Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 22
SomaticSeq Documentation
Li Tai Fang / li_tai.fang@roche.com
January 21, 2019
Contents
1 Introduction 2
1.1 Dependencies ............................................. 2
1.2 Dockerimages ............................................ 2
2 How to run SomaticSeq 3
2.1 SomaticSeqTrainingMode ..................................... 3
2.2 SomaticSeqPredictionMode .................................... 4
2.3 ConsensusMode ........................................... 4
3 Use SomaticSeq as a Python library 5
3.1 somaticseq.somaticseq modules . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 5
3.1.1 Module:run_somaticseq .................................. 5
3.1.2 Module: somatic_vcf2tsv and single_sample_vcf2tsv . . . . . . . . . . . . . . . . . . 6
3.1.3 Module:SSeq_tsv2vcf.................................... 7
3.2 somaticseq.utilitiesmodules..................................... 7
3.2.1 Module: split_Bed_into_equal_regions . . . . . . . . . . . . . . . . . . . . . . . . . . 7
3.2.2 Module: lociCounterWithLabels . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
4 The step-by-step SomaticSeq Workow 7
4.1 Apply inclusion and exclusion regions . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 7
4.2 Combinethecallsets......................................... 8
4.3 Convert the VCF le into TSV le . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
4.4 Model Training or Mutation Prediction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
5 To run the dockerized somatic mutation callers 10
5.1 Location................................................ 10
5.2 Requirements............................................. 10
5.3 Examplecommands ......................................... 10
5.3.1 Single-threadedJobs..................................... 10
5.3.2 Multi-threadedJobs ..................................... 10
5.3.3 SomaticSeqTraining..................................... 11
5.3.4 SomaticSeqPrediction.................................... 11
5.3.5 Parameters .......................................... 11
5.3.6 What does the single-threaded command do . . . . . . . . . . . . . . . . . . . . . . . . 12
5.3.7 What does the multi-threaded command do . . . . . . . . . . . . . . . . . . . . . . . . 13
6 Use BAMSurgeon to create training data 13
6.1 Requirements............................................. 14
6.2 Three scenario to simulate somatic mutations . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
6.2.1 When you have sequencing replicates of normal samples . . . . . . . . . . . . . . . . . 14
6.2.2 This example mimicks DREAM Challenge . . . . . . . . . . . . . . . . . . . . . . . . . 15
6.2.3 Merge and then split the input tumor and normal BAM les . . . . . . . . . . . . . . 15
1
6.3 ParametersandOptions....................................... 16
6.3.1 –merge-bam / –split-bam / –indel-realign . . . . . . . . . . . . . . . . . . . . . . . . . 17
6.4 To create SomaticSeq classiers . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
7 Release Notes 17
8 Contact Us 22
1 Introduction
SomaticSeq is a exible post-somatic-mutation-calling algorithm for improved accuracy. It is compatible
with 10+ somatic mutation caller(s). Any combination of them can be used to obtain a combined call set
with sequencing features extracted into TSV and VCF les. In addition, SomaticSeq uses machine learn-
ing (adaptive boosting) to distinguish true mutations from false positives from that call set. The mutation
callers we have incorporated are MuTect/Indelocator/MuTect2, VarScan2, JointSNVMix, SomaticSniper,
VarDict, MuSE, LoFreq, Scalpel, Strelka, and TNscope. You may incorporate some or all of those callers
into your own pipeline with SomaticSeq.
The manuscript, An ensemble approach to accurately detect somatic mutations using Somat-
icSeq, was published in Genome Biology 2015, 16:197. The SomaticSeq project is located at
https://github.com/bioinform/somaticseq. There have been some major improvements in SomaticSeq since
that Genome Biology publication in 2015.
The wrapper script somaticseq/run_somaticseq.py and its parallelized cousin somaticseq_parallel.py can
1) train the call set into a classier, 2) predict high-condence somatic mutations from the call set based
on a pre-dened classier, or 3) default to consensus mode, i.e., extract sequencing features and output the
TSV and VCF les, and then label the calls (i.e., PASS, LowQual, or REJECT) based on majority vote of
the tools.
1.1 Dependencies
• Python 3, plus pysam (v0.14.1), numpy (v1.14.3), and scipy (v1.1.0). The versions in parentheses are
in our docker images and validated to work, though other versions should work, too.
• R, plus the ada package in R.
• BEDTools (if inclusion and/or an exclusion region les are supplied, and/or running somatic-
seq_parallel.py instead of somaticseq/run_somaticseq.py)
• Optional: dbSNP in VCF format (if you want to use dbSNP membership as a part of the training).
• At least one of MuTect/Indelocator/MuTect2, VarScan2, JointSNVMix2, SomaticSniper, VarDict,
MuSE, LoFreq, Scalpel, Strelka2, TNscope, and/or Platypus. Those are the tools we have incorpo-
rated in SomaticSeq. If there are other somatic tools that may be good addition to our list, please
make the suggestion to us.
1.2 Docker images
SomaticSeq and the somatic mutation callers that we routinely use were dockerized.
• SomaticSeq: https://hub.docker.com/r/lethalfang/somaticseq
• MuTect2: https://hub.docker.com/r/broadinstitute/gatk
• VarScan2: https://hub.docker.com/r/djordjeklisic/sbg-varscan2
• JointSNVMix2: https://hub.docker.com/r/lethalfang/jointsnvmix2
• SomaticSniper: https://hub.docker.com/r/lethalfang/somaticsniper
2

• VarDict: https://hub.docker.com/r/lethalfang/vardictjava
• MuSE: https://hub.docker.com/r/marghoob/muse
• LoFreq: https://hub.docker.com/r/marghoob/lofreq
• Scalpel: https://hub.docker.com/r/lethalfang/scalpel
• Strelka2: https://hub.docker.com/r/lethalfang/strelka
2 How to run SomaticSeq
The somaticseq/run_somaticseq.py calls a series of programs and procedures after you have run your in-
dividual somatic mutation callers, and somaticseq_parallel.py is a wrapper script that allows parallel pro-
cessing. Section 5 will teach you how to run those mutation callers that we have been dockerized. It also
includes ways to create semi-simulated training data that can be used to create SomaticSeq classiers. In
the next section, we will describe the workow in this wrapper script in detail.
Both paired and single modes are supported, although single mode is not as well validated scientically as
the paired mode. To see the required and optional input les and parameters to somaticseq_parallel.py:
1# See the g l oba l input parameters
somaticseq_parallel .py −−help
3
# Input par amete rs f o r p aired−sample mode ( i . e . , tumor−normal )
5somaticseq_parallel .py paired −−help
7# Inp ut par ame ter s f o r s i n g l e −sample mode
somaticseq_parallel .py single −−help
2.1 SomaticSeq Training Mode
To create SomaticSeq classiers, you need a VCF le containing true SNVs and a VCF le containing true
INDELs. There is also an option to include a list of regions to include and/or exclude from this exercise.
The exclusion or inclusion regions can be VCF or BED les. An inclusion region may be subset of the call
sets where you have validated their true/false mutation status, so that only those regions will be used for
training. An exclusion region can be regions where the “truth” is ambigious. All the variants in the truth
VCF les are assumed to be true positives. Every mutation call not in the truth VCF les is assumed to
be false positives (as long as the genomic coordiante is in inclusion region and not in exclusion region if
those regions are provided).
All the output VCF les from individual callers are optional. Those VCF les can be bgzipped if they
have .vcf.gz extensions. It is imperative that you will use the same parameter for prediction as you do for
training.
# An example command f o r SomaticSeq T ra ining .
2somaticseq_parallel .py \
−−somaticseq−train \
4−−output−d i r e c t o r y $OUTPUT_DIR \
−−genome−r e f e r e n c e GRCh38. f a \
6−−truth−snv t r u e P o s i t i v e s . snv . vc f \
−−truth−i n d e l t r u e P o s i t i v e s . i n d e l . vc f \
8−− inclusion−reg io n genome . bed \
−− exclusion−r eg ion b l a c k l i s t . bed \
10 −− threads 12 \
paired \
12 −−tumor−bam−f i l e tumor .bam \
−−normal−bam−f i l e matched_normal .bam \
14 −−mutect2−vc f MuTect2/ v ar ian ts . vcf \
−−varscan−snv VarScan2/ va r i an t s . snp . vc f \
16 −−varscan−i n d e l VarScan2/ v a r i a n ts . i n d e l . v c f \
3

−−jsm−v cf JointSNVMix2/ v ar ia nt s . snp . vc f \
18 −− somaticsniper−v cf SomaticSniper / var ia nt s . snp . vc f \
−− v ard ict−vc f VarDict/ v ar ian ts . v cf \
20 −−muse−vc f MuSE/ va ri an ts . snp . v cf \
−− lofreq−snv LoFreq/ va ri an ts . snp . v cf \
22 −− lofreq−i n d e l LoFreq/ v a r i a nt s . i n d e l . v cf \
−− scalpel−vc f S ca l p e l / v a r i a nt s . i n d e l . v c f \
24 −− strelka−snv St re lk a / var ia nt s . snv . v cf \
−− strelka−i n d e l S t re l k a / v a r i a nt s . i n d e l . v c f
For the command’s argument placement, caller output and bam les are input “after” paired or single op-
tion. Everything else goes before, e.g., reference, ground truths, resources such as dbSNP and COSMIC,
etc.
Parallel processing is achieved by splitting the inclusion BED le into a number of sub-BED les of equal
region sizes, named 1.th.input.bed, 2.th.input.bed, ..., n.th.input.bed. Then each process will be run using
each sub-BED le as the inclusion BED le. If there is no inclusion BED le in the command argument, it
will split the reference.fa.fai le instead.
SomaticSeq supports any combination of the somatic mutation callers we have incorporated into the work-
ow. SomaticSeq will run based on the output VCFs you have provided. It will train for SNV and/or
INDEL if you provide the truePositives.snv.vcf and/or truePositives.indel.vcf le(s) and invoke the
--somaticseq-train option. Otherwise, it will fall back to the simple caller consensus mode.
2.2 SomaticSeq Prediction Mode
Make sure the classiers (.RData les) are supplied, Without either of them, or it will fall back to the sim-
ple caller consensus mode.
1# The ∗. RData f i l e s ar e t r a in e d c l a s s i f i e r from th e t r a i n i n g mode .
somaticseq_parallel .py \
3−− classifier−snv Ensemble . sSNV . t sv . ntChange . C l a s s i f i e r . RData \
−− classifier−i ndel Ensemble . sINDEL. ts v . ntChange . C l a s s i f i e r . RData \
5−−output−d i r e c t o r y $OUTPUT_DIR \
−−genome−r e f e r e n c e GRCh38. f a \
7−− inclusion−reg io n genome . bed \
−− exclusion−r eg ion b l a c k l i s t . bed \
9−− threads 12 \
paired \
11 −−tumor−bam−f i l e tumor .bam \
−−normal−bam−f i l e matched_normal .bam \
13 −−mutect2−vc f MuTect2/ v ar ian ts . v cf \
−−varscan−snv VarScan2/ va r i an t s . snp . vc f \
15 −−varscan−i n d e l VarScan2/ v a r i a n ts . i n d e l . v c f \
−−jsm−v cf JointSNVMix2/ v ar ia nt s . snp . vc f \
17 −− somaticsniper−v cf SomaticSniper / var ia nt s . snp . vc f \
−− v ard ict−vc f VarDict/ v ar ian ts . v cf \
19 −−muse−vc f MuSE/ va ri an ts . snp . v cf \
−− lofreq−snv LoFreq/ va ri an ts . snp . v cf \
21 −− lofreq−i n d e l LoFreq/ v a r i a nt s . i n d e l . v cf \
−− scalpel−vc f S ca l p e l / v a r i a nt s . i n d e l . v c f \
23 −− strelka−snv St re lk a / var ia nt s . snv . v cf \
−− strelka−i n d e l S t re l k a / v a r i a nt s . i n d e l . v c f
2.3 Consensus Mode
Same as the commands previously, but without including classiers or invoking –somaticseq-train. With-
out those information, SomaticSeq will forgo machine learning, and fall back into a simple majority vote.
4

3 Use SomaticSeq as a Python library
Section 2 described how to use SomaticSeq as a standalone software, but SomaticSeq can also be treated
as a python library for your own software.
3.1 somaticseq.somaticseq modules
The directory somaticseq/somaticseq contain some of the critical modules of SomaticSeq, and many can be
used as a function of a library.
3.1.1 Module: run_somaticseq
The script somaticseq/somaticseq/run_somaticseq.py contains the module to convert individual VCF les
(each from a popular somatic mutation caller) to SomaticSeq TSV and VCF les.
The code to produce the .TSV and .VCF les described in Section 2, for example, would be something like
this:
2# Module i s l oc a te d s om at ic se q / so ma ti cse q /run_somaticseq . py
import somaticseq . somaticseq . run_somaticseq as run_somaticseq
4
run_somaticseq . runPaired ( o ut di r=’/PATH/TO/SomaticSeq ’ , r e f =’/PATH/TO/GRCh38. fa ’ , tbam=’/PATH/TO
/tumor . bwa .bam’ , nbam=’/PATH/TO/ normal . bwa . bam’ , tumor_name=’TUMOR’ , normal_name=’NORMAL’ ,
truth_snv=None , t ru th_ ind el=None , c l a s s i f i e r _ s n v=None , c l a s s i f i e r _ i n d e l=None , p ass_thre shold
=0.5 , lowqual_threshold =0.1 , hom_threshold =0.85 , het_ threshold =0.01 , dbsnp=’/PATH/TO/
dbSNP_138. hg38 . v c f . v cf ’ , cosmic =’/PATH/TO/COSMIC. v85 . vcf ’ , i n c l u s i o n =’/PATH/TO/Exon_Capture .
bed ’ , ex cl usi on =’/PATH/TO/ i gn or e . bed ’ , mutect=None , i nd el oc at or=None , mutect2=’/PATH/TO/
MuTect2 . vcf ’ , varscan_snv=None , varscan_indel=None , jsm=None , sn ip er=None , va rdi ct =’/PATH/TO/
VarDict . vcf ’ , muse=’/PATH/TO/MuSE. vcf ’ , lofreq_snv =’/PATH/TO/LoFreq . snv . v cf . gz ’ , lo f r e q _in d e l
=’/PATH/TO/LoFreq . i nd e l . v cf . gz ’ , s c a lp e l=None , stre lka_sn v=’/PATH/TO/ S tr el ka / r e s u l t s / v ar ia nt s
/somatic_ssnv . v cf . gz ’ , s tr el ka _i nd el =’/PATH/TO/ S tr elk a / r e s u l t s / va ri an ts / so matic_s indel . vc f . gz
’ , tnscope=None , platypus=None , min_mq=1, min_bq=5, min_caller =0.5 , somaticseq_train=False ,
ense mbleOut Prefix =’Ensemble . ’ , c on sensusO ut Prefix =’Consensus . ’ , c l a s s i f i e d O u t P r e f i x =’SSeq .
C l a s s i f i e d . ’ , keep_ inte rmed iate s=Fa ls e )
The parameters of ensembleOutPrex,consensusOutPrex, and classiedOutPrex will dictate the output
le names under outdir.
Likewise, the single sample mode to convert various individual VCF outputs would be something like this:
2import somaticseq . somaticseq . run_somaticseq as run_somaticseq
4run_somaticseq . run Si ng le ( o ut di r=’/PATH/TO/SomaticSeq ’ , r e f =’/PATH/TO/GRCh38. f a ’ , bam=’/PATH/TO/
tumor . bwa . bam’ , tumor_name=’TUMOR’ , truth_snv=None , tr uth_ ind el=None , c l a s s i f i e r _ s n v=None ,
c l a s s i f i e r _ i n d e l=None , pass_threshold =0.5 , lowqual_threshold =0.1 , hom_threshold =0.85 ,
het_threshold =0.01 , dbsnp=’/PATH/TO/dbSNP_138 . hg38 . vc f . vcf ’ , cosmic=’/PATH/TO/COSMIC. v85 . vcf
’ , i n c l u s i o n =’/PATH/TO/Exon_Capture . bed ’ , e xc l us i on =’/PATH/TO/ i g no r e . bed ’ , mutect=None ,
mutect2=’/PATH/TO/MuTect2 . v cf ’ , varscan=None , v ar d ic t =’/PATH/TO/VarDict . v cf ’ , l o f r e q =’/PATH/
TO/LoFreq . vcf ’ , s c a l p e l=None , s t r e l k a =’/PATH/TO/ S tr e lk a . v cf ’ , min_mq=1, min_bq=5, min_cal ler
=0.5 , somaticseq_train=False , ensembleOutPrefix=’Ensemble . ’ , consensusOutPrefix =’Consensus . ’ ,
c l a s s i f i e d O u t P r e f i x =’SSeq . C l a s s i f i e d . ’ , k eep_int ermedia te s=F al se )
Parameters:
• truth_snv/truth_indel: if present, then the variants in these VCF les will be considered true posi-
tives, and everything else will be considered false positive. If None, then nothing with regard to true
positive or false positive will be annotated.
• classier_snv/classier_indel: if present, then SomaticSeq prediction will be invoked to create ma-
chine learning classied VCF les. if None, only majority-vote consensus VCF les will be created.
5

• inclusion: bed le so only variants in it will be considered (requires BEDTools on execution path)
• exclusion: bed le so variants in it will be tossed out (requires BEDTools on the execution path)
• mutect/mutect2/varscan/jsm/vardict/muse/lofreq/strelka/scalpel/tnscope: output VCF les from
the callers. If None, then it assumes that tool was not used.
• min_caller: only output variants if at least N number of callers have called it. Since some LowQual
calls are considered 0.5, an input of 0.5 tells the function to also return variants even if it’s only
been called as a “LowQual” by a tool. However, it will still lter out variants that’s only been “RE-
JECTED” by a caller.
somaticseq_train: if True, and also if truth_snv or truth_indel are present, then it will create So-
maticSeq classiers. If False, then will not invoke training mode.
3.1.2 Module: somatic_vcf2tsv and single_sample_vcf2tsv
Another useful module is the command to extract SomaticSeq features for variants in any VCF le, and
output the results to a TSV le. The following function requires both tumor and normal BAM les, and
the reference genome. COSMIC, dbSNP, etc. are optional. None for any null inputs. min_mq = 0 for
this purpose. This is a lter to only output variants that has been called by a minimum number of tools
(which you may specify as VCF inputs such as mutect, varscan, etc.)
1
import somaticseq . somaticseq . somatic_vcf2tsv as somatic_vcf2tsv
3
somatic_vcf2tsv . v cf 2t sv ( is_ vc f=’/PATH/TO/ va ria nt s . vcf ’ , is_bed=None , is_pos=None , nbam_fn=’/
PATH/TO/normal .bam’ , tbam_fn=’/PATH/TO/tumor .bam’ , truth=None , cosmic=’/PATH/TO/COSMIC. v85 .
vcf ’ , dbsnp=’/PATH/TO/dbSNP_138 . hg38 . vcf . vcf ’ , mutect=None , varscan=None , jsm=None , snip er=
None , v ar dic t=None , muse=None , l o f re q=None , s c al p e l=None , s t re l ka=None , tnscope=None ,
platypus=None , dedup=True , min_mq=1, min_bq=5, m in_call er =0, r ef _f a =’/PATH/TO/GRCh38 . fa ’ ,
p_scale=None , o u t f i l e =’/PATH/TO/SomaticSeq . FeaturesExtracted . tsv ’ )
You may also extract sequencing info for any VCF le if you just have one bam le
1
import somaticseq . somaticseq . single_sample_vcf2tsv as single_sample_vcf2tsv
3
single_sample_vcf2tsv . vc f2 tsv ( is _vcf =’/PATH/TO/ va ria nt s . vcf ’ , is_bed=None , is_pos=None , bam_fn
=’/PATH/TO/tumor .bam’ , truth=None , cosmic=’/PATH/TO/COSMIC. v85 . vcf ’ , dbsnp=’/PATH/TO/
dbSNP_138. hg38 . vcf . vcf ’ , mutect=None , varscan=None , v ar dic t=None , muse=None , l o f re q=None ,
s c a l p e l=None , s t r e l k a=None , dedup=True , min_mq=1, min_bq=5, min_c aller =0, r ef _f a =’/PATH/TO/
GRCh38. fa ’ , p_scale=None , o u t f i l e =’/PATH/TO/SomaticSeq . FeaturesExtracted . tsv ’ )
Both somaticseq/somaticseq/somatic_vcf2tsv.py and somaticseq/somaticseq/single_sample_vcf2tsv.py
may also be run as standalone scripts. Invoke the script with -h to learn their usages.
Parameters:
• is_vcf: the VCF le serves as the input le, from which every variant will have its sequencing feature
extracted from the BAM le(s).
• mutect/varscan/jsm/sniper/vardict/muse/lofreq/scalpel/strelka/tnscope: VCF les from these tools.
If present, the function will extract information from these les such as if a variant is called by the
tool. If None, everything associated with that tool will be “nan” in the TSV le.
6

3.1.3 Module: SSeq_tsv2vcf
This module converts SomaticSeq’s TSV le (described in Sec. 3.1.2) to SomaticSeq VCF les.
1
import somaticseq . somaticseq . SSeq_tsv2vcf as SSeq_tsv2vcf
3
SSeq_tsv2vcf . t s v2 v c f ( tsv_fn =’/PATH/TO/ SomaticSeq . tsv ’ , vcf_fn =’/PATH/TO/SomaticSeq . vcf ’ , t o o l s
=[ ’MuTect2 ’ , ’ SomaticSniper ’ , ’ Str elk a ’ ] , pass_score =0.5 , lowqual_score =0.1 , hom_threshold
=0.85 , het _th reshold =0.01 , single_mode=False , paired_mode=True , normal_sample_name=’NORMAL’ ,
tumor_sample_name=’TUMOR’ , p r i n t _ r e je c t=True , phred _sca led=True )
Parameters:
• tools: A list of tools that were run, can only be selected from MuTect2, MuTect, VarScan2,
JointSNVMix2, SomaticSniper, VarDict, MuSE, LoFreq, Scalpel, Strelka, TNscope, and/or Platypus.
• print_reject: if False, will only print PASS and LowQual variants into VCF. If True, will print ev-
erything from TSV to VCF.
• phred_scaled: if True, will print Phred-scaled score in QUAL column (if the TSV was produced with
SomaticSeq prediction). If False, will print the 0-1 scale. If no SomaticSeq prediction was done, will
print 0.
3.2 somaticseq.utilities modules
3.2.1 Module: split_Bed_into_equal_regions
Given a .bed or a .fa.fai le, it will split the input region into N number of bed les, such that each bed
le has equal-sized regions in them.
3.2.2 Module: lociCounterWithLabels
Given a list of .bed les and a .fa.fai le, it will return a .bed le detailing which regions were contained
from which .bed inputs.
1
somaticseq / u t i l i t i e s / lociCounterWithLabels . py −f a i GRCh38. fa . f a i −beds 1 . bed 2 . bed 3 . bed −
l a b e l s 01 02 03 −out overlapping .bed
Parameters:
• labels: A list of labels to be written in 4th column of the output bed le. If absent, the 4th column
will be populated by the input bed le names.
4 The step-by-step SomaticSeq Workow
We’ll describe the workow here.
4.1 Apply inclusion and exclusion regions
This step may be needed for model training. BEDTools is invoked by SomaticSeq. An inclusion region
means we will only use calls inside these regions. An exclusion region means we do not care about calls
inside this region. DREAM Challenge had exclusion regions, e.g., blacklisted regions, etc. It is also a rou-
tine used to parallelize the process by splitting large regions into equal-sized (in terms of number of pairs)
regions, so that they can be processed in parallel.
7
4.2 Combine the call sets
We use vcfModier/getUniqueVcfPositions.py and bedtools sort to combine the VCF les from dierent
callers. For each caller output, intermediate VCF le(s) may be created to separate the SNVs and INDELs
calls, and also remove some REJECT calls to reduce le sizes.
The following scripts are used to modify original VCF outputs.
1vc fModi fier /modify_JointSNVMix2 . py
vc fM od if ie r /modify_MuTect2 . py
3vc fM od if ie r /modify_MuTect . py
vc fM od if ie r /modify_SomaticSniper . py
5vc fM od if ie r /modify_ssMuTect2 . py
vc fM od if ie r /modify _ssStre lka . py
7vc fM od if ie r /modify_Strelka . py
vc fM od if ie r /modify_VarDict . py
9vc fM od if ie r /modify_VarScan2 . py
modify_ssTOOL.py denotes it’s for single-sample mode.
JointSNVMix2 does not output VCF les. In our own workow, we convert its out-
put into a basic VCF le with an 2 awk one-liners, which you may see at utilities/dock-
ered_pipelines/mutation_callers/submit_JointSNVMix2.sh.
1# To avoid te xt f i l e s on the or der o f te rab yte s , t h i s awk one−l i n e r ke eps e n t r i e s where the
r ef e re n c e i s not ”N” , and the somatic p r o b a b i l i t i e s are at l e a s t 0 .9 5 .
awk −F” \ t ” ’NR!=1 && $4!=”N” && $10+$11 >=0.95’
3
# This awk one−l i n e r con ver ts the text f i l e in to a ba s ic VCF f i l e
5awk −F” \ t ” ’{ p r i nt $1 ” \ t ” $2 ”\ t . \ t ” $3 ”\ t ” $4 ”\ t . \ t . \tAAAB=” $10 ” ;AABB=” $11 ”\tRD :AD\ t ” $5
” : ” $6 ”\ t ” $7 ” : ” $8 } ’
7## The a c tu a l commands we ’ ve used in our workflow :
echo −e ’##f i l e f o r m a t=VCFv4. 1 ’ > u nsor ted . v cf
9echo −e ’##INFO=<ID=AAAB, Number=1,Type=Fl oat , D es cr ip ti o n=”P r ob ab i li t y o f J oi nt Genotype AA i n
Normal and AB in Tumor”>’ >> unsorted . vc f
echo −e ’##INFO=<ID=AABB, Number=1,Type=Floa t , D e sc r ip t io n=”P ro b a bi li ty o f Jo in t Genotype AA i n
Normal and BB in Tumor”>’ >> unsorted . v cf
11 echo −e ’##FORMAT=<ID=RD, Number=1,Type=In te ge r , D e sc ri p ti on=”Depth o f r e fe r en c e−supporting ba ses (
reads1 ) ”>’ >> unsorted . v cf
echo −e ’##FORMAT=<ID=AD, Number=1,Type=In te ge r , D es cr i pt io n=”Depth o f v ar ia nt −support ing bases (
reads2 ) ”>’ >> unsorted . v cf
13 echo −e ’#CHROM\tPOS\tID\tREF\tALT\tQUAL\tFILTER\tINFO\tFORMAT\tNORMAL\tTUMOR’ >> unsorted . vc f
15 python $PATH/TO/jsm . py c l a s s i f y joint_snv_mix_two genome .GRCh37. fa normal .bam tumor .bam tra in ed .
parameter . c f g /dev/ s td ou t | \
awk −F” \ t ” ’NR!=1 && $4!=”N” && $10+$11 >=0.95’ | \
17 awk −F” \ t ” ’{ p r i nt $1 ” \ t ” $2 ”\ t . \ t ” $3 ”\ t ” $4 ”\ t . \ t . \tAAAB=” $10 ” ;AABB=” $11 ”\tRD :AD\ t ” $5
” : ” $6 ”\ t ” $7 ” : ” $8 } ’ >> unsor ted . v cf
4.3 Convert the VCF le into TSV le
This script somaticseq/somatic_vcf2tsv.py works for all VCF les (requires input for two BAM les).
It extracts information from the BAM les, as well as some individual callers’ output VCF les. If the
ground truth VCF le is included, a called variant will be annotated as a true positive, and everything
will be annotated as a false positive. somaticseq/single_sample_vcf2tsv.py is used for single-sample mode.
At the end of this, Ensemble.sSNV.tsv and Ensemble.sINDEL.tsv are created.
All the options for somaticseq/somatic_vcf2tsv.py or somaticseq/single_sample_vcf2tsv.py can be found
by running.
1somat ic se q /so ma ti c_v cf 2t sv . py −h
somat ic se q /sin gl e_ sa mple_v cf 2t sv . py −h
8

Note: Do not worry if Python throws a warning like this.
RuntimeWarning : i n v a l i d v al ue encou nte red in double_scalars
2z = ( s −expected ) / np . s qrt ( n1∗n2∗(n1+n2+1)/ 12 .0 )
This is to tell you that scipy was attempting some statistical test with empty data. That’s usually due to
the fact that normal BAM le has no variant reads at that given position. That is why lots of values are
NaN for the normal.
4.4 Model Training or Mutation Prediction
You can use Ensemble.sSNV.tsv and Ensemble.sINDEL.tsv les either for model training (provided that
their mutation status is annotated with 0 or 1) or mutation prediction. This is done with stochastic boost-
ing algorithm we have implemented in R.
Model training:
# Training :
2r_ sc ri pt s /ada_model_builder_ntChange .R Ensemble .sSNV. ts v Consistent_Mates Inconsistent_Mates
r_ sc ri pt s /ada_model_builder_ntChange .R Ensemble .sINDEL. tsv Strelka_QSS Strelka_TQSS
Consistent_Mates Inconsistent_Mates
Ensemble.sSNV.tsv.ntChange.Classier.RData and Ensemble.sINDEL.tsv.ntChange.Classier.RData will
be created from model training. The arguments after Ensemble.sSNV.tsv and Ensemble.sINDEL.tsv tells
the builder script to ignore those features in training. These features do not improve accuracy in our data
sets (mostly WGS data, but they may help other data sets)
Mutation prediction:
1# Mutation p r edi c t i o n :
r_ sc ri pt /ada_model_predictor .R Ensemble .sSNV . tsv . C l a s s i f i e r . RData Ensemble . sSNV . tsv Trained .
sSNV . tsv
3r_ sc ri pt /ada_model_predictor .R Ensemble . sINDEL. ts v . C l a s s i f i e r . RData Ensemble . sINDEL . ts v Trained .
sINDEL. tsv
After mutation prediction, if you feel like it, you may convert Trained.sSNV.tsv and Trained.sINDEL.tsv
into VCF les. Use -tools to list ONLY the individual tools used to have appropriately annotated VCF
les. Accepted tools are MuTect2/MuTect/Indelocator, VarScan2, JointSNVMix2, SomaticSniper, Var-
Dict, MuSE, LoFreq, Scalpel, Strelka, and/or TNscope. To list a tool without having run it, the VCF will
be annotated as if the tool was run but did not identify that position as a somatic variant, which is proba-
bly undesireable.
1# Pr ob a bi l it y above 0.7 lab e l e d PASS (−pass 0 . 7 ) , and between 0 . 1 and 0 . 7 l a b e l ed LowQual (−low
0 . 1 ) :
# Use −a l l to i nc lu de REJECT c a l l s in the VCF f i l e
3# Use −phred t o c on ver t p r o b a b i l i t y v a lu e s ( between 0 t o 1 ) i n t o Phred s c a l e i n th e QUAL column
in the VCF f i l e
5somat ic se q /SS eq_ts v2vcf . py −ts v Trained . sSNV . ts v −vc f Trained . sSNV. v cf −pass 0 .7 −low 0 .1 −
t o o l s MuTect2 VarScan2 JointSNVMix2 S omatic Sni per VarDict MuSE LoFreq S tr e lk a −a l l −phred
7somat ic se q /SS eq_ts v2vcf . py −t s v Trained . sINDEL . t sv −vc f Trained . sINDEL. vc f −pass 0 .7 −low 0 .1 −
t o o l s MuTect2 VarScan2 VarDict LoFreq S c al p el S tr e lk a −a l l −phred
9

5 To run the dockerized somatic mutation callers
For your convenience, we have created a couple of scripts that can generate run script for the dockerized
somatic mutation callers.
5.1 Location
• somaticseq/utilities/dockered_pipelines/
5.2 Requirements
• Have internet connection, and able to pull and run docker images from docker.io
• Have cluster management system such as Sun Grid Engine, so that the ”qsub” command is valid
5.3 Example commands
5.3.1 Single-threaded Jobs
This is best suited for whole exome sequencing or less.
1# Example command to submit the run s c r i p t s f or each of the f o ll ow i ng somatic mutation c a l l e r s
$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines / submit_callers_singleThread . sh \
3−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam \
−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam \
5−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a \
−−output−d i r /ABSOLUTE/PATH/TO/RESULTS \
7−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vc f \
−−somaticseq−d i r /ABSOLUTE/PATH/TO/ SomaticSeq \
9−− a cti on echo \
−−mutect2 −− somaticsniper −− v ard i ct −−muse −− lofreq −− scalpel −− strelka −−somaticseq
The command shown above will create scripts for MuTect2, SomaticSniper, VarDict, MuSE, LoFreq,
Scalpel, and Strelka. Then, it will create the SomaticSeq script that merges those 7 callers. This command
defaults to majority-vote consensus.
Since it’s --aciton echo, it will echo the mutation caller scripts locations, but these scripts will not be
run. If you do --action qsub instead, then those mutation caller scripts will be qsub’ed. You’ll still need
to mantually run/submit the SomaticSeq script after all the caller jobs are done.
5.3.2 Multi-threaded Jobs
This is best suited for whole genome sequencing. This is same as above, except it will create 36 equal-size
regions in 36 bed les, and parallelize the jobs into 36 regions.
# Submitting mutation c a l l e r job s by s p l i t t i n g each job in to 36 even reg io ns .
2$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines / submit_callers_multiThreads . sh \
−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam \
4−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam \
−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a \
6−−output−d i r /ABSOLUTE/PATH/TO/RESULTS \
−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vc f \
8−− threads 36 \
−− a cti on echo \
10 −−mutect2 −− somaticsniper −− v ard i ct −−muse −− lofreq −− scalpel −− strelka −−somaticseq
10
5.3.3 SomaticSeq Training
Two classiers will be created (*.RData les), one for SNV and one for INDEL.
# Submitting mutation c a l l e r job s by s p l i t t i n g each job in to 36 even reg io ns .
2$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines / submit_callers_singleThread . sh \
−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam \
4−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam \
−−truth−snv /ABSOLUTE/PATH/TO/snvTruth . vc f \
6−−truth−i n d e l /ABSOLUTE/PATH/TO/ i nd el Tr ut h . v c f \
−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a \
8−−output−d i r /ABSOLUTE/PATH/TO/RESULTS \
−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vc f \
10 −−somaticseq−d i r /ABSOLUTE/PATH/TO/ SomaticSeq \
−− a cti on echo \
12 −−mutect2 −− somaticsniper −− v ard i ct −−muse −− lofreq −− scalpel −− strelka −−somaticseq −−somaticseq
−train
Notice the command includes –truth-snv and –truth-indel, and invokes somaticseq-train.
For multi-threaded job, you should not invoke somaticseq-train. Instead, you should combine all the En-
semble.sSNV.tsv and Ensemble.sINDEL.tsv les (separately), and then train on the combined les.
5.3.4 SomaticSeq Prediction
# Submitting mutation c a l l e r job s by s p l i t t i n g each job in to 36 even reg io ns .
2$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines / submit_callers_singleThread . sh \
−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam \
4−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam \
−− classifier−snv /ABSOLUTE/PATH/TO/Ensemble .sSNV. tsv . ntChange . C l a s s i f i e r . RData \
6−− classifier−i ndel /ABSOLUTE/PATH/TO/Ensemble . sINDEL . ts v . ntChange . C l a s s i f i e r . RData \
−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38 . f a \
8−−output−d i r /ABSOLUTE/PATH/TO/RESULTS \
−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vc f \
10 −−somaticseq−d i r /ABSOLUTE/PATH/TO/ SomaticSeq \
−− a cti on echo \
12 −−mutect2 −− somaticsniper −− v ard i ct −−muse −− lofreq −− scalpel −− strelka −−somaticseq
Notice the command includes –classier-snv and –classier-indel.
5.3.5 Parameters
−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam ( Required )
2−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample . bam ( Required )
−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/ human_reference . f a ( Required )
4−−dbsnp /ABSOLUTE/PATH/TO/dbsnp . vc f ( Required f o r MuSE and LoFreq )
−−cosmic /ABSOLUTE/PATH/TO/cosmic . v cf ( Optional )
6−− s e l e c t o r /ABSOLUTE/PATH/TO/Capture_region . bed ( Optional . Will assume whole
genome from the . f a i f i l e without i t . )
−− exclu de /ABSOLUTE/PATH/TO/ B la ck li st _r eg io n . bed ( Optional )
8−−min−a f ( Opt ional . The minimum VAF c u t o f f for VarDict and VarScan2 .
Def au lts are 0. 10 f o r VarScan2 and 0 .05 f o r VarDict ) .
−− a cti on qsub ( Optional : the command pre cedi ng the .cmd s c r i p t s . Defau lt i s
echo)
10 −− threads 36 ( Optional f o r multi Threads and i n v a l i d f o r singleThread : evenly
s p l i t the genome i n to 36 BED f i l e s . Default = 12) .
−−mutect2 ( O ptional f l a g to i nvo ke MuTect2)
12 −−varsc an2 ( Optional f l a g to invoke VarScan2 )
−−j oin tsnvmix2 ( O ptional f l a g to i nvo ke JointSNVMix2 )
14 −− s oma tic sn ipe r ( O ptional f l a g to i nvo ke SomaticSn iper )
−− v a rd ic t ( O ptional f l a g to i nvo ke VarDict )
16 −−muse ( O ptional f l a g to i nvo ke MuSE)
11

−− l o f r e q ( O ptional f l a g to i nvo ke LoFreq )
18 −− s c a l p e l ( Optional f l a g t o invok e S c al pe l )
−− s t r e l k a ( Option al f l a g to invoke S tr e lk a )
20 −−s omaticseq ( O ptional f l a g to i nvo ke SomaticSeq . This s c r i p t always be echo ’ ed ,
as i t should not be submitted until a l l the c a l l e r s above complete ) .
−−output−d i r /ABSOLUTE/PATH/TO/OUTPUT_DIRECTORY ( Requi red )
22 −−somaticseq−d i r SomaticSeq_Output_Directory ( Op tio nal . The d i r e c t o r y name o f th e
SomaticSeq output . Defau lt = SomaticSeq ) .
−−somaticseq−t r a i n ( Optional f l ag to invoke SomaticSeq to produce c l a s s i f i e r s i f
ground tru th VCF f i l e s are provided . Only recommended i n s in gl eT hr ea d mode , bec ause o th e rw is e
it ’ s b ett er to combine the output TSV f i l e s f i r s t , and then train classifiers .)
24 −−somaticseq−a ct ion ( Optional . What to do with the somaticseq . cmd . Defa ult i s echo .
Only do ”qsub” i f you have al re ad y completed a l l the mutation c a l l e r s , but want to run
SomaticSeq at a d i f f e r e n t s e t ti n g . )
−− classifier−snv Trained_sSNV_Classifier . RData ( Optional i f the re i s a c l a s s i f e r you
want to use )
26 −− classifier−i n d e l Trai ned_sIN DEL_Cla ssifier . RData ( O pti ona l i f ther e i s a c l a s s i f e r
you want to use )
−−truth−snv sSNV_ground_truth . vc f ( Optional i f the re i s a ground truth , and
everything e l s e w i l l be l ab el ed f a l s e positive)
28 −−
truth−indel sINDEL_ground_truth . v cf ( Optional i f th er e i s a ground truth , and
everything e l s e w i l l be l ab el ed f a l s e positive)
−−exome ( Opti onal f l a g f o r S tr e lka )
30 −− scalpel−two−pass ( Optional parameter for S cal p e l . Default = f a l s e . )
−−mutect2−arguments ( Extra parameters to pass onto Mutect2 , e . g . , −−mutect2−arguments
’−− initial_tumor_lod 3 .0 −−log_somatic_prior −5.0 −−min_base_quality_score 20 ’)
32 −−mutect2−filter−arguments ( Extra parameters to pass onto F ilt erM ute ct Cal ls )
−−varscan−arguments ( Extra pa ramet ers t o p ass onto VarScan2 )
34 −−varscan−pileup−arguments ( Extra p ara met ers t o p as s onto sam to ol s mpileup t ha t c r e a t e s p i l eu p
files f o r VarScan )
−−jsm−train−arguments ( Extra pa ram ete rs t o pass onto JointSNVMix2 ’ s t r a i n command)
36 −−jsm−classify−arguments ( Extra parameters to pass onto JointSNVMix2 ’ s c l a s s i f y command)
−− somaticsniper−arguments ( Extra parameters to pass onto SomaticSniper )
38 −− v ard ict−arguments ( Extra parameters to pass onto VarDict )
−−muse−arguments ( Extra parameters to pass onto MuSE)
40 −− lofreq−arguments ( Extra par amete rs t o p as s onto LoFreq )
−− scalpel−discovery−arguments ( Extra parameters to pass onto Sc alpel ’ s di sc ov er y command)
42 −− scalpel−export−arguments ( Extra parameters to pass onto S ca lp el ’ s export command)
−− strelka−config−arguments ( Extra parameters to pass onto S tre lk a ’ s c on fi g command)
44 −− strelka−run−arguments ( Extra parameters to pass onto S tr ekl a ’ s run command)
−−somaticseq−arguments ( Extra parameters to pass onto SomaticSeq . Wrapper . sh )
5.3.6 What does the single-threaded command do
• For each ag such as --mutect2, --jointsnvmix2, ...., --strelka, a run script ending with .cmd will
be created in /ABSOLUTE/PATH/TO/RESULTS/logs. By default, these .cmd scripts will only be
created, and their le path will be printed on screen. However, if you do “--action qsub”, then these
scripts will be submitted via the qsub command. The default action is “echo.”
–Each of these .cmd script correspond to a mutation caller you specied. They all use docker
images.
–We may improve their functionalities in the future to allow more tunable parameters. For the
initial releases, POC and reproducibility take precedence.
• If you do “--somaticseq,” the somaticseq script will be created in /ABSOLUTE/PATH/TO/RE-
SULTS/SomaticSeq/logs. However, it will not be submitted until you manually do so after each of
these mutation callers is nished running.
–In the future, we may create more sophisticated solution that will automatically solves these
dependencies. For the initial release, we’ll focus on stability and reproducibility.
12

• Due to the way those run scripts are written, the Sun Grid Engine’s standard error log will record
the time the task completes (i.e., Done at 2017/10/30 29:03:02), and it will only do so when the task
is completed with an exit code of 0. It can be a quick way to check if a task is done, by looking at
the nal line of the standard error log le.
5.3.7 What does the multi-threaded command do
It’s very similar to the single-threaded WES solution, except the job will be split evenly based on genomic
lengths.
• If you specied “--threads 36,” then 36 BED les will be created. Each BED le represents 1/36 of
the total base pairs in the human genome (obtained from the .fa.fai le, but only including 1, 2, 3,
..., MT, or chr1, chr2, ..., chrM contigs). They are named 1.bed, 2.bed, ..., 36.bed, and will be cre-
ated into /ABSOLUTE/PATH/TO/RESULTS/1, /ABSOLUTE/PATH/TO/RESULTS/2, ..., /AB-
SOLUTE/PATH/TO/RESULTS/36. You may, of course, specify any number. The default is 12.
• For each mutation callers you specify (with the exception of SomaticSniper), a script will be created
into /ABSOLUTE/PATH/TO/RESULTS/1/logs, /ABSOLUTE/PATH/TO/RESULTS/2/logs, etc.,
with partial BAM input. Again, they will be automatically submitted if you do “--action qsub.”
• Because SomaticSniper does not support partial BAM input (one would have to manually split the
BAMs in order to parallelize SomaticSniper this way), the above mentioned procedure is not applied
to SomaticSniper. Instead, a single-threaded script will be created (and potentially qsub’ed) into
/ABSOLUTE/PATH/TO/RESULTS/logs.
–However, because SomaticSniper is by far the fastest tool there, single-thread is doable even for
WGS. Even single-threaded SomaticSniper will likely nish before parallelized Scalpel. When
I benchmarked the DREAM Challenge Stage 3 by splitting it into 120 regions, Scalpel took 10
hours and 10 minutes to complete 1/120 of the data. SomaticSniper took a little under 5 hours
for the whole thing.
–After SomaticSniper nishes, the result VCF les will be split into each of the /ABSOLUTE/-
PATH/TO/RESULTS/1, /ABSOLUTE/PATH/TO/RESULTS/2, etc.
• JointSNVMix2 also does not support partial BAM input. Unlike SomaticSniper, it’s slow and takes
massive amount of memory. It’s not a good idea to run JointSNVMix2 on a WGS data. The only
way to do so is to manually split the BAM les and run each separately. We may do so in the fu-
ture, but JointSNVMix2 is a 5-year old that’s no longer being supported, so we probably won’t
bother.
• Like the single-threaded case, a SomaticSeq run script will also be created for each partition like
/ABSOLUTE/PATH/TO/RESULTS/1/SomaticSeq/logs, but will not be submitted until you do
so manually.
–For simplicity, you may wait until all the mutation calling is done, then run a command like
1fi n d /ABSOLUTE/PATH/TO/RESULTS −name ’ somaticseq∗.cmd’ −exec qsub {} \ ;
6 Use BAMSurgeon to create training data
For your convenience, we have created a couple of wrapper scripts that can generate the run script to cre-
ate training data using BAMSurgon at somaticseq/utilities/dockered_pipelines/bamSimulator. Descrip-
tions and example commands can be found in the README there.
13

This pipeline is used to spike in in silico somatic mutations into existing BAM les in order to create a
training set for somatic mutations.
After the in silico data are generated, you can use the somatic mutation pipeline on the training data to
generate the SomaticSeq classiers.
Classiers built on training data work if the training data is similar to the data you want to predict. Ide-
ally, the training data are sequenced on the same platform, same sample prep, and similar depth of cover-
age as the data of interest.
This method is based on BAMSurgeon, slightly modied into our own fork for some speedups.
The proper citation for BAMSurgeon is Ewing AD, Houlahan KE, Hu Y, et al. Combining tumor genome
simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat Methods.
2015;12(7):623-30.
6.1 Requirements
• Have internet connection, and able to pull and run docker images from docker.io
• Have cluster management system such as Sun Grid Engine, so that the ”qsub” command is valid
6.2 Three scenario to simulate somatic mutations
Which scenario to use depend on the data sets available to you.
6.2.1 When you have sequencing replicates of normal samples
This is our approach to dene high-condence somatic mutations in SEQC2 consortium’s cancer reference
samples, presented here.
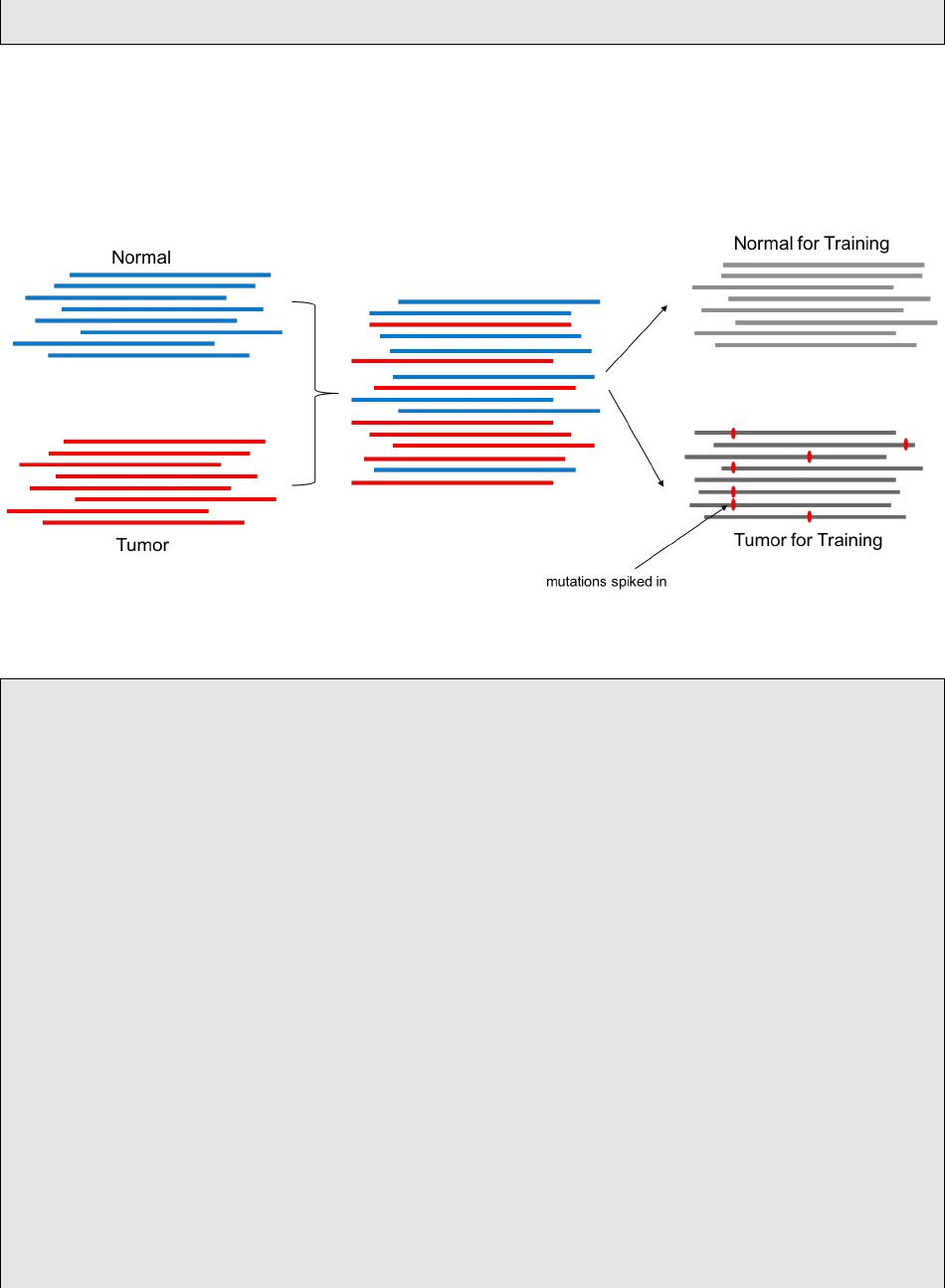
In this case, in silico mutations will be spiked into Replicate_002.bam. Since Replicate_002.bam and
Replicate_001.bam are otherwise the same sample, any mutations detected that you did not spike in are
false positives. The following command is a single-thread example.
1$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines /bamSimulator/BamSimulator_singleThread . sh \
−−genome−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38 . f a \
3−−tumor−bam−i n /ABSOLUTE/PATH/TO/ Replicate_001 .bam \
−−normal−bam−in /ABSOLUTE/PATH/TO/Replicate_002 .bam \
5−−tumor−bam−out syntheticTumor .bam \
−−normal−bam−out syntheticNormal .bam \
7−− split−proportion 0.5 \
−−num−snvs 20000 \
9−−num−i n d e l s 8000 \
−−min−va f 0.0 \
11 −−max−vaf 1.0 \
−− left−beta 2 \
13 −− right−beta 5 \
−−min−variant−read s 2 \
15 −−output−d i r /ABSOLUTE/PATH/TO/ t r a i n i n g S e t \
−− a cti on qsub
BamSimulator_*.sh creates semi-simulated tumor-normal pairs out of your input tumor-normal pairs. The
”ground truth” of the somatic mutations will be synthetic_snvs.vcf, synthetic_indels.vcf in the output
directory.
For multi-thread job (WGS), use BamSimulator_multiThreads.sh instead. See below for additional op-
tions and parameters.
A schematic of the BAMSurgeon simulation procedure
14

6.2.2 This example mimicks DREAM Challenge
DREAM Somatic Mutation Calling Challenge was an international competition to nd algorithms that
gave the most accurate performances.
In that case, a high-coverage BAM le is randomly split into two. One of which is designated normal, and
the other one is designated tumor where mutations will be spiked in. Like the previous example, any mu-
tations found between the designated tumor and designated normal are false positive, since not only are
they from the same sample, but also from the same sequencing run. This example will not capture false
positives as a result of run-to-run biases if they exist in your sequencing data. It will, however, still cap-
ture artefacts related to sequencing errors, sampling errors, mapping errors, etc.
$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines /bamSimulator/BamSimulator_multiThreads . sh \
2−−genome−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a −−tumor−bam−in /ABSOLUTE/PATH/TO/
highCoverageGenome .bam −−tumor−bam−out syntheticTumor .bam −−normal−bam−out syntheticNormal .
bam −− split−proportion 0.5 −−num−snvs 10000 −−num−i n d e l s 8000 −−num−sv s 1500 −−min−vaf 0. 0
−−max−vaf 1 .0 −− left−beta 2 −− right−beta 5 −−min−variant−reads 2 −−output−d i r /ABSOLUTE/PATH/
TO/trainingSet −−threads 24 −−a ct io n qsub −− split−bam −−indel−realign −−merge−output−bams
The –split-bem will randomly split the high coverage BAM le into two BAM les, one of which is desig-
nated normal and the other one designated tumor for mutation spike in. The –indel-realign is an option
that will perform GATK Joint Indel Realignment on the two BAM les. You may or may not invoke it de-
pending on your real data sets. The –merge-output-bams creates another script that will merge the BAM
and VCF les region-by-region. It will need to be run manually after all the spike in is done.
A schematic of the DREAM Challenge simulation procedure
6.2.3 Merge and then split the input tumor and normal BAM les
$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines /bamSimulator/BamSimulator_multiThreads . sh \
2−−genome−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a −−tumor−bam−in /ABSOLUTE/PATH/TO/Tumor_Sample .bam
−−normal−bam−in /ABSOLUTE/PATH/TO/Normal_Sample .bam −−tumor−bam−out syntheticTumor .bam −−
normal−bam−out syntheticNormal . bam −− split−proportion 0.5 −−num−snvs 30000 −−num−indels
10000 −−num−sv s 1500 −−min−vaf 0 . 0 −−max−vaf 1 . 0 −− left−beta 2 −− right−beta 5 −−min−variant−
15
reads 2 −−output−d i r /ABSOLUTE/PATH/TO/ t r a i n i n g S e t −− thre ads 24 −− act io n qsub −−merge−bam −−
split−bam −− indel−realign −−merge−output−bams
The --merge-bam will merge the normal and tumor BAM les into a single BAM le. Then, --split-bem
will randomly split the merged BAM le into two BAM les. One of which is designated normal, and one
of which is designated tumor. Synthetic mutations will then be spiked into the designated tumor to create
”real” mutations. This is the approach described in our 2017 AACR Abstract.
A schematic of the simulation procedure
6.3 Parameters and Options
−−genome−r e f e r e n c e /ABSOLUTE/PATH/TO/ human_reference . f a ( Required )
2−− s e l e c t o r /ABSOLUTE/PATH/TO/ capture_region . bed (BED f i l e to l i mi t where mutation s pi ke
in w i l l be attempted )
−−tumor−bam−i n Input BAM f i l e ( Required )
4−−normal−bam−in Input BAM f i l e ( Optional , but r eq ui red i f you want to merge i t with the tumor
input )
−−tumor−bam−out Output BAM f i l e f o r the des ig na te d tumor a f t e r BAMSurgeon mutation sp ike i n
6−−normal−bam−out Output BAM f i l e f o r the desi gnated normal i f −− split−bam i s chosen
−− split−pro po rt io n The f a c t i o n o f t o t a l re ad s d es gi na te d t o t he normal . ( Defaut = 0 . 5 )
8−−num−snvs Number of SNVs to sp ik e i n to the desig nate d tumor
−−num−i n d e l s Number o f INDELs to s p ik e i n to the de si gn at ed tumor
10 −−num−sv s Number of SVs to sp ik e in to the desi gnat ed tumor ( Default = 0)
−−min−depth Minimum depth where s pi ke in can take pl ac e
12 −−max−depth Maximum depth where s pi ke in can take pl ac e
−−min−va f Minimum VAF to si mul ate
14 −−max−vaf Maximum VAF to sim ulat e
−− left−beta Le ft beta of beta d i s t r i b u t i o n f o r VAF
16 −− right−beta Right bet a o f bet a d i s t r i b u t i o n f o r VAF
−−min−variant−read s Minimum number of variant−supporting reads f o r a s u c c e s s f u l sp ik e in
18 −−output−d i r Output d i r e c t o r y
−−merge−bam Flag to merge the tumor and normal bam f i l e input
20 −− split−bam Flag to s p l i t BAM f i l e f o r tumor and normal
−−clean−bam Flag to go through the BAM f i l e and remove r ea ds where more than 2 i d e n t i c a l
read names are present , or re ads where i t s read l en gth and CIGAR s t r i n g do not match . This
was necessary f o r some BAM f i l e s downloaded from TCGA. However , a proper pair−end BAM f i l e
should not have the same read name ap peari ng more than t wi ce . Use t h i s onl y when n ec es sa ry as
i t f i r s t s o r t s BAM f i l e by qname , goes through t he c le a n in g proc edure , then re−s o r t by
coordinates .
22 −− indel−r e a l i g n Conduct GATK Jo in t I nd el Realignment on the two output BAM f i l e s . I nst ea d o f
syntheticNormal .bam and syntheticTumor . bam, the f i n a l BAM f i l e s w i l l be syntheticNormal .
JointRealigned . bam and syntheticTumor . JointRe aligned .bam.
−−s ee d Random s eed . Pick any i n t e g e r for r e p r o d u c i b i l i t y purpos es .
16

24 −− th re ad s S p l i t th e BAM f i l e s ev en ly in N re gi on s , then pro ce ss each ( pa i r ) of sub−BAM
files in parallel .
−− a cti on The command p re ce din g the run s c r i p t cr ea ted i n to /ABSOLUTE/PATH/TO/
BamSurgeoned_SAMPLES/ l o g s . ”qsub” i s t o submit the s c r i p t in SGE system . Default = echo
6.3.1 –merge-bam / –split-bam / –indel-realign
If you have sequenced replicate normal, that’s the best data set for training. You can use one of the nor-
mal as normal, and designate the other normal (of the same sample) as tumor. Use --indel-realign to in-
voke GATK IndelRealign.
When you have a normal that’s roughly 2X the coverage as your data of choice, you can split that into
two halves. One designated as normal, and the other one designated as tumor. That DREAM Challenge’s
approach. Use --split-bam --indel-realign options.
Another approach is to merge the tumor and normal data, and then randomly split them as described
above. When you merge the tumor and normal, the real tumor mutations are relegated as germline or
noise, so they are considered false positives, because they are supposed to be evenly split into the desig-
nated normal. To take this approach, use --merge-bam --split-bam --indel-realign options.
Don’t use --indel-realign if you do not use indel realignment in your alignment pipeline.
In some BAM les, there are reads where read lengths and CIGAR strings don’t match. Spike in will fail
in these cases, and you’ll need to invoke --clean-bam to get rid of these problematic reads.
You can control and visualize the shape of target VAF distribution with python command:
1import s ci p y . s t a t s as s t a t s
import numpy as np
3import m at p lo t l ib . p yp lo t a s p l t
5le ftB et a , ri gthBeta = 2 ,5
minAF, maxAF = 0 ,1
7x = np . l i n s p a c e ( 0 , 1 , 1 01 )
y = s t a t s . bet a . pdf ( x , l ef t Be ta , rigthBeta , l oc = minAF, s c a l e = minAF + maxAF)
9_ = p l t . p l o t ( x , y )
6.4 To create SomaticSeq classiers
After the mutation simulation jobs are completed, you may create classiers with the training data with
the following command:
See our somatic mutation pipeline for more details.
1$PATH/TO/ somaticseq/ u t i l i t i e s / dockered_pipelines / submit_callers_multiThreads . sh \
−−output−d i r /ABSOLUTE/PATH/TO/ t r a i n i n g S e t / s om at ic Mu ta ti on Pi pe li ne \
3−−normal−bam /ABSOLUTE/PATH/TO/ t ra in in gS et / syntheticNormal .bam \
−−tumor−bam /ABSOLUTE/PATH/TO/ t ra in in gS et /syntheticTumor . bam \
5−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a \
−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vc f \
7−−thread 24 \
−−truth−snv /ABSOLUTE/PATH/TO/ t r a i n in g S e t / s yn th et ic _s nv s . v c f \
9−−truth−i nd e l /ABSOLUTE/PATH/TO/ t r ai n in g Se t / s y nt h et i c_ in d el s . l e f t A l i g n . v cf \
−− a cti on echo \
11 −−mutect2 −− somaticsniper −− v ard i ct −−muse −− lofreq −− strelka −−somaticseq
7 Release Notes
Make sure training and prediction use the same SomaticSeq version, or at least make sure the dierent
minor version changes do not change the results signicantly.
17
1. Version 1.0
Version used to generate data in the manuscript and Stage 5 of the ICGC-TCGA DREAM Somatic
Mutation Challenge, where SomaticSeq’s results were #1 for INDEL and #2 for SNV.
In the original manuscript, VarDict’s var2vcf_somatic.pl script was used to generate VarDict VCFs,
and subsequently “-lter somatic” was used for SSeq_merged.vcf2tsv.py. Since then (including
DREAM Challenge Stage 5), VarDict recommends var2vcf_paired.pl over var2vcf_somatic.pl, and
subsequently “-lter paired” was used for SSeq_merged.vcf2tsv.py. The dierence in SomaticSeq re-
sults, however, is pretty much negligible.
2. Version 1.1
Automated the SomaticSeq.Wrapper.sh script for both training and prediction mode. No change to
any algorithm.
3. Version 1.2
Have implemented the following improvement, mostly for indels:
• SSeq_merged.vcf2tsv.py can now accept pileup les to extract read depth and DP4 (reference
forward, reference reverse, alternate forward, and alternate reverse) information (mainly for in-
dels). Previously, that information can only be extracted from SAMtools VCF. Since the SAM-
tools or HaplotypeCaller generated VCFs hardly contain any indel information, this option im-
proves the indel model. The SomaticSeq.Wrapper.sh script is modied accordingly.
• Extract mapping quality (MQ) from VarDict output if this information cannot be found in
SAMtools VCF (also mostly benets the indel model).
• Indel length now positive for insertions and negative for deletions, instead of using the absolute
value previously.
4. Version 2.0
• Removed dependencies for SAMtools and HaplotypeCaller during feature extraction.
SSeq_merged.vcf2tsv.py extracts those information (plus more) directly from BAM les.
• Allow not only VCF le, but also BED le or a list of chromosome coordinate as input format
for SSeq_merged.vcf2tsv.py, i.e., use -mybed or -mypos instead of -myvcf.
• Instead of a separate step to annotate ground truth, that can be done directly by
SSeq_merged.vcf2tsv.py by supplying the ground truth VCF via -truth.
• SSeq_merged.vcf2tsv.py can annotate dbSNP and COSMIC information directly if BED le or
a list of chromosome coordinates are used as input in lieu of an annotated VCF le.
• Consolidated feature sets, e.g., removed some redunda Fixed a bug: if JointSNVMix2 is not in-
cluded, the values should be “NaN” instead of 0’s. This is to keep consistency with how we han-
dle all other callersnt feature sets coming from dierent resources.
5. Version 2.0.2
• Incorporated LoFreq.
• Used getopt to replace getopts in the SomaticSeq.Wrapper.sh script to allow long options.
6. Version 2.1.2
• Properly handle cases when multiple ALT’s are calls in the same position. The VCF les can
either contain multiple calls in the ALT column (i.e., A,G), or have multiple lines correspond-
ing to the same position (one line for each variant call). Some functions were signicantly re-
written to allow this.
• Incorporated Scalpel.
18
• Deprecated HaplotypeCaller and SAMTools dependencies completely as far as feature genera-
tion is concerned.
• The Wrapper script removed SnpSift/SnpE dependencies. Those information can be directly
obtained during the SSeq_merged.vcf2tsv.py step. Also removed some additional legacy steps
that has become useless since v2 (i.e., score_Somatic.Variants.py). Added a step to check the
correctness of the input. The v2.1 and 2.1.1 had some typos in the wrapper script, so only de-
scribing v2.1.2 here.
7. Version 2.2
• Added MuTect2 support.
8. Version 2.2.1
• InDel_3bp now stands for indel counts within 3 bps of the variant site, instead of exactly 3 bps
from the variant site as it was previously (likewise for InDel_2bp).
• Collapse MQ0 (mapping quality of 0) reads supporting reference/variant reads into a single
metric of MQ0 reads (i.e., tBAM_MQ0 and nBAM_MQ0). From experience, the number of
MQ0 reads is at least equally predictive of false positive calls, rather than distinguishing if
those MQ0 reads support reference or variant.
• Obtain SOR (Somatic Odds Ratio) from BAM les instead of VarDict’s VCF le.
• Fixed a typo in the SomaticSeq.Wrapper.sh script that did not handle inclusion region cor-
rectly.
9. Version 2.2.2
• Got around an occasional unexplained issue in then ada package were the SOR is sometimes
categorized as type, by forcing it to be numeric.
• Defaults PASS score from 0.7 to 0.5, and make them tunable in the SomaticSeq.Wrapper.sh
script (--pass-threshold and --lowqual-threshold).
10. Version 2.2.3
• Incorporated Strelka2 since it’s now GPLv3.
• Added another R script (ada_model_builder_ntChange.R) that uses nucleotide substitution
pattern as a feature. Limited experiences have shown us that it improves the accuracy, but it’s
not heavily tested yet.
• If a COSMIC site is labeled SNP in the COSMIC VCF le, if_cosmic and CNT will be labeled
as 0. The COSMIC ID will still appear in the ID column. This will not change any results be-
cause both of those features are turned o in the training R script.
• Fixed a bug: if JointSNVMix2 is not included, the values should be “NaN” instead of 0’s. This
is to keep consistency with how we handle all other callers.
11. Version 2.2.4
• Resolved a bug in v2.2.3 where the VCF les of Strelka INDEL and Scalpel clash on GATK
CombineVariants, by outputting a temporary VCF le for Strelka INDEL without the sample
columns.
• Caller classication: consider if_Scalpel = 1 only if there is a SOMATIC ag in its INFO.
12. Version 2.2.5
• Added a dockerle. Docker repo at https://hub.docker.com/r/lethalfang/somaticseq/.
19
• Ability to use vcfsort.pl instead of GATK CombineVariants to merge VCF les.
13. Version 2.3.0
• Moved some scripts to the utilities directory to clean up the clutter.
• Added the split_Bed_into_equal_regions.py to utilities, which will split a input BED le into
multiple BED les of equal size. This is to be used to parallelize large WGS jobs.
• Made compatible with MuTect2 from GATK4.
• Removed long options for the SomaticSeq.Wrapper.sh script because it’s more readable this
way.
• Added a script to add “GT” eld to Strelka’s VCF output before merging it with other VCF
les. That was what caused GATK CombineVariants errors mentioned in v2.2.4’s release notes.
• Added a bunch of scripts at utilities/dockered_pipelines that can be used to submit (requiring
Sun Grid Engine or equivalent) dockerized pipeline to a computing cluster.
14. Version 2.3.1
• Improve the automated run script generator at utilities/dockered_pipelines.
• No change to SomaticSeq algorithm
15. Version 2.3.2
• Added run script generators for dockerized BAMSurgeon pipelines at utilities/dock-
ered_pipelines/bamSurgeon
• Added an error message to r_scripts/ada_model_builder_ntChange.R when TrueVari-
ants_or_False don’t have both 0’s and 1’s. Other than this warning message change, no other
change to SomaticSeq algorithm.
16. Version 2.4.0
• Restructured the utilities scripts.
• Added the utilities/lter_SomaticSeq_VCF.py script that “demotes” PASS calls to LowQual
based on a set of tunable hard lters.
• BamSurgeon scripts invokes modied BamSurgeon script that splits a BAM le without the
need to sort by read name. This works if the BAM les have proper read names, i.e., 2 and
only 2 identical read names for each paired-end reads.
• No change to SomaticSeq algorithm
17. Version 2.4.1
• Updated some docker job scripts.
• Added a script that converts some items in the VCF’s INFO eld into the sample eld, to pre-
cipitate the need to merge multiple VCF les into a single multi-sample VCF, i.e., utilities/re-
format_VCF2SEQC2.py.
• No change to SomaticSeq algorithm
18. Version 2.5.0
• In modify_VJSD.py, get rid of VarDict’s END tag (in single sample mode) because it causes
problem with GATK CombineVariants.
• Added limited single-sample support, i.e., ssSomaticSeq.Wrapper.sh is the wrapper script. sin-
gleSample_callers_singleThread.sh is the wrapper script to submit single-sample mutation
caller scripts.
20
• Added run scripts for read alignments and post-alignment processing, i.e,. FASTQ →BAM, at
utilities/dockered_pipelines/alignments.
• Fixed a bug where the last two CD4 numbers were both alternate concordant reads in the out-
put VCF le, when the last number should’ve been alternate discordant reads.
• Changed the output le names from Trained.s(SNV|INDEL).vcf and
Untrained.s(SNV|INDEL).vcf to SSeq.Classied.s(SNV|INDE).vcf and
Consensus.s(SNV|INDEL).vcf. No change to the actual tumor-normal SomaticSeq algorithm.
• Added utilities/modify_VarDict.py to VarDict’s “complex” variant calls (e.g., GCA>TAC) into
SNVs when possible.
• Modied r_scripts/ada_model_builder_ntChange.R to allow you to ignore certain fea-
tures, e.g., r_scripts/ada_model_builder_ntChange.R Training_Data.tsv nBAM_REF_BQ
tBAM_REF_BQ SiteHomopolymer_Length ...
Everything after the input le are features to be ignored during training.
Also added r_scripts/ada_cross_validation.R.
19. Version 2.5.1
• Additional passable parameters options to pass extra parameters to somatic mutation callers.
Fixed a bug where the “two-pass” parameter is not passed onto Scalpel in multiThreads scripts.
• Ignore Strelka_QSS and Strelka_TQSS for indel training in the SomaticSeq.Wrapper.sh script.
20. Version 2.5.2
• Ported some pipeline scripts to singularities at utilities/singularities.
21. Version 2.6.0
• VarScan2_Score is no longer extracted from VarScan’s output. Rather, it’s now calculated di-
rectly using Fisher’s Exact Test, which reproduces VarScan’s output, but will have a real value
when VarScan2 does not output a particular variant.
• Incorporate TNscope’s output VCF into SomaticSeq, but did not incorporate TNscope caller
into the dockerized workow because we don’t have distribution license.
22. Version 2.6.1
• Optimized memory for singularity scripts.
• Updated utilities/bamQC.py and added utilities/trimSoftClippedReads.py (removed soft-
clipped bases on soft-clipped reads)
• Added some docker scripts at utilities/dockered_pipelines/QC
23. Version 2.7.0
• Added another feature: consistent/inconsistent calls for paired reads if the position is covered
by both forward and reverse reads. However, they’re excluded as training features in Somatic-
Seq.Wrapper.sh script for the time being.
• Change non-GCTA characters to N in VarDict.vcf le to make it conform to VCF le specica-
tions.
24. Version 2.7.1
• Without –gatk $PATH/TO/GenomeAnalysisTK.jar in the SomaticSeq.Wrapper.sh script, it
will use utilities/getUniqueVcfPositions.py and utilities/vcfsorter.pl to (in lieu of GATK3 Com-
bineVariants) to combine all the VCF les.
21
• Fixed bugs in the docker/singularities scripts where extra arguments for the callers are not cor-
rectly passed onto the callers.
25. Version 2.7.2
• Make compatible with .cram format
• Fixed a bug where Strelka-only calls are not considered by SomaticSeq.
26. Version 2.8.0
• The program is now designed to crash if the VCF le(s) are not sorted according to the .fasta
reference le.
27. Version 2.8.1
• Fixed a bug in the ssSomaticSeq.Wrapper.sh script (single-sample mode), where the SNV algo-
rithm weren’t looking for SNV VCF les during merging when using utilities/getUniqueVcfPo-
sitions.py, causing empty SNV les. For previous commands (invoking –gatk for CombineVari-
ants), the results have never changed.
28. Version 3.0.0
Refactored the codes.
• The wrapper scripts written in bash script (i.e., SomaticSeq.Wrapper.sh and ssSomatic-
Seq.Wrapper.sh) are replaced by somaticseq/run_somaticseq.py, though they’re still kept for
backward-compatibility.
• Allow parallel processing using somaticseq_parallel.py
29. Version 3.0.1
• Fixed a bug that didn’t handle Strelka/LoFreq indel calls correctly in somaticseq/com-
bine_callers.py module.
30. Version 3.1.0
• When splitting MuTect2 les into SNV and INDEL, make sure either the ref base or the alt
base (but not both) consists of a single base, i.e., discarding stu like GCAA>GCT.
• Fixed a bug introduced in v3.0.1 that caused the program to handle .vcf.gz les incorrectly.
• Incorporated Platypus into paired mode.
31. Version 3.1.1
• Fixed some bash scripts involved with single-sample multi-thread callers.
• vcfModier/splitVcf.py to handle multi-allelic calls better for indels.
8 Contact Us
For suggestions, bug reports, or technical support, please post in
https://github.com/bioinform/somaticseq/issues. The developers are alerted when issues are created
there. Alternatively, you may also email li_tai.fang@roche.com.
22