Silo Guide
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 358 [warning: Documents this large are best viewed by clicking the View PDF Link!]
- Chapter 1 Introduction to Silo
- Chapter 2 C and Fortran Functions
- 1 API Section Error Handling and Other Global Library Behavior
- DBErrfuncname
- DBErrno
- DBErrString
- DBShowErrors
- DBErrlvl
- DBErrfunc
- DBVariableNameValid
- DBVersion
- DBVersionDigits
- DBVersionGE
- DBSetAllowOverwrites
- DBGetAllowOverwrites
- DBSetAllowEmptyObjects
- DBGetAllowEmptyObjects
- DBForceSingle
- DBGetDatatypeString
- DBSetDataReadMask2
- DBGetDataReadMask2
- DBSetEnableChecksums
- DBGetEnableChecksums
- DBSetCompression
- DBGetCompression
- DBSetFriendlyHDF5Names
- DBGetFriendlyHDF5Names
- DBSetDeprecateWarnings
- DBGetDeprecateWarnings
- DB_VERSION_GE
- 2 API Section Files and File Structure
- DBRegisterFileOptionsSet
- DBUnregisterFileOptionsSet
- DBUnregisterAllFileOptionsSets
- DBSetUnknownDriverPriorities
- DBGetUnknownDriverPriorities
- DBCreate
- DBOpen
- DBClose
- DBGetToc
- DBFileVersion
- DBFileVersionDigits
- DBFileVersionGE
- DBVersionGEFileVersion
- DBSortObjectsByOffset
- DBMkDir
- DBSetDir
- DBGetDir
- DBCpDir
- DBGrabDriver
- DBUngrabDriver
- DBGetDriverType
- DBGetDriverTypeFromPath
- DBInqFile
- DBInqFileHasObjects
- _silolibinfo
- _hdf5libinfo
- _was_grabbed
- 3 API Section Meshes, Variables and Materials
- DBPutCurve
- DBGetCurve
- DBPutPointmesh
- DBGetPointmesh
- DBPutPointvar
- DBPutPointvar1
- DBGetPointvar
- DBPutQuadmesh
- DBGetQuadmesh
- DBPutQuadvar
- DBPutQuadvar1
- DBGetQuadvar
- DBPutUcdmesh
- DBPutUcdsubmesh
- DBGetUcdmesh
- DBPutZonelist
- DBPutZonelist2
- DBPutPHZonelist
- DBGetPHZonelist
- DBPutFacelist
- DBPutUcdvar
- DBPutUcdvar1
- DBGetUcdvar
- DBPutCsgmesh
- DBGetCsgmesh
- DBPutCSGZonelist
- DBGetCSGZonelist
- DBPutCsgvar
- DBGetCsgvar
- DBPutMaterial
- DBGetMaterial
- DBPutMatspecies
- DBGetMatspecies
- DBPutDefvars
- DBGetDefvars
- DBInqMeshname
- DBInqMeshtype
- 4 API Section Multi-Block Objects, Parallelism and Poor-Man’s Parallel I/O
- DBPutMultimesh
- DBGetMultimesh
- DBPutMultimeshadj
- DBGetMultimeshadj
- DBPutMultivar
- DBGetMultivar
- DBPutMultimat
- DBGetMultimat
- DBPutMultimatspecies
- DBGetMultimatspecies
- DBOpenByBcast
- PMPIO_Init
- PMPIO_CreateFileCallBack
- PMPIO_OpenFileCallBack
- PMPIO_CloseFileCallBack
- PMPIO_WaitForBaton
- PMPIO_HandOffBaton
- PMPIO_Finish
- PMPIO_GroupRank
- PMPIO_RankInGroup
- 5 API Section Part Assemblies, AMR, Slide Surfaces, Nodesets and Other Arbitrary Mesh Subsets
- 6 API Section Object Allocation, Free and IsEmpty
- 7 API Section Calculational and Utility
- 8 API Section Optlists
- 9 API Section User Defined (Generic) Data and Objects
- DBWrite
- DBWriteSlice
- DBReadVar
- DBReadVarSlice
- DBGetVar
- DBInqVarExists
- DBInqVarType
- DBGetVarByteLength
- DBGetVarDims
- DBGetVarLength
- DBGetVarType
- DBPutCompoundarray
- DBInqCompoundarray
- DBGetCompoundarray
- DBMakeObject
- DBFreeObject
- DBChangeObject
- DBClearObject
- DBAddDblComponent
- DBAddFltComponent
- DBAddIntComponent
- DBAddStrComponent
- DBAddVarComponent
- DBWriteComponent
- DBWriteObject
- DBGetObject
- DBGetComponent
- DBGetComponentType
- 10 API Section JSON Interface to Silo Objects
- 11 API Section Previously Undocumented Use Conventions
- 12 API Section Silo’s Fortran Interface
- 13 API Section Deprecated Functions
- 14 API Section Silo Library Header File
- 1 API Section Error Handling and Other Global Library Behavior
Lawrence
Livermore
National
Laboratory
Silo User’s Guide
Revision: July 2014
Version: 4.10 of the Silo Library
Document Release Number LLNL-SM-654357
This document was prepared as an account of work sponsored by an agency of the United States government. Neither
the United States government nor Lawrence Livermore National Security, LLC, nor any of their employees makes
any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or
usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe
privately owned rights. Reference herein to any specific commercial product, process, or service by trade name,
trademark, manufacturer, or otherwise does not necessarily constitute or imply its endorsement, recommendation, or
favoring by the United States government or Lawrence Livermore National Security, LLC. The views and opinions of
authors expressed herein do not necessarily state or reflect those of the United States government or Lawrence Liver-
more National Security, LLC, and shall not be used for advertising or product endorsement purposes.
ii
This work was performed under the auspices of the U.S. Department of Energy by Lawrence Livermore National
Laboratory in part under Contract W-7405-Eng-48 and in part under Contract DE-AC52-07NA27344.

Silo User’s Guide 1-1
Chapter 1 Introduction to Silo
1.1. Overview
Silo is a library which implements an application programing interface
(API) designed for reading and writing a wide variety of scientific data to
binary, disk files. The files Silo produces and the data within them can be
easily shared and exchanged between wholly independently developed
applications running on disparate computing platforms.
Consequently, the Silo API facilitates the development of general purpose
tools for processing scientific data. One of the more popular tools that pro-
cess Silo data files is the VisIt1 visualization tool.
Silo supports gridless (point) meshes, structured meshes, unstructured-zoo
and unstructured-arbitrary-polyhedral meshes, block structured AMR
meshes, constructive solid geometry (CSG) meshes as well as piecewise-
constant (e.g. zone-centered) and piecewise-linear (e.g. node-centered) vari-
ables defined on these meshes. In addition, Silo supports a wide array of
other useful objects to address various scientific computing applications’
needs.
Although the Silo library is a serial library, it has key features which enable
it to be applied quite effectively and scalably in parallel.
Architecturally, the library is divided into two main pieces; an upper-level
application programming interface (API) and a lower-level I/O implementa-
tion called a driver. Silo supports multiple I/O drivers, the two most com-
mon of which are the HDF5 (Hierarchical Data Format 5)2 and PDB
(Portable Data Base, a binary database file format developed at LLNL by
Stewart Brown) drivers. However, the reader should take care not to infer
1. VisIt can be obtained from http://www.llnl.gov/visit

1-2 Silo User’s Guide
from this that Silo can read any HDF5 file. It cannot. For the most part, Silo
is able to read only files that it has also written.
1.2. Where to Find Example Code
In the ‘tests’ directory within the Silo source release tarball, there are
numerous example C codes that demonstrate the use of Silo for writing var-
ious types of data. There are not as many examples of reading the data there.
If you are interested in point meshes, for example, you would search for
‘DBPutPointMesh’. Or, if you are interested in how to use some option like
DBOPT_CONSERVED, search for it within the C files in the tests direc-
tory.
1.3. Brief History and Background
Development of the Silo library began in the early 1990’s at Lawrence
Livermore National Laboratory to address a range of issues related to the
storage and exchange of data among a wide variety of scientific computing
applications and platforms.
In the early days of scientific computing, roughly 1950 - 1980, simulation
software development at many labs, like Livermore, invariably took the
form of a number of software “stovepipes”. Each big code effort included
sub-efforts to develop supporting tools for visualization, data differencing,
browsing and management.
Developers working in a particular stovepipe designed every piece of soft-
ware they wrote, simulation code and tools alike, to conform to a common
representation for the data. In a sense, all software in a particular stovepipe
was really just one big, monolithic application, typically held together by a
common, binary or ASCII file format.
Data exchanges across stovepipes were laborious and often achieved only
by employing one or more computer scientists whose sole task in life was to
write a conversion tool called a linker. Worse, each linker needed to be kept
it up to date as changes were made to one or the other codes that it linked. In
short, there was nothing but brute force data sharing and exchange. Further-
more, there was duplication of effort in the development of support tools for
each code.
Between 1980 and 2000, an important innovation emerged, the general pur-
pose I\O library. In fact, two variants emerged each working at a different
level of abstraction. One focused on the “objects” of computer science. That
is arrays, structs and linked lists (e.g. data structures). The other focused on
2. The National Center for Supercomputing Applications (NCSA) at the University
of Illinois at Urbana-Champaign (UIUC). The HDF5 software can be obtained
from http://hdf5.ncsa.uiuc.edu/HDF5/release/obtain5.html.
Architecture
Silo User’s Guide 1-3
the “objects” of computational modeling. That is structured and unstruc-
tured meshes with piecewise-constant and piecewise-linear fields. Exam-
ples of the former are CDF, HDF (HDF4 and HDF5) and PDBLib. Silo is an
example of the latter type of I/O library. At the same time, Silo makes use of
the former.
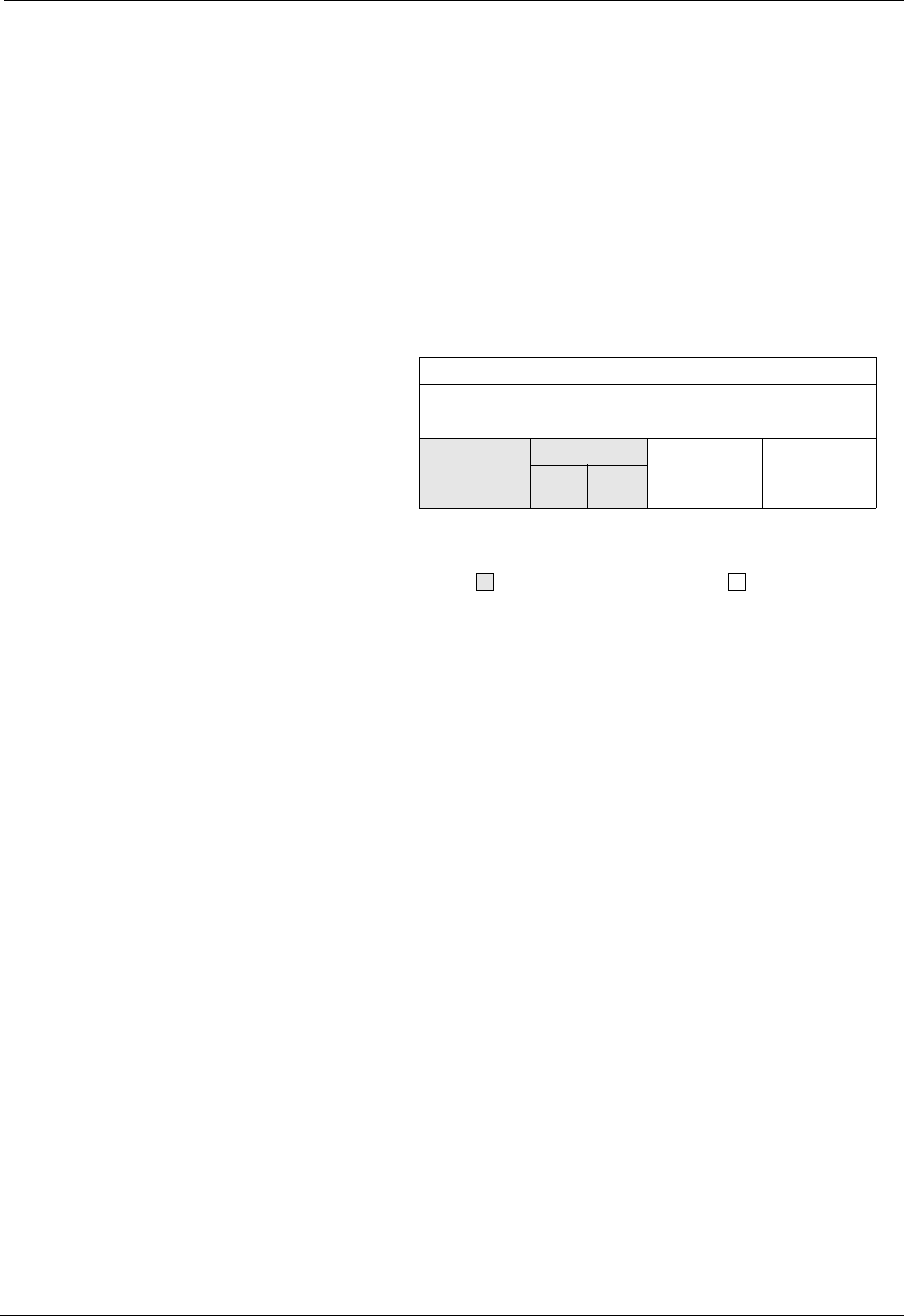
1.4. Silo Architecture
Silo has several drivers. Some are read-only and some are read-write. These
are illustrated in Figure 1-1:
Read/Write
Application
PDB
netcdf Taurus
Read only
HDF5
Silo-API
Drivers
Lite roper
Figure 1-1: Model of Silo Architecture.
Silo supports both read and write on the PDB (Portable Database) and
HDF5 drivers. In addition, Silo supports two different “flavors” of PDB
drivers. One known within Silo as “PDBLite” and is just called “PDB”
which is a very old version of PDB that was frozen into the Silo library in
1999. That is the default driver. The other flavor of PDB is known within
Silo as “PDB Proper” and can use a current release of the PDB library.
Although Silo can write and read PDB and HDF5 files, it cannot read just
any PDB or HDF5 file. It can read only PDB or HDF5 files that were also
written with Silo. Silo supports only read on the taurus and netcdf drivers.
The particular driver used to write data is chosen by an application when a
Silo file is created. It can be automatically determined by the Silo library
when a Silo file is opened.
1.4.1. Reading Silo Files
The Silo library has application-level routines to be used for reading mesh
and mesh-related data. These functions return compound C data structures
which represent data in a general way.
1.4.2. Writing Silo files
The Silo library contains application-level routines to be used for writing
mesh and mesh-related data into Silo files.

Computational Meshes Supported by Silo
1-4 Silo User’s Guide
In the C interface, the application provides a compound C data structure
representing the data. In the Fortran interface, the data is passed via individ-
ual arguments.
1.5. Terminology
Here is a short summary of some of the terms used throughout the Silo
interface and documentation. These terms are common to most computer
simulation environments.
Block This is the fundamental building block of a computational mesh. It
defines the nodal coordinates of one contiguous section of a mesh (also
known as a mesh-block).
Mesh A computational mesh, composed of one or more mesh-blocks. A mesh
can be composed of mesh-blocks of different types (quad, UCD) as well
as of different shapes.
Variable Data which are associated in some way with a computational mesh.
Variables usually represent values of some physics quantity (e.g., pres-
sure). Values are usually located either at the mesh nodes or at zone cen-
ters.
Material A physical material being modeled in a computer simulation.
Node A mathematical point. The fundamental building-block of a mesh or
zone.
Zone An area or volume of which meshes are comprised. Zones are polygons
or polyhedra with nodes as vertices (see “UCD 2-D and 3-D Cell
Shapes” on page 1-6.)
1.6. Computational Meshes Supported by Silo
Silo supports several classes, or types, of meshes. These are quadrilateral,
unstructured-zoo, unstructured-arbitrary, point, constructive solid geometry
(CSG), and adaptive refinement meshes.
1.6.3. Quadrilateral-Based Meshes and Related Data
A quadrilateral mesh is one which contains four nodes per zone in 2-D and
eight nodes per zone (four nodes per zone face) in 3-D. Quad meshes can be
either regular, rectilinear, or curvilinear, but they must be logically rectan-
gular (Fig. 1-2).
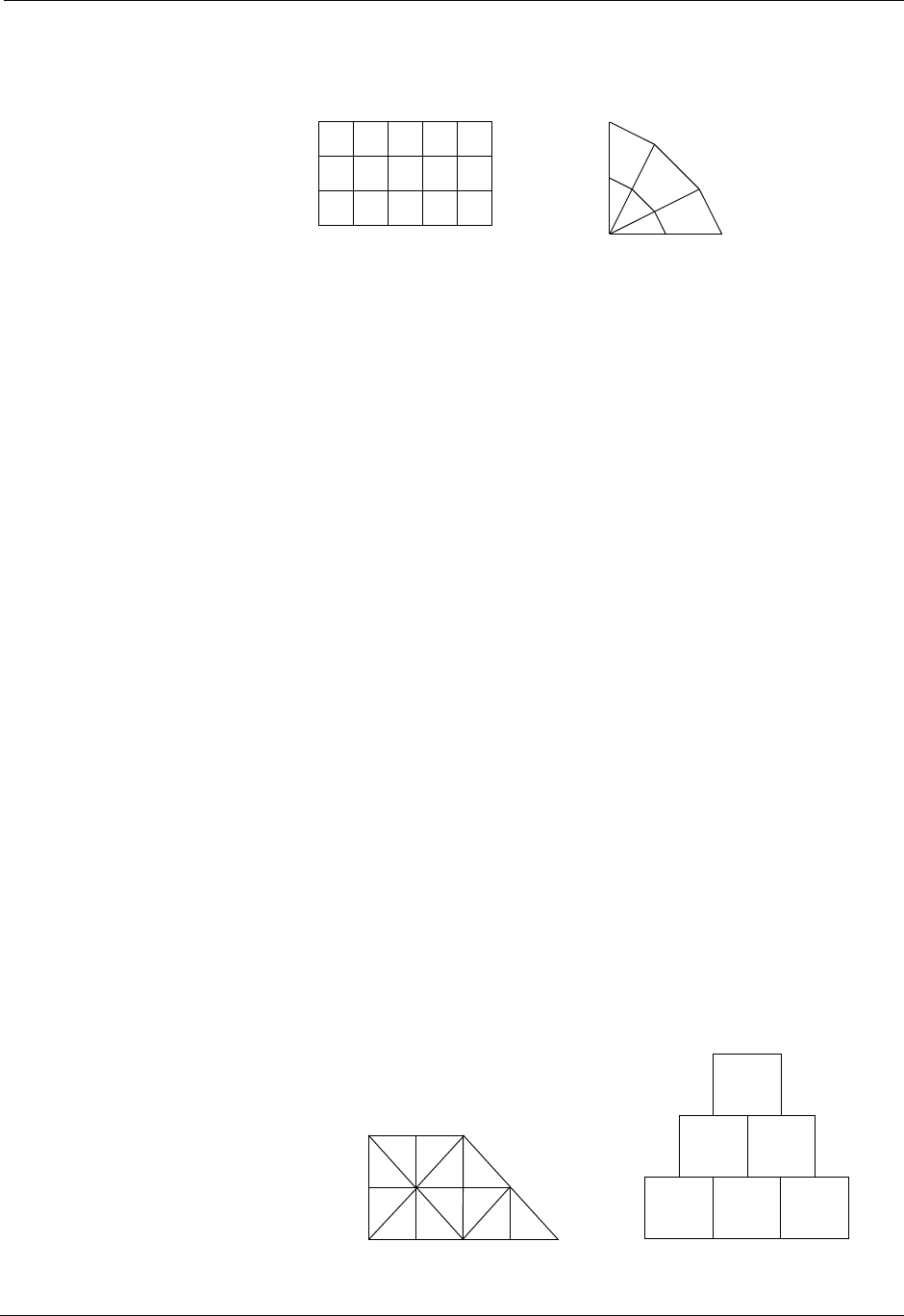
Rectilinear Curvilinear
X
Y
X = {0.0,1.0,2.0,3.0,
Y = {0.0,1.0,2.0,3.0}
X = {0.0,1.0,2.0,
0.0,0.8,1.6,
0.0,0.4,0.8,
0.0,0.0,0.0}
Y = {0.0,0.0,0.0,
0.0,0.4,0.8,
0.0,0.8,1.6,
0.0,1.0,2.0}
X
Y
4.0,5.0}
UCD Meshes
Silo User’s Guide 1-5
Figure 1-2: Examples of quadrilateral meshes.
1.6.4. UCD-Based Meshes and Related Data
An unstructured (UCD) mesh is a very general mesh representation; it is
composed of an arbitrary list of zones of arbitrary sizes and shapes. Most
meshes, including quadrilateral ones, can be represented as an unstructured
mesh (Fig. 1-4). Because of their generality, however, unstructured meshes
require more storage space and more complex algorithms.
In UCD meshes, the basic concept of zones (cells) still applies, but there is
no longer an implied connectivity between a zone and its neighbor, as with
the quadrilateral mesh. In other words, given a 2-D quadrilateral mesh zone
accessed by (i, j), one knows that this zone’s neighbors are (i-1,j), (i+1,j), (i,
j-1), and so on. This is not the case with a UCD mesh.
In a UCD mesh, a structure called a zonelist is used to define the nodes
which make up each zone. A UCD mesh need not be composed of zones of
just one shape (Fig. 1-5). Part of the zonelist structure describes the shapes
of the zones in the mesh and a count of how many of each zone shape
occurs in the mesh. The facelist structure is analogous to the zonelist struc-
ture, but defines the nodes which make up each zone face.
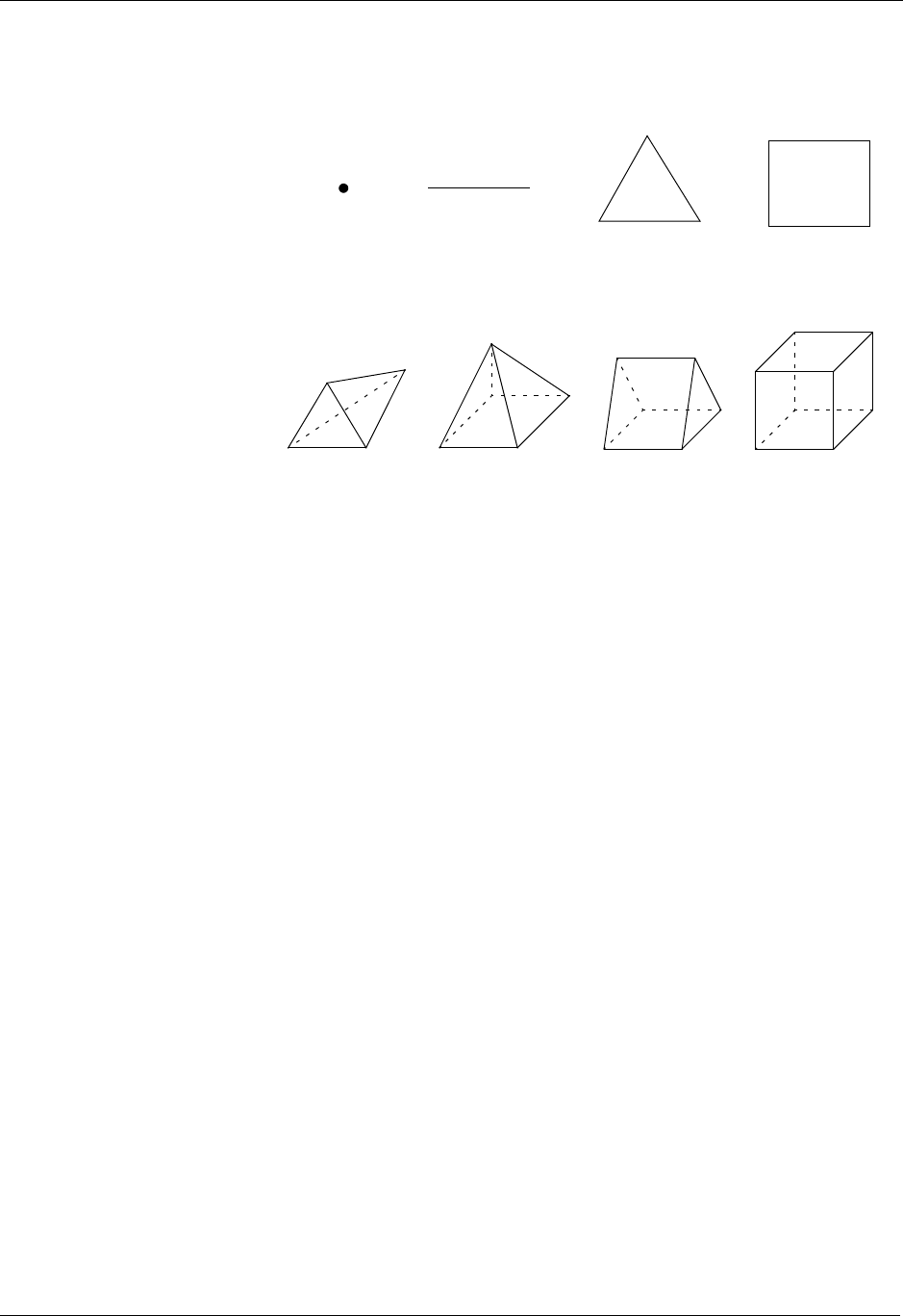
Silo Objects
1-6 Silo User’s Guide
Figure 1-3: Sample 2-D UCD Meshes
Tetrahedron Pyramid Prism Hexahedron
QuadrilateralTriangleLinePoint
Figure 1-4: UCD 2-D and 3-D Cell Shapes
1.6.5. Point Meshes and Related Data
A point mesh consists of a set of locations, or points, in space. This type of
mesh is well suited for representing random scalar data, such as tracer parti-
cles.
1.6.6. Constructive Solid Geometry (CSG) Meshes and Related Data
A constructive Solid Geometry mesh is constructed by boolean combina-
tions of solid model primitives such as spheres, cones, planes and quadric
surfaces. In a CSG mesh, a “zone” is a region defined by such a boolean
combination. CSG meshes support only zone-centered variables.
1.6.7. Block Structured, Adaptive Refinement Meshes (AMR) and Related
Data
Block structured AMR meshes are composed of a large number of Quad
meshes representing refinements of other quad meshes. The hierarchy of
refinement is characterized using a Mesh Region Grouping (MRG) tree.
1.7. Summary of Silo’s Computational Modeling
Objects
Objects are a grouping mechanism for maintaining related variables, dimen-
sions, and other data. The Silo library understands and operates on specific
types of objects including the previously described computational meshes

Silo Objects
Silo User’s Guide 1-7
and related data. The user is also able to define arbitrary objects for storage
of data if the standard Silo objects are not sufficient.
The objects are generalized representations for data commonly found in
physics simulations. These objects include:
Quadmesh A quadrilateral mesh. At a minimum, this must include the dimension
and coordinate data, but typically also includes the mesh’s coordinate
system, labelling and unit information, minimum and maximum
extents, and valid index ranges.
Quadvar A variable associated with a quadrilateral mesh. At a minimum, this
must include the variable’s data, centering information (node-centered
vs. zone centered), and the name of the quad mesh with which this vari-
able is associated. Additional information, such as time, cycle, units,
label, and index ranges can also be included.
Ucdmesh An unstructured mesh1. At a minimum, this must include the dimen-
sion, connectivity, and coordinate data, but typically also includes the
mesh’s coordinate system, labelling and unit information, minimum and
maximum extents, and a list of face indices.
Ucdvar A variable associated with a UCD mesh. This at a minimum must
include the variable’s data, centering information (node-centered vs.
zone-centered), and the name of the UCD mesh with which this variable
is associated. Additional information, such as time, cycle, units, and
label can also be included.
Pointmesh A point mesh. At a minimum, this must include dimension and coordi-
nate data.
Csgmesh A constructive solid geometry (CSG) mesh.
Csgvar A variable defined on a CSG mesh (always zone centered).
Defvar Defined variable representing an arithmetic expression involving other
variables.
Groupel Map Used in concert with an MRG tree to define subsetted regions of
meshes.
Multimat A set of materials. This object contains the names of the materials in the
set.
Multimatspecies A set of material species. This object contains the names of the material
species in the set.
Multimesh A set of meshes. This object contains the names of and types of the
meshes in the set.
Multivar Mesh variable data associated with a multimesh.
1. Unstructured cell data (UCD) is a term commonly used to denote an arbitrarily
connected mesh. Such a mesh is composed of vectors of coordinate values along
with an index array which identifies the nodes associated with each zone and/or
face. Zones may contain any number of nodes for 2-D meshes, and either four,
five, six, or eight nodes for 3-D meshes.

Silo Objects
1-8 Silo User’s Guide
Material Material information. This includes the number of materials present, a
list of valid material identifiers, and a zonal-length array which contains
the material identifiers for each zone.
Material species Extra material information. A material species is a type of a material.
They are used when a given material (i.e. air) may be made up of other
materials (i.e. oxygen, nitrogen) in differing amounts.
MRG Tree Mesh Region Grouping tree used to define various subset regions of
any of Silo’s mesh types.
Zonelist Zone-oriented connectivity information for a UCD mesh. This object
contains a sequential list of nodes which identifies the zones in the
mesh, and arrays which describe the shape(s) of the zones in the mesh.
PHZonelist Arbitrary, polyhedral extension of a zonelist.
Facelist Face-oriented connectivity information for a UCD mesh. This object
contains a sequential list of nodes which identifies the faces in the
mesh, and arrays which describe the shape(s) of the faces in the mesh. It
may optionally include arrays which provide type information for each
face.
Curve X versus Y data. This object must contain at least the domain and range
values, along with the number of points in the curve. In addition, a title,
variable names, labels, and units may be provided.
Variable Array data. This object contains, in addition to the data, the dimensions
and data type of the array. This object is not required to be associated
with a mesh.
1.7.8. Other Silo Objects
In addition to the objects listed in the previous section which are tailored to
the job of representing computational data from scientific computing appli-
cations. Silo supports a number of other objects useful to scientific comput-
ing applications. Some of the more useful ones are briefly summarized here.
Compound Array A compound array is an abstraction of a Fortran common block. It is
also somewhat like a C struct. It is a list of similarly typed by differently
named and sized (usually small in size) items that one often treats as a
group (particularly for I/O purposes).
Directory A silo file can be organized into directories in much the same way as a
UNIX filesystem.
Optlist An “options list” object used to pass additional options to various Silo
API functions.
Simple Variable A simple variable is just a named, multi-dimensional array of arbitrary
data.
User Defined Object A generic, user-defined object or arbitrary nature.

Silo Objects
Silo User’s Guide 1-9
1.8. Silo’s Fortran Interface
The Silo library is implemented in C. Nonetheless, a set of Fortran callable
wrappers have been written to make a majority of Silo’s functionality avail-
able to Fortran applications. These wrappers simply take the data that is
passed through a Fortran function interface, re-package it and call the
equivalent C function. However, there are a few limitations of the Fortran
interface.
1.8.9. Limitations of Fortran Interface
First, it is primarily a write-only interface. This means Fortran applications
can use the interface to write Silo files so that other tools, like VisIt, can
read them. However, for all but a few of Silo’s objects, only the functions
necessary to write the objects to a Silo file have been implemented in the
Fortran interface. This means Fortran applications cannot really use Silo for
restart file purposes.
Conceptually, the Fortran interface is identical to the C interface. To avoid
duplication of documentation, the Fortran interface is documented right
along with the C interface. However, because of differences in C and For-
tran argument passing conventions, there are key differences in the inter-
faces. Here, we use an example to outline the key differences in the
interfaces as well as the rules to be used to construct the Fortran interface
from the C.
1.8.10. Conventions used to construct the Fortran interface from C
In this section, we show an example of a C function in Silo and its equiva-
lent Fortran. We use this example to demonstrate many of the conventions
used to construct the Fortran interface from the C.
We describe these rules so that Fortran user’s can be assured of having up to
date documentation (which tends to always first come for the C interface)
but still be aware of key differences between the two.
A C function specification...
int DBAddRegionArray(DBmrgtree *tree, int nregn, const char **regn_names,
int info_bits, const char *maps_name, int nsegs, int *seg_ids, int *seg_lens,
int *seg_types, DBoptlist *opts)
The equivalent Fortran function...
integer function dbaddregiona(tree_id, nregn, regn_names, lregn_names,
type_info_bits, maps_name, lmaps_name, nsegs, seg_ids, seg_lens, seg_types,
optlist_id, status)
integer tree_id, nregn, lregn_names, type_info_bits, lmaps_name
integer nsegs, optlist_id, status
integer lregn_names(), seg_ids(), seg_lens(), seg_types()
character* maps_name
character*N regn_names

Silo Objects
1-10 Silo User’s Guide
l<strname> Wherever the C interface accepts a char*, the fortran interface accepts
two arguments; the character* argument followed by an integer argu-
ment indicating the string’s length. In the function specifications, it will
always be identified with an ell (‘l’) in front of the name of the charac-
ter* argument that comes before it. In the example above, this rule is
evident in the maps_name and lmaps_name arguments.
l<strname>s Wherever the C interface accepts an array of char* (e.g. char**), the
Fortran interface accepts a character*N followed by an array of lengths
of the strings. In the above example, this rule is evident by the
regn_names and lregn_names arguments. By default, N=32, but
the value for N can be changed, as needed by the dbset2dstrlen()
method.
<object>_id Wherever the C interface accepts a pointer to an abstract Silo object,
like the Silo database file handle (DBfile *) or, as in the example above,
a DBmrgtree*, the Fortran interface accepts an equivalent pointer_id. A
pointer_id is really an integer index into an internally maintained table
of pointers to Silo’s objects. In the above example, this rule is evident in
the tree_id aand optlist_id arguments.
data_ids Wherever the C interface accepts an array of void* (e.g. a void** argu-
ment), the Fortran interface accepts an array of integer pointer_ids. The
Fortran application may use the dbmkptr() function to a create the
pointer ids to populate this array. The above example does not demon-
strate this rule.
status Wherever the C interface returns integer error information in the return
value of the function, the Fortran interface accepts an extra integer
argument named status as the last argument in the list. The above exam-
ple demonstrates this rule.
Finally, there are a few function in Silo’s API that are unique to the Fortran
interface. Those functions are described in the section of the API manual
having to do with Fortran.
1.9. Using Silo in Parallel
Silo is a serial library. Nevertheless, it (as well as the tools that use it like
VisIt) has several features that enable its effective use in parallel with excel-
lent scaling behavior. However, using Silo effectively in parallel does
require an application to store its data to multiple Silo files; typically
between 8 and 64 depending on the number of concurrent I/O channels the
application has available.
The two features that enable Silo to be used effectively in parallel are its
ability to create separate namespaces (directories) within a single file and
the fact that a multi-block object can span multiple Silo files. With these fea-
tures, aparallel application can easily divide its processors into N groups
and write a separate Silo file for each group.

Silo Objects
Silo User’s Guide 1-11
Within a group, each processor in the group writes to its own directory
within the Silo file. One and only one processor has write access to the
group’s Silo file at any one time. So, I/O is serial within a group. However,
because each group has a separate Silo file to write to, each group has one
processor writing concurrently with other processors from other groups. So,
I/O is parallel across groups.
After all processors have created all their individual objects in various
directories within the each group’s Silo file, one processor is designated to
write multi-block objects. The multi-block objects serve as an assembly of
the names of all the individual objects written from various processors.
When N, the number of processor groups, is equal to one, I/O is effectively
serial. All the processors write their data to a single Silo file. When N is
equal to the number of processors, each processor writes its data to its own,
unique Silo file. Both of these extremes are bad for effective and scalable
parallel I/O. A good choice for N is the number of concurrent I/O channels
available to the application when it is actually running. For many parallel,
HPC platforms, this number is typically between 8 and 64.
This technique for using a serial I/O library effectively in parallel while
being able to tune the number of files concurrently being written to is affec-
tionately called Poor Man’s Parallel I/O (PMPIO).
There is a separate header file, pmpio.h, with a set of convenience methods
to support PMPIO-based parallel I/O with Silo. See “Multi-Block Objects,
Parallelism and Poor-Man’s Parallel I/O” on page 154 and See
“PMPIO_Init” on page 181 for more information.

Silo Objects
1-12 Silo User’s Guide

Silo User’s Guide 2-1
Chapter 2 C and Fortran Functions
2.1. C Interface Overview
This chapter documents the C and Fortran interface to the Silo library. The
C header file is “silo.h” and the Fortran header file is “silo.inc”
2.1.1. Optional Arguments
Many Silo functions have optional arguments. By optional, it is meant that a
dummy value can be supplied instead of an actual value. An argument to a
C function which the user does not want to provide, and which is docu-
mented as being optional, should be replaced with a NULL (as defined in
the file silo.h).
2.1.2. Using the Silo Option Parameter
Many of the functions take as one of their arguments a list of option-name/
option-value pairs. In this way additional information can be passed to a
function without having to change the function's interface. The following
sequence of function declarations outlines the procedure for creating and
populating such a list:
DBoptlist *DBMakeOptlist (int maxopts) /* Create a list with
maximum list length */
int DBAddOption ( /* Add an option to the list: */
DBoptlist *optlist, /* the list, */
int option_id, /* the option, */
void *option_value /* the option's value */
)

C Interface Overview
2-2 Silo User’s Guide
2.1.3. C Calling Sequence
The functions in the Silo output package should be called in a particular
order.
2.1.3.1. Write Sequence
Start by creating a Silo file, with DBCreate(), create any necessary directo-
ries, then call the remaining routines as needed for writing out the mesh,
material data, and any physics variables associated with the mesh.
Schematically, your program should look something like this:
DBCreate
DBMkdir
DBSetDir
DBPutQuadmesh
DBPutQuadvar1
DBPutQuadvar1
. . .
DBSetDir
DBMkdir
DBSetDir
DBPutZonelist
DBPutFacelist
DBPutUcdmesh
DBPutMaterial
DBPutUcdvar1
. . .
DBSetDir
DBClose
2.1.3.2. Example of C Calling Sequence for writing
The following C code is an example of the creation of a Silo file with just
one directory (the root):
#include <silo.h>
#include <string.h>
int main()
{
DBfile *file = NULL; /* The Silo file pointer */
char *coordnames[2]; /* Names of the coordinates */
float nodex[4]; /* The coordinate arrays */
float nodey[4];
float *coordinates[2]; /* The array of coordinate
arrays */
int dimensions[2]; /* The number of nodes in
each dimension */
/* Create the Silo file */

C Interface Overview
Silo User’s Guide 2-3
file = DBCreate(“sample.silo”, DB_CLOBBER, DB_LOCAL, NULL,
DB_PDB);
/* Name the coordinate axes ‘X’ and ‘Y’ */
coordnames[0] = strdup(“X”);
coordnames[1] = strdup(“Y”);
/* Give the x coordinates of the mesh */
nodex[0] = -1.1;
nodex[1] = -0.1;
nodex[2] = 1.3;
nodex[3] = 1.7;
/* Give the y coordinates of the mesh */
nodey[0] = -2.4;
nodey[1] = -1.2;
nodey[2] = 0.4;
nodey[3] = 0.8;
/* How many nodes in each direction? */
dimensions[0] = 4;
dimensions[1] = 4;
/* Assign coordinates to coordinates array */
coordinates[0] = nodex;
coordinates[1] = nodey;
/* Write out the mesh to the file */
DBPutQuadmesh(file, “mesh1”, coordnames, coordinates,
dimensions, 2, DB_FLOAT, DB_COLLINEAR, NULL);
/* Close the Silo file */
DBClose(file);
return (0);
}
2.1.3.3. Read Sequence
Start by opening the Silo file with DBOpen(), then change to the required
directory, and then read the mesh, material, and variables. Schematically,
your program should look something like this:
DBOpen
DBSetDir
DBGetQuadmesh
DBGetQuadvar1
DBGetQuadvar1
. . .

C Interface Overview
2-4 Silo User’s Guide
DBSetDir
DBGetUcdmesh
DBGetUcdvar1
DBGetMaterial
. . .
DBClose
2.2. Fortran Interface
Currently, C-callable functions exist for all routines, but Fortran-callable
functions exist for only a portion of the routines. The Fortran header file is
“silo.inc”.
2.2.4. Optional Arguments
The functions described below have optional arguments. By optional, it is
meant that a dummy value can be supplied instead of an actual value. An
argument to a Fortran function, which the user does not want to provide,
and which is documented as optional, should be replaced with the parameter
DB_F77NULL, which is defined in the file silo.inc.
2.2.5. Using the Silo Option Parameter
Many of the functions take as one of their arguments a list of option-name/
option-value pairs. In this way, additional information can be passed to a
function without having to change the function’s interface. The following
sequence of function declarations outlines the procedure for creating and
populating such a list:
integer function dbmkoptlist( ! Create a list:
maxopts, ! maximum list length
optlist_id ! list identifier
)
integer function dbaddiopt ( ! Add an integer option
! to the list:
optlist_id, ! the list
option_id, ! the option
int_value ! the option’s integer
! value
)
There also are functions for adding real and character option values to a list.
2.2.6. Fortran Calling Sequence
The functions in the Silo output package should be called in a particular
order. Start by creating a Silo file, with dbcreate(), create any necessary
directories, then call the remaining routines as needed for writing out the
mesh, material data, and any physics variables associated with the mesh.
Schematically, your program should look something like this:

C Interface Overview
Silo User’s Guide 2-5
dbcreate
dbmkdir
dbsetdir
dbputqm
dbputqv1
dbputqv1
dbputqv1
. . .
dbsetdir
dbmkdir
dbsetdir
dbputzl
dbputfl
dbputum
dbputmat
dbputuv1
. . .
dbsetdir
dbclose
2.3. Reading Silo Files
Silo functions that return Silo objects from an open file return a C struct
data structure defining the object. The most reliable source of information
on the C structure returned from each call is the silo header file, silo.h. For
reference, the header file for this version of Silo is attached as an appendix
to this manual.

C Interface Overview
2-6 Silo User’s Guide
1
Error Handling and Other Global Library Behavior..........................................7
DBErrfuncname . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
DBErrno . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
DBErrString . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
DBShowErrors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11
DBErrlvl . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
DBErrfunc. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 13
DBVariableNameValid. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
DBVersion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15
DBVersionDigits . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16
DBVersionGE. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
DBSetAllowOverwrites . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 18
DBGetAllowOverwrites . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 19
DBSetAllowEmptyObjects. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20
DBGetAllowEmptyObjects . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 21
DBForceSingle . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
DBGetDatatypeString. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 23
DBSetDataReadMask2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 24
DBGetDataReadMask2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 26
DBSetEnableChecksums . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27
DBGetEnableChecksums . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 28
DBSetCompression. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29
DBGetCompression . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 32
DBSetFriendlyHDF5Names . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33
DBGetFriendlyHDF5Names . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 34
DBSetDeprecateWarnings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
DBGetDeprecateWarnings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 36
DB_VERSION_GE . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 37
Files and File Structure.........................................................................................38
DBRegisterFileOptionsSet . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 40
DBUnregisterFileOptionsSet . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 44
DBUnregisterAllFileOptionsSets . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 45
DBSetUnknownDriverPriorities. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 46
DBGetUnknownDriverPriorities . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47
DBCreate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 48
DBOpen . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 53
DBClose . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 55
DBGetToc. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 56
DBFileVersion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 57
DBFileVersionDigits . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 58
DBFileVersionGE. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 59
DBVersionGEFileVersion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 60
DBSortObjectsByOffset . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 61
2
DBMkDir . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 62
DBSetDir . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 63
DBGetDir . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 64
DBCpDir. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 65
DBGrabDriver . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 66
DBUngrabDriver . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 67
DBGetDriverType . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 68
DBGetDriverTypeFromPath. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 69
DBInqFile . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 70
DBInqFileHasObjects. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 71
_silolibinfo . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 72
_hdf5libinfo . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 73
_was_grabbed . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 74
Meshes, Variables and Materials.........................................................................75
DBPutCurve . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 77
DBGetCurve . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 79
DBPutPointmesh. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 80
DBGetPointmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83
DBPutPointvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
DBPutPointvar1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86
DBGetPointvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 87
DBPutQuadmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 88
DBGetQuadmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 91
DBPutQuadvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 92
DBPutQuadvar1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 96
DBGetQuadvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 98
DBPutUcdmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
DBPutUcdsubmesh. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 107
DBGetUcdmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 108
DBPutZonelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 109
DBPutZonelist2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 110
DBPutPHZonelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 112
DBGetPHZonelist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 116
DBPutFacelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 117
DBPutUcdvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 119
DBPutUcdvar1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 122
DBGetUcdvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 124
DBPutCsgmesh. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 125
DBGetCsgmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 130
DBPutCSGZonelist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 131
DBGetCSGZonelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 136
DBPutCsgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 137
DBGetCsgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 139

3
DBPutMaterial . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 140
DBGetMaterial . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 144
DBPutMatspecies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 145
DBGetMatspecies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 148
DBPutDefvars. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 149
DBGetDefvars . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 151
DBInqMeshname . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 152
DBInqMeshtype . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 153
Multi-Block Objects, Parallelism and
Poor-Man’s Parallel I/O .....................................................................................154
DBPutMultimesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 156
DBGetMultimesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 161
DBPutMultimeshadj . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 162
DBGetMultimeshadj. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 165
DBPutMultivar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 166
DBGetMultivar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 170
DBPutMultimat. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 171
DBGetMultimat . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 174
DBPutMultimatspecies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 175
DBGetMultimatspecies. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 178
DBOpenByBcast. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 179
PMPIO_Init. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 181
PMPIO_CreateFileCallBack. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 184
PMPIO_OpenFileCallBack . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 185
PMPIO_CloseFileCallBack . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 186
PMPIO_WaitForBaton . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 187
PMPIO_HandOffBaton . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 188
PMPIO_Finish . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 189
PMPIO_GroupRank . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 190
PMPIO_RankInGroup . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 191
Part Assemblies, AMR, Slide Surfaces,
Nodesets and Other Arbitrary Mesh Subsets ...................................................192
DBMakeMrgtree. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 193
DBAddRegion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 197
DBAddRegionArray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 199
DBSetCwr. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 201
DBGetCwr . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 202
DBPutMrgtree. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 203
DBGetMrgtree . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 204
DBFreeMrgtree. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 205
4
DBMakeNamescheme . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 206
DBGetName . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 210
DBPutMrgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 211
DBGetMrgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 213
DBPutGroupelmap . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 214
DBGetGroupelmap . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 216
DBFreeGroupelmap . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 217
DBOPT_REGION_PNAMES . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 218
Object Allocation, Free and IsEmpty................................................................220
DBAlloc… . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 221
DBFree… . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 222
DBIsEmpty . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 223
Calculational and Utility.....................................................................................224
DBCalcExternalFacelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 225
DBCalcExternalFacelist2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 227
DBStringArrayToStringList . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 229
DBStringListToStringArray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 230
Optlists..................................................................................................................231
DBMakeOptlist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 232
DBAddOption. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 233
DBClearOption . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 234
DBGetOption . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 235
DBFreeOptlist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 236
DBClearOptlist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 237
User Defined (Generic) Data and Objects.........................................................238
DBWrite . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 239
DBWriteSlice . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 240
DBReadVar. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 242
DBReadVarSlice. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 243
DBGetVar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 244
DBInqVarExists . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 245
DBInqVarType . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 246
DBGetVarByteLength . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 248
DBGetVarDims . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 249
DBGetVarLength . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 250
5
DBGetVarType. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 251
DBPutCompoundarray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 252
DBInqCompoundarray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 253
DBGetCompoundarray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 254
DBMakeObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 255
DBFreeObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 256
DBChangeObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 257
DBClearObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 258
DBAddDblComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 259
DBAddFltComponent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 260
DBAddIntComponent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 261
DBAddStrComponent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 262
DBAddVarComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 263
DBWriteComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 264
DBWriteObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 265
DBGetObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 266
DBGetComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 267
DBGetComponentType . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 268
JSON Interface to Silo Objects ..........................................................................269
json-c extensions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 271
DBWriteJsonObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 272
DBGetJsonObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 273
Previously Undocumented Use Conventions ....................................................274
_visit_defvars . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 275
_visit_searchpath . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 276
_visit_domain_groups. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 277
AlphabetizeVariables . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 278
ConnectivityIsTimeVarying . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 279
MultivarToMultimeshMap_vars. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 280
MultivarToMultimeshMap_meshes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 281
Silo’s Fortran Interface ......................................................................................282
dbmkptr. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 283
dbrmptr . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 284
dbset2dstrlen . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 285
dbget2dstrlen. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 286
DBFortranAllocPointer. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 287
DBFortranAccessPointer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 288
DBFortranRemovePointer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 289
6
dbwrtfl. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 290
Deprecated Functions .........................................................................................291
Silo Library Header File.....................................................................................292

Silo User’s Guide 2-7
1 API Section Error Handling and Other Global Library
Behavior
The functions described in this section of the Silo Application Programming Interface (API) man-
ual, are those that effect behavior of the library, globally, for any file(s) that are or will be open.
These include such things as error handling, requiring Silo to do extra work to warn of and avoid
overwrites, to compute and warn of checksum errors and to compress data before writing it to disk.
The functions described here are...
DBErrfuncname . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 8
DBErrno . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
DBErrString . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 10
DBShowErrors . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 11
DBErrlvl . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 12
DBErrfunc. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 13
DBVariableNameValid. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
DBVersion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 15
DBVersionDigits . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 16
DBVersionGE. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
DBSetAllowOverwrites . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 18
DBGetAllowOverwrites . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 19
DBSetAllowEmptyObjects. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20
DBGetAllowEmptyObjects . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 21
DBForceSingle . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
DBGetDatatypeString. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 23
DBSetDataReadMask2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 24
DBGetDataReadMask2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 26
DBSetEnableChecksums . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 27
DBGetEnableChecksums . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 28
DBSetCompression. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 29
DBGetCompression . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 32
DBSetFriendlyHDF5Names . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 33
DBGetFriendlyHDF5Names . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 34
DBSetDeprecateWarnings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 35
DBGetDeprecateWarnings . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 36
DB_VERSION_GE . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 37

DBErrFunc
2-8 Silo User’s Guide
DBErrfuncname—Get name of error-generating function
Synopsis:
char const *DBErrfuncname (void)
Fortran Equivalent:
None
Returns:
DBErrfuncname returns a char const * containing the name of the function that generated the
last error. It cannot fail.
Description:
The DBErrfuncname function is used to find the name of the function that generated the last Silo
error. It is implemented as a macro. The returned pointer points into Silo private space and must
not be modified or freed.

DBErrno
Silo User’s Guide 2-9
DBErrno—Get internal error number.
Synopsis:
int DBErrno (void)
Fortran Equivalent:
integer function dberrno()
Returns:
DBErrno returns the internal error number of the last error. It cannot fail.
Description:
The DBErrno function is used to find the number of the last Silo error message. It is implemented
as a macro. The error numbers are not guaranteed to remain the same between different release
versions of Silo.

DBErrString
2-10 Silo User’s Guide
DBErrString—Get error message.
Synopsis:
char const *DBErrString (void)
Fortran Equivalent:
None
Returns:
DBErrString returns a char const * containing the last error message. It cannot fail.
Description:
The DBErrString function is used to find the last Silo error message. It is implemented as a macro.
The returned pointer points into Silo private space and must not be modified or freed.
DBShowErrors
Silo User’s Guide 2-11
DBShowErrors—Set the error reporting mode.
Synopsis:
void DBShowErrors (int level, void (*func)(char*))
Fortran Equivalent:
integer function dbshowerrors(level)
Arguments:
level Error reporting level. One of DB_ALL, DB_ABORT, DB_TOP, or DB_NONE.
func Function pointer to an error-handling function.
Returns:
DBShowErrors returns nothing (void). It cannot fail.
Description:
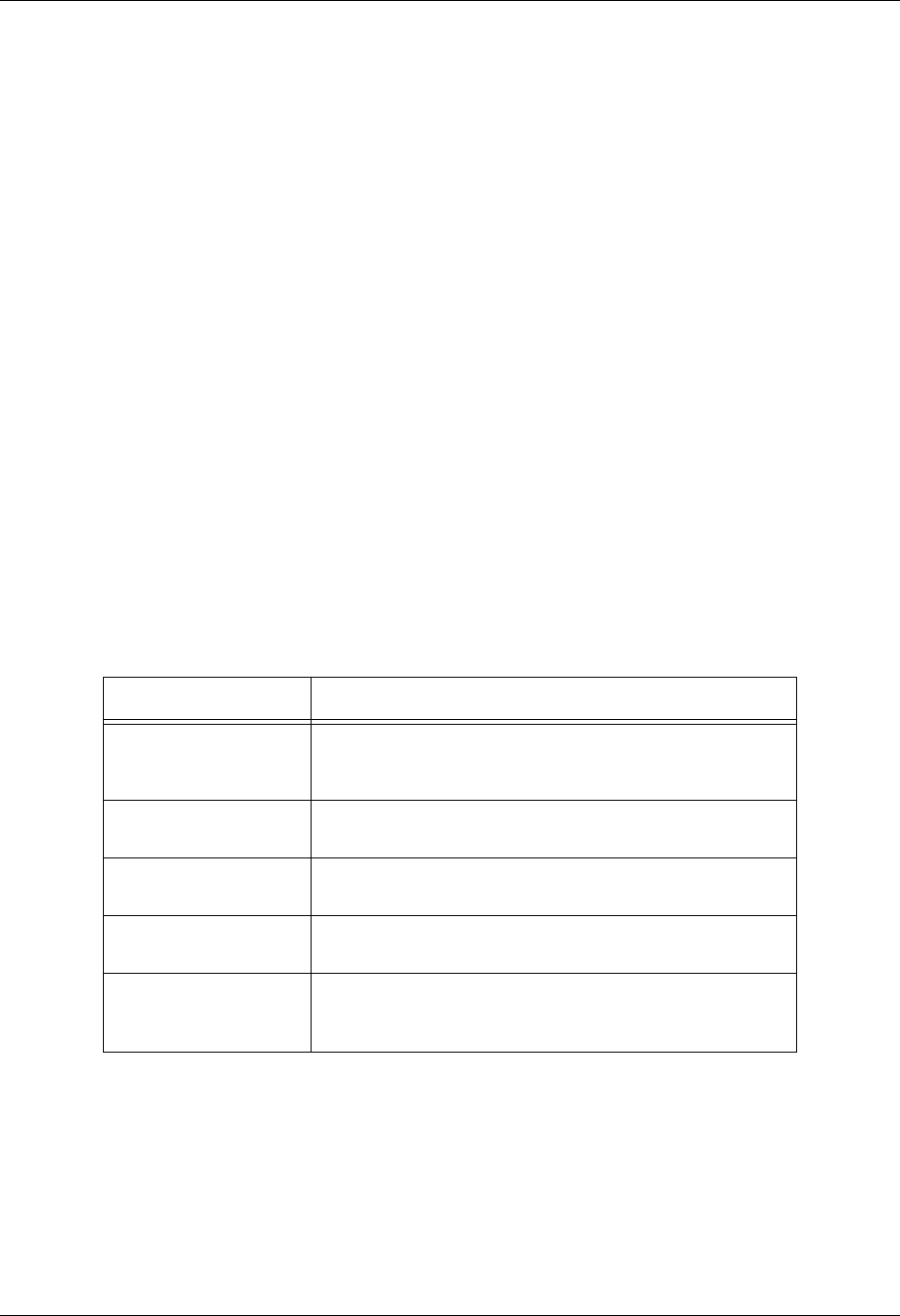
The DBShowErrors function sets the level of error reporting done by Silo when it encounters an
error. The following table describes the action taken upon error for different values of level.
Ordinarily, error reporting from the HDF5 library is disabled. However, DBShowErrors also influ-
ences the behavior of error reporting from the HDF5 library.
Error level value Error action
DB_ALL Show all errors, beginning with the (possibly internal) routine
that first detected the error and continuing up the call stack
to the application.
DB_ALL_AND_DRVR Same as DB_ALL execpt also show error messages gener-
ated by the underlying driver library (PDB or HDF5).
DB_ABORT Same as DB_ALL except abort is called after the error mes-
sage is printed.
DB_TOP (Default) Only the top-level Silo functions issue error mes-
sages.
DB_NONE The library does not handle error messages. The application
is responsible for checking the return values of the Silo func-
tions and handling the error.

DBShowErrors
2-12 Silo User’s Guide
DBErrlvl—Return current error level setting of the library
Synopsis:
int DBErrlvl(void)
Fortran Equivalent:
int dberrlvl()
Returns:
Returns current error level of the library. Cannot fail.

DBShowErrors
Silo User’s Guide 2-13
DBErrfunc—Get current error function set by DBShowErrors()
Synopsis:
void (*func)(char*) DBErrfunc(void);
Fortran Equivalent:
None
Description:
Returns the function pointer of the current error function set in the most recent previous call to
DBShowErrors().

DBVariableNameValid
2-14 Silo User’s Guide
DBVariableNameValid—check if character string represents a valid Silo variable name
Synopsis:
int DBValidVariableName(char const *s)
Fortran Equivalent:
None
Arguments:
sThe character string to check
Returns:
non-zero if the given character string represents a valid Silo variable name; zero otherwise
Description:
This is a convenience function for Silo applications to check whether a given variable name they
wish to use will be considered valid by Silo.
The only valid characters that can appear in a Silo variable name are all alphanumerics (e.g. [a-zA-
Z0-9]) and the underscore (e.g. ‘_’). If a candidate variable name contains any characters other
than these, that variable name is considered invalid. If that variable name is ever used in a call to
create an object in a Silo file, the call will fail with error E_INVALIDNAME.

DBVersion
Silo User’s Guide 2-15
DBVersion—Get the version of the Silo library.
Synopsis:
char const *DBVersion (void)
Fortran Equivalent:
None
Returns:
DBVersion returns the version as a character string.
Description:
The DBVersion function determines what version of the Silo library is being used and returns that
version in string form. The returned string should NOT be free’d by the caller.

DBVersionDigits
2-16 Silo User’s Guide
DBVersionDigits—Return the integer version digits of the library
Synopsis:
int DBVersionDigits(int *Maj, int *Min, int *Pat, int *Pre);
Fortran Equivalent:
None
Arguments:
Maj Pointer to returned major version digit
Min Pointer to returned minor version digit
Pat Pointer to returned patch version digit
Pre Pointer to returned pre-release version digit (if any)
Returns:
Returns 0 on success, -1 on failure..

DBVersionGE
Silo User’s Guide 2-17
DBVersionGE—Greater than or equal comparison for version of the Silo library
Synopsis:
int DBVersionGE(int Maj, int Min, int Pat)
Fortran Equivalent:
None
Arguments:
Maj Integer, major version number
Min Integer, minor version number
Pat Integer, patch version number
Returns:
One (1) if the library’s version number is greater than or equal to the version number specified by
Maj, Min, Pat arguments, zero (0) otherwise.
Description:
This function is the run-time equivalent of the DB_VERSION_GE macro.

DBSetAllowOverwrites
2-18 Silo User’s Guide
DBSetAllowOverwrites—Allow library to over-write existing objects in Silo files
Synopsis:
int DBSetAllowOverwrites(int allow)
Fortran Equivalent:
integer function dbsetovrwrt(allow)
Arguments:
allow Integer value controlling the Silo library’s overwrite behavior. A non-zero value
sets the Silo library to permit overwrites of existing objects. A zero value
disables overwrites. By default, Silo does NOT permit overwrites.
Returns:
Returns the previous setting of the value.
Description:
By default, Silo does not permit a caller to over-write existing objects in a Silo file. This is because
this kind of operation can often lead to corrupted files, particularly if the new object’s data does
not fit within the existing object’s space in the file.
However, there are often cases where a caller can ensure that the new object is the same size or
smaller and would like to over-write an existing object.

DBGetAllowOverwrites
Silo User’s Guide 2-19
DBGetAllowOverwrites—Get current setting for the allow overwrites flag
Synopsis:
int DBGetAllowOverwrites(void)
Fortran Equivalent:
integer function dbgetovrwrt()
Returns:
Returns the current setting for the allow overwrites flag
Description:
See DBSetAllowOverwrites for a description of the meaning of this flag

DBSetAllowEmptyObjects
2-20 Silo User’s Guide
DBSetAllowEmptyObjects—Permit the creation of empty silo objects
Synopsis:
int DBSetAllowEmptyObjects(int allow)
Fortran Equivalent:
integer function dbsetemptyok(allow)
Arguments:
allow Integer value indicating whether or not empty objects should be allowed to be
created in Silo files. A zero value prevents callers from creating empty objects
in Silo files. A non-zero value permits it. By default, the Silo library does NOT
permit callers to create empty objects.
Returns:
The previous setting of this value is returned.
Description:
For a long time, the “EMPTY” keyword convention (see “DBPutMultimesh” on page 156) was
sufficient for dealing with cases where callers needed to create multiple, related multi-block
objects with missing blocks. In fact, in many cases this convention was sufficient for combining
variables which by and large existed on different collections of blocks on a common multi-block
mesh.
More recently, the need has arisen for the Silo library to permit callers to instantiate within Silo
files “empty” objects; that is Silo objects with no problem-sized data associated with them. For
example, a point mesh with no points or a ucd variable with no variable arrays. This requirement
has been driven by the need to scale to larger problems and the use of nameschemes (see “DBMak-
eNamescheme” on page 206) in combination with meshes and variables with missing blocks.
Historically, such an operation has been considered an error by the Silo library and prevented.But,
that has been largely an overly cautious restriction in Silo to avert anticipated and not necessarily
any real problems. DBSetAllowEmptyObjects with a non-zero argument enables the Silo library to
by-pass these checks.

DBGetAllowEmptyObjects
Silo User’s Guide 2-21
DBGetAllowEmptyObjects—Get current setting for the allow empty objects flag
Synopsis:
int DBGetAllowEmptyobjets(void)
Fortran Equivalent:
integer function dbgetemptyok()
Arguments:
None
Description:
Get the current library setting for the allow empty objects flag.

DBForceSingle
2-22 Silo User’s Guide
DBForceSingle—Convert all datatype’d data read in read methods to type float
Synopsis:
int DBForceSingle(int force)
Fortran Equivalent:
None
Arguments:
force Flag to indicate if forcing should be set or not. Pass non-zero to force single
precision. Pass zero to NOT force single precision.
Returns:
Zero on success. -1 on failure
Description:
This setting is global to the whole library and effects subsequent read operations.
If force is non-zero, then any datatype'd arrays are converted on read from whatever their native
datatype is to float. A datatype'd array is an array that is part of some Silo object struct containing
a datatype member which indicates the type of data in the array. For example, a DBucdvar
struct has a datatype member to indicate the type of data in its var and mixvar arrays. Such
arrays will be converted on read if force here is non-zero. However, a DBmaterial object
struct is ALWAYS integer data. There is no datatype member for such an object and so its data
will NEVER be converted to float on read regardless of the force single status set here.
This function's original intention may have been to convert ONLY double precision arrays to sin-
gle precision. However, the PDB driver was apparently never designed that way and the PDB
driver's behavior sort of established the defacto meaning of DBForceSingle. Consequently, as
of Silo version 4.8 the HDF5 driver obeys these same semantics as well. Though, in fact the HDF5
driver was written to support the original intention of DBForceSingle and it worked in this
(buggy) fashion for many years before real problems with it were encountered.
This method is typically used by downstream, post-processing tools to reduce memory require-
ments. By default, Silo DOES NOT have single precision forcing enabled. When it is enabled,
only the methods that result in reading of floating point data from a Silo file are effected. Finally,
note that write methods are NOT effected.

DBForceSingle
Silo User’s Guide 2-23
DBGetDatatypeString—Return a string name for a given Silo datatype
Synopsis:
char *DBGetDatatypeString(int datatype)
Fortran Equivalent:
None
Arguments:
datatype One of the Silo datatypes (e.g. DB_INT, DB_FLOAT, DB_DOUBLE, etc.)
Returns:
A pointer to a newly allocated string representing the data type name. The caller must free the
returned string.
Description:
Obtain the string name of a given Silo datatype.

DBSetDataReadMask2
2-24 Silo User’s Guide
DBSetDataReadMask2—Set the data read mask
Synopsis:
unsigned long long DBSetDataReadMask2 (unsigned long long mask)
Fortran Equivalent:
None
Arguments:
mask The mask to use to read data. This is a bit vector of values that define whether
each data portion of the various Silo objects should be read.
Returns:
DBSetDataReadMask2 returns the previous data read mask.
Description:
DBSetDataReadMask2 replaces the now obsolete DBSetDataReadMask.
The DBSetDataReadMask2 allows the user to set the mask that’s used to read various large data
components within Silo objects.
Most Silo objects have a metadata portion and a data portion. The data portion is that part of the
object that consists of pointers to long arrays of data. These arrays are typically “problem sized”
but in any event require additional I/O to read. By default, the read mask is set to DBAll.
Setting the data read mask allows for a DBGet* call to return only part of the associated object’s
data. With the data read mask set to DBAll, the DBGet* functions return all of the information.
With the data read mask set to DBNone, they return only the metadata. The mask is a bit vector
specifying which part of the data model should be read.
A special case is found in the DBCalc flag. Sometimes data is not stored in the file, but is instead
calculated from other information. The DBCalc flag controls this behavior. If it is turned off, the
data is not calculated. If it is turned on, the data is calculated.
The values that DBSetDataReadMask takes as the mask parameter are binary-or’ed combinations
of the values shown in the following table:
Mask bit Meaning
DBAll All data values are read. This value is identical to specifying all of the
other mask bits or’ed together, setting all of the bit values to 1.
DBNone No data values are read. This value sets all of the bit values to 0.
DBCalc If data is calculable, calculate it. Otherwise, return NULL for that part.
DBMatMatnos Material numbers (matnos) read by DBGetMaterial.
DBMatMatnames Material names (matnames) read by DBGetMaterial.
DBSetDataReadMask2
Silo User’s Guide 2-25
Use the DBGetDataReadMask2 call to retrieve the current data read mask without setting one.
By default, the data read mask is set to DBAll. The data read mask effects only the read portion of
the Silo API.
DBMatMatlist Zone-by-zone material list read by DBGetMaterial.
DBMatMixList Mixed material information read by DBGetMaterial.
DBCurveArrays Data values of curves read by DBGetCurve.
DBPMCoords Coordinate arrays read by DBGetPointmesh.
DBPVData Var data arrays read by DBGetPointvar.
DBQMCoords Coordinate arrays read by DBGetQuadmesh.
DBQVData Var data arrays read by DBGetQuadvar.
DBUMCoords Coordinate arrays read by DBGetUcdmesh.
DBUMFacelist Facelists of UCD meshes read by DBGetUcdmesh.
DBUMZonelist Zonelists of UCD meshes read by DBGetUcdmesh.
DBUVData Var data arrays read by DBGetUcdvar.
DBFacelistInfo Nodelists and shape info read by DBGetFacelist.
DBZonelistInfo Nodelist and shape info read by DBGetZonelist.
DBUMGlobNodeNo Global node numbers read by DBGetUcdmesh
DBZonelistGlobZoneNo Global zone numbers read by DBGetUcdmesh
DBMatMatcolors Material colors read by DBGetMaterial and DBGetMultimat
DBMMADJNodelists Adjacency nodelists read by DBGetMultimeshadj
DBMMADJZonelists Adjacency zonelists read by DBGetMultimeshadj
DBCSGMBoundaryInfo Boundary list read by DBGetCsgmesh
DBCSGMZonelist Zonelist read by DBGetCsgmesh
DBCSGMBoundaryNames Boundary names read by DBGetCsgmesh
DBCSGVData Var data arrays read by DBGetCsgvar
DBCSGZonelistZoneNames Zone names read by DBGetCSGZonelist
DBCSGZonelistRegNames Region names read by DBGetCSGZonelist
DBPMGlobNodeNo Global node numbers read by DBGetPointmesh
DBPMGhostNodeLabels Ghost node labels read by DBGetPointmesh
DBQMGhostNodeLabels Ghost node labels read by DBGetQuadmesh
DBQMGhostZoneLabels Ghost zone lables read by DBGetQuadmesh
DBUMGhostNodeLabels Ghost node lables read by DBGetUcdmesh
DBZonelistGhostZoneLabels Ghost zone lables read by DBGetUcdmesh and/or DBGetZonelist
Mask bit Meaning

DBGetDataReadMask2
2-26 Silo User’s Guide
DBGetDataReadMask2—Get the current data read mask
Synopsis:
unsigned long long DBGetDataReadMask2 (void)
Fortran Equivalent:
None
Returns:
DBGetDataReadMask2 returns the current data read mask.
Description:
Note that DBGetDataReadMask2 replaces the now obsolete DBGetDataReadMask.
The DBGetDataReadMask2 allows the user to find out what mask is currently being used to read
the data within Silo objects.
See the documentation on DBSetDataReadMask2 for a complete description.

DBSetEnableChecksums
Silo User’s Guide 2-27
DBSetEnableChecksums—Set flag controlling checksum checks
Synopsis:
int DBSetEnableChecksums(int enable)
Fortran Equivalent:
integer function dbsetcksums(enable)
Arguments:
enable Integer value controlling checksum behavior of the Silo library. See description
for a complete explanation.
Returns:
Returns the previous setting for checksum behavior.
Description:
If checksums are enabled, whenever Silo writes data, it will compute checksums on the data in
memory and store these checksums with the data in the file. Note that during a write call, in no cir-
cumstance will Silo re-read data written to confirm it was written correctly (e.g. it gets back what
it wrote). In other words, Silo will not detect checksum errors on writes. It will detect checksum
errors only on reads, only if checksums were actually computed and stored with the data when it
was written and only when checksums are indeed enabled.
If checksums are enabled, whenever Silo reads data AND the data it is reading has checksums
stored in the file, it will compute and compare checksums. If the checksums computed on read do
not agree with the checksums stored in the file, the Silo call resulting in the data read will fail. The
error, E_CHECKSUM, will be set (See “DBShowErrors” on page 2-11). Note that because check-
sums are not checked on write, there is no foolproof way to detect whether a read has failed
because the data was corrupted when it was originally written or because the read itself has failed.
Checksum checks are supported ONLY on the HDF5 driver. The PDB driver DOES NOT support
checksum checks. Calling DBCreate() with checksumming enabled will fail if DB_PDB is
specified as the driver. If checksumming is enabled while any PDB file is opened, the request for
checksumming will be silently ignored by all attempts to write or read data from a PDB file.
In the HDF5 driver, only the data that winds up in HDF5 datasets in the file is checksumed. In
most applications, this represents more than 99% of all the data the client writes. However, it is
important to note that when checksuming is enabled, NOT ALL data written by Silo is check-
sumed. Various bits of metadata is not checksumed.
Finally, empirical results show that the resulting files are 1-5% larger and take about 1-5% longer
to write when checksumming is enabled. This is due primarily to the fact that a different class of
HDF5 dataset, called a chunked dataset, is required in order to enable checksumming.

DBGetEnableChecksums
2-28 Silo User’s Guide
DBGetEnableChecksums—Get current state of flag controlling checksumming
Synopsis:
int DBGetEnableChecksums(void)
Fortran Equivalent:
integer function dbgetcksums()
Returns:
Zero if checksumming is not currently enabled. Non-zero if checksumming is currently enabled.
Description:
This function returns the current setting for the library-global flag controlling checksumming
behavior.

DBSetCompression
Silo User’s Guide 2-29
DBSetCompression—Set compression options for succeeding writes of Silo data
Synopsis:n
int DBSetCompression(char const *options)
Fortran Equivalent:
integer function dbsetcompress(options, loptions)
Arguments:
options Character string containing the name of the compression method and various
parameters. The method set using the keyword, “METHOD=”. Any remaining
parameters are dependent on the compression method and are described below.
Returns:
Returns the previous value set for compression behavior.
Description:
Compression is currently supported only on the HDF5 driver.
Note that the responsibility for enabling compression falls only on the data producer. Any Silo cli-
ents attempting to read compressed data may do so without concern for whether the data in the file
is compressed or not. If the data is compressed, decompression will occur automatically during
read. This is true as long as the Silo library to which the client reading the data was compiled and
linked has the necessary decompression code. Because writer and reader need not be compiled and
linked to the same exact Silo library installation, each could be compiled with differing compres-
sion capabilities making it impossible to read data in some situations.
To the extent possible, the public installations of Silo on LLNL systems have all been enabled with
compatible compression features. However, because many application developers have taken to
creating their own installations of Silo, it is important to consider the effect of disabling (or
enabling) various compression features.
Compression features are controlled by an arbitrary string, whose contents are described in more
detail below. By default, the Silo library does not have compression enabled. A number of differ-
ent compression techniques are available. Some operate without regard to the type of data and
mesh being written. Others depend on the type of data and sometimes even the type of mesh.
Compression parameters global to all compression methods: There are two global parameters that
control behavior of compression algorithms. These must appear in the compression options string
before any compression-specific parameters.
The first is the error mode (“ERRMODE=<word>” which controls how the Silo library responds
when it encounters an error during compression and/or is unable to compress the data. The two
options are “FALLBACK” or “FAIL”. Including “ERRMODE=FALLBACK” in the compression
options string tells Silo that whenever compression fails, it should simply fallback to writing
uncompressed data. Including “ERRMODE=FAIL” in the compression options string tells Silo to
fail the write and return E_COMPRESSION error for the operation.

DBSetCompression
2-30 Silo User’s Guide
The second is the minimum compression ratio to be achieved by compressing the data. It is speci-
fied as “MINRATIO=<float>”. For example, including “MINRATIO=2.5” in the compression
options string tells Silo that all data must be compressed by at least a factor of 2.5:1. If it is unable
the compress by at least this amount, Silo will either fallback or fail the write depending on the
ERRMODE setting.
The remaining paragraphs describe compression algorithm specific options.
GZIP compression: is enabled using “METHOD=GZIP” in the options string. GZIP recognizes
the LEVEL=<int>, compression parameter. The compression level is an integer from 0 to 9, where
0 results in the fastest compression performance but at the expense of lower compression ratios.
Likewise, a level of 9 results in the slowest compression performance but with possibly better
compression ratios. If the “LEVEL=<int>” keyword does not appear in the options string or speci-
fies invalid values, the default is level one (1). The GZIP method of compression is applied inde-
pendently to float and integer data for all types of meshes and variables. It is also guaranteed to be
available to all Silo clients.
SZIP compression: is enabled using “METHOD=SZIP” in the options string. The SZIP compres-
sion algorithm is designed specifically for scientific data. SZIP recognizes the BLOCK=<int>, and
MASK={EC|NN} parameters. The BLOCK=<int>, takes an integer value from 0 to 32, which is a
blocking size and must be even and not greater than 32, with typical values being 8, 10, 16, or 32.
This parameter affects the compression ratio; the more values vary, the smaller this number should
be to achieve better performance. The MASK=EC, selects entropy coding method, this is best
suited for data that has been processed, working best for small numbers.MASK=NN, selects the
nearest neighbor coding method, preprocesses the data then applies the EC method as above. The
default parameters for SZIP compression are “METHOD=SZIP BLOCK=4 MASK=NN”. If in a
subsequent write operation (DBPutXXX, DBWrite, etc.) the value for BLOCK is bigger than the
total number of elements in a dataset, the write will fail. This means that you should take care not
to have compression turned on when doing small writes. To achieve optimal performance for SZIP
compression, it is recommended that one select a value for BLOCK that is an integral divisor of
the dataset’s fastest-changing dimension. Note that the SZIP compression encoder is licensed for
non-commercial use only while the decoder (e.g. decompression) is unlimited. Read more about
SZIP licensing at http://www.hdfgroup.org/doc_resource/SZIP/index.html. Note that SZIP decom-
pression is NOT guaranteed to be available to all Silo clients; only those for which the Silo library
was configured with SZIP compression capability enabled. Like GZIP, SZIP compression is
applied to float and integer data independently of the types of meshes and variables.
FPZIP compression: is enabled using “METHOD=FPZIP” in the options string. The FPZIP com-
pression algorithm was developed by Peter Lindstrom at LLNL and is also designed for high speed
compression of regular arrays of data. FPZIP recognizes the “LOSS=0|1|2|3” parameter which
specifies the amount of loss that is tolerable in the result in terms of quarters of full precision. For
example, “LOSS=3” indicates that a loss of 3/4 of full precision is tolerable (resulting in 8 bit
floats or 16 bit doubles). Note that for data being written from a double precision writer for down
stream visualization purposes, visualization tools such as VisIt often enforce single precision data.
Therefore, specifying a loss of 32 bits here for double precision data could have a dramatic impact
on compression and I/O performance with negligible effect in down stream visualization. If the
LOSS parameter is not specified, the default is “LOSS=0”. It is possible to build the Silo library
without FPZIP compression support. So, it is not always guaranteed to exist.
HZIP compression: is enabled using “METHOD=HZIP” in the options string. The HZIP compres-
sion algorithm was developed by Peter Lindstrom at LLNL and is designed for high-speed com-

DBSetCompression
Silo User’s Guide 2-31
pression of unstructured meshes of quad or hex elements and node-centered variables (it does not
yet support zone-centered variables) defined on a mesh. Before applying this compression method
to any given Silo mesh or variable object, the Silo library checks for compatibility with the con-
straints of the compression algorithm. If the mesh or variable object is compatible, the object will
be written with compression enabled. Otherwise, compression will be silently ignored. It is possi-
ble to build the Silo library without HZIP compression support. So, it is not always guaranteed to
exist.
Note that FPZIP and HZIP compression features are NOT available in a BSD Licensed release of
Silo library. They are available only in a Legacy licensed release of the Silo library.

DBGetCompression
2-32 Silo User’s Guide
DBGetCompression—Get current compression parameters
Synopsis:
char const *DBGetCompression()
Fortran Equivalent:
integer function dbgetcompress(options, loptions)
Arguments:
None
Returns:
NULL if no compress parameters have been set. A string of compression parameters if compres-
sion has been set
Description:
Obtain the current compression parameters. Caller should NOT free the returned string.

DBSetFriendlyHDF5Names
Silo User’s Guide 2-33
DBSetFriendlyHDF5Names—Set flag to indicate Silo should create friendly names for
HDF5 datasets
Synopsis:
int DBSetFriendlyHDF5Names(int enable)
Fortran Equivalent:
integer function dbsethdfnms(enable)
Arguments:
enable Flag to indicate if friendly names should be turned on (non-zero value) or off
(zero).
Returns:
Old setting for this flag
Description:
In versions of Silo prior to 4.8, the default behavior of the HDF5 driver was that it used HDF5 in a
way that made the data somewhat UNnatural to the user when viewed with HDF5 tools such as
h5ls, h5dump and hdfview as well as other tools that interact with the data via the HDF5 API. This
was not a problem for Silo but was a problem for these and other HDF5 tools.
DBSetFriendlyHDF5Names() was introduced as a way to address this issue so that the data
in an HDF5 file written by Silo looked more “natural”. Calling
DBSetFriendlyHDF5Names() with a value of one (‘1’) will result in additional HDF5 meta-
data being added to the file (in the form of soft links) with better names (and locations) for Silo
objects’ datasets. Note that creation of links does increase the file size somewhat. This affect is
less significant for larger files. It is also likely to have some negative but as yet to be investigated
effect on I/O performance
Calling DBSetFriendlyHDF5Names() with a value of two (‘2’) will foregoe the creation of
soft links and instead write the actual dataset data where those links would have been created (e.g.
the current working directory of the Silo file). This may be important for files consisting of a large
number of objects as it eliminates the creation of the /.silo group and subsequent very large
number of dataset objects in that one group.
In versions of Silo 4.8 and newer, the default behavior of the Silo library is to use mode ‘2’, that is
to create the datasets themselves there the links would have otherwise been created.
Notes:
If it was not obvious from the name, this method effects only the HDF5 driver.

DBGetFriendlyHDF5Names
2-34 Silo User’s Guide
DBGetFriendlyHDF5Names—Get setting for friendly HDF5 names flag
Synopsis:
int DBGetFriendlyHDF5Names()
Fortran Equivalent:
integer function dbgethdfnms()
Arguments:
None
Returns:
The current setting for the HDF5 friendly names flag.
Description:
See DBSetFriendlyHDF5Names().

DBSetDeprecateWarnings
Silo User’s Guide 2-35
DBSetDeprecateWarnings—Set maximum number of deprecate warnings Silo will
issue for any one function, option or convention
Synopsis:
int DBSetDeprecateWarnings(int max_count)
Fortran Equivalent:
integer function dbsetdepwarn(max_count)
Arguments:
max_count Maximum number of warnings Silo will issue for any single API function.
Returns:
The old maximum number of deprecate warnings
Description:
Some of Silo’s API functions have been deprecated. Some options on Silo objects have also been
deprecated. Finally, some conventional arrays, such as _visit_defvars, have been depre-
cated.
When an attempt to use a deprecated function, option or convention is detected, Silo will issue an
error message on stderr and proceed normally. The default number of error messages any given
deprecated function will report on stderr is 3. Note, this is on a per-deprecated function, option or
convention basis. If this number is decreased to zero by calling DBSetDeprecateWarn-
ings(0), no warnings will be generated on stderr. If it is increased, more warnings will be
issued.
Note that deprecated functions, options and conventions are guaranteed to operate correctly only
in the first release in which they became deprecated. In subsequent releases, they may be removed
entirely. So, it is wise to run your application for a while without turning off deprecation warnings
to get some inventory of functions that require attention.

DBGetDeprecateWarnings
2-36 Silo User’s Guide
DBGetDeprecateWarnings—Get maximum number of deprecated function warnings
Silo will issue
Synopsis:
int DBGetDeprecateWarnings()
Fortran Equivalent:
integer function dbgetdepwarn()
Arguments:
None
Returns:
The current maximum number of deprecate warnings
Description:

SILO_VERSION_GE
Silo User’s Guide 2-37
DB_VERSION_GE—Compile time macro to test silo version number
Synopsis:
DB_VERSION_GE(Maj,Min,Pat)
Arguments:
Maj Major version number digit
Min Minor version number digit. A zero is equivalent to no minor digit.
Pat Patch version number digit. A zero is equivalent to no patch digit.
Returns:
True (non-zero) if the combination of major, minor and patch digits results in a version number of
the Silo library that is greater (e.g. newer) than or equal to the version of the Silo library being
compiled against. False (zero), otherwise.
Description:
This macro is useful for writing version-specific code that interacts with the Silo library. Note,
however, that this macro appeared in version 4.6.1 of the Silo library and is not available in earlier
versions of the library.
As an example of use, the function DBSetDeprecateWarnings() was introduced in Silo version 4.6
and not available in earlier versions. You could use this macro like so...
#if DB_VERSION_GE(4,6,0)
DBSetDeprecateWarnings(0);
#endif

SILO_VERSION_GE
2-38 Silo User’s Guide
2 API Section Files and File Structure
If you are looking for information regarding how to use Silo from a parallel application, please See
“Multi-Block Objects, Parallelism and Poor-Man’s Parallel I/O” on page 154.
The Silo API is implemented on a number of different low-level drivers. These drivers control the
low-level file format Silo generates. For example, Silo can generate PDB (Portable DataBase) and
HDF5 formatted files. The specific choice of low-level file format is made at file creation time.
In addition, Silo files can themselves have directories. That is, within a single Silo file, one can
create directory hierarchies for storage of various objects. These directory hierarchies are analo-
gous to the Unix filesystem. Directories serve to divide the name space of a Silo file so the user
can organize content within a Silo file in a way that is natural to the application.
Note that the organization of objects into directories within a Silo file may have direct implications
for how these collections of objects are presented to users by post-processing tools. For example,
except for directories used to store multi-block objects (See “Multi-Block Objects, Parallelism and
Poor-Man’s Parallel I/O” on page 154.), VisIt will use directories in a Silo file to create submenus
within its Graphical User Interface (GUI). For example, if VisIt opens a Silo file with two directo-
ries called “foo” and “bar” and there are various meshes and variables in each of these directories,
then many of VisIt’s GUI menus will contain submenus named “foo” and “bar” where the objects
found in those directories will be placed in the GUI.
Silo also supports the concept of grabbing the low-level driver. For example, if Silo is using the
HDF5 driver, an application can obtain the actual HDF5 file id and then use the native HDF5 API
with that file id.
The functions described in this section of the interface are...
DBRegisterFileOptionsSet . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 40
DBUnregisterFileOptionsSet . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 44
DBUnregisterAllFileOptionsSets . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 45
DBSetUnknownDriverPriorities. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 46
DBGetUnknownDriverPriorities . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 47
DBCreate . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 48
DBOpen . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 53
DBClose . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 55
DBGetToc. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 56
DBFileVersion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 57
DBFileVersionDigits . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 58
DBFileVersionGE. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 59
DBVersionGEFileVersion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 60
DBSortObjectsByOffset . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 61
DBMkDir . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 62
DBSetDir . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 63
DBGetDir . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 64
DBCpDir. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 65
DBGrabDriver . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 66
DBUngrabDriver . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 67

SILO_VERSION_GE
Silo User’s Guide 2-39
DBGetDriverType . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 68
DBGetDriverTypeFromPath. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 69
DBInqFile . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 70
DBInqFileHasObjects. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 71
_silolibinfo . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 72
_hdf5libinfo . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 73
_was_grabbed . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 74

SILO_VERSION_GE
2-40 Silo User’s Guide
DBRegisterFileOptionsSet—Register a set of options for advanced control of the
low-level I/O driver
Synopsis:
int DBRegisterFileOptionsSet(const DBoptlist *opts)
Fortran Equivalent:
int dbregfopts(int optlist_id)
Arguments:
opts an options list object obtained from a DBMakeOptlist() call
Returns:
-1 on failure. Otherwise, the integer index of a registered file options set is returned.
Description:
File options sets are used in concert with the DB_HDF5_OPTS() macro in DBCreate or DBO-
pen calls to provide advanced and fine-tuned control over the behavior of the underlying driver
library and may be needed to affect memory usage and I/O performance as well as vary the behav-
ior of the underlying I/O driver away from its default mode of operation.
A file options set is nothing more than an optlist object (see “Optlists” on page 2-231), popu-
lated with file driver related options. A registered file options set is such an optlist that has
been registered with the Silo library via a call to this method, DBRegisterFileOptionsSet.
A maximum of 32 registered file options sets are currently permitted. Use DBUnregisterF-
ileOptionsSet to free up a slot in the list of registered file options sets.
Before a specific file options set may be used as part of a DBCreate or DBOpen call, the file
options set must be registered with the Silo library. In addition, the associated optlist object
should not be freed until after the last call to DBCreate or DBOpen in which it is needed.
Presently, the only options the Silo library defines are for the HDF5 driver. The table below
defines and describes the various options. A key option is the selection of the HDF5 Virtual File
Driver or VFD. See “DBCreate” on page 2-48 for a description of the available VFDs.
In the table of options below, some options are relevant to only a specific HDF5 VFD. Other
options effect the behavior of the HDF5 library as a whole, regardless of which underlying VFD is
used. This difference is notated in the scope column.
All of the options described here relate to options documented in the HDF5 library’s file access
property lists, http://www.hdfgroup.org/HDF5/doc/RM/RM_H5P.html. Therefore, rather than
duplicate a lot of the HDF5-specific documentation here, in most cases, we simply refer the reader
to the relevant sections of the HDF5 reference manual.
SILO_VERSION_GE
Silo User’s Guide 2-41
Note that all option names listed in left-most column of the table below have had their prefix
“DBOPT_H5_” removed to save space in the table. So, for example, the real name of the
CORE_ALLOC_INC option is DBOPT_H5_CORE_ALLOC_INC.
Option Name,
DBOPT_H5_... Scope Type Option Meaning Default Value
VFD VFD int Specifies which Virtual File Driver (VFD) the HDF5
library should use.
Set the integer value for this option to one of the
following values.
DB_H5VFD_DEFAULT, (use HDF5 default driver)
DB_H5VFD_SEC2 (use HDF5 sec2 driver)
DB_H5VFD_STDIO, (use HDF5 stdio driver)
DB_H5VFD_CORE, (use HDF5 core driver)
DB_H5VFD_LOG, (use HDF5 log river)
DB_H5VFD_SPLIT, (use HDF5 split driver)
DB_H5VFD_DIRECT, (use HDF5 direct i/o driver)
DB_H5VFD_FAMILY, (use HDF5 family driver)
DB_H5VFD_MPIO, (use HDF5 mpi-io driver)
DB_H5VFD_MPIP, (use HDF5 mpi posix driver)
DB_H5VFD_SILO, (use SILO BG/Q driver)
DB_H5VFD_FIC (use SILO file in core driver)
Many of the reamining options described in this
table apply to only certain of the above VFDs.
DB_H5VFD_DEFAULT
RAW_FILE_OPTS VFD int Applies only for the split VFD. Specifies a file
options set to use for the raw data file. May be any
value returned from a call to
DBRegisterFileOptionsSet() or can be any
one of the following pre-defined file options sets...
DB_FILE_OPTS_H5_DEFAULT_...
DEFAULT, SEC2, STDIO, CORE, LOG, SPLIT,
DIRECT, FAMILY, MPIO, MPIP, SILO.
See HDF5 reference manual for H5Pset_fapl_split
DB_FILE_OPTS_H5_DEFAULT
_DEFAULT
RAW_EXTENSION VFD char* Applies only for the split VFD. Specifies the file
extension/naming convention for raw data file. If
the string contains a ‘%s’ printf-like conversion
specifier, that will be replaced with the name of the
file passed in the DBCreate/DBOpen call. If the
string does NOT contain a ‘%s’ printf-like conver-
sion specifier, it is treated as an ‘extension’ which
is appended to the name of the file passed in
DBCreate/DBopen call.
See HDF5 reference manual for H5Pset_fapl_split
“-raw”
META_FILE_OPTS VFD int Same as DBOPT_H5_RAW_FILE_OPTS, above,
except for meta data file. See HDF5 reference
manual for H5Pset_fapl_split.
DB_FILE_OPTS_H5_DEFAULT
_CORE
META_EXTENSION VFD Same as DBOPT_H5_RAW_EXTENSION above,
except for meta data file. See HDF5 reference
manual for H5Pset_fapl_split.
“”
CORE_ALLOC_INC VFD int Applies only for core VFD. Specifies allocation
increment. See HDF5 reference manual for
H5Pset_fapl_core.
(1<<20)
CORE_NO_BACK_STORE VFD int Applies only for core VFD. Specifies whether or
not to store the file on close. See HDF5 reference
manual for H5Pset_fapl_core.
FALSE
LOG_NAME VFD char * Applies only for the log VFD. This is primarily a
debugging feature. Specifies name of the file to
which loggin data shall be stored. See HDF5
refrence manual for H5Pset_fapl_log.
“silo_hdf5_log.out”
SILO_VERSION_GE
2-42 Silo User’s Guide
LOG_BUF_SIZE VFD int Applies only for the log VFD. This is primarily a
debugging feature. Specifies size of the buffer to
which byte-for-byte HDF5 data type information is
written. See HDF5 refrence manual for
H5Pset_fapl_log.
0
META_BLOCK_SIZE GLOBAL int Applies the the HDF5 library as a whole (e.g. glob-
ally). Specifies the size of memory allocations the
library should use when allocating meta data. See
HDF5 reference manual for
H5Pset_meta_block_size.
0
SMALL_RAW_SIZE GLOBAL int Applies to the HDF5 library as a whole (e.g. glob-
ally). Specifies a threshold below which allocations
for raw data are aggregated into larger blocks
within HDF5. This can improve I/O performance by
reducing number of small I/O requests. Note, how-
ever, that with a block-oriented VFD such as the
Silo specific VFD, this parameter must be set to be
consistent with block size of the VFD. See the
HDF5 reference manual for
H5Pset_small_data_block_size.
0
ALIGN_MIN GLOBAL int Applies to the HDF5 library as a whole. Specified a
size threshold above which all datasets are
aligned in the file using the value specified in
ALIGN_VAL. See HDF5 reference manual for
H5Pset_alignment.
0
ALIGN_VAL GLOBAL int The alignment to be applied to datasets of size
greater than ALIGN_MIN. See HDF5 reference
manual for H5Pset_alignment.
0
DIRECT_MEM_ALIGN VFD int Applies only to the direct VFD. Specifies the align-
ment option. See the HDF5 reference manual for
H5Pset_fapl_direct.
0
DIRECT_BLOCK_SIZE VFD int Applies only to the direct VFD. Specifies the block
size the underlying filesystem is using. See the
HDF5 reference manual for H5Pset_fapl_direct.
DIRECT_BUF_SIZE Applies only to the direct VFD. Specifies a copy
buffer size. See the HDF5 reference manual for
H5Pset_fapl_direct.
MPIO_COMM
MPIO_INFO
MPIP_NO_GPFS_HINTS
SIEVE_BUF_SIZE GLOBAL int HDF5 sieve buf size. Only relevant if using either
compression and/or checksumming. See HDF5
reference manual for H5Pset_sieve_buf_size.
CACHE_NELMTS GLOBAL int HDF5 raw data chunk cache parameters. Only rel-
evant if using either compression and/or check-
summing. See the HDF5 reference manual for
H5Pset_cache.
CACHE_NBYTES HDF5 raw data chunk cache parameters. Only rel-
evant if using either compression and/or check-
summing. See the HDF5 reference manual for
H5Pset_cache.
CACHE_POLICY HDF5 raw data chunk cache parameters. Only rel-
evant if using either compression and/or check-
summing. See the HDF5 reference manual for
H5Pset_cache.
Option Name,
DBOPT_H5_... Scope Type Option Meaning Default Value
SILO_VERSION_GE
Silo User’s Guide 2-43
FAM_SIZE VFD int Size option for family VFD. See the HDF5 refer-
ence manual for H5Pset_fapl_family. The family
VFD is useful for handling files that would other-
wise be larger than 2Gigabytes on filesystems that
support a maximum file size of 2Gigabytes.
FAM_FILE_OPTS VFD int VFD options for each file in family VFD. See the
HDF5 reference manual for H5Pset_fapl_family.
The family VFD is useful for handling files that
would otherwise be larger than 2Gigabytes on file-
systems that support a maximum file size of
2Gigabytes.
USER_DRIVER_ID GLOBAL int Specify some user-defined VFD. Permtis applica-
tion to specify any user-defined VFD. See HDF5
reference manual for H5Pset_driver.
USER_DRIVER_INFO GLOBAL Specify user-defined VFD information struct. Per-
mtis application to specify any user-defined VFD.
See HDF5 reference manual for H5Pset_driver.
SILO_BLOCK_SIZE VFD int Block size option for Silo VFD. All I/O requests to/
from disk will occur in blocks of this size.
(1<<16)
SILO_BLOCK_COUNT VFD int Block count option for Silo VFD. This is the maxi-
mum number of blocks the Silo VFD will maintain
in memory at any one time.
32
SILO_LOG_STATS VFD int Flag to indicate if Silo VFD should gather I/O per-
formance statistics. This is primarily for debugging
and performance tuning of the Silo VFD.
0
SILO_USE_DIRECT VFD int Flag to indicate if Silo VFD should attempt to use
direct I/O. Tells the Silo VFD to use direct I/O
where it can. Note, if it cannot, this option will be
siliently ignored.
0
FIC_BUF VFD void* The buffer of bytes to be used as the “file in core”
to be opened in a DBOpen() call.
none
FIC_SIZE VFD int Size of the buffer of bytes to be used as the “file in
core” to be opened in a DBOpen() call.
none
Option Name,
DBOPT_H5_... Scope Type Option Meaning Default Value

SILO_VERSION_GE
2-44 Silo User’s Guide
DBUnregisterFileOptionsSet—Unregister a registered file options set
Synopsis:
int DBUnregisterFileOptionsSet(int opts_set_id)
Fortran Equivalent:
Arguments:
opts_set_id The identifer (obtained from a previous call to DBRegisterFileOptionsSet()) of
a file options set to unregister.
Returns:
Zero on success. -1 on failure.
Description:

SILO_VERSION_GE
Silo User’s Guide 2-45
DBUnregisterAllFileOptionsSets—Unregister all file options sets
Synopsis:
int DBUnregisterAllFileOptionsSets()
Fortran Equivalent:
Arguments:
None
Returns:
Zero on success, -1 on failure.
Description:

SILO_VERSION_GE
2-46 Silo User’s Guide
DBSetUnknownDriverPriorities—Set driver priorities for opening files with the
DB_UNKNOWN driver.
Synopsis:
static const int *DBSetUnknownDriverPriorities(int *driver_ids)
Fortran Equivalent:
None
Arguments:
driver_ids A -1 terminated list of driver ids such as DB_HDF5, DB_PDB,
DB_HDF5_CORE, or any driver id constructed with the DB_HDF5_OPTS()
macro.
Returns:
The previous
Description:
When opening files with DB_UNKNOWN driver, Silo iterates over drivers, trying each until it suc-
cessfuly opens a file.
This call can be used to affect the order in which driver ids are attempted and can improve behav-
ior and performance for opening files using DB_UNKNOWN driver.
If any of the driver ids specified in driver_ids is constructed using the DB_HDF5_OPTS()
macro, then the associated file options set must be registered with the Silo library.

SILO_VERSION_GE
Silo User’s Guide 2-47
DBGetUnknownDriverPriorities—Return the currently defined ordering of
drivers the DB_UNKNOWN driver will attempt.
Synopsis:
static const int *DBGetUnknownDriverPriorities(void)
Fortran Equivalent:
None
Description:

DBCreate
2-48 Silo User’s Guide
DBCreate—Create a Silo output file.
Synopsis:
DBfile *DBCreate (char *pathname, int mode, int target,
char *fileinfo, int filetype)
Fortran Equivalent:
integer function dbcreate(pathname, lpathname, mode, target,
fileinfo, lfileinfo, filetype, dbid)
returns created database file handle in dbid
Arguments:
pathname Path name of file to create. This can be either an absolute or relative path.
mode Creation mode. One of the predefined Silo modes: DB_CLOBBER or
DB_NOCLOBBER.
target Destination file format. One of the predefined types: DB_LOCAL, DB_SUN3,
DB_SUN4, DB_SGI, DB_RS6000, or DB_CRAY.
fileinfo Character string containing descriptive information about the file’s contents.
This information is usually printed by applications when this file is opened. If
no such information is needed, pass NULL for this argument.
filetype Destination file type. Applications typically use one of either DB_PDB, which
will create PDB files, or DB_HDF5, which will create HDF5 files. Other options
include DB_PDBP, DB_HDF5_SEC2, DB_HDF5_STDIO, DB_HDF5_CORE,
DB_HDF5_SPLIT or DB_FILE_OPTS(optlist_id) where
optlist_id is a registered file options set. For a description of the meaning
of these options as well as many other advanced features and control of
underlying I/O behavior, see “DBRegisterFileOptionsSet” on page 2-40.
Returns:
DBCreate returns a DBfile pointer on success and NULL on failure.
Description:
The DBCreate function creates a Silo file and initializes it for writing data.
Notes:
Silo supports two underlying drivers for storing named arrays and objects of machine independent
data. One is called the Portable DataBase Library (PDBLib or just PDB),
https://wci.llnl.gov/codes/pact/pdb.html and the other is Hierarchical Data Format, Version 5
(HDF5), http://www.hdfgroup.org/HDF5.
When Silo is configured with the --with-pdb-proper=<path-to-PACT> option, the Silo
library supports both the PDB driver that is built-in to Silo (which is actually an ancient version of
PACT’s PDB referred to internally as ‘PDB Lite’) identified with a filetype of DB_PDB and a
second variant of the PDB driver using a PACT installation (specified when Silo was configured)

DBCreate
Silo User’s Guide 2-49
with a filetype of DB_PDBP (Note the trailing ‘P’ for ‘PDB Proper’). PDB Proper is known to
give far superior performance than PDB Lite on BG/P and BG/L class systems and so is recom-
mended when using PDB driver on such systems.
For the HDF5 library, there are many more available options for fine tuned control of the underly-
ing I/O through the use of HDF5’s Virtual File Drivers (VFDs). For example, HDF5’s sec2 VFD
uses Unix Manual Section 2 I/O routines (e.g. create/open/read/write/close) while the
stdio VFD uses Standard I/O routines (e.g. fcreate/fopen/fread/fwrite/fclose).
Depending on the circumstances, the choice of VFD can have a profound impact on actual I/O per-
formance. For example, on BlueGene systems the customized Silo VFD (introduced to the Silo
library in Version 4.8) has demonstrated excellent performance compared to the default HDF5
VFD; sec2. The remaining paragraphs describe each of the available Virtual File Drivers as well as
parameters that govern their behavior.
DB_HDF5: From among the several VFDs that come pre-packaged with the HDF5 library, this
driver type uses whatever the HDF5 library defines as the default VFD. On non-Windows plat-
forms, this is the Section 2 (see below) VFD. On Windows platforms, it is a Windows specific
VFD.
DB_HDF5_SEC2: Uses the I/O system interface defined in section 2 of the Unix manual. That is
create, open, read, write, close. This is a VFD that comes pre-packaged with the HDF5 library. It
does little to no optimization of I/O requests. For example, two I/O requests that abutt in file
address space wind up being issued through the section 2 I/O routines as independent requests.
This can be disasterous for high latency filesystems such as might be available on BlueGene class
systems.
DB_HDF5_STDIO: Uses the Standard I/O system interface defined in Section 3 of the Unix man-
ual. That is fcreate, fopen, fread, fwrite, fclose. This is a VFD that comes pre-packaged with the
HDF5 library. It does little to no optimization of I/O requests. However, since it uses the stdio rou-
tines, it does benefit from whatever default buffering the implementation of the stdio interface on
the given platform provides. Because section 2 routines are unbuffered, the sec2 VFD typically
performs better when there are fewer, larger I/O requests while the stdio VFD performs better
when there are more, smaller requests. Unfortunately, the metric for what constitutes a “small” or
“large” request is system dependent. So, it helps to experiment with the different VFDs for the
HDF5 driver by running some typically sized use cases. Some results on the Luster file system for
tiny I/O requests (100’s of bytes) showed that the stdio VFD can perform 100x or more better than
the section 2. So, it pays to spend some time experimenting with this [Note: In future, it should be
possible to manipulate the buffer used for a given Silo file opened via the stdio VFD as one would
ordinarily do via such stdio calls as setvbuf(). However, due to limitations in the current
implementation, that is not yet possible. When and if that becomes possible, to use something
other than non-default stdio buffering, the Silo client will have to create and register an appropriate
file options set (see “DBRegisterFileOptionsSet” on page 2-40).]
DB_HDF5_CORE: Uses a memory buffer for the file with the option of either writing the resultant
buffer to disk or not. Conceptually, this VFD behaves more or less like a ramdisk. This is a VFD
that comes pre-packaged with the HDF5 library. I/O performance is optimal in the sense that only
a single I/O request for the entire file is issued to the underlying filesystem. However, this optimal-
ity comes at the expense of memory. The entire file must be capable of residing in memory. In
addition, releases of HDF5 library prior to 1.8.2 support the core VFD only when creating a new
file and not when open an existing file. Two parameters that govern behavior of the core VFD. The
allocation increment specifies the amount of memory the core VFD allocates, each time it needs to

DBCreate
2-50 Silo User’s Guide
increase the buffer size to accomodate the (possibly growing) file. The backing store indicates
whether the buffer should be saved to disk (if it has been changed) upon close. By default, using
DB_HDF5_CORE as the driver type results in an allocation incriment of 1 Megabyte and a back-
ing store option of TRUE, meaning it will store the file to disk upon close. To specify parameters
other than these default values, the Silo client will have to create and register an appropriate file
options set (see “DBRegisterFileOptionsSet” on page 2-40).
DB_HDF5_SPLIT: Splits HDF5 I/O operations across two VFDs. One VFD is used for all raw
data while the other VFD is used for everything else (e.g. meta data). For example, in Silo’s
DBPutPointvar() call, the data the caller passes in the vars argument is raw data. Everything else
including the object’s name, number of points, datatype, optlist options, etc. including all underly-
ing HDF5 metadata gets treated as meta data. This is a VFD that comes pre-packaged with the
HDF5 library. It results in two files being produced; one for the raw data and one for the meta data.
The reason this can be a benefit is that tiny bits of metadata intermingling with large raw data oper-
ations can degrade performance overall. Separating the datastreams can have a profound impact on
performance at the expense of two files being produced. Four parameters govern the behavior of
the split VFD. These are the VFD and filename extension for the raw and meta data, respectively.
By default, using DB_HDF5_SPLIT as the driver type results in Silo using sec2 and “-raw” as the
VFD and filename extension for raw data and core (default params) and “” (empty string) as the
VFD and extension for meta data. To specify parameters other than these default values, the Silo
client will have to create and register an appropriate file options set (see “DBRegisterFileOptions-
Set” on page 2-40).
DB_HDF5_FAMILY: Allows for the management of files larger than 2^32 bytes on 32-bit sys-
tems. The virtual file is decomposed into real files of size small enough to be managed on 32-bit
systems. This is a VFD that comes pre-packaged with the HDF5 library. Two parameters govern
the behavior of the family VFD. The size of each file in a family of files and the VFD used for the
individual files. By default, using DB_HDF5_FAMILY as the driver type results in Silo using a
size of 1 Gigabyte (1<<30) and the default VFD for the individual files. To specify parameters
other than these default values, the Silo client will have to create and register an appropriate file
options set (see “DBRegisterFileOptionsSet” on page 2-40).
DB_HDF5_LOG: While doing the I/O for HDF5 data, also collects detailed information regarding
VFD calls issued by the HDF5 library. The logging VFD writes detailed information regarding
VFD operations to a logfile. This is a VFD that comes pre-packaged with the HDF5 library. How-
ever, the logging VFD is a different code base than any other VFD that comes pre-packaged with
HDF5. So, while the logging information it produces is representative of the VFD calls made by
HDF5 library to the VFD interface, it is NOT representative of the actual I/O requests made by the
sec2 or stdio or other VFDs. Behavior of the logging VFD is governed by 3 parameters; the name
of the file to which log information is written, a set of flags which are or’d together to specify the
types of operations and information logged and, optionally, a buffer (which must be at least as
large as the actual file being written) which serves to map the kind of HDF5 data (there are about 8
different kinds) stores at each byte in the file. By default, using DB_HDF5_LOG as the driver type
results in Silo using a logfile name of “silo_hdf5_log.out”, flags of
H5FD_LOG_LOC_IO|H5FD_LOG_NUM_IO|H5FD_LOG_TIME_IO|H5FD_LOG_ALLOC and
a NULL buffer for the mapping information. To specify parameters other than these default val-
ues, the Silo client will have to create and register an appropriate file options set (see “DBRegister-
FileOptionsSet” on page 2-40). Users interested in this VFD should consult HDF5’s reference
manual for the meaning of the flags as well as how to interepret logging VFD output.

DBCreate
Silo User’s Guide 2-51
DB_HDF5_DIRECT: On systems that support the ‘O_DIRECT’ flag in section 2 create/open
calls, this VFD will use direct I/O. This VFD comes pre-packaged with the HDF5 library. Most
systems (both the system interfaces implementations for section 2 I/O as well as underlying file-
systems) do a lot of work to buffer and cache data to improve I/O performance. In some cases,
however, this extra work can actually get in the way of good performance, particularly when the
I/O operations are streaming like and large. Three parameters govern the behavior of the direct
VFD. The alignment specifies memory alignment requirement of raw data buffers. That generally
means that posix_memalign should be used to allocate any buffers you use to hold raw data
passed in calls to the Silo library. The block size indicates the underlying filesystem block size and
the copy buffer size gives the HDF5 library some additional flexibility in dealing with unaligned
requests. Few systems support the O_DIRECT flag and so this VFD is not often available in prac-
tice. However, when it is, using DB_HDF5_DIRECT as the driver type results in Silo using an
alignment of 4 kilobytes (1<<12), an alignment equal to the block size and a copy buffer size equal
to 256 times the blocksize.
DB_HDF5_SILO: This is a custom VFD designed specifically to address some of the performance
shortcommings of VFDs that come pre-packaged with the HDF5 library. The silo VFD is a very,
very simple, block-based VFD. It decomposes the file into blocks, keeps some number of blocks in
memory at any one time and issues I/O requests ONLY in whole blocks using section 2 I/O rou-
tines. In addition, it sets up some parameters that control HDF5 library’s allocation of meta data
and raw data such that each block winds up consisting primirily of either raw or meta data but not
both. It also disables meta data caching in HDF5 to reduce memory consumption of the HDF5
library to the bare minimum as there is no need for HDF5 to maintain cached metadata if it resides
in blocks kept in memory in the VFD. This is a suitable VFD for most scientific computing appli-
cations that are dumping either post-processing or restart files as applications that do that tend to
open the file, write a bunch of stuff from start to finish and close it or read a bunch of stuff from
start to finish and close it. Two parameters govern the behavior of the silo VFD; the block size and
the block count. The block size determines the size of individual blocks. All I/O requests will be
issued in whole blocks. The block count determines the number of blocks the silo VFD is permit-
ted to keep in memory at any one time. On BG/P class systems, good values are 1 Megabyte
(1<<20) block size and block count of 16 or 32. By default, the silo VFD uses a block size of 16
Kilobytes (1<<14) and a block count also of 16. To specify parameters other than these default val-
ues, the Silo client will have to create and register an appropriate file options set (see “DBRegister-
FileOptionsSet” on page 2-40).
DB_HDF5_MPIO and DB_HDF5_MPIOP: Although Silo itself DOES NOT support true parallel
I/O (e.g. multiple processors writing to the same file, concurrently), Silo can take advantage of any
performance capabilities which may be available in the MPI-IO implementation used by HDF5’s
mpio VFD. Two parameters govern the mpio VFD; the MPI communicator and an MPI_Info
object. By default, using DB_HDF5_MPIO as the driver type results in Silo using
MPI_COMM_SELF and an empty MPI_Info object. Note, because Silo is not designed to work
within the constraints of HDF5’s parallel interface, values for MPI_COMM_SELF (which lead to
a file per processor) are likely to result in deadlock and/or corrupted files.
Finally, both PDB and HDF5 support the concept of targeting output files. That is, a Sun IEEE file
can be created on the Cray, and vice versa. If creating files on a mainframe or other powerful com-
puter, it is best to target the file for the machine where the file will be processed. Because of the
extra time required to do the floating point conversions, however, one may wish to bypass the tar-
geting function by providing DB_LOCAL as the target.

DBCreate
2-52 Silo User’s Guide
In Fortran, an integer represent the file’s id is returned. That integer is then used as the database
file id in all functions to read and write data from the file.
Note that regardless of what type of file is created, it can still be read on any machine.
See notes in the documentation on DBOpen regarding use of the DB_UNKNOWN driver type.

DBOpen
Silo User’s Guide 2-53
DBOpen—Open an existing Silo file.
Synopsis:
DBfile *DBOpen (char *name, int type, int mode)
Fortran Equivalent:
integer function dbopen(name, lname, type, mode, dbid)
returns database file handle in dbid.
Arguments:
name Name of the file to open. Can be either an absolute or relative path.
type The type of file to open. One of the predefined types, typically DB_UNKNOWN,
DB_PDB, or DB_HDF5. However, there are other options as well as subtle but
important issues in using them. So, read description, below for more details.
mode The mode of the file to open. One of the values DB_READ or DB_APPEND.
Returns:
DBOpen returns a DBfile pointer on success and a NULL on failure.
Description:
The DBOpen function opens an existing Silo file. If the file type passed here is DB_UNKNOWN,
Silo will attempt to guess at the file type by iterating through the known types attempting to open
the file with each driver until it succeeds. This iteration does incur a small performance penalty. In
addition, use of DB_UNKNOWN can have other undesireable behavior described below. So, if at all
possible, it is best to open using a specific type. See DBGetDriverTypeFromPath() for a
function that uses cheap heuristics to determine the driver type given a candiate filename.
When writing general purpose code to read Silo files and you cannot know for certain ahead of
time what the correct driver to use is, there are a few options.
First, you can iterate over the available driver ids, calling DBOpen() using each one until one of
them succeds. But, that is exactly what the DB_UNKNOWN driver does so there is no need for a Silo
client to have to write that code. In addition, if you have a specific preference of order of drivers,
you can use DBSetUnknownDriverPriorities()to specify that ordering.
Undesireable behavior with DB_UNKNOWN can occur when the specified file can be successfully
opened using multiple of the available drivers and/or file options sets and it succceds with the
wrong one or one using options the caller neither expected or intended. See “DBSetUnknownDriv-
erPriorities” on page 2-46 for a way to specify the order of drivers tried by the DB_UNKNOWN
driver.
Indeed, in order to use a specific VFD (see “DBCreate” on page 2-48) in HDF5, it is necessary to
pass the specific DB_HDF5_XXX argument in this call or to set the unknown driver priorities
such that whatever specific HDF5 VFD(s) are desired are tried first before falling back to other,
perhaps less desirable ones.

DBOpen
2-54 Silo User’s Guide
The mode parameter allows a user to append to an existing Silo file. If a file is DBOpen’ed with a
mode of DB_APPEND, the file will support write operations as well as read operations.

DBClose
Silo User’s Guide 2-55
DBClose—Close a Silo database.
Synopsis:
int DBClose (DBfile *dbfile)
Fortran Equivalent:
integer function dbclose(dbid)
Arguments:
dbfile Database file pointer.
Returns:
DBClose returns zero on success and -1 on failure.
Description:
The DBClose function closes a Silo database.

DBGetToc
2-56 Silo User’s Guide
DBGetToc—Get the table of contents of a Silo database.
Synopsis:
DBtoc *DBGetToc (DBfile *dbfile)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
Returns:
DBGetToc returns a pointer to a DBtoc structure on success and NULL on error.
Description:
The DBGetToc function returns a pointer to a DBtoc structure, which contains the names of the
various Silo object contained in the Silo database. The returned pointer points into Silo private
space and must not be modified or freed. Also, calls to DBSetDir will free the DBtoc structure,
invalidating the pointer returned previously by DBGetToc.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBFileVersion
Silo User’s Guide 2-57
DBFileVersion—Version of the Silo library used to create the specified file
Synopsis:
char const *DBFileVersion(DBfile *dbfile)
Fortran Equivalent:
None
Arguments:
dbfile Database file handle
Returns:
A character string representation of the version number of the Silo library that was used to create
the Silo file. The caller should NOT free the returned string.
Description:
Note, that this is distinct from (e.g. can be the same or different from) the version of the Silo
library returned by the DBVersion() function.
DBFileVersion, here, returns the version of the Silo library that was used when DBCreate() was
called on the specified file. DBVersion() returns the version of the Silo library the executable is
currently linked with.
Most often, these two will be the same. But, not always. Also note that although is possible that a
single Silo file may have contents created within it from multiple versions of the Silo library, a call
to this function will return ONLY the version that was in use when DBCreate() was called; that is
when the file was first created.

DBFileVersionDigits
2-58 Silo User’s Guide
DBFileVersionDigits—Return integer digits of file version number
Synopsis:
int DBFileVersionDigits(const DBfile *dbfile,
int *maj, int *min, int *pat, int *pre)
Arguments:
dbfile Silo database file handle
maj Pointer to returned major version digit
min Pointer to returned minor version digit
pat Pointer to returned patch version digit
pre Pointer to returned pre-release version digit (if any)
Returns:
Zero on success. Negative value on failure.

DBFileVersionGE
Silo User’s Guide 2-59
DBFileVersionGE—Greater than or equal comparison for version of the Silo library a
given file was created with
Synopsis:
int DBFileVersionGE(DBfile *dbfile, int Maj, int Min, int Pat)
Fortran Equivalent:
None
Arguments:
dbfile Database file handle
Maj Integer major version number
Min Integer minor version number
Pat Integer patch version number
Returns:
One (1) if the version number of the library used to create the specified file is greater than or equal
to the version number specified by Maj, Min, Pat arguments, zero (0) otherwise. A negative value
is returned if a failure occurs.

DBVersionGEFileVersion
2-60 Silo User’s Guide
DBVersionGEFileVersion—Compare library version with file version
Synopsis:
int DBVersionGEFileVersion(const DBfile *dbfile)
Fortran Equivalent:
None
Arguments:
dbfile Silo database file handle obtained with a call to DBOpen
Returns:
Non-zero if the library version is greater than or equal to the file version. Zero otherwise.

DBSortObjectsByOffset
Silo User’s Guide 2-61
DBSortObjectsByOffset—Sort list of object names by order of offset in the file
Synopsis:
int DBSortObjectsByOffset(DBfile *, int nobjs,
const char *const *const obj_names, int *ordering)
Fortran Equivalent:
None
Arguments:
DBfile Database file pointer.
nobjs Number of object names in obj_names.
ordering Returned integer array of relative order of occurence in the file of each object.
For example, if ordering[i]==k, that means the object whose name is
obj_names[i] occurs kth when the objects are ordered according to offset at
which they exist in the file.
Returns:
0 on succes; -1 on failure. The only possible reason for failure is if the HDF5 driver is being used
to read the file and Silo is not compiled with HDF5 version 1.8 or later.
Description:
The intention of this function is to permit applications reading Silo files to order their reads in such
a way that objects are read in the order in which they occur in the file. This can have a postive
impact on I/O performance, particularly using a block-oriented VFD such as the Silo VFD as it can
reduce and/or eliminate unnecessary block pre-emption. The degree to which ordering reads
effects performance is not yet known.

DBMkDir
2-62 Silo User’s Guide
DBMkDir—Create a new directory in a Silo file.
Synopsis:
int DBMkDir (DBfile *dbfile, char const *dirname)
Fortran Equivalent:
integer function dbmkdir(dbid, dirname, ldirname, status)
Arguments:
dbfile Database file pointer.
dirname Name of the directory to create.
Returns:
DBMkDir returns zero on success and -1 on failure.
Description:
The DBMkDir function creates a new directory in the Silo file as a child of the current directory
(see DBSetDir). The directory name may be an absolute path name similar to “/dir/subdir”,
or may be a relative path name similar to “../../dir/subdir”.

DBSetDir
Silo User’s Guide 2-63
DBSetDir—Set the current directory within the Silo database.
Synopsis:
int DBSetDir (DBfile *dbfile, char const *pathname)
Fortran Equivalent:
integer function dbsetdir(dbid, pathname, lpathname)
Arguments:
dbfile Database file pointer.
pathname Path name of the directory. This can be either an absolute or relative path name.
Returns:
DBSetDir returns zero on success and -1 on failure.
Description:
The DBSetDir function sets the current directory within the given Silo database. Also, calls to
DBSetDir will free the DBtoc structure, invalidating the pointer returned previously by DBGet-
Toc. DBGetToc must be called again in order to obtain a pointer to the new directory’s DBtoc
structure.

DBGetDir
2-64 Silo User’s Guide
DBGetDir—Get the name of the current directory.
Synopsis:
int DBGetDir (DBfile *dbfile, char *dirname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
dirname Returned current directory name. The caller must allocate space for the returned
name. The maximum space used is 256 characters, including the NULL
terminator.
Returns:
DBGetDir returns zero on success and -1 on failure.
Description:
The DBGetDir function returns the name of the current directory.

DBGetDir
Silo User’s Guide 2-65
DBCpDir—Copy a directory hierarchy from one Silo file to another.
Synopsis:
int DBCpDir(DBfile *srcFile, const char *srcDir,
DBfile *dstFile, const char *dstDir)
Fortran Equivalent:
None
Arguments:
srcFile Source database file pointer.
srcDir Name of the directory within the source database file to copy.
dstFile Destination database file pointer.
dstDir Name of the top-level directory in the destination file. If an absolute path is
given, then all components of the path except the last must already exist.
Otherwise, the new directory is created relative to the current working directory
in the file.
Returns:
DBCpDir returns 0 on success, -1 on failure
Description:
DBCpDir copies an entire directory hierarchy from one Silo file to another.
Note that this function is available only on the HDF5 driver and only if the Silo library has been
compiled with HDF5 version 1.8 or later. This is because the implementation exploits functionality
available only in versions of HDF5 1.8 and later.

DBGrabDriver
2-66 Silo User’s Guide
DBGrabDriver—Obtain the low-level driver file handle
Synopsis:
void *DBGrabDriver(DBfile *file)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
Returns:
A void pointer to the low-level driver’s file handle on success. NULL(0) on failure.
Description:
This method is used to obtain the low-level driver’s file handle. For example, one can use it to
obtain the HDF5 file id. The caller is responsible for casting the returned pointer to a pointer to the
correct type. Use DBGetDriverType() to obtain information on the type of driver currently in use.
When the low-level driver’s file handle is grabbed, all Silo-level operations on the file are pre-
vented until the file is UNgrabbed. For example, after a call to DBGrabDriver, calls to functions
like DBPutQuadmesh or DBGetCurve will fail until the driver is UNgrabbed using DBUngrab-
Driver().
Notes:
As far as the integrity of a Silo file goes, grabbing is inherently dangerous. If the client is not care-
ful, one can easily wind up corrupting the file for the Silo library (though all may be ‘normal’ for
the underlying driver library). Therefore, to minimize the likelihood of corrupting the Silo file
while it is grabbed, it is recommended that all operations with the low-level driver grabbed be con-
fined to a separate sub-directory in the silo file. That is, one should not mix writing of Silo objects
and low-level driver objects in the same directory. To achieve this, before grabbing, create the
desired directory and descend into it using Silo’s DBMkDir() and DBSetDir() functions. Then,
grab the driver and do all the work with the low-level driver that is necessary. Finally, ungrab the
driver and immediately ascend out of the directory using Silo’s DBSetDir(“..”).
For reasons described above, if problems occur on files that have been grabbed, users will likely be
asked to re-produce the problem on a similar file that has NOT been grabbed to rule out the possi-
ble corruption from grabbing.

DBUngrabDriver
Silo User’s Guide 2-67
DBUngrabDriver—Ungrab the low-level file driver
Synopsis:
int DBUngrabDriver(DBfile *file, const void *drvr_hndl)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
drvr_hndl The low-level driver handle.
Returns:
The driver type on success, DB_UNKNOWN on failure.
Description:
This function returns the Silo file to an ungrabbed state, permitting ‘norma’ Silo calls to again pro-
ceed as normal.

DBGetDriverType
2-68 Silo User’s Guide
DBGetDriverType—Get the type of driver for the specified file
Synopsis:
int DBGetDriverType(const DBfile *file)
Fortran Equivalent:
None
Arguments:
file A Silo database file handle.
Returns:
DB_UNKNOWN for failure. Otherwise, the specified driver type is returned
Description:
This function returns the type of driver used for the specified file. If you want to ask this question
without actually opening the file, use DBGetDriverTypeFromPath

DBGetDriverTypeFromPath
Silo User’s Guide 2-69
DBGetDriverTypeFromPath—Guess the driver type used by a file with the given
pathname
Synopsis:
int DBGetDriverTypeFromPath(char const *path)
Fortran Equivalent:
None
Arguments:
path Path to a file on the filesystem
Returns:
DB_UNKNOWN on failure to determine type. Otherwise, the driver type such as DB_PDB,
DB_HDF5.
Description:
This function examines the first few bytes of the file for tell-tale signs of whether it is a PDB file
or an HDF5 file.
If it is a PDB file, it cannot distinguish between a file generated by DB_PDB driver and
DB_PDBP (PDB Proper) driver. It will always return DB_PDB for a PDB file.
If the file is an HDF5, the function is currently not implemented to distiniguish between various
HDF5 VFDs the file may have been generated with. It will always return DB_HDF5 for an HDF5
file.
Note, this function will determine only whether the underlying file is a PDB or HDF5 file. It will
not however, indicate whether the file is a PDB or HDF5 file that was indeed generated by Silo.
See “DBInqFile” on page 2-70 for a function that will indicate whether the file is indeed a Silo file.
Note, however, that DBInqFile is a more expensive operation.

DBInqFile
2-70 Silo User’s Guide
DBInqFile—Inquire if filename is a Silo file.
Synopsis:
int DBInqFile (char const *filename)
Fortran Equivalent:
integer function dbinqfile(filename, lfilename, is_file)
Arguments:
filename Name of file.
Returns:
DBInqFile returns 0 if filename is not a Silo file, a positive number if filename is a Silo file,
and a negative number if an error occurred.
Description:
The DBInqFile function is mainly used for its return value, as seen above.
Prior to version 4.7.1 of the Silo library, this function could return false positives when the file-
name referred to a PDB file that was NOT created by Silo. The reason for this is that all this func-
tion really did was check whether or not DBOpen would succeed on the file.
Starting in version 4.7.1 of the Silo library, this function will attempt to count the number of Silo
objects (not including directories) in the first non-empty directory it finds. If it cannot find any Silo
objects in the file, it will return zero (0) indicating the file is NOT a Silo file.
Because very early versions of the Silo library did not store anything to a Silo file to distinguish it
from a PDB file, it is conceivable that this function will return false negatives for very old, empty
Silo files. But, that case should be rare.
Similar problems do not exist for HDF5 files because Silo’s HDF5 driver has always stored infor-
mation in the HDF5 file which helps to distinguish it as a Silo file.

DBInqFileHasObjects
Silo User’s Guide 2-71
DBInqFileHasObjects—Determine if an open file has any Silo objects
Synopsis:
int DBInqFileHasObjects(DBfile *dbfile)
Fortran Equivalent:
None
Arguments:
dbfile The Silo database file handle
Description:
Examine an open file for existence of any Silo objects.

DBInqFileHasObjects
2-72 Silo User’s Guide
_silolibinfo—character array written by Silo to root directory indicating the Silo library
version number used to generate the file
Synopsis:
int n;
char vers[1024];
sprintf(vers, “silo-4.6”);
n = strlen(vers);
DBWrite(dbfile, “_silolibinfo”, vers, &n, 1, DB_CHAR);
Description:
This is a simple array variable written at the root directory in a Silo file that contains the Silo
library version string. It cannot be disabled.

DBInqFileHasObjects
Silo User’s Guide 2-73
_hdf5libinfo—character array written by Silo to root directory indicating the HDF5
library version number used to generate the file
Synopsis:
int n;
char vers[1024];
sprintf(vers, “hdf5-1.6.6”);
n = strlen(vers);
DBWrite(dbfile, “_hdf5libinfo”, vers, &n, 1, DB_CHAR);
Description:
This is a simple array variable written at the root directory in a Silo file that contains the HDF5
library version string. It cannot be disabled. Of course, it exists, only in files created with the
HDF5 driver.

DBInqFileHasObjects
2-74 Silo User’s Guide
_was_grabbed—single integer written by Silo to root directory whenever a Silo file has
been grabbed.
Synopsis:
int n=1;
DBWrite(dbfile, “_was_grabbed”, &n, &n, 1, DB_INT);
Description:
This is a simple array variable written at the root directory in a Silo whenever a Silo file has been
grabbed by the DBGrabDriver() function. It cannot be disabled.

DBInqFileHasObjects
Silo User’s Guide 2-75
3 API Section Meshes, Variables and Materials
If you are interested in learning how to deal with these objects in parallel, See “Multi-Block
Objects, Parallelism and Poor-Man’s Parallel I/O” on page 154.
This section of the Silo API manual describes all the high-level Silo objects that are sufficiently
self-describing as to be easily shared between a variety of applications.
Silo supports a variety of mesh types including simple 1D curves, structured meshes including
block-structured Adaptive Mesh Refinement (AMR) meshes, point (or gridless) meshes consisting
entirely of points, unstructured meshes consisting of the standard zoo of element types, fully arbi-
trary polyhedral meshes and Constructive Solid Geometry “meshes” described by boolean opera-
tions of primitive quadric surfaces.
In addition, Silo supports both piecewise constant (e.g. zone-centered) and piecewise-linear (e.g.
node-centered) variables (e.g. fields) defined on these meshes. Silo also supports the decomposi-
tion of these meshes into materials (and material species) including cases where multiple materials
are mixing within a single mesh element. Finally, Silo also supports the specification of expres-
sions representing derived variables.
The functions described in this section of the manual include...
DBPutCurve . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 77
DBGetCurve . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 79
DBPutPointmesh. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 80
DBGetPointmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 83
DBPutPointvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
DBPutPointvar1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 86
DBGetPointvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 87
DBPutQuadmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 88
DBGetQuadmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 91
DBPutQuadvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 92
DBPutQuadvar1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 96
DBGetQuadvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 98
DBPutUcdmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 99
DBPutUcdsubmesh. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 107
DBGetUcdmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 108
DBPutZonelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 109
DBPutZonelist2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 110
DBPutPHZonelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 112
DBGetPHZonelist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 116
DBPutFacelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 117
DBPutUcdvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 119
DBPutUcdvar1 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 122
DBGetUcdvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 124
DBPutCsgmesh. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 125
DBGetCsgmesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 130
DBPutCSGZonelist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 131

DBInqFileHasObjects
2-76 Silo User’s Guide
DBGetCSGZonelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 136
DBPutCsgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 137
DBGetCsgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 139
DBPutMaterial . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 140
DBGetMaterial . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 144
DBPutMatspecies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 145
DBGetMatspecies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 148
DBPutDefvars. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 149
DBGetDefvars . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 151
DBInqMeshname . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 152
DBInqMeshtype . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 153

DBPutCurve
Silo User’s Guide 2-77
DBPutCurve—Write a curve object into a Silo file
Synopsis:
int DBPutCurve (DBfile *dbfile, char const *curvename,
void const *xvals, void const *yvals, int datatype,
int npoints, DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputcurve(dbid, curvename, lcurvename, xvals,
yvals, datatype, npoints, optlist_id, status)
Arguments:
dbfile Database file pointer
curvename Name of the curve object
xvals Array of length npoints containing the x-axis data values. Must be NULL
when either DBOPT_XVARNAME or DBOPT_REFERENCE is used.
yvals Array of length npoints containing the y-axis data values. Must be NULL
when either DBOPT_YVARNAME or DBOPT_REFERENCE is used.
datatype Data type of the xvals and yvals arrays. One of the predefined Silo types.
npoints The number of points in the curve
optlist Pointer to an option list structure containing additional information to be
included in the compound array object written into the Silo file. Use NULL is
there are no options.
Returns:
DBPutCurve returns zero on success and -1 on failure.
Description:
The DBPutCurve function writes a curve object into a Silo file. A curve is a set of x/y points that
describes a two-dimensional curve.
Both the xvals and yvals arrays must have the same datatype.
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of this construct.
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_LABEL int Problem cycle value. 0
DBOPT_XLABEL char * Label for the x-axis NULL
DBOPT_YLABEL char * Label for the y-axis NULL
DBPutCurve
2-78 Silo User’s Guide
In some cases, particularly when writing multi-part silo files from parallel clients, it is convenient
to write curve data to something other than the “master” or “root” file. However, for a visualiza-
tion tool to become aware of such objects, the tool is then required to traverse all objects in all the
files of a multi-part file to find such objects. The DBOPT_REFERENCE option helps address this
issue by permitting the writer to create knowledge of a curve object in the “master” or “root” file
but put the actual curve object (the referenced object) wherever is most convenient. This output
option would be useful for other Silo objects, meshes and variables, as well. However, it is cur-
rently only available for curve objects.
DBOPT_XUNITS char * Character string defining the units for the
x-axis.
NULL
DBOPT_YUNITS char * Character string defining the units for the
y-axis
NULL
DBOPT_XVARNAME char * Name of the domain (x) variable. This is
the problem variable name, not the code
variable name passed into the xvals
argument.
NULL
DBOPT_YVARNAME char * Name of the domain (y) variable. This is
problem variable name, not the code vari-
able name passed into the yvals argu-
ment.
NULL
DBOPT_REFERENCE char * Name of the real curve object this object
references. The name can take the form of
‘<file:/path-to-curve-object>’ just as mesh
names in the DBPutMultiMesh call.
Note also that if this option is set, then the
caller must pass NULL for both xvals and
yvals arguments but must also pass valid
information for all other object attributes
including not only npoints and datatype
but also any options.
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_COORDSYS int Coordinate system. One of:
DB_CARTESIAN or DB_SPHERICAL
DB_CARTESIAN
DBOPT_MISSING_VALUE double Specify a numerical value that is intended
to represent “missing values” in the x or y
data arrays. Default is
DB_MISSING_VALUE_NOT_SET
DB_MISSING_VA
LUE_NOT_SET
Option Name
Value
Data Type Option Meaning Default Value

DBGetCurve
Silo User’s Guide 2-79
DBGetCurve—Read a curve from a Silo database.
Synopsis:
DBcurve *DBGetCurve (DBfile *dbfile, char const *curvename)
Fortran Equivalent:
integer function dbgetcurve(dbid, curvename, lcurvename, maxpts,
xvals, yvals, datatype, npts)
Arguments:
dbfile Database file pointer.
curvename Name of the curve to read.
Returns:
DBCurve returns a pointer to a DBcurve structure on success and NULL on failure.
Description:
The DBGetCurve function allocates a DBcurve data structure, reads a curve from the Silo data-
base, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutPointmesh
2-80 Silo User’s Guide
DBPutPointmesh—Write a point mesh object into a Silo file.
Synopsis:
int DBPutPointmesh (DBfile *dbfile, char const *name, int ndims,
void const * const coords[], int nels,
int datatype, DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputpm(dbid, name, lname, ndims, x, y, z, nels,
datatype, optlist_id, status)
void* x, y, z (if ndims<3, z=0 ok, if ndims<2, y=0 ok)
Arguments:
dbfile Database file pointer.
name Name of the mesh.
ndims Number of dimensions.
coords Array of length ndims containing pointers to coordinate arrays.
nels Number of elements (points) in mesh.
datatype Datatype of the coordinate arrays. One of the predefined Silo data types.
optlist Pointer to an option list structure containing additional information to be
included in the mesh object written into the Silo file. Typically, this argument is
NULL.
Returns:
DBPutPointmesh returns zero on success and -1 on failure.
Description:
The DBPutPointmesh function accepts pointers to the coordinate arrays and is responsible for
writing the mesh into a point-mesh object in the Silo file.
A Silo point-mesh object contains all necessary information for describing a mesh. This includes
the coordinate arrays, the number of dimensions (1,2,3,...) and the number of points.
DBPutPointmesh
Silo User’s Guide 2-81
Notes:
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of this construct.
Option Name
Data
Type Option Meaning
Default
Value
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_XLABEL char * Character string defining the label associ-
ated with the X dimension.
NULL
DBOPT_YLABEL char * Character string defining the label associ-
ated with the Y dimension.
NULL
DBOPT_ZLABEL char * Character string defining the label associ-
ated with the Z dimension.
NULL
DBOPT_NSPACE int Number of spatial dimensions used by this
mesh.
ndims
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0
DBOPT_XUNITS char * Character string defining the units associ-
ated with the X dimension.
NULL
DBOPT_YUNITS char * Character string defining the units associ-
ated with the Y dimension.
NULL
DBOPT_ZUNITS char * Character string defining the units associ-
ated with the Z dimension.
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_MRGTREE_NAME char * Name of the mesh region grouping tree to
be associated with this mesh.
NULL
DBOPT_NODENUM void* An array of length nnodes giving a global
node number for each node in the mesh.
By default, this array is treated as type int.
NULL
DBOPT_LLONGNZNUM int Indicates that the array passed for
DBOPT_NODENUM option is of long long
type instead of int.
0
DBOPT_LO_OFFSET int Zero-origin index of first non-ghost node.
All points in the mesh before this one are
considered ghost.
0
DBOPT_HI_OFFSET int Zero-origin index of last non-ghost node.
All points in the mesh after this one are
considered ghost.
nels-1
DBPutPointmesh
2-82 Silo User’s Guide
DBOPT_GHOST_NODE_LABELS char * Optional array of char values indicating
the ghost labeling
(DB_GHOSTTYPE_NOGHOST or
DB_GHOSTTYPE_INTDUP) of each point
NULL
DBOPT_ALT_NODENUM_VARS char ** A null terminated list of names of optional
array(s) or DBpointvar objects indicating
(multiple) alternative numbering(s) for
nodes.
NULL
The following optlist options have been deprecated. Instead use MRG trees
DBOPT_GROUPNUM int The group number to which this point-
mesh belongs.
-1 (not in
a group)
Option Name
Data
Type Option Meaning
Default
Value

DBGetPointmesh
Silo User’s Guide 2-83
DBGetPointmesh—Read a point mesh from a Silo database.
Synopsis:
DBpointmesh *DBGetPointmesh (DBfile *dbfile, char const *meshname)
Arguments:
dbfile Database file pointer.
meshname Name of the mesh.
Returns:
DBGetPointmesh returns a pointer to a DBpointmesh structure on success and NULL on failure.
Description:
The DBGetPointmesh function allocates a DBpointmesh data structure, reads a point mesh from
the Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutPointvar
2-84 Silo User’s Guide
DBPutPointvar—Write a vector/tensor point variable object into a Silo file.
Synopsis:
int DBPutPointvar (DBfile *dbfile, char const *name,
char const *meshname, int nvars, void const * cost vars[],
int nels, int datatype, DBoptlist const *optlist)
Fortran Equivalent:
None. See DBPutPointvar1
Arguments:
dbfile Database file pointer.
name Name of the variable set.
meshname Name of the associated point mesh.
nvars Number of variables supplied in vars array.
vars Array of length nvars containing pointers to value arrays.
nels Number of elements (points) in variable.
datatype Datatype of the value arrays. One of the predefined Silo data types.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. Typically, this argument
is NULL.
Returns:
DBPutPointvar returns zero on success and -1 on failure.
Description:
The DBPutPointvar function accepts pointers to the value arrays and is responsible for writing the
variables into a point-variable object in the Silo file.
A Silo point-variable object contains all necessary information for describing a variable associated
with a point mesh. This includes the number of arrays, the datatype of the variable, and the number
of points. This function should be used when writing vector or tensor quantities. Otherwise, it is
more convenient to use DBPutPointvar1.

DBPutPointvar
Silo User’s Guide 2-85
Notes:
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of this construct.
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_NSPACE int Number of spatial dimensions used by this
mesh.
ndims
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0
DBOPT_ASCII_LABEL int Indicate if the variable should be treated
as single character, ascii values. A value
of 1 indicates yes, 0 no.
0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_REGION_PNAMES char** A null-pointer terminated array of pointers
to strings specifying the pathnames of
regions in the mrg tree for the associated
mesh where the variable is defined. If
there is no mrg tree associated with the
mesh, the names specified here will be
assumed to be material names of the
material object associated with the mesh.
The last pointer in the array must be null
and is used to indicate the end of the list of
names. See
“DBOPT_REGION_PNAMES” on
page 218.
NULL
DBOPT_CONSERVED int Indicates if the variable represents a phys-
ical quantity that must be conserved under
various operations such as interpolation.
0
DBOPT_EXTENSIVE int Indicates if the variable represents a phys-
ical quantity that is extensive (as opposed
to intensive). Note, while it is true that any
conserved quantity is extensive, the con-
verse is not true. By default and histori-
cally, all Silo variables are treated as
intensive.
0
DBOPT_MISSING_VALUE double Specify a numerical value that is intended
to represent “missing values” variable data
array(s). Default is
DB_MISSING_VALUE_NOT_SET
DB_MISSING_VALUE_
NOT_SET

DBPutPointvar1
2-86 Silo User’s Guide
DBPutPointvar1—Write a scalar point variable object into a Silo file.
Synopsis:
int DBPutPointvar1 (DBfile *dbfile, char const *name,
char const *meshname, void const *var, int nels, int datatype,
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputpv1(dbid, name, lname, meshname, lmeshname,
var, nels, datatype, optlist_id, status)
Arguments:
dbfile Database file pointer.
name Name of the variable.
meshname Name of the associated point mesh.
var Array containing data values for this variable.
nels Number of elements (points) in variable.
datatype Datatype of the variable. One of the predefined Silo data types.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. Typically, this argument
is NULL.
Returns:
DBPutPointvar1 returns zero on success and -1 on failure.
Description:
The DBPutPointvar1 function accepts a value array and is responsible for writing the variable into
a point-variable object in the Silo file.
A Silo point-variable object contains all necessary information for describing a variable associated
with a point mesh. This includes the number of arrays, the datatype of the variable, and the number
of points. This function should be used when writing scalar quantities. To write vector or tensor
quantities, one must use DBPutPointvar.
See “DBPutPointvar” on page 84 to a description of the options accepted by this function.

DBGetPointvar
Silo User’s Guide 2-87
DBGetPointvar—Read a point variable from a Silo database.
Synopsis:
DBmeshvar *DBGetPointvar (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Name of the variable.
Returns:
DBGetPointvar returns a pointer to a DBmeshvar structure on success and NULL on failure.
Description:
The DBGetPointvar function allocates a DBmeshvar data structure, reads a variable associated
with a point mesh from the Silo database, and returns a pointer to that structure. If an error occurs,
NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutQuadmesh
2-88 Silo User’s Guide
DBPutQuadmesh—Write a quad mesh object into a Silo file.
Synopsis:
int DBPutQuadmesh (DBfile *dbfile, char const *name,
char const * const coordnames[], void const * const coords[],
int dims[], int ndims, int datatype, int coordtype,
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputqm(dbid, name, lname, xname, lxname, yname,
lyname, zname, lzname, x, y, z, dims, ndims,
datatype, coordtype, optlist_id, status)
void* x, y, z (if ndims<3, z=0 ok, if ndims<2, y=0 ok)
character* xname, yname, zname (if ndims<3, zname=0 ok, etc.)
Arguments:
dbfile Database file pointer.
name Name of the mesh.
coordnames Array of length ndims containing pointers to the names to be provided when
writing out the coordinate arrays. This parameter is currently ignored and can
be set as NULL.
coords Array of length ndims containing pointers to the coordinate arrays.
dims Array of length ndims describing the dimensionality of the mesh. Each value
in the dims array indicates the number of nodes contained in the mesh along
that dimension.
ndims Number of dimensions.
datatype Datatype of the coordinate arrays. One of the predefined Silo data types.
coordtype Coordinate array type. One of the predefined types: DB_COLLINEAR or
DB_NONCOLLINEAR. Collinear coordinate arrays are always one-
dimensional, regardless of the dimensionality of the mesh; non-collinear arrays
have the same dimensionality as the mesh.
optlist Pointer to an option list structure containing additional information to be
included in the mesh object written into the Silo file. Typically, this argument is
NULL.
Returns:
DBPutQuadmesh returns zero on success and -1 on failure.
Description:
The DBPutQuadmesh function accepts pointers to the coordinate arrays and is responsible for
writing the mesh into a quad-mesh object in the Silo file.

DBPutQuadmesh
Silo User’s Guide 2-89
A Silo quad-mesh object contains all necessary information for describing a mesh. This includes
the coordinate arrays, the rank of the mesh (1,2,3,...) and the type (collinear or non-collinear). In
addition, other information is useful and is therefore optionally included (row-major indicator,
time and cycle of mesh, offsets to ‘real’ zones, plus coordinate system type.)
Notes:
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of this construct.
Option Name
Data
Type Option Meaning Default Value
DBOPT_COORDSYS int Coordinate system. One of:
DB_CARTESIAN, DB_CYLINDRICAL,
DB_SPHERICAL, DB_NUMERICAL, or
DB_OTHER.
DB_OTHER
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_FACETYPE int Zone face type. One of the predefined
types: DB_RECTILINEAR or
DB_CURVILINEAR.
DB_RECTILINEAR
DBOPT_HI_OFFSET int * Array of length ndims which defines zero-
origin offsets from the last node for the
ending index along each dimension.
{0,0,...}
DBOPT_LO_OFFSET int * Array of ndims which defines zero-origin
offsets from the first node for the starting
index along each dimension.
{0,0,...}
DBOPT_XLABEL char * Character string defining the label associ-
ated with the X dimension.
NULL
DBOPT_YLABEL char * Character string defining the label associ-
ated with the Y dimension.
NULL
DBOPT_ZLABEL char * Character string defining the label associ-
ated with the Z dimension.
NULL
DBOPT_MAJORORDER int Indicator for row-major (0) or column-
major (1) storage for multidimensional
arrays.
0
DBOPT_NSPACE int Number of spatial dimensions used by this
mesh.
ndims
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0
DBOPT_PLANAR int Planar value. One of: DB_AREA or
DB_VOLUME.
DB_OTHER
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME dou-
ble
Problem time value. 0.0
DBOPT_XUNITS char * Character string defining the units associ-
ated with the X dimension.
NULL

DBPutQuadmesh
2-90 Silo User’s Guide
The options DB_LO_OFFSET and DB_HI_OFFSET should be used if the mesh being described
uses the notion of “phoney” zones (i.e., some zones should be ignored.) For example, if a 2-D
mesh had designated the first column and row, and the last two columns and rows as “phoney”,
then we would use: lo_off = {1,1} and hi_off = {2,2}.
DBOPT_YUNITS char * Character string defining the units associ-
ated with the Y dimension.
NULL
DBOPT_ZUNITS char * Character string defining the units associ-
ated with the Z dimension.
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_BASEINDEX int[3] Indicate the indices of the mesh within its
group.
0,0,0
DBOPT_MRGTREE_NAME char * Name of the mesh region grouping tree to
be associated with this mesh.
NULL
DBOPT_GHOST_NODE_LABELS char * Optional array of char values indicating
the ghost labeling
(DB_GHOSTTYPE_NOGHOST or
DB_GHOSTTYPE_INTDUP) of each
node
NULL
DBOPT_GHOST_ZONE_LABELS char * Optional array of char values indicating
the ghost labeling
(DB_GHOSTTYPE_NOGHOST or
DB_GHOSTTYPE_INTDUP) of each zone
NULL
DBOPT_ALT_NODENUM_VARS char
**
A null terminated list of names of optional
array(s) or DBquadvar objects indicating
(multiple) alternative numbering(s) for
nodes.
NULL
DBOPT_ALT_ZONENUM_VARS char
**
A null terminated list of names of optional
array(s) or DBquadvar objects indicating
(multiple) alternative numbering(s) for
zones.
NULL
The following options have been deprecated. Use MRG trees instead
DBOPT_GROUPNUM int The group number to which this quad-
mesh belongs.
-1 (not in a group)
Option Name
Data
Type Option Meaning Default Value

DBGetQuadmesh
Silo User’s Guide 2-91
DBGetQuadmesh—Read a quadrilateral mesh from a Silo database.
Synopsis:
DBquadmesh *DBGetQuadmesh (DBfile *dbfile, char const *meshname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
meshname Name of the mesh.
Returns:
DBGetQuadmesh returns a pointer to a DBquadmesh structure on success and NULL on failure.
Description:
The DBGetQuadmesh function allocates a DBquadmesh data structure, reads a quadrilateral mesh
from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutQuadvar
2-92 Silo User’s Guide
DBPutQuadvar—Write a vector/tensor quad variable object into a Silo file.
Synopsis:
int DBPutQuadvar (DBfile *dbfile, char const *name,
char const *meshname, int nvars,
char const * const varnames[], void const * const vars[],
int dims[], int ndims, void const * const mixvars[],
int mixlen, int datatype, int centering,
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputqv(dbid, vname, lvname, mname, lmname,
nvars, varnames, lvarnames, vars, dims, ndims, mixvar,
mixlen, datatype, centering, optlist_id, status)
varnames contains the names of the variables either in a matrix of characters form (if
fortran2DStrLen is non null) or in a vector of characters form (if fortran2DStrLen is null) with the
varnames length being found in the lvarnames integer array,
var is essentially a matrix of size <nvars> x <var-size> where var-size is determined by dims and
ndims. The first “row” of the var matrix is the first component of the quadvar. The second “row” of
the var matrix goes out as the second component of the quadvar, etc.
Arguments:
dbfile Database file pointer.
name Name of the variable.
meshname Name of the mesh associated with this variable (written with DBPutQuadmesh
or DBPutUcdmesh). If no association is to be made, this value should be NULL.
nvars Number of sub-variables which comprise this variable. For a scalar array, this is
one. If writing a vector quantity, however, this would be two for a 2-D vector
and three for a 3-D vector.
varnames Array of length nvars containing pointers to character strings defining the
names associated with each sub-variable.
vars Array of length nvars containing pointers to arrays defining the values
associated with each subvariable. For true edge- or face-centering (as opposed
to DB_EDGECENT centering when ndims is 1 and DB_FACECENT centering
when ndims is 2), each pointer here should point to an array that holds ndims
sub-arrays, one for each of the i-, j-, k-oriented edges or i-, j-, k-intercepting
faces, respectively. Read the description for more details.
dims Array of length ndims which describes the dimensionality of the data stored in
the vars arrays. For DB_NODECENT centering, this array holds the number of
nodes in each dimension. For DB_ZONECENT centering, DB_EDGECENT
centering when ndims is 1 and DB_FACECENT centering when ndims is 2,
this array holds the number of zones in each dimension. Otherwise, for
DB_EDGECENT and DB_FACECENT centering, this array should hold the

DBPutQuadvar
Silo User’s Guide 2-93
number of nodes in each dimension.
ndims Number of dimensions.
mixvars Array of length nvars containing pointers to arrays defining the mixed-data
values associated with each subvariable. If no mixed values are present, this
should be NULL.
mixlen Length of mixed data arrays, if provided.
datatype Datatype of the variable. One of the predefined Silo data types.
centering Centering of the subvariables on the associated mesh. One of the predefined
types: DB_NODECENT, DB_EDGECENT, DB_FACECENT or DB_ZONECENT.
Note that DB_EDGECENT centering on a 1D mesh is treated identically to
DB_ZONECENT centering. Likewise for DB_FACECENT centering on a 2D
mesh.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. Typically, this argument
is NULL.
Returns:
DBPutQuadvar returns zero on success and -1 on failure.
Description:
The DBPutQuadvar function writes a variable associated with a quad mesh into a Silo file. A
quad-var object contains the variable values.
For node- (or zone-) centered data, the question of which value in the vars array goes with which
node (or zone) is determined implicitly by a one-to-one correspondence with the multi-dimen-
sional array list of nodes (or zones) defined by the logical indexing for the associated mesh’s nodes
(or zones).
Edge- and face-centered data require a little more explanation. We can group edges according to
their logical orientation. In a 2D mesh of Nx by Ny nodes, there are (Nx-1)Ny i-oriented edges and
Nx(Ny-1) j-oriented edges. Likewise, in a 3D mesh of Nx by Ny by Nz nodes, there are
(Nx-1)NyNz i-oriented edges, Nx(Ny-1)Nz, j-oriented edges and NxNy(Nz-1) k-oriented edges.
Each group of edges is almost the same size as a normal node-centered variable. So, for conceptual
convenience we in fact treat them that way and treat the extra slots in them as phony data. So, in
the case of edge-centered data, each of the pointers in the vars argument to DBPutQuadvar is
interpreted to point to an array that is ndims times the product of nodal sizes (NxNyNz). The first
part of the array (of size NxNy nodes for 2D or NxNyNz nodes for 3D) holds the i-oriented edge
data, the next part the j-oriented edge data, etc.
A similar approach is used for face centered data. In a 3D mesh of Nx by Ny by Nz nodes, there
are Nx(Ny-1)(Nz-1) i-intercepting faces, (Nx-1)Ny(Nz-1) j-intercepting faces and (Nx-1)(Ny-
1)Nz k-intercepting faces. Again, just as for edge-centered data, each pointer in the vars array is
interpreted to point to an array that is ndims times the product of nodal sizes. The first part holds
the i-intercepting face data, the next part the j-interception face data, etc.
Unlike node- and zone-centered data, there does not necessarily exist in Silo an explicit list of
edges or faces. As an aside, the DBPutFacelist call is really for writing the external faces of a
mesh so that a downstream visualization tool need not have to compute them when it displays the
DBPutQuadvar
2-94 Silo User’s Guide
mesh. Now, requiring the caller to create explicit lists of edges and/or faces in order to handle
edge- or face-centered data results in unnecessary additional data being written to a Silo file. This
increases file size as well as the time to write and read the file. To avoid this, we rely upon implicit
lists of edges and faces.
Finally, since the zones of a one dimensional mesh are basically edges, the case of
DB_EDGECENT centering for a one dimensional mesh is treated identically to the DB_ZONECENT
case. Likewise, since the zones of a two dimensional mesh are basically faces, the DB_FACECENT
centering for a two dimensional mesh is treated identically to the DB_ZONECENT case.
Other information can also be included. This function is useful for writing vector and tensor fields,
whereas the companion function, DBPutQuadvar1, is appropriate for writing scalar fields.
Notes:
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of this construct.
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_COORDSYS int Coordinate system. One of:
DB_CARTESIAN, DB_CYLINDRICAL,
DB_SPHERICAL, DB_NUMERICAL, or
DB_OTHER.
DB_OTHER
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_FACETYPE int Zone face type. One of the predefined
types: DB_RECTILINEAR or
DB_CURVILINEAR.
DB_RECTILINEAR
DBOPT_LABEL char * Character string defining the label associ-
ated with this variable.
NULL
DBOPT_MAJORORDER int Indicator for row-major (0) or column-
major (1) storage for multidimensional
arrays.
0
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_UNITS char * Character string defining the units associ-
ated with this variable.
NULL
DBOPT_USESPECMF int Boolean (DB_OFF or DB_ON) value
specifying whether or not to weight the
variable by the species mass fraction
when using material species data.
DB_OFF
DBOPT_ASCII_LABEL int Indicate if the variable should be treated
as single character, ascii values. A value
of 1 indicates yes, 0 no.
0

DBPutQuadvar
Silo User’s Guide 2-95
DBOPT_CONSERVED int Indicates if the variable represents a phys-
ical quantity that must be conserved under
various operations such as interpolation.
0
DBOPT_EXTENSIVE int Indicates if the variable represents a phys-
ical quantity that is extensive (as opposed
to intensive). Note, while it is true that any
conserved quantity is extensive, the con-
verse is not true. By default and histori-
cally, all Silo variables are treated as
intensive.
0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_REGION_PNAMES char** A null-pointer terminated array of pointers
to strings specifying the pathnames of
regions in the mrg tree for the associated
mesh where the variable is defined. If
there is no mrg tree associated with the
mesh, the names specified here will be
assumed to be material names of the
material object associated with the mesh.
The last pointer in the array must be null
and is used to indicate the end of the list of
names. See
“DBOPT_REGION_PNAMES” on
page 218.
NULL
DBOPT_MISSING_VALUE double Specify a numerical value that is intended
to represent “missing values” variable data
array(s). Default is
DB_MISSING_VALUE_NOT_SET
DB_MISSING_VALUE_
NOT_SET
Option Name
Value
Data Type Option Meaning Default Value

DBPutQuadvar1
2-96 Silo User’s Guide
DBPutQuadvar1— Write a scalar quad variable object into a Silo file.
Synopsis:
int DBPutQuadvar1 (DBfile *dbfile, char const *name,
char const *meshname, void const *var, int const dims[],
int ndims, void const *mixvar, int mixlen, int datatype,
int centering, DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputqv1(dbid, name, lname, meshname, lmeshname,
var, dims, ndims, mixvar, mixlen, datatype,
centering, optlist_id, status)
Arguments:
dbfile Database file pointer.
name Name of the variable.
meshname Name of the mesh associated with this variable (written with DBPutQuadmesh
or DBPutUcdmesh.) If no association is to be made, this value should be NULL.
var Array defining the values associated with this variable. For true edge- or face-
centering (as opposed to DB_EDGECENT centering when ndims is 1 and
DB_FACECENT centering when ndims is 2), each pointer here should point to
an array that holds ndims sub-arrays, one for each of the i-, j-, k-oriented edges
or i-, j-, k-intercepting faces, respectively. Read the description for
DBPutQuadvar more details.
dims Array of length ndims which describes the dimensionality of the data stored in
the var array. For DB_NODECENT centering, this array holds the number of
nodes in each dimension. For DB_ZONECENT centering, DB_EDGECENT
centering when ndims is 1 and DB_FACECENT centering when ndims is 2,
this array holds the number of zones in each dimension. Otherwise, for
DB_EDGECENT and DB_FACECENT centering, this array should hold the
number of nodes in each dimension.
ndims Number of dimensions.
mixvar Array defining the mixed-data values associated with this variable. If no mixed
values are present, this should be NULL.
mixlen Length of mixed data arrays, if provided.
datatype Datatype of sub-variables. One of the predefined Silo data types.
centering Centering of the subvariables on the associated mesh. One of the predefined
types: DB_NODECENT, DB_EDGECENT, DB_FACECENT or DB_ZONECENT.
Note that DB_EDGECENT centering on a 1D mesh is treated identically to
DB_ZONECENT centering. Likewise for DB_FACECENT centering on a 2D
mesh.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. Typically, this argument

DBPutQuadvar1
Silo User’s Guide 2-97
is NULL.
Returns:
DBPutQuadvar1 returns zero on success and -1 on failure.
Description:
The DBPutQuadvar1 function writes a scalar variable associated with a quad mesh into a Silo file.
A quad-var object contains the variable values, plus the name of the associated quad-mesh. Other
information can also be included. This function should be used for writing scalar fields, and its
companion function, DBPutQuadvar, should be used for writing vector and tensor fields.
For edge- and face-centered data, please refer to the description for DBPutQuadvar for a more
detailed explanation.
Notes:
See “DBPutQuadvar” on page 92 for a description of options accepted by this function.

DBGetQuadvar
2-98 Silo User’s Guide
DBGetQuadvar—Read a quadrilateral variable from a Silo database.
Synopsis:
DBquadvar *DBGetQuadvar (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Name of the variable.
Returns:
DBGetQuadvar returns a pointer to a DBquadvar structure on success and NULL on failure.
Description:
The DBGetQuadvar function allocates a DBquadvar data structure, reads a variable associated
with a quadrilateral mesh from the Silo database, and returns a pointer to that structure. If an error
occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutUcdmesh
Silo User’s Guide 2-99
DBPutUcdmesh—Write a UCD mesh object into a Silo file.
Synopsis:
int DBPutUcdmesh (DBfile *dbfile, char const *name, int ndims,
char const * const coordnames[], void const * const coords[],
int nnodes, int nzones, char const *zonel_name,
char const *facel_name, int datatype,
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputum(dbid, name, lname, ndims, x, y, z, xname,
lxname, yname, lyname, zname, lzname, nnodes
nzones, zonel_name, lzonel_name, facel_name,
lfacel_name, datatype, optlist_id, status)
void *x,y,z (if ndims<3, z=0 ok, if ndims<2, y=0 ok)
character* xname,yname,zname (same rules)
Arguments:
dbfile Database file pointer.
name Name of the mesh.
ndims Number of spatial dimensions represented by this UCD mesh.
coordnames Array of length ndims containing pointers to the names to be provided when
writing out the coordinate arrays. This parameter is currently ignored and can
be set as NULL.
coords Array of length ndims containing pointers to the coordinate arrays.
nnodes Number of nodes in this UCD mesh.
nzones Number of zones in this UCD mesh.
zonel_name Name of the zonelist structure associated with this variable [written with
DBPutZonelist]. If no association is to be made or if the mesh is composed
solely of arbitrary, polyhedral elements, this value should be NULL. If a
polyhedral-zonelist is to be associated with the mesh, DO NOT pass the name of
the polyhedral-zonelist here. Instead, use the DBOPT_PHZONELIST option
described below. For more information on arbitrary, polyhedral zonelists, see
below and also see the documentation for DBPutPHZonelist.
facel_name Name of the facelist structure associated with this variable [written with
DBPutFacelist]. If no association is to be made, this value should be NULL.
datatype Datatype of the coordinate arrays. One of the predefined Silo data types.
optlist Pointer to an option list structure containing additional information to be
included in the mesh object written into the Silo file. See the table below for the
valid options for this function. If no options are to be provided, use NULL for
this argument.

DBPutUcdmesh
2-100 Silo User’s Guide
Returns:
DBPutUcdmesh returns zero on success and -1 on failure.
Description:
The DBPutUcdmesh function accepts pointers to the coordinate arrays and is responsible for writ-
ing the mesh into a UCD mesh object in the Silo file.
A Silo UCD mesh object contains all necessary information for describing a mesh. This includes
the coordinate arrays, the rank of the mesh (1,2,3,...) and the type (collinear or non-collinear.) In
addition, other information is useful and is therefore included (time and cycle of mesh, plus coor-
dinate system type).
A Silo UCD mesh may be composed of either zoo-type elements or arbitrary, polyhedral elements
or a mixture of both zoo-type and arbitrary, polyhedral elements. The zonelist (connectivity) infor-
mation for zoo-type elements is written with a call to DBPutZonelist. When there are only zoo-
type elements in the mesh, this is the only zonelist information associated with the mesh. However,
the caller can optionally specify the name of an arbitrary, polyhedral zonelist written with a call to
DBPutPHZonelist using the DBOPT_PHZONELIST option. If the mesh consists solely of arbi-
trary, polyhedral elements, the only zonelist associated with the mesh will be the one written with
the call to DBPutPHZonelist.
When a mesh is composed of both zoo-type elements and polyhedral elements, it is assumed that
all the zoo-type elements come first in the mesh followed by all the polyhedral elements. This has
implications for any DBPutUcdvar calls made on such a mesh. For zone-centered data, the vari-
able array should be organized so that values corresponding to zoo-type zones come first followed
by values corresponding to polyhedral zones. Also, since both the zoo-type zonelist and the poly-
hedral zonelist support hi- and lo- offsets for ghost zones, the ghost-zones of a mesh may consist
of zoo-type or polyhedral zones or a mixture of both.
Notes:
See the description of “DBCalcExternalFacelist” on page 2-225 or “DBCalcExternalFacelist2” on
page 2-227 for an automated way of computing the facelist needed for this call.
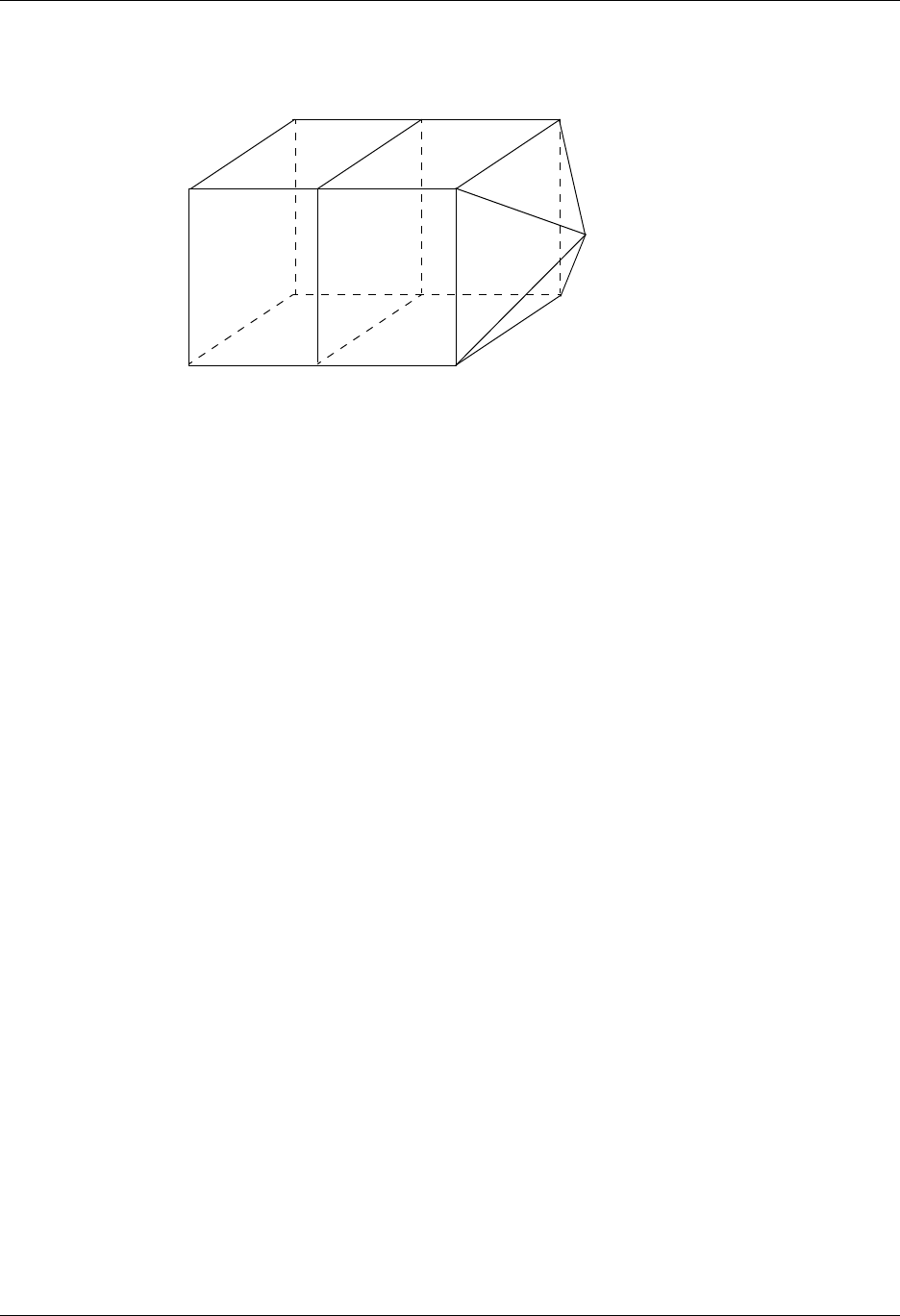
123
456
78 9
10 11 12
nnodes = 13
nzones = 3
nzshapes = 2
lznodelist = 2*8 + 1*5 = 21 zone nodes
nfaces = 13 external faces
nfshapes = 2 external face shapes
nftypes = 0
lfnodelist = 9*4 + 4*3 = 48 external face nodes
fnodelist = { 1,2,8,7 external face nodelist
2,3,9,8,
8,9,12,11,
5,6,12,11,...}
fshapesize = {4,3} external face shape sizes
fshapecnt = {9,4} external face shape counts
znodelist = { 7,10,11,8,1,4,5,2, zone nodelist
8,11,12,9,2,5,6,3,
zshapesize = {8,5} zone shape sizes
zshapecnt = {2,1} zone shape counts
x = {0,1,2,0,1,2,0,1,2,0,1,2,3}
y = {1,1,1,0,0,0,1,1,1,0,0,0,.5}
X
Y
Z
z = {1,1,1,1,1,1,0,0,0,0,0,0,.5}
fzoneno = {1,2,2,2,...}external face zone nos
13
3,9,12,6,13}
DBPutUcdmesh
Silo User’s Guide 2-101
Figure 0-1: Example usage of UCD zonelist and external facelist variables.
The order in which nodes are defined in the zonelist is important, especially for 3D cells. Nodes
defining a 2D cell should be supplied in either clockwise or counterclockwise order around the
DBPutUcdmesh
2-102 Silo User’s Guide
cell. The node, edge and face ordering and orientations for the predefined 3D cell types are illus-
trated below.
4
5
4
0
3
6
3
0
7
2
4
5
2
1
1
2
3
0
1
0
1
2
3
13
0
2
5
4
1
5
0
2
7
6
4
3
0
3
5
1
24
76
8
0
Node Order Edge Order Face Order
3
5
1
8
10
11
9
2
46
7
Tetrahedron
Pyramid
Prism
Hexahedron
0:012
1:023
2:031
3:132
0:0123
1:034
2:041
3:142
4:243
0:0123
1:034
2:0451
3:152
4:2543
0:0154
1:0321
2:0473
3:1265
4:2376
5:4567
Figure 0-2: Node, edge and face ordering for zoo-type UCD zone shapes.
Given the node ordering in the left-most column, there is indeed an algorithm for determining the
other orderings for each cell type.
For edges, each edge is identified by a pair of integer indices; the first being the “tail” of an arrow
oriented along the edge and the second being the “head” with the smaller node index always
placed first (at the tail). Next, the ordering of edges is akin to a lexicographic ordering of these
pairs of integers. This means that we start with the lowest node number of a cell shape, zero, and
find all edges with node zero as one of the points on the edge. Each such edge will have zero as its
tail. Since they all start with node 0 as the tail, we order these edges from smallest to largest “head”
node. Then we go to the next lowest node number on the cell that has edges that have yet to have

DBPutUcdmesh
Silo User’s Guide 2-103
been placed in the ordering. We find all the edges from that node (that have not already been
placed in the ordering) from smallest to largest “head” node. We continue this process until all the
edges on the cell have been placed in the ordering.
For faces, a similar algorithm is used. Starting with the lowest numbered node on a face, we enu-
merate the nodes over a face using the right hand rule for the normal to the face pointing away
from the innards of the cell. When one places the thumb of the right hand in the direction of this
normal, the direction of the fingers curling around it identify the direction we go to identify the
nodes of the face. Just as for edges, we start identifying faces for the lowest numbered node of the
cell (0). We find all faces that share this node. Of these, the face that enumerates the next lowest
node number as we traverse the nodes using the right hand rule, is placed first in the ordering.
Then, the face that has the next lowest node number and so on.
An example using arbitrary polyhedrons for some zones is illustrated in Figure 0-3 on page 104.
The nodes of a DB_ZONETYPE_POLYHEDRON are specified in the following fashion: First
specify the number of faces in the polyhedron. Then, for each face, specify the number of nodes in
the face followed by the nodes that make up the face. The nodes should be ordered such that they
are numbered in a counter-clockwise fashion when viewed from the outside (e.g. right-hand rules
yields an outward facing normal). For a fully arbitrarily connected mesh, see DBPutPHZonelist().
In addition, for a sequence of consecutive zones of type DB_ZONETYPE_POLYHEDRON in a
zonelist, the shapesize entry is taken to be the sum of all the associated positions occupied in the
nodelist data. So, for the example in Figure 0-3 on page 104, the shapesize entry for the
DB_ZONETYPE_POLYEDRON segment of the zonelist is ‘53’ because for the two arbitrary
polyhedral zones in the zonelist, 53 positions in the nodelist array are used.
123
456
78 9
10 11 12
nzones = 3
nzshapes = 2
lznodelist = 8 + 1 + 6 * 5 + 1 + 5 + 4 * 4 = 61
znodelist = {7,10,11,8,1,4,5,2,
X
Y
Z
13
6,
4,8,9,12,11,
4,9,3,6,12,
4,3,2,5,6,
4,2,8,11,5,
4,11,12,6,5,
4,9,8,2,3,
5,
4,9,12,6,3,
3,12,13,6,
3,9,13,12,
3,3,13,9,
3,6,13,3}
zshapetype = {DB_ZONETYPE_HEX,
DB_ZONETYPE_POLYHEDRON}
zshapesize = {8, 53}
zshapecnt = {1, 2}
DBPutUcdmesh
2-104 Silo User’s Guide
Figure 0-3: Example usage of UCD zonelist combining a hex and 2 polyhedra. This example is intended to illustrate
the representation of arbitrary polyhedra. So, although the two polyhedra represent a hex and pyramid which would
ordinarily be handled just fine by a ‘normal’ zonelist, they are expressed using arbitrary connectivity here.
DBPutUcdmesh
Silo User’s Guide 2-105
The following table describes the options accepted by this function:
Option Name
Data
Type Option Meaning Default Value
DBOPT_COORDSYS int Coordinate system. One of:
DB_CARTESIAN, DB_CYLINDRICAL,
DB_SPHERICAL, DB_NUMERICAL, or
DB_OTHER.
DB_OTHER
DBOPT_NODENUM void* An array of length nnodes giving a global
node number for each node in the mesh.
By default, this array is treated as type int.
NULL
DBOPT_LLONGNZNUM int Indicates that the array passed for
DBOPT_NODENUM option is of long long
type instead of int.
0
DBOPT_CYCLE int Problem cycle value 0
DBOPT_FACETYPE int Zone face type. One of the predefined
types: DB_RECTILINEAR or
DB_CURVILINEAR.
DB_RECTILINEAR
DBOPT_XLABEL char * Character string defining the label associ-
ated with the X dimension.
NULL
DBOPT_YLABEL char * Character string defining the label associ-
ated with the Y dimension.
NULL
DBOPT_ZLABEL char * Character string defining the label associ-
ated with the Z dimension.
NULL
DBOPT_NSPACE int Number of spatial dimensions used by this
mesh.
ndims
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0
DBOPT_PLANAR int Planar value. One of: DB_AREA or
DB_VOLUME.
DB_NONE
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME dou-
ble
Problem time value. 0.0
DBOPT_XUNITS char * Character string defining the units associ-
ated with the X dimension.
NULL
DBOPT_YUNITS char * Character string defining the units associ-
ated with the Y dimension.
NULL
DBOPT_ZUNITS char * Character string defining the units associ-
ated with the Z dimension.
NULL
DBOPT_PHZONELIST char * Character string holding the name for a
polyhedral zonelist object to be associated
with the mesh
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBPutUcdmesh
2-106 Silo User’s Guide
DBOPT_MRGTREE_NAME char * Name of the mesh region grouping tree to
be associated with this mesh.
NULL
DBOPT_TOPO_DIM int Used to indicate the topological dimension
of the mesh apart from its spatial dimen-
sion.
-1 (not specified)
DBOPT_TV_CONNECTIVTY int A non-zero value indicates that the con-
nectivity of the mesh varies with time
0
DBOPT_DISJOINT_MODE int Indicates if any elements in the mesh are
disjoint. There are two possible modes.
One is DB_ABUTTING indicating that ele-
ments abut spatially but actually reference
different node ids (but spatially equivalent
nodal positions) in the node list. The other
is DB_FLOATING where elements neither
share nodes in the nodelist nor abut spa-
tially.
DB_NONE
DBOPT_GHOST_NODE_LABELS char * Optional array of char values indicating
the ghost labeling
(DB_GHOSTTYPE_NOGHOST or
DB_GHOSTTYPE_INTDUP) of each point
NULL
DBOPT_ALT_NODENUM_VARS char
**
A null terminated list of names of optional
array(s) or DBpointvar objects indicating
(multiple) alternative numbering(s) for
nodes.
NULL
The following options have been deprecated. Use MRG trees instead
DBOPT_GROUPNUM int The group number to which this quad-
mesh belongs.
-1 (not in a group)
Option Name
Data
Type Option Meaning Default Value

DBPutUcdsubmesh
Silo User’s Guide 2-107
DBPutUcdsubmesh—Write a subset of a parent, ucd mesh, to a Silo file
Synopsis:
int DBPutUcdsubmesh(DBfile *file, const char *name,
const char *parentmesh, int nzones, const char *zlname,
const char *flname, DBoptlist const *opts)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
name The name of the ucd submesh object to create.
parentmesh The name of the parent ucd mesh this submesh is a portion of.
nzones The number of zones in this submesh.
zlname The name of the zonelist object.
fl [OPT] The name of the facelist object.
opts Additional options.
Returns:
A positive number on success; -1 on failure
Description:
DO NOT USE THIS METHOD.
It is an extremely limited, inefficient and soon to be retired way of trying to define subsets of
a ucd mesh. Instead, use a Mesh Region Grouping (MRG) tree. See “DBMakeMrgtree” on
page 193.

DBGetUcdmesh
2-108 Silo User’s Guide
DBGetUcdmesh—Read a UCD mesh from a Silo database.
Synopsis:
DBucdmesh *DBGetUcdmesh (DBfile *dbfile, char const *meshname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
meshname Name of the mesh.
Returns:
DBGetUcdmesh returns a pointer to a DBucdmesh structure on success and NULL on failure.
Description:
The DBGetUcdmesh function allocates a DBucdmesh data structure, reads a UCD mesh from the
Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutZonelist
Silo User’s Guide 2-109
DBPutZonelist—Write a zonelist object into a Silo file.
Synopsis:
int DBPutZonelist (DBfile *dbfile, char const *name, int nzones,
int ndims, int const nodelist[], int lnodelist, int origin,
int const shapesize[], int const shapecnt[], int nshapes)
Fortran Equivalent:
integer function dbputzl(dbid, name, lname, nzones, ndims,
nodelist, lnodelist, origin, shapesize,
shapecnt, nshapes, status)
Arguments:
dbfile Database file pointer.
name Name of the zonelist structure.
nzones Number of zones in associated mesh.
ndims Number of spatial dimensions represented by associated mesh.
nodelist Array of length lnodelist containing node indices describing mesh zones.
lnodelist Length of nodelist array.
origin Origin for indices in the nodelist array. Should be zero or one.
shapesize Array of length nshapes containing the number of nodes used by each zone
shape.
shapecnt Array of length nshapes containing the number of zones having each shape.
nshapes Number of zone shapes.
Returns:
DBPutZonelist returns zero on success or -1 on failure.
Description:
Do not use this method. Use DBPutZonelist2() instead.
The DBPutZonelist function writes a zonelist object into a Silo file. The name assigned to this
object can in turn be used as the zonel_name parameter to the DBPutUcdmesh function.
Notes:
See the write-up of DBPutUcdmesh for a full description of the zonelist data structures.

DBPutZonelist2
2-110 Silo User’s Guide
DBPutZonelist2—Write a zonelist object containing ghost zones into a Silo file.
Synopsis:
int DBPutZonelist2 (DBfile *dbfile, char const *name, int nzones,
int ndims, int const nodelist[], int lnodelist, int origin,
int lo_offset, int hi_offset, int const shapetype[],
int const shapesize[], int const shapecnt[], int nshapes,
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputzl2(dbid, name, lname, nzones, ndims,
nodelist, lnodelist, origin, lo_offset,
hi_offset, shapetype, shapesize, shapecnt,
nshapes, optlist_id, status)
Arguments:
dbfile Database file pointer.
name Name of the zonelist structure.
nzones Number of zones in associated mesh.
ndims Number of spatial dimensions represented by associated mesh.
nodelist Array of length lnodelist containing node indices describing mesh zones.
lnodelist Length of nodelist array.
origin Origin for indices in the nodelist array. Should be zero or one.
lo_offset The number of ghost zones at the beginning of the nodelist.
hi_offset The number of ghost zones at the end of the nodelist.
shapetype Array of length nshapes containing the type of each zone shape. See
description below.
shapesize Array of length nshapes containing the number of nodes used by each zone
shape.
shapecnt Array of length nshapes containing the number of zones having each shape.
nshapes Number of zone shapes.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. See the table below for
the valid options for this function. If no options are to be provided, use NULL
for this argument.
Returns:
DBPutZonelist2 returns zero on success or -1 on failure.

DBPutZonelist2
Silo User’s Guide 2-111
Description:
The DBPutZonelist2 function writes a zonelist object into a Silo file. The name assigned to this
object can in turn be used as the zonel_name parameter to the DBPutUcdmesh function.
The allowed shape types are described in the following table:
Type Description
DB_ZONETYPE_BEAM A line segment
DB_ZONETYPE_POLYGON A polygon where nodes are enumerated to form a polygon
DB_ZONETYPE_TRIANGLE A triangle
DB_ZONETYPE_QUAD A quadrilateral
DB_ZONETYPE_POLYHEDRON A polyhedron with nodes enumerated to form faces and
faces are enumerated to form a polyhedron
DB_ZONETYPE_TET A tetrahedron
DB_ZONETYPE_PYRAMID A pyramid
DB_ZONETYPE_PRISM A prism
DB_ZONETYPE_HEX A hexahedron
Notes:
The following table describes the options accepted by this function:
Option Name
Data
Type Option Meaning
Default
Value
DBOPT_ZONENUM void* Array of global zone numbers, one per
zone in this zonelist. By default, this is
assumed to be of type int.
NULL
DBOPT_LLONGNZNUM int Indicates that the array passed for
DBOPT_ZONENUM option is of long long
type instead of int.
0
DBOPT_GHOST_ZONE_LABELS char * Optional array of char values indicating
the ghost labeling
(DB_GHOSTTYPE_NOGHOST or
DB_GHOSTTYPE_INTDUP) of each zone
NULL
DBOPT_ALT_ZONENUM_VARS char ** A null terminated list of names of optional
array(s) or DBucdvar objects indicating
(multiple) alternative numbering(s) for
zones.
NULL
For a description of how the nodes for the allowed shapes are enumerated, see “DBPutUcdmesh”
on page 2-99

DBPutPHZonelist
2-112 Silo User’s Guide
DBPutPHZonelist—Write an arbitrary, polyhedral zonelist object into a Silo file.
Synopsis:
int DBPutPHZonelist (DBfile *dbfile, char const *name, int nfaces,
int const *nodecnts, int lnodelist, int const *nodelist,
char const *extface, int nzones, int const *facecnts,
int lfacelist, int const *facelist, int origin,
int lo_offset, int hi_offset, DBoptlist const *optlist)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Name of the zonelist structure.
nfaces Number of faces in the zonelist. Note that faces shared between zones should
only be counted once.
nodecnts Array of length nfaces indicating the number of nodes in each face. That is
nodecnts[i] is the number of nodes in face i.
lnodelist Length of the succeeding nodelist array.
nodelist Array of length lnodelist listing the nodes of each face. The list of nodes
for face i begins at index Sum(nodecnts[j]) for j=0...i-1.
extface An optional array of length nfaces where extface[i]!=0x0 means that
face i is an external face. This argument may be NULL.
nzones Number of zones in the zonelist.
facecnts Array of length nzones where facecnts[i] is number of faces for zone i.
lfacelist Length of the succeeding facelist array.
facelist Array of face ids for each zone. The list of faces for zone i begins at index
Sum(facecnts[j]) for j=0...i-1. Note, however, that each face is
identified by a signed value where the sign is used to indicate which ordering of
the nodes of a face is to be used. A face id >= 0 means that the node ordering as
it appears in the nodelist should be used. Otherwise, the value is negative
and it should be 1-complimented to get the face’s true id. In addition, the node
ordering for such a face is the opposite of how it appears in the nodelist. Finally,
node orders over a face should be specified such that a right-hand rule yields the
outward normal for the face relative to the zone it is being defined for.
origin Origin for indices in the nodelist array. Should be zero or one.
lo-offset Index of first real (e.g. non-ghost) zone in the list. All zones with index less than
(<) lo-offset are treated as ghost-zones.
hi-offset Index of last real (e.g. non-ghost) zone in the list. All zones with index greater
than (>) hi-offset are treated as ghost zones.
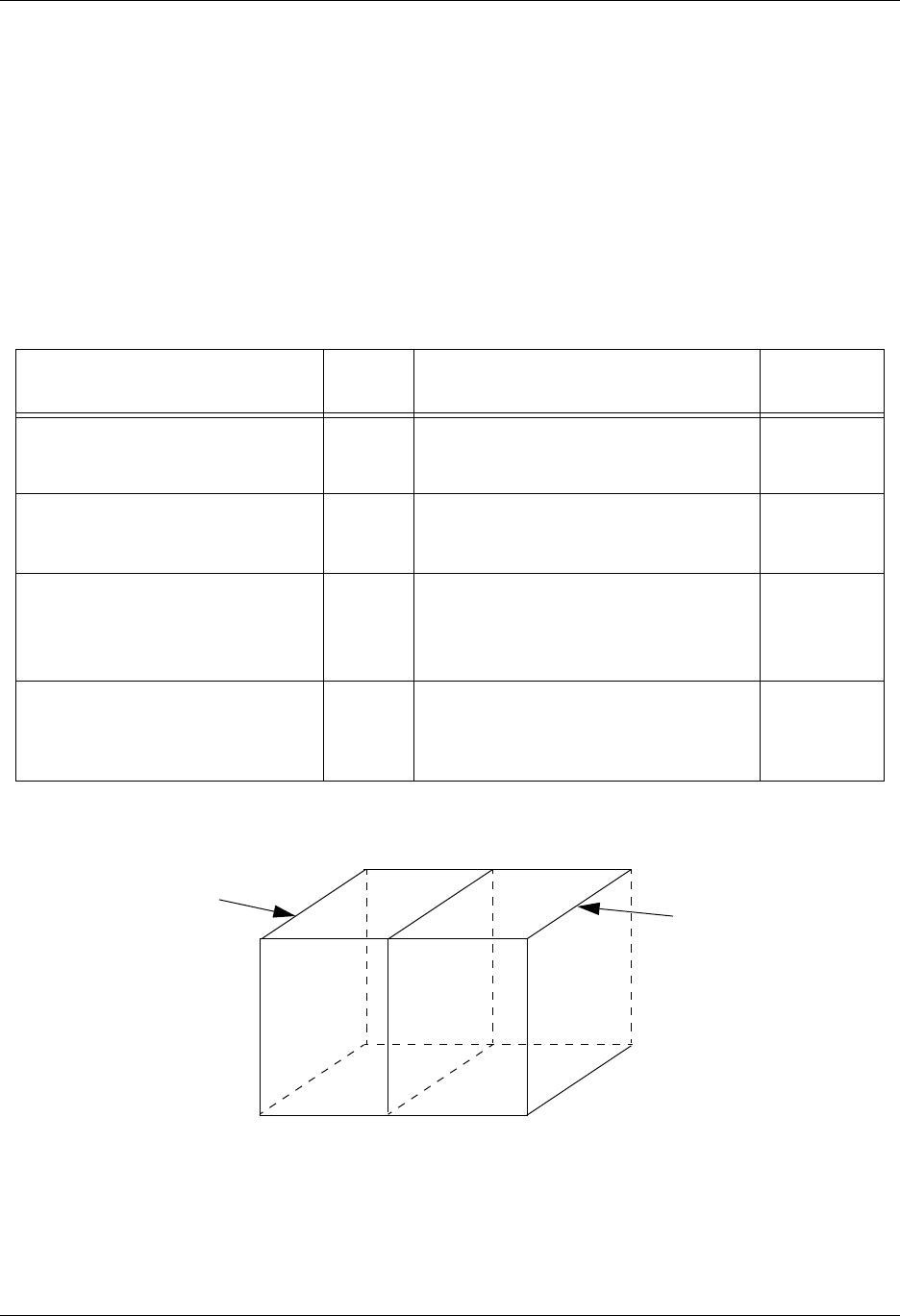
DBPutPHZonelist
Silo User’s Guide 2-113
Returns:
DBPutPHZonelist returns zero on success or -1 on failure.
Description:
The DBPutPHZonelist function writes a polyhedral-zonelist object into a Silo file. The name
assigned to this object can in turn be used as the parameter in the DBOPT_PHZONELIST option
for the DBPutUcdmesh function.
Notes:
The following table describes the options accepted by this function:
Option Name
Data
Type Option Meaning
Default
Value
DBOPT_ZONENUM void* Array of global zone numbers, one per
zone in this zonelist. By default, it is
assumed this array is of type int*.
NULL
DBOPT_LLONGNZNUM int Indicates that the array passed for
DBOPT_ZONENUM option is of long long
type instead of int.
0
DBOPT_GHOST_ZONE_LABELS char * Optional array of char values indicating
the ghost labeling
(DB_GHOSTTYPE_NOGHOST or
DB_GHOSTTYPE_INTDUP) of each zone
NULL
DBOPT_ALT_ZONENUM_VARS char ** A null terminated list of names of optional
array(s) or DBucdvar objects indicating
(multiple) alternative numbering(s) for
zones.
NULL
910 11
678
34 5
012
X
Z
Y
b
cd
ef
gh
ij
k
a
zone 0
zone 1
In interpreting the diagram above, numbers correspond to nodes while letters correspond to faces.
In addition, the letters are drawn such that they will always be in the lower, right hand corner of a

DBPutPHZonelist
2-114 Silo User’s Guide
face if you were standing outside the object looking towards the given face. In the example code
below, the list of nodes for a given face begin with the node nearest its corresponding letter.
For toplogically 2D meshes, two different approaches are possible for creating a polyhedral zonel-
ist. One is to simple have a single list of “faces” representing the polygons of the 2D mesh. The
other is to create an explicit list of “edges” and then define each polygon in terms of the edges it
comprises. Either is appropriate.

DBPutPHZonelist
Silo User’s Guide 2-115
#define NNODES 12
#define NFACES 11
#define NZONES 2
/* coordinate arrays */
float x[NNODES] = {0.0, 1.0, 2.0, 0.0, 1.0, 2.0, 0.0, 1.0, 2.0, 0.0, 1.0, 2.0};
float y[NNODES] = {0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 1.0, 1.0, 1.0, 1.0, 1.0, 1.0};
float z[NNODES] = {0.0, 0.0, 0.0, 1.0, 1.0, 1.0, 0.0, 0.0, 0.0, 1.0, 1.0, 1.0};
/* facelist where we enumerate the nodes over each face */
int nodecnts[NFACES] = {4,4,4,4,4,4,4,4,4,4,4};
int lnodelist = 4*NFACES;
/* a b c */
int nodelist[4*NFACES] = {1,7,6,0, 2,8,7,1 4,1,0,3,
/* d e f */
5,2,1,4, 3,9,10,4, 4,10,11,5,
/* g h i */
9,6,7,10, 10,7,8,11, 0,6,9,3,
/* j K */
1,7,10,4, 5,11,8,2};
/* zonelist where we enumerate the faces over each zone */
int facecnts[NZONES] = {6,6};
int lfacelist = 6*NZONES;
int facelist[6*NZONES] = {0,2,4,6,8,-9, 1,3,5,7,9,10};
Figure 0-4: Example of a polyhedral zonelist representation for two hexahedral elements.

DBGetPHZonelist
2-116 Silo User’s Guide
DBGetPHZonelist—Read a polyhedral-zonelist from a Silo database.
Synopsis:
DBphzonelist *DBGetPHZonelist (DBfile *dbfile,
char const *phzlname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
phzlname Name of the polyhedral-zonelist.
Returns:
DBGetPHZonelist returns a pointer to a DBphzonelist structure on success and NULL on failure.
Description:
The DBGetPHZonelist function allocates a DBphzonelist data structure, reads a polyhedral-zonel-
ist from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is
returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutFacelist
Silo User’s Guide 2-117
DBPutFacelist—Write a facelist object into a Silo file.
Synopsis:
int DBPutFacelist (DBfile *dbfile, char const *name, int nfaces,
int ndims, int const nodelist[], int lnodelist, int origin,
int const zoneno[], int const shapesize[],
int const shapecnt[], int nshapes, int const types[],
int const typelist[], int ntypes)
Fortran Equivalent:
integer function dbputfl(dbid, name, lname, ndims nodelist,
lnodelist, origin, zoneno, shapesize,
shapecnt, nshaps, types, typelist, ntypes,
status)
Arguments:
dbfile Database file pointer.
name Name of the facelist structure.
nfaces Number of external faces in associated mesh.
ndims Number of spatial dimensions represented by the associated mesh.
nodelist Array of length lnodelist containing node indices describing mesh faces.
lnodelist Length of nodelist array.
origin Origin for indices in nodelist array. Either zero or one.
zoneno Array of length nfaces containing the zone number from which each face
came. Use a NULL for this parameter if zone numbering info is not wanted.
shapesize Array of length nshapes containing the number of nodes used by each face
shape (for 3-D meshes only).
shapecnt Array of length nshapes containing the number of faces having each shape
(for 3-D meshes only).
nshapes Number of face shapes (for 3-D meshes only).
types Array of length nfaces containing information about each face. This
argument is ignored if ntypes is zero, or if this parameter is NULL.
typelist Array of length ntypes containing the identifiers for each type. This argument
is ignored if ntypes is zero, or if this parameter is NULL.
ntypes Number of types, or zero if type information was not provided.
Returns:
DBPutFacelist returns zero on success or -1 on failure.

DBPutFacelist
2-118 Silo User’s Guide
Description:
The DBPutFacelist function writes a facelist object into a Silo file. The name given to this object
can in turn be used as a parameter to the DBPutUcdmesh function.
Notes:
See the write-up of DBPutUcdmesh for a full description of the facelist data structures.

DBPutUcdvar
Silo User’s Guide 2-119
DBPutUcdvar—Write a vector/tensor UCD variable object into a Silo file.
Synopsis:
int DBPutUcdvar (DBfile *dbfile, char const *name,
char const *meshname, int nvars,
char const * const varnames[], void const * const vars[],
int nels, void const * const mixvars[], int mixlen,
int datatype, int centering, DBoptlist const *optlist)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Name of the variable.
meshname Name of the mesh associated with this variable (written with DBPutUcdmesh).
nvars Number of sub-variables which comprise this variable. For a scalar array, this is
one. If writing a vector quantity, however, this would be two for a 2-D vector
and three for a 3-D vector.
varnames Array of length nvars containing pointers to character strings defining the
names associated with each subvariable.
vars Array of length nvars containing pointers to arrays defining the values
associated with each subvariable.
nels Number of elements in this variable.
mixvars Array of length nvars containing pointers to arrays defining the mixed-data
values associated with each subvariable. If no mixed values are present, this
should be NULL.
mixlen Length of mixed data arrays (i.e., mixvars).
datatype Datatype of sub-variables. One of the predefined Silo data types.
centering Centering of the sub-variables on the associated mesh. One of the predefined
types: DB_NODECENT, DB_EDGECENT, DB_FACECENT,
DB_ZONECENT or DB_BLOCKCENT. See below for a discussion of
centering issues.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. See the table below for
the valid options for this function. If no options are to be provided, use NULL
for this argument.
Returns:
DBPutUcdvar returns zero on success and -1 on failure.

DBPutUcdvar
2-120 Silo User’s Guide
Description:
The DBPutUcdvar function writes a variable associated with an UCD mesh into a Silo file. Note
that variables can be node-centered, zone-centered, edge-centered or face-centered.
For node- (or zone-) centered data, the question of which value in the vars array goes with which
node (or zone) is determined implicitly by a one-to-one correspondence with the list of nodes in
the DBPutUcdmesh call (or zones in the DBPutZonelist or DBPutZonelist2 call). For
example, the 237th value in a zone-centered vars array passed here goes with the 237th zone in
the zonelist passed in the DBPutZonelist2 (or DBPutZonelist) call.
Edge- and face-centered data require a little more explanation. Unlike node- and zone-centered
data, there does not exist in Silo an explicit list of edges or faces. As an aside, the DBPutFacel-
ist call is really for writing the external faces of a mesh so that a downstream visualization tool
need not have to compute them when it displays the mesh. Now, requiring the caller to create
explicit lists of edges and/or faces in order to handle edge- or face-centered data results in unneces-
sary additional data being written to a Silo file. This increases file size as well as the time to write
and read the file. To avoid this, we rely upon implicit lists of edges and faces.
We define implicit lists of edges and faces in terms of a traversal of the zonelist structure of the
associated mesh. The position of an edge (or face) in its list is determined by the order of its first
occurrence in this traversal. The traversal algorithm is to visit each zone in the zonelist and, for
each zone, visit its edges (or faces) in local order. See Figure 0-2 on page 102. Because this tra-
versal will wind up visiting edges multiple times, the first time an edge (or face) is encountered is
what determines its position in the implicit edge (or face) list.
If the zonelist contains arbitrary polyhedra or the zonelist is a polyhedral zonelist (written with
DBPutPHZonelist), then the traversal algorithm involves visiting each zone, then each face for a
zone and finally each edge for a face.
Note that DBPutUcdvar() can also be used to define a block-centered variable on a multi-block
mesh by specifying a multi-block mesh name for the meshname and DB_BLOCKCENT for the
centering. This is useful in defining, for example, multi-block variable extents.
Other information can also be included. This function is useful for writing vector and tensor fields,
whereas the companion function, DBPutUcdvar1, is appropriate for writing scalar fields.
Notes:
The following table describes the options accepted by this function:
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_COORDSYS int Coordinate system. One of:
DB_CARTESIAN, DB_CYLINDRICAL,
DB_SPHERICAL, DB_NUMERICAL, or
DB_OTHER.
DB_OTHER
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_LABEL char * Character strings defining the label asso-
ciated with this variable.
NULL
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0

DBPutUcdvar
Silo User’s Guide 2-121
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_UNITS char * Character string defining the units associ-
ated with this variable.
NULL
DBOPT_USESPECMF int Boolean (DB_OFF or DB_ON) value
specifying whether or not to weight the
variable by the species mass fraction
when using material species data.
DB_OFF
DBOPT_ASCII_LABEL int Indicate if the variable should be treated
as single character, ascii values. A value
of 1 indicates yes, 0 no.
0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_REGION_PNAMES char** A null-pointer terminated array of pointers
to strings specifying the pathnames of
regions in the mrg tree for the associated
mesh where the variable is defined. If
there is no mrg tree associated with the
mesh, the names specified here will be
assumed to be material names of the
material object associated with the mesh.
The last pointer in the array must be null
and is used to indicate the end of the list of
names. See
“DBOPT_REGION_PNAMES” on
page 218.
NULL
DBOPT_CONSERVED int Indicates if the variable represents a phys-
ical quantity that must be conserved under
various operations such as interpolation.
0
DBOPT_EXTENSIVE int Indicates if the variable represents a phys-
ical quantity that is extensive (as opposed
to intensive). Note, while it is true that any
conserved quantity is extensive, the con-
verse is not true. By default and histori-
cally, all Silo variables are treated as
intensive.
0
DBOPT_MISSING_VALUE double Specify a numerical value that is intended
to represent “missing values” in the vari-
able data arrays. Default is
DB_MISSING_VALUE_NOT_SET
DB_MISSING_VALUE_
NOT_SET
Option Name
Value
Data Type Option Meaning Default Value

DBPutUcdvar1
2-122 Silo User’s Guide
DBPutUcdvar1—Write a scalar UCD variable object into a Silo file.
Synopsis:
int DBPutUcdvar1 (DBfile *dbfile, char const *name,
char const *meshname, void const *var, int nels,
void const *mixvar, int mixlen, int datatype, int centering,
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputuv1(dbid, name, lname, meshname, lmeshname,
var, nels, mixvar, mixlen, datatype,
centering, optlist_id, staus)
Arguments:
dbfile Database file pointer.
name Name of the variable.
meshname Name of the mesh associated with this variable (written with either
DBPutUcdmesh).
var Array of length nels containing the values associated with this variable.
nels Number of elements in this variable.
mixvar Array of length mixlen containing the mixed-data values associated with this
variable. If mixlen is zero, this value is ignored.
mixlen Length of mixvar array. If zero, no mixed data is present.
datatype Datatype of variable. One of the predefined Silo data types.
centering Centering of the sub-variables on the associated mesh. One of the predefined
types: DB_NODECENT, DB_EDGECENT, DB_FACECENT or
DB_ZONECENT.
optlist Pointer to an option list structure containing additional information to be
included in the variable object written into the Silo file. See the table below for
the valid options for this function. If no options are to be provided, use NULL
for this argument.
Returns:
DBPutUcdvar1 returns zero on success and -1 on failure.
Description:
DBPutUcdvar1 writes a variable associated with an UCD mesh into a Silo file. Note that variables
will be either node-centered or zone-centered. Other information can also be included. This func-
tion is useful for writing scalar fields, whereas the companion function, DBPutUcdvar, is appropri-
ate for writing vector and tensor fields.

DBGetUcdvar
2-124 Silo User’s Guide
DBGetUcdvar—Read a UCD variable from a Silo database.
Synopsis:
DBucdvar *DBGetUcdvar (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Name of the variable.
Returns:
DBGetUcdvar returns a pointer to a DBucdvar structure on success and NULL on failure.
Description:
The DBGetUcdvar function allocates a DBucdvar data structure, reads a variable associated with a
UCD mesh from the Silo database, and returns a pointer to that structure. If an error occurs, NULL
is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutCsgmesh
Silo User’s Guide 2-125
DBPutCsgmesh—Write a CSG mesh object to a Silo file
Synopsis:
DBPutCsgmesh(DBfile *dbfile, const char *name, int ndims,
int nbounds,
const int *typeflags, const int *bndids,
const void *coeffs, int lcoeffs, int datatype,
const double *extents, const char *zonel_name,
DBoptlist const *optlist);
Fortran Equivalent:
integer function dbputcsgm(dbid, name, lname, ndims, nbounds,
typeflags, bndids, coeffs, lcoeffs, datatype,
extents, zonel_name, lzonel_name, optlist_id,
status)
Arguments:
dbfile Database file pointer
name Name to associate with this DBcsgmesh object
ndims Number of spatial and topological dimensions of the CSG mesh object
nbounds Number of boundaries in the CSG mesh description.
typeflags Integer array of length nbounds of type information for each boundary. This is
used to encode various information about the type of each boundary such as, for
example, plane, sphere, cone, general quadric, etc as well as the number of
coefficients in the representation of the boundary. For more information, see the
description, below.
bndids Optional integer array of length nbounds which are the explicit integer
identifiers for each boundary. It is these identifiers that are used in expressions
defining a region of the CSG mesh. If the caller passes NULL for this argument,
a natural numbering of boundaries is assumed. That is, the boundary occurring
at position i, starting from zero, in the list of boundaries here is identified by
the integer i.
coeffs Array of length lcoeffs of coefficients used in the representation of each
boundary or, if the boundary is a transformed copy of another boundary, the
coefficients of the transformation. In the case where a given boundary is a
transformation of another boundary, the first entry in the coeffs entries for the
boundary is the (integer) identifier for the referenced boundary. Consequently, if
the datatype for coeffs is DB_FLOAT, there is an upper limit of about 16.7
million (2^24) boundaries that can be referenced in this way.
lcoeffs Length of the coeffs array.
datatype The data type of the data in the coeffs array.
zonel_name Name of CSG zonelist to be associated with this CSG mesh object
extents Array of length 2*ndims of spatial extents, xy(z)-minimums followed by
DBPutCsgmesh
2-126 Silo User’s Guide
xy(z)-maximums.
optlist Pointer to an option list structure containing additional information to be
included in the CSG mesh object written into the Silo file. Use NULL if there
are no options.
Returns:
DBPutCsgMesh returns zero on success and -1 on failure.
Description:
The word “mesh” in this function name is probably somewhat misleading because it suggests a
discretization of a domain into a “mesh”. In fact, a CSG (Constructive Solid Geometry) “mesh” in
Silo is a continuous, analytic representation of the geometry of some computational domain.
Nonetheless, most of Silo’s concepts for meshes, variables, materials, species and multi-block
objects apply equally well in the case of a CSG “mesh” and so that is what it is called, here. Pres-
ently, Silo does not have functions to discretize this kind of mesh. It has only the functions for stor-
ing and retrieving it. Nonetheless, a future version of Silo may include functions to discretize a
CSG mesh.
A CSG mesh is constructed by starting with a list of analytic boundaries, that is curves in 2D or
surfaces in 3D, such as planes, spheres and cones or general quadrics. Each boundary is defined by
an analytic expression (an equation) of the form f(x,y,z)=0 (or, in 2D, f(x,y)=0) in which the high-
est exponent for x, y or z is 2. That is, all the boundaries are quadratic (or “quadric”) at most.
The table below describes how to use the typeflags argument to define various kinds of bound-
aries in 3 dimensions.
typeflag
num-coeffs
coefficients and equation
DBCSG_QUADRIC_G 10
a0x2a1y2a2z2a3xy a4yz a5xz a6xa
7ya
8za
9
+++++++++ 0=
DBCSG_SPHERE_PR 4
xa
0
–
2ya
1
–
2za
2
–
2a3
2
–++ 0=
DBCSG_ELLIPSOID_PRRR 6
xa
0
–
2a3
2
ya
1
–
2a4
2
za
2
–
2a5
2
1–++ 0=
DBCSG_PLANE_G 4
a0xa
1ya
2za
3
+++ 0=
DBCSG_PLANE_X 1
xa
0
–0=
DBCSG_PLANE_Y 1
ya
0
–0=
DBCSG_PLANE_Z 1
za
0
–0=
DBCSG_PLANE_PN 6
xa
0
–a3ya
1
–a4za
2
–a5
++ 0=
DBCSG_PLANE_PPP 9
xa
0
–ya
1
–za
2
–
a3a0
–a
4a1
–a
5a2
–
a6a0
–a
7a1
–a
8a2
–
0=
DBCSG_CYLINDER_PNLR 8to be completed

DBPutCsgmesh
Silo User’s Guide 2-127
The table below defines an analogous set of typeflags for creating boundaries in two dimensions..
typeflag
num-coeffs
coefficients and equation
DBCSG_QUADRATIC_G 6
a0x2a1y2a2xy a3xa
4ya
5
++ +++ 0=
DBCSG_CIRCLE_PR 3
xa
0
–
2ya
1
–
2a2
2
–+0=
DBCSG_ELLIPSE_PRR 4
xa
0
–
2a2
2
ya
1
–
2a3
2
1–+0=
DBCSG_LINE_G 3
a0xa
1ya
2
++ 0=
DBCSG_LINE_X 1
xa
0
–0=
DBCSG_LINE_Y 1
ya
0
–0=
DBCSG_LINE_PN 4
xa
0
–a2ya
1
–a3
+0=
DBCSG_LINE_PP 4
a3a1
–
a2a0
–
----------------ya
1
–
xa
0
–
--------------–0=
DBCSG_BOX_XYXY 4to be completed
DBCSG_POLYGON_KP | K 2K to be completed
DBCSG_TRI_3P 6to be completed
DBCSG_QUAD_4P 8to be completed
By replacing the ‘=’ in the equation for a boundary with either a ‘<‘ or a ‘>’, whole regions in 2 or
3D space can be defined using these boundaries. These regions represent the set of all points that
satisfy the inequality. In addition, regions can be combined to form new regions by unions, inter-
sections and differences as well other operations (See DBPutCSGZonelist).
DBCSG_CYLINDER_PPR 7to be completed
DBCSG_BOX_XYZXYZ 6to be completed
DBCSG_CONE_PNLA 8to be completed
DBCSG_CONE_PPA to be completed
DBCSG_POLYHEDRON_KF | K 6K to be completed
DBCSG_HEX_6F 36 to be completed
DBCSG_TET_4F 24 to be completed
DBCSG_PYRAMID_5F 30 to be completed
DBCSG_PRISM_5F 30 to be completed
typeflag
num-coeffs
coefficients and equation
DBPutCsgmesh
2-128 Silo User’s Guide
In this call, only the analytic boundaries used in the expressions to define the regions are written.
The expressions defining the regions themselves are written in a separate call, DBPutCSG-
Zonelist.
If you compare this call to write a CSG mesh to a Silo file with a similar call to write a UCD mesh,
you will notice that the boundary list here plays a role similar to that of the nodal coordinates of a
UCD mesh. For the UCD mesh, the basic geometric primitives are points (nodes) and a separate
call, DBPutZonelist, is used to write out the information that defines how points (nodes) are
combined to form the zones of the mesh.
Similarly, here the basic geometric primitives are analytic boundaries and a separate call,
DBPutCSGZonelist, is used to write out the information that defines how the boundaries are
combined to form regions of the mesh.
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of the DBoptlist construct.
Option Name
Data
Type Option Meaning
Default
Value
DBOPT_CYCLE int Problem cycle value 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_XLABEL char * Character string defining the label associ-
ated with the X dimension.
NULL
DBOPT_YLABEL char * Character string defining the label associ-
ated with the Y dimension.
NULL
DBOPT_ZLABEL char * Character string defining the label associ-
ated with the Z dimension.
NULL
DBOPT_XUNITS char * Character string defining the units associ-
ated with the X dimension.
NULL
DBOPT_YUNITS char * Character string defining the units associ-
ated with the Y dimension.
NULL
DBOPT_ZUNITS char * Character string defining the units associ-
ated with the Z dimension.
NULL
DBOPT_BNDNAMES char ** Array of nboundaries character strings
defining the names of the individual
boundaries.
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_MRGTREE_NAME char * Name of the mesh region grouping tree to
be associated with this mesh.
NULL
DBOPT_TV_CONNECTIVTY int A non-zero value indicates that the con-
nectivity of the mesh varies with time
0
DBPutCsgmesh
Silo User’s Guide 2-129
DBOPT_DISJOINT_MODE int Indicates if any elements in the mesh are
disjoint. There are two possible modes.
One is DB_ABUTTING indicating that ele-
ments abut spatially but actually reference
different node ids (but spatially equivalent
nodal positions) in the node list. The other
is DB_FLOATING where elements neither
share nodes in the nodelist nor abut spa-
tially.
DB_NONE
DBOPT_ALT_NODENUM_VARS char ** A null terminated list of names of optional
array(s) or DBcsgvar objects indicating
(multiple) alternative numbering(s) for
boundaries.
NULL
The following options have been deprecated. Use MRG trees instead
DBOPT_GROUPNUM int The group number to which this quad-
mesh belongs.
-1 (not in a
group)
Option Name
Data
Type Option Meaning
Default
Value

DBGetCsgmesh
2-130 Silo User’s Guide
DBGetCsgmesh—Get a CSG mesh object from a Silo file
Synopsis:
DBcsgmesh *DBGetCsgmesh(DBfile *dbfile, const char *meshname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer
meshname Name of the CSG mesh object to read
Returns:
A pointer to a DBcsgmesh structure on success and NULL on failure.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutCSGZonelist
Silo User’s Guide 2-131
DBPutCSGZonelist—Put a CSG zonelist object in a Silo file.
Synopsis:
int DBPutCSGZonelist(DBfile *dbfile, const char *name, int nregs,
const int *typeflags,
const int *leftids, const int *rightids,
const void *xforms, int lxforms, int datatype,
int nzones, const int *zonelist,
DBoptlist *optlist);
Fortran Equivalent:
integer function dbputcsgzl(dbid, name, lname, nregs, typeflags,
leftids, rightids, xforms, lxforms, datatype,
nzones, zonelist, optlist_id, status)
Arguments:
dbfile Database file pointer
name Name to associate with the DBcsgzonelist object
nregs The number of regions in the regionlist.
typeflags Integer array of length nregs of type information for each region. Each entry
in this array is one of either DB_INNER, DB_OUTER, DB_ON, DB_XFORM,
DB_SWEEP, DB_UNION, DB_INTERSECT, and DB_DIFF.
The symbols, DB_INNER, DB_OUTER, DB_ON, DB_XFORM and DB_SWEEP
represent unary operators applied to the referenced region (or boundary). The
symbols DB_UNION, DB_INTERSECT, and DB_DIFF represent binary
operators applied to two referenced regions.
For the unary operators, DB_INNER forms a region from a boundary (See
DBPutCsgmesh) by replacing the ‘=’ in the equation representing the boundary
with ‘<‘. Likewise, DB_OUTER forms a region from a boundary by replacing
the ‘=’ in the equation representing the boundary with ‘>’. Finally, DB_ON
forms a region (of topological dimension one less than the mesh) by leaving the
‘=’ in the equation representing the boundary as an ‘=’. In the case of
DB_INNER, DB_OUTER and DB_ON, the corresponding entry in the leftids
array is a reference to a boundary in the boundary list (See DBPutCsgmesh).
For the unary operator, DB_XFORM, the corresponding entry in the leftids
array is a reference to a region to be transformed while the corresponding entry
in the rightids array is the index into the xform array of the row-by-row
coefficients of the affine transform.
The unary operator DB_SWEEP is not yet implemented.
leftids Integer array of length nregs of references to other regions in the regionlist or
boundaries in the boundary list (See DBPutCsgmesh). Each referenced region in

DBPutCSGZonelist
2-132 Silo User’s Guide
the leftids array forms the left operand of a binary expression (or single
operand of a unary expression) involving the referenced region or boundary.
rightids Integer array of length nregs of references to other regions in the regionlist.
Each referenced region in the rightids array forms the right operand of a
binary expression involving the region or, for regions which are copies of other
regions with a transformation applied, the starting index into the xforms array
of the row-by-row, affine transform coefficients. If for a given region no right
reference is appropriate, put a value of ‘-1’ into this array for the given region.
xforms Array of length lxforms of row-by-row affine transform coefficients for those
regions that are copies of other regions except with a transformation applied. In
this case, the entry in the leftids array indicates the region being copied and
transformed and the entry in the rightids array is the starting index into this
xforms array for the transform coefficients. This argument may be NULL.
lxforms Length of the xforms array. This argument may be zero if xforms is NULL.
datatype The data type of the values in the xforms array. Ignored if xforms is NULL.
nzones The number of zones in the CSG mesh. A zone is really just a completely
defined region.
zonelist Integer array of length nzones of the regions in the regionlist that form the
actual zones of the CSG mesh.
optlist Pointer to an option list structure containing additional information to be
included in this object when it is written to the Silo file. Use NULL if there are
no options.
Returns:
DBPutCSGZonelist returns zero on success and -1 on failure.
Description:
A CSG mesh is a list of curves in 2D or surfaces in 3D. These are analytic expressions of the
boundaries of objects that can be expressed by quadratic equations in x, y and z.
The zonelist for a CSG mesh is constructed by first defining regions from the mesh boundaries.
For example, given the boundary for a sphere, we can create a region by taking the inside
(DB_INNER) of that boundary or by taking the outside (DB_OUTER). In addition, regions can also
be created by boolean operations (union, intersect, diff) on other regions. The table below summa-
rizes how to construct regions using the typeflags argument.
op. symbol name type meaning
DBCSG_INNER unary specifies the region created by all points satisfying the equa-
tion defining the boundary with ‘<‘ replacing ‘=’.
left operand indicates the boundary, right operand ignored
DBCSG_OUTER unary specifies the region created by all points satisfying the equa-
tion defining the boundary with ‘>‘ replacing ‘=’.
left operand indicates the boundary, right operand ignored
DBPutCSGZonelist
Silo User’s Guide 2-133
However, not all regions in a CSG zonelist form the actual zones of a CSG mesh. Some regions
exist only to facilitate the construction of other regions. Only certain regions, those that are com-
pletely constructed, form the actual zones. Consequently, the zonelist for a CSG mesh involves
both a list of regions (as well as the definition of those regions) and then a list of zones (which are
really just completely defined regions).
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of the DBoptlist construct.
Option Name
Data
Type Option Meaning
Default
Value
DBOPT_REGNAMES char
**
Array of nregs character strings defining
the names of the individual regions.
NULL
DBOPT_ZONENAMES char** Array of nzones character strings defining
the names of individual zones.
NULL
DBOPT_ALT_ZONENUM_VARS char
**
A null terminated list of names of optional
array(s) or DBcsgvar objects indicating
(multiple) alternative numbering(s) for
zones.
NULL
DBCSG_ON unary specifies the region created by all points satisfying the equa-
tion defining the boundary.
left operand indicates the boundary, right operand ignored
DBCSG_UNION binary take the union of left and right operands
left and right operands indicate the regions
DBCSG_INTERSECT binary take the intersection of left and right operands
left and right operands indicate the regions
DBCSG_DIFF binary subtract the right operand from the left
left and right operands indicate the regions
DBCSG_COMPLIMENT unary take the compliment of the left operand,
left operand indicates the region, right operand ignored
DBCSG_XFORM unary to be implemented
DBCSG_SWEEP unary to be implemented
op. symbol name type meaning
front
top
side
DBPutCSGZonelist
2-134 Silo User’s Guide
Figure 0-5: A relatively simple object to represent as a CSG mesh. It models an A/C vent outlet for a 1994 Toyota
Tercel. It consists of two zones. One is a partially-spherical shaped ring housing (darker area). The other is a lens-
shaped fin used to direct airflow (lighter area).
The table below describes the contents of the boundary list (written in the DBPutCsgmesh call)
typeflags id coefficients name (optional)
DBCSG_SPHERE_PR 00.0, 0.0, 0.0, 5.0 “housing outer shell”
DBCSG_PLANE_X 1-2.5 “housing front”
DBCSG_PLANE_X 22.5 “housing back”
DBCSG_CYLINDER_PPR 30.0, 0.0, 0.0, 1.0, 0.0. 0.0, 3.0 “housing cavity”
DBCSG_SPHERE_PR 40.0, 0.0, 49.5, 50.0 “fin top side”
DBCSG_SPHERE_PR 50.0. 0.0, -49.5, 50.0 “fin bottom side”
The code below writes this CSG mesh to a silo file
int *typeflags={DBCSG_SPHERE_PR, DBCSG_PLANE_X, DBCSG_PLANE_X,
DBCSG_CYLINDER_PPR, DBCSG_SPHERE_PR, DBCSG_SPHERE_PR};
float *coeffs = {0.0, 0.0, 0.0, 5.0, 1.0, 0.0, 0.0, -2.5,
1.0, 0.0, 0.0, 2.5, 1.0, 0.0, 0.0, 0.0, 3.0,
0.0, 0.0, 49.5, 50.0, 0.0. 0.0, -49.5, 50.0};
DBPutCsgmesh(dbfile, “csgmesh”, 3, typeflags, NULL,
coeffs, 25, DB_FLOAT, “csgzl”, NULL);
DBPutCSGZonelist
Silo User’s Guide 2-135
The table below describes the contents of the regionlist, written in the DBPutCSGZonelist call.
typeflags regid leftids rightids notes
DBCSG_INNER 00-1 creates inner sphere region from boundary 0
DBCSG_INNER 11-1 creates front half-space region from boundary 1
DBCSG_OUTER 22-1 creates back half-space region from boundary 2
DBCSG_INNER 33-1 creates inner cavity region from boundary 3
DBCSG_INTERSE
CT
401cuts front of sphere by intersecting regions 0 &1
DBCSG_INTERSE
CT
542cuts back of sphere by intersecting regions 4 & 2
DBCSG_DIFF 653creates cavity in sphere by removing region 3
DBCSG_INNER 74-1 creates large sphere region for fin upper surface from boundary 4
DBCSG_INNER 85-1 creates large sphere region for fin lower surface from boundary 5
DBCSG_INTERSE
CT
978creates lens-shaped fin with razor edge protruding from sphere
housing by intersecting regions 7 & 8
DBCSG_INTERSE
CT
10 90cuts razor edge of lens-shaped fin to sphere housing
The table above creates 11 regions, only 2 of which form the actual zones of the CSG mesh. The 2
complete zones are for the spherical ring housing and the lens-shaped fin that sits inside it. They
are identified by region ids 6 and 10. The other regions exist solely to facilitate the construction.
The code to write this CSG zonelist to a silo file is given below.
int nregs = 11;
int *typeflags={DBCSG_INNER, DBCSG_INNER, DBCSG_OUTER, DBCSG_INNER,
DBCSG_INTERSECT, DBCSG_INTERSECT, DBCSG_DIFF,
DBCSG_INNER, DBCSG_INNER, DBCSG_INTERSECT,
DBCSG_INTERSECT};
int *leftids={0,1,2,3,0,4,5,4,5,7,9};
int *rightids={-1,-1,-1,-1,1,2,3,-1,-1,8,0};
int nzones = 2;
int *zonelist = {6, 10};
DBPutCSGZonelist(dbfile, “csgzl”, nregs, typeflags,
leftids, rightids, NULL, 0, DB_INT,
nzones, zonelist, NULL);

DBGetCSGZonelist
2-136 Silo User’s Guide
DBGetCSGZonelist—Read a CSG mesh zonelist from a Silo file
Synopsis:
DBcsgzonelist *DBGetCSGZonelist(DBfile *dbfile,
const char *zlname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer
zlname Name of the CSG mesh zonelist object to read
Returns:
A pointer to a DBcsgzonelist structure on success and NULL on failure.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutCsgvar
Silo User’s Guide 2-137
DBPutCsgvar—Write a CSG mesh variable to a Silo file
Synopsis:
int DBPutCsgvar(DBfile *dbfile, const char *vname,
const char *meshname, int nvars,
const char * const varnames[],
const void * const vars[], int nvals, int datatype,
int centering, DBoptlist const *optlist);
Fortran Equivalent:
integer function dbputcsgv(dbid, vname, lvname, meshname,
lmeshname, nvars, var_ids, nvals, datatype,
centering, optlist_id, status)
integer* var_ids (array of “pointer ids” created using dbmkptr)
Arguments:
dbfile Database file pointer
vname The name to be associated with this DBcsgvar object
meshname The name of the CSG mesh this variable is associated with
nvars The number of subvariables comprising this CSG variable
varnames Array of length nvars containing the names of the subvariables
vars Array of pointers to variable data
nvals Number of values in each of the vars arrays
datatype The type of data in the vars arrays (e.g. DB_FLOAT, DB_DOUBLE)
centering The centering of the CSG variable (DB_ZONECENT or DB_BNDCENT)
optlist Pointer to an option list structure containing additional information to be
included in this object when it is written to the Silo file. Use NULL if there are
no options
Description:
The DBPutCsgvar function writes a variable associated with a CSG mesh into a Silo file. Note that
variables will be either zone-centered or boundary-centered.
Just as UCD variables can be zone-centered or node-centered, CSG variables can be zone-centered
or boundary-centered. For a zone-centered variable, the value(s) at index i in the vars array(s) are
associated with the ith region (zone) in the DBcsgzonelist object associated with the mesh.
For a boundary-centered variable, the value(s) at index i in the vars array(s) are associated with the
ith boundary in the DBcsgbnd list associated with the mesh.

DBPutCsgvar
2-138 Silo User’s Guide
Other information can also be included via the optlist:
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_LABEL char * Character strings defining the label asso-
ciated with this variable.
NULL
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_UNITS char * Character string defining the units associ-
ated with this variable.
NULL
DBOPT_USESPECMF int Boolean (DB_OFF or DB_ON) value
specifying whether or not to weight the
variable by the species mass fraction
when using material species data.
DB_OFF
DBOPT_ASCII_LABEL int Indicate if the variable should be treated
as single character, ascii values. A value
of 1 indicates yes, 0 no.
0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_REGION_PNAMES char** A null-pointer terminated array of pointers
to strings specifying the pathnames of
regions in the mrg tree for the associated
mesh where the variable is defined. If
there is no mrg tree associated with the
mesh, the names specified here will be
assumed to be material names of the
material object associated with the mesh.
The last pointer in the array must be null
and is used to indicate the end of the list of
names. See
“DBOPT_REGION_PNAMES” on
page 218.
NULL
DBOPT_CONSERVED int Indicates if the variable represents a phys-
ical quantity that must be conserved under
various operations such as interpolation.
0
DBOPT_EXTENSIVE int Indicates if the variable represents a phys-
ical quantity that is extensive (as opposed
to intensive). Note, while it is true that any
conserved quantity is extensive, the con-
verse is not true. By default and histori-
cally, all Silo variables are treated as
intensive.
0
DBOPT_MISSING_VALUE double Specify a numerical value that is intended
to represent “missing values” in the x or y
data arrays. Default is
DB_MISSING_VALUE_NOT_SET
DB_MISSING_VALUE_
NOT_SET

DBGetCsgvar
Silo User’s Guide 2-139
DBGetCsgvar—Read a CSG mesh variable from a Silo file
Synopsis:
DBcsgvar *DBGetCsgvar(DBfile *dbfile, const char *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer
varname Name of CSG variable object to read
Returns:
A pointer to a DBcsgvar structure on success and NULL on failure.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutMaterial
2-140 Silo User’s Guide
DBPutMaterial—Write a material data object into a Silo file.
Synopsis:
int DBPutMaterial (DBfile *dbfile, char const *name,
char const *meshname, int nmat, int const matnos[],
int const matlist[], int const dims[], int ndims,
int const mix_next[], int const mix_mat[],
int const mix_zone[], void const *mix_vf, int mixlen,
int datatype, DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputmat(dbid, name, lname, meshname, lmeshname,
nmat, matnos, matlist, dims, ndims, mix_next,
mix_mat, mix_zone, mix_vf, mixlien, datatype,
optlist_id, status)
void* mix_vf
Arguments:
dbfile Database file pointer.
name Name of the material data object.
meshname Name of the mesh associated with this information.
nmat Number of materials.
matnos Array of length nmat containing material numbers.
matlist Array whose dimensions are defined by dims and ndims. It contains the
material numbers for each single-material (non-mixed) zone, and indices into
the mixed data arrays for each multi-material (mixed) zone. A negative value
indicates a mixed zone, and its absolute value is used as an index into the mixed
data arrays.
dims Array of length ndims which defines the dimensionality of the matlist
array.
ndims Number of dimensions in matlist array.
mix_next Array of length mixlen of indices into the mixed data arrays (one-origin).
mix_mat Array of length mixlen of material numbers for the mixed zones.
mix_zone Optional array of length mixlen of back pointers to originating zones. The
origin is determined by DBOPT_ORIGIN. Even if mixlen > 0, this argument
is optional.
mix_vf Array of length mixlen of volume fractions for the mixed zones. Note, this
can actually be either single- or double-precision. Specify actual type in
datatype.
mixlen Length of mixed data arrays (or zero if no mixed data is present). If mixlen >
0, then the “mix_” arguments describing the mixed data arrays must be non-
NULL.
DBPutMaterial
Silo User’s Guide 2-141
datatype Volume fraction data type. One of the predefined Silo data types.
optlist Pointer to an option list structure containing additional information to be
included in the material object written into the Silo file. See the table below for
the valid options for this function. If no options are to be provided, use NULL
for this argument.
Returns:
DBPutMaterial returns zero on success and -1 on failure.
Description:
Note that material functionality, even mixing materials, can now be handled, often more con-
veniently and efficiently, via a Mesh Region Grouping (MRG) tree. Users are encouraged to
consider an MRG tree as an alternative to DBPutMaterial(). See “DBMakeMrgtree” on
page 193.
The DBPutMaterial function writes a material data object into the current open Silo file. The min-
imum required information for a material data object is supplied via the standard arguments to this
function. The optlist argument must be used for supplying any information not requested
through the standard arguments.
Notes:
The following table describes the options accepted by this function. See the section titled “Using
the Silo Option Parameter” for details on the use of this construct.
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_LABEL char * Character string defining the label associ-
ated with material data.
NULL
DBOPT_MAJORORDER int Indicator for row-major (0) or column-
major (1) storage for multidimensional
arrays.
0
DBOPT_ORIGIN int Origin for mix_zone. Zero or one. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_MATNAMES char** Array of strings defining the names of the
individual materials.
NULL
DBOPT_MATCOLORS char** Array of strings defining the names of col-
ors to be associated with each material.
The color names are taken from the X win-
dows color database. If a color name
begins with a’#’ symbol, the remaining 6
characters are interpreted as the hexa-
decimal RGB value for the color.
NULL

DBPutMaterial
2-142 Silo User’s Guide
The model used for storing material data is the most efficient for VisIt, and works as follows:
One zonal array, matlist, contains the material number for a clean zone or an index into the
mixed data arrays if the zone is mixed. Mixed zones are marked with negative entries in
matlist, so you must take ABS(matlist[i]) to get the actual 1-origin mixed data index. All
indices are 1-origin to allow matlist to use zero as a material number.
The mixed data arrays are essentially a linked list of information about the mixed elements within
a zone. Each mixed data array is of length mixlen. For a given index i, the following information
is known about the i’th element:
mix_zone[i] The index of the zone which contains this element. The origin is determined by
DBOPT_ORIGIN.
mix_mat[i] The material number of this element
mix_vf[i] The volume fraction of this element
mix_next[i] The 1-origin index of the next material entry for this zone, else 0 if this is the
last entry.
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_ALLOWMAT0 int If set to non-zero, indicates that a zero
entry in the matlist array is actually not a
valid material number but is instead being
used to indicate an ‘unused’ zone.
0
Option Name
Value
Data Type Option Meaning Default Value
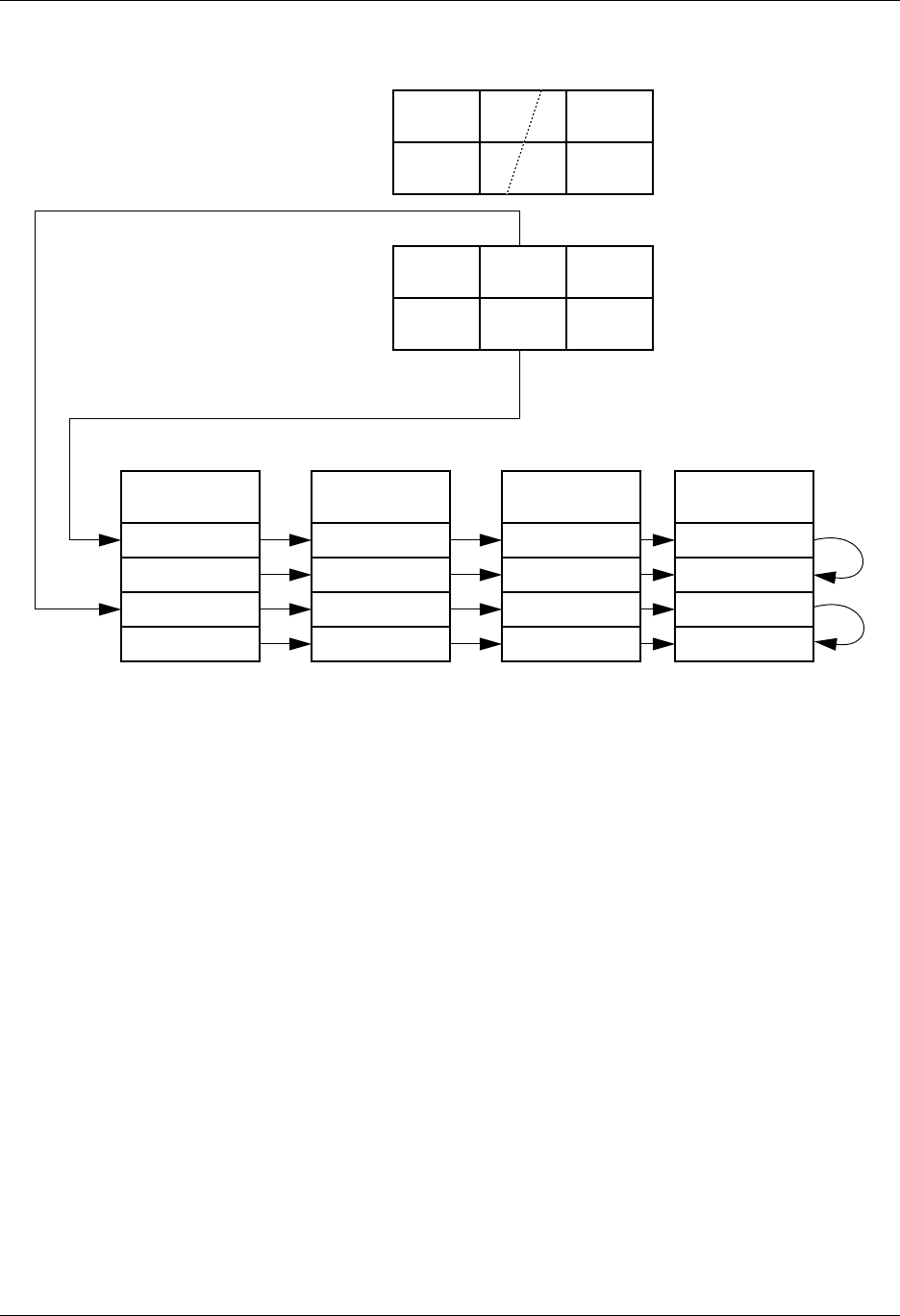
DBPutMaterial
Silo User’s Guide 2-143
.
1
11
1 2
2
2
2
Mesh ‘plot’
with material
numbers and
1
1
2
2
-1
-3 Corresponding
matlist array
mix_zone
1:
2:
3:
4:
2
2
5
5
mix_mat
1:
2:
3:
4:
1
2
1
2
mix_vf
1:
2:
3:
4:
.4
.6
.7
.3
mix_next
1:
2:
3:
4:
2
0
4
0
interface
Figure 0-6: Example using mixed data arrays for representing material information

DBGetMaterial
2-144 Silo User’s Guide
DBGetMaterial—Read material data from a Silo database.
Synopsis:
DBmaterial *DBGetMaterial (DBfile *dbfile, char const *mat_name)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
mat_name Name of the material variable to read.
Returns:
DBGetMaterial returns a pointer to a DBmaterial structure on success and NULL on failure.
Description:
The DBGetMaterial function allocates a DBmaterial data structure, reads material data from the
Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutMatspecies
Silo User’s Guide 2-145
DBPutMatspecies—Write a material species data object into a Silo file.
Synopsis:
int DBPutMatspecies (DBfile *dbfile, char const *name,
char const *matname, int nmat, int const nmatspec[],
int const speclist[], int const dims[], int ndims,
int nspecies_mf, void const *species_mf, int const mix_spec[],
int mixlen, int datatype, DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputmsp(dbid, name, lname, matname, lmatname,
nmat, nmatspec, speclist, dims, ndims,
species_mf, species_mf, mix_spec, mixlen,
datatype, optlist_id, status)
void *species_mf
Arguments:
dbfile Database file pointer.
name Name of the material species data object.
matname Name of the material object with which the material species object is associated.
nmat Number of materials in the material object referenced by matname.
nmatspec Array of length nmat containing the number of species associated with each
material.
speclist Array of dimension defined by ndims and dims of indices into the
species_mf array. Each entry corresponds to one zone. If the zone is clean,
the entry in this array must be positive or zero. A positive value is a 1-origin
index into the species_mf array. A zero can be used if the material in this
zone contains only one species. If the zone is mixed, this value is negative and is
used to index into the mix_spec array in a manner analogous to the mix_mat
array of the DBPutMaterial() call.
dims Array of length ndims that defines the shape of the speclist array.
ndims Number of dimensions in the speclist array.
nspecies_mf Length of the species_mf array.
species_mf Array of length nspecies_mf containing mass fractions of the material
species. Note, this can actually be either single or double precision. Specify type
in datatype argument.
mix_spec Array of length mixlen containing indices into the species_mf array.
These are used for mixed zones. For every index j in this array,
mix_list[j] corresponds to the DBmaterial structure’s material
mix_mat[j] and zone mix_zone[j].
mixlen Length of the mix_spec array.
datatype The datatype of the mass fraction data in species_mf. One of the predefined
DBPutMatspecies
2-146 Silo User’s Guide
Silo data types.
optlist Pointer to an option list structure containing additional information to be
included in the object written into the Silo file. Use a NULL if there are no
options.
Returns:
DBPutMatspecies returns zero on success and -1 on failure.
Description:
The DBPutMatspecies function writes a material species data object into a Silo file. The minimum
required information for a material species data object is supplied via the standard arguments to
this function. The optlist argument must be used for supplying any information not requested
through the standard arguments.
It is easiest to understand material species information by example. First, in order for a material
species object in Silo to have meaning, it must be associated with a material object. A material spe-
cies object by itself with no corresponding material object cannot be correctly interpreted.
So, suppose you had a problem which contains two materials, brass and steel. Now, neither brass
nor steel are themselves pure elements on the periodic table. They are instead alloys of other (pure)
metals. For example, common yellow brass is, nominally, a mixture of Copper (Cu) and Zinc (Zn)
while tool steel is composed primarily of Iron (Fe) but mixed with some Carbon (C) and a variety
of other elements.
For this example, lets suppose we are dealing with Brass (65% Cu, 35% Zn), T-1 Steel (76.3% Fe,
0.7% C, 18% W, 4% Cr,1% V) and O-1 Steel (96.2% Fe, 0.90% C,1.4% Mn, 0.50% Cr, 0.50% Ni,
0.50% W). Since T-1 Steel and O-1 Steel are composed of different elements, we wind up having to
represent each type of steel as a different material in the material object. So, the material object
would have 3 materials; Brass, T-1 Steel and O-1 Steel.
Brass is composed of 2 species, T-1 Steel, 5 species and O-1 Steel, 6. (Alternatively, one could opt
to characterize both T-1 Steel and O-1 Steel has having 7 species, Fe, C, Mn, Cr, Ni, W, V where
for T-1 Steel, the Mn and Ni components are always zero and for O-1 Steel the V component is
always zero. In that case, you would need only 2 materials in the associated material object.)
The material species object would be defined such that nmat=3 and nmatspec={2,5,6}. If
the composition of Brass, T-1 Steel and O-1 Steel is constant over the whole mesh, the
species_mf array would contain just 2 + 5 + 6 = 13 entries...
Brass (2
values)
T-1 Steel
(5 values starting at offset 3)
O-1 Steel
(6 values starting at offset 8)
species_mf .65 .35 .763 .007 .18 .04 .001 .962 .009 .014 .005 .005 .005
element Cu Zn Fe C W Cr VFe CMn Cr Ni W
1-origin index 12345678 910 11 12 13
If all of the zones in the mesh are clean (e.g. not mixing in material) and have the same composi-
tion of species, the speclist array would contain a ‘1’ for every Brass zone (1-origin indexing
would mean it would index species_mf[0]), a ‘3’ for every T-1 Steel zone and a ‘8’ for every O-1
Steel zone. However, if some cells had a Brass mixture with an extra 1% Cu, then you could create
DBPutMatspecies
Silo User’s Guide 2-147
another two entries at positions 14 and 15 in the species_mf array with the values 0.66 and
0.34, respectively, and the speclist array for those cells would point to ’14’ instead of ’1’.
The speclist entries indicate only where to start reading species mass fractions from the
species_mf array for a given zone. How do we know how many values to read? The associated
material object indicates which material is in the zone. The entry in the nmatspec array for that
material indicates how many mass fractions there are.
As simulations evolve, the relative mass fractions of species comprising each material vary away
from their nominal values. In this case, the species_mf array would grow to accommodate all
the variations of combinations of mass fraction for each material and the entries in the speclist
array would vary so that each zone would index the correct position in the species_mf array.
Finally, when zones contain mixing materials the speclist array needs to specify the
species_mf entries for each of the materials in the zone. In this case, negative values are
assigned to the speclist entries for these zones and the linked-list like structure of the associ-
ated material (e.g. mix_next, mix_mat, mix_vf, mix_zone args of the DBPutMaterial()
call) is used to traverse them.
Notes:
The following table describes the options accepted by this function:
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_MAJORORDER int Indicator for row-major (0) or column-
major (1) storage for multidimensional
arrays.
0
DBOPT_ORIGIN int Origin for arrays. Zero or one. 0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_SPECNAMES char** Array of strings defining the names of the
individual species. The length of this array
is the sum of the values in the nmatspec
argument to this function.
NULL
DBOPT_SPECCOLORS char** Array of strings defining the names of col-
ors to be associated with each species.
The color names are taken from the X win-
dows color database. If a color name
begins with a’#’ symbol, the remaining 6
characters are interpreted as the hexa-
decimal RGB value for the color. The
length of this array is the sum of the val-
ues in the nmatspec argument to this
function.
NULL

DBGetMatspecies
2-148 Silo User’s Guide
DBGetMatspecies—Read material species data from a Silo database.
Synopsis:
DBmatspecies *DBGetMatspecies (DBfile *dbfile,
char const *ms_name)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
ms_name Name of the material species data to read.
Returns:
DBGetMatspecies returns a pointer to a DBmatspecies structure on success and NULL on failure.
Description:
The DBGetMatspecies function allocates a DBmatspecies data structure, reads material species
data from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is
returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutDefvars
Silo User’s Guide 2-149
DBPutDefvars—Write a derived variable definition(s) object into a Silo file.
Synopsis:
int DBPutDefvars(DBfile *dbfile, const char *name, int ndefs,
const char * const names[], int const *types,
const char * const defns[], DBoptlist cost *optlist[]);
Fortran Equivalent:
integer function dbputdefvars(dbid, name, lname, ndefs, names,
lnames, types, defns, ldefns, optlist_id,
status)
character*N names (See “dbset2dstrlen” on page 285.)
character*N defns (See “dbset2dstrlen” on page 285.)
Arguments:
dbfile Database file pointer.
name Name of the derived variable definition(s) object.
ndefs number of derived variable definitions.
names Array of length ndefs of derived variable names
types Array of length ndefs of derived variable types such as
DB_VARTYPE_SCALAR, DB_VARTYPE_VECTOR,
DB_VARTYPE_TENSOR, DB_VARTYPE_SYMTENSOR,
DB_VARTYPE_ARRAY, DB_VARTYPE_MATERIAL,
DB_VARTYPE_SPECIES, DB_VARTYPE_LABEL
defns Array of length ndefs of derived variable definitions.
optlist Array of length ndefs pointers to option list structures containing additional
information to be included with each derived variable. The options available are
the same as those available for the respective variables.
Returns:
DBPutDefvars returns zero on success and -1 on failure.
Description:
The DBPutDefvars function is used to put definitions of derived variables in the Silo file. That is
variables that are derived from other variables in the Silo file or other derived variable definitions.
One or more variable definitions can be written with this function. Note that only the definitions of
the derived variables are written to the file with this call. The variables themselves are not in any
way computed by Silo.
If variable references within the defns strings do not have a leading slash (‘/’) (indicating an
absolute name), they are interpreted relative to the directory into which the Defvars object is writ-
ten. For the defns string, in cases where a variable’s name includes special characters (such as /
. { } [ ] + - = ), the entire variable reference should be bracketed by < and > characters.
DBPutDefvars
2-150 Silo User’s Guide
The interpretation of the defns strings written here is determined by the post-processing tool that
reads and interprets these definitions. Since in common practice that tool tends to be VisIt, the dis-
cussion that follows describes how VisIt would interpret this string.
The table below illustrates examples of the contents of the various array arguments to DBPutDe-
fvars for a case that defines 6 derived variables.
names types defns
0“totaltemp” DB_VARTYPE_SCALAR “nodet+zonetemp”
1“<stress/sz>” DB_VARTYPE_SCALAR “-<stress/sx>-<stress/sy>”
2“vel” DB_VARTYPE_VECTOR “{Vx, Vy, Vz}”
3“speed” DB_VARTYPE_SCALAR “magntidue(vel)”
4“dev_stress” DB_VARTYPE_TENSOR “{{<stress/sx>,<stress/txy>,<stress/txz>},
{ 0, <stress/sy>,<stress/tyz>},
{ 0, 0, <stress/sz>}}”
The first entry (0) defines a derived scalar variable named “totaltemp” which is the sum of vari-
ables whose names are “nodet” and “zonetemp”. The next entry (1) defines a derived scalar vari-
able named “sz” in a group of variables named “stress” (the slash character (‘/’) is used to group
variable names much the way file pathnames are grouped in Linux). Note also that the definition
of “sz” uses the special bracketing characters (‘<‘) and (‘>’) for the variable references due to the
fact that these variable references have a slash character (‘/’) in them.
The third entry (2) defines a derived vector variable named “vel” from three scalar variables
named “Vx”, “Vy”, and “Vz” while the fourth entry (3) defines a scalar variable, “speed” to be the
magnitude of the vector variable named “vel”. The last entry (4) defines a deviatoric stress tensor.
These last two cases demonstrate that derived variable definitions may reference other derived
variables.
The last few examples demonstrate the use of two operators, {}, and magnitude(). We call
these expression operators. In VisIt, there are numerous expression operators to help define
derived variables including such things as sqrt(), round(), abs(), cos(), sin(),
dot(), cross() as well as comparison operators, gt(), ge(), lt(), le(), eq(),
and the conditional if(). Furthermore, the list of expression operators in VisIt grows regularly.
Only a few examples are illustrated here. For a more complete list of the available expression
operators and their syntax, the reader is referred to the Expressions portion of the VisIt user’s man-
ual.

DBGetDefvars
Silo User’s Guide 2-151
DBGetDefvars—Get a derived variables definition object from a Silo file.
Synopsis:
DBdefvars DBGetDefvars(DBfile *dbfile, char const *name)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name The name of the DBdefvars object to read
Returns:
DBGetDefvars returns a pointer to a DBdefvars structure on success and NULL on failure.
Description:
The DBGetDefvars function allocates a DBdefvars data structure, reads the object from the Silo
database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBInqMeshname
2-152 Silo User’s Guide
DBInqMeshname—Inquire the mesh name associated with a variable.
Synopsis:
int DBInqMeshname (DBfile *dbfile, char const *varname,
char *meshname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Vari ab le n a m e.
meshname Returned mesh name. The caller must allocate space for the returned name. The
maximum space used is 256 characters, including the NULL terminator.
Returns:
DBInqMeshname returns zero on success and -1 on failure.
Description:
The DBInqMeshname function returns the name of a mesh associated with a mesh variable. Given
the name of a variable to access, one must call this function to find the name of the mesh before
calling DBGetQuadmesh or DBGetUcdmesh.
DBInqMeshtype
Silo User’s Guide 2-153
DBInqMeshtype—Inquire the mesh type of a mesh.
Synopsis:
int DBInqMeshtype (DBfile *dbfile, char const *meshname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
meshname Mesh name.
Returns:
DBInqMeshtype returns the mesh type on success and -1 on failure.
Description:
The DBInqMeshtype function returns the type of the given mesh. The value returned is described
in the following table:
Mesh Type Returned Value
Multi-Block DB_MULTIMESH
UCD DB_UCDMESH
Pointmesh DB_POINTMESH
Quad (Collinear) DB_QUAD_RECT
Quad (Non-Collinear) DB_QUAD_CURV
CSG DB_CSGMESH
DBInqMeshtype
2-154 Silo User’s Guide
4 API Section Multi-Block Objects, Parallelism and
Poor-Man’s Parallel I/O
Individual pieces of mesh created with a number of DBPutXxxmesh() calls can be assembled
together into larger, multi-block objects. Likewise for variables and materials defined on these
meshes.
In Silo, multi-block objects are really just lists of all the individual pieces of a larger, coherent
object. For example, a multi-mesh object is really just a long list of object names, each name being
the string passed as the name argument to a DBPutXxxmesh() call.
A key feature of multi-block object is that references to the individual pieces include the option of
specifying the name of the Silo file in which a piece is stored. This option is invoked when the
colon operator (‘:’) appears in the name of an individual piece. All characters before the colon
specify the name of a Silo file. All characters after a colon specify the directory path within the file
where the object lives.
The fact that multi-block objects can reference individual pieces that reside in different Silo files
means that Silo, a serial I/O library, can be used very effectively and scalably in parallel without
resorting to writing a file per processor. The “technique” used to affect parallel I/O in this manner
with Silo is affectionately called Poor Man’s Parallel I/O (PMPIO).
A separate convenience interface, PMPIO, is provided for this purpose. The PMPIO interface pro-
vides almost all of the functionality necessary to use Silo in a Poor Man’s Parallel way. The appli-
cation is required to implement a few callback functions. The PMPIO interface is described at the
end of this section.
The functions described in this section of the manual include...
DBPutMultimesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 156
DBGetMultimesh . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 161
DBPutMultimeshadj . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 162
DBGetMultimeshadj. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 165
DBPutMultivar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 166
DBGetMultivar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 170
DBPutMultimat. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 171
DBGetMultimat . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 174
DBPutMultimatspecies . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 175
DBGetMultimatspecies. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 178
DBOpenByBcast. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 179
PMPIO_Init. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 181
PMPIO_CreateFileCallBack. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 184
PMPIO_OpenFileCallBack . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 185
PMPIO_CloseFileCallBack . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 186
PMPIO_WaitForBaton . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 187
PMPIO_HandOffBaton . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 188
PMPIO_Finish . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 189
PMPIO_GroupRank . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 190

DBPutMultimesh
2-156 Silo User’s Guide
DBPutMultimesh—Write a multi-block mesh object into a Silo file.
Synopsis:
int DBPutMultimesh (DBfile *dbfile, char const *name, int nmesh,
char const * const meshnames[], int const meshtypes[],
DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputmmesh(dbid, name, lname, nmesh, meshnames,
lmeshnames, meshtypes, optlist_id, status)
character*N meshnames (See “dbset2dstrlen” on page 285.)
Arguments:
dbfile Database file pointer.
name Name of the multi-block mesh object.
nmesh Number of meshes pieces (blocks) in this multi-block object.
meshnames Array of length nmesh containing pointers to the names of each of the mesh
blocks written with a DBPut<whatever>mesh() call. See below for
description of how to populate meshnames when the pieces are in different
files as well as DBOPT_MB_FILE/BLOCK_NS options to use a printf-style
namescheme for large nmesh in lieu of explicitly enumerating them here.
meshtypes Array of length nmesh containing the type of each mesh block such as
DB_QUAD_RECT, DB_QUAD_CURV, DB_UCDMESH, DB_POINTMESH, and
DB_CSGMESH. Be sure to see description, below, for
DBOPT_MB_BLOCK_TYPE option to use single, constant value when all pieces
are the same type.
optlist Pointer to an option list structure containing additional information to be
included in the object written into the Silo file. Use a NULL if there are no
options.
Returns:
DBPutMultimesh returns zero on success and -1 on failure.
Description:
The DBPutMultimesh function writes a multi-block mesh object into a Silo file. It accepts as input
the names of the various sub-meshes (blocks) which are part of this mesh.
The meshe blocks may be stored in different sub-directories within a Silo file and, optionally, even
in different Silo files altogether. So, the name of each mesh block is specified using its full Silo
path name. The full Silo pathname is the form...
[<silo-filename>:]<path-to-mesh>

DBPutMultimesh
Silo User’s Guide 2-157
The existence of a colon (‘:’) anywhere in meshnames[i] indicates that the ith mesh block
name is specified using both the Silo filename and the path in the file. All characters before the
colon are the Silo file pathname within the filesystem on which the file(s) reside. Use whatever
slash character (‘\’ for Windows or ‘/’ for Unix) is appropriate for the underlying filesystem in this
part of the string only. Silo will automatically handle changes in the slash character in this part of
the string if this data is ever read on a different filesystem. All characters after the colon are the
path of the object within the Silo file and must use only the ‘/’ slash character.
Use the keyword “EMPTY” for any block for which the associated mesh object does not exist.
This convention is often convenient in cases where there are many related multi-block objects and/
or that evolve in time in such a way that some blocks do not exist for some times.
The individual mesh names referenced here CANNOT be the names of other multi-block meshes.
In other words, it is not valid to create a multi-mesh that references other multi-meshes.
For example, in the case where the are 6 blocks to be assembled into a larger mesh named ‘multi-
mesh’ in the file ‘foo.silo’ and the blocks are stored in three files as in the figure below,
/mesh1
/dir1/mesh2
/dir1/mesh1
file “foo.silo”
/dir2/mesh2
/a/b/c/mesh1
/mesh2
file “bar.silo”
/multi-mesh
file “gorfo.silo”
“/mesh1”
“/dir1/mesh2”
“bar.silo:/dir1/mesh1”
“bar.silo:/dir2/mesh2”
“gorfo.silo:/a/b/c/mesh1”
“gorfo.silo:/mesh2”
Figure 0-7: Strings for multi-block objects.
the array of strings to be passed as the meshnames argument of DBPutMultimesh are illustrated.
Note that the two pieces of mesh that are in the same file as the multi-mesh object itself, ‘multi-
mesh’, do NOT require the colon and filename option. Only those pieces of the multi-mesh object
that are in different files from the one the multi-block object itself resides in require the colon and
filename option.
You may pass NULL for the meshnames argument and instead use the namescheme options,
DBOPT_MB_FILE_NS and DBOPT_MB_BLOCK_NS described in the table of options, below.
This is particularly important for meshes consisting of O(105) or more blocks because it saves sub-
stantial memory and I/O time. See documentation on “DBMakeNamescheme” on page 2-206 for
how to specify nameschemes.
Note, however, that with the DBOPT_MB_FILE/BLOCK_NS options, you are specifying only the
string that a reader will later use in a call to DBMakeNamescheme() to create a namescheme
object suitable for generating the meshnames and not the namescheme object itself.
For convenience, two namescheme options are supported. One namescheme maps block numbers
to filenames. The other maps block numbers to object names. A reader is required to then combine
both to generate the complete block name for each mesh block. Optionally and where appropriate,
one can specify a block namescheme only. External array references may be used in the name-
schemes. Any such array names found in the namescheme are assumed to be the names of simple,
1D, integer arrays written with a DBWrite() call and existing in the same directory as the multi-
block object . Finally, keep in mind that in the nameschemes, blocks are numbered starting from
zero.
DBPutMultimesh
2-158 Silo User’s Guide
If you are using the namescheme options and have EMPTY blocks, since the meshnames argu-
ment is NULL, you can use the DBOPT_MB_EMPTY_COUNT/LIST options to explicitly enumer-
ate any empty blocks instead of having to incroporate them into your nameschemes.
Similarly, when the mesh consists of blocks of all the same type, you may pass NULL for the
meshtypes argumnt and instead use the DBOPT_MB_BLOCK_TYPE option to specify a single,
constant block type for all blocks. This option can result in important savings for large numbers of
blocks.
Finally, note that what is described here for the mulitmesh object in the way of name for the indi-
vidual blocks applies to all multi-block objects (e.g. DBPutMulti<whatever>).
Notes:
The following table describes the options accepted by this function:
Option Name
Value
Data Type Option Meaning
Default
Value
DBOPT_BLOCKORIGIN int The origin of the block numbers. 1
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_EXTENTS_SIZEaint Number of values in each extent tuple 0
DBOPT_EXTENTSadouble* Pointer to an array of length nmesh *
DBOPT_EXTENTS_SIZE doubles where
each group of DBOPT_EXTENTS_SIZE
doubles is an extent tuple for the mesh
coordinates (see below).
DBOPT_EXTENTS_SIZE must be set for
this option to work correctly.
NULL
DBOPT_ZONECOUNTSaint* Pointer to an array of length nmesh indi-
cating the number of zones in each block.
NULL
DBOPT_HAS_EXTERNAL_ZONESaint* Pointer to an array of length nmesh indi-
cating for each block whether that block
has zones external to the whole multi-
mesh object. A non-zero value at index i
indicates block i has external zones. A
value of 0 (zero) indicates it does not.
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_MRGTREE_NAME char * Name of the mesh region grouping tree to
be associated with this multimesh.
NULL
DBOPT_TV_CONNECTIVTY int A non-zero value indicates that the con-
nectivity of the mesh varies with time.
0
DBPutMultimesh
Silo User’s Guide 2-159
DBOPT_DISJOINT_MODE int Indicates if any elements in the mesh are
disjoint. There are two possible modes.
One is DB_ABUTTING indicating that ele-
ments abut spatially but actually reference
different node ids (but spatially equivalent
nodal positions) in the node list. The other
is DB_FLOATING where elements neither
share nodes in the nodelist nor abut spa-
tially.
DB_NONE
DBOPT_TOPO_DIM int Used to indicate the topological dimension
of the mesh apart from its spatial dimen-
sion.
-1 (not
specified)
DBOPT_MB_BLOCK_TYPE int Constant block type for all blocks (not speci-
fied)
DBOPT_MB_FILE_NS char* Multi-block file namescheme. This is a
namescheme, indexed by block number,
to generate filename in which each block
is stored.
NULL
DBOPT_MB_BLOCK_NS char* Multi-block block namescheme. This is a
namescheme, indexed by block number,
used to generate names of each block
object apart from the file in which it may
reside.
NULL
DBOPT_MB_EMPTY_LIST int* When namescheme options are used,
there is no meshnames argument in which
to use the keyword ‘EMPTY’ for empty
blocks. Instead, the empty blocks can be
enumerated here, indexed from zero.
NULL
DBOPT_MB_EMPTY_COUNT int Number of entries in the argument to
DBOPT_MB_EMPTY_LIST
0
The options specified below have been deprecated. Use Mesh Region Group (MRG) trees instead.
DBOPT_GROUPORIGIN int The origin of the group numbers. 1
DBOPT_NGROUPS int The total number of groups in this mul-
timesh object.
0
DBOPT_ADJACENCY_NAMEachar * Name of a multi-mesh, nodal adjacency
object written with a call to adj.
NULL
DBOPT_GROUPINGS_SIZE int Number of integer entries in the associ-
ated groupings array
0
DBOPT_GROUPINGS int * Integer array of length specified by
DBOPT_GROUPINGS_SIZE containing
information on how different mesh blocks
are organized into, possibly hierarchical,
groups. See below for detailed discussion.
NULL
DBOPT_GROUPINGS_NAMES char ** Optional set of names to be associated
with each group in the groupings array
NULL
Option Name
Value
Data Type Option Meaning
Default
Value

DBPutMultimesh
2-160 Silo User’s Guide
There is a class of options for DBMulti- objects that is VERY IMPORTANT in helping to acceler-
ate performance in down-stream post-processing tools. We call these Down-stream Performance
Options. In order of utility, these options are DBOPT_EXTENTS, DBOPT_MIXLENS and
DBOPT_MATLISTS and DBOPT_ZONECOUNTS. Although these options are creating redun-
dant data in the Silo database, the data is stored in a manner that is far more convenient to down-
stream applications that read Silo databases. Therefore, the user is strongly encouraged to make
use of these options.
Regarding the DBOPT_EXTENTS option, see the notes for DBPutMultivar. Note, however, that
here the extents are for the coordinates of the mesh.
Regarding the DBOPT_ZONECOUNTS option, this option will help down-stream post-processing
tools to select an appropriate static load balance of blocks to processors.
Regarding the DBOPT_HAS_EXTERNAL_ZONES option, this option will help down-stream post-
processing tools accelerate computation of external boundaries. When a block is known not to
contain any external zones, it can be quickly skipped in the computation. Note that while false pos-
itives can negatively effect only performance during downstream external boundary calculations,
false negatives will result in serious errors.
In other words, it is ok for a block that does not have external zones to be flagged as though it
does. In this case, all that will happen in down-stream post-processing tools is that work to com-
pute external faces that could have been avoided will be wasted. However, it is not ok for a block
that has external zones to be flagged as though it does not. In this case, down-stream post-process-
ing tools will skip boundary computation when it should have been computed.
Three options, DBOPT_GROUPINGS_SIZE, DBOPT_GROUPINGS are deprecated. Instead, use
MRG trees to handle grouping. Also, see notes regarding _visit_domain_groups variable conven-
tion.
a. Indicates a Down-stream Performance Option. See notes below.

DBGetMultimesh
Silo User’s Guide 2-161
DBGetMultimesh—Read a multi-block mesh from a Silo database.
Synopsis:
DBmultimesh *DBGetMultimesh (DBfile *dbfile, char const *meshname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
meshname Name of the multi-block mesh.
Returns:
DBGetMultimesh returns a pointer to a DBmultimesh structure on success and NULL on failure.
Description:
The DBGetMultimesh function allocates a DBmultimesh data structure, reads a multi-block mesh
from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutMultimeshadj
2-162 Silo User’s Guide
DBPutMultimeshadj—Write some or all of a multi-mesh adjacency object into a Silo
file.
Synopsis:
int DBPutMultimeshadj(DBfile *dbfile, char const *name,
int nmesh, int const *mesh_types, int const *nneighbors,
int const *neighbors, int const *back,
int const *nnodes, int const * const nodelists[],
int const *nzones, int const * const zonelists[],
DBoptlist const *optlist)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Name of the multi-mesh adjacency object.
nmesh The number of mesh pieces in the corresponding multi-mesh object. This value
must be identical in repeated calls to DBPutMultimeshadj.
mesh_types Integer array of length nmesh indicating the type of each mesh in the
corresponding multi-mesh object. This array must be identical to that which is
passed in the DBPutMultimesh call and in repeated calls to
DBPutMultimeshadj.
nneighbors Integer array of length nmesh indicating the number of neighbors for each
mesh piece. This array must be identical in repeated calls to
DBPutMultimeshadj.
In the argument descriptions to follow, let
nneighbors[i
i0=
k
=
. That is, let
Sk
be the sum of the first k entries in the nneighbors array.
neighbors Array of
Snmesh
integers enumerating for each mesh piece all other mesh pieces
that neighbor it. Entries from index
Sk
to index
Sk1+1–
enumerate the
neighbors of mesh piece k. This array must be identical in repeated calls to
DBPutMultimeshadj.
back Array of
Snmesh
integers enumerating for each mesh piece, the local index of that
mesh piece in each of its neighbors lists of neighbors. Entries from index
Sk
to
index
Sk1+1–
enumerate the local indices of mesh piece k in each of the
neighbors of mesh piece k. This argument may be NULL. In any case, this array
must be identical in repeated calls to DBPutMultimeshadj.
nnodes Array of
Snmesh
integers indicating for each mesh piece, the number of nodes
that it shares with each of its neighbors. Entries from index
Sk
to index
Sk1+1–
indicate the number of nodes that mesh piece k shares with each of its
neighbors. This array must be identical in repeated calls to DBPutMultimeshadj.
This argument may be NULL.

DBPutMultimeshadj
Silo User’s Guide 2-163
nodelists Array of
Snmesh
pointers to arrays of integers. Entries from index
Sk
to index
Sk1+1–
enumerate the nodes that mesh piece k shares with each of its
neighbors. The contents of a specific nodelist array depend on the types of
meshes that are neighboring each other (See description below). nodelists[m]
may be NULL even if nnodes[m] is non-zero. See below for a description of
repeated calls to DBPutMultimeshadj. This argument must be NULL if nnodes
is NULL.
nzones Array of
Snmesh
integers indicating for each mesh piece, the number of zones
that are adjacent with each of its neighbors. Entries from index
Sk
to index
Sk1+1–
indicate the number of zones that mesh piece k has adjacent to each of
its neighbors. This array must be identical in repeated calls to
DBPutMultimeshadj. This argument may be NULL.
zonelists Array of
Snmesh
pointers to arrays of integers. Entries from index
Sk
to index
Sk1+1–
enumerate the zones that mesh piece k has adjacent with each of its
neighbors. The contents of a specific zonelist array depend on the types of
meshes that are neighboring each other (See description below). zonelists[m]
may be NULL even if nzones[m] is non-zero. See below for a description of
repeated calls to DBPutMultimeshadj. This argument must be NULL if nzones
is NULL.
optlist Pointer to an option list structure containing additional information to be
included in the object written into the Silo file. Use a NULL if there are no
options.
Description:
Note that the functionality this object provides is now more efficiently and conveniently han-
dled via a Mesh Region Grouping (MRG) tree. Users are encouraged to use MRG trees as an
alternative to DBPutMultimeshadj(). See “DBMakeMrgtree” on page 193.
DBPutMultimeshadj is another Down-stream Performance Option (See “DBPutMultimesh” on
page 2-156). It is an alternative to including ghost-zones (See “DBPutMultimesh” on page 2-156)
in the mesh and can therefore help to reduce file size, particularly for unstructured meshes.
A multi-mesh adjacency object informs down-stream, post-processing tools such as VisIt how
nodes and/or zones, should be shared between neighboring mesh pieces to eliminate post-process-
ing discontinuity artifacts along the boundaries between the pieces. If neither this information is
provided nor ghost zones are stored in the file, post-processing tools must then infer this informa-
tion from global node or zone ids (if they exist) or, worse, by matching coordinates which is a
time-consuming process.
DBPutMultimeshadj is used to indicate how various mesh pieces in a multi-mesh object abut by
specifying for each mesh piece, the nodes it shares with other mesh pieces and/or the zones is has
adjacent to other mesh pieces. Note the important distinction in how nodes and zones are classi-
fied here. Nodes are shared between mesh pieces while zones are merely adjacent between mesh
pieces. In a call to DBPutMultimeshadj, a caller may write information for either shared nodes or
adjacent zones, or both.
In practice, applications tend to use the same mesh type for every mesh piece. Thus, for ucd and
point meshes, the nodelist (or zonelist) arrays will consists of pairs of integers where the first of
the pair identifies a node (or zone) in the given mesh while the second identifies the shared node
DBPutMultimeshadj
2-164 Silo User’s Guide
(or adjacent zone) in a neighbor. Likewise, for quad meshes, the nodelist (or zonelist) arrays will
consists of 15 integers the first 6 of which identify a slab of nodes (or zones) in the given quad
mesh. The second set of 6 integers identify the slab of shared nodes (or zones) in a neighbor quad
mesh and the last 3 integers indicate the orientation of the neighbor quad mesh relative to the given
quad mesh. For example the entries (1,2,3) for these 3 integers mean that all axes are aligned. The
entries (-2,1,3) mean that the -J axis of the neighbor mesh piece aligns with the +I axis of the given
mesh piece, the +I axis of the neighbor mesh piece aligns with the +J axis of the given mesh piece,
and the +K axes both align the same way.
The specific contents of a given nodelist array depend on the types of meshes between which it
enumerates shared nodes. The table below describes the contents of nodelist array m given the dif-
ferent mesh types that it may enumerate shared nodes for.
Neighbor mesh type
DB_POINT or DB_UCD DB_QUAD
Given mesh type
DB_POINT or DB_UCD
nnodes[m] pairs of integers
nnodes[m]+6 integers.
The first nnodes[m] integers
identify the nodes in the given
point or ucd mesh.
The next 6 integers identify ijk
bounds of the corresponding
nodes in the quad mesh neigh-
bor.
DB_QUAD
6+nnodes[m] integers.
The first 6 integers identify ijk
bounds of the nodes in the given
quad mesh.
The last nnodes[m] integers
identify the nodes in the neighbor
point or ucd mesh.
15 integers
The first set of 6 integers identify
ijk bounds of nodes in the given
quad mesh.
The second set of 6 integers
identify ijk bounds of nodes in the
neighbor quad mesh
The next 3 integers specify the
orientation of the neighbor quad
mesh relative to the given mesh.
This function is designed so that it may be called multiple times, each time writing a different por-
tion of multi-mesh adjacency information to the object. On the first call, space is allocated in the
Silo file for the entire object. The required space is determined by the contents of all but the nodel-
ists (and/or zonelists) arrays. The contents of the nodelists (and/or zonelists) arrays are the only
arguments that are permitted to vary from call to call and then they may vary only in which entries
are NULL and non-NULL. Whenever an entry is NULL and the corresponding entry in nnodes (or
nzones) array is non-zero, the assumption is that the information is provided in some other call to
DBPutMultimeshadj.

DBGetMultimeshadj
Silo User’s Guide 2-165
DBGetMultimeshadj—Get some or all of a multi-mesh nodal adjacency object
Synopsis:
DBmultimeshadj *DBGetMultimeshadj(DBfile *dbfile,
char const *name,
int nmesh, int const *mesh_pieces)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer
name Name of the multi-mesh nodal adjacency object
nmesh Number of mesh pieces for which nodal adjacency information is being
obtained. Pass zero if you want to obtain all nodal adjacency information in a
single call.
mesh_pieces Integer array of length nmesh indicating which mesh pieces nodal adjacency
information is desired for. May pass NULL if nmesh is zero.
Returns:
A pointer to a fully or partially populated DBmultimeshadj object or NULL on failure.
Description:
DBGetMultimeshadj returns a nodal adjacency object. This function is designed so that it may be
called multiple times to obtain information for different mesh pieces in different calls. The nmesh
and mesh_pieces arguments permit the caller to specify for which mesh pieces adjacency informa-
tion shall be obtained.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutMultivar
2-166 Silo User’s Guide
DBPutMultivar—Write a multi-block variable object into a Silo file.
Synopsis:
int DBPutMultivar (DBfile *dbfile, char const *name, int nvar,
char const * const varnames[], int const vartypes[],
DBoptlist const *optlist);
Fortran Equivalent:
integer function dbputmvar(dbid, name, lname, nvar, varnames,
lvarnames, vartypes, optlist_id, status)
character*N varnames (See “dbset2dstrlen” on page 285.)
Arguments:
dbfile Database file pointer.
name Name of the multi-block variable.
nvar Number of variables associated with the multi-block variable.
varnames Array of length nvar containing pointers to the names of the variables written
with DBPut<whatever>var() call. See “DBPutMultimesh” on page 2-156
for description of how to populate varnames when the pieces are in different
files as well as DBOPT_MB_BLOCK/FILE_NS options to use a printf-style
namescheme for large nvar in lieu of explicitly enumerating them here.
vartypes Array of length nvar containing the types of the variables such as
DB_POINTVAR, DB_QUADVAR, or DB_UCDVAR. See “DBPutMultimesh” on
page 2-156, for DBOPT_MB_BLOCK_TYPE option to use single, constant value
when all pieces are the same type.
optlist Pointer to an option list structure containing additional information to be
included in the object written into the Silo file. Use a NULL if there are no
options.
Returns:
DBPutMultivar returns zero on success and -1 on failure.
Description:
The DBPutMultivar function writes a multi-block variable object into a Silo file.
Notes:
The following table describes the options accepted by this function:
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_BLOCKORIGIN int The origin of the block numbers. 1
DBOPT_CYCLE int Problem cycle value. 0

DBPutPointmesh
Silo User’s Guide 2-167
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_EXTENTS_SIZEaint Number of values in each extent tuple 0
DBOPT_EXTENTSadouble* Pointer to an array of length nvar *
DBOPT_EXTENTS_SIZE doubles where
each group of DBOPT_EXTENTS_SIZE
doubles is an extent tuple (see below).
DBOPT_EXTENTS_SIZE must be set for
this option to work correctly.
NULL
DBOPT_MMESH_NAME char * Name of the multimesh this variable is
associated with. Note, this option is very
important as down-stream post process-
ing tools are otherwise required to guess
as to the mesh a given variable is associ-
ated with. Sometimes, the tools can guess
wrong.
NULL
DBOPT_TENSOR_RANK int Specify the variable type; one of either
DB_VARTYPE_SCALAR,
DB_VARTYPE_VECTOR
DB_VARTYPE_TENSOR,
DB_VARTYPE_SYMTENSOR,
DB_VARTYPE_ARRAY
DB_VARTYPE_LABEL
DB_VARTYPE
_SCALAR
DBOPT_REGION_PNAMES char** A null-pointer terminated array of pointers
to strings specifying the pathnames of
regions in the mrg tree for the associated
mesh where the variable is defined. If
there is no mrg tree associated with the
mesh, the names specified here will be
assumed to be material names of the
material object associated with the mesh.
The last pointer in the array must be null
and is used to indicate the end of the list of
names.See
“DBOPT_REGION_PNAMES” on
page 218.
NULL
DBOPT_CONSERVED int Indicates if the variable represents a phys-
ical quantity that must be conserved under
various operations such as interpolation.
0
Option Name
Value
Data Type Option Meaning Default Value
DBPutPointmesh
2-168 Silo User’s Guide
Regarding the DBOPT_EXTENTS option, an extent tuple is a tuple of the variable’s minimum
value(s) followed by the variable’s maximum value(s). If the variable is a single, scalar variable,
each extent tuple will be 2 values of the form {min,max}. Thus, DBOPT_EXTENTS_SIZE will be
2. If the variable consists of nvars subvariables (e.g. the nvars argument in any of DBPutPoint-
var, DBPutQuadvar, DBPutUcdvar is greater than 1), then each extent tuple is 2*nvars values of
each subvariable’s minimum value followed by each subvariable’s maximum value. In this case,
DBOPT_EXTENTS_SIZE will be 2*nvars.
For example, if we have a multi-var object of a 3D velocity vector on 2 blocks, then
DBOPT_EXTENTS_SIZE will be 2*3=6 and the DBOPT_EXTENTS array will be an array of
2*6 doubles organized as follows...
DBOPT_EXTENSIVE int Indicates if the variable represents a phys-
ical quantity that is extensive (as opposed
to intensive). Note, while it is true that any
conserved quantity is extensive, the con-
verse is not true. By default and histori-
cally, all Silo variables are treated as
intensive.
0
DBOPT_MB_BLOCK_TYPE int Constant block type for all blocks (not specified)
DBOPT_MB_FILE_NS char* Multi-block file namescheme. This is a
namescheme, indexed by block number,
to generate filename in which each block
is stored.
NULL
DBOPT_MB_BLOCK_NS char* Multi-block block namescheme. This is a
namescheme, indexed by block number,
used to generate names of each block
object apart from the file in which it may
reside.
NULL
DBOPT_MB_EMPTY_LIST int* When namescheme options are used,
there is no varnames argument in which
to use the keyword ‘EMPTY’ for empty
blocks. Instead, the empty blocks can be
enumerated here, indexed from zero.
NULL
DBOPT_MB_EMPTY_COUNT int Number of entries in the argument to
DBOPT_MB_EMPTY_LIST
0
DBOPT_MISSING_VALUE double Specify a numerical value that is intended
to represent “missing values” in the x or y
data arrays. Default is
DB_MISSING_VALUE_NOT_SET
DB_MISSING_
VALUE_NOT_
SET
The options below have been deprecated. Use MRG trees instead.
DBOPT_GROUPORIGIN int The origin of the group numbers. 1
DBOPT_NGROUPS int The total number of groups in this mul-
timesh object.
0
a. Indicates a Down-stream Performance Option. See notes for DBPutMultimesh.
Option Name
Value
Data Type Option Meaning Default Value

DBPutPointmesh
Silo User’s Guide 2-169
{Vx_min_0, Vy_min_0, Vz_min_0, Vx_max_0, Vy_max_0, Vz_max_0,
Vx_min_1, Vy_min_1, Vz_min_1, Vx_max_1, Vy_max_1, Vz_max_1}
Note that if ghost zones are present in a block, the extents must be computed such that they include
contributions from data in the ghost zones. On the other hand, if a variable has mixed components,
that is component values on materials mixing within zones, then the extents should NOT include
contributions from the mixed variable values.

DBGetMultivar
2-170 Silo User’s Guide
DBGetMultivar—Read a multi-block variable definition from a Silo database.
Synopsis:
DBmultivar *DBGetMultivar (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Name of the multi-block variable.
Returns:
DBGetMultivar returns a pointer to a DBmultivar structure on success and NULL on failure.
Description:
The DBGetMultivar function allocates a DBmultivar data structure, reads a multi-block variable
from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutMultimat
Silo User’s Guide 2-171
DBPutMultimat—Write a multi-block material object into a Silo file.
Synopsis:
int DBPutMultimat (DBfile *dbfile, char const *name, int nmat,
char const * const matnames[], DBoptlist const *optlist)
Fortran Equivalent:
integer function dbputmmat(dbid, name, lname, nmat, matnames,
lmatnames, optlist_id, status)
Arguments:
dbfile Database file pointer.
name Name of the multi-material object.
nmat Number of material blocks provided.
matnames Array of length nmat containing pointers to the names of the material block
objects, written with DBPutMaterial(). See “DBPutMultimesh” on page 2-156
for description of how to populate matnames when the pieces are in different
files as well as DBOPT_MB_BLOCK/FILE_NS options to use a printf-style
namescheme for large nmat in lieu of explicitly enumerating them here.
optlist Pointer to an option list structure containing additional information to be
included in the object written into the Silo file. Use a NULL if there are no
options
Returns:
DBPutMultimat returns zero on success and -1 on error.
Description:
The DBPutMultimat function writes a multi-material object into a Silo file.
Notes:
The following table describes the options accepted by this function:
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_BLOCKORIGIN int The origin of the block numbers. 1
DBOPT_NMATNOS int Number of material numbers stored in the
DBOPT_MATNOS option.
0
DBOPT_MATNOS int * Pointer to an array of length
DBOPT_NMATNOS containing a complete
list of the material numbers used in the
Multimat object. DBOPT_NMATNOS must
be set for this to work correctly.
NULL
DBPutMultimat
2-172 Silo User’s Guide
DBOPT_MATNAMES char** Pointer to an array of length
DBOPT_NMATNOS containing a complete
list of the material names used in the Mul-
timat object. DBOPT_NMATNOS must be
set for this to work correctly.
NULL
DBOPT_MATCOLORS char** Array of strings defining the names of col-
ors to be associated with each material.
The color names are taken from the X win-
dows color database. If a color name
begins with a’#’ symbol, the remaining 6
characters are interpreted as the hexa-
decimal RGB value for the color.
DBOPT_NMATNOS must be set for this to
work correctly.
NULL
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_MIXLENSaint* Array of nmat ints which are the values of
the mixlen arguments in each of the indi-
vidual block’s material objects.
DBOPT_MATCOUNTSaint* Array of nmat counts indicating the num-
ber of materials actually in each block.
NULL
DBOPT_MATLISTSaint* Array of material numbers in each block.
Length is the sum of values in
DBOPT_MATCOUNTS. DBOPT_MATCOUNTS
must be set for this option to work cor-
rectly.
NULL
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_ALLOWMAT0 int If set to non-zero, indicates that a zero
entry in the matlist array is actually not a
valid material number but is instead being
used to indicate an ‘unused’ zone.
0
DBOPT_MMESH_NAME char * Name of the multimesh this material is
associated with. Note, this option is very
important as down-stream post process-
ing tools are otherwise required to guess
as to the mesh a given material is associ-
ated with. Sometimes, the tools can guess
wrong.
NULL
DBOPT_MB_FILE_NS char* Multi-block file namescheme. This is a
namescheme, indexed by block number,
to generate filename in which each block
is stored.
NULL
Option Name
Value
Data Type Option Meaning Default Value

DBPutMultimat
Silo User’s Guide 2-173
Regarding the DBOPT_MIXLENS option, this option will help down-stream post-processing tools
to select an appropriate load balance of blocks to processors. Material mixing and material inter-
face reconstruction have a big effect on cost of certain post-processing operations.
Regarding the DBOPT_MATLISTS options, this option will give down-stream post-processing
tools better knowledge of how materials are distributed among blocks.
DBOPT_MB_BLOCK_NS char* Multi-block block namescheme. This is a
namescheme, indexed by block number,
used to generate names of each block
object apart from the file in which it may
reside.
NULL
DBOPT_MB_EMPTY_LIST int* When namescheme options are used,
there is no varnames argument in which
to use the keyword ‘EMPTY’ for empty
blocks. Instead, the empty blocks can be
enumerated here, indexed from zero.
NULL
DBOPT_MB_EMPTY_COUNT int Number of entries in the argument to
DBOPT_MB_EMPTY_LIST
0
The options below have been deprecated. Use MRG trees instead.
DBOPT_GROUPORIGIN int The origin of the group numbers. 1
DBOPT_NGROUPS int The total number of groups in this mul-
timesh object.
0
a. Indicates a Down-stream Performance Option. See notes for DBPutMultimesh.
Option Name
Value
Data Type Option Meaning Default Value

DBGetMultimat
2-174 Silo User’s Guide
DBGetMultimat—Read a multi-block material object from a Silo database
Synopsis:
DBmultimat *DBGetMultimat (DBfile *dbfile, char const *name)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer
name Name of the multi-block material object
Returns:
DBGetMultimat returns a pointer to a DBmultimat structure on success and NULL on failure.
Description:
The DBGetMultimat function allocates a DBmultimat data structure, reads a multi-block material
from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.
DBPutMultimatspecies
Silo User’s Guide 2-175
DBPutMultimatspecies—Write a multi-block species object into a Silo file.
Synopsis:
int DBPutMultimatspecies (DBfile *dbfile, char const *name,
int nspec, char const * const specnames[],
DBoptlist const *optlist)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Name of the multi-block species structure.
nspec Number of species objects provided.
specnames Array of length nspec containing pointers to the names of each of the species.
See “DBPutMultimesh” on page 2-156 for description of how to populate
specnames when the pieces are in different files as well as
DBOPT_MB_BLOCK/FILE_NS options to use a printf-style namescheme for
large nspec in lieu of explicitly enumerating them here.
optlist Pointer to an option list structure containing additional information to be
included in the object written into the Silo file. Use a NULL if there are no
options.
Returns:
DBPutMultimatspecies returns zero on success and -1 on failure.
Description:
The DBPutMultimatspecies function writes a multi-block material species object into a Silo file. It
accepts as input descriptions of the various sub-species (blocks) which are part of this mesh.
Notes:
The following table describes the options accepted by this function:
Option Name
Value
Data Type Option Meaning Default Value
DBOPT_BLOCKORIGIN int The origin of the block numbers. 1
DBOPT_MATNAME char * Character string defining the name of the
multi-block material with which this object
is associated.
NULL
DBOPT_NMAT int The number of materials in the associated
material object.
0
DBPutMultimatspecies
2-176 Silo User’s Guide
DBOPT_NMATSPEC int * Array of length DBOPT_NMAT containing
the number of material species associated
with each material. DBOPT_NMAT must be
set for this to work correctly.
NULL
DBOPT_CYCLE int Problem cycle value. 0
DBOPT_TIME float Problem time value. 0.0
DBOPT_DTIME double Problem time value. 0.0
DBOPT_HIDE_FROM_GUI int Specify a non-zero value if you do not
want this object to appear in menus of
downstream tools
0
DBOPT_SPECNAMES char** Array of strings defining the names of the
individual species. DBOPT_NMATSPEC
must be set for this to work correctly. The
length of this array is the sum of the val-
ues in the argument to the
DBOPT_NMATSPEC option.
NULL
DBOPT_SPECCOLORS char** Array of strings defining the names of col-
ors to be associated with each species.
The color names are taken from the X win-
dows color database. If a color name
begins with a’#’ symbol, the remaining 6
characters are interpreted as the hexa-
decimal RGB value for the color.
DBOPT_NMATSPEC must be set for this to
work correctly. The length of this array is
the sum of the values in the argument to
the DBOPT_NMATSPEC option.
NULL
DBOPT_MB_FILE_NS char* Multi-block file namescheme. This is a
namescheme, indexed by block number,
to generate filename in which each block
is stored.
NULL
DBOPT_MB_BLOCK_NS char* Multi-block block namescheme. This is a
namescheme, indexed by block number,
used to generate names of each block
object apart from the file in which it may
reside.
NULL
DBOPT_MB_EMPTY_LIST int* When namescheme options are used,
there is no varnames argument in which
to use the keyword ‘EMPTY’ for empty
blocks. Instead, the empty blocks can be
enumerated here, indexed from zero.
NULL
DBOPT_MB_EMPTY_COUNT int Number of entries in the argument to
DBOPT_MB_EMPTY_LIST
0
The options below have been deprecated. Use MRG trees instead.
Option Name
Value
Data Type Option Meaning Default Value

DBPutMultimatspecies
Silo User’s Guide 2-177
DBOPT_GROUPORIGIN int The origin of the group numbers. 1
DBOPT_NGROUPS int The total number of groups in this mul-
timesh object.
0
Option Name
Value
Data Type Option Meaning Default Value

DBGetMultimatspecies
2-178 Silo User’s Guide
DBGetMultimatspecies—Read a multi-block species from a Silo database.
Synopsis:
DBmultimesh *DBGetMultimatspecies (DBfile *dbfile,
char const *name)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Name of the multi-block material species.
Returns:
DBGetMultimatspecies returns a pointer to a DBmultimatspecies structure on success and NULL
on failure.
Description:
The DBGetMultimatspecies function allocates a DBmultimatspecies data structure, reads a multi-
block material species from the Silo database, and returns a pointer to that structure. If an error
occurs, NULL is returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBOpenByBcast
Silo User’s Guide 2-179
DBOpenByBcast—Specialized, read-only open method for parallel applications needing all
processors to read all (or most of) a given Silo file
Synopsis:
DBfile *DBOpenByBcast(char const *filename, MPI_Comm comm,
int rank_of_root)
Fortran Equivalent:
None
Arguments:
filename name of the Silo file to open
comm MPI communicator to use for the broadcast operation
rank_of_root MPI rank of the processor in the communicator comm that shall serve as the
root of the broadcast (typically 0).
Returns:
A Silo database file handle just as returned from DBOpen or DBCreate except that the file is read-
only. Available only for reading Silo files produced via the HDF5 driver.
Description:
This is an experimental interface! It is not fully integrated into the Silo library.
In many parallel applications, there is a master or root file that all processors need all (or most of)
the information from in order to bootstrap opening a larger collection of Silo files (similar to
PMPIO)
This method is provided to perform the operation in a way that is friendly to the underlying filesys-
tem by opening the file on a single processor using the HDF5 file-in-core feature and then broad-
casting the “file” buffer to all processors which then turn around and open the buffer as a file. In
this way, the application can avoid many processors interacting with and potentially overwhelming
the filesystem.
But, there are some important limitations too. First, it works only for reading Silo files. Next, the
entire Silo file is loaded into a buffer in memory and the broadcast in its entirety to all other pro-
cessors. If only some processors need only some of the data from the file, then there is potentially
a lot of memory and communication wasted for parts of the file not used on various processors.
When the file is closed with DBClose() all memory used by the file is released.
This method is not compiled into libsilo[h5].a. Instead, you are required to obtain the bcastopen.c
source file (which is installed to the include dir of the Silo install point) and compile it into your
application and then include a line of this form...
extern DBfile *DBOpenByBcast(char const *, MPI_Comm, int);

DBOpenByBcast
2-180 Silo User’s Guide
in your application.
Note that you can find an example of its use in the Silo source release “tests” directory in the
source file “bcastopen_test.c”

PMPIO_Init
Silo User’s Guide 2-181
PMPIO_Init—Initialize a Poor Man’s Parallel I/O interaction with the Silo library
Synopsis:
PMPIO_baton_t *PMPIO_Init(int numFiles, PMPIO_iomode_t ioMode,
MPI_Comm mpiComm, int mpiTag,
PMPIO_CreateFileCallBack createCb,
PMPIO_OpenFileCallBack openCb,
PMPIO_CloseFileCallBack closeCB,
void *userData)
Fortran Equivalent:
None
Arguments:
numFiles The number of individual Silo files to generate. Note, this is the number of
parallel I/O streams that will be running simultaneously during I/O. A value of 1
cause PMPIO to behave serially. A value equal to the number of processors
causes PMPIO to create a file-per-processor. Both values are unwise. For most
parallel HPC platforms, values between 8 and 64 are appropriate.
ioMode Choose one of either PMPIO_READ or PMPIO_WRITE. Note, you can not use
PMPIO to handle both read and write in the same interaction.
mpiComm The MPI communicator you would like PMPIO to use when passing the tiny
baton messages it needs to coordinate access to the underlying Silo files. See
documentation on MPI for a description of MPI communicators.
mpiTag The MPI message tag you would like PMPIO to use when passing the tiny baton
messages it needs to coordinate access to the underlying Silo files.
createCb The file creation callback function. This is a function you implement that
PMPIO will call when the first processor in each group needs to create the Silo
file for the group. It is needed only for PMPIO_WRITE operations. If default
behavior is acceptable, pass PMPIO_DefaultCreate here.
openCb The file open callback function. This is a function you implement that PMPIO
will call when the second and subsequent processors in each group need to open
a Silo file. It is needed for both PMPIO_READ and PMPIO_WRITE operations.
If default behavior is acceptable, pass PMPIO_DefaultOpen here.
closeCb The file close callback function. This is a function you implement that PMPIO
will call when a processor in a group needs to close a Silo file. If default
behavior is acceptable, pass PMPIO_DefaultClose here.
userData [OPT] Arbitrary user data that will be passed back to the various callback
functions. Pass NULL(0) if this is not needed.
Returns:
A pointer to a PMPIO_baton_t object to be used in subsequent PMPIO calls on success. NULL on
failure.

PMPIO_Init
2-182 Silo User’s Guide
Description:
The PMPIO interface was designed to be separate from the Silo library. To use it, you must
include the PMPIO header file, pmpio.h, after the MPI header file, mpi.h, in your applica-
tion. This interface was designed to work with any serial library and not Silo specifically. For
example, these same routines can be used with raw HDF5 or PDB files if so desired.
The PMPIO interface decomposes a set of P processors into N groups and then provides access, in
parallel, to a separate Silo file per group. This is the essence of Poor Man’s Parallel I/O.
For PMPIO_WRITE operations, each processor in a group creates its own Silo sub-directory
within the Silo file to write its data to. At any one moment, only one processor from each group
has a file open for writing. Hence, the I/O is serial within a group. However, because a processor in
each of the N groups is writing to its own Silo file, simultaneously, the I/O is parallel across
groups.
The number of files, N, can be chosen wholly independently of the total number of processors per-
mitting the application to tune N to the underlying filesystem. If N is set to 1, the result will be
serial I/O to a single file. If N is set to P, the result is one file per processor. Both of these are poor
choices.
Typically, one chooses N based on the number of available I/O channels. For example, a parallel
application running on 2,000 processors and writing to a filesystem that supports 8 parallel I/O
channels could select N=8 and achieve nearly optimum I/O performance and create only 8 Silo
files.
On every processor, the sequence of PMPIO operations takes the following form...
PMPIO_baton_t *bat = PMPIO_Init(...);
dbFile = (DBfile *) PMPIO_WaitForBaton(bat, ...);
/* local work (e.g. DBPutXXX() calls) for this processor */
.
.
.
PMPIO_HandOffBaton(bat, ...);
PMPIO_Finish(bat);
For a given PMPIO group of processors, only one processor in the group is in the “local work”
block of the above code. All other processors have either completed it or are waiting their prede-
cessor to finish. However, every PMPIO group will have one processor working in the “local
work” block, concurrently, to different files.
After PMPIO_Finish(), there is still one final step that PMPIO DOES NOT HELP with. That
is the creation of the multi-block objects that reference the individual pieces written by all the pro-
cessors with DBPutXXX calls in the “local work” part of the above sequence. It is the applica-
tion’s responsibility to correctly assembly the names of all these pieces and then create the multi-
block objects that reference them. Ordinarily, the application designates one processor to write
these multi-block objects and one of the N Silo files to write them to. Again, this last step is not
something PMPIO will help with.

PMPIO_Init
Silo User’s Guide 2-183
Poor Man’s Parallel I/O is a simple and effective I/O strategy that has been used by codes like
Ale3d and SAMRAI for many years and has shown excellent scaling behavior. A drawback of this
approach is, of course, that multiple files are generated. However, when used appropriately, this
number of files is typically small (e.g. 8 to 64). In addition, our experience has been that concur-
rent, parallel I/O to a single file which also supports sufficient variation in size, shape and pattern
of I/O requests from processor to processor is a daunting challenge to perform scalably. So, while
Poor Man’s Parallel I/O is not truly concurrent, parallel I/O, it has demonstrated that it is not only
highly flexible and highly scalable but also very easy to implement and for these reasons, often a
superior choice to true, concurrent, parallel I/O.

PMPIO_CreateFileCallBack
2-184 Silo User’s Guide
PMPIO_CreateFileCallBack—The PMPIO file creation callback
Synopsis:
typedef void *(*PMPIO_CreateFileCallBack)(const char *fname,
const char *dname, void *udata);
Fortran Equivalent:
None
Arguments:
fname The name of the Silo file to create.
dname The name of the directory within the Silo file to create.
udata A pointer to any additional user data. This is the pointer passed as the
userData argument to PMPIO_Init().
Returns:
A void pointer to the created file handle.
Description:
This defines the PMPIO file creation callback interface.
Your implementation of this file creation callback should minimally do the following things.
For PMPIO_WRITE operation, your implementation should DBCreate() a Silo file of name
fname, DBMkDir() a directory of name dname for the first processor of a group to write to and
DBSetDir() to that directory.
For PMPIO_READ operations, your implementation of this callback is never called.
The PMPIO_DefaultCreate function does only the minimal work, returning a void pointer to
the created DBfile Silo file handle.

PMPIO_OpenFileCallBack
Silo User’s Guide 2-185
PMPIO_OpenFileCallBack—The PMPIO file open callback
Synopsis:
typedef void *(*PMPIO_OpenFileCallBack)(const char *fname,
const char *dname, PMPIO_iomode_t iomode, void *udata);
Fortran Equivalent:
None
Arguments:
fname The name of the Silo file to open.
dname The name of the directory within the Silo file to work in.
iomode The iomode of this PMPIO interaction. This is the value passed as ioMode
argument to PMPIO_Init().
udate A pointer to any additional user data. This is the pointer passed as the
userData argument to PMPIO_Init().
Returns:
A void pointer to the opened file handle that was.
Description:
This defines the PMPIO open file callback.
Your implementation of this open file callback should minimally do the following things.
For PMPIO_WRITE operations, it should DBOpen() the Silo file named fname, DBMkDir() a
directory named dname and DBSetDir() to directory dname.
For PMPIO_READ operations, it should DBOpen() the Silo file named fname and then
DBSetDir() to the directory named dname.
The PMPIO_DefaultOpen function does only the minimal work, returning a void pointer to the
opened DBfile Silo handle.

PMPIO_CloseFileCallBack
2-186 Silo User’s Guide
PMPIO_CloseFileCallBack—The PMPIO file close callback
Synopsis:
typedef void (*PMPIO_CloseFileCallBack)(void *file, void *udata);
Fortran Equivalent:
None
Arguments:
file void pointer to the file handle (DBfile pointer).
udata A pointer to any additional user data. This is the pointer passed as the
userData argument to PMPIO_Init().
Returns:
None
Description:
This defines the PMPIO close file callback interface.
Your implementation of this callback function should simply close the file. It us up to the imple-
mentation to know the correct time of the file handle passed as the void pointer file.
The PMPIO_DefaultClose function simply closes the Silo file.

PMPIO_WaitForBaton
Silo User’s Guide 2-187
PMPIO_WaitForBaton—Wait for exclusive access to a Silo file
Synopsis:
void *PMPIO_WaitForBaton(PMPIO_baton_t *bat,
const char *filename, const char *dirname)
Fortran Equivalent:
None
Arguments:
bat The PMPIO baton handle obtained via a call to PMPIO_Init().
filename The name of the Silo file this processor will create or open.
dirname The name of the directory within the Silo file this processor will work in.
Returns:
NULL (0) on failure. Otherwise, for PMPIO_WRITE operations the return value is whatever the
create or open file callback functions return. For PMPIO_READ operations, the return value is
whatever the open file callback function returns.
Description:
All processors should call this function as the next PMPIO function to call following a call to
PMPIO_Init().
For all processors that are the first processors in their groups, this function will return immediately
after having called the file creation callback specified in PMPIO_Init(). Typically, this callback
will have created a file with the name filename and a directory in the file with the name
dirname as well as having set the current working directory to dirname.
For all processors that are not the first in their groups, this call will block, waiting for the processor
preceding it to finish its work on the Silo file for the group and pass the baton to the next proces-
sor.
A typical naming convention for filename is something like “my_file_%03d.silo” where the
“%03d” is replaced with the group rank (See “PMPIO_GroupRank” on page 190.) of the proces-
sor. Likewise, a typical naming convention for dirname is something like “domain_%03d” where
the “%03d” is replaced with the rank-in-group (See “PMPIO_RankInGroup” on page 191.) of the
processor.

PMPIO_HandOffBaton
2-188 Silo User’s Guide
PMPIO_HandOffBaton—Give up all access to a Silo file
Synopsis:
void PMPIO_HandOffBaton(const PMPIO_baton_t *bat, void *file)
Fortran Equivalent:
None
Arguments:
bat The PMPIO baton handle obtained via a call to PMPIO_Init().
file A void pointer to the Silo DBfile object.
Returns:
None
Description:
When a processor has completed all its work on a Silo file, it gives up access to the file by calling
this function. This has the effect of closing the Silo file and then passing the baton to the next pro-
cessor in the group.

PMPIO_Finish
Silo User’s Guide 2-189
PMPIO_Finish—Finish a Poor Man’s Parallel I/O interaction with the Silo library
Synopsis:
void PMPIO_Finish(PMPIO_baton *bat)
Fortran Equivalent:
None
Arguments:
bat The PMPIO baton handle obtained via a call to PMPIO_Init().
Returns:
None.
Description:
After a processor has finished a PMPIO interaction with the Silo library, call this function to free
the baton object associated with the interaction.

PMPIO_GroupRank
2-190 Silo User’s Guide
PMPIO_GroupRank—Obtain ‘group rank’ of a processor
Synopsis:
int PMPIO_GroupRank(const PMPIO_baton_t *bat, int rankInComm)
Fortran Equivalent:
None
Arguments:
bat The PMPIO baton handle obtained via a call to PMPIO_Init().
rankInComm Rank of processor in the MPI communicator passed in PMPIO_Init() for
which group rank is to be queried.
Returns:
The ‘group rank’ of the queiried processor. In other words, the group number of the queried pro-
cessor, indexed from zero.
Description:
This is a convenience function to help applications identify which PMPIO group a given processor
belongs to.

PMPIO_RankInGroup
Silo User’s Guide 2-191
PMPIO_RankInGroup—Obtain the rank of a processor within its PMPIO group
Synopsis:
int PMPIO_RankInGroup(const PMPIO_baton_t *bat, int rankInComm)
Fortran Equivalent:
None
Arguments:
bat The PMPIO baton handle obtained via a call to PMPIO_Init().
rankInComm Rank of the processor in the MPI communicator used in PMPIO_Init() to be
queried.
Returns:
The rank of the queried processor within its PMPIO group.
Description:
This is a convenience function for applications to determine which processor a given processor is
within its PMPIO group.

PMPIO_RankInGroup
2-192 Silo User’s Guide
5 API Section Part Assemblies, AMR, Slide Surfaces,
Nodesets and Other Arbitrary Mesh Subsets
This section of the API manual describes Mesh Region Grouping (MRG) trees and Groupel Maps.
MRG trees describe the decomposition of a mesh into various regions such as parts in an assembly,
materials (even mixing materials), element blocks, processor pieces, nodesets, slide surfaces,
boundary conditions, etc. Groupel maps describe the, problem sized, details of the subsetted
regions. MRG trees and groupel maps work hand-in-hand in efficiently (and scalably) characteriz-
ing the various subsets of a mesh.
MRG trees are associated with (e.g. bound to) the mesh they describe using the
DBOPT_MRGTREE_NAME optlist option in the DBPutXxxmesh() calls. MRG trees are used
both to describe a multi-mesh object and then again, to describe individual pieces of the multi-
mesh.
In addition, once an MRG tree has been defined for a mesh, variables to be associated with the
mesh can be defined on only specific subsets of the mesh using the DBOPT_REGION_PNAMES
optlist option in the DBPutXxxvar() calls.
Because MRG trees can be used to represent a wide variety of subsetting functionality and because
applications have still to gain experience using MRG trees to describe their subsetting applica-
tions, the methods defined here are design to be as free-form as possible with few or no limitations
on, for example, naming conventions of the various types of subsets. It is simply impossible to
know a priori all the different ways in which applications may wish to apply MRG trees to con-
struct subsetting information.
For this reason, where a specific application of MRG trees is desired (to represent materials for
example), we document the naming convention an application must use to affect the representa-
tion.
The functions described in this section of the API manual are...
DBMakeMrgtree. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 193
DBAddRegion . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 197
DBAddRegionArray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 199
DBSetCwr. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 201
DBGetCwr . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 202
DBPutMrgtree. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 203
DBGetMrgtree . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 204
DBFreeMrgtree. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 205
DBMakeNamescheme . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 206
DBGetName . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 210
DBPutMrgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 211
DBGetMrgvar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 213
DBPutGroupelmap . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 214
DBGetGroupelmap . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 216
DBFreeGroupelmap . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 217
DBOPT_REGION_PNAMES . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 218
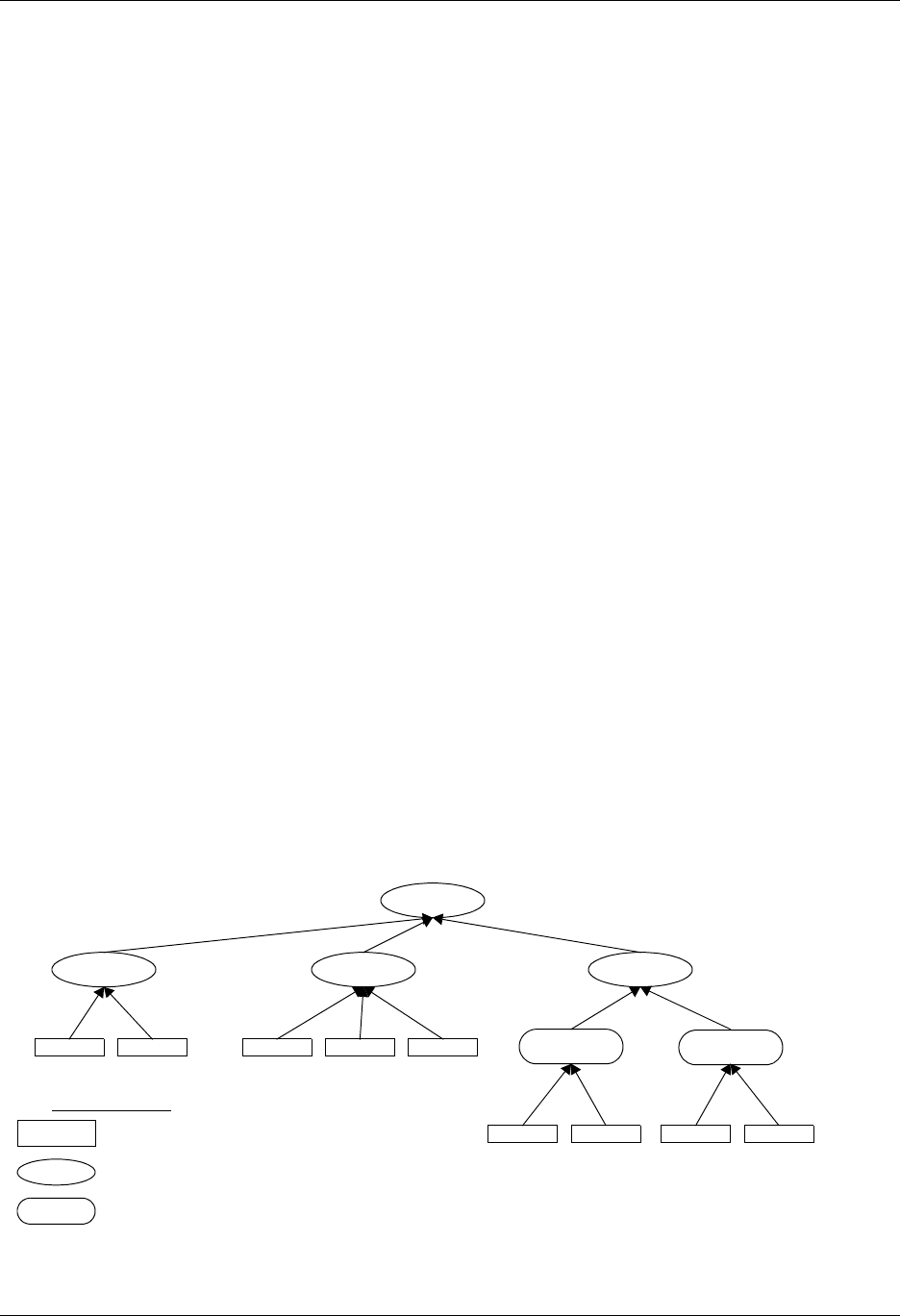
DBMakeMrgtree
Silo User’s Guide 2-193
DBMakeMrgtree—Create and initialize an empty mesh region grouping tree
Synopsis:
DBmrgtree *DBMakeMrgtree(int mesh_type, int info_bits,
int max_children, DBoptlist const *opts)
Fortran Equivalent:
integer function dbmkmrgtree(mesh_type, info_bits, max_children,
optlist_id, tree_id)
returns handle to newly created tree in tree_id.
Arguments:
mesh_type The type of mesh object the MRG tree will be associated with. An example
would be DB_MULTIMESH, DB_QUADMESH, DB_UCDMESH.
info_bits UNUSED
max_children Maximum number of immediate children of the root.
opts Additional options
Returns:
A pointer to a new DBmrgtree object on success and NULL on failure
Description:
This function creates a Mesh Region Grouping Tree (MRG) tree used to define different regions in
a mesh.
An MRG tree is used to describe how a mesh is composed of regions such as materials, parts in an
assembly, levels in an adaptive refinement hierarchy, nodesets, slide surfaces, boundary condi-
tions, as well as many other kinds of regions. An example is shown in Figure 0-8 on page 193.
mesh
nodesets materials assembly
wingsfueselage
ns_a ns_b copper zinc iron
left rightcabin cockpit
region-only node
grouping-only node
region & grouping node
node-type legend
Figure 0-8: Example of MRGTree
DBMakeMrgtree
2-194 Silo User’s Guide
In a multi-mesh setting, an MRG tree describing all of the subsets of the mesh is associated with
the top-level multimesh object. In addition, separate MRG trees representing the relevant portions
of the top-level MRG tree are also associated with each block.
MRG trees can be used to describe a wide variety of subsets of a mesh. In the paragraphs below,
we outline the use of MRG trees to describe a variety of common subsetting scenarios. In some
cases, a specific naming convention is required to fully specify the subsetting scenario.
The paragraphs below describe how to utilize an MRG tree to describe various common kinds of
decompositions and subsets.
Multi-Block Grouping (obsoletes DBOPT_GROUPING options for DBPutMultimesh,
_visit_domain_groups convention)
A multi-block grouping is the assignment of the blocks of a multi-block mesh (e.g. the mesh
objects created with DBPutXxxmesh() calls and enumerated by name in a DBPutMultimesh() call)
to one of several groups. Each group in the grouping represents several blocks of the multi-block
mesh. Historically, support for such a grouping in Silo has been limited in a couple of ways. First,
only a single grouping could be defined for a multi-block mesh. Second, the grouping could not be
hierarchically defined. MRG trees, however, support both multiple groupings and hierarchical
groupings.
In the MRG tree, define a child node of the root named “groupings.” All desired groupings shall be
placed under this node in the tree.
For each desired grouping, define a groupel map where the number of segments of the map is
equal to the number of desired groups. Map segment i will be of groupel type DB_BLOCKCENT
and will enumerate the blocks to be assigned to group i. Next, add regions (either an array of
regions or one at a time) to the MRG tree, one region for each group and specify the groupel map
name and other map parameters to be associated with these regions.
mesh
groupings
grouping A grouping B
side top bottom front skinny fat
Multiple Groupings
mesh
groupings
bottom front
Single, Hierarchical Grouping
groupel map object
groupel map segments refs.
interior exterior
deep shallow faces edges
Figure 0-9: Examples of MRG trees for single and multiple groupings.
In the diagram above, for the multiple grouping case, two groupel map objects are defined; one for
each grouping. For the ‘A’ grouping, the groupel map consists of 4 segments (all of which are of

DBMakeMrgtree
Silo User’s Guide 2-195
groupel type DB_BLOCKCENT) one for each grouping in ‘side’, ‘top’, ‘bottom’ and ‘front.’ Each
segment identifies the blocks of the multi-mesh (at the root of the MRG tree) that are in each of the
4 groups. For the ‘B’ grouping, the groupel map consists of 2 segments (both of type
DB_BLOCKCENT), for each grouping in ‘skinny’ and ‘fat’. Each segment identifies the blocks of
the multi-mesh that are in each group.
If, in addition to defining which blocks are in which groups, an application wishes to specify spe-
cific nodes and/or zones of the group that comprise each block, additional groupel maps of type
DB_NODECENT or DB_ZONECENT are required. However, because such groupel maps are speci-
fied in terms of nodes and/or zones, these groupel maps need to be defined on an MRG tree that is
associated with an individual mesh block. Nonetheless, the manner of representation is analogous.
Multi-Block Neighbor Connectivity (obsoletes DBPutMultimeshadj):
Multi-block neighbor connectivity information describes the details of how different blocks of a
multi-block mesh abut with shared nodes and/or adjacent zones. For a given block, multi-block
neighbor connectivity information lists the blocks that share nodes (or have adjacent zones) with
the given block and then, for each neighboring block, also lists the specific shared nodes (or adja-
cent zones).
If the underlying mesh type is structured (e.g. DBPutQuadmesh() calls were used to create the
individual mesh blocks), multi-block neighbor connectivity information can be scalably repre-
sented entirely at the multi-block level in an MRG tree. Otherwise, it cannot and it must be repre-
sented at the individual block level in the MRG tree. This section will describe both scenarios.
Note that these scenarios were previously handled with the now deprecated DBPutMultimeshadj()
call. That call, however, did not have favorable scalaing behavior for the unstructured case.
The first step in defining multi-block connectivity information is to define a top-level MRG tree
node named “neighbors.” Underneath this point in the MRG tree, all the information identifying
multi-block connectivity will be added.
Next, create a groupel map with number of segments equal to the number of blocks. Segment i of
the map will by of type DB_BLOCKCENT and will enumerate the neighboring blocks of block i.
Next, in the MRG tree define a child node of the root named “neighborhoods”. Underneath this
node, define an array of regions, one for each block of the multiblock mesh and associate the grou-
pel map with this array of regions.
For the structured grid case, define a second groupel map with number of segments equal to the
number of blocks. Segment i of the map will be of type DB_NODECENT and will enumerate the
slabs of nodes block i shares with each of its neighbors in the same order as those neighbors are
listed in the previous groupel map. Thus, segment i of the map will be of length equal to the num-
ber of neighbors of block i times 6 (2 ijk tuples specifying the lower and upper bounds of the slab
of shared nodes).
For the unstructured case, it is necessary to store groupel maps that enumerate shared nodes
between shared blocks on MRG trees that are associated with the individual blocks and NOT the
multi-block mesh itself. However, the process is otherwise the same.
In the MRG tree to be associated with a given mesh block, create a child of the root named “neigh-
bors.” For each neighboring block of the given mesh block, define a groupel map of type
DB_NODECENT, enumerating the nodes in the current block that are shared with another block
(or of type DB_ZONECENT enumerating the nodes in the current block that abut another block).

DBMakeMrgtree
2-196 Silo User’s Guide
Underneath this node in the MRG tree, create a region representing each neighboring block of the
given mesh block and associate the appropriate groupel map with each region.
Multi-Block, Structured Adaptive Mesh Refinement:
In a structured AMR setting, each AMR block (typically called a “patch” by the AMR commu-
nity), is written by a DBPutQuadmesh() call. A DBPutMultimesh() call groups all these
blocks together, defining all the individual blocks of mesh that comprise the complete AMR mesh.
An MRG tree, or portion thereof, is used to define which blocks of the multi-block mesh comprise
which levels in the AMR hierarchy as well as which blocks are refinements of other blocks.
First, the grouping of blocks into levels is identical to multi-block grouping, described previously.
For the specific purpose of grouping blocks into levels, a different top-level node in the MRG
needs to be defined named “amr-levels.” Below this node in the MRG tree, there should be a set of
regions, each region representing a separate refinement level. A groupel map of type
DB_BLOCKCENT with number of segments equal to number of levels needs to be defined and
associated with each of the regions defined under the “amr-levels’ region. The ith segment of the
map will enumerate those blocks that belong to the region representing level i. In addition, an
MRG variable defining the refinement ratios for each level named “amr-ratios” must be defined on
the regions defining the levels of the AMR mesh.
For the specific purpose of identifying which blocks of the multi-block mesh are refinements of a
given block, another top-level region node is added to the MRG tree called “amr-refinements”.
Below the “amr-refinements” region node, an array of regions representing each block in the
multi-block mesh should be defined. In addition, define a groupel map with a number of segments
equal to the number of blocks. Map segment i will be of groupel type DB_BLOCKCENT and will
define all those blocks which are immediate refinements of block i. Since some blocks, with finest
resolution do not have any refinements, the map segments defining the refinements for these
blocks will be of zero length.

DBAddRegion
Silo User’s Guide 2-197
DBAddRegion—Add a region to an MRG tree
Synopsis:
int DBAddRegion(DBmrgtree *tree, char const *reg_name,
int info_bits, int max_children, char const *maps_name,
int nsegs, int const *seg_ids, int const *seg_lens,
int const *seg_types, DBoptlist const *opts)
Fortran Equivalent:
integer function dbaddregion(tree_id, reg_name, lregname,
info_bits, max_children, maps_name,
lmaps_name, nsegs, seg_ids, seg_lens,
seg_types, optlist_id, status)
Arguments:
tree The MRG tree object to add a region to.
reg_name The name of the new region.
info_bits UNUSED
max_children Maximum number of immediate children this region will have.
maps_name [OPT] Name of the groupel map object to associate with this region. Pass
NULL if none.
nsegs [OPT] Number of segments in the groupel map object specified by the
maps_name argument that are to be associated with this region. Pass zero if
none.
seg_ids [OPT] Integer array of length nsegs of groupel map segment ids. Pass NULL (0)
if none.
seg_lens [OPT] Integer array of length nsegs of groupel map segment lengths. Pass
NULL (0) if none.
seg_types [OPT] Integer array of length nsegs of groupel map segment element types. Pass
NULL (0) if none. These types are the same as the centering options for
variables; DB_ZONECENT, DB_NODECENT, DB_EDGECENT,
DB_FACECENT and DB_BLOCKCENT (for the blocks of a multimesh)
opts [OPT] Additional options. Pass NULL (0) if none.
Returns:
A positive number on success; -1 on failure
Description:
Adds a single region node to an MRG tree below the current working region (See “DBSetCwr” on
page 201.).
DBAddRegion
2-198 Silo User’s Guide
If you need to add a large number of similarly purposed region nodes to an MRG tree, consider
using the more efficient DBAddRegionArray() function although it does have some limita-
tions with respect to the kinds of groupel maps it can reference.
A region node in an MRG tree can represent either a specific region, a group of regions or both all
of which are determined by actual use by the application.
Often, a region node is introduced to an MRG tree to provide a separate namespace for regions to
be defined below it. For example, to define material decompositions of a mesh, a region named
“materials” is introduced as a top-level region node in the MRG tree. Note that in so doing, the
region node named “materials” does NOT really represent a distinct region of the mesh. In fact, it
represents the union of all material regions of the mesh and serves as a place to define one, or
more, material decompositions.
Because MRG trees are a new feature in Silo, their use in applications is not fully defined and the
implementation here is designed to be as free-form as possible, to permit the widest flexibility in
representing regions of a mesh. At the same time, in order to convey the semantic meaning of cer-
tain kinds of information in an MRG tree, a set of pre-defined region names is described below.
Region Naming Convention Meaning
“materials” Top-level region below which material decomposition information is
defined. There can be multiple material decompositions, if so desired.
Each such decomposition would be rooted at a region named
“material_<name>” underneath the “materials” region node.
“groupings” Top-level region below which multi-block grouping information is
defined. There can be multiple groupings, if so desired. Each such
grouping would be rooted at a region named “grouping_<name>”
underneath the “groupings” region node.
“amr-levels” Top-level region below which Adaptive Mesh Refinement level group-
ings are defined.
“amr-refinements” Top-level region below which Adaptive Mesh Refinment refinement
information is defined. This where the information indicating which
blocks are refinements of other blocks is defined.
“neighbors” Top-level region below which multi-block adjacency information is
defined.
When a region is being defined in an MRG tree to be associated with a multi-block mesh, often the
groupel type of the maps associated with the region are of type DB_BLOCKCENT.

DBAddRegionArray
Silo User’s Guide 2-199
DBAddRegionArray—Efficiently add multiple, like-kind regions to an MRG tree
Synopsis:
int DBAddRegionArray(DBmrgtree *tree, int nregn,
char const * const *regn_names, int info_bits,
char const *maps_name, int nsegs, int const *seg_ids,
int const *seg_lens, int const *seg_types,
DBoptlist const *opts)
Fortran Equivalent:
integer function dbaddregiona(tree_id, nregn, regn_names,
lregn_names, info_bits, maps_name, lmaps_name, nsegs
seg_ids, seg_lens, seg_types, optlist_id, status)
Arguments:
tree The MRG tree object to add the regions to.
nregn The number of regions to add.
regn_names This is either an array of nregn pointers to character string names for each
region or it is an array of 1 pointer to a character string specifying a printf-style
naming scheme for the regions. The existence of a percent character (‘%’) (used
to introduce conversion specifications) anywhere in regn_names[0] will
indicate the latter mode.The latter mode is almost always preferable, especially
if nergn is large (say more than 100). See below for the format of the printf-
style naming string.
info_bits UNUSED
maps_name [OPT] Name of the groupel maps object to be associated with these regions.
Pass NULL (0) if none.
nsegs [OPT] The number of groupel map segments to be associated with each region.
Note, this is a per-region value. Pass 0 if none.
seg_ids [OPT] Integer array of length nsegs*nregn groupel map segment ids. The
first nsegs ids are associated with the first region. The second nsegs ids are
associated with the second region and so fourth. In cases where some regions
will have fewer than nsegs groupel map segments associated with them, pass -
1 for the corresponding segment ids. Pass NULL (0) if none.
seg_lens [OPT] Integer array of length nsegs*nregn indicating the lengths of each of
the groupel maps. In cases where some regions will have fewer than nsegs
groupel map segments associated with them, pass 0 for the corresponding
segment lengths. Pass NULL (0) if none.
seg_types [OPT] Integer array of length nsegs*nregn specifying the groupel types of
each segment. In cases where some regions will have fewer than nsegs
groupel map segments associated with them, pass 0 for the corresponding
segment lengths. Pass NULL (0) if none.
opts [OPT] Additional options. Pass NULL (0) if none.

DBAddRegionArray
2-200 Silo User’s Guide
Returns:
A positive number on success; -1 on failure
Description:
Use this function instead of DBAddRegion() when you have a large number of similarly purposed
regions to add to an MRG tree AND you can deal with the limitations of the groupel maps associ-
ated with these regions.
The key limitation of the groupel map associated with a region created with DBAddRegionArray()
array and a groupel map associated with a region created with DBAddRegion() is that every region
in the region array must reference nseg map segments (some of which can of course be of zero
length).
Adding a region array is a substantially more efficient way to add regions to an MRG tree than
adding them one at a time especially when a printf-style naming convention is used to specify the
region names.
The existence of a percent character (‘%’) anywhere in regn_names[0] indicates that a printf-style
namescheme is to be used. The format of a printf-style namescheme to specify region names is
described in the documentation of DBMakeNamescheme() (See “DBMakeNamescheme” on
page 206.)
Note that the names of regions within an MRG tree are not required to obey the same variable
naming conventions as ordinary Silo objects (See “DBVariableNameValid” on page 14.) except
that MRG region names can in no circumstance contain either a semi-colon character (‘;’) or a
new-line character (‘\n’).

DBSetCwr
Silo User’s Guide 2-201
DBSetCwr—Set the current working region for an MRG tree
Synopsis:
int DBSetCwr(DBmrgtree *tree, char const *path)
Fortran Equivalent:
integer function dbsetcwr(tree, path, lpath)
Arguments:
tree The MRG tree object.
path The path to set.
Returns:
Positive, depth in tree, on success, -1 on failure.
Description:
Sets the current working region of the MRG tree. The concept of the current working region is
completely analogous to the current working directory of a filesystem.
Notes:
Currently, this method is limited to settings up or down the MRG tree just one level. That is, it will
work only when the path is the name of a child of the current working region or is “..”. This limita-
tion will be relaxed in the next release.

DBGetCwr
2-202 Silo User’s Guide
DBGetCwr—Get the current working region of an MRG tree
Synopsis:
char const *GetCwr(DBmrgtree *tree)
Arguments:
tree The MRG tree.
Returns:
A pointer to a string representing the name of the current working region (not the full path name,
just current region name) on success; NULL (0) on failure.
Description:

DBPutMrgtree
Silo User’s Guide 2-203
DBPutMrgtree—Write a completed MRG tree object to a Silo file
Synopsis:
int DBPutMrgtree(DBfile *file, const char const *name,
char const *mesh_name, DBmrgtree const *tree,
DBoptlist const *opts)
Fortran Equivalent:
int dbputmrgtree(dbid, name, lname, mesh_name, lmesh_name,
tree_id, optlist_id, status)
Arguments:
file The Silo file handle
name The name of the MRG tree object in the file.
mesh_name The name of the mesh the MRG tree object is associated with.
tree The MRG tree object to write.
opts [OPT] Additional options. Pass NULL (0) if none.
Returns:
Positive or zero on success, -1 on failure.
Description:
After using DBPutMrgtree to write the MRG tree to a Silo file, the MRG tree object itself must be
freed using DBFreeMrgtree().

DBGetMrgtree
2-204 Silo User’s Guide
DBGetMrgtree—Read an MRG tree object from a Silo file
Synopsis:
DBmrgtree *DBGetMrgtree(DBfile *file, const char *name)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle
name The name of the MRG tree object in the file.
Returns:
A pointer to a DBmrgtree object on success; NULL (0) on failure.
Description:
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBFreeMrgtree
Silo User’s Guide 2-205
DBFreeMrgtree—Free the memory associated by an MRG tree object
Synopsis:
void DBFreeMrgtree(DBmrgtree *tree)
Fortran Equivalent:
integer function dbfreemrgtree(tree_id)
Arguments:
tree The MRG tree object to free.
Returns:
None.
Description:
Frees all the memory associated with an MRG tree.

DBMakeNamescheme
2-206 Silo User’s Guide
DBMakeNamescheme—Create a DBnamescheme object for on-demand name generation
Synopsis:
DBnamescheme *DBMakeNamescheme(const char *ns_str, ...)
Fortran Equivalent:
None
Arguments:
ns_str The namescheme string as described below.
... The remaining arguments take one of three forms depending on how the caller
wants external array references, if any are present in the format substring of
ns_str to be handled.
In the first form, the format substring of ns_str involves no externally
referenced arrays and so there are no additional arguments other than the
ns_str string itself.
In the second form, the caller has all externally referenced arrays needed in the
format substring of ns_str already in memory and simply passes their
pointers here as the remaining arguments in the same order in which they appear
in the format substring of ns_str. The arrays are bound to the returned
namescheme object and should not be freed until after the caller is done using
the returned namescheme object. In this case, DBFreeNamescheme() does
not free these arrays and the caller is required to explicitly free them.
In the third form, the caller makes a request for the Silo library to find in a given
file, read and bind to the returned namescheme object all externally referenced
arrays in the format substring of ns_str. To achieve this, the caller passes a 3-
tuple of the form...
“(void*) 0, (DBfile*) file, (char*) mbobjpath”
as the remaining arguments. The initial (void*)0 is required. The
(DBfile*)file is the database handle of the Silo file in which all externally
referenced arrays exist. The third (char*)mbobjpath, which may be 0/
NULL, is the path within the file, either relative to the file’s current working
directory, or absolute, at which the multi-block object holding the ns_str was
found in the file. All necessary externally referenced arrays must exist within
the specified file using either relative paths from multi-block object’s home
directory or the file’s current working directory or absolute paths. In this case
DBFreeNamescheme() also frees memory associated with these arrays.
Description:
A namescheme defines a mapping between the non-negative integers (e.g. the natural numbers)
and a sequence of strings such that each string to be associated with a given integer (n) can be gen-
erated from printf-style formatting involving simple expressions. Nameschemes are most often
used to define names of regions in region arrays or to define names of multi-block objects.

DBMakeNamescheme
Silo User’s Guide 2-207
The format of a printf-style namescheme is as follows. The first character of fmt is treated as
delimiter character definition. Wherever this delimiter character appears (except as the first char-
acter), this will indicate the end of one substring within fmt and the beginning of a next substring.
The delimiter character cannot be any of the characters used in the expression language (see
below) for defining expressions to generate names of a namescheme.
The first substring of fmt (that is the characters from position 1 to the first delimiter character)
will contain the complete printf-style format string. The remaining substrings will contain simple
expressions, one for each conversion specifier found in the format substring, which when evalu-
ated will be used as the corresponding argument in an sprintf call to generate the actual name,
when and if needed, on demand.
The expression language for building up the arguments to be used along with the printf-style for-
mat string is pretty simple.
It supports the ‘+’, ‘-’, ‘*’, ‘/’, ‘%’ (modulo), ‘|’, ‘&’, ‘^’ integer operators and a variant of the
question-mark-colon operator, ‘? : :’ which requires an extra, terminating colon.
It supports grouping via ‘(‘ and ‘)’ characters.
It supports grouping of characters into arbitrary strings via the string (single quote) characters ‘’’
and ‘’’. Any characters appearing between enclosing single quotes are treated as a literal string
suitable for an argument to be associated with a %s-type conversion specifier in the format string.
It supports references to external, integer valued arrays introduced via a ‘#’ character appearing
before an array’s name and external, string valued arrays introduced via a ‘$’ character appearing
before an array’s name.
Finally, the special operator ‘n’ appearing in an expression represents a natural number within the
sequence of names (zero-origin index). See below for some examples.
Except for singly quoted strings which evaluate to a literal string suitable for output via a %s type
conversion specifier, and $-type external array references which evaluate to an external string, all

DBMakeNamescheme
2-208 Silo User’s Guide
other expressions are treated as evaluating to integer values suitable for any of the integer conver-
sion specifiers (%[ouxXdi]) which may be used in the format substring..
fmt Interpretation
“|slide_%s|(n%2)?’master’:’slave’:” The delimiter character is ‘|’. The format substring is “slide_%s”. The
expression substring for the argument to the first (and only in this
case) conversion specifier (%s) is “(n%2)?’master’:’slave’:” When this
expression is evaluated for a given region, the region’s natural num-
ber will be inserted for ‘n’. The modulo operation with 2 will be applied.
If that result is non-zero, the ?:: expression will evaluate to ‘master’.
Otherwise, it will evaluate to ‘slave’. Note the terminating colon for the
?:: operator. This naming scheme might be useful for an array of
regions representing, alternately, master and slave sides of slide sur-
faces.
Note also for the ?:: operator, the caller can assume that only the sub-
expression corresponding to the whichever half of the operator is sat-
isfied is actually evaluated.
“Hblock_%02dx%02dHn/16Hn%16” The delimiter character is ‘H’. The format substring is
‘block_%02dx%02d”. The expression substring for the argument to
the first conversion specifier (%02d) is “n/256”. The expression sub-
string for the argument to the second conversion specifier (also
%02d) is “n%16”. When this expression is evaluated, the region’s nat-
ural number will be inserted for ‘n’ and the div and mod operators will
be evaluated. This naming scheme might be useful for a region array
of 256 regions to be named as a 2D array of regions with names like
“block_09x11”
“@domain_%03d@n” The delimiter character is ‘@’. The format substring is
“domain_%03d”. The expression substring for the argument to the
one and only conversion specifier is ‘n’. When this expression is eval-
uated, the region’s natural number is inserted for ‘n’. This results in
names like “domain_000”, “domain_001”, etc.

DBMakeNamescheme
Silo User’s Guide 2-209
Use DBFreeNamescheme() to free up the space associated with a namescheme.
Also note that there are numerous examples of nameschemes in “tests/nameschemes.c” in the Silo
source release tarball.
“@domain_%03d@n+1” This is just like the case above except that region names begin with
“domain_001” instead of “domain_000”. This might be useful to deal
with different indexing origins; Fortran vs. C.
“|foo_%03dx%03d|#P[n]|#U[n%4]” The delimiter character is ‘|’. The format substring is
“foo_%03dx%03d”. The expression substring for the first argument is
an external array reference ‘#P[n]’ where the index into the array is
just the natural number, n. The expression substring for the second
argument is another external array reference, ‘#U[n%4]’ where the
index is an expression ‘n%4’ on the natural number n.
If the caller is handling externally referenced arrays explicitly, because
‘P’ is the first externally referenced array in the format string, a pointer
to ‘P’ must be the first to appear in the varargs list of additional args to
DBMakeNamescheme. Similarly, because ‘U’ appears as the second
externally referenced array in the format string, a pointer to ‘U’ must
appear second in the varargs as in
DBMakeNamescheme(‘|foo_%03dx%03d|#P[n]|#U[n%4]’, p, u);
Alternatively, if the caller wants the Silo library to find ‘P’ and ‘U’ in a
Silo file, read the arrays from the file and bind them into the name-
scheme automatically, then ‘P’ and ‘U’ must be Silo arrays in the cur-
rent working directory of the file that is passed in as the 2-tuple “(int)
0, (DBfile *) dbfile’ in the varargs to DBMakeNamescheme as in
DBMakeNamescheme(“|foo_%03dx%03d|#P[n]|#U[n%4]”, 0, dbfile,
0);
fmt Interpretation

DBGetName
2-210 Silo User’s Guide
DBGetName—Generate a name from a DBnamescheme object
Synopsis:
char const *DBGetName(DBnamescheme *ns, int natnum)
Fortran Equivalent:
None
Arguments:
natnum Natural number of the entry in a namescheme to be generated. Must be greater
than or equal to zero.
Returns:
A string representing the generated name. If there are problems with the namescheme, the string
could be of length zero (e.g. the first character is a null terminator).
Description:
Once a namescheme has been created via DBMakeNamescheme, this function can be used to gen-
erate names at will from the namescheme. The caller must NOT free the returned string.
Silo maintains a tiny circular buffer of (32) names constructed and returned by this function so that
multiple evaluations in the same expression do not wind up overwriting each other. A call to
DBGetName(0,0) will free up all memory associated with this tiny circular buffer.

DBPutMrgvar
Silo User’s Guide 2-211
DBPutMrgvar—Write variable data to be associated with (some) regions in an MRG tree
Synopsis:
int DBPutMrgvar(DBfile *file, char const *name,
char const *mrgt_name,
int ncomps, char const * const *compnames,
int nregns, char const * const *reg_pnames,
int datatype, void const * const *data,
DBoptlist const *opts)
Fortran Equivalent:
integer function dbputmrgv(dbid, name, lname, mrgt_name,
lmrgt_name, ncomps, compnames, lcompnames,
nregns, reg_names, lreg_names, datatype,
data_ids, optlist_id, status)
character*N compnames (See “dbset2dstrlen” on page 285.)
character*N reg_names (See “dbset2dstrlen” on page 285.)
int* data_ids (use dbmkptr to get id for each pointer)
Arguments:
file Silo database file handle.
name Name of this mrgvar object.
tname name of the mrg tree this variable is associated with.
ncomps An integer specifying the number of variable components.
compnames [OPT] Array of ncomps pointers to character strings representing the names of
the individual components. Pass NULL(0) if no component names are to be
specified.
nregns The number of regions this variable is being written for.
reg_pnames Array of nregns pointers to strings representing the pathnames of the regions
for which the variable is being written. If nregns>1 and
reg_pnames[1]==NULL, it is assumed that reg_pnames[i]=NULL for
all i>0 and reg_pnames[0] contains either a printf-style naming
convention for all the regions to be named or, if reg_pnames[0] is found to
contain no printf-style conversion specifications, it is treated as the pathname of
a single region in the MRG tree that is the parent of all the regions for which
attributes are being written.
data Array of ncomps pointers to variable data. The pointer, data[i] points to an
array of nregns values of type datatype.
opts Additional options.
Returns:
Zero on success; -1 on failure.
DBPutMrgvar
2-212 Silo User’s Guide
Description:
Sometimes, it is necessary to associate variable data with regions in an MRG tree. This call allows
an application to associate variable data with a bunch of different regions in one of several ways all
of which are determined by the contents of the reg_pnames argument.
Variable data can be associated with all of the immediate children of a given region. This is the
most common case. In this case, reg_pnames[0] is the name of the parent region and
reg_pnames[i] is set to NULL for all i>0.
Alternatively, variable data can be associated with a bunch of regions whose names conform to a
common, printf-style naming scheme. This is typical of regions created with the DBPutRegion-
Array() call. In this case, reg_pnames[0] is the name of the parent region and
reg_pnames[i] is set to NULL for all i>0 and, in addition, reg_pnames[0] is a specially
formatted, printf-style string, for naming the regions. See “DBAddRegionArray” on page 199. for
a discussion of the regn_names argument format.
Finally, variable data can be associated with a bunch of arbitrarily named regions. In this case,
each region’s name must be explicitly specified in the reg_pnames array.
Because MRG trees are a new feature in Silo, their use in applications is not fully defined and the
implementation here is designed to be as free-form as possible, to permit the widest flexibility in
representing regions of a mesh. At the same time, in order to convey the semantic meaning of cer-
tain kinds of information in an MRG tree, a set of pre-defined MRG variables is descirbed below.
Variable Naming Convention Meaning
“amr-ratios” An integer variable of 3 components defining the refinement ratios (rx,
ry, rz) for an AMR mesh. Typically, the refinement ratios can be speci-
fied on a level-by-level basis. In this case, this variable should be
defined for nregns=<# of levels> on the level regions underneath the
“amr-levels” grouping. However, if refinment ratios need to be defined
on an individual patch basis instead, this variable should be defined
on the individual patch regions under the “amr-refinements” group-
ings.
“ijk-orientations” An integer variable of 3 components defined on the individual blocks
of a multi-block mesh defining the orientations of the individual blocks
in a large, ijk indexing space (Ares convention)
“<var>-extents” A double precision variable defining the block-by-block extents of a
multi-block variable. If <var>==”coords”, then it defines the spatial
extents of the mesh itself. Note, this convention obsoletes the
DBOPT_XXX_EXTENTS options on DBPutMultivar/DBPutMultimesh
calls.
Don’t forget to associate the resulting region variable object(s) with the MRG tree by using the
DBOPT_MRGV_ONAMES and DBOPT_MRGV_RNAMES options in the DBPutMrgtree() call.

DBGetMrgvar
Silo User’s Guide 2-213
DBGetMrgvar—Retrieve an MRG variable object from a silo file
Synopsis:
DBmrgvar *DBGetMrgvar(DBfile *file, char const *name)
Fortran Equivalent:
None
Arguments:
file Silo database file handle.
name The name of the region variable object to retrieve.
Returns:
A pointer to a DBmrgvar object on success; NULL (0) on failure.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBPutGroupelmap
2-214 Silo User’s Guide
DBPutGroupelmap—Write a groupel map object to a Silo file
Synopsis:
int DBPutGroupelmap(DBfile *file, char const *name,
int num_segs, int const *seg_types, int const *seg_lens,
int const *seg_ids, int const * const *seg_data,
void const * const *seg_fracs, int fracs_type,
DBoptlist const *opts)
Fortran Equivalent:
integer function dbputgrplmap(dbid, name, lname, num_segs,
seg_types, seg_lens, seg_ids, seg_data_ids,
seg_fracs_ids, fracs_type, optlist_id, status)
integer* seg_data_ids (use dbmkptr to get id for each pointer)
integer* seg_fracs_ids (use dbmkptr to get id for each pointer)
Arguments:
file The Silo database file handle.
name The name of the groupel map object in the file.
nsegs The number of segments in the map.
seg_types Integer array of length nsegs indicating the groupel type associated with each
segment of the map; one of DB_BLOCKCENT, DB_NODECENT,
DB_ZONECENT, DB_EDGECENT, DB_FACECENT.
seg_lens Integer array of length nsegs indicating the length of each segment
seg_ids [OPT] Integer array of length nsegs indicating the identifier to associate with
each segment. By default, segment identifiers are 0...negs-1. If default
identifiers are sufficient, pass NULL (0) here. Otherwise, pass an explicit list of
integer identifiers.
seg_data The groupel map data, itself. An array of nsegs pointers to arrays of integers
where array seg_data[i] is of length seg_lens[i].
seg_fracs [OPT] Array of nsegs pointers to floating point values indicating fractional
inclusion for the associated groupels. Pass NULL (0) if fractional inclusions are
not required. If, however, fractional inclusions are required but on only some of
the segments, pass an array of pointers such that if segment i has no fractional
inclusions, seg_fracs[i]=NULL(0). Fractional inclusions are useful for,
among other things, defining groupel maps involving mixing materials.
fracs_type [OPT] data type of the fractional parts of the segments. Ignored if seg_fracs
is NULL (0).
opts Additional options
Returns:
Zero on success; -1 on failure.
DBPutGroupelmap
Silo User’s Guide 2-215
Description:
By itself, an MRG tree is not sufficient to fully characterize the decomposition of a mesh into var-
ious regions. The MRG tree serves only to identify the regions and their relationships in gross
terms. This frees MRG trees from growing linearly (or worse) with problem size.
All regions in an MRG tree are ultimately defined, in detail, by enumerating a primitive set of
Grouping Elements (groupels) that comprise the regions. A groupel map is the object used for this
purpose. The problem-sized information needed to fully characterize the regions of a mesh is
stored in groupel maps.
The grouping elements or groupels are the individual pieces of mesh which, when enumerated,
define specific regions.
For a multi-mesh object, the groupels are whole blocks of the mesh. For Silo’s other mesh types
such as ucd or quad mesh objects, the groupels can be nodes (0d), zones (2d or 3d depending on
the mesh dimension), edges (1d) and faces (2d).
The groupel map concept is best illustrated by example. Here, we will define a groupel map for the
material case illustrated in Figure 0-6 on page 143.
1
11
1 2
2
2
2
Mesh ‘plot’
with material
numbers and
interface (zone #’s
012
345
in lower left)
03
14
25
14
.7 .4
.3 .6
(NULL)
(NULL)
seg_data[0]
seg_data[1]
seg_data[2]
seg_data[3]
seg_fracs[0]
seg_fracs[1]
seg_fracs[2]
seg_fracs[3]
num_segs = 4;
seg_types[] = {DB_ZONECENT, DB_ZONECENT, DB_ZONECENT, DB_ZONECENT};
seg_lens[] = {2,2,2,2};
material “1”
material “2”
seg_ids[] ={1,1,2,2}; /* material numbers used as ids */
Figure 0-10: Example of using groupel map for (mixing) materials.
In the example in the above figure, the groupel map has the behavior of representing the clean and
mixed parts of the material decomposition by enumerating in alternating segments of the map, the
clean and mixed parts for each successive material.

DBGetGroupelmap
2-216 Silo User’s Guide
DBGetGroupelmap—Read a groupel map object from a Silo file
Synopsis:
DBgroupelmap *DBGetGroupelmap(DBfile *file, char const *name)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
name The name of the groupel map object to read.
Returns:
A pointer to a DBgroupelmap object on success. NULL (0) on failure.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBFreeGroupelmap
Silo User’s Guide 2-217
DBFreeGroupelmap—Free memory associated with a groupel map object
Synopsis:
void DBFreeGroupelmap(DBgroupelmap *map)
Fortran Equivalent:
None
Arguments:
map Pointer to a DBgroupel map object.
Returns:
None
Description:

DBFreeGroupelmap
2-218 Silo User’s Guide
DBOPT_REGION_PNAMES—option for defining variables on specific regions of a mesh
Synopsis:
DBOPT_REGION_PNAMES char** A null-pointer terminated array of pointers
to strings specifying the pathnames of
regions in the mrg tree for the associated
mesh where the variable is defined. If
there is no mrg tree associated with the
mesh, the names specified here will be
assumed to be material names of the
material object associated with the mesh.
The last pointer in the array must be null
and is used to indicate the end of the list of
names.
NULL
All of Silo’s DBPutXxxvar() calls support the DBOPT_REGION_PNAMES option to specify
the variable on only some region(s) of the associated mesh. However, the use of the option has
implications regarding the ordering of the values in the vars[] arrays passed into the
DBPutXxxvar() functions. This section explains the ordering requirements.
Ordinarily, when the DBOPT_REGION_PNAMES option is not being used, the order of the values
in the vars arrays passed here is considered to be one-to-one with the order of the nodes (for
DB_NODECENT centering) or zones (for DB_ZONECENT centering) of the associated mesh. How-
ever, when the DBOPT_REGION_PNAMES option is being used, the order of values in the
vars[] is determined by other conventions described below.
If the DBOPT_REGION_PNAMES option references regions in an MRG tree, the ordering is one-
to-one with the groupel’s identified in the groupel map segment(s) (of the same groupel type as the
variable’s centering) associated with the region(s); all of the segment(s), in order, of the groupel
map of the first region, then all of the segment(s) of the groupel map of the second region, and so
on. If the set of groupel map segments for the regions specified include the same groupel multiple
times, then the vars[] arrays will wind up needing to include the same value, multiple times.
The preceding ordering convention works because the ordering is explicitly represented by the
order in which groupels are identified in the groupel maps. However, if the
DBOPT_REGION_PNAMES option references material name(s) in a material object created by a
DBPutMaterial() call, then the ordering is not explicitly represented. Instead, it is based on a
traversal of the mesh zones restricted to the named material(s). In this case, the ordering conven-
tion requires further explanation and is described below.
For DB_ZONECENT variables, as one traverses the zones of a mesh from the first zone to the last,
if a zone contains a material listed in DBOPT_REGION_PNAMES (wholly or partially), that zone
is considered in the traversal and placed conceptually in an ordered list of traversed zones. In addi-
tion, if the zone contains the material only partially, that zone is also placed conceptually in an
ordered list of traversed mixed zones. In this case, the values in the vars[] array must be one-to-
one with this traversed zones list. Likewise, the values of the mixvars[] array must be one-to-
one with the traversed mixed zones list. However, in the special case that the list of materials spec-
ified in DBOPT_REGION_PNAMES is of size one (1), an additional optimization is supported.
For the special case that the list of materials defined in DBOPT_REGION_PNAMES is of size one
(1), the requirement to specify separate values for zones containing the material only partially in

DBFreeGroupelmap
Silo User’s Guide 2-219
the mixvars[] array is removed. In this case, if the mixlen arg is zero (0) in the cooresponding
DBPutXXXvar() call, only the vars[] array, which is one-to-one with (all) traversed zones
containing the material either cleanly or partially, will be used. The reason this works is that in the
single material case, there is only ever one zonal variable value per zone regardless of whether the
zone contains the material cleanly or partially.
For DB_NODECENT variables, the situation is complicated by the fact that materials are zone-cen-
tric but the variable being defined is node-centered. So, an additional level of local traversal over a
zone’s nodes is required. In this case, as one traverses the zones of a mesh from the first zone to the
last, if a zone contains a material listed in DBOPT_REGION_PNAMES (wholly or partially), then
that zone’s nodes are traversed according to the ordering specified in “Node, edge and face order-
ing for zoo-type UCD zone shapes.” on page 2-102. On the first encounter of a node, that node is
considered in the traversal and placed conceptually in an ordered list of traversed nodes. The val-
ues in the vars[] array must be one-to-one with this traversed nodes list. Because we are not
aware of any cases of node-centered variables that have mixed material components, there is no
analogous traversed mixed nodes list.
For DBOPT_EDGECENT and DBOPT_FACECENT variables, the traversal is handled similarly.
That is, the list of zones for the mesh is traversed and for each zone found to contain one of the
materials listed in DBOPT_REGION_PNAMES, the zone’s edge’s (or face’s) are traversed in local
order specified in “Node, edge and face ordering for zoo-type UCD zone shapes.” on page 2-102.
For Quad meshes, there is no explicit list of zones (or nodes) comprising the mesh. So, the notion
of traversing the zones (or nodes) of a Quad mesh requires further explanation. If the mesh’s nodes
(or zones) were to be traversed, which would be the first? Which would be the second?
Unless the DBOPT_MAJORORDER option was used, the answer is that the traversal is identical to
the standard C programming language storage convention for multi-dimensional arrays often
called row-major storage order. That is, was we traverse through the list of nodes (or zones) of a
Quad mesh, we encounter first node with logical index [0,0,0], then [0,0,1], then
[0,0,2]...[0,1,0]...etc. A traversal of zones would behave similarly. Traversal of edges or faces of a
quad mesh would follow the description with “DBPutQuadvar” on page 2-92.

DBFreeGroupelmap
2-220 Silo User’s Guide
6 API Section Object Allocation, Free and IsEmpty
This section describes methods to allocate and initialize many of Silo’s objects. The functions
described here are...
DBAlloc… . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 221
DBFree… . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 222
DBIsEmpty . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 223

DBAlloc…
Silo User’s Guide 2-221
DBAlloc…—Allocate and initialize a Silo structure.
Synopsis:
DBcompoundarray *DBAllocCompoundarray (void)
DBcsgmesh *DBAllocCsgmesh (void)
DBcsgvar *DBAllocCsgvar (void)
DBcurve *DBAllocCurve (void)
DBcsgzonelist *DBAllocCSGZonelist (void)
DBdefvars *DBAllocDefvars (void)
DBedgelist *DBAllocEdgelist (void)
DBfacelist *DBAllocFacelist (void)
DBmaterial *DBAllocMaterial (void)
DBmatspecies *DBAllocMatspecies (void)
DBmeshvar *DBAllocMeshvar (void)
DBmultimat *DBAllocMultimat (void)
DBmultimatspecies *DBAllocMultimatspecies (void)
DBmultimesh *DBAllocMultimesh (void)
DBmultimeshadj *DBAllocMultimeshadj (void)
DBmultivar *DBAllocMultivar (void)
DBpointmesh *DBAllocPointmesh (void)
DBquadmesh *DBAllocQuadmesh (void)
DBquadvar *DBAllocQuadvar (void)
DBucdmesh *DBAllocUcdmesh (void)
DBucdvar *DBAllocUcdvar (void)
DBzonelist *DBAllocZonelist (void)
DBphzonelist *DBAllocPHZonelist (void)
DBnamescheme *DBAllocNamescheme(void);
DBgroupelmap *DBAllocGroupelmap(int, DBdatatype);
Fortran Equivalent:
None
Returns:
These allocation functions return a pointer to a newly allocated and initialized structure on success
and NULL on failure.
Description:
The allocation functions allocate a new structure of the requested type, and initialize all values to
NULL or zero. There are counterpart functions for freeing structures of a given type (see
DBFree….

DBFree…
2-222 Silo User’s Guide
DBFree…—Release memory associated with a Silo structure.
Synopsis:
void DBFreeCompoundarray (DBcompoundarray *x)
void DBFreeCsgmesh (DBcsgmesh *x)
void DBFreeCsgvar (DBcsgvar *x)
void DBFreeCSGZonelist (DBcsgzonelist *x)
void DBFreeCurve(DBcurve *);
void DBFreeDefvars (DBdefvars *x)
void DBFreeEdgelist (DBedgelist *x)
void DBFreeFacelist (DBfacelist *x)
void DBFreeMaterial (DBmaterial *x)
void DBFreeMatspecies (DBmatspecies *x)
void DBFreeMeshvar (DBmeshvar *x)
void DBFreeMultimesh (DBmultimesh *x)
void DBFreeMultimeshadj (DBmultimeshadj *x)
void DBFreeMultivar (DBmultivar *x)
void DBFreeMultimat(DBmultimat *);
void DBFreeMultimatspecies(DBmultimatspecies *);
void DBFreePointmesh (DBpointmesh *x)
void DBFreeQuadmesh (DBquadmesh *x)
void DBFreeQuadvar (DBquadvar *x)
void DBFreeUcdmesh (DBucdmesh *x)
void DBFreeUcdvar (DBucdvar *x)
void DBFreeZonelist (DBzonelist *x)
void DBFreePHZonelist (DBphzonelist *x)
void DBFreeNamescheme(DBnamescheme *);
void DBFreeMrgvar(DBmrgvar *mrgv);
void DBFreeMrgtree(DBmrgtree *tree);
void DBFreeGroupelmap(DBgroupelmap *map);
Arguments:
xA pointer to a structure which is to be freed. Its type must correspond to the type
in the function name.
Fortran Equivalent:
None
Returns:
These free functions return zero on success and -1 on failure.
Description:
The free functions release the given structure as well as all memory pointed to by these structures.
This is the preferred method for releasing these structures. There are counterpart functions for allo-
cating structures of a given type (see DBAlloc…). The functions will not fail if a NULL pointer is
passed to them.

DBIsEmpty
Silo User’s Guide 2-223
DBIsEmpty—Query a object returned from Silo for “emptiness”
Synopsis:
int DBIsEmptyCurve(DBcurve const *curve);
int DBIsEmptyPointmesh(DBpointmesh const *msh);
int DBIsEmptyPointvar(DBpointvar const *var);
int DBIsEmptyMeshvar(DBmeshvar const *var);
int DBIsEmptyQuadmesh(DBquadmesh const *msh);
int DBIsEmptyQuadvar(DBquadvar const *var);
int DBIsEmptyUcdmesh(DBucdmesh const *msh);
int DBIsEmptyFacelist(DBfacelist const *fl);
int DBIsEmptyZonelist(DBzonelist const *zl);
int DBIsEmptyPHZonelist(DBphzonelist const *zl);
int DBIsEmptyUcdvar(DBucdvar const *var);
int DBIsEmptyCsgmesh(DBcsgmesh const *msh);
int DBIsEmptyCSGZonelist(DBcsgzonelist const *zl);
int DBIsEmptyCsgvar(DBcsgvar const *var);
int DBIsEmptyMaterial(DBmaterial const *mat);
int DBIsEmptyMatspecies(DBmatspecies const *spec);
Arguments:
xPointer to a silo object structure to be queried
Description:
These functions return non-zero if the object is indeed empty and zero otherwise. When DBSetAl-
lowEmptyObjects() is enabled by a writer, it can produce objects in the file which contain useful
metadata but no “problems-sized” data. These methods can be used by a reader to determine if an
object read from a file is empty.

DBIsEmpty
2-224 Silo User’s Guide
7 API Section Calculational and Utility
This section of the API manual describes functions that can be used to compute things such as
Facelists as well as utility functions to, for example, catentate an array of strings into a single
string for simple output with DBWrite().
DBCalcExternalFacelist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 225
DBCalcExternalFacelist2 . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 227
DBStringArrayToStringList . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 229
DBStringListToStringArray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 230

DBCalcExternalFacelist
Silo User’s Guide 2-225
DBCalcExternalFacelist—Calculate an external facelist for a UCD mesh.
Synopsis:
DBfacelist *DBCalcExternalFacelist (int nodelist[], int nnodes,
int origin, int shapesize[],
int shapecnt[], int nshapes, int matlist[],
int bnd_method)
Fortran Equivalent:
integer function dbcalcfl(nodelist, nnodes, origin, shapesize,
shapecnt, nshapes, matlist, bnd_method,
flid)
returns the pointer-id of the created object in flid.
Arguments:
nodelist Array of node indices describing mesh zones.
nnodes Number of nodes in associated mesh.
origin Origin for indices in the nodelist array. Should be zero or one.
shapesize Array of length nshapes containing the number of nodes used by each zone
shape.
shapecnt Array of length nshapes containing the number of zones having each shape.
nshapes Number of zone shapes.
matlist Array containing material numbers for each zone (else NULL).
bnd_method Method to use for calculating external faces. See description below.
Returns:
DBCalcExternalFacelist returns a DBfacelist pointer on success and NULL on failure.
Description:
The DBCalcExternalFacelist function calculates an external facelist from the zonelist and zone
information describing a UCD mesh. The calculation of the external facelist is controlled by the
bnd_method parameter as defined in the table below:
bnd_method Meaning
0Do not use material boundaries when computing external faces. The
matlist parameter can be replaced with NULL.
1In addition to true external faces, include faces on material boundaries
between zones. Faces get generated for both zones sharing a common
face. This setting should not be used with meshes that contain mixed
material zones. If this setting is used with meshes with mixed material
zones, all faces which border a mixed material zone will be include. The
matlist parameter must be provided.

DBCalcExternalFacelist2
Silo User’s Guide 2-227
DBCalcExternalFacelist2—Calculate an external facelist for a UCD mesh
containing ghost zones.
Synopsis:
DBfacelist *DBCalcExternalFacelist2 (int nodelist[], int nnodes,
int low_offset, int hi_offset, int origin,
int shapetype[], int shapesize[],
int shapecnt[], int nshapes, int matlist[],
int bnd_method)
Fortran Equivalent:
None
Arguments:
nodelist Array of node indices describing mesh zones.
nnodes Number of nodes in associated mesh.
lo_offset The number of ghost zones at the beginning of the nodelist.
hi_offset The number of ghost zones at the end of the nodelist.
origin Origin for indices in the nodelist array. Should be zero or one.
shapetype Array of length nshapes containing the type of each zone shape. See
description below.
shapesize Array of length nshapes containing the number of noes used by each zone
shape.
shapecnt Array of length nshapes containing the number of zones having each shape.
nshapes Number of zone shapes.
matlist Array containing material numbers for each zone (else NULL).
bnd_method Method to use for calculating external faces. See description below.
Returns:
DBCalcExternalFacelist2 returns a DBfacelist pointer on success and NULL on failure.
DBCalcExternalFacelist2
2-228 Silo User’s Guide
Description:
The DBCalcExternalFacelist2 function calculates an external facelist from the zonelist and zone
information describing a UCD mesh. The calculation of the external facelist is controlled by the
bnd_method parameter as defined in the table below:
bnd_method Meaning
0Do not use material boundaries when computing external faces. The
matlist parameter can be replaced with NULL.
1In addition to true external faces, include faces on material boundaries
between zones. Faces get generated for both zones sharing a com-
mon face. This setting should not be used with meshes that contain
mixed material zones. If this setting is used with meshes with mixed
material zones, all faces which border a mixed material zone will be
included. The matlist parameter must be provided.
The allowed shape types are described in the following table:
Type Description
DB_ZONETYPE_BEAM A line segment
DB_ZONETYPE_POLYGON A polygon where nodes are enumerated to form a polygon
DB_ZONETYPE_TRIANGLE A triangle
DB_ZONETYPE_QUAD A quadrilateral
DB_ZONETYPE_POLYHED
RON
A polyhedron with nodes enumerated to form faces and
faces are enumerated to form a polyhedron
DB_ZONETYPE_TET A tetrahedron
DB_ZONETYPE_PYRAMID A pyramid
DB_ZONETYPE_PRISM A prism
DB_ZONETYPE_HEX A hexahedron
For a description of how the nodes for the allowed shapes are enumerated, see “DBPutUcdmesh”
on page 2-99.

DBCalcExternalFacelist2
Silo User’s Guide 2-229
DBStringArrayToStringList—Utility to catentate a group of strings into a single,
semi-colon delimited string.
Synopsis:
void DBStringArrayToStringList(char const * const *strArray,
int n, char **strList, int *m)
Fortran Equivalent:
None
Arguments:
strArray Array of strings to catenate together. Note that it can be ok if some entries in
strArray are the empty string, “” or NULL (0).
nThe number of entries in strArray. Passing -1 here indicates that the function
should count entries in strArray until reaching the first NULL entry. In this
case, embedded NULLs (0s) in strArray are, of course, not allowed.
strList The returned catenated, semi-colon separated, single, string.
mThe returned length of strList.
Description:
This is a utility function to facilitate writing of an array of strings to a file. This function will take
an array of strings and catenate them together into a single, semi-colon delimited list of strings.
Some characters are NOT permitted in the input strings. These are ‘\n’, ‘\0’ and ‘;’ characters.
This function can be used together with DBWrite() to easily write a list of strings to the a Silo data-
base.

DBCalcExternalFacelist2
2-230 Silo User’s Guide
DBStringListToStringArray—Given a single, semi-colon delimited string, de-
catenate it into an array of strings.
Synopsis:
char **DBStringListToStringArray(char *strList, int n,
int handleSlashSwap, int skipFirstSemicolon)
Fortran Equivalent:
None
Arguments:
strList A semi-colon separated, single string. Note that this string is modified by the
call. If the caller doesn’t want this, it will have to make a copy before calling.
nThe expected number of individual strings in strList. Pass -1 here if you
have no aprior knowledge of this number. Knowing the number saves an
additional pass over strList.
handleSlashSwap a boolean to indicate if slash characters should be swapped as per
differences in windows/linux filesystems.
This is specific to Silo’s internal handling of strings used in multi-block objects.
So, you should pass zero (0) here.
skipFirstSemicolon a boolean to indicate if the first semicolon in the string should be
skipped.
This is specific to Silo’s internal usage for legacy compatibility. You should
pass zero (0) here.
Description:
This function performs the reverse of DBStringArrayToStringList.

DBCalcExternalFacelist2
Silo User’s Guide 2-231
8 API Section Optlists
Many Silo functions take as a last argument a pointer to an Options List or optlist. This is intended
to permit the Silo API to grow and evolve as necessary without requiring substantial changes to the
API itself.
In the documentation associated with each function, the list of available options and their meaning
is described.
This section of the manual describes only the functions to create and manage options lists. These
are...
DBMakeOptlist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 232
DBAddOption. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 233
DBClearOption . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 234
DBGetOption . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 235
DBFreeOptlist. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 236
DBClearOptlist . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 237

DBMakeOptlist
2-232 Silo User’s Guide
DBMakeOptlist—Allocate an option list.
Synopsis:
DBoptlist *DBMakeOptlist (int maxopts)
Fortran Equivalent:
integer function dbmkoptlist(maxopts, optlist_id)
returns created optlist pointer-id in optlist_id
Arguments:
maxopts Initial maximum number of options expected in the optlist. If this maximum is
exceeded, the library will silently re-allocate more space using the golden-rule.
Returns:
DBMakeOptlist returns a pointer to an option list on success and NULL on failure.
Description:
The DBMakeOptlist function allocates memory for an option list and initializes it. Use the func-
tion DBAddOption to populate the option list structure, and DBFreeOptlist to free it.
In releases of Silo prior to 4.10, if the caller accidentally added more options to an optlist than it
was originally created for, an error would be generated. However, in version 4.10, the library will
silently just re-allocate the optlist to accommodate more options.

DBAddOption
Silo User’s Guide 2-233
DBAddOption—Add an option to an option list.
Synopsis:
int DBAddOption (DBoptlist *optlist, int option, void *value)
Fortran Equivalent:
integer function dbaddcopt (optlist_id, option, cvalue, lcvalue)
integer function dbaddcaopt (optlist_id, option, nval, cvalue,
lcvalue)
integer function dbadddopt (optlist_id, option, dvalue)
integer function dbaddiopt (optlist_id, option, ivalue)
integer function dbaddropt (optlist_id, option, rvalue)
integer ivalue, optlist_id, option, lcvalue, nval
double precision dvalue
real rvalue
character*N cvalue (See “dbset2dstrlen” on page 285.)
Arguments:
optlist Pointer to an option list structure containing option/value pairs. This structure is
created with the DBMakeOptlist function.
option Option definition. One of the predefined values described in the table in the
notes section of each command which accepts an option list.
value Pointer to the value associated with the provided option description. The data
type is implied by option.
Returns:
DBAddOption returns a zero on success and -1 on failure.
Description:
The DBAddOption function adds an option/value pair to an option list. Several of the output func-
tions accept option lists to provide information of an optional nature.
In releases of Silo prior to 4.10, if the caller accidentally added more options to an optlist than it
was originally created for, an error would be generated. However, in version 4.10, the library will
silently just re-allocate the optlist to accommodate more options.

DBClearOption
2-234 Silo User’s Guide
DBClearOption—Remove an option from an option list
Synopsis:
int DBClearOption(DBoptlist *optlist, int optid)
Fortran Equivalent:
None
Arguments:
optlist The option list object for which you wish to remove an option
optid The option id of the option you would like to remove
Returns:
DBClearOption returns zero on success and -1 on failure.
Description:
This function can be used to remove options from an option list. If the option specified by optid
exists in the given option list, that option is removed from the list and the total number of options
in the list is reduced by one.
This method can be used together with DBAddOption to modify an existing option in an option
list. To modify an existing option in an option list, first call DBClearOption for the option to be
modified and then call DBAddOption to re-add it with a new definition.
There is also a function to query for the value of an option in an option list, DBGetOption.

DBGetOption
Silo User’s Guide 2-235
DBGetOption—Retrieve the value set for an option in an option list
Synopsis:
void *DBGetOption(DBoptlist *optlist, int optid)
Fortran Equivalent:
None
Arguments:
optlist The optlist to query
optid The option id to query the value for
Returns:
Returns the pointer value set for a given option or NULL if the option is not defined in the given
option list.
Description:
This function can be used to query the contents of an optlist. If the given optlist has an
option of the given optid, then this function will return the pointer associated with the given
optid. Otherwise, it will return NULL indicating the optlist does not contain an option with
the given optid.

DBFreeOptlist
2-236 Silo User’s Guide
DBFreeOptlist—Free memory associated with an option list.
Synopsis:
int DBFreeOptlist (DBoptlist *optlist)
Fortran Equivalent:
integer function dbfreeoptlist(optlist_id)
Arguments:
optlist Pointer to an option list structure containing option/value pairs. This structure is
created with the DBMakeOptlist function.
Returns:
DBFreeOptlist returns a zero on success and -1 on failure.
Description:
The DBFreeOptlist function releases the memory associated with the given option list. The indi-
vidual option values are not freed.
DBFreeOptlist will not fail if a NULL pointer is passed to it.

DBClearOptlist
Silo User’s Guide 2-237
DBClearOptlist—Clear an optlist.
Synopsis:
int DBClearOptlist (DBoptlist *optlist)
Fortran Equivalent:
None
Arguments:
optlist Pointer to an option list structure containing option/value pairs. This structure is
created with the DBMakeOptlist function.
Returns:
DBClearOptlist returns zero on success and -1 on failure.
Description:
The DBClearOptlist function removes all options from the given option list.

DBClearOptlist
2-238 Silo User’s Guide
9 API Section User Defined (Generic) Data and Objects
If you want to create data that other applications (not written by you or someone working closely
with you) can read and understand, these are NOT the right functions to use. That is because the
data that these functions create is not self-describing and inherently non-shareable.
However, if you need to write data that only you (or someone working closely with you) will read
such as for restart purposes, the functions described here may be helpful. The functions described
here allow users to read and write arbitrary arrays of raw data as well as user-defined Silo objects.
These include...
DBWrite . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 239
DBWriteSlice . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 240
DBReadVar. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 242
DBReadVarSlice. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 243
DBGetVar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 244
DBInqVarExists . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 245
DBInqVarType . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 246
DBGetVarByteLength . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 248
DBGetVarDims . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 249
DBGetVarLength . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 250
DBGetVarType. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 251
DBPutCompoundarray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 252
DBInqCompoundarray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 253
DBGetCompoundarray . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 254
DBMakeObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 255
DBFreeObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 256
DBChangeObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 257
DBClearObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 258
DBAddDblComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 259
DBAddFltComponent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 260
DBAddIntComponent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 261
DBAddStrComponent. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 262
DBAddVarComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 263
DBWriteComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 264
DBWriteObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 265
DBGetObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 266
DBGetComponent . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 267
DBGetComponentType . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 268

DBWrite
Silo User’s Guide 2-239
DBWrite—Write a simple variable.
Synopsis:
int DBWrite (DBfile *dbfile, char const *varname, void const *var,
int const *dims, int ndims, int datatype)
Fortran Equivalent:
integer function dbwrite(dbid, varname, lvarname, var, dims,
ndims, datatype
Arguments:
dbfile Database file pointer.
varname Name of the simple variable.
var Array defining the values associated with the variable.
dims Array of length ndims which describes the dimensionality of the variable.
Each value in the dims array indicates the number of elements contained in the
variable along that dimension.
ndims Number of dimensions.
datatype Datatype of the variable. One of the predefined Silo data types.
Returns:
DBWrite returns zero on success and -1 on failure.
Description:
The DBWrite function writes a simple variable into a Silo file.

DBWriteSlice
2-240 Silo User’s Guide
DBWriteSlice—Write a (hyper)slab of a simple variable
Synopsis:
int DBWriteSlice (DBfile *dbfile, char const *varname,
void const *var, int datatype, int const *offset,
int cost *length, int const *stride, int const *dims,
int ndims)
Fortran Equivalent:
integer function dbwriteslice(dbid, varname, lvarname, var,
datatype, offset, length, stride, dims, ndims)
Arguments:
dbfile Database file pointer.
varname Name of the simple variable.
var Array defining the values associated with the slab.
datatype Datatype of the variable. One of the predefined Silo data types.
offset Array of length ndims of offsets in each dimension of the variable. This is the
0-origin position from which to begin writing the slice.
length Array of length ndims of lengths of data in each dimension to write to the
variable. All lengths must be positive.
stride Array of length ndims of stride steps in each dimension. If no striding is
desired, zeroes should be passed in this array.
dims Array of length ndims which describes the dimensionality of the entire
variable. Each value in the dims array indicates the number of elements
contained in the entire variable along that dimension.
ndims Number of dimensions.
Returns:
DBWriteSlice returns zero on success and -1 on failure.
Description:
The DBWriteSlice function writes a slab of data to a simple variable from the data provided in the
var pointer. Any hyperslab of data may be written.
The size of the entire variable (after all slabs have been written) must be known when the
DBWriteSlice function is called. The data in the var parameter is written into the entire variable
using the location specified in the offset, length, and stride parameters. The data that
makes up the entire variable may be written with one or more calls to DBWriteSlice.
The minimum length value is 1 and the minimum stride value is one.
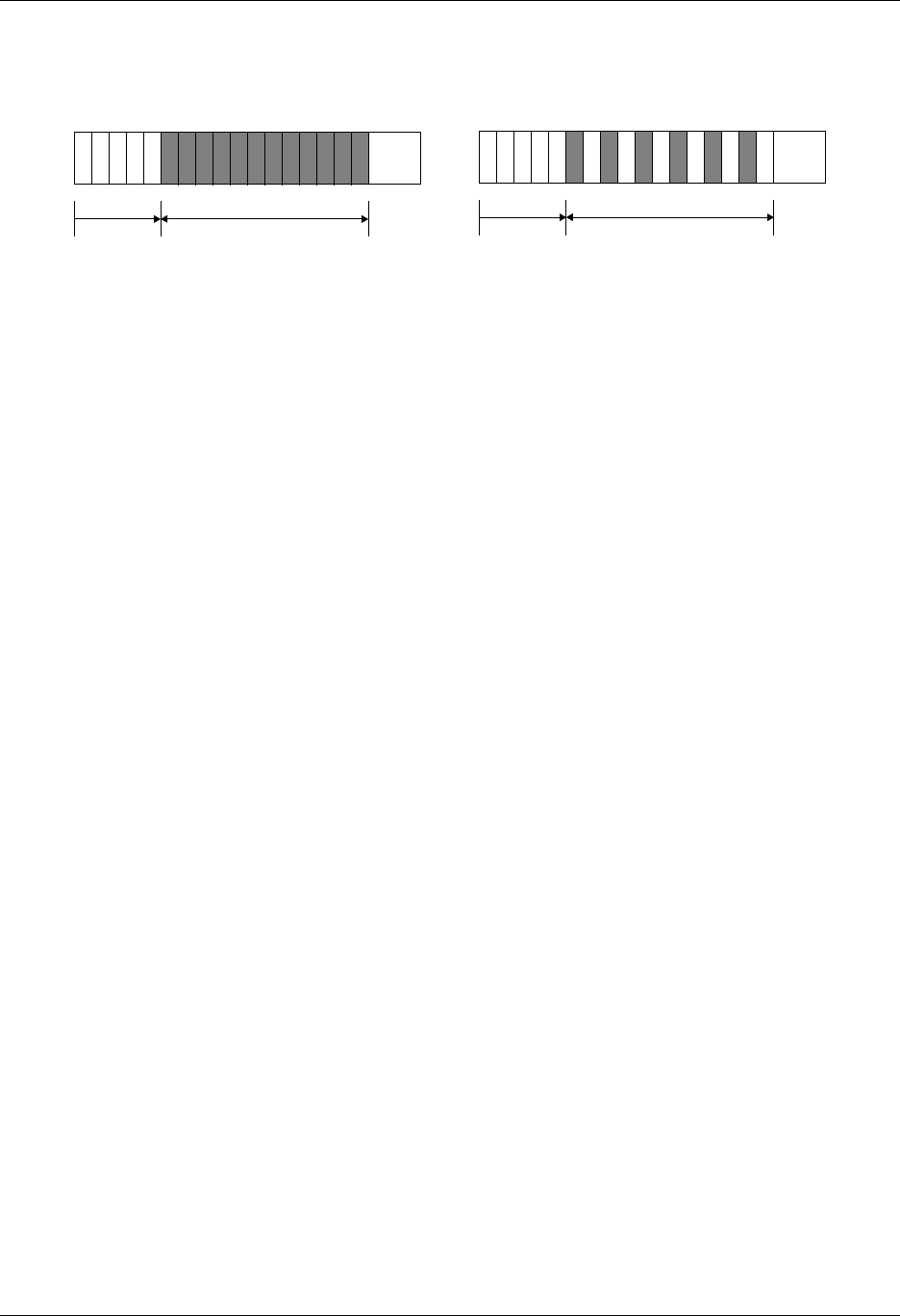
DBWriteSlice
Silo User’s Guide 2-241
A one-dimensional array slice:
Offset = 5 Length = 12
Stride = 1
Offset = 5 Length = 12
Stride = 2
Figure 0-11: Array slice

DBReadVar
2-242 Silo User’s Guide
DBReadVar—Read a simple Silo variable.
Synopsis:
int DBReadVar (DBfile *dbfile, char const *varname, void *result)
Fortran Equivalent:
integer function dbrdvar(dbid, varname, lvarname, ptr)
Arguments:
dbfile Database file pointer.
varname Name of the simple variable.
result Pointer to memory into which the variable should be read. It is up to the
application to provide sufficient space in which to read the variable.
Returns:
DBReadVar returns zero on success and -1 on failure.
Description:
The DBReadVar function reads a simple variable into the given space.
Notes:
See DBGetVar for a memory-allocating version of this function.

DBReadVarSlice
Silo User’s Guide 2-243
DBReadVarSlice—Read a (hyper)slab of data from a simple variable.
Synopsis:
int DBReadVarSlice (DBfile *dbfile, char const *varname,
int const *offset, int const *length, int const *stride,
int ndims, void *result)
Fortran Equivalent:
integer function dbrdvarslice(dbid, varname, lvarname, offset,
length, stride, ndims, ptr)
Arguments:
dbfile Database file pointer.
varname Name of the simple variable.
offset Array of length ndims of offsets in each dimension of the variable. This is the
0-origin position from which to begin reading the slice.
length Array of length ndims of lengths of data in each dimension to read from the
variable. All lengths must be positive.
stride Array of length ndims of stride steps in each dimension. If no striding is
desired, zeroes should be passed in this array.
ndims Number of dimensions in the variable.
result Pointer to location where the slice is to be written. It is up to the application to
provide sufficient space in which to read the variable.
Returns:
DBReadVarSlice returns zero on success and -1 on failure.
Description:
The DBReadVarSlice function reads a slab of data from a simple variable into a location provided
in the result pointer. Any hyperslab of data may be read.
Note that the minimum length value is 1 and the minimum stride value is one.
A one-dimensional array slice:
Offset = 5 Length = 12
Stride = 1
Offset = 5 Length = 12
Stride = 2
Figure 0-12: Array slice

DBGetVar
2-244 Silo User’s Guide
DBGetVar—Allocate space for, and return, a simple variable.
Synopsis:
void *DBGetVar (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Name of the variable
Returns:
DBGetVar returns a pointer to newly allocated space on success and NULL on failure.
Description:
The DBGetVar function allocates space for a simple variable, reads the variable from the Silo data-
base, and returns a pointer to the new space. If an error occurs, NULL is returned. It is up to the
application to cast the returned pointer to the correct data type.
Notes:
See DBReadVar and DBReadVar1 for non-memory allocating versions of this function.

DBInqVarExists
Silo User’s Guide 2-245
DBInqVarExists—Queries variable existence
Synopsis:
int DBInqVarExists (DBfile *dbfile, char const *name);
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Object name.
Returns:
DBInqVarExists returns non-zero if the object exists in the file. Zero otherwise.
Description:
The DBInqVarExists function is used to check for existence of an object in the given file.
If an object was written to a file, but the file has yet to be DBClose’d, the results of this function
querying that variable are undefined.
DBInqVarType
2-246 Silo User’s Guide
DBInqVarType—Return the type of the given object
Synopsis:
DBObjectType DBInqVarType (DBfile *dbfile, char const *name);
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
name Object name.
Returns:
DBInqVarType returns the DBObjectType corresponding to the given object.
Description:
The DBInqVarType function returns the DBObjectType of the given object. The value returned is
described in the following table:
Object Type Returned Value
Invalid object or the object was
not found in the file.
DB_INVALID_OBJECT
Quadmesh DB_QUADMESH
Quadvar DB_QUADVAR
UCD mesh DB_UCDMESH
UCD variable DB_UCDVAR
CSG mesh DB_CSGMESH
CSG variable DB_CSGVAR
Multiblock mesh DB_MULTIMESH
Multiblock variable DB_MULTIVAR
Multiblock material DB_MULTIMAT
Multiblock material species DB_MULTIMATSPECIES
Material DB_MATERIAL
Material species DB_MATSPECIES
Facelist DB_FACELIST
Zonelist DB_ZONELIST
Polyhedral-Zonelist DB_PHZONELIST
DBInqVarType
Silo User’s Guide 2-247
The function will signal an error if the given name does not exist in the file.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.
CSG-Zonelist DB_CSGZONELIST
Edgelist DB_EDGELIST
Curve DB_CURVE
Pointmesh DB_POINTMESH
Pointvar DB_POINTVAR
Defvars DB_DEFVARS
Compound array DB_ARRAY
Directory DB_DIR
Other variable (one written out
using DBWrite.)
DB_VARIABLE
User-defined DB_USERDEF
Object Type Returned Value

DBGetVarByteLength
2-248 Silo User’s Guide
DBGetVarByteLength—Return the byte length of a simple variable.
Synopsis:
int DBGetVarByteLength (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Vari ab le n a m e.
Returns:
DBGetVarByteLength returns the length of the given simple variable in bytes on success and -1 on
failure.
Description:
The DBGetVarByteLength function returns the length of the requested simple variable, in bytes.
This is useful for determining how much memory to allocate before reading a simple variable with
DBReadVar. Note that this would not be a concern if one used the DBGetVar function, which allo-
cates space itself.

DBGetVarByteLength
Silo User’s Guide 2-249
DBGetVarDims—Get dimension information of a variable in a Silo file
Synopsis:
int DBGetVarDims(DBfile *file, const char const *name, int
maxdims, int *dims)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
name The name of the Silo object to obtain dimension information for.
maxdims The maximum size of dims.
dims An array of maxdims integer values to be populated with the dimension
information returned by this call.
Returns:
The number of dimensions on success; -1 on failure
Description:
This function will populate the dims array up to a maximum of maxdims values with dimension
information of the specified Silo variable (object) name. The number of dimensions is returned as
the function’s return value.

DBGetVarLength
2-250 Silo User’s Guide
DBGetVarLength—Return the number of elements in a simple variable.
Synopsis:
int DBGetVarLength (DBfile *dbfile, char const *varname)
Fortran Equivalent:
integer function dbinqlen(dbid, varname, lvarname, len)
Arguments:
dbfile Database file pointer.
varname Vari ab le n a m e.
Returns:
DBGetVarLength returns the number of elements in the given simple variable on success and -1 on
failure.
Description:
The DBGetVarLength function returns the length of the requested simple variable, in number of
elements. For example a 16 byte array containing 4 floats has 4 elements.

DBGetVarType
Silo User’s Guide 2-251
DBGetVarType—Return the Silo datatype of a simple variable.
Synopsis:
int DBGetVarType (DBfile *dbfile, char const *varname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
varname Vari ab le n a m e.
Returns:
DBGetVarType returns the Silo datatype of the given simple variable on success and -1 on failure.
Description:
The DBGetVarType function returns the Silo datatype of the requested simple variable. For exam-
ple, DB_FLOAT for float variables.
Notes:
This only works for simple Silo variables (those written using DBWrite or DBWriteSlice). To
query the type of other variables, use DBInqVarType instead.

DBPutCompoundarray
2-252 Silo User’s Guide
DBPutCompoundarray—Write a Compound Array object into a Silo file.
Synopsis:
int DBPutCompoundarray (DBfile *dbfile, char const *name,
char const * const elemnames[], int const *elemlengths,
int nelems, void const *values, int nvalues, int datatype,
DBoptlist const *optlist);
Fortran Equivalent:
integer function dbputca(dbid, name, lname, elemnames, lelemnames,
elemlengths, nelems, values, datatype,
optlist_id, status)
character*N elemnames (See “dbset2dstrlen” on page 285.)
Arguments:
dbfile Database file pointer
name Name of the compound array structure.
elemnames Array of length nelems containing pointers to the names of the elements.
elemlengths Array of length nelems containing the lengths of the elements.
nelems Number of simple array elements.
values Array whose length is determined by nelems and elemlengths containing
the values of the simple array elements.
nvalues Total length of the values array.
datatype Data type of the values array. One of the predefined Silo types.
optlist Pointer to an option list structure containing additional information to be
included in the compound array object written into the Silo file. Use NULL is
there are no options.
Returns:
DBPutCompoundarray returns zero on success and -1 on failure.
Description:
The DBPutCompoundarray function writes a compound array object into a Silo file. A compound
array is an array whose elements are simple arrays. All of the simple arrays have elements of the
same data type, and each have a name.
Often, an application will partition a block of memory into named pieces, but write the block to a
database as a single entity. Fortran common blocks are used in this way. The compound array
object is an abstraction of this partitioned memory block.

DBInqCompoundarray
Silo User’s Guide 2-253
DBInqCompoundarray—Inquire Compound Array attributes.
Synopsis:
int DBInqCompoundarray (DBfile *dbfile, char const *name,
char ***elemnames, int *elemlengths,
int *nelems, int *nvalues, int *datatype)
Fortran Equivalent:
integer function dbinqca(dbid, name, lname, maxwidth, nelems,
nvalues, datatype)
Arguments:
dbfile Database file pointer.
name Name of the compound array.
elemnames Returned array of length nelems containing pointers to the names of the array
elements.
elemlengths Returned array of length nelems containing the lengths of the array elements.
nelems Returned number of array elements.
nvalues Returned number of total values in the compound array.
datatype Datatype of the data values. One of the predefined Silo data types.
Returns:
DBInqCompoundarray returns zero on success and -1 on failure.
Description:
The DBInqCompoundarray function returns information about the compound array. It does not
return the data values themselves; use DBGetCompoundarray instead.

DBGetCompoundarray
2-254 Silo User’s Guide
DBGetCompoundarray—Read a compound array from a Silo database.
Synopsis:
DBcompoundarray *DBGetCompoundarray (DBfile *dbfile,
char const *arrayname)
Fortran Equivalent:
integer function dbgetca(dbid, name, lname, lelemnames, elemnames,
elemlengths, nelems, values, nvalues,
datatype)
Arguments:
dbfile Database file pointer.
arrayname Name of the compound array.
Returns:
DBGetCompoundarray returns a pointer to a DBcompoundarray structure on success and NULL
on failure.
Description:
The DBGetCompoundarray function allocates a DBcompoundarray structure, reads a compound
array from the Silo database, and returns a pointer to that structure. If an error occurs, NULL is
returned.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBMakeObject
Silo User’s Guide 2-255
DBMakeObject—Allocate an object of the specified length and initialize it.
Synopsis:
DBobject *DBMakeObject (char const *objname, int objtype,
int maxcomps)
Fortran Equivalent:
None
Arguments:
objname Name of the object.
objtype Type of object. One of the predefined types: DB_QUADMESH,
DB_QUAD_RECT, DB_QUAD_CURV, DB_DEFVARS, DB_QUADVAR,
DB_UCDMESH, DB_UCDVAR, DB_POINTMESH, DB_POINTVAR,
DB_CSGMESH, DB_CSGVAR, DB_MULTIMESH, DB_MULTIVAR,
DB_MULTIADJ, DB_MATERIAL, DB_MATSPECIES, DB_FACELIST,
DB_ZONELIST, DB_PHZONELIST, DB_EDGELIST, DB_CURVE,
DB_ARRAY, or DB_USERDEF.
maxcomps Initial maximum number of components needed for this object. If this number is
exceeded, the library will silently re-allocate more space using the golden rule.
Returns:
DBMakeObject returns a pointer to the newly allocated and initialized object on success and
NULL on failure.
Description:
The DBMakeObject function allocates space for an object of maxcomps components.
In releases of the Silo library prior to 4.10, if a DBobject ever had more components added to it
than the maxcomps it was created with, an error would be generated and the operation to add a
component would fail. However, starting in version 4.10, the maxcomps argument is used only for
the initial object creation. If a caller attempts to add more than this number of components to an
object, Silo will simply re-allocate the object to accomodate the additional components.

DBFreeObject
2-256 Silo User’s Guide
DBFreeObject—Free memory associated with an object.
Synopsis:
int DBFreeObject (DBobject *object)
Fortran Equivalent:
None
Arguments:
object Pointer to the object to be freed. This object is created with the DBMakeObject
function.
Returns:
DBFreeObject returns zero on success and -1 on failure.
Description:
The DBFreeObject function releases the memory associated with the given object. The data asso-
ciated with the object’s components is not released.
DBFreeObject will not fail if a NULL pointer is passed to it.

DBFreeObject
Silo User’s Guide 2-257
DBChangeObject—Overwrite an existing object in a Silo file with a new object
Synopsis:
int DBChangeObject(DBfile *file, DBobject *obj)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
obj The new DBobject object (which knows its name) to write to the file.
Returns:
Zero on succes; -1 on failure
Description:
DBChangeObject writes a new DBobject object to a file, replacing the object in the file with the
same name.

DBClearObject
2-258 Silo User’s Guide
DBClearObject—Clear an object.
Synopsis:
int DBClearObject (DBobject *object)
Fortran Equivalent:
None
Arguments:
object Pointer to the object to be cleared. This object is created with the
DBMakeObject function.
Returns:
DBClearObject returns zero on success and -1 on failure.
Description:
The DBClearObject function clears an existing object. The number of components associated with
the object is set to zero.

DBAddDblComponent
Silo User’s Guide 2-259
DBAddDblComponent—Add a double precision floating point component to an object.
Synopsis:
int DBAddDblComponent (DBobject *object, char const *compname,
double d)
Fortran Equivalent:
None
Arguments:
object Pointer to an object. This object is created with the DBMakeObject function.
compname The component name.
dThe value of the double precision floating point component.
Returns:
DBAddDblComponent returns zero on success and -1 on failure.
Description:
The DBAddDblComponent function adds a component of double precision floating point data to
an existing object.

DBAddFltComponent
2-260 Silo User’s Guide
DBAddFltComponent—Add a floating point component to an object.
Synopsis:
int DBAddFltComponent (DBobject *object, char const *compname,
double f)
Fortran Equivalent:
None
Arguments:
object Pointer to an object. This object is created with the DBMakeObject function.
compname The component name.
fThe value of the floating point component.
Returns:
DBAddFltComponent returns zero on success and -1 on failure.
Description:
The DBAddFltComponent function adds a component of floating point data to an existing object.

DBAddIntComponent
Silo User’s Guide 2-261
DBAddIntComponent—Add an integer component to an object.
Synopsis:
int DBAddIntComponent (DBobject *object, char const *compname,
int i)
Fortran Equivalent:
None
Arguments:
object Pointer to an object. This object is created with the DBMakeObject function.
compname The component name.
iThe value of the integer component.
Returns:
DBAddIntComponent returns zero on success and -1 on failure.
Description:
The DBAddIntComponent function adds a component of integer data to an existing object.

DBAddStrComponent
2-262 Silo User’s Guide
DBAddStrComponent—Add a string component to an object.
Synopsis:
int DBAddStrComponent (DBobject *object, char const *compname,
char const *s)
Fortran Equivalent:
None
Arguments:
object Pointer to the object. This object is created with the DBMakeObject function.
compname The component name.
sThe value of the string component. Silo copies the contents of the string.
Returns:
DBAddStrComponent returns zero on success and -1 on failure.
Description:
The DBAddStrComponent function adds a component of string data to an existing object.

DBAddVarComponent
Silo User’s Guide 2-263
DBAddVarComponent—Add a variable component to an object.
Synopsis:
int DBAddVarComponent (DBobject *object, char const *compname,
char const *vardata)
Fortran Equivalent:
None
Arguments:
object Pointer to the object. This object is created with the DBMakeObject function.
compname Component name.
vardata Name of the variable object associated with the component (see Description).
Returns:
DBAddVarComponent returns zero on success and -1 on failure.
Description:
The DBAddVarComponent function adds a component of the variable type to an existing object.
The variable in vardata is stored verbatim into the object. No translation or typing is done on
the variable as it is added to the object.

DBWriteComponent
2-264 Silo User’s Guide
DBWriteComponent—Add a variable component to an object and write the associated
data.
Synopsis:
int DBWriteComponent (DBfile *dbfile, DBobject *object,
char const *compname, char const *prefix,
char const *datatype, void const *var, int nd,
long const *count)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
object Pointer to the object.
compname Component name.
prefix Path name prefix of the object.
datatype Data type of the component’s data. One of: “short”, “integer”, “long”, “float”,
“double”, “char”.
var Pointer to the component’s data.
nd Number of dimensions of the component.
count An array of length nd containing the length of the component in each of its
dimensions.
Returns:
DBWriteComponent returns zero on success and -1 on failure.
Description:
The DBWriteComponent function adds a component to an existing object and also writes the com-
ponent’s data to a Silo file.

DBWriteObject
Silo User’s Guide 2-265
DBWriteObject—Write an object into a Silo file.
Synopsis:
int DBWriteObject (DBfile *dbfile, DBobject const *object,
int freemem)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
object Object created with DBMakeObject and populated with DBAddFltComponent,
DBAddIntComponent, DBAddStrComponent, and DBAddVarComponent.
freemem If non-zero, then the object will be freed after writing.
Returns:
DBWriteObject returns zero on success and -1 on failure.
Description:
The DBWriteObject function writes an object into a Silo file. This is a user-defined object that
consists of various components. They are used when the basic Silo structures are not sufficient.

DBWriteObject
2-266 Silo User’s Guide
DBGetObject—Read an object from a Silo file as a generic object
Synopsis:
DBobject *DBGetObject(DBfile *file, char const *objname)
Fortran Equivalent:
None
Arguments:
file The Silo database file handle.
objname The name of the object to get.
Returns:
On success, a pointer to a DBobject struct containing the object’s data. NULL on failure.
Description:
Each of the object Silo supports has corresponding methods to both write them to a Silo database
file (DBPut...) and get them from a file (DBGet...).
However, Silo objects can also be accessed as generic objects through the generic object interface.
This is recommended only for objects that were written with DBWriteObject() method.
Notes:
For the details of the data structured returned by this function, see the Silo library header file,
silo.h, also attached to the end of this manual.

DBGetComponent
Silo User’s Guide 2-267
DBGetComponent—Allocate space for, and return, an object component.
Synopsis:
void *DBGetComponent (DBfile *dbfile, char const *objname,
char const *compname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
objname Object name.
compname Component name.
Returns:
DBGetComponent returns a pointer to newly allocated space containing the component value on
success, and NULL on failure.
Description:
The DBGetComponent function allocates space for one object component, reads the component,
and returns a pointer to that space. If either the object or component does not exist, NULL is
returned. It is up to the application to cast the returned pointer to the appropriate type.
DBGetComponentType
2-268 Silo User’s Guide
DBGetComponentType—Return the type of an object component.
Synopsis:
int DBGetComponentType (DBfile *dbfile, char const *objname,
char const *compname)
Fortran Equivalent:
None
Arguments:
dbfile Database file pointer.
objname Object name.
compname Component name.
Returns:
The values that are returned depend on the component’s type and how the component was written
into the object. The component types and their corresponding return values are listed in the table
below.
Component Type Return value
Integer DB_INT
Float DB_FLOAT
Double DB_DOUBLE
String DB_CHAR
Variable DB_VARIABLE
all others DB_NOTYPE
Description:
The DBGetComponentType function reads the component’s type and returns it. If either the object
or component does not exist, DB_NOTYPE is returned. This function allows the application to
process the component without having to know its type in advance.
DBGetComponentType
Silo User’s Guide 2-269
10 API Section JSON Interface to Silo Objects
WARNING: JSON support in Silo is experimental. The interface may be dramatically re-
worked, eliminated or replaced with something like Conduit. The Silo library must be con-
figured with --enable-json option to enable these JSON support functions. When this option
is enabled, the json-c library is compiled with Silo and installed to a json sub-directory at the
same install point as the Silo library. In addition, applications using Silo’s JSON interface
will have to link with the json-c library
(-I<silo-install>/json/include -L<silo-install>/json/lib -ljson).
JSON stands for JavaScript Object Notation. You can learn more about JSON at json.org. You can
learn more about the json-c library at https://github.com/json-c/json-c/wiki.
Silo’s JSON interface consists of two parts. The first part is just the json-c library interface which
includes methods such as json_object_new_int() which creates a new integer valued
JSON object and json_object_to_json_string() which returns an ascii string represen-
tation of a JSON object as well as many other methods. This interface is documented with the
json-c library and is not documented here.
The second part is some extensions to the json-c library we have defined for the purposes of pro-
viding a higher performance JSON interface for Silo objects. This includes the definition of a new
JSON object type; a pointer to an external array. This is called an extptr object and is actually a
specific assemblage of the following 4 JSON sub-objects.
Member name JSON type Meaning
“datatype” json_type_int An integer value representing one of the Silo types
DB_FLOAT, DB_INT, DB_DOUBLE, etc.
“ndims” json_type_int number of dimensions in the external array
“dims” json_type_array array of json_type_ints indicating size in each dimension
“ptr” json_type_string The ascii hexidecimal representation of a void* pointer hold-
ing the data of the array
The extpr object is used for all Silo data representing problem-sized array data. For example, it is
used to hold coordinate data for a mesh object, or variable data for a variable object or nodelist
data for a zonelist object.
Another extension of JSON we have defined for Silo is a binary format for serialized JSON
objects and methods to serialize and unserialize a JSON object to a binary buffer. Although JSON
implementations other than json-c also define a binary format (see for example, BSON) we have
defined one here as an extension to json-c. Silo’s binary format can be used, for example, by a par-
allel application to conveniently send Silo objects between processors by serializing to a binary
buffer at the sender and then unserializing at the receiver.
Any application wishing to use the JSON Silo interface must include the silo_json.h header file.
In this section we describe only those methods we have defined beyond those that come with the
json-c library. The functions in this part of the library are
json-c extensions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 271

DBGetComponentType
2-270 Silo User’s Guide
DBWriteJsonObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 272
DBGetJsonObject . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 273

DBGetComponentType
Silo User’s Guide 2-271
json-c extensions—Extensions to json-c library to support Silo
Synopsis:
/* Create/delete extptr object */
json_object* json_object_new_extptr(void *p, int ndims,
int const *dims, int datatype);
void json_object_extptr_delete(json_object *jso);
/* Inspect various members of an extptr object */
int json_object_is_extptr(json_object *obj);
int json_object_get_extptr_datatype(json_object *obj);
int json_object_get_extptr_ndims(json_object *obj);
int json_object_get_extptr_dims_idx(json_object *obj, int idx);
void* json_object_get_extptr_ptr(json_object *obj);
/* binary serialization */
int json_object_to_binary_buf(json_object *obj, int flags,
void **buf, int *len);
json_object* json_object_from_binary_buf(void *buf, int len);
/* Read/Write raw binary data to a file */
int json_object_to_binary_file(char const *filename,
json_object *obj);
json_object* json_object_from_binary_file(char const *filename);
/* Fix extptr members that were ascii-fied via standard json
string serialization */
void json_object_reconstitute_extptrs(json_object *o);
Fortran Equivalent:
None
Description:
As described in the introduction to this Silo API section, Silo defines a new JSON object type
called an extptr object. It is a pointer to an external array of data. Because the json-c library Silo
uses permits us to override the delete method for a JSON object, if you use the standard json-c
method of deleting a JSON object, json_object_put(), it will have the effect of deleting any
external arrays referenced by extptr objects.
Note that the binary serialization defined here can be UN-serialized only by this (Silo) implemen-
tation of JSON. If you serialize to a standard JSON string using the json-c library’s
json_object_to_json_string() the resulting serialization can be correctly interpreted
by *any* JSON implementation. However, in so doing, all extptr objects (which are unique to
Silo) are converted to the standard JSON array type. All performance advantages of extptr objects
are lost. They can, however, be re-constituted after UN-serializing a standard JSON string by the
json_object_reconstitute_extprs() method.

DBGetComponentType
2-272 Silo User’s Guide
DBWriteJsonObject—Write a JSON object to a Silo file
Synopsis:
DBWriteJsonObject(DBfile *db, json_object *jobj)
Fortran Equivalent:
None
Arguments:
db Silo database file handle
jobj JSON object pointer
Description:
This call takes a JSON object pointer and writes the object to a Silo file.
If the object is constructed so as to match one of Silo’s standard objects (any Silo object ordinarily
written with a DBPutXXX() call), then the JSON object will be written to the file such that any
Silo reader calling the matching DBGetXXX() method will successfully read the object. In other
words, it is possible to use this method to write first-class Silo objects to a file such as a ucd-mesh
or a quad-var, etc. All that is required is that the JSON object be constructed in such a way that it
holds all the metadata members Silo requires/uses for that specific object. See documentation for
the companion DBGetJsonObject().
Note that because there is no char const *name argument to this method, the JSON object
itself must indicate the name of the object. This is done by defining a string valued member with
key “silo_name”.

DBGetComponentType
Silo User’s Guide 2-273
DBGetJsonObject—Get an object from a Silo file as a JSON object
Synopsis:
json_object *DBGetJsonObject(DBfile *db, char const *name)
Fortran Equivalent:
None
Arguments:
db Silo database file handle
name Name of object to read
Description:
This method will read an object from a Silo file and return it as a JSON object. It can read *any*
Silo object from a Silo file including objects written to the file using DBPutXXX().
Note, however, that any problem-sized data associate with the object is returned as extptr sub-
objects. See introduction to this API section for a description of extptr objects.

DBGetComponentType
2-274 Silo User’s Guide
11 API Section Previously Undocumented Use Conventions
Silo is a relatively old library. It was originally developed in the early 1990’s. Over the years, a
number of use conventions have emerged and taken root and are now firmly entrenched in a vari-
ety of applications using Silo.
This section of the API manual simply tries to enumerate all these conventions and their meanings.
In a few cases, a long-standing use convention has been subsumed by the recent introduction of
formalized Silo objects or options to implement the convention. These cases are documented and
the user is encouraged to use the formal Silo approach.
Since everything documented in this section of the Silo API is a convention on the use of Silo,
where one would ordinarily see a function call prototype, instead example call(s) to the Silo that
implement the convention are described.
_visit_defvars . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 275
_visit_searchpath . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 276
_visit_domain_groups. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 277
AlphabetizeVariables . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 278
ConnectivityIsTimeVarying . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 279
MultivarToMultimeshMap_vars. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 280
MultivarToMultimeshMap_meshes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 281

_visit_defvars
Silo User’s Guide 2-275
_visit_defvars—convention for derived variable definitions
Synopsis:
int n;
char defs[1024];
sprintf(defs, “foo scalar x+y;bar vector {x,y,z};”
“gorfo scalar sqrt(x)”;
n = strlen(defs);
DBWrite(dbfile, “_visit_defvars”, defs, &n, 1, DB_CHAR);
Description:
Do not use this convention. Instead See “DBPutDefvars” on page 149.
_visit_defvars is an array of characters. The contents of this array is a semi-colon separated
list of derived variable expressions of the form
<name of derived variable> <space> <name of type> <space> <definition>
If an array of characters by this name exists in a Silo file, its contents will be used to populate the
post-processor’s derived variables. For VisIt, this would mean VisIt’s expression system.
This was also known as the “_meshtv_defvars” convention too.
This named array of characters can be written at any subdirectory in the Silo file.

_visit_searchpath
2-276 Silo User’s Guide
_visit_searchpath—directory order to search when opening a Silo file
Synopsis:
int n;
char dirs[1024];
sprintf(dirs, “nodesets;slides;”);
n = strlen(dirs);
DBWrite(dbfile, “_visit_searchpath”, dirs, &n, 1, DB_CHAR);
Description:
When opening a Silo file, an application is free to traverse directories in whatever order it wishes.
The _visit_searchpath convention is used by the data producer to control how downstream,
post-processing tools traverse a Silo file’s directory hierarchy.
_visit_searchpath is an array of characters representing a semi-colon separated list of
directory names. If a character array of this name is found at any directory in a Silo file, the direc-
tories it lists (which are considered to be relative to the directory in which this array is found
unless the directory names begin with a slash ‘/’) and only those directories are searched in the
order they are specified in the list.

_visit_domain_groups
Silo User’s Guide 2-277
_visit_domain_groups—method for grouping blocks in a multi-block mesh
Synopsis:
int domToGroupMap[16];
int j;
for (j = 0; j < 16; j++) domToGroupMap[j] = j%4;
DBWrite(dbfile, “_visit_domain_groups”, domToGroupMap,
&j, 1, DB_INT);
Description:
Do not use this convention. Instead use Mesh Region Grouping (MRG) trees. See “DBMak-
eMrgtree” on page 193.
_visit_domain_groups is an array of integers equal in size to the number of blocks in an
associated multi-block mesh object specifying, for each block, a group the block is a member of.
In the example above, there are 16 blocks assigned to 4 groups.

AlphabetizeVariables
2-278 Silo User’s Guide
AlphabetizeVariables—flag to tell post-processor to alphabetize variable lists
Synopsis:
int doAlpha = 1;
int n = 1;
DBWrite(dbfile, “AlphabetizeVariables”, &doAlpha, &n, 1, DB_INT);
Description:
The AlphabetizeVariables convention is a simple integer value which, if non-zero, indi-
cates that the post-processor should alphabetize its variable lists. In VisIt, this would mean that
various menus in the GUI, for example, are constructed such that variable names placed near the
top of the menus come alphabetically before variable names near the bottom of the menus. Other-
wise, variable names are presented in the order they are encountered in the database which is often
the order they were written to the database by the data producer.

ConnectivityIsTimeVarying
Silo User’s Guide 2-279
ConnectivityIsTimeVarying—flag telling post-processor if connectivity of
meshes in the Silo file is time varying or not
Synopsis:
int isTimeVarying = 1;
int n = 1;
DBWrite(dbfile, “ConnectivityIsTimeVarying”, &isTimeVarying, &n,
1, DB_INT);
Description:
The ConnectivityIsTimeVarying convention is a simple integer flag which, if non-zero,
indicates to post-processing tools that the connectivity for the mesh(s) in the database varies with
time. This has important performance implications and should only be specified if indeed it is nec-
essary as, for instance, in post-processors that assume connectivity is NOT time varying. This is an
assumption made by VisIt and the ConnectivityIsTimeVarying convention is a way to tell
VisIt to NOT make this assumption.

MultivarToMultimeshMap_vars
2-280 Silo User’s Guide
MultivarToMultimeshMap_vars—list of multivars to be associated with
multimeshes
Synopsis:
int len;
char tmpStr[256];
sprintf(tmpStr, "d;p;u;v;w;hist;mat1");
len = strlen(tmpStr);
DBWrite(dbfile, "MultivarToMultimeshMap_vars", tmpStr, &len, 1,
DB_CHAR);
Description:
Do not use this convention. Instead use the DBOPT_MMESH_NAME optlist option for a
DBPutMultivar() call to associate a multimesh with a multivar.
The MultivarToMultimeshMap_vars use convention goes hand-in-hand with the
MultivarToMultimeshMap_meshes use convention. The _vars portion is an array of
characters defining a semi-colon separated list of multivar object names to be associated with
multi-mesh names. The _mesh portion is an array of characters defining a semi-colon separated
list of associated multimesh object names. This convention was introduced to deal with a short-
coming in Silo where multivar objects did not know the multimesh object they were associated
with. This has since been corrected by the DBOPT_MMESH_NAME optlist option for a DBPut-
Multivar() call.

MultivarToMultimeshMap_meshes
Silo User’s Guide 2-281
MultivarToMultimeshMap_meshes—list of multimeshes to be associated with
multivars
Synopsis:
int len;
char tmpStr[256];
sprintf(tmpStr, "mesh1;mesh1;mesh1;mesh1;mesh1;mesh1;mesh1");
len = strlen(tmpStr);
DBWrite(dbfile, "MultivarToMultimeshMap_meshes", tmpStr, &len, 1,
DB_CHAR);
Description:
See “MultivarToMultimeshMap_vars” on page 280.

MultivarToMultimeshMap_meshes
2-282 Silo User’s Guide
12 API Section Silo’s Fortran Interface
The functions described in this section are either unique to the Fortran interface or facilitate the
mixing of C/C++ and Fortran within a single application interacting with a Silo file. Note that
when Silo was originally written, the vision was that only visualization/post-processing tools
would ever attempt to read the contents of Silo files. Therefore, the Fortran interface has never
included all the companion functions to read objects. That said, it is possible to write simple for-
tran callable wrappers to the C functions much like the write interface already implemented. Have
a look in the source file silo_f.c for examples.
The functions described here are...
dbmkptr. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 283
dbrmptr . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 284
dbset2dstrlen . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 285
dbget2dstrlen. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 286
DBFortranAllocPointer. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 287
DBFortranAccessPointer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 288
DBFortranRemovePointer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 289
dbwrtfl. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 290

dbmkptr
Silo User’s Guide 2-283
dbmkptr—create a pointer-id from a pointer
Synopsis:
integer function dbmkptr(void p)
Arguments:
ppointer for which a pointer-id is needed
Returns:
the integer pointer id to associate with the pointer
Description:
In cases where the C interface returns to the application a pointer to an abstract Silo object, in the
Fortran interface an integer pointer-id is created and returned instead. In addition, in cases where
the C interface would accept an array of pointers, such as in DBPutCsgvar(), the Fortran inter-
face accepts an array of pointer-ids. This function is used to create a pointer-id from a pointer.
A table of pointers is maintained internally in the Fortran wrapper library. The pointer-id is simply
the index into this table where the associated object’s pointer actually is. The caller can free up
space in this table using dbrmptr()

dbrmptr
2-284 Silo User’s Guide
dbrmptr—remove an old and no longer needed pointer-id
Synopsis:
integer function dbrmptr(ptr_id)
Arguments:
ptr_id the pointer-id to remove
Returns:
always 0

dbset2dstrlen
Silo User’s Guide 2-285
dbset2dstrlen—Set the size of a ‘row’ for pointers to ‘arrays’ of strings
Synopsis:
integer function dbset2dstrlen(int len)
integer len
Arguments:
len The length to set
Returns:
Returns the previously set value.
Description:
A number of functions in the Fortran interface take a char* argument that is really treated inter-
nally in the Fortran interface as a 2D array of characters. Calling this function allows the caller to
specify the length of the rows in this 2D array of characters. If necessary, this setting can be varied
from call to call.
The default value is 32 characters.

dbget2dstrlen
2-286 Silo User’s Guide
dbget2dstrlen—Get the size of a ‘row’ for pointers to ‘arrays’ of character strings
Synopsis:
integer function dbget2dstrlen()
Arguments:
None
Returns:
The current setting for the 2D string length.

DBFortranAllocPointer
Silo User’s Guide 2-287
DBFortranAllocPointer—Facilitates accessing C objects through Fortran
Synopsis:
int DBFortranAllocPointer (void *pointer)
Arguments:
pointer A pointer to a Silo object for which a Fortran identifier is needed
Returns:
DBFortranAllocPointer returns an integer that Fortran code can use to reference the given Silo
object.
Description:
The DBFortranAllocPointer function allows programs written in both C and Fortran to access the
same data structures. Many of the routines in the Fortran interface to Silo use an “object id”, an
integer which refers to a Silo object. DBFortanAllocPointer converts a pointer to a Silo object into
an integer that Fortran code can use. In some ways, this function is the inverse of DBFortranAc-
cessPointer.
The integer that DBFortranAllocPointer returns is used to index a table of Silo object pointers.
When done with the integer, the entry in the table may be freed for use later through the use of
DBFortranRemovePointer.
See “DBFortranAccessPointer” on page 2-288 and “DBFortranRemovePointer” on page 2-289 for
more information about how to use Silo objects in code that uses C and Fortran together.

DBFortranAccessPointer
2-288 Silo User’s Guide
DBFortranAccessPointer—Access Silo objects created through the Fortran Silo
interface.
Synopsis:
void *DBFortranAccessPointer (int value)
Arguments:
value The value returned by a Silo Fortran function, referencing a Silo object.
Returns:
DBFortranAccessPointer returns a pointer to a Silo object (which must be cast to the appropriate
type) on success, and NULL on failure.
Description:
The DBFortranAccessPointer function allows programs written in both C and Fortran to access the
same data structures. Many of the routines in the Fortran interface to Silo return an “object id”, an
integer which refers to a Silo object. DBFortranAccessPointer converts this integer into a C
pointer so that the sections of code written in C can access the Silo object directly.
See “DBFortranAllocPointer” on page 2-287 and “DBFortranRemovePointer” on page 2-289 for
more information about how to use Silo objects in code that uses C and Fortran together.

DBFortranRemovePointer
Silo User’s Guide 2-289
DBFortranRemovePointer—Removes a pointer from the Fortran-C index table
Synopsis:
void DBFortranRemovePointer (int value)
Arguments:
value An integer returned by DBFortranAllocPointer
Returns:
Nothing
Description:
The DBFortranRemovePointer function frees up the storage associated with Silo object pointers as
allocated by DBFortranAllocPointer.
Code that uses both C and Fortran may make use of DBFortranAllocPointer to allocate space in a
translation table so that the same Silo object may be referenced by both languages. DBFortranAc-
cessPointer provides access to this Silo object from the C side. Once the Fortran side of the code is
done referencing the object, the space in the translation table may be freed by calling DBFortran-
RemovePointer.
See “DBFortranAccessPointer” on page 2-288 and “DBFortranAllocPointer” on page 2-287 for
more information about how to use Silo objects in code that uses C and Fortran together.

DBFortranRemovePointer
2-290 Silo User’s Guide
dbwrtfl—Write a facelist object referenced by its object_id to a silo file
Synopsis:
dbwrtfl(dbid, name, lname, object_id, status)
Arguments:
dbid The identifier for the Silo database to write the object to.
name The name to be assigned to the object in the file.
lname The length of the name argument.
object_id The identifier for the facelist object, obtained via dbcalcfl.
status Return value indicating success or failure of the operation; 0 on success, -1 on
failure.
Returns:
Nothing
Description:
This function is designed to go hand-in-hand with dbcalcfl, the function used to calculate an
external facelist. When dbcalcfl is called, an object identifier is returned in object_id for
the newly created facelist. This call can then be used to write that facelist object to a Silo database.

DBFortranRemovePointer
Silo User’s Guide 2-291
13 API Section Deprecated Functions
The following functions were deprecated from Silo in version 4.6. Attempts to call these methods
in later versions may still succeed. However, deprecation warnings will be generated on stderr
(See “DBSetDeprecateWarnings” on page 35.). There is no guarantee that these methods will exist
in later versions of Silo.
DBGetComponentNames
DBGetAtt (completely removed in version 4.10)
DBListDir (completely removed in version 4.10)
DBReadAtt (completely removed in version 4.10)
DBGetQuadvar1 (completely removed in version 4.10)
DBcontinue (completely removed in version 4.10)
DBPause (completely removed in version 4.10)
DBPutZonelist
DBPutUcdsubmesh
DBErrFunc (use DBErrfunc instead)
DBSetDataReadMask (use DBSetDataReadMask2 instead)
DBGetDataReadMask (use DBGetDataReadMask2 instead)

DBFortranRemovePointer
2-292 Silo User’s Guide
14 API Section Silo Library Header File
We include the contents of the Silo header file here including a description of all DBxxx object
structs that are returned in DBGetXXX() calls as well as all other constant and symbols defined by
the library.

Page 1
/*
Copyright (c) 1994 - 2010, Lawrence Livermore National Security, LLC.
LLNL-CODE-425250.
All rights reserved.
This file is part of Silo. For details, see silo.llnl.gov.
Redistribution and use in source and binary forms, with or without
modification, are permitted provided that the following conditions
are met:
* Redistributions of source code must retain the above copyright
notice, this list of conditions and the disclaimer below.
* Redistributions in binary form must reproduce the above copyright
notice, this list of conditions and the disclaimer (as noted
below) in the documentation and/or other materials provided with
the distribution.
* Neither the name of the LLNS/LLNL nor the names of its
contributors may be used to endorse or promote products derived
from this software without specific prior written permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS
"AS IS" AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT
LIMITED TO, THE IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR
A PARTICULAR PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL LAWRENCE
LIVERMORE NATIONAL SECURITY, LLC, THE U.S. DEPARTMENT OF ENERGY OR
CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL,
EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO,
PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR
PROFITS; OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF
LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING
NEGLIGENCE OR OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS
SOFTWARE, EVEN IF ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.
This work was produced at Lawrence Livermore National Laboratory under
Contract No. DE-AC52-07NA27344 with the DOE.
Neither the United States Government nor Lawrence Livermore National
Security, LLC nor any of their employees, makes any warranty, express
or implied, or assumes any liability or responsibility for the
accuracy, completeness, or usefulness of any information, apparatus,
product, or process disclosed, or represents that its use would not
infringe privately-owned rights.
Any reference herein to any specific commercial products, process, or
services by trade name, trademark, manufacturer or otherwise does not
necessarily constitute or imply its endorsement, recommendation, or
favoring by the United States Government or Lawrence Livermore
National Security, LLC. The views and opinions of authors expressed
herein do not necessarily state or reflect those of the United States
Government or Lawrence Livermore National Security, LLC, and shall not
be used for advertising or product endorsement purposes.
*/

Page 2
/*
* SILO Public header file.
*
* This header file defines public constants and public prototypes.
* Before including this file, the application should define
* which file formats will be used.
*
* WARNING: The `#define' statements in this file are used when
* generating the Fortran include file `silo.inc'. Any
* such symbol that should not be an integer parameter
* in the Fortran include file should have the text
* `NO_FORTRAN_DEFINE' on the same line. #define statements
* that define macros (or any value not beginning with
* one of [a-zA-Z0-9_]) are ignored.
*/
#ifndef SILO_H
#define SILO_H
#include <stdio.h> /* for FILE* datatype for filters */
#include <silo_exports.h>
/* Why these Void Const Pointer (VCP) typedefs? (see below)... */
typedef void const * DBVCP1_t; /* single level array */
typedef void const * DBVCP2_t; /* double level array */
typedef void const * DBVCP3_t; /* triple level array */
/* And not these...
typedef void const * DBVCP1_t; single level array
typedef void const * const * DBVCP2_t; double level array
typedef void const * const * const * DBVCP3_t; triple level array
Ideally, the later typedefs would be used in the Silo API wherever a caller
needs to pass a single, double or triple level array of arbitrary type.
However, for multi-level arrays (e.g. more than a single star*), if the caller
doesn't explicitly cast a non-void pointer to the exact type including the
const qualifiers, the later definitions create obscure and non-intuitive
compiler
warnings for C callers and outright errors for C++ callers.
Basically, as counter-intuitive as it sounds neither C nor C++ compilers can
handle
passing something like a 'double **' as an argument to a function expecting a
'void const * const *'.
A good reference on this issue for C is http://c-faq.com/ansi/
constmismatch.html
A good reference for C++ is
http://www.embedded.com/electronics-blogs/programming-pointers/4025641/
Qualifiers-in-multilevel-pointers
Therefore, we introduce the DBVCPX_t typedefs, where 'X' is 1, 2 or 3, to
indicate the number of levels in the array the Silo library is expecting the

Page 3
caller to pass. Even though the typedef resolves to a 'void const *' for all
X, the underlying implementation will wind up treating it as a 'void const *'
for
X=1 (e.g. DBVCP1_t), a 'void const * const *' for X=2 (e.g. DBVCP2_t), etc.
*/
/* For the char-specific case of a constant array of strings, to facilitate
explicit casts */
typedef char const * const * DBCAS_t;
#ifndef FALSE
#define FALSE 0
#endif
#ifndef TRUE
#define TRUE 1
#endif
/* In the definitions for parts of the _B16 version numbers, below,
we use leading '0x0' to deal with possible blank minor and/or patch
version number. The _B16 variants are not really for Silo client
consumption. We use them here just allow for use of '0x0' leading
number to work-around issues with possible blanks. */
/* Major release number of silo library. */
#define _SILO_VERS_MAJ_@SILO_VERS_MAJ@
#ifdef _SILO_VERS_MAJ_
#define SILO_VERS_MAJ 0
#else
#define SILO_VERS_MAJ @SILO_VERS_MAJ@
#endif
/* Minor release number of silo library. Can be empty. */
#define _SILO_VERS_MIN_@SILO_VERS_MIN@
#ifdef _SILO_VERS_MIN_
#define SILO_VERS_MIN 0
#else
#define SILO_VERS_MIN @SILO_VERS_MIN@
#endif
/* Patch release number of silo library. Can be empty. */
#define _SILO_VERS_PAT_@SILO_VERS_PAT@
#ifdef _SILO_VERS_PAT_
#define SILO_VERS_PAT 0
#else
#define SILO_VERS_PAT @SILO_VERS_PAT@
#endif
/* Pre-release release number of silo library. Can be empty. */
#define _SILO_VERS_PRE_@SILO_VERS_PRE@
#ifdef _SILO_VERS_PRE_
#define SILO_VERS_PRE 0
#else
#define SILO_VERS_PRE @SILO_VERS_PRE@

Page 4
#endif
/* The symbol Silo uses to enforce link-time
header/object version compatibility */
#define SILO_VERS_TAG @SILO_VERS_TAG@
/* Useful macro for comparing Silo versions (and DB_ alias) */
#define SILO_VERSION_GE(Maj,Min,Pat) \
(((SILO_VERS_MAJ==Maj) && (SILO_VERS_MIN==Min) &&
(SILO_VERS_PAT>=Pat)) || \
((SILO_VERS_MAJ==Maj) && (SILO_VERS_MIN>Min)) || \
(SILO_VERS_MAJ>Maj))
#define DB_VERSION_GE(Maj,Min,Pat) SILO_VERSION_GE(Maj,Min,Pat)
/*-------------------------------------------------------------------------
* Drivers. This is a list of every driver that a user could use. Not all of
* them are necessarily compiled into the library. However, users are free
* to try without getting compilation errors. They are listed here so that
* silo.h doesn't have to be generated every time the library is recompiled.
*--------------------------------------------------------------------------*/
#define DB_NETCDF 0
#define DB_PDB 2 /* PDB Lite */
#define DB_TAURUS 3
#define DB_UNKNOWN 5
#define DB_DEBUG 6
#define DB_HDF5X 7
#define DB_PDBP 1 /* PDB Proper */
/* DO NOT USE. Obsoleted ways of specifying different HDF5 vfds */
#define DB_HDF5_SEC2_OBSOLETE 0x100
#define DB_HDF5_STDIO_OBSOLETE 0x200
#define DB_HDF5_CORE_OBSOLETE 0x300
#define DB_HDF5_MPIO_OBSOLETE 0x400
#define DB_HDF5_MPIOP_OBSOLETE 0x500
/* symbols for various HDF5 vfds */
#define DB_H5VFD_DEFAULT 0
#define DB_H5VFD_SEC2 1
#define DB_H5VFD_STDIO 2
#define DB_H5VFD_CORE 3
#define DB_H5VFD_LOG 4
#define DB_H5VFD_SPLIT 5
#define DB_H5VFD_DIRECT 6
#define DB_H5VFD_FAMILY 7
#define DB_H5VFD_MPIO 8
#define DB_H5VFD_MPIP 9
#define DB_H5VFD_SILO 10
#define DB_H5VFD_FIC 11 /* File Image in Core */
/* Macro for defining various HDF5 vfds as 'type' arg in create/open.
The 11 bit shift is to avoid possible collision with older versions
of Silo header file where VFDs where specified in bits 8-11. Their
obsoleted values are listed above. */
#define DB_HDF5_OPTS(OptsId) (DB_HDF5X|((OptsId&0x3F)<<11))

Page 5
/* Monikers for default file options sets */
/* We just make the default options sets the same as the vfd is */
#define DB_FILE_OPTS_H5_DEFAULT_DEFAULT DB_H5VFD_DEFAULT
#define DB_FILE_OPTS_H5_DEFAULT_SEC2 DB_H5VFD_SEC2
#define DB_FILE_OPTS_H5_DEFAULT_STDIO DB_H5VFD_STDIO
#define DB_FILE_OPTS_H5_DEFAULT_CORE DB_H5VFD_CORE
#define DB_FILE_OPTS_H5_DEFAULT_LOG DB_H5VFD_LOG
#define DB_FILE_OPTS_H5_DEFAULT_SPLIT DB_H5VFD_SPLIT
#define DB_FILE_OPTS_H5_DEFAULT_DIRECT DB_H5VFD_DIRECT
#define DB_FILE_OPTS_H5_DEFAULT_FAMILY DB_H5VFD_FAMILY
#define DB_FILE_OPTS_H5_DEFAULT_MPIO DB_H5VFD_MPIO
#define DB_FILE_OPTS_H5_DEFAULT_MPIP DB_H5VFD_MPIP
#define DB_FILE_OPTS_H5_DEFAULT_SILO DB_H5VFD_SILO
#define DB_FILE_OPTS_LAST DB_FILE_OPTS_H5_DEFAULT_SILO
/* note: no possible *default* settings for DB_H5VFD_FIC */
/* Various default HDF5 driver options. Users can define their own
sets of options using DBRegisterFileOptionsSets(). */
#define DB_HDF5 DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_DEFAULT)
#define DB_HDF5_SEC2 DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_SEC2)
#define DB_HDF5_STDIO DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_STDIO)
#define DB_HDF5_CORE DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_CORE)
#define DB_HDF5_LOG DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_LOG)
#define DB_HDF5_SPLIT DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_SPLIT)
#define DB_HDF5_DIRECT DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_DIRECT)
#define DB_HDF5_FAMILY DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_FAMILY)
#define DB_HDF5_MPIO DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_MPIO)
#define DB_HDF5_MPIOP DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_MPIP)
#define DB_HDF5_MPIP DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_MPIP)
#define DB_HDF5_SILO DB_HDF5_OPTS(DB_FILE_OPTS_H5_DEFAULT_SILO)
/*-------------------------------------------------------------------------
* Other library-wide constants.
*-------------------------------------------------------------------------*/
#define DB_NFILES 256 /*Max simultaneously open files */
#define DB_NFILTERS 32 /*Number of filters defined */
/*-------------------------------------------------------------------------
* Constants. All of these constants are always defined in the application.
* Each group of constants defined here are small integers used as an index
* into an array. Many of the groups have a total count of items in the
* group that will be used for array allocation and error checking--don't
* forget to increment the value when adding a new item to a constant group.
*-------------------------------------------------------------------------
*/
/* The following identifiers are for use with the DBDataReadMask() call. They
* specify what portions of the data beyond the metadata is allocated
* and read. Note that since these values are only necessary when reading
* and since the Fortran interface is primarily a write interface, it is not
* necessary for these symbols to appear in the silo.inc file. However, the
* reason they DO NOT APPEAR there inspite of the fact that DO NOT HAVE the
* 'NO_FORTRAN_DEFINE' text appearing on each line is that the mkinc script

Page 6
* requires an underscore in the symbol name for it to appear in silo.inc. */
#define DBAll 0xffffffffffffffff
#define DBNone 0x0000000000000000
#define DBCalc 0x0000000000000001
#define DBMatMatnos 0x0000000000000002
#define DBMatMatlist 0x0000000000000004
#define DBMatMixList 0x0000000000000008
#define DBCurveArrays 0x0000000000000010
#define DBPMCoords 0x0000000000000020
#define DBPVData 0x0000000000000040
#define DBQMCoords 0x0000000000000080
#define DBQVData 0x0000000000000100
#define DBUMCoords 0x0000000000000200
#define DBUMFacelist 0x0000000000000400
#define DBUMZonelist 0x0000000000000800
#define DBUVData 0x0000000000001000
#define DBFacelistInfo 0x0000000000002000
#define DBZonelistInfo 0x0000000000004000
#define DBMatMatnames 0x0000000000008000
#define DBUMGlobNodeNo 0x0000000000010000
#define DBZonelistGlobZoneNo 0x0000000000020000
#define DBMatMatcolors 0x0000000000040000
#define DBCSGMBoundaryInfo 0x0000000000080000
#define DBCSGMZonelist 0x0000000000100000
#define DBCSGMBoundaryNames 0x0000000000200000
#define DBCSGVData 0x0000000000400000
#define DBCSGZonelistZoneNames 0x0000000000800000
#define DBCSGZonelistRegNames 0x0000000001000000
#define DBMMADJNodelists 0x0000000002000000
#define DBMMADJZonelists 0x0000000004000000
#define DBPMGlobNodeNo 0x0000000008000000
#define DBPMGhostNodeLabels 0x0000000010000000
#define DBQMGhostNodeLabels 0x0000000020000000
#define DBQMGhostZoneLabels 0x0000000040000000
#define DBUMGhostNodeLabels 0x0000000080000000
#define DBZonelistGhostZoneLabels 0x0000000100000000
/* Definitions for COORD_TYPE */
/* Placed before DBObjectType enum because the
DB_QUAD_CURV and DB_QUAD_RECT symbols are
sometimes used as DBObjectType */
#define DB_COLLINEAR 130
#define DB_NONCOLLINEAR 131
#define DB_QUAD_RECT DB_COLLINEAR
#define DB_QUAD_CURV DB_NONCOLLINEAR
#ifdef __cplusplus
extern "C" {
#endif
/* Objects that can be stored in a data file */
typedef enum {
DB_INVALID_OBJECT= -1, /*causes enum to be signed, do not remove,

Page 7
space before minus sign necessary for
lint*/
DB_QUADRECT = DB_QUAD_RECT,
DB_QUADCURV = DB_QUAD_CURV,
DB_QUADMESH=500,
DB_QUADVAR=501,
DB_UCDMESH=510,
DB_UCDVAR=511,
DB_MULTIMESH=520,
DB_MULTIVAR=521,
DB_MULTIMAT=522,
DB_MULTIMATSPECIES=523,
DB_MULTIBLOCKMESH=DB_MULTIMESH,
DB_MULTIBLOCKVAR=DB_MULTIVAR,
DB_MULTIMESHADJ=524,
DB_MATERIAL=530,
DB_MATSPECIES=531,
DB_FACELIST=550,
DB_ZONELIST=551,
DB_EDGELIST=552,
DB_PHZONELIST=553,
DB_CSGZONELIST=554,
DB_CSGMESH=555,
DB_CSGVAR=556,
DB_CURVE=560,
DB_DEFVARS=565,
DB_POINTMESH=570,
DB_POINTVAR=571,
DB_ARRAY=580,
DB_DIR=600,
DB_VARIABLE=610,
DB_MRGTREE=611,
DB_GROUPELMAP=612,
DB_MRGVAR=613,
DB_USERDEF=700
} DBObjectType;
/* Data types */
typedef enum {
DB_INT=16,
DB_SHORT=17,
DB_LONG=18,
DB_FLOAT=19,
DB_DOUBLE=20,
DB_CHAR=21,
DB_LONG_LONG=22,
DB_NOTYPE=25 /*unknown type */
} DBdatatype;
/* Flags for DBCreate */
#define DB_CLOBBER 0
#define DB_NOCLOBBER 1
/* Flags for DBOpen */

Page 8
#define DB_READ 1
#define DB_APPEND 2
/* Target machine for DBCreate */
#define DB_LOCAL 0
#define DB_SUN3 10
#define DB_SUN4 11
#define DB_SGI 12
#define DB_RS6000 13
#define DB_CRAY 14
#define DB_INTEL 15
/* Options */
#define DBOPT_FIRST 260
#define DBOPT_ALIGN 260
#define DBOPT_COORDSYS 262
#define DBOPT_CYCLE 263
#define DBOPT_FACETYPE 264
#define DBOPT_HI_OFFSET 265
#define DBOPT_LO_OFFSET 266
#define DBOPT_LABEL 267
#define DBOPT_XLABEL 268
#define DBOPT_YLABEL 269
#define DBOPT_ZLABEL 270
#define DBOPT_MAJORORDER 271
#define DBOPT_NSPACE 272
#define DBOPT_ORIGIN 273
#define DBOPT_PLANAR 274
#define DBOPT_TIME 275
#define DBOPT_UNITS 276
#define DBOPT_XUNITS 277
#define DBOPT_YUNITS 278
#define DBOPT_ZUNITS 279
#define DBOPT_DTIME 280
#define DBOPT_USESPECMF 281
#define DBOPT_XVARNAME 282
#define DBOPT_YVARNAME 283
#define DBOPT_ZVARNAME 284
#define DBOPT_ASCII_LABEL 285
#define DBOPT_MATNOS 286
#define DBOPT_NMATNOS 287
#define DBOPT_MATNAME 288
#define DBOPT_NMAT 289
#define DBOPT_NMATSPEC 290
#define DBOPT_BASEINDEX 291 /* quad meshes for node and zone */
#define DBOPT_ZONENUM 292 /* ucd meshes for zone */
#define DBOPT_NODENUM 293 /* ucd/point meshes for node */
#define DBOPT_BLOCKORIGIN 294
#define DBOPT_GROUPNUM 295
#define DBOPT_GROUPORIGIN 296
#define DBOPT_NGROUPS 297
#define DBOPT_MATNAMES 298
#define DBOPT_EXTENTS_SIZE 299
#define DBOPT_EXTENTS 300

Page 9
#define DBOPT_MATCOUNTS 301
#define DBOPT_MATLISTS 302
#define DBOPT_MIXLENS 303
#define DBOPT_ZONECOUNTS 304
#define DBOPT_HAS_EXTERNAL_ZONES 305
#define DBOPT_PHZONELIST 306
#define DBOPT_MATCOLORS 307
#define DBOPT_BNDNAMES 308
#define DBOPT_REGNAMES 309
#define DBOPT_ZONENAMES 310
#define DBOPT_HIDE_FROM_GUI 311
#define DBOPT_TOPO_DIM 312 /* TOPOlogical DIMension */
#define DBOPT_REFERENCE 313 /* reference object */
#define DBOPT_GROUPINGS_SIZE 314 /* size of grouping array */
#define DBOPT_GROUPINGS 315 /* groupings array */
#define DBOPT_GROUPINGNAMES 316 /* array of size determined by
number of groups of names of groups. */
#define DBOPT_ALLOWMAT0 317 /* Turn off material numer "0" warnings*/
#define DBOPT_MRGTREE_NAME 318
#define DBOPT_REGION_PNAMES 319
#define DBOPT_TENSOR_RANK 320
#define DBOPT_MMESH_NAME 321
#define DBOPT_TV_CONNECTIVITY 322
#define DBOPT_DISJOINT_MODE 323
#define DBOPT_MRGV_ONAMES 324
#define DBOPT_MRGV_RNAMES 325
#define DBOPT_SPECNAMES 326
#define DBOPT_SPECCOLORS 327
#define DBOPT_LLONGNZNUM 328
#define DBOPT_CONSERVED 329
#define DBOPT_EXTENSIVE 330
#define DBOPT_MB_FILE_NS 331
#define DBOPT_MB_BLOCK_NS 332
#define DBOPT_MB_BLOCK_TYPE 333
#define DBOPT_MB_EMPTY_LIST 334
#define DBOPT_MB_EMPTY_COUNT 335
#define DBOPT_MB_REPR_BLOCK_IDX 336
#define DBOPT_MISSING_VALUE 337
#define DBOPT_ALT_ZONENUM_VARS 338
#define DBOPT_ALT_NODENUM_VARS 339
#define DBOPT_GHOST_NODE_LABELS 340
#define DBOPT_GHOST_ZONE_LABELS 341
#define DBOPT_LAST 499
/* Options relating to virtual file drivers */
#define DBOPT_H5_FIRST 500
#define DBOPT_H5_VFD 500
#define DBOPT_H5_RAW_FILE_OPTS 501
#define DBOPT_H5_RAW_EXTENSION 502
#define DBOPT_H5_META_FILE_OPTS 503
#define DBOPT_H5_META_EXTENSION 504
#define DBOPT_H5_CORE_ALLOC_INC 505
#define DBOPT_H5_CORE_NO_BACK_STORE 506
#define DBOPT_H5_META_BLOCK_SIZE 507

Page 10
#define DBOPT_H5_SMALL_RAW_SIZE 508
#define DBOPT_H5_ALIGN_MIN 509
#define DBOPT_H5_ALIGN_VAL 510
#define DBOPT_H5_DIRECT_MEM_ALIGN 511
#define DBOPT_H5_DIRECT_BLOCK_SIZE 512
#define DBOPT_H5_DIRECT_BUF_SIZE 513
#define DBOPT_H5_LOG_NAME 514
#define DBOPT_H5_LOG_BUF_SIZE 515
#define DBOPT_H5_MPIO_COMM 516
#define DBOPT_H5_MPIO_INFO 517
#define DBOPT_H5_MPIP_NO_GPFS_HINTS 518
#define DBOPT_H5_SIEVE_BUF_SIZE 519
#define DBOPT_H5_CACHE_NELMTS 520
#define DBOPT_H5_CACHE_NBYTES 521
#define DBOPT_H5_CACHE_POLICY 522
#define DBOPT_H5_FAM_SIZE 523
#define DBOPT_H5_FAM_FILE_OPTS 524
#define DBOPT_H5_USER_DRIVER_ID 525
#define DBOPT_H5_USER_DRIVER_INFO 526
#define DBOPT_H5_SILO_BLOCK_SIZE 527
#define DBOPT_H5_SILO_BLOCK_COUNT 528
#define DBOPT_H5_SILO_LOG_STATS 529
#define DBOPT_H5_SILO_USE_DIRECT 530
#define DBOPT_H5_FIC_SIZE 531
#define DBOPT_H5_FIC_BUF 532
#define DBOPT_H5_LAST 599
/* Error trapping method */
#define DB_TOP 0 /*default--API traps */
#define DB_NONE 1 /*no errors trapped */
#define DB_ALL 2 /*all levels trap (traceback) */
#define DB_ABORT 3 /*abort() is called */
#define DB_SUSPEND 4 /*suspend error reporting temporarily */
#define DB_RESUME 5 /*resume normal error reporting */
#define DB_ALL_AND_DRVR 6 /*DB_ALL + driver error reporting */
/* Errors */
#define E_NOERROR 0 /*No error */
#define E_BADFTYPE 1 /*Bad file type */
#define E_NOTIMP 2 /*Callback not implemented */
#define E_NOFILE 3 /*No data file specified */
#define E_INTERNAL 5 /*Internal error */
#define E_NOMEM 6 /*Not enough memory */
#define E_BADARGS 7 /*Bad argument to function */
#define E_CALLFAIL 8 /*Low-level function failure */
#define E_NOTFOUND 9 /*Object not found */
#define E_TAURSTATE 10 /*Taurus: database state error */
#define E_MSERVER 11 /*SDX: too many connections */
#define E_PROTO 12 /*SDX: protocol error */
#define E_NOTDIR 13 /*Not a directory */
#define E_MAXOPEN 14 /*Too many open files */
#define E_NOTFILTER 15 /*Filter(s) not found */
#define E_MAXFILTERS 16 /*Too many filters */
#define E_FEXIST 17 /*File already exists */

Page 11
#define E_FILEISDIR 18 /*File is actually a directory */
#define E_FILENOREAD 19 /*File lacks read permission. */
#define E_SYSTEMERR 20 /*System level error occured. */
#define E_FILENOWRITE 21 /*File lacks write permission. */
#define E_INVALIDNAME 22 /* Variable name is invalid */
#define E_NOOVERWRITE 23 /*Overwrite not permitted */
#define E_CHECKSUM 24 /*Checksum failed */
#define E_COMPRESSION 25 /*Compression failed */
#define E_GRABBED 26 /*Low level driver enabled */
#define E_NOTREG 27 /*The dbfile pointer is not resitered. */
#define E_CONCURRENT 28 /*File is opened multiply and not all read-
only. */
#define E_DRVRCANTOPEN 29 /*Driver cannot open the file. */
#define E_BADOPTCLASS 30 /*Optlist contains options for wrong class */
#define E_NOTENABLEDINBUILD 31 /*feature not enabled in this build */
#define E_MAXFILEOPTSETS 32 /*Too many file options sets */
#define E_NOHDF5 33 /*HDF5 driver not available */
#define E_EMPTYOBJECT 34 /*Empty object not currently permitted*/
#define E_OBJBUFFULL 35 /*No more temp. buffer space for object */
#define E_NERRORS 50
/* Definitions for MAJOR_ORDER */
#define DB_ROWMAJOR 0
#define DB_COLMAJOR 1
/* Definitions for CENTERING */
#define DB_NOTCENT 0
#define DB_NODECENT 110
#define DB_ZONECENT 111
#define DB_FACECENT 112
#define DB_BNDCENT 113 /* for CSG meshes only */
#define DB_EDGECENT 114
#define DB_BLOCKCENT 115 /* for 'block-centered' data on multimeshs
*/
/* Definitions for COORD_SYSTEM */
#define DB_CARTESIAN 120
#define DB_CYLINDRICAL 121 /* x,r; y,theta; z,height; 2D variant is
eqiv. to poloar */
#define DB_SPHERICAL 122 /* x,r; y,theta; z,phi; 2D variant is
equiv. to polar */
#define DB_NUMERICAL 123
#define DB_OTHER 124
/* Definitions for ZONE FACE_TYPE */
#define DB_RECTILINEAR 100
#define DB_CURVILINEAR 101
/* Definitions for PLANAR */
#define DB_AREA 140
#define DB_VOLUME 141
/* Definitions for flag values */
#define DB_ON 1000

Page 12
#define DB_OFF -1000
/* Definitions for disjoint flag */
#define DB_ABUTTING 142
#define DB_FLOATING 143
/* Definitions for derived variable types */
#define DB_VARTYPE_SCALAR 200
#define DB_VARTYPE_VECTOR 201
#define DB_VARTYPE_TENSOR 202
#define DB_VARTYPE_SYMTENSOR 203
#define DB_VARTYPE_ARRAY 204
#define DB_VARTYPE_MATERIAL 205
#define DB_VARTYPE_SPECIES 206
#define DB_VARTYPE_LABEL 207
/* Definitions for ghost labels */
#define DB_GHOSTTYPE_NOGHOST ((char) 0x00)
#define DB_GHOSTTYPE_INTDUP ((char) 0x01)
/* Definitions for CSG boundary types
Designed so low-order 16 bits are unused.
The last few characters of the symbol are intended
to indicate the representational form of the surface type
G = generalized form (n values, depends on surface type)
P = point (3 values, x,y,z in 3D, 2 values in 2D x,y)
N = normal (3 values, Nx,Ny,Nz in 3D, 2 values in 2D Nx,Ny)
R = radius (1 value)
A = angle (1 value in degrees)
L = length (1 value)
X = x-intercept (1 value)
Y = y-intercept (1 value)
Z = z-intercept (1 value)
K = arbitrary integer
F = planar face defined by point-normal pair (6 values)
*/
#define DBCSG_QUADRIC_G 0x01000000
#define DBCSG_SPHERE_PR 0x02010000
#define DBCSG_ELLIPSOID_PRRR 0x02020000
#define DBCSG_PLANE_G 0x03000000
#define DBCSG_PLANE_X 0x03010000
#define DBCSG_PLANE_Y 0x03020000
#define DBCSG_PLANE_Z 0x03030000
#define DBCSG_PLANE_PN 0x03040000
#define DBCSG_PLANE_PPP 0x03050000
#define DBCSG_CYLINDER_PNLR 0x04000000
#define DBCSG_CYLINDER_PPR 0x04010000
#define DBCSG_BOX_XYZXYZ 0x05000000
#define DBCSG_CONE_PNLA 0x06000000
#define DBCSG_CONE_PPA 0x06010000
#define DBCSG_POLYHEDRON_KF 0x07000000
#define DBCSG_HEX_6F 0x07010000

Page 13
#define DBCSG_TET_4F 0x07020000
#define DBCSG_PYRAMID_5F 0x07030000
#define DBCSG_PRISM_5F 0x07040000
/* Definitions for 2D CSG boundary types */
#define DBCSG_QUADRATIC_G 0x08000000
#define DBCSG_CIRCLE_PR 0x09000000
#define DBCSG_ELLIPSE_PRR 0x09010000
#define DBCSG_LINE_G 0x0A000000
#define DBCSG_LINE_X 0x0A010000
#define DBCSG_LINE_Y 0x0A020000
#define DBCSG_LINE_PN 0x0A030000
#define DBCSG_LINE_PP 0x0A040000
#define DBCSG_BOX_XYXY 0x0B000000
#define DBCSG_ANGLE_PNLA 0x0C000000
#define DBCSG_ANGLE_PPA 0x0C010000
#define DBCSG_POLYGON_KP 0x0D000000
#define DBCSG_TRI_3P 0x0D010000
#define DBCSG_QUAD_4P 0x0D020000
/* Definitions for CSG Region operators */
#define DBCSG_INNER 0x7F000000
#define DBCSG_OUTER 0x7F010000
#define DBCSG_ON 0x7F020000
#define DBCSG_UNION 0x7F030000
#define DBCSG_INTERSECT 0x7F040000
#define DBCSG_DIFF 0x7F050000
#define DBCSG_COMPLIMENT 0x7F060000
#define DBCSG_XFORM 0x7F070000
#define DBCSG_SWEEP 0x7F080000
/* definitions for MRG Tree traversal flags */
#define DB_PREORDER 0x00000001
#define DB_POSTORDER 0x00000002
#define DB_FROMCWR 0x00000004
/* Miscellaneous constants */
#define DB_F77NULL (-99) /*Fortran NULL pointer */
#define DB_F77NULLSTRING "NULLSTRING" /* FORTRAN STRING */
/*-------------------------------------------------------------------------
* Index selection macros
*-------------------------------------------------------------------------
*/
#define I4D(s,i,j,k,l) (l)*s[3]+(k)*s[2]+(j)*s[1]+(i)*s[0]
#define I3D(s,i,j,k) (k)*s[2]+(j)*s[1]+(i)*s[0]
#define I2D(s,i,j) (j)*s[1]+(i)*s[0]
/* Note we should not use MAX_DOUBLE here as its definition could be different
from one machine to the next. We need something that is a constant across
all machines we will operate on. And, yes, it means this particular number
cannot be used for any real data values or for the missing data value. I
think the risk of problems is minimal. Also, we want a zero in the file
to represent the fact that a missing value is NOT SET. Nonetheless, we

Page 14
want Silo clients to be able to specify that zero represents the missing
value. So, we adjust the interpretation of missing values on write and
on read to/from the file. On write, NOT_SET in mem gets mapped to zero in
the file and zero in mem gets mapped to NOT_SET in the file. On read, this
mapping is reversed. */
#define DB_MISSING_VALUE_NOT_SET ((double) (1.0e+308))
/*-------------------------------------------------------------------------
* Structures (just the public parts).
*-------------------------------------------------------------------------
*/
/*
* Database table of contents for the current directory only.
*/
typedef struct DBtoc_ {
char **curve_names;
int ncurve;
char **multimesh_names;
int nmultimesh;
char **multimeshadj_names;
int nmultimeshadj;
char **multivar_names;
int nmultivar;
char **multimat_names;
int nmultimat;
char **multimatspecies_names;
int nmultimatspecies;
char **csgmesh_names;
int ncsgmesh;
char **csgvar_names;
int ncsgvar;
char **defvars_names;
int ndefvars;
char **qmesh_names;
int nqmesh;
char **qvar_names;
int nqvar;
char **ucdmesh_names;
int nucdmesh;
char **ucdvar_names;

Page 15
int nucdvar;
char **ptmesh_names;
int nptmesh;
char **ptvar_names;
int nptvar;
char **mat_names;
int nmat;
char **matspecies_names;
int nmatspecies;
char **var_names;
int nvar;
char **obj_names;
int nobj;
char **dir_names;
int ndir;
char **array_names;
int narray;
char **mrgtree_names;
int nmrgtree;
char **groupelmap_names;
int ngroupelmap;
char **mrgvar_names;
int nmrgvar;
} DBtoc;
/*----------------------------------------------------------------------------
* Database Curve Object
*--------------------------------------------------------------------------
*/
typedef struct DBcurve_ {
/*----------- X vs. Y (Curve) Data -----------*/
int id; /* Identifier for this object */
int datatype; /* Datatype for x and y (float, double) */
int origin; /* '0' or '1' */
char *title; /* Title for curve */
char *xvarname; /* Name of domain (x) variable */
char *yvarname; /* Name of range (y) variable */
char *xlabel; /* Label for x-axis */
char *ylabel; /* Label for y-axis */
char *xunits; /* Units for domain */
char *yunits; /* Units for range */
void *x; /* Domain values for curve */

Page 16
void *y; /* Range values for curve */
int npts; /* Number of points in curve */
int guihide; /* Flag to hide from post-processor's GUI */
char *reference; /* Label to reference object */
int coord_sys; /* To indicate other coordinate systems */
double missing_value; /* Value to indicate var data is invalid/
missing */
} DBcurve;
typedef struct DBdefvars_ {
int ndefs; /* number of definitions */
char **names; /* [ndefs] derived variable names */
int *types; /* [ndefs] derived variable types */
char **defns; /* [ndefs] derived variable definitions */
int *guihides; /* [ndefs] flags to hide from
post-processor's GUI */
} DBdefvars;
typedef struct DBpointmesh_ {
/*----------- Point Mesh -----------*/
int id; /* Identifier for this object */
int block_no; /* Block number for this mesh */
int group_no; /* Block group number for this mesh */
char *name; /* Name associated with this mesh */
int cycle; /* Problem cycle number */
char *units[3]; /* Units for each axis */
char *labels[3]; /* Labels for each axis */
char *title; /* Title for curve */
void *coords[3]; /* Coordinate values */
float time; /* Problem time */
double dtime; /* Problem time, double data type */
/*
* The following two fields really only contain 3 elements. However, silo
* contains a bug in PJ_ReadVariable() as called by DBGetPointmesh() which
* can cause three doubles to be stored there instead of three floats.
*/
float min_extents[6]; /* Min mesh extents [ndims] */
float max_extents[6]; /* Max mesh extents [ndims] */
int datatype; /* Datatype for coords (float, double) */
int ndims; /* Number of computational dimensions */
int nels; /* Number of elements in mesh */
int origin; /* '0' or '1' */
int guihide; /* Flag to hide from post-processor's GUI */
void *gnodeno; /* global node ids */
char *mrgtree_name; /* optional name of assoc. mrgtree object */
int gnznodtype; /* datatype for global node/zone ids */
char *ghost_node_labels;
char **alt_nodenum_vars;
} DBpointmesh;
/*----------------------------------------------------------------------------
* Multi-Block Mesh Object

Page 17
*--------------------------------------------------------------------------
*/
typedef struct DBmultimesh_ {
/*----------- Multi-Block Mesh -----------*/
int id; /* Identifier for this object */
int nblocks; /* Number of blocks in mesh */
int ngroups; /* Number of block groups in mesh */
int *meshids; /* Array of mesh-ids which comprise mesh */
char **meshnames; /* Array of mesh-names for meshids */
int *meshtypes; /* Array of mesh-type indicators [nblocks] */
int *dirids; /* Array of directory ID's which contain blk
*/
int blockorigin; /* Origin (0 or 1) of block numbers */
int grouporigin; /* Origin (0 or 1) of group numbers */
int extentssize; /* size of each extent tuple */
double *extents; /* min/max extents of coords of each block */
int *zonecounts; /* array of zone counts for each block */
int *has_external_zones; /* external flags for each block */
int guihide; /* Flag to hide from post-processor's GUI */
int lgroupings; /* size of groupings array */
int *groupings; /* Array of mesh-ids, group-ids, and counts */
char **groupnames; /* Array of group-names for groupings */
char *mrgtree_name;/* optional name of assoc. mrgtree object */
int tv_connectivity;
int disjoint_mode;
int topo_dim; /* Topological dimension; max of all blocks.
*/
char *file_ns; /* namescheme for files (in lieu of meshnames)
*/
char *block_ns; /* namescheme for block objects (in lieu of
meshnames) */
int block_type; /* constant block type for all blocks (in lieu
of meshtypes) */
int *empty_list; /* list of empty block #'s (option for
namescheme) */
int empty_cnt; /* size of empty list */
int repr_block_idx; /* index of a 'representative' block */
char **alt_nodenum_vars;
char **alt_zonenum_vars;
char *meshnames_alloc; /* original alloc of meshnames as string
list */
} DBmultimesh;
/*----------------------------------------------------------------------------
* Multi-Block Mesh Adjacency Object
*--------------------------------------------------------------------------
*/
typedef struct DBmultimeshadj_ {
/*----------- Multi-Block Mesh Adjacency -----------*/
int nblocks; /* Number of blocks in mesh */
int blockorigin; /* Origin (0 or 1) of block numbers */
int *meshtypes; /* Array of mesh-type indicators [nblocks] */
int *nneighbors; /* Array [nblocks] neighbor counts */

Page 18
int lneighbors;
int *neighbors; /* Array [lneighbors] neighbor block numbers
*/
int *back; /* Array [lneighbors] neighbor block back */
int totlnodelists;
int *lnodelists; /* Array [lneighbors] of node counts shared */
int **nodelists; /* Array [lneighbors] nodelists shared */
int totlzonelists;
int *lzonelists; /* Array [lneighbors] of zone counts adjacent
*/
int **zonelists; /* Array [lneighbors] zonelists adjacent */
} DBmultimeshadj;
/*----------------------------------------------------------------------------
* Multi-Block Variable Object
*--------------------------------------------------------------------------
*/
typedef struct DBmultivar_ {
/*----------- Multi-Block Variable -----------*/
int id; /* Identifier for this object */
int nvars; /* Number of variables */
int ngroups; /* Number of block groups in mesh */
char **varnames; /* Variable names */
int *vartypes; /* variable types */
int blockorigin; /* Origin (0 or 1) of block numbers */
int grouporigin; /* Origin (0 or 1) of group numbers */
int extentssize; /* size of each extent tuple */
double *extents; /* min/max extents of each block */
int guihide; /* Flag to hide from post-processor's GUI */
char **region_pnames;
char *mmesh_name;
int tensor_rank; /* DB_VARTYPE_XXX */
int conserved; /* indicates if the variable should be
conserved
under various operations such as interp. */
int extensive; /* indicates if the variable reprsents an
extensiv
physical property (as opposed to intensive)
*/
char *file_ns; /* namescheme for files (in lieu of meshnames)
*/
char *block_ns; /* namescheme for block objects (in lieu of
meshnames) */
int block_type; /* constant block type for all blocks (in lieu
of meshtypes) */
int *empty_list; /* list of empty block #'s (option for
namescheme) */
int empty_cnt; /* size of empty list */
int repr_block_idx; /* index of a 'representative' block */
double missing_value; /* Value to indicate var data is invalid/
missing */

Page 19
char *varnames_alloc; /* original alloc of varnames as string
list */
} DBmultivar;
/*-------------------------------------------------------------------------
* Multi-material
*-------------------------------------------------------------------------
*/
typedef struct DBmultimat_ {
int id; /* Identifier for this object */
int nmats; /* Number of materials */
int ngroups; /* Number of block groups in mesh */
char **matnames; /* names of constiuent DBmaterial objects */
int blockorigin; /* Origin (0 or 1) of block numbers */
int grouporigin; /* Origin (0 or 1) of group numbers */
int *mixlens; /* array of mixlen values in each mat */
int *matcounts; /* counts of unique materials in each block */
int *matlists; /* list of materials in each block */
int guihide; /* Flag to hide from post-processor's GUI */
int nmatnos; /* global number of materials over all pieces
*/
int *matnos; /* global list of material numbers */
char **matcolors; /* optional colors for materials */
char **material_names; /* optional names of the materials */
int allowmat0; /* Flag to allow material "0" */
char *mmesh_name;
char *file_ns; /* namescheme for files (in lieu of meshnames)
*/
char *block_ns; /* namescheme for block objects (in lieu of
meshnames) */
int *empty_list; /* list of empty block #'s (option for
namescheme) */
int empty_cnt; /* size of empty list */
int repr_block_idx; /* index of a 'representative' block */
char *matnames_alloc; /* original alloc of matnames as string
list */
} DBmultimat;
/*-------------------------------------------------------------------------
* Multi-species
*-------------------------------------------------------------------------
*/
typedef struct DBmultimatspecies_ {
int id; /* Identifier for this object */
int nspec; /* Number of species */
int ngroups; /* Number of block groups in mesh */
char **specnames; /* Species object names */
int blockorigin; /* Origin (0 or 1) of block numbers */
int grouporigin; /* Origin (0 or 1) of group numbers */
int guihide; /* Flag to hide from post-processor's GUI */
int nmat; /* equiv. to nmatnos of a DBmultimat */
int *nmatspec; /* equiv. to matnos of a DBmultimat */
char **species_names; /* optional names of the species */
char **speccolors; /* optional colors for species */

Page 20
char *file_ns; /* namescheme for files (in lieu of meshnames)
*/
char *block_ns; /* namescheme for block objects (in lieu of
meshnames) */
int *empty_list; /* list of empty block #'s (option for
namescheme) */
int empty_cnt; /* size of empty list */
int repr_block_idx; /* index of a 'representative' block */
char *specnames_alloc; /* original alloc of matnames as string
list */
} DBmultimatspecies;
/*----------------------------------------------------------------------
* Definitions for the FaceList, ZoneList, and EdgeList structures
* used for describing UCD meshes.
*----------------------------------------------------------------------
*/
#define DB_ZONETYPE_BEAM 10
#define DB_ZONETYPE_POLYGON 20
#define DB_ZONETYPE_TRIANGLE 23
#define DB_ZONETYPE_QUAD 24
#define DB_ZONETYPE_POLYHEDRON 30
#define DB_ZONETYPE_TET 34
#define DB_ZONETYPE_PYRAMID 35
#define DB_ZONETYPE_PRISM 36
#define DB_ZONETYPE_HEX 38
typedef struct DBzonelist_ {
int ndims; /* Number of dimensions (2,3) */
int nzones; /* Number of zones in list */
int nshapes; /* Number of zone shapes */
int *shapecnt; /* [nshapes] occurences of each shape */
int *shapesize; /* [nshapes] Number of nodes per shape */
int *shapetype; /* [nshapes] Type of shape */
int *nodelist; /* Sequent lst of nodes which comprise zones
*/
int lnodelist; /* Number of nodes in nodelist */
int origin; /* '0' or '1' */
int min_index; /* Index of first real zone */
int max_index; /* Index of last real zone */
/*--------- Optional zone attributes ---------*/
int *zoneno; /* [nzones] zone number of each zone */
void *gzoneno; /* [nzones] global zone number of each zone */
int gnznodtype; /* datatype for global node/zone ids */
char *ghost_zone_labels;
char **alt_zonenum_vars;
} DBzonelist;
typedef struct DBphzonelist_ {

Page 21
int nfaces; /* Number of faces in facelist (aka
"facetable") */
int *nodecnt; /* Count of nodes in each face */
int lnodelist; /* Length of nodelist used to construct faces
*/
int *nodelist; /* List of nodes used in all faces */
char *extface; /* boolean flag indicating if a face is
external */
int nzones; /* Number of zones in this zonelist */
int *facecnt; /* Count of faces in each zone */
int lfacelist; /* Length of facelist used to construct zones
*/
int *facelist; /* List of faces used in all zones */
int origin; /* '0' or '1' */
int lo_offset; /* Index of first non-ghost zone */
int hi_offset; /* Index of last non-ghost zone */
/*--------- Optional zone attributes ---------*/
int *zoneno; /* [nzones] zone number of each zone */
void *gzoneno; /* [nzones] global zone number of each zone */
int gnznodtype; /* datatype for global node/zone ids */
char *ghost_zone_labels;
char **alt_zonenum_vars;
} DBphzonelist;
typedef struct DBfacelist_ {
/*----------- Required components ------------*/
int ndims; /* Number of dimensions (2,3) */
int nfaces; /* Number of faces in list */
int origin; /* '0' or '1' */
int *nodelist; /* Sequent list of nodes comprise faces */
int lnodelist; /* Number of nodes in nodelist */
/*----------- 3D components ------------------*/
int nshapes; /* Number of face shapes */
int *shapecnt; /* [nshapes] Num of occurences of each shape
*/
int *shapesize; /* [nshapes] Number of nodes per shape */
/*----------- Optional type component---------*/
int ntypes; /* Number of face types */
int *typelist; /* [ntypes] Type ID for each type */
int *types; /* [nfaces] Type info for each face */
/*--------- Optional node attributes ---------*/
int *nodeno; /* [lnodelist] node number of each node */
/*----------- Optional zone-reference component---------*/
int *zoneno; /* [nfaces] Zone number for each face */
} DBfacelist;
typedef struct DBedgelist_ {
int ndims; /* Number of dimensions (2,3) */
int nedges; /* Number of edges */

Page 22
int *edge_beg; /* [nedges] */
int *edge_end; /* [nedges] */
int origin; /* '0' or '1' */
} DBedgelist;
typedef struct DBquadmesh_ {
/*----------- Quad Mesh -----------*/
int id; /* Identifier for this object */
int block_no; /* Block number for this mesh */
int group_no; /* Block group number for this mesh */
char *name; /* Name associated with mesh */
int cycle; /* Problem cycle number */
int coord_sys; /* Cartesian, cylindrical, spherical */
int major_order; /* 1 indicates row-major for multi-d arrays */
int stride[3]; /* Offsets to adjacent elements */
int coordtype; /* Coord array type: collinear,
* non-collinear */
int facetype; /* Zone face type: rect, curv */
int planar; /* Sentinel: zones represent area or volume?
*/
void *coords[3]; /* Mesh node coordinate ptrs [ndims] */
int datatype; /* Type of coordinate arrays (double,float) */
float time; /* Problem time */
double dtime; /* Problem time, double data type */
/*
* The following two fields really only contain 3 elements. However, silo
* contains a bug in PJ_ReadVariable() as called by DBGetQuadmesh() which
* can cause three doubles to be stored there instead of three floats.
*/
float min_extents[6]; /* Min mesh extents [ndims] */
float max_extents[6]; /* Max mesh extents [ndims] */
char *labels[3]; /* Label associated with each dimension */
char *units[3]; /* Units for variable, e.g, 'mm/ms' */
int ndims; /* Number of computational dimensions */
int nspace; /* Number of physical dimensions */
int nnodes; /* Total number of nodes */
int dims[3]; /* Number of nodes per dimension */
int origin; /* '0' or '1' */
int min_index[3]; /* Index in each dimension of 1st
* non-phoney */
int max_index[3]; /* Index in each dimension of last
* non-phoney */
int base_index[3]; /* Lowest real i,j,k value for this block
*/
int start_index[3]; /* i,j,k values corresponding to original
* mesh */
int size_index[3]; /* Number of nodes per dimension for
* original mesh */
int guihide; /* Flag to hide from post-processor's GUI */
char *mrgtree_name; /* optional name of assoc. mrgtree object */
char *ghost_node_labels;

Page 23
char *ghost_zone_labels;
char **alt_nodenum_vars;
char **alt_zonenum_vars;
} DBquadmesh;
typedef struct DBucdmesh_ {
/*----------- Unstructured Cell Data (UCD) Mesh -----------*/
int id; /* Identifier for this object */
int block_no; /* Block number for this mesh */
int group_no; /* Block group number for this mesh */
char *name; /* Name associated with mesh */
int cycle; /* Problem cycle number */
int coord_sys; /* Coordinate system */
int topo_dim; /* Topological dimension. */
char *units[3]; /* Units for variable, e.g, 'mm/ms' */
char *labels[3]; /* Label associated with each dimension */
void *coords[3]; /* Mesh node coordinates */
int datatype; /* Type of coordinate arrays (double,float) */
float time; /* Problem time */
double dtime; /* Problem time, double data type */
/*
* The following two fields really only contain 3 elements. However, silo
* contains a bug in PJ_ReadVariable() as called by DBGetUcdmesh() which
* can cause three doubles to be stored there instead of three floats.
*/
float min_extents[6]; /* Min mesh extents [ndims] */
float max_extents[6]; /* Max mesh extents [ndims] */
int ndims; /* Number of computational dimensions */
int nnodes; /* Total number of nodes */
int origin; /* '0' or '1' */
DBfacelist *faces; /* Data structure describing mesh faces */
DBzonelist *zones; /* Data structure describing mesh zones */
DBedgelist *edges; /* Data struct describing mesh edges
* (option) */
/*--------- Optional node attributes ---------*/
void *gnodeno; /* [nnodes] global node number of each node */
/*--------- Optional zone attributes ---------*/
int *nodeno; /* [nnodes] node number of each node */
/*--------- Optional polyhedral zonelist ---------*/
DBphzonelist *phzones; /* Data structure describing mesh zones */
int guihide; /* Flag to hide from post-processor's GUI */
char *mrgtree_name; /* optional name of assoc. mrgtree object */
int tv_connectivity;
int disjoint_mode;
int gnznodtype; /* datatype for global node/zone ids */
char *ghost_node_labels;
char **alt_nodenum_vars;

Page 24
} DBucdmesh;
/*----------------------------------------------------------------------------
* Database Mesh-Variable Object
*---------------------------------------------------------------------------
*/
typedef struct DBquadvar_ {
/*----------- Quad Variable -----------*/
int id; /* Identifier for this object */
char *name; /* Name of variable */
char *units; /* Units for variable, e.g, 'mm/ms' */
char *label; /* Label (perhaps for editing purposes) */
int cycle; /* Problem cycle number */
int meshid; /* Identifier for associated mesh (Deprecated
Sep2005) */
void **vals; /* Array of pointers to data arrays */
int datatype; /* Type of data pointed to by 'val' */
int nels; /* Number of elements in each array */
int nvals; /* Number of arrays pointed to by 'vals' */
int ndims; /* Rank of variable */
int dims[3]; /* Number of elements in each dimension */
int major_order; /* 1 indicates row-major for multi-d arrays */
int stride[3]; /* Offsets to adjacent elements */
int min_index[3]; /* Index in each dimension of 1st
* non-phoney */
int max_index[3]; /* Index in each dimension of last
* non-phoney */
int origin; /* '0' or '1' */
float time; /* Problem time */
double dtime; /* Problem time, double data type */
/*
* The following field really only contains 3 elements. However, silo
* contains a bug in PJ_ReadVariable() as called by DBGetQuadvar() which
* can cause three doubles to be stored there instead of three floats.
*/
float align[6]; /* Centering and alignment per dimension */
void **mixvals; /* nvals ptrs to data arrays for mixed zones
*/
int mixlen; /* Num of elmts in each mixed zone data
* array */
int use_specmf; /* Flag indicating whether to apply species
* mass fractions to the variable. */
int ascii_labels;/* Treat variable values as ASCII values
by rounding to the nearest integer in
the range [0, 255] */
char *meshname; /* Name of associated mesh */
int guihide; /* Flag to hide from post-processor's GUI */
char **region_pnames;

Page 25
int conserved; /* indicates if the variable should be
conserved
under various operations such as interp. */
int extensive; /* indicates if the variable reprsents an
extensiv
physical property (as opposed to intensive)
*/
int centering; /* explicit centering knowledge; should agree
with alignment. */
double missing_value; /* Value to indicate var data is invalid/
missing */
} DBquadvar;
typedef struct DBucdvar_ {
/*----------- Unstructured Cell Data (UCD) Variable -----------*/
int id; /* Identifier for this object */
char *name; /* Name of variable */
int cycle; /* Problem cycle number */
char *units; /* Units for variable, e.g, 'mm/ms' */
char *label; /* Label (perhaps for editing purposes) */
float time; /* Problem time */
double dtime; /* Problem time, double data type */
int meshid; /* Identifier for associated mesh (Deprecated
Sep2005) */
void **vals; /* Array of pointers to data arrays */
int datatype; /* Type of data pointed to by 'vals' */
int nels; /* Number of elements in each array */
int nvals; /* Number of arrays pointed to by 'vals' */
int ndims; /* Rank of variable */
int origin; /* '0' or '1' */
int centering; /* Centering within mesh (nodal or zonal) */
void **mixvals; /* nvals ptrs to data arrays for mixed zones
*/
int mixlen; /* Num of elmts in each mixed zone data
* array */
int use_specmf; /* Flag indicating whether to apply species
* mass fractions to the variable. */
int ascii_labels;/* Treat variable values as ASCII values
by rounding to the nearest integer in
the range [0, 255] */
char *meshname; /* Name of associated mesh */
int guihide; /* Flag to hide from post-processor's GUI */
char **region_pnames;
int conserved; /* indicates if the variable should be
conserved
under various operations such as interp. */
int extensive; /* indicates if the variable reprsents an
extensiv
physical property (as opposed to intensive)
*/

Page 26
double missing_value; /* Value to indicate var data is invalid/
missing */
} DBucdvar;
typedef struct DBmeshvar_ {
/*----------- Generic Mesh-Data Variable -----------*/
int id; /* Identifier for this object */
char *name; /* Name of variable */
char *units; /* Units for variable, e.g, 'mm/ms' */
char *label; /* Label (perhaps for editing purposes) */
int cycle; /* Problem cycle number */
int meshid; /* Identifier for associated mesh (Deprecated
Sep2005) */
void **vals; /* Array of pointers to data arrays */
int datatype; /* Type of data pointed to by 'val' */
int nels; /* Number of elements in each array */
int nvals; /* Number of arrays pointed to by 'vals' */
int nspace; /* Spatial rank of variable */
int ndims; /* Rank of 'vals' array(s) (computatnl rank)
*/
int origin; /* '0' or '1' */
int centering; /* Centering within mesh (nodal,zonal,other)
*/
float time; /* Problem time */
double dtime; /* Problem time, double data type */
/*
* The following field really only contains 3 elements. However, silo
* contains a bug in PJ_ReadVariable() as called by DBGetPointvar() which
* can cause three doubles to be stored there instead of three floats.
*/
float align[6]; /* Alignmnt per dimension if
* centering==other */
/* Stuff for multi-dimensional arrays (ndims > 1) */
int dims[3]; /* Number of elements in each dimension */
int major_order; /* 1 indicates row-major for multi-d arrays */
int stride[3]; /* Offsets to adjacent elements */
/*
* The following two fields really only contain 3 elements. However, silo
* contains a bug in PJ_ReadVariable() as called by DBGetUcdmesh() which
* can cause three doubles to be stored there instead of three floats.
*/
int min_index[6]; /* Index in each dimension of 1st
* non-phoney */
int max_index[6]; /* Index in each dimension of last
non-phoney */
int ascii_labels;/* Treat variable values as ASCII values
by rounding to the nearest integer in
the range [0, 255] */
char *meshname; /* Name of associated mesh */
int guihide; /* Flag to hide from post-processor's GUI */

Page 27
char **region_pnames;
int conserved; /* indicates if the variable should be
conserved
under various operations such as interp. */
int extensive; /* indicates if the variable reprsents an
extensiv
physical property (as opposed to intensive)
*/
double missing_value; /* Value to indicate var data is invalid/
missing */
} DBmeshvar;
typedef DBmeshvar DBpointvar; /* better named alias for pointvar */
typedef struct DBmaterial_ {
/*----------- Material Information -----------*/
int id; /* Identifier */
char *name; /* Name of this material information block */
int ndims; /* Rank of 'matlist' variable */
int origin; /* '0' or '1' */
int dims[3]; /* Number of elements in each dimension */
int major_order; /* 1 indicates row-major for multi-d arrays */
int stride[3]; /* Offsets to adjacent elements in matlist */
int nmat; /* Number of materials */
int *matnos; /* Array [nmat] of valid material numbers */
char **matnames; /* Array of material names */
int *matlist; /* Array[nzone] w/ mat. number or mix index */
int mixlen; /* Length of mixed data arrays (mix_xxx) */
int datatype; /* Type of volume-fractions (double,float) */
void *mix_vf; /* Array [mixlen] of volume fractions */
int *mix_next; /* Array [mixlen] of mixed data indeces */
int *mix_mat; /* Array [mixlen] of material numbers */
int *mix_zone; /* Array [mixlen] of back pointers to mesh */
char **matcolors; /* Array of material colors */
char *meshname; /* Name of associated mesh */
int allowmat0; /* Flag to allow material "0" */
int guihide; /* Flag to hide from post-processor's GUI */
} DBmaterial;
typedef struct DBmatspecies_ {
/*----------- Species Information -----------*/
int id; /* Identifier */
char *name; /* Name of this matspecies information block
*/
char *matname; /* Name of material object with which the
* material species object is associated. */
int nmat; /* Number of materials */
int *nmatspec; /* Array of lngth nmat of the num of material
* species associated with each material. */
int ndims; /* Rank of 'speclist' variable */
int dims[3]; /* Number of elements in each dimension of the
* 'speclist' variable. */
int major_order; /* 1 indicates row-major for multi-d arrays */

Page 28
int stride[3]; /* Offsts to adjacent elmts in 'speclist' */
int nspecies_mf; /* Total number of species mass fractions. */
void *species_mf; /* Array of length nspecies_mf of mass
* frations of the material species. */
int *speclist; /* Zone array of dimensions described by ndims
* and dims. Each element of the array is an
* index into one of the species mass fraction
* arrays. A positive value is the index in
* the species_mf array of the mass fractions
* of the clean zone's material species:
* species_mf[speclist[i]] is the mass
fraction
* of the first species of material matlist[i]
* in zone i. A negative value means that the
* zone is a mixed zone and that the array
* mix_speclist contains the index to the
* species mas fractions: -speclist[i] is the
* index in the 'mix_speclist' array for zone
* i. */
int mixlen; /* Length of 'mix_speclist' array. */
int *mix_speclist; /* Array of lgth mixlen of 1-orig indices
* into the 'species_mf' array.
* species_mf[mix_speclist[j]] is the index
* in array species_mf' of the first of the
* mass fractions for material
* mix_mat[j]. */
int datatype; /* Datatype of mass fraction data. */
int guihide; /* Flag to hide from post-processor's GUI */
char **specnames; /* Array of species names; length is sum of
nmatspec */
char **speccolors; /* Array of species colors; length is sum of
nmatspec */
} DBmatspecies;
typedef struct DBcsgzonelist_ {
/*----------- CSG Zonelist -----------*/
int nregs; /* Number of regions in regionlist */
int origin; /* '0' or '1' */
int *typeflags; /* [nregs] type info about each region */
int *leftids; /* [nregs] left operand region refs */
int *rightids; /* [nregs] right operand region refs */
void *xform; /* [lxforms] transformation coefficients */
int lxform; /* length of xforms array */
int datatype; /* type of data in xforms array */
int nzones; /* number of zones */
int *zonelist; /* [nzones] region ids (complete regions) */
int min_index; /* Index of first real zone */
int max_index; /* Index of last real zone */
/*--------- Optional zone attributes ---------*/

Page 29
char **regnames; /* [nregs] names of each region */
char **zonenames; /* [nzones] names of each zone */
char **alt_zonenum_vars;
} DBcsgzonelist;
typedef struct DBcsgmesh_ {
/*----------- CSG Mesh -----------*/
int block_no; /* Block number for this mesh */
int group_no; /* Block group number for this mesh */
char *name; /* Name associated with mesh */
int cycle; /* Problem cycle number */
char *units[3]; /* Units for variable, e.g, 'mm/ms' */
char *labels[3]; /* Label associated with each dimension */
int nbounds; /* Total number of boundaries */
int *typeflags; /* nbounds boundary type info flags */
int *bndids; /* optional, nbounds explicit ids */
void *coeffs; /* coefficients in the representation of
each boundary */
int lcoeffs; /* length of coeffs array */
int *coeffidx; /* array of nbounds offsets into coeffs
for each boundary's coefficients */
int datatype; /* data type of coeffs data */
float time; /* Problem time */
double dtime; /* Problem time, double data type */
double min_extents[3]; /* Min mesh extents [ndims] */
double max_extents[3]; /* Max mesh extents [ndims] */
int ndims; /* Number of spatial & topological dimensions
*/
int origin; /* '0' or '1' */
DBcsgzonelist *zones; /* Data structure describing mesh zones */
/*--------- Optional boundary attributes ---------*/
char **bndnames; /* [nbounds] boundary names */
int guihide; /* Flag to hide from post-processor's GUI */
char *mrgtree_name; /* optional name of assoc. mrgtree object */
int tv_connectivity;
int disjoint_mode;
char **alt_nodenum_vars;
} DBcsgmesh;
typedef struct DBcsgvar_ {
/*----------- CSG Variable -----------*/
char *name; /* Name of variable */
int cycle; /* Problem cycle number */
char *units; /* Units for variable, e.g, 'mm/ms' */
char *label; /* Label (perhaps for editing purposes) */
float time; /* Problem time */
double dtime; /* Problem time, double data type */

Page 30
void **vals; /* Array of pointers to data arrays */
int datatype; /* Type of data pointed to by 'vals' */
int nels; /* Number of elements in each array */
int nvals; /* Number of arrays pointed to by 'vals' */
int centering; /* Centering within mesh (nodal or zonal) */
int use_specmf; /* Flag indicating whether to apply species
* mass fractions to the variable. */
int ascii_labels;/* Treat variable values as ASCII values
by rounding to the nearest integer in
the range [0, 255] */
char *meshname; /* Name of associated mesh */
int guihide; /* Flag to hide from post-processor's GUI */
char **region_pnames;
int conserved; /* indicates if the variable should be
conserved
under various operations such as interp. */
int extensive; /* indicates if the variable reprsents an
extensiv
physical property (as opposed to intensive)
*/
double missing_value; /* Value to indicate var data is invalid/
missing */
} DBcsgvar;
/*-------------------------------------------------------------------------
* A compound array is an array whose elements are simple arrays. A simple
* array is an array whose elements are all of the same primitive data
* type: float, double, integer, long... All of the simple arrays of
* a compound array have elements of the same data type.
*-------------------------------------------------------------------------
*/
typedef struct DBcompoundarray_ {
int id; /*identifier of the compound array */
char *name; /*name of te compound array */
char **elemnames; /*names of the simple array elements */
int *elemlengths; /*lengths of the simple arrays */
int nelems; /*number of simple arrays */
void *values; /*simple array values */
int nvalues; /*sum reduction of `elemlengths' vector */
int datatype; /*simple array element data type */
} DBcompoundarray;
typedef struct DBoptlist_ {
int *options; /* Vector of option identifiers */
void **values; /* Vector of pointers to option values */
int numopts; /* Number of options defined */
int maxopts; /* Total length of option/value arrays */
} DBoptlist;
#define DB_MAX_H5_OBJ_VALS 64

Page 31
typedef struct DBobject_ {
char *name;
char *type; /* Type of group/object */
char **comp_names; /* Array of component names */
char **pdb_names; /* Array of internal (PDB) variable names */
int ncomponents; /* Number of components */
int maxcomponents; /* Max number of components */
/* fields below are a hack for HDF5 driver to handle
customization of 'standard' objects */
char h5_vals[DB_MAX_H5_OBJ_VALS*3*sizeof(double)];
int h5_offs[DB_MAX_H5_OBJ_VALS];
int h5_sizes[DB_MAX_H5_OBJ_VALS];
int h5_types[DB_MAX_H5_OBJ_VALS];
char *h5_names[DB_MAX_H5_OBJ_VALS];
} DBobject;
typedef struct _DBmrgtnode {
char *name;
int narray;
char **names;
int type_info_bits;
int max_children;
char *maps_name;
int nsegs;
int *seg_ids;
int *seg_lens;
int *seg_types;
int num_children;
struct _DBmrgtnode **children;
/* internal stuff to support updates, i/o, etc. */
int walk_order;
struct _DBmrgtnode *parent;
} DBmrgtnode;
typedef void (*DBmrgwalkcb)(DBmrgtnode const *tnode, int nat_node_num, void
*data);
typedef struct _DBmrgtree {
char *name;
char *src_mesh_name;
int src_mesh_type;
int type_info_bits;
int num_nodes;
DBmrgtnode *root;
DBmrgtnode *cwr;
char **mrgvar_onames;
char **mrgvar_rnames;
} DBmrgtree;
typedef struct _DBmrgvar {

Page 32
char *name;
char *mrgt_name;
int ncomps;
char **compnames;
int nregns;
char **reg_pnames;
int datatype;
void **data;
} DBmrgvar ;
typedef struct _DBgroupelmap {
char *name;
int num_segments;
int *groupel_types;
int *segment_lengths;
int *segment_ids;
int **segment_data;
void **segment_fracs;
int fracs_data_type;
} DBgroupelmap;
#if !defined(DB_MAX_EXPSTRS) /* NO_FORTRAN_DEFINE */
#define DB_MAX_EXPSTRS 8 /* NO_FORTRAN_DEFINE */
#endif
typedef struct _DBnamescheme
{
char *fmt; /* orig. format string */
char const **fmtptrs; /* ptrs into first (printf) part of fmt for each
conversion spec. */
int fmtlen; /* len of first part of fmt */
int ncspecs; /* # of conversion specs in first part of fmt */
char delim; /* delimiter char used for parts of fmt */
int nembed; /* number of last embedded string encountered
(used in eval process) */
char *embedstrs[DB_MAX_EXPSTRS]; /* ptrs to copies of embedded strings
(used in eval process) */
int arralloc; /* flag indicating if Silo allocated the arrays or
not */
int narrefs; /* number of array refs in conversion specs */
char **arrnames; /* array names used by array refs */
void **arrvals; /* pointer to actual array data assoc. with each
name */
int *arrsizes; /* size of each array (only needed for
deallocating external arrays of strings) */
char **exprstrs; /* expressions to be evaluated for each conv.
spec. */
} DBnamescheme;
typedef struct DBfile *___DUMMY_TYPE; /* Satisfy ANSI scope rules */
/*
* All file formats are now anonymous except for the public properties
* and public methods.

Page 33
*/
typedef struct DBfile_pub {
/* Public Properties */
char *name; /*name of file */
int type; /*file type */
DBtoc *toc; /*table of contents */
int dirid; /*directory ID */
int fileid; /*unique file id [0,DB_NFILES-1] */
int pathok; /*driver handles paths in names */
int Grab; /*drive has access to low-level interface */
void *GrabId; /*pointer to low-level driver descriptor */
char *file_lib_version; /* version of lib file was created with
*/
/* Public Methods */
int (*close)(struct DBfile *);
int (*exist)(struct DBfile *, char const *);
int (*newtoc)(struct DBfile *);
DBObjectType (*inqvartype)(struct DBfile *, char const *);
int (*uninstall)(struct DBfile *);
DBobject *(*g_obj)(struct DBfile *, char const *);
int (*c_obj)(struct DBfile *, DBobject const *, int);
int (*w_obj)(struct DBfile *, DBobject const *, int);
void *(*g_comp)(struct DBfile *, char const *, char const *);
int (*g_comptyp)(struct DBfile *, char const *, char const *);
int (*w_comp)(struct DBfile *, DBobject *, char const *, char
const *,
char const *, void const *, int, long const *);
int (*write) (struct DBfile *, char const *, void const *, int
const *, int, int);
int (*writeslice)(struct DBfile *, char const *array_name, void
const *data,
int datatype, int const *offsets, int const *lens, int
const *stides,
int const *dims, int ndim);
int (*g_dir)(struct DBfile *, char *);
int (*mkdir)(struct DBfile *, char const *);
int (*cd)(struct DBfile *, char const *);
int (*r_var)(struct DBfile *, char const *, void *);
int (*module)(struct DBfile *, FILE *);
int (*r_varslice)(struct DBfile *, char const *, int const *,
int const *, int const *,
int, void *);
int (*g_compnames)(struct DBfile *, char const *, char ***,
char ***);
DBcompoundarray *(*g_ca)(struct DBfile *, char const *);
DBcurve *(*g_cu)(struct DBfile *, char const *);
DBdefvars *(*g_defv)(struct DBfile *, char const *);
DBmaterial *(*g_ma)(struct DBfile *, char const *);
DBmatspecies *(*g_ms)(struct DBfile *, char const *);
DBmultimesh *(*g_mm)(struct DBfile *, char const *);
DBmultivar *(*g_mv)(struct DBfile *, char const *);
DBmultimat *(*g_mt)(struct DBfile *, char const *);

Page 34
DBmultimatspecies *(*g_mms)(struct DBfile *, char const *);
DBpointmesh *(*g_pm)(struct DBfile *, char const *);
DBmeshvar *(*g_pv)(struct DBfile *, char const *);
DBquadmesh *(*g_qm)(struct DBfile *, char const *);
DBquadvar *(*g_qv)(struct DBfile *, char const *);
DBucdmesh *(*g_um)(struct DBfile *, char const *);
DBucdvar *(*g_uv)(struct DBfile *, char const *);
DBfacelist *(*g_fl)(struct DBfile *, char const *);
DBzonelist *(*g_zl)(struct DBfile *, char const *);
void *(*g_var)(struct DBfile *, char const *);
int (*g_varbl)(struct DBfile *, char const *); /*byte length
*/
int (*g_varlen)(struct DBfile *, char const *); /*nelems */
int (*g_vardims)(struct DBfile*, char const *, int, int *); /
*dims*/
int (*g_vartype)(struct DBfile *, char const *);
int (*i_meshname)(struct DBfile *, char const *, char *);
int (*i_meshtype)(struct DBfile *, char const *);
int (*p_ca)(struct DBfile *dbfile, char const *name, char const
* const *elemnames,
int const *elemlens, int nelems, void const *values,
int nvalues,
int datatype, DBoptlist const *);
int (*p_cu)(struct DBfile *dbfile, char const *name, void const
*xvals,
void const *yvals, int datatype, int npts, DBoptlist
const *opts);
int (*p_defv)(struct DBfile *dbfile, char const *name, int
ndefs,
char const * const *names, int const *types, char const
* const *defns,
DBoptlist const * const *opts);
int (*p_fl)(struct DBfile *dbfile, char const *name, int
nfaces, int ndims,
int const *nodelist, int lnodelist, int origin, int
const *zoneno,
int const *shapesize, int const *shapecnt, int nshapes,
int const *types,
int const *typelist, int ntypes);
int (*p_ma)(struct DBfile *dbfile, char const *name, char const
*meshname,
int nmat, int const *matnos, int const *matlist, int
const *dims,
int ndims, int const *mix_next, int const *mix_mat, int
const *mix_zone,
DBVCP1_t mix_vf, int mixlen, int datatype, DBoptlist
const *);
int (*p_ms)(struct DBfile *, char const *, char const *, int,
int const *, int const *,
int const *, int, int, DBVCP1_t, int const *, int, int,
DBoptlist const *);
int (*p_mm)(struct DBfile *, char const *, int, char const *
const *, int const *,
DBoptlist const *);

Page 35
int (*p_mv)(struct DBfile *, char const *, int, char const *
const *, int const *,
DBoptlist const *);
int (*p_mt)(struct DBfile *, char const *, int, char const *
const *, DBoptlist const *);
int (*p_mms)(struct DBfile *, char const *, int, char const *
const *, DBoptlist const *);
int (*p_pm)(struct DBfile *, char const *, int, DBVCP2_t, int,
int, DBoptlist const *);
int (*p_pv)(struct DBfile *, char const *, char const *, int,
DBVCP2_t, int,
int, DBoptlist const *);
int (*p_qm)(struct DBfile *, char const *, char const * const
*, DBVCP2_t, int const *,
int, int, int, DBoptlist const *);
int (*p_qv)(struct DBfile *, char const *, char const *, int,
char const * const *, DBVCP2_t,
int const *, int, DBVCP2_t, int, int, int, DBoptlist
const *);
int (*p_um)(struct DBfile *, char const *, int, char const *
const *, DBVCP2_t,
int, int, char const *, char const *, int, DBoptlist
const *);
int (*p_sm)(struct DBfile *, char const *, char const *,
int, char const *, char const *, DBoptlist const *);
int (*p_uv)(struct DBfile *, char const *, char const *, int,
char const * const *,
DBVCP2_t, int, DBVCP2_t, int, int, int, DBoptlist const
*);
int (*p_zl)(struct DBfile *, char const *, int, int, int const
*, int, int,
int const *, int const *, int);
int (*p_zl2)(struct DBfile *, char const *, int, int, int const
*, int, int,
int, int, int const *, int const *, int const *, int,
DBoptlist const *);
DBphzonelist *(*g_phzl)(struct DBfile *, char const *);
int (*p_phzl)(struct DBfile *, char const *, int, int const *,
int, int const *,
char const *, int, int const *, int, int const *, int,
int, int, DBoptlist const *);
int (*p_csgzl)(struct DBfile *, char const *, int, int const *,
int const *,
int const *, void const *, int, int, int, int const *,
DBoptlist const *);
DBcsgzonelist *(*g_csgzl)(struct DBfile *, char const *);
int (*p_csgm)(struct DBfile *, char const *, int, int, int
const *, int const *,
void const *, int, int, double const *, char const *,
DBoptlist const *);
DBcsgmesh *(*g_csgm)(struct DBfile *, char const *);
int (*p_csgv)(struct DBfile *, char const *, char const *, int,
char const * const *, void const * const *, int, int,
int, DBoptlist const *);

Page 36
DBcsgvar *(*g_csgv)(struct DBfile *, char const *);
DBmultimeshadj *(*g_mmadj)(struct DBfile *, char const *, int, int const
*);
int (*p_mmadj)(struct DBfile *, char const *, int, int const *,
int const *,
int const *, int const *, int const *, int const *
const *, int const *,
int const * const *, DBoptlist const *optlist);
int (*p_mrgt)(struct DBfile *dbfile, char const *name, char
const *mesh_name,
DBmrgtree const *tree, DBoptlist const *opts);
DBmrgtree *(*g_mrgt)(struct DBfile *, char const *name);
int (*p_grplm)(struct DBfile *dbfile, char const *map_name, int
num_segments,
int const *groupel_types, int const *segment_lengths,
int const *segment_ids,
int const * const *segment_data, void const *
const *segment_fracs,
int fracs_data_type, DBoptlist const *opts);
DBgroupelmap *(*g_grplm)(struct DBfile *dbfile, char const *name);
int (*p_mrgv)(struct DBfile *dbfile, char const *name, char
const *mrgt_name,
int ncomps, char const * const *compnames, int nregns,
char const * const *reg_pnames, int datatype, void
const * const *data,
DBoptlist const *opts);
DBmrgvar *(*g_mrgv)(struct DBfile *dbfile, char const *name);
int (*free_z)(struct DBfile *, char const *);
int (*cpdir)(struct DBfile *, char const *, struct DBfile *,
char const *);
int (*sort_obo)(struct DBfile *dbfile, int nobjs, char const
*const *obj_names, int *ranks);
} DBfile_pub;
typedef struct DBfile {
DBfile_pub pub;
/*private part follows per device driver */
} DBfile;
typedef void (*DBErrFunc_t)(char *);
/*-------------------------------------------------------------------------
* Public global variables.
*-------------------------------------------------------------------------
*/
SILO_API extern int DBDebugAPI; /*file desc for debug messages, or
zero */
SILO_API extern int db_errno; /*error number of last error */
SILO_API extern char db_errfunc[]; /*name of erring function */
#ifndef DB_MAIN
SILO_API extern DBfile *(*DBOpenCB[])(char const *, int, int);
SILO_API extern DBfile *(*DBCreateCB[])(char const *, int, int, int, char
const *);

Page 37
SILO_API extern int (*DBFSingleCB[])(int);
#endif
#define SILO_VSTRING_NAME "_silolibinfo"
#define SILO_VSTRING PACKAGE_VERSION
SILO_API extern int SILO_VERS_TAG;
#define SiloCheckVersion SILO_VERS_TAG = 1
/*
* SILO API FUNCTIONS
*/
/* Error handling and other global library behavior */
SILO_API extern void DBShowErrors(int, DBErrFunc_t);
SILO_API extern char const * DBErrString(void);
SILO_API extern char const * DBErrFuncname(void);
SILO_API extern DBErrFunc_t DBErrfunc(void);
SILO_API extern int DBErrno(void);
SILO_API extern int DBErrlvl(void);
/* Designed to prevent accidental use of old interface by forcing a human
readable compile time error */
#define DBSetDataReadMask(A)
,DBSetDataReadMask_is_replaced_with_DBSetDataReadMask2_using_unsigned_long_lon
g
#define DBGetDataReadMask()
,DBGetDataReadMask_is_replaced_with_DBGetDataReadMask2_using_unsigned_long_lon
g
SILO_API extern unsigned long long DBSetDataReadMask2(unsigned long long);
SILO_API extern unsigned long long DBGetDataReadMask2(void);
SILO_API extern char * DBGetDatatypeString(int datatype);
SILO_API extern int DBSetAllowOverwrites(int allow);
SILO_API extern int DBGetAllowOverwrites(void);
SILO_API extern int DBSetAllowEmptyObjects(int allow);
SILO_API extern int DBGetAllowEmptyObjects(void);
SILO_API extern int DBSetEnableChecksums(int enable);
SILO_API extern int DBGetEnableChecksums(void);
SILO_API extern void DBSetCompression(char const *);
SILO_API extern char const * DBGetCompression(void);
SILO_API extern int DBSetFriendlyHDF5Names(int enable);
SILO_API extern int DBGetFriendlyHDF5Names(void);
SILO_API extern int DBSetDeprecateWarnings(int max);
SILO_API extern int DBGetDeprecateWarnings();
SILO_API extern int const * DBSetUnknownDriverPriorities(int const
*);
SILO_API extern int const * DBGetUnknownDriverPriorities();
SILO_API extern int DBRegisterFileOptionsSet(DBoptlist
const *opts);
SILO_API extern int DBUnregisterFileOptionsSet(int
opts_set_id);
SILO_API extern void DBUnregisterAllFileOptionsSets();
SILO_API extern char const * DBVersion(void);
SILO_API extern int DBVersionDigits(int *Maj, int *Min, int
*Pat, int *Pre);
SILO_API extern int DBVersionGE(int Maj, int Min, int Pat);

Page 38
SILO_API extern int DBVariableNameValid(char const *s);
SILO_API extern int DBForceSingle(int);
/* Functions involving files, file structure and file inquiries */
SILO_API extern DBfile * DBOpenReal(char const *name, int
dbtype, int mode);
SILO_API extern DBfile * DBCreateReal(char const *name, int
mode, int targ, char const *info, int dbtype);
SILO_API extern int DBInqFileReal(char const *name);
/*
* The above functions are the 'Real' implementations of their macro
counterparts (below).
* These are the functions by which client code first gets into Silo. They are
separated
* out because they do a link-time header/library version check for us. It
works because
* we don't advertise the 'Real' functions and instead encourage clients to
use the macro
* counterparts (below). The macros wind up creating a reference to the Silo
vesion tag
* which is resolved only when the client is linked with a library that
defines the
* associated version symbol.
*/
#define DBOpen(NM, DR, MD) (SiloCheckVersion, DBOpenReal(NM, DR,
MD))
#define DBCreate(NM, MD, TG, NF, DR) (SiloCheckVersion, DBCreateReal(NM, MD,
TG, NF, DR))
#define DBInqFile(NM) (SiloCheckVersion, DBInqFileReal(NM))
SILO_API extern int DBClose(DBfile *);
SILO_API extern DBtoc * DBGetToc(DBfile *);
SILO_API extern int DBNewToc(DBfile *);
SILO_API extern void * DBGrabDriver(DBfile *);
SILO_API extern int DBUngrabDriver(DBfile *, void const *);
SILO_API extern int DBGetDriverType(DBfile const *);
SILO_API extern int DBGetDriverTypeFromPath(char const *);
SILO_API extern int DBVersionGEFileVersion(DBfile const
*dbfile);
SILO_API extern char const * DBFileVersion(DBfile const *dbfile);
SILO_API extern int DBFileVersionDigits(DBfile const
*dbfile, int *Maj, int *Min, int *Pat, int *Pre);
SILO_API extern int DBFileVersionGE(DBfile const *dbfile,
int Maj, int Min, int Pat);
SILO_API extern int DBGetDir(DBfile *, char *);
SILO_API extern int DBSetDir(DBfile *, char const *);
#define DBMkdir DBMkDir
SILO_API extern int DBMkDir(DBfile *, char const *);
SILO_API extern int DBCpDir(DBfile *dbfile, char const
*srcDir,
DBfile *dstFile, char const
*dstDir);
SILO_API extern int DBGuessHasFriendlyHDF5Names(DBfile *f);
SILO_API extern int DBInqVarExists(DBfile *, char const *);
SILO_API extern int DBUninstall(DBfile *);

Page 39
SILO_API extern int DBFreeCompressionResources(DBfile
*dbfile, char const *meshname);
SILO_API extern int DBSortObjectsByOffset(DBfile *, int
nobjs, char const * const *obj_names, int *ranks);
SILO_API extern int DBFilters(DBfile *, FILE *);
SILO_API extern int DBFilterRegistration(char const *, int
(*init) (DBfile *, char *),
int (*open) (DBfile *, char *));
SILO_API extern int DBInqFileHasObjects(DBfile *);
/* Object Allocation, Free and IsEmpty functions */
SILO_API extern DBcompoundarray * DBAllocCompoundarray(void);
SILO_API extern DBcurve * DBAllocCurve(void);
SILO_API extern DBdefvars * DBAllocDefvars(int);
SILO_API extern DBmultimesh * DBAllocMultimesh(int);
SILO_API extern DBmultimeshadj * DBAllocMultimeshadj(int);
SILO_API extern DBmultivar * DBAllocMultivar(int);
SILO_API extern DBmultimat * DBAllocMultimat(int);
SILO_API extern DBmultimatspecies * DBAllocMultimatspecies(int);
SILO_API extern DBcsgmesh * DBAllocCsgmesh(void);
SILO_API extern DBquadmesh * DBAllocQuadmesh(void);
SILO_API extern DBpointmesh * DBAllocPointmesh(void);
SILO_API extern DBmeshvar * DBAllocMeshvar(void);
SILO_API extern DBucdmesh * DBAllocUcdmesh(void);
SILO_API extern DBcsgvar * DBAllocCsgvar(void);
SILO_API extern DBquadvar * DBAllocQuadvar(void);
SILO_API extern DBucdvar * DBAllocUcdvar(void);
SILO_API extern DBzonelist * DBAllocZonelist(void);
SILO_API extern DBphzonelist * DBAllocPHZonelist(void);
SILO_API extern DBcsgzonelist * DBAllocCSGZonelist(void);
SILO_API extern DBedgelist * DBAllocEdgelist(void);
SILO_API extern DBfacelist * DBAllocFacelist(void);
SILO_API extern DBmaterial * DBAllocMaterial(void);
SILO_API extern DBmatspecies * DBAllocMatspecies(void);
SILO_API extern DBnamescheme * DBAllocNamescheme(void);
SILO_API extern DBgroupelmap * DBAllocGroupelmap(int, DBdatatype);
SILO_API extern void DBFreeMatspecies(DBmatspecies *);
SILO_API extern void DBFreeMaterial(DBmaterial *);
SILO_API extern void DBFreeFacelist(DBfacelist *);
SILO_API extern void DBFreeEdgelist(DBedgelist *);
SILO_API extern void DBFreeZonelist(DBzonelist *);
SILO_API extern void DBFreePHZonelist(DBphzonelist *);
SILO_API extern void DBFreeCSGZonelist(DBcsgzonelist *);
SILO_API extern void DBResetUcdvar(DBucdvar *);
SILO_API extern void DBFreeUcdvar(DBucdvar *);
SILO_API extern void DBResetQuadvar(DBquadvar *);
SILO_API extern void DBFreeCsgvar(DBcsgvar *);
SILO_API extern void DBFreeQuadvar(DBquadvar *);
SILO_API extern void DBFreeUcdmesh(DBucdmesh *);
SILO_API extern void DBFreeMeshvar(DBmeshvar *);
SILO_API extern void DBFreePointvar(DBpointvar *);
SILO_API extern void DBFreePointmesh(DBpointmesh *);
SILO_API extern void DBFreeQuadmesh(DBquadmesh *);

Page 40
SILO_API extern void DBFreeCsgmesh(DBcsgmesh *);
SILO_API extern void DBFreeDefvars(DBdefvars*);
SILO_API extern void DBFreeMultimesh(DBmultimesh *);
SILO_API extern void DBFreeMultimeshadj(DBmultimeshadj *);
SILO_API extern void DBFreeMultivar(DBmultivar *);
SILO_API extern void DBFreeMultimat(DBmultimat *);
SILO_API extern void DBFreeMultimatspecies(DBmultimatspecies
*);
SILO_API extern void DBFreeCompoundarray(DBcompoundarray *);
SILO_API extern void DBFreeCurve(DBcurve *);
SILO_API extern void DBFreeNamescheme(DBnamescheme *);
SILO_API extern void DBFreeMrgvar(DBmrgvar *mrgv);
SILO_API extern void DBFreeMrgtree(DBmrgtree *tree);
SILO_API extern void DBFreeGroupelmap(DBgroupelmap *map);
SILO_API extern int DBIsEmptyCurve(DBcurve const *curve);
SILO_API extern int DBIsEmptyPointmesh(DBpointmesh const
*msh);
SILO_API extern int DBIsEmptyPointvar(DBpointvar const
*var);
SILO_API extern int DBIsEmptyMeshvar(DBmeshvar const *var);
SILO_API extern int DBIsEmptyQuadmesh(DBquadmesh const
*msh);
SILO_API extern int DBIsEmptyQuadvar(DBquadvar const *var);
SILO_API extern int DBIsEmptyUcdmesh(DBucdmesh const *msh);
SILO_API extern int DBIsEmptyFacelist(DBfacelist const
*fl);
SILO_API extern int DBIsEmptyZonelist(DBzonelist const
*zl);
SILO_API extern int DBIsEmptyPHZonelist(DBphzonelist const
*zl);
SILO_API extern int DBIsEmptyUcdvar(DBucdvar const *var);
SILO_API extern int DBIsEmptyCsgmesh(DBcsgmesh const *msh);
SILO_API extern int DBIsEmptyCSGZonelist(DBcsgzonelist
const *zl);
SILO_API extern int DBIsEmptyCsgvar(DBcsgvar const *var);
SILO_API extern int DBIsEmptyMaterial(DBmaterial const
*mat);
SILO_API extern int DBIsEmptyMatspecies(DBmatspecies const
*spec);
/* User-defined (generic) Data and Object functions */
SILO_API extern int DBGetObjtypeTag(char const *);
SILO_API extern DBobject * DBMakeObject(char const *, int, int);
SILO_API extern int DBFreeObject(DBobject *);
SILO_API extern int DBClearObject(DBobject *);
SILO_API extern int DBAddVarComponent(DBobject *, char
const *, char const *);
SILO_API extern int DBAddIntComponent(DBobject *, char
const *, int);
SILO_API extern int DBAddFltComponent(DBobject *, char
const *, double);
SILO_API extern int DBAddDblComponent(DBobject *, char
const *, double);

Page 41
SILO_API extern int DBAddStrComponent(DBobject *, char
const *, char const *);
SILO_API extern int DBGetComponentNames(DBfile *, char
const *, char ***, char ***);
SILO_API extern DBobject * DBGetObject(DBfile *, char const *);
SILO_API extern int DBChangeObject(DBfile *, DBobject const
*);
SILO_API extern int DBWriteObject(DBfile *, DBobject const
*, int);
SILO_API extern void * DBGetComponent(DBfile *, char const *,
char const *);
SILO_API extern int DBGetComponentType(DBfile *, char const
*, char const *);
SILO_API extern int DBWriteComponent(DBfile *, DBobject *,
char const *, char const *, char const *,
void const *, int, long const *);
SILO_API extern int DBWrite(DBfile *, char const *, void
const *, int const *, int, int);
SILO_API extern int DBWriteSlice(DBfile *dbfile, char const
*array_name,
void const * data, int datatype,
int const *offsets,
int const *lengths, int const
*strides, int const *dims,
int ndims);
SILO_API extern int DBRead(DBfile *, char const *, void *);
SILO_API extern int DBReadVar(DBfile *, char const *, void
*);
SILO_API extern int DBReadVarSlice(DBfile *, char const *,
int const *, int const *, int const *, int, void *);
SILO_API extern DBcompoundarray * DBGetCompoundarray(DBfile *, char const
*);
SILO_API extern int DBInqCompoundarray(DBfile *, char const
*, char ***, int **, int *, int *, int *);
SILO_API extern void * DBGetVar(DBfile *, char const *);
SILO_API extern int DBGetVarByteLength(DBfile *, char const
*);
SILO_API extern int DBGetVarLength(DBfile *, char const *);
SILO_API extern int DBGetVarDims(DBfile *, char const *,
int, int *);
SILO_API extern int DBGetVarType(DBfile *, char const *);
SILO_API extern DBObjectType DBInqVarType(DBfile *, char const *);
/* Curve, Mesh, Variable and Material functions */
SILO_API extern DBcurve * DBGetCurve(DBfile *, char const *);
SILO_API extern DBdefvars * DBGetDefvars(DBfile *, char const *);
SILO_API extern DBmaterial * DBGetMaterial(DBfile *, char const *);
SILO_API extern DBmatspecies * DBGetMatspecies(DBfile *, char const
*);
SILO_API extern DBpointmesh * DBGetPointmesh(DBfile *, char const *);
SILO_API extern DBmeshvar * DBGetPointvar(DBfile *, char const *);
SILO_API extern DBquadmesh * DBGetQuadmesh(DBfile *, char const *);
SILO_API extern DBquadvar * DBGetQuadvar(DBfile *, char const *);
SILO_API extern DBucdmesh * DBGetUcdmesh(DBfile *, char const *);

Page 42
SILO_API extern DBucdvar * DBGetUcdvar(DBfile *, char const *);
SILO_API extern DBcsgmesh * DBGetCsgmesh(DBfile *, char const *);
SILO_API extern DBcsgvar * DBGetCsgvar(DBfile *, char const *);
SILO_API extern DBcsgzonelist * DBGetCSGZonelist(DBfile *, char const
*);
SILO_API extern DBfacelist * DBGetFacelist(DBfile *, char const *);
SILO_API extern DBzonelist * DBGetZonelist(DBfile *, char const *);
SILO_API extern DBphzonelist * DBGetPHZonelist(DBfile *, char const
*);
SILO_API extern int DBInqMeshname(DBfile *, char const *,
char *);
SILO_API extern int DBInqMeshtype(DBfile *, char const *);
SILO_API extern int DBPutCompoundarray(DBfile *dbfile, char
const *name, char const * const *elemnames,
int const *elemlens, int nelems,
void const *values, int nvalues, int datatype,
DBoptlist const *);
SILO_API extern int DBPutCurve(DBfile *dbfile, char const *
name, void const * xvals,
void const * yvals, int datatype,
int npts, DBoptlist const * opts);
SILO_API extern int DBPutDefvars(DBfile *dbfile, char const
*name, int, char const * const *names,
int const *types, char const *
const *defns, DBoptlist const * const *opts);
SILO_API extern int DBPutFacelist(DBfile *dbfile, char
const *, int nfaces, int ndims, int const *nodelist,
int lnodelist, int origin, int
const *zoneno, int const *shapesize,
int const *shapecnt, int nshapes,
int const *types, int const *typelist, int ntypes);
SILO_API extern int DBPutMaterial(DBfile *dbfile, char
const *name, char const *meshname, int nmat,
int const *matnos, int const
*matlist, int const *dims, int ndims,
int const *mix_next, int const
*mix_mat, int const *mix_zone, DBVCP1_t mix_vf,
int mixlen, int datatype, DBoptlist
const *opts);
SILO_API extern int DBPutMatspecies(struct DBfile *dbfile,
char const *name, char const *matnam,
int nmat, int const *nmatspec, int
const *speclist, int const *dims,
int ndims, int nspecies_mf,
DBVCP1_t species_mf, int const *mix_speclist,
int mixlen, int datatype, DBoptlist
const *optlist);
SILO_API extern int DBPutPointmesh(DBfile *, char const *,
int, DBVCP2_t, int, int, DBoptlist const *);
SILO_API extern int DBPutPointvar(DBfile *, char const *,
char const *, int, DBVCP2_t, int, int,
DBoptlist const *);
SILO_API extern int DBPutPointvar1(DBfile *, char const *,
char const *, DBVCP1_t, int, int,

Page 43
DBoptlist const *);
SILO_API extern int DBPutQuadmesh(DBfile *, char const *,
char const * const *, DBVCP2_t, int const *, int,
int, int, DBoptlist const *);
SILO_API extern int DBPutQuadvar(DBfile *, char const *,
char const *, int, char const * const *, DBVCP2_t,
int const *, int, DBVCP2_t, int,
int, int, DBoptlist const *);
SILO_API extern int DBPutQuadvar1(DBfile *, char const *,
char const *, DBVCP1_t, int const *, int,
DBVCP1_t, int, int, int, DBoptlist
const *);
SILO_API extern int DBPutUcdmesh(DBfile *, char const *,
int, char const * const *, DBVCP2_t, int,
int, char const *, char const *,
int, DBoptlist const *);
SILO_API extern int DBPutUcdsubmesh(DBfile *, char const *,
char const *, int,
char const *, char const *,
DBoptlist const *);
SILO_API extern int DBPutUcdvar(DBfile *, char const *,
char const *, int, char const * const *, DBVCP2_t,
int, DBVCP2_t, int, int, int,
DBoptlist const *);
SILO_API extern int DBPutUcdvar1(DBfile *, char const *,
char const *, DBVCP1_t, int, DBVCP1_t,
int, int, int, DBoptlist const *);
SILO_API extern int DBPutZonelist(DBfile *, char const *,
int, int, int const *, int, int,
int const *, int const *, int);
SILO_API extern int DBPutZonelist2(DBfile *, char const *,
int, int, int const *, int, int,
int, int, int const *, int const *,
int const *, int, DBoptlist const *);
SILO_API extern int DBPutPHZonelist(DBfile *, char const *,
int, int const *, int, int const *, char const *,
int, int const *, int, int const *,
int, int, int, DBoptlist const *);
SILO_API extern int DBPutCsgmesh(DBfile *, char const *,
int, int, int const *, int const *,
void const *, int, int, double
const *, char const *, DBoptlist const *);
SILO_API extern int DBPutCSGZonelist(DBfile *, char const
*, int, int const *,
int const *, int const *, void
const *, int, int, int, int const *,
DBoptlist const *);
SILO_API extern int DBPutCsgvar(DBfile *, char const *,
char const *, int, char const * const *,
DBVCP2_t, int, int, int, DBoptlist
const *);
/* Part Assemblies, AMR, Slide Surfaces, Nodesets and Other Arbitrary Mesh
Subsets */

Page 44
SILO_API extern void DBPrintMrgtree(DBmrgtnode *tnode, int
walk_order, void *data);
SILO_API extern void DBLinearizeMrgtree(DBmrgtnode *tnode,
int walk_order, void *data);
SILO_API extern void DBWalkMrgtree(DBmrgtree const *tree,
DBmrgwalkcb cb, void *wdata, int traversal_order);
SILO_API extern DBmrgtree * DBMakeMrgtree(int source_mesh_type, int
mrgtree_info, int max_root_descendents,
DBoptlist *opts);
SILO_API extern int DBAddRegion(DBmrgtree *tree, char const
*region_name, int type_info_bits,
int max_descendents, char const
*maps_name, int nsegs, int const *seg_ids,
int const *seg_sizes, int const
*seg_types, DBoptlist const *opts);
SILO_API extern int DBAddRegionArray(DBmrgtree *tree, int
nregn, char const * const *regn_names,
int type_info_bits, char const
*maps_name, int nsegs, int const *seg_ids,
int const *seg_sizes, int const
*seg_types, DBoptlist const *opts);
SILO_API extern int DBSetCwr(DBmrgtree *tree, char const
*path);
SILO_API extern char const * DBGetCwr(DBmrgtree *tree);
SILO_API extern int DBPutMrgtree(DBfile *dbfile, char const
*mrg_tree_name, char const *mesh_name,
DBmrgtree const *tree, DBoptlist
const *opts);
SILO_API extern int DBPutMrgvar(DBfile *dbfile, char const
*name, char const *mrgt_name,
int ncomps, char const
* const *compnames, int nregns,
char const * const *reg_pnames, int
datatype, DBVCP2_t data,
DBoptlist const *opts);
SILO_API extern int DBPutGroupelmap(DBfile *dbfile, char
const *map_name, int num_segments,
int const *groupel_types, int const
*segment_lengths, int const *segment_ids,
int const * const *segment_data,
DBVCP2_t segment_fracs,
int fracs_data_type, DBoptlist
const *opts);
SILO_API extern DBmrgtree * DBGetMrgtree(DBfile *dbfile, char const
*mrg_tree_name);
SILO_API extern DBgroupelmap * DBGetGroupelmap(DBfile *dbfile, char
const *name);
SILO_API extern DBmrgvar * DBGetMrgvar(DBfile *dbfile, char const
*name);
SILO_API extern DBnamescheme * DBMakeNamescheme(char const *fmt, ...);
SILO_API extern char const * DBGetName(DBnamescheme const *ns, int
natnum);
/* Multi-block objects and parallel I/O */

Page 45
SILO_API extern DBmultimesh * DBGetMultimesh(DBfile *, char const *);
SILO_API extern DBmultimeshadj * DBGetMultimeshadj(DBfile *, char const
*, int, int const *);
SILO_API extern DBmultivar * DBGetMultivar(DBfile *, char const *);
SILO_API extern DBmultimat * DBGetMultimat(DBfile *, char const *);
SILO_API extern DBmultimatspecies * DBGetMultimatspecies(DBfile *, char
const *);
SILO_API extern int DBPutMultimesh(DBfile *, char const *,
int, char const * const *, int const *,
DBoptlist const *);
SILO_API extern int DBPutMultimeshadj(DBfile *, char const
*, int, int const *, int const *,
int const *, int const *, int const
*, int const * const *, int const *,
int const * const *, DBoptlist
const *optlist);
SILO_API extern int DBPutMultivar(DBfile *, char const *,
int, char const * const *, int const *,
DBoptlist const *);
SILO_API extern int DBPutMultimat(DBfile *, char const *,
int, char const * const *, DBoptlist const *);
SILO_API extern int DBPutMultimatspecies(DBfile *, char
const *, int, char const * const *, DBoptlist const *);
/* Option lists */
SILO_API extern DBoptlist * DBMakeOptlist(int);
SILO_API extern int DBClearOptlist(DBoptlist *);
SILO_API extern int DBFreeOptlist(DBoptlist *);
SILO_API extern int DBAddOption(DBoptlist *, int, void *);
SILO_API extern void * DBGetOption(DBoptlist const *, int);
SILO_API extern int DBClearOption(DBoptlist *, int);
/* Calculational and Utility methods */
SILO_API extern int DBAnnotateUcdmesh(DBucdmesh *);
SILO_API extern DBfacelist * DBCalcExternalFacelist(int *, int, int,
int *, int *, int, int *, int);
SILO_API extern DBfacelist * DBCalcExternalFacelist2(int *, int,
int, int, int, int *, int *, int *, int, int *, int);
SILO_API extern char * DBJoinPath(char const *, char const *);
SILO_API extern void DBStringArrayToStringList(char const *
const *strArray, int n, char **strList, int *m);
SILO_API extern char ** DBStringListToStringArray(char const
*strList, int *n, int skipSemicolonAtIndexZero);
SILO_API extern int DBIsDifferentDouble(double a, double b,
double abstol, double reltol, double reltol_eps);
SILO_API extern int DBIsDifferentLongLong(long long a, long
long b, double abstol, double reltol, double reltol_eps);
/* Fortran interface functions */
SILO_API extern void * DBFortranAccessPointer(int value);
SILO_API extern int DBFortranAllocPointer(void *pointer);
SILO_API extern void DBFortranRemovePointer(int value);
SILO_API extern char * _db_safe_strdup(const char *);

Page 46
#ifdef __cplusplus
}
#endif
#undef NO_FORTRAN_DEFINE
#endif /* !SILO_H */


