum. Since you will apply your results from the damping study to your study of driven
oscillations and resonance, you should leave the wave driver attached to the pulley for your
damping study even though it is not in use.
The wave driver can be connected to the DAQ via the banana jacks on the upper right
of the 850 interface (see Fig. i.1). The rotation sensor plugs into one of the four “Passport”
sockets on the bottom row of the 850 interface. Capstone will automatically detect and
configure the rotation sensor, but you should configure output channel 1 to be an Output
Voltage Current Sensor, which will allow you to monitor the phase of the drive signal.
1This chapter was contributed by Priscilla Yitong Zhao (2013).
85
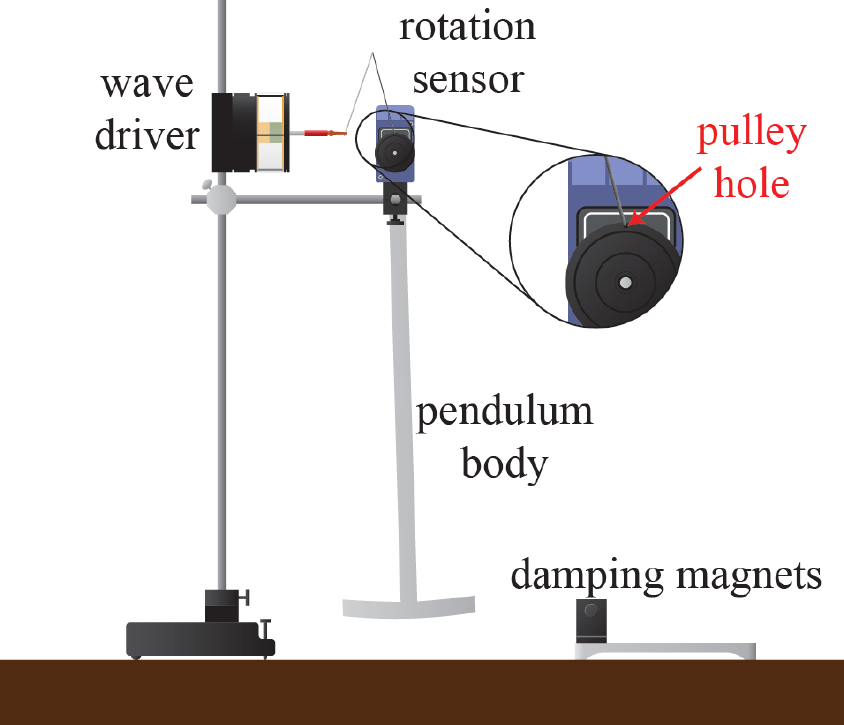
Figure 6.1: A physical pendulum for studying damping regimes and resonance. For small
oscillations, the pendulum is a harmonic oscillator, and its angular position can be measured
in real-time using a rotation sensor. A wave driver coupled to the pendulum by a torsion
spring (shaped like a Λ, shown above) can be used to study damped, driven oscillations. The
damping magnets can be moved into place and their spacing changed to provide a tunable
velocity-dependent damping force.
86
Damping will once again be provided by eddy-current heating due to the motion of the
oscillator. Unlike Experiment 5, however, this time it is the conductor that will be moving,
and the magnets will be stationary. By moving the magnet stand so that the pendulum
bottom passes midway between the two damping magnets, the amount of damping can be
tuned by adjusting the spacing between the magnet poles and the bottom of the pendulum
body.
You will acquire time traces for underdamped, overdamped, and critically-damped mo-
tion.
6.1 Theoretical Background
6.1.1 Undriven oscillations of a damped physical pendulum
A pendulum that is made of a rigid object (as opposed to a point-mass on an ideal flexible
string) is known as a physical pendulum. It is no more or less physical than a ball on a rope,
but this terminology is used nonetheless. The physical pendulum in this experiment is a sort
of anchor-shaped piece of aluminum alloy cut from sheet metal, and will be characterized by
a moment of inertia Iabout the rotation axis.
If we call the angular deviation of the pendulum from its equilibrium configuration θ, any
nonzero value of θwill lift the center of mass of the pendulum above its equilibrium height. If
the pendulum has a total mass Mand the location of its center of mass is a distance lbelow
the rotation axis, the restoring torque due to gravity will be given by τgrav =−Mgl sin(θ).
The spring attached at the top also exerts a torque on the pendulum about its rotation
axis. For an angular displacement θ, the spring’s length is changed by an amount propor-
tional to sin(θ). The torque this exerts on the pendulum is proportional to this force times
cos(θ), resulting in a restoring torque that we may write as τspring =−Bsin(θ) cos(θ).
Last, the magnet assembly at the bottom of the pendulum will exert a damping torque
on the pendulum, turning the angular kinetic energy into heat via eddy-currents in the
pendulum blade. This damping force is proportional to the tangential velocity of the blade,
so the damping torque will be proportional to the angular velocity, which we will write as
τdamping =−b˙
θ. Putting these three terms together gives us an equation of motion for the
pendulum that is rather complicated:
I¨
θ=X
i
τi
=−Mgl sin(θ)−Bsin(θ) cos(θ)−b˙
θ(6.1)
For small angular displacements (θ1 rad), we may expand the trig functions and keep
the first terms only, thereby invoking a small angle approximation (sin(θ)≈θ, cos(θ)≈1):
I¨
θ≈ −(Mgl +B)θ−b˙
θ
=−kθ −b˙
θ(6.2)
87
where we have defined a constant k≡Mgl +B.
If we compare Eq. 6.2 to Eq. 5.2, we see that this is exactly the same equation of motion as
we had in Experiment 5 with the replacement x→θ,m→Iand different physical definitions
of kand b(with different dimensions). We may therefore write down the solutions directly
by making these simple substitutions in Eq. 5.6
θ(t) = Aeiqk
I−b2
4I2t×e−bt/2I(6.3)
=Aeiωdamped t×e−t/τ .(6.4)
with the analogous definitions of the oscillatory frequency2and damping time:
ωdamped ≡sk
I−b2
4I2=sω2
o−1
τ2(6.5)
and
τ≡2I
b(6.6)
with the undamped resonance frequency ωo≡qk/I > 0.
The motion (described by the real part of θ) is again a sinusoidal oscillation at frequency
ωdamped that has an amplitude that exponentially decays with time constant τ. We identify
three regimes of damping in this system:
•Underdamped motion occurs when there is still oscillation of the pendulum for zero
initial velocity, meaning θchanges sign at least once. This occurs when the oscillation
frequency is faster than the damping rate: ωo>1
τ, which guarantees that ωdamped is
purely real. A good example of underdamped motion can be seen in Fig. 5.3.
•Overdamped motion occurs when ωo<1
τ. Looking at Eq. 6.5 shows us that in this
case, ωdamped will be purely imaginary, which means the angular displacement of the
pendulum for zero initial velocity (Eq. 6.4) will be described by a pure exponential
decay with no oscillations.
•Critically-damped motion is the crossover regime between these two cases, given by
ωo=1
τ. For critical damping, the system will return to its equilibrium state faster
than either overdamped or underdamped motion. Automobile suspension systems, the
dash pots on the doors to the lab, and earthquake suppression systems are all examples
of engineering tasks where one would want this feature of critical damping.
6.1.2 Driven oscillations
If the wave driver is driven by the analog output of the 850 interface, there will be a new
term in the equation of motion for this physical pendulum that oscillates with the driving
2Here, as elsewhere in this manual, we use ωas a symbol for angular frequency (units of rad/s or just
s−1) and fas a symbol for cyclic frequency (units of Hz).
88

current, which we will assume is proportional to cos(ωdt) for drive frequency ωd. In the
small-angle approximation, we have
I¨
θ=−kθ −b˙
θ+Ccos(ωdt).(6.7)
The solution to Eq. 6.7 consists of the sum of two terms, known as the complimentary
and particular solutions:
θ(t) = Acθc(t) + Apθp(t) (6.8)
where the complimentary solution is the one we have from undriven oscillations
θc(t)≡eiωdamped t×e−t/τ (6.9)
and the particular solution is given by [4]
θp(t)≡1
r(ω2
o−ω2
d)2+2ωd
τ2cos (ωdt−φ(ωd)) (6.10)
where
φ(ωd)≡arctan 2ωd
τ(ω2
o−ω2
d)!.(6.11)
The complimentary solution describes all of the transient behavior of the system, which
depends upon things such as initial conditions but does not depend upon the driving term
in any way. This behavior is called transient because as t→ ∞,θc→0. The timescale over
which the transient behavior dies out is τfor critical and underdamping. For this experiment,
this means that when you make a change to the system by, for instance, changing the drive
frequency, you must wait at least τbefore interpreting the behavior of the system as being
dominated by the particular solution. You should consider this when trying to decide how
to set the damping rate for the second half of this experiment.
6.1.3 Resonance
The particular solution, on the other hand, describes the steady-state behavior of the system
under a constant-amplitude, sinusoidal driving torque. The pendulum will oscillate at exactly
the drive frequency ωd, but with an amplitude and phase that depend upon the relationship
between ωdand ωdamped. Fig. 6.2 shows the calculated amplitude of the steady-state behavior
for a series of oscillators with the same undamped resonance frequency but different levels
of damping.
The drive frequency at which the amplitude is maximized is called the resonant frequency
of the damped, driven system, ωR. To find ωR, we can differentiate the amplitude of the
response in Eq. 6.10 with respect to ωdand solve for ωdsuch that this is zero, which gives
us the resonant frequency of a damped and driven harmonic oscillator:
ωR=sω2
o−21
τ2.(6.12)
89
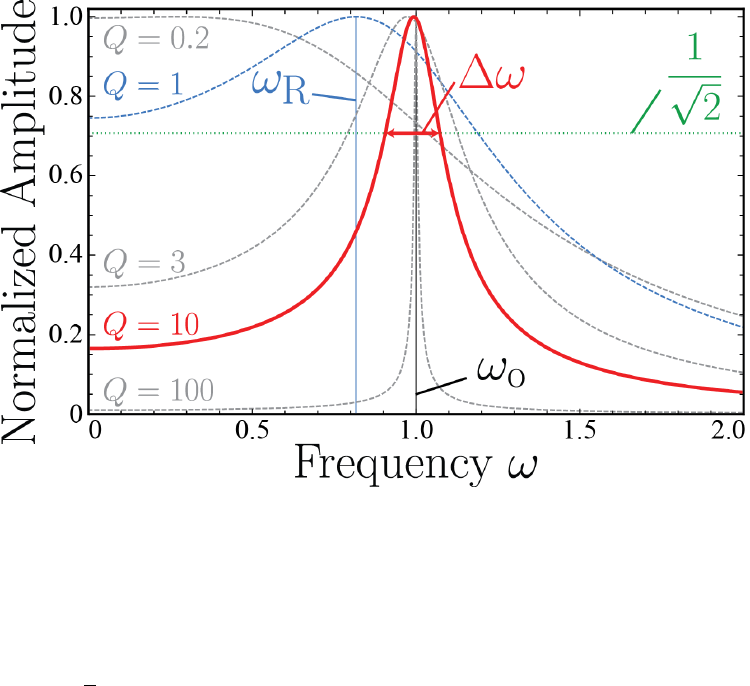
Figure 6.2: Amplitude response of damped resonators. The amplitudes are normalized to
set the peak amplitude equal to 1, and the undamped resonance frequency for all curves
is ωo= 1. Low-Qoscillators show resonant frequencies ωRthat are significantly different
from ωo. As the quality factor increases, the resonance position converges to ωR=ωo. The
resonance width ∆ωis shown for a Q= 10 resonance, where ∆ωis the full width when the
amplitude is 1/√2 times its maximum value.
90
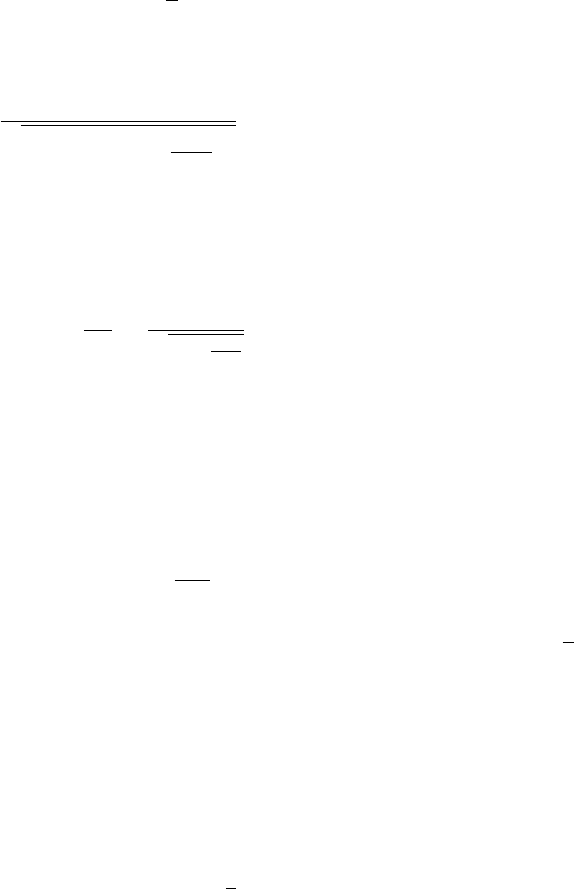
Comparing this to Eq. 6.5, we see that the resonant frequency can be close to ωdamped, and
in many situations they are used interchangeably. In the limit τ1/ωo, both of these
resonant frequencies converge to ωo, and you will find that they are often assumed to be
equal (especially for Q≥10 or so, see below).
We can define the quality factor Qof this system to be
Q≡1
2τωR(6.13)
to rewrite our particular solution as
θp(t)≡1
r(ω2
o−ω2
d)2+ωRωd
Q2cos (ωdt−φ(ωd)) (6.14)
For an oscillator with a high Q-factor (Q1), the damping time τis many oscilla-
tion periods long. As discussed above, in this case we see that ωRand ωdamped are both
approximately equal to ωo. In fact, we can rewrite Eq. 6.12 in terms of Qas
ωR
ωo
=1
q1 + 1
2Q2
.(6.15)
This factor goes to 1 very quickly as Qis increased. For instance, an oscillator with a modest
quality factor of Q= 3 has a resonance frequency that is only about 3% slower than the
undamped case.
For these cases where the damping is very slight and the Qis therefore substantially
greater than 1, it can be shown that the Q-factor is the ratio of the resonant frequency to
the width of the resonance curve
Q≈ωo
∆ω(6.16)
where ∆ωis the full width of the resonance, defined to be the frequency range over which
the amplitude response (the amplitude in Eq. 6.14) is greater than or equal to 1/√2 of its
maximum value. (This will be familiar to you if you did the Extra Credit from Experiment
5.)
Last, the phase term described in Eq. 6.11 is shown in Fig. 6.3 for the same oscillators as
the amplitudes in Fig. 6.2. This plot has been constrained to positive values of φto show that
the phase changes smoothly through resonance, a general property of damped oscillators.
Far below undamped resonance, the phase of the response is almost exactly the same as the
drive, but begins to increase as undamped resonance is approached. Right on undamped
resonance (ωd=ωo), the phase lag is exactly φ=π
2, regardless of the quality factor. Past
this point, the phase lag continues to increase, asymptotically approaching φ=πat infinite
frequency.
From a practical perspective, if we are tasked with trying to locate the (un-
damped) resonant frequency ωo(a common situation in physics and engineering),
looking at the phase response is going to be far more sensitive than using the
91
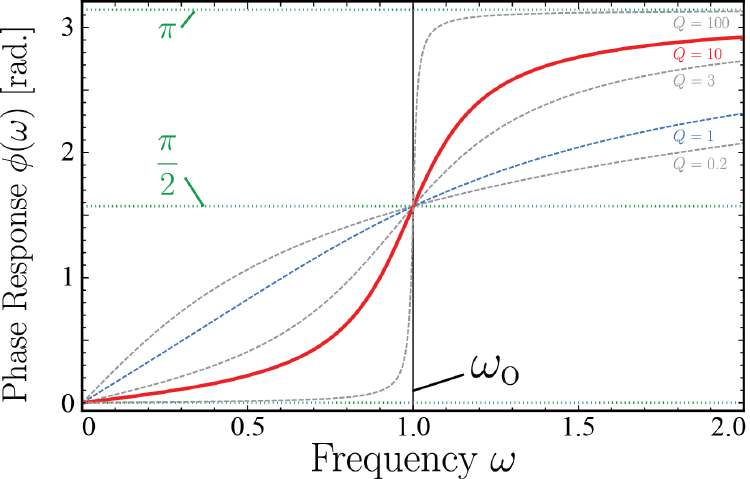
Figure 6.3: Phase response of damped resonators. The phase response is the difference
between the phase of the driving force and that of the displacement. Curves are shown for
the same parameters as Fig. 6.2. For driving frequencies below undamped resonance, the
response is said to be “in phase with the drive,” with the lag increasing as the drive frequency
increases. At the undamped resonant frequency, the response is out of phase from the drive
by π/2, regardless of Q. Past this point, the response is said to be “out of phase with the
drive” and the phase lag continues to increase with frequency, saturating at φ=π.
92
amplitude response. The reason is that in the immediate neighborhood of resonance,
the amplitude response levels out and does not depend sensitively on frequency, whereas
the phase response has a maximum slope at this point. The amplitude response therefore
becomes insensitive to frequency right where you need it the most, while the sensitivity of
the phase to frequency is maximized here.
6.2 Procedure
To set up the apparatus, start by connecting the spring on the wave driver to the wheel on
the pendulum rotation sensor as shown in Fig. 6.1. You will leave this connected for the
entire experiment. To make sure the pendulum swings freely for all parts of the experiment,
set the heights of the posts and clamps so that the pendulum blade can swing freely a couple
of inches above the table so that the blade is well-centered at the height of the magnets. In
order to center the angular readings about the equilibrium position, uncheck Zero Sensor
Measurements at Start in the options for the rotary motion sensor and click Zero Sensor
Now when the pendulum is in its equilibrium position.
6.2.1 Harmonic motion with damping
To investigate the various regimes of damping, you will be pulling back the pendulum to
some starting position and letting it go, recording the angular displacement vs. time using
the rotation sensor via the DAQ. Remember, the small-angle approximation we used in the
theory section is only valid for angles right around θ= 0, so be sure to set your starting
position no more than one blade length from the equilibrium position.
You may find it useful to have a photogate attached to the 850 to record the starting
condition when you let go of the pendulum. To do this, position the photogate such that
the pendulum blade just unblocks it when motion begins.
The amplitude sampling rate should be set so that you get enough data points to clearly
see which damping regime you are in without recording lots of points for the same value
of the angle. You should play around with this in Scope mode to choose an appropriate
sampling rate. You will be recording angle vs. time for different values of the spacing between
the magnets.
Record traces of the pendulum angle vs. time for the case where the magnets are removed
entirely, as well as 4 or five gaps between 50 mm and 10 mm. You will want to get a clear
set of data for underdamped motion and a clear set for overdamped motion for your report.
Next, determine the magnet spacing to achieve critical damping. Remember, you are
looking for the minimum damping such that the angular velocity of the pendulum does not
change sign. Take steps at least as small as 1 mm for this, and save a good set of data
showing critical damping for your report. Fig. 6.4 shows some example data for all three
regimes. Note that you may need to record where θ= 0 is for each data set to plot them on
the same axes.
93
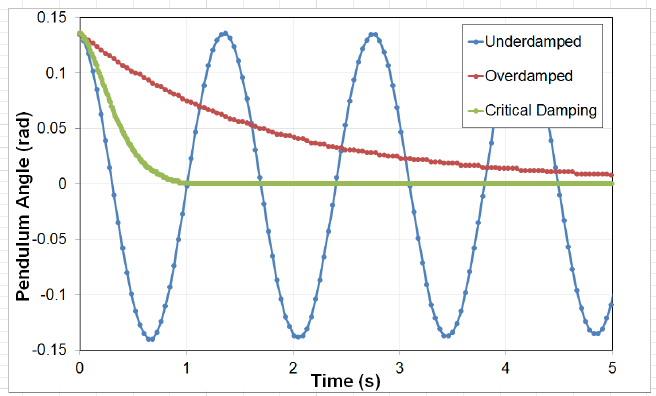
Figure 6.4: Illustration of damped motion in all three regimes of damping. All three plots
were obtained with the same initial displacement and no initial velocity. The critically-
damped trace clearly converges to θ= 0 the fastest.
6.2.2 Harmonic motion with driving
For the second part of the experiment, you will be examining the amplitude and phase
response of the driven, damped harmonic oscillator. For this whole section you will want to
set the magnet gap to something with fairly low damping. Remember, you are looking to
add just enough damping so that you don’t have to wait too long for the system to reach
steady state, but not so much that the oscillation amplitude is difficult to detect. You will
benefit from trying to use your data from the previous section to set the magnet so that
the free undriven oscillations damped out in about 5 to 10 seconds. Record a good trace of
displacement vs. time for this setting of the magnet in undriven conditions just like you did
for the previous part of the experiment.
You will be using the wave driver to produce a sinusoidal driving torque on the pendulum
using the Signal Generator function. This is physically accomplished by attaching the
banana plugs from the wave driver into Output 1 of the 850. The drive function will be a
Sine function, and you should probably keep the drive voltage below 5 V or so. The frequency
range you will be exploring will be very near the frequency of the undriven, undamped
oscillator, so take a look at your data from the first part with no damping magnets to
estimate the undamped frequency.
First, you will locate the driven, damped oscillator resonance ωR, which you may assume
is equal to ωosince you are working with low damping. As discussed, the easiest way to do
this is by looking at the phase response. To accomplish this, you will be making parametric
plots known as Lissajous figures. What I mean by “parametric plot” is that you will be
plotting two separate quantities that are related by a parameter, which in our case will be
94
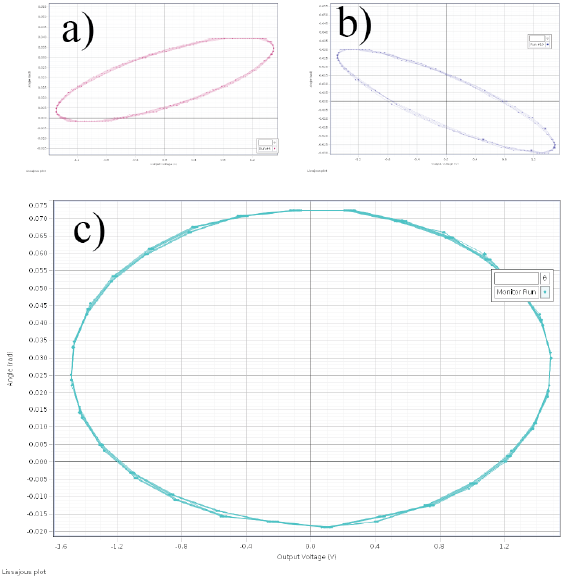
Figure 6.5: Parametric plots of the driving and response of a damped harmonic oscillator
showing Lissajous figures. These screen shots from Capstone show the measured pendulum
angle on the vertical axes and the drive voltage on the horizontal axes for drive frequencies
(a) below, (b) above, and (c) right on resonance.
time. You will be plotting Angle on the y-axis and Output Voltage on the x-axis of a
Graph display. You will also need to record data for several oscillations after the system
reaches steady-state for your analysis, so now is a good time to set up a Table for doing
that.
Your parametric plot will show both the drive voltage and the system response (the
angle) on one set of axes. For steady-state behavior, this plot will look like a closed curve.
You will need to play around with scaling the axes to try to make the extent of the shape on
the x-axis be about the same distance as the extent on the y-axis so that you can clearly see
the difference between a perfect circle and a slight ellipse. Fig. 6.5 shows an example screen
shot of the parametric plot.
To find ωR, you are looking for the drive frequency such that the steady-state behavior
looks like a symmetric circle on the parametric plot, such as shown in Fig. 6.5(c). You will
find that this technique is incredibly sensitive to the proximity to resonance, and even a
small deviation from resonance will be visible as a slightly-tilted ellipse. The direction of the
tilt tells you which direction you need to go to find resonance, as shown in Fig. 6.5(a) and
95
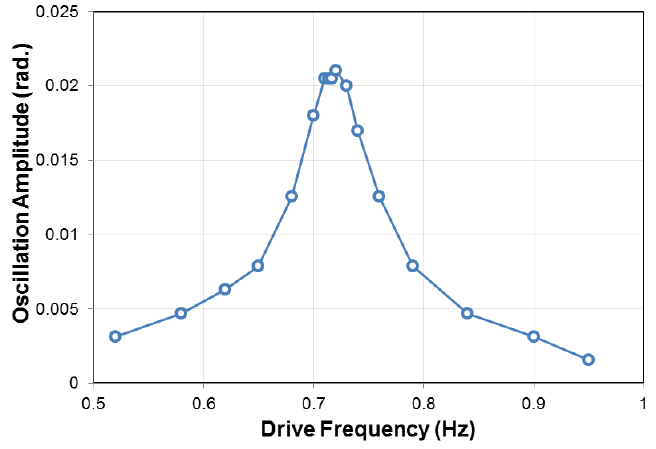
Figure 6.6: Measured oscillation amplitude spectrum of a damped, driven oscillator. The
points are connected by straight lines, showing the Lorentzian shape of the resonance as
predicted in Fig. 6.2.
(b). Estimate an uncertainty in your measurement of ωRbased on how small a deviation
you think you can make without being able to tell a difference in the figure. You will need to
save data for plotting Lissajous figures in your report for driving the system below, above,
and right on resonance.
Last, you will examine the amplitude response of the oscillator as a function of frequency.
It is by design that this lab manual is not going to provide you with detailed instructions
for how to go about this; you are expected to come up with an appropriate experiment on
your own (or with your lab partner) that you think makes sense. Record the amplitude of
the pendulum oscillations in steady-state for at least 10 different drive frequencies. You are
trying to make a figure that allows you to see the shape of the resonance curve, which will
allow you to find its height and estimate its width. An example is shown in Fig. 6.6 with the
data points connected by straight lines, which shows a Lorentzian shape similar to Fig. 6.2.
6.3 Analysis
For undriven damped motion, you recorded time traces of the angular displacement. From
your trace that was taken with no magnets present, you should be able to determine the
undamped oscillation frequency ωo. Make a plot clearly showing three time traces, one in
each regime of damping. You can assign an uncertainty to the critical damping gap between
the magnets based on how well you can measure the gap and how much change it would
96

take for you to say definitively that the damping has changed regimes.
For the driven, damped oscillator, plot your time trace of the undriven oscillator at the
magnet gap you used for the rest of the lab. Using the same technique you employed in
Experiment 5, find the damping time τby analyzing how much the amplitude decreases as
a function of time for undriven oscillations. Assign an uncertainty to your measurement of
τbased on how well you think you were able to measure the change in amplitude.
You recorded a measurement of the driven resonant frequency ωRthat was obtained
by looking at the Lissajous plots. Combine this with your measurement of τto report a
measured value for Qvia Eq. 6.13. Plot three Lissajous figures for your report showing the
shape below, on, and above resonance.
You have one more way to determine Q, which you will compare to this method. This
second method to measure Qis to examine the amplitude response to apply Eq. 6.16. To do
this, plot your amplitude spectrum and determine the peak height (which should occur at
ωR). It is sometimes helpful to connect the data points with straight lines for this analysis.
Using your plot, come up with some way to estimate the full width of the resonance where
the amplitude is 1/√2 times its maximum (∆ω), such as depicted in Fig. 6.2. You are
unlikely to have two data points at exactly the right place, so you will need to make an
educated guess about this width based on the plot and assign an uncertainty to it. You
have been assigning uncertainties in this course for many labs now, and you should be able
to come up with a way to do this that effectively reflects how well you really measured this
quantity. Use your measurement of ∆ωto calculate Q, and think about how much you trust
this measurement compared to your other method for determining Q.
6.4 Report guidelines
This week you will NOT turn in a lab report, nor will there be a worksheet
this week. Your presentation of this week’s experiment will be part of a double
credit lab report on resonance that will cover both Experiments 6 and 7. If you
do the extra credit, please save it for your lab report next week and include it
as if it were a normal part of your experiment. Even though there is no report
due next week, students are encouraged to make sure they can be prepared to
include everything they did this week.
1. Some items to consider including in your next report: It will be up to you to
form a coherent story in the lab report due next week. You must decide what graphs,
tables and other information should be included. Some general concepts for this lab
are
•Regimes of damping
•Resonance
•Q-factor and line widths
97

2. Extra Credit In the driven resonance part of this experiment, you estimated ∆ωby
using a plot that probably did not have two data points right at 1/√2 of the maximum
amplitude. Find these two frequencies experimentally by moving the drive frequency
around until the amplitude is at 1/√2 of its maximum value. The difference between
these two frequencies should be ∆ω. Plot Lissajous figures at both frequencies and
describe what they have in common. Use your new measured value for ∆ωto determine
Q. Further, since the frequency of the peak of your amplitude spectrum is ωR(even
when ωRand ωoare not approximately equal), this gives you yet a third method
for determining Q. The damped, driven resonance frequency ωRmust be halfway
between the two frequencies you found for achieving 1/√2 of the maximum amplitude.
Combine this with Eq. 6.15 to get a third measurement of Qand compare all of your
measurements and methods. Which do you believe is the most accurate, and why?
6.5 Epilogue
It is remarkable how much physics can be understood by modeling systems as harmonic oscil-
lators. The Lorentzian line shape shows up in everything from nuclear scattering to molecular
spectroscopy and plays an important role in almost every field of physics. Understanding
the fundamentals, such as the phase change across resonance and how the shape changes
with damping will be helpful for many advanced courses and research work in engineering
and physics.
98

Experiment 7: Waves on a Vibrating
String
In Experiments 3, 5, and 6 you studied systems that exhibited simple harmonic oscillations,
where the part of the system that was oscillating was often modeled as being a single point-
mass. Non-rigid, spatially-extensive systems can also exhibit simple harmonic motion, where
each point in space has some property that oscillates periodically in time and is somehow
related to the oscillations of its neighboring points. Phenomena such as these exhibit wave
behavior, and wave phenomena show up in everything from light to sound to the quantum
mechanical nature of probability amplitudes. In Experiment 7, you will be examining waves
on a stretched, massive string with clamped boundary conditions. You will measure the
group velocity of traveling waves on this string and predict and observe the behavior of
many modes of standing-wave oscillation. Last, you will investigate one of the curious
features of interference, where the addition of an intermediate boundary condition can be
used to suppress certain modes while leaving others effectively unchanged. Information about
laser safety can be found in Appendix A and should be read if you do not know how to safely
use a low-power visible laser.
7.1 Procedure
To configure the DAQ for this experiment, you will
need to be able to read the signal from a light sensor
(analog channel) and to send a signal to the wave
driver via output channel 1 (the red and black banana
plugs). The polarity of the plugs does not matter.
In order to excite and monitor wave behavior, we
will be using transverse motion of a thick, elastic
string about a meter long. The tension in the string is an important parameter for us
to control in a known way, so while one end of the string will be held in a clamp (shown in
Fig. 7.1a and b), the other will pass over a pulley wheel (Fig. 7.1a and c) and weights can
be hung from this end. The tension will be set by T= (m1+m2)g, where m1and m2are
the mass of the hanging weight and the portion of the string that is hanging from the pulley
99
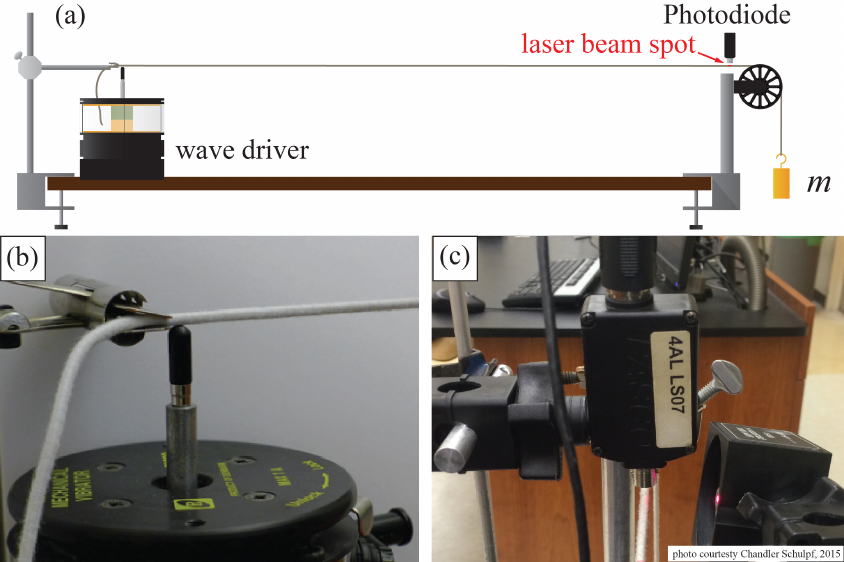
Figure 7.1: Apparatus for observing the vibrational motion of a massive string. (a,b) A
wave driver excites vibrations near the clamp, which can be detected when (c) a laser beam
scatters light into a photodetector at the other end. The tension in the string is controlled
by hanging known masses from a pulley that defines the other boundary.
100

(which has a non-negligible mass!).
The wave driver will excite vibrations in the string by moving the actuator in the vertical
direction. The actuator tip should just barely touch the string when the actuator is not
running, and should be positioned 1-2 mm from the clamp, as shown in Fig. 7.1b. You may
need to move the wave driver on the table until the driven waves are in the vertical plane
and do not have a significant horizontal component. It is recommended that you do not
put the string into the notch on the tip of the actuator, but rather just use the tip to push
against the string.
You will be measuring the displacement of the string in the vertical direction by scattering
laser light into a photodiode (also called a photodetector, light sensor, or simply “detector”).
The laser spot should be positioned so that it illuminates the top third or so of the string
when the string is not moving. The photodetector should be positioned directly above the
laser spot very close to the string to minimize the effect of ambient light getting into the
detector. In this way, if that section of the string moves downward, it scatters less of the
laser light into the detector and the signal decreases, with the converse being true if this
string moves upward. A photodiode gain setting of 10 seems to work well for this, but feel
free to experiment.
The laser beam should illuminate the string fairly close to the pulley (see Fig. 7.1c). For
Part 1 of this experiment (measuring the wave speed), it is recommended that you set the
beam (and photodetector) 6-8 cm from the pulley apex. For Part 2 and Part 3 (standing
waves), you will likely obtain the best results if the beam and photodiode detector are about
1 cm away from the pulley apex.
Setting the laser spot and detector positions correctly can be tricky, and it is worth your
time to make sure you do this well to get high-quality data. It is recommended that you
use Scope mode to monitor the Light Sensor while using Signal Generator to run the
wave driver in a sinusoid at 4-10 Hz with 1-2 V of output. For measuring the wave speed
(Part 1), adjust the laser beam and detector until the resulting signal is clearly visible above
the background noise. For the standing waves sections of this experiment (Parts 2 and 3),
you will also want to make sure that when you drive a standing wave on resonance with a
sinusoidal drive, the output signal also looks like a nice, symmetric sinusoid, which indicates
that the response is linear. Figure i.5 in Chapter i shows an example of non-sinusoidal
response, where an asymmetry is clearly visible. By moving the laser spot, detector, and
adjusting the drive amplitude, you should be able to get a signal that looks more like a
sinusoid.
7.1.1 Part 1: wave speed
In the first part of this experiment, you will measure the wave speed for several values of the
tension by hanging different weights from the string, but be sure to read below regarding
length measurements before changing weights. The wave velocity is predicted by a simple
model to be given by
v=sT
µ(7.1)
101

where Tis the tension and µis the linear mass density of the stretched string. Since
the string stretches under tension, its linear mass density will change depending
upon the amount of weight that is hung from the pulley.
As a first step to tackle this mass density issue, measure the mass Mof the string, and
be sure to subtract off the amount that will be hanging limply from the back of the clamp
since this portion will not stretch when the weights are applied and plays no role in the
experiment.
Example 7.1 A student has a string with total unstretched length of 2 m and total
mass M. After setting up the experiment, the student measures that there is 40 cm
of excess string hanging off the backside of the clamp. The mass of the string involved
in the experiment (this includes the portion hanging down to the weights since this
section stretches as well) is therefore given by
M=M1.6 m
2 m ,(7.2)
which is M= 0.8× M.
When tension is applied to the string, it will stretch, and the height of the knot holding
the end of the string to the hanging masses will change. The linear mass density is therefore
the mass Mdivided by total stretched length of the string from the clamp, over the pulley,
all the way to the knot at the top of the weights. This length will change when the tension
is changed, so be sure to keep track of it to note the changing linear mass density of the
string. You should also try to estimate the mass of the portion of the string that hangs from
the pulley to see if it needs to be included in your calculation of the tension, or if it is small
enough to be neglected.
You will be measuring the wave speed for 3 different values of the tension T, so it is
probably a good idea to go ahead and measure the stretched linear mass densities for 3
different values of the weight. For the second part of the experiment, you will want a good
bit of tension on the string, so choose at least one of your 3 weights to have a mass of 350 g
or more and then use this for Parts 2 and 3.
Conceptually, the wave speed measurement will proceed when you create a sharp rising
edge with the Signal Generator driving the wave driver and measure the time it takes the
resulting excitation to bounce back and forth between the pulley and the clamp. There
will be a signal on the photodiode (some sort of wiggle) every time the pulse arrives at the
photodiode. If the pulse travels down to the clamp and back in some time ∆t, the speed will
be the distance the wave traveled (call it 2L) divided by ∆t.
To take your wave speed measurement, select Continuous mode in the Controls palette.
Figure 7.2 shows Signal Generator settings appropriate for measuring the wave speed. Set
the signal generator to Auto, which will initiate the signal generator only when you start
recording data. You can create a fast rising edge by changing the Waveform from Sine to
Square. You will want the frequency to be quite low to give you lots of time to see the wave
reflecting back and forth on the string, so it is recommended that you set the Frequency to
102
Figure 7.2: Capstone settings for creating a fast rising edge to measure the wave speed.
0.2 Hz or so (the exact frequency does not matter). The recommended Amplitude is about 1
V. Last, set the Voltage Offset to be the same voltage as you used for the Amplitude so that
you don’t excite a pulse before you can record a rising edge.
You can visualize your results by opening a Graph session from the Displays palette.
You will be plotting two signals simultaneously (the wave driver voltage and the photodiode
signal) to see when the pulse is launched. To set this up, select Light Intensity (%) for the
y-axis and plot Time (s) on the x-axis. To add another y-axis, mouse over the plot area
to wake up the menu bar and click on the “Add new y-axis to active plot area” button .
The new axis will appear on the right side of the plot, and you can set it to plot Output
Voltage, Ch01 (V), as shown in Fig. 7.3. When you click Record on the Controls palette, the
photodiode signal will be fairly flat and the voltage should be zero until the rising edge, at
which point the voltage will jump to twice the amplitude setting and there will be a series of
peaks on the photodiode signal for each time the wave gets near the laser beam spot. Click
Stop once the voltage goes back to zero. Figure 7.3 shows a screen shot from Capstone with
an example signal.
The size and shape of the wiggles you get will probably look very different from everyone
else’s, but the only thing that matters is that you see some sort of signal that repeats itself a
few times. You can open a Table and save your data in Excel for your analysis and plotting.
This may involve quite a bit of data, but hopefully not too much to take home with you.
As always, be sure the table is displaying enough digits for you to get your data without
introducing nasty rounding errors. Save data for all 3 values of the tension so that you can
determine the wave speed during your analysis. Remember to measure the length between
the pulley and the clamp so that you know how far the pulse travels between successive
arrivals at the laser spot.
103
Figure 7.3: Capstone screen shot showing the launch of a pulse that makes many round-trips
along the string.
Figure 7.4: Close-up of the signal from a pulse reflecting back and forth from the clamp
and pulley. The wave speed can me measured by identifying some feature that repeats and
measuring the time duration between repetitions. In this case, the sharp downward spike
would probably be a good candidate. The uncertainty in this measurement can be estimated
from the width of the spike.
104
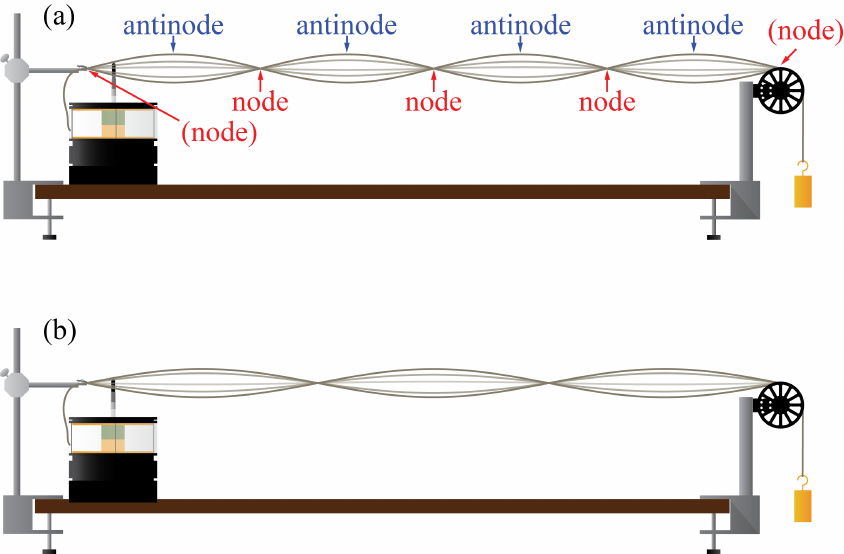
Figure 7.5: Transverse standing waves on a string. The positions where the displacement
amplitude is zero are called nodes, while the maxima are called antinodes. In contrast, a
normal mode of oscillation in this system is an entire standing wave. The n= 4 mode is
shown in (a), while an entirely different mode (with a correspondingly different frequency)
is shown in (b). The mode in (b) corresponds to n= 3.
7.1.2 Part 2: standing waves
For the second part of this experiment, you will be observing standing waves in the vibrating
string, as shown in Fig. 7.5. You will probably get the best results if you use your heaviest
weight from the first part of the experiment, and be sure to re-position the laser spot and
photodiode and optimize the linearity of the signal as mentioned above (driving the wave
driver with a sine wave should give you a nice sine wave on the photodiode signal).
Since both ends of the string have fixed boundary conditions, the string will support a
series of harmonics of the fundamental standing wave mode. Note: this resonance frequency
will probably be less than 20 Hz, so please do not try to turn on the wave driver at 1
kHz, which will primarily produce a painful noise. When driving the fundamental mode
on resonance, the string will oscillate with large amplitude (probably clearly visible by eye)
and will not have any nodes between end points. We can assign an integer n= 1 to this
mode. There will be an entire series of harmonics associated with this motion for other
integer values of n. Find the resonant frequency for this fundamental mode by monitoring
the amplitude of the photodiode signal and finding the frequency for which this amplitude
105
is maximized. Be sure to make a measurement that is as precise as possible in order to
accurately predict the frequency at which higher values of nwill also be normal modes of
the string. By using the DAQ, it should be possible for you to nail this down to at least
3 digits of precision (10s of mHz) using this “maximum amplitude” method. Record this
frequency for your records.
Before moving on to find the frequencies of higher-order modes, recall that you learned
last week how to find the undamped resonant frequency ωoof an oscillator using the phase
information of the signal. You created parametric plots of Lissajous figures such as Fig. 6.5,
and the resonant frequency was the frequency when the figure was most symmetric. Using
what you recall from last week about how to plot the system response (for this week, this
is the photodiode signal) vs. the drive signal (in this case, the wave driver output), set up
a parametric plot of your system that will allow you to identify resonant frequencies by
observing the symmetry of the waveform. You may assume ωR≈ωosince the damping time
in this system is many oscillation periods. Do this for the fundamental (n= 1) mode, and
see if you measure the same resonant frequency as you did with the “maximum amplitude”
method above. Be sure to record not just the resonant frequency, but also an uncertainty in
your measurement of this frequency. How far can you move the frequency before you notice
that the Lissajous figure is no longer symmetric?
Using your parametric plot, record the resonant frequencies for a series of harmonics of
this fundamental frequency (each harmonic is called a normal mode of oscillation). The nth
normal mode will have a resonant frequency that is ntimes the fundamental frequency, so
you should use your measured fundamental frequency to predict where to find each harmonic.
The nth mode will have nanti-nodes in it (nodes are positions with minimum amplitude, anti-
nodes are positions with maximum amplitude). Split up the measurement of the frequencies
of the harmonics with your parter up to n= 9. For instance, if there are two of you, one can
measure the frequencies of modes n= 1,3,5,7,9 and the other can measure n= 1,2,4,6,8.
If there are 3 of you sharing an apparatus, each should choose 5 harmonics at random from
n= 2 ···9.
For the higher frequency modes, you may find that it is helpful to turn up the signal
generator amplitude. If you have trouble finding those signals, you may have the laser and
photodiode too far from the pulley, so moving them closer may help. Try to find the 30th
harmonic (you should be able to predict where you think it will occur) and record what you
observe.
7.1.3 Part 3: boundary effects
The last part of the experiment is an investigation of intermediate boundary conditions and
interference. Using your measured resonant frequencies for n=2, 4, and 5, drive these modes
at some fixed drive amplitude and record the amplitude of the photodiode oscillations for
each one (Scope mode is good for this). The “amplitude” you record can simply be the
peak-to-peak size of the photodiode signal (because this number will be proportional to the
actual wave amplitude for transverse displacement of the string).
For all of the normal modes, the pulley apex and the clamp are always nodes, but for n= 2
106

and higher, there are also nodes between these endpoints. Drive the fundamental (n= 2)
mode with the same drive amplitude you just used when you measured its amplitude. Using
a ring stand and a post, constrain the motion of the string at the node that is in the middle
of the string for n= 2. You should be able to do this without substantially altering this
mode (the signal level should be the same). Record the new signal amplitude you measure
for this mode once you have optimized the position of the post to maximize it. Without
moving the post, drive the n= 4 mode with the same drive amplitude you used before, and
compare your new measured signal amplitude to the old one. Do the same (without moving
the post!) for n= 5 and record what you find.
7.2 Analysis
7.2.1 Part 1 analysis
To measure the wave speed from your data, for each tension, plot the photodiode signal
vs. time, as shown in Fig. 7.4. The excitation itself is likely to be a somewhat complicated
set of wiggles on the signal, but the important thing is not the shape of the signal so much
as it is identifying some feature that repeats itself. Each time the feature repeats, it will be
slightly smaller than the previous time, so you will have to make a decision about how far
out in time you want to look. Since you know the distance the excitation travels between
repetitions, measure the wave speed by dividing this distance by the time delay. Estimate an
uncertainty in your measurement based on how well you feel you can localize the excitation
feature in time and how well you know the distance traveled.
7.2.2 Part 2 analysis
The frequency of the nth normal mode is predicted to be given by
f(n) = nv
2L(7.3)
where vis the wave speed (predicted and measured for this tension in Part 1) and Lis
the distance between the clamp and pulley apex. Using your measured value for the wave
speed from Part 1, calculate a predicted value for the frequency of the fundamental mode.
Using Eq. 7.3, calculate predicted values for the higher-order modes whose frequencies you
measured. Compare your results.
7.3 Report guidelines
Write a full, formal lab report that covers Experiments 6 and 7.
1. Cover Sheet
•experiment number and title
107
•your name
•the date the lab was performed
•your lab section (for instance, “Monday 3pm”)
•your TA’s name
•your lab partners’ names
•the word count of your abstract
2. Guidelines
By this point in Physics 4AL, your experience in writing lab reports should enable you
to write a lab report without detailed guidelines. You will be responsible for developing
a coherent narrative to connect each of the results you obtained and deciding which
figures, plots, tables and analysis will be appropriate to accomplish this. You should
look back over your old returned reports and guidelines from earlier in this manual to
guide you through this process. If you did the extra credit from this week and/or last,
please find a way to include it in your lab report as if it were just another part of your
experiment.
3. Extra Credit In experiments 6 and 7, you used Lissajous figures to find the resonance
frequency of a couple of harmonic oscillators. Using this method for your standing
waves, try to find out what the highest mode you can find is. Can you see the n= 60
mode? Report your determination of each mode starting from n= 1 and going as high
in nas you can. State the distance you have between your laser spot on the string
and the point where the string touches the pulley. Calculate the frequency of the first
mode that will have a node at this location, and describe why this may make your
data taking for modes near this one difficult.
7.4 Epilogue
Throughout this course, you have investigated a series of different mechanical phenomena
in a laboratory setting. Many of the effect observed in this setting, however, manifest
themselves in many different contexts, including electricity and magnetism, atomic physics,
nuclear physics, astrophysics, communications, particle physics, et cetera. The material in
this course is therefore far more general than mechanics, and it is hoped that working with
your hands to investigate these phenomena will help to prepare you to have a more intuitive
understanding of these concepts as you move forward.
108
Appendix A
Laser safety.
109

Laser Safety
Lab Hazard Awareness Information
501 Westwood Plaza, 4th Floor • Los Angeles, CA 90095
Phone: 310-825-5689 • Fax: 310-825-7076 • www.ehs.ucla.edu
Provides information to lab workers/visitors who are NOT authorized laser users about hazards existing at this
location.
Location Knudsen 1238, 2115, 2122, 2136
LASERS
• Lasers may only be operated in these labs by personnel who have received permission to use them from a suitably
qualified class trainer.
• In order to maintain and protect equipment, personnel, and research, no worker, except for trained personnel, can
touch move or interfere with any laser equipment, materials or processes.
• Personnel should never use class-supplied laser pointers as presentation pointers in the classroom.
• Personnel should never introduce a shiny or mirrored surface in front of laser beam that might cause uncontrolled
specular reflection.
• Classroom laser personnel should not move or adjust laser equipment set up by any other class laser operator
unless specifically instructed to do so. This includes not detaching the lasers from any stands or moving the stands in
any way that could create a hazard for other classroom occupants.
• Laser equipment should be turned on only when necessary for presenting the demonstration.
• Laser equipment, whether powered by an external power supply or batteries, should be turned off following
completion of a given classroom demonstration so as to prevent inadvertent exposure during class dismissal.
EYE HAZARDS
• This lab uses and operates lasers that create light radiation which, if of sufficient power, can cause biological damage
to the eyes. Eye damage caused from lasers includes retinal burns, glaucoma, cataracts and total loss of vision.
• Personnel should never stare into a laser or look directly into a beam and never stand or sit at the same level as the
beam path.
• Laser operators must make sure that any laser beam points away from other members of the class.
• The person primarily responsible for the class training can order all personnel to leave the room if necessary for any
reason
Laser Safety
LASER LITE; A Quick Overview of Laser Safety 06/09
501 Westwood Plaza, 4th Floor • Los Angeles, CA 90095
Phone: 310-825-5689 • Fax: 310-825-7076 • www.ehs.ucla.edu
THE PRIMARY PURPOSE
OF THE UCLA LASER
SAFETY PROGRAM IS TO
AVOID GETTING A
LASER BEAM IN
YOUR EYE
NEVER STARE DIRECTLY
AT A LASER BEAM,
EVEN WITH PROTECTIVE
EYEWEAR
LASER LIGHT
• Maintains strength over long distances
• Produces significant eye hazards at relatively low levels
• Concentrates to a very high intensity when focused by a lens
• Occurs in the Visible and Non-visible spectrum
BIOLOGICAL HAZARDS
• Can be laser beam and non-beam related
• Can occur at all wavelengths
• Occur mainly in the eyes and skin
MOST INJURIES
• Affect the eyes
• Occur during alignment
• Result from operator error
PROTECTION
• Eyewear must be worn for Class 3B and Class 4 laser use
• Eyewear must meet ANSI standards and be marked with
Optical Density and Wavelength
• Appropriate skin protection is required for Class 4 lasers
LASER CLASSIFICATION TABLE
Class 1 / 1M
Maximum power output is a few microwatts. Visible spectrum output
Considered incapable of producing hazardous eye/skin exposure unless viewed with collecting
optics (1M). Does not apply to open Class 1 enclosures containing higher-class lasers
Class 2 / 2M
Maximum power output is < 1 mW. Visible spectrum output
Considered incapable of producing hazardous eye/skin exposure within the time period of human
eye aversion response (0.25 s).unless viewed with collecting optics (2M)
Class 3R
Maximum power output is 1 mW - < 5 mW. Visible and non-visible spectrum
Potentially hazardous under some direct and specular reflection viewing condition if the eye is
appropriately focused and stable or if viewed with collecting optics.
Class 3B
Maximum power output is 5 mW - < 500 mW Visible and non-visible spectrum
Presents a potential eye hazard for intrabeam (direct) or specular (mirror-like) conditions.
Presents a significant skin hazard by long-term diffuse (scatter) exposure if higher powered and
operating in 200 – 280 nm UVC ranges
Class 4
Maximum power output is > 500 mW. Visible and non-visible spectrum
Presents potential acute hazards to the eye and skin for all intrabeam and diffused conditions
Potential hazard for fire (ignition), explosion and emissions from target or process materials.
EYE DAMAGE - WAVELENGTHS
WAVELENGTH
AREA OF DAMAGE
PATHOLOGICAL EFFECT
180 - 315 nm
(Ultraviolet UV-B, UVC)
CORNEA; Deep-ultraviolet light causes
accumulating damage, even at very low power
Photokeratitis; Inflammation of the
cornea, similar to sunburn
315 - 400 nm
(Ultraviolet UV-A)
CORNEA and LENS
Photochemical cataract; Clouding
of the lens
400 - 780 nm
(Visible)
RETINA; Visible light is focused on the retina
Photochemical damage; Damage
to retina and retinal burns
780 - 1400 nm
(Near Infrared)
RETINA; Near IR light is not absorbed by iris
and is focused on the retina
Thermal damage to cataract and
retinal burns
1400 - 3000 nm
(Infrared)
CORNEA and LENS; IR light is absorbed by
transparent parts of eye before reaching the
retina
Aqueous flare; Protein in aqueous
humor, cataract, corneal burn
3000 – 10000 nm
(Far Infrared)
CORNEA
Corneal burn
Laser Safety
LASER LITE; A Quick Overview of Laser Safety 06/09
501 Westwood Plaza, 4th Floor • Los Angeles, CA 90095
Phone: 310-825-5689 • Fax: 310-825-7076 • www.ehs.ucla.edu
LASER OPERATION GUIDELINES
OPERATORS MUST;
• Review Standard Operating Procedures, operating and safety instructions and
laboratory-specific laser instructions
• Be trained in laser safety and specific laser procedures
• Observe all written procedures, safety rules and properly use appropriate
PPE
• Be authorized by the Principal Investigator
• Be directly supervised by a person knowledgeable in laser safety
• Wear appropriate PPE, follow safety procedures and SOP’s
• Never circumvent Administrative or Engineering safety controls
• Know the location and use of the power kill switch and fire extinguisher
• Use the buddy system when working with high voltage equipment
• Not wear reflective metal jewelry when working with laser beams
• Never stare directly into a beam even with eye protection; use indirect viewing
• Give sufficient attention to non-beam hazards to prevent possible injury and
illness
• Be aware of plasma and collateral radiation
• Notify supervisor immediately of potentially hazardous conditions, personal
injury, or property damage
IN EACH LAB, PERSONNEL MUST BE ABLE TO OBTAIN:
• Training on equipment, procedures and emergency procedures
• Safety Equipment that is sufficient in numbers for lab staff, appropriate for the
equipment in use and in good operating condition
• Standard Operational Procedures for the safe use of all equipment
• Information from the PI or Lab Manager about potential equipment hazards
EMERGENCY INSTRUCTIONS
1. Shut the laser off immediately and remove the interlock key. If not possible,
alert everyone to exit the laboratory and be the last to leave the laboratory
2. If there is a fire, get everyone out of the laboratory immediately. At the same
time shout “FIRE” loudly and frequently. Turn on a fire alarm. Do not try to
“fight” the fire from inside the laboratory; do it from the doorway to maintain an
escape route
3. In the event of MAJOR injury, Summon Medical Assistance. Call 911 from a
campus phone or 310-825-1491 from a cell phone
4. Call Security and/or Fire Department (911) as necessary
5. Call EHS Hotline (59797) to report the incident
Note; ALL incidents must be reported to the Laser Safety Officer
6. Contact the PI and/or Lab Manager and describe the emergency
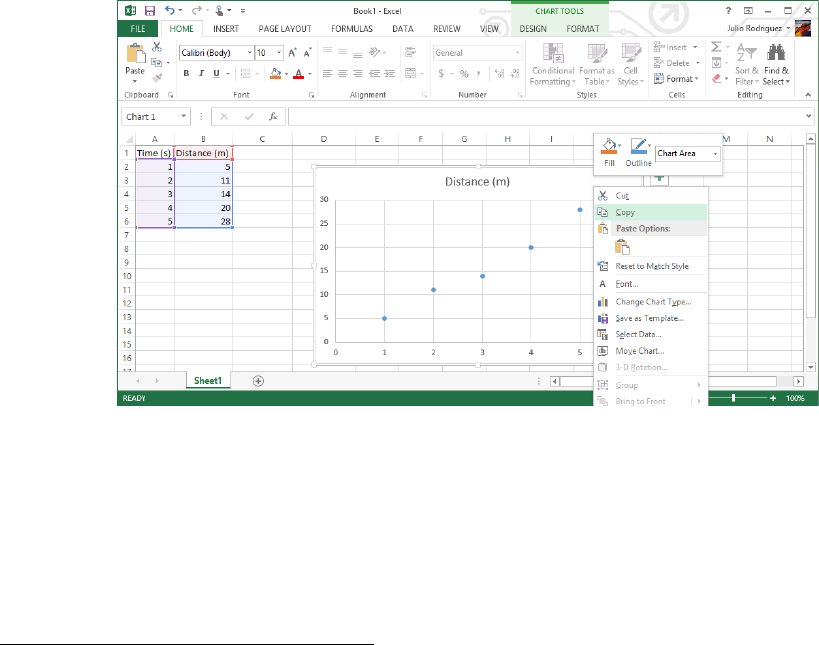
Appendix B
B.1 Guide for Making and Inserting Plots1
B.1.1 Obtaining Graph data from Excel
In MS Excel simply click on the white space in your graph, this will select the entire graph.
You can then use the ctrl+c keyboard shortcut to copy the graph. Alternatively you can
right click on the graphs white space and click “copy” from the menu. Simply paste this in
Word in the normal way (ctrl+p) or right click and selecting “paste.”
B.1.2 Formatting the Graph
Now that the graph is in MS Word, you need to format the graph according to the rules
agreed upon by the grading staff. Every graph needs these three things: Axis labels, units,
and a title. Additionally, graphs with multiple datasets require some way of determining
1This section was contributed by Julio S. Rodriguez, Jr. (2014) and revised by Anthony Ransford (2016).
113
what the data represents (either an explanation or a legend), and some graphs require a
trendline with an equation. Many of these things can be set up in Excel beforehand and will
be copied over. You may also generate them directly in Word. The lab manual has a pretty
good explanation of how to do all of this in Excel and the procedure is largely the same in
word. The labels, units, and trendline should appear on the graph while title and possibly
the trendline equation (with error) should appear in the caption. If you use a legend you do
not need an explanation of what the data points represent, if you do not have a legend, you
should include such an explanation in the caption.
Axis Labels
There is an easy way to add axis labels. Click on the whitespace in your graph and on
the ribbon, a few new tabs will open up under the group heading “Chart Tools.” On the
“Design” tab there are some preset layouts. In general, you should pick the preset with
axis labels (see Figure B.1). You can click on the axis labels to edit their content. I will
be working with the assumption that the legend is not necessary since we will have a very
descriptive caption. Simply click on the legend and press the “delete” key on the keyboard.
Figure B.1: There are templates in Excel that have axis labels by default, which is a good
way to make sure axes don’t end up in a report without labels.
Adding the caption
This is probably the trickiest part of the whole process. The caption function in Word is
great in that it auto-numbers things for you, and lets you use different numberings for figure
114
and tables. The cross-reference tool adds to its functionality, because changing the caption
numbering order will not mess up the numbering in the text of your paper. Before we add a
caption, we need to change the nature of the graph. Right now Word is treating our graph
like an object that takes up its own line, but we want Word to treat the graph as an object
embedded in the text (like the image on the right). Click on your graph and this time on
the ribbon click on the Format tab under the Chart Tools heading. Here click the Wrap
Text button and on the Square option (on older versions of Word, the image for this button
looks like a dog instead of an arched curve). Move your graph where you want it to appear
in your paper and then add the caption. Right click on the whitespace of your graph and
click the insertcaption option. All kinds of options come up, but the default settings are
okay for graphs. This will insert the caption as a text box, but we need the caption to be
associated with the graph. To do this: If the border and sizing handles on the textbox are
not already showing, click on the textbox (anywhere inside). While holding down ctrl, click
on the graph. This should select both objects. Release ctrl and right click on either object,
then select “Group >Group.”
These two objects will now be treated as a unit; moving one of them will move the other
one. Reformat the text in the text box to match rest of your document. You may choose
to format the title of the graph in bold font for clarity, but this is not strictly required.
To add equations, you can either type things out and use the “symbol” function to include
any non-standard symbols, or you can use the equation editor (alt = or insert >equation).
A common convention is to italicize variables and leave units un-italicized in equations,
remember to leave a space between the units and variables if the two are being multiplied.
When referencing you graphs or images in your paper, you should refer to them by their
designation. To make this as painless as possible, you may want to use the cross-reference
tool in Word. If you change the order of your graphs, insert another graph before one you
have already referenced, or anything else along those lines, this tool will keep all of your
numbers in order. On the ribbon under the references tab, click “Cross-reference.” Under
“Reference Type,” select “Figure,” and under, “Insert Reference To,” select “Only Label and
115
Number.” You can leave on the “hyperlink” function if you wish. Select which figure you
wish to make reference to in the box labeled “For which caption.” This inserts a reference
to your chart. For example, Fig. B.2 shows what a proper graph should look like. If you
insert any addition graphs or change the order, select your entire document and right click
any of the selected text and click “Update Field.”
Figure B.2: The blue dots on this graph represent data points taken from the Fakemeter 5000
while on a bicycle ride though Imaginary Park. Distance as a function of time was calculated
using the regression tool in Microsoft Excel 365 in order to find the average velocity of the
cyclist. The line of best fit is represented by D=t×(5.5±0.5) m
s−(1 ±2) m.
116
Appendix C
To help you figure out how to write bibliography entries in the style of Nature formatting,
a long example of bibliography formatting from Nature is reproduced here. This example
shows entries for published journal articles, journal articles in press, and published books.
These are page images taken from the paper reporting sequencing of the human genome
[2].
117

because of the availability of tissues from all developmental time
points. A challenge will be to de®ne the gene-speci®c patterns of
alternative splicing, which may affect half of human genes. Existing
collections of ESTs and cDNAs may allow identi®cation of the most
abundant of these isoforms, but systematic exploration of this
problem may require exhaustive analysis of cDNA libraries from
multiple tissues or perhaps high-throughput reverse transcription±
PCR studies. Deep understanding of gene function will probably
require knowledge of the structure, tissue distribution and abun-
dance of these alternative forms.
Large-scale identi®cation of regulatory regions
The one-dimensional script of the human genome, shared by
essentially all cells in all tissues, contains suf®cient information to
provide for differentiation of hundreds of different cell types, and
the ability to respond to a vast array of internal and external
in¯uences. Much of this plasticity results from the carefully orche-
strated symphony of transcriptional regulation. Although much has
been learned about the cis-acting regulatory motifs of some speci®c
genes, the regulatory signals for most genes remain uncharacterized.
Comparative genomics of multiple vertebrates offers the best hope
for large-scale identi®cation of such regulatory sites440. Previous
studies of sequence alignment of regulatory domains of ortho-
logous genes in multiple species has shown a remarkable
correlation between sequence conservation, dubbed `phylogenetic
footprints'441, and the presence of binding motifs for transcription
factors. This approach could be particularly powerful if combined
with expression array technologies that identify cohorts of genes
that are coordinately regulated, implicating a common set of cis-
acting regulatory sequences442±445. It will also be of considerable
interest to study epigenetic modi®cations such as cytosine methyla-
tion on a genome-wide scale, and to determine their biological
consequences446,447. Towards this end, a pilot Human Epigenome
Project has been launched448,449.
Sequencing of additional large genomes
More generally, comparative genomics allows biologists to peruse
evolution's laboratory notebookÐto identify conserved functional
features and recognize new innovations in speci®c lineages. Deter-
mination of the genome sequence of many organisms is very
desirable. Already, projects are underway to sequence the genomes
of the mouse, rat, zebra®sh and the puffer®shes T. nigroviridis and
Takifugu rubripes. Plans are also under consideration for sequencing
additional primates and other organisms that will help de®ne key
developments along the vertebrate and nonvertebrate lineages.
To realize the full promise of comparative genomics, however, it
needs to become simple and inexpensive to sequence the genome of
any organism. Sequencing costs have dropped 100-fold over the last
10 years, corresponding to a roughly twofold decrease every 18
months. This rate is similar to `Moore's law' concerning improve-
ments in semiconductor manufacture. In both sequencing and
semiconductors, such improvement does not happen automatically,
but requires aggressive technological innovation fuelled by major
investment. Improvements are needed to move current dideoxy
sequencing to smaller volumes and more rapid sequencing
times, based upon advances such as microchannel technology.
More revolutionary methods, such as mass spectrometry, single-
molecule sequencing and nanopore approaches76, have not yet
been fully developed, but hold great promise and deserve strong
encouragement.
Completing the catalogue of human variation
The human draft genome sequence has already allowed the identi-
®cation of more than 1.4 million SNPs, comprising a substantial
proportion of all common human variation. This program should
be extended to obtain a nearly complete catalogue of common
variants and to identify the common ancestral haplotypes present in
the population. In principle, these genetic tools should make it
possible to perform association studies and linkage disequilibrium
studies376 to identify the genes that confer even relatively modest risk
for common diseases. Launching such an intense era of human
molecular epidemiology will also require major advances in the cost
ef®ciency of genotyping technology, in the collection of carefully
phenotyped patient cohorts and in statistical methods for relating
large-scale SNP data to disease phenotype.
From sequence to function
The scienti®c program outlined above focuses on how the genome
sequence can be mined for biological information. In addition, the
sequence will serve as a foundation for a broad range of functional
genomic tools to help biologists to probe function in a more
systematic manner. These will need to include improved techniques
and databases for the global analysis of: RNA and protein expres-
sion, protein localization, protein±protein interactions and chemi-
cal inhibition of pathways. New computational techniques will be
needed to use such information to model cellular circuitry. A full
discussion of these important directions is beyond the scope of this
paper.
Concluding thoughts
The Human Genome Project is but the latest increment in a
remarkable scienti®c program whose origins stretch back a hundred
years to the rediscovery of Mendel's laws and whose end is nowhere
in sight. In a sense, it provides a capstone for efforts in the past
century to discover genetic information and a foundation for efforts
in the coming century to understand it.
We ®nd it humbling to gaze upon the human sequence now
coming into focus. In principle, the string of genetic bits holds long-
sought secrets of human development, physiology and medicine. In
practice, our ability to transform such information into under-
standing remains woefully inadequate. This paper simply records
some initial observations and attempts to frame issues for future
study. Ful®lling the true promise of the Human Genome Project will
be the work of tens of thousands of scientists around the world, in
both academia and industry. It is for this reason that our highest
priority has been to ensure that genome data are available rapidly,
freely and without restriction.
The scienti®c work will have profound long-term consequences
for medicine, leading to the elucidation of the underlying molecular
mechanisms of disease and thereby facilitating the design in many
cases of rational diagnostics and therapeutics targeted at those
mechanisms. But the science is only part of the challenge. We
must also involve society at large in the work ahead. We must set
realistic expectations that the most important bene®ts will not be
reaped overnight. Moreover, understanding and wisdom will be
required to ensure that these bene®ts are implemented broadly and
equitably. To that end, serious attention must be paid to the many
ethical, legal and social implications (ELSI) raised by the accelerated
pace of genetic discovery. This paper has focused on the scienti®c
achievements of the human genome sequencing efforts. This is not
the place to engage in a lengthy discussion of the ELSI issues, which
have also been a major research focus of the Human Genome
Project, but these issues are of comparable importance and could
appropriately ®ll a paper of equal length.
Finally, it is has not escaped our notice that the more we learn
about the human genome, the more there is to explore.
``We shall not cease from exploration. And the end of all our
exploring will be to arrive where we started, and know the place for
the ®rst time.''ÐT. S. Eliot450 M
Received 7 December 2000; accepted 9 January 2001.
1. Correns, C. Untersuchungen u
Èber die Xenien bei Zea mays. Berichte der Deutsche Botanische
Gesellschaft 17, 410±418 (1899).
2. De Vries, H. Sur la loie de disjonction des hybrides. Comptes Rendue Hebdemodaires, Acad. Sci. Paris
130, 845±847 (1900).
3. von Tschermack, E. Uber Ku
Ènstliche Kreuzung bei Pisum sativum. Berichte der Deutsche Botanische
Gesellschaft 18, 232±239. (1900).
4. Sanger, F. et al. Nucleotide sequence of bacteriophage FX174 DNA. Nature 265, 687±695 (1977).
5. Sanger, F. et al. The nucleotide sequence of bacteriophage FX174. J Mol Biol 125, 225±246 (1978).
articles
914 NATURE
|
VOL 409
|
15 FEBRUARY 2001
|
www.nature.com
© 2001 Macmillan Magazines Ltd

6. Sanger, F., Coulson, A. R., Hong, G. F., Hill, D. F. & Petersen, G. B. Nucleotide-sequence of
bacteriophage Lambda DNA. J. Mol. Biol. 162, 729±773 (1982).
7. Fiers, W. et al. Complete nucleotide sequence of SV40 DNA. Nature 273, 113±120 (1978).
8. Anderson, S. et al. Sequence and organization of the human mitochondrial genome. Nature 290,
457±465 (1981).
9. Botstein, D., White, R. L., Skolnick, M. & Davis, R. W. Construction of a genetic linkage map in man
using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314±331 (1980).
10. Olson, M. V. et al. Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl Acad.
Sci. USA 83, 7826±7830 (1986).
11. Coulson, A., Sulston, J., Brenner, S. & Karn, J. Toward a physical map of the genome of the nematode
Caenorhabditis elegans.Proc. Natl Acad. Sci. USA 83, 7821±7825 (1986).
12. Putney, S. D., Herlihy, W. C. & Schimmel, P. A new troponin T and cDNA clones for 13 different
muscle proteins, found by shotgun sequencing. Nature 302, 718±721 (1983).
13. Milner, R. J. & Sutcliffe, J. G. Gene expression in rat brain. Nucleic Acids Res. 11, 5497±5520 (1983).
14. Adams, M. D. et al. Complementary DNA sequencing: expressed sequence tags and human genome
project. Science 252, 1651±1656 (1991).
15. Adams, M. D. et al. Initial assessment of human gene diversity and expression patterns based upon
83 million nucleotides of cDNA sequence. Nature 377, 3±174 (1995).
16. Okubo, K. et al. Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of
gene expression. Nature Genet. 2, 173±179 (1992).
17. Hillier, L. D. et al. Generation and analysis of 280,000 human expressed sequence tags. Genome Res.
6, 807±828 (1996).
18. Strausberg, R. L., Feingold, E. A., Klausner, R. D. & Collins, F. S. The mammalian gene collection.
Science 286, 455±457 (1999).
19. Berry, R. et al. Gene-based sequence-tagged-sites (STSs) as the basis for a human gene map. Nature
Genet. 10, 415±423 (1995).
20. Houlgatte, R. et al. The Genexpress Index: a resource for gene discovery and the genic map of the
human genome. Genome Res. 5, 272±304 (1995).
21. Sinsheimer, R. L. The Santa Cruz WorkshopÐMay 1985. Genomics 5, 954±956 (1989).
22. Palca, J. Human genomeÐDepartment of Energy on the map. Nature 321, 371 (1986).
23. National Research Council Mapping and Sequencing the Human Genome (National Academy Press,
Washington DC, 1988).
24. Bishop, J. E. & Waldholz, M. Genome (Simon and Schuster, New York, 1990).
25. Kevles, D. J. & Hood, L. (eds) The Code of Codes: Scienti®c and Social Issues in the Human Genome
Project (Harvard Univ. Press, Cambridge, Massachusetts, 1992).
26. Cook-Deegan, R. The Gene Wars: Science, Politics, and the Human Genome (W. W. Norton & Co.,
New York, London, 1994).
27. Donis-Keller, H. et al. A genetic linkage map of the human genome. Cell 51, 319±337 (1987).
28. Gyapay, G. et al. The 1993±94 Genethon human genetic linkage map. Nature Genet. 7, 246±339
(1994).
29. Hudson, T. J. et al. An STS-based map of the human genome. Science 270, 1945±1954 (1995).
30. Dietrich, W. F. et al. A comprehensive genetic map of the mouse genome. Nature 380, 149±152
(1996).
31. Nusbaum, C. et al. A YAC-based physical map of the mouse genome. Nature Genet. 22, 388±393
(1999).
32. Oliver, S. G. et al. The complete DNA sequence of yeast chromosome III. Nature 357, 38±46 (1992).
33. Wilson, R. et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans.
Nature 368, 32±38 (1994).
34. Chen, E. Y. et al. The human growth hormone locus: nucleotide sequence, biology, and evolution.
Genomics 4, 479±497 (1989).
35. McCombie, W. R. et al. Expressed genes, Alu repeats and polymorphisms in cosmids sequenced
from chromosome 4p16.3. Nature Genet. 1, 348±353 (1992).
36. Martin-Gallardo, A. et al. Automated DNA sequencing and analysis of 106 kilobases from human
chromosome 19q13.3. Nature Genet. 1, 34±39 (1992).
37. Edwards, A. et al. Automated DNA sequencing of the human HPRT locus. Genomics 6, 593±608
(1990).
38. Marshall, E. A strategy for sequencing the genome 5 years early. Science 267, 783±784 (1995).
39. Project to sequence human genome moves on to the starting blocks. Nature 375, 93±94 (1995).
40. Shizuya, H. et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in
Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA 89, 8794±8797 (1992).
41. Burke, D. T., Carle, G. F. & Olson, M. V. Cloning of large segments of exogenous DNA into yeast by
means of arti®cial chromosome vectors. Science 236, 806±812 (1987).
42. Marshall, E. A second private genome project. Science 281, 1121 (1998).
43. Marshall, E. NIH to produce a `working draft' of the genome by 2001. Science 281, 1774±1775
(1998).
44. Pennisi, E. Academic sequencers challenge Celera in a sprint to the ®nish. Science 283, 1822±1823
(1999).
45. Bouck, J., Miller, W., Gorrell, J. H., Muzny, D. & Gibbs, R. A. Analysis of the quality and utility of
random shotgun sequencing at low redundancies. Genome Res. 8, 1074±1084 (1998).
46. Collins, F. S. et al. New goals for the U. S. Human Genome Project: 1998±2003. Science 282, 682±689
(1998).
47. Sanger, F. & Coulson, A. R. A rapid method for determining sequences in DNA by primed synthesis
with DNA polymerase. J. Mol. Biol. 94, 441±448 (1975).
48. Maxam, A. M. & Gilbert, W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA 74, 560±
564 (1977).
49. Anderson, S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids
Res. 9, 3015±3027 (1981).
50. Gardner, R. C. et al. The complete nucleotide sequence of an infectious clone of cauli¯ower mosaic
virus by M13mp7 shotgun sequencing. Nucleic Acids Res. 9, 2871±2888 (1981).
51. Deininger, P. L. Random subcloning of sonicated DNA: application to shotgun DNA sequence
analysis. Anal. Biochem. 129, 216±223 (1983).
52. Chissoe, S. L. et al. Sequence and analysis of the human ABL gene, the BCR gene, and regions
involved in the Philadelphia chromosomal translocation. Genomics 27, 67±82 (1995).
53. Rowen, L., Koop, B. F. & Hood, L. The complete 685-kilobase DNA sequence of the human beta T
cell receptor locus. Science 272, 1755±1762 (1996).
54. Koop, B. F. et al. Organization, structure, and function of 95 kb of DNA spanning the murine T-cell
receptor C alpha/C delta region. Genomics 13, 1209±1230 (1992).
55. Wooster, R. et al. Identi®cation of the breast cancer susceptibility gene BRCA2. Nature 378, 789±792
(1995).
56. Fleischmann, R. D. et al. Whole-genome random sequencing and assembly of Haemophilus
in¯uenzae Rd. Science 269, 496±512 (1995).
57. Lander, E. S. & Waterman, M. S. Genomic mapping by ®ngerprinting random clones: a
mathematical analysis. Genomics 2, 231±239 (1988).
58. Weber, J. L. & Myers, E. W. Human whole-genome shotgun sequencing. Genome Res. 7, 401±409
(1997).
59. Green, P. Against a whole-genome shotgun. Genome Res. 7, 410±417 (1997).
60. Venter, J. C. et al. Shotgun sequencing of the human genome. Science 280, 1540±1542 (1998).
61. Venter, J. C. et al. The sequence of the human genome. Science 291, 1304±1351 (2001).
62. Smith, L. M. et al. Fluorescence detection in automated DNA sequence analysis. Nature 321, 674±
679 (1986).
63. Ju, J. Y., Ruan, C. C., Fuller, C. W., Glazer, A. N. & Mathies, R. A. Fluorescence energy-transfer dye-
labeled primers for DNA sequencing and analysis. Proc. Natl Acad. Sci. USA 92, 4347±4351 (1995).
64. Lee, L. G. et al. New energy transfer dyes for DNA sequencing. Nucleic Acids Res. 25, 2816±2822 (1997).
65. Rosenblum, B. B. et al. New dye-labeled terminators for improved DNA sequencing patterns.
Nucleic Acids Res. 25, 4500±4504 (1997).
66. Metzker, M. L., Lu, J. & Gibbs, R. A. Electrophoretically uniform ¯uorescent dyes for automated
DNA sequencing. Science 271, 1420±1422 (1996).
67. Prober, J. M. et al. A system for rapid DNA sequencing with ¯uorescent chain-terminating
dideoxynucleotides. Science 238, 336±341 (1987).
68. Reeve, M. A. & Fuller, C. W. A novel thermostable polymerase for DNA sequencing. Nature 376,
796±797 (1995).
69. Tabor, S. & Richardson, C. C. Selective inactivation of the exonuclease activity of bacteriophage T7
DNA polymerase by in vitro mutagenesis. J. Biol. Chem. 264, 6447±6458 (1989).
70. Tabor, S. & Richardson, C. C. DNA sequence analysis with a modi®ed bacteriophage T7 DNA
polymeraseÐeffect of pyrophosphorolysis and metal ions. J. Biol. Chem. 265, 8322±8328 (1990).
71. Murray, V. Improved double-stranded DNA sequencing using the linear polymerase chain reaction.
Nucleic Acids Res. 17, 8889 (1989).
72. Guttman, A., Cohen, A. S., Heiger, D. N. & Karger, B. L. Analytical and micropreparative ultrahigh
resolution of oligonucleotides by polyacrylamide-gel high-performance capillary electrophoresis.
Anal. Chem. 62, 137±141 (1990).
73. Luckey, J. A. et al. High-speed DNA sequencing by capillary electrophoresis. Nucleic Acids Res. 18,
4417±4421 (1990).
74. Swerdlow, H., Wu, S., Harke, H. & Dovichi, N. J. Capillary gel-electrophoresis for DNA sequencingÐ
laser-induced ¯uorescence detection with the sheath ¯ow cuvette. J. Chromatogr. 516, 61±67 (1990).
75. Meldrum, D. Automation for genomics, part one: preparation for sequencing. Genome Res. 10,
1081±1092 (2000).
76. Meldrum, D. Automation for genomics, part two: sequencers, microarrays, and future trends.
Genome Res. 10, 1288±1303 (2000).
77. Ewing, B. & Green, P. Base-calling of automated sequencer traces using phred. II. Error probabilities.
Genome Res. 8, 186±194 (1998).
78. Ewing, B., Hillier, L., Wendl, M. C. & Green, P. Base-calling of automated sequencer traces using
phred. I. Accuracy assessment. Genome Res. 8, 175±185 (1998).
79. Bentley, D. R. Genomic sequence information should be released immediately and freely in the
public domain. Science 274, 533±534 (1996).
80. Guyer, M. Statement on the rapid release of genomic DNA sequence. Genome Res. 8, 413 (1998).
81. Dietrich, W. et al. A genetic map of the mouse suitable for typing intraspeci®c crosses. Genetics 131,
423±447 (1992).
82. Kim, U. J. et al. Construction and characterization of a human bacterial arti®cial chromosome
library. Genomics 34, 213±218 (1996).
83. Osoegawa, K. et al. Bacterial arti®cial chromosome libraries for mouse sequencing and functional
analysis. Genome Res. 10, 116±128 (2000).
84. Marra, M. A. et al. High throughput ®ngerprint analysis of large-insert clones. Genome Res. 7, 1072±
1084 (1997).
85. Marra, M. et al. A map for sequence analysis of the Arabidopsis thaliana genome. Nature Genet. 22,
265±270 (1999).
86. The International Human Genome Mapping Consortium. A physical map of the human genome.
Nature 409, 934±941 (2001).
87. Zhao, S. et al. Human BAC ends quality assessment and sequence analyses. Genomics 63, 321±332
(2000).
88. Mahairas, G. G. et al. Sequence-tagged connectors: A sequence approach to mapping and scanning
the human genome. Proc. Natl Acad. Sci. USA 96, 9739±9744 (1999).
89. Tilford, C. A. et al. A physical map of the human Y chromosome. Nature 409, 943±945 (2001).
90. Bentley, D. R. et al. The physical maps for sequencing human chromosomes 1, 6, 9, 10, 13, 20 and X.
Nature 409, 942±943 (2001).
91. Montgomery, K. T. et al. A high-resolution map of human chromosome 12. Nature 409, 945±946
(2001).
92. Bru
Èls, T. et al. A physical map of human chromosome 14. Nature 409, 947±948 (2001).
93. Hattori, M. et al. The DNA sequence of human chromosome 21. Nature 405, 311±319 (2000).
94. Dunham, I. et al. The DNA sequence of human chromosome 22. Nature 402, 489±495 (1999).
95. Cox, D. et al. Radiation hybrid map of the human genome. Science (in the press).
96. Osoegawa, K. et al. An improved approach for construction of bacterial arti®cial chromosome
libraries. Genomics 52, 1±8 (1998).
97. The International SNP Map Working Group. A map of human genome sequence variation
containing 1.42 million single nucleotide polymorphisms. Nature 409, 928±933 (2001).
98. Collins, F. S., Brooks, L. D. & Chakravarti, A. A DNA polymorphism discovery resource for research
on human genetic variation. Genome Res. 8, 1229±1231 (1998).
99. Stewart, E. A. et al. An STS-based radiation hybrid map of the human genome. Genome Res. 7, 422±
433 (1997).
100. Deloukas, P. et al. A physical map of 30,000 human genes. Science 282, 744±746 (1998).
101. Dib, C. et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites.
articles
NATURE
|
VOL 409
|
15 FEBRUARY 2001
|
www.nature.com 915
© 2001 Macmillan Magazines Ltd

Nature 380, 152±154 (1996).
102. Broman, K. W., Murray, J. C., Shef®eld, V. C., White, R. L. & Weber, J. L. Comprehensive human
genetic maps: individual and sex-speci®c variation in recombination. Am. J. Hum. Genet. 63, 861±
869 (1998).
103. The BAC Resource Consortium. Integration of cytogenetic landmarks into the draft sequence of the
human genome. Nature 409, 953±958 (2001).
104. Kent, W. J. & Haussler, D. GigAssembler: an algorithm for the initial assembly of the human working
draft . Technical Report UCSC-CRL-00-17 (Univ. California at Santa Cruz, Santa Cruz, California,
2001).
105. Morton, N. E. Parameters of the human genome. Proc. Natl Acad. Sci. USA 88, 7474±7476 (1991).
106. Podugolnikova, O. A. & Blumina, M. G. Heterochromatic regions on chromosomes 1, 9, 16, and Yin
children with some disturbances occurring during embryo development. Hum. Genet. 63, 183±188
(1983).
107. Lundgren, R., Berger, R. & Kristoffersson, U. Constitutive heterochromatin C-band polymorphism
in prostatic cancer. Cancer Genet. Cytogenet. 51, 57±62 (1991).
108. Lee, C., Wevrick, R., Fisher, R. B., Ferguson-Smith, M. A. & Lin, C. C. Human centromeric DNAs.
Hum. Genet. 100, 291±304 (1997).
109. Riethman, H. C. et al. Integration of telomere sequences with the draft human genome sequence.
Nature 409, 953±958 (2001).
110. Pruit, K. D. & Maglott, D. R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids
Res. 29, 137±140 (2001).
111. Wolfsberg, T. G., McEntyre, J. & Schuler, G. D. Guide to the draft human genome. Nature 409, 824±
826 (2001).
112. Hurst, L. D. & Eyre-Walker, A. Evolutionary genomics: reading the bands. Bioessays 22, 105±107
(2000).
113. Saccone, S. et al. Correlations between isochores and chromosomal bands in the human genome.
Proc. Natl Acad. Sci. USA 90, 11929±11933 (1993).
114. Zoubak, S., Clay, O. & Bernardi, G. The gene distribution of the human genome. Gene 174, 95±102
(1996).
115. Gardiner, K. Base composition and gene distribution: critical patterns in mammalian genome
organization. Trends Genet. 12, 519±524 (1996).
116. Duret, L., Mouchiroud, D. & Gautier, C. Statistical analysis of vertebrate sequences reveals that long
genes are scarce in GC-rich isochores. J. Mol. Evol. 40, 308±317 (1995).
117. Saccone, S., De Sario, A., Della Valle, G. & Bernardi, G. The highest gene concentrations in the
human genome are in telomeric bands of metaphase chromosomes. Proc. Natl Acad. Sci. USA 89,
4913±4917 (1992).
118. Bernardi, G. et al. The mosaic genome of warm-blooded vertebrates. Science 228, 953±958
(1985).
119. Bernardi, G. Isochores and the evolutionary genomics of vertebrates. Gene 241, 3±17 (2000).
120. Fickett, J. W., Torney, D. C. & Wolf, D. R. Base compositional structure of genomes. Genomics 13,
1056±1064 (1992).
121. Churchill, G. A. Stochastic models for heterogeneous DNA sequences. Bull. Math. Biol. 51, 79±94
(1989).
122. Bird, A., Taggart, M., Frommer, M., Miller, O. J. & Macleod, D. A fraction of the mouse genome that
is derived from islands of nonmethylated, CpG-rich DNA. Cell 40, 91±99 (1985).
123. Bird, A. P. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 3, 342±347 (1987).
124. Chan, M. F., Liang, G. & Jones, P. A. Relationship between transcription and DNA methylation. Curr.
Top. Microbiol. Immunol. 249, 75±86 (2000).
125. Holliday, R. & Pugh, J. E. DNA modi®cation mechanisms and gene activity during development.
Science 187, 226±232 (1975).
126. Larsen, F., Gundersen, G., Lopez, R. & Prydz, H. CpG islands as gene markers in the human genome.
Genomics 13, 1095±1107 (1992).
127. Tazi, J. & Bird, A. Alternative chromatin structure at CpG islands. Cell 60, 909±920 (1990).
128. Gardiner-Garden, M. & Frommer, M. CpG islands in vertebrate genomes. J. Mol. Biol. 196, 261±282
(1987).
129. Antequera, F. & Bird, A. Number of CpG islands and genes in human and mouse. Proc. Natl Acad.
Sci. USA 90, 11995±11999 (1993).
130. Ewing, B. & Green, P. Analysis of expressed sequence tags indicates 35,000 human genes. Nature
Genet. 25, 232±234 (2000).
131. Yu, A. Comparison of human genetic and sequence-based physical maps. Nature 409, 951±953 (2001).
132. Kaback, D. B., Guacci, V., Barber, D. & Mahon, J. W. Chromosome size-dependent control of meiotic
recombination. Science 256, 228±232 (1992).
133. Riles, L. et al. Physical maps of the 6 smallest chromosomes of Saccharomyces cerevisiae at a
resolution of 2.6-kilobase pairs. Genetics 134, 81±150 (1993).
134. Lynn, A. et al. Patterns of meiotic recombination on the long arm of human chromosome 21.
Genome Res. 10, 1319±1332 (2000).
135. Laurie, D. A. & Hulten, M. A. Further studies on bivalent chiasma frequency in human males with
normal karyotypes. Ann. Hum. Genet. 49, 189±201 (1985).
136. Roeder, G. S. Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600±2621 (1997).
137. Wu, T.-C. & Lichten, M. Meiosis-induced double-strand break sites determined by yeast chromatin
structure. Science 263, 515±518 (1994).
138. Gerton, J. L. et al. Global mapping of meiotic recombination hotspots and coldspots in the yeast
Saccharomyces cerevisiae.Proc. Natl Acad. Sci. USA 97, 11383±11390 (2000).
139. Li, W. -H. Molecular Evolution (Sinauer, Sunderland, Massachusetts, 1997).
140. Gregory, T. R. & Hebert, P. D. The modulation of DNA content: proximate causes and ultimate
consequences. Genome Res. 9, 317±324 (1999).
141. Hartl, D. L. Molecular melodies in high and low C. Nature Rev. Genet. 1, 145±149 (2000).
142. Smit, A. F. Interspersed repeats and other mementos of transposable elements in mammalian
genomes. Curr. Opin. Genet. Dev. 9, 657±663 (1999).
143. Prak, E. L. & Haig, H. K. Jr Mobile elements and the human genome. Nature Rev. Genet. 1, 134±144
(2000).
144. Okada, N., Hamada, M., Ogiwara, I. & Ohshima, K. SINEs and LINEs share common 39sequences: a
review. Gene 205, 229±243 (1997).
145. Esnault, C., Maestre, J. & Heidmann, T. Human LINE retrotransposons generate processed
pseudogenes. Nature Genet. 24, 363±367 (2000).
146. Wei, W. et al. Human L1 retrotransposition: cis-preference vs. trans-complementation. Mol. Cell.
Biol. 21, 1429±1439 (2001)
147. Malik, H. S., Henikoff, S. & Eickbush, T. H. Poised for contagion: evolutionary origins of the
infectious abilities of invertebrate retroviruses. Genome Res. 10, 1307±1318 (2000).
148. Smit, A. F. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6, 743±
748 (1996).
149. Clark, J. B. & Tidwell, M. G. A phylogenetic perspective on P transposable element evolution in
Drosophila. Proc. Natl Acad. Sci. USA 94, 11428±11433 (1997).
150. Haring, E., Hagemann, S. & Pinsker, W. Ancient and recent horizontal invasions of Drosophilids by
P elements. J. Mol. Evol. 51, 577±586 (2000).
151. Koga, A. et al. Evidence for recent invasion of the medaka ®sh genome by the Tol2 transposable
element. Genetics 155, 273±281 (2000).
152. Robertson, H. M. & Lampe, D. J. Recent horizontal transfer of a mariner transposable element
among and between Diptera and Neuroptera. Mol. Biol. Evol. 12, 850±862 (1995).
153. Simmons, G. M. Horizontal transfer of hobo transposable elements within the Drosophila
melanogaster species complex: evidence from DNA sequencing. Mol. Biol. Evol. 9, 1050±1060
(1992).
154. Malik, H. S., Burke, W. D. & Eickbush, T. H. The age and evolution of non-LTR retrotransposable
elements. Mol. Biol. Evol. 16, 793±805 (1999).
155. Kordis, D. & Gubensek, F. Bov-B long interspersed repeated DNA (LINE) sequences are present in
Vipera ammodytes phospholipase A2 genes and in genomes of Viperidae snakes. Eur. J. Biochem. 246,
772±779 (1997).
156. Jurka, J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet.
16, 418±420 (2000).
157. Sarich, V. M. & Wilson, A. C. Generation time and genome evolution in primates. Science 179,
1144±1147 (1973).
158. Smit, A. F., Toth, G., Riggs, A. D., & Jurka, J. Ancestral, mammalian-wide subfamilies of LINE-1
repetitive sequences. J. Mol. Biol. 246, 401±417 (1995).
159. Lim, J. K. & Simmons, M. J. Gross chromosome rearrangements mediated by transposable elements
in Drosophila melanogaster.Bioessays 16, 269±275 (1994).
160. Caceres, M., Ranz, J. M., Barbadilla, A., Long, M. & Ruiz, A. Generation of a widespread Drosophila
inversion by a transposable element. Science 285, 415±418 (1999).
161. Gray, Y. H. It takes two transposons to tango: transposable-element-mediated chromosomal
rearrangements. Trends Genet. 16, 461±468 (2000).
162. Zhang, J. & Peterson, T. Genome rearrangements by nonlinear transposons in maize. Genetics 153,
1403±1410 (1999).
163. Smit, A. F. Identi®cation of a new, abundant superfamily of mammalian LTR-transposons. Nucleic
Acids Res. 21, 1863±1872 (1993).
164. Cordonnier, A., Casella, J. F. & Heidmann, T. Isolation of novel human endogenous retrovirus-like
elements with foamy virus-related pol sequence. J. Virol. 69, 5890±5897 (1995).
165. Medstrand, P. & Mager, D. L. Human-speci®c integrations of the HERV-K endogenous retrovirus
family. J. Virol. 72, 9782±9787 (1998).
166. Myers, E. W. et al. A whole-genome assembly of Drosophila. Science 287, 2196±2204 (2000).
167. Petrov, D. A., Lozovskaya, E. R. & Hartl, D. L. High intrinsic rate of DNA loss in Drosophila.Nature
384, 346±349 (1996).
168. Li, W. H., Ellsworth, D. L., Krushkal, J., Chang, B. H. & Hewett-Emmett, D. Rates of nucleotide
substitution in primates and rodents and the generation-time effect hypothesis. Mol. Phylogenet.
Evol. 5, 182±187 (1996).
169. Goodman, M. et al. Toward a phylogenetic classi®cation of primates based on DNA evidence
complemented by fossil evidence. Mol. Phylogenet. Evol. 9, 585±598 (1998).
170. Kazazian, H. H. Jr & Moran, J. V. The impact of L1 retrotransposons on the human genome. Nature
Genet. 19, 19±24 (1998).
171. Malik, H. S. & Eickbush, T. H. NeSL-1, an ancient lineage of site-speci®c non-LTR retrotransposons
from Caenorhabditis elegans.Genetics 154, 193±203 (2000).
172. Casavant, N. C. et al. The end of the LINE?: lack of recent L1 activity in a group of South American
rodents. Genetics 154, 1809±1817 (2000).
173. Meunier-Rotival, M., Soriano, P., Cuny, G., Strauss, F. & Bernardi, G. Sequence organization and
genomic distribution of the major family of interspersed repeats of mouse DNA. Proc. Natl Acad. Sci.
USA 79, 355±359 (1982).
174. Soriano, P., Meunier-Rotival, M. & Bernardi, G. The distribution of interspersed repeats is
nonuniform and conserved in the mouse and human genomes. Proc. Natl Acad. Sci. USA 80, 1816±
1820 (1983).
175. Goldman, M. A., Holmquist, G. P., Gray, M. C., Caston, L. A. & Nag, A. Replication timing of genes
and middle repetitive sequences. Science 224, 686±692 (1984).
176. Manuelidis, L. & Ward, D. C. Chromosomal and nuclear distribution of the HindIII 1.9-kb human
DNA repeat segment. Chromosoma 91, 28±38 (1984).
177. Feng, Q., Moran, J. V., Kazazian, H. H. Jr & Boeke, J. D. Human L1 retrotransposon encodes a
conserved endonuclease required for retrotransposition. Cell 87, 905±916 (1996).
178. Jurka, J. Sequence patterns indicate an enzymatic involvement in integration of mammalian
retroposons. Proc. Natl Acad. Sci. USA 94, 1872±1877 (1997).
179. Arcot, S. S. et al. High-resolution cartography of recently integrated human chromosome 19-speci®c
Alu fossils. J. Mol. Biol. 281, 843±856 (1998).
180. Schmid, C. W. Does SINE evolution preclude Alu function? Nucleic Acids Res. 26, 4541±4550 (1998).
181. Chu, W. M., Ballard, R., Carpick, B. W., Williams, B. R. & Schmid, C. W. Potential Alu function:
regulation of the activity of double-stranded RNA-activated kinase PKR. Mol. Cell. Biol. 18, 58±68
(1998).
182. Li, T., Spearow, J., Rubin, C. M. & Schmid, C. W. Physiological stresses increase mouse short
interspersed element (SINE) RNA expression in vivo. Gene 239, 367±372 (1999).
183. Liu, W. M., Chu, W. M., Choudary, P. V. & Schmid, C. W. Cell stress and translational inhibitors
transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 23, 1758±
1765 (1995).
184. Filipski, J. Correlation between molecular clock ticking, codon usage ®delity of DNA repair,
chromosome banding and chromatin compactness in germline cells. FEBS Lett. 217, 184±186
(1987).
185. Sueoka, N. Directional mutation pressure and neutral molecular evolution. Proc. Natl Acad. Sci.
articles
916 NATURE
|
VOL 409
|
15 FEBRUARY 2001
|
www.nature.com
© 2001 Macmillan Magazines Ltd

USA 85, 2653±2657 (1988).
186. Wolfe, K. H., Sharp, P. M. & Li, W. H. Mutation rates differ among regions of the mammalian
genome. Nature 337, 283±285 (1989).
187. Bains, W. Local sequence dependence of rate of base replacement in mammals. Mutat. Res. 267, 43±
54 (1992).
188. Mathews, C. K. & Ji, J. DNA precursor asymmetries, replication ®delity, and variable genome
evolution. Bioessays 14, 295±301 (1992).
189. Holmquist, G. P. & Filipski, J. Organization of mutations along the genome: a prime determinant of
genome evolution. Trends Ecol. Evol. 9, 65±68 (1994).
190. Eyre-Walker, A. Evidence of selection on silent site base composition in mammals: potential
implications for the evolution of isochores and junk DNA. Genetics 152, 675±683 (1999).
191. The International SNP Map Working Group. An SNP map of the human genome generated by
reduced representation shotgun sequencing. Nature 407, 513±516 (2000).
192. Bohossian, H. B., Skaletsky, H. & Page, D. C. Unexpectedly similar rates of nucleotide substitution
found in male and female hominids. Nature 406, 622±625 (2000).
193. Skowronski, J., Fanning, T. G. & Singer, M. F. Unit-length LINE-1 transcripts in human
teratocarcinoma cells. Mol. Cell. Biol. 8, 1385±1397 (1988).
194. Boissinot, S., Chevret, P. & Furano, A. V. L1 (LINE-1) retrotransposon evolution and ampli®cation
in recent human history. Mol. Biol. Evol. 17, 915±928 (2000).
195. Moran, J. V. Human L1 retrotransposition: insights and peculiarities learned from a cultured cell
retrotransposition assay. Genetica 107, 39±51 (1999).
196. Kazazian, H. H. Jr et al. Haemophilia A resulting from de novo insertion of L1 sequences represents a
novel mechanism for mutation in man. Nature 332, 164±166 (1988).
197. Sheen, F.-m. et al. Reading between the LINEs: Human genomic variation introduced by LINE-1
retrotransposition. Genome Res. 10, 1496±1508 (2000).
198. Dombroski, B. A., Mathias, S. L., Nanthakumar, E., Scott, A. F. & Kazazian, H. H. Jr Isolation of an
active human transposable element. Science 254, 1805±1808 (1991).
199. Holmes, S. E., Dombroski, B. A., Krebs, C. M., Boehm, C. D. & Kazazian, H. H. Jr A new
retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric
insertion. Nature Genet. 7, 143±148 (1994).
200. Sassaman, D. M. et al. Many human L1 elements are capable of retrotransposition. Nature Genet. 16,
37±43 (1997).
201. Dombroski, B. A., Scott, A. F. & Kazazian, H. H. Jr Two additional potential retrotransposons
isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl
Acad. Sci. USA 90, 6513±6517 (1993).
202. Kimberland, M. L. et al. Full-length human L1 insertions retain the capacity for high frequency
retrotransposition in cultured cells. Hum. Mol. Genet. 8, 1557±1560 (1999).
203. Moran, J. V. et al. High frequency retrotransposition in cultured mammalian cells. Cell 87, 917±927
(1996).
204. Moran, J. V., DeBerardinis, R. J. & Kazazian, H. H. Jr Exon shuf¯ing by L1 retrotransposition. Science
283, 1530±1534 (1999).
205. Pickeral, O. K., Makalowski, W., Boguski, M. S. & Boeke, J. D. Frequent human genomic DNA
transduction driven by LINE-1 retrotransposition. Genome Res. 10, 411±415 (2000).
206. Miki, Y. et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon
cancer. Cancer Res. 52, 643±645 (1992).
207. Branciforte, D. & Martin, S. L. Developmental and cell type speci®city of LINE-1 expression in
mouse testis: implications for transposition. Mol. Cell. Biol. 14, 2584±2592 (1994).
208. Trelogan, S. A. & Martin, S. L. Tightly regulated, developmentally speci®c expression of the ®rst
open reading frame from LINE-1 during mouse embryogenesis. Proc. Natl Acad. Sci. USA 92, 1520±
1524 (1995).
209. Jurka, J. & Kapitonov, V. V. Sectorial mutagenesis by transposable elements. Genetica 107, 239±248
(1999).
210. Fraser, M. J., Ciszczon, T., Elick, T. & Bauser, C. Precise excision of TTAA-speci®c lepidopteran
transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from
two species of Lepidoptera. Insect Mol. Biol. 5, 141±151 (1996).
211. Brosius, J. Genomes were forged by massive bombardments with retroelements and retrosequences.
Genetica 107, 209±238 (1999).
212. Kruglyak, S., Durrett, R. T., Schug, M. D. & Aquadro, C. F. Equilibrium distribution of microsatellite
repeat length resulting from a balance between slippage events and point mutations. Proc. Natl Acad.
Sci. USA 95, 10774±10778 (1998).
213. Toth, G., Gaspari, Z. & Jurka, J. Microsatellites in different eukaryotic genomes: survey and analysis.
Genome Res. 10, 967±981 (2000).
214. Ellegren, H. Heterogeneous mutation processes in human microsatellite DNA sequences. Nature
Genet. 24, 400±402 (2000).
215. Ji, Y., Eichler, E. E., Schwartz, S. & Nicholls, R. D. Structure of chromosomal duplicons and their role
in mediating human genomic disorders. Genome Res. 10, 597±610 (2000).
216. Eichler, E. E. Masquerading repeats: paralogous pitfalls of the human genome. Genome Res. 8, 758±
762 (1998).
217. Mazzarella, R. & D. Schlessinger, D. Pathological consequences of sequence duplications in the
human genome. Genome Res. 8, 1007±1021 (1998).
218. Eichler, E. E. et al. Interchromosomal duplications of the adrenoleukodystrophy locus: a
phenomenon of pericentromeric plasticity. Hum. Mol. Genet. 6, 991±1002 (1997).
219. Horvath, J. E., Schwartz, S. & Eichler, E. E. The mosaic structure of human pericentromeric DNA: a
strategy for characterizing complex regions of the human genome. Genome Res. 10, 839±852 (2000).
220. Brand-Arpon, V. et al. A genomic region encompassing a cluster of olfactory receptor genes and a
myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13-q21 and
3p13. Genomics 56, 98±110 (1999).
221. Arnold, N., Wienberg, J., Ermert, K. & Zachau, H. G. Comparative mapping of DNA probes derived
from the V kappa immunoglobulin gene regions on human and great ape chromosomes by
¯uorescence in situ hybridization. Genomics 26, 147±150 (1995).
222. Eichler, E. E. et al. Duplication of a gene-rich cluster between 16p11.1 and Xq28: a novel
pericentromeric-directed mechanism for paralogous genome evolution. Hum. Mol. Genet. 5, 899±
912 (1996).
223. Potier, M. et al. Two sequence-ready contigs spanning the two copies of a 200-kb duplication on
human 21q: partial sequence and polymorphisms. Genomics 51, 417±426 (1998).
224. Regnier, V. et al. Emergence and scattering of multiple neuro®bromatosis (NF1)-related sequences
during hominoid evolution suggest a process of pericentromeric interchromosomal transposition.
Hum. Mol. Genet. 6, 9±16 (1997).
225. Ritchie, R. J., Mattei, M. G. & Lalande, M. A large polymorphic repeat in the pericentromeric region
of human chromosome 15q contains three partial gene duplications. Hum. Mol. Genet. 7, 1253±
1260 (1998).
226. Trask, B. J. et al. Members of the olfactory receptor gene family are contained in large blocks of DNA
duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet. 7, 13±26
(1998).
227. Trask, B. J. et al. Large multi-chromosomal duplications encompass many members of the olfactory
receptor gene family in the human genome. Hum. Mol. Genet. 7, 2007±2020 (1998).
228. van Deutekom, J. C. et al. Identi®cation of the ®rst gene (FRG1) from the FSHD region on human
chromosome 4q35. Hum. Mol. Genet. 5, 581±590 (1996).
229. Zachau, H. G. The immunoglobulin kappa locusÐorÐwhat has been learned from looking closely
at one-tenth of a percent of the human genome. Gene 135, 167±173 (1993).
230. Zimonjic, D. B., Kelley, M. J., Rubin, J. S., Aaronson, S. A. & Popescu, N. C. Fluorescence in situ
hybridization analysis of keratinocyte growth factor gene ampli®cation and dispersion in evolution
of great apes and humans. Proc. Natl Acad. Sci. USA 94, 11461±11465 (1997).
231. van Geel, M. et al. The FSHD region on human chromosome 4q35 contains potential coding regions
among pseudogenes and a high density of repeat elements. Genomics 61, 55±65 (1999).
232. Horvath, J. E. et al. Molecular structure and evolution of an alpha satellite/non-alpha satellite
junction at 16p11. Hum. Mol. Genet. 9, 113±123 (2000).
233. Guy, J. et al. Genomic sequence and transcriptional pro®le of the boundary between pericentromeric
satellites and genes on human chromosome arm 10q. Hum. Mol. Genet. 9, 2029±2042 (2000).
234. Reiter, L. T., Murakami, T., Koeuth, T., Gibbs, R. A. & Lupski, J. R. The human COX10 gene is
disrupted during homologous recombination between the 24 kb proximal and distal CMT1A-REPs.
Hum. Mol. Genet. 6, 1595±1603 (1997).
235. Amos-Landgraf, J. M. et al. Chromosome breakage in the Prader-Willi and Angelman syndromes
involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am. J.
Hum. Genet. 65, 370±386 (1999).
236. Christian, S. L., Fantes, J. A., Mewborn, S. K., Huang, B. & Ledbetter, D. H. Large genomic duplicons
map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-
q13). Hum. Mol. Genet. 8, 1025±1037 (1999).
237. Edelmann, L., Pandita, R. K. & Morrow, B. E. Low-copy repeats mediate the common 3-Mb deletion
in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 64, 1076±1086 (1999).
238. Shaikh, T. H. et al. Chromosome 22-speci®c low copy repeats and the 22q11.2 deletion syndrome:
genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 9, 489±501 (2000).
239. Francke, U. Williams-Beuren syndrome: genes and mechanisms. Hum. Mol. Genet. 8, 1947±1954
(1999).
240. Peoples, R. et al. A physical map, including a BAC/PAC clone contig, of the Williams-Beuren
syndrome-deletion region at 7q11.23. Am. J. Hum. Genet. 66, 47±68 (2000).
241. Eichler, E. E., Archidiacono, N. & Rocchi, M. CAGGG repeats and the pericentromeric duplication
of the hominoid genome. Genome Res. 9, 1048±1058 (1999).
242. O'Keefe, C. & Eichler, E. in Comparative Genomics: Empirical and Analytical Approaches to Gene
Order Dynamics, Map Alignment and the Evolution of Gene Families (eds Sankoff, D. & Nadeau, J.)
29±46 (Kluwer Academic, Dordrecht, 2000).
243. Lander, E. S. The new genomics: Global views of biology. Science 274, 536±539 (1996).
244. Eddy, S. R. Noncoding RNA genes. Curr. Op. Genet. Dev. 9, 695±699 (1999).
245. Ban, N., Nissen, P., Hansen, J., Moore, P. B. & Steitz, T. A. The complete atomic structure of the large
ribosomal subunit at 2.4 angstrom resolution. Science 289, 905±920 (2000).
246. Nissen, P., Hansen, J., Ban, N., Moore, P. B. & Steitz, T. A. The structural basis of ribosome activity in
peptide bond synthesis. Science 289, 920±930 (2000).
247. Weinstein, L. B. & Steitz, J. A. Guided tours: from precursor snoRNA to functional snoRNP. Curr.
Opin. Cell Biol. 11, 378±384 (1999).
248. Bachellerie, J.-P. & Cavaille, J. in Modi®cation and Editing of RNA (ed. Benne, H. G. a. R.) 255±272
(ASM, Washington DC, 1998).
249. Burge, C. & Sharp, P. A. Classi®cation of introns: U2-type or U12-type. Cell 91, 875±879 (1997).
250. Brown, C. J. et al. The Human Xist geneÐanalysis of a 17 kb inactive X-speci®c RNA that contains
conserved repeats and is highly localized within the nucleus. Cell 71, 527±542 (1992).
251. Kickhoefer, V. A., Vasu, S. K. & Rome, L. H. Vaults are the answer, what is the question? Trends Cell
Biol. 6, 174±178 (1996).
252. Hatlen, L. & Attardi, G. Proportion of the HeLa cell genome complementary to the transfer RNA and
5S RNA. J. Mol. Biol. 56, 535±553 (1971).
253. Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences
and sequences of tRNA genes. Nucleic Acids Res. 26, 148±153 (1998).
254. Long, E. O. & Dawid, I. B. Repeated genes in eukaryotes. Annu. Rev. Biochem. 49, 727±764 (1980).
255. Crick, F. H. Codon±anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548±555 (1966).
256. Guthrie, C. & Abelson, J. in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene
Expression (eds Strathern, J. & Broach J.) 487±528 (Cold Spring Harbor Laboratory Press,
Cold Spring Harbor, New York, 1982).
257. Soll, D. & RajBhandary, U. (eds) tRNA: Structure, Biosynthesis, and Function (ASM, Washington DC,
1995).
258. Ikemura, T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol.
Evol. 2, 13±34 (1985).
259. Bulmer, M. Coevolution of codon usage and transfer-RNA abundance. Nature 325, 728±730 (1987).
260. Duret, L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal
translation of highly expressed genes. Trends Genet. 16, 287±289 (2000).
261. Sharp, P. M. & Matassi, G. Codon usage and genome evolution. Curr. Opin. Genet. Dev. 4, 851±860
(1994).
262. Buckland, R. A. A primate transfer-RNA gene cluster and the evolution of human chromosome 1.
Cytogenet. Cell Genet. 61, 1±4 (1992).
263. Gonos, E. S. & Goddard, J. P. Human tRNA-Glu genes: their copy number and organization. FEBS
Lett. 276, 138±142 (1990).
264. Sylvester, J. E. et al. The human ribosomal RNA genes: structure and organization of the complete
repeating unit. Hum. Genet. 73, 193±198 (1986).
articles
NATURE
|
VOL 409
|
15 FEBRUARY 2001
|
www.nature.com 917
© 2001 Macmillan Magazines Ltd

265. Sorensen, P. D. & Frederiksen, S. Characterization of human 5S ribosomal RNA genes. Nucleic Acids
Res. 19, 4147±4151 (1991).
266. Timofeeva, M. et al. [Organization of a 5S ribosomal RNA gene cluster in the human genome].
Mol. Biol. (Mosk.) 27, 861±868 (1993).
267. Little, R. D. & Braaten, D. C. Genomic organization of human 5S rDNA and sequence of one tandem
repeat. Genomics 4, 376±383 (1989).
268. Maden, B. E. H. The numerous modi®ed nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic
Acid Res. Mol. Biol. 39, 241±303 (1990).
269. Tycowski, K. T., You, Z. H., Graham, P. J. & Steitz, J. A. Modi®cation of U6 spliceosomal RNA is
guided by other small RNAs. Mol. Cell 2, 629±638 (1998).
270. Pavelitz, T., Liao, D. Q. & Weiner, A. M. Concerted evolution of the tandem array encoding primate
U2 snRNA (the RNU2 locus) is accompanied by dramatic remodeling of the junctions with ¯anking
chromosomal sequences. EMBO J. 18, 3783±3792 (1999).
271. Lindgren, V., Ares, A., Weiner, A. M. & Francke, U. Human genes for U2 small nuclear RNA map to a
major adenovirus 12 modi®cation site on chromosome 17. Nature 314, 115±116 (1985).
272. Van Arsdell, S. W. & Weiner, A. M. Human genes for U2 small nuclear RNA are tandemly repeated.
Mol. Cell. Biol. 4, 492±499 (1984).
273. Gao, L. I., Frey, M. R. & Matera, A. G. Human genes encoding U3 snRNA associate with coiled
bodies in interphase cells and are clustered on chromosome 17p11. 2 in a complex inverted repeat
structure. Nucleic Acids Res. 25, 4740±4747 (1997).
274. Hawkins, J. D. A survey on intron and exon lengths. Nucleic Acids Res. 16, 9893±9908 (1988).
275. Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol.
268, 78±94 (1997).
276. Labeit, S. & Kolmerer, B. Titins: giant proteins in charge of muscle ultrastructure and elasticity.
Science 270, 293±296 (1995).
277. Sterner, D. A., Carlo, T. & Berget, S. M. Architectural limits on split genes. Proc. Natl Acad. Sci. USA
93, 15081±15085 (1996).
278. Sun, Q., Mayeda, A., Hampson, R. K., Krainer, A. R. & Rottman, F. M. General splicing factor SF2/
ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 7, 2598±
2608 (1993).
279. Tanaka, K., Watakabe, A. & Shimura, Y. Polypurine sequences within a downstream exon function as
a splicing enhancer. Mol. Cell. Biol. 14, 1347±1354 (1994).
280. Carlo, T., Sterner, D. A. & Berget, S. M. An intron splicing enhancer containing a G-rich repeat
facilitates inclusion of a vertebrate micro-exon. RNA 2, 342±353 (1996).
281. Burset, M., Seledtsov, I. A. & Solovyev, V. V. Analysis of canonical and non-canonical splice sites in
mammalian genomes. Nucleic Acids Res. 28, 4364±4375 (2000).
282. Burge, C. B., Padgett, R. A. & Sharp, P. A. Evolutionary fates and origins of U12-type introns.
Mol. Cell 2, 773±785 (1998).
283. Mironov, A. A., Fickett, J. W. & Gelfand, M. S. Frequent alternative splicing of human genes. Genome
Res. 9, 1288±1293 (1999).
284. Hanke, J. et al. Alternative splicing of human genes: more the rule than the exception? Trends Genet.
15, 389±390 (1999).
285. Brett, D. et al. EST comparison indicates 38% of human mRNAs contain possible alternative splice
forms. FEBS Lett. 474, 83±86 (2000).
286. Dunham, I. The gene guessing game. Yeast 17, 218±224 (2000).
287. Lewin, B. Gene Expression (Wiley, New York, 1980).
288. Lewin, B. Genes IV 466±481 (Oxford Univ. Press, Oxford, 1990).
289. Smaglik, P. Researchers take a gamble on the human genome. Nature 405, 264 (2000).
290. Fields, C., Adams, M. D., White, O. & Venter, J. C. How many genes in the human genome? Nature
Genet. 7, 345±346 (1994).
291. Liang, F. et al. Gene index analysis of the human genome estimates approximately 120,000 genes.
Nature Genet. 25, 239±240 (2000).
292. Roest Crollius, H. et al. Estimate of human gene number provided by genome-wide analysis using
Tetraodon nigroviridis DNA sequence. Nature Genet. 25, 235±238 (2000).
293. The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform
for investigating biology. Science 282, 2012±2018 (1998).
294. Rubin, G. M. et al. Comparative genomics of the eukaryotes. Science 287, 2204±2215 (2000).
295. Green, P. et al. Ancient conserved regions in new gene sequences and the protein databases. Science
259, 1711±1716 (1993).
296. Fraser, A. G. et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA
interference. Nature 408, 325±330 (2000).
297. Mott, R. EST_GENOME: a program to align spliced DNA sequences to unspliced genomic DNA.
Comput. Appl. Biosci. 13, 477±478 (1997).
298. Florea, L., Hartzell, G., Zhang, Z., Rubin, G. M. & Miller, W. A computer program for aligning a
cDNA sequence with a genomic DNA sequence. Genome Res. 8, 967±974 (1998).
299. Bailey, L. C. Jr, Searls, D. B. & Overton, G. C. Analysis of EST-driven gene annotation in human
genomic sequence. Genome Res. 8, 362±376 (1998).
300. Birney, E., Thompson, J. D. & Gibson, T. J. PairWise and SearchWise: ®nding the optimal alignment
in a simultaneous comparison of a protein pro®le against all DNA translation frames. Nucleic Acids
Res. 24, 2730±2739 (1996).
301. Gelfand, M. S., Mironov, A. A. & Pevzner, P. A. Gene recognition via spliced sequence alignment.
Proc. Natl Acad. Sci. USA 93, 9061±9066 (1996).
302. Kulp, D., Haussler, D., Reese, M. G. & Eeckman, F. H. A generalized hidden Markov model for the
recognition of human genes in DNA. ISMB 4, 134±142 (1996).
303. Reese, M. G., Kulp, D., Tammana, H. & Haussler, D. GenieÐgene ®nding in Drosophila
melanogaster.Genome Res. 10, 529±538 (2000).
304. Solovyev, V. & Salamov, A. The Gene-Finder computer tools for analysis of human and model
organisms genome sequences. ISMB 5, 294±302 (1997).
305. Guigo, R., Agarwal, P., Abril, J. F., Burset, M. & Fickett, J. W. An assessment of gene prediction
accuracy in large DNA sequences. Genome Res. 10, 1631±1642 (2000).
306. Hubbard, T. & Birney, E. Open annotation offers a democratic solution to genome sequencing.
Nature 403, 825 (2000).
307. Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 28, 263±266 (2000).
308. Birney, E. & Durbin, R. Using GeneWise in the Drosophila annotation experiment. Genome Res. 10,
547±548 (2000).
309. The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium.
Functional annotation of a full-length mouse cDNA collection. Nature 409, 685±690 (2001).
310. Basrai, M. A., Hieter, P. & Boeke, J. D. Small open reading frames: beautiful needles in the haystack.
Genome Res. 7, 768±771 (1997).
311. Janin, J. & Chothia, C. Domains in proteins: de®nitions, location, and structural principles. Methods
Enzymol. 115, 420±430 (1985).
312. Ponting, C. P., Schultz, J., Copley, R. R., Andrade, M. A. & Bork, P. Evolution of domain families.
Adv. Protein Chem. 54, 185±244 (2000).
313. Doolittle, R. F. The multiplicity of domains in proteins. Annu. Rev. Biochem. 64, 287±314 (1995).
314. Bateman, A. & Birney, E. Searching databases to ®nd protein domain organization. Adv. Protein
Chem. 54, 137±157 (2000).
315. Futreal, P. A. et al. Cancer and genomics. Nature 409, 850±852 (2001).
316. Nestler, E. J. & Landsman, D. Learning about addiction from the human draft genome. Nature 409,
834±835 (2001).
317. Tupler, R., Perini, G. & Green, M. R. Expressing the human genome. Nature 409, 832±835 (2001).
318. Fahrer, A. M., Bazan, J. F., Papathanasiou, P., Nelms, K. A. & Goodnow, C. C. A genomic view of
immunology. Nature 409, 836±838 (2001).
319. Li, W. -H., Gu, Z., Wang, H. & Nekrutenko, A. Evolutionary analyses of the human genome. Nature
409, 847±849 (2001).
320. Bock, J. B., Matern, H. T., Peden, A. A. & Scheller, R. H. A genomic perspective on membrane
compartment organization. Nature 409, 839±841 (2001).
321. Pollard, T. D. Genomics, the cytoskeleton and motility. Nature 409, 842±843 (2001).
322. Murray, A. W. & Marks, D. Can sequencing shed light on cell cycling? Nature 409, 844±846 (2001).
323. Clayton, J. D., Kyriacou, C. P. & Reppert, S. M. Keeping time with the human genome. Nature 409,
829±831 (2001).
324. Chervitz, S. A. et al. Comparison of the complete protein sets of worm and yeast: orthology and
divergence. Science 282, 2022±2028 (1998).
325. Aravind, L. & Subramanian, G. Origin of multicellular eukaryotesÐinsights from proteome
comparisons. Curr. Opin. Genet. Dev. 9, 688±694 (1999).
326. Attwood, T. K. et al. PRINTS-S: the database formerly known as PRINTS. Nucleic Acids Res. 28, 225±
227 (2000).
327. Hofmann, K., Bucher, P., Falquet, L. & Bairoch, A. The PROSITE database, its status in 1999. Nucleic
Acids Res. 27, 215±219 (1999).
328. Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs. Nucleic Acids Res. 25, 3389±3402 (1997).
329. Wolf, Y. I., Kondrashov, F. A. & Koonin, E. V. No footprints of primordial introns in a eukaryotic
genome. Trends Genet. 16, 333±334 (2000).
330. Brunner, H. G., Nelen, M., Breake®eld, X. O., Ropers, H. H. & van Oost, B. B. A. Abnormal behavior
associated with a point mutation in the structural gene for monoamine oxidase A. Science 262, 578±
580 (1993).
331. Cases, O. et al. Aggressive behavior and altered amounts of brain serotonin and norepinephrine in
mice lacking MAOA. Science 268, 1763±1766 (1995).
332. Brunner, H. G. et al. X-linked borderline mental retardation with prominent behavioral disturbance:
phenotype, genetic localization, and evidence for disturbed monoamine metabolism. Am. J. Hum.
Genet. 52, 1032±1039 (1993).
333. Deckert, J. et al. Excess of high activity monoamine oxidase A gene promoter alleles in female
patients with panic disorder. Hum. Mol. Genet. 8, 621±624 (1999).
334. Smith, T. F. & Waterman, M. S. Identi®cation of common molecular subsequences. J. Mol. Biol. 147,
195±197 (1981).
335. Tatusov, R. L., Koonin, E. V. & Lipman, D. J. A genomic perspective on protein families. Science 278,
631±637 (1997).
336. Ponting, C. P., Aravind, L., Schultz, J., Bork, P. & Koonin, E. V. Eukaryotic signalling domain
homologues in archaea and bacteria. Ancient ancestry and horizontal gene transfer. J. Mol. Biol. 289,
729±745 (1999).
337. Zhang, J., Dyer, K. D. & Rosenberg, H. F. Evolution of the rodent eosinophil-associated Rnase gene
family by rapid gene sorting and positive selection. Proc. Natl Acad. Sci. USA 97, 4701±4706 (2000).
338. Shashoua, V. E. Ependymin, a brain extracellular glycoprotein, and CNS plasticity. Ann. NY Acad.
Sci. 627, 94±114 (1991).
339. Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P. & Bork, P. SMART: a web-based tool for the study
of genetically mobile domains. Nucleic Acids Res. 28, 231±234 (2000).
340. Koonin, E. V., Aravind, L. & Kondrashov, A. S. The impact of comparative genomics on our
understanding of evolution. Cell 101, 573±576 (2000).
341. Bateman, A., Eddy, S. R. & Chothia, C. Members of the immunoglobulin superfamily in bacteria.
Protein Sci. 5, 1939±1941 (1996).
342. Sutherland, D., Samakovlis, C. & Krasnow, M. A. Branchless encodes a Drosophila FGF homolog
that controls tracheal cell migration and the pattern of branching. Cell 87, 1091±1101 (1996).
343. Warburton, D. et al. The molecular basis of lung morphogenesis. Mech. Dev. 92, 55±81 (2000).
344. Fuchs, T., Glusman, G., Horn-Saban, S., Lancet, D. & Pilpel, Y. The human olfactory subgenome:
from sequence to structure to evolution. Hum. Genet. 108, 1±13 (2001).
345. Glusman, G. et al. The olfactory receptor gene family: data mining, classi®cation and nomenclature.
Mamm. Genome 11, 1016±1023 (2000).
346. Rouquier, S. et al. Distribution of olfactory receptor genes in the human genome. Nature Genet. 18,
243±250 (1998).
347. Sharon, D. et al. Primate evolution of an olfactory receptor cluster: Diversi®cation by gene
conversion and recent emergence of a pseudogene. Genomics 61, 24±36 (1999).
348. Gilad, Y. et al. Dichotomy of single-nucleotide polymorphism haplotypes in olfactory receptor genes
and pseudogenes. Nature Genet. 26, 221±224 (2000).
349. Gearhart, J. & Kirschner, M. Cells, Embryos, and Evolution (Blackwell Science, Malden, Massachu-
setts, 1997).
350. Barbazuk, W. B. et al. The syntenic relationship of the zebra®sh and human genomes. Genome Res.
10, 1351±1358 (2000).
351. McLysaght, A., Enright, A. J., Skrabanek, L. & Wolfe, K. H. Estimation of synteny conservation and
genome compaction between puffer®sh (Fugu) and human. Yeast 17, 22±36 (2000).
352. Trachtulec, Z. et al. Linkage of TATA-binding protein and proteasome subunit C5 genes in mice and
humans reveals synteny conserved between mammals and invertebrates. Genomics 44, 1±7 (1997).
articles
918 NATURE
|
VOL 409
|
15 FEBRUARY 2001
|
www.nature.com
© 2001 Macmillan Magazines Ltd

353. Nadeau, J. H. Maps of linkage and synteny homologies between mouse and man. Trends Genet. 5,
82±86 (1989).
354. Nadeau, J. H. & Taylor, B. A. Lengths of chromosomal segments conserved since divergence of man
and mouse. Proc. Natl Acad. Sci. USA 81, 814±818 (1984).
355. Copeland, N. G. et al. A genetic linkage map of the mouse: current applications and future prospects.
Science 262, 57±66 (1993).
356. DeBry, R. W. & Seldin, M. F. Human/mouse homology relationships. Genomics 33, 337±351 (1996).
357. Nadeau, J. H. & Sankoff, D. The lengths of undiscovered conserved segments in comparative maps.
Mamm. Genome 9, 491±495 (1998).
358. Thomas, J. W. et al. Comparative genome mapping in the sequence-based era: early experience with
human chromosome 7. Genome Res. 10, 624±633 (2000).
359. Pletcher, M. T. et al. Chromosome evolution: The junction of mammalian chromosomes in the
formation of mouse chromosome 10. Genome Res. 10, 1463±1467 (2000).
360. Novacek, M. J. Mammalian phylogeny: shaking the tree. Nature 356, 121±125 (1992).
361. O'Brien, S. J. et al. Genome maps 10. Comparative genomics. Mammalian radiations. Wall chart.
Science 286, 463±478 (1999).
362. Romer, A. S. Vertebrate Paleontology (Univ. Chicago Press, Chicago and New York, 1966).
363. Paterson, A. H. et al. Toward a uni®ed genetic map of higher plants, transcending the monocot-dicot
divergence. Nature Genet. 14, 380±382 (1996).
364. Jenczewski, E., Prosperi, J. M. & Ronfort, J. Differentiation between natural and cultivated
populations of Medicago sativa (Leguminosae) from Spain: analysis with random ampli®ed
polymorphic DNA (RAPD) markers and comparison to allozymes. Mol. Ecol. 8, 1317±1330 (1999).
365. Ohno, S. Evolution by Gene Duplication (George Allen and Unwin, London, 1970).
366. Wolfe, K. H. & Shields, D. C. Molecular evidence for an ancient duplication of the entire yeast
genome. Nature 387, 708±713 (1997).
367. Blanc, G., Barakat, A., Guyot, R., Cooke, R. & Delseny, M. Extensive duplication and reshuf¯ing in
the arabidopsis genome. Plant Cell 12, 1093±1102 (2000).
368. Paterson, A. H. et al. Comparative genomics of plant chromosomes. Plant Cell 12, 1523±1540 (2000).
369. Vision, T., Brown, D. & Tanksley, S. The origins of genome duplications in Arabidopsis.Science 290,
2114±2117 (2000).
370. Sidow, A. & Bowman, B. H. Molecular phylogeny. Curr. Opin. Genet. Dev. 1, 451±456 (1991).
371. Sidow, A. & Thomas, W. K. A molecular evolutionary framework for eukaryotic model organisms.
Curr. Biol. 4, 596±603 (1994).
372. Sidow, A. Gen(om)e duplications in the evolution of early vertebrates. Curr. Opin. Genet. Dev. 6,
715±722 (1996).
373. Spring, J. Vertebrate evolution by interspeci®c hybridisationÐare we polyploid? FEBS Lett. 400, 2±8
(1997).
374. Skrabanek, L. & Wolfe, K. H. Eukaryote genome duplicationÐwhere's the evidence? Curr. Opin.
Genet. Dev. 8, 694±700 (1998).
375. Hughes, A. L. Phylogenies of developmentally important proteins do not support the hypothesis of
two rounds of genome duplication early in vertebrate history. J. Mol. Evol. 48, 565±576 (1999).
376. Lander, E. S. & Schork, N. J. Genetic dissection of complex traits. Science 265, 2037±2048 (1994).
377. Horikawa, Y. et al. Genetic variability in the gene encoding calpain-10 is associated with type 2
diabetes mellitus. Nature Genet. 26, 163±175 (2000).
378. Hastbacka, J. et al. The diastrophic dysplasia gene encodes a novel sulfate transporter: positional
cloning by ®ne-structure linkage disequilibrium mapping. Cell 78, 1073±1087 (1994).
379. Tischkoff, S. A. et al. Global patterns of linkage disequilibrium at the CD4 locus and modern human
origins. Science 271, 1380±1387 (1996).
380. Kidd, J. R. et al. Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus PAH,
in a global representation of populations. Am. J. Hum. Genet. 63, 1882±1899 (2000).
381. Mateu, E. et al. Worldwide genetic analysis of the CFTR region. Am. J. Hum. Genet. 68, 103±117
(2001).
382. Abecasis, G. R. et al. Extent and distribution of linkage disequilibrium in three genomic regions.
Am. J. Hum. Genet. 68, 191±197 (2001).
383. Taillon-Miller, P. et al. Juxtaposed regions of extensive and minimal linkage disequilibrium in Xq25
and Xq28. Nature Genet. 25, 324±328 (2000).
384. Martin, E. R. et al. SNPing away at complex diseases: analysis of single-nucleotide polymorphisms
around APOE in Alzheimer disease. Am. J. Hum. Genet. 67, 383±394 (2000).
385. Collins, A., Lonjou, C. & Morton, N. E. Genetic epidemiology of single-nucleotide polymorphisms.
Proc. Natl Acad. Sci. USA 96, 15173±15177 (1999).
386. Dunning, A. M. et al. The extent of linkage disequilibrium in four populations with distinct
demographic histories. Am. J. Hum. Genet. 67, 1544±1554 (2000).
387. Rieder, M. J., Taylor, S. L., Clark, A. G. & Nickerson, D. A. Sequence variation in the human
angiotensin converting enzyme. Nature Genet. 22, 59±62 (1999).
388. Collins, F. S. Positional cloning moves from perditional to traditional. Nature Genet. 9, 347±350
(1995).
389. Nagamine, K. et al. Positional cloning of the APECED gene. Nature Genet. 17, 393±398 (1997).
390. Reuber, B. E. et al. Mutations in PEX1 are the most common cause of peroxisome biogenesis
disorders. Nature Genet. 17, 445±448 (1997).
391. Portsteffen, H. et al. Human PEX1 is mutated in complementation group 1 of the peroxisome
biogenesis disorders. Nature Genet. 17, 449±452 (1997).
392. Everett, L. A. et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene
(PDS). Nature Genet. 17, 411±422 (1997).
393. Coffey, A. J. et al. Host response to EBV infection in X-linked lymphoproliferative disease results
from mutations in an SH2-domain encoding gene. Nature Genet. 20, 129±135 (1998).
394. Van Laer, L. et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5.
Nature Genet. 20, 194±197 (1998).
395. Sakuntabhai, A. et al. Mutations in ATP2A2, encoding a Ca2+ pump, cause Darier disease. Nature
Genet. 21, 271±277 (1999).
396. Gedeon, A. K. et al. Identi®cation of the gene (SEDL) causing X-linked spondyloepiphyseal
dysplasia tarda. Nature Genet. 22, 400±404 (1999).
397. Hurvitz, J. R. et al. Mutations in the CCN gene family member WISP3 cause progressive
pseudorheumatoid dysplasia. Nature Genet. 23, 94±98 (1999).
398. Laberge-le Couteulx, S. et al. Truncating mutations in CCM1, encoding KRIT1, cause hereditary
cavernous angiomas. Nature Genet. 23, 189±193 (1999).
399. Sahoo, T. et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause
cerebral cavernous malformations (CCM1). Hum. Mol. Genet. 8, 2325±2333 (1999).
400. McGuirt, W. T. et al. Mutations in COL11A2 cause non-syndromic hearing loss (DFNA13). Nature
Genet. 23, 413±419 (1999).
401. Moreira, E. S. et al. Limb-girdle muscular dystrophy type 2G is caused by mutations in the gene
encoding the sarcomeric protein telethonin. Nature Genet. 24, 163±166 (2000).
402. Ruiz-Perez, V. L. et al. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental
dysostosis. Nature Genet. 24, 283±286 (2000).
403. Kaplan, J. M. et al. Mutations in ACTN4, encoding alpha-actinin-4, cause familial focal segmental
glomerulosclerosis. Nature Genet. 24, 251±256 (2000).
404. Escayg, A. et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with
GEFS+2. Nature Genet. 24, 343±345 (2000).
405. Sacksteder, K. A. et al. Identi®cation of the alpha-aminoadipic semialdehyde synthase gene, which is
defective in familial hyperlysinemia. Am. J. Hum. Genet. 66, 1736±1743 (2000).
406. Kalaydjieva, L. et al. N-myc downstream-regulated gene 1 is mutated in hereditary motor and
sensory neuropathy-Lom. Am. J. Hum. Genet. 67, 47±58 (2000).
407. Sundin, O. H. et al. Genetic basis of total colourblindness among the Pingelapese islanders. Nature
Genet. 25, 289±293 (2000).
408. Kohl, S. et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor
cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21.
Hum. Mol. Genet. 9, 2107±2116 (2000).
409. Avela, K. et al. Gene encoding a new RING-B-box-coiled-coil protein is mutated in mulibrey
nanism. Nature Genet. 25, 298±301 (2000).
410. Verpy, E. et al. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear
sensory hair cells, underlies usher syndrome type 1C. Nature Genet. 26, 51±55 (2000).
411. Bitner-Glindzicz, M. et al. A recessive contiguous gene deletion causing infantile hyperinsulinism,
enteropathy and deafness identi®es the usher type 1C gene. Nature Genet. 26, 56±60 (2000).
412. The May-Hegglin/Fetchner Syndrome Consortium. Mutations in MYH9 result in the May-Hegglin
anomaly, and Fechtner and Sebastian syndromes. Nature Genet. 26, 103±105 (2000).
413. Kelley, M. J., Jawien, W., Ortel, T. L. & Korczak, J. F. Mutation of MYH9, encoding non-muscle
myosin heavy chain A, in May-Hegglin anomaly. Nature Genet. 26, 106±108 (2000).
414. Kirschner, L. S. et al. Mutations of the gene encoding the protein kinase A type I-aregulatory
subunit in patients with the Carney complex. Nature Genet. 26, 89±92 (2000).
415. Lalwani, A. K. et al. Human nonsyndromic hereditary deafness DFNA17 is due to a mutation in
non-muscle myosin MYH9. Am. J. Hum. Genet. 67, 1121±1128 (2000).
416. Matsuura, T. et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia
type 10. Nature Genet. 26, 191±194 (2000).
417. Delettre, C. et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is
mutated in dominant optic atrophy. Nature Genet. 26, 207±210 (2000).
418. Pusch, C. M. et al. The complete form of X-linked congenital stationary night blindness is caused by
mutations in a gene encoding a leucine-rich repeat protein. Nature Genet. 26, 324±327 (2000).
419. The ADHR Consortium. Autosomal dominant hypophosphataemic rickets is associated with
mutations in FGF23. Nature Genet. 26, 345±348 (2000).
420. Bomont, P. et al. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat
family, is mutated in giant axonal neuropathy. Nature Genet. 26, 370±374 (2000).
421. Tullio-Pelet, A. et al. Mutant WD-repeat protein in triple-A syndrome. Nature Genet. 26, 332±335
(2000).
422. Nicole, S. et al. Perlecan, the major proteoglycan of basement membranes, is altered in patients with
Schwartz-Jampel syndrome (chondrodystrophic myotonia). Nature Genet. 26, 480±483 (2000).
423. Rogaev, E. I. et al. Familial Alzheimer's disease in kindreds with missense mutations in a gene on
chromosome 1 related to the Alzheimer's disease type 3 gene. Nature 376, 775±778 (1995).
424. Sherrington, R. et al. Cloning of a gene bearing missense mutations in early-onset familial
Alzheimer's disease. Nature 375, 754±760 (1995).
425. Olivieri, N. F. & Weatherall, D. J. The therapeutic reactivation of fetal haemoglobin. Hum. Mol.
Genet. 7, 1655±1658 (1998).
426. Drews, J. Research & development. Basic science and pharmaceutical innovation. Nature Biotechnol.
17, 406 (1999).
427. Drews, J. Drug discovery: a historical perspective. Science 287, 1960±1964 (2000).
428. Davies, P. A. et al. The 5-HT3B subunit is a major determinant of serotonin-receptor function.
Nature 397, 359±363 (1999).
429. Heise, C. E. et al. Characterization of the human cysteinyl leukotriene 2 receptor. J. Biol. Chem. 275,
30531±30536 (2000).
430. Fan, W. et al. BACE maps to chromosome 11 and a BACE homolog, BACE2, reside in the obligate
Down Syndrome region of chromosome 21. Science 286, 1255a (1999).
431. Saunders, A. J., Kim, T. -W. & Tanzi, R. E. BACE maps to chromosome 11 and a BACE homolog,
BACE2, reside in the obligate Down Syndrome region of chromosome 21. Science 286, 1255a (1999).
432. Firestein, S. The good taste of genomics. Nature 404, 552±553 (2000).
433. Matsunami, H., Montmayeur, J. P. & Buck, L. B. A family of candidate taste receptors in human and
mouse. Nature 404, 601±604 (2000).
434. Adler, E. et al. A novel family of mammalian taste receptors. Cell 100, 693±702 (2000).
435. Chandrashekar, J. et al. T2Rs function as bitter taste receptors. Cell 100, 703±711 (2000).
436. Hardison, R. C. Conserved non-coding sequences are reliable guides to regulatory elements. Trends
Genet. 16, 369±372 (2000).
437. Onyango, P. et al. Sequence and comparative analysis of the mouse 1-megabase region orthologous
to the human 11p15 imprinted domain. Genome Res. 10, 1697±1710 (2000).
438. Bouck, J. B., Metzker, M. L. & Gibbs, R. A. Shotgun sample sequence comparisons between mouse
and human genomes. Nature Genet. 25, 31±33 (2000).
439. Marshall, E. Public-private project to deliver mouse genome in 6 months. Science 290, 242±243 (2000).
440. Wasserman, W. W., Palumbo, M., Thompson, W., Fickett, J. W. & Lawrence, C. E. Human-mouse
genome comparisons to locate regulatory sites. Nature Genet. 26, 225±228 (2000).
441. Tagle, D. A. et al. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago
crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic
footprints. J. Mol. Biol. 203, 439±455 (1988).
442. McGuire, A. M., Hughes, J. D. & Church, G. M. Conservation of DNA regulatory motifs and
discovery of new motifs in microbial genomes. Genome Res. 10, 744±757 (2000).
articles
NATURE
|
VOL 409
|
15 FEBRUARY 2001
|
www.nature.com 919
© 2001 Macmillan Magazines Ltd
Bibliography
[1] Philip R. Bevington and D. Kieth Robinson. Data Reduction and Error Analysis for
the Physical Sciences. McGraw-Hill, Inc., 2nd edition, 1992.
[2] International Human Genome Sequencing Consortium. Initial sequencing and analysis
of the human genome. Nature, 409:860, 2001.
[3] T. Kessler, C. Hagemann, C. Grebing, T. Legero, U. Sterr, F. Riehle, M. J. Martin,
L. Chen, and J. Ye. A sub-40-mHz-linewidth laser based on a silicon single-crystal
optical cavity. Nature Photonics, 6:687, 2012.
[4] Jerry B. Marion and Stephen T. Thornton. Classical Dynamics of Particles and Systems.
Saunders College Publishing, 4th edition, 1995.
[5] William J. Murray, 1984.
[6] Tony Reichhardt. NASA reworks its sums after Mars fiasco. Nature, 401:517, 1999.
[7] T. Rosenband, D. B. Hume, P. O. Schmidt, C. W. Chou, A. Brusch, L. Lorini, W. H.
Oskay, R. E. Drullinger, T. M. Fortier, J. E. Stalnaker, S. A. Diddams, W. C. Swann,
N. R. Newbury, W. M. Itano, D. J. Wineland, and J. C. Bergquist. Frequency ratio of
Al+and Hg+single-ion optical clocks; metrology at the 17th decimal place. Science,
319:1808, 2008.
[8] Barry N. Taylor and Ambler Thompson. NIST special publication 330: The Inter-
national System of Units (SI). Technical report, National Institute of Standards and
Technology, 2008. Available online at http://physics.nist.gov/Pubs/SP330/sp330.pdf.
[9] John R. Taylor. An Introduction to Error Analysis. University Science Books, Sausalito,
California, 2nd edition, 1997.
[10] Menglan Zeng and Donald H. Shields. Nonlinear thermal expansion and contraction of
asphalt concrete. Can. J. Civ. Eng., 26:26, 1999.
124