Installation Instructions Softwares For RNAseq Session March 2019 Windows
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 18

Steps to download softwares for
windows for RNAseq workshop
Mohak Sharda
Nitish Malhotra

Installing MobaXterm

Type in google search bar
Click on the link.
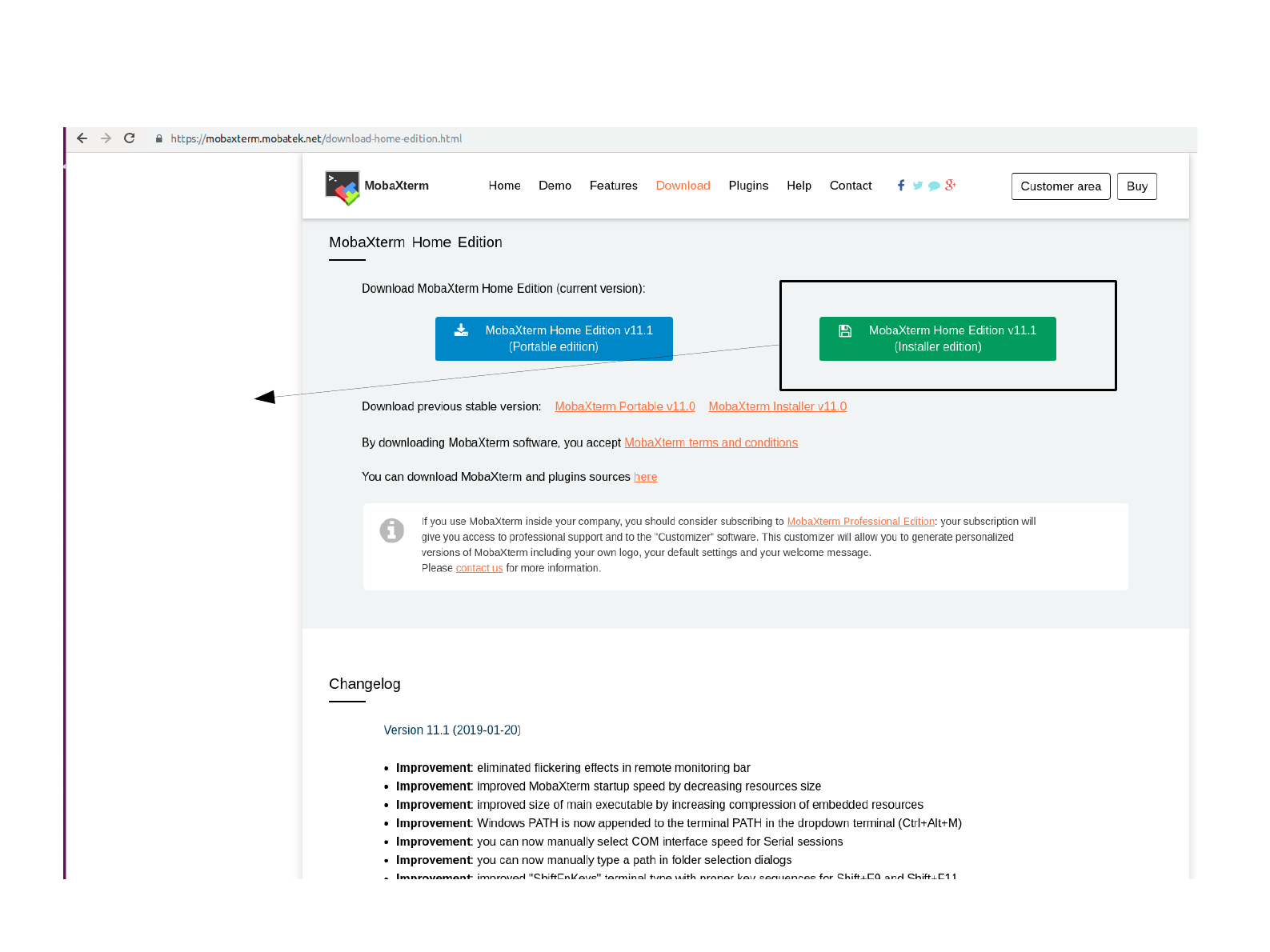
Download the compressed file
Click on the link
on the right only.

Open the .zip file and extract to a
desired folder

Open the MobaXterm setup from the
folder where you extracted your .zip file
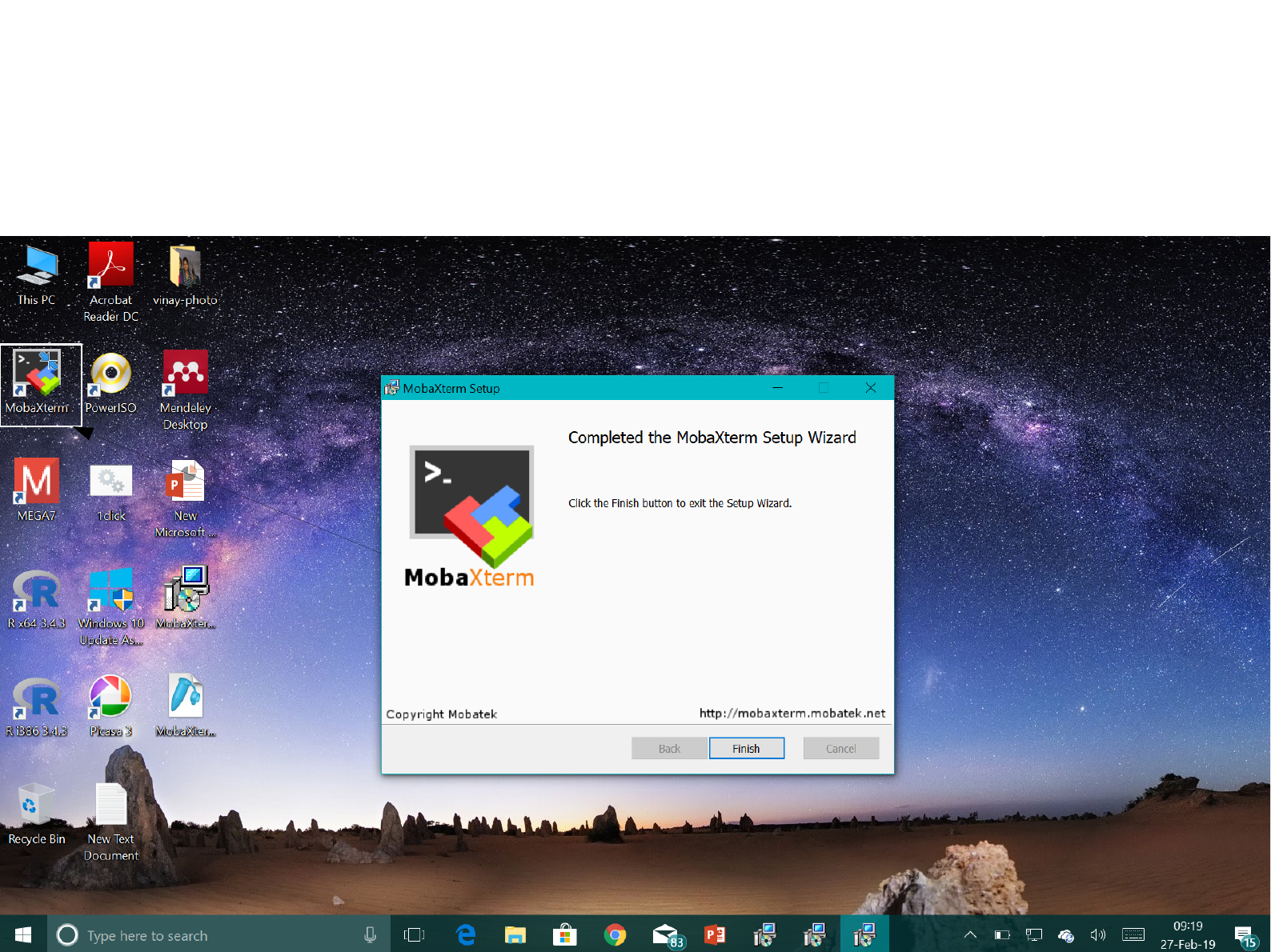
A shortcut of MobaXterm will be created
after finishing the setup
(all default settings)

Installing R
If you do not have R installed on your windows, please
follow the instructions ahead
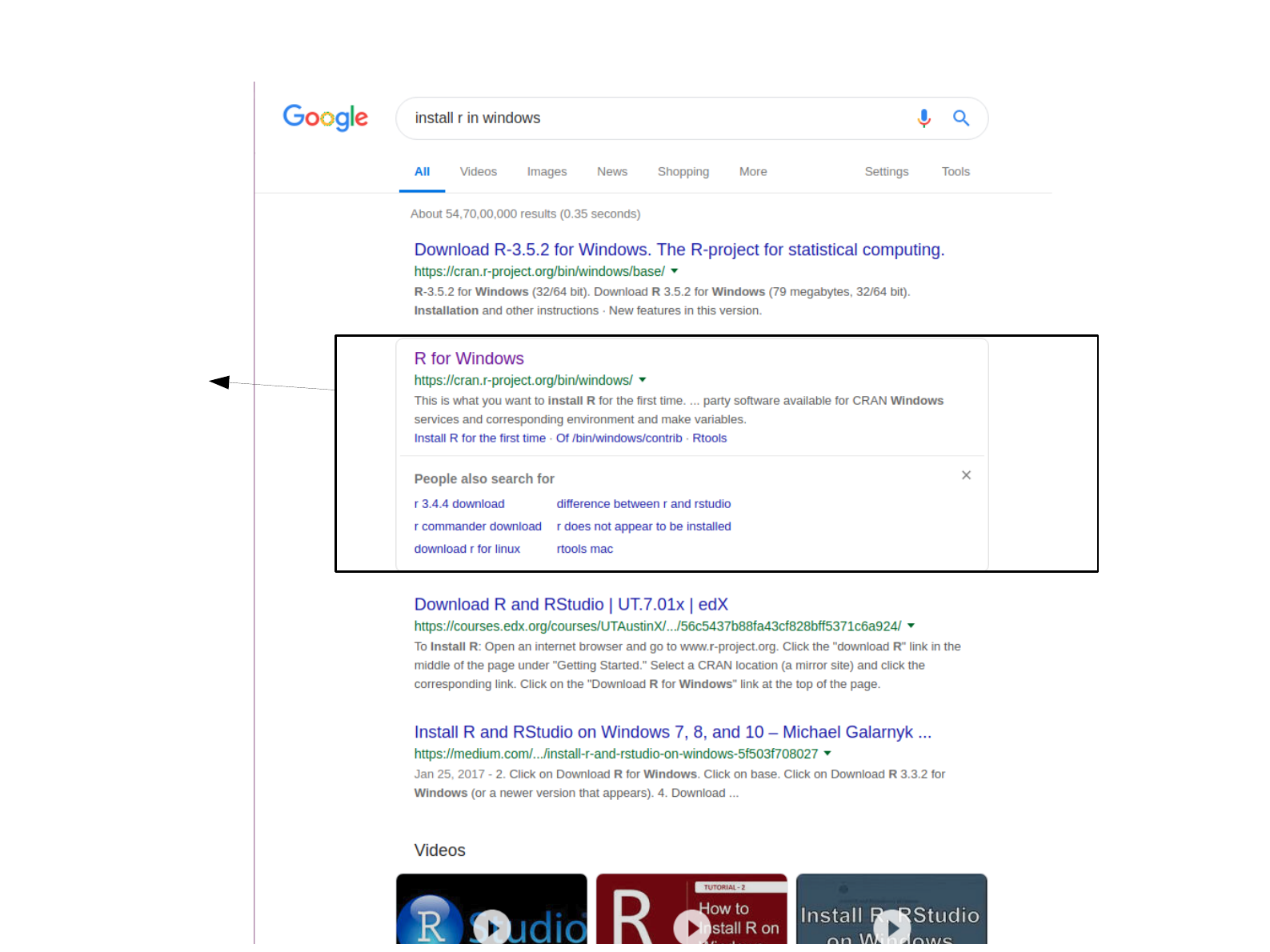
Type in google search bar
Click on the
link
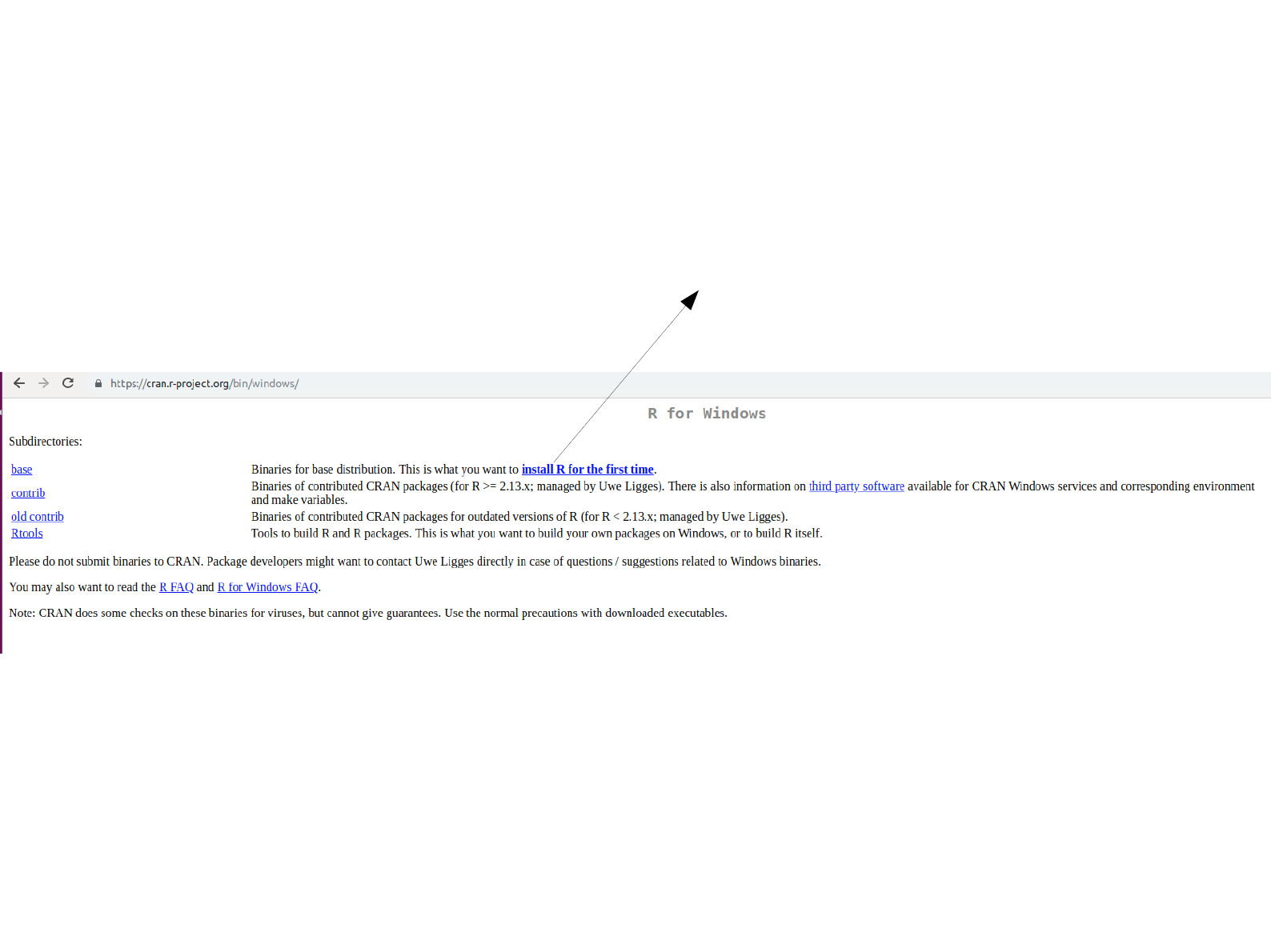
Open this hyperlink.
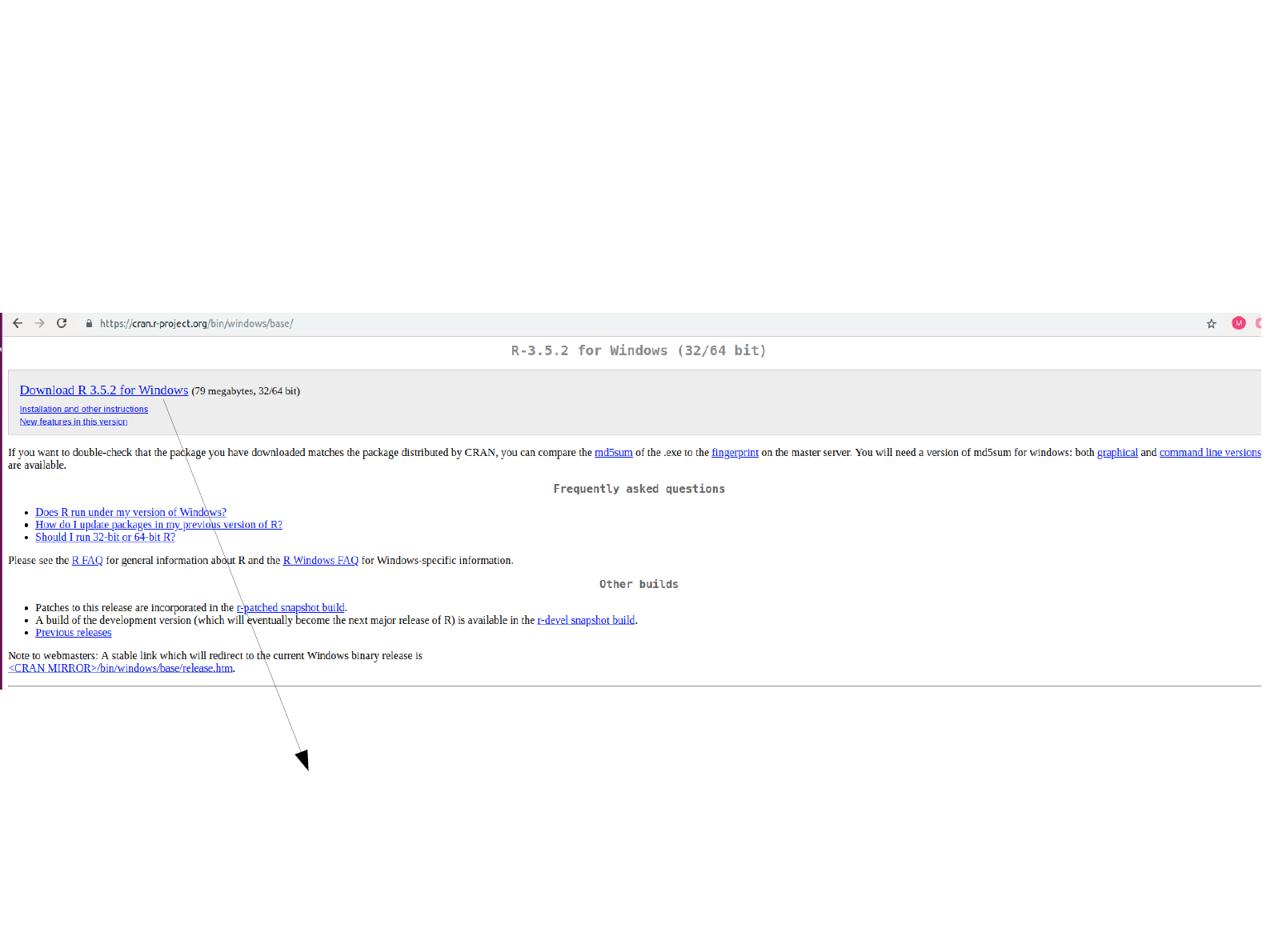
Download the hyperlink. Follow the instructions with
default settings to install R as previously done for
MobaXterm installation.

Installing Rstudio
In order for Rstudio to work, you need to have R already
installed on your system. Please go back to the previous
section and install R before proceeding.
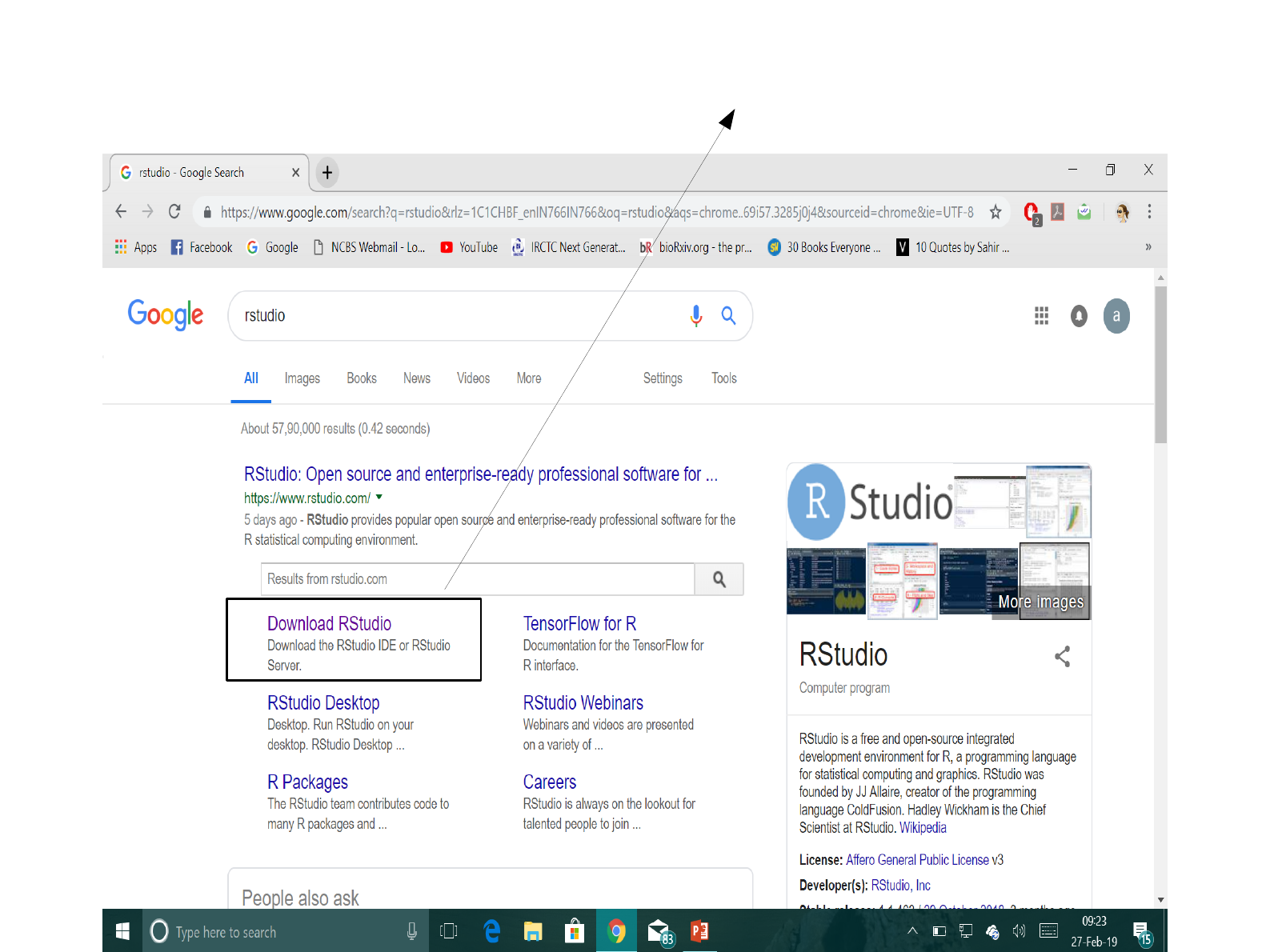
Click on the link highlighted
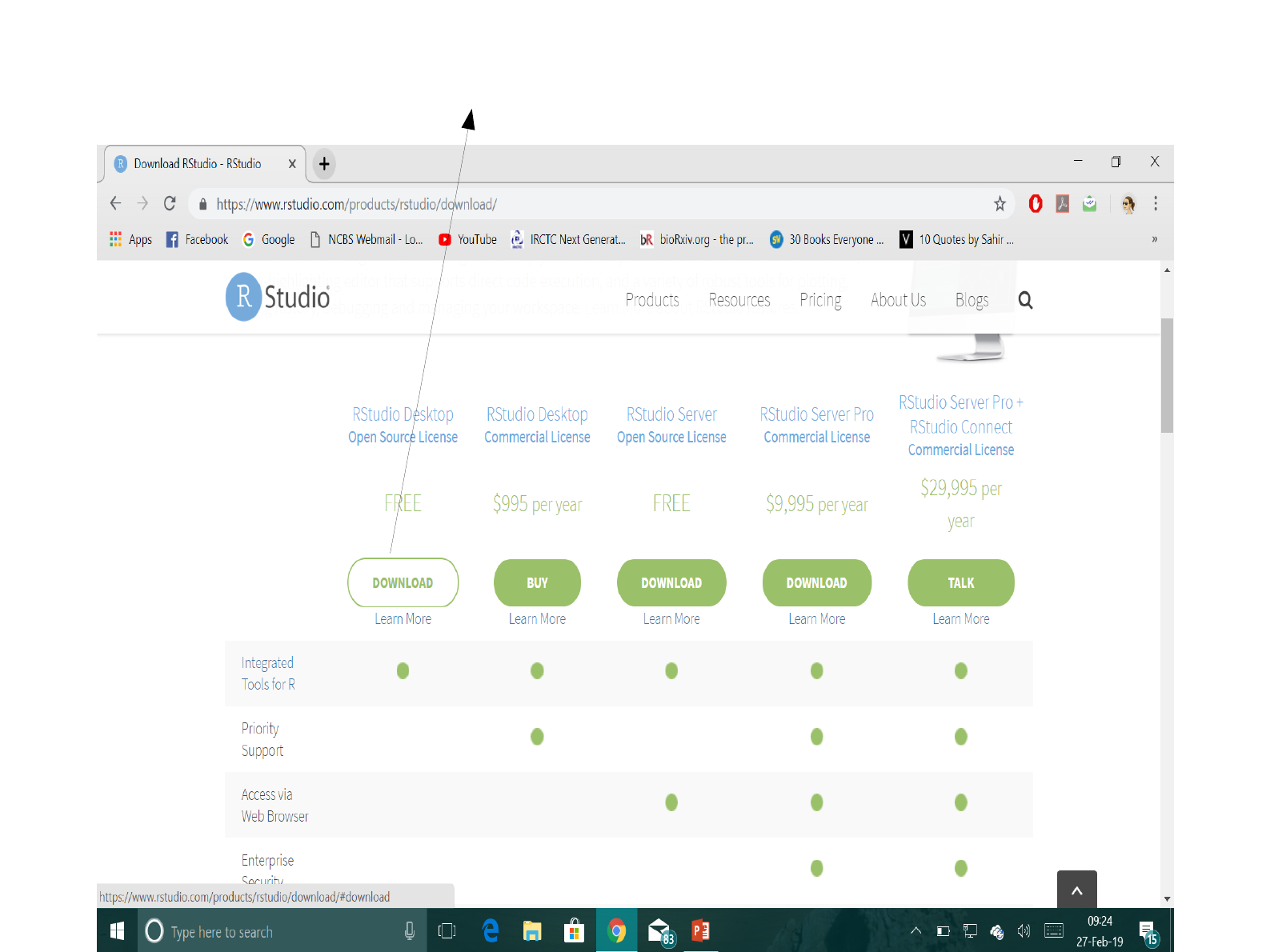
Click on the non-green fill “Download” option
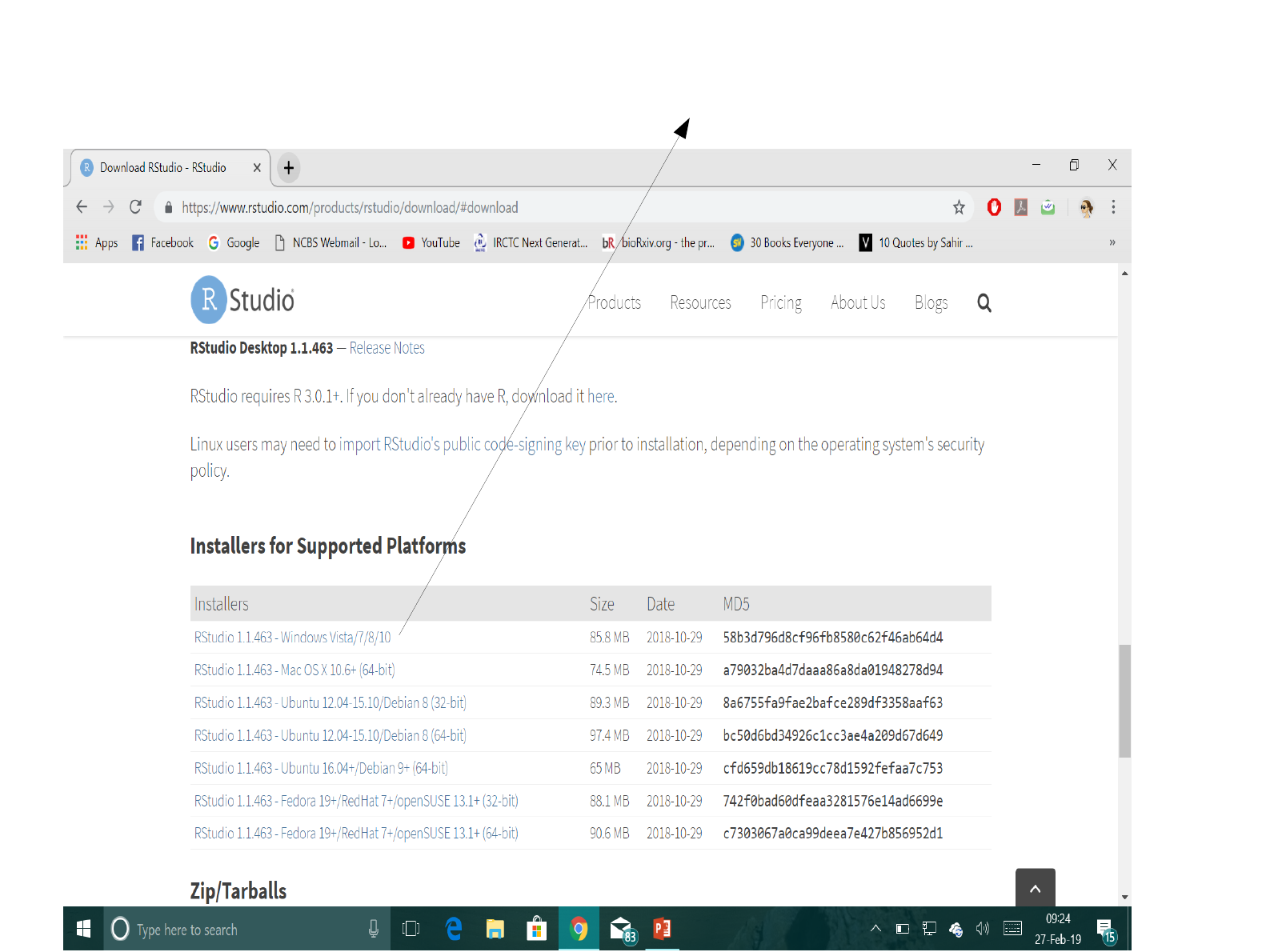
Click on the first link to download
Rstudio package on your system
Follow the instructions with default settings to complete the installation.

Installation of certain packages in R using Rstudio
Type the commands in blue and then press Ctrl+Enter in the Console. Wait for the commands to
complete, before the next “>” appears on console.
Note: Just pressing Enter alone will not execute the command. Press “Ctrl” and “Enter” at the
same time to execute the command.
Type commands in the console
1
2
3
4

Similarly, install statmod using console.
> install.packages(“statmod”)
The following packages will be installed in R for carrying out the
di"erential gene expression analysis:
1) edgeR
2) gplots
3) ape
4) statmod
See you all at the workshop!