Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 16
- Introduction
- To use SomaticSeq.Wrapper.sh
- The step-by-step SomaticSeq Workflow
- To run the dockerized somatic mutation callers
- Use BAMSurgeon to create training set
- Release Notes
- Version 1.0
- Version 1.1
- Version 1.2
- Version 2.0
- Version 2.0.2
- Version 2.1.2
- Version 2.2
- Version 2.2.1
- Version 2.2.2
- Version 2.2.3
- Version 2.2.4
- Version 2.2.5
- Version 2.3.0
- Version 2.3.1
- Version 2.3.2
- Version 2.4.0
- Version 2.4.1
- Version 2.5.0
- Version 2.5.1
- Version 2.5.2
- Version 2.6.0
- Version 2.6.1
- Version 2.7.0
- Version 2.7.1
- Version 2.7.2
- Version 2.8.0
- Contact Us
SomaticSeq Documentation
Li Tai Fang
June 23, 2018
1 Introduction
SomaticSeq is a exible post-somatic-mutation-calling workow for improved accuracy. We have incorporated
multiple somatic mutation caller(s) to obtain a combined call set, and then it uses machine learning to
distinguish true mutations from false positives from that call set. We have incorporated the following
somatic mutation caller: MuTect/Indelocator/MuTect2, VarScan2, JointSNVMix, SomaticSniper, VarDict,
MuSE, LoFreq, Scalpel, Strelka, and TNscope. You may incorporate some or all of those callers into your
own pipeline with SomaticSeq.
The manuscript, An ensemble approach to accurately detect somatic mutations using Somat-
icSeq, is published in Genome Biology 2015, 16:197. The SomaticSeq project is located at
http://bioinform.github.io/somaticseq/. The data described in the manuscript is also described at
http://bioinform.github.io/somaticseq/data.html. There have been some major improvements since the pub-
lication.
SomaticSeq.Wrapper.sh is a bash script that calls a series of scripts to combine the output of the somatic
mutation caller(s), after the somatic mutation callers are run. Then, depending on what R scripts are fed to
SomaticSeq.Wrapper.sh, it will either 1) train the call set into a classier, 2) predict high-condence somatic
mutations from the call set based on a pre-dened classier, or 3) simply label the calls (i.e., PASS, LowQual,
or REJECT) based on majority vote of the tools.
1.1 Dependencies
• Python 3, plus regex, pysam, numpy, and scipy libraries. All the .py scripts are written in Python 3.
• R, plus the ada package in R.
• BEDTools (if there is an inclusion and/or an exclusion region le)
• Optional: dbSNP and COSMIC les in VCF format (if you want to use dbSNP features as a part of
the training).
• At least one of MuTect/Indelocator/MuTect2, VarScan2, JointSNVMix2, SomaticSniper, VarDict,
MuSE, LoFreq, Scalpel, Strelka2 and/or TNscope. Those are the tools we have incorporated in So-
maticSeq. If there are other somatic tools that may be good addition to our list, please make the
suggestion to us.
1.2 Docker repos
SomaticSeq and most somatic mutation callers we have incorporated are now dockerized. The exceptions
are VarScan2 and TNscope because we do not have distribution rights.
• SomaticSeq is dockerized at https://hub.docker.com/r/lethalfang/somaticseq/.
• MuTect2 (tested with GATK4): https://hub.docker.com/r/broadinstitute/gatk
• VarScan2 (untested): https://hub.docker.com/r/djordjeklisic/sbg-varscan2/
1

• JointSNVMix2: https://hub.docker.com/r/lethalfang/jointsnvmix2/
• SomaticSniper: https://hub.docker.com/r/lethalfang/somaticsniper/
• VarDict: https://hub.docker.com/r/lethalfang/vardictjava/
• MuSE: https://hub.docker.com/r/marghoob/muse/
• LoFreq: https://hub.docker.com/r/marghoob/lofreq/
• Scalpel: https://hub.docker.com/r/lethalfang/scalpel/
• Strelka2: https://hub.docker.com/r/lethalfang/strelka/
2 To use SomaticSeq.Wrapper.sh
The SomaticSeq.Wrapper.sh is a wrapper script that calls a series of programs and procedures after you
have run your individual somatic mutation callers. In the next section, we will describe the workow in more
detail, so you may not be dependent on this wrapper script. You can either modify this wrapper script or
create your own workow.
2.1 To train data set into a classier
To create a trained classier, ground truth les are required for the data sets. There is also an option to
include a list of regions to include and/or exclude. The exclusion or inclusion regions can be VCF or BED
les. An inclusion region may be subset of the call sets where you have validated their true/false mutation
status, so that only those regions will be used for training. An exclusion region can be regions where the
“truth” is ambigious.
All the output VCF les from individual callers are optional. Those VCF les can be bgzipped if they
have .vcf.gz extensions. It is imperative that the parameters used for the training and prediction are identical.
1# For t r a i ni n g , t ru th f i l e and th e c o r r e c t R s c r i p t ar e r e qu i r ed .
3SomaticSeq . Wrapper . sh \
−−mutect MuTect/ v aria nts . snp . v cf \
5−−mutect2 MuTect2/ v arian ts . vc f \
−− i n d e l o c a t o r I n de l o ca t o r / v a r ia n t s . i n d e l . v c f \
7−−varscan−snv VarScan2/ va r ian t s . snp . v c f \
−−varscan−i n d e l VarScan2/ v a r i a n ts . i n d e l . v c f \
9−−jsm JointSNVMix2/ v ar ian ts . snp . v cf \
−− s ni per SomaticSniper/ v ar ia nt s . snp . v cf \
11 −− v ard ic t VarDict/ v ar ia nts . v cf \
−−muse MuSE/ va ri an ts . snp . v cf \
13 −− lofreq−snv LoFreq/ va ri an ts . snp . v cf \
−− lofreq−i n d e l LoFreq/ v a r i a n ts . i n d e l . v c f \
15 −− s c a l p e l S ca lp e l / v ar ia nt s . i n d el . v cf \
−− strelka−snv St rel ka / v ar ia nts . snv . v cf \
17 −− strelka−i n d e l S t re l k a / v a r i a nt s . i n d e l . v c f \
−−tnscope TNscope . vc f . gz \
19 −−normal−bam matched_normal .bam \
−−tumor−bam tumor .bam \
21 −−ada−r−script ada_model_builder .R \
−−genome−r e f e r e n c e human_b37 . f a s t a \
23 −−cosmic cosmic . b37 . v71 . vcf \
−−dbsnp dbSNP. b37 . v141 . v c f \
25 −−exclusion−r eg ion ig no r e . bed \
−− inclusion−r eg ion va l ida t ed . bed
27 −−truth−snv truth . snp . v cf \
−−truth−i n d e l tr ut h . i n d e l . v c f \
29 −−output−d i r $OUTPUT_DIR
2

SomaticSeq.Wrapper.sh supports any combination of the somatic mutation callers we have incorporated
into the workow. SomaticSeq will run based on the output VCFs you have provided. It will train for SNV
and/or INDEL if you provide the truth.snp.vcf and/or truth.indel.vcf le(s) as well as the proper R script
(ada_model_builder.R). Otherwise, it will fall back to the simple caller consensus mode.
2.2 To predict somatic mutation based on trained classiers
Make sure the classiers and the proper R script (ada_model_predictor.R) are supplied, Without either of
them, it will fall back to the simple caller consensus mode.
1# The ∗. RData f i l e s a re t r a in e d c l a s s i f i e r from the t r a i n i n g mode .
SomaticSeq . Wrapper . sh \
3−−mutect MuTect/ v ar ian ts . snp . vc f \
−−mutect2 MuTect2/ v ar ian ts . v cf \
5−− i n d e l o c a t o r I n de l o ca t o r / v a r ia n t s . i n d e l . v c f \
−−varscan−snv VarScan2/ va r ian t s . snp . v c f \
7−−varscan−i n d e l VarScan2/ v a r i a n ts . i n d e l . v c f \
−−jsm JointSNVMix2/ v ar ian ts . snp . v cf \
9−− s ni per SomaticSniper/ v ar ia nt s . snp . v cf \
−− v ard ic t VarDict/ v ar ia nts . v cf \
11 −−muse MuSE/ v ar ian ts . snp . v cf \
−− lofreq−snv LoFreq/ va ri an ts . snp . v cf \
13 −− lofreq−i n d e l LoFreq/ v a r i a n ts . i n d e l . v c f \
−− s c a l p e l S ca lp e l / v ar ia nt s . i n d el . v cf \
15 −− strelka−snv St rel ka / v ar ia nts . snv . v cf \
−− strelka−i n d e l S t re l k a / v a r i a nt s . i n d e l . v c f \
17 −−tnscope TNscope . vc f . gz \
−−normal−bam matched_normal .bam \
19 −−tumor−bam tumor .bam \
−−ada−r−s c r i p t ada_model_predictor .R \
21 −−genome−r e f e r e n c e human_b37 . f a s t a \
−−cosmic cosmic . b37 . v71 . vcf \
23 −−dbsnp dbSNP. b37 . v141 . v c f \
−−snpeff−d i r $PATH/TO/DIR/ s n p S i f t \
25 −−exclusion−r eg ion ig no r e . bed \
−− inclusion−r eg ion va l ida t ed . bed
27 −− classifier−snv sSNV . C l a s s i f i e r . RData \
−− classifier−i nd el sINDEL. C l a s s i f i e r . RData \
29 −−pass−thr esh ol d 0. 5 \
−−lowqual−th re sh o ld 0. 1 \
31 −−output−d i r $OUTPUT_DIR
If both MuTect2 and MuTect/Indelocator VCF les are provided, the script is written such that it will
use MuTect2 and ignore MuTect.
2.3 To classify based on simple majority vote
Same as the command previously, but not including the R script or the ground truth les. Without those
information, SomaticSeq will fall back into a simple majority vote.
3 The step-by-step SomaticSeq Workow
We’ll describe the workow here, so you may modify the workow and/or create your own workow instead
of using the wrapper script described previously.
3.1 Combine the call sets
We use utilities/getUniqueVcfPositions.py and vcfsorter.pl to combine the VCF les from dierent callers.
The intermediate VCF les were modied to separate SNVs and INDELs.
3
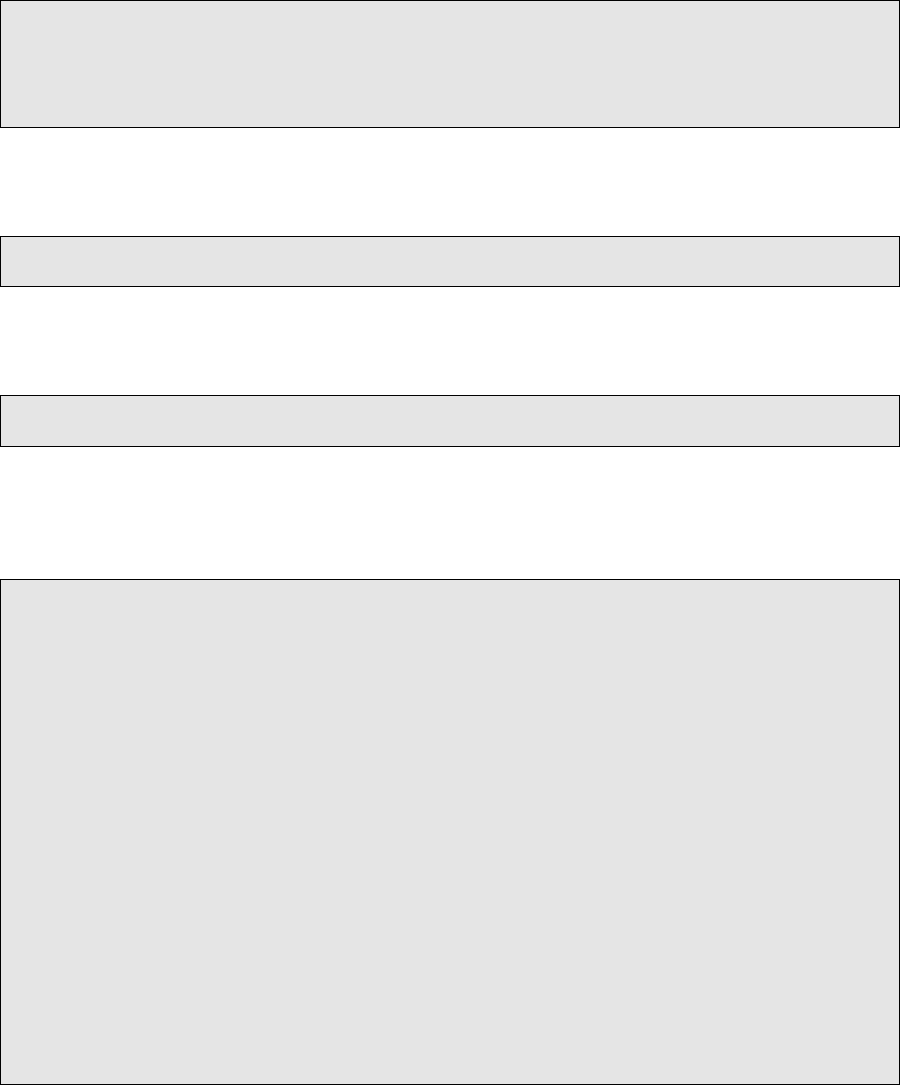
VCF modications were previously done to primarily make them compatible with GATK CombineVari-
ants. We no longer depend on CombineVariants, so some of the steps are legacy.
1. Modify MuTect and/or Indelocator output VCF les. Since MuTect’s output VCF do not always put
the tumor and normal samples in the same columns, the script will determine that information based
on either the BAM les (the header has sample name information), or based on the sample information
that you tell it, and then determine which column belongs to the normal, and which column belongs
to the tumor.
# Modify MuTect and I nd eloca to r ’ s output VCF based on BAM f i l e s
2modify_MuTect . py −i n f i l e input . vc f −o u t f i l e output . vc f −nbam normal .bam −tbam tumor .bam
4# Based on the sample name you supply :
modify_MuTect . py −i n f i l e input . vc f −o u t f i l e output . vc f −nsm NormalSampleName −tsm
TumorSampleName
2. For MuTect2, this script will split multi-allelic records into one variant per line in the VCF le. This
is to make thing easier for the SSeq_merged.vcf2tsv.py script later.
1# Based on the sample name you supply :
modify_MuTect2 . py −i n f i l e MuTect2 . F i l t e r e d . v cf −snv mutect . snp . vc f −i n d e l mutect . i n d e l . v c f
3. Modify VarScan’s output VCF les to be rigorously concordant to VCF format standard, and to attach
the tag ’VarScan2’ to somatic calls.
# Do i t f o r both t he SNV and i n d e l
2modify_VJSD . py −method VarScan2 −i n f i l e input . vc f −o u t f i l e output . vc f
4. JointSNVMix2 does not output VCF les. In our own workow, we have already converted its text
le into a basic VCF le with an 2 awk one-liners, which you may see in the Run_5_callers directory,
which are:
# To avoid t ex t f i l e s in the order of teraby tes , th i s awk one−l i n e r keeps e n t r i e s where t he
r ef e re n c e i s not ”N” , and the somatic p r o b a b i l i t i e s a re at l e a s t 0 . 9 5 .
2awk −F”\ t ” ’NR!=1 && $4!=”N” && $10+$11>=0.95’
4# This awk one−l i n e r con verts the tex t f i l e in t o a ba s i c VCF f i l e
awk −F”\ t ” ’{ p r i nt $1 ”\ t ” $2 ”\ t . \ t ” $3 ”\ t ” $4 ”\ t . \ t . \tAAAB=” $10 ” ;AABB=” $11 ”\tRD:AD\
t ” $5 ” : ” $6 ”\ t ” $7 ” : ” $8 } ’
6
## The a c tu al commands we ’ ve used i n our workflow :
8echo −e ’##f i l e f o r m a t=VCFv4. 1 ’ > un sort ed . v cf
echo −e ’##INFO=<ID=AAAB, Number=1,Type=Float , D es c ri pt i on=”Pr o ba b i li ty o f J oi nt Genotype AA
in Normal and AB in Tumor”>’ >> unsorted . v cf
10 echo −e ’##INFO=<ID=AABB, Number=1,Type=Float , D es cr ip ti o n=”P r ob a bi l i ty o f Jo in t Genotype AA
in Normal and BB in Tumor”>’ >> unsorted . v cf
echo −e ’##FORMAT=<ID=RD, Number=1,Type=I nt eg er , D es c ri pt io n=”Depth o f r ef e re n ce −supporting
bases ( reads1 ) ”>’ >> unsorted . v cf
12 echo −e ’##FORMAT=<ID=AD, Number=1,Type=In te ge r , D e sc r ip ti o n=”Depth o f v ari an t−supporting
bases ( reads2 ) ”>’ >> unsorted . v cf
echo −e ’#CHROM\tPOS\tID\tREF\tALT\tQUAL\tFILTER\tINFO\tFORMAT\tNORMAL\tTUMOR’ >> unsorted .
vcf
14
python $PATH/TO/jsm . py c l a s s i f y joint_snv_mix_two genome .GRCh37. f a normal . bam tumor .bam
tr ain e d . parameter . c f g /dev/ stdout | \
16 awk −F”\ t ” ’NR!=1 && $4!=”N” && $10+$11 >=0.95 ’ | \
awk −F”\ t ” ’{ p r i nt $1 ”\ t ” $2 ”\ t . \ t ” $3 ”\ t ” $4 ”\ t . \ t . \tAAAB=” $10 ” ;AABB=” $11 ”\tRD:AD\
t ” $5 ” : ” $6 ”\ t ” $7 ” : ” $8 } ’ >> u nso rted . v cf
4
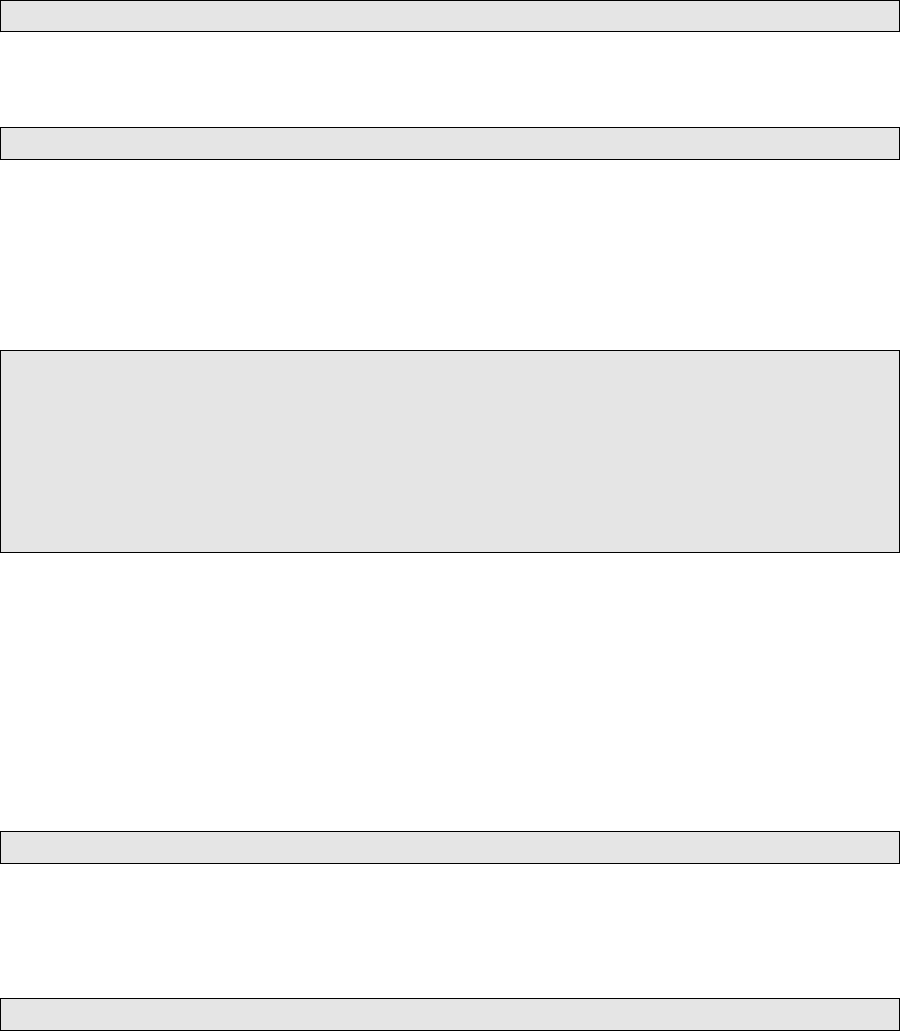
After that, you’ll also want to sort the VCF le. Now, to modify that basic VCF into something that
will be compatible with other VCF les under GATK CombineVariants:
1modify_VJSD . py −method JointSNVMix2 −i n f i l e input . vc f −o u t f i l e output . vc f
5. Modify SomaticSniper’s output:
1modify_VJSD . py −method SomaticSniper −i n f i l e input . vc f −o u t f i l e output . vc f
6. VarDict has both SNV and indel, plus some other variants in the same VCF le. Our script will
create two les, one for SNV and one for indel, while everything else is ignored for now. By default,
LikelySomatic/StrongSomatic and PASS calls will be labeled VarDict. However, in our SomaticSeq
paper, based on our experience in DREAM Challenge, we implemented two custom lters to relax the
VarDict tagging criteria.
1# De fault VarDict tag gin g c r i t e r i a , only PASS ( and L ik ely or Strong Somatic ) :
modify_VJSD . py −method VarDict −i n f i l e intp ut . v cf −o u t f i l e output . vc f
3
# When running VarDict , i f var2vcf_paired . pl i s used to g ene rat e the VCF f i l e , you may relax
the tagging c r i t e r i a with −f i l t e r pa ire d
5modify_VJSD . py −method VarDict −i n f i l e intp ut . v cf −o u t f i l e output . vc f −f i l t e r paire d
7# When running VarDict , i f var2vcf_somatic . pl i s used to ge ner ate the VCF f i l e , you may
rela x the tagging c r i t e r i a with −f i l t e r somatic
modify_VJSD . py −method VarDict −i n f i l e intp ut . v cf −o u t f i l e output . vc f −f i l t e r somatic
In the SomaticSeq paper, -lter somatic was used because var2vcf_somatic.pl was used to generate
VarDict’s VCF les. In the SomaticSeq.Wrapper.sh script, however, -lter paired is used because
VarDict authors have since recommended var2vcf_paired.pl script to create the VCF les. While there
are some dierences (dierent stringencies in some lters) in what VarDict labels as PASS between
the somatic.pl and paired.pl scripts, the dierence is miniscule after applying our custom lter (which
relaxes the lter, resulting in a dierence about 5 calls out of 15,000).
The output les will be snp.output.vcf and indel.output.vcf.
7. MuSE was not a part of our analysis in the SomaticSeq paper. We have implemented it later.
modify_VJSD . py −method MuSE −i n f i l e input . vc f −o u t f i l e output . vc f
8. LoFreq and Scalpel do not require modication. LoFreq has no sample columns anyway.
9. Add “GT” eld to sample columns to make it compatible with GATK CombineVariants.
1modify_Strelka .py −i n f i l e somatic . snvs . vc f . gz −o u t f i l e s t r al ka . snv . vc f
10. Finally, with the VCF les modied, you may combine them with GATK CombineVariants: one for
SNV and one for indel separately. There is no particular reason to use GATK CombineVariants.
Other combiners should also work. The only useful thing here is to combine the calls and GATK
CombineVariants does it well and pretty fast.
5
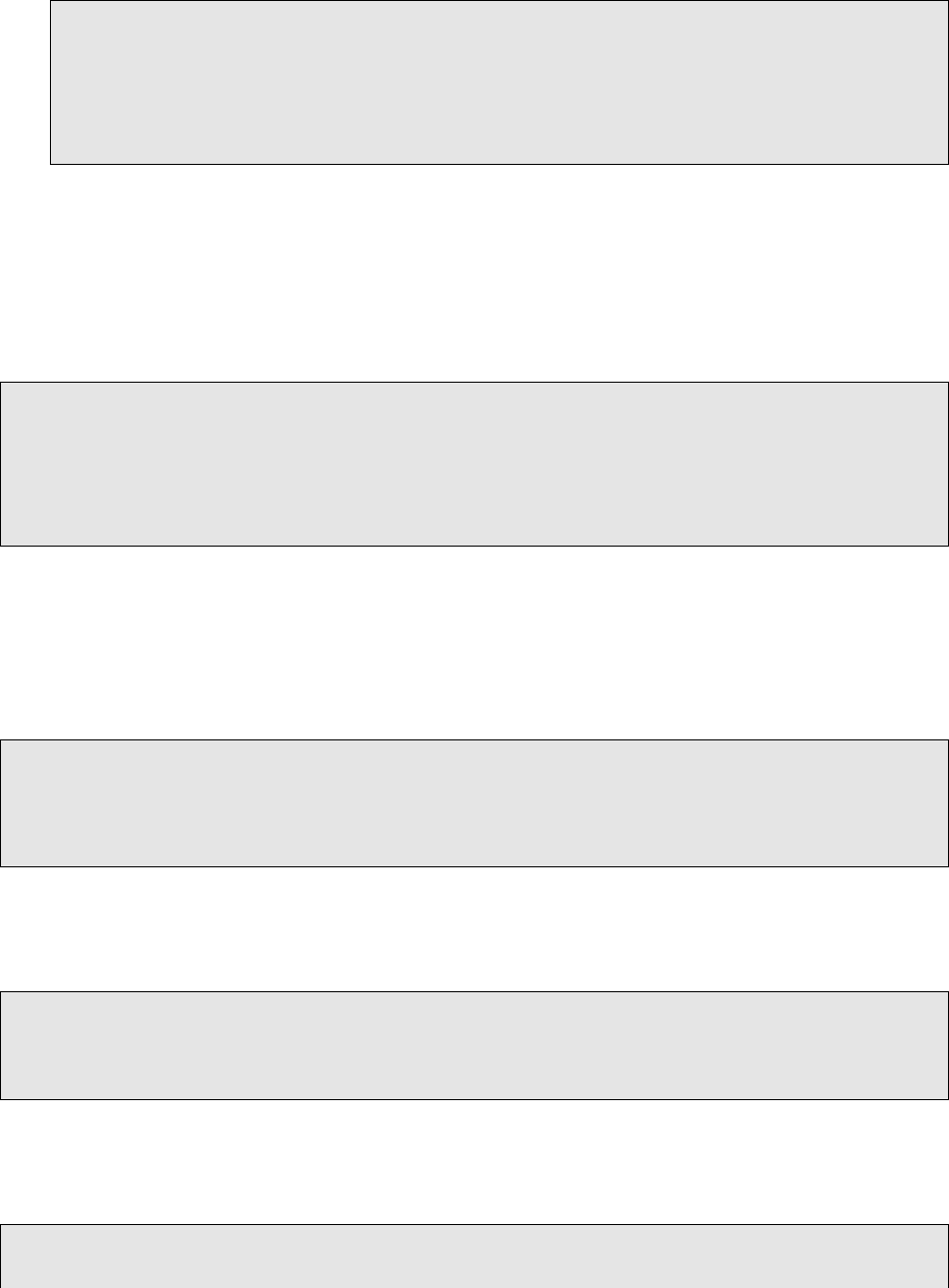
1# Combine the VCF f i l e s f o r SNV. Any or a l l of the VCF f i l e s may be prese nt .
#−nt 6 means to use 6 thr eads in p a r a l l e l
3java −j a r $PATH/TO/GenomeAnalysisTK . j a r −T CombineVariants −R genome .GRCh37. f a −nt 6 −−
setKey n u l l −−genotypemergeoption UNSORTED −V mutect . vcf −V varscan . snp . vcf −V
jointsnvmix . v c f −V snp . v a r dic t . v cf −V muse . vcf −−out CombineVariants . snp . v cf
java −j a r $PATH/TO/GenomeAnalysisTK . j a r −T CombineVariants −R genome .GRCh37. f a −nt 6 −−
setKey n u l l −−genotypemergeoption UNSORTED −V indelocator . vcf −V varscan . snp . vc f −V
i n d e l . v a r di c t . v c f −−out CombineVariants . i n d e l . v c f
3.2 For model training: process and annotate the VCF les (union of call sets)
This step may be needed for model training. The workow in SomaticSeq.Wrapper.sh allows for inclusion
and exclusion region. An inclusion region means we will only use calls inside these regions. An exclusion
region means we do not care about calls inside this region. DREAM Challenge had exclusion regions, e.g.,
blacklisted regions, etc.
# In the DREAM_Stage_3 d i rec t or y , we have i ncl ude d an exc l u s i o n re gi o n BED f i l e as an example
2# Th is command u se s BEDtools t o r i d o f a l l c a l l s i n the e x cl u s i on r eg io n
intersectBed −header −a BINA_somatic . snp . vc f −b ig n or e . bed −v > somatic . snp . p ro cesse d . v cf
4intersectBed −header −a BINA_somatic . i n d e l . v c f −b i gno re . bed −v > s oma tic . i n d e l . p ro ce s se d . v c f
6# A l t er n a ti v e ly ( or both ) , t h i s command u se s BEDtools t o keep only c a l l s i n the i n c l u s i o n r eg io n
intersectBed −header −a BINA_somatic . snp . vc f −b i n c l u s i o n . bed > soma tic . snp . p ro ce ss ed . v cf
8intersectBed −header −a BINA_somatic . i n d e l . v c f −b i n c l u s i o n . bed > so mat ic . i n d e l . p ro ce ss ed . v c f
3.3 Convert the VCF le, annotated or otherwise, into a tab separated le
This script works for all VCF les. It extracts information from BAM les as well as some VCF les created
by the individual callers. If the ground truth VCF le is included, a called variant will be annotated as a
true positive, and everything will be annotated as a false positive.
1# SNV
SSeq_merged . v c f 2t s v . py −r e f genome . GRCh37. f a −myvcf somatic . snp . pro cesse d . v cf −truth Ground . tr uth
. snp . vc f −mutect MuTect/ va r i a nts . snp . v c f . gz −varscan VarScan2/ var i a n ts . snp . vcf −jsm JSM2/
va r i a nts . vc f −s ni pe r SomaticSniper / var ia nt s . vc f −va r d i ct VarDict/snp . va r i a n ts . v cf −muse MuSE/
va r i a nts . vc f −l o f r e q LoFreq/ v ar ia nt s . snp . vc f −s t r e l k a S t re l ka / v a ri a nt s . snp . v cf −dedup −tbam
tumor .bam −nbam normal .bam −o u t f i l e Ensemble . sSNV. t sv
That was for SNV, and indel is almost the same thing. After version 2.1, we have replaced all information
from SAMtools and HaplotypeCaller with information directly from the BAM les. The accuracy dierences
are negligible with signicant improvement in usability and resource requirement.
# INDEL:
2SSeq_merged . v c f 2t s v . py −r e f genome . GRCh37. f a −myvcf som atic . i n d e l . p r oc e ss e d . v cf −truth Ground .
t ru th . i n d e l . v c f −varscan VarScan2/ va r i a nts . snp . vc f −v a r di c t VarDict / i n d e l . v a r i a nt s . v c f −
l o f r e q LoFreq/ v ar ia nt s . i n d el . v cf −s c a l p e l S c al pe l / v ar i an t s . i n d el . v cf −s t r e l k a S tr e lk a /
v a r ia n t s . i n d e l . v c f −tbam tumor .bam −nbam normal .bam −dedup −o u t f i l e Ensemble . sINDEL . t sv
At the end of this, Ensemble.sSNV.tsv and Ensemble.sINDEL.tsv are created.
All the options for SSeq_merged.vcf2tsv.py are listed here. They can also be displayed by running
SSeq_merged.vcf2tsv.py --help.
2−myvcf Input VCF f i l e o f the merged c a l l s [REQUIRED]
−r e f Genome r e f e r e n c e f a / f a s t a f i l e [REQUIRED]
6
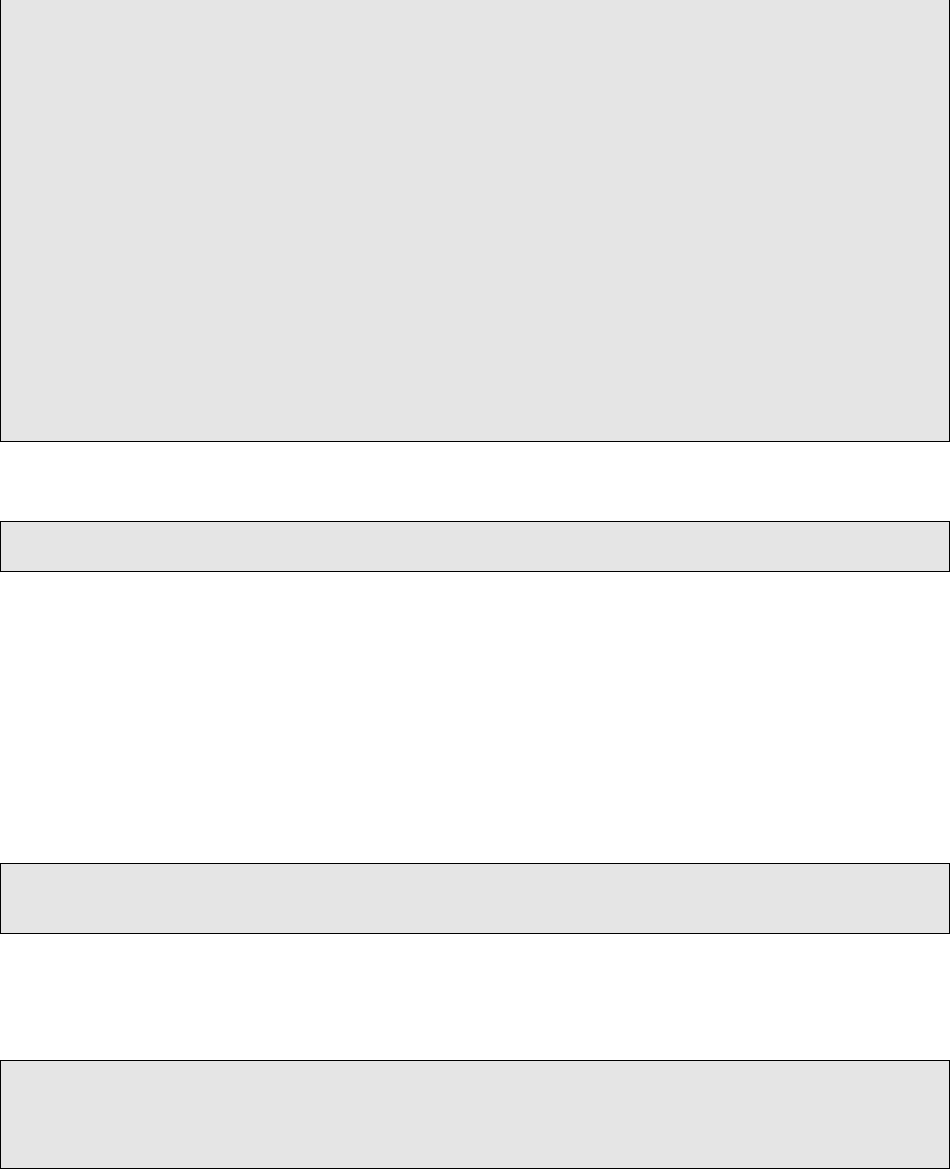
4−nbam BAM f i l e of the matched normal sample [REQUIRED]
−tbam BAM f i l e of the tumor sample [REQUIRED]
6−r e f Genome r e f e r e n c e f a / f a s t q f i l e [REQUIRED]
−t ru th Ground t ru th VCF f i l e . Every o th er p o s i t i o n i s a F al se P o s i t i v e .
8−dbsnp dbSNP VCF f i l e
−cosmic COSMIC VCF f i l e
10 −mutect VCF f i l e from e i t h e r MuTect2 , MuTect, or I n de lo c at o r
−sn i p e r VCF f i l e from SomaticSniper
12 −varscan VCF f i l e from VarScan2
−jsm VCF f i l e from Bina ’ s workflow that con ta ins JointSNVMix2
14 −va r d ict VCF f i l e that con ta in s only SNV or only INDEL from VarDict
−muse VCF f i l e from MuSE
16 −l o f r e q VCF f i l e from LoFreq
−s c a l p e l VCF f i l e from S ca l pe l
18 −s t r e l k a VCF f i l e from S t re l ka
−dedup A f l a g to co ns id er only primary r ead s
20 −minMQ Minimum mapping q u alit y for reads to be c on si de re d ( Default = 1)
−minBQ Minimum base q u a l i t y f o r reads to be con si de re d ( De fault = 5)
22 −mi n c all e r Minimum number of caller classification f o r a c a l l to be c on si de re d ( Use 0 .5 to
co ns ide r some LowQual c a l l s . Default = 0) .
−scale The op ti on s a re phred , f ra c t io n , o r None , t o c on ve rt numbers t o Phred s c a l e or
f r a c t i o n a l s c a le . ( d e fa ul t = None , i . e . , no co nv er si on )
24 −o u t f i l e Output TSV f i l e name
Note: Do not worry if Python throws a warning like this.
1RuntimeWarning : i n v a l i d v al ue enco unte red in double_scalars
z = ( s −expected ) / np . sq rt ( n1∗n2∗(n1+n2+1)/ 12 .0)
This is to tell you that scipy was attempting some statistical test with empty data. That’s usually due
to the fact that normal BAM le has no variant reads at that given position. That is why lots of values are
NaN for the normal.
3.4 Model Training or Mutation Prediction
You can use Ensemble.sSNV.tsv and Ensemble.sINDEL.tsv les either for model training (provided that their
mutation status is annotated with 0 or 1) or mutation prediction. This is done with stochastic boosting
algorithm we have implemented in R.
Model training:
# Training :
2r_ sc ri pt /ada_model_builder .R Ensemble .sSNV. t sv
r_ sc ri pt /ada_model_builder .R Ensemble . sINDEL . t sv
Ensemble.sSNV.tsv.Classier.RData and Ensemble.sINDEL.tsv.Classier.RData will be created from
model training.
Mutation prediction:
1# Mutation p r e d i c t i o n :
r_ sc ri pt /ada_model_predictor .R Ensemble . sSNV. t sv . C l a s s i f i e r . RData Ensemble . sSNV . t sv Trained .
sSNV . ts v
3r_ sc ri pt /ada_model_predictor .R Ensemble . sINDEL . t sv . C l a s s i f i e r . RData Ensemble .sINDEL . tsv Trained .
sINDEL . tsv
After mutation prediction, if you feel like it, you may convert Trained.sSNV.tsv and Trained.sINDEL.tsv
into VCF les. Use -tools to list ONLY the individual tools used to have appropriately annotated VCF
les. Accepted tools are CGA (for MuTect/Indelocator), VarScan2, JointSNVMix2, SomaticSniper, VarDict,
7

MuSE, LoFreq, and/or Scalpel. To list a tool without having run it, the VCF will be annotated as if the
tool was run but did not identify that position as a somatic variant.
1# Pr o bab i lit y above 0 .7 la b e l e d PASS (−pa ss 0 . 7 ) , and between 0 . 1 and 0 . 7 l a b el e d LowQual (−low
0 . 1 ) :
# Use −a l l to i nc lu de REJECT c a l l s i n the VCF f i l e
3# Use −phred t o co nv er t p r o b a b i l i t y v al u e s ( between 0 t o 1) i n to Phred s c a l e i n t he QUAL column
in the VCF f i l e
5SSeq_tsv2vcf .py −ts v Trained . sSNV. tsv −vc f Trained .sSNV . vcf −pass 0.7 −low 0 . 1 −t o o l s CGA
VarScan2 JointSNVMix2 SomaticSniper VarDict MuSE LoFreq S t r el k a −a l l −phred
7SSeq_tsv2vcf .py −ts v Tr ained . sINDEL . t sv −vc f Trained . sINDEL. v cf −pass 0.7 −low 0. 1 −t o o l s CGA
VarScan2 VarDict LoFreq S c a l p el St r e l ka −a l l −phred
4 To run the dockerized somatic mutation callers
For your convenience, we have created a couple of scripts that can generate run script for the dockerized
somatic mutation callers.
4.1 Location
• somaticseq/utilities/dockered_pipelines/
4.2 Requirements
• Have internet connection, and able to pull and run docker images from docker.io
• Have cluster management system such as Sun Grid Engine, so that the ”qsub” command is valid
4.3 Example commands
For single-threaded jobs, best suited for whole exome sequencing or less.
1# Example command to submit the run s c r i p t s f or each o f the fo l lo wi n g somatic mutation c a l l e r s
u t i l i t i e s / docke red _pipelines / singleThread / submit_callers_singleThread . sh \
3−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam \
−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam \
5−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a \
−−output−d i r /ABSOLUTE/PATH/TO/RESULTS \
7−− s e l e c t o r /ABSOLUTE/PATH/TO/Exome_Capture .GRCh38. bed \
−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vcf \
9−−a c ti o n qsub \
−−mutect2 −−jointsnvmix2 −−somaticsniper −−vardict −−muse −− lofreq −− scalpel −− strelka −−
somaticseq
For multi-threaded jobs, best suited for whole genome sequencing.
# Submitting mutation c a l l e r jo bs by s p l i t t i n g each job in to 36 even r eg ion s .
2u t i l i t i e s / docke red _pipelines /multiThreads/ submit_callers_multiThreads . sh \
−−normal−bam /ABSOLUTE/PATH/TO/normal_sample .bam \
4−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam \
−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/GRCh38. f a \
6−−output−d i r /ABSOLUTE/PATH/TO/RESULTS \
−−dbsnp /ABSOLUTE/PATH/TO/dbSNP.GRCh38. vcf \
8−−thr eads 36 \
−−a c ti o n qsub \
10 −−mutect2 −−somaticsniper −− v ard i c t −−muse −− lofreq −− scalpel −− strelka −−somaticseq
8

4.3.1 Parameters
−−normal−bam /ABSOLUTE/PATH/TO/normal_sample . bam ( Required )
2−−tumor−bam /ABSOLUTE/PATH/TO/tumor_sample .bam ( Required )
−−human−r e f e r e n c e /ABSOLUTE/PATH/TO/human_reference . f a ( Required )
4−−dbsnp /ABSOLUTE/PATH/TO/dbsnp . v cf ( Required f o r MuSE and LoFreq )
−−cosmic /ABSOLUTE/PATH/TO/cosmic . vc f ( Optional )
6−− s e l e c t o r /ABSOLUTE/PATH/TO/Capture_region . bed ( Optional . Will assume whole
genome from the . f a i f i l e without i t . )
−−exc lude /ABSOLUTE/PATH/TO/ B la ck li st _r eg io n . bed ( Optional )
8−−min−a f ( Opt ional . The minimum VAF c u t o f f for VarDict and VarScan2 .
Def au lt s are 0 .1 0 f o r VarScan2 and 0.0 5 f o r VarDict ) .
−−a c ti o n qsub ( Optional : the command pr ec edi ng the . cmd s c r i p t s . De faul t i s
echo)
10 −−thr eads 36 ( Optional f o r multiThreads and i n v a l i d f o r singleThread : evenly
s p l i t the genome i nt o 36 BED f i l e s . Default = 12) .
−−mutect2 ( Optional f l a g to i nvoke MuTect2)
12 −−va rscan2 ( Optional f l a g to in vok e VarScan2 )
−−joi ntsnvm ix2 ( O ptional f l a g to invok e JointSNVMix2 )
14 −−s oma tic sn ipe r ( O ptional f l a g to invok e SomaticSniper )
−− v ar di ct ( O ptional f l a g to invok e VarDict )
16 −−muse ( O ptional f l a g to invok e MuSE)
−− l o f r e q ( O ptional f l a g to invok e LoFreq )
18 −− s c a l p e l ( Optional f l a g to inv oke S ca lp e l )
−− s t r e l k a ( O pti onal f l a g to i nvo ke S t re lk a )
20 −−somati cseq ( O ptional f l a g to invok e SomaticSeq . This s c r i p t always be echo ’ ed ,
as i t should not be submitted until a l l the c a l l e r s above complete ) .
−−output−d i r /ABSOLUTE/PATH/TO/OUTPUT_DIRECTORY ( Requi red )
22 −−somaticseq−d i r SomaticSeq_Output_Directory ( Opt iona l . The d i r e c t o r y name o f t he
SomaticSeq output . Defa ult = SomaticSeq ) .
−−somaticseq−t r a in ( Optional f l a g to invoke SomaticSeq to produce c l a s s i f i e r s i f
ground truth VCF f i l e s are provided . Only recommended in s in gl eT hr ea d mode , beca use o th e rw i se
it ’ s be tter to combine the output TSV f i l e s f i r s t , and then train classifiers .)
24 −−somaticseq−act ion ( Optional . What to do with the somaticseq .cmd. Defa ult i s echo .
Only do ”qsub” i f you have a lr ea dy completed a l l the mutation c a l l e r s , but want to run
SomaticSeq at a d i f f e r e n t se t t in g . )
−− classifier−
snv Trained_sSNV_Classifier . RData ( Optional i f the re i s a c l a s s i f e r you
want to use )
26 −− classifier−i n d e l Traine d_sINDE L_Class ifier . RData ( Opt ion al i f ther e i s a c l a s s i f e r
you want to use )
−−truth−snv sSNV_ground_truth . v cf ( Optional i f there i s a ground truth , and
everything e l s e w i l l be l ab el ed f a l s e positive)
28 −−truth−i n d e l sINDEL_ground_truth . v c f ( O ptio nal i f t he re i s a ground truth , and
everything e l s e w i l l be l ab el ed f a l s e positive)
−−exome ( O ptional f l a g f o r St r e l ka )
30 −− scalpel−two−pass ( Optional parameter for S c a l p e l . Default = f a l s e . )
−−mutect2−arguments ( Extra parameters to pass onto Mutect2 , e . g . , −−mutect2−arguments
’−− initial_tumor_lod 3 .0 −−log_somatic_prior −5.0 −−min_base_quality_score 20 ’)
32 −−mutect2−filter−arguments ( Extra parameters to pass onto Fi lte rMu tec tCa lls )
−−varscan−arguments ( Extra pa ramet ers t o pass onto VarScan2 )
34 −−varscan−pileup−arguments ( Extra p arame ter s t o p as s onto s amtoo ls mpileup that c r e a t e s p il e u p
files f o r VarScan )
−−jsm−train−arguments ( Extra par ame ters t o p as s onto JointSNVMix2 ’ s t r a i n command)
36 −−jsm−classify−arguments ( Extra parameters to pass onto JointSNVMix2 ’ s c l a s s i f y command)
−−somaticsniper−arguments ( Extra parameters to pass onto SomaticSniper )
38 −−v ard ic t−arguments ( Extra parameters to pass onto VarDict )
−−muse−arguments ( Extra parameters to pass onto MuSE)
40 −− lofreq−arguments ( Extra pa ramet ers t o pass onto LoFreq )
−− scalpel−discovery−arguments ( Extra parameters to pass onto Sc al pe l ’ s d is co v er y command)
42 −− scalpel−export−arguments ( Extra parameters to pass onto Scalpe l ’ s export command)
−− strelka−config−arguments ( Extra parameters to pass onto S trelk a ’ s c on f ig command)
44 −− strelka−run−arguments ( Extra parameters to pass onto Str ek la ’ s run command)
−−somaticseq−arguments ( Extra parameters to pass onto SomaticSeq . Wrapper . sh )
9
4.3.2 What does the single-threaded command do
• For each ag such as --mutect2, --jointsnvmix2, ...., --strelka, a run script ending with .cmd will
be created in /ABSOLUTE/PATH/TO/RESULTS/logs. By default, these .cmd scripts will only be
created, and their le path will be printed on screen. However, if you do “--action qsub”, then these
scripts will be submitted via the qsub command. The default action is “echo.”
–Each of these .cmd script correspond to a mutation caller you specied. They all use docker
images.
–We may improve their functionalities in the future to allow more tunable parameters. For the
initial releases, POC and reproducibility take precedence.
• If you do “--somaticseq,” the somaticseq script will be created in /ABSOLUTE/PATH/TO/RESULT-
S/SomaticSeq/logs. However, it will not be submitted until you manually do so after each of these
mutation callers is nished running.
–In the future, we may create more sophisticated solution that will automatically solves these
dependencies. For the initial release, we’ll focus on stability and reproducibility.
• Due to the way those run scripts are written, the Sun Grid Engine’s standard error log will record the
time the task completes (i.e., Done at 2017/10/30 29:03:02), and it will only do so when the task is
completed with an exit code of 0. It can be a quick way to check if a task is done, by looking at the
nal line of the standard error log le.
4.3.3 What does the multi-threaded command do
It’s very similar to the single-threaded WES solution, except the job will be split evenly based on genomic
lengths.
• If you specied “--threads 36,” then 36 BED les will be created. Each BED le represents 1/36
of the total base pairs in the human genome (obtained from the .fa.fai le, but only including 1,
2, 3, ..., MT, or chr1, chr2, ..., chrM contigs). They are named 1.bed, 2.bed, ..., 36.bed, and will
be created into /ABSOLUTE/PATH/TO/RESULTS/1, /ABSOLUTE/PATH/TO/RESULTS/2, ...,
/ABSOLUTE/PATH/TO/RESULTS/36. You may, of course, specify any number. The default is 12.
• For each mutation callers you specify (with the exception of SomaticSniper), a script will be created
into /ABSOLUTE/PATH/TO/RESULTS/1/logs, /ABSOLUTE/PATH/TO/RESULTS/2/logs, etc.,
with partial BAM input. Again, they will be automatically submitted if you do “--action qsub.”
• Because SomaticSniper does not support partial BAM input (one would have to manually split the
BAMs in order to parallelize SomaticSniper this way), the above mentioned procedure is not applied
to SomaticSniper. Instead, a single-threaded script will be created (and potentially qsub’ed) into
/ABSOLUTE/PATH/TO/RESULTS/logs.
–However, because SomaticSniper is by far the fastest tool there, single-thread is doable even for
WGS. Even single-threaded SomaticSniper will likely nish before parallelized Scalpel. When I
benchmarked the DREAM Challenge Stage 3 by splitting it into 120 regions, Scalpel took 10
hours and 10 minutes to complete 1/120 of the data. SomaticSniper took a little under 5 hours
for the whole thing.
–After SomaticSniper nishes, the result VCF les will be split into each of the /ABSOLUTE/-
PATH/TO/RESULTS/1, /ABSOLUTE/PATH/TO/RESULTS/2, etc.
• JointSNVMix2 also does not support partial BAM input. Unlike SomaticSniper, it’s slow and takes
massive amount of memory. It’s not a good idea to run JointSNVMix2 on a WGS data. The only way
to do so is to manually split the BAM les and run each separately. We may do so in the future, but
JointSNVMix2 is a 5-year old that’s no longer being supported, so we probably won’t bother.
10

• Like the single-threaded case, a SomaticSeq run script will also be created for each partition like
/ABSOLUTE/PATH/TO/RESULTS/1/SomaticSeq/logs, but will not be submitted until you do so
manually.
–For simplicity, you may wait until all the mutation calling is done, then run a command like
1fi n d /ABSOLUTE/PATH/TO/RESULTS −name ’somaticseq∗.cmd’ −exec qsub {} \ ;
5 Use BAMSurgeon to create training set
For your convenience, we have created a couple of wrapper scripts that can generate the run script to create
training data using BAMSurgon at somaticseq/utilities/dockered_pipelines/bamSimulator. Descriptions
and example commands can be found in the README there.
5.1 Requirements
• Have internet connection, and able to pull and run docker images from docker.io
• Have cluster management system such as Sun Grid Engine, so that the ”qsub” command is valid
6 Release Notes
Make sure training and prediction use the same version. Otherwise the prediction is not valid.
6.1 Version 1.0
Version used to generate data in the manuscript and Stage 5 of the ICGC-TCGA DREAM Somatic Mutation
Challenge, where SomaticSeq’s results were #1 for INDEL and #2 for SNV.
In the original manuscript, VarDict’s var2vcf_somatic.pl script was used to generate VarDict VCFs,
and subsequently “-lter somatic” was used for SSeq_merged.vcf2tsv.py. Since then (including DREAM
Challenge Stage 5), VarDict recommends var2vcf_paired.pl over var2vcf_somatic.pl, and subsequently “-
lter paired” was used for SSeq_merged.vcf2tsv.py. The dierence in SomaticSeq results, however, is pretty
much negligible.
6.2 Version 1.1
Automated the SomaticSeq.Wrapper.sh script for both training and prediction mode. No change to any
algorithm.
6.3 Version 1.2
Have implemented the following improvement, mostly for indels:
• SSeq_merged.vcf2tsv.py can now accept pileup les to extract read depth and DP4 (reference forward,
reference reverse, alternate forward, and alternate reverse) information (mainly for indels). Previously,
that information can only be extracted from SAMtools VCF. Since the SAMtools or HaplotypeCaller
generated VCFs hardly contain any indel information, this option improves the indel model. The
SomaticSeq.Wrapper.sh script is modied accordingly.
• Extract mapping quality (MQ) from VarDict output if this information cannot be found in SAMtools
VCF (also mostly benets the indel model).
• Indel length now positive for insertions and negative for deletions, instead of using the absolute value
previously.
11
6.4 Version 2.0
• Removed dependencies for SAMtools and HaplotypeCaller during feature extraction.
SSeq_merged.vcf2tsv.py extracts those information (plus more) directly from BAM les.
• Allow not only VCF le, but also BED le or a list of chromosome coordinate as input format for
SSeq_merged.vcf2tsv.py, i.e., use -mybed or -mypos instead of -myvcf.
• Instead of a separate step to annotate ground truth, that can be done directly by
SSeq_merged.vcf2tsv.py by supplying the ground truth VCF via -truth.
• SSeq_merged.vcf2tsv.py can annotate dbSNP and COSMIC information directly if BED le or a list
of chromosome coordinates are used as input in lieu of an annotated VCF le.
• Consolidated feature sets, e.g., removed some redunda Fixed a bug: if JointSNVMix2 is not included,
the values should be “NaN” instead of 0’s. This is to keep consistency with how we handle all other
callersnt feature sets coming from dierent resources.
6.5 Version 2.0.2
• Incorporated LoFreq.
• Used getopt to replace getopts in the SomaticSeq.Wrapper.sh script to allow long options.
6.6 Version 2.1.2
• Properly handle cases when multiple ALT’s are calls in the same position. The VCF les can either
contain multiple calls in the ALT column (i.e., A,G), or have multiple lines corresponding to the same
position (one line for each variant call). Some functions were signicantly re-written to allow this.
• Incorporated Scalpel.
• Deprecated HaplotypeCaller and SAMTools dependencies completely as far as feature generation is
concerned.
• The Wrapper script removed SnpSift/SnpE dependencies. Those information can be directly obtained
during the SSeq_merged.vcf2tsv.py step. Also removed some additional legacy steps that has become
useless since v2 (i.e., score_Somatic.Variants.py). Added a step to check the correctness of the input.
The v2.1 and 2.1.1 had some typos in the wrapper script, so only describing v2.1.2 here.
6.7 Version 2.2
• Added MuTect2 support.
6.8 Version 2.2.1
• InDel_3bp now stands for indel counts within 3 bps of the variant site, instead of exactly 3 bps from
the variant site as it was previously (likewise for InDel_2bp).
• Collapse MQ0 (mapping quality of 0) reads supporting reference/variant reads into a single metric of
MQ0 reads (i.e., tBAM_MQ0 and nBAM_MQ0). From experience, the number of MQ0 reads is at
least equally predictive of false positive calls, rather than distinguishing if those MQ0 reads support
reference or variant.
• Obtain SOR (Somatic Odds Ratio) from BAM les instead of VarDict’s VCF le.
• Fixed a typo in the SomaticSeq.Wrapper.sh script that did not handle inclusion region correctly.
12
6.9 Version 2.2.2
• Got around an occasional unexplained issue in then ada package were the SOR is sometimes categorized
as type, by forcing it to be numeric.
• Defaults PASS score from 0.7 to 0.5, and make them tunable in the SomaticSeq.Wrapper.sh script
(--pass-threshold and --lowqual-threshold).
6.10 Version 2.2.3
• Incorporated Strelka2 since it’s now GPLv3.
• Added another R script (ada_model_builder_ntChange.R) that uses nucleotide substitution pattern
as a feature. Limited experiences have shown us that it improves the accuracy, but it’s not heavily
tested yet.
• If a COSMIC site is labeled SNP in the COSMIC VCF le, if_cosmic and CNT will be labeled as 0.
The COSMIC ID will still appear in the ID column. This will not change any results because both of
those features are turned o in the training R script.
• Fixed a bug: if JointSNVMix2 is not included, the values should be “NaN” instead of 0’s. This is to
keep consistency with how we handle all other callers.
6.11 Version 2.2.4
• Resolved a bug in v2.2.3 where the VCF les of Strelka INDEL and Scalpel clash on GATK Com-
bineVariants, by outputting a temporary VCF le for Strelka INDEL without the sample columns.
• Caller classication: consider if_Scalpel = 1 only if there is a SOMATIC ag in its INFO.
6.12 Version 2.2.5
• Added a dockerle. Docker repo at https://hub.docker.com/r/lethalfang/somaticseq/.
• Ability to use vcfsort.pl instead of GATK CombineVariants to merge VCF les.
6.13 Version 2.3.0
• Moved some scripts to the utilities directory to clean up the clutter.
• Added the split_Bed_into_equal_regions.py to utilities, which will split a input BED le into multiple
BED les of equal size. This is to be used to parallelize large WGS jobs.
• Made compatible with MuTect2 from GATK4.
• Removed long options for the SomaticSeq.Wrapper.sh script because it’s more readable this way.
• Added a script to add “GT” eld to Strelka’s VCF output before merging it with other VCF les.
That was what caused GATK CombineVariants errors mentioned in v2.2.4’s release notes.
• Added a bunch of scripts at utilities/dockered_pipelines that can be used to submit (requiring Sun
Grid Engine or equivalent) dockerized pipeline to a computing cluster.
6.14 Version 2.3.1
• Improve the automated run script generator at utilities/dockered_pipelines.
• No change to SomaticSeq algorithm
13
6.15 Version 2.3.2
• Added run script generators for dockerized BAMSurgeon pipelines at utilities/dock-
ered_pipelines/bamSurgeon
• Added an error message to r_scripts/ada_model_builder_ntChange.R when TrueVariants_or_False
don’t have both 0’s and 1’s. Other than this warning message change, no other change to SomaticSeq
algorithm.
6.16 Version 2.4.0
• Restructured the utilities scripts.
• Added the utilities/lter_SomaticSeq_VCF.py script that “demotes” PASS calls to LowQual based
on a set of tunable hard lters.
• BamSurgeon scripts invokes modied BamSurgeon script that splits a BAM le without the need to
sort by read name. This works if the BAM les have proper read names, i.e., 2 and only 2 identical
read names for each paired-end reads.
• No change to SomaticSeq algorithm
6.17 Version 2.4.1
• Updated some docker job scripts.
• Added a script that converts some items in the VCF’s INFO eld into the sample eld, to pre-
cipitate the need to merge multiple VCF les into a single multi-sample VCF, i.e., utilities/refor-
mat_VCF2SEQC2.py.
• No change to SomaticSeq algorithm
6.18 Version 2.5.0
• In modify_VJSD.py, get rid of VarDict’s END tag (in single sample mode) because it causes problem
with GATK CombineVariants.
• Added limited single-sample support, i.e., ssSomaticSeq.Wrapper.sh is the wrapper script. singleSam-
ple_callers_singleThread.sh is the wrapper script to submit single-sample mutation caller scripts.
• Added run scripts for read alignments and post-alignment processing, i.e,. FASTQ →BAM, at utili-
ties/dockered_pipelines/alignments.
• Fixed a bug where the last two CD4 numbers were both alternate concordant reads in the output VCF
le, when the last number should’ve been alternate discordant reads.
• Changed the output le names from Trained.s(SNV|INDEL).vcf and Untrained.s(SNV|INDEL).vcf to
SSeq.Classied.s(SNV|INDE).vcf and Consensus.s(SNV|INDEL).vcf. No change to the actual tumor-
normal SomaticSeq algorithm.
• Added utilities/modify_VarDict.py to VarDict’s “complex” variant calls (e.g., GCA>TAC) into SNVs
when possible.
• Modied r_scripts/ada_model_builder_ntChange.R to allow you to ignore certain features, e.g.,
r_scripts/ada_model_builder_ntChange.R Training_Data.tsv nBAM_REF_BQ tBAM_REF_BQ
SiteHomopolymer_Length ...
Everything after the input le are features to be ignored during training.
Also added r_scripts/ada_cross_validation.R.
14
6.19 Version 2.5.1
• Additional passable parameters options to pass extra parameters to somatic mutation callers. Fixed a
bug where the “two-pass” parameter is not passed onto Scalpel in multiThreads scripts.
• Ignore Strelka_QSS and Strelka_TQSS for indel training in the SomaticSeq.Wrapper.sh script.
6.20 Version 2.5.2
• Ported some pipeline scripts to singularities at utilities/singularities.
6.21 Version 2.6.0
• VarScan2_Score is no longer extracted from VarScan’s output. Rather, it’s now calculated directly
using Fisher’s Exact Test, which reproduces VarScan’s output, but will have a real value when VarScan2
does not output a particular variant.
• Incorporate TNscope’s output VCF into SomaticSeq, but did not incorporate TNscope caller into the
dockerized workow because we don’t have distribution license.
6.22 Version 2.6.1
• Optimized memory for singularity scripts.
• Updated utilities/bamQC.py and added utilities/trimSoftClippedReads.py (removed soft-clipped bases
on soft-clipped reads)
• Added some docker scripts at utilities/dockered_pipelines/QC
6.23 Version 2.7.0
• Added another feature: consistent/inconsistent calls for paired reads if the position is covered by both
forward and reverse reads. However, they’re excluded as training features in SomaticSeq.Wrapper.sh
script for the time being.
• Change non-GCTA characters to N in VarDict.vcf le to make it conform to VCF le specications.
6.24 Version 2.7.1
• Without –gatk $PATH/TO/GenomeAnalysisTK.jar in the SomaticSeq.Wrapper.sh script, it will use
utilities/getUniqueVcfPositions.py and utilities/vcfsorter.pl to (in lieu of GATK3 CombineVariants) to
combine all the VCF les.
• Fixed bugs in the docker/singularities scripts where extra arguments for the callers are not correctly
passed onto the callers.
6.25 Version 2.7.2
• Make compatible with .cram format
• Fixed a bug where Strelka-only calls are not considered by SomaticSeq.
6.26 Version 2.8.0
• The program will now throw an exception and crash if the VCF le(s) are not sorted according to the
.fasta reference le.
15