Octopus User Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 42
- Overview
- Introduction
- What’s in this manual?
- Availability
- License and copyright
- Further assistance
- Introduction
- Variant calling
- Hybrid mapping based variant calling
- Haplotype based variant calling
- Local variant phasing
- Installation
- System Requirements
- Hardware
- Required Software
- Optional software
- Downloading
- Building
- Easy install with Python
- Building with CMake
- Debug builds
- Running tests
- Getting started
- Basic usage
- Required arguments
- Optional arguments
- Reporting bugs
- Requesting new features
- Calling models
- Individual
- Population
- Trio
- Cancer
- Polyclone
- Examples
- Calling germline variants in a single sample
- Calling variants in a targeted exome panel
- Ignoring decoy contigs from a whole genome run
- Calling germline variants in a population
- Calling de novo mutations in a trio
- Calling somatic mutations in a tumour-normal pair
- HLA genotyping
- Calling variants in haploid organism
- Running in multithread mode
- Using a configuration file
- Random forest filtering
- Best practices
- Reference selection
- Read mapping
- Read preprocessing
- Variant calling
- Variant call filtering
- Command line reference
- General
- Read pre-processing
- Variant generation
- Haplotype generation
- Calling
- Trio
- Cancer
- POLYCLONE
- Phasing
- Call filtering
- VARIANT FILTERING
- Measure reference
- Threshold filtering
- Random forest filtering
- Training random forests
- Output format
- Performance optimisation
- Execution time
- Memory consumption
- Multithreading
- Variant generation
- Haplotype generation and phasing
- Calling model selection and parametrisation
- Troubleshooting
- Building
- Why are the requirements so strict?
- CMake chooses a bad compiler
- Compilation fails
- Linking fails
- Boost libraries fail to link
- Compilation has lots of #pragma warnings
- Runtime
- Segmentation fault
- Execution is slow
- Execution delays after initialising calling components in threaded mode
- Run hangs in decoy contigs
- Behaviour
- No calls are reported
- Regions are skipped because of too many haplotypes
- A call changes when a different input region is given
- Why doesn’t octopus report genotype likelihoods?
- Why do octopus VCF files contain * and .?
- SNP accuracy improves in fast mode
- Calling performance is worse with assembler
- Contact
- Appendix
- Installing requirements
- OS X
- Ubuntu
- Variant generation
- Haplotype generation
- Phasing
- Glossary
Octopus User Manual
Version 0.5.3 beta
1 January 2019
University of Oxford!
! of !1 42

OVERVIEW!6
Introduction!6 ........................................................................................................................
What’s in this manual?!6 ........................................................................................................
Availability!6 ..........................................................................................................................
License and copyright!6 .........................................................................................................
Further assistance!6 ..............................................................................................................
INTRODUCTION!7
Variant calling!7 .....................................................................................................................
Hybrid mapping based variant calling!7 ..................................................................................
Haplotype based variant calling!7 ...........................................................................................
Local variant phasing!7 ..........................................................................................................
INSTALLATION!8
System Requirements!8 ................................................................................................
Hardware!8 ...........................................................................................................................
Required Software!8 ..............................................................................................................
Optional software!9 ...............................................................................................................
Downloading!9 ..............................................................................................................
Building!9 .....................................................................................................................
Easy install with Python!9 ......................................................................................................
Building with CMake!10 .........................................................................................................
Debug builds!10 ....................................................................................................................
RUNNING TESTS!10
GETTING STARTED!11
Basic usage!11 ......................................................................................................................
Required arguments!11 .........................................................................................................
Optional arguments!11 ..........................................................................................................
Reporting bugs!11 .................................................................................................................
Requesting new features!12 ...................................................................................................
CALLING MODELS!13
Individual!13 ..........................................................................................................................
Population!13 ........................................................................................................................
Trio!13 ...................................................................................................................................
Cancer!13 .............................................................................................................................
! of !2 42

Polyclone!13 .........................................................................................................................
EXAMPLES!14
Calling germline variants in a single sample!14 .......................................................................
Calling variants in a targeted exome panel!14 .........................................................................
Ignoring decoy contigs from a whole genome run!14 ..............................................................
Calling germline variants in a population!15 ............................................................................
Calling de novo mutations in a trio!15 .....................................................................................
Calling somatic mutations in a tumour-normal pair!15 .............................................................
HLA genotyping!15 ................................................................................................................
Calling variants in haploid organism!15 ...................................................................................
Running in multithread mode!16 .............................................................................................
Using a configuration file!16 ...................................................................................................
Random forest filtering!16 ......................................................................................................
BEST PRACTICES!17
Reference selection!17 ..........................................................................................................
Read mapping!17 ..................................................................................................................
Read preprocessing!17 ..........................................................................................................
Variant calling!17 ...................................................................................................................
Variant call filtering!17 ............................................................................................................
COMMAND LINE REFERENCE!18
General!18 ....................................................................................................................
Read Pre-Processing!20 ................................................................................................
Variant Generation!22 ....................................................................................................
Haplotype Generation!23 ...............................................................................................
Calling!24 .....................................................................................................................
Trio!25 ..........................................................................................................................
Cancer!25 .....................................................................................................................
Polyclone!26 .................................................................................................................
Phasing!26 ...................................................................................................................
Call Filtering!26 .............................................................................................................
VARIANT FILTERING!28
Measure reference!28 ............................................................................................................
Threshold filtering!30 .............................................................................................................
! of !3 42

Random forest filtering!30 ......................................................................................................
Training random forests!31 .....................................................................................................
OUTPUT FORMAT!32
PERFORMANCE OPTIMISATION!33
Execution time!33 ..................................................................................................................
Memory consumption!33 .......................................................................................................
Multithreading!34 ...................................................................................................................
Variant generation!34 .............................................................................................................
Haplotype generation and phasing!35 ....................................................................................
Calling model selection and parametrisation!35 ......................................................................
TROUBLESHOOTING!36
Building!36 ...................................................................................................................
Why are the requirements so strict?!36 ..................................................................................
CMake chooses a bad compiler!36 ........................................................................................
Compilation fails!36 ...............................................................................................................
Linking fails!36 ......................................................................................................................
Boost libraries fail to link!37 ...................................................................................................
Compilation has lots of #pragma warnings!37 ........................................................................
Runtime!37 ...................................................................................................................
Segmentation fault!37 ............................................................................................................
Execution is slow!37 ..............................................................................................................
Execution delays after initialising calling components in threaded mode!37 ..............................
Run hangs in decoy contigs!37 ..............................................................................................
Behaviour!38 ................................................................................................................
No calls are reported!38 ........................................................................................................
Regions are skipped because of too many haplotypes!38 .......................................................
A call changes when a different input region is given!38 ..........................................................
Why doesn’t octopus report genotype likelihoods?!38 ............................................................
Why do octopus VCF files contain * and .?!38 ........................................................................
SNP accuracy improves in fast mode!39 ................................................................................
Calling performance is worse with assembler!39 .....................................................................
CONTACT!40
APPENDIX!40
Installing Requirements!40 .............................................................................................
! of !4 42

OS X!40 ................................................................................................................................
Ubuntu!40 .............................................................................................................................
Variant Generation!41 ....................................................................................................
Haplotype Generation!42 ...............................................................................................
Phasing!42 ...................................................................................................................
GLOSSARY!42
! of !5 42
OCTOPUS USER MANUAL
OVERVIEW
Introduction
Octopus is a command line tool that detects genetic variation from high-throughput sequencing data
(reads) relative to a reference sequence. The tool must be provided with an indexed FASTA reference file and
one or more SAM format mapped and aligned read files, and will produce a set of phased variants in the
VCF format. Octopus is able to call single nucleotide variants (SNVs) and small indels (< 2000bp), and can be
used detect and classify germline, somatic, or de novo mutations across multiple samples.
What’s in this manual?
This is a user manual intended for novice to advanced users to get octopus running optimally. It is not
intended to give a detailed description of the algorithms implemented in octopus (although some pertinent
details are given in the appendix to help understand some parameters), nor is it a technical manual for
software developers. Please refer to the octopus paper and developer manual for detailed descriptions of
these topics.
Availability
Octopus is hosted on Github.
License and copyright
Octopus is distributed under the MIT license. The copyright holder is the Daniel Cooke
(daniel.cooke@well.ox.ac.uk) who reserves the right to change the license terms.
Further assistance
•There is additional documentation on the Github wiki.
•For general discussion, please use the octopus Gitter chat.
•For bugs and feature requests, please use the octopus issue tracker.
•Other questions can be directed to Daniel Cooke: daniel.cooke@well.ox.ac.uk.!
! of !6 42
OCTOPUS USER MANUAL

INTRODUCTION
Variant calling
Variant calling is an inference task; the aim is to report the underlying genome of the sample under
consideration, given a set of indirect observations of the samples genome (reads). This is statistical problem as
the underlying read data is noisy due to the sequencing process (errors can be introduced in the library
preparation and sequencing itself). In practise, not all inferred genetic information is reported as the vast
majority of information is conserved amongst populations. Instead only differences compared to a reference
sequence for the population are reported, these are called variants.
Hybrid mapping based variant calling
Mapping based variant callers require preprocessed input from a read mapper. A read mapper takes raw
sequencing reads and attempts to determine the origin of each read independently relative to a reference
genome. Most mappers will subsequently align the read around the mapped location.
The variant calling task is much simplified when read mapping information is available as the domain of
possible variants is significantly reduced. However, with mapping based approaches, the overall accuracy of
the caller may be bounded by the accuracy of the mapper and alignment algorithm; the mapping stage itself
can be viewed as a variant calling process as the mapper must also account for deviations from the reference
sequence due to real variation and sequencing errors.
The other method is to avoid using a mapper entirely; just take the raw reads and assemble them into full
contigs. Such approaches do exist, and usually empty De Bruin graphs and similar algorithms, but are usually
underpowered compared to mapping based approaches. There is also a significant computational overhead
attached to assembly based approaches.
Experience is showing that the best overall solution is a hybrid approach where read mapping information is
used, but only partially. Reads mapping within a certain genomic interval are locally reassembled and then
aligned to the assembled contig. The idea being that the read mapper may be wrong, but it is unlikely to be
very wrong; the true read origin is unlikely to be far way from the mapped location.
Haplotype based variant calling
Haplotype based variant callers attempt to jointly genotype more than one genomic position simultaneously.
This is in contrast to traditional positional based variant calling that only genotype a single location at once.
The advantage of haplotype based is that the space of possible errors increases exponentially with haplotype
length, while the space of true haplotypes remains constant in the number of samples and organism ploidy. It
is therefore much easier to classify true variation and sequencing errors.
Local variant phasing
Phasing refers to assigning called alleles to a particular haplotype; calls are phased if information indicating
which called alleles occur on the same haplotype is provided. It is only possible to fully reconstruct a samples
genome if phased calls are generated. Octopus is able to generate phased calls - the phase information is
provided in the final call set.
! of !7 42
OCTOPUS USER MANUAL
INSTALLATION
This section gives detailed instructions on how to obtain, build, and install octopus. Please refer to the
troubleshooting section for common installation problems not addressed here.
SYSTEM REQUIREMENTS
Octopus is mostly written in C++, and therefore requires the source code to be compiled for the target
machine architecture with a C++ compiler. You will need to consult your operating systems technical
documentation to determine a suitable C++ compiler.
Hardware
In principle octopus can run on any machine capable of compiling a C++14 program. However, given the
complex numerical algorithms involved in running octopus the following guidelines are offered :
1
Most modern desktop and laptop computers should satisfy these requirements. The user should understand
that hardware requirements will vary greatly depending on the use-case and workload. For example, calling
many high coverage samples will require far greater memory than a single low coverage sample. Octopus is
fully multithreaded, and to achieve reasonable runtime performance on large tasks it is recommended to make
multiple processor cores available.
Octopus requires SSE2 hardware support.
Required Software
•A C++14 compliant compiler with SSE2 support
2
•An implementation of the C++14 standard library
3
•Boost 1.65 or greater
•htslib 1.4 or greater
•CMake 3.9 or greater
Technology
Minimum
Recommended
Processor
Intel Core i5
32 x Intel Core i5
Memory
8GB
16GB
Disk
500GB
1TB
These guidelines are based on running octopus on a single high coverage (~50x) human sample.
1
GCC 6.2.1 and below have bugs which affect octopus; only use GCC 6.3 and above. LLVM Clang 3.8 has been tested and compiles.
2
Visual Studios and Intel C++ compilers have not been tested.
It is highly recommended to use the compilers native C++ standard library implementation: libstdc++ for GCC and libc++ for Clang.
3
! of !8 42
OCTOPUS USER MANUAL

Optional software
•Git 2.5 or greater
•Python3
Instructions on obtaining the requirements on OS X and Ubuntu are given in the Appendix.
DOWNLOADING
Octopus is distributed via the project hosting website Github. There are two ways to obtain a copy of the
source code from Github:
1. Visit the octopus Github webpage and click the Clone or download box. This will download a zip file
named octopus-master.zip containing the octopus source code. Move the zip file to a suitable location,
unzip it, and rename the folder to octopus.
2. Open a command line terminal and move to a directory where you would like octopus to be downloaded,
then execute the git command:"
"
$ git clone https://github.com/luntergroup/octopus.git!
!
The octopus source code will be downloaded into a folder named octopus.
BUILDING
Once the source code is obtained there are two methods to create an executable for your target machine,
both require CMake to generate a native makefile which is used by a native build-tool to build the final
executable. It is highly recommended to do an out of source build.
Easy install with Python
In the scripts directory there is a Python3 script install.py which will execute all the necessary build
steps:
$ ./scripts/install.py
Which will install into the octopus bin directory. To install into a different location (e.g. /usr/local/bin) use:
$ ./scripts/install.py --prefix /usr/local/bin
If CMake is not able to find a suitable C++ compiler, it may be necessary to explicitly specify where such a
compiler exists:
$ ./scripts/install.py --cxx_compiler /path/to/compiler/cpp
On some systems, you may also need to specify a C compiler which is the same version as your C++
compiler, this can be done with the c_compiler option, e.g.:
$ ./scripts/install.py --cxx_compiler g++-7 --c_compiler gcc-7
! of !9 42
OCTOPUS USER MANUAL

The installation script can also be used to install all dependencies, including a suitable compiler:
$ ./scripts/install.py --install-dependencies
Building with CMake
It is also possible to build the source directly with CMake:
$ cd build
$ cmake .. && make install
$ cmake -DCMAKE_INSTALL_PREFIX=/usr/local/bin ..
$ cmake -DCMAKE_CXX_COMPILER=g++-4.2 ..
Using the python script is recommended however as it ensures an out of source build.
Debug builds
It is possible to build octopus with debug information. This is only recommended for debugging and will
hopefully not be needed for users. To do so, add the command --sanitize to the Python install script.
RUNNING TESTS
If you downloaded a developmental version of octopus, it is good practise to run all the packaged tests before
using any of the tools for production work. The release versions are guaranteed to have passed all tests. To
install octopus for testing and run the tests use:
$ test/install.py
Like the other install script this command can also be supplied with a compiler.
! of !10 42
OCTOPUS USER MANUAL

GETTING STARTED
Once successfully installed octopus is ready to run. This section is for novice users who want a gentle
introduction to variant calling. Advanced users should consult the command line reference section for detailed
descriptions of specific features.
Basic usage
Octopus is a command line tool and must be executed from a command terminal. The simplest octopus run is
without any arguments:
$ octopus
Which will report a user error informing there are missing required arguments! You can request a reminder of all
required and optional parameters with the --help command:
$ octopus --help
This will display a similar table to the command reference below.
Required arguments
Only two command line argument are required. First, the reference genome to use for analysis specified with
--reference; -R, which must be given a path to a FASTA file containing the reference genome. A FASTA
index file with the same name, but extension .fai is also required to exist in the same directory as the given
reference.
Second, a list of read file (BAM or CRAM format) paths must be supplied. These can either be supplied directly
with the --reads; -I option, or with the --reads-file; -i option, which must be given a path which
itself contains a list of paths to read files. These two options can also be used conjunctively, any duplicate files
will be ignored. Each read file must have an associated index file that exists in the same directory as the read
file (.bai for BAM and .crai for CRAM).
Optional arguments
Octopus has many optional arguments that affect accuracy and runtime performance. The default parameters
have been chosen with human germline sequence data mapped with BWA-MEM in mind; many users will find
the default arguments offer adequate performance on human samples. For non human data samples, the
default parameters may not offer good performance, especially for non-diploid organisms, and users are
advised to carefully read the available options. Even for users only interested in human samples, it is
recommended they briefly acquaint themselves with the available options. A detailed description of all
command line options can be found later in this manual.
Reporting bugs
Octopus is currently in pre-release, so it is likely that some bugs will be present. If you encounter a bug, please
first check the octopus issue tracker to make sure it is not already reported. Also, if you're using a tagged
! of !11 42
OCTOPUS USER MANUAL
release build, please check closed issues and newer releases before reporting the bug as it may already have
been fixed!
Once a bug has been verified, try if possible to find a minimal verifiable example (MVE); that is, the least
amount of data that triggers the bug. The first step to finding an MVE is usually to locate the approximate
genomic region where the bug is triggered, and then calling with smaller targeted regions to try to pinpoint the
problem. Usually this task is easier when running in a single thread, however, this can be time consuming if
calling over large amount of data, in which case you could try running with multiple threads and with the --
debug command which should help indicate where the issue occurred.
Once an MVE is found, recompile octopus in sanitize mode which adds significant debugging information
to the executable. Any errors will be reported to stderr should be recorded, and sent along with octopus's
own debug log to the octopus issue tracker.
Requesting new features
Feedback is very welcome! Please start by suggesting a new feature on the octopus forum and then if well
received make an official feature requests to the octopus issue tracker.
! of !12 42
OCTOPUS USER MANUAL

CALLING MODELS
Octopus provides a framework for genotyping samples given different states of knowledge about those
samples, such as different sample biology or experimental method, expressed via a calling model. A calling
model serves two purposes: firstly, it defines the type of calls and inferences that should be made (e.g.
somatic or de novo classification), and secondly, it defines a probability model to calculate posterior
probabilities for the given call types. Octopus currently has five calling models, which are briefly discussed
below.
Individual
The individual calling model is the simplest, it is intended to model a single healthy individual with known
chromosome copy number (ploidy) for all chromosomes (or contigs). The advantage of having a bespoke
model for an individual is that the genotype posterior distribution can be calculated exactly.
Population
The population calling model is intended for genotyping multiple unrelated samples from a population. Like the
individual model, it is assumed each sample is healthy with a known chromosome copy number. The first
advantage of calling samples jointly, as apposed to calling each sample individually and then merging the
results, is that power is increased to call common variation. This is particularly true for low coverage data. The
second advantage is that genotyping samples jointly allows a consistent call set to be produced; merging
independent call-sets can be very challenging.
Trio
The trio calling model is used to genotype a family consisting of a mother, father, and offspring. All members of
the trio are assumed to be healthy with known chromosome copy numbers. However, unlike the population
model, the trio model explicitly models the relationship between the samples, and is therefore able to classify
de novo mutations in the child.
Cancer
The cancer model is used to genotype tumours from a single individual. All tumours are assumed to be
metastasis from the same primary tumour (or the primary itself). The model can be used to classify somatic
mutations, and infer local copy number changes around called mutations. Unlike the other calling models, the
chromosome copy number of each tumour is not assumed to be known, however, if a normal sample with
known chromosome copy number is also present the classification power of the model is increased.
Polyclone
The polyclone calling model is designed for calling variants in a mixed haploid sample where the number and
mixture frequency of clones is unknown. An application is calling variants in bacterium samples which could
contain more than one isolate due to contamination, mixed infection, or in-host evolution. The number of
clones is automatically inferred from the data.!
! of !13 42
OCTOPUS USER MANUAL

EXAMPLES
This section contains some common use-case examples to get started. Please refer to the following sections
for more information regarding calling models and parameters. Note all of the examples in this section use the
default output mode (standard output) for brevity, to write to a file just add the --output; -o command.
Calling germline variants in a single sample
As previously described, octopus has two distinct models for germline variant calling - one for a single
individual and another for populations. Fortunately there is no concern for the user as the appropriate model is
selected automatically:
$ octopus -R human.fa -I NA12878.bam
Assuming the file NA12878.bam contains a single sample, this will use the individual calling model. Octopus
does not care how many samples are actually in a read file, so if the input read file contains multiple samples
but only a single sample is required for analysis, the name of the sample is required as input:
$ octopus -R human.fa -I multi_sample.bam --samples NA12878
Calling variants in a targeted exome panel
All octopus calling models can be supplied with a list of target intervals to analyse, for a small number of
regions the option --regions; -T can be used:
$ octopus -R human.fa -I NA12878.bam -T 22:35,799,116-35,799,685
This option can be used multiple times, or can be supplied with a space separated list of arguments. However,
for longer target interval lists it may be easier to create a file which lists the regions (one per line) and pass this
to octopus using the --regions-file; -t option:
$ octopus -R human.fa -I NA12878.bam -t exome-panel.txt
Note these options can be used in conjunction, and there is no need to worry about duplicates or overlaps -
octopus will resolve this internally.
Ignoring decoy contigs from a whole genome run
There is a useful option, --skip-regions; -K, that serves as the converse of the --regions command;
it informs octopus not to analyse the given options. The main utility of this is for ignoring decoy contigs or
centromeres in whole genome runs. There is a homologous command, --skip-regions-file; -k,
which takes a path to a file containing regions to ignore:
$ octopus -R human.fa -I NA12878.bam -k human-decoy.txt
It is possible to use all the region specific commands in conjunction to get fine grain control over which regions
to call.
! of !14 42
OCTOPUS USER MANUAL

Calling germline variants in a population
The population model is the default for more than a single sample, so just supply a list of samples:
$ octopus -R human.fa -I NA12878.bam NA12891.bam
For larger sample sets, it is usually better to have the read paths in a file:
$ octopus -R human.fa -i reads.txt
Calling de novo mutations in a trio
To call germane and de novo mutation in a trio, just specify --maternal-sample; -M and --paternal-
sample; -F:
$ octopus -R human.fa -i ceu_trio.txt -M NA12892 -F NA12892
The child is automatically deduced. The trio can also be specified with a PED file:
!$ octopus -R human.fa -i ceu_trio.txt --pedigree ceu_trio.ped
Calling somatic mutations in a tumour-normal pair
To call germline and somatic variants in tumour samples, supply either a normal sample:
$ octopus -R human.fa -I normal.bam tumour.bam -N normal
If a normal sample is unavailable, tumour only calling can be invoked by explicitly selecting the cancer calling
model:
$ octopus -R human.fa -I tumour1.bam tumour2.bam -C cancer
HLA genotyping
Octopus is able to call very long haplotypes, especially in variant dense regions, which makes it an ideal tool
for calling HLA haplotypes. By default octopus will not make maximally long haplotypes - and therefore phase
regions - due to the computational complexity involved in such optimisation, and the diminishing return of very
long haplotypes. But in the HLA, longer haplotypes are desired, which can be achieved using the --
phasing-level; -l command:
$ octopus -R human.fa -I NA12878.bam -t hla-regions.txt -l aggressive
It may also be beneficial to increase the default value of --max-haplotypes to 256 or 512.
Calling variants in haploid organism
The default parameters are set with human sequence data in mind, for non-human samples it is
recommended to adjust the options for the organism being analysed. For haploid organisms such as bacteria
and viruses, the most important parameter to change is --organism-ploidy; -P which sets the default
ploidy to use. Depending on the organism, it may also be important to adjust the variant priors: --snp-
heterozygosity and --indel-heterozygosity:
! of !15 42
OCTOPUS USER MANUAL

$ octopus -R ecoli.fa -I ecoli.bam -P 1 --snp_heterozygosity 0.01
To call variants in a haploid sample which potentially contains an unknown mix of multiple clones (e.g. bacteria
or viral samples), specify the polyclone calling model.
$ octopus -R H37Rv.fa -I mycobacterium_tuberculosis.bam -C polyclone
Running in multithread mode
By default all octopus runs execute using a single thread, but it is trivial to use multiple threads using the --
threads command:
$ octopus -R human.fa -I NA12878.bam --threads
This is the recommended approach to multithreading with octopus, but the command also takes an optional
number of threads to use, which must be specified immediately after the command:
$ octopus -R human.fa -I NA12878.bam --threads=4
The former form is recommended because it allows octopus to optimise thread usage, and also enables the
use of specific multithreaded algorithms.
Using a configuration file
Octopus allows all command line options to be specified using a configuration file, which some users may
prefer is the same configuration us used often. The configuration file is just a text file with each line containing
a option=value pair:
$ octopus --config my_octopus_config.txt
Random forest filtering
To use random forest filtering just specify the --forest-file option for germline calls and the --
somatic-forest-file option for somatic calls:
$ octopus -R human.fa -I NA12878.bam --forest-file germline.forest
! of !16 42
OCTOPUS USER MANUAL

BEST PRACTICES
This section gives some brief advise on best practise workflow from FASTQ to VCF.
Reference selection
Use the latest possible reference genome for your sample. Reference assemblies are often updated to reflect
resolutions of complex loci, or to add decoy sequence which reduces mapping issues and improve calling
quality.
Read mapping
Octopus requires mapped and aligned reads in the SAM format.The quality of the mapping software is
therefore an essential part of the variant calling process. While the performance of mappers can vary
considerably depending on the type of sequencing data used, BWA-MEM (default settings) is recommended
as it is widely used, well tested, and has been shown to perform well on a wide range of data - in particular
human genetic data.
Read preprocessing
Octopus does not require any read preprocessing after mapping, such as duplicate marking, indel realignment,
or base quality recalibration. Unlikely other variant callers, octopus is unlikely to benefit from such techniques
as reads are preprocessed internally, indels are essentially realigned during calling, and base quality scores are
also internally manipulated depending on sequence context. However, if you're data is already preprocessed,
octopus should perform equally well with this data.
Variant calling
Ensure the correct calling model is selected for the type of data to be analysed. Look at the private parameters
for the chosen calling model and verify the defaults are reasonable. At the very least, check the ploidy
assumptions are correct. Once the calling model is appropriately configured, consult the performance
optimisation section to help tune other calling parameters.
Variant call filtering
This version of octopus provides random forest and threshold based variant call filtering. We recommend using
the random forest for germline and somatic calling, and the default filter expressions for trio calling.
! of !17 42
OCTOPUS USER MANUAL

COMMAND LINE REFERENCE
This section contains a description of each command line option. The commands are separated into sections
which roughly correspond to different area of concern. At the end of each section a detailed explanation of any
non-trivial commands is given.
Some options have so called default implicit values, that is, they are be default disabled, but can be enabled
with the implicit default value by just specifying the option name. Implicit options are labelled as =(default
implicit value).
Entries with a red border are currently placeholders and are not yet implemented, they are included
to give an indication of what will be available in the first official release. Entries with an orange
border are currently implemented, but are likely to change before the first official release.
GENERAL
Command
Description
Default value
--reference, -R
The reference genome to use for analysis. Must match
the reference genome used to map reads against.
None
--reads, -I
The read files to use for analysis. Can be specified
multiple times and given a space separated list of
argument.
None
--reads-file, -i
A path that contains a list of read file paths, one per
line, to use for analysis.
None
--regions, -T
A list of genomic intervals to analyse.
All regions
present in the
reference
index.
--regions-file, -t
A path to a file that contains a list of genomic intervals
to analyse. Must have one region per line. BED format
is accepted.
None
--skip-regions, -K
A list of genomic intervals that should be ignored.
None
--skip-regions-file, k
A path to a file that contains a list of genomic intervals
that should be ignored. Must have one region per line.
BED format is accepted.
None
! of !18 42
OCTOPUS USER MANUAL
--one-based-indexing
Reads all user input regions using one-based indexing
rather than zero based.
No
--samples, -S
A list of samples to analyse, which must be a subset of
those samples in the reads.
All samples
found in the
reads.
--samples-file, -s
A path to a file containing a list of samples to analyse.
None
--pedigree
PED file containing sample pedigree. Only currently
used by trio calling model.
None
--fast
Disables various algorithmic features to significantly
reduce runtime, at the cost of worse calling accuracy.
Equivalent to -a off -l minimal -x 50.
Off
--very-fast
Disables various algorithmic features to significantly
reduce runtime, at the cost of worse calling accuracy.
Equivalent to --fast --inactive-flank-
scoring off.
Off
--threads
Enables multithreading. If not supplied with an
argument (recommended), the number of threads is
automatically determined. Otherwise the number of
threads is limited to the given number.
Disabled.
=(automatic)
--working-directory, -w
Any path given to octopus will be relative to the
working directory, unless the path is already valid.
None
--temp-directory-prefix
Prefix name of octopus temporary directory used
during calling (created in working directory).
octopus-temp
--max-reference-cache-
footprint, -X
The maximum amount of memory available to cache
reference sequence. Caching reference sequence
reduces file IO.
500MB
--target-read-buffer-
footprint, -B
The recommended amount of memory available for
buffering read data. This is not a strict limit.
6GB
--target-working-memory
Target maximum working memory for analysis. This is
not a strict limit, but may disable certain memory
intensive optimisations.
None
--max-open-read-files
Limits the number of open read files to the given
number. Note each read file also has an index which is
not accounted for.
250
--contig-output-order
Which order should contigs appear in the final output?
Possible values are: lexicographicalAscending,
lexicographicalDescending, contigSizeAscending,
contigSizeDescending, asInReference,
asInReferenceReversed.
asInReferenceI
ndex
Command
Description
Default value
! of !19 42
OCTOPUS USER MANUAL

READ PRE-PROCESSING
--sites-only
Remove genotype calls and associated information
from final VCF output.
No
--regenotype
A VCF file specifying sites to regenotype; only calls
listed in this files will appear in the output.
None
--legacy
Outputs a more conventional VCF file in addition to the
standard octopus format.
Off
--debug
Writes verbose debug information to a log file. Can be
supplied with a path.
Off
=(octopus_de
bug.log)
--trace
Writes very verbose debug information to a log file. For
maintainer use only. Can be supplied with a path.
Off
=(octopus_tra
ce.log)
--version
Displays the current version number and other meta
information.
None
--config
A configuration file that contains values for some or all
of the options listed here.
None
--bamout
Output realigned BAM files. Full path for single sample
calling, output directory for multi-sample calling.
None
--split-bamout
Output realigned split BAM files. Output prefix for
single sample calling, directory for multi-sample calling.
None
Command
Description
Default value
Command
Description
Default value
--read-transforms
Use to turn off all read transformations. Reads can still
be filtered.
On
--soft-clip-masking
Use to turn off soft clip masking (assigning base quality
zero) of soft clipped read flanks.
On
--mask-tails
Set this many tail base qualities of all reads to zero.
None
--mask-low-quality-tails
Masks (assigns base quality zero) the tail given number
of bases of each read.
No
=(3)
--mask-soft-clipped-
boundries
Masks (assigns base quality zero) to the soft clipped
flanks of reads, plus an additional number of given
bases.
No
! of !20 42
OCTOPUS USER MANUAL

--adapter-masking
Prevents read bases that are considered likely adapter
contaminants, as determined by octopuses native
adapter contamination detector, from being masked
(assigned base quality zero). This command is
redundant unless the command --allow-adapter-
contaminated-reads is also used.
On
--overlap-masking
Prevents masking (assigning base quality zero) of read
bases that overlap (w.r.t mapping location) of other
segments within the reads template. For paired-end
reads, this usually refers to the reads mate. Only one
corresponding base of each read is masked; the other
is left untouched.
On
--read-filtering
Prevents any read from being quality control filtered,
this does not affect downsampling.
On
--consider-unmapped-reads
Turns off filtering of reads marked as unmapped. Note
this is not the same as reads with mapping quality
zero.
No
--min-mapping-quality
Discards reads with mapping quality less than this
before calling.
20
--good-base-quality
The base quality threshold to use for the the options
--min-good-base-fraction and --min-good-
bases.
20
--min-good-base-fraction
The maximum fraction of bases below --min-good-
base-quality before the read is discarded.
Off
--min-good-bases
The minimum number of bases equal to or above --
min-good-base-quality before a read is
considered.
20
--allow-qc-fails
Prevents removal of reads marked as QC failed.
No
--min-read-length
Discards reads with less bases than this.
None
--max-read-length
Discards reads with more bases than this.
None
--allow-marked-duplicates
Prevents removal of reads pre-marked as duplicates.
No
--allow-octopus-
duplicates
Prevents removal of reads that octopuses native
duplicate detector marks as duplicates.
No
--allow-secondary-
alignmenets
Allows reads marked as being secondary alignments.
Yes
--allow-suplementary-
alignments
Allows reads marked as being supplementary
alignments.
Yes
Command
Description
Default value
! of !21 42
OCTOPUS USER MANUAL

VARIANT GENERATION
--no-reads-with-unmapped-
segments
Filter reads where one or more segments in the reads
template are marked as unmapped. For paired-end
reads, this usually refers to the read mate.
No
--no-reads-with-distance-
segments
Filter reads that have template segments mapped to a
different contig. For paired end reads, this usually refers
to the read mate.
No
--no-adapter-
contaminated-reads
Prevents removal of reads that are likely to contain
adapter contamination, as determined by octopuses
native adapter contamination detector.
No
--disable-downsampling
Turns off all downsampling. Reads may still be filtered.
No
--downsample-above
Trigger downsampling of a sample when the read
depth in a region is above this value.
500
--downsample-target
Once a region has been flagged for downsampling, try
to remove reads in the region to achieve this level of
coverage. Must be greater than --downsample-
above.
400
Command
Description
Default value
Command
Description
Default value
--raw-cigar-candidate-
generator, -g
Turn on or off the raw cigar variant candidate generator
to propose candidate variants.
On
--repeat-candidate-
generator
Turn on or off the repeat candidate generator to
propose candidate variants.
On
--assembly-candidate-
generator, -a
Turn on or off the local reassembler generator to
propose candidate variants.
On
--source-candidates
Consider all sites in the given VCF format as candidate
variants. This differs from the option --regenotype
as the final call set is not required to be a subset of
these calls.
None
--max-variant-size
The maximum variant size (w.r.t genomic interval span)
that any candidate variant generator may propose.
2,000
--min-supporting-reads
Overrides the default raw cigar generator and applies a
simple threshold inclusion predicate based on the
number of observed reads. Observations must have
base quality greater than that indicated in --min-
base-quality.
2
--min-base-quality
The minimum base quality a read base must have before it is
considered as supporting a variant.
20
! of !22 42
OCTOPUS USER MANUAL
HAPLOTYPE GENERATION
--kmer-sizes
Default k-mer sizes to use for assembly.
10 15 20
--num-fallback-kmers
The number of fallback k-mer sizes to try if the default
sizes fail to provide a valid graph.
10
--fallback-kmer-gap
The gap size of fallback k-mers.
10
--max-region-to-assemble
The maximum region size that will be used for local
reassembly. Larger sizes may result in larger structural
variation being found, but reduces sensitivity to smaller
variation.
400
--max-assemble-region-
overlap
The maximum number of bases assembly windows are
allowed to overlap. A higher overlap may increase
sensitivity but increase runtime.
200
--assemble-all
Forces local reassembly of all genomic regions.
No
--assembler-mask-base-
quality
Mismatching bases with quality less than this will be
masked as reference before being threaded into the
assembly graph.
10
--min-kmer-prune
The minimum number of k-mer observations to keep
the k-mer in the graph after pruning.
2
--max-bubbles
The maximum number of bubbles to extract from the
assembly graph.
30
--min-bubble-score
The minimum bubble score to extract from the
assembly graph.
2
Command
Description
Default value
Command
Description
Default value
--max-haplotypes, -x
The maximum number of haplotypes that can be used
to generate candidate genotypes. If the haplotype
generator proposes more haplotypes than this then the
excess will be filtered.
200
--haplotype-holdout-
threshold
If a region contains more haplotypes than this, then a
subset of alternative alleles will be temporarily removed
(held out) and only be analysed once some haplotypes
have been discarded.
2,500
--haplotype-overflow
The maximum number of haplotypes a region may
have before the region is unconditionally skipped
(without attempting to hold out alternative alleles).
200,000
--max-holdout-depth
The maximum number attempts to hold out alternative
alleles in a region before the region is skipped.
20
! of !23 42
OCTOPUS USER MANUAL

The option --max-haplotype is a target for the haplotype generator as well as a strict limit for the caller; the
haplotype generator will attempt to satisfy the request, but if it fails to do so, the caller will filter the generated
haplotype set to this limit.
CALLING
--extension-level
Level of haplotype extension. Possible values are
conservative, normal, optimistic, and aggressive.
Normal
--haplotype-extension-
threshold, -e
Haplotypes with posterior probability (of occurrence in
the sample set) can be removed before haplotype
extension.
100
--dedup-haplotypes-with-
prior-model
Deduplicate haplotypes using mutation prior model,
rather than naive method.
Yes
--protect-reference-
haplotype
Never filter the reference haplotype.
Yes
Command
Description
Default value
Command
Description
Default value
--caller, -C
Which calling model to use.
individual or
population
--organism-ploidy, -P
The autosome ploidy for the analysed organism. All
contigs will have this ploidy unless marked otherwise.
2
--contig-ploidies, -p
Assigns ploidies to contigs, overriding the default
organism ploidy.
Y=1 chrY=1
MT=1 chrM=1
--snp-heterozygosity
The SNP heterozygosity in the sample population.
0.001
--snp-heterozygosity-
stdev
The SNP heterozygosity standard deviation in the
sample population.
0.01
--indel-heterozygosity
The INDEL heterozygosity in the sample population.
0.0001
--min-variant-posterior
The minimum posterior probability (QUAL) for a variant
to be reported.
2
--use-uniform-genotype-
priors
Use uniform genotype priors.
No
--use-independent-
genotype-priors
Use independent genotype priors for joint calling.
No
--model-posterior
Calculate model posteriors for every call.
Off
--inactive-flank-scoring
Use to disable calculation to account for flank
mismatches in HMM routine.
On
--model-mapping-quality
Use read mapping quality in read likelihood calculation.
Yes
! of !24 42
OCTOPUS USER MANUAL

TRIO
CANCER
--max-genotypes
Maximum number of genotypes to consider. Currently
only used by cancer and polyclone calling models.
5,000
--max-joint-genotypes
Maximum number of joint genotype vectors that can
be considered (applicable to population and trio calling
models).
1,000,000
--sequence-error-model
The sequencing error model to use for read likelihood
calculation. Possible values are hiseq and x10.
hiseq
--max-vb-seeds
Maximum number of seeds to use in Variational Bayes
inference.
12
--refcall
Report reference confidence calls.
Off
--min-refcall-posterior
The minimum posterior probability (QUAL) for a
reference allele to be reported.
2
Command
Description
Default value
Command
Description
Default value
--maternal-sample; -M
Which of the given samples is the mother in the trio.
None
--paternal-sample; -F
Which of the given samples is the father in the trio.
None
--denovo-snv-mutation-
rate
The germline snv de novo mutation rate.
1.38 x 10-8
--denovo-indel-mutation-
rate
The germline indel de novo mutation rate.
10-9
--min-denovo-posterior
The minimum posterior probability (phred scale) to emit
a de novo mutation call.
3
--denovos-only
Only report DENOVO mutations.
No
Command
Description
Default value
--normal-sample; -N
Which of the given samples is the normal.
None
--max-somatic-haplotypes
Maximum number of somatic haplotypes to consider.
2
--somatic-snv-mutation-
rate
The somatic SNV mutation rate for the cancer to be
analysed.
10-4
--somatic-indel-
mutation-rate
The somatic INDEL mutation rate for the cancer to be
analysed.
10-5
! of !25 42
OCTOPUS USER MANUAL

POLYCLONE
PHASING
CALL FILTERING
--min-expected
-somatic-frequency
The minimum expected somatic allele frequency in
the sample.
0.03
--min-credible
-somatic-frequency
The minimum inferred somatic allele frequency that
will be emitted.
0.01
--credible-mass
Mass of the posterior allele frequency distribution to
use when calculating allele frequency.
0.9
--tumour-germline-
concentration
Dirichlet concentration parameter for tumour germline
haplotypes.
1.5
--normal-contamination-
risk
The risk level that the normal contains contamination
from the tumour. Possible values: low, high.
low
--min-somatic-posterior
The minimum posterior probability an allele is somatic
to be reported.
0.5
--somatics-only
Only report SOMATIC mutations.
No
Command
Description
Default value
Command
Description
Default value
--max-clones
The maximum number of clones to try use when
calling subclonal variants.
3
--min-clone-frequency
Minimum expected clone frequency in the sample.
0.01
Command
Description
Default value
--phasing-level, -l
The level of phasing. Possible values are: minimal,
conservative, moderate, normal, and aggressive.
Normal
--min-phase-score
The minimum phase score (phred scale) a potential
phase set may have to be called.
10
Command
Description
Default value
--call-filtering, -f
Use to enable Call Set Refinement (CSR).
On
! of !26 42
OCTOPUS USER MANUAL
--filter-expression
Boolean expression to use to filter calls. Current
version only supports OR operations and the measure
name must appear on the left hand side of the
comparator.
QUAL < 10 |
MQ < 10 | MP
< 10 | AF <
0.05 | SB >
0.98 | BQ <
15 | DP < 1
--somatic-filter-
expression
Filter expression for somatic calls.
QUAL < 2 |
GQ < 20 | MQ
< 30 | SB >
0.9 | SD > 0.9
| BQ < 20 |
DP < 3 | MF >
0.2 | NC > 1 |
FRF > 0.5
--denovo-filter-
expression
Filter expression for de novo calls.
QUAL < 50 |
PP < 40 | GQ
< 20 | MQ <
30 | AF < 0.1 |
SB > 0.95 |
BQ < 20 | DP
< 10 | DC > 1
| MF > 0.2 |
FRF > 0.5 |
MP < 30 |
MQ0 > 2
--refcall-filter-
expression
Filter expression for homozygous reference calls.
QUAL < 2 |
GQ < 20 | MQ
< 10 | DP <
10 | MF > 0.2
--use-calling-reads-for-
filtering
Use the reads used for calling for filtering. Otherwise
filtering reads will use default read filters and
transforms.
No
--keep-unfiltered-calls
If variant call filtering is turned on, also keep a copy of
unfiltered calls.
No
--training-annotations
Emits CSR measures to the output VCF in INFO fields.
None
--filter-vcf
Run CSR filtering on this octopus VCF without calling.
None
--forest-file
Trained ranger forest to use for germline variant
filtering.
None
--somatic-forest-file
Trained ranger forest to use for somatic variant filtering.
None
Command
Description
Default value
! of !27 42
OCTOPUS USER MANUAL
VARIANT FILTERING
Variant filtering is used to remove false positive calls that may be introduced due to systematic errors in
sequencing or mapping. Ideally, these sources of errors would be fully modelled by the data likelihood model,
but capturing all types of error at this stage is extremely difficult. A number of approaches to variant filtering
have been proposed, including simple threshold based approaches and sophisticated methods using machine
learning. However, all approaches first require defining a set of statistics, or measures, that will be used to
classify calls as passing or failing in one way or another. These quality of these statistics will ultimately decide
the accuracy of variant filtering, regardless of the actual methodology implemented. The default read filter has
been chosen to minimise the chance of filtering true positives, whilst eliminating high quality false positives. To
achieve very high specificity, it may be necessary to increase the stringency of the filter conditions.
Not all measures available in Octopus are computed during the calling phase, hence some filter expressions
require re-access to the read data. The main reason for this is that it may be beneficial to relax the read filtering
constraints used for calling compared to filtering. For example, during calling it is usually advisable to filter
reads with mapping quality less than 20 as these reads are a common source of false positives. To use them
during the calling step would likely increase the false positive rate considerably and increase computation time
as more candidates would need to be considered. However, these reads are useful for filtering as they indicate
the region where the reads are mapped is likely to contain mapping artefacts.
Measure reference
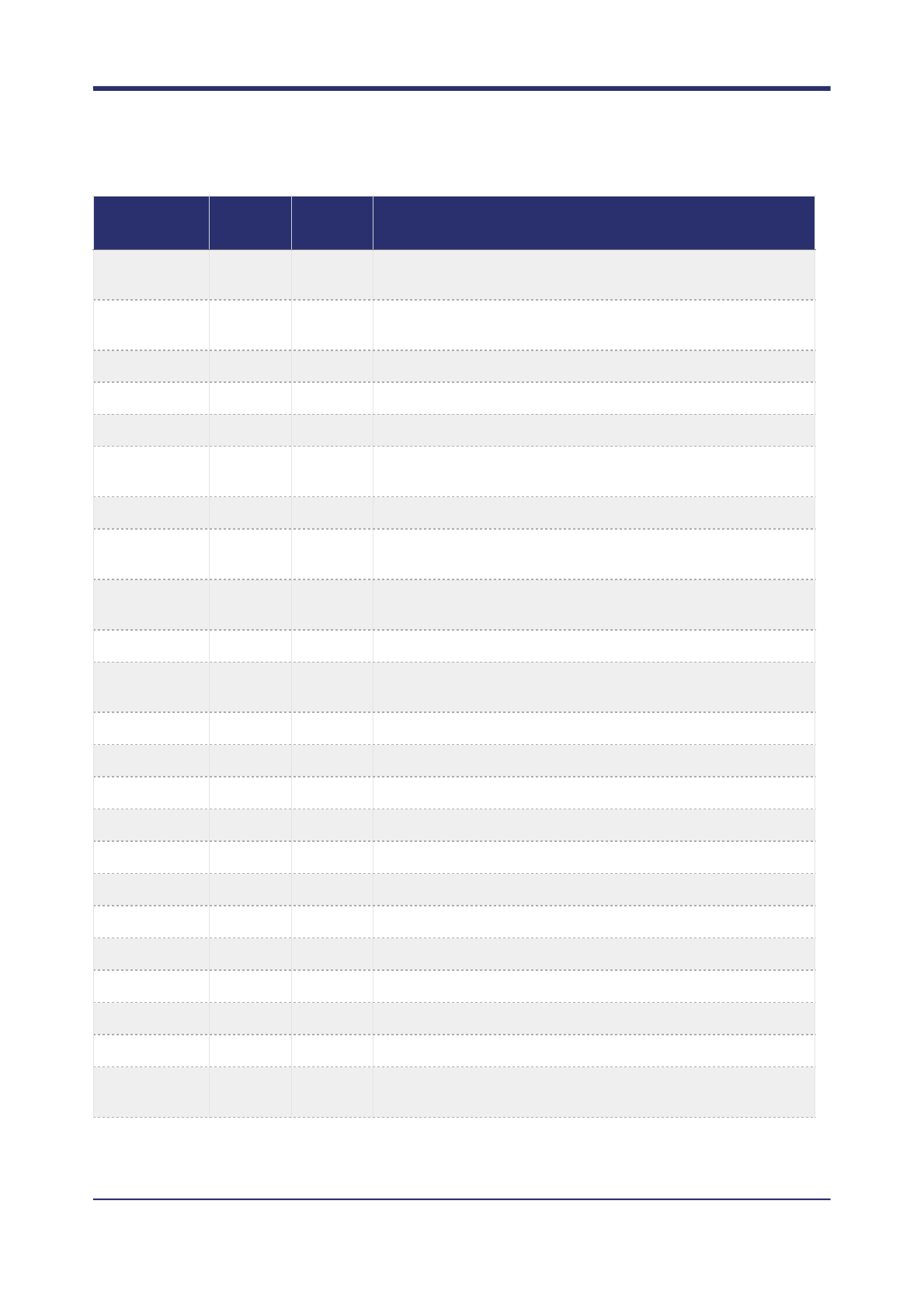
Below is a list of all available measures:
Measure
name
Requires
reads
Sample
specific?
Description
AC
No
No
Number of ALT alleles called.
AD
Yes
Yes
Minor empirical allele depth.
AF
Yes
Yes
Minor empirical allele frequency.
ARF
Yes
Yes
Fraction of reads overlapping the call that cannot be assigned to
a unique haplotype.
BMC
Yes
Yes
Number of base mismatches at variant position in reads
supporting variant haplotype.
BMF
Yes
Yes
Fraction of reads with base mismatches at variant position.
BQ
Yes
Yes
Median base quality of read bases supporting ALT alleles.
CC
No
No
Classification confidence: PP / QUAL
CRF
Yes
No
Fraction of reads supporting ALT alleles that are soft clipped.
DC
Yes
Yes
Number of reads supporting a de novo haplotype in the normal.
DENOVO
No
No
Is the call DENOVO?
! of !28 42
OCTOPUS USER MANUAL
DP
Yes
Yes
Number of reads overlapping the call. This is recalculated for
filtering so may be higher than the calling depth.
FRF
Yes
No
Fraction of reads overlapping the call that were filtered for
calling.
GC
No
No
GC content around the call.
GQ
No
Yes
The sample GQ.
GQD
Yes
Yes
Genotype Quality by Depth: GQ / DP
MC
Yes
Yes
Number of allele mismatches at variant position in reads
supporting variant haplotype.
MF
Yes
Yes
Fraction of reads with mismatches at variant position.
MP
No
No
The model posterior is the probability the model Octopus used
for calling is true, compared to other possible models.
MQ
Yes
No
Root Mean Squared (RMQ) mapping quality of reads
overlapping the call.
MQ0
Yes
No
Number of reads overlapping the call with mapping quality zero.
MQD
Yes
No
Maximum pairwise difference in median mapping qualities of
reads supporting each haplotype.
MRC
Yes
Yes
Number of reads supporting the call that appear misaligned.
NC
Yes
Yes
Number of reads supporting a somatic haplotype in the normal.
PP
No
Yes
The calls Posterior Probability.
PPD
Yes
Yes
Posterior Probability by Depth: PP / DP
QUAL
No
No
The calls quality score.
QD
No
No
Quality by Depth: QUAL / DP
REFCALL
No
No
Are all samples homozygous reference?
REB
Yes
Yes
Bias of variants at end (head or tail) of reads.
RSB
Yes
Yes
Bias of variant side in supporting reads.
RTB
Yes
Yes
Bias of variants at tail of reads.
SB
Yes
Yes
Strand bias of reads based on haplotype support.
SD
Yes
Yes
Strand bias of reads overlapping the site; probability mass in
tails of Beta distribution.
Measure
name
Requires
reads
Sample
specific?
Description
! of !29 42
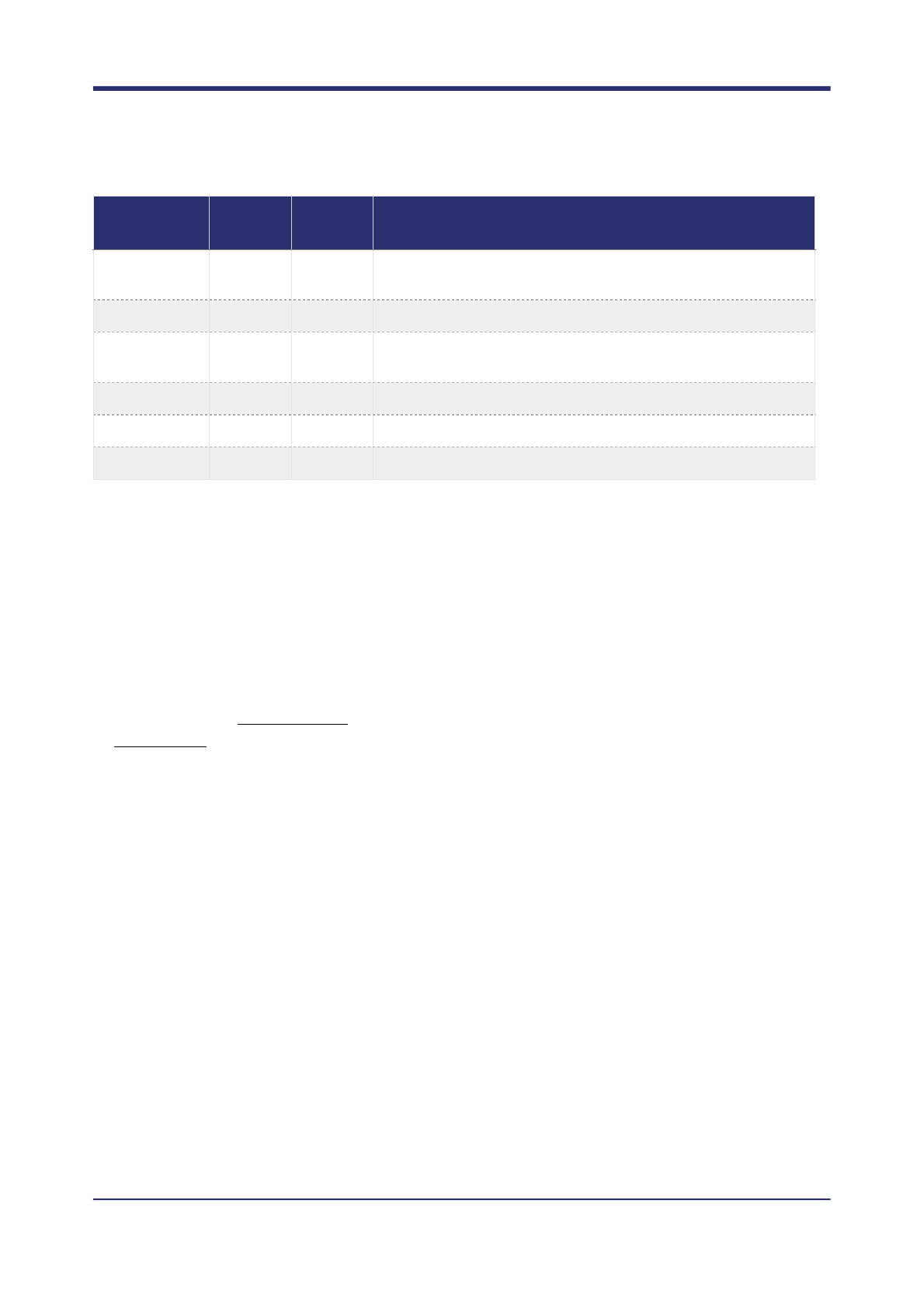
Threshold filtering
Octopus currently provides simple threshold based filtering. A number of measures that can be used to define
a Boolean filter expression. Currently, Octopus only supports expressions with OR operations and the less
than (<) and greater than (>) comparators. Furthermore, measure name must appear on the left hand side of
each condition in the expression.
Random forest filtering
Random forest filtering is a more flexible and powerful method to filter variant calls than threshold filtering, if
sufficient training data is available. Octopus supports random forest filtering for forests that have been trained
with the open source Ranger package. Pre-trained forests for germline and somatic variant calling are available
on Google Cloud. These can be downloaded manually, or automatically by adding the --download-
forests command to the Python installer:
$ ./scripts/install.py --download-forests
The forest files (ending in .forest) will be downloaded to the /resources/forests directory in the top level
source directory.
Octopus currently allows two random forests to be used: one for germline variants (--forest-file), and
another for somatic variants (--somatic-forest-file). In principle, it would be possible to just use one
forest that has been trained to cover all call types, but it is usually preferable to train separate forests when
there is known structure in the data, and sufficient training examples are available.
To apply random forest filtering to typical germline calling add the --forest-file option:
$ octopus -R human.fa -I NA12878.bam --forest-file germline.forest
To filter germline and somatic calls, both the --forest-file and --somatic-forest-file options
need to be given:
$ octopus -C cancer -R human.fa -I tumour.bam \
SF
Yes
Yes
Maximum fraction of reads supporting ALT alleles that are
supplementary.
SHC
Yes
Yes
Number of called somatic haplotypes.
SMQ
Yes
Yes
Median mapping quality of reads assigned to called somatic
haplotypes.
SOMATIC
No
Yes
Does the sample have somatic mutations?
STR_LENGTH
No
Yes
Length of overlapping STR.
STR_PERIOD
No
Yes
Period of overlapping STR.
Measure
name
Requires
reads
Sample
specific?
Description
! of !30 42

--forest-file germline.forest --somatic-forest-file somatic.forest
The pre-trained germline forest has been trained on various whole-genome replicates of NA12878, while the
somatic forest has been trained on synthetic whole genome tumour data. The coverages of the training data is
typical for WGS (10-60X), and may not be suitable for extremely high depth sequencing (e.g. amplicon).
Training random forests
Octopus expects ranger random forests that have been trained on all available measures. The full list is:
AC AD AF ARF BQ CC CRF DP FRF GC GQ GQD NC MC MF MP MRC MQ MQ0 MQD PP PPD QD
QUAL REFCALL REB RSB RTB SB SD SF SHC SMQ SOMATIC STR_LENGTH STR_PERIOD
Ranger expects a text file (either csv, tsv, or space separated) containing values for each measure (in the order
above), and a binary variable in the final column labelled (TP) indicating if the measures in the row originate
from a true or false call. Each row should therefore contain measures from a single sample. In order to
generate this file, Octopus needs to produce a VCF file annotated with each of these measures, which is
requested with the --training-annotations option provided with a list of of measures, or for convincing,
just "forest":
$ octopus -R human.fa -I NA12878.bam -o octopus.NA12878.annotated.vcf.gz
\ --training-annotations forest
Ranger supports several random forest types, Octopus requires the probability classification variants which is
specified with the --probability option. There are various other parameters that control the random forest
that can be specified. We have generally found a forest containing 100-500 trees, and a minimum node size of
5-20 works well.
There are two Python3 scripts in the /scripts directory: train_random_forest.py and
train_somatic_random_forest.py - in the top level source tree that can be used to train ranger
forests.!
! of !31 42
OUTPUT FORMAT
Octopus outputs variants using a simple but rich VCF format. Although the format is fully compliant with the
VCF specification (version 4.3), some users may find it unfamiliar, and some tools will fail to fully parse all
variants. Variant call output is challenging; the output should be consistent but succinct, however, many tools
use representations that is one or the other. For example, records such as the following are not uncommon:
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA12878
1 102738191 . ATTATTTAT A . . . GT 1/0
1 102738191 . ATTATTTATTTAT A . . . GT 1/0
The problem with this representation is that the two records are not consistent as both records infer the
reference allele at the same position, but the site is heterozygous non-reference for two different deletions. The
site can be consistently by joining both records, such as:
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA12878
1 102738191 . ATTATTTATTTAT ATTAT,A . . . GT 1/2
But this representation rapidly becomes unmanageable (and unreadable) as the length and number of
overlapping alleles increases. Octopus solves this issue by making use of two additional symbols:
•The asterisk symbol (*) in the ALT field is specified in the VCF specification as "The ‘*’ allele is reserved to
indicate that the allele is missing due to a upstream deletion".
• The dot symbol (.) in the GT field is specified in the VCF specification as "If a call cannot be made for a
sample at a given locus, ‘.’ should be specified for each missing allele in the GT fields".
Using these two symbols, octopus would represent the above site like:
#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA12878
1 102738191 . ATTATTTAT A,* . . . GT 1|2
1 102738191 . ATTATTTATTTAT A . . . GT .|1
There are three important observations here:
•The records are phased, so must be considered together.
•The first record, which is always contains the shorter allele (i.e. the one which ends first along the
reference sequence), specifies the allele on the first haplotype is the deletion of the length given in the ALT
field, while the other haplotype is non-reference, but is specified in a later record.
•The second record, specifies the deletion given in the ALT field on the other haplotype than the one in the
previous record. And a missing allele on the other haplotype.
When read sequentially, these observations suggest a single unique genotype for the sample at the site.
Although it may seem odd to specify the first allele in the second genotype as missing, this is only true without
the context of the first record, and crucially does not contradict the first record. Both records are consistent
when considered together.
To support tools unable to process octopus's default representation (e.g. RTG Tools), octopus has the --
legacy command line option which produces an additional VCF file using the first format in the example.
! of !32 42
PERFORMANCE OPTIMISATION
Octopus is a sophisticated program with many parameters. The default values for these parameters have been
chosen with an emphasis on calling accuracy, while keeping runtime reasonable. They should provide good all
round performance for most users. However, if octopus is not performing adequately on your particular
dataset it may be due to non-optimal parameterisation. Generally, there is a direct tradeoff between calling
accuracy and runtime resource consumption (memory and CPU time).
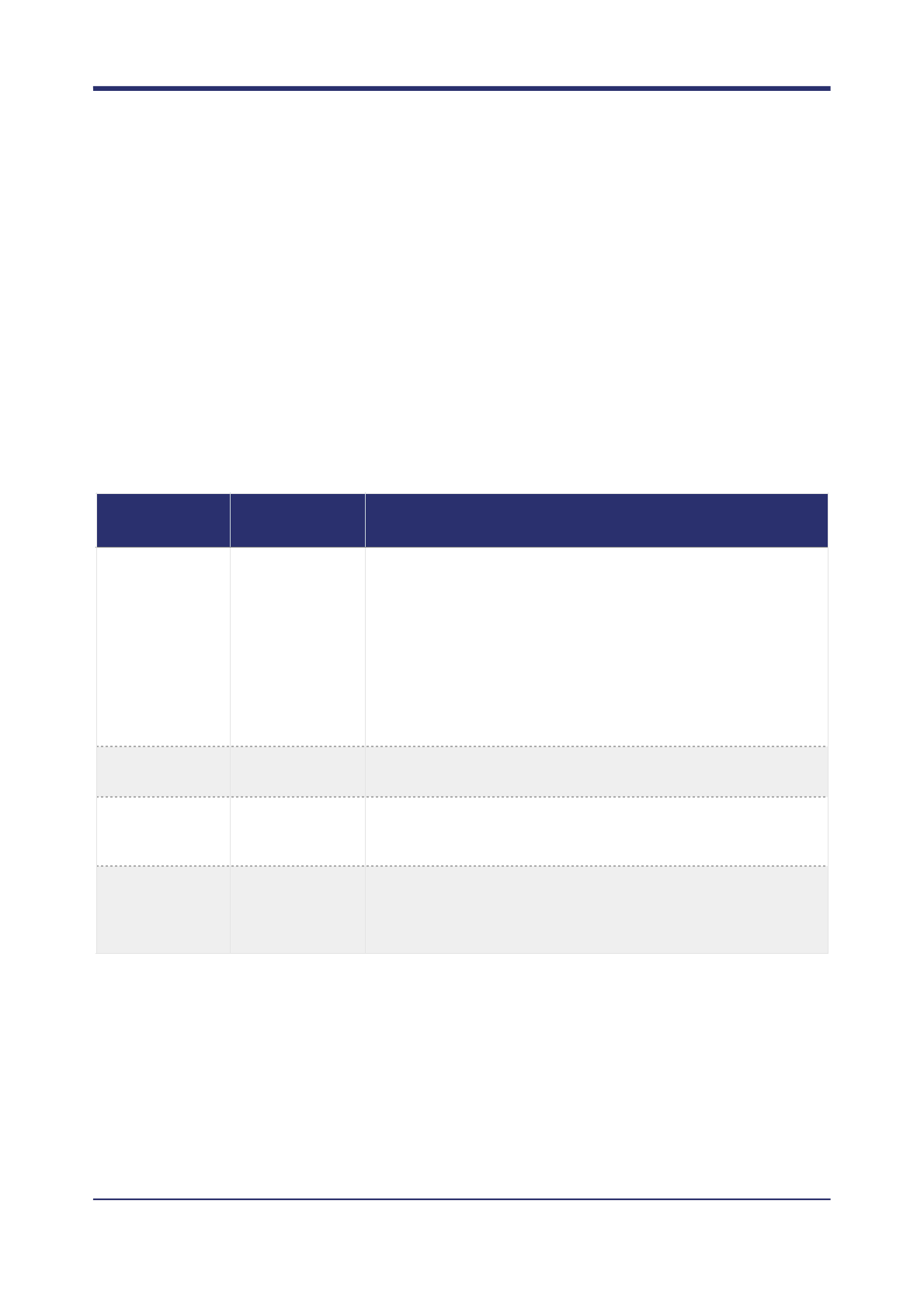
Execution time
By default octopus favours slower, more accurate variant calling. If accuracy is not critical, in particular around
highly polymorphic and complex indel regions, it is possible to achieve significant reductions in runtime by
altering the behaviour of certain components. In summary, the main components of interest are:
There are two convenience command line options --fast and --very-fast that can be used that
automatically adjust these parameters to achieve exceptional runtimes.
Memory consumption
Memory consumption will naturally fluctuate during an run depending on the complexity of the region currently
being analysed, however, by far the main source of memory consumption in octopus is from buffering read
data. While the size of the read buffer is not directly controllable, the user is able to hint at the maximum buffer
size with the --target-read-buffer-footprint option. As this is only a hint, it can be ignored, but in
Component
Associated
commands
Explanation
Variant generation
-a, --kmer-
sizes, --min-
bubble-score
The number of candidate variants that must be considered directly
affects runtime complexity. In general, more sensitive variant
generation will result in more accurate results but longer runtimes.
However, it is important to be aware generating too many
candidate variants may actually decrease accuracy as the model
may become overwhelmed.
Variant generation using local reassembly is also inherently
computational expensive, and the proportion of sites that must be
assembled will directly affect runtime.
Haplotype
generation
-x
The number of haplotypes considered has a direct impact on
calling accuracy and runtime.
Phasing
-l
Phasing requires potentially multiple marginalisation across the
entire genotype posterior distribution. This can be costly when
there are many genotypes, or many candidate variants.
Calling model
Model dependent
(see below)
Some calling models have computationally complex inference
procedures. For example, the cancer calling model implements an
iterative Variational Bayes algorithm. These algorithms usually have
convergence and performance limits which can affect runtime.
! of !33 42

most cases the request is satisfied. The exception to this is when multithreading is used, where there is a
greater possibility of the requested limit being exceeded. In this case it is recommended to set the target
around 20% less than the true limit.
Another source of memory consumption if reference caching which can be used to improve runtime execution
speed as it reduces the need to read from disk. A moderate default reference cache size is used, which can
be adjusted by the user.
Finally, significant memory usage can be observed if unreasonable calling parameters are chosen. For
example, setting --max-haplotypes above 5,000 is likely to lead to memory explosion. Octopus may issue
a warning in such cases, but will try to satisfy the users request.
The --target-working-memory option can be specified which may limit peak memory use. Although this
is not a strict limit, some algorithms have certain optimisations that require high memory use that may be
disabled when using this option. The Variational Bayes algorithm is an example.
Multithreading
The default behaviour is to run using only a single thread. To enable octopuses built in multithreading
capabilities, the --threads command must be used. If an argument is provided to this command then
octopus will use this many threads, if no argument is given to this command then octopus will automatically
determine the number of threads to use. It is highly recommended not to specify the number of threads, as
octopus can probably optimise thread usage better than the user, and it also enables use of specialised
multithreaded algorithms which are not available when the thread count is restricted.
When running a multithreaded job, it is also important to consider the sizes of the reference cache and read
buffer which are set with the -X and -B commands respectively. Octopus will always try to respect these
buffer sizes; even when using multiple threads. In doing so, a small read buffer size can see each thread being
assigned little work, and a small reference cache can lead to cache thrashing. While a larger reference cache
size will always improve runtime performance, increasing the read buffer sizes too much can lead to large
workloads for each thread which is undesirable as overall thread throughput can decrease. The optimal
balance is highly dependent on the data, most critically the depth of coverage. It is recommended the user
experiment with different buffer sizes to find an optimal throughput. As a rough guide it is recommended to at
least double the default reference cache and read buffer sizes when using multithreading, and quadruple the
default sizes when running on a machine with more than 100 cores.
Variant generation
Octopus uses two default candidate variant generators; the raw cigar generator and the local reassembly
generator. While is almost never a good reason to disable the raw cigar generator, it is worth considering the
benefit of leaving the assembler enabled, which can bring significant runtime overheads. The main reason to
have the assembler enabled is to resolve larger indels and complex variation. As a rule of thumb, the
assembler will have less impact with greater read length and decreasing sample diversity. For samples with
high diversity, it is recommended to leave the assembler on.
! of !34 42

Haplotype generation and phasing
Haplotype generation and phasing are closely related; longer haplotypes will produce longer range variant
phasing, and in general, longer haplotypes usually result in more accurate variant calls. Therefore it is important
to recognise that adjusting the level of phasing will also impact the quality of variant calling.
By default phasing is medium range. This means octopus will perform haplotype extension when possible, but
will stop if too much time is spent in a particular region, or if there are too many haplotypes supported by the
data. Experimentation has shown this to provide the best overall calling accuracy while also giving accurate
phasing in the vast majority of cases. For human data, the exception is the HLA which may benefit from higher
phase levels. In general, increasing the phasing level beyond the default level will not usually improve variant
call accuracy, and may decrease it, unless the number of allowed haplotypes is also increased, as the risk of
pruning a true haplotype increases with each haplotype extension.
Calling model selection and parametrisation
The most important consideration to make before variant calling is the calling model to use. Using the
population calling model on tumour samples will not result in high quality calls (or classified somatic mutations).
In most instances, if all relevant sample information is correctly entered to the command line then octopus will
automatically select the appropriate calling model. However, there may be exceptions (e.g. tumour only
calling), so it is important to check which calling model was invoked when execution begins, or explicitly set
the calling model.
Once the appropriate calling model has been chosen, it is vital to consider the parameters specific to that
model. For example, the de novo mutation rate is specific to the trio calling model. The more accurately the
calling model is parametrised, the more accurate the calls will be. It is therefore worth spending time reviewing
the parameters.
Some parameters are common to all calling models. Of these, the copy number directives are the most
important. In addition to choosing a default organism ploidy, octopus also allows copy number specification of
individual chromosomes, so it is important to set correct copy number for sex chromosomes if applicable.
! of !35 42

TROUBLESHOOTING
BUILDING
Why are the requirements so strict?
Octopus uses some advances features of the latest official C++ standard (C++14) and therefore requires a
mature C++ compiler. The other requirements have been set to recent versions because these are the versions
used to develop and test octopus, and we cannot guarantee that earlier versions will perform as well. In
particular CMake introduces improved interprocedural optimisation in version 3.9, whilst Boost 1.65 contains
bug fixes and improvements which octopus uses. This also reduces the burden on future releases to be
backwards compatible with ageing tools, which allows focus on new features and improvements.
CMake chooses a bad compiler
As the previous issue explains, some compilers advertised as being C++14 compliant are not due to compiler
bugs. Unfortunately, CMake cannot recognise this and will happily select a buggy compiler if you have one in a
standard location on your system. If you have installed another working compiler in a non-standard location,
you need to tell CMake how to find it with the CMAKE_CXX_COMPILER command:
$ cmake -D CMAKE_CXX_COMPILER=/path/to/compiler/cpp ..
Or if you’re using the Python install script (recommended):
$ ./scripts/install.py --cxx_compiler=/path/to/compiler/cpp
Compilation fails
Octopus uses advanced C++14, and therefore requires a robust C++14 compiler and standard library
implementation. Many of the compiler versions advertised as being C++14 compliant have bugs that prevent
compilation. The only compilers that have currently been shown to work are Clang 3.8 and GCC 6.2.
Linking fails
Ensure the correct compiler driver was selected: using Clang this means selecting clang++ and not clang:
$ ./scripts/install.py --cxx_compiler=/path/to/clang/bin/clang++
Similarly for GCC use g++ rather than gcc:
$ ./scripts/install.py --cxx_compiler=/path/to/gcc/bin/g++
On some systems, you may also need to specify a C compiler which is the same version as your C++
compiler, in which case you can also specify this with the c_compiler option:
$ ./install.py --cxx_compiler=g++-7 --c_compiler=gcc-7
If the issue is specifically with linking against Boost then see the next issue.
! of !36 42

Boost libraries fail to link
First ensure that Boost was built using the same compiler used to compile octopus - using different compilers
will often cause linking problems.
Second, if the required Boost libraries are not installed in a standard location on your system, and you have
not set appropriate environment variables, you may see an error message when running octopus (even after a
successful build). To resolve this you just need to set the appropriate environment variable:
$ export LD_LIBRARY_PATH=/path/to/boost/lib
before executing octopus.
Compilation has lots of #pragma warnings
This is a Boost issue that was introduced in Boost 1.60, and should be fixed in future releases of Boost. This
issue does not affect octopus.
RUNTIME
Segmentation fault
Sorry! Please see the advise under ‘reporting bugs’.
Execution is slow
The best way to improve runtime is to use multithreading with the --threads command. If this does not offer
adequate performance then consider switching off the assembler with the -a command or try the command
--fast. Please see the section Execution time under performance optimisation.
Execution delays after initialising calling components in threaded mode
This is due to the way tasks are distributed to threads: the challenge is to split the genome into chunks (or
tasks) that are independent in the sense that the union of the calls produced by calling each independently is
the same as calling them together. Clearly this is always true for different contigs, but it is non-trivial to detect
independent tasks for a single contig. In addition, if tasks are too small or too large, runtime performance is not
optimal. Octopus solves this problem by focusing on creating tasks that will lead good thread usage, while
relying on a conflict detection algorithm to find non-independent tasks after calling has completed. This
algorithm requires all tasks for a given contig to be known before any resolutions can be made, and therefore
all tasks for a contig must be generated before calling can begin.
Run hangs in decoy contigs
Decoy contigs often have very high coverage (above 20,000X) and therefore the downsampling will likely
occur. The downsampling algorithm octopus implements is non-trivial as it attempts to keep an even coverage
across the downsampled region to avoid any bias. Unfortunately in very high coverage regions this can be very
slow. We are working on improving this, but for now we suggest either increasing the downsampling
thresholds, or removing these decoy contigs from analysis completely (using the option --skip-regions(-
file).
! of !37 42

BEHAVIOUR
No calls are reported
Try turning off read filters with the command --read-filtering off. If this results in calls then one or
more of the filters is probably over-filtering, which usually implies a quirk of the read mapper that octopus does
not recognise. In such cases it is best to find which filter is triggering (the --debug command can be used to
help) and disable it.
Regions are skipped because of too many haplotypes
This warning can occur in very complex regions and is not normally a problem as it is usually reflective of bad
read mappings. However, it is possible to force octopus to call these regions by increasing the --max-
haplotypes and --haplotype-overflow command line options.
A call changes when a different input region is given
Haplotype based variant callers gain power by jointly calling adjacent alleles, hence the result for one position
is dependent on other nearby positions. If two target regions differ then different haplypes may be proposed
for each, which can affect the overall result for a position even if it is present in both target regions, and even if
no other calls result from the difference. Octopus tries to avoid this problem as much as possible by
considering regions beyond those requested, but this cannot eliminate the problem entirely - the only complete
solution is to call entire contigs.
Why doesn’t octopus report genotype likelihoods?
Octopus calculates the likelihood of the reads given a haplotype while records in the VCF file are alleles. It is
non-trivial to calculate the genotype likelihood w.r.t an allele given the genotype likelihoods w.r.t haplotypes as
the calculation must also condition on all other alleles used to compute the haplotypes likelihoods. This is not
the case for posteriors, which can be marginalised in a trivial way, as all required information regarding other
alleles is already present in the single posterior value.
While we appreciate users like to see genotype likelihoods. We feel we can offer users more accurate results
by providing flexible priors, and reporting exact genotype posteriors in the final output.
Why do octopus VCF files contain * and .?
Octopus uses advanced parts of the VCF specification to archive a consistent genotyping over all samples.
The problem appears because primarily because multi-allelic sites must either be represented on a single line
or split into multiple records. For shorter alleles the former option is fine, but the latter option is better for longer
alleles (otherwise records containing 100+ bases to represent a SNP occur). The principle problem with
splitting records like this is that inconstancies can arise if the records are treated independently, for example,
consider a simple example:
!#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA12878
1 100 . A C,G . . . GT 1|2
Now suppose we split the record into two separate records:
! of !38 42

#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA12878
1 100 . A C . . . GT 1|0
1 100 . A G . . . GT 0|1!
There is an inconstancy here! The first record states the genotype is C|A while the second claims it is A|G! The
problem is that the reference is overrepresented in the genotype call. What we should be saying here is that
the genotype cannot be fully represented due to a conflict with another call, which is exactly what octopus
does!
"#CHROM POS ID REF ALT QUAL FILTER INFO FORMAT NA12878
1 100 . A C . . . GT 1|.
1 100 . A G . . . GT .|1
Note: octopus would actual represent this call as a multi-allelic record (as in the first case), this example is just
intended to demonstrate the behaviour.
Now for the “*” which can appear as an ALT allele. This is somewhat like the dot used to indicate an
interaction, but specifically denotes an overlap with a deletion.
Unfortunately, many other variant callers - and therefore downstream analysis tools - do not output consistent
calls. In order to force octopus to output an additional potentially inconsistent VCF file which may work better
with downstream tools, just add the option --legacy.
SNP accuracy improves in fast mode
In fast mode octopus turns off haplotype extension, which means there is no haplotype posterior based
filtering. Haplotype extension is usually good; it can help resolve complex regions and allows long range
phasing. However, in regions where the data cannot be modelled correctly (e.g. if many reads are miss-
mapped), it can lead to significant portions of the overall posterior distribution being removed. This causes
false positive calls to be given far higher posterior (and hence QUAL) than if no extension was used. This is
especially true for SNPs.
Many of these high quality false positives will filtered with the default model filters, but it is possible for some to
pass these filters if too many reasonable haplotypes are removed before the model selection is applied. We are
working on ways to improve this, one idea is to do a double pass of any complex regions to improve
resolution. If this is causing a significant problem for you now, you can help alleviate the issue by increasing the
posterior filter threshold (--haplotype-extension-threshold), or simply turn off haplotype extension by
setting --phasing-level to minimal.
Calling performance is worse with assembler
The assembler often proposes many complex candidate variants which increases the number of haplotypes
considerably. This can force octopus to rely on its haplotype filtering algorithms more than without the
assembler, and while these algorithms often perform well, they are not foolproof. In addition, many gold
standard reference sets will not contain real complex variants of the type the assembler is able to propose, as
these are challenging to validate, and many variant callers are not even capable of calling such variants. It is
advised the user interest such results with a degree of caution.
! of !39 42

CONTACT
•Author and maintainer: Daniel Cooke (dcooke@well.ox.ac.uk).
•Author: Gerton Lunter (gerton.lunter@well.ox.ac.uk).
APPENDIX
INSTALLING REQUIREMENTS
OS X
On OS X, Clang is recommended, and all requirements can be installed with the package manager
Homebrew:
$ brew update
$ brew install git
$ brew install --with-clang llvm
$ brew install boost
$ brew install cmake
$ brew install python@3
$ brew install htslib
If you already have any of these packages installed on your system with Homebrew the command will fail, but
you can update to the latest version by using brew upgrade instead of brew install.
Ubuntu
On Ubuntu, most requirements can be obtained with apt-get. GCC 7 is recommended as this will simplify
installing Boost. To obtain the requirements execute:
$ sudo add-apt-repository ppa:ubuntu-toolchain-r/test
$ sudo apt-get update && sudo apt-get upgrade
$ sudo apt-get install gcc-7
$ sudo apt-get install git-all
$ sudo apt-get install python3
Test GCC was successfully installed by typing
$ g++-7 --version
As only htslib 1.2.1 is available using apt-get, htslib 1.4 must be installed manually:
$ sudo apt-get install autoconf
$ git clone https://github.com/samtools/htslib.git
$ cd htslib
$ autoheader
$ autoconf
! of !40 42
$ ./configure
$ make && sudo make install
CMake is also easy to install:
$ sudo apt-get purge cmake
$ mkdir ~/temp && cd ~/temp
$ wget https://cmake.org/files/v3.9/cmake-3.9.6.tar.gz
$ tar -xzvf cmake-3.9.6.tar.gz
$ cd cmake-3.9.6/
$ ./bootstrap
$ make -j4
$ sudo make install
$ cmake --version
Boost can be installed as follows:
$ wget -O boost_1_65_1.tar.gz http://sourceforge.net/projects/boost/
files/boost/1.65.1/boost_1_65_1.tar.gz/download
$ tar xzvf boost_1_65_1.tar.gz && cd boost_1_65_1
$ sudo apt-get update
$ sudo apt-get install build-essential g++ python-dev autotools-dev
libicu-dev build-essential libbz2-dev
$ ./bootstrap.sh --prefix=/usr/
$ ./b2
$ sudo ./b2 install
VARIANT GENERATION
Genotype calls are made by generating a set of candidate variants and weighing up the evidence for each. The
space of all possible variants is infinite, so heuristics must be employed to generate the candidate set. The
performance of the variant generator is critical as the final calls will be a subset of the generated candidate
variant set.
Octopus can make use of multiple independent variant generators and take the union of the result of each.
The two default variant generators are the raw cigar generator and the local reassembly generator. The
raw cigar generator proposes a candidate whenever a mismatch, as indicated in a reads cigar string, satisfies
some condition. The default condition used is dependent on the calling model selected, but can be modified
by the user. The local reassembly generator constructs a De Bruin graph of all reads in a genomic interval,
threaded around the reference sequence, and then extracts paths through the graph which deviate from the
single reference path.
Currently the only other non-default generator is the source file generator which simply extracts previously
made calls from a VCF format file. We are working on a generator that will extract known variants from online
databases.
! of !41 42

HAPLOTYPE GENERATION
The haplotype generator takes the set of candidate variants generated by the variant generators and
constructs sets of candidate haplotypes. The advantage of having these two stages independent is that
haplotypes can then by dynamically manipulated after construction. In particular, it allows a haplotype to be
extended conditionally on the posterior probability that the haplotype is present in the union of all sample
genotypes.
PHASING
Phasing takes place after variants have been called - the algorithm uses the regions spanned by the calls and
the inferred genotype posterior distribution to find optimal phase regions. It proceeds by arranging the called
regions into groups such that the entropy of marginal posterior distributions between groups is maximised. In
other words, if the posterior distribution assigns similar mass to different phasing of the same variant calls,
then the true phasing is unknown, but if one particular phasing carries most of the posterior mass, then there
is strong evidence that phasing is the true physical phasing.
GLOSSARY
Allele A particular instance of a genomic region, that is, a nucleotide sequence and a genomic region.
Genomic region/interval A coordinate range with respect to a reference genome. All genomic regions must
specify the contig (chromosome) and begin and end positions in the reference.
Genotype A collection of haplotypes or alleles of known cardinality.
Haplotype An ordered set of alleles which occur on the same chromosome in the sample.
! of !42 42