EcoPlate™ Biolog Ecoplate Instructions
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 2
Microbial Community Analysis
EcoPlate™
INTRODUCTION
Microbial communities provide useful information about
environmental change. Microorganisms are present in
virtually all environments and are typically the first organisms
to react to chemical and physical changes in the environment.
Because they are near the bottom of the food chain, changes in
microbial communities are often a precursor to changes in the
health and viability of the environment as a whole.
The Biolog EcoPlate™ (Figure 1) was created specifically for
community analysis and microbial ecological studies. It was
originally designed at the request of a group of microbial
ecologists that had been using the Biolog GN MicroPlate,
but wanted a panel that provided replicate sets of tests1.
Community analysis using Biolog MicroPlates was originally
described in 1991 by J. Garland and A. Mills2. They and other
researchers found that by inoculating Biolog GN MicroPlates
with a mixed population of microorganisms and measuring the
community metabolism over time, they could ascertain
characteristics of that community. This approach, called
community–level physiological profiling, or CLPP, has been
demonstrated to be effective at distinguishing spatial and
temporal changes in microbial communities. In applied
ecological research EcoPlates are used as both an assay of the
stability of a normal population and to detect and assess
changes following the onset of an environmental variable.
Studies have been done in diverse applications of microbial
ecology and have demonstrated the fundamental utility of
EcoPlates in detecting population changes in soil, water,
wastewater, activated sludge, compost, and industrial waste.
The utility of the information has been documented in
hundreds of publications using Biolog technology to analyze
microbial communities. A bibliography of publications is
posted on the Biolog website at
www.biolog.com/bibliography.php.
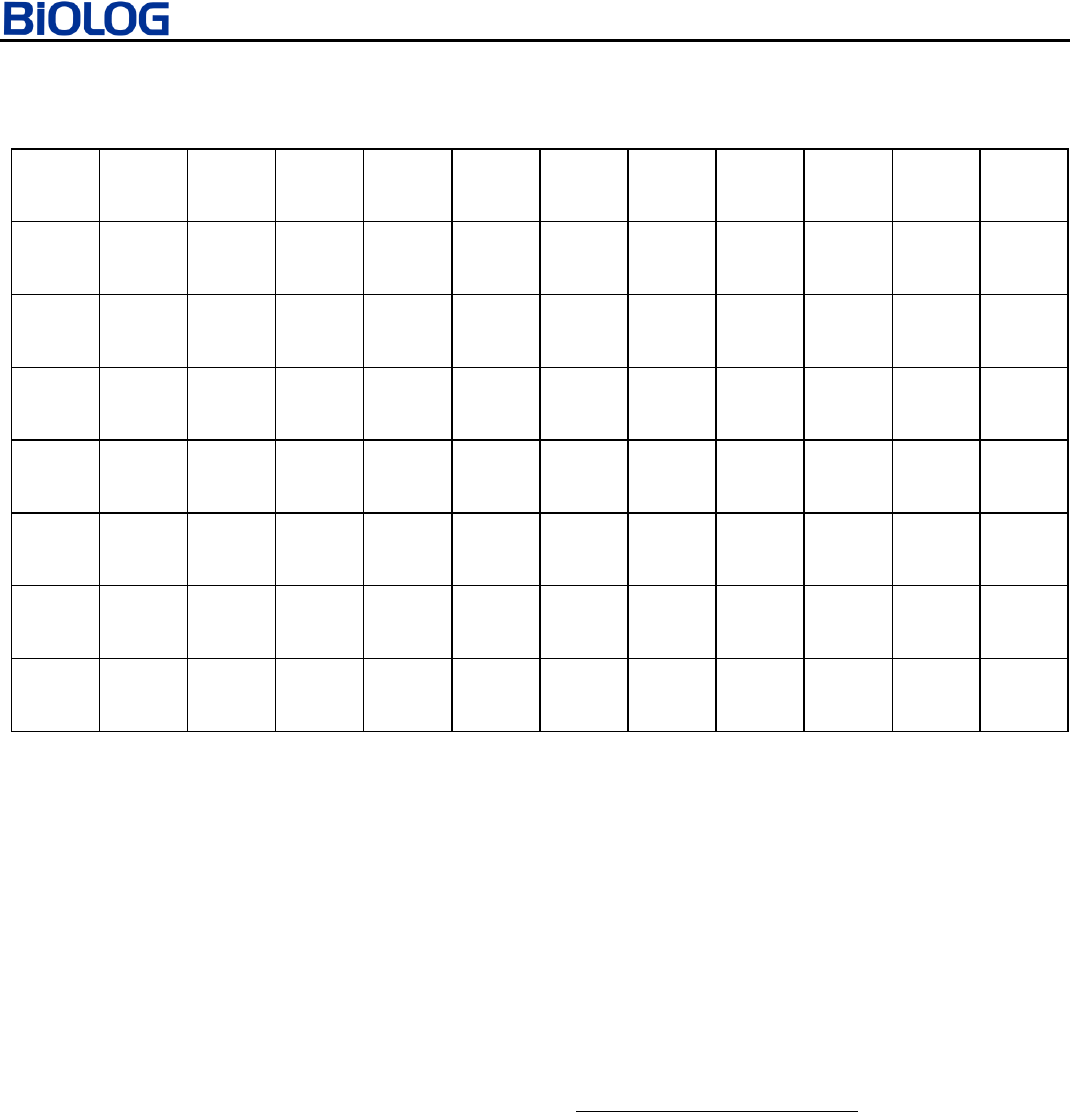
ECOPLATE
The EcoPlate contains 31 carbon sources that are useful for
community analysis. These 31 carbon sources are repeated 3
times to give the scientist more replicates of the data.
Communities of microorganisms will give a characteristic
reaction pattern called a metabolic fingerprint. From a single
EcoPlate, these fingerprint reaction patterns rapidly and easily
characterize the community.
A1
Water
A2
-Methyl-D-
Glucoside
A3
D-Galactonic
Acid
-Lactone
A4
L-Arginine
A5
Water
A6
-Methyl-D-
Glucoside
A7
D-Galactonic
Acid
-Lactone
A8
L-Arginine
A9
Water
A10
-Methyl-D-
Glucoside
A11
D-Galactonic
Acid
-Lactone
A12
L-Arginine
B1
Pyruvic Acid
Methyl Ester
B2
D-Xylose
B3
D-
Galacturonic
Acid
B4
L-Asparagine
B5
Pyruvic Acid
Methyl Ester
B6
D-Xylose
B7
D-
Galacturonic
Acid
B8
L-Asparagine
B9
Pyruvic Acid
Methyl Ester
B10
D-Xylose
B11
D-
Galacturonic
Acid
B12
L-Asparagine
C1
Tween 40
C2
i-Erythritol
C3
2-Hydroxy
Benzoic Acid
C4
L-
Phenylalanine
C5
Tween 40
C6
i-Erythritol
C7
2-Hydroxy
Benzoic Acid
C8
L-
Phenylalanine
C9
Tween 40
C10
i-Erythritol
C11
2-Hydroxy
Benzoic Acid
C12
L-
Phenylalanine
D1
Tween 80
D2
D-Mannitol
D3
4-Hydroxy
Benzoic Acid
D4
L-Serine
D5
Tween 80
D6
D-Mannitol
D7
4-Hydroxy
Benzoic Acid
D8
L-Serine
D9
Tween 80
D10
D-Mannitol
D11
4-Hydroxy
Benzoic Acid
D12
L-Serine
E1
-
Cyclodextrin
E2
N-Acetyl-D-
Glucosamine
E3
-Amino
Butyric Acid
E4
L-Threonine
E5
-
Cyclodextrin
E6
N-Acetyl-D-
Glucosamine
E7
-Amino
Butyric Acid
E8
L-Threonine
E9
-
Cyclodextrin
E10
N-Acetyl-D-
Glucosamine
E11
-Amino
Butyric Acid
E12
L-Threonine
F1
Glycogen
F2
D-
Glucosaminic
Acid
F3
Itaconic Acid
F4
Glycyl-L-
Glutamic Acid
F5
Glycogen
F6
D-
Glucosaminic
Acid
F7
Itaconic Acid
F8
Glycyl-L-
Glutamic Acid
F9
Glycogen
F10
D-
Glucosaminic
Acid
F11
Itaconic Acid
F12
Glycyl-L-
Glutamic Acid
G1
D-Cellobiose
G2
Glucose-1-
Phosphate
G3
-Keto
Butyric Acid
G4
Phenylethyl-
amine
G5
D-Cellobiose
G6
Glucose-1-
Phosphate
G7
-Keto
Butyric Acid
G8
Phenylethyl-
amine
G9
D-Cellobiose
G10
Glucose-1-
Phosphate
G11
-Keto
Butyric Acid
G12
Phenylethyl-
amine
H1
-D-Lactose
H2
D,L--
Glycerol
Phosphate
H3
D-Malic Acid
H4
Putrescine
H5
-D-Lactose
H6
D,L--
Glycerol
Phosphate
H7
D-Malic Acid
H8
Putrescine
H9
-D-Lactose
H10
D,L--
Glycerol
Phosphate
H11
D-Malic Acid
H12
Putrescine
FIGURE 1. Carbon Sources in EcoPlate

Microbial Community Analysis
21124 Cabot Blvd. Hayward, CA 94545 Telephone: 510-785-2564 Fax: 510-782-4639 www.biolog.com
EcoPlate, MicroPlate, Phenotype MicroArray and MicroStation are trademarks; OmniLog is a registered trademark of Biolog, Inc., Hayward, CA
Part# 00A 012, Rev. D, June 2018
The community reaction patterns are typically analyzed at
defined time intervals over 2 to 5 days. The changes in the
pattern are compared and analyzed using statistical analysis
software. The most popular method of analysis of the data is
via Principle Components Analysis (PCA) of average well
color development (AWCD) data, but alternative methods
may also offer advantages311. The changes observed in the
fingerprint pattern provide useful data about the microbial
population changes over time.
TYPICAL PROCEDURE3
STEP 1: Environmental samples are inoculated directly into
EcoPlates either as aqueous samples or after suspension (soil,
sludge, sediment, etc…).
STEP 2: The EcoPlates are incubated and analyzed at defined
time intervals.
STEP 3: The community-level physiological profile is
assessed for key characteristics:
o Pattern development (similarity)
o Rate of color change in each well (activity)
o Richness of well response (diversity)
Formation of purple color occurs when the microbes can
utilize the carbon source and begin to respire. The respiration
of the cells in the community reduces a tetrazolium dye that is
included with the carbon source.
The reaction patterns are most effectively analyzed using the
MicroStation™ System or an OmniLog® Instrument configured
for Phenotype MicroArray™ Analysis, which is especially
useful when reading a large number of plates, or when kinetic
analysis is required. However, any good microplate reader can
be used to provide optical density (OD590) values.
Statistical analysis of the data is typically performed using
standard software packages. Some researchers have found
that PCA provides greater resolution than other methods of
statistical analysis11.
EcoPlates: Catalog No. 1506 (10/box)
REFERENCES
[1] A new set of substrates proposed for community
characterization in environmental samples. H. Insam,
p. 260-261, In: Microbial Communities. Functional
versus structural approaches, H. Insam and A.
Rangger, editors, 1997, Springer.
[2] Classification and characterization of heterotrophic
microbial communities on the basis of patterns of
community level sole-carbon-source utilization. J.L.
Garland, A.L. Mills, Applied and Environmental
Microbiology, 1991, v.57, p. 2351-2359.
[3] Analysis and interpretation of community-level
physiological profiles in microbial ecology. J.L.
Garland, Federation of European Microbiological
Societies, Microbiology Ecology, 1997, v. 24, p289-
300.
[4] Community analysis by Biolog: curve integration for
statistical analysis of activated sludge microbial
habitats, J.B. Guckert, G.J. Carr, T.D. Johnson, B.G.
Hamm, D.H. Davidson, Y. Kumagai, Journal of
Microbiological Methods, 1996, v. 27:2-3, p. 183-
187.
[5] Statistical analysis of the time-course of Biolog
substrate utilization. C.A. Hackett, B.S. Griffiths,
Journal of Microbiological Methods, 1997, v. 30, p.
63-69.
[6] Statistical comparisons of community catabolic
profiles. E. Glimm, H. Heuer, B. Engelen, K. Smalla,
H. Backhaus, Journal of Microbiological Methods,
1997, v. 30, p. 71-80.
[7] Application of multivariate analysis of variance and
related techniques in soil studies with substrate
utilization tests, W. Hitzl, M. Henrich, M. Kessel,
and H. Insam, Journal of Microbiological Methods,
1997, v. 30, p. 81-89.
[8] Using the Gini coefficient with BIOLOG substrate
utilization data to provide an alternative quantitative
measure for comparing bacterial soil communities,
B.D. Harch, R.L. Correll, W. Meech, C.A. Kirkby,
and C.E. Pankhurst, Journal of Microbiological
Methods, 1997, v. 30, p. 91-101.
[9] Monitoring soil bacteria with community-level
physiological profiles using Biolog EcoPlates in the
Netherlands and Europe, Michiel Rutgers, Marja
Wouterse, Sytske M. Drost, Anton M. Breure,
Christian Mulder, Dorothy Stone, Rachel E.
Creamer, Anne Winding and Jaap Bloem, Applied
Soil Ecology, 2016, v. 97, p. 23-35.
[10] Community-level physiological profiling. K.P.
Weber and R. L. Legge, p. 263-281, In:
Bioremediation, Methods in Microbial Ecology v.
599, S.P. Cummings, editor, 2010, Springer.
[11] Defining soil quality in terms of microbial
community structure. M. Firestone, T. Balser, D.
Herman, Annual Reports of Research Projects, UC
Berkeley, 1997