Module Guide
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 24
flowPsi Module Guide
May 31, 2019
Contents
1 Introduction 1
1.1 Loci FVM Module Facilities . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1
1.2 flowPsi facilitiesandvariables............................ 3
2 Custom Output 6
3 Custom Boundary Condition Input 8
4 Custom Initial Conditions 10
5 Surface Integration 11
6 Volume Integration 13
7 Adding a Volume Source 14
8 Scalar Transport 16
8.1 TracerVariableExample............................... 17
8.2 The Menter one equation turbulence model . . . . . . . . . . . . . . . . . . . . 17
8.2.1 Implementation................................ 18

1 Introduction
This module guide provides a set of tutorials for creating add on modules for the flowPsi
solver. It is assumed that the reader has some basic understanding of the Loci programming
model. To gain that understanding please work through the tutorial provided with the Loci
framework. This document and examples will extend that tutorial to describe how to create
common types of extensions to the flowPsi solver. In general modules are loaded into flowPsi by
first making sure that the directory that contains the module is either in the LD LIBRARY PATH
or the LOCI MODULE PATH environment variable. Then the modules can be loaded into your
flowPsi case by adding a loadModule: followed by the module name at the top of the vars file.
The Makefile provided in these example directories are easy to use and can be copied for your
module development. The Makefile is designed such that all files that end in with the .loci
postfix will be compiled into a module. The file only needs to be edited to provide the module
name and the FLOWPSI BASE directory that points to where the flowPsi solver is installed.
1.1 Loci FVM Module Facilities
Before we begin discussing building modules for flowPsi lets review some of the basic facilities
that Loci provides for finite volume solvers that flowPsi utilizes. First, flowPsi uses the face
based data-structure that includes the face2node map that provides an ordered listing of the
nodes that form each face in the mesh as well as the cl and cr maps that provide the maps
from the face to the left and right cells. Faces on the boundary always have normals pointing
out of the domain and therefore the cl map points to the cell next to the boundary while the
cr cell may point to a ghost cell (in flowPsi, ghost cells are only used to implement periodic
boundary conditions). The ci and ref maps are only defined for boundary faces where ci
refers to the adjacent cell inside the mesh and ref refers to the reference surface that forms the
facets of that boundary. The pmap map associates the periodic boundary face with its periodic
pair. The cell to face maps upper,lower, and boundary map is used to form matrices that are
based on the face connectivity where upper refers to the faces that form the upper and lower
triangular matrix entries and the boundary map identifies boundary faces of the cell.
In addition to these data structure elements, the Loci FVM module provides some generic
rules for computing grid metrics such as face areas and cell volumes. Additionally the module
provides some basic facilities for computing gradients and interpolating solutions from cells
to nodes for plot output. These facilities generally will need the cell center values and the
boundary face values. The boundary face values will have the naming convention of having a
fpostfix annotation denoting that it is a face value. A table that summarizes that facilities
provided by the FVM module are provided in table 1.
1

Table 1: Table of Loci FVM provided utilities
Supplied by Loci FVM facilities
Variable Type Description
Mesh Data Structures
ci map cell next to boundary face
cl map cell on left side of face
cr map cell on right side of face
ref map map from boundary faces to surface
face2node multiMap map from face to nodes
pmap map periodic matching face
upper multiMap map from cell to upper faces
lower multiMap map from cell to lower faces
boundary map multiMap map from cell to boundary faces
Mesh Metrics
pos vector node positions
cellcenter vector cell centroid location
facecenter vector face centroid location
area composite face area and normal
vol scalar cell volume
gridvol parameter total grid volume
Parametric Rules
grads(X) vector gradient of scalar variable X
gradv3d(X) tensor gradient of vector variable X
gradv(X) generic vector gradient of mixture vector X
grads f(X) vector face gradient of scalar X
gradv3d f(X) tensor face gradient of vector X
gradv f(X) generic vector gradient of mixture vector X
cell2node(X) scalar cell to node interpolation for scalar X
cell2node v3d(X) vector cell to node interpolation for vector X
cell2nodeMax(X) scalar nodal maximum of Xfor connected cells
cell2nodeMin(X) scalar nodal minimum of Xfor connected cells
L1Norm(X) parameter volume integrated L1norm of cell variable X
L2Norm(X) parameter volume integrated L2norm of cell variable X
LinfNorm(X) parameter volume integrated L∞norm of cell variable X
fileNumber(X) integer file numbering for variable X
2
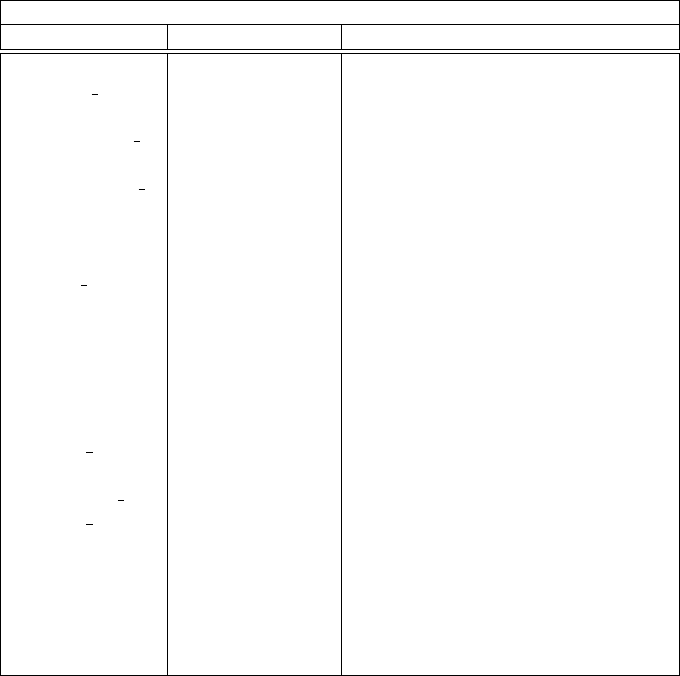
Table 2: Table of flowPsi Fluid Properties
flowPsi fluid properties variables
Variable Type Description
rho scalar fluid density
rho f scalar fluid density at face
temperature scalar fluid temperature
temperature f scalar fluid temperature at face
gagePressure scalar fluid pressure reference to Pambient
gagePressure f scalar fluid gage pressure at face
Pambient parameter reference pressure
pressure scalar fluid pressure
uvector fluid velocity in cell
u f vector fluid velocity at face
divu scalar divergence of u
vort vector fluid vorticity
vortMag scalar fluid vorticity magnitude
strainRate scalar strain rate magnitude, S
tau symmetric tensor fluid stress
mu scalar fluid laminar viscosity
mu f scalar fluid laminar viscosity at face
kconduct scalar fluid laminar conductivity
kconduct f scalar fluid laminar conductivity at face
us n scalar face grid velocity dot normal
us vector face grid velocity
Cp parameter fluid Cp
gamma parameter fluid isentropic index, γ
Rtilde parameter fluid gas constant
tmu scalar turbulent viscosity
soundSpeed scalar local speed of sound
1.2 flowPsi facilities and variables
When flowPsi runs the only the conservative variables (and gage pressure) are carried from
time-step to time-step. All other variables that are derived from these conservative variables
are only computed during the Newton iteration. Thus most of the computations in flowPsi
happen as part of the Newton iteration step. This section outlines some of the facilities that
are available during that step. Table 2 gives that generic fluid properties that are available.
Solver specific variables that can be replaced through priority rules or used in control are shown
in table 3 while the parametric utilities are described in table 4. These tables are provided to
give a brief survey of the kinds of data that is available for interfacing to flowPsi modules.
3

Table 3: Table of flowPsi solver specific variables
flowPsi solver specific variables
Variable Type Description
p0Ref scalar total pressure BC input
T0Ref scalar total temperature BC input
massFluxRef scalar mass flux BC input
temperatureRef scalar temperature BC input
gagePressureRef scalar gagePressure BC input
uRef vector fluid velocity BC input
Twall scalar viscous wall BC temperature
qwall scalar viscous wall BC heat flux
wallVelocity vector viscous wall tangential velocity
src general vector source array for mass, momentum, and energy
iflux Array(5) inviscid flux
vflux Array(4) viscous flux
scalar mdot scalar mass flux through face
srcJ matrix block diagonal jacobian term of src
fjp matrix block jacobian of flux from left
fjm matrix block jacobian of flux from right
min cell2noslip map Map from cell to closest viscous wall
dist noslip scalar distance from cell to nonslip wall
dt parameter timestep value in seconds
stime parameter simulation time in seconds
ncycle parameter simulation step
newtonFinished conditional Newton iteration complete
do restart conditional output restart files
do output conditional text output
do plot conditional plot file output
do boundary plot conditional boundary plot file output
4
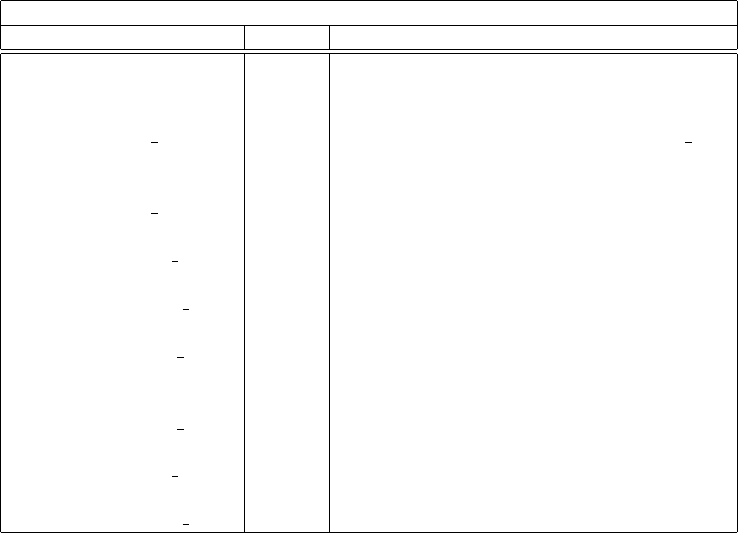
Table 4: Table of flowPsi parametric variables
flowPsi parametric variables
Variable Type Description
scalarTransport(X,C) generic build transport of scalar X, enabled by C
scalarTransportP(X,C) generic like above for positive scalar X
scalarMean(X) scalar time averaged cell value X
scalarMean f(X) scalar time averaged boundary face value X f
scalarVariance(X) scalar time variance of variable X
vect3dMean(X) vector time averaged vector mean
vect3dMean f(X) vector time averaged boundary vector mean
vect3dVariance(X) vector time variance of vector X
vect3dVariance f(X) vector time variance of boundary vector
vect3dCoVariance(X) vector co-variance of vector X
vect3dCoVariance f(X) vector co-variance of boundary vector x
scalarFavreMean(X) scalar Favre average of variable X
scalarFavreMean f(X) scalar Favre average of boundary variable
scalarFavreVar(X) scalar Favre weighted variance
vect3dFavreMean(X) vector vector Favre weighted mean
vect3dFavreMean f(X) vector vector Favre weighted boundary mean
vect3dFavreVar(X) vector vector Favre weighted variance
vect3dFavreVar f(X) vector vector Favre weighted variance boundary
vect3dFavreCoVar(X) vector vector Favre weighted co-variance
vect3dFavreCoVar f(X) vector vector Favre weighted co-variance boundary
5

2 Custom Output
One of the more common needs for add-on modules is the development of derived variable com-
putations. While it is possible to compute derived variables directly in post-processing software,
these computations can sometimes be misleading due to the fact that they are performed on
interpolated data which may introduce errors in the derived computations. So it is sometimes
useful to perform computations directly on the cell values before interpolation in order to get
more satisfactory results. For this example we will consider the output of a variable that can be
used for numerical schlieren imaging. This is done by computing the logarithm of the density
gradient. So in this example we will show how one goes about computing and outputting such
a variable in a module.
The first step to this is to copy the Makefile provided in the examples which will automatically
compile all .loci files in the directory. The only change that is needed is to set the MODULE NAME
variable in the Makefile to the name that you wish to use in the loadModule line of the vars
file. In this case we will call the module schlieren, or
# What is the name of the module given to loadModule
MODULE_NAME = schlieren
At this point we can begin editing our Loci program which we will conveniently name schlieren.loci.
At the beginning of the file we need to include some files that will be needed to access the fa-
cilities we will be using. This is accomplished with the following includes:
// Include Loci system
#include <Loci.h>
// defines types used by flowPsi
#include "flowTypes.h"
// imports flowPsi IO functionality
#include "flowPsiIO.h"
// defines Loci types for flowPsi variables
$include "flowPsi.lh"
It is convenient but not necessary to add the code in the C++ namespace called flowPsi.
Alternatively, we can add the code in our own namespace but add code to tell C++ to use
variables from the flowPsi namespace such as:
namespace schlieren {
using namespace flowPsi ;
We now need to compute the variable that we are going to plot. Since flowPsi computes
unknowns at cell centers this will be where we perform the computations. However, to improve
nodal interpolation accuracy we also need to compute the target variables for all boundary
faces as well. Since these variables are not part of the flowPsi infrastructure we will need to
define their types first:
// define types for cell and boundary face values
$type loggradrho store<double> ;
$type loggradrho_f store<double> ;
6

Then write rules to define how to compute these variables as well. These computations will use
the scalar gradient parametric rules provided by Loci’s FVM module. The rules will be defined
as:
// To interpolate to the nodal values we need both cell values and boundary
// face values. Thus we have two rules to compute the value of interest
$rule pointwise(loggradrho<-grads(rho)) {
$loggradrho = log10(max(norm($grads(rho)),1e-4)) ;
}
$rule pointwise(loggradrho_f<-grads_f(rho)) {
$loggradrho_f = log10(max(norm($grads_f(rho)),1e-4)) ;
}
At this point we have defined the variable that we wish to compute but as they are defined
now they will not be computed because flowPsi will never request these variables. Instead we
will want to tell flowPsi that these variables are ones that we need plot files. For this, flowPsi
provides a macro called OUTPUT SCALAR that interfaces with the flowPsi plotting facilities. It
will allow this variable to be written out when requested by placing the given variable name in
the plot output variable in the vars file. This is accomplished with the line:
OUTPUT_SCALAR("cell2node(loggradrho)", schlieren) ;
This line tells the code to write out the variable cell2node(loggradrho) as a variable named
schlieren. Note that the variable that we wrote out is not the variable that we computed.
This is because volume data is plotted from nodal values and since flowPsi computes its degrees
of freedom at the cells, an interpolation function is needed to compute the nodal values. The
cell2node parametric rules provided by the Loci FVM module performs this function. This
parametric rule will access the variables that we defined earlier. Also this macro will enable
requesting this variable on cutting plane and isosurface output from flowPsi as well.
In addition to volume data, we can also output boundary facet data. This is provided with the
macro OUTPUT BNDRY SCALAR. So if we wish to output the schlieren variable for the boundary
facets we can add the following line to or loci file:
OUTPUT_BNDRY_SCALAR("loggradrho_f",schlierenb,"ci") ;
The first argument of this macro is the variable that we wish to plot, while the second is the
name of the variable (hear with a b appended to differentiate boundary facet data from the
nodal data). Finally, the last argument defines the set of boundaries that we wish to write this
data. Since all boundary faces have the attribute ci this will write out for all boundary faces.
Also not, for outputting 3D vector data replace the SCALAR with VECTOR in the above macros.
7

3 Custom Boundary Condition Input
Most of the boundary conditions provided by flowPsi can be customized. Typically the inputs
for each boundary face are assigned to a Ref variables that can be overridden by modules to
change the basic behavior. For this example we will demonstrate this in a simple boundary
condition where the pressure is perturbed by a sinusoidal signal. In this particular case we will
override the definition of the gageP ressureRef variable so that instead of being the constant
defined in the boundary condition, it will instead be a time varying value. Note, it is possible
to also change the boundary condition to be reactive to the solution itself. However, in these
cases the stability of the resulting boundary condition cannot be guaranteed.
To add this capability to an inflow boundary condition we want to be able to add a specification
to each boundary where we want to add the perturbation that will give the amplitude and
frequency of the disturbance. By default the flowPsi boundary condition checking code will
not allow these parameters to be added without generating error messages. Therefore it is
necessary to register these new boundary condition arguments with flowPsi. This checker for
the boundary conditions are defined in the read grid.h header file which will need to be
included. The checker for these two new variables is added through the following code:
// Here we create a checking class that inherits from BC_Check that is defined
// in readGrid.h
class pwave_check : public BC_Check {
string error_message ;
public:
// List which boundary conditions this checker is for
std::string boundaryConditions() { return "inflow,supersonicInflow,farfield" ; }
// List which variables this checker will verify
std::string variablesChecked() { return "amplitude,frequency" ; }
// Check for the existence of amplitude and frequency
// in our implementation these are optional, but if amplitude is
// given then frequency must also be given
bool checkOptions(const options_list& bc_options) {
error_message = "" ;
bool check = true ;
if(bc_options.optionExists("amplitude")) {
if(!check_scalar_units(bc_options,"amplitude","Pa")) {
error_message += "Pressure units needed for ’amplitude’ " ;
check = false ;
}
if(!bc_options.optionExists("frequency")) {
error_message += "’amplitude’ must also specify ’frequency’ " ;
check = false ;
}
}
return check ;
}
// Print out error messsage. Used if checker fails to generate error
// message to the user. Usually the error message is remembered from
// when the checker was run.
8

std::ostream &ErrorMessage(std::ostream &s) {
}
} ;
// The register_BC template registers this object in the boundary checker
// database. The names are not important only that we create the object
// so that it is registered.
register_BC<pwave_check> register_BC_pwave_check ;
Next we will need to extract the values for amplitude and pressure from the BC options variable
that is defined for each boundary surface. For this we will use the constraint that is created
by the FVM grid reader that indicates what parameter are defined in the BC options variable.
This will ensure that the variables will only be extracted for the boundary conditions where
these arguments are specified. The rules that extract these variables are as follows:
$type amplitude_BC store<real> ;
$rule pointwise(amplitude_BC<-BC_options),constraint(amplitude_BCoption) {
$BC_options.getOptionUnits("amplitude","Pa",$amplitude_BC) ;
}
$type frequency_BC store<real> ;
$rule pointwise(frequency_BC<-BC_options),constraint(frequency_BCoption) {
$BC_options.getOption("frequency",$frequency_BC) ;
}
We can now compute the time varying prescribed pressure and override the gagePressureRef
variable that will be used by the inflow boundary condition.
$type pgDelta_BC store<real> ;
$rule pointwise(pgDelta_BC<-stime,amplitude_BC,frequency_BC) {
$pgDelta_BC = $amplitude_BC*sin($stime*$frequency_BC*2.*M_PI) ;
}
// Override the reference condtions for the faces that adds in the
// perturbation. This will be used instead of the default settings
$rule pointwise(distrbance::gagePressureRef<-
ref->(gagePressureRef_BC,pgDelta_BC)) {
$gagePressureRef = $ref->$gagePressureRef_BC + $ref->$pgDelta_BC ;
}
Other variables can also be overridden using a similar process. For a complete list of the Ref
variables that can be overridden see table 3.
9
4 Custom Initial Conditions
// For the initial conditons we define temperature_ic, gagePressure_ic, and
// initial velocity (u_ic)
$rule pointwise(temperature_ic,gagePressure_ic,u_ic<-
Rtilde,cellcenter,Pambient),constraint(geom_cells) {
const double x = $cellcenter.x ;
const double y = $cellcenter.y ;
double coef = 3.5 ;
double u = -1.*coef*cos(x)*sin(y) ;
double v = coef*sin(x)*cos(y) ;
double Pref = $Pambient ;
if(Pref < 10)
Pref = 1e5 ;
double rho = 1.0 ;
double p0 = Pref-$Pambient ;
double pg = p0-rho*0.25*coef*coef*(cos(2.*x)+cos(2.*y)) ;
double P = pg+$Pambient ;
double T = P/($Rtilde*rho) ;
$gagePressure_ic = pg ;
$temperature_ic = T ;
$u_ic = vect3d(u,v,0.0) ; ;
}
10

5 Surface Integration
In this example we consider how to add a module that can integrate a function over the boundary
surfaces of the mesh. In this example we will compute the area averaged temperature over all of
the boundary surfaces of the mesh. In general, Loci is quite flexible and will allow a wide range
of possible strategies for this. In effect, an integration is simply a reduction with the summation
operator over some set of entities that defines the surface of interest. How does one identify
the set of interest? Actually one could identify the set of interest in many different ways.
First, one could use the assigned boundary condition type. For example, one could integrate
over the set defined by the constraint viscousWall BC which would include all boundaries
that were assigned the viscousWall boundary condition. Alternatively, the user could define
a new boundary condition argument that would allow a user to mark that boundary with a
special integrate flag (say integrate) and then use a constraint of ref->integrate BCoption
to determine the set. In this example we use the parametric facility to integrate over each
boundary surface separately. To do this we use the fact that for each boundary a parametric
variable will be created in by the grid reader of the form boundaryName(X) where Xis replaced
by each boundary surface name. This is a string parameter which is defined over the set of
faces that form that boundary surface. For our mean temperature we will use an area weighted
average, so at first we will compute the total area for each boundary with the rule:
// This sets up the initial value of the boundary area before summing over
// faces. This needs to be set to zero as this is the indentity of the
// summation operator
$rule unit(boundaryArea_X<-boundaryName(X)),parametric(boundaryName(X)) {
$boundaryArea_X = 0 ;
}
// Add the areas from each face. In some cases, for example when
// running in axisymmetric mode the area might be zero, so the max
// function is there to keep that case from generating a divide by zero
// condition.
$rule apply(boundaryArea_X<-area,boundaryName(X))[Loci::Summation],
parametric(boundaryName(X)) {
// note, the area is the sada component of the area input, the n component
// is the normal vector
join($boundaryArea_X,max($area.sada,1e-13)) ;
}
In this set of parametric rules the boundaryArea X variable will be created for each boundary
condition with the Xreplaced by each boundary name. Now we can compute the weighted
temperature with the following rules:
// Now we will compute the area weighted average temperature
$type boundaryMeanTemp_X param<real> ;
// set the initial value for the mean temp
$rule unit(boundaryMeanTemp_X<-boundaryName(X)),parametric(boundaryName(X)) {
$boundaryMeanTemp_X = 0 ;
}
11
// Now compute the weighted sum of the of the temperature. The temperature
// at the boundary face is given by temperature_f
$rule apply(boundaryMeanTemp_X<-area,temperature_f,boundaryArea_X,boundaryName(X))[Loci::Summation],
parametric(boundaryName(X)) {
double weight = max($area.sada,1e-13)/$boundaryArea_X ;
join($boundaryMeanTemp_X,weight*$temperature_f) ;
}
Now we can output the integrated value. Note, that although this appears to be one rule, one
will be instantiated for each boundary surface. In the end this will generate output for each
boundary.
// Now output to stdout the average temperature. Do this output only
// when the code is outputting boundary plot files.
// Note, since this is only output once, we use the prelude and
// because we are in the prelude we must use the dereference operatpr
// ’*’ to access the computed parameters. The $[Once] tells Loci that
// you only want to output this once even when running in parallel where
// there would be multiple instances of execution
$rule pointwise(OUTPUT<-boundaryMeanTemp_X,boundaryName(X)),
conditional(do_boundary_plot),
parametric(boundaryName(X)), prelude {
$[Once] {
cout << "Boundary " << *$boundaryName(X) << " has mean temperature of "
<< *$boundaryMeanTemp_X << " Kelvin" << endl ;
}
} ;
//^ This semicolon is needed because we are not following the prelude
// section with a compute section.
12

6 Volume Integration
In this example we will show how to perform a volume integration over each named component
of the mesh. This will work similarly to the surface integration except that we will use the
volumeTag(X) parametric variable instead. For this example we will compute the integrated
fluid kinetic energy by simply adding up the kinetic energy integrated over each cell. The basic
approach is as follows:
// KEComponent_X is the integrated kinetic energy for component X. This
// rule is parametric so one of these variables will be created for each
// component.
$type KEComponent_X param<double> ;
$rule unit(KEComponent_X),constraint(volumeTag(X)),
parametric(volumeTag(X)) {
$KEComponent_X = 0 ;
}
// Here we integrate the kinetic energy over each cell which by use
// of the midpoint rule is simply vol*rho*0.5*dot(u,u), then the sum
// is the total kinetic energy
$rule apply(KEComponent_X<-vol,rho,u)[Loci::Summation],
constraint(volumeTag(X)),parametric(volumeTag(X)) {
double kecell = 0.5*$vol*$rho*dot($u,$u) ;
join($KEComponent_X,kecell) ;
}
Then KEComponent X can be written out to the screen in a similar fashion as the previous
example.
13
7 Adding a Volume Source
Another common type of addition to the solver is to add a new source term to the equations.
In this example we add a body force to return momentum that is lost to viscous walls for
the purpose of computing channel flow type of simulations. When using the implicit solver a
jacobian may need to also be computed depending on the stiffness of the source term. In flowPsi
there are two different possible jacobian types that will need to be supported to support all
implicit formulations. By default the code uses jacobians with respect to the primitive variables
density and gage pressure, but in modes using the local preconditioner it will use temperature
and gage pressure instead. If you want to support both modes of operation you will need to
define both types of jacobians.
For the channel flow case we first need to find out how much momentum was lost through the
fluid interaction with the viscous walls. For this we utilize a convenience variable (AllViscousBCs)
that contains all viscous boundary conditions including viscousWall and wallLaw. For the
integration of the momentum flux we consider both the inviscid flux, iflux, and the viscous
flux vflux. Since these variables contain mass, momentum, and energy fluxes, we need to
extract just the part that we are going to use. For the inviscid flux we skip the first item as
this is the mass flux. For the viscous flux the first item is the momentum flux as there is no
mass flux in the variable vflux
$type viscousWallMomentumFlux param<vect3d> ;
$rule unit(viscousWallMomentumFlux),constraint(UNIVERSE) {
$viscousWallMomentumFlux = vect3d(0,0,0) ;
}
// Note viscous and inviscid flux have already been integrated over the
// boundary facet area. We just need to sum up their contributions
$rule apply(viscousWallMomentumFlux<-iflux)[Loci::Summation],
constraint(AllViscousBCs) {
const int mi = 1
// extract inviscid momentum flux
vect3d mflux($iflux[mi+0],$iflux[mi+1],$iflux[mi+2]) ;
join($viscousWallMomentumFlux,mflux) ;
}
$rule apply(viscousWallMomentumFlux<-vflux,qvi)[Loci::Summation],
constraint(AllViscousBCs) {
const int mi = 0 ;
// extract viscous momentum flux
vect3d mflux($vflux[mi+0],$vflux[mi+1],$vflux[mi+2]) ;
join($viscousWallMomentumFlux,mflux) ;
}
Now we can compute the total force that we need to add back into the system to restore the lost
momentum. To do this we also integrate the total mass in the volume using a similar unit/apply
rule combination to compute the variable massSum. The body force is then computed for each
cell based on the local density. We also add a term to the energy equation to account for
the energy added to the system due to this addition (since we do not wish this energy to be
deducted from the internal energy of the fluid). Adding the resulting body force to the solver
then is implemented as:
14
//add friction loss to the source term
$rule apply(src<-dragAccel,rho,vol,cellcenter,u)[Loci::Summation] {
const int mi = 1 ;
const int mj = mi + 1 ;
const int mk = mi + 2 ;
const int ei = 4 ;
$src[mi] += $dragAccel.x*$rho*$vol ;
$src[mj] += $dragAccel.y*$rho*$vol ;
$src[mk] += $dragAccel.z*$rho*$vol ;
$src[ei] += $dragAccel.x*$u.x*$rho*$vol ;
}
The jacobians in flowPsi are computed with respect to the primitive variables gage pressure,
temperature, and velocity. The jacobians for the diagonal blocks are stored in the variable
srcJ. For this simple source term the jacobians are straightforward to implement which is
shown below:
$rule apply(srcJ<-rho,gagePressure,temperature,Pambient,u,dragAccel,vol)[Loci::Summation],
constraint(vol) {
const int mi = 1 ;
const int mj = mi + 1 ;
const int mk = mi + 2 ;
const int ei = 4 ;
const real drdt = -$rho/$temperature ;
const real drdp = $rho/($gagePressure+$Pambient) ;
real coefP = drdp*$vol ;
real coefT = drdt*$vol ;
real rho_vol = $rho*$vol ;
$srcJ[mi][4] += $dragAccel.x*coefP ;
$srcJ[mj][4] += $dragAccel.y*coefP ;
$srcJ[mk][4] += $dragAccel.z*coefP ;
$srcJ[ei][4] += dot($dragAccel,$u)*coefP ;
$srcJ[mi][0] += $dragAccel.x*coefT ;
$srcJ[mj][0] += $dragAccel.y*coefT ;
$srcJ[mk][0] += $dragAccel.z*coefT ;
$srcJ[ei][0] += dot($dragAccel,$u)*coefT ;
$srcJ[ei][mi] += $dragAccel.x*rho_vol ;
$srcJ[ei][mj] += $dragAccel.y*rho_vol ;
$srcJ[ei][mk] += $dragAccel.z*rho_vol ;
}
15
8 Scalar Transport
The flowPsi code provides an infrastructure that makes it simple to add new variables that
will be transported with the fluid flow. These variables makes it simple to add new models
to accompany the basic fluid flow. In this section we will discuss two examples. First the
development of a simple tracer variable that can inject spatially distributed sine waves that can
be convected with the flow and a second which goes through the implementation of a simple
one equation turbulence model. One advantage of using the scalar transport facilities is that
these facilities include interfaces with the restart, overset, and time integration facilities of the
flowPsi solver. Thus the scalar transport facilities gives a comprehensive way of adding new
variables to the flow solver.
Before we begin, we will have a quick review of the implementation of the scalar transport
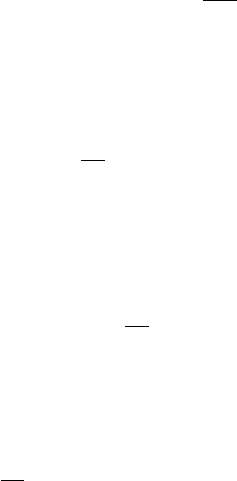
equations. The scalar transport equation in conservative form is written for a convected scalar,
φas ∂ρφ
∂t +∇ · (ρuφ) = ∇ · (λ∇φ) + Sφ,(1)
where ρis the fluid density, uis the fluid velocity vector, λis a diffusion coefficient, and Sφ
is a prescribed source term. We note that by substituting φ= 1 into the above equation one
recovers the global continuity equation. We discretize the above equation in time and space to
arrive at the expression
V
∆t(1 + ψ)(φn+1ρn+1 −φnρn) + ψ(φnρn−φn−1ρn−1)= (2)
−Xφn+1
u˙mf+XDφf+Sφ,
where φuis an upwinded extrapolation of φbased on the sign of the face mass flux ˙mand Dφf
is the numerical diffusion flux for scalar φ. Note that the discrete continuity equation is given
by
V
∆t(1 + ψ)(ρn+1 −ρn) + ψ(ρn−ρn−1)=−X˙mf.(3)
To improve the diagonal dominance of the scheme we employ a trick where we recognize that
Eq. (3) is zero when the system of equation is solved, thus we multiply Eq. (3) by φn+1 and
subtract it from Eq. (3) to arrive at the following expression:
V
∆t(1 + ψ)φn+1 −φnρn+φnρn−φn−1ρn−1ψ−ρn−ρn−1φn+1ψ−(4)
hφn+1 X˙mf−Xφn+1
u˙mf+XDφf+Sφi=Lφ(φn+1)=0.
The above operator, Lφ, is solved using a Newton method concurrently with the fluid equations.
These facilities can be enable to track the convection and diffusion of any arbitrary scalar. The
facility is provided by the parametric variables scalarTransport(X,C) or scalarTransportP(X,C)
where the first version is for variables that can be of any sign while the second is for variables
that are always positive. The first Xargument to the parameter is the name of the scalar to be
transported, while the second Cargument is a constraint that can be used to activate/deactivate
the scalar integration. Accessing this variable causes an entire iterative infrastructure to be
constructed that solves the scalar transport equations which can then be augmented with addi-
tional source terms to create a wide variety of models. An arbitrary number of scalar transport
equations can be constructed.
16

8.1 Tracer Variable Example
The first example that we have is simply the convection of a tracer variable that will simply
be convected with the flow. This will allow us to show how to set up the boundary and initial
conditions are setup. For this we will create the scalar transport for the variable tracer.
To setup flowPsi to transport the variable we first need to instantiate the scalar transport
equations. This is accomplished with the line:
$rule pointwise(OUTPUT<-scalarTransport(tracer,UNIVERSE)) { }
Note that this rule will never actually execute. Instead, the existence of this rule will cause the
parametric scalarTransport variable to be instantiated for the variable tracer. Setting the
second argument to UNIVERSE ensures that this scalar transport model will be always active.
To complete the scalar transport equations we need to provide the state of the tracer variable
at the boundaries. For this we define the boundary variable tracer bc. In general we will
define the variable for whatever our scalar name is followed by the bc postfix. In this case we
will use a spatially sinusoidal function for boundaries that we mark as having the tracer and set
the function to zero otherwise. By default the boundary condition will use simple upwinding
to determine the value of tracer f. To define this we simply use the rules:
$type tracer_bc store<real> ;
$rule pointwise(tracer_bc),constraint(ref->BC_options) {
$tracer_bc = 0 ;
}
$rule pointwise(marker::tracer_bc<-facecenter,tracerNormal,tracerPt,tracerInterval),constraint(ref->tracer_BCoption) {
real r = 2.*M_PI*dot($facecenter-$tracerPt,$tracerNormal)/$tracerInterval ;
$tracer_bc = cos(r) ;
}
Finally we need to output the tracer so that we can see where the tracer advected in the
simulation. This is accomplished with the line:
OUTPUT_SCALAR("cell2node(tracer)",tracer) ;
That is all that is needed to add the most basic type of scalar transport equation. Next we will
show how to develop a model with more comprehensive set of parts: a turbulence model.
8.2 The Menter one equation turbulence model
The Menter one equation turbulence model[1] is formally derived from the k−model after
eliminating kand from the equations by using the definition of the eddy viscosity and by
assuming that the turbulent shear stress is proportional to the turbulent kinetic energy. The
model solves the following single equation for the undamped eddy viscosity ˜νt:
17
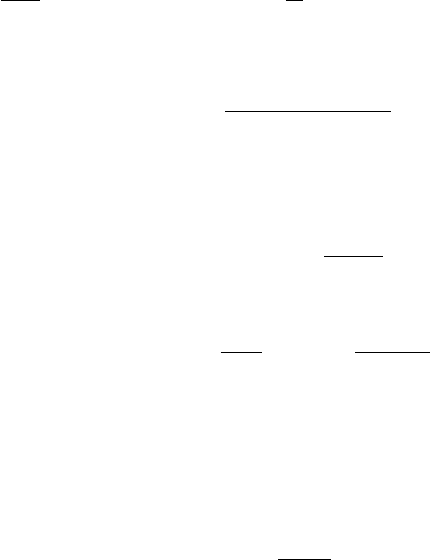
∂ρ˜νt
∂t +∇ · (ρ~u˜νt) = ∇ · ρν+˜νt
σ∇˜νt+ρc1D1˜νtS−ρc2E1e(5)
where Sis the strain rate defined by:
S=q(∇~u)·(∇~u +∇~uT) (6)
The modified destruction term E1eis defined as:
E1e=c3EBB tanh Ek−
c3EBB ,(7)
where
Ek−= ˜ν2
t1
LV K 2
= ˜ν2
t∇S· ∇S
S2(8)
involves the inverse of the von Karman length scale, and EBB is the Baldwin-Barth destruction
term:
EBB =∇˜νt· ∇˜νt.(9)
The damping terms D1and D2are defined as:
D1=νt+ν
˜νt+ν(10)
D2= 1 −e−(˜νt/A+κν)2,(11)
and the model constants are:
c1= 0.144, c2= 1.86, c3= 7, κ = 0.41, σ = 1, A+= 13 .(12)
The damped eddy viscosity is obtained from:
νt=D2˜νt.(13)
The boundary conditions for ˜νtat a solid wall are ˜νt= 0. The initial and free-stream conditions
are ˜νt∞≤ν∞. The default initial and free-stream value for ˜νtis 5 x 10−9m2/s. Note that the
boundary conditions are given in terms of the undamped eddy viscosity (˜νt) rather than νt.
8.2.1 Implementation
Examining equation (5) we note that the scalar transport will implement the first three terms.
We will need to define the diffusion coefficient and the last two terms to implement the turbu-
lence model. First we invoke the scalar transport of the variable nuTM which represents the ˜νt
in the equations. This is accomplished with the following code:
18

$rule pointwise(OUTPUT<-scalarTransportP(nuTM,menter)) { }
Here we constraint the model by the variable menter which will be defined as part of the model
as the set of constants that are used in the model. For the initial conditions we will extract
nu t from the initial conditions. This is extracted with the following rules:
$type nuTM_ic store<real> ;
$type icRegionInfo blackbox<ICparsedInitRegion> ;
$rule pointwise(nuTM_ic<-icRegionInfo,cellcenter),
constraint(geom_cells,menter) {
double nu_tm ;
$icRegionInfo.defaultState.get_nu_t(nu_tm) ;
$nuTM_ic = nu_tm ;
}
To complete the basic transport of nuTM we need to setup the boundary conditions for the
model For this we define:
$type nuTM_bcVal store<real> ;
// extract the boundary condition from BC_options
$rule pointwise(nuTM_bcVal<-BC_options) {
real nuTM = 5e-9 ;
if($BC_options.optionExists("nu_t"))
$BC_options.getOption("nu_t",nuTM) ;
$nuTM_bcVal = nuTM ;
}
// set boundary condition
$rule pointwise(nuTM_bc<- ref->nuTM_bcVal) {
$nuTM_bc = $ref->$nuTM_bcVal ;
}
This does not complete the boundary condition however, as the default is to use upwinding to
define the boundary face values. However for a boundary such as a viscous wall the boundary
value for nuTM f must be set explicitly. This is accomplished with the priority override rule
that will replace the upwind definition with this Dirichlet one:
$rule pointwise(noslip::nuTM_f),constraint(viscousWall_BC) {
$nuTM_f = 0.0 ;
}
Now we need to define the scalar diffusion term which is the third term in equation (5). This
will be defined by a face value for the diffusion coefficient which will have a name derived from
the scalar variable name by appending nu f. The diffusion term is thus implemented with the
following rules:
$type nuTM_nu_f store<real> ;
19

$rule pointwise(nuTM_nu_f<-rho_f,nuTM_f,mu_f,menter) {
const real sigma = $menter.sigma ;
$nuTM_nu_f = ($mu_f+$rho_f*$nuTM_f/sigma) ;
}
// The diffusion coefficient needs nuTM_f at all faces, but the
// infrastructure only defines them at the boundary.
// For interior faces we set the face value to be simply the
// reciprocal volume weighted average of the cell values
$rule pointwise(nuTM_f<-(cr,cl)->(vol,nuTM)) {
real vrvl = 1.0/($cr->$vol+$cl->$vol) ;
$nuTM_f = ($cr->$vol*$cl->$nuTM+$cl->$vol*$cr->$nuTM)*vrvl ;
}
Now we have completely defined the first three terms of equation (5). Now what remains is
to define the destruction and production terms. To add these terms we will use a unit rule
to combine them with the previous terms in the variable nuTM src. First lets consider the
production term which which can be added using the rules:
$type nuTM_prod store<real> ;
$rule pointwise(nuTM_prod<-nuTM,D1,rho,strainRate,vol,menter) {
const real c1 = $menter.c1 ;
const real P_k = $rho*c1*$D1*$nuTM*$strainRate ;
$nuTM_prod = P_k ;
}
//=======================================
// add integrated production term to rhs
//=======================================
$rule apply(nuTM_src<-nuTM_prod,vol)[Loci::Summation] {
$nuTM_src += $nuTM_prod*$vol ;
}
where strainRate is provide by the solver and D1 is the production damping function which
is defined by the rule:
$type D1 store<real> ;
$rule pointwise(D1<-nuTM,tmu,mu,rho) {
const real nut = $tmu/$rho ;
const real nu = $mu/$rho ;
$D1 = (nut+nu)/($nuTM+nu) ;
}
Note, the turbulent viscosity (tmu) has not yet been defined but will be shortly. The order
that rules appear in the file do not matter. Note, we have not considered the jacobian of the
production term. In general the sign of the production jacobian is destabilizing and as a general
rule it is desirable to leave this term out of an implicit implementation.
Next we will consider the last destruction term. This can be implemented using the following
rules:
20

$rule pointwise(nuTM_dest<-nuTM,strainRate,grads(nuTM),grads(strainRate),rho,menter) {
const real c3 = $menter.c3 ;
const real Ebb = dot($grads(nuTM),$grads(nuTM)) ;
const real Sdot = dot($grads(strainRate),$grads(strainRate)) ;
const real S2 = $strainRate*$strainRate ;
const real Eke = pow($nuTM,2)*Sdot/(S2+EPSILON) ;
real E1e = c3*Ebb*tanh(Eke/(c3*Ebb+EPSILON)) ;
const real c2 = $menter.c2 ;
$nuTM_dest = $rho*c2*E1e ;
}
//======================================
// Add integrated destruction term rhs
//======================================
$rule apply(nuTM_src<-nuTM_dest,vol)[Loci::Summation] {
join($nuTM_src, -$nuTM_dest*$vol) ;
}
One point to note on this is that we need the gradient of strainRate to implement this
term. For this implementation we are simply using the least squares gradient provided by the
FVM module to compute this. However, we will run into a problem here because the gradient
computation will require a value for strainRate at the face which is not provided by the solver.
For expediency we will assume that strainRate is relatively constant at the boundaries and
just copy from the cell to the face. This is accomplished with the rule
$rule pointwise(strainRate_f<-ci->strainRate) {
$strainRate_f = $ci->$strainRate ;
}
This implementation is OK in a pinch, but more sophisticated methods for computing this would
be suggested for a production level implementation. In particular the recursive application of
least squares gradients can be sensitive to numerical noise and the treatment of the boundary
values may lead to spurious results. For our limited tests this implementation seems to produce
satisfactory results.
For the destruction term it can be helpful to include a jacobian term. However, due to the
gradients of nuTM used in the destruction term, the exact jacobian can be expensive and tedious
to derive. For this reason we use a diagonalized approximation that can stabilize the solver
with a reasonable cost. The diagonal term of the jacobian is in the variable nuTM srcJ. Our
approximation is implemented using the following rule
//=================================================
// Apply an approximate destruction term jacobian
//=================================================
$type nuTM_srcJ store<real> ;
$rule apply(nuTM_srcJ<-nuTM_dest,vol,nuTM)[Loci::Summation] {
join($nuTM_srcJ, -$vol*2.*$nuTM*($nuTM_dest/pow($nuTM,2))) ;
}
21
Now all that remains is defining the turbulent viscosity which is computed from the scalar
variable we are solving combined with a the damping term D2. This is defined using the rules:
// nuTM damping function
$type D2 store<real> ;
$rule pointwise(D2<-nuTM,rho,mu,menter) {
const real aplus = $menter.aplus ;
const real kappa = $menter.kappa ;
const real nu = $mu/$rho ;
const real a1 = pow($nuTM/(aplus*kappa*nu),2) ;
$D2 = 1.-exp(-a1) ;
}
//=========================================================
// Define turbulent viscosity for mean flow solver to use
//=========================================================
$rule pointwise(menter::tmu<-nuTM,rho,D2) {
$tmu = $rho*$D2*$nuTM ;
}
And with this we complete the implementation of the turbulence model.
References
[1] F R. Menter. Eddy viscosity transport and equations and their relation to the k-model.
J. of Fluids Engineering, 119:876–884, 1997.
22