BrainNet Viewer Manual Brain Net
BrainNet_Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 38
BRAINNET VIEWER MANUAL
A visualization tool for brain network
Mingrui Xia
mingruixia@gmail.com; mxia@bnu.edu.cn
Released on April 3, 2017, Ver 1.6
1
BrainNet Viewer User Manual 1.6, April, 2017
Contents
1 Introduction ................................................................................................................. 2
2 Installation ................................................................................................................... 3
2.1 Run BrainNet Viewer on a PC with Matlab ............................................................................... 3
2.2 Run BrainNet Viewer on a PC without Matlab .......................................................................... 3
3 Pictures ......................................................................................................................... 5
4 Load Files ..................................................................................................................... 7
4.1 Load a surface file ...................................................................................................................... 7
4.2 Load a node file ......................................................................................................................... 9
4.3 Load an edge file ...................................................................................................................... 10
4.4 Load a volume file ................................................................................................................... 11
5 Visualize option .......................................................................................................... 13
5.1 Layout panel ............................................................................................................................ 13
5.2 Global panel ............................................................................................................................. 16
5.3 Surface panel ........................................................................................................................... 17
5.4 Node panel .............................................................................................................................. 18
5.5 Edge panel ............................................................................................................................... 21
5.6 Volume panel ........................................................................................................................... 24
5.7 Image panel ............................................................................................................................. 27
6 Menu .......................................................................................................................... 29
6.1 Files .......................................................................................................................................... 29
6.2 Option ...................................................................................................................................... 30
6.3 Visualize ................................................................................................................................... 30
6.4 Tools ......................................................................................................................................... 30
6.5 Help.......................................................................................................................................... 31
7 Toolbar ....................................................................................................................... 32
7.1 Load Files & Save as Image ...................................................................................................... 32
7.2 Print & Zoom ........................................................................................................................... 32
7.3 Move, Rotate & Get position ................................................................................................... 33
7.4 Standard view .......................................................................................................................... 33
7.5 Demo ....................................................................................................................................... 34
8 Command line ............................................................................................................ 35
References ..........................................................................................................................37

2
BrainNet Viewer User Manual 1.6, April, 2017
1 Introduction
Please cite as ‘... was/were visualized with the BrainNet Viewer (Xia et al., 2013,
http://www.nitrc.org/projects/bnv/)’ while using the package to make publicized pictures.
Reference: Xia M, Wang J, He Y (2013) BrainNet Viewer: A Network Visualization Tool for Human Brain
Connectomics. PLoS ONE 8: e68910.
BrainNet Viewer is a brain network visualization tool, which can help researchers to visualize structural
and functional connectivity patterns from different levels in a quick, easy and flexible way. It would be
greatly appreciated if you have any suggestions about the package or manual.
BrainNet Viewer is developed using MATLAB (The MathWorks Inc., Natick, MA, US) as a programming
language, with a user-friendly GUI, under a 64-bit Windows (Microsoft Corp., Redmond, WA, US)
environment. The toolbox includes functions of Statistical Parametric Mapping 8 (SPM,
http://www.fil.ion.ucl.ac.uk/spm/) for loading NIfTI, Analyze format and GIfTI surface files (*.nii; *.img;
*.gii). This toolbox has been successfully tested under a variety of operating systems with MATLAB
installed, including Windows (XP, 7, 8, 10 and Server versions), Linux (Ubuntu and CentOS) and Mac OS
in both 32- and 64-bit versions.
BrainNet Viewer is mainly developed by Mingrui Xia.
Contact information:
Mingrui Xia: mingruixia@gmail.com; mxia@bnu.edu.cn
Yong He: yong.h.he@gmail.com; yong.he@bnu.edu.cn
Copyright © 2017 Dr. Yong He’s Lab, National Key Laboratory of Cognitive Neuroscience and Learning,
IDG/McGovern Institute for Brain Research, Beijing Key Laboratory of Brain Imaging and Connectomics,
Beijing Normal University, Beijing, 100875, China
3
BrainNet Viewer User Manual 1.6, April, 2017
2 Installation
2.1 Run BrainNet Viewer on a PC with Matlab
1) Run Matlab. (A version of R2010b or above is recommended)
2) Add BrainNet Viewer path to Matlab search path:
Click ‘File’ in Matlab menu -> Click ‘Set Path’ -> Click ‘Add with Subfolders…’ button in the popup
dialog -> Select the ‘BrainNet Viewer’ folder on the machine -> Click ‘OK’ button -> Click ‘Save’
Button.
3) Run BrainNet.m:
Type ‘BrainNet’ in the command window of Matlab.
2.2 Run BrainNet Viewer on a PC without Matlab
Please contact us if you need standalone version. It cannot be found on the NITRC due to the large size
of Matlab Components Runtime.
Install Matlab Components Runtime (MCRInstall.exe for Windows OS, or MCRInstaller.bin for Linux and
Mac OS, ~200MB) using default settings.
Restart your computer (strongly recommended).
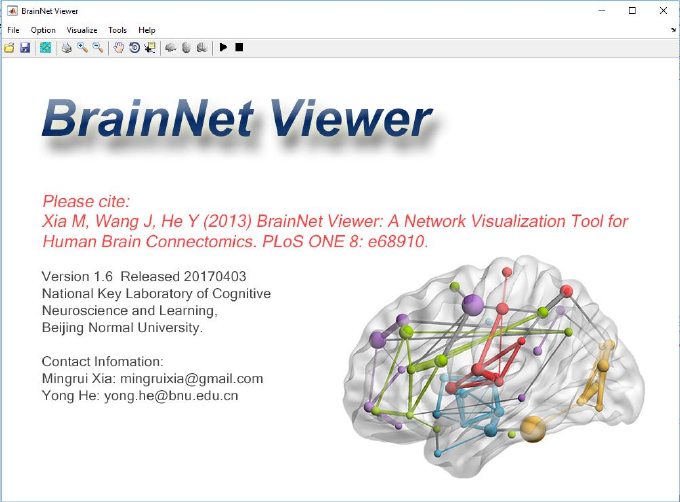
Run BrainNet.exe for Windows OS, run_BrainNet.sh for Linux or BrainNet.app for Mac OS, it should
take about one minute to start. You can find the interface below (Fig. 1) after successfully running the
BrainNet Viewer.
4
BrainNet Viewer User Manual 1.6, April, 2017
Fig. 1 The main window of BrainNet Viewer
5
BrainNet Viewer User Manual 1.6, April, 2017
3 Pictures
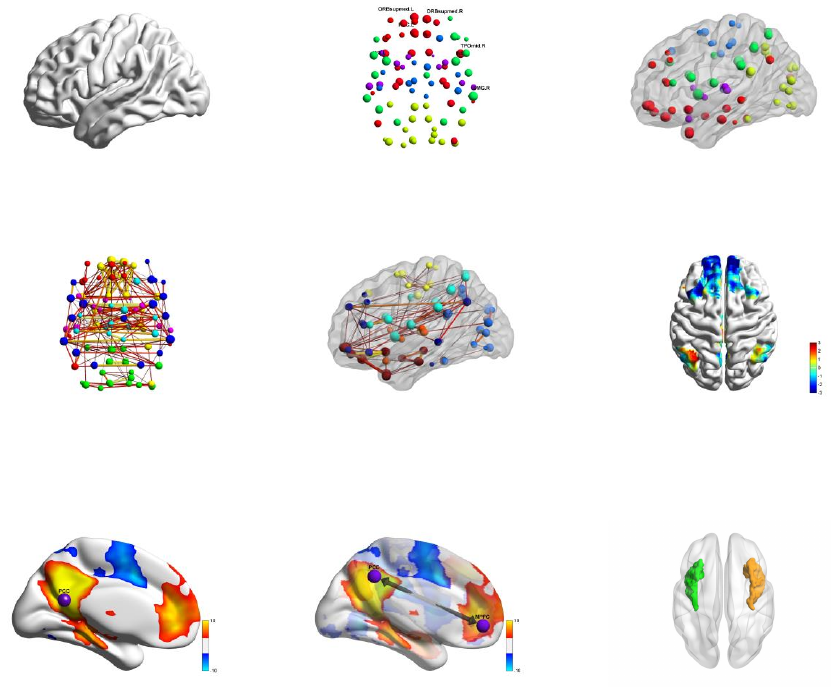
BrainNet Viewer doesn’t require to load surface, node, edge and volume file together. Instead, the
following combinations are acceptable and different combinations will generate different network
pictures (Fig. 2):
1) Brain surface: load brain surface file only. See section 4.1 for file preparation and section 5.3 for
visualization options.
2) Nodes: load node file only. See section 4.2 for file preparation and section 5.4 for visualization
options.
3) Brain surface and nodes: load both brain surface and node files. See sections 4.1 and 4.2 for file
preparation and section 5.3 and 5.4 for visualization options.
4) Nodes and edges: load both node and edge files. See sections 4.2 and 4.3 for file preparation and
section 5.4 and 5.5 for visualization options.
5) Brain surface, nodes and edges: load brain surface, node and edge files together. See sections 4.1
to 4.3 for file preparation and section 5.3 to 5.5 for visualization options.
6) Volume mapping to surface: load brain surface and volume files. See section 4.1 and 4.4 for file
preparation and section 5.3 and 5.6 for visualization options.
7) Volume mapping to surface with node: load brain surface, node and volume files. See section 4.1,
4.2 and 4.4 for file preparation and section 5.3, 5.4 and 5.6 for visualization options.
8) Volume mapping to surface with node and edge: load brain surface, node, edge and volume files.
See section 4.1 to 4.4 for file preparation and section 5.3 to 5.6 for visualization options.
9) ROI cluster drawing in volume: load brain surface and volume files. See section 4.1 and 4.4 for file
preparation and section 5.3 and 5.6 for visualization options.
6
BrainNet Viewer User Manual 1.6, April, 2017
1) Brain Surface
2) Nodes
3) Surface & Nodes
4) Nodes & Edges
5) Surface, Nodes &
Edges
6) Surface mapping
7) Surface mapping &
node
8) Surface mapping
with node & edge
9) ROI in Volume
Fig. 2 Brain network pictures from different file combinations
7
BrainNet Viewer User Manual 1.6, April, 2017
4 Load Files
To draw a brain network, some kinds of files such as brain surface, node file or edge file should be
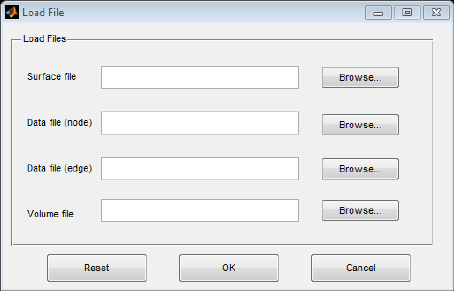
loaded in the first step. Click ‘Load File’ button on the toolbar or ‘File\Load File’ in the menu to open
Load File dialog shown below (Fig. 3). Select files to draw required graph.
In BrainNet Viewer, we provided several brain surface templates and example files (which were made
from various brain parcellation methods) including (1) Colin brain, smoothed Colin brain, Colin brain
with cerebellum, ICBM152 brain (MNI/Talaraich), smoothed ICBM152 brain (MNI/Talaraich),
hemispheres of ICBM152, hemispheres of smoothed ICBM152 brain surface and a cerebellum surface
in the folder ‘.\Data\SurfTemplate’; and (2) node and edge files for Automated Anatomical Labeling
(AAL, 90 regions) (Tzourio-Mazoyer et al., 2002), Brodmann areas (82 regions) (Brodmann, 1909),
Desikan-Killiany Atlas (68 regions) (Desikan et al., 2006), Harvard-Oxford Atlas (HOA, 112 regions)
(Smith et al., 2004), ROIs defined by Dosenbach et al.(160 ROIs) (Dosenbach et al., 2010), ROIs defined
by Fair et al. (34 ROIs) (Fair et al., 2009), LONI Probabilistic Brain Atlas (40 regions) (Shattuck et al.,
2008), ROIs defined by Power et al. (264 ROIs) (Power et al., 2011) and others (e.g., customized ROIs by
users) in the folder ‘.\Data\ExampleFiles’.
Fig. 3 Load File dialog
4.1 Load a surface file
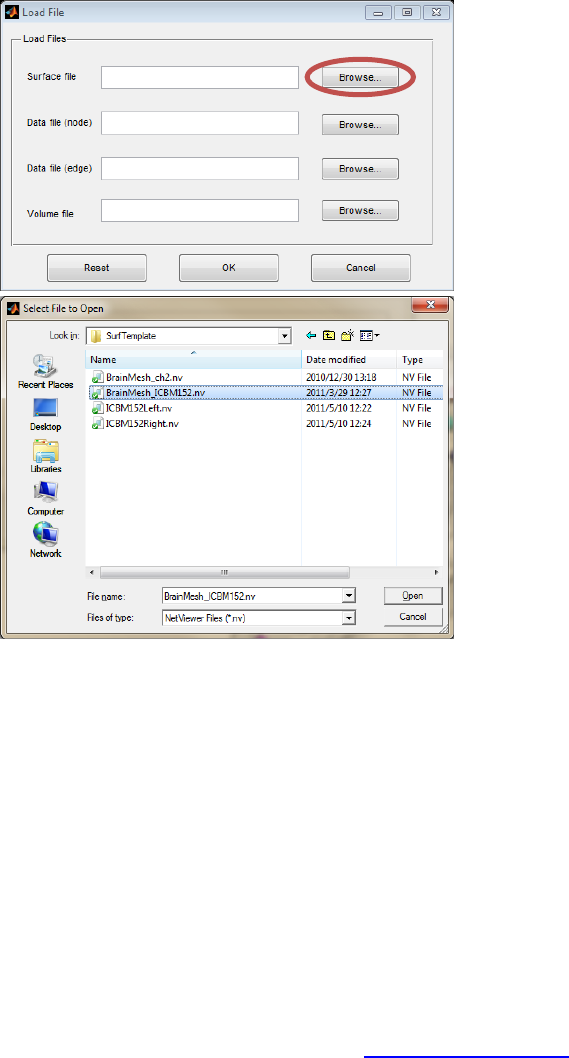
Click the ‘Browse…’ button next to the ‘Surface file’ in the ‘Load File’ dialog, and then select the
required brain surface file in the popup dialog. BrainNet Viewer provides several brain surfaces based
on two different brain templates, ICBM152 (.\Data\SurfTemplate\BrainMesh_ICBM152.nv) and Colin27
(.\Data\SurfTemplate\BrainMesh_ch2.nv), and separate hemisphere surfaces
8
BrainNet Viewer User Manual 1.6, April, 2017
(.\Data\SurfTemplate\ICBM152Left.nv, ICBM152Right.nv). In the below example, the ICBM152 template
is selected (Fig. 4). Note that the surface with a name ended with ‘tal’ is in the Talaraich space,
otherwise is in the MNI space.
Fig. 4 Select brain surface (ICBM152 is selected)
The information below is about the definition of the surface file. Usually, you don’t need to generate a
new surface file. Please read the file if interested or if you want to make a surface by yourself. The brain
surface file is defined as an ASCII text file with suffix ‘nv’ and contains four fields:
1) Vertex number;
2) Vertex coordinate;
3) Triangle faces number;
4) Index of vertex making up the triangles.
The ICBM152 brain surface was provided by Prof. Alan Evans (Montreal Neurological Institute, McGill
University) and the Colin27 brain surface was made by BrainVISA (http://brainvisa.info/). We
transferred and merged the original bilateral hemisphere files into one ‘.nv’ file. A surface merge tool is
provided in the tools menu (see more details in section 6.4 ‘Menus\Tool’).
9
BrainNet Viewer User Manual 1.6, April, 2017
BrainNet Viewer supports a number of different surface formats, including the ‘*.pial’ files generated
by FreeSurfer, (only hemisphere mesh), the ‘*.mesh’ files generated by BrainVISA ‘*.g’ files generated
by BrainWorks software, ‘*.obj’ files, ‘*.gii’ files of GIfTI surface and ‘*.mz3’ of Surf Ice.
4.2 Load a node file
The file represents the information from nodes obtained from the AAL90, Brodmann82,
Desikan-Killiany68, HOA112, Dos160, Fair34, LPBA40, Power264 and others (e.g., customized ROIs by
users). All files are stored in folder ‘.\Data\ExampleFiles\’, corresponding to its template name,
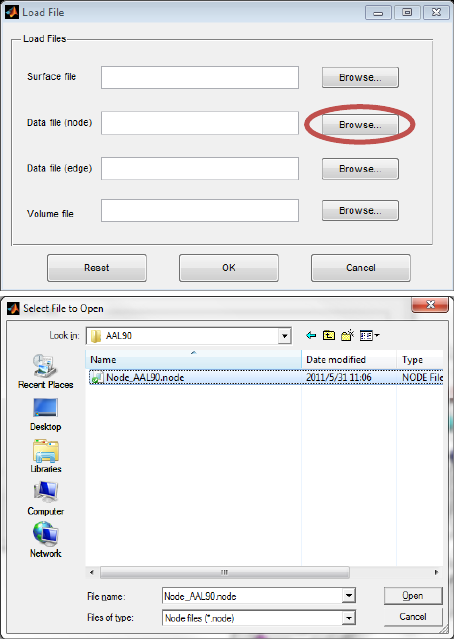
respectively. Click the ‘Browse…’ button next to ‘Data file (node)’ in the Load File dialog and select the
required node file. The AAL90 node file is selected as an example in Fig. 5.
Fig. 5 Select node file (AAL90 is selected)
The node file is defined as an ASCII text file with the suffix ‘node’. In the node file, there are 6 columns:
columns 1-3 represent node coordinates, column 4 represents node colors, column 5 represents node
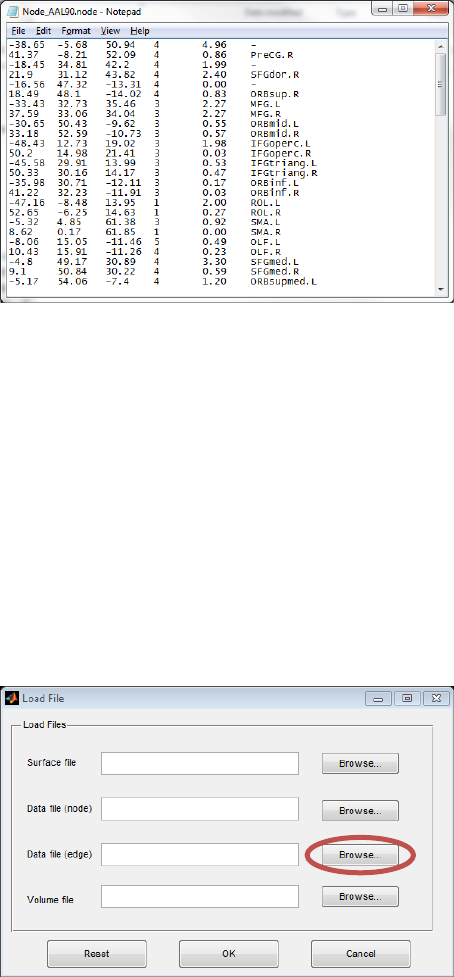
10
BrainNet Viewer User Manual 1.6, April, 2017
sizes, and the last column represents node labels. Please note, a symbol ‘-‘ (no ‘’) in column 6 means no
labels and blank characters in label would cause error. The user may put the modular information of
the nodes into column 4, like ‘1, 2, 3…’ or other information to be shown by color. Column 5 could be
set as nodal degree, centrality, T-value, etc. to emphasize nodal differences by size. The generation of
node file are flexible depending on different requirements.
Fig. 6 Node file (AAL90)
4.3 Load an edge file
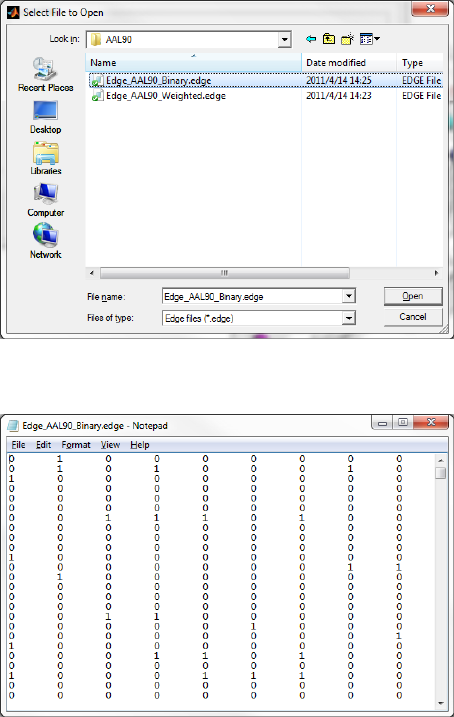
The brain edge file is defined as an ASCII text file with suffix ‘edge’, representing a connectivity (e.g.,
correlations) matrix among the ROIs, which could be either weighted or binarized, and therefore, the
dimensions of the matrix must correspond to the number of nodes. AAL90, Brodmann82, HOA112,
Dos160, Fair34, LPBA40 and other (e.g., customized ROIs by users) files are provided, and each file is in
the folder ‘.\Data\ExampleFiles\’ corresponding to its template name.
11
BrainNet Viewer User Manual 1.6, April, 2017
Fig. 7 Select an edge file (AAL90 binary file is selected)
Fig. 8 Edge file (AAL90, Binarized)
Both node and edge files can be generated/edited with text editors or Excel.
Notably, the size of the matrix should correspond to the number of nodes and comments can be
added with # at the beginning of the line in surface, node and edge files.
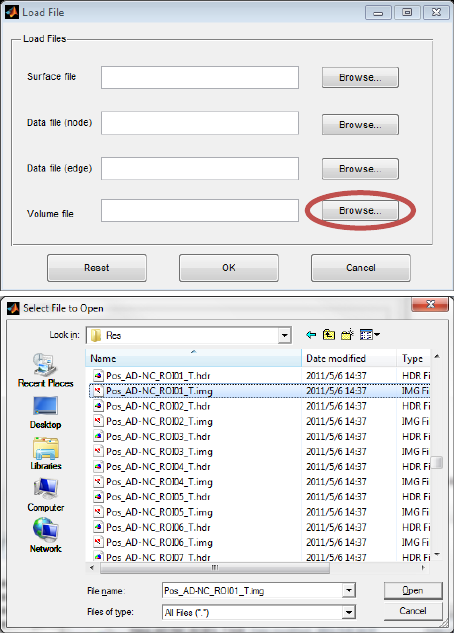
4.4 Load a volume file
This function lets users map the volume data to the brain surface. The volume file should be a 3D NIFTI
format, which could be T-map, Z-map, atlas or any other volume data, either paired files or nii file are
accepted. Besides, a text file containing an n × 1 vector is also accepted, in which n equals to the
vertex number of the brain surface (81924 vertexes in ICBM whole brain surface). The principle of
volume mapping is to transfer the vertex coordinates on the brain surface to the volume space, and
assign vertices with corresponding values by using different algorithms. The principle of ROI drawing is
to reconstruct voxels with same index in the image file to 3D volume.
12
BrainNet Viewer User Manual 1.6, April, 2017
Fig. 9 Volume file (a paired NIFTI file of T-test Map is selected)
13
BrainNet Viewer User Manual 1.6, April, 2017
5 Visualize option
The option panel has three parts (Fig. 10). The list box on the left includes ‘Layout’, ‘Global, ‘Surface’,
‘Node’, ‘Edge’, ’Volume’ and ‘Image,’ which represent different aspects of the figure. The main panel on
the right shows the detailed options of each part; click the text in the list box to change the panel.
There are six buttons on the bottom of the panel: use the ‘Load’ and ‘Save’ to acquire or save options
as a .mat file; ‘Reset’ to return all parameters to their original state; ‘OK’ to draw graph and close
option panel; ‘Apply’ to draw graph but keep option panel and ‘Cancel’ to exit the panel without
changes.
Fig. 10 Option panel
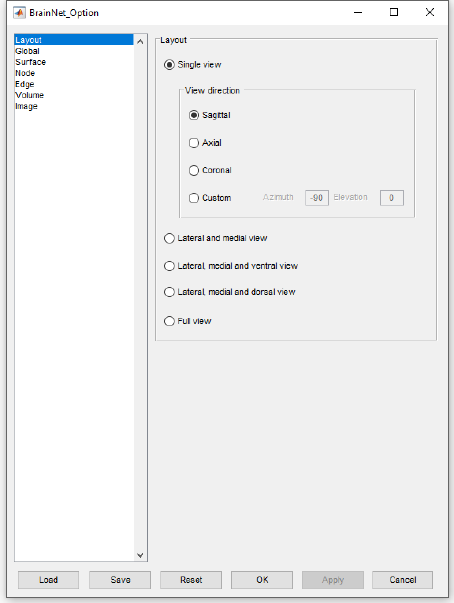
5.1 Layout panel
The layout panel (Figure 10) is primarily responsible for setting the output view of the brain model, in
which five types of views are provided:
Single view: Show only one brain model in the figure.
14
BrainNet Viewer User Manual 1.6, April, 2017
Sagittal Shows the brain in sagittal view (left side).
Axial Shows the brain in axial view (dorsal side).
Coronal Shows the brain in coronal view (frontal side).
Custom Shows the brain in a custom viewpoint, defined by azimuth and elevation (see more detail of
function ‘view’ in Matlab help).
Lateral and medial view: Show lateral and medial side of each hemisphere in the figure. The top row
contains lateral view of left and right hemispheres while the bottom row contains medial view of left
and right hemispheres.
Lateral, medial and ventral view: Show lateral, medial and ventral side of each hemisphere in the
figure. The top row contains lateral view of left and right hemispheres, the middle row contains medial
view of left and right hemispheres and the bottom row contains ventral view of left and right
hemispheres.
Lateral, medial and dorsal view: Show lateral and medial side of each hemisphere and the dorsal side
of the brain in the figure. The top row contains lateral view of left and right hemispheres, the bottom
row contains medial view of left and right hemispheres and the whole-brain is displayed from the
dorsal side in the center.
Full view: Show six or eight (depending on whether the brain surface can be divided into left and right
hemispheres) brain models. In the six brain mode, the top row from left to right are left side, top side
and frontal side, while the bottom row from left to right are right side, bottom side and back side. In
the eight brain mode, the first row from left to right are lateral view of left hemisphere, top side, lateral
view of right hemisphere, the second row from left to right are medial view of left hemisphere, bottom
side, medial view of right hemisphere, and the third row are frontal side and back side. See Fig. 11.
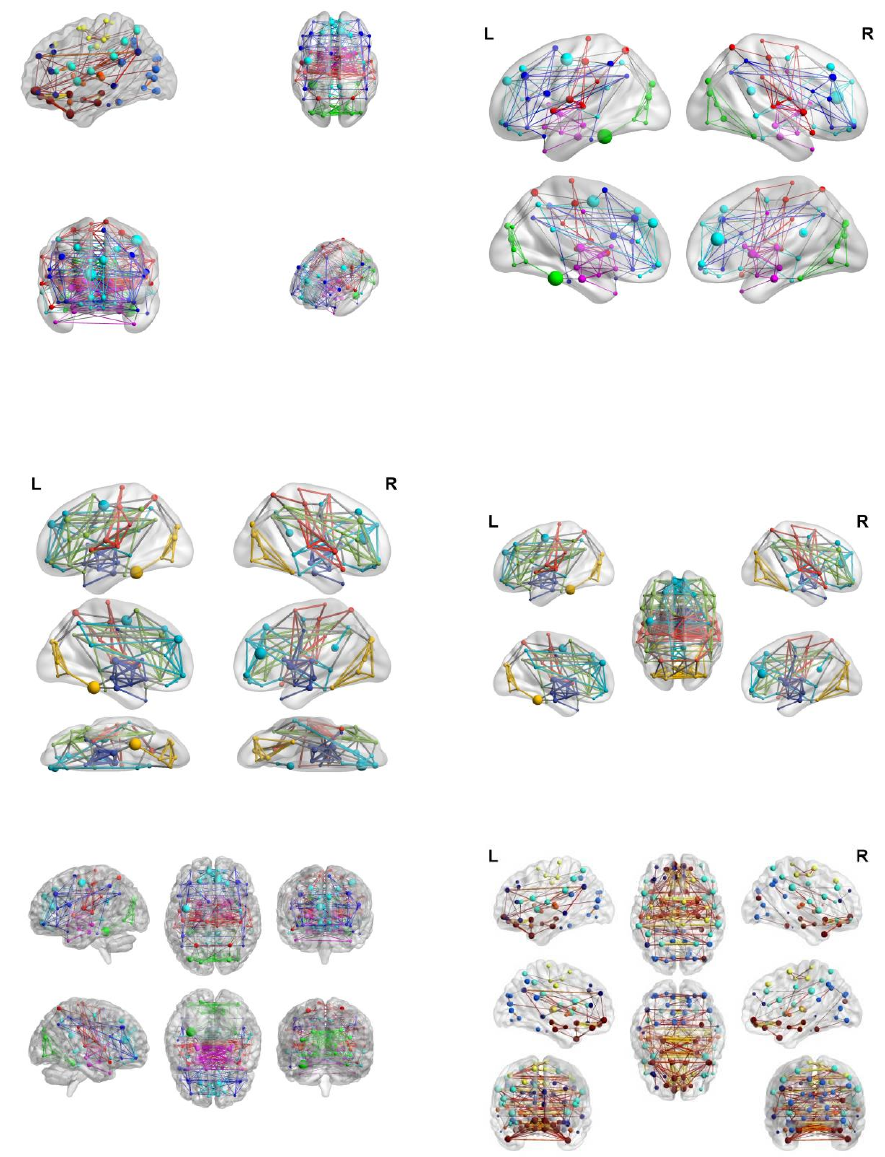
15
BrainNet Viewer User Manual 1.6, April, 2017
Single view: Sagittal
Single view: Axial
Single view: Coronal
Single view: Custom,
Az -130, El 30
Lateral and medial view
Lateral, medial and ventral view
Lateral, medial and dorsal view
Full View, six brain
Full View, eight brain
Fig. 11 Different layouts
16
BrainNet Viewer User Manual 1.6, April, 2017
5.2 Global panel
The global panel provides several different choices for the adjustment of the global figure, particularly
the display properties of these objects. (Fig 12)
Background Color: Change the color of the background. Right-click on the color square right beside the
text ‘Background Color’, and select the desired color in the popup dialog.
Object Material: Provide four manners to define material of the mode in figure, ‘Shiny’,
‘Dull’(default), ’Metal’ and ‘Custom’ which the ambient, diffuse, and specular can be freely defined.
Shading properties: Set color shading properties, ‘Flat’, ‘Faceted’ and ‘Interp’.
Flat, each triangle of the mesh has a constant color, appropriate for atlas or ROI display.
Faceted, show edges of the mesh.
Interp, varies the color of triangle by interpolating the colormap, appropriate for functional connectivity,
ALFF, ReHo or any volume with continuous data (default).
Lighting algorithm: Set lighting algorithm, ‘Flat’, ‘Gouraud’, ‘Phong’ and ‘None’.
Flat, produces uniform lighting across each of the faces of the object.
Gouraud, calculates the vertex normals and interpolates linearly across the triangles.
Phong, interpolates the vertex normals across each face and calculates the reflectance at each pixel.
(Better but costly than Gouraud, default)
None, turn off light.
Light direction: Set where the light comes from, ‘Headlight’, ‘Right’ (default) and ‘Left’.
Renderer: Set the render method, ‘OpenGL’ (default) and ‘zbuffer’. Texts displayed are sometimes
upside down with some type of AMD ATI graphic cards when using OpenGL mode. Turn this option to
zbuffer would solve this problem. However, the image is saved with texts in right direction.
Graph detail: Set the level of object detail by adjusting the numbers of vertex of nodes and edges when
drawing a graph theoretical network figure, ‘High’ (default), ’Moderate’ and ‘Low’
Mark left and right: Display marker ‘L’ and ‘R’ in the figure.
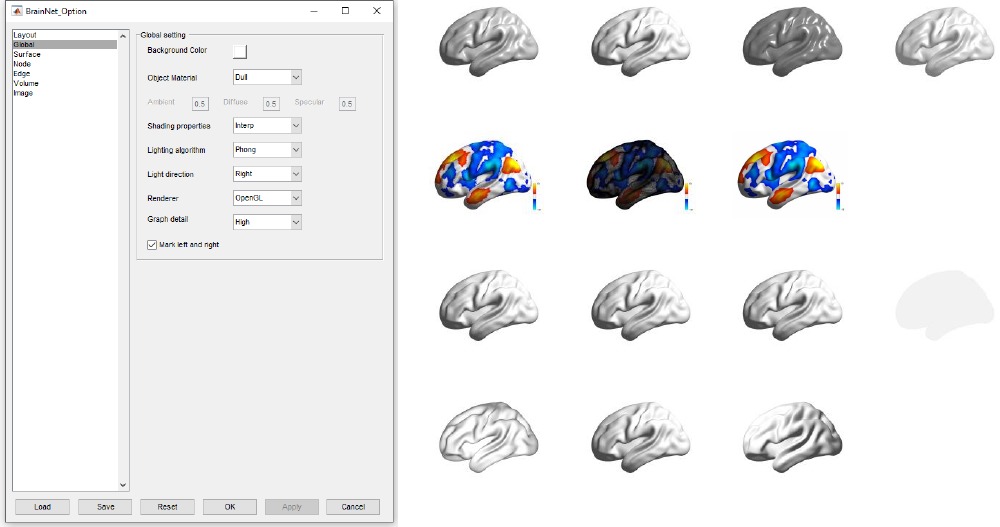
17
BrainNet Viewer User Manual 1.6, April, 2017
Shiny
Dull
Metal
Custom (0.5, 0.5, 0.5)
Object material
Flat
Faceted
Interp
Shading properties
Flat
Gouraud
Phong
None
Lighting algorithm
Headlight
Right
Left
Light direction
Fig. 12 Global panel
5.3 Surface panel
The surface panel is available for adjusting the properties of the brain surface.
Color: right-click the color square and select required color in the popup dialog to change color of the
brain surface.
Opacity: drag the slider bar or enter a number range from 0~1 in the edit box to change the
transparency of the brain surface.
Double Brain: click to display two brain models in one figure, usually used to display the relationship
between nodes in two time points. To display such figure, please arrange node and edge files as follow.
Node file: duplicate the node information and adjust with your own data at the end of the file. The first
half would be placed in the brain model on the left and the last half would be placed in the brain model
on the right. For instance, an original AAL90 node file includes 90 rows; they will be shown on the left.
Then copy them and paste as the row 91 to 180, this part would be shown on the right.
Edge file: the edge file includes intra and inter brain association matrix. For instance, the original AAL90
edge file includes a 90×90 matrix. In the double brain model, please arrange an edge file with 180×

18
BrainNet Viewer User Manual 1.6, April, 2017
180 matrix, in which the matrix (1:90, 1:90) and (91:180, 91:180) are intra connections of each brain,
and (1:90, 91:180) and (91:180, 1:90) are inter connections between the two brains.
Fig. 13 Surface panel and double brain model
5.4 Node panel
The node panel is developed with four zones to select node drawing, set labels, and adjust the node
size and color, respectively. All settings are dependent on the nodal information in the nodal file.
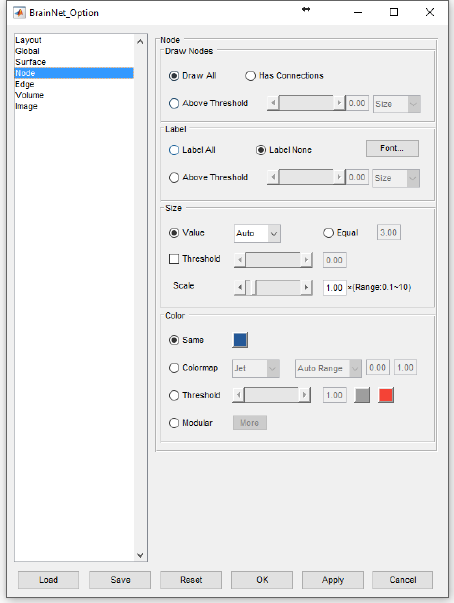
19
BrainNet Viewer User Manual 1.6, April, 2017
Fig. 14 Node panel
Draw nodes:
This function is used to decide which nodes are to be drawn.
Draw All: draw all nodes in the file.
Has Connections: only draw those nodes have connections in the edge file.
Above Threshold: set a threshold of color or size corresponding to column 4 or column 5 to draw those
nodes with higher value than the threshold.
Nodal label:
This panel is used to control the nodal label. Three options are available: ‘Label All’, ‘Label None’ or by a
threshold that only label those nodes with higher value than the threshold on size or color. Click the
‘Font…’ button to change the font of the labels in the popup dialog.
Nodal size:
There are two ways to set the size of the nodes:

20
BrainNet Viewer User Manual 1.6, April, 2017
Value: use the value in column 5 in the node file. In this manner, you can choose ‘Auto’ to arrange the
sizes of nodes to a proper range (radius: 2-7) by their value automatically, or choose ‘Raw’ to use the
original value in column 5 in the node file. When a threshold is selected, the nodes below the threshold
will be a small size (radius: 1), while those above the threshold will display by their Auto/Raw size. Drag
the slider bar or enter the threshold in the edit box. The range must be the same as that in column 5 in
the node file.
Equal: set all nodes to an equal size ignoring the size value in the file, and the size can be defined in the
edit box.
Scale: the volume ratio option is used to adjust the size of all nodes together, and the scale factor
ranges from 0.1 to 10.
Nodal color:
This panel provides four ways to control nodal color:
Same: to use the same color for all nodes ignoring the color index in the file, right-click the color square
and select the required color from the popup dialog.
Colormap: use a color map to display the value of the nodes from low end to high
end corresponding to column 4 in the node file. 13 kinds of color maps and
custom colormap can be selected (see the right picture for detail). The custom
colormap is a n*3 matrix, in which the columns represent the red, green and blue
channels, respectively, and the range of the values should be 0 to 1 (e.g., [0 0 0]
represents black while [1 1 1] represents white). Colormap range can be set as
auto or fixed. In auto mode, the minimum and maximum value in column 4 of
node file will be used while in fixed mode, users are allowed to input any values
they want.
Threshold: to binarize the color by a given threshold, drag the slider bar or enter the threshold in the
edit box, but the range must be the same as the range stated in column 4 of the node file. The nodes
with higher value will have one fixed color, while the nodes with lower value will have another fixed
color. Right-click the color square to select the color – the left one represents the higher value color
while the right one represents the lower value color.
Modular: modular color can be used to display different nodal colors for different modules. Set the
values of column 4 as ‘1, 2, 3…’ corresponding to modular 1, modular 2… in the node file. The
maximum number of modules is21 at present. Click to open the modular color dialog, and the left
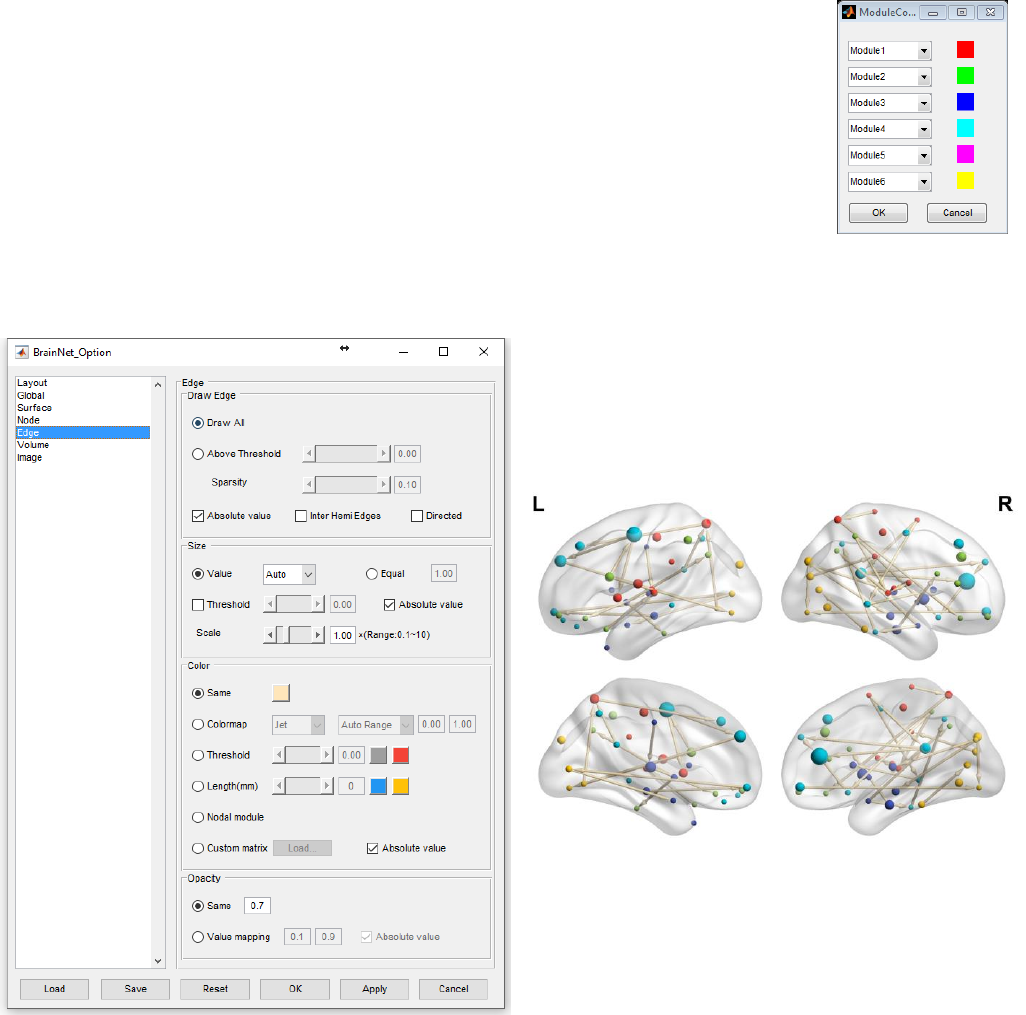
21
BrainNet Viewer User Manual 1.6, April, 2017
picture will display six modules with their color on the right. Click the popup menus
on the left to select other modules in the list and the color square will change to the
corresponding one. Right-click the color square to change color as described above.
5.5 Edge panel
The edge panel is similar to the node panel, with three parts that separately control edge extraction,
edge size and edge color.
Fig. 15 Edge panel and directed random network
Draw Edge:
This panel is used to extract edge information from the correlation matrix contained in the edge file,
and to decide whether all or parts of them are to be drawn.
Draw All: extract and draw all edges (all values not equal to zero) in the correlation matrix.
Threshold: extract the edge above a threshold. This threshold can be set as a value in the matrix or in
22
BrainNet Viewer User Manual 1.6, April, 2017
the sparsity of the matrix.
Absolute value: use absolute value to extract edges from the matrix.
Inter Hemi Edges: extract edges that travel across two hemispheres.
Directed: draw edges with direction.
Note that BrainNet Viewer will treat the value zero (0) in the matrix as a null edge, and only the right
upper triangle of the matrix will be considered in undirected mode. Always remember to change the
threshold when a weighted matrix is loaded, or it will draw the full connection among the nodes, which
would require a lot of time.
Edge size:
There are two ways to set the size of edges (here, size means the radius of the edge);
Value: employ the correlation matrix value in the edge file. In this manner, you can choose ‘Auto’ to
assign the edge sizes a proper range (radius: 0.3-1.5) by their value automatically or choose ‘Raw’ to
use the original value of the correlation matrix in the edge file. When a threshold is selected, the edges
with values lower than the threshold will have a fixed, smaller size while the edges above threshold will
be shown as Auto/Raw size. Drag the slider bar or enter the threshold into the edit box, but the range
must be the same as the correlation matrix in the edge file.
Equal: set all edges to an equal size, and the size can be defined in the edit box.
Scale: the scale option is used to adjust the size of all edges together. The scale factor ranges from 0.1
to 10.
Absolute value: use absolute value in matrix to calculate edge radius.
Edge color:
This panel provides six ways to control edge color:
Same: adopt the same color for all edges, right-click the color square and select the required color from
the popup dialog.
Colormap: use a colormap to render the value of the edge from low to high corresponding to the values
of the correlation matrix in the edge file. 13 kinds of colormaps and custom colormap, same as the
23
BrainNet Viewer User Manual 1.6, April, 2017
nodal colormaps can be selected. The custom colormap is a n*3 matrix, in which the columns represent
the red, green and blue channels, respectively, and the range of the values should be 0 to 1 (e.g., [0 0 0]
represents black while [1 1 1] represents white). Colormap range can be set as auto or fixed. In auto
mode, the minimum and maximum value edge file will be used while in fixed mode, users are allowed
to input any values they want.
Threshold: binarize the color by a given threshold, drag the slider bar or enter the threshold into the
edit box. The range must be the same as the correlation matrix in the edge file. Right-click the color
square to select colors – the left one represents the lower value while the right one represents the
higher value.
Length: binarize the color by a given threshold of Euclidean distance between two nodes (mm). The
edges with longer length have one fixed color, while the shorter ones have another fixed color. Drag the
slider bar or enter the threshold in the edit box; the threshold can range from zero to 100. Right-click
the color square to select colors, the left one represents the higher value while the right one represents
the lower value.
Nodal module: assign edge color according to the color of nodes it links. If two nodes of the edge have
same color, the edge will be set as the same color. If the two nodes are with different color, the edge
will be colored gray.
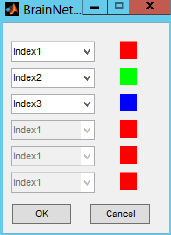
Custom matrix: load a matrix with same size to the network, in which the edges can be assigned with
color indices. For instance, edges in the network (n×n) were required to be
rendered with three different colors. Do follows: i) generate a matrix (n×n); ii)
assign each edge with an index value (1, 2 or 3) and save the matrix as a text file; iii)
load this file in this panel and set the color for each index in the popup window.
Absolute value: use absolute value in matrix to calculate edge color.
Edge opacity:
There are two ways to set the opacity of edges;
Same: set all edges with same opacity, default = 0.7;
Mapping: scale the edges’ opacity according to their values. The minimum and maximum opacity can
be set in the panel.
24
BrainNet Viewer User Manual 1.6, April, 2017
Absolute value: use absolute value in matrix to calculate edge opacity.
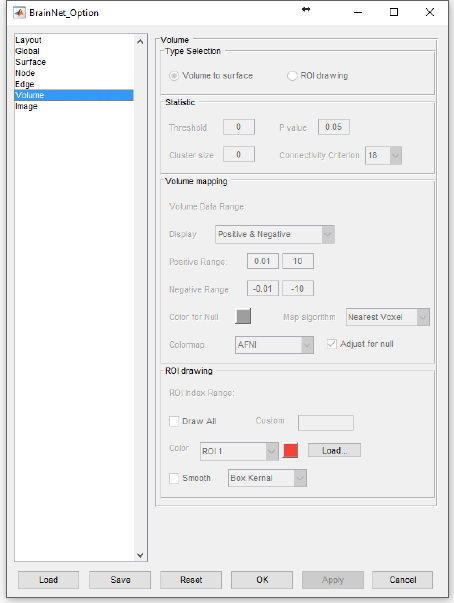
5.6 Volume panel
The volume panel is set to control the volume-to-surface mapping and draw ROI clusters with brain
surface. The volume file could be a T-map, Z-map, an atlas image etc.
Fig. 16 Volume panel
Type Selection: select to map volume to brain surface or to draw ROI volume in brain surface.
Statistic zone:
Threshold: set a threshold that only using the voxels with a value higher than the threshold or lower the
negative threshold.
P value: work with the statistical volumes generated by SPM, REST or DPABI.
Cluster size: set a threshold to select and display clusters have larger voxel number.
25
BrainNet Viewer User Manual 1.6, April, 2017
Connectivity Criterion: set connectivity criterion for voxel connections. 6 for surface connected, 18 for
edge connected and 26 for corner connected.
Volume mapping zone:
Volume Data Range: show the minimum and maximum values of the volume file.
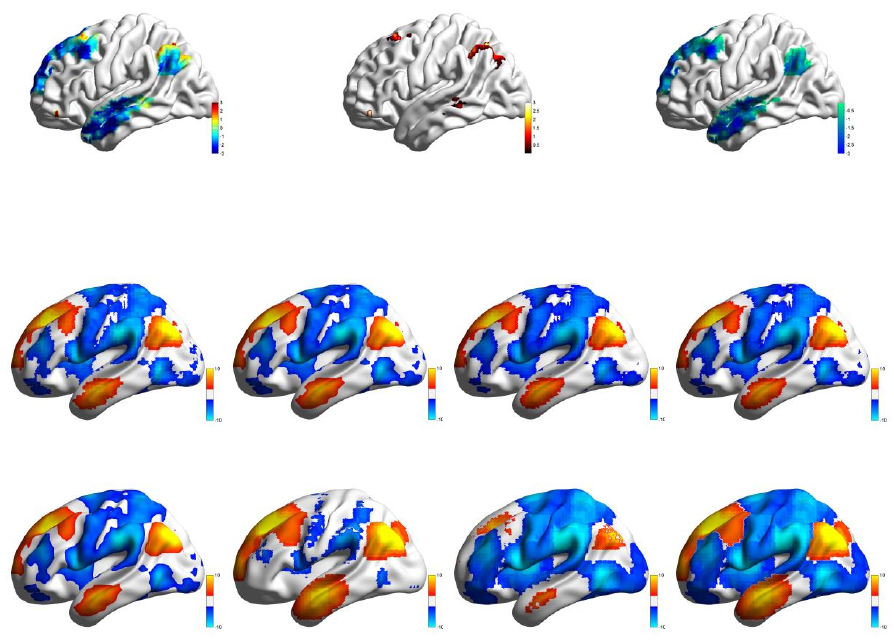
Display: contain three mapping manner, ‘Positive & Negative’, ‘Positive only’ and ‘Negative only’.
‘Positive & Negative’ sets the colorbar range from the minimum negative value to the maximum
positive value, and ‘Positive only’ and ‘Negative only’ just set the range of the colorbar in positive value
or negative value separately.
Positive Range and Negative Range: set the range of the color bar. The edit boxes on the left define the
value near zero on the color bar, while the right ones define the value away from zero. Take the above
picture as an example. When ‘Positive & Negative’ is chosen, the color bar would be arranged from -3
to 3, and -0.01 to 0.01 would be set as the null value range; if ‘Positive only’ is selected, the color bar
would be arranged from 0.01 to 3, any value below 0.01 would be set as a null value; and if ‘Negative
only’ is selected, the color bar would be arranged from -0.01 to -3, and any value above -0.01 would be
set as a null value (see Fig. 17).
Color for Null: define the color for null value part on the surface. Right-click the color square and select
required color.
Adjust for Null: when this option is selected, the colormap will be adjusted for null value vertex.
Specifically, in Positive & Negative mode, the vertex with value between high end of negative interval
and low end of positive interval will be set as color for null; in only positive mode, the vertex with value
below the low end of positive interval will be set as color for null; and in only negative mode, the vertex
with value larger than the low end of positive interval will be set as color for null.
Colormap: provide 24 kinds of colormaps including custom colormap. The custom colormap is a n*3
matrix, in which the columns represent the red, green and blue channels, respectively, and the range of
the values should be 0 to 1 (e.g., [0 0 0] represents black while [1 1 1] represents white).
Map algorithm: eight mapping algorithms are provided to determine the vertex values in BrainNet
Viewer: ‘Nearest Voxel’, assign the vertex with the value of the voxel in volume that is nearest to it,
suitable to display an atlas or mask; ‘Average Vertex’, assign the vertex with the value of the voxel in
volume that is nearest to it, and then average the vertex across its neighbors (high time consumption);
‘Average Voxel’, assign the vertex with average value of the voxel and its neighbors in volume that is
26
BrainNet Viewer User Manual 1.6, April, 2017
nearest to it; ‘Gaussian’, the volume first employs convolutions with a Gaussian kernel and then
assigns the vertex with the value of the voxel in volume that is nearest to it; ‘Interpolated’, the
coordinate of the vertex is determined in the volume space, and a trilinear interpolate method is then
used across its neighbors to calculate the value; ‘Maximum Voxel’, assign the vertex with the maximum
value of the voxel and its neighbors in volume that is nearest to it; ‘Minimum Voxel’, assign the vertex
with the minimum value of the voxel and its neighbors in volume that is nearest to it; ‘Extremum Voxel’,
assign the vertex with the extremum value of the voxel and its neighbors in volume that is nearest to it.
Positive & Negative
Colormap: Jet
Positive only
Colormap: Hot
Negative only
Colormap: Winter
Display
Nearest voxel
Average vertex
Average voxel
Gaussian
Interpolate
Maximum voxel
Minimum voxel
Extremum voxel
Map algorithm
Fig. 17 Volume mapping
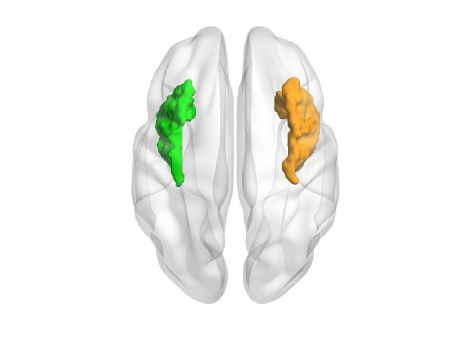
ROI drawing zone: (Please ensure your volume data is arranged with natural number index)
ROI Index Range: show the minimum and maximum index of the volume file, number 0 is out of use.
Draw All: construct and draw each ROI volume in sequence according to their index.
27
BrainNet Viewer User Manual 1.6, April, 2017
Custom: input the index number of ROIs, these ROIs will be selected to reconstruct and draw.
Color: set the color of each ROI volume. The ‘Load…’ button next to the color button allow users to load
a custom-defined color table for each ROI. This color table should be save as a ‘.txt’ file including n*3
matrix, where n corresponds to the number of ROIs and the columns represent the red, green and blue
channels, respectively. The range of the values should be 0 to 1 (e.g., [0 0 0] represents black while [1 1
1] represents white).
Smooth: smooth the surface of ROI volume. Two smooth kernel, ‘Box’ and ‘Gaussian’ are provided.
Please see the help for function ‘smooth3’ in Matlab for further information.
Fig. 18 ROI volume drawing
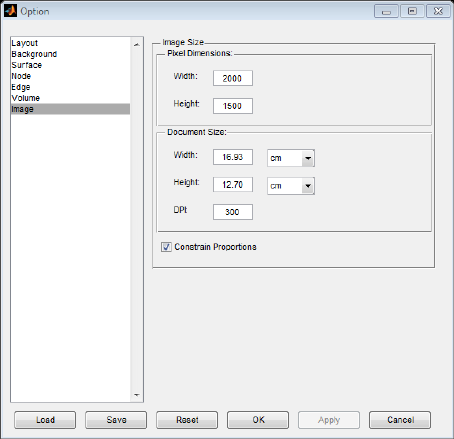
5.7 Image panel
In the image panel, the configurations are related to the size and resolution of the output images. The
width and height of the image can be adjusted in pixel dimensions for screen display or in real units
(centimeter or inch) for document use. The resolution of the output image can also be modified in dots
per inch (DPI). (Fig. 19).
28
BrainNet Viewer User Manual 1.6, April, 2017
Fig. 19 Image panel
29
BrainNet Viewer User Manual 1.6, April, 2017
6 Menu
6.1 Files
Load files:
Click to open load files panel (for more details, see Section ‘Load Files’).
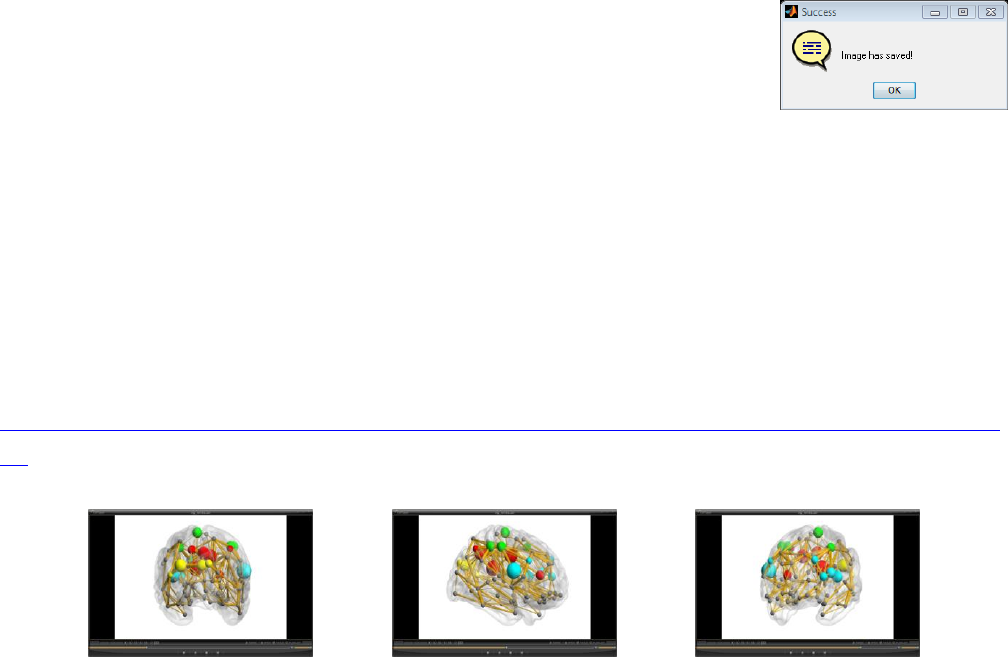
Save Image:
After visualization, click here to save the present figure as an image. At
present, TIFF, BMP, EPS, JPEG and PNG image formats are supported. The
parameters of the image such as pixel dimension, document size and dpi can
be adjusted in the ‘Option panel\Image’. After the image is saved, a message
box appears (see right picture).
Save Movie:
This function helps users to save a demonstration movie for network visualization. It produces a 12
seconds long, 30 FPS, 735×534, avi file in which the brain network rotates clockwise in a circle, one
degree per frame. This operation will take about 10 minutes. Please drink a cup of coffee to wait before
playing the movie. Note that this function should only be used in the ‘Single view’ layout. Pictures
below show different frames at different times. For an example, see
http://www.nitrc.org/docman/view.php/504/1023/Demo%20Video%20of%20Brain%20Network%20(14
M)
3s
6s
9s
Fig. 20 Frames of the network movie
Exit: Click to exit BrainNet Viewer.
30
BrainNet Viewer User Manual 1.6, April, 2017
6.2 Option
Option: Click to open the option panel (see more details in section ‘Visualize Option’).
Load Option: Load a previously saved visualize option file.
Save Option: Save current visualize option as a *.mat file.
Colormap Editor: Call colomap editor to edit colormap manually.
Apply Colormap: Apply edited colormap by colormap editor to all graphs in figure.
Save Colormap: save colormap as a text file. The saved colormap can be used by copy its text into
custom colormap in option panel.
6.3 Visualize
Redraw:
Clear figure and redraw network using the data and option last loaded.
Clear Figure:
Remove brain network and display the default information of BrainNet Viewer.
6.4 Tools
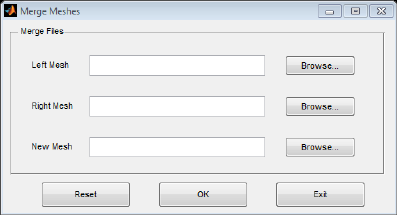
Merge Mesh:
This tool is used to merge the left and right hemisphere surface files extracted from FreeSurfer (*.pial)
or BrainVISA (*.mesh) from two separate files into one BrainNet Viewer surface template file (*.nv), or
to convert a one hemisphere surface file to a BrainNet Viewer surface template file (*.nv). When both
‘Left Mesh’ and ‘Right Mesh’ files are selected, the new mesh will combine two hemisphere files into
one file. If only one of the input files is selected, the new mesh file will convert only that hemisphere
file (Fig .21).
31
BrainNet Viewer User Manual 1.6, April, 2017
Fig. 21 Merge Meshes tool
6.5 Help
Manual:
Open this manual for help.
About:
Show version, author and contact information of BrainNet Viewer in a dialog.

32
BrainNet Viewer User Manual 1.6, April, 2017
7 Toolbar
The toolbar (Fig. 22) provides frequently-used and interaction commands to operate the brain network
graph, most of them are not included in the menu.
Fig. 22 Toolbar
7.1 Load Files & Save as Image
These two commands are included in menu, see details in section ‘Load Files’, and section
‘Menu\File\Save Images’.
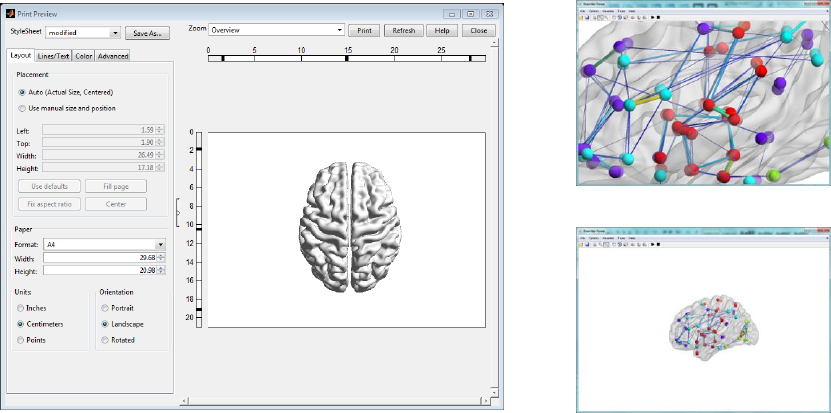
7.2 Print & Zoom
The Print command lets users print the current graph conveniently. A print panel like the one below will
pop up after the Print button is clicked.
The zoom in and zoom out functions help to observe the local or global status of the brain network.
33
BrainNet Viewer User Manual 1.6, April, 2017
Print panel
Zoom in & Zoom out
Fig. 23 Print panel and Zoom function
7.3 Move, Rotate & Get position
Click the ‘Move’ button and drag the brain anywhere in the window.
When the ‘Rotate’ button is pressed, hold left button of the mouse and move mouse to rotate the brain.
When rotate button is deselected, the light cam in the window will re-render the brain model
depending on the current orientation.
Click the ‘Get position’ button, and then click on the surface of the brain to display the coordinates and
value of the vertex on the surface, and it also provides the corresponding brain region labels in terms of
AAL and Brodmann atlases. Right click anywhere in the figure window, and select ‘Delete All Datatips’
to remove all coordinate labels.
7.4 Standard view
Shortcuts for three standard views, sagittal, axial and coronal, are available to quickly observe networks
from different standard views. These buttons should only be used for ‘Single view’ visualized brain
networks. Click twice to see the opposite side of the brain.

34
BrainNet Viewer User Manual 1.6, April, 2017
Sagittal View
Axial View
Coronal View
Fig. 24 Standard views
7.5 Demo
Press the black triangle button to make the brain rotate clockwise until the black square button is
pressed. This function only works for ‘Single View’ visualizations.
35
BrainNet Viewer User Manual 1.6, April, 2017
8 Command line
Considering the growing requirements for batched brain connectome figure mapping, such as dynamic
brain functional connectomes, the functionality to generate brain network figures in the command line
is provided. The function is called according to the following command line:
BrainNet_MapCfg(filename1, filename2…);
where the variables of filenames can be any one of the brain surface, node, edge and volume files.
Once the files are loaded, BrainNet Viewer draws the graphs with default configurations. For instance, a
command line of
BrainNet_MapCfg('BrainMesh_ICBM152.nv','Node_AAL90.node');
will draw the brain surface of 'BrainMesh_ICBM152.nv' and nodes in 'Node_AAL90.node' files using
default settings.
A pre-saved configuration file can also be included in this command line. For example, the command
line
BrainNet_MapCfg('BrainMesh_ICBM152_smoothed.nv','OneSample_T.nii','Cfg.mat');
would map the volume ‘OneSample_T.nii’ onto brain surface 'BrainMesh_ICBM152_smoothed.nv' using
the settings pre-saved in the ‘Cfg.mat’ file.
The command line also supports exporting the brain network figure as image file. The names of the
required image files are added to the command line
BrainNet_MapCfg('Node_AAL90.node','Edge_AAL90_Binary.edge', 'Net.jpg');
Using this command, BrainNet Viewer draws a network in which the node information is obtained from
'Node_AAL90.node' and the edge information is obtained from 'Edge_AAL90_Binary.edge' using default
settings, and this figure will be saved as a JPEG image as 'Net.jpg'. The order of these inputted
filenames is exchangeable, and the combinations of files are similar to the GUI version.

36
BrainNet Viewer User Manual 1.6, April, 2017
Acknowledgements
We thank the following colleagues for their kind helps during BrainNet Viewer developing and manual
revising:
Dr. G. Gong, Dr. N. Shu, Mr. T. Xie, Mr. Q. Lin, Dr. M. Cao, Mr. X. Wang, Mr. C. Tang, Mr. Z. Xu, and Ms. Y.
Xie, Beijing Normal University, China;
Prof. A.C. Evans, McGill University, Canada;
Dr. J. Wang, Hangzhou Normal University, China;
Dr. C. Yan, Chinese Academy of Sciences, China;
Dr. Z. Dai, Sun Yat-sen University, China;
Ms. J. Zhang, University College London, UK;
Mr. P. Clark, Pennsylvania State University, USA;
Mr. M. Ghasemi, Tarbiat Modares University, Iran.
We also thank the developers of the following softwares and toolboxes whose source codes or file
formats were referenced during our package developing:
Matlab: www.mathworks.com/products/matlab/
SurfStat: www.math.mcgill.ca/keith/surfstat/
FreeSurfer: http://surfer.nmr.mgh.harvard.edu/
BrainVISA: http://brainvisa.info/
SPM: www.fil.ion.ucl.ac.uk/spm/
REST: www.restfmri.net/
Surf Ice: www.nitrc.org/projects/surfice/
This project was supported by the Natural Science Foundation (Grant Nos. 81030028, 30870667,
81401479 and 81671767), the National Science Fund for Distinguished Young Scholars (Grant No.
81225012) and Beijing Natural Science Foundation (Grant No. Z111107067311036 and 7102090)
37
BrainNet Viewer User Manual 1.6, April, 2017
References
Brodmann K (1909) Vergleichende lokalisationslehre der grobhirnrinde. Barth: Leipzig.
Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP,
Hyman BT, Albert MS, Killiany RJ (2006) An automated labeling system for subdividing the
human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage
31:968-980.
Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, Nelson SM, Wig GS, Vogel AC,
Lessov-Schlaggar CN, Barnes KA, Dubis JW, Feczko E, Coalson RS, Pruett JR, Jr., Barch DM,
Petersen SE, Schlaggar BL (2010) Prediction of individual brain maturity using fMRI. Science
329:1358-1361.
Fair DA, Cohen AL, Power JD, Dosenbach NU, Church JA, Miezin FM, Schlaggar BL, Petersen SE (2009)
Functional brain networks develop from a "local to distributed" organization. PLoS Comput Biol
5:e1000381.
Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, Vogel AC, Laumann TO, Miezin FM,
Schlaggar BL, Petersen SE (2011) Functional network organization of the human brain. Neuron
72:665-678.
Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga
AW (2008) Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage
39:1064-1080.
Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De
Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM,
Matthews PM (2004) Advances in functional and structural MR image analysis and
implementation as FSL. Neuroimage 23 Suppl 1:S208-219.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M
(2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical
parcellation of the MNI MRI single-subject brain. Neuroimage 15:273-289.