PDF Rnews 2003 2
PDF Rnews_2003-2 CRAN - Contents of R News
User Manual: PDF CRAN: R News
Open the PDF directly: View PDF ![]() .
.
Page Count: 39
- Editorial
- R Help Desk
- Integrating grid Graphics Output with Base Graphics Output
- A New Package for the General Error Distribution
- Web-based Microarray Analysis using Bioconductor
- Sweave, Part II: Package Vignettes
- R Foundation News
- Recent Events
- Book Reviews
- Changes in R 1.8.0
- Changes on CRAN
- Crossword Solution
- Correction to ``Building Microsoft Windows Versions of R and R packages under Intel Linux''
News
The Newsletter of the R Project Volume 3/2, October 2003
Editorial
by Friedrich Leisch
Welcome to a new issue of R News, focussing on
graphics and user interfaces. Paul Murell’s grid
graphics system offers a modern infrastructure for R
graphics. As of R-1.8.0 it is part of the base distribu-
tion but, until now, grid graphics functions could not
be mixed with traditional (base) plotting functions.
In this newsletter Paul describes his gridBase pack-
age which allows mixing of grid and base graphics.
Marc Schwartz, known as frequent poster of answers
on the mailing list r-help, has been invited to con-
tribute to the R Help Desk. He presents “An Intro-
duction to Using R’s Base Graphics”.
The Bioconductor column was written by Colin
Smith and shows a web-based interface for microar-
ray analysis. Angelo Mineo describes normalp, a
new package for the general error distribution. In the
second part of my mini-series on Sweave I demon-
strate how users can interact with package vignettes
and how package authors can add such documents
to their packages.
To close the circle, a version of Paul’s article is a
vignette in package gridBase, so if you want to play
with the code examples, the easiest way is to use R’s
vignette tools. We hope to see vignettes used more
frequently in the future, as they are a simple, effec-
tive way of delivering code examples to users.
R 1.8.0 was released more than two weeks ago.
The list of new features is long; see “Changes in R”
for detailed release information. I will draw atten-
tion to a “minor” but rather emotional point that has
sparked heated dicussions on the mailing lists in the
past. Release 1.8.0 marks the end of the use of the un-
derscore as an assignment operator in the R dialect of
the S language. That is, x_1 is now a syntax error.
A new column in R News lists the new members
of the R Foundation for Statistical Computing. R has
become a mature and valuable tool and we would
like to ensure its continued development and the de-
velopment of future innovations in software for sta-
tistical and computational research. We hope to at-
tract sufficient funding to make these goals realities.
Listing members in our newsletter is one (small) way
of thanking them for their support.
Friedrich Leisch
Technische Universität Wien, Austria
Friedrich.Leisch@R-project.org
Contents of this issue:
Editorial ...................... 1
RHelpDesk .................... 2
Integrating grid Graphics Output
with Base Graphics Output . . . . . . . . . . 7
A New Package for the General Error Distri-
bution ...................... 13
Web-based Microarray Analysis using Biocon-
ductor ...................... 17
Sweave, Part II: Package Vignettes . . . . . . . 21
R Foundation News . . . . . . . . . . . . . . . . 25
RecentEvents ................... 26
BookReviews ................... 28
Changes in R 1.8.0 . . . . . . . . . . . . . . . . . 29
Changes on CRAN . . . . . . . . . . . . . . . . 35
Crossword Solution . . . . . . . . . . . . . . . . 38
Correction to “Building Microsoft Windows
Versions of R and R packages under Intel
Linux” ...................... 39

Vol. 3/2, October 2003 2
R Help Desk
An Introduction to Using R’s Base Graphics
Marc Schwartz
Preface
As the use of R grows dramatically, an increas-
ingly diverse base of users will begin their explo-
ration of R’s programmatic approach to graphics.
Some new users will start without prior experience
generating statistical graphics using coded functions
(ie. they may have used GUI based “point-and-click”
or “drag-and-drop” graphic processes) and/or they
may be overwhelmed by the vast array (pardon
the pun) of graphic and plotting functions in R.
This transition can not only present a steep learning
curve, but can perhaps, by itself, become a barrier to
using R entirely, which would be an unfortunate out-
come.
R has essentially two separate core plotting en-
vironments in the default (base plus ‘recommended
package’) installation. The first is the extensive set
of base graphic functions and the second is the com-
bination of the grid (Murrell,2002) and lattice pack-
ages (Sarkar,2002), which together provide for ex-
tensive Trellis conditioning plots and related stan-
dardized functionality. For the purpose of this intro-
duction, I shall focus exclusively on the former.
The key advantages of a programmatic plotting
approach are much finer control over the plotting
process and, importantly, reproducibility. Days,
weeks or even months later, you can return to re-
use your same code with the same data to achieve
the same output. Ultimately, productivity is also en-
hanced because, once created, a single plotting func-
tion can be called quickly, generating one or an entire
series of graphics in a largely automated fashion.
R has a large number of “high” and “low” level
plotting functions that can be used, combined and
extended for specific purposes. This extensibility en-
ables R to meet a wide spectrum of needs, as demon-
strated by the number of contributed packages on
CRAN that include additional specialized plotting
functionality.
The breadth of base plotting functions is usually
quite satisfactory for many applications. In conjunc-
tion with R’s innate ability to deal with data in vec-
torized structures and by using differing ‘methods’,
one can further reduce the need for lengthy, repeti-
tive and complex code. In many cases, entire data
structures (ie. a linear model object) can be passed as
a single argument to a single plotting function, creat-
ing a default plot or series of plots.
Further, where default plot settings are perhaps
inappropriate for a given task, these can be ad-
justed to your liking and/or disabled. The base
graphic can be enhanced by using various lower
level plotting functions to add data points, lines,
curves, shapes, titles, legends and text annotations.
Formatted complex mathematical formulae (Murrell
and Ihaka,2000;Ligges,2002) can also be included
where required.
If a graphics ‘device’ is not explicitly opened
by the user, R’s high level plotting functions will
open the default device (see ?Devices) specified by
options("device"). In an interactive session, this is
typically the screen. However, one can also open an
alternative device such as a bitmap (ie. PNG/JPEG) or
a PostScript/PDF file for publishing and/or presen-
tation. I will focus on using the screen here, since
the particulars concerning other devices can be plat-
form specific. Note that if you intend to create plots
for output to something other than the screen, then
you must explicitly open the intended device. Dif-
ferences between the screen and the alternate device
can be quite significant in terms of the resultant plot
output. For example, you can spend a lot of time cre-
ating the screen version of a plot, only to find out it
looks quite different in a PostScript file,
Various parameters of the figure and plot regions
within a device can be set in advance by the use of
the par() function before calling the initial plot func-
tion. Others can be set as named arguments to the
plot functions. Options set by par() affect all graph-
ics; options set in a graphics call affect only that call.
(See ?par and ?plot.default for some additional
details).
It is possible to divide the overall graphic device
into a row/column grid of figures and create individ-
ual plots within each grid section (ie. a matrix of scat-
terplots like a pairs() plot) or create a graphic that
contains different plot types (ie. a scatterplot with
boxplots placed on the x and y axes). For more in-
formation, see ?layout,?split.screen and graphic
parameters ‘mfcol’ and ‘mfrow’ in ?par.
For additional details regarding graphic devices,
parameters and other considerations, please review
“Graphical Procedures” (Ch. 12) in “An Introduc-
tion to R” (Venables, Smith and R Core,2003) and
“Graphics” (Ch. 4) in “Modern Applied Statistics
with S” (Venables and Ripley,2002).
Let’s Get Plotting
In this limited space, it is not possible to cover all
the combinations and permutations possible with R’s
base graphics functionality (which could be a thick
book in its own right). Thus, I will put forth a fi-
nite set of practical examples that cover a modest
range of base plots and enhancements. For each plot,
R News ISSN 1609-3631
Vol. 3/2, October 2003 3
we will create some simple data to work with, cre-
ate a basic plot using a standard function to demon-
strate default behavior and then enhance the base
plot with additional detail. The included graphic for
each will show the final result. I recommend that you
consult the R help system for each function (using
?FunctionName) to better understand the syntax of
each function call and how each argument impacts
the resultant output.
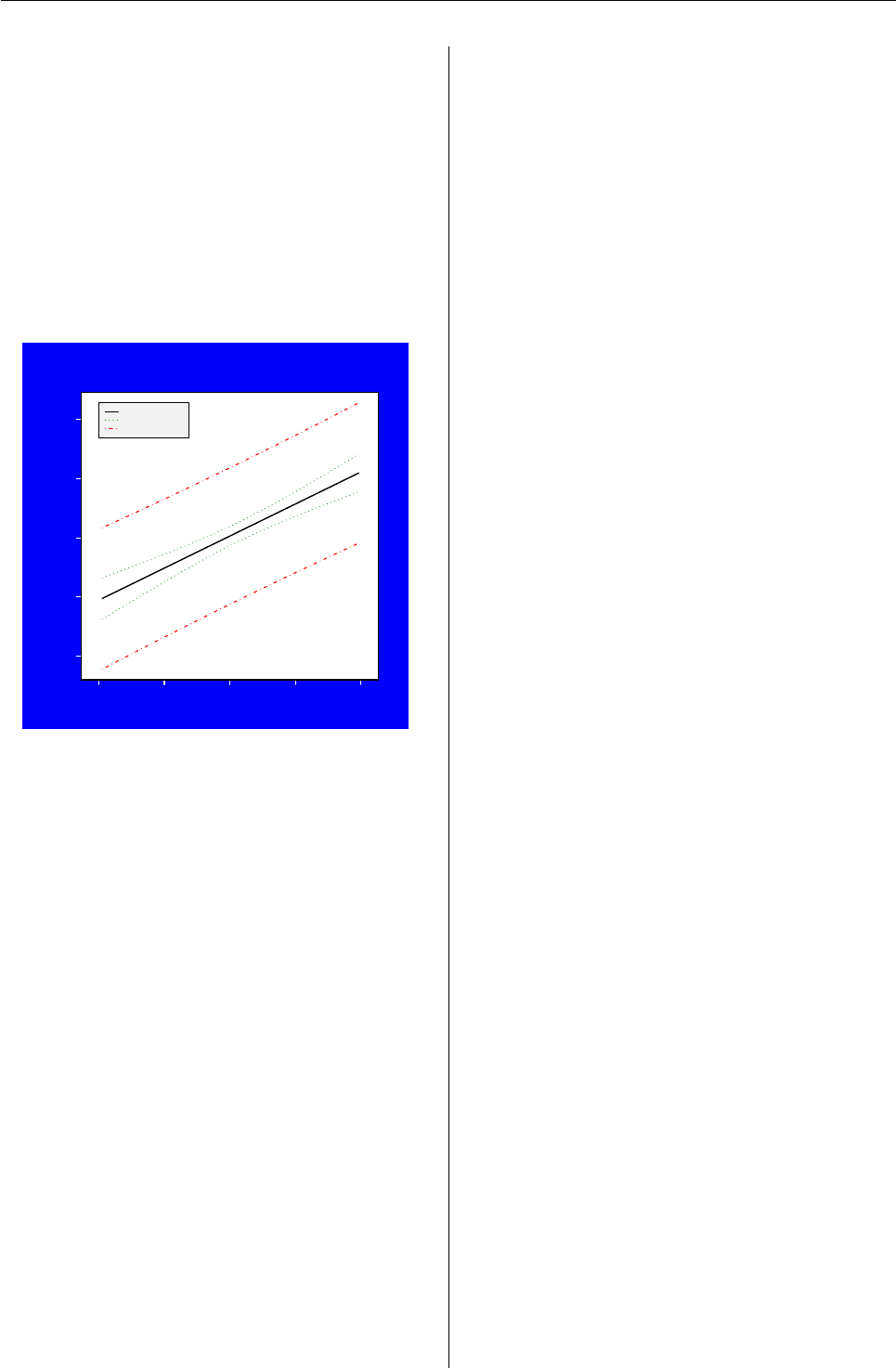
Scatterplot with a regression line and con-
fidence / prediction intervals
Linear Regression Plot
x vals
y vals
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
−2 −1 0 1 2
−4
−2
0
2
4
Fitted Line
Confidence Bands
Prediction Bands
The plot() function is a generic graphing func-
tion that can accept of variety of data structures
through specific defined ‘methods’. Frequently,
these arguments are numeric vectors representing
the two-dimensional (x,y) coordinate pairs of points
and/or lines to display. If you want to get a feel
for the breadth of plotting methods available use
methods(plot).
In the next example we first create a series of sim-
ple plots (not shown) then create the more complex
scatterplot shown above. To do this we create an x-y
scatterplot using type = "n" so that the axis ranges
are established, but nothing is plotted initially. We
then add the data points, the axes, a fitted regression
line, and confidence and prediction intervals for the
regression model:
# Create our data
set.seed(1)
x <- runif(50, -2, 2)
set.seed(2)
y <- x + rnorm(50)
# Create the model object
mod <- lm(y ~ x)
# Plot the data and add a regression line
# using default plot() behavior
plot(x, y)
abline(mod)
# Plot the model object, going through a
# sequence of diagnostic plots. See ?plot.lm
plot(mod)
# Create prediction values and confidence limits
# using a new dataframe of x values, noting the
# colnames need to match your model term names.
newData <- data.frame(x = seq(min(x), max(x),
by = (max(x) - min(x)) / 49))
pred.lim <- predict(mod, newdata = newData,
interval = "prediction")
conf.lim <- predict(mod, newdata = newData,
interval = "confidence")
# Function to color plot region
color.pr <- function(color = "white")
{
usr <- par("usr")
if (par("xlog"))
usr[1:2] <- 10 ^ usr[1:2]
if (par("ylog"))
usr[3:4] <- 10 ^ usr[3:4]
rect(usr[1], usr[3], usr[2], usr[4],
col = color)
}
# Color the plot background
par(bg = "blue")
# Define margins to enable space for labels
par(mar = c(5, 6, 5, 3) + 0.1)
# Create the plot. Do not plot the data points
# and axes to allow us to define them our way
plot(x, y, xlab = "x vals", ylab = "y vals",
type = "n", col.lab = "yellow", font.lab = 2,
cex.lab = 1.5, axes = FALSE, cex.main = 2,
main = "Linear Regression Plot",
col.main = "yellow", xlim = c(-2.1, 2.1),
ylim = range(y, pred.lim, na.rm = TRUE))
# Color the plot region white
color.pr("white")
# Plot the data points
points(x, y, pch = 21, bg = "yellow", cex=1.25)
# Draw the fitted regression line and the
# prediction and confidence intervals
matlines(newData$x, pred.lim, lty = c(1, 4, 4),
lwd = 2, col = c("black", "red", "red"))
matlines(newData$x, conf.lim, lty = c(1, 3, 3),
lwd = 2, col = c("black", "green4", "green4"))
# Draw the X and Y axes, repectively
axis(1, at = -2:2, col = "white",
col.axis = "white", lwd = 2)
axis(2, at = pretty(range(y), 3), las = 1,
col = "white", col.axis = "white", lwd = 2)
# Draw the legend
legend(-2, max(pred.lim, na.rm = TRUE),
legend = c("Fitted Line", "Confidence Bands",
"Prediction Bands"),
lty = c(1, 3, 4), lwd = 2,
col = c("black", "green4", "red"),
horiz = FALSE, cex = 0.9, bg = "gray95")
# Put a box around the plot
box(lwd = 2)
R News ISSN 1609-3631
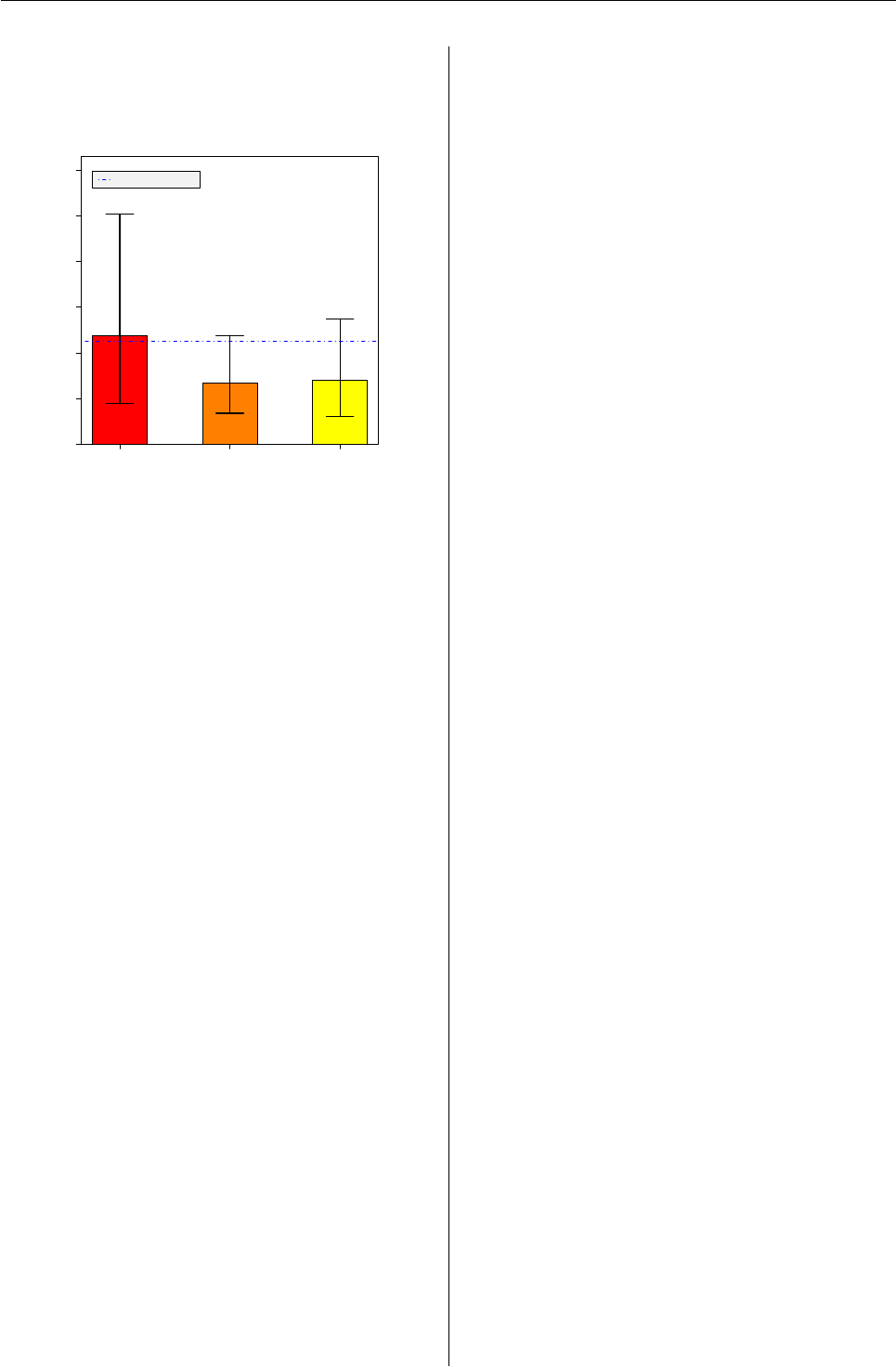
Vol. 3/2, October 2003 4
Barplot with confidence intervals and ad-
ditional annotation
0.0%
2.0%
4.0%
6.0%
8.0%
10.0%
12.0%
% Incidence (+/−95% CI)
A B C
126 409 284
p = 0.8285 p = 0.0931 p = 0.1977
4.8%
2.7% 2.8%
Benchmark Value: 4.5%
Incidence of Event By Group
Total N = 819
barplot() can draw essentially three types of
plots with either vertical or horizontal bars (using
the argument horiz = TRUE / FALSE). The first is
a series of individual bars where the height argu-
ment (which defines the bar values) is a simple vec-
tor. The second is a series of stacked multi-segment
bars where height is a matrix and beside = FALSE.
The third is a series of grouped bars where height is
a matrix and beside = TRUE. In the second and third
cases, each column of the matrix height represents
either the values of the bar segments in each stacked
bar, or the values of the individual bars in each bar
group, respectively.
barplot() returns either a vector or a matrix
(when beside = TRUE) of bar midpoints that can be
assigned to a variable (ie. mp <- barplot(...)). You
can use this information to locate bar midpoints for
text and/or line placement. To locate the midpoint
of bar groups, use colMeans(mp) to enable the place-
ment of a bar group label.
Here we will create a vertical barplot, with each
of the three bars representing a proportion. We will
add binomial confidence intervals and p values from
binom.test() using a ‘benchmark’ value that will be
plotted. We will label the y axis with percentages
(prop * 100), add bar values above the top of each
bar and put sample sizes centered below each bar un-
der the x axis.
# Create our data
A <- data.frame(Event = c(rep("Yes", 6),
rep("No", 120)), Group = "A")
B <- data.frame(Event = c(rep("Yes", 11),
rep("No", 398)), Group = "B")
C <- data.frame(Event = c(rep("Yes", 8),
rep("No", 276)), Group = "C")
BarData <- rbind(A, B, C)
attach(BarData)
# Create initial ’default’ barplots
barplot(table(Group))
barplot(table(Group), horiz = TRUE)
barplot(table(Event, Group))
barplot(table(Event, Group), beside = TRUE)
# Let’s get our summary data from the dataframe
table.data <- table(Event, Group)
# Get sample sizes
n <- as.vector(colSums(table.data))
# Get number of "Yes" events
events <- as.vector(table.data["Yes", ])
# Proportion of "Yes" events
prop.events <- events / n
# Group names from table dimnames
Group.Names <- dimnames(table.data)$Group
# Define our benchmark value
benchmark <- 0.045
# Get binomial confidence limits and p values
stats <- mapply(binom.test, x = events, n = n,
p = benchmark)
# ci[, 1] = lower and ci[, 2] = upper
ci <- matrix(unlist(stats["conf.int", ]),
ncol = 2, byrow = TRUE)
p.val <- unlist(stats["p.value", ])
# Define Y axis range to include CI’s and
# space for a legend in the upper LH corner
YMax <- max(ci[, 2]) * 1.25
# Define margins to enable space for labels
par(mar = c(5, 6, 5, 3) + 0.1)
# Do the barplot, saving bar midpoints in MidPts
MidPts <- barplot(prop.events, space = 1,
axes = FALSE,axisnames = FALSE,
ylim = c(0, YMax))
# Define formatted Y axis labels using
# axTicks() and draw the Y Axis and label
YLabels <- paste(formatC(axTicks(2) * 100,
format = "f", digits = 1),
"%", sep = "")
YAxisLab <- "% Incidence (+/-95% CI)"
axis(2, labels = YLabels, at = axTicks(2),
las = 1)
mtext(YAxisLab, side = 2, adj = 0.5,
line = 4.5, cex = 1.1, font = 2)
# Draw the X axis using Group Names at bar
# midpoints
axis(1, labels = Group.Names, at = MidPts,
font = 2, cex.axis = 1.25)
# Draw Sample Sizes and p Values below Group
# Names
mtext(n, side = 1, line = 2, at = MidPts,
cex = 0.9)
p.val.text <- paste("p = ",
formatC(p.val, format = "f", digits = 4),
sep = "")
mtext(p.val.text, side = 1, line = 3,
at = MidPts, cex = 0.9)
# Place formatted bar values above the left edge
# of each bar so that CI lines do not go through
# numbers. Left edge = MidPts - (’width’ / 2)
bar.vals <- paste(formatC(
prop.events * 100, format = "f", digits=1),
"%", sep = "")
text(MidPts - 0.5, prop.events, cex = 0.9,
labels = bar.vals, adj = c(0, -0.5), font=1)
R News ISSN 1609-3631
Vol. 3/2, October 2003 5
# Draw confidence intervals, first drawing
# vertical line segments and then upper and
# lower horizontal boundary segments
segments(MidPts, ci[, 1], MidPts, ci[, 2],
lty = "solid", lwd = 2)
segments(MidPts - 0.25, ci[, 1],
MidPts + 0.25, ci[, 1], lty = "solid", lwd=2)
segments(MidPts - 0.25, ci[, 2],
MidPts + 0.25, ci[, 2], lty = "solid", lwd=2)
# Plot benchmark line
abline(h = benchmark, lty = "dotdash",
lwd = 2, col = "blue")
# Draw legend
legend(1, YMax * 0.95, lty = "dotdash",
legend = "Benchmark Value: 4.5%", lwd = 2,
col = "blue", horiz = FALSE, cex = 0.9,
bg = "gray95")
# Draw title and sub-title
mtext("Incidence of Event By Group", side = 3,
line = 3, cex = 1.5, font = 2)
mtext(paste("Total N = ", sum(n), sep = ""),
side = 3, line = 1, cex = 1, font = 2)
# Put box around plot
box()
detach(BarData)
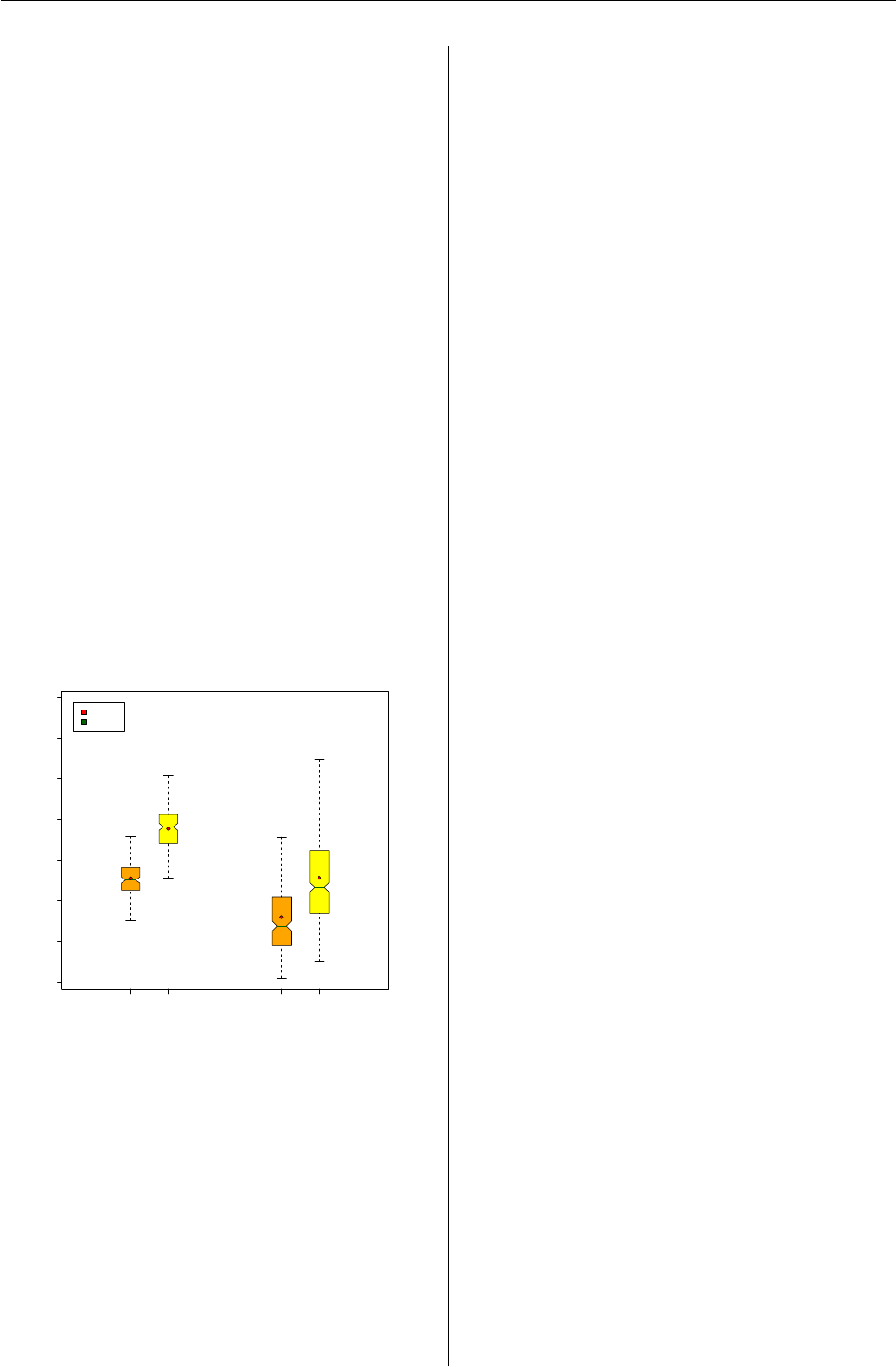
Paired Boxplots with outliers colored and
median / mean values labeled
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
0
2
4
6
8
10
12
14
A1 B1 A2 B2
135 175 250 500
5.1
7.5
3.2
5.1
5.0
7.6
2.7
4.6
Distribution of ’Measure’ by ’Group’
Mean
Median
J.W. Tukey’s Box-Whisker plots (Tukey,1977) are
a quick and easy way to visually review and com-
pare the distributions of continuous variables. For
some descriptive information on the structure and
interpretation of these plots including additional ref-
erences, see ?boxplot.stats.
Here we will generate continuous measures in
four groups. We will generate default plots and then
enhance the layout of the plot to visually group the
data and to annotate it with key labels.
# Create our data
set.seed(1)
A1 <- data.frame(Group = "A1",
Measure = rnorm(135, 5))
set.seed(2)
A2 <- data.frame(Group = "A2",
Measure = rgamma(250, 3))
set.seed(3)
B1 <- data.frame(Group = "B1",
Measure = rnorm(175, 7.5))
set.seed(4)
B2 <- data.frame(Group = "B2",
Measure = rgamma(500, 5))
BPData <- rbind(A1, A2, B1, B2)
attach(BPData)
# Create default boxplots
boxplot(Measure)
boxplot(Measure, horizontal = TRUE)
boxplot(Measure ~ Group)
# Adjust Group factor levels to put A1 / B1
# and A2 / B2 pairs together
Group <- factor(Group,
levels = c("A1", "B1", "A2", "B2"))
# Show default boxplot with re-grouping
boxplot(Measure ~ Group)
# Define that boxplot midpoints to separate
# the pairs of plots
at <- c(1.25, 1.75, 3.25, 3.75)
# Draw boxplot, returning boxplot stats in S
# which will contain summary data for each Group.
# See ?boxplot.stats
S <- boxplot(Measure ~ Group, boxwex = 0.25,
col = c("orange", "yellow"), notch = TRUE,
at = at, axes = FALSE)
# Draw thicker green lines for median values
# When notch = TRUE, median width = boxwex / 2
segments(at - 0.0625, S$stats[3, ],
at + 0.0625, S$stats[3, ],
lwd = 2, col = "darkgreen")
# Get Group means and plot them using a
# diamond plot symbol
means <- by(Measure, Group, mean)
points(at, means, pch = 23, cex = 0.75,
bg = "red")
# Color outlier values using x,y positions from S
points(at[S$group], S$out, pch = 21, bg="blue")
# Draw Y axis, rotating labels to horiz
axis(2, las = 1)
# Draw X Axis Group Labels
axis(1, at = at, labels = S$names,
cex.axis = 1.5, font.axis = 2)
mtext(S$n, side = 1, at = at, line = 3)
# Draw Mean values to the left edge of each
# boxplot
text(at - 0.125, means, labels = formatC(
means, format = "f", digits = 1),
pos = 2, cex = 0.9, col = "red")
# Draw Median values to the right edge of
# each boxplot
text(at + 0.125, S$stats[3, ],
labels = formatC(S$stats[3, ], format = "f",
digits = 1),
pos = 4, cex = 0.9, col = "darkgreen")
# Draw a box around plot
box()
# Add title and legend
title("Distribution of ’Measure’ by ’Group’",
R News ISSN 1609-3631

Vol. 3/2, October 2003 6
cex.main = 1.5)
legend(0.5, max(Measure),
legend = c("Mean", "Median"),
fill = c("red", "darkgreen"))
detach(BPData)
Additional Resources
For additional information on using R’s plotting
functionality, see: Venables, Smith and R Core (2003);
Venables and Ripley (2002); Fox (2002); Dalgaard
(2002). In addition, Uwe Ligges’ recent R News ar-
ticle (Ligges,2003) provides excellent insights into
how best to utilize R’s documentation and help re-
sources.
If you are in need of expert guidance on creating
analytic graphics, such as the pros and cons of using
particular graphic formats and their impact on the
interpretation of your data, two critically important
references are “Visualizing Data” (Cleveland,1993)
and “The Elements of Graphing Data” (Cleveland,
1994).
Bibliography
Cleveland, W. S. (1993): Visualizing Data. Summit,
NJ: Hobart Press. 6
Cleveland, W. S. (1994): The Elements of Graphing
Data. Summit, NJ: Hobart Press, revised edition.
6
Dalgaard, P. (2002): Introductory Statistics with R.
New York: Springer-Verlag. 6
Fox, J. (2002): An R and S-PLUS Companion to Applied
Regression. Thousand Oaks: Sage. 6
Ligges, U. (2002): R Help Desk – Automation of
Mathematical Annotation in Plots. R News, 2 (3),
32–34. ISSN 1609-3631. URL http://CRAN.
R-project.org/doc/Rnews/.2
Ligges, U. (2003): R Help Desk – Getting Help – R’s
Help Facilities and Manuals. R News, 3 (1), 26–28.
ISSN 1609-3631. URL http://CRAN.R-project.
org/doc/Rnews/.6
Murrell, P. (2002): The grid Graphics Package. R
News, 2 (2), 14–19. ISSN 1609-3631. URL http:
//CRAN.R-project.org/doc/Rnews/.2
Murrell, P. and Ihaka, R. (2000): An Approach to Pro-
viding Mathematical Annotation in Plots. Journal
of Computational and Graphical Statistics, 9 (3), 582–
599. 2
Sarkar, D. (2002): Lattice: An Implementation of Trel-
lis Graphics in R. R News, 2 (2), 19–23. ISSN
1609-3631. URL http://CRAN.R-project.org/
doc/Rnews/.2
Tukey, J. (1977): Exploratory Data Analysis. Reading,
MA: Addison-Wesley. 5
Venables, W. N. and Ripley, B. D. (2002): Modern Ap-
plied Statistics with S. New York: Springer-Verlag,
4th edition. 2,6
Venables, W. N., Smith, D. M. and the R De-
velopment Core Team (2003): An Introduction
to R. URL http://CRAN.R-project.org/doc/
manuals.html.2,6
Marc Schwartz
MedAnalytics, Inc., Minneapolis, Minnesota, USA
MSchwartz@MedAnalytics.com
R News ISSN 1609-3631
Vol. 3/2, October 2003 7
Integrating grid Graphics Output
with Base Graphics Output
by Paul Murrell
Introduction
The grid graphics package (Murrell,2002) is much
more powerful than the standard R graphics system
(hereafter “base graphics”) when it comes to com-
bining and arranging graphical elements. It is pos-
sible to create a greater variety of graphs more easily
with grid (see, for example, Deepayan Sarkar’s lat-
tice package (Sarkar,2002)). However, there are very
many plots based on base graphics (e.g., biplots), that
have not been implemented in grid, and the task of
reimplementing these in grid is extremely daunting.
It would be nice to be able to combine the ready-
made base plots with the sophisticated arrangement
features of grid.
This document describes the gridBase package
which provides some support for combining grid
and base graphics output.
Annotating base graphics
using grid
The gridBase package provides the baseViewports()
function, which supports adding grid output to a
base graphics plot. This function creates a set of grid
viewports that correspond to the current base plot.
These allow simple operations such as adding lines
and text using grid’s units to locate them relative to
a wide variety of coordinate systems, or something
more complex involving pushing further grid view-
ports.
baseViewports() returns a list of three grid view-
ports. The first corresponds to the base “inner” re-
gion. This viewport is relative to the entire device;
it only makes sense to push this viewport from the
“top level” (i.e., only when no other viewports have
been pushed). The second viewport corresponds to
the base “figure” region and is relative to the inner
region; it only makes sense to push it after the “in-
ner” viewport has been pushed. The third viewport
corresponds to the base “plot” region and is relative
to the figure region; it only makes sense to push it af-
ter the other two viewports have been pushed in the
correct order.
0246810
one
two
three
four
five
six
seven
eight
nine
ten
Figure 1: Annotating a base plot with grid.text().
A simple application of this facility involves
adding text to the margins of a base plot at an arbi-
trary orientation. The base function mtext() allows
text to be located in terms of a number of lines away
from the plot region, but only at rotations of 0 or 90
degrees. The base text() function allows arbitrary
rotations, but only locates text relative to the user co-
ordinate system in effect in the plot region (which is
inconvenient for locating text in the margins of the
plot). By contrast, the grid function grid.text() al-
lows arbitrary rotations and can be used in any grid
viewport. In the following code we first create a base
plot, leaving off the tick labels.
> midpts <- barplot(1:10, axes = FALSE)
> axis(2)
> axis(1, at = midpts, labels = FALSE)
Next we use baseViewports() to create grid view-
ports that correspond to the base plot and we push
those viewports1.
> vps <- baseViewports()
> par(new = TRUE)
> push.viewport(vps$inner, vps$figure,
+ vps$plot)
Finally, we draw rotated labels using grid.text()
(and pop the viewports to clean up after ourselves).
The final plot is shown in Figure 1.
1The par(new=TRUE) is necessary currently because the first grid action will try to move to a new page; it should be possible to remove
this step in future versions of R.
R News ISSN 1609-3631
Vol. 3/2, October 2003 8
> grid.text(c("one", "two", "three",
+ "four", "five", "six", "seven",
+ "eight", "nine", "ten"),
+ x = unit(midpts, "native"),
+ y = unit(-1, "lines"), just = "right",
+ rot = 60)
> pop.viewport(3)
The next example is a bit more complicated be-
cause it involves embedding grid viewports within
a base graphics plot. The dataset is a snapshot of
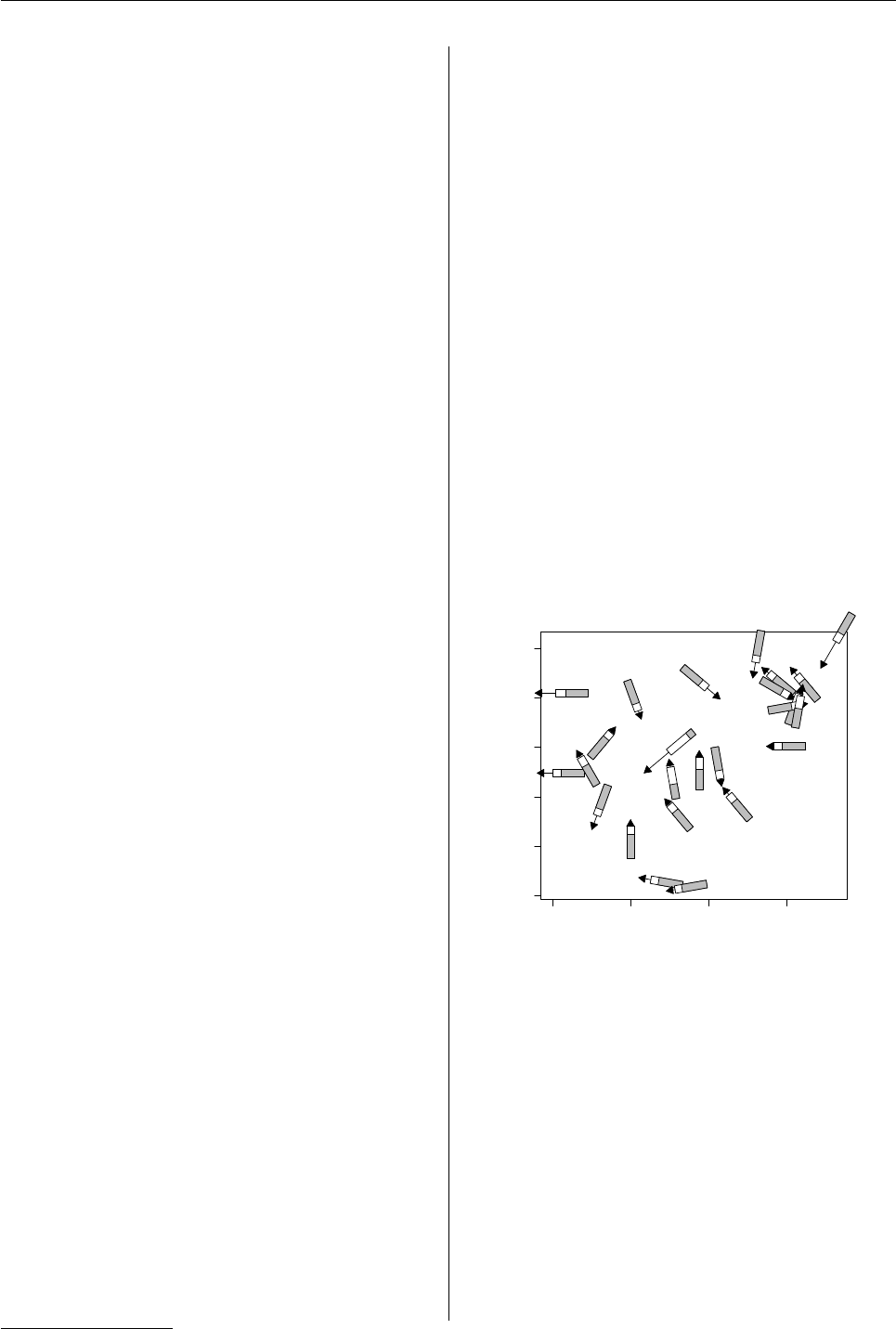
wind speed, wind direction, and temperature at sev-
eral weather stations in the South China Sea, south
west of Japan2.grid is used to produce novel plot-
ting symbols for a standard base plot.
First of all, we need to define the novel plotting
symbol. This consists of a dot at the data location,
with a thermometer extending “below” and an ar-
row extending “above”. The thermometer is used to
encode temperature and the arrow is used to indicate
wind speed (both scaled to [0, 1]).
> novelsym <- function(speed,
+ temp, width = unit(3, "mm"),
+ length = unit(0.5, "inches")) {
+ grid.rect(height = length,
+ y = 0.5, just = "top",
+ width = width,
+ gp = gpar(fill = "white"))
+ grid.rect(height = temp *
+ length, y = unit(0.5,
+ "npc") - length, width = width,
+ just = "bottom",
+ gp = gpar(fill = "grey"))
+ grid.arrows(x = 0.5,
+ y = unit.c(unit(0.5, "npc"),
+ unit(0.5, "npc") +
+ speed * length),
+ length = unit(3, "mm"),
+ type = "closed",
+ gp = gpar(fill = "black"))
+ grid.points(unit(0.5, "npc"),
+ unit(0.5, "npc"), size = unit(2,
+ "mm"), pch = 16)
+ }
Now we read in the data and generate a base plot,
but plot no points.
> chinasea <- read.table("chinasea.txt",
+ header = TRUE)
> plot(chinasea$lat, chinasea$long,
+ type = "n", xlab = "latitude",
+ ylab = "longitude",
+ main = "China Sea ...")
Now we use baseViewports() to align a grid view-
port with the plot region, and draw the symbols by
creating a grid viewport per (x,y)location (we rotate
the viewport to represent the wind direction). The fi-
nal plot is shown in Figure 2.
> speed <- 0.8 * chinasea$speed/14 +
+ 0.2
> temp <- chinasea$temp/40
> vps <- baseViewports()
> par(new = TRUE)
> push.viewport(vps$inner, vps$figure,
+ vps$plot)
> for (i in 1:25) {
+ push.viewport(viewport(
+ x = unit(chinasea$lat[i],
+ "native"),
+ y = unit(chinasea$long[i],
+ "native"),
+ angle = chinasea$dir[i]))
+ novelsym(speed[i], temp[i])
+ pop.viewport()
+ }
> pop.viewport(3)
22 23 24 25
119.5 120.0 120.5 121.0 121.5 122.0
China Sea Wind Speed/Direction and Temperature
latitude
longitude
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
Figure 2: Using grid to draw novel symbols on a
base plot.
Embedding base graphics plots in
grid viewports
gridBase provides several functions for adding base
graphics output to grid output. There are three func-
tions that allow base plotting regions to be aligned
with the current grid viewport; this makes it possi-
ble to draw one or more base graphics plots within a
grid viewport. The fourth function provides a set of
2Obtained from the CODIAC web site: http://www.joss.ucar.edu/codiac/codiac-www.html. The file chinasea.txt is in the grid-
Base/doc directory.
R News ISSN 1609-3631
Vol. 3/2, October 2003 9
graphical parameter settings so that base par() set-
tings can be made to correspond to some of3the cur-
rent grid graphical parameter settings.
The first three functions are gridOMI(),
gridFIG(), and gridPLT(). They return the appro-
priate par() values for setting the base “inner”, “fig-
ure”, and “plot” regions, respectively.
The main usefulness of these functions is to allow
you to create a complex layout using grid and then
draw a base plot within relevant elements of that lay-
out. The following example uses this idea to create
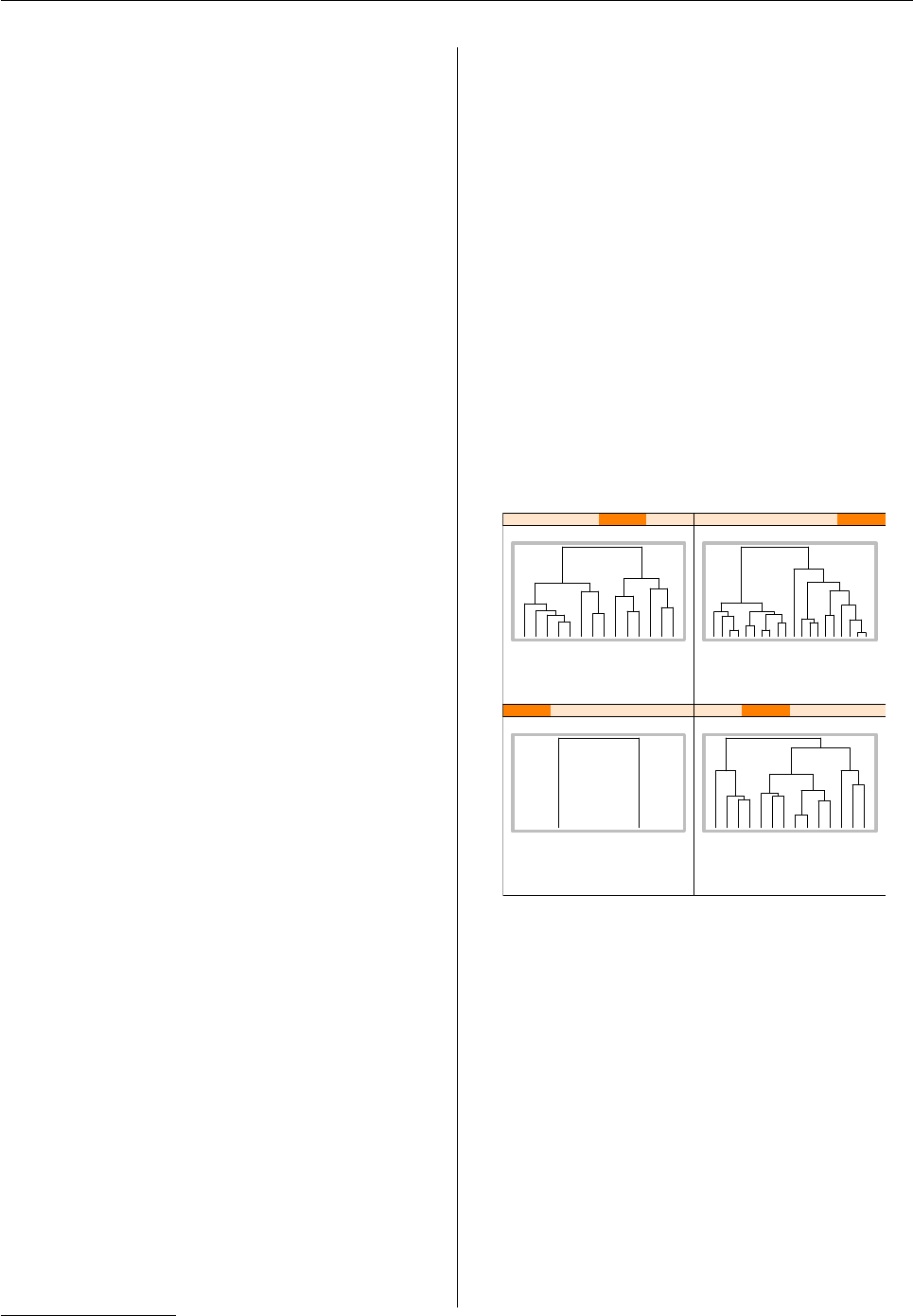
alattice plot where the panels contain dendrograms
drawn using base graphics functions4.
First of all, we create a dendrogram and cut it into
four subtrees5.
> library(mva)
> data(USArrests)
> hc <- hclust(dist(USArrests),
+ "ave")
> dend1 <- as.dendrogram(hc)
> dend2 <- cut(dend1, h = 70)
Now we create some dummy variables which corre-
spond to the four subtrees.
> x <- 1:4
> y <- 1:4
> height <- factor(round(unlist(
+ lapply(dend2$lower,
+ attr, "height"))))
Next we define a lattice panel function to draw the
dendrograms. The first thing this panel function
does is push a viewport that is smaller than the view-
port lattice creates for the panel; the purpose is to en-
sure there is enough room for the labels on the den-
drogram. The space variable contains a measure of
the length of the longest label. The panel function
then calls gridPLT() and makes the base plot region
correspond to the viewport we have just pushed. Fi-
nally, we call the base plot() function to draw the
dendrogram (and pop the viewport we pushed)6.
> space <- max(unit(rep(1, 50),
+ "strwidth",
+ as.list(rownames(USArrests))))
> dendpanel <- function(x, y,
+ subscripts, ...) {
+ push.viewport(viewport(y = space,
+ width = 0.9, height = unit(0.9,
+ "npc") - space,
+ just = "bottom"))
+ grid.rect(gp = gpar(col = "grey",
+ lwd = 5))
+ par(plt = gridPLT(), new = TRUE,
+ ps = 10)
+ plot(dend2$lower[[subscripts]],
+ axes = FALSE)
+ pop.viewport()
+ }
Finally, we draw a lattice xyplot, using lattice to set
up the arrangement of panels and strips and our
panel function to draw a base dendrogram in each
panel. The final plot is shown in Figure 3.
> library(lattice)
> xyplot(y ~ x | height, subscripts = TRUE,
+ xlab = "", ylab = "",
+ strip = function(...) {
+ strip.default(style = 4,
+ ...)
+ }, scales = list(draw = FALSE),
+ panel = dendpanel)
Florida
North Carolina
39 44 45 55
California
Maryland
Arizona
New Mexico
Delaware
Alabama
Louisiana
Illinois
New York
Michigan
Nevada
Alaska
Mississippi
South Carolina
39 44 45 55
Washington
Oregon
Wyoming
Oklahoma
Virginia
Rhode Island
Massachusetts
New Jersey
Missouri
Arkansas
Tennessee
Georgia
Colorado
Texas
39 44 45 55
Idaho
Nebraska
Kentucky
Montana
Ohio
Utah
Indiana
Kansas
Connecticut
Pennsylvania
Hawaii
West Virginia
Maine
South Dakota
North Dakota
Vermont
Minnesota
Wisconsin
Iowa
New Hampshire
39 44 45 55
Figure 3: Adding base dendrograms to a lattice plot.
The gridPLT() function is useful for embedding
just the plot region of a base graphics function (i.e.,
without labels and axes; another example of this us-
age is given in the next section). If labelling and axes
are to be included it will make more sense to use
gridFIG(). The gridOMI() function has pretty much
the same effect as gridFIG() except that it allows for
the possibility of embedding multiple base plots at
once. In the following code, a lattice plot is placed
alongside base diagnostic plots arranged in a 2-by-2
array.
We use the data from page 93 of “An Introduc-
tion to Generalized Linear Models” (Annette Dob-
son, 1990).
3Only lwd,lty,col are available yet. More should be available in future versions.
4Recall that lattice is built on grid so the panel region in a lattice plot is a grid viewport.
5the data and cluster analysis are copied from the example in help(plot.dendrogram).
6The grid.rect() call is just to show the extent of the extra viewport we pushed.
R News ISSN 1609-3631

Vol. 3/2, October 2003 10
> counts <- c(18, 17, 15, 20,
+ 10, 20, 25, 13, 12)
> outcome <- gl(3, 1, 9)
> treatment <- gl(3, 3)
We create two regions using grid viewports; the left
region is for the lattice plot and the right region is
for the diagnostic plots. There is a middle column of
1cm to provide a gap between the two regions.
> push.viewport(viewport(
+ layout = grid.layout(1,
+ 3, widths = unit(rep(1,
+ 3), c("null", "cm",
+ "null")))))
We draw a lattice plot in the left region.
> push.viewport(viewport(
+ layout.pos.col = 1))
> library(lattice)
> bwplot <- bwplot(counts ~ outcome |
+ treatment)
> print(bwplot, newpage = FALSE)
> pop.viewport()
We draw the diagnostic plots in the right region.
Here we use gridOMI() to set the base inner re-
gion and par(mfrow) and par(mfg) to insert multi-
ple plots7. The final plot is shown in Figure 4.
> push.viewport(viewport(layout.pos.col = 3))
> glm.D93 <- glm(counts ~ outcome +
+ treatment, family = poisson())
> par(omi = gridOMI(), mfrow = c(2,
+ 2), new = TRUE)
> par(cex = 0.5, mar = c(5, 4,
+ 1, 2))
> par(mfg = c(1, 1))
> plot(glm.D93, caption = "",
+ ask = FALSE)
> pop.viewport(2)
Notice that because there is only ever one cur-
rent grid viewport, it only makes sense to use one
of gridOMI(),gridFIG(), or gridPLT(). In other
words, it only makes sense to align either the inner
region, or the figure region, or the plot region with
the current grid viewport.
A more complex example
We will now look at a reasonably complex exam-
ple involving embedding base graphics within grid
viewports which are themselves embedded within a
base plot. This example is motivated by the follow-
ing problem8:
I am looking at a way of plotting a se-
ries of pie charts at specified locations on
an existing plot. The size of the pie chart
would be proportion to the magnitude of
the total value of each vector (x) and the
values in x are displayed as the areas of
pie slices.
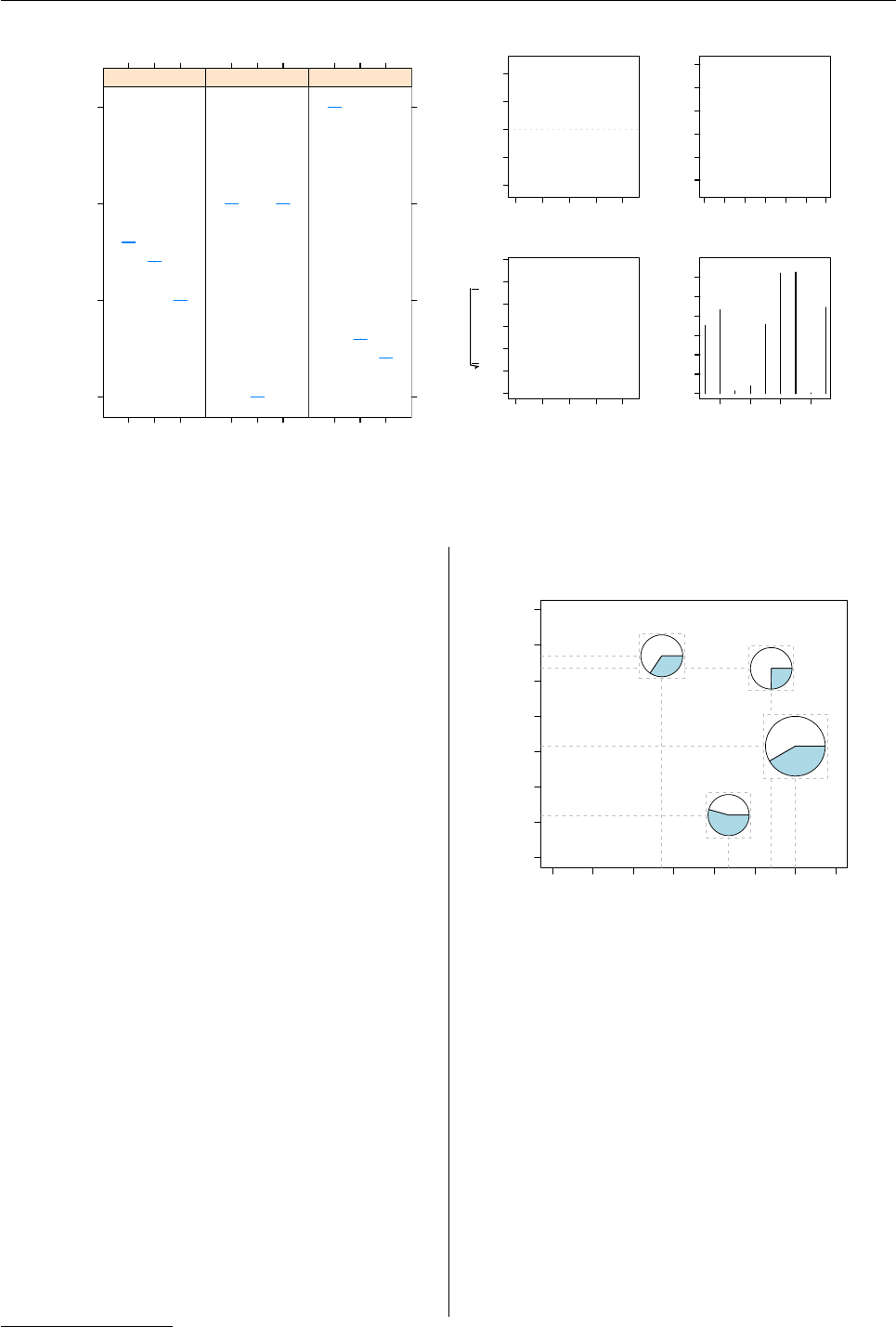
First of all, we construct some fake data, consist-
ing of four (x,y)values, and four (z1,z2)values :
> x <- c(0.88, 1, 0.67, 0.34)
> y <- c(0.87, 0.43, 0.04, 0.94)
> z <- matrix(runif(4 * 2), ncol = 2)
Before we start any plotting, we save the current
par() settings so that at the end we can “undo” some
of the complicated settings that we need to apply.
> oldpar <- par(no.readonly = TRUE)
Now we do a standard base plot of the (x,y)values,
but do not plot anything at these locations (we’re just
setting up the user coordinate system).
> plot(x, y, xlim = c(-0.2, 1.2),
+ ylim = c(-0.2, 1.2), type = "n")
Now we make use of baseViewports. This will cre-
ate a list of grid viewports that correspond to the
inner, figure, and plot regions set up by the base
plot. By pushing these viewports, we establish a grid
viewport that aligns exactly with the plot region cre-
ated by the base plot, including a (grid) “native” co-
ordinate system that matches the (base) user coordi-
nate system9.
> vps <- baseViewports()
> par(new = TRUE)
> push.viewport(vps$inner, vps$figure,
+ vps$plot)
> grid.segments(x0 = unit(c(rep(0,
+ 4), x), rep(c("npc", "native"),
+ each = 4)), x1 = unit(c(x,
+ x), rep("native", 8)), y0 = unit(c(y,
+ rep(0, 4)), rep(c("native",
+ "npc"), each = 4)), y1 = unit(c(y,
+ y), rep("native", 8)),
+ gp = gpar(lty = "dashed",
+ col = "grey"))
Before we draw the pie charts, we need to perform
a couple of calculations to determine their size. In
this case, we specify that the largest pie will be 1"in
diameter and the others will be a proportion of that
size based on ∑iz.i/max (∑iz.i)
7We use par(mfrow) to specify the 2-by-2 array and par(mfg) to start at position (1, 1)in the array.
8This description is from an email to R-help from Adam Langley, 18 July 2003
9The grid.segments call is just drawing some dashed lines to show that the pie charts we end up with are centred correctly at the
appropriate (x,y)locations.
R News ISSN 1609-3631
Vol. 3/2, October 2003 11
counts
●
●
●
10
15
20
25
1 2 3
1
●
●
●
1 2 3
2
●
●
●
1 2 3
3
2.6 2.7 2.8 2.9 3.0
−1.0 −0.5 0.0 0.5 1.0
Predicted values
Residuals
●
●
●●
●
●
●
●
●
6
9
2
●
●
●
●
●
●
●
●
●
−1.5 −0.5 0.5 1.0 1.5
−1.0 0.0 0.5 1.0 1.5
Theoretical Quantiles
Std. deviance resid.
6
9
2
2.6 2.7 2.8 2.9 3.0
0.0 0.2 0.4 0.6 0.8 1.0 1.2
Predicted values
Std. deviance resid.
●
●
●
●
●
●
●
●
●
6
9
2
2 4 6 8
0.0 0.1 0.2 0.3 0.4 0.5 0.6
Obs. number
Cook’s distance
7
6
9
Figure 4: Drawing multiple base plots within a grid viewport.
> maxpiesize <- unit(1, "inches")
> totals <- apply(z, 1, sum)
> sizemult <- totals/max(totals)
We now enter a loop to draw a pie at each (x,y)loca-
tion representing the corresponding (z1,z2)values.
The first step is to create a grid viewport at the (x,y)
location, then we use gridPLT() to set the base plot
region to correspond to the grid viewport. With that
done, we can use the base pie function to draw a pie
chart within the grid viewport10.
> for (i in 1:4) {
+ push.viewport(viewport(x = unit(x[i],
+ "native"), y = unit(y[i],
+ "native"), width = sizemult[i] *
+ maxpiesize, height = sizemult[i] *
+ maxpiesize))
+ grid.rect(gp = gpar(col = "grey",
+ fill = "white", lty = "dashed"))
+ par(plt = gridPLT(), new = TRUE)
+ pie(z[i, ], radius = 1,
+ labels = rep("", 2))
+ pop.viewport()
+ }
Finally, we clean up after ourselves by popping the
grid viewports and restoring the initial par settings.
> pop.viewport(3)
> par(oldpar)
The final plot is shown in Figure 5.
−0.2 0.0 0.2 0.4 0.6 0.8 1.0 1.2
−0.2 0.0 0.2 0.4 0.6 0.8 1.0 1.2
x
y
Figure 5: Base pie charts drawn within grid view-
ports, which are embedded within a base plot.
Problems and limitations
The functions provided by the gridBase package al-
low the user to mix output from two quite different
graphics systems and there are limits to how much
the systems can be combined. It is important that
users are aware that they are mixing two not wholly
compatible systems (which is why these functions
are provided in a separate package) and it is of course
important to know what the limitations are:
• The gridBase functions attempt to match grid
10We draw a grid.rect with a dashed border just to show the extent of each grid viewport. It is crucial that we again call par(new=TRUE)
so that we do not move on to a new page.
R News ISSN 1609-3631

Vol. 3/2, October 2003 12
graphics settings with base graphics settings
(and vice versa). This is only possible under
certain conditions. For a start, it is only possi-
ble if the device size does not change. If these
functions are used to draw into a window, then
the window is resized, the base and grid set-
tings will almost certainly no longer match and
the graph will become a complete mess. This
also applies to copying output between devices
of different sizes.
• It is not possible to embed base graphics output
within a grid viewport that is rotated.
• There are certain base graphics functions which
modify settings like par(omi) and par(fig)
themselves (e.g., coplot()). Output from these
functions may not embed properly within grid
viewports.
•grid output cannot be saved and restored so
any attempts to save a mixture of grid and base
output are likely to end in disappointment.
Summary
The functions in the gridBase package provide a sim-
ple mechanism for combining base graphics output
with grid graphics output for static, fixed-size plots.
This is not a full integration of the two graphics sys-
tems, but it does provide a useful bridge between the
existing large body of base graphics functions and
the powerful new features of grid.
Availability
The grid package is now part of the base distri-
bution of R (from R version 1.8.0). Additional in-
formation on grid is available from: http://www.
stat.auckland.ac.nz/~paul/grid/grid.html. The
gridBase package is available from CRAN (e.g.,
http://cran.us.r-project.org).
Bibliography
P. Murrell. The grid graphics package. R News, 2(2):
14–19, June 2002. URL http://CRAN.R-project.
org/doc/Rnews/.7
D. Sarkar. Lattice. R News, 2(2):19–23, June 2002. URL
http://CRAN.R-project.org/doc/Rnews/.7
Paul Murrell
University of Auckland, NZ
paul@stat.auckland.ac.nz
R News ISSN 1609-3631
Vol. 3/2, October 2003 13
A New Package for the General Error
Distribution
The normalp package
Angelo M. Mineo
Introduction
The General Error Distribution, whose first formula-
tion could be ascribed to the Russian mathematician
Subbotin (1923), is a general distribution for random
errors. To derive this random error distribution, Sub-
botin extended the two axioms used by Gauss to de-
rive the usual normal (Gaussian) error distribution,
by generalizing the first one. Subbotin used the fol-
lowing axioms:
1. The probability of an error εdepends only on
the greatness of the error itself and can be ex-
pressed by a function ϕ(ε)with continuous
first derivative almost everywhere.
2. The most likely value of a quantity, for which
direct measurements xiare available, must not
depend on the adopted unit of measure.
In this way Subbotin obtains the probability distribu-
tion with the following density function:
ϕ(ε) = mh
2Γ(1/m)·exp[−hm|ε|m]
with −∞<ε<+∞,h>0 and m≥1. This dis-
tribution is also known as Exponential Power Distri-
bution and it has been used, for example, by Box and
Tiao (1992) in Bayesian inference. In the Italian sta-
tistical literature, a different parametrization of this
distribution has been derived by Lunetta (1963), who
followed the procedure introduced by Pearson (1895)
to derive new probability distributions, solving this
differential equation
dlog f
dx =p·log f−log a
x−c
and obtaining a distribution with the following prob-
ability density function
f(x) = 1
2σpp1/pΓ(1+1/p)·exp −|x−µ|p
pσp
p!
with −∞<x<+∞and −∞<µ<+∞,σp>0
and p≥1. This distribution is known as the order
pnormal distribution (Vianelli, 1963). It is easy to
see how this distribution is characterized by three
parameters: µis the location parameter, σpis the
scale parameter and pis the structure parameter. By
changing the structure parameter p, we can recog-
nize some known probability distribution: for ex-
ample, for p=1 we have the Laplace distribution,
for p=2 we have the normal (Gaussian) distribu-
tion, for p→+∞we have the uniform distribution.
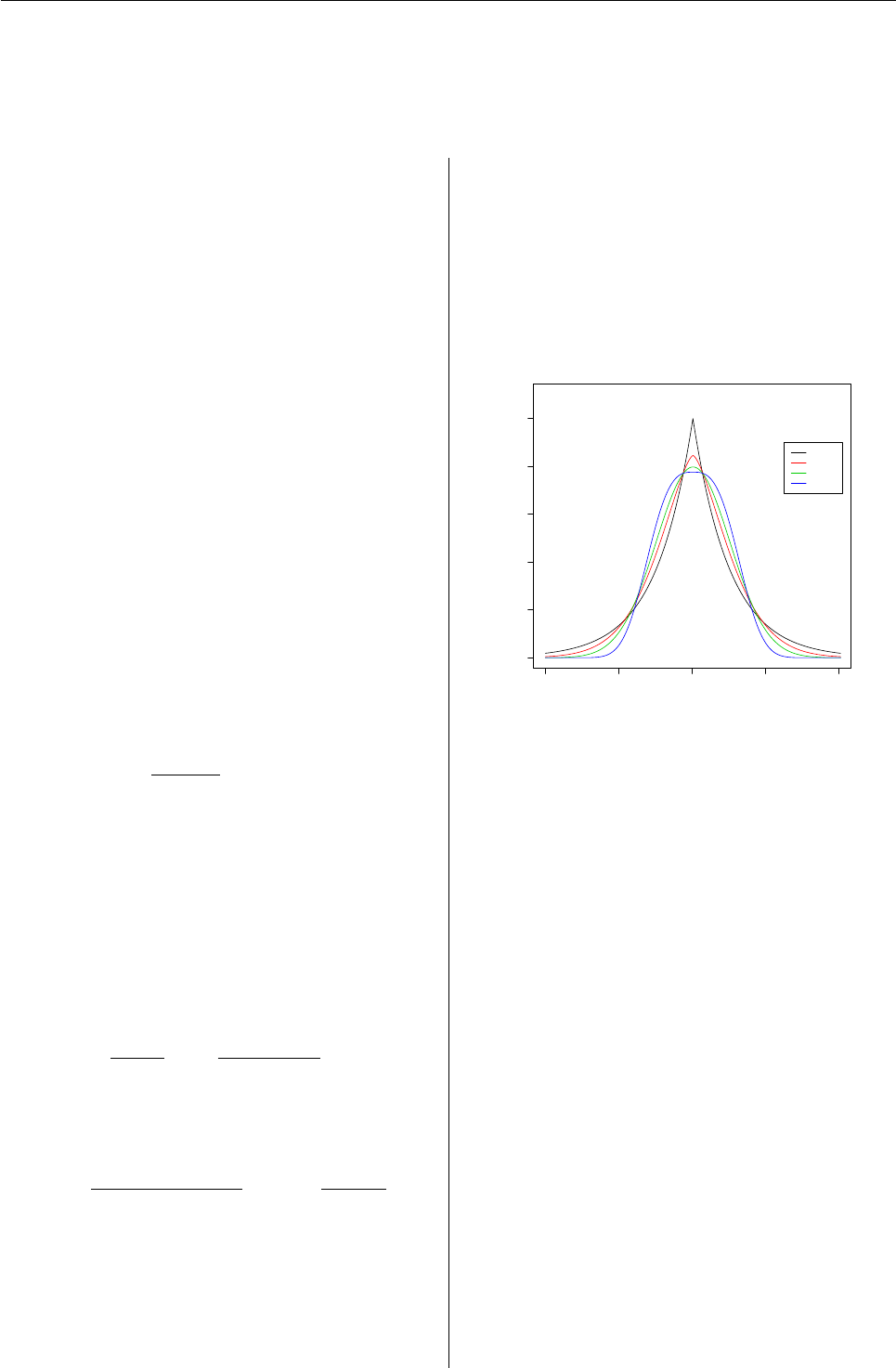
A graphical description of some normal of order p
curves is in figure 1 (this plot has been made with
the command graphnp() of the package normalp).
−4 −2 0 2 4
0.0 0.1 0.2 0.3 0.4 0.5
x
f(x)
p= 1
p= 1.5
p= 2
p= 3
Figure 1: Normal of order pcurves.
In this paper we present the main functions of the
package normalp and some examples of their use.
The normalp functions
The package contains the four classical functions
dealing with the computation of the density func-
tion, the distribution function, the quantiles and the
generation of pseudo-random observations from an
order pnormal distribution. Some examples related
to the use of these commands are the following:
> dnormp(3, mu = 0, sigmap = 1, p = 1.5,
+ log = FALSE)
[1] 0.01323032
> pnormp(0.5, mu = 2, sigmap = 3, p = 1.5)
[1] 0.3071983
> qnormp(0.3071983, mu = 2, sigmap = 3,
+ p = 1.5)
[1] 0.5
> rnormp(6, mu = 2, sigmap = 5, p = 2.5)
[1] 3.941597 -1.943872 -2.498598
[4] 1.869880 6.709037 14.873287
In case of generation of pseudo-random numbers we
have implemented two methods: one, faster, based
R News ISSN 1609-3631
Vol. 3/2, October 2003 14
on the relationship linking an order pnormal distri-
bution and a gamma distribution (see Lunetta, 1963),
and one based on the generalization of the Marsaglia
(1964) method to generate pseudo-random numbers
from a normal distribution. Chiodi (1986) describes
how the representation of the order pnormal distri-
bution as a generalization of a normal (Gaussian) dis-
tribution can be used for simulation of random vari-
ates.
Another group of functions concerns the estima-
tion of the order pnormal distribution parameters.
To estimate the structure parameter p, an estimation
method based on an index of kurtosis is used; in par-
ticular, the function estimatep() formulates an esti-
mate of pbased on the index VI given by
VI =õ2
µ1
=pΓ(1/p)Γ(3/p)
Γ(2/p).
by comparing its theoretical value and the empirical
value computed on the sample. For a comparison
between this estimation method and others based on
the likelihood approach see Mineo (2003). With the
function kurtosis() it is possible to compute the
theoretical values of, besides VI,β2and βpgiven by
β2=µ4
µ2
2
=Γ(1/p)Γ(5/p)
[Γ(3/p)]2
βp=√µ2p
µ2
p
=p+1
Moreover, it is possible to compute the empirical val-
ues of these indexes given by
c
VI =qn∑n
i=1(xi−M)2
∑n
i=1|xi−M|
ˆ
β2=n∑n
i=1(xi−M)4
[∑n
i=1(xi−M)2]2
ˆ
βp=n∑n
i=1|xi−M|2p
[∑n
i=1|xi−M|p]2.
Concerning the estimation of the location param-
eter µand the scale parameter σp, we have used
the maximum likelihood method, conditional on
the estimate of pthat we obtain from the function
estimatep(). The function we have to use in this
case is paramp(). We have implemented also a func-
tion simul.mp(), that allows a simulation study to
verify the behavior of the estimators used for the es-
timation of the parameters µ,σpand p. The com-
pared estimators are: the arithmetic mean and the
maximum likelihood estimator for the location pa-
rameter µ, the standard deviation and the maximum
likelihood estimator for the scale parameter σp; for
the structure parameter pwe used the estimation
method implemented by estimatep(). Through the
function plot.simul.mp() it is possible to see graph-
ically the behavior of the estimators. A possible use
of the function simul.mp() is the following:
> res <- simul.mp(n = 30, m = 1000, mu = 2,
+ sigmap = 3, p = 3)
> res
Mean Mp Sd
Mean 1.9954033 1.9991151 2.60598964
Variance 0.2351292 0.2849199 0.08791664
Sp p
Mean 2.9348828 3.415554
Variance 0.5481126 7.753024
N. samples with a difficult convergence: 26
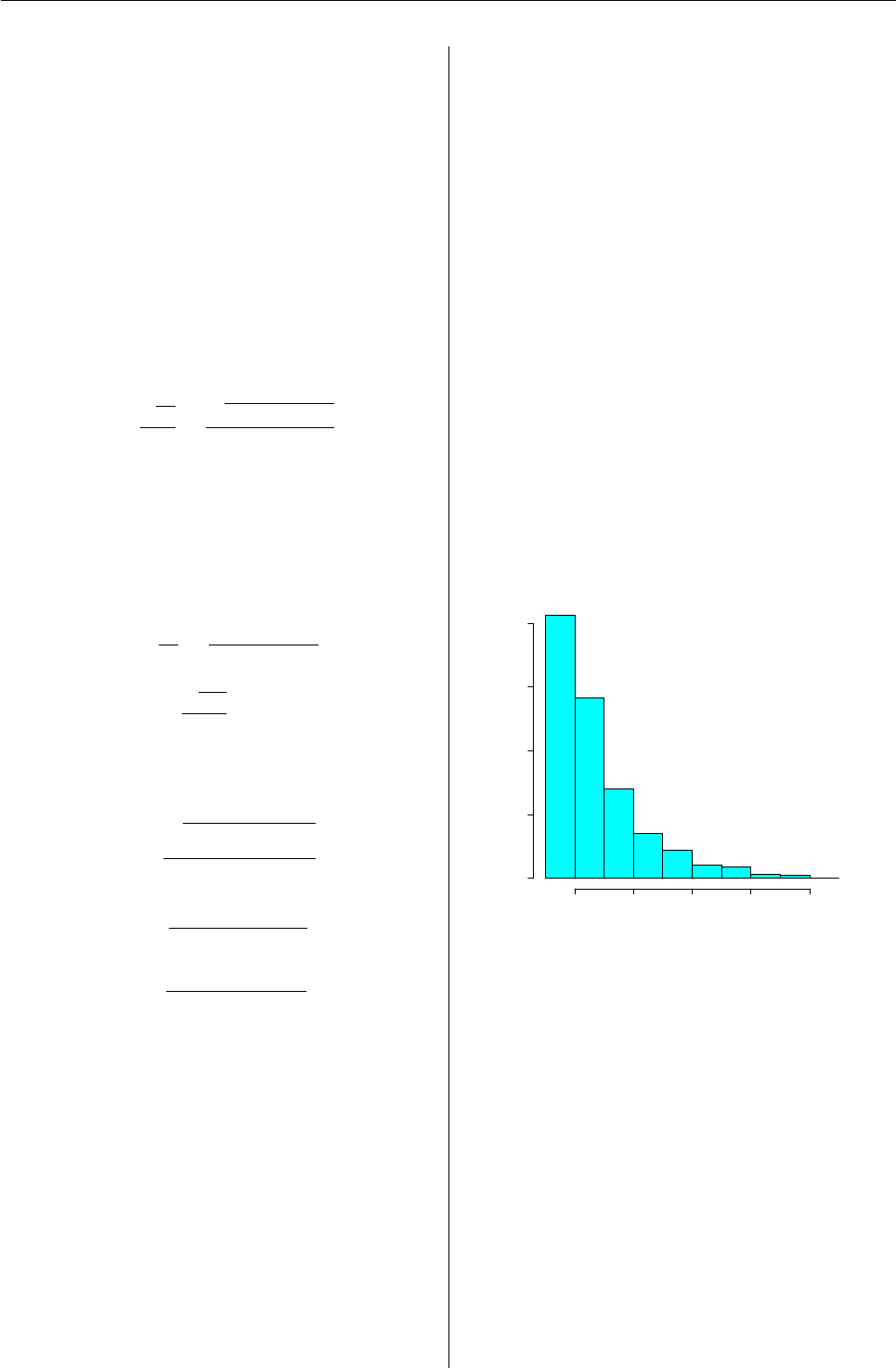
> plot(res)
The command plot(res) will produce an histogram
for every set of estimates created by the function
simul.mp(). In figure 2 we have the histogram for
ˆ
p. For more details see Mineo (1995-a).
Density
2 4 6 8 10
0.0 0.1 0.2 0.3 0.4
Figure 2: Histogram of ˆ
pobtained with the com-
mand plot.simul.mp(res).
It is also possible to estimate linear regression
models when we make the hypothesis of random er-
rors distributed according to an order pnormal dis-
tribution. The function we have to use in this case
is lmp(), which we can use like the function lm()
from the base package. In fact, the function lmp()
returns a list with all the most important results
drawn from a linear regression model with errors
distributed as a normal of order pcurve; moreover,
it returns an object that can form the argument of the
functions summary.lmp() and plot.lmp(): the func-
tion summary.lmp() returns a summary of the main
obtained results, while the function plot.lmp() re-
turns a set of graphs that in some way reproduces the
analysis of residuals that usually we conduct when
R News ISSN 1609-3631

Vol. 3/2, October 2003 15
we estimate a linear regression model with errors
distributed as a normal (Gaussian) distribution.
14 16 18 20
6 7 8 9 10 11
space
distance
Figure 3: Plot of the data considered in the data
frame cyclist.
To show an example of use of these functions, we
considered a data set reported in Devore (2000). In
this data set (see figure 3) the distance between a cy-
clist and a passing car (variable distance) and the
distance between the centre line and the cyclist in the
bike lane (variable space) has been recorded for each
of ten streets; by considering the variable distance
as a dependent variable and the variable space as an
independent variable, we produce the following ex-
ample:
> data(ex12.21, package = "Devore5")
> res <- lmp(distance ~ space,
+ data = ex12.21)
> summary(res)
Call:
lmp(formula = distance ~ space,
data = ex12.21)
Residuals:
Min 1Q Median 3Q Max
-0.7467 -0.5202 0.0045 0.3560 0.8363
Coefficients:
(Intercept) space
-2.4075 0.6761
Estimate of p
1.353972
Power deviation of order p: 0.6111
> plot(res)
In figure 4 we show one of the four graphs that we
have obtained with the command plot(res).
7 8 9 10 11
−0.5 0.0 0.5
Fitted values
Residuals
Residuals vs Fitted
lmp(formula = Car ~ Center, data = cyclist)
Figure 4: First graph obtained by using the command
plot.lmp(res).
Also for a linear regression model with errors
distributed as an order pnormal distribution we
have implemented a set of functions that allow a
simulation study to test graphically the suitabil-
ity of the estimators used. The main function is
simul.lmp(); besides this function, we have im-
plemented the functions summary.simul.lmp() and
plot.simul.lmp() that allow respectively to visual-
ize a summary of the results obtained from the func-
tion simul.lmp() and to show graphically the be-
havior of the produced estimates. A possible use of
these functions is the following:
> res <- simul.lmp(10, 500, 1, data = 1.5,
+ int = 1, sigmap = 1, p = 3, lp = FALSE)
> summary(res)
Results:
(intercept) x1
Mean 0.9959485 1.497519
Variance 0.5104569 1.577187
Sp p
Mean 0.90508039 3.196839
Variance 0.04555003 11.735883
Coefficients: (intercept) x1
1.0 1.5
Formula: y ~ +x1
Number of samples: 500
Value of p: 3
N. of samples with problems on convergence 10
> plot(res)
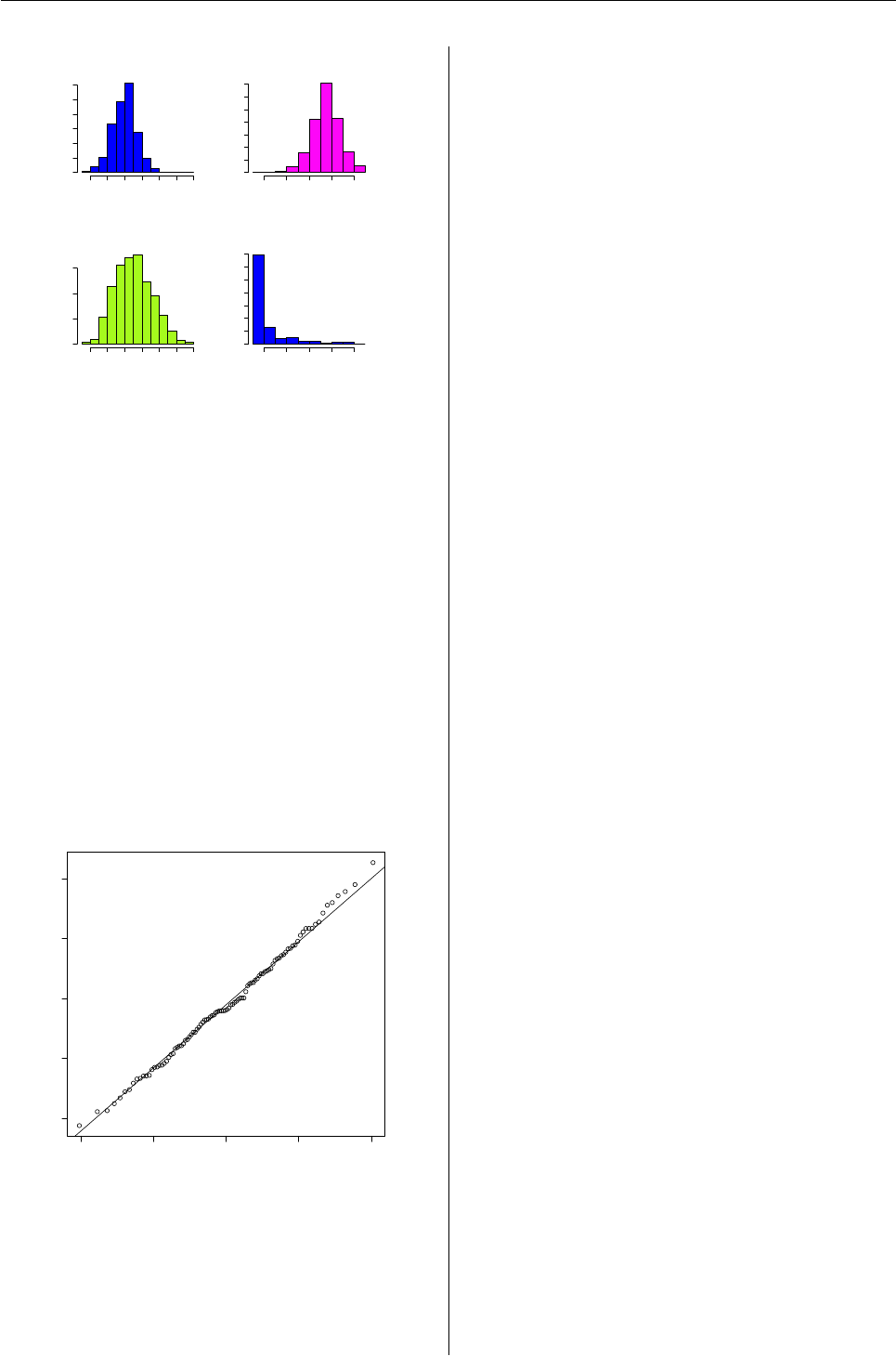
In figure 5 it is showed the result of plot(res). For
more details see Mineo (1995-b).
R News ISSN 1609-3631
Vol. 3/2, October 2003 16
Histogram of intercept
Density
−1012345
0.0 0.2 0.4 0.6
Histogram of x1
Density
−4 −2 0 2 4
0.00 0.10 0.20 0.30
Histogram of Sp
Density
0.4 0.6 0.8 1.0 1.2 1.4 1.6
0.0 0.5 1.0 1.5
Histogram of p
Density
2 4 6 8 10
0.0 0.2 0.4 0.6
Figure 5: Graphs obtained with the command
plot.simul.lmp(res).
Besides the described functions, we have im-
plemented two graphical functions. The command
graphnp() allows visualization of up to five different
order pnormal distributions: this is the command
used to obtain the plot in figure 1. The command
qqnormp() allows drawing a Quantile-Quantile plot
to check graphically if a set of observations follows a
particular order pnormal distribution. Close to this
function is the command qqlinep() that sketches a
line passing through the first and the third quartile
of the theoretical order pnormal distribution, line
sketched on a normal of order pQ-Q plot derived
with the command qqnormp(). In figure 6 there is a
graph produced by using these two functions.
−2 −1 0 1 2
−2 −1 0 1 2
Theoretical Quantiles
Sample Quantiles
p= 3
Figure 6: Normal of order pQ-Q plot.
Conclusion
In this article we have described the use of the new
package normalp, that implements some useful com-
mands where we have observations drawn from an
order pnormal distribution, known also as general
error distribution. The implemented functions deal
essentially with estimation problems for linear re-
gression models, besides some commands that gen-
eralize graphical tools already implemented in the
package base, related to observations distributed as
a normal (Gaussian) distribution. In the next future
we shall work on the computational improvement of
the code and on the implementation of other com-
mands to make this package still more complete.
Bibliography
G.E.P. Box and G.C. Tiao. Bayesian inference in statis-
tical analysis. Wiley, New York, 1992. First edition
for Addison-Wesley, 1973.
M. Chiodi. Procedures for generating pseudo-
random numbers from a normal distribution of or-
der p (p>1). Statistica Applicata, 1:7-26, 1986.
J.L. Devore. Probability and Statistics for Engineering
and the Sciences (5th edition). Duxbury, California,
2000.
G. Lunetta. Di una generalizzazione dello schema
della curva normale. Annali della Facoltà di Econo-
mia e Commercio dell’Università di Palermo, 17:237-
244, 1963.
G. Marsaglia and T.A. Bray. A convenient method for
generating normal variables. SIAM rev., 6:260-264,
1964.
A.M. Mineo. Stima dei parametri di intensità e di
scala di una curva normale di ordine p (p incog-
nito). Annali della Facoltà di Economia dell’Università
di Palermo (Area Statistico-Matematica), 49:125-159,
1995-a.
A.M. Mineo. Stima dei parametri di regressione
lineare semplice quando gli errori seguono una
distribuzione normale di ordine p (p incognito).
Annali della Facoltà di Economia dell’Università di
Palermo (Area Statistico-Matematica), 49:161-186,
1995-b.
A.M. Mineo. On the estimation of the structure pa-
rameter of a normal distribution of order p. To ap-
pear on Statistica, 2003.
K. Pearson. Contributions to the mathematical the-
ory of evolution. II. Skew variation in homo-
geneous material. Philosophical Transactions of the
Royal Society of London (A), 186:343-414, 1895.
M.T. Subbotin. On the law of frequency of errors.
Matematicheskii Sbornik, 31:296-301, 1923.
S. Vianelli. La misura della variabilità condizionata
in uno schema generale delle curve normali di fre-
quenza. Statistica, 23:447-474, 1963.
Angelo M. Mineo
University of Palermo, Italy
elio.mineo@dssm.unipa.it
R News ISSN 1609-3631

Vol. 3/2, October 2003 17
Web-based Microarray Analysis using
Bioconductor
by Colin A. Smith
Introduction
The Bioconductor project is an open source effort
which leverages R to develop infrastructure and al-
gorithms for the analysis of biological data, in par-
ticluar microarray data. Many features of R, includ-
ing a package-based distribution model, rapid proto-
typing, and selective migration to high performance
implementations lend themselves to the distributed
development model which the Bioconductor project
uses. Users also benefit from the flexible command
line environment which allows integration of the
available packages in unique ways suited to individ-
ual problems.
However, features to one individual may be road-
blocks to another. The use of microarrays for gene
expression profiling and other applications is grow-
ing rapidly. Many biologists who perform these ex-
periments lack the programming experience of the
typical R user and would strongly object to using a
command line interface for their analysis.
Here we present an overview of a web-based in-
terface that attempts to address some of the difficul-
ties facing individuals wishing to use Bioconductor
for their microarray data analysis. It is distributed as
the webbioc package available on the Bioconductor
web site at http://www.bioconductor.org/.
Target audience and interface goals
While targeted at many user types, the web interface
is designed for the lowest common denominator of
microarray users, e.g. a biologist with little computer
savvy and basic, but not extensive, statistical knowl-
edge in areas pertinant to microarray analysis. Note
that although this is the lowest level user targeted by
the web interface, this interface also caters to power
users by exposing finer details and allows flexibility
within the preconstructed workflows.
This article presents only the first version of a
Bioconductor web interface. With luck, more R and
Perl hackers will see fit to add interfaces for more as-
pects of microarray analysis (e.g. two-color cDNA
data preprocessing). To help maintain quality and
provide future direction, a number of user interface
goals have been established.
•Ease of use. Using the web interface, the user
should not need to know how to use either
a command line interface or the R language.
Depending on the technical experience of the
user, R tends to have a somewhat steep learn-
ing curve. The web interface has a short learn-
ing curve and should be usable by any biolo-
gist.
•Ease of installation. After initial installation by a
system administrator on a single system, there
is no need to install additional software on user
computers. Installing and maintaining an R in-
stallation with all the Bioconductor packages
can be a daunting task, often suited to a system
administrator. Using the web interface, only
one such installation needs to be maintained.
•Discoverability. Graphical user interfaces are
significantly more discoverable than command
line interfaces. That is, a user browsing around
a software package is much more likely to dis-
cover and use new features if they are graphi-
cally presented. Additionally, a unified user in-
terface for the different Bioconductor packages
can help add a degree to cohesiveness to what
is otherwise a disparate collection of functions,
each with a different interface. Ideally, a user
should be able to start using the web interface
without reading any external documentation.
•Documentation. Embedding context-sensitive
online help into the interface helps first-time
users make good decisions about which statis-
tical approaches to take. Because of its power,
Bioconductor includes a myriad of options for
analysis. Helping the novice statistician wade
through that pool of choices is an important as-
pect of the web interface.
Another aspect of the target audience is the de-
ployment platform. The web interface is written in
Perl, R, and shell scripts. It requires a Unix-based
operating system. Windows is not supported. It
also uses Netpbm and optionally PBS. For further in-
formation, see the webbioc vignette at http://www.
bioconductor.org/viglistingindex.html.
User-visible implementation deci-
sions
There are a number of existing efforts to create web
interfaces for R, most notably Rweb, which presents
the command line environment directly to the user.
See http://www.math.montana.edu/Rweb/. The Bio-
conductor web interface, on the other hand, entirely
abstracts the command line away from the user. This
results in an entirely different set of design decisions.
R News ISSN 1609-3631
Vol. 3/2, October 2003 18
The first choice made was the means by which
data is input and handled within the system. In an
R session, data is instantiated as variables which the
user can use and manipulate. However, in the web
interface, the user does not see variables associated
with an R session but rather individual files which
hold datasets, such as raw data, preprocessed data,
and analysis result tables.
Different stages of microarray analysis are di-
vided into individual modules. Each module leads
the user through a series of steps to gather process-
ing parameters. When ready, the system creates
an R script which it either runs locally or submits
to a computer cluster using the Portable Batch Sys-
tem. Any objects to be passed to another module are
saved in individual files.
Another decision which directly impacts the user
experience is that the system does not maintain ac-
counts for individual users. Instead, it uses the con-
cept of a uniquely identified session. When a user
first starts using the web interface, a session is cre-
ated which holds all the uploaded and processed
data. The system provides the user with a session
token comprised of a random string of letters and
numbers. The token allows the user to return to their
session at a future date.
This offers a number of advantages: 1) At the
discretion of the local system administrator, the web
analysis resource can be offered as either a public or
a private resource. Such a restriction can be made at
the web-server level rather than the code level. 2) It
allows rapid development of the web interface with-
out being bogged down in the implementation or in-
tegration of a user infrastructure. 3) As opposed to
having no session whatsoever, this allows a user to
input data only once. Raw data files are often quite
large. Uploading multiple copies of such datasets for
each change in parameters is not desirable.
Lastly, the web interface brings the idea of design-
by-contract used in the Bioconductor project down
to the package level. That is, individual interface
modules are responsible for a specific stage or type
of analysis. Modules may take the user through any
number of steps as long as they use standard input
and output formats. This allows the system to grow
larger and more powerful over time without making
individual components more complex than is neces-
sary to fulfill their function.
Analysis workflow
The web interface is currently limited to process-
ing data from microarray experiments based on the
Affymetrix GeneChip platform. It does however
handle an entire workflow going from raw intensity
values through to annotated lists of differentially ex-
pressed genes.
Affymetrix microarray data comes in the form of
CEL files containing intensity values for individual
probes. Because all processing is done server-side,
that data must first be transferred with the Upload
Manager. While raw data files can each be over ten
megabytes, today’s fast ethernet networks provide
very acceptable performance, with file transfers tak-
ing only a few seconds.
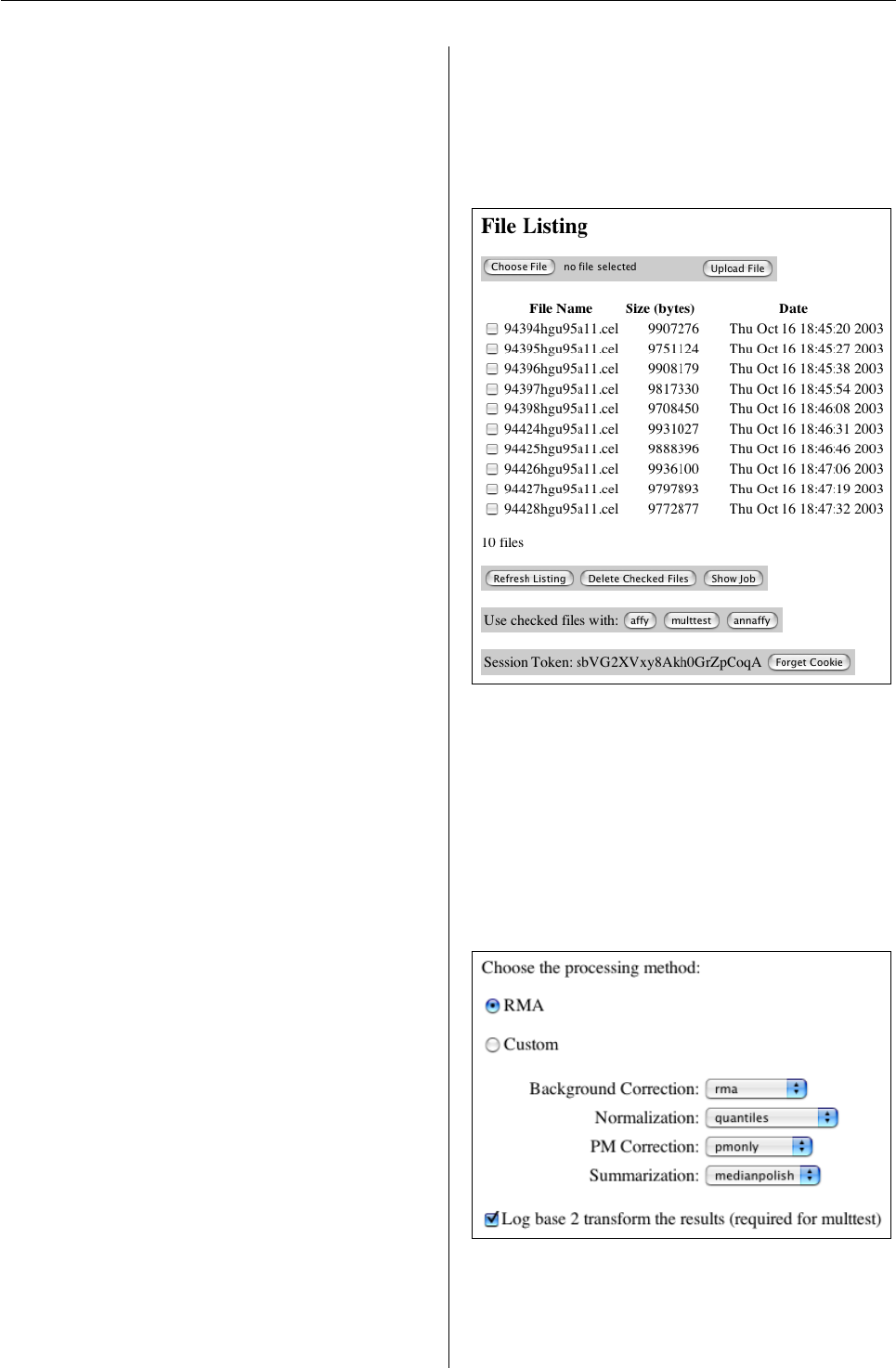
Figure 1: Upload Manager
Affymetrix preprocessing is handled by the affy
Bioconductor package. The core functionality of that
package is captured by only a handful of functions
and thus lends itself to a simple web interface. The
user may choose either the built-in high performance
function for RMA or a custom expression measure.
The custom expression measure also uses additional
plug-in methods from the vsn and gcrma packages,
which leverage the modular design of affy.
Figure 2: Affymetrix Data Preprocessing
There are a number of methods for identifying
differentailly expressed genes. The web interface
currently uses basic statistical tests (t-tests, F-tests,
R News ISSN 1609-3631

Vol. 3/2, October 2003 19
Figure 3: Differential Expression and Multiple Testing
etc.) combined with multiple testing procedures for
error control of many hypotheses. These are im-
plemented in the multtest package. Additionally,
the web interface automatically produces a number
of diagnostic plots common to microarray analysis.
Those include M vs. A (log fold change vs. overall
expression) and normal quantile-quantile plot.
The web interface completes the workflow by
producing tables with integrated results and meta-
data annotation. Where appropriate, the annota-
tion links to other online databases including a chro-
mosome viewer, PubMed abstracts, Gene Ontology
trees, and biochemical pathway schematics. The
metadata presentation is handled by annaffy, an-
other Bioconductor package.
In addition to presenting metadata, the web inter-
face provides facilities for searching that metadata.
For instance, it is trivial to map a set of GenBank
accession numbers onto a set of Affymetrix probe-
set ids or find all genes in a given Gene Ontology
branch. This assists biologists in making specific
hypotheses about differential gene expression while
maintining strong control over the error rate.
Lastly, because the web interface stores inter-
mediate data as R objects, users of Bioconductor
through either the command line or web interface
can easily exchange data back and forth. Data ex-
change is currently limited to exprSet objects, which
is the standard class for microarray data in Biocon-
ductor. Future development of the interface should
yield more data exchange options enabling novel col-
laboration between R users and non-users alike.
Final thoughts
An important consideration worthy of discussion is
the inherent lack of flexibility in graphical user inter-
faces. The R command line interface does not box
one into pre-scripted actions in the way that the web
interface does. It allows one to exercise much more
creativity in analysis and take more non-linear ap-
proaches. In the GUI trivial questions may by impos-
sible to answer simply because of unforeseen limita-
tions.
There are, however, a number of strengths in the
web interface beyond what is available in R. The
aforementioned interface goals are good examples of
this. Additionally, the web interface can help reduce
errors by distilling long series of typed commands
into simple point-and-click options. All actions and
parameters are tracked in a log for verification and
quality control.
Secondly, the web interface easily integrates into
existing institutional bioinformatics resources. The
web has been widely leveraged to bring univer-
R News ISSN 1609-3631
Vol. 3/2, October 2003 20
Figure 4: Annotated Results and Online Database Links
sally accessible interfaces to common command-line
bioinformatics tools. The system presented here can
sit right next to those tools on a web site. Because it
already uses PBS for dispatching computational jobs,
the web interface can take advantage of existing com-
puter clusters built for genomic search tools, such as
BLAST, and can scale to many simultaneous users.
The web interface has been deployed and is cur-
rently in use by two research groups. One group is
split between institutions located in different states.
They use common session tokens and collaborate by
sharing data and analysis results over the web.
Lastly, Bioconductor has implementations of a
number of algorithms not otherwise freely available.
Some newer algorithms have been exclusively imple-
mented in Bioconductor packages. The web interface
helps bring such innovations to the mainstream. It
may even wet the appetite of some users, convincing
them to take the plunge and learn R.
Colin A. Smith
NASA Center for Computational Astrobiology and Fun-
damental Biology
webbioc@colinsmith.org
R News ISSN 1609-3631

Vol. 3/2, October 2003 21
Sweave, Part II: Package Vignettes
Reading, writing and interacting with R package
primers in Sweave format.
by Friedrich Leisch
This is the second article in a two-part miniseries on
Sweave (Leisch,2002a), a tool that allows embed-
ding of R code in L
A
T
EX documents so that code, re-
sults, and descriptions are presented in a consistent
way. The first article (Leisch,2002b) introduced the
Sweave file format and the R functions to process it,
and demonstrated how to use Sweave as a report-
ing tool for literate statistical practice. This article
will concentrate on how to use files in Sweave format
as primers or manuals for R packages, so that users
have direct access to the code examples shown and
so that all code can be checked automatically for syn-
tax errors and for consistency of the usage descrip-
tion with the implementation.
R package documentation
The main vehicle for documenting R packages are
the help files, which are the sources, written in R doc-
umentation (Rd) format, of what you see after calling
help() on a topic. These files, which are divided into
sections, typically contain code in just two sections:
usage and examples. All examples in the R help files
are, by default, required to be executable such that
the user can copy & paste the code to a running R
process using
• the mouse,
• keyboard shortcuts if running R inside Emacs
with ESS, or
• R’s example() function.
Examples should be flagged as non-executable only
if there are good reasons for this, e.g. because they
require user interactivity like identify() and hence
cannot be executed in batch mode.
The tools for package quality control, available
through the R CMD check1command, test if all the
examples in the package can be successfully exe-
cuted. Furthermore, the code in the usage section is
compared with the actual implementation to check
for inconsistencies and for missing documentation.
The Rd file format was designed for refer-
ence documentation on single R objects (functions,
classes, data sets, . . . ). It is not intended for demon-
strating the interaction of multiple functions in a
package. For this task we have developed the con-
cept of package vignettes — short to medium-sized
documents explaining parts or all of the functionality
of a package in a more informal tone than the strict
format of reference help pages.
Reading vignettes
Books like Venables and Ripley (2002) that describe
the use of S for data analysis typically contain a mix-
ture of documentation, code, and output. Short doc-
uments in this style are ideally suited to explaining
the functionality of a package to new users. The di-
rectory ‘inst/doc’ of an R source package may con-
tain such package documentation in any format, al-
though we recommend PDF files because of their
platform independence.
We call a user guide located in ‘inst/doc’ a vi-
gnette only when it is a document from which the
user can extract the R code and interact with it.
Sweave is the only format for such documents that
is currently supported by R; there may be others in
the future. In short: every vignette is a user guide,
but not every user guide is a vignette.
Command line interface
Starting with R version 1.8.0 there is support in base
R for listing and viewing package vignettes. The
vignette() function works similar to data() and
demo(). If no argument is given, a list of all vignettes
in all installed packages is returned — see the exam-
ple R session in Figure 1.
Do not be surprised if this list is rather short or
even empty on your computer. At present only a
few of the packages on CRAN have vignettes. For
Bioconductor we have made package vignettes a re-
quirement and thus all Bioconductor packages pro-
vide one or more vignettes.
Following the title of each vignette listed in Fig-
ure 1, you will see in parenthesis a list of the formats
that are available. In Figure 1all the vignettes are
available in both source and PDF format. To view
the strucchange-intro vignette, all you need to do
is to issue
R> vignette("strucchange-intro")
and the PDF file is opened in your favorite PDF
reader (exactly which PDF reader depends on the
platform that you use). If the source file for a vignette
is available, one can easily extract the code shown
in the vignette, although we have not yet fully auto-
mated the procedure. First we get the full path to the
vignette directory
R> vigdir =
+ system.file("doc", package="strucchange")
and then we examine the names of the files it con-
tains
1R CMD xxx is Rcmd xxx in the Windows version of R.
R News ISSN 1609-3631

Vol. 3/2, October 2003 22
R > vig net te ()
Vig ne tte s in pac kag e ’ An nBuild er ’:
An nB ui lde r An nB ui lde r Basi c ( sou rce , pdf )
HowTo An nB ui lde r HowT o ( sou rce , pdf )
Vi gn et te s in pac ka ge ’ B iobase ’:
Bio ba se Bio ba se Pri mer ( source , pdf )
Bi oc ond uc tor Howto B io co ndu ct or ( s ource , pdf )
HowTo Ho wTo HowTo ( source , pdf )
esA pp ly esA pp ly I nt ro duc tio n ( source , p df )
...
Vig nette s in package ’ strucchange ’:
strucchange - intro str uc ch an ge : An R Pac kag e for T est ing for
Str uc tural C hang e in Linear R eg ression Models
(source , pdf)
...
Figure 1: Usage of vignette() to list available package vignattes.
R> list.files(vigdir)
[1] "00Index.dcf"
[2] "strucchange-intro.R"
[3] "strucchange-intro.Rnw"
[4] "strucchange-intro.pdf"
[5] "strucchange-intro.tex"
File ‘strucchange-intro.Rnw’ is the original Sweave
file, ‘strucchange-intro.R’ has the extracted R code for
all code chunks and could now be executed using
source() or opened in your favorite editor. If the
‘.R’ file is not available, we can create it in the cur-
rent working directory by
R> library("tools")
R> vig = listFilesWithType(vigdir, "vignette")
R> Stangle(vig[1])
Writing to file strucchange-intro.R
where listFilesWithType() returns the full path to
all files in vigdir that have type "vignette", i.e., an
extension marking them as Sweave files.
Graphical user interface
The simplest way to access vignettes is probably
through the HTML help system. If you execute
help.start() and click your way to a package con-
taining vignettes, then you will see, at the beginning
of the package’s table of contents, a link to an index
of all the available vignettes. In addition there is a
link to the directory containing the vignettes so that,
for example, you could use your browser to examine
the source files.
A more advanced interface to package vignettes
is available in the Bioconductor package tkWid-
gets, available from http://www.bioconductor.org.
Function vExplorer() lists all available vignettes in
a nice point & click menu. For example, after select-
ing the strucchange vignette the upper left window
shown in Figure 2is opened. The PDF version of
the vignette can be opened by clicking on the “View
PDF” button. Each code chunk of the vignette has
a button on the left side of the window, clicking on
the button shows the code in the "R Source Code"
text field. The code can be executed and the resulting
output is shown in the “Results of Execution” area.
The most powerful feature of this kind of inter-
face is that the S code in the source code field can
be modified by the user, e.g., to try variations of the
pre-fabricated examples. To modify the example, one
simply edits the code in the "R Source Code" area and
presses the "Execute Code" button again.
Dynamic statistical documents and their user in-
terfaces are an open research area, see also Buttrey
et al. (2001) and Sawitzki (2002) for other approaches.
Writing vignettes
Once the Sweave file is written (we cannot do that for
you), it is almost trivial to include it in an R package
and make it available to users by the tools described
above. Say you have written a file ‘foo.Rnw’ to be
used as a vignette for package foo. First you need to
add some meta-information to the file along the lines
of
% \VignetteIndexEntry{An R Package for ...}
% \VignetteDepends{foo, bar, ...}
% \VignetteKeyword{kwd1}
% \VignetteKeyword{kwd2}
R News ISSN 1609-3631
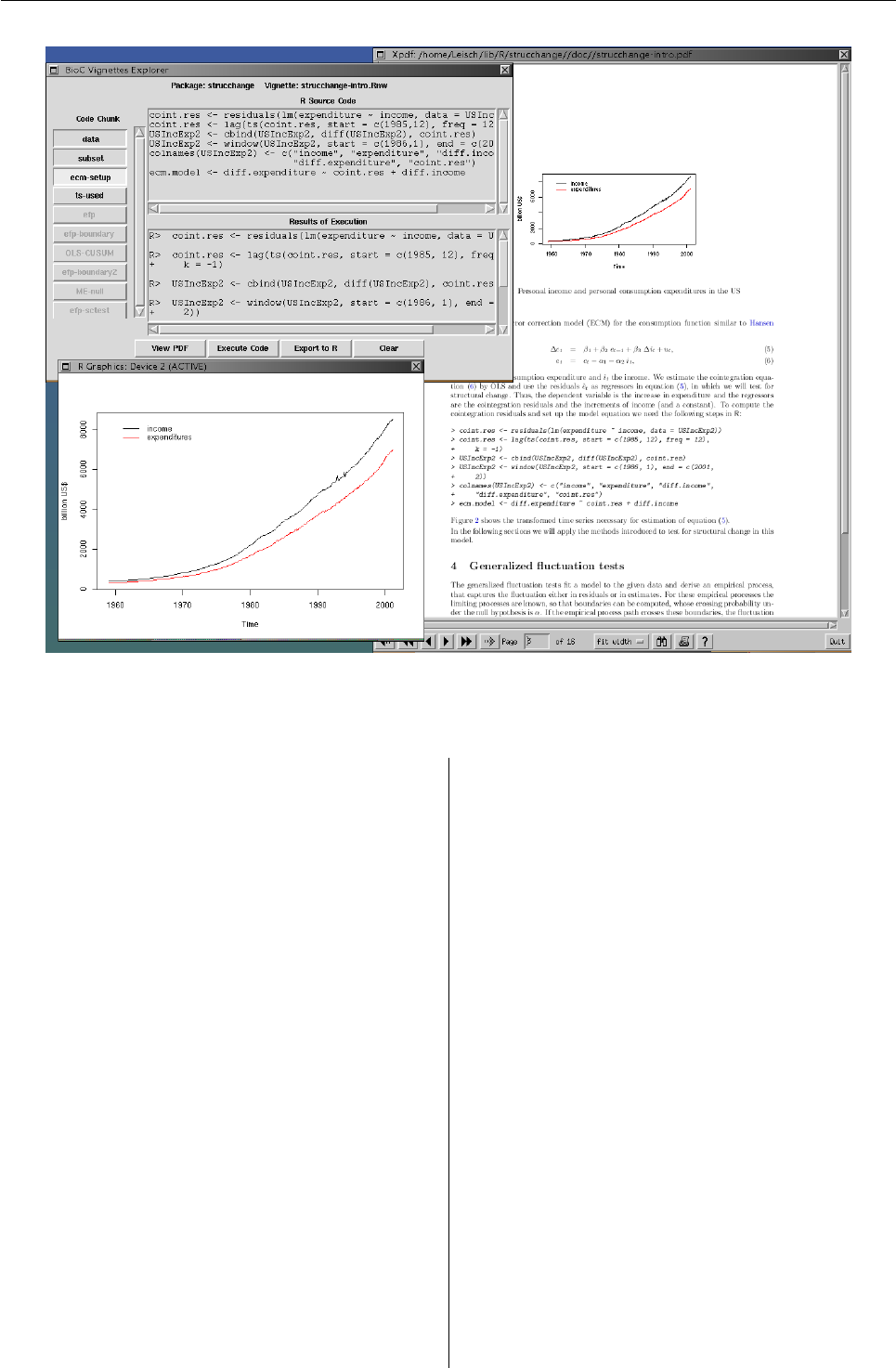
Vol. 3/2, October 2003 23
Figure 2: Screenshot of vExplorer() showing the vignette from package strucchange: main controls for code
chunk execution (upper left), currently active R graphics window (lower left) and a pdf viewer (right).
All of these should be in L
A
T
EX comments as we have
not defined them as proper L
A
T
EX commands. The in-
dex entry is used for the listings of vignette() or
vExplorer(); frequently it is the same as the title
of the document (or an abbreviated version thereof).
Note that it is directly used in text and HTML files
and hence should not contain any T
EX markup. The
dependency information is analogous to the Depends
field of a package ‘DESCRIPTION’ file and lists pack-
ages needed to execute the code in the vignette. The
list of \VignetteXXX meta-information specifications
will probably get longer in the near future, especially
for versioning etc.
Once this is done all you have to do is create
a subdirectory ‘inst/doc’ in yor package source tree
and copy ‘foo.Rnw’ to it. All the rest is taken care of
by the R package management system, e.g.
•R CMD check will extract the code from the vi-
gnette and test that it can be executed success-
fully.
•R CMD build will run Sweave() and pdflatex
on the vignette to create the PDF version.
• The package installation mechanism creates an
index of all vignettes in the package and links
it into the HTML help system.
Note that even code chunks with option
eval=FALSE are tested by R CMD check; if you want
to include code that should not be tested in a vi-
gnette, move it to a normal L
A
T
EX verbatim environ-
ment. The reason for this policy is because users
should be able to rely on code examples being exe-
cutable exactly as shown in the vignette.
By including the PDF version in the package
sources it is not necessary that the vignettes can be
compiled at install time, i.e., the package author can
use private L
A
T
EX extensions or BibTeX files. Only the
R code inside the vignettes is part of the checking
procedure; typesetting manuals is not part of pack-
age quality control.
For more details see the manual “Writing R Ex-
tensions”, which features a section on package vi-
gnettes.
In general it is assumed that package authors
run R CMD build on their machine (and may safely
assume that only they do that). R CMD check
on the other hand should be runnable by ev-
erybody, e.g., CRAN runs a check on all 250+
packages on a daily basis (the results are avail-
able at http://cran.r-project.org/src/contrib/
checkSummary.html). Bioconductor has opted for a
stricter policy such that even building packages (in-
cluding running latex on vignettes) should be re-
R News ISSN 1609-3631

Vol. 3/2, October 2003 24
producible on every machine which has the neces-
sary tools installed.
Acknowledgements
vignette() and most of R CMD check were written
by Kurt Hornik. vExplorer() and its helper func-
tions were written by Jeff Gentry and Jianhua Zhang
as part of the Bioconductor project. I want to thank
them and Robert Gentleman for helpful ideas and
discussions.
Bibliography
S. E. Buttrey, D. Nolan, and D. T. Lang. An environ-
ment for creating interactive statistical documents.
In E. J. Wegman, A. Braverman, A. Goodman, and
P. Smyth, editors, Computing Science and Statistics,
volume 33. Interface Foundation of North Amer-
ica, Fairfax Station, VA, USA, 2001. 22
F. Leisch. Sweave: Dynamic generation of statistical
reports using literate data analysis. In W. Härdle
and B. Rönz, editors, Compstat 2002 — Proceedings
in Computational Statistics, pages 575–580. Physika
Verlag, Heidelberg, Germany, 2002a. URL http:
//www.ci.tuwien.ac.at/~leisch/Sweave. ISBN
3-7908-1517-9. 21
F. Leisch. Sweave, part I: Mixing R and L
A
T
EX. R
News, 2(3):28–31, December 2002b. URL http:
//CRAN.R-project.org/doc/Rnews/.21
G. Sawitzki. Keeping statistics alive in documents.
Computational Statistics, 17:65–88, 2002. 22
W. N. Venables and B. D. Ripley. Modern Applied
Statistics with S. Fourth Edition. Springer, 2002. URL
http://www.stats.ox.ac.uk/pub/MASS4/. ISBN
0-387-95457-0.
21
Friedrich Leisch
Institut für Statistik & Wahrscheinlichkeitstheorie
Technische Universität Wien, Austria
Friedrich.Leisch@R-project.org
R News ISSN 1609-3631

Vol. 3/2, October 2003 25
R Foundation News
by Bettina Grün and Friedrich Leisch
New benefactors
• Department of Statistics, Brigham Young
University, Utah, USA
• Institute of Mathematical Statistics (IMS),
Ohio, USA
• MedAnalytics, Inc., Minnesota, USA
New supporting institutions
• Astra Zeneca R&D Mölndal, Mölndal, Sweden
• Baxter AG, Vienna, Austria
• Boehringer Ingelheim Austria GmbH, Vienna,
Austria
• Department of Economics, Stockholm Univer-
sity, Sweden
• Department of Statistics, University of
Wisconsin-Madison, Wisconsin, USA
• Lehrstuhl für Rechnerorientierte Statistik und
Datenanalyse, University of Augsburg, Ger-
many
• Spotfire, Massachusetts, USA
New supporting members
Klaus Abberger (Germany)
Luc Anselin (USA)
Anestis Antoniadis (France)
Carlos Enrique Carleos Artime (Spain)
Ricardo Azevedo (USA)
Pierluigi Ballabeni (Switzerland)
Saghir Bashir (UK)
Marcel Baumgartner (Switzerland)
Hans Werner Borchers (Germany)
Rollin Brant (Canada)
Alessandra R. Brazzale (Italy)
Karl W. Broman (USA)
Robert Burrows (USA)
Federico Calboli (Italy)
Charles M. Cleland (USA)
Jorge de la Vega Góngora (Mexico)
Jan de Leeuw (USA)
Ramón Diaz-Uriarte (Spain)
Dubravko Doli´c (Germany)
Dirk Eddelbuettel (USA)
Stephen Eglen (UK)
John Fox (Canada)
Simon Gatehouse (Australia)
Stefano Guazzetti (Italy)
Frank Harrell (USA)
Pascal Heus (USA)
Paul Hewson (UK)
Giles Heywood (UK)
Johannes Hüsing (Germany)
Rafael Irizarry (USA)
David A. James (USA)
Landon Jensen (USA)
Diego Kuonen (Switzerland)
Manuel Castéjon Limas (Spain)
Greg Louis (Canada)
Clifford E. Lunneborg (USA)
John Marsland (UK)
Andrew D. Martin (USA)
Gordon M. Morrison (UK)
Rashid Nassar (USA)
Vadim Ogranovich (USA)
John C. Paolillo (USA)
Thomas Petzoldt (Germany )
Bernhard Pfaff (Germany)
Jonas Ranneby (USA)
Gus Rashid (USA)
Greg Ridgeway (USA)
Jeffrey Rosenthal (Canada)
Claude Rubinson (USA)
Ingo Ruczinski (USA)
Erik A. Sauleau (France)
Martin Schlather (Germany)
Michael Scroggie (Australia)
Frederik von Ameln (Switzerland)
Scott R. Waichler (USA)
Rainer Walke (Germany)
Ko-Kang Kevin Wang (New Zealand)
Stefan Werner (Finland)
Victor D. Zurkowski (Canada)
New ordinary members
Roger Bivand (Norway)
Bill Venables (Australia)
Bettina Grün & Friedrich Leisch
Technische Universität Wien, Austria
Bettina.Gruen@ci.tuwien.ac.at
Friedrich.Leisch@R-project.org
R News ISSN 1609-3631

Vol. 3/2, October 2003 26
Recent Events
Statistical Computing 2003 at
Reisensburg
The Reisensburg meeting has become a regular at-
traction for those interested in Computational Statis-
tics. It is organized by three special interest groups
(Computational Statistics of the Biometric Society -DR,
Statistical Analysis Systems of the German Association
of Medical Informatics, Biometry and Epidemiology
GMDS, and Classification and Data Analysis in the bio-
sciences of the Gesellschaft für Klassifikation GfKl)
and it takes place near Ulm, Germany, in beautiful
Reisensburg castle, situated above the river Danube.
The main topics of this conference are fixed one
year in advance by the members of the working
groups. The organizers take great care that there is
sufficient time for discussion after the talks and at
the famous Reisensburg bartizan round tables.
Recent developments of statistical software has
been a major topic of previous meetings. Merits and
miseries of the various packages were discussed in
depth. This has changed. Discussion of the large
packages played a minor role this year, and R was
featured in many presentations. F. Bretz, T. Hothorn
and P. Westfall gave an overview on the multcomp
package for multiple comparisons. F. Leisch intro-
duced flexmix, a framework for fitting discrete mix-
tures of regression models.
As we all know, a lot has still to be done in R
to support advanced visualization and interactivity.
A. Zeileis, D. Meyer and K. Hornik demonstrated
visualizations using mosaic plots in R . S. Urbanek
showed Java based interactive graphics for R and
H. Hoffmann demonstrated what can be done in
other environments, using visualizations for condi-
tional distributions as an example and tools derived
from the Augsburg Dada collection.
The list of speakers and topics is too long
to be repeated here in full. Program and ab-
stracts are available from http://www.dkfz.de/
biostatistics/Reisensburg2003/.
The 2003 meeting highlighted the analysis of ge-
nomic data. Robert Gentleman presented a keynote
session on exploring and visualizing genomic data.
Subsequent sessions covered methodological as-
pects, in particular techniques for combining clas-
sifiers or variables and methods related to machine
learning. On the more applied side, topics included,
among others, C. Ittrich and A. Benner addressing
the role of microarrays in clinical trials, and U. Mans-
mann discussing simulation techniques for microar-
ray experiments. E. Brunner (Göttingen) used the
opportunity to demonstrate how classical statistical
analysis of the design of experiments may be applied
in this field to give concise answers instead of vague
conjectures.
High dimensional observations, combined with
very low sample sizes, are a well known peculiar-
ity of genomic data. Another peculiarity comes from
the strong dependence between the observed data.
The data refer to gene activities, and these are only
an aspect of the metabolic and regulatory dynam-
ics of a cell. Little is known about how to include
the knowledge of metabolic pathways and the re-
sulting dependencies in a statistical analysis. Us-
ing statistical inference from genomic data to identify
metabolic or regulatory structures is largely an open
task. F. Markowetz and R. Spang studied the effect
of perturbations on reconstructiing network struc-
ture; C. Becker and S. Kuhnt addressed robustness
in graphical modelling. From the application side,
A. v. Heydebreck reported on estimation of onco-
geneic tree models and W. Huber talked about iden-
tification of protein domain combinations.
The next Reisensburg working conference will
take place 2004, June 27.-30. By the time you read this
article, the call for papers should have been issued.
The main topics will be: applications of machine
learning; statistical analyis of graphs/networks; sta-
tistical software; bioinformatics; exploration of large
data sets.
Till then, working groups in cooperation with
the special research unit in Erlangen will organize
a workshop on Ensemble Learning, Erlangen 2004,
Jan. 23.-24. Stay tuned, and see http://www.imbe.
med.uni-erlangen.de/links/EnsembleWS/.
Günther Sawitzki
Universität Heidelberg
gs@statlab.uni-heidelberg.de
Statistical Inference, Computing
and Visualization for Graphs
On August 1–2, 2003, a workshop on using graphs
in statistical data analysis took place at Stanford
University. Quoting the workshop homepage
at http://www.research.att.com/~volinsky/
Graphs/Workshop.html “Graphs have become an in-
creasingly popular way of representing data in many dif-
ferent domains, including telecommunications research,
genomics and bioinformatics, epidemiology, computer
networks and web connectivity, social networks, mar-
keting and statistical graphical models. Analyzing these
data effectively depends on contributions from the areas of
data representation, algorithms, visualization (both static
and interactive), statistical modeling (including graphical
models) and inference. Each of these areas has its own
language for describing graphs, and its own favorite tools
and methods. Our goal for the workshop is to explore
R News ISSN 1609-3631

Vol. 3/2, October 2003 27
synergies that may exist between these different areas of
research.”
It was very interesting to see the number of dif-
ferent areas of applied data analysis in which graphs
(structures with nodes and edges) are used. There
are differences, most notably the sizes of the graphs,
ranging from a dozen nodes to several millions,
which has an impact on “natural” and efficient com-
putations. However, we also identified common-
alities, and having a central infrastructure in R for
representing graphs and performing common oper-
ations will certainly help to prevent reinventing the
wheel several times.
The Bioconductor project has started to provide
this infrastructure with the graph package and in-
terfaces to standard libraries for graph computations
and visualization (Rgraphviz,RBGL, . . . ). Develop-
ment versions of ggobi also have support for graphs
that can be tightly linked to R. If you are interested
to learn more about the workshop: you can down-
load the slides for any of the presentations from the
workshop homepage.
Finally, I want to thank the organizers for the
great job they did in organizing the workshop; both
the scientific program and the social atmosphere
made it a pleasure to participate.
Friedrich Leisch
Technische Universität Wien, Austria
Friedrich.Leisch@R-project.org
JSM 2003
At the 2003 Joint Statistical Meetings in San Fran-
cisco, an invited session was organized that is of par-
ticular interest to the R community. Jan de Leeuw
from University of California, Los Angeles, led off
the session with the a talk on “The State of Statis-
tical Software” (http://gifi.stat.ucla.edu/pub/
jsm03.pdf). He began with a overview of types
of statistical software one might use for activities
such as consulting, teaching and research providing
some history and thoughts for the future along the
way. Luke Tierney from University of Iowa, spoke
on “Some New Language Features of R” (http://
www.stat.uiowa.edu/~luke/talks/jsm03.pdf) fo-
cussing on namespaces, code analysis tools, excep-
tion handling and byte compilation. Duncan Tem-
ple Lang from Bell Laboratories spoke on “Con-
necting Scientific Software” (http://cm.bell-labs.
com/stat/duncan/Talks/JSM2003). The talk dealt
with connecting other software packages to R, with
particular attention to R DCOM services. The dis-
cussant, Wolfgang Hartmann from SAS, provided an
industry perspective (see http://www.cmat.pair.
com/wolfgang/jsm03.pdf) comparing the features
of different software, commercial and open-source,
with specific attention to R.
Balasubramanian Narasimhan
Stanford University, CA, USA
naras@stat.stanford.edu
gR 2003
On 17-20th September 2003, Aalborg University
hosted a workshop bringing together people from
many communities working with graphical models.
The common interest is development of a package
for R, supporting the use of graphical models for
data analysis. The workshop followed up on the gR
initiative described by Steffen Lauritzen in R News
2/3.
The workshop provided a kaleidoscope of ap-
plications as well as insight in experiences deal-
ing with practical graphical models. The applica-
tions presented were from the areas of epidemiol-
ogy, geostatistics, genetics, bioinformatics and ma-
chine learning.
The wide range of applications and methodology
showed that a unifying software package for graphi-
cal models must be widely extensible and flexible —
utilizing a variety of data formats, model specifica-
tions and estimation algorithms. The package should
also provide an attractive user interface that aids in
working with complex models interactively.
Development of a gR-package is evolving at
many levels. Some ’old’ stand-alone programs
are being ported as R-packages (CoCoR, BRugs),
some are being interfaced (mimR, JAGS, BugsR),
while others have been developed in R (ggm, deal,
GRAPPA).
Experiences from other existing packages can in-
spire the gR project. For example, the Bayes Net
Toolbox for Matlab includes many features that gR
will include. Intel is currently re-implementing the
Bayes Net Toolbox in C++ (called Probability Net-
work Library, PNL) and plan a December 2003 re-
lease, expected to be open source. An R interface to
PNL could be a possibility.
During the workshop an outline of a package gr-
base with basic elements was discussed and thought
to become a common ground for extensions. Impor-
tant features were to separate data, model and infer-
ence. The grbase package will include
• support for a variety of data formats, eg. as a
list of cases, a dataframe or a database connec-
tion. It should also be possible to work with a
model without data.
• a general model language capable of specify-
ing eg. (block-) recursive graphical models and
BUGS models.
• a variety of representation forms for graphs, eg.
using/extending the graph package from bio-
conductor.
R News ISSN 1609-3631

Vol. 3/2, October 2003 28
• a graphics system, for interactively working
with models. For example using R-Tcl/Tk,
Rggobi or the R-interface to Graphviz.
• an analyzing unit that combines data and
model with the possibility of using different in-
ference algorithms in the analyzing step.
A minimal version of grbase is planned for January
2004.
An invited session concerned with the gR de-
velopments is being planned for the Joint Statistical
Meeting in Toronto, 8-12 August 2004.
See http://www.math.auc.dk/gr/gr2003/ for
more information about the workshop and related
links, including links to the aforementioned soft-
ware.
Acknowledgments The gR-2003 workshop was
supported by the Danish National Research Founda-
tion Network in Mathematical Physics and Stochas-
tics - MaPhySto. The Danish activities of the gR
project are supported by the Danish Natural Science
Research Council.
Claus Dethlefsen
Aalborg University, Denmark
dethlef@math.auc.dk
Book Reviews
John Maindonald and John Braun:
Data Analysis and Graphics Using
R — An Example-based Approach
Cambridge University Press, Cambridge, United
Kingdom, 2003
362 pages, ISBN 0-521-81336-0
http://cbis.anu.edu/DAAG/
http://www.stats.uwo.ca/DAAG/
The aim of the book is to describe the ideas and
concepts of many statistical methodologies, that are
widely used in applications, by demonstrating the
use of R on a number of examples. Most examples in
the book use genuine data collected by the authors in
their combined several decades of statistical consult-
ing experience. The authors see the book as a com-
panion to other books that include more mathemati-
cal treatments of the relevant theory, and they avoid
mathematical notation and mathematical description
of statistical methods. The book is aimed at both sci-
entists and students interested in practical data anal-
ysis. Data and new R functions used in the book are
included in the DAAG package available from the
authors’ web sites and through the Comprehensive
R Archive Network (CRAN).
The book begins with a nice summary of the con-
tents of the twelve chapters of the book. Chapter 1,
A Brief Introduction to R, provides enough informa-
tion on using R to get the reader started. Chapter 2,
Style of Data Analysis, demonstrates with many ex-
amples the use of R to carry out basic exploratory
data analysis involving both graphical and numeri-
cal summaries of data. The authors not only describe
how to create graphs and plots but also show the
reader what to look for in the data summaries and
how to interpret the summaries in the context of each
particular example. Chapter 3, Statistical Models, de-
scribes the authors’ view on the importance of mod-
els as a framework for statistical analysis. Chapter 4,
An Introduction to Formal Inference, introduces the ba-
sic ideas of random sampling and sampling distribu-
tions of statistics necessary to understand confidence
intervals and hypothesis testing. It also includes
chi-square tests for contingency tables and one-way
ANOVA.
The next several chapters demonstrate the use of
R to analyze data using linear models. Chapter 5,
Regression with a Single Predictor, Chapter 6, Multi-
ple Linear Regression, Chapter 7, Exploiting the Linear
Model Framework, and Chapter 8, Logistic Regression
and Other Generalized Linear Models, use increasingly
complex models to lead the reader through several
examples of practical data analysis.
The next three chapters discuss more specialized
topics that arise frequently in practice. Chapter 9,
Multi-level Models, Time Series, and Repeated Measures,
goes through examples that use more complicated
error structures than examples found in previous
chapters. Chapter 10, Tree-based Classification and Re-
gression Trees, provides an introduction to tree-based
regression and classification modeling. Chapter 11,
Multivariate Data Exploration and Discrimination, de-
scribes both principle components analysis and dis-
criminant analysis.
The final chapter, Chapter 12, The R System — Ad-
ditional Topics, is a far more detailed introduction to
R than that contained in the initial chapters. It is also
intended as a reference to the earlier chapters.
The book is a primer on the nuts-and-bolts use
of R for the types of statistical analysis that arise
commonly in statistical practice, and it also teaches
the reader to think statistically when interpreting the
results of an analysis. The strength of the book is
in the extensive examples of practical data analysis
with complete examples of the R code necessary to
carry out the analyses. Short R commands appear
on nearly every page of the book and longer R code
examples appear frequently as footnotes.
R News ISSN 1609-3631

Vol. 3/2, October 2003 29
I would strongly recommend the book to scien-
tists who have already had a regression or a linear
models course and who wish to learn to use R. How-
ever, my recommendation has a few caveats. The
first chapter of the book takes the reader through an
introduction to R that has the potential to be a little
frustrating for a reader with no prior R experience.
For example, the first plotting command given is
plot(ACT ~ Year, data=ACTpop, pch=16)
The meaning of the pch=16 option is described and
the option data=ACTpop is self evident, but the syn-
tax ACT ~ Year is not explained and is potentially
confusing to an R beginner who does not automat-
ically translate ~into “is modeled by”. Page 5 gives
the advice to create a new workspace before exper-
imenting with R functions, but provides no details
on how one actually does this. Most examples of R
code in the book do contain adequate descriptions,
but there are a number of exceptions.
A second caveat is that the descriptions of statis-
tical methods are an adequate refresher, but are inad-
equate as a primary source of information. The au-
thors indicate clearly that the book is meant to com-
plement other books in the presentation of, and the
mathematical description of, statistical methods. I
agree that the book would not work well as a stand-
alone text book for a course on statistical modeling.
However, it is also not short and I would hesitate to
require students to buy it in addition to another com-
prehensive textbook. The scope of the book is greater
than simply serving as a companion book for teach-
ing R.
Despite my hesitation to use this book in teach-
ing, I give it a strong recommendation to the scien-
tist or data analyst who wishes an easy-to-read and
an understandable reference on the use of R for prac-
tical data analysis.
Bret Larget
University of Wisconsin—Madison
larget@stat.wisc.edu
Changes in R 1.8.0
by the R Core Team
MacOS changes
• As from this release there is only one R port
for the Macintosh, which runs only on MacOS
X. (The ‘Carbon’ port has been discontinued,
and the ‘Darwin’ port is part of the new ver-
sion.) The current version can be run either as a
command-line application or as an ‘Aqua’ con-
sole. There is a ‘Quartz’ device quartz(), and
the download and installation of both source
and binary packages is supported from the
Aqua console. Those CRAN and BioC pack-
ages which build under MacOS X have binary
versions updated daily.
User-visible changes
• The defaults for glm.control(epsilon=1e-8,
maxit=25) have been tightened: this will pro-
duce more accurate results, slightly slower.
• sub, gsub, grep, regexpr, chartr, tolower, toup-
per, substr, substring, abbreviate and strsplit
now handle missing values differently from
"NA".
• Saving data containing name space references
no longer warns about name spaces possibly
being unavailable on load.
• On Unix-like systems interrupt signals now set
a flag that is checked periodically rather than
calling longjmp from the signal handler. This is
analogous to the behavior on Windows. This
reduces responsiveness to interrupts but pre-
vents bugs caused by interrupting computa-
tions in a way that leaves the system in an in-
consistent state. It also reduces the number of
system calls, which can speed up computations
on some platforms and make R more usable
with systems like Mosix.
Changes to the language
• Error and warning handling has been mod-
ified to incorporate a flexible condition han-
dling mechanism. See the online documen-
tation of tryCatch() and signalCondition().
Code that does not use these new facilities
should remain unaffected.
• A triple colon operator can be used to access
values of internal variables in a name space (i.e.
a:::b is the value of the internal variable bin
name space a).
• Non-syntactic variable names can now be
specified by inclusion between backticks
Like This . The deparse() code has been
changed to output non-syntactical names with
this convention, when they occur as operands
in expressions. This is controlled by a backtick
R News ISSN 1609-3631

Vol. 3/2, October 2003 30
argument, which is by default TRUE for com-
posite expressions and FALSE for single sym-
bols. This should give minimal interference
with existing code.
• Variables in formulae can be quoted by
backticks, and such formulae can be used
in the common model-fitting functions.
terms.formula() will quote (by backticks)
non-syntactic names in its "term.labels" at-
tribute. [Note that other code using terms ob-
jects may expect syntactic names and/or not
accept quoted names: such code will still work
if the new feature is not used.]
New features
• New function bquote() does partial substitu-
tion like LISP backquote.
•capture.output() takes arbitrary connections
for file argument.
•contr.poly() has a new scores argument to
use as the base set for the polynomials.
•cor() has a new argument method =
c("pearson","spearman","kendall")’ as
cor.test() did forever. The two rank based
measures do work with all three missing value
strategies.
• New utility function cov2cor() Cov -> Corr
matrix.
•cut.POSIXt() now allows ‘breaks’ to be more
general intervals as allowed for the ‘by’ argu-
ment to seq.POSIXt().
•data() now has an envir argument.
•det() uses an LU decomposition and LA-
PACK. The method argument to det() no
longer has any effect.
•dev.control() now accepts enable as well as
inhibit. (Wishlist PR#3424)
•*,-and /work more generally on "difftime"
objects, which now have a diff() method.
•dt(*, ncp = V) is now implemented, thanks
to Claus Ekstroem.
•dump() only quotes object names in the file
where necessary.
•eval() of a promise forces the promise
•file.path() now returns an empty character
vector if given at least one zero-length argu-
ment.
•format() and hence print() make an effort to
handle corrupt data frames, with a warning.
•format.info() now also works with ‘nsmall’
in analogy with format.default().
•gamma(n) is very slightly more precise for inte-
ger n in 11:50.
•?and help() will accept more un-quoted ar-
guments, e.g. NULL.
• The ?operator has new forms for querying
documentation on S4 methods. See the online
documentation.
• New argument frame.plot = axes (==
TRUE) for filled.contour().
• New argument fixed = TRUE for grep() and
regexpr() to avoid the need to escape strings
to match.
•grep(x, ..., value = TRUE) preserves
names of x.
•hist.POSIXt() can now pass arguments to
hist.default()
•legend() and symbols() now make use of
xy.coords() and accept a wider range of co-
ordinate specifications.
• Added function library.dynam.unload() to
call dyn.unload() on a loaded DLL and tidy
up. This is called for all the standard packages
in namespaces with DLLs if their namespaces
are unloaded.
•lm(singular.ok = FALSE) is now imple-
mented.
• Empty lm() and glm() fits are now handled
by the normal code: there are no methods for
classes "lm.null" and "glm.null". Zero-rank
fits are handled consistently.
•make.names() has improvements, and there
is a new auxiliary function make.unique().
(Based on code contributed by Tom Minka,
since converted to a .Internal function.) In
particular make.names() now recognises that
names beginning with a dot are valid and that
reserved words are not.
•methods() has a print method which as-
terisks functions which are not user-visible.
methods(class = "foo") now lists non-
visible functions, and checks that there is a
matching generic.
•model.matrix() now warns when it removes
the response from the rhs of the formula: that
this happens is now documented on its help
page.
R News ISSN 1609-3631

Vol. 3/2, October 2003 31
• New option "locatorBell" to control the con-
firmation beep during the use of locator()
and identify().
• New option("scipen") provides some user
control over the printing of numbers in fixed-
point or exponential notation. (Contributed by
David Brahm.)
•plot.formula() now accepts horizontal=TRUE
and works correctly when boxplots are pro-
duced. (Wishlist PR#1207) The code has been
much simplified and corrected.
•polygon() and rect() now interpret density <
0 or NA to mean filling (by colour) is desired:
this allows filling and shading to be mixed in
one call, e.g. from legend().
• The predict() methods for classes lm, glm,
mlm and lqs take a ‘na.action’ argument that
controls how missing values in ‘newdata’ are
handled (and defaults to predicting NA). [Pre-
viously the value of getOption("na.action")
was used and this by default omitted cases
with missing values, even if set to ‘na.exclude’.]
•print.summary.glm() now reports omit-
ted coefficients in the same way as
print.summary.lm(), and both show them as
NAs in the table of coefficients.
•print.table() has a new argument
‘zero.print’ and is now documented.
•rank(x, na.last = "keep") now preserves
NAs in ‘x’, and the argument ‘ties.method’ al-
lows to use non-averaging ranks in the pres-
ence of ties.
•read.table()’s ’as.is’ argument can be charac-
ter, naming columns not to be converted.
•rep() is now a generic function, with de-
fault, POSIXct and POSIXlt methods. For ef-
ficiency, the base code uses rep.int() rather
than rep() where possible.
• New function replicate() for repeated eval-
uation of expression and collection of results,
wrapping a common use of sapply() for sim-
ulation purposes.
•rev() is now a generic function, with default
and dendrogram methods.
•serialize() and unserialize() functions are
available for low-level serialization to connec-
tions.
•socketSelect() allows waiting on multiple
sockets.
•sort(method = "quick", decreasing =
TRUE) is now implemented.
•sort.list() has methods "quick" (a wrapper
for sort(method = "quick", index.return
= TRUE) and "radix" (a very fast method for
small integers). The default "shell" method
works faster on long vectors with many ties.
•stripchart() now has ‘log’, ‘add’ and ‘at’ ar-
guments.
•strsplit(x, *) now preserves names() but
won’t work for non-character ‘x’ anymore
formerly used as.character(x), destroying
names(x).
•textConnection() now has a local argument
for use with output connections. local = TRUE
means the variable containing the output is as-
signed in the frame of the caller.
• Using UseMethod() with more than two argu-
ments now gives a warning (as R-lang.texi has
long claimed it did).
• New function vignette() for viewing or list-
ing vignettes.
•which.min(x) and which.max(x) now pre-
serve names.
•xy.coords() coerces "POSIXt" objects to
"POSIXct", allowing lines, etc. to be added to
plot.POSIXlt() plots.
•.Machine has a new entry, sizeof.pointer.
•.Random.seed is only looked for and stored in
the user’s workspace. Previously the first place
a variable of that name was found on the search
path was used.
• Subscripting for data.frames has been rational-
ized:
–Using a single argument now ignores any
‘drop’ argument (with a warning). Previ-
ously using ‘drop’ inhibited list-like sub-
scripting.
–adf$name <- value now checks for the
correct length of ‘value’, replicating a
whole number of times if needed.
–adf[j] <- value and adf[[j]] <-
value did not convert character vectors to
factors, but adf[,j] <- value did. Now
none do. Nor is a list ‘value’ coerced to
a data frame (thereby coercing character
elements to factors).
–Where replicating the replacement value
a whole number of times will produce
the right number of values, this is always
done (rather than some times but not oth-
ers).
R News ISSN 1609-3631

Vol. 3/2, October 2003 32
–Replacement list values can include
NULL elements.
–Subsetting a data frame can no longer pro-
duce duplicate column names.
–Subsetting with drop=TRUE no longer
sometimes drops dimensions on matrix or
data frame columns of the data frame.
–Attributes are no longer stripped when re-
placing part of a column.
–Columns added in replacement opera-
tions will always be named, using the
names of a list value if appropriate.
–as.data.frame.list() did not cope with
list names such as ‘check.rows’, and for-
matting/printing data frames with such
column names now works.
–Row names in extraction are still made
unique, but without forcing them to be
syntactic names.
–adf[x] <- list() failed if x was of
length zero.
• Setting dimnames to a factor now coerces to
character, as S does. (Earlier versions of R used
the internal codes.)
• When coercion of a list fails, a meaningful error
message is given.
• Adding to NULL with [[ ]] generates a list if
more than one element is added (as S does).
• There is a new command-line flag ‘--args’
that causes the rest of the command line to
be skipped (but recorded in commandArgs() for
further processing).
• S4 generic functions and method dispatch have
been modified to make the generic functions
more self-contained (e.g., usable in apply-type
operations) and potentially to speed dispatch.
• The data editor is no longer limited to 65535
rows, and will be substantially faster for large
numbers of columns.
• Standalone Rmath now has a get_seed func-
tion as requested (PR#3160).
• GC timing is not enabled until the first call
to gc.time(); it can be disabled by call-
ing gc.time(FALSE). This can speed up the
garbage collector and reduce system calls on
some platforms.
Standard packages
• New package ’mle’. This is a simple package to
find maximum likelihood estimates, and per-
form likelihood profiling and approximate con-
fidence limits based upon it. A well-behaved
likelihood function is assumed, and it is the re-
sponsibility of the user to gauge the applicabil-
ity of the asymptotic theory. This package is
based on S4 methods and classes.
• Changes in package ’mva’:
–factanal() now returns the test statis-
tic and P-value formerly computed in the
print method.
–heatmap() has many more arguments,
partly thanks to Wolfgang Huber and
Andy Liaw.
–Arguments ‘unit’ and ‘hmin’ of
plclust() are now implemented.
–prcomp() now accepts complex matrices,
and there is biplot() method for its out-
put (in the real case).
–dendrograms are slightly better docu-
mented, methods working with "label",
not "text" attribute. New rev() method
for dendrograms.
–plot.dendrogram() has an explicit
‘frame.plot’ argument defaulting to
FALSE (instead of an implicit one default-
ing to TRUE).
• Changes in package ’tcltk’:
–The package is now in a namespace. To
remove it you will now need to use
unloadNamespace("tcltk").
–The interface to Tcl has been made much
more efficient by evaluating Tcl com-
mands via a vector of Tcl objects rather
than by constructing the string represen-
tation.
–An interface to Tcl arrays has been intro-
duced.
–as.tclObj() has gained a ‘drop’ argu-
ment to resolve an ambiguity for vectors
of length one.
• Changes in package ’tools’:
–Utilities for testing and listing files, ma-
nipulating file paths, and delimited pat-
tern matching are now exported.
–Functions
checkAssignFuns()
checkDocArgs()
checkMethods()
R News ISSN 1609-3631

Vol. 3/2, October 2003 33
have been renamed to
checkReplaceFuns()
checkDocFiles()
checkS3methods()
to given better descriptions of what they
do.
–R itself is now used for analyzing the
markup in the \usage sections. Hence in
particular, replacement functions or S3 re-
placement methods are no longer ignored.
–checkDocFiles() now also determines
’over-documented’ arguments which are
given in the \arguments section but not in
\usage.
–checkDocStyle() and checkS3Methods()
now know about internal S3 generics and
S3 group generics.
–S4 classes and methods are included in the
QC tests. Warnings will be issued from
undoc() for classes and methods defined
but not documented. Default methods
automatically generated from nongeneric
functions do not need to be documented.
–New (experimental) functions
codocClasses()
codocData()
for code/documentation consistency
checking for S4 classes and data sets.
• Changes in package ’ts’:
–arima.sim() now checks for inconsis-
tent order specification (as requested in
PR#3495: it was previously documented
not to).
–decompose() has a new argument ‘filter’.
–HoltWinters() has new arguments ‘op-
tim.start’ and ‘optim.control’, and returns
more components in the fitted values. The
plot method allows ‘ylim’ to be set.
–plot.ts() has a new argument ‘nc’ con-
trolling the number of columns (with de-
fault the old behaviour for plot.mts).
–StructTS() now allows the first value of
the series to be missing (although it is bet-
ter to omit leading NAs). (PR#3990)
Using packages
•library() has a pos argument, controlling
where the package is attached (defaulting to
pos=2 as before).
•require() now maintains a list of required
packages in the toplevel environment (typ-
ically, .GlobalEnv). Two features use this:
detach() now warns if a package is detached
that is required by an attached package, and
packages that install with saved images no
longer need to use require() in the .First as
well as in the main source.
• Packages with name spaces can now be in-
stalled using ‘--save’.
• Packages that use S4 classes and methods
should now work with or without saved im-
ages (saved images are still recommended for
efficiency), writing setMethod(), etc. calls
with the default for argument ‘where’. The
topenv() function and sys.source() have
been changed correspondingly. See the online
help.
• Users can specify in the DESCRIPTION file the
collation order for files in the R source directory
of a package.
R documentation format
• New logical markup commands for empha-
sizing (\strong) and quoting (\sQuote and
\dQuote) text, for indicating the usage of an
S4 method (\S4method), and for indicating spe-
cific kinds of text (\acronym,\cite,\command,
\dfn,\env,\kbd,\option,\pkg,\samp,\var).
• New markup \preformatted for pre-
formatted blocks of text (like example but
within another section). (Based on a contri-
bution by Greg Warnes.)
• New markup \concept for concept index en-
tries for use by help.search().
• Rdconv now produces more informative out-
put from the special \method{GENERIC}{CLASS}
markup for indicating the usage of S3 methods,
providing the CLASS info in a comment.
•\dontrun sections are now marked within
comments in the user-readable versions of the
converted help pages.
•\dontshow is now the preferred name for
\testonly.
Installation changes
• The zlib code in the sources is used unless the
external version found is at least version 1.1.4
(up from 1.1.3).
• The regression checks now have to be passed
exactly, except those depending on recom-
mended packages (which cannot be assumed
to be present).
R News ISSN 1609-3631

Vol. 3/2, October 2003 34
• The target make check-all now runs R CMD
check on all the recommended packages (and
not just runs their examples).
• There are new macros DYLIB_* for building
dynamic libraries, and these are used for the
dynamic Rmath library (which was previously
built as a shared object).
• If a system function log1p is found, it is tested
for accuracy and if inadequate the substitute
function in src/nmath is used, with name
remapped to Rlog1p. (Apparently needed on
OpenBSD/NetBSD.)
C-level facilities
• There is a new installed header file
R_ext/Parse.h which allows R_ParseVector to
be called by those writing extensions. (Note
that the interface is changed from that used
in the unexported header Parse.h in earlier
versions, and is not guaranteed to remain un-
changed.)
• The header R_ext/Mathlib.h has been re-
moved. It was replaced by Rmath.h in R 1.2.0.
• PREXPR has been replaced by two macros,
PREXPR for obtaining the expression and
PRCODE for obtaining the code for use in
eval. The macro BODY_EXPR has been added
for use with closures. For a closure with a
byte compiled body, the macro BODY_EXPR
returns the expression that was compiled; if the
body is not compiled then the body is returned.
This is to support byte compilation.
• Internal support for executing byte compiled
code has been added. A compiler for produc-
ing byte compiled code will be made available
separately and should become part of a future
R release.
• On Unix-like systems calls to the popen() and
system() C library functions now go through
R_popen and R_system. On Mac OS X these
suspend SIGALRM interrupts around the li-
brary call. (Related to PR#1140.)
Utilities
• R CMD check accepts "ORPHANED" as pack-
age maintainer. Package maintainers can now
officially orphan a package, i.e., resign from
maintaining a package.
• R CMD INSTALL (Unix only) is now ’safe’: if
the attempt to install a package fails, leftovers
are removed. If the package was already in-
stalled, the old version is restored.
• R CMD build excludes possible (obsolete) data
and vignette indices in DCF format (and hence
also no longer rebuilds them).
• R CMD check now tests whether file names are
valid across file systems and supported oper-
ating system platforms. There is some support
for code/documentation consistency checking
for data sets and S4 classes. Replacement func-
tions and S3 methods in \usage sections are no
longer ignored.
• R CMD Rdindex has been removed.
Deprecated & defunct
• The assignment operator ‘_’ has been removed.
•printNoClass() is defunct.
• The classic MacOS port is no longer supported,
and its files have been removed from the
sources.
• The deprecated argument ’white’ of parse()
has been removed.
• Methods pacf/plot.mts() have been re-
moved and their functionality incorporated
into pacf.default/plot.ts().
•print.coefmat() is deprecated in favour of
printCoefmat() (which is identical apart from
the default for na.print which is changed from
"" to "NA", and better handling of the 0-rank
case where all coefficients are missing).
•codes() and codes<-() are deprecated, as al-
most all uses misunderstood what they actu-
ally do.
• The use of multi-argument return() calls is
deprecated: use a (named) list instead.
• anovalist.lm (replaced in 1.2.0) is now depre-
cated.
•-and Ops methods for POSIX[cl]t objects are
removed: the POSIXt methods have been used
since 1.3.0.
•glm.fit.null(),lm.fit.null() and
lm.wfit.null() are deprecated.
• Classes "lm.null" and "glm.null" are deprecated
and all of their methods have been removed.
• Method weights.lm(), a copy of
weights.default(), has been removed.
•print.atomic() is now deprecated.
• The back-compatibility entry point Rf_log1p in
standalone Rmath has been removed.
R News ISSN 1609-3631

Vol. 3/2, October 2003 35
Changes on CRAN
by Kurt Hornik and Friedrich Leisch
New contributed packages
DAAG various data sets used in examples and exer-
cises in the book Maindonald, J.H. and Braun,
W.J. (2003) "Data Analysis and Graphics Using
R". By John Maindonald and W. John Braun.
Devore6 Data sets and sample analyses from Jay
L. Devore (2003), "Probability and Statistics
for Engineering and the Sciences (6th ed)",
Duxbury. Original by Jay L. Devore, modifi-
cations by Douglas Bates.
Hmisc The Hmisc library contains many functions
useful for data analysis, high-level graph-
ics, utility operations, functions for computing
sample size and power, importing datasets, im-
puting missing values, advanced table making,
variable clustering, character string manipula-
tion, conversion of S objects to LaTeX code, and
recoding variables. By Frank E Harrell Jr, with
contributions from many other users.
HyperbolicDist This package includes the basic
functions for the hyperbolic distribution: prob-
ability density function, distribution function,
quantile function, a routine for generating ob-
servations from the hyperbolic, and a function
for fitting the hyperbolic distribution to data.
By David Scott.
VaR A set of methods for calculation of Value at Risk
(VaR). By Talgat Daniyarov.
bim Functions to sample and interpret Bayesian
QTL using MCMC. By Brian S. Yandell, Hao
Wu.
boolean A procedure for testing Boolean hypothe-
ses. By Bear F. Braumoeller, Jacob Kline.
cat Analysis of categorical-variable with missing
values. Original by Joseph L. Schafer. Ported
to R by Ted Harding and Fernando Tusell.
classPP PP Indices using class information. By Eun-
kyung Lee.
clines Calculates contour lines. By Paul Murrell.
diptest Compute Hartigan’s dip test statistic for
unimodality. By Martin Maechler, based
on Fortran and S-plus from Dario Ringach
(NYU.edu).
eha A package for survival and event history analy-
sis. By Göran Broström.
emme2 This package includes functions to read and
write to an EMME/2 databank. By Ben Stabler.
exactLoglinTest Monte Carlo and MCMC goodness
of fit tests for log-linear models. By Brian Caffo.
flexmix FlexMix implements a general framework
for finite mixtures of regression models using
the EM algorithm. FlexMix provides the E-
step and all data handling, while the M-step
can be supplied by the user to easily define
new models. Existing drivers implement mix-
tures of standard linear models, generalized
linear models and model-based clustering. By
Friedrich Leisch.
forward Forward search approach to robust analy-
sis in linear and generalized linear regression
models. By Originally written for S-Plus by:
Kjell Konis and Marco Riani. Ported to R by
Luca Scrucca.
fpc Fuzzy and crisp fixed point cluster analysis
based on Mahalanobis distance and linear re-
gression fixed point clusters. Semi-explorative,
semi-model-based clustering methods, operat-
ing on n*p data, do not need prespecification of
number of clusters, produce overlapping clus-
ters. Discriminant projections separate groups
optimally, used to visualize the separation of
groupings. Corresponding plot methods. Clus-
terwise linear regression by normal mixture
modeling. By Christian Hennig.
ftnonpar The package contains R-functions to per-
form the methods in nonparametric regression
and density estimation, described in Davies, P.
L. and Kovac, A. (2001) Local Extremes, Runs,
Strings and Multiresolution (with discussion)
Annals of Statistics. 29. p1-65 Davies, P. L.
and Kovac, A. (2003) Densities, Spectral Den-
sities and Modality Davies, P. L. (1995) Data
features. Statistica Neerlandica 49,185-245. By
Laurie Davies and Arne Kovac.
ggm Functions for defining directed acyclic graphs
and undirected graphs, finding induced graphs
and fitting Gaussian Markov models. By Gio-
vanni M. Marchetti.
gridBase Integration of base and grid graphics. By
Paul Murrell.
its The its package contains an S4 class for handling
irregular time series. By Portfolio & Risk Advi-
sory Group, Commerzbank Securities.
linprog This package can be used to solve Linear
Programming / Linear Optimization problems
R News ISSN 1609-3631

Vol. 3/2, October 2003 36
by using the simplex algorithm. By Arne Hen-
ningsen.
lme4 Fit linear and generalized linear mixed-effects
models. By Douglas Bates, and Saikat DebRoy.
lmeSplines Add smoothing spline modelling capa-
bility to nlme. Fit smoothing spline terms
in Gaussian linear and nonlinear mixed-effects
models. By Rod Ball.
logistf Firth’s bias reduced logistic regression ap-
proach with penalized profile likelihood based
confidence intervals for parameter estimates.
By Meinhard Ploner, Daniela Dunkler, Harry
Southworth, Georg Heinze.
mapdata Supplement to maps package, providing
the larger and/or higher-resolution databases.
Original S code by Richard A. Becker and Allan
R. Wilks. R version by Ray Brownrigg.
maps Display of maps. Projection code and larger
maps are in separate packages (mapproj and
mapdata). Original S code by Richard A.
Becker and Allan R. Wilks. R version by
Ray Brownrigg. Enhancements by Thomas P
Minka.
maptools Set of tools for manipulating and reading
geographic data, in particular ESRI shapefiles.
By Nicholas J. Lewin-Koh, modified by Roger
Bivand; C code used from shapelib ().
merror N methods are used to measure each of n
items. This data is used to estimate the accu-
racy and precision of the methods. Maximum
likelihood estimation is used for the precision
estimates. By Richard A. Bilonick.
mmlcr Mixed-mode latent class regression (also
known as mixed-mode mixture model regres-
sion or mixed-mode mixture regression mod-
els) which can handle both longitudinal and
one-time responses, although it is created with
longitudinal data in mind. By Steve Buyske.
mvnormtest Generalization of Shapiro-Wilk test for
multivariate variables. By Slawomir Jarek.
negenes Estimating the number of essential genes in
a genome on the basis of data from a random
transposon mutagenesis experiment, through
the use of a Gibbs sampler. By Karl W Broman.
nlmeODE This package combines the odesolve and
nlme packages for mixed-effects modelling us-
ing differential equations. By Christoffer W.
Tornoe.
nortest Five omnibus tests for the composite hy-
pothesis of normality. By Juergen Gross.
nprq Nonparametric and sparse quantile regression
methods. By Roger Koenker and Pin Ng.
orientlib Representations, conversions and display
of orientation SO(3) data. See the orientlib help
topic for details. By Duncan Murdoch.
pps The pps package contains functions to select
samples using PPS (probability proportional
to size) sampling. It also includes a function
for stratified simple random sampling, a func-
tion to compute joint inclusion probabilities for
Sampford’s method of PPS sampling, and a few
utility functions. By Jack G. Gambino.
prabclus Distance based parametric bootstrap tests
for clustering, mainly thought for presence-
absence data (clustering of species distribution
maps). Jaccard and Kulczynski distance mea-
sures, clustering of MDS scores, and nearest
neighbor based noise detection (R port of Byers
and Raftery’s (1998) "NNclean"). Main func-
tions are prabtest (for testing), prabclust (for
clustering), prabinit (for preparing the data)
and NNclean (for noise detection). The help-
pages for prabtest and prabclust contain simple
standard executions. By Christian Hennig.
psy Kappa, ICC, Cronbach alpha, screeplot, PCA
and related methods. By Bruno Falissard.
rqmcmb2 Markov Chain Marginal Bootstrap for
Quantile Regression. A resampling method for
inference in quantile regression. Suitable for
modest to large data sets. By Maria Kochergin-
sky, Xuming He.
sca Simple Component Analysis often provides
much more interpretable components than
Principal Components (PCA) without losing
too much. By Valentin Rousson and Martin
Maechler.
seacarb Calculates parameters of the seawater car-
bonate system. By Aurelien Proye and Jean-
Pierre Gattuso.
seao Software for simple evolutionary algorithms.
For all factors (genes) included, one can set the
lowest and highest values as well as the num-
ber of levels (alleles) or the step. An initial gen-
eration can be calculated in several ways and
following generations are calculated based on
a parent generation which can be constructed
using other, already calculated generations or
new generations (as long as the format is ok).
By Kurt Sys.
seao.gui Graphical interface for seao-package. All
functions can be called seperately, but there’s
also a function which can call all other func-
tions. The functions called with this graphical
R News ISSN 1609-3631

Vol. 3/2, October 2003 37
interface hasn’t the same flexibility of the func-
tions called from the command-line. This may
change in the future, although I doubt that. . . .
By Kurt Sys.
segmented Functions to estimate break-points of
segmented relationships in regression models
(GLMs). By Vito M. R. Muggeo.
shapefiles Functions to read and write ESRI shape-
files. By Ben Stabler.
shapes Routines for the statistical analysis of
shapes. In particular, the package provides
routines for procrustes analysis, displaying
shapes and principal components, testing for
mean shape difference, thin-plate spline trans-
formation grids and edge superimposition
methods. By Ian Dryden.
simpleboot Simple bootstrap routines. By Roger D.
Peng.
smoothSurv This package contains primarily a
function to fit a regression model with possi-
bly right, left or interval censored observations
and with the error distrbution expressed as a
mixture of G-splines. Core part of the computa-
tion is done in compiled C++ written using the
Scythe Statistical Libary Version 0.3. By Arnost
Komarek.
tapiR Tools for accessing online UK House of Com-
mons voting data, and datasets for the parlia-
ments 1992-97, 1997-2001 and 2001-now. By
David Firth and Arthur Spirling.
udunits This package provides an R interface to
the Unidata udunits library routines, which
can convert quantities between various units.
Units are indicated by human-readable strings,
such as "m/s", "J", "kg", or "in". Routines
for converting any quantity in known units to
other compatible units are provided. Of partic-
ular use are the time and calendar conversion
routines. Calendar dates are given with units
such as "days since 1900-01-01", for example.
Values with this unit can be converted to nor-
mal, readable calendar dates. This will let you
find that "32018 days since 1900-01-01" is ac-
tually 31 Aug 1987. These routines follow the
library’s C interface, so consult that section of
Unidata’s udunits manual for reference. Here
are some example formatted units strings that
can be used: "10 kilogram.meters/seconds2",
"10 kg-m/sec2", "(PI radian)2", "degF", "degC",
"100rpm", "geopotential meters", "33 feet wa-
ter". Note that the udunits library must already
be installed on your machine for this package
to work. By David Pierce.
Other changes
• Package grid is a base package in R 1.8.0.
• Package GeneSOM was renamed to som.
Kurt Hornik
Wirtschaftsuniversität Wien, Austria
Kurt.Hornik@R-project.org
Friedrich Leisch
Technische Universität Wien, Austria
Friedrich.Leisch@R-project.org
R News ISSN 1609-3631
Vol. 3/2, October 2003 38
Crossword Solution
by Barry Rowlingson
Unfortunately nobody got the crossword in the last
issue of R News (Vol. 3/1) exactly right, but one of
my clues had an alternate solution which could not
be eliminated as “wrong”. I’ll therefore draw a name
from the RNG hat:
And the winner is:
> sample(c("Rolf","Saikat","Simon"))[1]
[1] "Simon"
So a well-travelled (to the DSC-03 and back) 50
Euro note will be on its way to Simon Fear.
The solution, with some explanations, is
also available at http://www.maths.lancs.ac.uk/
~rowlings/Crossword/.
Barry Rowlingson
Lancaster University, UK
B.Rowlingson@lancaster.ac.uk
R I W C U A D U
OWNE R SH I P B R I A N
B T I A D A D R
E R R A T UM A ND A NT E
R A E B T V OP
T A NH B E NE F A CTOR
GE D R L E
ENT R A P SDO U G L A S
NT E E OE
T H R E A D SA F E JOHN
L I F Y L B K T
E X P OR T SANAN O V A
M L A T T T UB
A P E R M E L E M E NT A L
NY E M D S S E
R News ISSN 1609-3631

Vol. 3/2, October 2003 39
Correction to “Building Microsoft
Windows Versions of R and R packages
under Intel Linux”
by Jun Yan and A.J. Rossini
Unfortunately, due to an inexcusable oversight on
our part, we failed to be crystal clear in our article
Yan and Rossini (2003) that all the described steps
and the Makefile were summarized from several doc-
uments in the R sources (R Development Core Team,
2003).
These documents are INSTALL,readme.package,
and Makefile under the directory src/gnuwin32/ in
the R source. We intended to automate and illustrate
those steps by presenting an explicit example, hop-
ing that it might save people’s time. However, confu-
sion has been caused and inquiries have been raised
to the R-help mailing list. We apologize for the con-
fusion and claim sole responsibility. In addition, we
clarify that the final credit should go to the R Devel-
opment Core Team.
Bibliography
R Development Core Team. R: A language and envi-
ronment for statistical computing. R Foundation for
Statistical Computing, Vienna, Austria, 2003. URL
http://www.R-project.org. ISBN 3-900051-00-3.
39
J. Yan and A. Rossini. Building Microsoft Windows
versions of R and R packages under Intel Linux.
R News, 3(1):15–17, June 2003. URL http://CRAN.
R-project.org/doc/Rnews/.
39
Jun Yan
University of Iowa, U.S.A.
jyan@stat.uiowa.edu
A.J. Rossini
University of Washington, U.S.A.
rossini@u.washington.edu
Editor-in-Chief:
Friedrich Leisch
Institut für Statistik und Wahrscheinlichkeitstheorie
Technische Universität Wien
Wiedner Hauptstraße 8-10/1071
A-1040 Wien, Austria
Editorial Board:
Douglas Bates and Thomas Lumley.
Editor Programmer’s Niche:
Bill Venables
Editor Help Desk:
Uwe Ligges
Email of editors and editorial board:
firstname.lastname @R-project.org
R News is a publication of the R Foundation for Sta-
tistical Computing, communications regarding this
publication should be addressed to the editors. All
articles are copyrighted by the respective authors.
Please send submissions to regular columns to the
respective column editor, all other submissions to
the editor-in-chief or another member of the edi-
torial board (more detailed submission instructions
can be found on the R homepage).
R Project Homepage:
http://www.R-project.org/
This newsletter is available online at
http://CRAN.R-project.org/doc/Rnews/
R News ISSN 1609-3631