BIGSTICK Manual
BIGSTICK_Manual
User Manual: Pdf
Open the PDF directly: View PDF ![]() .
.
Page Count: 100
BIGSTICK: A flexible configuration-interaction
shell-model code 1
Calvin W. Johnson2, W. Erich Ormand3, Kenneth S. McElvain4, and Hongzhang Shan5
December 4, 2017
1UCRL number: LLNL-SM-739926
2Department of Physics, San Diego State University, 5500 Campanile Drive, San
Diego CA 92182-1233
3Lawrence Livermore National Laboratory, P.O. Box 808, L-414, Livermore, CA
94551
4Department of Physics, University of California, Berkeley 366 Leconte Hall MC
7300, Berkeley, CA 94720
5Computational Research Division, Lawrence Berkeley Laboratory, Berkeley, CA,
94720
Abstract
We present BIGSTICK, a flexible configuration-interaction open-source shell-
model code for the many-fermion problem. Written mostly in Fortran 90 with
some later extensions, BIGSTICK utilizes a factorized on-the-fly algorithm for
computing many-body matrix elements, and has both MPI (distributed mem-
ory) and OpenMP (shared memory) parallelization, and can run on platforms
ranging from laptops to the largest parallel supercomputers. It uses a flexible
yet efficient many-body truncation scheme, and reads input files in multiple for-
mats, allowing one to tackle both phenomenological (major valence shell space)
and ab initio (the so-called no-core shell model) calculations. BIGSTICK can
generate energy spectra, static and transition one-body densities, and expecta-
tion values of scalar operators. Using the built-in Lanczos algorithm one can
compute transition probability distributions and decompose wave functions into
components defined by group theory.
This manual provides a general guide to compiling and running BIGSTICK,
which comes with numerous sample input files, as well as some of the basic
theory underlying the code. The code also comes with an incomplete “inside
guide” which provides more details into the inner workings.
This code is distributed under the MIT Open Source License. The source
code and sample inputs are found at github.com/cwjsdsu/BigstickPublick.
Contents
1 Introduction 4
1.1 Expectations of users . . . . . . . . . . . . . . . . . . . . . . . . . 5
1.2 How to cite and copyright notices/licenses . . . . . . . . . . . . . 5
1.2.1 LAPACK copyright notice . . . . . . . . . . . . . . . . . . 6
1.3 Reporting bugs and other issues . . . . . . . . . . . . . . . . . . . 7
1.4 A brief history of BIGSTICK, and acknowledgements . . . . . . . 8
2 How we solve the many-body problem 9
2.1 Matrix formulation of the Schr¨odinger equation . . . . . . . . . . 10
2.2 Representation of the basis . . . . . . . . . . . . . . . . . . . . . 12
2.2.1 Factorization of the basis . . . . . . . . . . . . . . . . . . 14
2.3 The Lanczos algorithm and computational cost . . . . . . . . . . 17
2.4 Representation of the Hamiltonian . . . . . . . . . . . . . . . . . 18
2.5 An incomplete survey of other codes . . . . . . . . . . . . . . . . 19
3 Getting started with BIGSTICK 22
3.1 What can BIGSTICK do?....................... 22
3.2 Downloading and compiling the code . . . . . . . . . . . . . . . . 23
3.3 Requiredinputfiles.......................... 24
3.4 Runningthecode........................... 24
3.5 Somesampleruns .......................... 29
3.6 Typicalruntimes........................... 29
4 Using BIGSTICK, in detail 30
4.1 Overview of input files . . . . . . . . . . . . . . . . . . . . . . . . 30
4.2 Defining the model space . . . . . . . . . . . . . . . . . . . . . . 31
4.2.1 Particle-hole conjugation . . . . . . . . . . . . . . . . . . 35
4.2.2 Truncation of the many-body space . . . . . . . . . . . . 36
4.2.3 Advanced truncation options . . . . . . . . . . . . . . . . 38
4.2.4 How to handle ‘different’ proton-neutron spaces . . . . . . 39
4.3 Interactionfiles............................ 40
4.3.1 Scaling and autoscaling . . . . . . . . . . . . . . . . . . . 41
4.3.2 Proton-neutron and other isospin-breaking formats . . . . 42
4.3.3 MFDn formatinput...................... 45
1
4.3.4 Three-body forces . . . . . . . . . . . . . . . . . . . . . . 46
4.4 Primary runtime options . . . . . . . . . . . . . . . . . . . . . . . 47
4.4.1 Autoinput........................... 47
4.4.2 Standard or normal runs . . . . . . . . . . . . . . . . . . . 47
4.4.3 One-body density matrices and occupations . . . . . . . . 48
4.4.4 Other primary options . . . . . . . . . . . . . . . . . . . . 49
4.5 Diagonalization options . . . . . . . . . . . . . . . . . . . . . . . 50
4.5.1 Convergence ......................... 52
4.6 Secondary runtime options . . . . . . . . . . . . . . . . . . . . . . 53
4.6.1 Expectation value . . . . . . . . . . . . . . . . . . . . . . 54
4.6.2 Applying a one-body transition operator . . . . . . . . . . 55
4.6.3 Applying a two-body body scalar operator . . . . . . . . . 56
4.6.4 Generating strength function distributions . . . . . . . . . 56
4.6.5 Overlap or dot product of wave functions . . . . . . . . . 57
4.7 Outputfiles.............................. 58
4.7.1 Secondary files . . . . . . . . . . . . . . . . . . . . . . . . 59
4.7.2 Diagnosticfiles ........................ 59
4.8 Memoryusage ............................ 59
5 Applications 61
5.1 One-body density matrices . . . . . . . . . . . . . . . . . . . . . 61
5.1.1 Symmetries of density matrix elements . . . . . . . . . . . 63
5.1.2 Particle occupations from densities . . . . . . . . . . . . . 63
5.1.3 Strengths from density matrix elements . . . . . . . . . . 63
5.1.4 Sample case: spin-flip . . . . . . . . . . . . . . . . . . . . 65
5.1.5 Charge-changing transitions . . . . . . . . . . . . . . . . . 66
5.1.6 Sample case: 19F....................... 67
5.2 Strength function option . . . . . . . . . . . . . . . . . . . . . . . 67
5.2.1 Decomposition . . . . . . . . . . . . . . . . . . . . . . . . 67
5.2.2 Transition strength function distributions: the basics . . . 70
5.2.3 Transitions with good angular momentum . . . . . . . . . 72
5.2.4 Gamow-Teller with strength function option . . . . . . . 74
6 A peek behind the curtain 77
6.1 Anormalrun............................. 77
6.2 Explicit representation of the basis . . . . . . . . . . . . . . . . . 78
6.3 Explicit representation of the Hamiltonian . . . . . . . . . . . . . 80
7 Lanczos algorithm 81
7.1 Standard Lanczos algorithm . . . . . . . . . . . . . . . . . . . . . 81
7.2 Thick-restart Lanczos . . . . . . . . . . . . . . . . . . . . . . . . 83
7.3 Can I restart standard Lanczos? . . . . . . . . . . . . . . . . . . 85
2
8 Parallel computing and timing 86
8.1 MPI .................................. 87
8.1.1 Fragments........................... 87
8.1.2 Opbundles and optypes . . . . . . . . . . . . . . . . . . . 88
8.1.3 Modeling ........................... 89
8.2 OpenMP................................ 89
8.3 Timing................................. 90
8.3.1 Modetimes.......................... 90
8.3.2 Timing for parallel runs . . . . . . . . . . . . . . . . . . . 91
A Matrix elements and operators 92
A.1 Reduced matrix elements . . . . . . . . . . . . . . . . . . . . . . 92
A.2 The Hamiltonian and other operators in second quantization . . 93
A.3 Symmetries of matrix elements . . . . . . . . . . . . . . . . . . . 94
B Highlighted references 95
3
Chapter 1
Introduction
There are many approaches to the quantum many-body problem. BIGSTICK is
a configuration-interaction many-fermion code, written in Fortran 90. It solves
for low-lying eigenvalues of the Hamiltonian of a many fermion system; it does
this by creating a basis of many-body states of Slater determinants (actually, the
occupation representation of Slater determinants). The Slater determinants are
antisymmetrized products of single-particle states with good angular momen-
tum, typically derived from some shell-model-like potential; hence we call this
a shell-model basis. The Hamiltonian is assumed to be rotationally invariant
and to conserve parity, and is limited to two- and, optionally, three-body forces.
Otherwise no assumptions are made about the form of the single-particle states
or of the Hamiltonian.
The capabilities of BIGSTICK will be detailed below, but in addition to calcu-
lating the energy spectra and occupation-space wavefunctions, it can compute
particle occupations, expectation values of operators, and static and transition
densities and strengths. Most of the applications to date have been in low-
energy nuclear physics, but in principle any many-fermion system with two
fixed ‘species’ and rotational symmetry can be addressed by BIGSTICK, such
as the electronic structure of atoms and cold fermionic gases in a spherically
symmetric trap; although we have yet to publish papers, we have carried out
demonstration calculations for such systems, with ‘spin-up’ and ‘spin-down’ re-
placing ‘proton’ and ‘neutron.’ We apologize to any atomic physicist who will
have to translate our terminology.
In this next chapter we review the basic many-body problem. Chapter 2
outlines the configuration-interaction method and discusses in broad strokes
the principles of the algorithms in BIGSTICK. Chapter 3 gives an introduction
to how to compile and run BIGSTICK, while Chapter 4 goes into running the
code more detail. If you are interested in running BIGSTICK immediately,
go directly to Chapter 3.
In this manual we do not give substantial information on the inner workings
of the code. With the distribution you will find an as-yet incomplete Inside
Guide which provide many details. In addition, the code itself is heavily com-
4
mented. While internal information in BIGSTICK is highly compressed through
factorization, a technique outlined in Chapter 2, it is possible to get out explicit
representations of the many-body basis states and the many-body Hamiltonian
matrix; see Chapter 6. Chapter 7 discusses our use of the Lanczos algorithm.
Finally, parallel capabilities of the code is discussed in Chapter 8.
1.1 Expectations of users
Who do we expect to use BIGSTICK, and how do we expect them to use it?
We designed BIGSTICK to be run on a variety of platforms, from laptops to
leadership-class supercomputers. We also imagined, and tried to design, BIGSTICK
for a spectrum of users, with various expectations of them.
A crucial point for any and all users: BIGSTICK requires at least two
kinds of input files to run, a description of the single-particle space and a
file of interaction matrix elements. While we supply with the distribution a
number of example input files. it is important for both novice and routine users
to understand that such examples are just the beginning and not the sum of
nuclear physics. In general it is up to the user to provide interaction
files. We can use the .int interaction files usable by NuShell/NuShellX as
well as the interaction files used by MFDn.
It is also equally important to not ask BIGSTICK to be smarter than you are.
While BIGSTICK employs many error traps to avoid or at least flag the most
common mistakes, the principle of “garbage in, garbage out” still applies.
While this manual provides a fairly comprehensive introduction to running
BIGSTICK, it is not a detailed tutorial in configuration-interaction methods, the
atomic or nuclear shell models, or to basic nuclear physics. We expect the reader
to, above all, be comfortable with non-relativistic quantum mechanics (i.e., to
fully understand the Schr¨odinger equation and with Dirac’s bra-ket notation),
and to be fluent of the ideas and terminology of the shell model, especially the
nuclear shell model, and to understand the basic principles of configuration-
interaction methods. We review the latter in the opening of Chapter 2, so that
is a good place to start to check your level of comfort. We suggest additional
references in Appendix B.
1.2 How to cite and copyright notices/licenses
If you successfully use BIGSTICK in your research, please use the following cita-
tions:
•C. W. Johnson, W. E. Ormand, and P. G. Krastev, Comp. Phys. Comm.
184, 2761-2774 (2013). (You can also find this article at arXiv:1303.0905.)
•C. W. Johnson,W. E. Ormand, K. S. McElvain, and H. Z. Shan, UCRL number
LLNL-SM-739926, arXiv:XXXXX (this report)
The first paper, Johnson et al. [2013], in particular discusses the underlying
factorized on-the-fly algorithm. This documents focuses instead on how to run
5
BIGSTICK.
This code is distributed under the MIT Open Source License:
Copyright (c) 2017 Lawrence Livermore National Security and the San Diego
State University Research Foundation.
Permission is hereby granted, free of charge, to any person obtaining a copy
of this software and associated documentation files (the ”Software”), to deal in
the Software without restriction, including without limitation the rights to use,
copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the
Software, and to permit persons to whom the Software is furnished to do so,
subject to the following conditions:
The above copyright notice and this permission notice shall be included in
all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED ”AS IS”, WITHOUT WARRANTY OF
ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO
THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTIC-
ULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE
AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM,
DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CON-
TRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CON-
NECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS
IN THE SOFTWARE.
1.2.1 LAPACK copyright notice
We use LAPACK subroutines in our code. The following are the LAPACK
copyright notices.
Copyright (c) 1992-2013 The University of Tennessee and The University of
Tennessee Research Foundation. All rights reserved.
Copyright (c) 2000-2013 The University of California Berkeley. All rights
reserved.
Copyright (c) 2006-2013 The University of Colorado Denver. All rights re-
served.
Additional copyrights may follow
Redistribution and use in source and binary forms, with or without modifi-
cation, are permitted provided that the following conditions are met:
- Redistributions of source code must retain the above copyright notice, this list
of conditions and the following disclaimer.
- Redistributions in binary form must reproduce the above copyright notice,
this list of conditions and the following disclaimer listed in this license in the
documentation and/or other materials provided with the distribution.
6
- Neither the name of the copyright holders nor the names of its contributors
may be used to endorse or promote products derived from this software without
specific prior written permission.
The copyright holders provide no reassurances that the source code provided
does not infringe any patent, copyright, or any other intellectual property rights
of third parties. The copyright holders disclaim any liability to any recipient
for claims brought against recipient by any third party for infringement of that
parties intellectual property rights.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS
AND CONTRIBUTORS ”AS IS” AND ANY EXPRESS OR IMPLIED WAR-
RANTIES, INCLUDING, BUT NOT LIMITED TO, THE IMPLIED WAR-
RANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR
PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT
OWNER OR CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT,
INCIDENTAL, SPECIAL, EXEMPLARY, OR CONSEQUENTIAL DAMAGES
(INCLUDING, BUT NOT LIMITED TO, PROCUREMENT OF SUBSTI-
TUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS; OR
BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THE-
ORY OF LIABILITY, WHETHER IN CONTRACT, STRICT LIABILITY, OR
TORT (INCLUDING NEGLIGENCE OR OTHERWISE) ARISING IN ANY
WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF ADVISED OF
THE POSSIBILITY OF SUCH DAMAGE.
1.3 Reporting bugs and other issues
If you run into trouble, first read this manual. Most issues are caused by mistakes
in setting up input files, in particular inconsistencies between the single-particle
space defined and the interaction file(s). Second, please read the output carefully:
we have striven to write detailed error traps and often BIGSTICK will notify the
user of problems. Try running the sample cases and make sure they run to
correct completion and that you understand the inputs.
If, having exhausted all the resources presented here, you still have a prob-
lem, you may send your issue to Calvin Johnson, cjohnson@mail.sdsu.edu.
In particular send a copy of the entire output written to screen, which often
contains important clues, the input files, and all output files with the extensions
.res,.log and .bigstick. Although we hope to be able to help, we cannot
guarantee it.
As discussed elsewhere, BIGSTICK is developed for Linux and Linux-like envi-
ronments such as Mac OS X. We have made no attempt to adapt to a Windows
environment. Although it has a user-friendly menu-driven interface, it still as-
sumes a reasonable facility with many-body physics and in particular low-energy
nuclear physics.
Development of BIGSTICK is ongoing. We hope to release future versions of
the code as additional major capabilities come on line.
7
1.4 A brief history of BIGSTICK, and acknowl-
edgements
In 1997, when two of us (Ormand and Johnson) were both at Louisiana State
University, we decided to write our own many-fermion configuration-interactionI
code christened REDSTICK, English for Baton Rouge. Over the next decade
REDSTICK evolved and improved. Most important were the addition of three-
body forces and parallelization. As it approached the ten-year mark, we noticed
certain limitations, particularly in the set-up, and starting in 2007 we began
developing new algorithms.
By this time, Ormand had moved to Lawrence Livermore National Labora-
tory and Johnson had left for San Diego State University. Working first with
a student (Hai Ah Nam) and later a postdoc (Plamen Krastev) at San Diego
State University, we carefully studied bottlenecks in parallelization in the ap-
plication of the Hamiltonian. These studies led us to break up the application
of the Hamiltonian by basis sectors, which had two useful outcomes. First,
we rewrote our central application routines using simple arrays rather than the
derived types used in REDSTICK; this gave a speed-up of nearly a factor of 2.
Second, applying the Hamiltonian by quantum numbers allowed a more trans-
parent factorization of the Hamiltonian and better parallelization.
With these improvements and dramatic speed-ups, we had an entirely new
code, BIGSTICK.
Starting around 2014, through the good graces of Wick Haxton we teamed up
with UC Berkeley and Lawrence Berkeley Laboratory, and especially Haxton’s
graduate student Ken McElvain. Ken’s background in the computer industry
proved invaluable, and he was able to tweak the existing code into fantastic per-
formance, especially with regards to parallelism. Hongzhang Shan of Lawerence
Berkeley wrote an improved algorithm for using OpenMP in matvec operations.
In addition to the help of Hai Ah Nam and Plamen Krastev, we would also
like to thank Esmond Ng, Chao Yang, and Sam Williams, of Lawrence Berkeley
National Laboratory, James Vary and Pieter Maris of Iowa State University,
and many other colleagues who have provided helpful discussions, suggestions,
feedback and insight over the years. Jordan Fox helped find some bugs in this
most recent version, and Stephanie Lauber helped find typos and confusing
statements in this manual.
Over the years our primary research funding has come through the U.S.
Department of Energy, which has directly and often indirectly supported the
development of BIGSTICK. We are deeply grateful for this support. Support
for this project came primarily from the U.S. Department of Energy, in the
form of grants Grant DE-FG02-96ER40985, DE-FG52-03NA00082, DE-FG02-
03ER41272, as well as Louisiana State University, Lawrence Livermore National
Laboratory, San Diego State University, University of California, Berkeley, and
Lawrence Berkeley National Laboratory.
8
Chapter 2
How we solve the
many-body problem
In this chapter we discuss the principles of configuration-interaction (CI) many-
body calculations [Shavitt, 1998, Brussard and Glaudemans, 1977, Brown and
Wildenthal, 1988, Caurier et al., 2005, Cook, 1998, Jensen, 2017, Weiss, 1961,
L¨owdin, 1955, Sherrill and Schaefer, 1999], including some different classes of
CI codes, and give an overview of its application in BIGSTICK. Configuration-
interaction is sometimes called the interacting shell model, as (a) one typically
builds the many-body basis from spherical shell-model single particle states and
(b) to distinguish from the non-interacting shell model, sometimes also called
the independent particle model.
The key points here are:
•We represent the many-body Schr¨odinger equation as a matrix eigenvalue
problem, typically with very large basis dimensions. BIGSTICK can com-
pute problems with dimensions up to ∼107on a laptop, up to ∼108on
a desktop machine, and up to ∼1010 on parallel supercomputers.
•The large-basis-dimension eigenvalue problem has two computational bar-
riers. The first is how to solve the eigenvalue problem itself, especially
given that we almost never need all of the eigenvalues. The second is,
despite the fact the matrix is typically very sparse, the amount of data
required is still huge.
•We address the first problem by using the Lanczos algorithm, which effi-
ciently yields the low-lying eigenpairs.
•We address the second by not explicitly storing all the non-zero matrix
elements, but instead invoking a on-the-fly algorithm. This on-the-fly algo-
rithm, first implemented in the Strasbourg group’s code ANTOINE [Caurier
and Nowacki, 1999], exploits the fact that the interaction only acts on two-
9

or three- particles at a time. The on-the-fly algorithm can be thought of
as partially looping over spectator particles.
•The on-the-fly algorithm explicitly depends upon the existence of two
species of particles, for example protons and neutrons, or in the case of
atoms, spin-up and spin-down electrons, so that both the many-body basis
and the action of the Hamiltonian can be factorized into two components.
This factorization is guided by additive/multiplicative quantum numbers,
such as M, the z-component of angular momentum, and parity. This fac-
torization efficiently and losslessly “compresses” information; we outline
the basic concepts below.
•In order to implement many-body truncations, we have an additional ad-
ditive pseudo-quantum number, which we call W. This allows a general,
though not infinitely flexible, ability to truncate the basis. We discuss
these truncations below, but include for example n-particle, n-hole trun-
cations and the Nmax truncation typical of the no-core shell model.
With these efficiencies we can run both “phenomenological” and ab initio
or no-core shell model calculations, on machines ranging from laptops to super-
computers. Although we do not discuss it in depth in this document, we rely
heavily upon both factorization and use of quantum numbers in parallelization.
2.1 Matrix formulation of the Schr¨odinger equa-
tion
The basic goal is to solve the non-relativistic many-body Schr¨odinger equation
for Aidentical fermions of mass M,
A
X
i=1 −∇2
i
2M+X
i<j
V(~ri−~rj)
Ψ(~r1, ~r2, . . . , ~rA) = EΨ(~r1, ~r2, . . . , ~rA),(2.1)
which we often will write using the more compact Dirac bra-ket notation
ˆ
H|Ψi=E|Ψi.(2.2)
Already even Eq. (2.1) is simplified, as it leaves out explicit spin degrees of
freedom, and the potential here is purely local and two-body. BIGSTICK can
handle nonlocal interactions without blinking. BIGSTICK can also use three-
body forces, although the latter ups computational demands by nearly two
orders of magnitude, and in the current release the three-body forces are not
optimized.
The basic idea of configuration interaction is to expand the wavefunction in
some convenient many-body basis {|αi}:
|Ψi=X
α
cα|αi(2.3)
10
Then, if the basis states are orthonormal, hα|βi=δα,β , the Schr¨odinger equation
becomes a matrix eigenvalue equation
X
β
Hα,βcβ=Ecα.(2.4)
Because we typically deal with many-fermion systems, the wavefunction |Ψi
is completely antisymmetric under interchange of any two particles,
Ψ(~r1, ~r2, . . . , ~ri, . . . , ~rj, . . .) = −Ψ(~r1, ~r2, . . . , ~rj. . . , ~ri, . . .).(2.5)
(One can use configuration-interaction methods for many-boson systems, but
then the basis states would be totally symmetric, and a completely separate
code would be required.) A useful many-body basis are therefore Slater deter-
minants, which are antisymmetrized products of single-particle wavefunctions.
(As we will note several times in this manual, it is often important to distin-
guish between single-particle states and many-body states, as well as between,
for example, two-body matrix elements and many-body matrix elements.)
We do not explicitly use Slater determinants but rather the occupation rep-
resentation of Slater determinants using fermion creation and annihilation oper-
ators, also known as second quantization. We assume the reader is comfortable
with Slater determinants and the algebra of fermion operators, and therefore
give only a terse exposition in order to be clear about our terminology.
Suppose we have a set of Nssingle-particle states, φi(~r) where idescribes
each unique state by its quantum numbers. BIGSTICK assumes single-particle
states with rotational symmetry, and the available quantum numbers are n,
l,j, and m. Here lis the orbital angular momentum, jis the total angular
momentum, and mis the zcomponent of total angular momentum. nis the
‘radial’ quantum number; it distinguishes different states with the same angular
momentum quantum numbers but a different radial wavefunction. It plays no
other internal role in BIGSTICK, though it is relevant to calculating the value
of matrix element input into the code. BIGSTICK can use single-particle states
with arbitrary radial components, as long as they orthonormal; it is up to the
user to keep track of what radial wavefunction is being assumed. In many cases,
for example in so-called no-core shell model (NCSM) calculations, one uses a
harmonic oscillator basis, but that is by no means mandatory. In the same way,
lreally only gives the parity of each single-particle state.
Once a single-particle basis is defined, second quantization allows us to define
many-body states. Starting with a fermion vacuum state |0i, the operator ˆa†
i
creates the single fermion state φi. Then the many-body state
ˆa†
i1ˆa†
i2ˆa†
i3. . . |0i(2.6)
is the occupation representation of the Slater determinant of the single particle
states φi1, φi2, . . .. For succinctness we will refer to such many-body states as
‘Slater determinants’ even when we mean the occupation representation.
11
Using a one-body operator such as the kinetic energy ˆ
Tcan be written using
second quantization:
ˆ
T=X
ij
Tij ˆa†
iˆaj,(2.7)
where Tij =hi|ˆ
T|ji=Rφ∗
iˆ
T φjis the one-body matrix element of the operator;
the actual value is determined through the integral sketched above. Two-body
operators, e.g. interactions between two particles, can be similarly represented,
though with two annihilation operators followed by two creation operators. It
is useful to note that all BIGSTICK and similar CI codes read in are numerical
values of the matrix elements. This means the actual form of the single-particle
wavefunctions is hidden (although BIGSTICK, like nearly all other nuclear CI
codes, requires single-particle states to have good angular momentum).
The many-body matrix elements are thus exercises in fermion second quan-
tization algebra: Hαβ =hα|ˆ
H|βiwhere the basis states |αi,|βiand the Hamil-
tonian operator ˆ
Hare all expressed using creation and annihilation operators,
given exactly in Appendix A.2.
2.2 Representation of the basis
The occupation representation is a natural one for the computer as a single
particle state can either be occupied or unoccupied, represented by a 1 or a 0.
Thus the state
ˆa†
2ˆa†
4ˆa†
5ˆa†
8|0i
can be represented by the bit string
01011001
as the single particle states 2,4, 5 and 8 are occupied and the rest unoccupied. Of
course, consistency in ordering is important as one has to pay careful attention
to phases.
In the 1970s Whitehead and collaborators used bit manipulation for fast
calculation of matrix elements in the occupation scheme [Whitehead et al., 1977].
The basic idea is simple: consider a creation operator, say ˆa†
4, applied to some
Slater determinant represented by a bit string. If the 4th bit is 0, then the
action of applying ˆa†
4is to create a 1 in its place:
ˆa†
3|110001i=|110101i,
while if it is already occupied, then the state vanishes in a puff of digital smoke:
ˆa†
3|100101i= 0.
Similarly an annihilation operator such as ˆa2will destroy a state if the second
bit is empty
ˆa2|100111i= 0,
12
but will replace a 1 bit with a 0,
ˆa2|110101i=−|100101i.
The minus sign arises, of course, from fermion anticommutation relations. In
this way one can almost trivially find the action of, say, a two-body operator on
a state:
ˆa†
3ˆa†
5ˆa4ˆa1|1101011i=−ˆa†
3ˆa†
5|0100011i=−|01101011i.
Then one can search through the basis to find out what state |01101011iis.
In general we work with Hamiltonians which are rotationally invariant. This
means one can find eigenstates of the Hamiltonian which are also simultaneous
eigenstates of total angular momentum ˆ
J2=ˆ
J2
x+ˆ
Jy,2+ˆ
J2
zand of (by conven-
tion) ˆ
Jz, that is,
ˆ
H|Ψi=E|Ψi;
ˆ
J2|Ψi= ¯h2J(J+ 1)|Ψi;
ˆ
Jz|Ψi= ¯hM |Ψi.
We say such states ‘have good angular momentum.’ It is important to note that
Egenerally depends upon J, that is, except for special cases (usually involving
additional symmetries) states with different Jare not degenerate, for a given
value of Jthe value of Edoes not depend upon M. In practical terms, what this
means is that the Hamiltonian is block-diagonal in J; it is also block-diagonal
in M, but the blocks for the same Jbut different Mhave the exact same
eigenvalues.
Of course, whether or not the Hamiltonian is explicitly block diagonal de-
pends upon the choice of basis. We call these different choices basis ‘schemes.’
BIGSTICK, like most nuclear CI codes, constructs its many-body basis states
using single-particle states which also have good angular momentum, i.e., have
eigenvalues j(j+ 1) and mfor ˆ
J2and ˆ
Jz, respectively. (Here and hereafter we
set ¯h= 1.) The addition of total angular momentum is nontrivial, requiring
Clebsch-Gordan coefficient, but as ˆ
Jzis the generator of an Abelian subgroup,
any product of single-particle states each with good mihas good total M=
m1+m2+. . ..
What this means is it is both possible and easy to construct individual Slater
determinants which have good M(i.e., are eigenstates of ˆ
Jz). These will almost
never be states also of good J. But because ˆ
Hcommutes with both ˆ
Jzand
ˆ
J2, if we take all states of a given Mand diagonalize ˆ
H, the eigenstates will be
guaranteed to also have good J(barring ‘accidental’ degeneracies that rarely
occur). Taking states of fixed Mis called an M-scheme basis. It is the simplest
shell-model basis.
But the M-scheme isn’t the only choice. As mentioned above, one can also
make the many-body Hamiltonian matrix explicitly diagonal in Jas well as
M. This is a J-scheme basis. Such bases are significantly smaller in dimension,
typically an order of magnitude smaller than the M-scheme. Of course, there
are obvious costs. Almost always a state with good Jmust ultimately be a
13
superposition of M-scheme Slater determinants. This means both the J-scheme
basis states, and the many-body matrix elements in this basis, are more costly
to calculate.
(Historically, in chemical and atomic physics one used configuration state
functions with good angular momentum, which we would call the J-scheme.
The use of simple Slater determinants in chemical and atomic physics seems
to have been introduced by Knowles and Handy [1984] apparently unaware of
Whitehead’s innovation.)
One can go even further. Many nuclei exhibit strong rotational bands, which
can be reproduced using the group SU(3). If the nuclear Hamiltonian commuted
with the Casimir operators of SU(3), or nearly so, then the Hamiltonian would
be block diagonal in the irreps of SU(3), or nearly so, and SU(3) would be
dynamical symmetry. One can imagine other group structures as well.
Because of this, some groups use group-theoretical bases, also called symmetry-
adapted bases, such as a SU(3)-scheme basis [Draayer et al., 2012], based upon
calculations which suggest that nuclear wavefunction are dominated by a few
group irreps. The SU (3)-scheme is just like the Jonly more so: the basis is more
compact, but the basis states and the many-body matrix elements even more
complicated to derive. On the other hand, the SU (3)-scheme makes the origin
of rotational motion more transparent and potentially offers a more compact
representation and understanding of the wavefunctions. Each of these schemes
offer advantages and disadvantages.
2.2.1 Use of quantum numbers: factorization of the basis
One of the advantages of the M-scheme is that despite the fact it is the least
compact of basis schemes, it can be represented very efficiently with factoriza-
tion. Factorization is an idea used throughout BIGSTICK, and is most easily
illustrated in the basis.
We work in the M-scheme, which means every many-body basis state has
the same definite value of M. If we have an even number of particles, Mis an
integer, while for odd numbers it will be a half-integer (1/2,3/2,−5/2, etc.).
Internally BIGSTICK doubles these so they can be represented by even or odd
integers, respectively.
Each basis state, however, is a simple tensor product of a proton Slater de-
terminant and a neutron Slater determinant. Because the mquantum numbers
are additive, we have the total M=Mp+Mn, the sum of proton and neutron
M-values.
Absent other constraints, every proton Slater determinant with Mpnot only
can but must be combined with every neutron Slater determinant with Mn=
M−Mp; this, in part, guarantees that rotational invariance is respected and
that the final eigenstates will have good total J. This in turn leads to a shortcut.
Consider the case of the 27Al nucleus, using the sd valence space. This
assumes five valence protons and six valence neutrons above a frozen 16O core.
The total dimension of the many-body space is 80,115, but this is constructed
using only 792 five-proton states and 923 six-neutron states.
14
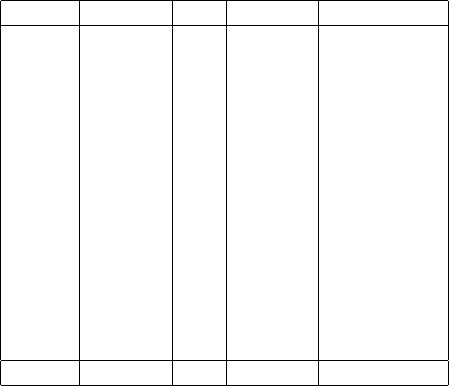
Table 2.1: Decomposition of the M-scheme basis for 5 protons and 6 neutrons
in the sd valence space (27Al), with total M=Mp+Mn+ 1/2. Here “pSD”
= proton Slater determinant and “nSD” = neutron Slater determinant, while
“combined” refers to the combined proton+neutron many-body basis states.
The subset of the basis labeled by fixed Mp(and thus fixed Mn) we label a
’sector’ of the basis.
Mp# pSDs Mn# nSDs # combined
+13/2 3 -6 9 27
+11/2 11 -5 21 231
+9/2 28 -4 47 1316
+7/2 51 -3 76 3876
+5/2 80 -2 109 8720
+3/2 104 -1 128 13,312
+1/2 119 0 142 16,898
-1/2 119 +1 128 15,232
-3/2 104 +2 109 11,336
-5/2 80 +3 76 6080
-7/2 51 +4 47 2444
-9/2 28 +5 21 588
-11/2 11 +6 9 99
-13/2 3 +7 1 3
Total 792 923 80,115
The reader will note that 792 ×923 = 731016 80115. Indeed, not every
five-proton state can be combined with every six-neutron state. The restric-
tion is due to conserving certain additive quantum numbers, and this restric-
tion turns out to limit usefully the nonzero matrix elements of the many-body
Hamiltonian, which we will discuss more in the next section.
For our example, we chose total M= +1/2 (though we could have chosen
a different half-integer value). This basis requires that Mp+Mn=M; and for
some given Mp,every proton Slater determinant with that Mpcombines with
every neutron Slater determinant with Mn=M−Mp. This is illustrated in
Table 2.1, which shows how the many-body basis is constructed from 792 proton
Slater determinants and 923 neutron Slater determinants. Note we are “miss-
ing” a neutron Slater determinant; the lone Mn=−7 state has no matching
(or ‘conjugate’) proton Slater determinants.
As a point of terminology, we divide up the basis (and thus any wavefunction
vectors) into sectors, each of which is labeled by Mp, and any additional quan-
tum numbers such as parity Πp; that is, all the basis states constructed with
the same Mp(Πp, etc.) belong to the same basis ‘sector’ and have contiguous
indices. Basis sectors are also useful for grouping operations of the Hamiltonian,
as described below, and can be the basis for distributing vectors across many
processors, although because sectors are of different sizes this creates nontrivial
issues for load balancing.
15
While we can represent the 80,115 basis states of 27Al in the sd with 792
proton Slater determinants and 923 neutron Slater determinants, the storage is
even more impressive for large systems. For example, in the pf shell, 60Zn, with
10 valence protons and 10 valence neutrons, has for M= 0 a basis dimension of
2.3 billion. But these are represented by ∼185,000 proton Slater determinants
and the same number of neutron Slater determinants. (In principle with self-
conjugate systems N=Zsystems one could gain further savings by keeping
only one set of Slater determinants. Because that is a small number of nuclides,
we chose not to do so.) The savings are not as dramatic for no-core shell model
calculations with Nmax truncation. For example, 12C in a basis or Nmax = 10
has a basis dimension of 7.8 billion, constructed from 1.8 million each proton
and neutron Slater determinants. The reason for the lessened efficiency is the
many-body truncation.
We note that factorization not only provides dramatic lossless compression
of data, it also accelerates the set up of data. In the set up phase of any CI
code, one of the major tasks is searching through long series of bitstrings and,
when one uses quantum numbers to organize the data, sorting. Factorization
improves this by reducing the length of lists to be searched and sorted. Our
second level of factorization further reduces those lists, making searches and
sorts even faster.
While factorization of the Hamiltonian was, to the best of our knowledge,
pioneered by Caurier and Nowacki [1999] in the code ANTOINE and adopted as
well by EICODE [Toivanen, 2006], NuShell/NuShellX [Brown and Rae, 2014],
and KSHELL [Shimizu, 2013] (and possibly others we are unaware of), BIGSTICK
has uniquely implemented a second level of factorization. Because most users
never see this level, we direct those interested to our paper for more details.
BIGSTICK does provide some information about this. In normal runs, as well
as in modeling runs, you will see
.... Building basis ...
Information about basis:
there are 27 sectors 1
there are 27 sectors 2
38760 SDs for species 1
184722 SDs for species 2
Total basis = 501113392
.... Basis built ...
The above example is for 56Fe in the pf shell with M= 0. The sectors are
the subsets of the proton and neutron Slater determinants (‘SDs’) with fixed
quantum number M, parity, and optionally W. Here ‘species 1’ refers to protons
and ‘species 2’ refers to neutrons.
16
2.3 The Lanczos algorithm and computational
cost
With bit manipulation allowing one to quickly calculate matrix elements, one
could address much larger spaces, spaces so large they were not amenable
to complete diagonalization, e.g., through the Householder algorithm [Parlett,
1980, Press et al., 1992]. But in nuclear structure configuration-interaction one
almost never wants all the eigensolutions; instead one typically just wants the
low-lying states. Thus Whitehead et al. [1977] introduced another innovation:
use of the Lanczos algorithm to find the extremal eigenstates.
The Lanczos algorithm is a subspecies of Arnoldi algorithms. We describe
the Lanczos algorithm in Chapter 7, but the key idea is that, starting from
an initial vector often called the pivot, one iteratively constructs a sequence of
orthonormal basis vectors that form a Krylov subspace, as well as the elements
of the Hamiltonian in that subspace. The genius of Arnoldi/Lanczos algorithms
is that they use the matrix to be diagonalized to construct the basis vectors; by
applying the Hamiltonian matrix to a given basis vector one constructs, after
orthogonalization, the next basis vector. One can show via the classical theory
of moments that the extremal eigenvalues of the Hamiltonian in the Krylov
subspace quickly converge to those of the full space. Although it depends upon
the model space, the Hamiltonian, and the choice of pivot (starting vector to
kick off the Lanczos iterations), one can often reach a converged ground state
energy in as few as twenty Lanczos iterations, and the lowest five states in as
few as 100 iterations.
Now let us think about the computational cost of carrying out CI, both
in terms of operations (time) and memory (storage). Before doing so let us
highlight a key point. Most often in discussing CI one cites the basis dimen-
sion. But, as we will argue below, the real measure of the computational cost is
the number of nonzero matrix elements. Now, for any given scheme, the num-
ber of nonzero matrix elements scales with the basis dimension. However, for
different schemes the proportionality is different: J-scheme is denser than M-
scheme; furthermore, even within the same basis scheme, different truncations
have different densities, e.g., the NCSM is much denser than ‘phenomenological’
calculations. Therefore, for absolute comparison of the computational cost of a
problem, the number of nonzero matrix elements is a much better measure than
basis dimension.
That said, let us look at the computational cost of matvec:
wα=X
β
Hα,βvβ.
Let the dimension of the vector space be N. If the many-body matrix His
a fully dense (but real, symmetric) matrix, the above matvec requires N2
operations as well as storage of N2many-body matrix elements. However, His
almost never fully dense. This can be most easily understood in the M-scheme,
where the fundamental occupation-space basis states can be represented as raw
17
bit strings. A two-body interaction can at most shift two bits. Therefore if
two basis states |αiand |βidiffer by more than two bits, the matrix element
between them must be zero. A typical ‘sparsity’ of M-scheme Hamiltonians is
2×10−6, that is, only two out of every million many-body matrix elements is
nonzero. (Three-body forces, naturally, lead to denser matrices, roughly two
orders of magnitude denser.)
If one has a basis dimension of a million, then there are roughly a million
nonzero matrix elements; because one needs not only the value of the matrix
element but some index to local it in the matrix, in single precision this requires
roughly 8 megabytes of memory. When one goes up to a basis dimension of one
billion, however, this number goes up by (103)2to 8 terabytes! Reading 8 Tb
of data even from fast solid state disks is a very slow proposition. If one stores
the matrix elements in core memory across many processors, as the code MFDn
does, this requires a minimum of many hundreds if not thousands of processors.
2.4 Representation of the Hamiltonian
As discussed above, M-scheme configuration-interaction calculations require a
many-body vector space of very large dimensions, and the many-body Hamil-
tonian matrix, while very sparse, still in large cases nonetheless the nonzero
matrix elements end requiring a huge amount of data.
If you were to examine closely, say, the bit representation of the basis states,
or the nonzero matrix elements, you’d find something confounding: quite a lot of
data is repeated, over and over. The same proton bit strings (which we generally
call proton Slater determinants, although technically they are representations of
said determinants) are repeated many times, sometimes many millions of times
or more, and the same for the neutron bit strings (neutron Slater determinants).
In the same way, the same values appear, thousands and millions of times, in the
non-zero many-body matrix elements, though with both positive and negative
values.
This redundancy can not only be understood, it can be turned to our ad-
vantage through factorization, both of the basis and of matvec operations.
The idea is similar to the factorization of the basis. Any two-body Hamil-
tonian can be split into forces that act only on protons, forces that act only
on neutrons, and interactions between protons and neutrons. Consider forces
acting only on protons; in a factorized basis, the neutrons are spectators. If
we write our basis states as a simple tensor product between a proton ‘Slater
determinant’ |ipiand and a neutron Slater determinant, |jni, so that the basis
state |αi=|ipi|jnithe pure proton Hamiltonian matrix element is
hα|ˆ
Hpp|α0i=hip|ˆ
Hpp|i0
piδjn,j0
n.(2.8)
We therefore only have to store the proton matrix element hip|ˆ
Hpp|i0
pi, and can
trivially loop over the neutron Slater determinants. You can see how you could
get dozens, hundreds, or thousands of matrix elements with the same value,
18
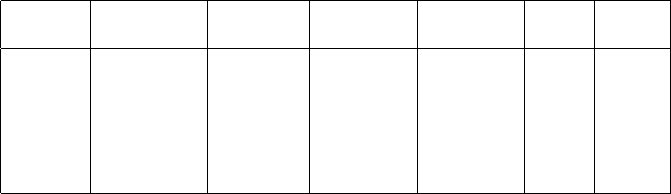
Table 2.2: Number of one- and two-body ‘jumps’ and storage requirements for
representative atomic nuclei in different model spaces (described in Appendix
B). For storage of nonzero matrix elements (penultimate column) we assume
each many-body matrix element is stored by a 4-byte real number and its lo-
cation encoded by a single 4-byte integer. Storage of a single jump (initial and
final Slater determinants for a species, and matrix element and phase) requires
13 bytes. All storage (final two columns) are in gigabytes (GB).
Nuclide space basis # 1-body # 2-body Store Store
dim jumps jumps m.e.s jumps
28Si sd 9.4×1044.8×1047.6×1030.2 0.002
52Fe pf 1.1×1084.0×1068.5×106700 0.16
56Ni pf 1.1×1091.5×1074.0×1079800 0.6
4He Nmax = 22 9 ×1075.3×1084.7×1099300 69
12CNmax = 8 6 ×1086×1083×1095200 45
13CNmax = 6 3.8×1077×1073×108210 4.3
just with different jn. Furthermore, because the neutron Slater determinants
are frozen, the quantum numbers cannot change, which severely restricts the
action of the proton-only Hamiltonian. The matrix elements hip|ˆ
Hpp|i0
piare
called jumps and we only need to store them (and know of the neutron indices
jnover which to loop).
For proton-neutron interactions the action is more complicated but the same
basic ideas hold: one stores separate proton jumps and neutron jumps and
reconstructs the value of the matrix element. Table 2.2 shows the storage for
nonzero matrix elements and of jumps needed for a number of representative
nuclei, in both phenomenological and NCSM calculations. You can see there
are at least two orders of magnitude difference. Thus, for example, 52Fe, which
would require 700 Gb of storage for just the nonzero matrix elements, only needs
less than a Gb for storage in factorization (in this particular case, storage of
the Lanczos vectors is much higher burden) and thus can be run on an ordinary
desktop computer.
The price one pays, of course, is the factorized reconstruct-on-the-fly algo-
rithm is much more complicated.
2.5 An incomplete survey of other codes
While this manual is about BIGSTICK, it is appropriate to put it in the context of
other (nuclear) configuration-interaction codes. One can broadly classify them
by (a) basis scheme, (b) representation and storage of many-body matrix el-
ements, (c) rank of interactions (i.e., two-body only or two- and three-body
forces), (d) parallelism, if any, and finally (e) general area of applicability, e.g.,
primarily to phenomenological spaces, which usually means a frozen core, and
interactions, or to ab initio no-core shell model calculations. Please keep in
mind that most of these codes are unpublished or have only partial informa-
19
tion published, and that many of the details have been gleaned from private
conversations; information on some codes, such as the powerful Japanese code
MSHELL, do not seem to be available. We apologize for any accidental misrep-
resentations. All of these codes have powerful capabilities and have made and
are making significant contributions to many-body physics.
Among the very earliest codes was the Oak Ridge-Rochester code from the
1950s and 1960s, which fully diagonalized the Hamiltonian after computing
the many-body J-scheme matrix elements via coefficients of fractional parent-
age. It was succeeded by the Whitehead (Glasgow) code and its descendents,
which used bit manipulation to compute the many-body matrix elements in the
M-scheme, and solved for low-lying eigenstates using the Lanczos algorithm,
ANTOINE [Caurier and Nowacki, 1999], MFDn [Sternberg et al., 2008], and KSHELL
[Shimizu, 2013] are also M-scheme codes. Examples of J-scheme codes in nu-
clear physics include OXBASH [Brown et al., 1985] and its successors NuShell
and NuShellX [Brown and Rae, 2014], NATHAN [Caurier et al., 1999], and EICODE
[Toivanen, 2006]. There have also been attempts to use group theory to con-
struct so-called symmetry-guided bases. The main effort is in SU(3) [Draayer
et al., 2012]. Although this approach is very promising, only time will tell for
sure if the advantages gained by group theory will outweigh the technical difficul-
ties needed to implement, for although the bases are small, they are significantly
denser, and furthermore the group theory is very challenging.
Regarding access to the many-body Hamiltonian matrix element, the Oak
Ridge-Rochester and Whitehead codes stored matrix elements on disk, as do
OXBASH and NuShell. The very successful code MFDn [Sternberg et al., 2008],
used primarily but not exclusively for no-core shell model calculations, stores the
many-body Hamiltonian matrix elements in RAM, much faster to access than
storing on disk, but for all but the most modest of problems requires distribution
across hundreds or thousands of nodes on a parallel computer spread across
many MPI processes.
Factorization methods, pioneered in ANTOINE [Caurier and Nowacki, 1999],
have been used in several other major CI codes: NATHAN[Caurier et al., 1999],
EICODE [Toivanen, 2006], NuShellX [Brown and Rae, 2014], KSHELL [Shimizu,
2013], and our own unpublished codes REDSTICK (so named because it was orig-
inated at Louisiana State University, located in Baton Rouge), and of course
BIGSTICK. Factorization has also been used in nuclear structure physics as a
gateway to approximation schemes [Andreozzi and Porrino, 2001, Papenbrock
and Dean, 2003, Papenbrock et al., 2004, Papenbrock and Dean, 2005]. The
codes most widely used by people beyond their authors have been OXBASH and its
successor NuShell/NuShellX, and ANTOINE. Because of their wide use, and be-
cause one of us (Ormand) heavily used OXBASH, the default formats for our input
.sps and .int files are heavily modeled upon the OXBASH/NuShell/NuShellX
formats.
Like BIGSTICK,MFDn has been parallelized with both MPI and OpenMP and
has carried out some of the largest supercomputer runs in the field. NuShellX
has only OpenMP parallelization. The parallelization of other codes is unknown.
In closing, we note that besides configuration-interaction there are many
20
other approaches to the many-body problem, such as the coupled cluster method,
the Green’s function Monte Carlo method, the in-medium similarity renormal-
ization group, density functional methods, and so on, each with their own ad-
vantages and disadvantages. There are also methods closely related to configu-
ration interaction, such as the ‘Monte Carlo shell model,’ the ‘shell-model Monte
Carlo,’ generator-coordinate codes, and the importance truncation shell model.
The main weakness of configuration interaction is that it is not size extensive,
which means unlinked diagrams must be cancelled and thus the dimensionality
of the problem grows exponentially with particle number and/or single-particle
basis. The advantages of CI is: it is fully microscopic; its connection to the
many-body Schr¨odinger equation (2.1) is pedagogically transparent; it gener-
ates excited states as easily as it does the ground state; it can handle even and
odd numbers of particle equally well and works well far from closed shells; and
finally places no restriction on the form of either the single-particle basis or on
the interaction (i.e., local and nonlocal forces are handled equally well, because
the occupation space basis is intrinsically nonlocal to begin with).
21
Chapter 3
Getting started with
BIGSTICK
BIGSTICK is a configuration-interaction many-fermion code, written in Fortran
90. It solves for low-lying eigenvalues of the Hamiltonian of a many fermion
system. The Hamiltonian is assumed to be rotationally invariant and to con-
serve parity, and is limited to two-body (and three-body, in progress) forces.
Otherwise few assumptions are made.
BIGSTICK allows for two species of fermions, such as protons and neutrons.
BIGSTICK is flexible, able to work with ”no-core” systems and phenomenolog-
ical valence systems alike, and can compute the electronic structure of single
atoms or cold fermionic gases (in which cases the two species are interpreted as
”spin-up” and ”spin-down”). BIGSTICK has a flexible many-body truncation
scheme that covers many common truncations. For nuclei it can assume isospin
symmetry or break isospin conservation. Interaction matrix elements must be
pre-computed by a third-party program and stored as a file, but BIGSTICK
accepts a variety of matrix element formats.
3.1 What can BIGSTICK do?
BIGSTICK can:
•compute the ground state energies and low-lying excitation spectra, in-
cluding angular momentum and, if relevant, isospin, of many-body sys-
tems with a rotationally invariant Hamiltonian; wave functions are also
generated;
•compute expectation values of scalar one- and two-body operators;
•compute one-body densities, including transition densities, among the low-
lying levels, which allows one to calculate transition rates, life times, mo-
ments, etc.;
22
•compute transition strength probabilities or strength functions for one-
body transition operators, useful when one needs to model transitions to
many excited states;
•use the strength function capability to decompose the wave function by
the eigenvalues of an operator, such as the Casimir of some group.
Along with this, one can ask, what are BIGSTICK’s limitations? This largely
depends upon the computer used and the many-body system. In low-energy
nuclear structure physics, which is the main focus of our research, one can eas-
ily run on a laptop any nuclide in the phenomenological sd space, and on a
workstation reach most nuclides in the phenomenological pf space. Although
dimensionality is not the most important determination of computational bur-
den, one can generally run cases of dimension up to a few million or even tens of
millions, if one is patient, on a laptop, a few hundred million on a workstation,
and a few billion on a parallel supercomputer.
As always, of course, much of the limitations depend upon the user. Al-
though we provide a few example input files, it is generally up to the user
to provide files for the model space, the interaction, and codes to postprocess
density matrices into transitions. (We do provide some tools for this.)
3.2 Downloading and compiling the code
BIGSTICK was developed for UNIX/Linux/MacOSX systems. We made no effort
to adapt it to running under Microsoft Windows.
To get BIGSTICK, download it from GitHub:
git clone https://www.github.com/cwjsdsu/BigstickPublick/
Your distribution includes a makefile. We have developed BIGSTICK to com-
pile and run successfully with Intel’s ifort compiler and GNU gfortran. You may
need to edit the makefile to put in the correct compiler and/or if you wish to use
for example LAPACK libraries. We have written the code to require minimal
special compile flags.
For example,
PROMPT> make serial
makes a serial version of the code with the Intel ifort compiler by default. Several
other options are:
PROMPT> make openmp →an OpenMP parallel version using ifort
PROMPT> make gfortran →a serial version using gfortran
PROMPT> make gfortran-openmp →an OpenMP version using gfortran
and so on. To see all the options encoded into the makefile,
PROMPT> make help
23
Each of these generates an executable with the nonstandard extension .x,
chosen to make deletion easy: bigstick.x, bigstick-openmp.x, bigstick-
-mpi.x, and bigstick-mp-omp.x. There are options for compiler on a number
of supercomputers. Please keep in mind, however, that compilers and compile
flags on supercomputers are a Red Queen’s Race, and it is up to the user to
tune the makefile for any given configuration.
Libraries. In routine operations, BIGSTICK uses the Lanczos algorithm to
reduce the Hamiltonian matrix to a truncated tridiagonal matrix whose eigen-
values approximate the extremal eigenvalues of the full matrix. This requires an
eigensolver for the tridiagonal. For modest cases, one can also choose to fully di-
agonalize the Hamiltonian, using a Householder algorithm. (In practice we find
this can be done quickly for basis dimensions up to a few thousand, and with
patience can be done up to a basis dimension ∼104.) For both cases we use the
LAPACK routine DSYEV, which solves the real-valued, double-precision symmet-
ric eigenvalue problem. The actual matrix elements are given in single-precision,
but we found when the density of eigenvalues is high, double-precision gives us
better values for observables, including angular momentum Jand isospin T.
Although in principle one could link to a library containing DSYEV, in practice
this is highly platform dependent. Also, except for special cases where one is
fully diagonalizing very large matrices and are impatient, the call to DSYEV is
a tiny fraction of the time. Hence we supply an unmodified copy of DSYEV and
required LAPACK routines, and there is no need to call any libraries.
3.3 Required input files
In order to solve the many-body Schr¨odinger equation, BIGSTICK requires at
least two inputs:
(a) A description of the single-particle space, usually through a file with exten-
sion .sps (although if one is running a no-core shell model calculation, there is
an option to generate this automatically); and
(b) A file containing the matrix elements of the interaction, in the form of
single-particle energies and two-body matrix elements (and, optionally, three-
body matrix elements).
We supply several example cases for both inputs, including some commonly
used spaces and interactions. But in general it is the user’s duty to supply
these input files and, importantly, to make sure they are consistent with each
other, i.e., to make sure the ordering of single-particle orbits in the .sps file
is consistent with those in the interaction file. We describe the file formats in
detail in Chapter 4
3.4 Running the code
BIGSTICK has a simple interactive input. It can also be run by pipelining the
input into the code.
24
To run:
PROMPT>bigstick.x
(we recommend you keep the source code, the executable, and the input
data files in separate directories, and make sure the executable is in your path).
We use the nonstandard extension .x to denote executables.
First up is a preamble, with the version number, information on parallel
processes, and a reminder for citations:
BIGSTICK: a CI shell-model code
Version 7.8.1 Aug 2017
by C. W. Johnson, W. E. Ormand,
K. S. McElvain, and H.Z. Shan
For reference please cite:
C. W. Johnson, W. E. Ormand, and P. G. Krastev
Comp. Phys. Comm. 184, 2761-2774 (2013)
[also found in arXiv:1303.0905]
This code distributed under Open Source License GPL.v2
Running on NERSC_HOST: none
scratch_dir (*.wfn,...): .
Number of MPI processors = 1
NUM_THREADS = 1
Next and most important is the main menu:
**************************************
* *
* OPTIONS (choose 1) *
* (i) Input automatically read from "autoinput.bigstick" file *
* (note: autoinput.bigstick file created with each nonauto run) *
* (n) Compute spectrum (default); (ns) to suppress eigenvector write up *
* (d) Densities: Compute spectrum + all one-body densities *
* (dx) Densities: Compute one-body densities from previous run *
* (x) eXpectation value of an operator (from previous wfn) *
* (?) Print out all options *
* *
**************************************
Enter choice
The most common choice is “n”for a normal run.
To facilitate batch runs or multiple runs with similar inputs, each time BIG-
STICK runs it creates a file autoinput.bigstick. This file can be edited;
choosing “i” from the initial menu will direct BIGSTICK to read all subsequent
commands from that file.
Next up:
25
Enter output name (enter "none" if none)
If you want your results stored to files, enter something like Si28run1.
The code will then create the following files:
Si28run1.res : text file of eigenenergies and timing information.
Si28run1.wfn: a binary file (not human readable) file of the wavefunctions for
post-processing or for other runs, e.g. “x” expectation values, etc.
Si28run1.log: a logfile of the run, useful for tracking the exact conditions
under which the run happened, as well as diagnosing problems.
Other files generated but not need by most users:
Si28run1.lcoef : text file of Lanczos coefficients;
timinginfo.bigstick and timingdata.bigstick: files on internal timing;
distodata.bigstick: a file contain information on distribution of work across
MPI processes;
and others used primarily by the authors for diagonsing behavior.
If you enter “none,” the .bigstick files will be created but no results file
(.res) and no wavefunction file (.wfn).
Enter file with s.p. orbit information (.sps)
(Enter "auto" to autofill s.p. orbit info )
This provides information about the single-particle space. A typical answer
might be sd, which tells BIGSTICK to open the file sd.sps, and read in informa-
tion about the sd valence space. (Please be aware that in most cases one does
not enter the extension, such as .sps or .int.) The auto option can only be
used for “no-core” nuclear shell-model calculations.
Enter # of protons, neutrons
These are the valence protons and neutrons. So, for example, if one wants to
compute 24Mg, which has 12 protons and 12 neutrons, but the sd single-particle
space assumes a closed 16O core, so one has 4 valence protons and 4 valence
neutrons. For other kinds of fermions, see the appendix.
Enter 2 x Jz of system
BIGSTICK is a “M-scheme” code, meaning the many-body basis states have fixed
total M=Jz(as opposed to J-scheme codes such as NuShell which the basis
has fixed total J). You must enter an integer which is twice the desired value
of M. If there are an even number of particles, this is usually 0. For an odd
number of nucleons, you must enter an odd integer, typically ±1. Because the
Hamiltonian is rotationally invariant, the results should not change for a value
±M. One can choose a non-minimal Mif, for example, you are interested in
high-spin states.
Enter parity +/- :
26
In addition to fixed M,BIGSTICK has fixed parity. BIGSTICK automatically
determines if more than one kind of parity is allowed and asks for the parity.
The sd space, for example, has only positive parity states, and so this input is
automatically skipped.
If you would like to compute both parities, enter ‘0’. (At the current time,
this is necessary if you want to compute parity-changing transitions, as for any
transition calculations BIGSTICK must work in the same basis.)
Would you like to truncate ? (y/n)
In some cases it is possible to truncate the many-body space, discussed in
detail in section 4.2.2.
BIGSTICK will then generate the basis; in most cases this takes only a fraction
of a second. BIGSTICK will print out some information about the basis, which
you can generally ignore.
The next item is to read in the matrix elements of the Hamiltonian.
Enter interaction file name (.int)
(Enter END to stop )
You can enter in a number of interaction files. The format for the interaction
files will be discussed below.
Enter scaling for spes, A,B,X ( (A/B)^X ) for TBMEs
(If B or X = 0, then scale by A )
After the interactions files have been read in, BIGSTICK sets up the jump arrays
for reconstructing the matrix elements on the fly. After that, the eigensolver
menu comes up:
/ ------------------------------------------------------------------------\
| |
| DIAGONALIZATION OPTIONS (choose one) |
| |
| (ex) Exact and full diagonalization (use for small dimensions only) |
| |
| (ld) Lanczos with default convergence (STANDARD) |
| (lf) Lanczos with fixed (user-chosen) iterations |
| (lc) Lanczos with user-defined convergence |
| |
| (td) Thick-restart Lanczos with default convergence |
| (tf) Thick-restart Lanczos with fixed iterations |
| (tc) Thick-restart Lanczos with user-defined convergence |
| (tx) Thick-restart Lanczos targeting states near specified energy |
| |
| (sk) Skip Lanczos (only used for timing set up) |
| |
\ ------------------------------------------------------------------------/
27
As noted, the standard choice is ‘ld’ for default Lanczos. Other options are
discussed later.
ld
Enter nkeep, max # iterations for lanczos
(nkeep = # of states printed out )
Except for very small cases, BIGSTICK does not find all the eigenvalues. In-
stead it uses the Lanczos algorithm (introduced by Whitehead et al to nuclear
physics) to find the low-lying eigenstates. The variable nkeep is the number of
targeted eigenpairs; typical values are 5-10. One can either set a fixed number
of iterations, typically 100-300, or set a maximal number of iterations and allow
BIGSTICK to stop sooner using a test for convergence (discussed in detail below).
BIGSTICK will then carry out the Lanczos iterations, printing out interme-
diate eigenvalues. The final result, which if a output file name was chose is also
written to the .res file, looks like
State E Ex J T
1 -149.77950 0.00000 0.000 0.000
2 -147.78011 1.99939 2.000 0.000
3 -144.92743 4.85207 4.000 0.000
4 -144.11148 5.66801 0.000 0.000
5 -142.72124 7.05826 3.000 0.000
This is fairly self-explanatory. Eis the absolute energy, Ex the excitation energy
relative to the first state, and Jand Tare the total angular momentum and
isospin, respectively. Even though only Mis fixed, because the Hamiltonian
commutes with ˆ
J2the final states will have good J. Lack of good Jmost likely
signals lack of convergence, or states degenerate in energy but with different
J). Lack of good Jcan also signal an error in the input file (specifically, a
disallowed Jfor a particular set of orbits; we have written error traps to catch
such a problem), or, lastly and only infrequently, a bug in the code itself.
If the input matrix elements respect isospin, then Tshould also be a good
quantum number. BIGSTICK allows one to read in isospin-breaking matrix ele-
ments, discussed in more detail in section 4.3.2.
BIGSTICK can also compute one-body density matrix elements at the end of
a run; choose option din the initial menu. The format and conventions for the
density matrices are in section 4.4.3.
The wavefunctions are saved to a .wfn file, unless you choose option ns
in the initial menu. BIGSTICK can then post-process the files, for example
computing the expectation value of a scalar (Hamiltonian-like) operator, section
4.6.1; compute overlap between wavefunctions from two different runs, section ;
or apply a non-scalar transition operator to a wavefunction and then compute
the strength distribution of that transition, sections 4.6.2, 5.2.2, and 5.2.3.
28
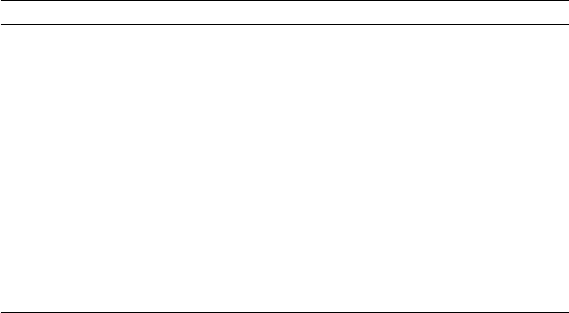
Nuclide space dim # ops min. store run time
24Mg sd 28,503 8.6M 34 Mb 5 s
48Cr pf 2 M 1.5 B 6 Gb 15 min
51Mn pf 44M 41B 160 Gb 9 hr
56Fe pf 500M 0.9 T 3.6 Tb 6 d
60Zn pf 2.3B 5T 20 Tb 35 d
12CNmax 6 32M 41 B 160 Gb 7 hr
6Li Nmax 12 49M 180 B 700 Gb 30 hr
12CNmax 8 594M 1.2T 5 Tb 8 d
16ONmax 8 1B 2 T 8 Tb 14 d
10BNmax 10 1.7B 5 T 20 Tb 35 d
6Li Nmax 16 800M 7T 27 Tb 46 d
Table 3.1: ‘Typical’ run times for various nuclides, running in serial for 150
Lanczos iterations. Here ‘min. store’ is an estimate of the minimal storage
required for nonzero matrix elements.
3.5 Some sample runs
In the directory SampleRuns that should be found in your distribution, you will
find various examples of runs, along with sample outputs to check the code is
working correctly.
3.6 Typical run times
In this section we survey ‘typical’ run times for calculations using BIGSTICK. Of
course, these depend upon the clock-speed of your chip as well as the compiler,
as well as how much parallelism you are exploiting. As we show below, BIGSTICK
does scale well in parallel mode.
Table 3.1 gives, for a variety of nuclides, the dimensionality of the space, the
number of operations (which is approximately though not exactly the number
of nonzero matrix elements), the minimal storage which would be required to
store the nonzero matrix elements, and finally an approximate run time, as-
suming 150 Lanczos iterations on a serial machine. The actual time may vary
a lot, depending on clock speed and how efficiently the operations are actually
processed. Parallelism, of course, can speed up the wall clock times considerably.
Empirically, one finds that the number of nonzero matrix element (here,
operations) generally scales like (dim)1.25 for two-body interactions, and ≈
(dim)1.5for three-body forces.
29
Chapter 4
Using BIGSTICK, in detail
BIGSTICK has two basic modes. It can calculate many-body spectra and wave
functions, and it can process those wave functions in several ways. In order to
generate the low-lying spectrum and wave functions, you need to, first, define
the model space, and second, provide an interaction.
4.1 Overview of input files
BIGSTICK uses three classes of externally generated files. Mandatory are: files
which define the single-particle space, and files for interaction matrix elements.
Optionally, BIGSTICK can also use files for one-body transition matrix elements.
Here we briefly summarize those files, and in later sections give more details.
Files which define the single-particle space have the extension either .sps or
.sp. When prompted, the user only supplies the name, not the extension, i.e.,
if the file is sd.sps only enter sd.BIGSTICK will automatically search for both
sd.sps and, if not found, then sd.sp. These files can assume isospin symmetry
or separate proton-neutron orbits, but at this time, BIGSTICK requires that the
proton and neutron single-particle spaces initially be the same. BIGSTICK can
however truncate the proton and neutron spaces differently.
If the user is carrying out a ‘no-core shell-model’ calculation where the single-
particle orbits are assumed to occur in a default order, BIGSTICK has an ‘auto’
option for defining the single-particle space and no input file is required.
BIGSTICK accepts two classes of files for interaction matrix elements. The
default format is derived from OXBASH/NuShell. It can be in isospin-conserving
format or in explicit proton-neutron format. Be aware that the latter has two
possibilities for normalization of the proton-neutron states. These files are used
primarily though not exclusively for phenomenological spaces and interactions.
All files with this format must end in the extension .int, and as with the single-
particle files, one enters only the name, i.e., if the file is usda.int one enters
in only usda. If the file is in isospin-conserving format, you only need to enter
the name of the file. If the file is in proton-neutron format, you must first tell
30
BIGSTICK the normalization convention, see section 4.3.2. These files have broad
options for scaling the magnitudes of matrix elements, see section 4.3.1.
BIGSTICK also accepts files in a format readable by the MFDn code. Here one
must enter in the full name of the file, even if it has the extension .int, so that
if the file is TBME.int you enter TBME.int not TBME; this signals to BIGSTICK
to expect the MFDn format. Go to section 4.3.3 for more details.
Finally, BIGSTICK can apply a one-body operator to a wave function in order
to generate a transition strength function. These have extension .opme. These
are defined in section 4.6.2, with advanced instruction and examples in sections
5.2.2 and 5.2.3.
While we supply sample files of these various formats, in general it is the
responsibility of the user to generate or obtain input files.
All other files BIGSTICK needs, such as wave function files with extension
.wfn, must been generated by a run of BIGSTICK itself.
4.2 Defining the model space
A many-body model space is defined by a single-particle space, the valence Zand
N, a total Mvalue, a total parity (if applicable), and, optionally, truncations on
that model space. Note that if you are carrying out what we call a secondary
option, which starts from an existing wave function as stored in a .wfn file,
BIGSTICK will automatically read from that file the information on the basis.
You only need to define the model space when carrying out a ‘primary’ option.
The single-particle space is defined one or two ways. Either read in a file
defining the single-particle space, or, for so-called no-core shell model calcula-
tions, automatically generate the basis in a pre-defined form, using the autofill
or ‘auto’ option.
For consistency, we generally refer to orbits as single-particle spaces labeled
by angular momentum jbut not jz, while states are labled by both jand jz.
Our default format for defining the single-particle space are derived from the
format for OXBASH/NuShell/NuShellX files. A typical file is the sd.sps file:
! sd-shell
iso
3
0.0 2.0 1.5 2
0.0 2.0 2.5 2
1.0 0.0 0.5 2
There is no particular formatting (spacing) to this file. Any header lines starting
with ! or # are skipped over. The first non-header line denotes about the isospin
symmetry or lack thereof. iso denotes the single-particle space for both species
is the same; one can still read in isospin breaking interactions. The second line
(3 in the example above) is the number of single-particle orbits. The quantum
numbers for the single-particle orbits as listed are: n, l, j, w; the first three
numbers are real or integers, jis a real number. nis the radial quantum
31
number, which play no role in BIGSTICK except to distinguish between different
states. lis the orbit angular momentum and jis the total angular momentum;
for the case of nucleons j=l±1/2. In BIGSTICK the most important quantum
number is j;lis used internally only to derive the parity of each state.
While for most applications jis a half-integer, i.e., 0.5, 1.5, 2.5, etc., it can
also be integer. In that case l=jand one should intepret ‘protons’ and ‘neu-
trons’ as ‘spin-up’ and ‘spin-down.’ One can compute the electronic structure
of isolated atoms, for example.
While nand lare not internally significant for BIGSTICK, they aid the human-
readability of the .sps files; in addition, they can be invaluable as input to other
code computing desired matrix elements.
BIGSTICK automatically unpacks each orbit to arrive at the 2j+ 1 single-
particle states with different jz.
The last ‘quantum number,’ w, is the weight factor, used for many-body
truncations, described in Section 4.2.2. It must be a nonnegative integer.
BIGSTICK can handle any set of single-particle orbits; the only requirement
is that each one have a unique set of n, l, j. (Although nand lare written above
as real numbers, for historical reasons, they must have integer values. jcan
take either half-integer values or integer values with l=j; this latter we refer
to as LS-coupling and is discussed in detail later on. All the j-values in a .sps
file must be consistent, that is, all half-integer or all integer.)
For example, one could have a set of l= 0, j = 1/2 states:
iso
4
0 0 0.5 0
1 0 0.5 0
2 0 0.5 0
3 0 0.5 0
As of the current version of BIGSTICK, one cannot define completely inde-
pendent proton and neutron spaces. One can however specify two variations.
One can have pns instead of iso, which originally signalled to BIGSTICK to
anticipate isospin-breaking interactions. This option is nearly obsolete. A more
useful option is wpn, where protons and neutrons can have different weights:
wpn
3 3
0.0 2.0 1.5 3
0.0 2.0 2.5 2
1.0 0.0 0.5 2
0.0 2.0 1.5 3
0.0 2.0 2.5 2
1.0 0.0 0.5 3
While the proton and neutron orbits can have different weights, at this time the
sets of quantum numbers must be the same and they must be listed in the same
32
order. In the example above, we have proton 0d3/2, 0d5/2, and 1s1/2, and then
the same for neutrons. Only the wvalues can be different.
The ordering of the single particle orbits is important and must be consistent
with the input interaction files. If one uses our default-format interaction files,
one must supply a .sps file.
It is possible to set environmental variables so that BIGSTICK automatically
searches for .sps files in a different directory:
You can set a path to a standard repository of .sps/.sp files
by using the environmental variable BIG_SPS_DIR.
Just do :
export BIG_SPS_DIR = (directory name)
export BIG_SPS_DIR=/Users/myname/sps_repo
Currently BIG_SPS_DIR is not set
While we recommend the default format, we also allow for NuShell/NuShellX-
compatible .sp files, which have a similar format. Like our default format, they
also come in isospin-symmetric and proton-neutron format. An annotated ex-
ample of the former is
! fp.sp
t ! isospin-symmetric
40 20 ! A, Z of core
4 ! number of orbits
1 4 ! number of species, orbits per species
1 1 3 7 ! index, n, l, 2 x j
2213
3135
4211
As with the default format, BIGSTICK will skip over any header lines starting
with ! or #. The next line, t, denotes isospin symmetry. (Note that, however,
because BIGSTICK requires the proton and neutron spaces to be the same, one
does not need this option, and independent of the form of the single-particle
space file one can read in interaction matrix elements in either isospin-conserving
or -breaking format.) The next line, 40 20 are the Aand Zof the core; these
are not actually needed but are inherited.
The third non-header line, here 4denotes the number of indexed orbits.
The fourth non-header line, 1 4, tells us there is just one ‘kind’ of particle
with 4 orbits. The next four lines are the orbits themselves, with the orbital
index, radial quantum number n, orbital angular momentum l, and twice the
total angular momentum j. Here ndistinguishes between different orbits which
otherwise have the same land j. In this example, nstarts at 1, while in our
other example nstarts at 0. This makes no difference for BIGSTICK’s workings.
This can be contrasted with the pn option for the same space, which has
separate indices for protons and neutrons.
! fppn.sp
33
pn
40 20
8
244
1137
2213
3135
4211
5137
6213
7135
8211
The main differentce are in the third and fourth lines. There are a total of 8
orbits labeled, among two kinds or ‘species’ of particles, each with 4 orbits. The
first 4 orbits are attributed to protons and the the next 4 to neutrons. While
BIGSTICK accepts both formats, in practical terms it does not make a difference.
At this time BIGSTICK does not allow for fully independent proton and neutron
spaces, and the ordering of proton and neutron orbits must be the same. (We
hope to install the capability for more flexible spaces in the future.)
Notice that the NuShell-compatible .sp format does not include the weight-
ing number w, which is assumed to be zero. Hence no many-body truncations
are possible with these files.
If, instead, one uses an MFDn-formatted interaction file, one can use the
autofill option for defining the single-particle states, by entering auto in place
of the name of the .sps file:
Enter file with s.p. orbit information (.sps)
(Enter "auto" to autofill s.p. orbit info )
auto
Enter maximum principle quantum number N
(starting with 0s = 0, 0p = 1, 1s0d = 2, etc. )
The autofill option creates a set of single-particle orbits assuming a harmonic
oscillator, in the following order: 0s1/2,0p1/2,0p3/2,1s1/2,0d3/2,0d5/2, etc., that
is, for given N, in order of increasing j, up to the maximal value N. It also
associates a value wequal to the principal quantum number of that orbit, e.g.,
2n+l, so that Nabove is the maximal principal quantum number. So, for
example, if one choose the principle quantum number N= 5 this includes up
to the 2p-1f-0hshells, which will looks like
iso
21
0.0 0.0 0.5 0
0.0 1.0 0.5 1
0.0 1.0 1.5 1
1.0 0.0 0.5 2
34
0.0 2.0 1.5 2
0.0 2.0 2.5 2
...
1.0 3.0 3.5 5
0.0 5.0 4.5 5
0.0 5.0 5.5 5
4.2.1 Particle-hole conjugation
BIGSTICK constructs the many-body basis states by listing the occupied particle
states. Because the available single-particle space is finite, one can alternately
list the unoccupied hole states. Such a representation can be advantageous if
the single-particle space is more than half-filled, which only happens in phe-
nomenological spaces: while the dimension of the Lanczos basis is unchanged,
because of our jump technology the matrix elements can take much more space
and memory. To understand this, , consider diagonal matrix elements, hα|ˆ
V|αi
which are a sum over occupied states:
hα|ˆ
V|αi=X
a,b∈α
V(ab, ab).
The number of terms in the sum is quadratic in the number of ‘particles’ in the
system. Switching to holes can dramatically decrease the terms in this sum: if
one has 12 single-particle states, for example, having two holes rather than ten
particles makes a difference of a factor of 25! The overall scaling is not so simple,
of course, for off-diagonal matrix elements (quickly: matrix elements of the form
PbV(ab, cb), a 6=c, that is, between two states which differ only by one particle,
go linearly in the number of particles, while those V(ab, cd), a 6=c, b 6=d, that
is, between two states which differ by two particles, are independent of the
number of particles), in large model spaces one can see a big difference. In
particular cases with a large excess of neutrons, so that we have a small number
of protons but nearly fill the neutron space, can lead to enormous slow downs,
as well as requiring many more jumps. Here transformation from particles to
holes make for much greater efficiency. In order to obtain the same spectra and
observables (density matrices), the matrix elements must be transformed via a
Pandya transformation.
How to invoke particle-hole conjugation: When you are asked to enter the
number of particles, you are told the maximum number of particles:
Enter # of valence protons (max 12 ), neutrons (max 12)
Simply enter the number of holes as a negative number, i.e.,
-2 -5
BIGSTICK will automatically carry out the Pandya transformation:
2 proton holes = 10 protons
5 neutron holes = 7 neutrons
35
You can conjugate protons, or neutrons, or both. If you enter the maximum
number of particles in a space, BIGSTICK will automatically regard it as zero
holes. Calculation of density matrices works correctly with particle-hole conju-
gation.
When written to file, hole numbers are also written as negative integers as
a flag, and when post-processing, BIGSTICK will correctly interpret them.
We find there is little significant performance difference in spaces with up
to about 20 single particle states, i.e. the pf shell, but beyond 20 the timing
difference can become quite dramatic.
4.2.2 Truncation of the many-body space
Given a defined single-particle space, the basis states have fixed total Mand
fixed parity. If we allow all such states, we have a full configuration many-body
space. Sometimes, motivated either by physics or computational tractability,
one wants to further truncate this many-body space. BIGSTICK allows a flexible
scheme for truncating the many-body space which encompasses many, though
not all, truncations schemes. We truncate the many-body space based upon sin-
gle particle occupations. One could truncate based upon many-body quantum
numbers, such as from non-Abelian groups (e.g., SU(2) for the J-scheme, or the
symmetry-adapted SU(3) scheme), but that is beyond the scope our algorithms.
Each single-particle orbit is assigned a weight factor w. This is read in from
the .sps file or if the autofill option is used, is equal to the harmonic oscillator
principal quantum number. wmust be a nonnegative integer. If all orbits have
the same wthen no truncation is possible and BIGSTICK does not query about
truncations.
wis treated as an addititive quantum number: each basis state has a total
Wwhich is the sum of the individual ws of the occupied states. Because wis
assigned to an orbit, it does not violate angular momentum or parity, and the
total Wis the same for all many-body basis states that are members of the same
configuration, e.g., (0d5/2)2(1s1/2)1(0d3/2)1. Typically one assigns the same w
to equivalent proton and neutron orbits (in principle one could assign different
ws, which would break isospin, but we haven’t explored this in depth).
Given the basis parameters, the single-particle orbits and their assigned ws
and the number of protons and neutrons, BIGSTICK computes the minimum and
maximum total Wpossible. The difference between these two is the maximal
excitation:
Would you like to truncate ? (y/n)
y
Max excite = 20
Max excite you allow
The user chooses any integer between 0 and “Max excite.” BIGSTICK then
creates all states with total Wup to this excitation.
This scheme encompasses two major trunction schemes. The first kind of
truncation is called a particle-hole truncation in nuclear physics, or sometimes
36
Inert core
Excluded
Limited valence
All valence
single particle states
Figure 4.1: Segregation of single-particle space. ’Inert core’ has all states filled.
‘Excluded’ disallows any occupied states. ‘All valence’ can have states up to the
number of valence particles filled, while ‘Limited valence’ can only have fewer
states filled (e.g. one, two, three...). See text for discussion. Figure taken from
Johnson et al. [2013].
n-particle, n-hole; in atomic physics (and occasionally in nuclear physics), one
uses the notation ‘singles,’ ‘doubles,’ ‘triples,’ etc. To understand this truncation
scheme, begin by considering a space of single-particle states, illustrated in
Figure 4.1. Any single-particle space can be partitioned into four parts. In the
first part, labeled ‘inert core’, the states are all filled and remain filled. In the
fourth and final part, labeled ‘excluded,’ no particles are allowed. Both the core
and excluded parts of the single-particle space need not be considered explicitly,
only implicitly. In some cases there is no core.
More important are the second and third sections, labeled ‘all valence’ and
‘limited valence’, respectively. The total number of particles in these combined
sections is fixed at Nv, and this is the valence or active space.
The difference between the ‘limited valence’ and the ‘all valence’ spaces is
that only some maximal number Nl< Nvof particles are allowed in the ’limited
valence’ space. So, for example, suppose we have four valence particles, but only
allow at most two particles into the ’limited valence’ space. In this case the ‘all
valence’ might contain four, three, or two particles, while the ’limited valence’
space might have zero, one, or two particles. In more standard language, Nl= 1
is called ‘one-particle, one-hole’ or ‘singles’, while Nl= 2 is called ‘two-particle,
two-hole’ or ’doubles’, and so on. There are no other restrictions aside from
global restrictions on quantum numbers such as parity and M.
The second truncation is commonly used in no-core shell model calculations,
where center-of-mass considerations weigh heavily. For all but the lightest sys-
tems, one must work in the laboratory frame, that is, the wavefunction is a
function of laboratory coordinates, Ψ = Ψ(r1, r2, r3, . . .). It is only the relative
degrees of freedom that are relevant, however, so ideally one would like to be
37
able to factorize this into relative and center-of-mass motion:
Ψ(r1, r2, r3, . . .) = Ψrel(~r1−~r2, ~r1−~r3, . . .)×ΨCM(~
RCM) (4.1)
(note that we have only sketched this factorization). In a harmonic oscillator
basis and with a translationally invariant interaction, one can achieve this fac-
torization exactly, if the many-body basis is truncated as follows [Palumbo,
1967, Palumbo and Prosperi, 1968, Gloeckner and Lawson, 1974]:
•In the non-interacting harmonic oscillator, each single-particle state has
an energy ei= ¯hΩ(Ni+ 3/2). Here Niis the principal quantum number, which
is 0 for the 0sshell, 1 for the 0pshell, 2 for the 1s-0dshell, and so on. The
frequency Ω of the harmonic oscillator is a parameter but its numerical value
plays no role in the basis truncation.
•We can then assign to each many-body state a non-interacting energy
ENI =Piei, the sum of the individual non-interacting energies of each particle.
There will be some minimum Emin and all subsequent non-interacting energies
will come in steps of ¯hΩ–in fact for states of the same parity, in steps of 2¯hΩ.
•Now choose some Nmax, and allow only states with non-interacting energy
ENI ≤Emin +Nmax¯hΩ. In practice, restricting states to the same parity means
that the ‘normal’ parity will have ENI =Emin,Emin + 2¯hΩ, Emin + 4¯hΩ, . . .,
Emin +Nmax¯hΩ, while ‘abnormal’ parity will have EN I =Emin + ¯hΩ, Emin +
3¯hΩ, . . .,Emin +Nmax¯hΩ.
This is sometimes called the Nmax truncation, the N¯hΩ truncation, or simply
the energy truncation. It is more complicated than the previous ‘particle-hole’
truncation. We identify with each principal quantum number Nia major shell;
for a 4¯hΩ we can excite four particles each up one shell, one particle up four
shells, two particles each up two shells, one particle up one shell and another
up three shells, and so on. While complicated, such a truncation allows us to
guarantee the center-of-mass wavefunction is a simple Gaussian.
More generally, one can adjust the truncation scheme further, based upon
skillful choice of single-particle ws. The assigned ws need not be contiguous;
the only requirement is that they be nonnegative.
4.2.3 Advanced truncation options
All truncation is based upon the wweight factors. In most applications, both
protons and neutron orbits have the same weights, and one typically truncates
equally. A more general truncation scheme is possible.
First, as discussed in section 4.2, it is possible for proton and neutron orbits
to have different values of w, if the .sps file has the ‘wpn’ format:
wpn
3 3
0.0 2.0 1.5 3
0.0 2.0 2.5 2
1.0 0.0 0.5 2
0.0 2.0 1.5 3
38
0.0 2.0 2.5 2
1.0 0.0 0.5 3
The dimensions of the proton and neutron orbits must be the same, as the order
of all the quantum numbers besides w.
Second, when asked for the truncation,
Would you like to truncate ? (y/n/?=more information)
choosing ‘p’ allows different truncation on protons and neutrons:
Max excite for sum, protons, neutrons?
(must be less than or equal to 8 4 4, respectively )
That is, the maximum values of Wp+Wn,Wp, and Wn, respectively. If you do
not choose this option, then the limits are the same for all three.
4.2.4 How to handle ‘different’ proton-neutron spaces
As of the current version, BIGSTICK cannot handle different proton-neutron
spaces. You can, however trick it into behaving that way, with a small cost.
Both involve deft usage of the truncation and, in many cases, of particle-hole
truncation.
Let’s consider two toy cases. First, suppose the proton and neutron spaces
are entirely separate. For example, let’s suppose valence protons occupy only
the 0f7/2space and valence neutrons only the 1p3/2. The .sps file can look like:
iso
2
0.0 3.0 3.5 0
1.0 1.0 1.5 1
By choosing a Max excite of zero, you will assure no particles are excited
out of the 0f7/2into the 1p3/2. (It is your responsiblity to set up the correct
interaction file. You do not have to include cross-shell matrix elements if they
are not needed; however if they are included, they will induce an effective single-
particle energy so choose wisely.)
A slightly more complex case: suppose you want protons active in 0f7/2,
1p3/2and 1p3/2, and neutrons in 1p3/2, 1p3/2, and 0f5/2. Set up the .sps file
wpn
4 4
0.0 3.0 3.5 0
1.0 1.0 1.5 0
1.0 1.0 0.5 0
0.0 3.0 2.5 1
0.0 3.0 3.5 0
1.0 1.0 1.5 1
1.0 1.0 0.5 1
0.0 3.0 2.5 1
39
It is required that the proton and neutron orbits be the same, though the weight
factors wis the last column can differ. Again, choosing Max excite of zero will
keep the protons and neutrons in their respective valence spaces. If the valence
spaces are significantly different, we strongly recommend utilizing particle-hole
conjugation for the neutrons.
One can make the truncations even more complex, for example allow a few
protons to be excited but no neutrons, by careful usage of the options provided.
4.3 Interaction files
After the model space is defined, BIGSTICK needs interaction matrix elements.
All matrix elements are defined in the one-, two-, or possibly three-body-space.
BIGSTICK’s job is to embed these matrix elements into a many-body space and
solve the eigenvalue problem. (Because three-body interaction files are highly
specialized, we do not discuss their format.)
The default format for two-body interaction file is derived from OXBASH/NuShell
and always ends in the extension .int. When entering the name of the file, only
enter the name, not the extension, i.e., usdb not usdb.int; otherwise BIGSTICK
will misinterpret the file.
! Brown-Richter USDB interaction
63 2.1117 -3.9257 -3.2079
2 2 2 2 1 0 -1.3796
2 2 2 1 1 0 3.4987
2 2 1 1 1 0 1.6647
2 2 1 3 1 0 0.0272
2 2 3 3 1 0 -0.5344
2 1 2 1 1 0 -6.0099
2 1 1 1 1 0 0.1922
2 1 1 3 1 0 1.6231
2 1 3 3 1 0 2.0226
1 1 1 1 1 0 -1.6582
1 1 1 3 1 0 -0.8493
1 1 3 3 1 0 0.1574
...
There is no specific spacing for this file. BIGSTICK will skip any header lines
starting with ! or #. The first line is
Ntbme spe(1) spe(2) spe(3) ...
where Ntbme is the number of two-body matrix elements (TBMEs) in the
rest of the file, and spe(i) is the single-particle energy of the ith orbit. Only 10
single particle energies are on each line.
The rest of the file are the two-body matrix elements. This is defined as
VJT (ab, cd) = hab;J T |V|cd;JT i,(4.2)
40

where a, b, c, d label orbits, as ordered in the .sps file or as created by the
autofill option; Jand Tare the total angular momentum and total isospin of
the two-body states |ab;JT i, which are normalized. This follows the convention
of Brussaard and Glaudemans. Each matrix element is read in as
a b c d J T VJT (ab, cd)
For input purposes, the order of a, b, c, d is not important (as long as one
has the correct phase), nor is the ordering of the TBMEs themselves. When
reading in the file, BIGSTICK automatically stores the matrix element according
to internal protocols, appropriately taking care of any relevant phases.
Matrix elements that are zero can be left out, as long as Ntbme correctly
gives the number of TBMEs in the file. More than one file can be read in; enter
end to tell BIGSTICK you are finished reading in files.
4.3.1 Scaling and autoscaling
Empirical studies with phenomenological interactions have found best agree-
ment with experiment if one scales the two-body matrix elements with mass
number A. (There is some justification based upon the scaling of harmonic
oscillator wave functions with A). A standard scaling factor is
Aref
Ax
(4.3)
where Aref is the reference mass number (typically Aof the frozen core +2, as it
is fit to the interaction of two particles above the frozen core), Ais the mass of
the desired nucleus, and xis an exponent, typically around 1/3. To accomodate
this scaling, when reading in the default format, BIGSTICK requests
Enter scaling for spes, A,B,X ( (A/B)^X ) for TBMEs
(If B or X = 0, then scale by A )
Typically the single particle energies are unscaled, but we allow for it. A typical
entry, for example for the USDA/B interactions [Brown and Richter, 2006],
would be
1 18 24 0.3
Here the single particle energies are unscaled, the core has mass number 16 and
hence the reference mass is 18, the target mass in this case has mass number
24, and the exponent is 0.3. Whoever provides the interaction has to provide
the exponent. If unsure, just enter
1, 1, 1, 1
Many files used with NuShell have autoscaling. For example, for the USDA/B
file, the first lines are
41
! 1=d3/2 2=d5/2 3=s1/2
! the first line has the three single-particle energies
! the - sign tells oxbash that the tbme have a mass dependence of the form
! [18/(16+n)]^0.3 where n is the number of valence particles
-63 1.9798 -3.9436 -3.0612 16.0000 18.0000 0.30000
A negative integer for the number of two-body matrix elements (here, -63)
initiates autoscaling. The next three numbers are the single-particle energies,
and the next numbers are Acore, the reference mass, and the exponent. If
BIGSTICK encounters a negative integer for the number of two-body matrix
elements, it autoscale the two-body matrix elements as described above. To
turn off autoscaling, change -63 to 63.
Keep in mind that not all interactions will be scaled. Ab initio interactions
are almost never scaled, and ‘phenomenological’ interactions depend on how
they were derived and fit. See your interaction provider for more information.
4.3.2 Proton-neutron and other isospin-breaking formats
Often one needs to break isospin. There are three modifications of the default
format which break isospin. In addition, ab initio inputs in the MFDn format,
described in section 4.3.3, also generally break isospin.
The most robust format, which we recommend, is the explicit proton-neutron
formalism. Here one has separate labels for proton and neutron orbits; however,
at this time the proton and neutron orbits must have the same quantum
numbers and be listed in the same order. For example, one might label
the proton orbits 1 = 0d3/2, 2 = 0d5/2, and 3 = 1s1/2. Then the neutron orbits
must be 4 = 0d3/2, 5 = 0d5/2, and 6 = 1s1/2.
While BIGSTICK generally allows for arbitrary order, for the proton-neutron
matrix elements the proton labels must be in the first and third columns and
neutron labels in the second and fourth columns, that is, for VJ(ab, cd), aand
cmust be proton labels and b, d must be neutron labels. With twice as many
defined orbits, one must also provide separate proton and neutron single particle
energies. As an example, here is part of the file of the p-shell Cohen-Kurath
matrix elements with good isospin:
! ORDER IS: 1 = 1P1/2 2 = 1P3/2
15 2.419 1.129
1 1 1 1 0 1 0.2440000
1 1 1 1 1 0 -4.2921500
2 1 1 1 1 0 1.2047000
2 1 2 1 1 0 -6.5627000
2 1 2 1 1 1 0.7344000
2 1 2 1 2 0 -4.0579000
2 1 2 1 2 1 -1.1443000
2 2 1 1 0 1 -5.0526000
and here is an excerpt in proton-neutron formalism
42

34 2.4190 1.1290 2.4190 1.1290
1 3 1 3 0 1 0.24400
1 1 1 1 0 1 0.24400
3 3 3 3 0 1 0.24400
1 3 1 3 1 1 -4.29215
1 3 1 4 1 1 -0.85185
1 3 2 3 1 1 0.85185
1 3 2 4 0 1 -5.05260
1 1 2 2 0 1 -5.05260
3 3 4 4 0 1 -5.05260
1 3 2 4 1 1 1.76980
1 4 1 4 1 1 -2.91415
1 2 1 2 1 1 0.73440
3 4 3 4 1 1 0.73440
In no case are headers required, but they do help as a check for the definition of
the orbits. BIGSTICK automatically checks that angular momentum and parity
selections are not violated. In the explicit proton neutron format Tis given in
the sixth column but not actually used.
There is one more question of convention one must deal with: the normal-
ization of the two-body states in the definition of matrix elements. All formats
assume two-proton and two-neutron states are normalized, and states with good
isospin are normalized. Files set up for NuShellX, however, have unnormalized
proton-neutron states.
BIGSTICK can read in default-format proton-neutron interactions with ei-
ther normalized (‘xpn’ or explicit proton-neutron) or unnormalized (‘upn’ or
unnormalized proton-neutron) conventions. In both cases the files also include
proton-proton and neutron-neutron matrix elements, with normalized states.
The relationship between the two is
Vxpn
J(aπbν, cπdν) = 2
p(1 + δab)(1 + δcd)Vupn
J(aπbν, cπdν) (4.4)
Here we have marked the orbits a, c as proton and b, d as neutron, but the
Kronecker-δs refer only to the quantum numbers n, l, j. For example, in the sd
shell, with the labels mentioned above,
Vxpn
J(16,25) = √2Vupn
J(16,25)
because proton orbit 1 (0d3/2) and neutron orbit 6 (1s1/2)are different, but
proton orbit 2 and neutron orbit 5 are both d5/2
Unfortunately it is up the user to know whether or not the file uses normal-
ized or unnormalized proton-neutron states. If the file was originally produced
for use with NuShellX, it is almost certainly the latter.
(This arises out of the conversion of normalized isospin wave function to
normalized proton-neutron wave functions and the result matrix elements. One
43

finds
Vpn
J(ab, cd) = p(1 + δab)(1 + δcd)
2Viso
J,T =0(ab, cd) + Viso
J,T =1(ab, cd),(4.5)
but the unnormalized convention yields the simpler
Vupn
J(ab, cd) = Viso
J,T =0(ab, cd) + Viso
J,T =1(ab, cd).(4.6)
While our preference is for the former, given the prominence of the latter through
NuShellX we include it as an option.)
In order to read in proton-neutron matrix elements in the default format,
you must first tell BIGSTICK to expect it.
For xpn/upn formats, you MUST specify the format.
In this format proton and neutron orbits are sequential and do not overlap,
E.g., proton orbits are 1,2,3 and neutron orbits are 4,5,6.
FOR NOW despite the distinct numbering the proton and neutron orbits
must encompass the same space.
NOTE: upn format is typical for TBME files distributed with NuShell;
xpn/upn files must have the name XXX.int, but enter XXX when requested.
As with default-format isospin-conserving files, the file name must be xxxx.int,
but the user enters in just ‘xxxx’.
Also as with default-format isospin-conserving files, after entering the name
of the file, the user is prompted for scaling. For maximal flexibility, there are two
layers of possible scaling. The first is the standard phenomenological scaling:
Enter global scaling for spes, A,B,X ( (A/B)^X ) for TBMEs
(If B or X = 0, then scale by A )
1 18. 24. 0.3
These scalings are applied to all single particle energies and to all two-body
matrix elements. In addition, one can enter in separate scaling factors for pro-
tons single-particle energies, neutron single-particle energies, proton-proton two-
body matrix elements, neutron-neutron two-body matrix elements, and finally
proton-neutron two-body matrix elements:
Enter individual scaling for: proton spes, neutron spes, pp TBMEs, nn TBMEs, p
n TBMES
(If not sure, just enter 1 1 1 1 1 )
There are two alternate formats for isospin-breaking files which build upon
the default format. These involve reading in separate files for proton-proton,
neutron-neutron, and proton-neutron, or for isoscalar, isovector, and isotensor
components. There are some tricky issues of definition, however. Thus we do not
actively support these alternative formats, instead recommending the explicit
proton-neutron format, whether normalized or unnormalized
44

One can mix all of these different formats. You can read in an isospin-
conserving file, a proton-neutron format file, and so on, in any order. To stop
reading in interaction files, enter ‘end’ at the prompt.
4.3.3 MFDn format input
Another major configuration-interaction code is MFDn (Many-Fermion-Dynamics,
nuclear version) out of Iowa State University [Sternberg et al., 2008]. While
within MFDn there are several variations on conventions, we describe here the
most common conventions.
Unlike the default format, to read in an MFDn-format file, you must enter
the entire name, including any extensions. This signals to BIGSTICK to prepare
to read in an an MFDn-format file. BIGSTICK will treat a file TBME.int very
differently if you answer ‘TBME’ versus ‘TBME.int’ for the file name. MFDn-format
files are almost always for ab initio or so-called no-core shell model calculations,
and almost always assume a harmonic oscillator basis.
The input file first line is
nTBME (other stuff which are not needed)
where nTBME is the number of TBMEs in the file. For example
2056271 13 14 20.0000 2.0000
The only number BIGSTICK requires is the first one. The fourth number, 20.000,
is ¯hΩ in MeV, but not all codes generate this information.
The MFDn format does not include explicit single-particle energies. Subsquent
lines are of the form
a b c d J T Trel Hrel Vcoul V
or, more commonly,
a b c d J T Trel Hrel Vcoul Vpn Vpp Vnn
Here all matrix elements are of the form hab;JT |V|cd;JT i, that is the ma-
trix element between normalized two-body states with a,b,c,d labels of single
particle orbits, J(and, optionally, T) are total angular momentum and isospin
of the coupled two-body states. The isospin Tis not really used.
Now for the matrix element. Trel is the relative kinetic energy, that is
ˆ
Trel =X
i<j
(~pi−~pj)2
2M,(4.7)
These matrix elements are computed in a harmonic oscillator basis for ¯hΩ =
1MeV , and A= 2, and thus must be rescaled correctly for the A-body system,
that is, must be multiplied by 2¯hΩ/A.
Now to the final matrix elements. The actual Hamiltonian one wants is
ˆ
H=ˆ
Trel +ˆ
Vrel +βc.m.(ˆ
Hcm −3
2¯hΩ).(4.8)
45
Here ˆ
Hcm is the center-of-mass Hamiltonian, used to push up spurious states via
the Palumbo-Lawson-Glocke method [Palumbo, 1967, Palumbo and Prosperi,
1968, Gloeckner and Lawson, 1974]:
ˆ
Hcm =P2
cm
2AmN
+1
2AmNΩ2R2
cm.(4.9)
where mNis the nucleon mass and
~
Rcm =1
AX
i
~ri,~
Pcm =¯h
iX
i
~
∇i.(4.10)
These have the correct commutation relation, that is, [ ~
Rcm,~
Pcm] = i¯h, so that
~
Pcm is the conjugate momentum to ~
Rcm.
It is useful to separate ˆ
Hcm into one- and two-body parts:
ˆ
Hcm =1
2AmNX
i
p2
i+1
2AmNΩ2X
i
r2
i+
1
2AmNX
i6=j
~pi·~pj+1
2AmNΩ2X
i6=j
~ri·~rj(4.11)
The first two terms are the single particle energies, with values ¯hΩ(N+ 3/2)/A,
with Nthe principal quantum number, and the second two terms is ˆ
Hrel ׯhΩ/A.
BIGSTICK automatically accounts for all the factors, as long as you provide the
correct ¯hΩ as shown below:
When you select an MFDn-format file, you will be prompted for the following:
For MFD-formatted input choose one of the following :
(I) No isospin breaking
(P) Explicit proton-neutron formalism
(C) Isospin breaking only through adding Coulomb
Almost always you should select option ‘p’.
In order to use an ab initio file, you need to enter in the value of ¯hΩ for
both the kinetic energy term and the center-of-mass Hamiltonian to push up
spurious states:
Enter oscillator frequency (in MeV) and center-of-mass strength
You should know the frequency at which the file was created. The second term
is βc.m.. Typical values of βc.m. are 1-10.
4.3.4 Three-body forces
While BIGSTICK has a validated capability for three-body forces, it is not op-
timized for large calculations; the main issue is storage of the large number of
matrix elements. If you have the capability to generate three-body forces, please
46

contact us, cjohnson@mail.sdsu.edu. We do not have the codes or capability
to generate three-body forces for users.
In your distribution three-body forces are likely disabled. They can be
re-enabled by setting the logical flag threebodycheck = .true. in module
flags3body in the file bmodules 3body.f90. If this flag is enabled, BIGSTICK
will query if you want to use three-body forces:
Do you want 3-body forces (y/n) ?
If you answer ‘n,’ BIGSTICK will proceed with just 2-body forces. If you answer
‘y,’ BIGSTICK will ask for the name of the file. Actually using three-body forces
is complicated and beyond the scope of this current manual.
You can, however, use two-body forces in three-body mode (the matrix ele-
ments are multiplied internally by ( ˆ
N−2)/(A−1) to turn them into genuine
three-body forces), by answering ‘none’ to the question of the name of the file
of three-body forces. Most users will not be interested in this.
4.4 Primary runtime options
Here we outline the major run time options, although some issues are discussed
in more detail elsewhere. The main menu can be divided into two categories,
primary and secondary. We discuss secondary options, which require results
of a previous run, in section 4.6. In most primary runs one solves the matrix
eigenvalue problem, which invokes the diagonalization options menu, discussed
in section 4.5.
4.4.1 Autoinput
* (i) Input automatically read from "autoinput.bigstick" file *
* (note: autoinput.bigstick file created with each nonauto run) *
Each time BIGSTICK runs, it writes the user’s responses to a file autoinput.bigstick.
This file can be edited or used as the basis of a batch file. The autoinput option,
‘i,’ will read in the autoinput.bigstick instead of taking responses from the
user.
4.4.2 Standard or normal runs
* (n) Compute spectrum (default); (ns) to suppress eigenvector write up *
The normal run, ‘n,’ will generate the low-lying eigenspectrum and wave func-
tions. This is the most common option. Two variations on it are ns which will
compute the eigenspectrum and the Jand Tvalues, but not write the wave-
functions to a file, and ne, which will only compute eigenenergies. These latter
options can save on time and file storage, but in most cases are not necessary.
47
4.4.3 One-body density matrices and occupations
One of the most important options for BIGSTICK is to generate the one-body
density matrices, defined as
ρfi
K(a†b) = [K]−1hJf||(a†b)K||Jii(4.12)
where we use the choice of reduced matrix elements from Edmonds [1996],
hJf||OK||Jii= [Jf](JfMf, KM|JiMi)−1hJfMf|OKM|JiMii(4.13)
The advantage of this definition of the density matrix is that the reduced matrix
element of a general, non-scalar one-body operator is just the density matrix ×
the reduced matrix elements of that operator, that is
hΨf, Jf|| ˆ
OK||Ψi, Jii=X
ab ha|| ˆ
OK||biρfi
K(a†b) (4.14)
where a, b are labels for single-particle orbits, and ha|| ˆ
OK||biare the reduced
one-body matrix matrix elements for the operator ˆ
Owith angular momentum
K.
BIGSTICK has a number of options to generate density matrices.
* (d) Densities: Compute spectrum + all one-body densities *
* (dx[m]) Densities: Compute one-body densities from previous run (.wfn) *
* optional m enables mathematica output *
The density matrix option ‘d’ runs just like the normal option, except at the
end of the run it generates the one-body density matrices, which we describe
more fully in section . If the interaction file has good isospin, then the one-body
density matrices will be coupled up to good isospin. If the interaction file breaks
isospin, the density matrices will be in proton-neutron format. If you use an
interaction with good isospin but want the density matrices in proton-neutron
format, use the option ‘dp.’
Two variations are the option ‘dx,’ which reads in a previously computed
wave function file and from it computes the one-body density matrices, and
‘dxm’, which does the same but generates the density matrices in a format read-
able by Mathematica. Technically these options are secondary.
* (p) Compute spectrum + single-particle occupations; (ps) to suppress wfn*
* (occ) single-particle occupations (from previous wfn) *
A restricted version of the one-body densities are the single-particle occupations.
In principle given the former one can compute the latter, as described in section
5.1.2, but for convenience we give an option to do this directly. Option ‘p’ does
this as in the normal option, but also writes the single-particle occupations to
the .res file. Option ’ps’ does the same but does not write the wave functions
to a file on disk. Finally, the secondary option ‘occ’ reads in an existing wave
function file and generates the single-particle occupations.
48
Single particle state quantum numbers
ORBIT : 1 2 3
N : 0 0 1
J : 3 5 1
L : 2 2 0
State E Ex J T
1 -62.78960 0.00000 2.500 1.500
p occ: 0.136 1.590 0.275
n_occ: 0.353 4.299 0.347
A detailed discussion of using one-body density matrices to get transition
probabilities can be found in Chapter 5.1.
4.4.4 Other primary options
* (m) print information for Modeling parallel distribution *
The modeling option ‘m’ is useful for seeing if enough nodes and memory can be
allocated for a large parallel run. See also section 8.1.3. No interaction files are
read in and no diagonalization is carried out.
There are a couple ofadditional options most users are unlikely to use but
which we mention nontheless.
* (f) Self-consistent mean-field approximation (prepare pivot) *
* (c) Compute traces *
The ‘f’ or self-consistent mean-field option is an attempt to quickly get an
good starting pivot for the ground state. Essentially what it does is replace the
full Hamiltonian by a one-body approximation. On each iteration, BIGSTICK
computes the one-body density matrices and generates an effective, in general
non-scalar one-body potential by coupling the two-body force to those one-body
densities . This process is much faster than applying the full two-body inter-
action. It is inspired by but not the same as the Hartree-Fock approximation,
which assumes a single Slater determinant.
The resulting approximate states are written to a .wfn file, which can be
read back in as a pivot using the option ‘np.’ In our experiments this pivot
speeds up convergence to the ground state but not excited states. More work
especially on multishell systems have yet to be done. We have not fully tested
this option in parallel.
BIGSTICK can also compute the trace of the Hamiltonian and the trace of
the Hamiltonian squared, using the option ‘c.’ Specifically, it computes the
centroid, which is the trace divided by the dimension, and the width, which is
the square root of the variance.
49
for dimension 28503, centroid = -43.544457, width = 11.298695
(saved to file trace.bigstick )
As shown above, the results are written to the file trace.bigstick. We caution
users that in particular computing the width can be time consuming. We have
not fully tested this option in parallel.
4.5 Diagonalization options
After the interactions files have been read in, BIGSTICK sets up the jump arrays
for reconstructing the matrix elements on the fly. After that, the eigensolver
menu comes up:
/ ------------------------------------------------------------------------\
| |
| DIAGONALIZATION OPTIONS (choose one) |
| |
| (ex) Exact and full diagonalization (use for small dimensions only) |
| |
| (ld) Lanczos with default convergence (STANDARD) |
| (lf) Lanczos with fixed (user-chosen) iterations |
| (lc) Lanczos with user-defined convergence |
| |
| (td) Thick-restart Lanczos with default convergence |
| (tf) Thick-restart Lanczos with fixed iterations |
| (tc) Thick-restart Lanczos with user-defined convergence |
| (tx) Thick-restart Lanczos targeting states near specified energy |
| |
| (sk) Skip Lanczos (only used for timing set up) |
| |
\ ------------------------------------------------------------------------/
The full diagonalization option, ‘ex,’ creates the entire Hamiltonian matrix,
stores it in memory, and solves it using the Householder algorithm as imple-
mented in the LAPACK routine DSYEV. As such, it should not be used except
for relatively small dimensions. On workstations one can solve up to ∼103in a
few or tens of minutes. We have solved up to ∼104, but that can take hours. In
principle MPI versions of Householder exist, but we have not installed one, as
one seldom has need for all eigensolutions of a very large matrix. (If you do not
need wave functions or the angular momenta, choosing option ‘ne’ will speed
this up dramatically, as DSYEV will run faster if one wants only eigenvalues.)
Under this option you can choose how many low-lying states, or all of them
if you wish, to keep. These get written to file.
The primary eigensolver is the Lanczos algorithm, described in Chapter 7.
Most of the time you will use option ‘ld’, the default Lanczos choice. Here you
get asked
50
Enter nkeep, max # iterations for lanczos
(nkeep = # of states printed out )
BIGSTICK will run until the standard convergence criterion, see section 4.5.1, is
satisfied, or until the maximum number of iterations is exceeded. The latter
must be specified to reserve memory for the Lanczos vectors. Although the
Lanczos and related Arnoldi algorithms are among the most studied in applied
mathematics, there is no simple, robust rule for the number of iterations needed.
For phenomenological spaces, the ground state will often converge in under 50
iterations, the first 5 states in 100 to 150 iterations, and so on. For no-core shell
model calculations, the time to convergence is usualy longer.
The default convergence check is discussed in the next section, 4.5.1. If you
want a fixed number of iterations without checking convergence, choose ‘lf.’ If
you want finer control over convergence, choose ‘lc,’ discussed in 4.5.1.
The Lanczos vectors are stored in memory. For large-dimension cases, es-
pecially on a laptop or desktop, one can run out of memory just storing these
vectors. Alternately, if one needs a large number of converged states, after
a number of iterations reorthogonalization actually starts to take more time
than matvec. A robust alternative is the thick-restart Lanczos algorithm [Wu
and Simon, 2000], which requires fewer vectors stored in memory but requires
more iterations. While standard Lanczos finds the lowest Nkeep eigensolutions
with Niter iterations, thick-restart has three numbers: Nkeep < Nthick < Niter.
As described more fully in section 7.2, after Niter iterations the approximate
Hamiltonian is diagonalized, and Nthick of these eigenvectors are kept for restart-
ing. This process is repeated until convergence or until a maximum number of
restarts has been exhausted.
td
Enter # of states to keep, number of iterations before restarting
5 50
Enter max # of restarts
10
As with standard Lanczos, the values chosen usually come with experience. We
find we usually want Nthick >∼3×Nkeep, and we take Niter as large as practical.
Specifically, BIGSTICK chooses
Nthick = max(3Nkeep, Nkeep + 5),
as long as this is not larger than Niter.
If you want a fixed number of iterations, choose ‘tf’, and you will be
prompted for Nkeep,Niter, and then Nthick:
tf
Enter # of states to keep, # of iterations before thick-restart
5 50
Enter # of vectors to keep after thick-restart
51

(Typical value would be 20 )
( Must be between 5 and 50 )
If you want to control the convergence, choose ‘tc’ and you will be prompted
for convergence choices much as for standard Lanczos.
Finally, ‘tx’ is an experimental mode attempting to find highly excited
states. It modifies thick-restart by choosing eigenpairs in the vincinity of a
selected absolute energy. In our experience the convergence is not very good,
but it does yield an eigenvector with very strong overlaps with the true eigen-
vectors in the vicinity of the target energy.
Option ‘sk’ is only for testing timing of set-up to this point.
4.5.1 Convergence
As BIGSTICK iterates, it checks for convergence. Every ten iterations it prints
to screen the current Nkeep lowest eigenvalues and the convergence criterion:
1 -135.86073
2 -133.92904
3 -131.25354
4 -131.02439
5 -129.53058
80 iterations
(energy convergence 0.70356 > criterion 0.00100)
1 -135.86073
2 -133.92904
3 -131.25354
4 -131.02439
5 -129.53059
90 iterations
(energy convergence 0.30353 > criterion 0.00100)
As far as we can tell from the literature, there is no robustly ideal convergence
criterion for general Lanczos. Our default convergence is on energy: BIGSTICK
takes the sum of the absolute value of the differences in energy between the
current iteration and one previous, not only for the Nkeep lowest energies but
also for the next 5, and divides by the square root of Nkeep + 5, that is,
δconv ≡PNkeep +5
i=1 Enew
i−Eold
i
pNkeep + 5 (4.15)
The reason for testing additional eigenvalues is to avoid the problem of
plunging eigenvalues, well known to occur in Lanzos Whitehead et al. [1977]; it
happens when by accident a low-lying state has a tiny overlap with the pivot or
52
initial vector. We divided not by the number of energies compared but by the
square root, because for a large number of energies one outlier could get washed
out by many small deviations. For our purposes this has worked well enough,
but it is not rigorously tuned.
If you want a different criterion, choose ‘lc’ on the diagonalization menu.
One then gets a series of questions with which to tune the convergence, including
comparing eigenvectors rather than eigenvalues:
Enter nkeep, max # iterations for lanczos
5 150
Enter how many ADDITIONAL states for convergence test
( Default= 5 ; you may choose 0 )
10
Enter one of the following choices for convergence control :
(0) Average difference in energies between one iteration and the last;
(1) Max difference in energies between one iteration and the last;
(2) Average difference in wavefunctions between one iteration and the last;
(3) Min difference in wavefunctions between one iteration and the last;
2
Enter desired tolerance
(default tol = 0.100E-04 )
Similar options are available for thick-restart Lanczos.
4.6 Secondary runtime options
Once BIGSTICK has generated wave function, it can further process the wave
functions in secondary options. We discuss those options in detail here. Some
of these were already mentioned in section 4.4. All of these options will ask for
the name of a previously generated .wfn file.
Enter input name of .wfn file
You do not have to read in a .sps or similar file to define the model space; from
the information in the .wfn file, BIGSTICK reconstucts the basis. Depending
upon the option, you may be asked to enter names of appropriate files, such as
interaction files.
We list these in the order they are presented in the menu, but the most
important and commonly used option are ‘x,’ expectation value of a scalar
operator (section 4.6.1), ‘o,’ apply a one-body non-scalar transition operator
(section 4.6.2 and Chapter 5), and ‘s,’ the strength function option (section 5.2
and Chapter 5).
* (np) Compute spectrum starting from prior pivot *
53
Option ‘np’ allows you to choose a pivot, or initial starting vector, from a
previously generated wave function. This might be useful, for example, if you
wanted to try to get states of a particular quantum number such as J, although
we do not currently have the capability to enforce this condition, and even if it is
an exact quantum number numerical noise will allow states with other quantum
numbers to creep in. Alternately, you can generate a guess of the ground state
by the self-consistent mean-field option ‘f.’ Our experiments suggest this latter
does lead to faster convergence of the ground state, but not excited states.
A related and more widely useful option is the strength function option ‘s’
discussed below and in section .
* (dx[m]) Densities: Compute one-body densities from previous run (.wfn) *
* optional m enables mathematica output
Options ‘dx’ and ‘dxm’ read in a previously generated wave function and compute
the one-body density matrices. The latter provides a Mathematica-friendly file
format. More details about one-body density matrices are found in section 5.1.
* (occ) single-particle occupations (from previous wfn) *
This option computes the single-particle occupations from a previously gener-
ated wave function file.
4.6.1 Expectation value
* (x) eXpectation value of a scalar Hamiltonian (from previous wfn) *
The option ‘x’ allows you to compute the expectation value of a operator, which
may have one-, two- (and in principle, three-) body components. It must be an
angular momentum scalar and thus is treated as a Hamiltonian, and is read in
exactly as Hamiltonians, along with standard requests for scaling information.
The results are written both to screen and, if an output name is given, to the
.res file.
STATE E J T^2 <H > (norm)
1 -92.7790 -0.0000 0.0000 499.287061 1.00000
2 -91.1196 2.0000 0.0000 488.826115 1.00000
3 -88.4779 2.0000 0.0000 509.922118 1.00000
4 -87.9781 4.0000 0.0000 452.853182 1.00000
The reason the norm of the vector is given is that after applying a one-body
transition operator, as described in the next section (4.6.2), the wave function
vector may no longer be normalized.
54
4.6.2 Applying a one-body transition operator
One of BIGSTICK’s important capabilities is to take a set of previously generated
wave functions and apply a non-scalar one-body operator to them:
* (o) Apply a one-body (transition) operator to previous wfn and write out*
* (oo) Apply a one-body (transition) operator with enforced orthogonality *
* (that is, forced resulting pivot to be orthogonal to starting pivot *
* to eliminate transition strength to original state) *
If you choose this option, you will be asked for the name of the input .wfn file as
well as the name of an output .wfn file. Then you will be asked for a file with
the reduced matrix elements of the operator, which must have the extension
.opme:
Enter name of .opme file
Here is an annotated example .opme file:
! header: Gamow-Teller-like
iso ! assumes isospin
3 ! # of single particle orbits
1 0 2 1.5 ! index, n, l, j of orbits
2 0 2 2.5
3 1 0 0.5
1 1 ! J, T of transition
1 1 -2.68328 ! a, b < a ||| O ||| b >
1 2 5.36656
2 1 -5.36656
2 2 5.01996
3 3 4.24264
The only formatting is the the first non-header line, here iso, must be flush
against the left. The file must contain the single-particle orbits, and BIGSTICK
checks against the orbits used to build the wave function. After the list of
orbits, the Jand Tof the operator come, and then the the non-zero reduced
matrix elements. Here, assuming isospin is a good quantum number, we have
doubly-reduced matrix elements. Although there is a symmetry ha||| ˆ
O|||bi=
(−1)ja−jbhb||| ˆ
O|||ai, at this time BIGSTICK requires both elements.
Many transition operators do not preserve isospin. Therefore BIGSTICK can
read in operators in an explicit proton-neutron symmetry:
! M1 matrix elements in the sd shell
pns
3
1 0 2 1.5
2 0 2 2.5
3 1 0 0.5
1 2
55
1 1 0.1568000 1.4481392 ! a b proton m.e. neutron m.e.
1 2 3.4710987 -2.8962784
2 1 -3.4710987 2.8962784
2 2 6.7871771 -2.7092204
3 3 3.3425579 -2.2897091
The code pns in the first non-header line signals that the matrix elements are
in proton-neutron formalism, with the same list of orbital quantum numbers for
protons and neutrons. The 2 in the 5th line also signals that one is breaking
isospin. Thus in the list of reduced (in Jonly) matrix elements, the columns
are for protons and neutrons, respectively.
BIGSTICK will read in all the wave functions |Ψiifrom the initial wave func-
tion file, and write ˆ
O|Ψiiin the final wave function file. These wave functions
will generally not be normalized and will not have good angular momentum or
isospin. More on this elsewhere.
Currently, both the initial and final wave functions must be in the same
basis. Thus, there are no explicit charge-changing transitions. To handle charge-
changing transitions, one must use an interaction with good isospin and exploit
isospin rotation, described in section 5.1.5. For transitions which change parity,
one must use a basis with both parities, option 0in the parity-selection menu.
Because transition operators are not in general unitary, the result wave func-
tion vectors are not normalized. This information is important, as it tells us
about total transition strengths, also known as the non-energy-weighted sum
rule.
4.6.3 Applying a two-body body scalar operator
* (a) Apply a scalar Hamiltonian to a previous wfn and write out *
Option ‘a’ works very similar to option ‘o:’ one reads in a previously-generated
file of wave functions, applies an operator to each wave function, and writes the
results to another file. The difference is here the operator must be a one-plus-
two-body scalar operator, that is, like a Hamiltonian. The files are read in the
same as a Hamiltonian, along with scaling, and so on.
4.6.4 Generating strength function distributions
One of the most powerful and most useful capabilities is ‘s,’ the strength func-
tion distribution option. We give an overview here, with many more details of
application in section 5.2.
Like all secondary options, the strength function option starts by prompting
the user for a previously generated .wfn file. The user is then prompted for
a Hamiltonian or Hamiltonian-like interaction file or files. Next the user must
enter the number of iterations for Lanczos:
Fixed iterations ONLY:
Enter nkeep, # iterations for lanczos
(nkeep = # of states printed out )
56
The only way to make this option work is through standard Lanczos. The num-
ber of results to keep and the number of iteration depends upon the application;
see section 5.2. Finally, the user must choose from the input file the pivot or
starting vector, a key decision:
There are 5 wavefunctions
5 states
STATE E J <H >
1 -92.7790 -0.0000 0.0000
2 -91.1196 2.0000 0.0000
3 -88.4779 2.0000 0.0000
4 -87.9781 4.0000 0.0000
5 -87.4348 3.0000 0.0000
Which do you want as pivot?
What happens next is that Lanczos runs normally, produces eigenvalues and
eigenvectors and writes them to file. It also additionally computes the overlap
of the pivot with each of the eigenstates, that is, |hf|pivoti|2:
Energy Strength
______ ________
17.30356 0.00007
49.98777 0.00123
110.94935 0.00956
184.33815 0.01945
249.75355 0.01641
301.54676 0.08014
383.34766 0.05833
428.29023 0.14090
498.95403 0.04282
534.87775 0.17398
588.87306 0.45712
______ ________
This is the strength function or strength distribution. If the starting vector has
a norm different from one, this is noted
0.99999999895896363 = total input strength
and this is included in the strengths. The usefulness of this capability cannot
be overestimated, and is discussed in depth in section 5.2.
4.6.5 Overlap or dot product of wave functions
* (v) Overlap of initial states with final states *
57
The output eigenstates are written as vectors. Although most users are unlikely
to need to use this, using the ‘v’ option BIGSTICK can compute the dot product
between two such wave functions, including from different files. From the first
file, you must choose a specific state:
Which do you want as initial state?
A second file is then opened (you can reopen the first file), and the intial state
is dotted against each of them. The results are written to the file overlap.dat:
Initial state = 1
state E J T <i|f> |<i|f>|^2
1 -87.10445 -0.0 0.0 0.99885 0.99771
2 -85.60214 2.0 0.0 -0.00000 0.00000
3 -82.98830 2.0 0.0 0.00000 0.00000
4 -82.73201 4.0 0.0 0.00000 0.00000
5 -82.03407 3.0 0.0 -0.00000 0.00000
6 -81.22187 4.0 0.0 -0.00000 0.00000
7 -79.76617 -0.0 0.0 -0.02326 0.00054
We found this option useful in validating other capabilities, such as the strength
function capability.
4.7 Output files
BIGSTICK generates a number of output files. These fall into two broad cat-
egories. The most important output files have a name supplied by the user,
e.g., mg24 followed by an extension, e.g. .res or .wfn. Other files, which are
not needed by most casual users, have the same standard name upon each run,
ending in .bigstick.
Results. The most important file are the results files, which have an exten-
sion .res. When you initiate BIGSTICK, after the main menu choice, BIGSTICK
almost always asks you for the name of the output files:
Enter output name (enter "none" if none)
If you enter “none” then several files are suppressed, in particular the results
file.
The results file generally contains the output spectrum, e.g.
State E Ex J T
1 -41.39657 0.00000 0.000 0.000
2 -39.58581 1.81077 2.000 0.000
3 -37.08646 4.31012 4.000 0.000
4 -34.46430 6.93227 0.000 0.000
5 -33.56871 7.82786 2.000 -0.000
58
It also may contain one-body density matrices, or the results of strength function
runs.
Wave functions. The .wfn file contains wave function information, stored
in binary (or “unformatted.”) In addition to containing the wave function vec-
tors, it has a header which contains enough information the basis can be recre-
ated.
Autoinput. On each run BIGSTICK generates a file autoinput.bigstick.
This saves the various input when run from the terminal. This is useful if one
is making small tweaks to run, or to use as the basis for input directives. To
use the autoinput file directly from terminal, choose ‘i’ at the opening menu.
Log file. The .log file summarizes information about the run, such as the
date, time, BIGSTICK version number, dimensions, parallelization (number of
MPI processes and OpenMP threads), internal flag settings, and so on. While
not needed by the casual user, they are useful to document the exact conditions
under which a particular result ran and for debugging. If no output name is
specified, this file is named logfile.bigstick.
4.7.1 Secondary files
BIGSTICK generates some intermediate files which are not needed for ordinary
runs but in some cases can be useful. The most useful of these are the .lcoef
files, which in an ordinary Lanczos run contains the Lanczos coefficients αi, βi.
If no output name is specified, this file is called lanczosvec.lcoef.
4.7.2 Diagnostic files
BIGSTICK also generates a number of diagnostic files, primarily for development,
tuning, and debugging.
timingdata.bigstick contains the time spent in different matvec modes
(SPE, PP, etc) on each MPI process.
distrodata.bigstick contains the type and size of jumps stored on each
MPI process.
4.8 Memory usage
The motivation for BIGSTICK’s on-the-fly algorithm is to save memory over
storing the nonzero many-body matrix elements. Despite this, BIGSTICK can
still be quite memory-hungry. The main sinks of memory are: the Lanczos
vectors themselves, the jumps factorizing the many-body matrix elements, and
the uncoupled two-body matrix elements. Which dominates depends upon the
59

system. For example, in large phenomenological calculations, the main memory
usage is from Lanczos vectors. In no-core shell model calculations, it is typically
the jumps, except for very light systems (A= 3,4) where for large spaces the
uncoupled matrix elements actually dominate.
BIGSTICK gives a report on memory usage. It also has some default caps on
memory and will halt if these are violated. The default caps can be changed by
the user.
Both in normal runs and in modeling runs, BIGSTICK produces a report:
RAM for 2 lanczos vector fragments (max) : 3923.728 Mb
RAM for jumps in storage (total) : 2353.914 Mb
Max RAM for local storage of jumps : 177.069 Mb
RAM for uncoupled two-body matrix elements : 0.017 Mb
The RAM report above is for the initial and final Lanczos vectors in matvec. In
order to reorthogonalize, BIGSTICK also stores all Lanczos vectors. When run
in MPI, these Lanczos vectors are distributed across many MPI processes:
Enter max number of Lanczos iterations
150
Assuming max memory per node to store Lanczos vectors 16.00000 Gb
Storage of Lanczos vectors distributed up across 128 nodes
Memory per node = 10.74658 Gb
The default memory caps can all be found in the module flagger in the file
bmodules flags.f90. The most important ones are
real :: maxjumpmemory_default = 16.0 ! in Gb
real :: maxlanczosstorage1 = 16.000 ! in Gb
These can be changed, though of course BIGSTICK must be recompiled.
60

Chapter 5
Applications
In this chapter we discuss in more detail applications of BIGSTICK, specifically
one-body density matrices and one-body transition strengths.
5.1 One-body density matrices
BIGSTICK can be directed to compute the reduced one-body density matrices,
ρfi
K(ab)≡1
p(2K+ 1) DΨfhˆca†⊗˜cbiKΨiE(5.1)
where we use reduced matrix elements as defined in Appendix A.1. We use this
particular definition because the reduced matrix element of a generic one-body
operator is the sum of products of the density matrix elements and the reduced
matrix elements, namely,
hΨf|| ˆ
OK||Ψii=X
ab
ρfi
K(ab)ha|| ˆ
OK||bi.(5.2)
It’s important to note that ha|| ˆ
OK||biare matrix elements between single-
particle states, while the density matrices are matrix elements between many-
body states. While some many-body codes compute the many-body matrix ele-
ments for specific operators, such as E2, M1, and so on, we chose for BIGSTICK to
produce one-body density matrices, allowing the user to compute the transition
matrix elements for any one-body operator.
For systems with good isospin one can also define “doubly-reduced” matrix
elements, that is, reduced in both angular momentum and isospin:
ρfi
K,T (ab)≡1
p(2K+ 1)(2T+ 1) hΨf|||hˆca†⊗˜cbiK,T |||Ψii(5.3)
When you choose the density matrix option, BIGSTICK will write to the .res
file (but not to screen) the density matrices, e.g.:
61
Initial state # 2 E = -85.60214 2xJ, 2xT = 4 0
Final state # 1 E = -87.10445 2xJ, 2xT = 0 0
Jt = 2, Tt = 0 1
1 1 -0.01957 0.00000
1 2 0.18184 0.00000
1 3 0.09721 0.00000
2 1 -0.18184 0.00000
2 2 -0.35744 0.00000
2 3 -0.26323 0.00000
3 1 -0.09721 0.00000
3 2 -0.26323 0.00000
The first two lines are the labels and energies of the initial and final wave-
functions; Jt and Tt are the angular momentum and isospin of the one-body
operator, and, for example,
1 3 0.09721 0.00000
1 is the label of the first single-particle orbit, 3 the label of the second (as defined
by the input .sps file), and the two real numbers are the T= 0,1 one-body
density matrix elements, that is,
Ψ2hˆa†
1⊗˜a3iJ=2,T =0
Ψ1= 0.09721
while that for T= 1 is, here, zero.
BIGSTICK has two options for densities. The option dwill compute one-
body densities with good isospin, where the output looks like (this example is
23Ne in the sd shell with the USDB interaction):
Initial state # 1 E = -62.78960 2xJ, 2xT = 5 3
Final state # 1 E = -62.78960 2xJ, 2xT = 5 3
Jt = 0, Tt = 0 1
1 1 0.84730 0.28105
2 2 8.32846 2.85633
3 3 1.52286 0.13245
Alternately, there is the option dp which puts the density matrix elements into
explicit proton-neutron form.
Initial state # 1 E = -62.78960 2xJ, 2xT = 5 3
Final state # 1 E = -62.78960 2xJ, 2xT = 5 3
Jt = 0, proton neutron
1 1 0.16625 0.43288
2 2 1.58968 4.29943
3 3 0.47558 0.60124
62
5.1.1 Symmetries of density matrix elements
A useful symmetry relation is
ρif
K,T (ba) = (−1)ja−jb+Ji−Jf+Ti−Tfρfi
K,T (ab).(5.4)
5.1.2 Particle occupations from densities
Particle occupations are the average number of particles in single-particle orbit
for a given wave function. Although there is an option, ‘p,’ to compute the orbit
occupation, you can also extract this information from the diagonal one-body
density matrices. The total number of particles in orbit ais
n(a) = [ja]
[Ji]ρii
K=0(a†a) (5.5)
If your densities are in proton-neutron format, you can extract the proton and
neutron occupations separately. If you have your densities in isospin formalism,
you can extract the total number of protons and neutrons in an orbit
nπ(a) + nν(a) = [ja][1/2]
[Ji][Ti]ρii
K=0,T =0(a†a) (5.6)
To separately extract proton and neutron occupation one must take careful
account of the Clebsch-Gordan coefficients. One must have f=i, so that
Jf=Ji=J0and Tf=Ti=T0, as well as considering only a=b. Furthermore,
the answer depends upon Tz= (Z−N)/2 (using the notation [x] = √2x+ 1)
nπ(a) = 1
2[ja]
[J0] [T0]
1
2 ρii
K=0,T =0(a†a) + Tz√3
pT0(T0+ 1)ρii
K,T =1(a†a)!,(5.7)
nν(a) = 1
2[ja]
[J0] [T0]
1
2 ρii
K=0,T =0(a†a)−Tz√3
pT0(T0+ 1)ρii
K,T =1(a†a)!.(5.8)
5.1.3 Strengths from density matrix elements
Given some transition operator ˆ
OKcarrying definite angular momentum K, the
transition strength between an initial and final state is just the square of the
matrix element:
hJfMf|ˆ
OKM |JiMii
2
.
This is the matrix element that goes into Fermi’s golden rule for decay and
transition rates.
But in most experimental situations we cannot pick out specific values of
Mi,f (unless we are doing an experiment with polarization). The final result
must then average over initial states and sum over final states, that is,
1
2Ji+ 1 X
MiX
MfhJfMf|ˆ
OKM |JiMii
2
.(5.9)
63
In most cases there is also implicitly a sum over M. (If not, the final result
will be different.) Now we can use the Wigner-Eckart theorem to rewrite the
average/sum as:
1
2Ji+ 1 X
MfX
MiX
M|(JiMi, KM|JfMf)|2(Jf|| ˆ
OK||Ji)
2
.(5.10)
Now we can use the the selection rule Mf=Mi+Mfto eliminate the sum over
Mfand the orthogonality of the Clebsch-Gordan coefficients to sum over Mi
and MX
MiX
M|(JiMi, KM|JfMi+Mf)|2= 1 (5.11)
Thus we get the result in terms of reduced matrix elements,
1
(2Ji+ 1) X
MiX
MfX
M(JfMf|ˆ
OKM |JiMi)
2
=1
(2Ji+ 1) (Jf|| ˆ
OK||Ji)
2
,(5.12)
As one often calls 1
2Ji+ 1 (Jf|| ˆ
OK||Ji)
2
,(5.13)
the B-value, written B(O) (for example, B(GT ) for Gamow-Teller, B(E2) for
electric quadrupole, etc.), this says the strength for an operator is B(O).
In the BIGSTICK code and most other shell-model codes, we compute tran-
sition strengths using transition density matrix elements: the doubly reduced
transition matrix element for a one-body operator ˆ
OK,T of angular momentum
rank Kand isospin rank Tis
hΨf||| ˆ
OK,T |||Ψii=X
ab
ρfi
K,T (ab)ha||| ˆ
OK,T |||bi.(5.14)
Although the default output is doubly-reduced matrix elements, the defi-
nition of B-values do not sum or average over ‘orientations’ in isospin space,
because Tz= (Z−N)/2 is fixed. Hence we have to account for that by un-
doing the Wigner-Eckart reduction in isospin, so that, for non-charge changing
transitions (e.g., γ-transitions),
B(O:i→f) = 1
2Ji+ 1 (Ψf:Jf|| ˆ
OJ||ΨiJi)
2
=1
2Ji+ 1 (Ψf:JfTf||| ˆ
OJ,T |||ΨiJiTi)
2
×|(TiTz, T 0|TfTz)|2
2Tf+ 1 .(5.15)
Note the last line uses the result of Eq. (5.14).
64

5.1.4 Sample case: spin-flip
Let’s consider a couple of simple cases, both in the sd shell with the USDB
interaction [Brown and Richter, 2006]. Let’s consider the spin-flip operator
~σ = 2~
S, which has the following doubly-reduced matrix elements:
One-body matrix element value
h0d3/2|||~σ|||0d3/2i-2.19089
h0d3/2|||~σ|||0d5/2i4.38178
h0d5/2|||~σ|||0d3/2i-4.38178
h0d5/2|||~σ|||0d5/2i4.09878
h1s1/2|||~σ|||1s1/2i3.46410
The nuclide 19F, which has only one valence proton and two valence neutrons,
has, with appropriate scaling of the matrix elements, the low-lying spectrum:
State E Ex J T
1 -23.86096 0.00000 0.500 0.500
2 -23.78367 0.07729 2.500 0.500
3 -22.09059 1.77037 1.500 0.500
4 -21.26237 2.59858 4.500 0.500
5 -19.25724 4.60371 6.500 0.500
The density matrix from the second state (J= 5/2) to the third (J= 3/2)
state is, up to some overall phases,
Initial state # 2 E = -23.78367 2xJ, 2xT = 5 1
Final state # 3 E = -22.09059 2xJ, 2xT = 3 1
Jt = 1, Tt = 0 1
1 1 -0.08640 -0.01635
1 2 0.44978 -0.36112
1 3 0.01255 -0.09014
2 1 -0.16826 0.08815
2 2 -0.00280 -0.35352
3 1 -0.03483 0.07521
3 3 0.28978 -0.20874
Because the vector of Pauli matrices ~σ carries one unit of angular momentum
and no isospin, we only use the (Tt= 0) set of matrix elements (column second
from the right ). BIGSTICK also generates the transition matrix elements for Jt
=2, 3, and 4, not shown. Applying (5.15), we get a B(σ: 2 →3)=1.2609
A second case is 20Ne. The ground state is at -40.4723 MeV, which the first
J= 1, T = 0 state, state #25, is at -27.8364 MeV (or 12.636 MeV excitation
energy). The density matrix is
Initial state # 1 E = -40.47233 2xJ, 2xT = 0 0
Final state # 25 E = -27.83635 2xJ, 2xT = 2 0
Jt = 1, Tt = 0 1
1 1 0.00069 0.00000
65

1 2 0.14575 0.00000
1 3 -0.10567 0.00000
2 1 0.18722 0.00000
2 2 -0.04822 0.00000
3 1 -0.02309 0.00000
3 3 0.28308 0.00000
and B(σ: 1 →25) = 0.3597.
5.1.5 Charge-changing transitions
Charge-changing transition such as Gamow-Teller are a little more subtle. If
we have isospin-conserving interactions, so that our initial and final states have
good isospin, we can use isospin rotation so that we don’t have to change basis.
If we want to have a transition
A
ZXN→A
Z±1YN∓1,
that is, from some initial Tz,i = (Z−N)/2 to some final Tz,f = (Z−N)/2±1,
we must work in the basis with the smaller Tz; then both initial and final states
will be somewhere in the spectrum. What we want to calculate is
hΨf:Jf, TfTz,f || ˆ
O||Ψi:Ji, TiTz,ii
2
,
but what we can actually calculate with BIGSTICK is
hΨf:Jf, TfTz|| ˆ
O||Ψi:Ji, TiTzi
2
.
Fortunately this can be accomplished with only a small modification of the
above procedure:
B(O:i→f) = 1
2Ji+ 1 (Ψf:Jf|| ˆ
OJ||ΨiJi)
2
=1
2Ji+ 1 (Ψf:JfTf||| ˆ
OJ,T |||ΨiJiTi)
2
×|(TiTz,i, T ±1|TfTz,f )|2
2Tf+ 1 ,(5.16)
where the difference between Eq. (5.15) and (5.16) is in the isospin Clebsch-
Gordan. There is, however, one more subtle point in treating the isospin rais-
ing/lowering operator, τ±. If one treats τas a rank-1 spherical tensor in isospin
space, one can show that
τ±=1
√2τ1,±1.(5.17)
Therefore, formally, in the above calculations, we are actually using 2−1/2τ1,0
in our calculation, and then rotating to a charge-changing transition.
It’s always good to have a way to check calculations, and in the case of
Gamow-Teller it’s the Ikeda sum rule, which says
X
f
B(~στ+:i→f)−B(~στ−:i→f) = 3(N−Z) (5.18)
66

independent of the initial state i. Here the isospin raising operator τ+changes a
neutron into a proton and hence is the operator for β−decay, while the isospin
lowering operator τ−changes a proton into a neutron and hence is the operator
for β+decay. This assume our convention that protons are isospin ‘up’ and
neutrons isospin ‘down;’ many authors have opposite conventions.
5.1.6 Sample case: 19F
Let’s calculate the Gamow-Teller B-value. The matrix elements for Gamow-
Teller are the same as for ~σ as shown above except multiplied by p3/2. Then
using the Tt= 1 one-body density matrix elements, we get B(GT: 2 →3) =
1.3990.
For 20Ne, there is at J= 1, T = 1 state at -29.3066 MeV (or 11.166 MeV
excitation energy, state #15); the density matrix is
Initial state # 1 E = -40.47233 2xJ, 2xT = 0 0
Final state # 15 E = -29.30659 2xJ, 2xT = 2 2
Jt = 1, Tt = 0 1
1 1 0.00000 0.05163
1 2 0.00000 0.09951
1 3 0.00000 -0.03397
2 1 0.00000 0.18236
2 2 0.00000 0.32717
3 1 0.00000 -0.03311
3 3 0.00000 -0.08363
Here B(GT) = 0.1654, for either β+ or β−.
5.2 Strength function option
One important capability of BIGSTICK is using the Lanczos algorithm to effi-
ciently compute transition strength function distributions and to decompose a
wavefunction using a scalar operator, or option ‘s’ in the main menu.
5.2.1 Decomposition
We’ll start with decomposition of a wavefunction using a scalar operator [John-
son, 2015], because operationally it is the most straightfoward. Suppose you
have a wavefunction, |Ψi, which you have previously computed using BIGSTICK
and have stored in a .wfn file; further suppose you have some operator ˆ
Owhich
is an angular momentum scalar, which in turn means its matrix elements can be
stored in a file just like a Hamiltonian. This operator ˆ
Oin turn has eigenpairs,
ˆ
O|Φωi=ω|Φωi.(5.19)
We can always expand |Ψiinto the eigenstates of ˆ
O:
|Ψi=X
ω
cω|Φωi(5.20)
67
and the fraction of the wavefunction |Ψilabeled by ωis simply
|hΦω|Ψi|2=|cω|2.
This is particularly useful when ˆ
Ois the Casimir of some group or subgroup,
such as total orbital angular momentum ˆ
L2or total spin ˆ
S2. In that case we
say we decompose the wavefunction |Ψiinto its L- or S- components.
BIGSTICK can carry out this decomposition easily. What you need is, first,
a previously computed wavefunction in some .wfn file, and a file or files which
contain the matrix elements of the decomposing operator.
To do this:
1. From the initial menu choose the option ‘s’:
* (s) Strength function (using starting pivot ) *
.
.
.
Enter choice
s
Note: the pivot is the starting vector; here it is the wavefunction you wish
to decompose. BIGSTICK can only decompose one wavefunction at a time.
2. Enter name of file containing the wavefunction to be decomposed (i.e.,
the pivot):
Compute strength function distribution using previous wfn
Enter input name of .wfn file
mg24
Here the choice of the wavefunction file is mg24.wfn; you do not include the
extension. At this point, BIGSTICK reads in some information from the .wfn
file:
testing magic number 31415926 31415926
dimbasischeck= 28503
Valence Z, N = 4 4
Single particle space :
N L 2xJ
023
025
101
Total # of orbits = 3
2 x Jz = 0
The ‘magic number’ is a test of internal consistency to make sure, first,
BIGSTICK is correctly reading the file (in particular if the binary file was created
68
on a different platform) and also between different versions of BIGSTICK if the
information protocol has changed.
From this information BIGSTICK reconstructs the basis and checks the di-
mensions agree. It then asks for the output file:
Enter output name (enter "none" if none)
After this, BIGSTICK will make the standard inquiries for the Hamiltonian.
When decomposing a wavefunction, the ‘Hamiltonian’ is actually an angular
momentum scalar which is a Casimir of the group or sub-group; for example, it
could be ˆ
S2or ˆ
L2. Such files are in in the same format as any interaction file.
While we provide a sample operator, it is up to the user to generate these files.
After the interaction file(s) have been read in, you must enter in the number
of iterations and number of states to keep (BIGSTICK automatically chooses a
fixed-iteration run for Lanczos):
Enter nkeep, # iterations for lanczos
(nkeep = # of states printed out )
Exactly how many iterations to to choose requires some knowledge of the group,
or, specifically, knowledge of the eigenvalues of the Casimir, and, in many cases,
a few trials. Remember that the irreps of the group are labled by the eigenvalues
of the Casimir, which means the eigenvalues are highly degenerate. The number
of iterations needed should be no greater than the number of distinct eigenvalues.
So, for example, if one has 8 nucleons and is decomposing via spin, the values
of Scan be 0,1,2,3,or 4. Therefore the number of iterations should be no
more than 4 (because one wants a total dimension of 5). Often one can use
fewer iterations. If you use too many iterations, you will get duplication of
eigenvalues or, worse, unconverged duplicate eigenvalues.
Finally, BIGSTICK will print out a list of the starting states in the pivot file,
and their energies and Jand Tvalues, and ask you to choose a pivot:
There are 5 wavefunctions
5 states
STATE E J <H >
1 -92.7790 -0.0000 0.0000
2 -91.1196 2.0000 0.0000
3 -88.4779 2.0000 0.0000
4 -87.9781 4.0000 0.0000
5 -87.4348 3.0000 0.0000
Which do you want as pivot?
Hence if you want to decompose the J= 3 state, enter 5.
Immediately after reading in the pivot, BIGSTICK will print out the norm of
the input pivot (that generally does play a role in this kind of decomposition,
but will in transition strength functions):
69
0.999999998379788 = total input strength
Often the norm or total input strength is far different from 1.
After carrying out the specified Lanczos iterations, the result will look some-
thing like this, depending on how many iterations:
Energy Strength
______ ________
0.00000 0.63545
2.00000 0.33880
6.00000 0.02515
12.00000 0.00059
20.00000 0.00000
______ ________
The ‘energies’ on the left are the eigenvalues of the operator you are using to
decompose the wavefunction, here ˆ
S2. Hence the J= 3 state (or state 5 in
the above example), is 63.5%S= 0, 33.9%S= 1, and so on. These results are
written to the standard .res file.
5.2.2 Transition strength function distributions: the ba-
sics
Often we want the transition function between two states, that is hΨf|ˆ
O|Ψii
2
where ˆ
Ois some one-body transition operator, for example the E2 or M1 tran-
sition operator. (As always, we assume the reader is familiar with these con-
cepts.) If one only wants one or two transitions, one can compute those using
the one-body density matrices, which we describe above in 5.1.3.
But sometimes we want many transitions to many final (or ‘daughter’) states
from a single initial (‘parent’) state, for example if we want to profile ‘giant’
resonances. We can do this using BIGSTICK in three to four steps. The first
step is to generate and write to file an initial wavefunction.
The second step is to apply a one-body operator, ˆ
O. The matrix elements
of the one-body operator must be stored in file with extension .opme, with the
format defined in the next section. To apply a one-body operator, choose option
‘o’ at the opening menu:
* (o) Apply a one-body (transition) operator to previous wfn....
BIGSTICK will then ask for the name of the input .wfn file and an output name,
required here. After reconstructing the basis from the information in the input
.wfn file, it will ask:
Enter name of .opme file
The matrix elements of the one-body operator are read in from a file with
extension .opme (‘operator matrix element’). While we distribute some sample
70
.opme files with BIGSTICK, in general it is up to the user to generate such files.
The format of such files are
iso ! indicating good isospin
3 ! # of single-particle orbits
1 0 2 1.5 ! index of orbit, n, l, and j
2 0 2 2.5
3 1 0 0.5
1 0 ! J and T of operator
1 1 -2.19089 ! a, b < a ||| O ||| b >
1 2 4.38178
2 1 -4.38178
2 2 4.09878
3 3 3.46410
BIGSTICK first checks the list and order of single-particle orbits agrees with that
of the read-in wavefunction. BIGSTICK will then read in the matrix elements of
the one-body operator and apply it to each wavefunction stored in the input
.wfn file and write them to a new output .wfn file.
The final step is to run BIGSTICK again, this time with the strength function
option ‘s,’ using the wavefunction generated in the second step as input. This
time, when BIGSTICK asks for the interaction file name, you should use the
same file(s) to generate the initial state, because you are diagonalizing the
Hamiltonian.
As with decomposition, BIGSTICK will now carry out a fixed number of
iterations and print out the transition strength. Because it includes the norm
of the input pivot, these strengths can be greater than 1.
An important question is that of convergence. As you probably know, in the
Lanczos algorithm the extremal eigenpairs converge first, with interior eigenpairs
converging later. This is true as well for the strengths described above: the
extremal strengths will converge quickly to strengths (and eigenenergies) of
extremal levels, but interior strengths will often not be converged; instead they
will be some sort of ‘local average’ of strengths.
What looks like a bug is actually a feature. In practice one often doesn’t need
each and every strength to be fully converged. Instead we only need integrals
over the strengths to be converged, and this does happen. While we can only
refer the reader to Caurier et al. [2005] and references therein, we can state that
the moments of the distributions of strengths do converge. In fact, if one carries
out NLanczos iterations, one has ∼2Nmoments of the distribution correctly.
Hence often only thirty or fifty iterations suffice.
As an example, consider 20Ne in the sd shell with the USDB interaction. If
we apply the σoperator, with the matrix elements given above, and then apply
the strength function option s, the output will look something like:
0.38353481218057317 = total strength
35 iterations
71

Energy Strength
______ ________
-40.47233 0.00000
-38.72564 0.00000
-36.29706 0.00000
-33.77148 0.00000
-32.88892 0.00000
-30.18655 0.00000
-27.83635 0.11991
-25.73419 0.00003
-25.49045 0.17314
-24.73655 0.01326
-22.86589 0.00027
-22.24522 0.01542
Notices the strength at -27.836 MeV (which is the J;T= 1; 0 state) is 0.11991;
using the Clesbsch-Gordan coefficients gives a factor of 3, or a total strength of
0.3597, which agrees with our previous result.
There is a small bug in this lovely ointment: it assumes we have treated
angular momentum (and isospin) correctly, a topic we now turn to.
5.2.3 Transition strength functions with good angular mo-
mentum
In the prior subsection we glided over questions of angular momentum, which
we treat more carefully here. An important question is correct calculation of
the B-values, as defined in Eq. (5.13) above, which assume an average over
final states and a sum over final states. But what we computed in the previous
section was
|hΨf|ˆ
O|Ψii|2
where the states have fixed Mand fixed Tz, because, as currently written, both
initial and final wavefunctions must be in the same basis. (We plan at a later
date to write a separate tool which will allow one to apply and operator from a
wavefunction in one basis to a wavefunction in a different basis.) If you want the
applied operator to change parity, then both parities must be included in the
basis (option 0 when entering parity in the initial calculation of wavefunctions).
To extract the B-value, one has to invoke the Wigner-Eckart theorem:
B(O:i→f) = 1
2Ji+ 1 (Ψf:Jf|| ˆ
OJ||ΨiJi)
2
=
2Jf+ 1
(2Ji+ 1)
hΨf:JfM|ˆ
OJ0|Ψi:JiMi
(JiM, J0|JfM)
2
.(5.21)
If one has Ji= 0 (which can only happen if M= 0), then the B-value is
72
straightforward to calculate:
B(O:i→f) = (2J+ 1)|hΨf:J0|ˆ
OJ0|Ψi: 00i|2.
It’s more complicated with Ji6= 0; in that case the triangle rule, |Ji−J| ≤ Jf≤
Ji+Jis in effect, and in fact the state produced by BIGSTICK,
ˆ
OK|Ψi:Jii
will be an admixture of states of different Jf. Thus one needs an additional
step, of projecting out states of good angular momentum after applying the
transition operator but before carrying out the strength function option via
Lanczos. Fortunately we already know how to do this via decomposition as
discussed in 5.2.1.
Therefore, to properly carry out calculation of strength functions, you will
need (a) files for the interaction, (b) a file for the one-body transition operator,
and (c) files with matrix elements of ˆ
J2and, separately, T2(you only need
the latter if your transition operator has isospin rank 1). Then carry out the
following steps:
(1) With your interaction use option (n) or similar option, generate a .wfn
file containing an initial state;
(2) Use option (o) to apply the one-body operator (note this will be applied
to every wavefunction in the file);
(3) If your initial state has Ji6= 0, you will need to filter out a state of good
Jffor every possible Jf; in fact what you will do will be to decompose the state
from step (2) into its components Jf. Here you use option (s). If there are N
possible values of Jfyou only need to do N−1 iterations.
If your transition operator has T= 1 and your initial Ti6= 0, you will need
to further decompose into possible Tfstates.
(4) Finally, use option (s) again, but this time with the original interaction,
to get the strength function distribution. You will have to apply the Wigner-
Eckart theorem as in Eq. (5.21), but now that step (3) guarantees a definite
value of Jf(and, if needed, Tf), you can carry this out.
You will have to repeat for each possible final value of Jf.
Here is an example using 19F: if we choose the J= 5/2 state (state #2) as
the pivot and apply ~σ, and then use the strength function option with ˆ
J2,
1.3726179231928042 = total strength
3 iterations
Energy Strength
______ ________
3.75000 1.13861
8.75000 0.05630
15.75000 0.17771
______ ________
73

This means 83% of the pivot has J= 3/2 and only 4.08% has J= 5/2. Next
we run the strength function again on the second state using the original (usdb)
interaction:
1.0000000433422094 = total strength
35 iterations
Energy Strength
______ ________
-23.86096 0.00000
-23.78367 0.00000
-22.09059 0.66440
-21.26237 0.00000
-19.25723 0.00000
Note that the wavefunction is normalized when it is read in. (By the way,
the zeroes show up because although there is no strength to them, or very
little, roundoff error allows them to grow during Lanczos. This is the same
phenomenon which forces us to orthogonalize new Lanczos vectors against old
ones.) We have to multiply 0.66440 ×1.13861 = 0.756 to get the ‘raw’ strength,
which here is |hJfM|~σ|JiMi|2, then we have to follow Eq. (5.21):
0.756 ×2(3/2) + 1
2(5/2) + 1 ×1
|(5/2 1/2,1 0|3/2 1/2)|2= 0.756 ×4
6×1
2/5= 1.260
which agrees with our previous result! Note that you have to do each step with
care; if don’t scale the two-body matrix elements, you will get different results.
5.2.4 Gamow-Teller with strength function option
Charge-changing transitions such as Gamow-Teller are a straightforward gen-
eralization but require even more care. Here one transitions from a state with
some initial Tz,i to some final Tz,f =Tz,i ±1. Because, as of the time of this
writing, BIGSTICK requires the same initial and final basis, we have to choose
Tz,0= min (abs(Tz,i),abs(Tz,f )) and invoke isospin rotation. Typically you will
have to filter both Jand T. The B-value is given by
B(O:i→f) = 1
2Ji+ 1 hΨf:Jf, TfTz,f || ˆ
OJ||Ψi:Ji, TiTz,ii
2
=
2Jf+ 1
(2Ji+ 1)
hΨf:JfM, TfTz,0|ˆ
O1 0,1 0|Ψi:JiM, TiTz,0i
(JiM, J0|JfM)
2
(5.22)
×
(TiTz,i,1±1|TfTz,f )
(TiTz,0,1 0|TfTz,0)
2
where the last line uses the isospin Wigner-Eckart theorem to transform from
the isospin frame the calculation is carried out in, to the physically desired
isospin frame.
74
We use BIGSTICK’s strength function option sto compute the matrix element
|hΨf:JfM, TfTz,0|ˆ
O1 0,1 0|Ψi:JiM, TiTz,0i|2. This is slightly involved. We
give now a detailed example, again with 19F. After applying the Gamow-Teller
operator, we filter out first with ˆ
J2, using the second state as the pivot:
0.72792934307990387 = total strength
3 iterations
Energy Strength
______ ________
3.75000 0.61870
8.75000 0.01998
15.75000 0.08925
______ ________
and then we filter this with ˆ
T2(applied to the first state, that is, the one with
J= 3/2
0.99999997896552772 = total strength
3 iterations
Energy Strength
______ ________
0.75000 0.77481
1.10978 0.00000
3.75000 0.22519
______ ________
and then finally applying the strength function with the usdb interaction:
1.0000000877719881 = total strength
35 iterations
Energy Strength
______ ________
-23.86096 0.00000
-23.78367 0.00000
-22.09059 0.87875
-21.26237 0.00000
-19.25722 0.00000
Thus in this example the ‘raw’ transition strength is that for the J;T=
5/2; 1/2→3/2; 1/2 which is
0.61870 ×0.77481 ×0.87875 = 0.42125.
This now has to be converted to a Gamow-Teller B-value by Eq. (5.22):
B(GT ) = 2·3
2+ 1
2·5
2+ 1 ×0.42125
5
2
1
2,1 0|3
2
1
2
2×1
2−1
2,1 1|1
2
1
2
1
2+1
2,1 0|1
2
1
2
2
75
A good exercise is to compute low-lying transitions two ways, first with
density matrices, and then via the strength function option, to confirm they
agree with each other. For Gamow-Teller, one can and should verify results by
using the Ikeda sum rule.
One can also use other operators for projection, for example, using center-
of-mass to project out nonspurious states in no-core shell model calculations.
We plan to later allow two-body transition operators, but as of version 7.8.1
these options have not yet been installed.
76

Chapter 6
A peek behind the curtain
Although this manual details how to use BIGSTICK, it only outlines the algo-
rithms and program. The distribution includes an Inside Guide which, while
incomplete, contains many more details on the code, and the source code itself is
heavily commented. Even so, it is a complex program, with more than seventy
Fortran files and on the order of 70,000 lines. Several files are for specialized
applications most users will not care about, as well as for features slated for
obsolescence.
There are ways to get BIGSTICK to present more information about its inner
workings, as well as ways to extert more control over the algorithm. A number
of logical flags turn behaviors on and off. The most important flags, and some
default settings, are found in bmodules flags.f90. Some additional flags for
output are in the module io in bmodules main.f90, and other flags can be
found elsewhere. To detail all the possibilities would expand this already long
manual by a significant amount.
In this chapter we outline the major steps BIGSTICK takes in carrying out
a ‘normal’ run, as well as telling the curious user how to print out an explicit
representation of the basis and of the many-body Hamiltonian matrix.
6.1 A normal run
Here are the steps BIGSTICK carries out in a ‘normal’ run, that is, setting up a
many-body Hamiltonian and finding the low-lying extremal eigensolutions.
•BIGSTICK sets up the basis.
•BIGSTICK counts up the number of jumps (data needed for constructing
the Hamiltonian on-the-fly).
•BIGSTICK gathers the interaction data.
•If running in parallel, BIGSTICK computes the distribution
77
•BIGSTICK generates the data needed (on a specific MPI process if running
in parallel), specifically the jumps and the decoupled matrix elements.
•BIGSTICK sets up storage for the Lanczos vectors.
•BIGSTICK begins Lanczos iterations.
•Upon completion of Lanczos, BIGSTICK constructs the low-lying eigenvec-
tors. It resets the jumps and computes angular momentum and isospin
as expectation values. Eigenvalues and eigenvectors are written to file. If
density matrices requested, BIGSTICK resets jumps for one-body operators
and computes.
•Upon finishing, BIGSTICK reports on timing and closes down.
6.2 Explicit representation of the basis
Through factorization and other tricks, BIGSTICK only implicitly stores the basis
and the Hamiltonian. In actual operations, BIGSTICK stores information on
pieces of Slater determinants, which we call “haikus.” Haikus are organized by
quantum numbers, as are the action of single-fermion creation and annihilation
operators on the haikus. These latter we called “hops” and from them we
construct jumps, and from jumps we construct many-body matrix elements,
and so on. Once the hops are created the haikus are not needed, and once the
jumps are constructed the hops are not needed.
It can be useful, however, to have explicit representations of both the basis
and of the many-body Hamiltonian. In standard runs, the final eigenvectors
are written to file with extension .wfn. These files are unformatted to save
space. Furthermore the detailed basis information is not saved; instead any
basis BIGSTICK is constructed in a standard order, and when reading a .wfn file
BIGSTICK swiftly reconstructs the basis.
From the main menu, however, the option ‘t’ will write out both the basis
and the eigenvectors in explicit, human-readable form, to a file with extension
.trwfn (originally written as an input to Petr Navratil’s density code TRDENS).
Here is an annotated example output from the p-shell, using the Cohen-
Kurath interaction. First is a header describing the nucleus:
4 ! valence Z
4 ! valence N
ckpot ! name of INTERACTION FILE
19.45492 ! HW (approx) 12
1 ! # of majors shells
12 1 ! total p+n s.p.s, # shells core
0 ! Nmax (excitations)
51 ! # of many-body configurations = basis dimension
1 ! parity, +
0 ! 2 x Jz
78
5 ! # of eigenstates kept
Next is a list of the eigenenergies, and numerical J and T values; the latter are
written as real numbers. Reading across we have E,J, and T.
-71.04467 3.6173283E-06 -4.1723251E-07
-66.39703 2.000000 -3.8743019E-07
-58.59552 1.000001 -4.4703484E-07
-57.57795 3.6235542E-06 -4.1723251E-07
-57.54143 4.000000 -2.9802322E-07
Then comes a list of the single particle state quantum numbers. Reading across
we have label, n(number of radial nodes), l, 2 ×j, 2 ×jz, and 2 ×tz:
1011-11
2013-11
3013-31
401111
501311
601331
7 0 1 1 -1 -1
8 0 1 3 -1 -1
9 0 1 3 -3 -1
10 0 1 1 1 -1
11 0 1 3 1 -1
12 0 1 3 3 -1
So single-particle state 1 is a 0p1/2with jz=−1/2 and is a proton, single-
particle state 2 is 0p3/2with jz=−1/2 and is a proton, etc.
Finally we have a listing of the 51 many-body basis states and their ampli-
tudes for the first five eigenstates:
1 2 3 5 8 10 11 12
0.1989746094 0.1647298932 -0.0000000709 -0.0788139924 -0.1036236882
1 2 3 5 7 10 11 12
0.0805789307 0.0938614532 0.0855044648 -0.1246829554 -0.0399925224
1 2 3 4 8 10 11 12
-0.0805789307 -0.0938614309 0.0855044946 0.1246829703 0.0399925113
1 2 3 4 7 10 11 12
-0.0582427122 -0.0433882587 0.0000000068 0.0952372476 0.0186970048
1 2 3 6 7 8 11 12
0.0825415403 -0.0342168659 -0.0919694155 -0.0390651748 0.1794791222
1 2 3 6 7 8 10 12
0.0697834268 -0.0333072022 0.0155492499 -0.1079809889 0.1385308802
1 2 3 6 9 10 11 12
-0.1513192952 0.0582505427 -0.0000000329 0.0562667921 -0.3108672500
2 3 4 5 7 8 11 12
0.0560085103 -0.0361775197 -0.0000000029 -0.0111581217 0.1036224812
...
79

So the first many-body basis states has occupied single particle states 1,2,3,5
(protons) and 8,10, 11, and 12 (neutrons). The five real numbers following are
the amplitudes for this basis state for the five eigenstates whose eigenenergies
are given above. You can see an example of factorization: the basis states 1 and
2 have the same proton occupancies loop over neutron occupancies; while basis
states 3 and 4 have the same proton occupancies (but different from basis states
1 and 2) and loops over the same neutron occupancies as basis states 1 and 2.
By adding up the jzvalues, the proton “Slater determinants” have Mp=−2
and the neutron Slater determinants have Mn= +2. The next basis states,
number 5 through 7, have Mp=−Mn=−1. In this way BIGSTICK builds up
the basis. As shown in the example, in constructing the basis via factorization,
the innermost loop is over neutron Slater determinants while the outer is over
protons.
We recommend against using this option on a regular basis, because writing
this information to a file is slow, and BIGSTICK does not have postprocessing
options for this format. Nonetheless it can be useful for understanding what is
going on, and could be a basis for a user’s own post-processing code.
6.3 Explicit representation of the Hamiltonian
The many-body Hamiltonian can also be explicitly generated. If from the Lanc-
zos menu you choose ‘ex’ for exact or full diagonalization, the entire Hamiltonian
is created and explicitly stored and solve using the LAPACK routine DSYEV. This
occurs in the routine exactdiag p in the file blanczos main.f90. If the basis
dimension is less than 100, BIGSTICK will automatically write out the Hamilto-
nian matrix elements to a file ham.dat. This occurs around line 1246; the user
can edit this part of the code to control the dimension cutoff for writing out
(in general we do not encourage writing out for large dimensions) as well as the
format.
80
Chapter 7
Lanczos algorithm
Today the most common way to find all the eigenpairs of a Hermitian (here,
real and symmetric) matrix is first reduce the matrix to tridiagonal form via a
sequence of unitary transformations, the Householder algorithm[Parlett, 1980,
Press et al., 1992], and then solve the resulting tridiagonal matrix via QL de-
composition with implicit shifts (or so we’ve been told). But for the very large
dimensions of standard CI calculations, one neither can extract all eigenpairs
nor does one want to: beyond a certain point one expects intruder many-body
states to enter and eventually dominate, rendering our calculated states lack-
ing physical meaning. (For the novice: an intruder state is a state outside the
designated model space. There is no simple way, at least for the non-expert, to
determine where we expect intruders to dominate.)
Instead we turn to the Lanczos and related algorithms [Parlett, 1980, Press
et al., 1992, Whitehead et al., 1977]. Lanczos is part of a family of so-called
Arnoldi algorithms, which iteratively construct a new orthonormal basis, the
Krylov subspace. In this new basis the Hamiltonian is tridiagonal, but unlike
the Householder algorithm, one does not need to fully carry out the transfor-
mation. The Lanczos algorithm is simple, beautiful, and powerful, though like
all algorithms it is not without its own limitations.
7.1 Standard Lanczos algorithm
The Lanczos algorithm is exceedingly straightforward. We will summarize it
here, though we will not explicate it in detail. Starting from some initial vector
|v1i, called the pivot, one iteratively generates a sequence of orthonormal vectors
81

{|vii, i = 1, k},hvi|vji=δij :
ˆ
H|v1i=α1|v1i+β1|v2i
ˆ
H|v2i=β1|v1i+α2|v2i+β2|v3i
ˆ
H|v3i=β2|v2i+α3|v3i+β3|v4i
. . .
ˆ
H|vii=βi−1|vi−1i+αi|vii+βi|vi+1i
. . .
ˆ
H|vki=βk−1|vk−1i+αk|vki
(7.1)
Each iteration generates a new Lanczos vector. If we stop at the k−1th iter-
ation, we have kLanczos vectors and a k-dimension Krylov subspace. Using
orthonormality of the vectors, one can show that in this basis, the Hamiltonian
is tridiagonal:
Hi,i =hvi|ˆ
H|vii=αi,(7.2)
Hi,i+1 =Hi+1,i =hvi|ˆ
H|vi+1i=βi.(7.3)
and all other matrix elements are zero.
The specific steps for creating the next Lanczos vectors are straightforward:
(1) |wii=ˆ
H|viiInitial matvec on vector i;
(2) αi=hvi|wiidot product to get αi;
(3) |wii←|wii − αi|viiorthogonalize against initial vector i;
(4) If i > 1|wii←|wii−βi−1|vi−1iorthogonalize against prior vector i-1;
(5) βi=phwi|wiifind norm to get βi;
(6) |vi+1i=β−1
i|wiiNormalize to get i+1th Lanczos vector.
If we had perfect arithmetic, this would be sufficient: the new Lanczos vector
|vi+1iwould be guaranteed to be orthogonal to all previous vectors. But we
don’t have perfect arithmetic, and due to round-off noise, small components of
prior Lanczos vectors will creep in and eventuall grow exponentially.
This requires us to enforce orthogonality against all prior Lanczos vectors:
(4)(alt.) For j = 1 to i-1 : |wii←|wii−|vjihvj|wii
If one does not reorthogonalize, eventualy one gets ‘ghost eigenvalues’, or
repetitions of the same eigenvalues. It is this need for reorthogonalization that
keeps Lanczos from supplanting Householder as the go-to algorithm for full
tridiagonalization of Hermitian matrices.
There has been much discussion and experimentation around partial re-
orthogonalization, but no one clearly successful recipe. BIGSTICK fully reorthog-
onalizes against all prior vectors; in most cases (a few hundred iterations) re-
orthogonalization work does not overwhelm matvec work.
You might notice that if one extended the for loop in our alternate step
(4), we would already get step (3). Because of finite arithmetic, order matters.
We find better results if we first compute αiand then orthogonalize against all
other vectors, rather than as a last step.
It is possible for a user to experiment with these fine tweaks in BIGSTICK. The
Lanczos iterations are found in subroutine lanczos p in file blanczoslib1.f90.
82
One can estimate the workload from reorthogonalization. Again, let Nbe
the dimension of the vector space. Each projection requires a dot product and
a subtraction, or about 2Noperations. For kLanczos vectors, one has k−1
iterations. For the jth iteration one orthogonalizes against jvectors or 2Nj
operations; thus for k−1 iterations one has 2N
Obviously with full reorthogonalization full Lanczos transformation to tridi-
agonal form becomes expensive; hence the dominance of the Householder algo-
rithm for complete diagonalization.
7.2 Thick-restart Lanczos
Sometimes there is insufficient storage for the number of Lanczos vectors re-
quired for convergence. An alternative is the thick-restart Lanczos[Wu and
Simon, 2000]. In standard Lanczos there are essentially two dimensions, Nkeep,
the number of converged states desired, and Niter, the number of iterations
(typically kabove). But one must store Niter + 1 Lanczos vector, which can be
prohibitive. For thick-restart Lanczos, there is an additional dimension Nthick,
with Nkeep < Nthick < Niter, and is the dimension of the Krylov subspace when
restarting. In otherwords, one iterative creates a subspace of dimension Niter +1,
but then truncates down to dimension Nthick, and then adds additional vectors
back up to a subspace of dimension Niter + 1, truncate back down, and repeat
until convergence. The advantage is that Niter, and the consequent number of
vectors stored, is much smaller than would be needed for standard Lanczos.
Thick-restart Lanczos follows this basic outline:
1. Start with some initial Lanczos pivot vector |v1ias usual.
2. Carry out kLanczos iterations so that you have k+ 1 Lanczos vectors
|vii, i = 1, k + 1, and a truncated k+ 1 ×k+ 1 Hamiltonian matrix Tk+1.
3. Diagonalize the k×ksubmatrix Tk.
4. From the eigenpairs of step (3), choose the Nthick lowest states. These
will form the “new” Lanczos eigenvectors. In addition, keep |vk+1iand
use this as the restarting vector for Lanczos.
5. Now restart Lanczos, but instead of starting with |v1i, start with |vk+1i
which is our new |vNthick+1i.
6. Iterate until you have again k+ 1 Lanczos vectors and an truncated k+
1×k+ 1 Hamiltonian matrix Tk+1. This new matrix will no longer be
tridiagonal, but it will have a simple form, given below.
Now let’s describe this in more detail. Suppose we have carried out kLanczos
iterations, so that we have a total of k+1 vectors |vii, including the pivot. Then
83
the transformed Hamiltonian, which is the Hamiltonian in the basis {|vii}, looks
like
Tk+1 =
α1β10 0 . . . 0
β1α2β20. . . 0
0β2α3β3. . . 0
.
.
.
0 0 . . . αk−1βk−10
0 0 . . . βk−1αkβk
0 0 . . . 0βkαk+1
(7.4)
Suppose, however, we only diagonalize Tk, that is, stopping at the kth column
and row, with Lbeing the k×kunitary matrix of eigenvectors, that is,
k
X
j=1 Tkij Ljµ =Liµ ˜
Eµ,(7.5)
for µ= 1, k. Here ˜
Eµare the approximate eigenenergies. If we apply the unitary
transform Lto the first kvectors, that is, introducing
|v0
µi=
k
X
i=1 |viiLi,µ, µ = 1, k (7.6)
and |v0
k+1i=|vk+1ithe transformed matrix, which is the Hamiltonian the basis
{|v0
ii}, now becomes
˜
E10 0 0 . . . βkLk1
0˜
E20 0 . . . βkLk2
0 0 ˜
E30. . . βkLk3
.
.
.
0 0 . . . ˜
Ek−10βkLk,k−1
0 0 . . . 0˜
EkβkLkk
βkLk1βkLk2. . . βkLk,k−1βkLkk αk+1
(7.7)
The key to thick-restart Lanczos is to judiciously truncate this. If you want,
as is the usual case, to get the lowest Nkeep states, truncate to some Nthick (with
Nkeep < Nthick < k) vectors, that is, to
|v0
1i,|v0
2i,|v0
3i, . . . |v0
Nthick i
plus the last Lanczos vector, |vk+1i, then the truncated Hamiltonian looks like
˜
E10 0 0 . . . βkLk1
0˜
E20 0 . . . βkLk2
0 0 ˜
E30. . . βkLk3
.
.
.
0 0 . . . ˜
ENthick−10βkLkNthick−1
0 0 . . . 0˜
ENthick βkLkNthick
βkLk1βkLk2. . . βkLk,Nthick −1βkLkNthick αk+1
(7.8)
84
Now declare |vk+1ito be the new |v0
Nthick+1iand start the Lanczos iterations
on it:
H|v0
Nthick+1i= (7.9)
βkLk1|v0
1i+βkLk2|v0
2i+. . . +αNthick+1|v0
Nthick+1i+βNthick+1|v0
Nthick+2i
This first step is not a tridiagonal relation; furthermore , although our new
|v0
Nthick+1iis the same as our old |vk+1i, and our new αNthick+1 is the same as
the old αk+1, the new vector |v0
Nthick+2iis not the same as |vk+1iwould have
been had we continued the previous iteration, although the former contains the
latter as a component, because the iteration step is different.
Now one continues iterations Nthick + 2, Nthick + 3, . . . , k + 1. Then one diag-
onalizes the approximate Tkagain,although it is no longer a pure tridiagonal,
and in fact looks like:
˜
E10 0 0 . . . βkLk10. . .
0˜
E20 0 . . . βkLk20. . .
0 0 ˜
E30. . . βkLk30. . .
.
.
....
0 0 . . . ˜
ENthick−10βkLkNthick−10. . .
0 0 . . . 0˜
ENthick βkLkNthick 0
βkLk1βkLk2. . . βkLk,Nthick −1βkLkNthick αNthick+1 βNthick+1
0 0 . . . 0 0 βNthick+1 αNthick+2
.
.
....
,
and restarts as above, repeating under convergence.
This thick-restart algorithm requires more matvec multiplications than stan-
dard Lanczos, because information is thrown away at each restart, but the stor-
age and reorthogonalization of Lanczos vectors can be greatly reduced. There is
no recommended value of Nthick or k, but one should take kas large as practical,
and “typical” values of Nthick ≈3Nkeep or so.
Although the usual application to thick restart is to find low-lying states, it
is conceivable to choose a slice of excited energy and to converge excited states.
This will be investigated.
7.3 Can I restart standard Lanczos?
The standard Lanzos algorithm is an iterative algorithm. In principle, if you
found the desired eigenpairs had not converged under the chosen number of
iterations, you could pick up and restart. To do this you would need the Lanczos
vectors created so far and the Lanczos coefficients.
Although in prior version BIGSTICK wrote the Lanczos vectors to disk, in
its current version stores all Lanczos vectors in RAM. In MPI parallelization
the Lanczos vectors are stored across multiple processes. Therefore right now
the restart option has been turned off. It is possible in future versions we may
restore it, although it is not a high priority.
85
Chapter 8
Parallel computing and
timing
BIGSTICK can run many non-trivial problems on modest desktop or even lap-
top computers. Because problems grow exponentially, however, single-processor
calculations quickly reach limits. To overcome these limits we invoke parallel
processing.
Although many parts of the set up portion of the code have been paral-
lelized, by far the most time-consuming part of the code is the matrix-vector
multiplication, followed by reorthogonalization, and it is these two portions it
is most important to parallelize.
For very large calculations, one needs to distribute both matvec operations
(work load balance) and data (memory load balance). Operations are paral-
lelized using both MPI (distributed memory) and OpenMP (shared memory)
while data can only be distributed with MPI.
When BIGSTICK starts, it tells you how many MPI processes and how many
OpenMP threads per process it is using:
Number of MPI processors = 512
NUM_THREADS = 8
This information is also written to the .log file. BIGSTICK does not have any
special requirements for setting up parallel runs, although to run in parallel one
must use an executable compiled with parallel options, i.e. bigstick-mpi.x
compiled with make mpi,bigstick-opemp.x compiled with make openmp, or
the hybrid bigstick-omp-mpi.x compiled with make openmp-mpi. Any user
who wishes to use the parallel capability should already have some idea about
submitting parallel jobs. For example, to set up the number of OpenMP threads
on a desktop machine you typically
PROMPT>export OMP_NUM_THREADS=8
and to submit an MPI job you may do
86
PROMPT>mpirun -n 512 bigstick-mpi.x
Of course, the details will depend upon the local environment. Unfortunately, in
our experience supercomputers do not have a uniform job submission protocol.
8.1 MPI
To carry out a Lanczos iteration, which includes a matvec followed by reorthog-
onalization, one needs the following data:
•an initial vector;
•an final vector;
•jump information used for on-the-fly construction of the many-body matrix
elements; and
•the uncoupled two- (or, optionally, three-) body matrix used in construction
of the many-body matrix elements;
•previously computed Lanczos vectors (used for calculation of the Lanczos
coefficients, for reorthogonalization and, ultimately, construction of the final
eigenvectors which represent wavefunctions).
In large calculations some or all of these may need to be distributed via MPI.
To compute the distribution efficiently, BIGSTICK goes throught the setup in
two stages. First, it calculates the number of operations in each matvec, that
is, in
vfinal
i=X
j
Hij vinitial
j,(8.1)
each update
vfinal
i←vfinal
i+Hij vinitial
j.(8.2)
Because of factorization, BIGSTICK does not have to actually generate every
operation. BIGSTICK then generates the distribution, and each MPI process
creates locally the data it needs.
BIGSTICK attemps to distribute the operations across MPI processes as
evenly as possible. The operations are constructed from jumps, but the ra-
tio of operations to jumps is not fixed. We find it helpful to think of matvec
operations as represented by the area of a rectangle, and the sides of the rectan-
gle representing the jumps. If the rectangle is nearly square, the reconstruction
is efficient, but if one has a long, thin rectangle in either dimension, one requires
considerably more storage of jumps relative to the number of operations. Oc-
casionally, an equitable distribution of operations will, on some small subset of
MPI processes, so many jumps they cannot be stored. In that case, BIGSTICK
will distribute those jumps over multiple MPI processes; this of course leads to
a load imbalance, but is necessary so as not to exhaust memory.
8.1.1 Fragments
If the basis dimension is so large both initial and final vectors cannot be con-
tained in core memory, they must be broken into fragments. When running in
87
MPI, or in modeling mode, BIGSTICK will ask for this automatically:
Enter desired limit on fragment size for breaking Lanczos vectors
(Largest un-splittable block = 16602236 )
( Default = 200000000 )
( Enter 0 to use default )
Somewhat counterintuitively, BIGSTICK achieves best efficiency when the frag-
ments are as large as possible. The reason for this is that the factorization
principle behind BIGSTICK works most efficiently when combining large conju-
gate data.
Fragments are generally combinations of contiguous sectors (a portion of
the vector which is labeled by the proton quantum numbers), although, because
the lengths of sectors can vary significantly, in some cases a fragment can be
comprised of a single sector. In the most extreme cases BIGSTICK will seek to
divide a sector into two new ‘sectors,’ although there are limitations to how
finely this can be done. Otherwise BIGSTICK attempts to make the fragments
as similar in size as practical.
If you run BIGSTICK in MPI mode, it will ask for the fragment size. The
fragment size is approximately the length of the initial and final vectors stored
on a given MPI process (because of the way the code chunks data, BIGSTICK
actually allows for a small overrun). Choosing a value of 0 will select the default
value, currently around 200 million, which is actually on the small size. Because
Lanczos vectors are stored in single precision, this requires roughly 1.6 Gb of
RAM for the initial and final vector fragments. On many machines you can
choose this to be larger.
Matvec operations are now defined from an initial fragment (of a Lanczos
vector) to a final fragment (of a Lanczos vector). This work will generally be
spread across mulitple MPI processes; hence one needs nproc (the number of
MPI processes) ≥nfragments2(the number of fragments). In fact, BIGSTICK
will complain if nproc <2×nfragments2.
8.1.2 Opbundles and optypes
The operations are organized by a derived type (Fortran’s designation for a
bundle of data, very much like a struct in C) called opbundles, or bundles of
operations. Opbundles are the ‘natural’ way to divide up work in BIGSTICK. Op-
bundles orchestrate the application of matvec operations, and BIGSTICK provide
information about opbundles. Most users will not need this information.
Each opbundle has an associated ‘optype,’ which classifies the physical origin
of the matrix elements being reconstructed. For example, the ‘PP’ optype is for
interactions betwen two protons, with neutrons as spectators. There are also
NN and PN optypes, and for three-body forces, PPP, NNN, PNN, and PPN.
Finally there has been an optype SPE for single-particle energies and related
single-particle potentials. However this has been absorbed into PP, PN, and
NN optypes. We do this by multiplying any one-body term by ( ˆ
N−1)/(A−1),
with ˆ
Nthe number operator and Athe (valence) mass number. In the same
88
way, if one runs with three-body forces, any two-body forces are subsumed into
three-body by multiplying the two-body operators by ( ˆ
N−2)(A−2).
Optypes signal different operations and invoke different methods of recon-
structing the matrix elements. PP optypes use proton ‘two-body jumps’ and
loop over spectator neutron Slater determinants, NN optypes use neutron two-
body jumps and loop over spectator proton Slater determinants, and PN op-
types use both proton one-body jumps and neutron one-body jumps. Not only
do these invoke different subroutines, the time per operation is different for
different optype, because the loops are different, and may be different on differ-
ent machines. This information in turn is used to calculation the distribution
of work. Information on the timing of these operations is found in the file
timinginfo.bigstick. In many cases, by carrying out a short run to establish
the time per operation, written to timinginfo.bigstick, and then running the
desired run using this information, can lead to significantly greater efficiency.
Unfortunately, the time per operation is not as fixed for an optype as we
originally hope, and detailed investigations show a great deal of fluctuations.
We are still investigating this issue and attempt to arrive at better weighting
and distribution algorithms.
8.1.3 Modeling
One menu option BIGSTICK offers is modeling, or choice ‘m’ on the main menu.
This will run mostly like a normal run, with the following differences:
•No interaction file information will be requested (although if three-body
forces are enables, it will ask if you want to model the use of three-body
forces);
•Prompt for mandatory information on fragments;
•Prompt for mandatory information on the number of MPI processes; in-
formation on the number of OpenMP threads is not needed;
•Prompt for the number of Lanczos vectors.
You can model a run using a different number of MPI processes than the
modelled number.
The modeling option will calculate the distribution of work and data. This
is useful because you can find out if the number of MPI processes requested is
insufficient, or if BIGSTICK can find a distribution solution at all. (In some rare
cases the algorithm currently fails.)
8.2 OpenMP
BIGSTICK uses OpenMP where it can, in particular in matvec. Unfortunately
due to the nature of the problem, there are limitations to the speedup form
OMP. Because the matrix elements are very sparse, one tends to lose locality.
89

Modern computers have at least three levels of storage: disk storage, RAM
storage, and cache storage. These three kinds of memory are increasingly close
to the CPU and thus are increasingly faster; they are also increasing smaller in
size. When data is fetched from disk or even from RAM, the CPU also fetches
nearby data and leaves it in the cache. If the program needs that cached data
next, it is handily nearby and thus faster to be accessed. Because of the highly
nonlocal nature of the data, however, BIGSTICK has trouble reaching maximum
efficiency. While we continue to work on this issue, by the very nature of the
sparse matrix this is difficult. Some of the work we have carried out is described
in Shan et al. [2015, 2017].
8.3 Timing
In order to improve efficiency, BIGSTICK contains a number of built-in vari-
ables and routines for tracking and reporting timing. When running in serial,
BIGSTICK uses the FORTRAN routines date and time or cpu time. Unfortu-
nately these do not provide very accurate timing, on the order of 0.01 second,
so some information is not accurate. When running in MPI, BIGSTICK uses
BMPI Wtime, which is much more accurate.
BIGSTICK will give an estimate of the time to run,
Approximate time per iterations estimated : 2112 sec, or 35.2 min
but keep in mind this is a rough estimate. This uses information in timinginfo.bigstick
which contains timing from previous runs. If you previously ran a similar prob-
lem, this estimate is likely reliable, but if the problem changes, or if you are
using the default assumption, when timinginfo.bigstick does not exist, then
the results may vary.
8.3.1 Mode times
The main timing in BIGSTICK is to measure the amount of time the code spends
in various modes of operation, i.e., in generating the basis, computing jumps,
matvec (matrix-vector multiplication), reorthognalization, and so on. At the
end of a run, BIGSTICK prints out the culmulative time. These times are written
to the terminal as well as the .res results file. The output looks something like
this:
Total time to run : 58.7889999998733
Time to compute basis : 3.999999724328518E-003
Time to count up jumps : 1.099999994039536E-002
Time to decouple matrix elements : 1.999999862164259E-003
Time to compute jumps : 1.899999985471368E-002
Time to compute lanczos : 42.0250000003725
Time total in H mat-vec multiply : 30.9959999998100
Time to apply sp energies : 4.599999962374568E-002
90
Time in pn : 13.1739999908023
Time in pn(back) : 8.17700000526384
Time in 2-body (pp) : 2.46199999982491
Time in 2-body (pp)(back) : 2.45600000442937
Time in 2-body (nn) : 4.66199999954551
Time in reorthogonalization : 10.8760000029579
Time to compute J^2, T^2 : 9.499999973922968E-002
Time in applyobs : 0.950999999884516
Time spent diagonalizing. : 7.299999939277768E-002
8.3.2 Timing for parallel runs
In addition to timing various modes during a run, BIGSTICK provides timing data
useful for load balancing MPI parallel runs. As discussed elsewhere, BIGSTICK
attempts to distribute work across MPI processes by counting up the number of
operations and distributing the work. Operations are managed by opbundles,
and each opbundle is associated with a particular Hamiltonian mode: proton-
proton (PP), neutron-neutron (NN), proton-neutron (PN), and so on. Therefore
BIGSTICK tracks the time spent on each MPI process, on each Hamiltonian mode
on each MPI process, and finally on each opbundle.
91

Appendix A
Matrix elements and
operators
A.1 Reduced matrix elements
The Wigner-Eckart theorem states that a matrix element which depends upon
Jzis proportional to a Clebsch-Gordan coefficient, that is,
hJfMf|ˆ
OKM |JiMii= [Jf]−1(JiMi, KM|JfMf)(Jf|| ˆ
OK||Ji) (A.1)
= (−1)Jf−MfJfK Ji
−MfMKMi(Jf|| ˆ
OK||Ji)
where (Jf|| ˆ
OK||Ji) is the reduced matrix element, which encapuslates the funda-
mental matrix element independent of orientation, and which in 5.1.3 is related
to a sum over all orientations.
Eq. (A.1) can also be thought of as the definition of the reduced matrix
element (and the Wigner-Eckart theorem a statement that this definition is
consistent using any set of Ms). Note that it is possible to have a variant defini-
tion with different pre-factors, that is, the phase and factors like p2Jf+ 1 are
conventions. Only the Clebsch-Gordan coefficients are dictated by the theorem.
The choices of (A.1), taken from Edmonds [1996] are the most widely used ones.
The Wigner-Eckart theorem applies not just to angular momentum but any
SU(2) algebra; hence one can reduce in isospin as well, and a doubly-reduced
matrix element follows naturally:
hJfMf;TfMT f |ˆ
OKM;T MT|JiMi;TiMT ii= (A.2)
(JiMi, KM|JfMf)
[Jf]
(TiMT i, T MT|TfMT f )
[Tf](Jf, TF|| ˆ
OK,T ||Ji, Ti).
92

A.2 The Hamiltonian and other operators in sec-
ond quantization
Here we carefully define our operators in second quantization, that is, using
fermion creation and annihilation operators and coupled up to good angular
momentum. To denote generic operators ˆα, ˆ
βcoupled up to good total angular
momentum Jand total z-component M, we use the notation
(ˆα׈
β)JM =X
mα,mβ
(jαmα, jβmβ|JM )ˆαjαmαˆ
βjβmβ,(A.3)
where (jαmα, jβmβ|JM ) is a Clebsch-Gordan coefficient (here and throughout
we use the conventions of Edmonds [1996]).
Hence we can define the general fermion pair creation operator
ˆ
A†
JM (ab) = (ˆa†×ˆ
b†)JM (A.4)
with two particles in orbits aand b. We also introduce the time-reverse of
A†
JM (ab), the pair annihilation operator,
˜
AJM (cd) = −(˜cט
d)JM (A.5)
Here we use the standard convention ˜cmc= (−1)jc+mcˆc−mc, where mcis the
z-component of angular momentum. An alternate notation is
ˆ
AJM (cd) = ˆ
A†
JM (cd)†
= (−1)J+M˜
AJ,−M(cd) (A.6)
With this we can write down a standard form for any one- plus two-body
Hamiltonian or Hamiltonian-like operator, which are angular momentum scalars.
To simplify we use
ˆ
H=X
ab
eab ˆnab
+1
4X
abcd
ζabζcd X
J
VJ(ab, cd)X
M
ˆ
A†
JM (ab)ˆ
AJM (cd),(A.7)
where ˆnab =Pmˆa†
mˆ
bmand ζab =√1 + δab. Here VJ(ab, cd) = hab;J|ˆ
V|cd;Ji
is the matrix element of the purely two-body part of ˆ
Hbetween normalized
two-body states with good angular momentum J; because it is a scalar it is in-
dependent of the z-component M. To make our results as broadly interpretable
as possible, we also write this as
X
ab
eab[ja]ˆa†×˜
b0,0
+1
4X
abcd
ζabζcd X
J
VJ(ab, cd) [J]ˆ
A†
J(ab)ט
AJ(cd)0,0(A.8)
93

where we use the notatation [x] = √2x+ 1, which some authors write as ˆx;
we use the former to avoid getting confused with operators which always are
denoted by either ˆaor ˜a.
Finally we also can introduce one-body transition operators with good an-
gular momentum rank Kand z-component of angular momentum M,
ˆ
FK,M =X
ab
Fab
1
[K]ˆa†×˜
bK,M (A.9)
Here Fab =ha|| ˆ
FK||biis the reduced one-body matrix element.
A.3 Symmetries of matrix elements
Two-body matrix elements satisfy the following symmetries:
VJ(ab, cd) = −(−1)ja+jb+JVJ(ba, cd) (A.10)
=−(−1)jc+jd+JVJ(ab, dc) = (−1)ja+jb+jc+jdVJ(ba, dc).
Including isospin,
VJT (ab, cd) = −(−1)ja+jb+J+1+TVJT (ba, cd) (A.11)
=−(−1)jc+jd+J+1+TVJT (ab, dc)=(−1)ja+jb+jc+jdVJT (ba, dc).
Because we assume real-valued matrix elements, VJT (ab, cd) = VJT (cd, ab). Al-
though internally BIGSTICK has a specified order for storing matrix elements,
the code can read in matrix elements in any order and with the indices a, b, c, d
in any order.
Non-scalar spherical tensors should satisfy [Edmonds, 1996]:
ˆ
FKM †
= (−1)Mˆ
FK,−M.(A.12)
For non-charge-changing transitions, Eq. (A.12) implies Fab = (−1)ja−jbF∗
ba.
94
Appendix B
Highlighted references
There are a number of books and review articles on the configuration-interaction
shell model. We focus on those in nuclear physics. One of the best, but nowa-
days difficult to get, is Brussard and Glaudemans [1977]. Some other useful
references, in historical order, are [De-Shalit and Talmi, 2013], Towner [1977],
Lawson and Lawson [1980] (thorough, but be aware his phase conventions differ
from most others), Talmi [1993], Heyde [1994], Suhonen [2007], and others. A
particular useful review article touching on many of the ideas here Caurier et al.
[2005]; the review article Brown and Wildenthal [1988] is older but has useful
information on applications of the shell model. The no-core shell model and
other ab initio methods are a rapidly evolving field, but good overviews of the
topic are Navr´atil et al. [2000] and Barrett et al. [2013].
For angular momentum coupling a widely used reference is the slim volume
by Edmonds [1996]. If you can’t find what you need in Edmonds, you can
almost certainly find it in Varshalovich et al. [1988]. Sadly, neither are good
pedagogical introductions to the topic of angular momentum algebra.
95
Bibliography
F. Andreozzi and A. Porrino. J. Phys. G: Nucl. Part. Phys, 27:845, 2001.
B. R. Barrett, P. Navr´atil, and J. P. Vary. Ab initio no core shell model. Progress
in Particle and Nuclear Physics, 69:131–181, 2013.
B. Brown, A. Etchegoyen, and W. Rae. Computer code OXBASH: the Oxford
University-Buenos Aires-MSU shell model code. Michigan State University
Cyclotron Laboratory Report No. 524, 1985.
B. A. Brown and W. D. M. Rae. The Shell-Model Code NuShellX@MSU.
Nuclear Data Sheets, 120:115–118, 2014.
B. A. Brown and W. A. Richter. New “usd” hamiltonians for the sd shell. Phys.
Rev. C, 74:034315, Sep 2006.
B. A. Brown and B. H. Wildenthal. Status of the nuclear shell model. Annual
Review of Nuclear and Particle Science, 38:29–66, 1988.
P. Brussard and P. Glaudemans. Shell-model applications in nuclear spec-
troscopy. North-Holland Publishing Company, Amsterdam, 1977.
E. Caurier and F. Nowacki. Present status of shell model techniques. Acta
Physica Polonica B, 30:705–714, 1999.
E. Caurier, G. Mart´ınez-Pinedo, F. Nowacki, A. Poves, J. Retamosa, and A. P.
Zuker. Full 0¯hω shell model calculation of the binding energies of the 1f7/2
nuclei. Phys. Rev. C, 59:2033–2039, Apr 1999.
E. Caurier, G. Martinez-Pinedo, F. Nowacki, A. Poves, and A. P. Zuker. The
shell model as a unified view of nuclear structure. Reviews of Modern Physics,
77:427–488, 2005.
D. B. Cook. Handbook of computational quantum chemistry. Oxford University
Press, 1998.
A. De-Shalit and I. Talmi. Nuclear shell theory, volume 14. Academic Press,
2013.
96
J. Draayer, T. Dytrych, K. Launey, and D. Langr. Symmetry-adapted no-
core shell model applications for light nuclei with qcd-inspired interactions.
Progress in Particle and Nuclear Physics, 67(2):516–520, 2012.
A. R. Edmonds. Angular momentum in quantum mechanics. Princeton Univer-
sity Press, 1996.
D. Gloeckner and R. Lawson. Spurious center-of-mass motion. Physics Letters
B, 53(4):313–318, 1974.
K. L. Heyde. The nuclear shell model. Springer, 1994.
F. Jensen. Introduction to computational chemistry. John Wiley & Sons, 2017.
C. W. Johnson. Spin-orbit decomposition of ab initio nuclear wave functions.
Phys. Rev. C, 91:034313, Mar 2015.
C. W. Johnson, W. E. Ormand, and P. G. Krastev. Factorization in large-scale
many-body calculations. Computer Physics Communications, 184:2761–2774,
2013.
P. Knowles and N. Handy. A new determinant-based full configuration interac-
tion method. Chemical physics letters, 111(4-5):315–321, 1984.
R. Lawson and R. Lawson. Theory of the nuclear shell model. Clarendon Press
Oxford, 1980.
P.-O. L¨owdin. Quantum theory of many-particle systems. i. physical interpre-
tations by means of density matrices, natural spin-orbitals, and convergence
problems in the method of configurational interaction. Phys. Rev., 97:1474–
1489, Mar 1955.
P. Navr´atil, J. Vary, and B. Barrett. Large-basis ab initio no-core shell model
and its application to 12 c. Physical Review C, 62(5):054311, 2000.
F. Palumbo. Intrinsic motion and translational invariance in shell-model calcu-
lations. Nuclear Physics A, 99(1):100–112, 1967.
F. Palumbo and D. Prosperi. Effects of translational invariance violation in
particle-hole calculations. application to 208pb. Nuclear Physics A, 115(2):
296–308, 1968.
T. Papenbrock and D. J. Dean. Factorization of shell-model ground states.
Phys. Rev. C, 67:051303, May 2003.
T. Papenbrock and D. J. Dean. Density matrix renormalization group and
wavefunction factorization for nuclei. Journal of Physics G: Nuclear and
Particle Physics, 31(8):S1377, 2005.
T. Papenbrock, A. Juodagalvis, and D. J. Dean. Solution of large scale nuclear
structure problems by wave function factorization. Phys. Rev. C, 69:024312,
Feb 2004.
97
B. N. Parlett. The symmetric eigenvalue problem, volume 7. SIAM, 1980.
W. H. Press, B. Flannery, S. Teukolsky, and W. Vetterling. Numerical recipes
in fortran. Cambridge university press, 1992.
H. Shan, S. Williams, C. Johnson, K. McElvain, and W. E. Ormand. Parallel im-
plementation and performance optimization of the configuration-interaction
method. In Proceedings of the International Conference for High Performance
Computing, Networking, Storage and Analysis, page 9. ACM, 2015.
H. Shan, S. Williams, C. Johnson, and K. McElvain. A locality-based
threading algorithm for the configuration-interaction method. 2017. URL
http://escholarship.org/uc/item/9sf515zf.
I. Shavitt. The history and evolution of configuration interaction. Molecular
Physics, 94:3–17, 1998.
C. D. Sherrill and H. F. Schaefer. The configuration interaction method: Ad-
vances in highly correlated approaches. Advances in quantum chemistry, 34:
143–269, 1999.
N. Shimizu. Nuclear shell-model code for massive parallel computation,” kshell”.
arXiv preprint arXiv:1310.5431, 2013.
P. Sternberg, E. Ng, C. Yang, P. Maris, J. Vary, M. Sosonkina, and H. V. Le.
Accelerating configuration interaction calculations for nuclear structure. The
Proceedings of the 2008 ACM/IEEE Conference on Supercomputing, 2008.
J. Suhonen. From Nucleons to Nucleus: Concepts of Microscopic Nuclear The-
ory. Springer Science & Business Media, 2007.
I. Talmi. Simple models of complex nuclei. CRC Press, 1993.
J. Toivanen. Efficient matrix-vector products for large-scale nuclear shell-model
calculations. arXiv preprint arXiv:nucl-th/0610028, 2006.
I. S. Towner. A shell model description of light nuclei. Clarendon Press Oxford,
1977.
D. A. Varshalovich, A. N. Moskalev, and V. K. Khersonskii. Quantum theory
of angular momentum. World scientific, 1988.
A. W. Weiss. Configuration interaction in simple atomic systems. Phys. Rev.,
122:1826–1836, Jun 1961.
R. R. Whitehead, A. Watt, B. J. Cole, and I. Morrison. Computational methods
for shell model calculations. Advances in Nuclear Physics, 9:123–176, 1977.
K. Wu and H. Simon. Thick-restart lanczos method for large symmetric eigen-
value problems. SIAM Journal on Matrix Analysis and Applications, 22(2):
602–616, 2000.
98