Detecting And Avoiding Likely False Positive Findings A Practical Guide: False‐positive – Guide Forstmeier Et Al 2
Detecting%20and%20avoiding%20likely%20false%E2%80%90positive%20findings%20%E2%80%93%20a%20practical%20guide%20Forstmeier_et_al-2
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 28

Biol. Rev. (2017), 92, pp. 1941–1968. 1941
doi: 10.1111/brv.12315
Detecting and avoiding likely false-positive
findings – a practical guide
Wolfgang Forstmeier1∗, Eric-Jan Wagenmakers2and Timothy H. Parker3
1Department of Behavioural Ecology and Evolutionary Genetics, Max Planck Institute for Ornithology, 82319 Seewiesen, Germany
2Department of Psychology, University of Amsterdam, PO Box 15906, 1001 NK Amsterdam, The Netherlands
3Department of Biology, Whitman College, Walla Walla, WA 99362, U.S.A.
ABSTRACT
Recently there has been a growing concern that many published research findings do not hold up in attempts to
replicate them. We argue that this problem may originate from a culture of ‘you can publish if you found a significant
effect’. This culture creates a systematic bias against the null hypothesis which renders meta-analyses questionable and
may even lead to a situation where hypotheses become difficult to falsify. In order to pinpoint the sources of error
and possible solutions, we review current scientific practices with regard to their effect on the probability of drawing a
false-positive conclusion. We explain why the proportion of published false-positive findings is expected to increase with
(i) decreasing sample size, (ii) increasing pursuit of novelty, (iii) various forms of multiple testing and researcher flexibility,
and (iv) incorrect P-values, especially due to unaccounted pseudoreplication, i.e. the non-independence of data points
(clustered data). We provide examples showing how statistical pitfalls and psychological traps lead to conclusions that
are biased and unreliable, and we show how these mistakes can be avoided. Ultimately, we hope to contribute to a
culture of ‘you can publish if your study is rigorous’. To this end, we highlight promising strategies towards making
science more objective. Specifically, we enthusiastically encourage scientists to preregister their studies (including a priori
hypotheses and complete analysis plans), to blind observers to treatment groups during data collection and analysis,
and unconditionally to report all results. Also, we advocate reallocating some efforts away from seeking novelty and
discovery and towards replicating important research findings of one’s own and of others for the benefit of the scientific
community as a whole. We believe these efforts will be aided by a shift in evaluation criteria away from the current
system which values metrics of ‘impact’ almost exclusively and towards a system which explicitly values indices of
scientific rigour.
Key words: confirmation bias, HARKing, hindsight bias, overfitting, P-hacking, power, preregistration, replication,
researcher degrees of freedom, Type I error.
CONTENTS
I. Introduction ..............................................................................................1942
II. Problems .................................................................................................1943
(1) The argument of Ioannidis and some extensions .....................................................1943
(2) Multiple testing in all of its manifestations ............................................................1946
(a) The temptation of selective reporting .............................................................1948
(b) Cryptic multiple testing during stepwise model simplification .....................................1949
(c) A priori hypothesis testing versus HARKing: Does it matter? .........................................1949
(d) Researcher degrees of freedom: (1) stopping rules ................................................1950
(e) Researcher degrees of freedom: (2) flexibility in analysis ..........................................1951
(3) Incorrect P-values ....................................................................................1952
(a) Pseudoreplication at the individual level ..........................................................1953
(b) Pseudoreplication due to genetic relatedness ......................................................1954
(c) Pseudoreplication due to spatial and temporal autocorrelation ...................................1955
(d) Pseudoreplication renders P-curve analysis invalid ...............................................1956
* Address for correspondence (Tel.: +49-8157-932346; E-mail: forstmeier@orn.mpg.de).
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
This is an open access article under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in any medium, provided the original
work is properly cited.

1942 W. Forstmeier and others
(4) Errors in interpretation of patterns ...................................................................1956
(a) Overinterpretation of apparent differences .......................................................1956
(b) Misinterpretation of measurement error ..........................................................1957
(5) Cognitive biases ......................................................................................1958
III. Solutions .................................................................................................1959
(1) Need for replication and rigorous assessment of context dependence .................................1959
(a) Obstacles to replication ...........................................................................1959
(b) Overcoming the obstacles .........................................................................1960
(c) Interpretation of differences in findings ...........................................................1960
(d) Is the world more complex or less complex than we think? .......................................1961
(2) Collecting evidence for the null and the elimination of zombie hypotheses ...........................1961
(3) Making science more objective .......................................................................1962
(a) Why should I preregister my next study? .........................................................1962
(b) Badges make good scientific practice visible ......................................................1962
(c) Blinding during data collection and analysis ......................................................1963
(d) Objective reporting of non-registered studies .....................................................1963
(e) Concluding recommendations for funding agencies ..............................................1963
IV. Conclusions ..............................................................................................1964
V. Glossary ..................................................................................................1964
VI. Acknowledgements .......................................................................................1966
VII. References ................................................................................................1966
I. INTRODUCTION
Several research fields appear to be in crisis of confidence
(McNutt, 2014; Nuzzo, 2014, 2015; Horton, 2015; Parker
et al., 2016) as evidence emerges that the majority of published
research findings cannot be replicated (Ioannidis, 2005;
Pereira & Ioannidis, 2011; Prinz, Schlange & Asadullah,
2011; Begley & Ellis, 2012; Open Science Collaboration,
2015). According to a recent survey in Nature (Baker, 2016),
52% of researchers believe that there is ‘a significant crisis’,
38% see ‘a slight crisis’, and only 3% see ‘no crisis’. This
suggests that many scientists are starting to contemplate the
following key questions: (i) to what extent are the findings in
my field reliable? (ii) How shall I judge the existing literature?
(iii) Can I distinguish findings that are likely false from those
that are likely true? (iv) How can I avoid building my own
research project on earlier findings that are false? (v)How
do I avoid repeating the mistakes that others seem to have
made? (vi) Which statistical approaches minimize my risk of
drawing false conclusions?
This review has the goal of providing guidance towards
answering these important questions. This requires a good
understanding of some basic statistical principles. To serve
as a practical guide for those less experienced or less versed
in statistics, we make an effort to explain basic concepts in
an easily accessible way (see also the Glossary in Section V),
and we choose a conversational style of writing to motivate
the reader to work through this important material. We have
compiled a collection of common pitfalls and illustrate them
with accessible examples. Our hope is that these examples
will prime our readers to recognize weaknesses or mistakes
when they critically examine the literature or review
manuscripts and help them avoid these mistakes when they
design their own studies and analyse their own data.
Our examples originate from our own research
experiences in behavioural ecology and evolutionary
genetics, but the same statistical issues occur across a wide
range of probabilistic scientific disciplines such as ecology,
physiology, neuroscience, medical sciences, and psychology.
Statistical analyses have been important in biology since the
development of tools like analysis of variance in the early
decades of the 20th century (Fisher, 1925), and statistical tools
remain essential and continue to proliferate (e.g. advanced
Bayesian statistics) across the biological sciences. Yet, no
matter whether you are running a simple t-test or a restricted
maximum likelihood animal model, there is always a risk
of getting it wrong [for examples of mistakes that lead
to over-confidence see Hadfield et al. (2010) and Valcu &
Valcu (2011)]. Hence, our first point is that there are some
common mistakes in the use of statistical tools and that these
mistakes often lead to nominal significance (P<0.05), yet
the P-value is often incorrect and (frequently) too small,
thereby contributing to false-positive claims in the literature.
Our second point is more philosophical but is a complement
to our first point. Statistically significant findings typically
seem more interesting than non-significant findings and are
thus easier to publish. This has created our current scientific
culture of actively seeking statistical significance, often with
practices that lead to misleading results. We hence try to raise
the general awareness of psychological biases that we need
to keep in check in order to ensure an objective reporting of
research outcomes. We believe that these two issues explain
much of the current crisis in science, and that we need to
rethink critically some of our common research practices.
The pitfalls we outline are unlikely to be equally serious
in all fields of science, so we want to avoid creating the false
impression that all current science is fundamentally flawed.
Our radical critique of the current research culture may leave
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1943
some readers frustrated and depressed, because it will be evi-
dent that making real scientific progress is much harder than
iconic research papers seem to suggest. However, instead
of frustration and depression we hope to offer optimism. We
invite our readers to be among the first to implement new
standards that will dramatically improve the reliability and
objectivity of research. This should be appealing and exciting
not only because researchers would like to have confidence
in the reliability of their own work, but also because new
tools allow them to signal the reliability of their research
findings to others. As this signalling (Gintis, Smith & Bowles,
2001) becomes more widespread, it will be harder for others
to cut corners and present results that are likely wrong.
Section II of our article outlines the existing problems.
We begin by reviewing the statistical parameters (prior
probability, realized αand β) that determine which
proportion of the published positive findings will be
false-positives (Section II.1). We show that unaccounted-for
multiple testing is a major source of false-positive findings,
and we present examples that illustrate how easily this
source of error creeps into our research if we fail to
develop a clear predetermined research plan. Flexibility in
defining and testing our hypotheses, combined with selective
reporting of apparent cases of success hence leads to a high
risk of publishing false-positive findings (Section II.2). This
risk increases further if we fail to acknowledge that the
data points we collected may not be independent of each
other. P-values derived from such pseudoreplicated data
will often mislead us into seeing patterns where none exist
(Section II.3). Sections II.2 and II.3 make up the bulk of
the present article because there are quite a few statistical
pitfalls to avoid. False-positive conclusions can also arise from
over-interpretation of differences or from misinterpretation
of measurement error, which we address in Section II.4.
Finally, we briefly touch on cognitive biases that render it
difficult to collect and interpret data objectively (Section II.5).
Section III focuses on possible solutions. Only a few
research fields have developed rigorous methodology that
limits the extent of false-positive reporting and ensures
that negative results are just as likely to get published as
positive results; consequently, many scientific disciplines
face a literature where it is difficult to distinguish likely
truth from falsehood. We therefore highlight the need for
rigorous replication studies (Section III.1) that help eliminate
hypotheses that are likely to be false (Section III.2). We then
conclude by discussing novel methods, like preregistration of
studies, which promote greater objectivity and less bias in
what gets reported in scientific publications (Section III.3).
II. PROBLEMS
(1) The argument of Ioannidis and some extensions
Approximately 10 years ago John Ioannidis famously
explained ‘Why Most Published Research Findings Are
False’ (Ioannidis, 2005). Although the title is somewhat
misleading (Ioannidis did not actually prove that most
findings are false), understanding his argument is essential
for an intuitive feeling of how likely it is that any published
positive finding is true or false. It is therefore worth following
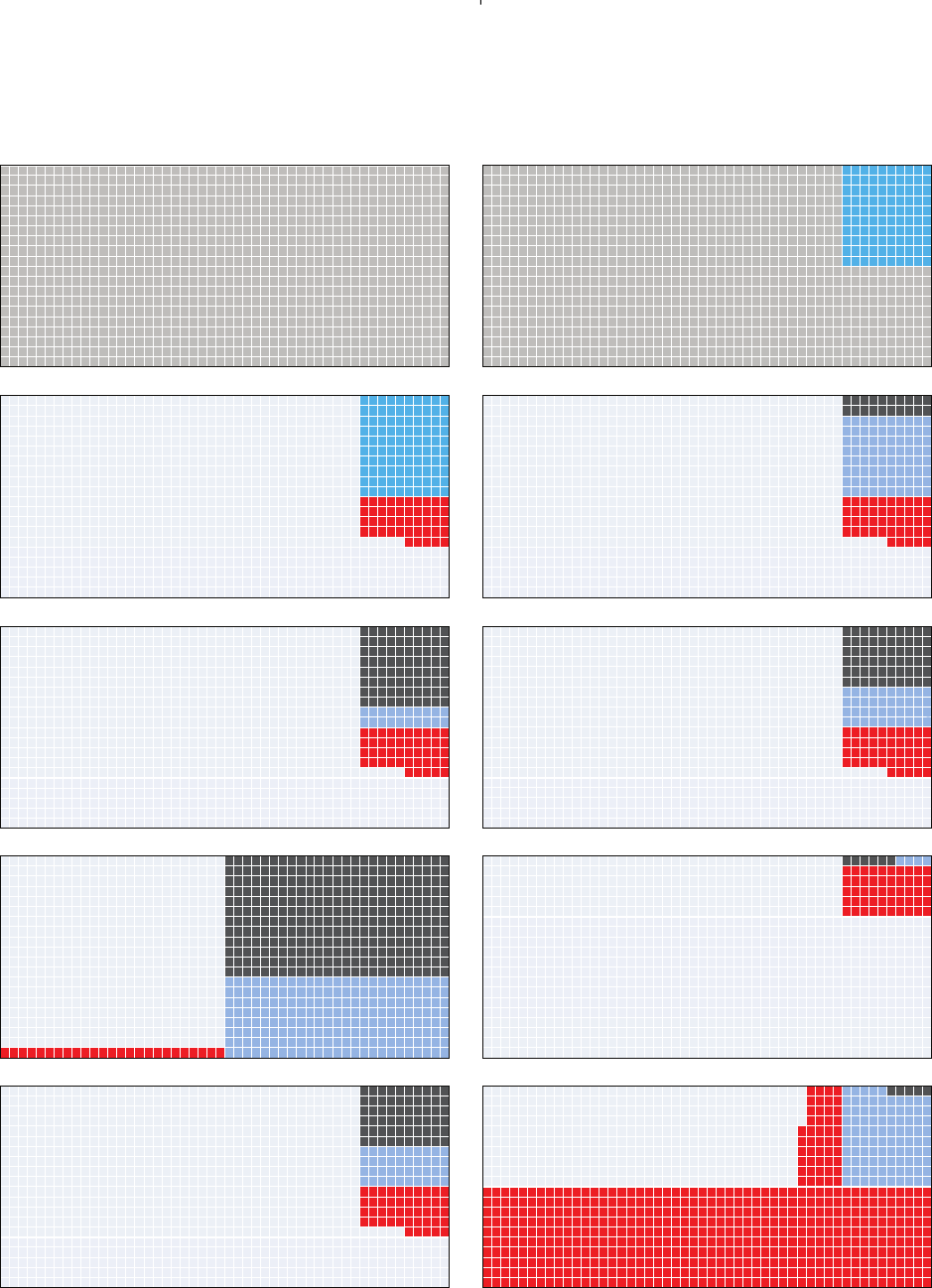
every step of the argument that we illustrate in Fig. 1 (see
also Lakens & Evers, 2014).
Consider a thousand hypotheses H1 that we might wish to
test (Fig. 1A). Many of these may not be true, so let us start
with a scenario where only 10% of the hypotheses at hand are
in fact true (Fig. 1B). This proportion of hypotheses being
true is often described with the symbol π(here π=0.1).
When testing the 900 hypotheses that are not true (dark grey
in Fig. 1B), we allow for 5% false-positive findings if we set
our significance threshold at α=0.05 (the accepted level of
making Type I errors). This means we will obtain 45 (i.e.
900 ×0.05) false-positive answers (red in Fig. 1C), where we
state that our data provide significant support for the hypoth-
esis H1 (or more formally speaking of ‘evidence against the
null hypothesis H0’) even though that hypothesis H1 is false
(and H0 is true). Now, when testing the 100 true hypotheses,
we will sometimes fall short of the significance threshold, i.e.
cases where we would conclude that the data do not support
that hypothesis H1, although it is true and H0 is false (a
false-negative or Type II error). The frequency with which
our test of the empirical data falls short of reaching signifi-
cance despite the hypothesis H1 being true is known as the
probability β(the probability of making a Type II error). The
probability βdepends on sample size (and effect size). When
the data set is very large, the risk of falling short of significance
is small, so we speak of the study having high statistical power
(which is defined as 1 −β). In our example in Fig. 1D, we
have a large sample size and hence a high power (80%) to sup-
port 80 out of the 100 true hypotheses correctly. In this case, β
will be 20%, leading to 20 false-negative conclusions shown in
black (i.e. where we reject the hypothesis despite it being true).
Here is the essential point of Ioannidis’ argument
(Ioannidis, 2005): when we consider only the subset of
positive outcomes, where a hypothesis H1 has been supported
by the data (the 45 red and the 80 blue cases in
Fig. 1D), 36% (i.e. 45/(45 +80)) will not be true. This
is the fraction of positive research findings (where data
provided significant support for a hypothesis) that are false.
This is also known as the false-positive report probability
(FPRP =(α(1 −π)/[α(1 −π)+(1 −β)π]). Notably, this
fraction is much higher than 5%. This highlights the fact
that a 5% false positive rate (i.e. setting αat 0.05) does not
mean that only 5% of significant research findings are false.
The situation may get worse. In many studies, sample sizes
are low, resulting in statistical power that is often as low as
20% (Møller & Jennions, 2002; Smith, Hardy & Gammell,
2011; Button et al., 2013; Parker et al., 2016). In this situation
we will have 80 instead of 20 cases of false-negative results
(black in Fig. 1E). If we then consider the positive outcomes
only, we observe that 69% of the significant research findings
are false [the red out of the red plus blue fraction in Fig. 1E;
45/(45 +20) =0.69]. This disturbingly high proportion is
what made Ioannidis (2005) claim that most findings are false.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
1944 W. Forstmeier and others
1000 hypotheses
10%
true
5% false-
positives
36% false
20% power
69% false
40% power
53% false
Unlikely hypotheses (1 in 10)
Likely hypotheses (1 in 2)
11% false
Highly unlikely hypotheses (1 in 100)
93% false
53% false
Unlikely hypotheses (1 in 10)
85% false
Unlikely hypotheses (1 in 10)
effective α= 60.7%
(B)(A)
(D)(C)
(F)(E)
(H)(G)
(J)(I)
10%
true 80%
power
40% power
40% power
40% power
95% power
Fig. 1. Different scenarios of testing 1000 hypotheses, of which a limited proportion is true. The colours in panels (B and C) refer to
hypotheses that are actually true (bright blue) or false (dark grey). The colours in panels (C–J) indicate false-positive findings (Type I
error; red), true positive findings (pale blue), false-negative findings (Type II error; black), and true negative findings (light grey). For
details see the main text. Illustration adopted and extended from http://www.economist.com/blogs/graphicdetail/2013/10/daily-
chart-2.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1945
For the following calculations, we will settle for an
intermediate sample size (larger than is typical in ecology
and evolution), which gives us a statistical power of 40%.
Under this condition, 53% of the positive findings will be
false (Fig. 1F). Now, it is essential to remember that we
started with a scenario where only 10% of the hypotheses
were actually true. That is, we were testing moderately
unlikely hypotheses to begin with (Fig. 1F). If, in contrast,
you are working in a research area where people mostly
test hypotheses that are likely (every second hypothesis being
actually true), the proportion of false-positive reports is quite
small (Fig. 1G). We would obtain only 25 false positive reports
(red in Fig. 1G), but as many as 200 true positives (blue in
Fig. 1G). In this case, readers of publications that present
positive findings will not often be misled (11% false). If,
however, a research field is testing highly unlikely hypotheses
(only one in a hundred being true) nearly all positive reports
will be incorrect (93% false, Fig. 1H).
To illustrate one final point, let us return to a situation
with moderately unlikely hypotheses (10% true) and still
intermediate power (1 −β=40%), which is shown in Fig. 1I.
Let us add a new dimension, which was brought up in
a seminal publication of Simmons, Nelson & Simonsohn
(2011). They stated that researchers actually have so much
flexibility in deciding how to analyse their data that this
flexibility allows them to coax statistically significant results
from nearly any data set [for similar insights see Barber
(1976), De Groot (1956/2014), Feynman (1974) and Gelman
& Loken (2014)].
Simmons et al. (2011) called this flexibility ‘researcher
degrees of freedom’. We will address these researcher degrees
of freedom in detail below, and we will give a range of
illustrative examples. For now, imagine that researchers have
to make many arbitrary decisions in data analysis, and if they
are trying hard (even unintentionally through self-deception)
to provide positive evidence for their hypothesis, at every
arbitrary step they may always go for the option that produces
the lowest P-value (‘significance seeking’). Using simulations,
Simmons et al. (2011) show that the combination of always
choosing the better option in four consecutive arbitrary steps
(each of which seems of minor importance, e.g. analysing
yearlings and adults together versus separately) adds up to
a dramatic effect of raising the α-level from α=0.050 to
0.607. That means, if we systematically chose the option
that reduces the P-value in each of the four steps, we will
be able to present an effect of interest as being statistically
significant (P<0.05) in 607 out of 1000 cases in which no
real effect exists (hence the formulation ‘allows presenting
anything as significant’). If this scenario of raising αto 60.7%
is applied to Ioannidis (2005) calculations, we would see 535
false positives (red in Fig. 1J) compared to approximately 95
true positives (blue in Fig 1J; note that this latter number is
a rough guess and not based on simulations), which would
mean that about 85% of all positive findings would be false.
According to the calculations illustrated in Fig. 1, the
proportion of false-positive reports (out of all positive reports)
will be highest for: (i) fields with mostly underpowered studies
(small sample size); (ii) fields with unlikely hypotheses (driven
by pursuit of novelty); (iii) fields that poorly guard against
raising the level of α(significance seeking).
More can be said about each of these influential factors:
1 A comparison between Fig. 1D,E illustrates why low
power produces relatively more false-positive findings.
The absolute number of false positives stays the same
(always 45 red cells), but we see fewer correct positives
(20 rather than 80 blue cells) as power drops from 0.80
to 0.20. Hence the proportion of positive findings that
are correct is decreasing. If you want to carry out your
own calculations to see how the statistical power in
your experiment depends on sample size, you will find
suitable calculator tools online (e.g. GPower; Faul et al.,
2009), but they will always ask you about the size of
the effect that you wish to detect. This is hard to know
a priori. In the fields of ecology and evolution observed
effect sizes are typically small (e.g. r=0.19; Møller &
Jennions, 2002), which is still likely an overestimate
(Hereford, Hansen & Houle, 2004; Parker et al., 2016).
Hence, large sample sizes are required to detect such
effects (required N=212, for detecting r=0.19 with
80% power). While studies in animal behaviour have a
reasonable power of around 70% for detecting a large
effect of r=0.5, the power for detecting an effect of
r=0.19 lies only around 15–20% [own calculations
using GPower 3.1 (Faul et al., 2009) based on results of
Jennions & Møller (2003) and Smith et al. (2011)].
2 The relative proportion of false-positive reports is most
strongly influenced by how likely one’s hypothesis is to
begin with (compare Fig. 1G with 1H). However, this
quantity may be difficult to gauge. Most researchers
would probably think (or at least hope) that they
are testing relatively likely hypotheses (much closer to
Fig. 1G than 1H). However, people’s impressions may
be deceiving. The existing literature is heavily biased
towards stories of success (Parker et al., 2016), with
84% of all publications finding support for their initial
hypotheses (Fanelli, 2010). As we will see in Section
II.2, this figure is far from an objective representation of
all hypothesis tests that have been conducted, because
null findings (non-significant results) are less likely to
get published (Rosenthal, 1979; Simonsohn et al., 2014),
and because various common data-analysis practices
increase the rate of false positives as well as the average
strength of reported effects among those results that are
published (Anderson, Martinson & De Vries, 2007;
Simmons et al., 2011; John, Loewenstein & Prelec,
2012; Parker et al., 2016). Even without problematic
data analysis, some (false) positive evidence can emerge
for any hypothesis (Fig. 1H). Thus, just because we see
support for a theory in the literature does not mean we
should assume that our hypothesis, which is based on
this theory, is likely to be true. Finally, one should realize
that high-impact journals are always on the lookout
for the most novel and surprising research findings.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1946 W. Forstmeier and others
Thus when researchers find evidence for surprising
hypotheses (Fig. 1H) and manage to secure publication
in these high-impact journals, other researchers may
be tempted to test increasingly far-fetched (non-trivial,
surprising) hypotheses. This could push a research field
into an arms race that comes at the expense of tests for
less-surprising hypotheses.
3 There are so many different ways in which the α-level
can be raised above the conventional threshold of 5%
(Fig. 1J) that this will keep us busy for most of this
review. Conceptually it is helpful to distinguish between
two problems. First (treated in Section II.2), there is
the issue of multiple hypothesis testing that comes in
various forms and can sometimes be deceivingly cryptic
(Parker et al., 2016). Here it is important to keep track
of the extent of multiple testing. This may allow us to
adjust α-levels accordingly, so that P-values can still
be interpreted in a meaningful way. Second (treated
in Section II.3), there are many ways of carrying
out statistical tests incorrectly which often will yield
highly significant P-values that are misleading and
incorrect to an extent that cannot be adjusted for.
The probably most important source of error here is
the non-independence of data points (Milinski, 1997),
which is typically referred to as pseudoreplication
(independent data points are considered as proper
replicates, while non-independent data points are
considered as pseudoreplicates) or as clustered data
(Weissgerber et al., 2016).
Table 1 provides an overview of the statistical and
psychological issues that will be addressed herein together
with a collection of possible solutions.
(2) Multiple testing in all of its manifestations
In this chapter we will focus on how multiple testing and
selective attention or reporting lead to inflated rates of Type
I error. If researchers were forced to report the outcome of
every single statistical test that they conduct, every obtained
P-value could be taken at face value. With αset at 0.05,
for each hypothesis H1 that is not true we would only have
a 5% risk of drawing a false-positive conclusion. However,
as soon as reporting becomes conditional on the outcome
(typically: positive findings being more likely to get reported)
or when we focus our attention on the promising outcomes
(ignoring or forgetting about negative outcomes), the risk of
a false-positive conclusion is much higher than 5% (e.g. 53%
in Fig. 1F).
When the total number of statistical tests conducted is
known (e.g. 10 tests), then it is possible to calculate the
probability of obtaining at least one significant result by
chance alone (1 −0.9510 =40%), and it is possible to adjust
α-levels (0.05/10 =0.005) for each test to ensure that the
probability of making one or several Type I errors remains
at about 5% (1 −0.99510 =4.9%). This adjustment is known
as the classical Bonferroni correction (Dunn, 1961). While
using such a strict α-threshold is effective in limiting Type
I errors, it inevitably will increase the number of Type II
errors (i.e. true effects that are discarded because they do not
pass this threshold). Hence, if you are more worried about
making Type II errors than about making Type I errors,
you may well discard the Bonferroni correction (Nakagawa,
2004), or go for less-strict methods of correction based on
false-discovery rate (Benjamini & Hochberg, 1995; Pike,
2011). Yet, whenever we allow our Type I error rate to rise
in the interest of keeping the Type II error rate low, we
will produce many false positives and thus need to seek to
replicate these exploratory findings (Pike, 2011). For instance,
if your aim is to discover a new treatment for a disease, you
want to make sure that you do not miss out on something
potentially interesting (and hence limit Type II errors). This is
the exploratory part of science. It is essential and important.
However, once you identified a potential treatment, you
should be interested in making sure that you are right so
as not to waste money or even cause harm, and hence you
want to reduce Type I errors. This is the confirmatory part
of science, the proper testing of a priori hypotheses.
Adjustments of αto multiple testing are typically called for
when researchers present large tables containing numerous
statistical tests, of which only a small fraction reaches
significance (P<0.05). Such tables elicit skepticism in
experienced researchers, who rightly worry that the content
of the entire table may be consistent with the null hypothesis.
As a pre-emptive response to such skepticism, authors may
avoid presenting too many non-significant results alongside
their positive findings. A threshold of P<0.05 seems fairly
reasonable when only a few P-values are shown and these
P-values mostly lie below the 0.05 threshold. By contrast,
referees may request a more stringent threshold when many
non-significant results are presented alongside, because the
long list clearly reveals the extent of multiple testing.
Problematically, when authors are free to choose which
results to present in their publication, it becomes impossible
to judge the appropriate statistical significance of the findings.
When, for instance, the authors highlight a single significant
finding from a pool of 10 tests they report, this inspires much
less confidence in that finding than if it had arisen from
a single planned test. This is a serious dilemma. Justified
skepticism from reviewers creates an incentive for reduced
transparency in scientific publications, thereby lowering the
overall utility of the reported work. This problem could
be mitigated if reviewers and editors would acknowledge
and appreciate the greater scientific value of a paper that
comprehensively reports all outcomes of a study compared
to the minimalistic presentation of a single finding.
There is compelling evidence that many tests do, in
fact, go unreported. As mentioned above, across scientific
disciplines, 84% of all studies present positive support for
their key hypothesis (Fanelli, 2010). Such a high success
rate is impossible to obtain without selective reporting or
biased attention that de-emphasizes non-significant findings
or likely a combination of both (see Fig. 1G). Even if all tested
hypotheses were true (which they are not), a statistical power
of 84% (rarely ever achieved) would be required to yield
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1947
Table 1. Collection of problems and possible solutions
Section Problems Solutions
II.1 •Small sample size (e.g. data hard to obtain) •Acknowledge preliminary nature
•Multi-laboratory collaborations
II.1 •Novelty seeking •Regard ‘surprising’ findings sceptically prior to replication
II.2a•Multiple testing and selective reporting (e.g. due
to too much trust in hypotheses, hindsight bias,
pressure from referees)
•Avoid excessive testing (think before data exploration)
•Keep track of number of tests conducted and report all tests
•Bonferroni correction, false-discovery rate or emphasize
preliminary nature of findings
•Average effect sizes across conceptually similar tests
•Referees and editors promote comprehensive and unbiased
reporting
II.2b•Multiple testing within models (stepwise model
simplification)
•Report the initial full model
•Global test of full model against null
•Test a pre-determined subset of models
•Average effects of individual variables across models
II.2b•Overfitting of models (inflated significance) •Keep N>3kfor correct P-values, where kis number of
parameters to be estimated (N>8kfor reliable parameter
estimates)
II.2c•HARKing (hypothesizing after the results are
known) and hindsight bias
•Preregister hypotheses
•Keep track of number of tests conducted
•Comprehensive reporting
II.2d•Data collection ends with reaching P<0.05 •Declare stopping rule
•Adjust P-value for multiple testing
II.2d•Discarding ‘unsuccessful’ experiments until an
experiment ‘works’
•Complete reporting of all experiments
II.2e•Arbitrary decision in analysis (e.g. selective
removal of outliers) are taken conditional on
reaching significance (confirmation bias)
•Make decisions apriori(preregistration)
•Ask colleagues to make decisions for you
•Blinding yourself during data analysis
•Specification-curve analysis: try all versions to examine
robustness of findings
II.3 •Non-independence of data points (e.g. related
individuals, temporal and spatial autocorrela-
tion)
•Test for non-independence, autocorrelation
•Fit grouping variables as random effects (intercepts, slopes,
space, time, pedigrees)
•Run analysis at the level where independence is met
•Balance experiments for confounding effects
II.3c•Overdispersed data •Transform data
•Control for overdispersion (random effects, quasi-likelihood)
II.4a•Over-interpretation of apparent differences •Test significance of interaction term
•Test context dependence in follow-up study
II.4b•Misinterpretation of regression to the mean •Avoid allocating individuals to different treatment groups
according to phenotype
•Set up a control group
•Model the expected effect
II.5 •Confirmation bias in data collection •Blinding observers to treatment groups
III.1 •Lack of close replication studies •Regard unreplicated findings as preliminary
•Preferentially cite confirmatory replication studies as the most
convincing evidence
•Replicate own findings
•Replicate important foundational studies as part of new
research
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
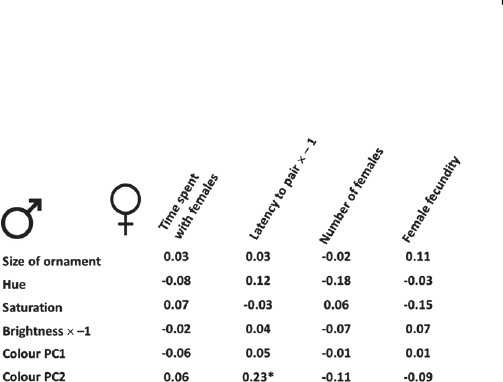
1948 W. Forstmeier and others
Fig. 2. A fictional table of correlation coefficients between
measures of male ornamentation and measures of male success
in pairing with females. The asterisk highlights a significant
correlation. Some parameters were multiplied by −1, such that
positive correlations indicate higher mating success for more
ornamented males.
this rate of success. Hence, this means that most disciplines
presumably sit on a huge pile of ‘failed’ experiments and
unpublished null results that are inaccessible because they
are hidden in the file-drawers of the experimenters [known
as the ‘file-drawer problem’ (Rosenthal, 1979)].
In the following we will discuss various forms of multiple
testing by giving typical examples to increase principle
awareness of problematic situations.
(a)The temptation of selective reporting
Imagine you study mate choice in species xy, and you would
like to understand why males of species xy have a colourful
plumage ornament that is absent in females. Hence, on the
side of males, you measure the size of the ornament as well
as its colour in terms of hue, saturation, and brightness, and
you also summarize the measures of the reflectance spectra
in two principal component scores. To assess female choice,
you measure how much time females spend close to each
male, the latency for males to secure a female partner, the
number of females each male sires offspring with, and the
number of eggs laid by females after pairing with a male of a
given ornamentation. You then look for positive correlations
between the degree of male ornamentation and their success
in attracting and pairing with females (Fig. 2).
The longer you look at this table of correlations with
one association being significant, the more tempting it
may become to convince yourself that, maybe, principal
component analysis actually represents the most objective
way of summarizing complex colour information, and that
maybe the latency to pair is the most meaningful measure
of male pairing success in this study species. Surely this
significant finding must be a true positive effect, since why
else would males have evolved these beautiful colours. Also
the use of Bonferroni correction has often been criticized
(Nakagawa, 2004) for being too conservative and leading to
many false-negative outcomes (Type II errors). Hence, we
might be tempted to publish only the association of ‘latency
to pair’ with ‘Colour PC1’ and ‘Colour PC2’ without
mentioning the remaining 22 null results (focussing on PC2
only without reporting on PC1 would be too extreme). We
might not even perceive this as unscientific conduct because
we have convinced ourselves of the biological and statistical
logic behind our ‘discovery’. As we convince ourselves that
the biology is right, we presumably feel an obligation to
share our discovery. Thus our personal focus on discovery
motivates us to publish this as a positive finding. Humans
are highly efficient at finding post-hoc justifications for their
choices (Trivers, 2011) if those choices produce a more
desirable outcome [positive results are likely easier to publish
than null findings (Franco, Malhotra & Simonovits, 2014)].
When we selectively report only 2 out of the 24 correlations
shown in Fig. 2, we often forget that the remaining 22
correlations actually represented equally valid tests of our
hypothesis that greater ornamentation enhances mating
success. A more objective approach would be to average
the 24 correlation coefficients to yield an estimate of the
overall effect of ornamentation. This can be done because all
variables were coded in such a way that high values always
refer to increased ornamentation and increased mating
success, meaning that positive correlations count as support
for the hypothesis. In our example, the average correlation
between ornamentation and mating success is exactly zero
(the mean of all positive and negative correlations is r=0.00).
Hence, if we started with the aim of objectively quantifying
something (rather than discovering something) we should
face less of a risk of misleading ourselves and our colleagues
and of having wasted efforts for the short-term benefit of
possibly publishing in a higher-ranking journal.
This hypothetical case clearly shows that ‘data do not
always speak for themselves’. Without knowing the context
of why the author decided to focus on principle component
analysis and on ‘latency to pair’, we cannot judge the
statistical significance of the finding. We will explore other
examples of deceiving statistical results below.
The literature on sexual selection acting on ornamental
traits is plagued by this problem of potential selective
reporting (not every study is biased, but there is no label
that would identify unbiased reporting). Since we have no
way of telling the extent of reporting bias, it is not clear
how we could draw a general conclusion about the strength
of sexual selection on ornaments from several decades of
work (Parker, 2013). This illustrates how inefficient research
can sometimes be if it fails to ensure maximal objectivity in
reporting. Meta-analyses that summarize all published effects
are not able to take into account these arbitrary decisions
made by authors (Ferguson & Heene, 2012). Although any
meta-analytic summary would certainly reveal a strong effect
of ornaments on mating success, it is unclear whether or to
what extent this is evidence for a theory as opposed to
evidence of selective reporting driven by a theory. There
are probably more than a few research areas where we
might benefit from a new round of empirical investigation
in which all results were made available. If we all begin now
with studies adhering to a standard of unbiased reporting
and we make such studies identifiable with the use of
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1949
badges (see Section III.3b), in a few years’ time we could
conduct meta-analysis comparing studies with and without
such badges to confirm or refute our past work.
(b)Cryptic multiple testing during stepwise model simplification
A table like that shown in Fig. 2 immediately reminds
researchers that they have to be aware of the issue of multiple
testing. A much less obvious form of multiple testing happens
when researchers fit complex models to explain variation in a
dependent variable by a combination of multiple predictors.
This has been termed ‘cryptic multiple hypotheses testing’
(Forstmeier & Schielzeth, 2011).
Imagine you are trying to explain variation in a variable
of interest with a set of six possible predictors. Besides the
six main effects that you are interested in, there is also the
possibility that any pair of two predictors might interact with
each other in influencing the dependent variable. To explore
all these possibilities you start by fitting a rather complex full
model where the dependent variable is a function of 6 predic-
tors plus their 15 two-way interactions, and you then carry
out a standard procedure of model simplification where, at
each step, you always delete the least significant term from
the model until you have only significant predictors (main
effects or interactions) left in the minimal model. Such exten-
sive data exploration minimizes the risk that you overlook a
potentially complex combination of factors that affects your
variable of interest. However, this widespread procedure
(recommended by some standard statistical textbooks, e.g.
Crawley, 2002) comes with a very high risk of Type I error.
In a simulation study it was shown (Forstmeier & Schielzeth,
2011), that when all null hypotheses are true (using randomly
generated data), the chance of finding at least one significant
effect lies close to 70%). This means that most of the time you
will be able to present a significant minimal model that seems
to reveal an interesting pattern [see also Mundry & Nunn
(2009) and Whittingham et al. (2006)]. Many researchers
seem unaware that they have actually examined 21 different
hypotheses at once, and that a Bonferroni correction of
setting αto 0.05/21 =0.0024 would be required to keep the
false-positive rate at the desired 5%.
This Bonferroni correction works reliably as long as the
full model was built on a reasonably sized data set. However,
when sample size becomes low relative to the number of
parameters to be estimated, then the estimation of model
fit and P-values becomes highly unreliable. This happens
because a small number of data points can often be explained
almost perfectly by a combination of predictors selected from
a relatively large pool of predictors. For instance, if the same
6 main effects and their 15 two-way interactions are fitted
to only 30 data points, the resulting minimal models are
often excessively significant. As many as 26% of the minimal
models cross even the Bonferroni-corrected threshold of
0.05/21 =0.0024, such that a much stricter correction to
0.05/286 =0.00017 would be required to ensure that only
5% of the minimal models pass that threshold. In other words,
running through such an automated assessment of your six
predictors and their two-way interactions by step-wise model
simplification is expected to give you P-values that are as low
as you would get from always picking the most significant
among an incredible 286 hypothesis tests.
Surely, this is an extreme case where P-values are no
longer correct (and not adjustable by Bonferroni correction)
because they are derived from an over-fitted model. Simula-
tions (Forstmeier & Schielzeth, 2011) revealed that P-values
begintobecomeexcessivelysmalloncetherearefewerthan
three data points per predictor (N<3kwith kbeing the num-
ber of parameters to be estimated). Regarding this result from
the other side, the observation that P-values were correct
(adjustable by Bonferroni correction) as long as there were
more than three data points per parameter, does not imply
that this sample size is sufficient in all respects. Statisticians
often recommend that at least eight data points per estimated
parameter should be available (e.g. N>8k+50; Field, 2005),
and they would consider the over-fitting of models described
in the previous paragraph a ‘statistical crime’. However,
when screening the literature in the field of ecology and
evolution, Forstmeier & Schielzeth (2011) found that authors
rarely described the initial full model that they had fitted.
This means that the extent of multiple testing and of possible
over-fitting could often not be reconstructed. Out of 50 stud-
ies examined, 28 used models with two or more predictors,
6 of which fitted between 6 and 17 effects, and 3 of which
violated the rule to not over-fit (N<3k). Moreover, and most
strikingly, none of the 28 studies considered any adjustment
of P-values for multiple testing (e.g. Bonferroni correction).
In some fields, iterative model building of the sort
we just described has become less common, but what
has replaced it is often not substantially better (Mundry,
2011). The replacement is typically a process by which
researchers develop a set of ‘plausible models’ and evaluate
them with measures of overall model fit (e.g. likelihood
ratio) or fit accounting for the number of predictors [e.g.
Akaike Information Criterion (AIC), or Bayesian Information
Criterion (BIC)]. Researchers may then assess parameter
estimates or tests of significance for individual predictors only
in the ‘best’ model. Just as with an iterative procedure, it is
unreasonable to assess the statistical significance of individual
variables in the ‘best’ model without correction for multiple
comparisons. Similarly, assessing the strength of effects in
only the ‘best’ model is also likely to produce an inflated effect.
(c)A priori hypothesis testing versus HARKing: Does it matter?
The above approach of exploratory data analysis means that
a fairly large number of hypotheses get tested in a very short
time (i.e. without careful thinking about specific hypotheses
considered plausible) and this comes with a high risk of
drawing a false-positive conclusion if we only report on the
subset of significant predictors. In fact, such exploratory
analysis could be seen as an act of generating hypotheses
rather than as an act of testing hypotheses, because you
only start thinking about the respective hypothesis once you
have discovered a significant association. This approach is
not wrong per se, as long as you are aware and honest
about the fact that the hypothesis was derived from the data.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1950 W. Forstmeier and others
The problem starts where researchers fail to acknowledge
this. The psychologist Norbert Kerr called this ‘HARKing’
(hypothesising after the results are known; Kerr, 1998).
Yet, does it really matter in terms of likelihood of a positive
finding being true whether we thought of the hypothesis a
priori (i.e. before data inspection) and then use the data to test
that hypothesis, or whether we came across the hypothesis
only after having explored the data and having focused on
only significant effects to begin with? Intuitively, we would
probably think that a priori hypothesis testing is less prone to
yield mistakes than ‘fishing for significance’ and HARKing,
but is that intuition correct?
In both cases we would use exactly the same data set (and
arrive at the same P-values), so for any given hypothesis,
the statistical outcome appears exactly the same. Although
this is true for any given hypothesis, fishing, HARKing,
and hindsight bias often produce hypotheses that researchers
never would have deduced from theory. Hindsight bias or the
‘knew-it-all-along’ effect (Fischhoff, 1975) is the phenomenon
that, after having seen the results of data analysis, these
results appear logical, inevitable, and in line with what we
must have predicted before. Hindsight bias is particularly
dangerous because we overestimate the plausibility of our
hypothesis (which in fact is a post hoc explanation, a hypothesis
that was derived from the data, not one that we had a priori).
Sometimes it is easy to spot unlikely hypotheses that were
derived from the data. When the title of a publication starts
with ‘Complex patterns of ...’ and the main finding of
the study consists of a difficult interaction between several
explanatory variables, then this complex hypothesis may well
have been derived from the data.
However, data exploration is not fundamentally a bad
thing. In fact when conducted transparently, it is very useful.
It may allow you to discover something for which theory has
not even been developed yet, or you may actually correctly
identify a complex pattern of interactions for which theory is
too simplistic. Yet, the main problem with data exploration is
that we normally do not keep track of the number of tests that
we have conducted or would have been willing to entertain,
so there exists no objective way of correcting for the extent
of multiple testing (De Groot, 1956/2014). Once a discovery
has been made (P<0.05) and a plausible explanation has
been found, it is very easy to deceive oneself into thinking
that one actually had that hypothesis in mind before starting
the exploration, and nothing seems wrong with writing up a
publication saying ‘here we test the hypothesis that ...’.
The failings of this approach were explained long ago
by De Groot (1956/2014) and they are strongly linked
to points we have already made. In exploratory analyses,
we are open to an array of possible relationships and
resulting interpretations. As the array of possible detectable
relationships expands, the likelihood that we might detect
false relationships expands as well. Of course we may well
also detect real relationships, but at this stage, we cannot
distinguish what is false from what is real. We have generated
a suite of hypotheses with our data exploration, and next we
(or others in the years to come) need to gather additional
data. With the new data, we should conduct only the very
limited set of analyses designed to test the hypotheses derived
from the exploratory work. Thus in this second round of
data collection and analyses, we can operate with a much
lower probability of detecting false positives. In other words,
we test hypotheses rather than just generate them.
In some fields it is common practice to masquerade
exploratory analyses as confirmatory hypothesis testing
because exploratory work is often perceived as inferior
or old-fashioned. In the distant past, data exploration
was presented in the Results section, and its subjective
interpretation was given in the Discussion section. Then
biologists adopted (or at least pretended to adopt) Popper’s
idea about hypothesis testing, and in the process started
to move their data-derived (post hoc) hypotheses to the
Introduction so they could pretend they were testing a priori
hypotheses. Unsurprisingly, when we ‘test’ a hypothesis with
the same data that generated that hypothesis, it tends to be
confirmed. In other words, you simply cannot ‘cherry-pick’
the hypothesis you wanted to test after having seen the
outcome of statistical analyses (Fig. 3).
(d)Researcher degrees of freedom: (1) stopping rules
As promised earlier, we now return to the issue of ‘researcher
degrees of freedom’ (Simmons et al., 2011), which refers to
researchers’ flexibility in how to collect and how to analyse
their data. One striking issue regards stopping rules for data
collection. How do you decide that you have enough data?
Say you are trying to test whether females of species xy
prefer males that sing with a lower-pitched voice. Initially,
you do not know how large such an effect might be, so
you start by collecting data on 10 females, after which
you conduct a simple regression test (pitch predicts female
response to song). Now let’s say you obtained a trend
in the expected direction (slight preference for low-pitch
voice), but the effect does not reach significance. You might
suspect that the effect is real but small, and that you lacked
statistical power to reach significance. You then collect data
on another 10 females and then conduct your regression
test again with all 20 females. Although the rationale behind
such a sampling design seems perfectly understandable, the
risk of making a Type I error has just risen from 5% to
approximately 7.7 (Simmons et al., 2011). This is because
you gave the data two chances of reaching significance.
Since the first data set is included in the second, these are
not two fully independent chances (which would yield 9.75%
false positives; 1 −(0.95 ×0.95) =0.0975), so the combined
risk of drawing a false-positive lies somewhere between 5
and 10% (this risk can be estimated from simulations). Thus,
when decisions about sample size are not made a priori,
and data sets are subject to iterative tests for significance as
data accumulate, you must correct for multiple testing. The
more often you stop data collection to check for significance,
the greater your risk of a false positive. It is important
to remember here that your decision to collect data on
another 10 females was conditional on the first outcome.
If you had obtained a statistically significant effect at the
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
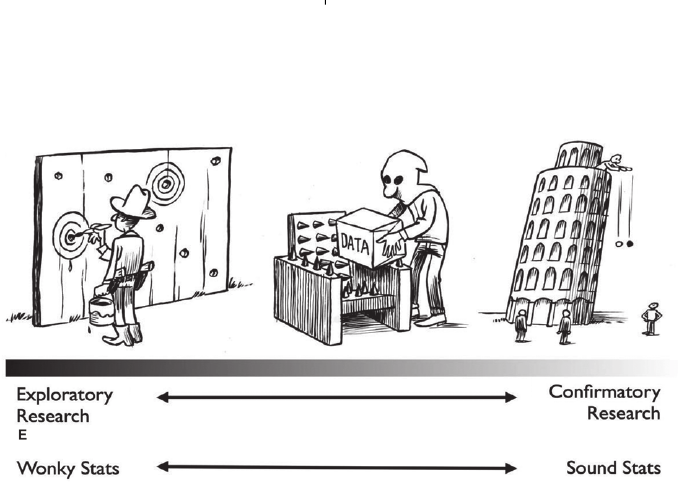
Avoiding false-positive findings 1951
Fig. 3. The graded di stinction between exploratory, hypothesis-generating research and confirmatory, hypothesis-testing research
(Wagenmakers et al., 2012). On the right side of the continuum, a purely confirmatory test is conducted. The test is transparent,
relevant hypotheses have been explicated beforehand, and a data analysis plan is present. This exemplifies the scenario of
hypothesis-testing research. For this type of research – and only for this type of research – statistical tests have their intended
meaning. On the left side of the continuum, a purely exploratory test takes place. The ‘Texas sharpshooter’ first fires at a fence,
and then proceeds to draw the targets around the bullet holes. There is no prediction here – there is only postdiction. This scenario
exemplifies the scenario of hypothesis-generating research. For this type of research, the resulting statistical tests (invented exclusively
for hypothesis-testing research) are misleading, or, in Ben Goldacre’s terms, ‘wonky’. In between the two extremes lies a continuum
where research is conducted that is partially confirmatory, typified by a degree of data massaging – in the figure, the data are
‘tortured until they confess’. The statistical results are partially wonky. Unfortunately, it is far too easy to make the mistake of
masquerading hypothesis generation as hypothesis testing. Most researchers, including the authors, admit to having done this (John
et al., 2012), either because of ignorance of the problem or because of self-deception [see Fischhoff (1975) and Trivers (2011)]. Figure
courtesy of Dirk-Jan Hoek.
first try, you would presumably not have collected more
data, but rather you would have concluded that the effect
seemed to be large because it reached significance with
only 10 females. In a scenario in which reaching statistical
significance always triggers an end to data gathering, there
is never an opportunity to discover whether a larger sample
size might eliminate significance.
In a worst-case scenario where you keep testing after every
sample until the expected effect reaches significance, you
are certain to find the effect eventually, since P-values will
undergo a random walk (Rouder et al., 2009) and will at
some point cross the 5% threshold (unadjusted for multiple
testing). Our own (unpublished) simulations with randomly
generated numbers show that you can expect to cross the
threshold of significance within the first N=100 in about 3 of
10 attempts (hence α=0.3 rather than α=0.05), although
if you are willing to continue to sample indefinitely, you will
eventually reach statistical significance in every single case
(Armitage, McPherson & Rowe, 1969). Hence, continued
sampling and a stopping rule based on reaching significance
unambiguously elevates Type I error rates and thus we expect
this to be one of the many factors leading to false positives
in the literature. Fortunately, as researchers are increasingly
becoming aware of this problem, it is slowly becoming good
practice to specify one’s stopping rule for sample size in the
methods section of a publication.
In a wider context, the same issue of multiple testing
applies to situations where researchers discard one or two
initial experiments that were ‘unsuccessful’ (for instance
because of a putative confounding factor that was not yet
controlled) and then have full trust in the first experiment
that yields the desired result.
(e)Researcher degrees of freedom: (2) flexibility in analysis
When analysing data, we face a wide variety of rather
arbitrary decisions that we have to make, such as: (i) should
I include covariate xin the model as a possible confounding
factor, and should xbe log-transformed or should I subdivide
it into categories (and how many)? (ii) Should I include or
exclude a particular outlier or an influential data point (high
leverage)? (iii) Should I transform the dependent variable
to approximate normality better, and which transformation
should I choose? (iv) Should I add baseline measures taken
before the start of the experiment as a covariate into the
model in order to remove some noise in the data? (v) Should
I control for sex as a fixed effect or also model a sex by
treatment interaction term? (vi) Should I exclude individuals
from the analysis for which the number of observations is
low? (vii) Should I remove a third treatment category that
seems unaffected by the treatment or should I lump it with
the control group?
With all these decisions to make, there is again a risk
of trying several versions (multiple testing) and of favouring
the version that renders the more interesting story (selective
reporting). Often, we may subconsciously favour the version
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1952 W. Forstmeier and others
that minimizes the P-value for the effect of interest because we
convince ourselves that this version must be the correct one or
the most powerful one. Since we often believe that an effect of
interest exists (and we designed the experiment to reveal the
effect), we tend to have greater trust in analyses that confirm
our belief. This powerful component of human nature
is called confirmation bias, and it has been documented
in a wide array of settings (Nickerson, 1998). Obviously,
confirmation bias can render our science highly subjective
unless we make all these arbitrary decisions a priori (if possible)
or at least blind to the outcome. By contrast, exploratory
analyses that are presented as confirmatory are always a
threat to objectivity. Unfortunately, full disclosure of post-hoc
decision making may often be quite challenging and requires
substantial conscientiousness, but increased awareness of the
issue is a first step towards mastering this challenge.
In an unpublished manuscript, A. Gelman & E. Loken
called this ‘the garden of forking paths’, which nicely
illustrates that there may be a near-endless diversity of
combinations of decision variants. Simonsohn, Simmons
& Nelson (2015) hence suggested an automated routine
of going through all possible combinations of identified
decisions in terms of their influence on the effect of interest
(the effect at the heart of the ‘story’ of a publication; see also
Steegen et al., 2016). Simonsohn et al. (2015) call this routine
‘Specification-Curve Analysis’ (SCA), and they demonstrate
its utility using the example of a recent study (Jung et al.,
2014) that led to some controversy about subjectivity in
decision making. In that study there were seven decisions to
be made, some of which had more than two options to choose
from, leading to a total of 3 ×6×2×2×2×4×3=1728
possible ways of analysis. SCA shows that only 37 out of these
1728 versions (2%) yield significant support for the prediction
that Jung et al. (2014) evaluated and confirmed in their
publication. A particularly problematic aspect of researcher
flexibility is the decision to remove outliers after having seen
their influence on the P-value. Selective removal of outliers
has a high potential of generating biased results (Holman et al.,
2016), so the removal of data points is generally discouraged.
Publications should always explain the reasons behind any
attrition (loss of data points between study initiation and
data analysis) and should discuss whether the missed samples
might have led to biased results. Up to this point we have
been dealing with the problem of multiple testing in all kinds
of versions, and we have seen that this problem can be
addressed by (i) limiting the number of tests conducted, and
(ii) adjusting α-thresholds to keep the false-positive rate at
some desired level. In all cases we assumed P-values to be
calculated correctly. Yet, in the following chapter we will see
that P-values are often incorrect (often too small), deceiving
us into over-confidence in our result.
(3) Incorrect P-values
P-values indicate how often chance alone will produce a
pattern of at least the strength observed in the experiment.
Accordingly, for any given sample size, if P=0.05 we might
still be sceptical whether the pattern could have arisen by
chance, but if P=0.0001 we will probably be much more
confident that we have discovered a true effect. However, this
confidence is only justified if the statistical test that yielded
the P-value was applied appropriately in the first place, but
not if the data violated the assumptions that underlie the
test. Statistical tests may have many underlying assumptions
(e.g. normally distributed residuals), although many of these
assumptions can be violated without drastic effects on the
P-values. One assumption, however, is crucial for P-values,
and that is the independence of data points. If data points do
not represent true independent replicates but are grouped in
clusters (‘clustered data’; Weissgerber et al., 2016), we speak
of pseudoreplication, and this may lead to over-optimistically
low P-values (Hurlbert, 1984). As we will see below, some
kind of structure in the data leading to non-independence
is ubiquitous. However, such structure only becomes a
problem for testing the significance of a predictor of interest
(e.g. treatment effect), if the samples are non-independent
with respect to the predictor. The latter is what defines
pseudoreplication.
There are many sources of non-independence of data:
repeated measures from the same individual, measures
from individuals that are closely genetically related to each
other, and measures from species that are related through
phylogeny are all non-independent of each other. Variation
in space, for instance in territory quality, may introduce
non-independence of measurements. The occurrence of a
disease may vary not only in space, but also in time, just like
data on daily weather or minute-by-minute data on whether
a bird is singing will show temporal non-independence.
All these dependencies lead to problems in P-value esti-
mation, the full extent of which is truly unknown. From own
experiences as reviewer or editor of manuscripts we gained
the impression that a substantial proportion of submitted
manuscripts contain analyses that are clearly incorrect, and
that the rate at which referees spot and eliminate these mis-
takes is not sufficiently high to ensure that the published liter-
ature would not contain numerous errors. Surely, awareness
of the pseudoreplication issue is well developed in some areas
like experimental design (Hurlbert, 1984; Milinski, 1997;
Ruxton & Colegrave, 2010) or phylogenetically controlled
analysis (Felsenstein, 1985; Freckleton, Harvey & Pagel,
2002). However, in some other fields, non-independence of
data has been overlooked for an extended period of time
because dependencies may be deceivingly cryptic (Schielzeth
& Forstmeier, 2009; Hadfield et al., 2010; Valcu & Kempe-
naers, 2010) and it seems likely that more such problems
will get highlighted and become better known in the
future.
Generally we feel that there is insufficient recognition
of the extent to which incorrect P-values resulting from
pseudoreplication have contributed to the current reliability
crisis. We therefore provide a practical introduction into
some aspects of pseudoreplication, starting from the most
basic principle that most readers will already be familiar with
and then exploring some less-obvious and more-specialized
examples. Those who feel sufficiently versed in statistics could
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
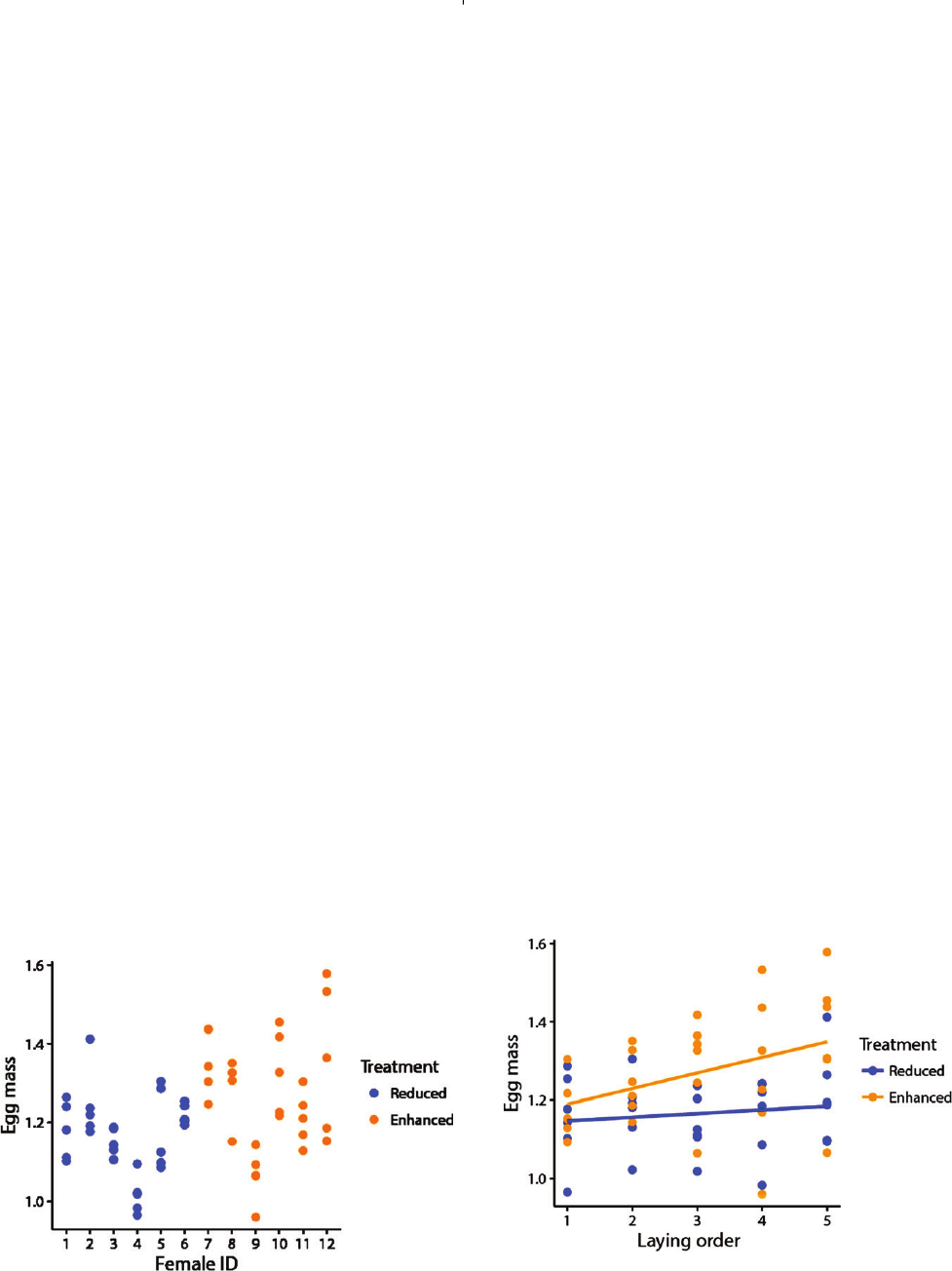
Avoiding false-positive findings 1953
skip the remainder of Section III.3, while the less experienced
may want to go through the examples that we provide.
(a)Pseudoreplication at the individual level
Imagine an experiment where you want to test whether
females lay larger eggs when mated to an attractive
male compared to an unattractive male (differential
allocation hypothesis; Sheldon, 2000). For that purpose you
experimentally enhance or reduce the ornamentation of
males of species xy, and you measure the size of the eggs that
females lay when paired to such males. You have 6 females,
each of which you pair to a different male with enhanced
ornamentation, and 6 different females each assigned to a
different male with reduced ornamentation, and for each of
the 12 females you measure the size of 5 eggs (60 eggs in
total; see Fig. 4). The five eggs that come from the same
female are obviously not independent of each other (i.e. they
are pseudoreplicates with respect to the treatment) and this is
problematic, because females are rather consistent in laying
eggs of a certain size (Fig. 4).
If this non-independence is ignored, and you test the 30
eggs from ‘enhanced’ against the 30 eggs from ‘reduced’, you
will get a highly misleading P=0.002 in this case [R-code:
glm(egg_mass∼treatment)]. Note that this P-value would
be correct if you had had 30 independent females in each
treatment group and if you had measured only one egg from
each of them. In the present case, you can either eliminate
pseudoreplication at the level of individuals by calculating
mean egg size per female and testing the six ‘enhanced’
means against the six ‘reduced’ means which yields P=0.10
[R-code: glm(mean_of_egg_mass∼treatment)], or you can
account statistically for the non-independence by fitting
female identity (ID) as a random effect (‘random intercepts’)
in your model [R-code using the lme4 package (Bates et al.,
2014): lmer(egg_mass∼treatment+(1|female_ID))], which
Fig. 4. Pseudoreplication at the individual level: different
intercepts. Fictional data on egg mass of 5 eggs from each
of 12 females, half of which were assigned to a male with
experimentally enhanced ornamentation and half to a male
with reduced ornaments. Individual females differ in their mean
egg mass (12 different intercepts).
should yield about the same P-value as the first option
(here P=0.07). Thus, it is important to acknowledge that, in
this example, the effective sample size is 6 females rather than
30 eggs per group. Therefore, always make sure to choose
the correct unit of analysis (where independence is ensured),
or make sure to identify sources of non-independence and
to model them correctly as random effects (watch out for
repeated measures on the same individual). Cases of such
overt pseudoreplication have become rare in the literature,
but they still persist in some research areas.
However, there is a risk of making another mistake,
less often spotted. After obtaining a non-significant result
(P=0.07) from testing the a priori hypothesis that females
would lay larger eggs for ‘enhanced’ males, it is tempting
to use the data set for further exploratory analysis. Maybe
the treatment effect will come out more clearly if we also
consider the order in which the five eggs of each female have
been laid (laying order).
In our example, egg mass typically increases from the
first to the fifth egg (Fig. 5). We do not know the function
(the adaptive value) of this increase, but we could speculate
that it mitigates competitive conditions for the last-hatching
chicks. We also notice that the increase in egg mass over
the laying sequence appears to be steeper for the ‘enhanced’
group (Fig. 5). We therefore test whether the treatment
interacts with laying order in its effect on egg mass [R-code
using the lme4 package: lmer(egg_mass∼treatment*
laying_order+(1|female_ID))], and indeed the interaction
term seems significant (P=0.042). This specification of the
model has been widely used, but it is in fact incorrect and
may yield 30% false-positive outcomes for the treatment
by laying order interaction term (Schielzeth & Forstmeier,
2009). So where is the mistake?
Fig. 5. Pseudoreplication at the individual level: different
slopes. Fictional data on egg mass from Fig. 4, but this time
plotted against the order in which the five eggs from each
female were laid. Egg mass appears to increase more steeply in
the ‘enhanced’ group (compared to the ‘reduced’ group), but
statistical testing requires specification of female-specific slopes
(six ‘enhanced’ versus six ‘reduced’ slopes).
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
1954 W. Forstmeier and others
Note that we have shifted our interest from testing for a
treatment main effect (Fig. 4) to testing for a treatment by
laying-order interaction, i.e. a difference in slopes between
treatments (Fig. 5). The former requires modelling of
individual-specific intercepts [R-code lme4: (1|female_ID)],
to acknowledge correctly that we are actually testing only
six versus six intercepts. The latter, testing for a difference
in slopes, requires modelling of individual-specific slopes
[R-code lme4: (laying_order|female_ID)], to acknowledge
correctly that we are actually testing only six versus six
slopes. Again, the mistake is to think that you could use
all 30 eggs from ‘enhanced’ for calculating the ‘enhanced’
slope (and the other 30 for the ‘reduced’ slope) as
if they were fully independent of each other. In fact,
each female has its own slope that you could calculate
and then do a t-test of six versus six slope estimates,
and, given the small sample size, this will rarely reach
significance. Indeed, when the full model is specified
correctly [R-code using lme4: lmer(egg_mass∼treatment*
laying_order+(laying_order|female_ID))] the P-value for
the treatment by laying-order interaction is clearly
non-significant (P=0.22).
Again, make it clear to yourself which hypothesis you are
testing (a difference in slopes), and what the independent
units are (female-specific slopes) for testing that hypothesis.
As in the earlier example (Fig. 4) where you had the option of
eliminating pseudoreplication by calculating mean egg mass
of each female, you here have the same option of calculating
a slope for each female and doing the testing on the derived
statistic. Yet in reality, not every female will lay exactly five
eggs, so the derived statistic (mean or slope) will vary in its
precision among females (most uncertain for females that lay
only two eggs). In that case, the method of choice is to fit
a model with the appropriate random-effects structure that
accounts for all non-independence in the data that is relevant
for hypothesis testing.
(b)Pseudoreplication due to genetic relatedness
In the previous section we focussed on repeated measures
within individuals that were obviously not independent of
each other. In this section we consider only one measurement
per individual, but focus on how individuals can be
non-independent of each other because of kinship (Hadfield
et al., 2010). This is most often a problem in observational
studies, and less so in experimental studies, because the latter
allow us to e.g. split up a pair of brothers into the two
treatment groups (making the confounding structure in the
data independent of the predictor of interest).
When working with animals that you breed yourself in
captivity, you rapidly begin to realize that individuals are not
independent of each other. Not surprisingly, pairs of siblings
tend to be more similar to each other when compared
to less-related individuals (Burley & Bartels, 1990). For
instance, you may find that across all individuals there
is a significant positive correlation (r=0.68, P=0.021)
between two phenotypic traits, here male body mass and
male courtship rate (Fig. 6).
Fig. 6. Pseudoreplication at the family level. Fictional data
showing the rate of male courtship (N=11 males) as a function
of male body mass (r=0.68, P=0.021). Note that significance
may be overestimated if the three males on the right come from
the same family that happens to carry alleles for high mass and
high courtship rate.
However, the statistical significance of that relationship
may have been overestimated if the 11 data points are
non-independent. Both phenotypic traits (mass and courtship
rate) are partly genetically inherited, and it might be the
case that the three males with the highest body mass and
highest courtship rate are three brothers. If this is the case,
we need to fit family identity as a random effect into the
model [R-code lme4: lmer(courtship∼mass+(1|family_ID))]
and let’s say that the other eight males come from eight
independent families. This changes the P-value for the effect
of mass on courtship rate from P=0.021 to 0.084. This is
still a trend, but one that is more likely to have come about
by chance, and so we should not have too much confidence
that we will observe the same pattern in other males. Some
researchers may argue that correcting for pseudoreplication
is misguided in this case since it could mask a real relationship
among these individuals. If the goal were only to describe the
pattern in this population of 11 males, then we would agree.
However, if we wanted to predict the likely pattern in other
populations or in the species in general, we want to avoid
being misled by chance associations driven by relatedness
among individuals in our sample (in which case P=0.084 is
a more realistic estimate of the probability that the observed
pattern arose by chance alone).
In the above example, relatedness may lead to inflated
significance because both the dependent variable (courtship
rate) and the predictor (body mass) are partly genetically
inherited. This confounding effect gets even larger when
the predictor is inherited entirely, as the next example
will show. Let’s say we study male courtship rate in zebra
finches (Taeniopygia guttata) in relation to alleles at genes that
are good candidates for affecting courtship rate (so-called
phenotype–genotype associations). We find that a particular
allele (ESR1_10) at the oestrogen receptor locus (ESR1)
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
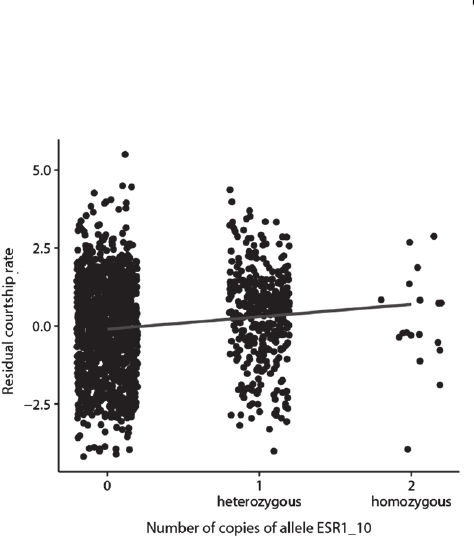
Avoiding false-positive findings 1955
Fig. 7. Average courtship rate (corrected for
between-generation differences) of 1556 male zebra finches as
a function of the number of ESR1_10 alleles they carry. Jitter
was added to the x-axis in order to increase the visibility of data
points. The regression line (y=0.4x−0.1) indicates by how
much courtship rate increases per gene copy, explaining 1.4%
of the total variance.
was associated with increased male courtship (Forstmeier,
Mueller & Kempenaers, 2010).
If we assume statistical independence of the 1556 males
from our captive zebra finch population (comprising
seven generations of birds) and model courtship rate as
a function of the genotype as illustrated in Fig. 7 we
obtain a remarkably significant P-value of P<10–15
[R-code: glm(courtship∼ESR1_10_copies)]. If we fit family
identity as a random effect into this model [R-code lme4:
lmer(courtship∼ESR1_10_copies+(1|family_ID))], where
family_ID groups together all full-brothers that come from
the same parents within each generation, the model yields
P<10–9, still a remarkably significant effect. However,
this coding of families accounts for non-independence of
brothers within generations, but ignores that both alleles
and behaviour are passed on longitudinally from father
to sons, making the corresponding family groups similar
in terms of alleles and behaviour. Hence, to account
for all genetic relationships, we need to fit the entire
seven-generations pedigree as a random effect into a
so-called ‘animal model’ [R-code for the pedigreemm
package (Vazquez et al., 2010; Bates & Vazquez, 2014):
pedigreemm(courtship∼ESR1_10_copies+(1|animal))].
Soberingly, this analysis yields P=0.015 for the effect of
this allele on courtship rate. Maybe this effect is still real,
but the exceedingly high confidence we had from the initial
analyses was unwarranted. As an aside, another lesson here
is that P-values are not useful for indicating strength of
effect. P-values here varied dramatically based on effective
sample size, but any effect of these alleles on courtship rate
was always weak as indicated by the r2value (0.014).
What is most disturbing about the problem of
non-independence driven by relatedness is that the problem
would be much harder to fix when studying a population
of animals in the wild (if relatedness is also high there). If
pedigree information is unavailable, we could genotype each
individual at say 10000 single nucleotide polymorphism
(SNP) markers, run a principal component analysis over
these data, and fit the principal components as fixed effects
to control for patterns of relatedness in this wild population
(Price et al., 2006). If we went through this trouble, we might
discover that a promising-looking phenotype–genotype
association has entirely evaporated in terms of its statistical
significance. This would no doubt be very disappointing, but
learning that this pattern is unreliable should save us from
wasting money on other expensive follow-up projects that
would have been built on an unreliable foundation.
(c)Pseudoreplication due to spatial and temporal autocorrelation
Above we have considered the case that individuals yield
non-independent data points because they are influenced by
the same genetic effects (shared alleles). Besides the effects of
genetics, individual phenotypes are influenced by numerous
environmental factors. Such environmental factors typically
vary in space and time, so individuals that are close to each
other in space and time will often share the same effects and
hence will be more similar to each other (non-independent).
Any such shared influences will create spatial and temporal
autocorrelation in the data. To examine those, you may want
to consider sorting your data either by time or in space and
checking the extent to which the preceding measurement
predicts the following one (this can be easily done by copying
your y-variable column into a new column but shifted down
by one row, and then quantifying the correlation between
the columns). However, remember that other confounding
factors (like repeated measures on the same individual) can
induce the illusion of temporal autocorrelation if subsequent
measures are typically from the same individual. If you
find autocorrelation in your data, you may want to consult
some of these references for methods of accounting for
non-independence (Cliff & Ord, 1981; Valcu & Kempenaers,
2010; Dale & Fortin, 2014).
When designing an experiment, it is always good
to consider the possibility of temporal and spatial
non-independence, because this will remind you to allocate
your treatments carefully (blocking is often better than
randomizing). You obviously should not locate all your
nutrient-enrichment plots in one field and your control plots
in another field. For the same reasons, you should not put the
cages holding the ‘enhanced’ males close to the room win-
dow and the ‘reduced’ males on the corridor side. Although
it may not be the case that daylight will affect egg size, this
will nevertheless put you into a situation where you lose
all power to detect a treatment effect, because you need to
control for the distance to the window as a covariate, which
will be strongly collinear with your treatment and hence the
two potential effects will be difficult to tease apart. Likewise,
putting all males of one treatment category into one aviary
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1956 W. Forstmeier and others
and all males of the other category into another aviary,
will leave you with N=1versus N =1, because you need to
control for the effect of aviary identity as a random effect.
Temporal non-independence of events also often leads to
the phenomenon of data being ‘overdispersed’. This means
that extreme values are more frequently observed than would
be expected from chance alone. For instance, subsequent
eggs are often more alike, not only in size (Fig. 5) but also
with regard to paternity. Studies of female promiscuity that
measure the proportion of eggs within a clutch that is sired by
an extra-pair male typically observe that extreme outcomes
(0 or 100% of extra-pair young) are more frequent than
expected by chance (Neuh¨
auser, Forstmeier & Bretz, 2001).
Such overdispersion in the data needs to be accounted for;
otherwise this will again result in anti-conservative P-values.
For this, the sequence of eggs does not have to be known, and
using clutch identity (unique code for every clutch) as a ran-
dom effect will typically help solve the issue. Hence, overdis-
persion may be easy to account for in cases where we under-
stand the source of non-independence (here clutch identity).
Another way of correcting for overdispersion is through the
use of quasi-likelihood models (Wedderburn, 1974).
A final mistake related to overdispersion that one can
sometimes observe is that measurements of latency (e.g. the
number of seconds that a bird takes to return to its nest) are
modelled as a Poisson trait (for count data). Rather obviously,
subsequent seconds are not statistically independent events,
and hence the data will typically be strongly overdispersed.
Also, when you compare different options, you will see that
P-values will strongly depend on whether you counted the
time in hours, minutes, seconds, or even milliseconds. An
example is shown in Fig. 8, where the latency to return to
the nest is analysed as a function of a measure of the bird’s
exploratory behaviour.
What is striking about this example (adopted and modified
from an unpublished study) is that a remarkably shallow
Fig. 8. Fictional data on the latency of 70 birds to return to
their nest after disturbance as a function of their exploratory
behaviour recorded in another test. The line is based on ordinary
least-squares regression on untransformed data.
regression line compared to the total range of variation is
accompanied by a P-value of <10–7.Inthisexample,each
minute gets evaluated as an independent event, and a few
very high values on the left side (up to 559 min) cause all
the apparent statistical significance. If the same data are
modelled (again wrongly) as counts of hours (ranging from
0 to 9), we arrive at P=0.078 although the data look
almost the same [R-code: glm(hours∼exploration, family =
‘poisson’)]. Latencies can often be transformed into a nice
normal distribution by taking the logarithm of the number of
minutes or seconds [here the choice does not really matter,
but note that ln(0) is not defined]. Modelling the natural
logarithm of the number of seconds as a Gaussian trait, we
obtain P=0.26, which fits the impression of a weak trend
given by Fig. 8 [R-code: glm(lnsec∼exploration)].
(d)Pseudoreplication renders P-curve analysis invalid
Simonsohn et al. (2014) recently suggested that one could
examine the subset of all published P-values that reach
significance (P<0.05) in order to find out whether a true
effect exists or not (referred to as ‘P-curve analysis’). In
the presence of a true effect, P-values between 0 and 0.01
should be more frequent than P-values between 0.04 and
0.05, i.e. there should be an excess of highly significant
P-values. Hence, right-skewed P-curves have been suggested
to be evidential for true effects (Jager & Leek, 2014;
Simonsohn et al., 2014). However, if genetic relatedness and
temporal or spatial autocorrelation are ubiquitous in real
data sets, and often lead to pseudoreplication that remains
unaccounted for (e.g. because relatedness is unknown),
then such pseudoreplication will cause an excess of overly
significant P-values that renders invalid such interpretation
of right-skewed P-curves as evidence for a true effect. The
assumptions of P-curve analyses are almost certainly seriously
violated in multiple other ways as well and so unfortunately
this briefly promising method for assessing biased reporting
cannot fulfil expectations (Gelman & O’Rourke, 2014;
Bishop & Thompson, 2016; Bruns & Ioannidis, 2016).
(4) Errors in interpretation of patterns
(a)Overinterpretation of apparent differences
Humans have a tendency readily to recognize patterns, even
where none exist. This may be partly enhanced by binary
thinking in terms of effect (if P<0.05) versus no effect (if
P>0.05). Accordingly, one can also find this as a common
mistake in the scientific literature: the title of a paper may
claim that ‘sexes differ in their response to a treatment’, but
the study only found that an effect was significant in males
and non-significant in females. This does not mean per se that
the sexes are actually different. Whether the difference itself
reaches significance has to be assessed by testing the sex by
treatment effect interaction term (Gelman & Stern, 2006).
Only if the P-value for that interaction passes the threshold of
P<0.05 can we conclude that the sexes differ significantly in
that treatment effect on whatever the dependent variable was.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.
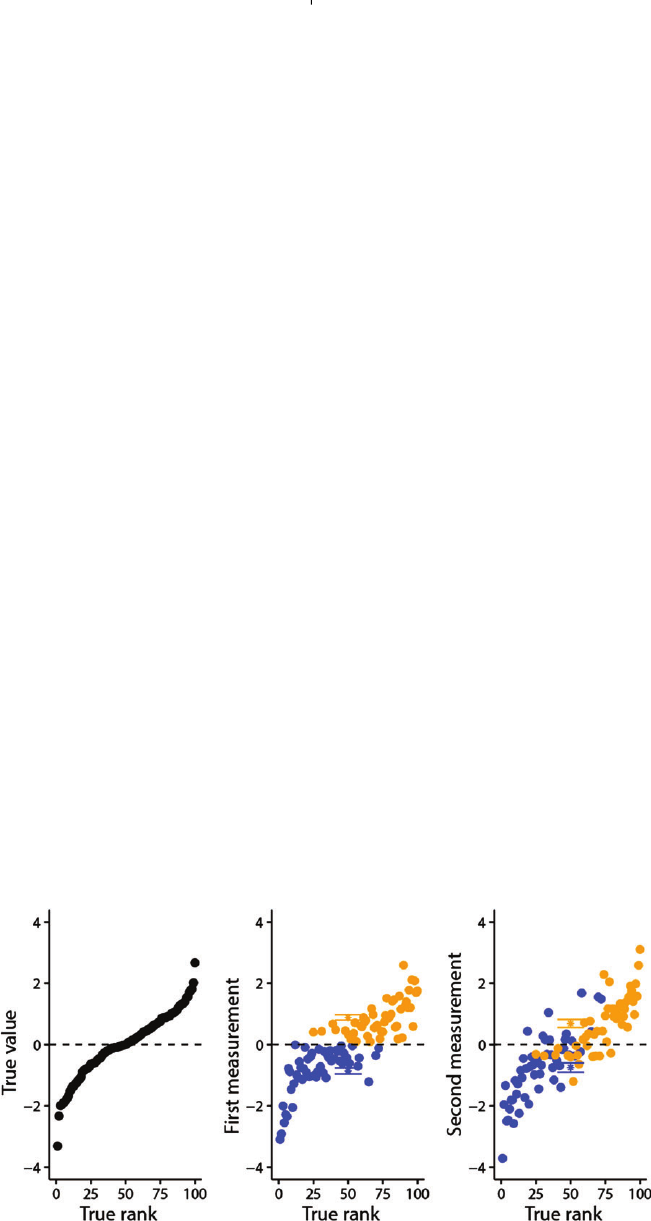
Avoiding false-positive findings 1957
Likewise, there is often a tendency to jump prematurely to
the conclusion that the findings of two studies are different.
Are they significantly different? Not very intuitively, a
parameter estimate from a replication study has a probability
of about one in six (16.6%) to fall outside the 95% confidence
interval of the estimate from the initial study (Cumming,
Williams & Fidler, 2004). This may come as a surprise,
because one may think that the 95% confidence interval
should contain 95% of the replication results (Cumming
et al., 2004). However, the 95% confidence interval is defined
in a way that it contains the (typically unknown) true value
of the parameter with a probability of 95%. And while the
true value is a fixed number, both the estimate from the first
study and the estimate from the second study come with
uncertainty. This means that either the first or the second
study could have yielded an unlikely (unusually extreme)
outcome, so the probability that they agree is lower than the
probability of one estimate agreeing with the fixed true value.
Again, a formal test for the study by effect interaction term
will inform you correctly about the probability of obtaining
such a difference between two studies by chance alone. So
make sure you are not over-interpreting a difference that
may not be real.
(b)Misinterpretation of measurement error
There is one final statistical phenomenon that we would
like to highlight: ‘regression to the mean’ (Barnett, van der
Pols & Dobson, 2005). Although it is not related to any of
the examples above, it is a sufficiently common trap and
has led to errors in a wide range of scientific disciplines
(Kelly & Price, 2005; Danchin, Wajnberg & Wagner, 2014).
Moreover, since the regression to the mean will consistently
produce a spurious but often significant effect, and since we
typically publish when encountering something significant,
one can readily find erroneous interpretations of this artefact
in the literature.
‘Regression to the mean’ is a phenomenon that results from
measurement error. Say we measure a group of individuals
once (e.g. we measure, with some error, the attractiveness of
individuals), and then divide them into two groups according
to the first measurement, namely those that lie above the
mean (attractive half) and those that lie below the mean
(unattractive half). If we then measure the individuals from
the two groups a second time, we can predict that the two
group averages will deviate less from the population mean
than in the first measure (hence ‘regression toward the mean’;
i.e. the attractive group will become less attractive, while the
unattractive group will become more attractive). Figure 9
illustrates the origin of this effect.
Regression to the mean leads to an apparent systematic
change in the phenotype of the individuals (on average, the
orange dots in Fig. 9 decreased and the blue dots increased
their trait values from first to second measurement). This
change has often been misinterpreted as resulting from an
experimental treatment that was also applied between the
first and the second measurement. When we know the
expected amount of measurement error that is inherent to
each measure (i.e. 1 −repeatability), we can make predictions
about the expected magnitude of regression toward the mean,
and we can test whether the experimental treatment had any
additional effects beyond this statistical artefact (Barnett et al.,
2005). However, in practice one should avoid such situations
whenever possible (Danchin et al., 2014). Hence, the rule
Fig. 9. The panel on the left shows the true trait values of 100 individuals sorted by their rank in trait values. In practice, such
true trait values are unknown, and we can only measure trait values with some measurement error (central panel). If we then assign
individuals into categories (‘below average’ in blue and ‘above average’ in orange) based on our first measurement, we make some
misassignments with respect to their true ranks (e.g. some with true rank >50 get assigned to the ‘below average’ group). A second
measurement on the same individuals will again approximate the true values with equal amount of error, but most of the previously
misassigned individuals and some of the correctly assigned ones will this time fall on the other side of the population average. As
a consequence, the means for the two groups (blue and orange asterisks) will move closer together (and closer to the population
average). Also note how the standard errors around the two group means (indicated by horizontal bars) increase from the first to the
second measurement because values can now vary over a wider range (no longer restricted by the ‘definition’ of having to lie above
or below the average).
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1958 W. Forstmeier and others
should be ‘never assign individuals to different treatments
according to their phenotype!’ If you cannot come up with a
better experimental design, you should at least be aware of the
phenomenon, i.e. you should expect that the more aggressive
individuals will become less aggressive when measured again,
and that the previously preferred option in a choice test will
become less preferred next time.
(5) Cognitive biases
Somewhat surprisingly, it appears that the human brain has
not evolved to maximize the objectivity of its judgements
(Haselton, Nettle & Murray, 2005). Accordingly, psycholo-
gists have described a near-endless list of cognitive biases that
influence our perception, reasoning and memory. In Table 2
we have compiled a selection of biases which should also
have an impact on the judgements made by scientists. Of
these, we have already discussed the hindsight bias in Section
II.2c, which makes it sometimes difficult to recall whether a
hypothesis was derived from the data or whether the test was
planned a priori. We further have touched on confirmation
bias in Section II.2ewhen discussing how wishful thinking
may influence our arbitrary decisions on how to analyse
our data. Another form of confirmation bias is worth
mentioning, namely that preconceptions may influence our
observations (‘observer bias’). In other words, if an observer
expects a treatment to produce a certain measureable effect,
the observer’s measurements may be unconsciously biased
towards detecting that effect. This bias can be minimized
by ‘blinding’ observers to the hypotheses being tested or to
the treatment categories of the individuals being measured.
However, blinding is rare. For example, in a collection of 79
studies of nest-mate recognition in ants, just 29% of the stud-
ies were conducted blind. This rarity of blinding appears to
have seriously impacted observations since non-blind studies
were much less likely (21%) to report aggression among
nest-mates than blind ones (73%), leading to a twofold
overestimation of effect size (van Wilgenburg & Elgar,
2013). In 83 pairs of evolutionary biology studies matched
for type of experiment, non-blind studies had substantially
larger effect sizes than blind studies (mean ±S.E. difference
in Hedges’ g=0.55 ±0.25), and the non-blind study had
a higher effect size than its matched blinded experiment in
significantly more cases (Holman et al., 2015). Comparisons
with much larger samples lend considerable support to these
observations. In 7644 papers identified via automated text
mining (from 4511 journals in the Open Access collection
of PubMed), the proportion of significant P-values in a paper
was significantly lower in blind than in non-blind papers
(Holman et al., 2015). Among a sample of 492 papers from
the disciplines of ecology, evolution, and behaviour pub-
lished in high-impact-factor journals in 2012, 248 presented
studies ‘that could have been influenced by observer bias’.
However, only 13% of these studies appeared to have
gathered data through a blind process (Kardish et al., 2015).
If we recognize our cognitive biases as fundamental to our
nature rather than as character flaws to be ashamed of, we
can structure our scientific endeavours in ways to minimize
their effects. We blind observers not because observers are
Table 2. A collection of cognitive biases that may hinder objectivity of researchers. Names and explanations were adopted from
Wikipedia (www.wikipedia.org) and inspired by a compilation of 175 cognitive biases by Buster Benson (https://betterhumans
.coach.me/cognitive-bias-cheat-sheet-55a472476b18)
Bias Explanation
Confirmation bias The tendency to search for, interpret, favour, and recall information in a way that confirms one’s
pre-existing beliefs or hypotheses, while giving disproportionately less consideration to alternative
possibilities
Selective perception The tendency not to notice and more quickly forget stimuli that cause emotional discomfort and
contradict our prior beliefs
Bias blind spot The cognitive bias of recognizing the impact of biases on the judgement of others, while failing to
see the impact of biases on one’s own judgment
Confabulation The production of fabricated, distorted or misinterpreted memories about oneself or the world,
without the conscious intention to deceive. This may help us in making sense of what we see
Clustering illusion The tendency to erroneously consider the inevitable ‘streaks’ or ‘clusters’ arising in small samples
from random distributions to be non-random
Illusion of validity A cognitive bias in which a person overestimates his or her ability to interpret and predict accurately
the outcome when analysing a set of data, in particular when the data analysed show a very
consistent pattern – that is, when the data ‘tell’ a coherent story
Belief bias The tendency to judge the strength of arguments based on the plausibility of their conclusion rather
than how strongly they support that conclusion. This is an error in reasoning, such as accepting
an invalid argument because it supports a conclusion that is plausible
Hindsight bias The inclination, after an event has occurred, to see the event as having been predictable, despite
there having been little or no objective basis for predicting it
Overconfidence effect A bias in which a person’s subjective confidence in his or her judgments is reliably greater than the
objective accuracy of those judgments
Appeal to novelty A fallacy in which one prematurely claims that an idea or proposal is correct or superior, exclusively
because it is new and modern
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1959
dishonest, but because we know that we all have a tendency
to see what we expect to see. We preregister analysis plans
only because we know that even people with the purest
conscious motives are more likely to choose the method that
produces the story that they most believe.
III. SOLUTIONS
(1) Need for replication and rigorous assessment of
context dependence
In face of the problems that we have outlined above
(multiple testing including researcher degrees of freedom,
pseudoreplication, selective reporting, and HARKing), it is
clear that we should be fastidiously sceptical consumers of
published scientific results. Given publication bias in favour
of positive results and given the rather soft criteria for
reaching significance, it is currently possible to find positive
‘evidence’ in the scientific literature for almost any possible
phenomenon. If you recognize this, you presumably will also
recognize that the extreme pursuit of novelty (by high-impact
journals) and researchers’ pursuit of impact (imposed by
employers and funding agencies) will contribute to an
ever-increasing body of false-positive claims that hampers
scientific progress (Ware & Munafo, 2015).
Hence, we need to promote scepticism of spectacular
but highly unlikely claims and turn attention to sorting out
all the false-positives from the true positives. How can this
be done? As we have seen, we can use a variety of clues
to identify findings that are less likely to be true, but what
we really need is a rigorous method of assessment in the
form of well-controlled and standardized attempts to closely
replicate previous studies (Nakagawa & Parker, 2015).
Further, institutions need to favour the publication of the
results of replication independent of their outcome. To see
the utility of unbiased replication attempts, we can look to the
recent coordinated effort to replicate 100 findings published
in top journals in the field of psychology (Open Science
Collaboration, 2015). This large-scale initiative found that
about 40% of the findings appear to hold up (reminding us
of Fig. 1F), while most of the remainder were contradicted
or not supported (but see Etz & Vandekerckhove, 2016).
The high statistical power of most of the replications lend
strength to the conclusion that many of the original studies
were either the result of error or were more dependent on
subtle differences in context than had been assumed.
(a)Obstacles to replication
In face of the request for novelty, researchers often address
a commonly asked question in a new species or with new
methods. However, in such quasi-replications (Palmer, 2000),
if results differ from the original, we are left to speculate
about why the outcomes differed and we will rarely be able
to identify the true reason for the difference in outcomes,
because quasi-replicates differ in so many aspects. Close
replication, by contrast, minimizes differences among studies
and facilitates the identification of plausible hypotheses to
explain divergent results. Unfortunately, close replications
are rare in many disciplines (Palmer, 2000; Kelly, 2006;
Drotar, 2010; Nakagawa & Parker, 2015). There are two
interrelated explanations for this. First, many researchers
have not yet come to appreciate the important role of
replication in developing robust scientific inference, and
second, the institutions that influence scientists’ choices do
not reward close replication (Nosek, Spies & Motyl, 2012).
Funding agencies and journal editors focus on novelty.
This is particularly hard to justify on the part of funders
since failing to invest in replication means failing to seek
robust answers to questions they already have made a
commitment to answering. If the answer truly was worth
paying for, then the replication should also be worth paying
for (Nakagawa & Parker, 2015). Promoting the funding of
replication studies would be relatively straightforward. Most
obviously, agencies could set aside funds for important and
well-justified replications. Agencies could also incentivize
replication by preferentially funding novel studies when
those studies rest on well-replicated foundations (Parker,
2013). They could also preferentially fund researchers whose
prior work has often been successfully replicated.
In the case of journals, pursuit of novelty may be harder
to curb, but there are paths to reducing the tyranny
of this pursuit. Journals seek novelty in part because of
the competition for impact factors. Studies which report
surprising (i.e. unlikely) findings are often highly cited and
thus contribute to the stature of the journal. Thus, ‘the more
surprising, the better’. This effect may be exacerbated by
the for-profit publishing industry. Fortunately, replications
can also be heavily cited. Recent attempts to replicate classic
studies in psychology have received citations at a much higher
rate than the average study in the journal in question. In
2014, the journal Social Psychology published an issue (issue 3,
May) devoted to replications of previously published studies.
As of 15 March 2016, the mean number of citations from
those 15 replications (of 15 different earlier studies) was 7.1
(median =4, with no articles having gone uncited). By the
same date in 2016, the average article from the previous
two issues (1 and 2 from 2014), including no replications,
had received 1.2 citations (median =1, with 5 of 12 articles
remaining uncited). Of course this may be in part due
to the current novelty of replication research (ironically).
However, robust, well-conducted replications of important
work will presumably attract considerable attention in the
future, especially as awareness grows about the importance
of replication in assessing validity of prior work. We expect
that as more journals explicitly invite replications (as some
are beginning to do), more researchers will come to recognize
their utility, and thus researchers will more often seek to cite
replications because of the strong inferences they facilitate.
Even without institutional obstacles, there are important
social obstacles to navigate. An attempt to replicate closely
someone else’s finding may be perceived as a personal attack
on the original researcher. The very act of replication implies
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1960 W. Forstmeier and others
insufficient confidence in the original findings, and in cases
of failure to confirm the original finding, the researcher
who published the original study may fear for his or her
reputation (although we expect this phenomenon to be less
common as replications become more frequent and failure
to get confirmed is recognized as normal). Journals almost
invariably ask the author of the previous study to review the
replication manuscript since he or she is the expert and is
directly concerned. This reviewer will often be predisposed to
be negative, sometimes trying to save his or her own results by
questioning the quality of the replication (e.g. making the case
that an incompetent person will often fail to get the correct
result; Bissell, 2013). Thus it may often be difficult to publish
a replication, especially when that replication contradicts
earlier work, and this is a strong disincentive to replicate.
(b)Overcoming the obstacles
A partial solution to this dilemma could come from
researchers replicating their own findings. This eliminates the
quality issue as well as issues related to dissimilarity in materi-
als or methods. A simple and cheap way of getting this started
was suggested by one of our colleagues, Jarrod Hadfield (Had-
field, 2015). He proposed that researchers running long-term
studies could publish addenda to their previous publications,
declaring in a one-page publication that their original finding
did or did not hold up in the data of the following years (after
the publication), and comparing the effect sizes between the
original data and the newer data. This would be a quick
way of producing another publication, and it would be enor-
mously helpful for the scientific field. This may also relax the
feeling of stigma when something does not hold up to future
evaluation. Admitting a failure to replicate could actually
be perceived as a signal of a researcher’s integrity and be
praised as a contribution to the scientific community. For
grant applications, funding agencies could even specifically
ask for visible signs of such integrity rather than exclusively
focussing on metrics of productivity and impact.
Apart from publishing addenda, how should we go about
conducting replication studies? First of all, the study should
be preregistered (see Section III.3) in order to solve two issues:
(i) preregistration of analysis plans takes out any researcher
degrees of freedom that would risk biasing the observed
effect size in the direction desired by the researcher (either
confirmation of the previous finding or clear refutation of it),
and (ii) preregistered studies that do not make it to the stage
of publication will still be accessible at a public repository,
documenting the attempt and hopefully also the reason for
failure. Besides being preregistered, replication studies should
attempt to match the previous study as closely as possible in
methods and materials and should aim for a larger sample
size. Incentives to preregister should be particularly strong for
replication studies since a study and analysis plan effectively
already exist, and preregistration will clearly signal to
reviewers and editors that the presentation of results has not
been altered in an attempt to achieve a particular outcome.
A particularly compelling option for publishing replica-
tions is through a process known as registered reports, a
format advocated by Chris Chambers (2013). Registered
reports involve preregistration, but with a registered report,
the researcher first submits the study and analysis plan to a
journal for review, potential revision of methods plans, and
preliminary acceptance prior to conducting the study. Thus
a proposed replication could be reviewed, and if judged mer-
itorious, provisionally accepted independent of results. This
would give the scientist who published the original study the
opportunity to recommend changes to methods of the repli-
cation before it was initiated, and thus increase the quality
of the replication while also reducing the opportunity for a
critique, spurious or genuine, of the quality of replication.
(c)Interpretation of differences in findings
When a replication fails to confirm the original result,
this is often interpreted as context dependence (e.g. ‘this
was a wetter year’, ‘this location contained more conifers’,
or ‘these animals were raised on a higher-protein diet’).
After all, we know that ecology and behaviour are highly
complex and we expect variability. However, in this situation
context dependence is simply an untested post-hoc hypothesis.
We cannot claim that divergent results stem from context
dependence without explicit testing with new data. As
explained in Section II.4a, it may be that the difference
in effect sizes observed in the two studies is no larger than
what one would expect from chance alone (sampling noise).
Meta-analysts (researchers who summarize effect sizes across
numerous studies) are very familiar with this idea, and
they quantify the extent of disagreement between studies as
‘heterogeneity’ in effect sizes. Since each observed effect size
is accompanied by a measure of uncertainty (for instance a
standard error or 95% confidence interval, both of which
depend on sample size), one can test statistically whether
there is significant heterogeneity in effect sizes. Such tests
are frequently significant, but does this observation provide
strong evidence for context dependence? Unfortunately not!
For tests of heterogeneity to provide insight into context
dependence, we need to minimize other sources of hetero-
geneity. One source of heterogeneity is publication bias in
favour of stronger effects, often facilitated by use of researcher
degrees of freedom to reach significance more often than
expected by chance. In an extreme example, two studies
could yield non-significant trends in opposite directions (a
small difference due to sampling noise), but those differences
could get amplified by ‘researcher degrees of freedom’ and
selective reporting, because each team of researchers (in good
faith of doing the right thing) feels obliged to emphasize the
outcome of their study by selecting the conditions where
trends approach or reach statistical significance. When pub-
lication bias is common, effect sizes often vary as a function
of sample size because as sample size declines, deriving a sta-
tistically significant effect requires larger effect sizes (Gelman
& Weakliem, 2009). This too can generate estimates of signif-
icant heterogeneity among studies in the absence of context
dependence. Thus an important step towards using tests for
heterogeneity as reasonable indicators of context dependence
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1961
is minimizing sources of bias, for instance through preregis-
tered studies. Heterogeneity can also arise due to differences
in study methods that are unrelated to hypothesized contex-
tual differences. If we are truly interested in context depen-
dence and its sources, then we should design replication stud-
ies explicitly to investigate context dependence. This means
systematically evaluating environmental variables that we
hypothesize may be driving context dependence using ‘repli-
cation batteries’, in which conditions hypothesized to drive
differences in results are manipulated while attempting to
hold other variables constant (Kelly, 2006). Post-hoc hypothe-
ses about context dependence are valid, but they remain
nothing more than hypotheses before studies have been
designed and implemented specifically to evaluate them.
(d)Is the world more complex or less complex than we think?
The tendency of many researchers to interpret all apparent
differences as important (see Section II.4a) and to attribute
these differences to context dependence by default has a
remarkable effect on the development of our world view.
Such world views become increasingly complex, emphasizing
the importance of context dependence which results in
hard-to-interpret interaction terms. By contrast, the sceptic
who emphasizes the large amount of sampling noise in most
sets of data which further may get inflated by researcher
flexibility, may often hold a nihilistic world view where most
or all effects are spurious. This is an interesting debate which
we only will be able to resolve by promoting transparency
and by replicating studies as rigorously as possible.
(2) Collecting evidence for the null and the
elimination of zombie hypotheses
Given the many false-positive findings in the literature, it is
the foremost goal of rigorous replication studies to validate or
reject previous findings. This will often mean that evidence
we collect contradicts the biological hypothesis. But can this
evidence lead to rejection of our biological hypothesis? After
all, we can never rule out the possibility that this hypothesis
might apply in some other context and that some unknown,
uncontrolled difference between our replication and the orig-
inal study led to our failure to replicate. In practice, however,
we think we can at least approximate rejection of the original
hypothesis. The key is multiple replications and some sort of
meta-analytical framework. For instance, when reporting a
result, we may calculate a 95% confidence interval around
our estimate, and then highlight that our study shows an
effect that is significantly smaller than this or that quantity.
We can calculate a weighted average of the two results, and
as we accumulate more independent replications, we gain
confidence in our average, and if this average approximates
zero, we become more confident that the hypothesis in ques-
tion is wrong, or at least of extremely narrow applicability
(Seguin & Forstmeier, 2012; Nakagawa & Parker, 2015).
Another excellent tool for drawing conclusions from a
series of replications is Bayesian statistics which allow us to
pitch two competing hypotheses (H0 and H1) against each
other and to evaluate which of them is better supported by
the data (Dienes, 2016). The Bayes factor (BF) can be used to
quantify how many times more likely the observed data are
if the hypothesis H1 is true rather than if H0 is true. A Bayes
factor of 1 means that the data are equally likely under both
hypotheses, so the data contribute no information towards
resolving the question of which hypothesis may be true.
Depending a bit on conventions, a Bayes factor larger than
3 is typically considered as substantial or moderate evidence
for H1, while a Bayes factor smaller than 1/3 represents
substantial or moderate evidence for the null hypothesis H0,
and these thresholds are approximately comparable to the
threshold of α=0.05 in significance testing (Dienes, 2016).
For the purpose of contrasting a specified hypothesis H1
(usually defined by the effect size reported in a previous study
that we want to replicate) against the null hypothesis H0,
Bayesian statistics hence allow us to assess whether the new
data from the replication study support either H1 or H0.
Of course Bayesian statistics do not guarantee that we will
resolve the issue (if 1/3 <BF <3). This lack of clarity may
come, for instance, if there is some truth to the published
finding, but the true effect size lies half-way between the one
that is published and the null (overestimation by a factor of
about two would make BF =1 a likely outcome).
In fact it is well known that published effect sizes tend
to be overestimates of true effects. It is frequently observed
that the first publications on a given topic report larger effect
sizes than later follow-up studies, leading to a general decline
in effect sizes over time (Jennions & Møller, 2002; Barto
& Rillig, 2012; Koricheva, Jennions & Lau, 2013). Equally
problematic is the observation that sample size and effect size
are typically negatively correlated in meta-analyses meaning
that only the studies with the largest sample size will yield
trustworthy effect-size estimates (Levine, Asada & Carpenter,
2009). Meta-analysts examine this phenomenon in so-called
funnel plots where effect sizes of studies are plotted in
relation to sample size (Pillemer & Light, 1984). Funnel
plots can be used to detect publication bias in meta-analyses
(Egger et al., 1997), but such tests should be regarded with
caution (Ioannidis & Trikalinos, 2007). Modest amounts of
publication bias, that may not be apparent from funnel plots,
can render the conclusion of a meta-analysis invalid (Scargle,
1999; Ferguson & Heene, 2012).
Besides many true effects being overestimated, it is
conceivable that some fields of research could exist for
extended periods of time in the complete absence of any
true effects of theoretical relevance. More than 40 years ago,
Greenwald (1975) complained that systematic ‘‘prejudice
against the null hypothesis’’ can lead to a dysfunctional system
of research and publication that allows untrue hypotheses
to persist undefeated, similar to what was recently proposed
for a substantial body of sexual selection research (Prum,
2010). More recently, and along the same lines, Ferguson &
Heene (2012) argued that our aversion to the null will result
in ‘a vast graveyard of undead theories’. We feel that it is
high time to overcome this aversion, and to remind ourselves
that researchers are supposed to be the unbiased referee in
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1962 W. Forstmeier and others
a game between H1 and H0, and that falsification of untrue
hypotheses lies at the heart of making scientific progress.
(3) Making science more objective
The weight of all these obstacles to objective science could
leave you frustrated and depressed. Scientists are often
unaware of many of the ways that science is not maximally
objective and the ways that scientific practices often serve
only the short-term interest of the scientist who is under
pressure to produce success stories in order to attract the
next grant. Fortunately, unscientific practices like fishing for
significance, HARKing, and many others are on their way
out, as a growing community becomes more aware of the
risks and better able to recognize the signs of bad practices.
Further, as discussions of these issues become more common,
we hope that more researchers will realize that sticking to
protocols that promote objectivity is the best strategy in
the long run. For instance, anyone who conducts a series
of studies in the same system will benefit from drawing
robust conclusions in their foundational work. Maximizing
objectivity, for instance through preregistering your studies,
should also bolster your reputation. Then, if one of your
findings is contradicted by later work you need not worry
about your reputation, and maybe more important, you can
feel good about the fact that you interpreted the data in the
most objective way. In this context, a recent commentary by
Markowetz (2015) is an interesting read. He lists five selfish
reasons why you should work reproducibly.
We have already touched on a couple of promising ways to
make science more reliable, and we add more details below.
(a)Why should I preregister my next study?
As noted above, preregistration of study plans solves the issues
of (i) HARKing, because you registered your hypotheses in
advance, (ii) researcher degrees of freedom, because you
registered your data-analysis strategy in advance, and (iii)
publication bias, because the added rigour makes nearly
every finding worth publishing (or otherwise the reason for
failure to publish gets documented in a public space). What
might hold you back from preregistering your next study?
1 I have checked the requirements, and it looks like a
substantial amount of work.
Preregistering your study may take you a couple of days, but
in the long run it will benefit you tremendously by forcing
you to think through your study plans very carefully. Likely
you will discover some weaknesses in your questions, study
design or analysis plan and you still have time to fix or
amend these issues before starting data collection. Also, the
preparatory work will make the data analysis easier since you
will already have a detailed plan, and it will make writing
your paper much easier, since you will already have written
your Methods section.
2 My colleagues are not preregistering their work, so why
should I?
If you are thinking of preregistering your study now, it is not
unlikely that you will be among the first in your field to do
so. Would you like to be able to say that you were among
the first to embrace this new tool that ensures objectivity?
Give it a try and you will likely discover that preregistering is
emotionally rewarding like the submission of a manuscript.
It also lends importance to your project.
3 I am worried that someone will steal my project idea.
No reason to worry. You can embargo your plans, and they
will only become publicly visible later.
4 I need more flexibility in study design.
Preregistration does not limit the freedom of the researcher;
it only documents the ideas and plans at any given time,
making the process maximally transparent. You always have
the possibility of modifying your study plans, and the exact
time of modification and reasons for modification will be
documented, probably still long before final data analysis
when researcher degrees of freedom would come into play.
The original study plan will always remain visible with its
date of registration, but making failed aspects of a plan
visible to others might also save them from repeating the
same mistake.
5 What if I make an unexpected discovery?
Having preregistered your study does not prevent you
from publishing any analysis you believe is interesting or
informative. All that preregistration does is clarify what tests
were formulated a priori and what tests were not. For instance,
you may subdivide your publication into a part that covers
the original analysis plan (the rigorous a priori testing part)
and a second part that explores the data post hoc and yields
unexpected discoveries.
If you have other questions, or want to start registering,
check out the website of the Center for Open Science
(https://cos.io/and specifically https://cos.io/prereg/). The
Center for Open Science is currently sponsoring a
‘Preregistration Challenge’ in which they will award
US$1000 to the first 1000 researchers to publish preregistered
studies. An alternative site that offers a maximally convenient
and hassle-free opportunity to preregister your study is
provided by AsPredicted (https://aspredicted.org/).
(b)Badges make good scientific practice visible
The Open Science Framework (https://osf .io/) has also
started an initiative to make good scientific practice
visible by awarding badges to studies that meet certain
criteria, currently including preregistration, the availability
of archived data, and the availability of detailed materials.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1963
Journals decide if they want to award badges to publications
that meet the criteria, and a few journals (mostly from
the field of psychology) have already started doing so.
Authors, when they submit their paper, apply for the
badge by declaring that the study complies with the
criteria. Editors or referees may check whether this is
true but they may also leave the responsibility with the
authors for making correct declarations (thereby minimizing
the additional workload for journals). There is already
evidence that awarding badges is effective in promoting
the sharing of data (Kidwell et al., 2016). Badges might
also facilitate evaluation of bias. For instance, badges could
make it possible to identify preregistered studies and thus to
compare effect sizes meta-analytically between preregistered
and non-preregistered studies. If you want to publish a
preregistered experiment, it may well make sense to contact
the editor-in-chief of the journal prior to submission to ask
whether they would be ready to give you a badge in case
of acceptance. Currently it seems that journal editors are
waiting to see whether they start receiving such requests
before deciding to join the list of journals that award badges
(see https://osf.io/tvyxz/wiki/5.%20Adoptions%20and%
20Endorsements/).
(c)Blinding during data collection and analysis
To rule out cognitive biases (Section II.5) such as the observer
effect during data collection or confirmation bias during data
analysis, it is generally a good idea to blind the researcher
(MacCoun & Perlmutter, 2015).
During data collection, blinding ourselves to treatment
groups provides an important protection against any intrinsic
subconscious biases that we may have (van Wilgenburg &
Elgar, 2013; Holman et al., 2015; MacCoun & Perlmutter,
2015). In all cases where data collection is not entirely
objective and free of observer effects, it is almost impossible
not to end up with bias. Despite the importance of observer
effects in many fields of science, strategies of observer blinding
are implemented in fewer studies than one would hope (van
Wilgenburg & Elgar, 2013; Holman et al., 2015; Kardish
et al., 2015).
During data analysis, blinding the researcher from the
effect of arbitrary decisions on statistical significance of the
hypothesis test is an important tool that ensures objectivity
(MacCoun & Perlmutter, in press). Hence, you should try
to take your choices of analysis variants before seeing their
effects on P-values. If this is not possible, you could ask
someone else to make these choices blindly (without regard
to outcome) for you, always going for the option that sounds
more reasonable based on criteria other than significance or
effect size.
(d)Objective reporting of non-registered studies
If you have not preregistered your hypotheses and analysis
plan, how can you reach comparable levels of objectivity
in your publication? First, you should explicitly distinguish
your a priori hypotheses (typically just a few, namely the
ones you designed the study for) from your exploratory data
analysis. Second, you should avoid selective reporting. For
any question or field of questions within your study, you
should attempt to consider all possibly relevant dependent
variables and predictors (see Fig. 2). If this makes the
manuscript too long, you can always put large tables of
results in an electronic supplementary file. You can then
report averaged effect sizes in the paper itself. Third, you
should avoid reporting estimates that are biased towards
significance by non-blind choice of ‘researcher degrees of
freedom’. If you cannot use blinding as described above,
you may want to consider using specification-curve analysis
(SCA) (Simonsohn et al., 2015) to go through all combinations
of analysis variants, and to average effect sizes across all
combinations. Currently, U. Simonsohn and co-workers are
planning to create an R-package that would make SCA easy
to carry out. A less-ambitious, but still noteworthy option has
been suggested by the same set of authors (Simmons, Nelson
& Simonsohn, 2012) who advocate ‘a 21 word solution’
of the problem in your Methods section. Specifically, your
Methods should say: ‘We report how we determined our
sample size, all data exclusions (if any), all manipulations,
and all measures in the study.’ This should take out a range of
researcher degrees of freedom, and should reduce the bias in
reported effects (see also http://www.researchtransparency
.org/). Note that it also covers the issue of specifying a
stopping rule for sample size.
Finally, when attempting to publish your manuscript, you
should emphasize its goal of yielding maximally objective
and unbiased estimates of effect sizes, as opposed to fishing,
HARKing and overselling. Remember that most effect sizes
are small and hence single studies usually lack the power
to detect them. Hence, any parameter estimate is worth
publishing, but only if it has been derived in an unbiased way.
(e)Concluding recommendations for funding agencies
In the future, what will scientists say when they look back on
our current organization of the scientific undertaking? We
think there are good reasons for calling this system inefficient
and wasteful.
The key problem is that we are expecting too much from
every single empirical study (despite knowing that most effect
sizes are small and hence power is very limited), meaning that
we set up the unrealistic expectation that each study should
yield a clear-cut conclusion by itself. This leads to a situation
where junior scientists complain when their laborious efforts
of data collection have not yielded a significant finding,
meaning that their work cannot be published. In response
senior scientists help with advice on alternative data analyses
designed to squeeze out something significant (see the torture
of data in Fig. 9). And there are more consequences of our
unrealistic expectations.
Weendupwithalargeamountofwastedeffortbecause
non-significant parameter estimates end up unpublished in
the so-called ‘file-drawer’. And, the studies that make it to
the publication stage often yield parameter estimates that
are biased upwards or are simply false positive, and these
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1964 W. Forstmeier and others
estimates therefore paint a distorted picture of the reality
that we set out to study. How absurd is a system in which
we measure an effect of interest with meticulous accuracy,
but then subject our measure to a self-imposed censorship by
only reporting it if it exceeded a certain strength?
It appears that this practice is currently ubiquitous across
a wide swathe of scientific disciplines (John et al., 2012),
despite the fact that it is fundamentally anti-scientific. We
believe that this practice could be stopped most effectively by
adequate measures from funding bodies. In an ideal world,
all measures of effect size would be reported (not necessarily
in peer-reviewed journals) and the respective raw data would
be made openly available (see also Morey et al., 2016).
Further, as preregistration became the norm, exploratory
studies would become more transparent and studies without
preregistration would come to be viewed as more provisional.
If funding bodies rewarded preregistration and unbiased
reporting practices along with intellectual merit rather than
rewarding only success in attracting citations, then we would
rapidly have a transition from a fairly dysfunctional to a
much more objective science.
Such a change in incentives would also be good news
for the above-mentioned junior scientists who would worry
about the rigour and merit of their experiments rather
than the outcome. They could move forward knowing that
well-designed tests of interesting ideas would make all their
parameter estimates valuable and publishable.
IV. CONCLUSIONS
(1) False-positive findings can easily arise when statistical
methods are applied incorrectly or when P-values
are interpreted without sufficient understanding of the
multiple-testing problem (false-positive report probability).
Incorrect P-values can arise from the over-fitting of
models or from a failure to control for pseudoreplication,
autocorrelation, or overdispersion. It is also essential to
understand the consequences of multiple testing that arise
from conditional stopping rules, from researcher flexibility
when choosing an analysis strategy post hoc, or from multiple
testing during the process of model selection.
(2) Psychological biases may also lead to false-positive
results. Researchers may be systematically biased against
the null hypothesis because positive findings are more
appealing than null results, and this bias may get amplified
by the selective interest of journals in discoveries. This
incentive is problematic because it can motivate a widespread
but unhelpful scientific practice: namely, extensive data
exploration in search for patterns that reach nominal
significance, followed by selective reporting of the most
interesting (significant) results, combined with depicting data
exploration as confirmatory testing of a priori hypotheses.
Researchers may often be unaware that such practice is
problematic, because hindsight bias can make many chance
findings appear plausible and in line with theory. Finally,
confirmation bias during data collection by non-blinded
observers may also contribute to biased results. Because of
these influences, ‘data do not speak for themselves’, but need
to be judged within the context of the procedures of data
collection, analysis, and presentation.
(3) Preregistration of hypotheses, research methods and
complete analysis plans solves many of these issues.
It prevents both HARKing and subjective choice of
analysis methods that is conditional on significance because
hypotheses and analysis plans have been specified in advance.
It also mitigates the problem of publication bias. Labelling
preregistered studies with badges may allow us to quantify
how much these biases contribute to overall effect-size
estimates.
(4) Non-preregistered studies should implement a strategy
of comprehensive and unbiased reporting that is not
conditional on significance. Researcher blinding during
analysis or specification-curve analysis are helpful techniques
that promote objectivity.
(5) Seeking novelty and discovery may be emotionally
rewarding, but in light of the currently low thresholds for
reaching nominal significance, isolated, unreplicated reports
of findings should be regarded as preliminary until confirmed
by rigorous replication studies. The term ‘evidence’ should
be used more cautiously (when there is consensus from
confirmatory tests) and the expression ‘as predicted’ should
maybe be limited to predictions that have been documented
or to those that strictly follow from theory. The crucial
second step from exploratory to confirmatory research
should be encouraged by funding bodies supporting rigorous
replication studies and by citation practices of researchers
who might want to prefer citing the rigorous confirmatory
over the initial exploratory study.
(6) A research system in which results are much more
likely to get reported if they reach statistical significance
violates scientific objectivity and is highly inefficient if our
interest lies in the quantification of effect sizes because
unbiased effect-size estimates are difficult to obtain in such
a system. Hence, researchers should recognize the value of
unbiased reporting and funding bodies should reward such
practice during the review process. The latter will have to
think of ways of assessing researcher performance in terms of
scientific rigour and integrity, because the current assessment
in terms of productivity and impact causes unwanted natural
selection pressure in favour of bad science (Smaldino &
McElreath, 2016).
V. GLOSSARY
Alpha probability = the accepted risk of drawing a
false-positive conclusion in a single statistical test, which
in most fields is arbitrarily set to α=5%.
A priori = typically before gathering data, but potentially just
before analysis or before seeing the data.
Attrition = reduction in sample size between study initiation
and data analysis, which might lead to a bias in the
results (e.g. when outliers are selectively removed).
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1965
Bayesian statistics = a statistical method that quantifies
uncertainty about parameters and models using the
laws of probability theory.
Bayes factor = a measure of predictive performance for two
competing models or hypotheses.
Beta probability = the accepted risk of drawing a
false-negative conclusion in a single statistical test (e.g.
β=20% when the statistical power equals 80%).
Bias = a systematic deviation from a true or representative
value (note how this differs from sampling noise which
causes a random deviation, with equal probability of
being too high or too low).
Bonferroni correction = adjustment of αthat allows us to
limit the risk of drawing one or more false-positive
conclusions from a whole series of tests.
Close (or exact) replication = an attempt to repeat an earlier
study with maximally similar methods.
Clustered data = non-independence of data points.
Collinearity = two or more predictors that are correlated
with each other, making it difficult to disentangle their
respective effects on the dependent variable.
Confirmatory testing = a planned test that seeks additional
evidence for a hypothesis (derived from theory or from
previous observations).
Exploratory analysis = a broad search for patterns in a given
data set with statistical or graphical methods.
False-negative finding = concluding an absence of effect
despite the opposite being true (failure to detect an
existing effect; =Type II error).
False-positive finding = concluding that an effect exists while
in fact it does not (=Type I error).
False-positive report probability (FPRP) = the probabil-
ity that a statistically significant finding is not
true (FPRP =(α(1 −π)/[α(1 −π)+(1 −β)π], with
α=Type I error probability, β=Type II error proba-
bility, π=the proportion of tested hypotheses that are
true).
File-drawer problem = studies with null-results (no signif-
icant effect) often do not get published and remain
hidden in the file-drawers of the researchers.
Fishing for significance = a range of strategies that can
be employed to increase the chance of obtaining a
statistically significant result.
HARKing = hypothesising after the results are known (see
Hindsight bias).
Heterogeneity in effect sizes = the degree to which estimates
of effects differ from one another (e.g. across a range
of studies). There is a range of statistical descriptors
of heterogeneity (Q,T2,I2) that come with different
properties and interpretation.
Hindsight bias = also known as ‘knew-it-all-along’ bias, this
is the tendency to underestimate the extent to which
outcomes were caused by noise, after these outcomes
have been observed. It can be a self-deceiving tendency
to believe, after seeing the data, that the result had
been predicted a priori when there was in fact no a priori
prediction.
Hypothesis testing = statistical analysis of data that usually
serves to reject a hypothesis.
Interaction term = two or more factors that interact with
each other rather than having effects that simply add up
(e.g. lifespan may be affected by smoking and gender,
but if the effect of smoking is larger in one sex than the
other, then the two factors interact in their effects on
lifespan).
Leverage = a data point that is an outlier with regard to the
dependent variable, which hence has a strong effect on
the position of a fitted regression line (such influential
data points are said to have high leverage).
Multiple hypotheses testing = the testing of several hypothe-
ses at once (which leads to a high probability of finding
at least one significant effect).
Outlier = a data point with an extreme value that has no
other data points nearby.
Overfitting = estimating too many parameters simultane-
ously from a limited number of data points, which
results in unreliable parameter estimates and P-values.
Pi (π) = symbol used for the proportion of hypotheses that
are in fact true.
P-hacking = another term for ‘fishing for significance’ (see
above).
P-value = probability that chance alone will produce an
effect (e.g. a correlation, a difference) as strong as or
stronger than the one observed in the data.
Post hoc = after analysis or after seeing the data.
Power (statistical power) = the probability that an existing
effect (of given size) will be detected (i.e. will reach
statistical significance) in a data set (of given size).
Power is defined as 1 −β, the probability of failing to
detect the effect.
Preregistration = submitting a document to a repository in
which one outlines the hypotheses, methods and analysis
strategies of a planned study before conducting the
study. This prevents post-hoc modification of hypotheses
(HARKing) and researcher flexibility in analysis and
thus reduces the risk of unreported multiple hypothesis
testing.
Pseudoreplication = independent data points represent
proper replicates, while non-independent data points
are referred to as pseudoreplicates. Such dependent
data points do not contribute as much information as
independent data points would. Most statistical tests
assume that data points are independent, and violating
this assumption leads to P-values that are too small.
Quasi-replication = replicating a study in a wider sense with
a different approach (e.g. different methods or different
species). In contrast to close replications, this leaves a
lot of room for interpreting differences in findings.
Researcher degrees of freedom = flexibility of the
researcher, who can choose how to analyse the
data, giving him/her the opportunity to select whatever
yields the desired outcome.
Sampling noise = random fluctuations in outcomes under
an identical data-generating process. Sampling noise
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1966 W. Forstmeier and others
arises from the fact that when sampling at random
from a population, we could have collected a different,
but equally random, sample leading to different
estimates.
Type I error = concluding that an effect exists while in fact
it does not (=false-positive finding).
Type II error = concluding an absence of effect despite the
opposite being true (failure to detect an existing effect;
=false-negative finding).
VI. ACKNOWLEDGEMENTS
We thank Martin Bulla, Malika Ihle, Bart Kempenaers,
Ulrich Knief, Holger Schielzeth and Isabel Winney for
critical comments on the manuscript, Malika Ihle and
Martin Bulla for help with figures, and Tim Coulson,
Jarrod Hadfield, Shinichi Nakagawa, and the other workshop
participants for inspiring and sometimes controversial
discussions. We also thank three referees for their very
constructive comments. We further thank the National
Science Foundation (DEB: 1548207), the Laura and John
Arnold Foundation and the Centre for Open Science
for supporting the workshop ‘Improving Inference in
Evolutionary Biology and Ecology’ which brought together
the authors and inspired the writing of this manuscript, which
was then supported by the Max Planck Society.
VII. REFERENCES
Anderson,M.S.,Martinson,B.C.&De Vries,R.(2007). Normative dissonance
in science: results from a national survey of US scientists. Journal of Empirical Research
on Human Research Ethics 2, 3–14.
Armitage,P.,McPherson,C.K.&Rowe,B.C.(
1969). Repeated significance
tests on accumulating data. Journal of the Royal Statistical Society, Series A (General) 132,
235– 244.
Baker,M.(
2016). Is there a reproducibility crisis? Nature 533, 452–454.
Barber,T.X.(
1976). Pitfalls in Human Research: Ten Pivotal Points. Pergamon Press Inc.,
New York.
Barnett,A.G.,van der Pols,J.C.&Dobson,A.J.(
2005). Regression to the
mean: what it is and how to deal with it. International Journal of Epidemiology 34,
215– 220.
Barto,E.K.&Rillig,M.C.(
2012). Dissemination biases in ecology: effect sizes
matter more than quality. Oikos 121, 228–235.
Bates,D.,Maechler,M.,Bolker,B.&Walker,S.(
2014). lme4: linear
mixed-effects models using Eigen and S4. R package, version 1.1-7.
Bates,D.&Vazquez,A.I.(
2014). pedigreemm: pedigree-based mixed-effects models.
R package, version 0.3-3, http://CRAN.R-project.org/package=pedigreemm
Accessed 15.4.2015.
Begley,C.G.&Ellis,L.M.(
2012). Raise standards for preclinical cancer research.
Nature 483, 531– 533.
Benjamini,Y.&Hochberg,Y.(
1995). Controlling the false discovery rate – a
practical and powerful approach to multiple testing. Journal of the Royal Statistical
Society, Series B (Methodological) 57, 289– 300.
Bishop,D.V.M.&Thompson,P.A.(
2016). Problems in using p-curve analysis and
text-mining to detect rate of p-hacking and evidential value. PeerJ 4, e1715.
Bissell,M.(
2013). The risks of the replication drive. Nature 503, 333–334.
Bruns,S.B.&Ioannidis,J.P.A.(
2016). P-Curve and p-Hacking in observational
research. PLoS One 11, e0149144.
Burley,N.&Bartels,P.J.(
1990). Phenotypic similarities of sibling zebra finches.
Animal Behaviour 39, 174–180.
Button,K.S.,Ioannidis,J.P.A.,Mokrysz,C.,Nosek,B.A.,Flint,J.,
Robinson,E.S.J.&Munafo,M.R.(
2013). Power failure: why small sample size
undermines the reliability of neuroscience. Nature Reviews. Neuroscience 14, 365 –376.
Chambers,C.D.(
2013). Registered reports: a new publishing initiative at cortex.
Cortex 49, 609– 610.
Cliff,A.D.&Ord,J.K.(
1981). Spatial Processes: Models & Applications. Pion, London.
Crawley,M.J.(
2002). Statistical Computing. An Introduction to Data Analysis using S-Plus.
Wiley, Chichester.
Cumming,G.,Williams,J.&Fidler,F.(
2004). Replication and researchers’
understanding of confidence intervals and standard error bars. Understanding Statistics
3, 299– 311.
Dale,M.R.T.&Fortin,M.-J.(
2014). Spatial Analysis: A Guide for Ecologists. Cambridge
University Press, Cambridge, UK.
Danchin,E.,Wajnberg,E.&Wagner,R.H.(
2014). Avoiding pitfalls in estimating
heritability with the common options approach. Scientific Reports 4, 3974.
De Groot,A.D.(
1956/2014). The meaning of ‘‘significance’’ for different types of
research. Translated and annotated by Eric-Jan Wagenmakers, Denny Borsboom,
Josine Verhagen, Rogier Kievit, Marjan Bakker, Angelique Cramer, Dora Matzke,
Don Mellenbergh, and Han L. J. van der Maas. Acta Psychologica 148, 188–194.
Dienes,Z.(
2016). How Bayes factor change scientific practice. Journal of Mathematical
Psychology 72, 78– 89.
Drotar,D.(
2010). Editorial: a call for replications of research in Pediatric Psychology
and guidance for authors. Journal of Pediatric Psychology 35, 801– 805.
Dunn,O.J.(
1961). Multiple comparisons among means. Journal of the American Statistical
Association 56, 52– 64.
Egger,M.,Smith,G.D.,Schneider,M.&Minder,C.(
1997). Bias in meta-analysis
detected by a simple, graphical test. British Medical Journal 315, 629– 634.
Etz,A.&Vandekerckhove,J.(
2016). A Bayesian perspective on the reproducibility
project: psychology. PLoS One 11, e0149794.
Fanelli,D.(
2010). ‘‘Positive’’ results increase down the hierarchy of the sciences.
PLoS One 5, e10068.
Faul, F., Erdfelder,E.,Buchner,A.&Lang,A.-G.(
2009). Statistical power
analyses using G*Power 3.1: tests for correlation and regression analyses. Behavior
Research Methods 41, 1149–1160.
Felsenstein,J.(
1985). Phylogenies and the comparative method. The American
Naturalist 125, 1–15.
Ferguson,C.J.&Heene,M.(
2012). A vast graveyard of undead theories: publication
bias and psychological science’s aversion to the null. Perspectives on Psychological Science
7, 555– 561.
Feynman,R.P.(
1974). Cargo cult science. Engineering and Science 37, 10 –13.
Field,A.(
2005). Discovering Statistics Using SPSS. Sage, London.
Fischhoff,B.(
1975). Hindsight not equal to foresight – effect of outcome knowledge
on judgment under uncertainty. Journal of Experimental Psychology: Human Perception and
Performance 1, 288– 299.
Fisher,R.A.(
1925). Statistical Methods for Research Workers. Genesis Publishing Pvt Ltd,
Guildford, UK.
Forstmeier,W.,Mueller,J.C.&Kempenaers,B.(
2010). A polymorphism in
the oestrogen receptor gene explains covariance between digit ratio and mating
behaviour. Proceedings of the Royal Society of London B: Biological Sciences 277, 3353–3361.
Forstmeier,W.&Schielzeth,H.(
2011). Cryptic multiple hypotheses testing in
linear models: overestimated effect sizes and the winner’s curse. Behavioral Ecology and
Sociobiology 65, 47– 55.
Franco,A.,Malhotra,N.&Simonovits,G.(
2014). Publication bias in the social
sciences: unlocking the file drawer. Science 345, 1502–1505.
Freckleton,R.P.,Harvey,P.H.&Pagel,M.(
2002). Phylogenetic analysis
and comparative data: a test and review of evidence. The American Naturalist 160,
712– 726.
Gelman,A.&Loken,E.(
2014). The statistical crisis in science. The American Scientist
102, 460– 465.
Gelman,A.&O’Rourke,K.(
2014). Discussion: difficulties in making inferences
about scientific truth from distributions of published p-values. Biostatistics 15, 18–23.
Gelman,A.&Stern,H.(
2006). The difference between ‘‘significant’’ and ‘‘not
significant’’ is not itself statistically significant. The American Statistician 60, 328–331.
Gelman,A.&Weakliem,D.(
2009). Of beauty, sex and power: statistical challenges
in estimating small effects. The American Scientist 97, 310– 316.
Gintis,H.,Smith,E.A.&Bowles,S.(
2001). Costly signaling and cooperation.
Journal of Theoretical Biology 213, 103–119.
Greenwald,A.G.(
1975). Consequences of prejudice against the null hypothesis.
Psychological Bulletin 82, 1– 20.
Hadfield,J.(
2015). There’s Madness in our Methods: improving infer-
ence in ecology and evolution. Methods.blog, https://methodsblog.wordpress.
com/2015/11/26/madness-in-our-methods Accessed 3.1.2016.
Hadfield,J.D.,Wilson,A.J.,Garant,D.,Sheldon,B.C.&Kruuk,L.E.B.
(2010). The misuse of BLUP in ecology and evolution. The American Naturalist 175,
116– 125.
Haselton,M.G.,Nettle,D.&Murray,D.R.(
2005). The evolution of cognitive
bias. In The Handbook of Evolutionary Psychology (ed. D. M. Buss), pp. 968–987. Wiley,
New York.
Hereford,J.,Hansen,T.F.&Houle,D.(
2004). Comparing strengths of directional
selection: how strong is strong? Evolution 58, 2133–2143.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

Avoiding false-positive findings 1967
Holman,L.,Head,M.L.,Lanfear,R.&Jennions,M.D.(2015). Evidence of
experimental bias in the Life Sciences: why we need blind data recording. PLoS
Biology 13, e1002190.
Holman,C.,Piper,S.K.,Grittner,U.,Diamantaras,A.A.,Kimmelman,
J., Siegerink,B.&Dirnagl,U.(
2016). Where have all the rodents gone? The
effects of attrition in experimental research on cancer and stroke. PLoS Biology 14,
e1002331.
Horton,R.(
2015). Offline: What is medicine’s 5 sigma? The Lancet 385, 1380.
Hurlbert,S.H.(
1984). Pseudoreplication and the design of ecological field
experiments. Ecological Monographs 54,187–211.
Ioannidis,J.P.A.(
2005). Why most published research findings are false. PLoS
Medicine 2, 696– 701.
Ioannidis,J.P.A.&Trikalinos,T.A.(
2007). The appropriateness of asymmetry
tests for publication bias in meta-analyses: a large survey. Canadian Medical Association
Journal 176, 1091– 1096.
Jager,L.R.&Leek,J.T.(
2014). An estimate of the science-wise false discovery rate
and application to the top medical literature. Biostatistics 15, 1– 12.
Jennions,M.D.&Møller,A.P.(
2002). Relationships fade with time: a meta-analysis
of temporal trends in publication in ecology and evolution. Proceedings of the Royal
Society of London B: Biological Sciences 269, 43–48.
Jennions,M.D.&Møller,A.P.(
2003). A survey of the statistical power of research
in behavioral ecology and animal behavior. Behavioral Ecology 14, 438– 445.
John,L.K.,Loewenstein,G.&Prelec,D.(
2012). Measuring the prevalence of
questionable research practices with incentives for truth telling. Psychological Science
23, 524– 532.
Jung,K.,Shavitt,S.,Viswanathan,M.&Hilbe,J.M.(
2014). Female hurricanes
are deadlier than male hurricanes. Proceedings of the National Academy of Sciences of the
United States of America 111, 8782–8787.
Kardish,M.R.,Mueller,U.G.,Amador-Vargas,S.,Dietrich,E.I.,Ma,R.,
Barrett,B.&Fang,C.-C.(
2015). Blind trust in unblinded observation in ecology,
evolution and behavior. Frontiers in Ecology and Evolution 3, 51.
Kelly,C.D.(
2006). Replicating empirical research in behavioral ecology: how and
why it should be done but rarely ever is. The Quarterly Review of Biology 81, 221–236.
Kelly,C.&Price,T.D.(
2005). Correcting for regression to the mean in behavior
and ecology. The American Naturalist 166, 700–707.
Kerr,N.L.(
1998). HARKing: hypothesizing after the results are known. Personality
and Social Psychology Review 2, 196–217.
Kidwell,M.C.,Lazarevi´
c,L.B.,Baranski,E.,Hardwicke,T.E.,
Piechowski,S.,Falkenberg, L.-S., Kennett,C.,Slowik,A.,Sonnleitner,
C., Hess-Holden,C.,Errington,T.M.,Fiedler,S.&Nosek,B.A.(
2016).
Badges to acknowledge open practices: a simple, low cost, effective method for
increasing transparency. PLoS Biology 14, e1002456.
Koricheva,J.,Jennions,M.D.&Lau,J.(
2013). Temporal trends in effect sizes:
causes, detection, & implications. In Handbook of Meta-analysis in Ecology and Evolution
(eds J. Gurevitch,J.Koricheva and K. Mengersen), pp. 237– 254. Princeton
University Press, Princeton.
Lakens,D.&Evers,E.R.K.(
2014). Sailing from the seas of chaos into the corridor
of stability: practical recommendations to increase the informational value of studies.
Perspectives on Psychological Science 9, 278–292.
Levine,T.,Asada,K.J.&Carpenter,C.(
2009). Sample sizes and effect sizes are
negatively correlated in meta-analyses: evidence and implications of a publication
bias against nonsignificant findings. Communication Monographs 76, 286–302.
MacCoun,R.&Perlmutter,S.(
2015). Hide results to seek the truth. Nature 526,
187– 189.
MacCoun,R.&Perlmutter,S.(
0000). Blind analysis as a correction for
confirmatory bias in physics and in psychology. In Psychological Science Under Scrutiny:
Recent Challenges and Proposed Solutions (eds S. O. Lilienfeld and I. Waldman),
pp. 589– 612. Wiley, New York.
Markowetz,F.(
2015). Five selfish reasons to work reproducibly. Genome Biology 16,1.
McNutt,M.(
2014). Reproducibility. Science 343,231–231.
Milinski,M.(
1997). How to avoid seven deadly sins in the study of behavior. Advances
in the Study of Behavior 26, 159–180.
Møller,A.P.&Jennions,M.D.(
2002). How much variance can be explained by
ecologists and evolutionary biologists? Oecologia 132, 492–500.
Morey,R.D.,Chambers,C.D.,Etchells,P.J.,Harris,C.R.,Hoekstra,R.,
Lakens,D.,Lewandowsky,S.,Morey,C.C.,Newman,D.P.,Schonbrodt,
F. D., Vanpaemel,W.,Wagenmakers,E.-J.&Zwaan,R.A.(
2016). The peer
reviewers’ openness initiative: incentivizing open research practices through peer
review. Royal Society Open Science 3, 150547.
Mundry,R.(
2011). Issues in information theory-based statistical inference – a
commentary from a frequentist’s perspective. Behavioral Ecology and Sociobiology 65,
57–68.
Mundry,R.&Nunn,C.L.(
2009). Stepwise model fitting and statistical inference:
turning noise into signal pollution. The American Naturalist 173, 119–123.
Nakagawa,S.(
2004). A farewell to Bonferroni: the problems of low statistical power
and publication bias. Behavioral Ecology 15, 1044–1045.
Nakagawa,S.&Parker,T.H.(
2015). Replicating research in ecology and evolution:
feasibility, incentives, and the cost-benefit conundrum. BMC Biology 13,88.
Neuh¨
auser,M.,Forstmeier,W.&Bretz,F.(
2001). The distribution of extra-pair
young within and among broods – a technique to calculate deviations from
randomness. Journal of Avian Biology 32, 358– 363.
Nickerson,R.S.(
1998). Confirmation bias: a ubiquitous phenomenon in many
guises. Review of General Psychology 2, 175– 220.
Nosek,B.A.,Spies,J.R.&Motyl,M.(
2012). Scientific utopia: II. restructuring
incentives and practices to promote truth over publishability. Perspectives on
Psychological Science 7, 615–631.
Nuzzo,R.(
2014). Statistical errors. Nature 506, 150– 152.
Nuzzo,R.(
2015). Foolong ourselves. Nature 526, 182– 185.
Open Science Collaboration (2015). Estimating the reproducibility of psychological
science. Science 349, aac4716.
Palmer,A.R.(
2000). Quasireplication and the contract of error: lessons from sex
ratios, heritabilities and fluctuating asymmetry. Annual Review of Ecology and Systematics
31, 441– 480.
Parker,T.H.(
2013). What do we really know about the signalling role of plumage
colour in blue tits? A case study of impediments to progress in evolutionary biology.
Biological Reviews 88, 511–536.
Parker,T.H.,Forstmeier,W.,Koricheva,J.,Fidler, F., Hadfield,J.D.,
Chee,Y.E.,Kelly,C.D.,Gurevitch,J.&Nakagawa,S.(
2016). Transparency
in ecology and evolution: real problems, real solutions. Trends in Ecology & Evolution
31, 711– 719.
Pereira,T.V.&Ioannidis,J.P.A.(
2011). Statistically significant meta-analyses of
clinical trials have modest credibility and inflated effects. Journal of Clinical Epidemiology
64, 1060– 1069.
Pike,N.(
2011). Using false discovery rates for multiple comparisons in ecology and
evolution. Methods in Ecology and Evolution 2, 278–282.
Pillemer,D.&Light,R.(
1984). Summing up: the science of reviewing research.Harvard
University Press, Cambridge.
Price,A.L.,Patterson,N.J.,Plenge,R.M.,Weinblatt,M.E.,Shadick,N.
A. & Reich,D.(
2006). Principal components analysis corrects for stratification in
genome-wide association studies. Nature Genetics 38, 904–909.
Prinz, F., Schlange,T.&Asadullah,K.(
2011). Believe it or not: how much can
we rely on published data on potential drug targets? Nature Reviews. Drug Discovery 10,
712.
Prum,R.O.(
2010). The Lande-Kirkpatrick mechanism is the null model of evolution
by intersexual selection: implications for meaning, honesty, and design in intersexual
signals. Evolution 64, 3085– 3100.
Rosenthal,R.(
1979). The file drawer problem and tolerance for null results.
Psychological Bulletin 86, 638– 641.
Rouder,J.N.,Speckman,P.L.,Sun,D.,Morey,R.D.&Iverson,G.(
2009).
Bayesian t tests for accepting and rejecting the null hypothesis. Psychonomic Bulletin
& Review 16, 225–237.
Ruxton,G.&Colegrave,N.(
2010). Experimental Design for the Life Sciences.Oxford
University Press, Oxford, UK.
Scargle,J.D.(
1999). Publication bias (the ‘‘File-Drawer Problem’’) in scientific
inference. arXiv preprint physics/9909033.
Schielzeth,H.&Forstmeier,W.(
2009). Conclusions beyond support:
overconfident estimates in mixed models. Behavioral Ecology 20, 416–420.
Seguin,A.&Forstmeier,W.(
2012). No band color effects on male courtship rate
or body mass in the zebra finch: four experiments and a meta-analysis. PLoS One 7,
e37785.
Sheldon,B.C.(
2000). Differential allocation: tests, mechanisms and implications.
Trends in Ecology & Evolution 15, 397–402.
Simmons,J.P.,Nelson,L.D.&Simonsohn,U.(
2011). False-positive psychology:
undisclosed flexibility in data collection and analysis allows presenting anything as
significant. Psychological Science 22, 1359–1366.
Simmons,J.P.,Nelson,L.D.&Simonsohn,U.(
2012). A 21 word solution.
Available at http://ssrn.com/abstract=2160588 Accessed 4.3.2014.
Simonsohn,U.,Nelson,L.D.&Simmons,J.P.(
2014). P-curve: a key to the
file-drawer. Journal of Experimental Psychology: General 143, 534 –547.
Simonsohn,U.,Simmons,J.P.&Nelson,L.D.(
2015). Specification curve:
descriptive and inferential statistics on all reasonable specifications. Manuscript
available at http://ssrn.com/abstract=2694998 Accessed 30.10.2015.
Smaldino,P.E.&McElreath,R.(
2016). The natural selection of bad science.
arXiv preprint arXiv:1605.09511.
Smith,D.R.,Hardy,I.C.W.&Gammell,M.P.(
2011). Power rangers: no
improvement in the statistical power of analyses published in Animal Behaviour.
Animal Behaviour 81, 347–352.
Steegen,S.,Tuerlinckx, F., Gelman,A.&Vanpaemel,W.(
2016). Increasing
transparency through a multiverse analysis. Perspectives on Psychological Science 11,
702– 712.
Trivers,R.(
2011). The Folly of Fools.Basic,NewYork.
Valcu,M.&Kempenaers,B.(
2010). Spatial autocorrelation: an overlooked concept
in behavioral ecology. Behavioral Ecology 21, 902–905.
Valcu,M.&Valcu,C.M.(
2011). Data transformation practices in biomedical
sciences. Nature Methods 8, 104–105.
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.

1968 W. Forstmeier and others
van Wilgenburg,E.&Elgar,M.A.(2013). Confirmation bias in studies of
nestmate recognition: a cautionary note for research into the behaviour of animals.
PLoS One 8, e53548.
Vazquez,A.I.,Bates,D.M.,Rosa,G.J.M.,Gianola,D.&Weigel,K.A.(
2010).
Technical note: an R package for fitting generalized linear mixed models in animal
breeding. Journal of Animal Science 88, 497–504.
Wagenmakers,E.-J.,Wetzels,R.,Borsboom,D.,van der Maas,H.L.J.&
Kievit,R.A.(
2012). An agenda for purely confirmatory research. Perspectives on
Psychological Science 7, 632–638.
Ware,J.J.&Munafo,M.R.(
2015). Significance chasing in research practice: causes,
consequences and possible solutions. Addiction 110,4–8.
Wedderburn,R.W.M.(
1974). Quasi-likelihood functions, generalized linear models,
and the Gauss-Newton method. Biometrika 61, 439–447.
Weissgerber,T.L.,Garovic,V.D.,Milin-Lazovic,J.S.,Winham,S.J.,
Obradovic,Z.,Trzeciakowski,J.P.&Milic,N.M.(
2016). Reinventing
biostatistics education for basic scientists. PLoS Biology 14, e1002430.
Whittingham,M.J.,Stephens,P.A.,Bradbury,R.B.&Freckleton,R.P.
(2006). Why do we still use stepwise modelling in ecology and behaviour? Journal of
Animal Ecology 75, 1182– 1189.
(Received 25 May 2016; revised 17 October 2016; accepted 19 October 2016; published online 23 November 2016)
Biological Reviews 92 (2017) 1941– 1968 ©2016 The Authors. Biological Reviews published by John Wiley & Sons Ltd on behalf of Cambridge Philosophical Society.