De Identification Of Free Text Medical Records Deid User Manual
DeidUserManual
DeidUserManual
DeidUserManual
DeidUserManual
DeidUserManual
User Manual: Pdf
Open the PDF directly: View PDF ![]() .
.
Page Count: 14

1
De-Identification of Free-Text Medical Records
User Manual
version 1.1
Written by
Ishna Neamatullah
September 5, 2006
Modified by
Li-wei Lehman May 15, 2009
Harvard-MIT Division of Health Sciences and Technology
Massachusetts Institute of Technology, Cambridge, MA 02139

2
Introduction
About this Document
This is the user’s manual for the de-identification software developed at the Harvard/MIT Division of Health
Sciences and Technology. It describes the dictionaries used by the software, file format used for various
input/output files, PHI tag types generated, and an overview of the top-level API of the software. This document
does not attempt to provide an exhaustive description of the software's purpose, structure or inner workings. For
these details consult references listed in the bibliography. Consult the README.txt file for a summary of the
software's installation and execution instructions.
About the De-Identification Software
The de-identification software is the product of a study at the Harvard-MIT Division of Health Science and
Technology (HST) to automatically de-identify confidential patient information from text medical records used
in intensive care units (ICUs). Patient records are a vital resource in medical research. Before such records can
be made available for research studies, protected health information (PHI) must be thoroughly scrubbed
according to HIPAA specifications to preserve patient confidentiality. Manual de-identification on large
databases tends to be prohibitively expensive, time-consuming and prone to error, making a computerized
algorithm an urgent need for large-scale de-identification purposes. We have developed an automated pattern-
matching de-identification algorithm that uses medical and hospital-specific information. The current version of
the algorithm has an overall recall/sensitivity of around 0.967 and a precision (or positive predictive value) of
0.748.

3
Software Installation and Execution
Platforms
Perl 5.8 or 5.10 under Fedora Core 10, Linux 2.6.27 (development and testing). The code is also expected to run
on Windows but have not been tested on that platform.
Code organization
The source code is contained in a single file (deid.pl). Each de-identification run can be configured using
deid.config. Associated dictionaries used for de-identification are in folders /lists and /dict. We suggest
extending or modifying all other lists and dictionaries to suit your particular needs. A comprehensive description
of all lists and dictionaries follows in the next section.
Installation and Execution
Please see the README.txt file.

4
File Formats
The input to the code needs to be a single text-file containing the gold standard corpus with an extension .text.
Format for id.text
Each record in the corpus starts with the following format:
START_OF_RECORD=<Patient-ID>||||<Record-Number>||||
The record must end with:
||||END_OF_RECORD
It is assumed that each patient has a unique patient ID, and each note has a unique record number for the patient.
Note that in this current release of the gold standard corpus, the record date is not supplied. A default date is
used in the perl code for date shifting when the record date is not supplied in the header. If you would like the
deid code to date shift the dates within the medical records properly, you need to supply a record date for each
record in the header as follows:
START_OF_RECORD=<Patient-ID>||||<Record-Number>||||<Record-Date>||||
The <Record-Date> should be in the format of <MM/DD/YYYY>.
Format for id.deid
The PHI location file (id.deid), containing all gold standard PHI locations, does not need to be passed into the
code as an argument. When the user requests performance statistics, the algorithm assumes that a PHI locations
file called <filename>.deid exists in the directory.
The format of this .deid is as follows:
Patient <Patient-ID><TAB>Note <Record-Number>
<PHI-Start><TAB><PHI-Start><TAB><PHI-End>
Patient <Patient-ID><TAB>Note <Record-Number>
<PHI-Start><TAB><PHI-Start><TAB><PHI-End>
An example follows for notes 1 and 2 for a patient with ID 1100:
Patient 1100 Note 1
12 12 15
24 24 29
Patient 1100 Note 2
10 10 18
245 245 251
310 310 312
Note: The first <PHI-Start> is the character index of the beginning of the word with the PHI. The second <PHI-
Start> is the index of the beginning of the PHI selection. In the Gold Standard corpus, they are the same number.
The third number <PHI-End> is the index of the last character selected as PHI.
Format for id.phi
This file has same format as id.deid.
Format for id.types
The PHI type/category file (id.types) contains the category of each PHI that appears in the gold standard corpus.
PHIs are classified into the following categories: PTName, PTNameInitial, HCPName, RelativeProxyName,

5
Location, Date, DateYear, Phone, etc. See Appendix B for a complete listing of the PHI types. Some of the
common PHI types are described as follows.
-PTName: Patient names (first, middle, or last names)
-PTNameInitial: Patient name initials
-RelativeProxyName: Names of patient's family members or proxies
-HCPName: Health Care Professional Names (doctors, nurses, hospital workers, etc.)
-Location: locations which include hospitals, company names, street addresses.
-Date: dates with day/month/year.
-DateYear: stand-alone year (without day or month).
-Phone: this includes telephone, pager and fax numbers.
The format of the PHI category file is as follows:
<Patient-ID> <Record-Number> <PHI-Start> <PHI-End> <Type>
where <Type> is one of the PHI categories.
Format for id.info
This file contains information on PHI locations and de-identification process for debugging purposes. The
format is <PHI-Start> <PHI-End> <PHI_Text> <PHI_TYPE>
The <PHI_Text> is the string from the text that corresponds to the PHI start end locations indicated in the first
two numbers. If it is preceded by #, it is NOT a PHI. This means that the string was considered by the deid
code as a potential PHI, but the algorithm ultimately decided that it's not a PHI. If it is not preceded by #, then it
is considered a PHI. In this case, the <PHI_Text> will be followed by the <PHI_TYPE> string.
6
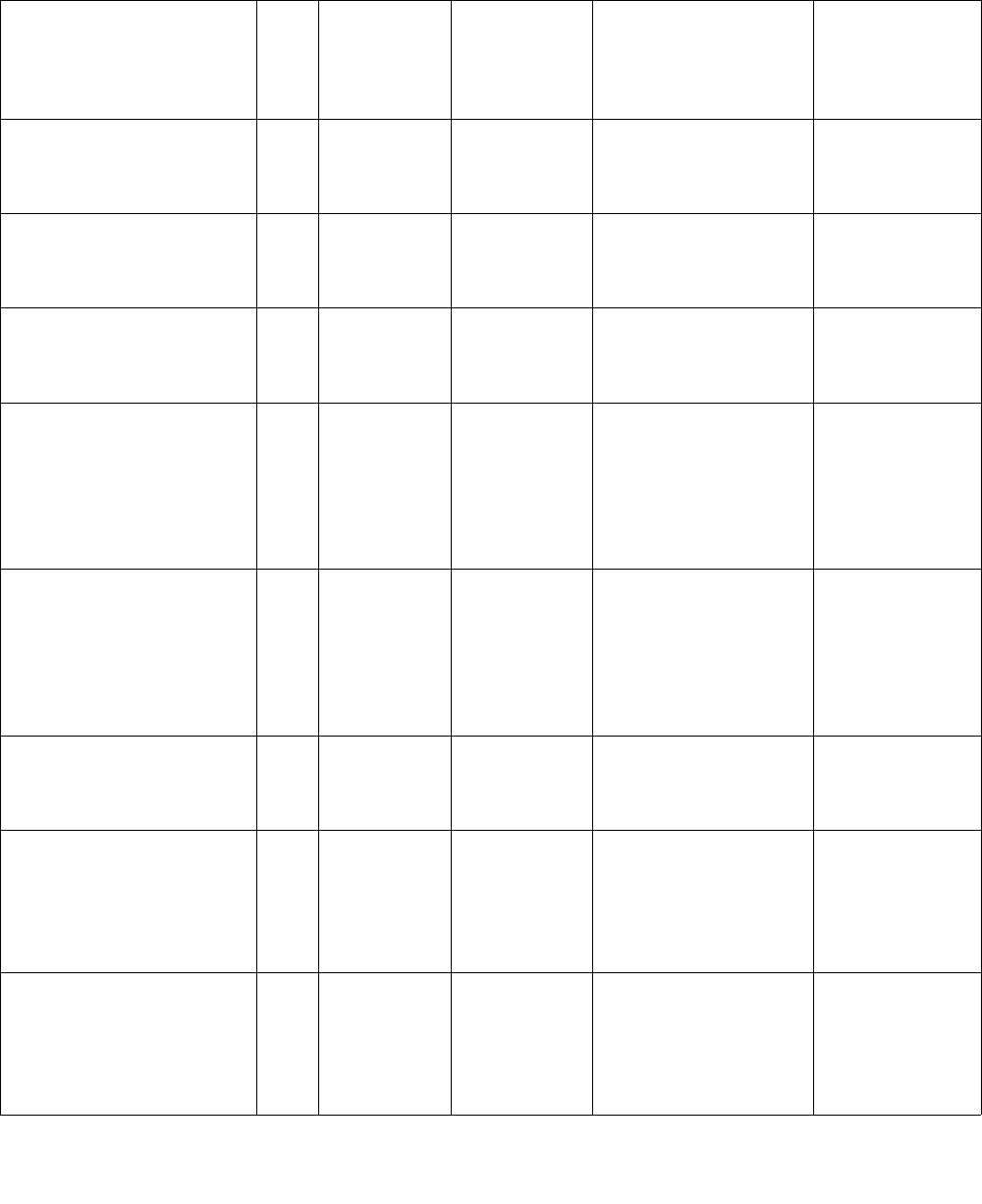
Dictionaries and Lists
The code uses multiple lists of known PHI and dictionaries of words and medical terms. We present the specific
format of each list/dictionary in the following table. Multi-word names are allowed in the dictionary unless
specified otherwise. In the case of multi-word names, the code will scan for the pattern in the note--all words in
the name must appear (in the order listed in the dictionary) for there to be a match.
Name of list/dictionary Dire
ctory
locat
ion
Description Format Use in code Notes
company_names_unambig.t
xt
/lists Unambiguous
names of
companies
1 name per
line
Each line is hashed and
scrubbed directly.
company_names_ambig.txt /lists Ambiguous
names of
companies
1 name per
line
Each line is hashed and
marked as potential
PHI.
countries_unambig.txt /lists Unambiguous
names of
countries
1 name per
line
Each line is hashed and
scrubbed directly.
doctor_first_names.txt /lists Unambiguous
first names of
doctors
(hospital-
specific)
1 name per
line
Each line is hashed and
scrubbed directly.
This file contains
a list of
unambiguous,
surrogate doctors'
names in gold
standard corpus.
doctor_last_names.txt /lists Unambiguous
last names of
doctors
(hospital-
specific)
1 name per
line
Each line is hashed and
scrubbed directly.
This file contains
a list of
unambiguous,
surrogate doctors'
names in gold
standard corpus.
ethnicities_unambig.txt /lists Unambiguous
names of
ethnicities
1 name per
line
Each line is hashed and
scrubbed directly.
female_names_unambig.txt /lists Unambiguous
female first
names
1 name per
line
Each line is hashed and
scrubbed directly. Also
used in name filter to
determine if a word is a
first name.
male_names_unambig.txt /lists Unambiguous
male first
names
1 name per
line
Each line is hashed and
scrubbed directly. Also
used in name filter to
determine if a word is a
first name.
7
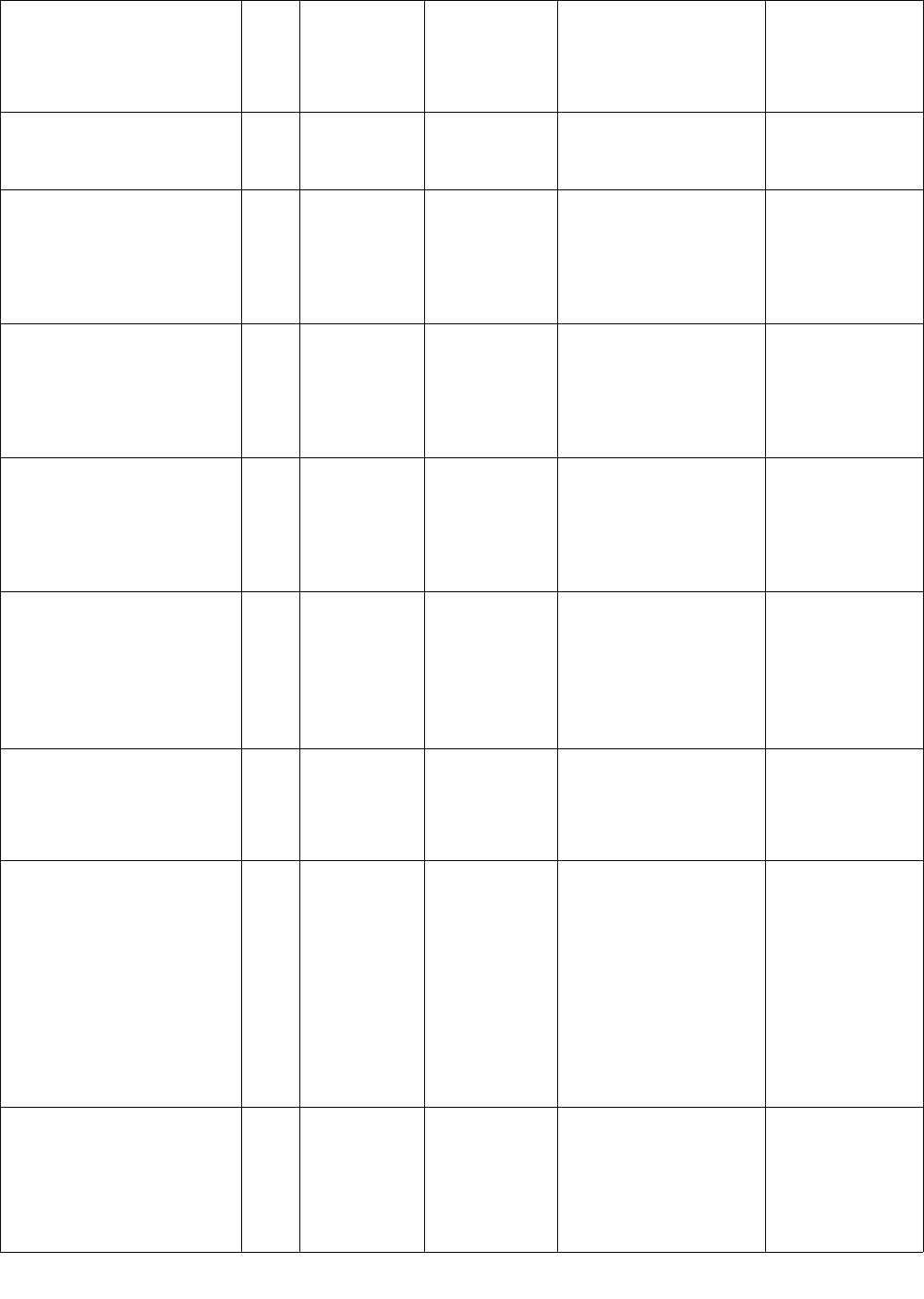
Name of list/dictionary Dire
ctory
locat
ion
Description Format Use in code Notes
last_names_unambig.txt /lists Unambiguous
last names
1 name per
line
Each line is hashed and
scrubbed directly.
female_names_ambig.txt /lists Ambiguous
female first
names
1 name per
line
Each line is hashed and
marked as potential
PHI. Also used in name
filter to determine if a
word is a first name.
male_names_ambig.txt /lists Ambiguous
male first
names
1 name per
line
Each line is hashed and
marked as potential
PHI. Also used in name
filter to determine if a
word is a first name.
last_names_ambig.txt /lists Ambiguous
last names
1 name per
line
Each line is hashed and
marked as potential
PHI. Also used in name
filter to determine if a
word is a last name.
female_names_popular.txt,
male_names_popular.txt,
last_names_popular.txt
/lists Popular
female/male
first names
and last
names
1 name per
line
Each line is hashed and
marked as potential
PHI. Also used in name
filter to determine if an
ambiguous name is also
a popular name.
last_name_prefixes.txt,
prefixes_unambig.txt
/lists Prefixes that
may appear
before a last
name
1 prefix per
line
Used to identify name
patterns.
locations_unambig.txt /lists Unambiguous
location
names
1 location
name per line
Each line is hashed and
scrubbed directly.
Multiple words are
scrubbed in the context
of the whole location
name, i.e. each word in
a multi-word location
will not be removed
when occurring in
isolation.
locations_ambig.txt /lists Ambiguous
location
names
1 location
name per line
Each line is hashed and
marked as potential
PHI. Multiple words are
scrubbed in the context
of the whole location
name, i.e. each word in
8
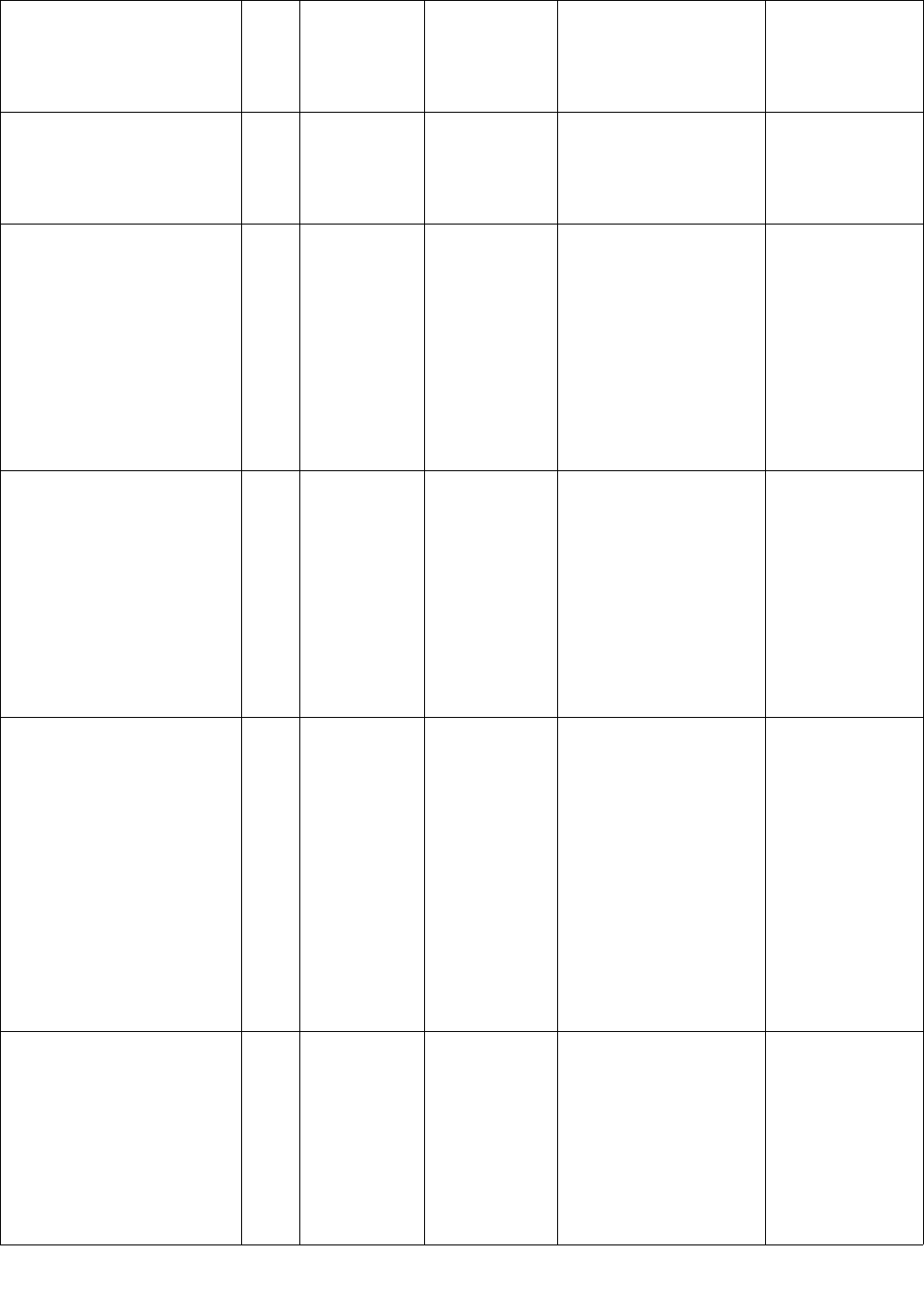
Name of list/dictionary Dire
ctory
locat
ion
Description Format Use in code Notes
a multi-word location
will not be removed
when occurring in
isolation.
local_places_unambig.txt /lists Towns and
cities around
the hospital
1 location
name per line
Each line is hashed and
scrubbed directly.
Multiple words are
scrubbed in the context
of the whole location
name, i.e. each word in
a multi-word location
will not be removed
when occurring in
isolation.
local_places_ambig.txt /lists Ambiguous
town and city
names around
the hospital
1 location
name per line
Each line is hashed and
marked as potential
PHI. Multiple words are
scrubbed in the context
of the whole location
name, i.e. each word in
a multi-word location
will not be removed
when occurring in
isolation.
pid_patientname.txt /lists Patient ID
(PID), patient
first names,
patient last
names
Per line: PID||||
firstname1
firstname2||||
lastname1
lastname2
Max 2 words
for first
names; max 2
words for last
names
For the PID of the
processed record, each
patient name (first or
last) is removed
directly.
This file contains
a list of surrogate
patient names
that appear in the
gold standard
corpus. For
names exceeding
4 words modify
existing code
segment at the
beginning of
deid() to extend
this functionality.
stripped_hospitals.txt /lists Unambiguous
hospital
names
1 name per
line
Each line is hashed and
scrubbed directly.
Multiple words are
scrubbed in the context
of the whole location
name, i.e. each word in
a multi-word location
will not be removed
when occurring in
9
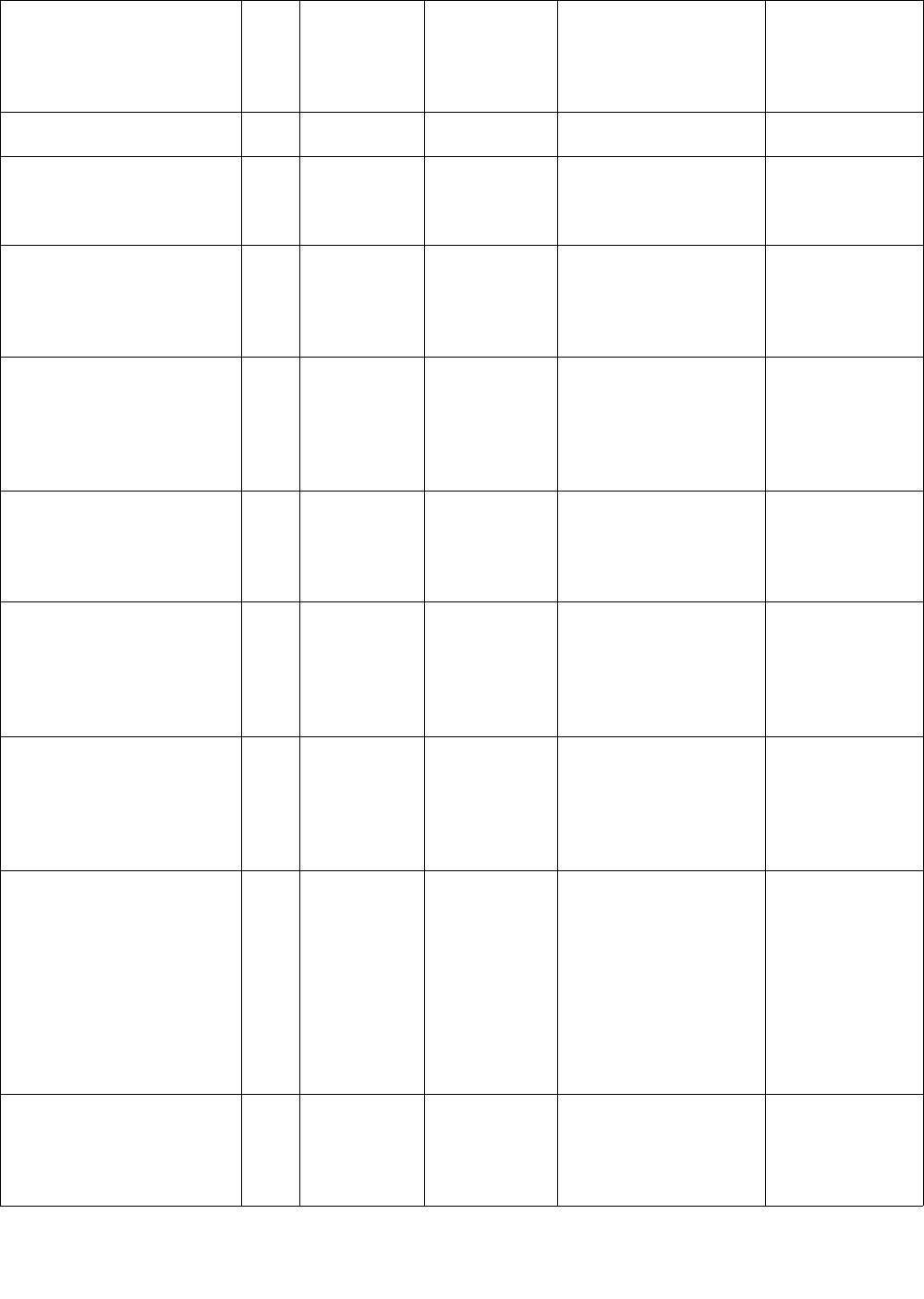
Name of list/dictionary Dire
ctory
locat
ion
Description Format Use in code Notes
isolation.
us_area_code.txt /lists US area
codes
1 area code
per line
Used to validate if
certain numeric patterns
are phone numbers.
us_states.txt,
us_states_abbre.txt,
more_us_state_abbreviation
s.txt
/lists US state
names
1 name per
line
Used to check for
zipcodes, and potential
locations.
commonest_words.txt /dict Words that
are very
common in
medical
records
1 word per
line
Used in multiple places
to check whether
possible names are
commonest words.
common_words.txt /dict Words that
are common
in medical
records
1 word per
line
Used in multiple places
to check whether
possible names are
common words.
sno_edited.txt /dict Medical
terms that
generally
should not be
removed
1 word per
line
Used in multiple places
to check whether
possible names are
medical terms.
medical_phrases.txt /dict Multi-word
medical
terms that
should not be
removed
Multiple
words per line
Used to check whether
possible names are part
of a medical phrase.
notes_common.txt /dict Really
common
words or
medical
terms
observed in
notes that
should not be
removed.
1 word per
line
Used to check for
potential names and
locations.
shift.txt Top-
level
direct
ory
PID to date
offset
mapping
PID||||Number
of days of
forward shift
Used in re-identifying
dates.

10
Bibliography
•Neamatullah I, Douglass M, Lehman LH, Reisner A, Villarroel M, Long WJ, Szolovits P,
Moody GB, Mark RG, and Clifford GD. Automated de-identification of free-text medical
records. BMC Med Inform Decis Mak 2008;8(32). URL http://www.biomedcentral.com/1472-
6947/8/32/.
•Neamatullah I. Automated De-Identification of Free-Text Medical Records. MEng Thesis,
Massachusetts Institute of Technology, Cambridge, MA, 2006.
•Douglass M. Computer-Assisted De-identification of Free-text Nursing Notes. MEng Thesis,
Massachusetts Institute of Technology, Cambridge, MA, USA, 2005.
•Douglass M, Clifford GD, Reisner A, Long WJ, Moody GB, Mark RG. De-identification
algorithm for free-text nursing notes. Computers in Cardiology, S6.2, 2005.
•Douglass M, Clifford GD, Reisner A, Moody GB, Mark RG. Computer-assisted de-
identification of free text in the MIMIC II database. Computers in Cardiology, M6.2, 2004.

11
APPENDIX A. De-Identification Code Description
In this Appendix, we document the major Perl subroutines in our de-identification software. The code
was implemented in Perl (version 5.8.8 and upgraded to version 5.10) and tested under Fedora Core 10,
Linux 2.6.27. De-identification involves scanning the entire text to identify PHI, classifying each item
of PHI based on the PHI categories, and replacing it with a PHI category tag (see Appendix B for the
PHI category tags used by the software).
An input configuration file is used to allow users to enable/disable the following filter types:
Name, SSN, URL, Email, Telephone, Unit Number (hospital patient identification number), Age (age
over 89), Location, Date, and U.S. State. There are also flags in the configuration file that can be used
to control whether certain dictionaries are to be loaded and used by the code for de-identification. The
dictionaries that can be enabled/disabled in the configuration file include: patient identification number
(PID) to patient name mapping, PID to date offset mapping (with a date shift value for each patient),
country names, company names, hospital names, location names, doctor names, U.S. city names, U.S.
state names, and ethnicity.
An overview of the main functions in the software follows in order of execution. The main function
responsible for de-identification is deid(), which calls findPHI() on each paragraph of text. findPHI()
scans through the paragraph of text and identifies PHI. After deid() has run each paragraph through
findPHI(), the software calls outputText() to create the de- and re-identified output of the paragraph.
We list the API of the major functions in the software below to provide an idea about its general
structure.
TOP MOST LEVEL OF CODE
Returns: None
Called by: Command at command prompt
Function synopsis:
The de-identification software initially sets the paths of lists and dictionaries in the working directory
that will be used in de-identification. Many of these lists and dictionaries are provided in a package
along with the software. The software declares arrays of context words that can be used to identify PHI.
The software calls the function setup() to create some lookup lists of known PHI in memory for fast
comparison with individual words during de-identification of text. It then calls the function deid() to
de-identify the text.
setup()
Arguments: None
Returns: None
Called by: Topmost level of code
Description: Creates some lookup lists to have in memory
Function synopsis:
The function sets up hashes of known PHI lists and dictionaries for direct identification of words in
text, e.g. last names, hospital names. The function preloads some PHI dictionaries into corresponding
arrays, e.g. locations, states, and generates associations between PHI in some lists and PHI categories.
deid()
Arguments: None
Returns: None

12
Called by: Topmost level of code
Description: This function reads in the text file to be de-identified, calls subroutine findPHI() to de-
identify text paragraph-by-paragraph, and outputs the de-identified text to a file.
Function synopsis:
The function opens the data file that contains the text to be de-identified. It reads in the data file
paragraph by paragraph so that items that extend over lines are not missed. It calls the function
findPHI() with each paragraph as the argument for the de-identification of the paragraph. The function
subsequently obtains and stores the PHI locations in the paragraph identified by findPHI(). The
function finally calls the function outputText() with hashes of identified PHI location to obtain the de-
identified text.
findPHI()
Arguments: paragraph of text
Returns: hash of PHI found
Called by: deid()
Description: Dispatched from the deid() function that perform de-identification. Reads in a paragraph
of text and runs the de-identification filters on it.
Function synopsis:
The function splits the data text into items demarcated by spaces. It performs an exact matching of each
item with lists of known PHI, e.g. proper names. The function then calls each filter function, e.g. name,
age, date filters, sequentially. The function returns a hash of approved PHI.
outputText()
Arguments: hash of PHI locations
Returns: None
Called by: deid()
Description: Creates the de-identified version of the text. Replaces dates with shifted dates, and other
PHI with their PHI types.
Function synopsis:
The function prints all the identified PHI locations to the output file. If a PHI is a date, the function
shifts the date and replaces it in the de-identified text. This shift may be a predetermined value or a
random value. If a PHI is not a date, the function replaces it in the de-identified text with a tag of the
PHI type. The function then prints the remaining non-PHI text to the de-identified text. Thus, the
function outputs a de-identified text file that is the original data file with all the identified PHI replaced
with PHI tags or shifted dates.
stat()
Arguments: filenames of file containing Gold Standard PHI locations and file containing PHI locations
of current de-identification run
Returns: None
Called by: Topmost level of code
Description: Calculates code performance statistics if comparison mode is set to 1 and if Gold Standard
is available.
Function Synopsis:
The function compares the PHI locations contained in the gold standard database and in the output
from the de-identification software, determines the recall and precision of the de-identification results,
and prints them on the screen.

13
APPENDIX B. PHI Tag Types
The de-identification algorithm replaces each PHI found in the medical notes with a PHI
category tag. In this section, we list the PHI tags defined in the code.
Name
The name filter replaces each name instance found in the medical notes with a PHI tag that indicates
the type of name replaced (e.g., first/last, female/male). In some cases, the pattern used to detect the
name is specified in parenthesis following the name type. For example, the tag [*** Name (PTitle)
***] indicates that the name matches patterns defined by plural titles such as “Drs.” and “Professors”.
Example name PHI tags are as follow.
[** Known patient firstname **] Name matched the patient’s first name listed in the dictionary.
[** Known patient lastname **] Name matched the patient’s last name in the dictionary.
[** Doctor First Name **] Doctor first name.
[** Doctor Last Name **] Doctor last name.
[** Female First Name (un) **] Unambiguous female first name.
[** Male First Name (un) **] Unambiguous male first name.
[** Name(LF) **] Last name followed by a comma and then a first name.
[** Name (MD) **] Doctor names followed by “MD”.
[** Name (PRE) **] Doctor name initial preceded by words such as “physician”, “PCP”, “provider”,
etc.
[** Name (NI) **] Names preceded by name indicators, such as “mother”, “brother”, “husband”.
[** Name (NameIs) **] Name preceded by the term “name is”.
[** Name Prefix (Prefixes) **] Name prefixes such as “de la”, or “van der”.
[** Last Name (Prefixes) **] Name preceded by prefixes such as “de la” or “van der”.
[** Name (STitle) **] Name followed by specific titles, such as “DR”, “MR” or “MS”.
[** Name (PTitle) **] Name followed by plural titles such as “Drs.” And “Professors”.
[** Name (NamePattern) **] Various name patterns that involve a first name, followed by an optional
1 or 2 middle initial(s), and then a last name.
Location
PHI category tags generated by the location filters include the following.
[** Street Address **] Street address.
[** Location **] Location in general, such as town, city names.
[** Location (Universities) **] University names.
[** Hospital **] Hospital names.
[** Wardname **] Hospital ward names.
[** PO BOX **] PO Box number.
[** State/Zipcode **] Zipcode preceded by state names.
[** State **] U.S. state names.
[** Country **] Country name.
[** Company **] Company name.
Telephone
The phone filter generates the following two types of PHI category tags.

14
[** Telephone/Fax **] Telephone or fax numbers.
[** Pager number **] Pager or beeper numbers.
Other
[** Social Security Number **] Social security numbers.
[** Medical Record Number **] Number associated with the medical record.
[** Unit Number **] Unique patient number.
[** Age over 90 **] Age equal to 90 or older.
[** E-mail address **] Email address.
[** URL **] Web URL address.
[** Holiday **] Holiday such as Christmas, Hanukah, Ramadan.
[** Ethnicity **] Words that indicate ethnicity or nationality, such as American, African, Spanish, etc.