MvdMlab Manual V0
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 46
MVDMLAB MANUAL

Copyright © 2018 van der Meer lab
www.vandermeerlab.org
Licensed under the Creative Commons BY-NC-SA License (the “License”), version 4.0; you may not use
this file except in compliance with the License. Briefly, you are free to share and adapt this material, un-
der the condition that you give appropriate credit, do not use the material for commercial purposes,
and distribute your contributions under the same terms. You may obtain a copy of the full License at
https://creativecommons.org/licenses/by-nc-sa/4.0/.
Inspired by similar lab manuals by others. Particularly helpful were the wonderful examples from the
Peelle and Aly labs.
Typeset in Palatino Linotype using L
A
T
EX and the tufte-book class.
Version 0.1, October 2018
Contents
Welcome! 7
About this manual 9
Research Using Animals 11
Values and Expectations 13
Expectations: everyone 13
Principal Investigator 14
Graduate Students 15
Research Technicians 15
Undergraduates 15
Code of Conduct 15
Doing good science 17
Failing fast and often, but learn from your mistakes 17
Documentation and taking good notes 18
Asking good questions 19
Reading papers 19
Communication 19
Lab space 21
General expectations 21
Experiment rooms 22
Fine Assembly Room (FAR) 23
4
Surgery and anteroom 23
Workshop 23
Shared facilities 23
Vivarium space 23
Offices 23
Electronic resources 25
Dropbox, Drive, Calendar 25
GitHub 25
Slack 25
Wiki 25
Mailing list 25
Computing 27
Lab computers 27
Skills 29
Software 30
Data management 33
Data storage 34
Data promotion 35
Data use cases 35
Animal care and recordkeeping 37
Animal ownership and responsibility 37
PBS and Dartmouth 39
Training required by Dartmouth 39
Important people and contact info 39
Dartmouth resources 39
Beyond the lab 41
Appendix: Subject email format template 43

Welcome!
I didn’t change my mind, it changed all by itself.
– Luna, Double Feature (1995)
The van der Meer lab brings together people who share an in-
terest in how the brain works. We aim to better understand how
learning, memory, and decision-making arise from the coordinated
activity of neurons. In the pursuit of this goal, we perform brain
surgery, design and construct strange mazes, painstakingly build
small devices so we can read the minds of rats, solder tiny circuit
boards to even tinier wires, write thousands of lines of computer
code, collect beautiful data, produce even more beautiful plots, and
many other activities.
Figure 1: Some strange mazes. (From
Emily Irvine’s shortcut experiment.)
Each of us brings their own particular blend of motivations for
engaging in these lab activities, but there are a few common threads:
curiosity about how the physical stuff of the brain gives rise to
thought and to behavior, a love for animals and computers alike, a
desire to help solve some of the big mysteries, and contributing to
human knowledge and ultimately a better world.
Figure 2: A small device for rat mind-
reading (by Andrew Alvarenga).
Our efforts are collaborative, not only within the lab, but also
with other labs in the department of Psychological & Brain Sciences
at Dartmouth. We share equipment, space, and interests with these
labs, and collaborate on joint projects and on creating a scientifically
exciting and supportive culture where everyone can thrive. We also
have joint projects with other labs around the world. We collaborate
because the problems we are trying to solve are hard, and because
learning from and working with others is one of the great joys of
working in science!
8
Working with others brings not only joy, but also expectations.
Coding and data analysis can be fun, but are full of pitfalls. Sharing
space and expensive equipment with others demands respect. There
is satisfaction in getting an experiment to work, but it can be a long
slog to get there. Perhaps most importantly of all, the use of animals
in research comes with practical as well as moral responsibilities. This
manual is intended to provide guidance on how to navigate these issues,
with particular focus on the van der Meer lab.
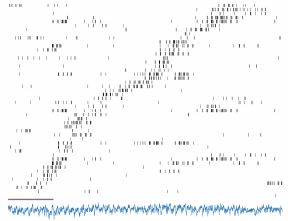
Figure 3: Some beautiful data, recorded
from R050’s hippocampus (by Alyssa
Carey). Vertical tick marks indicate
spikes (one row per neuron, sorted by
place field location); horizontal axis
indicates time; blue trace shows a local
field potential. Scale bar is 1s.
If you are new to the lab, I would like you to read this man-
ual through in its entirety. If you will not be working with animals
directly, it may seem like chapter on Animal Management does not
apply to you, but it will give you important insights into how the lab
operates.
Welcome. Let’s do some great science together!
–MvdM
About this manual
This document describes the principles that shape how the mem-
bers of the lab interact and work. Rather than dealing with the nuts
and bolts of how to get stuff done in the lab, the manual is about lab
philosophy, expectations, and resources. It is an introduction to the
lab. The lab manual is one of several classes (categories) of shared
documents in the lab, which are:
•Lab Manual. You are looking at it. It contains information that does
not change frequently. Only MvdM can change the lab manual,
but he wants to hear from you if you have thoughts of suggestions!
Periodically, we will review the manual as a group at lab meeting
to determine what needs updating.
• Private GitHub repositories1that contain protocols, i.e. documen- 1www.github.com/vandermeerlab.
Because these repositories are private,
you will not be able to see them unless
you are logged in and a member of the
vandermeerlab organization.
tation for experimental procedures2, and a separate repository for
2Examples include electrode plating,
histology, drive building, et cetera:
mvdmlab-protocols repository
behavioral tasks and associated computer code3. This documen-
3mvdmlab-tasks
tation is on GitHub so that we can say things like, “I used version
1.1of the histology protocol”, and so that we can track changes
to those protocols as well as the reasons for those changes. If you
perform procedures in the lab, you are expected to follow and to
contribute to these protocols; see section XXX for a more detailed
explanation.
• A lab wiki4that contains tutorials, guides, lists of useful links, et 4discovery.dartmouth.edu/~mvdm/wiki
cetera. The wiki is a more dynamic, easier to edit resource for
content that doesn’t rely as much on version control. The wiki
currently contains the lab’s MATLAB data analysis tutorials.
• We also have a lab Slack team5which is used for day to day com- 5mvdmlab.slack.com
munication. It hosts shared documents best described as “every-
thing else”, something that isn’t a protocol, task, or tutorial.
These different venues reflect an overall organization ranging
from content that is easy to create and easy to change (low overhead,
quick; Slack, wiki), to content that is moderately stable and takes a

10 mvdmlab manual
bit more effort to change (but easier to track; GitHub), to principles
that rarely change (lab manual). For instance, an idea for an exper-
iment may be initially discussed on Slack, lead to a draft protocol
shared there, and then get pushed to GitHub.
There are a number of other kinds of documents used in the lab
that, unlike the above categories, are not typically consulted by multi-
ple lab members. Most important among these are (1) animal records,
described in more detail in section XXX, and (2) lab notebooks, de-
scribed in section YYY.
I assume the lab manual and procedures on GitHub are accurate. This means
that you should follow all of the policies and procedures contained in the man-
ual and GitHub. If you notice something that seems to be wrong, please let
me know (for the lab manual) or change it yourself (if on GitHub). If there is
something in the lab manual or GitHub that you notice people aren’t doing,
please bring this up at lab meeting, or to me directly. Don’t assume this is
okay (it’s not).
Research Using Animals
We use animals in the lab, based on the conviction that doing so
accelerates scientific progess and provides a net benefit to humans,
and perhaps to animals, too. There are many examples of scientific
discoveries that were directly enabled by resarch in animals6. How- 6Some good examples include the
“Position statement of the Max Planck
Society concerning the use of animals
in experiments for basic research”,
https://www.mpg.de/10882259/MPG_
Whitepaper.pdf
ever, these successes do not mean we can experiment freely on any
animal we want. The use of animals for research carries a moral, sci-
entific, and legal obligation to only perform experiments where the
benefits outweigh the costs, where no suitable alternatives are avail-
able, and to care for our animals to the fullest extent possible. These
notions are often phrased as the “3R’s” (reduction, replacement,
refinement)7.7Some references on the 3Rs
More generally, research in animals demands that we do the best
job possible, and maximize the usefulness of the research outcomes.
This means that:
• We take care of our animals. If animals are comfortable and well
cared for, they will perform the behavioral tasks we want, be easy
to handle.
• We strive to design experiments so the outcomes will be as infor-
mative as possible.
• We aim to collect the highest quality data possible.
• For the data to be interpretable, good records need to be kept.
• Data is valuable. It needs to be protected, and be easy to use.
This means it needs to be organized and annotated in a specific
common format.
In sum, the fact we are working with animals shapes both our lab
philosophy and many practical aspects of working in the lab.
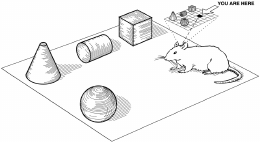
Figure 4: A cartoon rat with some
objects. Figure from Dudchenko et al.
Doing research with animals shapes not only the philosophy of the
lab, but also has pervasive consequences for day-to-day work. Later
12 mvdmlab manual
sections of this manual, such as XXX and YYY, will deal in more
detail with the nuts and bolts.
All of these components will be discussed on more detail in the
relevant sections. This chapter is intended to provide some key defi-
nitions, references, and principles.
Animal research at Dartmouth is overseen by the IACUC.
All work with animals (specifically, all procedures) needs to re-
ceive prior approval from the IACUC in the form of protocols8.8Protocol: document outlining the
scientific rationale, aims, and a set of
proposed experiments.
Animal care is provided by CCMR.
At a minimum, we are required to log all procedures9.9Procedure: Any action or change
beyond picking an animal up in the
colony room (e.g. for changing cages or
weighing) counts as a procedure
Apart from the moral obligation to care for our animals and en-
sure they can contribute to science, we often invest a tremendous
amount of time in each animal through behavioral training, con-
struction of chronic implants, surgery, and so on. If any one of these
procedures is not performed diligently, then your investment in all
the others may come to nothing.
Values and Expectations
The lab values personal and scientific integrity, respect for each other,
for animals, for equipment, and for data. We care about shaping and
maintaining an environment that fosters personal development, cu-
riosity, exploration, and the joy of discovery. We choose to work on
hard problems that require dedication, persistence, resourcefulness,
and resilience. We are humble. We support each other in when things
get tough, and celebrate each other’s successes. We value opportuni-
ties to learn from each other, set high standards but do not judge.
Holding these values implies the following:
Expectations: everyone
• Take initiative. If you are stuck, try something different. Ask
someone.
• Document. Anything you do more than once, or that you or
someone else might have to do again, should be written up as
aprotocol. Anything done with an animal is a procedure and
needs to be logged. Good recordkeeping is essential to doing good
science. Keep track of what you did with experiments and analy-
ses in your lab notebook.
• Communicate. Tell others what you are working on, what is work-
ing well and what could be improved. Post on updates on Slack
frequently.
• Contribute to community resources and space. If you notice some-
thing that can be improved, fix it, mention it on Slack, or open a
GitHub issue.
• Collaborate. Share your expertise. Make arrangements with others
to take care of their animals one weekend and reciprocate another.
14 mvdmlab manual
Offer to do code reviews, read each other’s manuscript, offer feed-
back on practive talks.
• Showing up. There will be days when it feels like you aren’t mak-
ing any progress and nothing is working. Don’t underestimate the
power of an iterative routine.
• Be a little obsessive.
• Be punctual.
• Take care of yourself. Know when it’s time to push, and when it’s
time to take a walk in Pine Park10.10 See here for a guide to some of
Hanover’s best trails, accessible from
the lab in the space of a lunch break.
• Tell others when you notice they did something well, or when they
made your life a little better today11.11 We have the octothorp, awarded
during lab meeting. But don’t stop
there!
• Keep at it. While brilliant ideas, creativity and skill certainly help,
the best predictor of success is how consistently you put in the
work.
Values: personal and scientific integrity; respect for animals, hu-
mans, equipment, data, space. Supportive and collaborative learning
atmosphere that fosters personal development, curiosity, exploration,
joy of discovery. For this to count, need persistence, resilience, and
hard work. This is only possible if you are pursuing something you
care about, and take care of yourself and those around you (so they
may reciprocate). Be kind. Non-judgmental, recognize that failing
often is part of science, changing your opinion is better than having
no opinion.
Well-being. Work hard but be kind to yourself and others.
Share your expertise, be respectful of others’ time.
Attend weekly lab meetings.
Principal Investigator
Have a vision for where the lab is going, and how to get there.
Provide an environment where great science can be done and
people can thrive.
Obtain funding.
Help train you.
values and expectations 15
Graduate Students
Make an original contribution to knowledge.
Research Technicians
Undergraduates
Onboarding process.
Letters of recommendation.
Funding sources for research and travel.
Code of Conduct
Short version: be nice. Science needs contributions from everyone.
There are well-documented barriers to participating that we want to
do our best to help eradicate.
Some specific things that work well:
Long version: laid out in Dartmouth policies.
Doing good science
This chapter provides some general strategies that effective scientists
use.
Failing fast and often, but learn from your mistakes
Science involves a lot of trial and error. You will be asking questions
whose answers are often not yet known, using combinations of tech-
niques and analyses that do not yet exist or have not been applied
to your particular question. So you want to find ways to learn from
errors as quickly as possible, and ways to avoid making them in the
first place.
The general principle to do this is to find ways to get feedback of-
ten. For projects that are at the idea stage, talk with your peers about
it, and solicit feedback from more senior colleagues. Find out what
the likely pitfalls and challenging steps are, and who has already
tried or even solved them before.
For projects that are experimental, the same idea applies: find
a setup in which the feedback cycle is as short as it can be. For in-
stance, if your project requires recording from a brain area that the
lab has not recorded from before, chronically implanting an animal
would take much longer until you find out if you hit, compared to
doing an acute (non-recovery) surgery.
Experiments often consist of multiple steps or components that all
need to work – failure in any component will make all the other steps
in vain12. This implies that you should find ways to get feedback 12 A typical example is that if you do
not follow aseptic surgical procedures,
an infection may cause your implant to
detach. Having built perfect electrodes
is then moot.
about whether each component is working correctly. For instance,
inspect your tetrodes under a microscope before attaching them to
the interface board.
For analysis projects, a similar logic again applies: formulate ex-
pectations about what the output of a given analysis step should look
18 mvdmlab manual
like.
For all of these processes, apply a hacker/startup mindset that
takes the quickest path towards a minimal working example or proof
of principle. At that point, if the results look promising, you should
clean up, improve and document things.
Often, things will not work. This is not always a problem – some-
times you just want to try something and only follow up if it looks
like things are going to work. However, some failures actually block
your way forward, and you will need to resolve them. Notes, as de-
scribed next, will help you (and others) identify what the problem
might be. Making the same mistake twice is not a good use of time
and resources.
Documentation and taking good notes
One way to avoid making the same mistake twice or to avoid making
them in the first place is to have good documentation.
A related but different reason to care about documentation is that
a desirable property of a scientific study is that it should be possible
for others to replicate the result.
Lab notebook
Protocols
Protocols are step-by-step instructions of procedures used in the lab.
Anything you expect to do again in the future should be written up
as a protocol. If you only do a thing once, you should still take notes!
Protocols are hosted on GitHub. Why?
What makes a good protocol? Use standard structure (purpose,
ingredients, equipment). Step-by-step instructions: it should be a
recipe, and algorithm. The main procedures should be clean and
minimal so that it is easy to get an overview, but adding copious
notes and explanations as footnotes/sidenotes/appendices are great.
Steps should be as easy as possible to follow, with checks/tests that
can be used to tell if things are working as expected. Pictures can be
very helpful here.
Some example protocols are:
doing good science 19
If you use a protocol, update it regularly, even if it is to say when it
was last used successfully (and by whom).
Asking good questions
Questions about science.
Experiments and analyses are ways of asking nature a question.
Your goal should never be to obtain a certain answer. You should
formulate expectations about possible answers and consider if those
would in fact inform the question you are asking.
Working models are good sources of questions.
Questions about computer code.
When troubleshooting computer code, aim to create a Minimal,
Complete, and Verifiable example. In doing so, you make it easier
for others to replicate the problem you are experiencing, helping
them help you. Moreover, often you’ll find that in the process of
creating such an example, you discover the source of the problem.
See this page on Stack Overflow13 for more detailed instructions. 13 www.stackoverflow.com, an excellent
resource for crowdsourced answers to
programming issues.
Reading papers
Inspectional, analytical, syntopical
Bibliography management
Find the classic papers and intellectual roots of the question you
are working on. If this doesn’t motivate or energize you at least from
time to time, ask yourself how much you actually care about your
question.
Communication
Writing well
Clear thesis
Structure of argument
Vomit draft, results packet
Iterating often
20 mvdmlab manual
Citing: first, best, and most recent (this is a good rule of thumb to
guide your reading, too.)
Simpler is better
See SOFTWARE SECTION for more info on recommendations.
Importance of visuals
Simpler is better
See SOFTWARE SECTION.
Lab space
Our lab space consists of the following:
Include map.
General expectations
In the lab, we assemble precision devices that will be surgically im-
planted in live animals. We perform behavioral experiments that are
sensitive to changes in many different conditions. The amount of
time invested in these procedures, and the fact that we are doing this
in live animals, demand careful attention. In addition, lab space is
subject to general lab safety requirements (link) and IACUC inspec-
tions (link).
The principle that guides the use of shared lab space is that any
lab member needs to be able to come in, find the items they need,
and get things done to a high standard. In addition, the condition in
which te lab spaces are kept should reflect pride taken in doing good
work – it will be seen and interpreted by others viewing our space.
Depending on the specific rooms this can mean somewhat different
things, discussed in more detail below, but there are some common
principles:
•Clean up after yourself. Use common sense about when to do this.
If you’re in the middle of something and stepping out to get some
lunch, it makes no sense to clear your work area. However, if
you’ve finished what you were working on, definitely clean up.
Before going home for the day is usually a good time to at least
clean a bit. See the specific room schedules for more specific clean-
ing requirements.
•Organize where things are stored. Any item in the lab has a home.
Some individual items are sufficiently important that they have
a dedicated place, such as “final cut scissors” (PICTURE). Some
22 mvdmlab manual
items don’t have a dedicated place but are a member of a more
general category, such as “BNC interconnects”. If you find an item
that does not have an obvious home, check on Slack if anyone has
suggestions, and/or create one for it. Creating a home for an item
can be as simple as labeling a piece of a shelf somewhere. Make
sure you tell everyone about it on Slack, in accordance with the
Communication Principle!
•Track stock levels. If something is running low, don’t just ignore
it. Post about it on Slack. Store parts that go with a certain piece
equipment with that equipment (for instance, by taping a ziploc
baggie to it).
•Label things. Any liquid and food containers MUST be labeled with
the contents and expiry date, if applicable. When you open a new
package of something, write on it when you opened it. When a
new thing comes in, write on it when it was received.
•Treat tools with respect. What may look like a cheap pair of scissors
is likely to cost at least $300. Use tools for their intended purpose,
and use the correct tool for the job. For instance, don’t use fine
scissors to make rough cuts into hard materials. Don’t use fine
forceps to hold metal parts. 14 14 Quick guide to forceps: #55 are finest.
•Be safe. The general lab safety training covers the basics, but spe-
cific spaces have hazards described below. First Aid kits are avail-
able in the FAR and in B101.
Experiment rooms
AKA “running rooms”
These are the main spaces that our rats interact with. That has a
few consequences:
• If you have a rat out, have the doors closed. Put a sign on the door
saying, “Experiment in Progress”.
• Don’t make changes to the layout of the room while you are run-
ning an experiment. Changing the cues and landmarks available
can change what strategy animals use to solve a behavioral task.
• Work to keep light levels, sound levels, and odor cues consistent.
• Any surface rats interact with needs to be cleanable. So, no card-
board, unsealed wood, and so on.
• Our IACUC protocols require cleaning apparatus at least weekly.
lab space 23
Cleanliness and organization is important, but don’t let that hold
you back in shaping your workspace to enable you do perform your
experiments well and efficiently. Tape procedures you are using
to the walls, print out the relevant atlas sections, make temporary
storage spaces for the tools you use frequently.
Fine Assembly Room (FAR)
Surgery and anteroom
If there is any single room that is especially important to be clean and
organized, this is it!
Workshop
Beware the Dremel and the belt sander. Wear gloves and goggles.
Shared facilities
Histology.
Vivarium space
Our room is...
Offices
Computing
Computing plays an increasingly important role in (neuro)science
and is central to many aspects of the lab’s workflow. The safety of
your hard-won data, the integrity and reproducibility of your anal-
yses and results, the ongoing development and sharing of the best
protocols and procedures, and the speed with which you can ac-
complish your goals all depend critically on correct use of the lab’s
computing resources.
Because many of these are shared (experimental machines) and/or
inherently collaborative (lab database, codebase) it is especially im-
portant to be aware of the issues below. Experience with some of the
more advanced concepts and tools is a highly valued skill in many
labs and workplaces; mastery of these will set you apart from many
of your peers.
Lab computers
The lab’s computers are a mixture of custom builds (by MvdM),
designed for a particular role, and general-purpose workstations
purchased from Dartmouth Computing Services. The different roles
determine the best way to use each machine. The roles are:
a) single-user machines in offices, one for each grad student/tech/postdoc
b) shared machine in B28, for use with NanoZ and high-power scopr
c) shared machine in B101, for use with the 3D printer d) shared
experimental equipment machines: for recording in B18
Single user machines (grad students and postdocs)
One of these will be exclusively yours to use during your tenure in
the lab. You are free to install software and change settings to suit
28 mvdmlab manual
your needs as long as any changes (1) do not interfere with the ma-
chine’s ability to support research, and (2) fall within the Dartmouth
Computing guidelines (link).
Single user machines have a boot drive (C:) for the operating sys-
tem and frequently used software. This is a fast SSD drive with lim-
ited space, so be mindful of what you install here. Absolutely do not
use this drive for any documents or data.
Save your work and data files on the data drive (D:), because this
is automatically backed up using RAID (i.e. there are actually two
“mirrored” hard disks which behave as one virtual disk). Run RaidX-
pert’s check utility periodically to ensure the health of the RAID
array.
Shared machines
Unlike the single user machines, when using the shared machine
you will need to consider how your actions affect other users. A few
guidelines:
• If need to save your work, create a folder with your name on the
Data drive. Do not leave work or data files on the desktop. If you
find anything that’s not supposed to be there on the desktop or
anywhere else, put it in the lost-and-found folder on the desktop.
• Do not install any software, or change any software and system
settings, without discussing with MvdM first. Do not delete any
files, but place them in lost-and-found instead.
• If you log in to web services, make sure you log out when you are
done.
Experimental machines
The above considerations for shared machines apply. In addition:
• If you acquire data for your project, use correct renaming and
backup procedure when you finish your acquisition session. This
is absolutely essential: data is expensive, and you individually and
the lab as a whole cannot afford to lose any of it, EVER. Do not
leave data in the acquisition folder.
• If you encounter a potential data file or folder that is not yours
and seems lost, make an effort to find out whose data it might be.
computing 29
• NEVER delete anything that looks like it might be data, unless you
are absolutely certain that it has been backed up correctly.
2. All machines, even the single-user ones, use the mvdmlab ac-
count.
3. Machines are distributed across physically different locations.
To facilitate communication and data sharing, we maintain a list with
machine names, MAC and IP addresses, and the network port they
are plugged in to. Remember to update this if anything changes.
To move data between machines, we use freeSSHd, Dropbox, and
WinVNC.
4. Web services accounts
gmail: mvdmlab@gmail.com dropbox: mvdmlab@gmail.com
skype: vandermeerlab (for contacting suppliers and other lab-related
business)
PRINTING
Please be considerate when using the printer. Keeping an elec-
tronic library of PDFs using an iPad, Dropbox and iAnnotate is a
worthwhile investment that will make it much easier to find papers
and your comments in the future, and will save a LOT of paper!
The recording computers both have local printers attached for
obtaining protocols and weight sheets.
6. If you see Windows updates to install on any machine, go ahead
and install them. If a computer needs rebooting because of updates
and you can do so safely, go ahead.
7. Use of University computers (which includes all lab computers)
is governed by the Guidelines on Use of Waterloo Computing and
Network Resources. These are generally common sense, and not
overly long so have a look.
Skills
Growth mindset.
Basic skills: learning about your filesystem, what makes sense to
store where
Learn to use the command line.
30 mvdmlab manual
Learn to code. MATLAB and Python are the most important. In
our subfield of neuroscience, MATLAB is still the most used, but
Python is gaining ground.
Good coding practices.
Style guide.
Other important software: reference manager, LaTeX if you plan to
write a paper or a thesis. Adobe Creative Suite for making figures.
How to ask good questions. Stack Overflow on how to create a
minimal, complete, and verifiable example.
Software
In general, you should feel free use whatever software is the best tool
for the job15, and you should feel free to do so. However, there are 15 ...where “best” is defined as the
intersection between the capabilities of
the software itself, suitability for the
task at hand, and the user’s ability to
use it correctly.
startup costs associated with learning how to use new software, and
the lab has built up expertise and resources in a number of software
packages that work well. Learning to use these software packages
will help you be productive in the lab, and will be applicable in other
settings as well.
spikes
MUA
LFPs position
SWR
events
left SWR
sequences
right SWR
sequences
left tuning
curves
right tuning
curves
left full
posterior
right full
posterior
L/R count
ratio
sequence
detection
decode
full data
exclude L/R
overlaps
encoding
model
candidate
detection
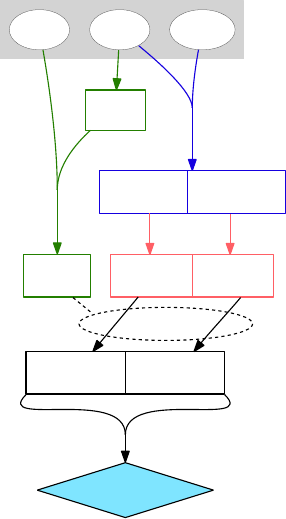
Figure 6: Example data analysis work-
flow, made with Graphviz
•Git is a tool for “collaborative version control”: a way to keep
track of changes made to files such as computer code, experi-
mental protocols or manuscrupt drafts, and to coordinate those
changes across multiple contributors.
•MATLAB is a scientific computing environment that includes a
programming language, libraries for performing specific tasks, a
development environment with a fully featured editor, debugger,
and interpreter, and many other features. It is widely used for data
analysis in neuroscience. Although it is not free, Dartmouth has a
site license that enables you to use it (download page). Python is
becoming increasingly popular, and is far more widely used than
MATLAB outside neuroscience. However, various toolboxes used
in the lab such as Chronux and FieldTrip are still MATLAB-only,
and MvdM is still a beginner in Python.
•Adobe Creative Suite is expensive, but produces superior docu-
ments. We use Illustrator to touch up and group together figure
panels for manuscripts based on raw output from MATLAB or
Graphviz, and to make posters. Photoshop is sometimes used to
process image files such as those from histology. Free alternatives
computing 31
such as Inkscape and GIMP exist, can do the job for simple docu-
ments, but I don’t (yet) recommend them for publication-quality
output.
•Graphviz produces schematics such as the one in Figure 6. Unlike
‘what-you-see-is-what-you-get” design software such as Photoshop
and Illustrator, the Graphviz tools create graphics from a plain text
source file. The source file contains a recipe for what the graphic
output should look like.
• L
A
T
EX. Unlike “what-you-see-is-what-you-get” content creation
programs such as Word, LaTeX creates documents from a plain
text source file. The source file contains instructions for what the
document should look like. Compared to Word, LaTeX does much
better with equations, produces more professional-looking docu-
ments16, and makes it much easier to maintain large documents. 16 Especially where typesetting features
such as spacing, kerning, and ligatures
are concerned.
Note that several of these require you to use the Command Line
(Terminal).
Operating Systems
Lab computers use Windows and/or Ubuntu Linux. There are a
few reasons for this: the data acquisition machines require Windows
(Neuralynx) or Windows/Linux (Open Ephys), and custom machine
builds are generally cheaper for PCs than for Apple Macs. Neverthe-
less, many lab members have Mac personal laptops that run OS X,
and most of the above software works fine on them. However, some
Neuralynx loaders only work on Windows, some codebase functions
have only been compiled to work on some OSes, and the lab only has
licenses for some software (Adobe) to work on Windows.
Data management
Data is valuable. After taking into account the number of hours
that went into training an animal, building a hyperdrive, preparing
for and performing surgery, painstakingly turning electrodes; animal
housing costs, costs of parts and consumables, and so on, the cost
of a single subject worth of data can run into tens of thousands of
dollars – and this is without the moral calculation of considering an
animal’s life. Data therefore needs to be treated with the greatest re-
spect: specifically, it needs to be managed so that it can be maximally
useful. The first requirement of data management is that data needs
to be available – i.e. backed up so that it can never be lost, and acces-
sible by those who need it. Our lab is committed to Open Science,
of which public data sharing is one component. Increasingly, jour-
nals and funders demand that data is made publicly available. The
second requirement of data management is that the data needs to be
organized such that it can be easily understood and used.
Before discussing how these two requirements – data storage
and data organization – are implemented in the lab, it is useful to
distinguish between different categories of data.
•Raw data is what gets saved on a data acquisition machine, such
as a running room computer connected to a Neuralynx record-
ing system, or a machine connected to a microscope. Depending
on the data you collect, a single session may yield many differ-
ent files, such as behavioral tracking data and neural recording
data, or a single file (an image). Raw data is only rarely suitable
for analysis beyond a few quick checks. At a minimum, freshly ac-
quired data sets typically must be annotated, and/or the files sys-
tematically renamed – for instance, with the ID of the experimental
subject and some information about recording locations – so that
the analyst can select which files to analyze, and combine results
across sessions and subjects. More complex pre-processing steps
include spike sorting (the process of assigning spike waveforms
34 mvdmlab manual
to putative single neurons to obtain their spike times), artefact
removal, and many others.
•Promoted data is data that is ready for analysis. Data files need
to be organized in a specific folder structure, named according
to the naming scheme, and supplied with annotations describing
the data. If applicable, various preprocessing steps may have been
carried out, for instance spike-sorting of raw voltage traces into
spike trains of putative single neurons, filtering of position data
to remove artifacts, and definition of trials on a behavioral tasks.
PDFs of handwritten notes and procedure logs, and images of his-
tology may also be included. A promoted data set typically does
not include the raw data. Promoted data should be sufficiently
well described and organized such that a competent reviewer or
collaborator can use it.
•InProcess data is data that is being worked on to move it from raw
to Promoted.
Data storage
Immediately after data is first acquired, do the following:
• Create and/or rename the new data folder according to the Lab
Data Naming Scheme.
• If applicable, compress any files that are large and compressible:
typical examples include Neuralynx .nvt (video tracking) files17.17 See section XXX for a quick primer on
data compression.
• Upload the data to the Incoming folder on the lab server. See sec-
tion XXX for instructions.
• Move the data out of the location where it is saved, into a folder
specific to your subject or experiment.
• Following completion of data collection from that subject or ex-
periment, verify that indeed all data is correct and present on the
server, and then delete it from the data acquisition machine.18 18 This is an important step, because
if there is no space available on a data
acquisition machine, we cannot acquire
more data!
The lab server uses a redundant data storage system that can
tolerate failure of a single hard drive without causing loss of data.
The contents of the datavault folder are periodically backed up to
“cold” offsite storage. However, it is good practive to also make your
own personal backup of your raw and promoted data. One good way
of doing this is to store that data on your desktop workstation, or
move it to an external HDD, just in case.
data management 35
Data promotion
What exactly is included in a promoted data set depends on the
specific experiment, but typical features include the following:
• Data should be organized and (re)named according to the lab Data
File Naming Scheme (link).
• Make sure raw and intermediate data files that are no longer
needed are removed.
•ExpKeys file.
• If applicable, task-specific metadata file(s).
• A description of the task/experiment.
• PDFs of relevant task notes and histology.
As you collect data, you should start drafting an example ExpKeys
file. Prior to starting data collection, you should have created a
protocol that describes the procedures used. You should use this
protocol as the basis for a description of the task/experiment.
Examples of nice promoted data with description include Alyssa’s
MotivationalT data (link) and Jimmie’s CueCoding data19.19 https://github.com/jgmaz/
vStrCueCoding
Data use cases
Some examples (“use cases”) that motivate careful data management
include:
• When analyzing your own data, you want to be able to easily
specify which subject(s) and session(s) you are working on.
• If you have a question that can be answered with someone else’s
data, you want to be able to “plug in” that data into your existing
workflow. When I (MvdM) joined the Redish lab, I ran a compar-
ison of dorsolateral striatum, ventral striatum, and hippocampus
data20. This was made possible through the use of a consistent 20 van der Meer et al. Neuron 2010
data formatting and annotation scheme.
• When publishing your work, it is the policy of the lab, and an
increasing number of journals and funders, that your data is made
publicly available. You want it to be easy to use and understand,
so that your colleagues are not annoyed with you and you don’t
have to answer their emails telling you things aren’t working.
36 mvdmlab manual
• Before you get to publishing, you will need to convince MvdM
of your results and conclusions. This likely invloves him running
your code on your data. That will only work if you’ve organized
your data correctly.
• MvdM is always working on applications to obtain research fund-
ing. Some of these applications, and the pilot data used to sup-
port them, are planned in advance; others are more spur-of-the-
moment, or even if planned, new insights can happen. Thus, there
is often a need to produce an analysis or figure in short order. This
is only possible, or at least made a lot easier, if the data is ready
for analysis..

Animal care and recordkeeping
Keeping detailed and accurate records on each of our animals is
an IACUC requirement (and indirectly a federal issue), as well as
scientifically important.
The main principles are that for any animal that is currently alive,
there must a binder (or a section in a binder) with that animal that
contains an up to date record of all procedures21 performed on that 21 What counts as a procedure: anything
beyond what’s required to change a
cage or weigh an animal. If in doubt,
log it anyway.
animal, as well as records of their weight.
Failure to log a procedure is a serious oversight that, if discovered, could have
real consequences, such as creating more work for everyone, or making our
IACUC renewals more painful. Don’t let it happen to you.
When performing invasive procedures (surgery) and when ad-
ministering drugs, CCMR requests that these are reported using a
special cage card22. Writing procedures on these cards is not suffi- 22 Picture of procedure cage card.
cient logging: you also need to log the procedure(s) in the animal’s
binder.
Once you euthanize and animal (or request it to be euthanized),
collect all weight and procedure logs, along with any additional
information (notes from behavior, for instance), scan them into a
single PDF23 and upload to the data vault. 23 The departmental copier/scanner is
great for this, you can feed it a pile of
documents and it can email you a PDF.
If you are working with animals, read the Animal Recordkeeping
Protocol (link), which provides detailed procedures for the above.
Animal ownership and responsibility
By default, CCMR staff will feed, water, and change cages for all our
animals. We pay them a per diem fee to do this.
By default, all animals receive ad lib24 food and water. 24 Ad libitem is the Latin phrase for,
literally, “to your libido”, i.e. as much as
desired. Incidentally, i.e. is Latin for id
est, “it is”.
38 mvdmlab manual
We can also request CCMR to feed rats 18g/day. This is a useful
amount that prevents rats from getting obese. To request this, use the
relevant cage card25.25 This one.
For any other food/water regimens, you’ll need to use the “Ex-
perimenter will feed/water” cage card. Doing so implies that the
experimenter listed takes ownership26 of that animal. 26 As the owner of an animal, you are
responsible for logging its weight
and all procedures. If the animal is
food and/or water-restricted, you are
responsible for supplying those things,
and logging that you have done so.
When animals first arrive, they are typically group-housed (multi-
ple animals per cage). They are named27 and start out with communal
27 Rats are named Rxxx, where xxx is
a3-digit counter that it incremented
with each new animal. Mice are named
Mxxx.
status. Communal animals do not have an owner yet. They are
weighed weekly by the Communal Animal Caretaker28. When com-
28 Note that to track weights of group-
housed animals, some kind of mark
needs to be used. The lab convention is
to write numbers 1,2,3etc. on the base
of the tail with a Sharpie. 1indicates
the animal with the lowest number.
Mouse tails are too small to write
numbers; stripes or rings can be used
instead.
munal animals are below 400g in weight, the Caretaker also handles
them weekly (about 5minutes per animal appears to be the sweet
spot, although you may want to take ownership of an animal that
will be implanted with a hyperdrive early on so that it can be han-
dled more). When communal animals reach 400g, the Caretaker
separates them into individual cages so that CCMR can feed them
18g/day.
These procedures are described in more detail in the Animal
Recordkeeping Protocol.
PBS and Dartmouth
Training required by Dartmouth
General lab safety training
Important people and contact info
PBS staff
CCMR
Dartmouth resources
Dick’s House
Graduate School
Postdoc Association
Workshops
Dartmouth design language and files
Beyond the lab
Resources on the internet
Industry vs. academia
Upper Valley
Appendix: Subject email format template
Subject line should read:
Appendix: Yearly evaluation forms
Graduate students
Staff
