XT800 JVI.03006 13.full
User Manual: XT800
Open the PDF directly: View PDF ![]() .
.
Page Count: 41

1
Rice Stripe Tenuivirus NSvc2 Glycoproteins Targeted to Golgi 1
Body by N-Terminal Transmembrane Domain and Adjacent 2
Cytosolic 24 Amino-Acids via COP I- and COP II-Dependent 3
Secretion Pathway 4
Min Yao 1 †, Xiaofan Liu 1 †, Shuo Li 2 †, Yi Xu 3 †, Yijun Zhou 2 *, Xueping Zhou 3,4 * and 5
Xiaorong Tao 1 * 6
7
1 Key Laboratory for the Integrated Management of Crop Diseases and Pests, Ministry of 8
Education, Department of Plant Pathology, Nanjing Agricultural University, Nanjing 210095, P. R. 9
China; 10
2 Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing 210014, P. R. 11
China; 12
3 State Key Laboratory of Rice Biology, Institute of Biotechnology, Zhejiang University, 13
Hangzhou 310029, P. R. China; 14
4 State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, 15
Chinese Academy of Agricultural Sciences, Beijing, P. R. China. 16
17
*Corresponding authors: Xiaorong Tao (taoxiaorong@njau.edu.cn); Xueping Zhou 18
(zzhou@zju.edu.cn) and Yijun Zhou (yjzhou@jass.ac.cn). 19
20
† These authors contributed equally to this study. 21
22
Running title: Requirements for Golgi targeting of RSV glycoproteins 23
24
Word count: Abstract, 203 25
Main body of the text, 4988 26
27
JVI Accepts, published online ahead of print on 3 January 2014
J. Virol. doi:10.1128/JVI.03006-13
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

2
Abstract 28
The NSvc2 glycoproteins encoded by Rice stripe tenuivirus (RSV) share many 29
characteristics common to the glycoproteins found among Bunyaviridae. Within this 30
viral family, glycoproteins targeting to the Golgi apparatus play a pivotal role in the 31
maturation of the enveloped spherical particles. RSV particles, however, adopt a long 32
filamentous morphology. Recently, RSV NSvc2 glycoproteins were shown to localize 33
exclusively to the ER in Sf9 insect cells. Here, we demonstrate that the 34
amino-terminal NSvc2 (NSvc2-N) targets to the Golgi apparatus in Nicotiana 35
benthamiana cells, whereas the carboxyl-terminal NSvc2 (NSvc2-C) accumulates in 36
the ER. Upon co-expression, NSvc2-N redirects NSvc2-C from the ER to the Golgi. 37
The NSvc2 glycoproteins move together with the Golgi stacks along the ER/actin 38
network. The targeting of the NSvc2 glycoproteins to the Golgi was strictly dependent 39
on functional anterograde traffic out of the ER to the Golgi or on a retrograde 40
transport route from the Golgi apparatus. The analysis of truncated and chimeric 41
NSvc2 proteins demonstrates that the Golgi targeting signal comprises amino acids 42
269-315 of NSvc2-N, encompassing the transmembrane domain and 24 adjacent 43
amino acids in the cytosolic tail. Our findings demonstrate for the first time that the 44
glycoproteins from an unenveloped Tenuivirus could target into Golgi bodies in plant 45
cells. 46
47
48
49
50
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

3
Importance 51
NSvc2 glycoprotein encoded by unenveloped Rice stripe tenuivirus (RSV) share 52
many characteristics in common with glycoprotein found among Bunyaviridae in 53
which all members have membrane-enveloped sphere particle. Recently, RSV NSvc2 54
glycoproteins were shown to localize exclusively to the ER in Sf9 insect cells. In this 55
study, we demonstrated that the RSV glycoproteins could target into Golgi in plant 56
cells. The targeting of NSvc2 glycoproteins to the Golgi was dependent on active 57
COP II or COP I. The Golgi targeting signal was mapped to the 23-amino-acids 58
transmembrane domain and the adjacent 24-amino-acids of the cytosolic tail of the 59
NSvc2-N. In light of the evidence from viruses in Bunyavidae that targeting into 60
Golgi is important for the viral particle assembly and vector transmission, we propose 61
that targeting of RSV glycoproteins into Golgi in plant cells represents a 62
physiologically relevant mechanism in the maturation of RSV particle complex for 63
insect vector transmission. 64
65
66
67
68
69
70
71
72
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

4
INTRODUCTION 73
Rice stripe virus (RSV) is the type member of the genus Tenuivirus (1). RSV has 74
caused severe damage to rice crops in China and is known to be transmitted by 75
Laodelphax striatellus in a persistent, circulative-propagative manner (2). The RSV 76
genome consists of four negative-sense single-stranded RNA segments, designated 77
RNA1, 2, 3 and 4, which encode seven ORFs using a negative or ambisense coding 78
strategy (3). RNA1 is negative sense and encodes an RNA-dependent RNA 79
polymerase (RdRp) (4). The other three segments adopt an ambisense coding strategy. 80
RNA2 encodes a 22.8 kDa protein (NSs2) from the viral RNA (vRNA) and a 94 kDa 81
protein (NSvc2) from the viral complementary RNA (vcRNA) (5). RNA3 encodes a 82
viral suppressor (NSs3, 23.9 kDa) from the vRNA (6) and a nucleocapsid protein 83
(NSvc3, 35 kDa) from the vcRNA (7, 8). RNA4 encodes a 20.5 kDa protein (NSs4) 84
from the vRNA and a movement protein (NSvc4, 32 kDa) from the vcRNA (9). 85
86
Based on phylogenetic relationship and their genome organization and gene 87
expression strategies, tenuiviruses are more closely related to the animal-infecting 88
viruses in the genus Phlebovirus of the family Bunyaviridae than they are to plant 89
tospoviruses (10). The NSvc2 protein encoded by RSV (hereinafter the NSvc2 90
glycoprotein) shares many characteristics in common with the glycoproteins found in 91
the Bunyaviridae family of viruses in which all members adopt an enveloped 92
spherical virion form (10). The glycoprotein encoded by the Bunyaviridae viruses is 93
processed into two proteins, Gn (the amino-terminal glycoprotein) and Gc (the 94
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

5
carboxyl-terminal glycoprotein), which together form the surface spikes of the mature 95
enveloped virion (11-14). The Gn protein of several viruses, including Uukuniemi 96
virus (UUKV) (15), the Punta toroviruses (16), and Rift valley fever virus (RVFV) 97
(17) in the genus Phlebovirus, as well as Tomato spotted wilt tospovirus (TSWV) (18), 98
has been shown to accumulate in the Golgi apparatus, while the Gc protein localizes 99
to the endoplasmic reticulum (ER). Upon co-expression, both glycoproteins localize 100
to the Golgi apparatus (16-19), suggesting that Gn can re-target Gc from the ER to the 101
Golgi. The targeting of the viral glycoproteins to the Golgi apparatus plays a pivotal 102
role in the maturation of the viral particles. The NSvc2 glycoprotein encoded by RSV 103
was predicted to be functionally similar to the glycoproteins found on other 104
Bunyaviridae viruses. RSV particles, however, adopt a long filamentous morphology 105
unenveloped (19, 20). The enveloped nature of Bunyaviridae versus the unenveloped 106
nature of Tenuivirus raises the question of what common or unique strategies have 107
evolved for them to form different morphology of viral particle. Zhao et al. (2012) 108
recently reported that the NSvc2 protein, or its two processing products, the 109
amino-terminus of NSvc2 (NSvc2-N) and the carboxyl-terminus of NSvc2 (NSvc2-C), 110
exclusively localized to the ER membrane in Spodoptera frugiperda (Sf9) insect cells 111
(21). It remains poorly understood whether the ER localization (the inability to target 112
to the Golgi apparatus) of the NSvc2 glycoproteins is the key step determining the 113
adoption of a long filamentous particle in RSV. It is also unknown why does a 114
nonenveloped teniuvirus encode glycoproteins. 115
116
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

6
RSV systemically infects Nicotiana benthamiana by mechanical inoculation (9, 22). 117
In this study, the subcellular targeting of the NSvc2 glycoproteins and the 118
requirements for their targeting were extensively characterized in N. benthamiana. 119
We demonstrated that the NSvc2-N glycoprotein alone is able to target to the Golgi 120
apparatus in N. benthamiana, whereas NSvc2-C localizes to the ER membrane in the 121
absence of NSvc2-N. Upon co-expression, NSvc2-N redirects NSvc2-C to the Golgi 122
apparatus. The NSvc2 glycoproteins were found to move together with the Golgi 123
stacks along the ER/actin network in N. benthamiana epidermal cells. Using 124
dominant-negative mutants, we demonstrated that the targeting of the NSvc2 proteins 125
from the ER to the Golgi was strictly dependent on COP I and COP II early secretion 126
pathways. The analysis of truncated and chimeric NSvc2 proteins demonstrated that 127
the Golgi targeting signal localized to amino acids 269-315, encompassing the 128
23-amino acid transmembrane domain and the 24 adjacent amino acids of the 129
cytosolic tail. Our findings provide novel insights into the cellular properties of RSV 130
glycoproteins in plant cells. 131
132
MATERIALS AND METHODS 133
Plasmid constructs and organelle markers 134
p1300S-NSvc2-N-YFP and p1300S-NSvc2-C-YFP. NSvc2-N and NSvc2-C were 135
amplified from total RNA isolated from rice infected by RSV using RT-PCR and the 136
primers XT746/XT747 and XT800/XT388 (Supplemental Table S1). The NSvc2-N 137
and NSvc2-C PCR fragments were digested with Kpn I and BamH I and inserted into 138
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

7
p1300S-YFP using the same restriction sites to obtain p1300S-NSvc2-N-YFP and 139
p1300S-NSvc2-C-YFP, respectively. 140
141
p1300S-NSvc2-Intron-YFP. A potato ST-LS1 intron (23) was inserted into the 142
AG/GT site at nucleotide (nt) position 1182 of NSvc2. The ST-LS1 intron, N-terminal 143
fragment (1182 nt) and C-terminal fragment (1423 nt) of NSvc2 were amplified using 144
the primers XT957/XT958, XT746/XT959 and XT960/XT388, respectively. The 145
three PCR fragments were mixed and amplified using XT746/XT388 to obtain 146
NSvc2-Intron, which was then digested with Kpn I and BamH I and inserted into 147
p1300S-YFP using the same restriction sites. 148
149
p1300S-NSvc2-N-46del-YFP and p1300S-NSvc2-N-63del-YFP. NSvc2-N 150
containing either a 46 or 63 amino acid deletion at the C-terminus was amplified 151
using the primer pairs XT746/XT807 or XT746/XT835, and the PCR products were 152
inserted into the Kpn I and BamH I sites of p1300S-YFP, respectively. 153
154
p1300S-SSNTMDNCTN-YFP, p1300S-SSNTMDNCTNdel46-YFP and 155
p1300S-SSNTMDNCTNdel63-YFP. The signal peptide (SSN), transmembrane domain 156
(TMDN) containing the full-length cytosolic domain (CTN), TMDN containing the 157
CTN with a 46 amino acid deletion and the TMDN with the CTN containing a 63 158
amino acid deletion at the C-terminus of NSvc2-N were amplified using the 159
corresponding primer pairs (XT746/XT837, XT836/XT747, XT836/XT807 and 160
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

8
XT836/XT835). The SSNTMDNCTN, SSNTMDNCTNdel46, and SSNTMDNCTNdel63 161
fragments were fused using overlap PCR and the primers XT746/XT747, 162
XT746/XT807 and XT746/XT835, and were inserted into the Kpn I and BamH I sites 163
of p1300S-YFP, respectively. 164
165
p1300S-NSvc-C(TMDNCTN)-YFP and p1300S-NSvc-C(TMD-CT-del46)-YFP. A 166
fragment of NSvc2-C lacking the TMDC and the CTC was amplified using the primers 167
XT800 and XT869. The TMDN fragment with the full-length CTN and the TMDN 168
fragment with the CTN containing a 46 amino acid deletion at the C-terminus of 169
NSvc2-N were amplified with the primer pairs XT747/XT868 and XT807/XT868. 170
They were then fused using overlap PCR and the primers XT800/XT747 and 171
XT800/XT807, respectively. The products of overlap PCR were digested with Kpn I 172
and BamH I and cloned into p1300S-YFP. 173
174
p1300S-CFP-Sec24 and p1300S-Arf1-CFP. The full-length Sec24 (AT3G07100) 175
and Arf1 genes were amplified using RT-PCR and the total RNA extracted from the 176
Col ecotype of Arabidopsis thaliana using the primers XT743/XT754 and 177
XT784/XT785, respectively. The Sec24 PCR fragments were digested with BamH I 178
and cloned into the Bgl II site of p1300S-CFP, while Arf1 was digested with BamH I 179
and cloned into the BamH I site of p1300S-CFP. 180
181
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

9
p1300S-Arf1 (T31N). To construct p1300S-ArfI (T31N), site-directed mutagenesis 182
was used to introduce the mutation into Arf1 using the primers XT784/XT795 and 183
XT794/XT785 and overlap PCR. The PCR product was digested with BamH I and 184
cloned into p1300S. 185
186
The ER marker mCherry-HDEL (24) and the Golgi marker Man49-mCherry (24) 187
were obtained from the Arabidopsis Biological Resource Center (ABRC). The Sar1 188
dominant-negative mutant construct Sar1 (H74L) was kindly provided by Professor 189
Taiyun Wei (25). 190
191
Plant material, transient expression and treatment 192
RSV (Jiangsu isolate) was collected from infected rice in a field in Nanjing and frozen 193
at -80°C until use. All transient expression experiments were performed using six- to 194
eight-week old N. benthamiana plants. Agrobacterium tumefaciens cells (C58C1 195
containing various RSV constructs and organelle markers) were grown using 196
kanamycin selection. The Agrobacterium cells were treated with infiltration buffer (10 197
mM MgCl2, 10 mM MES, pH 5.9, and 150 μM acetosyringone) for 3 hr at room 198
temperature before being infiltrated (OD600 = 0.5) into the abaxial surface of N. 199
benthamiana leaves. All agroinfiltrated plants were grown in growth chambers 200
(Model GXZ500D, Jiangnan Motor Factory, Ningbo, P. R. China) under a 16 h light/8 201
h dark cycle and a constant temperature of 25°C. The agroinfiltrated leaves were 202
examined for fluorescence expression between 24-72 hpi. When applicable, LatB 203
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

10
(Sigma) was infiltrated at a final concentration of 10 ȝM into N. benthamiana leaves 204
before fluorescence observation. 205
206
Confocal laser scanning microscopy 207
Leaf discs were dissected from the agroinfiltrated leaf area of N. benthamiana leaves 208
and mounted in water between two cover slips. Images and movies were captured 209
using a Carl Zeiss LSM 710 confocal laser scanning microscope and 20, 63 oil or 210
63 water immersion objective lenses. CFP fluorescence was excited at 405 nm and 211
emission captured at 440-470 nm, YFP were excited at 488 nm and emission captured 212
at 497-520 nm, and mCherry was excited at 561 nm and emission captured at 585-615 213
nm. Images were processed using the Zeiss 710 CLSM and Adobe Photoshop 214
programs (San Jose, CA, USA). Movies were edited using the Corel Video Studio Pro 215
X4 software (Ottawa, Ontario, Canada). 216
217
Western blot analysis 218
Plant leaves from N. benthamiana agroinfiltrated with NSvc2-N-YFP, NSvc2-C-YFP 219
and NSvc2-YFP constructs were ground in a 1:3 (w/v; 0.1 g/300 μL) ratio of 220
extraction buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 10% 221
glycerol, 0.1% Triton X-100 and 1plant protease inhibitor). After centrifugation for 222
10 min at 3,000 × g, the supernatant of the total protein preparation was separated by 223
SDS-polyacrylamide gel electrophoresis for immunoblot analysis. The blots were 224
probed with anti-YFP (Polyclonal antibody, 1:1,000 dilution; Biyuntian, Shanghai, 225
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

11
China) and visualized with AP conjugated Goat anti-rabbit secondary antibodies 226
(1:1,000 dilution; Biyuntian, Shanghai, China) followed by nitro-blue tetrazolium 227
(NBT) and 5-bromo-4-chloro-3'-indolyphosphate (BCIP) staining (ready-made 228
solutions; Shenggong, Shanghai, China). 229
230
For subcellular fractionations, the soluble and microsomal fractions were isolated 231
from N. benthamiana leaves agroinfiltrated with NSvc2-N-YFP, NSvc2-C-YFP and 232
NSvc2-YFP constructs as described by Peremyslov et al. (2004) (26). The antigens on 233
the membranes were blotted with anti-YFP (rabbit). It was detected by DyLight 234
680-coupled goat anti-rabbit antibodies (1:10,000 dilution; Pierce, IL USA) and then 235
visualized by Licor Odyssey scanner. 236
237
RESULTS 238
The NSvc2-N protein is targeted to the Golgi apparatus 239
N. benthamiana is an ideal plant species in which to assess the subcellular 240
localization of viral proteins. To characterize the subcellular target of the NSvc2 241
glycoproteins in plant cells, we first fused the yellow fluorescent protein (YFP) to the 242
C-terminus of NSvc2-N (Fig. 1) and then agroinfiltrated the construct into N. 243
benthamiana epidermal cells. Western blot analysis showed that NSvc2-N-YFP fusion 244
protein was expressed as a size of 68kDa protein (Fig. 2A), indicating a proper 245
expression of the NSvc2-N-YFP construct. To investigate the intracellular localization 246
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

12
of the NSvc2-N-YFP protein, we isolated soluble (S30) and microsomal (P30) protein 247
fractions from N. benthamiana leaves agroinfiltrated with NSvc2-N-YFP. We found 248
that NSvc2-N-YFP was localized exclusively in microsomal fractions that are known 249
to contain ER membrane structures and Golgi bodies (Fig. 2B). 250
251
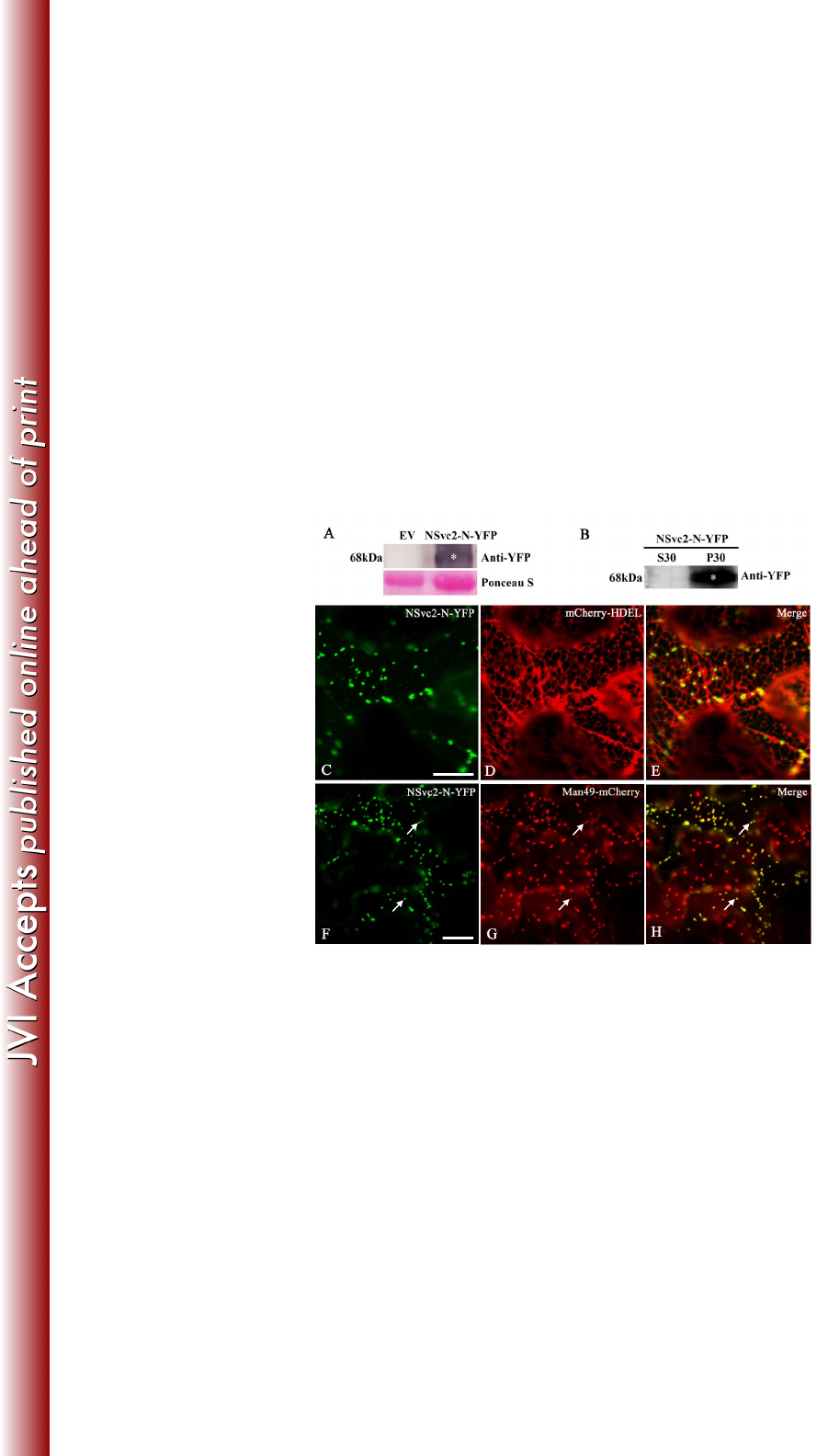
To further characterize the subcellular localization of NSvc2-N-YFP, the infiltrated 252
leaves were examined using Zeiss 710 confocal laser scanning microscopy. At 36 253
hours post-infiltration (hpi), NSvc2-N-YFP was observed as numerous small bodies 254
in the cortical cytoplasm of the cells (Fig. 2C). To determine whether NSvc2-N 255
accumulated in the ER membrane, we co-expressed the NSvc2-N-YFP protein with 256
the HDEL signal fused to the N-terminus of mCherry (mCherry-HDEL) in N. 257
benthamiana (24). The merge of NSvc2-N-YFP with mCherry-HDEL images 258
revealed that the NSvc2-N-YFP signal did not colocalize with the ER marker, while 259
those NSvc2-N-YFP punctate bodies were still associated with the ER membrane (Fig. 260
2C-E). 261
262
To determine whether the NSvc2-N-YFP bodies co-localized with the Golgi stacks, 263
we co-infiltrated the Golgi marker construct Man49-mCherry (24) with 264
NSvc2-N-YFP in N. benthamiana epidermal cells. At 36 hpi, we found that the 265
NSvc2-N-YFP bodies co-localized with the Golgi stacks (Fig. 2F-H), suggesting that 266
the NSvc2-N-YFP protein targets to the Golgi apparatus. We then examined the 267
NSvc2-N-YFP protein signal at three time points, 24, 48 and 72 hpi, and found that 268
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

13
NSvc2-N-YFP was targeted to the Golgi body as early as 24 hpi. 269
270
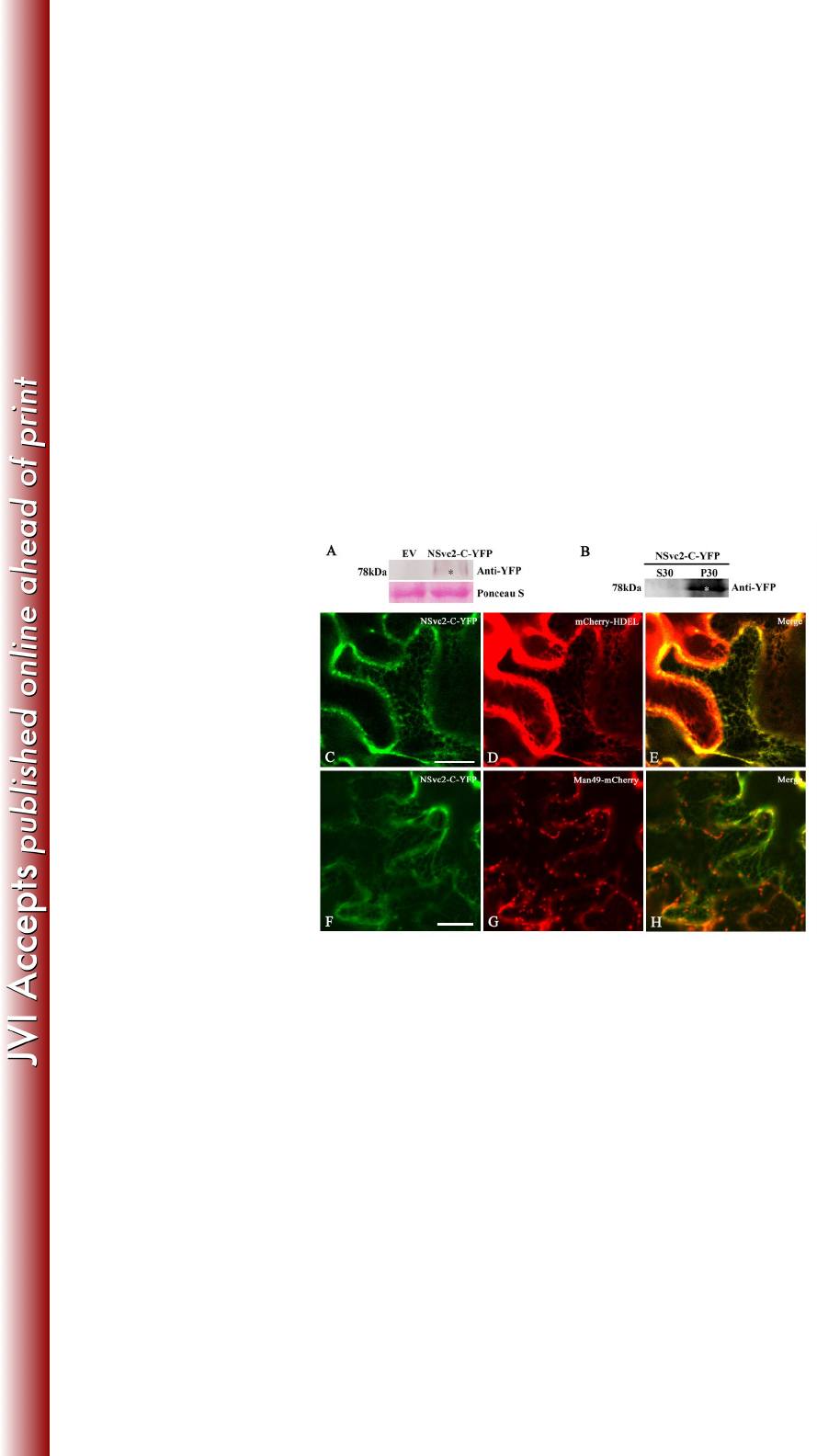
The NSvc2-C protein accumulates in the ER membrane 271
We also fused NSvc2-C protein with YFP at its C-terminus (Fig. 1) and infiltrated the 272
construct into N. benthamiana epidermal cells. Immunoblot analysis showed that 273
NSvc2-C-YFP protein expressed as 78kDa protein which is same as the predicted size 274
of NSvc2-C-YFP fusion protein (Fig. 3A). Fractionation analysis revealed that 275
NSvc2-C-YFP protein was localized only in the microsomal membrane fractions (Fig. 276
3B). To precisely define the intracellular distribution of NSvc2-C, the infiltrated 277
leaves were characterized using confocal laser scanning microscopy. The green 278
fluorescent signal of the NSvc2-C-YFP fusion protein appeared to be very weak, but 279
was still detectable in an ER-like network structure observed at 36 hpi (Fig. 3C). To 280
determine whether these fluorescent signals co-localized with the ER structure, the 281
cortical ER marker mCherry-HDEL was co-infiltrated with NSvc2-C-YFP. As shown 282
in Fig. 3C-E, the NSvc2-C-YFP protein co-localized with the ER membrane network. 283
284
To examine whether NSvc2-C-YFP accumulated in the Golgi stacks, we co-infiltrated 285
N. benthamiana cells with NSvc2-C-YFP and the Golgi marker Man49-mCherry. As 286
shown in Fig. 3F-H, no fluorescent signal associated with NSvc2-C-YFP was found to 287
accumulate in the Golgi apparatus. To confirm whether NSvc2-C-YFP exhibits any 288
accumulation in the Golgi stacks, we checked the fluorescent signal of NSvc2-C-YFP 289
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

14
at 24, 48 and 72 hpi. The NSvc2-C-YFP protein did not form any small bodies that 290
could target to the Golgi body at the three time points examined. These results suggest 291
that NSvc2-C-YFP was arrested in the ER in N. benthamiana. 292
293
The NSvc2-N protein recruits NSvc2-C from the ER to the Golgi apparatus 294
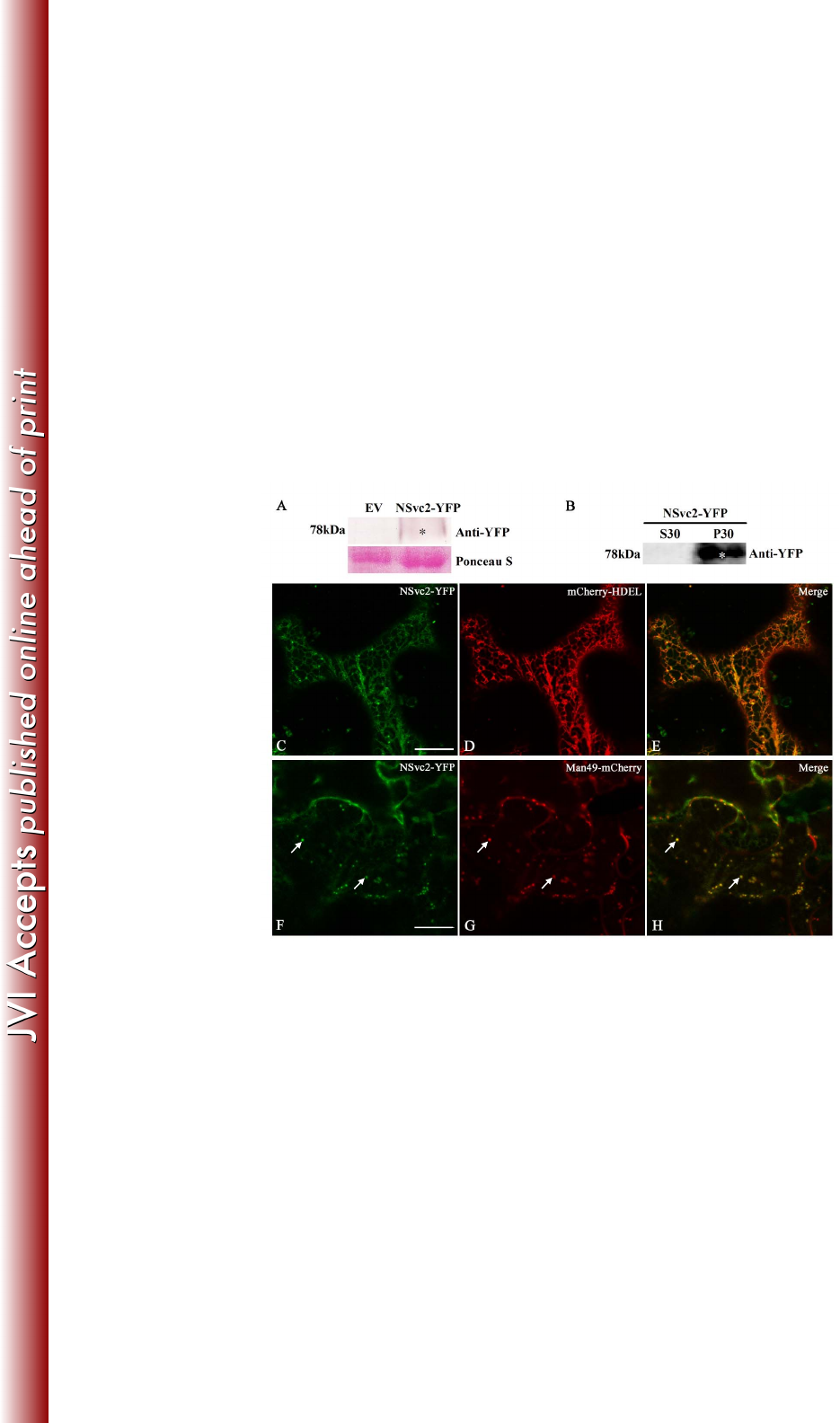
To determine the localization and trafficking of the NSvc2 glycoproteins when 295
expressed from their precursor, we fused YFP to the C-terminus of the NSvc2 296
precursor protein. However, the construct containing the full-length NSvc2 gene 297
cannot grow in E. coli cells, suggesting that the full-length NSvc2 gene is toxic to E. 298
coli. We therefore inserted a potato ST-LS1 intron (23) into the AG/GT site at 299
nucleotide (nt) position 1182 of NSvc2. The intron-containing construct, 300
NSvc2-Intron-YFP (Fig. 1), can successfully generate a green fluorescence signal in N. 301
benthamiana epidermal cells after agroinfiltration. Total RNA was then isolated from 302
infiltrated leaves and the NSvc2-Intron-YFP RT-PCR products were sequenced to 303
confirm that the intron had been precisely processed from the inserted site of NSvc2 304
(NSvc2-Intron-YFP is hereinafter referred to as NSvc2-YFP). Immunoblot analysis 305
showed that NSvc2-C-YFP has been efficiently processed from precursor protein 306
NSvc2-YFP and expressed as 78 kDa protein (Fig. 4A). The processed protein was 307
distributed exclusively in the microsomal fractions which are known to contain ER 308
membranes and Golgi bodies (Fig. 4B). 309
310
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

15
We then co-expressed NSvc2-YFP with the ER marker mCherry-HDEL in N. 311
benthamiana and the infiltrated leaves were examined using Zeiss confocal laser 312
scanning microscopy. Monitoring of NSvc2-C-YFP (NSvc2-C-YFP processed from 313
the NSvc2 precursor) showed that the fluorescent signal highlighted by NSvc2-C-YFP 314
co-localized in the ER network at 24-48 hpi. At 48-72 hpi, NSvc2-C-YFP began to 315
induce punctate structures along the ER membrane in the presence of NSvc2-N (Fig. 316
4C-E). To identify whether the newly formed bodies targeted to the Golgi apparatus, 317
we co-infiltrated N. benthamiana with NSvc2-YFP and the Golgi marker 318
Man49-mCherry. As shown in Fig. 4F-H, NSvc2-C-YFP bodies were indeed found to 319
be targeted to the Golgi apparatus. These results strongly suggest that NSvc2-N is 320
able to recruit NSvc2-C from the ER to the Golgi apparatus. 321
322
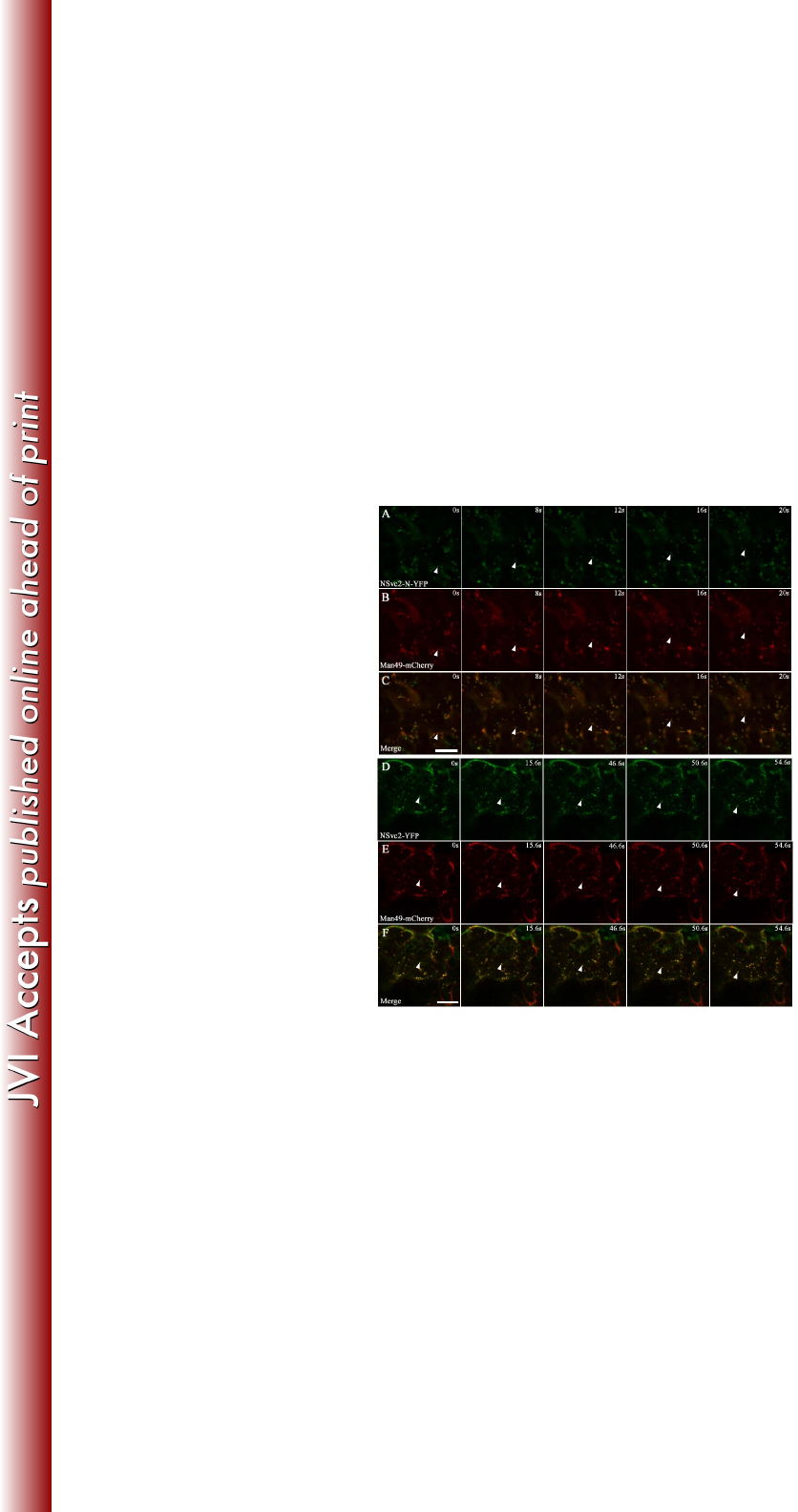
Targeted NSvc2 glycoproteins move together with the Golgi stacks in N. 323
benthamiana 324
In tobacco leaf cells, Golgi bodies traffic on an underlying ER track in an 325
actin-dependent manner (27, 28). To examine whether the targeted RSV NSvc2 326
glycoproteins move with the Golgi bodies, we utilized time-lapse confocal 327
microscopy to monitor the movement of NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP 328
(processed from the NSvc2-YFP precursor) in the presence of the Golgi marker. Fig. 329
5A-C and D-F show examples of the movement of the NSvc2-N-YFP and 330
NSvc2-N/NSvc2-C-YFP bodies with the Golgi stacks, and the arrows mark the 331
progressive movement of these bodies in each sequence. We found that both 332
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

16
NSvc2-N-YFP and NSvc2-N/NSvc2-C-YFP moved together with the Golgi bodies 333
(Fig. 5A-C and D-F; Supplemental Video S1 and S2). 334
335
To determine whether the movement of bodies labeled with NSvc2-N-YFP or 336
NSvc2-N/NSvc2-C-YFP is dependent on similar forces driving the movement of the 337
Golgi bodies, we treated agroinfiltrated leaves at 48 hpi with 10 μM latrunculin B, an 338
actin depolymerizing agent (29). After 3 h of chemical treatment, we found that 339
movement of the NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP as well as Golgi bodies 340
was completely inhibited. However, NSvc2-N-YFP, NSvc2-N/NSvc2-C-YFP and the 341
Golgi bodies remained co-localized (Supplemental Video S3 and S4). These data 342
suggest that the NSvc2-N-YFP or NSvc2-N/NSvc2-C-YFP bodies move together with 343
the Golgi stacks along the ER/actin network. 344
345
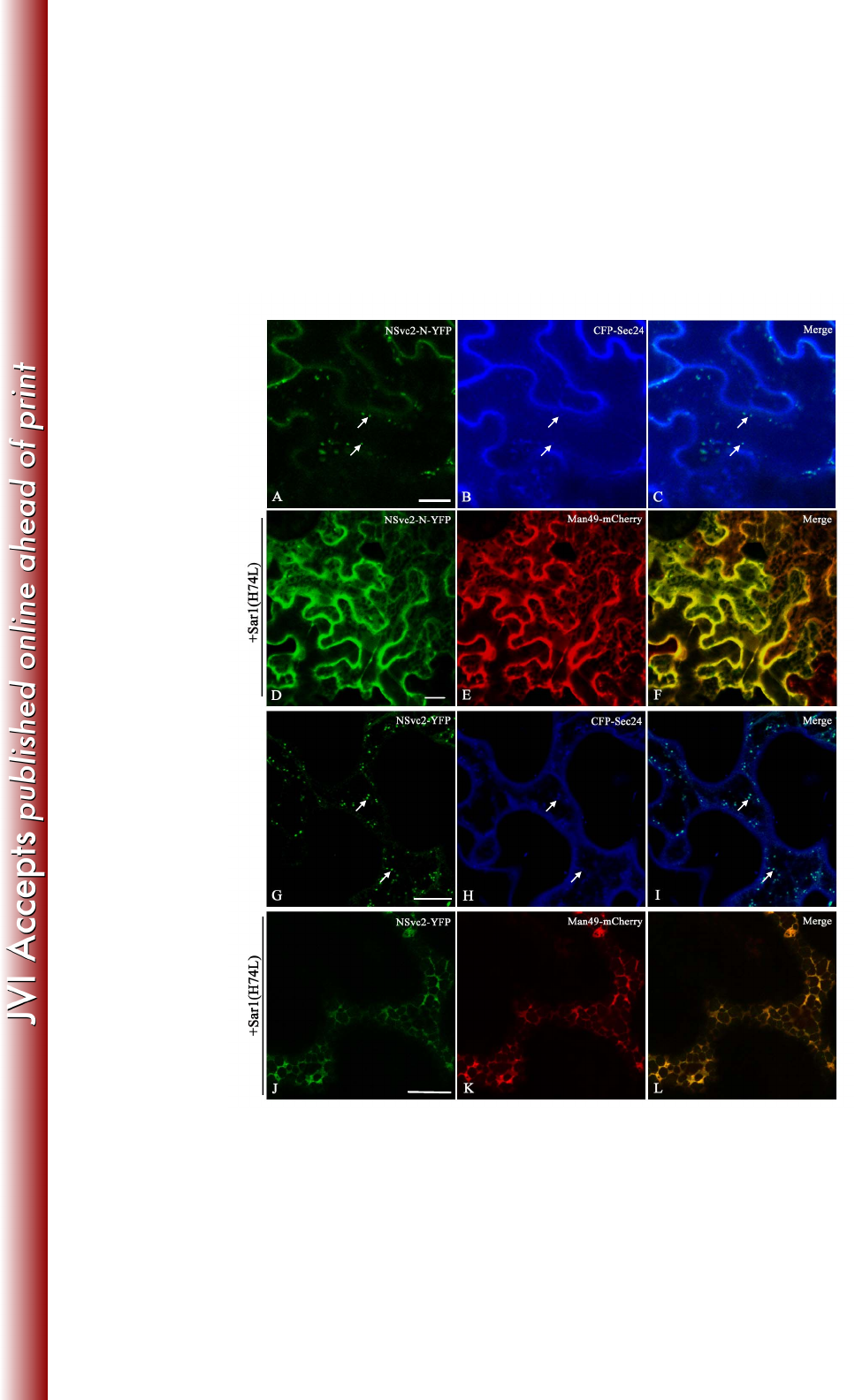
ER-to-Golgi targeting of NSvc2 glycoproteins is dependent on a functional COP 346
II complex 347
Given that the RSV NSvc2-N-YFP and NSvc2-YFP fusion proteins targeted to the 348
Golgi, we ask whether the Golgi targeting of viral glycoproteins results from traffic 349
out of the ER to the Golgi apparatus via ERES. To address this question, we 350
co-infiltrated an ERES-marker, CFP-Sec24 (30), with NSvc2-N-YFP or NSvc2-YFP 351
proteins into N. benthamiana leaf cells. As shown in Fig. 6A-C and G-I, the 352
NSvc2-N-YFP or NSvc2-YFP bodies co-localized with CFP-Sec24 fluorescence at 353
the ERES. These results suggest that NSvc2-N is able to redirect NSvc2-C from the 354
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

17
ER to the ERES, from where they subsequently co-migrate, most likely as a 355
heterodimer, to the Golgi apparatus. 356
357
The COP II complex is responsible for anterograde traffic out of the ER to the Golgi 358
apparatus (31). To test whether COPII vesicles are involved in ER-to-Golgi transport 359
of RSV NSvc2 glycoproteins, wild-type Sar1 or its dominant-negative mutant (H74L) 360
(32) was co-infiltrated with NSvc2-N-YFP or NSvc2-YFP together with the Golgi 361
marker Man49-mCherry into N. benthamiana. As shown in Fig. 6D-F and J-L, upon 362
co-expression of NSvc2-N-YFP or NSvc2-YFP with Sar1 (H74L), the florescence of 363
NSvc2-N-YFP or NSvc2-YFP, as well as of the Golgi bodies, was retrieved back to 364
the ER network, while co-expression with wild-type Sar1 did not cause the 365
NSvc2-N-YFP or NSvc2-YFP bodies to redistribute back to the ER (data not shown). 366
These results suggest that the accumulation of the RSV glycoproteins at the ERES and 367
in the Golgi bodies is dependent on a functional anterograde secretion pathway. 368
369
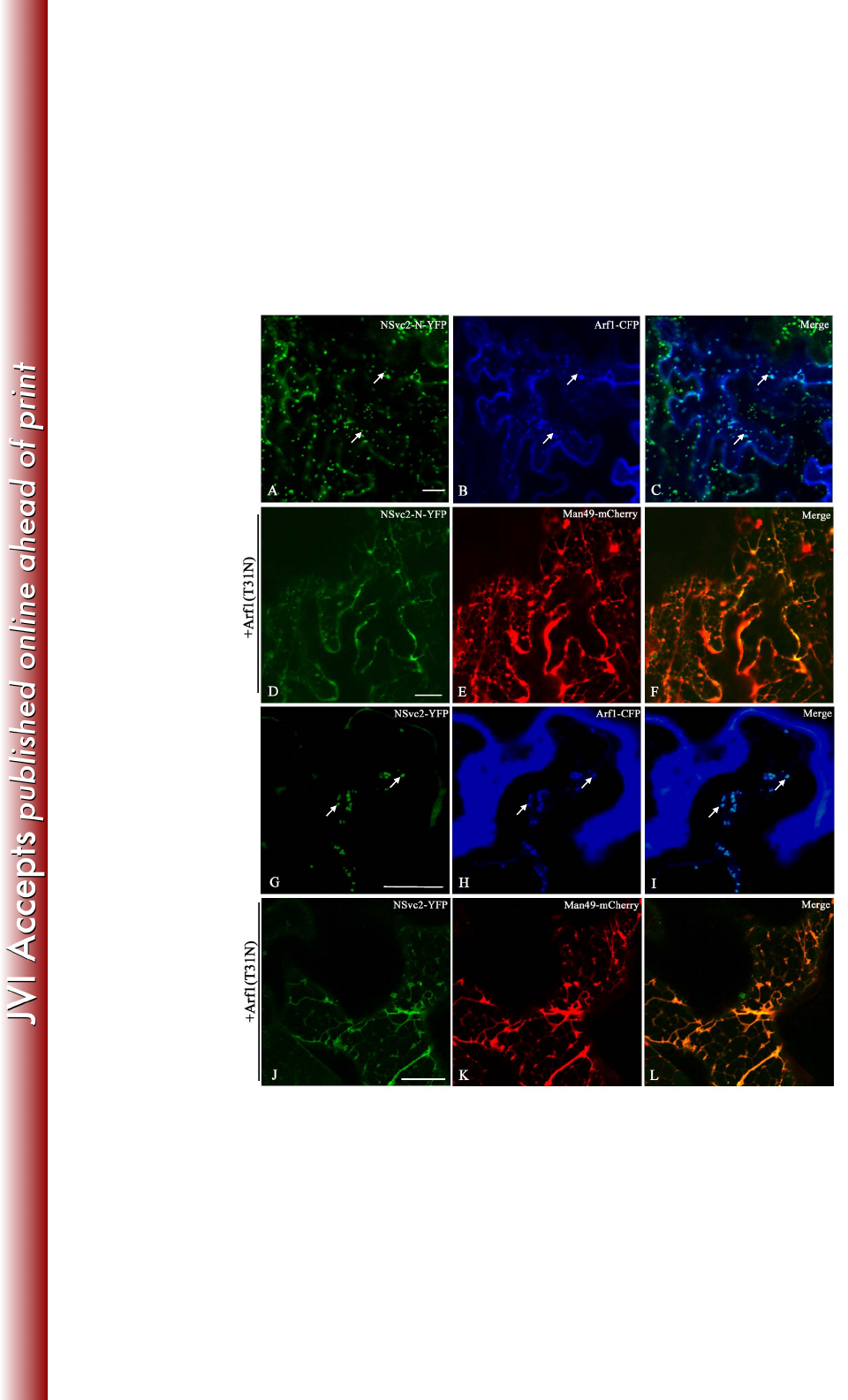
The accumulation of the NSvc2 glycoproteins at the Golgi bodies depends on 370
active COP I 371
To investigate whether the Golgi targeting of viral glycoproteins also involves 372
retrograde traffic, we co-infiltrated Arf1 tagged with CFP, a COP I vesicle marker 373
(33), with NSvc2-N-YFP or NSvc2-YFP in N. benthamiana. As shown in Fig. 7A-C 374
and G-I, the NSvc2-N-YFP or NSvc2-YFP bodies co-localized with COP I vesicles 375
labeled by Arf1-CFP. 376
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

18
377
To determine the dependency of the ER-to-Golgi transport of RSV NSvc2 378
glycoproteins on active COP I, wild-type Arf1 or Arf1 (T31N), a dominant-negative 379
mutant of COP I (33, 34), was co-infiltrated with NSvc2-N-YFP or NSvc2-YFP along 380
with the Golgi marker Man49-mCherry into N. benthamiana. We found that 381
NSvc2-N-YFP or NSvc2-YFP as well as Man49-mCherry labeled Golgi bodies 382
redistributed back to the ER membrane in the presence of the dominant-negative Arf1 383
(T31N) (Fig. 7D-E and J-L). However, the co-expression of wild-type Arf1 has no 384
such effect (data not shown). These data demonstrate that the Golgi targeting of RSV 385
glycoproteins is also dependent on an active retrograde export route. 386
387
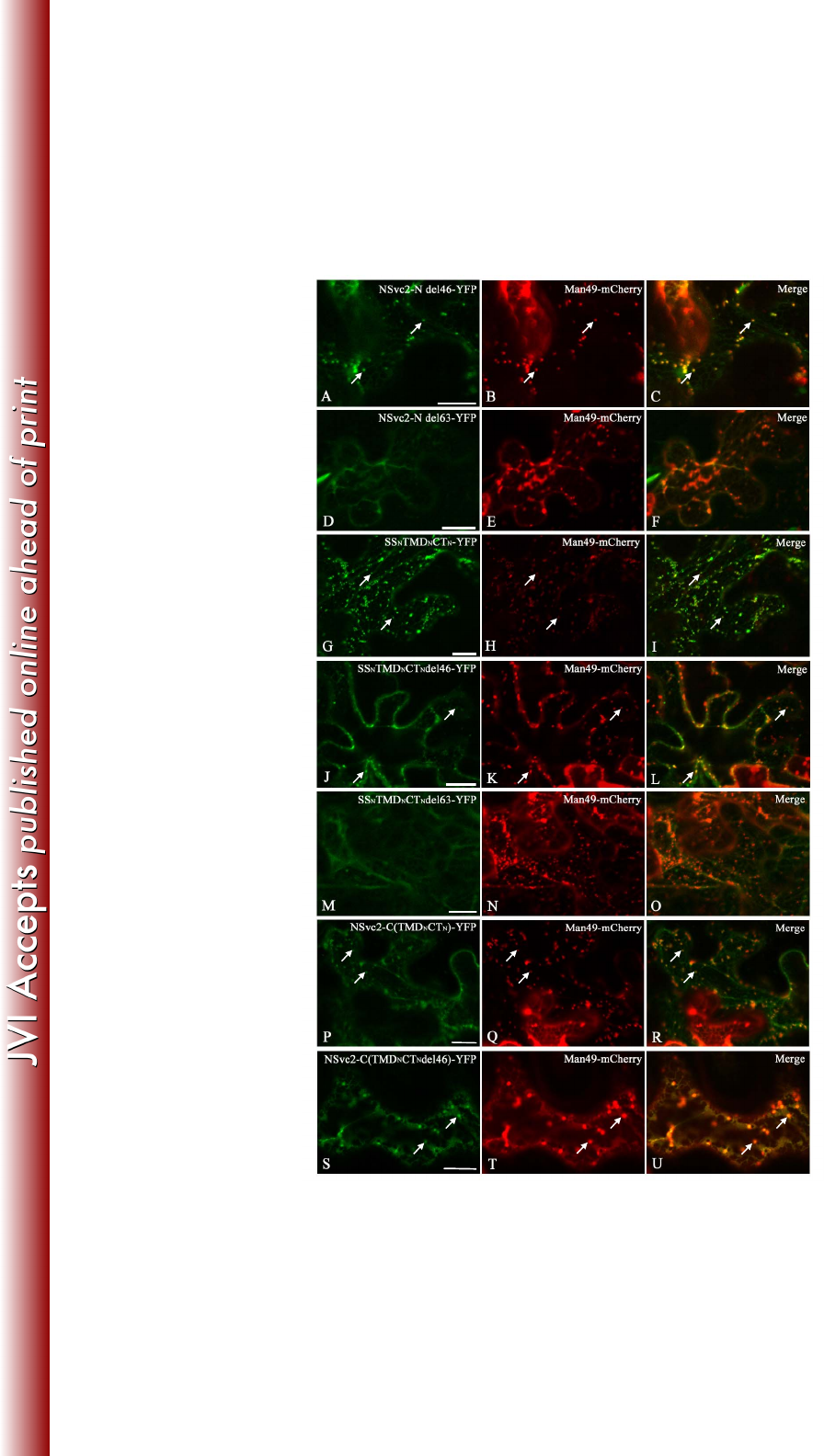
The Golgi targeting signal resides in a region of NSvc2-N encompassing a 388
transmembrane domain and the 24 adjacent amino acids of the cytosolic tail 389
Both the NSvc2-N-YFP and NSvc2-YFP expressed in N. benthamiana localized to 390
the Golgi complex, indicating that the Golgi retention signal resides in the N-terminus 391
of the NSvc2 protein. To map the domain responsible for the Golgi targeting of RSV 392
NSvc2-N, a truncated NSvc2-N del46-YFP protein, where 46 amino acids at the 393
C-terminal end of NSvc2-N within the cytosolic tail were deleted and fused with YFP 394
(Fig. 1), was constructed and transiently expressed in N. benthamiana. The 395
intracellular localization of this protein was determined by confocal fluorescence 396
analysis after 48 hpi. As illustrated in Fig. 8A-C, the truncated NSvc2-N del46-YFP 397
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

19
protein was still capable of targeting to the Golgi complex. Subsequently, 63 amino 398
acids of the C-terminal end of the NSvc2-N protein within the cytosolic tail were 399
deleted (Fig. 1). This truncated NSvc2-N del63-YFP protein was no longer targeted to 400
the Golgi apparatus (Fig. 8D-F), suggesting that the amino acids in the cytosolic tail 401
are required for entering into the Golgi. 402
403
To determine the minimum region required for Golgi targeting, the predicted 404
transmembrane domain (amino acids 269-291) and the entire cytosolic domain (amino 405
acids 292-361) of NSvc2-N were fused with its signal peptide sequence (amino acids 406
1-23) (Fig. 1). When this chimeric SSNTMDNCTN-YFP construct was expressed in N. 407
benthamiana leaf cells, we found that it accumulated in the Golgi apparatus (Fig. 408
8G-I). Subsequently, the transmembrane domain and the 24 adjacent amino acids 409
(CTdel46, amino acids 292-315) were fused with its signal peptide (Fig. 1). The 410
resulting SSNTMDNCTNdel46-YFP construct also localized to the Golgi apparatus 411
(Fig. 8J-L). Lastly, the transmembrane domain and the 7 adjacent amino acids 412
(CTdel63, amino acids 292-298) were fused with its signal peptide (Fig. 1). As shown 413
in Fig. 8M-O, this SSNTMDNCTNdel63-YFP construct was incapable of targeting to 414
the Golgi complex. These analyses suggest that both the transmembrane domain 415
(amino acids 269-291) and the 24 adjacent amino acids in the cytosolic tail of the 416
NSvc2-N protein are required for Golgi targeting. 417
418
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

20
To substantiate the observation that the Golgi retention signal is located within the 419
TMD and CT domains of NSvc2-N, the transmembrane domain (amino acids 269-291) 420
and the entire cytosolic domain (amino acids 292-361) of NSvc2-N were swapped 421
with those of NSvc2-C (Fig. 1). The resulting NSvc2-C(TMDNCTN)-YFP construct 422
was co-expressed with mCherry-HDEL and Man49-mCherry separately in N. 423
benthamiana. As shown in Fig. 8P-R, the chimeric NSvc2-C(TMDNCTN)-YFP 424
construct was capable of targeting to the Golgi apparatus, suggesting that the 425
transmembrane domain and the cytosolic domain of NSvc2-N was sufficient to direct 426
NSvc2-C-YFP to the Golgi complex (Fig. 8P-R). To analyze the requirement for the 427
Golgi targeting signal further, the transmembrane domain and the 24 adjacent amino 428
acids in the cytosolic domain of NSvc2-N were swapped with the corresponding 429
domain of NSvc2-C. As illustrated in Fig. 8S-U, this chimeric 430
NSvc2-C(TMDNCTNdel46)-YFP protein was also capable of localizing to the Golgi 431
apparatus. Taken together, these data suggest that the ER-to-Golgi targeting signal 432
resides in the C-terminal region (amino acids 269-315) of NSvc2-N, encompassing 433
the 23-amino-acids transmembrane domain and 24 adjacent amino acids in the 434
cytosolic tail. 435
436
DISCUSSION 437
In this study, using N. benthamiana as a model system we demonstrated here for the 438
first time that the glycoproteins from an unenveloped Tenuivirus could target into 439
Golgi bodies in plant cells. The RSV NSvc2-N glycoprotein alone targeted to the 440
Golgi apparatus, while the NSvc2-C glycoprotein accumulated in the ER membrane 441
in the absence of NSvc2-N. Upon co-expression, NSvc2-N was able to redirect 442
NSvc2-C from the ER to the Golgi apparatus. Using the Sar1 or Arf1 443
dominant-negative mutants, we demonstrated that the targeting of NSvc2 444
glycoproteins to the Golgi apparatus was dependent on an active COP I or COP II 445
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

21
secretion pathway. We further revealed that the Golgi targeting signal mapped to a 446
region of the NSvc2-N protein (amino acids 269-315) encompassing the 447
23-amino-acids transmembrane domain (TMD) and the adjacent 24 amino acids of the 448
cytosolic tail. 449
450
The targeting of viral glycoproteins to the Golgi apparatus plays a pivotal role in the 451
formation of enveloped spherical particles for the viruses (animal- and plant-infecting) 452
in the Bunyaviridae family (15, 17, 35-41). Although RSV particle adopt long 453
filamentous morphology (20, 21), the subcellular targeting to the Golgi apparatus 454
seems to be a conserved mechanism between the unenveloped Rice stripe tenuivirus 455
and the enveloped viruses in Bunyaviridae. Why RSV glycoproteins do not facilitate 456
the formation of an enveloped spherical particle remains to be extensively 457
investigated in the future. It is interesting to note that despite the common 458
glycoprotein characteristics shared by RSV and viruses in the Bunyaviridae, all of the 459
viruses in the Bunyaviridae have larger size of glycoproteins than are found in RSV. 460
461
For TSWV, the type member of Tospovirus which is the only genus containing 462
plant-infecting viruses in the family Bunyaviridae, the glycoproteins forming the 463
surface spikes of the mature viral particle play an important role in insect transmission 464
(42). The key step where the virus enters the insect midgut cells is mediated by these 465
glycoproteins (42). RSV particles must also enter the midgut cells of L. striatellus to 466
complete their circulative-propagative transmission. The RSV-encoded glycoproteins 467
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

22
were predicted to have a similar role in vector transmission. Although the NSvc2 468
protein was not detected in the filamentous RSV particle, this protein may function as 469
a bridge between the virus particle and recognition sites on the insect cell, as is seen, 470
for example, with helper component-proteinase (Hc-Pro) of potyvirus (43). The 471
targeting of RSV NSvc2 proteins to the Golgi apparatus could be an essential process 472
for glycoprotein modification and maturation, allowing the attachment of the RSV 473
RNP particle and subsequent vector transmission. 474
475
Zhao et al. (2012) reported that all of the RSV NSvc2 glycoproteins, including 476
NSvc2-N, NSvc2-C and the full-length NSvc2 localized exclusively to the ER 477
membrane in Sf9 insect cells (21). Our findings on the Golgi targeting of NSvc2 478
glycoproteins in N. benthamiana cells were different from those reported by Zhao et 479
al. (2012) in Sf9 insect cells. The RSV NSvc2 glycoproteins may have different 480
subcellular localization patterns in different systems. The NSvc2 glycoproteins target 481
to the Golgi apparatus in plant cells, while they were arrested in the ER membrane in 482
insect cells. These two different findings together lead to an interesting new concept 483
that acquisition of RSV viral particle from plant host by L. striatellus insect vector 484
may require glycoproteins which need to obtain glycosylation or similar modification 485
in the Golgi apparatus whereas transmission of RSV viral particle from insect vector 486
back into plant host may not require glycoproteins. 487
488
The leaf Golgi complex functions as a motile system that acquires products from a 489
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

23
relatively stationary ER system (28, 31). The glycoproteins of TSWV have shown to 490
target into Golgi body using a tobacco protoplast system (44). However, the 491
movement of the viral glycoproteins in the plant cell has not been shown previously. 492
We demonstrated in this study that the targeted NSvc2 glycoproteins moved together 493
with the Golgi stacks along the ER/actin network in N. benthamiana epidermal cells. 494
The movement of the NSvc2-N glycoprotein together with the Golgi stacks in the N. 495
benthamiana epidermal cells gives rise to an interesting hypothesis that the NSvc2-N 496
could be acting as a mobile system for picking up NSvc2-C from the ER and transport 497
it into the Golgi stacks. This hypothesis is consistent with the finding that the 498
NSvc2-N protein accumulated in the Golgi stacks as early as 24 hpi, whereas the 499
NSvc2-C protein alone remained consistently localized in the ER. NSvc2-C only 500
began to accumulate in the Golgi apparatus at 48 hpi in the presence of NSvc2-N. The 501
constant movement of NSvc2-N will continue to pick up NSvc2-C in the Golgi stacks 502
over time. 503
504
RSV NSvc2-N was able to facilitate NSvc2-C transport from the ER to the Golgi 505
apparatus. Export of proteins from the ER in plant cells has been suggested to occur 506
through different routes (45-48). For ER-to-Golgi transport, a widely accepted 507
pathway is based on the sequential action of COP II and COP I complexes (27). Our 508
results showed that RSV NSvc2-N and the NSvc2-N::NSvc2-C complex migrate to 509
the Golgi apparatus via the ERES and that Golgi targeting was strictly dependent on a 510
functional anterograde traffic out of the ER to the Golgi or a retrograde transport route 511
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

24
from the Golgi apparatus, as over-expression of Sar1 (H74L) and Arf1 (T31N) 512
aborted NSvc2-N as well as NSvc2-N::NSvc2-C complex trafficking to the Golgi. In 513
the mammalian system, it has been demonstrated that the COPII coat recognizes and 514
selects export cargo into ERES vesicles (49). Our finding that the targeting of NSvc2 515
protein into Golgi via ERES suggests that COPII machineries, such as Sar1 or 516
Sec23-Sec24 complex, may be involved in selecting NSvc2 glycoproteins to target 517
into Golgi. 518
519
For viruses in the Bunyaviridae family, intracellular maturation and budding in the 520
Golgi complex is mediated by the targeting and accumulation of the viral 521
glycoproteins in this cellular compartment (17, 18, 35, 38-40). Previous work has 522
shown that the Golgi targeting signal of the TSWV and BUNV glycoproteins resides 523
in the transmembrane domain of the Gn protein, allowing for sufficient ER-exit and 524
transport to the Golgi (35, 36). However, the Golgi localization signal of RVFV was 525
mapped to a 48-amino-acid region of Gn containing the transmembrane domain and 526
the adjacent 28 amino acids of the cytosolic tail (17). Although UUKV is also a 527
phlebovirus, the Golgi localization signal for the UUKV glycoproteins resides in the 528
cytosolic tail of Gn (15, 50). In this study, we have mapped the Golgi targeting signal 529
of RSV to a region encompassing the transmembrane domain and the 24 adjacent 530
amino acids of the cytosolic tail of the N-terminus of NSvc2. Although the 531
tenuiviruses has very close relationship to the phleboviruses, our finding support that 532
the Golgi targeting motif of the RSV glycoprotein is more closely related to that of 533
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

25
RVFV, instead of UUKV glycoprotein. 534
535
In summary, our results presented here reveal that Rice stripe tenuivirus glycoproteins 536
were able to target into Golgi apparatus in plant cells. Targeting of RSV glycoproteins 537
into Golgi apparatus is mediated by the N-Terminal transmembrane domain and the 538
adjacent cytosolic 24 amino-acids of NSvc2 in a COP I- and COP II-dependent 539
manner. In light of the evidence from viruses in Bunyavidae that targeting into Golgi 540
apparatus is important for the viral particle assembly and vector transmission, we 541
propose that targeting of RSV glycoproteins into Golgi apparatus in plant cells 542
represents a physiologically relevant mechanism in the maturation of RSV particle 543
complex for insect vector transmission. 544
545
ACKNOWLEDGMENTS 546
This work was financially supported by the Program for New Century Excellent 547
Talents in the University (NCET-12-0888), the National Natural Science Foundation 548
of China (31222045, 31171813 and 31170142), the Special Fund for Agro-scientific 549
Research in the Public Interest (201303021 and 201003031) and the National Program 550
on Key Basic Research Project of China (973 Program, 2014CB138400). We would 551
like to thank Professor Taiyun Wei for kindly providing the Sar1 (H74L) 552
dominant-negative mutant. We also thank three anonymous referees for their valuable 553
comments on earlier version of this paper. 554
555
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

26
REFERENCES 556
1. Tori y ama S. 2000. Rice stripe virus. Descriptions of Plant Viruses No. 375. 557
2. Falk BW, Tsai JH. 1998. Biology and molecular biology of viruses in the genus Tenuivirus. 558
Annu. Rev. Phytopathol. 36:139-163. 559
3. Ramirez BC, Haenni AL. 1994. Molecular biology of tenuiviruses, a remarkable group of 560
plant viruses. J. Gen. Virol. 75 ( Pt 3):467-475. 561
4. Toriyama S, Takahashi M, Sano Y, Shimizu T, Ishihama A. 1994. Nucleotide sequence of 562
RNA 1, the largest genomic segment of rice stripe virus, the prototype of the tenuiviruses. J. 563
Gen. Virol. 75 ( Pt 12):3569-3579. 564
5. Takahashi M, Toriyama S, Hamamatsu C, Ishihama A. 1993. Nucleotide sequence and 565
possible ambisense coding strategy of rice stripe virus RNA segment 2. J. Gen. Virol. 566
74:769-773. 567
6. Xiong R, Wu J, Zhou Y, Zhou X. 2009. Characterization and subcellular localization of an 568
RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 387:29-40. 569
7. Kakutani T, Hayano Y, Hayashi T, Minobe Y. 1991. Ambisense segment 3 of rice stripe 570
virus: the first instance of a virus containing two ambisense segments. J. Gen. Virol. 72 ( Pt 571
2):465-468. 572
8. Zhu Y, Hayakawa T, Toriyama S, Takahashi M. 1991. Complete nucleotide sequence of 573
RNA 3 of rice stripe virus: an ambisense coding strategy. J. Gen. Virol. 72:763-767. 574
9. Xiong RY, Wu JX, Zhou YJ, Zhou XP. 2008. Identification of a Movement Protein of the 575
Tenuivirus Rice Stripe Virus. J. Virol. 82:12304-12311. 576
10. Elliott RM. 1990. Molecular biology of the Bunyaviridae. J Gen Virol 71 ( Pt 3):501-522. 577
11. Elliott RM. 1996. The Bunyaviridae. New York, NY: Plenum Press. 578
12. Rusu M, Bonneau R, Holbrook MR, Watowich SJ, Birmanns S, Wriggers W, Freiberg 579
AN. 2012. An assembly model of rift valley Fever virus. Front. Microbiol. 3:254. 580
13. Goldbach R, Peters D. 1996. Molecular and biological aspects of tospoviruses. In The 581
Bunyaviridae, pp. 129–157. Edited by R. M. Elliott. New York, NY: Plenum Press. 582
14. Elliott RM. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3:572-577. 583
15. Andersson AM, Melin L, Persson R, Raschperger E, Wikstrom L, Pettersson RF. 1997. 584
Processing and membrane topology of the spike proteins G1 and G2 of Uukuniemi virus. J. 585
Virol. 71:218-225. 586
16. Matsuoka Y, Chen SY, Holland CE, Compans RW. 1996. Molecular determinants of Golgi 587
retention in the Punta Toro virus G1 protein. Arch. Biochem. Biophys. 336:184-189. 588
17. Gerrard SR, Nichol ST. 2002. Characterization of the Golgi Retention Motif of Rift Valley 589
Fever Virus GN Glycoprotein. J. Virol. 76:12200-12210. 590
18. Ribeiro D, Foresti O, Denecke J, Wellink J, Goldbach R, Kormelink RJM. 2008. Tomato 591
spotted wilt virus glycoproteins induce the formation of endoplasmic reticulum- and 592
Golgi-derived pleomorphic membrane structures in plant cells. J. Gen. Virol. 89:1811-1818. 593
19. Tori y ama S. 1982. Characterization of Rice Stripe Virus: a Heavy Component Carrying 594
Infectivity. J. Gen. Virol. 61:187-195. 595
20. Tori y ama S. 1986. An Rna-Dependent Rna-Polymerase Associated with the Filamentous 596
Nucleoproteins of Rice Stripe Virus. J. Gen. Virol. 67:1247-1255. 597
21. Zhao S, Zhang G, Dai X, Hou Y, Li M, Liang J, Liang C. 2012. Processing and intracellular 598
localization of rice stripe virus Pc2 protein in insect cells. Virology 429:148-154. 599
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

27
22. Yao M, Zhang T, Zhou T, Zhou Y, Zhou X, Tao X. 2012. Repetitive prime-and-realign 600
mechanism converts short capped RNA leaders into longer ones that may be more suitable for 601
elongation during rice stripe virus transcription initiation. J. Gen. Virol. 93:194-202. 602
23. Pang SZ, DeBoer DL, Wan Y, Ye GB, Layton JG, Neher MK, Armstrong CL, Fry JE, 603
Hinchee MAW, Fromm ME. 1996. An improved green fluorescent protein gene as a vital 604
marker in plants. Plant Physiol. 112:893-900. 605
24. Nelson BK, Cai X, Nebenfuhr A. 2007. A multicolored set of in vivo organelle markers for 606
co-localization studies in Arabidopsis and other plants. Plant J. 51:1126-1136. 607
25. Wei T, Wang A. 2008. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus 608
replication occurs at endoplasmic reticulum exit sites in a COPI- and COPII-dependent 609
manner. J. Virol. 82:12252-12264. 610
26. Peremyslov VV, Pan YW, Dolja VV. 2004. Movement protein of a closterovirus is a type III 611
integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 612
78:3704-3709. 613
27. DaSilva LLP, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F. 2004. 614
Endoplasmic reticulum export sites and golgi bodies behave as single mobile secretory units 615
in plant cells. Plant Cell 16:1753-1771. 616
28. Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. 1998. Stacks on 617
tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15:441-447. 618
29. Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS. 2009. The cauliflower mosaic 619
virus protein P6 forms motile inclusions that traffic along actin microfilaments and stabilize 620
microtubules. Plant Physiol. 149:1005-1016. 621
30. Hanton SL, Chatre L, Renna L, Matheson LA, Brandizzi F. 2007. De novo formation of 622
plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. 623
Plant Physiol. 143:1640-1650. 624
31. Nebenfuhr A, Staehelin LA. 2001. Mobile factories: Golgi dynamics in plant cells. Trends 625
Plant Sci. 6:160-167. 626
32. Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A. 2000. A dominant negative 627
mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi 628
apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23:517-525. 629
33. Stefano G, Renna L, Chatre L, Hanton SL, Moreau P, Hawes C, Brandizzi F. 2006. In 630
tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum 631
and of ER export sites depends on active COPI machinery. Plant J. 46:95-110. 632
34. Lee MH, Min MK, Lee YJ, Jin JB, Shin DH, Kim DH, Lee KH, Hwang I. 2002. 633
ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and 634
maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 635
129:1507-1520. 636
35. Shi X, Lappin DF, Elliott RM. 2004. Mapping the Golgi Targeting and Retention Signal of 637
Bunyamwera Virus Glycoproteins. J. Virol. 78:10793-10802. 638
36. Ribeiro D, Goldbach R, Kormelink R. 2009. Requirements for ER-Arrest and Sequential 639
Exit to the Golgi of Tomato Spotted Wilt Virus Glycoproteins. Traffic 10:664-672. 640
37. Chen SY, Compans RW. 1991. Oligomerization, transport, and Golgi retention of Punta Toro 641
virus glycoproteins. J. Virol. 65:5902-5909. 642
38. Chen SY, Matsuoka Y, Compans RW. 1991. Golgi complex localization of the Punta Toro 643
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

28
virus G2 protein requires its association with the G1 protein. Virology 183:351-365. 644
39. Ruusala A, Persson R, Schmaljohn CS, Pettersson RF. 1992. Coexpression of the 645
membrane glycoproteins G1 and G2 of Hantaan virus is required for targeting to the Golgi 646
complex. Virology 186:53-64. 647
40. Shi X, Elliott RM. 2002. Golgi Localization of Hantaan Virus Glycoproteins Requires 648
Coexpression of G1 and G2. Virology 300:31-38. 649
41. Kikkert M, Van Lent J, Storms M, Bodegom P, Kormelink R, Goldbach R. 1999. Tomato 650
spotted wilt virus particle morphogenesis in plant cells. J. Virol. 73:2288-2297. 651
42. Sin SH. 2005. Viral genetic determinants for thrips transmission of Tomato spotted wilt virus. 652
Proc. Nat. Acad. Sci. 102:5168-5173. 653
43. Maia IG, Haenni A, Bernardi F. 1996. Potyviral HC-Pro: a multifunctional protein. J. Gen. 654
Virol. 77:1335-1341. 655
44. Ribeiro D, Foresti O, Denecke J, Wellink J, Goldbach R, Kormelink RJ. 2008. Tomato 656
spotted wilt virus glycoproteins induce the formation of endoplasmic reticulum- and 657
Golgi-derived pleomorphic membrane structures in plant cells. J. Gen. Virol. 89:1811-1818. 658
45. Hanton SL, Matheson LA, Brandizzi F. 2006. Seeking a way out: export of proteins from 659
the plant endoplasmic reticulum. Trends Plant Sci. 11:335-343. 660
46. Oufattole M, Park JH, Poxleitner M, Jiang L, Rogers JC. 2005. Selective membrane 661
protein internalization accompanies movement from the endoplasmic reticulum to the protein 662
storage vacuole pathway in Arabidopsis. Plant Cell 17:3066-3080. 663
47. Takahashi H, Saito Y, Kitagawa T, Morita S, Masumura T, Tanaka K. 2005. A novel 664
vesicle derived directly from endoplasmic reticulum is involved in the transport of vacuolar 665
storage proteins in rice endosperm. Plant Cell Physiol. 46:245-249. 666
48. Tormakangas K, Hadlington JL, Pimpl P, Hillmer S, Brandizzi F, Teeri TH, Denecke J. 667
2001. A vacuolar sorting domain may also influence the way in which proteins leave the 668
endoplasmic reticulum. Plant Cell 13:2021-2032. 669
49. Barlowe C. 2003. Signals for COPII-dependent export from the ER: what's the ticket out? 670
Trends Cell Biol. 13:295-300. 671
50. Overby AK, Popov VL, Pettersson RF, Neve EPA. 2007. The Cytoplasmic Tails of 672
Uukuniemi Virus (Bunyaviridae) GN and GC Glycoproteins Are Important for Intracellular 673
Targeting and the Budding of Virus-Like Particles. J. Virol. 81:11381-11391. 674
675
676
677
678
679
680
681
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

29
FIGURE LEGENDS: 682
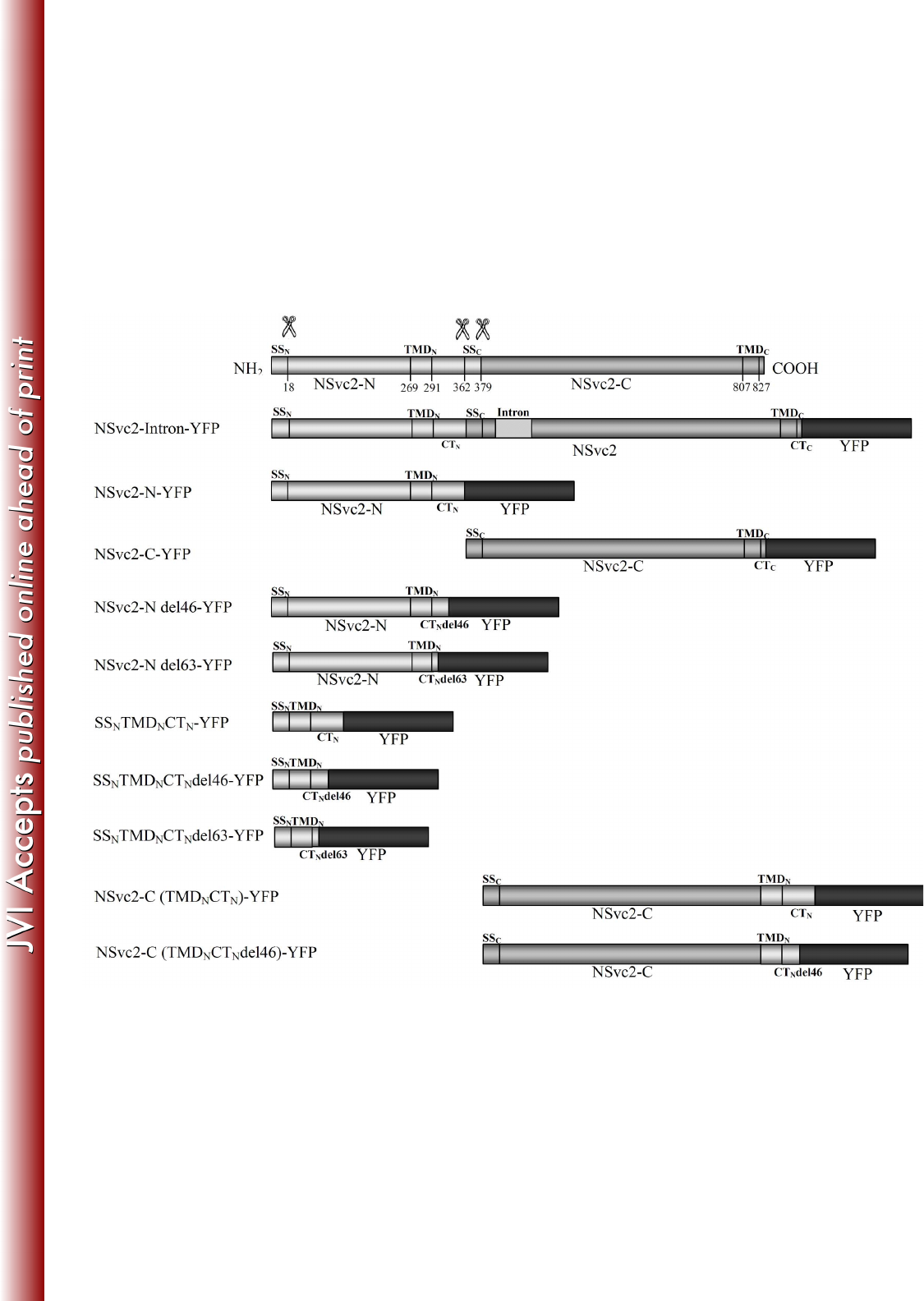
FIG 1 Schematic diagrams of the viral constructs used for expression analysis (the 683
glycoprotein constructs are aligned below the precursor). Predicted cleavage sites 684
(scissor symbols) and amino acid positions are indicated. SS, TMD and CT refer to 685
the signal sequence, the transmembrane domain and the cytosolic tail, respectively. 686
SSN and SSC refer to the SS of NSvc2-N and NSvc2-C, respectively. TMDN and 687
TMDC refer to the TMD of NSvc2-N and NSvc2-C, respectively. CTN and CTC refer 688
to the CT of NSvc2-N and NSvc2-C, respectively. An intron of the potato ST-LS1 689
was inserted at the nucleotide position of 1182 on NSvc2. In all constructs, the YFP 690
fluorophore was fused in frame at the site of the stop codon. 691
692
FIG 2 Subcellular localization of the NSvc2-N protein in Nicotiana benthamiana leaf 693
epidermal cells. (A) Immunoblot analysis of NSvc2-N-YFP fusion proteins expressed 694
by agroinfiltration in N. benthamiana leaves. The blots were probed using anti-YFP. 695
Empty vector (EV) was used as a negative control. Ponseau S was used as a loading 696
control. (B) Subcellular fractionation analysis of NSvc2-N-YFP fusion protein. The 697
soluble (S30) and microsomal (P30) fractions were isolated from agroinfiltrated 698
leaves of N. benthamiana. The membrane blots were probed using anti-YFP. (C-E) 699
The co-localization of the NSvc2-N-YFP (C) with the ER labeled by mCherry-HDEL 700
at 36 hpi (D). (E) Merged image of (C) and (D). (F-H) The co-localization of the 701
NSvc2-N-YFP (F) with the Golgi apparatus labeled by Man49-mCherry at 36 hpi (G). 702
The merged image illustrates the NSvc2-N protein targeted to the Golgi apparatus (H). 703
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

30
Scale bars, 20 μm. 704
705
FIG 3 Subcellular localization of the NSvc2-C protein in Nicotiana benthamiana leaf 706
epidermal cells. (A) Western blot analysis of NSvc2-C-YFP fusion proteins expressed 707
by agroinfiltration in N. benthamiana leaves. The blots were probed using anti-YFP. 708
Ponseau S was used as a loading control. Empty vector (EV) was used as a negative 709
control. (B) Subcellular distribution of NSvc2-C-YFP protein by fractionation 710
analysis. The soluble (S30) and microsomal (P30) fractions were isolated from 711
agroinfiltrated leaves of N. benthamiana. The membrane blots were probed using 712
anti-YFP. (C-E) The co-localization of the NSvc2-C-YFP (C) with the ER labeled by 713
mCherry-HDEL at 36 hpi (D). The merged image shows that NSvc2-C-YFP align 714
well with the ER membrane (E). (F-H) The co-localization of the NSvc2-C-YFP (F) 715
with the Golgi apparatus labeled by Man49-mCherry at 36 hpi (G). (H) Merged image 716
of (F) and (G). Scale bars, 20 μm. 717
718
FIG 4 Subcellular localization of NSvc2-YFP in Nicotiana benthamiana leaf 719
epidermal cells. (A) Immunoblot analysis of NSvc2-YFP fusion proteins (NSvc2-N 720
and NSvc2-C-YFP glycoproteins were processed from its common glycoprotein 721
precursor NSvc2-YFP) expressed by agroinfiltration in N. benthamiana leaves. The 722
membrane blots were probed using anti-YFP. Ponseau S was used as a loading control. 723
Empty vector (EV) was used as a negative control. (B) Subcellular distribution 724
analysis of NSvc2-YFP protein by fractionation. The soluble (S30) and microsomal 725
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

31
(P30) fractions were isolated from agroinfiltrated leaves of N. benthamiana. The 726
membrane blots were probed using anti-YFP. (C-E) Co-expression of the NSvc2-YFP 727
(NSvc2-C-YFP was processed from this glycoprotein precursor) (C) with 728
mCherry-HDEL (D) at 48 hpi. (E) Merged image of (C) and (D). (F-H) The 729
co-localization of the NSvc2-YFP (NSvc2-C-YFP was processed from the 730
glycoprotein precursor) (F) with the Golgi apparatus labeled by Man49-mCherry (G) 731
at 48 hpi. The merged image shows the NSvc2-C protein targeted to the Golgi 732
apparatus in the presence of NSvc2-N (H). Scale bars, 20 μm. 733
734
FIG 5 NSvc2 glycoproteins trafficking together with the Golgi stacks along the ER 735
track in Nicotiana benthamiana leaf epidermal cells. (A-C) Time-lapse confocal 736
images showing the movement of NSvc2-N-YFP (A) and the Golgi apparatus (B) 737
labeled by Man49-mCherry at the times indicated. The position of the tracked signal 738
is marked with an arrow. (C) Merged image of (A) and (B). (D-F) Time-lapse 739
confocal images showing the movement of NSvc2-YFP (NSvc2-N and NSvc2-C-YFP 740
were processed from the glycoprotein precursor NSvc2-YFP) (D) and the Golgi 741
apparatus (E) at the times indicated. The position of the tracked signal is marked with 742
an arrow. The merged images demonstrate that the NSvc2 proteins move together 743
with the Golgi apparatus along the ER track (F). Scale bars, 20 μm. 744
745
FIG 6 ER-to-Golgi targeting of RSV NSvc2 glycoproteins depends on a functional 746
COP II complex. (A-C) Confocal images of Nicotiana benthamiana epidermal cells 747
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

32
co-expressing NSvc2-N-YFP (A) and the COP II marker CFP-Sec24 at 36 hpi (B). (C) 748
Merged image of (A) and (B). The arrows mark co-localization of NSvc2-N-YFP 749
bodies with the ERES labeled with CFP-Sec24. (D-F) Co-expression of the 750
dominant-negative mutant Sar1 (H74L) causes the redistribution of NSvc2-N-YFP (D) 751
as well as the Golgi apparatus (E) back to the ER. (F) Merged image of (D) and (E). 752
(G-I) Cells co-expressing NSvc2-N and NSvc2-C-YFP (from their common precursor 753
NSvc2- YFP) (G) and the ERES labeled with CFP-Sec24 at 48 hpi (H). (I) Merged 754
image of (G) and (I). (J-L) Co-expression of the dominant-negative mutant Sar1 755
(H74L) inhibits the transport of NSvc2-N and NSvc2-C-YFP (co-expressed from their 756
common precursor NSvc2-YFP) to the Golgi complex. Scale bars, 20 μm. 757
758
FIG 7 ER-to-Golgi targeting of RSV NSvc2 glycoproteins depends on an active COP 759
I complex. (A-C) Confocal images of Nicotiana benthamiana epidermal cells 760
co-expressing NSvc2-N-YFP (A) and the COP I marker labeled with Arf1-CFP at 36 761
hpi (B). (C) Merged image of (A) and (B). (D-F) Co-expression of the 762
dominant-negative mutant Arf1 (T31N) led to the retention of NSvc2-N-YFP (D) as 763
well as the Golgi apparatus (E) in the ER at 48 hpi. (F) Merged image of (D) and (E). 764
(G-I) Cells co-expressing NSvc2-YFP (G) and the COP I marker labeled with 765
Arf1-CFP (H). (I) Merged image of (G) and (H). (J-L) Co-expression of the 766
dominant-negative Arf1 (T31N) blocks transport of NSvc2-N and NSvc2-C-YFP 767
(co-expressed from their common precursor NSvc2-YFP) to the Golgi complex. Scale 768
bars, 20 μm. 769
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from

33
770
FIG 8 Golgi targeting signal analysis of truncated and chimeric NSvc2-N proteins. 771
(A-U) Confocal images of Nicotiana benthamiana epidermal cells co-expressing 772
Man49-mCherry with the truncated or chimeric proteins NSvc2-N del46-YFP (A-C), 773
NSvc2-N del63-YFP (D-F), SSNTMDNCTN-YFP (G-I), SSnTMDN-CTNdel46-YFP 774
(J-L), SSNTMDNCTNdel63-YFP (M-O), NSvc2-C(TMDNCTN)-YFP (P-R), and 775
NSvc2-C(TMDNCTNdel46)-YFP (S-U), respectively, at 48 hpi. Scale bars, 20 μm. 776
777
778
779
780
781
782
783
on March 4, 2018 by guesthttp://jvi.asm.org/Downloaded from