Manual V0.2
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 55
EOS — A HEP
Program for Flavor Physics
User Manual
Danny van Dyk
Christoph Bobeth
Frederik Beaujean
version 0.2
May 20, 2018
Contents
Acknowledgments v
How to read this manual vii
1. Installation 3
1.1. Installingthedependencies .................................. 3
1.1.1. Overview......................................... 3
1.1.2. Linux: Installing the dependencies with APT (Debian, Ubuntu) . . . . . . . . . . 4
1.1.3. Linux: Installing Minuit2 and pmclib fromsource ................. 4
1.1.4. MacOSX: Installing the Dependencies with Homebrew and PyPi ......... 5
1.2. InstallingEOS .......................................... 6
1.2.1. MacOSXshortcuts .................................. 7
2. Usage 9
2.1. Browsing the List of Parameters, Observables and Constraints . . . . . . . . . . . . . . 9
2.2. EvaluatingObservables..................................... 10
2.3. Producing Random Parameter Samples . . . . . . . . . . . . . . . . . . . . . . . . . . . 11
2.4. Merging Markov Chains from multiple files . . . . . . . . . . . . . . . . . . . . . . . . . 15
2.5. Finding the Mode of a Probability Density . . . . . . . . . . . . . . . . . . . . . . . . . . 16
2.6. Bayesian Uncertainty Propagation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
2.7. PlottingRandomSamples................................... 19
3. Library Interface 21
3.1. Design .............................................. 21
3.2. CoreClasses........................................... 22
3.2.1. Class Parameters ................................... 22
3.2.2. Class Kinematics ................................... 23
3.2.3. Class Options ..................................... 23
3.2.4. Class Observable ................................... 23
4. Extending EOS 25
4.1. Howtoaddanewparameter ................................. 25
4.2. Howtoaddanewobservable................................. 26
4.3. Howtoaddanewconstraint ................................. 26
4.3.1. Type Amoroso ...................................... 27
4.3.2. Type Gaussian ..................................... 27
4.3.3. Type MultivariateGaussian ............................. 28
4.3.4. Type MultivariateGaussian(Covariance) .................... 28
5. Effective Field Theories 33
5.1. |∆B|=|∆S|= 1 ........................................ 34
5.2. |∆B|=|∆U|= 1 ........................................ 36
5.3. |∆B|=|∆C|= 1 ........................................ 36
iii
Acknowledgments
The creation and continued maintenance of EOS would not have been possible without the assistance
and encouragement of Gudrun Hiller.
Furthermore, we would like to extend our thanks to the following people whose input and support were
most helpful either the development or the maintenance of EOS, either through personal contributions
to the code, independent code review, or helpful suggestions: Thomas Blake, Marzia Bordone, Elena
Graverini, Daniel Greenwald, Nico Gubernari, Ahmet Kokulu, Christoph Langenbruch, David Leverton,
Nazila Mahmoudi, Ciaran McCreesh, Hideki Miyake, Bastian Müller, Konstantinos Petridis, Stefanie
Reichert, Martin Ritter, Denis Rosenthal, Alexander Shires, Rafael Silva Coutinho, Ismo Toijala, Chris-
tian Wacker .
Further development and maintenance of EOS and this manual is presently funded by the Deutsche
Forschungsgemeinschaft (DFG) within the Emmy-Noether Programme under contract ’DY 130/1-1’.
v
How to read this manual
To improve readability, the following concepts or entities discussed in this manual are highlighted.
shell specic Shell commands are monospaced-lowercase-and-grayed . Environment vari-
ables are $ALL_CAPS_DOLLAR_PREFIXED .
echo " Example commands are boxed with a gray background ."
# Comments are prefixed with a hash mark.
OS specic File names are light-blue, and /directories/end/with/a/slash//. Package
names try to follow the original typesetting and are green and bold.
File contents are boxed with a light - blue background .
EOS specic Obserservables,options and parameters are orange and adhere to a syntax
defined later in this manual. Classes are CamelCaseNoUnderscores. Methods are (almost al-
ways) lower_case_with_underscores.
1print ( 'Source code is boxed with a light orange back ground .')
2#Source code listings have line numbers.
vii
Software Documentation
1

1. Installation
The following instructions explain how to install EOS from source on a Linux-based operating system,
such as Debian or Ubuntu, 1or MacOS X.
1.1. Installing the dependencies
1.1.1. Overview
The dependencies can be roughly categorized as either system software, Python-related software, or
scientific software.
EOS requires the following system software:
g++ the GNU C++ compiler, in version 4.8.1 or higher;
autoconf the GNU tool for creating configure scripts, in version 2.69 or higher;
automake the GNU tool for creating makefiles, in version 1.14.1 or higher;
libtool the GNU tool for generic library support, in version 2.4.2 or higher;
pkg-config the freedesktop.org library helper, in version 0.28 or higher;
BOOST the BOOST C++ libraries (sub-libraries boost-filesystem and boost-system);
yaml-cpp a C++ YAML parser and interpreter library.
The Phython [1] interface to EOS requires the additional software:
python3 python3 interpreter in version 3.5 or higher, and required header files;
BOOST the BOOST C++ library boost-python for interfacing Python and C++;
h5py the Python interface to HDF5;
matplotlib the Python plotting library;
scipy the Python scientific library;
PyYAML the Python YAML parser and emitter library.
We recommend you install the above packages via your system’s software management system.
EOS requires the following scientific software:
GSL the GNU Scientific Library [2], in version 1.16 or higher;
1Other avors of Linux will work as well, however, note that we will exclusively use package names as they appear in the
Debian/Ubuntu apt databases.
3
1. Installation
HDF5 the Hierarchical Data Format version 5 (HDF5) [3], in version 1.8.11 or higher;
FFTW3 the C subroutine library for computing the discrete Fourier transform;
Minuit2 the physics analysis tool for function minimization, in version 5.28.00 or higher.
The optional (and highly recommended) Population Monte Carlo (PMC) sampling algorithm requires:
pmclib a free implementation of said algorithm [4], in version 1.01 or higher.
We recommend you install the above packages via your system’s software management system, ex-
cept for pmclib and Minuit2 in particular cases discussed below.
1.1.2. Linux: Installing the dependencies with APT (Debian, Ubuntu)
You can install the prerequisite software with the apt package management system on any Debian or
Ubuntu system as follows:
# for the 'System Software '
apt - get install g ++ aut oconf automake li btool pkg - config libboost - filesystem - dev \
libboost -system -dev libyaml -cpp - dev
# for the 'Python Software '
apt -get install python3 -dev libboost -python - dev python3 - h5py python3 - matplotlib python3 -scipy \
python3 - yaml
# for the 'Scientific Software '
apt -get install libgsl0 -dev libhdf5 - serial -dev libfftw3 -dev
Do not install Minuit2 via APT, since there is presently a bug in the Debian/Ubuntu packages for Mi-
nuit2, which prevents linking.
There are pre-built binary package files for pmclib and Minuit2 available for the Ubuntu long-term-
support varieties Xenial and Bionic via the Packagecloud web service. To use the EOS third-party
repository, create a new file eos.list within the directory /etc/apt/sources.list.d/ with the fol-
lowing contents:
deb http s :// p ack agec loud . io /eos /eos / ubuntu / DIST main
deb -src https :// p ack agec loud . io / eos /eos /ubuntu / DIST main
where you must replace DIST with either xenial or bionic, depending on your version of Ubuntu. Add
our repository’s GPG key by running
curl -L " https :// packagecloud .io/eos/eos / gpgkey" 2> /dev / null | apt -key add - &>/ dev/null
You can then install the binary packages through
apt - get update
apt - get install minuit2 libpmc
You can then proceed with the EOS installation as documented in section 1.2.
1.1.3. Linux: Installing Minuit2 and pmclib from source
To install Minuit2 from source, you need to disable the automatic support for OpenMP. Execute the
following commands:
mkdir / tmp/Minuit2
pushd / tmp/Minuit2
wget http :// www .cern .ch /ma thlibs /sw /5 _2 8_00 /Minui t2 /Minuit2 -5 .28.00. tar .gz
tar zxf Minuit2 -5.28.00. tar .gz
pushd Minuit2 -5.28.00
./ c onfigu re -- prefix =/ opt / pkgs /Minui t2 -5.28.0 0 -- disa bl e - op enmp
4

1.1. Installing the dependencies
make all
sudo make install
popd
popd
rm -R /tmp / Minuit2
Installation of pmclib for EOS’s purposes requires some modifications to the source code to make it
compatible with C++. Execute the following commands:
mkdir / tmp/libpmc
pushd / tmp/libpmc
wg et http :// www2 .iap .fr / users / kilbing e/ CosmoPMC / pm clib _v1 .01. tar . gz
tar zxf pmclib_v1 .01. tar .gz
pushd pm clib_v1 .01
./ waf configure --m64 --prefix =/opt/pkgs / pmclib -1.01
./ waf
sudo ./waf install
sudo find /opt /pkgs /pmclib -1.01/ include -name "*. h" \
-exec sed -i \
-e 's /# i nclude "e rrorl ist \. h "/ # include < pmctoo ls \/ e rrorl ist .h >/ ' \
-e 's/# include "io \. h"/# include <pm ctools \/ io .h >/ ' \
-e 's/# include "mvdens \.h "/# include < pmctool s\/ mvdens .h >/ ' \
-e 's/# include "maths \. h"/# include <pm ctools \/ maths .h >/ ' \
-e 's /# inc lude " ma ths_b ase \. h "/ # include < pmctools \/ math s_bas e.h >/ ' \
{} \;
sudo sed -i \
-e 's /^ double fmi n( double /\/\/&/ ' \
-e 's /^ double fma x( double /\/\/&/ ' \
/opt /pkgs /pmclib -1.01/ include /pmctools /maths .h
popd
popd
rm -R /tmp / libpmc
Note: The waf build script used by pmclib is written in Python2. On systems that use Python3 as the
default interpreter, you will see an error message similar to the following:
./ waf configure --m64 --prefix =/opt/pkgs / pmclib -1.01
/tmp/src / libpmc /pmclib_v1 .01/ wscript : error: Traceback (most recent call last):
File \
"/ tmp /src /libpmc /pmcli b_v1 .01/. waf3 -1.5.17 -496 be6 959 d6e0 cd4 06d5 f087 856 c4d7 9/ wafadmin /Utils .py" , \
line 198, in load_module
exec (compile (code , file_path ,'exec ') ,module .__dict__ )
File "/ tmp /src / libpmc /p mclib_v1 .01/ wscript ", line 130
except Exception ,e:
^
SyntaxError : invalid syntax
You can fixed this problem by replacing python with python2 in the very first line of the file waf in the
top-level pmclib directory.
You can then proceed with the EOS installation as documented in section 1.2.
1.1.4. MacOSX: Installing the Dependencies with Homebrew and PyPi
You can install most of the prerequisite software via Homebrew. You will need to make Homebrew
aware of EOS’ third-party repository as follows:
brew tap eos /eos
To install the packages, execute the following commands:
# for the 'System Software '
brew install autoc onf automake libtool pkg - config boost yaml - cpp
# for the 'Python Software '
brew install python3 boost -python3
# for the 'Scientific Software '
brew install gsl hdf5 minuit2 pmclib
5

1. Installation
Now you can use the pip3 command2to install the remaining packages from the PyPi package
index:
pip3 install h5py matplotlib scipy PyYAML
You can then proceed with the EOS installation as documented in section 1.2.
1.2. Installing EOS
You can obtain the most recent version of EOS is available from Github 3. To download it for the first
time, clone the repository via
git clon e -o eos -b master https :// g ithub .com /eos / eos . git
You must first create all the necessary build scripts via
cd eos
./ autogen.bash
You must specify where EOS will be installed by setting the $PREFIX environment variable.
1. If you are administrating the computer on which you install EOS, you may choose any prefix. We
recommend /usr/local/:
export PREFIX =/usr/ local
We strongly discourage using /usr/, which avoids conicts with your operating system’s pack-
age management software.
2. If you are not administrating the computer, you must install EOS into a directory below your
home directory or below any other directory for which you have the needed permissions. We
recommend $HOME/.local/:
export PREFIX=$HOME /. local /
Configuration
Next, you must configure the EOS build using the configure script.
1. If you want to use the EOS Python interface you must pass the --enable-python option to
configure . The default is --disable-python .
2. If you want to use the EOS PMC algorithm you must pass the --enable-pmc option to configure .
The default is --disable-pmc . If you installed pmclib from source to a non-standard location,
you must specify its installation prefix by passing on --with-pmc=PMC-INSTALLATION-PREFIX .
3. If you want to use ROOT’s internal copy of Minuit2 you must pass --with-minuit2=root to
configure .
4. Otherwise, if you have installed Minuit2 from source to a non-standard location you must specify
its installation prefix by passing on --with-minuit2=MINUIT2-INSTALLATION-PREFIX .
2Due to problems with the Python 3 installation provided by Mac OS X, we strongly recommend to use the Homebrew
provided pip3 instead via /usr/local/bin/pip3 .
3http://github.com/eos/eos
6
1.2. Installing EOS
The recommended configuration is achieved using
./ co nfigur e \
-- prefix =$PREFIX \
--enable -python \
--enable -pmc
If the configure script finds any problems with your system, it will complain loudly.
Building and installing
After successful configuration, build EOS using
make -j all
The -j option instructs make to use all available processors to parallelize the build process. We
strongly recommend testing the build by executing
make -j check VERBOSE =1
within the build directory. Please contact the authors if any test fails by opening an issue in the official
EOS Github repository. If all tests pass, install EOS using
make install # Use 'sudo make install ' if you install e.g. to '/usr/local '
# or a similarly privileged directory
If you installed EOS to a non-standard location (i.e.not /usr/local/), to use it from the command line
you must set up some environment variable. For BASH, which is the default Debian/Ubuntu shell, add
the following lines to $HOME/.bash_profile:
export PATH +=": $PREFIX / bin "
# uncomment the following line for Python support
# export PYTH ONPATH +=": $PRE FIX /lib /python3 .5/ site - packag es "
The last line should be uncommented only if you built EOS with Python support. Note that python3.5
piece must be replaced by the appropriate Python version with which EOS was built. You can deter-
mine the correct value using
python3 -c " import sys ; print (' python {0}.{1} '. format (sys .vers ion_i nfo [0] , sys . versi on_inf o [1]) )"
1.2.1. MacOS X short cuts
For users
The most recent development version of EOS (and all of its dependencies) can also be installed auto-
matically using Homebrew:
brew tap eos /eos
brew install --HEAD eos
For developers
If you intend to make changes to the EOS source code you can still use Homebrew. First, you should
clone the EOS repository into your home directory. We suggest
mk dir -p ~/ Repo sito ries / eos
cd ~/ Repos itor ies / eos
git clo ne ht tps :// github . com / eos / eos . git -b ma ster
# make and commit your changes
7
1. Installation
To setup your local for use with Homebrew you must “tap” the EOS “formula” as usual, and then edit
it:
brew tap eos /eos
brew edit eos
Change the line starting with head to look as follows:
head "/ Users / USERNAME / Repositories /eos /.git", :branch => " master ", :using => :git
where USERNAME is your username. You can determine it via whoami . Proceed to install the modified
EOS version via brew install --HEAD eos . To reinstall EOS after further changes to the source
code use brew reinstall --HEAD eos .
Note: Only changes that have been committed to the master branch of your local clone of the EOS
repository will be used for the installation.
8

2. Usage
EOS has been authored with several use cases in mind.
1. The evaluation of observables and further theoretical quantities in the field of avor physics. EOS
aspires to produce theory estimates of publication quality, and has produced such estimates in
the past.
2. The inference of parameters from experimental observations. For this task, EOS defaults to the
Bayesian framework of parameter inference.
3. The production of toy events for a variety of avor-physics-related processes using Monte Carlo
methods.
In the remainder of this chapter we document the usage of the existing EOS clients and scripts, to carry
out tasks corresponding to the above use cases. We assume further that only the built-in observables,
physics models and experimental constraints are used. The necessary steps to extend EOS with new
observables, physics models or constraints is discussed in chapter 4.
2.1. Browsing the List of Parameters, Observables and Constraints
EOS has understands parameters, observables, options, kinematic variables, and constraints. Pa-
rameters, observables and constraints share a common naming scheme, in order to allow categorize
these quantities more readily. The naming scheme is as follows: each name can be decomposed into
a prefix (before the :: separator), an optional tag (after the optional separator), and the short name
(between the separators); e.g.:
K^*::a_1_para@1GeV
Here Kˆ* indicated the association with the K∗meson, a_1_para is the short name for this object,
and the tag is present and reads 1GeV.
Parameters are scalar oating-point-valued inputs. They are real-valued numbers that can be set to
arbitrarily at run time. The full list of parameters known to EOS as well as their default values and
ranges can be browsed by running eos-list-parameters .
Observables are EOS-internal functions that take a set of parameters and return a single (real-valued)
number. These functions take a set of kinematic variables as their arguments. They include true ob-
servables in the physical sense (e.g. a branching ratio) but also pseudo observables such as hadronic
form factors. A full list of observables known to EOS can be listed by running eos-list-observables .
Options are used to modify the behavior of an observable at run time. Universal examples include
changing the physics model (via model) and changing form factor parametrization (via form-factors).
However, generally their names are specific to a single observable. There is presently no way to list
all possible options that pertain to a given observable.
9

2. Usage
Kinematic variables are used to provide numerical inputs to an observable that can be changed on a
per-observable level. As such, the names of kinematic variables are specific to each observable. The
required kinematic variables for each observable can be looked up using the eos-list-observables
client.
Constraints represent individual likelihood functions, either of an experimental measurement or a the-
oretical prediction. The list of built-in constraints is available through eos-list-constraints .
2.2. Evaluating Observables
Observables can be evaluated using eos-evaluate . It accepts the following command line argu-
ments:
--kinematics NAME VALUE
Within the scope of the next observable, declare a new kinematic variable with name NAME and
numerical value VALUE .
--range NAME MIN MAX POINTS
Within the scope of the next observable, declare a new kinematic variable with name NAME .
Subdivide the interval [ MIN ,MAX ] in POINTS subintervals, and evaluate the observable at each
subinterval boundary.
Note: More than one --range command can be issued per observable, but only one --range
command per kinematic variable.
--observable NAME
Add a new observable with name NAME to the list of observables that shall be evaluated. All
previously issued --kinematics and --range arguments apply, and will be used by the new
observable. The kinematics will be reset (i.e., all kinematic variables will be removed) in antici-
pation of the next --observable argument.
--vary NAME
Estimate the uncertainty based on variation of the parameter NAME , as if the parameter was
distributed like a univariate Gaussian.
--budget NAME
Create a new uncertainty budget, which encompasses all the subsequently issued --vary com-
mands (until the issue of a new --budget command). By default, the delta budget always
exists, and encompasses all variations.
As an example, we use the evaluation of the q2-integrated branching ratio B(¯
B0→π+µ−¯νµ), which is
addressable as B->pilnu::BR. For this example, we use the integration range
0GeV2≤q2≤12 GeV2.
Further, we use the BCL2008 [5] parametrization of the ¯
B→πform factor, as well as the Wolfen-
stein parametrization of the CKM matrix. The latter is achieved by choosing the physics model SM.
By default, EOS uses the most recent results of the UTfit collaboration’s fit of the CKM Wolfenstein
10

2.3. Producing Random Parameter Samples
parameter to data on tree-level decays. In this example, we evaluate the observable and estimate
parametric uncertainties based on the naive expectation of Gaussian uncertainty propagation. We
classify two budgets of parametric uncertainties: one for uncertainties pertaining to the form factors
(labeled ’FF’), and one for uncertainties pertaining to the CKM matrix elements (labeled ’CKM’).
Our intentions translate to the following call to eos-evaluate :
eos - evaluate \
-- kinematics s_min 0.0 \
-- kinematics s_max 12.0 \
-- observable "B -> pilnu ::BR ;l=mu ,form -factors =BCL2008" \
-- budget "FF" \
-- vary "B -> pi :: f_ +(0) @BCL2008 " \
-- vary "B -> pi :: b_ +^1 @ BCL2008 " \
-- vary "B -> pi :: b_ +^2 @ BCL2008 " \
-- budget "CKM" \
-- vary " CKM:: lambda " \
-- vary " CKM::A" \
-- vary " CKM:: rhobar " \
-- vary "CKM :: etabar "
The above call yields the following output:
# B-> pilnu ::BR : form - factors =BCL2008 ,l= mu
# s_min s_max central FF_min FF_max CKM_min CKM_max delta_min delta_max
0 12 0.000106816 3.00927e -05 1.46426e -05 9.14007e -06 9.70515e -06 3.14501e -05 1.75669e -05 \
( -29.4434% / +16.44 6%)
The output of calls to eos-evaluate is structured as follows:
• The first row names the observable at hand, as well as all active options.
• The second row contains column headers in the order:
–kinematics variables,
–the upper and lower uncertainty estimates for each individual uncertainty budget,
–the total upper and lower uncertainty estimates.
• The third row contains the result as described by the above columns. In addition, at the end of
the row the relative total uncertainties are given in parentheses.
The above structure repeats itself for every observable, as well as for each variation point of the kine-
matic variables as described by occurring --range arguments.
2.3. Producing Random Parameter Samples
For all the previously mentioned use cases (observable evaluation, Bayesian parameter inference, and
production of pseudo events) one requires to draw random samples from some arbitrary Probability
Density Function (PDF) P(⃗
θ). When using EOS, these random samples can be produced from Markov-
Chain Monte Carlo (MCMC) random walks, using the Metropolis-Hastings algorithm, by calls to the
eos-sample-mcmc client. In a second step, refined samples or samples for a very complicated setup,
can be obtained from an algorithm described in Ref. [6]. This algorithm uses an adaptive importance
sampling called Population Monte Carlo (PMC), implemented within the client eos-sample-pmc , and
requires a prior run of eos-sample-mcmc for initialization.
The follow command-line arguments are common to the eos-sample-mcmc and eos-sample-pmc
clients, as well as further clients described in subsequent sections:
11
2. Usage
--scan NAME --prior flat MIN MAX
--scan NAME [ABSMIN ABSMAX] --prior gaussian MIN CENTRAL MAX
--nuisance [...]
These arguments add a parameter to the statistical analysis, with either a at or a gaussian
prior. If ABSMIN and ABSMAX are specified, the prior will be cropped to this absolute interval.
The --scan and --nuisance arguments work identically, with one exception: --nuisance
declares the associated parameter as a nuisance parameter, which is agged in the HDF5 out-
put. The sampling algorithm treats nuisance parameters in the same way as scan parameters.
--constraint NAME
The named constraint from the internal database will be used as part of the likelihood. The
functional form of the likelihood, details such as correlations, and the required options for the
observables used will be automatically looked up. In order to browse the entries of the database,
use the eos-list-constraints client.
--fix NAME VALUE
The value of parameter NAME will be set to the supplied VALUE , and thus potentially deviate
from its default value.
The eos-sample-mcmc client further accepts the following arguments:
--seed [time|VALUE]
This argument sets the seed value for the Random Number Generator (RNG). Setting the seed
to a fixed numerical VALUE ensures being able to reproduce the results. This is important for
publication-quality usage of the client. If time is specified, the RNG is seeded with an integer
value based on the current time.
--prerun-min VALUE
For the prerun phase of the sampling algorithm, set the minimum number of steps to VALUE .
--prerun-max VALUE
For the prerun phase of the sampling algorithm, set the maximum number of steps to VALUE .
--prerun-update VALUE
For the prerun phase of the sampling algorithm, force an adaptation of the Markov chain’s pro-
posal function to its environment after every VALUE steps.
--store-prerun [0|1]
Either disable or enable storing of the prerun samples to the output file.
--output FILENAME
Use the file FILENAME to store the output, using the HDF5 file format. The resulting HDF5 file
follows the EOS-MCMC format, and can be accessed using, e.g., the eos.data.MCMCDataFile
Python class.
The eos-sample-pmc client additionally accepts the following command-line arguments:
12
2.3. Producing Random Parameter Samples
--seed [time|VALUE]
This argument sets the seed value for the RNG. Setting the seed to a fixed numerical VALUE
ensures being able to reproduce the results. This is important for publication-quality usage of
the client. If time is specified, the RNG is seeded with an interger value based on the current
time.
--hc-target-ncomponents N
When creating mixture components, create Ncomponents per existing MC group.
--hc-patch-length LENGTH
When clustering a group’s MCs onto the mixture components, cut the chains into patches of
LENGTH samples each.
--hc-skip-initial FRACTION
Skip the first FRACTION of all MCMC samples in the clustering step.
Note:FRACTION must be a decimal number between 0and 1.
--pmc-initialize-from-file HDF5FILE
Use the samples from a MCMC HDF5 output file HDF5FILE as generated with eos-sample-mcmc ,
in order to initialize the mixture density of the initial PMC step.
--pmc-group-by-r-value R
When forming groups of MCs from the initialization file, only add a chain to an existing group if
the chain’s R-value is less than R; create a new group otherwise.
--pmc-samples-per-component N
Set the number Nof samples that will be drawn per component and per update step of the PMC
run.
--pmc-final-samples N
Set the number Nof samples that will be drawn for the final step, i.e.: after the PMC updates
have converged.
--pmc-relative-std-deviation-over-last-step STD STEPS
If both perplexity and ESS have a standard deviation less than STD over the last STEPS updates,
declare convergence.
--pmc-ignore-ess [0|1]
Set whether convergence of the PMC updates shall be determined from the effective sample
size (ESS) in addition to the perplexity.
Default: Use the ESS.
--output FILENAME
Use the file FILENAME to store the output, using the HDF5 file format. The resulting HDF5 file
follows the EOS-PMC structure, and can be read using the eos.data.PMCDataFile Python class.
13

2. Usage
As an example, we define the a-priori PDF for a study of the decay ¯
B→π+µ−¯νµ. For the CKM Wolfen-
stein parameters, we use
λ= 0.22535 ±0.00065 , A = 0.807 ±0.020 ,
¯ρ= 0.128 ±0.055 ,¯η= 0.375 ±0.060 .
For the a-priori PDF, we use uniform distributions for the BCL2008 [5] parameters for the B→πform
factor fBπ
+. However, we construct a likelihood from the results of a recent study (IKMvD2016 [7])
of the form factor fBπ
+(q2)within Light-Cone Sum Rules (LCSRs). We now intend to draw random
numbers from the posterior PDF using EOS’ adaptive Metropolis-Hasting algorithm. During its prerun
phase, the algorithm adapts the chains’ proposal functions. As a consequence, the prerun samples
will in general not be distributed according to the posterior PDF. With a subequent PMC sampling run
in mind, we should demand at least 500 steps. In this simple problem, the Markov chains converge
(the prerun finishes) after a few thousand iterations; the adaption process should be executed after
every 500 steps.
Our intentions translate to the following call to eos-sample-mcmc :
eos - sample -mcmc \
--global -option model CKMScan \
-- global - option form - factors BCL2008 \
-- scan "CKM :: abs(V_ub )" 2e -3 5e -3 --prior flat \
-- scan "B->pi ::f_+(0) @BCL2008 " 0 1 -- prior flat \
-- scan "B->pi ::b_+^1 @BCL2008" -20 +20 -- prior flat \
-- scan "B->pi ::b_+^2 @BCL2008" -20 +20 -- prior flat \
-- constraint "B->pi:: f_+@IKMvD -2014" \
-- constraint "B^0->pi ^+ lnu ::BR@BaBar -2010 B" \
-- constraint "B^0->pi ^+ lnu ::BR@Belle -2010 A" \
-- constraint "B^0->pi ^+ lnu ::BR@BaBar -2012 D" \
-- constraint "B^0->pi ^+ lnu ::BR@Belle -2013 A" \
--prerun -min 500 \
--prerun -max 7500 \
--prerun -update 500 \
--prerun -only \
-- output / tmp/mcmc_prerun_btopi +ff.hdf5
# --chunks 10 \
# --chunk -size 1000 \
Optionally, if the prerun converges, the client can be used to perform a main run, in which the proposal
functions will be kept static. For such these main run samples, we wish for a total of 104, which we
artifically decompose into 10 chunks with 1000 samples each. While the sampling at hand will be quite
quick, sampling of computationally expensive functions should be done with small chunks so that the
progress of the computations can be monitored. The require options for these intentation are shown
above with a leading hash mark. Also, for a main run, the --prerun-only ag would need to be re-
moved.
We use the above call to eos-sample-mcmc in order to initialize a PMC run. We wish for 4mixture
components per MC group, and to skip 20% of the MCMC samples as part of the burn in. MC groups
will be created based on an R-value [8] threshold of 1.5. For each update, 500 samples per mixture
component shall be drawn, in order to produce 106samples in the final step. Convergence shall be
declared upon a standard deviation for the perplexity only, of 0.05 over the last 4update steps. The
call then reads:
eos - sample -pmc \
--global -option model CKMScan \
-- global - option form - factors BCL2008 \
-- scan "CKM :: abs(V_ub )" 2e -3 5e -3 --prior flat \
-- scan "B->pi ::f_+(0) @BCL2008 " 0 1 -- prior flat \
-- scan "B->pi ::b_+^1 @BCL2008" -20 +20 -- prior flat \
14
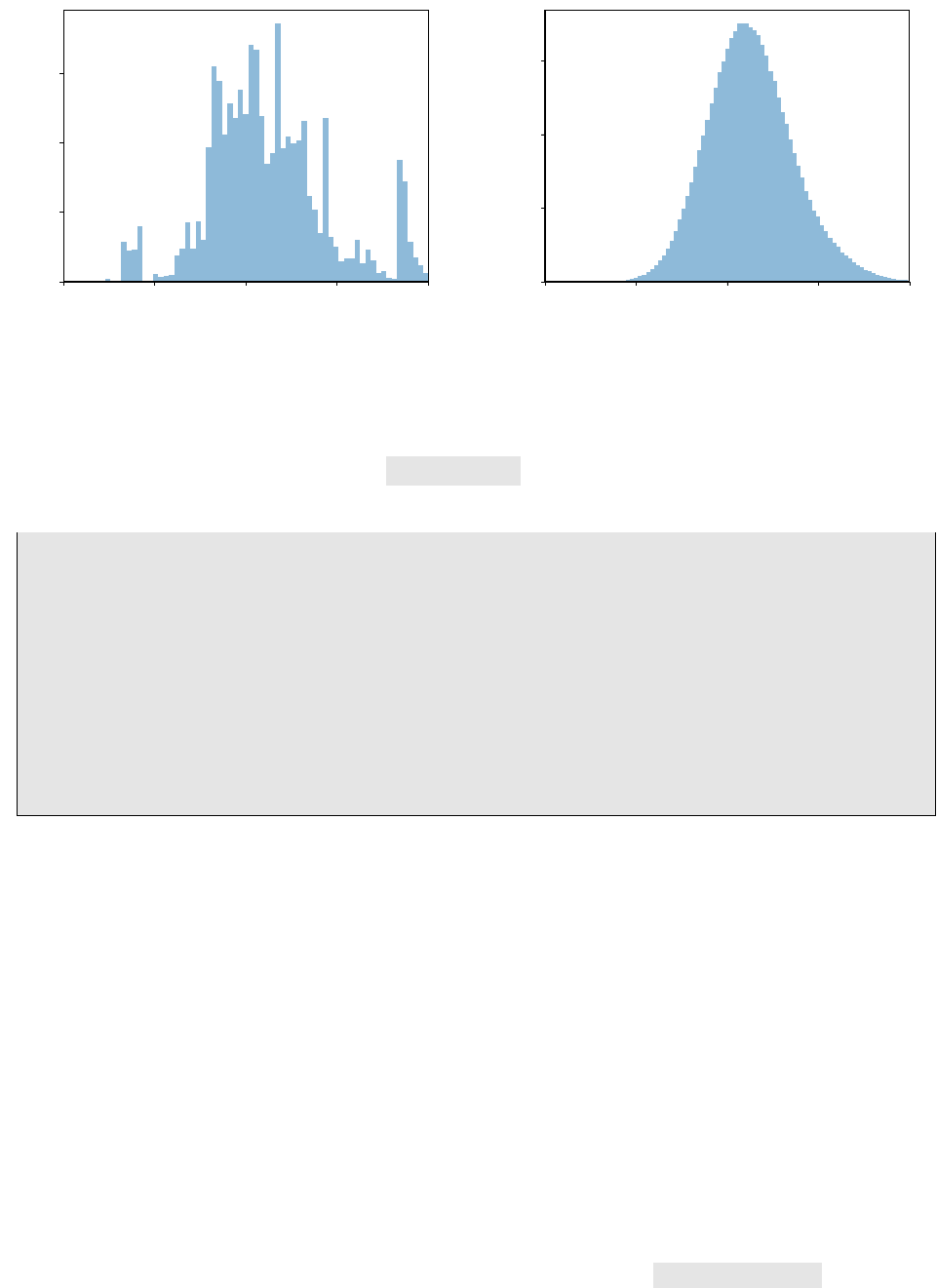
2.4. Merging Markov Chains from multiple files
0.0025 0.0030 0.0035 0.0040 0.0045
|Vub|
0
500
1000
1500
probability
EOS v0.1.1
(a) Histogram of the marginal posterior of |Vub |, using 4×
7500 samples. Despite the poor quality of these samples, they
can be used to initialize the PMC run as described in the text.
0.0025 0.0030 0.0035 0.0040 0.0045
|Vub|
0
500
1000
1500
probability
EOS v0.1.1
(b) Histogram of the marginal posterior of |Vub|, using 106
samples.
Figure 2.1.: Histograms of the parameter of interest |Vub|in the two example fits as described in sec-
tion 2.3, and plotted using the eos-plot-1d client; see section 2.7 for details.
-- scan "B -> pi :: b_ +^2 @ BCL2008 " -20 +20 -- prior flat \
-- constraint "B->pi:: f_+@IKMvD -2014" \
-- constraint "B^0->pi ^+ lnu ::BR@BaBar -2010 B" \
-- constraint "B^0->pi ^+ lnu ::BR@Belle -2010 A" \
-- constraint "B^0->pi ^+ lnu ::BR@BaBar -2012 D" \
-- constraint "B^0->pi ^+ lnu ::BR@Belle -2013 A" \
--pmc -initialize -from - file /tmp / mcmc_prerun_btopi +ff. hdf5 \
--hc -target - ncomponents 4 \
--hc -skip - initial 0.2 \
-- pmc -samples -per - component 500 \
-- pmc -group - by -r -value 1.5 \
-- pmc -final - samples 1000000 \
-- pmc -ignore -ess 1 \
-- pmc - relative - std - deviation - over - last - steps 0.05 4 \
-- output / tmp/pmc_monolithic_btopi+ff.hdf5
Both clients output copious amounts diagnostic data to the standard output, which include
• all information about the prior and the likelihood as specified on the command line (both clients);
• information about the convergence of the Markov chains within the parameter space, based on
the R-value criterion [8] (MCMC only);
• information about the convergence of the PMC run based on the perplexity and effective sam-
pling size (PMC only).
We display the outcome of both the MCMC (prerun) sampling as well as the PMC sampling steps in
figure 2.1.
2.4. Merging Markov Chains from multiple files
To run multiple Markov chains in parallel, one may use a compute cluster and submit independent
jobs, each of which creates its own output file with one or more chains inside. To initialize PMC, it is
necessary to have all Markov chains in a single HDF5 file. This is where eos-merge-mcmc comes into
play. It allows one to merge chains from multiple files into a single HDF5 file and optionally to apply a
15

2. Usage
cut such that chains that got stuck in irrelevant local modes in the prerun can be filtered out. Note that
chains both from the prerun and main run are copied into the output file. eos-merge-mcmc accepts
the following command-line arguments:
Any number of input files created by eos-sample-mcmc
--cut-off NUMBER
Skip chains whose maximum log posterior in the prerun is below the cut-off.
--input-file-list LIST
List input files in a text file, one file per line. If other input files are passed directly on the command
line, then those take precedence but all files will be merged.
--output FILENAME
The name of the output file in which the chains are stored.
2.5. Finding the Mode of a Probability Density
The mode, best-fit point, or simply the most-likely value, of some PDF P(⃗
θ)is regularly searched for
in physics analyses. EOS supplies the client eos-find-mode , which accepts the common set of
arguments describing the PDF as already discussed for the eos-sample-mcmc and eos-sample-pmc
clients, see section 2.3 for further information. In addition, it accepts the following command-line
arguments:
--starting-point { VALUE1 VALUE2 ... VALUEN }
Set the starting point for the maximization of the PDF P(⃗
θ)at θ= ( VALUE1 , . . . , VALUEN ).
--max-iterations INTEGER
The optimization algorithm is allowed to run at maximum NUMBER iterations.
--target-precision NUMBER
Attempt to determine the mode up to an uncertainty of NUMBER .
--output FILENAME
Write a YAML file with name FILENAME that contains the information of the mode in a machine-
readable format.
In order to illustrate the client’s usage, we use the same example as discussed in section 2.3. The
corresponding call then reads:
eos - find - mode \
--global -option model CKMScan \
-- global - option form - factors BCL2008 \
-- scan "CKM :: abs(V_ub )" 2e -3 5e -3 --prior flat \
-- nuisance "B -> pi :: f_ +(0) @B CL2008 " 0 1 -- prior flat \
-- nuisance "B -> pi :: b_ +^1 @BCL2008 " -20 +20 -- prior flat \
-- nuisance "B -> pi :: b_ +^2 @BCL2008 " -20 +20 -- prior flat \
-- constraint "B->pi:: f_+@IKMvD -2014" \
-- constraint "B^0->pi ^+ lnu ::BR@BaBar -2010 B" \
-- constraint "B^0->pi ^+ lnu ::BR@Belle -2010 A" \
-- constraint "B^0->pi ^+ lnu ::BR@BaBar -2012 D" \
-- constraint "B^0->pi ^+ lnu ::BR@Belle -2013 A" \
-- starting -point { 3.5e -3 0.31 0 0 } \
-- max - iterations 1000 \
--target -precision 1e -8 \
-- output /tmp/mode - btopipi - ff .yaml
16

2.6. Bayesian Uncertainty Propagation
The output starts with same diagnostic information on the composition of prior and likelihood as for
the sampling clients. The results are displayed in the last few lines, including
• the starting point for the mode-finding process;
• the coordinates of the best-fit point; and
• the log-posterior at the best-fit point.
# Starting optimization at ( 0.0035 0.31 0 0 )
# Found maximum at:
# ( 3.568778e-03 2.661032e-01 -2.670912e+00 2.231637e-02 )
# value = -3.101594e+02
2.6. Bayesian Uncertainty Propagation
EOS presently supports two ways to propagate theory uncertainties in the framework of Bayesian
statistics: First, by using prior PDFs that describes the nuisance parameters; second, by using samples
from a posterior PDF that have been obtained from running either eos-sample-mcmc or eos-sample-pmc .
It accepts the following command-line arguments:
--vary NAME --prior flat MIN MAX
--vary NAME [ABSMIN ABSMAX] --prior gaussian MIN CENTRAL MAX
These arguments add a parameter to the statistical analysis, with either a at or a Gaussian
prior. If ABSMIN and ABSMAX are specified, the prior will be cropped to this absolute interval.
--fix NAME VALUE
Fix parameter to a certain value. Use this to change default values for parameters that are not
varied.
--kinematics NAME VALUE
Within the scope of the next observable, declare a new kinematic variable with name NAME and
numerical value VALUE .
--observable NAME
Add a new observable with name NAME to the list of observables that shall be evaluated. All
previously issued --kinematics and --range arguments apply, and will be used by the new
obervable. The kinematics will be reset (i.e., all kinematic variables will be removed) in anticipa-
tion of the next --observable argument.
--global-option NAME VALUE
Set an option to a string value that applies to all following observables.
--samples NUMBER
Sets the number of samples that shall be produced per observable and worker thread.
--workers NUMBER
Sets the number of worker threads. Default: 4.
17

2. Usage
--parallel [0|1]
Activate multithreaded evaluation. Default: 1.
--mcmc-input HDF5FILE BEGIN END
Use the samples at index BEGIN up to index END from each chain in the file HDF5FILE . If
both preruns and main runs are present in the file, preference is give to the main run. To use all
samples, use BEGIN=0 and choose a value for END equal to or larger than the actual length of
any chain in the file.
Note: The file must have been produced by the eos-sample-mcmc client.
Note: If you have the HDF5 commandline tools available, use h5ls -r HDF5FILE to see the
number and length of the chains.
--mcmc-prefer-prerun [0|1]
Override the default setting to take samples from the prerun instead of the main run in case both
are present in the HDF5FILE . Default: 0.
--pmc-input HDF5FILE BEGIN END
Use the samples at index BEGIN up to index END from a named data set in the file HDF5FILE .
Note: The file must have been produced by the eos-sample-pmc client.
--pmc-sample-directory NAME
Use the named data set within the file specified with --pmc-input . Valid names are /data/X,
where X stands for either initial,final, or all numbers describing existing update steps that
were carried out in the PMC run.
Note: You should use /data/final unless you are debuging the PMC algorithm.
--output NAME
Set the output file name.
--store-parameters [0|1]
Store the parameter samples drawn from the prior into the output file. Has no effect with --mcmc-input
or --pmc-input . Default: 0.
--seed VALUE
Change the random number seed for generating parameter samples. Has no effect with --mcmc-input
or --pmc-input . By default a fixed seed is chosen corresponding to VALUE=0 . To get different
samples on each invocation, use VALUE=time .
As an example of the first way, we would like to predict the branching ratio for the decay B−→
τ−¯ντ. This prediction involves two parameters: First, the absolute value of Vub; and second, the value
of the B-meson decay constant fB. For the former, we choose the HFAG average of the inclusive
determination |Vub|= (4.45 ±0.26) ·10−3[9], while for the latter we use the FLAG average fB=
0.188±0.007 GeV [10]. Our intention translates to the following call to eos-propagate-uncertainty :
18

2.7. Plotting Random Samples
eos - propagate - uncert ainty \
--global -option model CKMScan \
-- global - option form - factors BCL2008 \
-- vary "CKM ::abs(V_ub )" 2e -3 5e -3 --prior gaussian 4.19e-3 4.45e-3 4.71e-3 \
-- vary "decay -constant :: B_u" 0.167 0.209 --prior gaussian 0.181 0.188 0.195 \
-- observable " B_u -> lnu :: BR ;l =tau " \
-- workers 4 \
-- samples 100000 \
-- output / tmp/unc_btotaunu . hdf5
As an exmple of the second way, we would like to use samples from the posterior PDF as obtained
in section 2.3 in order to predict the branching ratio of the differential decay ¯
B0→πe−¯νe, at various
points in the kinematic range as the decay to muons described previously. Our intention translates to
the following call to eos-propagate-uncertainty :
eos - propagate - uncert ainty \
-- kinematics s 0.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 0.25 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 0.50 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 0.75 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 1.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 1.50 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 2.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 2.50 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 3.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 3.50 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 4.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 6.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 7.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 8.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 9.00 --observable "B->pilnu :: dBR/ds;l=e,form -factors=BCL2008 ,model = CKMScan" \
-- kinematics s 10.00 --observable "B->pilnu :: dBR/ds;l=e ,form -factors = BCL2008 ,model=CKMScan " \
-- kinematics s 11.00 --observable "B->pilnu :: dBR/ds;l=e ,form -factors = BCL2008 ,model=CKMScan " \
-- kinematics s 12.00 --observable "B->pilnu :: dBR/ds;l=e ,form -factors = BCL2008 ,model=CKMScan " \
--pmc -input / tmp/pmc_monolithic_btopi +ff.hdf5 0 10000 \
-- pmc -sample - directo ry "/ data /final /" \
-- output / tmp/unc_btopilnu . hdf5
For both ways, the samples of the predictive distributions within the HDF5 files can be accessed using
the eos.data.UncertaintyDataFile Python class.
2.7. Plotting Random Samples
Once random samples have been obtained from either a posterior PDF (e.g. as described in sec-
tion 2.3) or a predictive PDF (e.g. as described in section 2.6), a visual inspection of the samples is the
next step. EOS provides scripts for this purpose, which plot histograms of either a marginalized 1D
(eos-plot-1d ) or heatmaps of 2D ( eos-plot-2d ) PDFs. Both scripts presently detect the output
format by inspection of the resepective HDF5 file name. Files containing MCMC samples should be
prefixed with mcmc_, while PMC sample files should be prefixed with pmc_monolithic_. Files contain-
ing samples from the uncertainty propagation should be prefixed with unc_.
The eos-plot-1d produces a 1D histogram of the samples for one parameter. It accepts the follow-
ing arguments:
HDF5FILE
The name of the HDF5 input file whose samples shall be plotted.
VAR
The name of the variable (either a Parameter or an Observable) whose density function shall
be plotted.
19
2. Usage
PDFFILE
The name of the PDF output file, into which the plot shall be saved.
--xmin XMIN ,--xmax XMAX
When specified, limit the plot range to the interval XMIN to XMAX . The default values are taken
from the description of the parameter within the HDF5 input file.
--kde [0|1]
When enabled, plots a univariate Kernel Density Estimate of the probability density based on the
available samples.
--kde-bandwidth KDEBANDWIDTH
When specified, multiplies the automatically determined KDE bandwidth parameter with KDEBANDWIDTH .
The eos-plot-2d produces a 2D heatmap of the samples for two parameters. It accepts the follow-
ing arguments
HDF5FILE
The name of the HDF5 input file whose samples shall be plotted.
XIDX
The numerical index for the parameter that shall be plotted on the xacis. XIDX starts with zero
YIDX
The numerical index for the parameter that shall be plotted on the yacis. YIDX starts with zero
PDFFILE
The name of the PDF output file, into which the plot shall be saved.
--xmin XMIN ,--xmax XMAX
When specified, limit the plot range on the xaxis to the interval XMIN to XMAX . The default
values are taken from the description of the parameter within the HDF5 input file.
--ymin YMIN ,--ymax YMAX
When specified, limit the plot range on the yaxis to the interval YMIN to YMAX . The default
values are taken from the description of the parameter within the HDF5 input file.
20

3. Library Interface
The rationale behind several of the design decisions of the EOS libraries are explained in section 3.1.
We document the core set of classes in section 3.2.
3.1. Design
To fulfill its intended use cases, the EOS libraries are designed by abiding to the follwing principles.
First, scalar quantities are treated as parameters. These include
• experimental inputs, such as particle masses and lifetimes;
• theoretically motivated quantities, such as quark masses (e.g.in the MS scheme) and the Wolfen-
stein parameters of the CKM matrix.
It is therefore straightforward to change a parameter’s role from one analysis to the next; e.g.from
being a nuisance parameter when producing theory estimates, to being a parameter of interest when
inferring knowedlge from experimental data. To differentiate between the various parameters, a nam-
ing scheme is put in place:
NAMESPACE::ID@TAG ,(3.1)
where the meta variables convey the following meaning:
NAMESPACE A short description of the context in which the parameter should be interpreted.
Possible namespaces include, but are not limited to: mass,decay-constants,
CKM, and others.
ID A descriptive handle for the parameter in its namespace.
TAG A forther pices of information that allows to distinguish between parameters with
the same ID.
Second, quantities that exhibit a functional dependence on either a parameter or a kinematic variable
are treated as plugins: An interface is created that allows implementing more than one realization of
the respective function. The actual implementations of this interface are then accesible via a factory
method.1Each implementation of a plugin can depend on its own subset of parameters. For clarity,
we use one of the hadronic form factor as an example. Consider the B→πvector form factor fBπ
+.
One possible parametrization of this form factor has been proposed in [5] (denotes as BCL2008). It is
achieved in terms of three parameters:
• the normalization fBπ
+(0), and
1A good example for such a modular design is found within the scope of hadronic matrix elements, in particular hadronic
form factors.
21

3. Library Interface
• two shape parameters bBπ,+
1and bBπ,+
2.
This parametrization is implemented within EOS as one of several options for a B→πform factor
parametrization. The relevant EOS parameters are contained in the namespace B->pi, and tagged for
the BCL2008 plugin:
B->pi::f_+(0)BCL2008 ,B->pi::b_+ˆ1BCL2008 ,and B->pi::b_+ˆ2BCL2008 .(3.2)
This removes the risk of namespace collisions among the various plugins’ parameters.
Observables that depend on the the B→πform factors can be implemented by relying on the
form factors interface, and leave the concrete parametrization to be a run-time option with name
form-factors. The BCL2008 parametrization can be selected by passing form-factors=BCL2008 to
the observable.
Third, data is only copied when explicitly requested. Copy constructors yield another object that points
to the same underlying data. This is achieved through extensive use of the Private Implementation
design pattern.2To make an independent copy, the EOS classes provide the clone() method.
3.2. Core Classes
At the core of the EOS API there are four classes:
Parameters which contains the whole set of Parameter known to EOS
Kinematics which contains the set of kinematic variables specific to one instance of Observable
Options which contains the set of options specific to one instance of Observable
Observable which encapsulates the numerical implemention of a single (pseudo-)observable
quantity.
3.2.1. Class Parameters
Implemented as a dictionary, with keys of type std:string and values of type Parameter. The default
set of parameters is loaded from the EOS-supplied YAML files. It can be loaded by using the static
Parameters::Defaults() method. Using the copy construtor does not yield an independent copy;
instead the second object uses the underlying data of the initial instance. To obtain an independent
copy with the same values as the present one, use the clone() method.
1Parame ters paramA = Para meter s :: D efaults () ;
2Parameters paramB(paramA );
3
4ObservablePtr obsA = Observable :: make("A", paramA , Kinematics{ }, Options { });
5ObservablePtr obsB = Observable :: make("A", paramB , Kinematics{ }, Options { });
You can access individual parameters via the array subscript operator. It takes a class QualifiedName
object as its key, which can be created implicitly from std::string or const char *. You can also
iterate over all parameters via the begin() and end() methods. In both cases, the underlying objects
are of class Parameter, which provides access to the current value via the evaluate() method.
2See https://cpppatterns.com/patterns/pimpl.html.
22
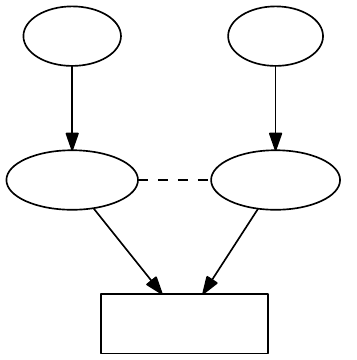
3.2. Core Classes
Parameter Data
paramA
depends
paramB
copy
depends
obsA
depends
obsB
depends
Figure 3.1.: Diagrammatic illustration that multiple observables can depend on the very same instance
of Parameters.
3.2.2. Class Kinematics
The class Kinematics is a dictionary from std::string-valued keys to KinematicVariable-valued
entries. Upon construction of an observable, a suitable instance of kinematics is bound to that ob-
servable. Acces to individual kinematic variables occurs through the array subscript operator. The
class is used for the run-time construction of observables. For this purpose, the variable names apply
only to a single observable and are not explicitly namespaced.
3.2.3. Class Options
The class Options is a dictionary from std::string-valued keys to std::string-valued entries. Upon
construction of an observable, a suitable instance of Options is bound to that observable. The under-
lying class’s constructor queries the options, and select the appropriate run-time behaviour.
Helper classes are available for repeated options-related task, e.g.the class SwitchOption.
3.2.4. Class Observable
The class Observable is an abstract base class. To create a new observable object, you must use the
static Observable::make() factory method. It requires three arguments:
• a class QualifiedName object n;
• a class Parameters object p; and
• a class Kinematics object k; and
• an class Options object o.
23
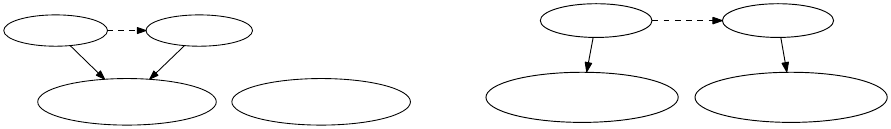
3. Library Interface
Observable A Observable A'
copy
Parameters A
mass::b(MSbar) = 4.20
Parameters B
mass::b(MSbar) = 4.16
(a) copy
Observable A Observable B
clone
Parameters A
mass::b(MSbar) = 4.20
Parameters B
mass::b(MSbar) = 4.16
(b) clone
Figure 3.2.: Diagrammatic illustration of the differences between copying an instance of Observable
via the copy-constructor, and cloning the observable via the clone method
To allow for more compact code, EOS’ internal observables are not implemented as individual classes.
Instead, related observables are bundled together into one class. Individual observable classes are
then created using the template <...> class ConcreteObservable. We refer to section 4.2 for de-
tails on how to extend EOS with the implementation of a new observable.
Note: Any changes to the Options object oafter construction of the observable do not affect the ob-
servable.
Note: All users of Observable must also support cloning. This allows to easily parallelize algorithms
acting on Observable.
24

4. Extending EOS
We discuss the three most common cases of extending EOS, by adding either a new parameter, a
new observable, or a new measurement. For extensions that do not fall in these categories please
contact the main authors, who will be happy to discuss your work with you. The most efficient way to
contribute new code or modifications to EOS is via a pull request in the main Github repository.1
4.1. How to add a new parameter
Parameters are stored in YAML files below eos/parameters/, including the default values and range,
as well as metadata and the origin thereof. Each file contains key-to-value maps:
• Keys starting and ending with an @sign are considered to be metadata pertaining to the entire
file.
• All other keys declare a new parameter.
The values in this map are the definitions of a parameters, and are also structured as maps. Valid
parameter definitions must include all of the following entries:
central the default central value;
min the minimal value for the default range;
max the maximal value for the default range.
Optional entries include:
comment a comment on the purpose;
latex the LaTeX command for the display;
unit the unit in which the parameter is expressed.
Example
We introduce two new parameters: mass::X@Example, and decay-constant::X@Example, standing in
for the mass and decay constant of a hypothetical particle X. We add the file eos/parameters/x.yaml
with contents:
'mass :: X@Example ' :
central: +1.0
min: +0.5
max: +1.5
unit : 'GeV '
latex : '$M_X$ '
comment: 'The mass of the unmotivated X particle '
'decay - constant :: X@ Examp le ' :
central: +0.10
1https://github.com/eos/eos
25
4. Extending EOS
min: +0.09
max: +0.11
unit : 'GeV '
latex : '$f_X$ '
comment: 'The decay constant of the unmotivated X particle '
4.2. How to add a new observable
An Observable is implemented in EOS as follows:
• The numerical code is implemented as the method double P::o(...) const, where Pis the
underlying class, and the dots stand in for any number of const double & arguments, includ-
ing zero. The class P must inherit from class ParameterUser. It must have one constructor
accepting a class Parameters and a class Options instance in that order. Schematically:
1#include <eos/utils/ parameters.hh >
2#include <eos/utils/ options.hh >
3
4class P :
5public ParameterUser
6{
7P(const Parameters \&, const Options \&};
8~P();
9
10 double obse rva ble _wi tho ut_ kin emat ics () const ;
11 double observable_with_one_kinematic_variable(const double &) const ;
12 };
• The method or methods are then associated with their names within the file eos/observable.cc
within the free-standing function make_observable_entries(). Within the existing map, new
entries are created through a call to make_observable(...). For a regular observable the ar-
guments are in order: the name of the observable; the pointer to its method; and the tuple
of std::strings that names the kinematic variables in the order the method expects them.
Schematically:
1#within eos/ observable .cc
2#[...]
3make _obs erva ble (" P:: ob serva ble1 " ,
4&P :: o bse rva ble _wi tho ut_ kine mat ics );
5make _obs erva ble (" P:: ob serva ble2 ( variab lena me )" ,
6&P::observable_with_one_kinematic_variable ,
7std :: m ake_ tuple (" vari able name ") );
8#[...]
4.3. How to add a new constraint
Constraints are stored in YAML files below eos/constraints/. New constraints can be added to
an existing file, or to an entirely new file. Each file contains key-to-value maps in which the top-level
keys declare a new constraint. The values in this map are the definitions of a constraint, and are also
structured as maps. Valid constraint definitions must at least include a type entry that governs the
functional form of the associated likelihood. EOS understands the following types of constraints:
•Amoroso,
•Gaussian,
26
4.3. How to add a new constraint
•MultivariateGaussian,
•MultivariateGaussian(Covariance).
4.3.1. Type Amoroso
Type Amoroso uses a likelihood arising from the PDF of the Amoroso distribution [11], a four-parameter
distribution. It is useful for the modeling of upper limits on as-of-yet undiscovered decays at given
probabilities. For example, the upper limits on the decay Bs→µ+µ−by the LHCb experiment prior to
the discovery of this decay are modeled using the Amoroso type of likelihood. The following keys are
required in the description of the constraint:
observable the qualified name for an Observable or a Parameter;
kinematics the map representation of the kinematic variables pertaining to the observable;
options the map representation of the options pertaining to the observable;
physical-limit the physical lower bound on the observable;
theta the θparameter of the Amoroso distribution;
alpha the αparameter of the Amoroso distribution;
beta the βparameter of the Amoroso distribution;
We strongly recommend contacting the EOS authors before adding your own constraint of type Amoroso.
4.3.2. Type Gaussian
Type Gaussian uses a univariate Gaussian or normally-distributed likelihood. The following keys are
required in the description of the constraint:
observable the qualified name of an Observable or a Parameter;
kinematics the map representation of the kinematic variables pertaining to the observable;
options the map representation of the options pertaining to the observable;
mean the mean of the distribution;
sigma-stat the map representation with keys hi and lo for the upper and lower statistical
uncertainty of the distribution, respectively;
sigma-sys the same as sigma-stat, but for the systematic uncertainty;
dof the degrees of freedom associated with this observation (should default to 1).
By default the uncertainties are symmetrized, and statistical and systematical uncertainties are added
in quadrature.
27
4. Extending EOS
4.3.3. Type MultivariateGaussian
Type MultivariateGaussian uses a multivariate Gaussian likelihood, parametrized in terms of its
mode and correlation matrix. The following keys are required in the description of the constraint:
dim the dimensionality of the multivariate distribution;
observable the list of size dim of qualified names for either Observables or Parameters;
kinematics the list of size dim of map representations of the kinematic variables pertaining
to each observable;
options the list of size dim of map representations of the options pertaining to each ob-
servable;
mean the list of size dim describing the mean of the distribution;
sigma-stat-hi the list of size dim for the upper statistical uncertainty of the distribution;
sigma-stat-lo the list of size dim for the lower statistical uncertainty of the distribution;
correlations the matrix of size dim ×dim for the statistical correlations;
sigma-sys the list of size dim for the systematic uncertainty;
dof the degrees of freedom associated with this observation (should default to dim).
By default the statistical and systematical uncertainties are added in quadrature, and the larger of the
upper and lower uncertainties are chosen as the standard deviation. The covariance matrix Σij is then
computed from the variances σiand the correlation matrix ρij as
Σij =σiσjρij .(4.1)
4.3.4. Type MultivariateGaussian(Covariance)
Type MultivariateGaussian(Covariance) uses a multivariate Gaussian likelihood, parametrized in
terms of its mode and covariance matrix. The following keys are required in the description of the
constraint:
dim the dimensionality of the multivariate distribution;
observable the list of size dim of qualified names for either Observables or Parameters;
kinematics the list of size dim of map representations of the kinematic variables pertaining
to each observable;
options the list of size dim of map representations of the options pertaining to each ob-
servable;
mean the list of size dim describing the mean of the distribution;
covariance the matrix of size dim ×dim for the covariance of the distribution;
dof the degrees of freedom associated with this observation (should default to dim).
28
4.3. How to add a new constraint
Example
We introduce a constraint to correlate the new parameters mass::X@Example, and decay-constant::X@Example
from section 4.1. The correlation is fixed at 50%, and the means and standard deviations reect the
previous one. We add the file eos/constraints/x.yaml with contents:
X:: mass +decay -constant@Example :
type : MultivariateGaussian
dim: 2
obser vabl es : [ 'mass :: X@Exam ple ' , ' decay - const ant :: X@Example ']
kinematics: [{} , {}]
options: [{} , {}]
means : [1.0, 0.1]
covari ance : [[0.2 5 , 2.5 e-3] , [2.5 e-3 , 1.0 e -4]]
dof: 2
29
Physics
31

5. Effective Field Theories
The electro-weak decays of hadrons (mesons and baryons) with masses much smaller than the
electro-weak scale of order of the W-boson mass, mW, can be efficiently described using effective
field theories (EFT) in the spirit of the well-known Fermi theory of the β-decay. This comprises prac-
tically all hadrons containing light quarks q= (u, d, s, c, b). In this context, specific avour-changing
higher-dimensional (dim > 4) operators arise in the standard model (SM) and it’s extensions, accom-
panied by effective couplings (Wilson coefficients) giving rise to the generic structure of the effective
Lagrangian
LEFT =LQCD×QED(light particles) +
iCi(µ)Oi+h.c. +. . . . (5.1)
The first term describes the SU (3)c×U(1)em gauge interactions of all light quarks, q, and leptons,
ℓ= (e, µ, τ), with QCD and QED gauge bosons. The second part are the aforementioned operators
Oiand effective couplings Ci, where the latter depend on a factorization scale µlow that is usually
of the order of the mass of the decaying hadron. The operators are composed out of light degrees
of freedom, i.e. fermions qand ℓ, as well as SU (3)cand U(1)em gauge bosons. Beyond the SM, it
is conceivable that in principle additional light degrees of freedom exist, which however must have
escaped direct detection so far. The Wilson coefficients are suppressed by inverse powers of the
electroweak scale or some new physics scale, depending on the dimension of their operators. Finally,
the dots denote higher-dimensional operators that have been neglected. In practical applications,
assuming the SM, they are suppressed at least by m2
b/m2
W∼0.004, where mbdenotes the bottom-
quark mass.
The EFT framework is sufficient to describe interactions at and below the scale µlow, including hadronic
binding effects due to strong interactions as well as electro-magnetic virtual and real corrections to
observables. The implementation of according observables in EOS is thus based on universal EFT La-
grangians. All respective short-distance interactions above µlow are fixed by the structure of the oper-
ators and the values of the Wilson coefficients. In the spirit of Fermi, the according Wilson coefficients
can be viewed as unknowns to be determined from experiment. This so-called model-independent ap-
proach implies the independence of all Wilson coefficients and correlation among observables arise
from the assumption of which operators are included, i.e. have non-vanishing Wilson coefficients at
the scale µlow (or some other).
The latter point is important in view of operator mixing (under QCD and QED), which is governed by
the anomalous dimension matrices (ADM) of the involved operators. In the case of operator mixing,
vanishing Wilson coefficients at some scale µ0can become non-zero at some other scale µ1, de-
pending on their mixing and the initial conditions of Wilson coefficients involved in the mixing. This is
summarized by the general solution of the renormalisation group equation (RGE)
Ci(µ1) =
j
[U(µ1, µ0)]ij Cj(µ0).(5.2)
The evolution matrix Udepends on the ADM’s of the operators, the strong and electro-magnetic cou-
plings, αsand αerespectively, and their respective RG-functions (beta-functions). Throughout Wilson
coefficients and couplings are MS-renormalized quantities.
33

5. Effective Field Theories
In the following we collect the conventions of the effective theories implemented in EOS for hadron
decays based on quark transitions
b→(d, s), . . . (5.3)
For an introduction to the topic, the reader is referred to the exhaustive review articles [12, 13].
The following abbreviations will be often used below for products of elements of the CKM quark mixing
matrix appearing in b-quark decays
λ(D)
U=VUbV∗
UD , D = (d, s), U = (u, c, t).(5.4)
5.1. |∆B|=|∆S|= 1
In this section we summarize the convention of the EFT for |∆B|=|∆S|= 1 decays covering transi-
tions
b→s+ (¯uu, ¯cc)current-current ,
b→s+ ¯qq QCD & QED penguin ,
b→s+ (γ, gluon)electro- & chromo-magnetic dipole ,
b→s+ (¯
ℓℓ, ¯νν)semi-leptonic ,
where the classification corresponds to the origin of the operators from decoupling Wand Zbosons
and the top quark in the SM. The basis contains several blocks that are however related via operator
mixing and renders the RGE of the Wilson coefficients non-trivial. The results for |∆B|=|∆D|= 1
transitions are obtained by the replacement s→d.
We start with the operators generated in the SM, where the initial Wilson coefficients and ADM’s are
known at several orders in perturbation theory. The most appropriate choice of basis for higher order
QCD calculations of ADM’s was given in [14, 15] with according extension for QED corrections in [16,
17]. The Lagrangian
LEFT =LQCD×QED(q;ℓ) + 4GF
√2λ(s)
u
2
i=1 Ci(Oc
i− Ou
i)
+4GF
√2λ(s)
t2
i=1 CiOc
i+
10
i=3 CiOi+
6
i=3 CiQOiQ +CbOb+h.c. .
(5.5)
incorporates unitarity of the CKM matrix λ(s)
u+λ(s)
c+λ(s)
t= 0. All Wilson coefficients Ciare evaluated
at µlow ∼mb. The up-quark sector ∼λ(s)
uis doubly-Cabibbo suppressed for b→stransitions and
leads to tiny CP-asymmetries in the SM.
The current-current (U=u, c) operators are defined as
OU
1= (¯sγµPLTaU)( ¯
UγµPLTab),OU
2= (¯sγµPLU)( ¯
UγµPLb),(5.6)
which arise already at tree-level from decoupling the W-boson in b→s+(¯uu, ¯cc)and mix into all other
operators, except the semi-leptonic O10. Here and below PL,R = (1∓γ5)/2 denote chirality projectors.
The QCD-penguin operators are
O3= (¯sγµPLb)
q
(¯qγµq),O5= (¯sγµνρPLb)
q
(¯qγµνρq),
O4= (¯sγµPLTab)
q
(¯qγµTaq),O6= (¯sγµνρPLTab)
q
(¯qγµνρTaq),
(5.7)
34

5.1. |∆B|=|∆S|= 1
where the sum extends over all q= (u, d, s, c, b)and Taare the generators of SU (3)c. Products of
several gamma matrices have been abbreviated γµνρ ≡γµγνγρ.
Analogous QED-penguin operators are defined as
O3Q= (¯sγµPLb)
q
Qq(¯qγµq),O5Q= (¯sγµνρPLb)
q
Qq(¯qγµνρq),
O4Q= (¯sγµPLTab)
q
Qq(¯qγµTaq),O6Q= (¯sγµνρPLTab)
q
Qq(¯qγµνρTaq),
(5.8)
where Qqdenotes the quark charges as multiples of e. Further, an additional operator has to be con-
sidered
Ob=−1
3(¯sγµPLb)(¯
bγµb) + 1
12(¯sγµνρPLb)(¯
bγµνρb),(5.9)
receiving contributions from electro-weak boxes. In four dimension it would correspond to (¯sγµPLb)(¯
bγµPLb),
however the above form allows to avoid traces with γ5to all orders in QCD.
The electro- and chromo-magnetic dipole operators
˜
O7=e
g2
s
[¯sσµν (mbPR+msPL)b]Fµν ,˜
O8=1
gs
[¯sσµν (mbPR+msPL)Tab]Ga
µν ,(5.10)
receive contributions from on-shell photon and gluon penguins. The appearing quark masses are
renormalized in the MS scheme.
In the SM there are two semi-leptonic operators
˜
Oℓ
9=e2
g2
s
(¯sγµPLb)(¯
ℓγµℓ),˜
Oℓ
10 =e2
g2
s
(¯sγµPLb)(¯
ℓγµγ5ℓ),(5.11)
describing b→s+¯
ℓℓ transitions. In this case the lepton charge Qℓhas been pulled into the definition
of the Wilson coefficient.
note definition of evanescent operators, without whom ADM’s and initial Wilson coefficients are mean-
ingless ...
For the tilde operators the normalization to (4π/gs)2, the QCD coupling, has been chosen for practical
reasons such that the leading SM 1-loop correction to the initial Wilson coefficients counts formally
as a strong correction rather then an electro-magnetic one. The initial Wilson coefficients are known
up to NNLO in QCD and NLO in EW corrections
Ci(µ0) = . . . (5.12)
(ai≡αi/(4π)) For practical applications, however, we use operators without a tilde, which are related
via
Oi=g2
s
(4π)2˜
Oi.(5.13)
The full basis of semi-leptonic operators is spanned by O9,10 as above, and additionally
Oℓ
9′=e2
(4π)2(¯sγµPRb)(¯
ℓγµℓ),Oℓ
10′=e2
(4π)2(¯sγµPRb)(¯
ℓγµγ5ℓ),(5.14)
Oℓ
S=e2
(4π)2(¯sPRb)(¯
ℓℓ),Oℓ
P=e2
(4π)2(¯sPRb)(¯
ℓγ5ℓ),(5.15)
Oℓ
S′=e2
(4π)2(¯sPLb)(¯
ℓℓ),Oℓ
P′=e2
(4π)2(¯sPLb)(¯
ℓγ5ℓ),(5.16)
Oℓ
T=e2
(4π)2(¯sσµν b)(¯
ℓσµν ℓ),Oℓ
T5=e2
(4π)2(¯sσµν b)(¯
ℓσµν γ5ℓ).(5.17)
35

5. Effective Field Theories
Quantity Qualified Name
i= 7,7′
Re Ci(µb)b->s::Re{ci}
Im Ci(µb)b->s::Im{ci}
i= 9,9′,10,10′, S, S′, P, P ′, T, T 5
Re C(ℓ)
i(µb)b->sℓℓ::Re{ci}
Im C(ℓ)
i(µb)b->sℓℓ::Im{ci}
i= 1 . . . 6,8,8′
Ci(µb)b->s::ci
Table 5.1.: Naming scheme for the |∆B|=|∆S|= 1 operators.
For studies of NP effects in these operators the values of the respective Wilson coefficients can be
changed within the EOS code at run time. For the changed to take effect, the observables must
be constructed with the option model set to WilsonScan. In that case, the coefficients Ciwith i∈
{7,7′,9,9′,10,10′, S, S′, P, P ′, T, T 5}are treated as complex-valued parameters. Since EOS parame-
ters are real-valued only, this implies that real and imaginary part of these coefficients can be adjusted
separately. The coefficients CjUwith j={1,2}are presently U-avour-universal, and are simply
considered as two independent coefficients Cj. The coefficients with j={1. . . 6,8}are treated as
real-valued parameters. The naming scheme for these coefficients is listed in table 5.1.
5.2. |∆B|=|∆U|= 1
The Lagrangian reads:
LEFT =LQCD×QED(q;ℓ)−4GF
√2Veff
ub
XCXOX.(5.18)
Note the change of sign compared to the journal version of [18], which has a wrong but inconsequential
global factor of −1. The operators are defined as
OV,L(R)≡¯uγµPL(R)b¯
ℓγµPLνℓ,
OS,L(R)≡¯uPL(R)b¯
ℓPLνℓ,
OT≡[¯uσµν b]¯
ℓσµν PLνℓ.
(5.19)
5.3. |∆B|=|∆C|= 1
The Lagrangian reads:
LEFT =LQCD×QED(q;ℓ)−4GF
√2Veff
cb
XCXOX.(5.20)
36

A. List of default parameters
The complete list of default parameters is given in this appendix, in a series of tables.
Qualified Name Parameter Description
b->s::c1 ???
b->s::c2 ???
b->s::c3 ???
b->s::c4 ???
b->s::c5 ???
b->s::c6 ???
b->s::Re{c7} ???
b->s::Im{c7} ???
b->s::c8 ???
b->see::Re{c9} ???
b->see::Im{c9} ???
b->see::Re{c10} ???
b->see::Im{c10} ???
b->smumu::Re{c9} ???
b->smumu::Im{c9} ???
b->smumu::Re{c10} ???
b->smumu::Im{c10} ???
b->s::Re{c7'} ???
b->s::Im{c7'} ???
b->s::c8' ???
b->see::Re{c9'} ???
b->see::Im{c9'} ???
b->see::Re{c10'} ???
b->see::Im{c10'} ???
b->smumu::Re{c9'} ???
b->smumu::Im{c9'} ???
b->smumu::Re{c10'} ???
b->smumu::Im{c10'} ???
b->see::Re{cS} ???
b->see::Im{cS} ???
b->see::Re{cS'} ???
b->see::Im{cS'} ???
b->see::Re{cP} ???
b->see::Im{cP} ???
b->see::Re{cP'} ???
39

A. List of default parameters
b->see::Im{cP'} ???
b->see::Re{cT} ???
b->see::Im{cT} ???
b->see::Re{cT5} ???
b->see::Im{cT5} ???
b->smumu::Re{cS} ???
b->smumu::Im{cS} ???
b->smumu::Re{cS'} ???
b->smumu::Im{cS'} ???
b->smumu::Re{cP} ???
b->smumu::Im{cP} ???
b->smumu::Re{cP'} ???
b->smumu::Im{cP'} ???
b->smumu::Re{cT} ???
b->smumu::Im{cT} ???
b->smumu::Re{cT5} ???
b->smumu::Im{cT5} ???
Table A.2.: The list of parameters describing the b->sqqbar,gamma,ll Wilson coefficients. See the EOS
manual for their physical definitions.
Qualified Name Parameter Description
HQET::lambda_1 λ1cf. [ALGH2001], Table 2, p. 13
HQET::lambda_2 λ2cf. [ALGH2001], Table 2, p. 13
B->B::mu_pi^2@1GeV µ2
π(1 GeV)cf. [BBMU2003], Eq. (19), p. 9
B->B::mu_G^2@1GeV µ2
G(1 GeV)cf. [BBMU2003], Eq. (17), p. 9
B->B::rho_D^3@1GeV ρ3
D(1 GeV)cf. [BBMU2003], between Eqs. (19),(20),
p. 9
B->B::rho_LS^3@1GeV ρ3
LS (1 GeV)cf. [BBMU2003], Eq. (20), p. 9
Lambda_b->Lambda_b::mu_pi^2@1GeV µ2
π(1 GeV)cf. [MvD2015]
Lambda_b->Lambda_b::rho_D^3@1GeV ρ3
D(1 GeV)cf. [MvD2015]
Table A.4.: The list of hadronic forward matrix elements of operators arising in the heavy quark expan-
sion. For the matrix elements µ2
π,G and similar, see [MTU2010] for the definition.
Qualified Name Parameter Description
CKM::A ACKM The Wolfenstein CKM parameter A
CKM::lambda λCKM The Wolfenstein CKM parameter λ
CKM::rhobar ¯ρCKM The Wolfenstein CKM parameter ¯ρ
40

CKM::etabar ¯ηCKM The Wolfenstein CKM parameter ¯η
CKM::abs(V_ud) |Vud|
CKM::arg(V_ud) arg Vud
CKM::abs(V_us) |Vus|
CKM::arg(V_us) arg Vus
CKM::abs(V_ub) |Vub|
CKM::arg(V_ub) arg Vub
CKM::abs(V_cd) |Vcd|
CKM::arg(V_cd) arg Vcd
CKM::abs(V_cs) |Vcs|
CKM::arg(V_cs) arg Vcs
CKM::abs(V_cb) |Vcb|
CKM::arg(V_cb) arg Vcb
CKM::abs(V_td) |Vtd|
CKM::arg(V_td) arg Vtd
CKM::abs(V_ts) |Vts|
CKM::arg(V_ts) arg Vts
CKM::abs(V_tb) |Vtb|
CKM::arg(V_tb) arg Vtb
Table A.6.: The list of parameters describing the CKM matrix. For the Wolfenstein parameters of the
CKM matrix see e.g. [UTFIT2013] for their definition.
41
43
Bibliography
[1] The Python Software Foundation, Python Language Reference, version 2.7.
[2] J. T. M. Galassi, et al., GNU Scientific Library.
[3] The HDF Group, The HDF5 library and data format.
[4] M. Kilbinger, et al., CosmoPMC.
[5] C. Bourrely, I. Caprini, and L. Lellouch, “Model-independent description of B→πℓ¯νdecays and
a determination of |Vub|”, Phys.Rev. D79, 013008 (2009), arXiv:0807.2722 [hep-ph].
[6] F. Beaujean, “A bayesian analysis of rare b decays with advanced monte carlo methods”, PhD
thesis (Fakultät für Physik, Technische Universität München, 2012).
[7] I. Sentitemsu Imsong, A. Khodjamirian, T. Mannel, and D. van Dyk, “Extrapolation and unitarity
bounds for the B→πform factor”, JHEP 02, 126 (2015), arXiv:1409.7816 [hep-ph].
[8] A. Gelman, and D. B. Rubin, “Inference from Iterative Simulation Using Multiple Sequences”,
Statist. Sci. 7, 457 (1992).
[9] Y. Amhis, et al., “Averages of b-hadron, c-hadron, and τ-lepton properties as of summer 2014”,
(2014), arXiv:1412.7515 [hep-ex].
[10] S. Aoki, et al., “Review of lattice results concerning low-energy particle physics”, Eur. Phys. J. C74,
2890 (2014), arXiv:1310.8555 [hep-lat].
[11] G. E. Crooks, “The Amoroso Distribution”, 2010.
[12] G. Buchalla, A. J. Buras, and M. E. Lautenbacher, “Weak decays beyond leading logarithms”,
Rev.Mod.Phys. 68, 1125 (1996), arXiv:hep-ph/9512380 [hep-ph].
[13] A. J. Buras, “Weak Hamiltonian, CP violation and rare decays”, 281 (1998), arXiv:hep-ph/9806471
[hep-ph].
[14] K. G. Chetyrkin, M. Misiak, and M. Munz, “Weak radiative Bmeson decay beyond leading loga-
rithms”, Phys.Lett. B400, 206 (1997), arXiv:hep-ph/9612313 [hep-ph].
[15] K. G. Chetyrkin, M. Misiak, and M. Munz, “|∆F|= 1 nonleptonic effective Hamiltonian in a simpler
scheme”, Nucl.Phys. B520, 279 (1998), arXiv:hep-ph/9711280 [hep-ph].
[16] C. Bobeth, P. Gambino, M. Gorbahn, and U. Haisch, “Complete NNLO QCD analysis of B→
Xsℓ+ℓ−and higher order electroweak effects”, JHEP 0404, 071 (2004), arXiv:hep-ph/0312090
[hep-ph].
[17] T. Huber, E. Lunghi, M. Misiak, and D. Wyler, “Electromagnetic logarithms in B→Xsℓ+ℓ−”,
Nucl.Phys. B740, 105 (2006), arXiv:hep-ph/0512066 [hep-ph].
[18] T. Feldmann, B. Müller, and D. van Dyk, “Analyzing b→utransitions in semileptonic ¯
Bs→K∗+(→
Kπ)ℓ−¯νℓdecays”, Phys. Rev. D92, 034013 (2015), arXiv:1503.09063 [hep-ph].
47