Bcl2fastq2 Conversion Software V2.18 User Guide V2 18 15051736 01
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 34

ILLUMINA PROPRIETARY
Document # 15051736 v01
April 2016
bcl2fastq2 Conversion v2.18
User Guide
For Research Use Only. Not for use in diagnostic procedures.
Introduction 3
Install bcl2fastq2 Conversion Software 5
BCL Conversion Input Files 7
Sample Sheet 13
Run BCL Conversion and Demultiplexing 16
BCL Conversion Output Files 21
Troubleshooting 28
Appendix: Installation Requirements 29
Revision History 31
Technical Assistance
This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"), and are intended solely for the
contractual use of its customer in connection with the use of the product(s) described herein and for no other purpose. This
document and its contents shall not be used or distributed for any other purpose and/or otherwise communicated, disclosed,
or reproduced in any way whatsoever without the prior written consent of Illumina. Illumina does not convey any license
under its patent, trademark, copyright, or common-law rights nor similar rights of any third parties by this document.
The instructions in this document must be strictly and explicitly followed by qualified and properly trained personnel in order
to ensure the proper and safe use of the product(s) described herein. All of the contents of this document must be fully read
and understood prior to using such product(s).
FAILURE TO COMPLETELY READ AND EXPLICITLY FOLLOW ALL OF THE INSTRUCTIONS CONTAINED HEREIN
MAY RESULT IN DAMAGE TO THE PRODUCT(S), INJURY TO PERSONS, INCLUDING TO USERS OR OTHERS, AND
DAMAGE TO OTHER PROPERTY.
ILLUMINA DOES NOT ASSUME ANY LIABILITY ARISING OUT OF THE IMPROPER USE OF THE PRODUCT(S)
DESCRIBED HEREIN (INCLUDING PARTS THEREOF OR SOFTWARE).
© 2016 Illumina, Inc. All rights reserved.
Illumina,24sure,BaseSpace,BeadArray,BlueFish,BlueFuse,BlueGnome,cBot,CSPro,CytoChip,DesignStudio,
Epicentre,ForenSeq,Genetic Energy,GenomeStudio,GoldenGate,HiScan,HiSeq,HiSeq X,Infinium,iScan,iSelect,
MiniSeq,MiSeq,MiSeqDx,MiSeq FGx,NeoPrep,NextBio,Nextera,NextSeq,Powered by Illumina,SureMDA,
TruGenome,TruSeq,TruSight,Understand Your Genome,UYG,VeraCode,verifi,VeriSeq, the pumpkin orange color,
and the streaming bases design are trademarks of Illumina, Inc. and/or its affiliate(s) in the U.S. and/or other countries. All
other names, logos, and other trademarks are the property of their respective owners.
Introduction
bcl2fastq2 Conversion Software v2.18 Guide 3
Introduction
The Illumina sequencing instruments generate per-cycle base call (BCL) files at the end of
the sequencing run. A majority of analysis applications use per-read FASTQ files as
input for analysis. You can use the bcl2fastq2 Conversion Software v2.18 to convert base
call(BCL) files from a sequencing run into FASTQ files.
Use this guide to install the bcl2fastq2 Conversion Software and run the BCL conversion
and demultiplexing process.
Supported Instruments
The bcl2fastq2 Conversion Software supports the following instruments:
}MiniSeq
}MiSeq
}NextSeq 500, 550
}HiSeq X
}HiSeq 2000, 2500, 3000, 4000
If your Illumina sequencing system runs a earlier software version of Real-Time Analysis
(RTA) than v1.18.54 and you want to convert BCL to FASTQ, install bcl2fastq v1.8.4, and
refer to the bcl2fastq Conversion User Guide Version v1.8.4 (part # 15038058) for instructions.
BCL Conversion and Demultiplexing Directory
The bcl2fastq2 Conversion Software performs BCL conversion and demultiplexing in a
single step. By default, the software puts the resulting demultiplexed compressed FASTQ
files in <run folder>/Data/Intensities/BaseCalls.
The software puts reads with undetermined indexes in files that begin with
Undetermined_S0_, unless the sample sheet specifies a sample ID or sample name for
reads without an index.
If the Sample_Project column is specified for a sample in the sample sheet, the FASTQ
files for that sample are placed in <run
folder>/Data/Intensities/BaseCalls/<Project>.
Multiple samples can use the same project directory. If the Sample_ID and Sample_
Name columns are specified but do not match, the FASTQ files are placed in an
additional sub-directory called <SampleId>.
BCLto FASTQ Conversion Process
The bcl2fastq2 Conversion Software converts the base calls in the per-cycle BCLfiles to
the per-read FASTQformat. As an option, the software can trim adapters and remove
Unique Molecular Identifier (UMI)bases from reads.
Adapter Trimming—The bcl2fastq2 Conversion Software checks whether a read extends
past the sample DNA insert and into the adapter sequence. The software uses an
approximate string matching algorithm to identify all or part of the adapter, and treats
the insertions and deletions as a single mismatch. If an adapter sequence is detected,
base calls matching the adapter and beyond the match are masked or removed in the
FASTQ file.
Unique Molecular Indentifiers (UMIs) Removal—UMIs are random k-mers attached to
the genomic DNAbefore polymerase chain reaction (PCR)amplification. After the UMI is
amplified with amplicons, the software can detect PCRduplicates and correct
amplification errors and can remove these bases and places them into the read name in
4Document # 15051736 v01
the FASTQfiles. Also, when the TrimUMI sample sheet setting is active, the software can
remove the bases from the reads.
Demultiplexing—First, the software reorganizes the FASTQ files based on the index
sequencing information. For best practices, avoid choosing indexes that differ by fewer
than 3 bases during sample preparation. Then, the software generates the statistics and
reports for the demultiplexed FASTQ files. Also, the software recalculates the base calling
analysis statistics and store the statistics in the InterOp folder. You can view the
statistics with the Sequencing Analysis Viewer (SAV) software from Illumina.
Output Files
}FASTQFiles
}InterOp Files
}ConversionStats File
}DemultiplexingStats File
}Adapter Trimming File
}FastqSummary and DemuxSummary
}HTML Reports
}JSONFile

Install bcl2fastq2 Conversion Software
bcl2fastq2 Conversion Software v2.18 Guide 5
Install bcl2fastq2 Conversion Software
You can download the bcl2fastq2 Conversion Software from the Downloads page on the
Illumina website.
For installation requirements, see Appendix: Installation Requirements on page 29.
Install from RPMPackage
You need to have access the root system to install.
1To install the RPM file, use the following command line:
yum install -y <rpm package-name>
The starting point for the bcl2fastq converter is the binary executable
/usr/local/bin/bcl2fastq.
2To install the RPM package in a user specified location, use the following command
line:
rpm --install --prefix <user specified directory>
<rpm package-name>
Install from Source
For installation, the directory locations are specified with the following environment
variables:
Variables Description
SOURCE Location of the bcl2fastq2 source code
BUILD Location of the build directory
INSTALL_DIR Location where the executable is installed
For example, the environment variables can be set as:
export TMP=/tmp
export SOURCE=${TMP}/bcl2fastq
export BUILD=${TMP}/bcl2fastq2-v2.18.x-build
export INSTALL_DIR=/usr/local/bcl2fastq2-v2.18.x
The build directory must be different from the source directory.
Follow these steps to install from source:
1Decompress and extract the source code.
cd ${TMP}
tar -xvzf path-to-tarball/bcl2fastq2-v2.18.x.tar.gz
This command creates a bcl2fastq sub-directory in the ${TMP} directory.
2Configure the build using the following commands:
mkdir ${BUILD}
cd ${BUILD}
${SOURCE}/src/configure --prefix=${INSTALL_DIR}
The commands in step 2 create a build directory. Move WHAT to that directory, and
then run the configuration in the directory.
The --prefix parameter provides the absolute path to the install the directory.
The command creates a sub-directory in the ${TMP} directory.
6Document # 15051736 v01
3Build the package using the following commands:
make
4Install the package using the following commands:
make install
Depending on the ${INSTALL_DIR} directory, you may need root privilege.

BCL Conversion Input Files
bcl2fastq2 Conversion Software v2.18 Guide 7
BCL Conversion Input Files
After sequencing, the instruments generate a BaseCalls directory, which contains the base
calls files (BCL), for demultiplexing.
For demultiplexing, the bcl2fastq2 Conversion Software requires the following input files:
Instrument Input Files
MiSeq and HiSeq 2000/2500 •BCL Files (*.bcl.gz)
•STATS Files
•FILTERFiles
•CONTROL Files
•Position Files
•RunInfo Files
•Config Files
•Sample Sheet Files (optional)
MiniSeq and NextSeq 500/550 •BCL Files (*bcl.bgzf)
•BCI Files
•FILTERFiles
•Position Files
•RunInfo Files
•Sample Sheet Files (optional)
HiSeq X and HiSeq 3000/4000 •BCL Files (*.bcl.gz)
•FILTERFiles
•Position Files
•RunInfo Files
•Sample Sheet Files (optional)
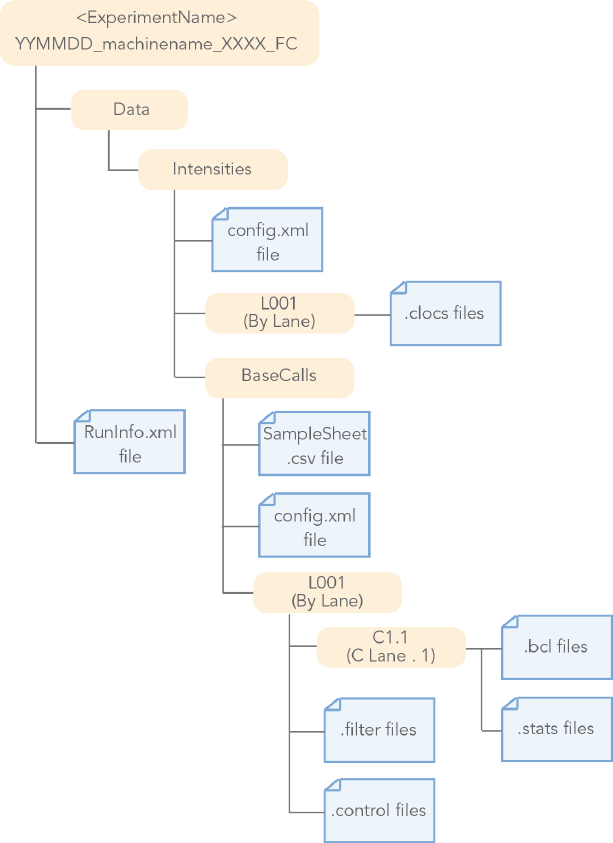
8Document # 15051736 v01
BCL Conversion Input Files Diagram
Figure 1 BCL Conversion Input Files from the MiSeq or HiSeq 2000/2500 System
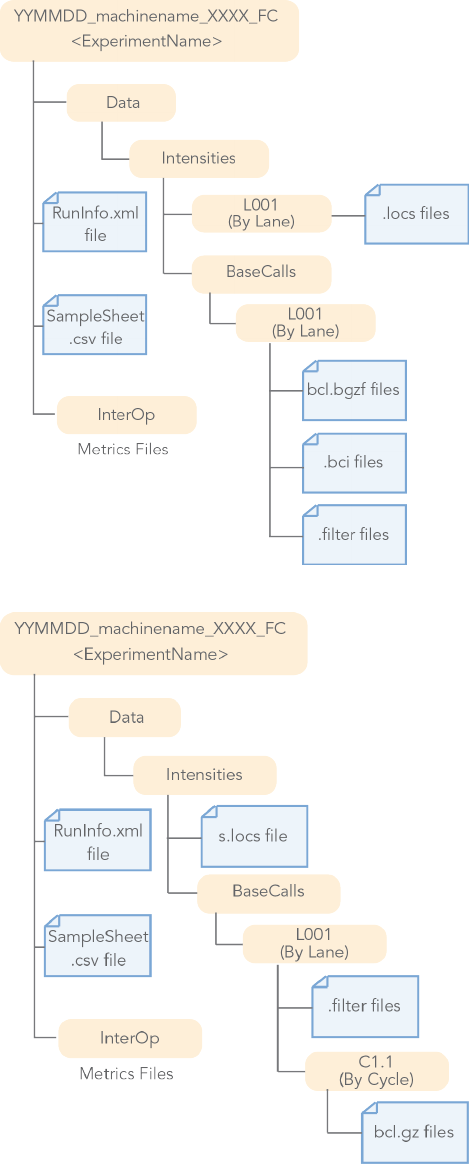
BCL Conversion Input Files
bcl2fastq2 Conversion Software v2.18 Guide 9
Figure 2 BCL Conversion Input Files from the MiniSeq or NextSeq System
Figure 3 BCL Conversion Input Files from the HiSeq X System
Folder and File Naming
The top-level run folder name is generated using 3 fields to identify the
<ExperimentName>, separated by underscores.

10 Document # 15051736 v01
The software generates the top-level run folder using 3 fields separated by underscores to
identify the <ExperimentName>.
Example:
YYMMDD_machinename_NNNN
For best practices, do not deviate from the run folder naming convention because doing
so can cause the software to stop.
}The first field is a six-digit number (YYMMDD) specifying the date of the run.
}The second field specifies the name of the sequencing machine. The field can consist
of any combination of upper or lower case letters, digits, or hyphens, but it cannot
contain any other characters or underscore.
}The third field is a four-digit specifies the experiment ID on that instrument. Each
instrument supplies a series of consecutively numbered experiment IDs from the on-
board sample tracking database or a LIMS.
For best practices, we recommend that you create unique names for the experiment or
sample IDs for each instrument to avoid naming conflicts.
For example, a run folder named 150108_instrument1_3147 indicates that the
experiment ID is 3147; the run is on instrument 1, and the date is on January 8, 2015
(YYMMDD). The date and instrument name specify a unique run folder for any number
of instruments.
Also, you can view the flow cell number in the run folder name.
Example:
YYMMDD_machinename_NNNN_FCYYY
When you publish the data to a public database, we recommend that you use a prefix for
each instrument with the identity of the sequencing center.
BCL Files
The BCL files are compressed with the gzip (*.gz) or the blocked GNU zip (*.bgzf) format.
The BaseCalls directory contains the BCL files. You can locate the files from the following
directory:
Data/Intensities/BaseCalls/L<lane>/C<Cycle>.1
Bytes Description Data type
Bytes 0–3 Number N of cluster Unsigned 32 bits
integer
Bytes 4–(N+3)
N—Cluster
index
Bits 0–1 are the bases, [A, C, G, T] for [0, 1, 2, 3]:
bits 2–7 are shifted by 2 bits and contain the quality
score.
All bits with 0 in a byte is reserved for no call.
Unsigned 8 bits
integer
Table 1 BCL File Format
BCI Files
The BCI (*.bci) files contain one record per tile for the sequencing run in binary format.
You can locate these files from the following directory:
<run directory>/Data/Intensities/BaseCalls/L<lane>

BCL Conversion Input Files
bcl2fastq2 Conversion Software v2.18 Guide 11
Bytes Description
Bytes 0–3 Tile number
Bytes 4–7 Number of clusters in the tile
Table 2 BCI File Format
STATS Files
The STATS file (*.stats) is a binary file that contains base calling statistics. You can locate
these files from the following directory:
Data/Intensities/BaseCalls/L00<lane>/C<cycle>.1
Start Description Data Type
Byte 0 Cycle number integer
Byte 4 Average Cycle Intensity double
Byte 12 Average intensity for A over all clusters with intensity for A double
Byte 20 Average intensity for C over all clusters with intensity for C double
Byte 28 Average intensity for G over all clusters with intensity for G double
Byte 36 Average intensity for T over all clusters with intensity for T double
Byte 44 Average intensity for A over clusters with base call A double
Byte 52 Average intensity for C over clusters with base call C double
Byte 60 Average intensity for G over clusters with base call G double
Byte 68 Average intensity for T over clusters with base call T double
Byte 76 Number of clusters with base call A integer
Byte 80 Number of clusters with base call C integer
Byte 84 Number of clusters with base call G integer
Byte 88 Number of clusters with base call T integer
Byte 92 Number of clusters with base call X integer
Byte 96 Number of clusters with intensity for A integer
Byte 100 Number of clusters with intensity for C integer
Byte 104 Number of clusters with intensity for G integer
Byte 108 Number of clusters with intensity for T integer
Table 3 Stats File Format
FILTER Files
The FILTER file (*.filter) is a binary file that contains the filter results. You can locate
these files from the following directory:
Data/Intensities/BaseCalls/L<lane>
Bytes Description
Bytes 0–3 Zero value (for backwards compatibility)
Bytes 4–7 Filter format version number
Bytes 8–11 Number of clusters
Bytes 12–(N+11)
N—cluster number
Unsigned 8 bits integer
Bit 0 is pass or failed filter
Table 4 Filter File Format
12 Document # 15051736 v01
CONTROL Files
The CONTROL (*.control) file is a binary files that contains the control results. You can
locate these files from the following directory:
<run directory>/Data/Intensities/BaseCalls/L00<lane>/
Bytes Description
Bytes 0–3 Zero value (for backwards compatibility)
Bytes 4–7 Format version number
Bytes 8–11 Number of clusters
Bytes 12–(2xN+11)
N—cluster index
The bit number indicates the following:
•Bit 0: always empty (0)
•Bit 1: was the read identified as a control?
•Bit 2: was the match ambiguous?
•Bit 3: did the read match the PhiX tag?
•Bit 4: did the read align to match the PhiX tag?
•Bit 5: did the read match the control index sequence?
•Bits 6,7: reserved for future use
•Bits 8..15: the report key for the matched record in the
controls.fasta file (specified by the REPORT_KEY metadata)
Table 5 Control File Format
CONFIG Files
The CONFIG (*config.xml) file records information specific to the generation of the
subfolders. The file contains a tag-value list that describes the cycle-image folders used to
generate each folder of intensity and sequence files. You can locate the file from the
following directory:
<run directory>/Data/Intensities/
The other CONFIG (*config.xml) file is in the BaseCalls directory, which contains the
meta-information on the base caller runs. You can locate the file from the following
directory:
<run directory>/Data/Intensities/BaseCalls/
Position Files
The BCL to FASTQ converter can use different types of position files.
The LOCS (*.locs) file is a binary file that contains the cluster positions. Additionally, the
*.clocs files are compressed versions of LOCS files.
The *_pos.txt files are text-based files with 2 columns and a number of rows equal to the
number of clusters. The first column is the X-coordinate and the second column is the Y-
coordinate. Each line has a <cr><lf> at the end.
You can locate these files from the following directory:
Data/Intensities/BaseCalls/L<lane>
RunInfo File
The RunInfo.xml file is located at the top-level run folder <run directory>. The file
contains information on the run, flow cell, and instrument IDs, date and read structure.
Also, the file provides the number of reads, the number of cycles per read, and the index
reads.

Sample Sheet
bcl2fastq2 Conversion Software v2.18 Guide 13
Sample Sheet
The sample sheet (*SampleSheet.csv) file provides information on the relationship
between samples and indexes during library creation. The sample sheet is optional and
is at the top-level run folder. When a sample sheet is not provided, all reads are assigned
to the default sample Undetermined_S0, which includes one file per lane per read.
Settings Section
The bcl2fastq2 Conversion Software uses the adapter settings for adapter trimming.
Setting Description
Adapter or TrimAdapter The adapter sequence to be trimmed. If an AdapterRead2 is
provided, this sequence is only used to trim Read 1.
AdapterRead2 or
TrimAdapterRead2
The adapter sequence to be trimmed in Read 2. If not
provided, the same sequence specified in Adapter is used.
MaskAdapter The adapter sequence to be masked rather than trimmed. If
MaskAdapterRead2 is provided, this sequence is only used to
mask Read 1.
MaskAdapterRead2 The adapter sequence to be masked in Read 2. If not provided,
the same sequence specified in MaskAdapter is used.
FindAdapterWithIndels 1 (default) or 0. If 1 (true), an approximate string matching
algorithm is used to identify the adapter, treating insertions
and deletions as a single mismatch (Myers 1999, J.ACM). If 0
(false), a sliding window algorithm is used, in which insertions
and deletions of bases inside the adapter sequence is not
tolerated.
Table 6 Adapter Specifications
Setting Description
Read1EndWithCycle The last cycle to use for Read 1.
Read2EndWithCycle The last cycle to use for Read 2.
Read1StartFromCycle The first cycle to use for Read 1.
Read2StartFromCycle The first cycle to use for Read 2.
Read1UMILength The length of the UMIused for Read 1.
Read2UMILength The length of the UMIused for Read 2.
Read1UMIStartFromCycle The first cycle to use for UMI in Read 1.
The cycle index is absolute and not affected by
Read1StartFromCycle. The software supports UMIs only
at the beginning or end of reads.
Read2UMIStartFromCycle The first cycle to use for UMI in Read 2.
The cycle index is absolute and not affected by
Read2StartFromCycle. The software currently supports
UMIs only at the beginning or end of reads.
TrimUMI 0 (default) or 1 (true). When TrimUMI setting is set to 1, the
software trims the UMI bases from Read 1 and Read 2.
ExcludeTiles Tiles to exclude. Separate tiles using a plus sign [+], or
specified as a range with a hyphen [-]. For
example,ExcludeTiles,1101+2201+1301-1306
meansskip tiles 1101, 2201, and 1301 through 1306.
ExcludeTilesLaneX Tiles to exclude for Lane X. For example,
ExcludeTilesLane6,1101–1108 means skip tiles 1101
through 1108 for lane 6 only.
Table 7 Cycle and Tile Specifications

14 Document # 15051736 v01
Setting Description
CreateFastqForIndexReads 0 (default) or 1. If 1 (true), generate FASTQ files for index
reads. Normally, these FASTQ files are not needed,
because demultiplexing is carried out automatically based
on the sample sheet. Also, the index sequence is already
placed in the sequence identifiers in the FASTQ files.
Generating FASTQ files is based on the following:
•The index read masks are specified from the --use-
bases-mask option.
•The RunInfo.xml file when the --use-bases-mask
option is not used.
ReverseComplement 0 (default) or 1. If 1 (true), all reads are reverse
complemented as they are written to FASTQ files. This
step is necessary in certain unusual cases (eg processing of
mate-pair data using BWA, which expects paired-end
data).
Table 8 FASTQ Specifications
DataSection
The bcl2fastq2 Conversion Software uses the information in the columns of the Data
section.
Column Description
Sample_Project The sample project name. The software creates a directory with
the specified sample project name and stores the FASTQ files
there. You can use multiple samples in the same project.
Lane When specified, the software generates FASTQ files for only the
samples with the specified lane number.
Sample_ID The sample ID.
Sample_Name The sample name.
index The index sequence.
index2 The index sequence for index 2.
If the Sample_ID and Sample_Name columns do not match, the FASTQ files are placed
in an additional sub-directory called <SampleId>.
You can use alphanumeric characters, hyphens [-], and underscores [_] for the Sample_
Project, Sample_ID, andSample_Name.
Sample Sheet Demultiplexing Scenarios
The Illumina Experiment Manager performs the following for sample sheet BCL
conversion and demultiplexing:
}All reads are placed in the Undetermined_S0 FASTQ files when there is no sample
sheet.
}All reads are placed in the Undetermined_S0 FASTQ files when there is a sample
sheet but no data section.
}All reads are placed in the sample FASTQ file as defined in the sample sheet when
there is a sample sheet and one sample has no indexes.
}When there is a sample sheet and the samples have indexes, the software performs
the following:
}Reads without a matching index are placed in the default Undetermined_S0
FASTQ files.

Sample Sheet
bcl2fastq2 Conversion Software v2.18 Guide 15
}Reads with a valid index are placed in the sample FASTQ file as defined in the
sample sheet.
For each sample, there is one file per lane per read number when reads exist for that
sample, lane, and read number.
NOTE
When the Lane column of the sample sheet Data section is populated, only those lanes are
converted. When the Lane column is not used, all lanes are converted.
Create a Sample Sheet with IEM
The Illumina Experiment Manager (IEM) software helps you create and edit sample
sheets for Illumina sequencers and analysis software. You can use IEM to create sample
sheets for any Illumina sequencer and for any Nextera or TruSeq libraries.
You can download EIMat support.illumina.com/sequencing/sequencing_
software/experiment_manager/downloads.html.
View the Illumina Experience Manager User Guide for creating a sample sheet.

16 Document # 15051736 v01
Run BCL Conversion and Demultiplexing
Use the following command to run the bcl2fastq2 Conversion Software :
nohup /usr/local/bin/bcl2fastq [options]
An example of a command with options:
nohup /usr/local/bin/bcl2fastq --runfolder-dir <RunFolder>
--output-dir <BaseCalls>
This command produces a set of FASTQ files in the BaseCalls directory. Reads with an
unresolved or erroneous index are placed in the Undetermined_S0 FASTQ files. By
default, --runfolder-dir is the current directory and --output-dir is the
Data/Intensities/BaseCalls sub-directory of the run folder.
NOTE
To generate a log file for a problematic bcl2fastq run, use the -l or --min-log-level
DEBUG option. By default, bcl2fastq generates a log file with logging level INFO.
BCL2FASTQ Options
The main command line options are the --runfolder-dir and --output-dir. For
command line options that have a corresponding sample sheet setting, the value passed
on the command line overwrites the value found in the sample sheet.
Option Description
-R, --runfolder-dir Path to run folder directory
Default: ./
-o, --output-dir Path to demultiplexed output
Default: <runfolder-
dir>/Data/Intensities/BaseCalls/
Table 9 Main Options
You can use the following advanced options for non-default settings or for customized
settings.
Option Description
-i, --input-dir Path to input directory
Default: <runfolder-dir>/Data/Intensities/BaseCalls/
--intensities-dir Path to intensities directory
If intensities directory is specified, then the input
directory must also be specified.
Default: <input-dir>/../
--interop-dir Path to demultiplexing statistics directory
Default: <runfolder-dir>/InterOp/
--stats-dir Path to human-readable demultiplexing statistics
directory
Default: <runfolder-dir>/Stats/
--reports-dir Path to reporting directory
Default: <runfolder-dir>/Reports/
--sample-sheet Path to sample sheet, so you can specify the location
and name of the sample sheet, if different from
default.
Default: <runfolder-dir>/SampleSheet.csv
Table 10 Directory Options

Run BCL Conversion and Demultiplexing
bcl2fastq2 Conversion Software v2.18 Guide 17
For processing, if your computing platform supports threading, the software manages the
threads by the following defaults:
}4 threads for reading the data
}4 threads for writing the data
}20% for demultiplexing data
}100% for processing demultiplexed data
The file i/o threads spend most of their time sleeping, and so take little processing time.
The processing of demultiplexed data is allocated 1 thread per CPU to make sure that
there are no idle CPUs, resulting in more threads than CPUs by default. You can use the
following options to provide control on threading. If, for example, you share your
computing resources with colleagues and wish to limit your usage, these options are
useful.
Option Description
-r, --loading-
threads
Number of threads used for loading BCL data.
Default depends on architecture.
-d,
--demultiplexing-
threads
Number of threads used for demultiplexing,
Default depends on architecture.
-p,
--processing-threads
Number of threads used for processing demultiplexed data.
Default depends on architecture.
-w,
--writing-threads
Number of threads used for writing FASTQ data. This number
must not be higher than number of samples.
Default depends on architecture.
Table 11 Processing Options
If you want to use these options to assign multiple threads, consider the following:
}The most CPU demanding stage is the processing step (-p option). Assign this step
the most threads.
}The second most CPU demanding stage is the demultiplexing step (-d option).
Assign this step the second highest number of threads. Tests indicate 20% of
processing time is used for demultiplexing a HiSeq X run.
}Reading and writing stages are lightweight and do not need many threads. This
consideration is especially important for a local hard drive where too many threads
mean too many parallel read write actions giving suboptimal performance.
}Use one thread per CPU core plus a little more to supply CPU with work. This
method prevents CPUs being idle due to a thread being blocked while waiting for
another thread.
}The number of threads depends on the data. If you specify more writing threads than
samples, the extra threads do no work but can cost time due to context switching.
Option Description
--adapter-stringency The minimum match rate that would trigger the masking or
trimming process. This value is calculated as MatchCount /
(MatchCount + MismatchCount) and ranges from 0 to 1, but it is
not recommended to use any value <0.5, as this value would
introduce too many false positives. The default value for this
parameter is 0.9, meaning that only reads with >90% sequence
identity with the adapter are trimmed.
Default: 0.9
Table 12 Behavioral Options

18 Document # 15051736 v01
Option Description
--aggregated-tiles This flag tells the converter about the structure of the input files.
Accepted values:
AUTO Automatically detects the tile setting
YES Tiles are aggregated into single input file
NO There are separate input files for individual tiles
Default: AUTO
--barcode-mismatches Number of allowed mismatches per index
Multiple entries, comma delimited allowed. Each entry is
applied to the corresponding index; last entry applies to all
remaining indexes.
Default: 1. Accepted values: 0, 1 or 2.
--create-fastq-for-
index-reads
Create FASTQ files also for Index Reads.
Generating FASTQ files is based on the following:
•The index read masks are specified from the --use-bases-mask
option.
•The RunInfo.xml file when the --use-bases-mask option is not
used.
--ignore-missing-
bcls
Missing or corrupt BCL files are ignored. Assumes 'N'/'#' for
missing calls
--ignore-missing-
filter
Missing or corrupt filter files are ignored. Assumes Passing
Filter for all clusters in tiles where filter files are missing.
--ignore-missing-
positions
Missing or corrupt positions files are ignored. If corresponding
position files are missing, bcl2fastq writes unique coordinate
positions in FASTQ header.
--ignore-missing-
controls
Missing or corrupt control files are ignored. Missing controls: 0
--minimum-trimmed-
read-length
Minimum read length after adapter trimming. bcl2fastq trims
the adapter from the read down to the value of this parameter.
If there is more adapter match below this value, then those
bases are masked, not trimmed (replaced by N rather than
removed).
Default: 35
--mask-short-
adapter-reads
This option applies when a read is trimmed to below the length
specified by the --minimum-trimmed-read-length option
(default of 35). These parameters specify the following
behavior:
If the number of bases left after adapter trimming is less than -
-minimum-trimmed-read-length, force the read length to
be equal to --minimum-trimmed-read-length by masking
adapter bases (replace with Ns) that fall below this length.
If the number of ACGT bases left after this process falls below -
-mask-short-adapter-reads, mask all bases, resulting in a
read with --minimum-trimmed-read-length number of Ns.
Default: 22
--tiles The --tiles argument takes a regular expression to select for
processing only a subset of the tiles available in the flow cell.
This argument can be specified multiple times, one time for each
regular expression. Examples:
To select all the tiles ending with 5 in all lanes:
--tiles [0–9][0–9][0–9]5
To select tile 2 in lane 1 and all the tiles in the other lanes:
--tiles s_1_0002 --tiles s_[2–8]

Run BCL Conversion and Demultiplexing
bcl2fastq2 Conversion Software v2.18 Guide 19
Option Description
--use-bases-mask The --use-bases-mask string specifies how to use each cycle.
An nmeans ignore the cycle.
AY(or y) means use the cycle.
An Imeans use the cycle for the Index Read.
A number means that the previous character is repeated that
many times.
An asterisk [*] means that the previous character is repeated
until the end of this read or index (length according to the
RunInfo.xml).
The read masks are separated with commas: ,
The format for dual indexing is as follows: --use-bases-mask
Y*,I*,I*,Y* or variations thereof as specified.
You can also specify the --use-bases-mask multiple times for
separate lanes, like this way:
--use-bases-mask 1:y*,i*,i*,y* --use-bases-mask
y*,n*,n*,y*
Where the 1: means: Use this setting for lane 1. In this case, the
second --use-bases-mask parameter is used for all other
lanes.
If this option is not specified, the mask is determined from the
'RunInfo.xml file in the run directory. If it cannot do this
determination, supply the --use-bases-mask.
When the --use-bases-mask option is specified, the number
of index cycles and the length of index in the sample sheet
should match.
--with-failed-reads Include all clusters in the output, even clusters that are non-PF.
These clusters would have been excluded by default.
--write-fastq-
reverse-complement
Generate FASTQ files containing reverse complements of actual
data.
--no-bgzf-
compression
Turn off BGZF compression, and use GZIP for FASTQ files.
BGZF compression allows downstream applications to
decompress in parallel. This parameter is available in case a
consumer of FASTQ data cannot handle all standard GZIP
formats.
--fastq-compression-
level
Zlib compression level (1–9) used for FASTQ files.
Default: 4
--no-lane-splitting Do not split FASTQ files by lane.
--find-adapters-
with-sliding-window
Find adapters with simple sliding window algorithm. Insertions
and deletions of bases inside the adapter sequence are not
handled.
NOTE
Do not use the --no-lane-splitting option if you want to upload the resulting FASTQ
files to BaseSpace. The FASTQ files generated from the --no-lane-splitting option
are not compatible with the BaseSpace file uploader. Files generated without this option
(the default setting) are compatible for upload to BaseSpace.
Option Description
-h,
--help
Produce help message and exit
-v,
--version
Print program version information
Table 13 General Options

20 Document # 15051736 v01
Option Description
-l,
--min-log-level
Minimum log level
Recognized values: NONE, FATAL, ERROR, WARNING, INFO,
DEBUG, TRACE
To generate a log file for a problematic bcl2fastq2 run, use the -
lor --min-log-level DEBUG option.
Default: INFO

BCL Conversion Output Files
bcl2fastq2 Conversion Software v2.18 Guide 21
BCL Conversion Output Files
The bcl2fastq2 Conversion Software provides the following output files: output directory
has the following characteristics:
}FASTQFiles
}InterOp Files
}ConversionStats File
}DemultiplexingStats File
}AdapterTrimming File
}FastqSummary and DemuxSummary
}HTML Reports
}JSONFile
FASTQ Files
The bcl2fastq2 Conversion Software converts *.bcl, *.bcl.gz, and *.bcl.bgzf files into
FASTQ files, which can be used as input for secondary analysis. When there is no
sample sheet, the software generates a Undetermined_S0 FASTQ file for each lane and
read number combination.
FASTQ File Names
FASTQ files are named with the sample name and the sample number. The sample
number is a numeric assignment based on the order that the sample is listed for the run.
For example:
Data\Intensities\BaseCalls\samplename_S1_L001_R1_001.fastq.gz
}samplename—The sample name listed for the sample. If a sample name is not
provided, the file name includes the sample ID.
}S1—The sample number based on the order that samples are listed for the run
starting with 1. In this example, S1 indicates that this sample is the first sample
listed for the run.
NOTE
Reads that cannot be assigned to any sample are written to a FASTQ file for sample
number 0, and excluded from downstream analysis.
}L001—The lane number.
}R1—The read. In this example, R1 means Read 1. For a paired-end run, a file from
Read 2 includes R2 in the file name. When generated, the Index Reads are I1 or I2.
}001—The last segment is always 001.
FASTQ files are compressed in the GNU zip format, as indicated by *.gz in the file name.
FASTQ files can be uncompressed using tools such as gzip (command-line) or 7-zip
(GUI).
FASTQ File Format
FASTQ file is a text-based file format that contains base calls and quality values per read.
Each record contains 4 lines:
}The identifier
}The sequence
}A plus sign (+)
}The quality scores in an ASCII encoded format

22 Document # 15051736 v01
The identifier is formatted as:
@Instrument:RunID:FlowCellID:Lane:Tile:X:Y:UMI
ReadNum:FilterFlag:0:SampleNumber
Example:
@SIM:1:FCX:1:15:6329:1045 1:N:0:2
TCGCACTCAACGCCCTGCATATGACAAGACAGAATC
+
<>;##=><9=AAAAAAAAAA9#:<#<;<<<????#=
Identifiers Description
@Each sequence identifier line starts with @.
instrument The instrument ID.
run number The run number on the instrument.
flowcell ID The flowcell ID.
lane The lane number.
tile The tile number.
x_pos The X coordinate of the cluster.
y_pos The Y coordinate of the cluster.
UMI [Optional] The Unique Molecular Identifiers (UMIs) are
restricted to A/T/G/C/N. The UMIsequences for Read 1 and
Read 1 are separated by a plus sign (+) when the UMIs are
specified in the sample sheet.
read Read 1—Single read.
Read 2—Paired-end read.
is filtered Y—The read is filtered.
N—The read is not filtered.
control number 0—No control bits are turned on.
Even number—Control bits are turned on.
index The Index reads are restricted to A/T/G/C/N.
Table 14 Identifiers Table
FASTQ Compression
FASTQ files are compressed in the GNU zip format, as indicated by *.gz in the file name.
FASTQ files can be uncompressed using tools such as gzip (command-line) or 7-zip
(GUI).
The BGZF variant facilitates parallel decompression of the FASTQ files by downstream
applications. If a downstream application cannot handle the BGZF variant, it can be
turned off with the --no-bgzf-compression command line.
FASTQ Control Values
When the read is identified as a control value, the number is greater than 0 and the
value specifies the type of control. When the read is not identified as a control, the 10th
column is 0.
The value is the decimal representation of a bit-wise encoding scheme. The scheme bit 0
has a decimal value of 1; bit 1 has a value of 2, bit 2 has a value of 4, and so on.
Quality Scores
A quality score, or Q-score, is a prediction of the probability of an incorrect base call. A
higher Q-score implies that a base call is more reliable.

BCL Conversion Output Files
bcl2fastq2 Conversion Software v2.18 Guide 23
Based on the Phred scale, the Q-score serves as a compact way to communicate small
error probabilities. Given a base call, X, the probability that X is not true, P(~X), results in
a quality score, Q(X), according to the relationship:
Q(X) = -10 log10(P(~X))
where P(~X) is the estimated error probability.
The following table shows the relationship between the quality score and error
probability.
Quality Score Q(X) Error Probability P(~X)
Q40 0.0001 (1 in 10,000)
Q30 0.001 (1 in 1,000)
Q20 0.01 (1 in 100)
Q10 0.1 (1 in 10)
For more information on the Phred quality score, see en.wikipedia.org/wiki/Phred_
quality_score.
During the sequencing run, base call quality scores are calculated after cycle 25 and
results are recorded in base call (*.bcl) files, which contain the base call and quality score
per cycle.
Quality Scores Encoding
In FASTQ files, quality scores are encoded into a compact form, which uses only 1 byte
per quality value. In this encoding, the quality score is represented as the character with
an ASCII code equal to its value + 33. The following table demonstrates the relationship
between the encoding character, its ASCII code, and the quality score represented.
NOTE
When Q-score binning is in use, the subset of Q-scores applied by the bins is displayed.

24 Document # 15051736 v01
Symbol ASCII Code Q-
Score
! 33 0
" 34 1
# 35 2
$ 36 3
% 37 4
& 38 5
' 39 6
( 40 7
) 41 8
* 42 9
+ 43 10
, 44 11
- 45 12
. 46 13
/ 47 14
0 48 15
1 49 16
2 50 17
3 51 18
4 52 19
5 53 20
Symbol ASCII Code Q-
Score
6 54 21
7 55 22
8 56 23
9 57 24
: 58 25
; 59 26
< 60 27
= 61 28
> 62 29
? 63 30
@ 64 31
A 65 32
B 66 33
C 67 34
D 68 35
E 69 36
F 70 37
G 71 38
H 72 39
I 73 40
Table 15 ASCII Characters Encoding Q-scores 0–40
InterOp Files
You can locate the InterOp files in the directory: <run directory>/InterOp. The
directory contains binary files used by the Sequencing Analysis Viewer (SAV) software to
summarize various analysis metrics, such as cluster density, intensities, quality scores,
and overall run quality.
The index metrics are stored in the IndexMetricsOut.bin file, which has the following
binary format:
Byte 0: file version (1)
Bytes (variable length): record:
}2 bytes: lane number (unint16)
}2 bytes: tile number (unint16)
}2 bytes: read number (unint16)
}2 bytes: number of bytes Y for index name (unint16)
BCL Conversion Output Files
bcl2fastq2 Conversion Software v2.18 Guide 25
}Y bytes: index name string (string in UTF8Encoding)
}4 bytes: # clusters identified as index (uint32)
}2 bytes: number of bytes V for sample name (unint16)
}V bytes: sample name string (string in UTF8Encoding)
}2 bytes: number of bytes W for sample project (unint16)
}W bytes: sample project string (string in UTF8Encoding)
ConversionStats File
You can locate the ConversionStats.xml file in the directory: <run
directory>/Stats/, or in the directory specified by the --stats-dir option.
The file contains the following information per tile:
}Raw Cluster Count
}Read number
}YieldQ30
}Yield
}QualityScore Sum
The file contains the following information per lane:
}Lane Number
DemultiplexingStats File
You can locate the DemultiplexingStats.xml file in the directory: <run
directory>/Stats/, or in the directory specified by the --stats-dir option. The file
contains the following information per lane, barcode, and sample, project.
Also, the file contains the following information for flow cell:
}Barcode Count
}PerfectBarcode Count
}OneMismatchBarcode Count
AdapterTrimming File
The AdapterTrimming file is a text-based file format that contains a statistic summary of
adapter trimming for the FASTQ file. You can locate the file in the <run
directory>/Stats/ or in the directory specified by the --stats-dir option.
The file contains the following information:
}Lane
}Read
}Project
}Sample ID
}Sample Name
}Sample Number
}TrimmedBases
}PercentageOfBased (being trimmed)
Also, the file contains the fraction of reads with untrimmed bases for each sample, lane,
and read number.

26 Document # 15051736 v01
FastqSummaryF1L#
The FastqSummaryF1L#.txt file (the #indicates the lane number) contains the number of
raw and passed filter reads for each sample number and tile.
DemuxSummaryF1L#
The DemuxSummaryF1L#.txt file (the #indicates the lane number) contains the
percentage of each tile that each sample makes up. The file also contains a list of the
1,000 most common unknown barcode sequences.
HTML Report
The HTML reports are generated from data in the DemultiplexingStats.xml and
ConversionStats.xml files. You can locate the reports in the directory: <run
directory>/Reports/html/, or in the directory specified by the --reports-dir
option.
The Flowcell Summary contains the following information:
}Clusters (Raw)
}Clusters (PF)
}Yield (MBases)
NOTE
For HiSeq X, HiSeq 4000, and HiSeq 3000, the number of raw clusters is actually the
number of wells on the flow cell that could potentially be seeded. The value is the same in
all cases.
The Lane Summary provides the following information for each project, sample, and
index sequence specified in the sample sheet:
}Lane #
}Clusters (Raw)
}% of the Lane
}% Perfect Barcode
}% One Mismatch
}Clusters (Filtered)
}Yield
}% PF Clusters
}%Q30 Bases
}Mean Quality Score
The Top Unknown Barcodes table in the HTML report provides the count and sequence
for the 10 most common unmapped bar codes in each lane.
JSON File
The Java Script Object Notification (JSON)file contains the *.json file extension. The
format for the JSON file makes it easier to parse the output data. The data in the
JSONfile are a combination of all the following files:
}InterOP
}ConversionStats
}DemultiplexingStats
}Adapter Trimming
}FastqSummary and DemuxSummary
BCL Conversion Output Files
bcl2fastq2 Conversion Software v2.18 Guide 27
}HTMLReport
28 Document # 15051736 v01
Troubleshooting
}If the bcl2fastq2 Conversion Software fails to complete a run, it could be missing an
input file or have a corrupt file. View the log file for missing or corrupt files. The
exact wording of the file status reported varies depending on the nature of the file
corruption. If the problem is the BCL file, launch the --ignore-missing-bcls
option. See BCLAdvanced Options.
}If there is a high percentage of reads assigned as undetermined, view the Top
Unknown Barcodes table in the HTMLreport on the index sequence.
}If the bcl2fastq2 Conversion Software has problems processing Small RNAsamples,
use the --minimum-trim-read-length 20 and --mask-short-adapter-
reads 20 command line instead of the default settings.

Appendix: Installation Requirements
bcl2fastq2 Conversion Software v2.18 Guide 29
Appendix: Installation Requirements
The bcl2fastq2 Conversion Software requires the following components:
Component Requirements
Network
Infrastructure
1 Gigabit minimum.
Server Infrastructure Single multiprocessor or multicore computer running Linux.
Analysis Computer Run software on the Linux operating systems only.
Memory 32 GB RAM.
Software We recommend the RedHat Enterprise Linux 5 platform. The
following software is required:
•zlib
•librt
•libpthread
The following software are required to build the bcl2fastq2
Conversion Software :
•gcc 4.7 (with support for C++11)
•boost 1.54
•CMake 2.8.9
•zlib
•librt
•libpthread
Notes

Revision History
bcl2fastq2 Conversion Software v2.18 Guide 31
Revision History
Part# Revision Date Description of Change
15051736 G July 2015 Updated to software requirements, gcc version.
15051736 F June 2015 Updated to support bcl2fastq2 v2.17.
15051736 01 April 2016 •Updated to support bcl2fastq2 v2.18.
•Reformatted the User Guide to Illumina style standards.
•Added JSON file and input files list for MiniSeq.
•Revised BCL2FASTQ options and sample sheet settings.
Notes

Technical Assistance
bcl2fastq2 Conversion Software v2.18 Guide
Technical Assistance
For technical assistance, contact Illumina Technical Support.
Website www.illumina.com
Email techsupport@illumina.com
Table 16 Illumina General Contact Information
Region Contact Number Region Contact Number
North America 1.800.809.4566 Japan 0800.111.5011
Australia 1.800.775.688 Netherlands 0800.0223859
Austria 0800.296575 New Zealand 0800.451.650
Belgium 0800.81102 Norway 800.16836
China 400.635.9898 Singapore 1.800.579.2745
Denmark 80882346 Spain 900.812168
Finland 0800.918363 Sweden 020790181
France 0800.911850 Switzerland 0800.563118
Germany 0800.180.8994 Taiwan 00806651752
Hong Kong 800960230 United Kingdom 0800.917.0041
Ireland 1.800.812949 Other countries +44.1799.534000
Italy 800.874909
Table 17 Illumina Customer Support Telephone Numbers
Safety data sheets (SDSs)—Available on the Illumina website at
support.illumina.com/sds.html.
Product documentation—Available for download in PDF from the Illumina website. Go
to support.illumina.com, select a product, then select Documentation & Literature.

Illumina
5200 Illumina Way
San Diego, California 92122 U.S.A.
+1.800.809.ILMN (4566)
+1.858.202.4566 (outside North America)
techsupport@illumina.com
www.illumina.com