PDF Rnews 2005 1
PDF Rnews_2005-1 CRAN: R News
User Manual: PDF CRAN - Contents of R News
Open the PDF directly: View PDF ![]() .
.
Page Count: 72
- Editorial
- Internationalization Features of R 2.1.0
- Packages and their Management in R 2.1.0
- Recent Changes in grid Graphics
- hoa: An R package bundle for higher order likelihood inference
- Fitting linear mixed models in R
- Using R for Statistical Seismology
- Literate programming for creating and maintaining packages
- CRAN Task Views
- Using Control Structures with Sweave
- The Value of R for Preclinical Statisticians
- Recreational mathematics with R: introducing the ``magic'' package
- Programmer's Niche
- Book Review of Julian J. Faraway: Linear Models with R
- R Foundation News
- Changes in R
- Changes on CRAN
- Events
News
The Newsletter of the R Project Volume 5/1, May 2005
Editorial
by Douglas Bates
This edition of R News is the work of a new editorial
team and I would particularly like to recognize my
associate editors, Paul Murrell and Torsten Hothorn,
who stepped up and did a superb job of handling
most of the submissions whilst I was otherwise oc-
cupied.
This issue accompanies the release of R 2.1.0 and
we begin with three articles describing new facili-
ties in this release. For many users the most notable
change with this release is the addition of interna-
tionalization and localization capabilities, described
in our feature article by Brian Ripley, who did most
of the work in adding these facilities to R. The next
article, also written by Brian Ripley, describes the
new package management structure in R 2.1.0 then
Paul Murrell updates us on changes in the standard
package grid.
Following these articles are three articles on con-
tributed packages beginning with Alessandra Braz-
zale’s description of the hoa bundle for statistical
inference using higher-order asymptotics. By con-
tributing the hoa bundle to CRAN Alessandra has
extended to five years the association of R with win-
ners of the Chambers Award for outstanding work
on statistical software by a student. She received the
2001 Chambers Award for her work on this software.
The 2002 Chambers Award winner, Simon Urbanek,
is now a member of the R Development Core Team.
The work of the 2003 winner, Daniel Adler for RGL,
the 2004 winner, Deepayan Sarkar for Lattice, and
the recently-announced 2005 winner, Markus Helbig
for JGR, are all intimately connected with R.
The next section of this newsletter is three articles
on tools for working with R. We follow these with an
article on experiences using R within a large phar-
maceutical company. This article is part of a contin-
uing series on the use of R in various settings. We
welcome contributions to this series. Finally, an ar-
ticle on magic squares and John Fox’s guest appear-
ance in the Programmer’s Niche column, where he
discusses a short but elegant function for producing
textual representations of numbers, put the focus on
programming in R, which is where the focus should
be, shouldn’t it?
Douglas Bates
University of Wisconsin – Madison, U.S.A.
bates@R-project.org
Contents of this issue:
Editorial ...................... 1
Internationalization Features of R 2.1.0 . . . . . 2
Packages and their Management in R 2.1.0 . . . 8
Recent Changes in grid Graphics . . . . . . . . 12
hoa: An R package bundle for higher order
likelihood inference . . . . . . . . . . . . . . 20
Fitting linear mixed models in R . . . . . . . . 27
Using R for Statistical Seismology . . . . . . . . 31
Literate programming for creating and main-
taining packages . . . . . . . . . . . . . . . . 35
CRAN Task Views . . . . . . . . . . . . . . . . . 39
Using Control Structures with Sweave . . . . . 40
The Value of R for Preclinical Statisticians . . . 44
Recreational mathematics with R: introducing
the “magic” package . . . . . . . . . . . . . . 48
Programmer’s Niche . . . . . . . . . . . . . . . 51
Book Review of
Julian J. Faraway: Linear Models with R . . . 56
R Foundation News . . . . . . . . . . . . . . . . 57
ChangesinR.................... 57
Changes on CRAN . . . . . . . . . . . . . . . . 67
Events........................ 71
Vol. 5/1, May 2005 2
Internationalization Features of R 2.1.0
by Brian D. Ripley
R 2.1.0 introduces a range of features to make it pos-
sible or easier to use Rin languages other than En-
glish or American English: this process is known as
internationalization, often abbreviated to i18n (since
18 letters are omitted). The process of adapting to
a particular language, currency and so on is known
as localization (abbreviated to l10n), and R 2.1.0 ships
with several localizations and the ability to add oth-
ers.
What is involved in supporting non-English lan-
guages? The basic elements are
1. The ability to represent words in the language.
This needs support for the encoding used for the
language (which might be OS-specific) as well
as support for displaying the characters, both
at the console and on graphical devices.
2. Manipulation of text in the language, by for
example grep(). This may sound straightfor-
ward, but earlier versions of Rworked with
bytes and not characters: at least two bytes are
needed to represent Chinese characters.
3. Translation of key components from English to
the user’s own language. Currently Rsupports
the translation of diagnostic messages and the
menus and dialogs of the Windows and MacOS
X GUIs.
There are other aspects to internationalization, for
example the support of international standard paper
sizes rather than just those used in North America,
different ways to represent dates and times,1mone-
tary amounts and the representation of numbers.2
Rhas ‘for ever’ had some support for Western
European languages, since several early members of
the core team had native languages other than En-
glish. We have been discussing more comprehen-
sive internationalization for a couple of years, and a
group of users in Japan have been producing a mod-
ified version of Rwith support for Japanese. During
those couple of years OS support for international-
ization has become much more widespread and reli-
able, and the increasing prevalence of UTF-83locales
as standard in Linux distributions made greater in-
ternationalization support more pressing.
How successfully your Rwill support non-
English languages depends on the level of operating
system support, although as always the core team
tries hard to make facilities available across all plat-
forms. Windows NT took internationalization seri-
ously from the mid 1990s, but took a different route
(16-bit characters) from most later internationaliza-
tion efforts. (We still have Rusers running the obso-
lete Windows 95/98/ME OSes, which had language-
specific versions.) MacOS X is a hybrid of compo-
nents which approach internationalization in differ-
ent ways but the Rmaintainers for that platform
have managed to smooth over many of its quirks.
Recent Linux and other systems using glibc have
extensive support. Commercial Unixes have varying
amounts of support, although Solaris was a pioneer
and often the model for Open Source implementa-
tions.
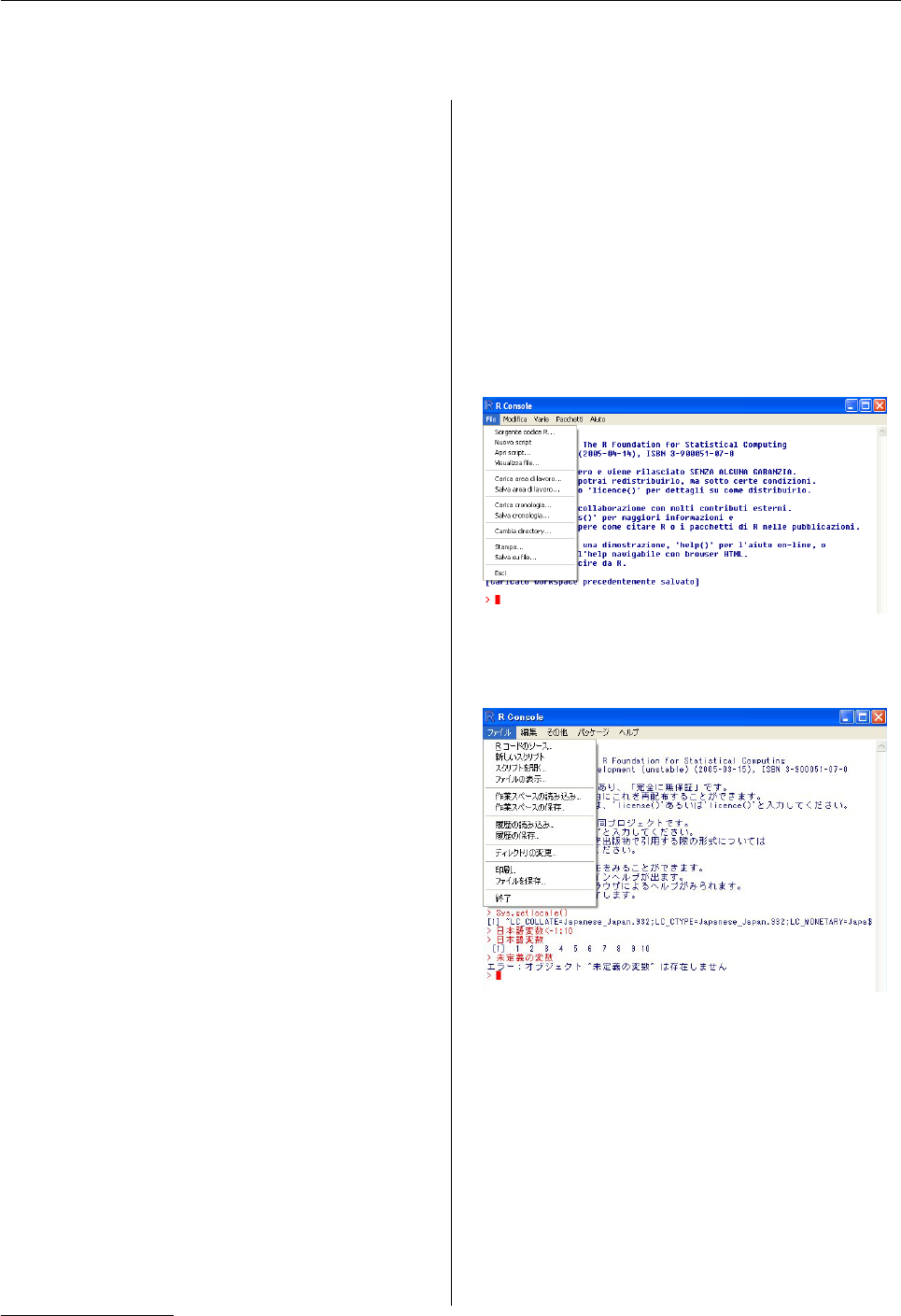
Figure 1: Part of the Windows console in an Italian
locale.
Figure 2: Part of the Windows console in Japanese.
Note the use of Japanese variable names, that the last
line is an error message in Japanese, as well as the
difference in width between English and Japanese
characters.
Hopefully, if your OS has enough support to al-
low you to work comfortably in your preferred lan-
guage, it will have enough support to allow you to
use Rin that language. Figures 1and 2show ex-
amples of internationalization for the Windows GUI:
note how the menus are translated, variable names
are accepted in the preferred language, and error
1e.g. the use of 12-hour or 24-hour clock.
2for example, the use of ,for the decimal point, and the use of ,or .or nothing as the thousands separator.
3see below.
R News ISSN 1609-3631

Vol. 5/1, May 2005 3
messages are translated too. The Japanese screen-
shot shows another difference: the Japanese charac-
ters are displayed at twice the width of the English
ones, and cursor movement has to take this into ac-
count.
The rest of this article expands on the concepts
and then some of the issues they raise for Rusers and
Rprogrammers. Quite a lot of background is needed
to appreciate what users of very different languages
need from the internationalization of R.
Locales
Alocale is a specification of the user-specific environ-
ment within which an operating system is running
an application program such as R. What exactly this
covers is OS-specific, but key components are
• The set of ‘legal’ characters the user might in-
put: this includes which characters are alpha-
betic and which are numeric. This is the C lo-
cale category LC_CTYPE.
• How characters are sorted: the C locale cate-
gory LC_COLLATE. Even where languages share
a character set they may sort words differently:
it comes as a surprise to English-speaking vis-
itors to Denmark to find ‘Aa’ sorted at the end
of the alphabet.
• How to represent dates and times (C locale cat-
egory LC_TIME). The meaning of times also de-
pends on the timezone which is sometimes re-
garded as part of the locale (and sometimes
not).
• Monetary formatting (C locale category
LC_MONETARY).
• Number formatting (C locale category
LC_NUMERIC).
• The preferred language in which to communi-
cate with the user (for some systems, C locale
category LC_MESSAGES).
• How characters in that language are encoded.
How these are specified is (once again) OS-
specific. The first four categories can be set by
Sys.setlocale(): the initial settings are taken
from the OS and can be queried by calling
Sys.getlocale() (see Figure 2). On Unix-like OSes
the settings are taken from environment variables
with the names given, defaulting to the value of
LC_ALL and then of LANG. If none of these are set, the
value is likely to be Cwhich is usually implemented
as the settings appropriate in the USA. Other aspects
of the locale are reported by Sys.localeconv().
Other OSes have other ways of specifying the lo-
cale: for example both Windows and MacOS X have
listboxes in their user settings. This is becoming
more common: when I select a ‘Language’ at the lo-
gin screen for a session in Fedora Core 3 Linux I am
actually selecting a locale, not just my preferred lan-
guage.
Rdoes not fully support LC_NUMERIC, and is un-
likely ever to. Because the comma is used as the sep-
arator in argument lists, it would be impossible to
parse expressions if it were also allowed as the deci-
mal point. We do allow the decimal point to be spec-
ified in scan(),read.table() and write.table().
Sys.setlocale() does allow LC_NUMERIC to be set,
with a warning and with inconsistent behaviour.
For the R user
The most important piece of advice is to specify your
locale when asking questions, since Rwill behave
differently in different locales. We have already seen
experienced Rusers insisting on R-help that R has
been broken when it seems they had changed locales.
To be sure, quote the output of Sys.getlocale() and
localeToCharset().
For the package writer
Try not to write language-specific code. A pack-
age which was submitted to CRAN with variables
named a~nos worked for the author and the CRAN
testers, but not on my Solaris systems. Use only
ASCII characters4for your variable names, and as far
as possible for your documentation.
Languages and character sets
What precisely do we mean by ‘the preferred lan-
guage’? Once again, there are many aspects to con-
sider.
• The language, such as ‘Chinese’ or ‘Spanish’.
There is an international standard ISO 6395for
two-letter abbreviations for languages, as well
as some three-letter ones. These are pretty stan-
dard, except that for Hebrew which has been
changed.
• Is this Spanish as spoken in Spain or in Latin
America?
• ISO 639 specifies no as ‘Norwegian’, but Nor-
way has two official languages, Bokmål and
Nynorsk. Which is this?6
4Digits and upper- and lower-case A–Z, without accents.
5http://www.w3.org/WAI/ER/IG/ert/iso639.htm or http://www.loc.gov/standards/iso639-2/englangn.html
6Bokmål, usually, with nn for Nynorsk and nb for Bokmål specifically.
R News ISSN 1609-3631

Vol. 5/1, May 2005 4
• The same language can be written in different
ways. The most prominent example is Chinese,
which has Simplified and Traditional orthogra-
phy, and most readers can understand only one
of them.
• Spelling. The Unix spell program has the fol-
lowing comment in the BUGS section of its man
page:
‘British spelling was done by an
American.’
but that of itself is an example of cultural impe-
rialism. The nations living on the British Isles
do not consider themselves to speak ‘British
English’ and do not agree on spelling: for ex-
ample in general Chambers’ dictionary (pub-
lished in Scotland) prefers ‘-ise’ and the Oxford
English Dictionary prefers ‘-ize’.
To attempt to make the description more precise, the
two-letter language code is often supplemented by a
two-letter ‘country or territory’ code from ISO 31667.
So, for example
pt_BR is Portuguese as written in Brazil.
zh_CN is (simplified) Chinese as written in most
of mainland China, and zh_TW is (traditional)
Chinese as written in Taiwan (which is writ-
ten in the same way as zh_HK used in Hong
Kong).
en_US is American.
en_GB is English as written somewhere (unspeci-
fied) in the UK.
We need to specify the language for at least three
purposes:
1. To delimit the set of characters which can be
used, and which of those are ‘legal’ in object
names.
2. To know the sorting order.
3. To decide what language to respond in.
In addition, we might need to know the direction of
the language, but currently Ronly supports left-to-
right processing of character strings.
The first two are part of the locale specification.
Specifying the language to be used for messages is
a little more complicated: a Nynorsk speaker might
prefer Nynorsk then Bokmål then generic Norwe-
gian then English, which can be expressed by set-
ting LANGUAGE=nn:nb:no (since generic ‘English’ is
the default).
Encodings
Computers work with bits and bytes, and encoding
is the act of representing characters in the language
as bytes, and vice versa. The earliest encodings (e.g.
for Telex) just represented capital letters and num-
bers but this expanded to ASCII, which has 92 print-
able characters (upper- and lower-case letters, digits
and punctuation) plus control codes, represented as
bytes with the topmost bit as 0. This is also the ISO
646 international standard and those characters are
included in most8other encodings.
Bytes allow 256 rather than 128 different charac-
ters, and ISO 646 has been extended into the bytes
with topmost bit as 1, in many different ways. There
is a series of standards ISO 8859-1 to ISO 8859-15
for such extensions, as well as many vendor-specific
ones (such as the ‘WinAnsi’ and other code pages).
Most languages have less than the 186 or more char-
acters9that an 8-bit encoding provides. The problem
is that given a sequence of bytes there is no way to
know that it is in an 8-bit encoding let alone which
one.
The CJK10 languages have tens of thousands of
characters. There have been many ways to repre-
sent such character sets, for example using shift se-
quences (like the shift, control, alt and altgr keys
on your keyboard) to shift between ‘pages’ of char-
acters. Windows uses a two-byte encoding for the
ideographs, with the ‘lead byte’ with topmost bit 1
followed by a ‘trail byte’. So the character sets used
for CJK languages on Windows occupy one or two
bytes.
A problem with such schemes is their lack of ex-
tensibility. What should we do when a new character
comes along, notably the Euro? Most commonly, re-
place something which is thought to be uncommon
(the generic currency symbol in ISO 8859-1 was re-
placed by the Euro in ISO 8859-15, amongst other
changes).
Unicode
Being able to represent one language may not be
enough: if for example we want to represent per-
sonal names we would like to be able to do so equally
for American, Chinese, Greek, Arabic and Maori11
speakers. So the idea emerged of a universal encod-
ing, to represent all the characters we might like to
7http://www.iso.org/iso/en/prods-services/iso3166ma/index.html.
8In a common Japanese encoding, backslash is replaced by Yen. As this is the encoding used for Windows Japanese fonts, file paths
look rather peculiar in a Japanese version of Windows.
9the rest are normally allocated to ‘space’ and control bytes
10Chinese, Japanese, Korean: known to Windows as ‘East Asian’. The CJK ideographs were also used for Vietnamese until the early
twentieth century.
11language code mi. Maori is usually encoded in ISO 8859-13, along with Lithuanian and Latvian!
R News ISSN 1609-3631

Vol. 5/1, May 2005 5
print—all human languages, mathematical and mu-
sical notation and so on.
This is the purpose of Unicode12, which allows up
to 232 different characters, although it is now agreed
that only 221 will ever be prescribed. The human
languages are represented in the ‘basic multilingual
plane’ (BMP), the first 216 characters, and so most
characters you will encounter have a 4-hex-digit rep-
resentation. Thus U+03B1 is alpha, U+05D0 is aleph
(the first letter of the Hebrew alphabet) and U+30C2
is the Katakana letter ‘di’.
If we all used Unicode, would everyone be
happy? Unfortunately not, as sometimes the same
character can be written in various ways. The
Unicode standard allows ‘only’ 20992 slots for CJK
ideographs, whereas the Taiwanese national stan-
dard defines 48711. The result is that different fonts
have different variants for e.g. U+8FCE and a variant
may be recognizable to only some of the users.
Nevertheless, Unicode is the best we have, and a
decade ago Windows NT adopted the BMP of Uni-
code as its native encoding, using 16-bit characters.
However, Unix-alike OSes expect nul (the zero byte)
to terminate a string such as a file path, and peo-
ple looked for ways to represent Unicode characters
as sequences of a variable number of bytes. By far
the most successful such scheme is UTF-8, which has
over the last couple of years become the de facto stan-
dard for Linux distributions. So when I select
English (UK) British English
as my language at the login screen for a session in Fe-
dora Core 3 Linux, I am also selecting UTF-8 as my
encoding.
In UTF-8, the 7-bit ASCII characters are repre-
sented as a single byte. All other characters are rep-
resented as two or more bytes all with topmost bit 1.
This introduces a number of implementation issues:
• Each character is represented by a variable
number of bytes, from one up to six (although
it is likely at most four will ever be used).
• Some bytes can never occur in UTF-8.
• Many byte sequences are not valid in UTF-8.
The major implementation issue in internationaliz-
ing Rhas been to support such multi-byte character
sets (MBCSs). There are other examples, including
EUC-JP used for Japanese on Unix-alikes and the
one- or two-byte encodings used for CJK on older
versions of Windows.
As UTF-8 locales can support multiple languages,
which are allowed for the ‘legal’ characters in Rob-
ject names? This may depend on the OS, but in all
the examples we know of all characters which would
be alphanumeric in at least one human language are
allowed. So for example ~n and ¨u are allowed in lo-
cale en_US.utf8 on Linux, just as they were in the
Latin-1 locale en_US (whereas on Windows the differ-
ent Latin-1 locales have different sets of ‘legal’ char-
acters, indeed different in different versions of Win-
dows for some languages). We have decided that
only the ASCII numbers will be regarded as numeric
in R.
Sorting orders in Unicode are handled by an OS
service: this is a very intricate subject so do not ex-
pect too much consistency between different imple-
mentations (which includes the same language in
different encodings).
Fonts
Being able to represent alpha, aleph and the
Katakana letter ‘di’ is one thing: being able to dis-
play them is another, and we want to be able to dis-
play them both at the console and in graphical out-
put. Effectively all fonts available cover quite small
subsets of Unicode, although they may be usable in
combinations to extend the range.
Rconsoles normally work in monospaced fonts.
That means that all printable ASCII characters have
the same width. Once we move to Unicode, char-
acters are normally categorized into three widths,
one (ASCII, Greek, Arabic, . . . ), two (most CJK
ideographs) and zero (the ‘combining characters’ of
languages such as Thai). Unfortunately there are two
conventions for CJK characters, with some fonts hav-
ing e.g. Katakana single-width and some (including
that used in Figure 2) double-width.
Which characters are available in a font can seem
capricious: for example the one I originally used to
test translations had directional single quotes but not
directional double quotes.
If displaying another language is not working in
a UTF-8 locale the most likely explanation is that the
font used is incomplete, and you may need to install
extra OS components to overcome this.
Font support for Rgraphics devices is equally dif-
ficult and has currently only been achieved on Win-
dows and on comprehensively equipped X servers.
In particular, postscript() and pdf() are restricted
to Western and Eastern European languages as the
commonly-available fonts are (and some are re-
stricted to ISO 8859-1).
For the R user
Now that Ris able to accept input in different encod-
ings, we need a way to specify the encoding. Con-
nections allow you to specify the encoding via the
encoding argument, and the encoding for stdin can
be set when reading from a file by the command-
12There is a more formal definition in the ISO 10646 standard, but for our purposes Unicode is a more precisely constrained version of
the same encoding. There are various versions of Unicode, currently 4.0.1—see http://www.unicode.org.
R News ISSN 1609-3631

Vol. 5/1, May 2005 6
line argument --encoding. Also, source() has an
encoding argument.
It is important to make sure the encoding is cor-
rect: in particular a UTF-8 file will be valid input in
most locales, but will only be interpreted properly
in a UTF-8 locale or if the encoding is specified. So
to read a data file produced on Windows containing
the names of Swiss students in a UTF-8 locale you
will need one of
A <- read.table(file("students",
encoding="latin1"))
A <- read.table(file("students",
encoding="UCS-2LE"))
the second if this is a ‘Unicode’ Windows file.13
In a UTF-8 locale, characters can be entered as
e.g. \u8fce or \u{8fce} and non-printable charac-
ters will be displayed by print() in the first of these
forms. Further, \U can be used with up to 8 hex digits
for characters not in the BMP.
For the R programmer
There is a common assumption that one byte = one
character = one display column. As from R 2.1.0
a character can use more than one byte and extend
over more than one column when printed. The func-
tion nchar() now has a type argument allowing
the number of bytes, characters or columns to be
found—note that this defaults to bytes for backwards
compatibility.
Switching between encodings
Ris able to support input in different encodings by
using on-the-fly translation between encodings. This
should be an OS service, and for modern glibc-
based systems it is. Even with such systems it can
be frustratingly difficult to find out what encod-
ings they support, and although most systems accept
aliases such as ISO_8859-1,ISO8859-1,ISO_8859_1,
8859_1 and LATIN-1, it is quite possible to find that
with three systems there is no name that they all ac-
cept. One solution is to install GNU libiconv: we
have done so for the Windows port of R, and Apple
have done so for MacOS X.
We have provided R-level support for changing
encoding via the function iconv(), and iconvlist()
will (if possible) list the supported encodings.
Quote symbols
ASCII has a quotation mark ", apostrophe ’and
grave accent ‘, although some fonts14 represent
the latter two as right and left quotes respectively.
Unicode provides a wide variety of quote sym-
bols, include left and right single (U+2018, U+2019)
and double (U+201C, U+201D) quotation marks. R
makes use of these for sQuote() and dQuote() in
UTF-8 locales.
Other languages use other conventions for quota-
tions, for example low and mirrored versions of the
right quotation mark (U+201A, U+201B), as well as
guillemets (U+00AB, U+00BB; something like ,).
Looking at translations of error messages in other
GNU projects shows little consistency between na-
tive writers of the same language, so we have made
no attempt to define language-specific versions of
sQuote() and dQuote().
Translations
Rcomes with a lot of documentation in English.
There is a Spanish translation of An Introduction to
Ravailable from CRAN, and an Italian introduction
based on it as well as links to Japanese translations
of An Introduction to R, other manuals and some help
pages.
R 2.1.0 facilitates the translation of messages from
‘English’ into other languages: it will ship with
a complete translation into Japanese and of many
of the messages into Brazilian Portuguese, Chinese,
German and Italian. Updates of these translations
and others can be added at a later date by translation
packages.
Which these ‘messages’ are was determined by
those (mainly me) who marked up the code for pos-
sible translation. We aimed to make all but the most
esoteric warning and error messages available for
translation, together with informational messages
such as the welcome message and the menus and di-
alogs of the Windows and MacOS X consoles. There
are about 4200 unique messages in command-line R
plus a few hundred in the GUIs.
One beneficial side-effect even for English read-
ers has been much improvement in the consistency
of the messages, as well as some re-writing where
translators found the messages unclear.
For the R user
All the end user needs to do is to ensure that the de-
sired translations are installed and that the language
is set correctly. To test the latter, try it and see: very
likely the internal code will be able to figure out the
correct language from the locale name, and the wel-
come message (and menus if appropriate) will ap-
pear in the correct language. If not, try setting the
environment variable LANGUAGE and if that does not
13This will probably not work unless the first two bytes are removed from the file. Windows is inclined to write a ‘Unicode’ file starting
with a Byte Order Mark 0xFEFE, whereas most Unix-alikes do not recognize a BOM. It would be technically easy to do so but apparently
politically impossible.
14most likely including the one used to display this document.
R News ISSN 1609-3631

Vol. 5/1, May 2005 7
help, read the appropriate section of the R Installation
and Administration manual.
Unix-alike versions of Rcan be built without sup-
port for translation, and it is possible that some OSes
will provide too impoverished an environment to de-
termine the language from the locale or even for the
translation support to be compiled in.
There is a pseudo-language en@quot for which
translations are supplied. This is intended for use
in UTF-8 locales, and makes use of the Unicode di-
rectional single and double quotation marks. This
should be selected via the LANGUAGE environment
variable.
For package writers
Any package can have messages marked for trans-
lation: see Writing R Extensions. Traditionally mes-
sages have been paste-d together, and such mes-
sages can be awkward or impossible to translate into
languages with other word orders. We have added
support for specifying messages via C-like format
strings, using the Rfunction gettextf. A typical us-
age is
stop(gettextf(
"autoloader did not find ’%s’ in ’%s’",
name, package),
domain = NA)
I chose this example as the Chinese translation re-
verses15 the order of the two variables. Using
gettextf() marks the format (the first argument)
for translation and then passes the arguments to
sprintf() for C-like formatting. The argument
domain = NA is passed to stop() as the message re-
turned by gettextf() will already be translated (if
possible) and so needs no further translation. As the
quotation marks are included in the format, trans-
lators can use other conventions (and the pseudo-
language en@quot will).
Plurals are another source of difficulties for trans-
lators. Some languages have no plural forms, others
have ‘singular’ and ‘plural’ as in English, and oth-
ers have up to four forms. Even for those languages
with just ‘singular’ and ‘plural’ there is the question
of whether zero is singular or plural, which differs
by language. There is quite general support for plu-
rals in the R and C functions ngettextf and a small
number16 of examples in the R sources.
See Writing R Extensions for more details.
For would-be translators
Additional translations and help completing the cur-
rent ones would be most welcome. Experience has
shown that it is helpful to have a small translation
team to cross-check each other’s work, discuss id-
iomatic usage and to share the load. Translation will
be an ongoing process as Rcontinues to grow.
Please see the documents linked from
http://developer.r-project.org.
Future directions
At this point we need to gain more experience
with internationalization infrastructure. We know it
works with the main Rplatforms (recent Linux, Win-
dows, MacOS X) and Solaris and for a small set of
quite diverse languages.
We currently have no way to allow Windows
users to use many languages in one session, as Win-
dows’ implementation of Unicode is not UTF-8 and
the standard C interface on Windows does not allow
UTF-8 as an encoding. We plan on making a ‘Uni-
code’ (in the Windows sense) implementation of R
2.2.0 that uses UTF-8 internally and 16-bit characters
to interface with Windows. It is likely that such a ver-
sion will only be available for NT-based versions of
Windows.
It is hoped to support a wider range of encodings
on the postscript() and pdf() devices.
The use of encodings in documentation remains
problematic, including in this article. Texinfo is
used for the Rmanuals but does not currently sup-
port even ISO 8859-1 correctly. We have started with
some modest support for encodings in .Rd files, and
may be able to do more.
Acknowledgements
The work of the Japanese group (especially Ei-ji
Nakama and Masafumi Okada) supplied valuable
early experience in internationalization.
The translations have been made available by
translation teams and in particular some enthusiastic
individuals. The infrastructure for translation is pro-
vided by GNU gettext, suitably bent to our needs
(for Ris a large and complex project, and in particu-
lar extensible).
Brian D. Ripley
University of Oxford, UK
ripley@stats.ox.ac.uk
15using ’%2$s’ and ’%1$s’ in the translation to refer to the second and first argument respectively.
16currently 22, about 0.5% of the messages.
R News ISSN 1609-3631

Vol. 5/1, May 2005 8
Packages and their Management in R 2.1.0
by Brian D. Ripley
R 2.1.0 introduces changes that make packages and
their management considerably more flexible and
less tied to CRAN.
What is a package?
The idea of an Rpackage was based quite closely on
that of an Slibrary section: a collection of Rfunc-
tions with associated documentation and perhaps
compiled code that is loaded from a library1by the
library() command. The Help Desk article (Ligges,
2003) provides more background.
Such packages still exist, but as from R 2.1.0
there are other types of Rextensions, distinguished
by the Type: field in the ‘DESCRIPTION’ file: the
classic package has Type: Package or no Type:
field. The Type: field tells R CMD INSTALL and
install.packages() what to do with the extension,
which is still distributed as a tarball or zip file con-
taining a directory of the same name as the package.
This allows new types of extensions to be added
in the future, but R 2.1.0 knows what to do with two
other types.
Translation packages
with Type: Translation. These are used to add or
update translations of the diagnostic messages pro-
duced by Rand the standard packages. The con-
vention is that languages are known by their ISO
639 two-letter (or less commonly three-letter) codes,
possibly with a ‘country or territory’ modifier. So
package Translation-sl should contain translations
into Slovenian, and Translation-zhTW translations
into traditional Chinese as used in Taiwan. Writing
R Extensions describes how to prepare such a pack-
age: it contains compiled translations and so all R
CMD INSTALL and install.packages() need to do is
to create the appropriate subdirectories and copy the
compiled translations into place.
Frontend packages
with Type: Frontend. These are only supported as
source packages on Unix, and are used to install al-
ternative front-ends such as the GNOME console,
previously part of the Rdistribution and now in
CRAN package gnomeGUI. For this type of package
the installation runs configure and the make and so
allows maximal flexibility for the package writer.
Source vs binary
Initially Rpackages were distributed in source
form as tarballs. Then the Windows binary
packages came along (distributed as Zip files)
and later Classic MacOS and MacOS X packages.
As from R 2.1.0 the package management func-
tions all have a type argument defaulting to the
value of getOption("pkgType"), with known values
"source","win.binary" and "mac.binary". These
distributions have different file extensions: .tar.gz,
.zip and .tgz respectively.
This allows packages of each type to be manip-
ulated on any platform: for example, whereas one
cannot install Windows binary packages on Linux,
one can download them on Linux or Solaris, or find
out if a MacOS X binary distribution is available.
Note that install.packages on MacOS X can
now install either "source" or "mac.binary" pack-
age files, the latter being the default. Similarly,
install.packages on Windows can now install ei-
ther "source" or "win.binary" package files. The
only difference between the platforms is the default
for getOption("pkgType").
Repositories
The package management functions such
as install.packages,update.packages and
packageStatus need to know where to look for pack-
ages. Function install.packages was originally
written to access a CRAN mirror, but other reposito-
ries of packages have been created, notably that of
Bioconductor. Function packageStatus was the first
design to allow multiple repositories.
We encourage people with collections of pack-
ages to set up a repository. Apart from the CRAN
mirrors and Bioconductor there is an Omegahat
repository, and I have a repository of hard-to-
compile Windows packages to supplement those
made automatically. We anticipate university de-
partments setting up repositories of support soft-
ware for their courses. A repository is just a tree of
files based at a URL: details of the format are in Writ-
ing R Extensions. It can include one, some or all of
"source","win.binary" and "mac.binary" types in
separate subtrees.
Specifying repositories
Specifying repositories has become quite compli-
cated: the (sometimes conflicting) aims were to be
intuitive to new users, very flexible and as far as pos-
sible backwards compatible.
1a directory containing subdirectories of installed packages
R News ISSN 1609-3631
Vol. 5/1, May 2005 9
As before, the place(s) to look for files are speci-
fied by the contriburl argument. This is a charac-
ter vector of URLs, and can include file:// URLs
if packages are stored locally, for example on a dis-
tribution CD-ROM. Different URLs in the vector can
be of different types, so one can mix a distribution
CD-ROM with a CRAN mirror.
However, as before, most people will not spec-
ify contriburl directly but rather the repos argu-
ment that replaces the CRAN argument.2The function
contrib.url is called to convert the base URLs in
repos into URLs pointing to the correct subtrees of
the repositories. For example
> contrib.url("http://base.url",
type = "source")
[1] "http://base.url/src/contrib"
> contrib.url("http://base.url",
type = "win.binary")
[1] "http://base.url/bin/windows/contrib/2.1"
> contrib.url("http://base.url",
type = "mac.binary")
[1] "http://base.url/bin/macosx/2.1"
This is of course hidden from the average user.
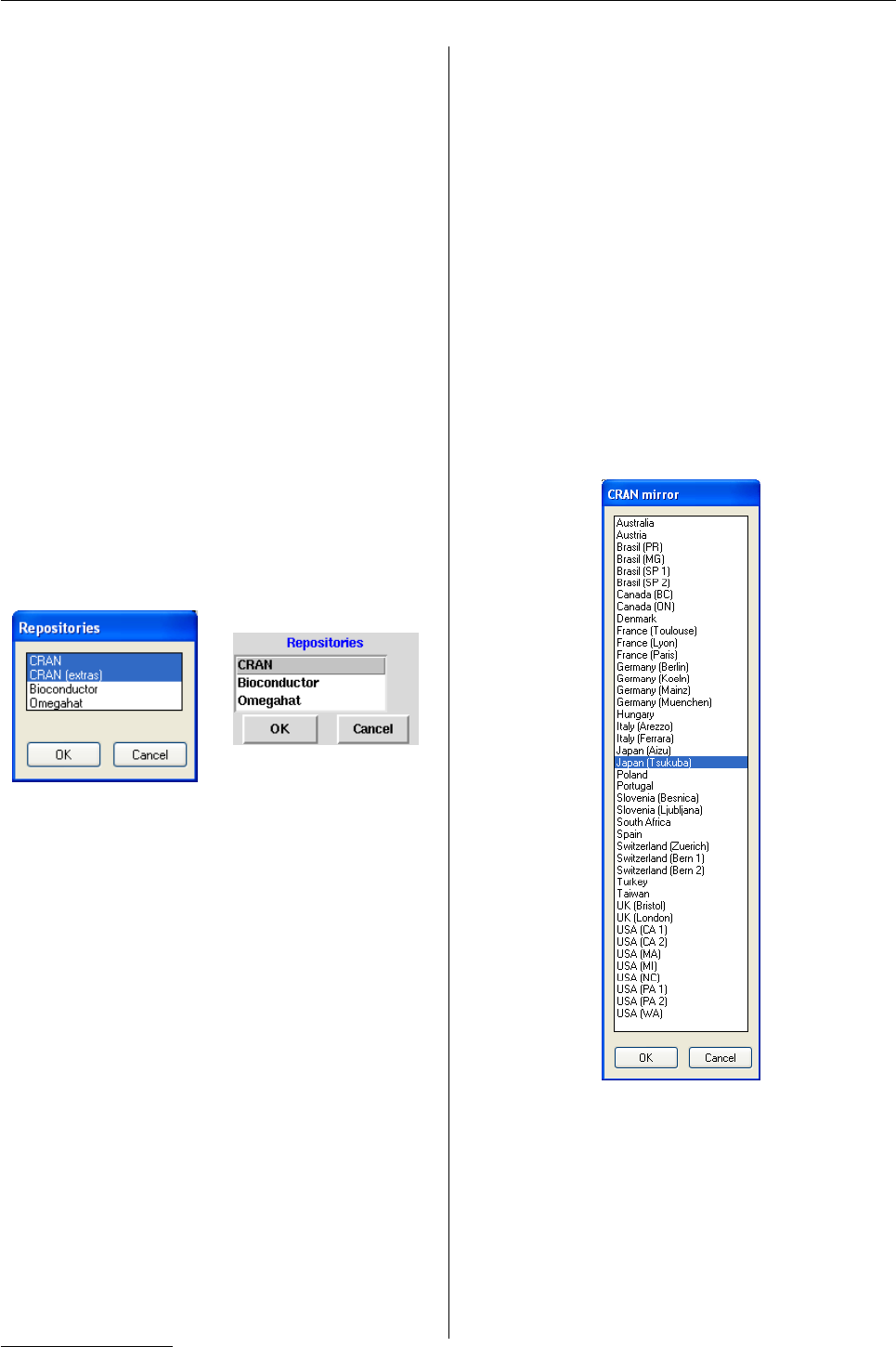
Figure 1: The listboxes from setRepositories() on
Windows (left) and Unix (right).
The repos argument of the package management
functions defaults to getOption("repos"). This has
a platform-specific default, and can be set by calling
setRepositories. On most systems this uses a list-
box widget, as shown in figure 1. Where no GUI is
available, a text list is used such as
> setRepositories()
--- Please select repositories for use
in this session ---
1: + CRAN
2: + CRAN (extras)
3: Bioconductor
4: Omegahat
Enter one or more numbers separated by spaces
1: 1 4
The list of repositories and which are selected by de-
fault can be customized for a user or for a site: see
?setRepositories. We envisaged people distribut-
ing a CD-ROM containing Rand a repository, with
setRepositories() customized to include the local
repository by default.
Suppose a package exists on more than one
repository?
Since multiple repositories are allowed, how is this
resolved? The rule is to select the latest version of
the package, from the first repository on the list that
is offering it. So if a user had
> getOption("repos")
[1] "file://d:/R/local"
[2] "http://cran.us.r-project.org"
packages would be fetched from the local repository
if they were current there, and from a US CRAN mir-
ror if there is an update available there.
Figure 2: Selecting a CRAN mirror on Windows.
No default CRAN
Windows users are already used to selecting a CRAN
mirror. Now ‘factory-fresh’ Rdoes not come with a
default CRAN mirror. On Unix the default is in fact
> getOption("repos")
CRAN
"@CRAN@"
2This still exists but has been deprecated and will be removed in R 2.2.0.
R News ISSN 1609-3631

Vol. 5/1, May 2005 10
Whenever @CRAN@ is encountered in a repository
specification, the function chooseCRANmirror is
called to ask for a mirror (and can also be called di-
rectly and from a menu item on the Windows GUI).
The Windows version is shown in Figure 2: there are
MacOS X and Tcl/Tk versions, as well as a text-based
version using menu.3
Experienced users can avoid being asked for a
mirror by setting options("repos") in their ‘.Rpro-
file’ files: examples might be
options(repos=
c(CRAN="http://cran.xx.r-project.org"))
on Unix-alikes and
options(repos=
c(CRAN="http://cran.xx.r-project.org",
CRANextra=
"http://www.stats.ox.ac.uk/pub/RWin"))
on Windows.
Although not essential, it is helpful to have a
named vector of repositories as in this example: for
example setRepositories looks at the names when
setting its initial selection.
Installing packages
Users of the Windows GUI will be used to selecting
packages to be installed from a scrollable list box.
This now works on all platforms from the command
line if install.packages is called with no pkgs (first)
argument. Packages from all the selected reposito-
ries are listed in alphabetical order, and the packages
from bundles are listed4individually in the form
MASS (VR).
The dependencies argument introduced in
R 2.0.0 can be used to install a package and its de-
pendencies (and their dependencies . . . ), and Rwill
arrange to install the packages in a suitable order.
The dependencies argument can be used to select
only those dependencies that are essential, or to in-
clude those which are suggested (and might only be
needed to run some of the examples).
Installing all packages
System administrators often want to install ‘all’ pack-
ages, or at least as many as can be installed on their
system: a standard Linux system lacks the additional
software needed to install about 20 and a handful can
only be installed on Windows or on Linux.
The new function new.packages() is a helpful
way to find out which packages are not installed of
those offered by the repositories selected. If called as
new.packages(ask = "graphics") a list box is used
to allow the user to select packages to be installed.
Figure 3: Selecting amongst uninstalled packages on
Linux.
Updating packages
The only real change in update.packages is the ask
argument. This defaults to ask = TRUE where as be-
fore the user is asked about each package (but with a
little more information and the option to quit). Other
options are ask = FALSE to install all updates, and
ask = "graphics" to use a listbox (similar to fig-
ure 3) to de-select packages (as by default all the up-
dates are selected).
Looking at repositories
The function CRAN.packages was (as it name im-
plies) designed to look at a single CRAN mirror. It
has been superseded by available.packages which
can look at one or more repositories and by default
looks at those specified by getOption("repos")
for package type (source/binary) specified by
getOption("pkgType"). This returns a matrix
with rather more columns than before, including
"Repository", the dependencies and the contents of
bundles.
Function packageStatus is still available but has
been re-implemented on top of functions such as
available.packages. Its summary method allows
a quick comparison of the installed packages with
those offered by the selected repositories. However,
its print method is concise: see figure 4.
Bibliography
U. Ligges. R help desk: Package management. R
News, 3(3):37–39, December 2003. URL http://
CRAN.R-project.org/doc/Rnews/.8
Brian D. Ripley
University of Oxford, UK
ripley@stats.ox.ac.uk
3menu has been much enhanced for R 2.1.0.
4provided the repository has been set up to supply the information.
R News ISSN 1609-3631

Vol. 5/1, May 2005 11
> (ps <- packageStatus())
--- Please select a CRAN mirror for use in this session ---
Number of installed packages:
ok upgrade unavailable
c:/R/library 34 1 0
c:/R/rw2010/library 12 0 13
Number of available packages (each package/bundle counted only once):
installed
http://www.sourcekeg.co.uk/cran/bin/windows/contrib/2.1 31
http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/2.1 4
not installed
http://www.sourcekeg.co.uk/cran/bin/windows/contrib/2.1 421
http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/2.1 8
> summary(ps)
Installed packages:
-------------------
*** Library c:/R/library
$ok
[1] "abind" "acepack" "akima" "ash" "car"
...
$upgrade
[1] "XML"
$unavailable
NULL
*** Library c:/R/rw2010/library
$ok
[1] "base" "datasets" "graphics" "grDevices" "grid" "methods" "splines" "stats" "stats4"
[10] "tcltk" "tools" "utils"
$upgrade
NULL
$unavailable
[1] "boot" "cluster" "foreign" "KernSmooth" "lattice"
...
Available packages:
-------------------
(each package appears only once)
*** Repository http://www.sourcekeg.co.uk/cran/bin/windows/contrib/2.1
$installed
[1] "abind" "acepack" "akima" "ash" "car"
...
$"not installed"
[1] "accuracy" "adapt" "ade4"
...
[421] "zicounts"
*** Repository http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/2.1
$installed
[1] "GLMMGibbs" "xgobi" "XML" "yags"
$"not installed"
[1] "gsl" "hdf5" "ncdf" "RDCOMClient" "rgdal" "RNetCDF" "survnnet" "udunits"
> upgrade(ps)
XML :
0.97-0 at c:/R/library
0.97-3 at http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/2.1
Update (y/N)? y
Figure 4: packageStatus on a Windows machine. The voluminous summary has been edited to save space.
R News ISSN 1609-3631

Vol. 5/1, May 2005 12
Recent Changes in grid Graphics
by Paul Murrell
Introduction
The grid graphics package provides an alternative
graphics system to the “traditional” S graphics sys-
tem that is provided by the graphics package.
The majority of high-level plotting functions (func-
tions that produce whole plots) that are currently
available in the base packages and in add-on pack-
ages are built on the graphics package, but the lattice
package (Sarkar,2002), which provides high-level
Trellis plots, is built on grid.
The grid graphics package can be useful for cus-
tomising lattice plots, creating complex arrange-
ments of several lattice plots, or for producing
graphical scenes from scratch.
The basic features of grid were described in an R
News article in June 2002 (Murrell,2002). This arti-
cle provides an update on the main features that have
been added to grid since then. This article assumes
a familiarity with the basic grid ideas of units and
viewports.
Changes to grid
The most important organisational change is that
grid is now a “base” package, so it is installed as part
of the standard R installation. In other words, pack-
age writers can assume that grid is available.
The two main areas where the largest changes have
been made to grid are viewports and graphical ob-
jects. The changes to viewports are described in the
next section and summarised in Table 1at the end of
the article. A separate section describes the changes
to graphical objects with a summary in Table 2at the
end of the article.
Changes to viewports
grid provides great flexibility for creating regions on
the page to draw in (viewports). This is good for be-
ing able to locate and size graphical output on the
page in very sophisticated ways (e.g., lattice plots),
but it is bad because it creates a complex environ-
ment when it comes to adding further output (e.g.,
annotating a lattice plot).
The first change to viewports is that they are now
much more persistent; it is possible to have any num-
ber of viewports defined at the same time.
There are also several new functions that provide a
consistent and straightforward mechanism for nav-
igating to a particular viewport when several view-
ports are currently defined.
A viewport is just a rectangular region that provides
a geometric and graphical context for drawing. The
viewport provides several coordinate systems for lo-
cating and sizing output, and it can have graphical
parameters associated with it to affect the appear-
ance of any output produced within the viewport.
The following code describes a viewport that occu-
pies the right half of the graphics device and within
which, unless otherwise specified, all output will be
drawn using thick green lines. A new feature is that a
viewport can be given a name. In this case the view-
port is called "rightvp". The name will be important
later when we want to navigate back to this view-
port.
> vp1 <- viewport(x=0.5, width=0.5,
just="left",
gp=gpar(col="green", lwd=3),
name="rightvp")
The above code only creates a description of a view-
port; a corresponding region is created on a graph-
ics device by pushing the viewport on the device, as
shown below.1
> pushViewport(vp1)
Now, all drawing occurs within the context defined
by this viewport until we change to another view-
port. For example, the following code draws some
basic shapes, all of which appear in the right half of
the device, with thick green lines (see the right half
of Figure 1). Another new feature is that a name
can be associated with a piece of graphical output.
In this case, the rectangle is called "rect1", the line
is called "line1", and the set of circles are called
"circles1". These names will be used later in the
section on “Changes to graphical objects”.
> grid.rect(name="rect1")
> r <- seq(0, 2*pi, length=5)[-5]
> x <- 0.5 + 0.4*cos(r + pi/4)
> y <- 0.5 + 0.4*sin(r + pi/4)
> grid.circle(x, y, r=0.05,
1The function used to be called push.viewport().
R News ISSN 1609-3631
Vol. 5/1, May 2005 13
name="circles1")
> grid.lines(x[c(2, 1, 3, 4)],
y[c(2, 1, 3, 4)],
name="line1")
There are two ways to change the viewport. You can
pop the viewport, in which case the region is perma-
nently removed from the device, or you can navigate
up from the viewport and leave the region on the de-
vice. The following code demonstrates the second
option, using the upViewport() function to revert to
using the entire graphics device for output, but leav-
ing a viewport called "rightvp" still defined on the
device.
> upViewport()
Next, a second viewport is defined to occupy the left
half of the device, this viewport is pushed, and some
output is drawn within it. This viewport is called
"leftvp" and the graphical output is associated with
the names "rect2","lines2", and "circles2". The
output from the code examples so far is shown in Fig-
ure 1.
> vp2 <- viewport(x=0, width=0.5,
just="left",
gp=gpar(col="blue", lwd=3),
name="leftvp")
> pushViewport(vp2)
> grid.rect(name="rect2")
> grid.circle(x, y, r=0.05,
name="circles2")
> grid.lines(x[c(3, 2, 4, 1)],
y[c(3, 2, 4, 1)],
name="line2")
●●
● ●
●●
● ●
Figure 1: Some simple shapes drawn in some simple
viewports.
There are now three viewports defined on the device;
the function current.vpTree() shows all viewports
that are currently defined.2
> current.vpTree()
viewport[ROOT]->(
viewport[leftvp],
viewport[rightvp])
There is always a ROOT viewport representing the en-
tire device and the two viewports we have created
are both direct children of the ROOT viewport. We
now have a tree structure of viewports that we can
navigate around. As before, we can navigate up from
the viewport we just pushed and, in addition, we can
navigate down to the previous viewport. The func-
tion downViewport() performs the navigation to a
viewport lower down the viewport tree. It requires
the name of the viewport to navigate to. In this case,
we navigate down to the viewport called "rightvp".
> upViewport()
> downViewport("rightvp")
It is now possible to add further output within the
context of the viewport called "rightvp". The fol-
lowing code draws some more circles, this time ex-
plicitly grey (but still with thick lines; see Figure 2).
The name "circles3" is associated with these extra
circles.
> x2 <- 0.5 + 0.4*cos(c(r, r+pi/8, r-pi/8))
> y2 <- 0.5 + 0.4*sin(c(r, r+pi/8, r-pi/8))
> grid.circle(x2, y2, r=0.05,
gp=gpar(col="grey"),
name="circles3")
> upViewport()
●●
● ●
●●
● ●
●
●
●
●
●
●
●
●
●
●
●
●
Figure 2: Navigating between viewports and anno-
tating.
What this simple demonstration shows is that it is
possible to define a number of viewports during
drawing and leave them in place for others to add
2The output of current.vpTree() has been reformatted to make the structure more obvious; the natural output is all just on a single
line.
R News ISSN 1609-3631
Vol. 5/1, May 2005 14
further output. A more sophisticated example is now
presented using a lattice plot.3
The following code creates a simple lattice plot. The
"trellis" object created by the histogram() func-
tion is stored in a variable, hist1, so that we can
use it again later; printing hist1 produces the plot
shown in Figure 3.4
> hist1 <- histogram(
par.settings=list(fontsize=list(text=8)),
rnorm(500), type = "density",
panel=
function(x, ...) {
panel.histogram(x, ...)
panel.densityplot(x,
col="brown",
plot.points=FALSE)
})
> trellis.par.set(canonical.theme("pdf"))
> print(hist1)
The lattice package uses grid to produce output and
it defines lots of viewports to draw in. In this case,
there are six viewports created, as shown below.
> current.vpTree()
viewport[ROOT]->(
viewport[plot1.toplevel.vp]->(
viewport[plot1.],
viewport[plot1.panel.1.1.off.vp],
viewport[plot1.panel.1.1.vp],
viewport[plot1.strip.1.1.off.vp],
viewport[plot1.xlab.vp],
viewport[plot1.ylab.vp]))
rnorm(500)
Density
−2 0 2
0.0
0.1
0.2
0.3
0.4
Figure 3: A simple lattice plot.
What we are going to do is add output to the lattice
plot by navigating to a couple of the viewports that
lattice set up and draw a border around the viewport
to show where it is. We will use the frame() function
defined below to draw a thick grey rectangle around
the viewport, then a filled grey rectangle at the top-
left corner, and the name of the viewport in white
within that.
> frame <- function(name) {
grid.rect(gp=gpar(lwd=3, col="grey"))
grid.rect(x=0, y=1,
height=unit(1, "lines"),
width=1.2*stringWidth(name),
just=c("left", "top"),
gp=gpar(col=NA, fill="grey"))
grid.text(name,
x=unit(2, "mm"),
y=unit(1, "npc") -
unit(0.5, "lines"),
just="left",
gp=gpar(col="white"))
}
The viewports used to draw the lattice plot con-
sist of a single main viewport called (in this case)
"plot1.toplevel.vp", and a number of other view-
ports within that for drawing the various compo-
nents of the plot. This means that the viewport
tree has three levels (including the top-most ROOT
viewport). With a multipanel lattice plot, there can
be many more viewports created, but the naming
scheme for the viewports uses a simple pattern and
3This low-level grid mechanism is general and available for any graphics output produced using grid including lattice output. An
additional higher-level interface has also been provided specifically for lattice plots (e.g., the trellis.focus() function).
4The call to trellis.par.set() sets lattice graphical parameters so that the colours used in drawing the histogram are those used on
a PDF device. This explicit control of the lattice “theme” makes it easier to exactly reproduce the histogram output in a later example.
R News ISSN 1609-3631
Vol. 5/1, May 2005 15
there are lattice functions provided to generate the
appropriate names (e.g., trellis.vpname()).
When there are more than two levels, there are two
ways to specify a particular low-level viewport. By
default, downViewport() will search within the chil-
dren of a viewport, so a single viewport name will
work as before. For example, the following code an-
notates the lattice plot by navigating to the viewport
"plot1.panel.1.1.off.vp" and drawing a frame to
show where it is. This example also demonstrates the
fact that downViewport() returns a value indicating
how many viewports were descended. This “‘depth”
can be passed to upViewport() to ensure that the cor-
rect number of viewports are ascended after the an-
notation.
> depth <-
downViewport("plot1.panel.1.1.off.vp")
> frame("plot1.panel.1.1.off.vp")
> upViewport(depth)
Just using the final destination viewport name can be
ambiguous if more than one viewport in the view-
port tree has the same name, so it is also possible
to specify a viewport path. A viewport path is a list
of viewport names that must be matched in order
from parent to child. For example, this next code
uses an explicit viewport path to frame the view-
port "plot1.xlab.vp" that is a child of the viewport
"plot1.toplevel.vp".
> depth <-
downViewport(vpPath("plot1.toplevel.vp",
"plot1.xlab.vp"))
> frame("plot1.xlab.vp")
> upViewport(depth)
It is also possible to use a viewport name or view-
port path in the vp argument to drawing functions, in
which case the output will occur in the named view-
port. In this case, the viewport path is strict, which
means that the full path must be matched start-
ing from the context in which the grob was drawn.
The following code adds a dashed white line to the
borders of the frames. This example also demon-
strates that viewport paths can be specified as ex-
plicit strings, with a "::" path separator.
The final output after all of these annotations is
shown in Figure 4.
> grid.rect(
gp=gpar(col="white", lty="dashed"),
vp="plot1.toplevel.vp::plot1.panel.1.1.off.vp")
> grid.rect(
gp=gpar(col="white", lty="dashed"),
vp="plot1.toplevel.vp::plot1.xlab.vp")
rnorm(500)
Density
−2 0 2
0.0
0.1
0.2
0.3
plot1.panel.1.1.off.vp
plot1.xlab.vp
Figure 4: Annotating the simple lattice plot.
Changes to graphical objects
All grid drawing functions create graphical objects
representing the graphical output. This is good be-
cause it makes it possible to manipulate these objects
to modify the current scene, but there needs to be a
coherent mechanism for working with these objects.
There are several new functions, and some existing
functions have been made more flexible, to provide
a consistent and straightforward mechanism for ac-
cessing, modifying, and removing graphical objects.
All grid functions that produce graphical output,
also create a graphical object, or grob, that represents
the output, and there are functions to access, modify,
and remove these graphical objects. This can be used
as an alternative to modifying and rerunning the R
code to modify a plot. In some situations, it will be
the only way to modify a low-level feature of a plot.
Consider the output from the simple viewport exam-
ple (shown in Figure 2). There are seven pieces of
output, each with a corresponding grob: two rect-
angles, "rect1" and "rect2"; two lines, "line1"
and "line2"; and three sets of circles, "circles1",
"circles2", and "circles3". A particular piece
of output can be modified by modifying the corre-
sponding grob; a particular grob is identified by its
name.
The names of all (top-level) grobs in the current
scene can be obtained using the getNames() func-
tion.5
5Only introduced in R version 2.1.0; a less efficient equivalent for version 2.0.0 is to use grid.get().
R News ISSN 1609-3631
Vol. 5/1, May 2005 16
> getNames()
[1] "rect1" "circles1" "line1" "rect2"
[5] "circles2" "line2" "circles3"
The following code uses the grid.get() function to
obtain a copy of all of the grobs. The first argument
specifies the names(s) of the grob(s) that should be
selected; the grep argument indicates whether the
name should be interpreted as a regular expression;
the global argument indicates whether to select just
the first match or all possible matches.
> grid.get(".*", grep=TRUE, global=TRUE)
(rect[rect1], circle[circles1], lines[line1],
rect[rect2], circle[circles2], lines[line2],
circle[circles3])
The value returned is a gList; a list of one or more
grobs. This is a useful way to obtain copies of one or
two objects representing some portion of a scene, but
it does not return any information about the context
in which the grobs were drawn, so, for example, just
drawing the gList is unlikely to reproduce the orig-
inal output (for that, see the grid.grab() function
below).
The following code makes use of the grid.edit()
function to change the colour of the grey circles to
black (see Figure 5). In general, most arguments pro-
vided in the creation of the output are available for
editing (see the documentation for individual func-
tions). It should be possible to modify the gp argu-
ment for all grobs.
> grid.edit("circles3", gp=gpar(col="black"))
●●
● ●
●●
● ●
●
●
●
●
●
●
●
●
●
●
●
●
Figure 5: Editing grid output.
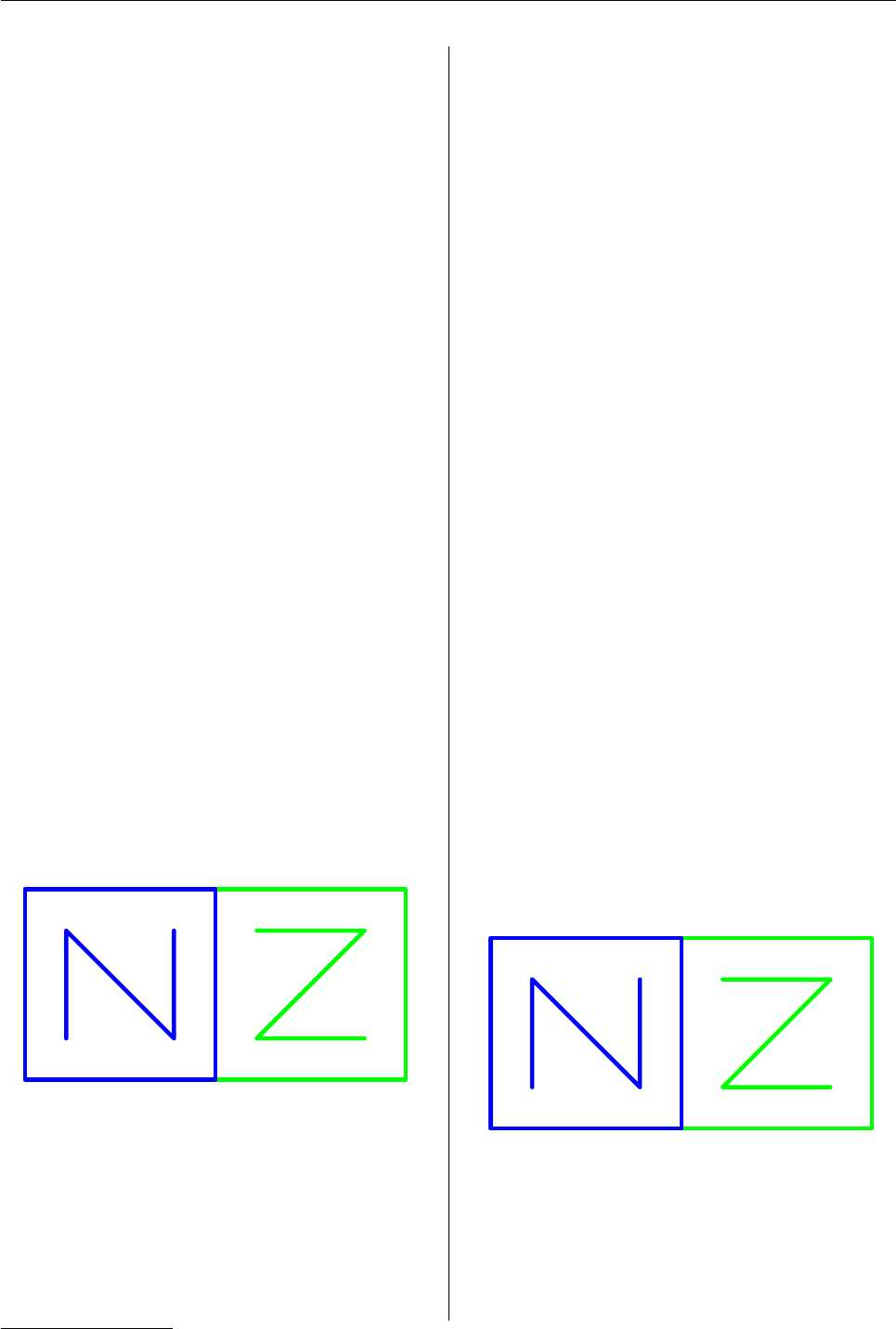
The following code uses grid.remove() to delete all
of the grobs whose names end with a "2" – all of the
blue output (see Figure 6).
> grid.remove(".+2$", grep=TRUE, global=TRUE)
●●
● ●
●
●
●
●
●
●
●
●
●
●
●
●
Figure 6: Deleting grid output.
These manipulations are possible with any grid out-
put. For example, in addition to lots of viewports, a
lattice plot creates a large number of grobs and all of
these can be accessed, modified, and deleted using
the grid functions (an example is given at the end of
the article).
Hierarchical graphical objects
Basic grobs simply consist of a description of some-
thing to draw. As shown above, it is possible to get
a copy of a grob, modify the description in a grob,
and/or remove a grob.
It is also possible to create and work with more com-
plicated graphical objects that have a tree structure,
where one graphical object is the parent of one or
more others. Such a graphical object is called a
gTree. The functions described so far all work with
gTrees as well, but there are some additional func-
tions just for creating and working with gTrees.
In simple usage, a gTree just groups several grobs
together. The following code uses the grid.grab()
function to create a gTree from the output in Figure
6.
> nzgrab <- grid.grab(name="nz")
If we draw this gTree on a new page, the output is
exactly the same as Figure 6, but there is now only
one graphical object in the scene: the gTree called
"nz".
> grid.newpage()
> grid.draw(nzgrab)
> getNames()
[1] "nz"
R News ISSN 1609-3631

Vol. 5/1, May 2005 17
This gTree has four children; the four original grobs
that were “grabbed”. The function childNames()
prints out the names of all of the child grobs of a
gTree.
> childNames(grid.get("nz"))
[1] "rect1" "circles1" "line1"
[4] "circles3"
AgTree contains viewports used to draw its child
grobs as well as the grobs themselves, so the origi-
nal viewports are also available.
> current.vpTree()
viewport[ROOT]->(
viewport[leftvp],
viewport[rightvp])
The grid.grab() function works with any output
produced using grid, including lattice plots, and
there is a similar function grid.grabExpr(), which
will capture the output from an expression.6The fol-
lowing code creates a gTree by capturing an expres-
sion to draw the histogram in Figure 3.
> histgrab <- grid.grabExpr(
{ trellis.par.set(canonical.theme("pdf"));
print(hist1) },
name="hist", vp="leftvp")
The grid.add() function can be used to add further
child grobs to a gTree. The following code adds the
histogram gTree as a child of the gTree called "nz".
An important detail is that the histogram gTree was
created with vp="leftvp"; this means that the his-
togram gets drawn in the viewport called "leftvp"
(i.e., in the left half of the scene). The output now
looks like Figure 7.
> grid.add("nz", histgrab)
There is still only one graphical object in the scene,
the gTree called "nz", but this gTree now has five
children: two lots of circles, one line, one rectangle,
and a lattice histogram (as a gTree).
> childNames(grid.get("nz"))
[1] "rect1" "circles1" "line1"
[4] "circles3" "hist"
The functions for accessing, modifying, and re-
moving graphical objects all work with hierarchical
graphical objects like this. For example, it is pos-
sible to remove a specific child from a gTree using
grid.remove().
When dealing with a scene that includes gTrees, a
simple grob name can be ambiguous because, by de-
fault, grid.get() and the like will search within the
children of a gTree to find a match.
Just as you can provide a viewport path in
downViewport() to unambiguously specify a partic-
ular viewport, it is possible to provide a grob path in
grid.get(),grid.edit(), or grid.remove() to un-
ambiguously specify a particular grob.
Agrob path is a list of grob names that must be
matched in order from parent to child. The following
code demonstrates the use of the gPath() function to
create a grob path. In this example, we are going to
modify one of the grobs that were created by lattice
when drawing the histogram. In particular, we are
going to modify the grob called "plot1.xlab" which
represents the x-axis label of the histogram.
In order to specify the x-axis label unambiguously,
we construct a grob path that identifies the grob
"plot1.xlab", which is a child of the grob called
"hist", which itself is a child of the grob called "nz".
That path is used to modify the grob which repre-
sents the xaxis label on the histogram. The xaxis la-
bel is moved to the far right end of its viewport and
is drawn using a bold-italic font face (see Figure 8).
> grid.edit(gPath("nz", "hist", "plot1.xlab"),
x=unit(1, "npc"), just="right",
gp=gpar(fontface="bold.italic"))
Summary
When a scene is created using the grid graphics sys-
tem, a tree of viewport objects is created to represent
the drawing regions used in the construction of the
scene and a list of graphical objects is created to rep-
resent the actual graphical output.
Functions are provided to view the viewport tree and
navigate within it in order to add further output to a
scene.
Other functions are provided to view the graphical
objects in a scene, to modify those objects, and/or
remove graphical objects from the scene. If graphi-
cal objects are modified or removed, the scene is re-
drawn to reflect the changes. Graphical objects can
also be grouped together in a tree structure and dealt
6The function grid.grabExpr() is only available in R version 2.1.0; the grid.grab() function could be used instead by explicitly
opening a new device, drawing the histogram, and then capturing it.
R News ISSN 1609-3631
Vol. 5/1, May 2005 18
rnorm(500)
Density
−2 0 2
0.0
0.1
0.2
0.3
0.4
Figure 7: Grabbing grid output.
rnorm(500)
Density
−2 0 2
0.0
0.1
0.2
0.3
0.4
Figure 8: Editing a gTree.
R News ISSN 1609-3631

Vol. 5/1, May 2005 19
Table 1: Summary of the new and changed functions in R 2.0.0 relating to viewports.
Function Description
pushViewport() Create a region for drawing on the cur-
rent graphics device.
popViewport() Remove the current drawing region
(does not delete any output).
upViewport() Navigate to parent drawing region (but
do not remove current region).
downViewport() Navigate down to named drawing re-
gion.
current.viewport() Return the current viewport.
current.vpTree() Return the current tree of viewports that
have been created on the current device.
vpPath() Create a viewport path; a concatenated
list of viewport names.
viewport(..., name=) A viewport can have a name associated
with it.
Table 2: Summary of the new and changed functions since R 2.0.0 relating to graphical objects (some functions
only available since R 2.1.0).
Function Description
grid.get() Return a single grob or a gList of grobs.
grid.edit() Modify one or more grobs and (option-
ally) redraw.
grid.remove() Remove one or more grobs and (option-
ally) redraw.
grid.add() Add a grob to a gTree.
grid.grab() Create a gTree from the current scene.
grid.grabExpr() Create a gTree from an R expression
(only available since R 2.1.0).
gPath() Create a grob path; a concatenated list of
grob names.
getNames() List the names of the top-level grobs in
the current scene (only available since R
2.1.0).
childNames() List the names of the children of a gTree.
grid.rect(..., name=) Grid drawing functions can associate a
name with their output.
R News ISSN 1609-3631

Vol. 5/1, May 2005 20
with as a single unit.
Bibliography
P. Murrell. The grid graphics package. R News, 2(2):
14–19, June 2002. URL http://CRAN.R-project.
org/doc/Rnews/.12
D. Sarkar. Lattice. R News, 2(2):19–23, June 2002. URL
http://CRAN.R-project.org/doc/Rnews/.12
Paul Murrell
University of Auckland, New Zealand
pmurrell@auckland.ac.nz
hoa: An R package bundle for higher
order likelihood inference
by Alessandra R. Brazzale
Introduction
The likelihood function represents the basic ingre-
dient of many commonly used statistical methods
for estimation, testing and the calculation of confi-
dence intervals. In practice, much application of like-
lihood inference relies on first order asymptotic re-
sults such as the central limit theorem. The approx-
imations can, however, be rather poor if the sample
size is small or, generally, when the average infor-
mation available per parameter is limited. Thanks
to the great progress made over the past twenty-
five years or so in the theory of likelihood inference,
very accurate approximations to the distribution of
statistics such as the likelihood ratio have been devel-
oped. These not only provide modifications to well-
established approaches, which result in more accu-
rate inferences, but also give insight on when to rely
upon first order methods. We refer to these develop-
ments as higher order asymptotics.
One intriguing feature of the theory of higher or-
der likelihood asymptotics is that relatively simple
and familiar quantities play an essential role. The
higher order approximations discussed in this paper
are for the significance function, which we will use
to set confidence limits or to calculate the p-value
associated with a particular hypothesis of interest.
We start with a concise overview of the approxi-
mations used in the remainder of the paper. Our
first example is an elementary one-parameter model
where one can perform the calculations easily, cho-
sen to illustrate the potential accuracy of the proce-
dures. Two more elaborate examples, an application
of binary logistic regression and a nonlinear growth
curve model, follow. All examples are carried out us-
ing the R code of the hoa package bundle.
Basic ideas
Assume we observed nrealizations y1,...,ynof in-
dependently distributed random variables Y1,...,Yn
whose density function f(yi;θ)depends on an un-
known parameter θ. Let `(θ) = ∑n
i=1log f(yi;θ)
denote the corresponding log likelihood and ˆ
θ=
argmaxθ`(θ)the maximum likelihood estimator. In
almost all applications the parameter θis not scalar
but a vector of length d. Furthermore, we may
re-express it as θ= (ψ,λ), where ψis the d0-
dimensional parameter of interest, about which we
wish to make inference, and λis a so-called nuisance
parameter, which is only included to make the model
more realistic.
Confidence intervals and p-values can be com-
puted using the significance function p(ψ;ˆ
ψ) =
Pr(ˆ
Ψ≤ˆ
ψ;ψ)which records the probability left of
the observed “data point” ˆ
ψfor varying values of
the unknown parameter ψ(Fraser,1991). Exact elim-
ination of λ, however, is possible only in few special
cases (Severini,2000, Sections 8.2 and 8.3). A com-
monly used approach is to base inference about ψon
the profile log likelihood `p(ψ) = `(ψ,ˆ
λψ), which we
obtain from the log likelihood function by replacing
the nuisance parameter with its constrained estimate
ˆ
λψobtained by maximising `(θ) = `(ψ,λ)with re-
spect to λfor fixed ψ. Let jp(ψ) = −∂2`p(ψ)/∂ψ∂ψ>
denote the observed information from the profile log
likelihood. Likelihood inference for scalar ψis typi-
cally based on the
• Wald statistic, w(ψ) = jp(ˆ
ψ)1/2(ˆ
ψ−ψ);
• likelihood root,
r(ψ) = sign(ˆ
ψ−ψ)2{`p(ˆ
ψ)−`p(ψ)}1/2;
or
• score statistic, s(ψ) = jp(ˆ
ψ)−1/2d`p(ψ)/dψ.
Under suitable regularity conditions on f(y;θ), all of
these have asymptotic standard normal distribution
R News ISSN 1609-3631
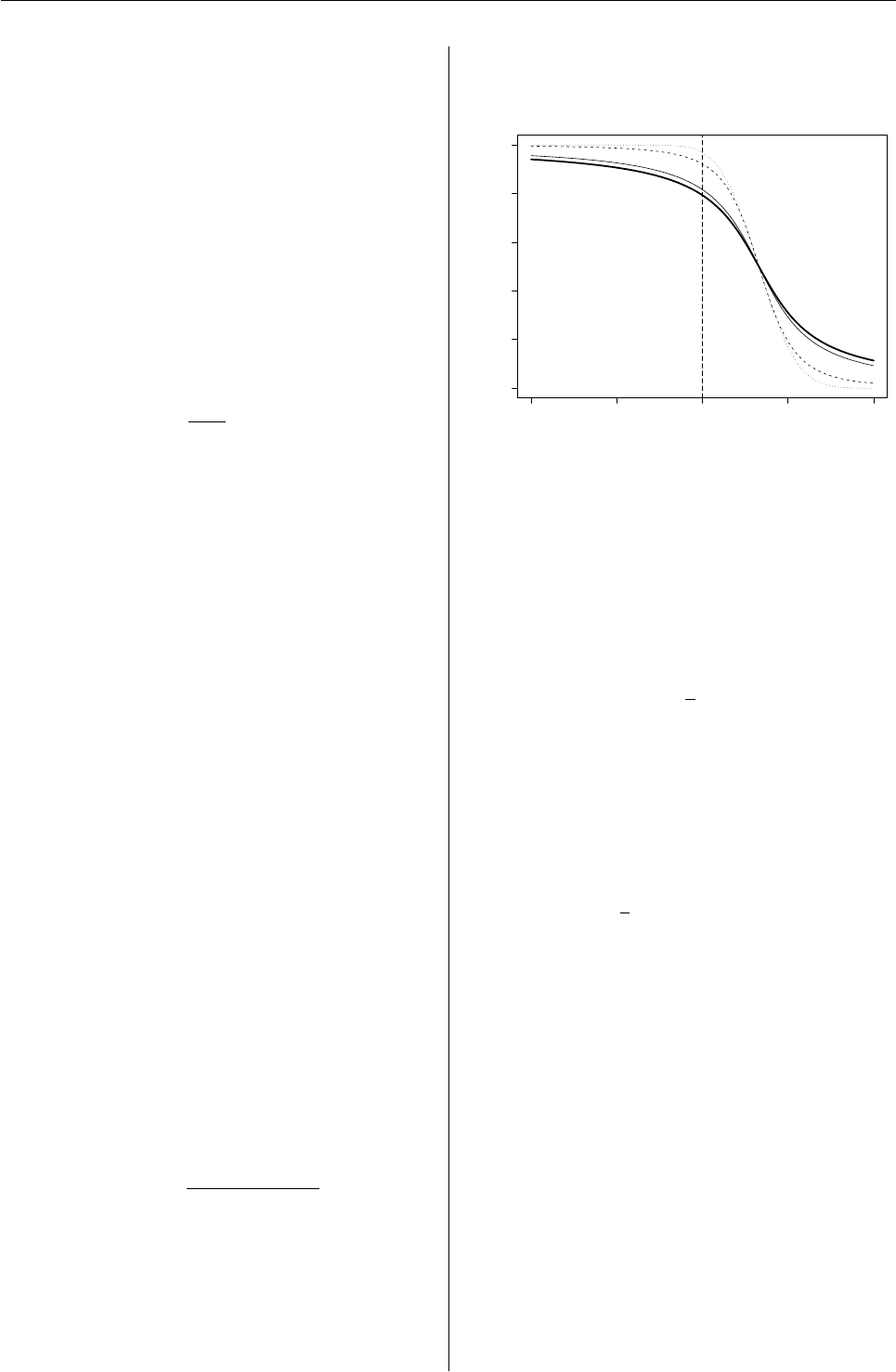
Vol. 5/1, May 2005 21
up to the first order. Using any of the above statis-
tics we can approximate the significance function by
Φ{w(ψ)},Φ{r(ψ)}or Φ{s(ψ)}. When d0>1, we
may use the quadratic forms of the Wald, likelihood
root and score statistics whose finite sample distribu-
tion is χ2
d0with d0degrees of freedom up to the sec-
ond order. We refer the reader to Chapters 3 and 4 of
Severini (2000) for a review of first order likelihood
inference.
Although it is common to treat `p(ψ)as if it were
an ordinary log likelihood, first order approxima-
tions can give poor results, particularly if the dimen-
sion of λis high and the sample size small. An im-
portant variant of the likelihood root is the modified
likelihood root
r∗(ψ) = r(ψ) + 1
r(ψ)log {q(ψ)/r(ψ)}, (1)
where q(ψ)is a suitable correction term. Expres-
sion (1) is a higher order pivot whose finite sample
distribution is standard normal up to the third or-
der. As it was the case for its first order counter-
part r, the significance function is approximated by
Φ{r∗(ψ)}, and there is a version of r∗for multidi-
mensional ψ(Skovgaard,2001, Section 3.1). More
details about the computation of the q(ψ)correction
term are given in the Appendix.
It is sometimes useful to decompose the modified
likelihood root as
r∗(ψ) = r(ψ) + rinf(ψ) + rnp(ψ),
where rinf is the information adjustment and rnp is the
nuisance parameter adjustment. The first term accounts
for non normality of r, while the second compensates
rfor the presence of the nuisance parameter λ.Pierce
and Peters (1992, Section 3) discuss the behaviour of
these two terms in the multiparameter exponential
family context. They find that while rnp is often ap-
preciable, the information adjustment rinf has typi-
cally a minor effect, provided the ψ-specific informa-
tion jp(ˆ
ψ)is not too small relative to the dimension
of λ.
A simple example
Suppose that a sample y1,...,ynis available from
the Cauchy density
f(y;θ) = 1
π{1+ (y−θ)2}. (2)
The maximum likelihood estimate ˆ
θof the unknown
location parameter θis the value which maximises
the log likelihood function
`(θ;y) = −
n
∑
i=1
log{1+ (yi−θ)2}.
For n=1, we obtain the exact distribution of ˆ
θ=y
from (2) as F(ˆ
θ;θ) = F(y;θ) = π−1arctan(y−θ).
−4 −2 0 2 4
0.0
0.2
0.4
0.6
0.8
1.0
θ
significance function
Figure 1: Significance functions for the location pa-
rameter of a Cauchy distribution when y=1.32:
exact (bold), Wald pivot (dotted), r(dashed) and r∗
(solid). The vertical dashed line corresponds to the
null hypothesis θ=0.
Assume that y=1.32 was observed. In Figure 1
we compare the exact significance function p(θ;y) =
Pr(Y≤y;θ)(bold line) to the two first order approx-
imations obtained from the Wald statistic
w(θ) = √2(y−θ),
(dotted line), and from the likelihood root
r(θ) = sign(ˆ
θ−θ)h2 log{1+ (y−θ)2}i1/2,
(dashed line). We also show the third order approx-
imation Φ{r∗(θ)}(solid line). Since this is a location
model and there is no nuisance parameter, the statis-
tic q(θ)in (1) is the score statistic
s(θ) = √2(y−θ)/{1+ (y−θ)2}
(see formula (6) in the Appendix). The vertical
dashed line corresponds to the null hypothesis that
θ=0. The exact p-value is 0.413, while the first
and third order approximations yield 0.0619 (Wald),
0.155 (r) and 0.367 (r∗). The r∗statistic is strikingly
accurate, while the first order approximations are
very poor. This is surprising if we consider that the
score function is not monotonic in yand that only
one observation is available. Figure 2summarises
the R code used to generate Figure 1and obtain the
above p-values.
Suppose now that we observed a sample of size
n=15 from the Student t distribution with 3 de-
grees of freedom. It is no longer possible to derive
the exact distribution of the maximum likelihood es-
timator ˆ
θ, but we may use the code provided in the
marg package of the hoa package bundle to compute
the p-values for testing the significance of the loca-
tion parameter.
R News ISSN 1609-3631

Vol. 5/1, May 2005 22
## likelihood pivots
> wald.stat <- function(theta, y) {
+ sqrt(2) * (y - theta) }
> lik.root <- function(theta, y) {
+ sign(y - theta) * sqrt( 2 * log(1 + (y - theta)^2) ) }
> score.stat <- function(theta, y) {
+ ( sqrt(2) * (y - theta) )/( 1 + (y - theta)^2 ) }
> rstar <- function(theta, y) {
+ lik.root(theta, y) + 1/lik.root(theta, y) * log( score.stat(theta, y)/lik.root(theta, y) ) }
## significance functions : Figure 1
> theta.seq <- seq(-4, 4, length = 100)
> par( las = 1, mai = c(0.9, 0.9, 0.2, 0.2) )
> plot( theta.seq, pcauchy( q = 1.32 - theta.seq ), type = "l", lwd = 2, ylim = c(0,1),
+ xlab = expression(theta), ylab = "significance function", cex.lab = 1.5, cex.axis = 1.5 )
> lines( theta.seq, pnorm( wald.stat(theta.seq, 1.32) ), lty = "dotted" )
> lines( theta.seq, pnorm( lik.root(theta.seq, 1.32) ), lty = "dashed" )
> lines( theta.seq, pnorm( rstar(theta.seq, 1.32) ), lty = "solid" )
> abline( v = 0, lty = "longdash" )
## p-values
> 2 * ( min( tp <- pt(1.32, df = 1), 1 - tp ) )
> 2 * ( min( tp <- pnorm( wald.stat(0, 1.32) ), 1 - tp ) )
> 2 * ( min( tp <- pnorm( lik.root(0, 1.32) ), 1 - tp ) )
> 2 * ( min( tp <- pnorm( rstar(0, 1.32) ), 1 - tp ) )
Figure 2: R code for drawing Figure 1and calculating the p-values for testing the significance of the location
parameter θof a Cauchy distribution when y=1.32.
## simulated data
> library(marg)
> set.seed(321)
> y <- rt(n = 15, df = 3)
> y.rsm <- rsm(y ~ 1, family = student(3))
> y.cond <- cond(y.rsm, offset = 1)
> summary(y.cond, test = 0)
The previous set of instructions yields the p-values
0.282 (Wald), 0.306 (r) and 0.354 (r∗). The difference
between first order and higher order approximations
is slightly smaller than it was the case before. For this
particular model a sample size of n=15 still does
not provide enough information on the scalar param-
eter θto wipe out completely the effect of higher or-
der corrections.
Higher order asymptotics in R
hoa is an R package bundle which implements higher
order inference for three widely used model classes:
logistic regression, linear non normal models and
nonlinear regression with possibly non homoge-
neous variance. The corresponding code is organ-
ised in three packages, namely cond,marg and nlreg.
We already saw a (very elementary) application of
the marg code. The two examples which follow will
give a glimpse of the use of the routines in cond
and nlreg. Attention is restricted to the calculation
of p-values and confidence intervals, although sev-
eral routines for accurate point estimation and model
checking are also available. The hoa bundle includes
a fourth package, called sampling, which we will not
discuss here. It implements a Metropolis-Hastings
sampler which can be used to simulate from the
conditional distribution of the higher order statistics
considered in marg.
The hoa package bundle will soon be available on
CRAN. More examples of applications, and gener-
ally of the use of likelihood asymptotics, are given in
Brazzale et al. (to appear).
Example 1: Binary data
Collet (1998) gives a set of binary data on the pres-
ence of a sore throat in a sample of 35 patients un-
dergoing surgery during which either of two devices
was used to secure the airway. In addition to the
variable of interest, device type (1=tracheal tube or
0=laryngeal mask), there are four further explana-
tory variables: the age of the patient in years, an in-
dicator variable for sex (1=male, 0=female), an indi-
cator variable for lubricant use (1=yes, 0=no) and
the duration of the surgery in minutes. The obser-
vations form the data frame airway which is part of
the hoa bundle.
A natural starting point for the analysis is a logis-
tic regression model with success probability of the
R News ISSN 1609-3631
Vol. 5/1, May 2005 23
form
Pr(Y=1; β) = exp(x>β)
1+exp(x>β),
where xrepresents the explanatory variables associ-
ated with the binary response Y(1=sore throat and
0=no sore throat). The following set of instructions
fits this model to the data with all five explanatory
variables included.
## binomial model fit
> airway.glm <- glm( formula(airway),
+ family = binomial, data = airway )
> summary( airway.glm )
[omitted]
Coefficients:
Estimate Std. Error z value Pr(>|z
(Intercept) -2.75035 2.08914 -1.316 0.18
age 0.02245 0.03763 0.597 0.55
sex1 0.32076 1.01901 0.315 0.75
lubricant1 0.08448 0.97365 0.087 0.93
duration 0.07183 0.02956 2.430 0.01
type1 -1.62968 0.94737 -1.720 0.08
[omitted]
The coefficient of device type is only marginally sig-
nificant.
As in the previous example we may wonder
whether the sample size is large enough to allow us
to rely upon first order inference. For the airway data
we have n=35 and p=5, so we might expect
higher order corrections to the usual approximations
to have little effect. We can check this using the rou-
tines in the cond package.
## higher order inference
> library(cond)
> airway.cond <- cond( airway.glm,
+ offset = type1 )
> summary( airway.cond )
# produces 95% confidence intervals
As our model is a canonical exponential family,
the correction term q(ψ)in (1) involves the Wald
statistic w(ψ)plus parts of the observed informa-
tion matrix (see formula (5) in the Appendix). The
95% confidence intervals obtained from the Wald
pivot and from the likelihood root are respectively
(−3.486, 0.227)and (−3.682, 0.154). The third or-
der statistic r∗yields a 95% confidence interval of
(−3.130, 0.256). First and third order results are
rather different, especially with respect to the lower
bound.
−5 −4 −3 −2 −1 0 1 2
−3
−2
−1
0
1
2
3
coefficient of type
pivot
Figure 3: airway data analysis: profile plots of the
pivots w(ψ)(dashed line), r(ψ)(solid line) and r∗(ψ)
(bold line), where ψis the coefficient of the covariate
device type.
Figure 3plots the profiles of the first and third order
pivots w(ψ)(dashed line), r(ψ)(solid line) and r∗(ψ)
(bold line). The correction term q(ψ)is particularly
significant for values of ψbelonging to the lower half
of the confidence interval. The nuisance parameter
correction is rnp =0.51, while rinf =0.059 is about
ten times smaller.
Example 2: Nonlinear regression
A simple illustration of nonlinear regression is Exam-
ple 7.7 of Davison and Hinkley (1997), which refers
to the calcium data of package boot. This data set
records the calcium uptake (in nmoles/mg) of cells y
as a function of time x(in minutes), after being sus-
pended in a solution of radioactive calcium. There
are 27 observations in all. The model is
yi=β0{1−exp(−β1xi)}+σiεi, (3)
where β0and β1are unknown regression coefficients
and the error term εi∼N(0, 1)is standard normal.
We complete the definition of model (3) by allowing
the response variance σ2
i=σ2(1+xi)γto depend
nonlinearly on the time covariate through the two
variance parameters γand σ2.
Figure 4gives the R code for the analysis. The
variables cal and time represent respectively the cal-
cium uptake and suspension time. Model (3) is fitted
by maximum likelihood using the nlreg routine of
package nlreg, which yields ˆ
β0=4.317 (s.e. 0.323),
ˆ
β1=0.207 (s.e. 0.036), ˆγ=0.536 (s.e. 0.320), and
log ˆσ2=−2.343 (s.e. 0.634). Note that the base-
line variance σ2is fitted on the logarithmic scale.
This does not affect inference based on the rand r∗
statistics, which are parametrisation invariant, and
ensures positive values forσ2when the Wald statistic
1Technical details are omitted from the output.
R News ISSN 1609-3631
Vol. 5/1, May 2005 24
> library(boot)
> library(nlreg)
## maximum likelihood fit
> calcium.nl <- nlreg( cal ~ b0 * (1 - exp(-b1 * time)), weights = ~ (1 + time)^g,
+ data = calcium, start = c(b0 = 4, b1 = 0.1, g = 0) )
> summary( calcium.nl ) # yields estimates and standard errors
## pivot profiling for \gamma
> calcium.prof <- profile( calcium.nl, offset = g )
> summary( calcium.prof )
Two-sided confidence intervals for g
lower upper
r* - Fr (0.95) -0.14340 1.191
r* - Sk (0.95) -0.14320 1.190
r (0.95) -0.12441 1.154
Wald (0.95) -0.08991 1.163
Estimate Std. Error
g 0.5364 0.3196
[omitted]
## inference on proportion of maximum
> calcium.nl <- nlreg( cal ~ b0 * (1 - exp(- log(1 + exp(psi)) * time / 15)),
+ data = calcium, start = c(b0 =4.3, psi =2) )
> calcium.prof <- profile( calcium.nl, offset = psi )
> calcium.sum <- summary( calcium.prof )
> exp(calcium.sum$CI) / (1 + exp(calcium.sum$CI)) # 95% confidence intervals for \pi
## profile and contour plots : Figure 5
> calcium.prof <- profile( calcium.nl )
> par( las = 1, mai = c(0.5, 0.5, 0.2, 0.2) )
> contour( calcium.prof, alpha = 0.05, cl1 = "black", cl2 = "black", lwd2 = 2 )
Figure 4: calcium uptake data analysis: R code for model fitting and pivot profiling for different parameters
of interest (variance parameter γand “proportion of maximum” π).
is used. Figure 4shows also how to use the profile
method of the nlreg package to set various first and
higher order 95% confidence intervals for the vari-
ance parameter γ.1A difficulty we had not to face in
the previous two examples is that it is no longer pos-
sible to calculate the correction term in (1) exactly.
The profile function implements two slightly dif-
ferent versions of the higher order pivot r∗which we
obtain by using the two approximations of q(ψ)dis-
cussed in the Appendix. The four statistics agree in
letting us question the heterogeneity of the response
variance.
Davison and Hinkley (1997, p. 356) consider not
only inference on the nonlinear mean function, but
also on other aspects of the model such as the
“proportion of maximum”, π=1−exp(−β1x).
For x=15 minutes they give the estimate ˆπ=
0.956 and the associated 95% bootstrap confidence
interval (0.83, 0.98). We may obtain the corre-
sponding first and higher order likelihood analogues
by reparametrizing the mean response curve into
(π,β0)and re-running the whole analysis. This time
we assume that the response variance is homoge-
neous. Because of the constraint that πmust lie
in the interval (0, 1), we actually fit the model for
ψ=log{π/(1−π)}and back-transform to the orig-
inal scale by π=exp(ψ)/{1+exp(ψ)}. This yields
the intervals (0.87, 0.99)and (0.88, 0.99)for respec-
tively the Wald and likelihood root statistics and
(0.87, 0.99)for both versions of r∗, which is in agree-
ment with the bootstrap simulation.
The profile method of the nlreg package pro-
vides also all elements needed to display graphi-
cally a fitted nonlinear model, as done for instance
in Figure 5with respect to the second model fit. The
contour method of the nlreg package represents,
in fact, an enhanced version of the original algo-
rithm by Bates and Watts (1988, Chapter 6), to which
we refer the reader for the interpretation of these
plots. The dashed, solid and bold lines represent re-
spectively the Wald pivot, the likelihood root rand
Skovgaard’s (1996) approximation to the r∗statistic
R News ISSN 1609-3631
Vol. 5/1, May 2005 25
3.5 4.5 5.5
−3
−2
−1
0
1
2
3
b0
−3 −1 1 3
−3
−2
−1
0
1
2
3
3.5 4.5 5.5
2
3
4
5
−3 −1 1 3
−3
−2
−1
0
1
2
3
3.5 4.5 5.5
−2.0
−1.5
−1.0
−0.5
2 3 4 5
−3
−2
−1
0
1
2
3
psi
−3 −1 1 3
−3
−2
−1
0
1
2
3
2345
−2.0
−1.5
−1.0
−0.5
−2.0 −1.0
−3
−2
−1
0
1
2
3
logs
Figure 5: calcium uptake data analysis: profile plots and profile pair sketches for the parameters β0,ψand
logσ2using the Wald statistic (dashed), the likelihood root r(solid) and Skovgaard’s (1996) approximation to
r∗(bold).
(see the Appendix). The bivariate contour plots in
the lower triangle are plotted on the original scale,
whereas the ones in the upper triangle are on the
rscale. Figure 5highlights different aspects of the
model fit. First, the maximum likelihood estimate
of log σ2is biased downwards, which we can tell
from the fact the corresponding r∗profile is shifted
to the right of r. Otherwise, there does not seem to
be a huge difference between first and higher order
methods as the corresponding profiles and contours
are not too different. The finite sample estimates of
β0and ψare strongly correlated, while they are al-
most independent of log ˆσ2. The contours of r(ψ)
and r∗(ψ)are close to elliptic which indicates that
the log likelihood function is not too far from being
quadratic. A further indication for a small curvature
effect due to parametrisation is that the contours on
the original and on the rscale look similar.
Acknowledgments
I am in debt with Ruggero Bellio, Anthony Davison
and Nancy Reid for the many helpful discussions on
and around higher order likelihood asymptotics. I
would like to thank two anonymous referees whose
suggestions and comments helped improving a pre-
vious version of this paper.
Appendix: q(ψ)correction term
In this appendix we give the general expression of
the correction term q(ψ)in (1) and the explicit for-
mulae for two special model classes, that is, linear ex-
ponential families and regression-scale models. We
will furthermore discuss two ways of approximating
q(ψ)in case we cannot calculate it explicitly.
Basic expression
Let `(θ) = `(ψ,λ)be the log likelihood function,
ˆ
θ= ( ˆ
ψ,ˆ
λ)the maximum likelihood estimator of the
d-dimensional parameter θ= (ψ,λ), and j(θ) =
−∂2`(θ)/∂θ∂θ>the d×dobserved information ma-
trix. Denote ˆ
λψthe constrained maximum likeli-
hood estimator of the nuisance parameter λgiven
the value of the scalar parameter of interest ψ. Write
jλλ(θ)the corner of j(θ) = j(ψ,λ)which corre-
sponds to λ, and ˆ
θψ= (ψ,ˆ
λψ).
R News ISSN 1609-3631

Vol. 5/1, May 2005 26
The basic expression for q(ψ)is
q(ψ) = |`;ˆ
θ(ˆ
θ)−`;ˆ
θ(ˆ
θψ)`λ>;ˆ
θ(ˆ
θψ)|
|jλλ(ˆ
θψ)||j(ˆ
θ)|1/2, (4)
where |·| indicates determinant (Severini,2000, Sec-
tion 7.4.1). The d×dmatrix appearing in the numer-
ator of q(ψ)consists of a column vector formed using
so-called sample space derivatives
`;ˆ
θ(θ) = ∂`(θ;ˆ
θ|a)
∂ˆ
θ,
and a d×(d−1)matrix of mixed derivatives
`λ>;ˆ
θ=∂2`(ψ,λ;ˆ
θ|a)
∂λ>∂ˆ
θ.
The former are defined as the derivatives of the log
likelihood function `(ψ,λ;ˆ
θ|a)with respect to the
maximum likelihood estimator ˆ
θ; mixed derivatives
furthermore involve differentiation with respect to
the whole parameter θor parts of it (Severini,2000,
Section 6.2.1). Note that to do so, the data vector
has to be re-expressed as y= ( ˆ
θ,a), where arepre-
sents the observed value of an ancillary statistic upon
which we condition.
Approximations
Exact computation of the sample space derivatives
involved in expression (4) requires that we are able to
write the data vector yas a function of the maximum
likelihood estimator ˆ
θand of an ancillary statistic a.
This is, with few exceptions, only feasible for linear
exponential families and transformation models, in
which cases the q(ψ)term involves familiar likeli-
hood quantities. If the reference model is a full rank
exponential family with ψand λtaken as canonical
parameters, the correction term
q(ψ) = w(ψ)|jλλ (ˆ
θ)|/|jλλ(ˆ
θψ)|1/2(5)
depends upon the Wald statistic. In case of a
regression-scale model, that is, of a linear regression
model with non necessarily normal errors,
q(ψ) = s(ψ)|jλλ (ˆ
θψ)|/|jλλ (ˆ
θ)|1/2(6)
involves the score statistic. Here, ψis linear in (β,σ)
and the nuisance parameter λis taken linear in βand
ξ=logσ, where βand σrepresent respectively the
regression coefficients and the scale parameter.
In general, the calculation of the sample space
derivatives `;ˆ
θ(θ)and mixed derivatives `λ>;ˆ
θ(θ)
may be difficult or impossible. To deal with this dif-
ficulty, several approximations were proposed. For
a comprehensive review we refer the reader to Sec-
tion 6.7 of Severini (2000). Here we will mention
two of them. A first approximation, due to Fraser
et al. (1999), is based upon the idea that in order to
differentiate the likelihood function `(θ;ˆ
θ|a)on the
surface in the n-dimensional sample space defined
by conditioning on awe need not know exactly the
transformation from yto (ˆ
θ,a), but only the dvec-
tors which are tangent to this surface (Severini,2000,
Section 6.7.2). Skovgaard (1996) on the other hand
suggests to approximate the sample space and mixed
derivatives by suitable covariances of the log like-
lihood and of the score vector (Severini,2000, Sec-
tion 6.7.3). While the first approximation maintains
the third order accuracy of r∗, we lose one degree
when following Skovgaard’s (1996) approach. See
Sections 7.5.3 and 7.5.4 of Severini (2000) for the de-
tails.
The hoa package
The expressions of q(ψ)implemented in the hoa
package bundle are: i) (5) and (6) for respectively the
cond and marg packages (logistic and linear non nor-
mal regression), and ii) the two approximations dis-
cussed above for the nlreg package (nonlinear het-
eroscedastic regression). The formulae are given in
Brazzale et al. (to appear). The nlreg package also
implements Skovgaard’s (2001, Section 3.1) multipa-
rameter extension of the modified likelihood root.
The implementation of the cond and nlreg packages
is discussed in Brazzale (1999) and Bellio and Braz-
zale (2003).
Bibliography
D. M. Bates and D. G. Watts. Nonlinear Regression
Analysis and Its Applications. Wiley, New York,
1988. 24
R. Bellio and A. R. Brazzale. Higher-order asymp-
totics unleashed: Software design for nonlinear
heteroscedastic models. Journal of Computational
and Graphical Statistics, 12:682–697, 2003. 26
A. R. Brazzale. Approximate conditional inference in
logistic and loglinear models. Journal of Computa-
tional and Graphical Statistics, 8:653–661, 1999. 26
A. R. Brazzale, A. C. Davison, and N. Reid. Applied
Asymptotics. Cambridge University Press, Cam-
bridge, to appear. 22,26
D. Collet. Binary data. In P. Armitage and T. Colton,
editors, Encyclopedia of Biostatistics. John Wiley &
Sons, Chichester, 1998. 22
A. C. Davison and D. V. Hinkley. Bootstrap Methods
and Their Application. Cambridge University Press,
Cambridge, 1997. 23,24
D. A. S. Fraser. Statistical inference: Likelihood to
significance. Journal of the American Statistical Asso-
ciation, 86:258–265, 1991. 20
R News ISSN 1609-3631

Vol. 5/1, May 2005 27
D. A. S. Fraser, N. Reid, and J. Wu. A simple general
formula for tail probabilities for frequentist and
Bayesian inference. Biometrika, 86:249–264, 1999.
26
D. A. Pierce and D. Peters. Practical use of higher-
order asymptotics for multiparameter exponential
families. Journal of the Royal Statistical Society Series
B, 54:701–737, 1992. 21
T. A. Severini. Likelihood Methods in Statistics. Oxford
University Press, Oxford, 2000. 20,21,26
I. M. Skovgaard. An explicit large-deviation approx-
imation to one-parameter tests. Bernoulli, 2:145–
165, 1996. 26
I. M. Skovgaard. Likelihood asymptotics. Scandina-
vian Journal of Statistics, 28:3–32, 2001. 21
Alessandra R. Brazzale
Institute of Biomedical Engineering, Italian National Re-
search Council
alessandra.brazzale@isib.cnr.it
Fitting linear mixed models in R
Using the lme4 package
by Douglas Bates
The lme function, which fits linear mixed models of
the form described in Pinheiro and Bates (2000), has
been available in the required R package nlme for
several years. Recently my colleagues and I have
been developing another R package, called lme4, and
its lmer function which provides more flexible fitting
of linear mixed models and also provides extensions
to generalized linear mixed models.
The good news for lme users is that the lmer func-
tion fits a greater range of models, is more reliable,
and is faster than the lme function. The bad news
is that the model specification has been changed
slightly. The purpose of this article is to introduce
lmer, to describe how it can be used to fit linear
mixed models and to highlight some of the differ-
ences between lmer and lme.
Linear mixed models
Before describing how to fit linear mixed models I
will describe what they are. In building a statisti-
cal model for experimental or observational data we
often want to characterize the dependence of a re-
sponse, such as a patient’s heart rate, on one or more
covariates, such as the patient identifier, whether the
patient is in the treatment group or the control group,
and the time under treatment. We also want to char-
acterize the “unexplained” variation in the response.
Empirical models (i.e., models that are derived from
the data itself, not from external assumptions on the
mechanism generating the data) are generally cho-
sen to be linear in the parameters as these are much
simpler to use than are nonlinear models.
Some of the available covariates may be repeat-
able in the sense that (conceptually, at least) we can
obtain new observations with the same values of the
covariate as in the current study. For example, we
can recruit a new patient to the study and assign
this patient to the treatment group or to the control
group. We can then observe this patient’s heart rate
at one minute, five minutes, etc. after treatment.
Hence we would regard both the treatment factor
and the time after treatment covariate as repeatable.
A factor is repeatable if the set of possible levels
of the factor is fixed and each of these levels is itself
repeatable. In most studies we would not regard the
patient identifier factor (or, more generally, the “sub-
ject” factor or any other factor representing an exper-
imental unit) as being repeatable. Instead we regard
the subjects in the study as a random sample from
the population of interest.
Our goals in modeling repeatable covariates and
non-repeatable covariates are different. With a
repeatable covariate we want to characterize the
change in the response between different levels and
for this we use fixed-effects terms that represent, say,
the typical rate of change of the response with respect
to time under treatment or the difference between
a typical response in the treatment and the control
groups. For a non-repeatable covariate we want to
characterize the variation induced in the response by
the different levels of the covariate and for this we
use random-effects terms.
A statistical model that incorporates both fixed-
effects terms and random-effects terms is called a
mixed-effects model or, more simply, a mixed model.
Single grouping factor
As indicated above, a random effect is associated
with a grouping factor, which would be the patient
identifier in our example, and possibly with other co-
variates. We specify a random-effects term in lmer by
a linear model term and a grouping factor separated
by ‘|’, which we would read as “given” or “condi-
tional on”. That is, a random effect is a linear model
term conditional on the level of the grouping factor.
Because the precedence of ‘|’ as an operator
R News ISSN 1609-3631

Vol. 5/1, May 2005 28
is lower than most other operators used in linear
model formulae, the entire random-effects expres-
sion should be enclosed in parentheses.
Many models for longitudinal data (repeated
measurements over time on each of several subjects)
incorporate random effects associated with a single
grouping factor. Consider the HR (heart rate) data
from the SASmixed package
> data("HR", package = "SASmixed")
> names(HR)
[1] "Patient" "Drug" "baseHR"
[4] "HR" "Time"
Initially we fit a linear mixed model with fixed-
effects terms for the base heart rate, the time since
administration of the medication, the type of drug
and the time/drug interaction. The random effect as-
sociated with the patient is a simple additive shift.
> (fm1 <- lmer(HR ~ baseHR + Time *
+ Drug + (1 | Patient), HR))
Linear mixed-effects model fit by REML
Formula: HR ~ baseHR+Time*Drug+(1|Patient)
Data: HR
AIC BIC logLik MLdeviance REMLdeviance
790.7 815.8 -386.3 791.9 772.7
Random effects:
Groups Name Variance Std.Dev.
Patient (Intercept) 44.5 6.67
Residual 29.8 5.46
# of obs: 120, groups: Patient, 24
Fixed effects:
Estimate Std. Error DF
(Intercept) 33.962 9.931 113
baseHR 0.588 0.118 113
Time -10.698 2.421 113
Drugb 3.380 3.784 113
Drugp -3.778 3.802 113
Time:Drugb 3.512 3.424 113
Time:Drugp 7.501 3.424 113
t value Pr(>|t|)
(Intercept) 3.42 0.00087
baseHR 4.97 2.5e-06
Time -4.42 2.3e-05
Drugb 0.89 0.37358
Drugp -0.99 0.32244
Time:Drugb 1.03 0.30717
Time:Drugp 2.19 0.03050
The first few lines of the output state that the
model has been fit using the REML (restricted or
residual maximum likelihood) criterion and indicate
the formula and the data set used. The next line pro-
vides several measures of the quality of the fit in-
cluding Akaike’s Information Criterion, Schwartz’s
Bayesian Information Criterion, the log-likelihood
(actually the log-restricted likelihood because REML
is being used) and the ML and REML versions of the
deviance.
The estimates of the variance components are
given next. Note that the column headed Std.Dev. is
not the standard error of the estimate of the variance
component. It is simply the square root of the esti-
mated variance and is included because many peo-
ple (and I am one) find it easier to interpret an es-
timated standard deviation instead of an estimated
variance.
Finally the estimates for the fixed-effects coef-
ficients are summarized. If we wish to consider
the significance of terms rather than individual co-
efficients we can use a single-argument call to the
anova generic which provides the sequential sums of
squares and the corresponding F tests.
> anova(fm1)
Analysis of Variance Table
Df Sum Sq Mean Sq Denom
baseHR 1 746 746 113
Time 1 753 753 113
Drug 2 87 43 113
Time:Drug 2 143 72 113
F value Pr(>F)
baseHR 25.05 2.1e-06
Time 25.28 1.9e-06
Drug 1.46 0.237
Time:Drug 2.40 0.095
At present the denominator degrees of freedom
shown in the coefficient table and in the analysis of
variance table are upper bounds. In a sense there
is no “correct” denominator degrees of freedom be-
cause the F test is always an approximation for these
models. But, even taking this into account, it is still
the case that the denominator degrees of freedom for
the Drug term should be lower than shown. The rea-
son that these degrees of freedom are not more ac-
curately approximated at present is because it is dif-
ficult to decide exactly how this should be done for
the general models described below.
Before changing the fixed effects terms in the
model we may wish to examine models with more
general specifications of the random effects, such as
both a random intercept and a random slope (with
respect to time) for each patient.
> VarCorr(fm2 <- lmer(HR ~ baseHR +
+ Time * Drug + (Time | Patient),
+ HR))
Groups Name Variance Std.Dev.
Patient (Intercept) 60.6 7.79
Time 37.8 6.15
Residual 24.4 4.94
Corr
-0.563
R News ISSN 1609-3631

Vol. 5/1, May 2005 29
> head(model.matrix(~Time, HR), n = 3)
(Intercept) Time
1 1 0.01667
2 1 0.08333
3 1 0.25000
> head(ranef(fm2)$Patient, n = 3)
(Intercept) Time
201 0.871 4.04733
202 -9.341 6.79574
203 5.005 -0.07822
> anova(fm1, fm2)
Data: HR
Models:
fm1:HR ~ baseHR+Time*Drug+(1|Patient)
fm2:HR ~ baseHR+Time*Drug+(Time|Patient)
Df AIC BIC logLik Chisq Chi Df
fm1 9 810 835 -396
fm2 11 810 841 -394 3.77 2
Pr(>Chisq)
fm1
fm2 0.15
To save space I summarized this fitted model us-
ing VarCorr which describes only the estimates of
the variance components. Notice that the expression
Time generates a model matrix (model.matrix) with
two columns, (Intercept) and Time, so there are
two random effects associated with each patient.
The distribution of the random effects is a bivariate
normal distribution with mean zero and a positive-
definite 2 ×2 variance-covariance matrix. The esti-
mates of the two variances and the estimated corre-
lation are given in the output.
The ranef generic function returns the BLUPs
(Best Linear Unbiased Predictors) of the random ef-
fects and anova provides a likelihood ratio test when
given multiple model fits to the same data set. As
described in Pinheiro and Bates (2000), the p-value
calculated for this test will be conservative (i.e. it is
an upper bound on the true p-value) because the pa-
rameter space is bounded and in the null hypothesis
one of the parameters is at the boundary.
Multiple random-effects expres-
sions
As shown above, the estimated variance-covariance
matrix from a random-effects expression, such as
(Time|Patient), for which the model matrix has
multiple columns is a general, positive-definite, sym-
metric matrix. (“General” in the sense that there are
no constraints imposed on this matrix other than its
being positive-definite.)
Occasionally we wish to impose further con-
straints such as independence of random effects as-
sociated with the same grouping factor. For exam-
ple we can fit a model with an intercept and slope
for each patient but assuming independence of these
random effects with
> VarCorr(fm3 <- lmer(HR ~ baseHR +
+ Time * Drug + (1 | Patient) +
+ (Time - 1 | Patient), HR))
Groups Name Variance Std.Dev.
Patient (Intercept) 47.9 6.92
Patient Time 25.0 5.00
Residual 25.6 5.06
> anova(fm1, fm3, fm2)
Data: HR
Models:
fm1: HR~baseHR+Time*Drug+(1|Patient)
fm3: HR~baseHR+Time*Drug+(1|Patient)
+(Time-1|Patient)
fm2: HR~baseHR+Time*Drug+(Time|Patient)
Df AIC BIC logLik Chisq Chi Df
fm1 9 810 835 -396
fm3 10 811 839 -396 0.84 1
fm2 11 810 841 -394 2.93 1
Pr(>Chisq)
fm1
fm3 0.358
fm2 0.087
The fitted model fm3 has a random intercept and a
random slope for each patient, as does fm2, but in fm3
these random effects are assumed to be independent
within patient. Hence fm3 uses one fewer degree of
freedom than does fm2.
In general the random effects from each expres-
sion are modeled as independent. If the model ma-
trix from the ith random-effects expression has qi
columns and the grouping factor has kilevels (this
number is well-defined because unused levels are
dropped during construction of the model frame) the
corresponding random effects vector has length kiqi
and could be arranged as a ki×qimatrix as shown in
the ranef output above. The rows share a common
ki×kivariance-covariance matrix. Different rows are
independent.
Nested and non-nested grouping factors
We have seen an example of multiple random-effects
expressions for the same grouping factor. It is more
common to create random-effects expressions asso-
ciated with different grouping factors. For example
in a multicenter trial we may have observations over
time on each of several patients at each of several in-
stitutions and it could be appropriate to use random
effects for both patient and institution.
If each patient is observed at only one institution
we say that the levels of patient are nested within the
R News ISSN 1609-3631

Vol. 5/1, May 2005 30
levels of institution; otherwise the factors are non-
nested. Notice that it only takes one patient migrat-
ing between institutions to cause the grouping fac-
tors to lose the nesting property. This may not be a
primary concern in the analysis of multicenter trials
but it is an important issue in the analysis of student
test scores over time where it is quite common to
have some portion of the students observed at mul-
tiple schools. In these cases analysis of the data as
if students were nested within schools is at best an
approximation and at worst quite inappropriate.
A major distinction between lme and lmer is that
lme is optimized for nested grouping factors whereas
lmer handles nested and non-nested grouping fac-
tors equally easily. In lmer one simply provides mul-
tiple random-effects expressions and the determina-
tion of nested or non-nested is done in the code.
Consider the star data (Student Teacher
Achievement Ratio) from the mlmRev package. These
data are from a large study - 26,796 observations
on a total of 11,598 students in 80 schools. We in-
ferred teacher ids from characteristics of the teacher
and classroom, resulting in 1387 distinct teacher ids.
(This teacher count is not entirely accurate but it is a
reasonable approximation.)
We fit an initial model to the math test scores.
> VarCorr(fm4 <- lmer(math ~ gr +
+ sx + eth + cltype + (yrs |
+ id) + (yrs | sch), star))
Groups Name Variance Std.Dev.Corr
id (Intercept) 1079.2 32.85
yrs 22.9 4.78 -0.315
sch (Intercept) 292.8 17.11
yrs 56.8 7.53 -0.777
Residual 584.1 24.17
> anova(fm4)
Analysis of Variance Table
Df Sum Sq Mean Sq Denom
gr 3 1622511 540837 24566
sx 1 9641 9641 24566
eth 5 192909 38582 24566
cltype 2 63848 31924 24566
F value Pr(>F)
gr 925.9 < 2e-16
sx 16.5 4.9e-05
eth 66.0 < 2e-16
cltype 54.6 < 2e-16
It happens in this case that the grouping factors
id and sch are not nested but if they were nested
there would be no change in the model specification.
It is likely that the lmer function would converge
more rapidly if the grouping factors were nested
but even with the nonnested grouping factors in this
large study convergence is reasonably fast.
Specifying levels
Because nested and non-nested grouping factors are
expressed in exactly the same way, it is not possible
to use implicit nesting when specifying the levels of
the grouping factors. Implicit nesting occurs when
the levels of the “inner” grouping factor are reused
for different levels of the “outer” grouping factor.
For example, if the patients are numbered starting at
patient 1 for each institution then patient 1 at insti-
tution 1 is distinguished from patient 1 at institution
2 only if it is assumed that patient is nested within
institution.
For lmer each distinct experimental unit must
correspond to a distinct level in the corresponding
grouping factor. It is easy to create a new group-
ing factor with this property from implicitly nested
factors using the interaction operator ‘:’. The Pixel
data set in the MEMSS package has one grouping fac-
tor Dog and another factor Side. If we wish to fit a
model with random effects for “side within dog” we
must first create a dog/side factor as
> Pixel$DS <- with(Pixel,Dog:Side)[drop=TRUE]
In this case the subset expression [drop=TRUE] is
not needed but it is a good practice to use it when-
ever creating such a factor. The expression results
in any combinations of levels that does not occur in
the data being dropped from the list of levels in the
newly created factor.
Summary
The lmer function in the lme4 package fits linear
mixed models. There are vast changes in the inter-
nals of lmer relative to the earlier lme function and
we hope they will ensure that lmer is faster, more re-
liable, and easier to use than was lme. The biggest
impact on the user is the change in model specifica-
tion, which was made so as to clarify the model being
fit. The lmer function uses a single model formula in-
cluding random-effects expressions that specify both
a linear model term and a grouping factor and can be
used to fit models based on multiple nested or non-
nested grouping factors. It can also be used to fit
generalized linear mixed models but that is a topic
for another day.
Bibliography
J. C. Pinheiro and D. M. Bates. Mixed-Effects Models
in S and S-PLUS. Springer, 2000. 27,29
Douglas Bates
University of Wisconsin - Madison, U.S.A.
bates@wisc.edu
R News ISSN 1609-3631

Vol. 5/1, May 2005 31
Using R for Statistical Seismology
by Ray Brownrigg & David Harte
Statistical Seismology is a relatively new term used
to describe the application of statistical methodol-
ogy to earthquake data. The purpose is to raise
new questions about the physical earthquake pro-
cess, and to describe stochastic components not ex-
plained by physical models. Such stochastic quantifi-
cation allows one to test the validity of various phys-
ical hypotheses, and also makes probability forecasts
more feasible.
We describe here a suite of Rpackages, known
as the "Statistical Seismology Library" (SSLib). This
paper firstly introduces SSLib, providing a little his-
tory, and a description of its structure. Then the
various components of SSLib are described in more
detail and some examples are given. While the
packages were developed primarily to analyse earth-
quake data, two of the packages (Fractal and PtPro-
cess) have more general applicability.
For a general text on seismology, see Lay and Wal-
lace (1995). Further, a large collection of seismolog-
ical software can be found at http://orfeus.knmi.
nl/other.services/software.links.shtml.
Introduction to SSLib
The Statistical Seismology Library (SSLib) is a collec-
tion of earthquake hypocentral catalogues (3D loca-
tion of the rupture initiation point, time and mag-
nitude) and Rfunctions to manipulate, describe and
model event data contained in the catalogues. At this
stage, analyses include graphical data displays, fit-
ting of point process models, estimation of fractal di-
mensions, and routines to apply the M8 Algorithm.
While we have named it the "Statistical Seismology
Library", the type of analyses that are performed re-
ally only reflect the research interests of the authors.
Part of the rationale was to require our students and
collaborators to formally document their programs
so that others could determine what they were sup-
posed to do, and to be able to use them after they
have possibly moved on. Another aim was to make
some of our statistical methods and models more di-
rectly available to our seismologist and geophysicist
colleagues.
The library is divided into a number of Rpack-
ages. Details of these and source code can all be
found on the Statistical Seismology Library web
page (http://homepages.paradise.net.nz/david.
harte/SSLib/). Package names with a specifically
seismological character are prefixed by "ss" (e.g. the
New Zealand catalogue is named ssNZ), whereas
those with a more general statistical interest are not
(e.g. PtProcess and Fractal). A reference manual
(standard Rformat) for each package can also be
found on the web page, along with a Users Guide
that contains examples and shows how the different
packages relate to each other (Harte,2005e).
SSLib was started in 1996 as an S-PLUS library
(Harte,1998). After being ported to the Rlanguage
in 1999, development of SSLib switched to using the
Rimplementation. At this time, SSLib was only avail-
able on the Unix and Linux platforms, but in 2004 a
Windows version was released.
Earthquake Catalogues
Generally, users will want to use their own earth-
quake catalogues. However SSLib does contain
various earthquake catalogues including the New
Zealand catalogue and the PDE catalogue (Pre-
liminary Determinations of Epicentres) which is
a worldwide catalogue collated by the US Geo-
logical Survey. Further details of the catalogues
available in SSLib can be found on the SSLib
web page (see http://homepages.paradise.net.
nz/david.harte/SSLib/).
The earthquake catalogues are based on raw data
available from the World Wide Web. These data are
generally collected by national seismological obser-
vatories. The raw data appears in many different for-
mats, but the SSLib input routines coerce these dif-
ferent formats into a single common structure. This
allows for both a uniformity in analysis, and the abil-
ity to make comparisons between the different cata-
logues. Nevertheless, the data structure within the
catalogue packages allows for the inclusion of any
number of extra data fields; see Harte (2005e) for fur-
ther details.
Catalogue Manipulation Utilities
The ssBase package (Harte,2005b) provides cata-
logue preparation and data manipulation functions.
These include functions for date/time formatting,
data summaries, printing and data subsetting. This
package also contains other functions of a general na-
ture used by the other packages.
A catalogue can be subsetted on any combination
of location, time range, depth range or magnitude
range, with the location being rectangular (in the lat-
itude/longitude sense), circular (actually cylindrical
in 3 dimensions), spherical, or based on an arbitrary
polygon on the surface of the earth. Many of the
analysis functions will work directly with a subset
’object’.
R News ISSN 1609-3631
Vol. 5/1, May 2005 32
Exploratory Data Analyses
The ssEDA package (Harte,2005c) consists of func-
tions for exploratory data analysis. In particular
these can provide histograms of event depth or event
frequency (monthly or yearly), line plots of magni-
tude over time, maps of the locations of the epicen-
tres, and frequency-magnitude plots to determine a
b-value (see Figure 2) or to check the completeness of
a catalogue. Further, the epicentre maps can identify
different magnitudes and depths. Interactive graph-
ics can be used to identify individual events on epi-
central plots, and dynamic graphics can be used to
show 3-dimensional views with rotation and linked
plots.
172 173 174 175 176 177 178 179
−42 −41 −40 −39 −38 −37 −36 −35
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
● ●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
NZ Events Since 1965 With M ≥4.5
Figure 1: Epicentral plot of events from the NZ cata-
logue since 1965 with magnitude ≥4.5 (1777 events).
The Pacific tectonic plate to the east is subducting the
Australian plate to the west.
Figure 1shows a map of the epicentres of a sub-
set of events from the New Zealand catalogue. The
colour of the point depicts the depth of the event,
with the most shallow events at the red end of the
spectrum and the deep events at the blue end of the
spectrum. The Rcommands required to display this
plot follow.
library(ssNZ)
library(ssEDA)
b <- subset.rect(NZ, minday=julian(1, 1, 1965),
minmag=4.5, minlat=-42,
maxlat=-35, minlong=172,
maxlong=179, mindepth=40)
epicentres(b, depth=c(40, 60, 80, 100, 150,
200, Inf), mapname="nz",
criteria=FALSE, cex=0.8,
usr=c(172, 179, -42, -35))
title(expression(paste("NZ Events Since 1965
With ", M >= 4.5)))
Figure 2is a frequency-magnitude plot for the
same subset as used in Figure 1and displays the
Gutenberg-Richter law (Lay and Wallace,1995). This
law says that the logarithm of the cumulative num-
ber of events with magnitude greater than mis lin-
ear as a function of m. The R commands required
to present this graph (given the call to subset.rect
from the previous figure) follow.
freq.magnitude(b)
title("Frequency Magnitude Power-Law
Relationship")
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●
●●●
●
●
● ●
●
●
4.5 5.0 5.5 6.0 6.5
−3.0 −2.5 −2.0 −1.5 −1.0 −0.5 0.0
m
log10(Proportion of Events with Magnitude > m)
Frequency Magnitude Power−Law Relationship
Figure 2: Plot of events from the NZ catalogue since
1965 with magnitude ≥4.5 showing the Gutenberg-
Richter law. The absolute value of the slope is re-
ferred to as the b-value and is typically about one.
The interactive graphics is provided by the
epicentres.identify function through use of the
identify R function. The dynamic graphics is pro-
vided through the threeD function, which links to
an external viewer provided by the xgobi software
(Swayne et al.,1998) via the xgobi R function. Un-
fortunately xgobi is not easily usable on a Windows
system, so work is in progress to use ggobi (GGobi)
instead.
Point Process Modelling
The PtProcess package (Harte,2004), which can be
used independently of the other packages, provides
a framework for point process modelling. This in-
cludes parameter estimation, model evaluation and
R News ISSN 1609-3631
Vol. 5/1, May 2005 33
simulation. This package is likely to be of most inter-
est to a general statistical audience.
Our PtProcess package has a slightly different
emphasis to the point process packages spatstat
(available on CRAN) and ptproc (http://sandybox.
typepad.com/software/ptproc/index.html). Our
point process models are for events strictly ordered
by time, and are conditional on the history of the pro-
cess up to time t. This could be thought of as the
ground process (Daley and Vere-Jones,2003). Other
event characteristics could be added as marks, e.g.
earthquake event magnitude. The spatstat package
(Baddeley and Turner,2005) has an emphasis on the
spatial location of items, presumably at the same
point in time, e.g. tree locations or animals in a for-
est, etc. Emphasis is then on modelling the spatial
intensity. Here marks can be added, e.g. the particu-
lar species of tree. The ptproc package (Peng,2003) is
derived from our package, and has extended the pro-
cedure into multidimensional processes. However,
these additional variables are not treated as marks,
and hence provides an alternative direction to our in-
tended direction.
While the conditional intensity functions pro-
vided within the package have a distinctly seismo-
logical flavour, the general methodology and struc-
ture of the package is probably applicable to a rea-
sonably large class of point process models. The
models fitted are essentially marked point processes
(Daley and Vere-Jones,2003), where the mark dis-
tribution has been explicitly built into the condi-
tional intensity function. Our next task is to sepa-
rate the mark distribution and ground intensity func-
tion, and further generalise the framework so that
any number of mark distributions can be attached to
a given ground intensity function.
Currently the conditional intensity function is the
most basic "building block". The conditional inten-
sity function, λ(t|Ht), can be thought of as an instan-
taneous value of the Poisson rate parameter at time t
and is conditional on the history of the process up to
but not including t. We have given each conditional
intensity function a suffix ".cif". There are a num-
ber of "generic"-like functions which perform some
operation given an intensity function, for example,
simulate a process, perform a goodness of fit evalua-
tion, etc.
As an example, consider the ETAS (Epidemic
Type Aftershock Sequence) model. This assumes
that earthquake events behave in a similar way to
an epidemic, where each event reproduces a number
of aftershocks. The larger the event, the more after-
shocks that will be reproduced. If various criticality
conditions are satisfied, the aftershock sequence will
eventually die out. See Harte (2004) and Utsu and
Ogata (1997) for further technical details about the
ETAS model.
The package contains an example dataset pro-
vided by Yosihiko Ogata. These data were simulated
over the time interval (0, 800). Using these data and
approximate maximum likelihood solutions for the
parameters contained in p(the ETAS model contains
5 parameters), the conditional intensity function can
be plotted as follows (see Figure 3).
library(PtProcess)
data(Ogata)
p <- c(0.02, 70.77, 0.47, 0.002, 1.25)
ti <- seq(0, 800, 0.5)
plot(ti, log(etas.cif(Ogata, ti, p)),
ylab=expression(paste("log ", lambda(t))),
xlab="Time", type="l", xlim=c(0, 800),
main="Conditional Intensity Function")
0 200 400 600 800
−4 −3 −2 −1 0 1 2 3
Conditional Intensity Function
Time
log λ(t)
Figure 3: The spikes occur at event times and their
heights are proportional to the event magnitudes,
with an "Omori" decay (Lay and Wallace,1995) in the
rate after each event.
Further, the log-likelihood can be calculated as
follows.
pp.LL(Ogata, etas.cif, p, c(0, 800))
Using the vector pas initial values, maximum
likelihood parameter estimates can be calculated as
follows.
posterior <- make.posterior(Ogata, etas.cif,
c(0, 800))
neg.posterior <- function(params)
(-posterior(params))
p <- c(0.02, 70.77, 0.47, 0.002, 1.25)
z <- nlm(neg.posterior, p, hessian=TRUE,
iterlim=1000, typsize=p)
The function make.posterior creates a log-
likelihood function to be evaluated on the time inter-
val (0, 800), and also gives one the option of enforc-
ing constraints on the parameters (none here). We
R News ISSN 1609-3631

Vol. 5/1, May 2005 34
then create a negative posterior function because the
function nlm is a minimiser. The maximum likeli-
hood parameter estimates are contained within the
object z.
The creation of the posterior function was partly
done so that the function to be "optimised" within S-
PLUS had only one argument. This is not necessary
in R. Further, the use of priors has not been as useful
as was initially thought. Consequently, it is probably
best to revise the package so that the optimisation
works more directly on the pp.LL function.
One way to test for the goodness of fit is to cal-
culate the transformed residual process. This effec-
tively creates a new time variable which magnifies or
contracts the original process, assuming that the fit-
ted model is correct, in such a manner that the resul-
tant process is a homogeneous Poisson process with
rate parameter one. A plot of the transformed event
times versus the event number should roughly fol-
low the line x=y. Large deviations from this line
indicate a poorly fitting model. This can be achieved
with the following code.
tau <- pp.resid(Ogata, z$estimate, etas.cif)
n <- nrow(Ogata)
plot(1:n, tau, type="l", xlab="Event Number",
ylab="Transformed Time",
xlim=c(0, n), ylim=c(0, n))
abline(a=0, b=1, lty=2, col="red")
Using the maximum likelihood parameter esti-
mates, one can simulate the process over the subse-
quent interval (800, 1200), say. This is achieved as
follows.
x <- pp.sim(Ogata, z$estimate, etas.cif,
TT=c(800, 1200))
The object xcontains the original Ogata data,
with the new simulated events appended. One may
be interested in forecasting the time taken for an
event with magnitude ≥6, say, to occur after time
800. One would then perform many such simula-
tions, and determine the empirically simulated dis-
tribution for the given event of interest.
As can be seen, the conditional intensity function
is the essential ingredient in each of the above analy-
ses, and hence an entity like this is an essential com-
ponent in a more object oriented setup for these mod-
els. It is a relatively simple step to set this up in a
more object oriented manner, however, we have held
off with this until we have disentangled the condi-
tional intensity into its ground process and a general
number of mark distributions.
Other Analyses
The ssM8 package (Harte,2005d) implements the
Keilis-Borok & Kossobokov M8 algorithm (Keilis-
Borok and Kossobokov,1990). It is an empirically
based algorithm that attempts to deduce "times of
increased probability". We have been involved in
projects that have attempted to test the efficacy of
this algorithm.
The Fractal package (Harte,2005a) has been used
to calculate various fractal dimensions based on
earthquake hypocentral locations, for example, see
Harte (2001).
Problems and Future Development
We have already mentioned a number of extensions
to the Point Process package: separating the condi-
tional intensity into a ground intensity and a gen-
eral number of mark distributions, writing more ob-
ject oriented code, and determining if there is still
a need for the make.posterior function. To imple-
ment more object oriented code, the naming conven-
tions would clearly need to be changed.
There is a difference with the functions contained
in the chron package (Ripley and Hornik,2001;
Grothendieck and Petzoldt,2004) and our "date-
times" format in ssBase (Harte,2005b). We would
prefer to use the chron functions, however, we
would like the format.times function within chron
to have greater flexibility, including fractional num-
bers of seconds. For example, we would like the
ability to specify a times format as hh:mm:ss.s, or
hh:mm:ss.ss. Further, many historical earthquake
events do not have all date-time components avail-
able, and our "datetimes" format has a mechanism to
deal with this. Note that the POSIX functions are in-
appropriate, as the event times are generally stored
as UTC times.
A feature that we have included in all of our pack-
ages is a "changes" manual page. This documents all
recent changes made to the package with the date.
We have found this particularly useful when old
analyses have been repeated and different answers
are produced!
As noted earlier, the type of analyses included in
SSLib largely reflects our own research interests. This
also means that it is continually being changed and
augmented.
Bibliography
A. Baddeley and R. Turner. Spatstat: an Rpackage
for analyzing spatial point patterns. Journal of Sta-
tistical Software, 12(6):1–42, 2005. ISSN 1548-7660.
URL http://www.jstatsoft.org.33
D. Daley and D. Vere-Jones. An Introduction to the
Theory of Point Processes, volume I: Elementary
Theory and Methods. Springer-Verlag, New York,
second edition, 2003. 33
GGobi. GGobi data visualization system. http:
//www.ggobi.org/, 2003. 32
R News ISSN 1609-3631

Vol. 5/1, May 2005 35
G. Grothendieck and T. Petzoldt. Rhelp desk: Date
and time classes in R.R News, 4(1):29–32, 2004. 34
D. Harte. Documentation for the Statistical Seis-
mology Library. Technical Report 98-10, School
of Mathematical and Computing Sciences, Vic-
toria University of Wellington, Wellington, New
Zealand, 1998. 31
D. Harte. Multifractals: Theory and Applications. Chap-
man and Hall/CRC, Boca Raton, 2001. 34
D. Harte. Package PtProcess: Time Dependent Point
Process Modelling. Statistics Research Asso-
ciates, Wellington, New Zealand, 2004. URL
http://homepages.paradise.net.nz/david.
harte/SSLib/Manuals/pp.pdf.32,33
D. Harte. Package Fractal: Fractal Analysis. Statistics
Research Associates, Wellington, New Zealand,
2005a. URL http://homepages.paradise.net.
nz/david.harte/SSLib/Manuals/fractal.pdf.
34
D. Harte. Package ssBase: Base Functions
for SSLib. Statistics Research Associates,
Wellington, New Zealand, 2005b. URL
http://homepages.paradise.net.nz/david.
harte/SSLib/Manuals/base.pdf.31,34
D. Harte. Package ssEDA: Exploratory Data Analysis
for Earthquake Data. Statistics Research Asso-
ciates, Wellington, New Zealand, 2005c. URL
http://homepages.paradise.net.nz/david.
harte/SSLib/Manuals/eda.pdf.32
D. Harte. Package ssM8: M8 Earthquake Fore-
casting Algorithm. Statistics Research Asso-
ciates, Wellington, New Zealand, 2005d. URL
http://homepages.paradise.net.nz/david.
harte/SSLib/Manuals/m8.pdf.34
D. Harte. Users Guide for the Statistical Seismol-
ogy Library. Statistics Research Associates,
Wellington, New Zealand, 2005e. URL
http://homepages.paradise.net.nz/david.
harte/SSLib/Manuals/guide.pdf.31
V. Keilis-Borok and V. Kossobokov. Premonitory ac-
tivation of earthquake flow: algorithm M8.Phys.
Earth & Planet. Int., 61:73–83, 1990. 34
T. Lay and T. Wallace. Modern Global Seismology. Aca-
demic Press, San Diego, 1995. 31,32,33
R. Peng. Multi-dimensional point process models in
R.Journal of Statistical Software, 8(16):1–24, 2003.
ISSN 1548-7660. URL http://www.jstatsoft.
org.33
B. Ripley and K. Hornik. Date-time classes. R News,
1(2):8–11, 2001. 34
D. F. Swayne, D. Cook, and A. Buja. XGobi:
Interactive dynamic data visualization in the X
window system. Journal of Computational and
Graphical Statistics, 7(1):113–130, 1998. ISSN
1061-8600. URL http://www.research.att.com/
areas/stat/xgobi/.32
T. Utsu and Y. Ogata. Statistical analysis of seismic-
ity. In J. Healy, V. Keilis-Borok, and W. Lee, edi-
tors, Algorithms for Earthquake Statistics and Predic-
tion, pages 13–94. IASPEI, Menlo Park CA, 1997.
33
Ray Brownrigg
Victoria University of Wellington
ray@mcs.vuw.ac.nz
David Harte
Statistics Research Associates
david@statsresearch.co.nz
Literate programming for creating and
maintaining packages
Jonathan Rougier
Outline
I describe a strategy I have found useful for devel-
oping large packages with lots of not-obvious math-
ematics that needs careful documentation. The basic
idea is to combine the ‘noweb’ literate programming
tool with the Unix ‘make’ utility. The source of the
package itself has the usual structure, but with the
addition of a noweb directory alongside the Rand man
directories. For further reference below, the general
directory tree structure I adopt is
pkgName
zzu
u
u
u
u
u
u
u
u
u
%%
J
J
J
J
J
J
J
J
J
**
T
T
T
T
T
T
T
T
T
T
T
T
T
T
T
T
Rman, . . . inst
noweb
doc doc
where ‘. . . ’ denotes other optional directories such as
src and data.
The files in the noweb directory are the entry-point
R News ISSN 1609-3631

Vol. 5/1, May 2005 36
for all of the documented code in the package, in-
cluding *.R files, *.Rd files, low-level source code,
datasets, and so on. The noweb tool, under the con-
trol of the file ‘Makefile’ and the Unix make utility, is
used to strip the files in the noweb directory into the
appropriate places in the other directories. Other tar-
gets in ‘Makefile’ also handle things like building the
package, installing it to a specific location, cleaning
up and creating various types of documentation.
A complete example R package is available
at http://maths.dur.ac.uk/stats/people/jcr/
myPkg.tar.gz containing the code below, and a bit
more to make ‘myPkg’ into a proper R package.
Literate programming and noweb
The basic idea of literate programming is that when
writing complicated programs, it makes a lot of sense
to keep the code and the documentation of the code
together, in one file. This requires a mechanism for
treating the file one way to strip out and assemble the
code, and another way to create the documentation.
With such a mechanism in place, the functionality
of the code can be made much more transparent—
and the documentation much more informative—
because the code itself can be written in a modular or
‘bottom-up’ fashion, and then assembled in the cor-
rect order automatically.
I use the noweb literate programming tool. A
noweb file looks like a L
A
T
EX file with additional envi-
ronments delimited by <<name of environment>>=
and @. These define code chunks that ap-
pear specially-formatted in the documentation, and
which get stripped out and assembled into the ac-
tual code. Here is an example of a minimal noweb
file, called, say, ‘myDoc1.nw’.
\documentclass[a4paper]{article}
\usepackage{noweb}\noweboptions{smallcode}
\pagestyle{noweb}
\begin{document}
Here is some \LaTeX\ documentation. Followed
by the top-level source code.
<<R>>=
# The source of this file is noweb/myDoc1.nw
<<foo function>>
<<bar function>>
@
Now I can describe the [[foo]] function in more
detail, and then give the code.
<<foo function>>=
"foo" <- function(x) 2 * x^2 - 1
@
And here is the [[bar]] function.
<<bar function>>=
"bar" <- function(y) sqrt((1 + y) / 2)
@
% stuff for man page not shown
\end{document}
The [[name]] syntax is used by noweb to highlight
variable names. There are many additional features
of noweb that are useful to programmers. More infor-
mation can be found at the noweb homepage, http:
//www.eecs.harvard.edu/~nr/noweb/.
Tangle
noweb strips out the code chunks and assembles them
using the ‘notangle’ command. One useful feature is
that it can strip out chunks with specific identifiers
using the -R flag. Thus the command notangle -RR
myDoc1.nw will strip all the code chunks identified
directly or indirectly by <<R>>=, assembling them in
the correct order. The result of this command is the
file
# The source of this file is noweb/myDoc1.nw
"foo" <- function(x) 2 * x^2 - 1
"bar" <- function(y) sqrt((1 + y) / 2)
which is written to standard output. By redirecting
the standard output we can arrange for this file to go
in the appropriate place in the package directory tree.
So the full command in this case would be notangle
-RR myDoc1.nw > ../R/myDoc1.R. So as long as we
are consistent in always labelling the ‘top’ chunk of
the R code with an <<R>>= identifier, we can auto-
mate the process of getting the R code out.
If we want, we can do the same thing using
<<man>>= for the content of man pages, <<src>>= for
low-level code, <<data>>= for datasets, and so on.
Each of these can be stripped out and saved as a file
into the appropriate place in the package directory
tree.
Weave
To build the documentation, noweb uses the ‘noweave’
command, where it is useful to set the -delay option.
Again, this is written to the standard output, and can
be redirected to place the resulting L
A
T
EX file at the
appropriate place in the package tree. I put the L
A
T
EX
versions of the *.nw files in the directory noweb/doc.
Thus the appropriate command is noweave -delay
myDoc1.nw > doc/myDoc1.tex. This file contains
quite a lot of L
A
T
EX commands inserted by noweb to
help with cross-referencing and indexing, and so is
not particularly easy to read. But after using latex,
the result is a dvi file which integrates the L
A
T
EX doc-
umentation and the actual code, in a very readable
format.
R News ISSN 1609-3631

Vol. 5/1, May 2005 37
April 13, 2005 myDoc1.nw 1
Here is some L
A
T
E
X documentation. Followed by the top-level source code.
hRi≡
# The source of this file is noweb/myDoc1.nw
hfoo functioni
hbar functioni
Now I can describe the foo function in more detail, and then give the code.
hfoo functioni≡
"foo" <- function(x) 2 * x^2 - 1
And here is the bar function.
hbar functioni≡
"bar" <- function(y) sqrt((1 + y) / 2)
Similarity to ‘Sweave’
Many users of R will be familiar with Friedrich
Leisch’s ‘Sweave’ function, available in the tools
package. Sweave provides a means of embedding R
code into a report, such as a L
A
T
EX report, and then
automatically replacing this code with its output. It
uses the notangle functionality of a literate program-
ming tool like noweb, because the R code must be
stripped out, prior to evaluating it. The difference
is that when Sweave builds the resulting document it
does more than noweave would do, because it must
actually evaluate R on each code chunk and put the
output back into the document. In the literate pro-
gramming described in this article, all that notangle
does is wrap the code chunk in a L
A
T
EX command
to format it differently. Sweave can duplicate the
simpler behaviour of notangle with the arguments
echo=TRUE,eval=FALSE in each code chunk: see the
help file for the function ‘RWeaveLatex’ for more de-
tails.
Using make
Our starting point is a collection of *.nw files in the
noweb subdirectory. We could, by hand, issue a series
of notangle and noweave commands, each time we
update one or more of these files. Happily, the Unix
tool make can be used instead. make requires a file
of specifiers, often called ‘Makefile’, which describe
the kinds of things that can be made, and the com-
mands necessary to make them. Obvious candidates
for things we would like to make are the *.R and
*.Rd files that go into the Rand man subdirectories,
and the documentation that goes into the inst/doc
subdirectory.
The make utility is very powerful, and compli-
cated. I am not an expert at writing a ‘Makefile’,
but the following approach seems to work well. My
Makefile lives in the noweb directory, and all of the
path specifiers take this as their starting point.
Simple make targets
Suppose that we have compiled a list of files that
ought to be in the Rsubdirectory (say, ‘RFILES’), and
we want to use the command ‘make R’ to update
these files according to changes that have occurred
in the noweb/*.nw files; the argument Ris known as
a ‘target’ of the make command. We need the follow-
ing lines in Makefile:
# make R files in ../R
R: $(RFILES)
$(RFILES): %.R : %.nw
@echo ’Building R file for $<’
@notangle -RR $< > ../R/$@
@if test ‘egrep ’^<<man>>=’ $<‘ ; then \
notangle -Rman $< > ../man/$*.Rd ; fi
After the comment, prefixed by #, the first line states
that the command ‘make R’ depends on the files in
the list ‘RFILES’, which we have already compiled.
The next set of lines is executed for each component
of ‘RFILES’ in turn, conditionally on the *.R file being
older than the corresponding *.nw file. Where the *.R
file is out-of-date in this way, a message is printed,
notangle is called, and the *.R file is rebuilt and
placed in the Rsubdirectory; if there is a <<man>>=
entry, the *.Rd file is also rebuilt and placed in the
man subdirectory.
A ‘Makefile’ has its own syntax. Within a target
specification, the tags ‘$<’, ‘$@’ and ‘$*’ indicate the
origin file, the result file, and the file stem. Under
each initial line the collection of subsequent com-
mands is indented by tabbing, and the backslash at
the end of the line indicates that the next line is a con-
tinuation. The @at the start of each command line
suppresses the printing of the command to the stan-
dard output.
The attractive feature of make is that it only re-
builds files that are out-of-date, and it does this
all automatically using information in the file date-
stamps. This saves a lot of time with a large pack-
age in which there are many *.nw files in the noweb
directory. In my experience, however, one feels a
certain loss of control with so much automation.
The useful command ‘make --dry-run R’ will print
out the commands that will be performed when the
command ‘make R’ is given, but not actually do
them, which is a useful sanity check. The com-
mand ‘touch myDoc1.nw’ will change the date-stamp
on ‘myDoc1.nw’, so that all derived files such as
‘myDoc1.R’ will automatically be out-of-date, which
will force a rebuild even if ‘myDoc1.nw’ has not been
modified: this can be another useful sanity-check.
Suppose instead we want to rebuild the L
A
T
EX
documentation in the noweb/doc subdirectory, using
the command ‘make latex’. We need the following
lines in ‘Makefile’:
# make latex files in doc
latex: $(TEXFILES)
$(TEXFILES): %.tex : %.nw
@echo ’Building tex file for $<’
@noweave -delay $< > doc/$@
R News ISSN 1609-3631

Vol. 5/1, May 2005 38
@cd doc; \
latex $@ > /dev/null
where ‘TEXFILES’ is a list of files that ought to be in
the noweb/doc subdirectory. As before, each *.tex
file is only rebuilt if it is out-of-date relative to the
corresponding *.nw file. The resulting *.tex file is
put into noweb/doc, and then latex is called to create
the corresponding *.dvi file (rather lazily, the output
from latex is diverted to /dev/null and lost).
Slightly more complicated, suppose we want to
put pdf versions of the *.tex files into the subdirec-
tory inst/doc, using the command ‘make pdf’. We
need the following lines in ‘Makefile’:
# make pdf files in ../inst/doc
pdf: $(TEXFILES) $(PDFFILES)
$(PDFFILES): %.pdf : %.tex
@echo ’Building pdf file for $<’
@cd doc; \
pdflatex $< > /dev/null;
@mv doc/$@ ../inst/doc/
where ‘PDFFILES’ is a list of files that ought to be
in the inst/doc subdirectory. The new feature here
is that the command ‘make pdf’ depends on both
‘TEXFILES’ and ‘PDFFILES’. This means that each
component of ‘TEXFILES’ is updated, if necessary,
before the *.pdf file is rebuilt from the *.tex file.
To rebuild the *.pdf file, the pdflatex command is
called on the *.tex file in noweb/doc, and then the
result is moved to inst/doc.
Additional lines in the ‘Makefile’
The main role of additional lines in ‘Makefile’ is to
supply the lists ‘RFILES’, ‘TEXFILES’ and so on. The
following commands achieve this, and a little bit else
besides, and go at the start of ‘Makefile’:
## minimal Makefile
# look in other directories for files
vpath %.R ../R
vpath %.tex doc
vpath %.pdf ../inst/doc
# get lists of files
NWFILES = $(wildcard *.nw)
RFILES = $(NWFILES:.nw=.R)
TEXFILES = $(NWFILES:.nw=.tex)
PDFFILES = $(NWFILES:.nw=.pdf)
# here are the various targets for make
.PHONY: R latex pdf install
# Now insert the lines from above ...
The three ‘vpath’ lines are necessary to help make to
look in other directories than noweb for certain types
of file. Thus the *.R files are to be found in ../R,
the *.tex files in doc, and so on (all relative to the
noweb directory). The next four lines compile the
lists of various types of file: ‘NWFILES’ are found
in the noweb directory, ‘RFILES’ are ‘NWFILES’ files
with the ‘nw’ suffix replaced with a ‘R’ suffix, and so
on. For the final line, the .PHONY command is a bit of
defensive programming to identify the targets of the
make command.
Example
The file http://maths.dur.ac.uk/stats/people/
jcr/myPkg.tar.gz contains a small example of an R
package created using noweb and make, where there
is only one *.nw file in the noweb directory, namely
myDoc1.nw. The following commands illustrate the
steps outlined above (‘debreu’ is my computer):
debreu% make --dry-run R
make: Nothing to be done for ‘R’.
debreu% touch myDoc1.nw
debreu% make --dry-run R
echo ’Building R file for myDoc1.nw’
notangle -RR myDoc1.nw > ../R/myDoc1.R
if test ‘egrep ’^<<man>>=’ myDoc1.nw‘ ; then \
notangle -Rman myDoc1.nw > ../man/myDoc1.Rd ; fi
debreu% make R
Building R file for myDoc1.nw
debreu% make --dry-run pdf
echo ’Building tex file for myDoc1.nw’
noweave -delay myDoc1.nw > doc/myDoc1.tex
cd doc; \
latex myDoc1.tex > /dev/null
echo ’Building pdf file for myDoc1.tex’
cd doc; \
pdflatex myDoc1.tex > /dev/null;
mv doc/myDoc1.pdf ../inst/doc/
debreu% make pdf
Building tex file for myDoc1.nw
Building pdf file for myDoc1.tex
Initially the source file myDoc1.nw is up-to-date. I
touch it to make it more recent than the derived
files myDoc1.{R,Rd,tex,pdf}. The dry run shows
the commands that will be executed when I type
make R: in this case the file myDoc1.R is built and
moved to ../R. After issuing the command make R
the only output to screen is the comment ‘Building R
file for myDoc1.nw’. The dry run for make pdf shows
a more complicated set of operations, because before
myDoc1.pdf can be built, myDoc1.tex has to be built.
More complicated targets
The make command can also be used to perform more
complicated operations. Two that I have found use-
ful are building the package, and installing the pack-
age onto my $R_LIBS path. In both cases, these oper-
ations must first make sure that the files are all up-to-
date. For ‘make install’, for example, we might have:
R News ISSN 1609-3631

Vol. 5/1, May 2005 39
PKGNAME = myPkg
RLIBRARY = /tmp/
R=R
# install
install: $(RFILES) $(PDFFILES)
@echo ’Installing $(PKGNAME) at $(RLIBRARY)’
@cd ../..; \
$(R) CMD INSTALL -l $(RLIBRARY) $(PKGNAME)
where the variables ‘PKGNAME’, ‘RLIBRARY’ and ‘R’
are specified separately, so that it is easy to change
them for different packages, different locations on
$R_LIBS or different versions of R. These are best put
at the top of the ‘Makefile’. The target install should
be added to the .PHONY line.
Issuing the make install command when every-
thing is up-to-date gives:
debreu% make --dry-run install
echo ’Installing myPkg at /tmp/’
cd ../..; R CMD INSTALL -l /tmp/ myPkg
debreu% make install
Installing myPkg at /tmp/
* Installing *source* package ’myPkg’ ...
** R
** inst
** help
>>> Building/Updating help pgs for package ’myPkg’
Formats: text html latex example
* DONE (myPkg)
If some of the files were not up-to-date then they
would have been rebuilt from the original *.nw files
first.
Conclusion
The great thing about literate programming is that
there is no arguing with the documentation, since the
documentation actually includes the code itself, pre-
sented in a format that is easy to check. For those
of us who use L
A
T
EX, noweb is a very simple literate
programming tool. I have found using noweb to be a
good discipline when developing code that is mod-
erately complicated. I have also found that it saves
a lot of time when providing code for other people,
because the programmer’s notes and the documen-
tation become one and the same thing: I shudder to
think how I used to write the code first, and then doc-
ument it afterwards.
For large projects, for which the development of
an R package is appropriate, it is often possible to
break the tasks down into chunks, and assign each
chunk to a separate file. At this point the Unix make
utility is useful both for automating the processing
of the individual files, and also for speeding up this
process by not bothering with up-to-date files. The
‘Makefile’ that controls this process is almost com-
pletely generic, so that the one described above can
be used in any package which conforms to the out-
line given in the introduction, and which uses the
tags ‘R’, ‘man’ and so on to identify the type of code
chunk in each *.nw file.
Jonathan Rougier
Department of Mathematical Sciences, University of
Durham, UK
J.C.Rougier@durham.ac.uk
CRAN Task Views
by Achim Zeileis
With the fast-growing list of packages on CRAN (cur-
rently about 500), the following two problems be-
came more apparent over the last years:
1. When a new user comes to CRAN and is look-
ing for packages that are useful for a certain
task (e.g., econometrics, say), which of all the
packages should he/she look at as they might
contain relevant functionality?
2. If it is clear that a collection of packages is use-
ful for a certain task, it would be nice if the full
collection could be installed easily in one go.
The package ctv tries to address both problems by
providing infrastructure for maintained task views
on CRAN-style repositories. The idea is the follow-
ing: a (group of) maintainer(s) should provide: (a)
a list of packages that are relevant for a specific task
(which can be used for automatic installation) along
with (b) meta-information (from which HTML pages
can be generated) giving an overview of what each
package is doing. Both aspects of the task views are
equally important as is the fact that the views are
maintained. This should provide some quality con-
trol and also provide the meta-information in the jar-
gon used in the community that the task view ad-
dresses.
Using CRAN task views is very simple: the
HTML overviews are available at http://CRAN.
R-project.org/src/contrib/Views/ and the task
view installation tools are very similar to the package
installation tools. The list of views can be queried by
CRAN.views() that returns a list of "ctv" objects:
R> library(ctv)
R> x <- CRAN.views()
R> x
CRAN Task Views
R News ISSN 1609-3631

Vol. 5/1, May 2005 40
---------------
Name: Econometrics
Topic: Computational Econometrics
Maintainer: Achim Zeileis
Repository: http://cran.r-project.org
---------------
Name: Finance
Topic: Empirical Finance
Maintainer: Dirk Eddelbuettel
Repository: http://cran.r-project.org
---------------
Name: MachineLearning
Topic: Machine Learning&Statistical Learning
Maintainer: Torsten Hothorn
Repository: http://cran.r-project.org
---------------
Name: gR
Topic: gRaphical models in R
Maintainer: Claus Dethlefsen
Repository: http://cran.r-project.org
R> x[[1]]
CRAN Task View
--------------
Name: Econometrics
Topic: Computational Econometrics
Maintainer: Achim Zeileis
Repository: http://cran.r-project.org
Packages: bayesm, betareg, car*, Design,
dse, dynlm, Ecdat, fCalendar,
Hmisc, ineq, its, lmtest*, Matrix,
micEcon, MNP, nlme, quantreg,
sandwich*, segmented, sem,
SparseM, strucchange, systemfit,
tseries*, urca*, uroot, VR,
zicounts, zoo*
(* = core package)
Note that currently each CRAN task view is as-
sociated with a single CRAN-style repository (i.e.,
a repository which has in particular a src/contrib
structure), future versions of ctv should relax this
and make it possible to include packages from var-
ious repositories into a view, but this is not imple-
mented, yet.
A particular view can be installed subsequently
by either passing its name or the corresponding
"ctv" object to install.views():
R> install.views("Econometrics",
+ lib = "/path/to/foo")
R> install.views(x[[1]],
+ lib = "/path/to/foo")
An overview of these client-side tools is given on
the manual page of these functions.
Writing a CRAN task is also very easy: all infor-
mation can be provided in a single XML-based for-
mat called .ctv. The .ctv file specifies the name,
topic and maintainer of the view, has an information
section (essentially in almost plain HTML), a list of
the associated packages and further links. For exam-
ples see the currently available views in ctv and also
the vignette contained in the package. All it takes for
a maintainer to write a new task view is to write this
.ctv file, the rest is generated automatically when
the view is submitted to us. Currently, there are task
views available for econometrics, finance, machine
learning and graphical models in R—furthermore,
task views for spatial statistic and statistics in the so-
cial sciences are under development. But to make
these tools more useful, task views for other topics
are needed: suggestions for new task views are more
than welcome and should be e-mailed to me. Of
course, other general comments about the package
ctv are also appreciated.
Achim Zeileis
Wirtschaftsuniversität Wien, Austria
Achim.Zeileis@R-project.org
Using Control Structures with Sweave
Damian Betebenner
Sweave is a tool loaded by default with the utils
package that permits the integration of R/S with
L
A
T
EX. In one of its more prominant applications,
Sweave enables literate statistical practice—where
R/S source code is interwoven with correspond-
ing L
A
T
EX formatted documentation (R Development
Core Team,2005;Leisch,2002a,b). A particularly ele-
gant implementation of this is the vignette() func-
tion (Leisch,2003). Another, more pedestrian, use
of Sweave, which is the focus of this article, is the
batch processing of reports whose contents, includ-
ing figures and variable values, are dynamic in na-
ture. Dynamic forms are common on the web where
a base .html template is populated with user specific
data drawn, most often, from a database. The incor-
poration of repetitive and conditional control struc-
tures into the processed files allows for almost limit-
less possibilities to weave together output from R/S
within the confines of a L
A
T
EX document and produce
professional quality dynamic output.
R News ISSN 1609-3631

Vol. 5/1, May 2005 41
Motivation for the current project came as a result
of pilot data analyses for a state department of edu-
cation. Dissemination of the analyses to schools in
the state required the production of approximately
3,000 school reports documenting the results using
school specific numbers/text, tables, and figures.
The R/Sweave/L
A
T
EX combination was deployed in
various ways to produce prototype reports. This arti-
cle provides details of the process so that others with
similar document production needs can employ this
combination of tools.
Outline of Process
The production process is quite basic in its im-
plementation: Sweave a file using an appropriate
dataframe to create a .tex that includes appropri-
ate output. Most users of Sweave would have little
problem creating such a .tex file based upon a given
dataframe. Adding the capacity to deal with numer-
ous different dataframes is possible using basic con-
trol structures available in R. In general, there are two
files necessary for the mass production of reports:
template.Rnw A template file that provides the tem-
plate into which all the relevant R output is
placed.
master.R A master file that loops over all the re-
cipients of the template file, creates the sub-
set of data to be used within the template file,
Sweaves the template and saves the output to
a recipient specific .tex file.
In the following, the essential parts of each of these
files is discussed in detail:
master.R
The master.R file implements a control structure out-
side of the Sweave process. This allows one to create
a.tex file for each recipient. The key to the creation
of these files is the creation of a distinct outfile for
each unique recipient in the list. A basic master.R
file which performs this looks like:
load(the.data.frame)
recip.list <- unique(the.data.frame$recip)
for (name in recip.list){
subset.data <- subset(the.data.frame,
the.data.frame$recip==name)
outfile <- paste(name, "tex", sep=".")
Sweave("template.Rnw", output=outfile)
}
The process starts by sourcing master.R. This
loads a master data frame that contains the data for
all the potential report recipients. A list of recipi-
ents is then derived from that dataframe. A looping
structure is implemented that subsets the appropri-
ate data file from the master data frame. Crucially,
this subsetted data frame contains the recipient spe-
cific data that is used to construct the individualized
report. Finally, a recipient specific .tex file is created
by by Sweaving the template, template.Rnw.
Depending upon the number of recipients, it is
entirely possible for thousands of .tex files to be
generated. To avoid the files accumulating in the
same directory, one can easily modify the code to
place the output in other directories. For example,
if one wished to create a distinct directory for each
recipient, the following additions would suffice:
for (name in recip.list){
subset.data <- subset(the.data.frame,
the.data.frame$recip==name)
outfile <- paste(name, "tex", sep=".")
dir.create(name)
setwd(name)
Sweave(file.path("..", "template.Rnw"),
output=outfile)
setwd("..")
}
This basic control structure can be tweaked in many
ways. For example, a call can be made to a relational
database that pulls the relevant data into R for pro-
cessing. This is particularly attractive for those appli-
cations where data storage is cumbersome and better
administered with a relational database.
template.Rnw
The template, template.Rnw, is a standard Sweave
file containing L
A
T
EX markup together with docu-
mentation and code chunks. The file, as its name sug-
gests, is a template into which recipient specific num-
bers/text, tables, and figures can be included. One of
the greatest challenges to producing a template is en-
suring that it will produce the expected output given
the range of possible values it is expected to accept.
One of the easiest ways of providing recipient
specific output with the template.Rnw file is through
the liberal use of the \Sexpr function. For the afore-
mentioned school reports, the name of the school
was included throughout the text, tables, and figures
to provide a customized feel. In particular, the fancy-
hdr L
A
T
EX package was used and the school name was
included in the left header. Because school names,
and string expressions in general, vary widely in
their length, care must be exercised so that any place
that the \Sexpr is utilized, the string (or number) re-
sulting from \Sexpr will function in the L
A
T
EX docu-
ment for the range of values of the field that occur in
the data set.
The creation of recipient specific tables is a
frequently encountered (particularly with .html)
means of dynamic report generation. There are
a number of ways of including tables into the
R News ISSN 1609-3631

Vol. 5/1, May 2005 42
template.Rnw document. Perhaps the simplest way
is through the use of the xtable function within
David Dahl’s xtable package which produces L
A
T
EX
output for a variety of R objects. A basic code chunk
yielding .tex markup producing a table is given by
<<echo=FALSE,results=tex>>=
xtable(object, caption="table:caption",
label="table:label")
@
Though suitable for a variety of tasks, the func-
tion xtable is limited in its capacity to produce
complex tables (e.g., tables with multicolumn head-
ers). Another frequently encountered difficulty oc-
curs when tables being produced are long and possi-
bly extend over several pages. Such long tables are
not a problem with |.html| because the pages can
be of nearly infinite length. However, printed pages
must have the table broken into pieces according to
the space available on the page. The L
A
T
EX code pro-
duced by xtable will not yield acceptable results. To
circumvent this, one can use the latex.table func-
tion provided by Roger Koenker’s quantreg pack-
age together with the option for using the longtable
L
A
T
EX package. The latex.table function accepts
matrix objects and outputs results to a specified file.
As such, its incorporation into the Sweave document
requires a slightly different two part technique than
that used with xtable:
1. latex.table is called within a code chunk and
produces the required L
A
T
EX markup and saves
it to an output file.
<<echo=False>>=
latex.table(matrix,
file=paste(name, ":table.tex",
sep=""))
@
2. The L
A
T
EX markup for the table is inserted into
the document (n.b., outside of a code chunk) at
the appropriate place:
\begin{center}
\input{\Sexpr{name}:table.tex}
\end{center}
As with xtable, there are still limitations to
the amount of tabular complexity available with
latex.table. Multicolumn headings, multirow
headings, colored columns, etc. are all difficult if
not impossible to produce. Depending upon ones
willingness to program, however, all effects can ul-
timately be produced. Though tedious, frequent use
of Sexpr commands outside of code chunks or cat
commands within code chunks allows all L
A
T
EX com-
mands to be incorporated into the final .tex docu-
ment upon Sweave compilation.
In many instances, particularly with tables and
figures, it’s necessary to produce multiple tables or
figures within a single Sweave document based upon
particular attributes of the recipient. For example,
with the school project each school’s report con-
tained information pertinent to each grade in the
school. Thus, a table was produced for each grade
within the school. To implement this, a loop is
placed within template.Rnw. This procedure, given
in FAQ A.8 from Friedrich Leisch’s Sweave User
Manual (Leisch,2005), is useful is many situations.
A major difficulty with control structures within
the template is the variable nature of the output. In
the example with school reports, some rural schools
have grades 1 through 12. Thus, if a table or figure
is produced for each grade, one must take account
of the fact that so many tables or figures will result
in the .tex document. The simplest way to accom-
modate different numbers of loop iterations within
the template is to create a \newpage each time the
loop completes. Thus, for example, each figure for
each grade receives a page to itself. This doesn’t al-
ways look acceptable for especially parsimonious ta-
bles or figures. However, combining multiple tables
on a single page can often cause overfull hbox and
vbox situations to occur. A workaround to this diffi-
culty is to use the L
A
T
EX\scalebox and \resizebox
commands. These commands allow great flexibility
in shrinking output to fit a given page. In general,
a good deal of planning and experimentation is nec-
essary to produce a template that works well for all
situations.
The extent to which one wishes to make the re-
ports specific to a particular recipient can be ex-
tended far beyond expressing the recipient specific
information in tables and figures. For reports where
the recipient is an individual, text within the re-
port can be made gender specific through the use
of if else statements based upon values in the
subsetted data frame that the template is using to
construct the recipient specific report. Supposing
the gender code in the subsetted data frame is 1
for males and 2for females, one could incorporate
he/she (or him/her) pronouns into the text using a
simple if else statement such as
\Sexpr{if (data.frame$gender==1) "he"
else "she"}
The amount of customization is limited only by the
amount of work put into constructing the template.
One is not limited to the basic L
A
T
EX classes
(e.g., article.cls or report.cls) when produc-
ing the template. The beamer.cls (Tantau,2004) is
Sweave compatible and provides extensive function-
ality to produce rich, full color .pdf output with
extensive hyperlink capabilities. Figure 1presents
a two page example demonstrating some possibili-
ties of the R/L
A
T
EX/Sweave combination for dynamic
.pdf report generation using the beamer.cls. Other
R News ISSN 1609-3631
Vol. 5/1, May 2005 43
School Name Goes Here Title of Report Goes Here
School Name Goes Here
A Report on Student Achievement
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
R
E
A
L
L
Y
Z
O
O
M
I
N
G
I
N
!
Your Graphic Here
This prototype provides a picture of what is pos-
sible by using Sweave/L
A
T
E
X/R with the beamer.cls.
There are many possible ways to customize the re-
port. The easiest is to liberally use the Sexpr function
throughout the report. For example, one can easily
indicate the name of the recipient on the report using
Sexpr within the .Rnw template file. This allows one
a great deal of flexibility with regard to inputing user
specific data directly into the document.
The first page of the report might provide some
basic instructions for the person receiving the report.
These instructions are likely generic and don’t include
much customization. As one refines the template,
however, it is not difficult to produce increasingly com-
plicated reports. One just has to be careful that the
template works for all the data that is sent to it. One
problem encountered in the school project arose when
using Sexpr{school.name}in the template.Rnw file.
Some school names used the & character. Sexpr re-
turned that string including the & which caused the
pdfL
A
T
E
X stage to come to a halt. Weaving user spe-
cific information into the prose is not difficult using
the Sexpr command in combination with various R
functions. One just has to be careful.
Organization Producing Report Goes Here February 18, 2005 1 / 2
School Name Goes Here Title of Report Goes Here
The following tables provide data summarizing student achievement at
Insert School Name Here. One could additionally insert text to guide
the reader through the tables and figures that appear on the page.
Grade “Insert Grade Here” Reading
2004 Student Number of Percent of Multi-column header here Multi-Column header here
Performance Level Students School Total School District State School District State
Lowest Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic
Low Middle Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic
High Middle Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic
Highest Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic
Total Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic Dynamic
Lowest Low Middle High Middle Highest
0 10 20 30 40 50
A Colorful Barplot for Insert School Name Here
Organization Producing Report Goes Here February 18, 2005 2 / 2
Figure 1: A basic two-page example demonstrating dynamic .pdf using the R/L
A
T
EX/Sweave combination.
L
A
T
EX classes, packages and font families are avail-
able that can be combined to yield unlimited possi-
bilities.
Final Compilation
Upon Sweave completion, master.R yields numer-
ous .tex files that need to be compiled. For the
school project, approximately 3,000 .tex files were
created. In addition, there were, in most cases, three
.pdf figures created for each school. All of these files
can be compiled from within R using the texi2dvi
command available in the tools package. The fol-
lowing loop which can be placed into master.R ac-
complishes this task:
for (name in recip.list){
outfile <- paste(name, "tex", sep=".")
texi2dvi(outfile, pdf=TRUE, clean=TRUE)
}
texi2dvi will run L
A
T
EX and BIBT
EX the appropriate
number of times to resolve any reference and citation
dependencies. The result is compiled versions of all
the .tex files generated from the earlier loop contain-
ing the Sweave process. It is also possible to perform
this task outside of R using a script or batch file to
perform the same operations.
R News ISSN 1609-3631

Vol. 5/1, May 2005 44
Conclusions
For batch production of dynamic .pdf or .ps doc-
umentation, the R/Sweave/L
A
T
EX combination is
powerful and nearly limitless in its capabilities. A
major benefit to producing reports using this combi-
nation of tools is how closely Sweave integrates the
processing power of R with the typesetting capabil-
ity of L
A
T
EX. The key to producing dynamic reports
for a large number of recipients is the use of itera-
tive control structures in R. This article provides the
author’s “homebrew” code. Other, more elegant, so-
lutions are likely possible.
A next step after report generation is report distri-
bution. In theory, given the appropriate server con-
figuration, it should be possible to electronically dis-
tribute the reports to the appropriate recipient based
upon, for example, an email address contained in the
database. I would appreciate learning how others
have addressed this and similar problems.
Acknowledgments
I would like to thank Bill Oliver for introducing me
to R and for his gracious help. In addition, I would
like to thank one of the reviewers for providing par-
ticularly insightful improvements to my code. Fi-
nally, I would like to express gratitude to the devel-
opers/contributors of R, Sweave, and L
A
T
EX, without
their efforts none of this would be possible.
Bibliography
F. Leisch. Sweave: Dynamic generation of statistical
reports using literate data analysis. In W. Härdle
and B. Rönz, editors, Compstat 2002—Proceedings
in Computational Statistics, pages 575–580, Heidel-
berg, Germany, 2002a. Physika Verlag. ISBN 3-
7908-1517-9. URL http://www.ci.tuwien.ac.at/
~leisch/Sweave.40
F. Leisch. Sweave, Part I: Mixing R and L
A
T
EX. R News,
2(3):28–31, December 2002b. URL http://cran.
r-project.org/doc/Rnews/Rnews_2002-3.pdf.
40
F. Leisch. Sweave, Part II: Package Vignettes. R News,
3(2):21–24, October 2003. URL http://cran.
r-project.org/doc/Rnews/Rnews_2003-2.pdf.
40
F. Leisch. Sweave User Manual, 2005. 42
R Development Core Team. R: A language and envi-
ronment for statistical computing. R Foundation for
Statistical Computing, Vienna, Austria, 2005. URL
http://www.R-project.org. 3-900051-07-0. 40
T. Tantau. User’s Guide to the Beamer Class. Source-
forge.net, 2004. URL http://latex-beamer.
sourceforge.net.42
Damian Betebenner
Lynch School of Education
Educational Research, Measurement and Evaluation
Boston College
Damian.Betebenner@bc.edu
The Value of R for Preclinical Statisticians
Bill Pikounis and Andy Liaw
Merck & Co., Inc.
Participation on the R-help mailing list has shown R
to be widely used across organizations of all types
the world over. This article discusses one corner
where R has become an indispensable tool for effec-
tive statistical practice: a preclinical pharmaceutical
environment at the authors’ place of employment.
Preclinical is defined here as a portion of the
research and development process for prescription
medicines. In this article that portion is defined
to start with fundamental discovery, and to end up
where a potential product is first put into human
subjects in clinical trials. There is a wide variety of
knowledge sought across this spectrum, including:
biological targets believed to be important influences
on a given disease; chemical entities that affect such
targets; effectiveness and safety in animals; formu-
lation of product itself. This brief listing is only in-
tended to provide a broad overview. In reality, there
are many more areas that could be included based on
desired granularity. A complementary term of “non-
clinical,” which is also commonly used, is perhaps
more appropriate here; however, we will stay with
the phrase preclinical for brevity.
The hallmark diversity of preclinical research
presents unlimited opportunities for statisticians.
Data are collected everywhere in the process, and in
large quantities. We have the liberty to choose meth-
ods of processing and analysis. This is a different
environment than clinical biostatistics, where SAS is
the dominant tool for various reasons. Below we
present examples and discussion on how and why
we use R and find it invaluable.
R News ISSN 1609-3631

Vol. 5/1, May 2005 45
Daily operations
Our corporate-issued laptops run Windows XP and
we have a small cluster of dual-processor Opterons
that run 64-bit Linux for intensive jobs. The client-
server configuration of VNC (TightVNC ,2004) pro-
vides a very stable way to bridge the two environ-
ments onto one monitor. An X-window session ap-
pears as just another window on the Windows desk-
top. Samba (Samba Team ,1992-2005) sets up file
sharing so that a /home directory share under Linux
appears as a network drive in Windows. The auto-
cutsel (Witrant ,2004) utility eases cut-and-paste op-
erations between the two operating systems.
If we work on data that can be comfortably ex-
plored on our laptops, we stay there. Generating
reports for our research collaborators is helped by
the right-click cutting and default pasting of a Win-
dows metafile from the windows graphics device into
a Microsoft Office document. The internal Data Ed-
itor (see ?edit) is very useful for browsing a data
frame. Once a workspace has been saved in a Win-
dows folder, launching it from Explorer automati-
cally sets the container working directory. The over-
all R GUI console is thankfully unobtrusive, and we
well appreciate the separation of it from graphics de-
vice windows in SDI mode. We find it much more
pleasant to switch amongst desktop windows where
each stands equally on its own, rather than having to
search inside a set of windows within the same ap-
plication.
If an analysis task becomes too large (in mem-
ory or time) for the laptops, it is a simple matter
to transfer the R workspace image to a Linux box
and pick up right where we left off. Through the
aforementioned network drive mapping of a Linux
share, source code editing can be done in the same
Windows editor, and file operations of saving, copy-
ing, opening, etc. are transparently done through the
standard Windows GUI.
All these task-oriented details add up to in-
creased productivity. More importantly, however, is
the content of the environment itself. We point out
here that some of what we say in the rest of the ar-
ticle is not necessarily unique to R, and such ben-
efits can be found in the other implementation of
the S language, namely S-PLUS. In his “Exegeses
on Linear Models” talk (Venables ,2000), one exe-
gesis from Bill Venables that we particularly admire,
and paraphrase here, is that “the subject should dic-
tate what the program should do and not the other
way around.” Our work environment streams a con-
stant wealth of different data structures and types
of scientific questions to address, and we can exer-
cise our freedom to choose our strategies of analy-
sis. Comprehensive data analyses are only limited
by our own human capabilities, not by R. R enables
us to seamlessly move from data management to ex-
ploration to formal evaluations to communications
of results. No excuse remains to prevent the produc-
tion of good data graphs for the purposes of valu-
able study or presentation. The implementation of
modern methods allows us to use resampling, re-
sistance/robustness, and dimension reduction meth-
ods on a routine basis, to name a few. We work
with cutting edge science, which we firmly believe
deserves cutting edge data analysis. R promotes this
for us, with its virtues of flexibility, stability and effi-
ciency so clearly practiced by its R Core caretakers.
Professor Brian D. Ripley, in “How Computing
has Changed Statistics” (Ripley ,2004), aptly states
of R that “the only barrier to understanding how it
works, precisely, is skill.” We interpret the mean-
ing of “how it works” to be on multiple levels, and
the level most important for us is that a true, contin-
uous investment to learn its many functions, struc-
tures, and features pays off time and again. It helps
us bring value to scientific research, gain trust from
our collaborators, and stimulates the intellect.
Illustrations
Preclinical statisticians do not get nearly enough op-
portunities to provide input to design of experi-
ments. When we do get such an opportunity, we
are especially eager to make recommendations as
quickly as possible. A recent query involved a
crossover design with binary responses. An impor-
tant part of the design plan was to estimate the num-
ber of subjects needed to see a meaningful difference
between two conditions, where incidence rates were
low. Sample size determination tends to be the pre-
dominant aspect of such study design requests.
Estimating sample size is a process of several ap-
proximations, but with some effort such approxi-
mations do not substantially diminish its value. In
this example, tracking down software that could
be readily used, or finding a literature reference
where a recipe could be programmed, was not timely
enough. So a simulation-based approach was taken.
calcPower <- function(nsubj, pyn, pny, pnnyy,
numsim=1000) {
## A simple approach to calculate the power of
## McNemar’s matched-pair test for these inputs:
## nsubj = the number of subjects/pairs of
## measurements (msmts)
## (note that msmt1 comes from condition 1 and
## msmt2 comes from condition 2)
## pyn = p(msmt1 = yes & msmt2 = no)
## pny = p(msmt1 = no & msmt2 = yes)
## pnnyy = p(msmt1 != msmt2)
## numsim = Number of Simulations
## (at least 1000 recommended)
outcomes <- rmultinom(n=numsim, size=nsubj,
prob=c(pnn=pnnyy, pyy=0,
pny=pny, pyn=pyn))
tscompares <-
apply(outcomes, 2, function(x) {
R News ISSN 1609-3631

Vol. 5/1, May 2005 46
ts <- (((x[3] - x[4])^2) / (x[3] + x[4]))
if (!is.finite(ts)) ts <- 0
ts
})
mean(tscompares > qchisq(0.95, 1), na.rm=FALSE)
}
This simple-minded function evaluates how Mc-
Nemar’s test would behave for a given configuration
of underlying parameters, thereby providing an esti-
mate of its power to detect that difference. Recall that
small occurrence rates were expected. A grid of po-
tential configurations can be evaluated:
powerGrid <-
expand.grid(pyn=seq(0.01, 0.10, by=.01),
pny=c(0.001, 0.002, 0.005,
seq(0.01, 0.09, by=.01)))
powerGrid$pnnyy <- (1 - powerGrid$pyn -
powerGrid$pny)
powerGrid <- subset(powerGrid, pyn > pny)
powerGrid <- powerGrid[order(powerGrid$pyn,
powerGrid$pny),]
powerGrid$power <-
apply(powerGrid, 1,
function(x) calcPower(100, x[1],
x[2], x[3]))
From the resulting data frame powerGrid, the
context of the study objectives and available re-
sources, a variety of alternatives were explored and
recommended to the researcher. Ensuing discussions
then produced a design that was later successfully
implemented.
We realize that potential influences of period and
sequence were ignored in the above calculations, as
well as perhaps a more suitable parametric model
approach to the postulated analysis. But as we men-
tioned previously, we felt the approximations in the
approach were reasonable enough to address the
questions at hand.
Similar sample size scenarios we have encoun-
tered include (1) survival data with simple Type I
censoring; (2) single population binomial parame-
ter estimation; (3) inspection of device performance
within its specified tolerances; and (4) comparability
of old and new drug formulations. In all these sce-
narios the principal recipe of simulating power re-
mains the same as the example above; the key differ-
ences include the postulated underlying distribution
and the methodology of estimation or testing.
One could argue the drawback of computational
time that might be needed to generate sufficient grids
of sample size and power values. In the above ex-
ample, a nsim=10000 or 100000 would provide bet-
ter estimates of power and the computational time is
not prohibitive; we are talking minutes and at most
hours to get the needed results. Here is where our
small cluster of Linux servers, or simply scheduling
something to run overnight, takes advantage of what
computers are really good at.
Molecular Modeling
As another illustration, one particular area where
our group has been involved in is molecular mod-
eling. Specifically, we are referring to the problem
of predicting biological activities of small organic
molecules from their chemical “descriptors”. The
biological activities can be quantitative (e.g., per-
cent inhibition against a target enzyme) or qualita-
tive (“active” vs. “inactive”). The chemical descrip-
tors are properties/features of the molecules com-
puted from their structures. There are two possible
goals. One is simply prediction: Given a collection
of molecules with unknown biological activities, pre-
dict which ones are likely to be active. The other pos-
sible goal is interpretation: What chemical properties
or substructures are biologically relevant?
Our computational chemistry colleagues have
traditionally used techniques such as k-nearest
neighbors, partial least squares, and neural net-
works, etc. for such problems, mostly using tools
written in-house. A couple of years ago, we started
to convince our colleagues that more modern, pow-
erful tools such as random forests (Breiman,2001;
Svetnik et. al. ,2003) can be readily accessible
through R. We started working on linking Breiman
and Cutler’s Fortran code to R after we got tired
of using the Fortran code alone, since every lit-
tle change in the data or parameters required re-
compiling the source code. It would have been im-
possible to convince our colleagues to use the tool
in that form, no matter how powerful the tool may
be. As a result of making random forests available
in R, it has become an important component of the
methodologies utilized by our colleagues.
Currently, R is installed on the main computer
system for the computational chemists, who are in-
variably Unix based. Whatever R functionalities are
required, the development group (who are responsi-
ble for research and implementation of new method-
ologies) would wrap them in shell or Perl scripts.
These scripts are then used by the applications group
to support specific projects.
Delivery of Tools
Merck preclinical statisticians are outnumbered at
least ten to one by potential researchers to collaborate
with, and we have global sites that we strive to serve
since they have no access to local statisticians. As
briefly alluded to in the previous section, the avail-
ability of R to communicate with other software of-
fers great potential to serve our customers.
We have recently embarked on a COM-driven1
framework to take advantage of existing infrastruc-
1COM stands for Common Object Model, a Microsoft-driven “standard” for communications between software components.
R News ISSN 1609-3631

Vol. 5/1, May 2005 47
ture. For instance, virtually everyone at Merck runs
the same version of Windows XP and the same ver-
sion of Microsoft Excel. Use of R(D)COM (Baier
and Neuwirth ,2004) allows construction of an ap-
plication where the user interface is entirely in Ex-
cel; namely, input data storage, selection of data and
choices through Userforms, and formatted output.
The underlying work of the R engine is invisible to
the user as it provides calculations and the raw out-
put to Excel for the formatting and the organized pre-
sentation. This framework leverages Excel strengths
of formatting, familiarity, and ubiquity, and R pro-
vides numerical reliability and breadth of data ana-
lytic functionality. While we are currently in early
stages, the framework has demonstrated reliability
for widespread distribution.
Summary
There are several specific aspects that make R
uniquely suitable for us and our colleagues:
• Availability of modern methodologies;
• Flexibility for implementing new methods;
• Facilities to package added functionalities;
• Seamless integration with other software;
• Liberty to (try to) install on any platform.
The first point is discussed above. We add that col-
leagues in our department rely on R for their support
of genomic and proteomic research, where access to
(or the ability to implement) cutting-edge method-
ologies is crucial. The second through fourth points
are important for us as tool developers. The fact that
R is a full-featured language enables us to follow the
spirit of “turn ideas into software, quickly and faith-
fully” (Chambers ,1998). The packaging facility in
R lets us easily create, maintain, and distribute tools
and associated documentation. The same cannot re-
ally be said about most other statistical languages or
packages. The COM framework discussed above is
but one of many options for integration of R with
processes or GUIs, etc. The last point is important
not because R is free (as in beer), but because we are
not limited to run the software on whatever platform
a vendor chooses to support. As long as we can get
R to compile and pass all its tests, we are comfort-
able using it. As an example, when we bought our
first 64-bit machine (a dual Opteron 244 with 16GB
of RAM), the main motivation was to overcome the
memory limitation of a 32-bit platform. Because we
can build R from source, we readily built a 64-bit ver-
sion of R on that box.
This article focused on preclinical statistics use of
R. Nearly four years ago we organized our first in-
house introductory course on R, and about 20 people
attended, mostly from preclinical statistics and other
basic research departments that provide quantitative
analysis support to researchers. As a sign of the in-
creasing acceptance of R at Merck three years later,
attendance more than doubled and mostly included
clinical statisticians and SAS programmers. Results
based on the use of R by preclinical statisticians have
been included in regulatory responses and filings,
and no issues have arisen. We are not aware of R
use in the traditional core workflows of clinical trials
production work to date, but we do expect the use of
R within Merck to continue to increase in all areas,
including clinical.
In our small corner of the world that is preclinical
drug research & development, R is a guide towards
better statistical practice and helps expands our in-
fluence on scientific research. It is indispensable. If
R— the software and community — did not exist, we
would be wishing that something like it would come
along.
Footnote: The first author recently changed employ-
ment from Merck to Centocor. The preclinical/nonclinical
experiences with researchers there have been quite similar
to date.
Bibliography
T. Baier and E. Neuwirth (2004) R (D)COM
Server V1.35 URL http://cran.r-project.org/
contrib/extra/dcom/RSrv135.html 47
L. Breiman (2001) Random forests Machine Learning,
45, 5–32. 46
J. M. Chambers (1998) Programming with Data New
York: Springer-Verlag. 47
B. D. Ripley (2004) How Computing Has Changed
Statistics (and is changing. . .) Symposium in
Honour of David Cox’s 80th birthday, Neuchatel,
July 2004 URL http://www.stats.ox.ac.uk/
~ripley/Cox80.pdf 45
Samba URL http://www.samba.org/ 45
V. Svetnik, A. Liaw, C. Tong, C. Culberson,
R. P. Sheridan, and B. P. Feuston (2003) Random
Forest: A Classification and Regression Tool for
Compound Classification and QSAR Modeling J.
Chem. Inf. Comput. Sci. 43, 1047–1058. 46
TightVNC URL http://www.tightvnc.com/ 45
W. N. Venables (2000) Exegeses on Linear Models Pa-
per presented to the S-PLUS User’s Conference Wash-
ington, DC, 8-9th October, 1998 URL http://www.
stats.ox.ac.uk/pub/MASS3/Exegeses.pdf 45
M. Witrant (2004) autocutsel URL http://savannah.
nongnu.org/projects/autocutsel/ 45
R News ISSN 1609-3631
Vol. 5/1, May 2005 48
Recreational mathematics with R:
introducing the “magic” package
Creating and investigating magic squares with R
Robin K. S. Hankin
Preface
The R computer language (R Development Core
Team,2004) has been applied with a great deal of
success to a wide variety of statistical, physical, and
medical applications. Here, I show that R is an
equally superb research tool in the field of recre-
ational mathematics.
Recreational mathematics is easier to recognize
than define, but seems to be characterized by requir-
ing a bare minimum of “raw material”: complex no-
tation is not needed, and problems are readily com-
municated to the general public.
This is not to say that all problems of recreational
mathematics are trivial: one could argue that much
number theory is recreational in nature; yet attempts
to prove Fermat’s Last Theorem, or the search for
ever higher perfect numbers, have been the catalyst
for the development of many fruitful new areas of
mathematics.
The study of magic squares is also an example of
nontrivial recreational mathematics as the basic con-
cept is simple to grasp—yet there remain unsolved
problems in the field whose study has revealed deep
mathematical truths.
Here, I introduce the “magic” package, and show
that R is an excellent environment for the creation
and investigation of magic squares. I also show
that one’s appreciation of magic squares may be en-
hanced through computer tools such as R, and that
the act of translating ‘paper’ algorithms of the litera-
ture into R idiom can lead to new insight.
Magic squares
Magic squares have essentially zero practical use;
their fascination—like much of pure mathematics—
lies in the appeal of æsthetics and structure rather
than immediate usefulness.
The following definitions are almost universal:
• A semimagic square is one all of whose row sums
equal all its columnwise sums (i.e. the magic
constant).
• A magic square is a semimagic square with the
sum of both unbroken diagonals equal to the
magic constant.
• A panmagic square is a magic square all of whose
broken diagonals sum to the magic constant.
(all squares are understood to be n×nand to be
normal, that is, to comprise n2consecutive inte-
gers1). Functions is.semimagic(),is.magic(), and
is.panmagic() test for these properties.
A good place to start is the simplest—and by far
the most commonly encountered—magic square, lo
zhu:
> magic(3)
[,1] [,2] [,3]
[1,] 2 7 6
[2,] 9 5 1
[3,] 4 3 8
This magic square has been known since antiq-
uity (legend has it that the square was revealed to
humanity inscribed upon the shell of a divine turtle).
More generally, if consecutive numbers of a magic
square are joined by lines, a pleasing image is of-
ten obtained (figure 1, for example, shows a magic
square of order 7; when viewed in this way, the algo-
rithm for creating such a square should be immedi-
ately obvious).
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
Figure 1: Magic square of order 7 in graphical form
(obtained by magicplot(magic.2np1(3)))
1Most workers require the entries to start at 1, which is the convention here; but there are several instances where starting at 0 is far
more convenient. In any case, if xis magic, then x+n is magic for any integer n.
R News ISSN 1609-3631
Vol. 5/1, May 2005 49
Function magic() takes an integer argument n
and returns a normal magic square of size n×n.
There are eight equivalent forms for lo zhu or in-
deed any magic square, achieved by rotating and re-
flecting the matrix (Benson and Jacoby,1976); such
equivalence is tested by eq() or %eq%. Of these eight
forms, a magic square ais said to be in Frénicle’s
standard form if a[1,1]≤b[1,1] whenever a %eq% b,
and a[1,2]<a[2,1]. Function is.standard() tests
for this, and function as.standard() places a magic
square in standard form. Magic squares returned by
magic() are always in standard form.
A typical (paper) algorithm for placing magic
square ain standard form would be “rotate a
until a[1,1]<min(a[1,n],a[n,1],a[n,n]) then, if
a[1,2]>a[2,1], take the transpose”. I shall show
later that expressing such an algorithm in R leads to
new insight when considering magic hypercubes.
A wide variety of algorithms exists for calculating
magic squares. For a given order n, these algorithms
generally depend on nmodulo 4.
A typical paper algorithm for magic squares of
order n=4mwould go as follows.
Algorithm 1: in a square of order 4m,
shade the long major diagonal. Then
shade all major diagonals distant by a
multiple of 4 cells from the long diagonal.
Do the same with the minor diagonals.
Then, starting with “1” at the top left cor-
ner and proceeding from left to right and
top to bottom, count from 1 to n2, filling
in the shaded squares with the appropri-
ate number and omitting the unshaded
ones [figure 2]. Fill in the remaining (un-
shaded) squares in the same way, start-
ing at the lower right corner, moving left-
wards and upwards [figure 3].
Such paper algorithms are common in the litera-
ture but translating this one into code that uses R’s
vectorized tools effectively can lead to new insight.
The magicness of such squares may be proved by
considering the increasing and decreasing sequences
separately.
57
33
25
1
50
42
18
10
51
43
19
11
60
36
28
4
61
37
29
5
54
46
22
14
55
47
23
15
64
40
32
8
Figure 2: Half-completed magic square of order 8
57
16
24
33
25
48
56
1
7
50
42
31
39
18
10
63
6
51
43
30
38
19
11
62
60
13
21
36
28
45
53
4
61
12
20
37
29
44
52
5
3
54
46
27
35
22
14
59
2
55
47
26
34
23
15
58
64
9
17
40
32
41
49
8
Figure 3: Magic square of order 8
The interesting part of the above paper algo-
rithm lies in determining the pattern of shaded
and unshaded squares2. As the reader may
care to verify, parsing the algorithm into R id-
iom is not straightforward. An alternative,
readily computed in R, would be to recog-
nize that the repeating 4 ×4 cell a[2:5,2:5]
is kronecker(diag(2),matrix(1,2,2)) -> b say,
replicate it with kronecker(matrix(1,3,3),b) ->
g; then trim off the border by selecting only the
middle elements, in this case g[2:9,2:9]. Function
magic.4n() implements the algorithm for general m.
2If a <- matrix(1:(n*n),n,n), with jj a Boolean vector of length n2with TRUE corresponding to shaded squares, then with it is clear
that a[jj] <- rev(a[jj]) will return the above magic square.
R News ISSN 1609-3631

Vol. 5/1, May 2005 50
Magic hypercubes
One of the great strengths of R is its ability to han-
dle arbitrary dimensioned arrays in an efficient and
elegant manner.
Generalizing magic squares to magic hyper-
cubes (Hendricks,1973) is thus natural when work-
ing in R. The following definitions represent a gen-
eral consensus, but are far from universal:
• A semimagic hypercube has all “rook’s move”
sums equal to the magic constant (that is,
each ∑n
ir=1a[i1,i2,...,ir−1,ir,ir+1,...,id]with
1≤r≤dis equal to the magic constant for
all values of the other i’s).
• A magic hypercube is a semimagic hypercube
with the additional requirement that all 2d−1
long (ie extreme point-to-extreme point) diag-
onals sum correctly.
• A perfect magic hypercube is a magic hypercube
with all nonbroken diagonals summing cor-
rectly3.
• A pandiagonal hypercube is a perfect magic hy-
percube with all broken diagonals summing
correctly.
(a magic hypercube is understood to be of di-
mension rep(n,d) and normal). Functions
is.semimagichypercube(),is.magichypercube()
and is.perfect(a) test for the first three proper-
ties; the fourth is not yet implemented. Function
is.diagonally.correct() tests for correct summa-
tion of the 2d(sic) long diagonals.
Magic hypercubes of order 4n
Consider algorithm 1 generalized to a d-dimensional
hypercube. The appropriate generalization of
the repeating cell of the 8 ×8 magic square dis-
cussed above is not immediately obvious when
considering figure 2, but the R formalism (viz
kronecker(diag(2),matrix(1,2,2))) makes it
clear that the appropriate generalization is to replace
matrix(1,2,2) with array(1,rep(2,d)).
The appropriate generalization for diag(2) (call
it g) is not so straightforward, but one might be
guided by the following requirements:
• The dimension of gmust match the first argu-
ment to kronecker(), viz rep(2,d)
• The number of 0s must be equal to the number
of 1s: sum(g==1)==sum(g==0)
• The observation that diag(2) is equal to its
transpose would generalize to requiring that
aperm(g,K) be identical to gfor any permuta-
tion K.
These lead to specifying that g[i1,...,id] should
be zero if (i1,...,id)contains an odd number of 2s
and one otherwise.
One appropriate R idiom would be to de-
fine a function dimension(a,p) to be an inte-
ger matrix with the same dimensions as a, with
element (n1,n2, ..., nd)being np, then if jj =
∑d
i=1dimension(a,i), we can specify g=jj*0 and
then g[jj%%2==1] <- 1.
Another application of kronecker() gives a hy-
percube that is of extent 4m+2 in each of its ddimen-
sions, and this may be trimmed off as above to give
an array of dimensions rep(4m,d) using do.call()
and [<-. The numbers may be filled in exactly as for
the 2d case.
The resulting hypercube is magic, in the sense
defined above4, although it is not perfect; function
magichypercube.4n() implements the algorithm.
The ability to generate magic hypercubes of arbitrary
dimension greater than one is apparently novel.
Standard form for hypercubes
Consider again the paper definition for Frénicle’s
standard form of a magic square a: it is rotated so
that the smallest number appears at the top left; then
if a[1,2]<a[2,1], the transpose is taken.
When coding up such an algorithm in R with an
eye to generalizing it to arbitrarily high dimensional
hypercubes, it becomes apparent that “rotation” is
not a natural operation. The generalization used in
the package is directly suggested by R’s array capa-
bilities: it is a two-step process in which the first step
is to maneuver the smallest possible element to posi-
tion [1,1,...,1] using only operations that reverse
the index of some (or all) dimensions. An example
would be a <- a[1:n,n:1,1:n,n:1].
The second step is to use function aperm()
to ensure that the appropriate generalization of
a[1,2]<a[2,1], which would be
a[1,1,...,1,2] <a[1,...,2,1] <. . .
. . . <a[1,2,...,1] <a[2,1,...,1]
holds; the appropriate R idiom is a <-
aperm(a,order(-a[1+diag(d)]))).
This generalization of Frénicle’s standard form to
arbitrary dimensional hypercubes appears to be new;
it arises directly from the power and elegance of R’s
array handling techniques.
3This condition is quite restrictive; in the case of a tesseract, this would include subsets such as ∑n
i=1a[1, i,n−i+1, n]summing
correctly.
4If I had a rigorous proof of this, the margin might be too narrow for it.
R News ISSN 1609-3631

Vol. 5/1, May 2005 51
Conclusions
The R language is a natural environment for the in-
vestigation of magic squares and hypercubes; and
the discipline of translating published algorithms
into R idiom can yield new insight. These insights
include a new generalization of Frénicle’s standard
form to hypercubes, and also what appears to be the
first algorithm for generating magic hypercubes of
any dimension,
Insofar as magic squares and hypercubes are wor-
thy of attention, it is worth creating fast, efficient rou-
tines to carry out the “paper” algorithms of the liter-
ature. I hope that the magic package will continue to
facilitate the study of these fascinating objects.
Acknowledgements
I would like to acknowledge the many stimulating
and helpful comments made by the R-help list over
the years.
Bibliography
W. H. Benson and O. Jacoby. New recreations with
magic squares. Dover, 1976. 49
J. R. Hendricks. Magic tesseracts and N-dimensional
magic hypercubes. Journal of Recreational Mathe-
matics, 6(3):193–201, 1973. 50
R Development Core Team. R: A language and envi-
ronment for statistical computing. R Foundation for
Statistical Computing, Vienna, Austria, 2004. URL
http://www.R-project.org. ISBN 3-900051-07-0.
48
Robin Hankin
Southampton Oceanography Centre
European Way
Southampton
United Kingdom
SO14 3ZH
r.hankin@soc.soton.ac.uk
Programmer’s Niche
How Do You Spell That Number?
John Fox
Frank Duan recently posted a question to the r-help
mailing list asking how to translate numbers into
words. The program described in this column is a
cleaned-up and slightly enhanced version of my re-
sponse to his question. I found the problem to be an
interesting puzzle, and the solution uses several pro-
gramming techniques that demonstrate the flexibil-
ity of R, including its ability to manipulate character-
string data and to employ recursive function calls.
One intriguing aspect of the problem is that it re-
quired me to raise into consciousness my subcon-
scious knowledge about how numbers are spoken
and written in English. I was much more aware
of these conventions in the languages (French and
Spanish) that I had studied as a non-native speaker.
A bit later, I realized that there are variations among
English-speaking countries in the manner in which
numbers are spoken and written down. Because I
was born in the United States and have lived most
of my adult life in Canada, I’m terminally confused
about English spelling and usage. Canadian conven-
tions are an amalgam of American and British rules.
In any event, it didn’t take much time to see that
the numbers from one to nineteen are represented
by individual words; the numbers from twenty-one
to ninety-nine are formed as compound words, with
components for the tens and units digits — with the
exceptions of multiples of ten (twenty,thirty, etc.),
which are single words. The Chicago Manual of Style
tells me that these compound words should be hy-
phenated (but offered little additional useful advice
about how numbers are to be written out). Num-
bers from 100 to 999 are written by tacking on (at
the left) a phrase like “six hundred” — that is, com-
posed of a number from one to nine plus the suffix
hundred (and there is no hyphen). Above this point,
additional terms are added at the left, representing
multiples of powers of 1000. In American English
(and in Canada), the first few powers of 1000 have
the following names, to be used as suffixes:
10001thousand
10002million
10003billion
10004trillion
Thus, for example, the number 210,363,258 would
be rendered “two hundred ten million, three hun-
dred sixty-three thousand, two hundred fifty-eight.”
There really is no point in going beyond tril-
lions, because double-precision numbers can repre-
sent integers exactly only to about 15 decimal dig-
its, or hundreds of trillions. Of course, I could
allow numbers to be specified optionally by ar-
bitrarily long character strings of numerals (e.g.,
"210363258347237492310"), but I didn’t see a real
need to go higher than hundreds of trillions.
One approach to converting numbers to words
would be to manipulate the numbers as integers, but
R News ISSN 1609-3631

Vol. 5/1, May 2005 52
it seemed to me simpler to convert numbers to char-
acter strings of numerals, which could then be split
into individual characters: (1) larger integers can
be represented exactly as double-precision floating-
point numbers than as integers in R; (2) it is easier to
manipulate the individual numerals than to perform
repeated integer arithmetic to extract digits; and (3)
having the numerals in character form allows me to
take advantage of R’s ability to index vectors by ele-
ment names (see below).
I therefore defined the following function to con-
vert a number to a vector of characters containing the
numerals composing the number:
> makeDigits <- function(x)
+ strsplit(as.character(x), "")[[1]]
Here are some examples of the use of this func-
tion:
> makeDigits(123456)
[1] "1" "2" "3" "4" "5" "6"
> makeDigits(-123456)
[1] "-" "1" "2" "3" "4" "5" "6"
> makeDigits(1000000000)
[1] "1" "e" "+" "0" "9"
Notice the problems revealed by the second and
third examples: It’s necessary to make provision for
negative numbers, and R wants to render certain
numbers in scientific notation.1By setting the scipen
(“scientific notation penalty”) option to a large num-
ber, we can avoid the second problem:
> options(scipen=100)
> makeDigits(1000000000)
[1] "1" "0" "0" "0" "0" "0" "0" "0" "0" "0"
It also seemed useful to have a function that con-
verts a vector of numerals in character form back into
a number:
> makeNumber <- function(x)
+ as.numeric(paste(x, collapse=""))
> makeNumber(c("1", "2", "3", "4", "5"))
[1] 12345
Finally, by way of preparation, I constructed sev-
eral vectors of number words:
> ones <- c("zero", "one", "two", "three",
+ "four", "five", "six", "seven",
+ "eight", "nine")
> teens <- c("ten", "eleven", "twelve",
+ "thirteen", "fourteen", "fifteen",
+ "sixteen", " seventeen", "eighteen",
+ "nineteen")
> names(ones) <- names(teens) <- 0:9
> tens <- c("twenty", "thirty", "forty",
+ "fifty", "sixty", "seventy", "eighty",
+ "ninety")
> names(tens) <- 2:9
> suffixes <- c("thousand,", "million,",
+ "billion,", "trillion,")
Because the names of the elements of the first
three vectors are numerals, they can conveniently be
indexed; for example:
> ones["5"]
5
"five"
> teens["3"]
3
"thirteen"
> tens["7"]
7
"seventy"
The vector of suffixes includes a comma after
each word.
Figure 1shows a function for converting a single
integer to words; I’ve added line numbers to make it
easier to describe how the function works:
And here are some examples of its use, wrapping
long lines of output to fit on the page:
> number2words(123456789)
[1] "one hundred twenty-three million,
four hundred fifty-six thousand,
seven hundred eighty-nine"
> number2words(-123456789)
[1] "minus one hundred twenty-three million,
four hundred fifty-six thousand,
seven hundred eighty-nine"
> number2words(-123456000)
[1] "minus one hundred twenty-three million,
four hundred fifty-six thousand"
I believe that the first five lines of the function are
essentially self-explanatory. The rest of the function
probably requires some explanation, however:
[6] If the number is composed of a single digit,
then we can find the answer by simply in-
dexing into the vector ones; the function
as.vector is used to remove the name of
(i.e., the numeral labelling) the selected el-
ement.
[7-9] If the number is composed of two digits
and is less than or equal to 19, then we can
get the answer by indexing into teens with
the last digit (i.e., the second element of the
digits vector). If the number is 20 or larger,
then we need to attach the tens digit to the
ones digit, with a hyphen in between. If,
1I don’t want to mislead the reader: I discovered these and other problems the hard way, when they surfaced as bugs. The account
here is a reconstruction that avoids my missteps. I can honestly say, however, that it took me much longer to write this column explaining
how the program works than to write the original program. Moreover, in the process of writing up the program, I saw several ways to
improve it, especially in clarity — a useful lesson.
R News ISSN 1609-3631

Vol. 5/1, May 2005 53
[ 1] number2words <- function(x){
[ 2] negative <- x < 0
[ 3] x <- abs(x)
[ 4] digits <- makeDigits(x)
[ 5] nDigits <- length(digits)
[ 6] result <- if (nDigits == 1) as.vector(ones[digits])
[ 7] else if (nDigits == 2)
[ 8] if (x <= 19) as.vector(teens[digits[2]])
[ 9] else trim(paste(tens[digits[1]], "-", ones[digits[2]], sep=""))
[10] else if (nDigits == 3) {
[11] tail <- makeNumber(digits[2:3])
[12] if (tail == 0) paste(ones[digits[1]], "hundred")
[13] else trim(paste(ones[digits[1]], "hundred", number2words(tail)))
[14] }
[15] else {
[16] nSuffix <- ((nDigits + 2) %/% 3) - 1
[17] if (nSuffix > length(suffixes) || nDigits > 15)
[18] stop(paste(x, "is too large!"))
[19] pick <- 1:(nDigits - 3*nSuffix)
[20] trim(paste(number2words(makeNumber(digits[pick])),
[21] suffixes[nSuffix], number2words(makeNumber(digits[-pick]))))
[22] }
[23] if (negative) paste("minus", result) else result
[24] }
Figure 1: A function to convert a single integer into words.
however, the ones digit is 0, ones["0"] is
"zero", and thus we have an embarrassing
result such as "twenty-zero". More gener-
ally, the program can produce spurious hy-
phens, commas, spaces, and the strings ",
zero" and "-zero" in appropriate places.
My solution was to write a function to trim
these off:
trim <- function(text){
gsub("(^\ *)|((\ *|-|,\ zero|-zero)$)",
"", text)
}
The trim function makes use of R’s ability
to process “regular expressions.” See Lum-
ley (2003) for a discussion of the use of regu-
lar expressions in R.
[10-14] If the number consists of three digits,
then the first digit is used for hundreds, and
the remaining two digits can be processed as
an ordinary two-digit number; this is done
by a recursive call to number2words2— un-
less the last two digits are 0, in which case,
we don’t need to convert them into words.
The hundreds digit is then pasted onto the
representation of the last two digits, and the
result is trimmed. Notice that makeNumber
is used to put the last two digits back into a
number (called tail).
[15-22] Finally, if the number contains more than
three digits, we’re into the realm of thou-
sands, millions, etc. The computation on line
[16] determines with which power of 1000
we’re dealing. Then, if the number is not too
large, the appropriate digits are stripped off
from the left of the number and attached to
the proper suffix; the remaining digits to the
right are recomposed into a number and pro-
cessed with a recursive call, to be attached at
the right.
[23] If the original number was negative, the
word "minus" is pasted onto the front before
the result is returned.
The final function, called numbers2words (shown
in Figure 2), adds some bells and whistles: The vari-
ous vectors of names are defined locally in the func-
tion; the utility functions makeDigits,makeNumbers,
and trim, are similarly defined as local functions;
and the function number2words, renamed helper,
is also made local. Using a helper function rather
than a recursive call permits efficient vectorization,
via sapply, at the end of numbers2words. Were
2It’s traditional in S to use Recall for a recursive function call, but I’m not fond of this convention, and I don’t see an argument for it
here: It’s unlikely that number2words will be renamed, and in any event, it will become a local function in the final version of the program
(see below).
R News ISSN 1609-3631

Vol. 5/1, May 2005 54
numbers2words to call itself recursively, the local def-
initions of objects (such as the vector ones and the
function trim) would be needlessly recomputed at
each call, rather than only once. Because of R’s lex-
ical scoping, objects defined in the environment of
numbers2words are visible to helper. For more on
recursion in R, see Venables (2001).
numbers2words includes a couple of additional
features. First, according to the Oxford English Dic-
tionary, the definition of “billion” differs in the U.S.
and (traditionally) in Britain: “1. orig. and still
commonly in Great Britain: A million millions. (=
U.S. trillion.) ... 2. In U.S., and increasingly in
Britain: A thousand millions.” Thus, if the argument
billion is set to "UK", a different vector of suffixes
is used. Moreover, provision is made to avoid awk-
ward translations that repeat the word “million,”
such as “five thousand million, one hundred million,
... ,” which is instead, and more properly rendered as
“five thousand, one hundred million, ... .”
Second, Bill Venables tells me that outside of the
U.S., it is common to write or speak a number such
101 as “one hundred and one” rather than as “one
hundred one.” (Both of these phrases seem correct
to me, but as I said, I’m hopelessly confused about
international variations in English.) I have there-
fore included another argument, called and, which
is pasted into the number at the appropriate point.
By default, this argument set is to "" when billion
is "US" and to "and" when billion is "UK".
Some examples, again wrapping long lines of
output:
> numbers2words(c(1234567890123, -0123, 1000))
[1] "one trillion,
two hundred thirty-four billion,
five hundred sixty-seven million,
eight hundred ninety thousand,
one hundred twenty-three"
[2] "minus one hundred twenty-three"
[3] "one thousand"
> numbers2words(c(1234567890123, -0123, 1000),
+ billion="UK")
[1] "one billion,
two hundred and thirty-four thousand,
five hundred and sixty-seven million,
eight hundred and ninety thousand,
one hundred and twenty-three"
[2] "minus one hundred and twenty-three"
[3] "one thousand"
> numbers2words(c(1234567890123, -0123, 1000),
+ and="and")
[1] "one trillion,
two hundred and thirty-four billion,
five hundred and sixty-seven million,
eight hundred and ninety thousand,
one hundred and twenty-three"
[2] "minus one hundred and twenty-three"
[3] "one thousand"
Finally, a challenge to the reader: At present,
numbers2words rounds its input to whole numbers.
Modify the program so that it takes a digits argu-
ment (with default 0), giving the number of places to
the right of the decimal point to which numbers are
to be rounded, and then make provision for translat-
ing such numbers (e.g., 1234567.890) into words.
John Fox
Sociology, McMaster University
jfox@mcmaster.ca
Bibliography
T. Lumley. Programmer’s niche: Little bits of string.
R News, 3(3):40–41, December 2003. URL http:
//CRAN.R-project.org/doc/Rnews/.53
B. Venables. Programmer’s niche. R News, 1(1):27–
30, January 2001. URL http://CRAN.R-project.
org/doc/Rnews/.54
R News ISSN 1609-3631

Vol. 5/1, May 2005 55
numbers2words <- function(x, billion=c("US", "UK"),
and=if (billion == "US") "" else "and"){
billion <- match.arg(billion)
trim <- function(text){
gsub("(^\ *)|((\ *|-|,\ zero|-zero)$)", "", text)
}
makeNumber <- function(x) as.numeric(paste(x, collapse=""))
makeDigits <- function(x) strsplit(as.character(x), "")[[1]]
helper <- function(x){
negative <- x < 0
x <- abs(x)
digits <- makeDigits(x)
nDigits <- length(digits)
result <- if (nDigits == 1) as.vector(ones[digits])
else if (nDigits == 2)
if (x <= 19) as.vector(teens[digits[2]])
else trim(paste(tens[digits[1]], "-", ones[digits[2]], sep=""))
else if (nDigits == 3) {
tail <- makeNumber(digits[2:3])
if (tail == 0) paste(ones[digits[1]], "hundred")
else trim(paste(ones[digits[1]], trim(paste("hundred", and)),
helper(tail)))
}
else {
nSuffix <- ((nDigits + 2) %/% 3) - 1
if (nSuffix > length(suffixes) || nDigits > 15)
stop(paste(x, "is too large!"))
pick <- 1:(nDigits - 3*nSuffix)
trim(paste(helper(makeNumber(digits[pick])),
suffixes[nSuffix], helper(makeNumber(digits[-pick]))))
}
if (billion == "UK"){
words <- strsplit(result, " ")[[1]]
if (length(grep("million,", words)) > 1)
result <- sub(" million, ", ", ", result)
}
if (negative) paste("minus", result) else result
}
opts <- options(scipen=100)
on.exit(options(opts))
ones <- c("zero", "one", "two", "three", "four", "five", "six", "seven",
"eight", "nine")
teens <- c("ten", "eleven", "twelve", "thirteen", "fourteen", "fifteen",
"sixteen", " seventeen", "eighteen", "nineteen")
names(ones) <- names(teens) <- 0:9
tens <- c("twenty", "thirty", "forty", "fifty", "sixty", "seventy", "eighty",
"ninety")
names(tens) <- 2:9
suffixes <- if (billion == "US")
c("thousand,", "million,", "billion,", "trillion,")
else
c("thousand,", "million,", "thousand million,", "billion,")
x <- round(x)
if (length(x) > 1) sapply(x, helper) else helper(x)
}
Figure 2: A function to convert a vector of integers into a vector of strings containing word-equivalents of the
integers.
R News ISSN 1609-3631

Vol. 5/1, May 2005 56
Book Review of
Julian J. Faraway: Linear Models with R
Chapman & Hall/CRC, Boca Raton FL, USA, 2005
229 pages, ISBN 1-58488-425-8
http://www.stat.lsa.umich.edu/∼faraway/LMR
This book is useful to serve for the practical as-
pects of an advanced undergraduate linear regres-
sion course. Regarding the potential readership, the
Author writes in the preface that the book is “not in-
troductory” and that it “presumes some knowledge
of basic statistical theory and practice. Readers are
expected to know the essentials of statistical infer-
ence such as estimation, hypothesis testing and con-
fidence intervals. A basic knowledge of data anal-
ysis is presumed. Some linear algebra and calculus
are also required.” Thus this book is most suitable
for undergraduate statistics majors at least half way
through their degrees. The book needs to be accom-
panied by a theoretical book, such as Seber and Lee
(2003). A somewhat similar competitor to the book
is Fox (2002).
With a large number (16) of chapters in this small-
ish book, most of them are short and mention ideas
briefly. Of course, some chapters are on core practical
topics such as residual diagnostics, transformations,
weighted and generalized least squares, ANOVA
(factorial designs, block designs), ANCOVA, vari-
able selection techniques, and inference. A good fea-
ture of the book is that more ‘optional’ topics are
covered, include missing values, regression splines,
robust regression (M-estimation and least trimmed
squares), permutation tests, shrinkage methods such
as partial least squares, measurement error mod-
els including SIMEX, latin squares and iteratively
reweighted least squares. Most instructors are un-
likely to be familiar with all these topics, and the
book does a good job in giving a very gentle intro-
duction to these topics. The reader is usually referred
to references at the back for further reading.
The book has two small appendices. The first de-
scribes R installation, functions and data, while the
second is a quick introduction to R. The examples in
the book are based on R 1.9.0. In my copy there were
a few font problems, e.g., p.189 has (αβ)i j instead of
(αβ)i j.
The tone of the book is that of an informal tutorial
with some necessary theory interdispersed through-
out. Explanations are good overall, and the math-
ematical notation is quite standard. The Author’s
website serves as the repository for the book’s pack-
age, called faraway, and includes errata, R com-
mands and data sets (the package is also on CRAN.)
The Author writes “Data analysis cannot be learned
without actually doing it” and this is facilitated
by practical exercises (which are usually short and
sharp) at the end of each chapter. Solutions appear
unavailable.
The book centers on lm() and assumes familiar-
ity with the S language, e.g., I found it referred to “.”
in a formula without reminding or telling the reader
that it means all variables in the data frame other
than the response. The book is rather unsuitable as a
reference book, for example, summation constraints
J
∑
j=1
αj=0 is a popular parameterization for a factor
with Jlevels. The function contr.sum is alluded to
but not mentioned directly in the book, nor how to
switch between different parameterizations. Ideally,
a handy summary of topics such as the Wilkinson-
Rogers operators (e.g., *,/,:) and all the different
contrast options available should be put into an ap-
pendix for fast and easy reference. This would make
Chambers and Hastie (1991) less necessary for stu-
dents to buy or refer to. The book makes good and
frequent use of basic graphics with a liberal sprin-
kling of plots everywhere, and many examples.
Altogether, there are 27 data sets used in the book;
the large majority of these are small. Even though
the figures are in black and white, the reader is not
encouraged to use colors in the plotting—something
very useful on a computer screen. It would be good
also that the reader be reminded how to create PDF
files of figures (especially if the user uses Linux),
which is useful if the user writes reports.
The book has many good points but I picked up
a few bad points. For example, the use of extractor
functions is not always taken where possible, e.g.,
coef(fit) should be used instead of fit$coef. Also,
numerous examples use cut-and-paste from previ-
ous output rather than extracting quantities from ob-
jects, for example, p.20, 60.975 is used rather than
mdls$sigma. I would have liked the paragraph on
additive models expanded to make the reader aware
of how to check the linearity assumptions using lm()
and regression splines.
I thought there were a number of omissions.
The prediction problem should be mentioned (in
S-PLUS data-dependent terms such as scale(x),
poly(x, 2) and bs(x) give problems, whereas in
R, a data-dependent function called inside another
function are problematic, e.g., poly(scale(x), 2)).
Other omissions include not mentioning computa-
tional details such as the QR-algorithm and not us-
ing more advanced graphics such as those found in
the lattice package.
Although I agree that the book is “not in-
troductory”, neither is it advanced. For exam-
ple, a broken stick regression model is fitted in
R News ISSN 1609-3631

Vol. 5/1, May 2005 57
Section 7.2.1. Improvements to this example in-
clude using coef() instead of $coef, and using
the predict generic function, i.e., predict(gb,
data.frame(pop15=seq(20,48,by=1))). The two
(dotted) lines described in the text actually do not
show up on the plot. Furthermore, although it is in-
structional to create two basis functions lhs() and
rhs(), the reader is not informed that bs(pop15,
degree=1, df=1, knot=35) would be equivalent.
In conclusion, the book is quite suitable to serve
for the practical component of an advanced under-
graduate course in linear models to reasonably pre-
pared students. A sequel on generalized linear mod-
els and extensions would be a natural next step!
Bibliography
J. Fox. An R and S-Plus Companion to Applied Regres-
sion, 2002. Thousand Oaks, CA: Sage Publications.
56
G. A. F. Seber and A. J. Lee. Linear Regression Analysis,
2nd Edition, 2003. New York: Wiley. 56
Chambers, J. M. and Hastie, T. J. Statistical Models
in S, 1991. Pacific Grove, Calif.: Wadsworth &
Brooks/Cole. 56
Thomas Yee
University of Auckland, New Zealand
yee@stat.auckland.ac.nz
R Foundation News
by Bettina Grün
Donations and new members
Donations
Adelchi Azzalini (Italy)
BC Cancer Agency, Vancouver (Canada)
David W. Crawford (USA)
Peter L. Flom (USA)
Google Inc., Mountain View, California (USA)
Faculty of Economics, University of Groningen
(Netherlands)
Shigeru Mase (Japan)
Merck and Co., Inc. (USA)
Network Theory Ltd, Bristol (United Kingdom)
Stanford University, California (USA)
Douglas Wilson (Canada)
Ivo Welch (USA)
New benefactors
Burns Statistics Ltd., London, U.K.
Loyalty Matrix Inc., California, USA
Statisticon AB, Uppsala, Sweden
Merck and Co., Inc., USA
New supporting institutions
Baxter Healthcare Corp., California, USA
Department of Mathematics and Statistics, Utah
State University, USA
Department of Statistics, Iowa State University, USA
Dipartimento di Statistica, Università Ca’ Foscari di
Venezia, Italy
School of Economics and Finance, Victoria Univer-
sity of Wellington, New Zealand
New supporting members
Claudio Agostinelli (Italy)
Roger J. Bos (USA)
Dianne Cook (USA)
Gregor Gorjanc (Slovenia)
Ivan Kojadinovic (France)
Iago Mosqueira (Spain)
Jonas Ranstam (Sweden)
Christian Schulz (Gerrmany)
Mario Walter (Germany)
Bettina Grün
Technische Universität Wien, Austria
Bettina.Gruen@ci.tuwien.ac.at
Changes in R
by the R Core Team
User-visible changes
• box plots by boxplot() or bxp() now have the
median line three times the normal line width
in order to distinguish it from the quartile ones.
• Unix-alike versions of R can now be used in
UTF-8 locales on suitably equipped OSes. See
the internationalization section below.
R News ISSN 1609-3631

Vol. 5/1, May 2005 58
• The meaning of ’encoding’ for a connection has
changed: See the internationalization section
below.
• There has been some rationalization of the for-
mat of warning/error messages, to make them
easier to translate. Generally names of func-
tions and arguments are single-quoted, and
classes double-quoted.
• Reading text files with embedded "\" (as in
Windows file names) may now need to use
scan(* , allowEscapes = FALSE), see also
below.
New features
• %% now warns if its accuracy is likely to be
affected by lack of precision (as in 1e18 %%
11, the unrealistic expectation of PR#7409), and
tries harder to return a value in range when it
is.
•abbreviate() now warns if used with non-
ASCII chars, as the algorithm is designed for
English words.
• The default methods for add1() and drop1()
check for changes in the number of cases in use.
The "lm" and "glm" methods for add1() quoted
the <none> model on the original fitted values
when using (with a warning) a smaller set of
cases for the expanded models.
• Added alarm() function to generate a bell or
beep or visual alert.
•all/any() now attempt to coerce their argu-
ments to logical, as documented in the Blue
Book. This means e.g. any(list()) works.
• New functions for multivariate linear
models: anova.mlm(),SSD(),estVar(),
mauchley.test() (for sphericity).
vcov() now does something more sensible for
"mlm" class objects.
•as.data.frame.table() has a new argument
’responseName’ (contributed by Bill Venables).
•as.dist() and cophenetic() are now generic,
and the latter has a new method for objects of
class "dendrogram".
•as.ts() is now generic.
•binomial() has a new "cauchit" link (sug-
gested by Roger Koenker).
•chisq.test() has a new argument ’rescale.p’.
It is now possible to simulate (slowly) the P
value also in the 1D case (contributed by Rolf
Turner).
•choose(n,k) and lchoose(.) now also work
for arbitrary (real) n in accordance with the
general binomial theorem. choose(*,k) is
more accurate (and faster) for small k.
• Added colorRamp() and colorRampPalette()
functions for color interpolation.
•colSums()/rowSums() now allow arrays with a
zero-length extent (requested by PR#7775).
•confint() has stub methods for classes "glm"
and "nls" that invoke those in package MASS.
This avoids using the "lm" method for "glm"
objects if MASS is not attached.
confint() has a default method using asymp-
totic normality.
•contr.SAS() has been moved from the ’nlme’
package to the ’stats’ package.
• New function convertColors() maps between
color spaces. colorRamp() uses it.
• The cov() function in the non-Pearson cases
now ranks data after removal of missing val-
ues, not before. The pairwise-complete method
should now be consistent with cor.test. (Code
contributed by Shigenobu Aoki.)
• Added delayedAssign() function to replace
delay(), which is now deprecated.
•dir.create() has a new argument ’recursive’
serving the same purpose as Unix’s mkdir -p.
•do.call() now takes either a function or a
character string as its first argument. The sup-
plied arguments can optionally be quoted.
•duplicated() and unique() now accept "list"
objects, but are fast only for simple list objects.
•ecdf() now has jumps of the correct size (a
multiple of 1/n) if there are ties. (Wished by
PR#7292).
•eff.aovlist() assumed orthogonal contrasts
for any term with more than one degree of free-
dom: this is now documented and checked for.
Where each term only occurs in only one stra-
tum the efficiencies are all one: this is detected
and orthogonal contrasts are not required.
• New function encodeString() to encode char-
acter strings in the same way that printing
does.
R News ISSN 1609-3631

Vol. 5/1, May 2005 59
•file("clipboard") now work for reading the
primary selection on Unix-alikes with an active
X11 display. (It has long worked for reading
and writing under Windows.) The secondary
selection can also be read: see ?file.
file() now allows mode "w+b" as well as
"w+".
•file.append() has been tuned, including for
the case of appending many files to a single file.
• Functions flush.console() and select.list()
are now available on all platforms. There is a
Tcl/Tk-based version of select.list() called
tk_select.list() in package tcltk.
•gc() now reports maximum as well as current
memory use.
• A new function getGraphicsEvent() has been
added which will allow mouse or keyboard in-
put from a graphics device. (NB: currently only
the Windows screen device supports this func-
tion. This should improve before the 2.1.0 re-
lease.)
• New functions gray.colors()/grey.colors()
for gray color palettes.
•grep(),gsub(),sub() and regexpr() now al-
ways attempt to coerce their ’pattern’, ’x’, ’re-
placement’ and ’text’ arguments to character.
Previously this was undocumented but done
by [g]sub() and regexpr() for some values of
their other arguments. (Wish of PR#7742.)
•gsub/sub() have a new ’fixed’ method.
• New function hcl() for creating colors for a
given hue, chroma and luminance (i.e. percep-
tual hsv).
•isTRUE() convenience function to be used for
programming.
•kmeans() now returns an object of class
"kmeans" which has a print() method.
Two alternative algorithms have been imple-
mented.
If the number of centres is supplied, it has a
new option of multiple random starts.
• The limits on the grid size in layout() are now
documented, and have been raised somewhat
by using more efficient internal structures.
•legend() now accepts positioning by keyword,
e.g. "topleft", and can put a title within the
legend. (Suggested by Elizabeth Purdom in
PR#7400.)
•mahalanobis() now has a ’...’ argument which
is passed to solve() for computing the inverse
of the covariance matrix, this replaces the for-
mer ’tol.inv’ argument.
•menu() uses a multi-column layout if possible
for more than 10 choices.
menu(graphics = TRUE) is implemented
on most platforms via select.list() or
tk_select.list().
• New function message() in ’base’ for gener-
ating "simple" diagnostic messages, replacing
such a function in the ’methods’ package.
•na.contiguous() is now (S3) generic with first
argument renamed to ’object’.
• New function normalizePath() to find canon-
ical paths (and on Windows, canonical names
of components).
• The default in options("expressions") has
been increased to 5000, and the maximal set-
table value to 500000.
•p.adjust() has a new method "BY".
•pbeta() now uses a different algorithm for
large values of at least one of the shape pa-
rameters, which is much faster and is accurate
and reliable for very large values. (This affects
pbinom(),pf(),qbeta() and other functions
using pbeta at C level.)
•pch="." now by default produces a rectangle
at least 0.01" per side on high-resolution de-
vices. (It used to be one-pixel square even
on high-resolution screens and Windows print-
ers, but 1/72" on postscript() and pdf() de-
vices.) Additionally, the size is now scalable by
’cex’; see ?points and note that the details are
subject to change.
•pdf() now responds to the ’paper’ and ’page-
centre’ arguments. The default value of ’paper’
is "special" for backward-compatibility (this is
different from the default for postscript()).
•plot.data.frame() tries harder to produce
sensible plots for non-numeric data frames
with one or two columns.
• The predict() methods for "prcomp" and
"princomp" now match the columns of ’new-
data’ to the original fit using column names if
these are available.
• New function recordGraphics() to encapsu-
late calculations and graphics output together
on graphics engine display list. To be used with
care.
R News ISSN 1609-3631

Vol. 5/1, May 2005 60
• New function RSiteSearch() to query R-
related resources on-line (contributed by
Jonathan Baron and Andy Liaw).
•scan() arranges to share storage of duplicated
character strings read in: this can dramati-
cally reduce the memory requirements for large
character vectors which will subsequently be
turned into factors with relatively few levels.
For a million items this halved the time and re-
duced storage by a factor of 20.
scan() has a new argument ’allowEscapes’
(default TRUE) that controls when C-style es-
capes in the input are interpreted. Previously
only \n and \r were interpreted, and then only
within quoted strings when no separator was
supplied.
scan() used on an open connection now
pushes back on the connection its private
‘ungetc’ and so is safer to use to read partial
lines.
•scatter.smooth() and loess.smooth() now
handle missing values in their inputs.
•seq.Date() and seq.POSIXt() now allow ’to’
to be before ’from’ if ’by’ is negative.
•sprintf() has been enhanced to allow the
POSIX/XSI specifiers like "%2$6d", and also ac-
cepts "%x" and "%X".
sprintf() does limited coercion of its argu-
ments.
sprintf() accepts vector arguments and op-
erates on them in parallel (after re-cycling if
needed).
• New function strtrim() to trim character vec-
tors to a display width, allowing for double-
width characters in multi-byte character sets.
•subset() now has a method for matrices, sim-
ilar to that for data frames.
• Faster algorithm in summaryRprof().
•sunflowerplot() has new arguments ’col’ and
’bg’.
•sys.function() now has argument ’which’ (as
has long been presaged on its help page).
•Sys.setlocale("LC_ALL", ) now only sets
the locale categories which R uses, and
Sys.setlocale("LC_NUMERIC", ) now gives a
warning (as it can cause R to malfunction).
•unclass() is no longer allowed for environ-
ments and external pointers (since these cannot
be copied and so unclass() was destructive of
its argument). You can still change the "class"
attribute.
• File-name matching is no longer case-
insensitive with unz() connections, even on
Windows.
• New argument ’immediate.’ to warning() to
send an immediate warning.
• New convenience wrappers write.csv() and
write.csv2().
• There is a new version for write.table()
which is implemented in C. For simple matri-
ces and data frames this is several times faster
than before, and uses negligible memory com-
pared to the object size.
The old version (which no longer coerces a ma-
trix to a data frame and then back to a matrix)
is available for now as write.table0().
• The functions xinch(),yinch(), and xyinch()
have been moved from package ’grDevices’
into package ’graphics’.
• Plotmath now allows underline in expressions.
(PR#7286, contributed by Uwe Ligges.)
• BATCH on Unix no longer sets --gui="none"
as the X11 module is only loaded if needed.
• The X11 module (and the hence X11(),jpeg()
and png() devices and the X-based dataentry
editor) is now in principle available under all
Unix GUIs except --gui="none", and this is re-
flected in capabilities().
capabilities("X11") determines if an X
server can be accessed, and so is more likely to
be accurate.
• Printing of arrays now honours the ’right’ ar-
gument if there are more than two dimensions.
• Tabular printing of numbers now has headers
right-justified, as they were prior to version
1.7.0 (spotted by Rob Baer).
• Lazy-loading databases are now cached in
memory at first use: this enables R to run much
faster from slow file systems such as USB flash
drives. There is a small (less than 2Mb) increase
in default memory usage.
• The implicit class structure for numeric vectors
has been changed, so that integer/real vectors
try first methods for class "integer"/"double"
and then those for class "numeric".
The implicit classes for matrices and arrays
have been changed to be "matrix"/"array" fol-
lowed by the class(es) of the underlying vec-
tor.
R News ISSN 1609-3631

Vol. 5/1, May 2005 61
•splines::splineDesign() now allows the
evaluation of a B-spline basis everywhere in-
stead of just inside the "inner" knots, by setting
the new argument ‘outer.ok = TRUE’.
• Hashing has been tweaked to use half as much
memory as before.
• Readline is not used for tilde expansion when
R is run with --no-readline, nor from embed-
ded applications. Then " name" is no longer ex-
panded, but " " still is.
• The regular expression code is now based on
that in glibc 2.3.3. It has stricter conformance
to POSIX, so metachars such as + * may need
to be escaped where before they did not (but
could have been).
• New encoding ’TeXtext.enc’ improves the way
postscript() works with Computer Modern
fonts.
• Replacement in a non-existent column of a data
frame tries harder to create a column of the cor-
rect length and so avoid a corrupt data frame.
• For Windows and readline-based history, the
saved file size is re-read from R_HISTSIZE im-
mediately before saving.
• Collected warnings during start-up are now
printed before the initial prompt rather than af-
ter the first command.
• Changes to package ’grid’:
–preDrawDetails(),drawDetails(), and
postDrawDetails() methods are now
recorded on the graphics engine display
list. This means that calculations within
these methods are now run when a device
is resized or when output is copied from
one device to another.
–Fixed bug in grid.text() when ’rot’ ar-
gument has length 0. (privately reported
by Emmanuel Paradis)
–New getNames() function to return just
the names of all top-level grobs on the dis-
play list.
–Recording on the grid display list is
turned off within preDrawDetails(),
drawDetails(), and postDrawDetails()
methods.
–Grid should recover better from errors or
user-interrupts during drawing (i.e., not
leave you in a strange viewport or with
strange graphical parameter settings).
–New function grid.refresh() to redraw
the grid display list.
–New function grid.record() to capture
calculations with grid graphics output.
–grobWidth and grobHeight ("grobwidth"
and "grobheight" units) for primitives
(text, rects, etc, ...) are now calculated
based on a bounding box for the relevant
grob.
NOTE: this has changed the calculation of
the size of a scalar rect (or circle or lines).
–New arguments ’warn’ and ’wrap’ for
function grid.grab()
–New function grid.grabExpr() which
captures the output from an expression
(i.e., not from the current scene) without
doing any drawing (i.e., no impact on the
current scene).
–upViewport() now (invisibly) returns the
path that it goes up (suggested by Ross
Ihaka).
–The ’gamma’ gpar has been deprecated
(this is a device property not a property
of graphical objects; suggested by Ross
Ihaka).
–New ’lex’ gpar; a line width multiplier.
–grid.text() now handles any language
object as mathematical annotation (in-
stead of just expressions).
–plotViewport() has default value for
’margins’ argument (that match the de-
fault value for par(mar)).
–The ’extension’ argument to dataViewport()
can now be vector, in which case the first
value is used to extend the xscale and the
second value is used to extend the y scale.
(suggested by Ross Ihaka).
–All ’just’ arguments (for viewports, lay-
outs, rectangles, text) can now be numeric
values (typically between 0 [left] and 1
[right]) as well as character values ("left",
"right", ...).
For rectangles and text, there are addi-
tional ’hjust’ and ’vjust’ arguments which
allow numeric vectors of justification in
each direction (e.g., so that several pieces
of text can have different justifications).
(suggested by Ross Ihaka)
–New ’edits’ argument for grid.xaxis()
and grid.yaxis() to allow specification
of on-the-fly edits to axis children.
–applyEdit(x, edit) returns x if target of
edit (i.e., child specified by a gPath) can-
not be found.
–Fix for calculation of length of
max/min/sum unit. Length is now (cor-
rectly) reported as 1 (was reported as
length of first arg).
R News ISSN 1609-3631

Vol. 5/1, May 2005 62
–Viewport names can now be any string
(they used to have to be a valid R symbol).
–The ’label’ argument for grid.xaxis()
and grid.yaxis() can now also be a lan-
guage object or string vector, in which
case it specifies custom labels for the tick
marks.
Internationalization
• Unix-alike versions of R can now be used in
UTF-8 and other multi-byte locales on suit-
ably equipped OSes if configured with option
--enable-mbcs (which is the default). [The
changes to font handling in the X11 module
are based on the Japanization patches of Eiji
Nakama.]
Windows versions of R can be used in ‘East
Asian’ locales on suitable versions of Windows.
See the ’Internationalization’ chapter in the ’In-
stallation and Administration’ manual.
• New command-line flag --encoding to specify
the encoding to be assumed for stdin (but not
for a console).
• New function iconv() to convert charac-
ter vectors between encodings, on those
OSes which support this. See the new
capabilities("iconv").
• The meaning of ’encoding’ for a connection has
changed: it now allows any charset encoding
supported by iconv on the platform, and can
re-encode output as well as input.
As the new specification is a character string
and the old was numeric, this should not cause
incorrect operation.
• New function localeToCharset() to
find/guess encoding(s) from the locale name.
•nchar() returns the true number of bytes
stored (including any embedded nuls), this be-
ing 2 for missing values. It has an optional ar-
gument ’type’ with possible non-default values
"chars" and "width" to give the number of char-
acters or the display width in columns.
• Characters can be entered in hexadecimal as
e.g. \x9c, and in UTF-8 and other multibyte
locales as \uxxxx,\u{xxxx},\Uxxxxxxxx or
\U{xxxxxxxx}. Non-printable Unicode char-
acters are displayed C-style as \uxxxx or
\Uxxxxxxxx.
• LC_MONETARY is set to the locale, which af-
fects the result of Sys.localeconv(), but noth-
ing else in R itself. (It could affect add-on pack-
ages.)
•source() now has an ’encoding’ argument
which can be used to make it try out vari-
ous possible encodings. This is made use of
by example() which will convert (non-UTF-8)
Latin-1 example files in a UTF-8 locale.
•read/writeChar() work in units of characters,
not bytes.
•.C() now accepts an ENCODING= argument
where re-encoding is supported by the OS. See
‘Writing R Extensions’.
• delimMatch (tools) now reports match posi-
tions and lengths in units of characters, not
bytes. The delimiters can be strings, not just
single ASCII characters.
• .Rd files can indicate via a \encoding{} argu-
ment the encoding that should be assumed for
non-ASCII characters they contain.
• Phrases in .Rd files can be marked by \enc{}{}
to show a transliteration to ASCII for use in e.g.
text help.
• The use of ’pch’ in points() now allows for
multi-byte character sets: in such a locale a
glyph can either be specified as a multi-byte
single character or as a number, the Unicode
point.
• New function l10n_info() reports on aspects
of the locale/charset currently in use.
•scan() is now aware of double-byte locales
such as Shift-JIS in which ASCII characters can
occur as the second (’trail’) byte.
• Functions sQuote() and dQuote() use the Uni-
code directional quotes if in a UTF-8 locale.
• The infrastructure is now in place for C-level
error and warning messages to be translated
and used on systems with Native Language
Support. This has been used for the startup
message in English and to translate American-
isms such as ’color’ into English: translations
to several other languages are under way, and
some are included in this release.
See ’Writing R Extensions’ for how to make use
of this in a package: all the standard packages
have been set up to do translation, and the ’lan-
guage’ ’en@quot’ is implemented to allow Uni-
code directional quotes in a UTF-8 locale.
• R-level stop(),warning() and message()
messages can be translated, as can other mes-
sages via the new function gettext(). Tools
xgettext() and xgettext2pot() are provided
in package tools to help manage error mes-
sages.
gettextf() is a new wrapper to call sprintf()
using gettext() on the format string.
R News ISSN 1609-3631

Vol. 5/1, May 2005 63
• Function ngettext() allows the management
of singular and plural forms of messages.
Utilities
• New functions mirror2html() and
checkCRAN().
• R CMD check has a new option
’--use-valgrind’.
• R CMD check now checks that Fortran and C++
files have LF line endings, as well as C files.
It also checks Makevars[.in] files for portable
compilation flags.
• R CMD check will now work on a source tarball
and prints out information about the version of
R and the package.
•tools:::.install_package_code_files()
(used to collate R files when installing pack-
ages) ensures files are separated by a line feed.
•vignette() now returns an object of class "vi-
gnette" whose print() method opens the cor-
responding PDF file. The edit() method can
be used to open the code of the vignette in an
editor.
• R CMD INSTALL on Unix has a new option
’--build’ matching that on Windows, to pack-
age as tarball the installed package.
• R CMD INSTALL on Unix can now install bi-
nary bundles.
• R CMD build now changes src files to LF line
endings if necessary.
• R CMD build now behaves consistently be-
tween source and binary builds: in each case
it prepares a source directory and then either
packages that directory as a tarball or calls R
CMD INSTALL -build on the prepared sources.
This means that R CMD build --binary now
respects .Rbuildignore and will rebuild vi-
gnettes (unless the option --no-vignettes is
used). For the latter, it now installs the current
sources into a temporary library and uses that
version of the package/bundle to rebuild the
vignettes.
• R CMD build now reports empty directories in
the source tree.
• New function write_PACKAGES() in package
’tools’ to help with preparing local package
repositories. (Based on a contribution by Uwe
Ligges.) How to prepare such repositories is
documented in the ’R Installation and Admin-
istration’ manual.
•package.skeleton() adds a bit more to DE-
SCRIPTION.
• Sweave changes:
–\usepackage[nogin]{Sweave} in the
header of an Sweave file suppresses auto-
setting of graphical parameters such as
the width of the graphics output.
–The new \SweaveInput{} command
works similar to LaTeX’s \input{} com-
mand.
–Option value strip.white=all strips all
blank lines from the output of a code
chunk.
–Code chunks with eval=false are com-
mented out by Stangle() and hence no
longer tested by R CMD check.
Documentation
• File doc/html/faq.html no longer exists, and
doc/manual/R-FAQ.html (which has active
links to other manuals) is used instead. (If
makeinfo >= 4.7 is not available, the version on
CRAN is linked to.)
• Manual ’Writing R Extensions’ has further de-
tails on writing new front-ends for R using the
new public header files.
• There are no longer any restrictions on charac-
ters in the \name{} field of a .Rd file: in partic-
ular _ is supported.
C-level facilities
• There are new public C/C++ header files Rin-
terface.h and R_ext/RStartup.h for use with ex-
ternal GUIs.
• Added an onExit() function to graphics de-
vices, to be executed upon user break if non-
NULL.
• ISNAN now works even in C++ code that un-
defines the ’isnan’ macro.
• R_alloc’s limit on 64-bit systems has been
raised from just under 231 bytes (2Gb) to just
under 234 (16Gb), and is now checked.
• New math utility functions log1pmx(x),
lgamma1p(x),logspace_add(logx, logy),
and logspace_sub(logx, logy).
R News ISSN 1609-3631

Vol. 5/1, May 2005 64
Deprecated & defunct
• The aqua module for MacOS X has been re-
moved: --with-aqua now refers to the unbun-
dled Cocoa GUI.
• Capabilities "bzip2", "GNOME, "libz" and
"PCRE" are defunct.
• The undocumented use of UseMethod() with
no argument was deprecated in 2.0.1 and is
now regarded as an error.
• Capability "IEEE754" is deprecated.
• The ’CRAN’ argument to update.packages(),
old.packages(),new.packages(),
download.packages() and install.packages()
is deprecated in favour of ’repos’, which re-
places it as a positional argument (so this is
only relevant for calls with named args).
• The S3 methods for getting and setting names
of "dist" objects have been removed (as they
provided names with a different length from
the "dist" object itself).
• Option "repositories" is no longer used and so
not set.
•loadURL() is deprecated in favour of
load(url()).
•delay() is deprecated. Use delayAssign() in-
stead.
Installation changes
• New configure option --enable-utf8 to en-
able support for UTF-8 locales, on by default.
• R_XTRA_[CF]FLAGS are now used during
the configuration tests, and [CF]PICFLAGS if
--enable-R-shlib was specified. This ensures
that features such as inlining are only used if
the compilation flags specified support them.
(PR#7257)
• Files FAQ, RESOURCES, doc/html/resources.html
are no longer in the SVN sources but are made
by ’make dist’.
• The GNOME GUI is unbundled, now provided
as a package on CRAN.
• Configuring without having the recom-
mended packages is now an error unless
--with-recommended-packages=no (or equiv-
alent) is used.
• Configuring without having the X11 head-
ers and libraries is now an error unless
--with-x=no (or equivalent) is used.
• Configure tries harder to find a minimal set
of FLIBS. Under some circumstances this may
remove from R_LD_LIBRARY_PATH path el-
ements that ought to have specified in LD-
FLAGS (but were not).
• The C code for most of the graphics device
drivers and their afm files are now in package
grDevices.
• R is now linked against ncurses/termlib/termcap
only if readline is specified (now the default)
and that requires it.
• Makeinfo 4.7 or later is now required for build-
ing the HTML and Info versions of the manu-
als.
Installation changes
• There are new types of packages, identified by
the Type field in the DESCRIPTION file. For
example the GNOME console is now a sepa-
rate package (on CRAN), and translations can
be distributed as packages.
• There is now support of installing from within
R both source and binary packages on Ma-
cOS X and Windows. Most of the R func-
tions now have a ’type’ argument defaulting to
getOption("pkgType") and with possible val-
ues "source", "win.binary" and "mac.binary".
The default is "source" except under Windows
and the CRAN GUI build for MacOS X.
•install.packages() and friends now accept a
vector of URLs for ’repos’ or ’contriburl’ and
get the newest available version of a package
from the first repository on the list in which it is
found. The argument ’CRAN’ is still accepted,
but deprecated.
install.packages() on Unix can now install
from local .tar.gz files via repos = NULL (as has
long been done on Windows).
install.packages() no longer asks if down-
loaded packages should be deleted: they will
be deleted at the end of the session anyway
(and can be deleted by the user at any time).
If the repository provides the information,
install.packages() will now accept the name
of a package in a bundle.
If ’pkgs’ is omitted install.packages() will
use a listbox to display the available packages,
on suitable systems.
’dependencies’ can be a character vector to al-
low only some levels of dependencies (e.g. not
"Suggests") to be requested.
R News ISSN 1609-3631

Vol. 5/1, May 2005 65
• There is a new possible value
update.packages(ask="graphics") that uses
a widget to (de)select packages, on suitable sys-
tems.
• The option used is now getOption("repos")
not getOption("CRAN") and it is initially set to
a dummy value. Its value can be a character
vector (preferably named) giving one or sev-
eral repositories.
A new function chooseCRANmirror() will se-
lect a CRAN mirror. This is called automati-
cally if the contrib.url() encounters the ini-
tial dummy value of getOption("repos")
A new function setRepositories() can be
used to create getOption("repos") from a
(platform-specific) list of known repositories.
• New function new.packages() to report unin-
stalled packages available at the requested
repositories. This also reports incomplete bun-
dles. It will optionally install new packages.
• New function available.packages(), similar
to CRAN.packages() but for use with multiple
repositories. Both now only report packages
whose R version requirements are met.
•update.packages() and old.packages() have
a new option ’checkBuilt’ to allow packages in-
stalled under earlier versions of R to be up-
dated.
•remove.packages() can now remove bundles.
• The Contains: field of the DESCRIPTION file
of package bundles is now installed, so later
checks can find out if the bundle is complete.
•packageStatus() is now built on top of *.pack-
ages, and gains a ’method’ argument. It de-
faults to the same repositories as the other
tools, those specified by getOption("repos").
Bug fixes
• Configuring for Tcl/Tk makes use
of ${TK_LIB_SPEC} ${TK_LIBS} not
${TK_LIB_SPEC} ${TK_XLIBSW}, which is cor-
rect for recent versions of Tk, but conceivably
not for old tkConfig.sh files.
•detach() was not recomputing the S4 methods
for primitives correctly.
• Methods package now has class "expression"
partly fixed in basic classes, so S4 classes can
extend these (but "expression" is pretty broken
as a vector class in R).
• Collected warnings had messages with un-
needed trailing space.
• S4 methods for primitive functions must be ex-
ported from namespaces; this is now done au-
tomatically. Note that is.primitive() is now
in ’base’, not ’methods’.
• Package grid:
–Fixed bug in grid.text() when "rot" ar-
gument has length 0. (reported by Em-
manuel Paradis)
•.install_package_vignette_index() cre-
ated an index even in an empty ’doc’ directory.
• The print() method for factors now escapes
characters in the levels in the same way as they
are printed.
•str() removed any class from environment ob-
jects.
str() no longer interprets control characters
in character strings and factor levels; also no
longer truncates factor levels unless they are
longer than ’nchar.max’. Truncation of such
long strings is now indicated ”outside” the
string.
str(<S4.object>) was misleading for the case
of a single slot.
str() now also properly displays S4 class defi-
nitions (such as returned by getClass().
•print.factor(quote=TRUE) was not quoting
levels, causing ambiguity when the levels con-
tained spaces or quotes.
• R CMD check was confused by a trailing / on
a package name.
•write.table() was writing incorrect column
names if the data frame contained any matrix-
like columns.
•write.table() was not quoting row names for
a 0-column x.
•t(x)’s default method now also preserves
names(dimnames(x)) for 1D arrays ’x’.
•r <- a %*% b no longer gives names(dimnames(r))
== c("", "") unless one of aor bhas named
dimnames.
• Some .Internal functions that were supposed
to return invisibly did not. This was behind
PR#7397 and PR#7466.
•eval(expr, NULL, encl) now looks up vari-
ables in encl, as eval(expr, list(), encl) al-
ways did
• Coercing as.data.frame(NULL) to a pairlist
caused an error.
R News ISSN 1609-3631

Vol. 5/1, May 2005 66
•p.adjust(p, ..) now correctly works when
‘p’ contains NAs (or when it is of length 0 or
length 2 for method = "hommel").
• ’methods’ initialization was calling a function
intended for .Call() with .C().
•optim() needed a check that the objective func-
tion returns a value of length 1 (spotted by Ben
Bolker).
•X11() was only scaling its fonts to pointsize if
the dpi was within 0.5 of 100dpi.
•X11() font selection was looking for any sym-
bol font, and sometimes got e.g. bold italic if
the server has such a font.
•dpois(*, lambda=Inf) now returns 0 (or -Inf
for log).
• Using pch="" gave a square (pch=0)! Now it is
regarded as the same as NA, which was also
undocumented but omits the point.
• Base graphics now notices (ab)lines which have
a zero coordinate on log scale, and omits them.
(PR#7559)
•stop() and warning() now accept NULL as
they are documented to do (although this
seems of little use and is equivalent to "").
•weighted.mean() now checks the length of the
weight vector w.
•getAnywhere() was confused by names with
leading or trailing dots (spotted by Robert
McGehee)
•eval() was not handling values from return()
correctly.
•par(omd) is now of the form c(x1, x2, y1,
y2) to match the documentation and for S-
PLUS compatibility.
[Previously, par(omd) was of the form
c(bottom, left, top, right) like par(oma)
and par(omi)]
•formatC() did not check its ’flag’ argu-
ment, and could segfault if it was incorrect.
(PR#7686)
• Contrasts needed to be coerced to numeric (e.g.
from integer) inside model.matrix. (PR#7695)
•socketSelect() did not check for buffered in-
put.
• Reads on a non-blocking socket with no avail-
able data were not handled properly and could
result in a segfault.
• The "aovlist" method for se.contrast() failed
in some very simple cases that were effectively
not multistratum designs, e.g. only one treat-
ment occurring in only one stratum.
•pgamma() uses completely re-written algo-
rithms, and should work for all (even very
extreme) arguments; this is based on Morten
Welinder’s contribution related to PR#7307.
•dpois(10, 2e-308, log=TRUE) and similar
cases gave -Inf.
•x <- 2^(0:1000); plot(x, x^.9, type="l",
log="xy") and x <- 2^-(1070:170); plot(x,
x^.9, type="l", log="xy") now both work
•summary.lm() asked for a report on a reason-
able occurrence, but the check failed to take ac-
count of NAs.
•lm() was miscalculating ’df.residual’ for empty
models with a matrix response.
•summary.lm() now behaves more sensibly for
empty models.
•plot.window() was using the wrong sign
when adjusting xlim/ylim for positive ’asp’
and a reversed axis.
• If malloc() fails when allocating a large ob-
ject the allocator now does a gc and tries the
malloc() again.
•packageSlot() and getGroupMembers() are
now exported from the ’methods’ package
as they should from documentation and the
Green Book.
•rhyper() was giving numbers slightly too
small, due to a bug in the original algorithm.
(PR#7314)
•gsub() was sometimes incorrectly matching ^
inside a string, e.g. gsub("^12", "x", "1212")
was "xx".
•[g]sub(perl = TRUE) was giving random re-
sults for a 0-length initial match. (PR#7742)
•[g]sub was ignoring most 0-length matches,
including all initial ones. Note that sub-
stitutions such as gsub("[[:space:]]*", "
", ...) now work as they do in ’sed’
(whereas the effect was previously the same as
gsub("[[:space:]]+", " ", ...)). (In part
PR#7742)
• Promises are now evaluated when extracted
from an environment using $or [[ ]] .
•reshape(direction="wide") had some sort-
ing problems when guessing time points
(PR#7669)
R News ISSN 1609-3631

Vol. 5/1, May 2005 67
•par() set ’xaxp’ before ’xlog’ and ’yaxp’ before
’ylog’, causing PR#831.
• The logic in tclRequire() to check the avail-
ability of a Tcl package turned out to be falli-
ble. It now uses a try()-and-see mechanism
instead.
• Opening a unz() connection on a non-existent
file left a file handle in use.
• "dist" objects of length 0 failed to print.
• INSTALL and the libR try harder to find a tem-
porary directory (since there might be one left
over with the same PID).
•acf() could cause a segfault with some
datasets. (PR#7771)
•tan(1+LARGEi) now gives 0+1i rather than
0+NaNi (PR#7781)
•summary(data.frame(mat = I(matrix(1:8,
4)))) does not go into infinite recursion any-
more.
•writeBin() performed byte-swapping incor-
rectly on complex vectors, also swapping real
and imaginary parts. (PR#7778)
•read.table() sometimes discarded as blank
lines containing only white space, even if
sep=",".
Changes on CRAN
by Kurt Hornik
New contributed packages
AMORE A MORE flexible neural network package.
This package was born to release the TAO ro-
bust neural network algorithm to the R users.
It has grown and can be of interest for the users
wanting to implement their own training algo-
rithms as well as for those others whose needs
lie only in the “user space”. By Manuel Caste-
jón Limas, Joaquín B. Ordieres Meré, Eliseo P.
Vergara González, Francisco Javier Martínez de
Pisón Ascacibar, Alpha V. Pernía Espinoza, and
Fernando Alba Elías.
BHH2 Functions and data sets reproducing some
examples in “Statistics for Experimenters II” by
G. E. P. Box, J. S. Hunter, and W. C. Hunter,
2005, John Wiley and Sons. By Ernesto Barrios.
Bolstad Functions and data sets for the book “Intro-
duction to Bayesian Statistics” by W. M. Bol-
stad, 2004, John Wiley and Sons. By James Cur-
ran.
Ecdat Data sets from econometrics textbooks. By
Yves Croissant.
GDD Platform and X11 independent device for cre-
ating bitmaps (png, gif and jpeg). By Simon Ur-
banek.
GeneNT The package implements a two-stage algo-
rithm to screen co-expressed gene pairs with
controlled FDR and MAS. The packages also
constructs relevance networks and clusters co-
expressed genes (both similarly co-expressed
and transitively co-expressed). By Dongxiao
Zhu.
Geneland Detection of spatial structure from ge-
netic data. By Gilles Guillot.
HTMLapplets Functions inserting dynamic scatter-
plots and grids in documents generated by
R2HTML. By Gregoire Thomas.
IDPmisc The IDPmisc package contains different
high-level graphics functions for displaying
large datasets, brewing color ramps, draw-
ing nice arrows, creating figures with differ-
ently colored margins and plot regions, and
other useful goodies. By Andreas Ruckstuhl,
Thomas Unternährer, and Rene Locher.
LDheatmap Produces a graphical display, as a heat
map, of measures of pairwise linkage disequi-
libria between SNPs. Users may optionally in-
clude the physical locations or genetic map dis-
tances of each SNP on the plot. By Ji-Hyung
Shin, Sigal Blay, Jinko Graham, and Brad Mc-
Neney.
LogicReg Routines for Logic Regression. By Charles
Kooperberg and Ingo Ruczinski.
MEMSS Data sets and sample analyses from
“Mixed-effects Models in S and S-PLUS” by J.
Pinheiro and D. Bates, 2000, Springer. By Dou-
glas Bates.
MatchIt Select matched samples of the original
treated and control groups with similar covari-
ate distributions—can be used to match exactly
on covariates, to match on propensity scores,
or perform a variety of other matching proce-
dures. By Daniel Ho, Kosuke Imai, Gary King,
and Elizabeth Stuart.
R News ISSN 1609-3631

Vol. 5/1, May 2005 68
Matching Provides functions for multivariate and
propensity score matching and for finding op-
timal balance based on a genetic search algo-
rithm. A variety of univariate and multivariate
tests to determine if balance has been obtained
are also provided. By Jasjeet Singh Sekhon.
NORMT3 Evaluates the probability density func-
tion of the sum of the Gaussian and Student’s t
density on 3 degrees of freedom. Evaluates the
p.d.f. of the sphered Student’s tdensity func-
tion. Also evaluates the erf and erfc functions
on complex-valued arguments. By Guy Nason.
PK Estimation of pharmacokinetic parameters. By
Martin Wolfsegger.
ProbForecastGOP Probabilistic weather field fore-
casts using the Geostatistical Output Pertur-
bation method introduced by Gel, Raftery
and Gneiting (2004). By Yulia Gel, Adrian
E. Raftery, Tilmann Gneiting, and Veronica J.
Berrocal.
R.matlab Provides methods to read and write MAT
files. It also makes it possible to communi-
cate (evaluate code, send and retrieve objects
etc.) with Matlab v6 or higher running locally
or on a remote host. The auxiliary Java class
provides static methods to read and write Java
data types. By Henrik Bengtsson.
R.oo Methods and classes for object-oriented pro-
gramming in R with or without references. By
Henrik Bengtsson.
RGrace Mouse/menu driven interactive plotting
application. By M. Kondrin.
RII Estimation of the relative index of inequality
for interval-censored data using natural cubic
splines. By Jamie Sergeant.
ROCR ROC graphs, sensitivity/specificity curves,
lift charts, and precision/recall plots are popu-
lar examples of trade-off visualizations for spe-
cific pairs of performance measures. ROCR is
a flexible tool for creating cutoff-parametrized
2D performance curves by freely combining
two from over 25 performance measures (new
performance measures can be added using a
standard interface). By Tobias Sing, Oliver
Sander, Niko Beerenwinkel, and Thomas
Lengauer.
ResistorArray Electrical properties of resistor net-
works. By Robin K. S. Hankin.
Rlab Functions and data sets for the NCSU ST370
class. By Dennis D. Boos, Atina Dunlap Brooks,
and Douglas Nychka.
SciViews A bundle of packages to implement a full
reusable GUI API for R. Contains svGUI with
the main GUI features, svDialogs for the dialog
boxes, svIO for data import/export, svMisc
with miscellaneous supporting functions, and
svViews providing views and report features
(views are HTML presentations of the content
of R objects, combining text, tables and graphs
in the same document). By Philippe Grosjean
& Eric Lecoutre.
SemiPar Functions for semiparametric regression
analysis, to complement the book “Semipara-
metric Regression” by R. Ruppert, M. P. Wand,
and R. J. Carroll, 2003, Cambridge University
Press. By Matt Wand.
SeqKnn Estimate missing values sequentially from
the gene that had least missing rate in microar-
ray data. By Ki-Yeol Kim and Gwan-Su Yi.
UsingR Data sets to accompany the textbook “Us-
ing R for Introductory Statistics” by J. Verzani,
2005, Chapman & Hall/CRC. By John Verzani.
Zelig Everyone’s statistical software: an easy-to-use
program that can estimate, and help interpret
the results of, an enormous range of statisti-
cal models. By Kosuke Imai, Gary King, and
Olivia Lau.
adlift Adaptive Wavelet transforms for signal de-
noising. By Matt Nunes and Marina Popa.
alr3 Methods and data to accompany the textbook
“Applied Linear Regression” by S. Weisberg,
2005, Wiley. By Sanford Weisberg.
arules Provides the basic infrastructure for mining
and analyzing association rules and an inter-
face to the C implementations of Apriori and
Eclat by Christian Borgelt. By Bettina Gruen,
Michael Hahsler and Kurt Hornik.
bayesm Covers many important models used in
marketing and micro-econometrics applica-
tions. The package includes: Bayes Re-
gression (univariate or multivariate depen-
dent variable), Multinomial Logit (MNL) and
Multinomial Probit (MNP), Multivariate Pro-
bit, Multivariate Mixtures of Normals, Hi-
erarchical Linear Models with normal prior
and covariates, Hierarchical Multinomial Log-
its with mixture of normals prior and covari-
ates, Bayesian analysis of choice-based conjoint
data, Bayesian treatment of linear instrumental
variables models, and Analyis of Multivariate
Ordinal survey data with scale usage hetero-
geneity. By Peter Rossi and Rob McCulloch.
bitops Functions for Bitwise operations on integer
vectors. S original by Steve Dutky, initial R port
R News ISSN 1609-3631

Vol. 5/1, May 2005 69
and extensions by Martin Maechler. Revised
and modified by Steve Dutky.
boost Contains a collection of boosting methods,
these are ‘BagBoost’, ‘LogitBoost’, ‘AdaBoost’
and ‘L2Boost’, along with feature preselection
by the Wilcoxon test statistic. Moreover, meth-
ods for the simulation of data according to cor-
relation and mean structures of existing real
datasets are included. By Marcel Dettling.
changeLOS Change in length of hospital stay (LOS).
By Matthias Wangler and Jan Beyersmann.
clac Clust Along Chromosomes, a method to call
gains/losses in CGH array data. By Pei
Wang, with contributions from Balasubrama-
nian Narasimhan.
climatol Functions to fill missing data in climato-
logical (monthly) series and to test their homo-
geneity, plus functions to draw wind-rose and
Walter&Lieth diagrams. By José A. Guijarro.
clue CLUster Ensembles. By Kurt Hornik, with con-
tributions from Walter Boehm.
coin Conditional inference procedures for the gen-
eral independence problem including two-
sample, K-sample, correlation, censored, or-
dered and multivariate problems. By Torsten
Hothorn and Kurt Hornik, with contributions
by Mark van de Wiel and Achim Zeileis.
colorspace Carries out mapping between assorted
color spaces. By Ross Ihaka.
concor Concordance, providing “SVD by blocks”.
By R. Lafosse.
ctv Server-side and client-side tools for task views to
CRAN-style repositories. By Achim Zeileis and
Kurt Hornik.
cyclones Functions for locating local min-
ima/maxima. By Rasmus E. Benestad.
eco Fits parametric and nonparametric Bayesian
models for ecological inference in 2 by 2 ta-
bles. The models are fit using the Markov chain
Monte Carlo algorithms that are described in
Imai and Lu (2004). By Ying Lu and Kosuke
Imai.
edci Detection of edgepoints in images based on the
difference of two asymmetric M kernel estima-
tors. Linear and circular regression clustering
based on redescending M estimators. Detec-
tion of linear edges in images. By Tim Garlipp.
elasticnet Elastic net regularization and variable se-
lection. Also implements the sparse PCA algo-
rithm based on the elastic net/lasso. By Hui
Zou and Trevor Hastie.
ensembleBMA Uses Bayesian Model Averaging to
create probabilistic forecasts of ensembles us-
ing a mixture of normal distributions. By
Adrian E. Raftery, J. McLean Sloughter, and
Michael Polakowski.
epitools Basic tools for applied epidemiology. By
Tomas Aragon.
epsi Smoothing methods for images which are
based on a redescending M kernel estimator
which preserves edges and corners. By Tim
Garlipp.
far Modelizations and previsions functions for
Functional AutoRegressive processes using
nonparametric methods: functional kernel, es-
timation of the covariance operator in a sub-
space, . . . . By Damon Julien and Guillas Serge.
frailtypack Fit a shared gamma frailty model and
Cox proportional hazards model using a Penal-
ized Likelihood on the hazard function. Left
truncated, censored data and strata (max=2)
are allowed. Original Fortran routines by Vir-
ginie Rondeau. Modified Fortran routines, R
code and packaging by Juan R Gonzalez.
gcmrec Parameters estimation of the general semi-
parametric model for recurrent event data pro-
posed by Peña and Hollander. By Juan R. Gon-
zalez, Elizabeth H. Slate, and Edsel A Peña.
genalg R based genetic algorithm for binary and
floating point chromosomes. By Egon Wil-
lighagen.
gmp Multiple precision Arithmetic (prime numbers,
. . . ), “arithmetic without limitations” using the
C library gmp. By Antoine Lucas, Immanuel
Scholz, Rainer Boehme, and Sylvain Jasson.
gsl An R wrapper for the special functions and quasi
random number generators of the Gnu Scien-
tific Library (http://www.gnu.org/software/
gsl/). By Robin K. S. Hankin; qrng functions
by Duncan Murdoch.
hmm.discnp Fits hidden Markov models with dis-
crete non-parametric observation distributions
to data sets. Simulates data from such models.
By Rolf Turner and Limin Liu..
hopach The Hierarchical Ordered Partitioning and
Collapsing Hybrid (HOPACH) clustering algo-
rithm. By Katherine S. Pollard, with Mark J.
van der Laan.
intcox Implementation of the Iterated Convex Mino-
rant Algorithm for the Cox proportional haz-
ard model for interval censored event data. By
Volkmar Henschel, Christiane Heiss, and Ul-
rich Mansmann.
R News ISSN 1609-3631

Vol. 5/1, May 2005 70
irr Coefficients of Interrater Reliability and Agree-
ment for quantitative, ordinal and nominal
data: ICC, Finn-Coefficient, Robinson’s A,
Kendall’s W, Cohen’s Kappa, . . . . By Matthias
Gamer.
kknn Weighted k-Nearest Neighbors Classification
and Regression. By Klaus Schliep & Klaus
Hechenbichler.
ks Bandwidth matrices for kernel density estimators
and kernel discriminant analysis for bivariate
data. By Tarn Duong.
labdsv A variety of ordination and vegetation anal-
yses useful in analysis of datasets in commu-
nity ecology. Includes many of the common
ordination methods, with graphical routines to
facilitate their interpretation, as well as several
novel analyses. By David W. Roberts.
latticeExtra Generic function and standard methods
for Trellis-based displays. By Deepayan Sarkar.
ltm Analysis of multivariate Bernoulli data using la-
tent trait models (including the Rasch model)
under the Item Response Theory approach. By
Dimitris Rizopoulos.
maanova Analysis of N-dye Micro Array experi-
ment using mixed model effect. Containing
analysis of variance, permutation and boot-
strap, cluster and consensus tree. By Hao Wu,
with ideas from Gary Churchill, Katie Kerr and
Xiangqin Cui.
matlab Emulate MATLAB code using R. By P. Roe-
buck.
mcmc Functions for Markov chain Monte Carlo
(MCMC). By Charles J. Geyer.
meta Fixed and random effects meta-analysis. Func-
tions for tests of bias, forest and funnel plot. By
Guido Schwarzer.
micEcon Tools for microeconomic analysis and mi-
croeconomic modelling. By Arne Henningsen.
minpack.lm Provides R interface for two func-
tions from MINPACK library, solving nonlin-
ear least squares problem by modification of
the Levenberg-Marquardt algorithm. By Timur
V. Elzhov.
mlica An R code implementation of the maximum
likelihood (fixed point) algorithm of Hyvaeri-
nen, Karhuna and Oja for independent compo-
nent analysis. By Andrew Teschendorff.
mlmRev Data and examples from a multilevel mod-
elling software review as well as other well-
known data sets from the multilevel modelling
literature. By Douglas Bates.
mvoutlier Outlier detection using robust estima-
tions of location and covariance structure. By
Moritz Gschwandtner and Peter Filzmoser.
nice Get or set UNIX priority (niceness) of running
R process. By Charles J. Geyer.
ouch Fit and compare Ornstein-Uhlenbeck models
for evolution along a phylogenetic tree. By
Aaron A. King.
outliers A collection of some tests commonly used
for identifying outliers. By Lukasz Komsta.
perturb Evaluates collinearity by adding random
noise to selected variables. By John Hendrickx.
phpSerialize Serializes R objects for import by PHP
into an associative array. Can be used to build
interactive web pages with R. By Dieter Menne.
pls Multivariate regression by partial least squares
regression (PLSR) and principal components
regression (PCR). This package supersedes the
pls.pcr package. By Ron Wehrens and Bjørn-
Helge Mevik.
plsgenomics Provides routines for PLS-based ge-
nomic analyses. By Anne-Laure Boulesteix and
Korbinian Strimmer.
plugdensity Kernel density estimation with global
bandwidth selection via “plug-in”". By Eva
Herrmann (C original); R interface et cetera by
Martin Maechler.
polycor Computes polychoric and polyserial corre-
lations by quick “two-step” methods or ML,
optionally with standard errors; tetrachoric
and biserial correlations are special cases. By
John Fox.
ppc Sample classification of protein mass spectra by
peak probability contrasts. By Balasubrama-
nian Narasimhan, R. Tibshirani, and T. Hastie.
proto An object oriented system using prototype
or object-based (rather than class-based) object
oriented ideas. By Louis Kates and Thomas
Petzoldt.
pvclust Assessing the uncertainty in hierarchi-
cal cluster analysis. By Ryota Suzuki and
Hidetoshi Shimodaira.
qtlDesign Tools for the design of QTL experiments.
By Saunak Sen, Jaya Satagopan, and Gary
Churchill.
qvalue Q-value estimation for false discovery rate
control. By Alan Dabney and John D. Storey,
with assistance from Gregory R. Warnes.
R News ISSN 1609-3631

Vol. 5/1, May 2005 71
relsurv Various functions for regression in relative
survival. By Maja Pohar.
resper Two accept-and-reject algorithms to sample
from permutations. By Johannes Hüsing.
rstream Unified object oriented interface for multi-
ple independent streams of random numbers
from different sources. By Josef Leydold.
rwt Performing digital signal processing. By P. Roe-
buck, based on MATLAB extension by Rice
University’s DSP group.
seqinr Exploratory data analysis and data visualiza-
tion for biological sequence (DNA and protein)
data. By Delphine Charif and Jean Lobry.
spectrino Spectra organizer, visualization and data
extraction from within R. By Teodor Krastev.
stepwise A stepwise approach to identifying recom-
bination breakpoints in a sequence alignment.
By Jinko Graham, Brad McNeney, and Fran-
coise Seillier-Moiseiwitsch, R interface by Sigal
Blay.
survBayes Fits a proportional hazards model to time
to event data by a Bayesian approach. Right
and interval censored data and a log-normal
frailty term can be fitted. By Volkmar Henschel,
Christiane Heiss, Ulrich Mansmann.
tdist Computes the distribution of a linear combina-
tion of independent Student’s t-variables (with
small degrees of freedom, dff ≤100) and/or
standard Normal Zrandom variables. By Vik-
tor Witkovsky and Alexander Savin.
tweedie Maximum likelihood computations for
Tweedie families. By Peter Dunn.
uroot Unit root tests (KPSS, ADF, CH and HEGY)
and graphics for seasonal time series. By Javier
López-de-Lacalle & Ignacio Díaz-Emparanza.
vabayelMix Performs inference of a gaussian mix-
ture model within a bayesian framework us-
ing an optimal separable approximation to the
posterior density. The optimal posterior ap-
proximation is obtained using a variational ap-
proach. By Andrew Teschendorff.
verification Contains utilities for verification of dis-
crete and probabilistic forecasts. By the NCAR
Research Application Program.
zicounts Fits classical and zero-inflated count data
regression model as well as censored count
data regression. By S M Mwalili.
Other changes
• Packages CoCoAn,gpls, and multiv were
moved from the main CRAN section to the
Archive.
• Package Dopt was moved from the Devel sec-
tion of CRAN to the Archive.
• Package multidim had to be removed from
CRAN.
Kurt Hornik
Wirtschaftsuniversität Wien, Austria
Kurt.Hornik@R-project.org
Events
Chambers Workshop
A workshop entitled “40 Years of Statistical Comput-
ing and Beyond” was held at Bell Laboratories, Mur-
ray Hill, NJ, U.S.A. on April 29, 2005 to mark the oc-
casion of John Chambers’ retirement from the Labs
but not from active research. He is continuing his
record of innovation by becoming the first Emeritus
Member of Technical Staff in the history of Bell Labs.
R is an implementation of the S language and
John is the originator and principal designer of S.
Without John’s work on S there never would have
been an R. His patient responding to questions dur-
ing early development of R was crucial to its success
and, since 2000, he has been a member of the R De-
velopment Core Team.
The history and development of the S language
and of its implementation in R were featured in
many of the presentations at the workshop, espe-
cially in those by Rick Becker and Allan Wilks. Nat-
urally, the highlight of the day was John’s presen-
tation in which he first looked back on his 40 years
of involvement in statistical computing and the de-
velopment of languages for programming with data
and then gave some tantalizing glimpses into the fu-
ture. The agenda for the workshop can be seen at
http://stat.bell-labs.com.
DSC 2005
DSC 2005 – Directions in Statistical Computing – will
be held from the evening of August 12 through Au-
gust 15, in Seattle, Washington. This conference fol-
lows on from the successful DSC 1999, 2001, and 2003
conferences at the Vienna University of Technology.
R News ISSN 1609-3631

Vol. 5/1, May 2005 72
The workshop will focus on, but is not limited to,
open source statistical computing.
We are inviting abstracts for contributed
presentations on issues related to the devel-
opment of statistical computing and graphics
environments. Abstracts should be sent to
dsc2005@u.washington.edu, by May 15 and ques-
tions to tlumley@u.washington.edu. More informa-
tion is available at http://depts.washington.edu/
dsc2005/ and online registration will soon open at
that website.
Bioconductor User Conference
The 2005 Bioconductor User Conference will be
hosted at the Fred Hutchinson Cancer Research Cen-
ter in Seattle on August 16 and 17, following the
DSC. The two day meeting will consist of morn-
ing lectures and afternoon laboratories. Scheduled
speakers include: Michael Boutros, Eric Schadt, Srid-
har Ramaswamy, Rafael Irizarry, and Robert Gentle-
man. The conference web site can be found at http:
//www.bioconductor.org/meeting05. For more de-
tails contact biocworkshop@fhcrc.org.
Editor-in-Chief:
Douglas Bates
Department of Statistics
1300 University Ave University of Wisconsin
Madison, WI 53706
USA
Editorial Board:
Paul Murrell and Torsten Hothorn.
Editor Programmer’s Niche:
Bill Venables
Editor Help Desk:
Uwe Ligges
Email of editors and editorial board:
firstname.lastname @R-project.org
R News is a publication of the R Foundation for Sta-
tistical Computing, communications regarding this
publication should be addressed to the editors. All
articles are copyrighted by the respective authors.
Please send submissions to regular columns to the
respective column editor, all other submissions to
the editor-in-chief or another member of the edi-
torial board (more detailed submission instructions
can be found on the R homepage).
R Project Homepage:
http://www.R-project.org/
This newsletter is available online at
http://CRAN.R-project.org/doc/Rnews/
R News ISSN 1609-3631