AMICI Guide
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 527 [warning: Documents this large are best viewed by clicking the View PDF Link!]
- 1 About AMICI
- 2 License Conditions
- 3 How to contribute
- 4 Installation
- 5 Python Interface
- 6 MATLAB Interface
- 7 C++ Interface
- 8 FAQ
- 9 Namespace Documentation
- 10 Class Documentation
- 10.1 AmiException Class Reference
- 10.2 AmiVector Class Reference
- 10.3 AmiVectorArray Class Reference
- 10.4 BackwardProblem Class Reference
- 10.5 CvodeException Class Reference
- 10.6 ExpData Class Reference
- 10.7 ForwardProblem Class Reference
- 10.8 IDAException Class Reference
- 10.9 IntegrationFailure Class Reference
- 10.10 IntegrationFailureB Class Reference
- 10.11 Model Class Reference
- 10.12 Model_DAE Class Reference
- 10.13 Model_ODE Class Reference
- 10.14 NewtonFailure Class Reference
- 10.15 NewtonSolver Class Reference
- 10.16 NewtonSolverDense Class Reference
- 10.17 NewtonSolverIterative Class Reference
- 10.18 NewtonSolverSparse Class Reference
- 10.19 Constant Class Reference
- 10.20 Expression Class Reference
- 10.21 LogLikelihood Class Reference
- 10.22 ModelQuantity Class Reference
- 10.23 Observable Class Reference
- 10.24 ODEExporter Class Reference
- 10.25 ODEModel Class Reference
- 10.26 Parameter Class Reference
- 10.27 SigmaY Class Reference
- 10.28 State Class Reference
- 10.29 TemplateAmici Class Reference
- 10.30 ReturnData Class Reference
- 10.31 SBMLException Class Reference
- 10.32 SbmlImporter Class Reference
- 10.33 SetupFailure Class Reference
- 10.34 Solver Class Reference
- 10.35 SteadystateProblem Class Reference
- 11 File Documentation
- Index
AMICI
Generated by Doxygen 1.8.14

CONTENTS i
Contents
1 About AMICI 2
2 License Conditions 2
3 How to contribute 3
4 Installation 3
5 Python Interface 6
6 MATLAB Interface 8
7 C++ Interface 13
8 FAQ 14
9 Namespace Documentation 14
9.1 amici Namespace Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
9.2 amici.ode_export Namespace Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 75
9.3 amici.plotting Namespace Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 81
9.4 amici.sbml_import Namespace Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 82
10 Class Documentation 86
10.1 AmiException Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86
10.2 AmiVector Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 89
10.3 AmiVectorArray Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
10.4 BackwardProblem Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 105
10.5 CvodeException Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 111
10.6 ExpData Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 112
10.7 ForwardProblem Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 139
10.8 IDAException Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 147
10.9 IntegrationFailure Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 148
10.10IntegrationFailureB Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 150
10.11Model Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 151
Generated by Doxygen

CONTENTS 1
10.12Model_DAE Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 282
10.13Model_ODE Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 314
10.14NewtonFailure Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 344
10.15NewtonSolver Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 345
10.16NewtonSolverDense Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 352
10.17NewtonSolverIterative Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 355
10.18NewtonSolverSparse Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 359
10.19Constant Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 361
10.20Expression Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 362
10.21LogLikelihood Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 364
10.22ModelQuantity Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 365
10.23Observable Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 366
10.24ODEExporter Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 367
10.25ODEModel Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 372
10.26Parameter Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 385
10.27SigmaY Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 386
10.28State Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 388
10.29TemplateAmici Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 389
10.30ReturnData Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 390
10.31SBMLException Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 408
10.32SbmlImporter Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 408
10.33SetupFailure Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 423
10.34Solver Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 424
10.35SteadystateProblem Class Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 493
11 File Documentation 500
11.1 amici.cpp File Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 500
11.2 cblas.cpp File Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 502
11.3 interface_matlab.cpp File Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 502
11.4 spline.cpp File Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 504
11.5 symbolic_functions.cpp File Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 505
Generated by Doxygen

2 CONTENTS
Index 507
1 About AMICI
AMICI provides a multilanguage (Python, C++, Matlab) interface for the SUNDIALS solvers CVODES (for ordinary
differential equations) and IDAS (for algebraic differential equations). AMICI allows the user to read differential equa-
tion models specified as SBML and automatically compiles such models as .mex simulation files, C++ executables
or python modules. In contrast to the SUNDIALSTB interface, all necessary functions are transformed into native
C++ code, which allows for a significantly faster simulation. Beyond forward integration, the compiled simulation
file also allows for forward sensitivity analysis, steady state sensitivity analysis and adjoint sensitivity analysis for
likelihood based output functions.
The interface was designed to provide routines for efficient gradient computation in parameter estimation of bio-
chemical reaction models but is also applicable to a wider range of differential equation constrained optimization
problems.
Online documentation is available as github-pages.
Publications
Fröhlich, F., Kaltenbacher, B., Theis, F. J., & Hasenauer, J. (2017). Scalable
Parameter Estimation for Genome-Scale Biochemical Reaction Networks. Plos
Computational Biology, 13(1), e1005331. doi: 10.1371/journal.pcbi.1005331
Fröhlich, F., Theis, F. J., Rädler, J. O., & Hasenauer, J. (2017). Parameter
estimation for dynamical systems with discrete events and logical operations.
Bioinformatics, 33(7), 1049-1056. doi: 10.1093/bioinformatics/btw764
Current build status
2 License Conditions
Copyright (c) 2015-2018, Fabian Fröhlich, Jan Hasenauer, Daniel Weindl and Paul Stapor All rights reserved.
Redistribution and use in source and binary forms, with or without modification, are permitted provided that the
following conditions are met:
1. Redistributions of source code must retain the above copyright notice, this list of conditions and the following
disclaimer.
2. Redistributions in binary form must reproduce the above copyright notice, this list of conditions and the fol-
lowing disclaimer in the documentation and/or other materials provided with the distribution.
3. Neither the name of the copyright holder nor the names of its contributors may be used to endorse or promote
products derived from this software without specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS" AND ANY
EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WARRANTIES OF
MERCHANTABILITY AND FITNESS FOR A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL
THE COPYRIGHT HOLDER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SP←-
ECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO, PROCUREMENT
OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR BUSINESS INTERRUPTI←-
ON) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY,
OR TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS
SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
Generated by Doxygen

3 How to contribute 3
3 How to contribute
We are happy about contributions to AMICI in any form (new functionality, documentation, bug reports, ...).
Making code changes
When making code changes:
• Check if you agree to release your contribution under the conditions provided in LICENSE
• Start a new branch from master
• Implement your changes
• Submit a pull request
• Make sure your code is documented appropriately
–Run mtoc/makeDocumentation.m to check completeness of your documentation
• Make sure your code is compatible with C++11, gcc and clang
• when adding new functionality, please also provide test cases (see tests/cpputest/)
• Write meaningful commit messages
• Run all tests to ensure nothing got broken
–Run tests/cpputest/wrapTestModels.m followed by CI tests scripts/buildAll.sh
&& scripts/run-cpputest.sh
–Run tests/testModels.m
• When all tests are passing and you think your code is ready to merge, request a code review
Adding/Updating tests
To add new tests add a new corresponding python script (see, e.g., tests/example_dirac.py)
and add it to and run tests/generateTestConfigurationForExamples.sh To update test re-
sults replace tests/cpputest/expectedResults.h5 by tests/cpputest/writeResults.←-
h5.bak [ONLY DO THIS AFTER TRIPLE CHECKING CORRECTNESS OF RESULTS] Before replacing
the test results, confirm that only expected datasets have changed, e.g. using h5diff -v -r 1e-8
tests/cpputest/expectedResults.h5 tests/cpputest/writeResults.h5.bak |less
4 Installation
Availability
The sources for AMICI are accessible as
• Source tarball
• Source zip
• GIT repository on github
Generated by Doxygen

4 CONTENTS
Obtaining AMICI via the GIT versioning system
In order to always stay up-to-date with the latest AMICI versions, simply pull it from our GIT repository and recompile
it when a new release is available. For more information about GIT checkout their website
The GIT repository can currently be found at https://github.com/ICB-DCM/AMICI and a direct clone is
possible via
git clone https://github.com/ICB-DCM/AMICI.git AMICI
Installation
If AMICI was downloaded as a zip, it needs to be unpacked in a convenient directory. If AMICI was obtained via
cloning of the git repository, no further unpacking is necessary.
Dependencies
The MATLAB interface only depends on the symbolic toolbox, which is needed for model compilation, but not
simulation.
Symbolic Engine
The MATLAB interface requires the symbolic toolbox for model compilation. The symbolic toolbox is not required
for model simulation.
Math Kernel Library (MKL)
The python and C++ interfaces require a system installation of BLAS. AMICI has been tested with various native and
general purpose MKL implementations such as Accelerate, Intel MKL, cblas, openblas, atlas. The matlab interface
uses the MATLAB MKL, which requires no prior installation.
HDF5
The python and C++ interfaces provide routines to read and write options and results in hdf5 format. For the
python interface, the installation of hdf5 is optional, but for the C++ interace it is required. HDF can be installed
using package managers such as brew or apt:
brew install hdf5
or
apt-get install libhdf5-serial-dev
SWIG
The python interface requires SWIG, which has to be installed by the user. Swig can be installed using package
managers such as brew or apt:
brew install swig
or
apt-get install swig3.0
We note here that some linux package managers may provide swig executables as swig3.0, but installation as
swig is required. This can be fixed using, e.g., symbolic links:
mkdir -p ~/bin/ && ln -s $(which swig3.0) ~/bin/swig && export PATH=~/bin/:$PATH
Generated by Doxygen

4 Installation 5
python packages
The python interface requires the python packages pkgconfig and numpy to be installed before AMICI can be
installed. These can be installed via pip:
pip3 install pkgconfig numpy
MATLAB
To use AMICI from MATLAB, start MATLAB and add the AMICI/matlab direcory to the MATLAB path. To add all
toolbox directories to the MATLAB path, execute the matlab script
installAMICI.m
To store the installation for further MATLAB session, the path can be saved via
savepath
For the compilation of .mex files, MATLAB needs to be configured with a working C compiler. The C compiler needs
to be installed and configured via:
mex -setup c
For a list of supported compilers we refer to the mathworks documentation: mathworks.com Note that Microsoft
Visual Studio compilers are currently not supported.
python
To use AMICI from python, install the module and all other requirements using pip
pip3 install amici
You can now import it as python module:
import amici
C++
To use AMICI from C++, run the
./scripts/buildSundials.sh
./scripts/buildSuitesparse.sh
./scripts/buildAmici.sh
script to compile amici libary. The static library file can then be linked from
./build/libamici.a
In CMake-based packages, amici can be linked via
find_package(Amici)
Generated by Doxygen

6 CONTENTS
Dependencies
The MATLAB interface requires the Mathworks Symbolic Toolbox for model generation via amiwrap(...), but
not for execution of precompiled models. Currently MATLAB R2018a or newer is not supported (see https←-
://github.com/ICB-DCM/AMICI/issues/307)
The python interface requires python 3.6 or newer and cblas library to be installed. Windows installations via pip
are currently not supported, but users may try to install amici using the build scripts provided for the C++ interface
(these will by default automatically install the python module).
The C++ interface requires cmake and cblas to be installed.
The tools SUNDIALS and SuiteSparse shipped with AMICI do not require explicit installation.
AMICI uses the following packages from SUNDIALS:
CVODES: the sensitivity-enabled ODE solver in SUNDIALS. Radu Serban and Alan C. Hindmarsh. ASME 2005 In-
ternational Design Engineering Technical Conferences and Computers and Information in Engineering Conference.
American Society of Mechanical Engineers, 2005. PDF
IDAS
AMICI uses the following packages from SuiteSparse:
Algorithm 907: KLU, A Direct Sparse Solver for Circuit Simulation Problems. Timothy A. Davis, Ekanathan Pala-
madai Natarajan, ACM Transactions on Mathematical Software, Vol 37, Issue 6, 2010, pp 36:1 - 36:17. PDF
Algorithm 837: AMD, an approximate minimum degree ordering algorithm, Patrick R. Amestoy, Timothy A. Davis,
Iain S. Duff, ACM Transactions on Mathematical Software, Vol 30, Issue 3, 2004, pp 381 - 388. PDF
Algorithm 836: COLAMD, a column approximate minimum degree ordering algorithm, Timothy A. Davis, John R.
Gilbert, Stefan I. Larimore, Esmond G. Ng ACM Transactions on Mathematical Software, Vol 30, Issue 3, 2004, pp
377 - 380. PDF
5 Python Interface
In the following we will give a detailed overview how to specify models in Python and how to call the generated
simulation files.
Model Definition
This guide will guide the user on how to specify models in Python using SBML. For example implementations see
the examples in the python/examples directory.
SBML input
First, import an sbml file using the amici.sbml_import.SbmlImporter class:
import amici
sbmlImporter = amici.SbmlImporter(’model_steadystate_scaled.sbml’)
the sbml document as imported by libSBML is available as
sbml = sbmlImporter.sbml
Generated by Doxygen

5 Python Interface 7
Constants
parameters that should be considered constants can be specified in a list of strings specifying the respective SbmlId
of a parameter.
constantParameters=[’k4’]
Observables
assignment rules that should be considered as observables can extracted using the amici.assignment←-
Rules2observables function
observables = amici.assignmentRules2observables(sbml, filter=lambda variableId:
variableId.startswith(’observable_’) and not variableId.endswith(’_sigma’))
Standard Deviations
standard deviations can be specified as dictionaries ...
sigmas = {’observable_x1withsigma’: ’observable_x1withsigma_sigma’}
Model Compilation
to compile the sbml as python module, the user has to call the method amici.sbml_import.SbmlImporter.sbml2amici,
passing all the previously defined model specifications
sbmlImporter.sbml2amici(’test’, ’test’,
observables=observables,
constantParameters=constantParameters,
sigmas=sigma)
Model Simulation
currently the model folder has to be manually added to the python path
import sys
sys.path.insert(0, ’test’)
the compiled model can now be imported as python module
import test as modelModule
to obtain a model instance call the getModel() method. This model instance will be instantiated using the defautl
parameter values specified in the sbml.
model = modelModule.getModel()
then pass the simulation timepoints to amici.Model.setTimepoints
model.setTimepoints(np.linspace(0, 60, 60))
for simulation we need to generate a solver instance
solver = model.getSolver()
the model simulation can now be carried out using amici.runAmiciSimulation
rdata = amici.runAmiciSimulation(model, solver)
Generated by Doxygen
8 CONTENTS
6 MATLAB Interface
In the following we will give a detailed overview how to specify models in MATLAB and how to call the generated
simulation files.
Model Definition
This guide will guide the user on how to specify models in MATLAB. For example implementations see the examples
in the matlab/examples directory.
Header
The model definition needs to be defined as a function which returns a struct with all symbolic definitions and
options.
function [model] = example_model_syms()
Options
Set the options by specifying the respective field of the modelstruct
model.(fieldname) = value
The options specify default options for simulation, parametrisation and compilation. All of these options are optional.
field description default
.param default parametrisation 'log'/'log10'/'lin' 'lin'
.debug flag to compile with debug symbols false
.forward flag to activate forward sensitivities true
.adjoint flag to activate adjoint sensitivities true
When set to false, the fields 'forward' and 'adjoint' will speed up the time required to compile the model but also
disable the respective sensitivity computation.
States
Create the respective symbolic variables. The name of the symbolic variable can be chosen arbitrarily.
syms state1 state2 state3
Create the state vector containing all states:
model.sym.x = [ state1 state2 state3 ];
Parameters
Create the respective symbolic variables. The name of the symbolic variable can be chosen arbitrarily. Sensitivities
will be derived for all paramaters.
syms param1 param2 param3 param4 param5 param6
Create the parameters vector
model.sym.p = [ param1 param2 param3 param4 param5 param6 ];
Generated by Doxygen

6 MATLAB Interface 9
Constants
Create the respective symbolic variables. The name of the symbolic variable can be chosen arbitrarily. Sensitivities
with respect to constants will not be derived.
syms const1 const2
Create the parameters vector
model.sym.k = [ const1 const2 ];
Differential Equation
For time-dependent differential equations you can specify a symbolic variable for time. This needs to be denoted
by t.
syms t
Specify the right hand side of the differential equation for xdot
model.sym.xdot(1) = [ const1 - param1*state1 ];
model.sym.xdot(2) = [ +param2*state1 + dirac(t-param3) - const2*state2 ];
model.sym.xdot(3) = [ param4*state2 ];
or
model.sym.f(1) = [ const1 - param1*state1 ];
model.sym.f(2) = [ +param2*state1 + dirac(t-param3) - const2*state2 ];
model.sym.f(3) = [ param4*state2 ];
The specification of for xdot may depend on states, parameters and constants.
For DAEs also specify the mass matrix.
model.sym.M = [1, 0, 0;...
0, 1, 0;...
0, 0, 0];
The specification of M may depend on parameters and constants.
For ODEs the integrator will solve the equation ˙x=fand for DAEs the equations M·˙x=f. AMICI will decide
whether to use CVODES (for ODEs) or IDAS (for DAEs) based on whether the mass matrix is defined or not.
In the definition of the differential equation you can use certain symbolic functions. For a full list of available functions
see src/symbolic_functions.cpp.
Dirac functions can be used to cause a jump in the respective states at the specified time-point. This is typically
used to model injections, or other external stimuli. Spline functions can be used to model time/state dependent
response with unkown time/state dependence.
Initial Conditions
Specify the initial conditions. These may depend on parameters on constants and must have the same size as x.
model.sym.x0 = [ param4, 0, 0 ];
Generated by Doxygen

10 CONTENTS
Observables
Specify the observables. These may depend on parameters and constants.
model.sym.y(1) = state1 + state2;
model.sym.y(2) = state3 - state2;
In the definition of the observable you can use certain symbolic functions. For a full list of available functions see
src/symbolic_functions.cpp. Dirac functions in observables will have no effect.
Events
Specifying events is optional. Events are specified in terms of a trigger function, a bolus fuction and an output
function. The roots of the trigger function defines the occurences of the event. The bolus function defines the
change in the state on event occurences. The output function defines the expression which is evaluated and
reported by the simulation routine on every event occurence. The user can create events by constructing a vector
of objects of the class amievent.
model.sym.event(1) = amievent(state1 - state2,0,[]);
Events may depend on states, parameters and constants but not on observables.
For more details about event support see https://doi.org/10.1093/bioinformatics/btw764
Standard Deviation
Specifying standard deviations is optional. It only has an effect when computing adjoint sensitivities. It allows the
user to specify standard deviations of experimental data for observables and events.
Standard deviaton for observable data is denoted by sigma_y
model.sym.sigma_y(1) = param5;
Standard deviaton for event data is denoted by sigma_t
model.sym.sigma_t(1) = param6;
Both sigma_y and sigma_t can either be a scalar or of the same dimension as the observables / events function.
They can depend on time and parameters but must not depend on the states or observables. The values provided
in sigma_y and sigma_t will only be used if the value in D.Sigma_Y or D.Sigma_T in the user-provided
data struct is NaN. See simulation for details.
Objective Function
By default, AMICI assumes a normal noise model and uses the corresponding negative log-likelihood
J = 1/2*sum(((y_i(t)-my_ti)/sigma_y_i)^2 + log(2*pi*sigma_y^2)
as objective function. A user provided objective function can be specified in
model.sym.Jy
As reference see the default specification of this.sym.Jy in amimodel.makeSyms.
Generated by Doxygen

6 MATLAB Interface 11
SBML
AMICI can also import SBML models using the command SBML2AMICI. This will generate a model specification
as described above, which may be edited by the user to apply further changes.
Model Compilation
The model can then be compiled by calling amiwrap.m:
amiwrap(modelname,’example_model_syms’,dir,o2flag)
Here modelname should be a string defining the name of the model, dir should be a string containing
the path to the directory in which simulation files should be placed and o2flag is a flag indicating whether
second order sensitivities should also be compiled. The user should make sure that the previously defined
function ‘'example_model_syms’is in the user path. Alternatively, the user can also
call the function'example_model_syms'`
[model] = example_model_syms()
and subsequently provide the generated struct to amiwrap(...), instead of providing the symbolic function:
amiwrap(modelname,model,dir,o2flag)
In a similar fashion, the user could also generate multiple models and pass them directly to amiwrap(...)
without generating respective model definition scripts.
Model Simulation
After the call to amiwrap(...) two files will be placed in the specified directory. One is a modelname.mex and
the other is simulate_ modelname.m. The mex file should never be called directly. Instead the MATLAB script,
which acts as a wrapper around the .mex simulation file should be used.
The simulate_ modelname.m itself carries extensive documentation on how to call the function, what it returns and
what additional options can be specified. In the following we will give a short overview of possible function calls.
Integration
Define a time vector:
t = linspace(0,10,100)
Generate a parameter vector:
theta = ones(6,1);
Generate a constants vector:
kappa = ones(2,1);
Integrate:
sol = simulate_modelname(t,theta,kappa,[],options)
The integration status will be indicated by the sol.status flag. Negative values indicated failed integration. The
states will then be available as sol.x. The observables will then be available as sol.y. The event outputs will then
be available as sol.z. If no event occured there will be an event at the end of the considered interval with the final
value of the root function is stored in sol.rz.
Alternatively the integration can also be called via
[status,t,x,y] = simulate_modelname(t,theta,kappa,[],options)
The integration status will be indicated by the flag status . Negative values indicated failed integration. The states
will then be available as x. The observables will then be available as y. No event output will be given.
Generated by Doxygen

12 CONTENTS
Forward Sensitivities
Set the sensitivity computation to forward sensitivities and integrate:
options.sensi = 1;
options.sensi_meth = ’forward;
sol = simulate_modelname(t,theta,kappa,[],options)
The integration status will be indicated by the sol.status flag. Negative values indicate failed integration. The
states will be available as sol.x, with the derivative with respect to the parameters in sol.sx. The observables
will be available as sol.y, with the derivative with respect to the parameters in sol.sy. The event outputs will
be available as sol.z, with the derivative with respect to the parameters in sol.sz. If no event occured there
will be an event at the end of the considered interval with the final value of the root function stored in sol.rz, with
the derivative with respect to the parameters in sol.srz.
Alternatively the integration can also be called via
[status,t,x,y,sx,sy] = simulate_modelname(t,theta,kappa,[],options)
The integration status will be indicated by the status flag. Negative values indicate failed integration. The states will
then be available as x, with derivative with respect to the parameters in sx. The observables will then be available
as y, with derivative with respect to the parameters in sy. No event output will be given.
Adjoint Sensitivities
Set the sensitivity computation to adjoint sensitivities:
options.sensi = 1;
options.sensi_meth = ’adjoint’;
Define Experimental Data:
D.Y = [NaN(1,2)],ones(length(t)-1,2)];
D.Sigma_Y = [0.1*ones(length(t)-1,2),NaN(1,2)];
D.T = ones(1,1);
D.Sigma_T = NaN;
The NaN values in Sigma_Y and Sigma_T will be replaced by the specification in model.sym.sigma_y and
model.sym.sigma_t. Data points with NaN value will be completely ignored.
Integrate:
sol = simulate_modelname(t,theta,kappa,D,options)
The integration status will be indicated by the sol.status flag. Negative values indicate failed integration. The log-
likelihood will then be available as sol.llh and the derivative with respect to the parameters in sol.sllh.
Notice that for adjoint sensitivities no state, observable and event sensitivities will be available. Yet this approach
can be expected to be significantly faster for systems with a large number of parameters.
Generated by Doxygen

7 C++ Interface 13
Steady State Sensitivities
This will compute state sensitivities according to the formula sx
k=−∂f
∂x −1∂f
∂θk
In the current implementation this formulation does not allow for conservation laws as this would result in a singular
Jacobian.
Set the final timepoint as infinity, this will indicate the solver to compute the steadystate:
t = Inf;
Set the sensitivity computation to steady state sensitivities:
options.sensi = 1;
Integrate:
sol = simulate_modelname(t,theta,kappa,D,options)
The states will be available as sol.x, with the derivative with respect to the parameters in sol.sx. The observ-
ables will be available as sol.y, with the derivative with respect to the parameters in sol.sy. Notice that for
steady state sensitivities no event sensitivities will be available. For the accuracy of the computed derivatives it is
essential that the system is sufficiently close to a steady state. This can be checked by examining the right hand
side of the system at the final time-point via sol.diagnosis.xdot.
7 C++ Interface
The Python Interface and MATLAB Interface can translate the model definition into C++ code, which is then com-
piled into a .mex file or a python module. Advanced users can also use this code within stand-alone C/C++ ap-
plication for use in other environments (e.g. on high performance computing systems). This section will give a
short overview over the generated files and provide a brief introduction of how this code can be included in other
applications.
Generated model files
amiwrap.m and amici.SbmlImporter.sbml2amici write the model source files to ${AMICI_ROOT_←-
DIR}/models/${MODEL_NAME} by default. The content of a model source directory might look something like this
(given MODEL_NAME=model_steadystate):
CMakeLists.txt
hashes.mat
main.cpp
model_steadystate_deltaqB.cpp
model_steadystate_deltaqB.h
[... many more files model_steadystate_*.(cpp|h|md5|o) ]
wrapfunctions.cpp
wrapfunctions.h
model_steadystate.h
Generated by Doxygen

14 CONTENTS
Running a simulation
The entry function for running an AMICI simulation is runAmiciSimulation(...), declared in amici.h.
This function requires (i) a Model instance. For the example model_steadystate the respective class
is provided as Model_model_steadystate in model_steadystate.h. For convenience, the header
wrapfunctions.h defines a function getModel(), that returns an instance of that class. (ii) a Solver
instance. This solver instance needs to match the requirements of the model and can be generated using
model->getSolver(). (iii) optionally an ExpData instance, which contains any experimental data.
A scaffold for a standalone simulation program is generated in main.cpp in the model source directory. This
programm shows how to initialize the above-mentioned structs and how to obtain the simulation results.
Compiling and linking
The complete AMICI API is available through amici.h; this is the only header file that needs to be included.
hdf5.h provides some functions for reading and writing HDF5 files).
You need to compile and link ${AMICI_ROOT_DIR}/models/${MODEL_NAME}/∗.cpp,${AMICI_RO←-
OT_DIR}/src/∗.cpp, the SUNDIALS and the SUITESPARSE library, or use the CMake package configuration
from the build directory which tells CMake about all AMICI dependencies.
Along with main.cpp, a CMake file (CMakeLists.txt) will be generated automatically. The CMake file
shows the abovementioned library dependencies. These files provide a scaffold for a standalone simulation
program. The required numerical libraries are shipped with AMICI. To compile them, run ${AMICI_ROOT_←-
DIR}/scripts/run-tests.sh once. HDF5 libraries and header files need to be installed separately. More
information on how to run the compiled program is provided in main.cpp.
8 FAQ
Q: My model fails to build.
A: Remove the corresponding model directory located in AMICI/models/∗yourmodelname∗and compile again.
Q: It still does not compile.
A: Make an issue and we will have a look.
Q: I get an out of memory error while compiling my model on a Windows machine.
A: This may be due to an old compiler version. See issue #161 for instructions on how to install a new compiler.
Q: The simulation/sensitivities I get are incorrect.
A: There are some known issues, especially with adjoint sensitivities, events and DAEs. If your particular problem
is not featured in the issues list, please add it!
9 Namespace Documentation
9.1 amici Namespace Reference
The AMICI Python module (in doxygen this will also contain documentation about the C++ library)
Generated by Doxygen

9.1 amici Namespace Reference 15
Namespaces
•ode_export
The C++ ODE export module for python.
•plotting
Plotting related functions.
•sbml_import
The python sbml import module for python.
Classes
• class AmiException
amici exception handler class
• class AmiVector
• class AmiVectorArray
• class BackwardProblem
class to solve backwards problems.
• class CvodeException
cvode exception handler class
• class ExpData
ExpData carries all information about experimental or condition-specific data.
• class ForwardProblem
The ForwardProblem class groups all functions for solving the backwards problem. Has only static members.
• class IDAException
ida exception handler class
• class IntegrationFailure
integration failure exception for the forward problem this exception should be thrown when an integration failure
occured for this exception we can assume that we can recover from the exception and return a solution struct to
the user
• class IntegrationFailureB
integration failure exception for the backward problem this exception should be thrown when an integration failure
occured for this exception we can assume that we can recover from the exception and return a solution struct to the
user
• class Model
The Model class represents an AMICI ODE model. The model can compute various model related quantities based
on symbolically generated code.
• class Model_DAE
The Model class represents an AMICI DAE model. The model does not contain any data, but represents the state of
the model at a specific time t. The states must not always be in sync, but may be updated asynchroneously.
• class Model_ODE
The Model class represents an AMICI ODE model. The model does not contain any data, but represents the state of
the model at a specific time t. The states must not always be in sync, but may be updated asynchroneously.
• class NewtonFailure
newton failure exception this exception should be thrown when the steady state computation failed to converge for
this exception we can assume that we can recover from the exception and return a solution struct to the user
• class NewtonSolver
The NewtonSolver class sets up the linear solver for the Newton method.
• class NewtonSolverDense
The NewtonSolverDense provides access to the dense linear solver for the Newton method.
• class NewtonSolverIterative
The NewtonSolverIterative provides access to the iterative linear solver for the Newton method.
• class NewtonSolverSparse
Generated by Doxygen

16 CONTENTS
The NewtonSolverSparse provides access to the sparse linear solver for the Newton method.
• class ReturnData
class that stores all data which is later returned by the mex function
• class SetupFailure
setup failure exception this exception should be thrown when the solver setup failed for this exception we can assume
that we cannot recover from the exception and an error will be thrown
• class Solver
• class SteadystateProblem
The SteadystateProblem class solves a steady-state problem using Newton's method and falls back to integration on
failure.
Typedefs
• typedef double realtype
• typedef void(∗msgIdAndTxtFp) (const char ∗identifier, const char ∗format,...)
msgIdAndTxtFp
Enumerations
• enum BLASLayout {rowMajor = 101, colMajor = 102 }
• enum BLASTranspose {noTrans = 111, trans = 112, conjTrans = 113 }
• enum ParameterScaling {none,ln,log10 }
• enum SecondOrderMode {none,full,directional }
• enum SensitivityOrder {none,first,second }
• enum SensitivityMethod {none,forward,adjoint }
• enum LinearSolver {
dense = 1, band = 2, LAPACKDense = 3, LAPACKBand = 4,
diag = 5, SPGMR = 6, SPBCG = 7, SPTFQMR = 8,
KLU =9}
• enum InternalSensitivityMethod {simultaneous = 1, staggered = 2, staggered1 =3}
• enum InterpolationType {hermite = 1, polynomial =2}
• enum LinearMultistepMethod {adams = 1, BDF =2}
• enum NonlinearSolverIteration {functional = 1, newton =2}
• enum StateOrdering {AMD,COLAMD,natural }
• enum SteadyStateSensitivityMode {newtonOnly,simulationFSA }
• enum NewtonStatus {failed =-1, newt =1, newt_sim =2, newt_sim_newt =3 }
• enum mexRhsArguments {
RHS_TIMEPOINTS,RHS_PARAMETERS,RHS_CONSTANTS,RHS_OPTIONS,
RHS_PLIST,RHS_XSCALE_UNUSED,RHS_INITIALIZATION,RHS_DATA,
RHS_NUMARGS_REQUIRED = RHS_DATA, RHS_NUMARGS }
The mexFunctionArguments enum takes care of the ordering of mex file arguments (indexing in prhs)
Functions
• void printErrMsgIdAndTxt (const char ∗identifier, const char ∗format,...)
• void printWarnMsgIdAndTxt (const char ∗identifier, const char ∗format,...)
• std::unique_ptr<ReturnData >runAmiciSimulation (Solver &solver, const ExpData ∗edata, Model &model)
• void amici_dgemv (BLASLayout layout, BLASTranspose TransA, const int M, const int N, const double alpha,
const double ∗A, const int lda, const double ∗X, const int incX, const double beta, double ∗Y, const int incY)
• void amici_dgemm (BLASLayout layout, BLASTranspose TransA, BLASTranspose TransB, const int M, const
int N, const int K, const double alpha, const double ∗A, const int lda, const double ∗B, const int ldb, const
double beta, double ∗C, const int ldc)
Generated by Doxygen

9.1 amici Namespace Reference 17
• void amici_daxpy (int n, double alpha, const double ∗x, const int incx, double ∗y, int incy)
Compute y = a∗x + y.
• void setModelData (const mxArray ∗prhs[ ], int nrhs, Model &model)
setModelData sets data from the matlab call to the model object
• void setSolverOptions (const mxArray ∗prhs[ ], int nrhs, Solver &solver)
setSolverOptions solver options from the matlab call to a solver object
• ReturnDataMatlab ∗setupReturnData (mxArray ∗plhs[ ], int nlhs)
• std::unique_ptr<ExpData >expDataFromMatlabCall (const mxArray ∗prhs[ ], const Model &model)
• int checkFinite (const int N, const realtype ∗array, const char ∗fun)
• void unscaleParameters (const double ∗bufferScaled, const ParameterScaling ∗pscale, int n, double ∗buffer←-
Unscaled)
Remove parameter scaling according to the parameter scaling in pscale.
• void unscaleParameters (std::vector<double >const &bufferScaled, std::vector<ParameterScaling >const
&pscale, std::vector<double >&bufferUnscaled)
Remove parameter scaling according to the parameter scaling in pscale.
• double getUnscaledParameter (double scaledParameter, ParameterScaling scaling)
Remove parameter scaling according to scaling
• bool operator== (const Model &a, const Model &b)
• mxArray ∗getReturnDataMatlabFromAmiciCall (ReturnData const ∗rdata)
• mxArray ∗initMatlabReturnFields (ReturnData const ∗rdata)
• mxArray ∗initMatlabDiagnosisFields (ReturnData const ∗rdata)
•template<typename T >
void writeMatlabField0 (mxArray ∗matlabStruct, const char ∗fieldName, T fieldData)
•template<typename T >
void writeMatlabField1 (mxArray ∗matlabStruct, const char ∗fieldName, std::vector<T>fieldData, const int
dim0)
•template<typename T >
void writeMatlabField2 (mxArray ∗matlabStruct, const char ∗fieldName, std::vector<T>fieldData, int dim0,
int dim1, std::vector<int >perm)
•template<typename T >
void writeMatlabField3 (mxArray ∗matlabStruct, const char ∗fieldName, std::vector<T>fieldData, int dim0,
int dim1, int dim2, std::vector<int >perm)
•template<typename T >
void writeMatlabField4 (mxArray ∗matlabStruct, const char ∗fieldName, std::vector<T>fieldData, int dim0,
int dim1, int dim2, int dim3, std::vector<int >perm)
• double ∗initAndAttachArray (mxArray ∗matlabStruct, const char ∗fieldName, std::vector<mwSize >dim)
• void checkFieldNames (const char ∗∗fieldNames, const int fieldCount)
•template<typename T >
std::vector<T>reorder (const std::vector<T>input, const std::vector<int >order)
•template<typename T >
char ∗serializeToChar (T const &data, int ∗size)
•template<typename T >
TdeserializeFromChar (const char ∗buffer, int size)
•template<typename T >
std::string serializeToString (T const &data)
•template<typename T >
std::vector<char >serializeToStdVec (T const &data)
•template<typename T >
TdeserializeFromString (std::string const &serialized)
• bool operator== (const Solver &a, const Solver &b)
• int spline (int n, int end1, int end2, double slope1, double slope2, double x[ ], double y[ ], double b[ ], double
c[ ], double d[ ])
• double seval (int n, double u, double x[ ], double y[ ], double b[ ], double c[ ], double d[ ])
Evaluate the cubic spline function.
• double sinteg (int n, double u, double x[ ], double y[ ], double b[ ], double c[ ], double d[ ])
Generated by Doxygen

18 CONTENTS
• double log (double x)
• double dirac (double x)
• double heaviside (double x)
• double min (double a, double b, double c)
• double Dmin (int id, double a, double b, double c)
• double max (double a, double b, double c)
• double Dmax (int id, double a, double b, double c)
• double pos_pow (double base, double exponent)
• int isNaN (double what)
• int isInf (double what)
• double getNaN ()
• double sign (double x)
• double spline (double t, int num,...)
• double spline_pos (double t, int num,...)
• double Dspline (int id, double t, int num,...)
• double Dspline_pos (int id, double t, int num,...)
• double DDspline (int id1, int id2, double t, int num,...)
• double DDspline_pos (int id1, int id2, double t, int num,...)
• def runAmiciSimulation (model, solver, edata=None)
Convenience wrapper around amici.runAmiciSimulation (generated by swig)
• def ExpData (rdata, sigma_y, sigma_z)
Convenience wrapper for ExpData constructor.
• def runAmiciSimulations (model, solver, edata_list)
Convenience wrapper for loops of amici.runAmiciSimulation.
• def pysb2amici (model, output_dir=None, observables=None, constantParameters=None, sigmas=None, ver-
bose=False, assume_pow_positivity=False, compiler=None)
Generate AMICI C++ files for the model provided to the constructor.
• int dbl2int (const double x)
• char amici_blasCBlasTransToBlasTrans (BLASTranspose trans)
• std::vector<realtype >mxArrayToVector (const mxArray ∗array, int length)
•realtype getValueById (std::vector<std::string >const &ids, std::vector<realtype >const &values, std←-
::string const &id, const char ∗variable_name, const char ∗id_name)
local helper function to get parameters
• void setValueById (std::vector<std::string >const &ids, std::vector<realtype >&values, realtype value,
std::string const &id, const char ∗variable_name, const char ∗id_name)
local helper function to set parameters
• int setValueByIdRegex (std::vector<std::string >const &ids, std::vector<realtype >&values, realtype value,
std::string const ®ex, const char ∗variable_name, const char ∗id_name)
local helper function to set parameters via regex
Variables
•msgIdAndTxtFp errMsgIdAndTxt = &printErrMsgIdAndTxt
•msgIdAndTxtFp warnMsgIdAndTxt = &printWarnMsgIdAndTxt
• constexpr double pi = 3.14159265358979323846
• bool hdf5_enabled = False
boolean indicating if amici was compiled with hdf5 support
• bool has_clibs = False
boolean indicating if this is the full package with swig interface or the raw package without
•amici_path
absolute root path of the amici repository
•amiciSwigPath = os.path.join(amici_path, 'swig')
Generated by Doxygen

9.1 amici Namespace Reference 19
absolute path of the amici swig directory
•amiciSrcPath = os.path.join(amici_path, 'src')
absolute path of the amici source directory
•amiciModulePath = os.path.dirname(__file__)
absolute root path of the amici module
9.1.1 Detailed Description
The AMICI Python module provides functionality for importing SBML models and turning them into C++ Python
extensions.
Getting started:
creating a extension module for an SBML model:
import amici
amiSbml = amici.SbmlImporter(’mymodel.sbml’)
amiSbml.sbml2amici(’modelName’,’outputDirectory’)
using the created module (set python path)
import modelName
help(modelName)
9.1.2 Typedef Documentation
9.1.2.1 realtype
typedef double realtype
defines variable type for simulation variables (determines numerical accuracy)
Definition at line 51 of file defines.h.
9.1.2.2 msgIdAndTxtFp
typedef void(∗msgIdAndTxtFp) (const char ∗identifier, const char ∗format,...)
Parameters
identifier string with error message identifier
format string with error message printf-style format
... arguments to be formatted
Definition at line 159 of file defines.h.
9.1.3 Enumeration Type Documentation
Generated by Doxygen

20 CONTENTS
9.1.3.1 BLASLayout
enum BLASLayout [strong]
BLAS Matrix Layout, affects dgemm and gemv calls
Definition at line 54 of file defines.h.
9.1.3.2 BLASTranspose
enum BLASTranspose [strong]
BLAS Matrix Transposition, affects dgemm and gemv calls
Definition at line 60 of file defines.h.
9.1.3.3 ParameterScaling
enum ParameterScaling [strong]
modes for parameter transformations
Definition at line 67 of file defines.h.
9.1.3.4 SecondOrderMode
enum SecondOrderMode [strong]
modes for second order sensitivity analysis
Definition at line 74 of file defines.h.
9.1.3.5 SensitivityOrder
enum SensitivityOrder [strong]
orders of sensitivity analysis
Definition at line 81 of file defines.h.
Generated by Doxygen

9.1 amici Namespace Reference 21
9.1.3.6 SensitivityMethod
enum SensitivityMethod [strong]
methods for sensitivity computation
Definition at line 88 of file defines.h.
9.1.3.7 LinearSolver
enum LinearSolver [strong]
linear solvers for CVODES/IDAS
Definition at line 95 of file defines.h.
9.1.3.8 InternalSensitivityMethod
enum InternalSensitivityMethod [strong]
CVODES/IDAS forward sensitivity computation method
Definition at line 108 of file defines.h.
9.1.3.9 InterpolationType
enum InterpolationType [strong]
CVODES/IDAS state interpolation for adjoint sensitivity analysis
Definition at line 115 of file defines.h.
9.1.3.10 LinearMultistepMethod
enum LinearMultistepMethod [strong]
CVODES/IDAS linear multistep method
Definition at line 121 of file defines.h.
Generated by Doxygen

22 CONTENTS
9.1.3.11 NonlinearSolverIteration
enum NonlinearSolverIteration [strong]
CVODES/IDAS Nonlinear Iteration method
Definition at line 127 of file defines.h.
9.1.3.12 StateOrdering
enum StateOrdering [strong]
KLU state reordering
Definition at line 133 of file defines.h.
9.1.3.13 SteadyStateSensitivityMode
enum SteadyStateSensitivityMode [strong]
Sensitivity computation mode in steadyStateProblem
Definition at line 140 of file defines.h.
9.1.3.14 NewtonStatus
enum NewtonStatus [strong]
State in which the steady state computionat finished
Definition at line 146 of file defines.h.
9.1.4 Function Documentation
9.1.4.1 printErrMsgIdAndTxt()
void printErrMsgIdAndTxt (
const char ∗identifier,
const char ∗format,
... )
printErrMsgIdAndTxt prints a specified error message associated to the specified identifier
Generated by Doxygen
9.1 amici Namespace Reference 23
Parameters
in identifier error identifier
Type: char
in format string with error message printf-style format
... arguments to be formatted
Returns
void
Definition at line 130 of file amici.cpp.
9.1.4.2 printWarnMsgIdAndTxt()
void printWarnMsgIdAndTxt (
const char ∗identifier,
const char ∗format,
... )
printErrMsgIdAndTxt prints a specified warning message associated to the specified identifier
Parameters
in identifier warning identifier
Type: char
in format string with error message printf-style format
... arguments to be formatted
Returns
void
Definition at line 151 of file amici.cpp.
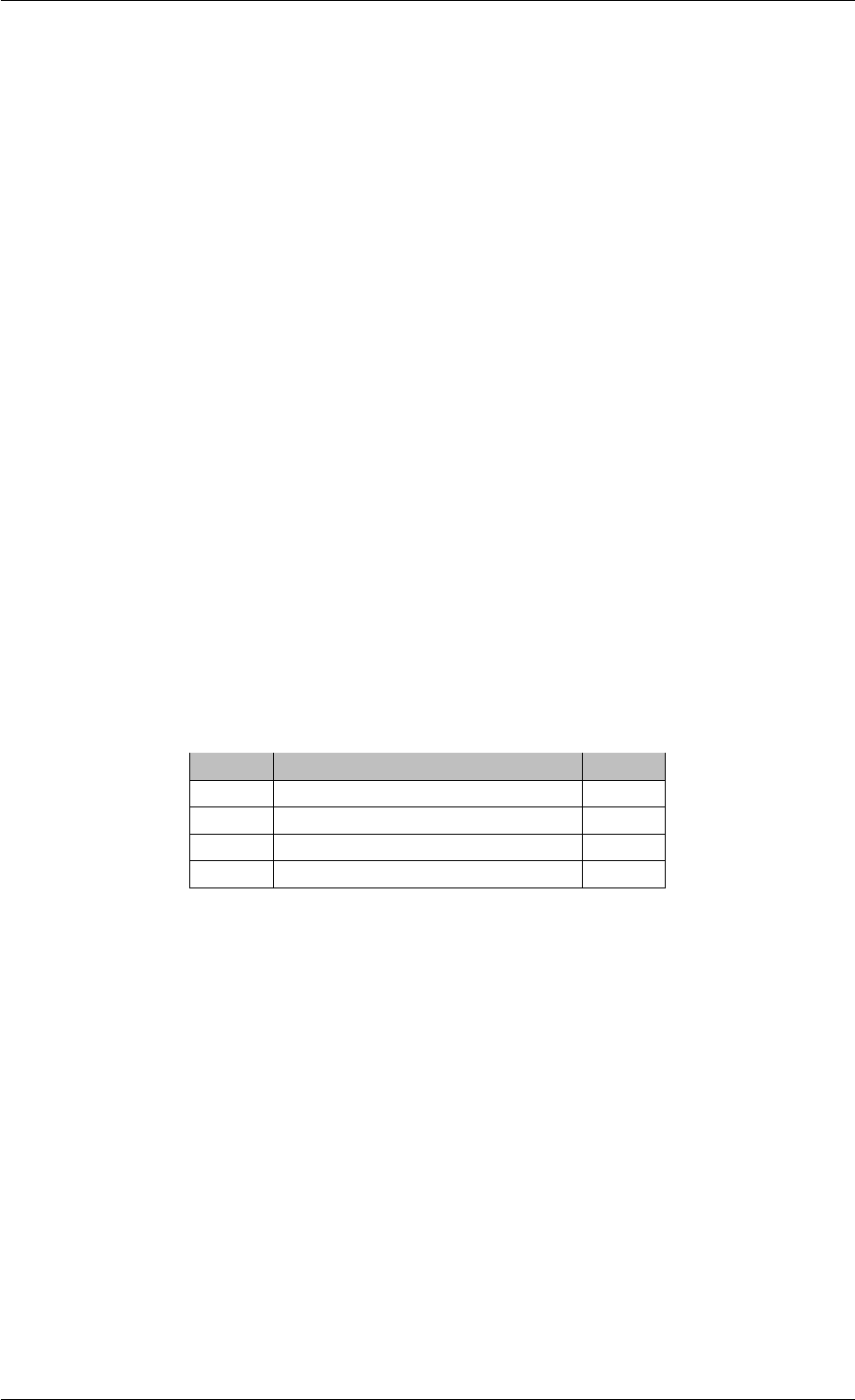
9.1.4.3 runAmiciSimulation() [1/2]
std::unique_ptr<ReturnData >runAmiciSimulation (
Solver &solver,
const ExpData ∗edata,
Model &model )
runAmiciSimulation is the core integration routine. It initializes the solver and runs the forward and backward prob-
lem.
Parameters
solver Solver instance
edata pointer to experimental data object
model model specification object
Generated by Doxygen
24 CONTENTS
Returns
rdata pointer to return data object
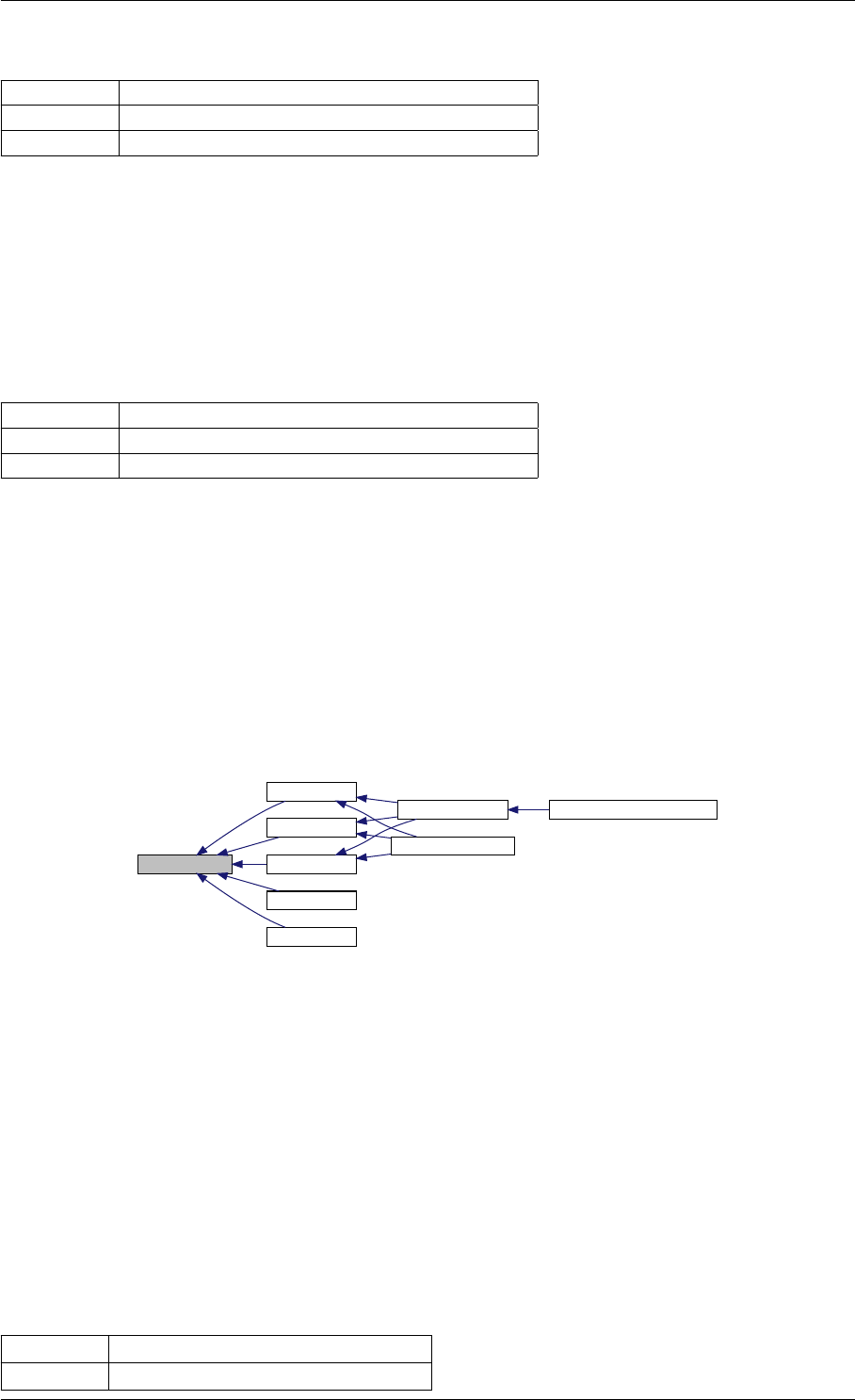
Definition at line 59 of file amici.cpp.
Here is the call graph for this function:
runAmiciSimulation
amici::ExpData::nt
amici::ExpData::getTimepoints
amici::ReturnData::
invalidate
amici::ReturnData::
invalidateLLH
amici::AmiException
::what
amici::AmiException
::getBacktrace
amici::ReturnData::
applyChainRuleFactorToSimulation
Results
getNaN
unscaleParameters getUnscaledParameter
Here is the caller graph for this function:
runAmiciSimulation
runAmiciSimulation
runAmiciSimulations
9.1.4.4 amici_dgemv()
void amici_dgemv (
BLASLayout layout,
BLASTranspose TransA,
const int M,
const int N,
const double alpha,
const double ∗A,
const int lda,
Generated by Doxygen

9.1 amici Namespace Reference 25
const double ∗X,
const int incX,
const double beta,
double ∗Y,
const int incY )
amici_dgemm provides an interface to the blas matrix vector multiplication routine dgemv. This routines computes
y = alpha∗A∗x + beta∗y with A: [MxK] B:[KxN] C:[MxN]
Parameters
in layout can be AMICI_BLAS_ColMajor or AMICI_BLAS_RowMajor.
in TransA flag indicating whether A should be transposed before multiplication
in Mnumber of rows in A
in Nnumber of columns in A
in alpha coefficient alpha
in Amatrix A
in lda leading dimension of A (m or n)
in Xvector X
in incX increment for entries of X
in beta coefficient beta
in,out Yvector Y
in incY increment for entries of Y
amici_dgemm provides an interface to the CBlas matrix vector multiplication routine dgemv. This routines computes
y = alpha∗A∗x + beta∗y with A: [MxN] x:[Nx1] y:[Mx1]
Parameters
layout always needs to be AMICI_BLAS_ColMajor.
TransA flag indicating whether A should be transposed before multiplication
Mnumber of rows in A
Nnumber of columns in A
alpha coefficient alpha
Amatrix A
lda leading dimension of A (m or n)
Xvector X
incX increment for entries of X
beta coefficient beta
Yvector Y
incY increment for entries of Y
Returns
void
Definition at line 73 of file cblas.cpp.
Generated by Doxygen

26 CONTENTS
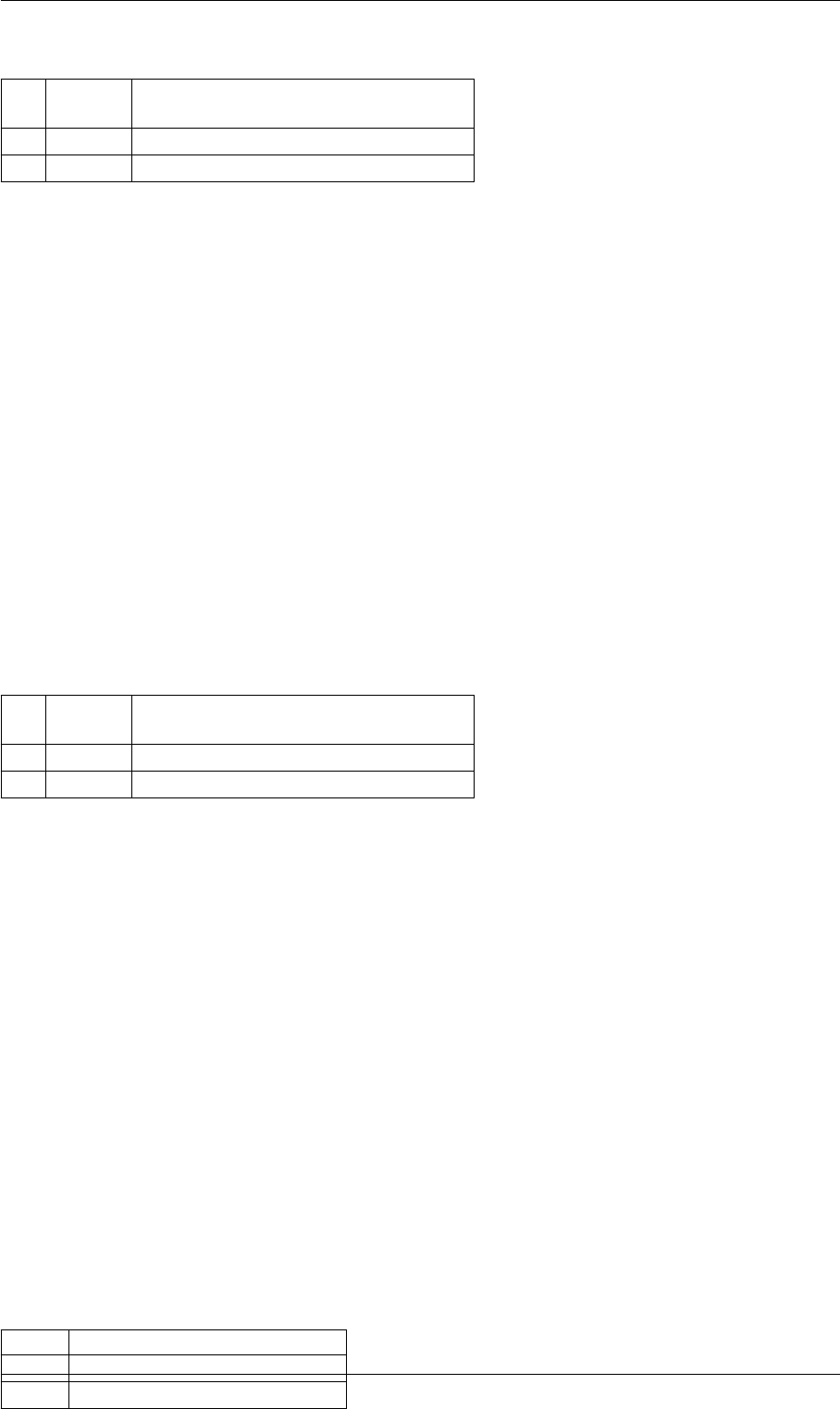
9.1.4.5 amici_dgemm()
void amici_dgemm (
BLASLayout layout,
BLASTranspose TransA,
BLASTranspose TransB,
const int M,
const int N,
const int K,
const double alpha,
const double ∗A,
const int lda,
const double ∗B,
const int ldb,
const double beta,
double ∗C,
const int ldc )
amici_dgemm provides an interface to the blas matrix matrix multiplication routine dgemm. This routines computes
C = alpha∗A∗B + beta∗C with A: [MxK] B:[KxN] C:[MxN]
Parameters
in layout can be AMICI_BLAS_ColMajor or AMICI_BLAS_RowMajor.
in TransA flag indicating whether A should be transposed before multiplication
in TransB flag indicating whether B should be transposed before multiplication
in Mnumber of rows in A/C
in Nnumber of columns in B/C
in Knumber of rows in B, number of columns in A
in alpha coefficient alpha
in Amatrix A
in lda leading dimension of A (m or k)
in Bmatrix B
in ldb leading dimension of B (k or n)
in beta coefficient beta
in,out Cmatrix C
in ldc leading dimension of C (m or n)
amici_dgemm provides an interface to the CBlas matrix matrix multiplication routine dgemm. This routines computes
C = alpha∗A∗B + beta∗C with A: [MxK] B:[KxN] C:[MxN]
Parameters
layout memory layout.
TransA flag indicating whether A should be transposed before multiplication
TransB flag indicating whether B should be transposed before multiplication
Mnumber of rows in A/C
Nnumber of columns in B/C
Knumber of rows in B, number of columns in A
alpha coefficient alpha
Amatrix A
lda leading dimension of A (m or k)
Bmatrix B
Generated by Doxygen
9.1 amici Namespace Reference 27
Parameters
ldb leading dimension of B (k or n)
beta coefficient beta
Cmatrix C
ldc leading dimension of C (m or n)
Returns
void
Definition at line 44 of file cblas.cpp.
Here is the caller graph for this function:
amici_dgemm
amici::Model::fsy
amici::Model::fsJy
amici::Model::fdJydp
amici::Model::fdJydx
amici::Model::fsJz
amici::Model::fdJzdp
amici::Model::fdJzdx
9.1.4.6 amici_daxpy()
void amici_daxpy (
int n,
double alpha,
const double ∗x,
const int incx,
double ∗y,
int incy )
Generated by Doxygen
28 CONTENTS
Parameters
nnumber of elements in y
alpha scalar coefficient of x
xvector of length n∗incx
incx x stride
yvector of length n∗incy
incy y stride
Definition at line 90 of file cblas.cpp.
9.1.4.7 setModelData()
void setModelData (
const mxArray ∗prhs[ ],
int nrhs,
Model &model )
Parameters
in prhs pointer to the array of input arguments
Type: mxArray
in nrhs number of elements in prhs
in,out model model to update
Definition at line 389 of file interface_matlab.cpp.
Here is the call graph for this function:
setModelData dbl2int
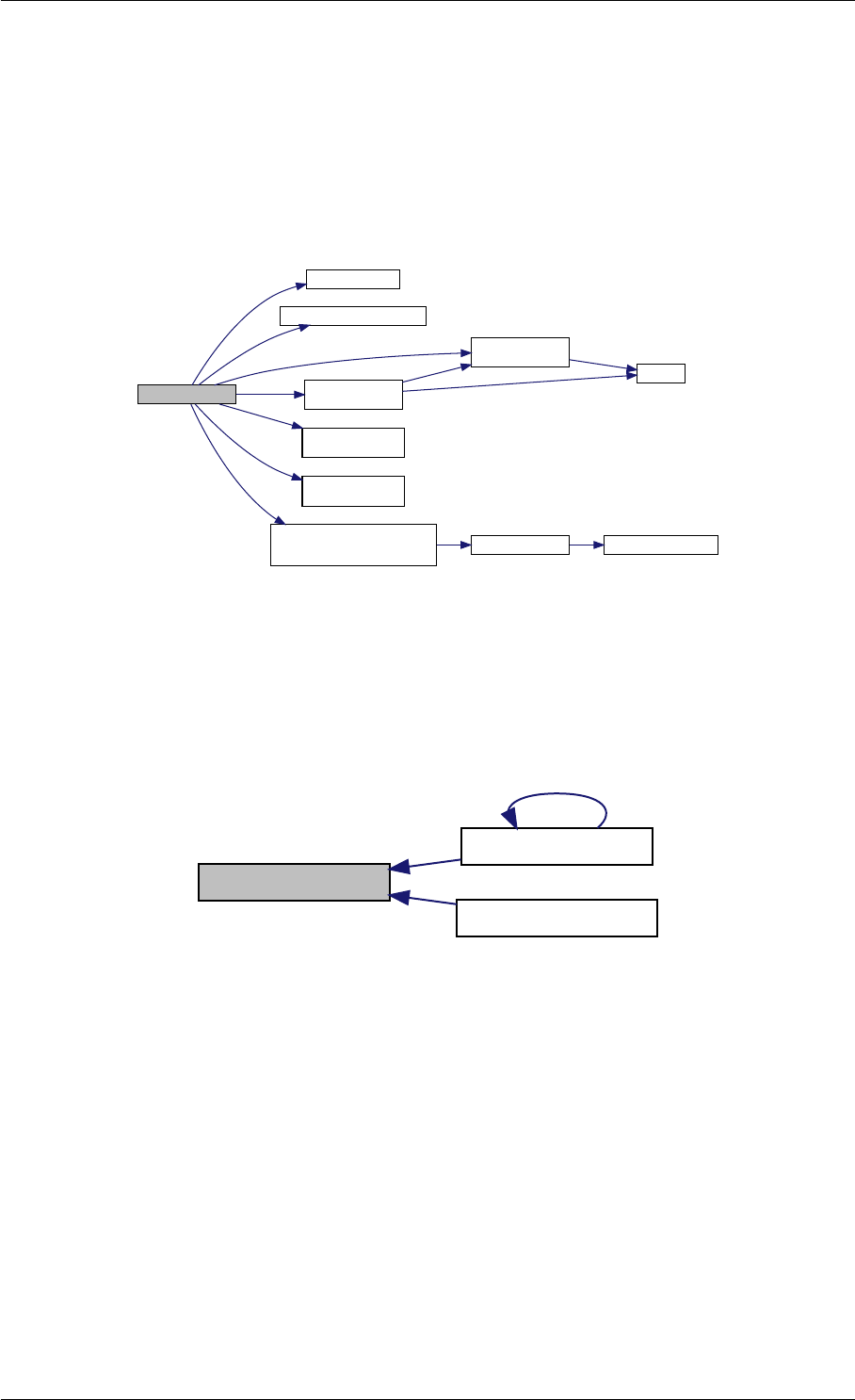
9.1.4.8 setSolverOptions()
void setSolverOptions (
const mxArray ∗prhs[ ],
int nrhs,
Solver &solver )
Generated by Doxygen
9.1 amici Namespace Reference 29
Parameters
in prhs pointer to the array of input arguments
Type: mxArray
in nrhs number of elements in prhs
in,out solver solver to update
Definition at line 304 of file interface_matlab.cpp.
Here is the call graph for this function:
setSolverOptions
amici::Solver::setAbsolute
Tolerance
amici::Solver::setRelative
Tolerance
amici::Solver::setAbsolute
ToleranceQuadratures
amici::Solver::setRelative
ToleranceQuadratures
amici::Solver::setMaxSteps
dbl2int
amici::Solver::setMaxSteps
BackwardProblem
amici::Solver::setLinear
MultistepMethod
amici::Solver::setNonlinear
SolverIteration
amici::Solver::setInterpolationType
amici::Solver::setLinear
Solver
amici::Solver::setSensitivity
Order
amici::Solver::setInternal
SensitivityMethod
amici::Solver::setSensitivity
Method
amici::Solver::setStateOrdering
amici::Solver::setStability
LimitFlag
amici::Solver::setNewton
Preequilibration
amici::Solver::setNewton
MaxSteps
amici::Solver::setNewton
MaxLinearSteps
amici::Solver::getMallocDone
amici::Solver::applyTolerances
amici::Solver::applySensitivity
Tolerances
amici::Solver::setSStolerances
amici::Solver::setMaxNum
Steps
amici::Solver::setMaxNum
StepsB
amici::Solver::setStabLimDet
amici::Solver::setStabLimDetB
Generated by Doxygen
30 CONTENTS
9.1.4.9 setupReturnData()
ReturnDataMatlab∗amici::setupReturnData (
mxArray ∗plhs[ ],
int nlhs )
setupReturnData initialises the return data struct
Parameters
in plhs user input
Type: mxArray
in nlhs number of elements in plhs
Type: mxArray
Returns
rdata: return data struct
Type:∗ReturnData
9.1.4.10 expDataFromMatlabCall()
std::unique_ptr<ExpData >expDataFromMatlabCall (
const mxArray ∗prhs[ ],
const Model &model )
expDataFromMatlabCall initialises the experimental data struct
Parameters
in prhs user input
Type:∗mxArray
Returns
edata: experimental data struct
Type:∗ExpData
expDataFromMatlabCall parses the experimental data from the matlab call and writes it to an ExpData class object
Parameters
prhs pointer to the array of input arguments
model pointer to the model object, this is necessary to perform dimension checks
Returns
edata pointer to experimental data object
Generated by Doxygen
9.1 amici Namespace Reference 31
Definition at line 179 of file interface_matlab.cpp.
Here is the call graph for this function:
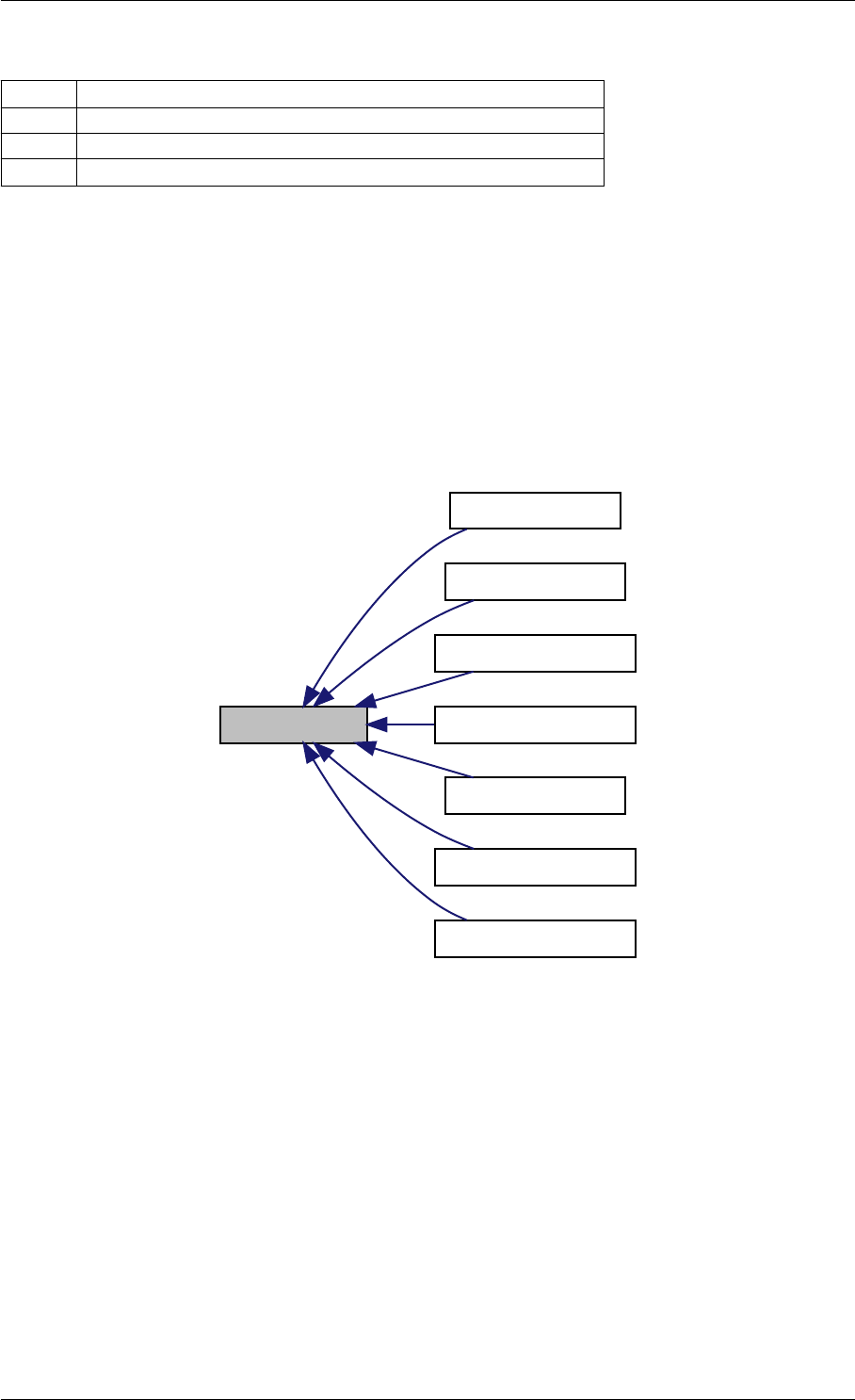
expDataFromMatlabCall
ExpData
mxArrayToVector
Here is the caller graph for this function:
expDataFromMatlabCall mexFunction
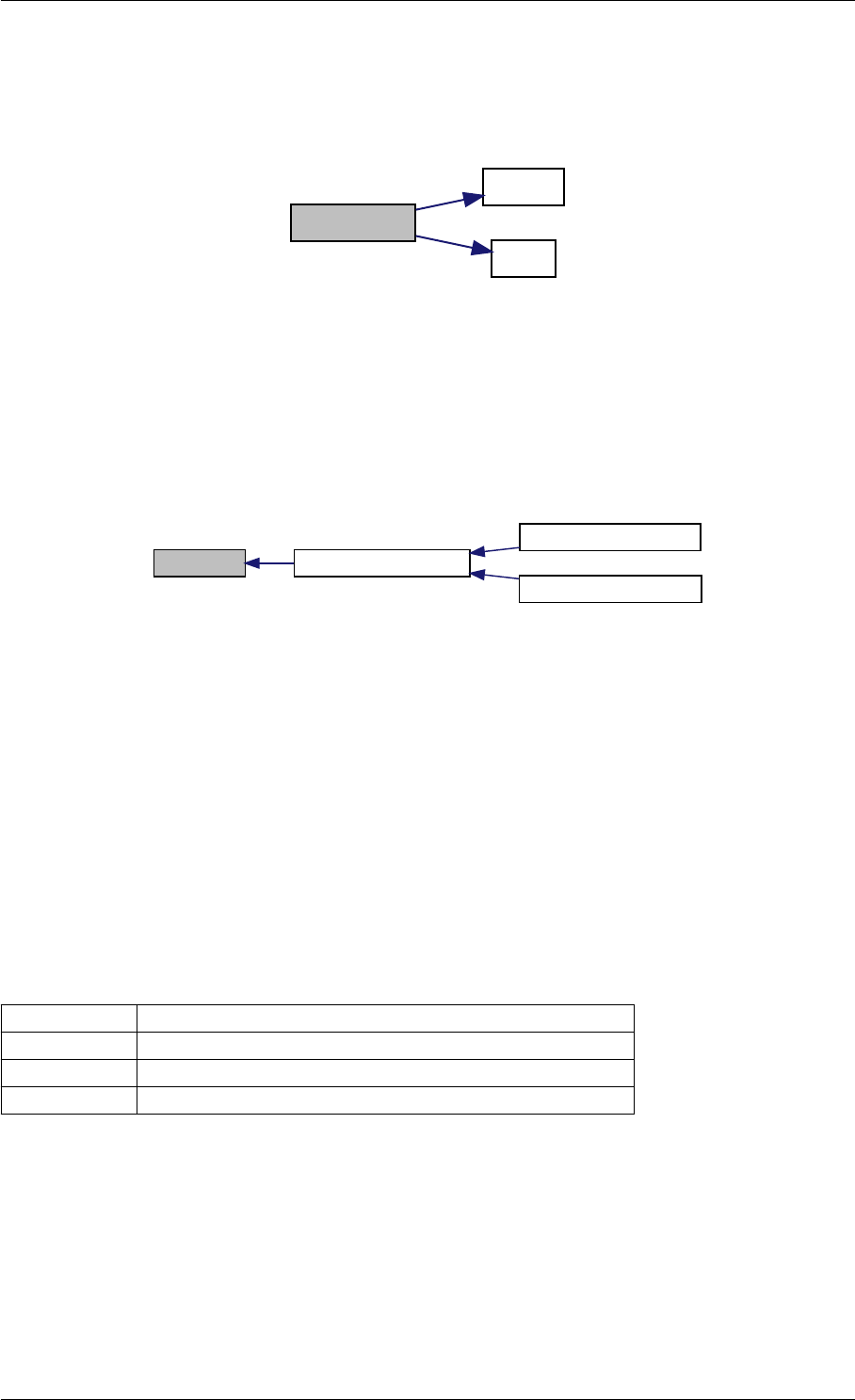
9.1.4.11 checkFinite()
int checkFinite (
const int N,
const realtype ∗array,
const char ∗fun )
Checks the values in an array for NaNs and Infs
Parameters
Nnumber of elements in array
array array
fun name of calling function
Returns
AMICI_RECOVERABLE_ERROR if a NaN/Inf value was found, AMICI_SUCCESS otherwise
Definition at line 17 of file misc.cpp.
Generated by Doxygen
32 CONTENTS
Here is the call graph for this function:
checkFinite
isNaN
isInf
Here is the caller graph for this function:
checkFinite amici::Model::checkFinite
amici::Model_DAE::fJDiag
amici::Model_ODE::fJDiag
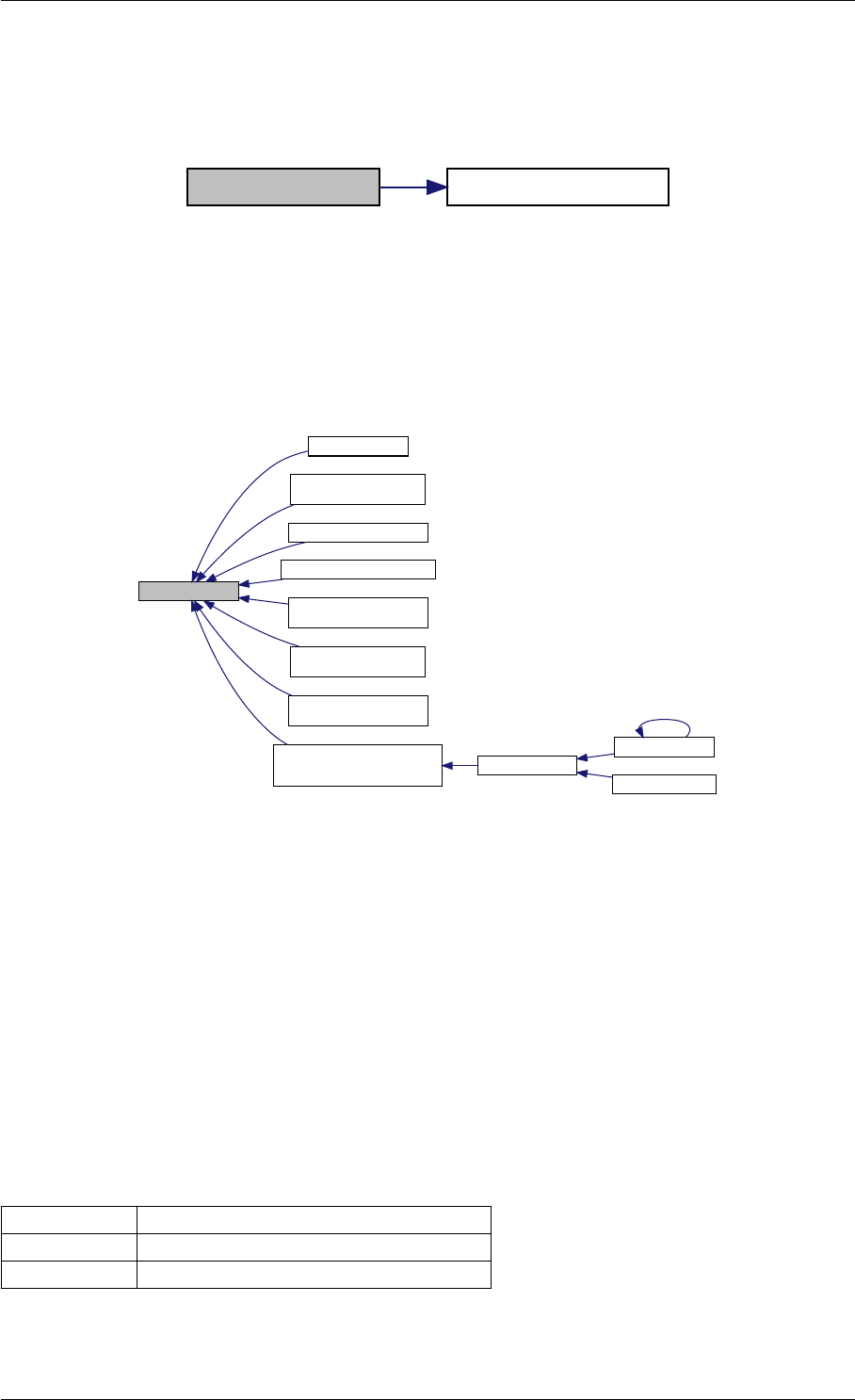
9.1.4.12 unscaleParameters() [1/2]
void unscaleParameters (
const double ∗bufferScaled,
const ParameterScaling ∗pscale,
int n,
double ∗bufferUnscaled )
Parameters
bufferScaled scaled parameters
pscale parameter scaling
nnumber of elements in bufferScaled, pscale and bufferUnscaled
bufferUnscaled unscaled parameters are written to the array
Returns
status flag indicating success of execution
Type: int
Definition at line 44 of file misc.cpp.
Generated by Doxygen
9.1 amici Namespace Reference 33
Here is the call graph for this function:
unscaleParameters getUnscaledParameter
Here is the caller graph for this function:
unscaleParameters
unscaleParameters
amici::Model::setParameter
Scale
amici::Model::setParameters
amici::Model::setParameterById
amici::Model::setParameters
ByIdRegex
amici::Model::setParameter
ByName
amici::Model::setParameters
ByNameRegex
amici::ReturnData::
applyChainRuleFactorToSimulation
Results
runAmiciSimulation
runAmiciSimulation
runAmiciSimulations
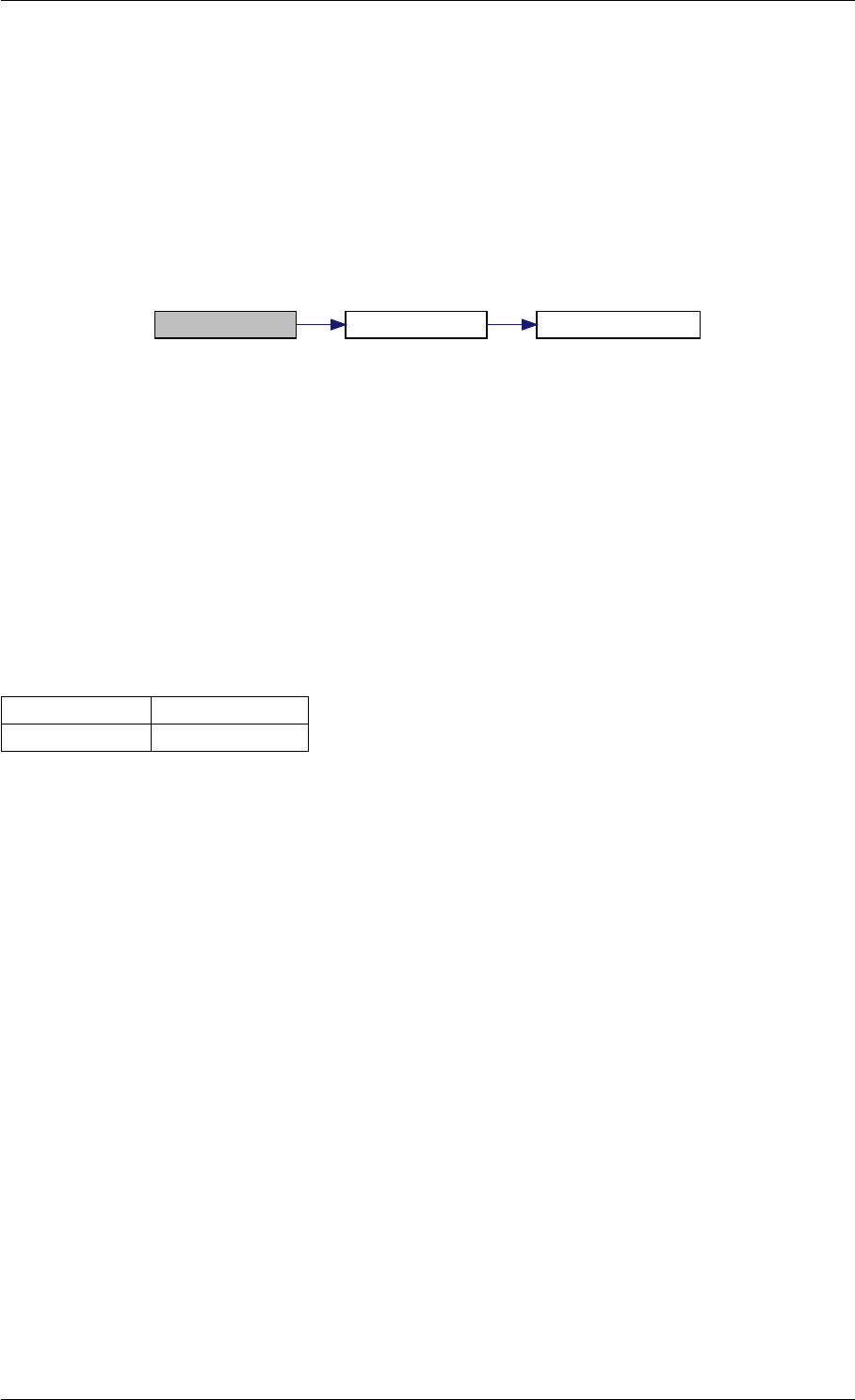
9.1.4.13 unscaleParameters() [2/2]
void unscaleParameters (
std::vector<double >const & bufferScaled,
std::vector<ParameterScaling >const & pscale,
std::vector<double >&bufferUnscaled )
All vectors must be of same length
Parameters
bufferScaled scaled parameters
pscale parameter scaling
bufferUnscaled unscaled parameters are written to the array
Generated by Doxygen
34 CONTENTS
Returns
status flag indicating success of execution
Type: int
Definition at line 52 of file misc.cpp.
Here is the call graph for this function:
unscaleParameters unscaleParameters getUnscaledParameter
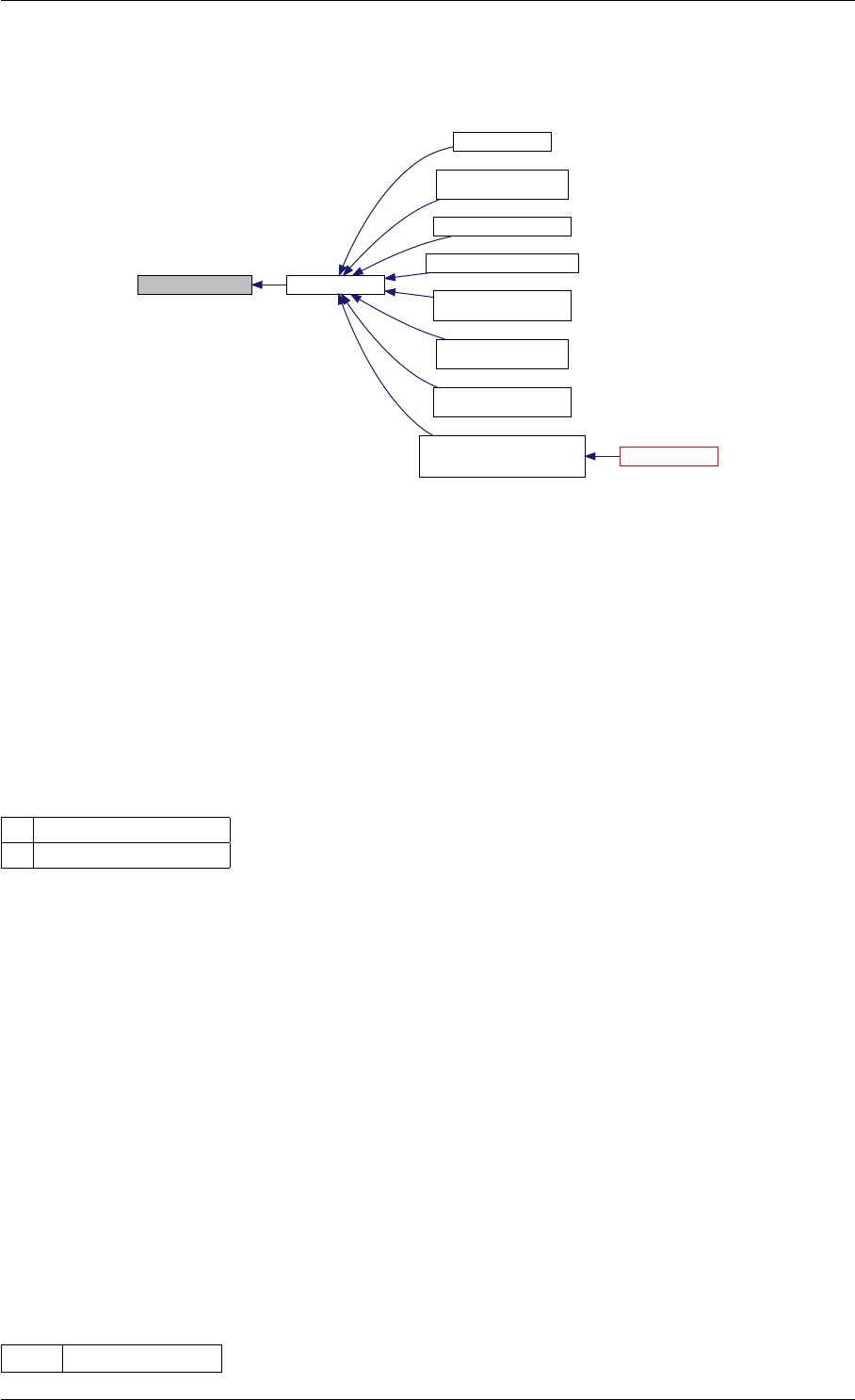
9.1.4.14 getUnscaledParameter()
double getUnscaledParameter (
double scaledParameter,
ParameterScaling scaling )
Parameters
scaledParameter scaled parameter
scaling parameter scaling
Returns
Unscaled parameter
Definition at line 32 of file misc.cpp.
Generated by Doxygen
9.1 amici Namespace Reference 35
Here is the caller graph for this function:
getUnscaledParameter unscaleParameters
unscaleParameters
amici::Model::setParameter
Scale
amici::Model::setParameters
amici::Model::setParameterById
amici::Model::setParameters
ByIdRegex
amici::Model::setParameter
ByName
amici::Model::setParameters
ByNameRegex
amici::ReturnData::
applyChainRuleFactorToSimulation
Results
runAmiciSimulation
9.1.4.15 operator==() [1/2]
bool operator== (
const Model &a,
const Model &b)
Parameters
afirst model instance
bsecond model instance
Returns
equality
Definition at line 1312 of file model.cpp.
9.1.4.16 getReturnDataMatlabFromAmiciCall()
mxArray ∗getReturnDataMatlabFromAmiciCall (
ReturnData const ∗rdata )
generates matlab mxArray from a ReturnData object
Parameters
rdata ReturnDataObject
Generated by Doxygen
36 CONTENTS
Returns
rdatamatlab ReturnDataObject stored as matlab compatible data
generates matlab mxArray from a ReturnData object
Parameters
rdata ReturnDataObject
Returns
rdatamatlab ReturnDataObject stored as matlab compatible data
Definition at line 7 of file returndata_matlab.cpp.
Here is the call graph for this function:
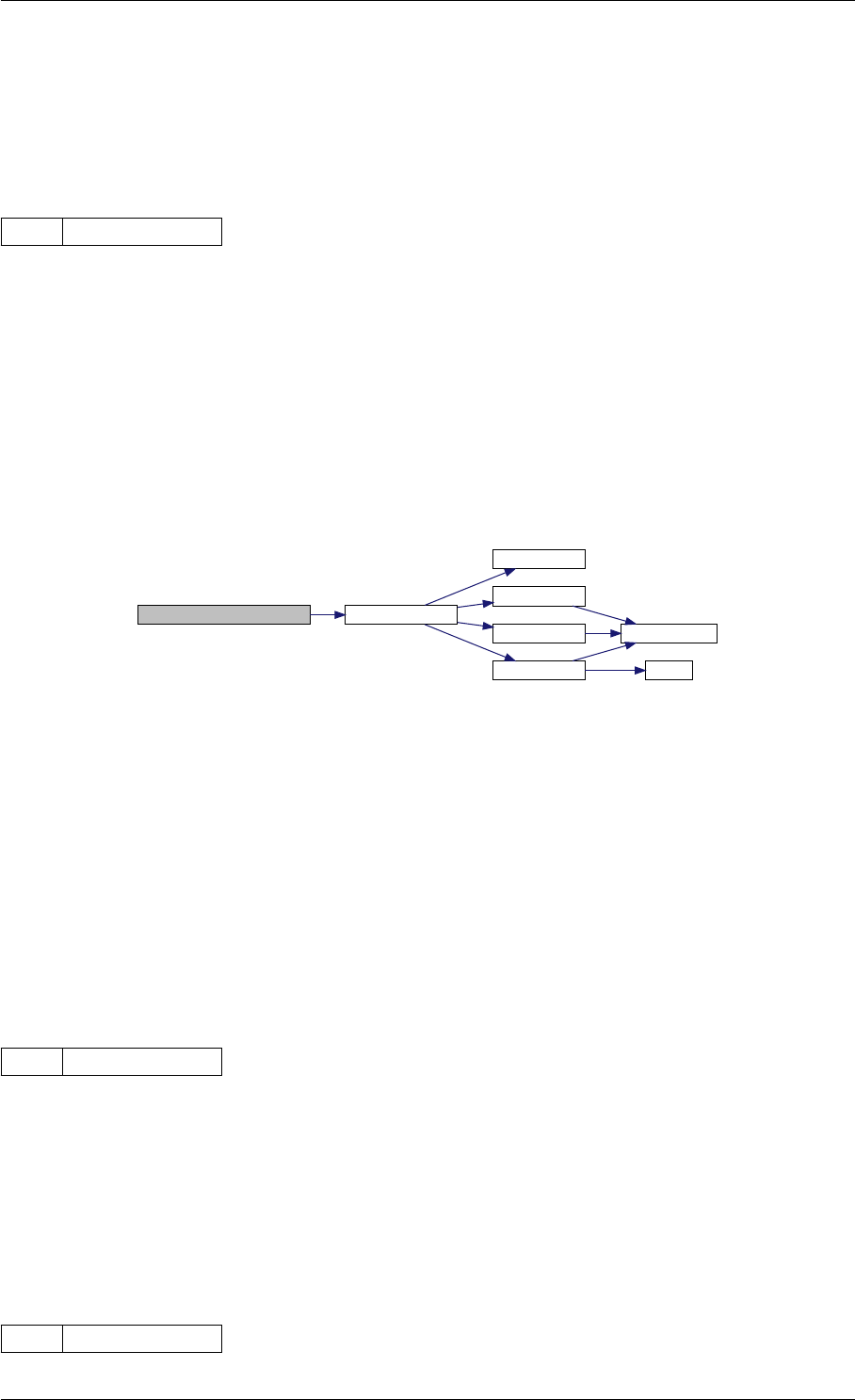
getReturnDataMatlabFromAmiciCall initMatlabReturnFields
checkFieldNames
writeMatlabField0
writeMatlabField1
writeMatlabField2
initAndAttachArray
reorder
9.1.4.17 initMatlabReturnFields()
mxArray ∗initMatlabReturnFields (
ReturnData const ∗rdata )
allocates and initialises solution mxArray with the corresponding fields
Parameters
rdata ReturnDataObject
Returns
Solution mxArray
allocates and initialises solution mxArray with the corresponding fields
Parameters
rdata ReturnDataObject
Generated by Doxygen
9.1 amici Namespace Reference 37
Returns
Solution mxArray
Definition at line 18 of file returndata_matlab.cpp.
Here is the call graph for this function:
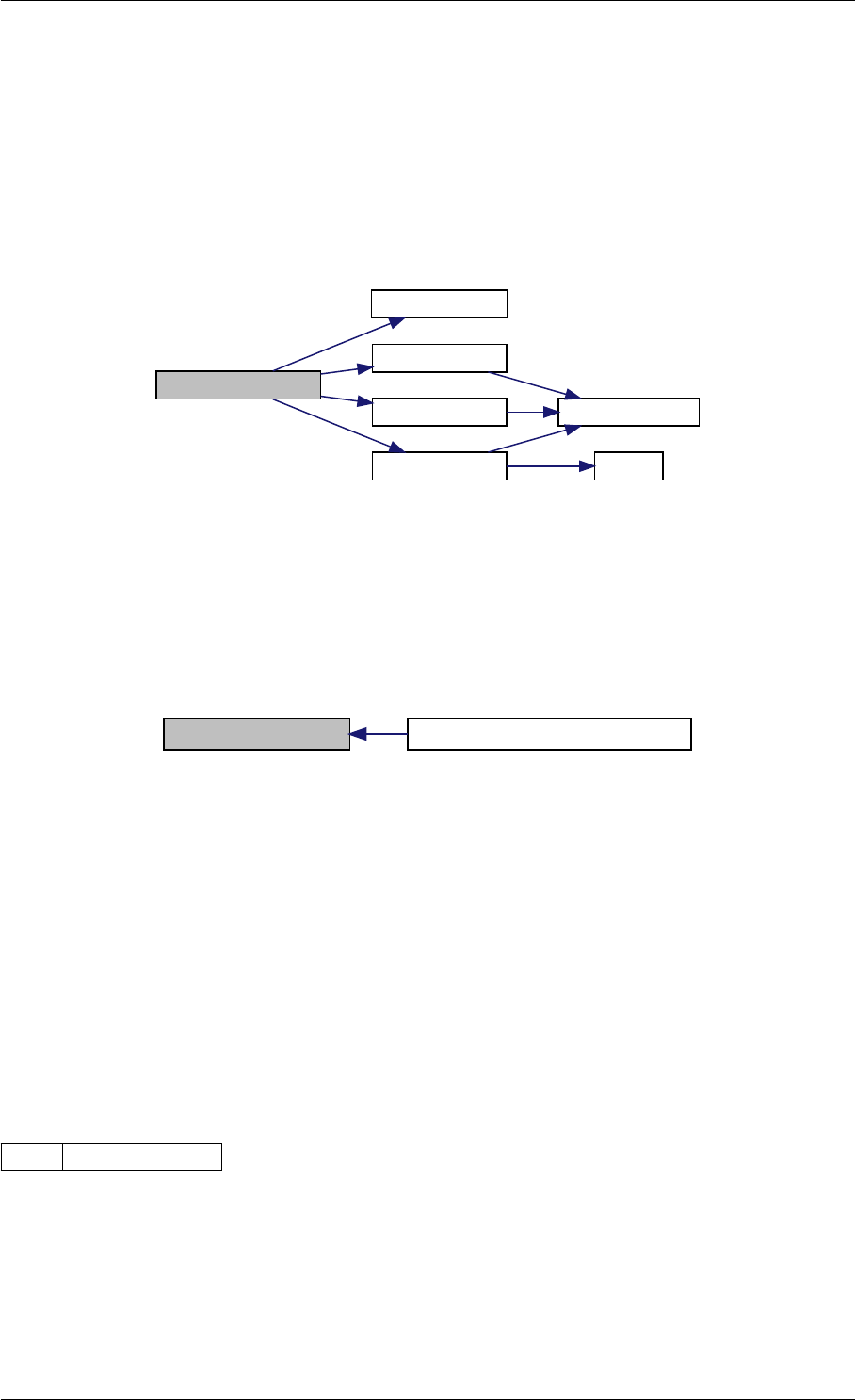
initMatlabReturnFields
checkFieldNames
writeMatlabField0
writeMatlabField1
writeMatlabField2
initAndAttachArray
reorder
Here is the caller graph for this function:
initMatlabReturnFields getReturnDataMatlabFromAmiciCall
9.1.4.18 initMatlabDiagnosisFields()
mxArray ∗initMatlabDiagnosisFields (
ReturnData const ∗rdata )
allocates and initialises diagnosis mxArray with the corresponding fields
Parameters
rdata ReturnDataObject
Returns
Diagnosis mxArray
allocates and initialises diagnosis mxArray with the corresponding fields
Generated by Doxygen
38 CONTENTS
Parameters
rdata ReturnDataObject
Returns
Diagnosis mxArray
Definition at line 116 of file returndata_matlab.cpp.
Here is the call graph for this function:
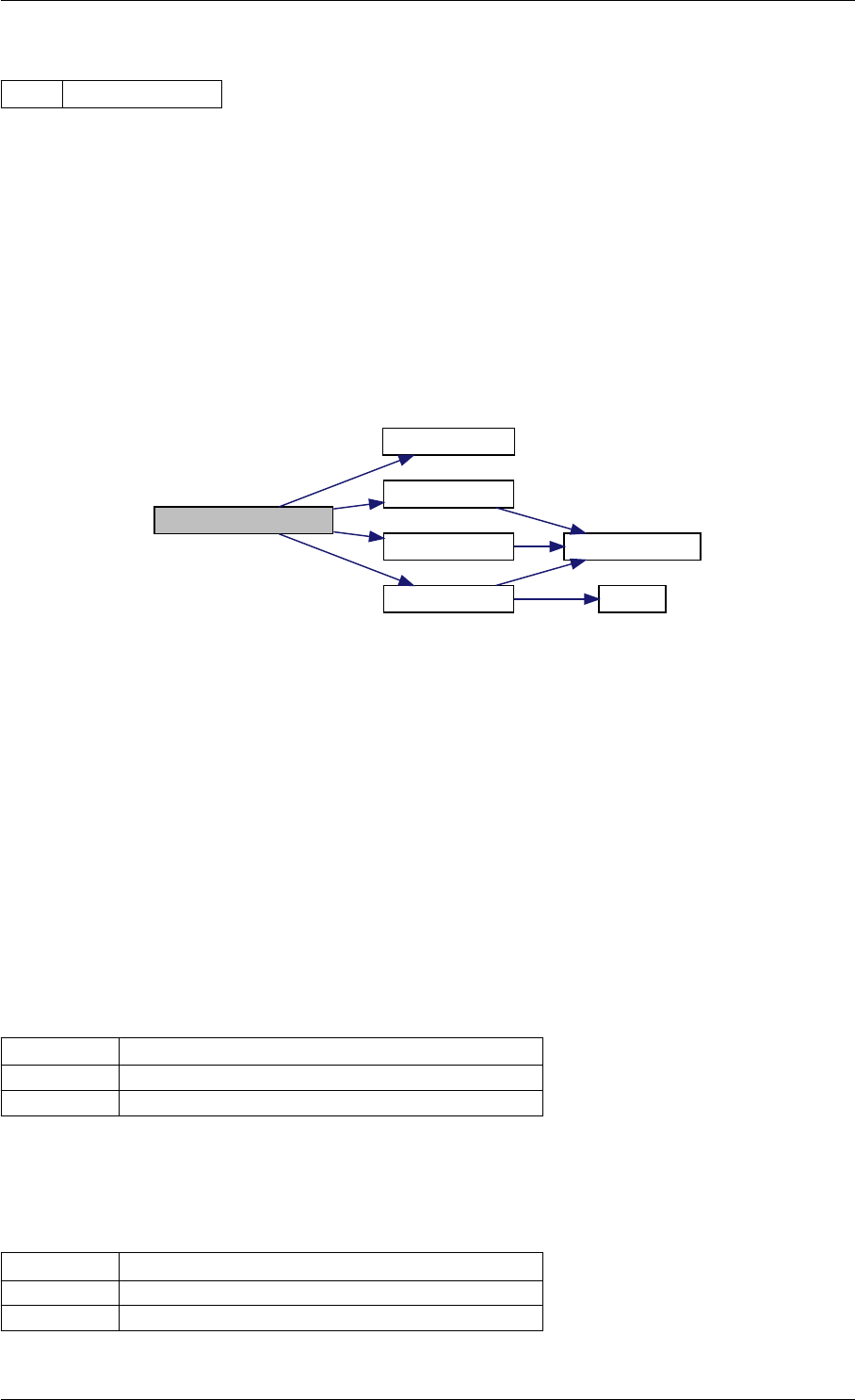
initMatlabDiagnosisFields
checkFieldNames
writeMatlabField1
writeMatlabField2
writeMatlabField0 initAndAttachArray
reorder
9.1.4.19 writeMatlabField0()
void writeMatlabField0 (
mxArray ∗matlabStruct,
const char ∗fieldName,
TfieldData )
initialise vector and attach to the field
Parameters
matlabStruct pointer of the field to which the vector will be attached
fieldName Name of the field to which the vector will be attached
fieldData Data wich will be stored in the field
initialise vector and attach to the field
Parameters
matlabStruct pointer of the field to which the vector will be attached
fieldName Name of the field to which the vector will be attached
fieldData Data wich will be stored in the field
Generated by Doxygen
9.1 amici Namespace Reference 39
Definition at line 181 of file returndata_matlab.cpp.
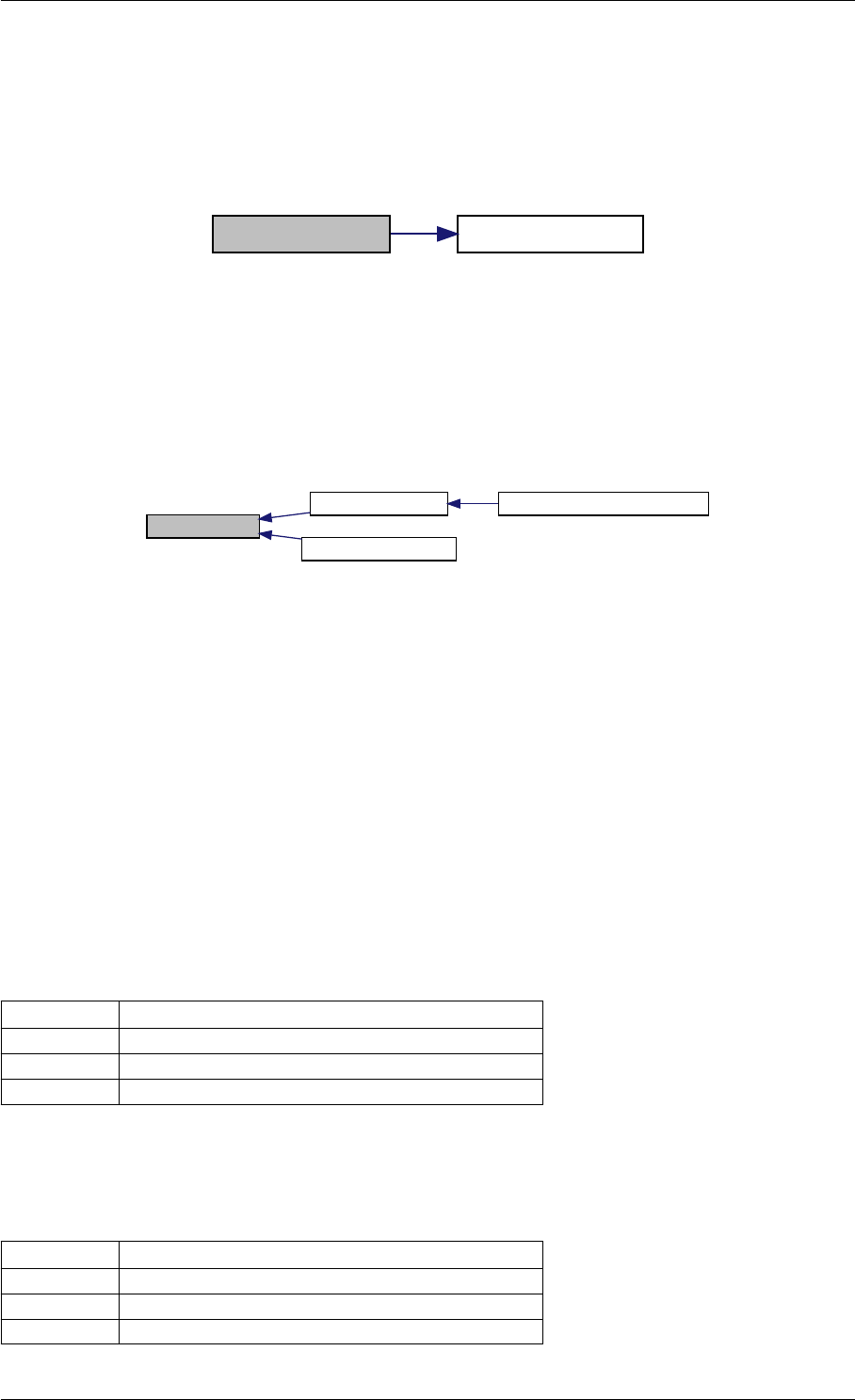
Here is the call graph for this function:
writeMatlabField0 initAndAttachArray
Here is the caller graph for this function:
writeMatlabField0
initMatlabReturnFields
initMatlabDiagnosisFields
getReturnDataMatlabFromAmiciCall
9.1.4.20 writeMatlabField1()
void writeMatlabField1 (
mxArray ∗matlabStruct,
const char ∗fieldName,
std::vector<T>fieldData,
const int dim0 )
initialise vector and attach to the field
Parameters
matlabStruct pointer of the field to which the vector will be attached
fieldName Name of the field to which the vector will be attached
fieldData Data wich will be stored in the field
dim0 Number of elements in the vector
initialise vector and attach to the field
Parameters
matlabStruct pointer of the field to which the vector will be attached
fieldName Name of the field to which the vector will be attached
fieldData Data wich will be stored in the field
dim0 Number of elements in the vector
Generated by Doxygen
40 CONTENTS
Definition at line 199 of file returndata_matlab.cpp.
Here is the call graph for this function:
writeMatlabField1 initAndAttachArray
Here is the caller graph for this function:
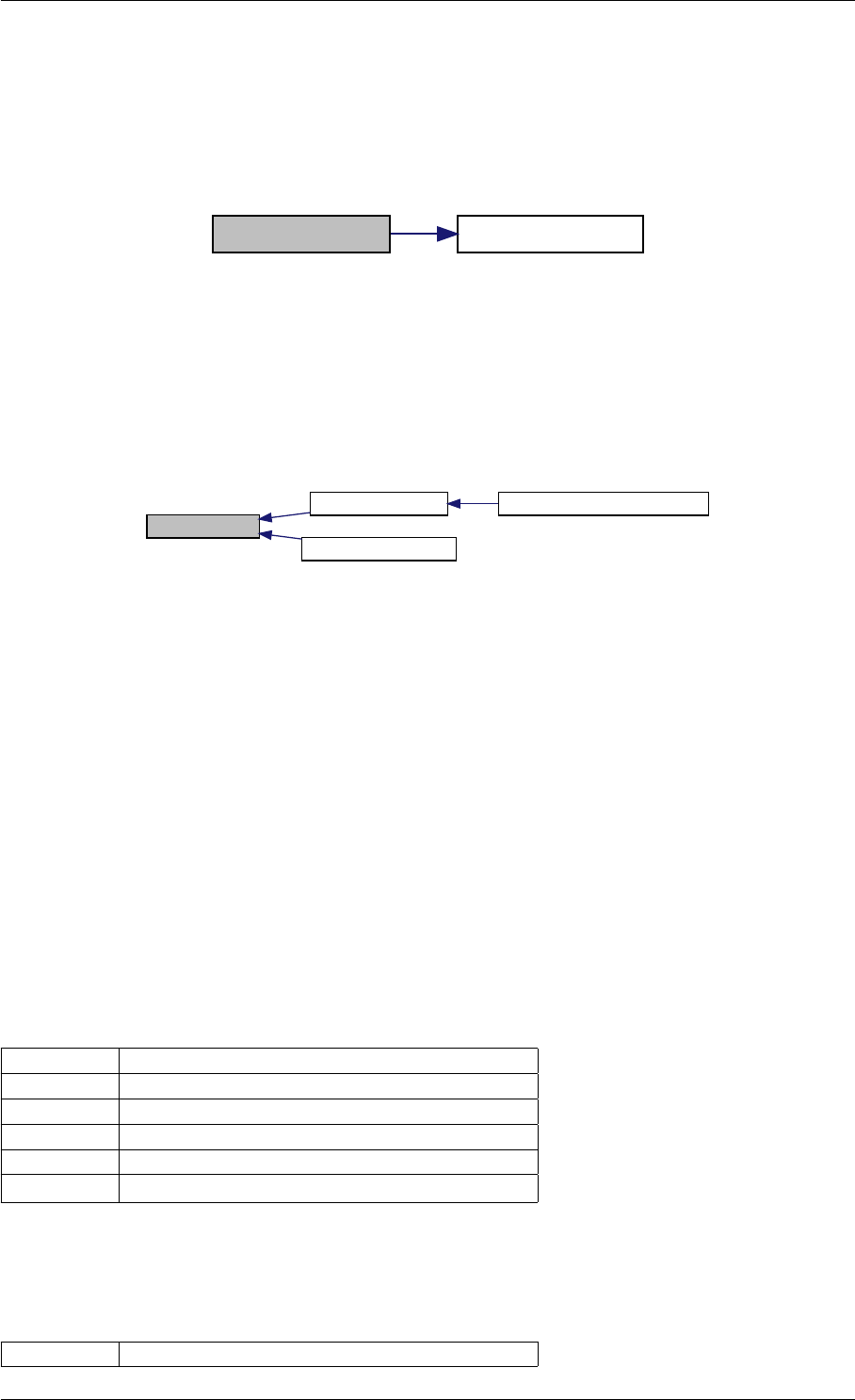
writeMatlabField1
initMatlabReturnFields
initMatlabDiagnosisFields
getReturnDataMatlabFromAmiciCall
9.1.4.21 writeMatlabField2()
void writeMatlabField2 (
mxArray ∗matlabStruct,
const char ∗fieldName,
std::vector<T>fieldData,
int dim0,
int dim1,
std::vector<int >perm )
initialise matrix, attach to the field and write data
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
fieldData Data wich will be stored in the field
dim0 Number of rows in the tensor
dim1 Number of columns in the tensor
perm reordering of dimensions (i.e., transposition)
initialise matrix, attach to the field and write data
Parameters
matlabStruct Pointer to the matlab structure
Generated by Doxygen
9.1 amici Namespace Reference 41
Parameters
fieldName Name of the field to which the tensor will be attached
fieldData Data wich will be stored in the field
dim0 Number of rows in the tensor
dim1 Number of columns in the tensor
perm reordering of dimensions (i.e., transposition)
Definition at line 221 of file returndata_matlab.cpp.
Here is the call graph for this function:
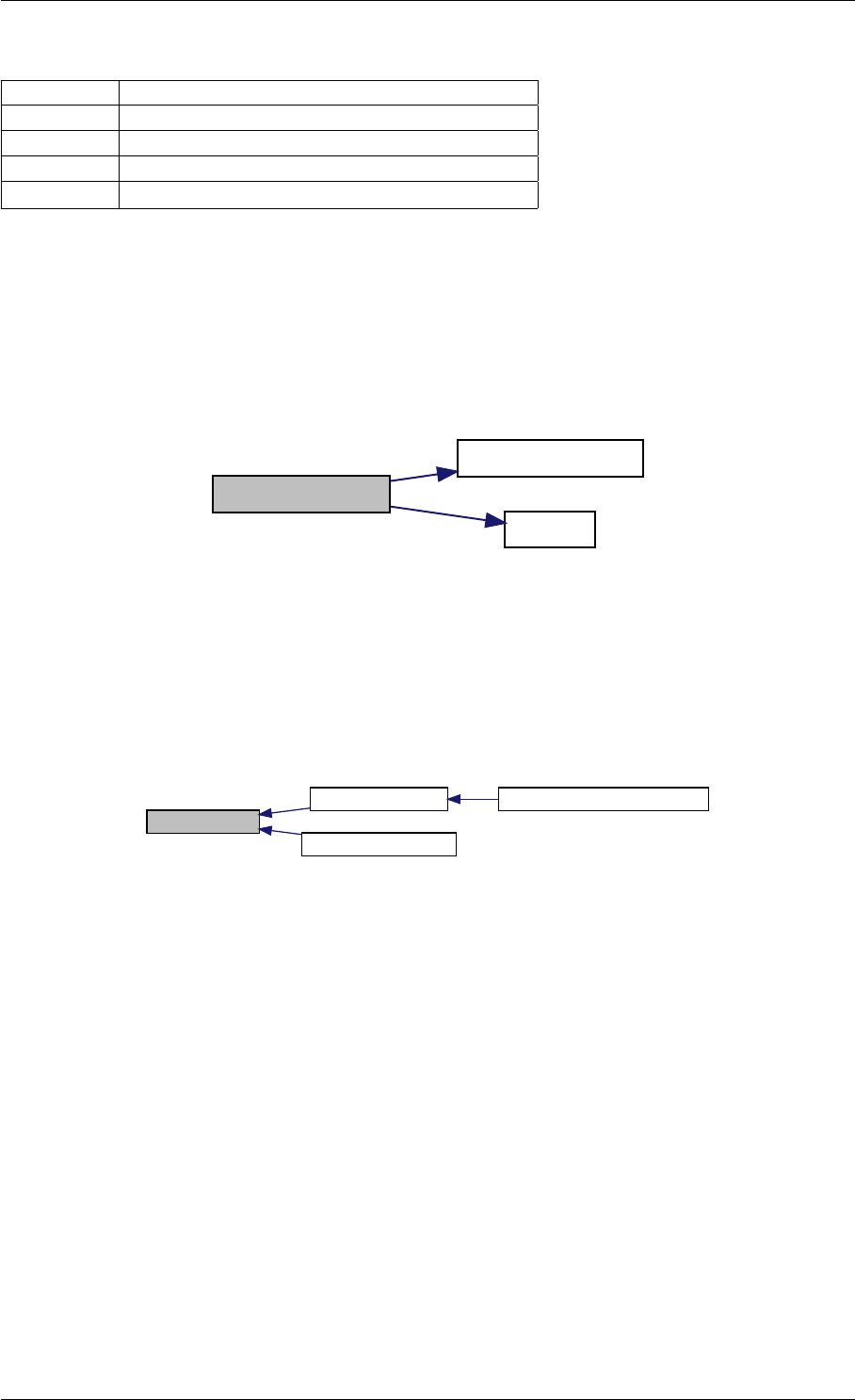
writeMatlabField2
initAndAttachArray
reorder
Here is the caller graph for this function:
writeMatlabField2
initMatlabReturnFields
initMatlabDiagnosisFields
getReturnDataMatlabFromAmiciCall
9.1.4.22 writeMatlabField3()
void writeMatlabField3 (
mxArray ∗matlabStruct,
const char ∗fieldName,
std::vector<T>fieldData,
int dim0,
int dim1,
int dim2,
std::vector<int >perm )
initialise 3D tensor, attach to the field and write data
Generated by Doxygen
42 CONTENTS
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
fieldData Data wich will be stored in the field
dim0 number of rows in the tensor
dim1 number of columns in the tensor
dim2 number of elements in the third dimension of the tensor
perm reordering of dimensions
initialise 3D tensor, attach to the field and write data
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
fieldData Data wich will be stored in the field
dim0 number of rows in the tensor
dim1 number of columns in the tensor
dim2 number of elements in the third dimension of the tensor
perm reordering of dimensions
Definition at line 254 of file returndata_matlab.cpp.
Here is the call graph for this function:
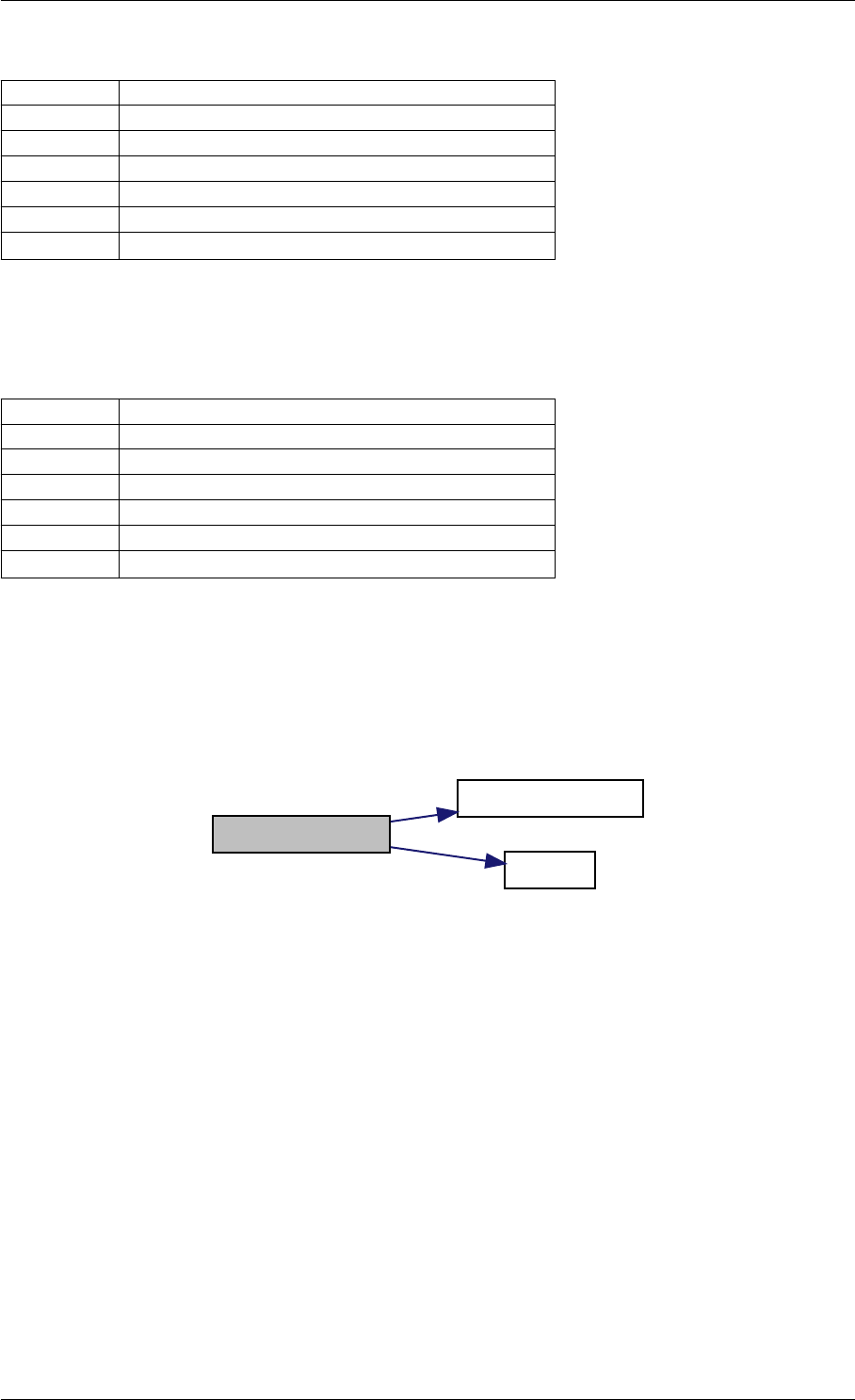
writeMatlabField3
initAndAttachArray
reorder
9.1.4.23 writeMatlabField4()
void writeMatlabField4 (
mxArray ∗matlabStruct,
const char ∗fieldName,
std::vector<T>fieldData,
int dim0,
int dim1,
int dim2,
int dim3,
std::vector<int >perm )
initialise 4D tensor, attach to the field and write data
Generated by Doxygen
9.1 amici Namespace Reference 43
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
fieldData Data wich will be stored in the field
dim0 number of rows in the tensor
dim1 number of columns in the tensor
dim2 number of elements in the third dimension of the tensor
dim3 number of elements in the fourth dimension of the tensor
perm reordering of dimensions
initialise 4D tensor, attach to the field and write data
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
fieldData Data wich will be stored in the field
dim0 number of rows in the tensor
dim1 number of columns in the tensor
dim2 number of elements in the third dimension of the tensor
dim3 number of elements in the fourth dimension of the tensor
perm reordering of dimensions
Definition at line 290 of file returndata_matlab.cpp.
Here is the call graph for this function:
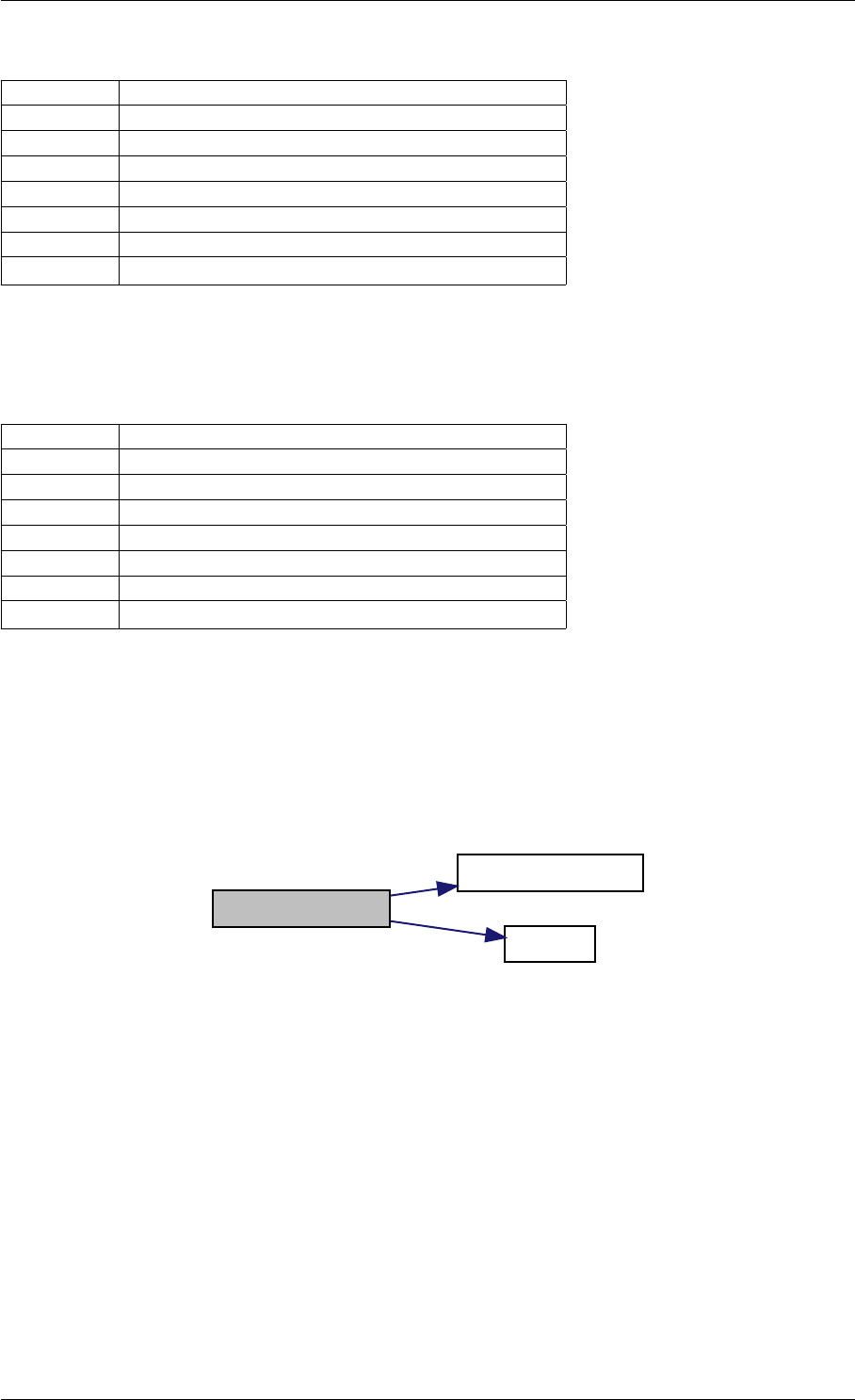
writeMatlabField4
initAndAttachArray
reorder
9.1.4.24 initAndAttachArray()
double ∗initAndAttachArray (
mxArray ∗matlabStruct,
const char ∗fieldName,
std::vector<mwSize >dim )
initialises the field fieldName in matlabStruct with dimension dim
Generated by Doxygen
44 CONTENTS
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
dim vector of field dimensions
Returns
Pointer to field data
initialises the field fieldName in matlabStruct with dimension dim
Parameters
matlabStruct Pointer to the matlab structure
fieldName Name of the field to which the tensor will be attached
dim vector of field dimensions
Returns
Pointer to field data
Definition at line 328 of file returndata_matlab.cpp.
Here is the caller graph for this function:
initAndAttachArray
writeMatlabField0
writeMatlabField1
writeMatlabField2
writeMatlabField3
writeMatlabField4
initMatlabReturnFields
initMatlabDiagnosisFields
getReturnDataMatlabFromAmiciCall
9.1.4.25 checkFieldNames()
void checkFieldNames (
const char ∗∗ fieldNames,
const int fieldCount )
checks whether fieldNames was properly allocated
Parameters
fieldNames array of field names
fieldCount expected number of fields in fieldNames
Generated by Doxygen
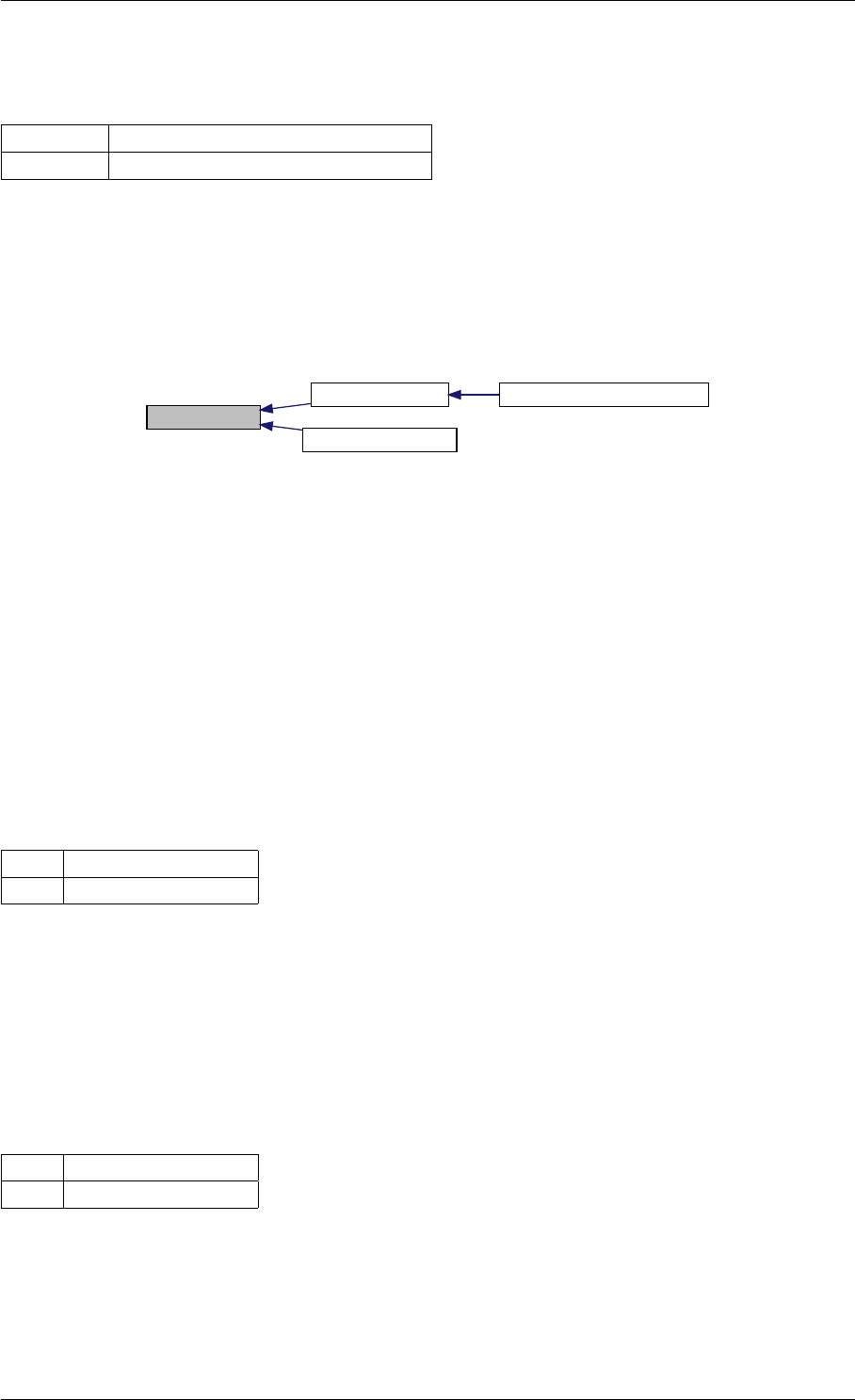
9.1 amici Namespace Reference 45
checks whether fieldNames was properly allocated
Parameters
fieldNames array of field names
fieldCount expected number of fields in fieldNames
Definition at line 349 of file returndata_matlab.cpp.
Here is the caller graph for this function:
checkFieldNames
initMatlabReturnFields
initMatlabDiagnosisFields
getReturnDataMatlabFromAmiciCall
9.1.4.26 reorder()
std::vector<T>reorder (
const std::vector<T>input,
const std::vector<int >order )
template function that reorders elements in a std::vector
Parameters
input unordered vector
order dimension permutation
Returns
Reordered vector
template function that reorders elements in a std::vector
Parameters
input unordered vector
order dimension permutation
Returns
Reordered vector
Definition at line 362 of file returndata_matlab.cpp.
Generated by Doxygen

46 CONTENTS
Here is the caller graph for this function:
reorder
writeMatlabField2
writeMatlabField3
writeMatlabField4
initMatlabReturnFields
initMatlabDiagnosisFields
getReturnDataMatlabFromAmiciCall
9.1.4.27 serializeToChar()
char∗amici::serializeToChar (
T const & data,
int ∗size )
Serialize object to char array
Parameters
data input object
size maximum char length
Returns
The object serialized as char
Definition at line 172 of file serialization.h.
9.1.4.28 deserializeFromChar()
T amici::deserializeFromChar (
const char ∗buffer,
int size )
Deserialize object that has been serialized using serializeToChar
Parameters
buffer serialized object
size length of buffer
Generated by Doxygen
9.1 amici Namespace Reference 47
Returns
The deserialized object
Definition at line 205 of file serialization.h.
9.1.4.29 serializeToString()
std::string amici::serializeToString (
T const & data )
Serialize object to string
Parameters
data input object
Returns
The object serialized as string
Definition at line 230 of file serialization.h.
9.1.4.30 serializeToStdVec()
std::vector<char>amici::serializeToStdVec (
T const & data )
Serialize object to std::vector<char>
Parameters
data input object
Returns
The object serialized as std::vector<char>
Definition at line 256 of file serialization.h.
9.1.4.31 deserializeFromString()
T amici::deserializeFromString (
std::string const & serialized )
Deserialize object that has been serialized using serializeToString
Generated by Doxygen
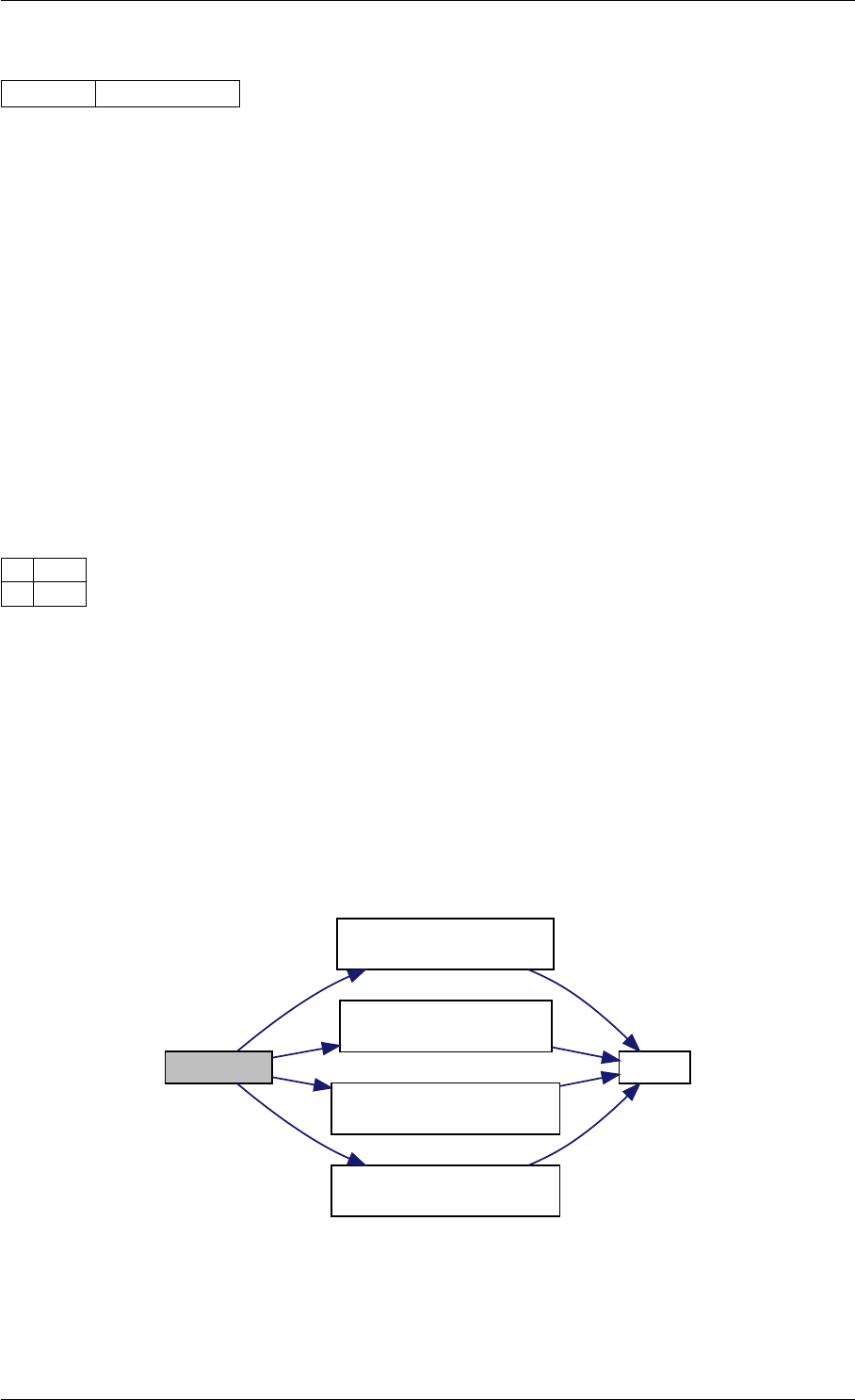
48 CONTENTS
Parameters
serialized serialized object
Returns
The deserialized object
Definition at line 283 of file serialization.h.
9.1.4.32 operator==() [2/2]
bool operator== (
const Solver &a,
const Solver &b)
Parameters
a
b
Returns
Definition at line 378 of file solver.cpp.
Here is the call graph for this function:
operator==
amici::Solver::getAbsolute
ToleranceSteadyState
amici::Solver::getRelative
ToleranceSteadyState
amici::Solver::getAbsolute
ToleranceSteadyStateSensi
amici::Solver::getRelative
ToleranceSteadyStateSensi
isNaN
Generated by Doxygen
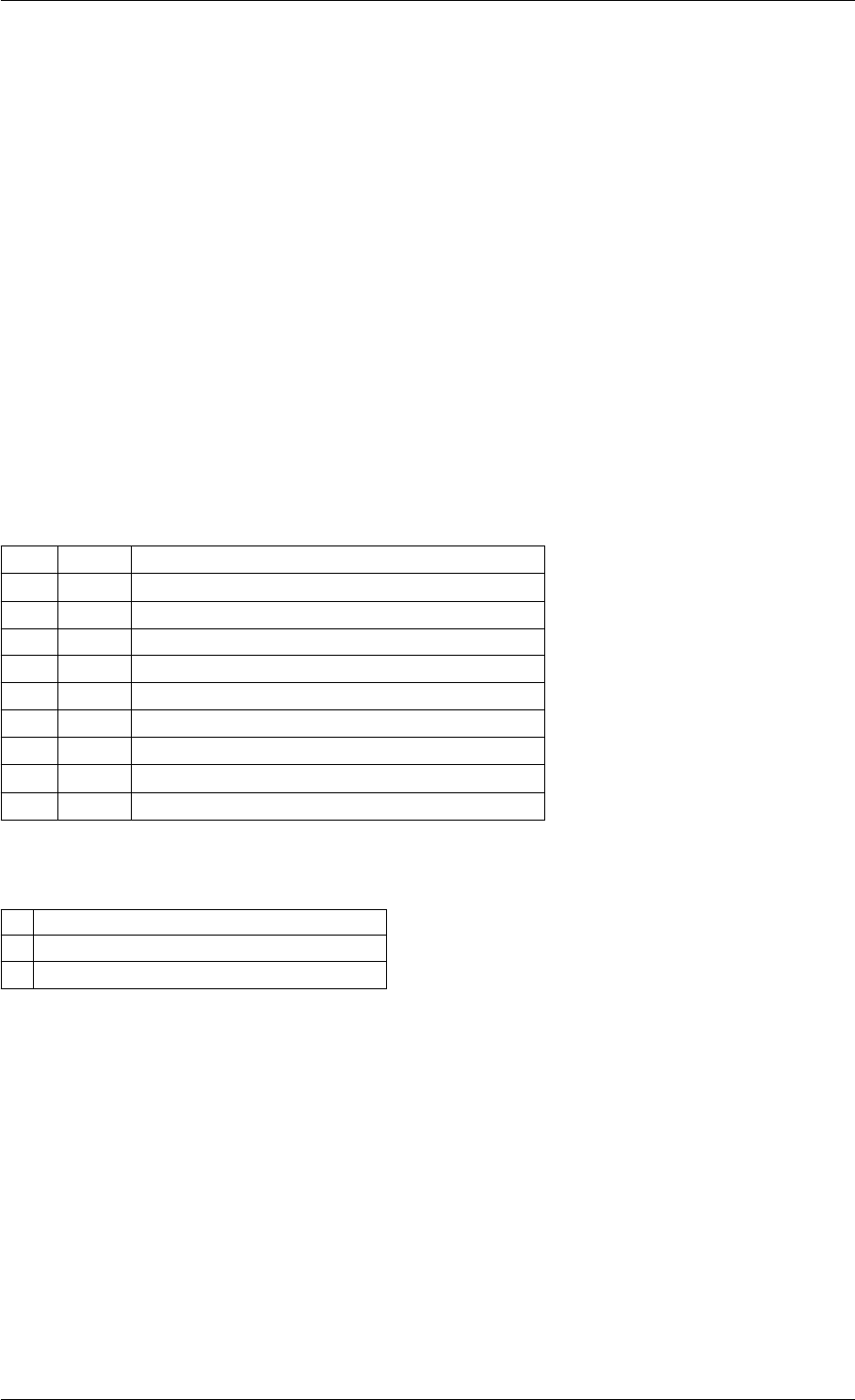
9.1 amici Namespace Reference 49
9.1.4.33 spline() [1/2]
int spline (
int n,
int end1,
int end2,
double slope1,
double slope2,
double x[ ],
double y[ ],
double b[ ],
double c[ ],
double d[ ] )
Evaluate the coefficients b[i], c[i], d[i], i = 0, 1, .. n-1 for a cubic interpolating spline
S(xx) = Y[i] + b[i] ∗w + c[i] ∗w∗∗2 + d[i] ∗w∗∗3 where w = xx - x[i] and x[i] <= xx <= x[i+1]
The n supplied data points are x[i], y[i], i = 0 ... n-1.
Parameters
in nThe number of data points or knots (n >= 2)
in end1 0: default condition 1: specify the slopes at x[0]
in end2 0: default condition 1: specify the slopes at x[n-1]
in slope1 slope at x[0]
in slope2 slope at x[n-1]
in x[ ] the abscissas of the knots in strictly increasing order
in y[ ] the ordinates of the knots
out b[ ] array of spline coefficients
out c[ ] array of spline coefficients
out d[ ] array of spline coefficients
Return values
0normal return
1less than two data points; cannot interpolate
2x[] are not in ascending order
Notes
• The accompanying function seval() may be used to evaluate the spline while deriv will provide the first deriva-
tive.
• Using p to denote differentiation y[i] = S(X[i]) b[i] = Sp(X[i]) c[i] = Spp(X[i])/2 d[i] = Sppp(X[i])/6 ( Derivative
from the right )
• Since the zero elements of the arrays ARE NOW used here, all arrays to be passed from the main program
should be dimensioned at least [n]. These routines will use elements [0 .. n-1].
• Adapted from the text Forsythe, G.E., Malcolm, M.A. and Moler, C.B. (1977) "Computer Methods for Mathe-
matical Computations" Prentice Hall
• Note that although there are only n-1 polynomial segments, n elements are requird in b, c, d. The elements
b[n-1], c[n-1] and d[n-1] are set to continue the last segment past x[n-1].
Generated by Doxygen
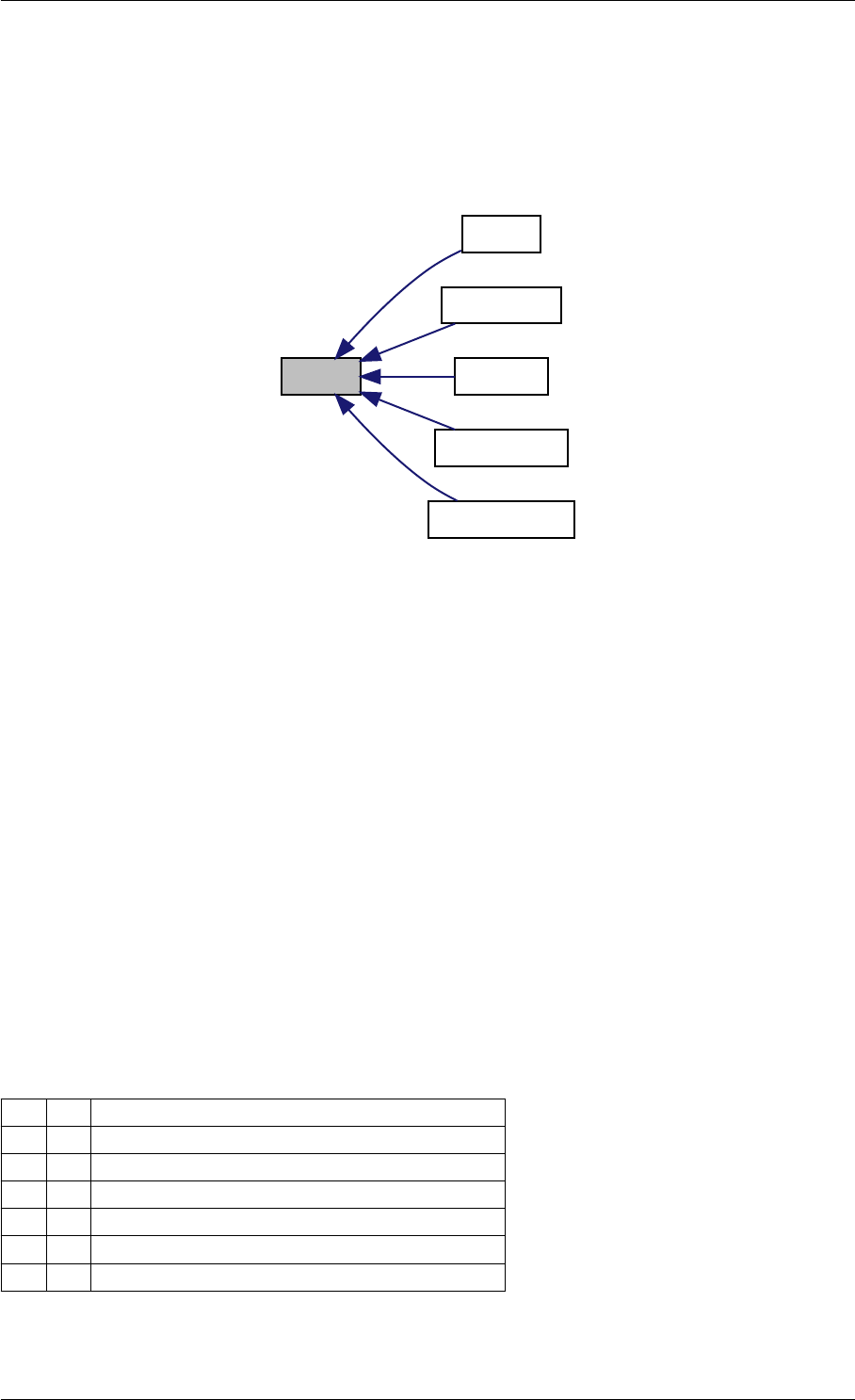
50 CONTENTS
Definition at line 16 of file spline.cpp.
Here is the caller graph for this function:
spline
spline
spline_pos
Dspline
Dspline_pos
DDspline_pos
9.1.4.34 seval()
double seval (
int n,
double u,
double x[ ],
double y[ ],
double b[ ],
double c[ ],
double d[ ] )
S(xx) = y[i] + b[i] ∗w + c[i] ∗w∗∗2 + d[i] ∗w∗∗3 where w = u - x[i] and x[i] <= u <= x[i+1] Note that Horner's rule is
used. If u <x[0] then i = 0 is used. If u >x[n-1] then i = n-1 is used.
Parameters
in nThe number of data points or knots (n >= 2)
in uthe abscissa at which the spline is to be evaluated
in x[ ] the abscissas of the knots in strictly increasing order
in y[ ] the ordinates of the knots
in barray of spline coefficients computed by spline().
in carray of spline coefficients computed by spline().
in darray of spline coefficients computed by spline().
Generated by Doxygen
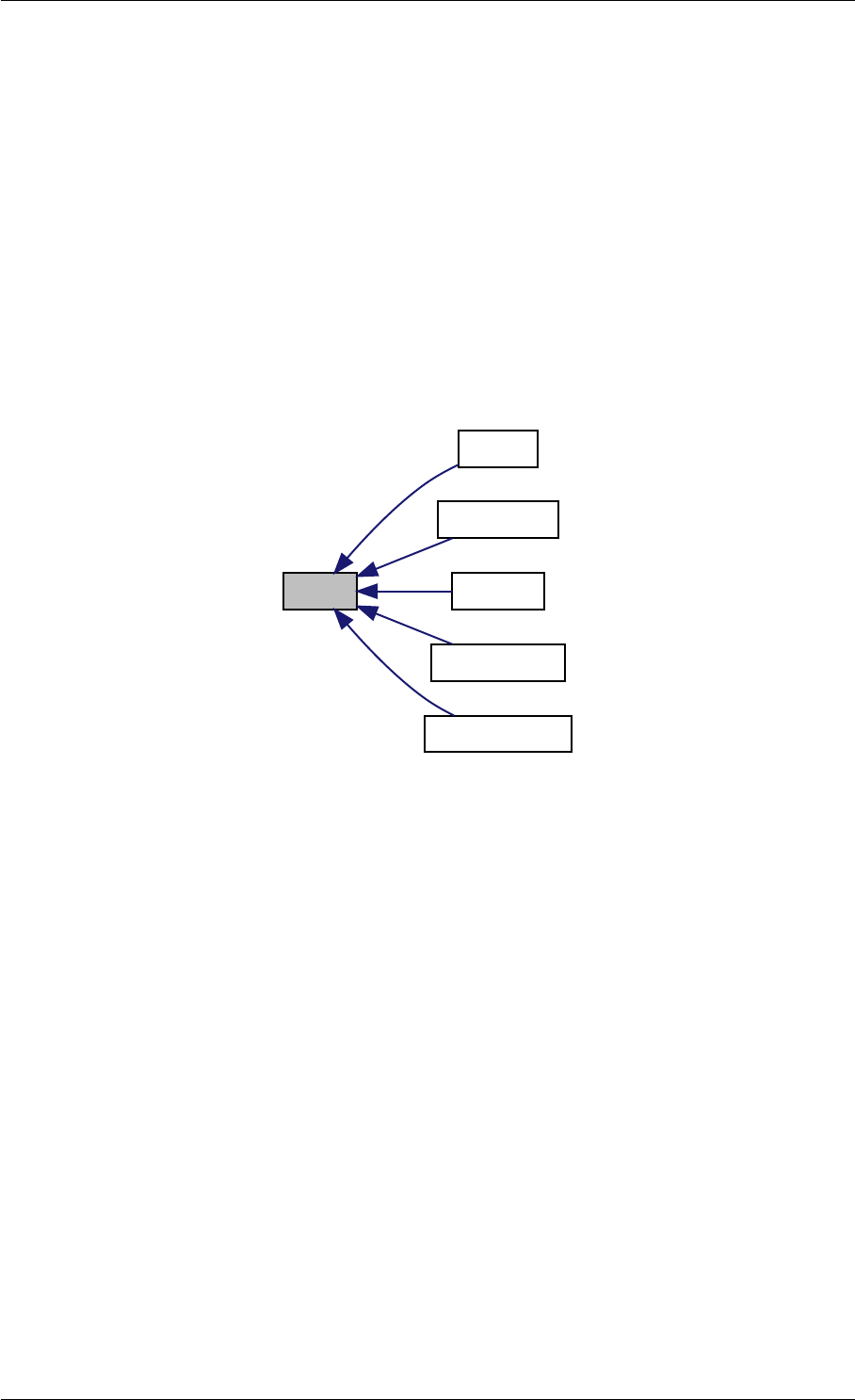
9.1 amici Namespace Reference 51
Returns
the value of the spline function at u
Notes
• If u is not in the same interval as the previous call then a binary search is performed to determine the proper
interval.
Definition at line 195 of file spline.cpp.
Here is the caller graph for this function:
seval
spline
spline_pos
Dspline
Dspline_pos
DDspline_pos
9.1.4.35 sinteg()
double sinteg (
int n,
double u,
double x[ ],
double y[ ],
double b[ ],
double c[ ],
double d[ ] )
Integrate the cubic spline function
S(xx) = y[i] + b[i] ∗w + c[i] ∗w∗∗2 + d[i] ∗w∗∗3 where w = u - x[i] and x[i] <= u <= x[i+1]
The integral is zero at u = x[0].
If u <x[0] then i = 0 segment is extrapolated. If u >x[n-1] then i = n-1 segment is extrapolated.
Generated by Doxygen
52 CONTENTS
Parameters
in nthe number of data points or knots (n >= 2)
in uthe abscissa at which the spline is to be evaluated
in x[ ] the abscissas of the knots in strictly increasing order
in y[ ] the ordinates of the knots
in barray of spline coefficients computed by spline().
in carray of spline coefficients computed by spline().
in darray of spline coefficients computed by spline().
Returns
the value of the spline function at u
Notes
• If u is not in the same interval as the previous call then a binary search is performed to determine the proper
interval.
Definition at line 256 of file spline.cpp.
9.1.4.36 log()
double log (
double x)
c implementation of log function, this prevents returning NaN values for negative values
Parameters
xargument
Generated by Doxygen
9.1 amici Namespace Reference 53
Returns
if(x>0) then log(x) else -Inf
Definition at line 62 of file symbolic_functions.cpp.
Here is the caller graph for this function:
log
spline_pos
Dspline_pos
DDspline_pos
9.1.4.37 dirac()
double dirac (
double x)
c implementation of matlab function dirac
Parameters
xargument
Returns
if(x==0) then INF else 0
Definition at line 77 of file symbolic_functions.cpp.
9.1.4.38 heaviside()
double heaviside (
double x)
c implementation of matlab function heaviside
Generated by Doxygen
54 CONTENTS
Parameters
xargument
Returns
if(x>0) then 1 else 0
Definition at line 92 of file symbolic_functions.cpp.
9.1.4.39 min()
double min (
double a,
double b,
double c)
c implementation of matlab function min
Parameters
avalue1
bvalue2
cbogus parameter to ensure correct parsing as a function
Returns
if(a <b) then a else b
Definition at line 149 of file symbolic_functions.cpp.
Here is the call graph for this function:
min max isNaN
Generated by Doxygen

9.1 amici Namespace Reference 55
9.1.4.40 Dmin()
double Dmin (
int id,
double a,
double b,
double c)
parameter derivative of c implementation of matlab function max
Generated by Doxygen
56 CONTENTS
Parameters
id argument index for differentiation
avalue1
bvalue2
cbogus parameter to ensure correct parsing as a function
Returns
id == 1: if(a >b) then 1 else 0
id == 2: if(a >b) then 0 else 1
Definition at line 191 of file symbolic_functions.cpp.
Here is the call graph for this function:
Dmin Dmax
9.1.4.41 max()
double max (
double a,
double b,
double c)
c implementation of matlab function max
Parameters
avalue1
bvalue2
cbogus parameter to ensure correct parsing as a function
Returns
if(a >b) then a else b
Definition at line 128 of file symbolic_functions.cpp.
Generated by Doxygen
9.1 amici Namespace Reference 57
Here is the call graph for this function:
max isNaN
Here is the caller graph for this function:
max min
9.1.4.42 Dmax()
double Dmax (
int id,
double a,
double b,
double c)
parameter derivative of c implementation of matlab function max
Parameters
id argument index for differentiation
avalue1
bvalue2
cbogus parameter to ensure correct parsing as a function
Returns
id == 1: if(a >b) then 1 else 0
id == 2: if(a >b) then 0 else 1
Definition at line 164 of file symbolic_functions.cpp.
Generated by Doxygen
58 CONTENTS
Here is the caller graph for this function:
Dmax Dmin
9.1.4.43 pos_pow()
double pos_pow (
double base,
double exponent )
specialized pow functions that assumes positivity of the first argument
Parameters
base base
exponent exponent
Returns
pow(max(base,0.0),exponen)
Definition at line 203 of file symbolic_functions.cpp.
9.1.4.44 isNaN()
int isNaN (
double what )
c++ interface to the isNaN function
Parameters
what argument
Returns
isnan(what)
Definition at line 35 of file symbolic_functions.cpp.
Generated by Doxygen
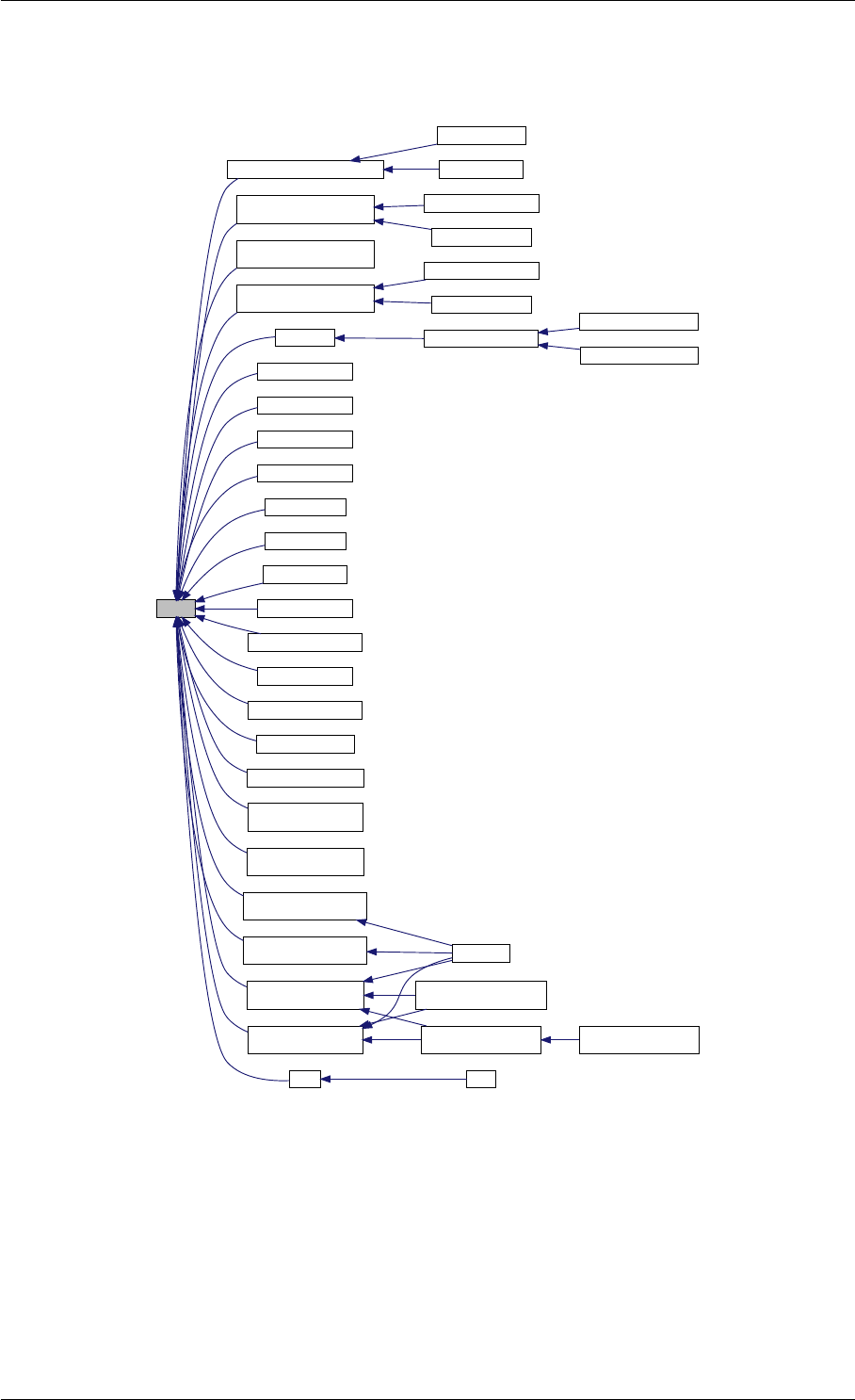
9.1 amici Namespace Reference 59
Here is the caller graph for this function:
isNaN
amici::ExpData::isSetObservedData
amici::ExpData::isSetObserved
DataStdDev
amici::ExpData::isSetObserved
Events
amici::ExpData::isSetObserved
EventsStdDev
checkFinite
amici::Model::fdJydp
amici::Model::fdJydx
amici::Model::fdJzdp
amici::Model::fdJzdx
amici::Model::fJy
amici::Model::fJz
amici::Model::fJrz
amici::Model::fdJydy
amici::Model::fdJydsigma
amici::Model::fdJzdz
amici::Model::fdJzdsigma
amici::Model::fdJrzdz
amici::Model::fdJrzdsigma
amici::Solver::getRelative
ToleranceSensi
amici::Solver::getAbsolute
ToleranceSensi
amici::Solver::getRelative
ToleranceSteadyState
amici::Solver::getAbsolute
ToleranceSteadyState
amici::Solver::getRelative
ToleranceSteadyStateSensi
amici::Solver::getAbsolute
ToleranceSteadyStateSensi
max
amici::Model::fres
amici::Model::fsres
amici::Model::fsigmay
amici::Model::fdsigmaydp
amici::Model::fsigmaz
amici::Model::fdsigmazdp
amici::Model::checkFinite
amici::Model_DAE::fJDiag
amici::Model_ODE::fJDiag
operator==
amici::SteadystateProblem
::checkConvergence
amici::SteadystateProblem
::createSteadystateSimSolver
amici::SteadystateProblem
::getSteadystateSimulation
min
9.1.4.45 isInf()
int isInf (
double what )
c++ interface to the isinf function
Generated by Doxygen
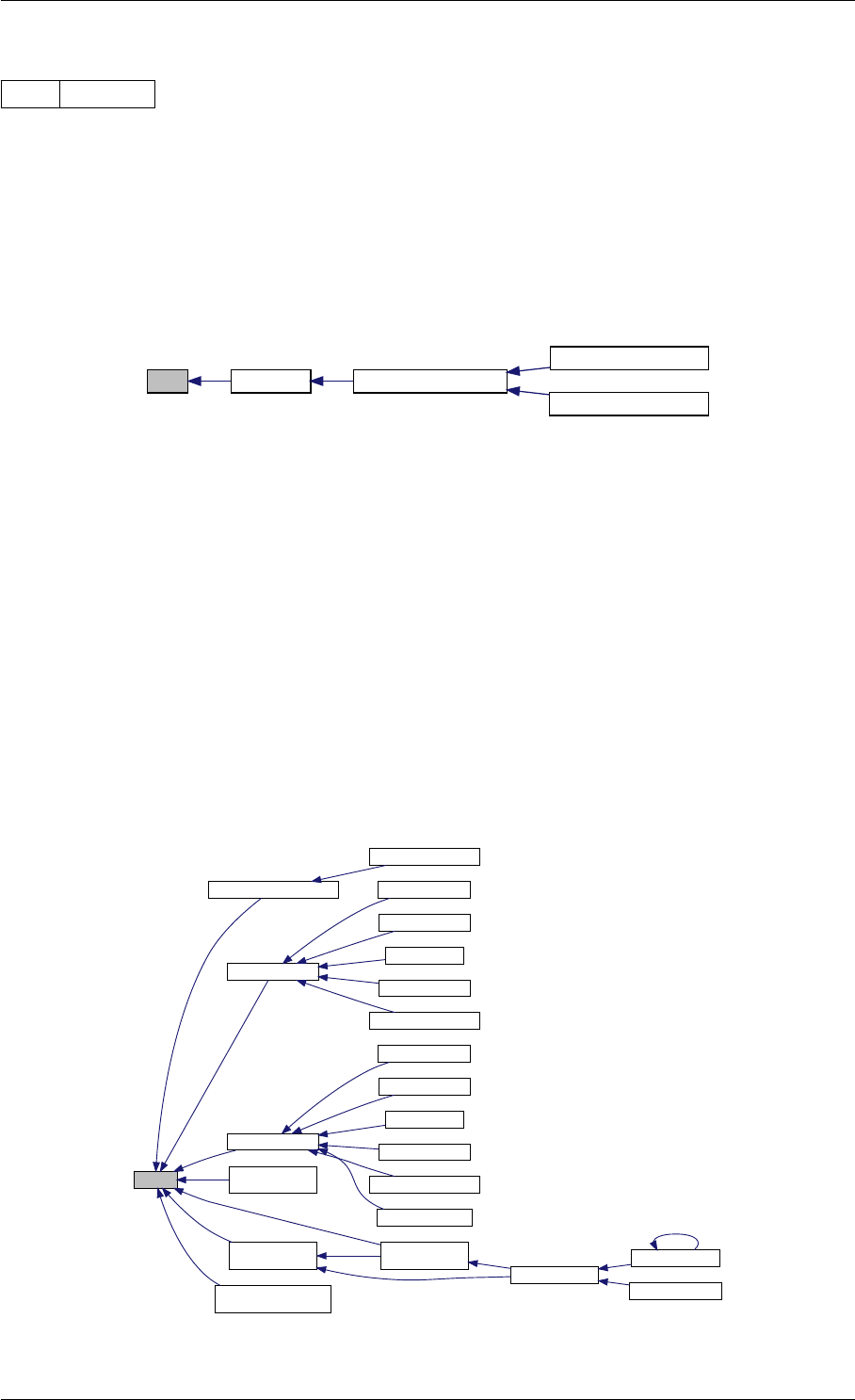
60 CONTENTS
Parameters
what argument
Returns
isnan(what)
Definition at line 44 of file symbolic_functions.cpp.
Here is the caller graph for this function:
isInf checkFinite amici::Model::checkFinite
amici::Model_DAE::fJDiag
amici::Model_ODE::fJDiag
9.1.4.46 getNaN()
double getNaN ( )
function returning nan
Returns
NaN
Definition at line 52 of file symbolic_functions.cpp.
Here is the caller graph for this function:
getNaN
amici::ExpData::setTimepoints
amici::Model::getmy
amici::Model::getmz
amici::ReturnData::
ReturnData
amici::ReturnData::
invalidate
amici::ReturnData::
invalidateLLH
amici::SteadystateProblem
::applyNewtonsMethod
amici::ExpData::ExpData
amici::Model::fdJydp
amici::Model::fdJydx
amici::Model::fJy
amici::Model::fdJydy
amici::Model::fdJydsigma
amici::Model::fdJzdp
amici::Model::fdJzdx
amici::Model::fJz
amici::Model::fdJzdz
amici::Model::fdJzdsigma
amici::Model::fdJrzdz
runAmiciSimulation
runAmiciSimulation
runAmiciSimulations
Generated by Doxygen
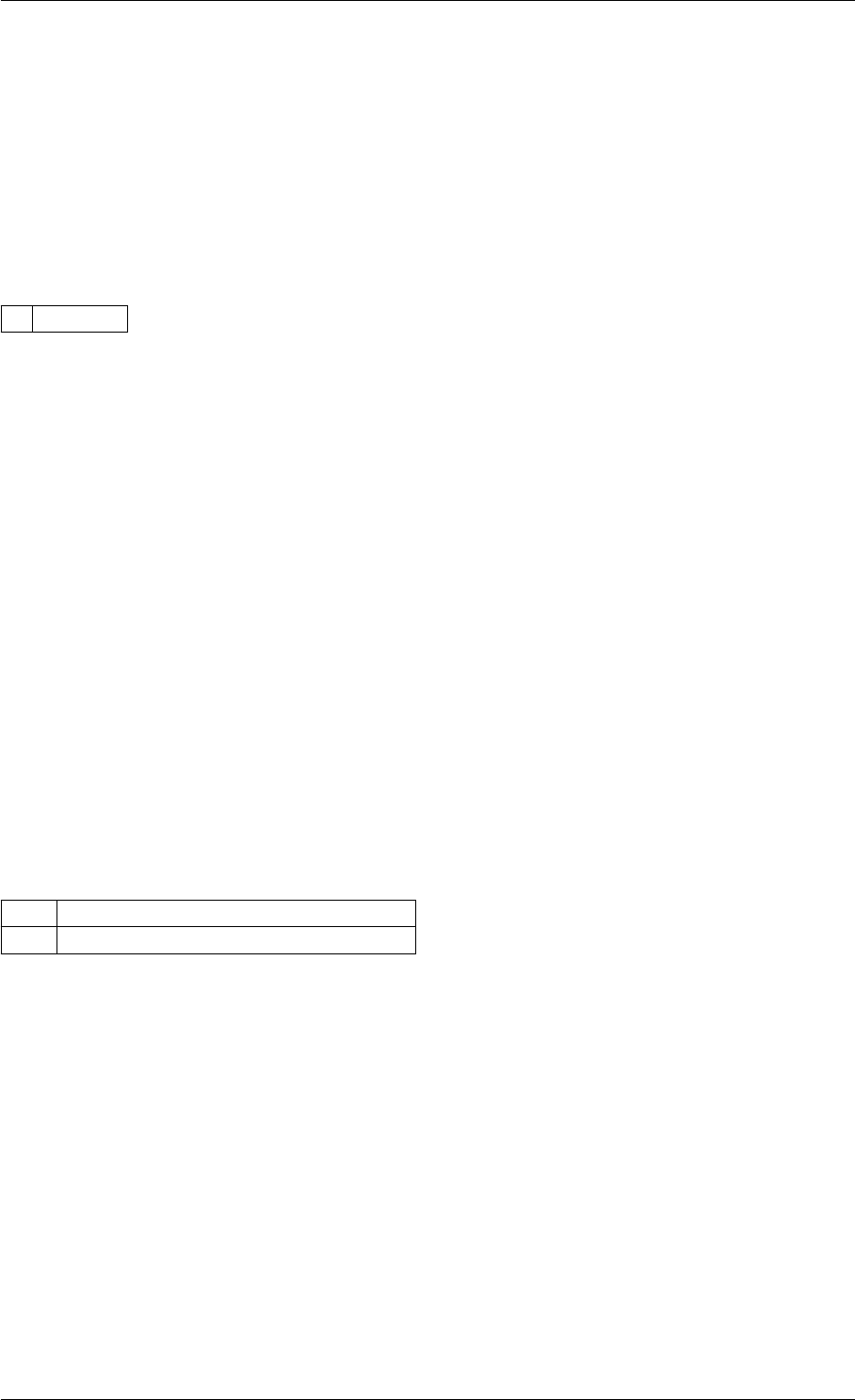
9.1 amici Namespace Reference 61
9.1.4.47 sign()
double sign (
double x)
c implementation of matlab function sign
Parameters
xargument
Returns
0
Definition at line 107 of file symbolic_functions.cpp.
9.1.4.48 spline() [2/2]
double spline (
double t,
int num,
... )
spline function, takes variable argument pairs (ti,pi) with ti: location of node i and pi: spline value at node i. the
last two arguments are always ss: flag indicating whether slope at first node should be user defined and dudt
user defined slope at first node. All arguments must be of type double.
Parameters
tpoint at which the spline should be evaluated
num number of spline nodes
Generated by Doxygen
62 CONTENTS
Returns
spline(t)
Definition at line 222 of file symbolic_functions.cpp.
Here is the call graph for this function:
spline
spline
seval
9.1.4.49 spline_pos()
double spline_pos (
double t,
int num,
... )
exponentiated spline function, takes variable argument pairs (ti,pi) with ti: location of node i and pi: spline value
at node i. the last two arguments are always ss: flag indicating whether slope at first node should be user defined
and dudt user defined slope at first node. All arguments must be of type double.
Parameters
tpoint at which the spline should be evaluated
num number of spline nodes
Generated by Doxygen
9.1 amici Namespace Reference 63
Returns
spline(t)
Definition at line 273 of file symbolic_functions.cpp.
Here is the call graph for this function:
spline_pos
log
spline
seval
9.1.4.50 Dspline()
double Dspline (
int id,
double t,
int num,
... )
derivation of a spline function, takes variable argument pairs (ti,pi) with ti: location of node i and pi: spline value
at node i. the last two arguments are always ss: flag indicating whether slope at first node should be user defined
and dudt user defined slope at first node. All arguments but id must be of type double.
Parameters
id index of node to which the derivative of the corresponding spline coefficient should be computed
tpoint at which the spline should be evaluated
num number of spline nodes
Returns
dsplinedp(t)
Definition at line 326 of file symbolic_functions.cpp.
Generated by Doxygen

64 CONTENTS
Here is the call graph for this function:
Dspline
spline
seval
9.1.4.51 Dspline_pos()
double Dspline_pos (
int id,
double t,
int num,
... )
derivation of an exponentiated spline function, takes variable argument pairs (ti,pi) with ti: location of node i and
pi: spline value at node i. the last two arguments are always ss: flag indicating whether slope at first node should
be user defined and dudt user defined slope at first node. All arguments but id must be of type double.
Parameters
id index of node to which the derivative of the corresponding spline coefficient should be computed
tpoint at which the spline should be evaluated
num number of spline nodes
Returns
dsplinedp(t)
Definition at line 382 of file symbolic_functions.cpp.
Generated by Doxygen
9.1 amici Namespace Reference 65
Here is the call graph for this function:
Dspline_pos
log
spline
seval
9.1.4.52 DDspline()
double DDspline (
int id1,
int id2,
double t,
int num,
... )
second derivation of a spline function, takes variable argument pairs (ti,pi) with ti: location of node i and pi: spline
value at node i. the last two arguments are always ss: flag indicating whether slope at first node should be user
defined and dudt user defined slope at first node. All arguments but id1 and id2 must be of type double.
Parameters
id1 index of node to which the first derivative of the corresponding spline coefficient should be computed
id2 index of node to which the second derivative of the corresponding spline coefficient should be computed
tpoint at which the spline should be evaluated
num number of spline nodes
Returns
ddspline(t)
Definition at line 448 of file symbolic_functions.cpp.
9.1.4.53 DDspline_pos()
double DDspline_pos (
int id1,
Generated by Doxygen
66 CONTENTS
int id2,
double t,
int num,
... )
derivation of an exponentiated spline function, takes variable argument pairs (ti,pi) with ti: location of node i and
pi: spline value at node i. the last two arguments are always ss: flag indicating whether slope at first node should
be user defined and dudt user defined slope at first node. All arguments but id1 and id2 must be of type double.
Parameters
id1 index of node to which the first derivative of the corresponding spline coefficient should be computed
id2 index of node to which the second derivative of the corresponding spline coefficient should be computed
tpoint at which the spline should be evaluated
num number of spline nodes
Returns
ddspline(t)
Definition at line 468 of file symbolic_functions.cpp.
Here is the call graph for this function:
DDspline_pos
log
spline
seval
9.1.4.54 runAmiciSimulation() [2/2]
def amici.runAmiciSimulation (
model,
solver,
edata = None )
Parameters
model Model instance
solver Solver instance, must be generated from Model.getSolver()
edata ExpData instance (optional)
Generated by Doxygen
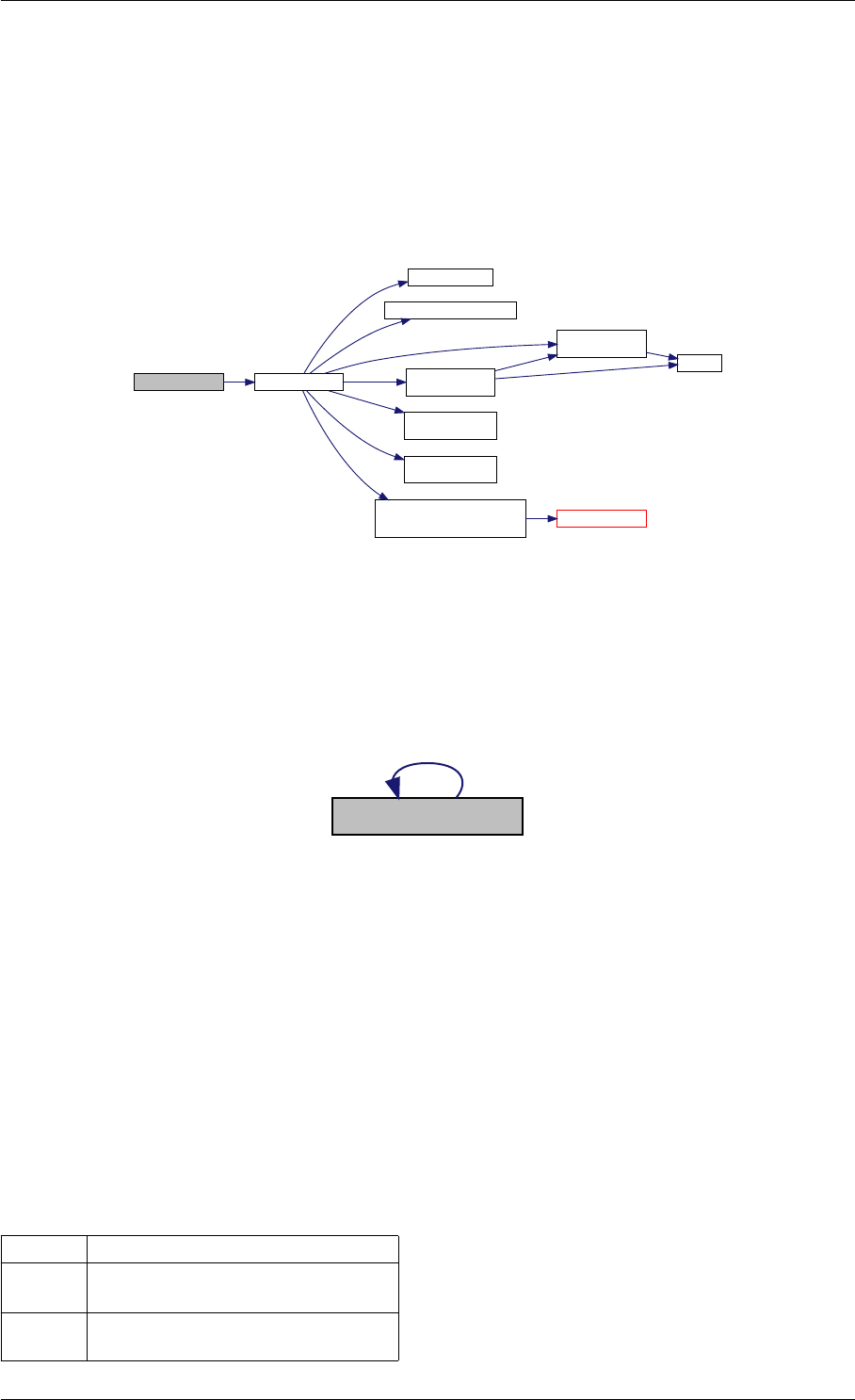
9.1 amici Namespace Reference 67
Returns
ReturnData object with simulation results
Definition at line 92 of file __init__.py.
Here is the call graph for this function:
runAmiciSimulation runAmiciSimulation
amici::ExpData::nt
amici::ExpData::getTimepoints
amici::ReturnData::
invalidate
amici::ReturnData::
invalidateLLH
amici::AmiException
::what
amici::AmiException
::getBacktrace
amici::ReturnData::
applyChainRuleFactorToSimulation
Results
getNaN
unscaleParameters
Here is the caller graph for this function:
runAmiciSimulation
9.1.4.55 ExpData()
def amici.ExpData (
rdata,
sigma_y,
sigma_z )
Parameters
rdata rdataToNumPyArrays output
sigma←-
_y
standard deviation for ObservableData
sigma←-
_z
standard deviation for EventData
Generated by Doxygen
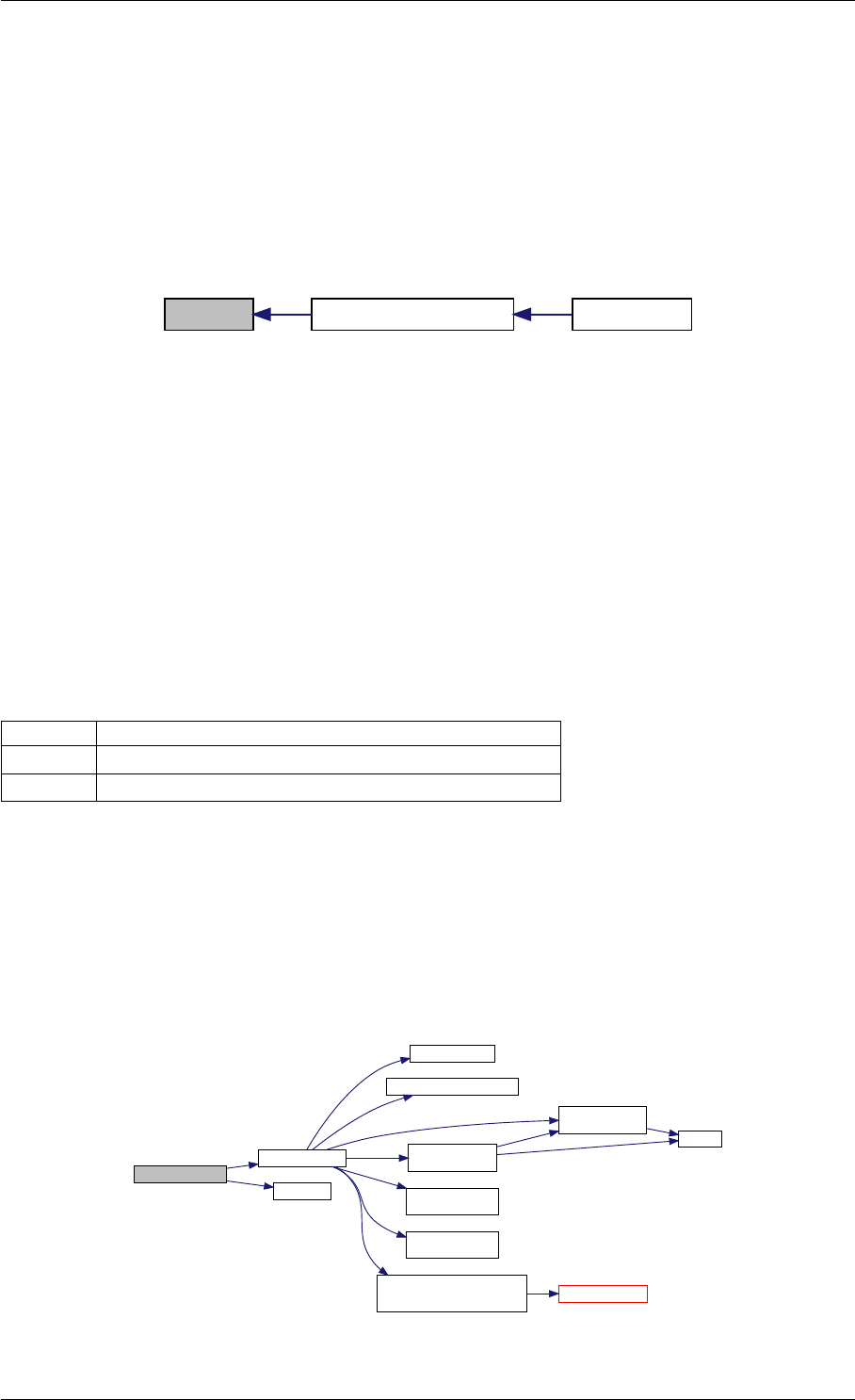
68 CONTENTS
Returns
ExpData Instance
Definition at line 112 of file __init__.py.
Here is the caller graph for this function:
ExpData expDataFromMatlabCall mexFunction
9.1.4.56 runAmiciSimulations()
def amici.runAmiciSimulations (
model,
solver,
edata_list )
Parameters
model Model instance
solver Solver instance, must be generated from Model.getSolver()
edata_list list of ExpData instances
Returns
list of ReturnData objects with simulation results
Definition at line 129 of file __init__.py.
Here is the call graph for this function:
runAmiciSimulations
runAmiciSimulation
pysb2amici
amici::ExpData::nt
amici::ExpData::getTimepoints
amici::ReturnData::
invalidate
amici::ReturnData::
invalidateLLH
amici::AmiException
::what
amici::AmiException
::getBacktrace
amici::ReturnData::
applyChainRuleFactorToSimulation
Results
getNaN
unscaleParameters
Generated by Doxygen

9.1 amici Namespace Reference 69
9.1.4.57 pysb2amici()
def amici.pysb2amici (
model,
output_dir = None,
observables = None,
constantParameters = None,
sigmas = None,
verbose = False,
assume_pow_positivity = False,
compiler = None )
Parameters
model pysb model, model.name will determine the name of the generated module
Type: pysb.Model
output_dir see sbml_import.setPaths()
Type: str
observables list of pysb.Expressions names that should be interpreted as observables
Type: list
sigmas list of pysb.Expressions names that should be interpreted as sigmas
Type: list
constantParameters list of pysb.Parameter names that should be interpreted as constantParameters
verbose more verbose output if True
Type: bool
assume_pow_positivity if set to true, a special pow function is used to avoid problems with state variables
that may become negative due to numerical errors
Type: bool
compiler distutils/setuptools compiler selection to build the python extension
Type: str
Returns
Definition at line 181 of file __init__.py.
Here is the caller graph for this function:
pysb2amici runAmiciSimulations
Generated by Doxygen
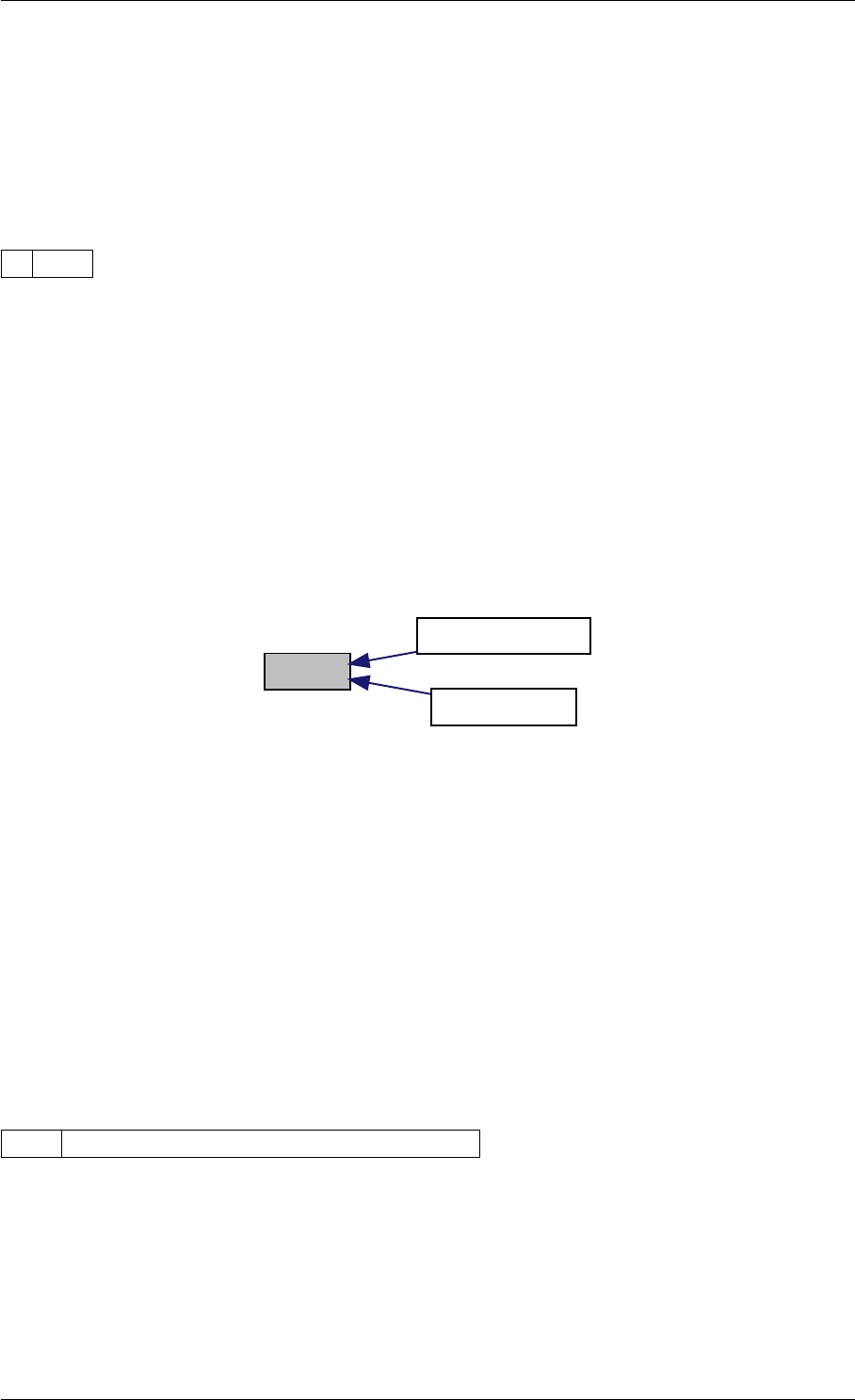
70 CONTENTS
9.1.4.58 dbl2int()
int dbl2int (
const double x)
conversion from double to int with checking for loss of data
Parameters
xinput
Returns
int_x casted value
Definition at line 298 of file interface_matlab.cpp.
Here is the caller graph for this function:
dbl2int
setSolverOptions
setModelData
9.1.4.59 amici_blasCBlasTransToBlasTrans()
char amici::amici_blasCBlasTransToBlasTrans (
BLASTranspose trans )
amici_blasCBlasTransToBlasTrans translates AMICI_BLAS_TRANSPOSE values to CBlas readable strings
Parameters
trans flag indicating transposition and complex conjugation
Returns
cblastrans CBlas readable CHAR indicating transposition and complex conjugation
Definition at line 52 of file interface_matlab.cpp.
Generated by Doxygen
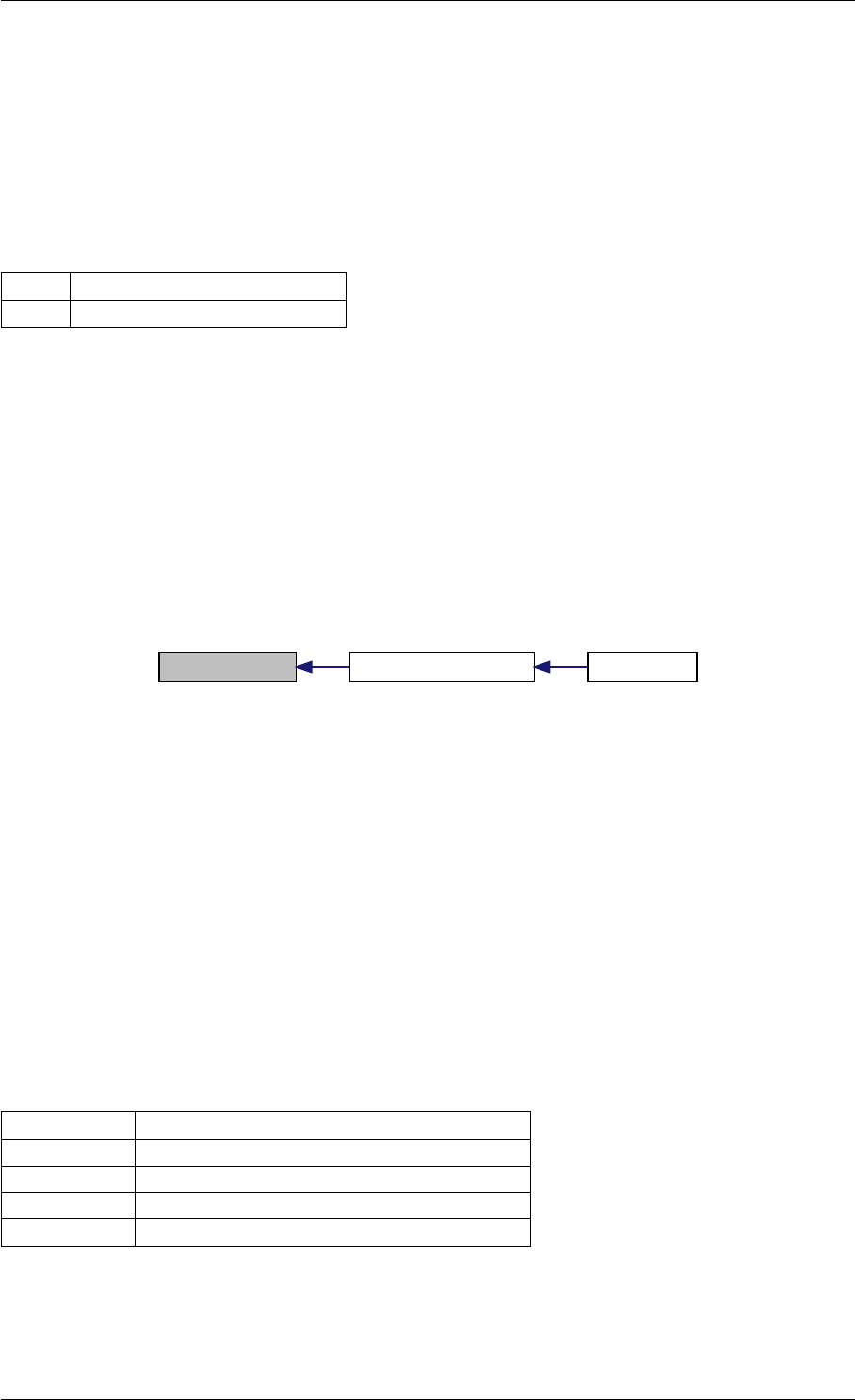
9.1 amici Namespace Reference 71
9.1.4.60 mxArrayToVector()
std::vector<realtype>amici::mxArrayToVector (
const mxArray ∗array,
int length )
conversion from mxArray to vector<realtype>
Parameters
array Matlab array to create vector from
length Number of elements in array
Returns
std::vector with data from array
Definition at line 166 of file interface_matlab.cpp.
Here is the caller graph for this function:
mxArrayToVector expDataFromMatlabCall mexFunction
9.1.4.61 getValueById()
realtype amici::getValueById (
std::vector<std::string >const & ids,
std::vector<realtype >const & values,
std::string const & id,
const char ∗variable_name,
const char ∗id_name )
Parameters
ids vector of name/ids of (fixed)Parameters
values values of the (fixed)Parameters
id name/id to look for in the vector
variable_name string indicating what variable we are lookin at
id_name string indicating whether name or id was specified
Returns
value of the selected parameter
Generated by Doxygen
72 CONTENTS
Definition at line 323 of file model.cpp.
Here is the caller graph for this function:
getValueById
amici::Model::getParameterById
amici::Model::getParameter
ByName
amici::Model::getFixedParameterById
amici::Model::getFixedParameter
ByName
9.1.4.62 setValueById()
void amici::setValueById (
std::vector<std::string >const & ids,
std::vector<realtype >&values,
realtype value,
std::string const & id,
const char ∗variable_name,
const char ∗id_name )
Parameters
ids vector of names/ids of (fixed)Parameters
values values of the (fixed)Parameters
value for the selected parameter
id name/id to look for in the vector
variable_name string indicating what variable we are lookin at
id_name string indicating whether name or id was specified
Definition at line 341 of file model.cpp.
Generated by Doxygen
9.1 amici Namespace Reference 73
Here is the caller graph for this function:
setValueById
amici::Model::setParameterById
amici::Model::setParameter
ByName
amici::Model::setFixedParameterById
amici::Model::setFixedParameter
ByName
9.1.4.63 setValueByIdRegex()
int amici::setValueByIdRegex (
std::vector<std::string >const & ids,
std::vector<realtype >&values,
realtype value,
std::string const & regex,
const char ∗variable_name,
const char ∗id_name )
Parameters
ids vector of names/ids of (fixed)Parameters
values values of the (fixed)Parameters
value for the selected parameter
regex string according to which names/ids are to be matched
variable_name string indicating what variable we are lookin at
id_name string indicating whether name or id was specified
Returns
number of matched names/ids
Definition at line 360 of file model.cpp.
Generated by Doxygen
74 CONTENTS
Here is the caller graph for this function:
setValueByIdRegex
amici::Model::setParameters
ByIdRegex
amici::Model::setParameters
ByNameRegex
amici::Model::setFixedParameters
ByIdRegex
amici::Model::setFixedParameters
ByNameRegex
9.1.5 Variable Documentation
9.1.5.1 errMsgIdAndTxt
msgIdAndTxtFp errMsgIdAndTxt = &printErrMsgIdAndTxt
errMsgIdAndTxt is a function pointer for printErrMsgIdAndTxt
Definition at line 45 of file amici.cpp.
9.1.5.2 warnMsgIdAndTxt
msgIdAndTxtFp warnMsgIdAndTxt = &printWarnMsgIdAndTxt
warnMsgIdAndTxt is a function pointer for printWarnMsgIdAndTxt
Definition at line 13 of file model_dae.h.
9.1.5.3 pi
constexpr double pi = 3.14159265358979323846
MS definition of PI and other constants
Definition at line 13 of file defines.h.
Generated by Doxygen

9.2 amici.ode_export Namespace Reference 75
9.1.5.4 amici_path
amici_path
Initial value:
1 = os.path.abspath(os.path.join(
2 os.path.dirname(__file__), ’..’,’..’))
Definition at line 55 of file __init__.py.
9.2 amici.ode_export Namespace Reference
The C++ ODE export module for python.
Classes
• class Constant
AConstant is a fixed variable in the model with respect to which sensitivites cannot be computed, abbreviated by k
• class Expression
An Expressions is a recurring elements in symbolic formulas.
• class LogLikelihood
ALogLikelihood defines the distance between measurements and experiments for a particular observable.
• class ModelQuantity
Base class for model components.
• class Observable
An Observable links model simulations to experimental measurements, abbreviated by y
• class ODEExporter
The ODEExporter class generates AMICI C++ files for ODE model as defined in symbolic expressions.
• class ODEModel
An ODEModel defines an Ordinay Differential Equation as set of ModelQuantities.
• class Parameter
AParameter is a free variable in the model with respect to which sensitivites may be computed, abbreviated by p
• class SigmaY
A Standard Deviation SigmaY rescales the distance between simulations and measurements when computing resid-
uals, abbreviated by sigmay
• class State
AState variable defines an entity that evolves with time according to the provided time derivative, abbreviated by x
• class TemplateAmici
Template format used in AMICI (see string.template for more details).
Functions
• def var_in_function_signature (name, varname)
Checks if the values for a symbolic variable is passed in the signature of a function.
• def getSymbolicDiagonal (matrix)
Get symbolic matrix with diagonal of matrix matrix.
• def applyTemplate (sourceFile, targetFile, templateData)
Load source file, apply template substitution as provided in templateData and save as targetFile.
• def ODEModel_from_pysb_importer (model, constants=None, observables=None, sigmas=None)
Creates an ODEModel instance from a pysb.Model instance.
Generated by Doxygen
76 CONTENTS
Variables
•pysb = None
pysb module, if this is None, pysb is not available
• dictionary functions
prototype for generated C++ functions, keys are the names of functions
• list sparse_functions
list of sparse functions
• list sensi_functions
list of sensitivity functions
• list multiobs_functions
list of multiobs functions
• dictionary symbol_to_type
indicates which type some attributes in ODEModel should have
9.2.1 Detailed Description
!/usr/bin/env python3
9.2.2 Function Documentation
9.2.2.1 var_in_function_signature()
def amici.ode_export.var_in_function_signature (
name,
varname )
Parameters
name name of the function
Type: str
varname name of the symbolic variable
Type: str
Generated by Doxygen
9.2 amici.ode_export Namespace Reference 77
Returns
Definition at line 279 of file ode_export.py.
Here is the caller graph for this function:
var_in_function_signature amici.ode_export.ODEModel.sym
Names
9.2.2.2 getSymbolicDiagonal()
def amici.ode_export.getSymbolicDiagonal (
matrix )
Parameters
matrix Matrix from which to return the diagonal
Type: symengine.DenseMatrix
Returns
A Symbolic matrix with the diagonal of matrix.
Exceptions
Exception The provided matrix was not square
Definition at line 2199 of file ode_export.py.
Here is the caller graph for this function:
getSymbolicDiagonal amici.ode_export.ODEModel.generate
BasicVariables
amici.ode_export.ODEModel.import
_from_sbml_importer
Generated by Doxygen
78 CONTENTS
9.2.2.3 applyTemplate()
def amici.ode_export.applyTemplate (
sourceFile,
targetFile,
templateData )
Parameters
sourceFile relative or absolute path to template file
Type: str
targetFile relative or absolute path to output file
Type: str
templateData template keywords to substitute (key is template variable without TemplateAmici.delimiter)
Type: dict
Returns
Definition at line 2234 of file ode_export.py.
Here is the call graph for this function:
applyTemplate ODEModel_from_pysb
_importer
Here is the caller graph for this function:
applyTemplate amici.ode_export.ODEExporter.compile
Model
9.2.2.4 ODEModel_from_pysb_importer()
def amici.ode_export.ODEModel_from_pysb_importer (
model,
constants = None,
observables = None,
sigmas = None )
Generated by Doxygen
9.2 amici.ode_export Namespace Reference 79
Parameters
model pysb model
Type: pysb.Model
constants list of Parameters that should be constants
Type: list
observables list of Expressions that should be observables
Type: list
sigmas dict with observable Expression name as key and sigma Expression name as expression
Type: list
Returns
New ODEModel instance according to pysbModel
Definition at line 2263 of file ode_export.py.
Here is the caller graph for this function:
ODEModel_from_pysb
_importer applyTemplate amici.ode_export.ODEExporter.compile
Model
9.2.3 Variable Documentation
9.2.3.1 functions
dictionary functions
signature: defines the argument part of the function signature, input variables should have a const flag
assume_pow_positivity: identifies the functions on which assume_pow_positivity will have an effect when specified
during model generation. generally these are functions that are used for solving the ODE, where negative values
may negatively affect convergence of the integration algorithm
sparse: specifies whether the result of this function will be stored in sparse format. sparse format means that the
function will only return an array of nonzero values and not a full matrix.
Definition at line 42 of file ode_export.py.
Generated by Doxygen

80 CONTENTS
9.2.3.2 sparse_functions
list sparse_functions
Initial value:
1 = [
2 function for function in functions
3if ’sparse’ in functions[function]
4and functions[function][’sparse’]
5 ]
Definition at line 248 of file ode_export.py.
9.2.3.3 sensi_functions
list sensi_functions
Initial value:
1 = [
2 function for function in functions
3if ’const int ip’ in functions[function][’signature’]
4and function is not ’sxdot’
5 ]
Definition at line 254 of file ode_export.py.
9.2.3.4 multiobs_functions
list multiobs_functions
Initial value:
1 = [
2 function for function in functions
3if ’const int iy’ in functions[function][’signature’]
4 ]
Definition at line 260 of file ode_export.py.
9.2.3.5 symbol_to_type
dictionary symbol_to_type
Initial value:
1 = {
2’species’: State,
3’parameter’: Parameter,
4’fixed_parameter’: Constant,
5’observable’: Observable,
6’sigmay’: SigmaY,
7’llhy’: LogLikelihood,
8’expression’: Expression,
9 }
Definition at line 537 of file ode_export.py.
Generated by Doxygen
9.3 amici.plotting Namespace Reference 81
9.3 amici.plotting Namespace Reference
Plotting related functions.
Functions
• def plotStateTrajectories (rdata, state_indices=None, ax=None)
Plot state trajectories.
• def plotObservableTrajectories (rdata, observable_indices=None, ax=None)
Plot observable trajectories.
9.3.1 Function Documentation
9.3.1.1 plotStateTrajectories()
def amici.plotting.plotStateTrajectories (
rdata,
state_indices = None,
ax = None )
Parameters
rdata AMICI simulation results as returned by amici.getSimulationResults()
state_indices Indices of states for which trajectories are to be plotted
ax matplotlib.axes.Axes instance to plot into
Returns
Definition at line 17 of file plotting.py.
9.3.1.2 plotObservableTrajectories()
def amici.plotting.plotObservableTrajectories (
rdata,
observable_indices = None,
ax = None )
Parameters
rdata AMICI simulation results as returned by amici.getSimulationResults()
observable_indices Indices of observables for which trajectories are to be plotted
ax matplotlib.axes.Axes instance to plot into
Generated by Doxygen

82 CONTENTS
Returns
Definition at line 42 of file plotting.py.
9.4 amici.sbml_import Namespace Reference
The python sbml import module for python.
Classes
• class SBMLException
• class SbmlImporter
The SbmlImporter class generates AMICI C++ files for a model provided in the Systems Biology Markup Language
(SBML).
Functions
• def getRuleVars (rules)
Extract free symbols in SBML rule formulas.
• def assignmentRules2observables (sbml_model, filter_function=lambda ∗_:True)
Turn assignment rules into observables.
• def constantSpeciesToParameters (sbml_model)
Convert constant species in the SBML model to constant parameters.
• def replaceLogAB (x)
Replace log(a, b) in the given string by ln(b)/ln(a)
• def l2s (inputs)
transforms an list into list of strings
Variables
• dictionary default_symbols
default dict for symbols
9.4.1 Detailed Description
!/usr/bin/env python3
9.4.2 Function Documentation
9.4.2.1 getRuleVars()
def amici.sbml_import.getRuleVars (
rules )
Generated by Doxygen
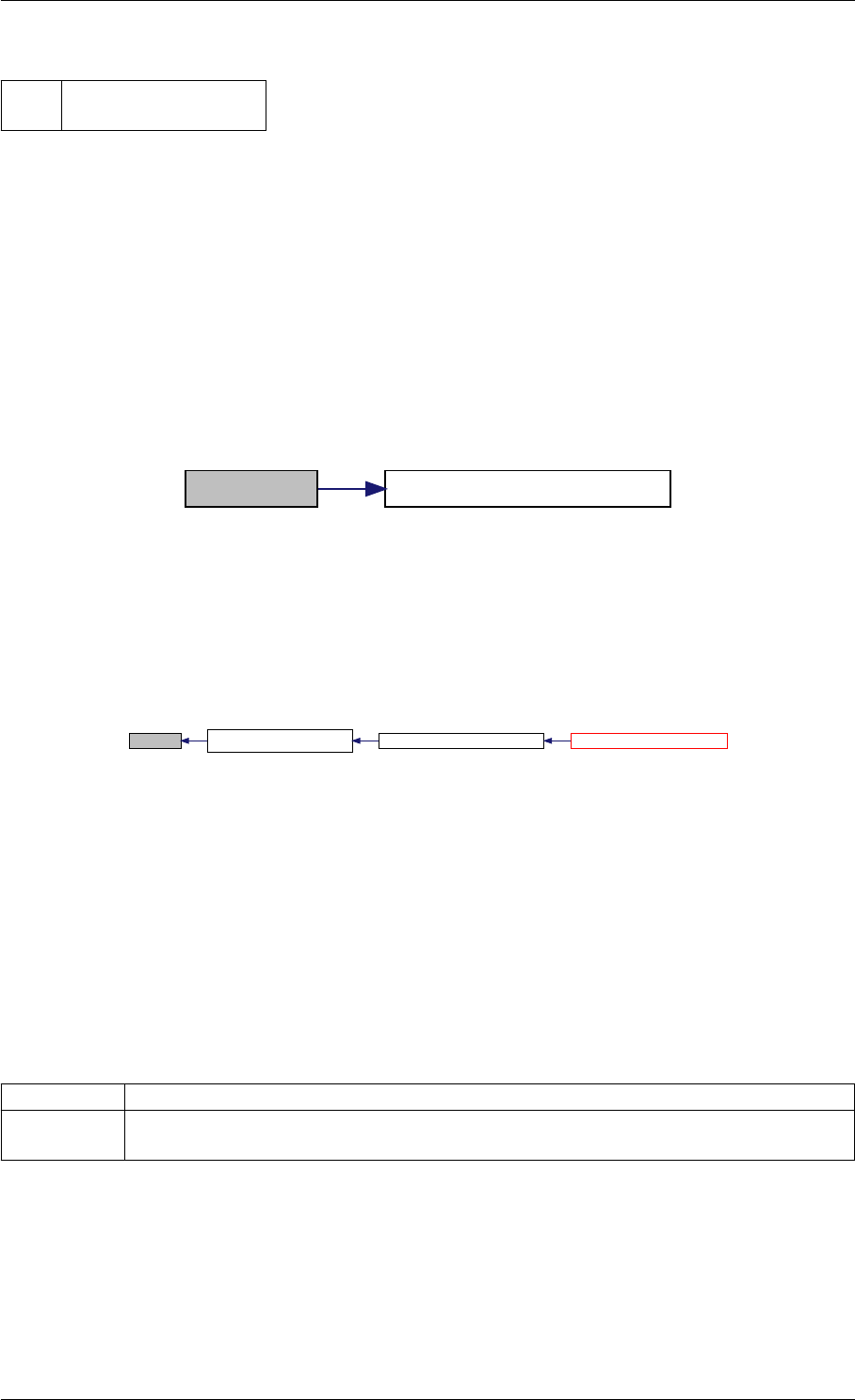
9.4 amici.sbml_import Namespace Reference 83
Parameters
rules sbml definitions of rules
Type: list
Returns
Tuple of free symbolic variables in the formulas all provided rules
Definition at line 992 of file sbml_import.py.
Here is the call graph for this function:
getRuleVars assignmentRules2observables
Here is the caller graph for this function:
getRuleVars amici.sbml_import.SbmlImporter.process
Rules amici.sbml_import.SbmlImporter.processSBML amici.sbml_import.SbmlImporter.sbml2amici
9.4.2.2 assignmentRules2observables()
def amici.sbml_import.assignmentRules2observables (
sbml_model,
filter_function = lambda ∗_: True )
Parameters
sbml_model an sbml Model instance
filter_function callback function taking assignment variable as input and returning True/False to indicate if the
respective rule should be turned into an observable
Returns
A dictionary(observableId:{ 'name': observableNamem, 'formula': formulaString })
Definition at line 1018 of file sbml_import.py.
Generated by Doxygen
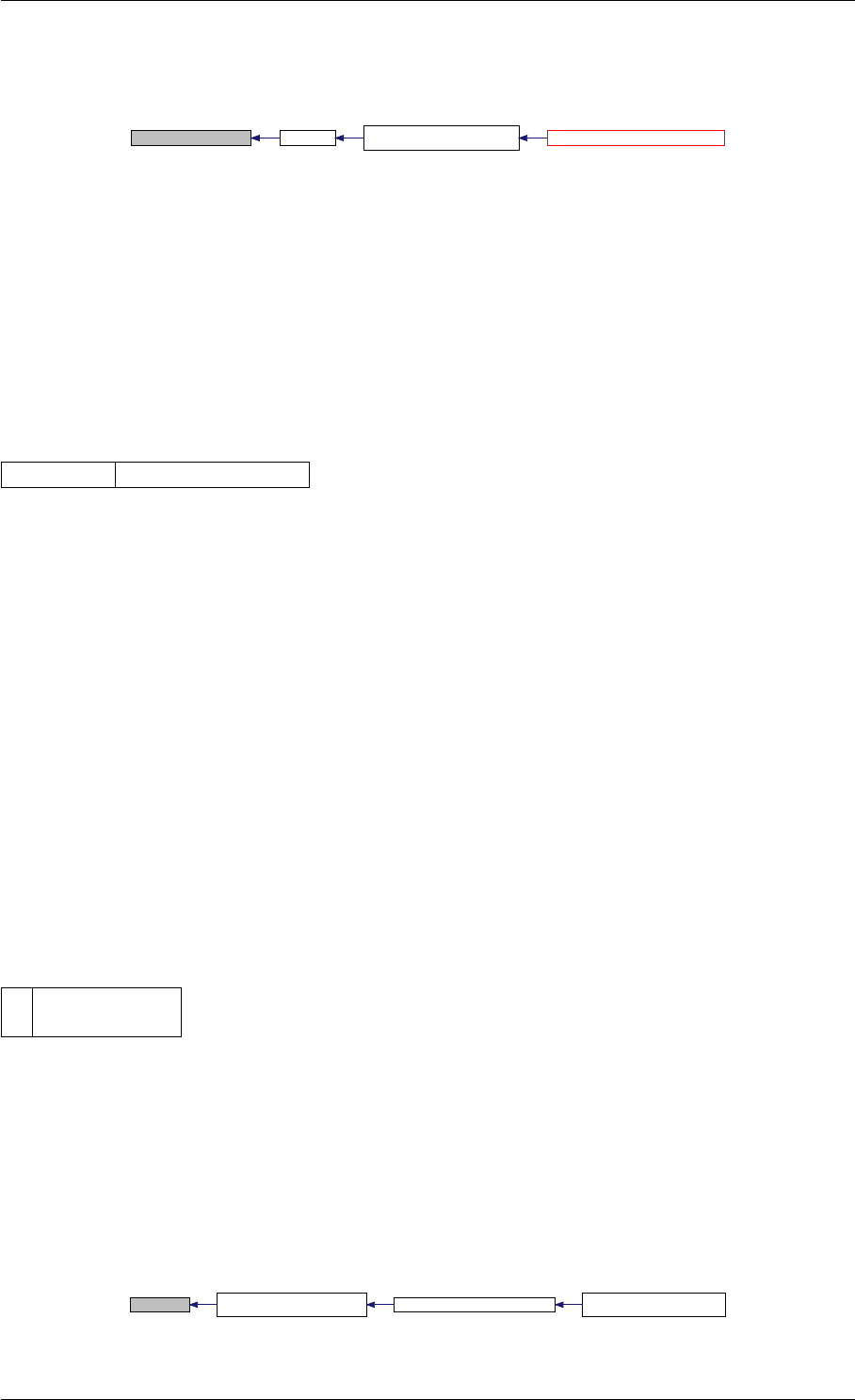
84 CONTENTS
Here is the caller graph for this function:
assignmentRules2observables getRuleVars amici.sbml_import.SbmlImporter.process
Rules amici.sbml_import.SbmlImporter.processSBML
9.4.2.3 constantSpeciesToParameters()
def amici.sbml_import.constantSpeciesToParameters (
sbml_model )
Parameters
sbml_model libsbml model instance
Returns
species IDs that have been turned into constants
Definition at line 1049 of file sbml_import.py.
9.4.2.4 replaceLogAB()
def amici.sbml_import.replaceLogAB (
x)
Works for nested parentheses and nested 'log's. This can be used to circumvent the incompatible argument order
between symengine (log(x, basis)) and libsbml (log(basis, x)).
Parameters
xstring to replace
Type: str
Returns
string with replaced 'log's
Definition at line 1097 of file sbml_import.py.
Here is the caller graph for this function:
replaceLogAB amici.sbml_import.SbmlImporter.process
Observables amici.sbml_import.SbmlImporter.sbml2amici amici.sbml_import.SbmlImporter.check
LibSBMLErrors
Generated by Doxygen
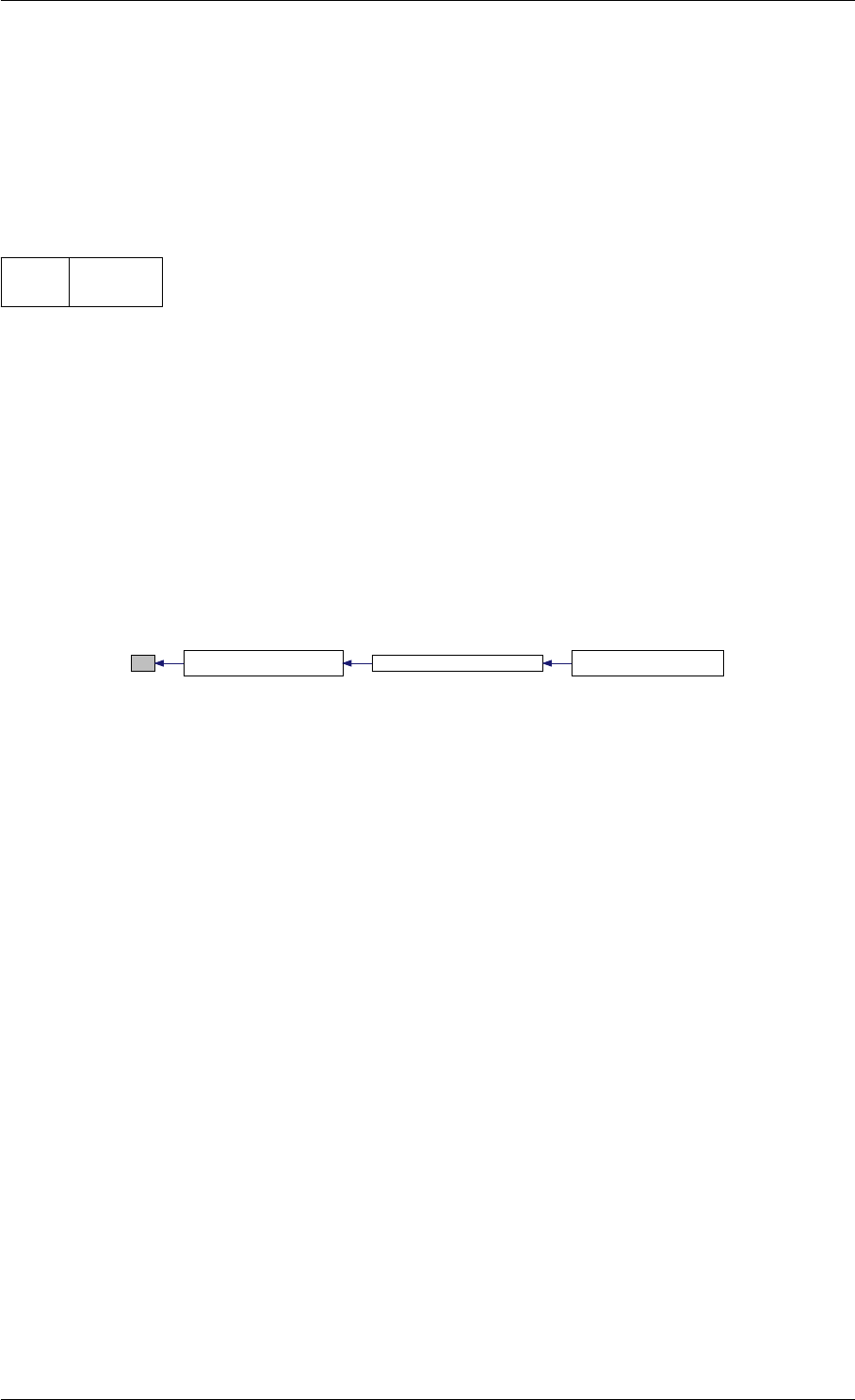
9.4 amici.sbml_import Namespace Reference 85
9.4.2.5 l2s()
def amici.sbml_import.l2s (
inputs )
Parameters
inputs objects
Type: list
Returns
list of str(object)
Definition at line 1143 of file sbml_import.py.
Here is the caller graph for this function:
l2s amici.sbml_import.SbmlImporter.process
Observables amici.sbml_import.SbmlImporter.sbml2amici amici.sbml_import.SbmlImporter.check
LibSBMLErrors
9.4.3 Variable Documentation
9.4.3.1 default_symbols
dictionary default_symbols
Initial value:
1 = {
2’species’: {},
3’parameter’: {},
4’fixed_parameter’: {},
5’observable’: {},
6’expression’: {},
7’sigmay’: {},
8’my’: {},
9’llhy’: {},
10 }
Definition at line 17 of file sbml_import.py.
Generated by Doxygen
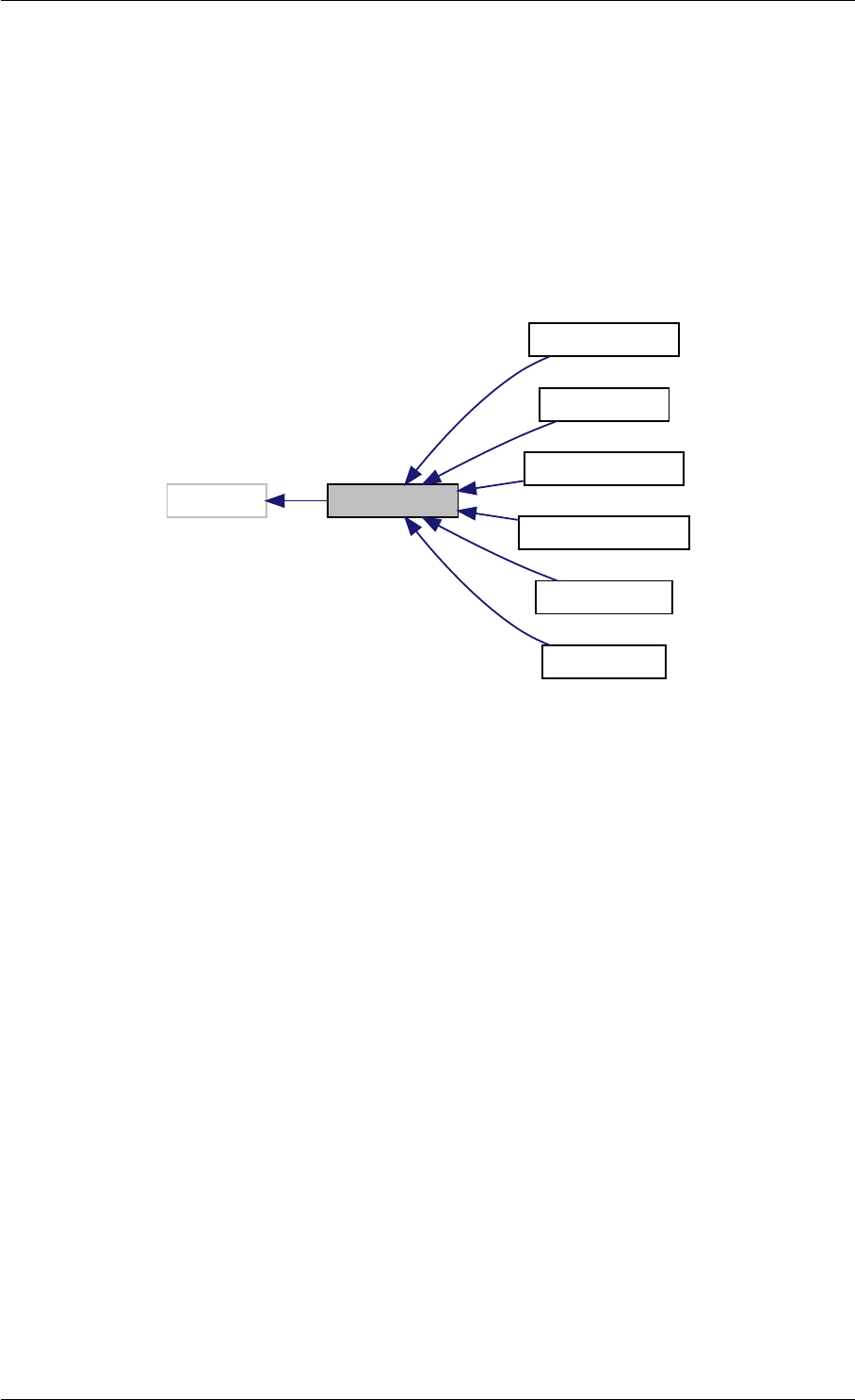
86 CONTENTS
10 Class Documentation
10.1 AmiException Class Reference
amici exception handler class
#include <exception.h>
Inheritance diagram for AmiException:
AmiException
CvodeException
IDAException
IntegrationFailure
IntegrationFailureB
NewtonFailure
SetupFailure
exception
Public Member Functions
•AmiException (char const ∗fmt,...)
•AmiException (const AmiException &old)
• const char ∗what () const throw ()
• const char ∗getBacktrace () const
• void storeBacktrace (const int nMaxFrames)
10.1.1 Detailed Description
has a printf style interface to allow easy generation of error messages
Definition at line 29 of file exception.h.
10.1.2 Constructor & Destructor Documentation
10.1.2.1 AmiException() [1/2]
AmiException (
char const ∗fmt,
... )
constructor with printf style interface
Generated by Doxygen
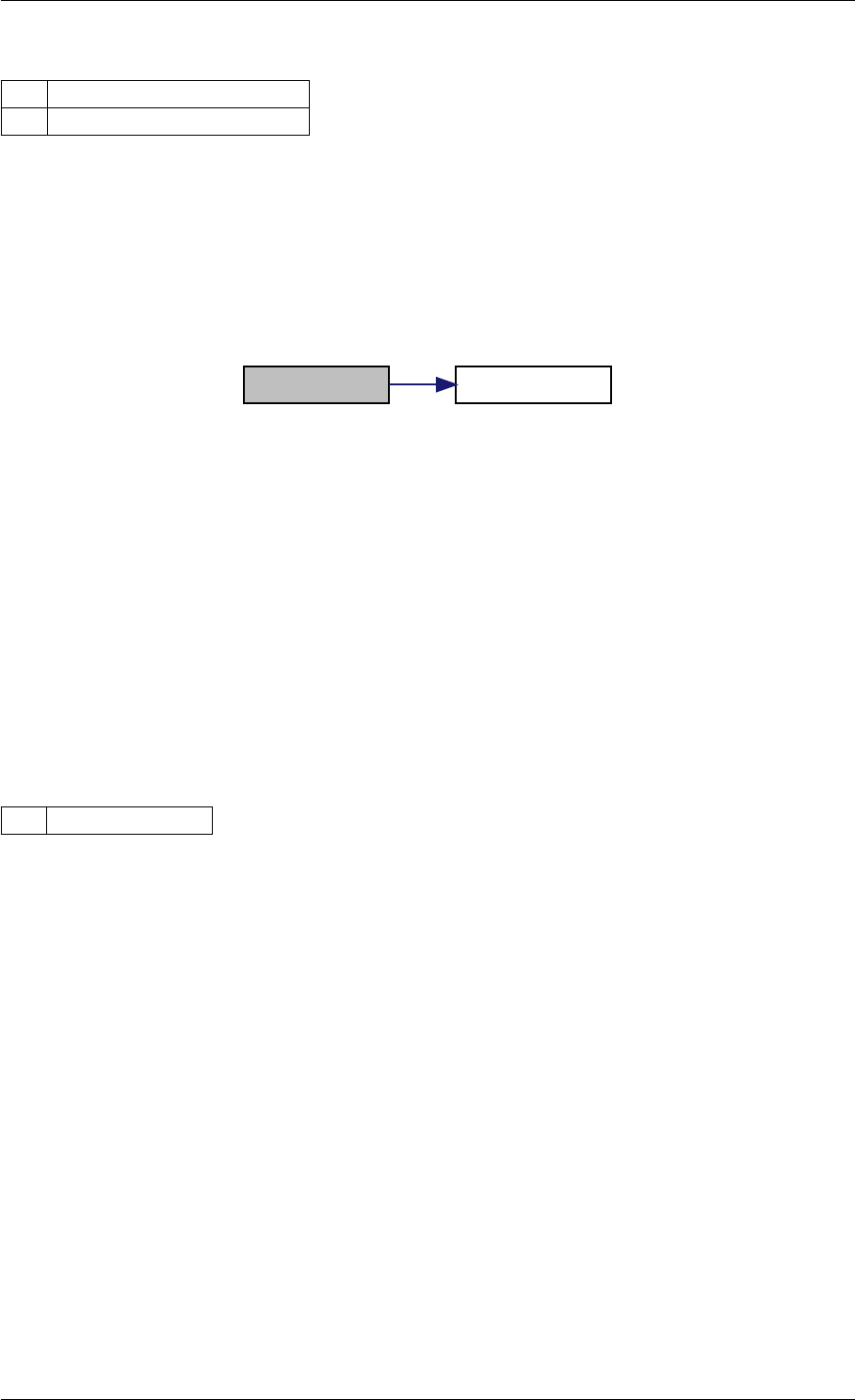
10.1 AmiException Class Reference 87
Parameters
fmt error message with printf format
... printf formating variables
Definition at line 31 of file exception.h.
Here is the call graph for this function:
AmiException storeBacktrace
10.1.2.2 AmiException() [2/2]
AmiException (
const AmiException &old )
copy constructor
Parameters
old object to copy from
Definition at line 43 of file exception.h.
10.1.3 Member Function Documentation
10.1.3.1 what()
const char∗what ( ) const throw )
override of default error message function
Generated by Doxygen
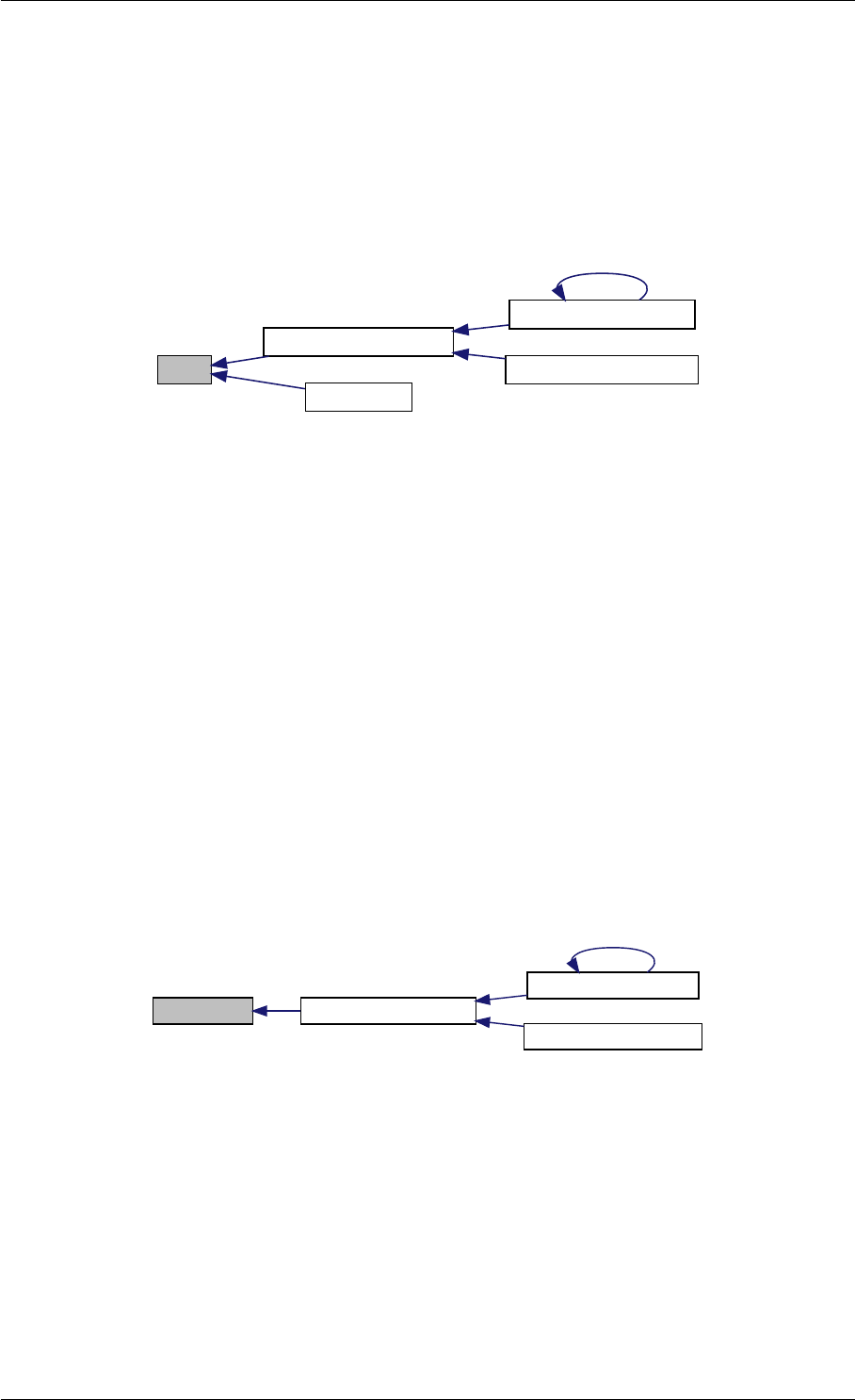
88 CONTENTS
Returns
msg error message
Definition at line 54 of file exception.h.
Here is the caller graph for this function:
what
amici::runAmiciSimulation
mexFunction
amici.runAmiciSimulation
amici.runAmiciSimulations
10.1.3.2 getBacktrace()
const char∗getBacktrace ( ) const
returns the stored backtrace
Returns
trace backtrace
Definition at line 61 of file exception.h.
Here is the caller graph for this function:
getBacktrace amici::runAmiciSimulation
amici.runAmiciSimulation
amici.runAmiciSimulations
10.1.3.3 storeBacktrace()
void storeBacktrace (
const int nMaxFrames )
stores the current backtrace
Generated by Doxygen
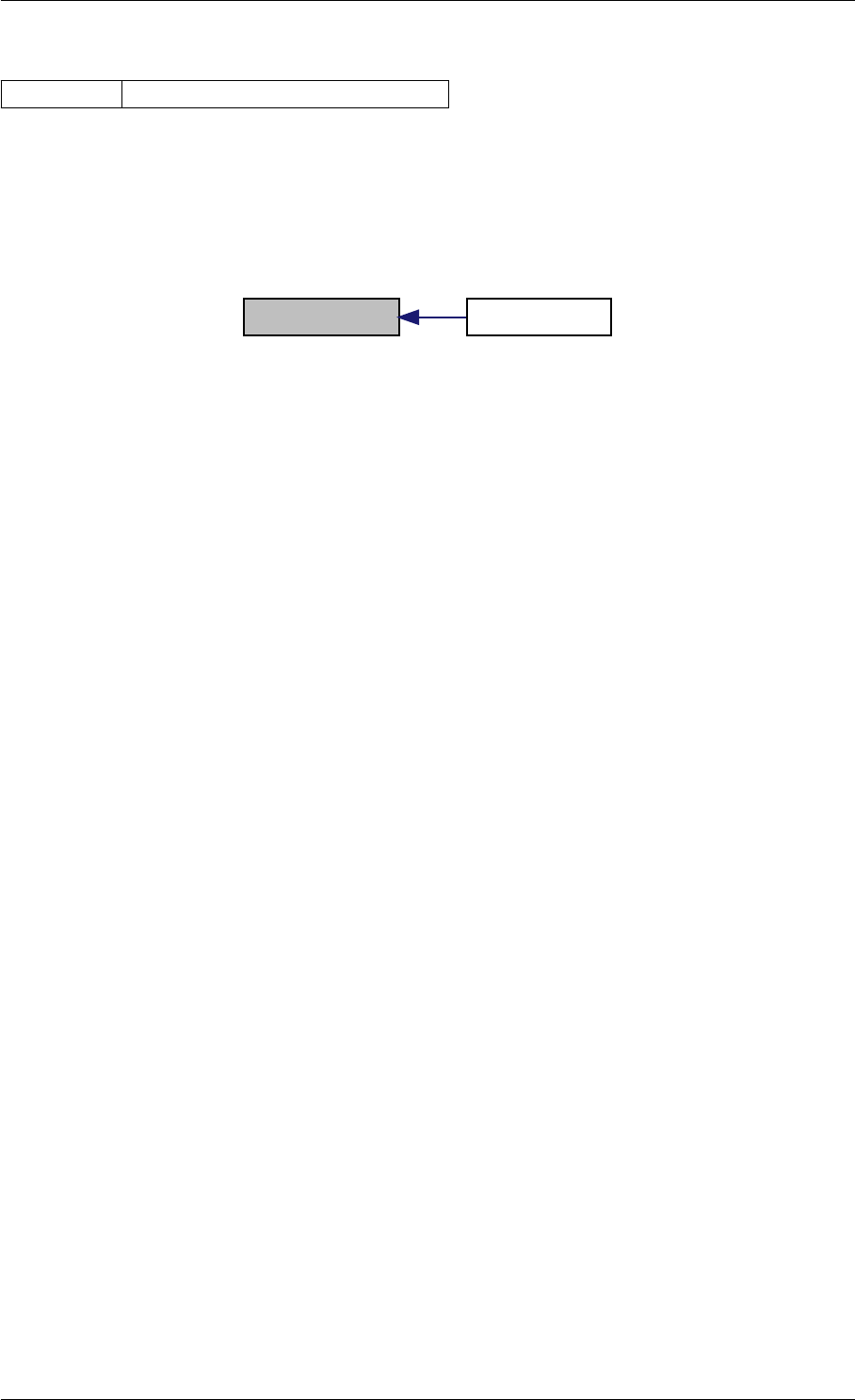
10.2 AmiVector Class Reference 89
Parameters
nMaxFrames number of frams to go back in stacktrace
Definition at line 68 of file exception.h.
Here is the caller graph for this function:
storeBacktrace AmiException
10.2 AmiVector Class Reference
#include <vector.h>
Public Member Functions
•AmiVector (const long int length)
•AmiVector (std::vector<realtype >rvec)
•AmiVector (const AmiVector &vold)
•AmiVector &operator= (AmiVector const &other)
•realtype ∗data ()
• const realtype ∗data () const
• N_Vector getNVector () const
• std::vector<realtype >const & getVector () const
• int getLength () const
• void reset ()
• void minus ()
• void set (realtype val)
•realtype &operator[ ] (int pos)
•realtype &at (int pos)
10.2.1 Detailed Description
AmiVector class provides a generic interface to the NVector_Serial struct
Definition at line 11 of file vector.h.
10.2.2 Constructor & Destructor Documentation
10.2.2.1 AmiVector() [1/3]
AmiVector (
const long int length )
Creates an std::vector<realtype>and attaches the data pointer to a newly created N_Vector_Serial. Using N←-
_VMake_Serial ensures that the N_Vector module does not try to deallocate the data vector when calling N_V←-
Destroy_Serial
Generated by Doxygen

90 CONTENTS
Parameters
length number of elements in vector
Returns
new AmiVector instance
Definition at line 23 of file vector.h.
10.2.2.2 AmiVector() [2/3]
AmiVector (
std::vector<realtype >rvec )
constructor from std::vector, copies data from std::vector and constructs an nvec that points to the data
Parameters
rvec vector from which the data will be copied
Returns
new AmiVector instance
Definition at line 34 of file vector.h.
10.2.2.3 AmiVector() [3/3]
AmiVector (
const AmiVector &vold )
copy constructor
Parameters
vold vector from which the data will be copied
Definition at line 43 of file vector.h.
10.2.3 Member Function Documentation
Generated by Doxygen
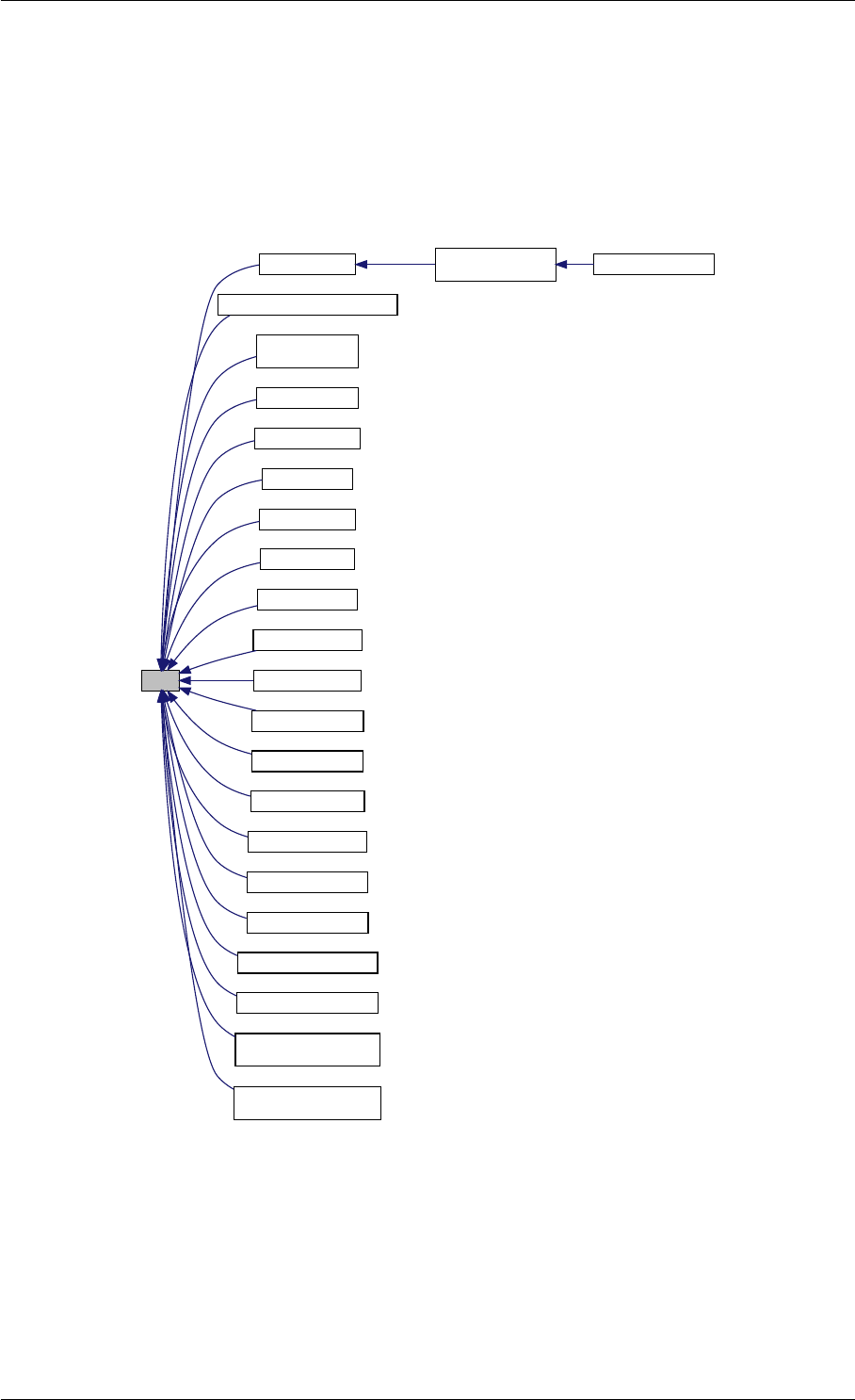
92 CONTENTS
Returns
pointer to data array
Definition at line 63 of file vector.h.
Here is the caller graph for this function:
data
amici::Model::fx0
amici::Model::fx0_fixedParameters
amici::Model::fsx0
_fixedParameters
amici::Model::fsx0
amici::Model::fstau
amici::Model::fz
amici::Model::fsz
amici::Model::frz
amici::Model::fsrz
amici::Model::fdzdp
amici::Model::fdzdx
amici::Model::fdrzdp
amici::Model::fdrzdx
amici::Model::fdeltax
amici::Model::fdeltasx
amici::Model::fdeltaxB
amici::Model::fdeltaqB
amici::Model_DAE::fJDiag
amici::Model_ODE::fJDiag
amici::NewtonSolverDense
::solveLinearSystem
amici::NewtonSolverSparse
::solveLinearSystem
amici::Model::initialize
States amici::Model::initialize
10.2.3.3 data() [2/2]
const realtype∗data ( ) const
const data accessor
Generated by Doxygen

10.2 AmiVector Class Reference 93
Returns
const pointer to data array
Definition at line 70 of file vector.h.
10.2.3.4 getNVector()
N_Vector getNVector ( ) const
N_Vector accessor
Returns
N_Vector
Definition at line 77 of file vector.h.
Generated by Doxygen
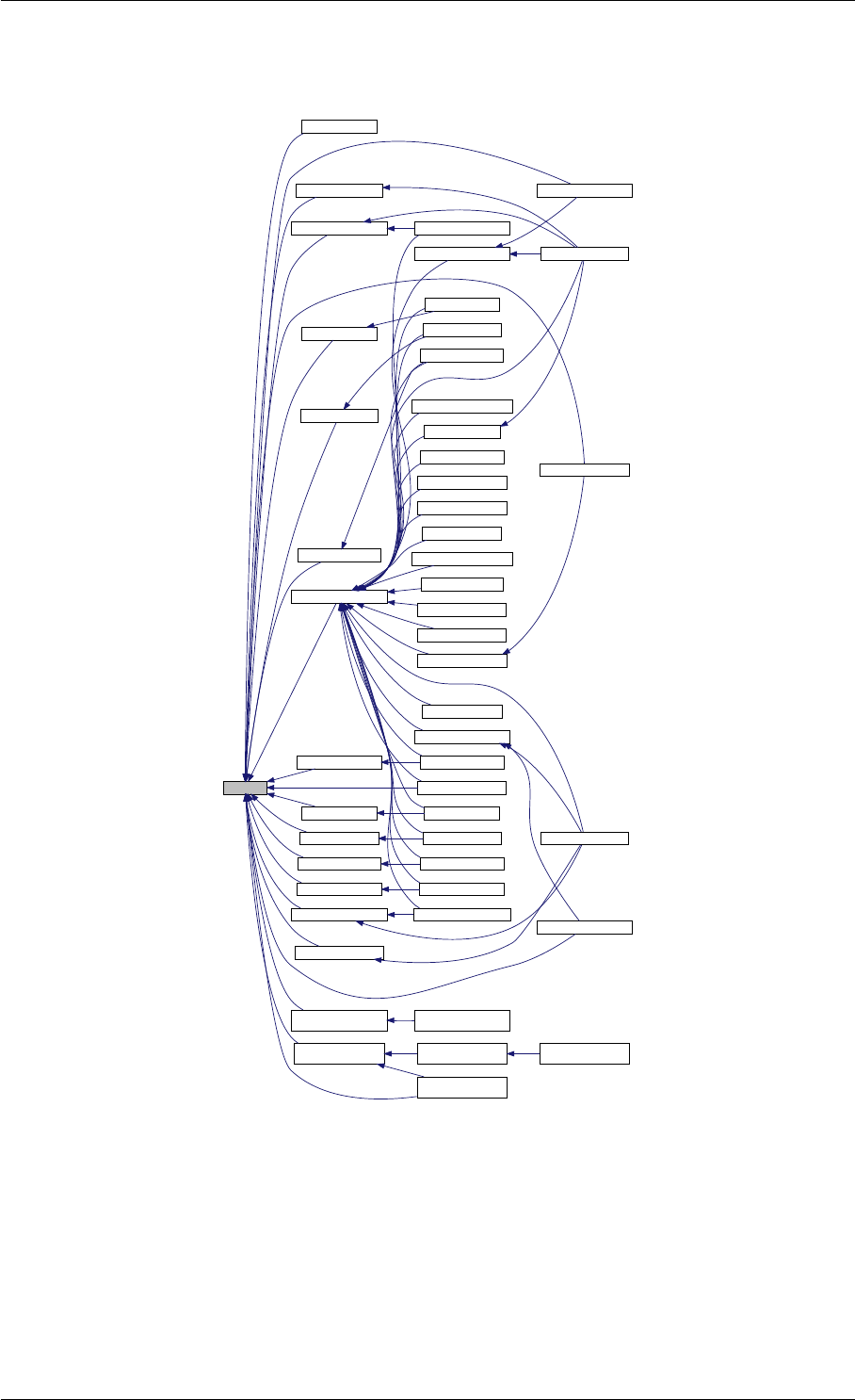
94 CONTENTS
Here is the caller graph for this function:
getNVector
amici::Model::fdeltasx
amici::Model::computeX_pos
amici::Model_DAE::fJDiag
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fJDiag
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::froot
amici::Model_DAE::fxdot
amici::Model_DAE::fsxdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::froot
amici::Model_ODE::fxdot
amici::Model_ODE::fsxdot
amici::NewtonSolverIterative
::linsolveSPBCG
amici::SteadystateProblem
::getWrmsNorm
amici::SteadystateProblem
::applyNewtonsMethod
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::froot
amici::Model_DAE::fxdot
amici::Model_DAE::fdxdotdp amici::Model_DAE::fsxdot
amici::Model_DAE::fM
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_DAE::fqBdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::froot
amici::Model_ODE::fxdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_ODE::fqBdot
amici::NewtonSolverIterative
::solveLinearSystem
amici::SteadystateProblem
::checkConvergence
amici::SteadystateProblem
::getSteadystateSimulation
10.2.3.5 getVector()
std::vector<realtype>const& getVector ( ) const
Vector accessor
Generated by Doxygen
10.2 AmiVector Class Reference 95
Returns
Vector
Definition at line 84 of file vector.h.
Here is the caller graph for this function:
getVector amici::ForwardProblem
::workForwardProblem
10.2.3.6 getLength()
int getLength ( ) const
returns the length of the vector
Generated by Doxygen
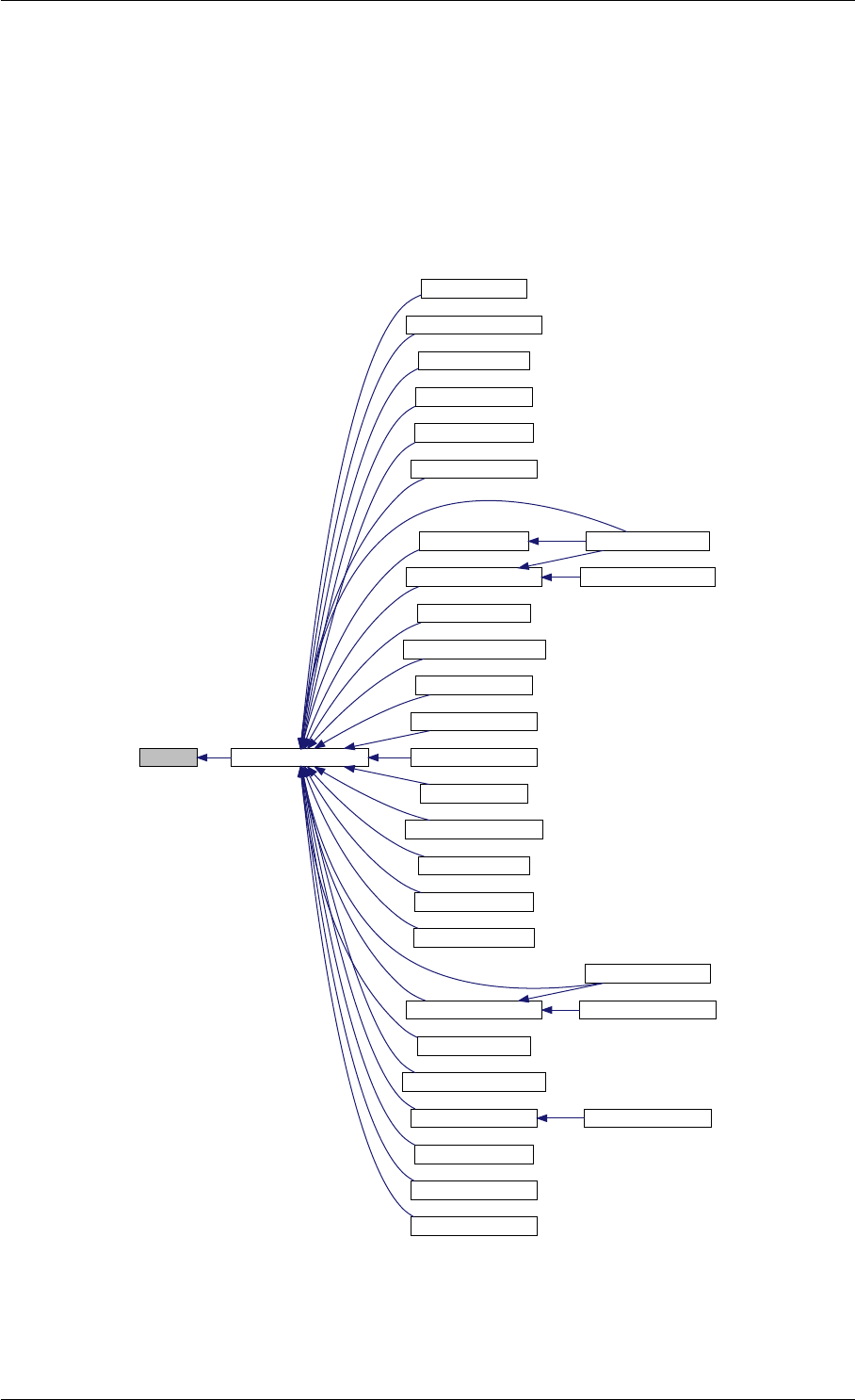
96 CONTENTS
Returns
length
Definition at line 91 of file vector.h.
Here is the caller graph for this function:
getLength amici::Model::computeX_pos
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::froot
amici::Model_DAE::fxdot
amici::Model_DAE::fJDiag
amici::Model_DAE::fdxdotdp
amici::Model_DAE::fsxdotamici::Model_DAE::fM
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_DAE::fqBdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::froot
amici::Model_ODE::fxdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_ODE::fqBdot
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fJDiag
Generated by Doxygen
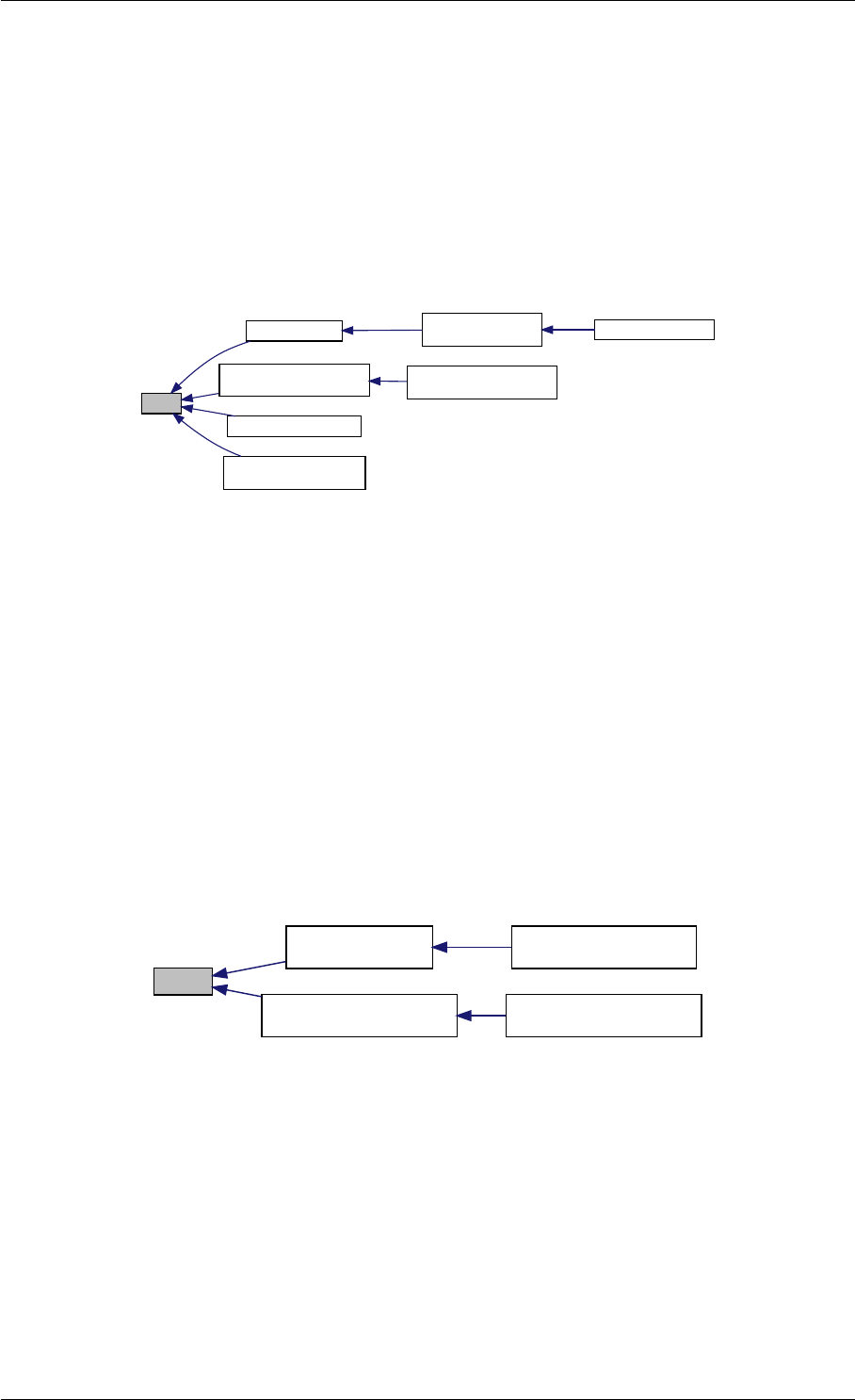
10.2 AmiVector Class Reference 97
10.2.3.7 reset()
void reset ( )
resets the Vector by filling with zero values
Definition at line 97 of file vector.h.
Here is the caller graph for this function:
reset
amici::Model::fx0
amici::NewtonSolverIterative
::linsolveSPBCG
amici::Solver::setupAMIB
amici::SteadystateProblem
::applyNewtonsMethod
amici::Model::initialize
States amici::Model::initialize
amici::NewtonSolverIterative
::solveLinearSystem
10.2.3.8 minus()
void minus ( )
changes the sign of data elements
Definition at line 103 of file vector.h.
Here is the caller graph for this function:
minus
amici::NewtonSolver
::getStep
amici::NewtonSolverIterative
::linsolveSPBCG
amici::SteadystateProblem
::applyNewtonsMethod
amici::NewtonSolverIterative
::solveLinearSystem
10.2.3.9 set()
void set (
realtype val )
sets all data elements to a specific value
Generated by Doxygen
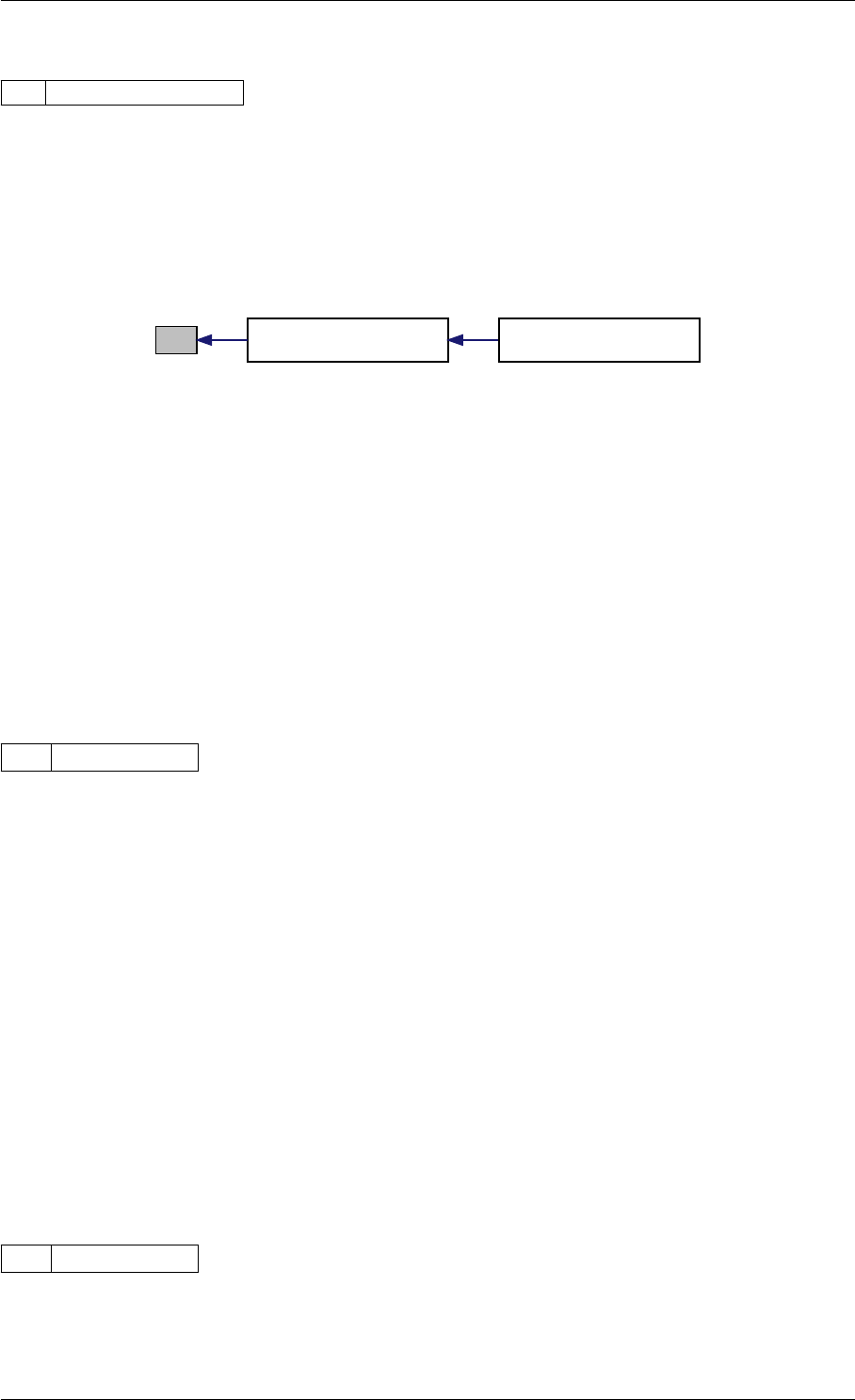
98 CONTENTS
Parameters
val value for data elements
Definition at line 112 of file vector.h.
Here is the caller graph for this function:
set amici::NewtonSolverIterative
::linsolveSPBCG
amici::NewtonSolverIterative
::solveLinearSystem
10.2.3.10 operator[]()
realtype& operator[ ] (
int pos )
accessor to data elements of the vector
Parameters
pos index of element
Returns
element
Definition at line 120 of file vector.h.
10.2.3.11 at()
realtype& at (
int pos )
accessor to data elements of the vector
Parameters
pos index of element
Generated by Doxygen
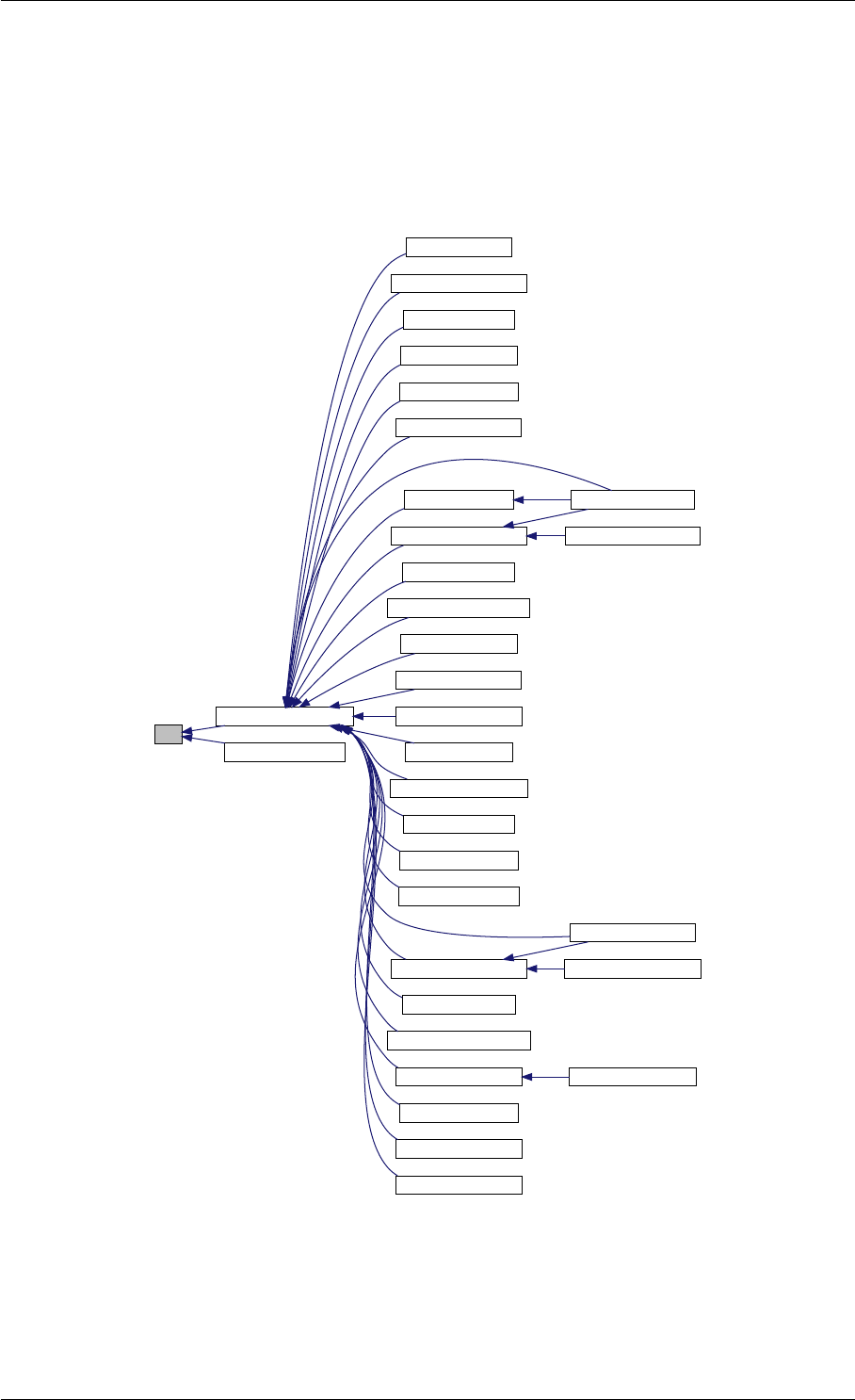
10.3 AmiVectorArray Class Reference 99
Returns
element
Definition at line 128 of file vector.h.
Here is the caller graph for this function:
at
amici::Model::computeX_pos
amici::Solver::setupAMIB
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::froot
amici::Model_DAE::fxdot
amici::Model_DAE::fJDiag
amici::Model_DAE::fdxdotdp
amici::Model_DAE::fsxdotamici::Model_DAE::fM
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_DAE::fqBdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::froot
amici::Model_ODE::fxdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_ODE::fqBdot
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fJDiag
10.3 AmiVectorArray Class Reference
#include <vector.h>
Generated by Doxygen

100 CONTENTS
Public Member Functions
•AmiVectorArray (long int length_inner, long int length_outer)
•AmiVectorArray (const AmiVectorArray &vaold)
•realtype ∗data (int pos)
• const realtype ∗data (int pos) const
•realtype &at (int ipos, int jpos)
• N_Vector ∗getNVectorArray ()
• N_Vector getNVector (int pos)
•AmiVector &operator[ ] (int pos)
• const AmiVector &operator[ ] (int pos) const
• int getLength () const
• void reset ()
10.3.1 Detailed Description
AmiVectorArray class. provides a generic interface to arrays of NVector_Serial structs
Definition at line 148 of file vector.h.
10.3.2 Constructor & Destructor Documentation
10.3.2.1 AmiVectorArray() [1/2]
AmiVectorArray (
long int length_inner,
long int length_outer )
creates an std::vector<realype>and attaches the data pointer to a newly created N_VectorArray using Clone←-
VectorArrayEmpty ensures that the N_Vector module does not try to deallocate the data vector when calling N_V←-
DestroyVectorArray_Serial
Parameters
length_inner length of vectors
length_outer number of vectors
Returns
New AmiVectorArray instance
Definition at line 159 of file vector.h.
10.3.2.2 AmiVectorArray() [2/2]
AmiVectorArray (
const AmiVectorArray &vaold )
copy constructor
Generated by Doxygen
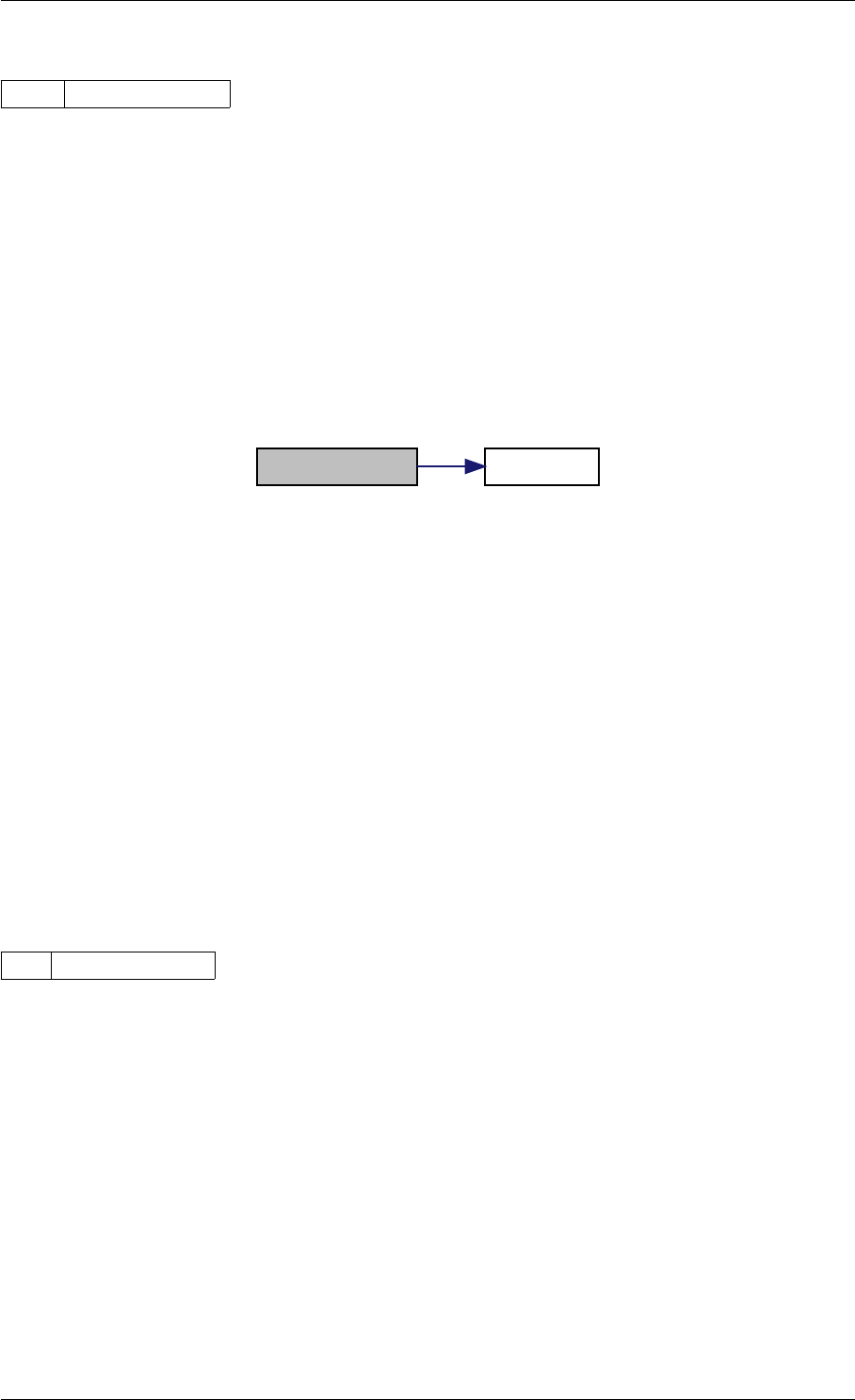
10.3 AmiVectorArray Class Reference 101
Parameters
vaold object to copy from
Returns
new AmiVectorArray instance
Definition at line 173 of file vector.h.
Here is the call graph for this function:
AmiVectorArray getLength
10.3.3 Member Function Documentation
10.3.3.1 data() [1/2]
realtype∗data (
int pos )
accessor to data of AmiVector elements
Parameters
pos index of AmiVector
Generated by Doxygen
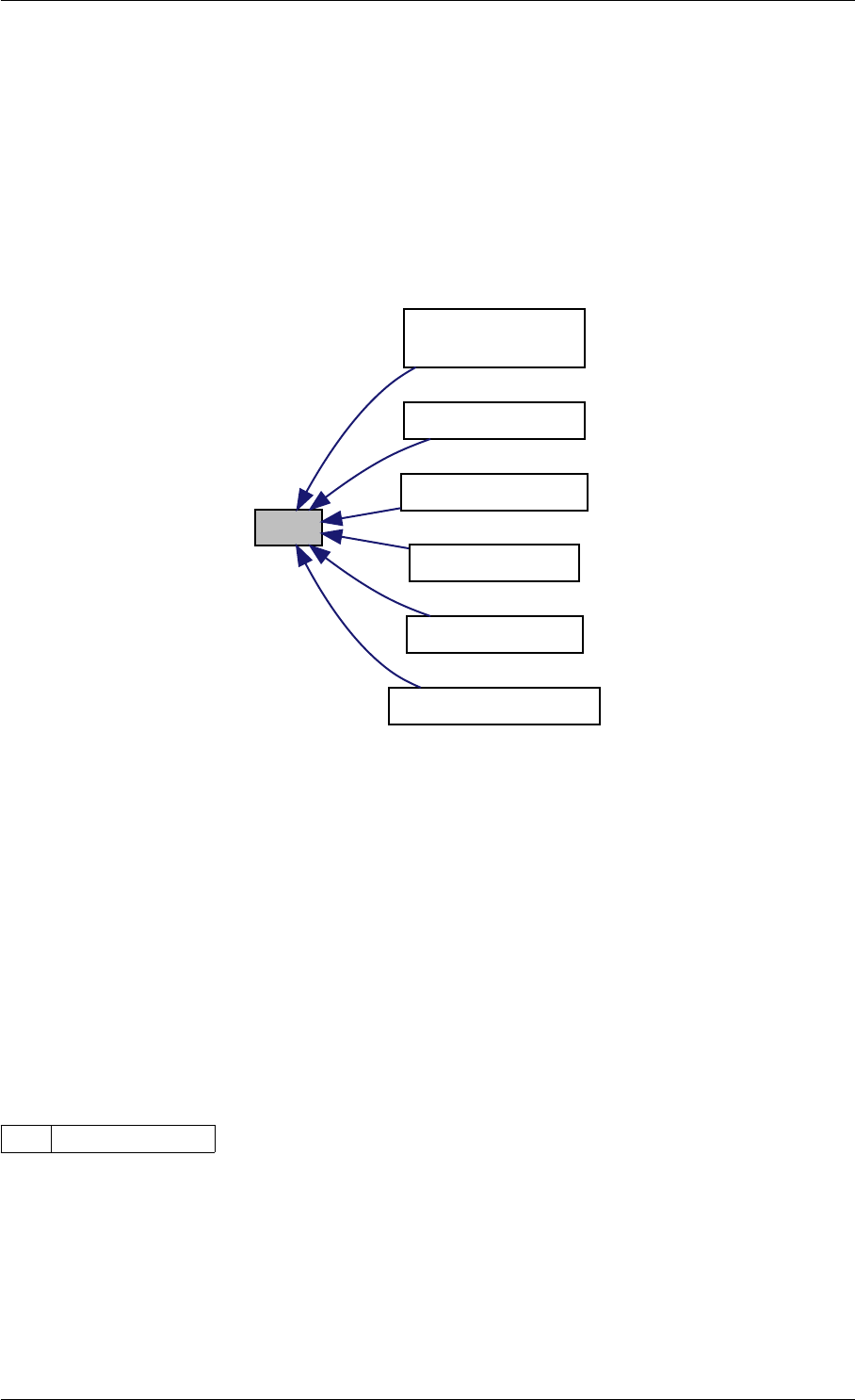
102 CONTENTS
Returns
pointer to data array
Definition at line 184 of file vector.h.
Here is the caller graph for this function:
data
amici::Model::fsx0
_fixedParameters
amici::Model::fsx0
amici::Model::fstau
amici::Model::fsz
amici::Model::fsrz
amici::Model::fdeltasx
10.3.3.2 data() [2/2]
const realtype∗data (
int pos ) const
const accessor to data of AmiVector elements
Parameters
pos index of AmiVector
Returns
const pointer to data array
Definition at line 192 of file vector.h.
Generated by Doxygen
10.3 AmiVectorArray Class Reference 103
10.3.3.3 at()
realtype& at (
int ipos,
int jpos )
accessor to elements of AmiVector elements
Parameters
ipos inner index in AmiVector
jpos outer index in AmiVectorArray
Returns
element
Definition at line 201 of file vector.h.
Here is the caller graph for this function:
at
amici::Model::fsJz
amici::NewtonSolver
::computeNewtonSensis
10.3.3.4 getNVectorArray()
N_Vector∗getNVectorArray ( )
accessor to NVectorArray
Returns
N_VectorArray
Definition at line 208 of file vector.h.
10.3.3.5 getNVector()
N_Vector getNVector (
int pos )
accessor to NVector element
Generated by Doxygen
104 CONTENTS
Parameters
pos index of corresponding AmiVector
Returns
N_Vector
Definition at line 216 of file vector.h.
10.3.3.6 operator[]() [1/2]
AmiVector& operator[ ] (
int pos )
accessor to AmiVector elements
Parameters
pos index of AmiVector
Returns
AmiVector
Definition at line 224 of file vector.h.
10.3.3.7 operator[]() [2/2]
const AmiVector& operator[ ] (
int pos ) const
const accessor to AmiVector elements
Parameters
pos index of AmiVector
Returns
const AmiVector
Definition at line 232 of file vector.h.
Generated by Doxygen
10.4 BackwardProblem Class Reference 105
10.3.3.8 getLength()
int getLength ( ) const
length of AmiVectorArray
Returns
length
Definition at line 239 of file vector.h.
Here is the caller graph for this function:
getLength AmiVectorArray
10.3.3.9 reset()
void reset ( )
resets every AmiVector in AmiVectorArray
Definition at line 244 of file vector.h.
Here is the caller graph for this function:
reset amici::Model::fsx0
10.4 BackwardProblem Class Reference
class to solve backwards problems.
#include <backwardproblem.h>
Generated by Doxygen

106 CONTENTS
Public Member Functions
• void workBackwardProblem ()
•BackwardProblem (const ForwardProblem ∗fwd)
•realtype gett () const
• int getwhich () const
• int ∗getwhichptr ()
•AmiVector ∗getxBptr ()
•AmiVector ∗getxQBptr ()
•AmiVector ∗getdxBptr ()
• std::vector<realtype >const & getdJydx () const
10.4.1 Detailed Description
solves the backwards problem for adjoint sensitivity analysis and handles events and data-points
Definition at line 23 of file backwardproblem.h.
10.4.2 Constructor & Destructor Documentation
10.4.2.1 BackwardProblem()
BackwardProblem (
const ForwardProblem ∗fwd )
Construct backward problem from forward problem
Parameters
fwd pointer to corresponding forward problem
Returns
new BackwardProblem instance
Definition at line 18 of file backwardproblem.cpp.
Generated by Doxygen
10.4 BackwardProblem Class Reference 107
Here is the call graph for this function:
BackwardProblem
amici::ForwardProblem
::getTime
amici::ForwardProblem
::getRootCounter
10.4.3 Member Function Documentation
10.4.3.1 workBackwardProblem()
void workBackwardProblem ( )
workBackwardProblem solves the backward problem. if adjoint sensitivities are enabled this will also compute
sensitivies workForwardProblem should be called before this is function is called
Definition at line 42 of file backwardproblem.cpp.
Here is the call graph for this function:
workBackwardProblem amici::Solver::getSensitivity
Order
10.4.3.2 gett()
realtype gett ( ) const
accessor for t
Generated by Doxygen
108 CONTENTS
Returns
t
Definition at line 32 of file backwardproblem.h.
Here is the caller graph for this function:
gett amici::Solver::setupAMIB
10.4.3.3 getwhich()
int getwhich ( ) const
accessor for which
Returns
which
Definition at line 39 of file backwardproblem.h.
Here is the caller graph for this function:
getwhich amici::Solver::setupAMIB
Generated by Doxygen
10.4 BackwardProblem Class Reference 109
10.4.3.4 getwhichptr()
int∗getwhichptr ( )
accessor for pointer to which
Returns
which
Definition at line 46 of file backwardproblem.h.
Here is the caller graph for this function:
getwhichptr amici::Solver::setupAMIB
10.4.3.5 getxBptr()
AmiVector∗getxBptr ( )
accessor for pointer to xB
Returns
&xB
Definition at line 53 of file backwardproblem.h.
Here is the caller graph for this function:
getxBptr amici::Solver::setupAMIB
Generated by Doxygen
110 CONTENTS
10.4.3.6 getxQBptr()
AmiVector∗getxQBptr ( )
accessor for pointer to xQB
Returns
&xQB
Definition at line 60 of file backwardproblem.h.
Here is the caller graph for this function:
getxQBptr amici::Solver::setupAMIB
10.4.3.7 getdxBptr()
AmiVector∗getdxBptr ( )
accessor for pointer to dxB
Returns
&dxB
Definition at line 67 of file backwardproblem.h.
Here is the caller graph for this function:
getdxBptr amici::Solver::setupAMIB
Generated by Doxygen
10.5 CvodeException Class Reference 111
10.4.3.8 getdJydx()
std::vector<realtype>const& getdJydx ( ) const
accessor for dJydx
Returns
dJydx
Definition at line 74 of file backwardproblem.h.
Here is the caller graph for this function:
getdJydx amici::Solver::setupAMIB
10.5 CvodeException Class Reference
cvode exception handler class
#include <exception.h>
Inheritance diagram for CvodeException:
CvodeException
AmiException
exception
Public Member Functions
•CvodeException (const int error_code, const char ∗function)
Generated by Doxygen
112 CONTENTS
10.5.1 Detailed Description
Definition at line 117 of file exception.h.
10.5.2 Constructor & Destructor Documentation
10.5.2.1 CvodeException()
CvodeException (
const int error_code,
const char ∗function )
constructor
Parameters
error_code error code returned by cvode function
function cvode function name
Definition at line 123 of file exception.h.
10.6 ExpData Class Reference
ExpData carries all information about experimental or condition-specific data.
#include <edata.h>
Public Member Functions
•ExpData ()
•ExpData (int nytrue, int nztrue, int nmaxevent)
•ExpData (int nytrue, int nztrue, int nmaxevent, std::vector<realtype >ts)
•ExpData (int nytrue, int nztrue, int nmaxevent, std::vector<realtype >ts, std::vector<realtype >
observedData, std::vector<realtype >observedDataStdDev, std::vector<realtype >observedEvents,
std::vector<realtype >observedEventsStdDev)
•ExpData (const Model &model)
•ExpData (const ReturnData &rdata, realtype sigma_y, realtype sigma_z)
•ExpData (const ReturnData &rdata, std::vector<realtype >sigma_y, std::vector<realtype >sigma_z)
•ExpData (const ExpData &other)
Copy constructor.
• const int nt () const
• void setTimepoints (const std::vector<realtype >&ts)
• std::vector<realtype >const & getTimepoints () const
•realtype getTimepoint (int it) const
• void setObservedData (const std::vector<realtype >&observedData)
• void setObservedData (const std::vector<realtype >&observedData, int iy)
• bool isSetObservedData (int it, int iy) const
Generated by Doxygen

10.6 ExpData Class Reference 113
• std::vector<realtype >const & getObservedData () const
• const realtype ∗getObservedDataPtr (int it) const
• void setObservedDataStdDev (const std::vector<realtype >&observedDataStdDev)
• void setObservedDataStdDev (const realtype stdDev)
• void setObservedDataStdDev (const std::vector<realtype >&observedDataStdDev, int iy)
• void setObservedDataStdDev (const realtype stdDev, int iy)
• bool isSetObservedDataStdDev (int it, int iy) const
• std::vector<realtype >const & getObservedDataStdDev () const
• const realtype ∗getObservedDataStdDevPtr (int it) const
• void setObservedEvents (const std::vector<realtype >&observedEvents)
• void setObservedEvents (const std::vector<realtype >&observedEvents, int iz)
• bool isSetObservedEvents (int ie, int iz) const
• std::vector<realtype >const & getObservedEvents () const
• const realtype ∗getObservedEventsPtr (int ie) const
• void setObservedEventsStdDev (const std::vector<realtype >&observedEventsStdDev)
• void setObservedEventsStdDev (const realtype stdDev)
• void setObservedEventsStdDev (const std::vector<realtype >&observedEventsStdDev, int iz)
• void setObservedEventsStdDev (const realtype stdDev, int iz)
• bool isSetObservedEventsStdDev (int ie, int iz) const
• std::vector<realtype >const & getObservedEventsStdDev () const
• const realtype ∗getObservedEventsStdDevPtr (int ie) const
Public Attributes
• const int nytrue
• const int nztrue
• const int nmaxevent
• std::vector<realtype >fixedParameters
• std::vector<realtype >fixedParametersPreequilibration
• std::vector<realtype >fixedParametersPresimulation
•realtype t_presim = 0
duration of pre-simulation if this is >0, presimualation will be performed from (model->t0 - t_presim) to model->t0
using the fixedParameters in fixedParametersPresimulation
Protected Member Functions
• void checkDataDimension (std::vector<realtype >input, const char ∗fieldname) const
• void checkEventsDimension (std::vector<realtype >input, const char ∗fieldname) const
• void checkSigmaPositivity (std::vector<realtype >sigmaVector, const char ∗vectorName) const
• void checkSigmaPositivity (realtype sigma, const char ∗sigmaName) const
Protected Attributes
• std::vector<realtype >ts
• std::vector<realtype >observedData
• std::vector<realtype >observedDataStdDev
• std::vector<realtype >observedEvents
• std::vector<realtype >observedEventsStdDev
10.6.1 Detailed Description
Definition at line 14 of file edata.h.
Generated by Doxygen

114 CONTENTS
10.6.2 Constructor & Destructor Documentation
10.6.2.1 ExpData() [1/8]
ExpData ( )
default constructor
Definition at line 14 of file edata.cpp.
10.6.2.2 ExpData() [2/8]
ExpData (
int nytrue,
int nztrue,
int nmaxevent )
constructor that only initializes dimensions
Parameters
nytrue
nztrue
nmaxevent
Definition at line 16 of file edata.cpp.
10.6.2.3 ExpData() [3/8]
ExpData (
int nytrue,
int nztrue,
int nmaxevent,
std::vector<realtype >ts )
constructor that initializes timepoints from vectors
Parameters
nytrue (dimension: scalar)
nztrue (dimension: scalar)
nmaxevent (dimension: scalar)
ts (dimension: nt)
Definition at line 21 of file edata.cpp.
Generated by Doxygen
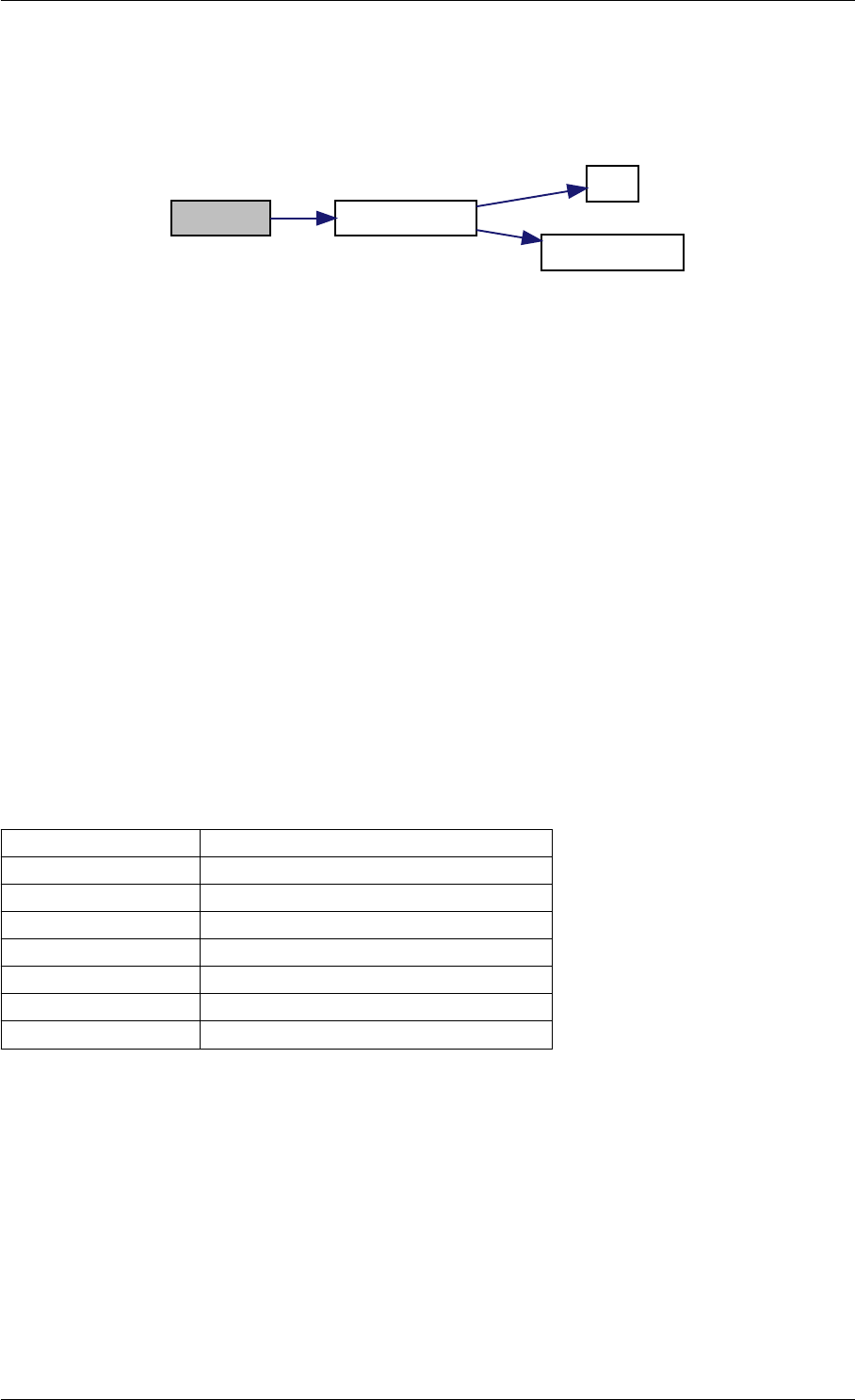
10.6 ExpData Class Reference 115
Here is the call graph for this function:
ExpData setTimepoints
nt
amici::getNaN
10.6.2.4 ExpData() [4/8]
ExpData (
int nytrue,
int nztrue,
int nmaxevent,
std::vector<realtype >ts,
std::vector<realtype >observedData,
std::vector<realtype >observedDataStdDev,
std::vector<realtype >observedEvents,
std::vector<realtype >observedEventsStdDev )
constructor that initializes timepoints and data from vectors
Parameters
nytrue (dimension: scalar)
nztrue (dimension: scalar)
nmaxevent (dimension: scalar)
ts (dimension: nt)
observedData (dimension: nt x nytrue, row-major)
observedDataStdDev (dimension: nt x nytrue, row-major)
observedEvents (dimension: nmaxevent x nztrue, row-major)
observedEventsStdDev (dimension: nmaxevent x nztrue, row-major)
Definition at line 28 of file edata.cpp.
Generated by Doxygen
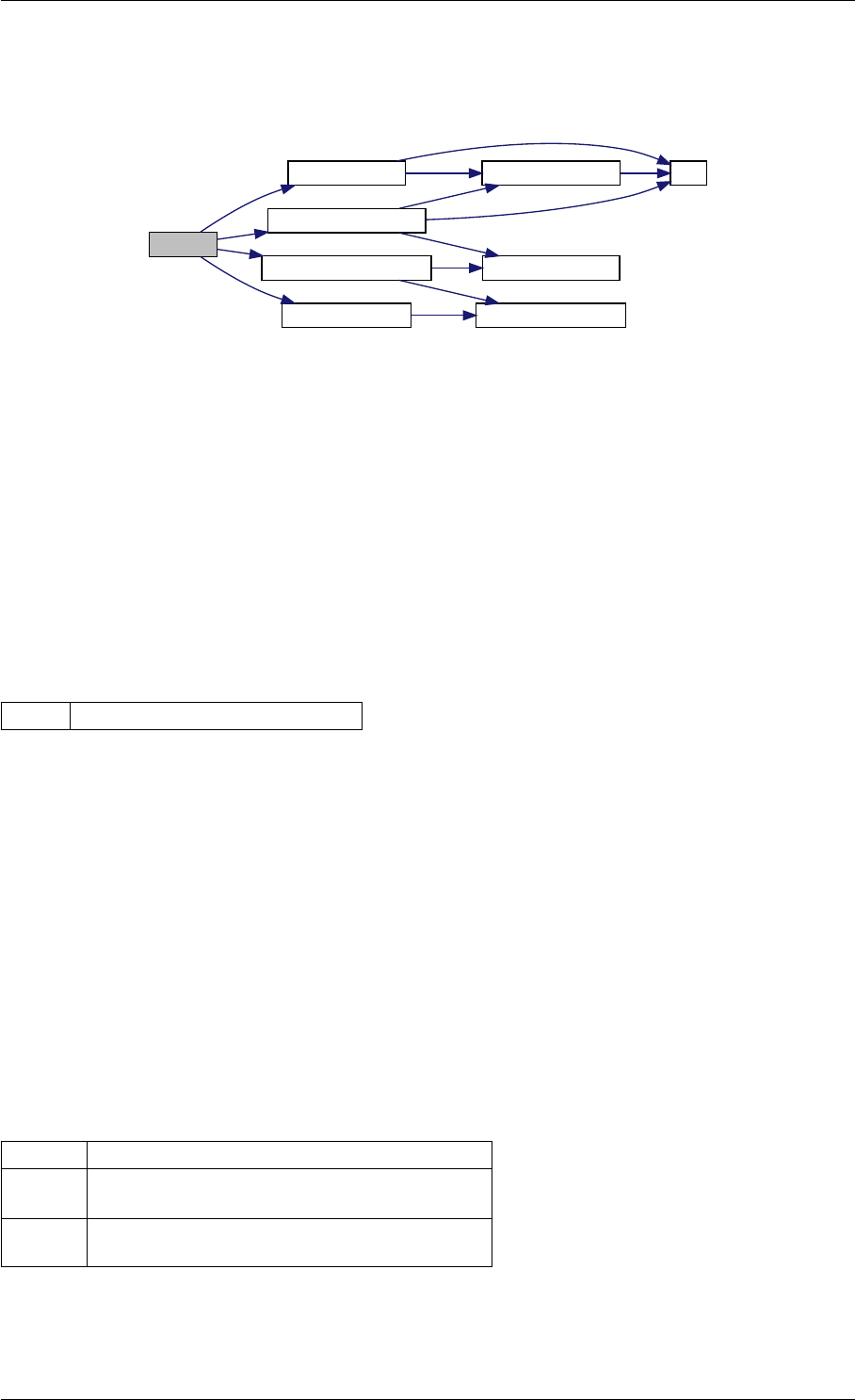
116 CONTENTS
Here is the call graph for this function:
ExpData
setObservedData
setObservedDataStdDev
setObservedEvents
setObservedEventsStdDev
checkDataDimension nt
checkSigmaPositivity
checkEventsDimension
10.6.2.5 ExpData() [5/8]
ExpData (
const Model &model )
constructor that initializes with Model
Parameters
model pointer to model specification object
Definition at line 42 of file edata.cpp.
10.6.2.6 ExpData() [6/8]
ExpData (
const ReturnData &rdata,
realtype sigma_y,
realtype sigma_z )
constructor that initializes with returnData, adds
Parameters
rdata return data pointer with stored simulation results
sigma←-
_y
scalar standard deviations for all observables
sigma←-
_z
scalar standard deviations for all event observables
Definition at line 56 of file edata.cpp.
Generated by Doxygen
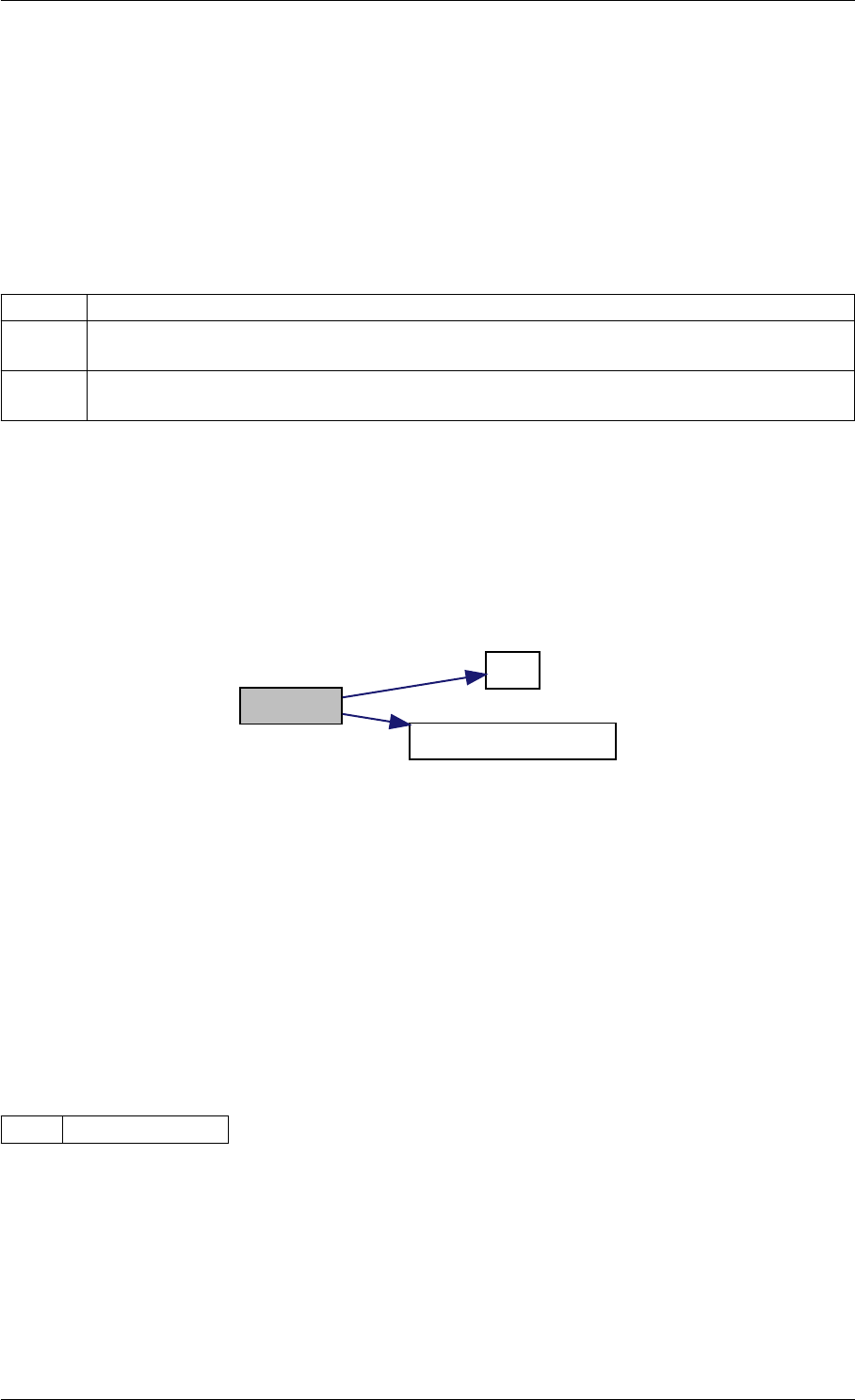
10.6 ExpData Class Reference 117
10.6.2.7 ExpData() [7/8]
ExpData (
const ReturnData &rdata,
std::vector<realtype >sigma_y,
std::vector<realtype >sigma_z )
constructor that initializes with returnData, adds
Parameters
rdata return data pointer with stored simulation results
sigma←-
_y
vector of standard deviations for observables (dimension: nytrue or nt x nytrue, row-major)
sigma←-
_z
vector of standard deviations for event observables (dimension: nztrue or nmaxevent x nztrue,
row-major)
Definition at line 61 of file edata.cpp.
Here is the call graph for this function:
ExpData
nt
checkSigmaPositivity
10.6.2.8 ExpData() [8/8]
ExpData (
const ExpData &other )
Parameters
other object to copy from
Definition at line 48 of file edata.cpp.
10.6.3 Member Function Documentation
Generated by Doxygen
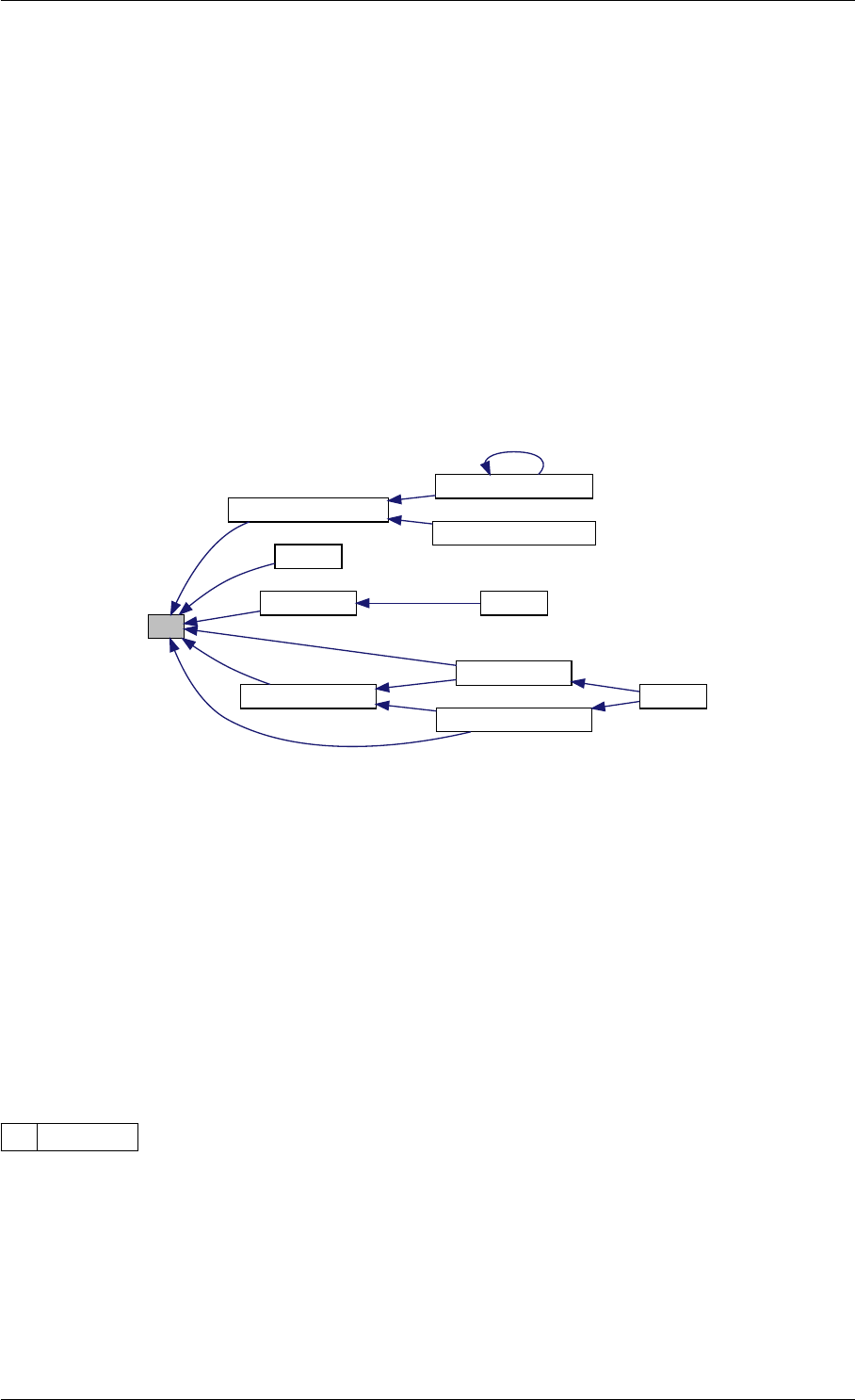
118 CONTENTS
10.6.3.1 nt()
const int nt ( ) const
number of timepoints
Returns
number of timepoints
Definition at line 110 of file edata.cpp.
Here is the caller graph for this function:
nt
amici::runAmiciSimulation
ExpData
setTimepoints
setObservedData
setObservedDataStdDev
checkDataDimension
amici.runAmiciSimulation
amici.runAmiciSimulations
ExpData
ExpData
10.6.3.2 setTimepoints()
void setTimepoints (
const std::vector<realtype >&ts )
set function that copies data from input to ExpData::ts
Parameters
ts timepoints
Definition at line 97 of file edata.cpp.
Generated by Doxygen
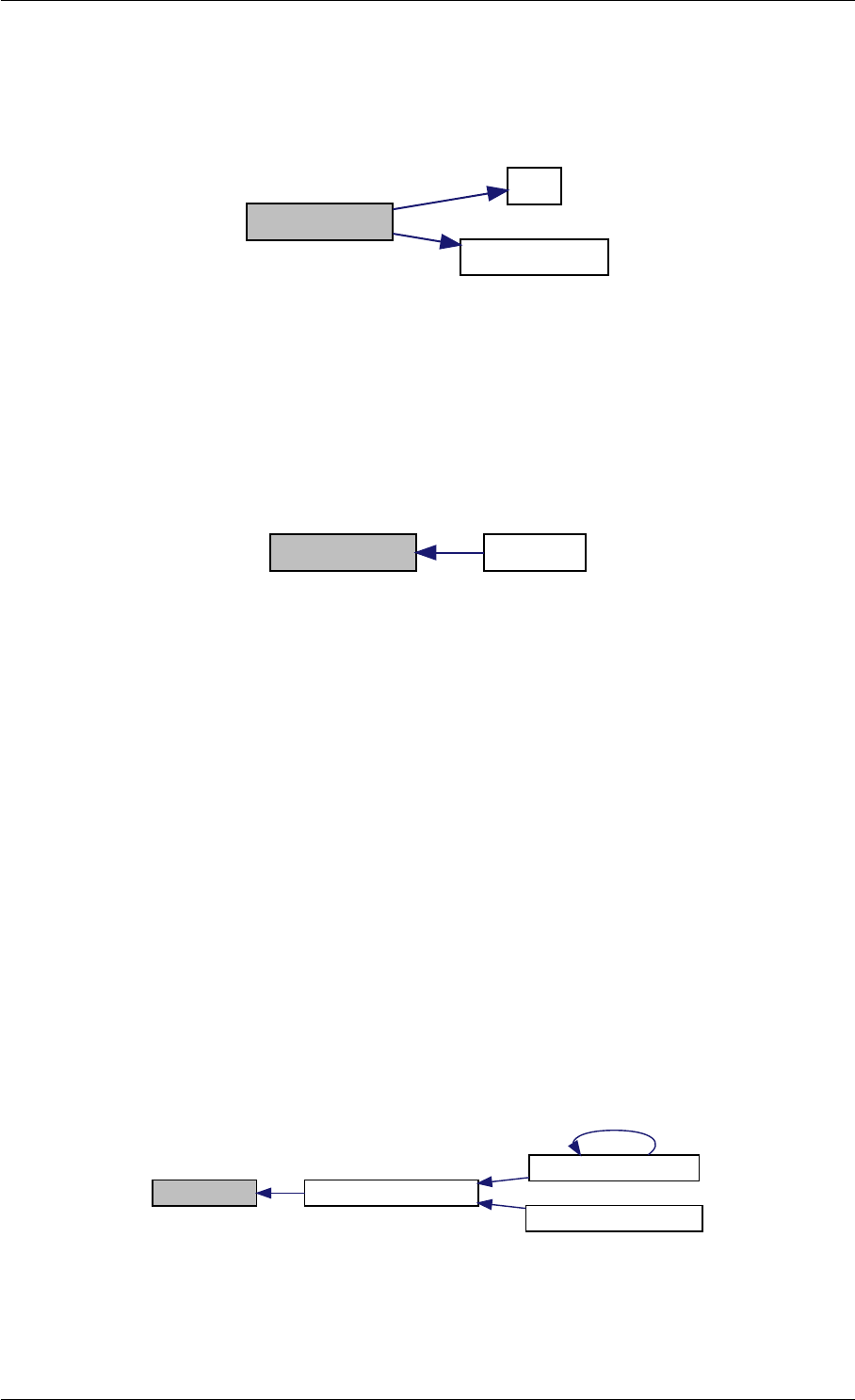
10.6 ExpData Class Reference 119
Here is the call graph for this function:
setTimepoints
nt
amici::getNaN
Here is the caller graph for this function:
setTimepoints ExpData
10.6.3.3 getTimepoints()
std::vector<realtype >const & getTimepoints ( ) const
get function that copies data from ExpData::ts to output
Returns
ExpData::ts
Definition at line 106 of file edata.cpp.
Here is the caller graph for this function:
getTimepoints amici::runAmiciSimulation
amici.runAmiciSimulation
amici.runAmiciSimulations
Generated by Doxygen

120 CONTENTS
10.6.3.4 getTimepoint()
realtype getTimepoint (
int it ) const
get function that returns timepoint at index
Parameters
it timepoint index
Returns
timepoint timepoint at index
Definition at line 114 of file edata.cpp.
10.6.3.5 setObservedData() [1/2]
void setObservedData (
const std::vector<realtype >&observedData )
set function that copies data from input to ExpData::my
Parameters
observedData observed data (dimension: nt x nytrue, row-major)
Definition at line 118 of file edata.cpp.
Here is the call graph for this function:
setObservedData
checkDataDimension
nt
Generated by Doxygen
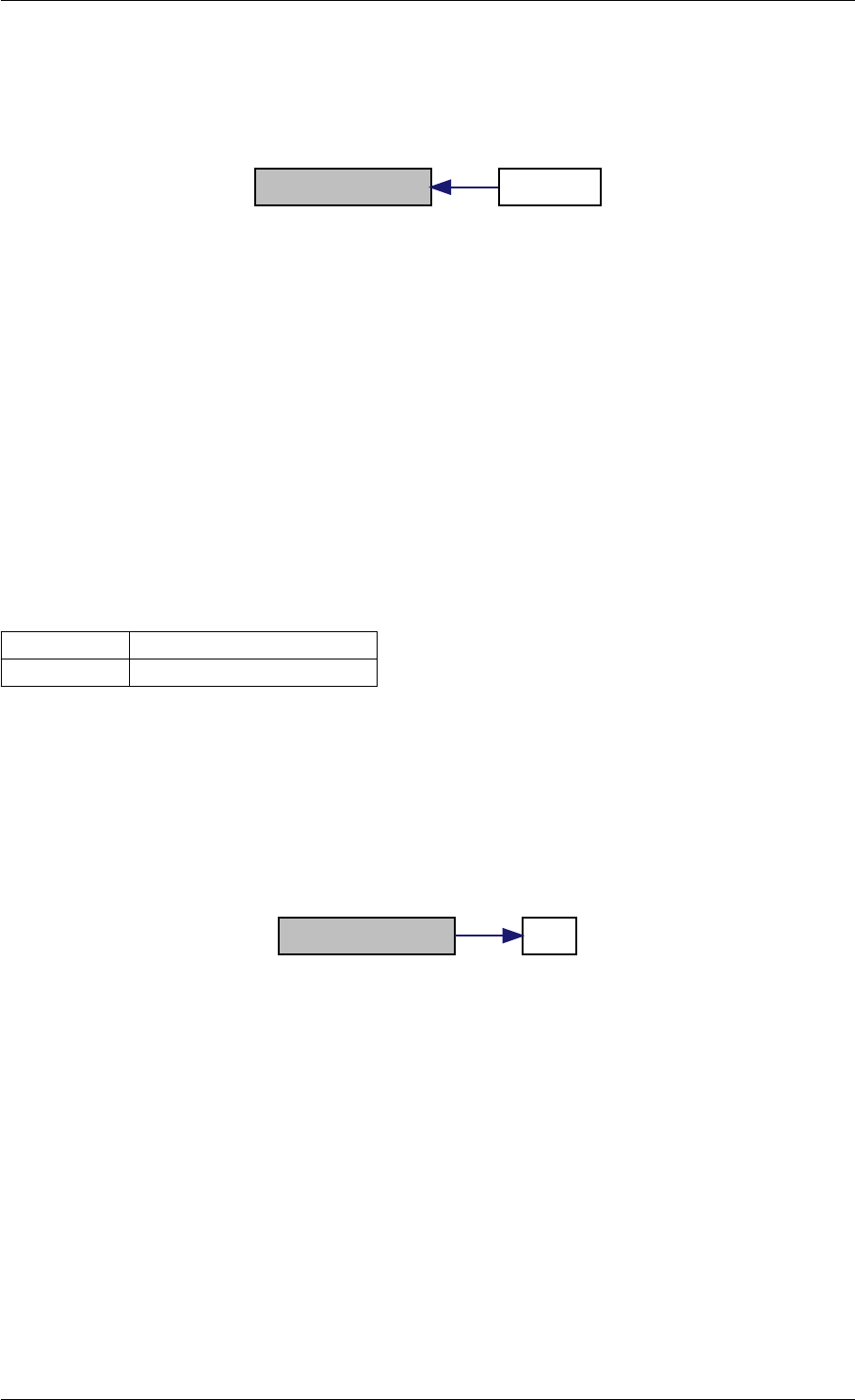
10.6 ExpData Class Reference 121
Here is the caller graph for this function:
setObservedData ExpData
10.6.3.6 setObservedData() [2/2]
void setObservedData (
const std::vector<realtype >&observedData,
int iy )
set function that copies observed data for specific observable
Parameters
observedData observed data (dimension: nt)
iy oberved data index
Definition at line 127 of file edata.cpp.
Here is the call graph for this function:
setObservedData nt
10.6.3.7 isSetObservedData()
bool isSetObservedData (
int it,
int iy ) const
get function that checks whether data at specified indices has been set
Generated by Doxygen
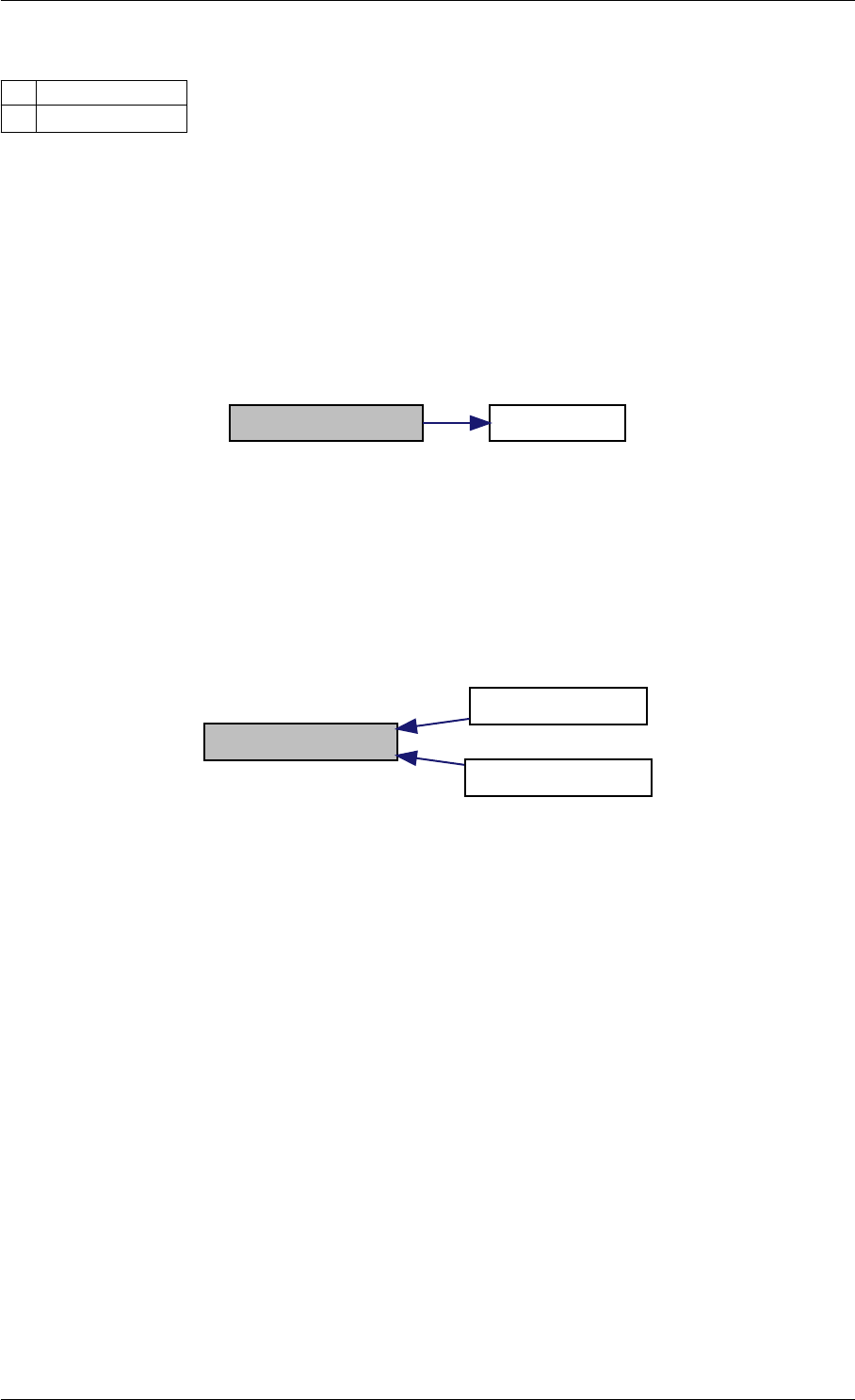
122 CONTENTS
Parameters
it time index
iy observable index
Returns
boolean specifying if data was set
Definition at line 135 of file edata.cpp.
Here is the call graph for this function:
isSetObservedData amici::isNaN
Here is the caller graph for this function:
isSetObservedData
amici::Model::fres
amici::Model::fsres
10.6.3.8 getObservedData()
std::vector<realtype >const & getObservedData ( ) const
get function that copies data from ExpData::observedData to output
Returns
observed data (dimension: nt x nytrue, row-major)
Definition at line 139 of file edata.cpp.
10.6.3.9 getObservedDataPtr()
const realtype ∗getObservedDataPtr (
int it ) const
get function that returns a pointer to observed data at index
Generated by Doxygen
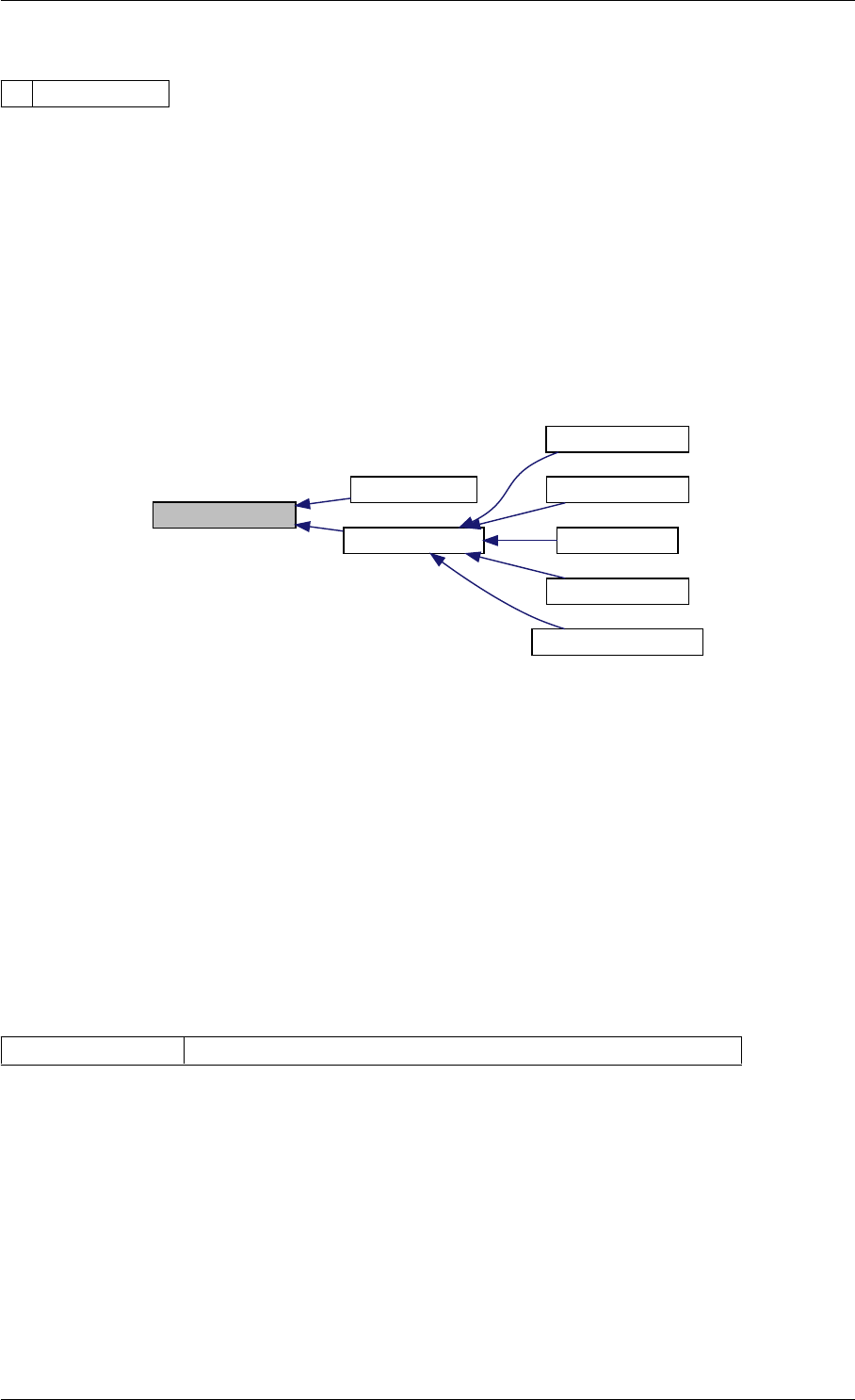
10.6 ExpData Class Reference 123
Parameters
it timepoint index
Returns
pointer to observed data at index (dimension: nytrue)
Definition at line 143 of file edata.cpp.
Here is the caller graph for this function:
getObservedDataPtr
amici::Model::fres
amici::Model::getmy
amici::Model::fdJydp
amici::Model::fdJydx
amici::Model::fJy
amici::Model::fdJydy
amici::Model::fdJydsigma
10.6.3.10 setObservedDataStdDev() [1/4]
void setObservedDataStdDev (
const std::vector<realtype >&observedDataStdDev )
set function that copies data from input to ExpData::observedDataStdDev
Parameters
observedDataStdDev standard deviation of observed data (dimension: nt x nytrue, row-major)
Definition at line 150 of file edata.cpp.
Generated by Doxygen
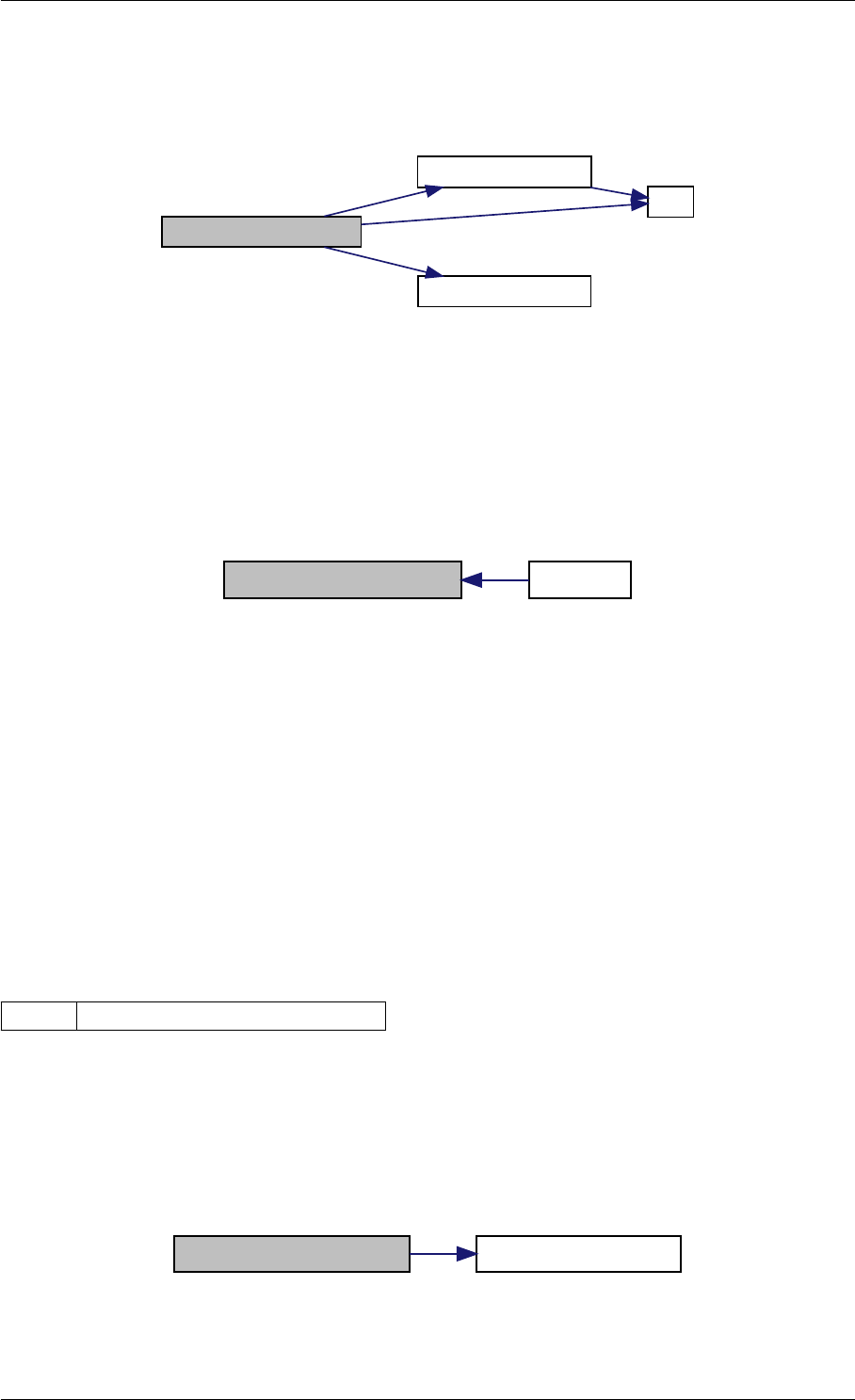
124 CONTENTS
Here is the call graph for this function:
setObservedDataStdDev
checkDataDimension
nt
checkSigmaPositivity
Here is the caller graph for this function:
setObservedDataStdDev ExpData
10.6.3.11 setObservedDataStdDev() [2/4]
void setObservedDataStdDev (
const realtype stdDev )
set function that sets all ExpData::observedDataStdDev to the input value
Parameters
stdDev standard deviation (dimension: scalar)
Definition at line 160 of file edata.cpp.
Here is the call graph for this function:
setObservedDataStdDev checkSigmaPositivity
Generated by Doxygen
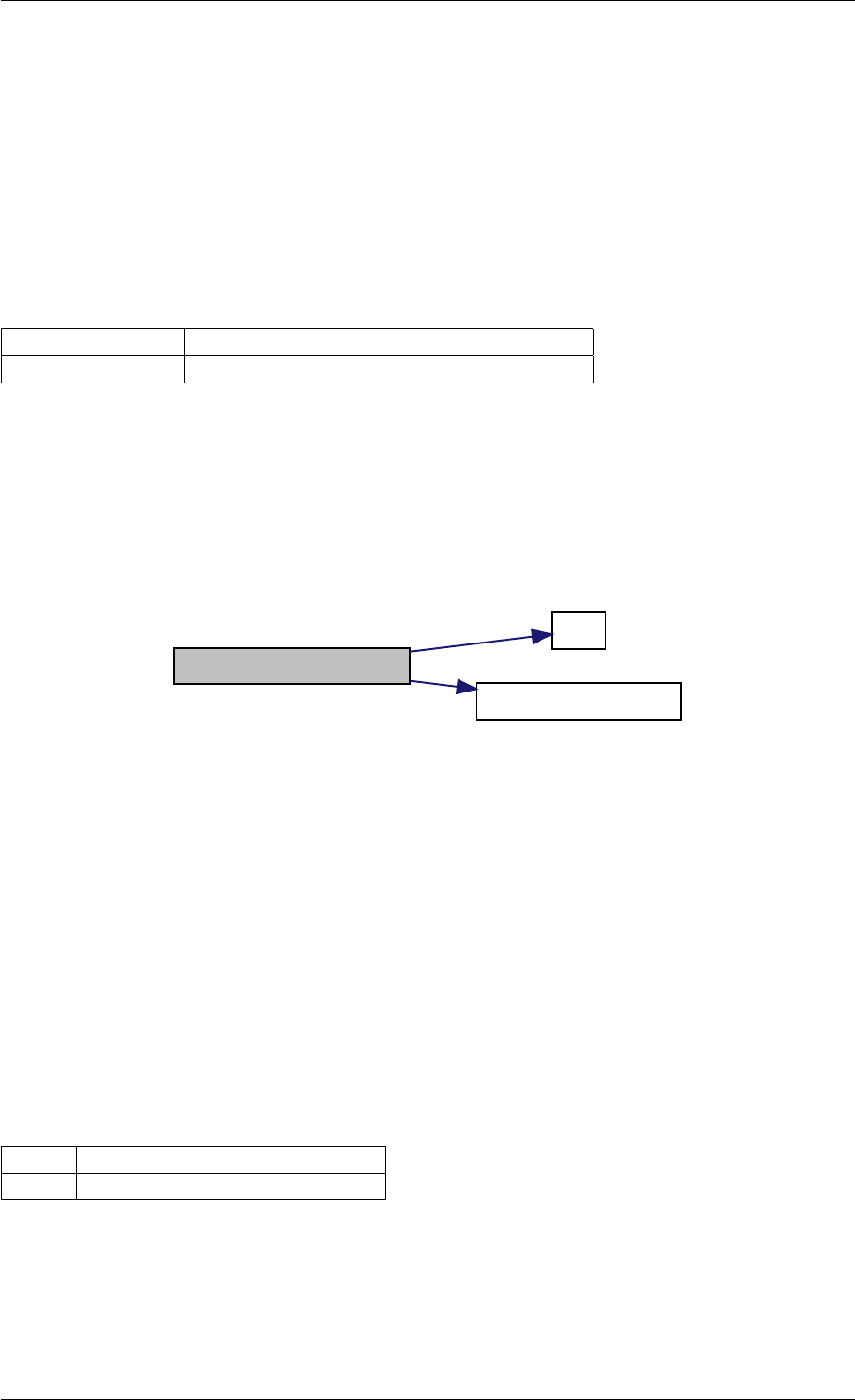
10.6 ExpData Class Reference 125
10.6.3.12 setObservedDataStdDev() [3/4]
void setObservedDataStdDev (
const std::vector<realtype >&observedDataStdDev,
int iy )
set function that copies standard deviation of observed data for specific observable
Parameters
observedDataStdDev standard deviation of observed data (dimension: nt)
iy observed data index
Definition at line 165 of file edata.cpp.
Here is the call graph for this function:
setObservedDataStdDev
nt
checkSigmaPositivity
10.6.3.13 setObservedDataStdDev() [4/4]
void setObservedDataStdDev (
const realtype stdDev,
int iy )
set function that sets all standard deviation of a specific observable to the input value
Parameters
stdDev standard deviation (dimension: scalar)
iy observed data index
Definition at line 174 of file edata.cpp.
Generated by Doxygen
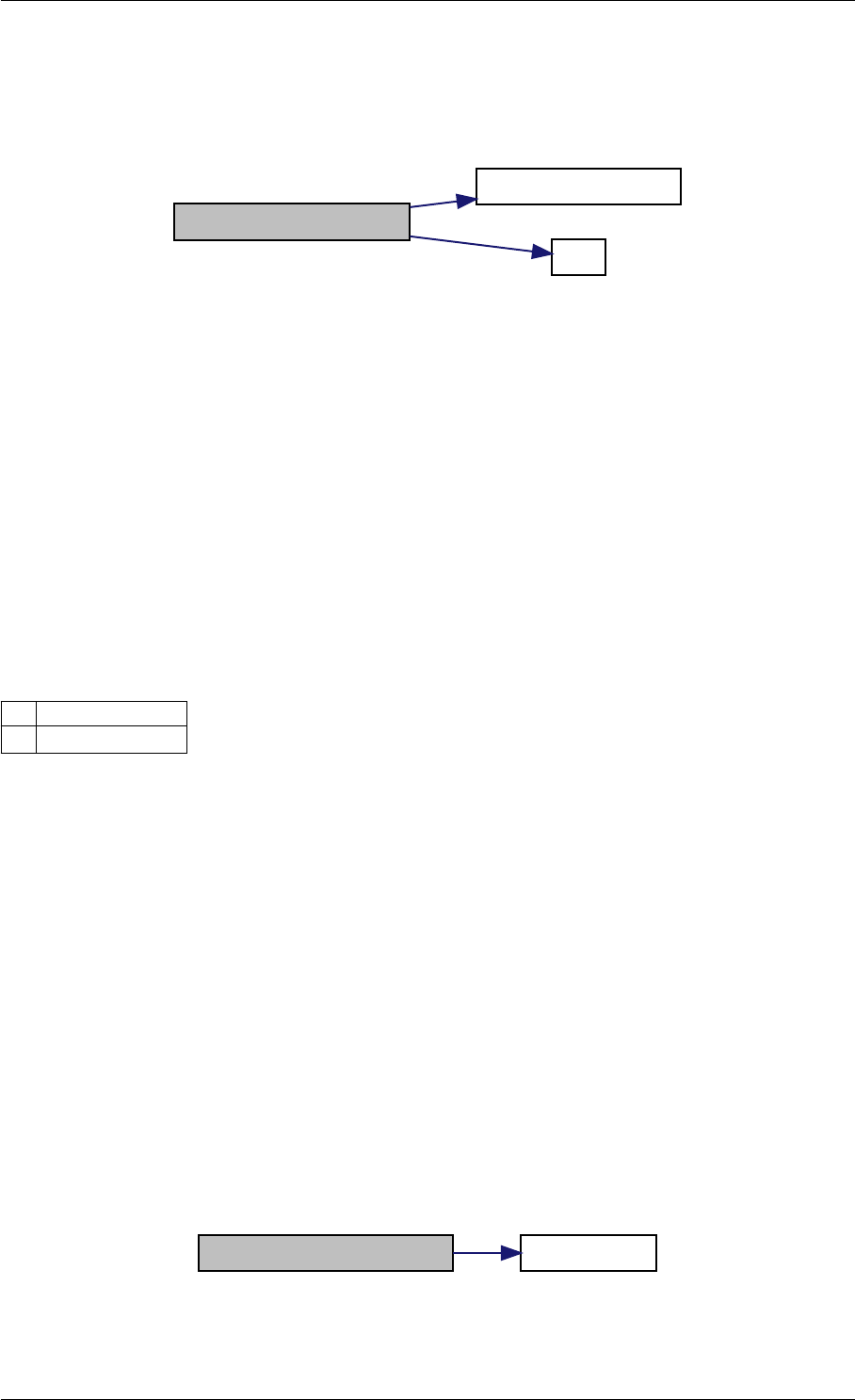
126 CONTENTS
Here is the call graph for this function:
setObservedDataStdDev
checkSigmaPositivity
nt
10.6.3.14 isSetObservedDataStdDev()
bool isSetObservedDataStdDev (
int it,
int iy ) const
get function that checks whether standard deviation of data at specified indices has been set
Parameters
it time index
iy observable index
Returns
boolean specifying if standard deviation of data was set
Definition at line 180 of file edata.cpp.
Here is the call graph for this function:
isSetObservedDataStdDev amici::isNaN
Generated by Doxygen
10.6 ExpData Class Reference 127
Here is the caller graph for this function:
isSetObservedDataStdDev
amici::Model::fsigmay
amici::Model::fdsigmaydp
10.6.3.15 getObservedDataStdDev()
std::vector<realtype >const & getObservedDataStdDev ( ) const
get function that copies data from ExpData::observedDataStdDev to output
Returns
standard deviation of observed data
Definition at line 184 of file edata.cpp.
10.6.3.16 getObservedDataStdDevPtr()
const realtype ∗getObservedDataStdDevPtr (
int it ) const
get function that returns a pointer to standard deviation of observed data at index
Parameters
it timepoint index
Returns
pointer to standard deviation of observed data at index
Definition at line 188 of file edata.cpp.
Generated by Doxygen
128 CONTENTS
Here is the caller graph for this function:
getObservedDataStdDevPtr amici::Model::fsigmay
10.6.3.17 setObservedEvents() [1/2]
void setObservedEvents (
const std::vector<realtype >&observedEvents )
set function that copies observed event data from input to ExpData::observedEvents
Parameters
observedEvents observed data (dimension: nmaxevent x nztrue, row-major)
Definition at line 195 of file edata.cpp.
Here is the call graph for this function:
setObservedEvents checkEventsDimension
Here is the caller graph for this function:
setObservedEvents ExpData
Generated by Doxygen

10.6 ExpData Class Reference 129
10.6.3.18 setObservedEvents() [2/2]
void setObservedEvents (
const std::vector<realtype >&observedEvents,
int iz )
set function that copies observed event data for specific event observable
Parameters
observedEvents observed data (dimension: nmaxevent)
iz observed event data index
Definition at line 204 of file edata.cpp.
10.6.3.19 isSetObservedEvents()
bool isSetObservedEvents (
int ie,
int iz ) const
get function that checks whether event data at specified indices has been set
Parameters
ie event index
iz event observable index
Returns
boolean specifying if data was set
Definition at line 213 of file edata.cpp.
Here is the call graph for this function:
isSetObservedEvents amici::isNaN
Generated by Doxygen
130 CONTENTS
10.6.3.20 getObservedEvents()
std::vector<realtype >const & getObservedEvents ( ) const
get function that copies data from ExpData::mz to output
Returns
observed event data
Definition at line 217 of file edata.cpp.
10.6.3.21 getObservedEventsPtr()
const realtype ∗getObservedEventsPtr (
int ie ) const
get function that returns a pointer to observed data at ieth occurence
Parameters
ie event occurence
Returns
pointer to observed event data at ieth occurence
Definition at line 221 of file edata.cpp.
Here is the caller graph for this function:
getObservedEventsPtr amici::Model::getmz
amici::Model::fdJzdp
amici::Model::fdJzdx
amici::Model::fJz
amici::Model::fdJzdz
amici::Model::fdJzdsigma
amici::Model::fdJrzdz
Generated by Doxygen
10.6 ExpData Class Reference 131
10.6.3.22 setObservedEventsStdDev() [1/4]
void setObservedEventsStdDev (
const std::vector<realtype >&observedEventsStdDev )
set function that copies data from input to ExpData::observedEventsStdDev
Parameters
observedEventsStdDev standard deviation of observed event data
Definition at line 228 of file edata.cpp.
Here is the call graph for this function:
setObservedEventsStdDev
checkEventsDimension
checkSigmaPositivity
Here is the caller graph for this function:
setObservedEventsStdDev ExpData
10.6.3.23 setObservedEventsStdDev() [2/4]
void setObservedEventsStdDev (
const realtype stdDev )
set function that sets all ExpData::observedDataStdDev to the input value
Parameters
stdDev standard deviation (dimension: scalar)
Generated by Doxygen
132 CONTENTS
Definition at line 238 of file edata.cpp.
Here is the call graph for this function:
setObservedEventsStdDev checkSigmaPositivity
10.6.3.24 setObservedEventsStdDev() [3/4]
void setObservedEventsStdDev (
const std::vector<realtype >&observedEventsStdDev,
int iz )
set function that copies standard deviation of observed data for specific observable
Parameters
observedEventsStdDev standard deviation of observed data (dimension: nmaxevent)
iz observed data index
Definition at line 243 of file edata.cpp.
Here is the call graph for this function:
setObservedEventsStdDev checkSigmaPositivity
10.6.3.25 setObservedEventsStdDev() [4/4]
void setObservedEventsStdDev (
const realtype stdDev,
int iz )
set function that sets all standard deviation of a specific observable to the input value
Generated by Doxygen
10.6 ExpData Class Reference 133
Parameters
stdDev standard deviation (dimension: scalar)
iz observed data index
Definition at line 252 of file edata.cpp.
Here is the call graph for this function:
setObservedEventsStdDev checkSigmaPositivity
10.6.3.26 isSetObservedEventsStdDev()
bool isSetObservedEventsStdDev (
int ie,
int iz ) const
get function that checks whether standard deviation of even data at specified indices has been set
Parameters
ie event index
iz event observable index
Returns
boolean specifying if standard deviation of event data was set
Definition at line 259 of file edata.cpp.
Here is the call graph for this function:
isSetObservedEventsStdDev amici::isNaN
Generated by Doxygen
134 CONTENTS
Here is the caller graph for this function:
isSetObservedEventsStdDev
amici::Model::fsigmaz
amici::Model::fdsigmazdp
10.6.3.27 getObservedEventsStdDev()
std::vector<realtype >const & getObservedEventsStdDev ( ) const
get function that copies data from ExpData::observedEventsStdDev to output
Returns
standard deviation of observed event data
Definition at line 266 of file edata.cpp.
10.6.3.28 getObservedEventsStdDevPtr()
const realtype ∗getObservedEventsStdDevPtr (
int ie ) const
get function that returns a pointer to standard deviation of observed event data at ieth occurence
Parameters
ie event occurence
Returns
pointer to standard deviation of observed event data at ieth occurence
Definition at line 270 of file edata.cpp.
Generated by Doxygen
10.6 ExpData Class Reference 135
Here is the caller graph for this function:
getObservedEventsStdDevPtr amici::Model::fsigmaz
10.6.3.29 checkDataDimension()
void checkDataDimension (
std::vector<realtype >input,
const char ∗fieldname ) const [protected]
checker for dimensions of input observedData or observedDataStdDev
Parameters
input vector input to be checked
fieldname name of the input
Definition at line 277 of file edata.cpp.
Here is the call graph for this function:
checkDataDimension nt
Here is the caller graph for this function:
checkDataDimension
setObservedData
setObservedDataStdDev
ExpData
Generated by Doxygen
136 CONTENTS
10.6.3.30 checkEventsDimension()
void checkEventsDimension (
std::vector<realtype >input,
const char ∗fieldname ) const [protected]
checker for dimensions of input observedEvents or observedEventsStdDev
Parameters
input vector input to be checked
fieldname name of the input
Definition at line 282 of file edata.cpp.
Here is the caller graph for this function:
checkEventsDimension
setObservedEvents
setObservedEventsStdDev
ExpData
10.6.3.31 checkSigmaPositivity() [1/2]
void checkSigmaPositivity (
std::vector<realtype >sigmaVector,
const char ∗vectorName ) const [protected]
checks input vector of sigmas for not strictly positive values
Parameters
sigmaVector vector input to be checked
vectorName name of the input
Definition at line 287 of file edata.cpp.
Generated by Doxygen
10.6 ExpData Class Reference 137
Here is the caller graph for this function:
checkSigmaPositivity
ExpData
setObservedDataStdDev
setObservedEventsStdDev
ExpData
10.6.3.32 checkSigmaPositivity() [2/2]
void checkSigmaPositivity (
realtype sigma,
const char ∗sigmaName ) const [protected]
checks input scalar sigma for not strictly positive value
Parameters
sigma input to be checked
sigmaName name of the input
Definition at line 292 of file edata.cpp.
10.6.4 Member Data Documentation
10.6.4.1 nytrue
const int nytrue
number of observables
Definition at line 309 of file edata.h.
10.6.4.2 nztrue
const int nztrue
number of event observables
Definition at line 311 of file edata.h.
Generated by Doxygen

138 CONTENTS
10.6.4.3 nmaxevent
const int nmaxevent
maximal number of event occurences
Definition at line 313 of file edata.h.
10.6.4.4 fixedParameters
std::vector<realtype>fixedParameters
condition-specific parameters of size Model::nk() or empty
Definition at line 316 of file edata.h.
10.6.4.5 fixedParametersPreequilibration
std::vector<realtype>fixedParametersPreequilibration
condition-specific parameters for pre-equilibration of size Model::nk() or empty. Overrides Solver::newton_preeq
Definition at line 319 of file edata.h.
10.6.4.6 fixedParametersPresimulation
std::vector<realtype>fixedParametersPresimulation
condition-specific parameters for pre-simulation of size Model::nk() or empty.
Definition at line 321 of file edata.h.
10.6.4.7 ts
std::vector<realtype>ts [protected]
observation timepoints (dimension: nt)
Definition at line 364 of file edata.h.
Generated by Doxygen

10.7 ForwardProblem Class Reference 139
10.6.4.8 observedData
std::vector<realtype>observedData [protected]
observed data (dimension: nt x nytrue, row-major)
Definition at line 367 of file edata.h.
10.6.4.9 observedDataStdDev
std::vector<realtype>observedDataStdDev [protected]
standard deviation of observed data (dimension: nt x nytrue, row-major)
Definition at line 369 of file edata.h.
10.6.4.10 observedEvents
std::vector<realtype>observedEvents [protected]
observed events (dimension: nmaxevents x nztrue, row-major)
Definition at line 372 of file edata.h.
10.6.4.11 observedEventsStdDev
std::vector<realtype>observedEventsStdDev [protected]
standard deviation of observed events/roots (dimension: nmaxevents x nztrue, row-major)
Definition at line 375 of file edata.h.
10.7 ForwardProblem Class Reference
The ForwardProblem class groups all functions for solving the backwards problem. Has only static members.
#include <forwardproblem.h>
Generated by Doxygen

140 CONTENTS
Public Member Functions
•ForwardProblem (ReturnData ∗rdata, const ExpData ∗edata,Model ∗model,Solver ∗solver)
• void workForwardProblem ()
•realtype getTime () const
•AmiVectorArray const & getStateSensitivity () const
•AmiVectorArray const & getStatesAtDiscontinuities () const
•AmiVectorArray const & getRHSAtDiscontinuities () const
•AmiVectorArray const & getRHSBeforeDiscontinuities () const
• std::vector<int >const & getNumberOfRoots () const
• std::vector<realtype >const & getDiscontinuities () const
• std::vector<int >const & getRootIndexes () const
• std::vector<realtype >const & getDJydx () const
• std::vector<realtype >const & getDJzdx () const
• int getRootCounter () const
•AmiVector ∗getStatePointer ()
•AmiVector ∗getStateDerivativePointer ()
•AmiVectorArray ∗getStateSensitivityPointer ()
•AmiVectorArray ∗getStateDerivativeSensitivityPointer ()
Public Attributes
•Model ∗model
•ReturnData ∗rdata
•Solver ∗solver
• const ExpData ∗edata
10.7.1 Detailed Description
Definition at line 23 of file forwardproblem.h.
10.7.2 Constructor & Destructor Documentation
10.7.2.1 ForwardProblem()
ForwardProblem (
ReturnData ∗rdata,
const ExpData ∗edata,
Model ∗model,
Solver ∗solver )
Constructor
Parameters
rdata pointer to ReturnData instance
edata pointer to ExpData instance
model pointer to Model instance
solver pointer to Solver instance
Generated by Doxygen
10.7 ForwardProblem Class Reference 141
Definition at line 42 of file forwardproblem.cpp.
10.7.3 Member Function Documentation
10.7.3.1 workForwardProblem()
void workForwardProblem ( )
workForwardProblem solves the forward problem. if forward sensitivities are enabled this will also compute sensi-
tivies
Definition at line 77 of file forwardproblem.cpp.
Here is the call graph for this function:
workForwardProblem
amici::Solver::setup
amici::ReturnData::
initializeObjectiveFunction
amici::Solver::getNewton
Preequilibration
amici::AmiVector::getVector
amici::Solver::getSensitivity
Method
amici::Solver::allocateSolver
amici::Solver::init
amici::Solver::applyTolerances
amici::Solver::setErrHandlerFn
amici::Solver::setUserData
amici::Solver::setMaxNum
Steps
amici::Solver::setStabLimDet
amici::Solver::rootInit
amici::Solver::initialize
LinearSolver
amici::Solver::getMallocDone
amici::Solver::setSStolerances
amici::Solver::dense
amici::Solver::setDenseJacFn
amici::Solver::band
amici::Solver::setBandJacFn
10.7.3.2 getTime()
realtype getTime ( ) const
accessor for t
Generated by Doxygen
142 CONTENTS
Returns
t
Definition at line 37 of file forwardproblem.h.
Here is the caller graph for this function:
getTime amici::BackwardProblem
::BackwardProblem
10.7.3.3 getStateSensitivity()
AmiVectorArray const& getStateSensitivity ( ) const
accessor for sx
Returns
sx
Definition at line 44 of file forwardproblem.h.
10.7.3.4 getStatesAtDiscontinuities()
AmiVectorArray const& getStatesAtDiscontinuities ( ) const
accessor for x_disc
Returns
x_disc
Definition at line 51 of file forwardproblem.h.
Generated by Doxygen

10.7 ForwardProblem Class Reference 143
10.7.3.5 getRHSAtDiscontinuities()
AmiVectorArray const& getRHSAtDiscontinuities ( ) const
accessor for xdot_disc
Returns
xdot_disc
Definition at line 58 of file forwardproblem.h.
10.7.3.6 getRHSBeforeDiscontinuities()
AmiVectorArray const& getRHSBeforeDiscontinuities ( ) const
accessor for xdot_old_disc
Returns
xdot_old_disc
Definition at line 65 of file forwardproblem.h.
10.7.3.7 getNumberOfRoots()
std::vector<int>const& getNumberOfRoots ( ) const
accessor for nroots
Returns
nroots
Definition at line 72 of file forwardproblem.h.
10.7.3.8 getDiscontinuities()
std::vector<realtype>const& getDiscontinuities ( ) const
accessor for discs
Returns
discs
Definition at line 79 of file forwardproblem.h.
Generated by Doxygen

144 CONTENTS
10.7.3.9 getRootIndexes()
std::vector<int>const& getRootIndexes ( ) const
accessor for rootidx
Returns
rootidx
Definition at line 86 of file forwardproblem.h.
10.7.3.10 getDJydx()
std::vector<realtype>const& getDJydx ( ) const
accessor for dJydx
Returns
dJydx
Definition at line 93 of file forwardproblem.h.
10.7.3.11 getDJzdx()
std::vector<realtype>const& getDJzdx ( ) const
accessor for dJzdx
Returns
dJzdx
Definition at line 100 of file forwardproblem.h.
Generated by Doxygen
10.7 ForwardProblem Class Reference 145
10.7.3.12 getRootCounter()
int getRootCounter ( ) const
accessor for iroot
Returns
iroot
Definition at line 107 of file forwardproblem.h.
Here is the caller graph for this function:
getRootCounter amici::BackwardProblem
::BackwardProblem
10.7.3.13 getStatePointer()
AmiVector∗getStatePointer ( )
accessor for pointer to x
Returns
&x
Definition at line 114 of file forwardproblem.h.
10.7.3.14 getStateDerivativePointer()
AmiVector∗getStateDerivativePointer ( )
accessor for pointer to dx
Returns
&dx
Definition at line 121 of file forwardproblem.h.
Generated by Doxygen

146 CONTENTS
10.7.3.15 getStateSensitivityPointer()
AmiVectorArray∗getStateSensitivityPointer ( )
accessor for pointer to sx
Returns
&sx
Definition at line 128 of file forwardproblem.h.
10.7.3.16 getStateDerivativeSensitivityPointer()
AmiVectorArray∗getStateDerivativeSensitivityPointer ( )
accessor for pointer to sdx
Returns
&sdx
Definition at line 135 of file forwardproblem.h.
10.7.4 Member Data Documentation
10.7.4.1 model
Model∗model
pointer to model instance
Definition at line 140 of file forwardproblem.h.
10.7.4.2 rdata
ReturnData∗rdata
pointer to return data instance
Definition at line 142 of file forwardproblem.h.
Generated by Doxygen
10.8 IDAException Class Reference 147
10.7.4.3 solver
Solver∗solver
pointer to solver instance
Definition at line 144 of file forwardproblem.h.
10.7.4.4 edata
const ExpData∗edata
pointer to experimental data instance
Definition at line 146 of file forwardproblem.h.
10.8 IDAException Class Reference
ida exception handler class
#include <exception.h>
Inheritance diagram for IDAException:
IDAException
AmiException
exception
Public Member Functions
•IDAException (const int error_code, const char ∗function)
10.8.1 Detailed Description
Definition at line 129 of file exception.h.
Generated by Doxygen
148 CONTENTS
10.8.2 Constructor & Destructor Documentation
10.8.2.1 IDAException()
IDAException (
const int error_code,
const char ∗function )
constructor
Parameters
error_code error code returned by ida function
function ida function name
Definition at line 135 of file exception.h.
10.9 IntegrationFailure Class Reference
integration failure exception for the forward problem this exception should be thrown when an integration failure
occured for this exception we can assume that we can recover from the exception and return a solution struct to the
user
#include <exception.h>
Inheritance diagram for IntegrationFailure:
IntegrationFailure
AmiException
exception
Public Member Functions
•IntegrationFailure (int code, realtype t)
Generated by Doxygen
10.9 IntegrationFailure Class Reference 149
Public Attributes
• int error_code
•realtype time
10.9.1 Detailed Description
Definition at line 144 of file exception.h.
10.9.2 Constructor & Destructor Documentation
10.9.2.1 IntegrationFailure()
IntegrationFailure (
int code,
realtype t)
constructor
Parameters
code error code returned by cvode/ida
ttime of integration failure
Definition at line 154 of file exception.h.
10.9.3 Member Data Documentation
10.9.3.1 error_code
int error_code
error code returned by cvode/ida
Definition at line 147 of file exception.h.
10.9.3.2 time
realtype time
time of integration failure
Definition at line 149 of file exception.h.
Generated by Doxygen
150 CONTENTS
10.10 IntegrationFailureB Class Reference
integration failure exception for the backward problem this exception should be thrown when an integration failure
occured for this exception we can assume that we can recover from the exception and return a solution struct to the
user
#include <exception.h>
Inheritance diagram for IntegrationFailureB:
IntegrationFailureB
AmiException
exception
Public Member Functions
•IntegrationFailureB (int code, realtype t)
Public Attributes
• int error_code
•realtype time
10.10.1 Detailed Description
Definition at line 166 of file exception.h.
10.10.2 Constructor & Destructor Documentation
10.10.2.1 IntegrationFailureB()
IntegrationFailureB (
int code,
realtype t)
constructor
Generated by Doxygen

10.11 Model Class Reference 151
Parameters
code error code returned by cvode/ida
ttime of integration failure
Definition at line 176 of file exception.h.
10.10.3 Member Data Documentation
10.10.3.1 error_code
int error_code
error code returned by cvode/ida
Definition at line 169 of file exception.h.
10.10.3.2 time
realtype time
time of integration failure
Definition at line 171 of file exception.h.
10.11 Model Class Reference
The Model class represents an AMICI ODE model. The model can compute various model related quantities based
on symbolically generated code.
#include <model.h>
Inheritance diagram for Model:
Model
Model_DAE Model_ODE
Generated by Doxygen

152 CONTENTS
Public Member Functions
•Model ()
•Model (const int nx, const int nxtrue, const int ny, const int nytrue, const int nz, const int nztrue, const int
ne, const int nJ, const int nw, const int ndwdx, const int ndwdp, const int nnz, const int ubw, const int lbw,
amici::SecondOrderMode o2mode, const std::vector<amici::realtype >&p, std::vector<amici::realtype >
k, const std::vector<int >&plist, std::vector<amici::realtype >idlist, std::vector<int >z2event)
•Model (Model const &other)
• virtual ∼Model ()
•Model &operator= (Model const &other)=delete
• virtual Model ∗clone () const =0
Clone this instance.
• virtual std::unique_ptr<Solver >getSolver ()=0
• virtual void froot (realtype t,AmiVector ∗x, AmiVector ∗dx, realtype ∗root)=0
• virtual void fxdot (realtype t,AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot)=0
• virtual void fsxdot (realtype t,AmiVector ∗x, AmiVector ∗dx, int ip, AmiVector ∗sx, AmiVector ∗sdx, AmiVector
∗sxdot)=0
• virtual void fJ (realtype t,realtype cj, AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, DlsMat J)=0
• virtual void fJSparse (realtype t,realtype cj, AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, SlsMat J)=0
• virtual void fJDiag (realtype t,AmiVector ∗Jdiag, realtype cj, AmiVector ∗x, AmiVector ∗dx)=0
• virtual void fdxdotdp (realtype t,AmiVector ∗x, AmiVector ∗dx)=0
• virtual void fJv (realtype t,AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, AmiVector ∗v, AmiVector ∗nJv,
realtype cj)=0
• void fx0 (AmiVector ∗x)
• void fx0_fixedParameters (AmiVector ∗x)
• virtual void fdx0 (AmiVector ∗x0, AmiVector ∗dx0)
• void fsx0 (AmiVectorArray ∗sx, const AmiVector ∗x)
• void fsx0_fixedParameters (AmiVectorArray ∗sx, const AmiVector ∗x)
• virtual void fsdx0 ()
• void fstau (const realtype t, const int ie, const AmiVector ∗x, const AmiVectorArray ∗sx)
• void fy (int it, ReturnData ∗rdata)
• void fdydp (const int it, ReturnData ∗rdata)
• void fdydx (const int it, ReturnData ∗rdata)
• void fz (const int nroots, const int ie, const realtype t, const AmiVector ∗x, ReturnData ∗rdata)
• void fsz (const int nroots, const int ie, const realtype t, const AmiVector ∗x, const AmiVectorArray ∗sx,
ReturnData ∗rdata)
• void frz (const int nroots, const int ie, const realtype t, const AmiVector ∗x, ReturnData ∗rdata)
• void fsrz (const int nroots, const int ie, const realtype t, const AmiVector ∗x, const AmiVectorArray ∗sx,
ReturnData ∗rdata)
• void fdzdp (const realtype t, const int ie, const AmiVector ∗x)
• void fdzdx (const realtype t, const int ie, const AmiVector ∗x)
• void fdrzdp (const realtype t, const int ie, const AmiVector ∗x)
• void fdrzdx (const realtype t, const int ie, const AmiVector ∗x)
• void fdeltax (const int ie, const realtype t, const AmiVector ∗x, const AmiVector ∗xdot, const AmiVector ∗xdot←-
_old)
• void fdeltasx (const int ie, const realtype t, const AmiVector ∗x, const AmiVectorArray ∗sx, const AmiVector
∗xdot, const AmiVector ∗xdot_old)
• void fdeltaxB (const int ie, const realtype t, const AmiVector ∗x, const AmiVector ∗xB, const AmiVector ∗xdot,
const AmiVector ∗xdot_old)
• void fdeltaqB (const int ie, const realtype t, const AmiVector ∗x, const AmiVector ∗xB, const AmiVector ∗xdot,
const AmiVector ∗xdot_old)
• void fsigmay (const int it, ReturnData ∗rdata, const ExpData ∗edata)
• void fdsigmaydp (const int it, ReturnData ∗rdata, const ExpData ∗edata)
• void fsigmaz (const realtype t, const int ie, const int ∗nroots, ReturnData ∗rdata, const ExpData ∗edata)
• void fdsigmazdp (const realtype t, const int ie, const int ∗nroots, ReturnData ∗rdata, const ExpData ∗edata)
Generated by Doxygen

10.11 Model Class Reference 153
• void fJy (const int it, ReturnData ∗rdata, const ExpData ∗edata)
• void fJz (const int nroots, ReturnData ∗rdata, const ExpData ∗edata)
• void fJrz (const int nroots, ReturnData ∗rdata, const ExpData ∗edata)
• void fdJydy (const int it, const ReturnData ∗rdata, const ExpData ∗edata)
• void fdJydsigma (const int it, const ReturnData ∗rdata, const ExpData ∗edata)
• void fdJzdz (const int nroots, const ReturnData ∗rdata, const ExpData ∗edata)
• void fdJzdsigma (const int nroots, const ReturnData ∗rdata, const ExpData ∗edata)
• void fdJrzdz (const int nroots, const ReturnData ∗rdata, const ExpData ∗edata)
• void fdJrzdsigma (const int nroots, const ReturnData ∗rdata, const ExpData ∗edata)
• void fsy (const int it, ReturnData ∗rdata)
• void fsz_tf (const int ∗nroots, const int ie, ReturnData ∗rdata)
• void fsJy (const int it, const std::vector<realtype >&dJydx, ReturnData ∗rdata)
• void fdJydp (const int it, const ExpData ∗edata, ReturnData ∗rdata)
• void fdJydx (std::vector<realtype >∗dJydx, const int it, const ExpData ∗edata, const ReturnData ∗rdata)
• void fsJz (const int nroots, const std::vector<realtype >&dJzdx, AmiVectorArray ∗sx, ReturnData ∗rdata)
• void fdJzdp (const int nroots, realtype t, const ExpData ∗edata, const ReturnData ∗rdata)
• void fdJzdx (std::vector<realtype >∗dJzdx, const int nroots, realtype t, const ExpData ∗edata, const
ReturnData ∗rdata)
• void initialize (AmiVector ∗x, AmiVector ∗dx)
• void initializeStates (AmiVector ∗x)
• void initHeaviside (AmiVector ∗x, AmiVector ∗dx)
• int nplist () const
number of paramaeters wrt to which sensitivities are computed
• int np () const
total number of model parameters
• int nk () const
number of constants
• const double ∗k() const
fixed parameters
• int nMaxEvent () const
Get nmaxevent.
• void setNMaxEvent (int nmaxevent)
Set nmaxevent.
• int nt () const
Get number of timepoints.
• std::vector<ParameterScaling >const & getParameterScale () const
Get ParameterScale for each parameter.
• void setParameterScale (ParameterScaling pscale)
Set ParameterScale for each parameter.
• void setParameterScale (const std::vector<ParameterScaling >&pscale)
Set ParameterScale for each parameter.
• std::vector<realtype >const & getParameters () const
Get the parameter vector.
• void setParameters (std::vector<realtype >const &p)
Sets the parameter vector.
• std::vector<realtype >const & getUnscaledParameters () const
Gets parameters with transformation according to ParameterScale applied.
• std::vector<realtype >const & getFixedParameters () const
Gets the fixedParameter member.
• void setFixedParameters (std::vector<realtype >const &k)
Sets the fixedParameter member.
•realtype getFixedParameterById (std::string const &par_id) const
Generated by Doxygen

154 CONTENTS
Get value of fixed parameter with the specified Id.
•realtype getFixedParameterByName (std::string const &par_name) const
Get value of fixed parameter with the specified name, if multiple parameters have the same name, the first parameter
with matching name is returned.
• void setFixedParameterById (std::string const &par_id, realtype value)
Set value of first fixed parameter with the specified id.
• int setFixedParametersByIdRegex (std::string const &par_id_regex, realtype value)
Set values of all fixed parameters with the id matching the specified regex.
• void setFixedParameterByName (std::string const &par_name, realtype value)
Set value of first fixed parameter with the specified name,.
• int setFixedParametersByNameRegex (std::string const &par_name_regex, realtype value)
Set value of all fixed parameters with name matching the specified regex,.
• std::vector<realtype >const & getTimepoints () const
Get the timepoint vector.
• void setTimepoints (std::vector<realtype >const &ts)
Set the timepoint vector.
• std::vector<bool >const & getStateIsNonNegative () const
gets flags indicating whether states should be treated as non-negative
• void setStateIsNonNegative (std::vector<bool >const &stateIsNonNegative)
sets flags indicating whether states should be treated as non-negative
• double t(int idx) const
Get timepoint for given index.
• std::vector<int >const & getParameterList () const
Get the list of parameters for which sensitivities are computed.
• void setParameterList (std::vector<int >const &plist)
Set the list of parameters for which sensitivities are computed.
• std::vector<realtype >const & getInitialStates () const
Get the initial states.
• void setInitialStates (std::vector<realtype >const &x0)
Set the initial states.
• std::vector<realtype >const & getInitialStateSensitivities () const
Get the initial states sensitivities.
• void setInitialStateSensitivities (std::vector<realtype >const &sx0)
Set the initial state sensitivities.
• double t0 () const
get simulation start time
• void setT0 (double t0)
set simulation start time
• int plist (int pos) const
entry in parameter list
• void requireSensitivitiesForAllParameters ()
Require computation of sensitivities for all parameters p [0..np[ in natural order.
• void fw (const realtype t, const N_Vector x)
Recurring terms in xdot.
• void fw (const realtype t, const realtype ∗x)
Recurring terms in xdot.
• void fdwdp (const realtype t, const N_Vector x)
Recurring terms in xdot, parameter derivative.
• void fdwdp (const realtype t, const realtype ∗x)
Recurring terms in xdot, parameter derivative.
• void fdwdx (const realtype t, const N_Vector x)
Generated by Doxygen

10.11 Model Class Reference 155
Recurring terms in xdot, state derivative.
• void fdwdx (const realtype t, const realtype ∗x)
Recurring terms in xdot, state derivative.
• void fres (const int it, ReturnData ∗rdata, const ExpData ∗edata)
• void fchi2 (const int it, ReturnData ∗rdata)
• void fsres (const int it, ReturnData ∗rdata, const ExpData ∗edata)
• void fFIM (const int it, ReturnData ∗rdata)
• void updateHeaviside (const std::vector<int >&rootsfound)
• void updateHeavisideB (const int ∗rootsfound)
•realtype gett (const int it, const ReturnData ∗rdata) const
• int checkFinite (const int N, const realtype ∗array, const char ∗fun) const
Check if the given array has only finite elements. If not try to give hints by which other fields this could be caused.
• virtual bool hasParameterNames () const
Reports whether the model has parameter names set.
• virtual std::vector<std::string >getParameterNames () const
Get names of the model parameters.
• virtual bool hasStateNames () const
Reports whether the model has state names set.
• virtual std::vector<std::string >getStateNames () const
Get names of the model states.
• virtual bool hasFixedParameterNames () const
Reports whether the model has fixed parameter names set.
• virtual std::vector<std::string >getFixedParameterNames () const
Get names of the fixed model parameters.
• virtual bool hasObservableNames () const
Reports whether the model has observable names set.
• virtual std::vector<std::string >getObservableNames () const
Get names of the observables.
• virtual bool hasParameterIds () const
Reports whether the model has parameter ids set.
• virtual std::vector<std::string >getParameterIds () const
Get ids of the model parameters.
•realtype getParameterById (std::string const &par_id) const
Get value of first model parameter with the specified id.
•realtype getParameterByName (std::string const &par_name) const
Get value of first model parameter with the specified name,.
• void setParameterById (std::string const &par_id, realtype value)
Set value of first model parameter with the specified id.
• int setParametersByIdRegex (std::string const &par_id_regex, realtype value)
Set all values of model parameters with ids matching the specified regex.
• void setParameterByName (std::string const &par_name, realtype value)
Set value of first model parameter with the specified name.
• int setParametersByNameRegex (std::string const &par_name_regex, realtype value)
Set all values of all model parameters with names matching the specified regex.
• virtual bool hasStateIds () const
Reports whether the model has state ids set.
• virtual std::vector<std::string >getStateIds () const
Get ids of the model states.
• virtual bool hasFixedParameterIds () const
Reports whether the model has fixed parameter ids set.
• virtual std::vector<std::string >getFixedParameterIds () const
Generated by Doxygen

156 CONTENTS
Get ids of the fixed model parameters.
• virtual bool hasObservableIds () const
Reports whether the model has observable ids set.
• virtual std::vector<std::string >getObservableIds () const
Get ids of the observables.
• void setSteadyStateSensitivityMode (const SteadyStateSensitivityMode mode)
sets the mode how sensitivities are computed in the steadystate simulation
•SteadyStateSensitivityMode getSteadyStateSensitivityMode () const
gets the mode how sensitivities are computed in the steadystate simulation
• void setReinitializeFixedParameterInitialStates (bool flag)
set whether initial states depending on fixedParmeters are to be reinitialized after preequilibration and presimulation
• bool getReinitializeFixedParameterInitialStates () const
get whether initial states depending on fixedParmeters are to be reinitialized after preequilibration and presimulation
Public Attributes
• const int nx
• const int nxtrue
• const int ny
• const int nytrue
• const int nz
• const int nztrue
• const int ne
• const int nw
• const int ndwdx
• const int ndwdp
• const int nnz
• const int nJ
• const int ubw
• const int lbw
• const SecondOrderMode o2mode
• const std::vector<int >z2event
• const std::vector<realtype >idlist
• std::vector<realtype >sigmay
• std::vector<realtype >dsigmaydp
• std::vector<realtype >sigmaz
• std::vector<realtype >dsigmazdp
• std::vector<realtype >dJydp
• std::vector<realtype >dJzdp
• std::vector<realtype >deltax
• std::vector<realtype >deltasx
• std::vector<realtype >deltaxB
• std::vector<realtype >deltaqB
• std::vector<realtype >dxdotdp
Generated by Doxygen

10.11 Model Class Reference 157
Protected Member Functions
• void initializeVectors ()
Set the nplist-dependent vectors to their proper sizes.
• virtual void fx0 (realtype ∗x0, const realtype t, const realtype ∗p, const realtype ∗k)
• virtual void fx0_fixedParameters (realtype ∗x0, const realtype t, const realtype ∗p, const realtype ∗k)
• virtual void fsx0_fixedParameters (realtype ∗sx0, const realtype t, const realtype ∗x0, const realtype ∗p, const
realtype ∗k, const int ip)
• virtual void fsx0 (realtype ∗sx0, const realtype t, const realtype ∗x0, const realtype ∗p, const realtype ∗k,
const int ip)
• virtual void fstau (realtype ∗stau, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗sx, const int ip, const int ie)
• virtual void fy (realtype ∗y, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h, const realtype ∗w)
• virtual void fdydp (realtype ∗dydp, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const int ip, const realtype ∗w, const realtype ∗dwdp)
• virtual void fdydx (realtype ∗dydx, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗w, const realtype ∗dwdx)
• virtual void fz (realtype ∗z, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h)
• virtual void fsz (realtype ∗sz, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const realtype ∗sx, const int ip)
• virtual void frz (realtype ∗rz, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h)
• virtual void fsrz (realtype ∗srz, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const realtype ∗sx, const int ip)
• virtual void fdzdp (realtype ∗dzdp, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const
realtype ∗k, const realtype ∗h, const int ip)
• virtual void fdzdx (realtype ∗dzdx, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const
realtype ∗k, const realtype ∗h)
• virtual void fdrzdp (realtype ∗drzdp, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const
realtype ∗k, const realtype ∗h, const int ip)
• virtual void fdrzdx (realtype ∗drzdx, const int ie, const realtype t, const realtype ∗x, const realtype ∗p, const
realtype ∗k, const realtype ∗h)
• virtual void fdeltax (realtype ∗deltax, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const int ie, const realtype ∗xdot, const realtype ∗xdot_old)
• virtual void fdeltasx (realtype ∗deltasx, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗w, const int ip, const int ie, const realtype ∗xdot, const realtype ∗xdot_old,
const realtype ∗sx, const realtype ∗stau)
• virtual void fdeltaxB (realtype ∗deltaxB, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const int ie, const realtype ∗xdot, const realtype ∗xdot_old, const realtype ∗xB)
• virtual void fdeltaqB (realtype ∗deltaqB, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const int ip, const int ie, const realtype ∗xdot, const realtype ∗xdot_old, const realtype
∗xB)
• virtual void fsigmay (realtype ∗sigmay, const realtype t, const realtype ∗p, const realtype ∗k)
• virtual void fdsigmaydp (realtype ∗dsigmaydp, const realtype t, const realtype ∗p, const realtype ∗k, const int
ip)
• virtual void fsigmaz (realtype ∗sigmaz, const realtype t, const realtype ∗p, const realtype ∗k)
• virtual void fdsigmazdp (realtype ∗dsigmazdp, const realtype t, const realtype ∗p, const realtype ∗k, const int
ip)
• virtual void fJy (realtype ∗nllh, const int iy, const realtype ∗p, const realtype ∗k, const realtype ∗y, const
realtype ∗sigmay, const realtype ∗my)
• virtual void fJz (realtype ∗nllh, const int iz, const realtype ∗p, const realtype ∗k, const realtype ∗z, const
realtype ∗sigmaz, const realtype ∗mz)
• virtual void fJrz (realtype ∗nllh, const int iz, const realtype ∗p, const realtype ∗k, const realtype ∗z, const
realtype ∗sigmaz)
Generated by Doxygen

158 CONTENTS
• virtual void fdJydy (realtype ∗dJydy, const int iy, const realtype ∗p, const realtype ∗k, const realtype ∗y, const
realtype ∗sigmay, const realtype ∗my)
• virtual void fdJydsigma (realtype ∗dJydsigma, const int iy, const realtype ∗p, const realtype ∗k, const realtype
∗y, const realtype ∗sigmay, const realtype ∗my)
• virtual void fdJzdz (realtype ∗dJzdz, const int iz, const realtype ∗p, const realtype ∗k, const realtype ∗z, const
realtype ∗sigmaz, const realtype ∗mz)
• virtual void fdJzdsigma (realtype ∗dJzdsigma, const int iz, const realtype ∗p, const realtype ∗k, const realtype
∗z, const realtype ∗sigmaz, const realtype ∗mz)
• virtual void fdJrzdz (realtype ∗dJrzdz, const int iz, const realtype ∗p, const realtype ∗k, const realtype ∗rz,
const realtype ∗sigmaz)
• virtual void fdJrzdsigma (realtype ∗dJrzdsigma, const int iz, const realtype ∗p, const realtype ∗k, const
realtype ∗rz, const realtype ∗sigmaz)
• virtual void fw (realtype ∗w, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h)
• virtual void fdwdp (realtype ∗dwdp, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗w)
• virtual void fdwdx (realtype ∗dwdx, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗w)
• void getmy (const int it, const ExpData ∗edata)
• void getmz (const int nroots, const ExpData ∗edata)
• const realtype ∗gety (const int it, const ReturnData ∗rdata) const
• const realtype ∗getx (const int it, const ReturnData ∗rdata) const
• const realtype ∗getsx (const int it, const ReturnData ∗rdata) const
• const realtype ∗getz (const int nroots, const ReturnData ∗rdata) const
• const realtype ∗getrz (const int nroots, const ReturnData ∗rdata) const
• const realtype ∗getsz (const int nroots, const int ip, const ReturnData ∗rdata) const
• const realtype ∗getsrz (const int nroots, const int ip, const ReturnData ∗rdata) const
• virtual bool isFixedParameterStateReinitializationAllowed () const
• N_Vector computeX_pos (N_Vector x)
computes nonnegative state vector according to stateIsNonNegative if anyStateNonNegative is set to false, i.e., all
entries in stateIsNonNegative are false, this function directly returns x, otherwise all entries of x are copied in to
x_pos_tmp and negative values are replaced by 0 where applicable
Protected Attributes
• SlsMat J= nullptr
• std::vector<realtype >my
• std::vector<realtype >mz
• std::vector<realtype >dJydy
• std::vector<realtype >dJydsigma
• std::vector<realtype >dJzdz
• std::vector<realtype >dJzdsigma
• std::vector<realtype >dJrzdz
• std::vector<realtype >dJrzdsigma
• std::vector<realtype >dzdx
• std::vector<realtype >dzdp
• std::vector<realtype >drzdx
• std::vector<realtype >drzdp
• std::vector<realtype >dydp
• std::vector<realtype >dydx
• std::vector<realtype >w
• std::vector<realtype >dwdx
• std::vector<realtype >dwdp
• std::vector<realtype >M
Generated by Doxygen

10.11 Model Class Reference 159
• std::vector<realtype >stau
• std::vector<realtype >h
• std::vector<realtype >unscaledParameters
• std::vector<realtype >originalParameters
• std::vector<realtype >fixedParameters
• std::vector<int >plist_
• std::vector<double >x0data
• std::vector<realtype >sx0data
• std::vector<realtype >ts
• std::vector<bool >stateIsNonNegative
• bool anyStateNonNegative = false
•AmiVector x_pos_tmp
• int nmaxevent = 10
• std::vector<ParameterScaling >pscale
• double tstart = 0.0
•SteadyStateSensitivityMode steadyStateSensitivityMode = SteadyStateSensitivityMode::newtonOnly
• bool reinitializeFixedParameterInitialStates = false
Friends
•template<class Archive >
void boost::serialization::serialize (Archive &ar, Model &r, const unsigned int version)
Serialize Model (see boost::serialization::serialize)
• bool operator== (const Model &a, const Model &b)
Check equality of data members.
10.11.1 Detailed Description
Definition at line 40 of file model.h.
10.11.2 Constructor & Destructor Documentation
10.11.2.1 Model() [1/3]
Model ( )
default constructor
Definition at line 43 of file model.h.
Generated by Doxygen

160 CONTENTS
10.11.2.2 Model() [2/3]
Model (
const int nx,
const int nxtrue,
const int ny,
const int nytrue,
const int nz,
const int nztrue,
const int ne,
const int nJ,
const int nw,
const int ndwdx,
const int ndwdp,
const int nnz,
const int ubw,
const int lbw,
amici::SecondOrderMode o2mode,
const std::vector<amici::realtype >&p,
std::vector<amici::realtype >k,
const std::vector<int >&plist,
std::vector<amici::realtype >idlist,
std::vector<int >z2event )
constructor with model dimensions
Parameters
nx number of state variables
nxtrue number of state variables of the non-augmented model
ny number of observables
nytrue number of observables of the non-augmented model
nz number of event observables
nztrue number of event observables of the non-augmented model
ne number of events
nJ number of objective functions
nw number of repeating elements
ndwdx number of nonzero elements in the x derivative of the repeating elements
ndwdp number of nonzero elements in the p derivative of the repeating elements
nnz number of nonzero elements in Jacobian
ubw upper matrix bandwidth in the Jacobian
lbw lower matrix bandwidth in the Jacobian
o2mode second order sensitivity mode
pparameters
kconstants
plist indexes wrt to which sensitivities are to be computed
idlist indexes indicating algebraic components (DAE only)
z2event mapping of event outputs to events
Definition at line 626 of file model.cpp.
Generated by Doxygen
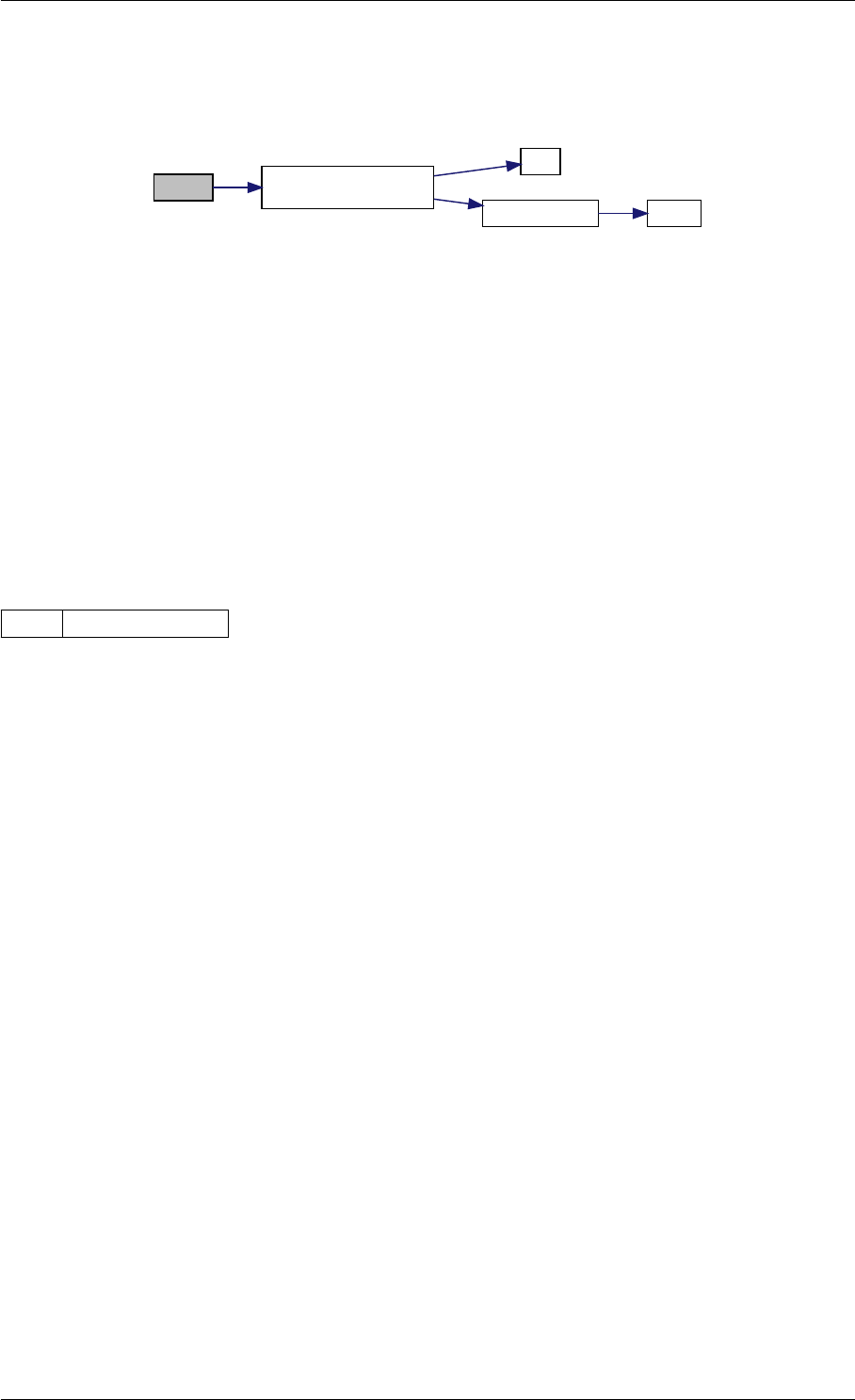
10.11 Model Class Reference 161
Here is the call graph for this function:
Model requireSensitivitiesForAll
Parameters
np
initializeVectors nplist
10.11.2.3 Model() [3/3]
Model (
Model const & other )
Copy constructor
Parameters
other object to copy from
Returns
Definition at line 703 of file model.cpp.
10.11.2.4 ∼Model()
∼Model ( ) [virtual]
destructor
Definition at line 762 of file model.cpp.
10.11.3 Member Function Documentation
10.11.3.1 operator=()
Model& operator= (
Model const & other ) [delete]
Copy assignment is disabled until const members are removed
Generated by Doxygen

162 CONTENTS
Parameters
other object to copy from
Returns
10.11.3.2 clone()
virtual Model∗clone ( ) const [pure virtual]
Returns
The clone
10.11.3.3 getSolver()
virtual std::unique_ptr<Solver>getSolver ( ) [pure virtual]
Retrieves the solver object
Returns
The Solver instance
Implemented in Model_ODE, and Model_DAE.
10.11.3.4 froot()
virtual void froot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
realtype ∗root ) [pure virtual]
Root function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
root array to which values of the root function will be written
Generated by Doxygen
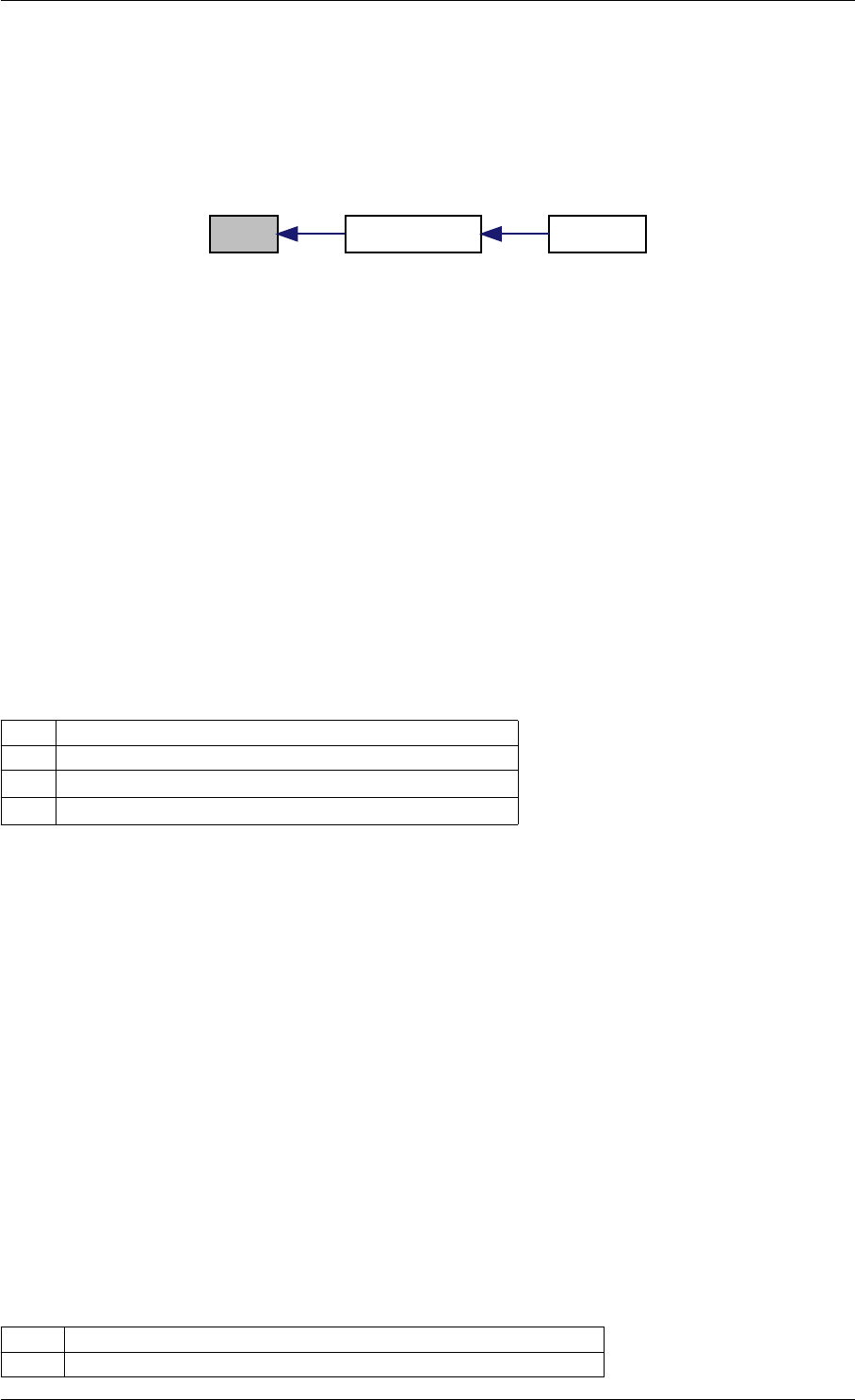
10.11 Model Class Reference 163
Implemented in Model_ODE, and Model_DAE.
Here is the caller graph for this function:
froot initHeaviside initialize
10.11.3.5 fxdot()
virtual void fxdot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot ) [pure virtual]
Residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
xdot array to which values of the residual function will be written
Implemented in Model_ODE, and Model_DAE.
10.11.3.6 fsxdot()
virtual void fsxdot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
int ip,
AmiVector ∗sx,
AmiVector ∗sdx,
AmiVector ∗sxdot ) [pure virtual]
Sensitivity Residual function
Parameters
ttime
xstate
Generated by Doxygen
164 CONTENTS
Parameters
dx time derivative of state (DAE only)
ip parameter index
sx sensitivity state
sdx time derivative of sensitivity state (DAE only)
sxdot array to which values of the sensitivity residual function will be written
Implemented in Model_ODE, and Model_DAE.
10.11.3.7 fJ()
virtual void fJ (
realtype t,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
DlsMat J) [pure virtual]
Dense Jacobian function
Parameters
ttime
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
Jdense matrix to which values of the jacobian will be written
Implemented in Model_ODE, and Model_DAE.
Here is the caller graph for this function:
fJ amici::NewtonSolverDense
::prepareLinearSystem
Generated by Doxygen
10.11 Model Class Reference 165
10.11.3.8 fJSparse()
virtual void fJSparse (
realtype t,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
SlsMat J) [pure virtual]
Sparse Jacobian function
Parameters
ttime
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
Jsparse matrix to which values of the Jacobian will be written
Implemented in Model_ODE, and Model_DAE.
Here is the caller graph for this function:
fJSparse amici::NewtonSolverSparse
::prepareLinearSystem
10.11.3.9 fJDiag()
virtual void fJDiag (
realtype t,
AmiVector ∗Jdiag,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx ) [pure virtual]
Diagonal Jacobian function
Parameters
ttime
Jdiag array to which the diagonal of the Jacobian will be written
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
Generated by Doxygen

166 CONTENTS
Returns
flag indicating successful evaluation
Implemented in Model_ODE, and Model_DAE.
Here is the caller graph for this function:
fJDiag amici::NewtonSolverIterative
::linsolveSPBCG
amici::NewtonSolverIterative
::solveLinearSystem
10.11.3.10 fdxdotdp()
virtual void fdxdotdp (
realtype t,
AmiVector ∗x,
AmiVector ∗dx ) [pure virtual]
parameter derivative of residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
Returns
flag indicating successful evaluation
Implemented in Model_ODE, and Model_DAE.
Here is the caller graph for this function:
fdxdotdp amici::NewtonSolver
::computeNewtonSensis
Generated by Doxygen
10.11 Model Class Reference 167
10.11.3.11 fJv()
virtual void fJv (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
AmiVector ∗v,
AmiVector ∗nJv,
realtype cj ) [pure virtual]
Jacobian multiply function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
vmultiplication vector (unused)
nJv array to which result of multiplication will be written
cj scaling factor (inverse of timestep, DAE only)
Implemented in Model_ODE, and Model_DAE.
Here is the caller graph for this function:
fJv amici::NewtonSolverIterative
::linsolveSPBCG
amici::NewtonSolverIterative
::solveLinearSystem
10.11.3.12 fx0() [1/2]
void fx0 (
AmiVector ∗x)
Initial states
Parameters
xpointer to state variables
Definition at line 783 of file model.cpp.
Generated by Doxygen
168 CONTENTS
Here is the call graph for this function:
fx0
amici::AmiVector::reset
amici::AmiVector::data
Here is the caller graph for this function:
fx0 initializeStates initialize
10.11.3.13 fx0_fixedParameters() [1/2]
void fx0_fixedParameters (
AmiVector ∗x)
Sets only those initial states that are specified via fixedParmeters
Parameters
xpointer to state variables
Definition at line 788 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 169
Here is the call graph for this function:
fx0_fixedParameters
getReinitializeFixedParameter
InitialStates
amici::AmiVector::data
10.11.3.14 fdx0()
void fdx0 (
AmiVector ∗x0,
AmiVector ∗dx0 ) [virtual]
Initial value for time derivative of states (only necessary for DAEs)
Parameters
x0 Vector with the initial states
dx0 Vector to which the initial derivative states will be written (only DAE)
Definition at line 801 of file model.cpp.
Here is the caller graph for this function:
fdx0 initialize
10.11.3.15 fsx0() [1/2]
void fsx0 (
AmiVectorArray ∗sx,
const AmiVector ∗x)
Initial value for initial state sensitivities
Generated by Doxygen
170 CONTENTS
Parameters
sx pointer to state sensitivity variables
xpointer to state variables
Definition at line 803 of file model.cpp.
Here is the call graph for this function:
fsx0
amici::AmiVectorArray
::reset
amici::AmiVectorArray
::data
amici::AmiVector::data
10.11.3.16 fsx0_fixedParameters() [1/2]
void fsx0_fixedParameters (
AmiVectorArray ∗sx,
const AmiVector ∗x)
Sets only those initial states sensitivities that are affected from fx0 fixedParmeters
Parameters
sx pointer to state sensitivity variables
xpointer to state variables
Definition at line 793 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 171
Here is the call graph for this function:
fsx0_fixedParameters
getReinitializeFixedParameter
InitialStates
amici::AmiVectorArray
::data
amici::AmiVector::data
10.11.3.17 fsdx0()
void fsdx0 ( ) [virtual]
Sensitivity of derivative initial states sensitivities sdx0 (only necessary for DAEs)
Definition at line 809 of file model.cpp.
10.11.3.18 fstau() [1/2]
void fstau (
const realtype t,
const int ie,
const AmiVector ∗x,
const AmiVectorArray ∗sx )
Sensitivity of event timepoint, total derivative
Parameters
tcurrent timepoint
ie event index
xpointer to state variables
sx pointer to state sensitivity variables
Definition at line 811 of file model.cpp.
Generated by Doxygen
172 CONTENTS
Here is the call graph for this function:
fstau
t
amici::AmiVector::data
amici::AmiVectorArray
::data
10.11.3.19 fy() [1/2]
void fy (
int it,
ReturnData ∗rdata )
Observables / measurements
Parameters
it timepoint index
rdata pointer to return data instance
Definition at line 818 of file model.cpp.
Here is the call graph for this function:
fy
fw
getx
t
Generated by Doxygen
10.11 Model Class Reference 173
10.11.3.20 fdydp() [1/2]
void fdydp (
const int it,
ReturnData ∗rdata )
partial derivative of observables y w.r.t. model parameters p
Parameters
it timepoint index
rdata pointer to return data instance
Definition at line 825 of file model.cpp.
Here is the call graph for this function:
fdydp fwgetx
fdwdp
t
10.11.3.21 fdydx() [1/2]
void fdydx (
const int it,
ReturnData ∗rdata )
partial derivative of observables y w.r.t. state variables x
Parameters
it timepoint index
rdata pointer to return data instance
Definition at line 846 of file model.cpp.
Generated by Doxygen
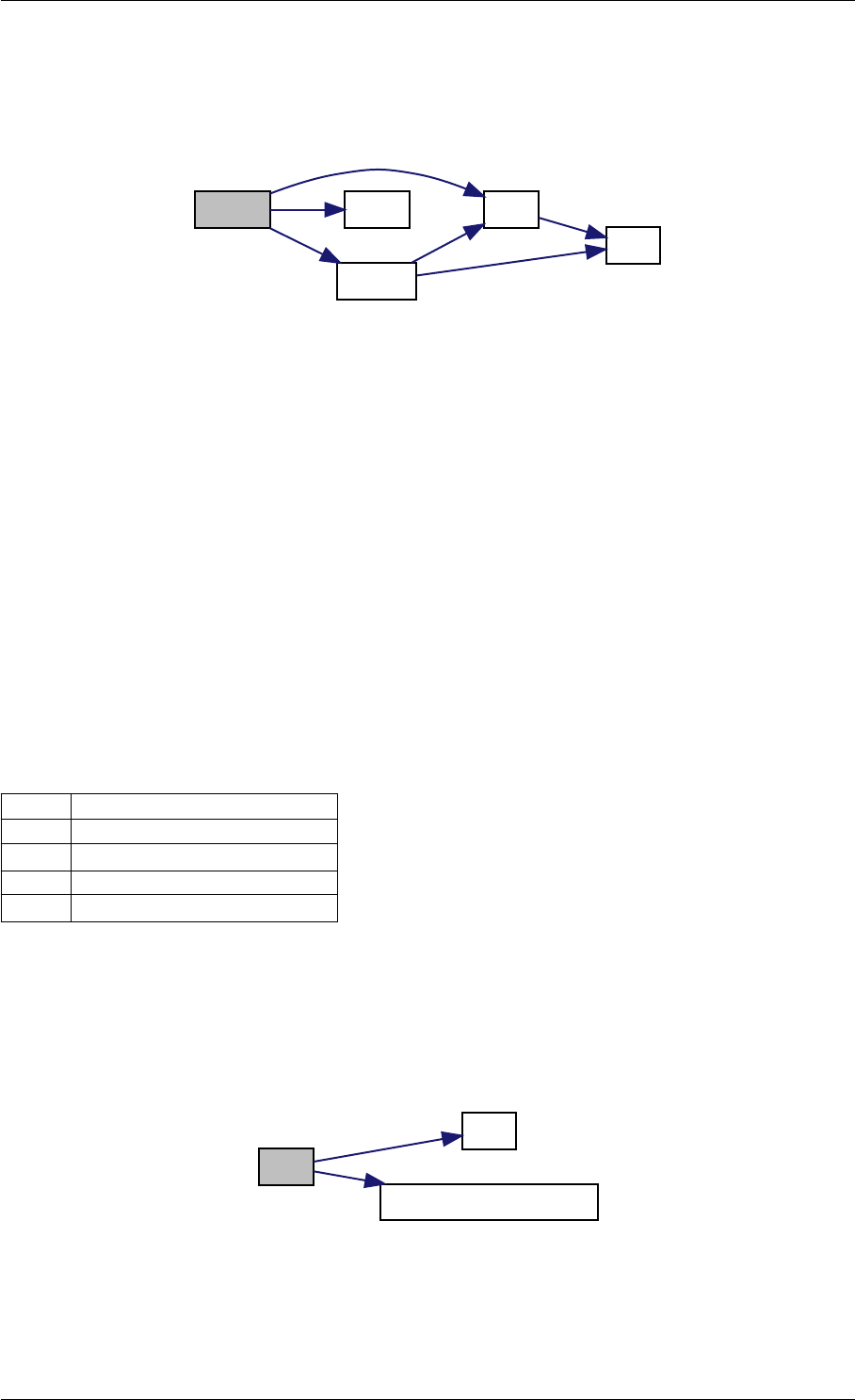
174 CONTENTS
Here is the call graph for this function:
fdydx fwgetx
fdwdx
t
10.11.3.22 fz() [1/2]
void fz (
const int nroots,
const int ie,
const realtype t,
const AmiVector ∗x,
ReturnData ∗rdata )
Event-resolved output
Parameters
nroots number of events for event index
ie event index
tcurrent timepoint
xcurrent state
rdata pointer to return data instance
Definition at line 856 of file model.cpp.
Here is the call graph for this function:
fz
t
amici::AmiVector::data
Generated by Doxygen
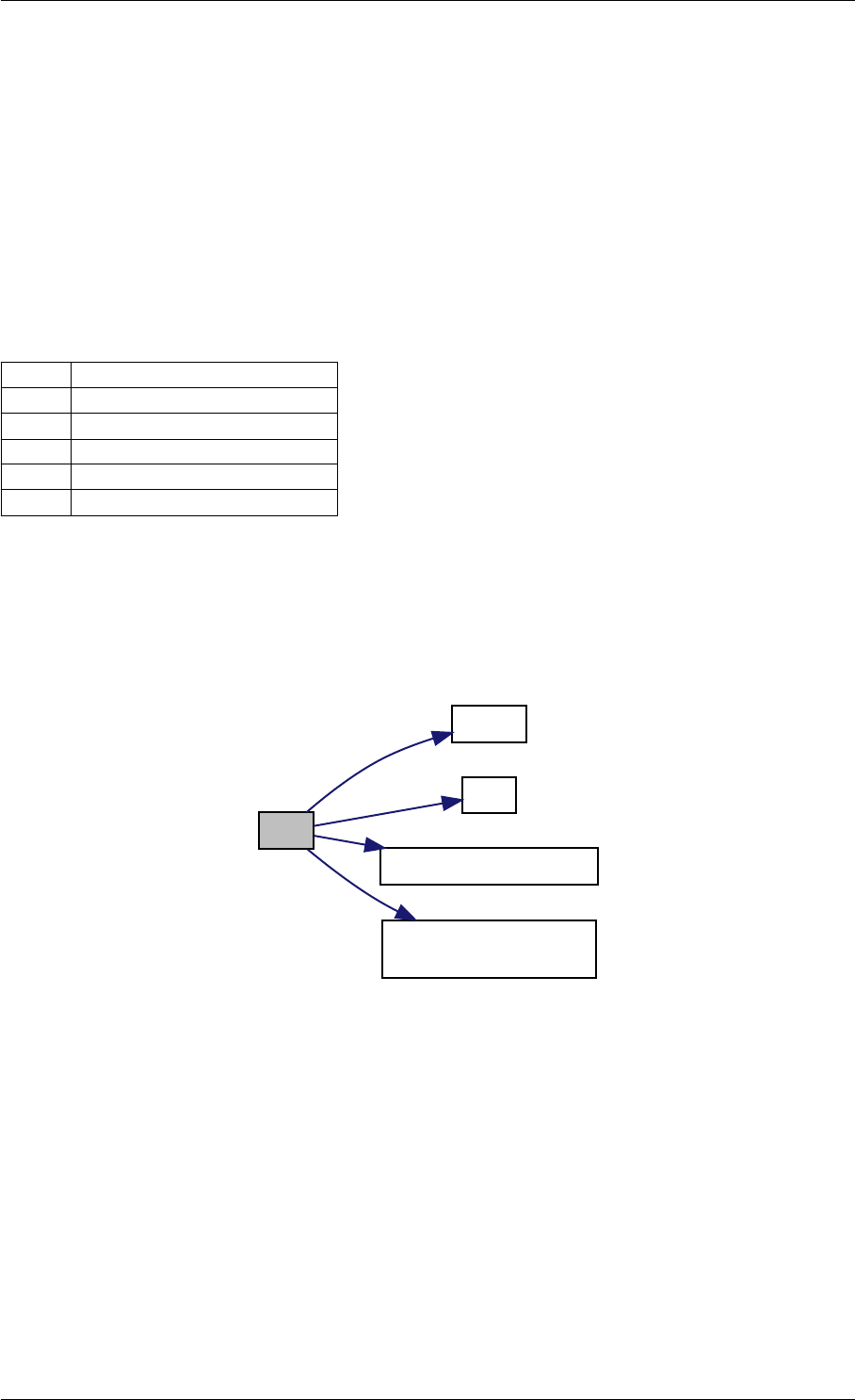
10.11 Model Class Reference 175
10.11.3.23 fsz() [1/2]
void fsz (
const int nroots,
const int ie,
const realtype t,
const AmiVector ∗x,
const AmiVectorArray ∗sx,
ReturnData ∗rdata )
Sensitivity of z, total derivative
Parameters
nroots number of events for event index
ie event index
tcurrent timepoint
xcurrent state
sx current state sensitivities
rdata pointer to return data instance
Definition at line 860 of file model.cpp.
Here is the call graph for this function:
fsz
nplist
t
amici::AmiVector::data
amici::AmiVectorArray
::data
10.11.3.24 frz() [1/2]
void frz (
const int nroots,
const int ie,
const realtype t,
const AmiVector ∗x,
ReturnData ∗rdata )
Event root function of events (equal to froot but does not include non-output events)
Generated by Doxygen
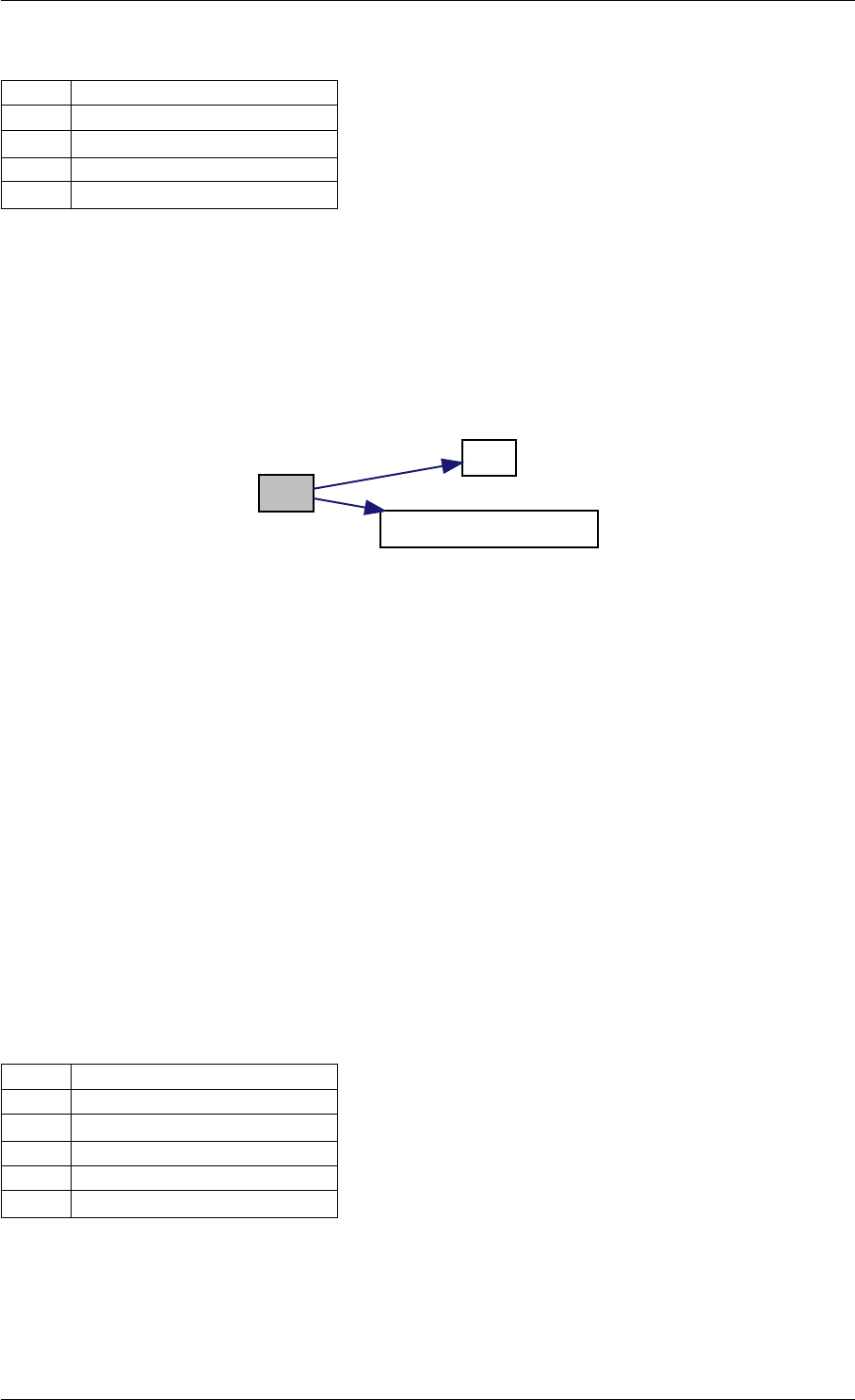
176 CONTENTS
Parameters
nroots number of events for event index
ie event index
tcurrent timepoint
xcurrent state
rdata pointer to return data instance
Definition at line 866 of file model.cpp.
Here is the call graph for this function:
frz
t
amici::AmiVector::data
10.11.3.25 fsrz() [1/2]
void fsrz (
const int nroots,
const int ie,
const realtype t,
const AmiVector ∗x,
const AmiVectorArray ∗sx,
ReturnData ∗rdata )
Sensitivity of rz, total derivative
Parameters
nroots number of events for event index
ie event index
tcurrent timepoint
xcurrent state
sx current state sensitivities
rdata pointer to return data instance
Definition at line 870 of file model.cpp.
Generated by Doxygen
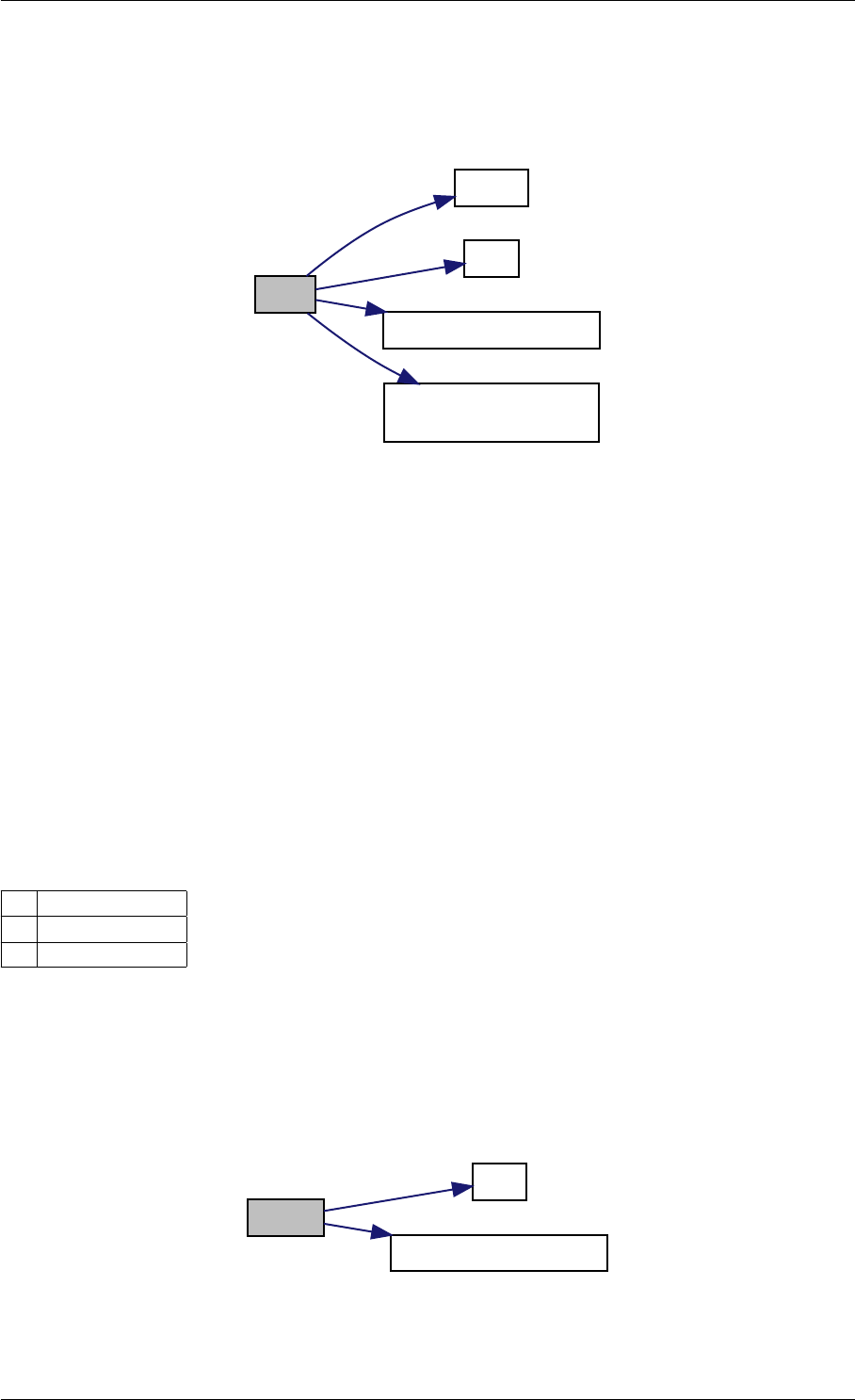
10.11 Model Class Reference 177
Here is the call graph for this function:
fsrz
nplist
t
amici::AmiVector::data
amici::AmiVectorArray
::data
10.11.3.26 fdzdp() [1/2]
void fdzdp (
const realtype t,
const int ie,
const AmiVector ∗x)
partial derivative of event-resolved output z w.r.t. to model parameters p
Parameters
ie event index
tcurrent timepoint
xcurrent state
Definition at line 876 of file model.cpp.
Here is the call graph for this function:
fdzdp
t
amici::AmiVector::data
Generated by Doxygen
178 CONTENTS
10.11.3.27 fdzdx() [1/2]
void fdzdx (
const realtype t,
const int ie,
const AmiVector ∗x)
partial derivative of event-resolved output z w.r.t. to model states x
Parameters
ie event index
tcurrent timepoint
xcurrent state
Definition at line 883 of file model.cpp.
Here is the call graph for this function:
fdzdx
t
amici::AmiVector::data
10.11.3.28 fdrzdp() [1/2]
void fdrzdp (
const realtype t,
const int ie,
const AmiVector ∗x)
Sensitivity of event-resolved root output w.r.t. to model parameters p
Parameters
ie event index
tcurrent timepoint
xcurrent state
Definition at line 888 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 179
Here is the call graph for this function:
fdrzdp
t
amici::AmiVector::data
10.11.3.29 fdrzdx() [1/2]
void fdrzdx (
const realtype t,
const int ie,
const AmiVector ∗x)
Sensitivity of event-resolved measurements rz w.r.t. to model states x
Parameters
ie event index
tcurrent timepoint
xcurrent state
Definition at line 896 of file model.cpp.
Here is the call graph for this function:
fdrzdx
t
amici::AmiVector::data
Generated by Doxygen
180 CONTENTS
10.11.3.30 fdeltax() [1/2]
void fdeltax (
const int ie,
const realtype t,
const AmiVector ∗x,
const AmiVector ∗xdot,
const AmiVector ∗xdot_old )
State update functions for events
Parameters
ie event index
tcurrent timepoint
xcurrent state
xdot current residual function values
xdot_old value of residual function before event
Definition at line 901 of file model.cpp.
Here is the call graph for this function:
fdeltax
t
amici::AmiVector::data
10.11.3.31 fdeltasx() [1/2]
void fdeltasx (
const int ie,
const realtype t,
const AmiVector ∗x,
const AmiVectorArray ∗sx,
const AmiVector ∗xdot,
const AmiVector ∗xdot_old )
Sensitivity update functions for events, total derivative
Parameters
ie event index
tcurrent timepoint
xcurrent state
sx current state sensitivity
xdot current residual function values
xdot_old value of residual function before event
Generated by Doxygen
10.11 Model Class Reference 181
Definition at line 907 of file model.cpp.
Here is the call graph for this function:
fdeltasx
fw
t
amici::AmiVector::getNVector
amici::AmiVector::data
amici::AmiVectorArray
::data
10.11.3.32 fdeltaxB() [1/2]
void fdeltaxB (
const int ie,
const realtype t,
const AmiVector ∗x,
const AmiVector ∗xB,
const AmiVector ∗xdot,
const AmiVector ∗xdot_old )
Adjoint state update functions for events
Parameters
ie event index
tcurrent timepoint
xcurrent state
xB current adjoint state
xdot current residual function values
xdot_old value of residual function before event
Definition at line 916 of file model.cpp.
Generated by Doxygen
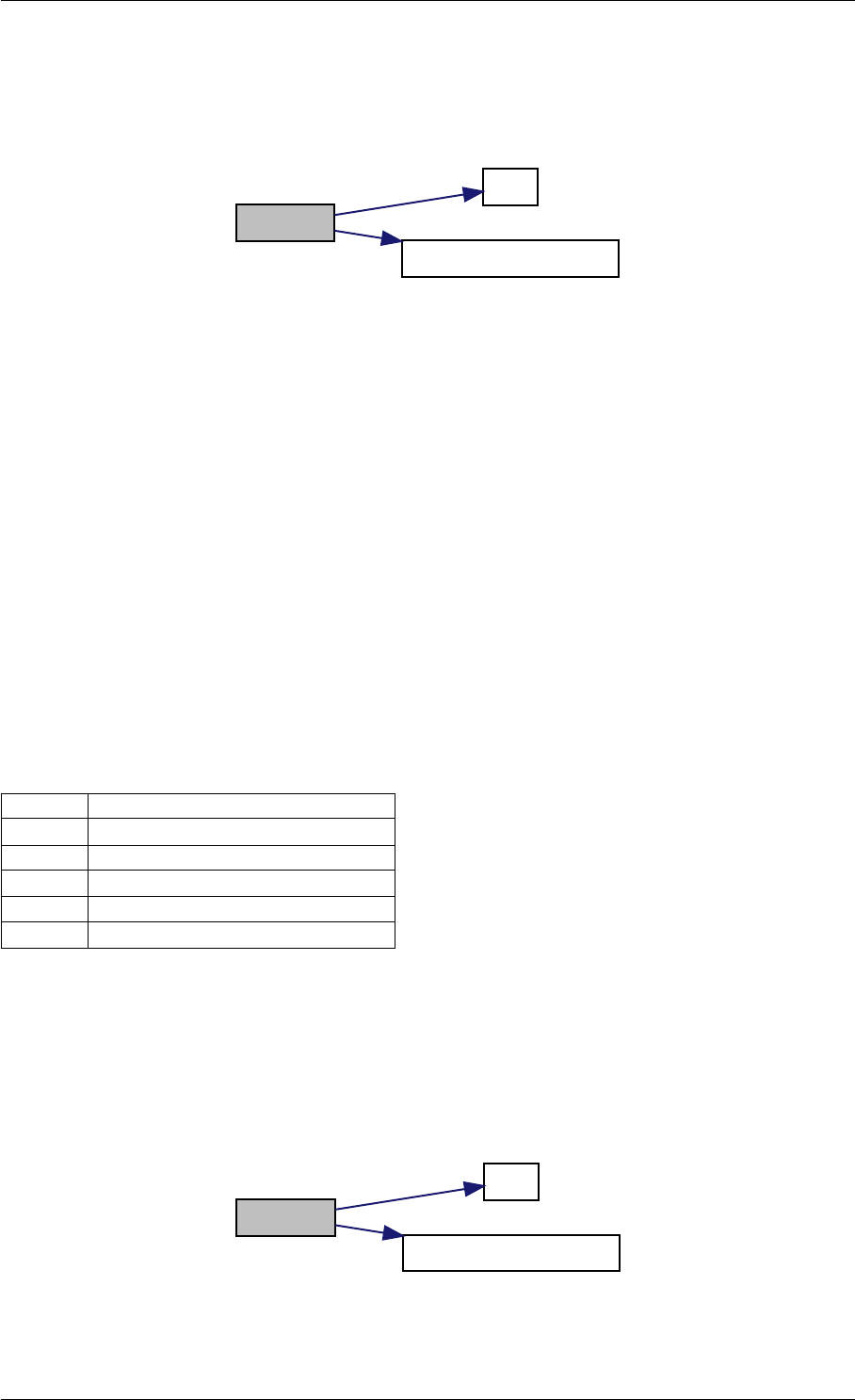
182 CONTENTS
Here is the call graph for this function:
fdeltaxB
t
amici::AmiVector::data
10.11.3.33 fdeltaqB() [1/2]
void fdeltaqB (
const int ie,
const realtype t,
const AmiVector ∗x,
const AmiVector ∗xB,
const AmiVector ∗xdot,
const AmiVector ∗xdot_old )
Quadrature state update functions for events
Parameters
ie event index
tcurrent timepoint
xcurrent state
xB current adjoint state
xdot current residual function values
xdot_old value of residual function before event
Definition at line 922 of file model.cpp.
Here is the call graph for this function:
fdeltaqB
t
amici::AmiVector::data
Generated by Doxygen
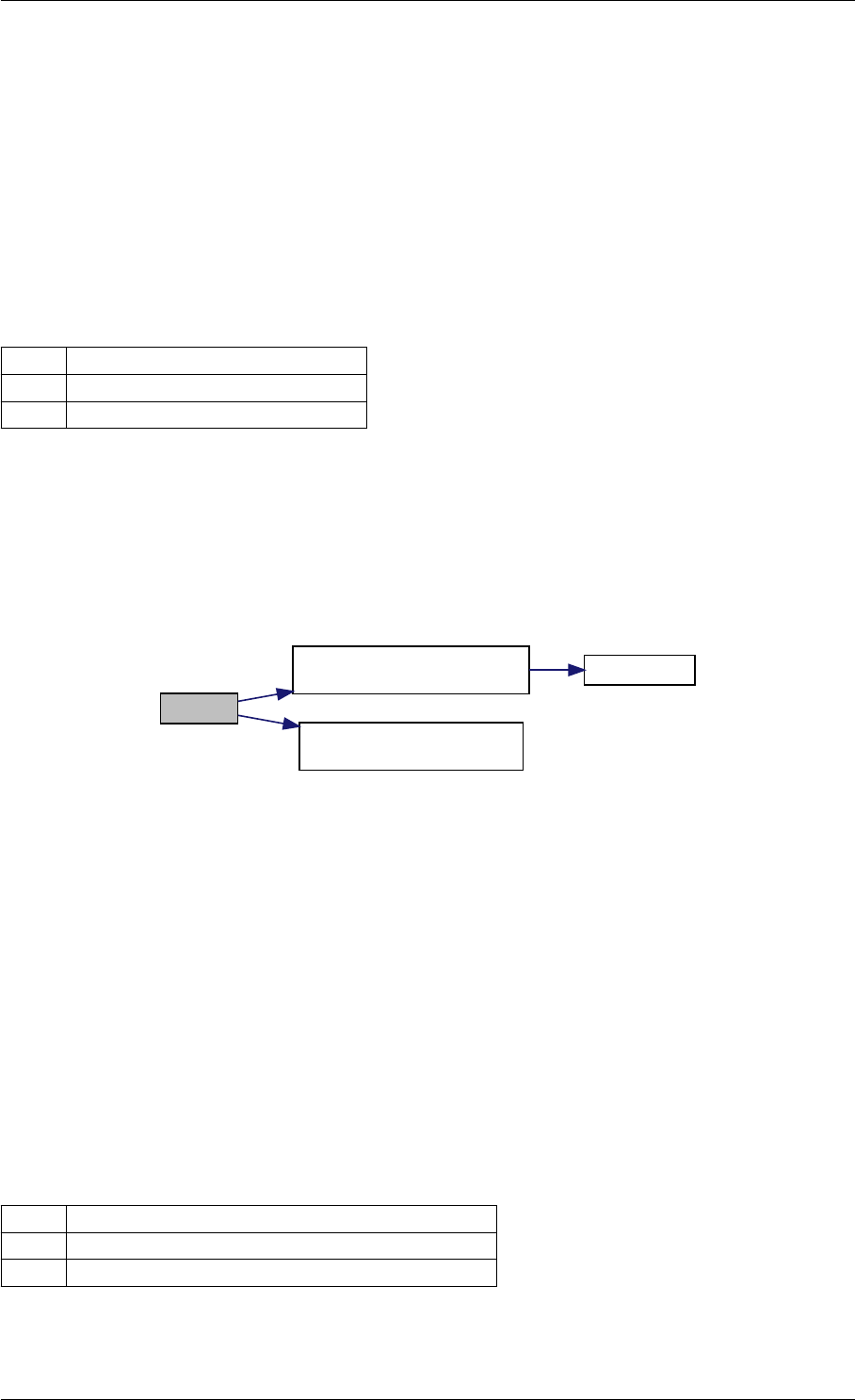
10.11 Model Class Reference 183
10.11.3.34 fsigmay() [1/2]
void fsigmay (
const int it,
ReturnData ∗rdata,
const ExpData ∗edata )
Standard deviation of measurements
Parameters
it timepoint index
edata pointer to experimental data instance
rdata pointer to return data instance
Definition at line 930 of file model.cpp.
Here is the call graph for this function:
fsigmay
amici::ExpData::isSetObserved
DataStdDev
amici::ExpData::getObserved
DataStdDevPtr
amici::isNaN
10.11.3.35 fdsigmaydp() [1/2]
void fdsigmaydp (
const int it,
ReturnData ∗rdata,
const ExpData ∗edata )
partial derivative of standard deviation of measurements w.r.t. model
Parameters
it timepoint index
rdata pointer to return data instance
edata pointer to ExpData data instance holding sigma values
Definition at line 949 of file model.cpp.
Generated by Doxygen
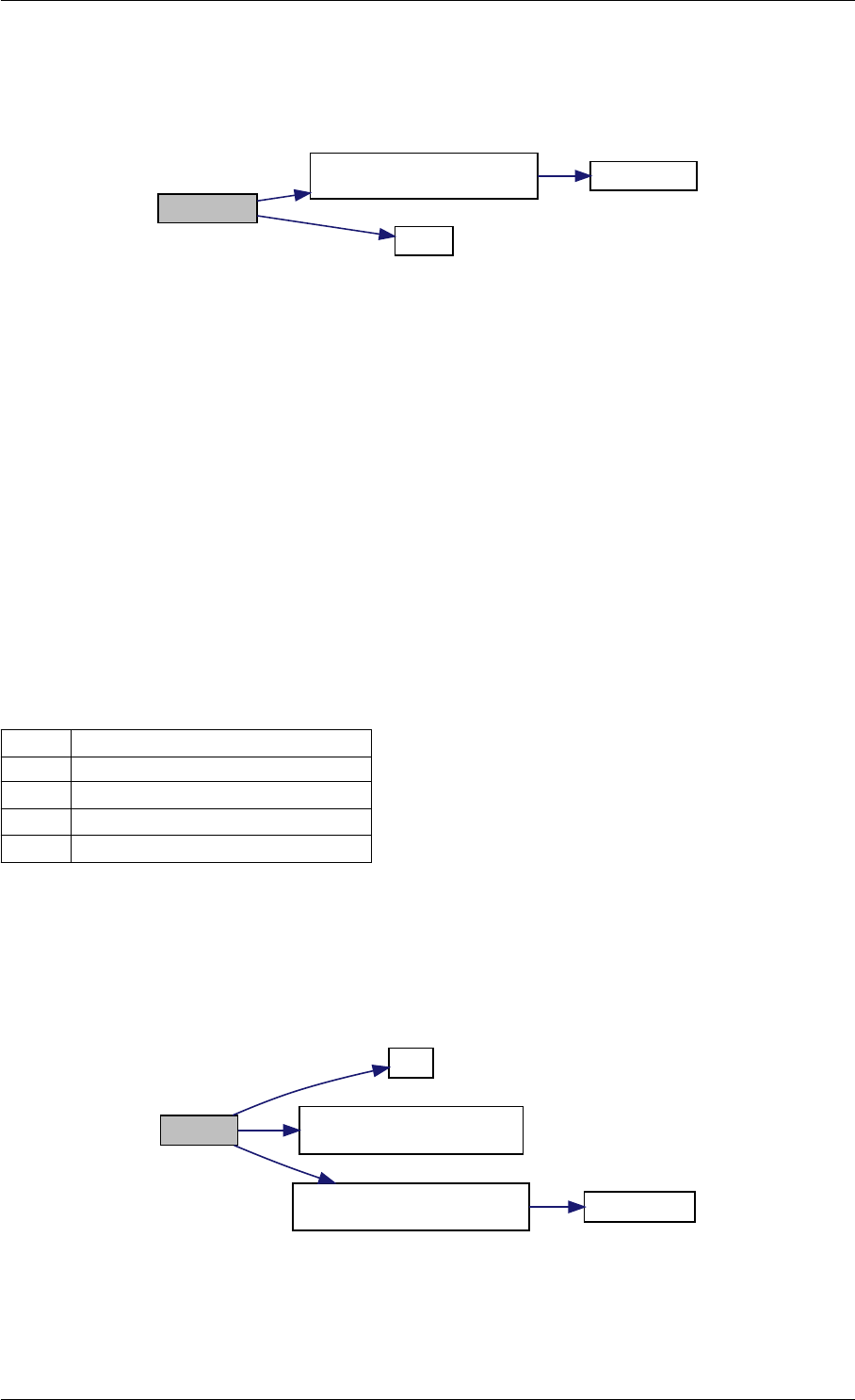
184 CONTENTS
Here is the call graph for this function:
fdsigmaydp
amici::ExpData::isSetObserved
DataStdDev
nplist
amici::isNaN
10.11.3.36 fsigmaz() [1/2]
void fsigmaz (
const realtype t,
const int ie,
const int ∗nroots,
ReturnData ∗rdata,
const ExpData ∗edata )
Standard deviation of events
Parameters
tcurrent timepoint
ie event index
nroots array with event numbers
edata pointer to experimental data instance
rdata pointer to return data instance
Definition at line 978 of file model.cpp.
Here is the call graph for this function:
fsigmaz
t
amici::ExpData::getObserved
EventsStdDevPtr
amici::ExpData::isSetObserved
EventsStdDev amici::isNaN
Generated by Doxygen
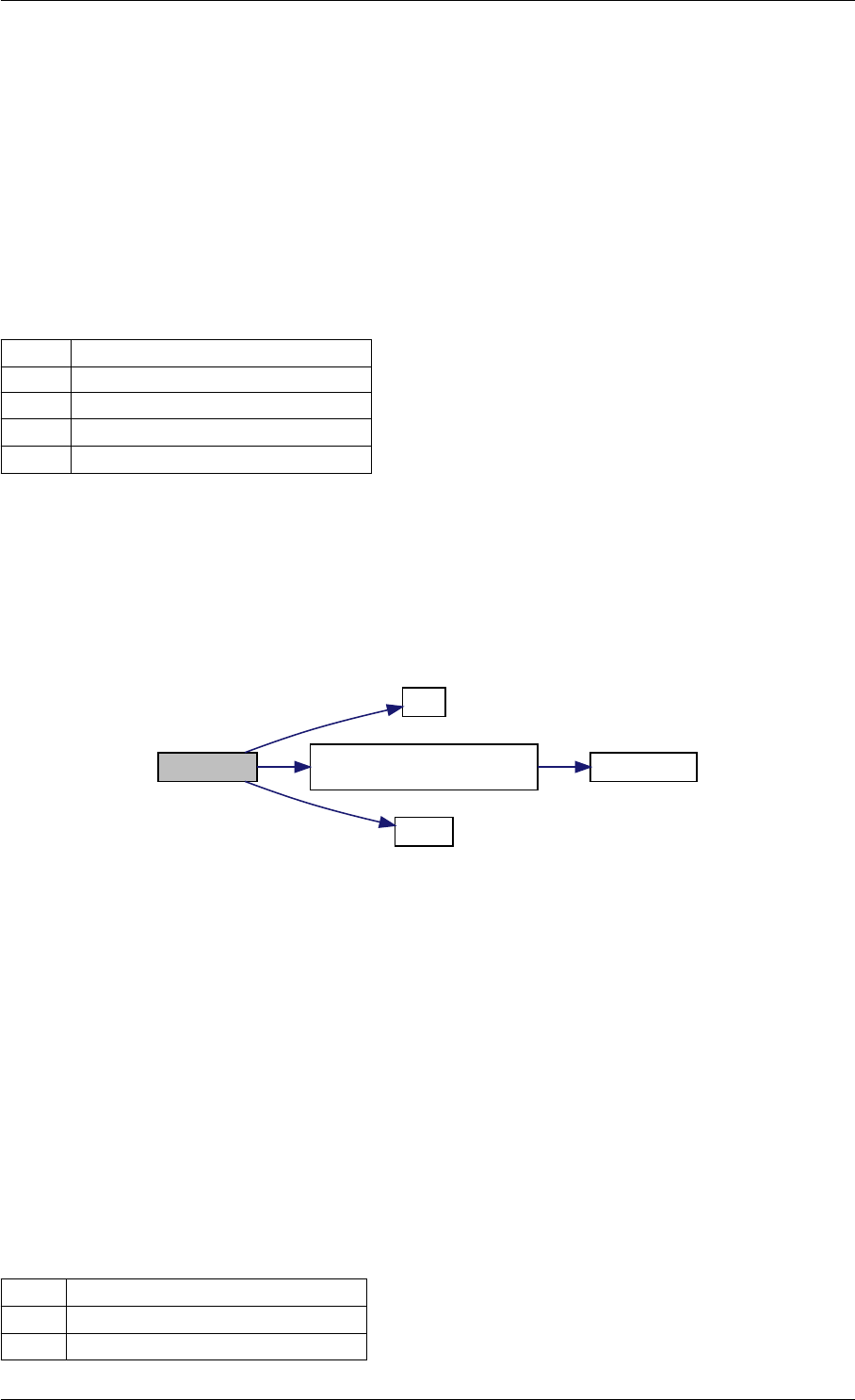
10.11 Model Class Reference 185
10.11.3.37 fdsigmazdp() [1/2]
void fdsigmazdp (
const realtype t,
const int ie,
const int ∗nroots,
ReturnData ∗rdata,
const ExpData ∗edata )
Sensitivity of standard deviation of events measurements w.r.t. model parameters p
Parameters
tcurrent timepoint
ie event index
nroots array with event numbers
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 996 of file model.cpp.
Here is the call graph for this function:
fdsigmazdp
t
amici::ExpData::isSetObserved
EventsStdDev
nplist
amici::isNaN
10.11.3.38 fJy() [1/2]
void fJy (
const int it,
ReturnData ∗rdata,
const ExpData ∗edata )
negative log-likelihood of measurements y
Parameters
it timepoint index
rdata pointer to return data instance
edata pointer to experimental data instance
Generated by Doxygen
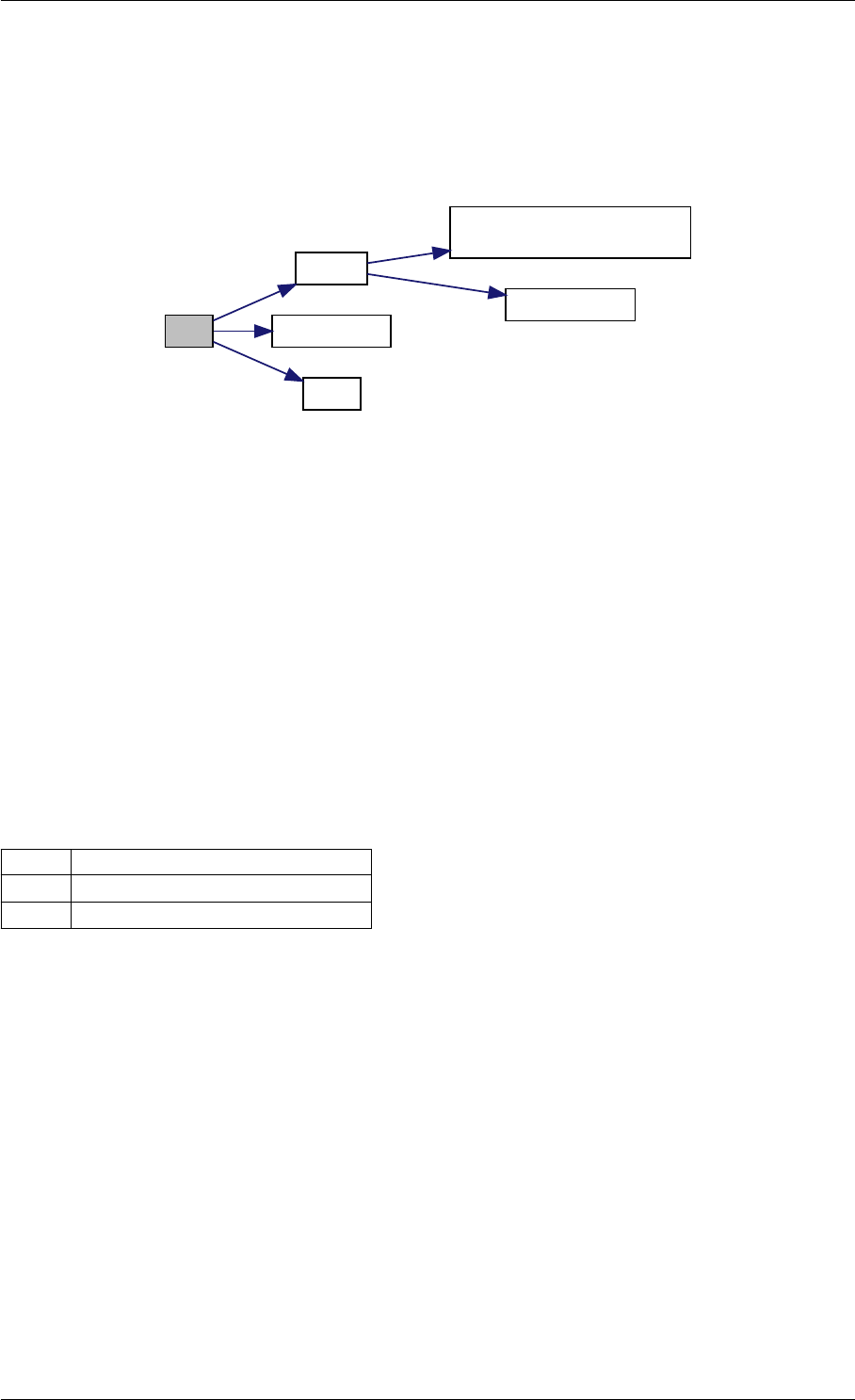
186 CONTENTS
Definition at line 1021 of file model.cpp.
Here is the call graph for this function:
fJy
getmy
amici::isNaN
gety
amici::ExpData::getObserved
DataPtr
amici::getNaN
10.11.3.39 fJz() [1/2]
void fJz (
const int nroots,
ReturnData ∗rdata,
const ExpData ∗edata )
negative log-likelihood of event-resolved measurements z
Parameters
nroots event index
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 1033 of file model.cpp.
Generated by Doxygen
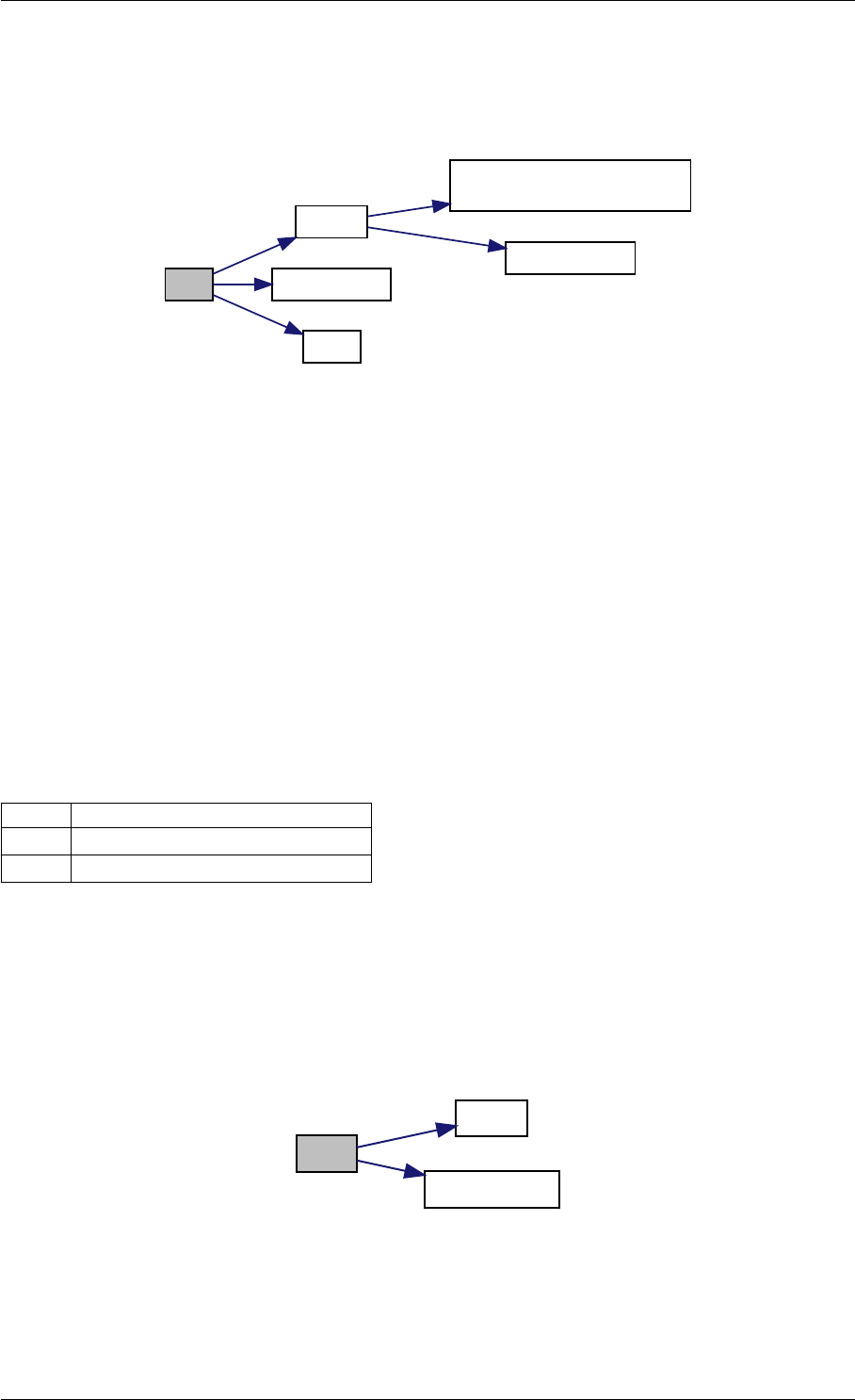
10.11 Model Class Reference 187
Here is the call graph for this function:
fJz
getmz
amici::isNaN
getz
amici::ExpData::getObserved
EventsPtr
amici::getNaN
10.11.3.40 fJrz() [1/2]
void fJrz (
const int nroots,
ReturnData ∗rdata,
const ExpData ∗edata )
regularization of negative log-likelihood with roots of event-resolved measurements rz
Parameters
nroots event index
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 1045 of file model.cpp.
Here is the call graph for this function:
fJrz
getrz
amici::isNaN
Generated by Doxygen
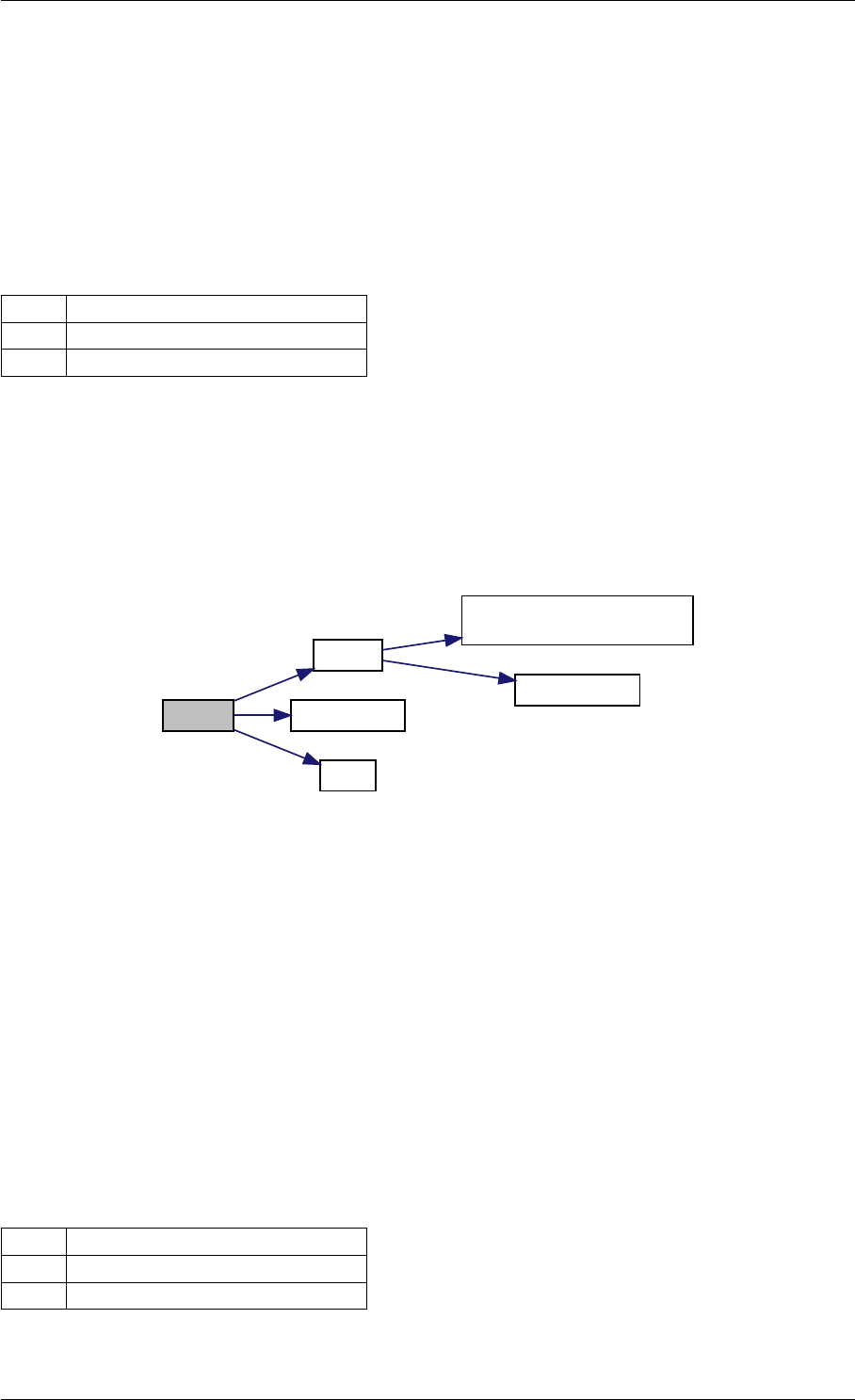
188 CONTENTS
10.11.3.41 fdJydy() [1/2]
void fdJydy (
const int it,
const ReturnData ∗rdata,
const ExpData ∗edata )
partial derivative of time-resolved measurement negative log-likelihood Jy
Parameters
it timepoint index
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 1057 of file model.cpp.
Here is the call graph for this function:
fdJydy
getmy
amici::isNaN
gety
amici::ExpData::getObserved
DataPtr
amici::getNaN
10.11.3.42 fdJydsigma() [1/2]
void fdJydsigma (
const int it,
const ReturnData ∗rdata,
const ExpData ∗edata )
Sensitivity of time-resolved measurement negative log-likelihood Jy w.r.t. standard deviation sigma
Parameters
it timepoint index
rdata pointer to return data instance
edata pointer to experimental data instance
Generated by Doxygen
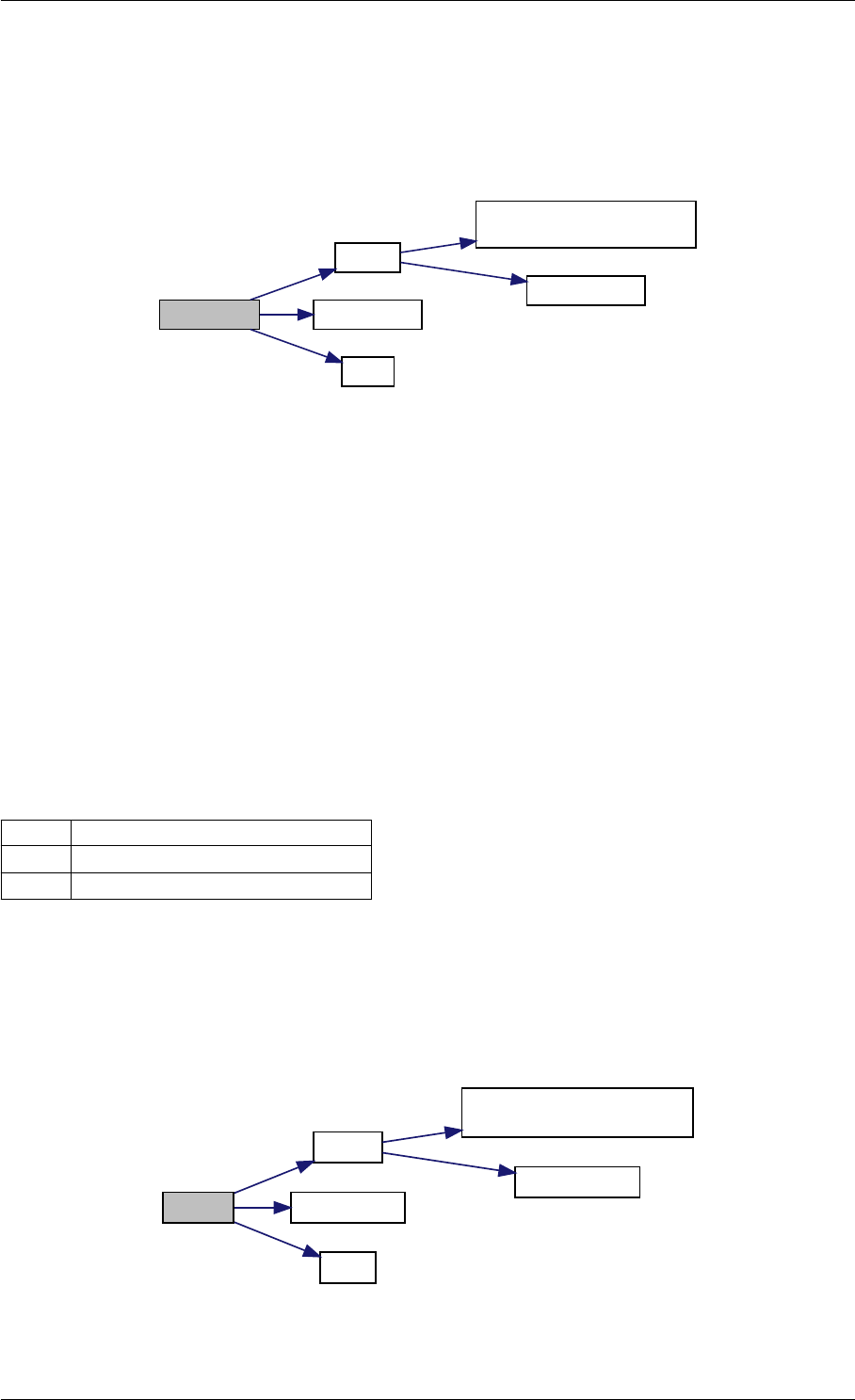
10.11 Model Class Reference 189
Definition at line 1077 of file model.cpp.
Here is the call graph for this function:
fdJydsigma
getmy
amici::isNaN
gety
amici::ExpData::getObserved
DataPtr
amici::getNaN
10.11.3.43 fdJzdz() [1/2]
void fdJzdz (
const int nroots,
const ReturnData ∗rdata,
const ExpData ∗edata )
partial derivative of event measurement negative log-likelihood Jz
Parameters
nroots event index
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 1097 of file model.cpp.
Here is the call graph for this function:
fdJzdz
getmz
amici::isNaN
getz
amici::ExpData::getObserved
EventsPtr
amici::getNaN
Generated by Doxygen
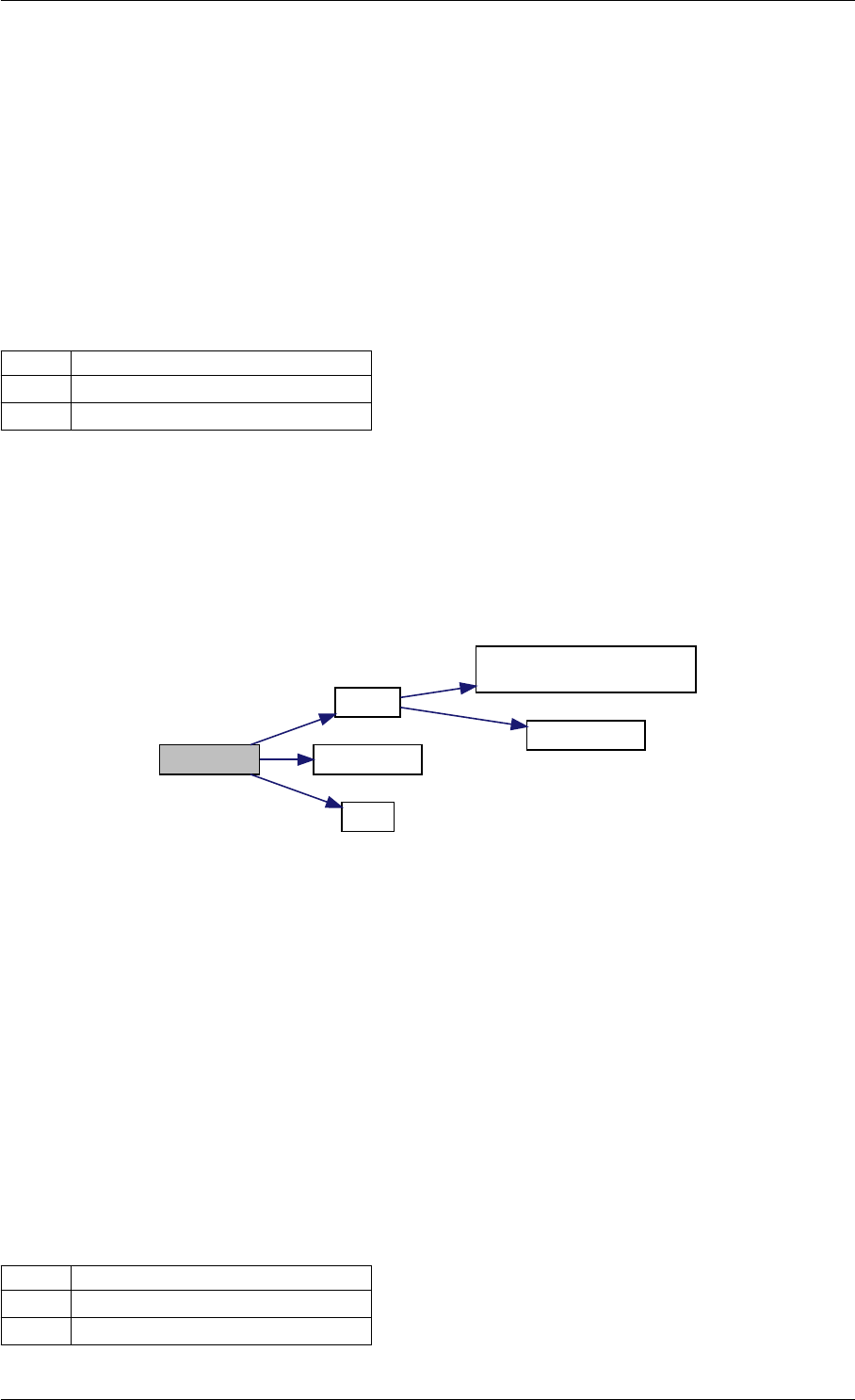
190 CONTENTS
10.11.3.44 fdJzdsigma() [1/2]
void fdJzdsigma (
const int nroots,
const ReturnData ∗rdata,
const ExpData ∗edata )
Sensitivity of event measurement negative log-likelihood Jz w.r.t. standard deviation sigmaz
Parameters
nroots event index
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 1108 of file model.cpp.
Here is the call graph for this function:
fdJzdsigma
getmz
amici::isNaN
getz
amici::ExpData::getObserved
EventsPtr
amici::getNaN
10.11.3.45 fdJrzdz() [1/2]
void fdJrzdz (
const int nroots,
const ReturnData ∗rdata,
const ExpData ∗edata )
partial derivative of event measurement negative log-likelihood Jz
Parameters
nroots event index
rdata pointer to return data instance
edata pointer to experimental data instance
Generated by Doxygen
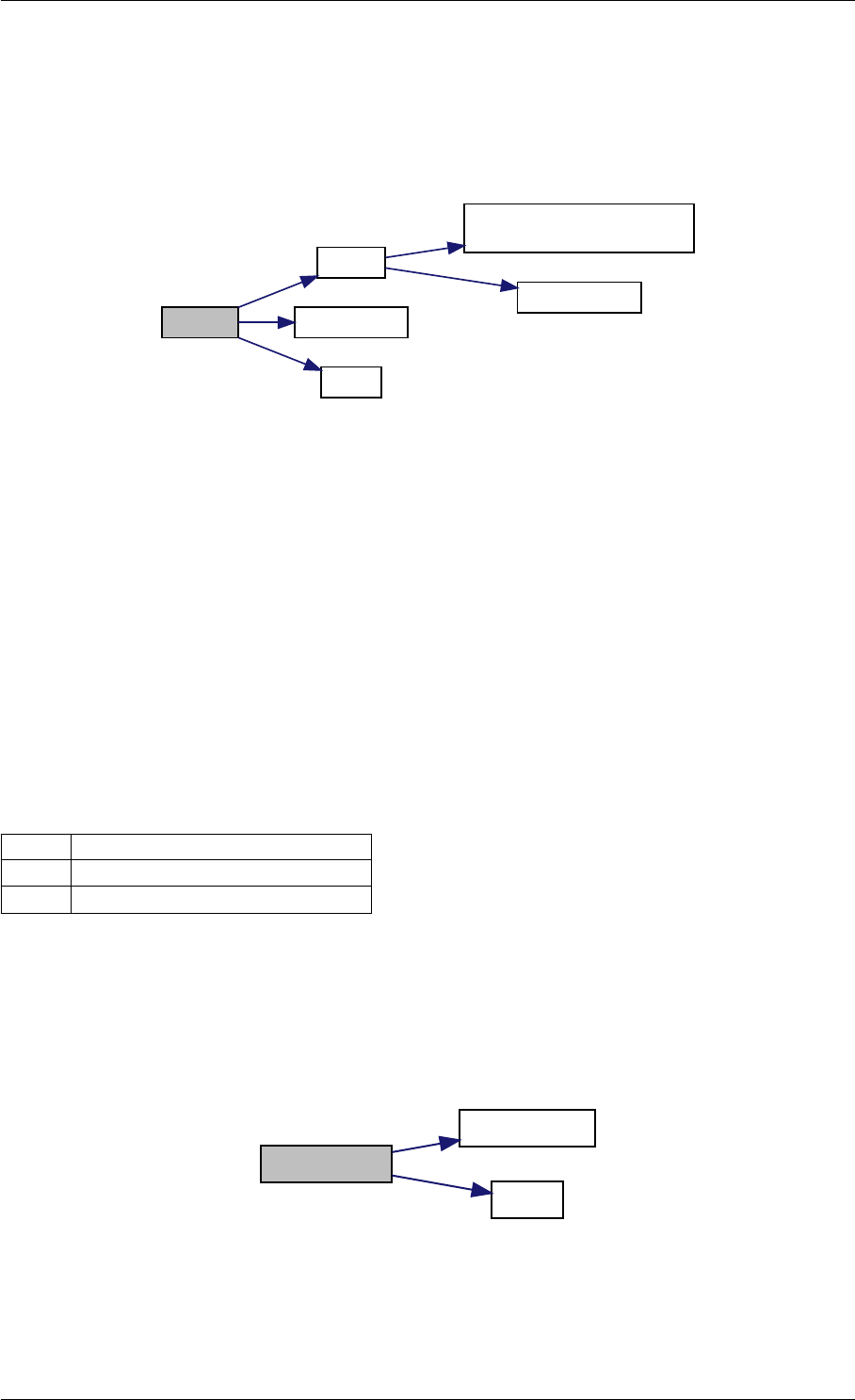
10.11 Model Class Reference 191
Definition at line 1119 of file model.cpp.
Here is the call graph for this function:
fdJrzdz
getmz
amici::isNaN
getrz
amici::ExpData::getObserved
EventsPtr
amici::getNaN
10.11.3.46 fdJrzdsigma() [1/2]
void fdJrzdsigma (
const int nroots,
const ReturnData ∗rdata,
const ExpData ∗edata )
Sensitivity of event measurement negative log-likelihood Jz w.r.t. standard deviation sigmaz
Parameters
nroots event index
rdata pointer to return data instance
edata pointer to experimental data instance
Definition at line 1130 of file model.cpp.
Here is the call graph for this function:
fdJrzdsigma
amici::isNaN
getrz
Generated by Doxygen
192 CONTENTS
10.11.3.47 fsy()
void fsy (
const int it,
ReturnData ∗rdata )
Sensitivity of measurements y, total derivative sy = dydx ∗sx + dydp
Parameters
it timepoint index
rdata pointer to return data instance
Definition at line 14 of file model.cpp.
Here is the call graph for this function:
fsy
nplist
amici::amici_dgemm
10.11.3.48 fsz_tf()
void fsz_tf (
const int ∗nroots,
const int ie,
ReturnData ∗rdata )
Sensitivity of z at final timepoint (ignores sensitivity of timepoint), total derivative
Parameters
nroots number of events for event index
ie event index
rdata pointer to return data instance
Definition at line 30 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 193
Here is the call graph for this function:
fsz_tf nplist
10.11.3.49 fsJy()
void fsJy (
const int it,
const std::vector<realtype >&dJydx,
ReturnData ∗rdata )
Sensitivity of time-resolved measurement negative log-likelihood Jy, total derivative
Parameters
it timepoint index
dJydx vector with values of state derivative of Jy
rdata pointer to return data instance
Definition at line 38 of file model.cpp.
Here is the call graph for this function:
fsJy
nplist
amici::amici_dgemm
10.11.3.50 fdJydp()
void fdJydp (
const int it,
Generated by Doxygen
194 CONTENTS
const ExpData ∗edata,
ReturnData ∗rdata )
Compute sensitivity of time-resolved measurement negative log-likelihood Jy w.r.t. parameters for the given time-
point. Add result to respective fields in rdata.
Parameters
it timepoint index
edata pointer to experimental data instance
rdata pointer to return data instance
Definition at line 67 of file model.cpp.
Here is the call graph for this function:
fdJydp
getmy
amici::isNaN
amici::amici_dgemm
amici::ExpData::getObserved
DataPtr
amici::getNaN
10.11.3.51 fdJydx()
void fdJydx (
std::vector<realtype >∗dJydx,
const int it,
const ExpData ∗edata,
const ReturnData ∗rdata )
Sensitivity of time-resolved measurement negative log-likelihood Jy w.r.t. state variables
Parameters
dJydx pointer to vector with values of state derivative of Jy
it timepoint index
edata pointer to experimental data instance
rdata pointer to return data instance
Definition at line 115 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 195
Here is the call graph for this function:
fdJydx
getmy
amici::isNaN
amici::amici_dgemm
amici::ExpData::getObserved
DataPtr
amici::getNaN
10.11.3.52 fsJz()
void fsJz (
const int nroots,
const std::vector<realtype >&dJzdx,
AmiVectorArray ∗sx,
ReturnData ∗rdata )
Sensitivity of event-resolved measurement negative log-likelihood Jz, total derivative
Parameters
nroots event index
dJzdx vector with values of state derivative of Jz
sx pointer to state sensitivities
rdata pointer to return data instance
Definition at line 134 of file model.cpp.
Here is the call graph for this function:
fsJz
nplist
amici::AmiVectorArray::at
amici::amici_dgemm
Generated by Doxygen
196 CONTENTS
10.11.3.53 fdJzdp()
void fdJzdp (
const int nroots,
realtype t,
const ExpData ∗edata,
const ReturnData ∗rdata )
Sensitivity of event-resolved measurement negative log-likelihood Jz w.r.t. parameters
Parameters
nroots event index
tcurrent timepoint
edata pointer to experimental data instance
rdata pointer to return data instance
Definition at line 168 of file model.cpp.
Here is the call graph for this function:
fdJzdp
getmz
amici::isNaN
amici::amici_dgemm
amici::ExpData::getObserved
EventsPtr
amici::getNaN
10.11.3.54 fdJzdx()
void fdJzdx (
std::vector<realtype >∗dJzdx,
const int nroots,
realtype t,
const ExpData ∗edata,
const ReturnData ∗rdata )
Sensitivity of event-resolved measurement negative log-likelihood Jz w.r.t. state variables
Parameters
dJzdx pointer to vector with values of state derivative of Jz
nroots event index
tcurrent timepoint
edata pointer to experimental data instance
rdata pointer to return data instance
Generated by Doxygen
10.11 Model Class Reference 197
Definition at line 204 of file model.cpp.
Here is the call graph for this function:
fdJzdx
getmz
amici::isNaN
amici::amici_dgemm
amici::ExpData::getObserved
EventsPtr
amici::getNaN
10.11.3.55 initialize()
void initialize (
AmiVector ∗x,
AmiVector ∗dx )
initialization of model properties
Parameters
xpointer to state variables
dx pointer to time derivative of states (DAE only)
Definition at line 227 of file model.cpp.
Here is the call graph for this function:
initialize
initializeStates
fdx0
initHeaviside
fx0
amici::AmiVector::reset
amici::AmiVector::data
froot
Generated by Doxygen
198 CONTENTS
10.11.3.56 initializeStates()
void initializeStates (
AmiVector ∗x)
initialization of initial states
Parameters
xpointer to state variables
Definition at line 238 of file model.cpp.
Here is the call graph for this function:
initializeStates fx0
amici::AmiVector::reset
amici::AmiVector::data
Here is the caller graph for this function:
initializeStates initialize
10.11.3.57 initHeaviside()
void initHeaviside (
AmiVector ∗x,
AmiVector ∗dx )
initHeaviside initialises the heaviside variables h at the intial time t0 heaviside variables activate/deactivate on event
occurences
Parameters
xpointer to state variables
dx pointer to time derivative of states (DAE only)
Generated by Doxygen
10.11 Model Class Reference 199
Definition at line 249 of file model.cpp.
Here is the call graph for this function:
initHeaviside froot
Here is the caller graph for this function:
initHeaviside initialize
10.11.3.58 nplist()
int nplist ( ) const
Returns
length of sensitivity index vector
Definition at line 266 of file model.cpp.
Generated by Doxygen
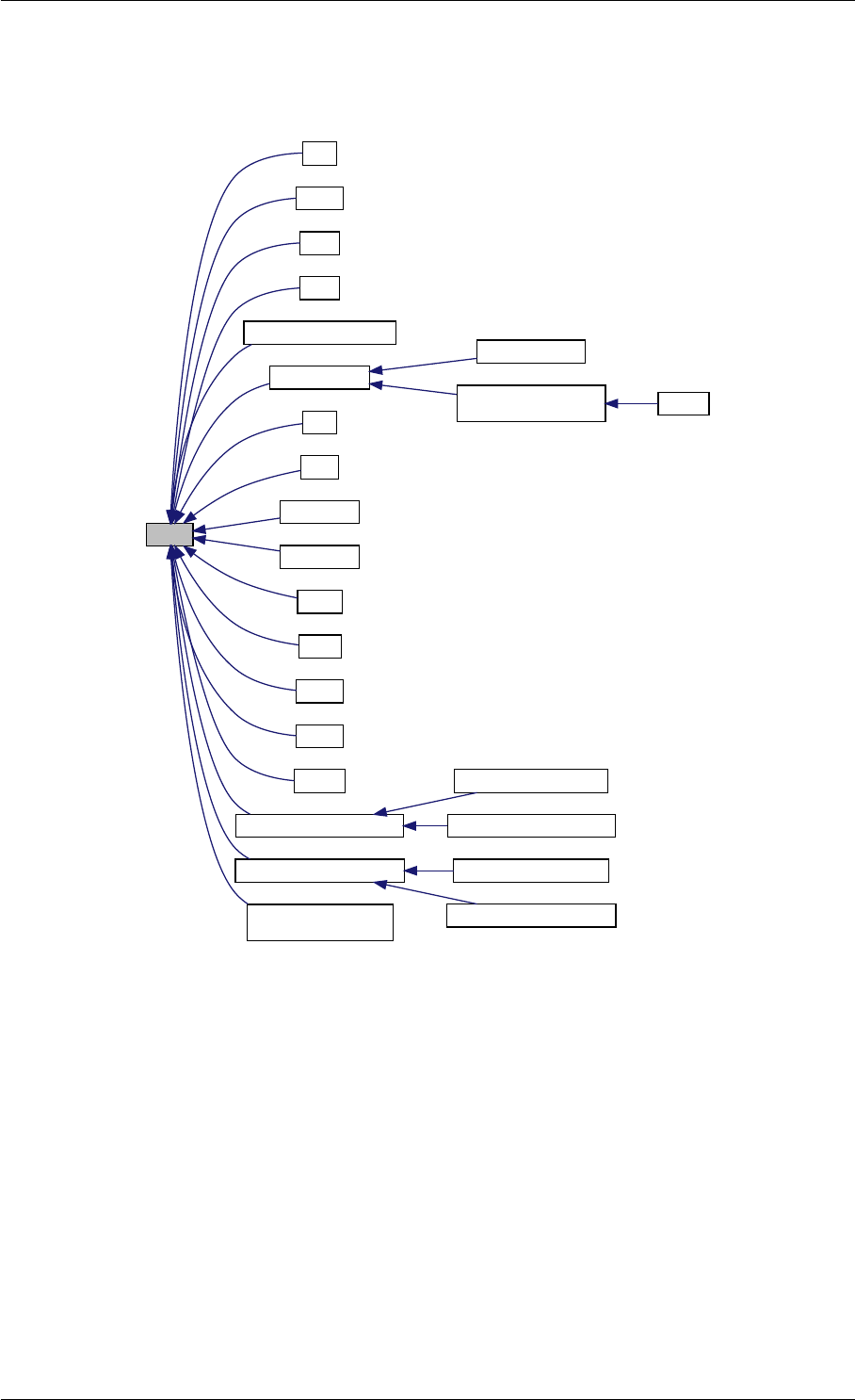
200 CONTENTS
Here is the caller graph for this function:
nplist
fsy
fsz_tf
fsJy
fsJz
setInitialStateSensitivities
initializeVectors
fsz
fsrz
fdsigmaydp
fdsigmazdp
fsres
fFIM
getsx
getsz
getsrz
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fdxdotdp
amici::NewtonSolver
::computeNewtonSensis
setParameterList
requireSensitivitiesForAll
Parameters Model
amici::Model_DAE::fsxdot
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fdxdotdp
10.11.3.59 np()
int np ( ) const
Returns
length of parameter vector
Definition at line 270 of file model.cpp.
Generated by Doxygen
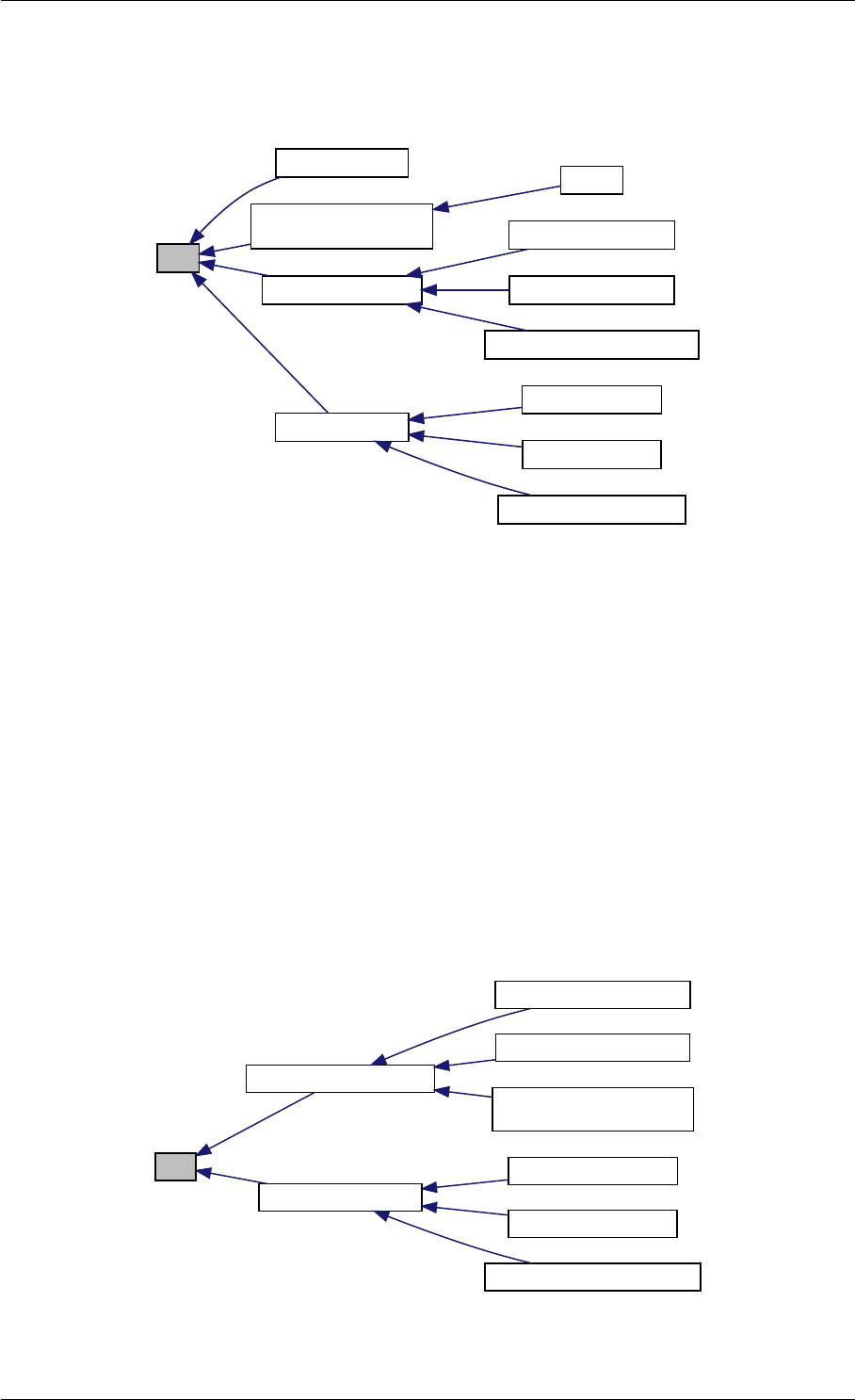
10.11 Model Class Reference 201
Here is the caller graph for this function:
np
setParameterList
requireSensitivitiesForAll
Parameters
hasParameterNames
hasParameterIds
Model
getParameterByName
setParameterByName
setParametersByNameRegex
getParameterById
setParameterById
setParametersByIdRegex
10.11.3.60 nk()
int nk ( ) const
Returns
length of constant vector
Definition at line 274 of file model.cpp.
Here is the caller graph for this function:
nk
hasFixedParameterNames
hasFixedParameterIds
getFixedParameterByName
setFixedParameterByName
setFixedParametersByName
Regex
getFixedParameterById
setFixedParameterById
setFixedParametersByIdRegex
Generated by Doxygen
202 CONTENTS
10.11.3.61 k()
const double ∗k ( ) const
Returns
pointer to constants array
Definition at line 278 of file model.cpp.
Here is the caller graph for this function:
k setFixedParameters
10.11.3.62 nMaxEvent()
int nMaxEvent ( ) const
Returns
maximum number of events that may occur for each type
Definition at line 282 of file model.cpp.
10.11.3.63 setNMaxEvent()
void setNMaxEvent (
int nmaxevent )
Parameters
nmaxevent maximum number of events that may occur for each type
Definition at line 286 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 203
10.11.3.64 nt()
int nt ( ) const
Returns
number of timepoints
Definition at line 290 of file model.cpp.
10.11.3.65 getParameterScale()
const std::vector<ParameterScaling >& getParameterScale ( ) const
Returns
vector of parameter scale
Definition at line 294 of file model.cpp.
10.11.3.66 setParameterScale() [1/2]
void setParameterScale (
ParameterScaling pscale )
Parameters
pscale scalar parameter scale for all parameters
Definition at line 298 of file model.cpp.
Here is the call graph for this function:
setParameterScale amici::unscaleParameters amici::getUnscaledParameter
10.11.3.67 setParameterScale() [2/2]
void setParameterScale (
const std::vector<ParameterScaling >&pscale )
Generated by Doxygen
204 CONTENTS
Parameters
pscale vector of parameter scales
Definition at line 304 of file model.cpp.
Here is the call graph for this function:
setParameterScale amici::unscaleParameters amici::getUnscaledParameter
10.11.3.68 getParameters()
std::vector<realtype >const & getParameters ( ) const
Returns
The user-set parameters (see also getUnscaledParameters)
Definition at line 310 of file model.cpp.
10.11.3.69 setParameters()
void setParameters (
std::vector<realtype >const & p)
Parameters
pvector of parameters
Definition at line 397 of file model.cpp.
Here is the call graph for this function:
setParameters amici::unscaleParameters amici::getUnscaledParameter
Generated by Doxygen
10.11 Model Class Reference 205
10.11.3.70 getUnscaledParameters()
const std::vector<realtype >& getUnscaledParameters ( ) const
Returns
unscaled parameters
Definition at line 459 of file model.cpp.
10.11.3.71 getFixedParameters()
const std::vector<realtype >& getFixedParameters ( ) const
Returns
vector of fixed parameters
Definition at line 463 of file model.cpp.
10.11.3.72 setFixedParameters()
void setFixedParameters (
std::vector<realtype >const & k)
Parameters
kvector of fixed parameters
Definition at line 489 of file model.cpp.
Here is the call graph for this function:
setFixedParameters k
10.11.3.73 getFixedParameterById()
realtype getFixedParameterById (
std::string const & par_id ) const
Generated by Doxygen
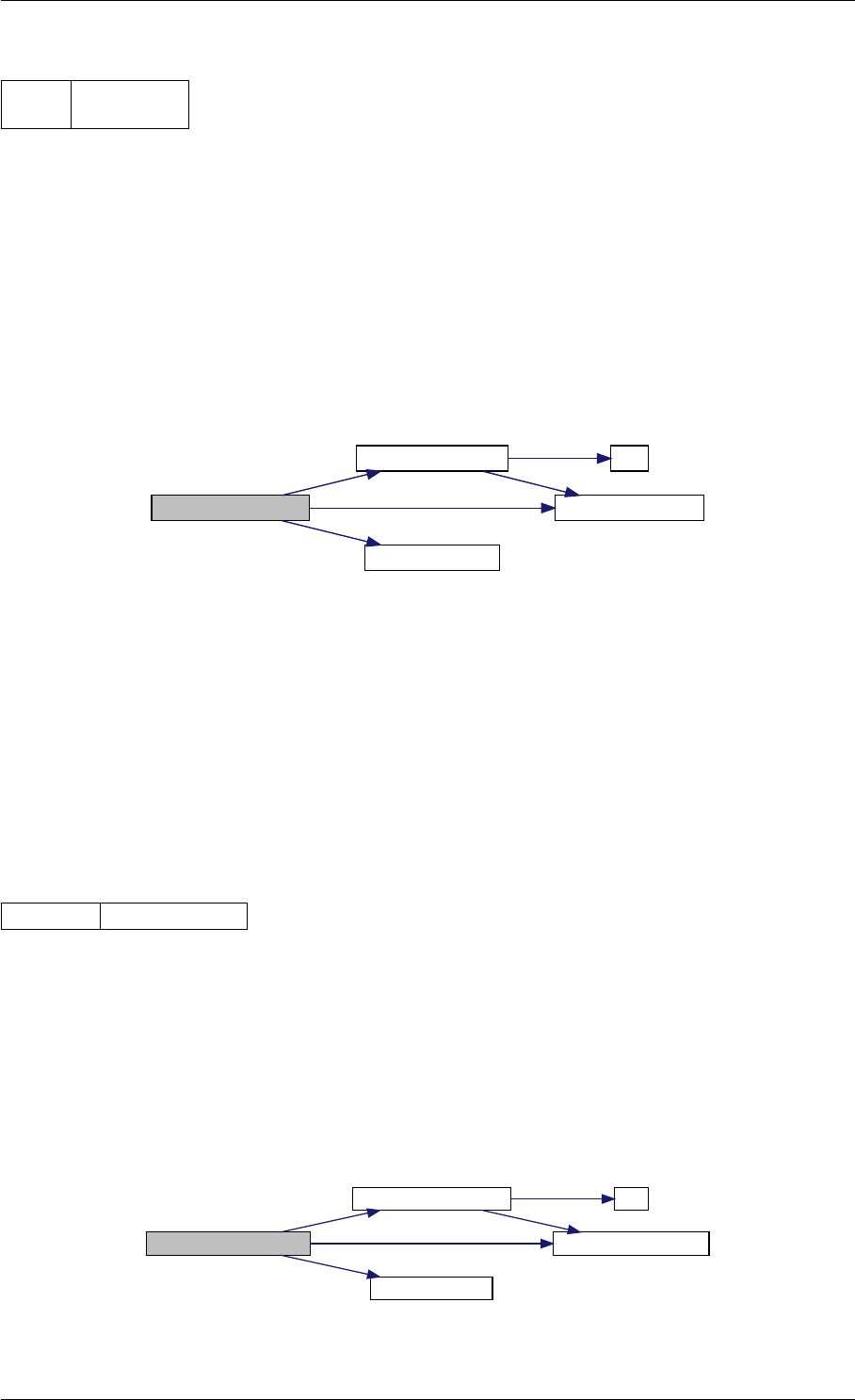
206 CONTENTS
Parameters
par←-
_id
parameter id
Returns
parameter value
Definition at line 467 of file model.cpp.
Here is the call graph for this function:
getFixedParameterById
hasFixedParameterIds
getFixedParameterIds
amici::getValueById
nk
10.11.3.74 getFixedParameterByName()
realtype getFixedParameterByName (
std::string const & par_name ) const
Parameters
par_name parameter name
Returns
parameter value
Definition at line 478 of file model.cpp.
Here is the call graph for this function:
getFixedParameterByName
hasFixedParameterNames
getFixedParameterNames
amici::getValueById
nk
Generated by Doxygen
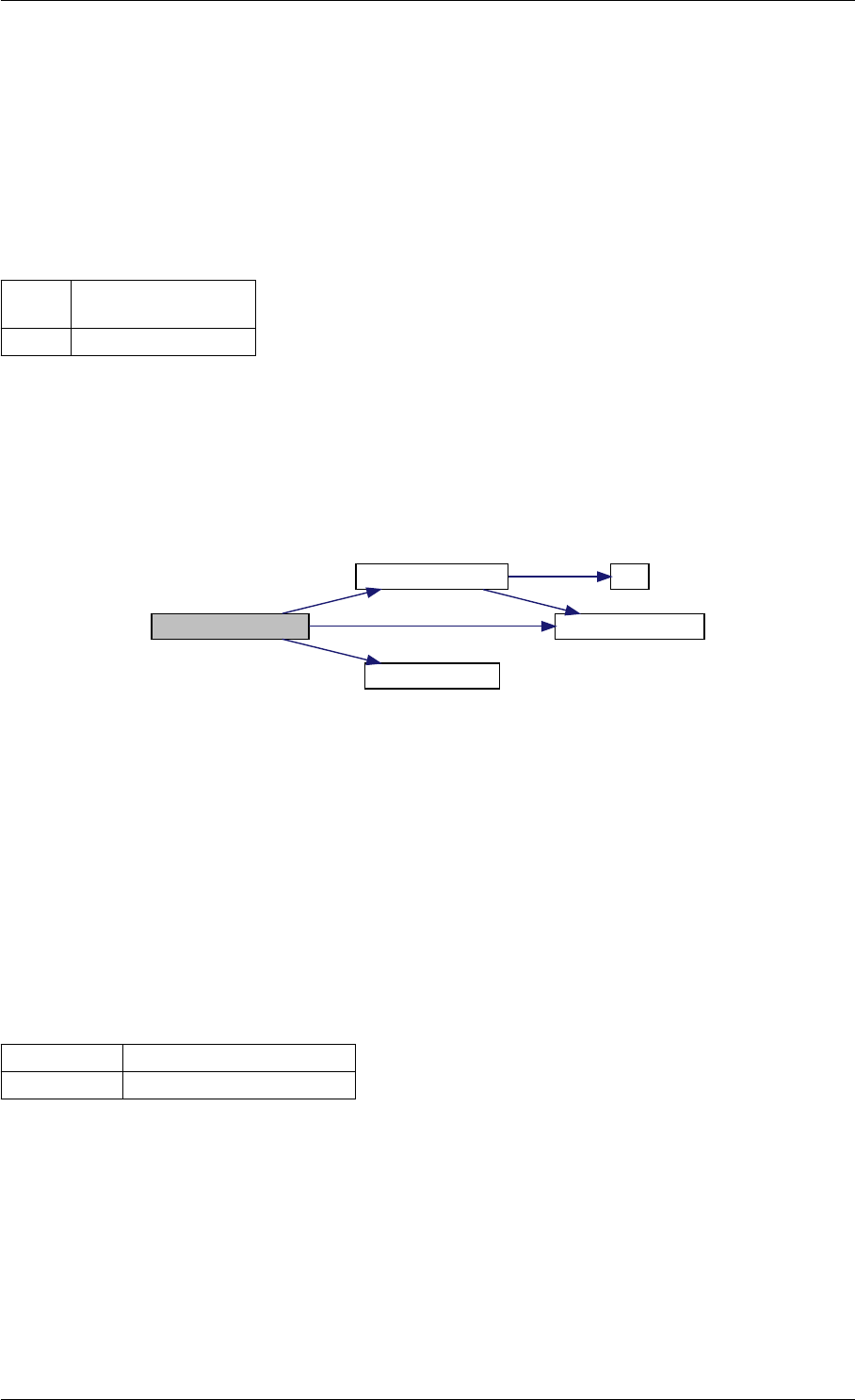
10.11 Model Class Reference 207
10.11.3.75 setFixedParameterById()
void setFixedParameterById (
std::string const & par_id,
realtype value )
Parameters
par←-
_id
fixed parameter id
value fixed parameter value
Definition at line 495 of file model.cpp.
Here is the call graph for this function:
setFixedParameterById
hasFixedParameterIds
getFixedParameterIds
amici::setValueById
nk
10.11.3.76 setFixedParametersByIdRegex()
int setFixedParametersByIdRegex (
std::string const & par_id_regex,
realtype value )
Parameters
par_id_regex fixed parameter name regex
value fixed parameter value
Returns
number of fixed parameter ids that matched the regex
Definition at line 507 of file model.cpp.
Generated by Doxygen
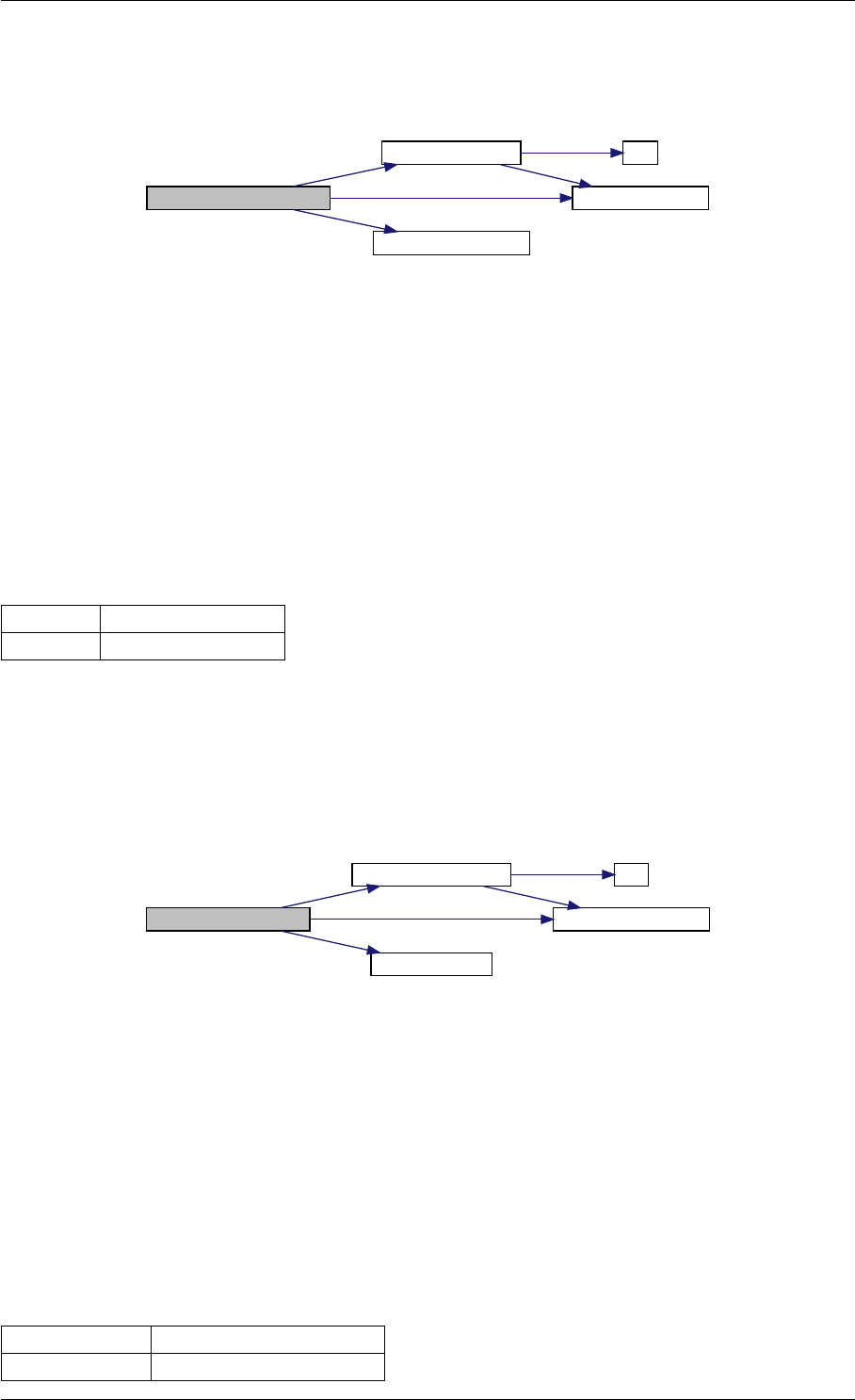
208 CONTENTS
Here is the call graph for this function:
setFixedParametersByIdRegex
hasFixedParameterIds
getFixedParameterIds
amici::setValueByIdRegex
nk
10.11.3.77 setFixedParameterByName()
void setFixedParameterByName (
std::string const & par_name,
realtype value )
Parameters
par_name fixed parameter id
value fixed parameter value
Definition at line 519 of file model.cpp.
Here is the call graph for this function:
setFixedParameterByName
hasFixedParameterNames
getFixedParameterNames
amici::setValueById
nk
10.11.3.78 setFixedParametersByNameRegex()
int setFixedParametersByNameRegex (
std::string const & par_name_regex,
realtype value )
Parameters
par_name_regex fixed parameter name regex
value fixed parameter value
Generated by Doxygen
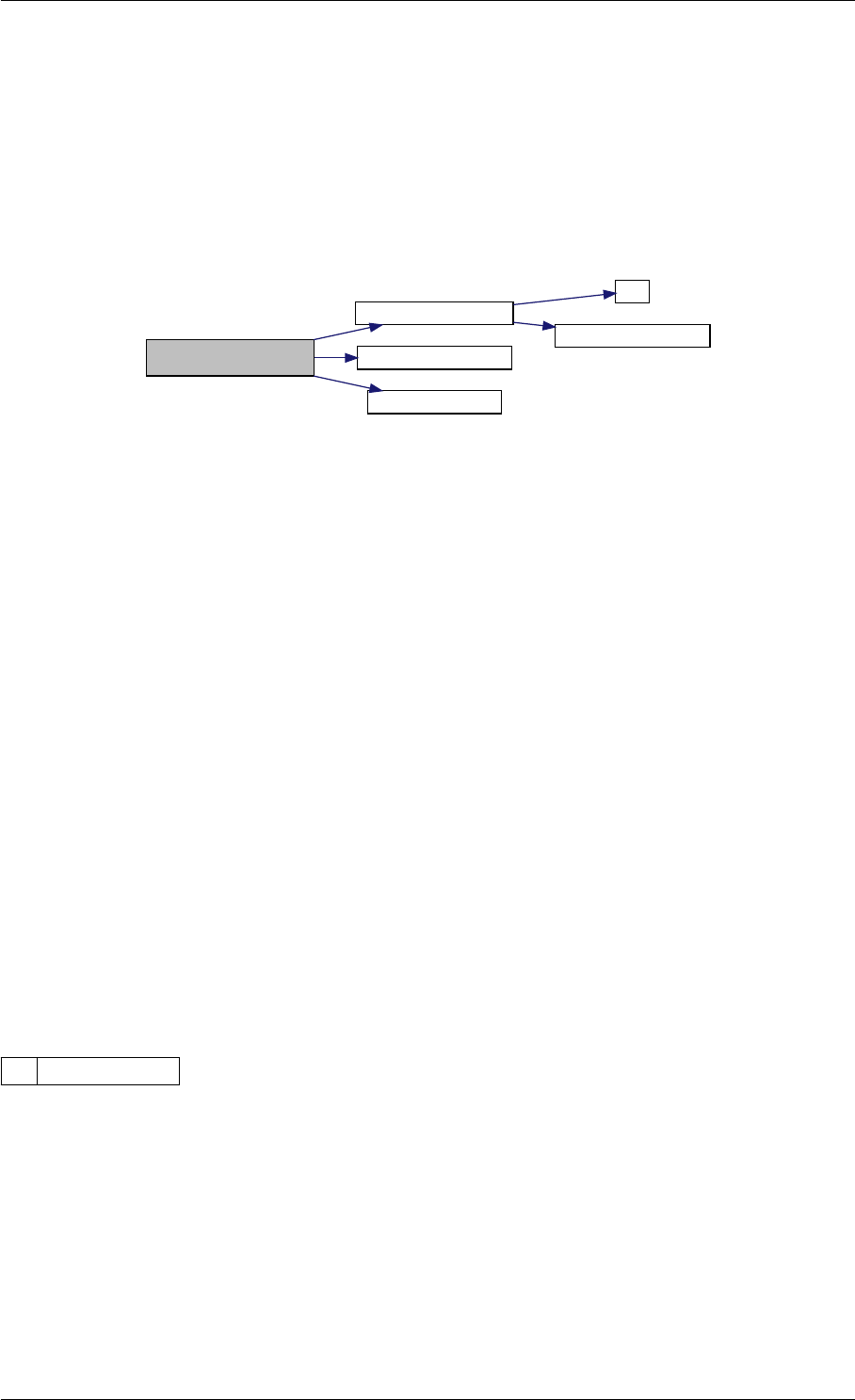
10.11 Model Class Reference 209
Returns
number of fixed parameter names that matched the regex
Definition at line 531 of file model.cpp.
Here is the call graph for this function:
setFixedParametersByName
Regex
hasFixedParameterNames
amici::setValueByIdRegex
getFixedParameterIds
nk
getFixedParameterNames
10.11.3.79 getTimepoints()
std::vector<realtype >const & getTimepoints ( ) const
Returns
timepoint vector
Definition at line 543 of file model.cpp.
10.11.3.80 setTimepoints()
void setTimepoints (
std::vector<realtype >const & ts )
Parameters
ts timepoint vector
Definition at line 547 of file model.cpp.
10.11.3.81 getStateIsNonNegative()
std::vector<bool >const & getStateIsNonNegative ( ) const
Generated by Doxygen
210 CONTENTS
Returns
vector of flags
Definition at line 553 of file model.cpp.
10.11.3.82 setStateIsNonNegative()
void setStateIsNonNegative (
std::vector<bool >const & stateIsNonNegative )
Parameters
stateIsNonNegative vector of flags
Definition at line 557 of file model.cpp.
10.11.3.83 t()
double t (
int idx ) const
Parameters
idx timepoint index
Returns
timepoint
Definition at line 570 of file model.cpp.
Generated by Doxygen
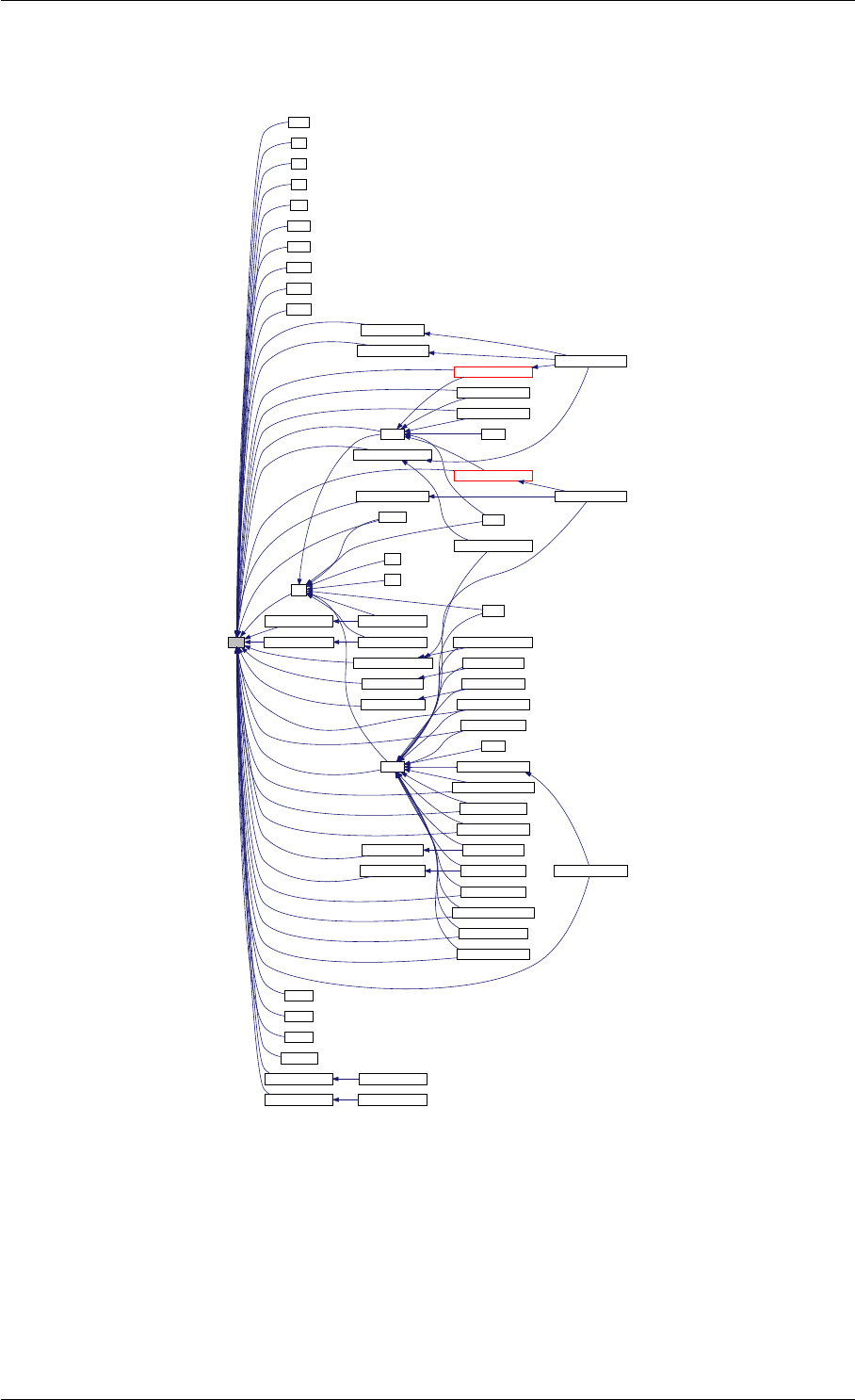
10.11 Model Class Reference 211
Here is the caller graph for this function:
t
fstau
fz
fsz
frz
fsrz
fdzdp
fdzdx
fdrzdp
fdrzdx
fdeltax
fdeltasx
fdeltaxB
fdeltaqB
fsigmaz
fdsigmazdp
fw
fdwdp
amici::Model_DAE::fdxdotdp
amici::Model_DAE::fqBdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fqBdot
fdwdx
amici::Model_DAE::fJDiag
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::froot
amici::Model_DAE::fxdot
amici::Model_DAE::fM
amici::Model_DAE::fsxdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::froot
amici::Model_ODE::fxdot
amici::Model_ODE::fsxdot
fy
fdydp
fdydx
fw
amici::Model_DAE::fxdot
amici::Model_ODE::fxdot
fdwdp
amici::Model_DAE::fsxdot
amici::Model_ODE::fsxdot
fdwdx
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::fJDiag
amici::Model_DAE::froot
amici::Model_ODE::froot
10.11.3.84 getParameterList()
const std::vector<int >& getParameterList ( ) const
Generated by Doxygen
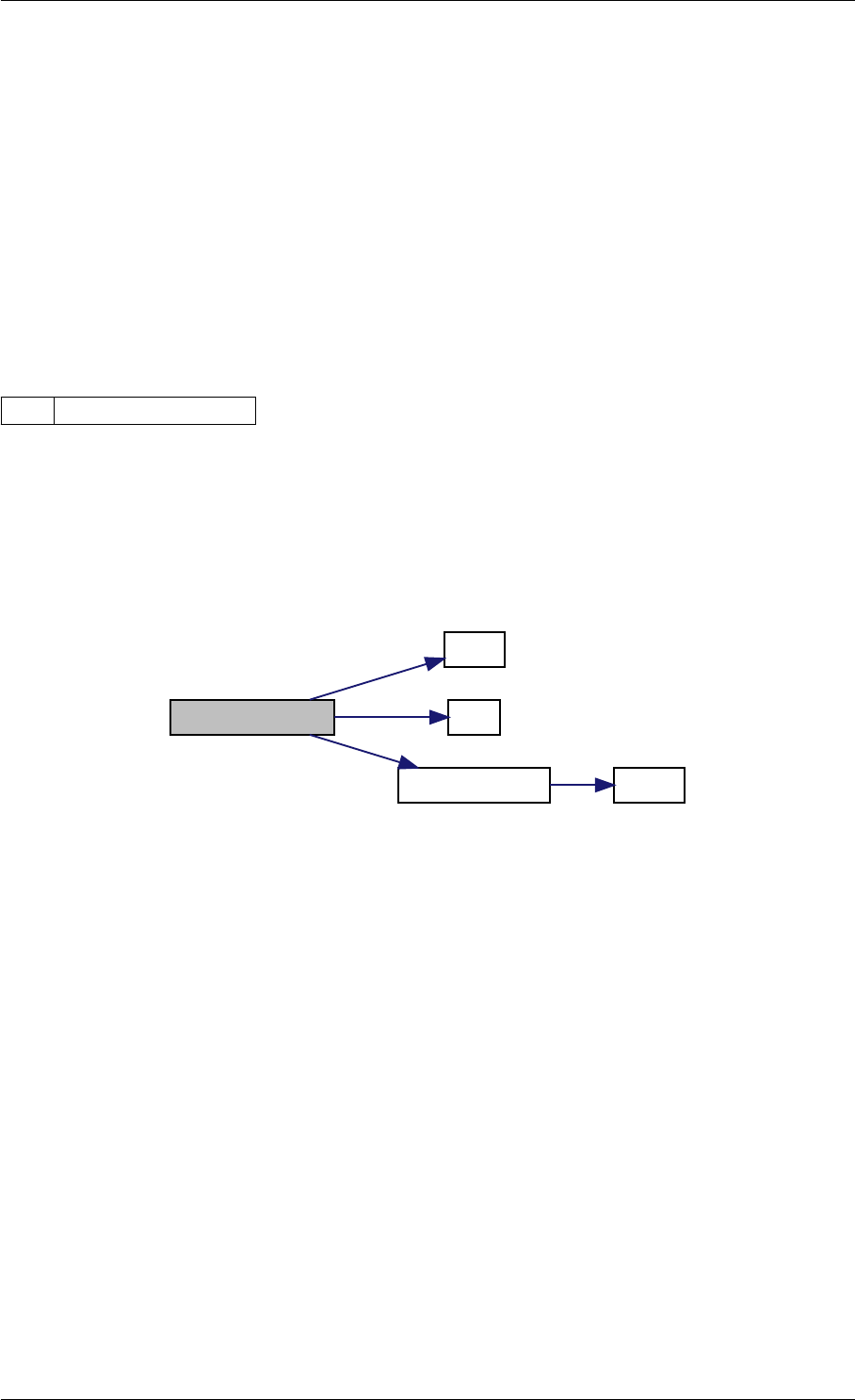
212 CONTENTS
Returns
list of parameter indices
Definition at line 574 of file model.cpp.
10.11.3.85 setParameterList()
void setParameterList (
std::vector<int >const & plist )
Parameters
plist list of parameter indices
Definition at line 578 of file model.cpp.
Here is the call graph for this function:
setParameterList
plist
np
initializeVectors nplist
10.11.3.86 getInitialStates()
std::vector<realtype >const & getInitialStates ( ) const
Returns
initial state vector
Definition at line 587 of file model.cpp.
10.11.3.87 setInitialStates()
void setInitialStates (
std::vector<realtype >const & x0 )
Generated by Doxygen
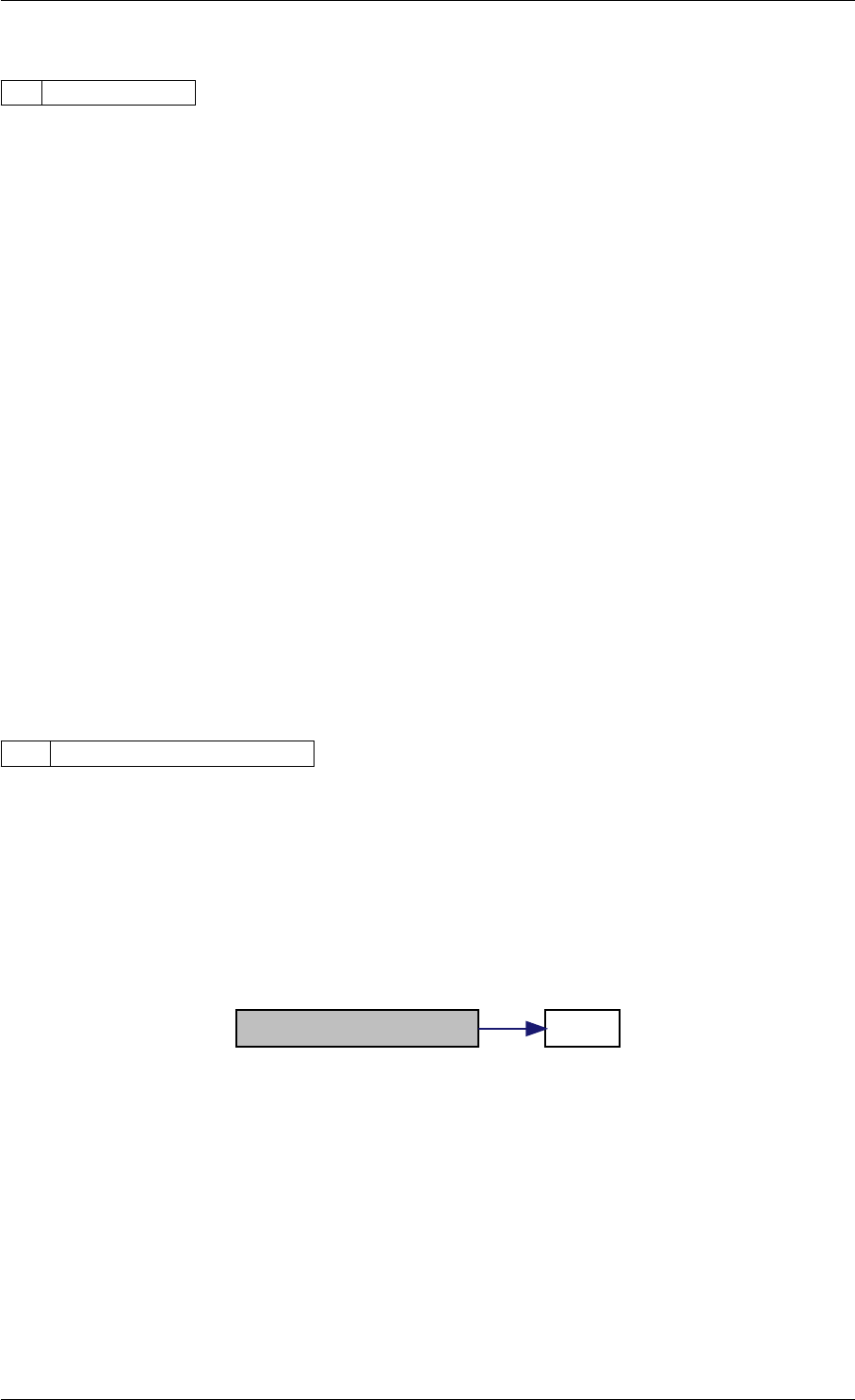
10.11 Model Class Reference 213
Parameters
x0 initial state vector
Definition at line 591 of file model.cpp.
10.11.3.88 getInitialStateSensitivities()
const std::vector<realtype >& getInitialStateSensitivities ( ) const
Returns
vector of initial state sensitivities
Definition at line 600 of file model.cpp.
10.11.3.89 setInitialStateSensitivities()
void setInitialStateSensitivities (
std::vector<realtype >const & sx0 )
Parameters
sx0 vector of initial state sensitivities
Definition at line 604 of file model.cpp.
Here is the call graph for this function:
setInitialStateSensitivities nplist
10.11.3.90 t0()
double t0 ( ) const
Generated by Doxygen
214 CONTENTS
Returns
simulation start time
Definition at line 613 of file model.cpp.
Here is the caller graph for this function:
t0 setT0
10.11.3.91 setT0()
void setT0 (
double t0 )
Parameters
t0 simulation start time
Definition at line 617 of file model.cpp.
Here is the call graph for this function:
setT0 t0
10.11.3.92 plist()
int plist (
int pos ) const
Parameters
pos index
Generated by Doxygen
10.11 Model Class Reference 215
Returns
entry
Definition at line 621 of file model.cpp.
Here is the caller graph for this function:
plist setParameterList
10.11.3.93 fw() [1/3]
void fw (
const realtype t,
const N_Vector x)
Parameters
ttimepoint
xVector with the states
Definition at line 1140 of file model.cpp.
Here is the call graph for this function:
fw t
Generated by Doxygen
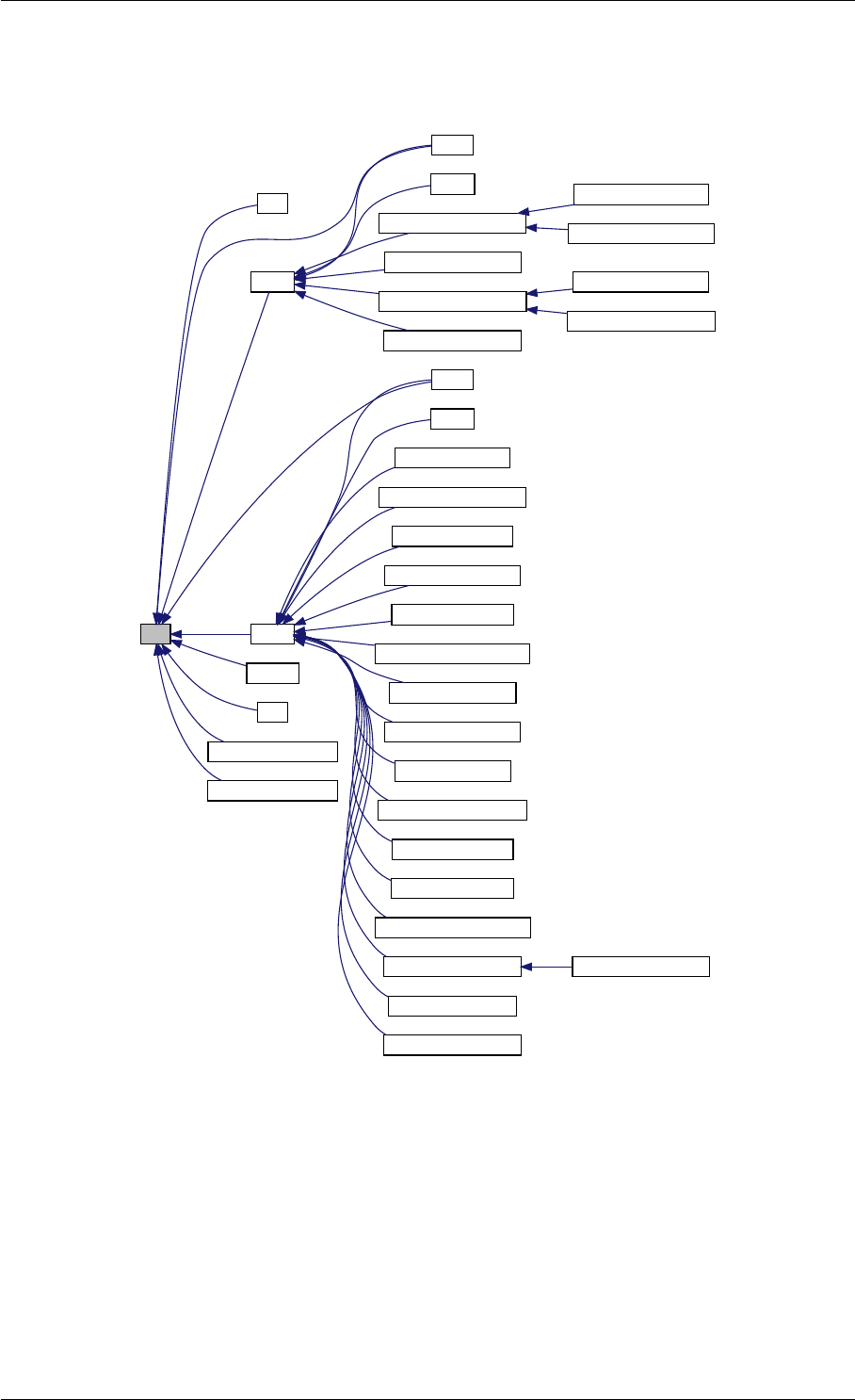
216 CONTENTS
Here is the caller graph for this function:
fw
fy
fdydp
fdydx
fdeltasx
fw
fdwdp
fdwdx
amici::Model_DAE::fxdot
amici::Model_ODE::fxdot
fdwdp
amici::Model_DAE::fdxdotdp
amici::Model_DAE::fqBdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fqBdot
amici::Model_DAE::fsxdot
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fdxdotdp
fdwdx
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::fJDiag
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_ODE::fJDiag
10.11.3.94 fw() [2/3]
void fw (
const realtype t,
const realtype ∗x)
Generated by Doxygen
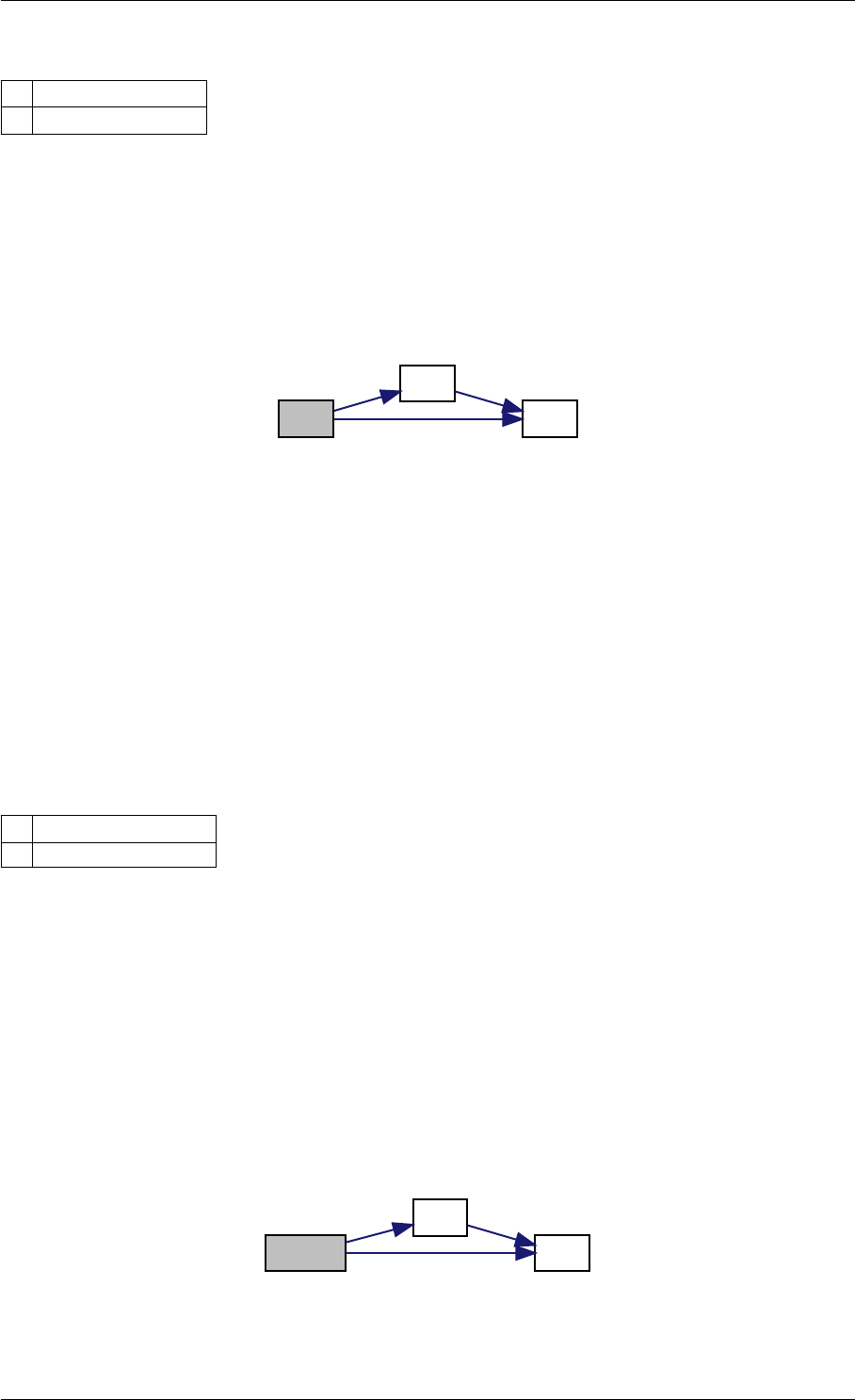
10.11 Model Class Reference 217
Parameters
ttimepoint
xarray with the states
Definition at line 1145 of file model.cpp.
Here is the call graph for this function:
fw
fw
t
10.11.3.95 fdwdp() [1/3]
void fdwdp (
const realtype t,
const N_Vector x)
Parameters
ttimepoint
xVector with the states
Definition at line 1150 of file model.cpp.
Here is the call graph for this function:
fdwdp
fw
t
Generated by Doxygen
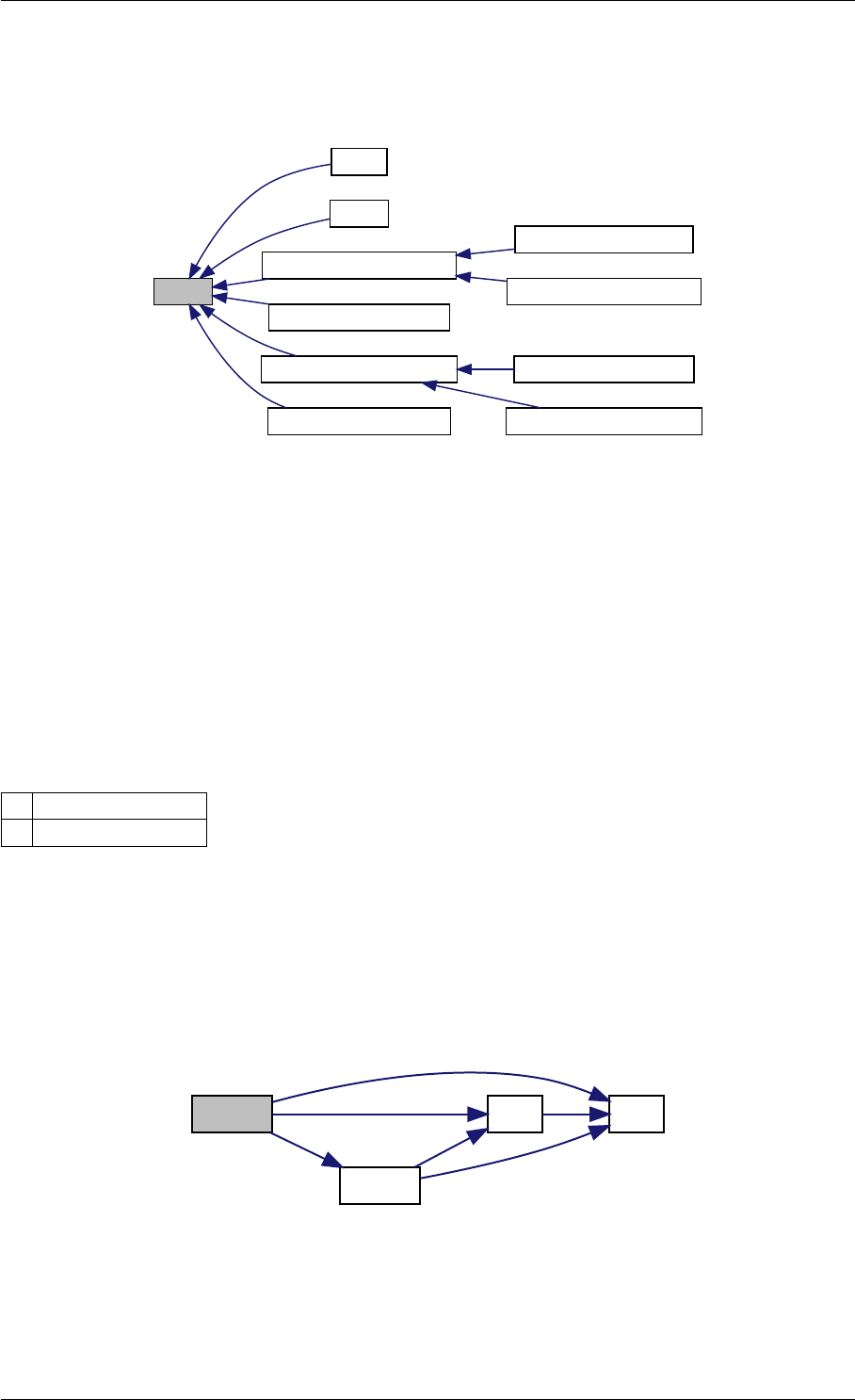
218 CONTENTS
Here is the caller graph for this function:
fdwdp
fdydp
fdwdp
amici::Model_DAE::fdxdotdp
amici::Model_DAE::fqBdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fqBdot
amici::Model_DAE::fsxdot
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fdxdotdp
10.11.3.96 fdwdp() [2/3]
void fdwdp (
const realtype t,
const realtype ∗x)
Parameters
ttimepoint
xarray with the states
Definition at line 1156 of file model.cpp.
Here is the call graph for this function:
fdwdp fw t
fdwdp
Generated by Doxygen
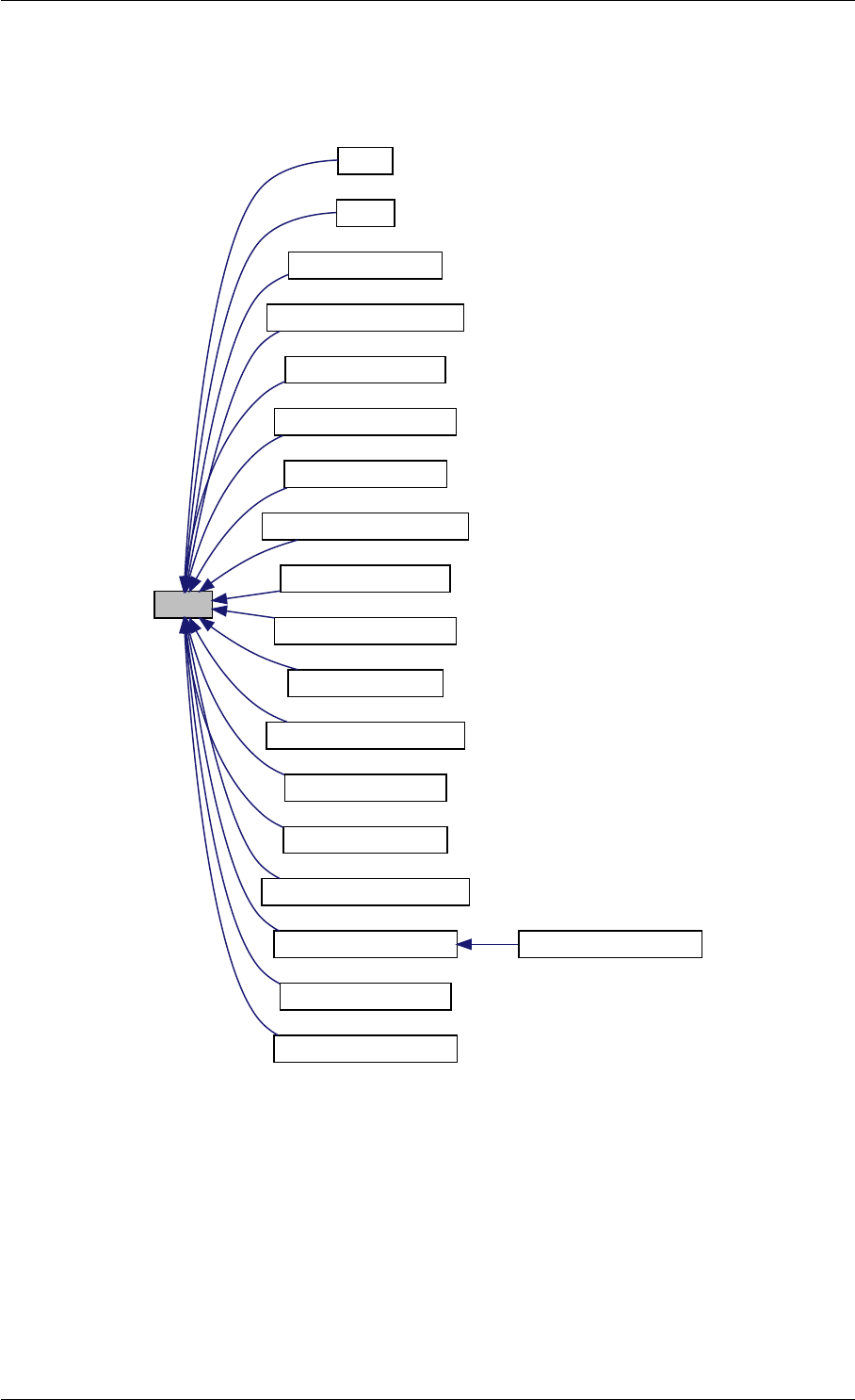
220 CONTENTS
Here is the caller graph for this function:
fdwdx
fdydx
fdwdx
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::fJDiag
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_ODE::fJDiag
10.11.3.98 fdwdx() [2/3]
void fdwdx (
const realtype t,
const realtype ∗x)
Generated by Doxygen
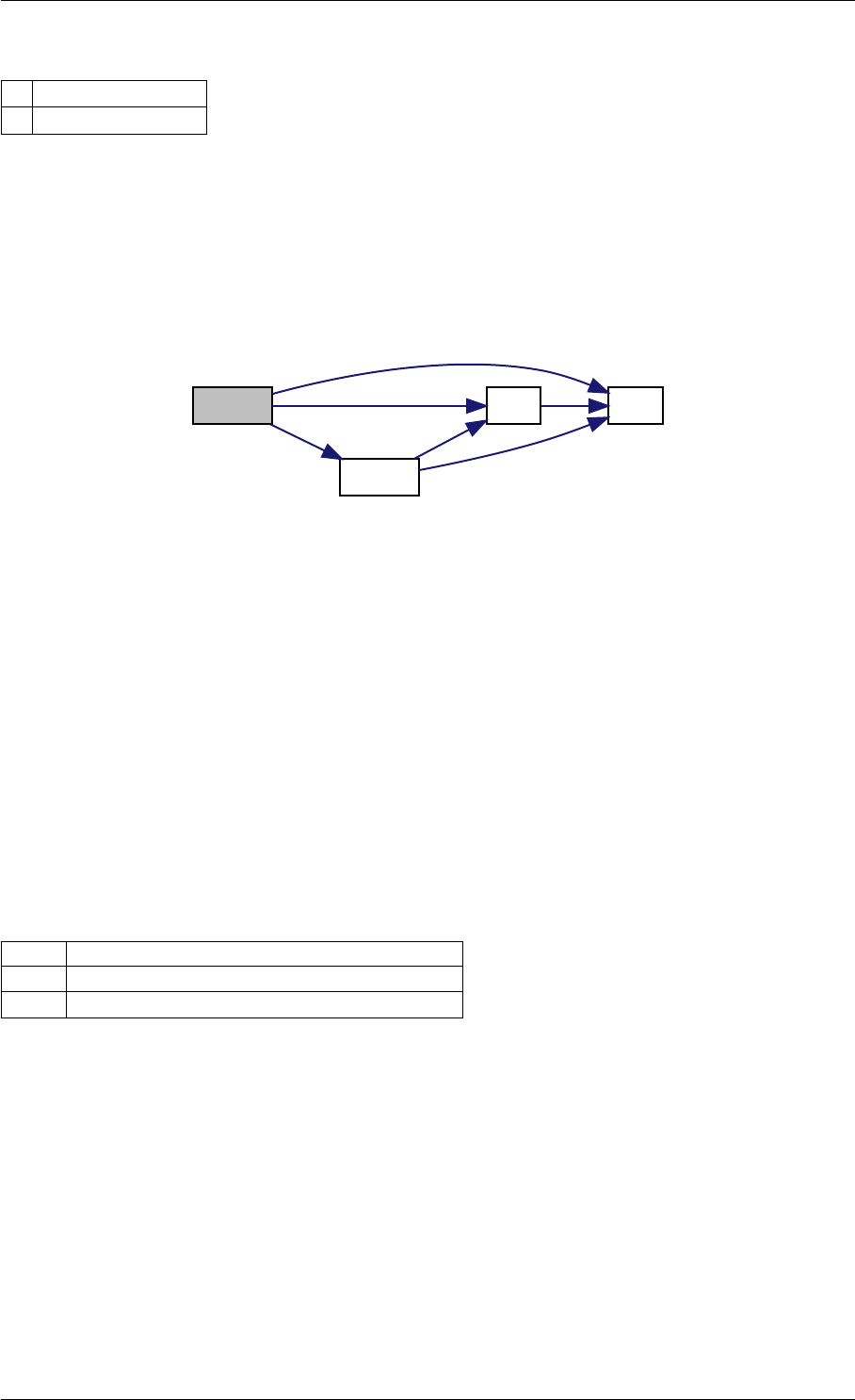
10.11 Model Class Reference 221
Parameters
ttimepoint
xarray with the states
Definition at line 1168 of file model.cpp.
Here is the call graph for this function:
fdwdx fw t
fdwdx
10.11.3.99 fres()
void fres (
const int it,
ReturnData ∗rdata,
const ExpData ∗edata )
residual function
Parameters
it time index
rdata ReturnData instance to which result will be written
edata ExpData instance containing observable data
Definition at line 1174 of file model.cpp.
Generated by Doxygen
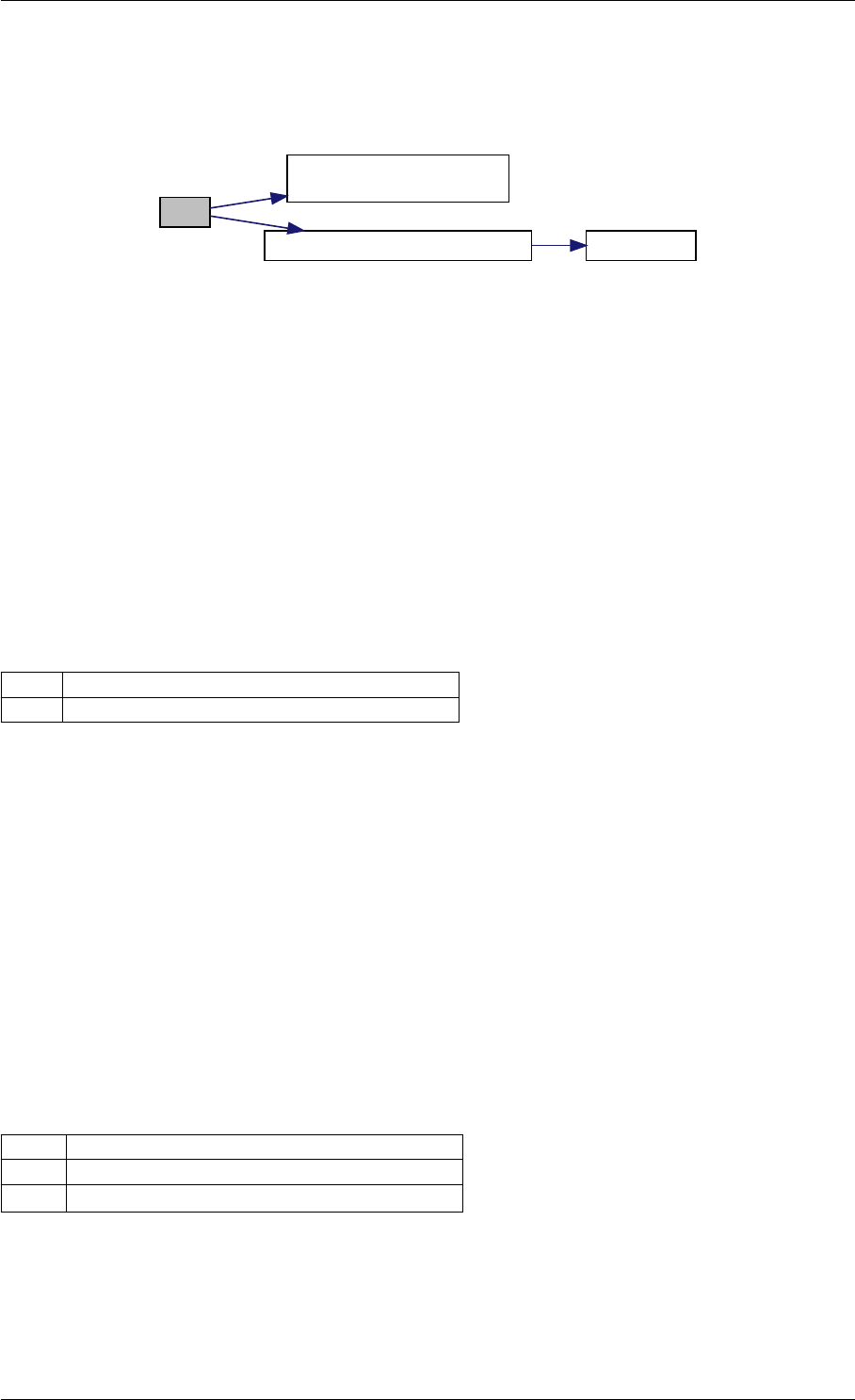
222 CONTENTS
Here is the call graph for this function:
fres
amici::ExpData::getObserved
DataPtr
amici::ExpData::isSetObservedData amici::isNaN
10.11.3.100 fchi2()
void fchi2 (
const int it,
ReturnData ∗rdata )
chi-squared function
Parameters
it time index
rdata ReturnData instance to which result will be written
Definition at line 1189 of file model.cpp.
10.11.3.101 fsres()
void fsres (
const int it,
ReturnData ∗rdata,
const ExpData ∗edata )
residual sensitivity function
Parameters
it time index
rdata ReturnData instance to which result will be written
edata ExpData instance containing observable data
Definition at line 1199 of file model.cpp.
Generated by Doxygen
10.11 Model Class Reference 223
Here is the call graph for this function:
fsres
amici::ExpData::isSetObservedData
nplist
amici::isNaN
10.11.3.102 fFIM()
void fFIM (
const int it,
ReturnData ∗rdata )
fisher information matrix function
Parameters
it time index
rdata ReturnData instance to which result will be written
Definition at line 1215 of file model.cpp.
Here is the call graph for this function:
fFIM nplist
10.11.3.103 updateHeaviside()
void updateHeaviside (
const std::vector<int >&rootsfound )
updateHeaviside updates the heaviside variables h on event occurences
Generated by Doxygen

224 CONTENTS
Parameters
rootsfound provides the direction of the zero-crossing, so adding it will give the right update to the heaviside
variables (zero if no root was found)
Definition at line 1231 of file model.cpp.
10.11.3.104 updateHeavisideB()
void updateHeavisideB (
const int ∗rootsfound )
updateHeavisideB updates the heaviside variables h on event occurences in the backward problem
Parameters
rootsfound provides the direction of the zero-crossing, so adding it will give the right update to the heaviside
variables (zero if no root was found)
Definition at line 1237 of file model.cpp.
10.11.3.105 gett()
realtype gett (
const int it,
const ReturnData ∗rdata ) const
get current timepoint from index
Parameters
it timepoint index
rdata pointer to return data instance
Returns
current timepoint
Definition at line 1264 of file model.cpp.
10.11.3.106 checkFinite()
int checkFinite (
const int N,
const realtype ∗array,
const char ∗fun ) const
Generated by Doxygen
10.11 Model Class Reference 225
Parameters
Nnumber of datapoints in array
array arrays of values
fun name of the fucntion that generated the values
Returns
AMICI_RECOVERABLE_ERROR if a NaN/Inf value was found, AMICI_SUCCESS otherwise
Definition at line 1292 of file model.cpp.
Here is the call graph for this function:
checkFinite amici::checkFinite
amici::isNaN
amici::isInf
Here is the caller graph for this function:
checkFinite
amici::Model_DAE::fJDiag
amici::Model_ODE::fJDiag
10.11.3.107 hasParameterNames()
virtual bool hasParameterNames ( ) const [virtual]
Generated by Doxygen
226 CONTENTS
Returns
boolean indicating whether parameter names were set
Definition at line 902 of file model.h.
Here is the call graph for this function:
hasParameterNames
np
getParameterNames
Here is the caller graph for this function:
hasParameterNames
getParameterByName
setParameterByName
setParametersByNameRegex
10.11.3.108 getParameterNames()
virtual std::vector<std::string>getParameterNames ( ) const [virtual]
Returns
the names
Definition at line 908 of file model.h.
Generated by Doxygen
10.11 Model Class Reference 227
Here is the caller graph for this function:
getParameterNames
getParameterByName
setParameterByName
setParametersByNameRegex
hasParameterNames
10.11.3.109 hasStateNames()
virtual bool hasStateNames ( ) const [virtual]
Returns
boolean indicating whether state names were set
Definition at line 914 of file model.h.
Here is the call graph for this function:
hasStateNames getStateNames
10.11.3.110 getStateNames()
virtual std::vector<std::string>getStateNames ( ) const [virtual]
Generated by Doxygen
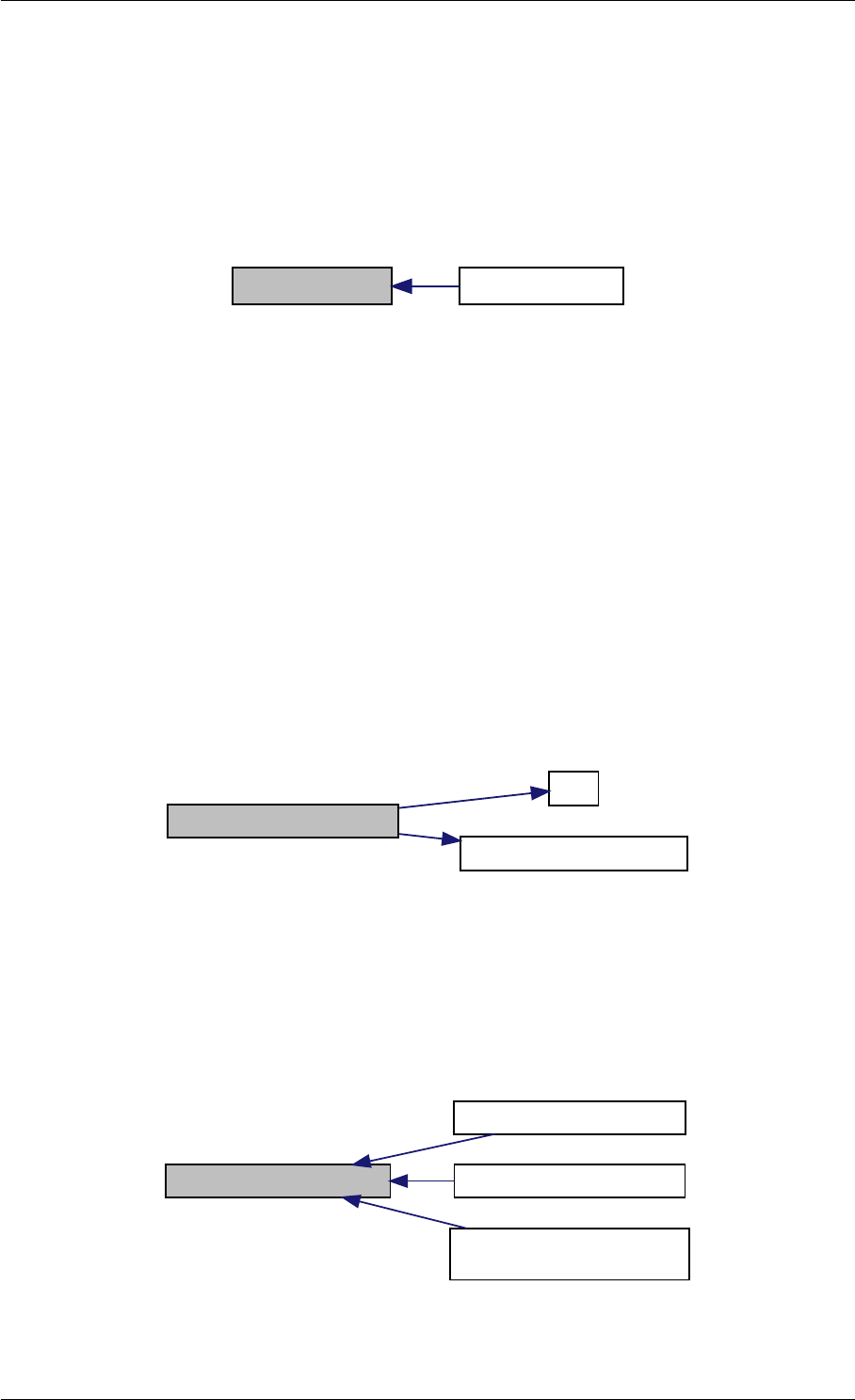
228 CONTENTS
Returns
the names
Definition at line 920 of file model.h.
Here is the caller graph for this function:
getStateNames hasStateNames
10.11.3.111 hasFixedParameterNames()
virtual bool hasFixedParameterNames ( ) const [virtual]
Returns
boolean indicating whether fixed parameter names were set
Definition at line 926 of file model.h.
Here is the call graph for this function:
hasFixedParameterNames
nk
getFixedParameterNames
Here is the caller graph for this function:
hasFixedParameterNames
getFixedParameterByName
setFixedParameterByName
setFixedParametersByName
Regex
Generated by Doxygen
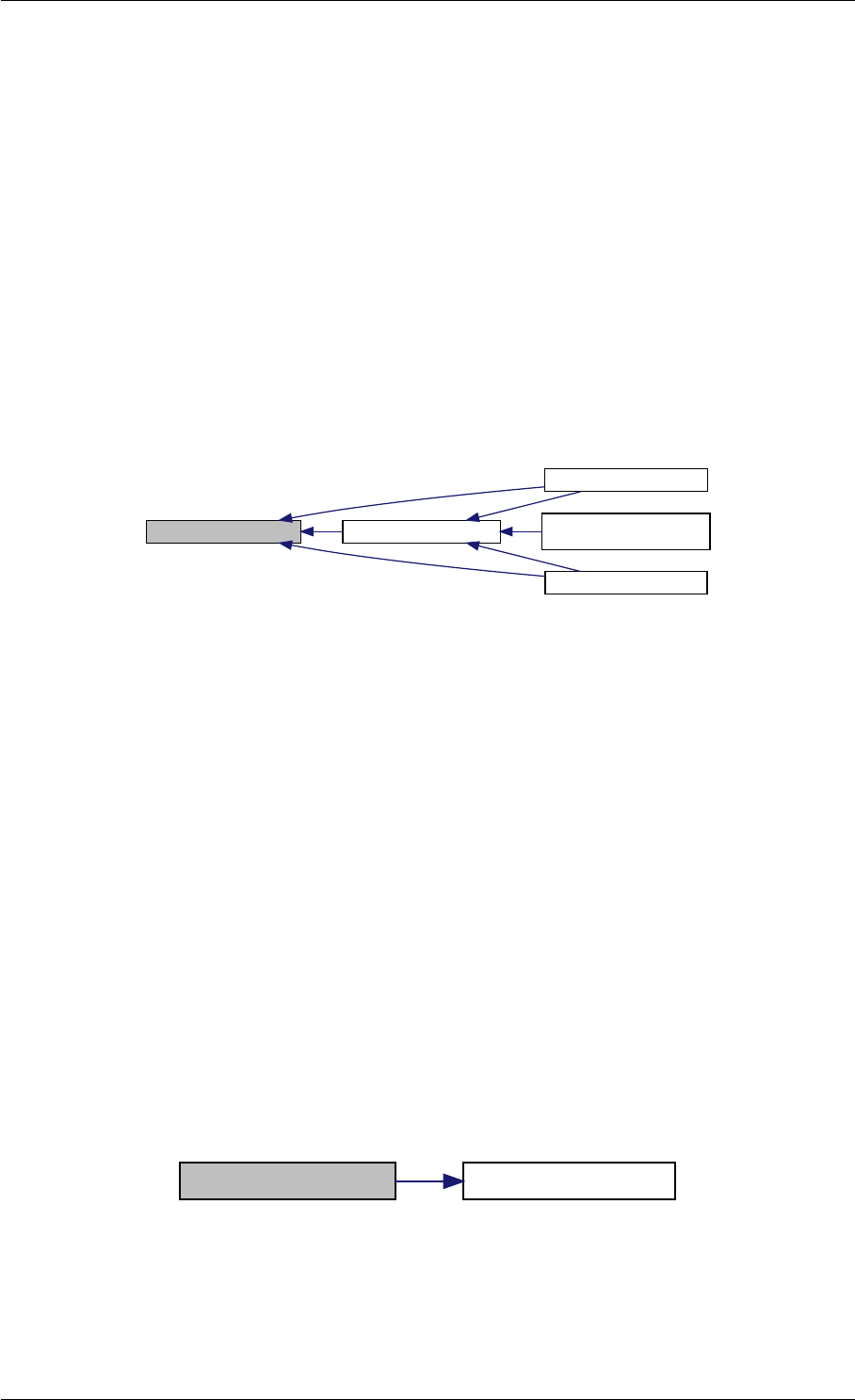
10.11 Model Class Reference 229
10.11.3.112 getFixedParameterNames()
virtual std::vector<std::string>getFixedParameterNames ( ) const [virtual]
Returns
the names
Definition at line 932 of file model.h.
Here is the caller graph for this function:
getFixedParameterNames
getFixedParameterByName
setFixedParameterByName
hasFixedParameterNames setFixedParametersByName
Regex
10.11.3.113 hasObservableNames()
virtual bool hasObservableNames ( ) const [virtual]
Returns
boolean indicating whether observabke names were set
Definition at line 938 of file model.h.
Here is the call graph for this function:
hasObservableNames getObservableNames
Generated by Doxygen
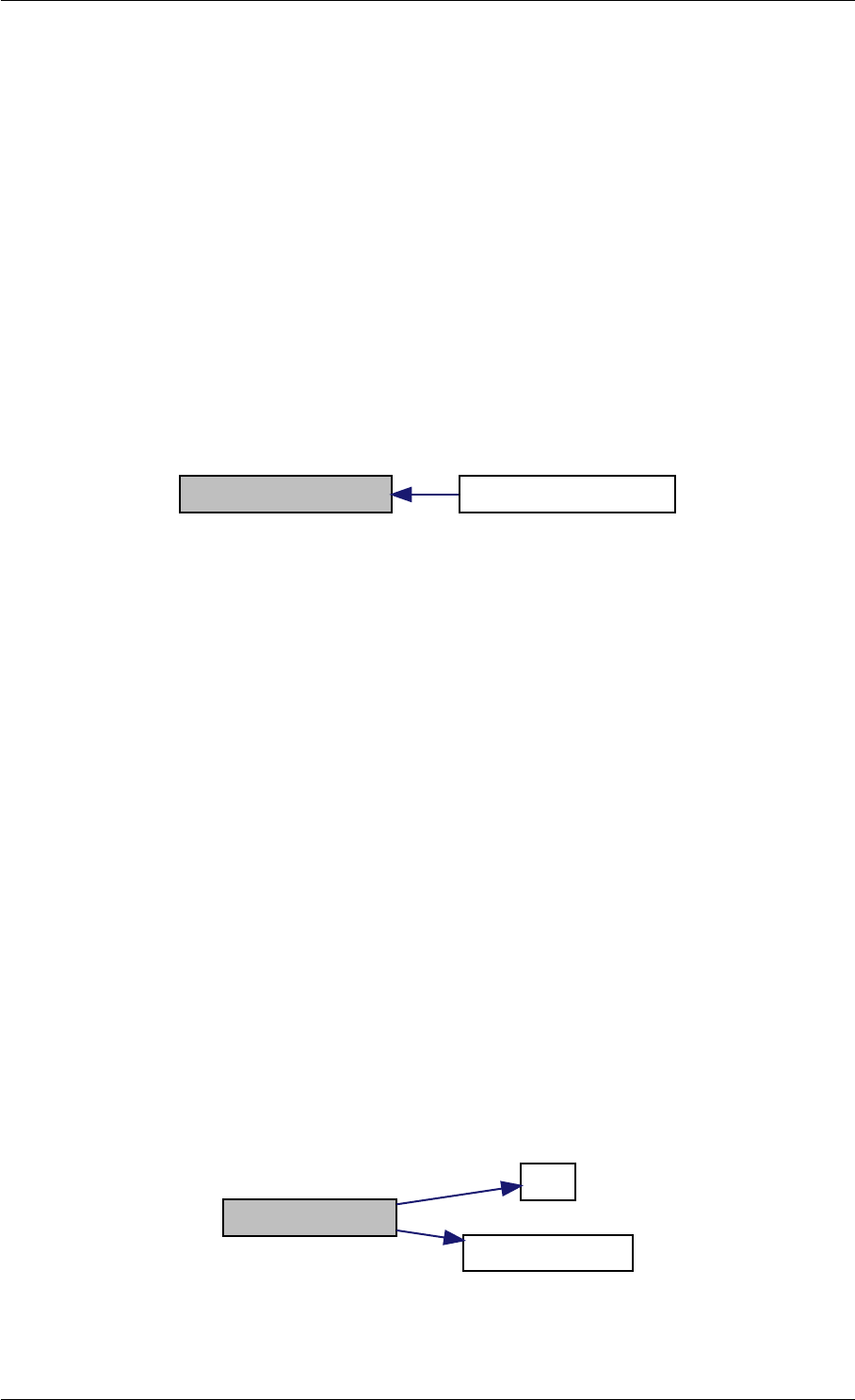
230 CONTENTS
10.11.3.114 getObservableNames()
virtual std::vector<std::string>getObservableNames ( ) const [virtual]
Returns
the names
Definition at line 944 of file model.h.
Here is the caller graph for this function:
getObservableNames hasObservableNames
10.11.3.115 hasParameterIds()
virtual bool hasParameterIds ( ) const [virtual]
Returns
boolean indicating whether parameter ids were set
Definition at line 950 of file model.h.
Here is the call graph for this function:
hasParameterIds
np
getParameterIds
Generated by Doxygen
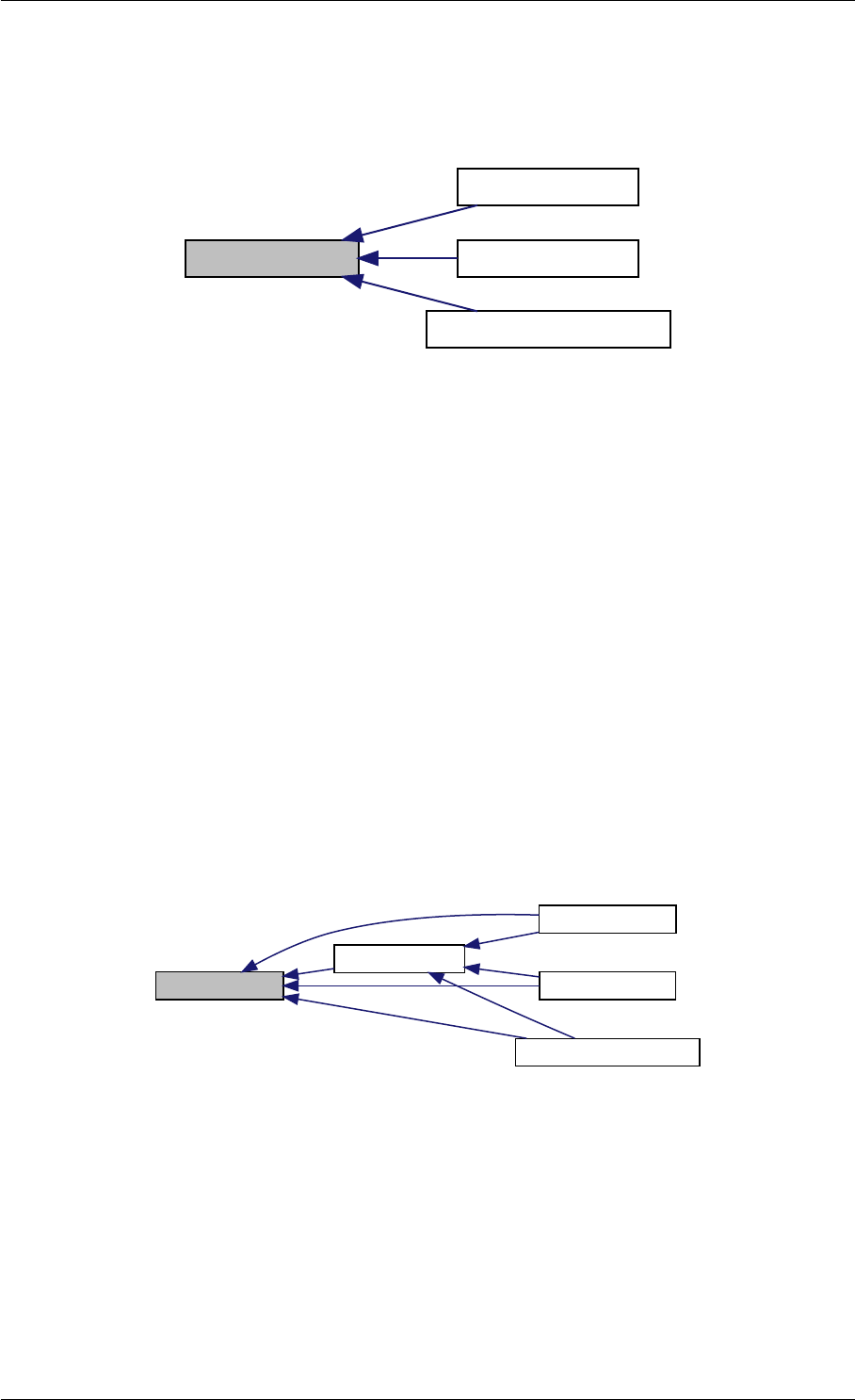
10.11 Model Class Reference 231
Here is the caller graph for this function:
hasParameterIds
getParameterById
setParameterById
setParametersByIdRegex
10.11.3.116 getParameterIds()
virtual std::vector<std::string>getParameterIds ( ) const [virtual]
Returns
the ids
Definition at line 956 of file model.h.
Here is the caller graph for this function:
getParameterIds
getParameterById
setParameterById
setParametersByIdRegex
hasParameterIds
10.11.3.117 getParameterById()
realtype getParameterById (
std::string const & par_id ) const
Generated by Doxygen
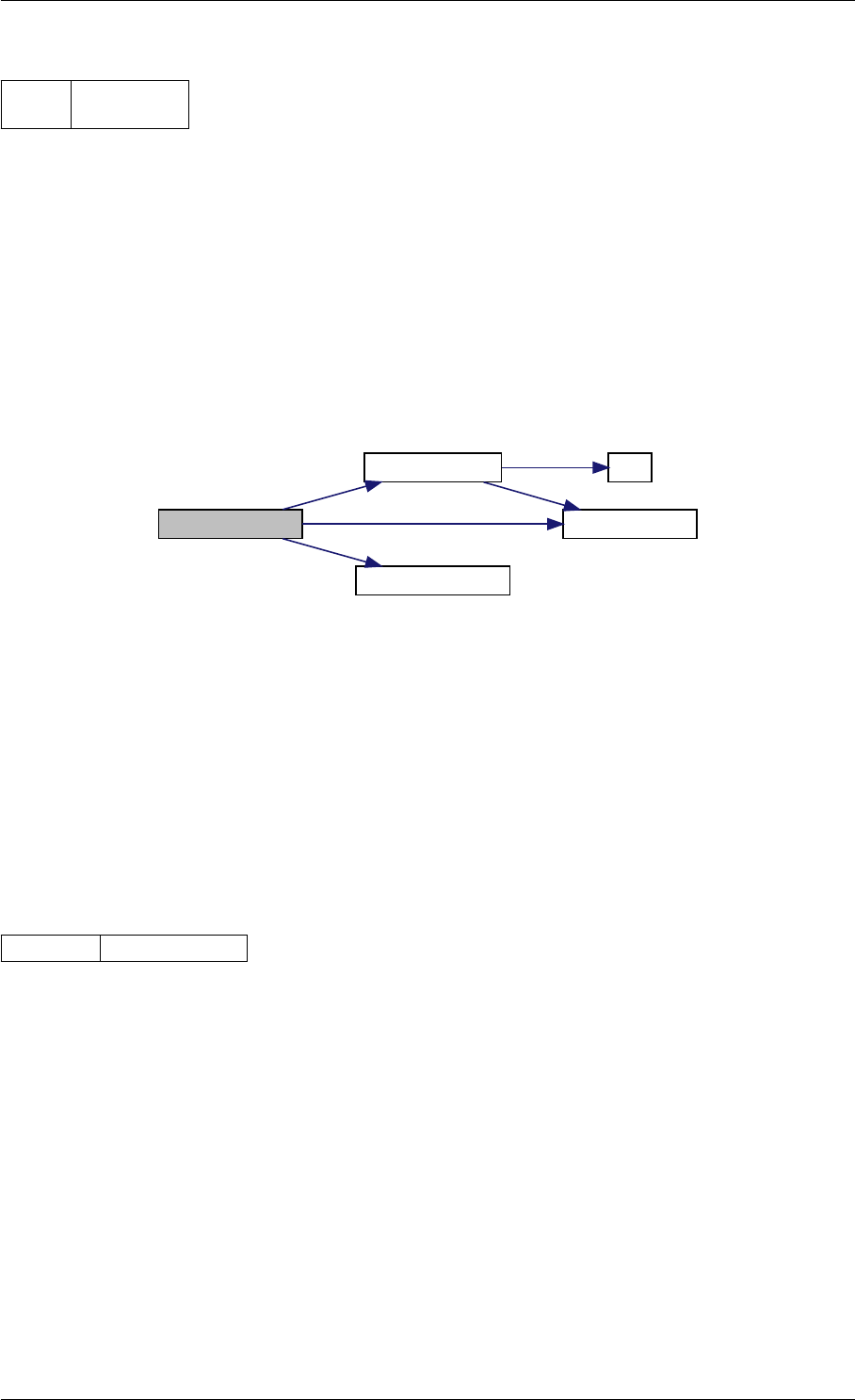
232 CONTENTS
Parameters
par←-
_id
parameter id
Returns
parameter value
Definition at line 381 of file model.cpp.
Here is the call graph for this function:
getParameterById
hasParameterIds
getParameterIds
amici::getValueById
np
10.11.3.118 getParameterByName()
realtype getParameterByName (
std::string const & par_name ) const
Parameters
par_name parameter name
Generated by Doxygen
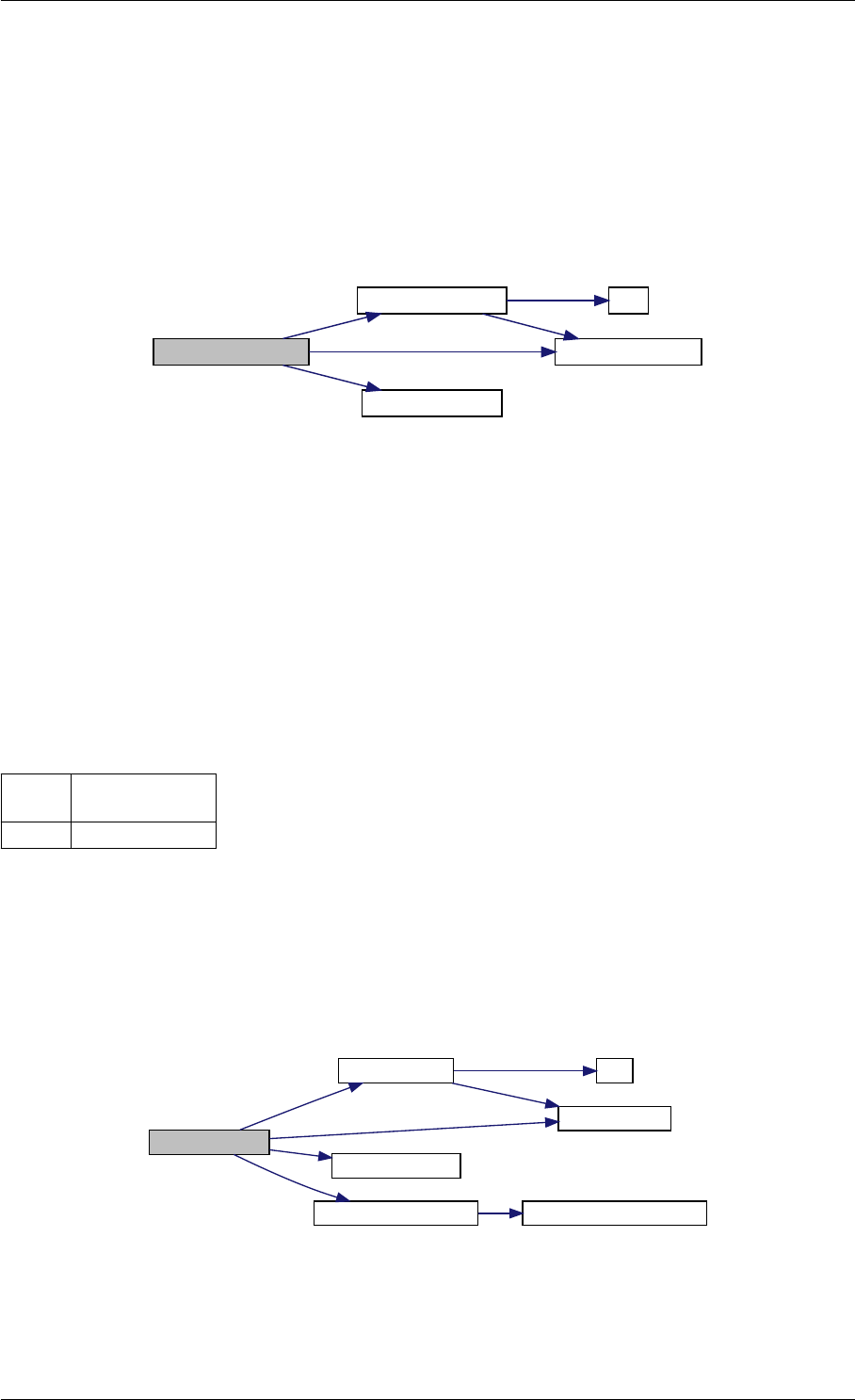
10.11 Model Class Reference 233
Returns
parameter value
Definition at line 387 of file model.cpp.
Here is the call graph for this function:
getParameterByName
hasParameterNames
getParameterNames
amici::getValueById
np
10.11.3.119 setParameterById()
void setParameterById (
std::string const & par_id,
realtype value )
Parameters
par←-
_id
parameter id
value parameter value
Definition at line 405 of file model.cpp.
Here is the call graph for this function:
setParameterById
hasParameterIds
getParameterIds
amici::setValueById
amici::unscaleParameters
np
amici::getUnscaledParameter
Generated by Doxygen
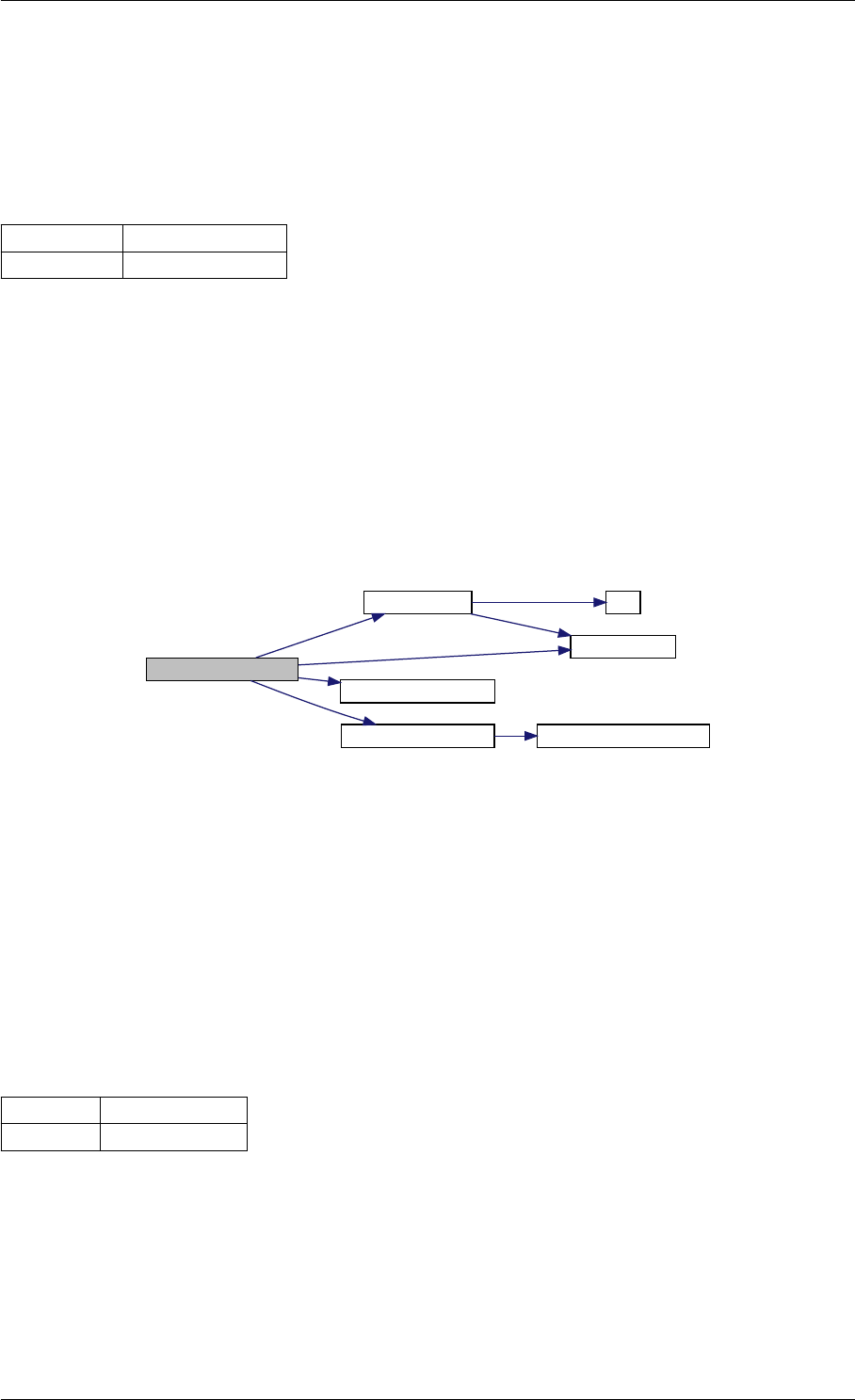
234 CONTENTS
10.11.3.120 setParametersByIdRegex()
int setParametersByIdRegex (
std::string const & par_id_regex,
realtype value )
Parameters
par_id_regex parameter id regex
value parameter value
Returns
number of parameter ids that matched the regex
Definition at line 418 of file model.cpp.
Here is the call graph for this function:
setParametersByIdRegex
hasParameterIds
getParameterIds
amici::setValueByIdRegex
amici::unscaleParameters
np
amici::getUnscaledParameter
10.11.3.121 setParameterByName()
void setParameterByName (
std::string const & par_name,
realtype value )
Parameters
par_name parameter name
value parameter value
Definition at line 431 of file model.cpp.
Generated by Doxygen
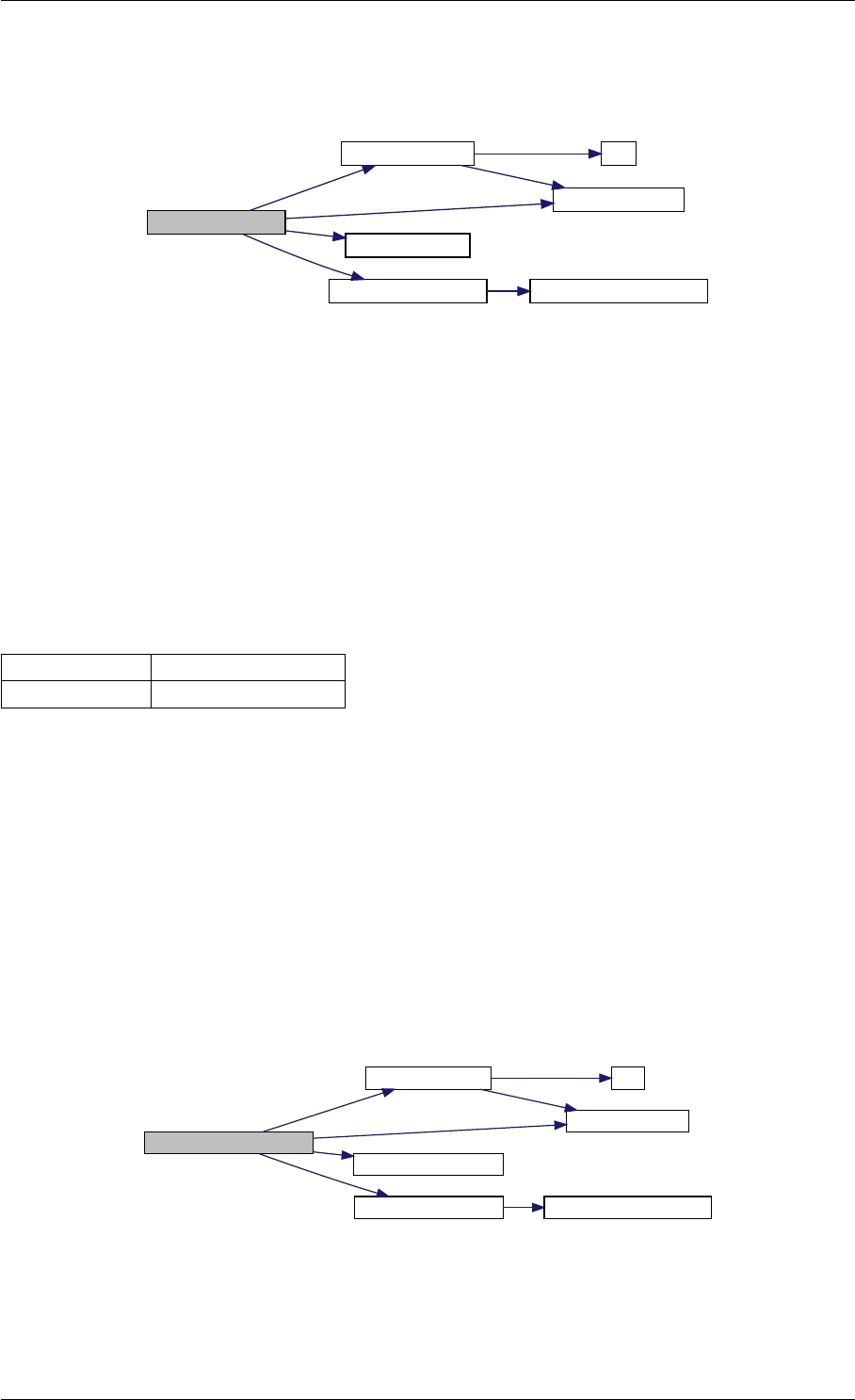
10.11 Model Class Reference 235
Here is the call graph for this function:
setParameterByName
hasParameterNames
getParameterNames
amici::setValueById
amici::unscaleParameters
np
amici::getUnscaledParameter
10.11.3.122 setParametersByNameRegex()
int setParametersByNameRegex (
std::string const & par_name_regex,
realtype value )
Parameters
par_name_regex parameter name regex
value parameter value
Returns
number of fixed parameter names that matched the regex
Definition at line 444 of file model.cpp.
Here is the call graph for this function:
setParametersByNameRegex
hasParameterNames
getParameterNames
amici::setValueByIdRegex
amici::unscaleParameters
np
amici::getUnscaledParameter
Generated by Doxygen
236 CONTENTS
10.11.3.123 hasStateIds()
virtual bool hasStateIds ( ) const [virtual]
Returns
Definition at line 1008 of file model.h.
Here is the call graph for this function:
hasStateIds getStateIds
10.11.3.124 getStateIds()
virtual std::vector<std::string>getStateIds ( ) const [virtual]
Returns
the ids
Definition at line 1014 of file model.h.
Here is the caller graph for this function:
getStateIds hasStateIds
Generated by Doxygen
10.11 Model Class Reference 237
10.11.3.125 hasFixedParameterIds()
virtual bool hasFixedParameterIds ( ) const [virtual]
Returns
boolean indicating whether fixed parameter ids were set
Definition at line 1022 of file model.h.
Here is the call graph for this function:
hasFixedParameterIds
nk
getFixedParameterIds
Here is the caller graph for this function:
hasFixedParameterIds
getFixedParameterById
setFixedParameterById
setFixedParametersByIdRegex
10.11.3.126 getFixedParameterIds()
virtual std::vector<std::string>getFixedParameterIds ( ) const [virtual]
Generated by Doxygen
238 CONTENTS
Returns
the ids
Definition at line 1028 of file model.h.
Here is the caller graph for this function:
getFixedParameterIds
getFixedParameterById
setFixedParameterById
setFixedParametersByIdRegex
setFixedParametersByName
Regex
hasFixedParameterIds
10.11.3.127 hasObservableIds()
virtual bool hasObservableIds ( ) const [virtual]
Returns
boolean indicating whether observale ids were set
Definition at line 1036 of file model.h.
Here is the call graph for this function:
hasObservableIds getObservableIds
Generated by Doxygen
10.11 Model Class Reference 239
10.11.3.128 getObservableIds()
virtual std::vector<std::string>getObservableIds ( ) const [virtual]
Returns
the ids
Definition at line 1042 of file model.h.
Here is the caller graph for this function:
getObservableIds hasObservableIds
10.11.3.129 setSteadyStateSensitivityMode()
void setSteadyStateSensitivityMode (
const SteadyStateSensitivityMode mode )
Parameters
mode steadyStateSensitivityMode
Definition at line 1050 of file model.h.
10.11.3.130 getSteadyStateSensitivityMode()
SteadyStateSensitivityMode getSteadyStateSensitivityMode ( ) const
Returns
flag value
Definition at line 1058 of file model.h.
10.11.3.131 setReinitializeFixedParameterInitialStates()
void setReinitializeFixedParameterInitialStates (
bool flag )
Generated by Doxygen
240 CONTENTS
Parameters
flag true/false
Definition at line 1067 of file model.h.
Here is the call graph for this function:
setReinitializeFixedParameter
InitialStates
isFixedParameterStateReinitialization
Allowed
10.11.3.132 getReinitializeFixedParameterInitialStates()
bool getReinitializeFixedParameterInitialStates ( ) const
Returns
flag true/false
Definition at line 1081 of file model.h.
Here is the caller graph for this function:
getReinitializeFixedParameter
InitialStates
fx0_fixedParameters
fsx0_fixedParameters
10.11.3.133 fx0() [2/2]
virtual void fx0 (
realtype ∗x0,
const realtype t,
const realtype ∗p,
const realtype ∗k) [protected], [virtual]
model specific implementation of fx0
Generated by Doxygen
10.11 Model Class Reference 241
Parameters
x0 initial state
tinitial time
pparameter vector
kconstant vector
Definition at line 1161 of file model.h.
10.11.3.134 fx0_fixedParameters() [2/2]
virtual void fx0_fixedParameters (
realtype ∗x0,
const realtype t,
const realtype ∗p,
const realtype ∗k) [protected], [virtual]
model specific implementation of fx0_fixedParameters
Parameters
x0 initial state
tinitial time
pparameter vector
kconstant vector
Definition at line 1171 of file model.h.
10.11.3.135 fsx0_fixedParameters() [2/2]
virtual void fsx0_fixedParameters (
realtype ∗sx0,
const realtype t,
const realtype ∗x0,
const realtype ∗p,
const realtype ∗k,
const int ip ) [protected], [virtual]
model specific implementation of fsx0_fixedParameters
Parameters
sx0 initial state sensitivities
tinitial time
x0 initial state
pparameter vector
kconstant vector
ip sensitivity index
Generated by Doxygen

242 CONTENTS
Definition at line 1182 of file model.h.
10.11.3.136 fsx0() [2/2]
virtual void fsx0 (
realtype ∗sx0,
const realtype t,
const realtype ∗x0,
const realtype ∗p,
const realtype ∗k,
const int ip ) [protected], [virtual]
model specific implementation of fsx0
Parameters
sx0 initial state sensitivities
tinitial time
x0 initial state
pparameter vector
kconstant vector
ip sensitivity index
Definition at line 1193 of file model.h.
10.11.3.137 fstau() [2/2]
virtual void fstau (
realtype ∗stau,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗sx,
const int ip,
const int ie ) [protected], [virtual]
model specific implementation of fstau
Parameters
stau total derivative of event timepoint
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
sx current state sensitivity
ip sensitivity index
ie event index
Generated by Doxygen
10.11 Model Class Reference 243
Definition at line 1208 of file model.h.
10.11.3.138 fy() [2/2]
virtual void fy (
realtype ∗y,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w) [protected], [virtual]
model specific implementation of fy
Parameters
ymodel output at current timepoint
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
wrepeating elements vector
Definition at line 1221 of file model.h.
10.11.3.139 fdydp() [2/2]
virtual void fdydp (
realtype ∗dydp,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip,
const realtype ∗w,
const realtype ∗dwdp ) [protected], [virtual]
model specific implementation of fdydp
Parameters
dydp partial derivative of observables y w.r.t. model parameters p
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
ip parameter index w.r.t. which the derivative is requested
wrepeating elements vector
dwdp Recurring terms in xdot, parameter derivative
Generated by Doxygen

244 CONTENTS
Definition at line 1236 of file model.h.
10.11.3.140 fdydx() [2/2]
virtual void fdydx (
realtype ∗dydx,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation of fdydx
Parameters
dydx partial derivative of observables y w.r.t. model states x
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
wrepeating elements vector
dwdx Recurring terms in xdot, state derivative
Definition at line 1250 of file model.h.
10.11.3.141 fz() [2/2]
virtual void fz (
realtype ∗z,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h) [protected], [virtual]
model specific implementation of fz
Parameters
zvalue of event output
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
Generated by Doxygen
10.11 Model Class Reference 245
Definition at line 1263 of file model.h.
10.11.3.142 fsz() [2/2]
virtual void fsz (
realtype ∗sz,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗sx,
const int ip ) [protected], [virtual]
model specific implementation of fsz
Parameters
sz Sensitivity of rz, total derivative
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
sx current state sensitivity
ip sensitivity index
Definition at line 1278 of file model.h.
10.11.3.143 frz() [2/2]
virtual void frz (
realtype ∗rz,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h) [protected], [virtual]
model specific implementation of frz
Parameters
rz value of root function at current timepoint (non-output events not included)
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
Generated by Doxygen
246 CONTENTS
Definition at line 1291 of file model.h.
10.11.3.144 fsrz() [2/2]
virtual void fsrz (
realtype ∗srz,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗sx,
const int ip ) [protected], [virtual]
model specific implementation of fsrz
Parameters
srz Sensitivity of rz, total derivative
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
sx current state sensitivity
hheavyside vector
ip sensitivity index
Definition at line 1306 of file model.h.
10.11.3.145 fdzdp() [2/2]
virtual void fdzdp (
realtype ∗dzdp,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip ) [protected], [virtual]
model specific implementation of fdzdp
Parameters
dzdp partial derivative of event-resolved output z w.r.t. model parameters p
ie event index
tcurrent time
Generated by Doxygen
10.11 Model Class Reference 247
Parameters
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
ip parameter index w.r.t. which the derivative is requested
Definition at line 1320 of file model.h.
10.11.3.146 fdzdx() [2/2]
virtual void fdzdx (
realtype ∗dzdx,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h) [protected], [virtual]
model specific implementation of fdzdx
Parameters
dzdx partial derivative of event-resolved output z w.r.t. model states x
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
Definition at line 1333 of file model.h.
10.11.3.147 fdrzdp() [2/2]
virtual void fdrzdp (
realtype ∗drzdp,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip ) [protected], [virtual]
model specific implementation of fdrzdp
Generated by Doxygen

248 CONTENTS
Parameters
drzdp partial derivative of root output rz w.r.t. model parameters p
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
ip parameter index w.r.t. which the derivative is requested
Definition at line 1347 of file model.h.
10.11.3.148 fdrzdx() [2/2]
virtual void fdrzdx (
realtype ∗drzdx,
const int ie,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h) [protected], [virtual]
model specific implementation of fdrzdx
Parameters
drzdx partial derivative of root output rz w.r.t. model states x
ie event index
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
Definition at line 1360 of file model.h.
10.11.3.149 fdeltax() [2/2]
virtual void fdeltax (
realtype ∗deltax,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ie,
Generated by Doxygen
10.11 Model Class Reference 249
const realtype ∗xdot,
const realtype ∗xdot_old ) [protected], [virtual]
model specific implementation of fdeltax
Parameters
deltax state update
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
ie event index
xdot new model right hand side
xdot_old previous model right hand side
Definition at line 1375 of file model.h.
10.11.3.150 fdeltasx() [2/2]
virtual void fdeltasx (
realtype ∗deltasx,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w,
const int ip,
const int ie,
const realtype ∗xdot,
const realtype ∗xdot_old,
const realtype ∗sx,
const realtype ∗stau ) [protected], [virtual]
model specific implementation of fdeltasx
Parameters
deltasx sensitivity update
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
wrepeating elements vector
ip sensitivity index
ie event index
xdot new model right hand side
xdot_old previous model right hand side
sx state sensitivity
stau event-time sensitivity
Generated by Doxygen
250 CONTENTS
Definition at line 1395 of file model.h.
10.11.3.151 fdeltaxB() [2/2]
virtual void fdeltaxB (
realtype ∗deltaxB,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ie,
const realtype ∗xdot,
const realtype ∗xdot_old,
const realtype ∗xB ) [protected], [virtual]
model specific implementation of fdeltaxB
Parameters
deltaxB adjoint state update
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
ie event index
xdot new model right hand side
xdot_old previous model right hand side
xB current adjoint state
Definition at line 1413 of file model.h.
10.11.3.152 fdeltaqB() [2/2]
virtual void fdeltaqB (
realtype ∗deltaqB,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip,
const int ie,
const realtype ∗xdot,
const realtype ∗xdot_old,
const realtype ∗xB ) [protected], [virtual]
model specific implementation of fdeltaqB
Generated by Doxygen
10.11 Model Class Reference 251
Parameters
deltaqB sensitivity update
tcurrent time
xcurrent state
pparameter vector
kconstant vector
hheavyside vector
ip sensitivity index
ie event index
xdot new model right hand side
xdot_old previous model right hand side
xB adjoint state
Definition at line 1431 of file model.h.
10.11.3.153 fsigmay() [2/2]
virtual void fsigmay (
realtype ∗sigmay,
const realtype t,
const realtype ∗p,
const realtype ∗k) [protected], [virtual]
model specific implementation of fsigmay
Parameters
sigmay standard deviation of measurements
tcurrent time
pparameter vector
kconstant vector
Definition at line 1442 of file model.h.
10.11.3.154 fdsigmaydp() [2/2]
virtual void fdsigmaydp (
realtype ∗dsigmaydp,
const realtype t,
const realtype ∗p,
const realtype ∗k,
const int ip ) [protected], [virtual]
model specific implementation of fsigmay
Generated by Doxygen
252 CONTENTS
Parameters
dsigmaydp partial derivative of standard deviation of measurements
tcurrent time
pparameter vector
kconstant vector
ip sensitivity index
Definition at line 1453 of file model.h.
10.11.3.155 fsigmaz() [2/2]
virtual void fsigmaz (
realtype ∗sigmaz,
const realtype t,
const realtype ∗p,
const realtype ∗k) [protected], [virtual]
model specific implementation of fsigmaz
Parameters
sigmaz standard deviation of event measurements
tcurrent time
pparameter vector
kconstant vector
Definition at line 1463 of file model.h.
10.11.3.156 fdsigmazdp() [2/2]
virtual void fdsigmazdp (
realtype ∗dsigmazdp,
const realtype t,
const realtype ∗p,
const realtype ∗k,
const int ip ) [protected], [virtual]
model specific implementation of fsigmaz
Parameters
dsigmazdp partial derivative of standard deviation of event measurements
tcurrent time
pparameter vector
kconstant vector
ip sensitivity index
Generated by Doxygen

10.11 Model Class Reference 253
Definition at line 1474 of file model.h.
10.11.3.157 fJy() [2/2]
virtual void fJy (
realtype ∗nllh,
const int iy,
const realtype ∗p,
const realtype ∗k,
const realtype ∗y,
const realtype ∗sigmay,
const realtype ∗my ) [protected], [virtual]
model specific implementation of fJy
Parameters
nllh negative log-likelihood for measurements y
iy output index
pparameter vector
kconstant vector
ymodel output at timepoint
sigmay measurement standard deviation at timepoint
my measurements at timepoint
Definition at line 1487 of file model.h.
10.11.3.158 fJz() [2/2]
virtual void fJz (
realtype ∗nllh,
const int iz,
const realtype ∗p,
const realtype ∗k,
const realtype ∗z,
const realtype ∗sigmaz,
const realtype ∗mz ) [protected], [virtual]
model specific implementation of fJz
Parameters
nllh negative log-likelihood for event measurements z
iz event output index
pparameter vector
kconstant vector
zmodel event output at timepoint
sigmaz event measurement standard deviation at timepoint
mz event measurements at timepoint
Generated by Doxygen

254 CONTENTS
Definition at line 1499 of file model.h.
10.11.3.159 fJrz() [2/2]
virtual void fJrz (
realtype ∗nllh,
const int iz,
const realtype ∗p,
const realtype ∗k,
const realtype ∗z,
const realtype ∗sigmaz ) [protected], [virtual]
model specific implementation of fJrz
Parameters
nllh regularization for event measurements z
iz event output index
pparameter vector
kconstant vector
zmodel event output at timepoint
sigmaz event measurement standard deviation at timepoint
Definition at line 1511 of file model.h.
10.11.3.160 fdJydy() [2/2]
virtual void fdJydy (
realtype ∗dJydy,
const int iy,
const realtype ∗p,
const realtype ∗k,
const realtype ∗y,
const realtype ∗sigmay,
const realtype ∗my ) [protected], [virtual]
model specific implementation of fdJydy
Parameters
dJydy partial derivative of time-resolved measurement negative log-likelihood Jy
iy output index
pparameter vector
kconstant vector
ymodel output at timepoint
sigmay measurement standard deviation at timepoint
my measurement at timepoint
Generated by Doxygen
10.11 Model Class Reference 255
Definition at line 1524 of file model.h.
10.11.3.161 fdJydsigma() [2/2]
virtual void fdJydsigma (
realtype ∗dJydsigma,
const int iy,
const realtype ∗p,
const realtype ∗k,
const realtype ∗y,
const realtype ∗sigmay,
const realtype ∗my ) [protected], [virtual]
model specific implementation of fdJydsigma
Parameters
dJydsigma Sensitivity of time-resolved measurement negative log-likelihood Jy w.r.t. standard deviation sigmay
iy output index
pparameter vector
kconstant vector
ymodel output at timepoint
sigmay measurement standard deviation at timepoint
my measurement at timepoint
Definition at line 1539 of file model.h.
10.11.3.162 fdJzdz() [2/2]
virtual void fdJzdz (
realtype ∗dJzdz,
const int iz,
const realtype ∗p,
const realtype ∗k,
const realtype ∗z,
const realtype ∗sigmaz,
const realtype ∗mz ) [protected], [virtual]
model specific implementation of fdJzdz
Parameters
dJzdz partial derivative of event measurement negative log-likelihood Jz
iz event output index
pparameter vector
kconstant vector
zmodel event output at timepoint
sigmaz event measurement standard deviation at timepoint
mz event measurement at timepoint
Generated by Doxygen
256 CONTENTS
Definition at line 1553 of file model.h.
10.11.3.163 fdJzdsigma() [2/2]
virtual void fdJzdsigma (
realtype ∗dJzdsigma,
const int iz,
const realtype ∗p,
const realtype ∗k,
const realtype ∗z,
const realtype ∗sigmaz,
const realtype ∗mz ) [protected], [virtual]
model specific implementation of fdJzdsigma
Parameters
dJzdsigma Sensitivity of event measurement negative log-likelihood Jz w.r.t. standard deviation sigmaz
iz event output index
pparameter vector
kconstant vector
zmodel event output at timepoint
sigmaz event measurement standard deviation at timepoint
mz event measurement at timepoint
Definition at line 1568 of file model.h.
10.11.3.164 fdJrzdz() [2/2]
virtual void fdJrzdz (
realtype ∗dJrzdz,
const int iz,
const realtype ∗p,
const realtype ∗k,
const realtype ∗rz,
const realtype ∗sigmaz ) [protected], [virtual]
model specific implementation of fdJrzdz
Parameters
dJrzdz partial derivative of event penalization Jrz
iz event output index
pparameter vector
kconstant vector
rz model root output at timepoint
sigmaz event measurement standard deviation at timepoint
Generated by Doxygen

10.11 Model Class Reference 257
Definition at line 1581 of file model.h.
10.11.3.165 fdJrzdsigma() [2/2]
virtual void fdJrzdsigma (
realtype ∗dJrzdsigma,
const int iz,
const realtype ∗p,
const realtype ∗k,
const realtype ∗rz,
const realtype ∗sigmaz ) [protected], [virtual]
model specific implementation of fdJrzdsigma
Parameters
dJrzdsigma Sensitivity of event penalization Jrz w.r.t. standard deviation sigmaz
iz event output index
pparameter vector
kconstant vector
rz model root output at timepoint
sigmaz event measurement standard deviation at timepoint
Definition at line 1595 of file model.h.
10.11.3.166 fw() [3/3]
virtual void fw (
realtype ∗w,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h) [protected], [virtual]
model specific implementation of fw
Parameters
wRecurring terms in xdot
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
Definition at line 1608 of file model.h.
Generated by Doxygen

258 CONTENTS
10.11.3.167 fdwdp() [3/3]
virtual void fdwdp (
realtype ∗dwdp,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w) [protected], [virtual]
model specific implementation of dwdp
Parameters
dwdp Recurring terms in xdot, parameter derivative
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
Definition at line 1620 of file model.h.
10.11.3.168 fdwdx() [3/3]
virtual void fdwdx (
realtype ∗dwdx,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w) [protected], [virtual]
model specific implementation of dwdx
Parameters
dwdx Recurring terms in xdot, state derivative
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
Definition at line 1632 of file model.h.
Generated by Doxygen
10.11 Model Class Reference 259
10.11.3.169 getmy()
void getmy (
const int it,
const ExpData ∗edata ) [protected]
create my slice at timepoint
Parameters
it timepoint index
edata pointer to experimental data instance
Definition at line 1244 of file model.cpp.
Here is the call graph for this function:
getmy
amici::ExpData::getObserved
DataPtr
amici::getNaN
Here is the caller graph for this function:
getmy
fdJydp
fdJydx
fJy
fdJydy
fdJydsigma
Generated by Doxygen

260 CONTENTS
10.11.3.170 getmz()
void getmz (
const int nroots,
const ExpData ∗edata ) [protected]
create mz slice at event
Parameters
nroots event occurence
edata pointer to experimental data instance
Definition at line 1268 of file model.cpp.
Here is the call graph for this function:
getmz
amici::ExpData::getObserved
EventsPtr
amici::getNaN
Generated by Doxygen
10.11 Model Class Reference 261
Here is the caller graph for this function:
getmz
fdJzdp
fdJzdx
fJz
fdJzdz
fdJzdsigma
fdJrzdz
10.11.3.171 gety()
const realtype ∗gety (
const int it,
const ReturnData ∗rdata ) const [protected]
create y slice at timepoint
Parameters
it timepoint index
rdata pointer to return data instance
Returns
y y-slice from rdata instance
Definition at line 1260 of file model.cpp.
Generated by Doxygen
262 CONTENTS
Here is the caller graph for this function:
gety
fJy
fdJydy
fdJydsigma
10.11.3.172 getx()
const realtype ∗getx (
const int it,
const ReturnData ∗rdata ) const [protected]
create x slice at timepoint
Parameters
it timepoint index
rdata pointer to return data instance
Returns
x x-slice from rdata instance
Definition at line 1252 of file model.cpp.
Here is the caller graph for this function:
getx
fy
fdydp
fdydx
Generated by Doxygen
10.11 Model Class Reference 263
10.11.3.173 getsx()
const realtype ∗getsx (
const int it,
const ReturnData ∗rdata ) const [protected]
create sx slice at timepoint
Parameters
it timepoint index
rdata pointer to return data instance
Returns
sx sx-slice from rdata instance
Definition at line 1256 of file model.cpp.
Here is the call graph for this function:
getsx nplist
10.11.3.174 getz()
const realtype ∗getz (
const int nroots,
const ReturnData ∗rdata ) const [protected]
create z slice at event
Parameters
nroots event occurence
rdata pointer to return data instance
Returns
z slice
Generated by Doxygen
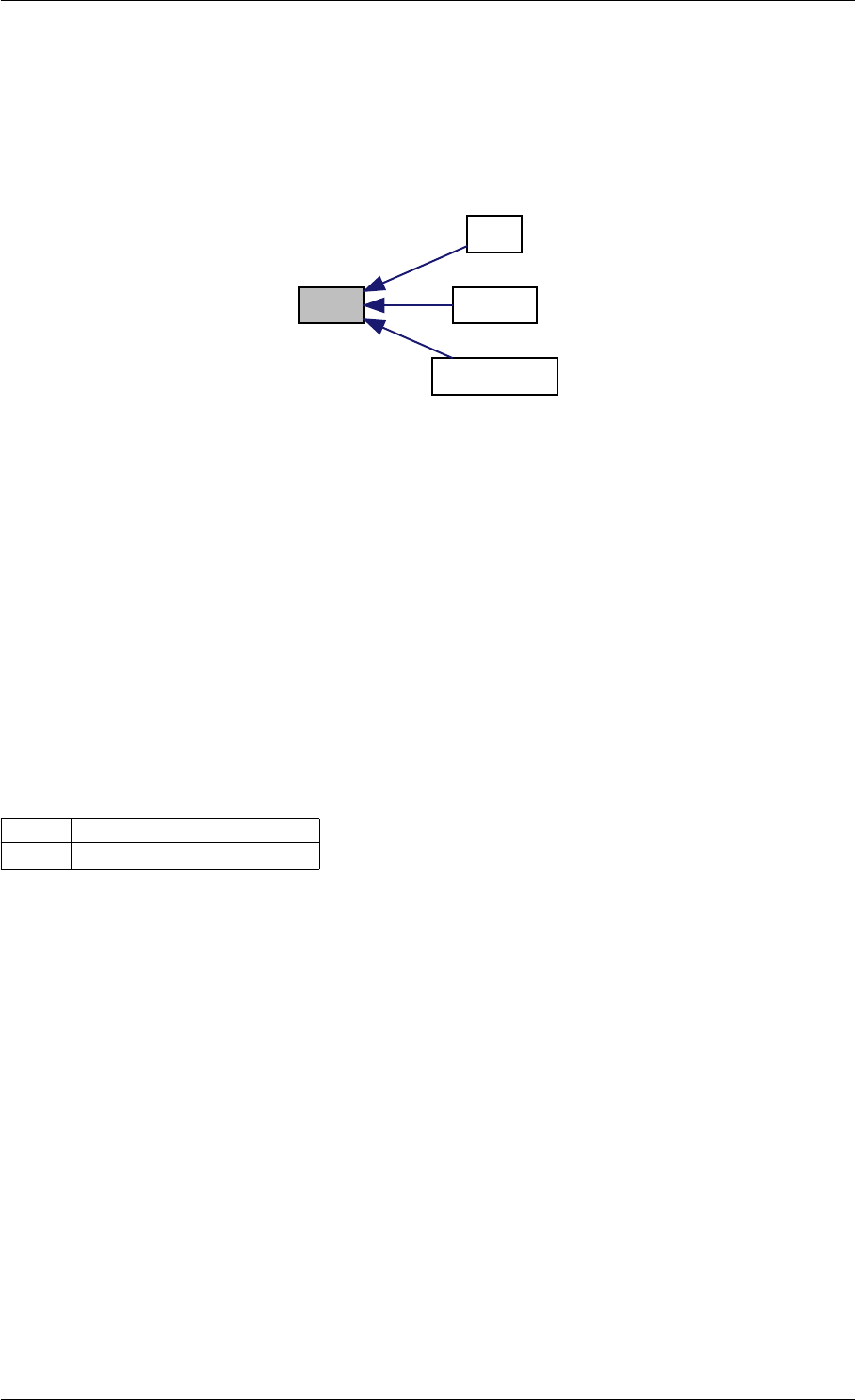
264 CONTENTS
Definition at line 1276 of file model.cpp.
Here is the caller graph for this function:
getz
fJz
fdJzdz
fdJzdsigma
10.11.3.175 getrz()
const realtype ∗getrz (
const int nroots,
const ReturnData ∗rdata ) const [protected]
create rz slice at event
Parameters
nroots event occurence
rdata pointer to return data instance
Returns
rz slice
Definition at line 1280 of file model.cpp.
Generated by Doxygen
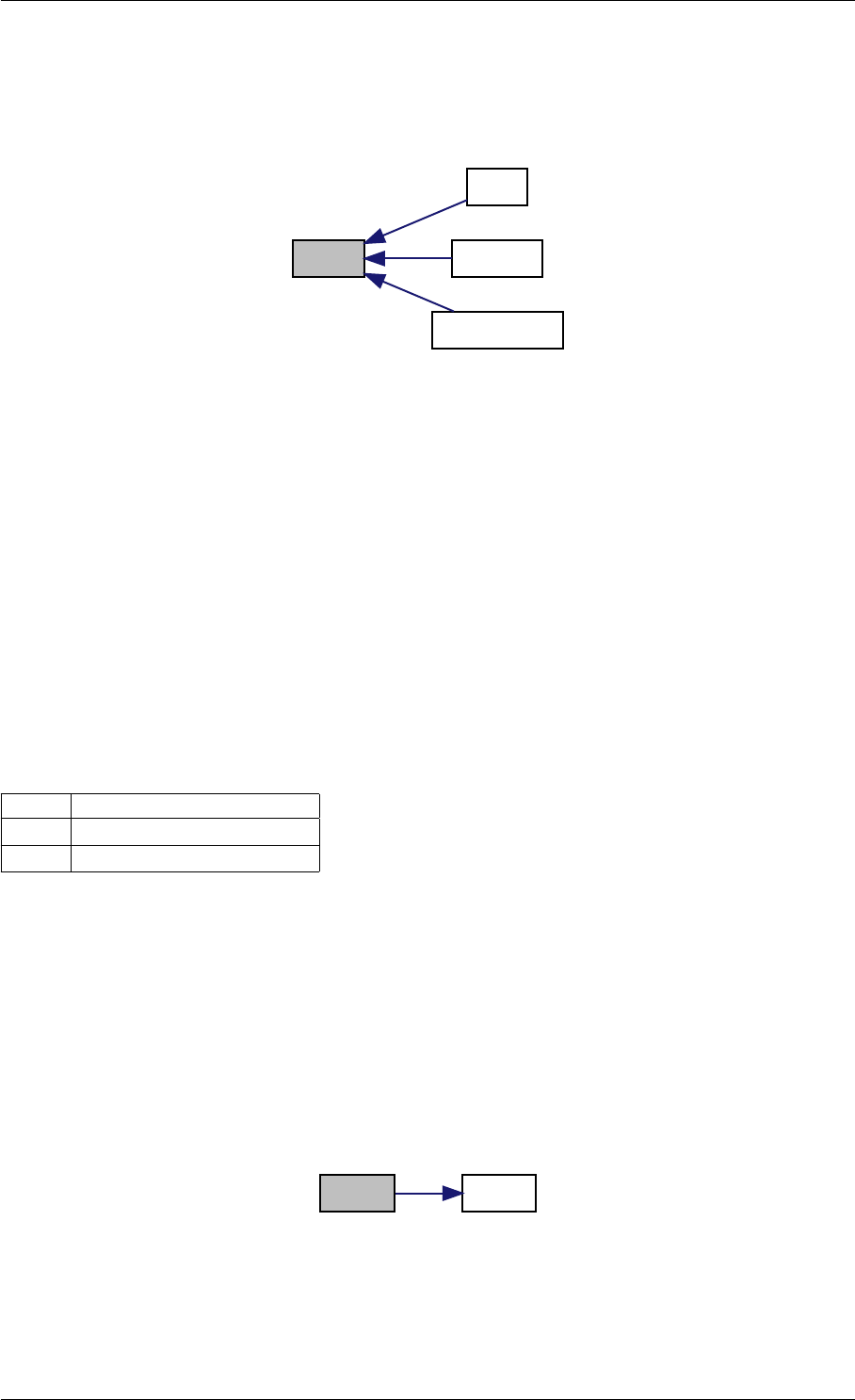
10.11 Model Class Reference 265
Here is the caller graph for this function:
getrz
fJrz
fdJrzdz
fdJrzdsigma
10.11.3.176 getsz()
const realtype ∗getsz (
const int nroots,
const int ip,
const ReturnData ∗rdata ) const [protected]
create sz slice at event
Parameters
nroots event occurence
ip sensitivity index
rdata pointer to return data instance
Returns
z slice
Definition at line 1284 of file model.cpp.
Here is the call graph for this function:
getsz nplist
Generated by Doxygen
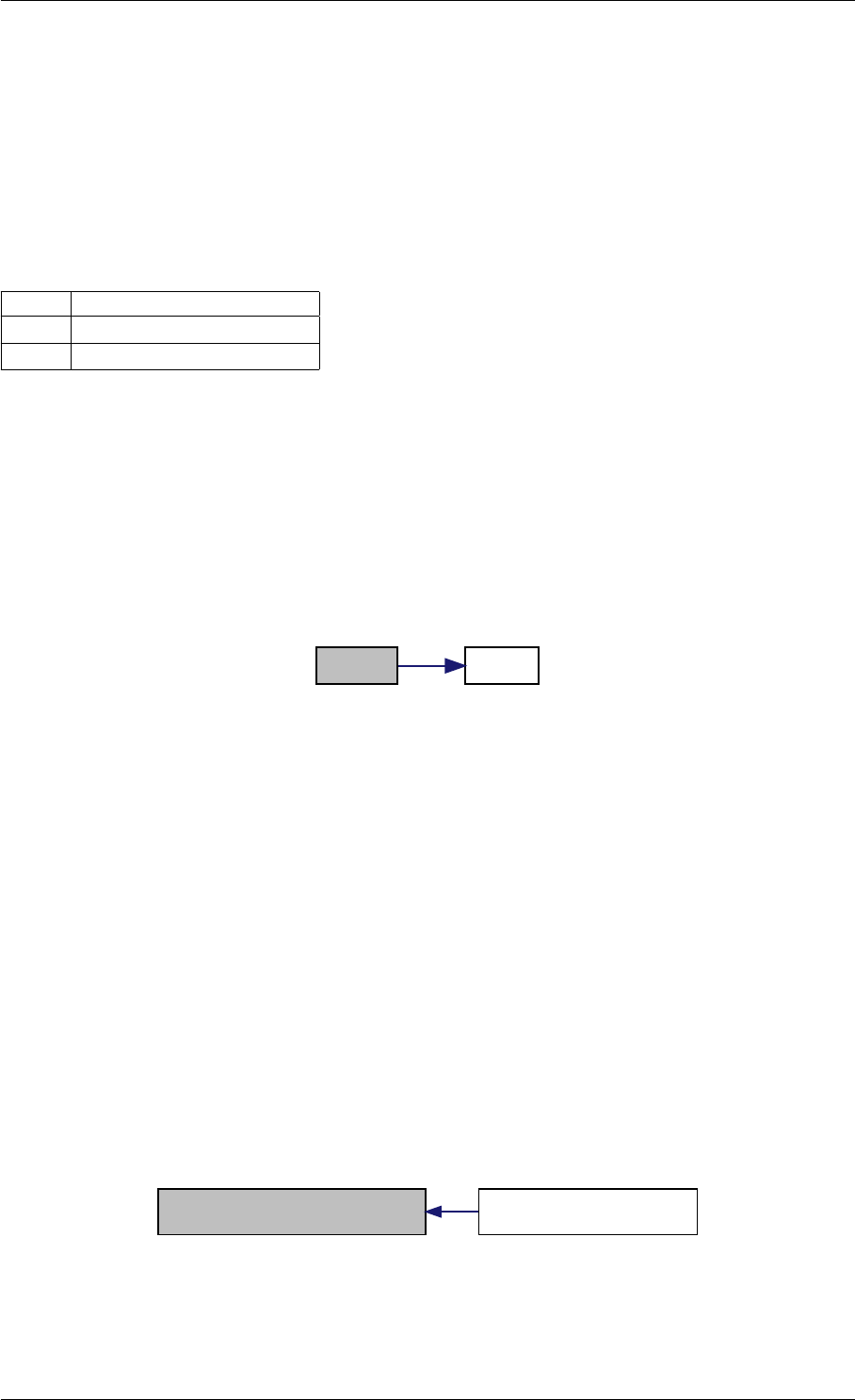
266 CONTENTS
10.11.3.177 getsrz()
const realtype ∗getsrz (
const int nroots,
const int ip,
const ReturnData ∗rdata ) const [protected]
create srz slice at event
Parameters
nroots event occurence
ip sensitivity index
rdata pointer to return data instance
Returns
rz slice
Definition at line 1288 of file model.cpp.
Here is the call graph for this function:
getsrz nplist
10.11.3.178 isFixedParameterStateReinitializationAllowed()
virtual bool isFixedParameterStateReinitializationAllowed ( ) const [protected], [virtual]
function indicating whether reinitialization of states depending on fixed parameters is permissible
Returns
flag inidication whether reinitialization of states depending on fixed parameters is permissible
Definition at line 1703 of file model.h.
Here is the caller graph for this function:
isFixedParameterStateReinitialization
Allowed
setReinitializeFixedParameter
InitialStates
Generated by Doxygen

10.11 Model Class Reference 267
10.11.3.179 computeX_pos()
N_Vector computeX_pos (
N_Vector x) [protected]
Parameters
xstate vector possibly containing negative values
Returns
state vector with negative values replaced by 0 according to stateIsNonNegative
Definition at line 1348 of file model.cpp.
Here is the call graph for this function:
computeX_pos
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
Generated by Doxygen
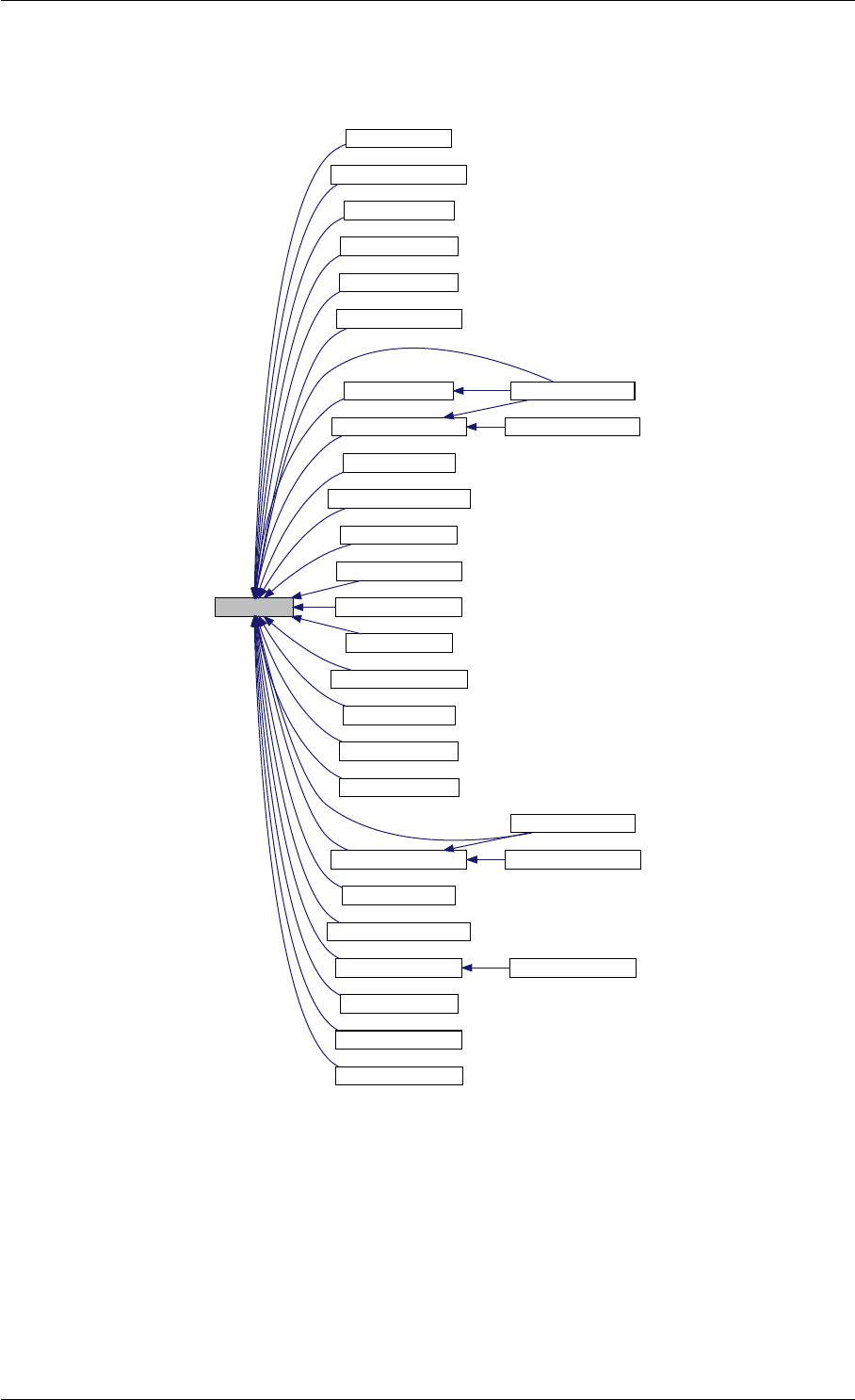
268 CONTENTS
Here is the caller graph for this function:
computeX_pos
amici::Model_DAE::fJ
amici::Model_DAE::fJSparse
amici::Model_DAE::fJv
amici::Model_DAE::froot
amici::Model_DAE::fxdot
amici::Model_DAE::fJDiag
amici::Model_DAE::fdxdotdp
amici::Model_DAE::fsxdotamici::Model_DAE::fM
amici::Model_DAE::fJB
amici::Model_DAE::fJSparseB
amici::Model_DAE::fJvB
amici::Model_DAE::fxBdot
amici::Model_DAE::fqBdot
amici::Model_ODE::fJ
amici::Model_ODE::fJSparse
amici::Model_ODE::fJv
amici::Model_ODE::froot
amici::Model_ODE::fxdot
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fsxdot
amici::Model_ODE::fJB
amici::Model_ODE::fJSparseB
amici::Model_ODE::fJDiag
amici::Model_ODE::fJvB
amici::Model_ODE::fxBdot
amici::Model_ODE::fqBdot
amici::Model_DAE::fdxdotdp
amici::Model_ODE::fdxdotdp
amici::Model_ODE::fJDiag
10.11.4 Friends And Related Function Documentation
10.11.4.1 boost::serialization::serialize
void boost::serialization::serialize (
Archive & ar,
Generated by Doxygen
10.11 Model Class Reference 269
Model &r,
const unsigned int version ) [friend]
Parameters
ar Archive to serialize to
rData to serialize
version Version number
10.11.4.2 operator==
bool operator== (
const Model &a,
const Model &b) [friend]
Parameters
afirst model instance
bsecond model instance
Returns
equality
Definition at line 1312 of file model.cpp.
10.11.5 Member Data Documentation
10.11.5.1 nx
const int nx
number of states
Definition at line 1086 of file model.h.
10.11.5.2 nxtrue
const int nxtrue
number of states in the unaugmented system
Definition at line 1088 of file model.h.
Generated by Doxygen

270 CONTENTS
10.11.5.3 ny
const int ny
number of observables
Definition at line 1090 of file model.h.
10.11.5.4 nytrue
const int nytrue
number of observables in the unaugmented system
Definition at line 1092 of file model.h.
10.11.5.5 nz
const int nz
number of event outputs
Definition at line 1094 of file model.h.
10.11.5.6 nztrue
const int nztrue
number of event outputs in the unaugmented system
Definition at line 1096 of file model.h.
10.11.5.7 ne
const int ne
number of events
Definition at line 1098 of file model.h.
Generated by Doxygen

10.11 Model Class Reference 271
10.11.5.8 nw
const int nw
number of common expressions
Definition at line 1100 of file model.h.
10.11.5.9 ndwdx
const int ndwdx
number of derivatives of common expressions wrt x
Definition at line 1102 of file model.h.
10.11.5.10 ndwdp
const int ndwdp
number of derivatives of common expressions wrt p
Definition at line 1104 of file model.h.
10.11.5.11 nnz
const int nnz
number of nonzero entries in jacobian
Definition at line 1106 of file model.h.
10.11.5.12 nJ
const int nJ
dimension of the augmented objective function for 2nd order ASA
Definition at line 1108 of file model.h.
Generated by Doxygen

272 CONTENTS
10.11.5.13 ubw
const int ubw
upper bandwith of the jacobian
Definition at line 1110 of file model.h.
10.11.5.14 lbw
const int lbw
lower bandwith of the jacobian
Definition at line 1112 of file model.h.
10.11.5.15 o2mode
const SecondOrderMode o2mode
flag indicating whether for sensi == AMICI_SENSI_ORDER_SECOND directional or full second order derivative will
be computed
Definition at line 1115 of file model.h.
10.11.5.16 z2event
const std::vector<int>z2event
index indicating to which event an event output belongs
Definition at line 1117 of file model.h.
10.11.5.17 idlist
const std::vector<realtype>idlist
flag array for DAE equations
Definition at line 1119 of file model.h.
Generated by Doxygen

10.11 Model Class Reference 273
10.11.5.18 sigmay
std::vector<realtype>sigmay
data standard deviation for current timepoint (dimension: ny)
Definition at line 1122 of file model.h.
10.11.5.19 dsigmaydp
std::vector<realtype>dsigmaydp
parameter derivative of data standard deviation for current timepoint (dimension: nplist x ny, row-major)
Definition at line 1124 of file model.h.
10.11.5.20 sigmaz
std::vector<realtype>sigmaz
event standard deviation for current timepoint (dimension: nz)
Definition at line 1126 of file model.h.
10.11.5.21 dsigmazdp
std::vector<realtype>dsigmazdp
parameter derivative of event standard deviation for current timepoint (dimension: nplist x nz, row-major)
Definition at line 1128 of file model.h.
10.11.5.22 dJydp
std::vector<realtype>dJydp
parameter derivative of data likelihood for current timepoint (dimension: nplist x nJ, row-major)
Definition at line 1130 of file model.h.
Generated by Doxygen

274 CONTENTS
10.11.5.23 dJzdp
std::vector<realtype>dJzdp
parameter derivative of event likelihood for current timepoint (dimension: nplist x nJ, row-major)
Definition at line 1132 of file model.h.
10.11.5.24 deltax
std::vector<realtype>deltax
change in x at current timepoint (dimension: nx)
Definition at line 1135 of file model.h.
10.11.5.25 deltasx
std::vector<realtype>deltasx
change in sx at current timepoint (dimension: nplist x nx, row-major)
Definition at line 1137 of file model.h.
10.11.5.26 deltaxB
std::vector<realtype>deltaxB
change in xB at current timepoint (dimension: nJ x nxtrue, row-major)
Definition at line 1139 of file model.h.
10.11.5.27 deltaqB
std::vector<realtype>deltaqB
change in qB at current timepoint (dimension: nJ x nplist, row-major)
Definition at line 1141 of file model.h.
Generated by Doxygen

10.11 Model Class Reference 275
10.11.5.28 dxdotdp
std::vector<realtype>dxdotdp
tempory storage of dxdotdp data across functions (dimension: nplist x nx, row-major)
Definition at line 1144 of file model.h.
10.11.5.29 J
SlsMat J = nullptr [protected]
Sparse Jacobian (dimension: nnz)
Definition at line 1709 of file model.h.
10.11.5.30 my
std::vector<realtype>my [protected]
current observable (dimension: nytrue)
Definition at line 1712 of file model.h.
10.11.5.31 mz
std::vector<realtype>mz [protected]
current event measurement (dimension: nztrue)
Definition at line 1714 of file model.h.
10.11.5.32 dJydy
std::vector<realtype>dJydy [protected]
observable derivative of data likelihood (dimension nJ x nytrue x ny, ordering = ?)
Definition at line 1716 of file model.h.
Generated by Doxygen

276 CONTENTS
10.11.5.33 dJydsigma
std::vector<realtype>dJydsigma [protected]
observable sigma derivative of data likelihood (dimension nJ x nytrue x ny, ordering = ?)
Definition at line 1718 of file model.h.
10.11.5.34 dJzdz
std::vector<realtype>dJzdz [protected]
event ouput derivative of event likelihood (dimension nJ x nztrue x nz, ordering = ?)
Definition at line 1721 of file model.h.
10.11.5.35 dJzdsigma
std::vector<realtype>dJzdsigma [protected]
event sigma derivative of event likelihood (dimension nJ x nztrue x nz, ordering = ?)
Definition at line 1723 of file model.h.
10.11.5.36 dJrzdz
std::vector<realtype>dJrzdz [protected]
event ouput derivative of event likelihood at final timepoint (dimension nJ x nztrue x nz, ordering = ?)
Definition at line 1725 of file model.h.
10.11.5.37 dJrzdsigma
std::vector<realtype>dJrzdsigma [protected]
event sigma derivative of event likelihood at final timepoint (dimension nJ x nztrue x nz, ordering = ?)
Definition at line 1727 of file model.h.
Generated by Doxygen

10.11 Model Class Reference 277
10.11.5.38 dzdx
std::vector<realtype>dzdx [protected]
state derivative of event output (dimension: nz ∗nx, ordering = ?)
Definition at line 1729 of file model.h.
10.11.5.39 dzdp
std::vector<realtype>dzdp [protected]
parameter derivative of event output (dimension: nz ∗nplist, ordering = ?)
Definition at line 1731 of file model.h.
10.11.5.40 drzdx
std::vector<realtype>drzdx [protected]
state derivative of event timepoint (dimension: nz ∗nx, ordering = ?)
Definition at line 1733 of file model.h.
10.11.5.41 drzdp
std::vector<realtype>drzdp [protected]
parameter derivative of event timepoint (dimension: nz ∗nplist, ordering = ?)
Definition at line 1735 of file model.h.
10.11.5.42 dydp
std::vector<realtype>dydp [protected]
parameter derivative of observable (dimension: nplist ∗ny, row-major)
Definition at line 1737 of file model.h.
Generated by Doxygen

278 CONTENTS
10.11.5.43 dydx
std::vector<realtype>dydx [protected]
state derivative of observable (dimension: ny ∗nx, ordering = ?)
Definition at line 1740 of file model.h.
10.11.5.44 w
std::vector<realtype>w [protected]
tempory storage of w data across functions (dimension: nw)
Definition at line 1742 of file model.h.
10.11.5.45 dwdx
std::vector<realtype>dwdx [protected]
tempory storage of sparse dwdx data across functions (dimension: ndwdx)
Definition at line 1744 of file model.h.
10.11.5.46 dwdp
std::vector<realtype>dwdp [protected]
tempory storage of sparse dwdp data across functions (dimension: ndwdp)
Definition at line 1746 of file model.h.
10.11.5.47 M
std::vector<realtype>M [protected]
tempory storage of M data across functions (dimension: nx)
Definition at line 1748 of file model.h.
Generated by Doxygen

10.11 Model Class Reference 279
10.11.5.48 stau
std::vector<realtype>stau [protected]
tempory storage of stau data across functions (dimension: nplist)
Definition at line 1750 of file model.h.
10.11.5.49 h
std::vector<realtype>h [protected]
flag indicating whether a certain heaviside function should be active or not (dimension: ne)
Definition at line 1754 of file model.h.
10.11.5.50 unscaledParameters
std::vector<realtype>unscaledParameters [protected]
unscaled parameters (dimension: np)
Definition at line 1757 of file model.h.
10.11.5.51 originalParameters
std::vector<realtype>originalParameters [protected]
orignal user-provided, possibly scaled parameter array (size np)
Definition at line 1760 of file model.h.
10.11.5.52 fixedParameters
std::vector<realtype>fixedParameters [protected]
constants (dimension: nk)
Definition at line 1763 of file model.h.
Generated by Doxygen

280 CONTENTS
10.11.5.53 plist_
std::vector<int>plist_ [protected]
indexes of parameters wrt to which sensitivities are computed (dimension nplist)
Definition at line 1766 of file model.h.
10.11.5.54 x0data
std::vector<double>x0data [protected]
state initialisation (size nx)
Definition at line 1769 of file model.h.
10.11.5.55 sx0data
std::vector<realtype>sx0data [protected]
sensitivity initialisation (size nx ∗nplist, ordering = ?)
Definition at line 1772 of file model.h.
10.11.5.56 ts
std::vector<realtype>ts [protected]
timepoints (size nt)
Definition at line 1775 of file model.h.
10.11.5.57 stateIsNonNegative
std::vector<bool>stateIsNonNegative [protected]
vector of bools indicating whether state variables are to be assumed to be positive
Definition at line 1778 of file model.h.
Generated by Doxygen

10.11 Model Class Reference 281
10.11.5.58 anyStateNonNegative
bool anyStateNonNegative = false [protected]
boolean indicating whether any entry in stateIsNonNegative is true
Definition at line 1781 of file model.h.
10.11.5.59 x_pos_tmp
AmiVector x_pos_tmp [protected]
temporary storage of positified state variables according to stateIsNonNegative
Definition at line 1784 of file model.h.
10.11.5.60 nmaxevent
int nmaxevent = 10 [protected]
maximal number of events to track
Definition at line 1787 of file model.h.
10.11.5.61 pscale
std::vector<ParameterScaling>pscale [protected]
parameter transformation of originalParameters (dimension np)
Definition at line 1790 of file model.h.
10.11.5.62 tstart
double tstart = 0.0 [protected]
starting time
Definition at line 1793 of file model.h.
Generated by Doxygen

282 CONTENTS
10.11.5.63 steadyStateSensitivityMode
SteadyStateSensitivityMode steadyStateSensitivityMode = SteadyStateSensitivityMode::newtonOnly
[protected]
flag indicating whether steadystate sensivities are to be computed via FSA when steadyStateSimulation is used
Definition at line 1797 of file model.h.
10.11.5.64 reinitializeFixedParameterInitialStates
bool reinitializeFixedParameterInitialStates = false [protected]
flag indicating whether reinitialization of states depending on fixed parameters is activated
Definition at line 1802 of file model.h.
10.12 Model_DAE Class Reference
The Model class represents an AMICI DAE model. The model does not contain any data, but represents the state
of the model at a specific time t. The states must not always be in sync, but may be updated asynchroneously.
#include <model_dae.h>
Inheritance diagram for Model_DAE:
Model_DAE
Model
Generated by Doxygen

10.12 Model_DAE Class Reference 283
Public Member Functions
•Model_DAE ()
•Model_DAE (const int nx, const int nxtrue, const int ny, const int nytrue, const int nz, const int nztrue, const
int ne, const int nJ, const int nw, const int ndwdx, const int ndwdp, const int nnz, const int ubw, const int lbw,
const SecondOrderMode o2mode, const std::vector<realtype >p, const std::vector<realtype >k, const
std::vector<int >plist, const std::vector<realtype >idlist, const std::vector<int >z2event)
• virtual void fJ (realtype t,realtype cj, AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, DlsMat J) override
• void fJ (realtype t,realtype cj, N_Vector x, N_Vector dx, N_Vector xdot, DlsMat J)
• void fJB (realtype t,realtype cj, N_Vector x, N_Vector dx, N_Vector xB, N_Vector dxB, DlsMat JB)
• virtual void fJSparse (realtype t,realtype cj, AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, SlsMat J) override
• void fJSparse (realtype t,realtype cj, N_Vector x, N_Vector dx, SlsMat J)
• void fJSparseB (realtype t,realtype cj, N_Vector x, N_Vector dx, N_Vector xB, N_Vector dxB, SlsMat JB)
• virtual void fJDiag (realtype t,AmiVector ∗JDiag, realtype cj, AmiVector ∗x, AmiVector ∗dx) override
• virtual void fJv (realtype t,AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, AmiVector ∗v, AmiVector ∗nJv,
realtype cj) override
• void fJv (realtype t, N_Vector x, N_Vector dx, N_Vector v, N_Vector Jv, realtype cj)
• void fJvB (realtype t, N_Vector x, N_Vector dx, N_Vector xB, N_Vector dxB, N_Vector vB, N_Vector JvB,
realtype cj)
• virtual void froot (realtype t,AmiVector ∗x, AmiVector ∗dx, realtype ∗root) override
• void froot (realtype t, N_Vector x, N_Vector dx, realtype ∗root)
• virtual void fxdot (realtype t,AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot) override
• void fxdot (realtype t, N_Vector x, N_Vector dx, N_Vector xdot)
• void fxBdot (realtype t, N_Vector x, N_Vector dx, N_Vector xB, N_Vector dxB, N_Vector xBdot)
• void fqBdot (realtype t, N_Vector x, N_Vector dx, N_Vector xB, N_Vector dxB, N_Vector qBdot)
• void fdxdotdp (const realtype t, const N_Vector x, const N_Vector dx)
• virtual void fdxdotdp (realtype t,AmiVector ∗x, AmiVector ∗dx) override
• void fsxdot (realtype t,AmiVector ∗x, AmiVector ∗dx, int ip, AmiVector ∗sx, AmiVector ∗sdx, AmiVector ∗sxdot)
override
• void fsxdot (realtype t, N_Vector x, N_Vector dx, int ip, N_Vector sx, N_Vector sdx, N_Vector sxdot)
• void fM (realtype t, const N_Vector x)
Mass matrix for DAE systems.
• virtual std::unique_ptr<Solver >getSolver () override
Protected Member Functions
• virtual void fJ (realtype ∗J, const realtype t, const realtype ∗x, const double ∗p, const double ∗k, const realtype
∗h, const realtype cj, const realtype ∗dx, const realtype ∗w, const realtype ∗dwdx)=0
• virtual void fJB (realtype ∗JB, const realtype t, const realtype ∗x, const double ∗p, const double ∗k, const
realtype ∗h, const realtype cj, const realtype ∗xB, const realtype ∗dx, const realtype ∗dxB, const realtype ∗w,
const realtype ∗dwdx)
• virtual void fJSparse (SlsMat JSparse, const realtype t, const realtype ∗x, const double ∗p, const double ∗k,
const realtype ∗h, const realtype cj, const realtype ∗dx, const realtype ∗w, const realtype ∗dwdx)=0
• virtual void fJSparseB (SlsMat JSparseB, const realtype t, const realtype ∗x, const double ∗p, const double
∗k, const realtype ∗h, const realtype cj, const realtype ∗xB, const realtype ∗dx, const realtype ∗dxB, const
realtype ∗w, const realtype ∗dwdx)
• virtual void fJDiag (realtype ∗JDiag, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype cj, const realtype ∗dx, const realtype ∗w, const realtype ∗dwdx)
• virtual void fJv (realtype ∗Jv, const realtype t, const realtype ∗x, const double ∗p, const double ∗k, const
realtype ∗h, const realtype cj, const realtype ∗dx, const realtype ∗v, const realtype ∗w, const realtype ∗dwdx)
• virtual void fJvB (realtype ∗JvB, const realtype t, const realtype ∗x, const double ∗p, const double ∗k, const
realtype ∗h, const realtype cj, const realtype ∗xB, const realtype ∗dx, const realtype ∗dxB, const realtype
∗vB, const realtype ∗w, const realtype ∗dwdx)
• virtual void froot (realtype ∗root, const realtype t, const realtype ∗x, const double ∗p, const double ∗k, const
realtype ∗h, const realtype ∗dx)
Generated by Doxygen

284 CONTENTS
• virtual void fxdot (realtype ∗xdot, const realtype t, const realtype ∗x, const double ∗p, const double ∗k, const
realtype ∗h, const realtype ∗dx, const realtype ∗w)=0
• virtual void fxBdot (realtype ∗xBdot, const realtype t, const realtype ∗x, const double ∗p, const double ∗k,
const realtype ∗h, const realtype ∗xB, const realtype ∗dx, const realtype ∗dxB, const realtype ∗w, const
realtype ∗dwdx)
• virtual void fqBdot (realtype ∗qBdot, const int ip, const realtype t, const realtype ∗x, const double ∗p, const
double ∗k, const realtype ∗h, const realtype ∗xB, const realtype ∗dx, const realtype ∗dxB, const realtype ∗w,
const realtype ∗dwdp)
• virtual void fdxdotdp (realtype ∗dxdotdp, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const int ip, const realtype ∗dx, const realtype ∗w, const realtype ∗dwdp)
• virtual void fsxdot (realtype ∗sxdot, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const int ip, const realtype ∗dx, const realtype ∗sx, const realtype ∗sdx, const realtype ∗w,
const realtype ∗dwdx, const realtype ∗M, const realtype ∗J, const realtype ∗dxdotdp)
• virtual void fM (realtype ∗M, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k)
Additional Inherited Members
10.12.1 Detailed Description
Definition at line 24 of file model_dae.h.
10.12.2 Constructor & Destructor Documentation
10.12.2.1 Model_DAE() [1/2]
Model_DAE ( )
default constructor
Definition at line 27 of file model_dae.h.
10.12.2.2 Model_DAE() [2/2]
Model_DAE (
const int nx,
const int nxtrue,
const int ny,
const int nytrue,
const int nz,
const int nztrue,
const int ne,
const int nJ,
const int nw,
const int ndwdx,
const int ndwdp,
const int nnz,
const int ubw,
const int lbw,
const SecondOrderMode o2mode,
const std::vector<realtype >p,
const std::vector<realtype >k,
const std::vector<int >plist,
const std::vector<realtype >idlist,
const std::vector<int >z2event )
constructor with model dimensions
Generated by Doxygen
10.12 Model_DAE Class Reference 285
Parameters
nx number of state variables
nxtrue number of state variables of the non-augmented model
ny number of observables
nytrue number of observables of the non-augmented model
nz number of event observables
nztrue number of event observables of the non-augmented model
ne number of events
nJ number of objective functions
nw number of repeating elements
ndwdx number of nonzero elements in the x derivative of the repeating elements
ndwdp number of nonzero elements in the p derivative of the repeating elements
nnz number of nonzero elements in Jacobian
ubw upper matrix bandwidth in the Jacobian
lbw lower matrix bandwidth in the Jacobian
o2mode second order sensitivity mode
pparameters
kconstants
plist indexes wrt to which sensitivities are to be computed
idlist indexes indicating algebraic components (DAE only)
z2event mapping of event outputs to events
Definition at line 53 of file model_dae.h.
10.12.3 Member Function Documentation
10.12.3.1 fJ() [1/3]
void fJ (
realtype t,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
DlsMat J) [override], [virtual]
Dense Jacobian function
Parameters
ttime
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
Jdense matrix to which values of the jacobian will be written
Generated by Doxygen
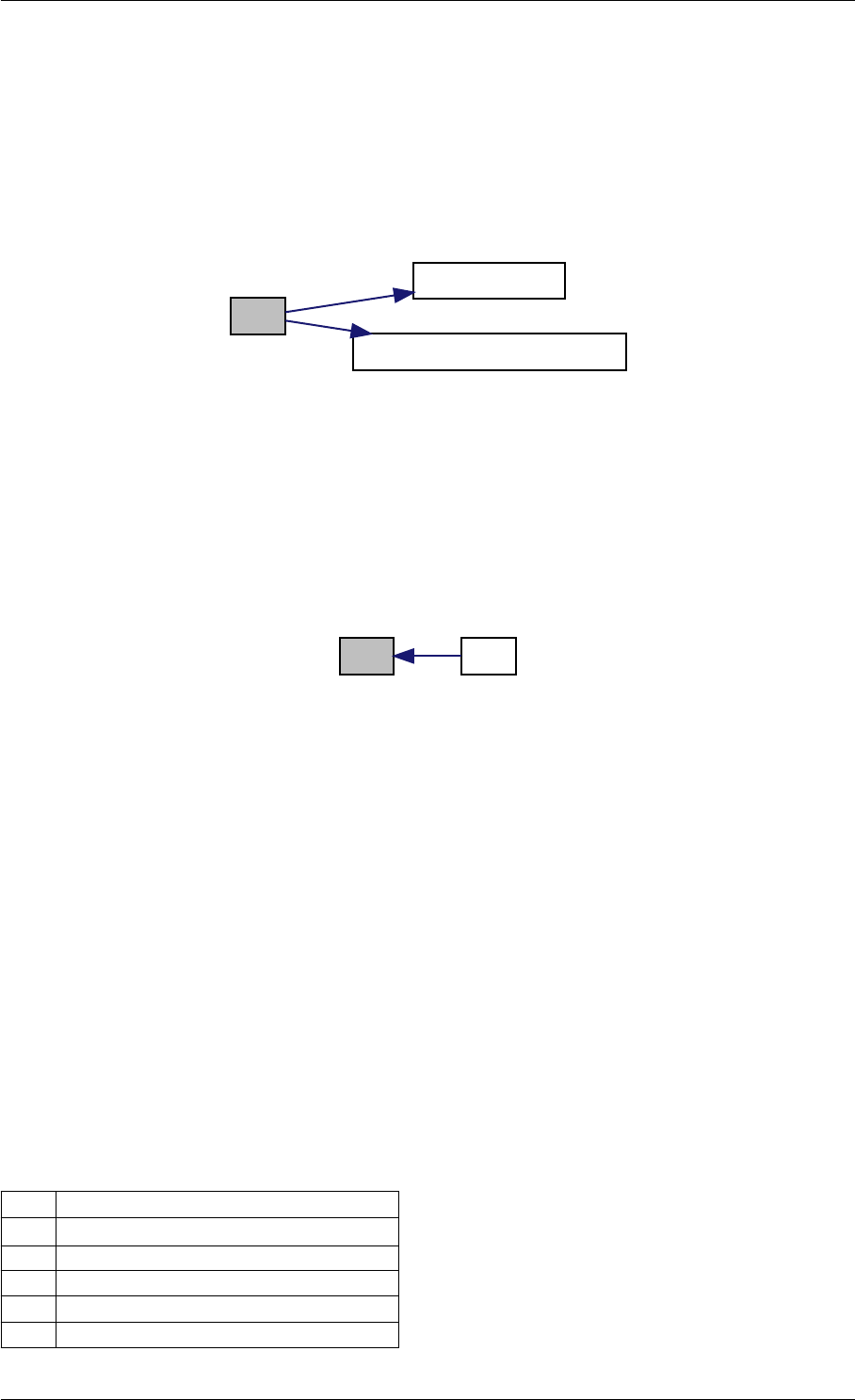
286 CONTENTS
Implements Model.
Definition at line 6 of file model_dae.cpp.
Here is the call graph for this function:
fJ
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fJ fJ
10.12.3.2 fJ() [2/3]
void fJ (
realtype t,
realtype cj,
N_Vector x,
N_Vector dx,
N_Vector xdot,
DlsMat J)
Jacobian of xdot with respect to states x
Parameters
ttimepoint
cj scaling factor, inverse of the step size
xVector with the states
dx Vector with the derivative states
xdot Vector with the right hand side
JMatrix to which the Jacobian will be written
Generated by Doxygen
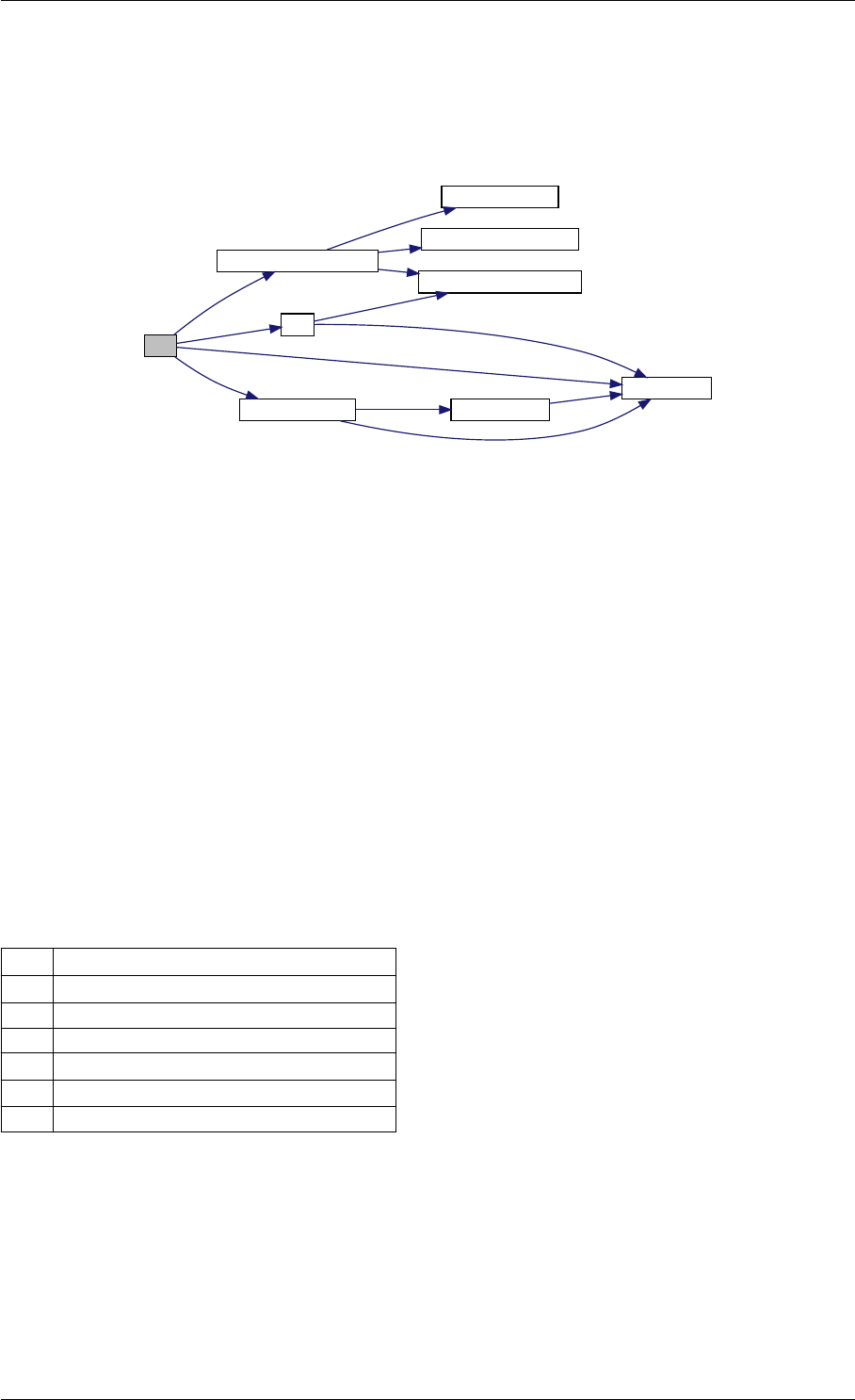
10.12 Model_DAE Class Reference 287
Definition at line 20 of file model_dae.cpp.
Here is the call graph for this function:
fJ
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
fJ
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.3 fJB() [1/2]
void fJB (
realtype t,
realtype cj,
N_Vector x,
N_Vector dx,
N_Vector xB,
N_Vector dxB,
DlsMat JB )
Jacobian of xBdot with respect to adjoint state xB
Parameters
ttimepoint
cj scaling factor, inverse of the step size
xVector with the states
dx Vector with the derivative states
xB Vector with the adjoint states
dxB Vector with the adjoint derivative states
JB Matrix to which the Jacobian will be written
Definition at line 166 of file model_dae.cpp.
Generated by Doxygen
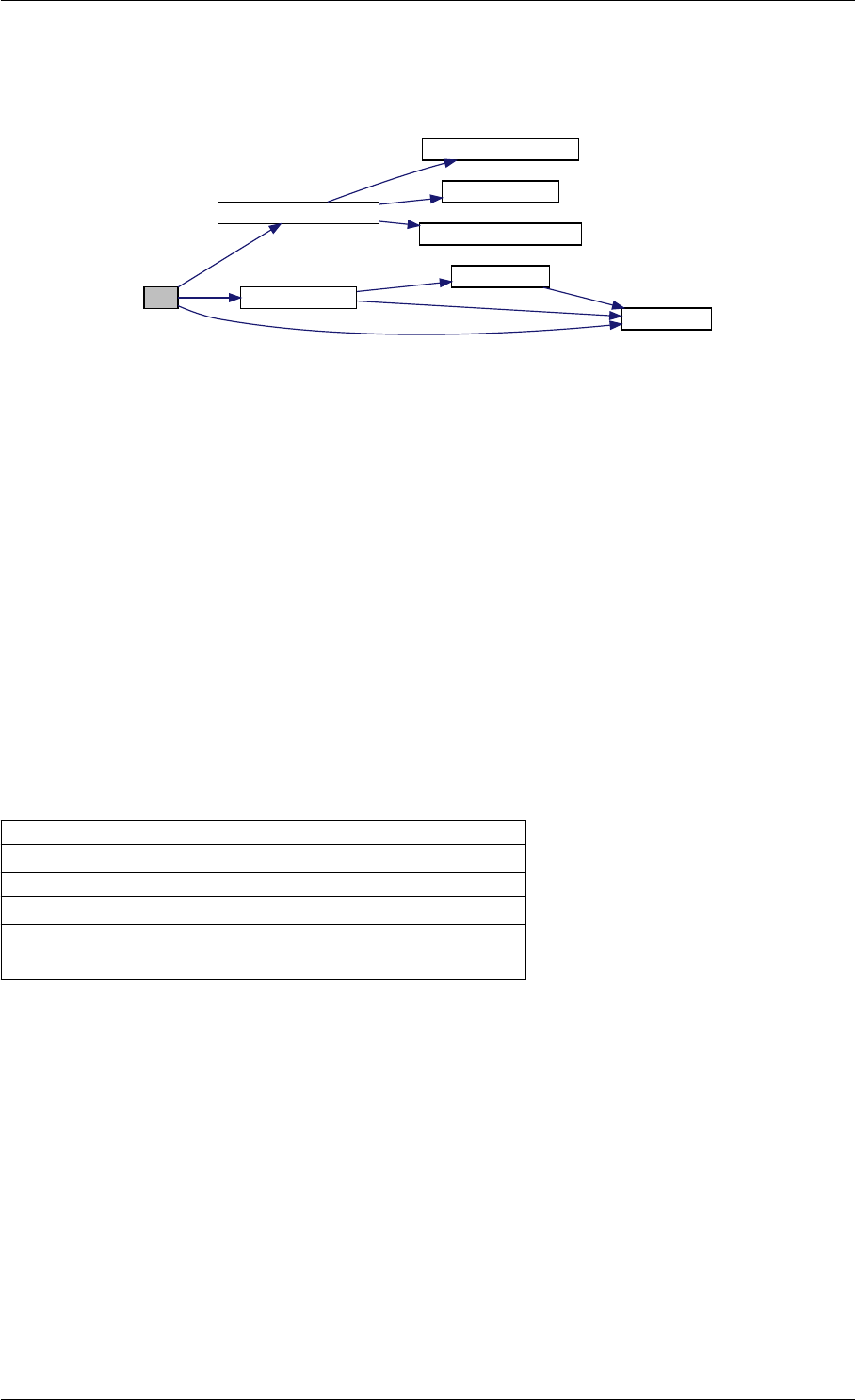
288 CONTENTS
Here is the call graph for this function:
fJB
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.4 fJSparse() [1/3]
void fJSparse (
realtype t,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
SlsMat J) [override], [virtual]
Sparse Jacobian function
Parameters
ttime
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
Jsparse matrix to which values of the Jacobian will be written
Implements Model.
Definition at line 29 of file model_dae.cpp.
Generated by Doxygen
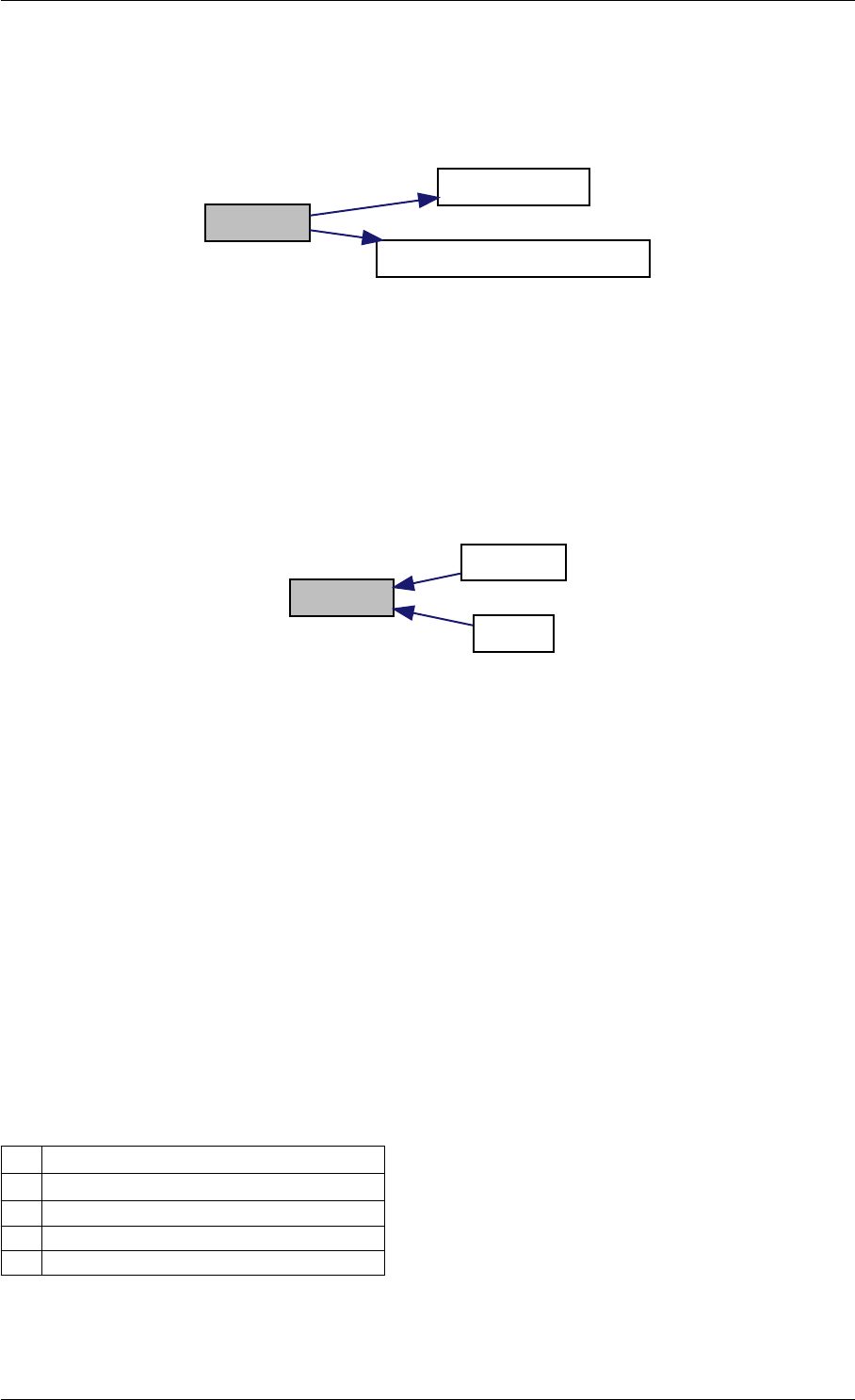
10.12 Model_DAE Class Reference 289
Here is the call graph for this function:
fJSparse
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fJSparse
fJSparse
fsxdot
10.12.3.5 fJSparse() [2/3]
void fJSparse (
realtype t,
realtype cj,
N_Vector x,
N_Vector dx,
SlsMat J)
J in sparse form (for sparse solvers from the SuiteSparse Package)
Parameters
ttimepoint
cj scalar in Jacobian (inverse stepsize)
xVector with the states
dx Vector with the derivative states
JMatrix to which the Jacobian will be written
Definition at line 41 of file model_dae.cpp.
Generated by Doxygen
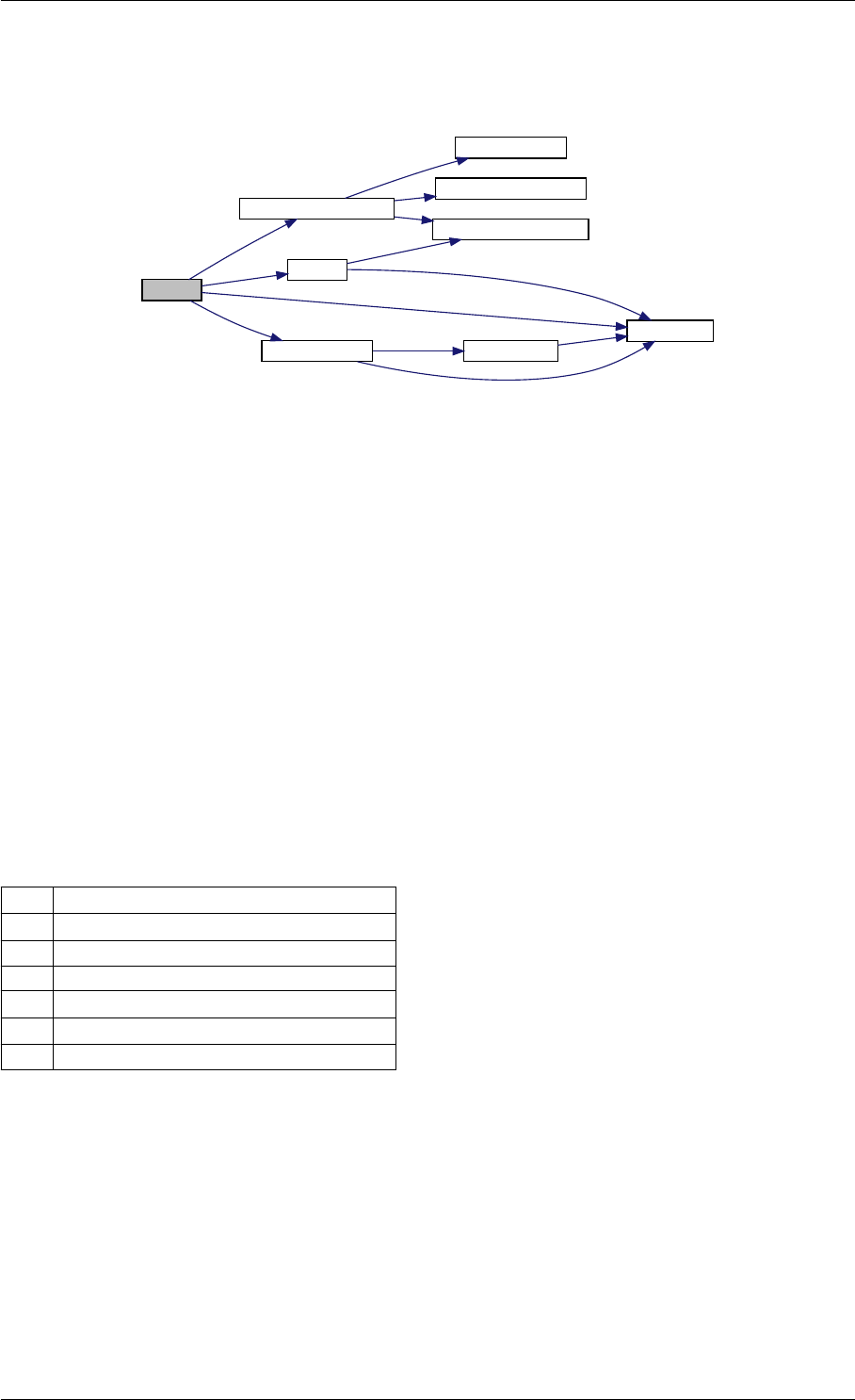
290 CONTENTS
Here is the call graph for this function:
fJSparse
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
fJSparse
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.6 fJSparseB() [1/2]
void fJSparseB (
realtype t,
realtype cj,
N_Vector x,
N_Vector dx,
N_Vector xB,
N_Vector dxB,
SlsMat JB )
JB in sparse form (for sparse solvers from the SuiteSparse Package)
Parameters
ttimepoint
cj scalar in Jacobian
xVector with the states
dx Vector with the derivative states
xB Vector with the adjoint states
dxB Vector with the adjoint derivative states
JB Matrix to which the Jacobian will be written
Definition at line 184 of file model_dae.cpp.
Generated by Doxygen

10.12 Model_DAE Class Reference 291
Here is the call graph for this function:
fJSparseB
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.7 fJDiag() [1/2]
void fJDiag (
realtype t,
AmiVector ∗JDiag,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx ) [override], [virtual]
diagonalized Jacobian (for preconditioning)
Parameters
ttimepoint
JDiag Vector to which the Jacobian diagonal will be written
cj scaling factor, inverse of the step size
xVector with the states
dx Vector with the derivative states
Returns
status flag indicating successful execution
Implements Model.
Definition at line 116 of file model_dae.cpp.
Generated by Doxygen
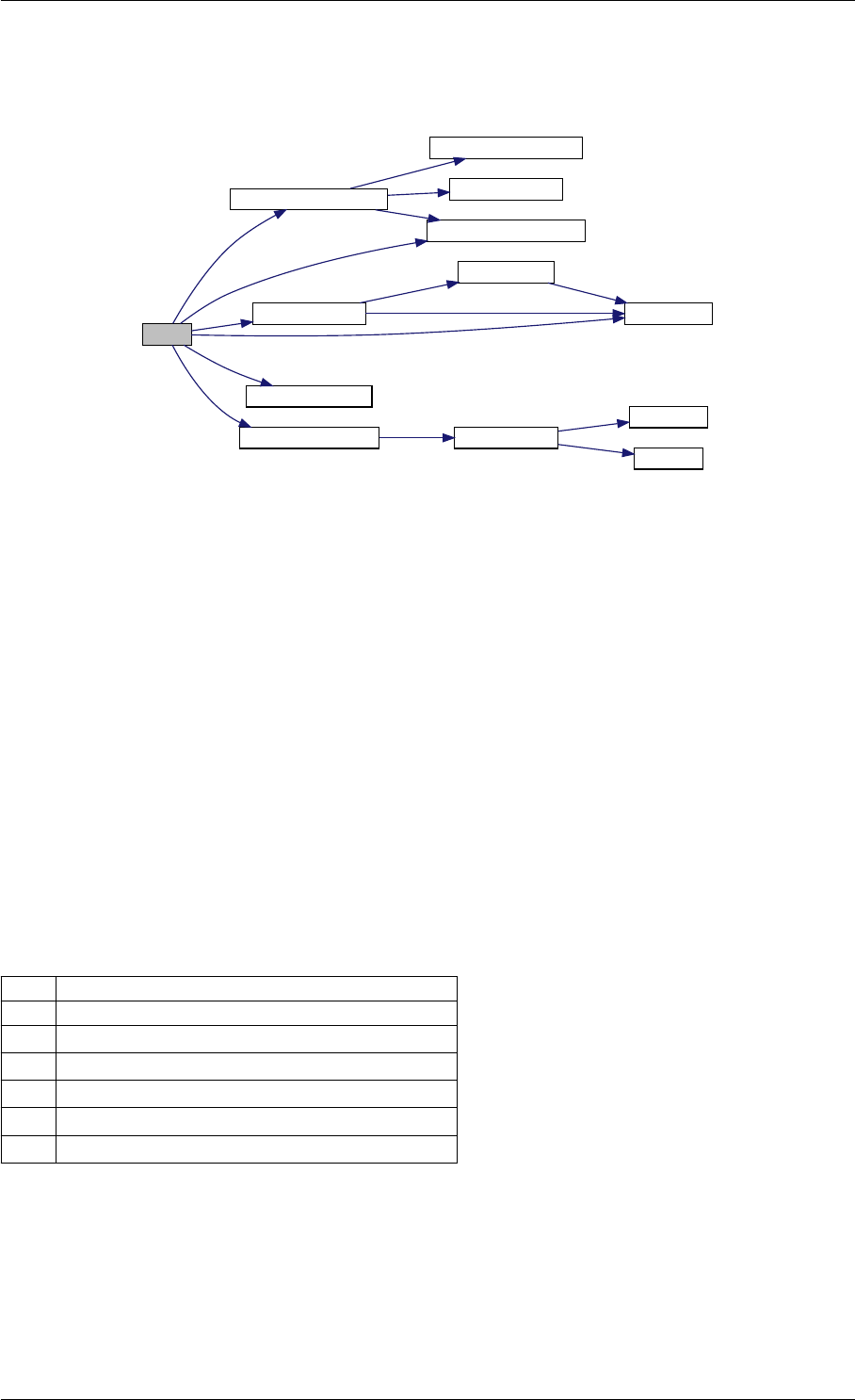
292 CONTENTS
Here is the call graph for this function:
fJDiag
amici::Model::computeX_pos
amici::AmiVector::getNVector
amici::Model::fdwdx amici::Model::t
amici::AmiVector::data
amici::Model::checkFinite
amici::AmiVector::getLength
amici::AmiVector::at
amici::Model::fw
amici::checkFinite
amici::isNaN
amici::isInf
10.12.3.8 fJv() [1/3]
void fJv (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
AmiVector ∗v,
AmiVector ∗nJv,
realtype cj ) [override], [virtual]
Jacobian multiply function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
vmultiplication vector (unused)
nJv array to which result of multiplication will be written
cj scaling factor (inverse of timestep, DAE only)
Implements Model.
Definition at line 49 of file model_dae.cpp.
Generated by Doxygen
10.12 Model_DAE Class Reference 293
Here is the call graph for this function:
fJv
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fJv fJv
10.12.3.9 fJv() [2/3]
void fJv (
realtype t,
N_Vector x,
N_Vector dx,
N_Vector v,
N_Vector Jv,
realtype cj )
Matrix vector product of J with a vector v (for iterative solvers)
Parameters
ttimepoint
Type: realtype
cj scaling factor, inverse of the step size
xVector with the states
dx Vector with the derivative states
vVector with which the Jacobian is multiplied
Jv Vector to which the Jacobian vector product will be written
Definition at line 64 of file model_dae.cpp.
Generated by Doxygen
294 CONTENTS
Here is the call graph for this function:
fJv
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
fJv
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.10 fJvB() [1/2]
void fJvB (
realtype t,
N_Vector x,
N_Vector dx,
N_Vector xB,
N_Vector dxB,
N_Vector vB,
N_Vector JvB,
realtype cj )
Matrix vector product of JB with a vector v (for iterative solvers)
Parameters
ttimepoint
xVector with the states
dx Vector with the derivative states
xB Vector with the adjoint states
dxB Vector with the adjoint derivative states
vB Vector with which the Jacobian is multiplied
JvB Vector to which the Jacobian vector product will be written
cj scalar in Jacobian (inverse stepsize)
Definition at line 204 of file model_dae.cpp.
Generated by Doxygen

10.12 Model_DAE Class Reference 295
Here is the call graph for this function:
fJvB
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.11 froot() [1/3]
void froot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
realtype ∗root ) [override], [virtual]
Root function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
root array to which values of the root function will be written
Implements Model.
Definition at line 73 of file model_dae.cpp.
Here is the call graph for this function:
froot
amici::Model::t
amici::AmiVector::getNVector
Generated by Doxygen
296 CONTENTS
Here is the caller graph for this function:
froot froot
10.12.3.12 froot() [2/3]
void froot (
realtype t,
N_Vector x,
N_Vector dx,
realtype ∗root )
Event trigger function for events
Parameters
ttimepoint
xVector with the states
dx Vector with the derivative states
root array with root function values
Definition at line 83 of file model_dae.cpp.
Here is the call graph for this function:
froot
amici::Model::computeX_pos
froot
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
Generated by Doxygen
10.12 Model_DAE Class Reference 297
10.12.3.13 fxdot() [1/3]
void fxdot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot ) [override], [virtual]
Residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
xdot array to which values of the residual function will be written
Implements Model.
Definition at line 90 of file model_dae.cpp.
Here is the call graph for this function:
fxdot
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fxdot fxdot
Generated by Doxygen
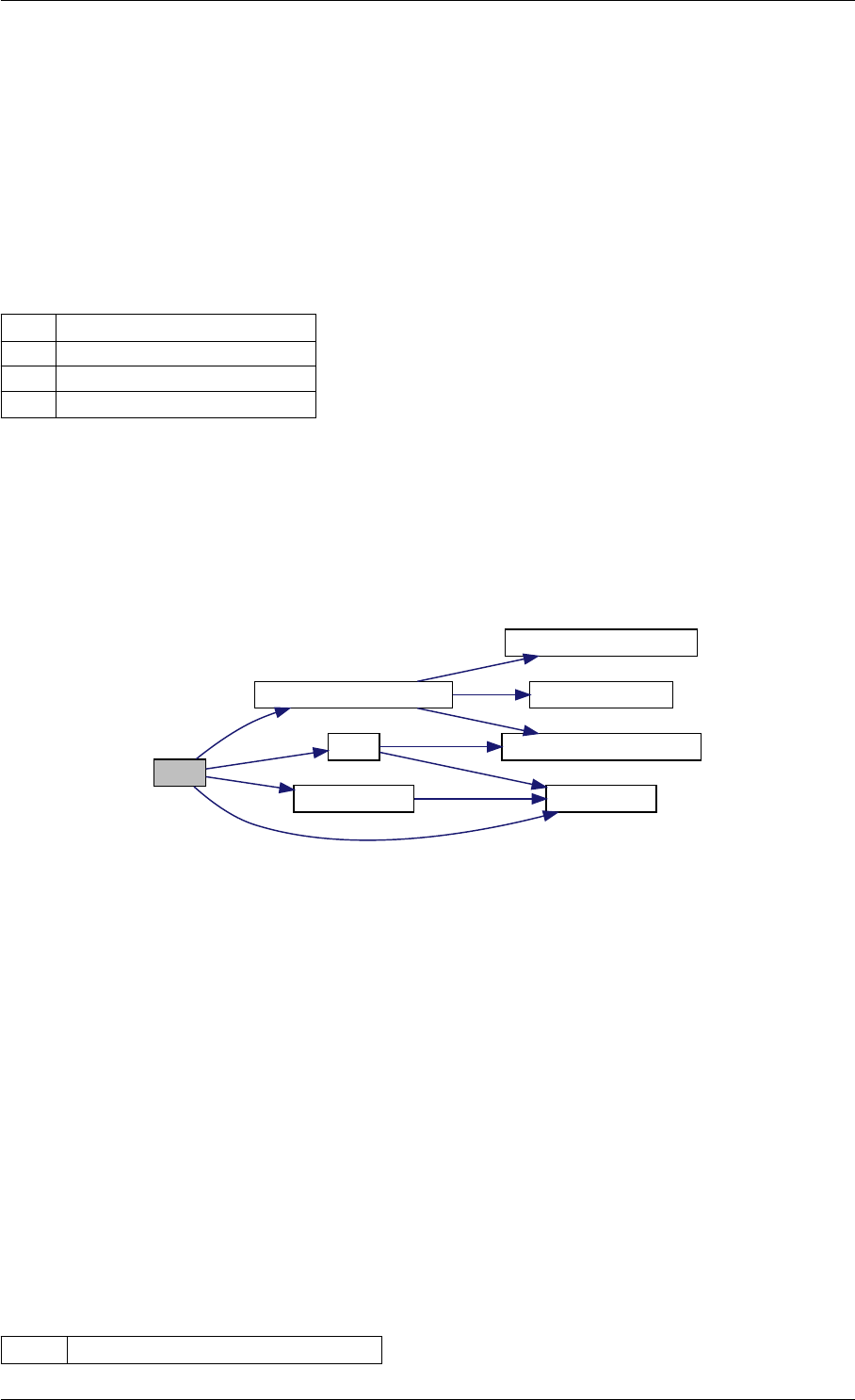
298 CONTENTS
10.12.3.14 fxdot() [2/3]
void fxdot (
realtype t,
N_Vector x,
N_Vector dx,
N_Vector xdot )
residual function of the DAE
Parameters
ttimepoint
xVector with the states
dx Vector with the derivative states
xdot Vector with the right hand side
Definition at line 100 of file model_dae.cpp.
Here is the call graph for this function:
fxdot
amici::Model::computeX_pos
amici::Model::fw amici::Model::t
fxdot
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
10.12.3.15 fxBdot() [1/2]
void fxBdot (
realtype t,
N_Vector x,
N_Vector dx,
N_Vector xB,
N_Vector dxB,
N_Vector xBdot )
Right hand side of differential equation for adjoint state xB
Parameters
ttimepoint
Generated by Doxygen
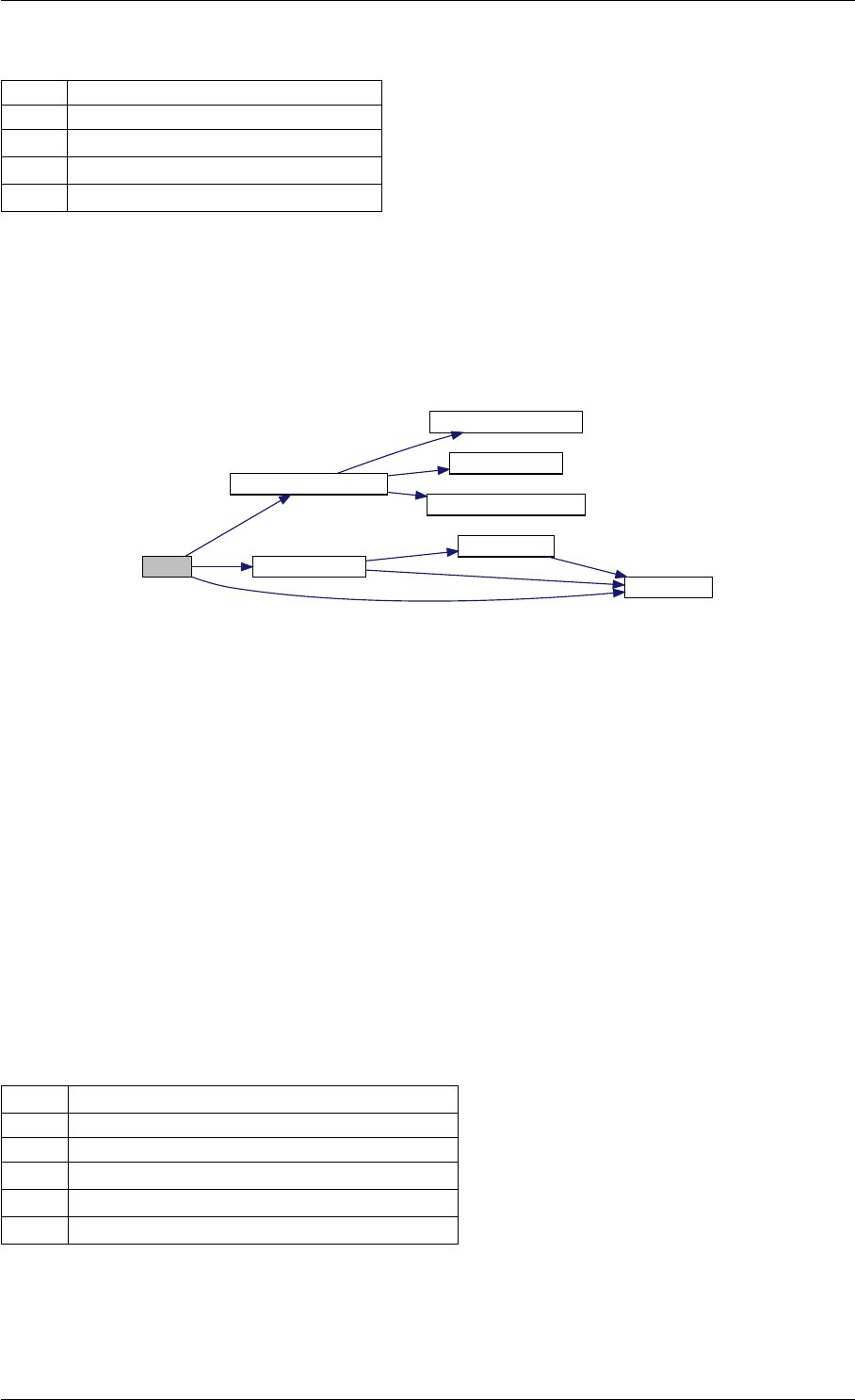
10.12 Model_DAE Class Reference 299
Parameters
xVector with the states
dx Vector with the derivative states
xB Vector with the adjoint states
dxB Vector with the adjoint derivative states
xBdot Vector with the adjoint right hand side
Definition at line 222 of file model_dae.cpp.
Here is the call graph for this function:
fxBdot
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.16 fqBdot() [1/2]
void fqBdot (
realtype t,
N_Vector x,
N_Vector dx,
N_Vector xB,
N_Vector dxB,
N_Vector qBdot )
Right hand side of integral equation for quadrature states qB
Parameters
ttimepoint
xVector with the states
dx Vector with the derivative states
xB Vector with the adjoint states
dxB Vector with the adjoint derivative states
qBdot Vector with the adjoint quadrature right hand side
Definition at line 240 of file model_dae.cpp.
Generated by Doxygen
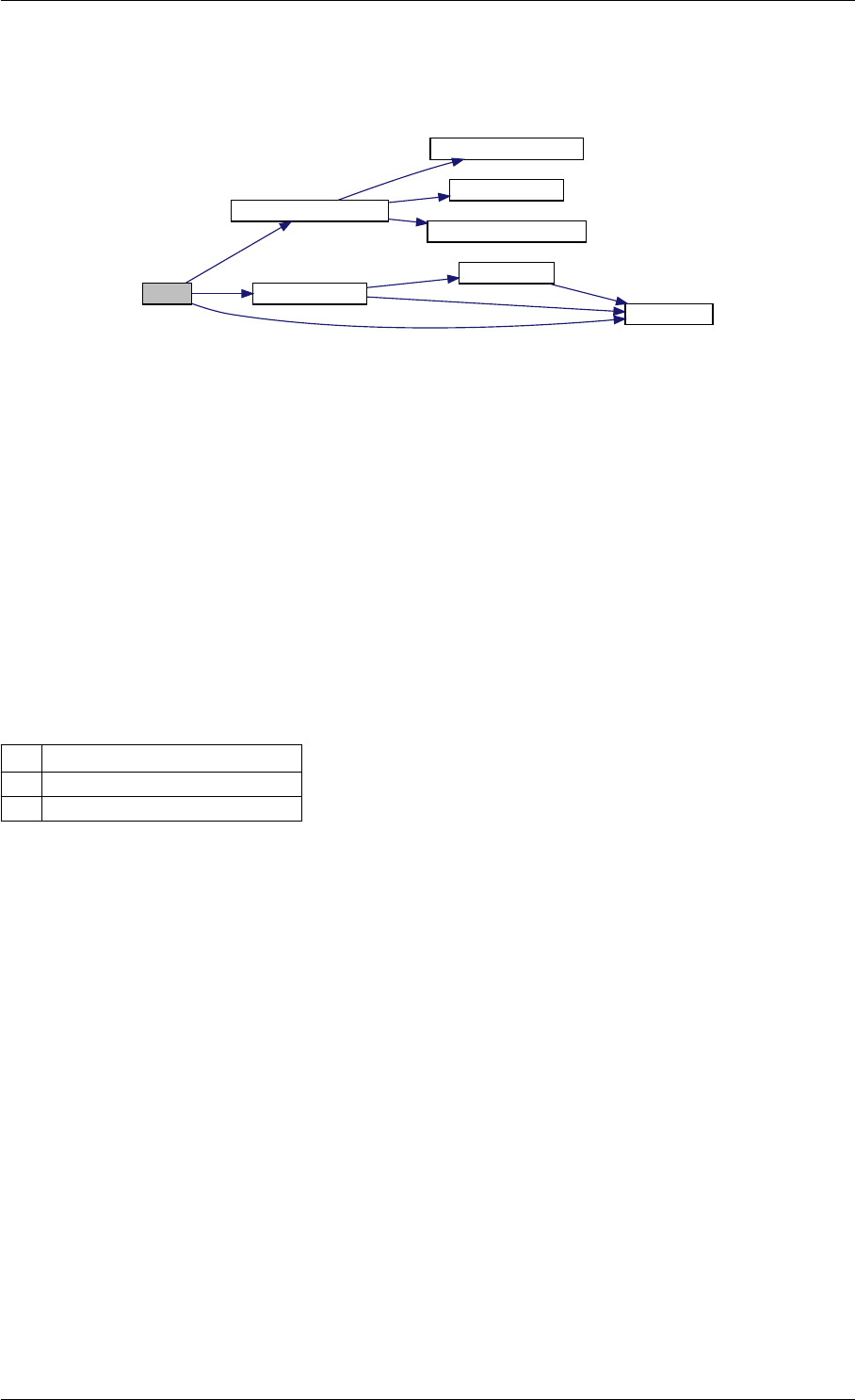
300 CONTENTS
Here is the call graph for this function:
fqBdot
amici::Model::computeX_pos
amici::Model::fdwdp
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.12.3.17 fdxdotdp() [1/3]
void fdxdotdp (
const realtype t,
const N_Vector x,
const N_Vector dx )
Sensitivity of dx/dt wrt model parameters p
Parameters
ttimepoint
xVector with the states
dx Vector with the derivative states
Returns
status flag indicating successful execution
Definition at line 133 of file model_dae.cpp.
Generated by Doxygen
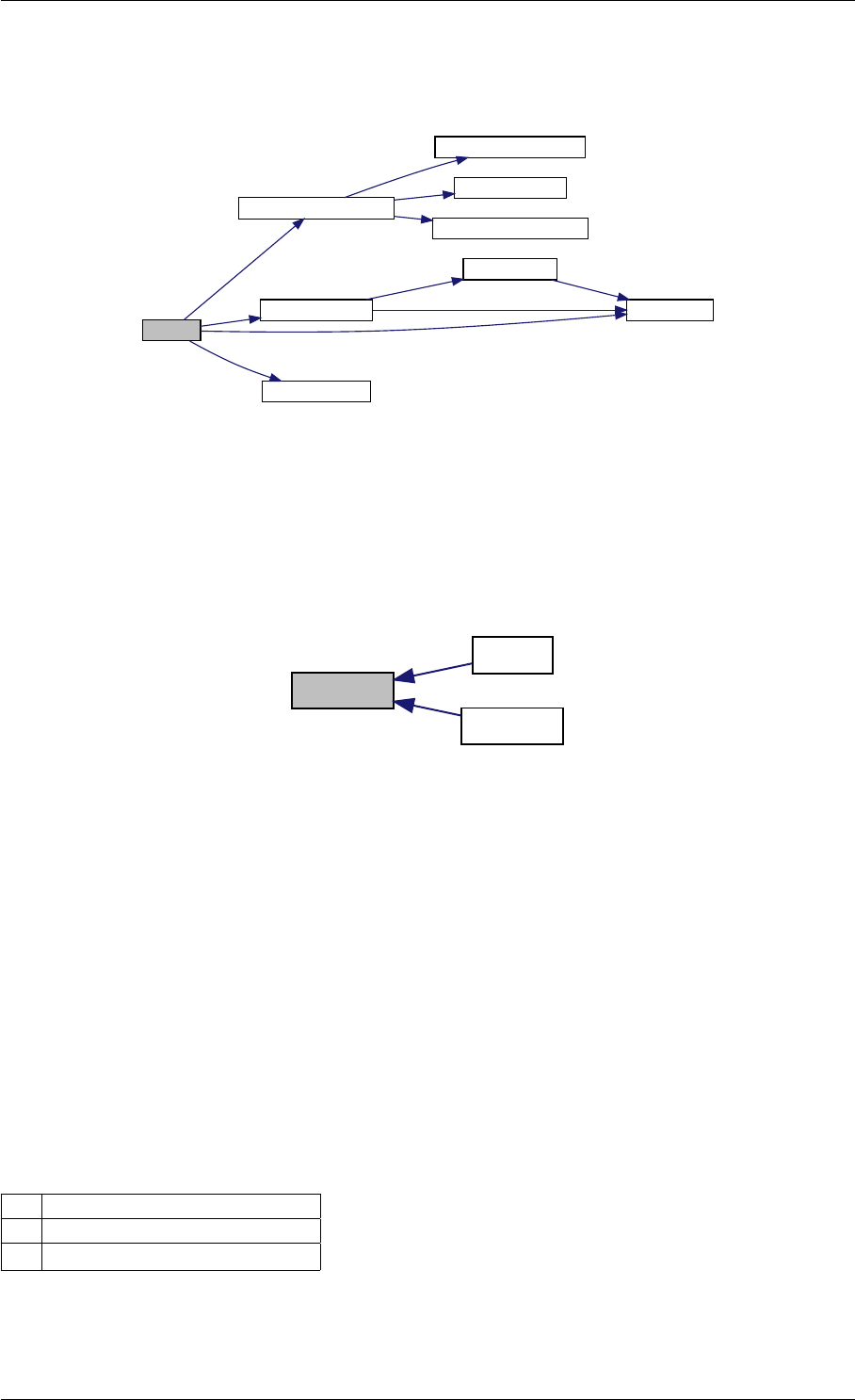
10.12 Model_DAE Class Reference 301
Here is the call graph for this function:
fdxdotdp
amici::Model::computeX_pos
amici::Model::fdwdp amici::Model::t
amici::Model::nplist
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
Here is the caller graph for this function:
fdxdotdp
fsxdot
fdxdotdp
10.12.3.18 fdxdotdp() [2/3]
virtual void fdxdotdp (
realtype t,
AmiVector ∗x,
AmiVector ∗dx ) [override], [virtual]
parameter derivative of residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
Generated by Doxygen
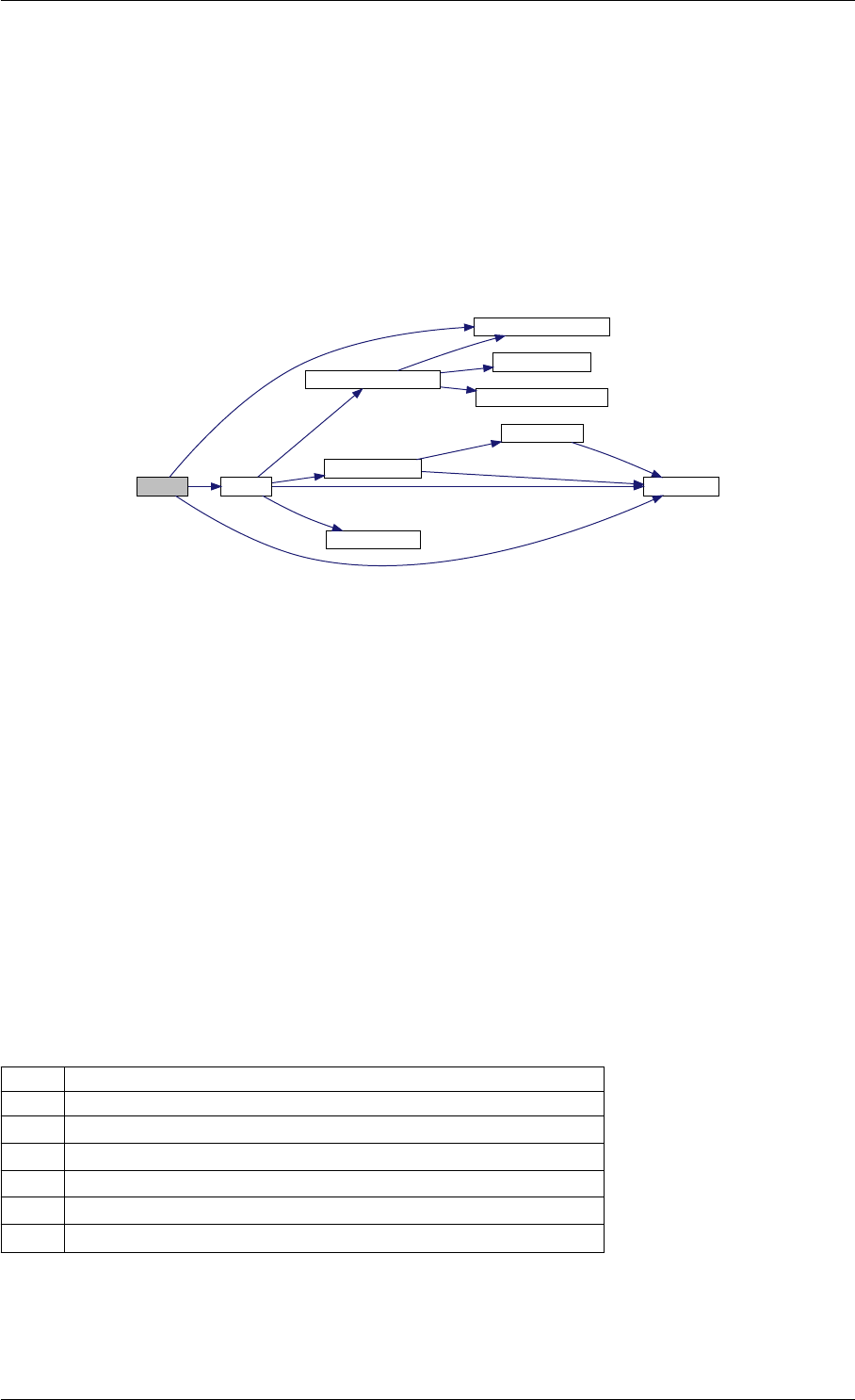
302 CONTENTS
Returns
flag indicating successful evaluation
Implements Model.
Definition at line 95 of file model_dae.h.
Here is the call graph for this function:
fdxdotdp fdxdotdp
amici::AmiVector::getNVector
amici::Model::t
amici::Model::computeX_pos
amici::Model::fdwdp
amici::Model::nplist
amici::AmiVector::getLength
amici::AmiVector::at
amici::Model::fw
10.12.3.19 fsxdot() [1/3]
void fsxdot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
int ip,
AmiVector ∗sx,
AmiVector ∗sdx,
AmiVector ∗sxdot ) [override], [virtual]
Sensitivity Residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
ip parameter index
sx sensitivity state
sdx time derivative of sensitivity state (DAE only)
sxdot array to which values of the sensitivity residual function will be written
Implements Model.
Definition at line 250 of file model_dae.cpp.
Generated by Doxygen
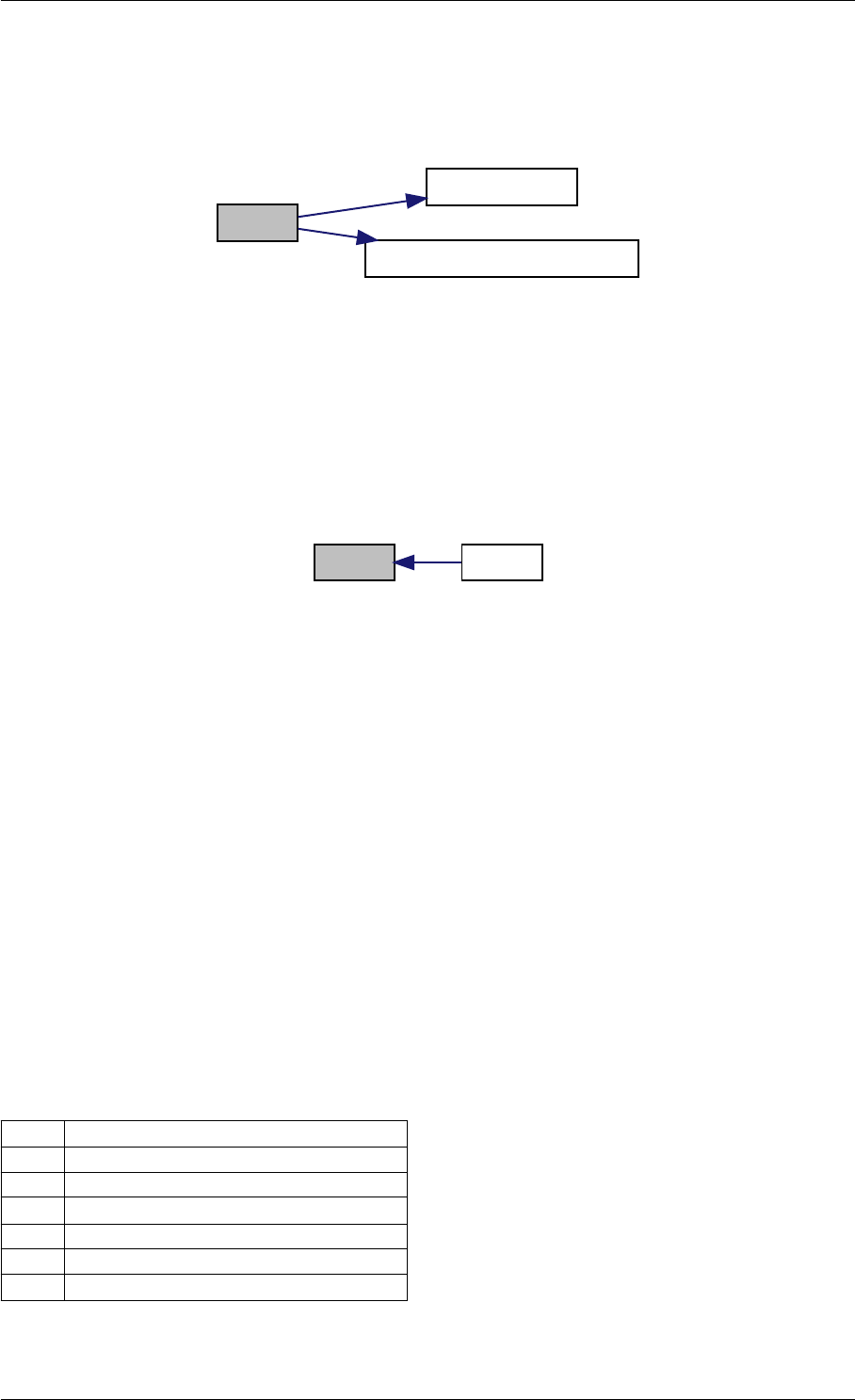
10.12 Model_DAE Class Reference 303
Here is the call graph for this function:
fsxdot
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fsxdot fsxdot
10.12.3.20 fsxdot() [2/3]
void fsxdot (
realtype t,
N_Vector x,
N_Vector dx,
int ip,
N_Vector sx,
N_Vector sdx,
N_Vector sxdot )
Right hand side of differential equation for state sensitivities sx
Parameters
ttimepoint
xVector with the states
dx Vector with the derivative states
ip parameter index
sx Vector with the state sensitivities
sdx Vector with the derivative state sensitivities
sxdot Vector with the sensitivity right hand side
Definition at line 266 of file model_dae.cpp.
Generated by Doxygen
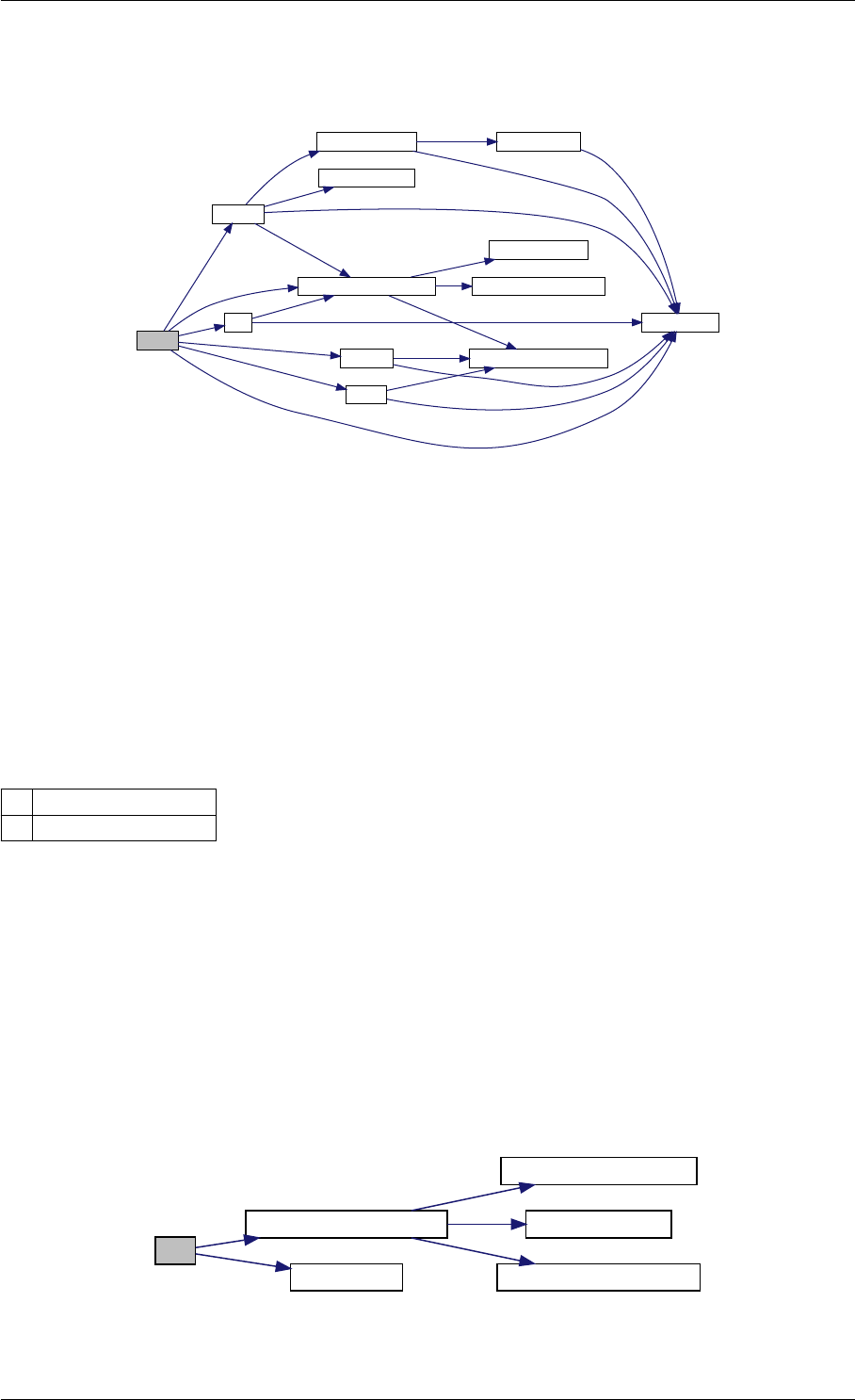
304 CONTENTS
Here is the call graph for this function:
fsxdot
amici::Model::computeX_pos
fM amici::Model::t
fdxdotdp
fJSparse
fsxdot
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fdwdp
amici::Model::nplist
amici::Model::fw
10.12.3.21 fM() [1/2]
void fM (
realtype t,
const N_Vector x)
Parameters
ttimepoint
xVector with the states
Definition at line 147 of file model_dae.cpp.
Here is the call graph for this function:
fM
amici::Model::computeX_pos
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
Generated by Doxygen

10.12 Model_DAE Class Reference 305
Here is the caller graph for this function:
fM fsxdot
10.12.3.22 getSolver()
std::unique_ptr<Solver >getSolver ( ) [override], [virtual]
Retrieves the solver object
Returns
The Solver instance
Implements Model.
Definition at line 153 of file model_dae.cpp.
10.12.3.23 fJ() [3/3]
virtual void fJ (
realtype ∗J,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗dx,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [pure virtual]
model specific implementation for fJ
Parameters
JMatrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
dx Vector with the derivative states
wvector with helper variables
dwdx derivative of w wrt x
Generated by Doxygen
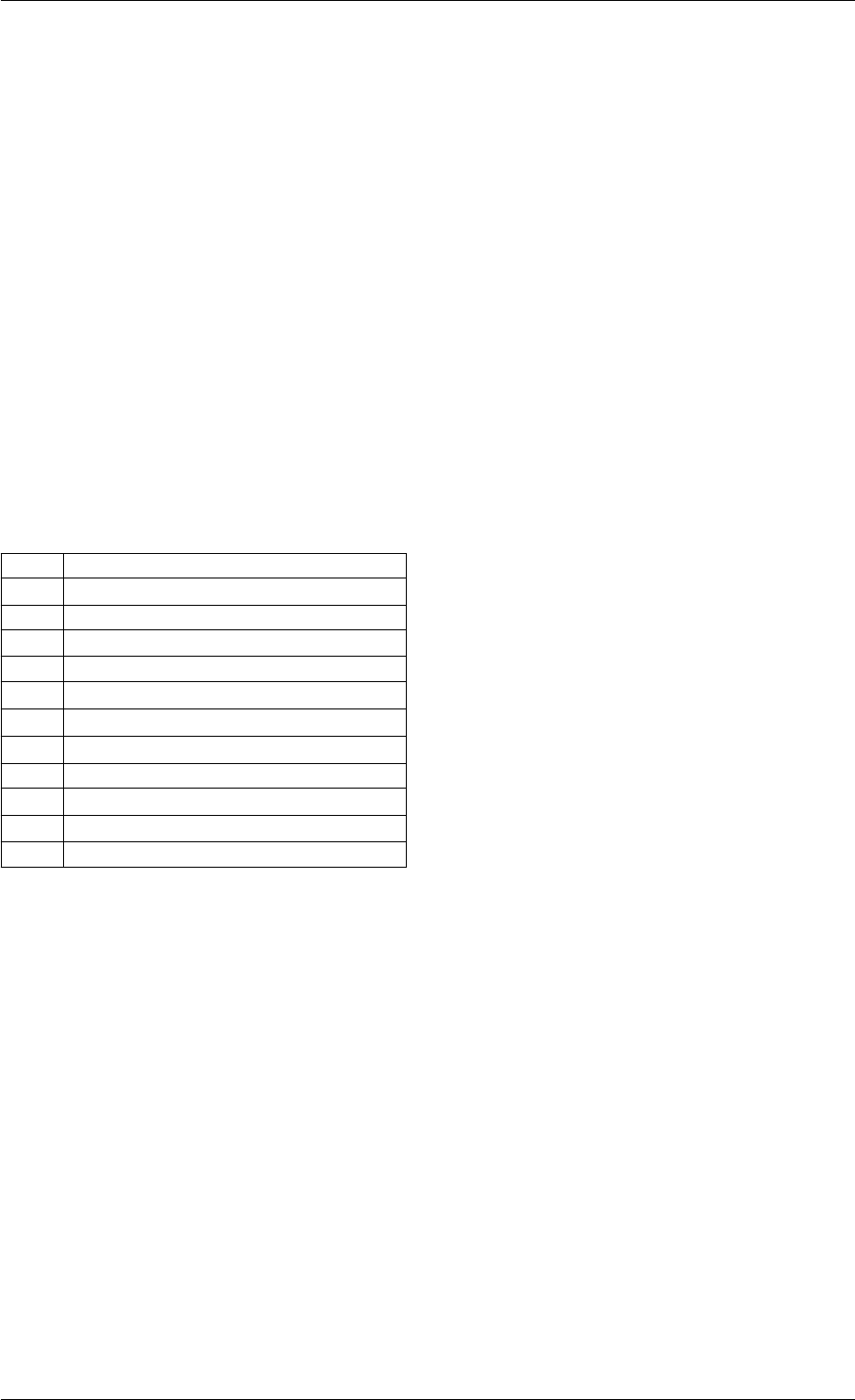
306 CONTENTS
10.12.3.24 fJB() [2/2]
virtual void fJB (
realtype ∗JB,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗xB,
const realtype ∗dx,
const realtype ∗dxB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJB
Parameters
JB Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
xB Vector with the adjoint states
dx Vector with the derivative states
dxB Vector with the adjoint derivative states
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 139 of file model_dae.h.
10.12.3.25 fJSparse() [3/3]
virtual void fJSparse (
SlsMat JSparse,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗dx,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [pure virtual]
model specific implementation for fJSparse
Generated by Doxygen
10.12 Model_DAE Class Reference 307
Parameters
JSparse Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
dx Vector with the derivative states
wvector with helper variables
dwdx derivative of w wrt x
10.12.3.26 fJSparseB() [2/2]
virtual void fJSparseB (
SlsMat JSparseB,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗xB,
const realtype ∗dx,
const realtype ∗dxB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJSparseB
Parameters
JSparseB Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
xB Vector with the adjoint states
dx Vector with the derivative states
dxB Vector with the adjoint derivative states
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 174 of file model_dae.h.
Generated by Doxygen
308 CONTENTS
10.12.3.27 fJDiag() [2/2]
virtual void fJDiag (
realtype ∗JDiag,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗dx,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJDiag
Parameters
JDiag array to which the Jacobian diagonal will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
dx Vector with the derivative states
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 192 of file model_dae.h.
10.12.3.28 fJv() [3/3]
virtual void fJv (
realtype ∗Jv,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗dx,
const realtype ∗v,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJv
Parameters
Jv Matrix vector product of J with a vector v
ttimepoint
Generated by Doxygen
10.12 Model_DAE Class Reference 309
Parameters
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
dx Vector with the derivative states
vVector with which the Jacobian is multiplied
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 210 of file model_dae.h.
10.12.3.29 fJvB() [2/2]
virtual void fJvB (
realtype ∗JvB,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype cj,
const realtype ∗xB,
const realtype ∗dx,
const realtype ∗dxB,
const realtype ∗vB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJvB
Parameters
JvB Matrix vector product of JB with a vector v
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
cj scaling factor, inverse of the step size
xB Vector with the adjoint states
dx Vector with the derivative states
dxB Vector with the adjoint derivative states
vB Vector with which the Jacobian is multiplied
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 231 of file model_dae.h.
Generated by Doxygen
310 CONTENTS
10.12.3.30 froot() [3/3]
virtual void froot (
realtype ∗root,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype ∗dx ) [protected], [virtual]
model specific implementation for froot
Parameters
root values of the trigger function
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
dx Vector with the derivative states
Definition at line 246 of file model_dae.h.
10.12.3.31 fxdot() [3/3]
virtual void fxdot (
realtype ∗xdot,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype ∗dx,
const realtype ∗w) [protected], [pure virtual]
model specific implementation for fxdot
Parameters
xdot residual function
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
dx Vector with the derivative states
Generated by Doxygen
10.12 Model_DAE Class Reference 311
10.12.3.32 fxBdot() [2/2]
virtual void fxBdot (
realtype ∗xBdot,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗dx,
const realtype ∗dxB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fxBdot
Parameters
xBdot adjoint residual function
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
dx Vector with the derivative states
dxB Vector with the adjoint derivative states
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 277 of file model_dae.h.
10.12.3.33 fqBdot() [2/2]
virtual void fqBdot (
realtype ∗qBdot,
const int ip,
const realtype t,
const realtype ∗x,
const double ∗p,
const double ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗dx,
const realtype ∗dxB,
const realtype ∗w,
const realtype ∗dwdp ) [protected], [virtual]
model specific implementation for fqBdot
Generated by Doxygen

312 CONTENTS
Parameters
qBdot adjoint quadrature equation
ip sensitivity index
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
dx Vector with the derivative states
dxB Vector with the adjoint derivative states
wvector with helper variables
dwdp derivative of w wrt p
Definition at line 297 of file model_dae.h.
10.12.3.34 fdxdotdp() [3/3]
virtual void fdxdotdp (
realtype ∗dxdotdp,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip,
const realtype ∗dx,
const realtype ∗w,
const realtype ∗dwdp ) [protected], [virtual]
model specific implementation of fdxdotdp
Parameters
dxdotdp partial derivative xdot wrt p
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
ip parameter index
dx Vector with the derivative states
wvector with helper variables
dwdp derivative of w wrt p
Definition at line 315 of file model_dae.h.
Generated by Doxygen
10.12 Model_DAE Class Reference 313
10.12.3.35 fsxdot() [3/3]
virtual void fsxdot (
realtype ∗sxdot,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip,
const realtype ∗dx,
const realtype ∗sx,
const realtype ∗sdx,
const realtype ∗w,
const realtype ∗dwdx,
const realtype ∗M,
const realtype ∗J,
const realtype ∗dxdotdp ) [protected], [virtual]
model specific implementation of fsxdot
Parameters
sxdot sensitivity rhs
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
ip parameter index
dx Vector with the derivative states
sx Vector with the state sensitivities
sdx Vector with the derivative state sensitivities
wvector with helper variables
dwdx derivative of w wrt x
Mmass matrix
Jjacobian
dxdotdp parameter derivative of residual function
Definition at line 337 of file model_dae.h.
10.12.3.36 fM() [2/2]
virtual void fM (
realtype ∗M,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k) [protected], [virtual]
model specific implementation of fM
Generated by Doxygen
314 CONTENTS
Parameters
Mmass matrix
ttimepoint
xVector with the states
pparameter vector
kconstants vector
Definition at line 351 of file model_dae.h.
10.13 Model_ODE Class Reference
The Model class represents an AMICI ODE model. The model does not contain any data, but represents the state
of the model at a specific time t. The states must not always be in sync, but may be updated asynchroneously.
#include <model_ode.h>
Inheritance diagram for Model_ODE:
Model_ODE
Model
Public Member Functions
•Model_ODE ()
•Model_ODE (const int nx, const int nxtrue, const int ny, const int nytrue, const int nz, const int nztrue, const
int ne, const int nJ, const int nw, const int ndwdx, const int ndwdp, const int nnz, const int ubw, const int lbw,
const SecondOrderMode o2mode, const std::vector<realtype >p, const std::vector<realtype >k, const
std::vector<int >plist, const std::vector<realtype >idlist, const std::vector<int >z2event)
•Model_ODE (Model_ODE const &other)
Copy constructor.
• virtual void fJ (realtype t,realtype cj, AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, DlsMat J) override
• void fJ (realtype t, N_Vector x, N_Vector xdot, DlsMat J)
• void fJB (realtype t, N_Vector x, N_Vector xB, N_Vector xBdot, DlsMat JB)
• virtual void fJSparse (realtype t,realtype cj, AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, SlsMat J) override
• void fJSparse (realtype t, N_Vector x, SlsMat J)
• void fJSparseB (realtype t, N_Vector x, N_Vector xB, N_Vector xBdot, SlsMat JB)
• void fJDiag (realtype t, N_Vector JDiag, N_Vector x)
• virtual void fJDiag (realtype t,AmiVector ∗Jdiag, realtype cj, AmiVector ∗x, AmiVector ∗dx) override
Generated by Doxygen

10.13 Model_ODE Class Reference 315
• virtual void fJv (realtype t,AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot, AmiVector ∗v, AmiVector ∗nJv,
realtype cj) override
• void fJv (N_Vector v, N_Vector Jv, realtype t, N_Vector x)
• void fJvB (N_Vector vB, N_Vector JvB, realtype t, N_Vector x, N_Vector xB)
• virtual void froot (realtype t,AmiVector ∗x, AmiVector ∗dx, realtype ∗root) override
• void froot (realtype t, N_Vector x, realtype ∗root)
• virtual void fxdot (realtype t,AmiVector ∗x, AmiVector ∗dx, AmiVector ∗xdot) override
• void fxdot (realtype t, N_Vector x, N_Vector xdot)
• void fxBdot (realtype t, N_Vector x, N_Vector xB, N_Vector xBdot)
• void fqBdot (realtype t, N_Vector x, N_Vector xB, N_Vector qBdot)
• void fdxdotdp (const realtype t, const N_Vector x)
• virtual void fdxdotdp (realtype t,AmiVector ∗x, AmiVector ∗dx) override
• void fsxdot (realtype t,AmiVector ∗x, AmiVector ∗dx, int ip, AmiVector ∗sx, AmiVector ∗sdx, AmiVector ∗sxdot)
override
• void fsxdot (realtype t, N_Vector x, int ip, N_Vector sx, N_Vector sxdot)
• virtual std::unique_ptr<Solver >getSolver () override
Protected Member Functions
• virtual void fJ (realtype ∗J, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h, const realtype ∗w, const realtype ∗dwdx)=0
• virtual void fJB (realtype ∗JB, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h, const realtype ∗xB, const realtype ∗w, const realtype ∗dwdx)
• virtual void fJSparse (SlsMat JSparse, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const realtype ∗w, const realtype ∗dwdx)=0
• virtual void fJSparseB (SlsMat JSparseB, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const realtype ∗xB, const realtype ∗w, const realtype ∗dwdx)
• virtual void fJDiag (realtype ∗JDiag, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗w, const realtype ∗dwdx)
• virtual void fJv (realtype ∗Jv, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h, const realtype ∗v, const realtype ∗w, const realtype ∗dwdx)
• virtual void fJvB (realtype ∗JvB, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h, const realtype ∗xB, const realtype ∗vB, const realtype ∗w, const realtype ∗dwdx)
• virtual void froot (realtype ∗root, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k, const
realtype ∗h)
• virtual void fxdot (realtype ∗xdot, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗w)=0
• virtual void fxBdot (realtype ∗xBdot, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const realtype ∗xB, const realtype ∗w, const realtype ∗dwdx)
• virtual void fqBdot (realtype ∗qBdot, const int ip, const realtype t, const realtype ∗x, const realtype ∗p, const
realtype ∗k, const realtype ∗h, const realtype ∗xB, const realtype ∗w, const realtype ∗dwdp)
• virtual void fdxdotdp (realtype ∗dxdotdp, const realtype t, const realtype ∗x, const realtype ∗p, const realtype
∗k, const realtype ∗h, const int ip, const realtype ∗w, const realtype ∗dwdp)
• virtual void fsxdot (realtype ∗sxdot, const realtype t, const realtype ∗x, const realtype ∗p, const realtype ∗k,
const realtype ∗h, const int ip, const realtype ∗sx, const realtype ∗w, const realtype ∗dwdx, const realtype ∗J,
const realtype ∗dxdotdp)
Additional Inherited Members
10.13.1 Detailed Description
Definition at line 23 of file model_ode.h.
Generated by Doxygen

316 CONTENTS
10.13.2 Constructor & Destructor Documentation
10.13.2.1 Model_ODE() [1/3]
Model_ODE ( )
default constructor
Definition at line 26 of file model_ode.h.
10.13.2.2 Model_ODE() [2/3]
Model_ODE (
const int nx,
const int nxtrue,
const int ny,
const int nytrue,
const int nz,
const int nztrue,
const int ne,
const int nJ,
const int nw,
const int ndwdx,
const int ndwdp,
const int nnz,
const int ubw,
const int lbw,
const SecondOrderMode o2mode,
const std::vector<realtype >p,
const std::vector<realtype >k,
const std::vector<int >plist,
const std::vector<realtype >idlist,
const std::vector<int >z2event )
constructor with model dimensions
Parameters
nx number of state variables
nxtrue number of state variables of the non-augmented model
ny number of observables
nytrue number of observables of the non-augmented model
nz number of event observables
nztrue number of event observables of the non-augmented model
ne number of events
nJ number of objective functions
nw number of repeating elements
ndwdx number of nonzero elements in the x derivative of the repeating elements
ndwdp number of nonzero elements in the p derivative of the repeating elements
nnz number of nonzero elements in Jacobian
ubw upper matrix bandwidth in the Jacobian
Generated by Doxygen
10.13 Model_ODE Class Reference 317
Parameters
lbw lower matrix bandwidth in the Jacobian
o2mode second order sensitivity mode
pparameters
kconstants
plist indexes wrt to which sensitivities are to be computed
idlist indexes indicating algebraic components (DAE only)
z2event mapping of event outputs to events
Definition at line 52 of file model_ode.h.
10.13.2.3 Model_ODE() [3/3]
Model_ODE (
Model_ODE const & other )
Parameters
other
Definition at line 66 of file model_ode.h.
10.13.3 Member Function Documentation
10.13.3.1 fJ() [1/3]
void fJ (
realtype t,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
DlsMat J) [override], [virtual]
Dense Jacobian function
Parameters
ttime
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
Jdense matrix to which values of the jacobian will be written
Generated by Doxygen
318 CONTENTS
Implements Model.
Definition at line 6 of file model_ode.cpp.
Here is the call graph for this function:
fJ
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fJ fJ
10.13.3.2 fJ() [2/3]
void fJ (
realtype t,
N_Vector x,
N_Vector xdot,
DlsMat J)
implementation of fJ at the N_Vector level, this function provides an interface to the model specific routines for the
solver implementation aswell as the AmiVector level implementation
Parameters
ttimepoint
xVector with the states
xdot Vector with the right hand side
JMatrix to which the Jacobian will be written
Definition at line 20 of file model_ode.cpp.
Generated by Doxygen
10.13 Model_ODE Class Reference 319
Here is the call graph for this function:
fJ
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
fJ
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.3 fJB() [1/2]
void fJB (
realtype t,
N_Vector x,
N_Vector xB,
N_Vector xBdot,
DlsMat JB )
implementation of fJB at the N_Vector level, this function provides an interface to the model specific routines for the
solver implementation
Parameters
ttimepoint
xVector with the states
xB Vector with the adjoint states
xBdot Vector with the adjoint right hand side
JB Matrix to which the Jacobian will be written
Definition at line 147 of file model_ode.cpp.
Generated by Doxygen
320 CONTENTS
Here is the call graph for this function:
fJB
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.4 fJSparse() [1/3]
void fJSparse (
realtype t,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
SlsMat J) [override], [virtual]
Sparse Jacobian function
Parameters
ttime
cj scaling factor (inverse of timestep, DAE only)
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
Jsparse matrix to which values of the Jacobian will be written
Implements Model.
Definition at line 28 of file model_ode.cpp.
Generated by Doxygen
10.13 Model_ODE Class Reference 321
Here is the call graph for this function:
fJSparse
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fJSparse
fJSparse
fsxdot
10.13.3.5 fJSparse() [2/3]
void fJSparse (
realtype t,
N_Vector x,
SlsMat J)
implementation of fJSparse at the N_Vector level, this function provides an interface to the model specific routines
for the solver implementation aswell as the AmiVector level implementation
Parameters
ttimepoint
xVector with the states
JMatrix to which the Jacobian will be written
Definition at line 40 of file model_ode.cpp.
Generated by Doxygen
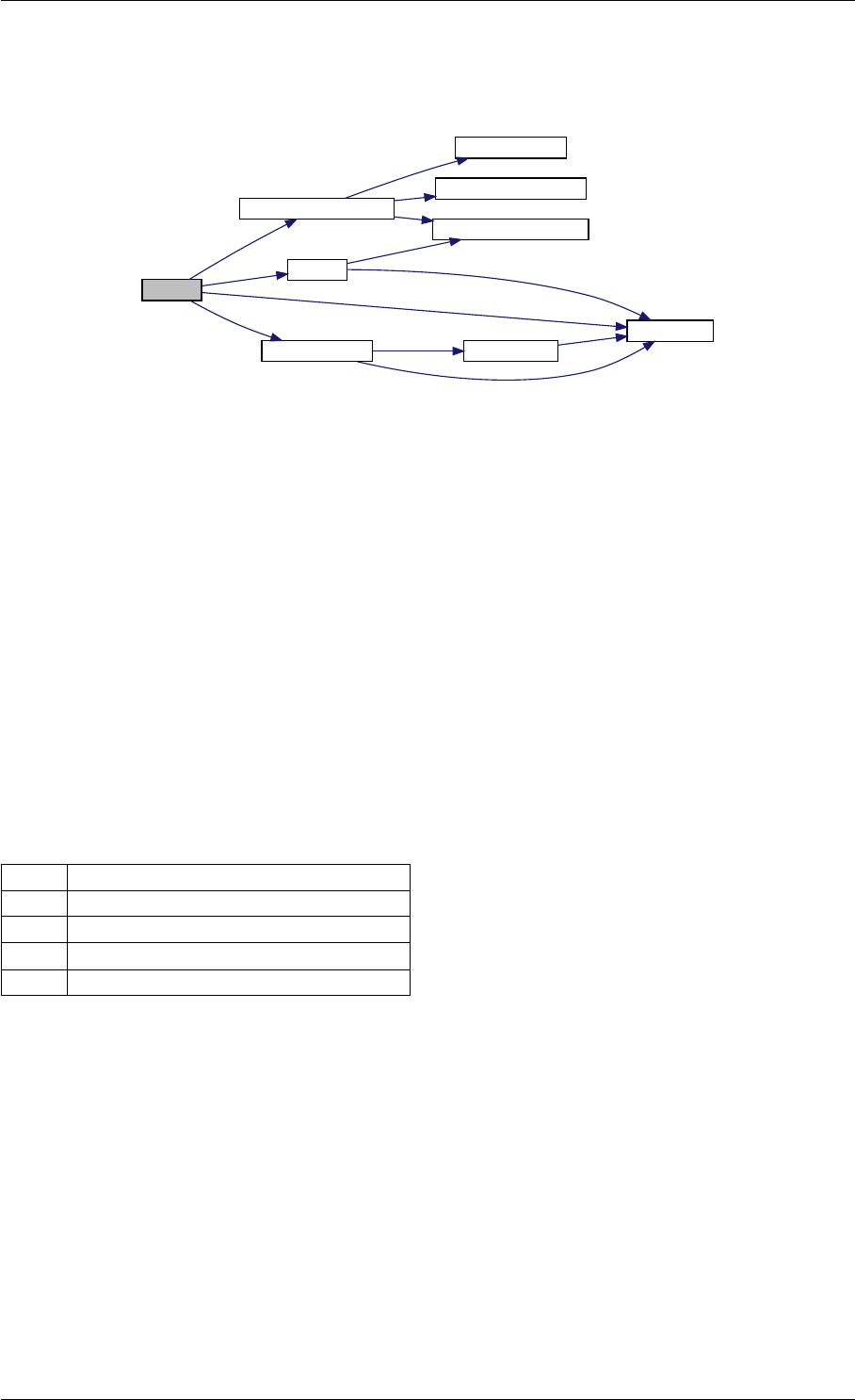
322 CONTENTS
Here is the call graph for this function:
fJSparse
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
fJSparse
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.6 fJSparseB() [1/2]
void fJSparseB (
realtype t,
N_Vector x,
N_Vector xB,
N_Vector xBdot,
SlsMat JB )
implementation of fJSparseB at the N_Vector level, this function provides an interface to the model specific routines
for the solver implementation
Parameters
ttimepoint
xVector with the states
xB Vector with the adjoint states
xBdot Vector with the adjoint right hand side
JB Matrix to which the Jacobian will be written
Definition at line 164 of file model_ode.cpp.
Generated by Doxygen
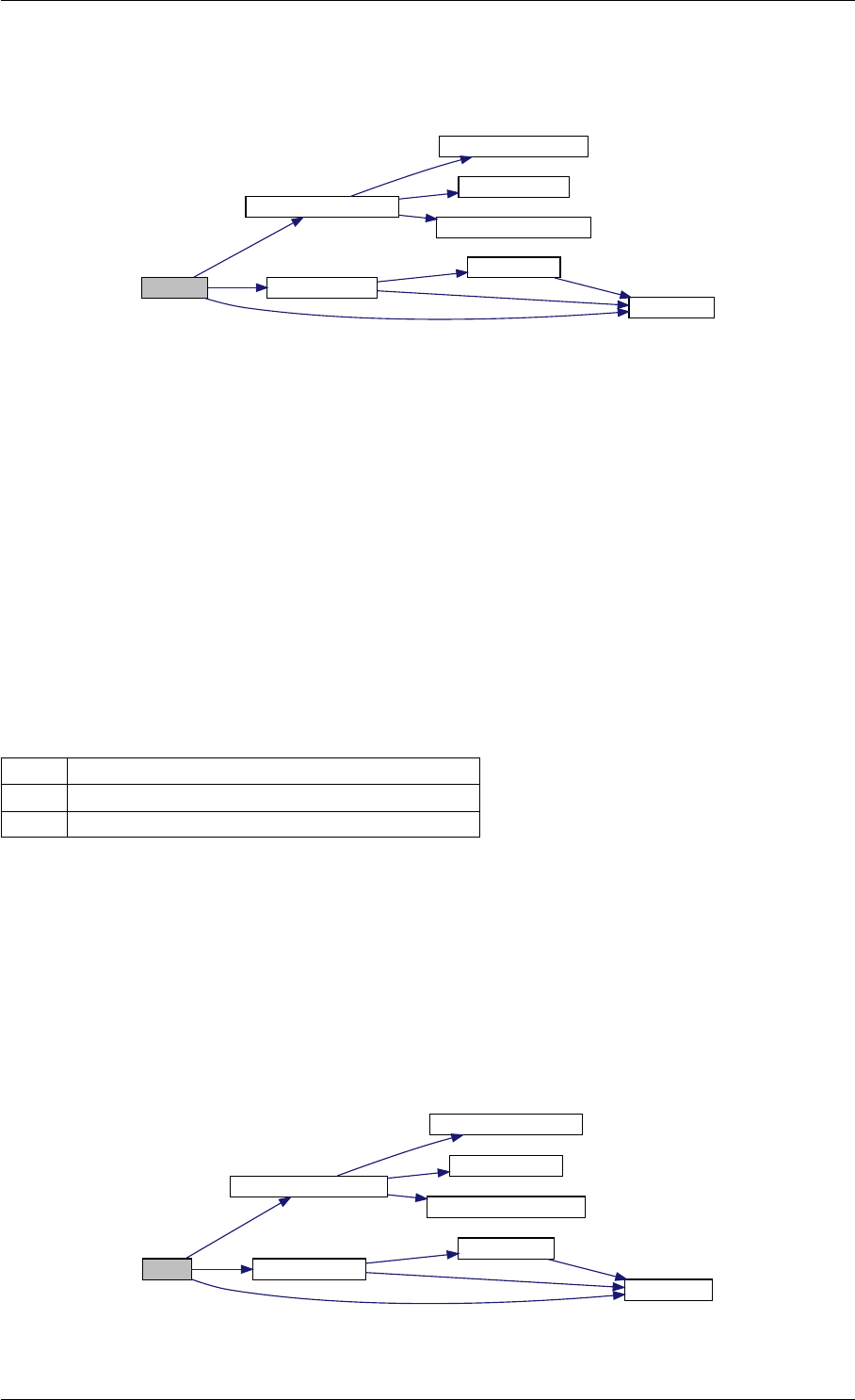
10.13 Model_ODE Class Reference 323
Here is the call graph for this function:
fJSparseB
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.7 fJDiag() [1/3]
void fJDiag (
realtype t,
N_Vector JDiag,
N_Vector x)
implementation of fJDiag at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation
Parameters
ttimepoint
JDiag Vector to which the Jacobian diagonal will be written
xVector with the states
Definition at line 178 of file model_ode.cpp.
Here is the call graph for this function:
fJDiag
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
Generated by Doxygen

324 CONTENTS
Here is the caller graph for this function:
fJDiag fJDiag
10.13.3.8 fJDiag() [2/3]
void fJDiag (
realtype t,
AmiVector ∗JDiag,
realtype cj,
AmiVector ∗x,
AmiVector ∗dx ) [override], [virtual]
diagonalized Jacobian (for preconditioning)
Parameters
ttimepoint
JDiag Vector to which the Jacobian diagonal will be written
cj scaling factor, inverse of the step size
xVector with the states
dx Vector with the derivative states
Returns
status flag indicating successful execution
Implements Model.
Definition at line 114 of file model_ode.cpp.
Generated by Doxygen
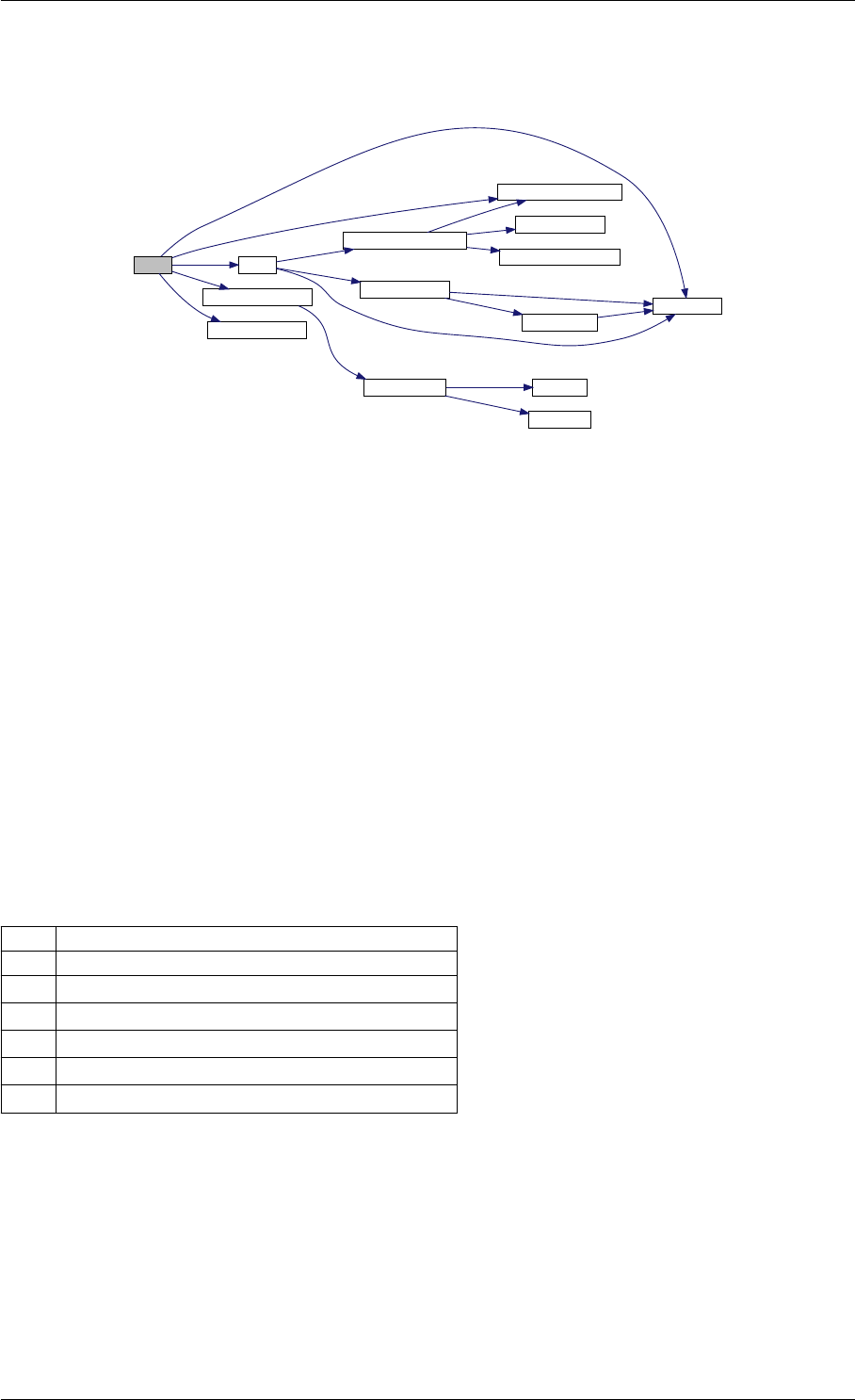
10.13 Model_ODE Class Reference 325
Here is the call graph for this function:
fJDiag fJDiag
amici::AmiVector::getNVector
amici::Model::t
amici::Model::checkFinite
amici::AmiVector::data
amici::Model::computeX_pos
amici::Model::fdwdx
amici::AmiVector::getLength
amici::AmiVector::at
amici::Model::fw
amici::checkFinite
amici::isNaN
amici::isInf
10.13.3.9 fJv() [1/3]
void fJv (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot,
AmiVector ∗v,
AmiVector ∗nJv,
realtype cj ) [override], [virtual]
Jacobian multiply function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
xdot values of residual function (unused)
vmultiplication vector (unused)
nJv array to which result of multiplication will be written
cj scaling factor (inverse of timestep, DAE only)
Implements Model.
Definition at line 48 of file model_ode.cpp.
Generated by Doxygen
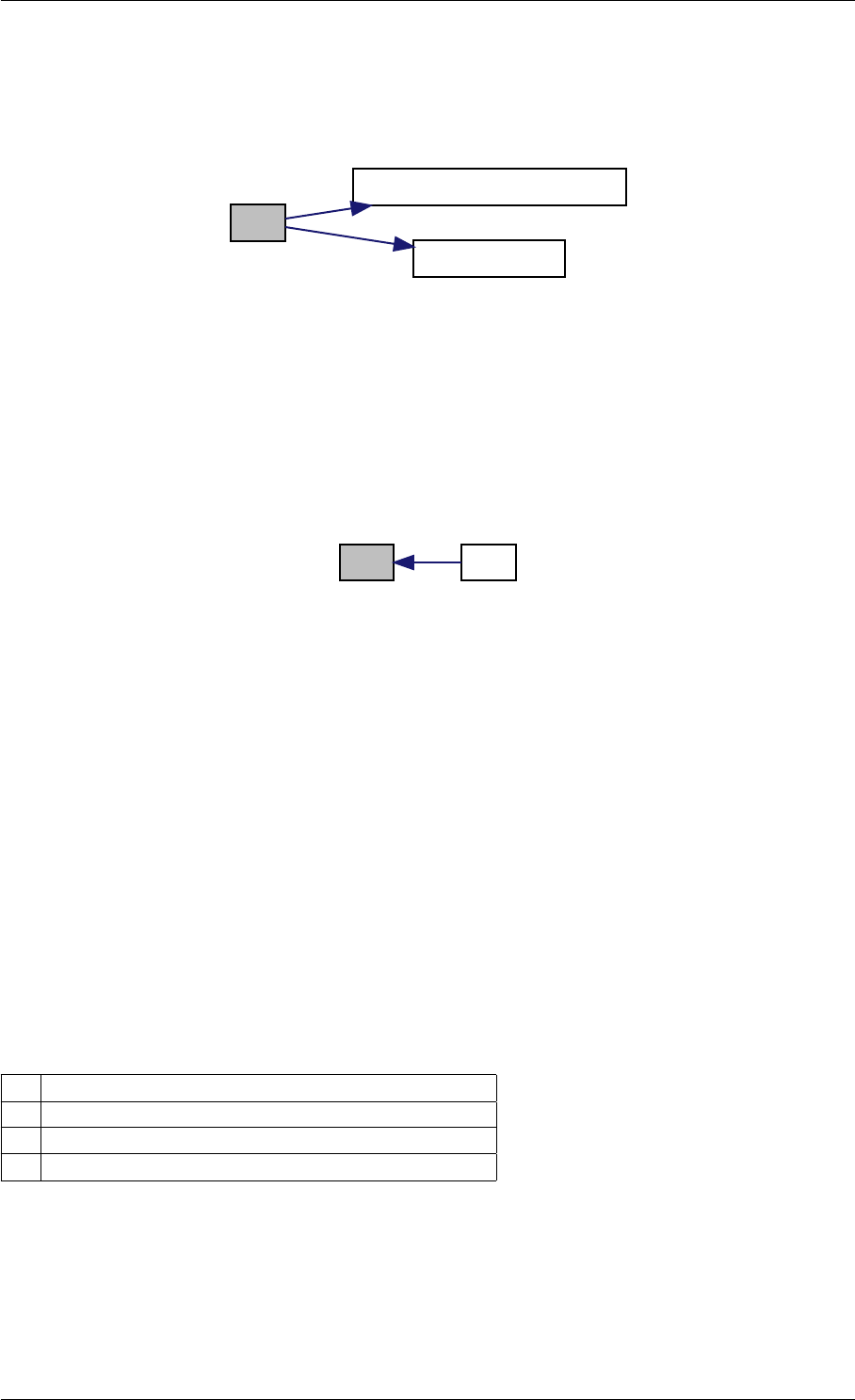
326 CONTENTS
Here is the call graph for this function:
fJv
amici::AmiVector::getNVector
amici::Model::t
Here is the caller graph for this function:
fJv fJv
10.13.3.10 fJv() [2/3]
void fJv (
N_Vector v,
N_Vector Jv,
realtype t,
N_Vector x)
implementation of fJv at the N_Vector level, this function provides an interface to the model specific routines for the
solver implementation aswell as the AmiVector level implementation
Parameters
ttimepoint
xVector with the states
vVector with which the Jacobian is multiplied
Jv Vector to which the Jacobian vector product will be written
Definition at line 62 of file model_ode.cpp.
Generated by Doxygen
10.13 Model_ODE Class Reference 327
Here is the call graph for this function:
fJv
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
fJv
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.11 fJvB() [1/2]
void fJvB (
N_Vector vB,
N_Vector JvB,
realtype t,
N_Vector x,
N_Vector xB )
implementation of fJvB at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation
Parameters
ttimepoint
xVector with the states
xB Vector with the adjoint states
vB Vector with which the Jacobian is multiplied
JvB Vector to which the Jacobian vector product will be written
Definition at line 195 of file model_ode.cpp.
Generated by Doxygen

328 CONTENTS
Here is the call graph for this function:
fJvB
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.12 froot() [1/3]
void froot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
realtype ∗root ) [override], [virtual]
Root function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
root array to which values of the root function will be written
Implements Model.
Definition at line 70 of file model_ode.cpp.
Here is the call graph for this function:
froot
amici::Model::t
amici::AmiVector::getNVector
Generated by Doxygen
10.13 Model_ODE Class Reference 329
Here is the caller graph for this function:
froot froot
10.13.3.13 froot() [2/3]
void froot (
realtype t,
N_Vector x,
realtype ∗root )
implementation of froot at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation aswell as the AmiVector level implementation
Parameters
ttimepoint
xVector with the states
root array with root function values
Definition at line 81 of file model_ode.cpp.
Here is the call graph for this function:
froot
amici::Model::computeX_pos
froot
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
Generated by Doxygen
330 CONTENTS
10.13.3.14 fxdot() [1/3]
void fxdot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
AmiVector ∗xdot ) [override], [virtual]
Residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
xdot array to which values of the residual function will be written
Implements Model.
Definition at line 87 of file model_ode.cpp.
Here is the call graph for this function:
fxdot
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fxdot fxdot
Generated by Doxygen
10.13 Model_ODE Class Reference 331
10.13.3.15 fxdot() [2/3]
void fxdot (
realtype t,
N_Vector x,
N_Vector xdot )
implementation of fxdot at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation aswell as the AmiVector level implementation
Parameters
ttimepoint
xVector with the states
xdot Vector with the right hand side
Definition at line 98 of file model_ode.cpp.
Here is the call graph for this function:
fxdot
amici::Model::computeX_pos
amici::Model::fw amici::Model::t
fxdot
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
10.13.3.16 fxBdot() [1/2]
void fxBdot (
realtype t,
N_Vector x,
N_Vector xB,
N_Vector xBdot )
implementation of fxBdot at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation
Parameters
ttimepoint
xVector with the states
xB Vector with the adjoint states
xBdot Vector with the adjoint right hand side
Generated by Doxygen
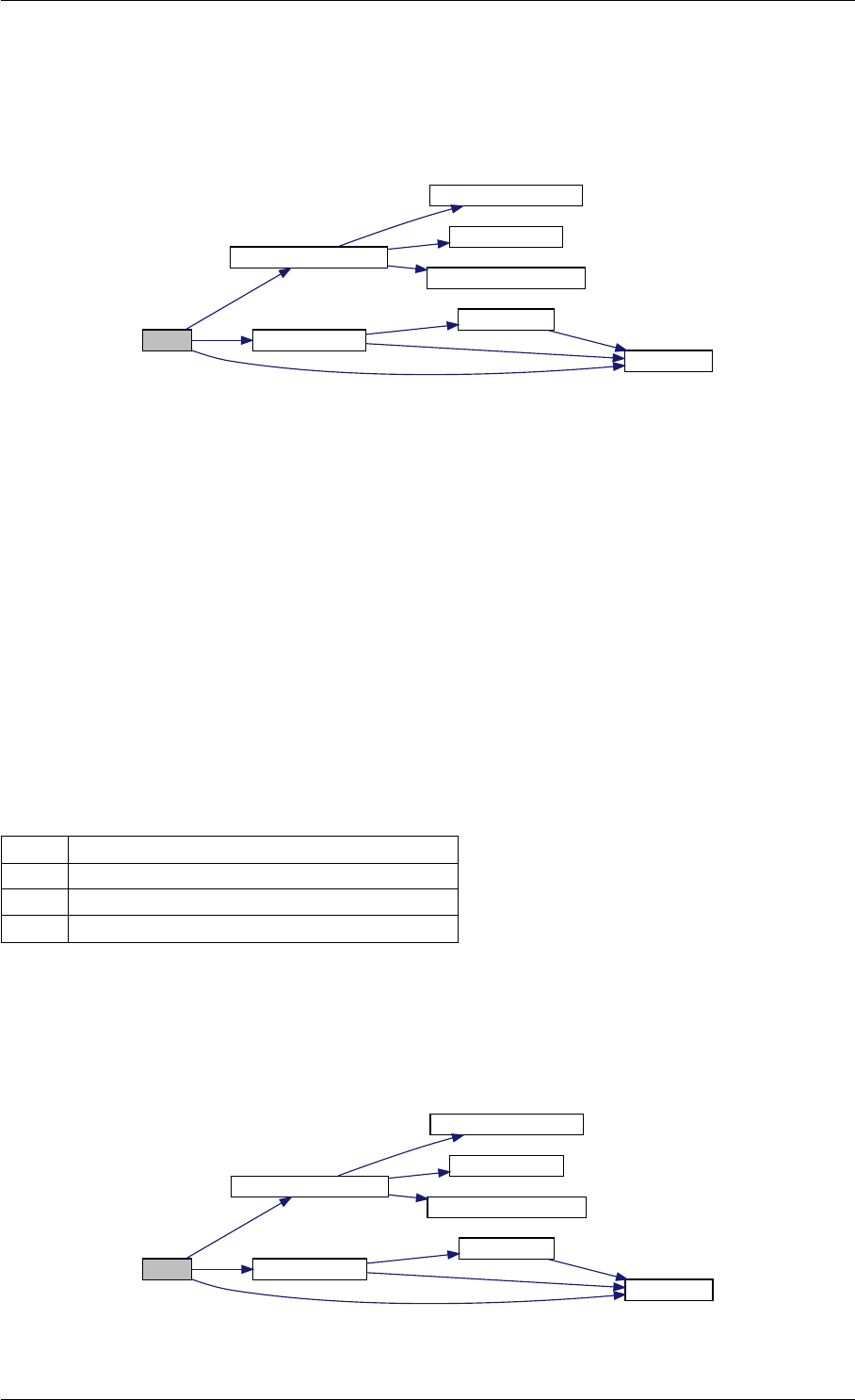
332 CONTENTS
Definition at line 210 of file model_ode.cpp.
Here is the call graph for this function:
fxBdot
amici::Model::computeX_pos
amici::Model::fdwdx
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
10.13.3.17 fqBdot() [1/2]
void fqBdot (
realtype t,
N_Vector x,
N_Vector xB,
N_Vector qBdot )
implementation of fqBdot at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation
Parameters
ttimepoint
xVector with the states
xB Vector with the adjoint states
qBdot Vector with the adjoint quadrature right hand side
Definition at line 225 of file model_ode.cpp.
Here is the call graph for this function:
fqBdot
amici::Model::computeX_pos
amici::Model::fdwdp
amici::Model::t
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
Generated by Doxygen
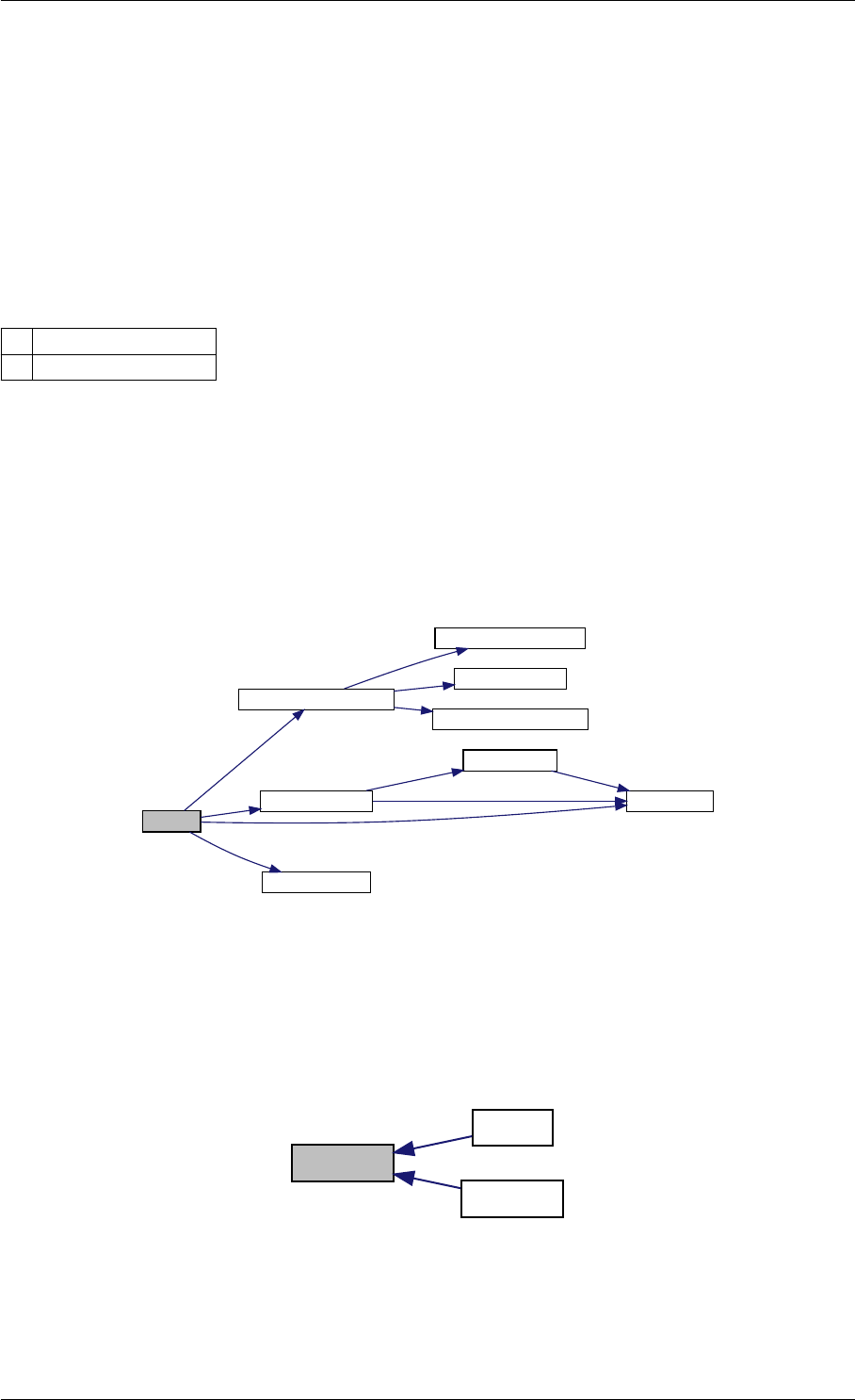
10.13 Model_ODE Class Reference 333
10.13.3.18 fdxdotdp() [1/3]
void fdxdotdp (
const realtype t,
const N_Vector x)
Sensitivity of dx/dt wrt model parameters p
Parameters
ttimepoint
xVector with the states
Returns
status flag indicating successful execution
Definition at line 126 of file model_ode.cpp.
Here is the call graph for this function:
fdxdotdp
amici::Model::computeX_pos
amici::Model::fdwdp amici::Model::t
amici::Model::nplist
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fw
Here is the caller graph for this function:
fdxdotdp
fsxdot
fdxdotdp
Generated by Doxygen
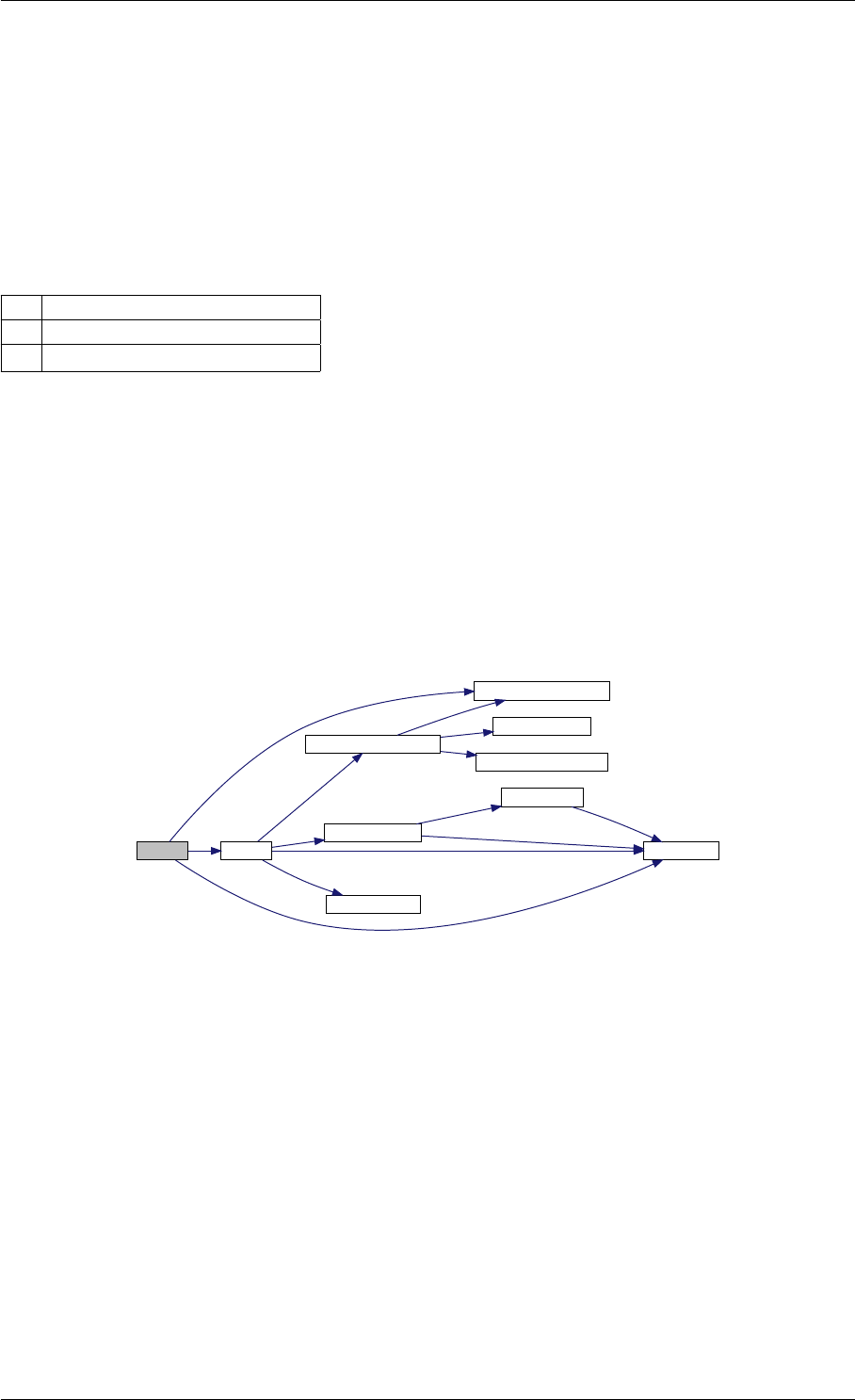
334 CONTENTS
10.13.3.19 fdxdotdp() [2/3]
virtual void fdxdotdp (
realtype t,
AmiVector ∗x,
AmiVector ∗dx ) [override], [virtual]
parameter derivative of residual function
Parameters
ttime
xstate
dx time derivative of state (DAE only)
Returns
flag indicating successful evaluation
Implements Model.
Definition at line 104 of file model_ode.h.
Here is the call graph for this function:
fdxdotdp fdxdotdp
amici::AmiVector::getNVector
amici::Model::t
amici::Model::computeX_pos
amici::Model::fdwdp
amici::Model::nplist
amici::AmiVector::getLength
amici::AmiVector::at
amici::Model::fw
10.13.3.20 fsxdot() [1/3]
void fsxdot (
realtype t,
AmiVector ∗x,
AmiVector ∗dx,
int ip,
AmiVector ∗sx,
AmiVector ∗sdx,
AmiVector ∗sxdot ) [override], [virtual]
Sensitivity Residual function
Generated by Doxygen
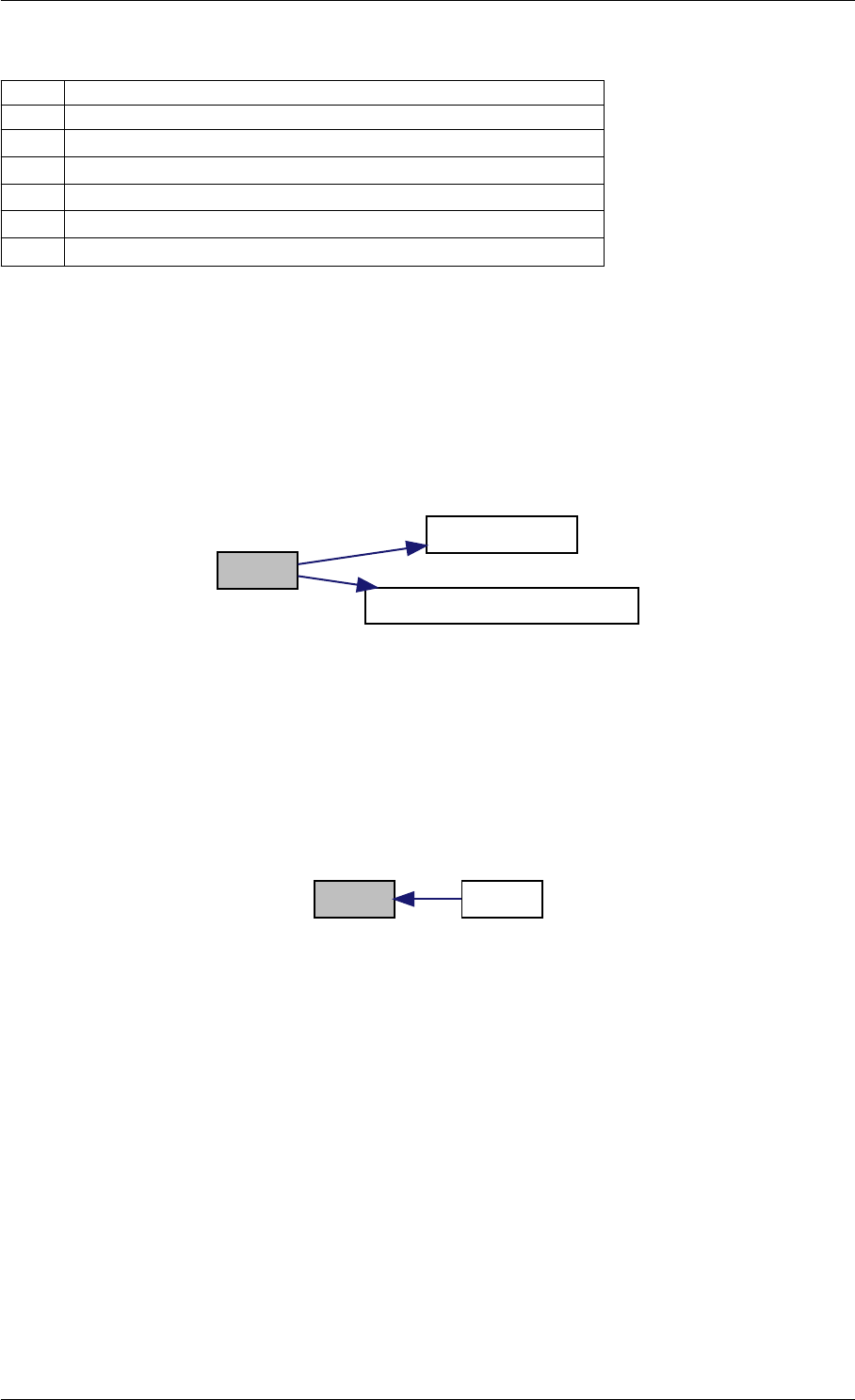
10.13 Model_ODE Class Reference 335
Parameters
ttime
xstate
dx time derivative of state (DAE only)
ip parameter index
sx sensitivity state
sdx time derivative of sensitivity state (DAE only)
sxdot array to which values of the sensitivity residual function will be written
Implements Model.
Definition at line 235 of file model_ode.cpp.
Here is the call graph for this function:
fsxdot
amici::Model::t
amici::AmiVector::getNVector
Here is the caller graph for this function:
fsxdot fsxdot
10.13.3.21 fsxdot() [2/3]
void fsxdot (
realtype t,
N_Vector x,
int ip,
N_Vector sx,
N_Vector sxdot )
implementation of fsxdot at the N_Vector level, this function provides an interface to the model specific routines for
the solver implementation
Generated by Doxygen
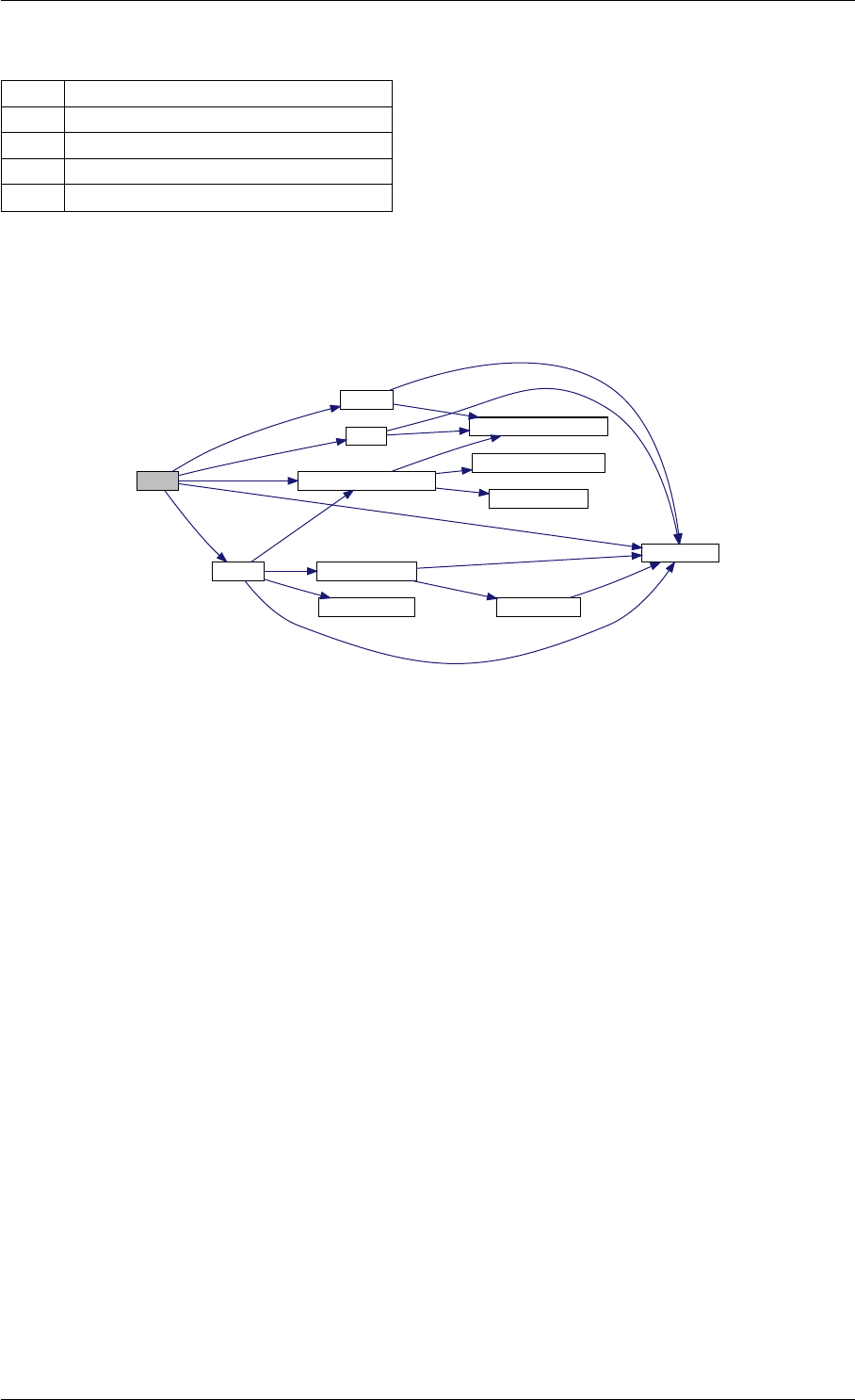
336 CONTENTS
Parameters
ttimepoint
xVector with the states
ip parameter index
sx Vector with the state sensitivities
sxdot Vector with the sensitivity right hand side
Definition at line 248 of file model_ode.cpp.
Here is the call graph for this function:
fsxdot amici::Model::computeX_pos
fdxdotdp
amici::Model::t
fJSparse
fsxdot
amici::AmiVector::getLength
amici::AmiVector::at
amici::AmiVector::getNVector
amici::Model::fdwdp
amici::Model::nplist amici::Model::fw
10.13.3.22 getSolver()
std::unique_ptr<Solver >getSolver ( ) [override], [virtual]
Retrieves the solver object
Returns
The Solver instance
Implements Model.
Definition at line 135 of file model_ode.cpp.
10.13.3.23 fJ() [3/3]
virtual void fJ (
realtype ∗J,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [pure virtual]
model specific implementation for fJ
Generated by Doxygen
10.13 Model_ODE Class Reference 337
Parameters
JMatrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
dwdx derivative of w wrt x
10.13.3.24 fJB() [2/2]
virtual void fJB (
realtype ∗JB,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJB
Parameters
JB Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 139 of file model_ode.h.
10.13.3.25 fJSparse() [3/3]
virtual void fJSparse (
SlsMat JSparse,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
Generated by Doxygen
338 CONTENTS
const realtype ∗h,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [pure virtual]
model specific implementation for fJSparse
Parameters
JSparse Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
dwdx derivative of w wrt x
10.13.3.26 fJSparseB() [2/2]
virtual void fJSparseB (
SlsMat JSparseB,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJSparseB
Parameters
JSparseB Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 167 of file model_ode.h.
10.13.3.27 fJDiag() [3/3]
virtual void fJDiag (
realtype ∗JDiag,
Generated by Doxygen
10.13 Model_ODE Class Reference 339
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJDiag
Parameters
JDiag Matrix to which the Jacobian will be written
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 182 of file model_ode.h.
10.13.3.28 fJv() [3/3]
virtual void fJv (
realtype ∗Jv,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗v,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJv
Parameters
Jv Matrix vector product of J with a vector v
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
vVector with which the Jacobian is multiplied
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 198 of file model_ode.h.
Generated by Doxygen
340 CONTENTS
10.13.3.29 fJvB() [2/2]
virtual void fJvB (
realtype ∗JvB,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗vB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fJvB
Parameters
JvB Matrix vector product of JB with a vector v
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
vB Vector with which the Jacobian is multiplied
wvector with helper variables
dwdx derivative of w wrt x
Definition at line 215 of file model_ode.h.
10.13.3.30 froot() [3/3]
virtual void froot (
realtype ∗root,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h) [protected], [virtual]
model specific implementation for froot
Parameters
root values of the trigger function
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
Generated by Doxygen

10.13 Model_ODE Class Reference 341
Definition at line 228 of file model_ode.h.
10.13.3.31 fxdot() [3/3]
virtual void fxdot (
realtype ∗xdot,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗w) [protected], [pure virtual]
model specific implementation for fxdot
Parameters
xdot residual function
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
wvector with helper variables
10.13.3.32 fxBdot() [2/2]
virtual void fxBdot (
realtype ∗xBdot,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗w,
const realtype ∗dwdx ) [protected], [virtual]
model specific implementation for fxBdot
Parameters
xBdot adjoint residual function
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
wvector with helper variables
dwdx derivative of w wrt x
Generated by Doxygen
342 CONTENTS
Definition at line 254 of file model_ode.h.
10.13.3.33 fqBdot() [2/2]
virtual void fqBdot (
realtype ∗qBdot,
const int ip,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const realtype ∗xB,
const realtype ∗w,
const realtype ∗dwdp ) [protected], [virtual]
model specific implementation for fqBdot
Parameters
qBdot adjoint quadrature equation
ip sensitivity index
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
xB Vector with the adjoint states
wvector with helper variables
dwdp derivative of w wrt p
Definition at line 271 of file model_ode.h.
10.13.3.34 fdxdotdp() [3/3]
virtual void fdxdotdp (
realtype ∗dxdotdp,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip,
const realtype ∗w,
const realtype ∗dwdp ) [protected], [virtual]
model specific implementation of fdxdotdp
Generated by Doxygen

10.13 Model_ODE Class Reference 343
Parameters
dxdotdp partial derivative xdot wrt p
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
ip parameter index
wvector with helper variables
dwdp derivative of w wrt p
Definition at line 287 of file model_ode.h.
10.13.3.35 fsxdot() [3/3]
virtual void fsxdot (
realtype ∗sxdot,
const realtype t,
const realtype ∗x,
const realtype ∗p,
const realtype ∗k,
const realtype ∗h,
const int ip,
const realtype ∗sx,
const realtype ∗w,
const realtype ∗dwdx,
const realtype ∗J,
const realtype ∗dxdotdp ) [protected], [virtual]
model specific implementation of fsxdot
Parameters
sxdot sensitivity rhs
ttimepoint
xVector with the states
pparameter vector
kconstants vector
hheavyside vector
ip parameter index
sx Vector with the state sensitivities
wvector with helper variables
dwdx derivative of w wrt x
Jjacobian
dxdotdp parameter derivative of residual function
Definition at line 306 of file model_ode.h.
Generated by Doxygen
344 CONTENTS
10.14 NewtonFailure Class Reference
newton failure exception this exception should be thrown when the steady state computation failed to converge for
this exception we can assume that we can recover from the exception and return a solution struct to the user
#include <exception.h>
Inheritance diagram for NewtonFailure:
NewtonFailure
AmiException
exception
Public Member Functions
•NewtonFailure (int code, const char ∗function)
Public Attributes
• int error_code
10.14.1 Detailed Description
Definition at line 201 of file exception.h.
10.14.2 Constructor & Destructor Documentation
10.14.2.1 NewtonFailure()
NewtonFailure (
int code,
const char ∗function )
constructor, simply calls AmiException constructor
Generated by Doxygen
10.15 NewtonSolver Class Reference 345
Parameters
function name of the function in which the error occured
code error code
Definition at line 209 of file exception.h.
10.14.3 Member Data Documentation
10.14.3.1 error_code
int error_code
error code returned by solver
Definition at line 204 of file exception.h.
10.15 NewtonSolver Class Reference
The NewtonSolver class sets up the linear solver for the Newton method.
#include <newton_solver.h>
Inheritance diagram for NewtonSolver:
NewtonSolver
NewtonSolverDense NewtonSolverIterative NewtonSolverSparse
Public Member Functions
•NewtonSolver (realtype ∗t,AmiVector ∗x,Model ∗model,ReturnData ∗rdata)
• void getStep (int ntry, int nnewt, AmiVector ∗delta)
• void computeNewtonSensis (AmiVectorArray ∗sx)
• virtual void prepareLinearSystem (int ntry, int nnewt)=0
• virtual void solveLinearSystem (AmiVector ∗rhs)=0
Static Public Member Functions
• static std::unique_ptr<NewtonSolver >getSolver (realtype ∗t,AmiVector ∗x,LinearSolver linsolType, Model
∗model,ReturnData ∗rdata, int maxlinsteps, int maxsteps, double atol, double rtol)
Generated by Doxygen
346 CONTENTS
Public Attributes
• int maxlinsteps = 0
• int maxsteps = 0
• double atol = 1e-16
• double rtol = 1e-8
Protected Attributes
•realtype ∗t
•Model ∗model
•ReturnData ∗rdata
•AmiVector xdot
•AmiVector ∗x
•AmiVector dx
10.15.1 Detailed Description
Definition at line 25 of file newton_solver.h.
10.15.2 Constructor & Destructor Documentation
10.15.2.1 NewtonSolver()
NewtonSolver (
realtype ∗t,
AmiVector ∗x,
Model ∗model,
ReturnData ∗rdata )
default constructor, initializes all members with the provided objects
Parameters
tpointer to time variable
xpointer to state variables
model pointer to the AMICI model object
rdata pointer to the return data object
Definition at line 18 of file newton_solver.cpp.
10.15.3 Member Function Documentation
Generated by Doxygen
10.15 NewtonSolver Class Reference 347
10.15.3.1 getSolver()
std::unique_ptr<NewtonSolver >getSolver (
realtype ∗t,
AmiVector ∗x,
LinearSolver linsolType,
Model ∗model,
ReturnData ∗rdata,
int maxlinsteps,
int maxsteps,
double atol,
double rtol ) [static]
Tries to determine the steady state of the ODE system by a Newton solver, uses forward intergration, if the Newton
solver fails, restarts Newton solver, if integration fails. Computes steady state sensitivities
Parameters
tpointer to time variable
xpointer to state variables
linsolType integer indicating which linear solver to use
model pointer to the AMICI model object
rdata pointer to the return data object
maxlinsteps maximum number of allowed linear steps per Newton step for steady state computation
maxsteps maximum number of allowed Newton steps for steady state computation
atol absolute tolerance
rtol relative tolerance
Returns
solver NewtonSolver according to the specified linsolType
Definition at line 36 of file newton_solver.cpp.
Here is the caller graph for this function:
getSolver amici::SteadystateProblem
::workSteadyStateProblem
10.15.3.2 getStep()
void getStep (
int ntry,
int nnewt,
AmiVector ∗delta )
Computes the solution of one Newton iteration
Generated by Doxygen
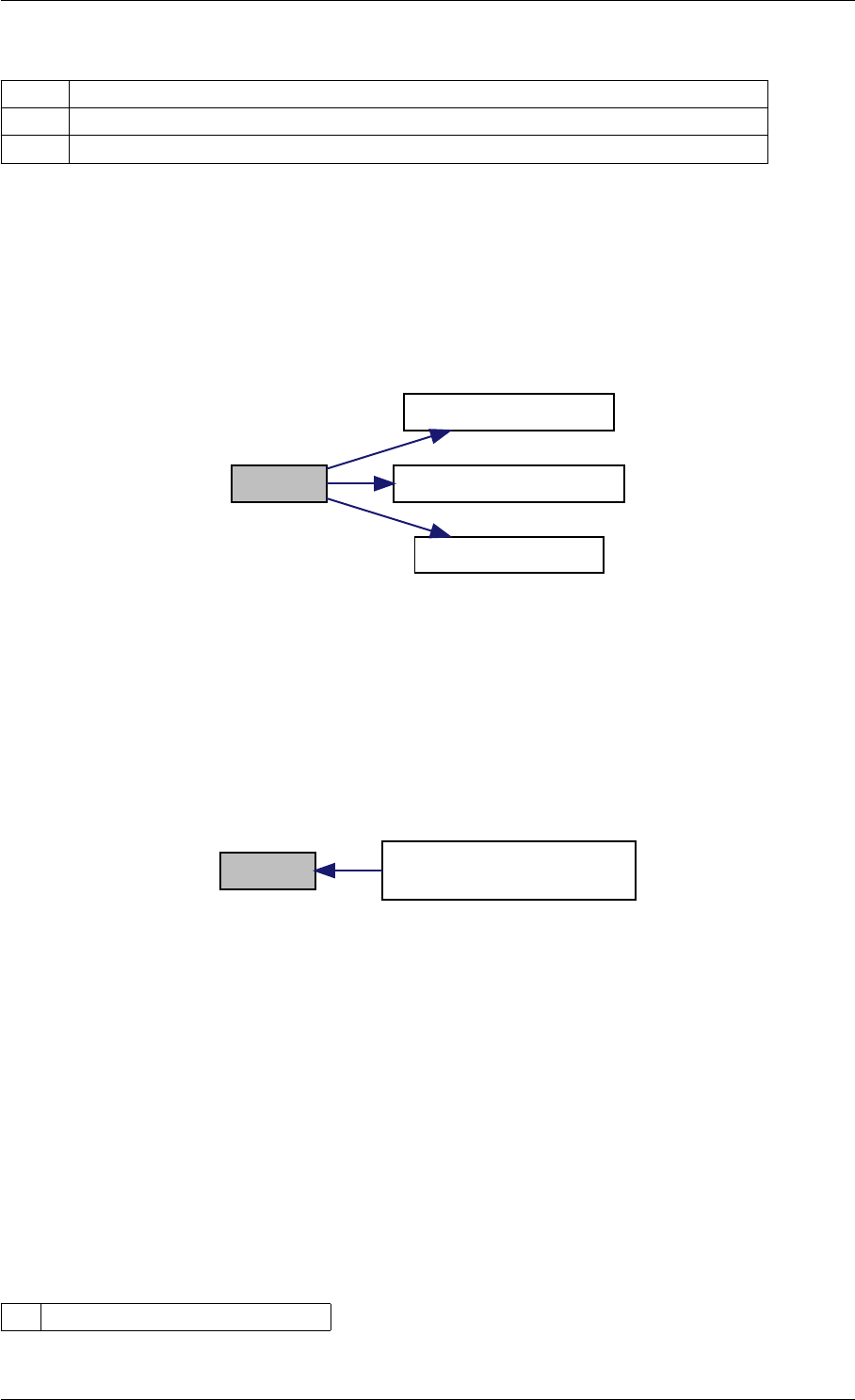
348 CONTENTS
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
delta containing the RHS of the linear system, will be overwritten by solution to the linear system
Definition at line 107 of file newton_solver.cpp.
Here is the call graph for this function:
getStep
prepareLinearSystem
amici::AmiVector::minus
solveLinearSystem
Here is the caller graph for this function:
getStep amici::SteadystateProblem
::applyNewtonsMethod
10.15.3.3 computeNewtonSensis()
void computeNewtonSensis (
AmiVectorArray ∗sx )
Computes steady state sensitivities
Parameters
sx pointer to state variable sensitivities
Generated by Doxygen
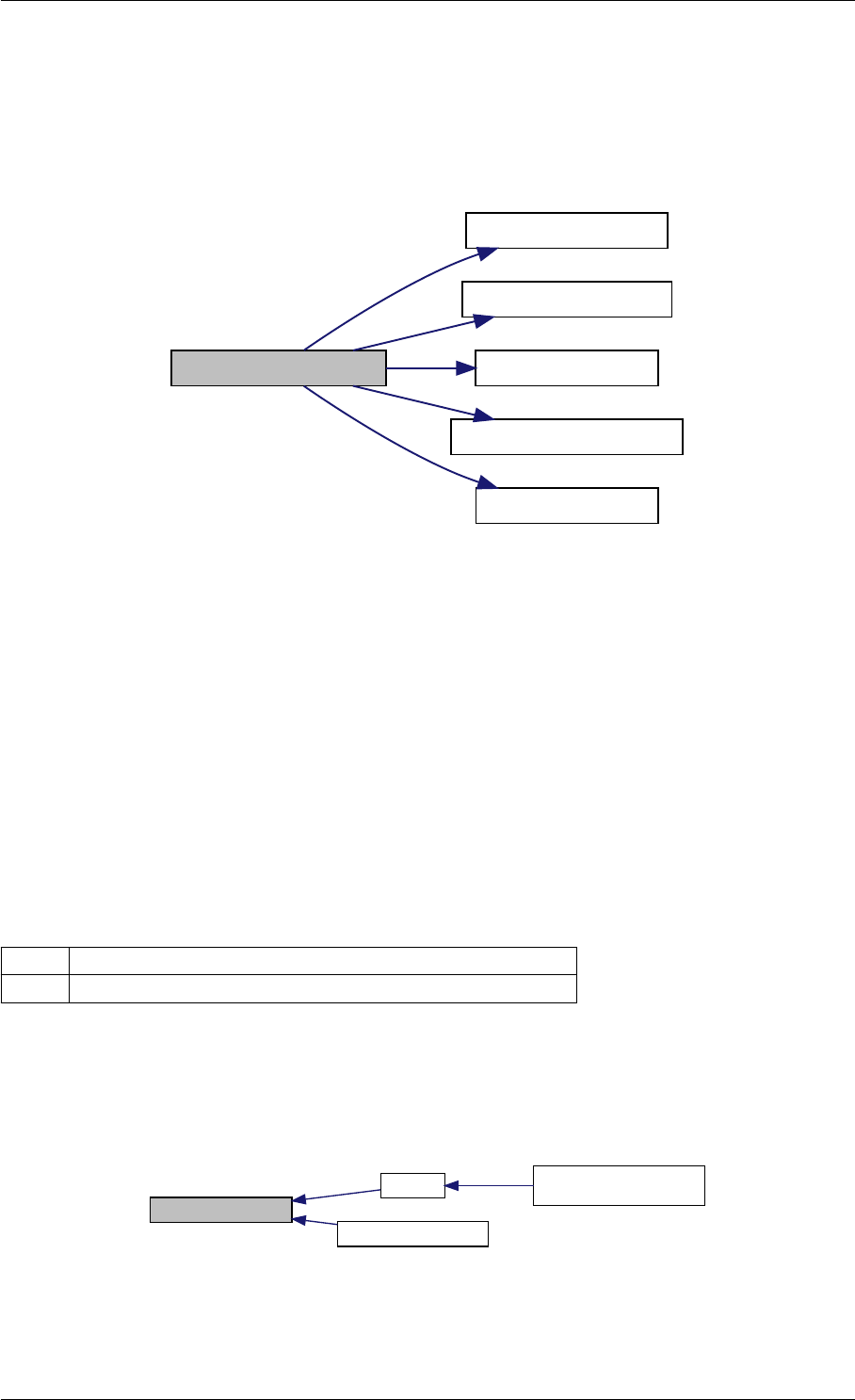
10.15 NewtonSolver Class Reference 349
Definition at line 127 of file newton_solver.cpp.
Here is the call graph for this function:
computeNewtonSensis
prepareLinearSystem
amici::Model::fdxdotdp
amici::Model::nplist
amici::AmiVectorArray::at
solveLinearSystem
10.15.3.4 prepareLinearSystem()
virtual void prepareLinearSystem (
int ntry,
int nnewt ) [pure virtual]
Writes the Jacobian for the Newton iteration and passes it to the linear solver
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Implemented in NewtonSolverIterative,NewtonSolverSparse, and NewtonSolverDense.
Here is the caller graph for this function:
prepareLinearSystem
getStep
computeNewtonSensis
amici::SteadystateProblem
::applyNewtonsMethod
Generated by Doxygen
350 CONTENTS
10.15.3.5 solveLinearSystem()
virtual void solveLinearSystem (
AmiVector ∗rhs ) [pure virtual]
Solves the linear system for the Newton step
Parameters
rhs containing the RHS of the linear system, will be overwritten by solution to the linear system
Implemented in NewtonSolverIterative,NewtonSolverSparse, and NewtonSolverDense.
Here is the caller graph for this function:
solveLinearSystem
getStep
computeNewtonSensis
amici::SteadystateProblem
::applyNewtonsMethod
10.15.4 Member Data Documentation
10.15.4.1 maxlinsteps
int maxlinsteps = 0
maximum number of allowed linear steps per Newton step for steady state computation
Definition at line 59 of file newton_solver.h.
10.15.4.2 maxsteps
int maxsteps = 0
maximum number of allowed Newton steps for steady state computation
Definition at line 61 of file newton_solver.h.
Generated by Doxygen

10.15 NewtonSolver Class Reference 351
10.15.4.3 atol
double atol = 1e-16
absolute tolerance
Definition at line 63 of file newton_solver.h.
10.15.4.4 rtol
double rtol = 1e-8
relative tolerance
Definition at line 65 of file newton_solver.h.
10.15.4.5 t
realtype∗t [protected]
time variable
Definition at line 69 of file newton_solver.h.
10.15.4.6 model
Model∗model [protected]
pointer to the AMICI model object
Definition at line 71 of file newton_solver.h.
10.15.4.7 rdata
ReturnData∗rdata [protected]
pointer to the return data object
Definition at line 73 of file newton_solver.h.
Generated by Doxygen

352 CONTENTS
10.15.4.8 xdot
AmiVector xdot [protected]
right hand side AmiVector
Definition at line 75 of file newton_solver.h.
10.15.4.9 x
AmiVector∗x [protected]
current state
Definition at line 77 of file newton_solver.h.
10.15.4.10 dx
AmiVector dx [protected]
current state time derivative (DAE)
Definition at line 79 of file newton_solver.h.
10.16 NewtonSolverDense Class Reference
The NewtonSolverDense provides access to the dense linear solver for the Newton method.
#include <newton_solver.h>
Inheritance diagram for NewtonSolverDense:
NewtonSolverDense
NewtonSolver
Generated by Doxygen
10.16 NewtonSolverDense Class Reference 353
Public Member Functions
•NewtonSolverDense (realtype ∗t,AmiVector ∗x,Model ∗model,ReturnData ∗rdata)
• void solveLinearSystem (AmiVector ∗rhs) override
• void prepareLinearSystem (int ntry, int nnewt) override
Additional Inherited Members
10.16.1 Detailed Description
Definition at line 88 of file newton_solver.h.
10.16.2 Constructor & Destructor Documentation
10.16.2.1 NewtonSolverDense()
NewtonSolverDense (
realtype ∗t,
AmiVector ∗x,
Model ∗model,
ReturnData ∗rdata )
default constructor, initializes all members with the provided objects and initializes temporary storage objects
Parameters
tpointer to time variable
xpointer to state variables
model pointer to the AMICI model object
rdata pointer to the return data object
Definition at line 152 of file newton_solver.cpp.
10.16.3 Member Function Documentation
10.16.3.1 solveLinearSystem()
void solveLinearSystem (
AmiVector ∗rhs ) [override], [virtual]
Solves the linear system for the Newton step
Generated by Doxygen
354 CONTENTS
Parameters
rhs containing the RHS of the linear system, will be overwritten by solution to the linear system
Solves the linear system for the Newton step
Parameters
rhs containing the RHS of the linear system, will be overwritten by solution to the linear system
Implements NewtonSolver.
Definition at line 191 of file newton_solver.cpp.
Here is the call graph for this function:
solveLinearSystem amici::AmiVector::data
10.16.3.2 prepareLinearSystem()
void prepareLinearSystem (
int ntry,
int nnewt ) [override], [virtual]
Writes the Jacobian for the Newton iteration and passes it to the linear solver
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Writes the Jacobian for the Newton iteration and passes it to the linear solver
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Implements NewtonSolver.
Definition at line 171 of file newton_solver.cpp.
Generated by Doxygen
10.17 NewtonSolverIterative Class Reference 355
Here is the call graph for this function:
prepareLinearSystem amici::Model::fJ
10.17 NewtonSolverIterative Class Reference
The NewtonSolverIterative provides access to the iterative linear solver for the Newton method.
#include <newton_solver.h>
Inheritance diagram for NewtonSolverIterative:
NewtonSolverIterative
NewtonSolver
Public Member Functions
•NewtonSolverIterative (realtype ∗t,AmiVector ∗x,Model ∗model,ReturnData ∗rdata)
• void solveLinearSystem (AmiVector ∗rhs)
• void prepareLinearSystem (int ntry, int nnewt)
• void linsolveSPBCG (int ntry, int nnewt, AmiVector ∗ns_delta)
Additional Inherited Members
10.17.1 Detailed Description
Definition at line 136 of file newton_solver.h.
10.17.2 Constructor & Destructor Documentation
Generated by Doxygen
356 CONTENTS
10.17.2.1 NewtonSolverIterative()
NewtonSolverIterative (
realtype ∗t,
AmiVector ∗x,
Model ∗model,
ReturnData ∗rdata )
default constructor, initializes all members with the provided objects
Parameters
tpointer to time variable
xpointer to state variables
model pointer to the AMICI model object
rdata pointer to the return data object
Definition at line 312 of file newton_solver.cpp.
10.17.3 Member Function Documentation
10.17.3.1 solveLinearSystem()
void solveLinearSystem (
AmiVector ∗rhs ) [virtual]
Solves the linear system for the Newton step
Parameters
rhs containing the RHS of the linear system, will be overwritten by solution to the linear system
Solves the linear system for the Newton step by passing it to linsolveSPBCG
Parameters
rhs containing the RHS of the linear system, will be overwritten by solution to the linear system
Implements NewtonSolver.
Definition at line 350 of file newton_solver.cpp.
Generated by Doxygen
10.17 NewtonSolverIterative Class Reference 357
Here is the call graph for this function:
solveLinearSystem linsolveSPBCG
amici::AmiVector::minus
amici::Model::fJDiag
amici::AmiVector::set
amici::AmiVector::getNVector
amici::AmiVector::reset
amici::Model::fJv
10.17.3.2 prepareLinearSystem()
void prepareLinearSystem (
int ntry,
int nnewt ) [virtual]
Writes the Jacobian for the Newton iteration and passes it to the linear solver
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Writes the Jacobian for the Newton iteration and passes it to the linear solver. Also wraps around getSensis for
iterative linear solver.
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Implements NewtonSolver.
Definition at line 329 of file newton_solver.cpp.
Generated by Doxygen
358 CONTENTS
10.17.3.3 linsolveSPBCG()
void linsolveSPBCG (
int ntry,
int nnewt,
AmiVector ∗ns_delta )
Iterative linear solver created from SPILS BiCG-Stab. Solves the linear system within each Newton step if iterative
solver is chosen.
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
ns_delta Newton step
Definition at line 363 of file newton_solver.cpp.
Here is the call graph for this function:
linsolveSPBCG
amici::AmiVector::minus
amici::Model::fJDiag
amici::AmiVector::set
amici::AmiVector::getNVector
amici::AmiVector::reset
amici::Model::fJv
Here is the caller graph for this function:
linsolveSPBCG solveLinearSystem
Generated by Doxygen
10.18 NewtonSolverSparse Class Reference 359
10.18 NewtonSolverSparse Class Reference
The NewtonSolverSparse provides access to the sparse linear solver for the Newton method.
#include <newton_solver.h>
Inheritance diagram for NewtonSolverSparse:
NewtonSolverSparse
NewtonSolver
Public Member Functions
•NewtonSolverSparse (realtype ∗t,AmiVector ∗x,Model ∗model,ReturnData ∗rdata)
• void solveLinearSystem (AmiVector ∗rhs) override
• void prepareLinearSystem (int ntry, int nnewt) override
Additional Inherited Members
10.18.1 Detailed Description
Definition at line 109 of file newton_solver.h.
10.18.2 Constructor & Destructor Documentation
10.18.2.1 NewtonSolverSparse()
NewtonSolverSparse (
realtype ∗t,
AmiVector ∗x,
Model ∗model,
ReturnData ∗rdata )
default constructor, initializes all members with the provided objects, initializes temporary storage objects and the
klu solver
Generated by Doxygen
360 CONTENTS
Parameters
tpointer to time variable
xpointer to state variables
model pointer to the AMICI model object
rdata pointer to the return data object
Definition at line 221 of file newton_solver.cpp.
10.18.3 Member Function Documentation
10.18.3.1 solveLinearSystem()
void solveLinearSystem (
AmiVector ∗rhs ) [override], [virtual]
Solves the linear system for the Newton step
Parameters
rhs containing the RHS of the linear system, will be overwritten by solution to the linear system
Solves the linear system for the Newton step
Parameters
rhs containing the RHS of the linear system,will be overwritten by solution to the linear system
Implements NewtonSolver.
Definition at line 278 of file newton_solver.cpp.
Here is the call graph for this function:
solveLinearSystem amici::AmiVector::data
Generated by Doxygen
10.19 Constant Class Reference 361
10.18.3.2 prepareLinearSystem()
void prepareLinearSystem (
int ntry,
int nnewt ) [override], [virtual]
Writes the Jacobian for the Newton iteration and passes it to the linear solver
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Writes the Jacobian for the Newton iteration and passes it to the linear solver
Parameters
ntry integer newton_try integer start number of Newton solver (1 or 2)
nnewt integer number of current Newton step
Implements NewtonSolver.
Definition at line 244 of file newton_solver.cpp.
Here is the call graph for this function:
prepareLinearSystem amici::Model::fJSparse
10.19 Constant Class Reference
AConstant is a fixed variable in the model with respect to which sensitivites cannot be computed, abbreviated by k
Inheritance diagram for Constant:
Constant
ModelQuantity
Generated by Doxygen

362 CONTENTS
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new Expression instance.
10.19.1 Detailed Description
Definition at line 484 of file ode_export.py.
10.19.2 Constructor & Destructor Documentation
10.19.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Parameters
identifier unique identifier of the Constant
Type: sympy.Symbol
name individual name of the Constant (does not need to be unique)
Type: str
value numeric value
Type: float
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 503 of file ode_export.py.
10.20 Expression Class Reference
An Expressions is a recurring elements in symbolic formulas.
Generated by Doxygen

10.20 Expression Class Reference 363
Inheritance diagram for Expression:
Expression
ModelQuantity
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new Expression instance.
10.20.1 Detailed Description
Specifying this may yield more compact expression which may lead to substantially shorter model compilation times,
but may also reduce model simulation time, abbreviated by w
Definition at line 431 of file ode_export.py.
10.20.2 Constructor & Destructor Documentation
10.20.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Parameters
identifier unique identifier of the Expression
Type: sympy.Symbol
name individual name of the Expression (does not need to be unique)
Type: str
value formula
Type: symengine.Basic
Generated by Doxygen
364 CONTENTS
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 449 of file ode_export.py.
10.21 LogLikelihood Class Reference
ALogLikelihood defines the distance between measurements and experiments for a particular observable.
Inheritance diagram for LogLikelihood:
LogLikelihood
ModelQuantity
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new Expression instance.
10.21.1 Detailed Description
The final LogLikelihood value in the simulation will be the sum of all specified LogLikelihood instances evaluated at
all timepoints, abbreviated by Jy
Definition at line 513 of file ode_export.py.
10.21.2 Constructor & Destructor Documentation
10.21.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Generated by Doxygen
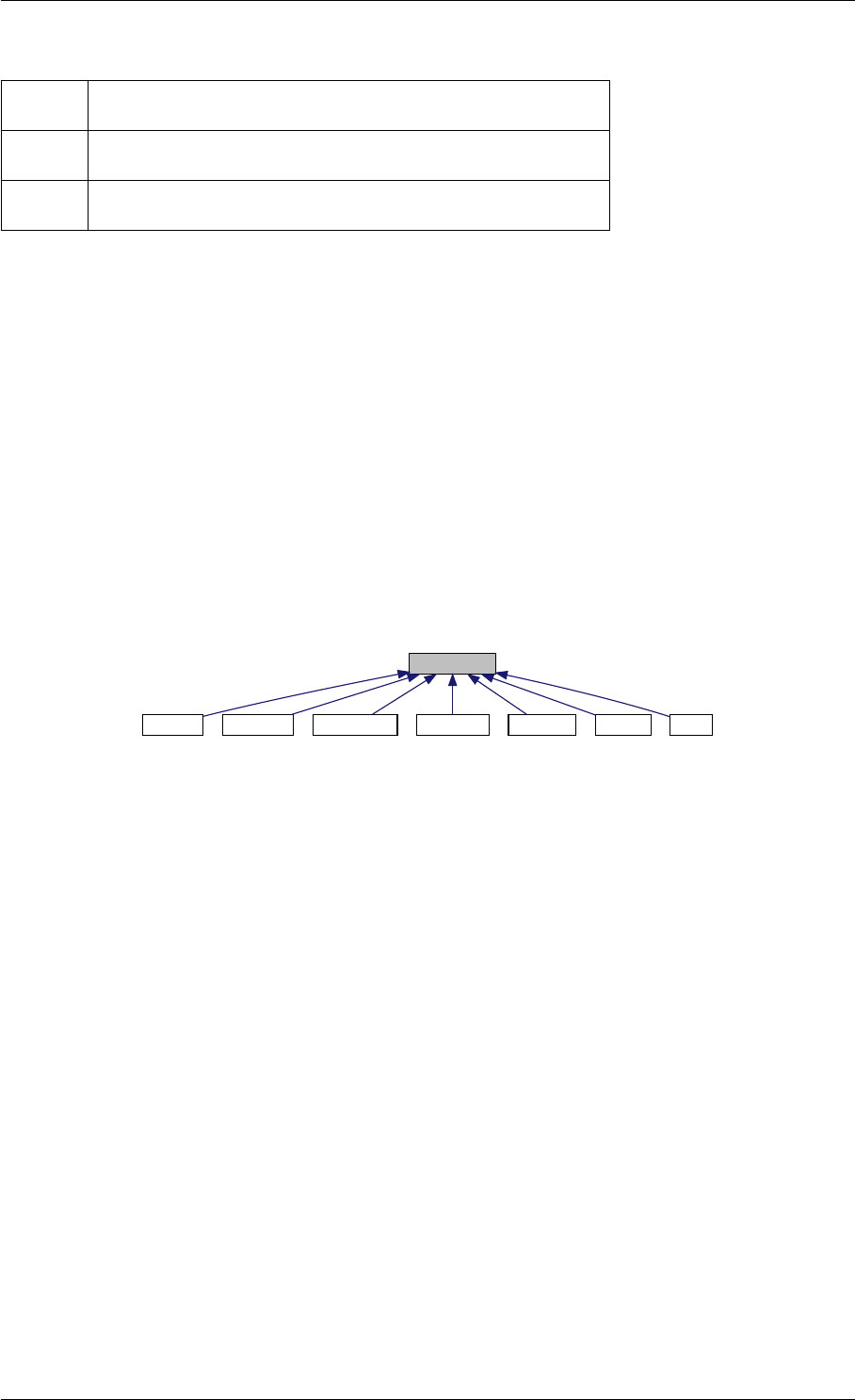
10.22 ModelQuantity Class Reference 365
Parameters
identifier unique identifier of the LogLikelihood
Type: sympy.Symbol
name individual name of the LogLikelihood (does not need to be unique)
Type: str
value formula
Type: symengine.Basic
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 533 of file ode_export.py.
10.22 ModelQuantity Class Reference
Base class for model components.
Inheritance diagram for ModelQuantity:
ModelQuantity
Constant Expression LogLikelihood Observable Parameter SigmaY State
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new ModelQuantity instance.
• def __repr__ (self)
Representation of the ModelQuantity object.
10.22.1 Detailed Description
Definition at line 292 of file ode_export.py.
10.22.2 Constructor & Destructor Documentation
10.22.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Generated by Doxygen

366 CONTENTS
Parameters
identifier unique identifier of the quantity
Type: sympy.Symbol
name individual name of the quantity (does not need to be unique)
Type: str
value either formula, numeric value or initial value
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 310 of file ode_export.py.
10.22.3 Member Function Documentation
10.22.3.1 __repr__()
def __repr__ (
self )
Returns
string representation of the ModelQuantity
Definition at line 337 of file ode_export.py.
10.23 Observable Class Reference
An Observable links model simulations to experimental measurements, abbreviated by y
Inheritance diagram for Observable:
Observable
ModelQuantity
Generated by Doxygen

10.24 ODEExporter Class Reference 367
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new Observable instance.
10.23.1 Detailed Description
Definition at line 377 of file ode_export.py.
10.23.2 Constructor & Destructor Documentation
10.23.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Parameters
identifier unique identifier of the Observable
Type: sympy.Symbol
name individual name of the Observable (does not need to be unique)
Type: str
value formula
Type: symengine.Basic
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 395 of file ode_export.py.
10.24 ODEExporter Class Reference
The ODEExporter class generates AMICI C++ files for ODE model as defined in symbolic expressions.
Generated by Doxygen

368 CONTENTS
Public Member Functions
• def __init__ (self, ode_model, outdir=None, verbose=False, assume_pow_positivity=False, compiler=None)
Generate AMICI C++ files for the ODE provided to the constructor.
• def generateModelCode (self)
Generates the native C++ code for the loaded model.
• def compileModel (self)
Compiles the generated code it into a simulatable module.
• def setPaths (self, output_dir)
Set output paths for the model and create if necessary.
• def setName (self, modelName)
Sets the model name.
Public Attributes
•outdir
see sbml_import.setPaths()
Type: str
•verbose
more verbose output if True
Type: bool
•assume_pow_positivity
if set to true, a special pow function is
•compiler
distutils/setuptools compiler selection to build the
•codeprinter
allows export of symbolic variables as C++ code
•modelName
name of the model that will be used for compilation
•modelPath
path to the generated model specific files
Type: str
•modelSwigPath
path to the generated swig files
Type: str
•model
ODE definition
Type:ODEModel.
•functions
carries C++ function signatures and other specifications
•allow_reinit_fixpar_initcond
indicates whether reinitialization of
10.24.1 Detailed Description
initial states depending on fixedParmeters is allowed for this model
Type: bool
Definition at line 1437 of file ode_export.py.
Generated by Doxygen
10.24 ODEExporter Class Reference 369
10.24.2 Constructor & Destructor Documentation
10.24.2.1 __init__()
def __init__ (
self,
ode_model,
outdir = None,
verbose = False,
assume_pow_positivity = False,
compiler = None )
Parameters
ode_model ODE definition
Type:ODEModel
outdir see sbml_import.setPaths()
Type: str
verbose more verbose output if True
Type: bool
assume_pow_positivity if set to true, a special pow function is used to avoid problems with state variables
that may become negative due to numerical errors
Type: bool
compiler distutils/setuptools compiler selection to build the python extension
Type: str
Returns
Definition at line 1530 of file ode_export.py.
10.24.3 Member Function Documentation
10.24.3.1 generateModelCode()
def generateModelCode (
self )
Generated by Doxygen
370 CONTENTS
Returns
Definition at line 1564 of file ode_export.py.
Here is the call graph for this function:
generateModelCode
_prepareModelFolder
_generateCCode
10.24.3.2 compileModel()
def compileModel (
self )
Generated by Doxygen
10.24 ODEExporter Class Reference 371
Returns
Definition at line 1577 of file ode_export.py.
Here is the call graph for this function:
compileModel
_compileCCode
_writeFunctionFile
_writeIndexFiles
_writeWrapfunctionsCPP
_writeWrapfunctionsHeader
_writeModelHeader
_writeCMakeFile
_writeSwigFiles
_writeModuleSetup
_getFunctionBody
_getSymLines
_getSparseSymLines
amici.ode_export.applyTemplate
_getSymbolNameInitializerList
_getSymbolIDInitializerList
amici.ode_export.ODEModel
_from_pysb_importer
10.24.3.3 setPaths()
def setPaths (
self,
output_dir )
Generated by Doxygen

372 CONTENTS
Parameters
output_dir relative or absolute path where the generated model code is to be placed. will be created if does
not exists.
Type: str
Returns
Definition at line 2165 of file ode_export.py.
10.24.3.4 setName()
def setName (
self,
modelName )
Parameters
modelName name of the model (must only contain valid filename characters)
Type: str
Returns
Definition at line 2184 of file ode_export.py.
10.25 ODEModel Class Reference
An ODEModel defines an Ordinay Differential Equation as set of ModelQuantities.
Public Member Functions
• def __init__ (self)
Create a new ODEModel instance.
• def import_from_sbml_importer (self, si)
Imports a model specification from a amici.SBMLImporter instance.
• def add_component (self, component)
Adds a new ModelQuantity to the model.
• def nx (self)
Number of states.
• def ny (self)
Number of Observables.
• def nk (self)
Generated by Doxygen

10.25 ODEModel Class Reference 373
Number of Constants.
• def np (self)
Number of Parameters.
• def sym (self, name)
Returns (and constructs if necessary) the identifiers for a symbolic entity.
• def sparsesym (self, name)
Returns (and constructs if necessary) the sparsified identifiers for a sparsified symbolic variable.
• def eq (self, name)
Returns (and constructs if necessary) the formulas for a symbolic entity.
• def sparseeq (self, name)
Returns (and constructs if necessary) the sparsified formulas for a sparsified symbolic variable.
• def colptr (self, name)
Returns (and constructs if necessary) the column pointers for a sparsified symbolic variable.
• def rowval (self, name)
Returns (and constructs if necessary) the row values for a sparsified symbolic variable.
• def val (self, name)
Returns (and constructs if necessary) the numeric values of a symbolic entity.
• def name (self, name)
Returns (and constructs if necessary) the names of a symbolic variable.
• def generateBasicVariables (self)
Generates the symbolic identifiers for all variables in ODEModel.variable_prototype.
• def symNames (self)
Returns a list of names of generated symbolic variables.
10.25.1 Detailed Description
This class provides general purpose interfaces to compute arbitrary symbolic derivatives that are necessary for
model simulation or sensitivity computation
derivative from a partial derivative call to enforce a partial
derivative in the next recursion. prevents infinite recursion
Definition at line 558 of file ode_export.py.
10.25.2 Constructor & Destructor Documentation
10.25.2.1 __init__()
def __init__ (
self )
Generated by Doxygen
374 CONTENTS
Returns
New ODEModel instance
Definition at line 713 of file ode_export.py.
Here is the call graph for this function:
__init__
amici::Solver::nx
nx
ny
sym _generateSymbol
10.25.3 Member Function Documentation
10.25.3.1 import_from_sbml_importer()
def import_from_sbml_importer (
self,
si )
Parameters
si imported SBML model
Type: amici.SbmlImporter
Returns
Definition at line 806 of file ode_export.py.
Generated by Doxygen
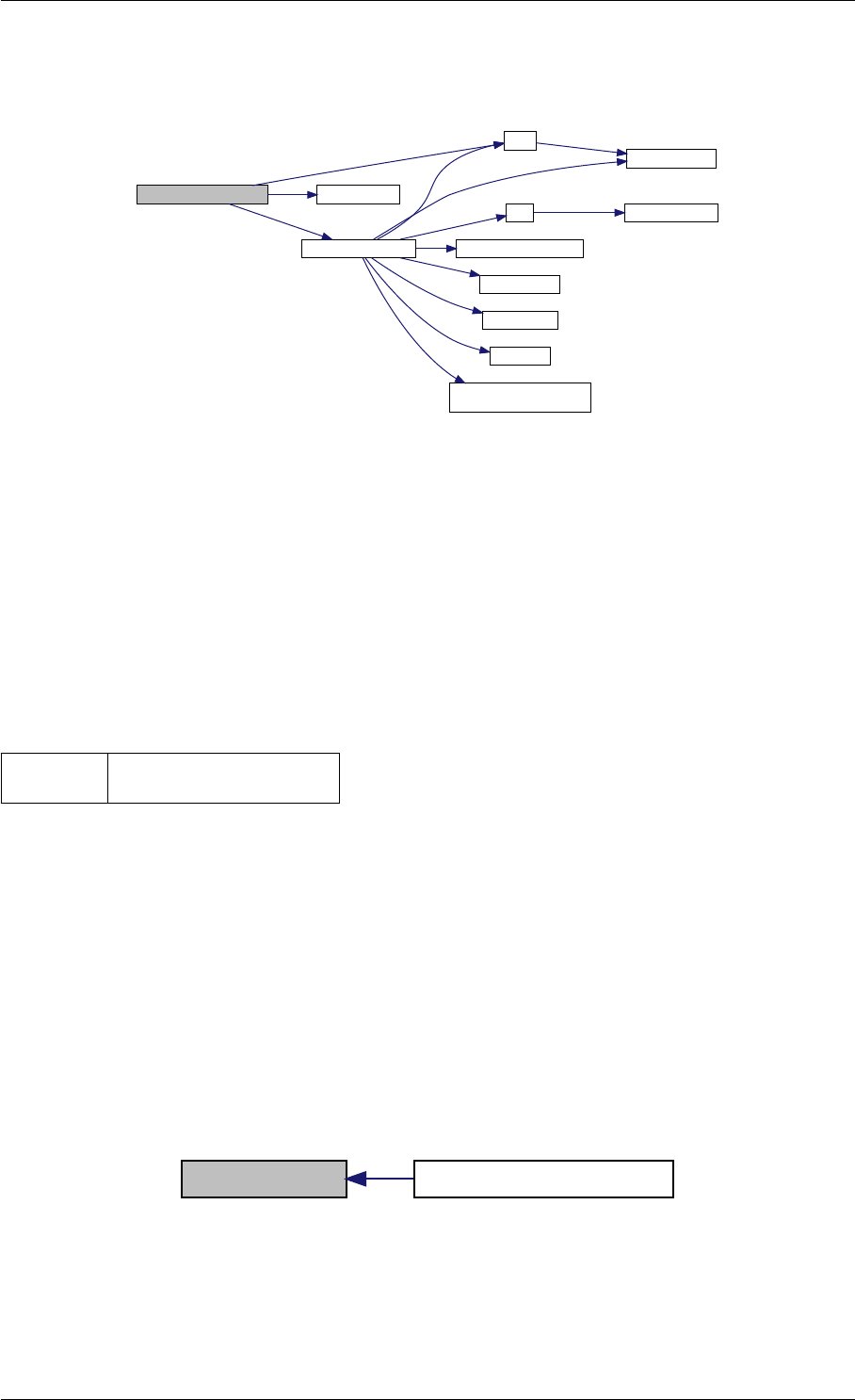
10.25 ODEModel Class Reference 375
Here is the call graph for this function:
import_from_sbml_importer
sym
add_component
generateBasicVariables
_generateSymbol
eq
_equationFromComponent
_totalDerivative
_multiplication
_derivative
amici.ode_export.getSymbolic
Diagonal
_computeEquation
10.25.3.2 add_component()
def add_component (
self,
component )
Parameters
component model quantity to be added
Type:ModelQuantity
Returns
Definition at line 839 of file ode_export.py.
Here is the caller graph for this function:
add_component import_from_sbml_importer
Generated by Doxygen
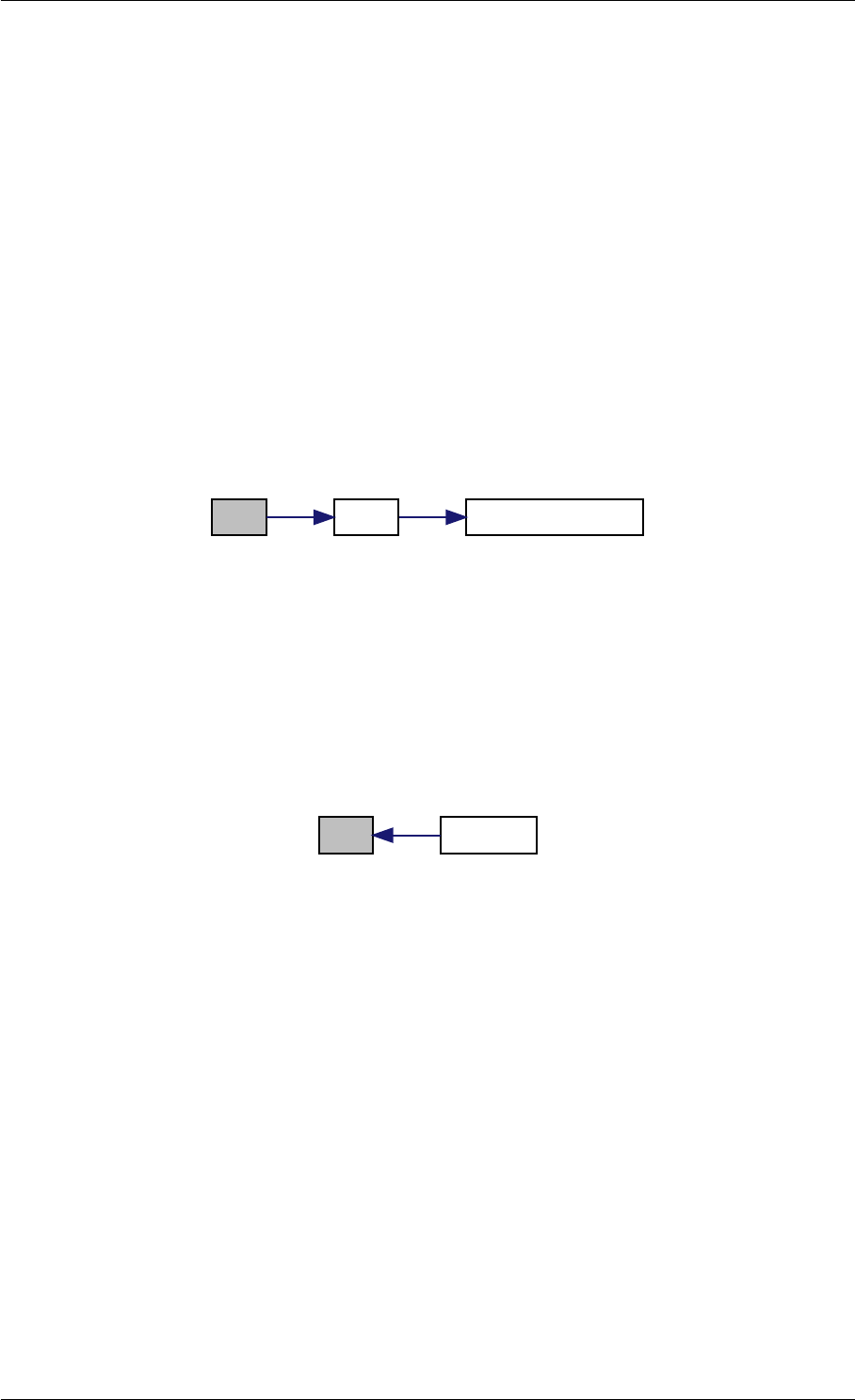
376 CONTENTS
10.25.3.3 nx()
def nx (
self )
Returns
number of state variable symbols
Definition at line 859 of file ode_export.py.
Here is the call graph for this function:
nx sym _generateSymbol
Here is the caller graph for this function:
nx __init__
10.25.3.4 ny()
def ny (
self )
Returns
number of observable symbols
Definition at line 872 of file ode_export.py.
Generated by Doxygen
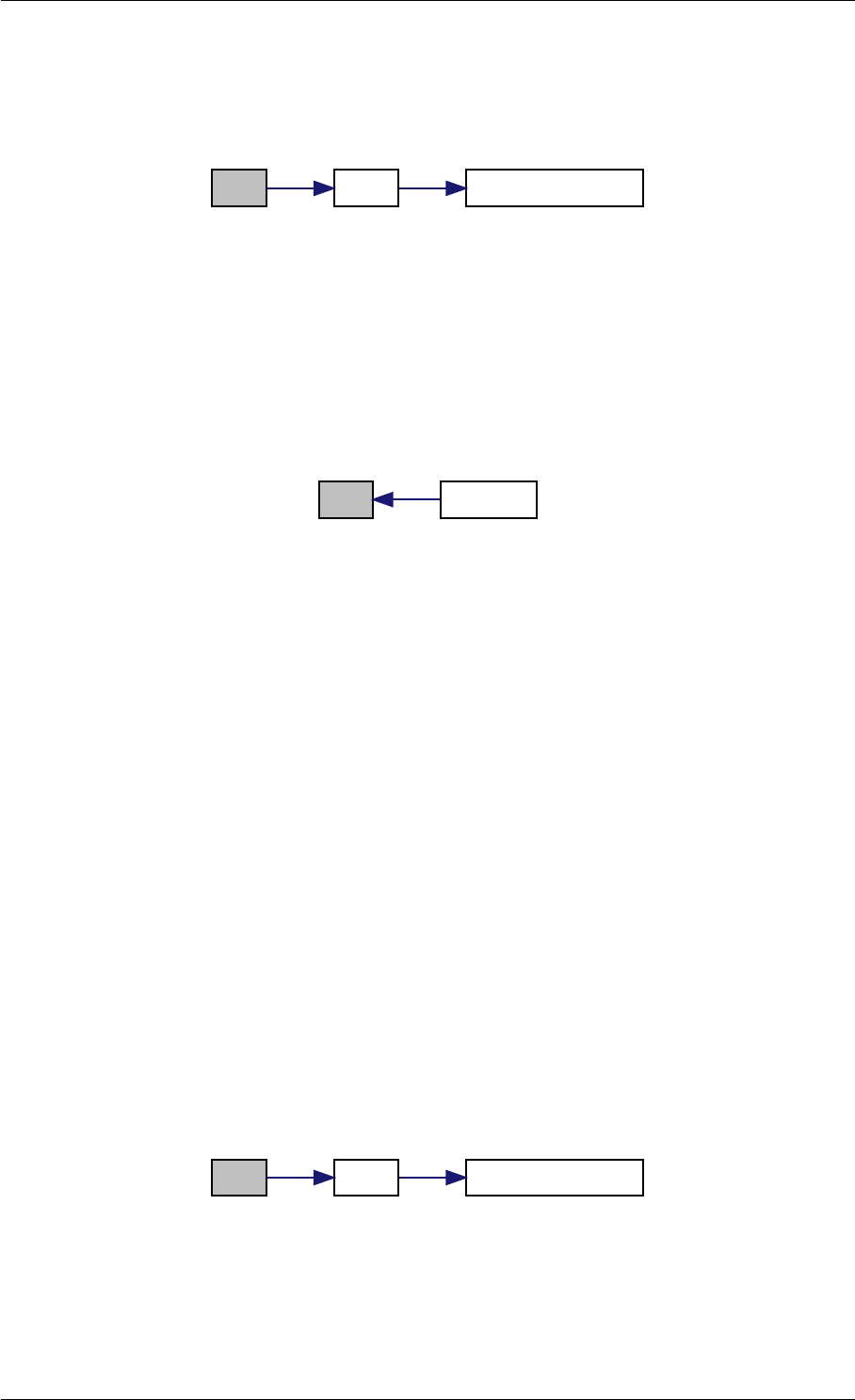
10.25 ODEModel Class Reference 377
Here is the call graph for this function:
ny sym _generateSymbol
Here is the caller graph for this function:
ny __init__
10.25.3.5 nk()
def nk (
self )
Returns
number of constant symbols
Definition at line 885 of file ode_export.py.
Here is the call graph for this function:
nk sym _generateSymbol
Generated by Doxygen
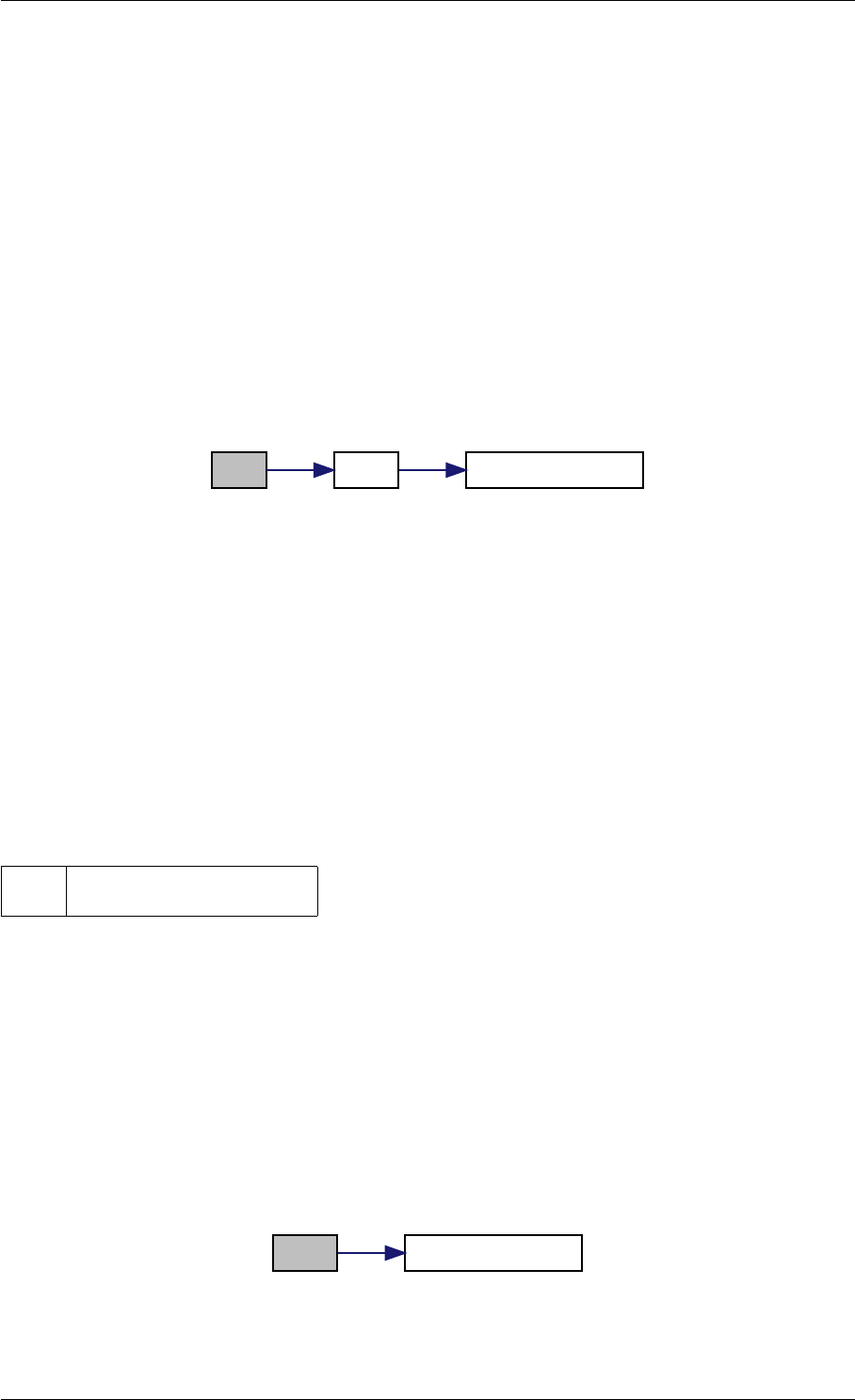
378 CONTENTS
10.25.3.6 np()
def np (
self )
Returns
number of parameter symbols
Definition at line 898 of file ode_export.py.
Here is the call graph for this function:
np sym _generateSymbol
10.25.3.7 sym()
def sym (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
containing the symbolic identifiers
Type: symengine.DenseMatrix
Definition at line 913 of file ode_export.py.
Here is the call graph for this function:
sym _generateSymbol
Generated by Doxygen
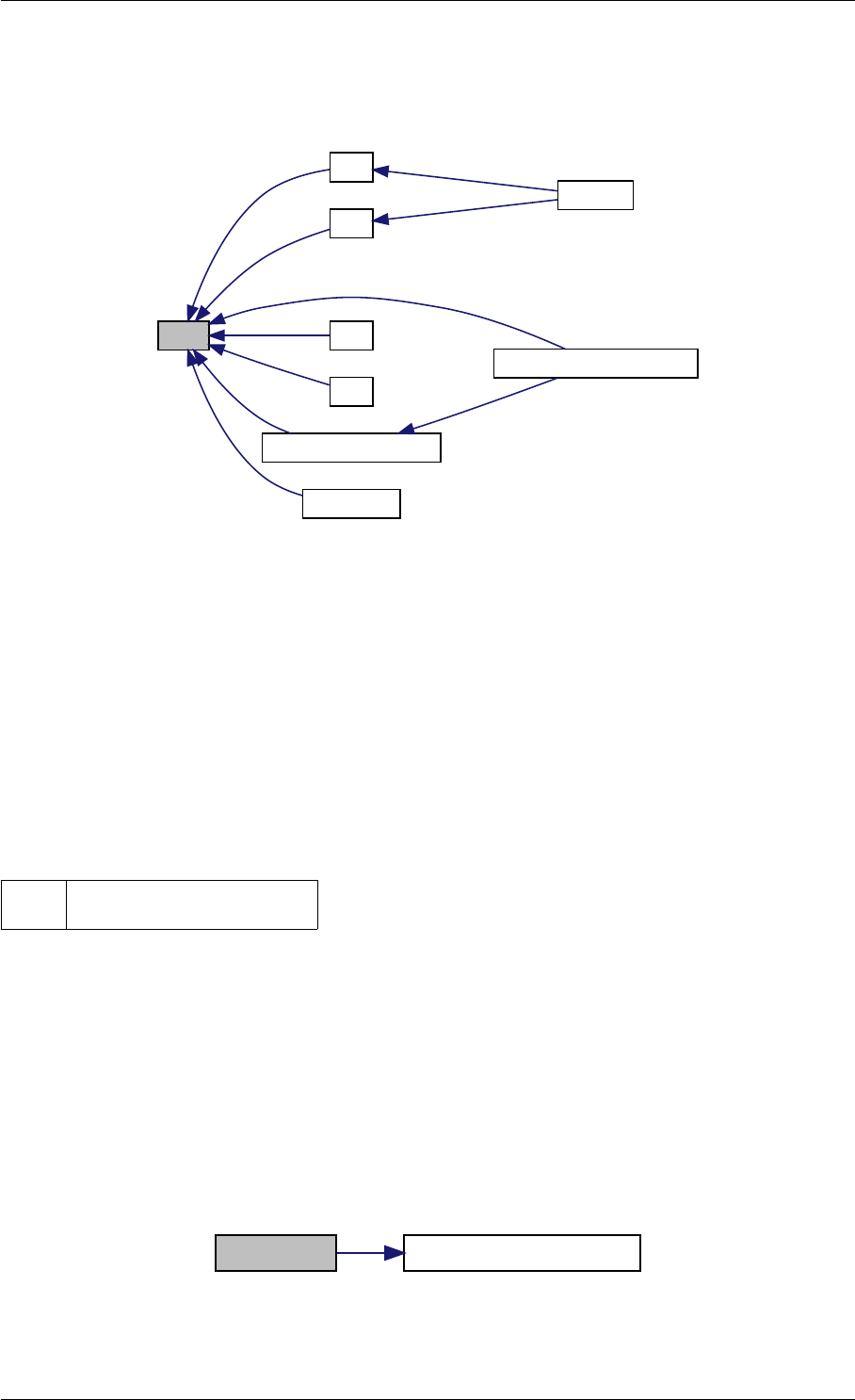
10.25 ODEModel Class Reference 379
Here is the caller graph for this function:
sym
import_from_sbml_importer
nx
ny
nk
np
generateBasicVariables
symNames
__init__
10.25.3.8 sparsesym()
def sparsesym (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
linearized symengine.DenseMatrix containing the symbolic identifiers
Definition at line 930 of file ode_export.py.
Here is the call graph for this function:
sparsesym _generateSparseSymbol
Generated by Doxygen
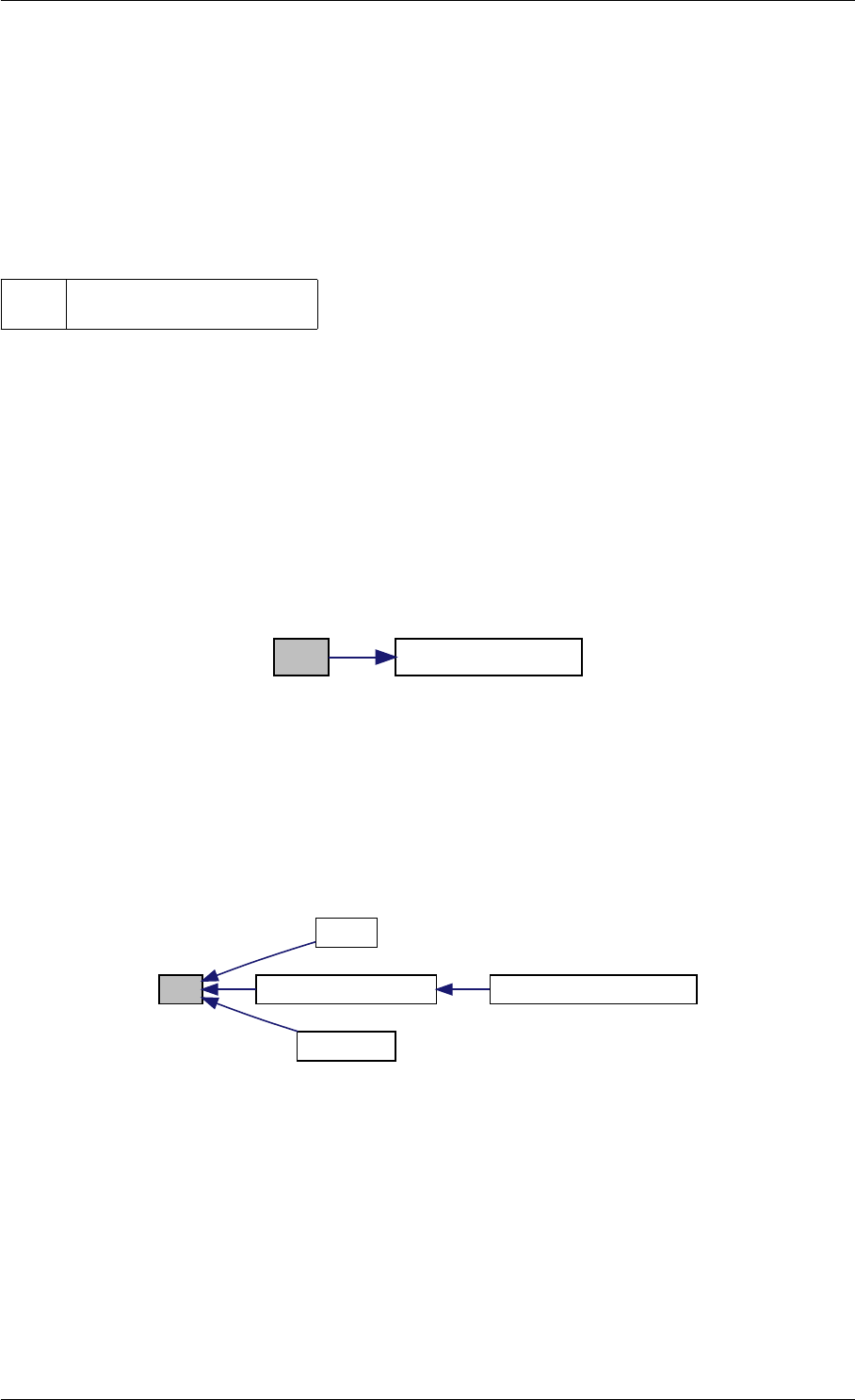
380 CONTENTS
10.25.3.9 eq()
def eq (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
symengine.DenseMatrix containing the symbolic identifiers
Definition at line 949 of file ode_export.py.
Here is the call graph for this function:
eq _computeEquation
Here is the caller graph for this function:
eq
name
generateBasicVariables
symNames
import_from_sbml_importer
10.25.3.10 sparseeq()
def sparseeq (
self,
name )
Generated by Doxygen
10.25 ODEModel Class Reference 381
Parameters
name name of the symbolic variable
Type: str
Returns
linearized symengine.DenseMatrix containing the symbolic formulas
Definition at line 966 of file ode_export.py.
Here is the call graph for this function:
sparseeq _generateSparseSymbol
10.25.3.11 colptr()
def colptr (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
symengine.DenseMatrix containing the column pointers
Definition at line 985 of file ode_export.py.
Here is the call graph for this function:
colptr _generateSparseSymbol
Generated by Doxygen
382 CONTENTS
10.25.3.12 rowval()
def rowval (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
symengine.DenseMatrix containing the row values
Definition at line 1004 of file ode_export.py.
Here is the call graph for this function:
rowval _generateSparseSymbol
10.25.3.13 val()
def val (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
list containing the numeric values
Definition at line 1023 of file ode_export.py.
Generated by Doxygen
10.25 ODEModel Class Reference 383
Here is the call graph for this function:
val _generateValue
10.25.3.14 name()
def name (
self,
name )
Parameters
name name of the symbolic variable
Type: str
Returns
list of names
Definition at line 1039 of file ode_export.py.
Here is the call graph for this function:
name
_generateName
_generateSparseSymbol
eq _computeEquation
Generated by Doxygen
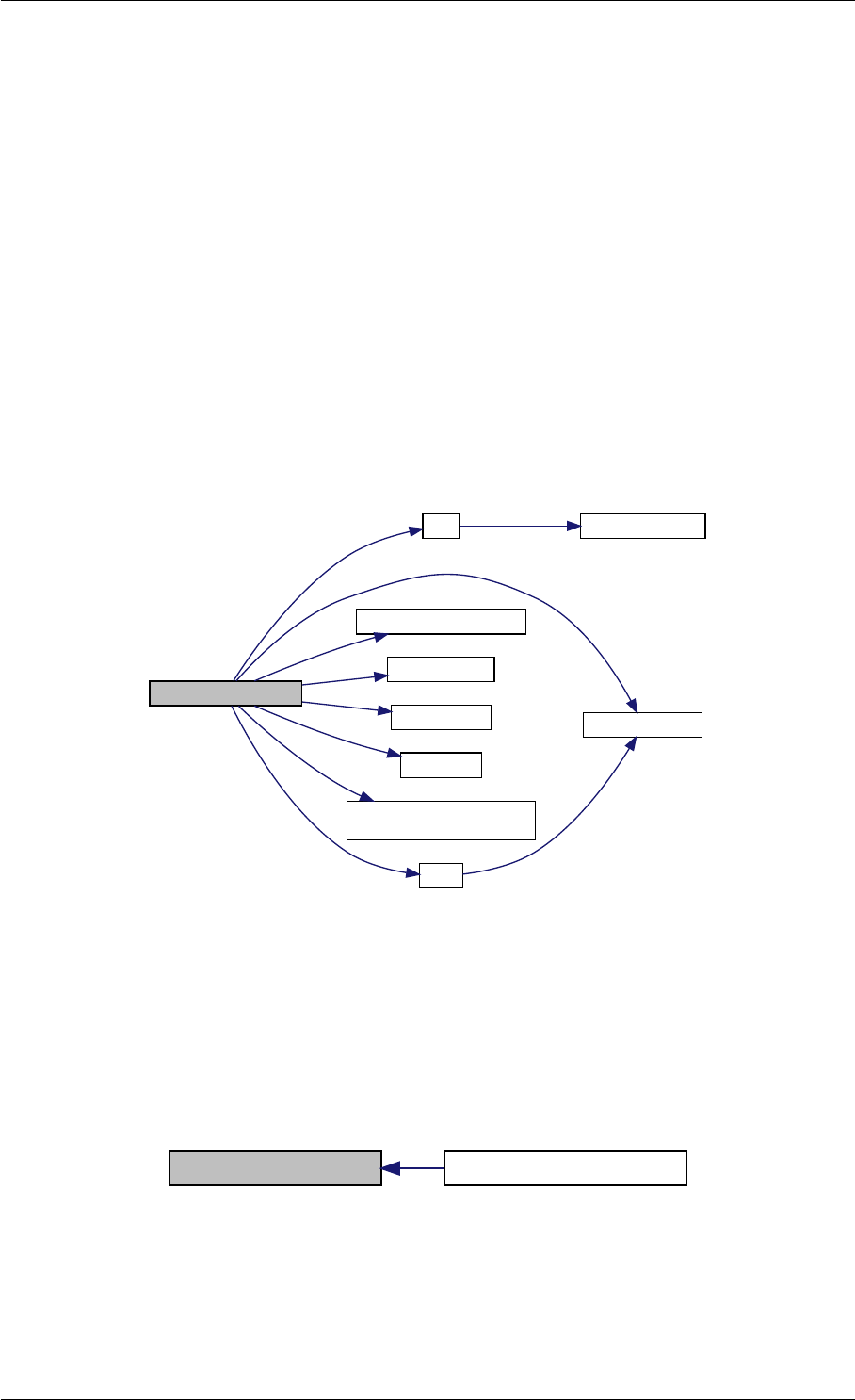
384 CONTENTS
10.25.3.15 generateBasicVariables()
def generateBasicVariables (
self )
Returns
Definition at line 1092 of file ode_export.py.
Here is the call graph for this function:
generateBasicVariables
_generateSymbol
eq
_equationFromComponent
_totalDerivative
_multiplication
_derivative
amici.ode_export.getSymbolic
Diagonal
sym
_computeEquation
Here is the caller graph for this function:
generateBasicVariables import_from_sbml_importer
Generated by Doxygen
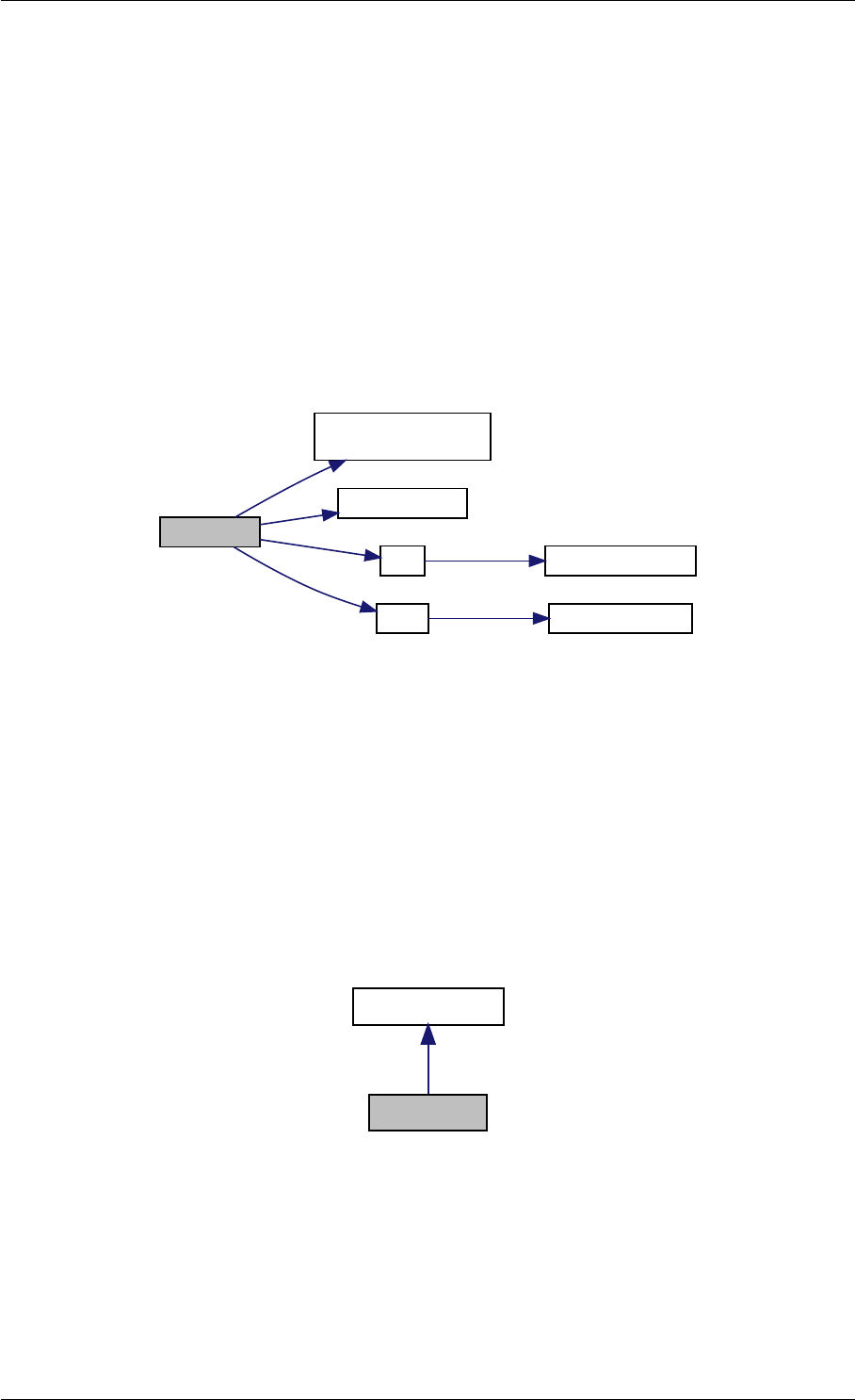
10.26 Parameter Class Reference 385
10.25.3.16 symNames()
def symNames (
self )
Returns
list of names
Definition at line 1216 of file ode_export.py.
Here is the call graph for this function:
symNames
amici.ode_export.var
_in_function_signature
_totalDerivative
eq
sym
_computeEquation
_generateSymbol
10.26 Parameter Class Reference
AParameter is a free variable in the model with respect to which sensitivites may be computed, abbreviated by p
Inheritance diagram for Parameter:
Parameter
ModelQuantity
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new Expression instance.
Generated by Doxygen

386 CONTENTS
10.26.1 Detailed Description
Definition at line 457 of file ode_export.py.
10.26.2 Constructor & Destructor Documentation
10.26.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Parameters
identifier unique identifier of the Parameter
Type: sympy.Symbol
name individual name of the Parameter (does not need to be unique)
Type: str
value numeric value
Type: float
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 476 of file ode_export.py.
10.27 SigmaY Class Reference
A Standard Deviation SigmaY rescales the distance between simulations and measurements when computing resid-
uals, abbreviated by sigmay
Generated by Doxygen

10.27 SigmaY Class Reference 387
Inheritance diagram for SigmaY:
SigmaY
ModelQuantity
Public Member Functions
• def __init__ (self, identifier, name, value)
Create a new Standard Deviation instance.
10.27.1 Detailed Description
Definition at line 403 of file ode_export.py.
10.27.2 Constructor & Destructor Documentation
10.27.2.1 __init__()
def __init__ (
self,
identifier,
name,
value )
Parameters
identifier unique identifier of the Standard Deviation
Type: sympy.Symbol
name individual name of the Standard Deviation (does not need to be unique)
Type: str
value formula
Type: symengine.Basic
Returns
ModelQuantity instance
Generated by Doxygen
388 CONTENTS
TypeError
is thrown if input types do not match documented types
Definition at line 422 of file ode_export.py.
10.28 State Class Reference
AState variable defines an entity that evolves with time according to the provided time derivative, abbreviated by x
Inheritance diagram for State:
State
ModelQuantity
Public Member Functions
• def __init__ (self, identifier, name, value, dt)
Create a new State instance.
10.28.1 Detailed Description
Definition at line 345 of file ode_export.py.
10.28.2 Constructor & Destructor Documentation
10.28.2.1 __init__()
def __init__ (
self,
identifier,
name,
value,
dt )
Extends ModelQuantity.__init__ by dt
Generated by Doxygen
10.29 TemplateAmici Class Reference 389
Parameters
identifier unique identifier of the state
Type: sympy.Symbol
name individual name of the state (does not need to be unique)
Type: str
value initial value
Type: symengine.Basic
dt time derivative
Type: symengine.Basic
Returns
ModelQuantity instance
TypeError
is thrown if input types do not match documented types
Definition at line 366 of file ode_export.py.
10.29 TemplateAmici Class Reference
Template format used in AMICI (see string.template for more details).
Inheritance diagram for TemplateAmici:
TemplateAmici
Template
Static Public Attributes
• string delimiter = 'TPL_'
delimiter that identifies template variables
Type: str
10.29.1 Detailed Description
Definition at line 2212 of file ode_export.py.
Generated by Doxygen

390 CONTENTS
10.30 ReturnData Class Reference
class that stores all data which is later returned by the mex function
#include <rdata.h>
Public Member Functions
•ReturnData ()
default constructor
•ReturnData (std::vector<realtype >ts, int np, int nk, int nx, int nxtrue, int ny, int nytrue, int nz, int nztrue,
int ne, int nJ, int nplist, int nmaxevent, int nt, int newton_maxsteps, std::vector<ParameterScaling >pscale,
SecondOrderMode o2mode,SensitivityOrder sensi,SensitivityMethod sensi_meth)
ReturnData.
•ReturnData (Solver const &solver, const Model ∗model)
• void initializeObjectiveFunction ()
initializeObjectiveFunction
• void invalidate (const realtype t)
• void invalidateLLH ()
• void applyChainRuleFactorToSimulationResults (const Model ∗model)
Public Attributes
• const std::vector<realtype >ts
• std::vector<realtype >xdot
• std::vector<realtype >J
• std::vector<realtype >z
• std::vector<realtype >sigmaz
• std::vector<realtype >sz
• std::vector<realtype >ssigmaz
• std::vector<realtype >rz
• std::vector<realtype >srz
• std::vector<realtype >s2rz
• std::vector<realtype >x
• std::vector<realtype >sx
• std::vector<realtype >y
• std::vector<realtype >sigmay
• std::vector<realtype >sy
• std::vector<realtype >ssigmay
• std::vector<realtype >res
• std::vector<realtype >sres
• std::vector<realtype >FIM
• std::vector<int >numsteps
• std::vector<int >numstepsB
• std::vector<int >numrhsevals
• std::vector<int >numrhsevalsB
• std::vector<int >numerrtestfails
• std::vector<int >numerrtestfailsB
• std::vector<int >numnonlinsolvconvfails
• std::vector<int >numnonlinsolvconvfailsB
• std::vector<int >order
• int newton_status = 0
Generated by Doxygen

10.30 ReturnData Class Reference 391
• double newton_cpu_time = 0.0
• std::vector<int >newton_numsteps
• std::vector<int >newton_numlinsteps
•realtype t_steadystate = NAN
•realtype wrms_steadystate = NAN
•realtype wrms_sensi_steadystate = NAN
• std::vector<realtype >x0
• std::vector<realtype >sx0
•realtype llh = 0.0
•realtype chi2 = 0.0
• std::vector<realtype >sllh
• std::vector<realtype >s2llh
• int status = 0
• const int np
• const int nk
• const int nx
• const int nxtrue
• const int ny
• const int nytrue
• const int nz
• const int nztrue
• const int ne
• const int nJ
• const int nplist
• const int nmaxevent
• const int nt
• const int newton_maxsteps
• std::vector<ParameterScaling >pscale
• const SecondOrderMode o2mode
• const SensitivityOrder sensi
• const SensitivityMethod sensi_meth
Friends
•template<class Archive >
void boost::serialization::serialize (Archive &ar, ReturnData &r, const unsigned int version)
Serialize ReturnData (see boost::serialization::serialize)
10.30.1 Detailed Description
NOTE: multidimensional arrays are stored in row-major order (FORTRAN-style)
Definition at line 28 of file rdata.h.
10.30.2 Constructor & Destructor Documentation
Generated by Doxygen

392 CONTENTS
10.30.2.1 ReturnData() [1/2]
ReturnData (
std::vector<realtype >ts,
int np,
int nk,
int nx,
int nxtrue,
int ny,
int nytrue,
int nz,
int nztrue,
int ne,
int nJ,
int nplist,
int nmaxevent,
int nt,
int newton_maxsteps,
std::vector<ParameterScaling >pscale,
SecondOrderMode o2mode,
SensitivityOrder sensi,
SensitivityMethod sensi_meth )
Parameters
ts see amici::Model::ts
np see amici::Model::np
nk see amici::Model::nk
nx see amici::Model::nx
nxtrue see amici::Model::nxtrue
ny see amici::Model::ny
nytrue see amici::Model::nytrue
nz see amici::Model::nz
nztrue see amici::Model::nztrue
ne see amici::Model::ne
nJ see amici::Model::nJ
nplist see amici::Model::nplist
nmaxevent see amici::Model::nmaxevent
nt see amici::Model::nt
newton_maxsteps see amici::Solver::newton_maxsteps
pscale see amici::Model::pscale
o2mode see amici::Model::o2mode
sensi see amici::Solver::sensi
sensi_meth see amici::Solver::sensi_meth
Definition at line 40 of file rdata.cpp.
Generated by Doxygen
10.30 ReturnData Class Reference 393
Here is the call graph for this function:
ReturnData amici::getNaN
10.30.2.2 ReturnData() [2/2]
ReturnData (
Solver const & solver,
const Model ∗model )
constructor that uses information from model and solver to appropriately initialize fields
Parameters
solver solver
model pointer to model specification object bool
Definition at line 22 of file rdata.cpp.
10.30.3 Member Function Documentation
10.30.3.1 invalidate()
void invalidate (
const realtype t)
routine to set likelihood, state variables, outputs and respective sensitivities to NaN (typically after integration failure)
Parameters
ttime of integration failure
Definition at line 117 of file rdata.cpp.
Generated by Doxygen
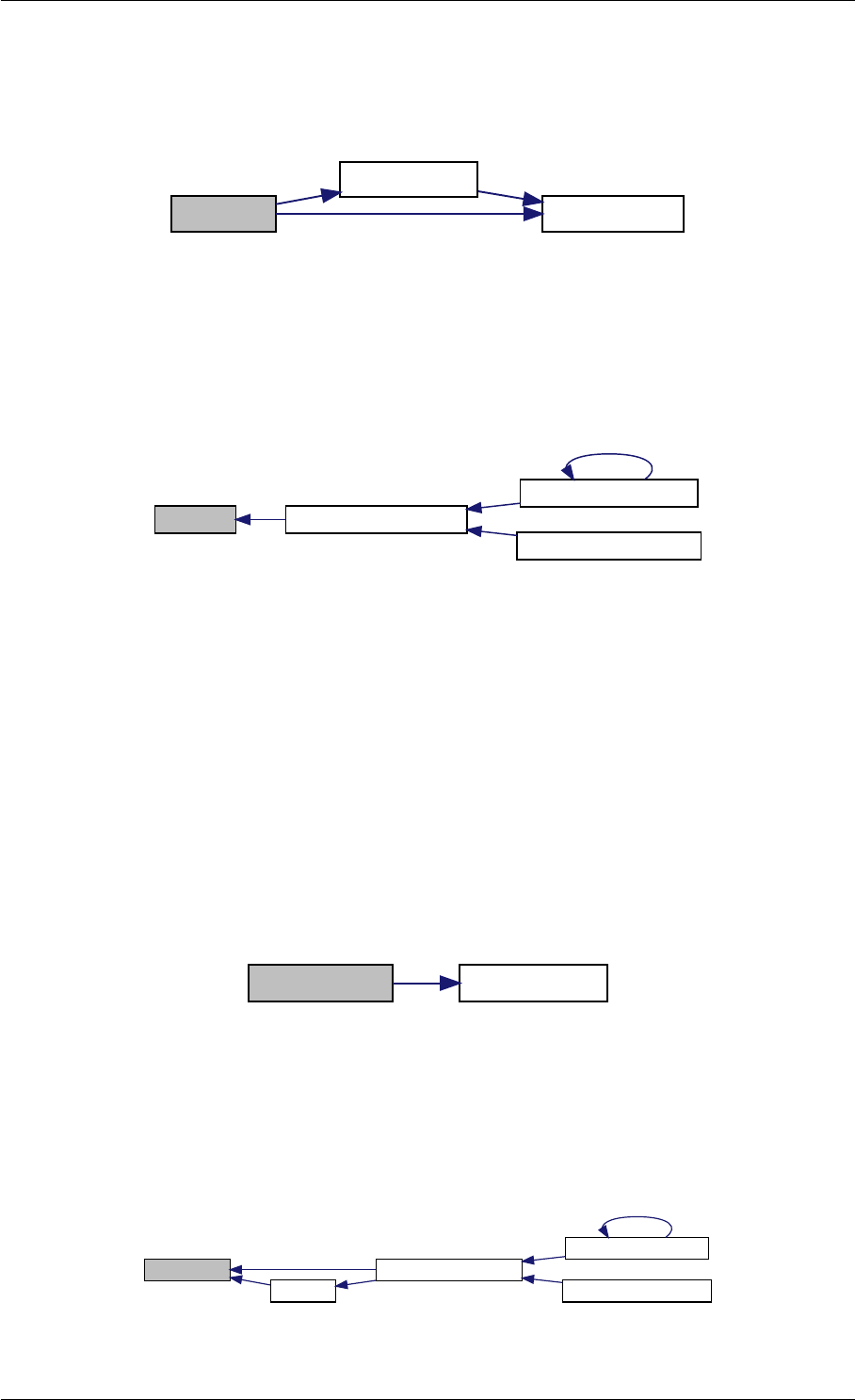
394 CONTENTS
Here is the call graph for this function:
invalidate
invalidateLLH
amici::getNaN
Here is the caller graph for this function:
invalidate amici::runAmiciSimulation
amici.runAmiciSimulation
amici.runAmiciSimulations
10.30.3.2 invalidateLLH()
void invalidateLLH ( )
routine to set likelihood and respective sensitivities to NaN (typically after integration failure)
Definition at line 156 of file rdata.cpp.
Here is the call graph for this function:
invalidateLLH amici::getNaN
Here is the caller graph for this function:
invalidateLLH amici::runAmiciSimulation
invalidate
amici.runAmiciSimulation
amici.runAmiciSimulations
Generated by Doxygen
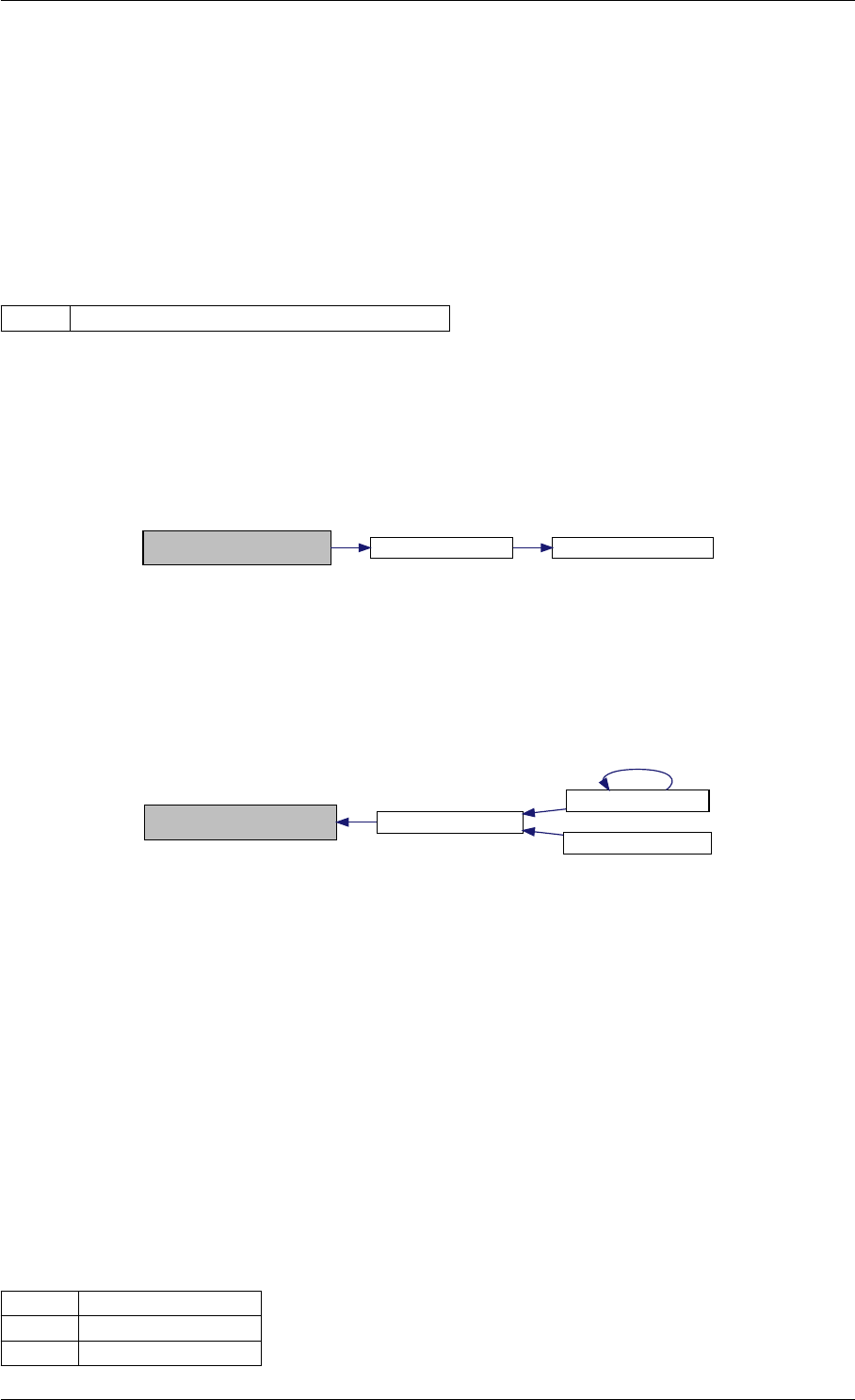
10.30 ReturnData Class Reference 395
10.30.3.3 applyChainRuleFactorToSimulationResults()
void applyChainRuleFactorToSimulationResults (
const Model ∗model )
applies the chain rule to account for parameter transformation in the sensitivities of simulation results
Parameters
model Model from which the ReturnData was obtained
Definition at line 168 of file rdata.cpp.
Here is the call graph for this function:
applyChainRuleFactorToSimulation
Results amici::unscaleParameters amici::getUnscaledParameter
Here is the caller graph for this function:
applyChainRuleFactorToSimulation
Results amici::runAmiciSimulation
amici.runAmiciSimulation
amici.runAmiciSimulations
10.30.4 Friends And Related Function Documentation
10.30.4.1 boost::serialization::serialize
void boost::serialization::serialize (
Archive & ar,
ReturnData &r,
const unsigned int version ) [friend]
Parameters
ar Archive to serialize to
rData to serialize
version Version number
Generated by Doxygen

396 CONTENTS
10.30.5 Member Data Documentation
10.30.5.1 ts
const std::vector<realtype>ts
timepoints (dimension: nt)
Definition at line 77 of file rdata.h.
10.30.5.2 xdot
std::vector<realtype>xdot
time derivative (dimension: nx)
Definition at line 80 of file rdata.h.
10.30.5.3 J
std::vector<realtype>J
Jacobian of differential equation right hand side (dimension: nx x nx, row-major)
Definition at line 84 of file rdata.h.
10.30.5.4 z
std::vector<realtype>z
event output (dimension: nmaxevent x nz, row-major)
Definition at line 87 of file rdata.h.
10.30.5.5 sigmaz
std::vector<realtype>sigmaz
event output sigma standard deviation (dimension: nmaxevent x nz, row-major)
Definition at line 91 of file rdata.h.
Generated by Doxygen

10.30 ReturnData Class Reference 397
10.30.5.6 sz
std::vector<realtype>sz
parameter derivative of event output (dimension: nmaxevent x nz, row-major)
Definition at line 95 of file rdata.h.
10.30.5.7 ssigmaz
std::vector<realtype>ssigmaz
parameter derivative of event output standard deviation (dimension: nmaxevent x nz, row-major)
Definition at line 99 of file rdata.h.
10.30.5.8 rz
std::vector<realtype>rz
event trigger output (dimension: nmaxevent x nz, row-major)
Definition at line 102 of file rdata.h.
10.30.5.9 srz
std::vector<realtype>srz
parameter derivative of event trigger output (dimension: nmaxevent x nz x nplist, row-major)
Definition at line 106 of file rdata.h.
10.30.5.10 s2rz
std::vector<realtype>s2rz
second order parameter derivative of event trigger output (dimension: nmaxevent x nztrue x nplist x nplist, row-
major)
Definition at line 110 of file rdata.h.
Generated by Doxygen

398 CONTENTS
10.30.5.11 x
std::vector<realtype>x
state (dimension: nt x nx, row-major)
Definition at line 113 of file rdata.h.
10.30.5.12 sx
std::vector<realtype>sx
parameter derivative of state (dimension: nt x nplist x nx, row-major)
Definition at line 117 of file rdata.h.
10.30.5.13 y
std::vector<realtype>y
observable (dimension: nt x ny, row-major)
Definition at line 120 of file rdata.h.
10.30.5.14 sigmay
std::vector<realtype>sigmay
observable standard deviation (dimension: nt x ny, row-major)
Definition at line 123 of file rdata.h.
10.30.5.15 sy
std::vector<realtype>sy
parameter derivative of observable (dimension: nt x nplist x ny, row-major)
Definition at line 127 of file rdata.h.
Generated by Doxygen

10.30 ReturnData Class Reference 399
10.30.5.16 ssigmay
std::vector<realtype>ssigmay
parameter derivative of observable standard deviation (dimension: nt x nplist x ny, row-major)
Definition at line 131 of file rdata.h.
10.30.5.17 res
std::vector<realtype>res
observable (dimension: nt∗ny, row-major)
Definition at line 134 of file rdata.h.
10.30.5.18 sres
std::vector<realtype>sres
parameter derivative of residual (dimension: nt∗ny x nplist, row-major)
Definition at line 138 of file rdata.h.
10.30.5.19 FIM
std::vector<realtype>FIM
fisher information matrix (dimension: nplist x nplist, row-major)
Definition at line 142 of file rdata.h.
10.30.5.20 numsteps
std::vector<int>numsteps
number of integration steps forward problem (dimension: nt)
Definition at line 145 of file rdata.h.
Generated by Doxygen

400 CONTENTS
10.30.5.21 numstepsB
std::vector<int>numstepsB
number of integration steps backward problem (dimension: nt)
Definition at line 148 of file rdata.h.
10.30.5.22 numrhsevals
std::vector<int>numrhsevals
number of right hand side evaluations forward problem (dimension: nt)
Definition at line 151 of file rdata.h.
10.30.5.23 numrhsevalsB
std::vector<int>numrhsevalsB
number of right hand side evaluations backwad problem (dimension: nt)
Definition at line 154 of file rdata.h.
10.30.5.24 numerrtestfails
std::vector<int>numerrtestfails
number of error test failures forward problem (dimension: nt)
Definition at line 157 of file rdata.h.
10.30.5.25 numerrtestfailsB
std::vector<int>numerrtestfailsB
number of error test failures backwad problem (dimension: nt)
Definition at line 160 of file rdata.h.
Generated by Doxygen

10.30 ReturnData Class Reference 401
10.30.5.26 numnonlinsolvconvfails
std::vector<int>numnonlinsolvconvfails
number of linear solver convergence failures forward problem (dimension: nt)
Definition at line 164 of file rdata.h.
10.30.5.27 numnonlinsolvconvfailsB
std::vector<int>numnonlinsolvconvfailsB
number of linear solver convergence failures backwad problem (dimension: nt)
Definition at line 168 of file rdata.h.
10.30.5.28 order
std::vector<int>order
employed order forward problem (dimension: nt)
Definition at line 171 of file rdata.h.
10.30.5.29 newton_status
int newton_status = 0
flag indicating success of Newton solver
Definition at line 174 of file rdata.h.
10.30.5.30 newton_cpu_time
double newton_cpu_time = 0.0
computation time of the Newton solver [s]
Definition at line 177 of file rdata.h.
Generated by Doxygen

402 CONTENTS
10.30.5.31 newton_numsteps
std::vector<int>newton_numsteps
number of Newton steps for steady state problem (length = 2)
Definition at line 180 of file rdata.h.
10.30.5.32 newton_numlinsteps
std::vector<int>newton_numlinsteps
number of linear steps by Newton step for steady state problem (length = newton_maxsteps ∗2)
Definition at line 183 of file rdata.h.
10.30.5.33 t_steadystate
realtype t_steadystate = NAN
time at which steadystate was reached in the simulation based approach
Definition at line 186 of file rdata.h.
10.30.5.34 wrms_steadystate
realtype wrms_steadystate = NAN
weighted root-mean-square of the rhs when steadystate was reached
Definition at line 190 of file rdata.h.
10.30.5.35 wrms_sensi_steadystate
realtype wrms_sensi_steadystate = NAN
weighted root-mean-square of the rhs when steadystate was reached
Definition at line 194 of file rdata.h.
Generated by Doxygen

10.30 ReturnData Class Reference 403
10.30.5.36 x0
std::vector<realtype>x0
preequilibration steady state found be Newton solver (dimension: nx)
Definition at line 197 of file rdata.h.
10.30.5.37 sx0
std::vector<realtype>sx0
preequilibration sensitivities found be Newton solver (dimension: nplist x nx, row-major)
Definition at line 200 of file rdata.h.
10.30.5.38 llh
realtype llh = 0.0
loglikelihood value
Definition at line 203 of file rdata.h.
10.30.5.39 chi2
realtype chi2 = 0.0
chi2 value
Definition at line 206 of file rdata.h.
10.30.5.40 sllh
std::vector<realtype>sllh
parameter derivative of loglikelihood (dimension: nplist)
Definition at line 209 of file rdata.h.
Generated by Doxygen

404 CONTENTS
10.30.5.41 s2llh
std::vector<realtype>s2llh
second order parameter derivative of loglikelihood (dimension: (nJ-1) x nplist, row-major)
Definition at line 213 of file rdata.h.
10.30.5.42 status
int status = 0
status code
Definition at line 216 of file rdata.h.
10.30.5.43 np
const int np
total number of model parameters
Definition at line 219 of file rdata.h.
10.30.5.44 nk
const int nk
number of fixed parameters
Definition at line 221 of file rdata.h.
10.30.5.45 nx
const int nx
number of states
Definition at line 223 of file rdata.h.
Generated by Doxygen

10.30 ReturnData Class Reference 405
10.30.5.46 nxtrue
const int nxtrue
number of states in the unaugmented system
Definition at line 225 of file rdata.h.
10.30.5.47 ny
const int ny
number of observables
Definition at line 227 of file rdata.h.
10.30.5.48 nytrue
const int nytrue
number of observables in the unaugmented system
Definition at line 229 of file rdata.h.
10.30.5.49 nz
const int nz
number of event outputs
Definition at line 231 of file rdata.h.
10.30.5.50 nztrue
const int nztrue
number of event outputs in the unaugmented system
Definition at line 233 of file rdata.h.
Generated by Doxygen

406 CONTENTS
10.30.5.51 ne
const int ne
number of events
Definition at line 235 of file rdata.h.
10.30.5.52 nJ
const int nJ
dimension of the augmented objective function for 2nd order ASA
Definition at line 237 of file rdata.h.
10.30.5.53 nplist
const int nplist
number of parameter for which sensitivities were requested
Definition at line 240 of file rdata.h.
10.30.5.54 nmaxevent
const int nmaxevent
maximal number of occuring events (for every event type)
Definition at line 242 of file rdata.h.
10.30.5.55 nt
const int nt
number of considered timepoints
Definition at line 244 of file rdata.h.
Generated by Doxygen

10.30 ReturnData Class Reference 407
10.30.5.56 newton_maxsteps
const int newton_maxsteps
maximal number of newton iterations for steady state calculation
Definition at line 246 of file rdata.h.
10.30.5.57 pscale
std::vector<ParameterScaling>pscale
scaling of parameterization (lin,log,log10)
Definition at line 248 of file rdata.h.
10.30.5.58 o2mode
const SecondOrderMode o2mode
flag indicating whether second order sensitivities were requested
Definition at line 250 of file rdata.h.
10.30.5.59 sensi
const SensitivityOrder sensi
sensitivity order
Definition at line 252 of file rdata.h.
10.30.5.60 sensi_meth
const SensitivityMethod sensi_meth
sensitivity method
Definition at line 254 of file rdata.h.
Generated by Doxygen

408 CONTENTS
10.31 SBMLException Class Reference
Inheritance diagram for SBMLException:
SBMLException
Exception
10.31.1 Detailed Description
Definition at line 13 of file sbml_import.py.
10.32 SbmlImporter Class Reference
The SbmlImporter class generates AMICI C++ files for a model provided in the Systems Biology Markup Language
(SBML).
Public Member Functions
• def __init__ (self, SBMLFile, check_validity=True)
Create a new Model instance.
• def reset_symbols (self)
Reset the symbols attribute to default values.
• def loadSBMLFile (self, SBMLFile)
Parse the provided SBML file.
• def checkLibSBMLErrors (self)
Checks the error log in the current self.sbml_doc.
• def sbml2amici (self, modelName, output_dir=None, observables=None, constantParameters=None, sig-
mas=None, verbose=False, assume_pow_positivity=False, compiler=None)
Generate AMICI C++ files for the model provided to the constructor.
• def processSBML (self, constantParameters=None)
Read parameters, species, reactions, and so on from SBML model.
• def checkSupport (self)
Check whether all required SBML features are supported.
• def processSpecies (self)
Get species information from SBML model.
• def processParameters (self, constantParameters=None)
Get parameter information from SBML model.
• def processCompartments (self)
Generated by Doxygen

10.32 SbmlImporter Class Reference 409
Get compartment information, stoichiometric matrix and fluxes from SBML model.
• def processReactions (self)
Get reactions from SBML model.
• def processRules (self)
Process Rules defined in the SBML model.
• def processVolumeConversion (self)
Convert equations from amount to volume.
• def processTime (self)
Convert time_symbol into cpp variable.
• def processObservables (self, observables, sigmas)
Perform symbolic computations required for objective function evaluation.
• def replaceInAllExpressions (self, old, new)
Replace 'old' by 'new' in all symbolic expressions.
• def cleanReservedSymbols (self)
Remove all reserved symbols from self.symbols.
• def replaceSpecialConstants (self)
Replace all special constants by their respective SBML csymbol definition.
Public Attributes
•check_validity
indicates whether the validity of the SBML document
•symbols
dict carrying symbolic definitions
Type: dict
•SBMLreader
the libSBML sbml reader [!not storing this will result
•sbml_doc
document carrying the sbml defintion [!not storing this
•sbml
sbml definition [!not storing this will result in a segfault!]
•speciesIndex
maps species names to indices
Type: dict
•speciesCompartment
compartment for each species
Type:
•constantSpecies
ids of species that are marked as constant
Type: list
•boundaryConditionSpecies
ids of species that are marked as boundary
•speciesHasOnlySubstanceUnits
flags indicating whether a species has
•speciesConversionFactor
conversion factors for every species
Type:
•compartmentSymbols
compartment ids
Type: symengine.DenseMatrix
•compartmentVolume
Generated by Doxygen

410 CONTENTS
numeric/symbnolic compartment volumes
Type:
•stoichiometricMatrix
stoichiometrix matrix of the model
Type:
•fluxVector
reaction kinetic laws
Type: symengine.DenseMatrix
10.32.1 Detailed Description
Definition at line 35 of file sbml_import.py.
10.32.2 Constructor & Destructor Documentation
10.32.2.1 __init__()
def __init__ (
self,
SBMLFile,
check_validity = True )
Parameters
SBMLFile Path to SBML file where the model is specified
Type: string
check_validity Flag indicating whether the validity of the SBML document should be checked
Type: bool
Returns
SbmlImporter instance with attached SBML document
Definition at line 143 of file sbml_import.py.
10.32.3 Member Function Documentation
10.32.3.1 reset_symbols()
def reset_symbols (
self )
Generated by Doxygen

10.32 SbmlImporter Class Reference 411
Returns
Definition at line 162 of file sbml_import.py.
Here is the caller graph for this function:
reset_symbols sbml2amici checkLibSBMLErrors
10.32.3.2 loadSBMLFile()
def loadSBMLFile (
self,
SBMLFile )
Parameters
SBMLFile path to SBML file
Type: str
Returns
Definition at line 175 of file sbml_import.py.
10.32.3.3 checkLibSBMLErrors()
def checkLibSBMLErrors (
self )
Returns
raises SBMLException if errors with severity ERROR or FATAL have
Exceptions
occured
Generated by Doxygen
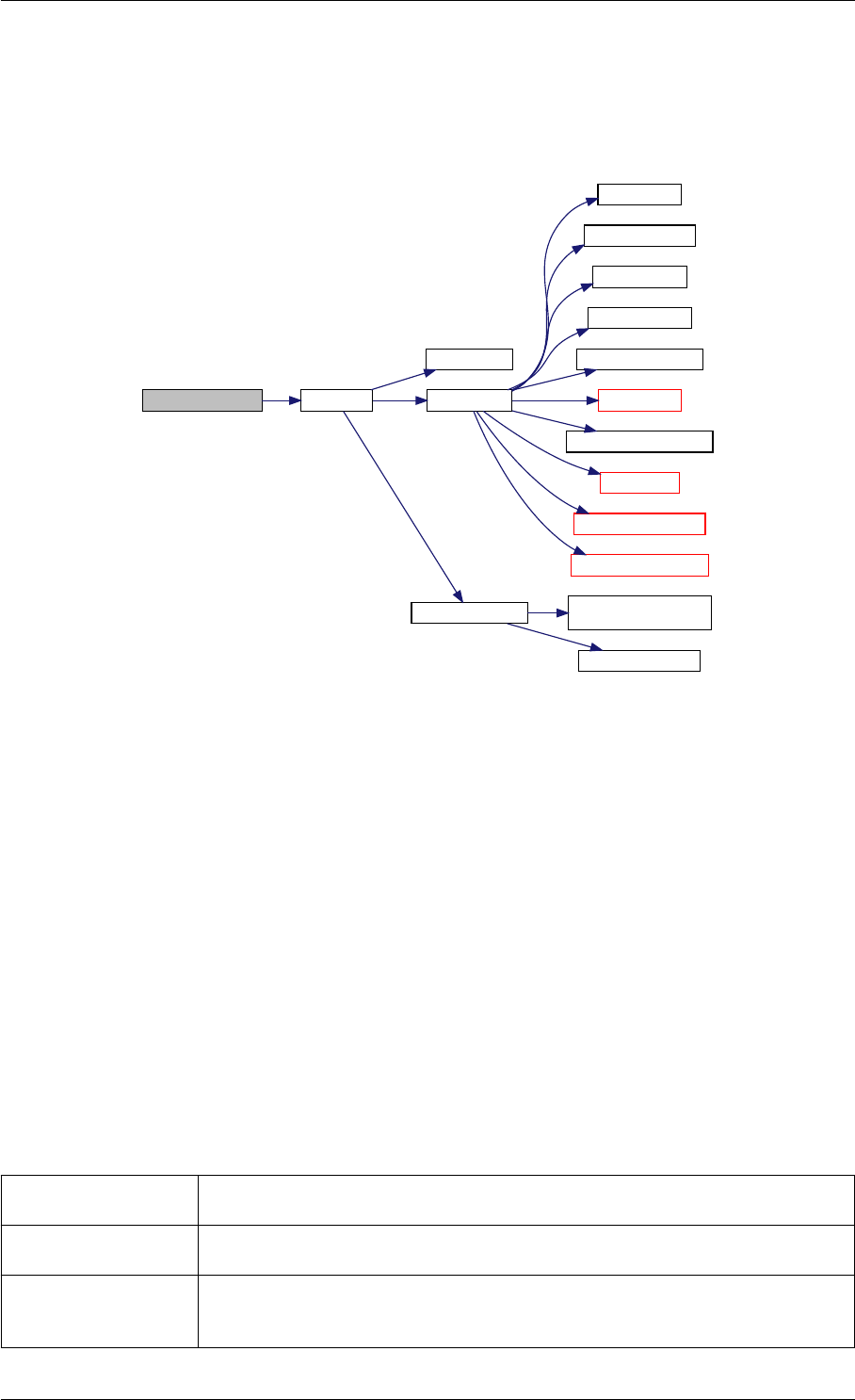
412 CONTENTS
Definition at line 216 of file sbml_import.py.
Here is the call graph for this function:
checkLibSBMLErrors sbml2amici
reset_symbols
processSBML
processObservables
checkSupport
processParameters
processSpecies
processReactions
processCompartments
processRules
processVolumeConversion
processTime
cleanReservedSymbols
replaceSpecialConstants
amici.sbml_import.replace
LogAB
amici.sbml_import.l2s
10.32.3.4 sbml2amici()
def sbml2amici (
self,
modelName,
output_dir = None,
observables = None,
constantParameters = None,
sigmas = None,
verbose = False,
assume_pow_positivity = False,
compiler = None )
Parameters
modelName name of the model/model directory
Type: str
output_dir see sbml_import.setPaths()
Type: str
observables dictionary( observableId:{'name':observableName (optional),
'formula':formulaString)}) to be added to the model
Type: dict
Generated by Doxygen
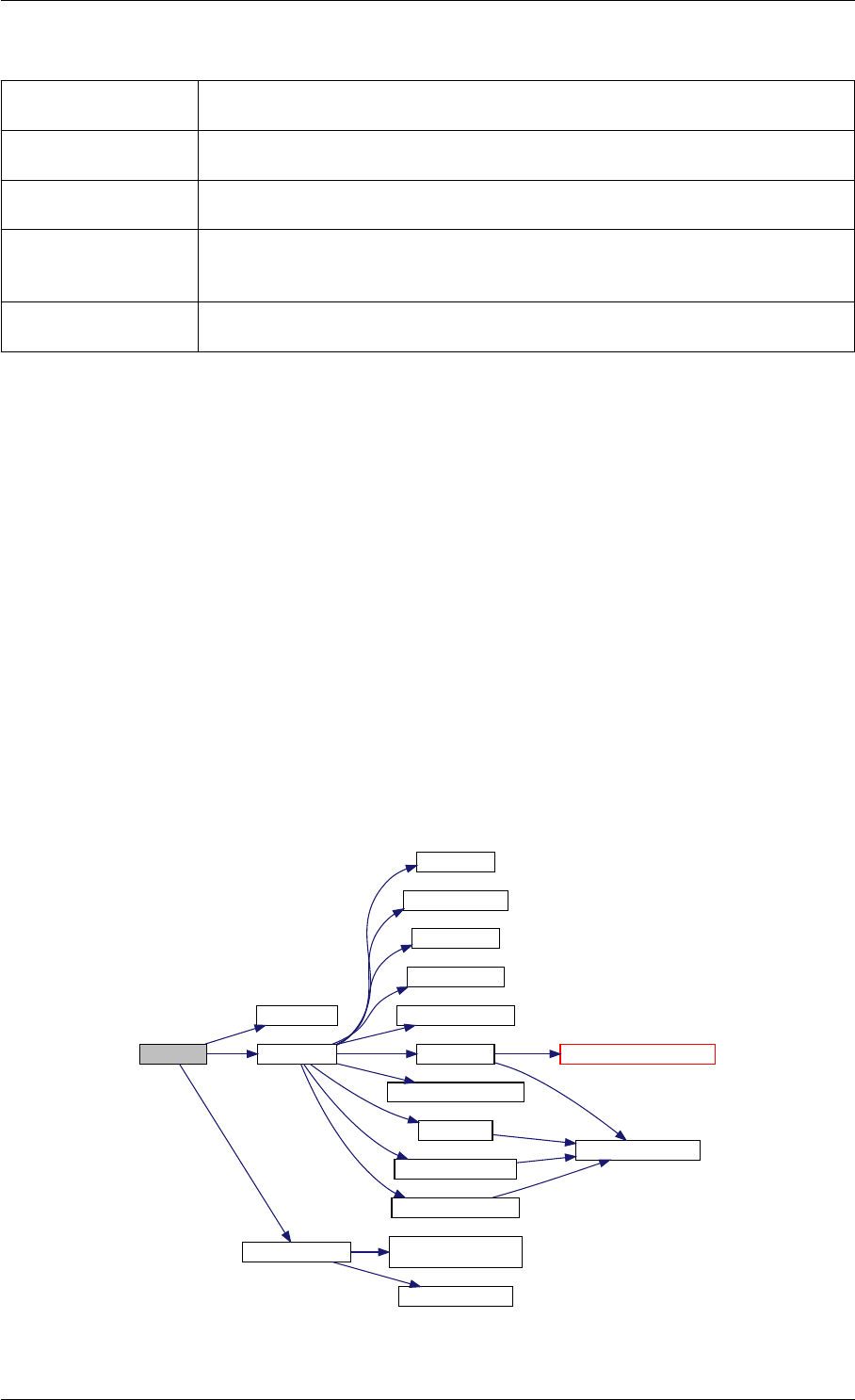
10.32 SbmlImporter Class Reference 413
Parameters
sigmas dictionary(observableId: sigma value or (existing) parameter name)
Type: dict
constantParameters list of SBML Ids identifying constant parameters
Type: dict
verbose more verbose output if True
Type: bool
assume_pow_positivity if set to true, a special pow function is used to avoid problems with state variables
that may become negative due to numerical errors
Type: bool
compiler distutils/setuptools compiler selection to build the python extension
Type: str
Returns
Definition at line 274 of file sbml_import.py.
Here is the call graph for this function:
sbml2amici
reset_symbols
processSBML
processObservables
checkSupport
processParameters
processSpecies
processReactions
processCompartments
processRules
processVolumeConversion
processTime
cleanReservedSymbols
replaceSpecialConstants
amici.sbml_import.getRuleVars
replaceInAllExpressions
amici.sbml_import.replace
LogAB
amici.sbml_import.l2s
Generated by Doxygen
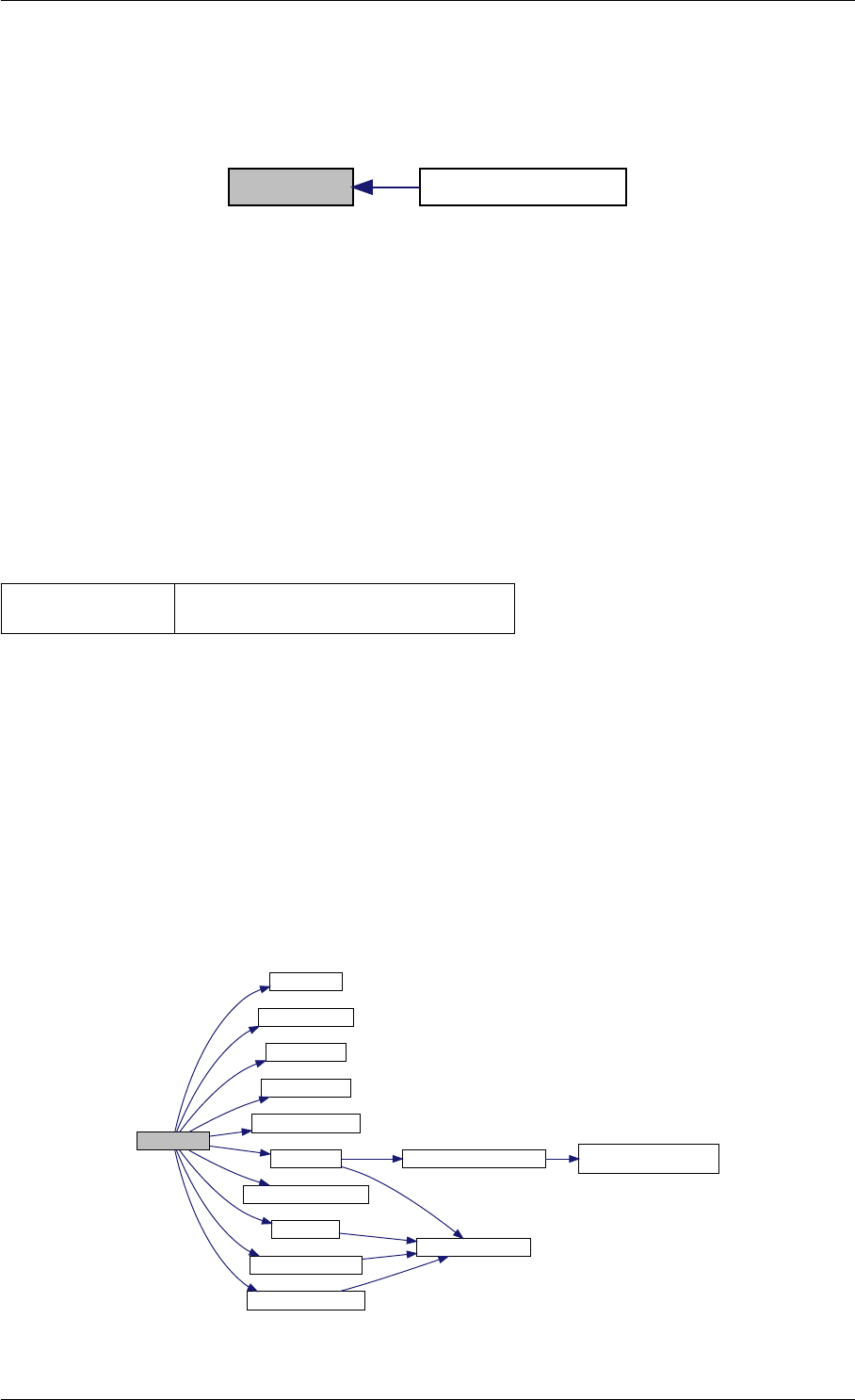
414 CONTENTS
Here is the caller graph for this function:
sbml2amici checkLibSBMLErrors
10.32.3.5 processSBML()
def processSBML (
self,
constantParameters = None )
Parameters
constantParameters SBML Ids identifying constant parameters
Type: list
Returns
Definition at line 312 of file sbml_import.py.
Here is the call graph for this function:
processSBML
checkSupport
processParameters
processSpecies
processReactions
processCompartments
processRules
processVolumeConversion
processTime
cleanReservedSymbols
replaceSpecialConstants
amici.sbml_import.getRuleVars
replaceInAllExpressions
amici.sbml_import.assignment
Rules2observables
Generated by Doxygen
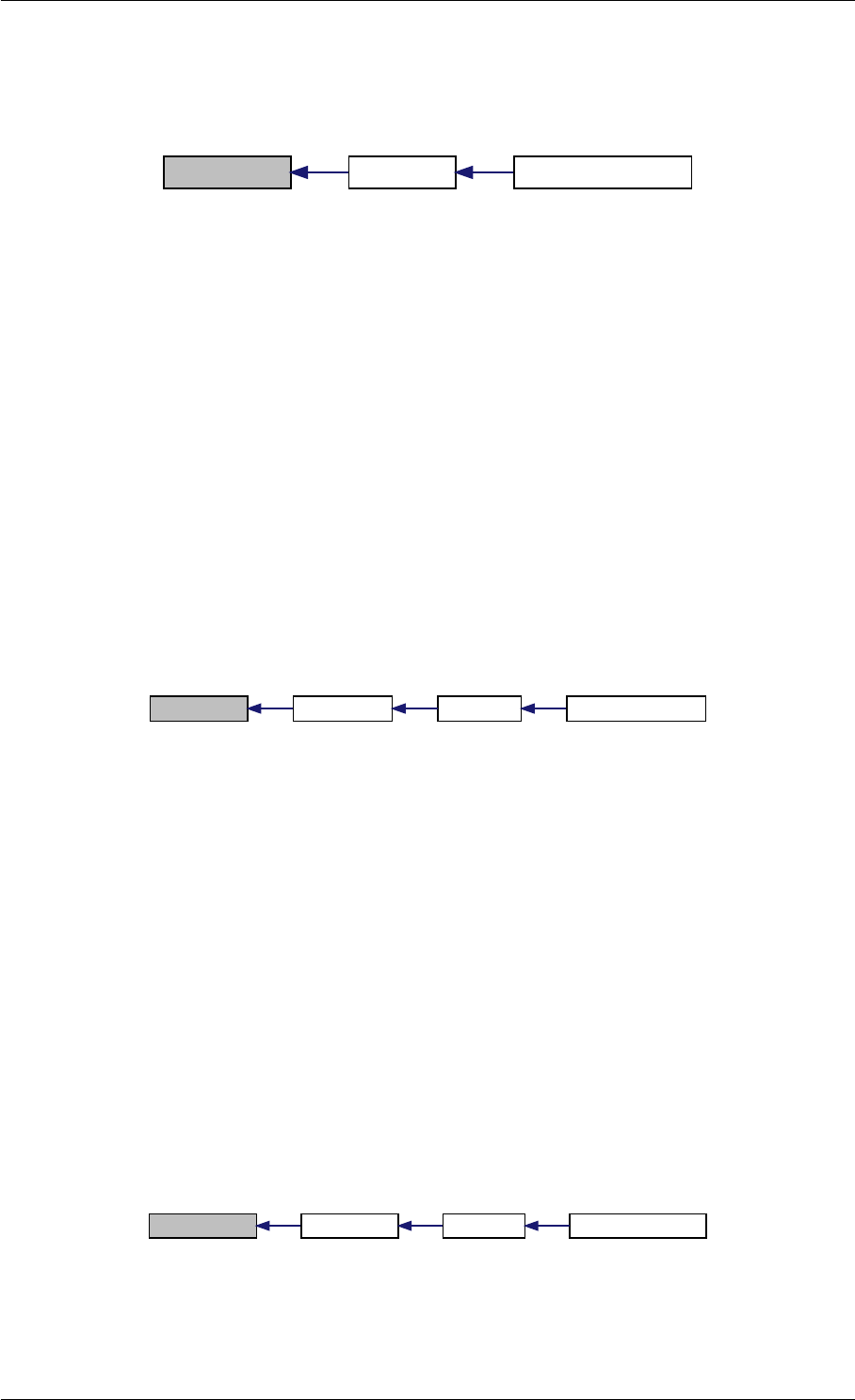
10.32 SbmlImporter Class Reference 415
Here is the caller graph for this function:
processSBML sbml2amici checkLibSBMLErrors
10.32.3.6 checkSupport()
def checkSupport (
self )
Returns
Definition at line 337 of file sbml_import.py.
Here is the caller graph for this function:
checkSupport processSBML sbml2amici checkLibSBMLErrors
10.32.3.7 processSpecies()
def processSpecies (
self )
Returns
Definition at line 374 of file sbml_import.py.
Here is the caller graph for this function:
processSpecies processSBML sbml2amici checkLibSBMLErrors
Generated by Doxygen

416 CONTENTS
10.32.3.8 processParameters()
def processParameters (
self,
constantParameters = None )
Generated by Doxygen
10.32 SbmlImporter Class Reference 417
Parameters
constantParameters SBML Ids identifying constant parameters
Type: list
Returns
Definition at line 466 of file sbml_import.py.
Here is the caller graph for this function:
processParameters processSBML sbml2amici checkLibSBMLErrors
10.32.3.9 processCompartments()
def processCompartments (
self )
Returns
Definition at line 543 of file sbml_import.py.
Here is the caller graph for this function:
processCompartments processSBML sbml2amici checkLibSBMLErrors
Generated by Doxygen
418 CONTENTS
10.32.3.10 processReactions()
def processReactions (
self )
Returns
Definition at line 577 of file sbml_import.py.
Here is the caller graph for this function:
processReactions processSBML sbml2amici checkLibSBMLErrors
10.32.3.11 processRules()
def processRules (
self )
Returns
Definition at line 689 of file sbml_import.py.
Here is the call graph for this function:
processRules
amici.sbml_import.getRuleVars
replaceInAllExpressions
amici.sbml_import.assignment
Rules2observables
Here is the caller graph for this function:
processRules processSBML sbml2amici checkLibSBMLErrors
Generated by Doxygen
10.32 SbmlImporter Class Reference 419
10.32.3.12 processVolumeConversion()
def processVolumeConversion (
self )
Returns
Definition at line 766 of file sbml_import.py.
Here is the caller graph for this function:
processVolumeConversion processSBML sbml2amici checkLibSBMLErrors
10.32.3.13 processTime()
def processTime (
self )
Returns
Definition at line 788 of file sbml_import.py.
Here is the call graph for this function:
processTime replaceInAllExpressions
Here is the caller graph for this function:
processTime processSBML sbml2amici checkLibSBMLErrors
Generated by Doxygen

420 CONTENTS
10.32.3.14 processObservables()
def processObservables (
self,
observables,
sigmas )
Generated by Doxygen
10.32 SbmlImporter Class Reference 421
Parameters
observables dictionary( observableId:{'name':observableName (optional), 'formula':formulaString)}) to be
added to the model
Type: dict
sigmas dictionary(observableId: sigma value or (existing) parameter name)
Type: dict
Returns
Definition at line 810 of file sbml_import.py.
Here is the call graph for this function:
processObservables
amici.sbml_import.replace
LogAB
amici.sbml_import.l2s
Here is the caller graph for this function:
processObservables sbml2amici checkLibSBMLErrors
10.32.3.15 replaceInAllExpressions()
def replaceInAllExpressions (
self,
old,
new )
Generated by Doxygen
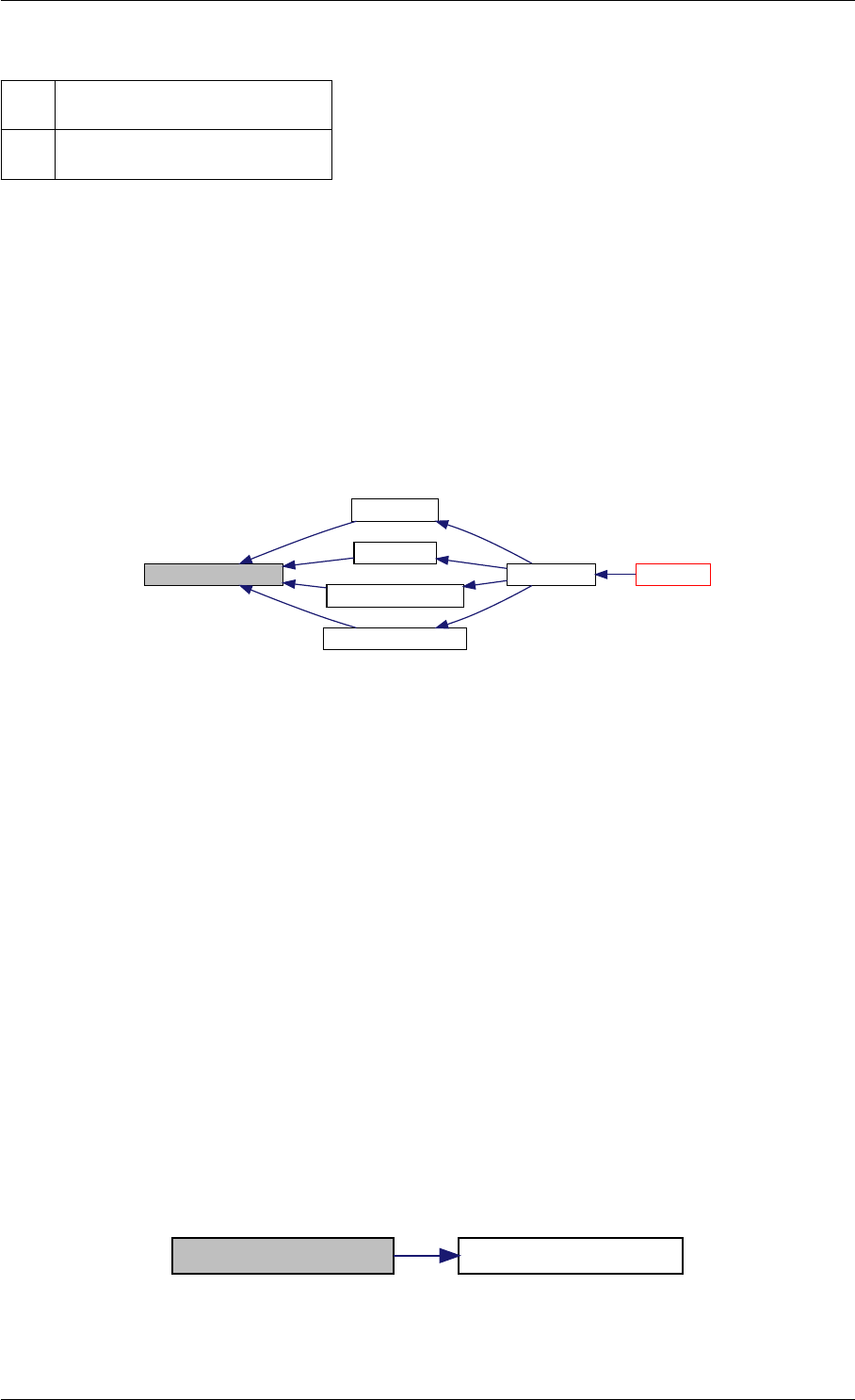
422 CONTENTS
Parameters
old symbolic variables to be replaced
Type: symengine.Symbol
new replacement symbolic variables
Type: symengine.Symbol
Returns
Definition at line 915 of file sbml_import.py.
Here is the caller graph for this function:
replaceInAllExpressions
processRules
processTime
cleanReservedSymbols
replaceSpecialConstants
processSBML sbml2amici
10.32.3.16 cleanReservedSymbols()
def cleanReservedSymbols (
self )
Returns
Definition at line 942 of file sbml_import.py.
Here is the call graph for this function:
cleanReservedSymbols replaceInAllExpressions
Generated by Doxygen
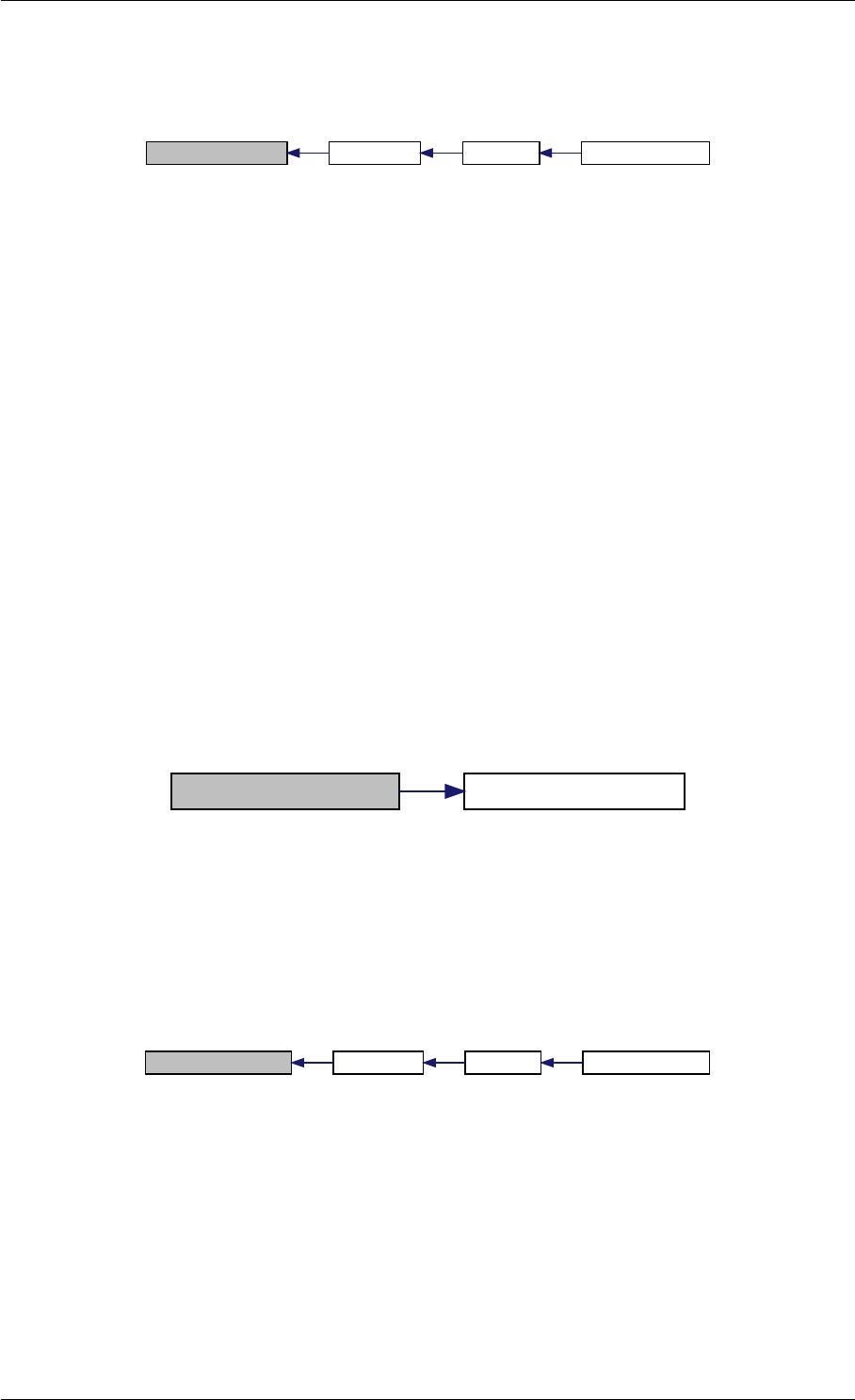
10.33 SetupFailure Class Reference 423
Here is the caller graph for this function:
cleanReservedSymbols processSBML sbml2amici checkLibSBMLErrors
10.32.3.17 replaceSpecialConstants()
def replaceSpecialConstants (
self )
Returns
Definition at line 963 of file sbml_import.py.
Here is the call graph for this function:
replaceSpecialConstants replaceInAllExpressions
Here is the caller graph for this function:
replaceSpecialConstants processSBML sbml2amici checkLibSBMLErrors
10.33 SetupFailure Class Reference
setup failure exception this exception should be thrown when the solver setup failed for this exception we can
assume that we cannot recover from the exception and an error will be thrown
#include <exception.h>
Generated by Doxygen
424 CONTENTS
Inheritance diagram for SetupFailure:
SetupFailure
AmiException
exception
Public Member Functions
•SetupFailure (const char ∗msg)
10.33.1 Detailed Description
Definition at line 188 of file exception.h.
10.33.2 Constructor & Destructor Documentation
10.33.2.1 SetupFailure()
SetupFailure (
const char ∗msg )
constructor, simply calls AmiException constructor
Parameters
msg
Definition at line 193 of file exception.h.
10.34 Solver Class Reference
#include <solver.h>
Generated by Doxygen

10.34 Solver Class Reference 425
Public Member Functions
•Solver (const Solver &other)
Solver copy constructor.
• virtual Solver ∗clone () const =0
Clone this instance.
• void setup (AmiVector ∗x, AmiVector ∗dx, AmiVectorArray ∗sx, AmiVectorArray ∗sdx, Model ∗model)
Initialises the ami memory object and applies specified options.
• void setupAMIB (BackwardProblem ∗bwd, Model ∗model)
• virtual void getSens (realtype ∗tret, AmiVectorArray ∗yySout) const =0
• void getDiagnosis (const int it, ReturnData ∗rdata) const
• void getDiagnosisB (const int it, ReturnData ∗rdata, int which) const
• virtual void getRootInfo (int ∗rootsfound) const =0
• virtual void reInit (realtype t0, AmiVector ∗yy0, AmiVector ∗yp0)=0
• virtual void sensReInit (AmiVectorArray ∗yS0, AmiVectorArray ∗ypS0)=0
• virtual void calcIC (realtype tout1, AmiVector ∗x, AmiVector ∗dx)=0
• virtual void calcICB (int which, realtype tout1, AmiVector ∗xB, AmiVector ∗dxB)=0
• virtual int solve (realtype tout, AmiVector ∗yret, AmiVector ∗ypret, realtype ∗tret, int itask)=0
• virtual int solveF (realtype tout, AmiVector ∗yret, AmiVector ∗ypret, realtype ∗tret, int itask, int ∗ncheckPtr)=0
• virtual void solveB (realtype tBout, int itaskB)=0
• virtual void setStopTime (realtype tstop)=0
• virtual void reInitB (int which, realtype tB0, AmiVector ∗yyB0, AmiVector ∗ypB0)=0
• virtual void getB (int which, realtype ∗tret, AmiVector ∗yy, AmiVector ∗yp) const =0
• virtual void getQuadB (int which, realtype ∗tret, AmiVector ∗qB) const =0
• virtual void quadReInitB (int which, AmiVector ∗yQB0)=0
• virtual void turnOffRootFinding ()=0
•SensitivityMethod getSensitivityMethod () const
• void setSensitivityMethod (SensitivityMethod sensi_meth)
setSensitivityMethod
• int getNewtonMaxSteps () const
getNewtonMaxSteps
• void setNewtonMaxSteps (int newton_maxsteps)
setNewtonMaxSteps
• bool getNewtonPreequilibration () const
getNewtonPreequilibration
• void setNewtonPreequilibration (bool newton_preeq)
setNewtonPreequilibration
• int getNewtonMaxLinearSteps () const
getNewtonMaxLinearSteps
• void setNewtonMaxLinearSteps (int newton_maxlinsteps)
setNewtonMaxLinearSteps
•SensitivityOrder getSensitivityOrder () const
returns the sensitvity order
• void setSensitivityOrder (SensitivityOrder sensi)
sets the sensitvity order
• double getRelativeTolerance () const
returns the relative tolerances for the forward & backward problem
• void setRelativeTolerance (double rtol)
sets the relative tolerances for the forward & backward problem
• double getAbsoluteTolerance () const
returns the absolute tolerances for the forward & backward problem
• void setAbsoluteTolerance (double atol)
Generated by Doxygen

426 CONTENTS
sets the absolute tolerances for the forward & backward problem
• double getRelativeToleranceSensi () const
returns the relative tolerances for the forward sensitivity problem
• void setRelativeToleranceSensi (double rtol)
sets the relative tolerances for the forward sensitivity problem
• double getAbsoluteToleranceSensi () const
returns the absolute tolerances for the forward sensitivity problem
• void setAbsoluteToleranceSensi (double atol)
sets the absolute tolerances for the forward sensitivity problem
• double getRelativeToleranceQuadratures () const
returns the relative tolerance for the quadrature problem
• void setRelativeToleranceQuadratures (double rtol)
sets the relative tolerance for the quadrature problem
• double getAbsoluteToleranceQuadratures () const
returns the absolute tolerance for the quadrature problem
• void setAbsoluteToleranceQuadratures (double atol)
sets the absolute tolerance for the quadrature problem
• double getRelativeToleranceSteadyState () const
returns the relative tolerance for the steady state problem
• void setRelativeToleranceSteadyState (double rtol)
sets the relative tolerance for the steady state problem
• double getAbsoluteToleranceSteadyState () const
returns the absolute tolerance for the steady state problem
• void setAbsoluteToleranceSteadyState (double atol)
sets the absolute tolerance for the steady state problem
• double getRelativeToleranceSteadyStateSensi () const
returns the relative tolerance for the sensitivities of the steady state problem
• void setRelativeToleranceSteadyStateSensi (double rtol)
sets the relative tolerance for the sensitivities of the steady state problem
• double getAbsoluteToleranceSteadyStateSensi () const
returns the absolute tolerance for the sensitivities of the steady state problem
• void setAbsoluteToleranceSteadyStateSensi (double atol)
sets the absolute tolerance for the sensitivities of the steady state problem
• int getMaxSteps () const
returns the maximum number of solver steps for the forward problem
• void setMaxSteps (int maxsteps)
sets the maximum number of solver steps for the forward problem
• int getMaxStepsBackwardProblem () const
returns the maximum number of solver steps for the backward problem
• void setMaxStepsBackwardProblem (int maxsteps)
sets the maximum number of solver steps for the backward problem
•LinearMultistepMethod getLinearMultistepMethod () const
returns the linear system multistep method
• void setLinearMultistepMethod (LinearMultistepMethod lmm)
sets the linear system multistep method
•NonlinearSolverIteration getNonlinearSolverIteration () const
returns the nonlinear system solution method
• void setNonlinearSolverIteration (NonlinearSolverIteration iter)
sets the nonlinear system solution method
•InterpolationType getInterpolationType () const
getInterpolationType
Generated by Doxygen

10.34 Solver Class Reference 427
• void setInterpolationType (InterpolationType interpType)
sets the interpolation of the forward solution that is used for the backwards problem
•StateOrdering getStateOrdering () const
sets KLU state ordering mode
• void setStateOrdering (StateOrdering ordering)
sets KLU state ordering mode (only applies when linsol is set to amici.AMICI_KLU)
• booleantype getStabilityLimitFlag () const
returns stability limit detection mode
• void setStabilityLimitFlag (booleantype stldet)
set stability limit detection mode
•LinearSolver getLinearSolver () const
getLinearSolver
• void setLinearSolver (LinearSolver linsol)
setLinearSolver
•InternalSensitivityMethod getInternalSensitivityMethod () const
returns the internal sensitivity method
• void setInternalSensitivityMethod (InternalSensitivityMethod ism)
sets the internal sensitivity method
Protected Member Functions
• virtual void init (AmiVector ∗x, AmiVector ∗dx, realtype t)=0
• virtual void binit (int which, AmiVector ∗xB, AmiVector ∗dxB, realtype t)=0
• virtual void qbinit (int which, AmiVector ∗qBdot)=0
• virtual void rootInit (int ne)=0
• virtual void sensInit1 (AmiVectorArray ∗sx, AmiVectorArray ∗sdx, int nplist)=0
• virtual void setDenseJacFn ()=0
• virtual void setSparseJacFn ()=0
• virtual void setBandJacFn ()=0
• virtual void setJacTimesVecFn ()=0
• virtual void setDenseJacFnB (int which)=0
• virtual void setSparseJacFnB (int which)=0
• virtual void setBandJacFnB (int which)=0
• virtual void setJacTimesVecFnB (int which)=0
• virtual void allocateSolver ()=0
• virtual void setSStolerances (double rtol, double atol)=0
• virtual void setSensSStolerances (double rtol, double ∗atol)=0
• virtual void setSensErrCon (bool error_corr)=0
• virtual void setQuadErrConB (int which, bool flag)=0
• virtual void setErrHandlerFn ()=0
• virtual void setUserData (Model ∗model)=0
• virtual void setUserDataB (int which, Model ∗model)=0
• virtual void setMaxNumSteps (long int mxsteps)=0
• virtual void setMaxNumStepsB (int which, long int mxstepsB)=0
• virtual void setStabLimDet (int stldet)=0
• virtual void setStabLimDetB (int which, int stldet)=0
• virtual void setId (Model ∗model)=0
• virtual void setSuppressAlg (bool flag)=0
• virtual void setSensParams (realtype ∗p, realtype ∗pbar, int ∗plist)=0
• virtual void getDky (realtype t, int k, AmiVector ∗dky) const =0
• virtual void adjInit ()=0
• virtual void allocateSolverB (int ∗which)=0
Generated by Doxygen

428 CONTENTS
• virtual void setSStolerancesB (int which, realtype relTolB, realtype absTolB)=0
• virtual void quadSStolerancesB (int which, realtype reltolQB, realtype abstolQB)=0
• virtual void dense (int nx)=0
• virtual void denseB (int which, int nx)=0
• virtual void band (int nx, int ubw, int lbw)=0
• virtual void bandB (int which, int nx, int ubw, int lbw)=0
• virtual void diag ()=0
• virtual void diagB (int which)=0
• virtual void spgmr (int prectype, int maxl)=0
• virtual void spgmrB (int which, int prectype, int maxl)=0
• virtual void spbcg (int prectype, int maxl)=0
• virtual void spbcgB (int which, int prectype, int maxl)=0
• virtual void sptfqmr (int prectype, int maxl)=0
• virtual void sptfqmrB (int which, int prectype, int maxl)=0
• virtual void klu (int nx, int nnz, int sparsetype)=0
• virtual void kluSetOrdering (int ordering)=0
• virtual void kluSetOrderingB (int which, int ordering)=0
• virtual void kluB (int which, int nx, int nnz, int sparsetype)=0
• virtual void getNumSteps (void ∗ami_mem, long int ∗numsteps) const =0
• virtual void getNumRhsEvals (void ∗ami_mem, long int ∗numrhsevals) const =0
• virtual void getNumErrTestFails (void ∗ami_mem, long int ∗numerrtestfails) const =0
• virtual void getNumNonlinSolvConvFails (void ∗ami_mem, long int ∗numnonlinsolvconvfails) const =0
• virtual void getLastOrder (void ∗ami_mem, int ∗order) const =0
• void initializeLinearSolver (const Model ∗model)
• void initializeLinearSolverB (const Model ∗model, const int which)
• virtual int nplist () const =0
• virtual int nx () const =0
• virtual const Model ∗getModel () const =0
• virtual bool getMallocDone () const =0
• virtual bool getAdjMallocDone () const =0
• virtual void ∗getAdjBmem (void ∗ami_mem, int which)=0
• void applyTolerances ()
• void applyTolerancesFSA ()
• void applyTolerancesASA (int which)
• void applyQuadTolerancesASA (int which)
• void applySensitivityTolerances ()
Static Protected Member Functions
• static void wrapErrHandlerFn (int error_code, const char ∗module, const char ∗function, char ∗msg, void
∗eh_data)
Protected Attributes
• std::unique_ptr<void, std::function<void(void ∗)> > solverMemory
• std::vector<std::unique_ptr<void, std::function<void(void ∗)>>>solverMemoryB
• bool solverWasCalled = false
•InternalSensitivityMethod ism = InternalSensitivityMethod::simultaneous
•LinearMultistepMethod lmm = LinearMultistepMethod::BDF
•NonlinearSolverIteration iter = NonlinearSolverIteration::newton
•InterpolationType interpType = InterpolationType::hermite
• int maxsteps = 10000
Generated by Doxygen
10.34 Solver Class Reference 429
Friends
•template<class Archive >
void boost::serialization::serialize (Archive &ar, Solver &r, const unsigned int version)
Serialize Solver (see boost::serialization::serialize)
• bool operator== (const Solver &a, const Solver &b)
Check equality of data members.
10.34.1 Detailed Description
Solver class. provides a generic interface to CVode and IDA solvers, individual realizations are realized in the
CVodeSolver and the IDASolver class.
Definition at line 38 of file solver.h.
10.34.2 Constructor & Destructor Documentation
10.34.2.1 Solver()
Solver (
const Solver &other )
Parameters
other
Definition at line 46 of file solver.h.
10.34.3 Member Function Documentation
10.34.3.1 clone()
virtual Solver∗clone ( ) const [pure virtual]
Returns
The clone
10.34.3.2 setup()
void setup (
AmiVector ∗x,
AmiVector ∗dx,
AmiVectorArray ∗sx,
AmiVectorArray ∗sdx,
Model ∗model )
Generated by Doxygen
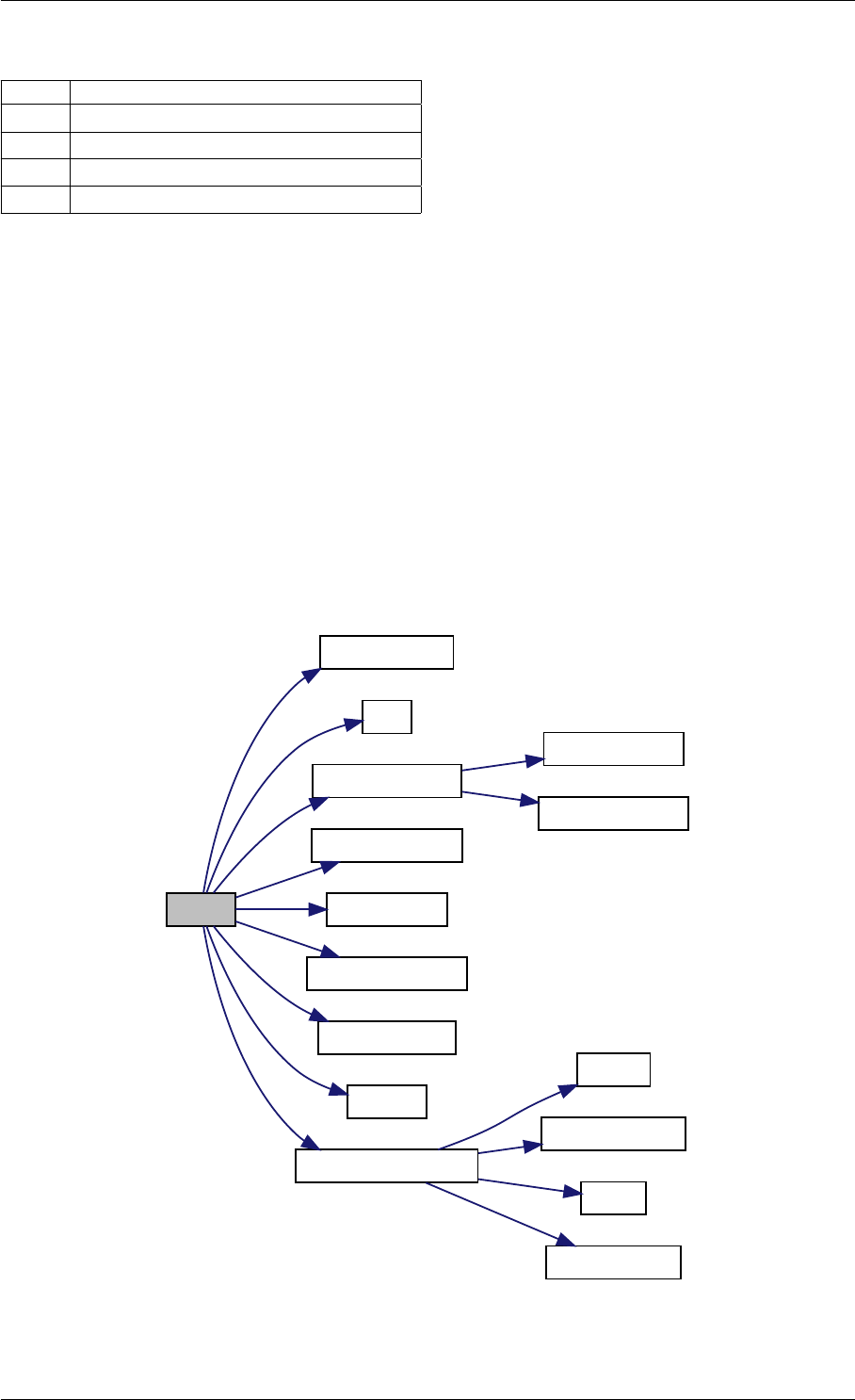
430 CONTENTS
Parameters
xstate vector
dx state derivative vector (DAE only)
sx state sensitivity vector
sdx state derivative sensitivity vector (DAE only)
model pointer to the model object
Definition at line 27 of file solver.cpp.
Here is the call graph for this function:
setup
allocateSolver
init
applyTolerances
setErrHandlerFn
setUserData
setMaxNumSteps
setStabLimDet
rootInit
initializeLinearSolver
getMallocDone
setSStolerances
dense
setDenseJacFn
band
setBandJacFn
Generated by Doxygen
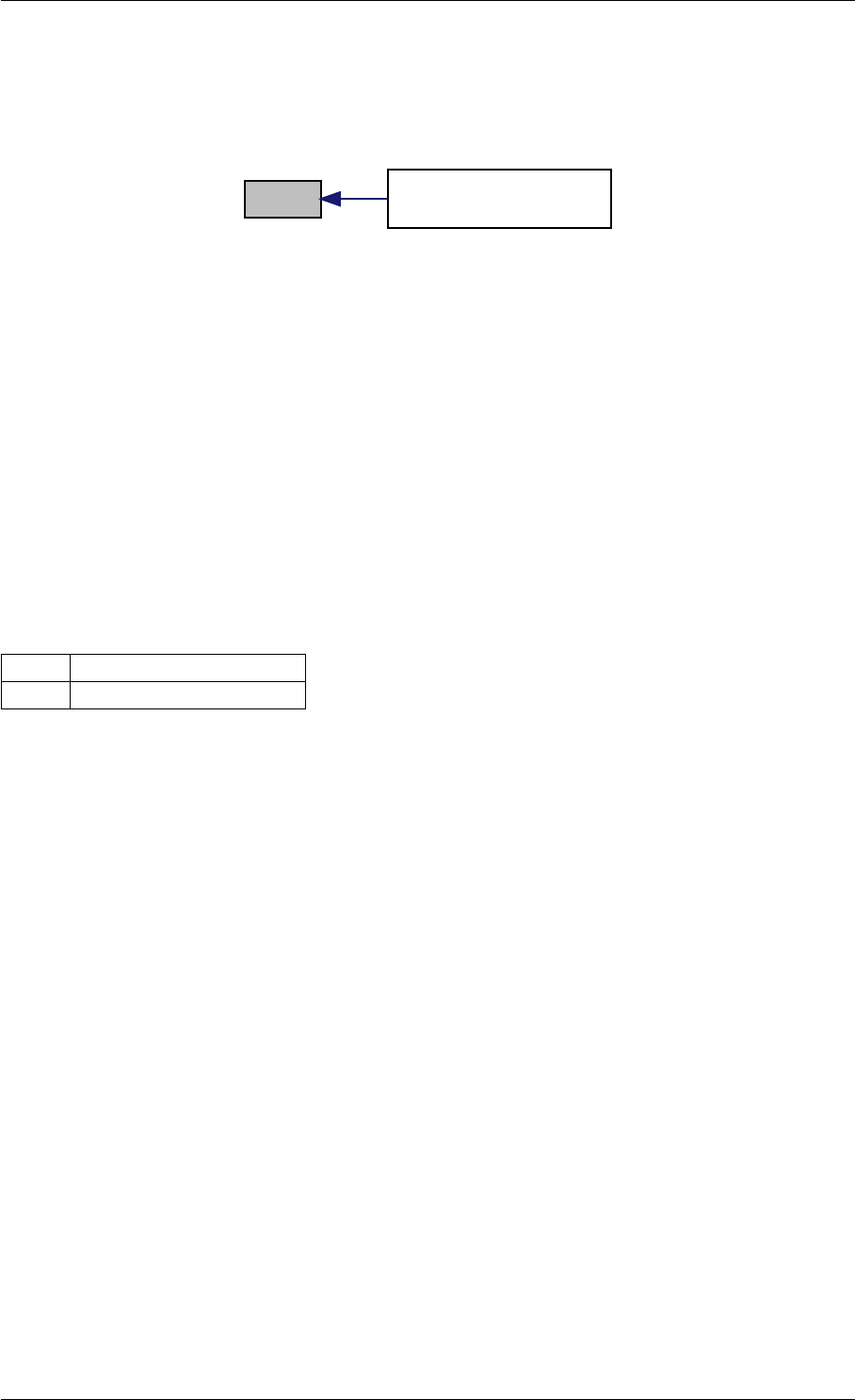
10.34 Solver Class Reference 431
Here is the caller graph for this function:
setup amici::ForwardProblem
::workForwardProblem
10.34.3.3 setupAMIB()
void setupAMIB (
BackwardProblem ∗bwd,
Model ∗model )
setupAMIB initialises the AMI memory object for the backwards problem
Parameters
bwd pointer to backward problem
model pointer to the model object
Definition at line 100 of file solver.cpp.
Generated by Doxygen
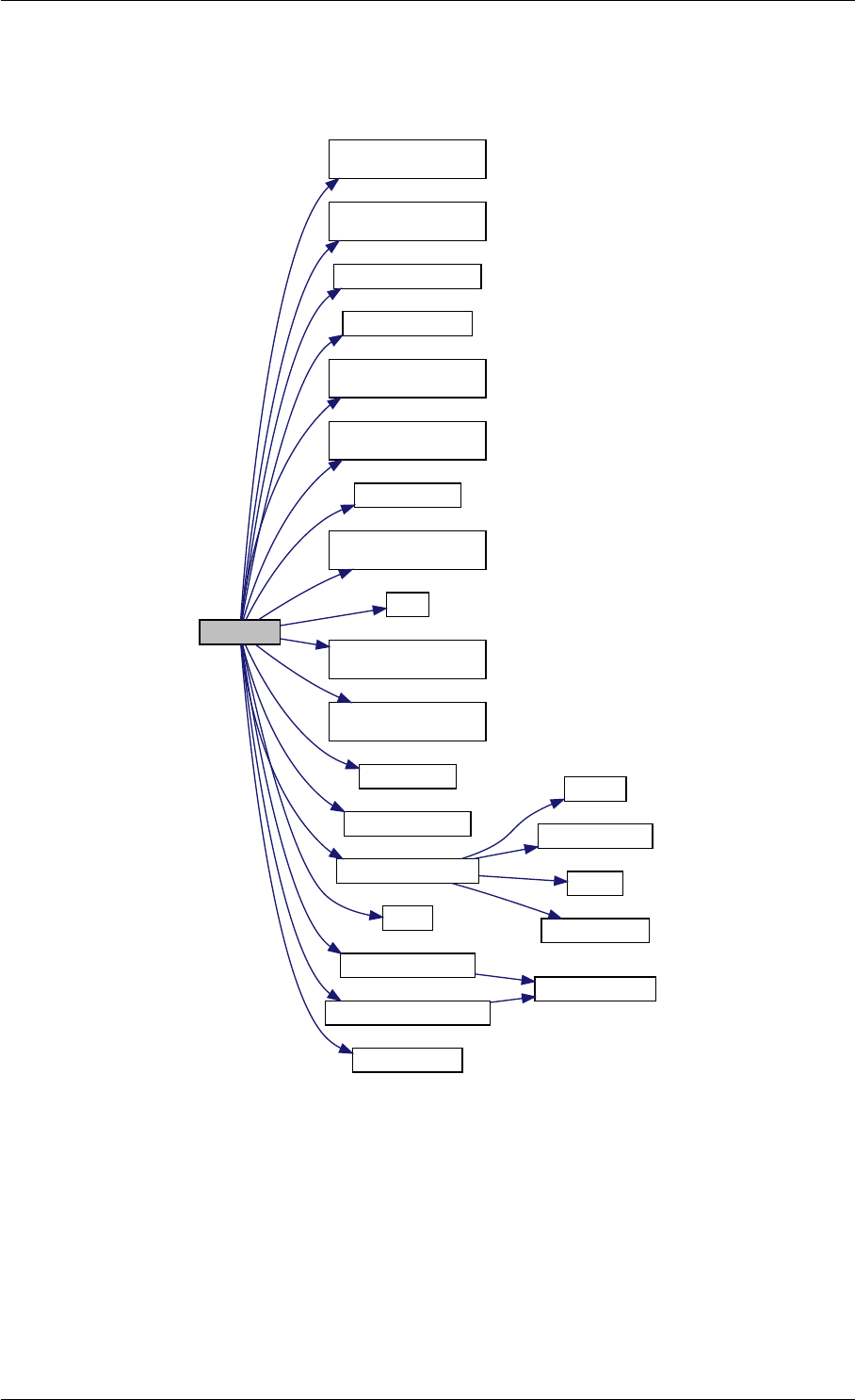
432 CONTENTS
Here is the call graph for this function:
setupAMIB
amici::BackwardProblem
::getdJydx
amici::BackwardProblem
::getxBptr
amici::AmiVector::reset
amici::AmiVector::at
amici::BackwardProblem
::getdxBptr
amici::BackwardProblem
::getxQBptr
allocateSolverB
amici::BackwardProblem
::getwhichptr
binit
amici::BackwardProblem
::getwhich
amici::BackwardProblem
::gett
setUserDataB
setMaxNumStepsB
initializeLinearSolverB
qbinit
applyTolerancesASA
applyQuadTolerancesASA
setStabLimDetB
denseB
setDenseJacFnB
bandB
setBandJacFnB
getAdjMallocDone
10.34.3.4 getSens()
virtual void getSens (
realtype ∗tret,
AmiVectorArray ∗yySout ) const [pure virtual]
getSens extracts diagnosis information from solver memory block and writes them into the return data instance
Generated by Doxygen
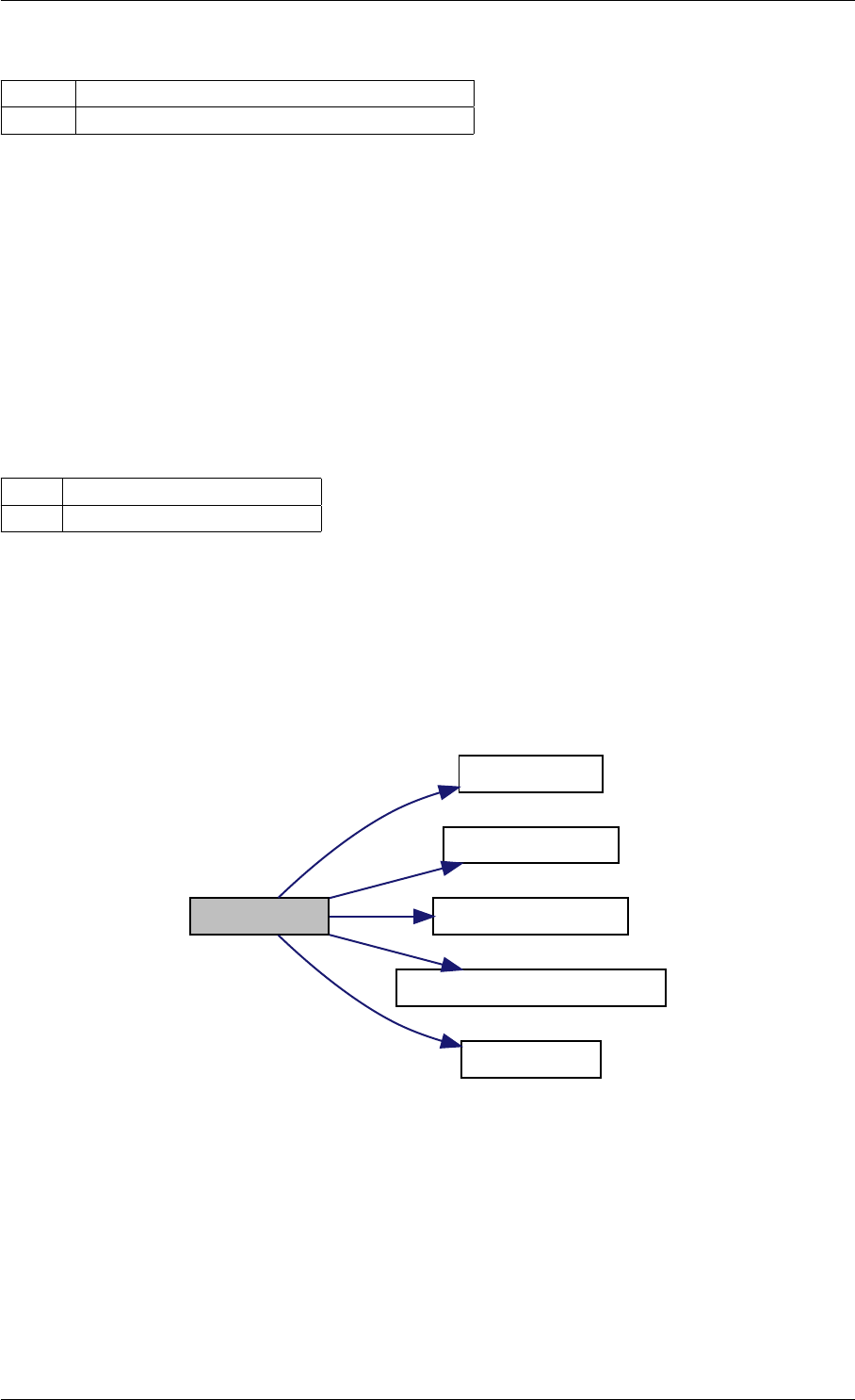
10.34 Solver Class Reference 433
Parameters
tret time at which the sensitivities should be computed
yySout vector with sensitivities
10.34.3.5 getDiagnosis()
void getDiagnosis (
const int it,
ReturnData ∗rdata ) const
getDiagnosis extracts diagnosis information from solver memory block and writes them into the return data object
Parameters
it time-point index
rdata pointer to the return data object
Definition at line 184 of file solver.cpp.
Here is the call graph for this function:
getDiagnosis
getNumSteps
getNumRhsEvals
getNumErrTestFails
getNumNonlinSolvConvFails
getLastOrder
10.34.3.6 getDiagnosisB()
void getDiagnosisB (
const int it,
Generated by Doxygen
434 CONTENTS
ReturnData ∗rdata,
int which ) const
getDiagnosisB extracts diagnosis information from solver memory block and writes them into the return data object
for the backward problem
Parameters
it time-point index
rdata pointer to the return data object
which identifier of the backwards problem
Definition at line 204 of file solver.cpp.
Here is the call graph for this function:
getDiagnosisB
getNumSteps
getNumRhsEvals
getNumErrTestFails
getNumNonlinSolvConvFails
10.34.3.7 getRootInfo()
virtual void getRootInfo (
int ∗rootsfound ) const [pure virtual]
getRootInfo extracts information which event occured
Parameters
rootsfound array with flags indicating whether the respective event occured
10.34.3.8 reInit()
virtual void reInit (
Generated by Doxygen
10.34 Solver Class Reference 435
realtype t0,
AmiVector ∗yy0,
AmiVector ∗yp0 ) [pure virtual]
ReInit reinitializes the states in the solver after an event occurence
Parameters
t0 new timepoint
yy0 new state variables
yp0 new derivative state variables (DAE only)
10.34.3.9 sensReInit()
virtual void sensReInit (
AmiVectorArray ∗yS0,
AmiVectorArray ∗ypS0 ) [pure virtual]
SensReInit reinitializes the state sensitivites in the solver after an event occurence
Parameters
yS0 new state sensitivity
ypS0 new derivative state sensitivities (DAE only)
10.34.3.10 calcIC()
virtual void calcIC (
realtype tout1,
AmiVector ∗x,
AmiVector ∗dx ) [pure virtual]
CalcIC calculates consistent initial conditions, assumes initial states to be correct (DAE only)
Parameters
tout1 next timepoint to be computed (sets timescale)
xinitial state variables
dx initial derivative state variables (DAE only)
10.34.3.11 calcICB()
virtual void calcICB (
int which,
realtype tout1,
Generated by Doxygen

436 CONTENTS
AmiVector ∗xB,
AmiVector ∗dxB ) [pure virtual]
CalcIBC calculates consistent initial conditions for the backwards problem, assumes initial states to be correct (DAE
only)
Parameters
which identifier of the backwards problem
tout1 next timepoint to be computed (sets timescale)
xB states of final solution of the forward problem
dxB derivative states of final solution of the forward problem (DAE only)
10.34.3.12 solve()
virtual int solve (
realtype tout,
AmiVector ∗yret,
AmiVector ∗ypret,
realtype ∗tret,
int itask ) [pure virtual]
Solve solves the forward problem until a predefined timepoint
Parameters
tout timepoint until which simulation should be performed
yret states
ypret derivative states (DAE only)
tret pointer to the time variable
itask task identifier, can be CV_NORMAL or CV_ONE_STEP
Returns
status flag indicating success of execution
Here is the caller graph for this function:
solve amici::SteadystateProblem
::getSteadystateSimulation
Generated by Doxygen

10.34 Solver Class Reference 437
10.34.3.13 solveF()
virtual int solveF (
realtype tout,
AmiVector ∗yret,
AmiVector ∗ypret,
realtype ∗tret,
int itask,
int ∗ncheckPtr ) [pure virtual]
SolveF solves the forward problem until a predefined timepoint (adjoint only)
Parameters
tout timepoint until which simulation should be performed
yret states
ypret derivative states (DAE only)
tret pointer to the time variable
itask task identifier, can be CV_NORMAL or CV_ONE_STEP
ncheckPtr pointer to a number that counts the internal checkpoints
Returns
status flag indicating success of execution
10.34.3.14 solveB()
virtual void solveB (
realtype tBout,
int itaskB ) [pure virtual]
SolveB solves the backward problem until a predefined timepoint (adjoint only)
Parameters
tBout timepoint until which simulation should be performed
itaskB task identifier, can be CV_NORMAL or CV_ONE_STEP
10.34.3.15 setStopTime()
virtual void setStopTime (
realtype tstop ) [pure virtual]
SetStopTime sets a timepoint at which the simulation will be stopped
Parameters
tstop timepoint until which simulation should be performed
Generated by Doxygen
438 CONTENTS
10.34.3.16 reInitB()
virtual void reInitB (
int which,
realtype tB0,
AmiVector ∗yyB0,
AmiVector ∗ypB0 ) [pure virtual]
ReInitB reinitializes the adjoint states after an event occurence
Parameters
which identifier of the backwards problem
tB0 new timepoint
yyB0 new adjoint state variables
ypB0 new adjoint derivative state variables (DAE only)
10.34.3.17 getB()
virtual void getB (
int which,
realtype ∗tret,
AmiVector ∗yy,
AmiVector ∗yp ) const [pure virtual]
getB returns the current adjoint states
Parameters
which identifier of the backwards problem
tret time at which the adjoint states should be computed
yy adjoint state variables
yp adjoint derivative state variables (DAE only)
10.34.3.18 getQuadB()
virtual void getQuadB (
int which,
realtype ∗tret,
AmiVector ∗qB ) const [pure virtual]
getQuadB returns the current adjoint states
Parameters
which identifier of the backwards problem
tret time at which the adjoint states should be computed
qB adjoint quadrature state variables Generated by Doxygen
10.34 Solver Class Reference 439
10.34.3.19 quadReInitB()
virtual void quadReInitB (
int which,
AmiVector ∗yQB0 ) [pure virtual]
ReInitB reinitializes the adjoint states after an event occurence
Parameters
which identifier of the backwards problem
yQB0 new adjoint quadrature state variables
10.34.3.20 turnOffRootFinding()
virtual void turnOffRootFinding ( ) [pure virtual]
turnOffRootFinding disables rootfinding
10.34.3.21 getSensitivityMethod()
SensitivityMethod getSensitivityMethod ( ) const
sensitivity method
Returns
method enum
Definition at line 471 of file solver.cpp.
Here is the caller graph for this function:
getSensitivityMethod amici::ForwardProblem
::workForwardProblem
10.34.3.22 setSensitivityMethod()
void setSensitivityMethod (
SensitivityMethod sensi_meth )
Generated by Doxygen
440 CONTENTS
Parameters
sensi_meth
Definition at line 475 of file solver.cpp.
Here is the caller graph for this function:
setSensitivityMethod amici::setSolverOptions
10.34.3.23 getNewtonMaxSteps()
int getNewtonMaxSteps ( ) const
Returns
Definition at line 479 of file solver.cpp.
Here is the caller graph for this function:
getNewtonMaxSteps amici::SteadystateProblem
::workSteadyStateProblem
10.34.3.24 setNewtonMaxSteps()
void setNewtonMaxSteps (
int newton_maxsteps )
Generated by Doxygen
10.34 Solver Class Reference 441
Parameters
newton_maxsteps
Definition at line 483 of file solver.cpp.
Here is the caller graph for this function:
setNewtonMaxSteps amici::setSolverOptions
10.34.3.25 getNewtonPreequilibration()
bool getNewtonPreequilibration ( ) const
Returns
Definition at line 489 of file solver.cpp.
Here is the caller graph for this function:
getNewtonPreequilibration amici::ForwardProblem
::workForwardProblem
10.34.3.26 setNewtonPreequilibration()
void setNewtonPreequilibration (
bool newton_preeq )
Generated by Doxygen
442 CONTENTS
Parameters
newton_preeq
Definition at line 493 of file solver.cpp.
Here is the caller graph for this function:
setNewtonPreequilibration amici::setSolverOptions
10.34.3.27 getNewtonMaxLinearSteps()
int getNewtonMaxLinearSteps ( ) const
Returns
Definition at line 497 of file solver.cpp.
Here is the caller graph for this function:
getNewtonMaxLinearSteps amici::SteadystateProblem
::workSteadyStateProblem
10.34.3.28 setNewtonMaxLinearSteps()
void setNewtonMaxLinearSteps (
int newton_maxlinsteps )
Parameters
newton_maxlinsteps
Generated by Doxygen
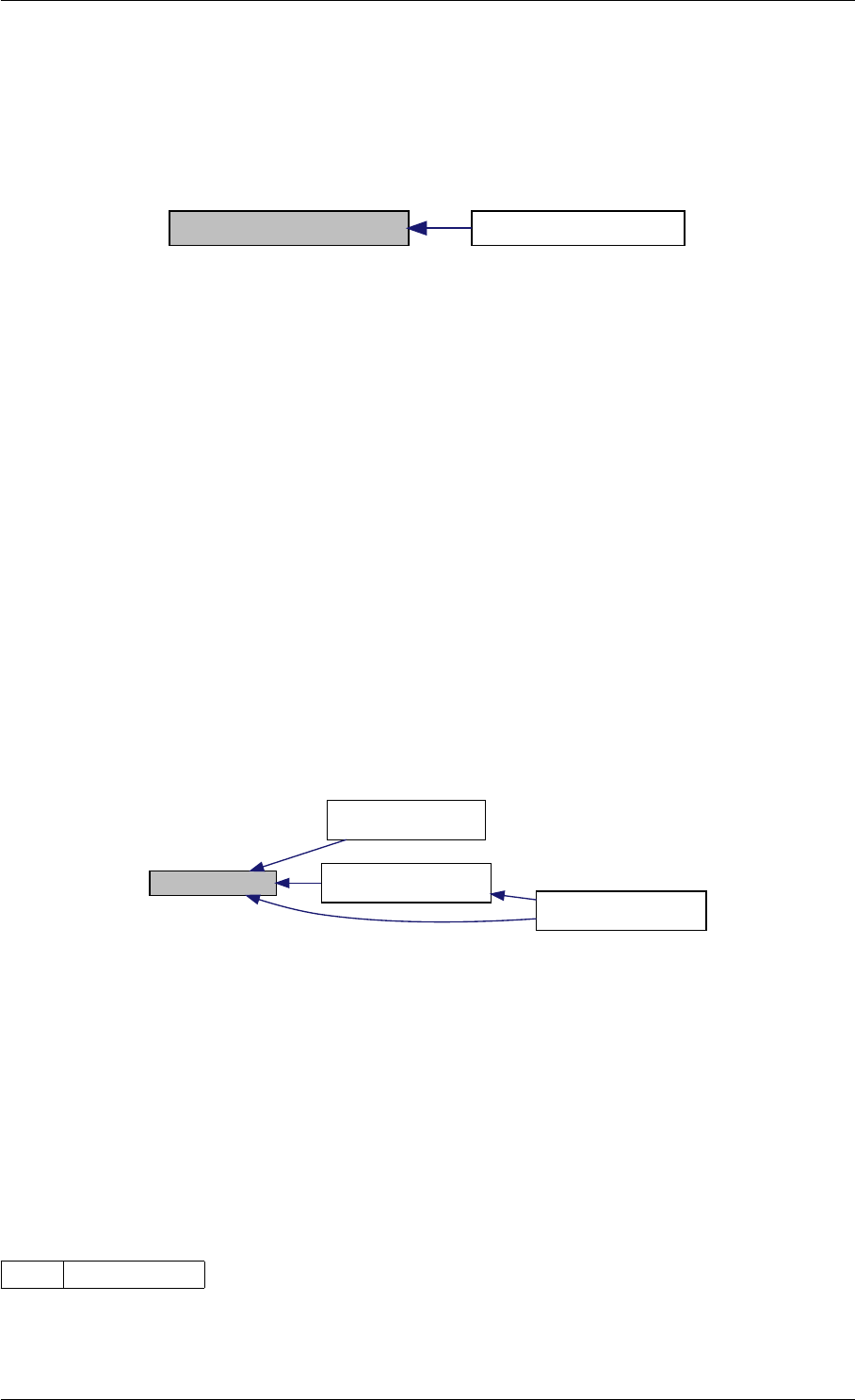
10.34 Solver Class Reference 443
Definition at line 501 of file solver.cpp.
Here is the caller graph for this function:
setNewtonMaxLinearSteps amici::setSolverOptions
10.34.3.29 getSensitivityOrder()
SensitivityOrder getSensitivityOrder ( ) const
Returns
sensitivity order
Definition at line 507 of file solver.cpp.
Here is the caller graph for this function:
getSensitivityOrder
amici::BackwardProblem
::workBackwardProblem
amici::SteadystateProblem
::checkConvergence
amici::SteadystateProblem
::getSteadystateSimulation
10.34.3.30 setSensitivityOrder()
void setSensitivityOrder (
SensitivityOrder sensi )
Parameters
sensi sensitivity order
Definition at line 511 of file solver.cpp.
Generated by Doxygen
444 CONTENTS
Here is the call graph for this function:
setSensitivityOrder
getMallocDone
applySensitivityTolerances
Here is the caller graph for this function:
setSensitivityOrder amici::setSolverOptions
10.34.3.31 getRelativeTolerance()
double getRelativeTolerance ( ) const
Returns
relative tolerances
Definition at line 518 of file solver.cpp.
Here is the caller graph for this function:
getRelativeTolerance
amici::SteadystateProblem
::workSteadyStateProblem
amici::SteadystateProblem
::createSteadystateSimSolver
Generated by Doxygen
10.34 Solver Class Reference 445
10.34.3.32 setRelativeTolerance()
void setRelativeTolerance (
double rtol )
Parameters
rtol relative tolerance (non-negative number)
Definition at line 522 of file solver.cpp.
Here is the call graph for this function:
setRelativeTolerance
getMallocDone
applyTolerances
applySensitivityTolerances
setSStolerances
Here is the caller graph for this function:
setRelativeTolerance amici::setSolverOptions
10.34.3.33 getAbsoluteTolerance()
double getAbsoluteTolerance ( ) const
Returns
absolute tolerances
Definition at line 534 of file solver.cpp.
Generated by Doxygen
446 CONTENTS
Here is the caller graph for this function:
getAbsoluteTolerance
amici::SteadystateProblem
::workSteadyStateProblem
amici::SteadystateProblem
::createSteadystateSimSolver
10.34.3.34 setAbsoluteTolerance()
void setAbsoluteTolerance (
double atol )
Parameters
atol absolute tolerance (non-negative number)
Definition at line 538 of file solver.cpp.
Here is the call graph for this function:
setAbsoluteTolerance
getMallocDone
applyTolerances
applySensitivityTolerances
setSStolerances
Here is the caller graph for this function:
setAbsoluteTolerance amici::setSolverOptions
Generated by Doxygen
10.34 Solver Class Reference 447
10.34.3.35 getRelativeToleranceSensi()
double getRelativeToleranceSensi ( ) const
Returns
relative tolerances
Definition at line 550 of file solver.cpp.
Here is the call graph for this function:
getRelativeToleranceSensi amici::isNaN
10.34.3.36 setRelativeToleranceSensi()
void setRelativeToleranceSensi (
double rtol )
Parameters
rtol relative tolerance (non-negative number)
Definition at line 554 of file solver.cpp.
Here is the call graph for this function:
setRelativeToleranceSensi
getMallocDone
applySensitivityTolerances
Generated by Doxygen
448 CONTENTS
10.34.3.37 getAbsoluteToleranceSensi()
double getAbsoluteToleranceSensi ( ) const
Returns
absolute tolerances
Definition at line 565 of file solver.cpp.
Here is the call graph for this function:
getAbsoluteToleranceSensi amici::isNaN
10.34.3.38 setAbsoluteToleranceSensi()
void setAbsoluteToleranceSensi (
double atol )
Parameters
atol absolute tolerance (non-negative number)
Definition at line 569 of file solver.cpp.
Here is the call graph for this function:
setAbsoluteToleranceSensi
getMallocDone
applySensitivityTolerances
Generated by Doxygen
10.34 Solver Class Reference 449
10.34.3.39 getRelativeToleranceQuadratures()
double getRelativeToleranceQuadratures ( ) const
Returns
relative tolerance
Definition at line 580 of file solver.cpp.
10.34.3.40 setRelativeToleranceQuadratures()
void setRelativeToleranceQuadratures (
double rtol )
Parameters
rtol relative tolerance (non-negative number)
Definition at line 584 of file solver.cpp.
Here is the caller graph for this function:
setRelativeToleranceQuadratures amici::setSolverOptions
10.34.3.41 getAbsoluteToleranceQuadratures()
double getAbsoluteToleranceQuadratures ( ) const
Returns
absolute tolerance
Definition at line 598 of file solver.cpp.
10.34.3.42 setAbsoluteToleranceQuadratures()
void setAbsoluteToleranceQuadratures (
double atol )
Generated by Doxygen
450 CONTENTS
Parameters
atol absolute tolerance (non-negative number)
Definition at line 602 of file solver.cpp.
Here is the caller graph for this function:
setAbsoluteToleranceQuadratures amici::setSolverOptions
10.34.3.43 getRelativeToleranceSteadyState()
double getRelativeToleranceSteadyState ( ) const
Returns
relative tolerance
Definition at line 616 of file solver.cpp.
Here is the call graph for this function:
getRelativeToleranceSteady
State amici::isNaN
Here is the caller graph for this function:
getRelativeToleranceSteady
State
amici::operator==
amici::SteadystateProblem
::checkConvergence
amici::SteadystateProblem
::createSteadystateSimSolver
amici::SteadystateProblem
::getSteadystateSimulation
Generated by Doxygen
10.34 Solver Class Reference 451
10.34.3.44 setRelativeToleranceSteadyState()
void setRelativeToleranceSteadyState (
double rtol )
Parameters
rtol relative tolerance (non-negative number)
Definition at line 620 of file solver.cpp.
10.34.3.45 getAbsoluteToleranceSteadyState()
double getAbsoluteToleranceSteadyState ( ) const
Returns
absolute tolerance
Definition at line 627 of file solver.cpp.
Here is the call graph for this function:
getAbsoluteToleranceSteady
State amici::isNaN
Here is the caller graph for this function:
getAbsoluteToleranceSteady
State
amici::operator==
amici::SteadystateProblem
::checkConvergence
amici::SteadystateProblem
::createSteadystateSimSolver
amici::SteadystateProblem
::getSteadystateSimulation
10.34.3.46 setAbsoluteToleranceSteadyState()
void setAbsoluteToleranceSteadyState (
double atol )
Generated by Doxygen
452 CONTENTS
Parameters
atol absolute tolerance (non-negative number)
Definition at line 631 of file solver.cpp.
10.34.3.47 getRelativeToleranceSteadyStateSensi()
double getRelativeToleranceSteadyStateSensi ( ) const
Returns
relative tolerance
Definition at line 638 of file solver.cpp.
Here is the call graph for this function:
getRelativeToleranceSteady
StateSensi amici::isNaN
Here is the caller graph for this function:
getRelativeToleranceSteady
StateSensi amici::operator==
10.34.3.48 setRelativeToleranceSteadyStateSensi()
void setRelativeToleranceSteadyStateSensi (
double rtol )
Generated by Doxygen
10.34 Solver Class Reference 453
Parameters
rtol relative tolerance (non-negative number)
Definition at line 642 of file solver.cpp.
10.34.3.49 getAbsoluteToleranceSteadyStateSensi()
double getAbsoluteToleranceSteadyStateSensi ( ) const
Returns
absolute tolerance
Definition at line 649 of file solver.cpp.
Here is the call graph for this function:
getAbsoluteToleranceSteady
StateSensi amici::isNaN
Here is the caller graph for this function:
getAbsoluteToleranceSteady
StateSensi amici::operator==
10.34.3.50 setAbsoluteToleranceSteadyStateSensi()
void setAbsoluteToleranceSteadyStateSensi (
double atol )
Generated by Doxygen
454 CONTENTS
Parameters
atol absolute tolerance (non-negative number)
Definition at line 653 of file solver.cpp.
10.34.3.51 getMaxSteps()
int getMaxSteps ( ) const
Returns
maximum number of solver steps
Definition at line 660 of file solver.cpp.
Here is the caller graph for this function:
getMaxSteps
amici::SteadystateProblem
::getSteadystateSimulation
amici::SteadystateProblem
::createSteadystateSimSolver
10.34.3.52 setMaxSteps()
void setMaxSteps (
int maxsteps )
Parameters
maxsteps maximum number of solver steps (non-negative number)
Definition at line 664 of file solver.cpp.
Generated by Doxygen
10.34 Solver Class Reference 455
Here is the call graph for this function:
setMaxSteps setMaxNumSteps
Here is the caller graph for this function:
setMaxSteps amici::setSolverOptions
10.34.3.53 getMaxStepsBackwardProblem()
int getMaxStepsBackwardProblem ( ) const
Returns
maximum number of solver steps
Definition at line 673 of file solver.cpp.
10.34.3.54 setMaxStepsBackwardProblem()
void setMaxStepsBackwardProblem (
int maxsteps )
Parameters
maxsteps maximum number of solver steps (non-negative number)
Definition at line 677 of file solver.cpp.
Generated by Doxygen
456 CONTENTS
Here is the call graph for this function:
setMaxStepsBackwardProblem setMaxNumStepsB
Here is the caller graph for this function:
setMaxStepsBackwardProblem amici::setSolverOptions
10.34.3.55 getLinearMultistepMethod()
LinearMultistepMethod getLinearMultistepMethod ( ) const
Returns
linear system multistep method
Definition at line 687 of file solver.cpp.
Here is the caller graph for this function:
getLinearMultistepMethod amici::SteadystateProblem
::createSteadystateSimSolver
10.34.3.56 setLinearMultistepMethod()
void setLinearMultistepMethod (
LinearMultistepMethod lmm )
Generated by Doxygen
10.34 Solver Class Reference 457
Parameters
lmm linear system multistep method
Definition at line 691 of file solver.cpp.
Here is the caller graph for this function:
setLinearMultistepMethod amici::setSolverOptions
10.34.3.57 getNonlinearSolverIteration()
NonlinearSolverIteration getNonlinearSolverIteration ( ) const
Returns
Definition at line 697 of file solver.cpp.
Here is the caller graph for this function:
getNonlinearSolverIteration amici::SteadystateProblem
::createSteadystateSimSolver
10.34.3.58 setNonlinearSolverIteration()
void setNonlinearSolverIteration (
NonlinearSolverIteration iter )
Parameters
iter nonlinear system solution method
Generated by Doxygen

458 CONTENTS
Definition at line 701 of file solver.cpp.
Here is the caller graph for this function:
setNonlinearSolverIteration amici::setSolverOptions
10.34.3.59 getInterpolationType()
InterpolationType getInterpolationType ( ) const
Returns
Definition at line 707 of file solver.cpp.
10.34.3.60 setInterpolationType()
void setInterpolationType (
InterpolationType interpType )
Parameters
interpType interpolation type
Definition at line 711 of file solver.cpp.
Here is the caller graph for this function:
setInterpolationType amici::setSolverOptions
Generated by Doxygen
10.34 Solver Class Reference 459
10.34.3.61 getStateOrdering()
StateOrdering getStateOrdering ( ) const
Returns
Definition at line 717 of file solver.cpp.
10.34.3.62 setStateOrdering()
void setStateOrdering (
StateOrdering ordering )
Parameters
ordering state ordering
Definition at line 721 of file solver.cpp.
Here is the caller graph for this function:
setStateOrdering amici::setSolverOptions
10.34.3.63 getStabilityLimitFlag()
int getStabilityLimitFlag ( ) const
Returns
stldet can be amici.FALSE (deactivated) or amici.TRUE (activated)
Definition at line 731 of file solver.cpp.
Generated by Doxygen
460 CONTENTS
Here is the caller graph for this function:
getStabilityLimitFlag amici::SteadystateProblem
::createSteadystateSimSolver
10.34.3.64 setStabilityLimitFlag()
void setStabilityLimitFlag (
booleantype stldet )
Parameters
stldet can be amici.FALSE (deactivated) or amici.TRUE (activated)
Definition at line 735 of file solver.cpp.
Here is the call graph for this function:
setStabilityLimitFlag
setStabLimDet
setStabLimDetB
Here is the caller graph for this function:
setStabilityLimitFlag amici::setSolverOptions
Generated by Doxygen
10.34 Solver Class Reference 461
10.34.3.65 getLinearSolver()
LinearSolver getLinearSolver ( ) const
Returns
Definition at line 748 of file solver.cpp.
Here is the caller graph for this function:
getLinearSolver
amici::SteadystateProblem
::workSteadyStateProblem
amici::SteadystateProblem
::createSteadystateSimSolver
10.34.3.66 setLinearSolver()
void setLinearSolver (
LinearSolver linsol )
Parameters
linsol
Definition at line 752 of file solver.cpp.
Here is the caller graph for this function:
setLinearSolver amici::setSolverOptions
Generated by Doxygen
462 CONTENTS
10.34.3.67 getInternalSensitivityMethod()
InternalSensitivityMethod getInternalSensitivityMethod ( ) const
Returns
internal sensitivity method
Definition at line 760 of file solver.cpp.
10.34.3.68 setInternalSensitivityMethod()
void setInternalSensitivityMethod (
InternalSensitivityMethod ism )
Parameters
ism internal sensitivity method
Definition at line 764 of file solver.cpp.
Here is the caller graph for this function:
setInternalSensitivityMethod amici::setSolverOptions
10.34.3.69 init()
virtual void init (
AmiVector ∗x,
AmiVector ∗dx,
realtype t) [protected], [pure virtual]
init initialises the states at the specified initial timepoint
Parameters
xinitial state variables
dx initial derivative state variables (DAE only)
tinitial timepoint
Generated by Doxygen
10.34 Solver Class Reference 463
Here is the caller graph for this function:
init setup amici::ForwardProblem
::workForwardProblem
10.34.3.70 binit()
virtual void binit (
int which,
AmiVector ∗xB,
AmiVector ∗dxB,
realtype t) [protected], [pure virtual]
binit initialises the adjoint states at the specified final timepoint
Parameters
which identifier of the backwards problem
xB initial adjoint state variables
dxB initial adjoint derivative state variables (DAE only)
tfinal timepoint
Here is the caller graph for this function:
binit setupAMIB
10.34.3.71 qbinit()
virtual void qbinit (
int which,
AmiVector ∗qBdot ) [protected], [pure virtual]
qbinit initialises the quadrature states at the specified final timepoint
Generated by Doxygen
464 CONTENTS
Parameters
which identifier of the backwards problem
qBdot initial adjoint quadrature state variables
Here is the caller graph for this function:
qbinit setupAMIB
10.34.3.72 rootInit()
virtual void rootInit (
int ne ) [protected], [pure virtual]
RootInit initialises the rootfinding for events
Parameters
ne number of different events
Here is the caller graph for this function:
rootInit setup amici::ForwardProblem
::workForwardProblem
10.34.3.73 sensInit1()
virtual void sensInit1 (
AmiVectorArray ∗sx,
AmiVectorArray ∗sdx,
int nplist ) [protected], [pure virtual]
SensInit1 initialises the sensitivities at the specified initial timepoint
Generated by Doxygen
10.34 Solver Class Reference 465
Parameters
sx initial state sensitivities
sdx initial derivative state sensitivities (DAE only)
nplist number parameter wrt which sensitivities are to be computed
10.34.3.74 setDenseJacFn()
virtual void setDenseJacFn ( ) [protected], [pure virtual]
SetDenseJacFn sets the dense Jacobian function Here is the caller graph for this function:
setDenseJacFn initializeLinearSolver setup amici::ForwardProblem
::workForwardProblem
10.34.3.75 setSparseJacFn()
virtual void setSparseJacFn ( ) [protected], [pure virtual]
SetSparseJacFn sets the sparse Jacobian function
10.34.3.76 setBandJacFn()
virtual void setBandJacFn ( ) [protected], [pure virtual]
SetBandJacFn sets the banded Jacobian function Here is the caller graph for this function:
setBandJacFn initializeLinearSolver setup amici::ForwardProblem
::workForwardProblem
10.34.3.77 setJacTimesVecFn()
virtual void setJacTimesVecFn ( ) [protected], [pure virtual]
SetJacTimesVecFn sets the Jacobian vector multiplication function
10.34.3.78 setDenseJacFnB()
virtual void setDenseJacFnB (
int which ) [protected], [pure virtual]
SetDenseJacFn sets the dense Jacobian function
Generated by Doxygen
466 CONTENTS
Parameters
which identifier of the backwards problem
Here is the caller graph for this function:
setDenseJacFnB initializeLinearSolverB setupAMIB
10.34.3.79 setSparseJacFnB()
virtual void setSparseJacFnB (
int which ) [protected], [pure virtual]
SetSparseJacFn sets the sparse Jacobian function
Parameters
which identifier of the backwards problem
10.34.3.80 setBandJacFnB()
virtual void setBandJacFnB (
int which ) [protected], [pure virtual]
SetBandJacFn sets the banded Jacobian function
Parameters
which identifier of the backwards problem
Here is the caller graph for this function:
setBandJacFnB initializeLinearSolverB setupAMIB
Generated by Doxygen
10.34 Solver Class Reference 467
10.34.3.81 setJacTimesVecFnB()
virtual void setJacTimesVecFnB (
int which ) [protected], [pure virtual]
SetJacTimesVecFn sets the Jacobian vector multiplication function
Parameters
which identifier of the backwards problem
10.34.3.82 wrapErrHandlerFn()
void wrapErrHandlerFn (
int error_code,
const char ∗module,
const char ∗function,
char ∗msg,
void ∗eh_data ) [static], [protected]
ErrHandlerFn extracts diagnosis information from solver memory block and writes them into the return data object
for the backward problem
Parameters
error_code error identifier
module name of the module in which the error occured
function name of the function in which the error occured
Type: char
msg error message
eh_data unused input
Definition at line 149 of file solver.cpp.
10.34.3.83 allocateSolver()
virtual void allocateSolver ( ) [protected], [pure virtual]
Create specifies solver method and initializes solver memory for the forward problem Here is the caller graph for
Generated by Doxygen
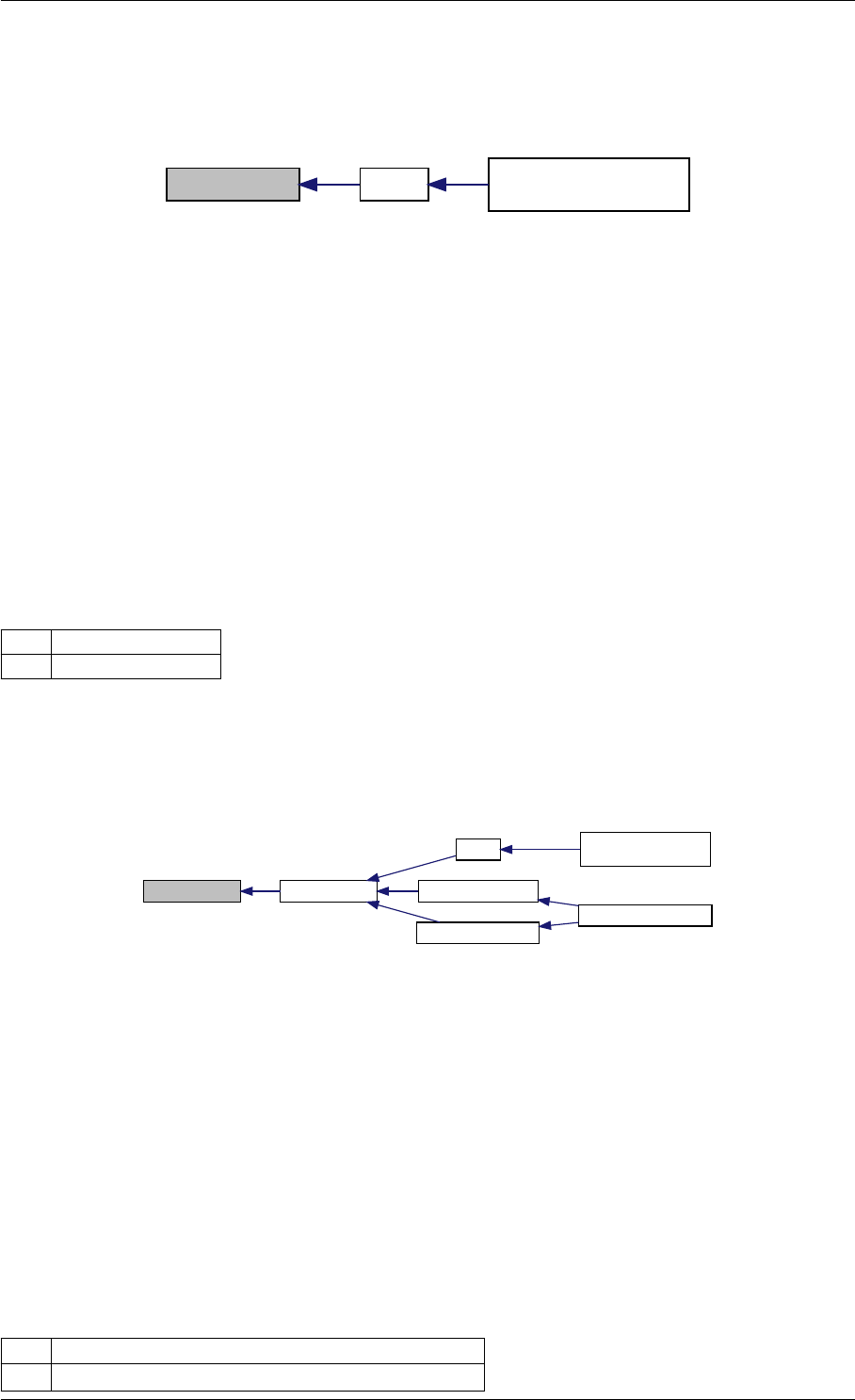
468 CONTENTS
this function:
allocateSolver setup amici::ForwardProblem
::workForwardProblem
10.34.3.84 setSStolerances()
virtual void setSStolerances (
double rtol,
double atol ) [protected], [pure virtual]
SStolerances sets scalar relative and absolute tolerances for the forward problem
Parameters
rtol relative tolerances
atol absolute tolerances
Here is the caller graph for this function:
setSStolerances applyTolerances
setup
setRelativeTolerance
setAbsoluteTolerance
amici::ForwardProblem
::workForwardProblem
amici::setSolverOptions
10.34.3.85 setSensSStolerances()
virtual void setSensSStolerances (
double rtol,
double ∗atol ) [protected], [pure virtual]
SensSStolerances activates sets scalar relative and absolute tolerances for the sensitivity variables
Parameters
rtol relative tolerances
atol array of absolute tolerances for every sensitivy variable
Generated by Doxygen
10.34 Solver Class Reference 469
10.34.3.86 setSensErrCon()
virtual void setSensErrCon (
bool error_corr ) [protected], [pure virtual]
SetSensErrCon specifies whether error control is also enforced for sensitivities for the forward problem
Parameters
error_corr activation flag
10.34.3.87 setQuadErrConB()
virtual void setQuadErrConB (
int which,
bool flag ) [protected], [pure virtual]
SetSensErrCon specifies whether error control is also enforced for the backward quadrature problem
Parameters
which identifier of the backwards problem
flag activation flag
10.34.3.88 setErrHandlerFn()
virtual void setErrHandlerFn ( ) [protected], [pure virtual]
SetErrHandlerFn attaches the error handler function (errMsgIdAndTxt) to the solver Here is the caller graph for this
function:
setErrHandlerFn setup amici::ForwardProblem
::workForwardProblem
10.34.3.89 setUserData()
virtual void setUserData (
Model ∗model ) [protected], [pure virtual]
SetUserData attaches the user data instance (here this is a Model) to the forward problem
Generated by Doxygen
470 CONTENTS
Parameters
model Model instance,
Here is the caller graph for this function:
setUserData setup amici::ForwardProblem
::workForwardProblem
10.34.3.90 setUserDataB()
virtual void setUserDataB (
int which,
Model ∗model ) [protected], [pure virtual]
SetUserDataB attaches the user data instance (here this is a Model) to the backward problem
Parameters
which identifier of the backwards problem
model Model instance,
Here is the caller graph for this function:
setUserDataB setupAMIB
10.34.3.91 setMaxNumSteps()
virtual void setMaxNumSteps (
long int mxsteps ) [protected], [pure virtual]
SetMaxNumSteps specifies the maximum number of steps for the forward problem
Generated by Doxygen
10.34 Solver Class Reference 471
Parameters
mxsteps number of steps
Here is the caller graph for this function:
setMaxNumSteps
setup
setMaxSteps
amici::ForwardProblem
::workForwardProblem
amici::setSolverOptions
10.34.3.92 setMaxNumStepsB()
virtual void setMaxNumStepsB (
int which,
long int mxstepsB ) [protected], [pure virtual]
SetMaxNumStepsB specifies the maximum number of steps for the forward problem
Parameters
which identifier of the backwards problem
mxstepsB number of steps
Here is the caller graph for this function:
setMaxNumStepsB
setupAMIB
setMaxStepsBackwardProblem amici::setSolverOptions
10.34.3.93 setStabLimDet()
virtual void setStabLimDet (
int stldet ) [protected], [pure virtual]
SetStabLimDet activates stability limit detection for the forward problem
Generated by Doxygen
472 CONTENTS
Parameters
stldet flag for stability limit detection (TRUE or FALSE)
Here is the caller graph for this function:
setStabLimDet
setup
setStabilityLimitFlag
amici::ForwardProblem
::workForwardProblem
amici::setSolverOptions
10.34.3.94 setStabLimDetB()
virtual void setStabLimDetB (
int which,
int stldet ) [protected], [pure virtual]
SetStabLimDetB activates stability limit detection for the backward problem
Parameters
which identifier of the backwards problem
stldet flag for stability limit detection (TRUE or FALSE)
Here is the caller graph for this function:
setStabLimDetB
setupAMIB
setStabilityLimitFlag amici::setSolverOptions
10.34.3.95 setId()
virtual void setId (
Model ∗model ) [protected], [pure virtual]
SetId specify algebraic/differential components (DAE only)
Generated by Doxygen
10.34 Solver Class Reference 473
Parameters
model model specification
10.34.3.96 setSuppressAlg()
virtual void setSuppressAlg (
bool flag ) [protected], [pure virtual]
SetId deactivates error control for algebraic components (DAE only)
Parameters
flag deactivation flag
10.34.3.97 setSensParams()
virtual void setSensParams (
realtype ∗p,
realtype ∗pbar,
int ∗plist ) [protected], [pure virtual]
SetSensParams specifies the scaling and indexes for sensitivity computation
Parameters
pparamaters
pbar parameter scaling constants
plist parameter index list
10.34.3.98 getDky()
virtual void getDky (
realtype t,
int k,
AmiVector ∗dky ) const [protected], [pure virtual]
getDky interpolates the (derivative of the) solution at the requested timepoint
Parameters
ttimepoint
kderivative order
dky interpolated solution
Generated by Doxygen
474 CONTENTS
10.34.3.99 adjInit()
virtual void adjInit ( ) [protected], [pure virtual]
AdjInit initializes the adjoint problem
10.34.3.100 allocateSolverB()
virtual void allocateSolverB (
int ∗which ) [protected], [pure virtual]
specifies solver method and initializes solver memory for the backward problem
Parameters
which identifier of the backwards problem
Here is the caller graph for this function:
allocateSolverB setupAMIB
10.34.3.101 setSStolerancesB()
virtual void setSStolerancesB (
int which,
realtype relTolB,
realtype absTolB ) [protected], [pure virtual]
SStolerancesB sets relative and absolute tolerances for the backward problem
Parameters
which identifier of the backwards problem
relTolB relative tolerances
absTolB absolute tolerances
Generated by Doxygen
10.34 Solver Class Reference 475
10.34.3.102 quadSStolerancesB()
virtual void quadSStolerancesB (
int which,
realtype reltolQB,
realtype abstolQB ) [protected], [pure virtual]
SStolerancesB sets relative and absolute tolerances for the quadrature backward problem
Parameters
which identifier of the backwards problem
reltolQB relative tolerances
abstolQB absolute tolerances
10.34.3.103 dense()
virtual void dense (
int nx ) [protected], [pure virtual]
Dense attaches a dense linear solver to the forward problem
Parameters
nx number of state variables
Here is the caller graph for this function:
dense initializeLinearSolver setup amici::ForwardProblem
::workForwardProblem
10.34.3.104 denseB()
virtual void denseB (
int which,
int nx ) [protected], [pure virtual]
DenseB attaches a dense linear solver to the backward problem
Parameters
which identifier of the backwards problem
nx number of state variables
Generated by Doxygen
476 CONTENTS
Here is the caller graph for this function:
denseB initializeLinearSolverB setupAMIB
10.34.3.105 band()
virtual void band (
int nx,
int ubw,
int lbw ) [protected], [pure virtual]
Band attaches a banded linear solver to the forward problem
Parameters
nx number of state variables
ubw upper matrix bandwidth
lbw lower matrix bandwidth
Here is the caller graph for this function:
band initializeLinearSolver setup amici::ForwardProblem
::workForwardProblem
10.34.3.106 bandB()
virtual void bandB (
int which,
int nx,
int ubw,
int lbw ) [protected], [pure virtual]
BandB attaches a banded linear solver to the backward problem
Generated by Doxygen
10.34 Solver Class Reference 477
Parameters
which identifier of the backwards problem
nx number of state variables
ubw upper matrix bandwidth
lbw lower matrix bandwidth
Here is the caller graph for this function:
bandB initializeLinearSolverB setupAMIB
10.34.3.107 diag()
virtual void diag ( ) [protected], [pure virtual]
Diag attaches a diagonal linear solver to the forward problem
10.34.3.108 diagB()
virtual void diagB (
int which ) [protected], [pure virtual]
DiagB attaches a diagonal linear solver to the backward problem
Parameters
which identifier of the backwards problem
10.34.3.109 spgmr()
virtual void spgmr (
int prectype,
int maxl ) [protected], [pure virtual]
DAMISpgmr attaches a scaled predonditioned GMRES linear solver to the forward problem
Parameters
prectype preconditioner type PREC_NONE, PREC_LEFT, PREC_RIGHT or PREC_BOTH
maxl maximum Kryloc subspace dimension
Generated by Doxygen

478 CONTENTS
10.34.3.110 spgmrB()
virtual void spgmrB (
int which,
int prectype,
int maxl ) [protected], [pure virtual]
DAMISpgmrB attaches a scaled predonditioned GMRES linear solver to the backward problem
Parameters
which identifier of the backwards problem
prectype preconditioner type PREC_NONE, PREC_LEFT, PREC_RIGHT or PREC_BOTH
maxl maximum Kryloc subspace dimension
10.34.3.111 spbcg()
virtual void spbcg (
int prectype,
int maxl ) [protected], [pure virtual]
Spbcg attaches a scaled predonditioned Bi-CGStab linear solver to the forward problem
Parameters
prectype preconditioner type PREC_NONE, PREC_LEFT, PREC_RIGHT or PREC_BOTH
maxl maximum Kryloc subspace dimension
10.34.3.112 spbcgB()
virtual void spbcgB (
int which,
int prectype,
int maxl ) [protected], [pure virtual]
SpbcgB attaches a scaled predonditioned Bi-CGStab linear solver to the backward problem
Parameters
which identifier of the backwards problem
prectype preconditioner type PREC_NONE, PREC_LEFT, PREC_RIGHT or PREC_BOTH
maxl maximum Kryloc subspace dimension
Generated by Doxygen

10.34 Solver Class Reference 479
10.34.3.113 sptfqmr()
virtual void sptfqmr (
int prectype,
int maxl ) [protected], [pure virtual]
Sptfqmr attaches a scaled predonditioned TFQMR linear solver to the forward problem
Parameters
prectype preconditioner type PREC_NONE, PREC_LEFT, PREC_RIGHT or PREC_BOTH
maxl maximum Kryloc subspace dimension
10.34.3.114 sptfqmrB()
virtual void sptfqmrB (
int which,
int prectype,
int maxl ) [protected], [pure virtual]
SptfqmrB attaches a scaled predonditioned TFQMR linear solver to the backward problem
Parameters
which identifier of the backwards problem
prectype preconditioner type PREC_NONE, PREC_LEFT, PREC_RIGHT or PREC_BOTH
maxl maximum Kryloc subspace dimension
10.34.3.115 klu()
virtual void klu (
int nx,
int nnz,
int sparsetype ) [protected], [pure virtual]
KLU attaches a sparse linear solver to the forward problem
Parameters
nx number of state variables
nnz number of nonzero entries in the jacobian
sparsetype sparse storage type, CSC_MAT for column matrix, CSR_MAT for row matrix
10.34.3.116 kluSetOrdering()
virtual void kluSetOrdering (
Generated by Doxygen
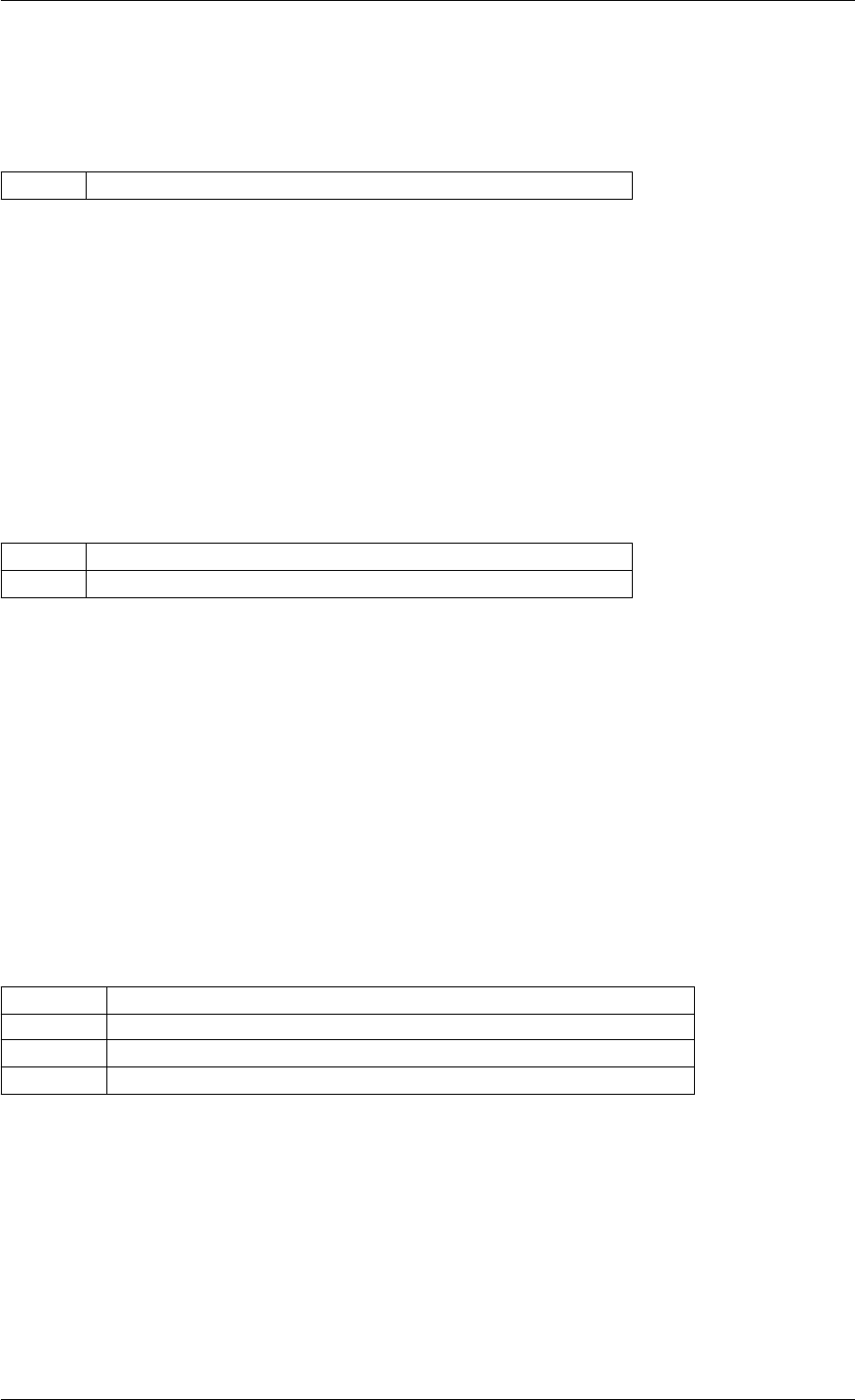
480 CONTENTS
int ordering ) [protected], [pure virtual]
KLUSetOrdering sets the ordering for the sparse linear solver of the forward problem
Parameters
ordering ordering algorithm to reduce fill 0:AMD 1:COLAMD 2: natural ordering
10.34.3.117 kluSetOrderingB()
virtual void kluSetOrderingB (
int which,
int ordering ) [protected], [pure virtual]
KLUSetOrderingB sets the ordering for the sparse linear solver of the backward problem
Parameters
which identifier of the backwards problem
ordering ordering algorithm to reduce fill 0:AMD 1:COLAMD 2: natural ordering
10.34.3.118 kluB()
virtual void kluB (
int which,
int nx,
int nnz,
int sparsetype ) [protected], [pure virtual]
KLUB attaches a sparse linear solver to the forward problem
Parameters
which identifier of the backwards problem
nx number of state variables
nnz number of nonzero entries in the jacobian
sparsetype sparse storage type, CSC_MAT for column matrix, CSR_MAT for row matrix
10.34.3.119 getNumSteps()
virtual void getNumSteps (
void ∗ami_mem,
long int ∗numsteps ) const [protected], [pure virtual]
getNumSteps reports the number of solver steps
Generated by Doxygen
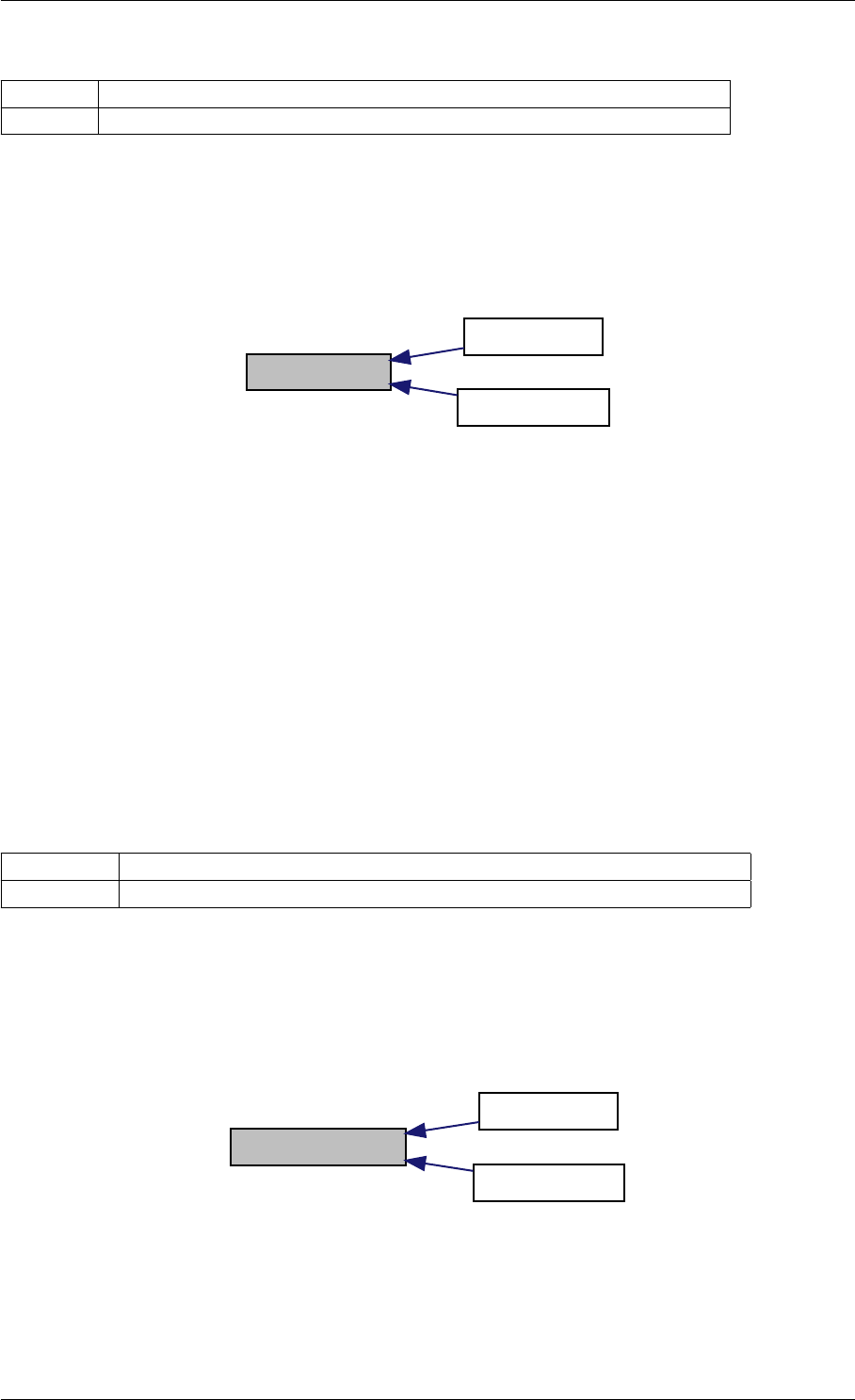
10.34 Solver Class Reference 481
Parameters
ami_mem pointer to the solver memory instance (can be from forward or backward problem)
numsteps output array
Here is the caller graph for this function:
getNumSteps
getDiagnosis
getDiagnosisB
10.34.3.120 getNumRhsEvals()
virtual void getNumRhsEvals (
void ∗ami_mem,
long int ∗numrhsevals ) const [protected], [pure virtual]
getNumRhsEvals reports the number of right hand evaluations
Parameters
ami_mem pointer to the solver memory instance (can be from forward or backward problem)
numrhsevals output array
Here is the caller graph for this function:
getNumRhsEvals
getDiagnosis
getDiagnosisB
Generated by Doxygen
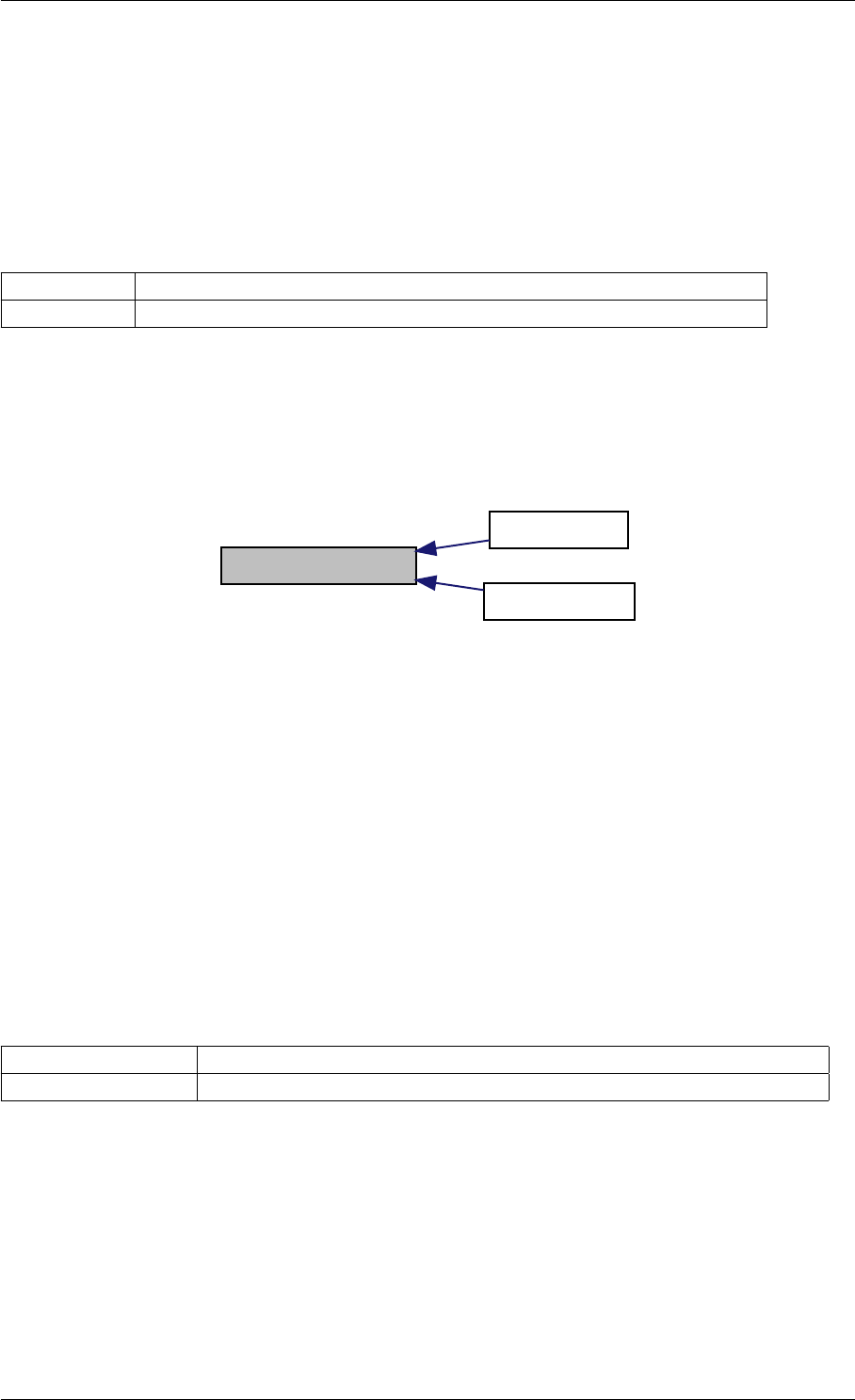
482 CONTENTS
10.34.3.121 getNumErrTestFails()
virtual void getNumErrTestFails (
void ∗ami_mem,
long int ∗numerrtestfails ) const [protected], [pure virtual]
getNumErrTestFails reports the number of local error test failures
Parameters
ami_mem pointer to the solver memory instance (can be from forward or backward problem)
numerrtestfails output array
Here is the caller graph for this function:
getNumErrTestFails
getDiagnosis
getDiagnosisB
10.34.3.122 getNumNonlinSolvConvFails()
virtual void getNumNonlinSolvConvFails (
void ∗ami_mem,
long int ∗numnonlinsolvconvfails ) const [protected], [pure virtual]
getNumNonlinSolvConvFails reports the number of nonlinear convergence failures
Parameters
ami_mem pointer to the solver memory instance (can be from forward or backward problem)
numnonlinsolvconvfails output array
Generated by Doxygen
10.34 Solver Class Reference 483
Here is the caller graph for this function:
getNumNonlinSolvConvFails
getDiagnosis
getDiagnosisB
10.34.3.123 getLastOrder()
virtual void getLastOrder (
void ∗ami_mem,
int ∗order ) const [protected], [pure virtual]
Reports the order of the integration method during the last internal step
Parameters
ami_mem pointer to the solver memory instance (can be from forward or backward problem)
order output array
Here is the caller graph for this function:
getLastOrder getDiagnosis
10.34.3.124 initializeLinearSolver()
void initializeLinearSolver (
const Model ∗model ) [protected]
initializeLinearSolver sets the linear solver for the forward problem
Generated by Doxygen
484 CONTENTS
Parameters
model pointer to the model object
Definition at line 227 of file solver.cpp.
Here is the call graph for this function:
initializeLinearSolver
dense
setDenseJacFn
band
setBandJacFn
Here is the caller graph for this function:
initializeLinearSolver setup amici::ForwardProblem
::workForwardProblem
10.34.3.125 initializeLinearSolverB()
void initializeLinearSolverB (
const Model ∗model,
const int which ) [protected]
Sets the linear solver for the backward problem
Parameters
model pointer to the model object
which index of the backward problem
Generated by Doxygen
10.34 Solver Class Reference 485
Definition at line 304 of file solver.cpp.
Here is the call graph for this function:
initializeLinearSolverB
denseB
setDenseJacFnB
bandB
setBandJacFnB
Here is the caller graph for this function:
initializeLinearSolverB setupAMIB
10.34.3.126 nplist()
virtual int nplist ( ) const [protected], [pure virtual]
Accessor function to the number of sensitivity parameters in the model stored in the user data
Returns
number of sensitivity parameters
Generated by Doxygen
486 CONTENTS
10.34.3.127 nx()
virtual int nx ( ) const [protected], [pure virtual]
Accessor function to the number of state variables in the model stored in the user data
Returns
number of state variables
Here is the caller graph for this function:
nx amici.ode_export.ODEModel.
__init__
10.34.3.128 getModel()
virtual const Model∗getModel ( ) const [protected], [pure virtual]
Accessor function to the model stored in the user data
Returns
user data model
10.34.3.129 getMallocDone()
virtual bool getMallocDone ( ) const [protected], [pure virtual]
checks whether memory for the forward problem has been allocated
Generated by Doxygen
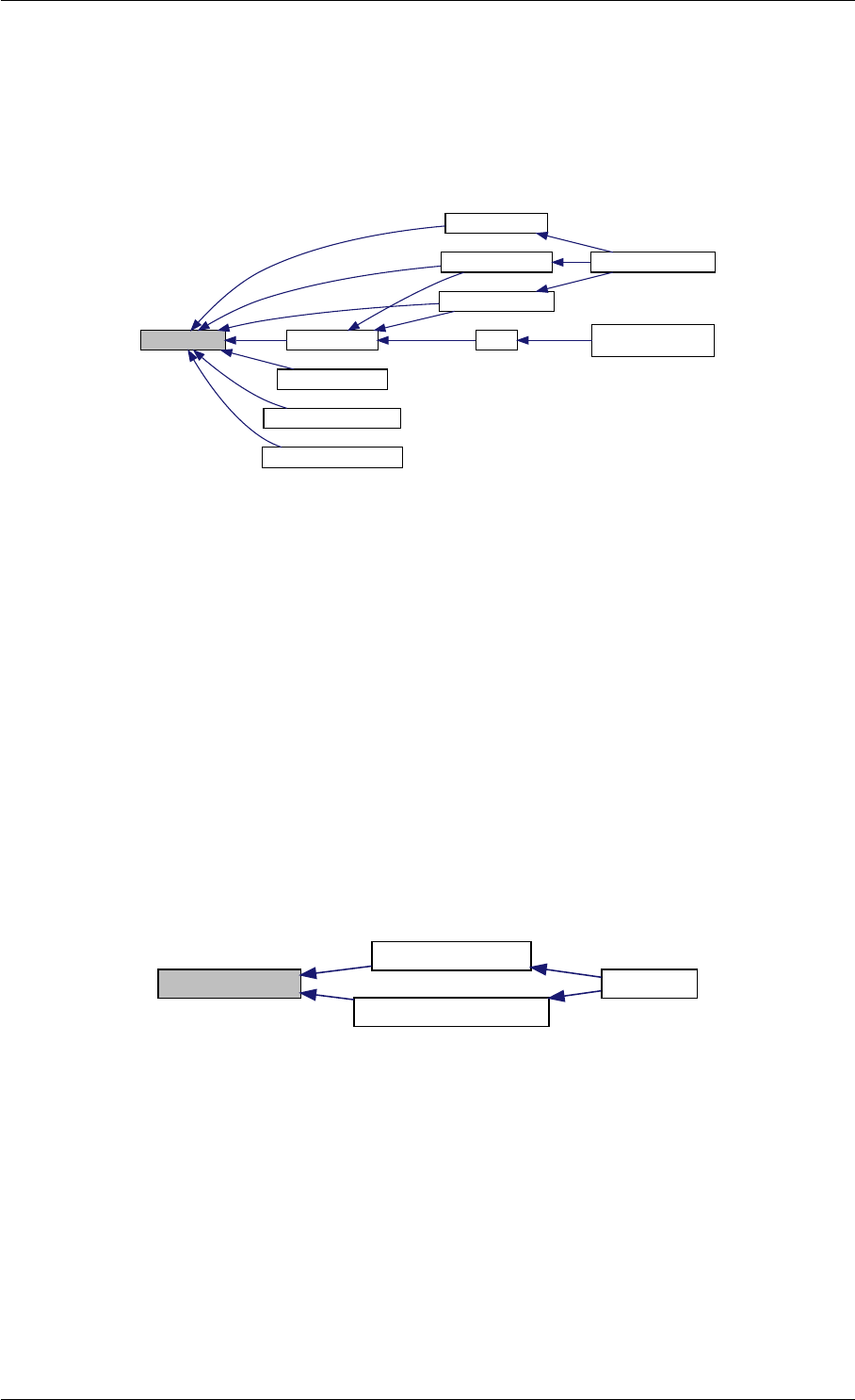
10.34 Solver Class Reference 487
Returns
solverMemory->(cv|ida)__MallocDone
Here is the caller graph for this function:
getMallocDone applyTolerances
setRelativeTolerance
setAbsoluteTolerance
applyTolerancesFSA
setSensitivityOrder
setRelativeToleranceSensi
setAbsoluteToleranceSensi
setup amici::ForwardProblem
::workForwardProblem
amici::setSolverOptions
10.34.3.130 getAdjMallocDone()
virtual bool getAdjMallocDone ( ) const [protected], [pure virtual]
checks whether memory for the backward problem has been allocated
Returns
solverMemory->(cv|ida)__adjMallocDone
Here is the caller graph for this function:
getAdjMallocDone
applyTolerancesASA
applyQuadTolerancesASA
setupAMIB
10.34.3.131 getAdjBmem()
virtual void∗getAdjBmem (
void ∗ami_mem,
int which ) [protected], [pure virtual]
getAdjBmem retrieves the solver memory instance for the backward problem
Generated by Doxygen
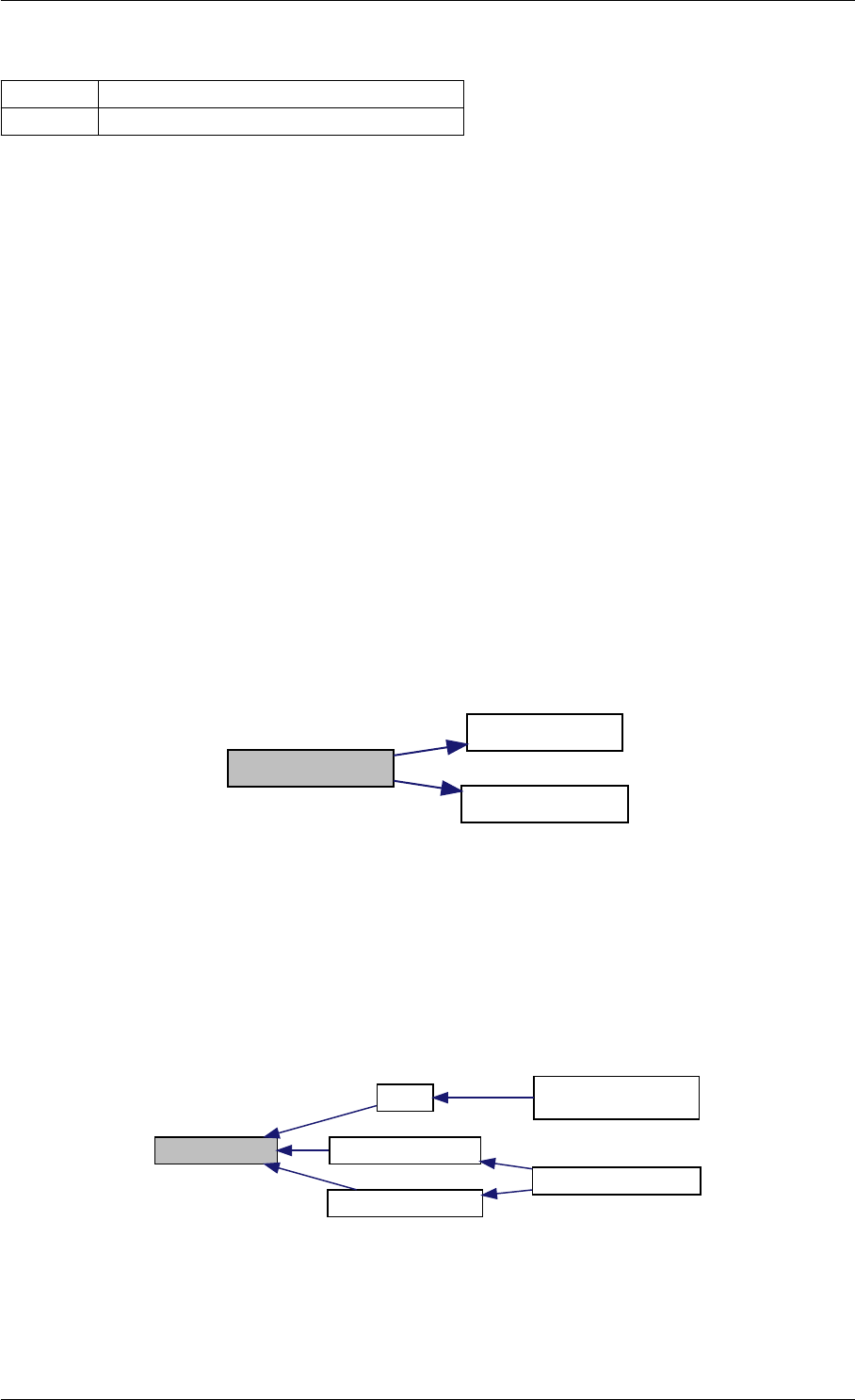
488 CONTENTS
Parameters
which identifier of the backwards problem
ami_mem pointer to the forward solver memory instance
Returns
ami_memB pointer to the backward solver memory instance
10.34.3.132 applyTolerances()
void applyTolerances ( ) [protected]
updates solver tolerances according to the currently specified member variables
Definition at line 408 of file solver.cpp.
Here is the call graph for this function:
applyTolerances
getMallocDone
setSStolerances
Here is the caller graph for this function:
applyTolerances
setup
setRelativeTolerance
setAbsoluteTolerance
amici::ForwardProblem
::workForwardProblem
amici::setSolverOptions
Generated by Doxygen
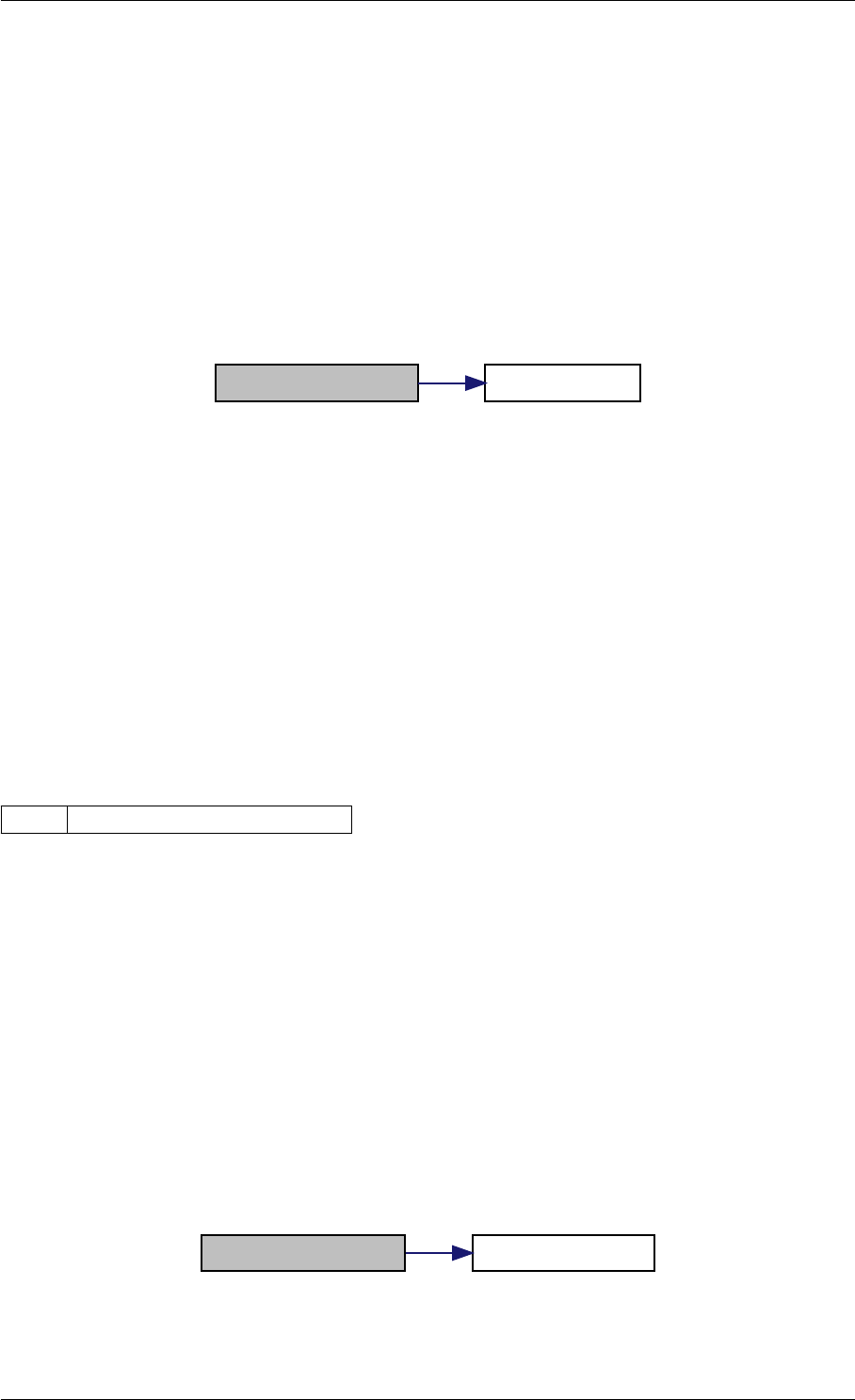
10.34 Solver Class Reference 489
10.34.3.133 applyTolerancesFSA()
void applyTolerancesFSA ( ) [protected]
updates FSA solver tolerances according to the currently specified member variables
Definition at line 415 of file solver.cpp.
Here is the call graph for this function:
applyTolerancesFSA getMallocDone
10.34.3.134 applyTolerancesASA()
void applyTolerancesASA (
int which ) [protected]
updates ASA solver tolerances according to the currently specified member variables
Parameters
which identifier of the backwards problem
Definition at line 429 of file solver.cpp.
Here is the call graph for this function:
applyTolerancesASA getAdjMallocDone
Generated by Doxygen
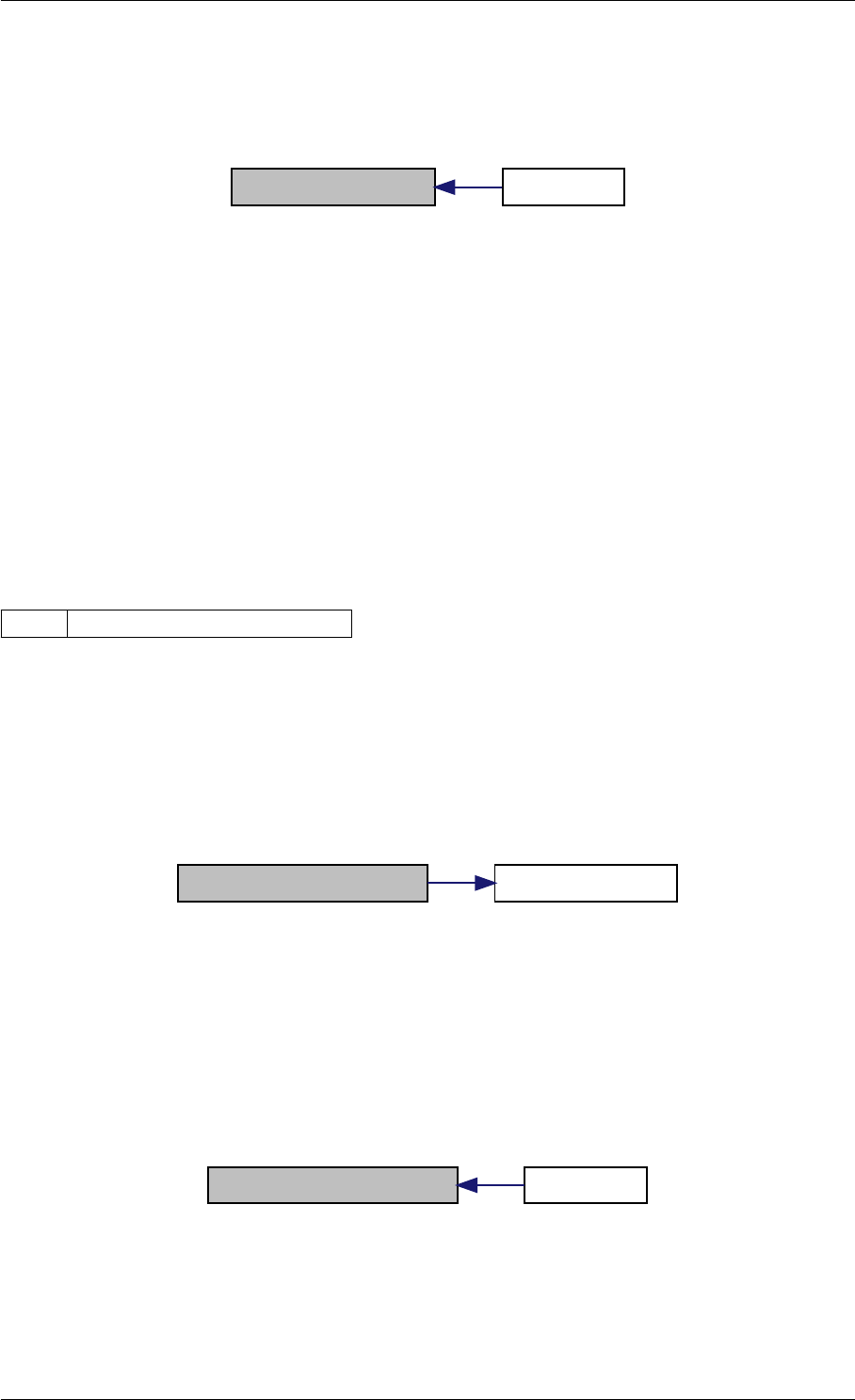
490 CONTENTS
Here is the caller graph for this function:
applyTolerancesASA setupAMIB
10.34.3.135 applyQuadTolerancesASA()
void applyQuadTolerancesASA (
int which ) [protected]
updates ASA quadrature solver tolerances according to the currently specified member variables
Parameters
which identifier of the backwards problem
Definition at line 440 of file solver.cpp.
Here is the call graph for this function:
applyQuadTolerancesASA getAdjMallocDone
Here is the caller graph for this function:
applyQuadTolerancesASA setupAMIB
Generated by Doxygen
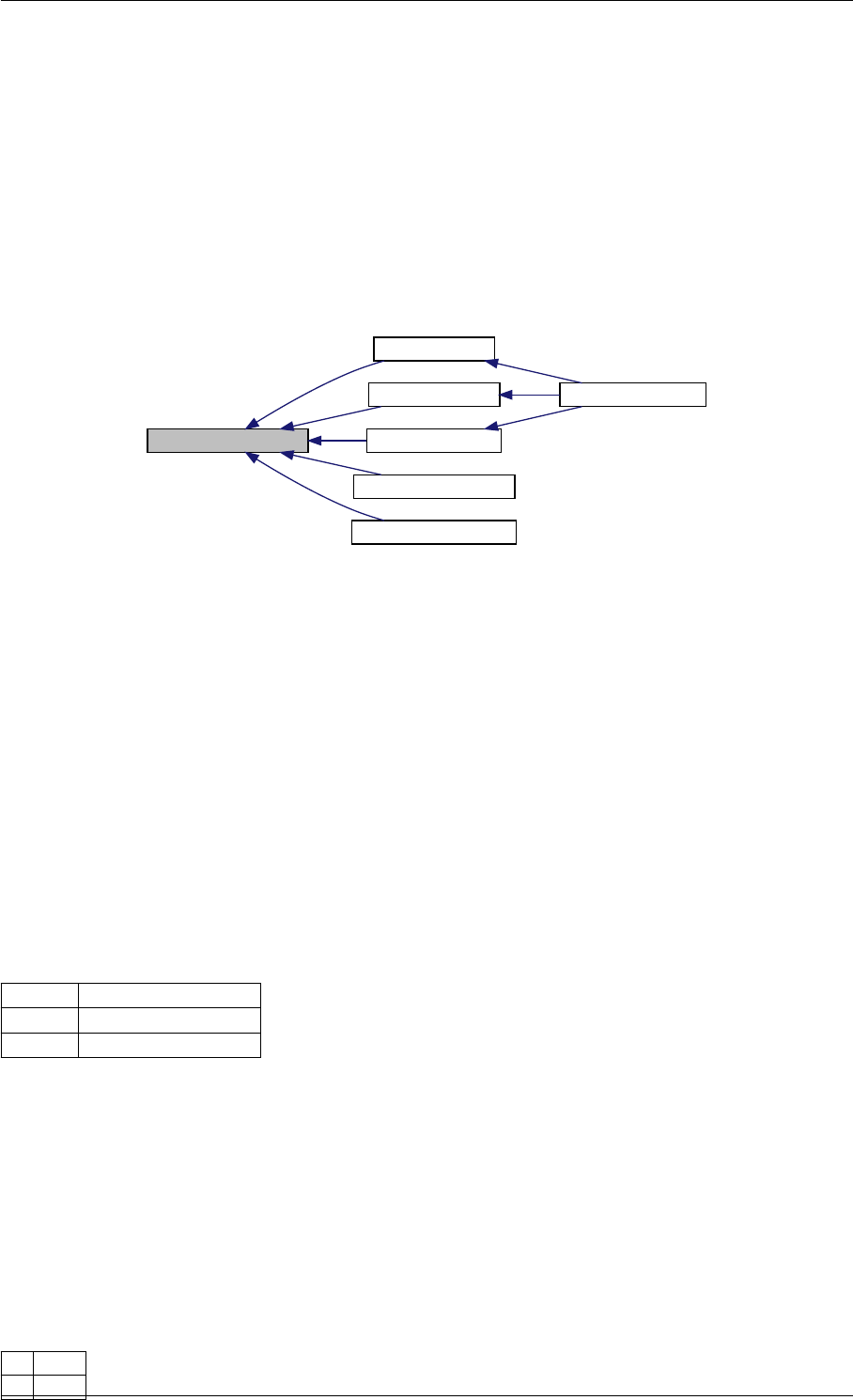
10.34 Solver Class Reference 491
10.34.3.136 applySensitivityTolerances()
void applySensitivityTolerances ( ) [protected]
updates all senstivivity solver tolerances according to the currently specified member variables
Definition at line 459 of file solver.cpp.
Here is the caller graph for this function:
applySensitivityTolerances
setSensitivityOrder
setRelativeTolerance
setAbsoluteTolerance
setRelativeToleranceSensi
setAbsoluteToleranceSensi
amici::setSolverOptions
10.34.4 Friends And Related Function Documentation
10.34.4.1 boost::serialization::serialize
void boost::serialization::serialize (
Archive & ar,
Solver &r,
const unsigned int version ) [friend]
Parameters
ar Archive to serialize to
rData to serialize
version Version number
10.34.4.2 operator==
bool operator== (
const Solver &a,
const Solver &b) [friend]
Parameters
a
b
Generated by Doxygen

492 CONTENTS
Returns
Definition at line 378 of file solver.cpp.
10.34.5 Member Data Documentation
10.34.5.1 solverMemory
std::unique_ptr<void, std::function<void(void ∗)> > solverMemory [protected]
pointer to solver memory block
Definition at line 1105 of file solver.h.
10.34.5.2 solverMemoryB
std::vector<std::unique_ptr<void, std::function<void(void ∗)>>>solverMemoryB [protected]
pointer to solver memory block
Definition at line 1108 of file solver.h.
10.34.5.3 solverWasCalled
bool solverWasCalled = false [protected]
flag indicating whether the solver was called
Definition at line 1111 of file solver.h.
10.34.5.4 ism
InternalSensitivityMethod ism = InternalSensitivityMethod::simultaneous [protected]
internal sensitivity method flag used to select the sensitivity solution method. Only applies for Forward Sensitivities.
Definition at line 1115 of file solver.h.
Generated by Doxygen

10.35 SteadystateProblem Class Reference 493
10.34.5.5 lmm
LinearMultistepMethod lmm = LinearMultistepMethod::BDF [protected]
specifies the linear multistep method.
Definition at line 1119 of file solver.h.
10.34.5.6 iter
NonlinearSolverIteration iter = NonlinearSolverIteration::newton [protected]
specifies the type of nonlinear solver iteration
Definition at line 1124 of file solver.h.
10.34.5.7 interpType
InterpolationType interpType = InterpolationType::hermite [protected]
interpolation type for the forward problem solution which is then used for the backwards problem.
Definition at line 1129 of file solver.h.
10.34.5.8 maxsteps
int maxsteps = 10000 [protected]
maximum number of allowed integration steps
Definition at line 1132 of file solver.h.
10.35 SteadystateProblem Class Reference
The SteadystateProblem class solves a steady-state problem using Newton's method and falls back to integration
on failure.
#include <steadystateproblem.h>
Public Member Functions
• void workSteadyStateProblem (ReturnData ∗rdata, Solver ∗solver, Model ∗model, int it)
•realtype getWrmsNorm (AmiVector const &x, AmiVector const &xdot, realtype atol, realtype rtol)
• bool checkConvergence (const Solver ∗solver, Model ∗model)
• void applyNewtonsMethod (ReturnData ∗rdata, Model ∗model, NewtonSolver ∗newtonSolver, int newton_try)
• void writeNewtonOutput (ReturnData ∗rdata, const Model ∗model, NewtonStatus newton_status, double
run_time, int it)
• void getSteadystateSimulation (ReturnData ∗rdata, Solver ∗solver, Model ∗model, int it)
• std::unique_ptr<CVodeSolver >createSteadystateSimSolver (Solver ∗solver, Model ∗model, realtype tstart)
•SteadystateProblem (realtype ∗t, AmiVector ∗x, AmiVectorArray ∗sx)
Generated by Doxygen
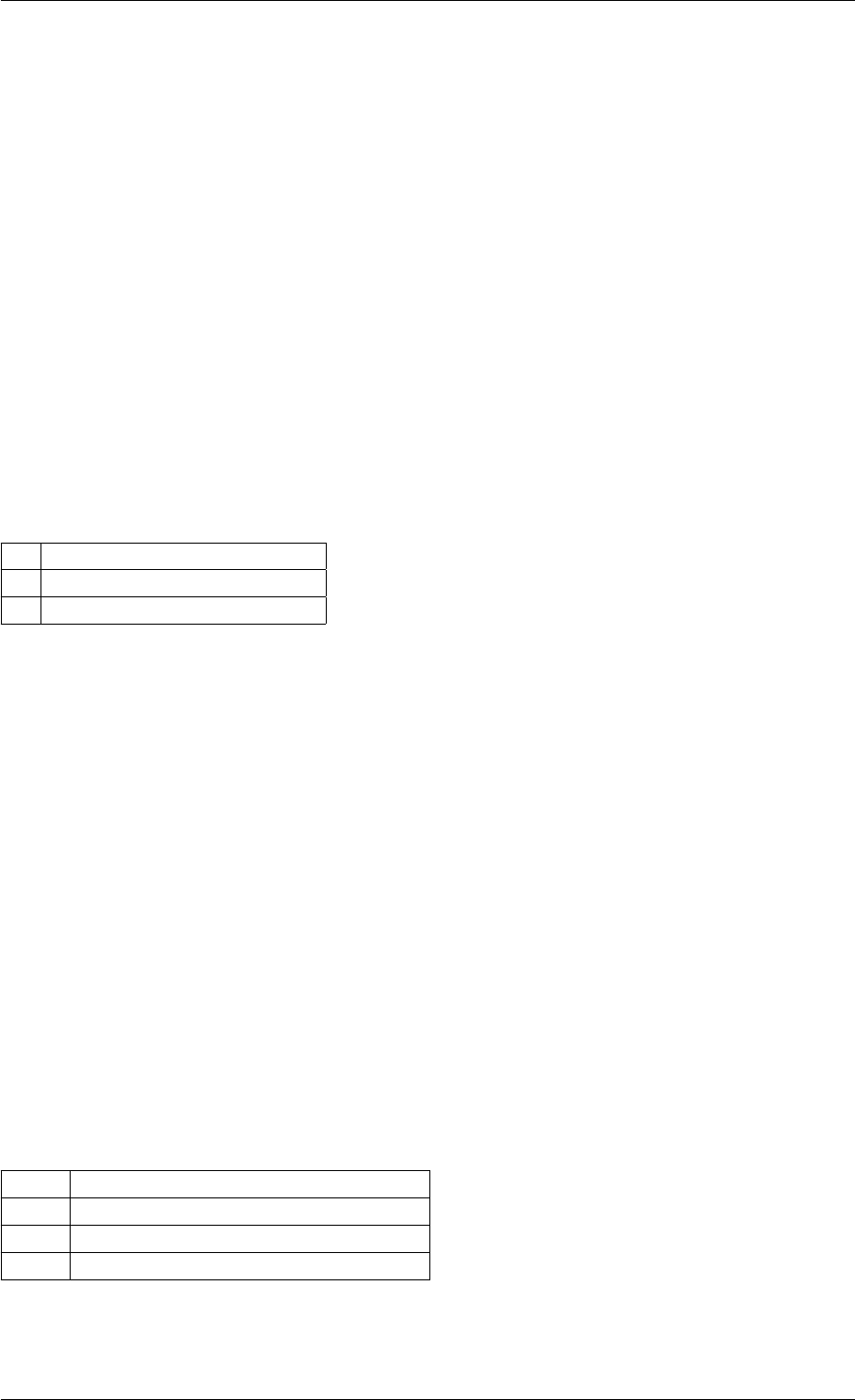
494 CONTENTS
10.35.1 Detailed Description
Definition at line 25 of file steadystateproblem.h.
10.35.2 Constructor & Destructor Documentation
10.35.2.1 SteadystateProblem()
SteadystateProblem (
realtype ∗t,
AmiVector ∗x,
AmiVectorArray ∗sx )
default constructor
Parameters
tpointer to time variable
xpointer to state variables
sx pointer to state sensitivity variables
Definition at line 108 of file steadystateproblem.h.
10.35.3 Member Function Documentation
10.35.3.1 workSteadyStateProblem()
void workSteadyStateProblem (
ReturnData ∗rdata,
Solver ∗solver,
Model ∗model,
int it )
Tries to determine the steady state of the ODE system by a Newton solver, uses forward intergration, if the Newton
solver fails, restarts Newton solver, if integration fails. Computes steady state sensitivities
Parameters
solver pointer to the AMICI solver object
model pointer to the AMICI model object
it integer with the index of the current time step
rdata pointer to the return data object
Definition at line 21 of file steadystateproblem.cpp.
Generated by Doxygen
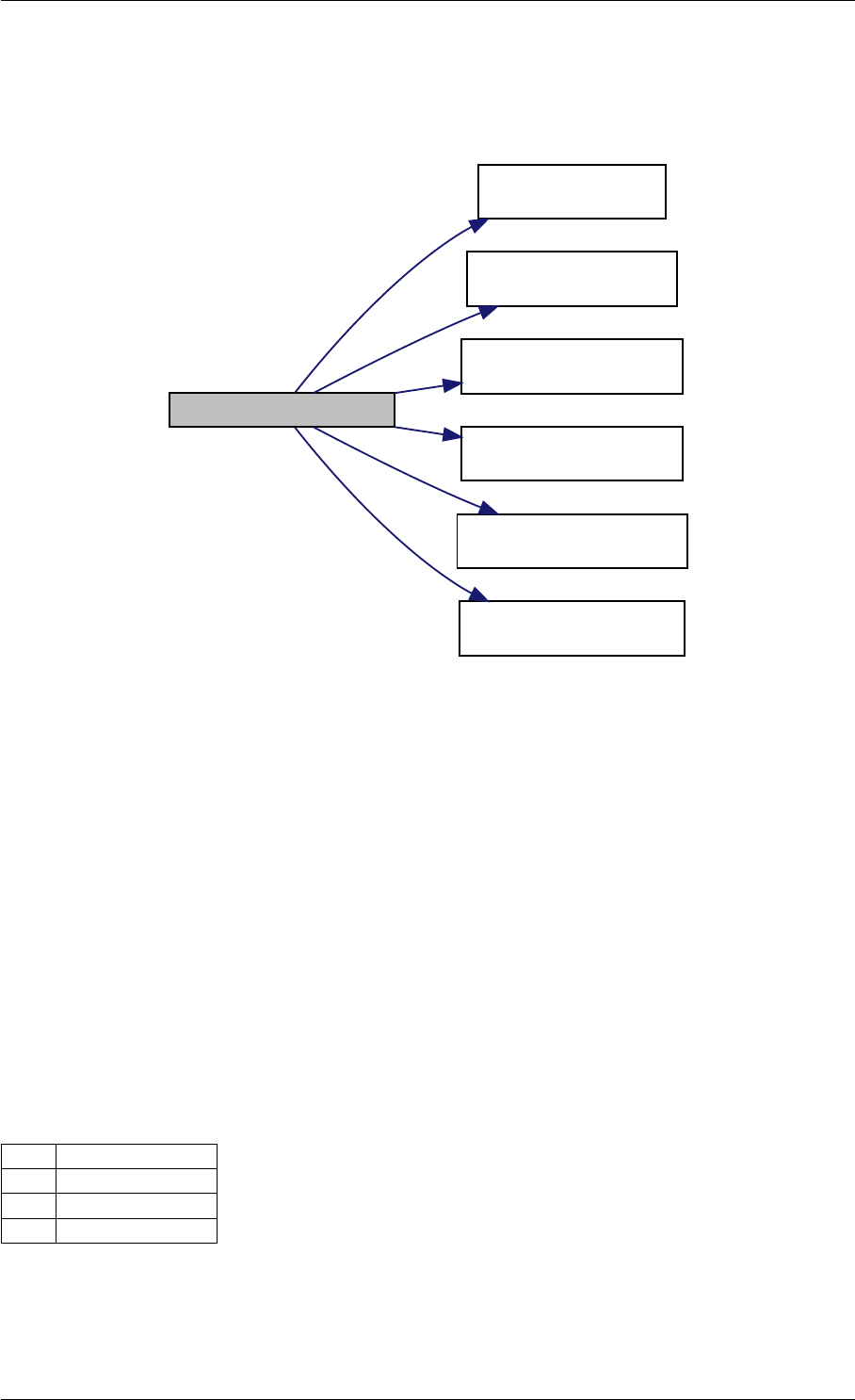
10.35 SteadystateProblem Class Reference 495
Here is the call graph for this function:
workSteadyStateProblem
amici::NewtonSolver
::getSolver
amici::Solver::getLinear
Solver
amici::Solver::getNewton
MaxLinearSteps
amici::Solver::getNewton
MaxSteps
amici::Solver::getAbsolute
Tolerance
amici::Solver::getRelative
Tolerance
10.35.3.2 getWrmsNorm()
realtype getWrmsNorm (
AmiVector const & x,
AmiVector const & xdot,
realtype atol,
realtype rtol )
Computes the weighted root mean square of xdot the weights are computed according to x: w_i = 1 / ( rtol ∗x_i +
atol )
Parameters
xcurrent state
xdot current rhs
atol absolute tolerance
rtol relative tolerance
Returns
root-mean-square norm
Generated by Doxygen
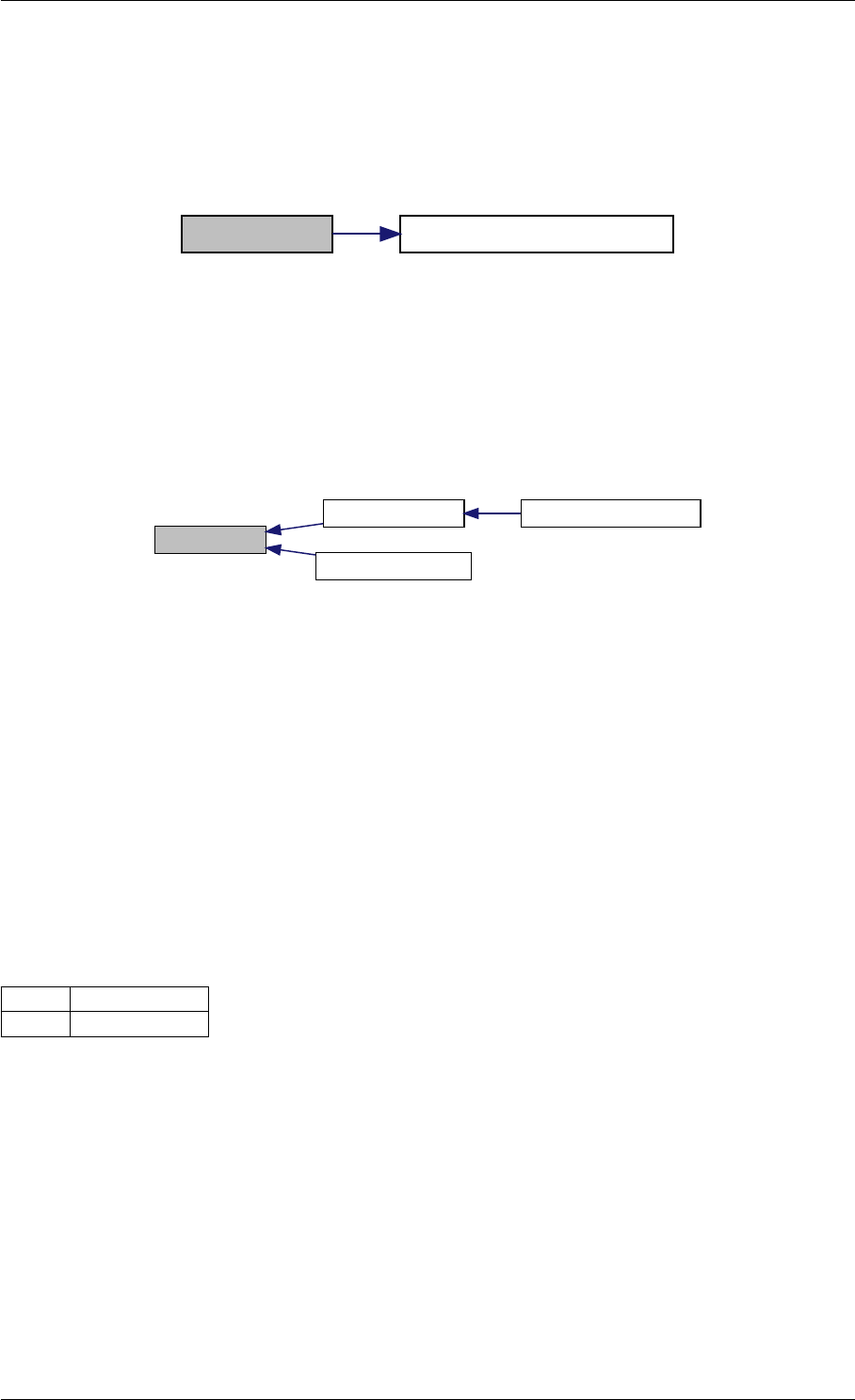
496 CONTENTS
Definition at line 95 of file steadystateproblem.cpp.
Here is the call graph for this function:
getWrmsNorm amici::AmiVector::getNVector
Here is the caller graph for this function:
getWrmsNorm
checkConvergence
applyNewtonsMethod
getSteadystateSimulation
10.35.3.3 checkConvergence()
bool checkConvergence (
const Solver ∗solver,
Model ∗model )
Checks convergence for state and respective sensitivities
Parameters
solver Solver instance
model instance
Returns
boolean indicating convergence
Definition at line 107 of file steadystateproblem.cpp.
Generated by Doxygen
10.35 SteadystateProblem Class Reference 497
Here is the call graph for this function:
checkConvergence
getWrmsNorm
amici::Solver::getAbsolute
ToleranceSteadyState
amici::Solver::getRelative
ToleranceSteadyState
amici::Solver::getSensitivity
Order
amici::AmiVector::getNVector
amici::isNaN
Here is the caller graph for this function:
checkConvergence getSteadystateSimulation
10.35.3.4 applyNewtonsMethod()
void applyNewtonsMethod (
ReturnData ∗rdata,
Model ∗model,
NewtonSolver ∗newtonSolver,
int newton_try )
Runs the Newton solver iterations and checks for convergence to steady state
Parameters
rdata pointer to the return data object
model pointer to the AMICI model object
newtonSolver pointer to the NewtonSolver object
Type:NewtonSolver
newton_try integer start number of Newton solver (1 or 2)
Definition at line 132 of file steadystateproblem.cpp.
Generated by Doxygen
498 CONTENTS
Here is the call graph for this function:
applyNewtonsMethod
amici::AmiVector::reset
getWrmsNorm amici::AmiVector::getNVector
amici::NewtonSolver
::getStep
amici::getNaN
amici::NewtonSolver
::prepareLinearSystem
amici::AmiVector::minus
amici::NewtonSolver
::solveLinearSystem
10.35.3.5 writeNewtonOutput()
void writeNewtonOutput (
ReturnData ∗rdata,
const Model ∗model,
NewtonStatus newton_status,
double run_time,
int it )
Stores output of workSteadyStateProblem in return data
Parameters
newton_status integer flag indicating when a steady state was found
run_time double coputation time of the solver in milliseconds
rdata pointer to the return data instance
model pointer to the model instance
it current timepoint index, <0 indicates preequilibration
Definition at line 221 of file steadystateproblem.cpp.
10.35.3.6 getSteadystateSimulation()
void getSteadystateSimulation (
ReturnData ∗rdata,
Solver ∗solver,
Model ∗model,
int it )
Forward simulation is launched, if Newton solver fails in first try
Generated by Doxygen
10.35 SteadystateProblem Class Reference 499
Parameters
solver pointer to the AMICI solver object
model pointer to the AMICI model object
rdata pointer to the return data object
it current timepoint index, <0 indicates preequilibration
Definition at line 246 of file steadystateproblem.cpp.
Here is the call graph for this function:
getSteadystateSimulation
checkConvergence
amici::Solver::getSensitivity
Order
amici::Solver::solve
amici::Solver::getMaxSteps
getWrmsNorm
amici::Solver::getAbsolute
ToleranceSteadyState
amici::Solver::getRelative
ToleranceSteadyState
amici::AmiVector::getNVector
amici::isNaN
10.35.3.7 createSteadystateSimSolver()
std::unique_ptr<CVodeSolver >createSteadystateSimSolver (
Solver ∗solver,
Model ∗model,
realtype tstart )
initialize CVodeSolver instance for preequilibration simulation
Parameters
solver pointer to the AMICI solver object
model pointer to the AMICI model object
tstart time point for starting Newton simulation
Returns
solver instance
Definition at line 273 of file steadystateproblem.cpp.
Generated by Doxygen
500 CONTENTS
Here is the call graph for this function:
createSteadystateSimSolver
amici::Solver::getLinear
MultistepMethod
amici::Solver::getNonlinear
SolverIteration
amici::Solver::getAbsolute
Tolerance
amici::Solver::getRelative
Tolerance
amici::Solver::getAbsolute
ToleranceSteadyState
amici::Solver::getRelative
ToleranceSteadyState
amici::Solver::getMaxSteps
amici::Solver::getStability
LimitFlag
amici::Solver::getLinear
Solver
amici::isNaN
11 File Documentation
11.1 amici.cpp File Reference
core routines for integration
#include "amici/amici.h"
#include "amici/backwardproblem.h"
#include "amici/forwardproblem.h"
#include "amici/misc.h"
#include <sundials/sundials_types.h>
#include <cvodes/cvodes.h>
#include <type_traits>
#include <cassert>
#include <cstdlib>
#include <cstring>
#include <cstdarg>
#include <memory>
Generated by Doxygen

11.1 amici.cpp File Reference 501
#include <cmath>
Include dependency graph for amici.cpp:
amici.cpp
amici/amici.h
cmath
cstdarg cstring
memoryamici/misc.h
amici/backwardproblem.h amici/forwardproblem.h
sundials/sundials_types.h cvodes/cvodes.h type_traits cassert cstdlib
amici/model.h
amici/exception.h
amici/defines.h
amici/symbolic_functions.h
amici/solver.h
amici/rdata.h amici/edata.h
amici/cblas.h
amici/vector.h
vector
sundials/sundials_direct.hsundials/sundials_sparse.h numeric
nvector/nvector_serial.h
functional
Namespaces
•amici
The AMICI Python module (in doxygen this will also contain documentation about the C++ library)
Macros
• #define _USE_MATH_DEFINES
• #define M_PI 3.14159265358979323846
Functions
• std::unique_ptr<ReturnData >runAmiciSimulation (Solver &solver, const ExpData ∗edata, Model &model)
• void printErrMsgIdAndTxt (const char ∗identifier, const char ∗format,...)
• void printWarnMsgIdAndTxt (const char ∗identifier, const char ∗format,...)
11.1.1 Macro Definition Documentation
11.1.1.1 _USE_MATH_DEFINES
#define _USE_MATH_DEFINES
MS definition of PI and other constants
Definition at line 23 of file amici.cpp.
11.1.1.2 M_PI
#define M_PI 3.14159265358979323846
define PI if we still have no definition
Definition at line 27 of file amici.cpp.
Generated by Doxygen
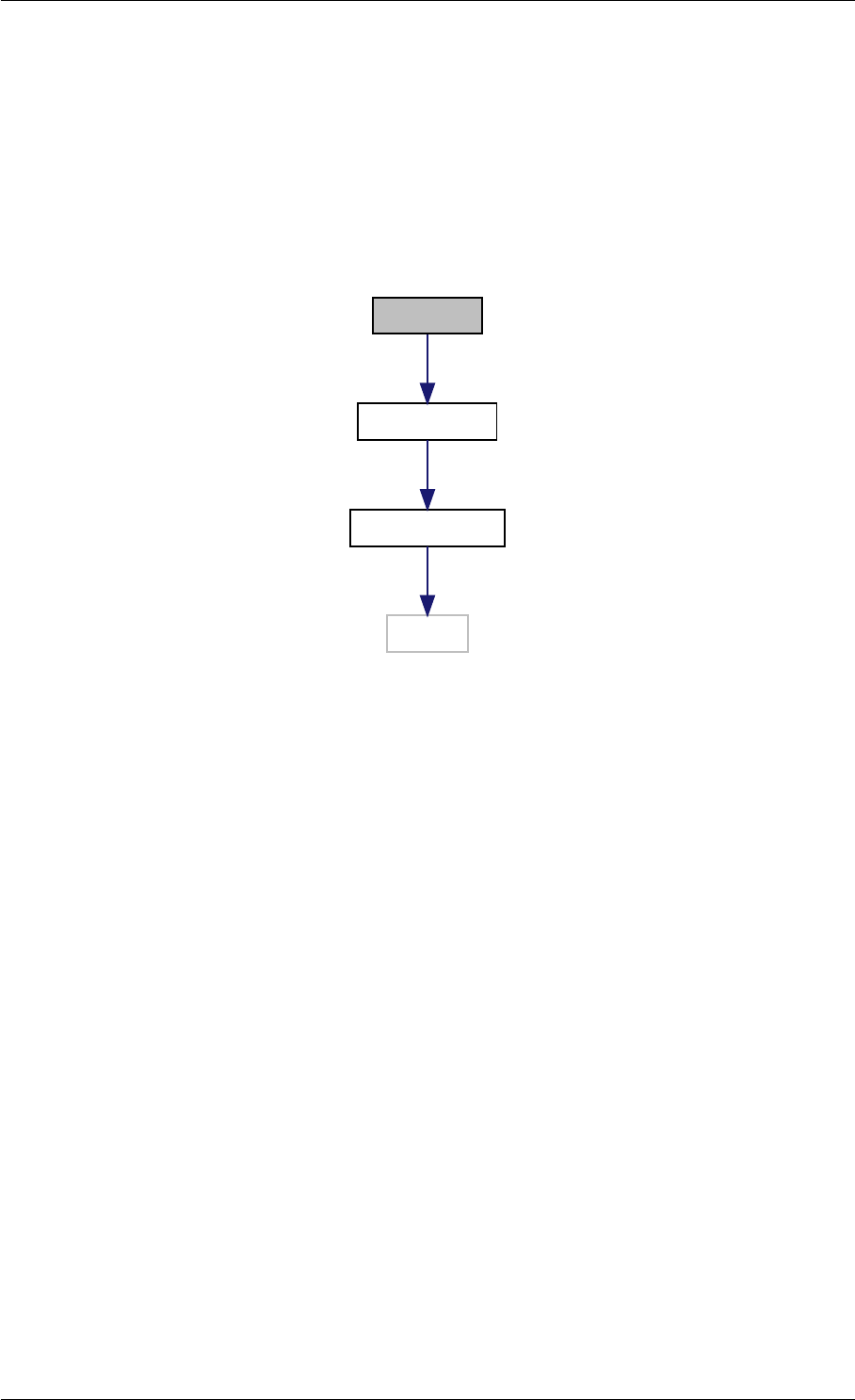
502 CONTENTS
11.2 cblas.cpp File Reference
BLAS routines required by AMICI.
#include "amici/cblas.h"
Include dependency graph for cblas.cpp:
cblas.cpp
amici/cblas.h
amici/defines.h
cmath
Namespaces
•amici
The AMICI Python module (in doxygen this will also contain documentation about the C++ library)
Functions
• void amici_dgemm (BLASLayout layout, BLASTranspose TransA, BLASTranspose TransB, const int M, const
int N, const int K, const double alpha, const double ∗A, const int lda, const double ∗B, const int ldb, const
double beta, double ∗C, const int ldc)
• void amici_dgemv (BLASLayout layout, BLASTranspose TransA, const int M, const int N, const double alpha,
const double ∗A, const int lda, const double ∗X, const int incX, const double beta, double ∗Y, const int incY)
• void amici_daxpy (int n, double alpha, const double ∗x, const int incx, double ∗y, int incy)
Compute y = a∗x + y.
11.3 interface_matlab.cpp File Reference
core routines for mex interface
#include "amici/interface_matlab.h"
#include "amici/model.h"
Generated by Doxygen
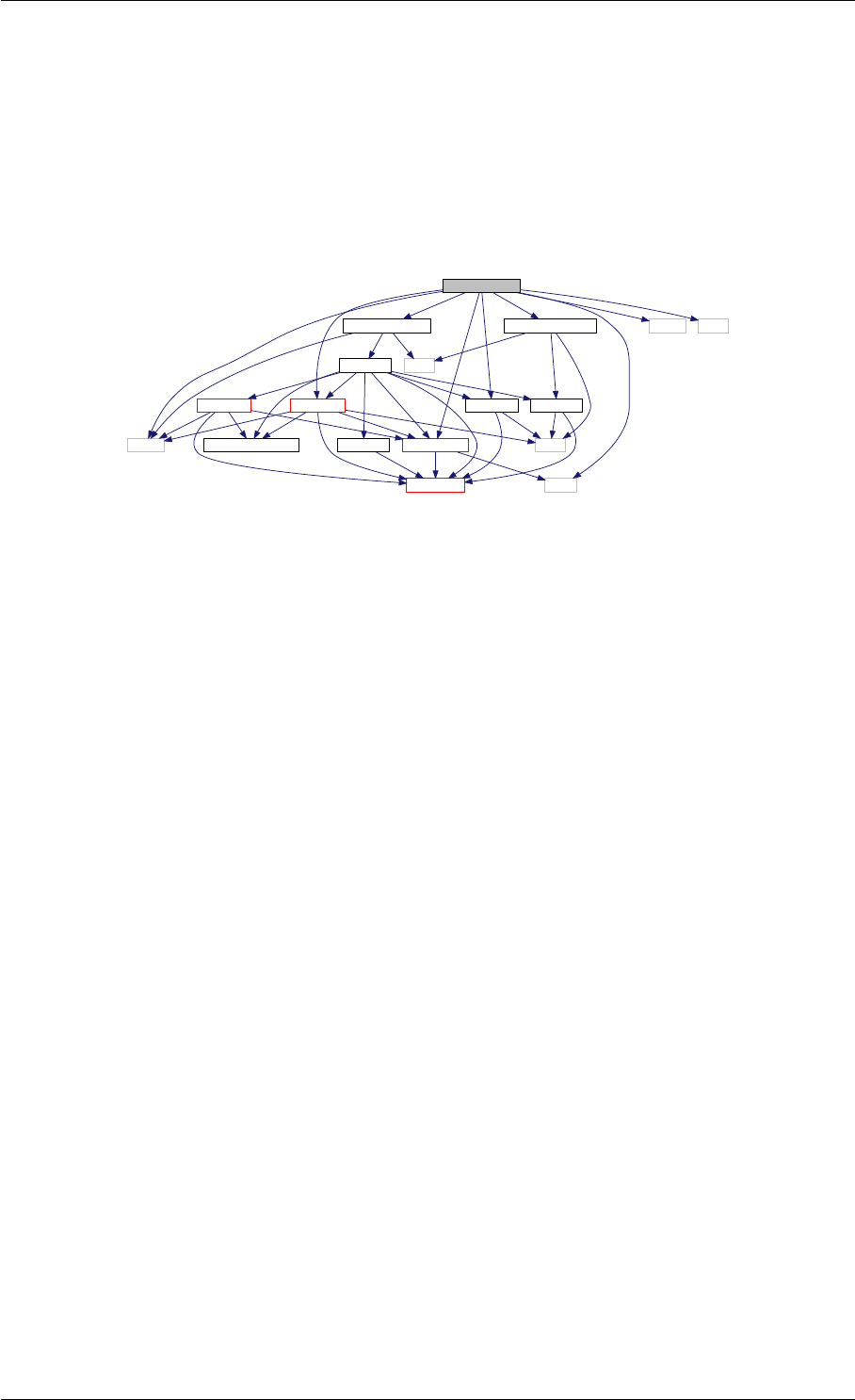
11.3 interface_matlab.cpp File Reference 503
#include "amici/exception.h"
#include "amici/edata.h"
#include "amici/returndata_matlab.h"
#include <assert.h>
#include <blas.h>
#include <cstring>
#include <memory>
Include dependency graph for interface_matlab.cpp:
interface_matlab.cpp
amici/interface_matlab.h
amici/model.h
amici/exception.h
cstring
memory
amici/edata.h
amici/returndata_matlab.h assert.h blas.h
amici/amici.h mex.h
amici/defines.h
amici/symbolic_functions.h
amici/solver.h amici/rdata.h
amici/cblas.h vector
Namespaces
•amici
The AMICI Python module (in doxygen this will also contain documentation about the C++ library)
Enumerations
• enum mexRhsArguments {
RHS_TIMEPOINTS,RHS_PARAMETERS,RHS_CONSTANTS,RHS_OPTIONS,
RHS_PLIST,RHS_XSCALE_UNUSED,RHS_INITIALIZATION,RHS_DATA,
RHS_NUMARGS_REQUIRED = RHS_DATA, RHS_NUMARGS }
The mexFunctionArguments enum takes care of the ordering of mex file arguments (indexing in prhs)
Functions
• int dbl2int (const double x)
• char amici_blasCBlasTransToBlasTrans (BLASTranspose trans)
• void amici_dgemm (BLASLayout layout, BLASTranspose TransA, BLASTranspose TransB, const int M, const
int N, const int K, const double alpha, const double ∗A, const int lda, const double ∗B, const int ldb, const
double beta, double ∗C, const int ldc)
• void amici_dgemv (BLASLayout layout, BLASTranspose TransA, const int M, const int N, const double alpha,
const double ∗A, const int lda, const double ∗X, const int incX, const double beta, double ∗Y, const int incY)
• void amici_daxpy (int n, double alpha, const double ∗x, const int incx, double ∗y, int incy)
Compute y = a∗x + y.
• std::vector<realtype >mxArrayToVector (const mxArray ∗array, int length)
• std::unique_ptr<ExpData >expDataFromMatlabCall (const mxArray ∗prhs[ ], const Model &model)
• void setSolverOptions (const mxArray ∗prhs[ ], int nrhs, Solver &solver)
setSolverOptions solver options from the matlab call to a solver object
• void setModelData (const mxArray ∗prhs[ ], int nrhs, Model &model)
setModelData sets data from the matlab call to the model object
• void mexFunction (int nlhs, mxArray ∗plhs[ ], int nrhs, const mxArray ∗prhs[ ])
Generated by Doxygen
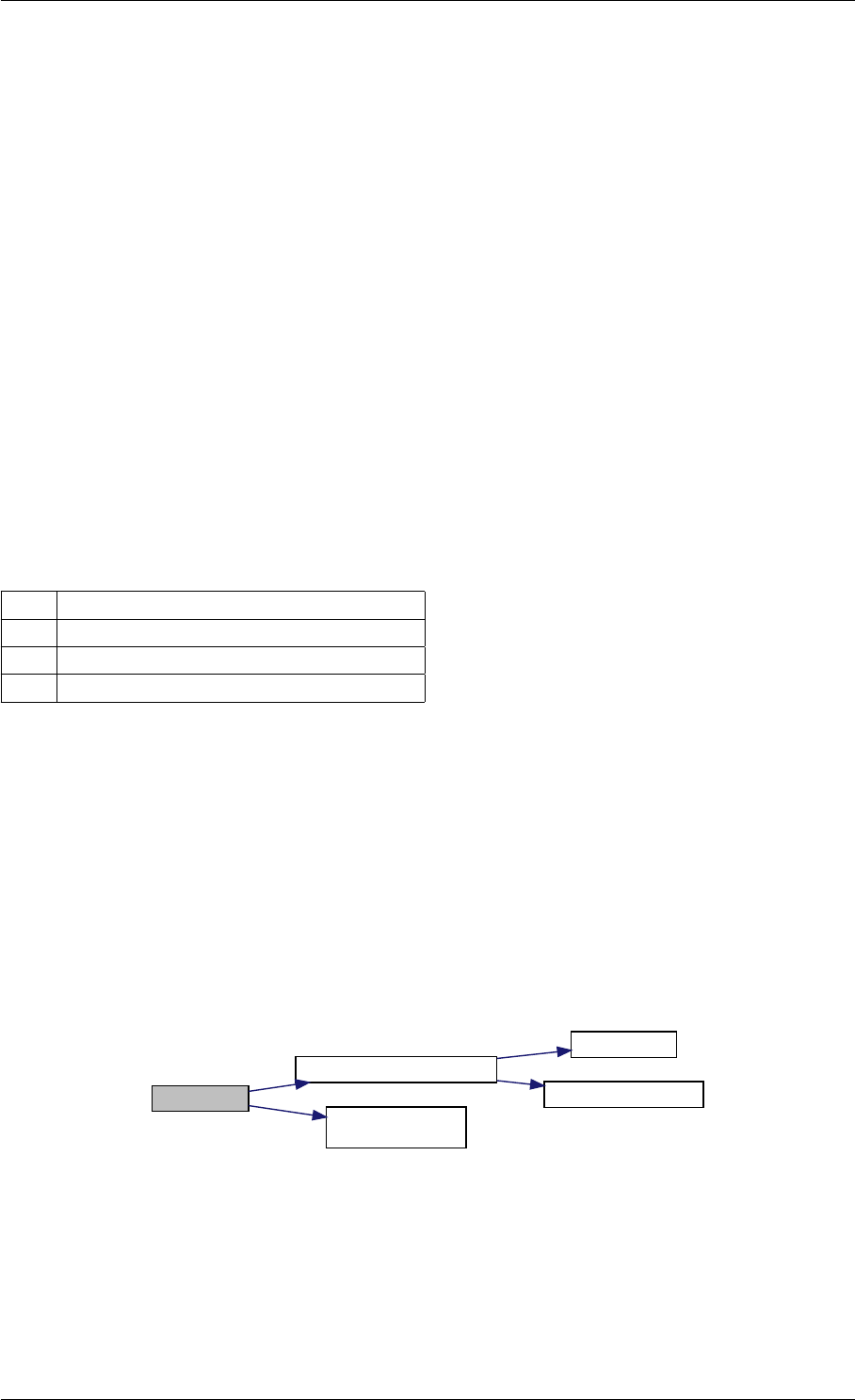
504 CONTENTS
11.3.1 Detailed Description
This file defines the fuction mexFunction which is executed upon calling the mex file from matlab
11.3.2 Function Documentation
11.3.2.1 mexFunction()
void mexFunction (
int nlhs,
mxArray ∗plhs[ ],
int nrhs,
const mxArray ∗prhs[ ] )
mexFunction is the main interface function for the MATLAB interface. It reads in input data (udata and edata) and
creates output data compound (rdata) and then calls the AMICI simulation routine to carry out numerical integration.
Parameters
nlhs number of output arguments of the matlab call
plhs pointer to the array of output arguments
nrhs number of input arguments of the matlab call
prhs pointer to the array of input arguments
Returns
void
Definition at line 525 of file interface_matlab.cpp.
Here is the call graph for this function:
mexFunction
amici::expDataFromMatlabCall
amici::AmiException
::what
amici.ExpData
amici::mxArrayToVector
11.4 spline.cpp File Reference
definition of spline functions
Generated by Doxygen
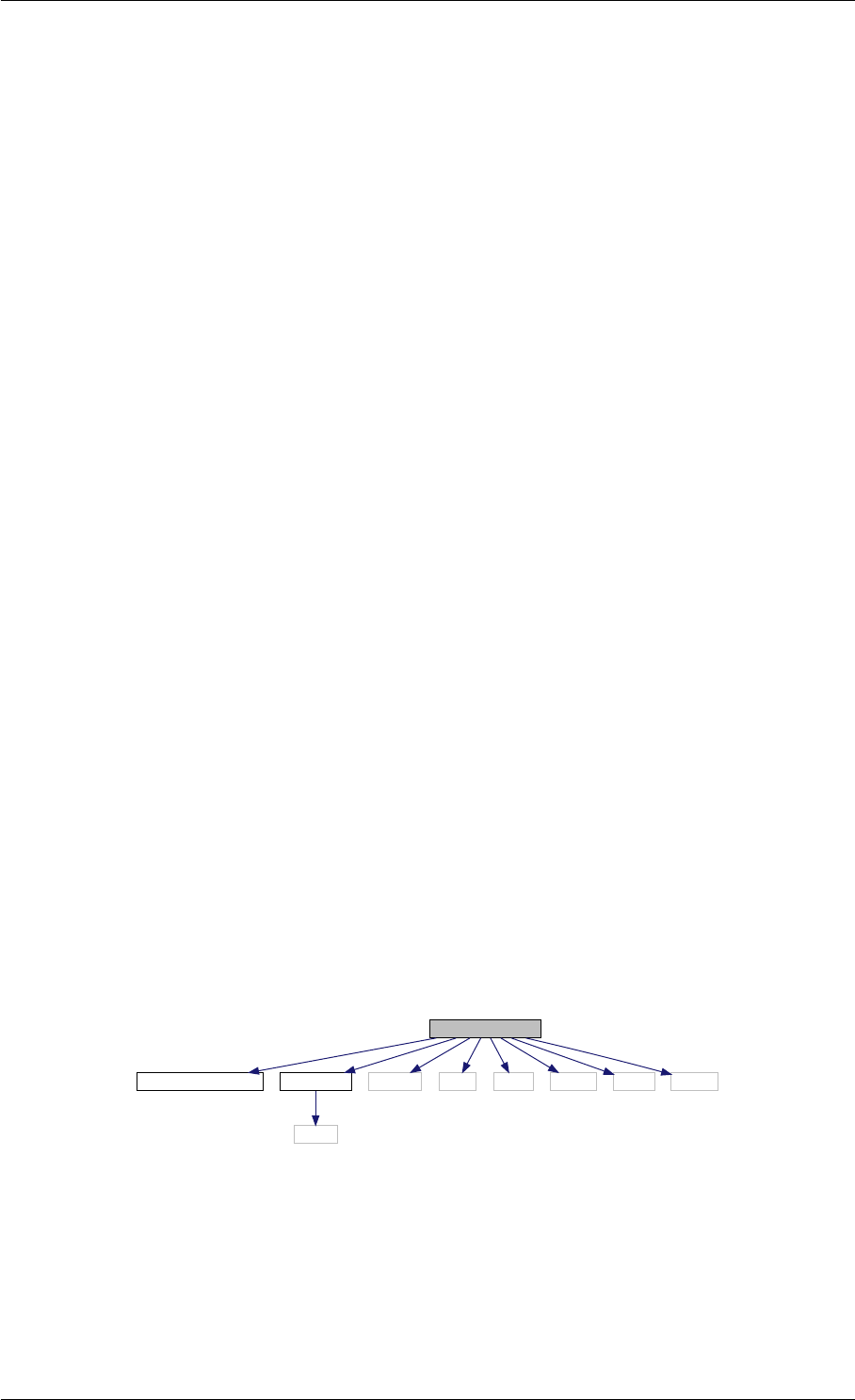
11.5 symbolic_functions.cpp File Reference 505
Namespaces
•amici
The AMICI Python module (in doxygen this will also contain documentation about the C++ library)
Functions
• int spline (int n, int end1, int end2, double slope1, double slope2, double x[ ], double y[ ], double b[ ], double
c[ ], double d[ ])
• double seval (int n, double u, double x[ ], double y[ ], double b[ ], double c[ ], double d[ ])
Evaluate the cubic spline function.
• double sinteg (int n, double u, double x[ ], double y[ ], double b[ ], double c[ ], double d[ ])
11.4.1 Detailed Description
Author
Peter & Nigel, Design Software, 42 Gubberley St, Kenmore, 4069, Australia.
11.5 symbolic_functions.cpp File Reference
definition of symbolic functions
#include "amici/symbolic_functions.h"
#include "amici/spline.h"
#include <algorithm>
#include <cfloat>
#include <cmath>
#include <cstdarg>
#include <cstdlib>
#include <alloca.h>
Include dependency graph for symbolic_functions.cpp:
symbolic_functions.cpp
amici/symbolic_functions.h amici/spline.h algorithm cfloat cmath cstdarg cstdlib alloca.h
math.h
Namespaces
•amici
The AMICI Python module (in doxygen this will also contain documentation about the C++ library)
Generated by Doxygen

506 CONTENTS
Functions
• int isNaN (double what)
• int isInf (double what)
• double getNaN ()
• double log (double x)
• double dirac (double x)
• double heaviside (double x)
• double sign (double x)
• double max (double a, double b, double c)
• double min (double a, double b, double c)
• double Dmax (int id, double a, double b, double c)
• double Dmin (int id, double a, double b, double c)
• double pos_pow (double base, double exponent)
• double spline (double t, int num,...)
• double spline_pos (double t, int num,...)
• double Dspline (int id, double t, int num,...)
• double Dspline_pos (int id, double t, int num,...)
• double DDspline (int id1, int id2, double t, int num,...)
• double DDspline_pos (int id1, int id2, double t, int num,...)
11.5.1 Detailed Description
This file contains definitions of various symbolic functions which
Generated by Doxygen
Index
_USE_MATH_DEFINES
amici.cpp, 501
__init__
amici::ode_export::Constant, 362
amici::ode_export::Expression, 363
amici::ode_export::LogLikelihood, 364
amici::ode_export::ModelQuantity, 365
amici::ode_export::ODEExporter, 369
amici::ode_export::ODEModel, 373
amici::ode_export::Observable, 367
amici::ode_export::Parameter, 386
amici::ode_export::SigmaY, 387
amici::ode_export::State, 388
amici::sbml_import::SbmlImporter, 410
__repr__
amici::ode_export::ModelQuantity, 366
∼Model
amici::Model, 161
add_component
amici::ode_export::ODEModel, 375
adjInit
amici::Solver, 474
allocateSolver
amici::Solver, 467
allocateSolverB
amici::Solver, 474
AmiException, 86
amici::AmiException, 86,87
AmiVector, 89
amici::AmiVector, 89,90
AmiVectorArray, 99
amici::AmiVectorArray, 100
amici, 14
amici_blasCBlasTransToBlasTrans, 70
amici_daxpy, 27
amici_dgemm, 25
amici_dgemv, 24
amici_path, 74
BLASLayout, 19
BLASTranspose, 20
checkFieldNames, 44
checkFinite, 31
DDspline, 65
DDspline_pos, 65
dbl2int, 69
deserializeFromChar, 46
deserializeFromString, 47
dirac, 53
Dmax, 57
Dmin, 54
Dspline, 63
Dspline_pos, 64
errMsgIdAndTxt, 74
ExpData, 67
expDataFromMatlabCall, 30
getNaN, 60
getReturnDataMatlabFromAmiciCall, 35
getUnscaledParameter, 34
getValueById, 71
heaviside, 53
initAndAttachArray, 43
initMatlabDiagnosisFields, 37
initMatlabReturnFields, 36
InternalSensitivityMethod, 21
InterpolationType, 21
isInf, 59
isNaN, 58
LinearMultistepMethod, 21
LinearSolver, 21
log, 52
max, 56
min, 54
msgIdAndTxtFp, 19
mxArrayToVector, 70
NewtonStatus, 22
NonlinearSolverIteration, 21
operator==, 35,48
ParameterScaling, 20
pi, 74
pos_pow, 58
printErrMsgIdAndTxt, 22
printWarnMsgIdAndTxt, 23
pysb2amici, 69
realtype, 19
reorder, 45
runAmiciSimulation, 23,66
runAmiciSimulations, 68
SecondOrderMode, 20
SensitivityMethod, 20
SensitivityOrder, 20
serializeToChar, 46
serializeToStdVec, 47
serializeToString, 47
setModelData, 28
setSolverOptions, 28
setValueById, 72
setValueByIdRegex, 73
setupReturnData, 29
seval, 50
sign, 61
sinteg, 51
spline, 48,61
spline_pos, 62
StateOrdering, 22
SteadyStateSensitivityMode, 22
unscaleParameters, 32,33
warnMsgIdAndTxt, 74
writeMatlabField0, 38
writeMatlabField1, 39

508 INDEX
writeMatlabField2, 40
writeMatlabField3, 41
writeMatlabField4, 42
amici.cpp, 500
_USE_MATH_DEFINES, 501
M_PI, 501
amici.ode_export, 75
amici.plotting, 81
amici.sbml_import, 82
amici::AmiException
AmiException, 86,87
getBacktrace, 88
storeBacktrace, 88
what, 87
amici::AmiVector
AmiVector, 89,90
at, 98
data, 91,92
getLength, 95
getNVector, 93
getVector, 94
minus, 97
operator=, 90
operator[], 98
reset, 96
set, 97
amici::AmiVectorArray
AmiVectorArray, 100
at, 102
data, 101,102
getLength, 104
getNVector, 103
getNVectorArray, 103
operator[], 104
reset, 105
amici::BackwardProblem
BackwardProblem, 106
getdJydx, 110
getdxBptr, 110
gett, 107
getwhich, 108
getwhichptr, 108
getxBptr, 109
getxQBptr, 109
workBackwardProblem, 107
amici::CvodeException
CvodeException, 112
amici::ExpData
checkDataDimension, 135
checkEventsDimension, 135
checkSigmaPositivity, 136,137
ExpData, 114–117
fixedParameters, 138
fixedParametersPreequilibration, 138
fixedParametersPresimulation, 138
getObservedData, 122
getObservedDataPtr, 122
getObservedDataStdDev, 127
getObservedDataStdDevPtr, 127
getObservedEvents, 129
getObservedEventsPtr, 130
getObservedEventsStdDev, 134
getObservedEventsStdDevPtr, 134
getTimepoint, 119
getTimepoints, 119
isSetObservedData, 121
isSetObservedDataStdDev, 126
isSetObservedEvents, 129
isSetObservedEventsStdDev, 133
nmaxevent, 137
nt, 117
nytrue, 137
nztrue, 137
observedData, 138
observedDataStdDev, 139
observedEvents, 139
observedEventsStdDev, 139
setObservedData, 120,121
setObservedDataStdDev, 123–125
setObservedEvents, 128
setObservedEventsStdDev, 130–132
setTimepoints, 118
ts, 138
amici::ForwardProblem
edata, 147
ForwardProblem, 140
getDJydx, 144
getDJzdx, 144
getDiscontinuities, 143
getNumberOfRoots, 143
getRHSAtDiscontinuities, 142
getRHSBeforeDiscontinuities, 143
getRootCounter, 144
getRootIndexes, 143
getStateDerivativePointer, 145
getStateDerivativeSensitivityPointer, 146
getStatePointer, 145
getStateSensitivity, 142
getStateSensitivityPointer, 145
getStatesAtDiscontinuities, 142
getTime, 141
model, 146
rdata, 146
solver, 146
workForwardProblem, 141
amici::IDAException
IDAException, 148
amici::IntegrationFailure
error_code, 149
IntegrationFailure, 149
time, 149
amici::IntegrationFailureB
error_code, 151
IntegrationFailureB, 150
time, 151
amici::Model
Generated by Doxygen

INDEX 509
∼Model, 161
anyStateNonNegative, 280
boost::serialization::serialize, 268
checkFinite, 224
clone, 162
computeX_pos, 266
dJrzdsigma, 276
dJrzdz, 276
dJydp, 273
dJydsigma, 275
dJydy, 275
dJzdp, 273
dJzdsigma, 276
dJzdz, 276
deltaqB, 274
deltasx, 274
deltax, 274
deltaxB, 274
drzdp, 277
drzdx, 277
dsigmaydp, 273
dsigmazdp, 273
dwdp, 278
dwdx, 278
dxdotdp, 274
dydp, 277
dydx, 277
dzdp, 277
dzdx, 276
fFIM, 223
fJDiag, 165
fJSparse, 164
fJrz, 187,254
fJv, 166
fJy, 185,253
fJz, 186,253
fchi2, 222
fdJrzdsigma, 191,257
fdJrzdz, 190,256
fdJydp, 193
fdJydsigma, 188,255
fdJydx, 194
fdJydy, 187,254
fdJzdp, 196
fdJzdsigma, 190,256
fdJzdx, 196
fdJzdz, 189,255
fdeltaqB, 182,250
fdeltasx, 180,249
fdeltax, 179,248
fdeltaxB, 181,250
fdrzdp, 178,247
fdrzdx, 179,248
fdsigmaydp, 183,251
fdsigmazdp, 184,252
fdwdp, 217,218,257
fdwdx, 218,220,258
fdx0, 169
fdxdotdp, 166
fdydp, 172,243
fdydx, 173,244
fdzdp, 177,246
fdzdx, 178,247
fixedParameters, 279
fJ, 164
fres, 221
froot, 162
frz, 175,245
fsJy, 193
fsJz, 195
fsdx0, 171
fsigmay, 183,251
fsigmaz, 184,252
fsres, 222
fsrz, 176,246
fstau, 171,242
fsx0, 169,242
fsx0_fixedParameters, 170,241
fsxdot, 163
fsy, 191
fsz, 174,245
fsz_tf, 192
fw, 215,216,257
fx0, 167,240
fx0_fixedParameters, 168,241
fxdot, 163
fy, 172,243
fz, 174,244
getFixedParameterById, 205
getFixedParameterByName, 206
getFixedParameterIds, 237
getFixedParameterNames, 229
getFixedParameters, 205
getInitialStateSensitivities, 213
getInitialStates, 212
getObservableIds, 238
getObservableNames, 229
getParameterById, 231
getParameterByName, 232
getParameterIds, 231
getParameterList, 211
getParameterNames, 226
getParameterScale, 203
getParameters, 204
getReinitializeFixedParameterInitialStates, 240
getSolver, 162
getStateIds, 236
getStateIsNonNegative, 209
getStateNames, 227
getSteadyStateSensitivityMode, 239
getTimepoints, 209
getUnscaledParameters, 204
getmy, 258
getmz, 259
getrz, 264
getsrz, 265
Generated by Doxygen

510 INDEX
getsx, 263
getsz, 265
gett, 224
getx, 262
gety, 261
getz, 263
h, 279
hasFixedParameterIds, 236
hasFixedParameterNames, 228
hasObservableIds, 238
hasObservableNames, 229
hasParameterIds, 230
hasParameterNames, 225
hasStateIds, 235
hasStateNames, 227
idlist, 272
initHeaviside, 198
initialize, 197
initializeStates, 197
isFixedParameterStateReinitializationAllowed, 266
J, 275
k, 202
lbw, 272
M, 278
Model, 159,161
my, 275
mz, 275
nMaxEvent, 202
ndwdp, 271
ndwdx, 271
ne, 270
nJ, 271
nk, 201
nmaxevent, 281
nnz, 271
np, 200
nplist, 199
nt, 202
nw, 270
nx, 269
nxtrue, 269
ny, 269
nytrue, 270
nz, 270
nztrue, 270
o2mode, 272
operator=, 161
operator==, 269
originalParameters, 279
plist, 214
plist_, 279
pscale, 281
reinitializeFixedParameterInitialStates, 282
setFixedParameterById, 207
setFixedParameterByName, 208
setFixedParameters, 205
setFixedParametersByIdRegex, 207
setFixedParametersByNameRegex, 208
setInitialStateSensitivities, 213
setInitialStates, 212
setNMaxEvent, 202
setParameterById, 233
setParameterByName, 234
setParameterList, 212
setParameterScale, 203
setParameters, 204
setParametersByIdRegex, 233
setParametersByNameRegex, 235
setReinitializeFixedParameterInitialStates, 239
setStateIsNonNegative, 210
setSteadyStateSensitivityMode, 239
setT0, 214
setTimepoints, 209
sigmay, 272
sigmaz, 273
stateIsNonNegative, 280
stau, 278
steadyStateSensitivityMode, 281
sx0data, 280
t, 210
t0, 213
ts, 280
tstart, 281
ubw, 271
unscaledParameters, 279
updateHeaviside, 223
updateHeavisideB, 224
w, 278
x0data, 280
x_pos_tmp, 281
z2event, 272
amici::Model_DAE
fJDiag, 291,307
fJSparse, 288,289,306
fJSparseB, 290,307
fJB, 287,306
fJv, 292,293,308
fJvB, 294,309
fdxdotdp, 300,301,312
fJ, 285,286,305
fM, 304,313
fqBdot, 299,311
froot, 295,296,310
fsxdot, 302,303,312
fxBdot, 298,311
fxdot, 296,297,310
getSolver, 305
Model_DAE, 284
amici::Model_ODE
fJDiag, 323,324,338
fJSparse, 320,321,337
fJSparseB, 322,338
fJB, 319,337
fJv, 325,326,339
fJvB, 327,340
fdxdotdp, 333,342
Generated by Doxygen

INDEX 511
fJ, 317,318,336
fqBdot, 332,342
froot, 328,329,340
fsxdot, 334,335,343
fxBdot, 331,341
fxdot, 329,330,341
getSolver, 336
Model_ODE, 316,317
amici::NewtonFailure
error_code, 345
NewtonFailure, 344
amici::NewtonSolver
atol, 350
computeNewtonSensis, 348
dx, 352
getSolver, 346
getStep, 347
maxlinsteps, 350
maxsteps, 350
model, 351
NewtonSolver, 346
prepareLinearSystem, 349
rdata, 351
rtol, 351
solveLinearSystem, 349
t, 351
x, 352
xdot, 351
amici::NewtonSolverDense
NewtonSolverDense, 353
prepareLinearSystem, 354
solveLinearSystem, 353
amici::NewtonSolverIterative
linsolveSPBCG, 357
NewtonSolverIterative, 355
prepareLinearSystem, 357
solveLinearSystem, 356
amici::NewtonSolverSparse
NewtonSolverSparse, 359
prepareLinearSystem, 360
solveLinearSystem, 360
amici::ReturnData
applyChainRuleFactorToSimulationResults, 395
boost::serialization::serialize, 395
chi2, 403
FIM, 399
invalidate, 393
invalidateLLH, 394
J, 396
llh, 403
ne, 405
newton_cpu_time, 401
newton_maxsteps, 406
newton_numlinsteps, 402
newton_numsteps, 401
newton_status, 401
nJ, 406
nk, 404
nmaxevent, 406
np, 404
nplist, 406
nt, 406
numerrtestfails, 400
numerrtestfailsB, 400
numnonlinsolvconvfails, 400
numnonlinsolvconvfailsB, 401
numrhsevals, 400
numrhsevalsB, 400
numsteps, 399
numstepsB, 399
nx, 404
nxtrue, 404
ny, 405
nytrue, 405
nz, 405
nztrue, 405
o2mode, 407
order, 401
pscale, 407
res, 399
ReturnData, 391,393
rz, 397
s2llh, 403
s2rz, 397
sensi, 407
sensi_meth, 407
sigmay, 398
sigmaz, 396
sllh, 403
sres, 399
srz, 397
ssigmay, 398
ssigmaz, 397
status, 404
sx, 398
sx0, 403
sy, 398
sz, 396
t_steadystate, 402
ts, 396
wrms_sensi_steadystate, 402
wrms_steadystate, 402
x, 397
x0, 402
xdot, 396
y, 398
z, 396
amici::SetupFailure
SetupFailure, 424
amici::Solver
adjInit, 474
allocateSolver, 467
allocateSolverB, 474
applyQuadTolerancesASA, 490
applySensitivityTolerances, 490
applyTolerances, 488
Generated by Doxygen

512 INDEX
applyTolerancesASA, 489
applyTolerancesFSA, 488
band, 476
bandB, 476
binit, 463
boost::serialization::serialize, 491
calcICB, 435
calcIC, 435
clone, 429
dense, 475
denseB, 475
diag, 477
diagB, 477
getAbsoluteTolerance, 445
getAbsoluteToleranceQuadratures, 449
getAbsoluteToleranceSensi, 447
getAbsoluteToleranceSteadyState, 451
getAbsoluteToleranceSteadyStateSensi, 453
getAdjBmem, 487
getAdjMallocDone, 487
getDiagnosis, 433
getDiagnosisB, 433
getDky, 473
getInternalSensitivityMethod, 461
getInterpolationType, 458
getLastOrder, 483
getLinearMultistepMethod, 456
getLinearSolver, 460
getMallocDone, 486
getMaxSteps, 454
getMaxStepsBackwardProblem, 455
getModel, 486
getNewtonMaxLinearSteps, 442
getNewtonMaxSteps, 440
getNewtonPreequilibration, 441
getNonlinearSolverIteration, 457
getNumErrTestFails, 481
getNumNonlinSolvConvFails, 482
getNumRhsEvals, 481
getNumSteps, 480
getQuadB, 438
getRelativeTolerance, 444
getRelativeToleranceQuadratures, 448
getRelativeToleranceSensi, 447
getRelativeToleranceSteadyState, 450
getRelativeToleranceSteadyStateSensi, 452
getRootInfo, 434
getSens, 432
getSensitivityMethod, 439
getSensitivityOrder, 443
getStabilityLimitFlag, 459
getStateOrdering, 458
getB, 438
init, 462
initializeLinearSolver, 483
initializeLinearSolverB, 484
interpType, 493
ism, 492
iter, 493
klu, 479
kluSetOrdering, 479
kluSetOrderingB, 480
kluB, 480
lmm, 492
maxsteps, 493
nplist, 485
nx, 485
operator==, 491
qbinit, 463
quadReInitB, 439
quadSStolerancesB, 474
reInit, 434
reInitB, 438
rootInit, 464
sensInit1, 464
sensReInit, 435
setAbsoluteTolerance, 446
setAbsoluteToleranceQuadratures, 449
setAbsoluteToleranceSensi, 448
setAbsoluteToleranceSteadyState, 451
setAbsoluteToleranceSteadyStateSensi, 453
setBandJacFn, 465
setBandJacFnB, 466
setDenseJacFn, 465
setDenseJacFnB, 465
setErrHandlerFn, 469
setId, 472
setInternalSensitivityMethod, 462
setInterpolationType, 458
setJacTimesVecFn, 465
setJacTimesVecFnB, 467
setLinearMultistepMethod, 456
setLinearSolver, 461
setMaxNumSteps, 470
setMaxNumStepsB, 471
setMaxSteps, 454
setMaxStepsBackwardProblem, 455
setNewtonMaxLinearSteps, 442
setNewtonMaxSteps, 440
setNewtonPreequilibration, 441
setNonlinearSolverIteration, 457
setQuadErrConB, 469
setRelativeTolerance, 444
setRelativeToleranceQuadratures, 449
setRelativeToleranceSensi, 447
setRelativeToleranceSteadyState, 450
setRelativeToleranceSteadyStateSensi, 452
setSStolerances, 468
setSStolerancesB, 474
setSensErrCon, 469
setSensParams, 473
setSensSStolerances, 468
setSensitivityMethod, 439
setSensitivityOrder, 443
setSparseJacFn, 465
setSparseJacFnB, 466
Generated by Doxygen

INDEX 513
setStabLimDet, 471
setStabLimDetB, 472
setStabilityLimitFlag, 460
setStateOrdering, 459
setStopTime, 437
setSuppressAlg, 473
setUserData, 469
setUserDataB, 470
setup, 429
setupAMIB, 431
solve, 436
solveB, 437
solveF, 436
Solver, 429
solverMemory, 492
solverMemoryB, 492
solverWasCalled, 492
spbcg, 478
spbcgB, 478
spgmr, 477
spgmrB, 478
sptfqmr, 478
sptfqmrB, 479
turnOffRootFinding, 439
wrapErrHandlerFn, 467
amici::SteadystateProblem
applyNewtonsMethod, 497
checkConvergence, 496
createSteadystateSimSolver, 499
getSteadystateSimulation, 498
getWrmsNorm, 495
SteadystateProblem, 494
workSteadyStateProblem, 494
writeNewtonOutput, 498
amici::ode_export
applyTemplate, 77
functions, 79
getSymbolicDiagonal, 77
multiobs_functions, 80
ODEModel_from_pysb_importer, 78
sensi_functions, 80
sparse_functions, 79
symbol_to_type, 80
var_in_function_signature, 76
amici::ode_export::Constant
__init__, 362
amici::ode_export::Expression
__init__, 363
amici::ode_export::LogLikelihood
__init__, 364
amici::ode_export::ModelQuantity
__init__, 365
__repr__, 366
amici::ode_export::ODEExporter
__init__, 369
compileModel, 370
generateModelCode, 369
setName, 372
setPaths, 371
amici::ode_export::ODEModel
__init__, 373
add_component, 375
colptr, 381
eq, 380
generateBasicVariables, 383
import_from_sbml_importer, 374
name, 383
nk, 377
np, 377
nx, 375
ny, 376
rowval, 382
sparseeq, 380
sparsesym, 379
sym, 378
symNames, 384
val, 382
amici::ode_export::Observable
__init__, 367
amici::ode_export::Parameter
__init__, 386
amici::ode_export::SigmaY
__init__, 387
amici::ode_export::State
__init__, 388
amici::plotting
plotObservableTrajectories, 81
plotStateTrajectories, 81
amici::sbml_import
assignmentRules2observables, 83
constantSpeciesToParameters, 84
default_symbols, 85
getRuleVars, 82
l2s, 85
replaceLogAB, 84
amici::sbml_import::SbmlImporter
__init__, 410
checkLibSBMLErrors, 411
checkSupport, 415
cleanReservedSymbols, 422
loadSBMLFile, 411
processCompartments, 417
processObservables, 419
processParameters, 415
processReactions, 417
processRules, 418
processSBML, 414
processSpecies, 415
processTime, 419
processVolumeConversion, 418
replaceInAllExpressions, 421
replaceSpecialConstants, 423
reset_symbols, 410
sbml2amici, 412
amici_blasCBlasTransToBlasTrans
amici, 70
Generated by Doxygen

514 INDEX
amici_daxpy
amici, 27
amici_dgemm
amici, 25
amici_dgemv
amici, 24
amici_path
amici, 74
anyStateNonNegative
amici::Model, 280
applyChainRuleFactorToSimulationResults
amici::ReturnData, 395
applyNewtonsMethod
amici::SteadystateProblem, 497
applyQuadTolerancesASA
amici::Solver, 490
applySensitivityTolerances
amici::Solver, 490
applyTemplate
amici::ode_export, 77
applyTolerances
amici::Solver, 488
applyTolerancesASA
amici::Solver, 489
applyTolerancesFSA
amici::Solver, 488
assignmentRules2observables
amici::sbml_import, 83
at
amici::AmiVector, 98
amici::AmiVectorArray, 102
atol
amici::NewtonSolver, 350
BLASLayout
amici, 19
BLASTranspose
amici, 20
BackwardProblem, 105
amici::BackwardProblem, 106
band
amici::Solver, 476
bandB
amici::Solver, 476
binit
amici::Solver, 463
boost::serialization::serialize
amici::Model, 268
amici::ReturnData, 395
amici::Solver, 491
calcICB
amici::Solver, 435
calcIC
amici::Solver, 435
cblas.cpp, 502
checkConvergence
amici::SteadystateProblem, 496
checkDataDimension
amici::ExpData, 135
checkEventsDimension
amici::ExpData, 135
checkFieldNames
amici, 44
checkFinite
amici, 31
amici::Model, 224
checkLibSBMLErrors
amici::sbml_import::SbmlImporter, 411
checkSigmaPositivity
amici::ExpData, 136,137
checkSupport
amici::sbml_import::SbmlImporter, 415
chi2
amici::ReturnData, 403
cleanReservedSymbols
amici::sbml_import::SbmlImporter, 422
clone
amici::Model, 162
amici::Solver, 429
colptr
amici::ode_export::ODEModel, 381
compileModel
amici::ode_export::ODEExporter, 370
computeNewtonSensis
amici::NewtonSolver, 348
computeX_pos
amici::Model, 266
Constant, 361
constantSpeciesToParameters
amici::sbml_import, 84
createSteadystateSimSolver
amici::SteadystateProblem, 499
CvodeException, 111
amici::CvodeException, 112
DDspline
amici, 65
DDspline_pos
amici, 65
dJrzdsigma
amici::Model, 276
dJrzdz
amici::Model, 276
dJydp
amici::Model, 273
dJydsigma
amici::Model, 275
dJydy
amici::Model, 275
dJzdp
amici::Model, 273
dJzdsigma
amici::Model, 276
dJzdz
amici::Model, 276
data
amici::AmiVector, 91,92
Generated by Doxygen

INDEX 515
amici::AmiVectorArray, 101,102
dbl2int
amici, 69
default_symbols
amici::sbml_import, 85
deltaqB
amici::Model, 274
deltasx
amici::Model, 274
deltax
amici::Model, 274
deltaxB
amici::Model, 274
dense
amici::Solver, 475
denseB
amici::Solver, 475
deserializeFromChar
amici, 46
deserializeFromString
amici, 47
diag
amici::Solver, 477
diagB
amici::Solver, 477
dirac
amici, 53
Dmax
amici, 57
Dmin
amici, 54
drzdp
amici::Model, 277
drzdx
amici::Model, 277
dsigmaydp
amici::Model, 273
dsigmazdp
amici::Model, 273
Dspline
amici, 63
Dspline_pos
amici, 64
dwdp
amici::Model, 278
dwdx
amici::Model, 278
dx
amici::NewtonSolver, 352
dxdotdp
amici::Model, 274
dydp
amici::Model, 277
dydx
amici::Model, 277
dzdp
amici::Model, 277
dzdx
amici::Model, 276
edata
amici::ForwardProblem, 147
eq
amici::ode_export::ODEModel, 380
errMsgIdAndTxt
amici, 74
error_code
amici::IntegrationFailure, 149
amici::IntegrationFailureB, 151
amici::NewtonFailure, 345
ExpData, 112
amici, 67
amici::ExpData, 114–117
expDataFromMatlabCall
amici, 30
Expression, 362
fFIM
amici::Model, 223
FIM
amici::ReturnData, 399
fJDiag
amici::Model, 165
amici::Model_DAE, 291,307
amici::Model_ODE, 323,324,338
fJSparse
amici::Model, 164
amici::Model_DAE, 288,289,306
amici::Model_ODE, 320,321,337
fJSparseB
amici::Model_DAE, 290,307
amici::Model_ODE, 322,338
fJB
amici::Model_DAE, 287,306
amici::Model_ODE, 319,337
fJrz
amici::Model, 187,254
fJv
amici::Model, 166
amici::Model_DAE, 292,293,308
amici::Model_ODE, 325,326,339
fJvB
amici::Model_DAE, 294,309
amici::Model_ODE, 327,340
fJy
amici::Model, 185,253
fJz
amici::Model, 186,253
fchi2
amici::Model, 222
fdJrzdsigma
amici::Model, 191,257
fdJrzdz
amici::Model, 190,256
fdJydp
amici::Model, 193
fdJydsigma
Generated by Doxygen

516 INDEX
amici::Model, 188,255
fdJydx
amici::Model, 194
fdJydy
amici::Model, 187,254
fdJzdp
amici::Model, 196
fdJzdsigma
amici::Model, 190,256
fdJzdx
amici::Model, 196
fdJzdz
amici::Model, 189,255
fdeltaqB
amici::Model, 182,250
fdeltasx
amici::Model, 180,249
fdeltax
amici::Model, 179,248
fdeltaxB
amici::Model, 181,250
fdrzdp
amici::Model, 178,247
fdrzdx
amici::Model, 179,248
fdsigmaydp
amici::Model, 183,251
fdsigmazdp
amici::Model, 184,252
fdwdp
amici::Model, 217,218,257
fdwdx
amici::Model, 218,220,258
fdx0
amici::Model, 169
fdxdotdp
amici::Model, 166
amici::Model_DAE, 300,301,312
amici::Model_ODE, 333,342
fdydp
amici::Model, 172,243
fdydx
amici::Model, 173,244
fdzdp
amici::Model, 177,246
fdzdx
amici::Model, 178,247
fixedParameters
amici::ExpData, 138
amici::Model, 279
fixedParametersPreequilibration
amici::ExpData, 138
fixedParametersPresimulation
amici::ExpData, 138
fJ
amici::Model, 164
amici::Model_DAE, 285,286,305
amici::Model_ODE, 317,318,336
fM
amici::Model_DAE, 304,313
ForwardProblem, 139
amici::ForwardProblem, 140
fqBdot
amici::Model_DAE, 299,311
amici::Model_ODE, 332,342
fres
amici::Model, 221
froot
amici::Model, 162
amici::Model_DAE, 295,296,310
amici::Model_ODE, 328,329,340
frz
amici::Model, 175,245
fsJy
amici::Model, 193
fsJz
amici::Model, 195
fsdx0
amici::Model, 171
fsigmay
amici::Model, 183,251
fsigmaz
amici::Model, 184,252
fsres
amici::Model, 222
fsrz
amici::Model, 176,246
fstau
amici::Model, 171,242
fsx0
amici::Model, 169,242
fsx0_fixedParameters
amici::Model, 170,241
fsxdot
amici::Model, 163
amici::Model_DAE, 302,303,312
amici::Model_ODE, 334,335,343
fsy
amici::Model, 191
fsz
amici::Model, 174,245
fsz_tf
amici::Model, 192
functions
amici::ode_export, 79
fw
amici::Model, 215,216,257
fx0
amici::Model, 167,240
fx0_fixedParameters
amici::Model, 168,241
fxBdot
amici::Model_DAE, 298,311
amici::Model_ODE, 331,341
fxdot
amici::Model, 163
Generated by Doxygen

INDEX 517
amici::Model_DAE, 296,297,310
amici::Model_ODE, 329,330,341
fy
amici::Model, 172,243
fz
amici::Model, 174,244
generateBasicVariables
amici::ode_export::ODEModel, 383
generateModelCode
amici::ode_export::ODEExporter, 369
getAbsoluteTolerance
amici::Solver, 445
getAbsoluteToleranceQuadratures
amici::Solver, 449
getAbsoluteToleranceSensi
amici::Solver, 447
getAbsoluteToleranceSteadyState
amici::Solver, 451
getAbsoluteToleranceSteadyStateSensi
amici::Solver, 453
getAdjBmem
amici::Solver, 487
getAdjMallocDone
amici::Solver, 487
getBacktrace
amici::AmiException, 88
getDJydx
amici::ForwardProblem, 144
getDJzdx
amici::ForwardProblem, 144
getDiagnosis
amici::Solver, 433
getDiagnosisB
amici::Solver, 433
getDiscontinuities
amici::ForwardProblem, 143
getDky
amici::Solver, 473
getFixedParameterById
amici::Model, 205
getFixedParameterByName
amici::Model, 206
getFixedParameterIds
amici::Model, 237
getFixedParameterNames
amici::Model, 229
getFixedParameters
amici::Model, 205
getInitialStateSensitivities
amici::Model, 213
getInitialStates
amici::Model, 212
getInternalSensitivityMethod
amici::Solver, 461
getInterpolationType
amici::Solver, 458
getLastOrder
amici::Solver, 483
getLength
amici::AmiVector, 95
amici::AmiVectorArray, 104
getLinearMultistepMethod
amici::Solver, 456
getLinearSolver
amici::Solver, 460
getMallocDone
amici::Solver, 486
getMaxSteps
amici::Solver, 454
getMaxStepsBackwardProblem
amici::Solver, 455
getModel
amici::Solver, 486
getNVector
amici::AmiVector, 93
amici::AmiVectorArray, 103
getNVectorArray
amici::AmiVectorArray, 103
getNaN
amici, 60
getNewtonMaxLinearSteps
amici::Solver, 442
getNewtonMaxSteps
amici::Solver, 440
getNewtonPreequilibration
amici::Solver, 441
getNonlinearSolverIteration
amici::Solver, 457
getNumErrTestFails
amici::Solver, 481
getNumNonlinSolvConvFails
amici::Solver, 482
getNumRhsEvals
amici::Solver, 481
getNumSteps
amici::Solver, 480
getNumberOfRoots
amici::ForwardProblem, 143
getObservableIds
amici::Model, 238
getObservableNames
amici::Model, 229
getObservedData
amici::ExpData, 122
getObservedDataPtr
amici::ExpData, 122
getObservedDataStdDev
amici::ExpData, 127
getObservedDataStdDevPtr
amici::ExpData, 127
getObservedEvents
amici::ExpData, 129
getObservedEventsPtr
amici::ExpData, 130
getObservedEventsStdDev
amici::ExpData, 134
Generated by Doxygen

518 INDEX
getObservedEventsStdDevPtr
amici::ExpData, 134
getParameterById
amici::Model, 231
getParameterByName
amici::Model, 232
getParameterIds
amici::Model, 231
getParameterList
amici::Model, 211
getParameterNames
amici::Model, 226
getParameterScale
amici::Model, 203
getParameters
amici::Model, 204
getQuadB
amici::Solver, 438
getRHSAtDiscontinuities
amici::ForwardProblem, 142
getRHSBeforeDiscontinuities
amici::ForwardProblem, 143
getReinitializeFixedParameterInitialStates
amici::Model, 240
getRelativeTolerance
amici::Solver, 444
getRelativeToleranceQuadratures
amici::Solver, 448
getRelativeToleranceSensi
amici::Solver, 447
getRelativeToleranceSteadyState
amici::Solver, 450
getRelativeToleranceSteadyStateSensi
amici::Solver, 452
getReturnDataMatlabFromAmiciCall
amici, 35
getRootCounter
amici::ForwardProblem, 144
getRootIndexes
amici::ForwardProblem, 143
getRootInfo
amici::Solver, 434
getRuleVars
amici::sbml_import, 82
getSens
amici::Solver, 432
getSensitivityMethod
amici::Solver, 439
getSensitivityOrder
amici::Solver, 443
getSolver
amici::Model, 162
amici::Model_DAE, 305
amici::Model_ODE, 336
amici::NewtonSolver, 346
getStabilityLimitFlag
amici::Solver, 459
getStateDerivativePointer
amici::ForwardProblem, 145
getStateDerivativeSensitivityPointer
amici::ForwardProblem, 146
getStateIds
amici::Model, 236
getStateIsNonNegative
amici::Model, 209
getStateNames
amici::Model, 227
getStateOrdering
amici::Solver, 458
getStatePointer
amici::ForwardProblem, 145
getStateSensitivity
amici::ForwardProblem, 142
getStateSensitivityPointer
amici::ForwardProblem, 145
getStatesAtDiscontinuities
amici::ForwardProblem, 142
getSteadyStateSensitivityMode
amici::Model, 239
getSteadystateSimulation
amici::SteadystateProblem, 498
getStep
amici::NewtonSolver, 347
getSymbolicDiagonal
amici::ode_export, 77
getTime
amici::ForwardProblem, 141
getTimepoint
amici::ExpData, 119
getTimepoints
amici::ExpData, 119
amici::Model, 209
getUnscaledParameter
amici, 34
getUnscaledParameters
amici::Model, 204
getValueById
amici, 71
getVector
amici::AmiVector, 94
getWrmsNorm
amici::SteadystateProblem, 495
getB
amici::Solver, 438
getdJydx
amici::BackwardProblem, 110
getdxBptr
amici::BackwardProblem, 110
getmy
amici::Model, 258
getmz
amici::Model, 259
getrz
amici::Model, 264
getsrz
amici::Model, 265
Generated by Doxygen

INDEX 519
getsx
amici::Model, 263
getsz
amici::Model, 265
gett
amici::BackwardProblem, 107
amici::Model, 224
getwhich
amici::BackwardProblem, 108
getwhichptr
amici::BackwardProblem, 108
getx
amici::Model, 262
getxBptr
amici::BackwardProblem, 109
getxQBptr
amici::BackwardProblem, 109
gety
amici::Model, 261
getz
amici::Model, 263
h
amici::Model, 279
hasFixedParameterIds
amici::Model, 236
hasFixedParameterNames
amici::Model, 228
hasObservableIds
amici::Model, 238
hasObservableNames
amici::Model, 229
hasParameterIds
amici::Model, 230
hasParameterNames
amici::Model, 225
hasStateIds
amici::Model, 235
hasStateNames
amici::Model, 227
heaviside
amici, 53
IDAException, 147
amici::IDAException, 148
idlist
amici::Model, 272
import_from_sbml_importer
amici::ode_export::ODEModel, 374
init
amici::Solver, 462
initAndAttachArray
amici, 43
initHeaviside
amici::Model, 198
initMatlabDiagnosisFields
amici, 37
initMatlabReturnFields
amici, 36
initialize
amici::Model, 197
initializeLinearSolver
amici::Solver, 483
initializeLinearSolverB
amici::Solver, 484
initializeStates
amici::Model, 197
IntegrationFailure, 148
amici::IntegrationFailure, 149
IntegrationFailureB, 150
amici::IntegrationFailureB, 150
interface_matlab.cpp, 502
mexFunction, 504
InternalSensitivityMethod
amici, 21
interpType
amici::Solver, 493
InterpolationType
amici, 21
invalidate
amici::ReturnData, 393
invalidateLLH
amici::ReturnData, 394
isFixedParameterStateReinitializationAllowed
amici::Model, 266
isInf
amici, 59
isNaN
amici, 58
isSetObservedData
amici::ExpData, 121
isSetObservedDataStdDev
amici::ExpData, 126
isSetObservedEvents
amici::ExpData, 129
isSetObservedEventsStdDev
amici::ExpData, 133
ism
amici::Solver, 492
iter
amici::Solver, 493
J
amici::Model, 275
amici::ReturnData, 396
k
amici::Model, 202
klu
amici::Solver, 479
kluSetOrdering
amici::Solver, 479
kluSetOrderingB
amici::Solver, 480
kluB
amici::Solver, 480
l2s
Generated by Doxygen

520 INDEX
amici::sbml_import, 85
lbw
amici::Model, 272
LinearMultistepMethod
amici, 21
LinearSolver
amici, 21
linsolveSPBCG
amici::NewtonSolverIterative, 357
llh
amici::ReturnData, 403
lmm
amici::Solver, 492
loadSBMLFile
amici::sbml_import::SbmlImporter, 411
log
amici, 52
LogLikelihood, 364
M
amici::Model, 278
M_PI
amici.cpp, 501
max
amici, 56
maxlinsteps
amici::NewtonSolver, 350
maxsteps
amici::NewtonSolver, 350
amici::Solver, 493
mexFunction
interface_matlab.cpp, 504
min
amici, 54
minus
amici::AmiVector, 97
Model, 151
amici::Model, 159,161
model
amici::ForwardProblem, 146
amici::NewtonSolver, 351
Model_DAE, 282
amici::Model_DAE, 284
Model_ODE, 314
amici::Model_ODE, 316,317
ModelQuantity, 365
msgIdAndTxtFp
amici, 19
multiobs_functions
amici::ode_export, 80
mxArrayToVector
amici, 70
my
amici::Model, 275
mz
amici::Model, 275
nMaxEvent
amici::Model, 202
name
amici::ode_export::ODEModel, 383
ndwdp
amici::Model, 271
ndwdx
amici::Model, 271
ne
amici::Model, 270
amici::ReturnData, 405
newton_cpu_time
amici::ReturnData, 401
newton_maxsteps
amici::ReturnData, 406
newton_numlinsteps
amici::ReturnData, 402
newton_numsteps
amici::ReturnData, 401
newton_status
amici::ReturnData, 401
NewtonFailure, 344
amici::NewtonFailure, 344
NewtonSolver, 345
amici::NewtonSolver, 346
NewtonSolverDense, 352
amici::NewtonSolverDense, 353
NewtonSolverIterative, 355
amici::NewtonSolverIterative, 355
NewtonSolverSparse, 359
amici::NewtonSolverSparse, 359
NewtonStatus
amici, 22
nJ
amici::Model, 271
amici::ReturnData, 406
nk
amici::Model, 201
amici::ReturnData, 404
amici::ode_export::ODEModel, 377
nmaxevent
amici::ExpData, 137
amici::Model, 281
amici::ReturnData, 406
nnz
amici::Model, 271
NonlinearSolverIteration
amici, 21
np
amici::Model, 200
amici::ReturnData, 404
amici::ode_export::ODEModel, 377
nplist
amici::Model, 199
amici::ReturnData, 406
amici::Solver, 485
nt
amici::ExpData, 117
amici::Model, 202
amici::ReturnData, 406
Generated by Doxygen

INDEX 521
numerrtestfails
amici::ReturnData, 400
numerrtestfailsB
amici::ReturnData, 400
numnonlinsolvconvfails
amici::ReturnData, 400
numnonlinsolvconvfailsB
amici::ReturnData, 401
numrhsevals
amici::ReturnData, 400
numrhsevalsB
amici::ReturnData, 400
numsteps
amici::ReturnData, 399
numstepsB
amici::ReturnData, 399
nw
amici::Model, 270
nx
amici::Model, 269
amici::ReturnData, 404
amici::Solver, 485
amici::ode_export::ODEModel, 375
nxtrue
amici::Model, 269
amici::ReturnData, 404
ny
amici::Model, 269
amici::ReturnData, 405
amici::ode_export::ODEModel, 376
nytrue
amici::ExpData, 137
amici::Model, 270
amici::ReturnData, 405
nz
amici::Model, 270
amici::ReturnData, 405
nztrue
amici::ExpData, 137
amici::Model, 270
amici::ReturnData, 405
o2mode
amici::Model, 272
amici::ReturnData, 407
ODEExporter, 367
ODEModel, 372
ODEModel_from_pysb_importer
amici::ode_export, 78
Observable, 366
observedData
amici::ExpData, 138
observedDataStdDev
amici::ExpData, 139
observedEvents
amici::ExpData, 139
observedEventsStdDev
amici::ExpData, 139
operator=
amici::AmiVector, 90
amici::Model, 161
operator==
amici, 35,48
amici::Model, 269
amici::Solver, 491
operator[]
amici::AmiVector, 98
amici::AmiVectorArray, 104
order
amici::ReturnData, 401
originalParameters
amici::Model, 279
Parameter, 385
ParameterScaling
amici, 20
pi
amici, 74
plist
amici::Model, 214
plist_
amici::Model, 279
plotObservableTrajectories
amici::plotting, 81
plotStateTrajectories
amici::plotting, 81
pos_pow
amici, 58
prepareLinearSystem
amici::NewtonSolver, 349
amici::NewtonSolverDense, 354
amici::NewtonSolverIterative, 357
amici::NewtonSolverSparse, 360
printErrMsgIdAndTxt
amici, 22
printWarnMsgIdAndTxt
amici, 23
processCompartments
amici::sbml_import::SbmlImporter, 417
processObservables
amici::sbml_import::SbmlImporter, 419
processParameters
amici::sbml_import::SbmlImporter, 415
processReactions
amici::sbml_import::SbmlImporter, 417
processRules
amici::sbml_import::SbmlImporter, 418
processSBML
amici::sbml_import::SbmlImporter, 414
processSpecies
amici::sbml_import::SbmlImporter, 415
processTime
amici::sbml_import::SbmlImporter, 419
processVolumeConversion
amici::sbml_import::SbmlImporter, 418
pscale
amici::Model, 281
amici::ReturnData, 407
Generated by Doxygen

522 INDEX
pysb2amici
amici, 69
qbinit
amici::Solver, 463
quadReInitB
amici::Solver, 439
quadSStolerancesB
amici::Solver, 474
rdata
amici::ForwardProblem, 146
amici::NewtonSolver, 351
reInit
amici::Solver, 434
reInitB
amici::Solver, 438
realtype
amici, 19
reinitializeFixedParameterInitialStates
amici::Model, 282
reorder
amici, 45
replaceInAllExpressions
amici::sbml_import::SbmlImporter, 421
replaceLogAB
amici::sbml_import, 84
replaceSpecialConstants
amici::sbml_import::SbmlImporter, 423
res
amici::ReturnData, 399
reset
amici::AmiVector, 96
amici::AmiVectorArray, 105
reset_symbols
amici::sbml_import::SbmlImporter, 410
ReturnData, 390
amici::ReturnData, 391,393
rootInit
amici::Solver, 464
rowval
amici::ode_export::ODEModel, 382
rtol
amici::NewtonSolver, 351
runAmiciSimulation
amici, 23,66
runAmiciSimulations
amici, 68
rz
amici::ReturnData, 397
s2llh
amici::ReturnData, 403
s2rz
amici::ReturnData, 397
SBMLException, 408
sbml2amici
amici::sbml_import::SbmlImporter, 412
SbmlImporter, 408
SecondOrderMode
amici, 20
sensInit1
amici::Solver, 464
sensReInit
amici::Solver, 435
sensi
amici::ReturnData, 407
sensi_functions
amici::ode_export, 80
sensi_meth
amici::ReturnData, 407
SensitivityMethod
amici, 20
SensitivityOrder
amici, 20
serializeToChar
amici, 46
serializeToStdVec
amici, 47
serializeToString
amici, 47
set
amici::AmiVector, 97
setAbsoluteTolerance
amici::Solver, 446
setAbsoluteToleranceQuadratures
amici::Solver, 449
setAbsoluteToleranceSensi
amici::Solver, 448
setAbsoluteToleranceSteadyState
amici::Solver, 451
setAbsoluteToleranceSteadyStateSensi
amici::Solver, 453
setBandJacFn
amici::Solver, 465
setBandJacFnB
amici::Solver, 466
setDenseJacFn
amici::Solver, 465
setDenseJacFnB
amici::Solver, 465
setErrHandlerFn
amici::Solver, 469
setFixedParameterById
amici::Model, 207
setFixedParameterByName
amici::Model, 208
setFixedParameters
amici::Model, 205
setFixedParametersByIdRegex
amici::Model, 207
setFixedParametersByNameRegex
amici::Model, 208
setId
amici::Solver, 472
setInitialStateSensitivities
amici::Model, 213
Generated by Doxygen

INDEX 523
setInitialStates
amici::Model, 212
setInternalSensitivityMethod
amici::Solver, 462
setInterpolationType
amici::Solver, 458
setJacTimesVecFn
amici::Solver, 465
setJacTimesVecFnB
amici::Solver, 467
setLinearMultistepMethod
amici::Solver, 456
setLinearSolver
amici::Solver, 461
setMaxNumSteps
amici::Solver, 470
setMaxNumStepsB
amici::Solver, 471
setMaxSteps
amici::Solver, 454
setMaxStepsBackwardProblem
amici::Solver, 455
setModelData
amici, 28
setNMaxEvent
amici::Model, 202
setName
amici::ode_export::ODEExporter, 372
setNewtonMaxLinearSteps
amici::Solver, 442
setNewtonMaxSteps
amici::Solver, 440
setNewtonPreequilibration
amici::Solver, 441
setNonlinearSolverIteration
amici::Solver, 457
setObservedData
amici::ExpData, 120,121
setObservedDataStdDev
amici::ExpData, 123–125
setObservedEvents
amici::ExpData, 128
setObservedEventsStdDev
amici::ExpData, 130–132
setParameterById
amici::Model, 233
setParameterByName
amici::Model, 234
setParameterList
amici::Model, 212
setParameterScale
amici::Model, 203
setParameters
amici::Model, 204
setParametersByIdRegex
amici::Model, 233
setParametersByNameRegex
amici::Model, 235
setPaths
amici::ode_export::ODEExporter, 371
setQuadErrConB
amici::Solver, 469
setReinitializeFixedParameterInitialStates
amici::Model, 239
setRelativeTolerance
amici::Solver, 444
setRelativeToleranceQuadratures
amici::Solver, 449
setRelativeToleranceSensi
amici::Solver, 447
setRelativeToleranceSteadyState
amici::Solver, 450
setRelativeToleranceSteadyStateSensi
amici::Solver, 452
setSStolerances
amici::Solver, 468
setSStolerancesB
amici::Solver, 474
setSensErrCon
amici::Solver, 469
setSensParams
amici::Solver, 473
setSensSStolerances
amici::Solver, 468
setSensitivityMethod
amici::Solver, 439
setSensitivityOrder
amici::Solver, 443
setSolverOptions
amici, 28
setSparseJacFn
amici::Solver, 465
setSparseJacFnB
amici::Solver, 466
setStabLimDet
amici::Solver, 471
setStabLimDetB
amici::Solver, 472
setStabilityLimitFlag
amici::Solver, 460
setStateIsNonNegative
amici::Model, 210
setStateOrdering
amici::Solver, 459
setSteadyStateSensitivityMode
amici::Model, 239
setStopTime
amici::Solver, 437
setSuppressAlg
amici::Solver, 473
setT0
amici::Model, 214
setTimepoints
amici::ExpData, 118
amici::Model, 209
setUserData
Generated by Doxygen

524 INDEX
amici::Solver, 469
setUserDataB
amici::Solver, 470
setValueById
amici, 72
setValueByIdRegex
amici, 73
setup
amici::Solver, 429
setupAMIB
amici::Solver, 431
SetupFailure, 423
amici::SetupFailure, 424
setupReturnData
amici, 29
seval
amici, 50
SigmaY, 386
sigmay
amici::Model, 272
amici::ReturnData, 398
sigmaz
amici::Model, 273
amici::ReturnData, 396
sign
amici, 61
sinteg
amici, 51
sllh
amici::ReturnData, 403
solve
amici::Solver, 436
solveLinearSystem
amici::NewtonSolver, 349
amici::NewtonSolverDense, 353
amici::NewtonSolverIterative, 356
amici::NewtonSolverSparse, 360
solveB
amici::Solver, 437
solveF
amici::Solver, 436
Solver, 424
amici::Solver, 429
solver
amici::ForwardProblem, 146
solverMemory
amici::Solver, 492
solverMemoryB
amici::Solver, 492
solverWasCalled
amici::Solver, 492
sparse_functions
amici::ode_export, 79
sparseeq
amici::ode_export::ODEModel, 380
sparsesym
amici::ode_export::ODEModel, 379
spbcg
amici::Solver, 478
spbcgB
amici::Solver, 478
spgmr
amici::Solver, 477
spgmrB
amici::Solver, 478
spline
amici, 48,61
spline.cpp, 504
spline_pos
amici, 62
sptfqmr
amici::Solver, 478
sptfqmrB
amici::Solver, 479
sres
amici::ReturnData, 399
srz
amici::ReturnData, 397
ssigmay
amici::ReturnData, 398
ssigmaz
amici::ReturnData, 397
State, 388
stateIsNonNegative
amici::Model, 280
StateOrdering
amici, 22
status
amici::ReturnData, 404
stau
amici::Model, 278
SteadyStateSensitivityMode
amici, 22
steadyStateSensitivityMode
amici::Model, 281
SteadystateProblem, 493
amici::SteadystateProblem, 494
storeBacktrace
amici::AmiException, 88
sx
amici::ReturnData, 398
sx0
amici::ReturnData, 403
sx0data
amici::Model, 280
sy
amici::ReturnData, 398
sym
amici::ode_export::ODEModel, 378
symNames
amici::ode_export::ODEModel, 384
symbol_to_type
amici::ode_export, 80
symbolic_functions.cpp, 505
sz
amici::ReturnData, 396
Generated by Doxygen

INDEX 525
t
amici::Model, 210
amici::NewtonSolver, 351
t0
amici::Model, 213
t_steadystate
amici::ReturnData, 402
TemplateAmici, 389
time
amici::IntegrationFailure, 149
amici::IntegrationFailureB, 151
ts
amici::ExpData, 138
amici::Model, 280
amici::ReturnData, 396
tstart
amici::Model, 281
turnOffRootFinding
amici::Solver, 439
ubw
amici::Model, 271
unscaleParameters
amici, 32,33
unscaledParameters
amici::Model, 279
updateHeaviside
amici::Model, 223
updateHeavisideB
amici::Model, 224
val
amici::ode_export::ODEModel, 382
var_in_function_signature
amici::ode_export, 76
w
amici::Model, 278
warnMsgIdAndTxt
amici, 74
what
amici::AmiException, 87
workBackwardProblem
amici::BackwardProblem, 107
workForwardProblem
amici::ForwardProblem, 141
workSteadyStateProblem
amici::SteadystateProblem, 494
wrapErrHandlerFn
amici::Solver, 467
writeMatlabField0
amici, 38
writeMatlabField1
amici, 39
writeMatlabField2
amici, 40
writeMatlabField3
amici, 41
writeMatlabField4
amici, 42
writeNewtonOutput
amici::SteadystateProblem, 498
wrms_sensi_steadystate
amici::ReturnData, 402
wrms_steadystate
amici::ReturnData, 402
x
amici::NewtonSolver, 352
amici::ReturnData, 397
x0
amici::ReturnData, 402
x0data
amici::Model, 280
x_pos_tmp
amici::Model, 281
xdot
amici::NewtonSolver, 351
amici::ReturnData, 396
y
amici::ReturnData, 398
z
amici::ReturnData, 396
z2event
amici::Model, 272
Generated by Doxygen

