Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 5

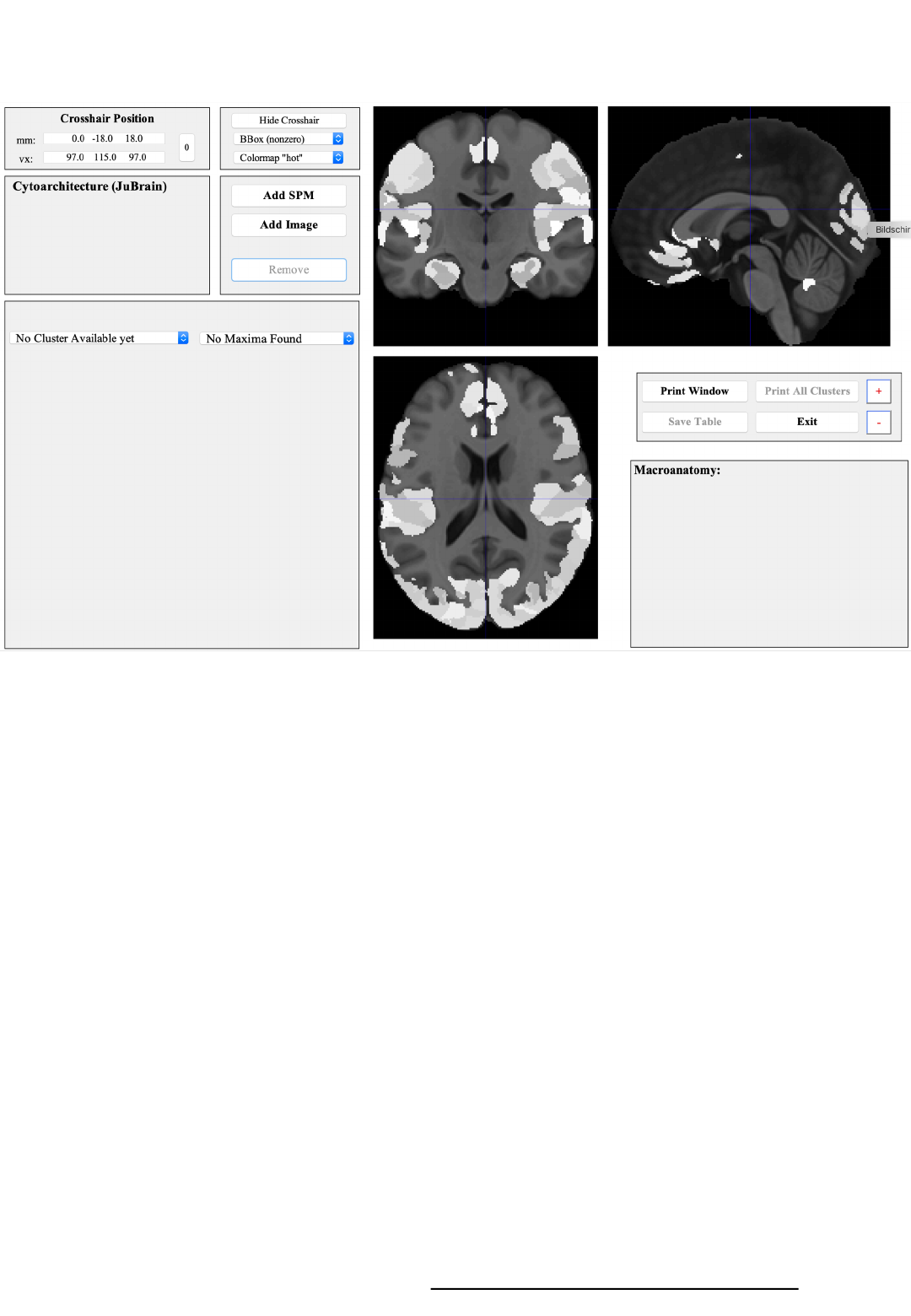
Atlas & Assignment Tool
1 2
3
5 4
6
7
8
1) Coordinate panel
Showing and changing the crosshair position
2) Figure settings
Modifying the display of the orthogonal sections
3) Orthogonal sections
Showing the MNI152 template in the background
with the JuBrain Maximum Probability Map in lighter greyx
5) JuBrain Voxel Assignment
Cytoarchitectonic information for the crosshair location
6) JuBrain Cluster assignment
Cytoarchitectonic information per overlay-cluster
+ Navigation through the overlay (SPM map, image)
8) Macroanatomical panel
Assignment of the crosshair position and the current overlay cluster
(if applicable) to the Harvard-Oxford macroanatomical atlas
4). Overlay controls
Load or remove an SPM map or an image overlay
7) Control panel
Various controls including the change of font size (+ / -buttons)
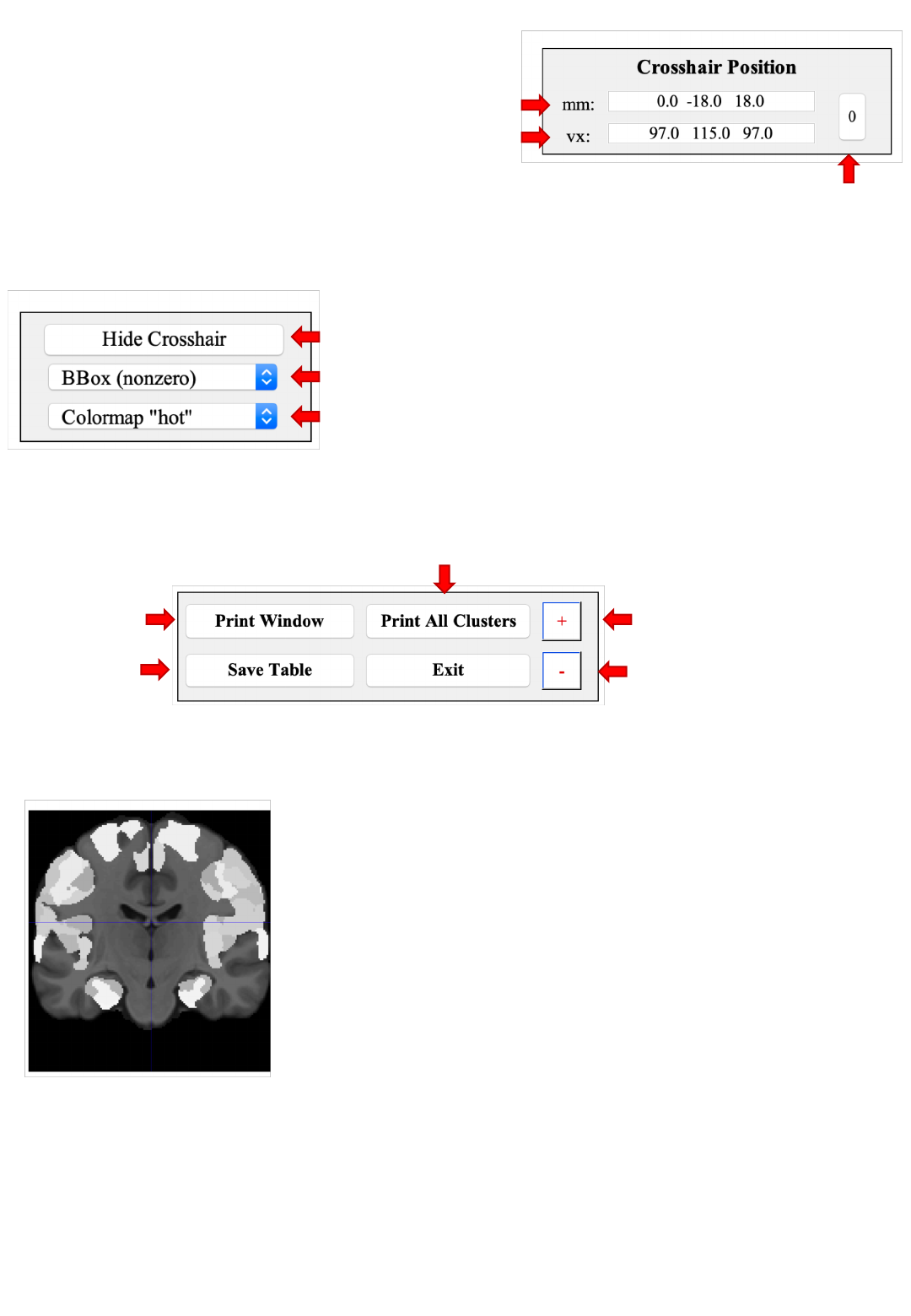
Coordinate panel
Cross-hair position in MNI152 (world) space
Cross-hair position in MPM voxel space
Move cross-hair
to origin
Changing the coordinates (confirmed by ↩)
moves the cross-hair and updates the voxel assignment
Toggle cross-hairs on / off (not affecting behavior)
Zoom in/our focused on the cross-hair location
Change the colormap of an overlay
(not affecting the atlas & template display)
Figure settings
Orthogonal sections
Overlays, e.g., findings from functional or structural imaging studies may be
overlayed on this map and are shown using the color-scale defined in the
figure settings panel.
Clicking on any of the three sections changes the cross-hair position to that
location. In this process, the coordinate panel, the JuBrain voxel assignment
and the macroanatomical information is updated.
In dark grey, the (non-linear) MNI152 template is
shown, light grey represents the assignment to
histological areas based on the JuBrain Maximum
Probability Map. This map assigns each voxel to the
most likely histological area at that position.
Control panel Cycle through all clusters and save a screen-shot
Increase font size
Decrease font size
Screen-
shot
Save
JuBrain
assignment for all clusters as tsv
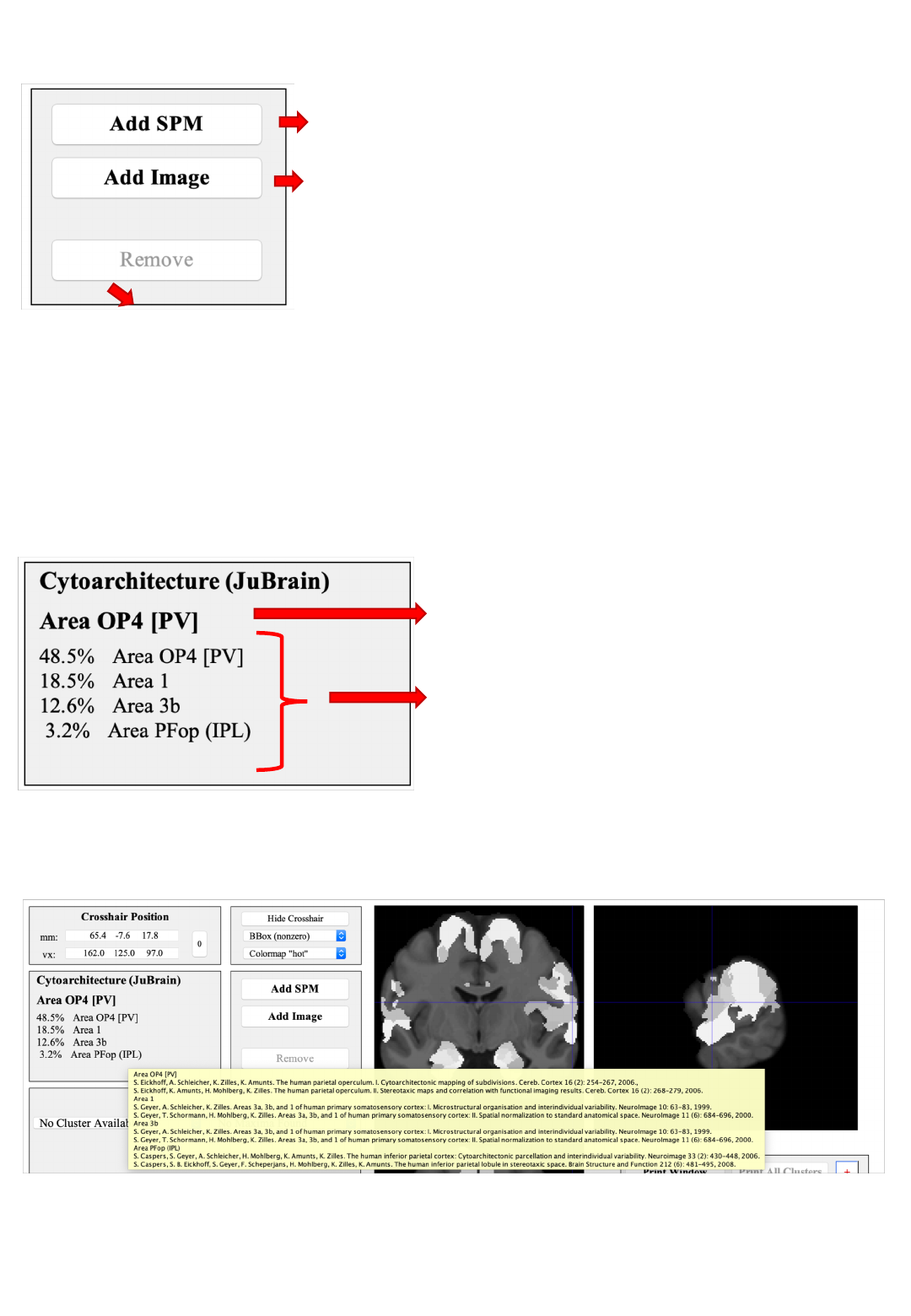
Overlay controls
Bring up the SPM contrast manager, allowing
you to define, evaluate and threshold contrasts
Remove the current overlay (only one overlay can be shown at any time)
Add an overlay image from a nifti-file.
Premultiply: Multiply all image values by a scalar
Hight threshold: Evaluated after pre-multiplication
Extent threshold: In voxels (native image space)
Overlays need to be in MNI152 space and are resampled to the template
resolution (1 mm isotropic)
JuBrain Voxel Assignment
“Hard” assignment based on the
Maximum Probability Map (MPM)
Full description: probabilities for all
histological area found at this position
Information in this panel pertain (only) to the voxel at the cross-hair position
Pointing the cursor on the name of a cytoarchitectonic area and keeping it there
for ~1 second reveals the references describing the cytoarchitectonic mapping
of this histological area.
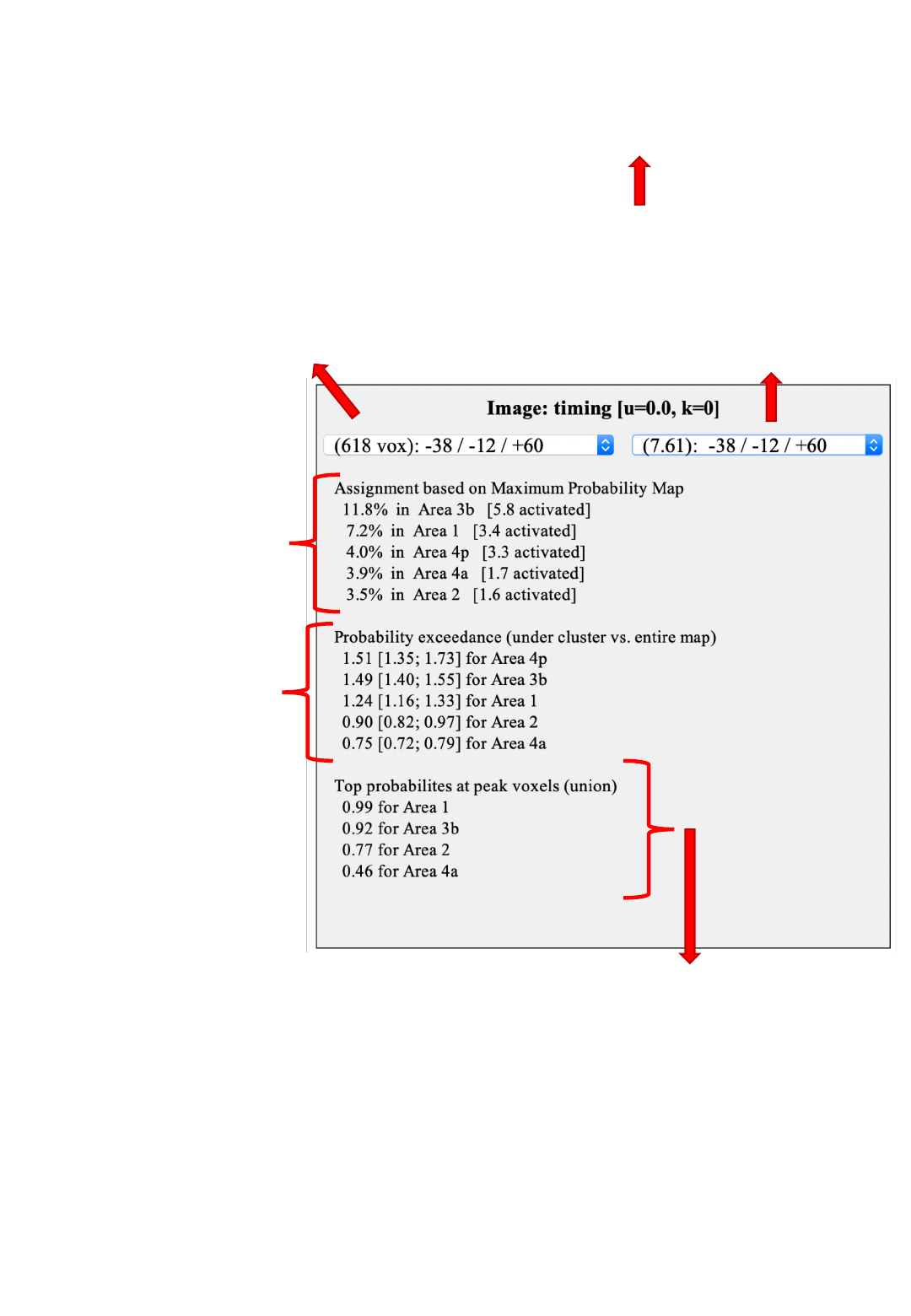
JuBrain Cluster assignment
Use this menu to jump to the
different clusters.
Coordinates indicate the location
of the cluster maximum
Use this menu to jump to the
different peaks (local maxima)
within a cluster.
Values in parenthesis indicate the
overlay value (e.g., T / Z statistic).
Comparison between
the current cluster and
the JuBrain MPM
Overlap is provided
relative to cluster and
area (in brackets) volume
(Eickhoff et al., 2005)
Average probability for
each JuBrain area
at the location of the
cluster relative to its full
probability map
Higher values indicate a
location more towards the
center of the area (cf.
Eickhoff et al., 2007)
This will update the JuBrain
Voxel Assignment panel for
information on maxima location
Union of the probability values for each area at the
location of the local maxima
High values indicate that at least one peak was most likely
located in the respective area give histological variability
The Macroanatomical panel follows the same layout
It is concurrently updated when a new cluster / maximum is selected