SDTMIG V3.1.2 SDTM Implementation Guide
User Manual: Pdf
Open the PDF directly: View PDF ![]() .
.
Page Count: 298 [warning: Documents this large are best viewed by clicking the View PDF Link!]
- Cover page
- Table of Contents
- 1 Introduction
- 2 Fundamentals of the SDTM
- 3 Submitting Data in Standard Format
- 4 Assumptions for Domain Models
- 4.1 General Assumptions for All Domains
- 4.1.1 General Domain Assumptions
- 4.1.1.1 Review Study Data Tabulation and Implementation Guide
- 4.1.1.2 Relationship to Analysis Datasets
- 4.1.1.3 Additional Timing Variables
- 4.1.1.4 Order of the Variables
- 4.1.1.5 CDISC Core Variables
- 4.1.1.6 Additional Guidance on Dataset Naming
- 4.1.1.7 Splitting Domains
- 4.1.1.8 Origin Metadata
- 4.1.1.9 Assigning Natural Keys in the Metadata
- 4.1.2 General Variable Assumptions
- 4.1.2.1 Variable-Naming Conventions
- 4.1.2.2 Two-Character Domain Identifier
- 4.1.2.3 Use of “Subject” and USUBJID
- 4.1.2.4 Case Use of Text in Submitted Data
- 4.1.2.5 Convention for Missing Values
- 4.1.2.6 Grouping Variables and Categorization
- 4.1.2.7 Submitting Free Text from the CRF
- 4.1.2.8 Multiple Values for a Variable
- 4.1.3 Coding and Controlled Terminology Assumptions
- 4.1.3.1 Types of Controlled Terminology
- 4.1.3.2 Controlled Terminology Text Case
- 4.1.3.3 Controlled Terminology Values
- 4.1.3.4 Use of Controlled Terminology and Arbitrary Number Codes
- 4.1.3.5 Storing Controlled Terminology for Synonym Qualifier Variables
- 4.1.3.6 Storing Topic Variables for General Domain Models
- 4.1.3.7 Use of “Yes” and “No” Values
- 4.1.4 Actual and Relative Time Assumptions
- 4.1.4.1 Formats for Date/Time Variables
- 4.1.4.2 Date/Time Precision
- 4.1.4.3 Intervals of Time and Use of Duration for --DUR Variables
- 4.1.4.4 Use of the “Study Day” Variables
- 4.1.4.5 Clinical Encounters and Visits
- 4.1.4.6 Representing Additional Study Days
- 4.1.4.7 Use of Relative Timing Variables
- 4.1.4.8 Date and Time Reported in a Domain Based on Findings
- 4.1.4.9 Use of Dates as Result Variables
- 4.1.4.10 Representing Time Points
- 4.1.5 Other Assumptions
- 4.1.5.1 Original and Standardized Results of Findings and Tests Not Done
- 4.1.5.2 Linking of Multiple Observations
- 4.1.5.3 Text Strings That Exceed the Maximum Length for General-Observation-Class Domain Variables
- 4.1.5.4 Evaluators in the Interventions and Events Observation Classes
- 4.1.5.5 Clinical Significance for Findings Observation Class Data
- 4.1.5.6 Supplemental Reason Variables
- 4.1.5.7 Presence or Absence of Pre-Specified Interventions and Events
- 4.1.1 General Domain Assumptions
- 4.1 General Assumptions for All Domains
- 5 Models for Special-Purpose Domains
- 6 Domain Models Based on the General Observation Classes
- 6.1 Interventions
- 6.2 Events
- 6.3 Findings
- 6.3.1 ECG Test Results — EG
- 6.3.2 Inclusion/Exclusion Criteria Not Met — IE
- 6.3.3 Laboratory Test Results — LB
- 6.3.4 Physical Examination — PE
- 6.3.5 Questionnaire — QS
- 6.3.6 Subject Characteristics — SC
- 6.3.7 Vital Signs — VS
- 6.3.8 Drug Accountability — DA
- 6.3.9 Microbiology Domains — MB and 1104HMS
- 6.3.10 Pharmacokinetics Domains — PC and PP
- 6.3.10.1 Assumptions for Pharmacokinetic Concentrations (PC) Domain Model
- 6.3.10.2 Examples for Pharmacokinetic Concentrations (PC) Domain Model
- 6.3.10.3 Assumptions for Pharmacokinetic Parameters (PP) Domain Model
- 6.3.10.4 Example for Pharmacokinetic Parameters (PP) Domain Model
- 6.3.10.5 Relating PP Records to PC Records
- 6.3.10.6 Conclusions
- 6.3.10.7 Suggestions for Implementing RELREC in the Submission of PK Data
- 6.4 Findings about Events or Interventions
- 7 Trial Design Datasets
- 7.1 Introduction
- 7.2 Trial Arms
- 7.2.1 Trial Arms Dataset — TA
- 7.2.2 Assumptions for TA Dataset
- 7.2.3 Trial Arms Examples
- 7.2.3.1 Example Trial 1, a Parallel Trial
- 7.2.3.2 Example Trial 2, a Crossover Trial
- 7.2.3.3 Example Trial 3, a Trial with Multiple Branch Points
- 7.2.3.4 Example Trial 4, Cycles of Chemotherapy
- 7.2.3.5 Example Trial 5, Cycles with Different Treatment Durations
- 7.2.3.6 Example Trial 6, Chemotherapy Trial with Cycles of Different Lengths
- 7.2.3.7 Example Trial 7, Trial with Disparate Arms
- 7.2.4 Issues in Trial Arms Datasets
- 7.3 Trial Elements
- 7.4 Trial Visits
- 7.5 Trial Inclusion/Exclusion Criteria
- 7.6 Trial Summary Information
- 7.7 How to Model the Design of a Clinical Trial
- 8 Representing Relationships and Data
- Appendices
- Appendix A: CDISC SDS Team *
- Appendix B: Glossary and Abbreviations
- Appendix C: Controlled Terminology
- Appendix C1: Controlled Terms or Format for SDTM Variables (see also 1437HAppendix C3: Trial Summary Codes)
- Appendix C2: Reserved Domain Codes
- Appendix C2a: Reserved Domain Codes under Discussion
- Appendix C3: Trial Summary Codes
- Appendix C4: Drug Accountability Test Codes
- Appendix C5: Supplemental Qualifiers Name Codes
- Appendix D: CDISC Variable-Naming Fragments
- Appendix E: Revision History
- Appendix F: Representations and Warranties, Limitations of Liability, and Disclaimers
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 1
Final November 12, 2008
Study Data Tabulation Model
Implementation Guide:
Human Clinical Trials
Prepared by the
CDISC Submission Data Standards Team
Notes to Readers
This is the implementation guide for Human Clinical Trials corresponding to Version 1.2 of the CDISC
Study Data Tabulation Model.
This Implementation Guide comprises version 3.1.2 (V3.1.2) of the CDISC Submission Data Standards
and domain models.
Revision History
Date
Version
Summary of Changes
2008-11-12
3.1.2 Final
Released version reflecting all changes and
corrections identified during comment period.
2007-07-25
3.1.2 Draft
Draft for comment.
2005-08-26
3.1.1 Final
Released version reflecting all changes and
corrections identified during comment period.
2004-07-14
3.1
Released version reflecting all changes and
corrections identified during comment periods.
Note: Please see 1570HAppendix F for Representations and Warranties, Limitations of Liability, and Disclaimers.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 2 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
CONTENTS
0H1 INTRODUCTION ................................................................................................... 1571H7
1H1.1 PURPOSE ............................................................................................................................................................. 1572H7
2H1.2 ORGANIZATION OF THIS DOCUMENT ................................................................................................................... 1573H7
3H1.3 RELATIONSHIP TO PRIOR CDISC DOCUMENTS ................................................................................................... 1574H8
4H1.4 HOW TO READ THIS IMPLEMENTATION GUIDE .................................................................................................... 1575H9
5H1.5 SUBMITTING COMMENTS .................................................................................................................................... 1576H9
6H2 FUNDAMENTALS OF THE SDTM ...................................................................... 1577H10
7H2.1 OBSERVATIONS AND VARIABLES ....................................................................................................................... 1578H10
8H2.2 DATASETS AND DOMAINS ................................................................................................................................. 1579H11
9H2.3 SPECIAL-PURPOSE DATASETS ........................................................................................................................... 1580H12
10H2.4 THE GENERAL OBSERVATION CLASSES ............................................................................................................. 1581H12
11H2.5 THE SDTM STANDARD DOMAIN MODELS ....................................................................................................... 1582H13
12H2.6 CREATING A NEW DOMAIN ............................................................................................................................... 1583H14
13H3 SUBMITTING DATA IN STANDARD FORMAT .................................................. 1584H16
14H3.1 STANDARD METADATA FOR DATASET CONTENTS AND ATTRIBUTES .................................................................. 1585H16
15H3.2 USING THE CDISC DOMAIN MODELS IN REGULATORY SUBMISSIONS — DATASET METADATA ....................... 1586H17
16H3.2.1.1 Primary Keys ....................................................................................................................................... 1587H19
17H3.2.1.2 CDISC Submission Value-Level Metadata .......................................................................................... 1588H20
18H3.2.2 Conformance........................................................................................................................................ 1589H20
19H4 ASSUMPTIONS FOR DOMAIN MODELS .......................................................... 1590H21
20H4.1 GENERAL ASSUMPTIONS FOR ALL DOMAINS .................................................................................................... 1591H21
21H4.1.1 General Domain Assumptions ............................................................................................................. 1592H21
22H4.1.1.1 Review Study Data Tabulation and Implementation Guide ................................................................. 1593H21
23H4.1.1.2 Relationship to Analysis Datasets ........................................................................................................ 1594H21
24H4.1.1.3 Additional Timing Variables ................................................................................................................ 1595H21
25H4.1.1.4 Order of the Variables .......................................................................................................................... 1596H21
26H4.1.1.5 CDISC Core Variables ......................................................................................................................... 1597H21
27H4.1.1.6 Additional Guidance on Dataset Naming ............................................................................................ 1598H22
28H4.1.1.7 Splitting Domains ................................................................................................................................ 1599H22
29H4.1.1.8 Origin Metadata ................................................................................................................................... 1600H25
30H4.1.1.9 Assigning Natural Keys in the Metadata ............................................................................................. 1601H26
31H4.1.2 General Variable Assumptions ............................................................................................................. 1602H28
32H4.1.2.1 Variable-Naming Conventions ............................................................................................................. 1603H28
33H4.1.2.2 Two-Character Domain Identifier ........................................................................................................ 1604H28
34H4.1.2.3 Use of ―Subject‖ and USUBJID .......................................................................................................... 1605H29
35H4.1.2.4 Case Use of Text in Submitted Data .................................................................................................... 1606H29
36H4.1.2.5 Convention for Missing Values ............................................................................................................ 1607H29
37H4.1.2.6 Grouping Variables and Categorization ............................................................................................... 1608H29
38H4.1.2.7 Submitting Free Text from the CRF..................................................................................................... 1609H31
39H4.1.2.8 Multiple Values for a Variable ............................................................................................................. 1610H33
40H4.1.3 Coding and Controlled Terminology Assumptions .............................................................................. 1611H35
41H4.1.3.1 Types of Controlled Terminology ........................................................................................................ 1612H35
42H4.1.3.2 Controlled Terminology Text Case ...................................................................................................... 1613H35
43H4.1.3.3 Controlled Terminology Values ........................................................................................................... 1614H35
44H4.1.3.4 Use of Controlled Terminology and Arbitrary Number Codes ............................................................ 1615H36
45H4.1.3.5 Storing Controlled Terminology for Synonym Qualifier Variables ..................................................... 1616H36
46H4.1.3.6 Storing Topic Variables for General Domain Models .......................................................................... 1617H36
47H4.1.3.7 Use of ―Yes‖ and ―No‖ Values ............................................................................................................. 1618H36

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 3
Final November 12, 2008
48H4.1.4 Actual and Relative Time Assumptions ............................................................................................... 1619H37
49H4.1.4.1 Formats for Date/Time Variables ......................................................................................................... 1620H37
50H4.1.4.2 Date/Time Precision ............................................................................................................................. 1621H38
51H4.1.4.3 Intervals of Time and Use of Duration for --DUR Variables ............................................................... 1622H39
52H4.1.4.4 Use of the ―Study Day‖ Variables ........................................................................................................ 1623H40
53H4.1.4.5 Clinical Encounters and Visits ............................................................................................................. 1624H41
54H4.1.4.6 Representing Additional Study Days ................................................................................................... 1625H41
55H4.1.4.7 Use of Relative Timing Variables ........................................................................................................ 1626H42
56H4.1.4.8 Date and Time Reported in a Domain Based on Findings ................................................................... 1627H44
57H4.1.4.9 Use of Dates as Result Variables.......................................................................................................... 1628H44
58H4.1.4.10 Representing Time Points .................................................................................................................... 1629H44
59H4.1.5 Other Assumptions ............................................................................................................................... 1630H47
60H4.1.5.1 Original and Standardized Results of Findings and Tests Not Done ................................................... 1631H47
61H4.1.5.2 Linking of Multiple Observations ........................................................................................................ 1632H50
62H4.1.5.3 Text Strings That Exceed the Maximum Length for General-Observation-Class Domain Variables .. 1633H50
63H4.1.5.4 Evaluators in the Interventions and Events Observation Classes......................................................... 1634H51
64H4.1.5.5 Clinical Significance for Findings Observation Class Data ................................................................. 1635H52
65H4.1.5.6 Supplemental Reason Variables ........................................................................................................... 1636H52
66H4.1.5.7 Presence or Absence of Pre-Specified Interventions and Events ......................................................... 1637H52
67H5 MODELS FOR SPECIAL-PURPOSE DOMAINS ................................................. 1638H54
68H5.1 DEMOGRAPHICS ............................................................................................................................................... 1639H54
69H5.1.1 Demographics — DM .......................................................................................................................... 1640H54
70H5.1.1.1 Assumptions for Demographics Domain Model.................................................................................. 1641H56
71H5.1.1.2 Examples for Demographics Domain Model ....................................................................................... 1642H57
72H5.2 COMMENTS....................................................................................................................................................... 1643H64
73H5.2.1 Comments — CO ................................................................................................................................ 1644H64
74H5.2.1.1 Assumptions for Comments Domain Model ....................................................................................... 1645H65
75H5.2.1.2 Examples for Comments Domain Model ............................................................................................. 1646H66
76H5.3 SUBJECT ELEMENTS AND VISITS ....................................................................................................................... 1647H67
77H5.3.1 Subject Elements — SE ....................................................................................................................... 1648H67
78H5.3.1.1 Assumptions for Subject Elements Domain Model ............................................................................. 1649H68
79H5.3.1.2 Examples for Subject Elements Domain Model .................................................................................. 1650H70
80H5.3.2 Subject Visits — SV ............................................................................................................................ 1651H72
81H5.3.2.1 Assumptions for Subject Visits Domain Model ................................................................................... 1652H73
82H5.3.2.2 Examples for Subject Visits Domain Model ........................................................................................ 1653H74
83H6 DOMAIN MODELS BASED ON THE GENERAL OBSERVATION CLASSES .... 1654H75
84H6.1 INTERVENTIONS ................................................................................................................................................ 1655H75
85H6.1.1 Concomitant Medications — CM ........................................................................................................ 1656H75
86H6.1.1.1 Assumptions for Concomitant Medications Domain Model................................................................ 1657H78
87H6.1.1.2 Examples for Concomitant Medications Domain Model ..................................................................... 1658H80
88H6.1.2 Exposure — EX ................................................................................................................................... 1659H82
89H6.1.2.1 Assumptions for Exposure Domain Model .......................................................................................... 1660H84
90H6.1.2.2 Examples for Exposure Domain Model ............................................................................................... 1661H85
91H6.1.3 Substance Use — SU ........................................................................................................................... 1662H89
92H6.1.3.1 Assumptions for Substance Use Domain Model ................................................................................. 1663H92
93H6.1.3.2 Example for Substance Use Domain Model ........................................................................................ 1664H93
94H6.2 EVENTS ............................................................................................................................................................ 1665H94
95H6.2.1 Adverse Events — AE ......................................................................................................................... 1666H94
96H6.2.1.1 Assumptions for Adverse Event Domain Model ................................................................................. 1667H97
97H6.2.1.2 Examples for Adverse Events Domain Model ................................................................................... 1668H100
98H6.2.2 Disposition — DS .............................................................................................................................. 1669H103
99H6.2.2.1 Assumptions for Disposition Domain Model .................................................................................... 1670H104
100H6.2.2.2 Examples for Disposition Domain Model ......................................................................................... 1671H106

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 4 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
101H6.2.3 Medical History — MH ..................................................................................................................... 1672H110
102H6.2.3.1 Assumptions for Medical History Domain Model ............................................................................. 1673H112
103H6.2.3.2 Examples for Medical History Domain Model .................................................................................. 1674H114
104H6.2.4 Protocol Deviations — DV ................................................................................................................ 1675H117
105H6.2.4.1 Assumptions for Protocol Deviations Domain Model ....................................................................... 1676H118
106H6.2.4.2 Examples for Protocol Deviations Domain Model ............................................................................ 1677H118
107H6.2.5 Clinical Events — CE ........................................................................................................................ 1678H119
108H6.2.5.1 Assumptions for Clinical Events Domain Model .............................................................................. 1679H121
109H6.2.5.2 Examples for Clinical Events Domain Model ................................................................................... 1680H122
110H6.3 FINDINGS ........................................................................................................................................................ 1681H124
111H6.3.1 ECG Test Results — EG .................................................................................................................... 1682H124
112H6.3.1.1 Assumptions for ECG Test Results Domain Model ........................................................................... 1683H127
113H6.3.1.2 Examples for ECG Test Results Domain Model ................................................................................ 1684H127
114H6.3.2 Inclusion/Exclusion Criteria Not Met — IE ...................................................................................... 1685H130
115H6.3.2.1 Assumptions for Inclusion/Exclusion Criteria Not Met Domain Model ........................................... 1686H131
116H6.3.2.2 Examples for Inclusion/Exclusion Not Met Domain Model .............................................................. 1687H132
117H6.3.3 Laboratory Test Results — LB .......................................................................................................... 1688H133
118H6.3.3.1 Assumptions for Laboratory Test Results Domain Model ................................................................. 1689H137
119H6.3.3.2 Examples for Laboratory Test Results Domain Model ...................................................................... 1690H137
120H6.3.4 Physical Examination — PE .............................................................................................................. 1691H140
121H6.3.4.1 Assumptions for Physical Examination Domain Model .................................................................... 1692H142
122H6.3.4.2 Examples for Physical Examination Domain Model ......................................................................... 1693H143
123H6.3.5 Questionnaire — QS .......................................................................................................................... 1694H144
124H6.3.5.1 Assumptions for Questionnaire Domain Model ................................................................................ 1695H147
125H6.3.5.2 Examples for Questionnaire Domain Model ..................................................................................... 1696H148
126H6.3.6 Subject Characteristics — SC ............................................................................................................ 1697H150
127H6.3.6.1 Assumptions for Subject Characteristics Domain Model .................................................................. 1698H151
128H6.3.6.2 Example for Subject Charactistics Domain Model ............................................................................ 1699H152
129H6.3.7 Vital Signs — VS ............................................................................................................................... 1700H153
130H6.3.7.1 Assumptions for Vital Signs Domain Model ..................................................................................... 1701H156
131H6.3.7.2 Example for Vital Signs Domain Model ............................................................................................ 1702H156
132H6.3.8 Drug Accountability — DA ............................................................................................................... 1703H158
133H6.3.8.1 Assumptions for Drug Accountability Domain Model ...................................................................... 1704H159
134H6.3.8.2 Examples for Drug Accountability Domain Model ........................................................................... 1705H160
135H6.3.9 Microbiology Domains — MB and MS ............................................................................................ 1706H161
136H6.3.9.1 Microbiology Specimen (MB) Domain Model .................................................................................. 1707H161
137H6.3.9.2 Assumptions for Microbiology Specimen (MB) Domain Model ...................................................... 1708H164
138HMicrobiology Susceptibility (MS) Domain Model ............................................................................................ 1709H165
139H6.3.9.3 Assumptions for Microbiology Susceptibility (MS) Domain Model ................................................. 1710H168
140H6.3.9.4 Examples for MB and MS Domain Models ....................................................................................... 1711H169
141H6.3.10 Pharmacokinetics Domains — PC and PP ......................................................................................... 1712H172
142H6.3.10.1 Assumptions for Pharmacokinetic Concentrations (PC) Domain Model........................................... 1713H176
143H6.3.10.2 Examples for Pharmacokinetic Concentrations (PC) Domain Model ................................................ 1714H176
144H6.3.10.3 Assumptions for Pharmacokinetic Parameters (PP) Domain Model ................................................. 1715H179
145H6.3.10.4 Example for Pharmacokinetic Parameters (PP) Domain Model ........................................................ 1716H179
146H6.3.10.5 Relating PP Records to PC Records .................................................................................................. 1717H181
147H6.3.10.6 Conclusions........................................................................................................................................ 1718H193
148H6.3.10.7 Suggestions for Implementing RELREC in the Submission of PK Data ........................................... 1719H193
149H6.4 FINDINGS ABOUT EVENTS OR INTERVENTIONS ................................................................................................ 1720H194
150H6.4.1 When to Use Findings About ............................................................................................................. 1721H194
151H6.4.2 Naming Findings About Domains ..................................................................................................... 1722H195
152H6.4.3 Variables Unique to Findings About .................................................................................................. 1723H195
153H6.4.4 Findings About (FA) Domain Model ................................................................................................. 1724H196
154H6.4.5 Assumptions for Findings About Domain Model .............................................................................. 1725H198
155H6.4.6 Findings About Examples .................................................................................................................. 1726H199

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 5
Final November 12, 2008
156H7 TRIAL DESIGN DATASETS .............................................................................. 1727H211
157H7.1 INTRODUCTION ............................................................................................................................................... 1728H211
158H7.1.1 Purpose of Trial Design Model .......................................................................................................... 1729H211
159H7.1.2 Definitions of Trial Design Concepts ................................................................................................ 1730H211
160H7.1.3 Current and Future Contents of the Trial Design Model .................................................................... 1731H213
161H7.2 TRIAL ARMS ................................................................................................................................................... 1732H214
162H7.2.1 Trial Arms Dataset — TA .................................................................................................................. 1733H214
163H7.2.2 Assumptions for TA Dataset .............................................................................................................. 1734H214
164H7.2.3 Trial Arms Examples ......................................................................................................................... 1735H215
165H7.2.3.1 Example Trial 1, a Parallel Trial ........................................................................................................ 1736H216
166H7.2.3.2 Example Trial 2, a Crossover Trial .................................................................................................... 1737H219
167H7.2.3.3 Example Trial 3, a Trial with Multiple Branch Points ....................................................................... 1738H223
168H7.2.3.4 Example Trial 4, Cycles of Chemotherapy ........................................................................................ 1739H226
169H7.2.3.5 Example Trial 5, Cycles with Different Treatment Durations ............................................................ 1740H230
170H7.2.3.6 Example Trial 6, Chemotherapy Trial with Cycles of Different Lengths .......................................... 1741H232
171H7.2.3.7 Example Trial 7, Trial with Disparate Arms ...................................................................................... 1742H235
172H7.2.4 Issues in Trial Arms Datasets ............................................................................................................. 1743H238
173H7.2.4.1 Distinguishing between Branches and Transitions ............................................................................ 1744H238
174H7.2.4.2 Subjects not Assigned to an Arm ....................................................................................................... 1745H238
175H7.2.4.3 Defining Epochs ................................................................................................................................ 1746H238
176H7.2.4.4 Rule Variables .................................................................................................................................... 1747H238
177H7.3 TRIAL ELEMENTS ........................................................................................................................................... 1748H239
178H7.3.1 Trial Elements Dataset — TE ............................................................................................................ 1749H239
179H7.3.2 Assumptions for TE Dataset .............................................................................................................. 1750H240
180H7.3.3 Trial Elements Examples ................................................................................................................... 1751H241
181H7.3.4 Trial Elements Issues ......................................................................................................................... 1752H242
182H7.3.4.1 Granularity of Trial Elements ............................................................................................................ 1753H242
183H7.3.4.2 Distinguishing Elements, Study Cells, and Epochs ........................................................................... 1754H242
184H7.3.4.3 Transitions between Elements ........................................................................................................... 1755H243
185H7.4 TRIAL VISITS .................................................................................................................................................. 1756H244
186H7.4.1 Trial Visits Dataset — TV .................................................................................................................. 1757H244
187H7.4.2 Assumptions for TV Dataset .............................................................................................................. 1758H244
188H7.4.3 Trial Visits Examples ......................................................................................................................... 1759H245
189H7.4.4 Trial Visits Issues ............................................................................................................................... 1760H246
190H7.4.4.1 Identifying Trial Visits ....................................................................................................................... 1761H246
191H7.4.4.2 Trial Visit Rules ................................................................................................................................. 1762H246
192H7.4.4.3 Visit Schedules Expressed with Ranges............................................................................................. 1763H247
193H7.4.4.4 Contingent Visits ................................................................................................................................ 1764H247
194H7.5 TRIAL INCLUSION/EXCLUSION CRITERIA ........................................................................................................ 1765H248
195H7.5.1 Trial Inclusion/Exclusion Criteria Dataset — TI ............................................................................... 1766H248
196H7.5.2 Assumptions for TI Dataset ............................................................................................................... 1767H248
197H7.5.3 Examples for Trial Inclusion/Exclusion Dataset Model .................................................................... 1768H249
198H7.6 TRIAL SUMMARY INFORMATION ..................................................................................................................... 1769H249
199H7.6.1 Trial Summary Dataset — TS ............................................................................................................ 1770H249
200H7.6.2 Assumptions for Trial Summary Dataset Model ................................................................................ 1771H250
201H7.6.3 Examples for Trial Summary Dataset Model ..................................................................................... 1772H251
202H7.7 HOW TO MODEL THE DESIGN OF A CLINICAL TRIAL ....................................................................................... 1773H254
203H8 REPRESENTING RELATIONSHIPS AND DATA .............................................. 1774H255
204H8.1 RELATING GROUPS OF RECORDS WITHIN A DOMAIN USING THE --GRPID VARIABLE ..................................... 1775H256
205H8.1.1 --GRPID Example ............................................................................................................................. 1776H256
206H8.2 RELATING PEER RECORDS .............................................................................................................................. 1777H257
207H8.2.1 RELREC Dataset ............................................................................................................................... 1778H257
208H8.2.2 RELREC Dataset Examples .............................................................................................................. 1779H258

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 6 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
209H8.3 RELATING DATASETS ...................................................................................................................................... 1780H259
210H8.3.1 RELREC Dataset Relationship Example ........................................................................................... 1781H259
211H8.4 RELATING NON-STANDARD VARIABLES VALUES TO A PARENT DOMAIN ......................................................... 1782H260
212H8.4.1 Supplemental Qualifiers: SUPPQUAL or SUPP-- Datasets .............................................................. 1783H261
213H8.4.2 Submitting Supplemental Qualifiers in Separate Datasets ................................................................. 1784H262
214H8.4.3 SUPP-- Examples .............................................................................................................................. 1785H262
215H8.4.4 When Not to Use Supplemental Qualifiers ........................................................................................ 1786H264
216H8.5 RELATING COMMENTS TO A PARENT DOMAIN ................................................................................................ 1787H265
217H8.6 HOW TO DETERMINE WHERE DATA BELONG IN THE SDTM ........................................................................... 1788H265
218H8.6.1 Guidelines for Determining the General Observation Class .............................................................. 1789H265
219H8.6.2 Guidelines for Forming New Domains .............................................................................................. 1790H266
220H8.6.3 Guidelines for Differentiating between Events, Findings, and Findings about Events ...................... 1791H266
221HAPPENDICES ............................................................................................................. 1792H269
222HAPPENDIX A: CDISC SDS TEAM *............................................................................................................................. 1793H269
223HAPPENDIX B: GLOSSARY AND ABBREVIATIONS .......................................................................................................... 1794H270
224HAPPENDIX C: CONTROLLED TERMINOLOGY ............................................................................................................... 1795H271
225HAppendix C1: Controlled Terms or Format for SDTM Variables (see also Appendix C3: Trial Summary Codes 1796H271
226HAppendix C2: Reserved Domain Codes ................................................................................................................ 1797H274
227HAppendix C2a: Reserved Domain Codes under Discussion .................................................................................. 1798H277
228HAppendix C3: Trial Summary Codes ..................................................................................................................... 1799H279
229HAppendix C4: Drug Accountability Test Codes ..................................................................................................... 1800H283
230HAppendix C5: Supplemental Qualifiers Name Codes............................................................................................ 1801H283
231HAPPENDIX D: CDISC VARIABLE-NAMING FRAGMENTS ............................................................................................. 1802H284
232HAPPENDIX E: REVISION HISTORY ............................................................................................................................... 1803H286
233HAPPENDIX F: REPRESENTATIONS AND WARRANTIES, LIMITATIONS OF LIABILITY, AND DISCLAIMERS ........................ 1804H298

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 7
Final November 12, 2008
1 Introduction
1.1 PURPOSE
This document comprises the CDISC Version 3.1.2 (V3.1.2) Study Data Tabulation Model Implementation Guide
for Human Clinical Trials (SDTMIG), which has been prepared by the Submissions Data Standards (SDS) team of
the Clinical Data Interchange Standards Consortium (CDISC). Like its predecessors, V3.1.2 is intended to guide the
organization, structure, and format of standard clinical trial tabulation datasets submitted to a regulatory authority
such as the US Food and Drug Administration (FDA). V3.1.2 supersedes all prior versions of the CDISC
Submission Data Standards.
The SDTMIG should be used in close concert with the current version of the CDISC Study Data Tabulation Model
(SDTM, available at http://www.cdisc.org/standards) that describes the general conceptual model for representing
clinical study data that is submitted to regulatory authorities and should be read prior to reading the SDTMIG.
V3.1.2 provides specific domain models, assumptions, business rules, and examples for preparing standard
tabulation datasets that are based on the SDTM.
Tabulation datasets, which are electronic listings of individual observations for a subject that comprise the essential
data reported from a clinical trial, are one of four types of data currently submitted to the FDA along with patient
profiles, listings, and analysis files. By submitting tabulations that conform to the standard structure, sponsors may
benefit by no longer having to submit separate patient profiles or listings with a product marketing application.
SDTM datasets are not intended to fully meet the needs supported by analysis datasets, which will continue to be
submitted separately in addition to the tabulations. Since July 2004, the FDA has referenced use of the SDTM in the
Study Data Specifications for the Electronic Common Technical Document, available at
235Hhttp://www.fda.gov/cder/regulatory/ersr/Studydata-v1.2.pdf.
The availability of standard submission data will provide many benefits to regulatory reviewers. Reviewers can be
trained in the principles of standardized datasets and the use of standard software tools, and thus be able to work
with the data more effectively with less preparation time. Another benefit of the standardized datasets is that they
will support 1) the FDA‘s efforts to develop a repository for all submitted trial data, and 2) a suite of standard review
tools to access, manipulate, and view the tabulations. Use of these data standards is also expected to benefit industry
by streamlining the flow of data from collection through submission, and facilitating data interchange between
partners and providers. Note that the SDTM represents an interchange standard, rather than a presentation format. It
is assumed that tabulation data will be transformed by software tools to better support viewing and analysis.
This document is intended for companies and individuals involved in the collection, preparation, and analysis of
clinical data that will be submitted to regulatory authorities.
1.2 ORGANIZATION OF THIS DOCUMENT
This document is organized into the following sections:
236HSection 1, 1805HIntroduction, provides an overall introduction to the V3.1.2 models and describes changes from
prior versions.
237HSection 2, 1806HFundamentals of the SDTM, recaps the basic concepts of the SDTM, and describes how this
implementation guide should be used in concert with the SDTM.
238HSection 3, 1807HSubmitting Data in Standard Format, explains how to describe metadata for regulatory
submissions, and how to assess conformance with the standards.
239HSection 4, 1808HAssumptions for Domain Models, describes basic concepts, business rules, and assumptions that
should be taken into consideration before applying the domain models.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 8 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
240HSection 5, 241HModels for Special-Purpose Domains, describes special-purpose domains, including
Demographics, Comments, Subject Visits, and Subject Elements.
242HSection 6, 1809HDomain Models Based on the General Observation Classes, provides specific metadata models
based on the three general observation classes, along with assumptions and example data.
243HSection 7, 1810HTrial Design Datasets, provides specific metadata models, assumptions, and examples.
244HSection 8, 245HRepresenting Relationships and Data, describes how to represent relationships between separate
domains, datasets, and/or records, and information to help sponsors determine where data belongs in the
SDTM.
1811HAppendices provide additional background material and describe other supplemental material relevant to
implementation.
1.3 RELATIONSHIP TO PRIOR CDISC DOCUMENTS
This document, together with the SDTM, represents the most recent version of the CDISC Submission Data Domain
Models. Since all updates are intended to be backward compatible the term ―V3.x‖ is used to refer to Version 3.1
and all subsequent versions. The most significant changes since the prior version, V3.1.1, include:
New domain models for Clinical Events and Findings About Events and Interventions (formerly Clinical
Findings in v3.1.2 Draft), and inclusion of previously posted domain models for Protocol Deviations, Drug
Accountability, pharmacokinetic data, and microbiology.
Additional assumptions and rules for representing common data scenarios and naming of datasets in
246HSection 4, including guidance on the use of keys and representing data with multiple values for a single
question.
Corrections and clarifications regarding the use of ISO 8601 date formats in 247HSection 4.1.4.
Additional guidance about how to address Findings data collected as a result of Events or Interventions,
and data submitted for pre-specified Findings and Events.
The use of new SDTM variables (Section 6.2 of the SDTM).
Implementation advice on the use of new timing variables, --STRTPT, --ENRTPT, --STTPT, and --ENTPT
(248HSection 4.1.4.7), and the new variable --OBJ (249HSection 6.4.3).
Listing of Qualifier variables from the same general observation class that would not generally be used in
the standard domains.
Several changes to the organization of the document, including the reclassification of Subject Elements
(SE) and Subject Visits (SV) as special-purpose domain datasets in 250HSection 5 (these were formerly included
as part of Trial Design), and moving data examples from a separate section (former Section 9) to locations
immediately following each domain model in 251HSection 5 and 252HSection 6.
Changes to the method for representing multiple RACE values in DM and SUPPDM with examples.
Removed the Origin column from domain models based on the three general classes since origins will need
to be defined by the sponsor in most cases. Definitions of origin metadata have been added.
A detailed list of changes between versions is provided in 253HAppendix E.
V3.1 was the first fully implementation-ready version of the CDISC Submission Data Standards that was directly
referenced by the FDA for use in human clinical studies involving drug products. However, future improvements
and enhancements such as V3.1.2 will continue to be made as sponsors gain more experience submitting data in this
format. Therefore, CDISC will be preparing regular updates to the implementation guide to provide corrections,
clarifications, additional domain models, examples, business rules, and conventions for using the standard domain
models. CDISC will produce further documentation for controlled terminology as separate publications, so sponsors
are encouraged to check the CDISC website (254Hwww.cdisc.org/standards/) frequently for additional information. See
255HSection 4.1.3 for the most up-to-date information on applying Controlled Terminology.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 9
Final November 12, 2008
1.4 HOW TO READ THIS IMPLEMENTATION GUIDE
This SDTM Implementation Guide (SDTMIG) is best read online, so the reader can benefit from the many
hyperlinks included to both internal and external references. The following guidelines may be helpful in reading this
document:
1. First, read the SDTM to gain a general understanding of SDTM concepts.
2. Next, read Sections 1-3 of this document to review the key concepts for preparing domains and submitting
data to regulatory authorities. Refer to the Glossary in 1812H Appendix B as necessary.
3. Read the 256HGeneral Assumptions for all Domains in 257HSection 4.
4. Review 258HSection 5 and 259HSection 6 in detail, referring back to Assumptions as directed (hyperlinks are
provided). Note the implementation examples for each domain to gain an understanding of how to apply
the domain models for specific types of data.
5. Read 260HSection 7 to understand the fundamentals of the Trial Design Model and consider how to apply the
concepts for typical protocols. New extensions to the trial design model will be published separately on the
CDISC website.
6. Review 261HSection 8 to learn advanced concepts of how to express relationships between datasets, records and
additional variables not specifically defined in the models.
7. Finally, review the 1813H Appendices as appropriate.
1.5 SUBMITTING COMMENTS
Comments on this document can be submitted through the 262HCDISC Discussion Board.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 10 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
2 Fundamentals of the SDTM
2.1 OBSERVATIONS AND VARIABLES
The V3.x Submission Data Standards are based on the SDTM‘s general framework for organizing clinical trials
information that is to be submitted to the FDA. The SDTM is built around the concept of observations collected
about subjects who participated in a clinical study. Each observation can be described by a series of variables,
corresponding to a row in a dataset or table. Each variable can be classified according to its Role. A Role determines
the type of information conveyed by the variable about each distinct observation and how it can be used. Variables
can be classified into five major roles:
Identifier variables, such as those that identify the study, subject, domain, and sequence number of the record
Topic variables, which specify the focus of the observation (such as the name of a lab test)
Timing variables, which describe the timing of the observation (such as start date and end date)
Qualifier variables, which include additional illustrative text or numeric values that describe the results or
additional traits of the observation (such as units or descriptive adjectives)
Rule variables, which express an algorithm or executable method to define start, end, and branching or looping
conditions in the Trial Design model
The set of Qualifier variables can be further categorized into five sub-classes:
Grouping Qualifiers are used to group together a collection of observations within the same domain. Examples
include --CAT and --SCAT.
Result Qualifiers describe the specific results associated with the topic variable in a Findings dataset. They
answer the question raised by the topic variable. Result Qualifiers are --ORRES, --STRESC, and --STRESN.
Synonym Qualifiers specify an alternative name for a particular variable in an observation. Examples include
--MODIFY and --DECOD, which are equivalent terms for a --TRT or --TERM topic variable, --TEST and
--LOINC which are equivalent terms for a --TESTCD.
Record Qualifiers define additional attributes of the observation record as a whole (rather than describing a
particular variable within a record). Examples include --REASND, AESLIFE, and all other SAE flag variables
in the AE domain; AGE, SEX, and RACE in the DM domain; and --BLFL, --POS, --LOC, --SPEC and --NAM
in a Findings domain
Variable Qualifiers are used to further modify or describe a specific variable within an observation and are only
meaningful in the context of the variable they qualify. Examples include --ORRESU, --ORNRHI, and
--ORNRLO, all of which are Variable Qualifiers of --ORRES; and --DOSU, which is a Variable Qualifier of
--DOSE.
For example, in the observation, ―Subject 101 had mild nausea starting on Study Day 6, ― the Topic variable value is
the term for the adverse event, ―NAUSEA‖. The Identifier variable is the subject identifier, ―101‖. The Timing
variable is the study day of the start of the event, which captures the information, ―starting on Study Day 6‖, while
an example of a Record Qualifier is the severity, the value for which is ―MILD‖. Additional Timing and Qualifier
variables could be included to provide the necessary detail to adequately describe an observation.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 11
Final November 12, 2008
2.2 DATASETS AND DOMAINS
Observations about study subjects are normally collected for all subjects in a series of domains. A domain is defined
as a collection of logically related observations with a common topic. The logic of the relationship may pertain to the
scientific subject matter of the data or to its role in the trial. Each domain is represented by a single dataset.
Each domain dataset is distinguished by a unique, two-character code that should be used consistently throughout
the submission. This code, which is stored in the SDTM variable named DOMAIN, is used in four ways: as the
dataset name, the value of the DOMAIN variable in that dataset, as a prefix for most variable names in that dataset,
and as a value in the RDOMAIN variable in relationship tables (263HSection 8).
All datasets are structured as flat files with rows representing observations and columns representing variables. Each
dataset is described by metadata definitions that provide information about the variables used in the dataset. The
metadata are described in a data definition document named ―define‖ that is submitted with the data to regulatory
authorities. (See the 264HCase Report Tabulation Data Definition Specification [define.xml], available at www.CDISC.org).
Define.xml specifies seven distinct metadata attributes to describe SDTM data:
The Variable Name (limited to 8 characters for compatibility with the SAS Transport format)
A descriptive Variable Label, using up to 40 characters, which should be unique for each variable in the dataset
The data Type (e.g., whether the variable value is a character or numeric)
The set of controlled terminology for the value or the presentation format of the variable (Controlled Terms or Format)
The Origin of each variable (see 265HSection 4.1.1.8)
The Role of the variable, which determines how the variable is used in the dataset. For the V3.x domain models,
Roles are used to represent the categories of variables such as Identifier, Topic, Timing, or the five types of
Qualifiers.
Comments or other relevant information about the variable or its data included by the sponsor as necessary to
communicate information about the variable or its contents to a regulatory agency.
Data stored in SDTM datasets include both raw (as originally collected) and derived values (e.g., converted into
standard units, or computed on the basis of multiple values, such as an average). The SDTM lists only the name,
label, and type, with a set of brief CDISC guidelines that provide a general description for each variable used for a
general observation class.
The domain dataset models included in 266HSection 5 and 267HSection 6 of this document provide additional information
about Controlled Terms or Format, notes on proper usage, and examples. Controlled terminology (CT) is now
represented one of four ways:
A single asterisk when there is no specific CT available at the current time, but the SDS Team expects that sponsors
may have their own CT and/or the CDISC Controlled Terminology Team may be developing CT.
A list of controlled terms for the variable when values are not yet maintained externally
The name of an external codelist whose values can be found via the hyperlinks in either the domain or 268HAppendix C1.
A common format such as ISO 8601
The CDISC Controlled Terminology team will be publishing additional guidance on use of controlled terminology
separately.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 12 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
2.3 SPECIAL-PURPOSE DATASETS
The SDTM includes three types of special-purpose datasets:
Domain datasets, consisting of Demographics (DM), Comments (CO), Subject Elements (SE), and Subject
Visits (SV)0F
1, all of which include subject-level data that do not conform to one of the three general
observation classes. These are described in 269HSection 5.
Trial Design Model (TDM) datasets, such as Trial Arms (TA) and Trial Elements (TE), which represent
information about the study design but do not contain subject data. These are described in 270HSection 7.
Relationship datasets, which include the RELREC and SUPP-- datasets described in 271HSection 8.
2.4 THE GENERAL OBSERVATION CLASSES
Most subject-level observations collected during the study should be represented according to one of the three
SDTM general observation classes: Interventions, Events, or Findings. The lists of variables allowed to be used in
each of these can be found in the STDM.
The Interventions class captures investigational, therapeutic and other treatments that are administered to the
subject (with some actual or expected physiological effect) either as specified by the study protocol (e.g.,
exposure to study drug), coincident with the study assessment period (e.g., concomitant medications), or
self-administered by the subject (such as use of alcohol, tobacco, or caffeine).
The Events class captures planned protocol milestones such as randomization and study completion, and
occurrences, conditions, or incidents independent of planned study evaluations occurring during the trial (e.g.,
adverse events) or prior to the trial (e.g., medical history).
The Findings class captures the observations resulting from planned evaluations to address specific tests or
questions such as laboratory tests, ECG testing, and questions listed on questionnaires.
In most cases, the choice of observation class appropriate to a specific collection of data can be easily determined
according to the descriptions provided above. The majority of data, which typically consists of measurements or
responses to questions usually at specific visits or time points, will fit the Findings general observation class.
Additional guidance on choosing the appropriate general observation class is provided in 272HSection 8.6.1.
General assumptions for use with all domain models and custom domains based on the general observation classes
are described in 273HSection 4 of this document; specific assumptions for individual domains are included with the
domain models.
1 SE and SV were included as part of the Trial Design Model in earlier versions of the SDTMIG.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 13
Final November 12, 2008
2.5 THE SDTM STANDARD DOMAIN MODELS
The following standard domains with their respective domain codes have been defined or referenced by the CDISC SDS
Team in this document. Note that other domain models may be posted separately for comment after this publication.
Special-Purpose Domains (defined in 274HSection 5):
Demographics — 1814HDM
Comments — 1815HCO
Subject Elements — 1816HSE
Subject Visits — 1817HSV
Interventions General Observation Class (defined in 275HSection 6.1):
Concomitant Medications — 1818HCM
Exposure — 1819HEX
Substance Use — 1820HSU
Events General Observation Class (defined in 276HSection 6.2):
Adverse Events — 1821H AE
Disposition — 1822HDS
Medical History — 1823HMH
Protocol Deviations — 1824HDV
Clinical Events — 1825HCE
Findings General Observation Class (defined in 277HSection 6.3):
ECG Test Results — 1826HEG
Inclusion/Exclusion Criterion Not Met — 1827HIE
Laboratory Test Results — 1828HLB
Physical Examination — 1829HPE
Questionnaires — 1830HQS
Subject Characteristics — 1831HSC
Vital Signs — 1832HVS
Drug Accountability — 1833HDA
Microbiology Specimen — 1834HMB
Microbiology Susceptibility Test — MS
PK Concentrations — 1836HPC
PK Parameters —PP
Findings About (defined in 280HSection 6.4)
Findings About — 281HFA
Trial Design Domains (defined in 282HSection 7):
Trial Arms — 283HTA
Trial Elements — 284HTE
Trial Visits — 1837HTV
Trial Inclusion/Exclusion Criteria — 285HTI
Trial Summary — 286HTS
Relationship Datasets (defined in 287HSection 8):
288HSupplemental Qualifiers — SUPPQUAL or
multiple SUPP-- datasets
Related Records — 289HRELREC
A sponsor should only submit domain datasets that were actually collected (or directly derived from the collected
data) for a given study. Decisions on what data to collect should be based on the scientific objectives of the study,
rather than the SDTM. Note that any data that was collected and will be submitted in an analysis dataset must also
appear in a tabulation dataset.
The collected data for a given study may use some or all of the SDS standard domains as well as additional custom
domains based on the three general observation classes. A list of standard domain codes for many commonly used
domains is provided in . Additional standard domain models will be published by CDISC as they are developed, and
sponsors are encouraged to check the CDISC website for updates.
These general rules apply when determining which variables to include in a domain:
The Identifier variables, STUDYID, USUBJID, DOMAIN, and --SEQ are required in all domains based on the
general observation classes. Other Identifiers may be added as needed.
Any Timing variables are permissible for use in any submission dataset based on a general observation class
except where restricted by specific domain assumptions.
Any additional Qualifier variables from the same general observation class may be added to a domain model
except where restricted by specific domain assumptions.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 14 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Sponsors may not add any other variables than those described in the preceding three bullets. The addition of
non-standard variables will compromise the FDA‘s abilities to populate the data repository and to use standard
tools. The SDTM allows for the inclusion of the sponsors non-SDTM variables using the Supplemental
Qualifiers special-purpose dataset structure, described in 290HSection 8.4. As the SDTM continues to evolve over
time, certain additional standard variables may be added to the general observation classes. Therefore, Sponsors
wishing to nominate such variables for future consideration should provide a rationale and description of the
proposed variable(s) along with representative examples to the CDISC Public Discussion Forum.
Standard variables must not be renamed or modified for novel usage. Their metadata should not be changed.
As long as no data was collected for Permissible variables, a sponsor is free to drop them and the corresponding
descriptions from the define.xml.
2.6 CREATING A NEW DOMAIN
This section describes the overall process for creating a custom domain, which must be based on one of the three
SDTM general observation classes. The number of domains submitted should be based on the specific requirements
of the study. Follow the process below to create a custom domain:
1. Confirm that none of the existing published domains will fit the need. A custom domain may only be
created if the data are different in nature and do not fit into an existing published domain.
Establish a domain of a common topic (i.e., where the nature of the data is the same), rather than by
a specific method of collection (e.g. electrocardiogram - EG). Group and separate data within the
domain using --CAT, --SCAT, --METHOD, --SPEC, --LOC, etc. as appropriate. Examples of
different topics are: microbiology, tumor measurements, pathology/histology, vital signs, and
physical exam results.
Do not create separate domains based on time, rather represent both prior and current observations
in a domain (e.g., CM for all non-study medications). Note that AE and MH are an exception to this
best practice because of regulatory reporting needs.
How collected data are used (e.g., to support analyses and/or efficacy endpoints) must not result in
the creation of a custom domain. For example, if blood pressure measurements are endpoints in a
hypertension study, they must still be represented in the VS (Vital Signs) domain as opposed to a
custom ―efficacy‖ domain. Similarly, if liver function test results are of special interest, they must
still be represented in the LB (Laboratory Tests) domain.
Data that were collected on separate CRF modules or pages may fit into an existing domain (such as
separate questionnaires into the QS domain, or prior and concomitant medications in the CM domain).
If it is necessary to represent relationships between data that are hierarchical in nature (e.g., a parent
record must be observed before child records), then establish a domain pair (e.g., MB/MS, PC/PP).
Note, domain pairs have been modeled for microbiology data (MB/MS domains) and PK data
(PC/PP domains) to enable dataset-level relationships to be described using RELREC. The domain
pair uses DOMAIN as an Identifier to group parent records (e.g., MB) from child records (e.g., MS)
and enables a dataset-level relationship to be described in RELREC. Without using DOMAIN to
facilitate description of the data relationships, RELREC, as currently defined could not be used
without introducing a variable that would group data like DOMAIN.
2. Check the Submission Data Standards area of the CDISC website (292Hhttp://www.cdisc.org/) for models added
after the last publication of the SDTMIG.
3. Look for an existing, relevant domain model to serve as a prototype. If no existing model seems
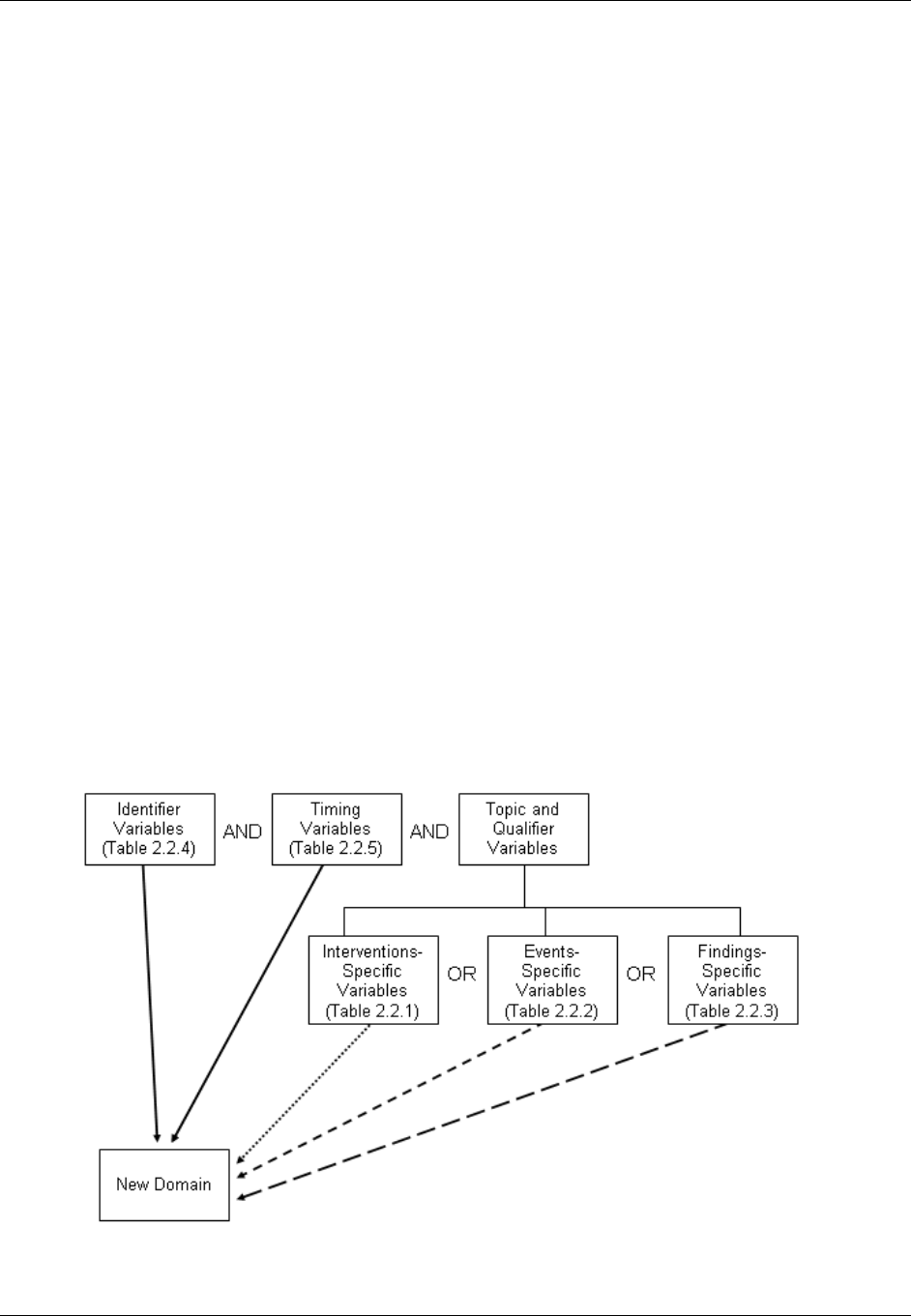
appropriate, choose the general observation class (Interventions, Events, or Findings) that best fits the data
by considering the topic of the observation The general approach for selecting variables for a custom
domain is as follows (also see Figure 2.6 below)
a. Select and include the required Identifier variables (e.g., STUDYID, DOMAIN, USUBJID, --SEQ)
and any permissible Identifier variables from SDTM Table 2.2.4.
b. Include the Topic variable from the identified general observation class (e.g., --TESTCD for
Findings) (SDTM table 2.2.1, SDTM table 2.2.2 or SDTM table 2.2.3).
c. Select and include the relevant Qualifier variables from the identified general observation class
(SDTM table 2.2.1, SDTM table 2.2.2 or SDTM table 2.2.3). Variables belonging to other general
observation classes must not be added.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 15
Final November 12, 2008
d. Select and include the applicable Timing variables (SDTM Table 2.2.5). Determine the domain
code. Check 293H Appendix C2 and 294HAppendix C2A for reserved two-character domain identifiers or
abbreviations. If one has not been assigned by CDISC, then the sponsor may select the unique
two-character domain code to be used consistently throughout the submission.
e. Apply the two-character domain code to the appropriate variables in the domain. Replace all
variable prefixes (shown in the models as two hyphens ―--―) with the domain code. If no domain
code exists in 295H Appendix C2 or 296HAppendix C2A for this data and if it desired to have this domain code
as part of CDISC controlled terminology then submit a request to add the new domain via the
CDISC website. Requests for new domain codes must include:
1) Two-letter domain code and description
2) Rationale for domain code
3) Domain model with assumptions
4) Examples
Upon receipt, the SDS Domain Code Subteam will review the package. If accepted, then the
proposal will be submitted to the SDS Team for review. Upon approval, a response will be sent to
the requestor and package processing will begin (i.e., prepare for inclusion in a next release of the
SDTM and SDTMIG, mapping concepts to BRIDG, and posting an update to the CDISC website). If
declined, then the Domain Code Subteam will draft a response for SDS Team review. Upon
agreement, the response will be sent to the requestor and also posted to the CDISC website.
f. Set the order of variables consistent with the order defined in SDTM Tables 2.2.1, 2.2.2, or 2.2.3,
depending upon the general observation class the custom domain is based on.
g. Adjust the labels of the variables only as appropriate to properly convey the meaning in the context
of the data being submitted in the newly created domain. Use title case for all labels (title case
means to capitalize the first letter of every word except for articles, prepositions, and conjunctions).
h. Ensure that appropriate standard variables are being properly applied by comparing the use of
variables in standard domains.
i. Describe the dataset within the define.xml document (see 297HSection 3.2).
j. Place any non-standard (SDTM) variables in a Supplemental Qualifier dataset. Mechanisms for
representing additional non-standard Qualifier variables not described in the general observation
classes and for defining relationships between separate datasets or records are described in 298HSection 8.4
of this document.
Figure 2.6. Creating a New Domain

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 16 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
3 Submitting Data in
Standard Format
3.1 STANDARD METADATA FOR DATASET CONTENTS AND ATTRIBUTES
The SDTMIG provides standard descriptions of some of the most commonly used data domains, with metadata
attributes. The descriptive metadata attributes that should be included in a define.xml as applied in the domain
models are:
The SDTMIG -standard variable name (standardized for all submissions, even though sponsors may be
using other variable names internally in their operational database)
The SDTMIG -standard variable label
Expected data types (the SDTMIG uses character or numeric to conform to the data types consistent with
SAS V5 transport file format, but define.xml allows for more descriptive data types, such as integer or
float)
The actual controlled terms and formats used by the sponsor (do not include the asterisk (*) included in the
CDISC domain models to indicate when controlled terminology applies)
The origin or source of the data (e.g., CRF, derived; see definitions in 299HSection 4.1.1.8)
The role of the variable in the dataset corresponding to the role in the SDTM if desired. Since these roles
are predefined for all standard domains that follow the general observation classes, they do not need to be
specified by sponsors in their define.xml for these domains.
Any Comments provided by the sponsor that may be useful to the Reviewer in understanding the variable
or the data in it.
In addition to these metadata attributes, the CDISC domain models include three other shaded columns that are not
sent to the FDA. These columns assist sponsors in preparing their datasets:
"CDISC Notes" is for notes to the sponsor regarding the relevant to the use of each variable
"Core" indicates how a variable is classified as a CDISC Core Variable (see 300HSection 4.1.1.5)
"References" provides references to relevant section of the SDTM or the SDTMIG.), and one to provide
references to relevant section of the SDTM or the SDTMIG.
The domain models in 301HSection 6 illustrate how to apply the SDTM when creating a specific domain dataset. In
particular, these models illustrate the selection of a subset of the variables offered in one of the general observation
classes along with applicable timing variables. The models also show how a standard variable from a general
observation class should be adjusted to meet the specific content needs of a particular domain, including making the
label more meaningful, specifying controlled terminology, and creating domain-specific notes and examples. Thus
the domain models demonstrate not only how to apply the model for the most common domains, but also give
insight on how to apply general model concepts to other domains not yet defined by CDISC.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 17
Final November 12, 2008
3.2 USING THE CDISC DOMAIN MODELS IN REGULATORY
SUBMISSIONS — DATASET METADATA
The define.xml that accompanies a submission should also describe each dataset that is included in the submission
and describe the natural key structure of each dataset. While most studies will include DM and a set of safety
domains based on the three general observation classes (typically including EX, CM, AE, DS, MH, IE, LB, and VS),
the actual choice of which data to submit will depend on the protocol and the needs of the regulatory reviewer.
Dataset definition metadata should include dataset filenames, descriptions, locations, structures, class, purpose, keys,
and comments as described below in Table 3.2.1.
In the event that no records are present in a dataset (e.g., a small PK study where no subjects took concomitant
medications), the empty dataset should not be submitted and should not be described in the define.xml document.
The annotated CRF will show the data that would have been submitted had data been received; it need not be re-
annotated to indicate that no records exist.
Table 3.2.1. SDTM Submission Dataset-Definition Metadata Example
Dataset
Description
Class
Structure
Purpose
1838HKeys*
Location
1839HDM
Demographics
Special Purpose
Domains
One record per subject
Tabulation
STUDYID,
USUBJID
dm.xpt
1840HCO
Comments
Special Purpose
Domains
One record per comment
per subject
Tabulation
STUDYID,
USUBJID,
COSEQ
co.xpt
1841HSE
Subject Elements
Special Purpose
Domains
One record per actual
Element per subject
Tabulation
STUDYID,
USUBJID,
ETCD,
SESTDTC
se.xpt
1842HSV
Subject Visits
Special Purpose
Domains
One record per actual visit
per subject
Tabulation
STUDYID,
USUBJID,
VISITNUM
sv.xpt
1843HCM
Concomitant
Medications
Interventions
One record per recorded
medication occurrence or
constant-dosing interval per
subject.
Tabulation
STUDYID,
USUBJID,
CMTRT,
CMSTDTC
cm.xpt
1844HEX
Exposure
Interventions
One record per constant
dosing interval per subject
Tabulation
STUDYID,
USUBJID,
EXTRT,
EXSTDTC
ex.xpt
1845HSU
Substance Use
Interventions
One record per substance
type per reported occurrence
per subject
Tabulation
STUDYID,
USUBJID,
SUTRT,
SUSTDTC
su.xpt
1846HAE
Adverse Events
Events
One record per adverse
event per subject
Tabulation
STUDYID,
USUBJID,
AEDECOD,
AESTDTC
ae.xpt
1847HDS
Disposition
Events
One record per disposition
status or protocol milestone
per subject
Tabulation
STUDYID,
USUBJID,
DSDECOD,
DSSTDTC
ds.xpt
1848HMH
Medical History
Events
One record per medical
history event per subject
Tabulation
STUDYID,
USUBJID,
MHDECOD
mh.xpt
1849HDV
Protocol
Deviations
Events
One record per protocol
deviation per subject
Tabulation
STUDYID,
USUBJID,
DVTERM,
DVSTDTC
dv.xpt
1850HCE
Clinical Events
Events
One record per event per
subject
Tabulation
STUDYID,
USUBJID,
CETERM,
CESTDTC
ce.xpt

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 18 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Dataset
Description
Class
Structure
Purpose
1838HKeys*
Location
1851HEG
ECG Test Results
Findings
One record per ECG
observation per time point
per visit per subject
Tabulation
STUDYID,
USUBJID,
EGTESTCD,
VISITNUM,
EGTPTREF,
EGTPTNUM
eg.xpt
1852HIE
Inclusion/
Exclusion Criteria
Not Met
Findings
One record per
inclusion/exclusion criterion
not met per subject
Tabulation
STUDYID,
USUBJID,
IETESTCD
ie.xpt
1853HLB
Laboratory Tests
Results
Findings
One record per analyte per
planned time point number
per time point reference per
visit per subject
Tabulation
STUDYID,
USUBJID,
LBTESTCD,
LBSPEC,
VISITNUM,
LBTPTREF,
LBTPTNUM
lb.xpt
1854HPE
Physical
Examination
Findings
One record per body system
or abnormality per visit per
subject
Tabulation
STUDYID,
USUBJID,
PETESTCD,
VISITNUM
pe.xpt
1855HQS
Questionnaires
Findings
One record per
questionnaire per question
per time point per visit per
subject
Tabulation
STUDYID,
USUBJID,
QSCAT,
QSTESTCD,
VISITNUM,
QSTPTREF,
QSTPTNUM
qs.xpt
1856HSC
Subject
Characteristics
Findings
One record per
characteristic per subject
Tabulation
STUDYID,
USUBJID,
SCTESTCD
sc.xpt
1857HVS
Vital Signs
Findings
One record per vital sign
measurement per time point
per visit per subject
Tabulation
STUDYID,
USUBJID,
VSTESTCD,
VISITNUM,
VSTPTREF,
VSTPTNUM
vs.xpt
1858HDA
Drug
Accountability
Findings
One record per drug
accountability finding per
subject
Tabulation
STUDYID,
USUBJID,
DATESTCD,
DADTC
da.xpt
1859HMB
Microbiology
Specimen
Findings
One record per
microbiology specimen
finding per time point per
visit per subject
Tabulation
STUDYID,
USUBJID,
MBTESTCD,
VISITNUM,
MBTPTREF,
MBTPTNUM
mb.xpt
1860HMS
Microbiology
Susceptibility Test
Findings
One record per
microbiology susceptibility
test (or other organism-
related finding) per
organism found in MB
Tabulation
STUDYID,
USUBJID,
MSTESTCD,
VISITNUM,
MSTPTREF,
MSTPTNUM
ms.xpt
1861HPC
Pharmacokinetic
Concentrations
Findings
One record per analyte per
planned time point number
per time point reference per
visit per subject"
Tabulation
STUDYID,
USUBJID,
PCTESTCD,
VISITNUM,
PCTPTREF,
PCTPTNUM
pc.xpt
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 19
Final November 12, 2008
Dataset
Description
Class
Structure
Purpose
1838HKeys*
Location
302HPP
Pharmacokinetic
Parameters
Findings
One record per PK
parameter per time-
concentration profile per
modeling method per
subject
Tabulation
STUDYID,
USUBJID,
PPTESTCD,
PPCAT,
VISITNUM,
PPTPTREF
pp.xpt
303HFA
Findings About
Events or
Interventions
Findings
One record per finding per
object per time point per
time point reference per
visit per subject
Tabulation
STUDYID,
USUBJID,
FATESTCD,
FAOBJ,
VISITNUM,
FATPTREF,
FATPTNUM
fa.xpt
304HTA
Trial Arms
Trial Design
One record per planned
Element per Arm
Tabulation
STUDYID,
ARMCD,
TAETORD
ta.xpt
305HTE
Trial Elements
Trial Design
One record per planned
Element
Tabulation
STUDYID,
ETCD
te.xpt
1862HTV
Trial Visits
Trial Design
One record per planned
Visit per Arm
Tabulation
STUDYID,
VISITNUM,
ARMCD
tv.xpt
306HTI
Trial Inclusion/
Exclusion Criteria
Trial Design
One record per I/E criterion
Tabulation
STUDYID,
IETESTCD
ti.xpt
307HTS
Trial Summary
Trial Design
One record per trial
summary parameter
value
Tabulation
STUDYID,
TSPARMCD,
TSSEQ
ts.xpt
1863HRELREC
Related Records
Special Purpose
Datasets
One record per related
record, group of records or
datasets
Tabulation
STUDYID,
RDOMAIN,
USUBJID,
IDVAR,
IDVARVAL,
RELID
relrec.xpt
308HSUPP--
**
Supplemental
Qualifiers for
[domain name]
Special-Purpose
Datasets
One record per IDVAR,
IDVARVAL, and QNAM
value per subject
Tabulation
STUDYID,
RDOMAIN,
USUBJID,
IDVAR,
IDVARVAL,
QNAM
supp--.xpt or
suppqual.xpt
* Note that the key variables shown in this table are examples only. A sponsor‘s actual key structure may be
different.
** Separate Supplemental Qualifier datasets of the form supp--.xpt are recommended. See 309HSection 8.4.
3.2.1.1 PRIMARY KEYS
310HTable 3.2.1 above shows examples of what a sponsor might submit as variables that comprise the primary key for
SDTM datasets. Since the purpose of this column is to aid reviewers in understanding the structure of a dataset,
sponsors should list all of the natural keys (see definition below) for the dataset. These keys should define uniqueness
for records within a dataset, and may define a record sort order. The naming of these keys should be consistent with
the description of the structure in the Structure column. For all the general-observation-class domains (and for some
special-purpose domains), the --SEQ variable was created so that a unique record could be identified consistently
across all of these domains via its use, along with STUDYID, USUBJID, DOMAIN. In most domains, --SEQ will be
a surrogate key (see definition below) for a set of variables which comprise the natural key. In certain instances, a
Supplemental Qualifier (SUPP--) variable might also contribute to the natural key of a record for a particular domain.
See 311Hassumption 4.1.1.9 for how this should be represented, and for additional information on keys.
A natural key is a piece of data (one or more columns of an entity) that uniquely identify that entity, and distinguish
it from any other row in the table. The advantage of natural keys is that they exist already, and one does not need to
introduce a new ―unnatural‖ value to the data schema. One of the difficulties in choosing a natural key is that just
about any natural key one can think of has the potential to change. Because they have business meaning, natural

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 20 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
keys are effectively coupled to the business, and they may need to be reworked when business 312Hrequirements change.
An example of such a change in clinical trials data would be the addition of a position or location that becomes a
key in a new study, but wasn‘t collected in previous studies.
A surrogate key is a single-part, artificially established identifier for a record. Surrogate key assignment is a special
case of derived data, one where a portion of the primary key is derived. A surrogate key is immune to changes in
business needs. In addition, the key depends on only one field, so it‘s compact. A common way of deriving surrogate
key values is to assign integer values sequentially. The --SEQ variable in the SDTM datasets is an example of a
surrogate key for most datasets; in some instances, however, --SEQ might be a part of a natural key as a replacement
for what might have been a key (e.g. a repeat sequence number) in the sponsor's database
3.2.1.2 CDISC SUBMISSION VALUE-LEVEL METADATA
In general, the CDISC V3.x Findings data models are closely related to normalized, relational data models in a
vertical structure of one record per observation. Since the V3.x data structures are fixed, sometimes information that
might have appeared as columns in a more horizontal (denormalized) structure in presentations and reports will
instead be represented as rows in an SDTM Findings structure. Because many different types of observations are all
presented in the same structure, there is a need to provide additional metadata to describe the expected differences
that differentiate, for example, hematology lab results from serum chemistry lab results in terms of data type,
standard units and other attributes.
For example, the Vital Signs data domain could contain subject records related to diastolic and systolic blood
pressure, height, weight, and body mass index (BMI). These data are all submitted in the normalized SDTM
Findings structure of one row per vital signs measurement. This means that there could be five records per subject
(one for each test or measurement) for a single visit or time point, with the parameter names stored in the Test
Code/Name variables, and the parameter values stored in result variables. Since the unique Test Code/Names could
have different attributes (i.e., different origins, roles, and definitions) there would be a need to provide value-level
metadata for this information.
The value-level metadata should be provided as a separate section of the Case Report Tabulation Data Definition
Specification (CRT-DDS). This information, which historically has been submitted as a pdf document named
―define.pdf‖, should henceforth be submitted in an XML format. For details on the CDISC specification for
submitting define.xml, see 313Hwww.cdisc.org/standards/
3.2.2 CONFORMANCE
Conformance with the SDTMIG Domain Models is minimally indicated by:
Following the complete metadata structure for data domains
Following SDTMIG domain models wherever applicable
Using SDTM-specified standard domain names and prefixes where applicable
Using SDTM-specified standard variable names
Using SDTM-specified variable labels for all standard domains
Using SDTM-specified data types for all variables
Following SDTM-specified controlled terminology and format guidelines for variables, when provided
Including all collected and relevant derived data in one of the standard domains, special-purpose datasets, or
general-observation-class structures
Including all Required and Expected variables as columns in standard domains, and ensuring that all
Required variables are populated
Ensuring that each record in a dataset includes the appropriate Identifier and, Timing variables, as well as a
Topic variable
Conforming to all business rules described in the CDISC Notes column and general and domain-specific
assumptions.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 21
Final November 12, 2008
4 Assumptions for Domain
Models
4.1 GENERAL ASSUMPTIONS FOR ALL DOMAINS
4.1.1 GENERAL DOMAIN ASSUMPTIONS
4.1.1.1 REVIEW STUDY DATA TABULATION AND IMPLEMENTATION GUIDE
Review the Study Data Tabulation Model as well as this Implementation Guide before attempting to use any of the
individual domain models. See the Case Report Tabulation Data Definition Specification (define.xml), available on
the CDISC website, for information about an xml representation of the define.xml document.
4.1.1.2 RELATIONSHIP TO ANALYSIS DATASETS
Specific guidance on preparing analysis datasets can be found in the CDISC Analysis Dataset Model General
Considerations document, available at 314Hwww.cdisc.org/standards/
4.1.1.3 ADDITIONAL TIMING VARIABLES
Additional Timing variables can be added as needed to a standard domain model based on the three general
observation classes except where discouraged in 315HAssumption 4.1.4.8 and specific domain assumptions. Timing
variables can be added to special-purpose domains only where specified in the SDTMIG domain model
assumptions. Timing variables cannot be added to SUPPQUAL datasets or to RELREC (described in 316HSection 8).
4.1.1.4 ORDER OF THE VARIABLES
The order of variables in the define.xml should reflect the order of variables in the dataset. The order of variables in
the CDISC domain models has been chosen to facilitate the review of the models and application of the models.
Variables for the three general observation classes should be ordered with Identifiers first, followed by the Topic,
Qualifier, and Timing variables. Within each role, variables are ordered as shown in Tables 2.2.1, 2.2.2, 2.2.3,
2.2.3.1, 2.2.4, and 2.2.5 of the SDTM.
4.1.1.5 CDISC CORE VARIABLES
The concept of core variable is used both as a measure of compliance, and to provide general guidance to sponsors.
Three categories of variables are specified in the ―Core‖ column in the domain models:
A Required variable is any variable that is basic to the identification of a data record (i.e., essential key
variables and a topic variable) or is necessary to make the record meaningful. Required variables must always
be included in the dataset and cannot be null for any record.
An Expected variable is any variable necessary to make a record useful in the context of a specific domain.
Expected variables may contain some null values, but in most cases will not contain null values for every record.
When no data has been collected for an expected variable, however, a null column should still be included in the
dataset, and a comment should be included in the define.xml to state that data was not collected.
A Permissible variable should be used in a domain as appropriate when collected or derived. Except where
restricted by specific domain assumptions, any SDTM Timing and Identifier variables, and any Qualifier
variables from the same general observation class are permissible for use in a domain based on that general
observation class. The Sponsor can decide whether a Permissible variable should be included as a column
when all values for that variable are null. The sponsor does not have the discretion to not submit permissible
variables when they contain data.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 22 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.1.6 ADDITIONAL GUIDANCE ON DATASET NAMING
SDTM datasets are normally named to be consistent with the domain code; for example, the Demographics dataset
(DM) is named dm.xpt (see 317H Appendix C2 for a list of standard and reserved domain codes). Exceptions to this rule
are described in 318HSection 4.1.1.7 for general-observation-class datasets and in 319HSection 8 for the RELREC and SUPP--
datasets.
In some cases, sponsors may need to define new custom domains other than those represented in the SDTMIG or
listed in 320HAppendix C2, and may be concerned that CDISC domain codes defined in the future will conflict with
those they choose to use. To eliminate any risk of a sponsor using a name that CDISC later determines to have a
different meaning, domain codes beginning with the letters X, Y, or Z have been reserved for the creation of custom
domains. Any letter or number may be used in the second position. Note the use of codes beginning with X, Y, or Z
is optional, and not required for custom domains.
4.1.1.7 SPLITTING DOMAINS
Sponsors may choose to split a domain of topically related information into physically separate datasets. In such
cases, one of two approaches should be implemented:
1) For a domain based on a general observation class, splitting should be according to values in --CAT (which
must not be null).
2) The Findings About (FA) domain (321HSection 6.4) can be split either by --CAT values (per the bullet above) or
relative to the parent domain of the value in --OBJ. For example, FACM would store Findings About CM
records. See 322HSection 6.4.2 for more details.
The following rules must be adhered to when splitting a domain into separate datasets to ensure they can be
appended back into one domain dataset:
1) The value of DOMAIN must be consistent across the separate datasets as it would have been if they had not
been split (e.g., QS, FA).
2) All variables that require a domain prefix (e.g., --TESTCD, --LOC) must use the value of DOMAIN as the
prefix value (e.g., QS, FA).
3) --SEQ must be unique within USUBJID for all records across all the split datasets. If there are 1000 records
for a USUBJID across the separate datasets, all 1000 records need unique values for --SEQ.
4) When relationship datasets (e.g., SUPPxx, FAxx, CO, RELREC) relate back to split parent domains, IDVAR
should generally be --SEQ. When IDVAR is a value other than --SEQ (e.g., --GRPID, --REFID, --SPID),
care should be used to ensure that the parent records across the split datasets have unique values for the
variable specified in IDVAR, so that related children records do not accidentally join back to incorrect parent
records.
5) Variables of the same name in separate datasets should have the same SAS Length attribute to avoid any
difficulties if the sponsor or FDA should decide to append datasets together.
6) Permissible variables included in one split dataset need not be included in all split datasets. Should the
datasets be appended in SAS, permissible variables not used in some split datasets will have null values in
the appended datasets. Care is advised, however, when considering variable order. Should a permissible
variable used in one (or more) split datasets not be included in the first dataset used in a SAS Set statement,
the order of variables could be compromised.
7) Split dataset names can be up to four characters in length. For example, if splitting by --CAT, then dataset
names would be the domain name plus up to two additional characters (e.g., QS36 for SF-36). If splitting
Findings About by parent domain, then the dataset name would be the domain name plus the two-character
domain code describing the parent domain code (e.g., FACM). The four-character dataset-name limitation
allows the use of a Supplemental Qualifier dataset associated with the split dataset.
8) Supplemental Qualifier datasets for split domains would also be split. The nomenclature would include the
additional one-to-two characters used to identify the split dataset (e.g., SUPPQS36, SUPPFACM). The value
of RDOMAIN in the SUPP-- datasets would be the two-character domain code (e.g., QS, FA).
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 23
Final November 12, 2008
9) In RELREC, if a dataset-level relationship is defined for a split Findings About domain, then RDOMAIN
may contain the four-character dataset name, as shown in the following example.
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
ABC
CM
CMSPID
ONE
1
ABC
FACM
FASPID
MANY
1
10) See the SDTM Metadata Implementation Guide for guidance on how to represent the metadata for a set of
split domain datasets in the define.xml.
Note that submission of split SDTM domains may be subject to additional dataset splitting conventions as defined
by regulators via technical specifications (e.g., Study Data Specifications) and/or as negotiated with regulatory
reviewers.
4.1.1.7.1 EXAMPLE OF SPLITTING QUESTIONNAIRES
This example shows the split QS domain data into three datasets: Clinical Global Impression (QSCG), Cornell Scale
for Depression in Dementia (QSCS) and Mini Mental State Examination (QSMM). Each dataset represents a subset
of the QS domain data and has only one value of QSCAT.
QS Domains
qscg.xpt (Clinical Global Impressions)
Row
STUDYID
DOMAIN
USUBJID
QSSEQ
QSSPID
QSTESTCD
QSTEST
QSCAT
1
CDISC01
QS
CDISC01.100008
1
CGI-
CGI-I
CGIGLOB
Global
Improvement
Clinical Global
Impressions
2
CDISC01
QS
CDISC01.100008
2
CGI-
CGI-I
CGIGLOB
Global
Improvement
Clinical Global
Impressions
3
CDISC01
QS
CDISC01.100014
1
CGI-
CGI-I
CGIGLOB
Global
Improvement
Clinical Global
Impressions
4
CDISC01
QS
CDISC01.100014
2
CGI-
CGI-I
CGIGLOB
Global
Improvement
Clinical Global
Impressions
Row
QSORRES
QSSTRESC
QSSTRESN
QSBLFL
VISITNUM
VISIT
VISITDY
QSDTC
QSDY
1
(cont)
No change
4
4
3
WEEK
2
15
2003-
05-13
15
2
(cont)
Much
Improved
2
2
10
WEEK
24
169
2003-
10-13
168
3
(cont)
Minimally
Improved
3
3
3
WEEK
2
15
2003-
10-31
17
4
(cont)
Minimally
Improved
3
3
10
WEEK
24
169
2004-
03-30
168
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 24 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
qscs.xpt (Cornell Scale for Depression in Dementia)
Row
STUDYID
DOMAIN
USUBJID
QSSEQ
QSSPID
QSTESTCD
QSTEST
QSCAT
1
CDISC01
QS
CDISC01.
100008
3
CSDD-01
CSDD01
Anxiety
Cornell Scale for
Depression in Dementia
2
CDISC01
QS
CDISC01.
100008
23
CSDD-01
CSDD01
Anxiety
Cornell Scale for
Depression in Dementia
3
CDISC01
QS
CDISC01.
100014
3
CSDD-01
CSDD01
Anxiety
Cornell Scale for
Depression in Dementia
4
CDISC01
QS
CDISC01.
100014
28
CSDD-06
CSDD06
Retardation
Cornell Scale for
Depression in Dementia
Row
QSORRES
QSSTRESC
QSSTRESN
QSBLFL
VISITNUM
VISIT
VISITDY
QSDTC
QSDY
1 (cont)
Severe
2
2
1
SCREEN
-13
2003-04-15
-14
2 (cont)
Severe
2
2
Y
2
BASELINE
1
2003-04-29
1
3 (cont)
Severe
2
2
1
SCREEN
-13
2003-10-06
-9
4 (cont)
Mild
1
1
Y
2
BASELINE
1
2003-10-15
1
qsmm.xpt (Mini Mental State Examination)
Row
STUDYID
DOMAIN
USUBJID
QSSEQ
QSSPID
QSTESTCD
QSTEST
QSCAT
1
CDISC01
QS
CDISC01.
100008
81
MMSE-A.1
MMSEA1
Orientation Time
Score
Mini Mental State
Examination
2
CDISC01
QS
CDISC01.
100008
88
MMSE-A.1
MMSEA1
Orientation Time
Score
Mini Mental State
Examination
3
CDISC01
QS
CDISC01.
100014
81
MMSE-A.1
MMSEA1
Orientation Time
score
Mini Mental State
Examination
4
CDISC01
QS
CDISC01.
100014
88
MMSE-A.1
MMSEA1
Orientation Time
score
Mini Mental State
Examination
Row
QSORRES
QSSTRESC
QSSTRESN
QSBLFL
VISITNUM
VISIT
VISITDY
QSDTC
QSDY
1 (cont)
4
4
4
1
SCREEN
-13
2003-04-15
-14
2 (cont)
3
3
3
Y
2
BASELINE
1
2003-04-29
1
3 (cont)
2
2
2
1
SCREEN
-13
2003-10-06
-9
4 (cont)
2
2
2
Y
2
BASELINE
1
2003-10-15
1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 25
Final November 12, 2008
SUPPQS Domains
suppqscg.xpt: Supplemental Qualifiers for QSCG
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
CDISC01
QS
CDISC01.
100008
QSCAT
Clinical Global
Impressions
QSLANG
Questionnaire
Language
GERMAN
CRF
2
CDISC01
QS
CDISC01.
100014
QSCAT
Clinical Global
Impressions
QSLANG
Questionnaire
Language
FRENCH
CRF
suppqscs.xpt: Supplemental Qualifiers for QSCS
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
CDISC01
QS
CDISC01.
100008
QSCAT
Cornell Scale
for Depression
in Dementia
QSLANG
Questionnaire
Language
GERMAN
CRF
2
CDISC01
QS
CDISC01.
100014
QSCAT
Cornell Scale
for Depression
in Dementia
QSLANG
Questionnaire
Language
FRENCH
CRF
suppqsmm.xpt: Supplemental Qualifiers for QSMM
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
CDISC01
QS
CDISC01.
100008
QSCAT
Mini Mental State
Examination
QSLANG
Questionnaire
Language
GERMAN
CRF
2
CDISC01
QS
CDISC01.
100014
QSCAT
Mini Mental State
Examination
QSLANG
Questionnaire
Language
FRENCH
CRF
4.1.1.8 ORIGIN METADATA
4.1.1.8.1 ORIGIN METADATA FOR VARIABLES
The Origin column of the define.xml is used to indicate where the data originated. Its purpose is to unambiguously
communicate to the reviewer whether data was collected on a CRF (and thus should be traceable to an annotated
CRF), derived (and thus traceable to some derivation algorithm), or assigned by some subjective process (and thus
traceable to some external evaluator). The SDTMIG defines the following controlled terms for specifying Origin:
CRF: The designation of ‖CRF‖ (along with a reference) as an origin in the define.xml means that data was
collected as part of a CRF and that there is an annotated CRF associated with the variable. Sponsors may specify
additional details about the origin that may be helpful to the Reviewer (e.g., electronic diary) in the Comments
section of the define.xml. An origin of ―CRF‖ includes information that is preprinted on the CRF (e.g.,
―RESPIRATORY SYSTEM DISORDERS‖ for MHCAT).
eDT: The designation of "eDT" as an origin in the define.xml means that the data are received via an electronic Data
Transfer (eDT) and usually does not have associated annotations. An origin of eDT refers to data collected via data
streams such as laboratory, ECG, or IVRS. Sponsors may specify additional details about the origin that may be
helpful to the Reviewer in the Comments section of the define.xml.
Derived: Derived data are not directly collected on the CRF but are calculated by an algorithm or reproducible rule,
which is dependent upon other data values. This algorithm is applied across all values and may reference other
SDTM datasets. The derivation is assumed to be performed by the Sponsor. This does not apply to derived lab test
results performed directly by labs (or by devices).

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 26 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Examples illustrating the distinction between collected and derived values include the following:
A value derived by an eCRF system from other entered fields has an origin of "Derived, " since the sponsor
controls the derivation.
A value derived from collected data by the sponsor, or a CRO working on their behalf, has an origin of
"Derived."
A value derived by an investigator and written/entered on a CRF has an origin of "CRF" (along with a
reference) rather than ―derived‖.
A value derived by a vendor (e.g., a central lab) according to their procedures is considered collected rather
than derived, and would have an origin of ―eDT‖.
Assigned: A value that is determined by individual judgment (by an evaluator other than the subject or investigator),
rather than collected as part of the CRF or derived based on an algorithm. This may include third party attributions
by an adjudicator. Coded terms that are supplied as part of a coding process (as in --DECOD) are considered to have
an Origin of ―Assigned‖. Values that are set independently of any subject-related data values in order to complete
SDTM fields such as DOMAIN and --TESTCD are considered to have an Origin of ―Assigned‖.
Protocol: A value that is defined as part of the Trial Design preparation (see 323HSection 7). An example would be
VSPOS (Vital Signs Position), which may be specified only in the protocol and not appear on a CRF.
The term ―Sponsor Defined‖ was used in earlier versions of the SDTMIG to advise the Sponsor to supply the
appropriate Origin value in the metadata. The text ―Sponsor Defined‖ was not intended to be used in the define.xml
and is no longer used in V3.1.2 and later.
4.1.1.8.2 ORIGIN METADATA FOR RECORDS
Sponsors are cautioned to recognize that an Origin of ―Derived‖ means that all values for that variable were derived,
and that ―CRF‖ (along with a reference) means that all were collected. In some cases, both collected and derived
values may be reported in the same field. For example, some records in a Findings dataset such as QS contain values
collected from the CRF and other records may contain derived values such as a total score. When both derived and
collected values are reported in a field, the value-level metadata origin will indicate at the test level if the value is
―Derived‖ or ―CRF‖ and the variable-level metadata origin will list all types for that variable separated by commas
(e.g., ―Derived, CRF‖).
4.1.1.9 ASSIGNING NATURAL KEYS IN THE METADATA
324HSection 3.2 indicates that a sponsor should include in the metadata the variables that contribute to the natural key for
a domain. The following examples are illustrations of how to do this, and include a case where a Supplemental
Qualifier variable is referenced because it forms part of the natural key.
Physical Examination (PE) domain example:
Sponsor A chooses the following natural key for the PE domain:
STUDYID, USUBJID, VISTNUM, PETESTCD
Sponsor B collects data in such a way that the location (PELOC) and method (PEMETHOD) variables need to be
included in the natural key to identify a unique row, but they do not collect a visit variable; instead they use the visit
date (PEDTC) to sequence the data. Sponsor B then defines the following natural key for the PE domain.
STUDYID, USUBJID, PEDTC, PETESTCD, PELOC, PEMETHOD
In certain instances a Supplemental Qualifier variable (i.e., a QNAM value, see 325HSection 8.4) might also contribute to
the natural key of a record, and therefore needs to be referenced as part of the natural key for a domain. The
important concept here is that a domain is not limited by physical structure. A domain may be comprised of more
than one physical dataset, for example the main domain dataset and its associated Supplemental Qualifiers dataset.
Supplemental Qualifiers variables should be referenced in the natural key by using a two-part name. The word
QNAM must be used as the first part of the name to indicate that the contributing variable exists in a dataset (and
this can be either a domain-specific SUPP-- dataset or the general SUPPQUAL dataset) and the second part is the

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 27
Final November 12, 2008
value of QNAM that ultimately becomes a column reference (e.g., QNAM.XVAR when the SUPP-- record has a
QNAM of ―XVAR‖) when the SUPPQUAL records are joined on to the main domain dataset.
Continuing with the PE domain example above, Sponsor B might have used ultrasound as a method of measurement
and might have collected additional information such as the makes and models of ultrasound equipment employed.
The sponsor considers the make and model information to be essential data that contributes to the uniqueness of the
test result, and so creates Supplemental Qualifier variables for make (QNAM=PEMAKE) and model
(QNAM=PEMODEL). The natural key is then defined as follows:
STUDYID, USUBJID, PEDTC, PETESTCD, PELOC, PEMETHOD, QNAM.PEMAKE, QNAM.PEMODEL
This approach becomes very useful in a Findings domain when a sponsor might choose to employ generic
--TESTCD values rather than compound --TESTCD values. The use of generic test codes helps to create distinct
lists of manageable controlled terminology for --TESTCD. In studies where multiple repetitive tests or
measurements are being made, for example in a rheumatoid arthritis study where repetitive measurements of bone
erosion in the hands and wrists might be made using both X-ray and MRI equipment, one approach to recording this
data might be to create an individual --TESTCD value for each measurement. Taking just the phalanges, a sponsor
might want to express the following in a test code in order to make it unique:
Left or Right hand
Phalange position (proximal / distal / middle)
Rotation of the hand
Method of measurement (X-ray / MRI)
Machine Make
Machine Model
Trying to encapsulate all of this information within a unique value of a --TESTCD is not a recommended approach
for the following reasons:
It results in the creation of a potentially large number of test codes
The eight-character values of --TESTCD becoming less intuitively meaningful
Multiple test codes are essentially representing the same test or measurement simply to accommodate
attributes of a test within the --TESTCD value itself (e.g., to represent a body location at which a
measurement was taken).
As a result, the preferred approach would be to use a generic (or simple) test code that requires associated qualifier
variables to fully express the test detail. Using this approach in the above example, a generic --TESTCD value might
be ―EROSION‖ and the additional components of the compound test codes discussed above would be represented in
a number of distinct qualifier variables. These may include domain variables (--LOC, --METHOD, etc.) and
Supplemental Qualifier variables (QNAM.MAKE, QNAM.MODEL, etc.). Expressing the natural key becomes very
important in this situation in order to communicate the variables that contribute to the uniqueness of a test.
If a generic --TESTCD was used the following variables would be used to fully describe the test. The test is
―EROSION‖, the location is ―Left MCP I‖, the method of measurement is ―Ultrasound‖, the make of the ultrasound
machine is ―ACME‖ and the model of the ultrasound machine is ―u 2.1‖. This domain includes both domain
variables and Supplemental Qualifier variables that contribute to the natural key of each row and to describe the
uniqueness of the test.
--TESTCD
--TEST
--LOC
--METHOD
QNAM.MAKE
QNAM.MODEL
EROSION
EROSION
LEFT MCP I
ULTRASOUND
ACME
U 2.1

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 28 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.2 GENERAL VARIABLE ASSUMPTIONS
4.1.2.1 VARIABLE-NAMING CONVENTIONS
SDTM variables are named according to a set of conventions, using fragment names (defined in 1864HAppendix D).
Variables with names ending in ―CD‖ are ―short‖ versions of associated variables that do not include the ―CD‖
suffix (e.g., --TESTCD is the short version of --TEST).
Values of --TESTCD must be limited to 8 characters, and cannot start with a number, nor can they contain characters
other than letters, numbers, or underscores. This is to avoid possible incompatibility with SAS V5 Transport files.
This limitation will be in effect until the use of other formats (such as XML) becomes acceptable to regulatory
authorities.
QNAM serves the same purpose as --TESTCD within supplemental qualifier datasets, and so values of QNAM are
subject to the same restrictions as values of --TESTCD.
Values of other ―CD‖ variables are not subject to the same restrictions as --TESTCD.
ETCD (the companion to ELEMENT) and TSPARMCD (the companion to TSPARM) are limited to 8
characters and do not have special character restrictions. These values should be short for ease of use in
programming, but it is not expected that they will need to serve as variable names.
ARMCD is limited to 20 characters and does not have special character restrictions. The maximum length
of ARMCD is longer than for other ―short‖ variables to accommodate the kind of values that are likely to
be needed for crossover trials. For example, if ARMCD values for a seven-period crossover were
constructed using two-character abbreviations for each treatment and separating hyphens, the length of
ARMCD values would be 20.
Variable descriptive names (labels), up to 40 characters, should be provided as data variable labels.
Use of variable names (other than domain prefixes), formats, decodes, terminology, and data types for the same type
of data (even for custom domains and Supplemental Qualifiers) should be consistent within and across studies
within a submission. Sponsors must use the predefined SDTM-standard labels in all standard domains.
4.1.2.2 TWO-CHARACTER DOMAIN IDENTIFIER
In order to minimize the risk of difficulty when merging/joining domains for reporting purposes, the two-character
Domain Identifier is used as a prefix in most variable names.
Special-Purpose domains (see 326HSection 5), Standard domains (see 327HSection 6), Trial Design domains (see 328HSection 7)
and Relationship datasets (see 329HSection 8) already specify the complete variable names, so no action is required.
When creating custom domains based on the General Observation Classes, sponsors must replace the -- (two
hyphens) prefix in the General Observation Class, Timing, and Identifier variables with the two-character Domain
Identifier (DOMAIN) variable value for that domain/dataset. The two-character domain code is limited to A to Z for
the first character, and A-Z, 0 to 9 for the 2nd character. No special characters are allowed for compatibility with SAS
version 5 transport files, and with file naming for the Electronic Common Technical Document (eCTD).
The philosophy applied to determine which variable names use a prefix was that all variable names are prefixed with
the Domain Identifier in which they originate except the following:
a. Required Identifiers (STUDYID, DOMAIN, USUBJID)
b. Commonly used grouping and merge Keys (VISIT, VISITNUM, VISITDY), and many of the variables
in trial design (such as ELEMENT and ARM)
c. All Demographics domain (DM) variables other than DMDTC and DMDY
d. All variables in RELREC and SUPPQUAL, and some variables in Comments and Trial Design datasets.
Required Identifiers are not prefixed because they are usually used as keys when merging/joining observations. The
--SEQ and the optional Identifiers --GRPID and --REFID are prefixed because they may be used as keys when
relating observations across domains.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 29
Final November 12, 2008
4.1.2.3 USE OF “SUBJECT” AND USUBJID
―Subject‖ should be used where applicable to generically refer to both ―patients‖ and ―healthy volunteers‖ in order
to be consistent with the recommendation in FDA guidance. The term ―Subject‖ should be used consistently in all
labels and comments. To identify a subject uniquely across all studies for all applications or submissions involving
the product, a unique identifier (USUBJID) should be assigned and included in all datasets.
The unique subject identifier (USUBJID) is required in all datasets containing subject-level data. USUBJID values
must be unique for each trial participant (subject) across all trials in the submission. This means that no two (or
more) subjects, across all trials in the submission, may have the same USUBJID. Additionally, the same person who
participates in multiple clinical trials (when this is known) must be assigned the same USUBJID value in all trials.
Sample Rows from individual study dm.xpt files for a same subject that participates first in ACME01 study, then
ACME14 study. Note that this is only one example of the possible values for USUBJID. CDISC does not
recommend any specific format for the values of USUBJID, only that the values need to be unique for all subjects in
the submission, and across multiple submissions for the same compound. Many sponsors concatenate values for the
Study, Site and Subject into USUBJID, but this is not a requirement. It is acceptable to use any format for
USUBJID, as long as the values are unique across all subjects per FDA guidance.
Study ACME01 dm.xpt
STUDYID
DOMAIN
USUBJID
SUBJID
SITEID
INVNAM
ACME01
DM
ACME01-05-001
001
05
John Doe
Study ACME14 dm.xpt
STUDYID
DOMAIN
USUBJID
SUBJID
SITEID
INVNAM
ACME14
DM
ACME01-05-001
017
14
Mary Smith
4.1.2.4 CASE USE OF TEXT IN SUBMITTED DATA
It is recommended that text data be submitted in upper case text. Exceptions may include long text data (such as
comment text); values of --TEST in Findings datasets (which may be more readable in title case if used as labels in
transposed views); and certain controlled terminology (see 330HSection 4.1.3.2) that are already in mixed case. The
Sponsor‘s define.xml may indicate as a general note or assumption whether case sensitivity applies to text data for
any or all variables in the dataset.
4.1.2.5 CONVENTION FOR MISSING VALUES
Missing values for individual data items should be represented by nulls. This is a change from previous versions of
the SDTMIG, which allowed sponsors to define their conventions for missing values. Conventions for representing
observations not done using the SDTM --STAT and --REASND variables are addressed in 331H Section 4.1.5.1.2 and the
individual domain models.
4.1.2.6 GROUPING VARIABLES AND CATEGORIZATION
Grouping variables are Identifiers and Qualifiers that group records in the SDTM domains/datasets such as the
--CAT (Category) and --SCAT (Subcategory) variables assigned by sponsors to categorize topic-variable values. For
example, a lab record with LBTEST = ―SODIUM‖ might have LBCAT = ―CHEMISTRY‖ and LBSCAT =
―ELECTROLYTES‖. Values for --CAT and --SCAT should not be redundant with the domain name or dictionary
classification provided by --DECOD and --BODSYS.
1. Hierarchy of Grouping Variables
STUDYID
DOMAIN
--CAT
--SCAT
USUBJID
--GRPID

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 30 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
2. How Grouping Variables Group Data
A. For the subject
1. All records with the same USUBJID value are a group of records that describe that subject.
B. Across subjects (records with different USUBJID values)
1. All records with the same STUDYID value are a group of records that describe that study
2. All records with the same DOMAIN value are a group of records that describe that domain
3. --CAT (Category) and --SCAT (Sub-category) values further subset groups within the domain.
Generally, --CAT/--SCAT values have meaning within a particular domain. However, it is
possible to use the same values for --CAT/--SCAT in related domains (e.g., MH and AE). When
values are used across domains, the meanings should be the same. Examples of where
--CAT/--SCAT may have meaning across domains/datasets:
a. Some limited cases where they will have meaning across domains within the same
general observation class, because those domains contain similar conceptual information.
Adverse Events (AE), Medical History (MH) and Clinical Events (CE), for example, are
conceptually the same data, the only differences being when the event started relative to
the study start and whether the event is considered a regulatory reportable adverse event
in the study. Neurotoxicities collected in Oncology trials both as separate Medical
History CRFs (MH domain) and Adverse Event CRFs (AE domain) could both
identify/collect ―Paresthesia of the left Arm.‖ In both domains, the --CAT variable could
have the value of NEUROTOXICITY.
b. Cases where multiple datasets are necessary to capture data in the same domain. As an
example, perhaps the existence, start and stop date of ―Paresthesia of the left Arm‖ is
reported as an Adverse Event (AE domain), but the severity of the event is captured at
multiple visits and recorded as Findings About (FA dataset). In both cases the --CAT
variable could have a value of NEUROTOXICITY.
c. Cases where multiple domains are necessary to capture data that was collected together and
have an implicit relationship, perhaps identified in the Related Records (RELREC) special
purpose dataset. Stress Test data collection, for example, may capture the following:
i. Information about the occurrence, start, stop, and duration of the test in an
Events or Interventions custom general observation class dataset
ii. Vital Signs recorded during the stress test (VS domain)
iii. Treatments (e.g., oxygen) administered during the stress test (in an Interventions
domain).
In such cases, the data collected during the stress tests recorded in three separate domains
may all have --CAT/--SCAT values (STRESS TEST) that identify this data was collected
during the stress test.
C. Within subjects (records with the same USUBJID values)
1. --GRPID values further group (subset) records within USUBJID. All records in the same domain with the
same --GRPID value are a group of records within USUBJID. Unlike --CAT and --SCAT, --GRPID values
are not intended to have any meaning across subjects and are usually assigned during or after data collection.
2. Although --SPID and --REFID are Identifier variables, usually not considered to be grouping variables, they
may have meaning across domains.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 31
Final November 12, 2008
3. Differences between Grouping Variables
A. The primary distinctions between --CAT/--SCAT and --GRPID are:
--CAT/--SCAT are known (identified) about the data before it is collected
--CAT/--SCAT values group data across subjects
--CAT/--SCAT may have some controlled terminology
--GRPID is usually assigned during or after data collection at the discretion of the Sponsor
--GRPID groups data only within a subject
--GRPID values are sponsor-defined, and will not be subject to controlled terminology.
Therefore, data that would be the same across subjects is usually more appropriate in --CAT/--SCAT, and
data that would vary across subjects is usually more appropriate in --GRPID. For example, a Concomitant
Medication administered as part of a known combination therapy for all subjects (Mayo Clinic Regimen
for example) would more appropriately use --CAT/--SCAT to identify the medication as part of that
regimen. Groups of medications taken to treat an SAE, recorded in/on the SAE collection, and could be
part of a different grouping of medications for each subject would more appropriately use --GRPID.
In domains based on the Findings general observation class, the --RESCAT variable can be used to categorize results
after the fact. --CAT and --SCAT by contrast, are generally pre-defined by the Sponsor or used by the investigator at
the point of collection, not after assessing the value of Findings results.
4.1.2.7 SUBMITTING FREE TEXT FROM THE CRF
Sponsors often collect free text data on a CRF to supplement a standard field. This often occurs as part of a list of
choices accompanied by ―Other, specify.‖ The manner in which these data are submitted will vary based on their
role.
4.1.2.7.1 “SPECIFY” VALUES FOR NON-RESULT QUALIFIER VARIABLES
When free-text information is collected to supplement a standard non-result Qualifier field, the free-text value
should be placed in the SUPP-- dataset described in 332HSection 8.4. When applicable, controlled terminology should be
used for SUPP-- field names (QNAM) and their associated labels (QLABEL) (see 333HSection 8.4 and 1865HAppendix C5).
For example, when a description of "Other Medically Important Serious Adverse Event" category is collected on a
CRF, the free text description should be stored in the SUPPAE dataset.
AESMIE=Y
SUPPAE QNAM=AESOSP, QLABEL= Other Medically Important SAE, QVAL=HIGH RISK FOR
ADDITIONAL THROMBOSIS
Another example is a CRF that collects reason for dose adjustment with additional free-text description:
Reason for Dose Adjustment (EXADJ)
Describe
Adverse event
_____________________
Insufficient response
_____________________
Non-medical reason
_____________________
The free text description should be stored in the SUPPEX dataset.
EXADJ=NONMEDICAL REASON
SUPPEX QNAM=EXADJDSC, QLABEL= Reason For Dose Adjustment, QVAL=PATIENT
MISUNDERSTOOD INSTRUCTIONS
Note that QNAM references the ―parent‖ variable name with the addition of ―OTH, ‖ one of the standard variable
naming fragments for ―Other‖ (see 1866HAppendix D). Likewise, the label is a modification of the parent variable label.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 32 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
When the CRF includes a list of values for a qualifier field that includes "Other" and the "Other" is supplemented
with a "Specify" free text field, then the manner in which the free text "Specify" value is submitted will vary based
on the sponsor's coding practice and analysis requirements. For example, consider a CRF that collects the
anatomical location of administration (EXLOC) of a study drug given as an injection:
Location of Injection
Right Arm Left Arm
Right Thigh Left Thigh
Other, Specify: _________________
An investigator has selected ―OTHER‖ and specified ―UPPER RIGHT ABDOMEN‖. Several options are available
for submission of this data:
1) If the sponsor wishes to maintain controlled terminology for the EXLOC field and limit the terminology to the 5
pre-specified choices, then the free text is placed in SUPPEX.
EXLOC
OTHER
QNAM
QLABEL
QVAL
EXLOCOTH
Other Location of Dose Administration
UPPER RIGHT ABDOMEN
2) If the sponsor wishes to maintain controlled terminology for EXLOC but will expand the terminology based on
values seen in the specify field, then the value of EXLOC will reflect the sponsor‘s coding decision and
SUPPEX could be used to store the verbatim text.
EXLOC
ABDOMEN
QNAM
QLABEL
QVAL
EXLOCOTH
Other Location of Dose Administration
UPPER RIGHT ABDOMEN
Note that the sponsor might choose a different value for EXLOC (e.g., UPPER ABDOMEN, TORSO)
depending on the sponsor's coding practice and analysis requirements.
3) If the sponsor does not require that controlled terminology be maintained and wishes for all responses to be
stored in a single variable, then EXLOC will be used and SUPPEX is not required.
EXLOC
UPPER RIGHT ABDOMEN
4.1.2.7.2 “SPECIFY” VALUES FOR RESULT QUALIFIER VARIABLES
When the CRF includes a list of values for a result field that includes "Other" and the "Other" is supplemented with
a "Specify" free text field, then the manner in which the free text "Specify" value is submitted will vary based on the
sponsor's coding practice and analysis requirements. For example, consider a CRF where the sponsor requests the
subject's eye color:
Eye Color
Brown Black
Blue Green
Other, specify: ________
An investigator has selected "OTHER" and specified "BLUEISH GRAY." As in the above discussion for non-result
Qualifier values, the sponsor has several options for submission:
1) If the sponsor wishes to maintain controlled terminology in the standard result field and limit the terminology to
the 5 pre-specified choices, then the free text is placed in --ORRES and the controlled terminology in
--STRESC.
SCTEST=Eye Color, SCORRES=BLUEISH GRAY, SCSTRESC=OTHER

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 33
Final November 12, 2008
2) If the sponsor wishes to maintain controlled terminology in the standard result field, but will expand the
terminology based on values seen in the specify field, then the free text is placed in --ORRES and the value of
--STRESC will reflect the sponsor's coding decision.
SCTEST=Eye Color, SCORRES=BLUEISH GRAY, SCSTRESC=GRAY
3) If the sponsor does not require that controlled terminology be maintained, the verbatim value will be copied to
--STRESC.
SCTEST=Eye Color, SCORRES=BLUEISH GRAY, SCSTRESC=BLUEISH GRAY
Note that rules for the use of ―Other, Specify‖ for the Result Qualifier variable, --OBJ, is discussed in 334HSection 6.4.3.
4.1.2.7.3 “SPECIFY” VALUES FOR TOPIC VARIABLES
Interventions: If a list of specific treatments is provided along with ―Other, Specify‖, --TRT should be populated with
the name of the treatment found in the specified text. If the sponsor wishes to distinguish between the pre-specified
list of treatments and those recorded under ―Other, Specify, ‖ the --PRESP variable could be used. For example:
Indicate which of the following concomitant medications was used to treat the subject‘s headaches:
Acetaminophen
Aspirin
Ibuprofen
Naproxen
Other: ______
If ibuprofen and diclofenac were reported, the CM dataset would include the following:
CMTRT=IBUPROFEN, CMPRESP=Y
CMTRT=DICLOFENAC, CMPRESP is null.
Events: ―Other, Specify‖ for Events may be handled similarly to Interventions. --TERM should be populated with
the description of the event found in the specified text and --PRESP could be used to distinguish between
pre-specified and free text responses.
Findings: ―Other, Specify‖ for tests may be handled similarly to Interventions. --TESTCD and --TEST should be
populated with the code and description of the test found in the specified text. If specific tests are not prespecified on
the CRF and the investigator has the option of writing free text for tests, then the name of the test would have to be
coded to ensure that all --TESTCD and --TEST values are controlled terminology and are not free text. For example,
a lab CRF has tests of Hemoglobin, Hematocrit and ―Other, specify‖. The value the investigator wrote for ―Other,
specify‖ is Prothrombin time with an associated result and units. The sponsor would submit the controlled
terminology for this test which is LBTESTCD = PT and LBTEST = Prothrombin Time.
4.1.2.8 MULTIPLE VALUES FOR A VARIABLE
4.1.2.8.1 MULTIPLE VALUES FOR AN INTERVENTION OR EVENT TOPIC VARIABLE
If multiple values are reported for a topic variable (i.e., --TRT in an Interventions general-observation-class dataset or
--TERM in an Events general-observation-class dataset), it is assumed that the sponsor will split the values into
multiple records or otherwise resolve the multiplicity as per the sponsor‘s standard data management procedures. For
example, if an adverse event term of ―Headache and Nausea‖ or a concomitant medication of ―Tylenol and Benadryl‖ is
reported, sponsors will often split the original report into separate records and/or query the site for clarification. By the
time of submission, the datasets should be in conformance with the record structures described in the SDTMIG. Note
that the Disposition dataset (DS) is an exception to the general rule of splitting multiple topic values into separate
records. For DS, one record for each disposition or protocol milestone is permitted according to the domain structure.
For cases of multiple reasons for discontinuation see 335HSection 6.2.2.1, Assumption 5 for additional information.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 34 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.2.8.2 MULTIPLE VALUES FOR A FINDINGS RESULT VARIABLE
If multiple result values (--ORRES) are reported for a test in a Findings class dataset, multiple records should be
submitted for that --TESTCD. Example:
EGTESTCD=RHYRATE, EGTEST=Rhythm and Rate, EGORRES=ATRIAL FIBRILLATION
EGTESTCD=RHYRATE, EGTEST=Rhythm and Rate, EGORRES=ATRIAL FLUTTER
Note that in this case, the sponsor‘s operational database may have a result-sequence variable as part of the natural key.
Some sponsors may elect to keep this variable in a Supplemental Qualifier record, while others may decide to use
--SPID or --SEQ to replace it. Dependent variables such as result Qualifiers should never be part of the natural key.
4.1.2.8.3 MULTIPLE VALUES FOR A NON-RESULT QUALIFIER VARIABLE
The SDTM permits one value for each Qualifier variable per record. If multiple values exist (e.g., due to a ―Check all that
apply‖ instruction on a CRF), then the value for the Qualifier variable should be ―MULTIPLE‖ and SUPP-- should be
used to store the individual responses. It is recommended that the SUPP-- QNAM value reference the corresponding
standard domain variable with an appended number or letter. In some cases, the standard variable name will be shortened
to meet the 8 character variable name requirement or it may be clearer to append a meaningful character string as shown in
the 2nd AE example below where the 1st 3 characters of the drug name are appended. Likewise the QLABEL value should
be similar to the standard label. The values stored in QVAL should be consistent with the controlled terminology
associated with the standard variable. See 336HSection 8.4 for additional guidance on maintaining appropriately unique QNAM
values. The following example includes selected variables from the ae.xpt and suppae.xpt datasets for a rash whose
locations are the face, neck, and chest.
AE Dataset
AETERM
AELOC
RASH
MULTIPLE
SUPPAE Dataset
QNAM
QLABEL
QVAL
AELOC1
Location of the Reaction 1
FACE
AELOC2
Location of the Reaction 2
NECK
AELOC3
Location of the Reaction 3
CHEST
In some cases, values for QNAM and QLABEL more specific than those above may be needed. For example, a
sponsor might conduct a study with two study drugs (e.g., open-label study of Abcicin + Xyzamin), and may require
the investigator assess causality and describe action taken for each drug for the rash:
AE Dataset
AETERM
AEREL
AEACN
RASH
MULTIPLE
MULTIPLE
SUPPAE Dataset
QNAM
QLABEL
QVAL
AERELABC
Causality of Abcicin
POSSIBLY RELATED
AERELXYZ
Causality of Xyzamin
UNLIKELY RELATED
AEACNABC
Action Taken with Abcicin
DOSE REDUCED
AEACNXYZ
Action Taken with Xyzamin
DOSE NOT CHANGED
In each of the above examples, the use of SUPPAE should be documented in the metadata and the annotated CRF.
The controlled terminology used should be documented as part of value-level metadata.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 35
Final November 12, 2008
If the sponsor has clearly documented that one response is of primary interest (e.g., in the CRF, protocol, or analysis
plan), the standard domain variable may be populated with the primary response and SUPP-- may be used to store
the secondary response(s). For example, if Abcicin is designated as the primary study drug in the example above:
AE Dataset
AETERM AEREL AEACN
RASH POSSIBLY RELATED DOSE REDUCED
SUPPAE Dataset
QNAM QLABEL QVAL
AERELX Causality of Xyzamin UNLIKELY RELATED
AEACNX Action Taken with Xyzamin DOSE NOT CHANGED
Note that in the latter case the label for standard variables AEREL and AEACN will have no indication that they
pertain to Abcicin. This association must be clearly documented in the metadata and annotated CRF.
4.1.3 CODING AND CONTROLLED TERMINOLOGY ASSUMPTIONS
PLEASE NOTE: Examples provided in the column “CDISC Notes” are only examples and not intended to imply
controlled terminology. Please check current controlled terminology at this link:
337H1561Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
4.1.3.1 TYPES OF CONTROLLED TERMINOLOGY
For SDTMIG V3.1.1 the presence of a single asterisk (*) or a double asterisk (**) in the “Controlled Terms or
Format” column indicated that a discrete set of values (controlled terminology) was expected for the variable. This
set of values was sponsor-defined in cases where standard vocabularies had not yet been defined (represented by *)
or from an external published source such as MedDRA (represented by **). For V3.1.2, controlled terminology is
now represented one of three ways:
• A single asterisk when there is no specific CT available at the current time, but the SDS Team expects
that sponsors may have their own CT and/or the CDISC Controlled Terminology Team may be
developing CT.
• A list of controlled terms for the variable when values are not yet maintained externally
• The name of an external codelist whose values can be found via the hyperlinks in either the domain or
Appendix C.
In addition, the “Controlled Terms or Format” column has been used to indicate a common format such as ISO
8601.
4.1.3.2 CONTROLLED TERMINOLOGY TEXT CASE
It is recommended that controlled terminology be submitted in upper case text for all cases other than those
described as exceptions below. Deviations to this rule should be described in the define.xml.
a. If the external reference for the controlled terminology is not in upper case then the data should
conform to the case prescribed in the external reference (e.g., MedDRA and LOINC).
b. Units, which are considered symbols rather than abbreviated text (e.g., mg/dL).
4.1.3.3 CONTROLLED TERMINOLOGY VALUES
The controlled terminology or a link to the controlled terminology should be included in the define.xml wherever
applicable. All values in the permissible value set for the study should be included, whether they are represented in
the submitted data or not. Note that a null value should not be included in the permissible value set. A null value is
implied for any list of controlled terms unless the variable is “Required” (see 338HSection 4.1.1.5).
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 36 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.3.4 USE OF CONTROLLED TERMINOLOGY AND ARBITRARY NUMBER CODES
Controlled terminology or decoded text should be used instead of arbitrary number codes in order to reduce
ambiguity for submission reviewers. For example, for concomitant medications, the verbatim term and/or dictionary
term should be presented, rather than numeric codes. Separate code values may be submitted as Supplemental
Qualifiers and may be necessary in analysis datasets.
4.1.3.5 STORING CONTROLLED TERMINOLOGY FOR SYNONYM QUALIFIER VARIABLES
For events such as AEs and Medical History, populate --DECOD with the dictionary‘s preferred term and
populate --BODSYS with the preferred body system name. If a dictionary is multi-axial, the value in
--BODSYS should represent the system organ class (SOC) used for the sponsor‘s analysis and summary
tables, which may not necessarily be the primary SOC.
For concomitant medications, populate CMDECOD with the drug's generic name and populate CMCLAS
with the drug class used for the sponsor‘s analysis and summary tables. If coding to multiple classes, follow
339Hassumption 4.1.2.8.1 or omit CMCLAS.
In either case, no other intermediate levels (e.g., MedDRA LLT, HLT, HLGT) or relationships should be stored in
the dataset. These may be provided in a Supplemental Qualifiers dataset (see 340HSection 8.4 and Appendix C5 for more
information). By knowing the dictionary and version used, the reviewer will be able to obtain intermediate levels in
a hierarchy (as in MedDRA), or a drug‘s ATC codes (as in WHO Drug). The sponsor is expected to provide the
dictionary name and version used to map the terms by utilizing the define.xml external codelist attributes.
4.1.3.6 STORING TOPIC VARIABLES FOR GENERAL DOMAIN MODELS
The topic variable for the Interventions and Events general-observation-class models is often stored as verbatim text.
For an Events domain, the topic variable is --TERM. For an Interventions domain, the topic variable is --TRT. For a
Findings domain, the topic variable, --TESTCD, should use Controlled Terminology (e.g., SYSBP for Systolic
Blood Pressure). If CDISC standard controlled terminology exists, it should be used; otherwise sponsors should
define their own controlled list of terms. If the verbatim topic variable in an Interventions or Event domain is
modified to facilitate coding, the modified text is stored in --MODIFY. In most cases (other than PE), the dictionary-
coded text is derived into --DECOD. Since the PEORRES variable is modified instead of the topic variable for PE,
the dictionary-derived text would be placed in PESTRESC. The variables used in each of the defined domains are:
Domain
Original Verbatim
Modified Verbatim
Standardized Value
AE
AETERM
AEMODIFY
AEDECOD
DS
DSTERM
DSDECOD
CM
CMTRT
CMMODIFY
CMDECOD
MH
MHTERM
MHMODIFY
MHDECOD
PE
PEORRES
PEMODIFY
PESTRESC
4.1.3.7 USE OF “YES” AND “NO” VALUES
Variables where the response is ―Yes‖ or ―No‖ (―Y‖ or ―N‖) should normally be populated for both ―Y‖ and ―N‖
responses. This eliminates confusion regarding whether a blank response indicates ―N‖ or is a missing value.
However, some variables are collected or derived in a manner that allows only one response, such as when a single
check box indicates ―Yes‖. In situations such as these, where it is unambiguous to only populate the response of
interest, it is permissible to only populate one value (―Y‖ or ―N‖) and leave the alternate value blank. An example of
when it would be acceptable to use only a value of ―Y‖ would be for Baseline Flag (--BLFL) variables, where ―N‖ is
not necessary to indicate that a value is not a baseline value.
Note: Permissible values for variables with controlled terms of ―Y‖ or ―N‖ may be extended to include ―U‖ or ―NA‖ if
it is the sponsor‘s practice to explicitly collect or derive values indicating ―Unknown‖ or ―Not Applicable‖ for that
variable.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 37
Final November 12, 2008
4.1.4 ACTUAL AND RELATIVE TIME ASSUMPTIONS
Timing variables (Table 2.2.5 of the SDTM) are an essential component of all SDTM subject-level domain datasets.
In general, all domains based on the three general observation classes should have at least one Timing variable. In
the Events or Interventions general observation class this could be the start date of the event or intervention. In the
Findings observation class where data are usually collected at multiple visits, at least one Timing variable must be
used.
The SDTMIG requires dates and times of day to be stored according to the international standard ISO 8601
(342Hhttp://www.iso.org). ISO 8601 provides a text-based representation of dates and/or times, intervals of time, and
durations of time.
4.1.4.1 FORMATS FOR DATE/TIME VARIABLES
An SDTM DTC variable may include data that is represented in ISO 8601 format as a complete date/time, a partial
date/time, or an incomplete date/time.
The SDTMIG template uses ISO 8601 for calendar dates and times of day, which are expressed as follows:
ο YYYY-MM-DDThh:mm:ss
where:
ο [YYYY] = four-digit year
ο [MM] = two-digit representation of the month (01-12, 01=January, etc.)
ο [DD] = two-digit day of the month (01 through 31)
ο [T] = (time designator) indicates time information follows
ο [hh] = two digits of hour (00 through 23) (am/pm is NOT allowed)
ο [mm] = two digits of minute (00 through 59)
ο [ss] = two digits of second (00 through 59)
Other characters defined for use within the ISO 8601 standard are:
ο [-] (hyphen): to separate the time Elements "year" from "month" and "month" from "day" and to represent
missing date components.
ο [:] (colon): to separate the time Elements "hour" from "minute" and "minute" from "second"
ο [/] (solidus): to separate components in the representation of date/time intervals
ο [P] (duration designator): precedes the components that represent the duration
ο NOTE: Spaces are not allowed in any ISO 8601 representations
Key aspects of the ISO 8601 standard are as follows:
• ISO 8601 represents dates as a text string using the notation YYYY-MM-DD.
• ISO 8601 represents times as a text string using the notation hh:mm:ss.
• The SDTM and SDTMIG require use of the ISO 8601 Extended format, which requires hyphen delimiters
for date components and colon delimiters for time components. The ISO 8601 basic format, which does not
require delimiters, should not be used in SDTM datasets.
• When a date is stored with a time in the same variable (as a date/time), the date is written in front of the
time and the time is preceded with “T” using the notation YYYY-MM-DDThh:mm:ss
(e.g. 2001-12-26T00:00:01).
Implementation of the ISO 8601 standard means that date/time variables are character/text data types. The SDS
fragment employed for date/time character variables is DTC.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 38 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.4.2 DATE/TIME PRECISION
The concept of representing date/time precision is handled through use of the ISO 8601 standard. According to ISO
8601, precision (also referred to by ISO 8601 as "completeness" or "representations with reduced accuracy") can be
inferred from the presence or absence of components in the date and/or time values. Missing components are
represented by right truncation or a hyphen (for intermediate components that are missing). If the date and time
values are completely missing the SDTM date field should be null. Every component except year is represented as
two digits. Years are represented as four digits; for all other components, one-digit numbers are always padded with
a leading zero.
The table below provides examples of ISO 8601 representation complete date and truncated date/time values using
ISO 8601 "appropriate right truncations" of incomplete date/time representations. Note that if no time component is
represented, the [T] time designator (in addition to the missing time) must be omitted in ISO 8601 representation.
Date and Time as
Originally Recorded
Precision
ISO 8601 Date/Time
1
December 15, 2003 13:14:17
Complete date/time
2003-12-15T13:14:17
2
December 15, 2003 13:14
Unknown seconds
2003-12-15T13:14
3
December 15, 2003 13
Unknown minutes and seconds
2003-12-15T13
4
December 15, 2003
Unknown time
2003-12-15
5
December, 2003
Unknown day and time
2003-12
6
2003
Unknown month, day, and time
2003
This date and date/time model also provides for imprecise or estimated dates, such as those commonly seen in
Medical History. To represent these intervals while applying the ISO 8601 standard, it is recommended that the
sponsor concatenate the date/time values (using the most complete representation of the date/time known) that
describe the beginning and the end of the interval of uncertainty and separate them with a solidus as shown in the
table below:
Interval of Uncertainty
ISO 8601 Date/Time
1
Between 10:00 and 10:30 on the Morning of December 15, 2003
2003-12-15T10:00/2003-12-15T10:30
2
Between the first of this year (2003) until "now" (February 15, 2003)
2003-01-01/2003-02-15
3
Between the first and the tenth of December, 2003
2003-12-01/2003-12-10
4
Sometime in the first half of 2003
2003-01-01/2003-06-30
Other uncertainty intervals may be represented by the omission of components of the date when these components
are unknown or missing. As mentioned above, ISO 8601 represents missing intermediate components through the
use of a hyphen where the missing component would normally be represented. This may be used in addition to
"appropriate right truncations" for incomplete date/time representations. When components are omitted, the
expected delimiters must still be kept in place and only a single hyphen is to be used to indicate an omitted
component. Examples of this method of omitted component representation are shown in the table below:
Date and Time as Originally
Recorded
Level of Uncertainty
ISO 8601 Date/Time
1
December 15, 2003 13:15:17
Complete date
2003-12-15T13:15:17
2
December 15, 2003 ??:15
Unknown hour with known minutes
2003-12-15T-:15
3
December 15, 2003 13:??:17
Unknown minutes with known date, hours,
and seconds
2003-12-15T13:-:17
4
The 15th of some month in 2003, time
not collected
Unknown month and time with known year
and day
2003---15
5
December 15, but can't remember the
year, time not collected
Unknown year with known month and day
--12-15
6
7:15 of some unknown date
Unknown date with known hour and
minute
-----T07:15

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 39
Final November 12, 2008
Note that Row 6 above where a time is reported with no date information represents a very unusual situation. Since
most data is collected as part of a visit, when only a time appears on a CRF, it is expected that the date of the visit
would usually be used as the date of collection.
Using a character-based data type to implement the ISO 8601 date/time standard will ensure that the date/time
information will be machine and human readable without the need for further manipulation, and will be platform
and software independent.
4.1.4.3 INTERVALS OF TIME AND USE OF DURATION FOR --DUR VARIABLES
4.1.4.3.1 INTERVALS OF TIME AND USE OF DURATION FOR --DUR VARIABLES
As defined by ISO 8601, an interval of time is the part of a time axis, limited by two time "instants" such as the
times represented in SDTM by the variables --STDTC and --ENDTC. These variables represent the two instants that
bound an interval of time, while the duration is the quantity of time that is equal to the difference between these time
points.
ISO 8601 allows an interval to be represented in multiple ways. One representation, shown below, uses two dates in
the format:
YYYY-MM-DDThh:mm:ss/YYYY-MM-DDThh:mm:ss
While the above would represent the interval (by providing the start date/time and end date/time to "bound" the
interval of time), it does not provide the value of the duration (the quantity of time).
Duration is frequently used during a review; however, the duration timing variable (--DUR) should generally be
used in a domain if it was collected in lieu of a start date/time (--STDTC) and end date/time (--ENDTC). If both
--STDTC and --ENDTC are collected, durations can be calculated by the difference in these two values, and need
not be in the submission dataset.
Both duration and duration units can be provided in the single --DUR variable, in accordance with the ISO 8601
standard. The values provided in --DUR should follow one of the following ISO 8601 duration formats:
PnYnMnDTnHnMnS or PnW
where:
[P] (duration designator): precedes the alphanumeric text string that represents the duration. NOTE: The
use of the character P is based on the historical use of the term "period" for duration.
[n] represents a positive -number or zero
[W] is used as week designator, preceding a data Element that represents the number of calendar weeks
within the calendar year (e.g., P6W represents 6 weeks of calendar time).
The letter "P" must precede other values in the ISO 8601 representation of duration. The ―n‖ preceding each letter
represents the number of Years, Months, Days, Hours, Minutes, Seconds, or the number of Weeks. As with the
date/time format, ―T‖ is used to separate the date components from time components.
Note that weeks cannot be mixed with any other date/time components such as days or months in duration expressions.
As is the case with the date/time representation in --DTC, --STDTC, or --ENDTC only the components of duration
that are known or collected need to be represented. Also, as is the case with the date/time representation, if no time
component is represented, the [T] time designator (in addition to the missing time) must be omitted in ISO 8601
representation.
ISO 8601 also allows that the "lowest-order components" of duration being represented may be represented in
decimal format. This may be useful if data are collected in formats such as "one and one-half years", "two and one-
half weeks", "one-half a week" or "one quarter of an hour" and the sponsor wishes to represent this "precision" (or
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 40 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
lack of precision) in ISO 8601 representation. Remember that this is ONLY allowed in the lowest-order (right-most)
component in any duration representation.
The table below provides some examples of ISO-8601-compliant representations of durations:
Duration as originally recorded
ISO 8601 Duration
2 Years
P2Y
10 weeks
P10W
3 Months 14 days
P3M14D
3 Days
P3D
6 Months 17 Days 3 Hours
P6M17DT3H
14 Days 7 Hours 57 Minutes
P14DT7H57M
42 Minutes 18 Seconds
PT42M18S
One-half hour
PT0.5H
5 Days 12¼ Hours
P5DT12.25H
4 ½ Weeks
P4.5W
Note that a leading zero is required with decimal values less than one.
4.1.4.3.2 INTERVAL WITH UNCERTAINTY
When an interval of time is an amount of time (duration) following an event whose start date/time is recorded (with
some level of precision, i.e. when one knows the start date/time and the duration following the start date/time), the
correct ISO 8601 usage to represent this interval is as follows:
YYYY-MM-DDThh:mm:ss/PnYnMnDTnHnMnS
where the start date/time is represented before the solidus [/], the "Pn…", following the solidus, represents a
―duration‖, and the entire representation is known as an ―interval‖. NOTE: This is the recommended representation
of elapsed time, given a start date/time and the duration elapsed.
When an interval of time is an amount of time (duration) measured prior to an event whose start date/time is
recorded (with some level of precision, i.e. where one knows the end date/time and the duration preceding that end
date/time), the syntax is:
PnYnMnDTnHnMnS/YYYY-MM-DDThh:mm:ss
where the duration, "Pn…", is represented before the solidus [/], the end date/time is represented following the
solidus, and the entire representation is known as an ―interval‖.
4.1.4.4 USE OF THE “STUDY DAY” VARIABLES
The permissible Study Day variables (--DY, --STDY, and --ENDY) describe the relative day of the observation
starting with the reference date as Day 1. They are determined by comparing the date portion of the respective
date/time variables (--DTC, --STDTC, and --ENDTC) to the date portion of the Subject Reference Start Date
(RFSTDTC from the Demographics domain).
The Subject Reference Start Date (RFSTDTC) is designated as Study Day 1. The Study Day value is incremented by
1 for each date following RFSTDTC. Dates prior to RFSTDTC are decremented by 1, with the date preceding
RFSTDTC designated as Study Day -1 (there is no Study Day 0). This algorithm for determining Study Day is
consistent with how people typically describe sequential days relative to a fixed reference point, but creates
problems if used for mathematical calculations because it does not allow for a Day 0. As such, Study Day is not
suited for use in subsequent numerical computations, such as calculating duration. The raw date values should be
used rather than Study Day in those calculations.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 41
Final November 12, 2008
All Study Day values are integers. Thus, to calculate Study Day:
--DY = (date portion of --DTC) - (date portion of RFSTDTC) + 1 if --DTC is on or after RFSTDTC
--DY = (date portion of --DTC) - (date portion of RFSTDTC) if --DTC precedes RFSTDTC
This algorithm should be used across all domains.
4.1.4.5 CLINICAL ENCOUNTERS AND VISITS
All domains based on the three general observation classes should have at least one timing variable. For domains in
the Events or Interventions observations classes, and for domains in the Findings observation class for which data
are collected only once during the study, the most appropriate timing variable may be a date (e.g., --DTC, --STDTC)
or some other timing variable. For studies that are designed with a prospectively defined schedule of visit-based
activities, domains for data that are to be collected more than once per subject (e.g., Labs, ECG, Vital Signs) are
expected to include VISITNUM as a timing variable.
Clinical encounters are described by the CDISC Visit variables. For planned visits, values of VISIT, VISITNUM,
and VISITDY must be those defined in the Trial Visits dataset, see 343HSection 7.4. For planned visits:
Values of VISITNUM are used for sorting and should, wherever possible, match the planned chronological
order of visits. Occasionally, a protocol will define a planned visit whose timing is unpredictable (e.g., one
planned in response to an adverse event, a threshold test value, or a disease event), and completely
chronological values of VISITNUM may not be possible in such a case.
There should be a one-to-one relationship between values of VISIT and VISITNUM.
For visits that may last more than one calendar day, VISITDY should be the planned day of the start of the visit.
Sponsor practices for populating visit variables for unplanned visits may vary across sponsors.
VISITNUM should generally be populated, even for unplanned visits, as it is expected in many Findings
domains, as described above. The easiest method of populating VISITNUM for unplanned visits is to assign
the same value (e.g., 99) to all unplanned visits, but this method provides no differentiation between the
unplanned visits and does not provide chronological sorting. Methods that provide a one-to-one relationship
between visits and values of VISITNUM, that are consistent across domains, and that assign VISITNUM
values that sort chronologically require more work and must be applied after all of a subject's unplanned visits
are known.
VISIT may be left null or may be populated with a generic value (e.g., "Unscheduled") for all unplanned
visits, or individual values may be assigned to different unplanned visits.
VISITDY should not be populated for unplanned visits, since VISITDY is, by definition, the planned study
day of visit, and since the actual study day of an unplanned visit belongs in a --DY variable.
The following table shows an example of how the visit identifiers might be used for lab data:
USUBJID
VISIT
VISITNUM
VISITDY
LBDY
001
Week 1
2
7
7
001
Week 2
3
14
13
001
Week 2 Unscheduled
3.1
17
4.1.4.6 REPRESENTING ADDITIONAL STUDY DAYS
The SDTM allows for --DTC values to be represented as study days (--DY) relative to the RFSTDTC reference start
date variable in the DM dataset, as described above in 344HSection 4.1.4.4. The calculation of additional study days
within subdivisions of time in a clinical trial may be based on one or more sponsor-defined reference dates not
represented by RFSTDTC. In such cases, the Sponsor may define Supplemental Qualifier variables and the
define.xml should reflect the reference dates used to calculate such study days. If the sponsor wishes to define ―day
within element‖ or ―day within epoch,‖ the reference date/time will be an element start date/time in the Subject
Elements dataset (345HSection 5.3.1).

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 42 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.4.7 USE OF RELATIVE TIMING VARIABLES
--STRF and --ENRF
The variables --STRF and --ENRF represent the timing of an observation relative to the sponsor-defined reference period
when information such as "BEFORE‖, ―PRIOR‖,‖ONGOING‖', or ―CONTINUING‖ is collected in lieu of a date and this
collected information is in relation to the sponsor-defined reference period. The sponsor-defined reference period is the
continuous period of time defined by the discrete starting point (RFSTDTC) and the discrete ending point (RFENDTC) for
each subject in the Demographics dataset.
--STRF is used to identify the start of an observation relative to the sponsor-defined reference period.
--ENRF is used to identify the end of an observation relative to the sponsor-defined reference period.
Allowable values for --STRF and --ENRF are ―BEFORE‖, ―DURING‖, ―DURING/AFTER‖, ―AFTER‖, and ―U‖ (for
unknown).
As an example, a CRF checkbox that identifies concomitant medication use that began prior to the study treatment
period would translate into CMSTRF = ―BEFORE‖ if selected. Note that in this example, the information collected
is with respect to the start of the concomitant medication use only and therefore the collected data corresponds to
variable CMSTRF, not CMENRF. Note also that the information collected is relative to the study treatment period,
which meets the definition of CMSTRF.
Some sponsors may wish to derive --STRF and --ENRF for analysis or reporting purposes even when dates are
collected. Sponsors are cautioned that doing so in conjunction with directly collecting or mapping data such as
―BEFORE‖, ―PRIOR", etc. to --STRF and --ENRF will blur the distinction between collected and derived values
within the domain. Sponsors wishing to do such derivations are instead encouraged to use supplemental variables or
analysis datasets for this derived data.
In general, sponsors are cautioned that representing information using variables --STRF and --ENRF may not be as
precise as other methods, particularly because information is often collected relative to a point in time or to a period
of time other than the one defined as the study reference period. SDTMIG V3.1.2 has attempted to address these
limitations by the addition of four new relative timing variables, which are described in the following paragraph.
Sponsors should use the set of variables that allows for accurate representation of the collected data. In many cases,
this will mean using these new relative timing variables in place of --STRF and --ENRF.
--STRTPT, --STTPT, --ENRTPT, and --ENTPT
While the variables --STRF and --ENRF are useful in the case when relative timing assessments are made coincident
with the start and end of the study reference period, these may not be suitable for expressing relative timing
assessments such as ―Prior‖ or ―Ongoing‖ that are collected at other times of the study. As a result, four new timing
variables have been added in V3.1.2 to express a similar concept at any point in time. The variables --STRTPT and
--ENRTPT contain values similar to --STRF and --ENRF, but may be anchored with any timing description or
date/time value expressed in the respective --STTPT and --ENTPT variables, and not be limited to the study reference
period. Unlike the variables --STRF and --ENRF, which for all domains are defined relative to one study reference
period, the timing variables --STRTPT, --STTPT, --ENRTPT, and --ENTPT are defined to be unique within a domain
only. Allowable values for --STRTPT and --ENRTPT are as follows:
If the reference time point corresponds to the date of collection or assessment:
Start values: an observation can start BEFORE that time point, can start COINCIDENT with that time
point, or it is unknown (U) when it started
End values: an observation can end BEFORE that time point, can end COINCIDENT with that time point, can
be known that it didn‘t end but was ONGOING, or it is unknown (U) at all when it ended or if it was ongoing.
AFTER is not a valid value in this case because it would represent an event after the date of collection.
If the reference time point is prior to the date of collection or assessment:
Start values: an observation can start BEFORE the reference point, can start COINCIDENT with the
reference point, can start AFTER the reference point, or it may not be known (U) when it started
End values: an observation can end BEFORE the reference point, can end COINCIDENT with the
reference point, can end AFTER the reference point, can be known that it didn‘t end but was ONGOING, or
it is unknown (U) when it ended or if it was ongoing.
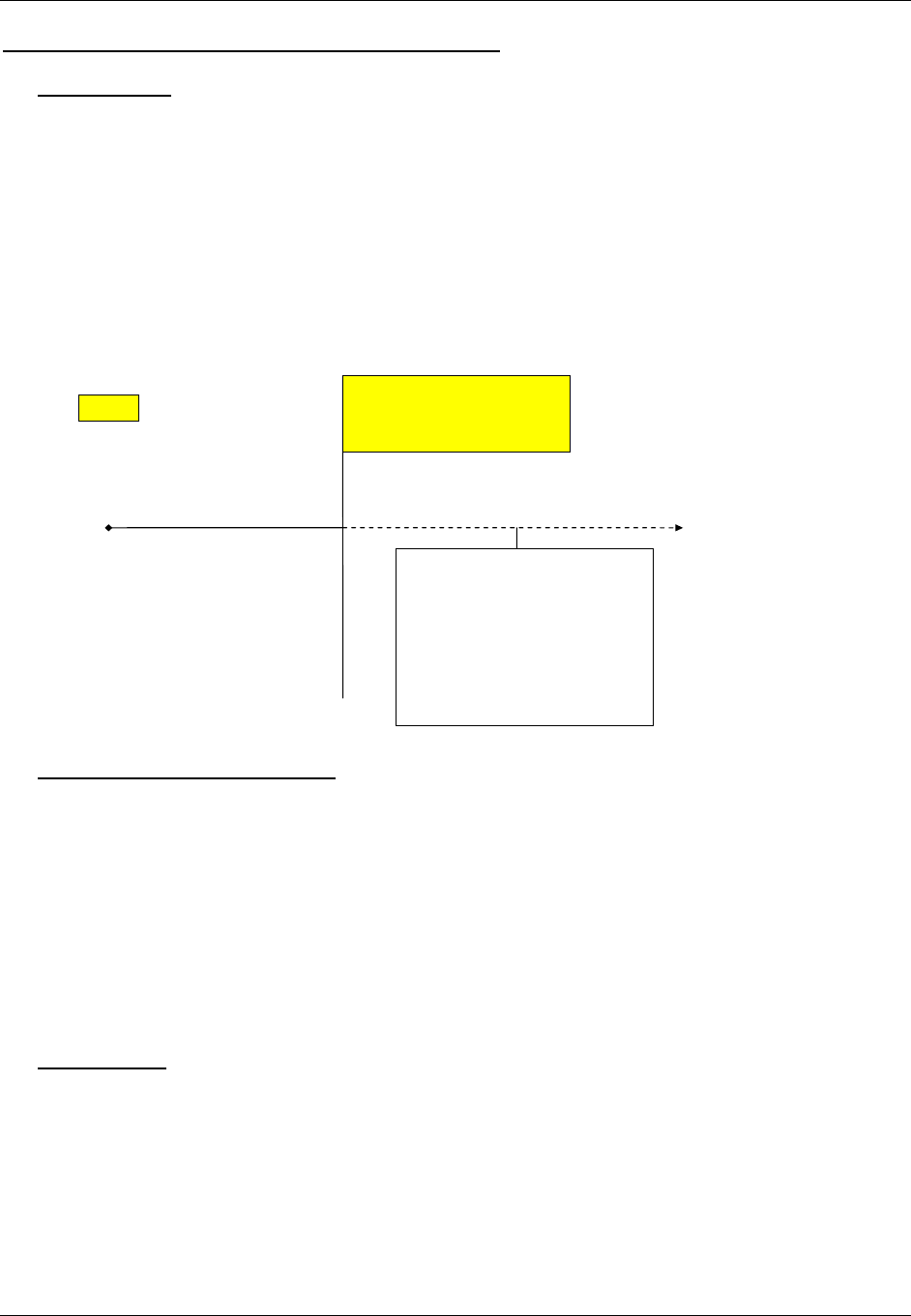
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 43
Final November 12, 2008
Examples of --STRTPT, --STTPT, --ENRTPT, and --ENTPT
1. Medical History
Assumptions:
CRF contains "Year Started" and check box for "Active"
"Date of Assessment" is collected
Example when "Active" is checked:
MHDTC = date of assessment value, ex. "2006-11-02"
MHSTDTC = year of condition start, e.g., "2002"
MHENRTPT = "ONGOING"
MHENTPT = date of assessment value, e.g., "2006-11-02"
Figure 4.1.4.7 Example of --ENRTPT and --ENTPT for Medical History
MHENTPT
Assessment Date and
Reference Time Point of
2006-11-02
MHDTC = 2006-11-02
MHSTDTC = 2002
MHENRTPT = ONGOING
MHENTPT = 2006-11-02
Medical event began in 2002 and
was ongoing at the reference time
point of 2006-11-02. The medical
event may or may not have ended
at any time after that.
2002
Prior and Concomitant Medications
Assumptions:
CRF contains "Start Date", "Stop Date", and check boxes for "Prior" if unknown or uncollected Start Date, and
"Continuing" if no Stop Date was collected. Prior refers to screening visit and Continuing refers to final study visit.
Example when both "Prior" and "Continuing" are checked:
CMSTDTC = [null]
CMENDTC = [null]
CMSTRTPT = "BEFORE"
CMSTTPT = screening date, e.g., "2006-10-21"
CMENRTPT = "ONGOING"
CMENTPT = final study visit date, e.g., "2006-11-02"
2. Adverse Events
Assumptions:
CRF contains "Start Date", "Stop Date", and "Outcome" with check boxes including "Continuing" and
"Unknown" (Continuing and Unknown are asked at the end of the subject's study participation)
No assessment date or visit information is collected
Example when "Unknown" is checked:
AESTDTC = start date, e.g., "2006-10-01"
AEENDTC = [null]
AEENRTPT = "U"
AEENTPT = final subject contact date, e.g., "2006-11-02"
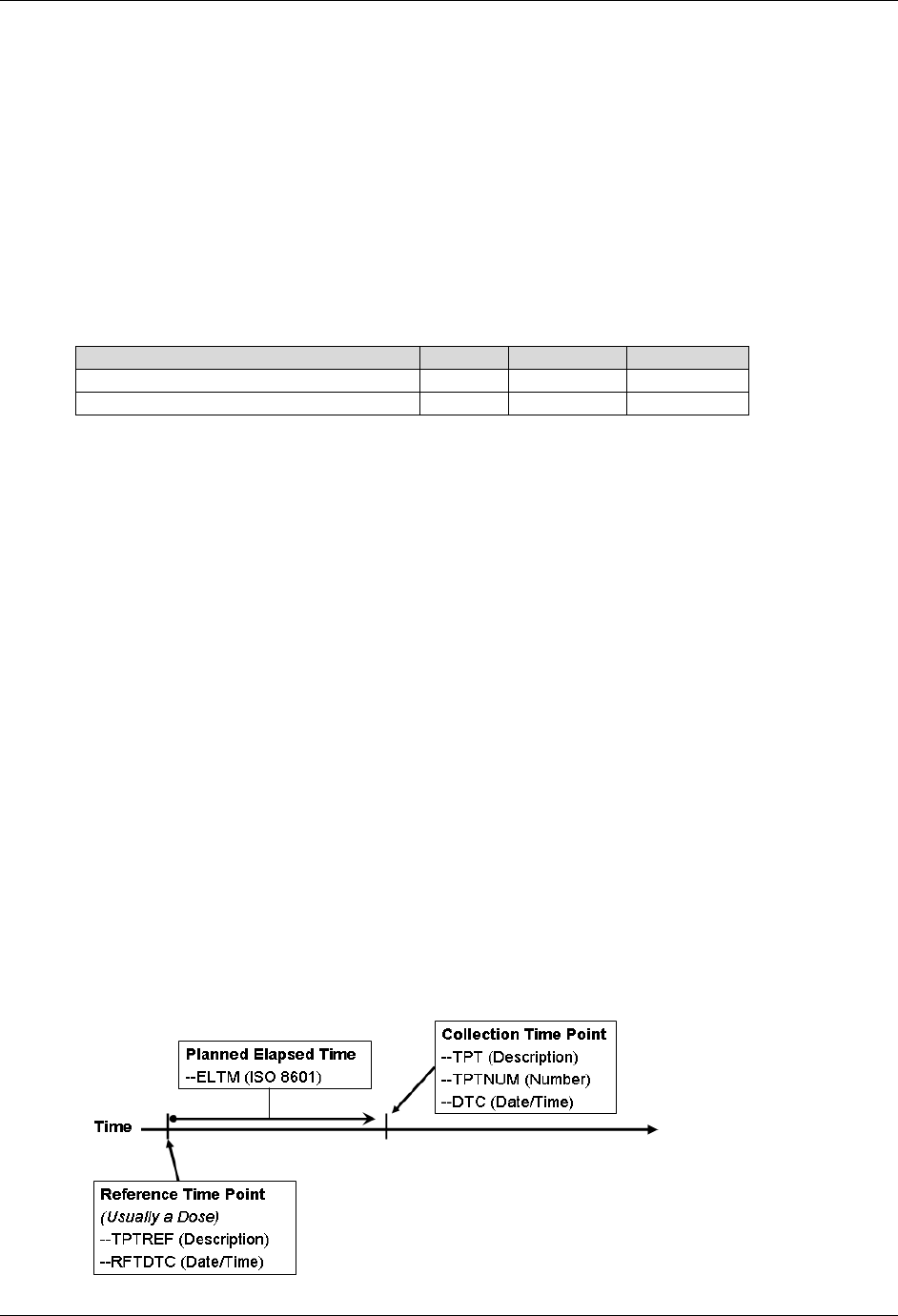
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 44 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.4.8 DATE AND TIME REPORTED IN A DOMAIN BASED ON FINDINGS
When the date/time of collection is reported in any domain, the date/time should go into the --DTC field (e.g., EGDTC for
Date/Time of ECG). For any domain based on the Findings general observation class, such as lab tests which are based on a
specimen, the collection date is likely to be tied to when the specimen or source of the finding was captured, not necessarily
when the data was recorded. In order to ensure that the critical timing information is always represented in the same variable,
the --DTC variable is used to represent the time of specimen collection. For example, in the LB domain the LBDTC variable
would be used for all single-point blood collections or spot urine collections. For timed lab collections (e.g., 24-hour urine
collections) the LBDTC variable would be used for the start date/time of the collection and LBENDTC for the end date/time
of the collection. This approach will allow the single-point and interval collections to use the same date/time variables
consistently across all datasets for the Findings general observation class. The table below illustrates the proper use of these
variables. Note that --STDTC is not used for collection dates over an interval, so is blank in the following table.
Collection Type
--DTC
--STDTC
--ENDTC
Single-Point Collection
X
Interval Collection
X
X
4.1.4.9 USE OF DATES AS RESULT VARIABLES
Dates are generally used only as timing variables to describe the timing of an event, intervention, or collection activity, but there may
be occasions when it may be preferable to model a date as a result (--ORRES) in a Findings dataset. Note that using a date as a result
to a Findings question is unusual and atypical, and should be approached with caution, but this situation may occasionally occur when
a) a group of questions (each of which has a date response) is asked and analyzed together; or b) the Event(s) and Intervention(s) in
question are not medically significant (often the case when included in questionnaires). Consider the following cases:
Calculated due date
Date of last day on the job
Date of high school graduation
One approach to modeling these data would be to place the text of the question in --TEST and the response to the
question, a date represented in ISO 8601 format, in --ORRES and --STRESC as long as these date results do not
contain the dates of medically significant events or interventions.
Again, use extreme caution when storing dates as the results of Findings. Remember, in most cases, these dates
should be timing variables associated with a record in an Intervention or Events dataset.
4.1.4.10 REPRESENTING TIME POINTS
Time points can be represented using the time point variables, --TPT, --TPTNUM, --ELTM, and the time point
anchors, --TPTREF (text description) and --RFTDTC (the date/time). Note that time-point data will usually have an
associated --DTC value. The interrelationship of these variables is shown in Figure 4.1.4.10 below.
Figure 4.1.4.10

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 45
Final November 12, 2008
Values for these variables for Vital Signs measurements taken at 30, 60, and 90 minutes after dosing would look like
the following.
VSTPTNUM
VSTPT
VSELTM
VSTPTREF
VSRFTDTC
VSDTC
1
30 MIN
PT30M
DOSE ADMINISTRATION
2006-08-01T08:00
2006-08-01T08:30
2
60 MIN
PT1H
DOSE ADMINISTRATION
2006-08-01T08:00
2006-08-01T09:01
3
90 MIN
PT1H30M
DOSE ADMINISTRATION
2006-08-01T08:00
2006-08-01T09:32
Note that the actual elapsed time is not an SDTM variable, but can be derived by an algorithm representing VSDTC-VSRFTDTC.
Values for these variables for Urine Collections taken pre-dose, and from 0-12 hours and 12-24 hours after dosing
would look like the following.
LBTPTNUM
LBTPT
LBELTM
LBTPTREF
LBRFTDTC
LBDTC
1
15 MIN PRE-DOSE
-PT15M
DOSE ADMINISTRATION
2006-08-01T08:00
2006-08-01T08:30
2
0-12 HOURS
PT12H
DOSE ADMINISTRATION
2006-08-01T08:00
2006-08-01T20:35
3
12-24 HOURS
PT24H
DOSE ADMINISTRATION
2006-08-01T08:00
2006-08-02T08:40
Note that the value in LBELTM represents the end of the interval at which the collection ends.
When time points are used, --TPTNUM is expected. Time points may or may not have an associated --TPTREF.
Sometimes, --TPTNUM may be used as a key for multiple values collected for the same test within a visit; as such,
there is no dependence upon an anchor such as --TPTREF, but there will be a dependency upon the VISITNUM. In
such cases, VISITNUM will be required to confer uniqueness to values of --TPTNUM.
If the protocol describes the scheduling of a dose using a reference intervention or assessment, then --TPTREF
should be populated, even if it does not contribute to uniqueness. The fact that time points are related to a reference
time point, and what that reference time point is, are important for interpreting the data collected at the time point.
Not all time points will require all three variables to provide uniqueness. In fact, in some cases a time point may be
uniquely identified without the use of VISIT, or without the use of --TPTREF, or, rarely, without the use of either
one. For instance:
A trial might have time points only within one visit, so that the contribution of VISITNUM to uniqueness is
trivial.
A trial might have time points that do not relate to any visit, such as time points relative to a dose of drug
self-administered by the subject at home.
A trial may have only one reference time point per visit, and all reference time points may be similar, so that
only one value of --TPTREF (e.g., "DOSE") is needed.
A trial may have time points not related to a reference time point. For instance, --TPTNUM values could be
used to distinguish first, second, and third repeats of a measurement scheduled without any relationship to
dosing.
For trials with many time points, the requirement to provide uniqueness using only VISITNUM, --TPTREF, and
--TPTNUM may lead to a scheme where multiple natural keys are combined into the values of one of these variables.
For instance, in a crossover trial with multiple doses on multiple days within each period, either of the following
options could be used. VISITNUM might be used to designate period, --TPTREF might be used to designate the day
and the dose, and --TPTNUM might be used to designate the timing relative to the reference time point. Alternatively,
VISITNUM might be used to designate period and day within period, --TPTREF might be used to designate the dose
within the day, and --TPTNUM might be used to designate the timing relative to the reference time point.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 46 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Option 1
VISIT VISITNUM --TPT --TPTNUM --TPTREF
PERIOD 1 3 PRE-DOSE 1 DAY 1, AM DOSE
1H 2
4H 3
PRE-DOSE 1 DAY 1, PM DOSE
1H 2
4H 3
PRE-DOSE 1 DAY 5, AM DOSE
1H 2
4H 3
PRE-DOSE 1 DAY 5, PM DOSE
1H 2
4H 3
PERIOD 2 4 PRE-DOSE 1 DAY 1, AM DOSE
1H 2
4H 3
PRE-DOSE 1 DAY 1, PM DOSE
1H 2
4H 3
Option 2
VISIT VISITNUM --TPT --TPTNUM --TPTREF
PERIOD 1, DAY 1 3 PRE-DOSE 1 AM DOSE
1H 2
4H 3
PRE-DOSE 1
PM DOSE
1H 2
4H 3
PERIOD 1, DAY 5 4 PRE-DOSE 1 AM DOSE
1H 2
4H 3
PRE-DOSE 1
PM DOSE
1H 2
4H 3
PERIOD 2, DAY 1 5 PRE-DOSE 1 AM DOSE
1H 2
4H 3
PRE-DOSE 1
PM DOSE
1H 2
4H 3
Within the context that defines uniqueness for a time point, which may include domain, visit, and reference time
point, there must be a one-to-relationship between values of --TPT and --TPTNUM. In other words, if domain, visit,
and reference time point uniquely identify subject data, then if two subjects have records with the same values of
DOMAIN, VISITNUM, --TPTREF, and --TPTNUM, then these records may not have different time point
descriptions in --TPT.
Within the context that defines uniqueness for a time point, there is likely to be a one-to-one relationship between
most values of --TPT and --ELTM. However, since --ELTM can only be populated with ISO 8601 periods of time
(as described in 346HSection 4.1.4.3), --ELTM may not be populated for all time points. For example, --ELTM is likely to
be null for time points described by text such as "pre-dose" or "before breakfast." When --ELTM is populated, if two
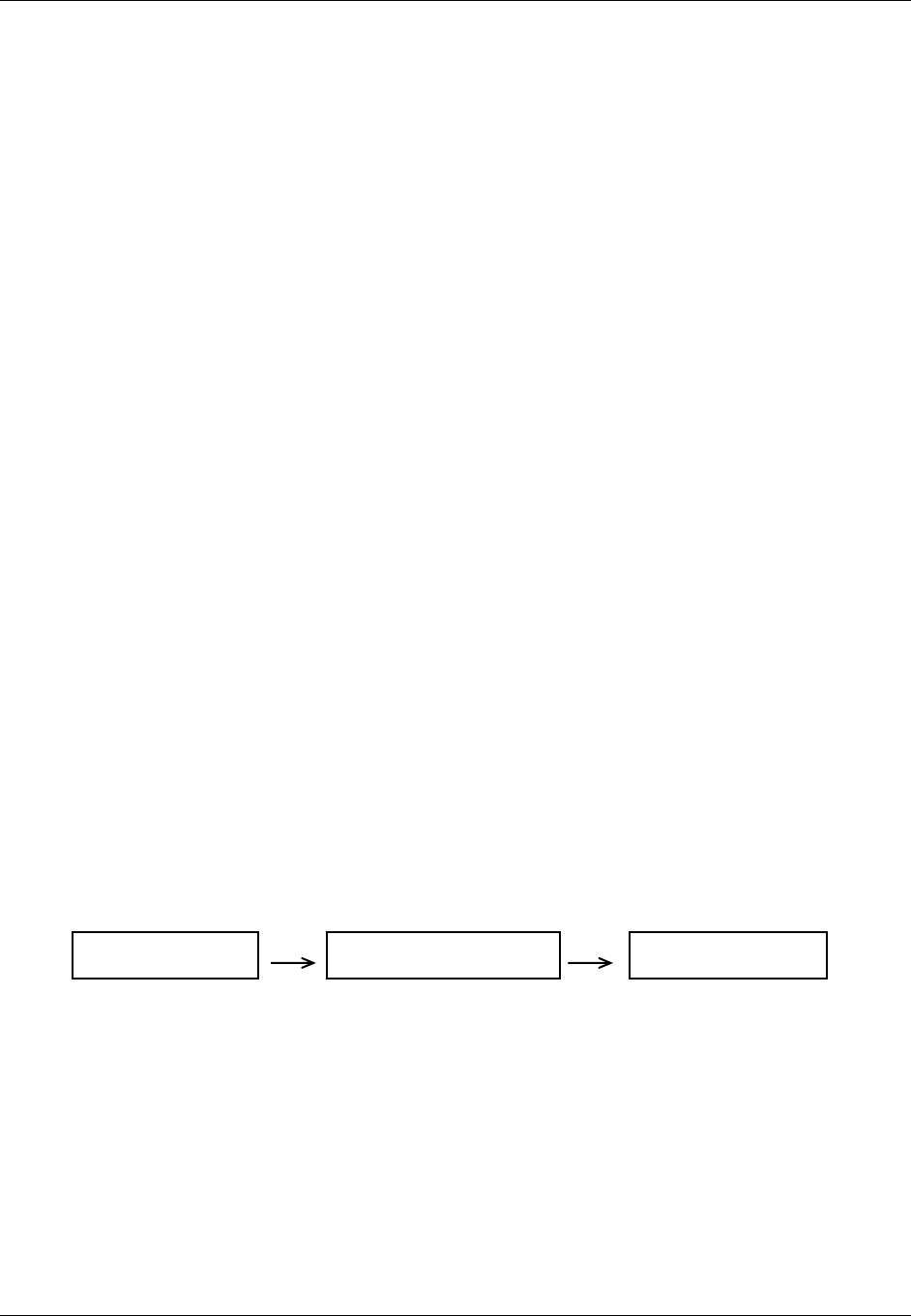
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 47
Final November 12, 2008
subjects have records with the same values of DOMAIN, VISITNUM, --TPTREF, and --TPTNUM, then these
records may not have different values in --ELTM.
When the protocol describes a time point with text such as "4-6 hours after dose" or "12 hours +/- 2 hours after
dose" the sponsor may choose whether and how to populate --ELTM. For example, a time point described as "4-6
hours after dose" might be associated with an --ELTM value of PT4H. A time point described as "12 hours +/- 2
hours after dose" might be associated with an --ELTM value of PT12H. Conventions for populating --ELTM should
be consistent (the examples just given would probably not both be used in the same trial). It would be good practice
to indicate the range of intended timings by some convention in the values used to populate --TPT.
Sponsors may, of course, use more stringent requirements for populating --TPTNUM, --TPT, and --ELTM. For
instance, a sponsor could decide that all time points with a particular --ELTM value would have the same values of
--TPTNUM and --TPT, across all visits, reference time points, and domains.
4.1.5 OTHER ASSUMPTIONS
4.1.5.1 ORIGINAL AND STANDARDIZED RESULTS OF FINDINGS AND TESTS NOT DONE
4.1.5.1.1 ORIGINAL AND STANDARDIZED RESULTS
The --ORRES variable contains the result of the measurement or finding as originally received or collected.
--ORRES is an expected variable and should always be populated, with two exceptions:
When --STAT = ―NOT DONE‖
--ORRES should generally not be populated for derived records
Derived records are flagged with the --DRVFL variable. When the derived record comes from more than one visit,
the sponsor must define the value for VISITNUM, addressing the correct temporal sequence. If a new record is
derived for a dataset, and the source is not eDT, then that new record should be flagged as derived. For example in
ECG data, if QTc Intervals are derived in-house by the sponsor, then the derived flag is set to ―Y‖. If the QTc
Intervals are received from a vendor the derived flag is not populated.
When --ORRES is populated, --STRESC must also be populated, regardless of whether the data values are character
or numeric. The variable, --STRESC, is derived either by the conversion of values in --ORRES to values with
standard units, or by the assignment of the value of --ORRES (as in the PE Domain, where --STRESC could contain
a dictionary-derived term). A further step is necessary when --STRESC contains numeric values. These are
converted to numeric type and written to --STRESN. Because --STRESC may contain a mixture of numeric and
character values, --STRESN may contain null values, as shown in the flowchart below.
--ORRES
(all original values)
--STRESC
(derive or copy all results)
--STRESN
(numeric results only)
When the original measurement or finding is a selection from a defined codelist, in general, the --ORRES and
--STRESC variables contain results in decoded format, that is, the textual interpretation of whichever code was
selected from the codelist. In some cases where the code values in the codelist are statistically meaningful
standardized values or scores, which are defined by sponsors or by valid methodologies such as SF36
questionnaires, the --ORRES variables will contain the decoded format, whereas, the --STRESC variables as well as
the --STRESN variables will contain the standardized values or scores.
Occasionally data that are intended to be numeric are collected with characters attached that cause the character-to-numeric
conversion to fail. For example, numeric cell counts in the source data may be specified with a greater than (>) or less than (<)
sign attached (e.g. >10, 000 or <1). In these cases the value with the greater than (>) or less than (<) sign attached should be
moved to the --STRESC variable, and --STRESN should be null. The rules for modifying the value for analysis purposes
should be defined in the analysis plan and only changed in the ADaM datasets. If the value in --STRESC has different units,
the greater than (>) or less than (<) sign should be maintained. An example is included in 347H Section 4.1.5.1.3, Rows 11 and 12.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 48 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
4.1.5.1.2 TESTS NOT DONE
When an entire examination (Laboratory draw, ECG, Vital Signs, or Physical Examination), or a group of tests
(hematology or urinalysis), or an individual test (glucose, PR interval, blood pressure, or hearing) is not done, and
this information is explicitly captured on the CRF with a yes/no or done/not done question, this information should
be presented in the dataset. The reason for the missing information may or may not have been collected. A sponsor
has two options; one is to submit individual records for each test not done or to submit one record for a group of
tests that were not done. See the examples below for submitting groups of tests not done.
If the data on the CRF is missing and yes/no or done/not done was not explicitly captured a record should not be
created to indicate that the data was not collected.
If a group of tests were not done:
--TESTCD should be --ALL
--TEST should be <Name of the Module>
--CAT should be <Name of Group of Tests>
--ORRES should be null
--STAT should be ―NOT DONE‖
--REASND, if collected, might be ‖Specimen lost‖
For example, if urinalysis is not done then:
LBTESTCD should be ―LBALL‖
LBTEST should be ―Labs Data‖
LBCAT should be "URINALYSIS"
LBORRES should be NULL
LBSTAT should be ―NOT DONE‖
LBREASND, if collected, might be ―Subject could not void‖
4.1.5.1.3 EXAMPLES OF ORIGINAL AND STANDARD UNITS AND TEST NOT DONE
The following examples are meant to illustrate the use of Findings results variables, and are not meant as
comprehensive domain examples. Certain required and expected variables are omitted, and the samples may
represent data for more than one subject.
Lab Data Examples
Numeric values that have been converted (Row 1) or copied (Row 3).
A character result that has been copied (Row 2).
A result of ―TRACE‖ shows ―TRACE‖ in LBSTRESC and LBSTRESN is null (Row 4).
Value of 1+ in LBORRES, 1+ in LBSTRESC and LBSTRESN is null (Row 5).
A result of ―BLQ‖ was collected. That value was copied to LBSTRESC and LBSTRESN is null. Note that
the standard units are populated by sponsor decision, but could be left null. (Row 6).
A result is missing because the observation was ―NOT DONE‖, as reflected in the --STAT variable; neither
LBORRES nor LBSTRESC are populated (Row 7).
A result is derived from multiple records such as an average of baseline measurements for a baseline value,
so LBDRVFL = Y (Row 8). Note that the original collected data are not shown in this example.
None of the scheduled tests were completed as planned (Row 9).
A category of tests was not completed as planned (Row 10).
Shows when LBSTRESC has been standardized and the less than (<) sign has been maintained (Row 11).
Shows when LBSTRESC has been standardized and the less than (<) sign has been maintained (Row 12).
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 49
Final November 12, 2008
Row
LBTESTCD
LBCAT
LBORRES
LBORRESU
LBSTRESC
LBSTRESN
LBSTRESU
LBSTAT
LBDRVFL
1
GLUC
CHEMISTRY
6.0
mg/dL
60.0
60.0
mg/L
2
BACT
URINALYSIS
MODERATE
MODERATE
3
ALT
CHEMISTRY
12.1
mg/L
12.1
12.1
mg/L
4
RBC
URINALYSIS
TRACE
TRACE
5
WBC
URINALYSIS
1+
1+
6
KETONES
CHEMISTRY
BLQ
mg/L
BLQ
mg/L
7
HCT
HEMATOLOGY
NOT DONE
8
MCHC
HEMATOLOGY
33.8
33.8
g/dL
Y
9
LBALL
NOT DONE
10
LBALL
HEMATOLOGY
NOT DONE
11
WBC
HEMATOLOGY
<4, 000
/mm3
<4,000
/mm3
12
BILI
CHEMISTRY
<0.1
mg/dL
<1.71
umol/L
The SDS Team realizes that for rows 4, 5, and 6, this change is not backward compatible, but the example has been
modified to reflect harmonization with ADaM and comments received during the review period. The changes are
directed at decreasing the amount of sponsor subjectivity in converting original results to standard results.
ECG Examples:
Numeric and character values that have been converted (Rows 2 and 3) or copied (Rows 1 and 4).
A result is missing because the test was ―NOT DONE‖, as reflected in the EGSTAT variable; neither
EGORRES nor EGSTRESC is populated (Row 5).
The overall interpretation is included as a new record (Row 6)
The entire ECG was not done (Row 7)
Row
EGTESTCD
EGORRES
EGORRESU
EGSTRESC
EGSTRESN
EGSTRESU
EGSTAT
EGDRVFL
1
QRSDUR
0.362
sec
0.362
0.362
sec
2
QTMEAN
221
msec
.221
.221
sec
3
QTCB
412
msec
.412
.412
sec
4
RHYMRATE
ATRIAL FLUTTER
ATRIAL FLUTTER
5
PRMEAN
NOT DONE
6
INTP
ABNORMAL
ABNORMAL
7
EGALL
NOT DONE
Vital Signs Example:
Numeric values that have converted (Rows 1 and 2).
A result is missing because the Vital Signs test was ―NOT DONE‖, as reflected in the VSSTAT variable;
neither VSORRES nor VSSTRESC is populated (Row 3).
The result is derived by having multiple records for one measurement (Rows 4 and 5), and the derived
value is recorded in a new row with the derived record flagged. (Row 6).
The entire examination was not done (Row 7).
Row
VSTESTCD
VSORRES
VSORRESU
VSSTRESC
VSSTRESN
VSSTRESU
VSSTAT
VSDRVFL
1
HEIGHT
60
IN
152
152
cm
2
WEIGHT
110
LB
50
50
kg
3
HR
NOT DONE
4
SYSBP
96
mmHg
96
96
mmHg
5
SYSBP
100
mmHg
100
100
mmHg
6
SYSBP
98
98
mmHg
Y
7
VSALL
NOT DONE
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 50 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Questionnaire Example:
Note that this is for a standard instrument for which no subjectivity is involved in representing the original result as
a numeric value.
A Character value that has been converted to a standard score (Rows 1, 5, and 6).
A result is derived from multiple records (Row 2). The records for the original collected results are not shown in
this example.
A result is missing because the observation was ―NOT DONE‖, as reflected in the QSSTAT variable;
neither QSORRES nor QSSTRESC is populated (Row 3).
The entire questionnaire was not done (Row 4).
Shows when a summary score in Row 7 is derived from the data in Rows 5 and 6 and QSORRES should not
be populated because the character values cannot be added to give a meaningful result (Rows 5, 6, and 7).
Row
QSTESTCD
QSTEST
QSORRES
QSSTRESC
QSSTRESN
QSSTAT
QSDRVFL
1
QS1
Health
VERY GOOD
4.4
4.4
2
QS2
Health Perceptions (0-100)
82
82
Y
3
QS1
Health
NOT DONE
4
QSALL
Questionnaire
NOT DONE
5
QSP10
Healthy As Anyone
MOSTLY TRUE
4
4
6
QSP11
Expect Health To Get Better
DEFINITELY TRUE
5
5
7
QSPSUM
Total of Scores
9
9
Y
4.1.5.2 LINKING OF MULTIPLE OBSERVATIONS
See 348HSection 8 for guidance on expressing relationships among multiple observations.
4.1.5.3 TEXT STRINGS THAT EXCEED THE MAXIMUM LENGTH FOR GENERAL-
OBSERVATION-CLASS DOMAIN VARIABLES
4.1.5.3.1 TEST NAME (--TEST) GREATER THAN 40 CHARACTERS
Sponsors may have test descriptions (--TEST) longer than 40 characters in their operational database. Since the
--TEST variable is meant to serve as a label for a --TESTCD when a Findings dataset is transposed to a more
horizontal format, the length of --TEST is normally limited to 40 characters to conform to the limitations of the SAS
V5 Transport format currently used for submission datasets. Therefore, sponsors have the choice to either insert the
first 40 characters or a text string abbreviated to 40 characters in --TEST. Sponsors should include the full
description for these variables in the study metadata in one of two ways:
If the annotated CRF contains the full text, provide a link to the annotated CRF page containing the full test
description in the define.xml Origin column for --TEST.
If the annotated CRF does not specify the full text, then create a pdf document to store full-text descriptions.
In the define.xml Comments column for --TEST insert a link to the full test description in the pdf.
The convention above should also be applied to the Qualifier Value Label (QLABEL) in Supplemental Qualifiers
(SUPP--) datasets. IETEST values in IE and TI are exceptions to the above 40-character rule and are limited to 200
characters since they are not expected to be transformed to a column labels. Values of IETEST that exceed 200
characters should be described in study metadata as per the convention above. For further details see IE domain
349HSection 6.3.2.1 Assumption 4 and TI domain 350HSection 7.5.2 Assumption 5.
4.1.5.3.2 TEXT STRINGS> 200 CHARACTERS IN OTHER VARIABLES
Some sponsors may collect data values longer than 200 characters for some variables. Because of the current
requirement for Version 5 SAS transport file format, it will not be possible to store those long text strings using only
one variable. Therefore, the SDTMIG has defined a convention for storing a long text string by using a combination
of the standard domain dataset and the Supplemental Qualifiers (SUPP--) datasets, which applies to all domains
based on a general observation class. Note that the Comments domain is not based on a general observation class
and has different rules. See 351HSection 5.2 for information on handling comment text more than 200 characters long.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 51
Final November 12, 2008
The first 200 characters of text should be stored in the standard domain variable and each additional 200 characters
of text should be stored as a record in the SUPP-- dataset (see 352HSection 8.4). In this dataset, the value for QNAM
should contain a sequential variable name, which is formed by appending a one-digit integer, beginning with 1, to
the original standard domain variable name. When splitting a text string into several records, the text should be split
between words to improve readability.
As an example, if there was a verbatim response for a Medical History Reported Term (MHTERM) of 500 characters
in length, the sponsor would put the first 200 characters of text in the standard domain variable and dataset (MHTERM
in MH), the next 200 characters of text as a first supplemental record in the SUPPMH dataset, and the final 100
characters of text as a second record in the SUPPMH dataset (see Example 1 below). Variable QNAM would have the
values MHTERM1 and MHTERM2 for these two records in SUPPMH, respectively, for this one particular text string.
Sponsors should place the text itself into variable QVAL and the label of the original standard domain variable into
variable QLABEL. In this case, IDVAR and IDVARVAL should be used in SUPPMH to relate the associated
supplemental text records to the parent record containing the first 200 characters of text in the standard domain.
In cases where the standard domain variable name is already 8 characters in length, sponsors should replace the last
character with a digit when creating values for QNAM. As an example, for Other Action Taken in Adverse Events
(AEACNOTH), values for QNAM for the SUPPAE records would have the values AEACNOT1, AEACNOT2, and so on.
Example 1: MHTERM with 500 characters.
suppmh.xpt
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
12345
MH
99-123
MHSEQ
6
MHTERM1
Reported Term
for the Medical
History
2nd 200
chars of text
CRF
12345
MH
99-123
MHSEQ
6
MHTERM2
Reported Term
for the Medical
History
last 100
chars of text
CRF
Example 2: AEACN with 400 characters.
suppae.xpt
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
12345
AE
99-123
AESEQ
4
AEACNOT1
Other Action
Taken
2nd 200
chars of text
CRF
The only exceptions to the above rules are Comments (CO) and TS (Trial Summary). Please see 353Hsection 5.2.1.1 for
Comments and 354HSection 7.6.1 for Trial Summary. NOTE: Only the Comments (CO) and Trial Summary (TS)
domains are allowed to add variables for the purpose of handling text exceeding 200 characters. All other domains
must use SUPPQUAL variables as noted in the examples above.
4.1.5.4 EVALUATORS IN THE INTERVENTIONS AND EVENTS OBSERVATION CLASSES
The observations recorded in the Findings class include the --EVAL qualifier because the observation may originate
from more than one source (e.g., an Investigator or Central Reviewer). For the Interventions and Events observation
classes, which do not include the --EVAL variable, all data are assumed to be attributed to the Principal Investigator.
The QEVAL variable can be used to describe the evaluator for any data item in a SUPP-- dataset (355HSection 8.4.1), but
is not required when the data are objective. For observations that have primary and supplemental evaluations of
specific qualifier variables, sponsors should put data from the primary evaluation into the standard domain dataset
and data from the supplemental evaluation into the Supplemental Qualifier datasets (SUPP--). Within each SUPP--
record, the value for QNAM should be formed by appending a ―1‖ to the corresponding standard domain variable
name. In cases where the standard domain variable name is already eight characters in length, sponsors should
replace the last character with a ―1‖ (incremented for each additional attribution). The following is an example of
how to represent the case where an adjudication committee evaluates an adverse event in SUPPAE. See 356HSection 8.4
for additional details on how to use SUPP--.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 52 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Note that QNAM takes on the value AERELNS1, as the corresponding standard domain variable AERELNST is
already eight characters in length. The adverse event data as determined by the primary investigator would reside in
the standard AE dataset.
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
12345
AE
99-123
AESEQ
3
AESEV1
Severity/
Intensity
MILD
CRF
ADJUDICATION
COMMITTEE
12345
AE
99-123
AESEQ
3
AEREL1
Causality
POSSIBLY
RELATED
CRF
ADJUDICATION
COMMITTEE
12345
AE
99-123
AESEQ
3
AERELNS1
Relationship
to Non-Study
Treatment
Possibly
related to
aspirin use
CRF
ADJUDICATION
COMMITTEE
4.1.5.5 CLINICAL SIGNIFICANCE FOR FINDINGS OBSERVATION CLASS DATA
For assessments of clinical significance when the overall interpretation is a record in the domain, use Supplemental
Qualifier (SUPP--) record (with QNAM = --CLSIG) linked to the record that contains the overall interpretation or a
particular result. An example would be a QNAM value of EGCLSIG in SUPPEG with a value of ―Y‖, indicating
that an ECG result of ATRIAL FIBRILLATION was clinically significant.
Separate from clinical significance are results of NORMAL or ABNORMAL, or lab values which are out of range.
Examples of the latter include the following:
An ECG test with EGTESTCD=INTP addresses the ECG as a whole should have a result or of NORMAL
or ABNORMAL. A record for EGTESTCD=INTP may also have a record in SUPPEG indicating whether
the result is clinically significant.
A record for a vital signs measurement (e.g., systolic blood pressure) or a lab test (e.g., hematocrit) that
contains a measurement may have a normal range and a normal range indicator. It could also have a
SUPP-- record indicating whether the result was clinically significant.
4.1.5.6 SUPPLEMENTAL REASON VARIABLES
The SDTM general observation classes include the --REASND variable to submit the reason an observation was not
collected. However, sponsors sometimes collect the reason that something was done. For the Interventions general
observation class, --INDC and --ADJ are available to indicate the reason for the intervention or for the dose adjustment.
For the Findings general observation class, if the sponsor collects the reason for performing a test or examination, it
should be placed in the SUPP-- dataset as described in 357HSection 8.4.1. The standard SUPP-- QNAM value of --REAS
should be used as described in 1867HAppendix C5. If multiple reasons are reported, refer to 358HSection 4.1.2.8.3.
For example, if the sponsor collects the reason that extra lab tests were done, the SUPP-- record might be populated
as follows:
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
12345
LB
99-123
LBSEQ
3
LBREAS
Reason Test or Examination
was Performed
ORIGINAL
SAMPLE LOST
CRF
4.1.5.7 PRESENCE OR ABSENCE OF PRE-SPECIFIED INTERVENTIONS AND EVENTS
Interventions (e.g., concomitant medications) and Events (e.g., medical history) can generally be collected in two
different ways, by recording either verbatim free text or the responses to a pre-specified list of treatments or terms.
Since the method of solicitation for information on treatments and terms may affect the frequency at which they are
reported, whether they were pre-specified may be of interest to reviewers. The --PRESP variable is used to indicate
whether a specific intervention (--TRT) or event (--TERM) was solicited. The --PRESP variable has controlled
terminology of Y (for ―Yes‖) or a null value. It is a permissible variable, and should only be used when the topic
variable values come from a pre-specified list. Questions such as ―Did the subject have any concomitant
medications?‖ or ―Did the subject have any medical history?‖ should not have records in SDTM domain because 1)
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 53
Final November 12, 2008
these are not valid values for the respective topic variables of CMTRT and MHTERM, and 2) records whose sole
purpose is to indicate whether or not a subject had records are not meaningful.
The --OCCUR variable is used to indicate whether a pre-specified intervention or event occurred or did not occur. It
has controlled terminology of Y and N (for ―Yes‖ and ―No‖). It is a permissible variable and may be omitted from
the dataset if no topic-variable values were pre-specified.
If a study collects both pre-specified interventions and events as well as free-text events and interventions, the value
of --OCCUR should be ―Y‖ or ―N‖ for all pre-specified interventions and events, and null for those reported as free-
text.
The --STAT and --REASND variables can be used to provide information about pre-specified interventions and
events for which there is no response (e.g., investigator forgot to ask). As in Findings, --STAT has controlled
terminology of NOT DONE.
Situation
Value of
--PRESP
Value of
--OCCUR
Value of
--STAT
Spontaneously reported event occurred
Pre-specified event occurred
Y
Y
Pre-specified event did not occur
Y
N
Pre-specified event has no response
Y
NOT DONE
Refer to the standard domains in the Events and Interventions General Observation Classes for additional
assumptions and examples.
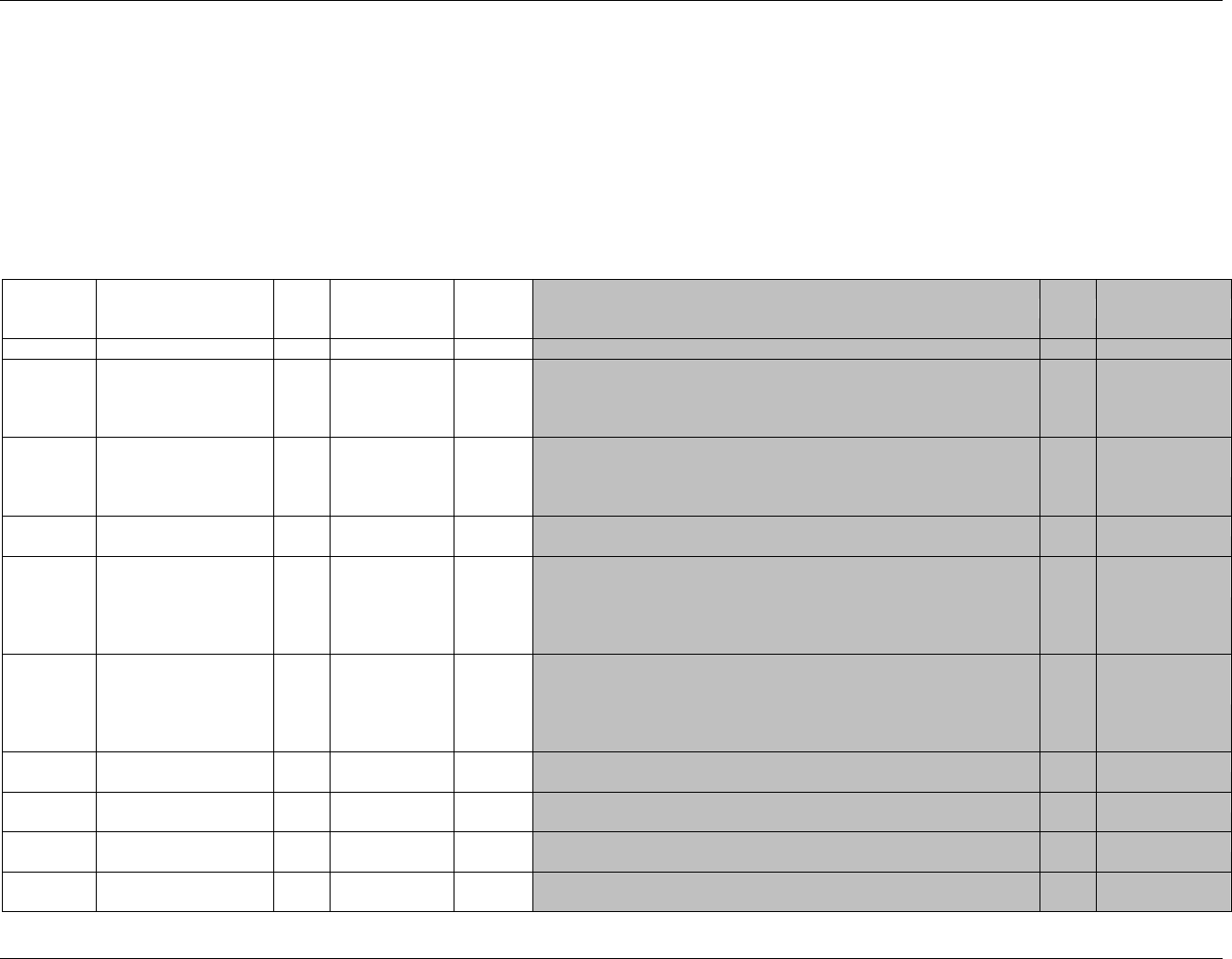
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 54 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
5 Models for Special-Purpose Domains
5.1 DEMOGRAPHICS
5.1.1 55BDEMOGRAPHICS — DM
dm.xpt, Demographics — Version 3.1.2. One record per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1868HDM
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
359H360HSDTMIG 4.1.2.2,
361HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product. This must be a unique
number, and could be a compound identifier formed by concatenating
STUDYID-SITEID-SUBJID.
Req
SDTM 2.2.4,
362H363HSDTMIG 4.1.2.3
SUBJID
Subject Identifier for the
Study
Char
Topic
Subject identifier, which must be unique within the study. Often the ID of
the subject as recorded on a CRF.
Req
RFSTDTC
Subject Reference Start
Date/Time
Char
ISO 8601
Record
Qualifier
Reference Start Date/time for the subject in ISO 8601 character format.
Usually equivalent to date/time when subject was first exposed to study
treatment. Required for all randomized subjects; will be null for all
subjects who did not meet the milestone the date requires, such as screen
failures or unassigned subjects.
Exp
364H
SDTM 2.2.5,
375HSDTMIG 4.1.4.1 508H
RFENDTC
Subject Reference End
Date/Time
Char
ISO 8601
Record
Qualifier
Reference End Date/time for the subject in ISO 8601 character format.
Usually equivalent to the date/time when subject was determined to have
ended the trial, and often equivalent to date/time of last exposure to study
treatment. Required for all randomized subjects; null for screen failures or
unassigned subjects.
Exp
365H
SDTM 2.2.5,
375HSDTMIG 4.1.4.1
SITEID
Study Site Identifier
Char
Record
Qualifier
Unique identifier for a site within a study.
Req
INVID
Investigator Identifier
Char
Record
Qualifier
An identifier to describe the Investigator for the study. May be used in
addition to SITEID. Not needed if SITEID is equivalent to INVID.
Perm
INVNAM
Investigator Name
Char
Synonym
Qualifier
Name of the investigator for a site.
Perm
BRTHDTC
Date/Time of Birth
Char
ISO 8601
Record
Qualifier
Date/time of birth of the subject.
Perm
366HSDTM 2.2.5,
375HSDTMIG 4.1.4.1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 55
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
AGE
Age
Num
Record
Qualifier
Age expressed in AGEU. May be derived from RFSTDTC and
BRTHDTC, but BRTHDTC may not be available in all cases (due to
subject privacy concerns).
Exp
AGEU
Age Units
Char
(1869HAGEU)
Variable
Qualifier
Units associated with AGE.
Exp
SEX
Sex
Char
(367HSEX)
Record
Qualifier
Sex of the subject.
Req
RACE
Race
Char
(368HRACE)
Record
Qualifier
Race of the subject. Sponsors should refer to ―Collection of Race and
Ethnicity Data in Clinical Trials‖ (FDA, September 2005) for guidance
regarding the collection of race
(369Hhttp://www.fda.gov/cder/guidance/5656fnl.htm) See Assumption below
regarding RACE.
Exp
ETHNIC
Ethnicity
Char
(1870HETHNIC)
Record
Qualifier
The ethnicity of the subject. Sponsors should refer to ―Collection of Race
and Ethnicity Data in Clinical Trials‖ (FDA, September 2005) for
guidance regarding the collection of ethnicity
(370Hhttp://www.fda.gov/cder/guidance/5656fnl.htm).
Perm
ARMCD
Planned Arm Code
Char
*
Record
Qualifier
ARMCD is limited to 20 characters and does not have special character
restrictions. The maximum length of ARMCD is longer than for other
―short‖ variables to accommodate the kind of values that are likely to be
needed for crossover trials. For example, if ARMCD values for a seven-
period crossover were constructed using two-character abbreviations for
each treatment and separating hyphens, the length of ARMCD values
would be 20.
Req
371HSDTMIG 4.1.2.1
ARM
Description of Planned
Arm
Char
*
Synonym
Qualifier
Name of the Arm to which the subject was assigned.
Req
372HSDTMIG 4.1.2.1,
373HSDTMIG 4.1.2.4
COUNTRY
Country
Char
(1871HCOUNTRY)
ISO 3166
Record
Qualifier
Country of the investigational site in which the subject participated in the
trial.
Req
DMDTC
Date/Time of Collection
Char
ISO 8601
Timing
Date/time of demographic data collection.
Perm
SDTM 2.2.5,
374HSDTMIG 4.1.4.1
DMDY
Study Day of Collection
Num
Timing
Study day of collection measured as integer days.
Perm
SDTM 2.2.5,
375HSDTMIG 4.1.4.1
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 56 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
5.1.1.1 ASSUMPTIONS FOR DEMOGRAPHICS DOMAIN MODEL
1. Investigator and site identification: Companies use different methods to distinguish sites and investigators. CDISC assumes that SITEID will always be
present, with INVID and INVNAM used as necessary. This should be done consistently and the meaning of the variable made clear in the define.xml.
2. Every subject in a study must have a subject identifier (SUBJID). In some cases a subject may participate in more than one study. To identify a subject
uniquely across all studies for all applications or submissions involving the product, a unique identifier (USUBJID) must be included in all datasets.
Subjects occasionally change sites during the course of a clinical trial. The sponsor must decide how to populate variables such as USUBJID, SUBJID
and SITEID based on their operational and analysis needs, but only one DM record should be submitted for the subject. The Supplemental Qualifiers
dataset may be used if appropriate to provide additional information.
3. Concerns for subject privacy suggest caution regarding the collection of variables like BRTHDTC. This variable is included in the Demographics model
in the event that a sponsor intends to submit it; however, sponsors should follow regulatory guidelines and guidance as appropriate.
4. The values of ARM and ARMCD in DM must match entries in the Trial Arms (TA) dataset, except for subjects who were not fully assigned to an Arm.
Subjects who did not receive the treatments to which they were assigned will still have the values of ARM and ARMCD to which they were assigned.
SE/DM Examples 1 and 2 in 376Hsection 5.3.1.2 show examples of subjects whose actual treatment did not match their planned treatment.
Some subjects may leave the trial before they can be assigned to an Arm, or, in the case of trials where Arm is assigned by two or more successive
allocation processes, may leave before the last of these processes. Such subjects will not be assigned to one of the planned Arms described in the Trial
Arms dataset, and must have special values of ARM and ARMCD assigned.
Data for screen failure subjects, if submitted, should be included in the Demographics dataset, with ARMCD = ―SCRNFAIL" and ARM = ―Screen
Failure‖. Sponsors may include a record in the Disposition dataset indicating when the screen failure event occurred. DM/SE Example 6 shows an
example of data submitted for a screen failure subject.
Some trial designs include Elements after screening but before Arm assignments are made, and so may have subjects who are not screen failures, but are
not assigned to an Arm. Subjects withdrawn from a trial before assignment to an Arm, if they are not screen failures, should have ARMCD =
"NOTASSGN" and ARM = "Not Assigned". Example 1872HTrial 1 in 377HSection 7.2.3.1, which includes a screening Epoch and a run-in Epoch before
randomization, is an example of such a trial; data for a subject who passed screening but was not randomized in this trial are shown in DM/SE Example 6.
In trials where Arm assignment is done by means of two or more allocation processes at separate points in time, subjects who drop out after the first
allocation process but before the last allocation process, should be assigned values of ARMCD that reflect only the allocation processes they
underwent. Example 1873HTrial 3, 378HSection 7.2.3.3, is such a trial. DM/SE Example 7 shows sample data for subjects in this trial.
5. When study population flags are included in SDTM, they are treated as Supplemental Qualifiers (see 379HSection 8.4) to DM and placed in the SUPPDM
dataset. Controlled terms for these subject-level population flags, (e.g., COMPLT, SAFETY, ITT and PPROT) are listed in Appendix C5. See ICH E9 for
more information and definitions. Note that the ADaM subject-level analysis dataset (ADSL) includes population flags; consult the ADaM
Implementation Guide for more information about these variables.
6. Submission of multiple race responses should be represented in the Demographics domain and Supplemental Qualifiers (SUPPDM) dataset as described
in 380Hassumption 4.1.2.8.3, Multiple Responses for a Non-Result Qualifier. If multiple races are collected then the value of RACE should be ―MULTIPLE‖
and the additional information will be included in the Supplemental Qualifiers dataset. Controlled terminology for RACE should be used in both DM and
SUPPDM so that consistent values are available for summaries regardless of whether the data are found in a column or row. If multiple races were
collected and one was designated as primary, RACE in DM should be the primary race and additional races should be reported in SUPPDM. When
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 57
Final November 12, 2008
additional free text information is reported about subject's RACE using ―Other, Specify‖, Sponsors should refer to 381HSection 4.1.2.7.1. If the race was
collected via an ―Other, Specify‖ field and the sponsor chooses not to map the value as described in the current FDA guidance (see CDISC Notes for RACE) then the
value of RACE should be ―OTHER‖. If a subject refuses to provide race information, the value of RACE could be ―UNKNOWN‖. Examples are provided below in
382HSection 5.1.1.2.
7. RFSTDTC, RFENDTC, and BRTHDTC represent date/time values, but they are considered to have a Record Qualifier role in DM. They are not
considered to be Timing Variables because they are not intended for use in the general observation classes.
8. Additional Permissible Identifier, Qualifier and Timing Variables
Only the following Timing variables are permissible and may be added as appropriate: VISITNUM, VISIT, VISITDY. The Record Qualifier DMXFN
(External File Name) is the only additional variable that may be added, which is adopted from the Findings general observation class, may also be used to
refer to an external file, such as a patient narrative.
5.1.1.2 EXAMPLES FOR DEMOGRAPHICS DOMAIN MODEL
Examples of using the DM domain for typical scenarios are provided below. Example 1 displays the all Required and Expected variables; in examples 2 - 6,
certain Required or Expected variables have been omitted in consideration of space and clarity. Example 1 is a general Demographics example showing typical
data recorded for a clinical trial. Examples 2 through 5 display various scenarios for representing race and ethnicity information. Example 6 shows the handling
of ARMCD for Subjects Withdrawn before Assignment to an Arm, and Example 7 shows the handling ARMCD for Subjects Withdrawn when assignment to an
Arm is Incomplete.
DM Example 1 – General Demographics
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
SUBJID
RFSTDTC
RFENDTC
SITEID
INVNAM
BIRTHDTC
AGE
AGEU
1
ABC123
DM
ABC12301001
001
2006-01-12
2006-03-10
01
JOHNSON, M
1948-12-13
57
YEARS
2
ABC123
DM
ABC12301002
002
2006-01-15
2006-02-28
01
JOHNSON, M
1955-03-22
50
YEARS
3
ABC123
DM
ABC12301003
003
2006-01-16
2006-03-19
01
JOHNSON, M
1938-01-19
68
YEARS
4
ABC123
DM
ABC12301004
004
01
JOHNSON, M
1941-07-02
5
ABC123
DM
ABC12302001
001
2006-02-02
2006-03-31
02
GONZALEZ, E
1950-06-23
55
YEARS
6
ABC123
DM
ABC12302002
002
2006-02-03
2006-04-05
02
GONZALEZ, E
1956-05-05
49
YEARS
Row
SEX
RACE
ETHNIC
ARMCD
ARM
COUNTRY
1 (cont)
M
WHITE
HISPANIC OR LATINO
A
Drug A
USA
2 (cont)
M
WHITE
NOT HISPANIC OR LATINO
P
Placebo
USA
3 (cont)
F
BLACK OR AFRICAN AMERICAN
NOT HISPANIC OR LATINO
P
Placebo
USA
4 (cont)
M
ASIAN
NOT HISPANIC OR LATINO
SCRNFAIL
Screen Failure
USA
5 (cont)
F
AMERICAN INDIAN OR ALASKA NATIVE
NOT HISPANIC OR LATINO
P
Placebo
USA
6 (cont)
F
NATIVE HAWAIIAN OR OTHER PACIFIC ISLANDERS
NOT HISPANIC OR LATINO
A
Drug A
USA
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 58 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
DM Example 2 – Single Race/Single Ethnicity Choice.
Sample CRF:
Ethnicity
Check one
Hispanic or Latino
Not Hispanic or Latino
Race
Check one
American Indian or Alaska Native
Asian
Black or African American
Native Hawaiian or Other Pacific Islander
White
Row 1: Subject 001 was Not-Hispanic and Asian.
Row 2: Subject 002 was Hispanic and White.
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
RACE
ETHNIC
1
ABC
DM
001
ASIAN
NOT HISPANIC OR LATINO
2
ABC
DM
002
WHITE
HISPANIC OR LATINO
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 59
Final November 12, 2008
DM Example 3 - Multiple Race Choices
In this example, the subject is permitted to check all applicable races.
Sample CRF:
Race
Check all that apply
American Indian or Alaska Native
Asian
Black or African American
Native Hawaiian or Other Pacific Islander
White
Other, Specify: ____________________
Row 1 (DM) and
Row 1 (SUPPDM): Subject 001 checked ―Other, Specify:‖ and entered ―Brazilian‖ as race.
Row 2 (DM) and
Rows 2, 3, 4, 5 (SUPPDM): Subject 002 checked three races, including an ―Other, Specify‖ value. The three values are reported in SUPPDM using QNAM
values RACE1 - RACE3. The specified information describing other race for is submitted in the same manner as subject 001.
Row 3 (DM): Subject 003 refused to provide information on race.
Row 4 (DM): Subject 004 checked ―Asian‖ as their only race.
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
RACE
1
ABC
DM
001
OTHER
2
ABC
DM
002
MULTIPLE
3
ABC
DM
003
4
ABC
DM
004
ASIAN
suppdm.xpt
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
ABC
DM
001
RACEOTH
Race, Other
BRAZILIAN
CRF
2
ABC
DM
002
RACE1
Race 1
BLACK OR AFRICAN AMERICAN
CRF
3
ABC
DM
002
RACE2
Race 2
AMERICAN INDIAN OR ALASKA NATIVE
CRF
4
ABC
DM
002
RACE3
Race 3
OTHER
CRF
5
ABC
DM
002
RACEOTH
Race, Other
ABORIGINE
CRF
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 60 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
DM Example 4: Mapping Predefined Races
In this example, the sponsor has chosen to map some of the predefined races to other races, specifically Japanese and Non-Japanese to Asian. Note: Sponsors
may choose not to map race data, in which case the previous examples should be followed.
Sample CRF
Race
Check One
American Indian or Alaska Native
Asian
Japanese
Non-Japanese
Black or African American
Native Hawaiian or Other Pacific Islander
White
Row 1 (DM), Row 1 (SUPPDM): Subject 001 checked ―Non-Japanese‖ which was mapped by the sponsor to ―Asian‖.
Row 2 (DM), Row 2 (SUPPDM): Subject 002 checked ―Japanese‖ which was mapped by the sponsor to ―Asian‖.
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
RACE
1
ABC
DM
001
ASIAN
2
ABC
DM
002
ASIAN
suppdm.xpt
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
ABC
DM
001
RACEOR
Original Race
NON-JAPANESE
CRF
2
ABC
DM
002
RACEOR
Original Race
JAPANESE
CRF
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 61
Final November 12, 2008
DM Example 5: Mapping “Other, Specify” Races.
In this example, the sponsor has chosen to map the values entered into the ―Other, Specify‖ field to one of the preprinted races.
Note: Sponsors may choose not to map race data, in which case the first two examples should be followed.
Sample CRF and Data:
Race
Check One
American Indian or Alaska Native
Asian
Black or African American
Native Hawaiian or Other Pacific Islander
White
Other, Specify: _____________________
Row 1 (DM), Row 1 (SUPPDM): Subject 001 checked ―Other, Specify‖ and entered ―Japanese‖ which was mapped to ―Asian‖ by the sponsor.
Row 2 (DM), Row 2 (SUPPDM): Subject 002 checked ―Other, Specify‖ and entered ―Swedish‖ which was mapped to ―White‖ by the sponsor.
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
RACE
1
ABC
DM
001
ASIAN
2
ABC
DM
002
WHITE
suppdm.xpt
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
ABC
DM
001
RACEOR
Original Race
JAPANESE
CRF
2
ABC
DM
002
RACEOR
Original Race
SWEDISH
CRF
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 62 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
DM/SE Example 6
The following examples illustrate values of ARMCD for subjects in Example 1874HTrial 1, described in 383HSection 7.2.3.1. The sponsor is submitting data on screen-
failure subjects.
Row 1: Subject 001 was randomized to Arm A. Rows 1-3 of SE dataset show that the subject completed all the Elements for Arm A.
Row 2: Subject 002 was randomized to Arm B. Rows 4-6 of SE dataset show that the subject completed all the Elements for Arm B.
Row 3: Subject 003 was a screen failure. Row 7 of SE dataset shows that they passed through only the Screen Element.
Row 4: Subject 004 withdrew during the Run-in Element. They were not considered a screen failure, but they were not randomized, so they have been given the
special ARMCD value NOTASSGN. Rows 8-9 of the SE dataset show the two Elements (Screen and Run-in) this subject passed through.
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
ARMCD
1
ABC
DM
001
A
2
ABC
DM
002
B
3
ABC
DM
003
SCRNFAIL
4
ABC
DM
004
NOTASSGN
se.xpt
Row
STUDYID
DOMAIN
USUBJID
SESEQ
ETCD
ELEMENT
SESTDTC
SEENDTC
1
ABC
SE
001
1
SCRN
Screen
2006-06-01
2006-06-07
2
ABC
SE
001
2
RI
Run-In
2006-06-07
2006-06-21
3
ABC
SE
001
3
A
Drug A
2006-06-21
2006-07-05
4
ABC
SE
002
1
SCRN
Screen
2006-05-03
2006-05-10
5
ABC
SE
002
2
RI
Run-In
2006-05-10
2006-05-24
6
ABC
SE
002
3
B
Drug B
2006-05-24
2006-06-07
7
ABC
SE
003
1
SCRN
Screen
2006-06-27
2006-06-30
8
ABC
SE
004
1
SCRN
Screen
2006-05-14
2006-05-21
9
ABC
SE
004
2
RI
Run-In
2006-05-21
2006-05-26
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 63
Final November 12, 2008
DM/SE Example 7:
The following example illustrates values of ARMCD for subjects in Example 1875HTrial 3, described in 384HSection 7.2.3.3.
Row 1: Subject 001 was randomized to Drug A. At the end of the Double Blind Treatment Epoch, they were assigned to Open Label A. Thus their ARMCD is
AA. Rows 1-3 of the SE dataset show that subject passed through all three Elements for the AA Arm.
Row 2: Subject 002 was randomized to Drug A. They were lost to follow-up during the Double Blind Epoch, so never reached the Open Label Epoch, when they
would have been assigned to either the Open Drug A or the Rescue Element. Their ARMCD is A. Note that A is not one of the Arm code values in the
Trial Arms dataset for this trial. See 385HSection 7.2.4.2 for more information on handling subjects who do not reach all branch points in the trial design.
Rows 4-5 of the SE dataset show the two Elements (Screen and Treatment A) the subject passed through.
dm.xpt
Row
STUDYID
DOMAIN
USUBJID
ARMCD
ARM
1
DEF
DM
001
AA
A-OPEN A
2
DEF
DM
002
A
A
se.xpt
Row
STUDYID
DOMAIN
USUBJID
SESEQ
ETCD
ELEMENT
SESTDTC
SEENDTC
1
DEF
SE
001
1
SCRN
Screen
2006-01-07
2006-01-12
2
DEF
SE
001
2
DBA
Treatment A
2006-01-12
2006-04-10
3
DEF
SE
001
3
OA
Open Drug A
2006-04-10
2006-07-05
4
DEF
SE
002
1
SCRN
Screen
2006-02-03
2006-02-10
5
DEF
SE
002
2
DBA
Treatment A
2006-02-10
2006-03-24
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 64 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
5.2 COMMENTS
5.2.1 COMMENTS — CO
co.xpt, Comments —Version 3.1.2,One record per comment per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1876HCO
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
386H387HSDTMIG 4.1.2.2,
388HSDTMIG
Appendix C2
RDOMAIN
Related Domain
Abbreviation
Char
*
Record
Qualifier
Two-character abbreviation for the domain of the parent record(s). Null for
comments collected on a general comments or additional information CRF page.
Perm
USUBJID
Unique Subject
Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all applications
or submissions involving the product.
Req
SDTM 2.2.4,
389H390HSDTMIG 4.1.2.3
COSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a domain.
May be any valid number.
Req
SDTM 2.2.4
IDVAR
Identifying Variable
Char
*
Record
Qualifier
Identifying variable in the parent dataset that identifies the record(s) to which the
comment applies. Examples AESEQ or CMGRPID. Used only when individual
comments are related to domain records. Null for comments collected on separate CRFs.
Perm
IDVARVAL
Identifying Variable
Value
Char
Record
Qualifier
Value of identifying variable of the parent record(s). Used only when individual
comments are related to domain records. Null for comments collected on separate CRFs.
Perm
COREF
Comment Reference
Char
Record
Qualifier
Sponsor-defined reference associated with the comment. May be the CRF page
number (e.g. 650), or a module name (e.g. DEMOG), or a combination of
information that identifies the reference (e.g. 650-VITALS-VISIT 2).
Perm
COVAL
Comment
Char
Topic
The text of the comment. Text over 200 characters can be added to additional
columns COVAL1-COVALn. See 391Hassumption 5.2.1.1.3.
Req
COEVAL
Evaluator
Char
*
Record
Qualifier
Used to describe the originator of the comment. Examples: CENTRAL, REVIEWER,
ADJUDICATION COMMITTEE, PRINCIPAL INVESTIGATOR.
Perm
CODTC
Date/Time of
Comment
Char
ISO 8601
Timing
Date/time of comment on dedicated comment form. Should be null if this is a
child record of another domain or if comment date was not collected.
Perm
392HSDTM 2.2.5,
375HSDTMIG 4.1.4.1
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 65
Final November 12, 2008
5.2.1.1 ASSUMPTIONS FOR COMMENTS DOMAIN MODEL
1. The Comments special-purpose domain provides a solution for submitting free-text comments related to data in one or more SDTM domains (as
described in 393HSection 8.5) or collected on a separate CRF page dedicated to comments. Comments are generally not responses to specific questions;
instead, comments usually consist of voluntary, free-text or unsolicited observations.
2. The CO dataset accommodates three sources of comments:
a. Those unrelated to a specific domain or parent record(s), in which case the values of the variables RDOMAIN, IDVAR and IDVARVAL are
null. CODTC should be populated if captured. See example, Rows 1.
b. Those related to a domain but not to specific parent record(s), in which case the value of the variable RDOMAIN is set to the DOMAIN code
of the parent domain and the variables IDVAR and IDVARVAL are null. CODTC should be populated if captured. See example, Row 2.
c. Those related to a specific parent record or group of parent records, in which case the value of the variable RDOMAIN is set to the DOMAIN
code of the parent record(s) and the variables IDVAR and IDVARVAL are populated with the key variable name and value of the parent
record(s). Assumptions for populating IDVAR and IDVARVAL are further described in 394HSection 8.5. CODTC should be null because the timing
of the parent record(s) is inherited by the comment record. See example, Rows 3-5.
3. When the comment text is longer than 200 characters, the first 200 characters of the comment will be in COVAL, the next 200 in COVAL1, and
additional text stored as needed to COVALn. See example, Rows 3-4.
4. Additional information about how to relate comments to parent SDTM records is provided in 395HSection 8.5.
5. The variable COREF may be null unless it is used to identify the source of the comment. See example, Rows 1 and 5.
6. Only following Identifier and Timing variables that are permissible and may be added as appropriate when comments are not related to other
domain records: COGRPID, COREFID, COSPID, VISIT, VISITNUM, VISITDY, TAETORD, CODY, COTPT, COTPTNUM, COELTM,
COTPTREF, CORFTDTC.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 66 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
5.2.1.2 EXAMPLES FOR COMMENTS DOMAIN MODEL
In the example below:
Row 1: Shows a comment unrelated to any specific domain or record, because it was collected on a separate comments page...
Row 2: Shows a comment related to a specific domain (PE in this example), but not to any specific record because it was collected on the
bottom of the PE page without any indication of specific records it applies to. COREF is populated with the text ―VISIT 7‖ to show
this comment came from the VISIT 7 PE page.
Rows 3-5: Show comments related to parent records in the AE, EX and VS domains.
Row 3 shows a comment related to a single AE record having it‘s AESEQ=7.
Row 4 shows a comment related to multiple EX records having their EXGRPID=―COMBO1‖.
Row 5 shows a comment related to multiple VS records having their VSGRPID=―VS2‖
Rows 6-8: Show three options for representing a comment unrelated to any specific general observation class record(s) because it was collected
on a separate comments page, but the page was associated with a specific visit.
Row 6 shows the comment related to the Subject Visit record in SV. The RDOMAIN variable is populated with SV (the Subject
Visits domain) and the variables IDVAR and IDVARVAL are populated with the key variable name and value of the parent
Subject-Visit record.
Row 7 shows the comment unrelated to any parent records, RDOMAIN, IDVAR and IDVARVAL are not populated. COREF is
populated to indicate that the comment reference is ―VISIT 4‖
Row 8 also shows the comment unrelated to any parent records, but instead of populating COREF, the VISIT Timing variable was
added to the CO dataset and populated with 4 to indicate Visit 4.
Row
STUDYID
DOMAIN
USUBJID
COSEQ
RDOMAIN
IDVAR
IDVARVAL
COREF
COVAL
COVAL1
COVAL2
COEVAL
VISIT
CODTC
1
1234
CO
AB-99
1
Comment text
PRINCIPAL
INVESTIGATOR
2003-11-08
2
1234
CO
AB-99
2
PE
VISIT 7
Comment text
PRINCIPAL
INVESTIGATOR
2004-01-14
3
1234
CO
AB-99
3
AE
AESEQ
7
PAGE 650
First 200
characters
Next 200
characters
Remaining text
PRINCIPAL
INVESTIGATOR
4
1234
CO
AB-99
4
EX
EXGRPID
COMBO1
PAGE 320-355
First 200
characters
Remaining text
PRINCIPAL
INVESTIGATOR
5
1234
CO
AB-99
5
VS
VSGRPID
VS2
Comment text
PRINCIPAL
INVESTIGATOR
6
1234
CO
AB-99
6
SV
VISITNUM
4
Comment Text
PRINCIPAL
INVESTIGATOR
7
1234
CO
AB-99
7
VISIT 4
Comment Text
PRINCIPAL
INVESTIGATOR
8
1234
CO
AB-99
8
Comment Text
PRINCIPAL
INVESTIGATOR
4

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 67
Final November 12, 2008
5.3 SUBJECT ELEMENTS AND VISITS
The Trial Elements, Trial Arms, and Trial Visits datasets in the Trial Design model describe the planned design of the study (see 396HSection 7.3, Section 7.2 and
Section 7.4), but it is also necessary to collect the corresponding actual data. Subject assignment to an Arm is reported in the ARM variable in Demographics.
Actual Elements and Visits data for each subject are described in two additional datasets:
The Subject Elements dataset (397HTable 5.3.1)
The Subject Visits dataset (398HTable 5.3.2).
5.3.1 SUBJECT ELEMENTS — SE
The Subject Elements dataset consolidates information about the timing of each subject‘s progress through the Epochs and Elements of the trial. For Elements
that involve study treatments, the identification of which Element the subject passed through (e.g., Drug X vs. placebo) is likely to derive from data in the
Exposure domain or another Interventions domain. The dates of a subject‘s transition from one Element to the next will be taken from the Interventions
domain(s) and from other relevant domains, according to the definitions (TESTRL values) in the Trial Elements dataset (see 399HSection 7.3).
The Subject Elements dataset is particularly useful for studies with multiple treatment periods, such as crossover studies. The Subject Elements dataset contains
the date/times at which a subject moved from one Element to another, so when the Trial Arms (400HSection 7.2), Trial Elements (401HSection 7.3), and Subject Elements
datasets are included in a submission, reviewers can relate all the observations made about a subject to that subject‘s progression through the trial.
Comparison of the --DTC of a finding observation to the Element transition dates (values of SESTDTC and SEENDTC) tells which Element the subject
was in at the time of the finding. Similarly, one can determine the Element during which an event or intervention started or ended.
―Day within Element‖ or ―day within Epoch‖ can be derived. Such variables relate an observation to the start of an Element or Epoch in the same way
that study day (--DY) variables relate it to the reference start date (RFSTDTC) for the study as a whole. See 402HSection 4.1.4.4
Having knowledge of Subject Element start and end dates can be helpful in the determination of baseline values.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 68 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
se.xpt, Subject Elements — Version 3.1.2. One record per actual Element per subject.
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1877HSE
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
403H404HSDTMIG 4.1.2.2,
405HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
406H407HSDTMIG 4.1.2.3
SESEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. Should be assigned to be consistent chronological order.
Req
SDTM 2.2.4
ETCD
Element Code
Char
*
Topic
1. ETCD (the companion to ELEMENT) is limited to 8 characters and does
not have special character restrictions. These values should be short for ease
of use in programming, but it is not expected that ETCD will need to serve as
a variable name.
2. If an encountered Element differs from the planned Element to the point
that it is considered a new Element, then use ―UNPLAN‖ as the value for
ETCD to represent this Element.
Req
408HSDTMIG 4.1.2.1
ELEMENT
Description of Element
Char
*
Synonym
Qualifier
The name of the Element. If ETCD has a value of ―UNPLAN‖ then
ELEMENT should be Null.
Perm
409HSDTMIG 4.1.2.1,
410HSDTMIG 4.1.2.4
SESTDTC
Start Date/Time of
Element
Char
ISO 8601
Timing
Start date/time for an Element for each subject.
Req
SDTM 2.2.5,
411HSDTMIG 4.1.4.1
SEENDTC
End Date/Time of
Element
Char
ISO 8601
Timing
End date/time for an Element for each subject.
Exp
SDTM 2.2.5,
412HSDTMIG 4.1.4.1
TAETORD
Planned Order of
Elements within Arm
Num
Timing
Number that gives the planned order of the Element within the subject's
assigned ARM.
Perm
EPOCH
Epoch
Char
*
Timing
Epoch associated with the Element in the planned sequence of Elements for
the ARM to which the subject was assigned
Perm
SDTM 2.2.5,
413HSDTMIG 7.1.2
SEUPDES
Description of Unplanned
Element
Char
Synonym
Qualifier
Description of what happened to the subject during this unplanned Element.
Used only if ETCD has the value of ―UNPLAN‖.
Perm
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
5.3.1.1 ASSUMPTIONS FOR SUBJECT ELEMENTS DOMAIN MODEL
1. Submission of the Subject Elements dataset is strongly recommended, as it provides information needed by reviewers to place observations in context within
the study. The Trial Elements and Trial Arms datasets should also be submitted, as they define the design and the terms referenced by the Subject Elements
dataset.
2. The Subject Elements domain allows the submission of data on the timing of the trial Elements a subject actually passed through in their participation in the
trial. Please read 414HSection 7.3, on the Trial Elements dataset and 415HSection 7.2, on the Trial Arms dataset, as these datasets define a trial's planned Elements, and
describe the planned sequences of Elements for the Arms of the trial.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 69
Final November 12, 2008
3. For any particular subject, the dates in the subject Elements table are the dates when the transition events identified in the Trial Elements table occurred. Judgment may be
needed to match actual events in a subject's experience with the definitions of transition events (the events that mark the starts of new Elements) in the Trial Elements table,
since actual events may vary from the plan. For instance, in a single dose PK study, the transition events might correspond to study drug doses of 5 and 10 mg. If a subject
actually received a dose of 7 mg when they were scheduled to receive 5 mg, a decision will have to be made on how to represent this in the SE domain.
4. If the date/time of a transition Element was not collected directly, the method used to infer the Element start date/time should be explained in the Comments
column of the define.xml.
5. Judgment will also have to be used in deciding how to represent a subject's experience if an Element does not proceed or end as planned. For instance, the
plan might identify a trial Element which is to start with the first of a series of 5 daily doses and end after 1 week, when the subject transitions to the next
treatment Element. If the subject actually started the next treatment Epoch (see 416HSection 7.1.2) after 4 weeks, the sponsor will have to decide whether to
represent this as an abnormally long Element, or as a normal Element plus an unplanned non-treatment Element.
6. If the sponsor decides that the subject's experience for a particular period of time cannot be represented with one of the planned Elements, then that period of
time should be represented as an unplanned Element. The value of ETCD for an unplanned Element is ―UNPLAN‖ and SEUPDES should be populated with
a description of the unplanned Element.
7. The values of SESTDTC provide the chronological order of the actual subject Elements. SESEQ should be assigned to be consistent with the chronological order. Note that
the requirement that SESEQ be consistent with chronological order is more stringent than in most other domains, where --SEQ values need only be unique within subject.
8. When TAETORD is included in the SE domain, it represents the planned order of an Element in an Arm. This should not be confused with the actual order of the
Elements, which will be represented by their chronological order and SESEQ. TAETORD will not be populated for subject Elements that are not planned for the
Arm to which the subject was assigned. Thus, TAETORD will not be populated for any Element with an ETCD value of ―UNPLAN‖. TAETORD will also not be
populated if a subject passed through an Element that, although defined in the TE dataset, was out of place for the Arm to which the subject was assigned. For
example, if a subject in a parallel study of Drug A vs. Drug B was assigned to receive Drug A, but received Drug B instead, then TAETORD would be left blank
for the SE record for their Drug B Element. If a subject was assigned to receive the sequence of Elements A, B, C, D, and instead received A, D, B, C, then the
sponsor would have to decide for which of these subject Element records TAETORD should be populated. The rationale for this decision should be documented
in the Comments column of the define.xml.
9. For subjects who follow the planned sequence of Elements for the Arm to which they were assigned, the values of EPOCH in the SE domain will match
those associated with the Elements for the subject's Arm in the Trial Arms dataset. The sponsor will have to decide what value, if any, of EPOCH to assign
SE records for unplanned Elements and in other cases where the subject's actual Elements deviate from the plan. The sponsor's methods for such decisions
should be documented in the define.xml, in the row for EPOCH in the SE dataset table.
10. Since there are, by definition, no gaps between Elements, the value of SEENDTC for one Element will always be the same as the value of SESTDTC for the next Element.
11. Note that SESTDTC is required, although --STDTC is not required in any other subject-level dataset. The purpose of the dataset is to record the Elements a
subject actually passed through. We assume that if it is known that a subject passed through a particular Element, then there must be some information on when
it started, even if that information is imprecise. Thus, SESTDTC may not be null, although some records may not have all the components (e.g., year, month,
day, hour, minute) of the date/time value collected.
12. The following Identifier variables are permissible and may be added as appropriate: --GRPID, --REFID, --SPID.
13. Care should be taken in adding additional Timing variables:
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 70 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The purpose of --DTC and --DY in other domains with start and end dates (Event and Intervention Domains) is to record the date and study day on
which data was collected. The starts and ends of elements are generally ―derived‖ in the sense that they are a secondary use of data collected
elsewhere, and it is not generally useful to know when those date/times were recorded.
--DUR could be added only if the duration of an element was collected, not derived.
It would be inappropriate to add the variables that support time points (--TPT, --TPTNUM, --ELTM, --TPTREF, and --RFTDTC), since the topic of
this dataset is Elements.
5.3.1.2 EXAMPLES FOR SUBJECT ELEMENTS DOMAIN MODEL
STUDYID and DOMAIN, which are required in the SE and DM domains, have not been included in the following examples, to improve readability.
Example 1
This example shows data for two subjects for a crossover trial with four Epochs.
Row 1: The record for the SCREEN Element for subject 789. Note that only the date of the start of the SCREEN Element was collected, while for the end
of the Element, which corresponds to the start of IV dosing, both date and time were collected.
Row 2: The record for the IV Element for subject 789. The IV Element started with the start of IV dosing and ended with the start of oral dosing, and full
date/times were collected for both.
Row 3: The record for the ORAL Element for subject 789. Only the date, and not the time, of start of Follow-up was collected.
Row 4: The FOLLOWUP Element for subject 789 started and ended on the same day. Presumably, the Element had a positive duration, but no times were
collected.
Rows 5-8: Subject 790 was treated incorrectly, as shown by the fact that the values of SESEQ and TAETORD do not match. This subject entered the IV
Element before the Oral Element, but the planned order of Elements for this subject was ORAL, then IV. The sponsor has assigned EPOCH
values for this subject according to the actual order of Elements, rather than the planned order. The correct order of Elements is the subject's
ARMCD, shown in Row 2 of the DM dataset.
se.xpt
Row
USUBJID
SESEQ
ETCD
SESTDTC
SEENDTC
SEUPDES
TAETORD
EPOCH
1
789
1
SCREEN
2006-06-01
2006-06-03T10:32
1
SCREEN
2
789
2
IV
2006-06-03T10:32
2006-06-10T09:47
2
FIRST TREATMENT
3
789
3
ORAL
2006-06-10T09:47
2006-06-17
3
SECOND TREATMENT
4
789
4
FOLLOWUP
2006-06-17
2006-06-17
4
FOLLOW-UP
5
790
1
SCREEN
2006-06-01
2006-06-03T10:14
1
SCREEN
6
790
2
IV
2006-06-03T10:14
2006-06-10T10:32
3
FIRST TREATMENT
7
790
3
ORAL
2006-06-10T10:32
2006-06-17
2
SECOND TREATMENT
8
790
4
FOLLOWUP
2006-06-17
2006-06-17
4
FOLLOW-UP
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 71
Final November 12, 2008
dm.xpt
Row
USUBJID
SUBJID
RFSTDTC
RFENDTC
SITEID
INVNAM
BIRTHDTC
AGE
AGEU
SEX
RACE
ETHNIC
ARMCD
ARM
COUNTRY
1
789
001
2006-06-03
2006-06-17
01
SMITH, J
1948-12-13
57
YEARS
M
WHITE
HISPANIC OR
LATINO
IO
IV-ORAL
USA
2
790
002
2006-06-03
2006-06-17
01
SMITH, J
1955-03-22
51
YEARS
M
WHITE
NOT
HISPANIC OR
LATINO
OI
ORAL-IV
USA
Example 2
The data below represent two subjects enrolled in Example 1878HTrial 3, described in 417HSection 7.2.3.3.
Rows 1-2: Subject 123 completed only two Elements of the trial. The double-blind treatment Epoch starts with the start of dosing, but in this trial only the
date, and not the time, of the start of dosing has been collected. Note that, for this subject, events that occurred on, or data collected on, 2006-06-
03 cannot be assigned to an Element or an Epoch on the basis of dates alone. When sponsors choose to collect only dates, they must deal with
such ambiguity in the algorithms they use to assign data to Elements or Epochs. Row 1 of the Demographics dataset shows that this subject has an
ARMCD value of A. See DM/SE 418HExample 6 in Section 5.1.1.2 for other examples of ARM and ARMCD values for this trial.
Rows 3-6: Subject 456 completed the trial, but received the wrong drug for the last 2 weeks of the double-blind treatment period. This has been represented
by treating the period when the subject received the wrong drug as an unplanned Element. Note that TAETORD, which represents the planned
order of Elements within an Arm, has not been populated for this unplanned Element. However, even though this Element was unplanned, the
sponsor assigned a value of DOUBLE BLIND TREATMENT to EPOCH. Row 2 of the Demographics dataset shows that the values of ARM and
ARMCD for this subject reflect their planned treatment, and are not affected by the fact that their treatment deviated from plan.
se.xpt
Row
USUBJID
SESEQ
ETCD
SESTDTC
SEENDTC
SEUPDES
TAETORD
EPOCH
1
123
1
SCRN
2006-06-01
2006-06-03
1
SCREEN
2
123
2
DBA
2006-06-03
2006-06-10
2
DOUBLE-BLIND TREATMENT
3
456
1
SCRN
2006-05-01
2006-05-03
1
SCREEN
4
456
2
DBA
2006-05-03
2006-05-31
2
DOUBLE-BLIND TREATMENT
5
456
3
UNPLAN
2006-05-31
2006-06-13
Drug B dispensed in error
DOUBLE-BLIND TREATMENT
6
456
4
RSC
2006-06-13
2006-07-30
3
OPEN-LABEL TREATMENT
dm.xpt
Row
USUBJID
SUBJID
RFSTDTC
RFENDTC
SITEID
INVNAM
BIRTHDTC
AGE
AGEU
SEX
RACE
ETHNIC
ARMCD
ARM
COUNTRY
1
123
012
2006-06-03
2006-06-10
01
JONES, D
1943-12-08
62
YEARS
M
ASIAN
HISPANIC
OR
LATINO
A
A
USA
2
456
103
2006-05-03
2006-07-30
01
JONES, D
1950-05-15
55
YEARS
F
WHITE
NOT
HISPANIC
OR
LATINO
AR
A-
Rescue
USA
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 72 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
5.3.2 SUBJECT VISITS — SV
The Subject Visits domain consolidates information about the timing of subject visits that is otherwise spread over domains that include the visit variables (VISITNUM and
possibly VISIT and/or VISITDY). Unless the beginning and end of each visit is collected, populating the Subject Visits dataset will involve derivations. In a simple case,
where, for each subject visit, exactly one date appears in every such domain, the Subject Visits dataset can be created easily, by populating both SVSTDTC and SVENDTC
with the single date for a visit. When there are multiple dates and/or date/times for a visit for a particular subject, the derivation of values for SVSTDTC and SVENDTC may
be more complex. The method for deriving these values should be consistent with the visit definitions in the Trial Visits dataset (see 419HSection 7.4). For some studies, a visit
may be defined to correspond with a clinic visit that occurs within one day, while for other studies, a visit may reflect data collection over a multi-day period.
The Subject Visits dataset provides reviewers with a summary of a subject‘s Visits. Comparison of an individual subject‘s SV dataset with the TV dataset (420HSection 7.4),
which describes the planned Visits for the trial, quickly identifies missed Visits and ―extra‖ Visits. Comparison of the values of STVSDY and SVENDY to VISIT and/or
VISITDY can often highlight departures from the planned timing of Visits.
sv.xpt, Subject Visits — Version 3.1.2,. One record per subject per actual visit.
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1879HSV
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
421H422HSDTMIG 4.1.2.2,
423HSDTMIG
Appendix C2
USUBJID
Unique Subject
Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
424H425HSDTMIG 4.1.2.3
VISITNUM
Visit Number
Num
Topic
1. Clinical encounter number. (Decimal numbering may be useful for
inserting unplanned visits.)
2. Numeric version of VISIT, used for sorting.
Req
SDTM 2.2.5,
426HSDTMIG 4.1.4.5,
427HSDTMIG 7.4
VISIT
Visit Name
Char
Synonym
Qualifier
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY as a text
description of the clinical encounter.
Perm
SDTM 2.2.5,
428HSDTMIG 4.1.4.5,
429HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the start of the visit based upon RFSTDTC in
Demographics.
Perm
SDTM 2.2.5,
430HSDTMIG 4.1.4.5,
431HSDTMIG 7.4
SVSTDTC
Start Date/Time of
Visit
Char
ISO 8601
Timing
Start date/time for a Visit.
Exp
SDTM 2.2.5,
432HSDTMIG 4.1.4.1
SVENDTC
End Date/Time of
Visit
Char
ISO 8601
Timing
End date/time of a Visit.
Exp
SDTM 2.2.5,
433HSDTMIG 4.1.4.1
SVSTDY
Study Day of Start of
Visit
Num
Timing
Study day of start of visit relative to the sponsor-defined RFSTDTC.
Perm
SDTM 2.2.5,
434HSDTMIG 4.1.4.4
SVENDY
Study Day of End of
Visit
Num
Timing
Study day of end of visit relative to the sponsor-defined RFSTDTC.
Perm
SDTM 2.2.5,
435HSDTMIG 4.1.4.4
SVUPDES
Description of
Unplanned Visit
Char
Synonym
Qualifier
Description of what happened to the subject during an unplanned visit.
Perm
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 73
Final November 12, 2008
5.3.2.1 ASSUMPTIONS FOR SUBJECT VISITS DOMAIN MODEL
1. The Subject Visits domain allows the submission of data on the timing of the trial visits a subject actually passed through in their participation in the
trial. Please read 436HSection 7.4 on the Trial Visits dataset, as the Trial Visits dataset defines the planned visits for the trial.
2. The identification of an actual visit with a planned visit sometimes calls for judgment. In general, data collection forms are prepared for particular visits,
and the fact that data was collected on a form labeled with a planned visit is sufficient to make the association. Occasionally, the association will not be
so clear, and the sponsor will need to make decisions about how to label actual visits. The sponsor's rules for making such decisions should be
documented in the define.xml document.
3. Records for unplanned visits should be included in the SV dataset. For unplanned visits, SVUPDES should be populated with a description of the reason
for the unplanned visit. Some judgment may be required to determine what constitutes an unplanned visit. When data are collected outside a planned
visit, that act of collecting data may or may not be described as a "visit." The encounter should generally be treated as a visit if data from the encounter
are included in any domain for which VISITNUM is included, since a record with a missing value for VISITNUM is generally less useful than a record
with VISITNUM populated. If the occasion is considered a visit, its date/times must be included in the SV table and a value of VISITNUM must be
assigned. See 437HSection 4.1.4.5 for information on the population of visit variables for unplanned visits.
4. VISITDY is the Planned Study Day of a visit. It should not be populated for unplanned visits.
5. If SVSTDY is included, it is the actual study day corresponding to SVSTDTC. In studies for which VISITDY has been populated, it may be desirable to
populate SVSTDY, as this will facilitate the comparison of planned (VISITDY) and actual (SVSTDY) study days for the start of a visit.
6. If SVENDY is included, it is the actual day corresponding to SVENDTC.
7. For many studies, all visits are assumed to occur within one calendar day, and only one date is collected for the Visit. In such a case, the values for
SVENDTC duplicate values in SVSTDTC. However, if the data for a visit is actually collected over several physical visits and/or over several days,
then SVSTDTC and SVENDTC should reflect this fact. Note that it is fairly common for screening data to be collected over several days, but for the
data to be treated as belonging to a single planned screening visit, even in studies for which all other visits are single-day visits.
8. Differentiating between planned and unplanned visits may be challenging if unplanned assessments (e.g., repeat labs) are performed during the time
period of a planned visit.
9. Algorithms for populating SVSTDTC and SVENDTC from the dates of assessments performed at a visit may be particularly challenging for screening
visits since baseline values collected at a screening visit are sometimes historical data from tests performed before the subject started screening for the
trial
10. The following Identifier variables are permissible and may be added as appropriate: --SEQ, --GRPID, --REFID, and --SPID.
11. Care should be taken in adding additional Timing variables:
If TAETORD and/or EPOCH are added, then the values must be those at the start of the visit.
The purpose of --DTC and --DY in other domains with start and end dates (Event and Intervention Domains) is to record the data on which data
was collected. It seems unnecessary to record the date on which the start and end of a visit were recorded.
--DUR could be added if the duration of a visit was collected.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 74 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
It would be inappropriate to add the variables that support time points (--TPT, --TPTNUM, --ELTM, --TPTREF, and --RFTDTC), since the topic
of this dataset is visits.
--STRF and --ENRF could be used to say whether a visit started and ended before, during, or after the study reference period, although this seems
unnecessary.
--STRTPT, --STTPT, --ENRTPT, and --ENTPT could be used to say that a visit started or ended before or after particular dates, although this
seems unnecessary.
5.3.2.2 EXAMPLES FOR SUBJECT VISITS DOMAIN MODEL
The data below represents the visits for a single subject.
Row 1: Data for the screening visit was actually gathered over the course of six days.
Row 2: The visit called DAY 1 actually started and ended as planned, on Day 1.
Row 3: The visit scheduled for Day 8 occurred one day early, on Day 7.
Row 4: The visit called WEEK 2 actually started and ended as planned, on Day 15,
Row 5: Shows an unscheduled visit. SVUPDES provides the information that this visit dealt with evaluation of an adverse event. Since this visit was not
planned, VISITDY was not populated. The sponsor chose not to populate VISIT. VISITNUM was populated, probably because the data collected at
this encounter is in a Findings domain such as EG, LB, or VS, in which VISIT is treated as an important timing variable.
Row 6: This subject had their last visit, a follow-up visit on study Day 26, eight days after the unscheduled visit, but well before the scheduled visit day of
71.
Row
STUDYID
DOMAIN
USUBJID
VISITNUM
VISIT
VISITDY
SVSTDTC
SVENDTC
SVSTDY
SVENDY
SVUPDES
1
123456
SV
101
1
SCREEN
-7
2006-01-15
2006-01-20
-6
-1
2
123456
SV
101
2
DAY 1
1
2006-01-21
2006-01-21
1
1
3
123456
SV
101
3
WEEK 1
8
2006-01-27
2006-01-27
7
7
4
123456
SV
101
4
WEEK 2
15
2006-02-04
2006-02-04
15
15
5
123456
SV
101
4.1
2006-02-07
2006-02-07
18
18
Evaluation of AE
6
123456
SV
101
8
FOLLOW-UP
71
2006-02-15
2006-02-15
26
26
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 75
Final November 12, 2008
6 Domain Models Based on the General
Observation Classes
6.1 INTERVENTIONS
6.1.1 CONCOMITANT MEDICATIONS — CM
cm.xpt, Concomitant Medications — Interventions, Version 3.1.2,. One record per recorded intervention occurrence or constant-dosing interval per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1880HCM
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
438H439HSDTMIG 4.1.2.2,
440HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
441H442HSDTMIG 4.1.2.3
CMSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
CMGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a subject.
Perm
SDTM 2.2.4
443HSDTMIG 4.1.2.6
CMSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Examples: a number pre-printed on the CRF
as an explicit line identifier or record identifier defined in the sponsor‘s
operational database. Example: line number on a concomitant medication page.
Perm
SDTM 2.2.4
CMTRT
Reported Name of Drug,
Med, or Therapy
Char
Topic
Verbatim medication name that is either pre-printed or collected on a
CRF.
Req
SDTM 2.2.1
CMMODIFY
Modified Reported Name
Char
Synonym
Qualifier
If CMTRT is modified to facilitate coding, then CMMODIFY will
contain the modified text.
Perm
SDTM 2.2.1,
444H445HSDTMIG 4.1.3.6
CMDECOD
Standardized Medication
Name
Char
*
Synonym
Qualifier
Standardized or dictionary-derived text description of CMTRT or
CMMODIFY. Equivalent to the generic medication name in WHO Drug.
The sponsor is expected to provide the dictionary name and
version used to map the terms utilizing the define.xml external
codelist attributes. If an intervention term does not have a decode value in
the dictionary then CMDECOD will be left blank.
Perm
SDTM 2.2.1,
446HSDTMIG 4.1.3.6
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 76 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
CMCAT
Category for Medication
Char
*
Grouping
Qualifier
Used to define a category of medications/treatments. Examples: PRIOR,
CONCOMITANT, ANTI-CANCER MEDICATION, or GENERAL
CONMED.
Perm
SDTM 2.2.1,
447HSDTMIG 4.1.2.6
CMSCAT
Subcategory for
Medication
Char
*
Grouping
Qualifier
A further categorization of medications/ treatment. Examples:
CHEMOTHERAPY, HORMONAL THERAPY, ALTERNATIVE THERAPY.
Perm
SDTM 2.2.1,
448HSDTMIG 4.1.2.6
CMPRESP
CM Pre-Specified
Char
(1881HNY)
Record
Qualifier
Used to indicate whether (Y/null) information about the use of a specific
medication was solicited on the CRF.
Perm
SDTM 2.2.1,
449H450HSDTMIG 4.1.2.7,
451HSDTMIG 4.1.5.7
CMOCCUR
CM Occurrence
Char
(1882HNY)
Record
Qualifier
When the use of specific medications is solicited, CMOCCUR is used to
indicate whether or not (Y/N) use of the medication occurred. Values are
null for medications not specifically solicited.
Perm
SDTM 2.2.1,
452HSDTMIG 4.1.5.7
CMSTAT
Completion Status
Char
(1883HND)
Record
Qualifier
Used to indicate that a question about a pre-specified medication was not
answered. Should be null or have a value of NOT DONE.
Perm
SDTM 2.2.1,
453H454HSDTMIG 4.1.5.1,
455HSDTMIG 4.1.5.7
CMREASND
Reason Medication Not
Collected
Char
Record
Qualifier
Describes the reason concomitant medication was not collected. Used in
conjunction with CMSTAT when value is NOT DONE.
Perm
SDTM 2.2.1,
456HSDTMIG 4.1.5.1,
457HSDTMIG 4.1.5.7
CMINDC
Indication
Char
Record
Qualifier
Denotes why a medication was taken or administered. Examples:
NAUSEA, HYPERTENSION.
Perm
SDTM 2.2.1,
458HSDTMIG 4.1.5.6
CMCLAS
Medication Class
Char
*
Variable
Qualifier
Drug class. May be obtained from coding. When coding to a single class,
populate with class value. If using a dictionary and coding to multiple
classes, then follow 459Hassumption 4.1.2.8.3 or omit CMCLAS.
Perm
SDTM 2.2.1,
460H461H462H463HSDTMIG 4.1.3.5
CMCLASCD
Medication Class Code
Char
*
Variable
Qualifier
Class code corresponding to CMCLAS. Drug class. May be obtained from
coding. When coding to a single class, populate with class code. If using a
dictionary and coding to multiple classes, then follow 464Hassumption
4.1.2.8.3 or omit CMCLASCD.
Perm
SDTM 2.2.1,
465H466H467H468HSDTMIG 4.1.3.5
CMDOSE
Dose per Administration
Num
Record
Qualifier
Amount of CMTRT taken.
Perm
SDTM 2.2.1
CMDOSTXT
Dose Description
Char
Record
Qualifier
Dosing amounts or a range of dosing information collected in text form.
Units may be stored in CMDOSU. Example: 200-400, 15-20.
Perm
SDTM 2.2.1
CMDOSU
Dose Units
Char
(469HUNIT)
Variable
Qualifier
Units for CMDOSE, CMDOSTXT, and CMDOSTOT. Examples: ng, mg,
or mg/kg.
Perm
SDTM 2.2.1,
470H471HSDTMIG 4.1.3.2
CMDOSFRM
Dose Form
Char
(1884HFRM)
Record
Qualifier
Dose form for CMTRT. Examples: TABLET, LOTION.
Perm
SDTM 2.2.1
CMDOSFRQ
Dosing Frequency per
Interval
Char
(472HFREQ)
Variable
Qualifier
Usually expressed as the number of repeated administrations of CMDOSE
within a specific time period. Examples: BID (twice daily), Q12H (every 12
hours).
Perm
SDTM 2.2.1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 77
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
CMDOSTOT
Total Daily Dose
Num
Record
Qualifier
Total daily dose of CMTRT using the units in CMDOSU. Total dose over
a period other than day could be recorded in a separate Supplemental
Qualifier variable. CMDOSTOT should be used in addition to CMDOSE,
and not in place of it.
Perm
SDTM 2.2.1
CMDOSRGM
Intended Dose Regimen
Char
Variable
Qualifier
Text description of the (intended) schedule or regimen for the
Intervention. Examples: TWO WEEKS ON, TWO WEEKS OFF.
Perm
SDTM 2.2.1
CMROUTE
Route of Administration
Char
(1885HROUTE)
Variable
Qualifier
Route of administration for CMTRT. Examples: ORAL,
INTRAVENOUS.
Perm
SDTM 2.2.1
CMSTDTC
Start Date/Time of
Medication
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
473HSDTMIG 4.1.4.1,
474H475HSDTMIG 4.1.4.3
CMENDTC
End Date/Time of
Medication
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
476HSDTMIG 4.1.4.1,
477HSDTMIG 4.1.4.3
CMSTDY
Study Day of Start of
Medication
Num
Timing
Study day of start of medication relative to the sponsor-defined
RFSTDTC.
Perm
SDTM 2.2.5,
478HSDTMIG 4.1.4.4,
479HSDTMIG 4.1.4.6
CMENDY
Study Day of End of
Medication
Num
Timing
Study day of end of medication relative to the sponsor-defined
RFSTDTC.
Perm
SDTM 2.2.5,
480HSDTMIG 4.1.4.4,
481HSDTMIG 4.1.4.6
CMDUR
Duration of Medication
Char
ISO 8601
Timing
Collected duration for a treatment episode. Used only if collected on the
CRF and not derived from start and end date/times.
Perm
SDTM 2.2.5,
482HSDTMIG 4.1.4.3
CMSTRF
Start Relative to Reference
Period
Char
(1886HSTENRF)
Timing
Describes the start of the medication relative to sponsor-defined reference
period. The sponsor-defined reference period is a continuous period of
time defined by a discrete starting point and a discrete ending point
(represented by RFSTDTC and RFENDTC in Demographics). If
information such as "PRIOR", "ONGOING", or "CONTINUING" was
collected, this information may be translated into CMSTRF.
Perm
SDTM 2.2.5,
483H484HSDTMIG 4.1.4.7
CMENRF
End Relative to Reference
Period
Char
(1887HSTENRF)
Timing
Describes the end of the medication relative to the sponsor-defined
reference period. The sponsor-defined reference period is a continuous
period of time defined by a discrete starting point and a discrete ending
point (represented by RFSTDTC and RFENDTC in Demographics). If
information such as "PRIOR", "ONGOING", or "CONTINUING" was
collected, this information may be translated into CMENRF.
Perm
SDTM 2.2.5,
485H486HSDTMIG 4.1.4.7
CMSTRTPT
Start Relative to Reference
Time Point
Char
BEFORE,
COINCIDENT,
AFTER, U
Timing
Identifies the start of the medication as being before or after the reference
time point defined by variable CMSTTPT.
Perm
SDTM 2.2.5,
487H488HSDTMIG 4.1.4.7
CMSTTPT
Start Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the reference
point referred to by CMSTRTPT. Examples: "2003-12-15" or "VISIT 1".
Perm
SDTM 2.2.5,
489HSDTMIG 4.1.4.7
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 78 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
CMENRTPT
End Relative to Reference
Time Point
Char
BEFORE,
COINCIDENT,
AFTER,
ONGOING, U
Timing
Identifies the end of the medication as being before or after the reference
time point defined by variable CMENTPT.
Perm
SDTM 2.2.5,
490HSDTMIG 4.1.4.7
CMENTPT
End Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the reference
point referred to by CMENRTPT. Examples: "2003-12-25" or "VISIT 2".
Perm
SDTM 2.2.5,
491HSDTMIG 4.1.4.7
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.1.1.1 ASSUMPTIONS FOR CONCOMITANT MEDICATIONS DOMAIN MODEL
1. CM Definition and Structure
a. CRF data that captures the Concomitant and Prior Medications/Therapies used by the subject. Examples are the Concomitant Medications/Therapies
given on an as-needed basis and the usual and background medications/therapies given for a condition.
b. The structure of the CM domain is one record per medication intervention episode, constant-dosing interval, or pre-specified medication assessment per
subject. It is the sponsor's responsibility to define an intervention episode. This definition may vary based on the sponsor's requirements for review and
analysis. The submission dataset structure may differ from the structure used for collection. One common approach is to submit a new record when there
is a change in the dosing regimen. Another approach is to collapse all records for a medication to a summary level with either a dose range or the highest
dose level. Other approaches may also be reasonable as long as they meet the sponsor's evaluation requirements.
2. Concomitant Medications Description and Coding
a. CMTRT captures the name of the Concomitant Medications/Therapy and it is the topic variable. It is a required variable and must have a value. CMTRT
should only include the medication/therapy name and should not include dosage, formulation, or other qualifying information. For example, ―ASPIRIN
100MG TABLET‖ is not a valid value for CMTRT. This example should be expressed as CMTRT= ―ASPIRIN‖, CMDOSE= ―100‖, CMDOSU= ―MG‖,
and CMDOSFRM= ―TABLET‖.
b. CMMODIFY should be included if the sponsor‘s procedure permits modification of a verbatim term for coding.
c. CMDECOD is the standardized medication/therapy term derived by the sponsor from the coding dictionary. It is expected that the reported term
(CMTRT) or the modified term (CMMODIFY) will be coded using a standard dictionary. The sponsor is expected to provide the dictionary name and
version used to map the terms utilizing the define.xml external codelist attributes.
3. Pre-specified Terms; Presence or Absence of Concomitant Medications
a. Information on concomitant medications is generally collected in two different ways, either by recording free text or using a pre-specified list of terms.
Since the solicitation of information on specific concomitant medications may affect the frequency at which they are reported, the fact that a specific
medication was solicited may be of interest to reviewers. CMPRESP and CMOCCUR are used together to indicate whether the intervention in CMTRT
was pre-specified and whether it occurred , respectively.
b. CMOCCUR is used to indicate whether a pre-specified medication was used. A value of Y indicates that the medication was used and N indicates that it was not.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 79
Final November 12, 2008
c. If a medication was not pre-specified the value of CMOCCUR should be null. CMPRESP and CMOCCUR is a permissible fields and may be omitted
from the dataset if all medications were collected as free text. Values of CMOCCUR may also be null for pre-specified medications if no Y/N response
was collected; in this case, CMSTAT = NOT DONE, and CMREASND could be used to describe the reason the answer was missing.
4. Additional Timing Variables
a. CMSTRTPT, CMSTTPT, CMENRTPT and CMENTPT may be populated as necessary to indicate when a medication was used relative to specified time
points. For example, assume a subject uses birth control medication. The subject has used the same medication for many years and continues to do so.
The date the subject began using the medication (or at least a partial date) would be stored in CMSTDTC. CMENDTC is null since the end date is
unknown (it hasn‘t happened yet). This fact can be recorded by setting CMENTPT=‖2007-04-30‖ (the date the assessment was made) and
CMENRTPT=‖ONGOING‖.
5. Additional Permissible Interventions Qualifiers
a. Any additional Qualifiers from the Interventions Class may be added to this domain.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 80 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.1.1.2 EXAMPLES FOR CONCOMITANT MEDICATIONS DOMAIN MODEL
Example 1: Spontaneous concomitant medications with dosing information
Sponsors collect the timing of concomitant medication use with varying specificity, depending on the pattern of use; the type, purpose, and importance of the
medication; and the needs of the study. It is often unnecessary to record every unique instance of medication use, since the same information can be conveyed
with start and end dates and frequency of use. If appropriate, medications taken as needed (intermittently or sporadically over a time period) may be reported
with a start and end date and a frequency of ―PRN‖.
The example below shows three subjects who took the same medication on the same day.
Rows 1-6: For the first subject (USUBJID=ABC-0001, each instance is recorded separately, and frequency (CMDOSFRQ) is ONCE.
Rows 7-9: For the second subject (USUBJID=ABC-0002, the second record (CMSEQ=2) shows that aspirin was taken twice on January 7th, so the frequency
is BID. The frequency is also included for the other daily records to avoid confusion.
Row 10: Records for the third subject are collapsed (this is shown as an example only, not as a recommendation) into a single entry that spans the relevant
time period, with a frequency of PRN. This approach assumes that knowing exactly when aspirin was used is not important for evaluating safety
and efficacy in this study.
Row
STUDYID
DOMAIN
USUBJID
CMSEQ
CMTRT
CMDOSE
CMDOSU
CMDOSFRQ
CMSTDTC
CMENDTC
1
ABC
CM
ABC-0001
1
ASPIRIN
100
MG
ONCE
2004-01-01
2004-01-01
2
ABC
CM
ABC-0001
2
ASPIRIN
100
MG
ONCE
2004-01-02
2004-01-02
3
ABC
CM
ABC-0001
3
ASPIRIN
100
MG
ONCE
2004-01-03
2004-01-03
4
ABC
CM
ABC-0001
4
ASPIRIN
100
MG
ONCE
2004-01-07
2004-01-07
5
ABC
CM
ABC-0001
5
ASPIRIN
100
MG
ONCE
2004-01-07
2004-01-07
6
ABC
CM
ABC-0001
6
ASPIRIN
100
MG
ONCE
2004-01-09
2004-01-09
7
ABC
CM
ABC-0002
1
ASPIRIN
100
MG
Q24H
2004-01-01
2004-01-03
8
ABC
CM
ABC-0002
2
ASPIRIN
100
MG
BID
2004-01-07
2004-01-07
9
ABC
CM
ABC-0002
3
ASPIRIN
100
MG
Q24H
2004-01-09
2004-01-09
10
ABC
CM
ABC-0003
1
ASPIRIN
100
MG
PRN
2004-01-01
2004-01-09
Example 2: Spontaneous concomitant medications without dosing information
The example below is for a study that has a particular interest in whether subjects use any anticonvulsant medications. The medication history, dosing, etc. are
not of interest; the study only asks for the anticonvulsants to which subjects are being exposed.
Row
STUDYID
DOMAIN
USUBJID
CMSEQ
CMTRT
CMCAT
1
ABC123
CM
1
1
LITHIUM
ANTI-CONVULSANT
2
ABC123
CM
2
1
VPA
ANTI-CONVULSANT
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 81
Final November 12, 2008
Example 3: Pre-specified concomitant medications using CMPRESP, CMOCCUR, CMSTAT, and CMREASND
Sponsors often are interested in whether subjects are exposed to specific concomitant medications, and collect this information using a checklist. The example
below is for a study that has a particular interest in the antidepressant medications that subjects use. For the study‘s purposes, the absence is just as important as
the presence of a medication. This can be clearly shown by using CMOCCUR.
In this example, CMPRESP shows that the subjects were specifically asked if they use any of three antidepressants (Zoloft, Prozac, or Paxil). The value of
CMOCCUR indicates the response to the pre-specified medication question. CMSTAT indicates whether the response was missing for a pre-specified
medication, and CMREASND shows the reason for missing response. The medication details (e.g., dose, frequency) were not of interest in this study.
Row 1: Medication was solicited on CRF and was taken.
Row 2: Medication use solicited in CRF and was not taken.
Row 3: Medication use solicited in CRF but data was not collected.
Row
STUDYID
DOMAIN
USUBJID
CMSEQ
CMTRT
CMPRESP
CMOCCUR
CMSTAT
CMREASND
1
ABC123
CM
1
1
ZOLOFT
Y
Y
2
ABC123
CM
1
2
PROZAC
Y
N
3
ABC123
CM
1
3
PAXIL
Y
NOT DONE
Didn't ask due to interruption
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 82 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.1.2 EXPOSURE — EX
ex.xpt, Exposure — Interventions, Version 3.1.2. One record per constant dosing interval per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1888HEX
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
492HSDTMIG 4.1.2.2,
493HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
494H495HSDTMIG 4.1.2.3
EXSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
EXGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4
496HSDTMIG 4.1.2.6
EXSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a CRF Page.
Perm
SDTM 2.2.4
EXTRT
Name of Actual Treatment
Char
Topic
Name of the intervention treatment — usually the verbatim name of the
investigational treatment given during the dosing period for the observation.
Req
SDTM 2.2.1
EXCAT
Category for Treatment
Char
*
Grouping
Qualifier
Used to define a category of related records. Example: COMPARATOR
CLASS.
Perm
SDTM 2.2.1,
497HSDTMIG 4.1.2.6
EXSCAT
Subcategory for Treatment
Char
*
Grouping
Qualifier
A further categorization of treatment.
Perm
SDTM 2.2.1,
498HSDTMIG 4.1.2.6
EXDOSE
Dose per Administration
Num
Record
Qualifier
Amount of EXTRT administered or given.
Exp
SDTM 2.2.1
EXDOSTXT
Dose Description
Char
Record
Qualifier
Dosing amounts or a range of dosing information collected in text form.
Example: 200-400.
Perm
SDTM 2.2.1
EXDOSU
Dose Units
Char
(499HUNIT)
Variable
Qualifier
Units for EXDOSE and EXDOSTOT. Examples: ng, mg, or mg/kg.
Exp
SDTM 2.2.1,
500H501HSDTMIG 4.1.3.2
EXDOSFRM
Dose Form
Char
(1889HFRM)
Record
Qualifier
Dose form for EXTRT. Examples: TABLET, LOTION.
Exp
SDTM 2.2.1
EXDOSFRQ
Dosing Frequency per
Interval
Char
(502HFREQ)
Variable
Qualifier
Usually expressed as the number of repeated administrations of EXDOSE
within a specific time period. Examples: BID (twice daily), Q4S (once
every four weeks), BIS (twice a week).
Perm
SDTM 2.2.1
EXDOSTOT
Total Daily Dose
Num
Record
Qualifier
Total daily dose of EXTRT using the units in EXDOSU. Total dose over a
period other than day could be recorded in a separate Supplemental
Qualifier variable.
Perm
SDTM 2.2.1
EXDOSRGM
Intended Dose Regimen
Char
Variable
Qualifier
Text description of the (intended) schedule or regimen for the
Intervention. Examples: TWO WEEKS ON, TWO WEEKS OFF.
Perm
SDTM 2.2.1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 83
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
EXROUTE
Route of Administration
Char
(1890HROUTE)
Variable
Qualifier
Route of administration for EXTRT. Examples: ORAL,
INTRAVENOUS.
Perm
SDTM 2.2.1
EXLOT
Lot Number
Char
Record
Qualifier
Lot Number of the EXTRT product.
Perm
SDTM 2.2.1
EXLOC
Location of Dose
Administration
Char
(503HLOC)
Record
Qualifier
Specifies location of administration. Example: LEFT ARM for a topical
application.
Perm
SDTM 2.2.1
EXTRTV
Treatment Vehicle
Char
*
Record
Qualifier
Describes vehicle used for treatment. Example: SALINE.
Perm
SDTM 2.2.1
EXVAMT
Treatment Vehicle
Amount
Num
Variable
Qualifier
Amount administered of the treatment vehicle indicated by EXTRTV
Perm
SDTM 2.2.1
EXVAMTU
Treatment Vehicle
Amount Units
Char
(504HUNIT)
Variable
Qualifier
Units of the treatment vehicle amount indicated by EXVAMT
Perm
SDTM 2.2.1
EXADJ
Reason for Dose
Adjustment
Char
*
Record
Qualifier
Describes reason or explanation of why a dose is adjusted – used only
when an adjustment is represented in EX.
Perm
SDTM 2.2.1
TAETORD
Order of Element within
Arm
Num
Timing
Number that gives the order of the Element within the Arm.
Perm
SDTM 2.2.5,
505HSDTMIG 5.3.1
EPOCH
Epoch
Char
*
Timing
Trial Epoch of the Exposure record. Examples: SCREENING,
TREATMENT PHASE, FOLLOW-UP
Perm
SDTM 2.2.5,
506HSDTMIG 7.1.2
EXSTDTC
Start Date/Time of
Treatment
Char
ISO 8601
Timing
The time when administration of the treatment indicated by EXTRT and
EXDOSE began.
Exp
SDTM 2.2.5,
507HSDTMIG 4.1.4.1,
508HSDTMIG 4.1.4.3
EXENDTC
End Date/Time of
Treatment
Char
ISO 8601
Timing
The time when administration of the treatment indicated by EXTRT and
EXDOSE ended.
Perm
SDTM 2.2.5,
509HSDTMIG 4.1.4.1,
510HSDTMIG 4.1.4.3
EXSTDY
Study Day of Start of
Treatment
Num
Timing
Study day of start of treatment relative to the sponsor-defined RFSTDTC.
Perm
SDTM 2.2.5,
511HSDTMIG 4.1.4.4,
512HSDTMIG 4.1.4.6
EXENDY
Study Day of End of
Treatment
Num
Timing
Study day of end of treatment relative to the sponsor-defined RFSTDTC.
Perm
SDTM 2.2.5,
513HSDTMIG 4.1.4.4,
514HSDTMIG 4.1.4.6
EXDUR
Duration of Treatment
Char
ISO 8601
Timing
Collected duration and unit of a treatment. Used only if collected on the
CRF and not derived from start and end date/times.
Perm
SDTM 2.2.5,
515HSDTMIG 4.1.4.3
EXTPT
Planned Time Point Name
Char
Timing
1. Text Description of time when a dose should be given.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See EXTPTNUM and EXTPTREF.
Examples: Start or 5 min post.
Perm
SDTM 2.2.5,
516H517HSDTMIG 4.1.4.10
EXTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of EXTPT to aid in sorting.
Perm
SDTM 2.2.5,
518H519HSDTMIG 4.1.4.10
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 84 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
EXELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Planned elapsed time (in ISO 8601 format) relative to the planned fixed
reference (EXTPTREF). This variable is useful where there are repetitive
measures. Not a clock time. Represented as an ISO duration.
Perm
SDTM 2.2.5,
520H521HSDTMIG 4.1.4.10
EXTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by EXELTM, EXTPTNUM,
and EXTPT. Examples: PREVIOUS DOSE, PREVIOUS MEAL.
Perm
SDTM 2.2.5,
522HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.1.2.1 ASSUMPTIONS FOR EXPOSURE DOMAIN MODEL
1. EX Definition
a. The Exposure domain model records the details of a subject‘s exposure to protocol-specified study treatment. Study treatment may be any intervention
that is prospectively defined as a test material within a study, and is typically but not always supplied to the subject. Examples include but are not
limited to placebo, active comparators, and investigational products. Treatments that are not protocol-specified should be recorded in the Concomitant
Medication (CM) domain.
b. This domain should contain one record per constant dosing interval per subject. "Constant dosing interval" is sponsor-defined, and may include any
period of time that can be described in terms of a known treatment given at a consistent dose and frequency. For example, for a study with once-a-week
administration of a standard dose for 6 weeks, exposure may be represented as one of the following:
If information about each dose is not collected, there would be a single record per subject, spanning the entire treatment phase
If the sponsor monitors each treatment administration and deviations in treatment or dose occur, there could be up to six records (one for
each weekly administration).
c. The Exposure domain is required for all studies that include investigational product. Exposure information may be directly or indirectly determined.
Regardless of how it is known, it must be represented using the Exposure domain, and the metadata should explain how it was populated. Common
methods for determining exposure (from most direct to least direct) include the following:
1) Actual observation of the administration of drug by the investigator
2) Automated dispensing device which records administrations
3) Subject recall (e.g., via diary)
4) Derived from drug accountability data (e.g., pill counts)
5) Derived from the protocol
2. Categorization and Grouping
a. EXCAT and EXSCAT may be used when appropriate to categorize treatments into categories and subcategories. For example, if a study contains
several active comparator medications, EXCAT may be set to ―ACTIVE COMPARATOR.‖ Such categorization may not be useful in most studies, so
these variables are permissible but not expected.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 85
Final November 12, 2008
3. Exposure Treatment Description
a. EXTRT captures the name of the investigational treatment and it is the topic variable. It is a required variable and must have a value. EXTRT should
only include the treatment name and should not include dosage, formulation or other qualifying information. For example, ―ASPIRIN 100MG
TABLET‖ is not a valid value for EXTRT. This example should be expressed as EXTRT= ―ASPIRIN‖, EXDOSE= ―100‖, EXDOSU= ―mg‖, and
EXDOSFRM= ―TABLET‖.
b. Doses of placebo should be represented as per Exposure Example 5 below.
4. Timing Variables
a. The timing of exposure to study treatment is captured by the start/end date and start/end time of each constant dosing interval. If the subject is only
exposed to study medication within a clinical encounter (e.g., if an injection is administered at the clinic), VISITNUM may be added to the domain as
an additional timing variable. VISITDY and VISIT would then also be permissible Qualifiers. However if the beginning and end of a constant dosing
interval is not confined within the time limits of a clinical encounter (e.g., if a subjects takes pills at home), then it is not appropriate to include
VISITNUM in the EX domain. This is because EX is designed to capture the timing of exposure to treatment, not the timing of dispensing treatment.
Furthermore, VISITNUM should not be used to indicate that treatment began at a particular visit and continued for a period of time. The SDTM does
not have any provision for recording "start visit" and "end visit" since such information is redundant with start date/time and end date/time.
5. Additional Interventions Qualifiers
b. The variables --PRESP, --OCCUR, --STAT, and --REASND from the Interventions general observation class would not generally be used in the EX
domain because EX should only contain medications received.
c. Other additional Qualifiers from the SDTM Interventions Class may be added to this domain.
6.1.2.2 EXAMPLES FOR EXPOSURE DOMAIN MODEL
Example 1:
This is an example of an Exposure dataset for a parallel-design study. In this example, subjects were randomized to one of three treatment groups: Drug A 40 mg
Q24H, Drug A 20 mg Q24H, or Drug B 150 mg BID. Drug C was assigned as supplemental therapy for the three groups. The study included 8 weeks of treatment,
with subjects remaining on the same treatment throughout the study. With respect to timing of doses, the sponsor only collected the start and stop dates of
uninterrupted periods of treatment. Note below that Subject 12345003 missed taking study medications on Study Days 23 and 24.
Row
STUDYID
DOMAIN
USUBJID
EXSEQ
EXTRT
EXDOSE
EXDOSU
EXDOSFRM
EXDOSFRQ
EXDOSTOT
EXROUTE
EXSTDTC
EXENDTC
EXSTDY
EXENDY
1
12345
EX
12345001
1
DRUG A
40
mg
TABLET
Q24H
40
ORAL
2002-01-10
2002-03-08
1
58
2
12345
EX
12345001
2
DRUG C
30
mg
CAPSULE
BID
60
ORAL
2002-01-10
2002-03-08
1
58
3
12345
EX
12345002
1
DRUG A
20
mg
TABLET
Q24H
20
ORAL
2002-01-10
2002-03-07
1
57
4
12345
EX
12345002
2
DRUG C
30
mg
CAPSULE
BID
60
ORAL
2002-01-10
2002-03-07
1
57
5
12345
EX
12345003
1
DRUG C
30
mg
CAPSULE
BID
60
ORAL
2002-01-11
2002-02-01
1
22
6
12345
EX
12345003
2
DRUG B
150
mg
TABLET
BID
300
ORAL
2002-01-11
2002-02-01
1
22
7
12345
EX
12345003
3
DRUG C
30
mg
CAPSULE
BID
60
ORAL
2002-02-04
2002-03-06
25
55
8
12345
EX
12345003
4
DRUG B
150
mg
TABLET
BID
300
ORAL
2002-02-04
2002-03-06
25
55
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 86 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 2:
This is an example of an Exposure dataset for a single crossover study comparing once daily oral administration of Drug A 20 mg capsules with Drug B 30 mg
coated tablets. Study drug was taken for 3 consecutive mornings 30 minutes prior to a standardized breakfast. There was a 6-day washout period between
treatments.
Row
STUDYID
DOMAIN
USUBJID
EXSEQ
EXGRPID
EXTRT
EXDOSE
EXDOSU
EXDOSFRM
EXDOSFRQ
1
56789
EX
56789001
1
1
DRUG A
20
mg
CAPSULE
Q24H
2
56789
EX
56789001
2
1
DRUG A
20
mg
CAPSULE
Q24H
3
56789
EX
56789001
3
1
DRUG A
20
mg
CAPSULE
Q24H
4
56789
EX
56789001
4
2
DRUG B
30
mg
TABLET, COATED
Q24H
5
56789
EX
56789001
5
2
DRUG B
30
mg
TABLET, COATED
Q24H
6
56789
EX
56789001
6
2
DRUG B
30
mg
TABLET, COATED
Q24H
7
56789
EX
56789003
1
1
DRUG B
30
mg
TABLET, COATED
Q24H
8
56789
EX
56789003
2
1
DRUG B
30
mg
TABLET, COATED
Q24H
9
56789
EX
56789003
3
1
DRUG B
30
mg
TABLET, COATED
Q24H
10
56789
EX
56789003
4
2
DRUG A
20
mg
CAPSULE
Q24H
11
56789
EX
56789003
5
2
DRUG A
20
mg
CAPSULE
Q24H
12
56789
EX
56789003
6
2
DRUG A
20
mg
CAPSULE
Q24H
Row
EXDOSTOT
EXROUTE
EXSTDTC
EXENDTC
EXSTDY
EXENDY
EXTPT
EXTPTREF
1 (cont)
20
ORAL
2002-07-01T07:30
2002-07-01T07:30
1
1
30 MINUTES PRIOR
STD BREAKFAST
2 (cont)
20
ORAL
2002-07-02T07:30
2002-07-02T07:30
2
2
30 MINUTES PRIOR
STD BREAKFAST
3 (cont)
20
ORAL
2002-07-03T07:32
2002-07-03T07:32
3
3
30 MINUTES PRIOR
STD BREAKFAST
4 (cont)
30
ORAL
2002-07-09T07:30
2002-07-09T07:30
9
9
30 MINUTES PRIOR
STD BREAKFAST
5 (cont)
30
ORAL
2002-07-10T07:30
2002-07-10T07:30
10
10
30 MINUTES PRIOR
STD BREAKFAST
6 (cont)
30
ORAL
2002-07-11T07:34
2002-07-11T07:34
11
11
30 MINUTES PRIOR
STD BREAKFAST
7 (cont)
30
ORAL
2002-07-03T07:30
2002-07-03T07:30
1
1
30 MINUTES PRIOR
STD BREAKFAST
8 (cont)
30
ORAL
2002-07-04T07:24
2002-07-04T07:24
2
2
30 MINUTES PRIOR
STD BREAKFAST
9 (cont)
30
ORAL
2002-07-05T07:24
2002-07-05T07:24
3
3
30 MINUTES PRIOR
STD BREAKFAST
10 (cont)
20
ORAL
2002-07-11T07:30
2002-07-11T07:30
9
9
30 MINUTES PRIOR
STD BREAKFAST
11 (cont)
20
ORAL
2002-07-12T07:43
2002-07-12T07:43
10
10
30 MINUTES PRIOR
STD BREAKFAST
12 (cont)
20
ORAL
2002-07-13T07:38
2002-07-13T07:38
11
11
30 MINUTES PRIOR
STD BREAKFAST

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 87
Final November 12, 2008
Example 3:
This is an example of an Exposure dataset for an open-label study examining the tolerability of different doses of Drug A. Study drug was taken daily for three
months. Dose adjustments were allowed as needed in response to tolerability or efficacy issues.
Row
STUDYID
DOMAIN
USUBJID
EXSEQ
EXTRT
EXDOSE
EXDOSU
EXDOSFRM
EXADJ
EXSTDTC
EXENDTC
1
37841
EX
37841001
1
DRUG A
20
mg
TABLET
2002-07-01
2002-10-01
2
37841
EX
37841002
1
DRUG A
20
mg
TABLET
2002-04-02
2002-04-21
3
37841
EX
37841002
2
DRUG A
15
mg
TABLET
Reduced due to toxicity
2002-04-22
2002-07-01
4
37841
EX
37841003
1
DRUG A
20
mg
TABLET
2002-05-09
2002-06-01
5
37841
EX
37841003
2
DRUG A
25
mg
TABLET
Increased due to
suboptimal efficacy
2002-06-02
2002-07-01
6
37841
EX
37841003
3
DRUG A
30
mg
TABLET
Increased due to
suboptimal efficacy
2002-07-02
2002-08-01

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 88 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 4:
This is an example of a titration Exposure dataset for a study that gradually increases dosage while simultaneously evaluating efficacy and toleration of the
treatment regimen. The schedule specifies that Drug A be administered twice daily starting with 100 mg for 3 days, then increase to 200 mg daily for 3 days, then
increase further in 100-mg increments every three days until signs of intolerance are noted or no improvement in efficacy is observed.
Row
STUDYID
DOMAIN
USUBJID
EXSEQ
EXGRPID
EXTRT
EXDOSE
EXDOSU
EXDOSFRM
EXDOSFRQ
1
70912
EX
23301996
1
1
DRUG A
100
mg
CAPSULE
BID
2
23301
EX
23301996
2
1
DRUG A
100
mg
CAPSULE
BID
3
23301
EX
23301996
3
1
DRUG A
100
mg
CAPSULE
BID
4
23301
EX
23301996
4
2
DRUG A
200
mg
CAPSULE
BID
5
23301
EX
23301996
5
2
DRUG A
200
mg
CAPSULE
BID
6
23301
EX
23301996
6
2
DRUG A
200
mg
CAPSULE
BID
7
23301
EX
23301996
7
1
DRUG A
300
mg
CAPSULE
BID
8
23301
EX
23301996
8
1
DRUG A
300
mg
CAPSULE
BID
9
23301
EX
23301996
9
1
DRUG A
300
mg
CAPSULE
BID
10
23301
EX
23301996
10
2
DRUG A
400
mg
CAPSULE
BID
11
23301
EX
23301996
11
2
DRUG A
400
mg
CAPSULE
BID
12
23301
EX
23301996
12
2
DRUG A
400
mg
CAPSULE
BID
Row
EXDOSTOT
EXROUTE
EXSTDTC
EXENDTC
EXSTDY
EXENDY
1 (cont)
200
ORAL
2004-07-01T07:30
2004-07-01T07:30
1
1
2 (cont)
200
ORAL
2004-07-02T07:30
2004-07-02T07:30
2
2
3 (cont)
200
ORAL
2004-07-03T07:32
2004-07-03T07:32
3
3
4 (cont)
400
ORAL
2004-07-09T07:30
2004-07-09T07:30
9
9
5 (cont)
400
ORAL
2004-07-10T07:30
2004-07-10T07:30
10
10
6 (cont)
400
ORAL
2004-07-11T07:34
2004-07-11T07:34
11
11
7 (cont)
600
ORAL
2004-07-01T07:30
2004-07-01T07:30
1
1
8 (cont)
600
ORAL
2004-07-02T07:30
2004-07-02T07:30
2
2
9 (cont)
600
ORAL
2004-07-03T07:32
2004-07-03T07:32
3
3
10 (cont)
800
ORAL
2004-07-09T07:30
2004-07-09T07:30
9
9
11 (cont)
800
ORAL
2004-07-10T07:30
2004-07-10T07:30
10
10
12 (cont)
800
ORAL
2004-07-11T07:34
2004-07-11T07:34
11
11
Example 5:
The table below presents data for a study comparing low dose aspirin to placebo. Two rows are shown: one for a subject receiving active study drug and one for a
subject receiving placebo.
USUBJID
EXSEQ
EXTRT
EXDOSE
EXDOSU
EXDOSEFRM
EXDOSFRQ
2008-039-001
1
Aspirin
81
mg
TABLET
QD
2008-039-002
1
Placebo
0
mg
TABLET
QD
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 89
Final November 12, 2008
6.1.3 SUBSTANCE USE — SU
su.xpt, Substance Use — Interventions, Version 3.1.2. One record per substance type per reported occurrence per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms or
Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1891HSU
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
523HSDTMIG 4.1.2.2,
524HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
525H526HSDTMIG 4.1.2.3
SUSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
SUGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4
527HSDTMIG 4.1.2.6
SUSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a Tobacco & Alcohol use CRF page.
Perm
SDTM 2.2.4
SUTRT
Reported Name of
Substance
Char
Topic
Substance name. Examples: Cigarettes, Coffee.
Req
SDTM 2.2.1
SUMODIFY
Modified Substance Name
Char
Synonym
Qualifier
If SUTRT is modified, then the modified text is placed here.
Perm
SDTM 2.2.1,
528HSDTMIG 4.1.3.6
SUDECOD
Standardized Substance
Name
Char
*
Synonym
Qualifier
Standardized or dictionary-derived text description of SUTRT or
SUMODIFY if the sponsor chooses to code the substance use. The
sponsor is expected to provide the dictionary name and version used to
map the terms utilizing the define.xml external codelist attributes.
Perm
SDTM 2.2.1,
529HSDTMIG 4.1.3.6
SUCAT
Category for Substance
Use
Char
*
Grouping
Qualifier
Used to define a category of related records. Examples: TOBACCO,
ALCOHOL, or CAFFEINE.
Perm
SDTM 2.2.1,
530HSDTMIG 4.1.2.6
SUSCAT
Subcategory for Substance
Use
Char
*
Grouping
Qualifier
A further categorization of substance use. Examples: CIGARS,
CIGARETTES, BEER, WINE
Perm
SDTM 2.2.1,
531HSDTMIG 4.1.2.6
SUPRESP
SU Pre-Specified
Char
(1892HNY)
Record
Qualifier
Used to indicate whether (Y/null) information about the use of a specific
substance was solicited on the CRF.
Perm
SDTM 2.2.1,
532HSDTMIG 4.1.2.7.3,
533HSDTMIG 4.1.5.7
SUOCCUR
SU Occurrence
Char
(1893HNY)
Record
Qualifier
When the use of specific substances is solicited, SUOCCUR is used to
indicate whether or not (Y/N) a particular pre-specified substance was
used. Values are null for substances not specifically solicited.
Perm
SDTM 2.2.1,
534HSDTMIG 4.1.5.7
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 90 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms or
Format
Role
CDISC Notes
Core
References
SUSTAT
Completion Status
Char
(1894HND)
Record
Qualifier
When the use of pre-specified substances is solicited, the completion status
indicates that there was no response to the question about the pre-specified
substance. When there is no pre-specified list on the CRF, then the completion
status indicates that substance use was not assessed for the subject.
Perm
SDTM 2.2.1,
535HSDTMIG 4.1.5.1,
536HSDTMIG 4.1.5.7
SUREASND
Reason Substance Use Not
Collected
Char
Record
Qualifier
Describes the reason substance use was not collected. Used in conjunction
with SUSTAT when value of SUSTAT is NOT DONE.
Perm
SDTM 2.2.1,
537HSDTMIG 4.1.5.1,
538HSDTMIG 4.1.5.7
SUCLAS
Substance Use Class
Char
*
Variable
Qualifier
Substance use class. May be obtained from coding. When coding to a
single class, populate with class value. If using a dictionary and coding to
multiple classes, 539Hthen follow 464Hassumption 4.1.2.8.3 or omit SUCLAS.
Perm
SDTM 2.2.1,
540H541H542H543HSDTMIG 4.1.3.5
SUCLASCD
Substance Use Class Code
Char
*
Variable
Qualifier
Code corresponding to SUCLAS. May be obtained from coding.
Perm
SDTM 2.2.1,
544H545H546HSDTMIG 4.1.3.5
SUDOSE
Substance Use
Consumption
Num
Record
Qualifier
Amount of SUTRT consumed.
Perm
SDTM 2.2.1
SUDOSTXT
Substance Use
Consumption Text
Char
Record
Qualifier
Substance use consumption amounts or a range of consumption
information collected in text form.
Perm
SDTM 2.2.1
SUDOSU
Consumption Units
Char
(547HUNIT)
Variable
Qualifier
Units for SUDOSE, SUDOSTXT, and SUDOSTOT. Examples:
OUNCES, CIGARETTE EQUIVALENTS, or GRAMS.
Perm
SDTM 2.2.1,
548HSDTMIG 4.1.3.2
SUDOSFRM
Dose Form
Char
*
Record
Qualifier
Dose form for SUTRT. Examples: INJECTABLE, LIQUID, or
POWDER.
Perm
SDTM 2.2.1
SUDOSFRQ
Use Frequency Per Interval
Char
(549HFREQ)
Variable
Qualifier
Usually expressed as the number of repeated administrations of SUDOSE
within a specific time period. Example: Q24H (every day)
Perm
SDTM 2.2.1
SUDOSTOT
Total Daily Consumption
Num
Record
Qualifier
Total daily use of SUTRT using the units in SUDOSU. If sponsor needs to
aggregate the data over a period other than daily, then the aggregated total
could be recorded in a Supplemental Qualifier variable.
Perm
SDTM 2.2.1
SUROUTE
Route of Administration
Char
(1895HROUTE)
Variable
Qualifier
Route of administration for SUTRT. Examples: ORAL,
INTRAVENOUS.
Perm
SDTM 2.2.1
SUSTDTC
Start Date/Time of
Substance Use
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
550HSDTMIG 4.1.4.1,
551HSDTMIG 4.1.4.3
SUENDTC
End Date/Time of
Substance Use
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
552HSDTMIG 4.1.4.1,
553HSDTMIG 4.1.4.3
SUSTDY
Study Day of Start of
Substance Use
Num
Timing
Study day of start of substance use relative to the sponsor-defined
RFSTDTC.
Perm
SDTM 2.2.5,
554HSDTMIG 4.1.4.4,
555HSDTMIG 4.1.4.6
SUENDY
Study Day of End of
Substance Use
Num
Timing
Study day of end of substance use relative to the sponsor-defined
RFSTDTC.
Perm
SDTM 2.2.5,
556HSDTMIG 4.1.4.4,
557HSDTMIG 4.1.4.6
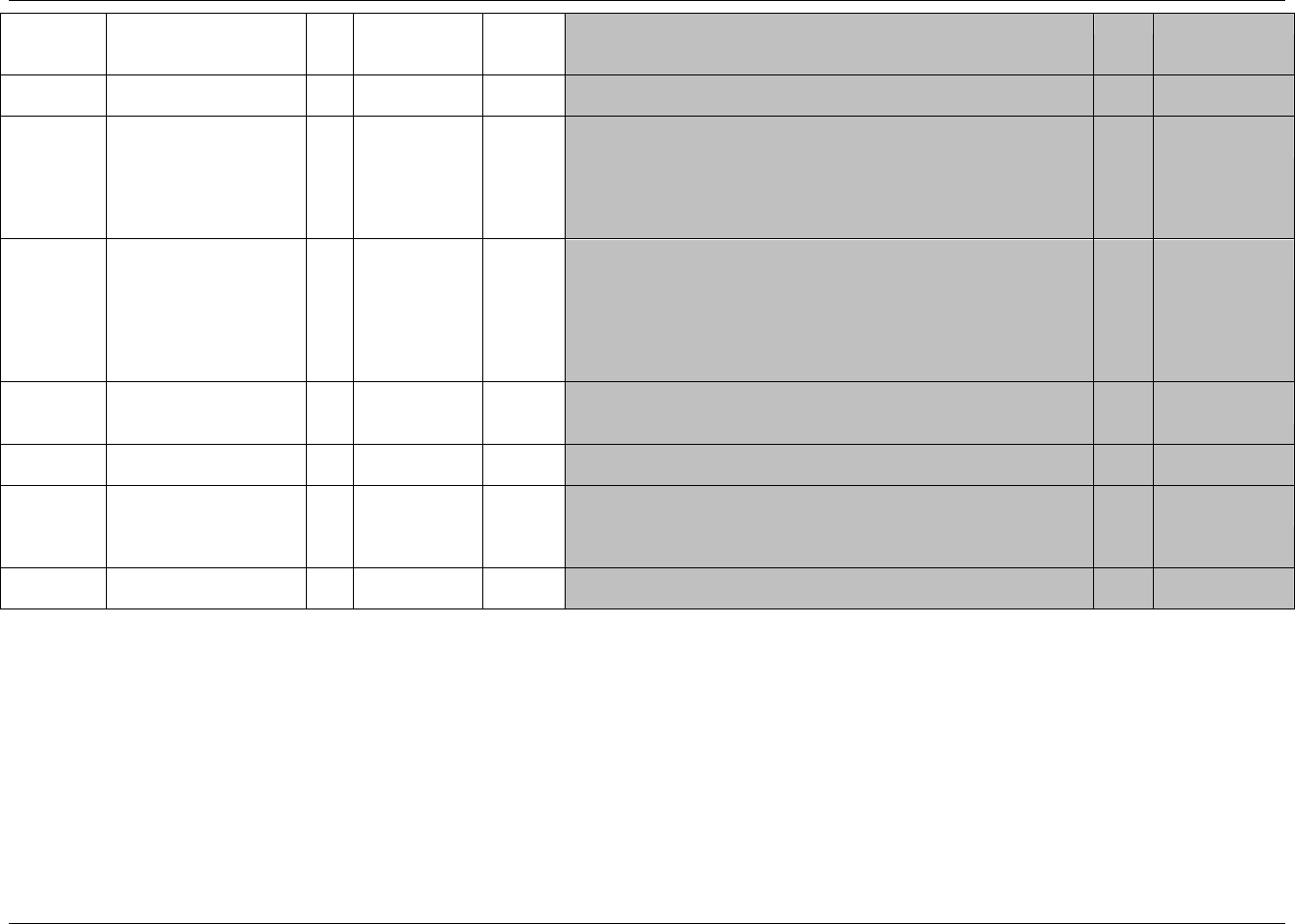
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 91
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms or
Format
Role
CDISC Notes
Core
References
SUDUR
Duration of Substance Use
Char
ISO 8601
Timing
Collected duration of substance use in ISO 8601 format. Used only if
collected on the CRF and not derived from start and end date/times.
Perm
SDTM 2.2.5,
558HSDTMIG 4.1.4.3
SUSTRF
Start Relative to Reference
Period
Char
(1896HSTENRF)
Timing
Describes the start of the substance use relative to the sponsor-defined
reference period. The sponsor-defined reference period is a continuous
period of time defined by a discrete starting point and a discrete ending
point (represented by RFSTDTC and RFENDTC in Demographics). If
information such as "PRIOR", "ONGOING", or "CONTINUING" was
collected, this information may be translated into SUSTRF.
Perm
SDTM 2.2.5,
559HSDTMIG 4.1.4.7
SUENRF
End Relative to Reference
Period
Char
(1897HSTENRF)
Timing
Describes the end of the substance use with relative to the sponsor-
defined reference period. The sponsor-defined reference period is a
continuous period of time defined by a discrete starting point and a
discrete ending point (represented by RFSTDTC and RFENDTC in
Demographics). If information such as "PRIOR", "ONGOING", or
"CONTINUING" was collected, this information may be translated into
SUENRF.
Perm
SDTM 2.2.5,
560HSDTMIG 4.1.4.7
SUSTRTPT
Start Relative to Reference
Time Point
Char
BEFORE,
COINCIDENT,
AFTER, U
Timing
Identifies the start of the substance as being before or after the reference
time point defined by variable SUSTTPT.
Perm
SDTM 2.2.5,
561HSDTMIG 4.1.4.7
SUSTTPT
Start Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the reference
point referred to by SUSTRTPT. Examples: "2003-12-15" or "VISIT 1".
Perm
SDTM 2.2.5,
562HSDTMIG 4.1.4.7
SUENRTPT
End Relative to Reference
Time Point
Char
BEFORE,
COINCIDENT,
AFTER,
ONGOING, U
Timing
Identifies the end of the substance as being before or after the reference
time point defined by variable SUENTPT.
Perm
SDTM 2.2.5,
563HSDTMIG 4.1.4.7
SUENTPT
End Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the reference
point referred to by SUENRTPT. Examples: "2003-12-25" or "VISIT 2".
Perm
SDTM 2.2.5,
564HSDTMIG 4.1.4.7
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 92 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.1.3.1 ASSUMPTIONS FOR SUBSTANCE USE DOMAIN MODEL
1. The intent of the domain is to capture substance use information that may be used to assess the efficacy and/or safety of therapies that look to mitigate the
effects of chronic substance use, or that could be used as covariates in other efficacy and/or safety analyses.
2. SU Definition
a. This information may be independent of planned study evaluations, or may be a key outcome (e.g., planned evaluation) of a clinical trial.
b. In many clinical trials, detailed substance use information as provided for in the domain model above may not be required (e.g., the only information
collected may be a response to the question ―Have you ever smoked tobacco?‖); in such cases, many of the Qualifier variables would not be submitted.
c. SU may contain responses to questions about use of pre-specified substances as well as records of substance use collected as free text.
3. Substance Use Description and Coding
a. SUTRT captures the verbatim or the pre-specified text collected for the substance. It is the topic variable for the SU dataset. SUTRT is a required
variable and must have a value.
b. SUMODIFY is a permissible variable and should be included if coding is performed and the sponsor‘s procedure permits modification of a verbatim
substance use term for coding. The modified term is listed in SUMODIFY. The variable may be populated as per the sponsor‘s procedures.
c. SUDECOD is the preferred term derived by the sponsor from the coding dictionary if coding is performed. It is a permissible variable. Where deemed
necessary by the sponsor, the verbatim term (SUTRT) should be coded using a standard dictionary such as WHO Drug. The sponsor is expected to
provide the dictionary name and version used to map the terms utilizing the define.xml external codelist attributes.
4. Additional Categorization and Grouping
a. SUCAT and SUSCAT should not be redundant with the domain code or dictionary classification provided by SUDECOD, or with SUTRT. That is,
they should provide a different means of defining or classifying SU records. For example, a sponsor may be interested in identifying all substances that
the investigator feels might represent opium use, and to collect such use on a separate CRF page. This categorization might differ from the
categorization derived from the coding dictionary.
b. SUGRPID may be used to link (or associate) different records together to form a block of related records within SU at the subject level (see 565HSection
4.1.2.6). It should not be used in place of SUCAT or SUSCAT.
5. Timing Variables
a. SUSTDTC and SUENDTC may be populated as required.
b. If substance use information is collected more than once within the CRF (indicating that the data are visit-based) then VISITNUM would be added to
the domain as an additional timing variable. VISITDY and VISIT would then be permissible variables.
6. Additional Permissible Interventions Qualifiers
a. Any additional Qualifiers from the Interventions Class may be added to this domain.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 93
Final November 12, 2008
6.1.3.2 EXAMPLE FOR SUBSTANCE USE DOMAIN MODEL
The example below illustrates how typical substance use data could be populated. Here, the CRF collected smoking data (smoking status: previous, current,
never; if a current or past smoker, how many packs per day; if a former smoker, what year did the subject quit) and current caffeine use (what caffeine drinks
have been consumed today; how many cups today). SUCAT allows the records to be grouped into smoking-related data and caffeine-related data. In this
example, the treatments are pre-specified on the CRF page so SUTRT does not require a standardized SUDECOD equivalent.
Row 1: Subject 1234005 is a 2-pack/day current smoker. ―Current‖ implies that smoking started sometime before the time the question was asked
(SUSTTPT = 2006-01-01, SUSTRTPT = BEFORE) and will end sometime after that date (SUENRTPT = ONGOING). See 566HSection 4.1.4.7 for the
use of these variables. Both the beginning and ending reference time points for this question are the date of the assessment.
Row 2: The same subject drank three cups of coffee on the day of the assessment.
Row 3: Subject 1234006 is a former smoker. The date the subject began smoking is unknown but we know that it was sometime before the assessment date.
This is shown by the values of SUSTTPT and SUSTRTPT (taken from the timing variables for all classes). The end date of smoking was collected
so SUENTPT and SUENRTPT are not populated. Instead, the end date is in SUENDTC.
Rows 4-5: The same subject drank tea (Row 4) and coffee (Row 5) on the day of the assessment.
Row 6: Subject 1234007 has missing data for the smoking questions. This is indicated by SUSTAT=NOT DONE. The reason is in SUREASND.
Row 7: The same subject also had missing data for all of the caffeine questions.
Not shown: Subject 1234008 has never smoked, so does not have a tobacco record. Alternatively, a row for the subject could have been included with
SUOCCUR=N and not populating the dosing and timing fields; the interpretation would be the same. The subject did not drink any caffeinated
drinks on the day of the assessment so does not have any caffeine records. Therefore this subject does not appear in the data.
Row
STUDYID
DOMAIN
USUBJID
SUSEQ
SUTRT
SUCAT
SUSTAT
SUREASND
SUDOSE
SUDOSU
SUDOSFRQ
1
1234
SU
1234005
1
CIGARETTES
TOBACCO
2
PACK
PER DAY
2
1234
SU
1234005
2
COFFEE
CAFFEINE
3
CUP
PER DAY
3
1234
SU
1234006
1
CIGARETTES
TOBACCO
1
PACK
PER DAY
4
1234
SU
1234006
2
TEA
CAFFEINE
1
CUP
PER DAY
5
1234
SU
1234006
3
COFFEE
CAFFEINE
2
CUP
PER DAY
6
1234
SU
1234007
1
CIGARETTES
TOBACCO
NOT
DONE
Subject left office before CRF
was completed
7
1234
SU
1234007
2
CAFFEINE
CAFFEINE
NOT
DONE
Subject left office before CRF
was completed
Row
SUSTDTC
SUENDTC
SUSTTPT
SUSTRTPT
SUENTPT
SUENRTPT
1 (cont)
2006-01-01
BEFORE
2006-01-01
ONGOING
2 (cont)
2006-01-01
2006-01-01
3 (cont)
2003
2006-03-15
BEFORE
4 (cont)
2006-03-15
2006-03-15
5 (cont)
2006-03-15
2006-03-15
6 (cont)
7 (cont)
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 94 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2 EVENTS
6.2.1 ADVERSE EVENTS — AE
ae.xpt, Adverse Events — Events, Version 3.1.2,. One record per adverse event per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1898HAE
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
567HSDTMIG 4.1.2.2,
568HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4
569H570HSDTMIG 4.1.2.3
AESEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
AEGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a subject.
Perm
SDTM 2.2.4
AEREFID
Reference ID
Char
Identifier
Internal or external identifier such as a serial number on an SAE reporting
form
Perm
SDTM 2.2.4
AESPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined identifier. It may be pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on an Adverse Events page.
Perm
SDTM 2.2.4
AETERM
Reported Term for the
Adverse Event
Char
Topic
Verbatim name of the event.
Req
SDTM 2.2.2,
571HSDTMIG 4.1.3.6
AEMODIFY
Modified Reported Term
Char
Synonym
Qualifier
If AETERM is modified to facilitate coding, then AEMODIFY will
contain the modified text.
Perm
SDTM 2.2.2,
572HSDTMIG 4.1.3.6
AEDECOD
Dictionary-Derived Term
Char
*
Synonym
Qualifier
Dictionary-derived text description of AETERM or AEMODIFY.
Equivalent to the Preferred Term (PT in MedDRA). The sponsor is
expected to provide the dictionary name and version used to map
the terms utilizing the define.xml external codelist attributes
Req
SDTM 2.2.2,
573H574H575HSDTMIG 4.1.3.5
576HSDTMIG 4.1.3.6
AECAT
Category for Adverse
Event
Char
*
Grouping
Qualifier
Used to define a category of related records. Example: BLEEDING,
NEUROPSYCHIATRIC.
Perm
SDTM 2.2.2,
577HSDTMIG 4.1.2.6
AESCAT
Subcategory for Adverse
Event
Char
*
Grouping
Qualifier
A further categorization of adverse event. Example: NEUROLOGIC.
Perm
SDTM 2.2.2,
578HSDTMIG 4.1.2.6
AEPRESP
Pre-Specified Adverse
Event
Char
(1899HNY)
Record
Qualifier
A value of ―Y‖ indicates that this adverse event was pre-specified on the
CRF. Values are null for spontaneously reported events (i.e., those
collected as free-text verbatim terms)
Perm
SDTM 2.2.2,
579HSDTMIG 4.1.2.7
580HSDTMIG 4.1.5.7
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 95
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or Format
Role
CDISC Notes
Core
References
AEBODSYS
Body System or Organ
Class
Char
*
Record
Qualifier
Dictionary derived. Body system or organ class used by the sponsor from
the coding dictionary (e.g., MedDRA). When using a multi-axial
dictionary such as MedDRA, this should contain the SOC used for the
sponsor‘s analyses and summary tables which may not necessarily be the
primary SOC.
Exp
SDTM 2.2.2,
581H582H583HSDTMIG 4.1.3.5
AELOC
Location of Event
Char
(584HLOC)
Record
Qualifier
Describes anatomical location relevant for the event (e.g., LEFT ARM for
skin rash).
Perm
SDTM 2.2.2
AESEV
Severity/Intensity
Char
(1900HAESEV)
Record
Qualifier
The severity or intensity of the event. Examples: MILD, MODERATE,
SEVERE.
Perm
SDTM 2.2.2
AESER
Serious Event
Char
(1901HNY)
Record
Qualifier
Is this a serious event?
Exp
SDTM 2.2.2
AEACN
Action Taken with Study
Treatment
Char
(1902HACN)
Record
Qualifier
Describes changes to the study treatment as a result of the event. AEACN
is specifically for the relationship to study treatment. AEACNOTH is for
actions unrelated to dose adjustments of study treatment. Examples of
AEACN values include ICH E2B values: DRUG WITHDRAWN, DOSE
REDUCED, DOSE INCREASED, DOSE NOT CHANGED,
UNKNOWN or NOT APPLICABLE
Exp
SDTM 2.2.2
AEACNOTH
Other Action Taken
Char
Record
Qualifier
Describes other actions taken as a result of the event that are unrelated to
dose adjustments of study treatment. Usually reported as free text.
Example: ―TREATMENT UNBLINDED. PRIMARY CARE
PHYSICIAN NOTIFIED.‖
Perm
SDTM 2.2.2
AEREL
Causality
Char
*
Record
Qualifier
Records the investigator's opinion as to the causality of the event
to the treatment. ICH E2A and E2B examples include NOT
RELATED, UNLIKELY RELATED, POSSIBLY RELATED,
RELATED. Controlled Terminology may be defined in the future.
Check with regulatory authority for population of this variable
Exp
SDTM 2.2.2
AERELNST
Relationship to Non-
Study Treatment
Char
Record
Qualifier
Records the investigator's opinion as to whether the event may have been
due to a treatment other than study drug. May be reported as free text.
Example: "MORE LIKELY RELATED TO ASPIRIN USE.‖.
Perm
SDTM 2.2.2
AEPATT
Pattern of Adverse Event
Char
*
Record
Qualifier
Used to indicate the pattern of the event over time. Examples:
INTERMITTENT, CONTINUOUS, SINGLE EVENT.
Perm
SDTM 2.2.2
AEOUT
Outcome of Adverse
Event
Char
(1903HOUT)
Record
Qualifier
Description of the outcome of an event.
Perm
SDTM 2.2.2
AESCAN
Involves Cancer
Char
(1904HNY)
Record
Qualifier
Was the serious event associated with the development of cancer?
Perm
SDTM 2.2.2
AESCONG
Congenital Anomaly or
Birth Defect
Char
(1905HNY)
Record
Qualifier
Was the serious event associated with congenital anomaly or birth defect?
Perm
SDTM 2.2.2
AESDISAB
Persist or Signif
Disability/Incapacity
Char
(1906HNY)
Record
Qualifier
Did the serious event result in persistent or significant
disability/incapacity?
Perm
SDTM 2.2.2
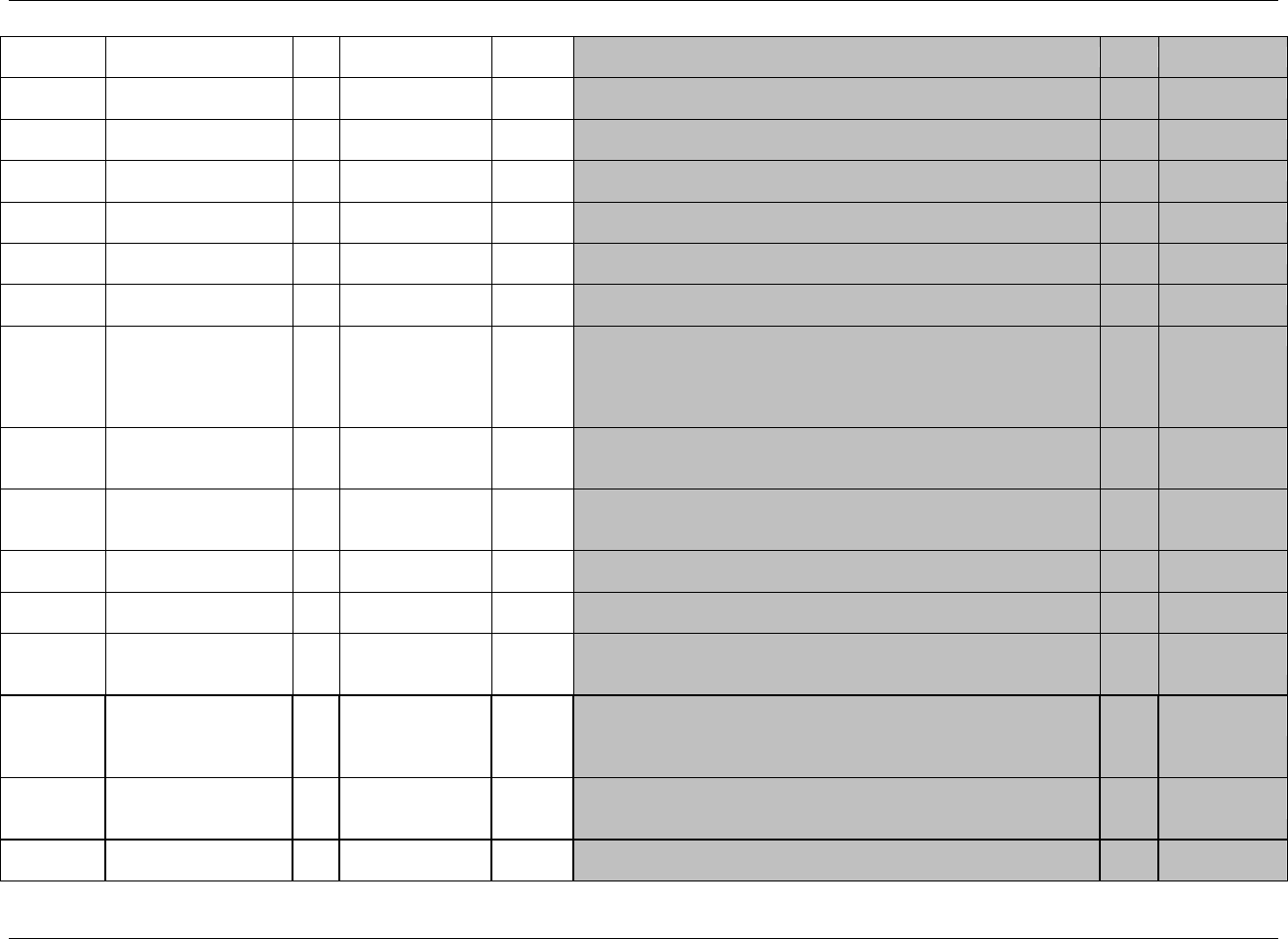
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 96 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or Format
Role
CDISC Notes
Core
References
AESDTH
Results in Death
Char
(1907HNY)
Record
Qualifier
Did the serious event result in death?
Perm
SDTM 2.2.2
AESHOSP
Requires or Prolongs
Hospitalization
Char
(1908HNY)
Record
Qualifier
Did the serious event require or prolong hospitalization?
Perm
SDTM 2.2.2
AESLIFE
Is Life Threatening
Char
(1909HNY)
Record
Qualifier
Was the serious event life threatening?
Perm
SDTM 2.2.2
AESOD
Occurred with Overdose
Char
(1910HNY)
Record
Qualifier
Did the serious event occur with an overdose?
Perm
SDTM 2.2.2
AESMIE
Other Medically
Important Serious Event
Char
(1911HNY)
Record
Qualifier
Do additional categories for seriousness apply?
Perm
SDTM 2.2.2
AECONTRT
Concomitant or
Additional Trtmnt Given
Char
(1912HNY)
Record
Qualifier
Was another treatment given because of the occurrence of the event?
Perm
SDTM 2.2.2
AETOXGR
Standard Toxicity Grade
Char
*
Record
Qualifier
Toxicity grade according to a standard toxicity scale such as Common
Terminology Criteria for Adverse Events v3.0 (CTCAE). Sponsor should
specify name of the scale and version used in the metadata (see 585HSection
6.2.1.1, Assumption 6d). If value is from a numeric scale, represent only
the number (e.g., ―2‖ and not ―Grade 2‖).
Perm
SDTM 2.2.2
AESTDTC
Start Date/Time of
Adverse Event
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
586HSDTMIG 4.1.4.1,
587HSDTMIG 4.1.4.2
AEENDTC
End Date/Time of
Adverse Event
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
588HSDTMIG 4.1.4.1;
589HSDTMIG 4.1.4.2
AESTDY
Study Day of Start of
Adverse Event
Num
Timing
Study day of start of adverse event relative to the sponsor-defined
RFSTDTC.
Perm
SDTM 2.2.5,
590HSDTMIG 4.1.4.4
AEENDY
Study Day of End of
Adverse Event
Num
Timing
Study day of end of event relative to the sponsor-defined RFSTDTC.
Perm
SDTM 2.2.5,
591HSDTMIG 4.1.4.4
AEDUR
Duration of Adverse
Event
Char
ISO 8601
Timing
Collected duration and unit of an adverse event. Used only if collected on
the CRF and not derived from start and end date/times. Example:
P1DT2H (for 1 day, 2 hours).
Perm
SDTM 2.2.5,
592HSDTMIG 4.1.4.3
AEENRF
End Relative to
Reference Period
Char
(STENRF)
Timing
Describes the end of the event relative to the sponsor-defined reference
period. The sponsor-defined reference period is a continuous period of
time defined by a discrete starting point (RFSTDTC) and a discrete
ending point (RFENDTC) of the trial.
Perm
SDTM 2.2.5,
593HSDTMIG 4.1.4.7
AEENRTPT
End Relative to
Reference Time Point
Char
BEFORE, AFTER,
COINCIDENT,
ONGOING, U
Timing
Identifies the end of the event as being before or after the reference time
point defined by variable AEENTPT.
Perm
SDTM 2.2.5,
594HSDTMIG 4.1.4.7
AEENTPT
End Reference Time
Point
Char
Timing
Description of date/time in ISO 8601 character format of the reference
point referred to by AEENRTPT. Examples: "2003-12-25" or "VISIT 2".
Perm
SDTM 2.2.5,
595HSDTMIG 4.1.4.7
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 97
Final November 12, 2008
6.2.1.1 ASSUMPTIONS FOR ADVERSE EVENT DOMAIN MODEL
1. AE Definition
The Adverse Events dataset includes clinical data describing "any untoward medical occurrence in a patient or clinical investigation subject administered a
pharmaceutical product and which does not necessarily have to have a causal relationship with this treatment" (ICH E2A). In consultation with regulatory
authorities, sponsors may extend or limit the scope of adverse event collection (e.g., collecting pre-treatment events related to trial conduct, not collecting
events that are assessed as efficacy endpoints). The events included in the AE dataset should be consistent with the protocol requirements. Adverse events
may be captured either as free text or via a pre-specified list of terms.
2. Adverse Event Description and Coding
a. AETERM captures the verbatim term collected for the event. It is the topic variable for the AE dataset. AETERM is a required variable and must have
a value.
b. AEMODIFY is a permissible variable and should be included if the sponsor‘s procedure permits modification of a verbatim term for coding. The
modified term is listed in AEMODIFY. The variable should be populated as per the sponsor‘s procedures.
c. AEDECOD is the preferred term derived by the sponsor from the coding dictionary. It is a required variable and must have a value. It is expected that
the reported term (AETERM) will be coded using a standard dictionary such as MedDRA. The sponsor is expected to provide the dictionary name and
version used to map the terms utilizing the define.xml external codelist attributes.
d. AEBODSYS is the system organ class from the coding dictionary associated with the adverse event by the sponsor. This value may differ from the
primary system organ class designated in the coding dictionary's standard hierarchy. It is expected that this variable will be populated.
e. Sponsors may include the values of additional levels from the coding dictionary's hierarchy (i.e., High-Level Group Term, High-Level Term, Lower-
Level Term) in the SUPPAE dataset as described in 1913HAppendix C5 (standard Supplemental Qualifier name codes) and 596HSection 8.4.
3. Additional Categorization and Grouping
a. AECAT and AESCAT should not be redundant with the domain code or dictionary classification provided by AEDECOD and AEBODSYS (i.e., they
should provide a different means of defining or classifying AE records). AECAT and AESCAT are intended for categorizations that are defined in
advance. For example, a sponsor may have a separate CRF page for AEs of special interest and then another page for all other AEs. AECAT and
AESCAT should not be used for after-the-fact categorizations such as clinically significant. In cases where a category of AEs of special interest
resembles a part of the dictionary hierarchy (e.g., "CARDIAC EVENTS"), the categorization represented by AECAT and AESCAT may differ from the
categorization derived from the coding dictionary.
b. AEGRPID may be used to link (or associate) different records together to form a block of related records at the subject level within the AE domain.
See 597HSection 4.1.2.6 for discussion of grouping variables.
4. Pre-Specified Terms; Presence or Absence of Events
a. Adverse events are generally collected in two different ways, either by recording free text or using a pre-specified list of terms. In the latter case, the
solicitation of information on specific adverse events may affect the frequency at which they are reported; therefore, the fact that a specific adverse
event was solicited may be of interest to reviewers. An AEPRESP value of ―Y‖ is used to indicate that the event in AETERM was pre-specified on the
CRF.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 98 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
b. If it is important to know which adverse events from a pre-specified list were not reported as well as those that did occur, these data should be
submitted in a Findings class dataset such as Findings About Events and Interventions (FA, 598HSection 6.4). A record should be included in that Findings
dataset for each pre-specified adverse-event term. Records for adverse events that actually occurred should also exist in the AE dataset with AEPRESP
set to ―Y.‖
c. If a study collects both pre-specified adverse events as well as free-text events, the value of AEPRESP should be ―Y‖ for all pre-specified events and
null for events reported as free-text. AEPRESP is a permissible field and may be omitted from the dataset if all adverse events were collected as free
text.
d. When adverse events are collected with the recording of free text, a record may be entered into the sponsor‘s data management system to indicate ―no
adverse events‖ for a specific subject. For these subjects, do not include a record in the AE submission dataset to indicate that there were no events.
Records should be included in the submission AE dataset only for adverse events that have actually occurred.
5. Timing Variables
a. Relative timing assessment ―Ongoing‖ is common in the collection of Adverse Event information. AEENRF may be used when this relative timing
assessment is made coincident with the end of the study reference period for the subject represented in the Demographics dataset (RFENDTC).
AEENRTPT with AEENTPT may be used when "Ongoing" is relative to another date such as the final safety follow-up visit date. See 599HSection 4.1.4.7.
b. Additional timing variables (such as AEDTC) may be used when appropriate.
6. Other Qualifier Variables
a. If categories of serious events are collected secondarily to a leading question, as in the example below, the values of the variables that capture reasons
an event is considered serious (i.e., AESCAN, AESCONG, etc.) may be null. For example, if Serious is answered ―No, ― the values for these variables
may be null. However, if Serious is answered ―Yes, ― at least one of them will have a ―Y‖ response. Others may be N or null, according to the
sponsor‘s convention.
Serious? [ ] Yes [ ] No
If yes, check all that apply: [ ] Fatal [ ] Life-threatening [ ] Inpatient hospitalization… [ ] etc.
On the other hand, if the CRF is structured so that a response is collected for each seriousness category, all category variables (e.g., AESDTH,
AESHOSP) would be populated and AESER would be derived.
b. The serious categories ―Involves cancer‖ (AESCAN) and ―Occurred with overdose‖ (AESOD) are not part of the ICH definition of a serious adverse
event, but these categories are available for use in studies conducted under guidelines that existed prior to the FDA‘s adoption of the ICH definition.
c. When a description of Other Medically Important Serious Adverse Events category is collected on a CRF, sponsors should place the description in the
SUPPAE dataset using the standard supplemental qualifier name code AESOSP as described in 600HSection 8.4 and Appendix C5.
d. In studies using toxicity grade according to a standard toxicity scale such as Common Terminology Criteria for Adverse Events v3.0 (CTCAE),
published by the NCI (National Cancer Institute) at 601Hhttp://ctep.cancer.gov/reporting/ctc.html)), AETOXGR should be used instead of AESEV. In most
cases, either AESEV or AETOXGR is populated but not both. There may be cases when a sponsor may need to populate both variables. The sponsor is
expected to provide the dictionary name and version used to map the terms utilizing the define.xml external codelist attributes
e. AE Structure
The structure of the AE domain is one record per adverse event per subject. It is the sponsor's responsibility to define an event. This definition may
vary based on the sponsor's requirements for characterizing and reporting product safety and is usually described in the protocol. For example, the

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 99
Final November 12, 2008
sponsor may submit one record that covers an adverse event from start to finish. Alternatively, if there is a need to evaluate AEs at a more granular
level, a sponsor may submit a new record when severity, causality, or seriousness changes or worsens. By submitting these individual records, the
sponsor indicates that each is considered to represent a different event. The submission dataset structure may differ from the structure at the time of
collection. For example, a sponsor might collect data at each visit in order to meet operational needs, but submit records that summarize the event and
contain the highest level of severity, causality, seriousness, etc. Examples of dataset structure:
1. One record per adverse event per subject for each unique event. Multiple adverse event records reported by the investigator are submitted
as summary records ―collapsed‖ to the highest level of severity, causality, seriousness, and the final outcome.
2. One record per adverse event per subject. Changes over time in severity, causality, or seriousness are submitted as separate events.
Alternatively, these changes may be submitted in a separate dataset based on the Findings About Events and Interventions model (see
602HSection 6.4).
3. Other approaches may also be reasonable as long as they meet the sponsor's safety evaluation requirements and each submitted record
represents a unique event. The domain-level metadata (See Section 3.2) should clarify the structure of the dataset.
7. Use of EPOCH and TAETORD
When EPOCH is included in the Adverse Event domain, it should be the epoch of the start of the adverse event. In other words, it should be based on
AESTDTC, rather than AEENDTC. The computational method for EPOCH in the define.xml should describe any assumptions made to handle cases
where an adverse event starts on the same day that a subject starts an epoch, if AESTDTC and SESTDTC are not captured with enough precision to
determine the epoch of the onset of the adverse event unambiguously. Similarly, if TAETORD is included in the Adverse Events domain, it should be
the value for the start of the adverse event, and the computational method in the define.xml should describe any assumptions.
8. Additional Events Qualifiers
The following Qualifiers would not be used in AE: --OCCUR, --STAT, and--REASND. They are the only Qualifiers from the SDTM Events Class not
in the AE domain. They are not permitted because the AE domain contains only records for adverse events that actually occurred. See Assumption 4b
above for information on how to deal with negative responses or missing responses to probing questions for pre-specified adverse events.
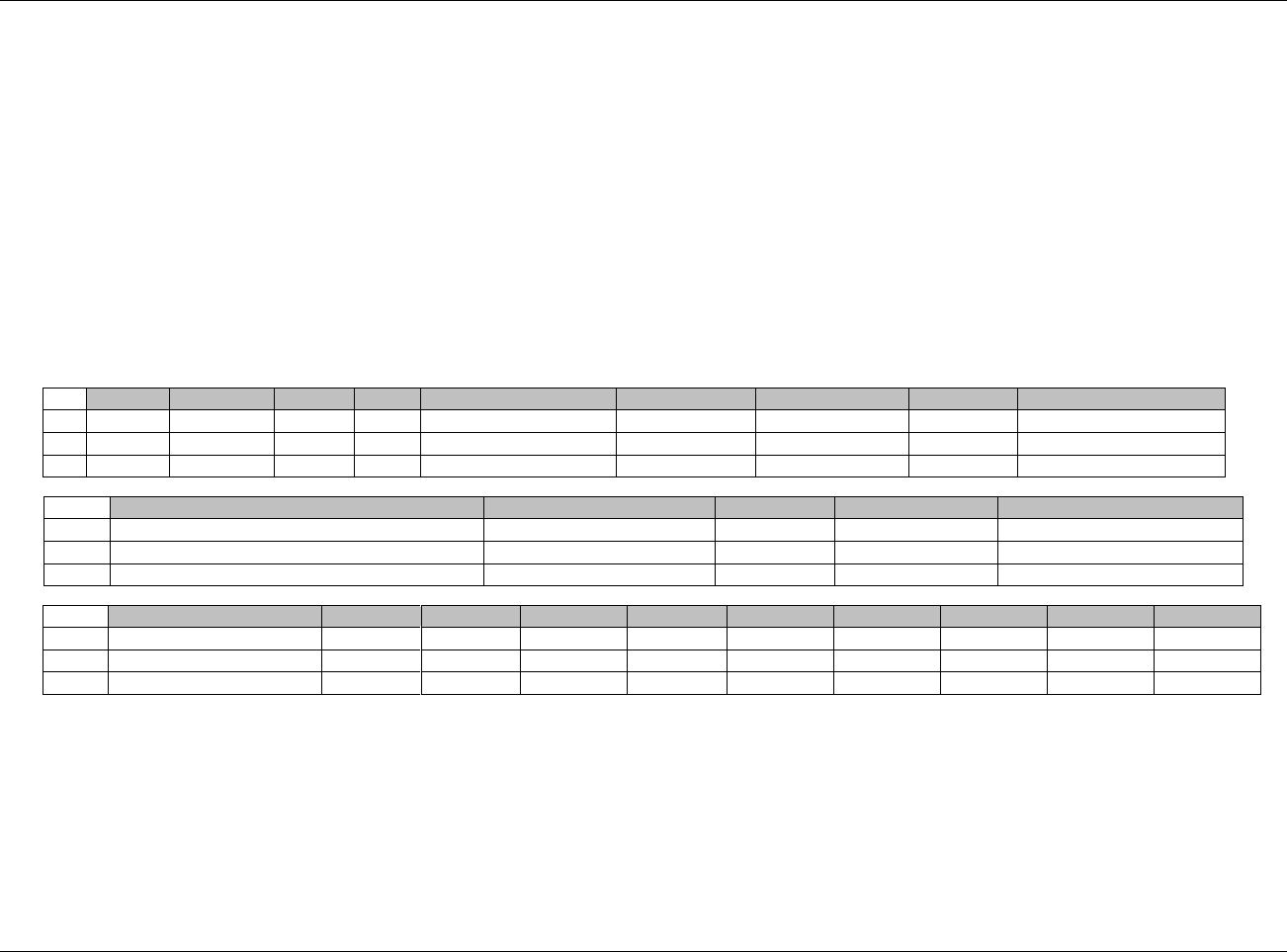
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 100 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.1.2 EXAMPLES FOR ADVERSE EVENTS DOMAIN MODEL
Example 1
This is an example of data from an AE CRF that collects AE terms as free text. The first study drug was administered to the subject on October 13, 2006 at 12:00.
Three AEs were reported. AEs were coded using MedDRA, and the sponsor‘s procedures include the possibility of modifying the reported term to aid in coding.
The CRF is structured so that seriousness category variables (e.g., AESDTH, AESHOSP) are checked only when AESER is answered ―Y.‖
Rows 1 and 2 Show the following:
-an example of modifying the reported term for coding purposes. The modified value is in AEMODIFY.
-an example of the overall seriousness question AESER answered with an ―N‖ and corresponding seriousness category variables (e.g.,
AESDTH, AESHOSP) left blank.
Row 3 Shows an example of the overall seriousness question AESER answered with a ―Y‖ and the relevant corresponding seriousness category
variables (AESHOSP and AESLIFE) answered with a ―Y‖. The other seriousness category variables are left blank. This row also shows an
example of AEENRF being populated because the AE was marked as ―Continuing‖ as of the end of the study reference period for the subject
(see 603HSection 4.1.4.7).
Row
STUDYID
DOMAIN
USUBJID
AESEQ
AETERM
AESTDTC
AEENDTC
AEMODIFY
AEDECOD
1
ABC123
AE
123101
1
POUNDING HEADACHE
2005-10-12
2005-10-12
HEADACHE
Headache
2
ABC123
AE
123101
2
BACK PAIN FOR 6 HOURS
2005-10-13T13:05
2005-10-13T19:00
BACK PAIN
Back pain
3
ABC123
AE
123101
3
PULMONARY EMBOLISM
2005-10-21
Pulmonary embolism
Row
AEBODSYS
AESEV
AESER
AEACN
AEREL
1 (cont)
Nervous system disorders
SEVERE
N
NOT APPLICABLE
DEFINITELY NOT RELATED
2 (cont)
Musculoskeletal and connective tissue disorders
MODERATE
N
DOSE REDUCED
PROBABLY RELATED
3 (cont)
Vascular disorders
MODERATE
Y
DOSE REDUCED
PROBABLY NOT RELATED
Row
AEOUT
AESCONG
AESDISAB
AESDTH
AESHOSP
AESLIFE
AESMIE
AESTDY
AEENDY
AEENRF
1 (cont)
RECOVERED/RESOLVED
-1
-1
2 (cont)
RECOVERED/RESOLVED
1
1
3 (cont)
RECOVERING/RESOLVING
Y
Y
9
AFTER
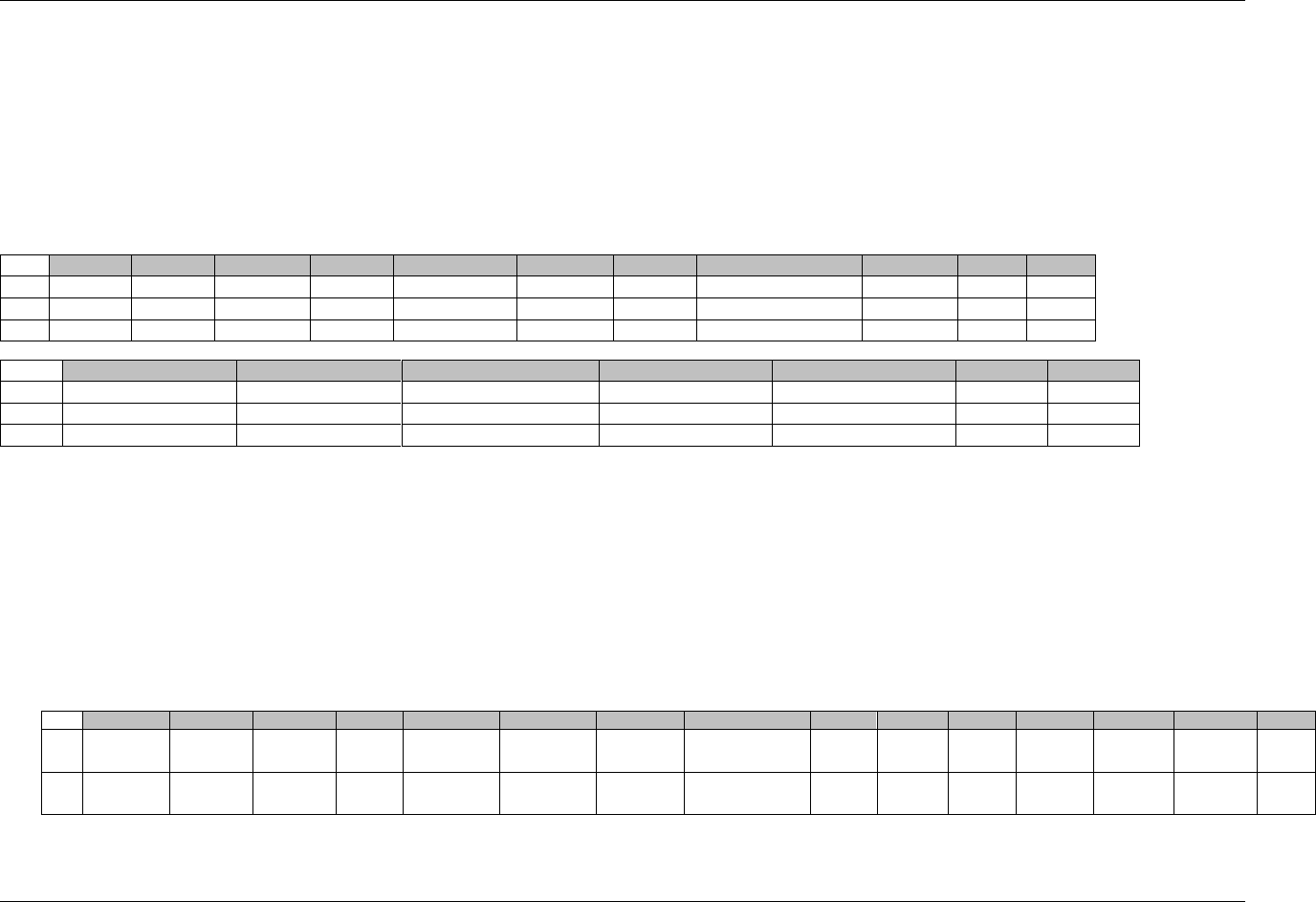
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 101
Final November 12, 2008
Example 2
In this example, a CRF module occurring at several visits asks whether or not nausea, vomiting, or diarrhea occurred. The responses to the probing questions
(Yes, No, or Not done) will be represented in the Findings About (FA) domain (see 604HSection 6.4). If ―Yes ‖ the investigator is instructed to complete the Adverse
Event CRF. In the Adverse Events dataset, data on AEs solicited by means of pre-specified on the CRF will have an AEPRESP value of Y. For AEs solicited by a
general question, AEPRESP will be null. RELREC may be used to relate AE records with FA records.
Rows 1 and 2 Show that nausea and vomiting were pre-specified on a CRF, as indicated by AEPRESP = ―Y‖. The subject did not experience diarrhea, so no
record for that term exists in the AE dataset.
Row 3 Shows an example of an AE (headache) that is not pre-specified on a CRF as indicated by a blank for AEPRESP
Row
STUDYID
DOMAIN
USUBJID
AESEQ
AETERM
AEDECOD
AEPRESP
AEBODSYS
AESEV
AESER
1
ABC123
AE
123101
1
NAUSEA
Nausea
Y
Gastrointestinal disorders
SEVERE
N
2
ABC123
AE
123101
2
VOMITING
Vomiting
Y
Gastrointestinal disorders
MODERATE
N
3
ABC123
AE
123101
3
HEADACHE
Headache
Nervous system disorders
MILD
N
Row
AEACN
AEREL
AEOUT
AESTDTC
AEENDTC
AESTDY
AEENDY
1 (cont)
DOSE REDUCED
RELATED
RECOVERED/RESOLVED
2005-10-12
2005-10-13
2
3
2 (cont)
DOSE REDUCED
RELATED
RECOVERED/RESOLVED
2005-10-13T13:00
2005-10-13T19:00
3
3
3 (cont)
DOSE NOT CHANGED
POSSIBLY RELATED
RECOVERED/RESOLVED
2005-10-21
2005-10-21
11
11
Example 3
In this example, a CRF module occurs only once and asks whether or not nausea, vomiting, or diarrhea occurred. In the context of this study, the conditions that
occurred are reportable as Adverse Events. No additional data about these events is collected. No other adverse event information is collected via general
questions. The responses to the probing questions (Yes, No, or Not done) will be represented in the Findings About (FA) domain (see 605HSection 6.4). Since all
adverse events must be submitted in AE dataset, this represents an unusual case; the AE dataset will be populated with the term and the flag indicating that it was
pre-specified, but timing information is limited to the date of collection, and other expected Qualifiers are not available. RELREC may be used to relate AE
records with FA records.
Rows 1 and 2 Subject was found to have experienced nausea and vomiting by means of the probing questions. The subject did not experience diarrhea, so no
record for that term exists in the AE dataset
Row
STUDYID
DOMAIN
USUBJID
AESEQ
AETERM
AEDECOD
AEPRESP
AEBODSYS
AESER
AEACN
AEREL
AESTDTC
AEENDTC
AEDTC
AEDY
1
ABC123
AE
123101
1
NAUSEA
Nausea
Y
Gastrointestinal
disorders
2005-10-29
19
2
ABC123
AE
123101
2
VOMITING
Vomiting
Y
Gastrointestinal
disorders
2005-10-29
19
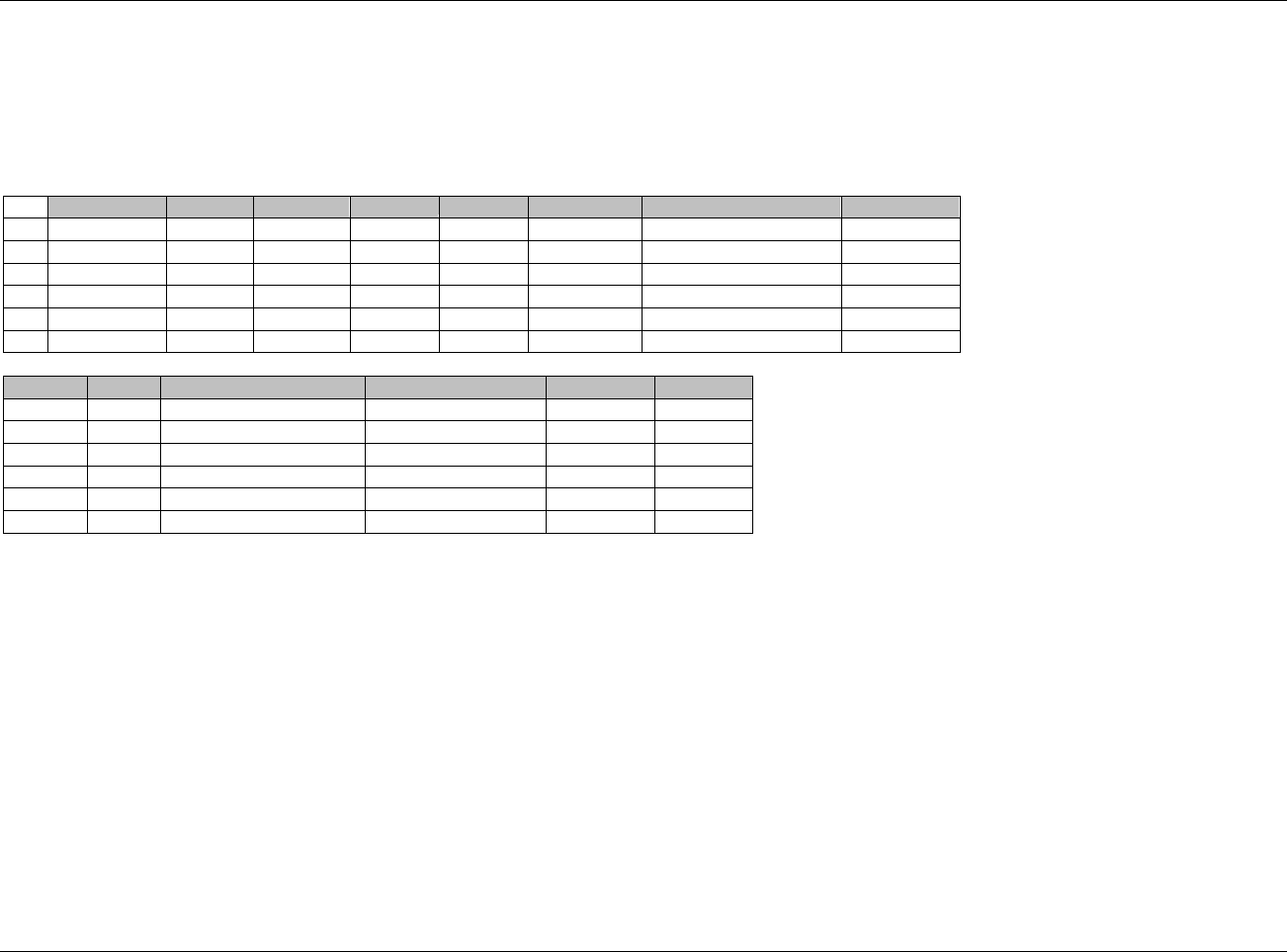
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 102 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 4
In this example, the investigator was instructed to create a new adverse-event record each time the severity of an adverse event changes. AEGRPID can be used
to identify the group of records related to a single event for a subject.
Row 1 Shows an adverse event of nausea, whose severity was moderate.
Rows 2-6 Show how AEGRPID can be used to identify the group of records related to a single event for a subject.
Row
STUDYID
DOMAIN
USUBJID
AESEQ
AEGRPID
AETERM
AEBODSYS
AESEV
1
ABC123
AE
123101
1
NAUSEA
Gastrointestinal disorders
MODERATE
2
ABC123
AE
123101
2
1
VOMITING
Gastrointestinal disorders
MILD
3
ABC123
AE
123101
3
1
VOMITING
Gastrointestinal disorders
SEVERE
4
ABC123
AE
123101
4
1
VOMITING
Gastrointestinal disorders
MILD
5
ABC123
AE
123101
5
2
DIARRHEA
Gastrointestinal disorders
SEVERE
6
ABC123
AE
123101
6
2
DIARRHEA
Gastrointestinal disorders
MODERATE
Row
AESER
AEACN
AEREL
AESTDTC
AEENDTC
1 (cont‘d)
N
DOSE NOT CHANGED
RELATED
2005-10-13
2005-10-14
2 (cont‘d)
N
DOSE NOT CHANGED
POSSIBLY RELATED
2005-10-14
2005-10-16
3 (cont‘d)
N
DOSE NOT CHANGED
POSSIBLY RELATED
2005-10-16
2005-10-17
4 (cont‘d)
N
DOSE NOT CHANGED
POSSIBLY RELATED
2005-10-17
2005-10-20
5 (cont‘d)
N
DOSE NOT CHANGED
POSSIBLY RELATED
2005-10-16
2005-10-17
6 (cont‘d)
N
DOSE NOT CHANGED
POSSIBLY RELATED
2005-10-17
2005-10-21
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 103
Final November 12, 2008
6.2.2 DISPOSITION — DS
ds.xpt, Disposition — Events, Version 3.1.2. One record per disposition status or protocol milestone per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1916HDS
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
606HSDTMIG 4.1.2.2,
607HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4
608H609HSDTMIG 4.1.2.3
DSSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
DSGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
610HSDTMIG 4.1.2.6
SDTM 2.2.4
DSREFID
Reference ID
Char
Identifier
Internal or external identifier.
Perm
SDTM 2.2.4
DSSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a Disposition page.
Perm
SDTM 2.2.4
DSTERM
Reported Term for the
Disposition Event
Char
Topic
Verbatim name of the event or protocol milestone. Some terms in
DSTERM will match DSDECOD, but others, such as ―Subject moved‖
will map to controlled terminology in DSDECOD, such as ―LOST TO
FOLLOW-UP.‖
Req
SDTM 2.2.2,
611HSDTMIG 4.1.3.6
DSDECOD
Standardized Disposition
Term
Char
(1917HNCOMPLT)
Synonym
Qualifier
Controlled terminology for the name of disposition event or protocol
milestone. Examples of protocol milestones: INFORMED CONSENT
OBTAINED, RANDOMIZED
Req
SDTM 2.2.2,
612H613H614HSDTMIG 4.1.3.5
DSCAT
Category for Disposition
Event
Char
(615HDSCAT)
Grouping
Qualifier
Used to define a category of related records. DSCAT is now an
―Expected‖ variable. DSCAT was permissible in SDTMIG 3.1.1 and
earlier versions. The change from ―permissible‖ to ―expected‖ is based on
the requirement to distinguish protocol milestones and/or other events
from disposition events. DSCAT may be null if there are only ―disposition
events‖; however, it is recommended that DSCAT always be populated.
Exp
SDTM 2.2.2,
616HSDTMIG 4.1.2.6
DSSCAT
Subcategory for
Disposition Event
Char
*
Grouping
Qualifier
A further categorization of disposition event.
Perm
SDTM 2.2.2,
617HSDTMIG 4.1.2.6
EPOCH
Epoch
Char
*
Timing
EPOCH may be used when DSCAT = ―DISPOSITION EVENT‖.
Examples: SCREENING, TREATMENT PHASE, FOLLOW-UP
Perm
SDTM 2.2.5,
618HSDTMIG 7.1.2
DSDTC
Date/Time of Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
619HSDTMIG 4.1.4.1
DSSTDTC
Start Date/Time of
Disposition Event
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
620HSDTMIG 4.1.4.1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 104 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
DSSTDY
Study Day of Start of
Disposition Event
Num
Timing
Study day of start of event relative to the sponsor-defined RFSTDTC.
Perm
SDTM 2.2.5,
621HSDTMIG 4.1.4.4
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.2.2.1 ASSUMPTIONS FOR DISPOSITION DOMAIN MODEL
1. DS Definition
The Disposition dataset provides an accounting for all subjects who entered the study and may include protocol milestones, such as randomization, as well as
the subject's completion status or reason for discontinuation for the entire study or each phase or segment of the study, including screening and post-treatment
follow-up. Sponsors may choose which disposition events and milestones to submit for a study. See ICH E3, Section 10.1 for information about disposition
events.
2. Categorization
a. DSCAT is used to distinguish between disposition events, protocol milestones and other events. The controlled terminology for DSCAT consists of
―DISPOSITION EVENT,‖ ―PROTOCOL MILESTONE,‖ and ―OTHER EVENT.‖
b. A ―DISPOSITION EVENT‖ describes whether a subject completed the study or portion of a study (Epoch) or the reason they did not complete. The
subject‘s disposition is often described for each study Epoch (e.g., screening, initial treatment, washout, cross-over treatment, follow-up).
c. A ―PROTOCOL MILESTONE‖ is a protocol-specified, ―point-in-time‖ event. The most common protocol milestones are ―INFORMED CONSENT
OBTAINED‖ and ―RANDOMIZED.‖
d. Other important events that occur during a trial, but are not driven by protocol requirements and are not captured in another Events or Interventions class
dataset, are classified as ―OTHER EVENT.‖ ―TREATMENT UNBLINDED‖ is an example of ―OTHER EVENT.‖
3. DS Description and Coding
a. DSTERM and DSDECOD are required. DSDECOD values are drawn from sponsor-defined controlled terminology. The controlled terminology will
depend on the value of DSCAT. When DSCAT="DISPOSITION EVENT", DSTERM contains either "COMPLETE" or, if the subject did not complete,
specific verbatim information about the disposition event.
b. When DSTERM = "COMPLETED", DSDECOD = "COMPLETED". When DSTERM contains verbatim text, DSDECOD will contain a standard term
from a controlled terminology list. For example, DSTERM = "Subject moved" might map to "LOST TO FOLLOW-UP" in the sponsor's controlled
terminology.
c. A sponsor may collect one disposition event for the trial as a whole, or they may collect disposition for each Epoch of the trial. When disposition is
collected for each Epoch, the variable EPOCH should be included in the DS dataset. When EPOCH is populated for disposition events (records with
DSCAT = DISPOSITION EVENT), EPOCH is the name of the Epoch for the disposition event described in the record. This is a subtly different
meaning from that of EPOCH when it is used in other general-observation-class domains, where EPOCH, as a Timing variable, is the name of the Epoch
during which --STDTC or --DTC falls. The values of EPOCH are drawn from the Trial Arms domain, 622HSection 7.2.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 105
Final November 12, 2008
d. When DSCAT="PROTOCOL MILESTONE", DSTERM and DSDECOD will contain the same value drawn from the sponsor's controlled terminology.
Examples of controlled terms include "INFORMED CONSENT OBTAINED" and "RANDOMIZED." EPOCH should not be populated when DSCAT =
―PROTOCOL MILESTONE‖.
e. Events that are not disposition or milestone related are classified as an ―OTHER EVENT‖ (see Assumption 2d above). If a reason for the OTHER
EVENT was collected, then the reason is in DSTERM. For example, treatment was unblinded due to investigator error. DSTERM = INVESTIGATOR
ERROR and DSDECOD = TREATMENT UNBLINDED. IF no reason was collected then DSTERM = DSDECOD.
4. Timing Variables
a. DSSTDTC is expected and is used for the date/time of the disposition event. Disposition events do not have start and end dates since disposition events
do not span an interval (e.g. randomization date) but occur at a single date/time (e.g., randomization date).
b. DSSTDTC documents the date/time that a protocol milestone, disposition event, or other event occurred. In the case of a disposition event, the reason
for not completing the referenced study Epoch may be related to an event or intervention reported in another dataset. DSSTDTC is the date/time that the
Epoch was completed and is not necessarily the same as the start or end date/time of the event or intervention that led to discontinuation. For example, a
subject reported severe vertigo on June 1, 2006 (AESTDTC). After ruling out other possible causes, the investigator decided to discontinue study
treatment on June 6, 2006 (DSSTDTC). The subject reported that the vertigo had resolved on June 8, 2006 (AEENDTC).
5. Reasons for Termination
a. ICH E3, Section 10.1 indicates that ―the specific reason for discontinuation‖ should be presented, and that summaries should be ―grouped by treatment
and by major reason.‖ The CDISC SDS Team interprets this guidance as requiring one standardized disposition term (DSDECOD) per disposition event.
If multiple reasons are reported, the sponsor should identify a primary reason and use that to populate DSTERM and DSDECOD. Additional reasons
should be submitted in SUPPDS. Example:
DSTERM= SEVERE NAUSEA
DSDECOD=ADVERSE EVENT
SUPPDS QNAM=DSTERM1
SUPPDS QLABEL= Reported Term for Disposition Event 1
SUPPDS QVAL=SUBJECT REFUSED FURTHER TREATMENT
SUPPDS QNAM=DSDECOD1
SUPPDS QLABEL= Standardized Disposition Term 1
SUPPDS QVAL=WITHDREW CONSENT
6. Additional Event Qualifiers
The following Qualifiers would generally not be used in DS: --PRESP, --OCCUR, --STAT, --REASND, --BODSYS, --LOC, --SEV, --SER, --ACN,
--ACNOTH, --REL, --RELNST, --PATT, --OUT, --SCAN, --SCONG, --SDISAB, --SDTH, --SHOSP, --SLIFE, --SOD, --SMIE, --CONTRT, --TOXGR.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 106 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.2.2 EXAMPLES FOR DISPOSITION DOMAIN MODEL
Example 1
In this example, a DS CRF collected multiple disposition events at different time points in the study indicated by EPOCH. There are also several protocol
milestones which are indicated by DSCAT = ―PROTOCOL MILESTONE‖. DSTERM is populated with controlled terminology with the same value as
DSDECOD except in the case when there is free text for DSTERM such as ―Subject moved‖. In this case, the controlled terminology is only in DSDECOD
(LOST TO FOLLOW-UP).
Rows 1-21: There are multiple disposition events and protocol milestones per subject. EPOCH is populated when DSCAT has a value of DISPOSITION
EVENT and null when DSCAT has value of PROTOCOL MILESTONE.
Rows 2, 4, 5: Subject 123101 has 3 records to indicate the completion of 3 stages of the study, which are screening, treatment phase, and follow-up. The study
also collected the protocol milestones of INFORMED CONSENT and RANDOMIZATION.
Row 7: Subject 123102 is a screen drop (also known as a screen failure). Screen drops are identified by a DSDECOD that is not equal to ―COMPLETED‖
for the SCREENING stage. This is an example of the submission of the verbatim reason for discontinuation in DSTERM. Also note that although
DSDECOD is ―PROTOCOL VIOLATION‖, this record represents the disposition event for the SCREENING stage and documents the reason for
not completing (―SUBJECT DENIED MRI PROCEDURE‖) and the corresponding date of discontinuation (DSSTDTC). A record describing the
protocol deviation event itself should appear in the DV dataset.
Rows 9, 11: Subject 123103 completed the screening stage but did not complete the treatment stage.
Row 11: The verbatim reason the subject dropped is in DSTERM (SUBJECT MOVED) and the controlled term is in DSDECOD (LOST TO FOLLOW-UP).
Row 16: Subject 123104 died in an automobile accident on October 29, 2003 (see DSSTDTC) after the completion of treatment, but prior to the completion
of follow-up. Note that the date of collection of the event information (DSDTC = October 31, 2003) was different from the date of the disposition
event.
Rows 20, 21: Subject 123105 discontinued study treatment due to an AE, but went on to complete the follow-up phase of the trial.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 107
Final November 12, 2008
Row
STUDYID
DOMAIN
USUBJID
DSSEQ
DSTERM
DSDECOD
DSCAT
EPOCH
DSDTC
DSSTDTC
1
ABC123
DS
123101
1
INFORMED CONSENT
OBTAINED
INFORMED CONSENT
OBTAINED
PROTOCOL MILESTONE
2003-09-21
2003-09-21
2
ABC123
DS
123101
2
COMPLETED
COMPLETED
DISPOSITION EVENT
SCREENING
2003-09-29
2003-09-29
3
ABC123
DS
123101
3
RANDOMIZED
RANDOMIZED
PROTOCOL MILESTONE
2003-09-30
2003-09-30
4
ABC123
DS
123101
4
COMPLETED
COMPLETED
DISPOSITION EVENT
TREATMENT
PHASE
2003-10-31
2003-10-31
5
ABC123
DS
123101
5
COMPLETED
COMPLETED
DISPOSITION EVENT
FOLLOW-UP
2003-11-15
2003-11-15
6
ABC123
DS
123102
1
INFORMED CONSENT
OBTAINED
INFORMED CONSENT
OBTAINED
PROTOCOL MILESTONE
2003-11-21
2003-11-21
7
ABC123
DS
123102
2
SUBJECT DENIED MRI
PROCEDURE
PROTOCOL VIOLATION
DISPOSITION EVENT
SCREENING
2003-11-22
2003-11-20
8
ABC123
DS
123103
1
INFORMED CONSENT
OBTAINED
INFORMED CONSENT
OBTAINED
PROTOCOL MILESTONE
2003-09-15
2003-09-15
9
ABC123
DS
123103
2
COMPLETED
COMPLETED
DISPOSITION EVENT
SCREENING
2003-09-22
2003-09-22
10
ABC123
DS
123103
3
RANDOMIZED
RANDOMIZED
PROTOCOL MILESTONE
2003-09-30
2003-09-30
11
ABC123
DS
123103
4
SUBJECT MOVED
LOST TO FOLLOW-UP
DISPOSITION EVENT
TREATMENT
PHASE
2003-10-31
2003-10-31
12
ABC123
DS
123104
1
INFORMED CONSENT
OBTAINED
INFORMED CONSENT
OBTAINED
PROTOCOL MILESTONE
2003-09-15
2003-09-15
13
ABC123
DS
123104
2
COMPLETED
COMPLETED
DISPOSITION EVENT
SCREENING
2003-09-22
2003-09-22
14
ABC123
DS
123104
3
RANDOMIZED
RANDOMIZED
PROTOCOL MILESTONE
2003-09-30
2003-09-30
15
ABC123
DS
123104
4
COMPLETED
COMPLETED
DISPOSITION EVENT
TREATMENT
PHASE
2003-10-15
2003-10-15
16
ABC123
DS
123104
5
AUTOMOBILE
ACCIDENT
DEATH
DISPOSITION EVENT
FOLLOW-UP
2003-10-31
2003-10-29
17
ABC123
DS
123105
1
INFORMED CONSENT
OBTAINED
INFORMED CONSENT
OBTAINED
PROTOCOL MILESTONE
2003-09-28
2003-09-28
18
ABC123
DS
123105
2
COMPLETED
COMPLETED
DISPOSITION EVENT
SCREENING
2003-10-02
2003-10-02
19
ABC123
DS
123105
3
RANDOMIZED
RANDOMIZED
PROTOCOL MILESTONE
2003-10-02
2003-10-02
20
ABC123
DS
123105
4
ANEMIA
ADVERSE EVENT
DISPOSITION EVENT
TREATMENT
PHASE
2003-10-17
2003-10-17
21
ABC123
DS
123105
5
COMPLETED
COMPLETED
DISPOSITION EVENT
FOLLOW-UP
2003-11-02
2003-11-02
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 108 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 2
In this example, the sponsor has chosen to simply submit whether or not the subject completed the study, so there is only one record per subject.
Row 1: Subject who completed the study
Rows 2, 3: Subjects who discontinued.
Row
STUDYID
DOMAIN
USUBJID
DSSEQ
DSTERM
DSDECOD
DSCAT
DSSTDTC
1
ABC456
DS
456101
1
COMPLETED
COMPLETED
DISPOSITION EVENT
2003-09-21
2
ABC456
DS
456102
1
SUBJECT TAKING STUDY MED
ERRATICALLY
PROTOCOL VIOLATION
DISPOSITION EVENT
2003-09-29
3
ABC456
DS
456103
1
LOST TO FOLLOW-UP
LOST TO FOLLOW-UP
DISPOSITION EVENT
2003-10-15
Example 3
Rows 1, 2: Subject completed the treatment and follow-up phase
Rows 3, 5: Subject did not complete the treatment phase but did complete the follow-up phase.
Row 4: Subject‘s treatment is unblinded. The date of the unblinding is represented in DSSTDTC. Maintaining the blind as per protocol is not considered
to be an event since there is no change in the subject‘s state.
Row
STUDYID
DOMAIN
USUBJID
DSSEQ
DSTERM
DSDECOD
DSCAT
EPOCH
DSSTDTC
1
ABC789
DS
789101
1
COMPLETED
COMPLETED
DISPOSITION EVENT
TREATMENT PHASE
2004-09-12
2
ABC789
DS
789101
2
COMPLETED
COMPLETED
DISPOSITION EVENT
FOLLOW-UP
2004-12-20
3
ABC789
DS
789102
1
SKIN RASH
ADVERSE
EVENT
DISPOSITION EVENT
TREATMENT PHASE
2004-09-30
4
ABC789
DS
789102
2
SUBJECT HAD
SEVERE RASH
TREATMENT
UNBLINDED
OTHER EVENT
TREATMENT PHASE
2004-10-01
5
ABC789
DS
789102
3
COMPLETED
COMPLETED
DISPOSITION EVENT
FOLLOW-UP
2004-12-28
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 109
Final November 12, 2008
Example 4
In this example, the CRF documents a link between the DS record and the AE record. This relationship is documented in the RELREC dataset.
Disposition (DS) Dataset
Row 1: Shows that Subject died of heart failure.
Row
STUDYID
DOMAIN
USUBJID
DSSEQ
DSTERM
DSDECOD
DSCAT
EPOCH
DSDTC
DSSTDTC
1
ABC123
DS
123102
1
Heart Failure
DEATH
DISPOSITION
EVENT
TREATMENT
PHASE
2003-09-29
2003-09-29
Adverse Event (AE) Dataset:
Row 1: Shows that Subject died due to heart failure.
Row
STUDYID
DOMAIN
USUBJID
AESEQ
AETERM
AESTDTC
AEENDTC
AEDECOD
AEBODSYS
AESEV
AESER
AEACN
1
ABC123
AE
123102
1
Heart Failure
2003-09-29
2003-09-29
HEART
FAILURE
CARDIOVASCULAR
SYSTEM
SEVERE
Y
NOT APPLICABLE
Row
AEREL
AEOUT
AESCAN
AESCONG
AESDISAB
AESDTH
AESHOSP
AESLIFE
AESOD
AESMIE
1 (cont)
DEFINITELY NOT RELATED
FATAL
N
N
N
Y
N
N
N
N
RELREC Dataset
Rows 1, 2: Show that the subject‘s disposition status that is related to the AE record.
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
1
ABC123
DS
123102
DSSEQ
1
1
2
ABC123
AE
123102
AESEQ
1
1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 110 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.3 MEDICAL HISTORY — MH
mh.xpt, Medical History — Events, Version 3.1.2. One record per medical history event per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1918HMH
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
623HSDTMIG 4.1.2.2,
624HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4
625H626HSDTMIG 4.1.2.3
MHSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
MHGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
627HSDTMIG 4.1.2.6,
SDTM 2.2.4
MHREFID
Reference ID
Char
Identifier
Internal or external medical history identifier.
Perm
SDTM 2.2.4
MHSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a Medical History page.
Perm
SDTM 2.2.4
MHTERM
Reported Term for the
Medical History
Char
Topic
Verbatim or preprinted CRF term for the medical condition or event.
Req
SDTM 2.2.2,
628HSDTMIG 4.1.3.6
MHMODIFY
Modified Reported Term
Char
Synonym
Qualifier
If MHTERM is modified to facilitate coding, then MHMODIFY will
contain the modified text.
Perm
SDTM 2.2.2,
629H630H631HSDTMIG 4.1.3.5
MHDECOD
Dictionary-Derived Term
Char
*
Synonym
Qualifier
Dictionary-derived text description of MHTERM or MHMODIFY.
Equivalent to the Preferred Term (PT in MedDRA). The sponsor is
expected to provide the dictionary name and version used to map
the terms utilizing the define.xml external codelist attributes
Perm
SDTM 2.2.2,
632H633H634HSDTMIG 4.1.3.5
MHCAT
Category for Medical
History
Char
*
Grouping
Qualifier
Used to define a category of related records. Examples: CARDIAC or
GENERAL
Perm
SDTM 2.2.2,
635HSDTMIG 4.1.2.6
MHSCAT
Subcategory for Medical
History
Char
*
Grouping
Qualifier
A further categorization of the condition or event.
Perm
SDTM 2.2.2,
636HSDTMIG 4.1.2.6
MHPRESP
Medical History Event Pre-
Specified
Char
(1919HNY)
Record
Qualifier
A value of ―Y‖ indicates that this medical history event was pre-specified
on the CRF. Values are null for spontaneously reported events (i.e., those
collected as free-text verbatim terms)
Perm
SDTM 2.2.2,
637H638HSDTMIG 4.1.2.7
639HSDTMIG 4.1.5.7
MHOCCUR
Medical History
Occurrence
Char
(1920HNY)
Record
Qualifier
Used when the occurrence of specific medical history conditions is
solicited to indicate whether or not (Y/N) a medical condition (MHTERM)
had ever occurred. Values are null for spontaneously reported events.
Perm
SDTM 2.2.2,
640HSDTMIG 4.1.5.7
MHSTAT
Completion Status
Char
(1921HND)
Record
Qualifier
The status indicates that the pre-specified question was not answered.
Perm
SDTM 2.2.2,
641HSDTMIG 4.1.5.1,
642HSDTMIG 4.1.5.7
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 111
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MHREASND
Reason Medical History
Not Collected
Char
Record
Qualifier
Describes the reason data for a pre-specified condition was not collected.
Used in conjunction with MHSTAT when value is NOT DONE.
Perm
SDTM 2.2.2
643HSDTMIG 4.1.5.1
644HSDTMIG 4.1.5.7
MHBODSYS
Body System or Organ
Class
Char
*
Record
Qualifier
Dictionary-derived. Body system or organ class that is involved in an
event or measurement from a standard hierarchy (e.g., MedDRA). When
using a multi-axial dictionary such as MedDRA, this should contain the
SOC used for the sponsor‘s analyses and summary tables which may not
necessarily be the primary SOC.
Perm
SDTM 2.2.2,
645H646H647HSDTMIG 4.1.3.5
MHDTC
Date/Time of History
Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5
648HSDTMIG 4.1.4.1
MHSTDTC
Start Date/Time of Medical
History Event
Char
ISO 8601
Timing
Perm
SDTM 2.2.5
649HSDTMIG 4.1.4.1
MHENDTC
End Date/Time of Medical
History Event
Char
ISO 8601
Timing
Perm
SDTM 2.2.5
650HSDTMIG 4.1.4.1
MHDY
Study Day of History
Collection
Num
Timing
1. Study day of medical history collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5
651HSDTMIG 4.1.4.4
MHENRF
End Relative to Reference
Period
Char
(STENRF)
Timing
Describes the end of the event relative to the sponsor-defined reference
period. The sponsor-defined reference period is a continuous period of
time defined by a discrete starting point and a discrete ending point
(represented by RFSTDTC and RFENDTC in Demographics)
Perm
SDTM 2.2.5
652HSDTMIG 4.1.4.7
MHENRTPT
End Relative to Reference
Time Point
Char
BEFORE,
AFTER,
COINCIDENT,
ONGOING, U
Timing
Identifies the end of the event as being before or after the reference time
point defined by variable MHENTPT.
Perm
SDTM 2.2.5
653HSDTMIG 4.1.4.7
MHENTPT
End Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the reference
point referred to by MHENRTPT. Examples: "2003-12-25" or "VISIT 2".
Perm
SDTM 2.2.5
654HSDTMIG 4.1.4.7
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 112 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.3.1 ASSUMPTIONS FOR MEDICAL HISTORY DOMAIN MODEL
1. MH Definition
a. The Medical History dataset generally includes the subject's prior and concomitant conditions at the start of the trial. Examples of subject medical
history information could include general medical history and gynecological history. Note that prior and concomitant medications should be
submitted in an appropriate dataset from the Interventions class (e.g., CM).
2. Medical History Description and Coding
a. MHTERM captures the verbatim term collected for the condition or event. It is the topic variable for the MH dataset. MHTERM is a required
variable and must have a value.
b. MHMODIFY is a permissible variable and should be included if the sponsor‘s procedure permits modification of a verbatim term for coding. The
modified term is listed in MHMODIFY. The variable should be populated as per the sponsor‘s procedures; null values are permitted.
c. If the sponsor codes the reported term (MHTERM) using a standard dictionary, then MHDECOD will be populated with the preferred term
derived from the dictionary. The sponsor is expected to provide the dictionary name and version used to map the terms utilizing the define.xml
external codelist attributes
d. MHBODSYS is the system organ class from the coding dictionary associated with the adverse event by the sponsor. This value may differ from
the primary system organ class designated in the coding dictionary's standard hierarchy.
e. Sponsors may include the values of additional levels from the coding dictionary's hierarchy (e.g., High Level Group Term, High Level Term,
Lower Level Term) in the SUPPMH dataset as described in 655HSection 8.4. See 1922HAppendix C5 for standard Supplemental Qualifier name codes.
f. If a CRF collects medical history by pre-specified body systems and the sponsor also codes reported terms using a standard dictionary, then
MHDECOD and MHBODSYS are populated using the standard dictionary. MHCAT and MHSCAT should be used for the pre-specified body
systems.
3. Additional Categorization and Grouping
a. MHCAT and MHSCAT may be populated with the sponsor's pre-defined categorization of medical history events, which are often pre-specified on
the CRF. Note that even if the sponsor uses the body system terminology from the standard dictionary, MHBODSYS and MHCAT may differ,
since MHBODSYS is derived from the coding system, while MHCAT is effectively assigned when the investigator records a condition under the
pre-specified category.
i. This categorization should not group all records (within the MH Domain) into one generic group such as ―Medical History‖ or ―General
Medical History‖ because this is redundant information with the domain code. If no smaller categorization can be applied, then it is not
necessary to include or populate this variable.
ii. Examples of MHCAT could include ―General Medical History― (see above assumption since if ―General Medical History‖ is an MHCAT
value then there should be other MHCAT values), ―Allergy Medical History, ― and ―Reproductive Medical History.‖
b. MHGRPID may be used to link (or associate) different records together to form a block of related records at the subject level within the MH
domain. It should not be used in place of MHCAT or MHSCAT, which are used to group data across subjects. For example, if a group of
syndromes reported for a subject were related to a particular disease then the MHGRPID variable could be populated with the appropriate text.
4. Pre-Specified Terms; Presence or Absence of Events
a. Information on medical history is generally collected in two different ways, either by recording free text or using a pre-specified list of terms. The
solicitation of information on specific medical history events may affect the frequency at which they are reported; therefore, the fact that a specific
medical history event was solicited may be of interest to reviewers. MHPRESP and MHOCCUR are used together to indicate whether the condition
in MHTERM was pre-specified and whether it occurred, respectively. A value of ―Y‖ in MHPRESP indicates that the term was pre-specified.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 113
Final November 12, 2008
b. MHOCCUR is used to indicate whether a pre-specified medical condition occurred; a value of Y indicates that the event occurred and N indicates
that it did not.
c. If a medical history event was reported using free text, the values of MHPRESP and MHOCCUR should be null. MHPRESP and MHOCCUR are
permissible fields and may be omitted from the dataset if all medical history events were collected as free text.
d. MHSTAT and MHREASND provide information about pre-specified medical history questions for which no response was collected. MHSTAT and
MHREASND are permissible fields and may be omitted from the dataset if all medications were collected as free text or if all pre-specified
conditions had responses in MHOCCUR.
Situation
Value of MHPRESP
Value of MHOCCUR
Value of MHSTAT
Spontaneously reported event occurred
Pre-specified event occurred
Y
Y
Pre-specified event did not occur
Y
N
Pre-specified event has no response
Y
NOT DONE
e. When medical history events are collected with the recording of free text, a record may be entered into the data management system to indicate ―no
medical history‖ for a specific subject or pre-specified body system category (e.g., Gastrointestinal). For these subjects or categories within subject,
do not include a record in the MH dataset to indicate that there were no events.
5. Timing Variables
a. Relative timing assessments such as ―Ongoing‖ or "Active" are common in the collection of Medical History information. MHENRF may be used
when this relative timing assessment is coincident with the start of the study reference period for the subject represented in the Demographics
dataset (RFSTDTC). MHENRTPT and MHENTPT may be used when "Ongoing" is relative to another date such as the screening visit date. See
examples below and 656HSection 4.1.4.7.
b. Additional timing variables (such as MHSTRF) may be used when appropriate.
6. Additional Events Qualifiers
The following Qualifiers would generally not be used in MH: --SER, --ACN, --ACNOTH, --REL, --RELNST, --OUT, --SCAN, --SCONG, --SDISAB,
--SDTH, --SHOSP, --SLIFE, --SOD, --SMIE.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 114 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.3.2 EXAMPLES FOR MEDICAL HISTORY DOMAIN MODEL
Example 1
In this example, a General Medical History CRF collects verbatim descriptions of conditions and events by body system (e.g., Endocrine, Metabolic) and asks
whether or not the conditions were ongoing at the time of the visit. Another CRF page is used for Cardiac history events and for primary diagnosis; this page does
not include the ongoing question.
Rows 1-3: MHSCAT displays the body systems specified on the General Medical History CRF. The reported events are coded using a standard dictionary.
MHDECOD and MHBODSYS display the preferred term and body system assigned through the coding process.
Rows 1-3: MHENRTPT has been populated based on the response to the "Ongoing " question on the General Medical History CRF. MHENTPT displays the
reference date for MHENRTPT - that is, the date the information was collected. If "Yes" is specified for Ongoing, MHENRTPT="ONGOING" if
"No" is checked, MHENRTPT="BEFORE." See 657HSection 4.1.4.7 for further guidance.
Row 4: MHCAT indicates that this record displays the primary diagnosis, "ISCHEMIC STROKE". This term was not coded.
Row 5: MHCAT indicates that this term was reported on the Cardiac Medical History page.
Row
STUDYID
DOMAIN
USUBJID
MHSEQ
MHTERM
MHDECOD
MHCAT
MHSCAT
MHBODSYS
MHSTDTC
MHENRTPT
MHENTPT
1
ABC123
MH
123101
1
ASTHMA
Asthma
GENERAL
MEDICAL
HISTORY
RESPIRATORY
Respiratory system
disorders
ONGOING
2004-09-18
2
ABC123
MH
123101
2
FREQUENT
HEADACHES
Headache
GENERAL
MEDICAL
HISTORY
CNS
Central and peripheral
nervous system
disorders
ONGOING
2004-09-18
3
ABC123
MH
123101
3
BROKEN LEG
Bone fracture
GENERAL
MEDICAL
HISTORY
OTHER
Musculoskeletal
system disorders
BEFORE
2004-09-18
4
ABC123
MH
123101
4
ISCHEMIC
STROKE
PRIMARY
DIAGNOSIS
2004-09-17T07:30
5
ABC123
MH
123101
5
CHF
Cardiac
failure
congestive
CARDIAC
MEDICAL
HISTORY
Cardiac disorders
2004-06
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 115
Final November 12, 2008
Example 2
This is an example of a medical history CRF where the history of specific (prespecified) conditions is solicited. The conditions are not coded using a standard
dictionary. The data are collected as part of the Screening visit.
Rows 1-10 MHPRESP values of ―Y‖ indicate that each condition was pre-specified on the CRF. The presence or absence of a condition is documented
with MHOCCUR. The data are collected as part of the Screening visit.
Rows 1-3, 7, 9: The absence of a condition is documented with MHOCCUR.
Rows 4-6, 8: The presence of a condition is documented with MHOCCUR.
Row 10: The question regarding ASTHMA was not asked. MHSTAT is used to indicate this and MHOCCUR is null.
Row
STUDYID
DOMAIN
USUBJID
MHSEQ
MHTERM
MHPRESP
MHOCCUR
MHSTAT
MHREASND
VISIT
VISITNUM
MHDTC
MHDY
1
ABC123
MH
101002
1
HISTORY OF EARLY
CORONARY ARTERY DISEASE
(<55 YEARS OF AGE)
Y
N
SCREEN
1
2006-04-22
-5
2
ABC123
MH
101002
2
CONGESTIVE HEART FAILURE
Y
N
SCREEN
1
2006-04-22
-5
3
ABC123
MH
101002
3
PERIPHERAL VASCULAR
DISEASE
Y
N
SCREEN
1
2006-04-22
-5
4
ABC123
MH
101002
4
TRANSIENT ISCHEMIC ATTACK
Y
Y
SCREEN
1
2006-04-22
-5
5
ABC123
MH
101002
5
ASTHMA
Y
Y
SCREEN
1
2006-04-22
-5
6
ABC123
MH
101003
1
HISTORY OF EARLY
CORONARY ARTERY DISEASE
(<55 YEARS OF AGE)
Y
Y
SCREEN
1
2006-05-03
-3
7
ABC123
MH
101003
2
CONGESTIVE HEART FAILURE
Y
N
SCREEN
1
2006-05-03
-3
8
ABC123
MH
101003
3
PERIPHERAL VASCULAR
DISEASE
Y
Y
SCREEN
1
2006-05-03
-3
9
ABC123
MH
101003
4
TRANSIENT ISCHEMIC ATTACK
Y
N
SCREEN
1
2006-05-03
-3
10
ABC123
MH
101003
5
ASTHMA
Y
NOT
DONE
FORGOT TO
ASK
SCREEN
1
2006-05-03
-3
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 116 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 3
In this example, three CRFs related to medical history are collected:
A General Medical History CRF collects descriptions of conditions and events by body system (e.g., Endocrine, Metabolic) and asks whether or not the
conditions were ongoing at study start. The reported events are coded using a standard dictionary.
A second CRF collects Stroke History.
A third CRF asks whether or not the subject had any of a list of 4 specific risk factors, with space for the investigator to write in other risk factors.
MHCAT is used to indicate the CRF from which the medical condition came.
Rows 1-3: MHSCAT displays the body systems specified on the General Medical History CRF. The reported events are coded using a standard dictionary.
Rows 1-3: MHENRF has been populated based on the response to the "Ongoing at Study Start" question on the General Medical History CRF. If "Yes" is specified,
MHENRF="DURING/AFTER;" if "No" is checked, MHENRF="BEFORE" See 658HSection 4.1.4.7 for further guidance on using --STRF and --ENRF.
Row 4: MHCAT indicates that this record displays Stroke History. This term is not coded.
Rows 1-4: MHPRESP and MHOCCUR are null for the conditions, which are not prespecified.
Rows 5-9: MHCAT indicates that these terms were reported on the RISK FACTORS page. These terms are not coded.
Rows 5-8: MHPRESP values of ―Y‖ indicate that each risk factor was pre-specified on the CRF. MHOCCUR is populated with Y or N corresponding to the
CRF response to the questions for the 4 pre-specified risk factors.
Row 9: MHPRESP and MHOCCUR are null for the other risk factor written in by the investigator as free text.
Row
STUDYID
DOMAIN
USUBJID
MHSEQ
MHTERM
MHDECOD
MHCAT
MHSCAT
MHPRESP
1
ABC123
MH
123101
1
ASTHMA
Asthma
GENERAL MEDICAL HISTORY
RESPIRATORY
2
ABC123
MH
123101
2
FREQUENT HEADACHES
Headache
GENERAL MEDICAL HISTORY
CNS
3
ABC123
MH
123101
3
BROKEN LEG
Bone fracture
GENERAL MEDICAL HISTORY
OTHER
4
ABC123
MH
123101
4
ISCHEMIC STROKE
STROKE HISTORY
5
ABC123
MH
123101
5
DIABETES
RISK FACTORS
Y
6
ABC123
MH
123101
6
HYPERCHOLESTEROLEMIA
RISK FACTORS
Y
7
ABC123
MH
123101
7
HYPERTENSION
RISK FACTORS
Y
8
ABC123
MH
123101
8
TIA
RISK FACTORS
Y
9
ABC123
MH
123101
9
MATERNAL FAMILY HX OF STROKE
RISK FACTORS
Row
MHOCCUR
MHBODSYS
MHSTDTC
MHENRF
1 (cont’d)
Respiratory system disorders
DURING/AFTER
2 (cont’d)
Central and peripheral nervous system disorders
DURING/AFTER
3 (cont’d)
Musculoskeletal system disorders
BEFORE
4 (cont’d)
2004-09-17T07:30
5 (cont’d)
Y
6 (cont’d)
Y
7 (cont’d)
Y
8 (cont’d)
N
9 (cont’d)
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 117
Final November 12, 2008
6.2.4 PROTOCOL DEVIATIONS — DV
dv.xpt, Protocol Deviations — Events, Version 3.1.2. One record per protocol deviation per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1923HDV
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
659HSDTMIG 4.1.2.2,
660HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4
661H662HSDTMIG 4.1.2.3
DVSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
DVREFID
Reference ID
Char
Identifier
Internal or external identifier.
Perm
SDTM 2.2.4
DVSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a CRF page.
Perm
SDTM 2.2.4
DVTERM
Protocol Deviation Term
Char
Topic
Verbatim name of the protocol deviation criterion. Example: IVRS
PROCESS DEVIATION - NO DOSE CALL PERFORMED. The DVTERM
values will map to the controlled terminology in DVDECOD, such as
TREATMENT DEVIATION.
Req
SDTM 2.2.2,
663HSDTMIG 4.1.3.6
DVDECOD
Protocol Deviation Coded
Term
Char
*
Synonym
Qualifier
Controlled terminology for the name of the protocol deviation. Examples:
SUBJECT NOT WITHDRAWN AS PER PROTOCOL, SELECTION
CRITERIA NOT MET, EXCLUDED CONCOMITANT MEDICATION,
TREATMENT DEVIATION.
Perm
SDTM 2.2.2,
664H665H666HSDTMIG 4.1.3.5
DVCAT
Category for Protocol
Deviation
Char
*
Grouping
Qualifier
Category of the protocol deviation criterion.
Perm
SDTM 2.2.2,
667HSDTMIG 4.1.2.6
DVSCAT
Subcategory for Protocol
Deviation
Char
*
Grouping
Qualifier
A further categorization of the protocol deviation.
Perm
SDTM 2.2.2,
668HSDTMIG 4.1.2.6
EPOCH
Epoch
Char
*
Timing
Epoch associated with the start date/time of the deviation. Examples:
TREATMENT PHASE, SCREENING, and FOLLOW-UP.
Perm
SDTM 2.2.5
669HSDTMIG 7.1.2
DVSTDTC
Start Date/Time of
Deviation
Char
ISO 8601
Timing
Start date/time of deviation represented in ISO 8601 character format.
Perm
SDTM 2.2.5
670HSDTMIG 4.1.4.1
DVENDTC
End Date/Time of
Deviation
Char
ISO 8601
Timing
End date/time of deviation represented in ISO 8601 character format.
Perm
SDTM 2.2.5
671HSDTMIG 4.1.4.1
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 118 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.4.1 ASSUMPTIONS FOR PROTOCOL DEVIATIONS DOMAIN MODEL
1. The DV domain is an Events model for collected protocol deviations and not for derived protocol deviations that are more likely to be part of analysis.
Events typically include what the event was, captured in --TERM (the topic variable), and when it happened (captured in its start and/or end dates). The
intent of the domain model is to capture protocol deviations that occurred during the course of the study (see ICH E3, Section 10.2). Usually these are
deviations that occur after the subject has been randomized or received the first treatment.
2. This domain should not be used to collect entry criteria information. Violated inclusion/exclusion criteria are stored in IE. The Deviations domain is for
more general deviation data. A protocol may indicate that violating an inclusion/exclusion criterion during the course of the study (after first dose) is a
protocol violation. In this case, this information would go into DV.
3. Additional Events Qualifiers
The following Qualifiers would generally not be used in DV: --PRESP, --OCCUR, --STAT, --REASND, --BODSYS, --LOC, --SEV, --SER, --ACN,
--ACNOTH, --REL, --RELNST, --PATT, --OUT, --SCAN, --SCONG, --SDISAB, --SDTH, --SHOSP, --SLIFE, --SOD, --SMIE, --CONTRT, --TOXGR.
6.2.4.2 EXAMPLES FOR PROTOCOL DEVIATIONS DOMAIN MODEL
Example 1
This is an example of data that was collected on a protocol-deviations CRF. The DVDECOD column is for controlled terminology whereas the DVTERM is free
text.
Rows 1 and 3: Show examples of a TREATMENT DEVIATION type of protocol deviation.
Row 2: Shows an example of a deviation due to the subject taking a prohibited concomitant mediation.
Rows 4: Shows an example of a medication that should not be taken during the study.
Row
STUDYID
DOMAIN
USUBJID
DVSEQ
DVTERM
DVDECOD
EPOCH
DVSTDTC
1
ABC123
DV
123101
1
IVRS PROCESS DEVIATION - NO DOSE
CALL PERFORMED.
TREATMENT DEVIATION
TREATMENT PHASE
2003-09-21
2
ABC123
DV
123103
1
DRUG XXX ADMINISTERED DURING
STUDY TREATMENT PERIOD
EXCLUDED CONCOMITANT
MEDICATION
TREATMENT PHASE
2003-10-30
3
ABC123
DV
123103
2
VISIT 3 DOSE <15 MG
TREATMENT DEVIATION
TREATMENT PHASE
2003-10-30
4
ABC123
DV
123104
1
TOOK ASPIRIN
PROHIBITED MEDS
TREATMENT PHASE
2003-11-30
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 119
Final November 12, 2008
6.2.5 CLINICAL EVENTS — CE
ce.xpt, Clinical Events — Events, Version 3.1.2. One record per event per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or
Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1924HCE
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4
672HSDTMIG 4.1.2.2,
673HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
674H675HSDTMIG 4.1.2.3
CESEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
CEGRPID
Group ID
Char
Identifier
Used to tie together a block of related records for a subject within a
domain.
Perm
SDTM 2.2.4
676HSDTMIG 2.1,
677HSDTMIG 4.1.2.6
CEREFID
Reference ID
Char
Identifier
Internal or external identifier.
Perm
SDTM 2.2.4
CESPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a CRF page.
Perm
SDTM 2.2.4
CETERM
Reported Term for the
Clinical Event
Char
Topic
Term for the medical condition or event. Most likely pre-printed on CRF.
Req
SDTM 2.2.2,
678HSDTMIG 4.1.3.6
CEDECOD
Dictionary-Derived Term
Char
*
Synonym
Qualifier
Controlled terminology for the name of the clinical event. The sponsor
is expected to provide the dictionary name and version used to
map the terms utilizing the define.xml external codelist attributes
Perm
SDTM 2.2.2,
679H680H681HSDTMIG 4.1.3.5
CECAT
Category for Clinical Event
Char
*
Grouping
Qualifier
Used to define a category of related records.
Perm
SDTM 2.2.2,
682HSDTMIG 4.1.2.6
CESCAT
Subcategory for Clinical
Event
Char
*
Grouping
Qualifier
A further categorization of the condition or event.
Perm
SDTM 2.2.2,
683HSDTMIG 4.1.2.6
CEPRESP
Clinical Event Pre-
Specified
Char
(1925HNY)
Record
Qualifier
Used to indicate whether the Event in CETERM was pre-specified. Value
is Y for pre-specified events, null for spontaneously reported events.
Perm
SDTM 2.2.2,
684H685HSDTMIG 4.1.2.7
686HSDTMIG 4.1.5.7
CEOCCUR
Clinical Event Occurrence
Char
(1926HNY)
Record
Qualifier
Used when the occurrence of specific events is solicited to indicate
whether or not a clinical event occurred. Values are null for spontaneously
reported events.
Perm
SDTM 2.2.2,
687HSDTMIG 4.1.5.7
CESTAT
Completion Status
Char
(1927HND)
Record
Qualifier
The status indicates that a question from a pre-specified list was not
answered.
Perm
SDTM 2.2.2,
688HSDTMIG 4.1.5.1,
689HSDTMIG 4.1.5.7
CEREASND
Reason Clinical Event Not
Collected
Char
Record
Qualifier
Describes the reason clinical event data was not collected. Used in
conjunction with CESTAT when value is NOT DONE.
Perm
SDTM 2.2.2,
690HSDTMIG 4.1.5.1
691HSDTMIG 4.1.5.7
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 120 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or
Format
Role
CDISC Notes
Core
References
CEBODSYS
Body System or Organ
Class
Char
*
Record
Qualifier
Dictionary-derived. Body system or organ class that is involved in an
event or measurement from a standard hierarchy (e.g., MedDRA). When
using a multi-axial dictionary such as MedDRA, this should contain the
SOC used for the sponsor‘s analyses and summary tables which may not
necessarily be the primary SOC.
Perm
SDTM 2.2.2,
692H693H694HSDTMIG 4.1.3.5
CESEV
Severity/Intensity
Char
*
Record
Qualifier
The severity or intensity of the event. Examples: MILD, MODERATE,
SEVERE
Perm
SDTM 2.2.2,
CEDTC
Date/Time of Event
Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
695HSDTMIG 4.1.4.1
CESTDTC
Start Date/Time of Clinical
Event
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
696HSDTMIG 4.1.4.1;
697HSDTMIG 4.1.4.2
CEENDTC
End Date/Time of Clinical
Event
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
698HSDTMIG 4.1.4.1;
699HSDTMIG 4.1.4.2
CEDY
Study Day of Event
Collection
Num
Timing
1. Study day of clinical event collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5,
700HSDTMIG 4.1.4.4
CESTRF
Start Relative to Reference
Period
Char
(1928HSTENRF)
Timing
Describes the start of the clinical event relative to the sponsor-defined
reference period. The sponsor-defined reference period is a continuous
period of time defined by a discrete starting point and a discrete ending
point (represented by RFSTDTC and RFENDTC in Demographics).
Perm
SDTM 2.2.5,
701HSDTMIG 4.1.4.7
CEENRF
End Relative to Reference
Period
Char
(1929HSTENRF)
Timing
Describes the end of the event relative to the sponsor-defined reference
period. The sponsor-defined reference period is a continuous period of
time defined by a discrete starting point and a discrete ending point
(represented by RFSTDTC and RFENDTC in Demographics).
Perm
SDTM 2.2.5,
702HSDTMIG 4.1.4.7
CESTRTPT
Start Relative to Reference
Time Point
Char
BEFORE, AFTER,
COINCIDENT, U
Timing
Identifies the start of the observation as being before or after the reference
time point defined by variable CESTTPT.
Perm
SDTM 2.2.5,
703HSDTMIG 4.1.4.7
CESTTPT
Start Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the sponsor-
defined reference point referred to by --STRTPT. Examples:
"2003-12-15" or "VISIT 1".
Perm
SDTM 2.2.5,
704HSDTMIG 4.1.4.7
CEENRTPT
End Relative to Reference
Time Point
Char
BEFORE, AFTER,
COINCIDENT,
ONGOING, U
Timing
Identifies the end of the event as being before or after the reference time
point defined by variable CEENTPT.
Perm
SDTM 2.2.5,
705HSDTMIG 4.1.4.7
CEENTPT
End Reference Time Point
Char
Timing
Description or date/time in ISO 8601 character format of the reference
point referred to by CEENRTPT. Examples: "2003-12-25" or "VISIT 2".
Perm
SDTM 2.2.5,
706HSDTMIG 4.1.4.7
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 121
Final November 12, 2008
6.2.5.1 ASSUMPTIONS FOR CLINICAL EVENTS DOMAIN MODEL
1. The intent of the domain model is to capture clinical events of interest that would not be classified as adverse events. The data may be data about
episodes of symptoms of the disease under study (often known as signs and symptoms), or about events that do not constitute adverse events in
themselves, though they might lead to the identification of an adverse event. For example, in a study of an investigational treatment for migraine
headaches, migraine headaches may not be considered to be adverse events per protocol. The occurrence of migraines or associated signs and symptoms
might be reported in CE. Other studies might track the occurrence of specific events as efficacy endpoints. For example, in a study of an investigational
treatment for prevention of ischemic stroke, all occurrences of TIA, stroke and death might be captured as clinical events and assessed as to whether
they meet endpoint criteria. Note that other information about these events may also be reported in other datasets. For example, the event leading to
death would be reported in AE and death would also be a reason for study discontinuation in DS.
2. CEOCCUR and CEPRESP are used together to indicate whether the event in CETERM was pre-specified, and whether it occurred. CEPRESP can be
used to separate records that correspond to probing questions for pre-specified events from those that represent spontaneously reported events, while
CEOCCUR contains the responses to such questions. The table below shows how these variables are populated in various situations.
Situation
Value of
CEPRESP
Value of
CEOCCUR
Value of CESTAT
Spontaneously reported event occurred
Pre-specified event occurred
Y
Y
Pre-specified event did not occur
Y
N
Pre-specified event has no response
Y
NOT DONE
3. The collection of write-in events on a Clinical Events CRF should be considered with caution. Sponsors must ensure that all adverse events are recorded
in the AE domain.
4. Timing Variables
a. Relative timing assessments "Prior" or ―Ongoing‖ are common in the collection of Clinical Event information. CESTRF or CEENRF may be used
when this timing assessment is relative to the study reference period for the subject represented in the Demographics dataset (RFENDTC).
CESTRTPT with CESTTPT, and/or CEENRTPT with CEENTPT may be used when "Prior" or "Ongoing" are relative to specific dates other than
the start and end of the study reference period. See 707HSection 4.1.4.7.
b. Additional Timing variables may be used when appropriate.
5. Additional Events Qualifiers
The following Qualifiers would generally not be used in CE: --SER, --ACN, --ACNOTH, --REL, --RELNST, --OUT, --SCAN, --SCONG,
--SDISAB, --SDTH, --SHOSP, --SLIFE, --SOD, --SMIE.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 122 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.2.5.2 EXAMPLES FOR CLINICAL EVENTS DOMAIN MODEL
Example 1
Assumptions:
CRF data are collected about pre-specified events that, in the context of this study, are not reportable as Adverse Events.
The data being collected includes ―event-like‖ timing and other Qualifiers.
Data are collected about pre-specified clinical events in a log independent of visits, rather than in a visit-based CRF module.
Note that data collected is the start date of the event, which is ―event-like‖ data.
No "yes/no" data on the occurrence of the event is collected.
CRF:
Record start dates of any of the following signs that occur.
Clinical Sign
Start Date
Rash
Wheezing
Edema
Conjunctivitis
Data:
Rows 1-3: Show records for clinical events for which start dates were recorded. Since conjunctivitis was not observed, no start date was recorded and there is no
CE record.
Row
STUDYID
DOMAIN
USUBJID
CESEQ
CETERM
CEPRESP
CEOCCUR
CESTDTC
1
ABC123
CE
123
1
Rash
Y
Y
2006-05-03
2
ABC123
CE
123
2
Wheezing
Y
Y
2006-05-03
3
ABC123
CE
123
3
Edema
Y
Y
2006-05-06
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 123
Final November 12, 2008
Example 2
Assumptions:
CRF includes both questions about pre-specified clinical events (events not reportable as AEs in the context of this study) and spaces for the investigator
to write in additional clinical events.
Note that data being collected are start and end dates, which are "event-like, " and severity, which is a Qualifier in the Events general observation class.
CRF:
Event
Yes
No
Date Started
Date Ended
Severity
Nausea
Vomit
Diarrhea
Other, Specify
Data:
Row 1: Shows a record for a response to the pre-specified clinical event "Nausea." The CEPRESP value of ―Y‖ indicates that there was a probing question,
and the response to the probe (CEOCCUR) was "Yes."
Row 2: Shows a record for a response to the pre-specified clinical event "Vomit." The CEPRESP value of ―Y‖ indicates that there was a probing question,
and the response to the question (CEOCCUR) was "No‖.
Row 3: Shows a record for the pre-specified clinical event "Diarrhea." A value of Y for CEPRESP indicates it was pre-specified. The CESTAT value of NOT
DONE indicates that there was either 1) no probing question (investigator error) or a probing question with no response.
Row 4: Shows a record for a write-in Clinical Event recorded in the "Other, Specify" space. Because this event was not pre-specified, CEPRESP and
CEOCCUR are null (708HSection 4.1.2.7 further information on populating the Topic variable when ―Other, specify‖ is used on the CRF).
Row
STUDYID
DOMAIN
USUBJID
CESEQ
CETERM
CEPRESP
CEOCCUR
CESTAT
CESEV
CESTDTC
CEENDTC
1
ABC123
CE
123
1
NAUSEA
Y
Y
MODERATE
2005-10-12
2005-10-15
2
ABC123
CE
123
2
VOMIT
Y
N
3
ABC123
CE
123
3
DIARRHEA
Y
NOT DONE
4
ABC123
CE
123
4
SEVERE HEAD
PAIN
SEVERE
2005-10-09
2005-10-11
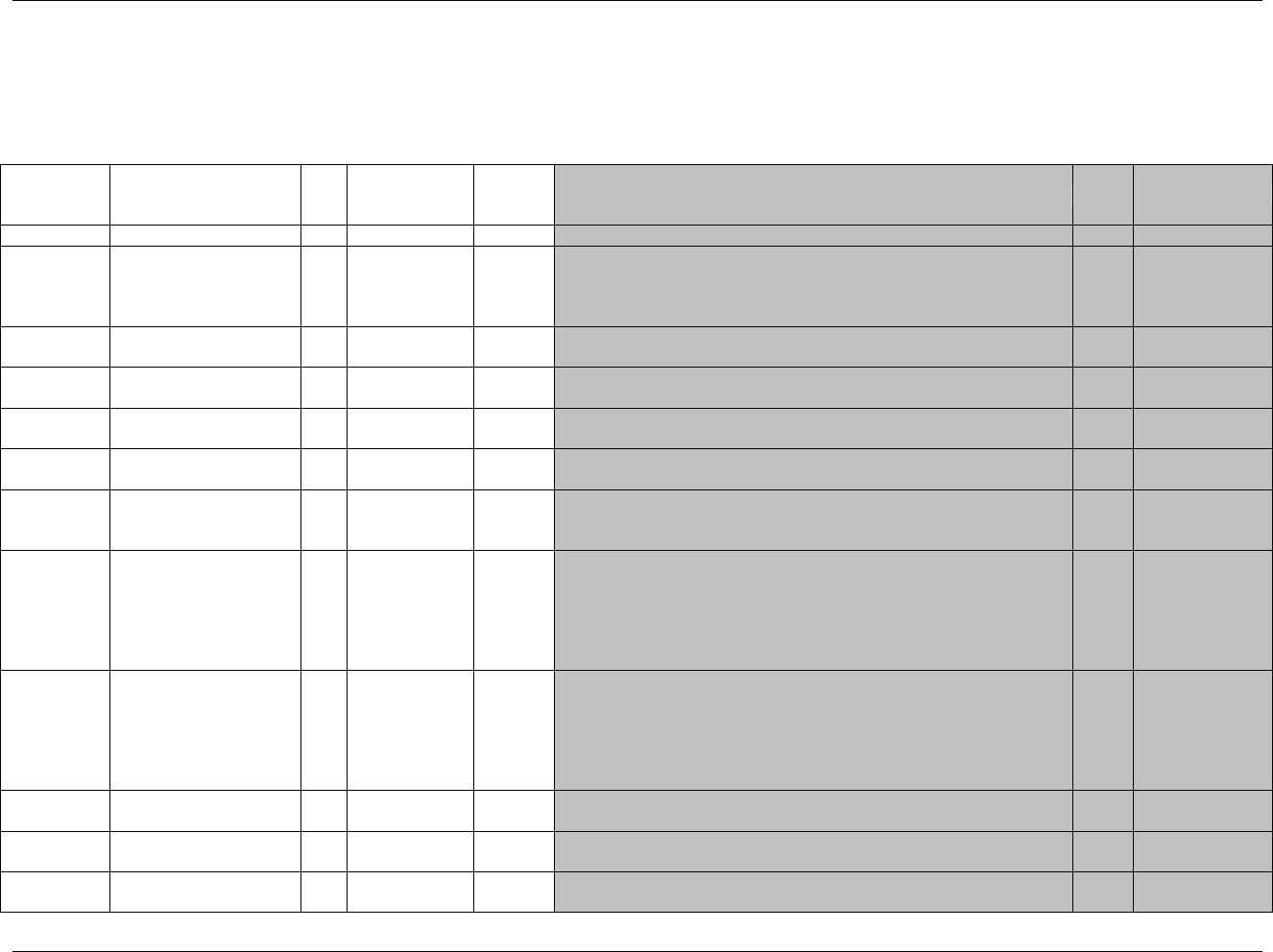
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 124 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3 FINDINGS
6.3.1 ECG TEST RESULTS — EG
eg.xpt, ECG — Findings, Version 3.1.2. One record per ECG observation per time point per visit per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1930HEG
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
709HSDTMIG 4.1.2.2,
710HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
711H712HSDTMIG 4.1.2.3
EGSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
EGGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
713HSDTMIG 4.1.2.6
EGREFID
ECG Reference ID
Char
Identifier
Internal or external ECG identifier. Example: UUID.
Perm
SDTM 2.2.4,
714HSDTMIG 4.1.2.6
EGSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor's operational database.
Example: Line number from the ECG page.
Perm
SDTM 2.2.4,
715HSDTMIG 4.1.2.6
EGTESTCD
ECG Test or Examination
Short Name
Char
(716HEGTESTCD)
Topic
Short name of the measurement, test, or examination described in
EGTEST. It can be used as a column name when converting a dataset
from a vertical to a horizontal format. The value in EGTESTCD cannot be
longer than 8 characters, nor can it start with a number (e.g., ―1TEST‖).
EGTESTCD cannot contain characters other than letters, numbers, or
underscores. Examples :PRMEAN, QTMEAN
Req
SDTM 2.2.3,
717H718HSDTMIG 4.1.1.9,
719HSDTMIG 4.1.2.1,
SDTMIG 4.1.5.5
720HSDTMIG
Appendix C1
EGTEST
ECG Test or Examination
Name
Char
(721HEGTEST)
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the measurement
or finding. The value in EGTEST cannot be longer than 40 characters.
Examples: Summary (Mean) PR Duration, Summary (Mean) QT Duration
Req
SDTM 2.2.3,
722HSDTMIG 4.1.2.1,
723HSDTMIG 4.1.2.4,
724HSDTMIG 4.1.5.3.1,
725HSDTMIG
Appendix C1
EGCAT
Category for ECG
Char
*
Grouping
Qualifier
Used to categorize ECG observations across subjects. Examples:
MEASUREMENT, FINDING, INTERVAL
Perm
SDTM 2.2.3,
726HSDTMIG 4.1.2.6
EGSCAT
Subcategory for ECG
Char
*
Grouping
Qualifier
A further categorization of the ECG.
Perm
SDTM 2.2.3,
727HSDTMIG 4.1.2.6
EGPOS
ECG Position of Subject
Char
(728HPOSITION)
Record
Qualifier
Position of the subject during a measurement or examination. Examples:
SUPINE, STANDING, SITTING.
Perm
SDTM 2.2.3
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 125
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
EGORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the ECG measurement or finding as originally received or
collected. Examples of expected values are 62 or 0.151 when the result is
an interval or measurement, or ―ATRIAL FIBRILLATION‖ or ―QT
PROLONGATION‖ when the result is a finding.
Exp
SDTM 2.2.3,
729H730HSDTMIG 4.1.5.1
EGORRESU
Original Units
Char
(731HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for EGORRES.
Examples: sec or msec.
Perm
SDTM 2.2.3,
732HSDTMIG 4.1.3.2,
733HSDTMIG 4.1.5.1
EGSTRESC
Character Result/Finding
in Std Format
Char
(734HEGSTRESC)
Result
Qualifier
Contains the result value for all findings, copied or derived from
EGORRES in a standard format or standard units. EGSTRESC should
store all results or findings in character format; if results are numeric, they
should also be stored in numeric format in EGSTRESN. For example, if a
test has results of ―NONE‖, ―NEG‖, and ―NEGATIVE‖ in EGORRES
and these results effectively have the same meaning, they could be
represented in standard format in EGSTRESC as ―NEGATIVE‖. For
other examples, see general assumptions. Additional examples of result
data: SINUS BRADYCARDIA, ATRIAL FLUTTER, ATRIAL
FIBRILLATION.
Exp
SDTM 2.2.3,
735HSDTMIG 4.1.3.6,
736HSDTMIG 4.1.5.1
EGSTRESN
Numeric Result/Finding in
Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from EGSTRESC. EGSTRESN should store all
numeric test results or findings.
Perm
SDTM 2.2.3,
737HSDTMIG 4.1.5.1
EGSTRESU
Standard Units Char (738HUNIT) Variable
Qualifier Standardized unit used for EGSTRESC or EGSTRESN. Perm SDTM 2.2.3,
739HSDTMIG 4.1.3.2,
740HSDTMIG 4.1.5.1
EGSTAT
Completion Status Char (1931HND) Record
Qualifier Used to indicate an ECG was not done, or an ECG measurement was not
taken. Should be null if a result exists in EGORRES. Perm SDTM 2.2.3,
741HSDTMIG 4.1.5.1,
742HSDTMIG 4.1.5.7,
743HSDTMIG
Appendix C1
EGREASND
Reason ECG Not
Performed Char Record
Qualifier Describes why a measurement or test was not performed. Examples:
BROKEN EQUIPMENT or SUBJECT REFUSED. Used in conjunction
with EGSTAT when value is NOT DONE.
Perm SDTM 2.2.3,
744HSDTMIG 4.1.5.1,
745HSDTMIG 4.1.5.7
EGXFN ECG External File Name Char Record
Qualifier File name and path for the external ECG Waveform file. Perm SDTM 2.2.3
EGNAM
Vendor Name Char Record
Qualifier Name or identifier of the laboratory or vendor who provided the test
results. Perm SDTM 2.2.3
EGLOC
Lead Location Used for
Measurement Char (746HLOC) Record
Qualifier The lead used for the measurement, examples, V1, V6, aVR, I, II, III. Perm SDTM 2.2.3,
747HSDTMIG 4.1.1.9
EGMETHOD
Method of ECG Test Char (749HEGMETHOD) Record
Qualifier Method of the ECG test. Examples: 12 LEAD STANDARD. Perm SDTM 2.2.3,
750HSDTMIG
Appendix C1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 126 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
EGBLFL
Baseline Flag
Char
(1932HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or
null.
Exp
SDTM 2.2.3,
751HSDTMIG 4.1.3.7,
752HSDTMIG
Appendix C1
EGDRVFL
Derived Flag
Char
(1933HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records which
represent the average of other records, or that do not come from the CRF, or are not
as originally collected or received are examples of records that would be derived
for the submission datasets. If EGDRVFL=Y, then EGORRES could be null, with
EGSTRESC, and (if numeric) EGSTRESN having the derived value.
Perm
SDTM 2.2.3,
753HSDTMIG 4.1.3.7,
754HSDTMIG 4.1.5.1,
755HSDTMIG
Appendix C1
EGEVAL
Evaluator
Char
*
Record
Qualifier
Role of the person who provided the evaluation. Used only for results that
are subjective (e.g., assigned by a person or a group). Should be null for
records that contain collected or derived data. Examples:
INVESTIGATOR, ADJUDICATION COMMITTEE, VENDOR.
Perm
SDTM 2.2.3,
756HSDTMIG 4.1.5.4
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
757HSDTMIG 4.1.4.5,
758HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
759HSDTMIG 4.1.4.5,
760HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
761HSDTMIG 4.1.4.5,
762HSDTMIG 7.4
EGDTC
Date/Time of ECG
Char
ISO 8601
Timing
Date of ECG.
Exp
SDTM 2.2.5,
763HSDTMIG 4.1.4.1,
764HSDTMIG 4.1.4.2,
765H766HSDTMIG 4.1.4.8
EGDY
Study Day of ECG
Num
Timing
1. Study day of the ECG, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm
SDTM 2.2.5,
767HSDTMIG 4.1.4.4,
768HSDTMIG 4.1.4.6
EGTPT
Planned Time Point Name
Char
Timing
1. Text Description of time when measurement should be taken.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See EGTPTNUM and EGTPTREF.
Examples: Start, 5 min post.
Perm
SDTM 2.2.5,
769HSDTMIG 4.1.4.10
EGTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of EGTPT to aid in sorting.
Perm
SDTM 2.2.5,
770HSDTMIG 4.1.4.10
EGELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Planned elapsed time (in ISO 8601) relative to a fixed time point reference
(EGTPTREF). Not a clock time or a date time variable. Represented as an ISO
8601 duration. Examples: ―-PT15M‖ to represent the period of 15 minutes
prior to the reference point indicated by EGTPTREF, or ―PT8H‖ to represent
the period of 8 hours after the reference point indicated by EGTPTREF.
Perm
SDTM 2.2.5,
771HSDTMIG 4.1.4.3
772HSDTMIG 4.1.4.10
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 127
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
EGTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by EGELTM, EGTPTNUM,
and EGTPT. Examples: PREVIOUS DOSE, PREVIOUS MEAL.
Perm
SDTM 2.2.5,
773HSDTMIG 4.1.4.10
EGRFTDTC
Date/Time of Reference
Time Point
Char
ISO 8601
Timing
Date/time of the reference time point, EGTPTREF.
Perm
SDTM 2.2.5,
774HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.1.1 ASSUMPTIONS FOR ECG TEST RESULTS DOMAIN MODEL
1. EG Definition: CRF data that captures interval measurements and summary information from an ECG. This domain captures ECG data collected on
the CRF or received from a central provider or vendor.
2. EGREFID is intended to store an identifier (e.g., UUID) for the associated ECG tracing. EGFXN is intended to store the name of and path to the ECG
waveform file when it is submitted.
3. The method for QT interval correction is specified in the test name by controlled terminology: EGTESTCD = QTCF and EGTEST = QTcF for
Fridericia‘s Correction Formula; EGTESTCD=QTCB and EGTEST = QTcB for Bazett's Correction Formula.
4. EGNRIND can be added to indicate where a result falls with respect to reference range defined by EGORNRLO and EGORNRHI. Examples: HIGH,
LOW. Clinical significance would be represented as described in 775HSection 4.1.5.5 as a record in SUPPEG with a QNAM of EGCLSIG (see also ECG
Example 1 below).
5. When QTCF and QTCB are derived by the sponsor, the derived flag (EGDRVFL) is set to Y. However, when the QTCF or QTCB is received from a
central provider or vendor, the value would go into EGORRES and EGDRVFL would be null (See 776HSection 4.1.1.8.1).
6. The following Qualifiers would not generally be used in EG: --MODIFY, --BODSYS, --SPEC, --SPCCND, --FAST, --SEV. It is recommended that
--LOINC not be used.
6.3.1.2 EXAMPLES FOR ECG TEST RESULTS DOMAIN MODEL
Example 1
Rows 1-6: Show how ECG measurements are represented.
Row 1: Shows a measurement of ventricular rate. The Supplemental Qualifier record related to this EG record, Row 1 in the SUPPEG dataset, has
QNAM = EGCLSIG and QVAL = "N". This indicates that the ventricular rate of 62 bpm was assessed as not being clinically significant. See
777HSection 4.1.5.5 for more on clinical significance.
Rows 2-4: Show the data in original units of measure in EGORRES, EGSTRESC, and EGSTRESN. See 778HSection 4.1.5.1 for additional examples for the
population of Result Qualifiers.
Row 2: The TEST " Summary (Mean) PR Duration ―has a result of 0.15 sec. The Supplemental Qualifier record related to this EG record, Row 2 in the
SUPPEG dataset, has QNAM = CLSIG and QVAL = "Y". This indicates that the PR interval of 0.15 sec was assessed as being clinically
significant. See 779HSection 4.1.5.5 for more on clinical significance.
Rows 2-10: Show how EGCAT could be used to group the intervals and the findings.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 128 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Rows 5-6: Show QTCB and QTCF. The data shows a ―Y‖ in the EGDRVFL column since these results are derived by the sponsor in this example. Note that
EGORRES is null for these derived records.
Rows 7-10: Show how ECG findings are represented.
Row 11: Shows a way of representing technical problems that are important to the overall understanding of the ECG, but which are not truly findings or
interpretations.
Row 12: The TEST "Interpretation" (i.e., the interpretation of the ECG strip as a whole), is "ABNORMAL ".
eg.xpt
Row
STUDYID
DOMAIN
USUBJID
EGSEQ
EGCAT
EGREFID
EGTESTCD
EGTEST
EGPOS
EGORRES
EGORRESU
1
XYZ
EG
XYZ-US-701-002
1
MEASUREMENT
334PT89
HRMEAN
Summary (Mean) Heart Rate
SUPINE
62
BEATS/MIN
2
XYZ
EG
XYZ-US-701-002
2
INTERVAL
334PT89
PRMEAN
Summary (Mean) PR Duration
SUPINE
0.15
sec
3
XYZ
EG
XYZ-US-701-002
3
INTERVAL
334PT89
QRSDUR
Summary (Mean) QRS
Duration
SUPINE
0.103
sec
4
XYZ
EG
XYZ-US-701-002
4
INTERVAL
334PT89
QTMEAN
Summary (Mean) QT Duration
SUPINE
0.406
sec
5
XYZ
EG
XYZ-US-701-002
5
INTERVAL
334PT89
QTCB
QTcB – Bazett's Correction
Formula
SUPINE
6
XYZ
EG
XYZ-US-701-002
6
INTERVAL
334PT89
QTCF
QTcF – Fridericia's Correction
Formula
SUPINE
7
XYZ
EG
XYZ-US-701-002
7
FINDING
334PT89
RHYRATE
Rhythm and Rate
SUPINE
ATRIAL FIBRILLATION
8
XYZ
EG
XYZ-US-701-002
8
FINDING
334PT89
RHYRATE
Rhythm and Rate
SUPINE
ATRIAL FLUTTER
9
XYZ
EG
XYZ-US-701-002
9
FINDING
334PT89
QTABN
QT Abnormalities
SUPINE
PROLONGED QT
10
XYZ
EG
XYZ-US-701-002
10
FINDING
334PT89
VCABN
Ventricular Conduction
Abnormalities
SUPINE
LEFT VENTRICULAR
HYPERTROPHY
11
XYZ
EG
XYZ-US-701-002
11
334PT89
TECHPROB
Technical Problems
SUPINE
INCORRECT ELECTRODE
PLACEMENT
12
XYZ
EG
XYZ-US-701-002
12
334PT89
INTP
Interpretation
SUPINE
ABNORMAL
Row
EGSTRESC
EGSTRESN
EGSTRESU
EGXFN
EGNAM
EGDRVFL
EGEVAL
VISITNUM
VISIT
EGDTC
EGDY
1 (cont)
62
62
BEATS/MIN
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
2 (cont)
150
150
msec
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
3 (cont)
103
103
msec
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
4 (cont)
406
406
msec
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
5 (cont)
469
469
msec
PQW436789-07.xml
Test Lab
Y
1
Screening 1
2003-04-15T11:58
-36
6 (cont)
446
446
msec
PQW436789-07.xml
Test Lab
Y
1
Screening 1
2003-04-15T11:58
-36
7 (cont)
ATRIAL FIBRILLATION
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
8 (cont)
ATRIAL FLUTTER
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
9 (cont)
PROLONGED QT
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 129
Final November 12, 2008
Row
EGSTRESC
EGSTRESN
EGSTRESU
EGXFN
EGNAM
EGDRVFL
EGEVAL
VISITNUM
VISIT
EGDTC
EGDY
10 (cont)
LEFT VENTRICULAR
HYPERTROPHY
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
11 (cont)
INCORRECT ELECTRODE
PLACEMENT
PQW436789-07.xml
Test Lab
1
Screening 1
2003-04-15T11:58
-36
12 (cont) ABNORMAL PRINCIPAL
INVESTIGA
TOR 1 Screening 1 2003-04-15T11:58 -36
suppeg.xpt
Row 1: Shows that the record in the EG dataset with value of EGSEQ of 1 has Supplemental Qualifier record indicating that the ventricular rate of 62 bpm was
assessed as not being clinically significant.
Row 2: Shows that the record in the EG dataset with value of EGSEQ of 2 has Supplemental Qualifier record indicating that the PR interval of 0.15 sec was
assessed as being clinically significant.
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
XYZ
EG
XYZ-US-701-002
EGSEQ
1
EGCLSIG
Clinically Significant
N
CRF
2
XYZ
EG
XYZ-US-701-002
EGSEQ
2
EGCLSIG
Clinically Significant
Y
CRF
Example 2
Example 2 shows results for one subject across multiple visits where only the overall assessment was collected. In addition the ECG done at Visit 4 was read by
the principal investigator and a cardiologist. In this example the EGGRPID is the same number and the EGSEQ increments by one.
Rows 1-5: Show that when an interpretation is collected the evaluator is stored in EGEVAL.
Row 2: Shows the record selected as Baseline.
Row 3: Shows a date/time in ISO 8601 representation where both the date and time were collected.
Rows 4-5: Show where EGGRPID is used to group related results.
Row
STUDYID
DOMAIN
USUBJID
EGSEQ
EGGRPID
EGTESTCD
EGTEST
EGPOS
EGORRES
EGSTRESC
EGSTRESN
1
ABC
EG
ABC-99-CA-456
1
1
INTP
Interpretation
SUPINE
NORMAL
NORMAL
2
ABC
EG
ABC-99-CA-456
2
2
INTP
Interpretation
SUPINE
ABNORMAL
ABNORMAL
3
ABC
EG
ABC-99-CA-456
3
3
INTP
Interpretation
SUPINE
ABNORMAL
ABNORMAL
4
ABC
EG
ABC-99-CA-456
4
4
INTP
Interpretation
SUPINE
ABNORMAL
ABNORMAL
5
ABC
EG
ABC-99-CA-456
5
4
INTP
Interpretation
SUPINE
ABNORMAL
ABNORMAL
Row
EGBLFL
EGEVAL
VISITNUM
VISIT
VISITDY
EGDTC
EGDY
1 (cont)
PRINCIPAL INVESTIGATOR
1
SCREEN I
-2
2003-11-26
-2
2 (cont)
Y
PRINCIPAL INVESTIGATOR
2
SCREEN II
-1
2003-11-27
-1
3 (cont)
PRINCIPAL INVESTIGATOR
3
DAY 10
10
2003-12-07T09:02
10
4 (cont)
PRINCIPAL INVESTIGATOR
4
DAY 15
15
2003-12-12
15
5 (cont)
CARDIOLOGIST
4
DAY 15
15
2003-12-12
15
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 130 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.2 INCLUSION/EXCLUSION CRITERIA NOT MET — IE
ie.xpt, Inclusion/Exclusion Criteria Not Met — Findings, Version 3.1.2. One record per inclusion/exclusion criterion not met per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1934HIE
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
780HSDTMIG 4.1.2.2,
781HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
782H783HSDTMIG 4.1.2.3
IESEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
IESPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Inclusion or Exclusion criteria number from CRF.
Perm
SDTM 2.2.4,
784HSDTMIG 4.1.2.6
IETESTCD
Inclusion/Exclusion
Criterion Short Name
Char
*
Topic
Short name of the criterion described in IETEST. The value in
IETESTCD cannot be longer than 8 characters, nor can it start with a
number (e.g.‖1TEST‖). IETESTCD cannot contain characters other than
letters, numbers, or underscores. Examples: IN01, EX01.
Req
SDTM 2.2.3,
SDTMIG 4.1.1.9
785H 787HSDTMIG 4.1.2.1
IETEST
Inclusion/Exclusion
Criterion
Char
Synonym
Qualifier
Verbatim description of the inclusion or exclusion criterion that was the
exception for the subject within the study. IETEST cannot be longer than
200 characters.
Req
SDTM 2.2.3,
788HSDTMIG 4.1.2.1,
789HSDTMIG 4.1.2.4,
790HSDTMIG 4.1.5.3.1
IECAT
Inclusion/Exclusion
Category
Char
(1935HIECAT)
Grouping
Qualifier
Used to define a category of related records across subjects.
Req
SDTM 2.2.3,
791HSDTMIG 4.1.2.6,
792HSDTMIG
Appendix C1
IESCAT
Inclusion/Exclusion
Subcategory
Char
*
Grouping
Qualifier
A further categorization of the exception criterion. Can be used to
distinguish criteria for a sub-study or for to categorize as a major or minor
exceptions. Examples: MAJOR, MINOR.
Perm
SDTM 2.2.3,
793HSDTMIG 4.1.2.6
IEORRES
I/E Criterion Original
Result
Char
(1936HNY)
Result
Qualifier
Original response to Inclusion/Exclusion Criterion question. Inclusion or
Exclusion criterion met?
Req
SDTM 2.2.3,
794H795HSDTMIG 4.1.5.1,
796HSDTMIG
Appendix C1
IESTRESC
I/E Criterion Result in Std
Format
Char
(1937HNY)
Result
Qualifier
Response to Inclusion/Exclusion criterion result in standard format.
Req
SDTM 2.2.3,
797HSDTMIG 4.1.3.6,
798HSDTMIG 4.1.5.1,
799HSDTMIG
Appendix C1

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 131
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Perm
SDTM 2.2.5,
800HSDTMIG 4.1.4.5,
801HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
802HSDTMIG 4.1.4.5,
803HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
804HSDTMIG 4.1.4.5,
805HSDTMIG 7.4
IEDTC
Date/Time of Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
806HSDTMIG 4.1.4.1,
807HSDTMIG 4.1.4.2,
808HSDTMIG 4.1.4.8
IEDY
Study Day of Collection
Num
Timing
1. Study day of collection of the inclusion/exclusion exceptions, measured
as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5,
809HSDTMIG 4.1.4.4,
810HSDTMIG 4.1.4.6
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.2.1 ASSUMPTIONS FOR INCLUSION/EXCLUSION CRITERIA NOT MET DOMAIN MODEL
1. IE Definition:
CRF data that captures inclusion and exclusion criteria exceptions per subject. All inclusion or exclusion criteria that are violated, should be stored
here, even if a sponsor has granted a waiver or if the subject was admitted by mistake. In cases where a CRF may allow a response of ―Not
Applicable‖ and this is checked, no criteria were violated, so these records would not be in IE.
2. The intent of the domain model is to collect responses to only those criteria that the subject did not meet, and not the responses to all criteria. The
complete list of Inclusion/Exclusion criteria can be found in the TI trial inclusion/exclusion criteria dataset described in 811HSection 7.5.
3. This domain should be used to document the exceptions to inclusion or exclusion criteria at the time that eligibility for study entry is determined
(e.g., at the end of a run-in period or immediately before randomization). This domain should not be used to collect protocol deviations/violations
incurred during the course of the study, typically after randomization or start of study medication. See 812HSection 6.2.4.1 for the DV events domain
model that is used to submit protocol deviations/violations.
4. IETEST is to be used only for the verbatim description of the inclusion or exclusion criteria. If the text is <200 characters, it goes in IETEST; if the
text is > 200 characters, put meaningful text in IETEST and describe the full text in the study metadata. See 813Hsection 4.1.5.3.2 for further
information.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 132 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
5. The following Qualifiers would not generally be used in IE: --MODIFY, --POS, --BODSYS, --ORRESU, --ORNRLO, --ORNRHI, --STRESN,
--STRESU, --STNRLO, --STNRHI, --STNRC, --NRIND, --RESCAT, --XFN, --NAM, --LOINC, --SPEC, --SPCCND, --LOC, --METHOD,
--BLFL, --FAST, --DRVFL, --TOX, --TOXGR, --SEV, --STAT.
6.3.2.2 EXAMPLES FOR INCLUSION/EXCLUSION NOT MET DOMAIN MODEL
This example shows records for three subjects; one with 2 inclusion/exclusion exceptions, and the others with one exception each. Subject XYZ-0007 failed
exclusion criterion number 17 and inclusion criterion 3, but was included in the trial. The other two subjects each failed inclusion criterion number 3, but were
also included in the trial.
Row
STUDYID
DOMAIN
USUBJID
IESEQ
IESPID
IETESTCD
IETEST
IECAT
IEORRES
IESTRESC
1
XYZ
IE
XYZ-0007
1
17
EXCL17
Ventricular Rate
EXCLUSION
Y
Y
2
XYZ
IE
XYZ-0007
2
3
INCL03
Acceptable mammogram from local radiologist?
INCLUSION
N
N
3
XYZ
IE
XYZ-0047
1
3
INCL03
Acceptable mammogram from local radiologist?
INCLUSION
N
N
4
XYZ
IE
XYZ-0096
1
3
INCL03
Acceptable mammogram from local radiologist?
INCLUSION
N
N
Row
VISITNUM
VISIT
VISITDY
IEDTC
IEDY
1 (cont)
1
WEEK -8
-56
1999-01-10
-58
2 (cont)
1
WEEK -8
-56
1999-01-10
-58
3 (cont)
1
WEEK -8
-56
1999-01-12
-56
4 (cont)
1
WEEK -8
-56
1999-01-13
-55
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 133
Final November 12, 2008
6.3.3 LABORATORY TEST RESULTS — LB
lb.xpt, Labs — Findings, Version 3.1.2. One record per lab test per time point per visit per subject, Tabulation
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1938HLB
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
814HSDTMIG 4.1.2.2,
815HSDTMIG Appendix
C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
816H817HSDTMIG 4.1.2.3
LBSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
LBGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
818HSDTMIG 4.1.2.6
LBREFID
Specimen ID
Char
Identifier
Internal or external specimen identifier. Example: Specimen ID.
Perm
SDTM 2.2.4,
819HSDTMIG 4.1.2.6
LBSPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on the Lab page.
Perm
SDTM 2.2.4,
820HSDTMIG 4.1.2.6
LBTESTCD
Lab Test or Examination
Short Name
Char
(1939HLBTESTCD)
Topic
Short name of the measurement, test, or examination described in
LBTEST. It can be used as a column name when converting a dataset from
a vertical to a horizontal format. The value in LBTESTCD cannot be
longer than 8 characters, nor can it start with a number (e.g.‖1TEST‖).
LBTESTCD cannot contain characters other than letters, numbers, or
underscores. Examples: ALT, LDH.
Req
SDTM 2.2.3,
SDTMIG 4.1.1.9
821H 823HSDTMIG 4.1.2.1,
824HSDTMIG Appendix
C1
LBTEST
Lab Test or Examination
Name
Char
(1940HLBTEST)
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the measurement
or finding. Note any test normally performed by a clinical laboratory is
considered a lab test. The value in LBTEST cannot be longer than 40
characters. Examples: Alanine Aminotransferase, Lactate Dehydrogenase.
Req
SDTM 2.2.3,
825HSDTMIG 4.1.2.1,
826HSDTMIG 4.1.2.4,
827HSDTMIG 4.1.5.3.1
828HSDTMIG Appendix
C1
LBCAT
Category for Lab Test
Char
*
Grouping
Qualifier
Used to define a category of related records across subjects. Examples:
such as HEMATOLOGY, URINALYSIS, CHEMISTRY.
Exp
SDTM 2.2.3,
829HSDTMIG 4.1.2.6
LBSCAT
Subcategory for Lab Test
Char
*
Grouping
Qualifier
A further categorization of a test category such as DIFFERENTIAL,
COAGULATON, LIVER FUNCTION, ELECTROLYTES.
Perm
SDTM 2.2.3,
830HSDTMIG 4.1.2.6
LBORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the measurement or finding as originally received or collected.
Exp
SDTM 2.2.3, 831H
832HSDTMIG 4.1.5.1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 134 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
LBORRESU
Original Units
Char
(833HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for LBORRES.
Example: g/L.
Exp
SDTM 2.2.3,
834HSDTMIG 4.1.3.2,
835HSDTMIG 4.1.5.1
LBORNRLO
Reference Range Lower
Limit in Orig Unit
Char
Variable
Qualifier
Lower end of reference range for continuous measurements in original
units. Should be populated only for continuous results.
Exp
SDTM 2.2.3
LBORNRHI
Reference Range Upper
Limit in Orig Unit
Char
Variable
Qualifier
Upper end of reference range for continuous measurements in original
units. Should be populated only for continuous results.
Exp
SDTM 2.2.3
LBSTRESC
Character Result/Finding
in Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from
LBORRES in a standard format or standard units. LBSTRESC should
store all results or findings in character format; if results are numeric, they
should also be stored in numeric format in LBSTRESN. For example, if a
test has results ―NONE‖, ―NEG‖, and ―NEGATIVE‖ in LBORRES and
these results effectively have the same meaning, they could be represented
in standard format in LBSTRESC as ―NEGATIVE‖. For other examples,
see general assumptions.
Exp
SDTM 2.2.3,
836H837HSDTMIG 4.1.5.1
LBSTRESN
Numeric Result/Finding
in Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from LBSTRESC. LBSTRESN should store all
numeric test results or findings.
Exp
SDTM 2.2.3,
838HSDTMIG 4.1.5.1
LBSTRESU
Standard Units
Char
(839HUNIT)
Variable
Qualifier
Standardized unit used for LBSTRESC or LBSTRESN.
Exp
SDTM 2.2.3,
840HSDTMIG 4.1.3.2,
841HSDTMIG 4.1.5.1
LBSTNRLO
Reference Range Lower
Limit-Std Units
Num
Variable
Qualifier
Lower end of reference range for continuous measurements for
LBSTRESC/LBSTRESN in standardized units. Should be populated only
for continuous results.
Exp
SDTM 2.2.3
LBSTNRHI
Reference Range Upper
Limit-Std Units
Num
Variable
Qualifier
Upper end of reference range for continuous measurements in standardized
units. Should be populated only for continuous results.
Exp
SDTM 2.2.3
LBSTNRC
Reference Range for Char
Rslt-Std Units
Char
Variable
Qualifier
For normal range values that are character in ordinal scale or if categorical
ranges were supplied (e.g., ―-1 to +1‖, ―NEGATIVE TO TRACE‖).
Perm
SDTM 2.2.3
LBNRIND
Reference Range
Indicator
Char
*
Variable
Qualifier
1. Indicates where the value falls with respect to reference range defined
by LBORNRLO and LBORNRHI, LBSTNRLO and LBSTNRHI, or by
LBSTNRC. Examples: NORMAL, ABNORMAL, HIGH, LOW.
2. Sponsors should specify in the study metadata (Comments column in the
define.xml) whether LBNRIND refers to the original or standard reference
ranges and results.
3. Should not be used to indicate clinical significance.
Exp
SDTM 2.2.3 8
LBSTAT
Completion Status
Char
(1941HND)
Record
Qualifier
Used to indicate exam not done. Should be null if a result exists in
LBORRES.
Perm
SDTM 2.2.3,
843HSDTMIG 4.1.5.1,
844HSDTMIG 4.1.5.7,
845HSDTMIG Appendix
C1
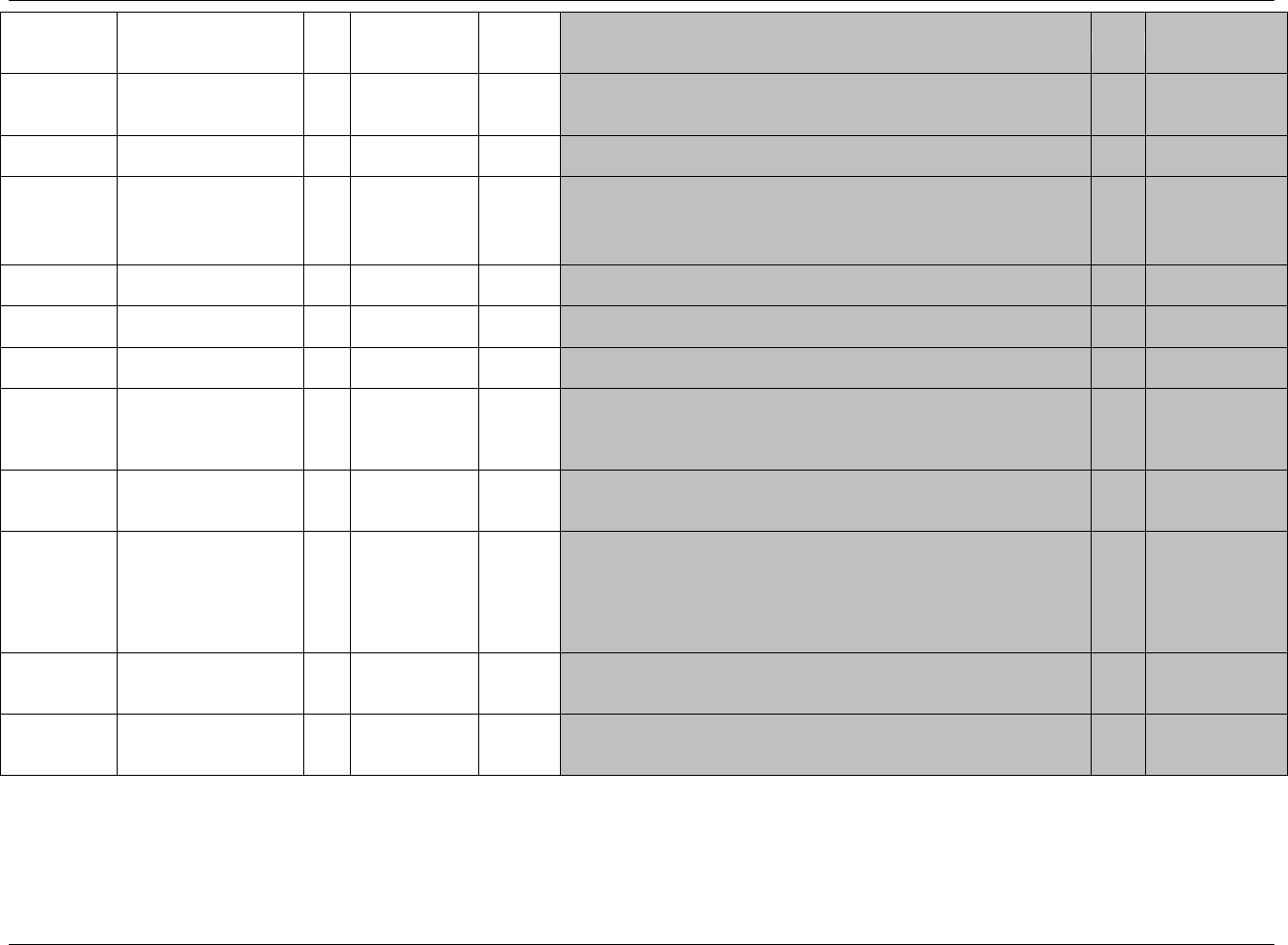
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 135
Final November 12, 2008
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
LBREASND
Reason Test Not Done
Char
Record
Qualifier
Describes why a measurement or test was not performed such as BROKEN
EQUIPMENT, SUBJECT REFUSED, or SPECIMEN LOST. Used in
conjunction with LBSTAT when value is NOT DONE.
Perm
SDTM 2.2.3,
846HSDTMIG 4.1.5.1,
847HSDTMIG 4.1.5.7
LBNAM
Vendor Name
Char
Record
Qualifier
The name or identifier of the laboratory that performed the test.
Perm
SDTM 2.2.3
LBLOINC
LOINC Code
Char
*
Synonym
Qualifier
1. Dictionary-derived LOINC Code for LBTEST.
2. The sponsor is expected to provide the dictionary name and
version used to map the terms utilizing the define.xml external
codelist attributes
Perm
SDTM 2.2.3,
848HSDTMIG 4.1.3.2
LBSPEC
Specimen Type
Char
*
Record
Qualifier
Defines the type of specimen used for a measurement. Examples: SERUM,
PLASMA, URINE.
Perm
SDTM 2.2.3
LBSPCCND
Specimen Condition
Char
*
Record
Qualifier
Free or standardized text describing the condition of the specimen e.g.
HEMOLYZED, ICTERIC, LIPEMIC etc.
Perm
SDTM 2.2.3
LBMETHOD
Method of Test or
Examination
Char
*
Record
Qualifier
Method of the test or examination. Examples: EIA (Enzyme
Immunoassay), ELECTROPHORESIS, DIPSTICK
Perm
SDTM 2.2.3
LBBLFL
Baseline Flag
Char
(1942HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or
null.
Exp
SDTM 2.2.3,
849HSDTMIG 4.1.3.7,
850HSDTMIG Appendix
C1
LBFAST
Fasting Status
Char
(1943HNY)
Record
Qualifier
Indicator used to identify fasting status such as Y, N, U, or null if not
relevant.
Perm
SDTM 2.2.3,
851HSDTMIG Appendix
C1
LBDRVFL
Derived Flag
Char
(1944HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records
that represent the average of other records, or do not come from the CRF,
or are not as originally received or collected are examples of records that
might be derived for the submission datasets. If LBDRVFL=Y, then
LBORRES may be null, with LBSTRESC, and (if numeric) LBSTRESN
having the derived value.
Perm
SDTM 2.2.3,
852HSDTMIG 4.1.3.7,
853HSDTMIG 4.1.5.1,
854HSDTMIG Appendix
C1
LBTOX
Toxicity
Char
*
Variable
Qualifier
Description of toxicity quantified by LBTOXGR. The sponsor is expected
to provide the name of the scale and version used to map the terms,
utilizing the define.xml external codelist attributes.
Perm
SDTM 2.2.3
LBTOXGR
Standard Toxicity Grade
Char
*
Variable
Qualifier
Records toxicity grade value using a standard toxicity scale (such as the
NCI CTCAE). If value is from a numeric scale, represent only the number
(e.g., ―2‖ and not ―Grade 2‖).
Perm
SDTM 2.2.3
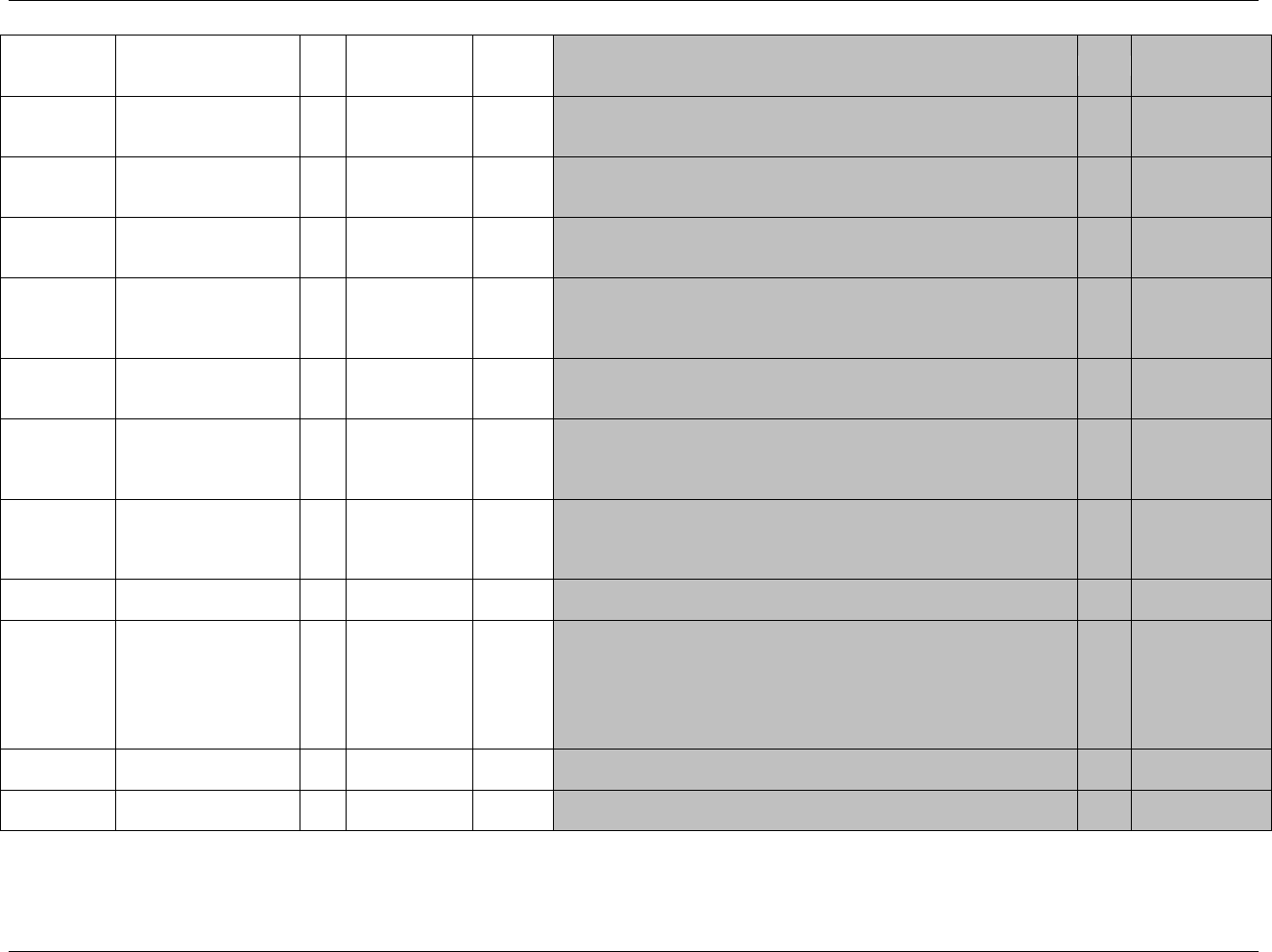
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 136 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
855HSDTMIG 4.1.4.5,
856HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter
2. May be used in addition to VISITNUM and/or VISITDY
Perm
SDTM 2.2.5,
857HSDTMIG 4.1.4.5,
858HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
859HSDTMIG 4.1.4.5,
860HSDTMIG 7.4
LBDTC
Date/Time of Specimen
Collection
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
861HSDTMIG 4.1.4.1,
SDTMIG 4.1.4.2
862HSDTMIG 4.1.4.8
LBENDTC
End Date/Time of
Specimen Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
863HSDTMIG 4.1.4.1,
864HSDTMIG 4.1.4.8
LBDY
Study Day of Specimen
Collection
Num
Timing
1. Study day of specimen collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5,
865HSDTMIG 4.1.4.4,
866HSDTMIG 4.1.4.6
LBTPT
Planned Time Point
Name
Char
Timing
1. Text Description of time when specimen should be taken.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See LBTPTNUM and LBTPTREF.
Examples: Start, 5 min post.
Perm
SDTM 2.2.5,
867HSDTMIG 4.1.4.10
LBTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of LBTPT to aid in sorting.
Perm
SDTM 2.2.5,
868HSDTMIG 4.1.4.10
LBELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Planned Elapsed time (in ISO 8601) relative to a planned fixed reference
(LBTPTREF). This variable is useful where there are repetitive measures. Not
a clock time or a date time variable. Represented as an ISO 8601 duration.
Examples: ―-PT15M‖ to represent the period of 15 minutes prior to the
reference point indicated by LBTPTREF, or ―PT8H‖ to represent the
period of 8 hours after the reference point indicated by LBTPTREF.
Perm
SDTM 2.2.5,
869HSDTMIG 4.1.4.3,
870HSDTMIG 4.1.4.10
LBTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by LBELTM, LBTPTNUM,
and LBTPT. Examples: PREVIOUS DOSE, PREVIOUS MEAL.
Perm
SDTM 2.2.5,
871HSDTMIG 4.1.4.10
LBRFTDTC
Date/Time of Reference
Time Point
Char
ISO 8601
Timing
Date/time of the reference time point, LBTPTREF.
Perm
SDTM 2.2.5,
872HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 137
Final November 12, 2008
6.3.3.1 ASSUMPTIONS FOR LABORATORY TEST RESULTS DOMAIN MODEL
1. LB Definition: This domain captures laboratory data collected on the CRF or received from a central provider or vendor
2. For lab tests that do not have continuous numeric results (e.g., urine protein as measured by dipstick, descriptive tests such as urine color), LBSTNRC could
be populated either with normal range values that are character in an ordinal scale (e.g., ―NEGATIVE to TRACE') or a delimited set of values that are
considered to be normal (e.g., ―YELLOW‖, ―AMBER‖). LBORNRLO, LBORNRHI, LBSTNRLO, and LBSTNRHI should be null for these types of tests.
3. LBNRIND can be added to indicate where a result falls with respect to reference range defined by LBORNRLO and LBORNRHI. Examples: HIGH, LOW.
Clinical significance would be represented as described in 873HSection 4.1.5.5 as a record in SUPPLB with a QNAM of LBCLSIG (see also LB Example 1
below).
4. For lab tests where the specimen is collected over time, i.e., 24-hour urine collection, the start date/time of the collection goes into LBDTC and the end
date/time of collection goes into LBENDTC. See 874HAssumption 4.1.4.8.
5. The following Qualifiers would not generally be used in LB: --BODSYS, --SEV.
6. A value derived by a central lab according to their procedures is considered collected rather than derived. See 875HSection 4.1.1.8.1.
6.3.3.2 EXAMPLES FOR LABORATORY TEST RESULTS DOMAIN MODEL
Example 1:
Row 1: Shows a value collected in one unit, but converted to selected standard unit. See 876HSection 4.1.5.1 for additional examples for the population of Result Qualifiers.
Rows 2-4: Show two records (rows 2 and 3) for Alkaline Phosphatase done at the same visit, one day apart. Row 4 shows how to create a derived record
(average of the records 2 and 3) and flagged derived (LBDRVFL = ―Y‖) and as the record to use as baseline (LBBLFL = ―Y‖).
Rows 6 and 7: Show a suggested use of the LBSCAT variable. It could be used to further classify types of tests within a laboratory panel (i.e., ―DIFFERENTIAL‖).
Row 9: Shows the proper use of the LBSTAT variable to indicate "NOT DONE", where a reason was collected when a test was not done.
Row 10: The subject had cholesterol measured. The normal range for this test is <200 mg/dL. Note that the sponsor has decided to make LBSTNRHI
=199 however another sponsor may have chosen a different value.
Row 12: Shows use of LBSTNRC for Urine Protein that is not reported as a continuous numeric result.
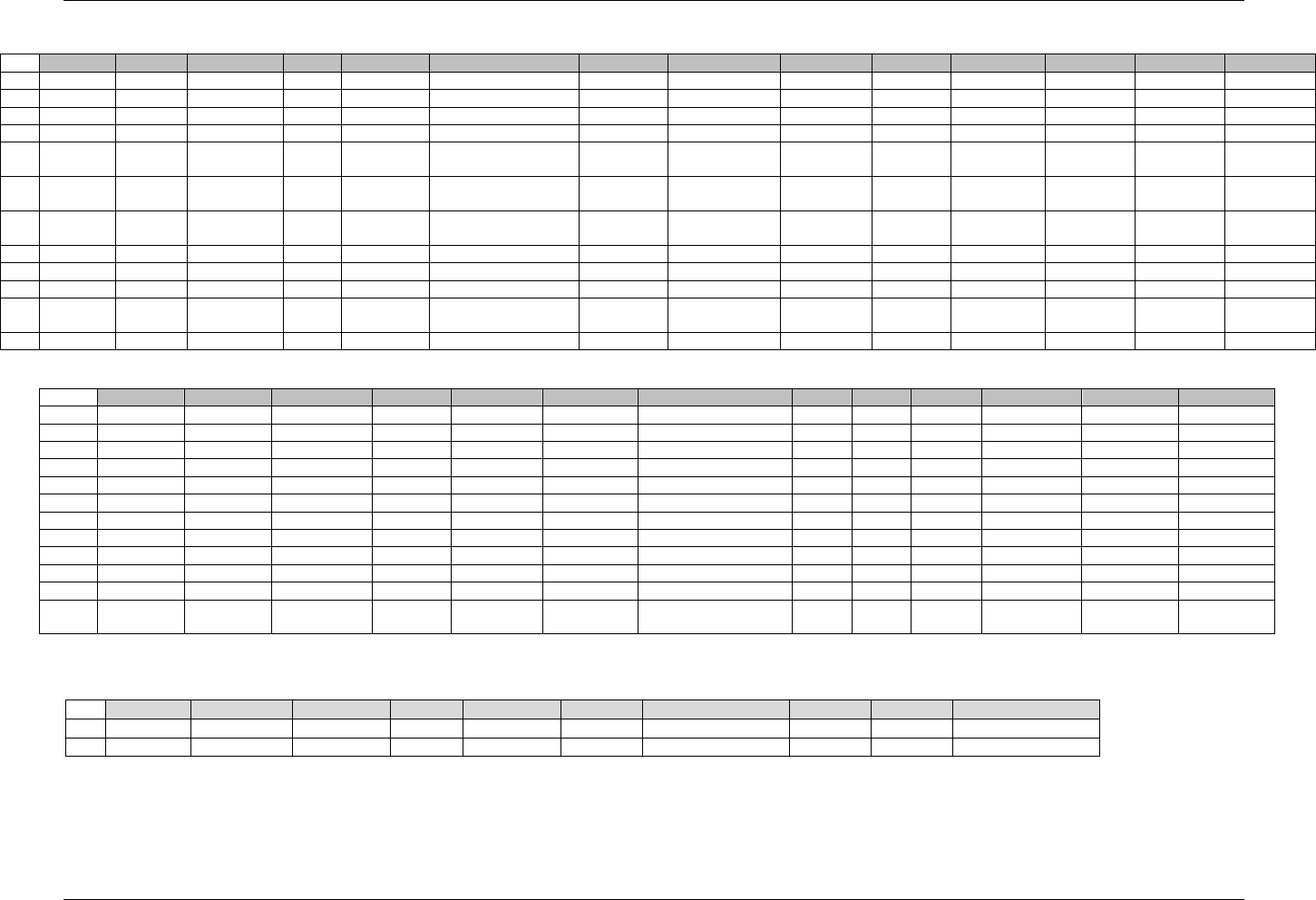
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 138 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
lb.xpt
Row
STUDYID
DOMAIN
USUBJID
LBSEQ
LBTESTCD
LBTEST
LBCAT
LBSCAT
LBORRES
LBORRESU
LBORNRLO
LBORNRHI
LBSTRESC
LBSTRESN
1
ABC
LB
ABC-001-001
1
ALB
Albumin
CHEMISTRY
30
g/L
35
50
3.0
3.0
2
ABC
LB
ABC-001-001
2
ALP
Alkaline Phosphatase
CHEMISTRY
398
IU/L
40
160
398
398
3
ABC
LB
ABC-001-001
3
ALP
Alkaline Phosphatase
CHEMISTRY
350
IU/L
40
160
350
350
4
ABC
LB
ABC-001-001
4
ALP
Alkaline Phosphatase
CHEMISTRY
374
374
5
ABC
LB
ABC-001-001
5
WBC
Leukocytes
HEMATOLO
GY
5.9
10^9/L
4
11
5.9
5.9
6
ABC
LB
ABC-001-001
6
LYMLE
Lymphocytes
HEMATOLO
GY
DIFFERENTIAL
6.7
%
25
40
6.7
6.7
7
ABC
LB
ABC-001-001
7
NEUT
Neutrophils
HEMATOLO
GY
DIFFERENTIAL
5.1
10^9/L
2
8
5.1
5.1
8
ABC
LB
ABC-001-001
8
PH
pH
URINALYSIS
7.5
5.0
9.0
7.5
9
ABC
LB
ABC-001-001
9
ALB
Albumin
CHEMISTRY
10
ABC
LB
ABC-001-001
10
CHOL
Cholesterol
CHEMISTRY
229
mg/dL
0
<200
229
229
11
ABC
LB
ABC-001-001
11
WBC
Leukocytes
HEMATOLO
GY
5.9
10^9/L
4
11
5.9
5.9
12
ABC
LB
ABC-001-001
12
PROT
Protein
URINALYSIS
MODERATE
MODERATE
Note that the use of 10^9 as a unit is not a standard representation.
Row
LBSTRESU
LBSTNRLO
LBSTNRHI
LBSTRNRC
LBNRIND
LB STAT
LBREASND
LBBLFL
LBFAST
LBDRVFL
VISITNUM
VISIT
LBDTC
1 (cont)
g/dL
3.5
5
LOW
Y
Y
1
Week 1
1999-06-19
2 (cont)
units/L
40
160
Y
1
Week 1
1999-06-19
3 (cont)
units/L
40
160
Y
1
Week 1
1999-06-20
4 (cont)
units/L
40
160
Y
Y
Y
1
Week 1
1999-06-19
5 (cont)
10^3/uL
4
11
Y
Y
1
Week 1
1999-06-19
6 (cont)
%
25
40
LOW
Y
Y
1
Week 1
1999-06-19
7 (cont)
10^9/L
2
8
Y
Y
1
Week 1
1999-06-19
8 (cont)
5.00
9.00
Y
Y
1
Week 1
1999-06-19
9 (cont)
NOT DONE
INSUFFICIENT SAMPLE
2
Week 2
1999-07-21
10 (cont)
mg/dL
0
199
2
Week 2
1999-07-21
11 (cont)
10^3/uL
4
11
Y
2
Week 2
1999-07-21
12 (cont)
NEGATIVE
to TRACE
ABNORMAL
2
Week 2
1999-07-21
supplb.xpt
Row 1, 6: The SUPPLB dataset example shows clinical significance assigned by the investigator for test results where LBNRIND (reference range indicator) is populated.
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
ABC
LB
ABC-001-001
LBSEQ
1
LBCLSIG
Clinical Significance
N
CRF
INVESTIGATOR
2
ABC
LB
ABC-001-001
LBSEQ
6
LBCLSIG
Clinical Significance
N
CRF
INVESTIGATOR
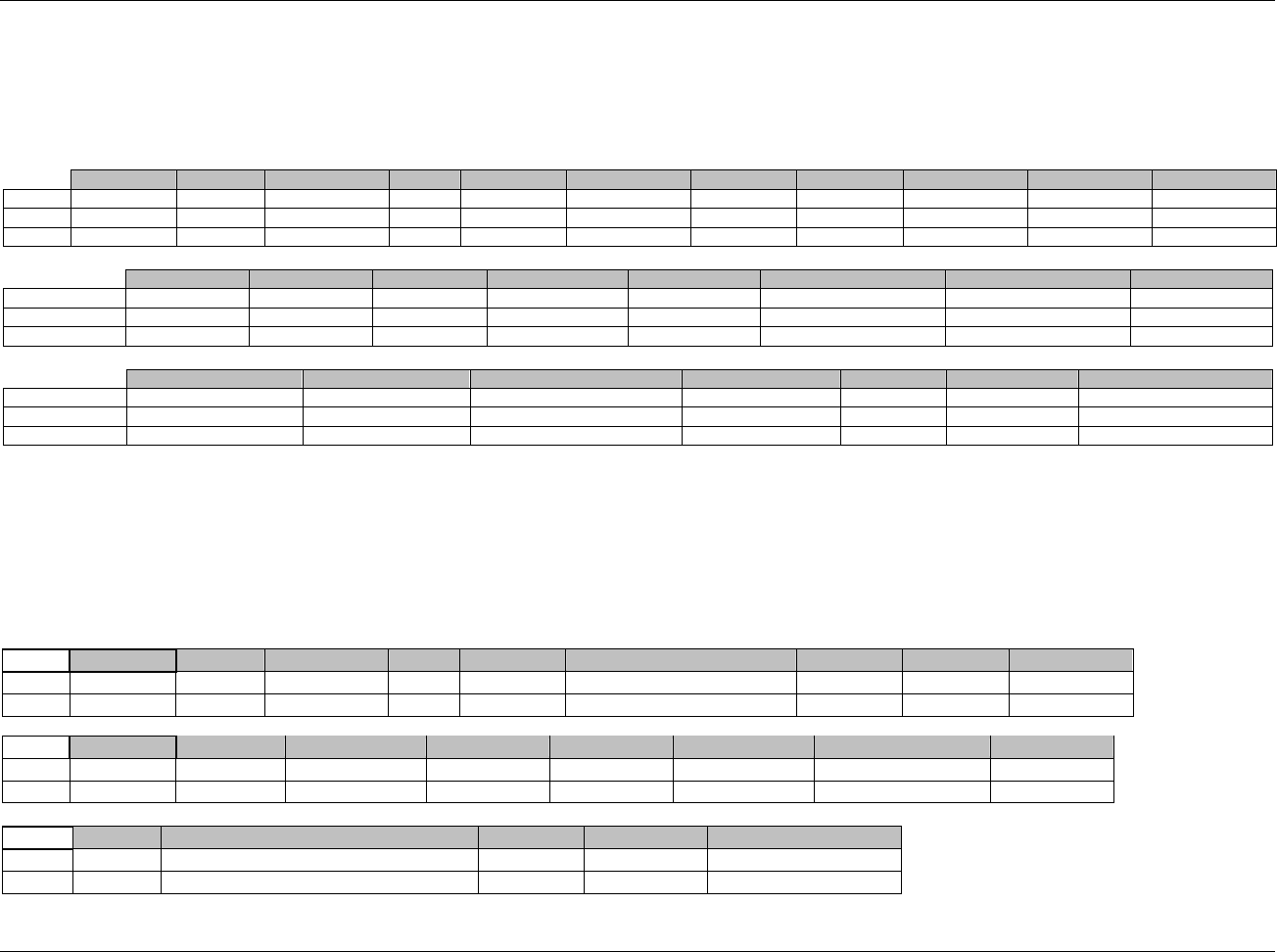
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 139
Final November 12, 2008
Example 2
Rows 1: Shows an example of a pre-dose urine collection interval (from 4 hours prior to dosing until 15 minutes prior to dosing) with a negative value for
LBELTM that reflects the end of the interval in reference to the fixed reference LBTPTREF, the date of which is recorded in LBRFTDTC.
Rows 2 and 3: Show an example of post-dose urine collection intervals with values for LBELTM that reflect the end of the intervals in reference to the fixed
reference LBTPTREF, the date of which is recorded in LBRFTDTC.
STUDYID
DOMAIN
USUBJID
LBSEQ
LBTESTCD
LBTEST
LBCAT
LBORRES
LBORRESU
LBORNRLO
LBORNRHI
Row 1
ABC
LB
ABC-001-001
1
GLUCOSE
Glucose
URINALYSIS
7
mg/dL
1
15
Row 2
ABC
LB
ABC-001-001
2
GLUCOSE
Glucose
URINALYSIS
11
mg/dL
1
15
Row 3
ABC
LB
ABC-001-001
3
GLUCOSE
Glucose
URINALYSIS
9
mg/dL
1
15
LBSTRESC
LBSTRESN
LBSTRESU
LBSTNRLO
LBSTNRHI
LBNRIND
VISIT
VISITNUM
Row 1 (cont)
0.38
0.38
mmol/L
0.1
0.8
NORMAL
INITIAL DOSING
2
Row 2 (cont)
0.61
0.61
mmol/L
0.1
0.8
NORMAL
INITIAL DOSING
2
Row 3 (cont)
0.5
0.5
mmol/L
0.1
0.8
NORMAL
INITIAL DOSING
2
LBDTC
LBENDTC
LBTPT
LBTPTNUM
LBELTM
LBTPTREF
LBRFTDTC
Row 1 (cont)
1999-06-19T04:00
1999-06-19T07:45
Pre-dose
1
-PT15M
Dosing
1999-06-19T08:00
Row 2 (cont)
1999-06-19T08:00
1999-06-19T16:00
0-8 hours after dosing
2
PT8H
Dosing
1999-06-19T08:00
Row 3 (cont)
1999-06-19T16:00
1999-06-20T00:00
8-16 hours after dosing
3
PT16H
Dosing
1999-06-19T08:00
Example 3:
This is an example of pregnancy test records, one with a result and one with no result because the test was not performed due to the subject being male.
Row 1: Shows an example of a pregnancy test record that returns a result of ―-― (negative sign) in LBORRES and is standardized to the text value
―NEGATIVE‖ in LBSTRESC
Row 2: Show an example of a pregnancy test that was not performed because the subject was male, and the sponsor felt it was necessary to report a record
documenting the reason why the test was not performed, rather then simply excluding the record.
Row
STUDYID
DOMAIN
USUBJID
LBSEQ
LBTESTCD
LBTEST
LBCAT
LBORRES
LBORRESU
1
ABC
LB
ABC-001-001
1
HCG
Choriogonadotropin Beta
CHEMISTRY
-
2
ABC
LB
ABC-001-002
1
HCG
Choriogonadotropin Beta
CHEMISTRY
Row
LBORNRLO
LBORNRHI
LBSTRESC
LBSTRESN
LBSTRESU
LBSTNRLO
LBSTRNHI
LBNRIND
1 (cont)
NEGATIVE
2 (cont)
Row
LBSTAT
LBREASND
VISIT
VISITNUM
LBDTC
1 (cont)
BASELINE
1
1999-06-19T04:00
2 (cont)
NOT DONE
NOT APPLICABLE (SUBJECT MALE)
BASELINE
1
1999-06-24T08:00
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 140 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.4 PHYSICAL EXAMINATION — PE
pe.xpt, Physical Examination — Findings, Version 3.1.2. One record per body system or abnormality per visit per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1945HPE
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
877HSDTMIG 4.1.2.2,
878HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
879H880HSDTMIG 4.1.2.3
PESEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
PEGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
881HSDTMIG 4.1.2.6
PESPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a CRF.
Perm
SDTM 2.2.4,
882HSDTMIG 4.1.2.6
PETESTCD
Body System Examined
Short Name
Char
*
Topic
Short name of the measurement, test, or examination described in
PETEST. It can be used as a column name when converting a dataset
from a vertical to a horizontal format. The value in PETESTCD cannot be
longer than 8 characters, nor can it start with a number (e.g.‖1TEST‖).
PETESTCD cannot contain characters other than letters, numbers, or
underscores.
Req
SDTM 2.2.3,
SDTMIG 4.1.1.9,
883H88SDTMIG 4.1.2.1
PETEST
Body System Examined
Char
*
Synonym
Qualifier
Verbatim term part of the body examined. The value in PETEST cannot
be longer than 40 characters. Examples: Cardiovascular and Respiratory.
For subject-level exam, value should be ―Physical Examination‖.
Req
SDTM 2.2.3,
886HSDTMIG 4.1.2.1,
887HSDTMIG 4.1.2.4,
888HSDTMIG 4.1.5.3.1
PEMODIFY
Modified Reported Term
Char
Synonym
Qualifier
If PEORRES is modified as part of a defined procedure, then PEMODIFY
will contain the modified text.
Perm
SDTM 2.2.3,
889HSDTMIG 4.1.3.6
PECAT
Category for Examination
Char
*
Grouping
Qualifier
Used to define a category of examination. Examples: GENERAL,
NEUROLOGICAL.
Perm
SDTM 2.2.3,
890HSDTMIG 4.1.2.6
PESCAT
Subcategory for
Examination
Char
*
Grouping
Qualifier
A further categorization of the examination. Used if needed to add further
detail to PECAT.
Perm
SDTM 2.2.3,
891HSDTMIG 4.1.2.6
PEBODSYS
Body System or Organ
Class
Char
Result
Qualifier
1. Body system or organ class ( MedDRA SOC) that is involved in a
measurement from the standard hierarchy (e.g., MedDRA).
Perm
SDTM 2.2.3,
892HSDTMIG 4.1.3.5

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 141
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
PEORRES
Verbatim Examination
Finding
Char
Result
Qualifier
Text description of any abnormal findings. If the examination was
completed and there were no abnormal findings, the value should be
NORMAL. If the examination was not performed on a particular body
system, or at the subject level, then the value should be null, and NOT
DONE should appear in PESTAT.
Exp
SDTM 2.2.3,
893HSDTMIG 4.1.3.6
PEORRESU
Original Units
Char
(894HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for PEORRES.
Perm
SDTM 2.2.3,
895HSDTMIG 4.1.3.2
PESTRESC
Character Result/Finding in
Std Format
Char
Result
Qualifier
If there are findings for a body system, then either the dictionary preferred
term (if findings are coded using a dictionary) or PEORRES (if findings
are not coded) should appear here. If PEORRES is null, PESTRESC
should be null
Exp
SDTM 2.2.3,
896HSDTMIG 4.1.3.6,
897HSDTMIG 4.1.5.1
PESTAT
Completion Status
Char
(1946HND)
Record
Qualifier
Used to indicate exam not done. Should be null if a result exists in
PEORRES.
Perm
SDTM 2.2.3,
898HSDTMIG 4.1.5.1,
899HSDTMIG 4.1.5.7,
900HSDTMIG
Appendix C1
PEREASND
Reason Not Examined
Char
Record
Qualifier
Describes why an examination was not performed or why a body system
was not examined. Example: SUBJECT REFUSED. Used in conjunction
with STAT when value is NOT DONE.
Perm
SDTM 2.2.3,
901HSDTMIG 4.1.5.1,
902HSDTMIG 4.1.5.7
PELOC
Location of Physical Exam
Finding
Char
(903HLOC)
Record
Qualifier
Can be used to specify where a physical exam finding occurred. Example:
LEFT ARM for skin rash.
Perm
SDTM 2.2.3
SDTMIG 4.1.1.9
PEMETHOD
Method of Test or
Examination
Char
*
Record
Qualifier
Method of the test or examination. Examples: XRAY, MRI.
Perm
SDTM 2.2.3
PEEVAL
Evaluator
Char
*
Record
Qualifier
Role of the person who provided the evaluation. Used only for results that
are subjective (e.g., assigned by a person or a group). Should be null for
records that contain collected or derived data. Examples:
INVESTIGATOR, ADJUDICATION COMMITTEE, VENDOR.
Perm
SDTM 2.2.3,
904HSDTMIG 4.1.5.4
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5
905HSDTMIG 4.1.4.5,
906HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
907HSDTMIG 4.1.4.5,
908HSDTMIG 7.4
VISITDY
Planned Study Day of Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
909HSDTMIG 4.1.4.5,
910HSDTMIG 7.4
PEDTC
Date/Time of Examination
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
911HSDTMIG 4.1.4.1,
912HSDTMIG 4.1.4.8
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 142 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
PEDY
Study Day of Examination
Num
Timing
1. Study day of physical exam, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm
SDTM 2.2.5,
913HSDTMIG 4.1.4.4,
914HSDTMIG 4.1.4.6
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.4.1 ASSUMPTIONS FOR PHYSICAL EXAMINATION DOMAIN MODEL
1. PE Definition: Data that captures findings about physical exams. This could be information about which body systems that were examined and specific
abnormalities were collected.
2. The PE domain provides an example where the result, PEORRES, is coded. This is in contrast to Events and Interventions domains (e.g., AE, CM, and MH),
in which the topic variable (AETERM, CMTRT, and EXTRT, respectively) is the one coded.
3. The following Qualifiers would not generally be used in PE: --XFN, --NAM, --LOINC, --FAST, --TOX, --TOXGR.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 143
Final November 12, 2008
6.3.4.2 EXAMPLES FOR PHYSICAL EXAMINATION DOMAIN MODEL
The example shows data for one subject collected at two different visits. In all of the records except 8 and 13 the data comes from the general physical
examination. In this case PECAT is "GENERAL". Additional data collected about an ophthalmologic examination is also added to this domain.
Row 1: Shows how PESTRESC is populated if result is ―NORMAL‖.
Row 2: Shows the proper use of the --STAT variable to indicate "NOT DONE", and when PEREASND is used to indicate why a body system
(PETEST) was not examined.
Rows 4-6: Show how PESPID is used to show the sponsor-defined identifier, which in this case is a CRF sequence number used for identifying
abnormalities within a body system.
Rows 4-7: Show how PESTRESC is populated if abnormality is dictionary coded.
Rows 8, 13: Show how the PECAT variable can be used to indicate a different type of physical examination. In this case, the ophthalmologic examination
may have been collected in a separate dataset in the operational database.
Row
STUDYID
DOMAIN
USUBJID
PESEQ
PESPID
PETESTCD
PETEST
PECAT
PELOC
PEBODSYS
1
ABC
PE
ABC-001-001
1
1
HEAD
Head
GENERAL
2
ABC
PE
ABC-001-001
2
1
RESP
Respiratory
GENERAL
3
ABC
PE
ABC-001-001
3
1
ENT
Ear/nose/throat
GENERAL
4
ABC
PE
ABC-001-001
4
1
SKIN
Skin
GENERAL
FACE
SKIN
5
ABC
PE
ABC-001-001
5
2
SKIN
Skin
GENERAL
HANDS
SKIN
6
ABC
PE
ABC-001-001
6
3
SKIN
Skin
GENERAL
LEFT ARM
SKIN
7
ABC
PE
ABC-001-001
7
1
CV
Cardiovascular
GENERAL
CARDIOVASCULAR
8
ABC
PE
ABC-001-001
8
1
FUNDOSCP
Fundoscopic
OPHTHAMOLOGIC
9
ABC
PE
ABC-001-001
9
1
RESP
Respiratory
GENERAL
10
ABC
PE
ABC-001-001
10
1
ENT
Ear/nose/throat
GENERAL
11
ABC
PE
ABC-001-001
11
1
NECK
Neck
GENERAL
12
ABC
PE
ABC-001-001
12
1
CARDIO
Cardiovascular
GENERAL
13
ABC
PE
ABC-001-001
13
1
FUNDOSCP
Fundoscopic
OPHTHAMOLOGIC
Row
PEORRES
PESTRESC
PESTAT
PEREASND
VISITNUM
VISIT
VISITDY
PEDTC
PEDY
1 (cont)
NORMAL
NORMAL
1
BASELINE
1
1999-06-06
-3
2 (cont)
NOT DONE
INVESTIGATOR ERROR
1
BASELINE
1
1999-06-06
-3
3 (cont)
NORMAL
NORMAL
1
BASELINE
1
1999-06-06
-3
4 (cont)
ACNE
ACNE NOS
1
BASELINE
1
1999-06-06
-3
5 (cont)
ALLERGIC REACTION
DERMATITIS
1
BASELINE
1
1999-06-06
-3
6 (cont)
SKINRASH
RASH
1
BASELINE
1
1999-06-06
-3
7 (cont)
HEART MURMUR
CARDIAC MURMUR
1
BASELINE
1
1999-06-06
-3
8 (cont)
NORMAL
NORMAL
1
BASELINE
1
1999-06-06
-3
9 (cont)
NORMAL
NORMAL
2
VISIT 1
45
1999-07-21
45
10 (cont)
NORMAL
NORMAL
2
VISIT 1
45
1999-07-21
45
11 (cont)
NORMAL
NORMAL
2
VISIT 1
45
1999-07-21
45
12 (cont)
NORMAL
NORMAL
2
VISIT 1
45
1999-07-21
45
13 (cont)
NORMAL
NORMAL
2
VISIT 1
45
1999-07-21
45
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 144 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.5 QUESTIONNAIRE — QS
qs.xpt, Questionnaires — Findings, Version 3.1.2. One record per questionnaire per question per time point per visit per subject, Tabulation
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1947HQS
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
915HSDTMIG 4.1.2.2,
916HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
917H918HSDTMIG 4.1.2.3
QSSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
QSGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
919HSDTMIG 4.1.2.6
QSSPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Question number on a questionnaire.
Perm
SDTM 2.2.4,
920HSDTMIG 4.1.2.6
QSTESTCD
Question Short Name
Char
*
Topic
Topic variable for QS. Short name for the value in QSTEST, which can be
used as a column name when converting the dataset from a vertical format
to a horizontal format. The value in QSTESTCD cannot be longer than 8
characters, nor can it start with a number (e.g.‖1TEST‖). QSTESTCD
cannot contain characters other than letters, numbers, or underscores.
Examples: COG01, GH1, PF1.
Req
SDTM 2.2.3,
SDTMIG 4.1.1.9
921H923HSDTMIG 4.1.2.1
QSTEST
Question Name
Char
Synonym
Qualifier
Verbatim name of the question or group of questions used to obtain the
measurement or finding. The value in QSTEST cannot be longer than 40
characters. Example: In General, How is Your Health?
Req
SDTM 2.2.3,
924HSDTMIG 4.1.2.1,
925HSDTMIG 4.1.2.4,
926HSDTMIG 4.1.5.3.1
QSCAT
Category of Question
Char
*
Grouping
Qualifier
Used to define a category of related records that will be meaningful to the
Reviewer. Examples: HAMILTON DEPRESSION SCALE, SF36, ADAS.
Req
SDTM 2.2.3,
927HSDTMIG 4.1.2.6
QSSCAT
Subcategory for Question
Char
*
Grouping
Qualifier
A further categorization of the questions within the category. Examples:
MENTAL HEALTH DOMAIN, DEPRESSION DOMAIN, WORD
RECALL.
Perm
SDTM 2.2.3,
928HSDTMIG 4.1.2.6
QSORRES
Finding in Original Units
Char
Result
Qualifier
Finding as originally received or collected (e.g. RARELY, SOMETIMES).
When sponsors apply codelist to indicate the code values are statistically
meaningful standardized scores, which are defined by sponsors or by
valid methodologies such as SF36 questionnaires, QSORRES will
contain the decode format, and QSSTRESC and QSSTRESN may contain
the standardized code values or scores.
Exp
SDTM 2.2.3,
929H930HSDTMIG 4.1.5.1

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 145
Final November 12, 2008
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
QSORRESU
Original Units
Char
(931HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for QSORRES,
such as minutes or seconds or the units associated with a visual analog
scale.
Perm
SDTM 2.2.3,
932HSDTMIG 4.1.3.2,
933HSDTMIG 4.1.5.1
QSSTRESC
Character Result/Finding
in Std Format
Char
Result
Qualifier
Contains the finding for all questions or sub-scores, copied or derived from
QSORRES in a standard format or standard units. QSSTRESC should store
all findings in character format; if findings are numeric, they should also be
stored in numeric format in QSSTRESN. If question scores are derived from
the original finding, then the standard format is the score. Examples: 0, 1.
When sponsors apply codelist to indicate the code values are statistically
meaningful standardized scores, which are defined by sponsors or by valid
methodologies such as SF36 questionnaires, QSORRES will contain the
decode format, and QSSTRESC and QSSTRESN may contain the
standardized code values or scores.
Exp
SDTM 2.2.3,
934H935HSDTMIG 4.1.5.1
QSSTRESN
Numeric Finding in
Standard Units
Num
Result
Qualifier
Used for continuous or numeric findings in standard format; copied in
numeric format from QSSTRESC. QSSTRESN should store all numeric
results or findings.
Perm
SDTM 2.2.3,
936HSDTMIG 4.1.5.1
QSSTRESU
Standard Units
Char
(937HUNIT)
Variable
Qualifier
Standardized unit used for QSSTRESC or QSSTRESN.
Perm
SDTM 2.2.3,
938HSDTMIG 4.1.3.2,
939HSDTMIG 4.1.5.1
QSSTAT
Completion Status
Char
(1948HND)
Record
Qualifier
Used to indicate a questionnaire or response to a questionnaire was not
done. Should be null if a result exists in QSORRES.
Perm
SDTM 2.2.3,
940HSDTMIG 4.1.5.1,
941HSDTMIG 4.1.5.7,
942HSDTMIG
Appendix C1
QSREASND
Reason Not Performed
Char
Record
Qualifier
Describes why a question was not answered. Used in conjunction with
QSSTAT when value is NOT DONE. Example: SUBJECT REFUSED.
Perm
SDTM 2.2.3,
943HSDTMIG 4.1.5.1,
944HSDTMIG 4.1.5.7
QSBLFL
Baseline Flag
Char
(1949HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or null.
Exp
SDTM 2.2.3,
945HSDTMIG 4.1.3.7,
946HSDTMIG
Appendix C1
QSDRVFL
Derived Flag
Char
(1950HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records that
represent the average of other records or questionnaire sub-scores that do not
come from the CRF are examples of records that would be derived for the
submission datasets. If QSDRVFL=Y, then QSORRES may be null with
QSSTRESC and (if numeric) QSSTRESN having the derived value.
Perm
SDTM 2.2.3,
947HSDTMIG 4.1.3.7,
948HSDTMIG 4.1.5.1,
949HSDTMIG
Appendix C1
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
950HSDTMIG 4.1.4.5,
951HSDTMIG 7.4
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 146 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
952HSDTMIG 4.1.4.5,
953HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
954HSDTMIG 4.1.4.5,
955HSDTMIG 7.4
QSDTC
Date/Time of Finding
Char
ISO 8601
Timing
Date of questionnaire.
Exp
SDTM 2.2.5,
956HSDTMIG 4.1.4.1,
SDTMIG 4.1.4.2
957HSDTMIG 4.1.4.8
QSDY
Study Day of Finding
Num
Timing
1. Study day of finding collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm
SDTM 2.2.5,
958HSDTMIG 4.1.4.4,
959HSDTMIG 4.1.4.6
QSTPT
Planned Time Point
Name
Char
Timing
1. Text Description of time when questionnaire should be administered.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See QSTPTNUM and QSTPTREF.
Perm
SDTM 2.2.5,
960HSDTMIG 4.1.4.10
QSTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of QSTPT to aid in sorting.
Perm
SDTM 2.2.5,
961HSDTMIG 4.1.4.10
QSELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Planned Elapsed time (in ISO 8601) relative to a planned fixed reference
(QSTPTREF). This variable is useful where there are repetitive measures.
Not a clock time or a date time variable. Represented as an ISO 8601
duration. Examples: ―-PT15M‖ to represent the period of 15 minutes
prior to the reference point indicated by QSTPTREF, or ―PT8H‖ to
represent the period of 8 hours after the reference point indicated by
QSTPTREF.
Perm
SDTM 2.2.5,
962HSDTMIG 4.1.4.3,
963HSDTMIG 4.1.4.10
QSTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by QSELTM, QSTPTNUM,
and QSTPT. Examples: PREVIOUS DOSE, PREVIOUS MEAL.
Perm
SDTM 2.2.5,
964HSDTMIG 4.1.4.10
QSRFTDTC
Date/Time of Reference
Time Point
Char
ISO 8601
Timing
Date/time of the reference time point, LBTPTREF.
Perm
SDTM 2.2.5,
965HSDTMIG 4.1.4.10
QSEVLINT
Evaluation Interval
Char
ISO 8601
Timing
Evaluation Interval associated with a QSTEST question represented in ISO
8601 character format. Example: "-P2Y" to represent an interval of 2 years
in the question "Have you experienced any episodes in the past 2 years?"
Perm
SDTM 2.2.5
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 147
Final November 12, 2008
6.3.5.1 ASSUMPTIONS FOR QUESTIONNAIRE DOMAIN MODEL
1. QS Definition: A written or electronic survey instrument comprised of a series of questions, designed to measure a specific item or set of items. Questionnaires
are research instruments that usually have a documented method of administration, a standard format for data collection, and documented methods for scoring,
analysis, and interpretation of results. A questionnaire is often analyzed by applying a numeric scoring system to question responses, where each question
response is assigned a specific numeric ―score‖ that can be totaled to give an overall score (and possibly sectional sub-scores). Questionnaire data may
include, but are not limited to subject reported outcomes and validated or non-validated questionnaires. The QS domain is not intended for use in submitting a
set of questions grouped on the CRF for convenience of data capture. Some diaries are vehicles for collecting data for a validated questionnaire while others
may simply facilitate capture of routine study data. When objective numeric data with result Qualifiers are collected in a questionnaire or diary format, the
sponsor should consider whether this data actually belongs in a separate (new or existing) domain. For example, if the subject records the number of
caffeinated beverages consumed each day in a diary, this information might be more appropriate for the Substance Use domain. The names of the
questionnaires should be described under the variable QSCAT in the questionnaire domain. These could be either abbreviations or longer names, at the
sponsor‘s discretion until controlled terminology is developed. For example, Alzheimer's Disease Assessment Scale (ADAS), SF-36 Health Survey (SF36),
Positive and Negative Syndrome Scale (PANSS).
2. Names of subcategories for groups of items/questions could be described under QSSCAT.
3. Derived information such as total scores and sub scores, etc., may be stored in the QS domain as derived records with appropriate category/subcategory names
(QSSCAT), item names (QSTEST), and results (QSSTRESC, QSSTRESN). Derived records should be flagged by QSDRVFL. Single score measurements or
results may go into questionnaire (e.g., APACHE Score, ECOG), but the sponsor should consider if the results should go into a more appropriate domain.
4. The following Qualifiers would not generally be used in QS: --POS, --BODSYS, --ORNRLO, --ORNRHI, --STNRLO, --STNRHI, --STRNC, --NRIND,
--RESCAT, --XFN, --LOINC, --SPEC, --SPCCND, --LOC, --METHOD, --FAST, --TOX, --TOXGR, --SEV.
5. The sponsor is expected to provide information about the version used for each validated questionnaire in the metadata (using the Comments column in the
define.xml ). This could be provided as value-level metadata for QSCAT. If more than one version of a questionnaire is used in a study, the version used for
each record should be specified in the Supplemental Qualifiers datasets, as described in 966HSection 8.4. The sponsor is expected to provide information about the
scoring rules in the metadata.
6. If the verbatim question text is > 40 characters, put meaningful text in QSTEST and describe the full text in the study metadata. See 967Hsection 4.1.5.3.1 for
further information.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 148 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.5.2 EXAMPLES FOR QUESTIONNAIRE DOMAIN MODEL
Example 1:
This is an example of data from a questionnaire from one subject at one visit with standard text answers that have an associated score. In this example the subject
answered all of the questions in Rows 1-4 and Rows 7-9. The standard text (e.g., very good) translates to a score of 4.4. The value of 4.4 is populated in both
QSSTRESN and QSSTRESC. Since this is the baseline data there is a flag in all records in QSBLFL. The values in Rows 5, 6, 10, and 11 are derived from previous
records and are flagged with a Y in QSDRVFL. The example shows how the textual answer is handled in the QSORRES variable, while the QSSTRESC and
QSSTRESN contain the standardized score value.
Rows 5, 6, 10, 11 Show derived records. Notice how QSORRES is blank for derived records because there is no corresponding text value for the numeric
value shown (see 968HSection 4.1.5.1).
Row
STUDYID
DOMAIN
USUBJID
QSSEQ
QSTESTCD
QSTEST
QSCAT
QSSCAT
1
STUDYX
QS
P0001
1
GH1
Health
SF36
GENERAL HEALTH
2
STUDYX
QS
P0001
2
GH11A
Sick a little easier
SF36
GENERAL HEALTH
3
STUDYX
QS
P0001
3
GH11B
Healthy as anybody
SF36
GENERAL HEALTH
4
STUDYX
QS
P0001
4
GH11C
Expect health to get worse
SF36
GENERAL HEALTH
5
STUDYX
QS
P0001
5
GH
SF-36 General health perceptions
SF36
GENERAL HEALTH
6
STUDYX
QS
P0001
6
GHINDEX
SF-36 General health perceptions (0-100)
SF36
GENERAL HEALTH
7
STUDYX
QS
P0001
7
RP4A
Phys. Health-cut down time spent
SF36
ROLE-PHYSICAL
8
STUDYX
QS
P0001
8
RP4B
Phys. Health-accomplished less
SF36
ROLE-PHYSICAL
9
STUDYX
QS
P0001
9
RP4C
Phys. Health-limit kind of work
SF36
ROLE-PHYSICAL
10
STUDYX
QS
P0001
10
RP
SF-36 Role-physical
SF36
ROLE-PHYSICAL
11
STUDYX
QS
P0001
11
RPINDEX
SF-36 Role-physical (0-100)
SF36
ROLE-PHYSICAL
Row
QSORRES
QSSTRESC
QSSTRESN
QSBLFL
QSDRVFL
VISITNUM
VISIT
QSDTC
QSDY
1 (cont)
VERY GOOD
4.4
4.4
Y
2
BASELINE
2001-03-28
-2
2 (cont)
MOSTLY FALSE
4
4
Y
2
BASELINE
2001-03-28
-2
3 (cont)
MOSTLY TRUE
4
4
Y
2
BASELINE
2001-03-28
-2
4 (cont)
DEFINITELY FALSE
5
5
Y
2
BASELINE
2001-03-28
-2
5 (cont)
21.4
21.4
Y
Y
2
BASELINE
2001-03-28
-2
6 (cont)
82
82
Y
Y
2
BASELINE
2001-03-28
-2
7 (cont)
NO
2
2
Y
2
BASELINE
2001-03-28
-2
8 (cont)
NO
2
2
Y
2
BASELINE
2001-03-28
-2
9 (cont)
NO
2
2
Y
2
BASELINE
2001-03-28
-2
10 (cont)
8
8
Y
Y
2
BASELINE
2001-03-28
-2
11 (cont)
100
100
Y
Y
2
BASELINE
2001-03-28
-2
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 149
Final November 12, 2008
Example 2:
This example shows data from one subject collected at one visit for a questionnaire with standard text answers.
Rows 1-10: Answers are not associated with a numeric score, so QSSTRESC is copied from QSORRES, and QSSTRESN is null. Notice that QSTPTNUM is
used to distinguish the same question being asked at various time points on the same date where no time was collected. For more information on
time points, see 969HSection 4.1.4.10. Note that QSTPTREF is not used in the examples because QSTPTNUM is being used only to organize the results
by type of question, and the timing to a reference point is not important in this study. In this study, QSTPTNUM is an arbitrary number, sponsor
defined to aid in sorting
Row 11: Shows a derived record. Notice how QSORRES is blank for derived records because there is no corresponding text value for the numeric value
shown (see 970HSection 4.1.5.1). The derived record, however, does have a derived value in QSSTRESN.
Row
STUDYID
DOMAIN
USUBJID
QSSEQ
QSTESTCD
QSTEST
QSCAT
QSSCAT
QSORRES
QSSTRESC
1
STUDYX
QS
P0001
1
COG01T02
ARM
ADAS
WORD RECALL
NO
NO
2
STUDYX
QS
P0001
2
COG01T02
ARM
ADAS
WORD RECALL
NO
NO
3
STUDYX
QS
P0001
3
COG01T02
ARM
ADAS
WORD RECALL
NO
NO
4
STUDYX
QS
P0001
4
COG01T03
BUTTER
ADAS
WORD RECALL
NO
NO
5
STUDYX
QS
P0001
5
COG01T03
BUTTER
ADAS
WORD RECALL
NO
NO
6
STUDYX
QS
P0001
6
COG01T03
BUTTER
ADAS
WORD RECALL
NO
NO
7
STUDYX
QS
P0001
7
COG01T04
CABIN
ADAS
WORD RECALL
NO
NO
8
STUDYX
QS
P0001
8
COG01T04
CABIN
ADAS
WORD RECALL
NO
NO
9
STUDYX
QS
P0001
9
COG01T04
CABIN
ADAS
WORD RECALL
NO
NO
10
STUDYX
QS
P0001
10
COG01T09
GRASS
ADAS
WORD RECALL
NO
NO
11
STUDYX
QS
P0001
11
COG01X
WORD RECALL
ADAS
WORD RECALL
9
Row
QSSTRESN
QSBLFL
QSDRVFL
VISITNUM
VISIT
VISITYDY
QSDTC
QSDY
QSTPTNUM
1 (cont)
1
SCREENING
-14
2001-03-20
-10
1
2 (cont)
1
SCREENING
-14
2001-03-20
-10
2
3 (cont)
1
SCREENING
-14
2001-03-20
-10
3
4 (cont)
1
SCREENING
-14
2001-03-20
-10
1
5 (cont)
1
SCREENING
-14
2001-03-20
-10
2
6 (cont)
1
SCREENING
-14
2001-03-20
-10
3
7 (cont)
1
SCREENING
-14
2001-03-20
-10
1
8 (cont)
1
SCREENING
-14
2001-03-20
-10
2
9 (cont)
1
SCREENING
-14
2001-03-20
-10
3
10 (cont)
1
SCREENING
-14
2001-03-20
-10
1
11 (cont)
9
Y
1
SCREENING
-14
2001-03-20
-10
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 150 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.6 SUBJECT CHARACTERISTICS — SC
sc.xpt, Subject Characteristics — Findings, Version 3.1.2. One record per characteristic per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1952HSC
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
971HSDTMIG 4.1.2.2,
972HSDTMIG
Appendix C2
USUBJID
Unique Subject
Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
973H974HSDTMIG 4.1.2.3
SCSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
SCGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
975HSDTMIG 4.1.2.6
SCSPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Perm
SDTM 2.2.4,
976HSDTMIG 4.1.2.6
SCTESTCD
Subject Characteristic
Short Name
Char
(977HSCCD)
Topic
Short name of the measurement, test, or examination described in
SCTEST. It can be used as a column name when converting a dataset from
a vertical to a horizontal format. The value in SCTESTCD cannot be
longer than 8 characters, nor can it start with a number (e.g.‖1TEST‖).
SCTESTCD cannot contain characters other than letters, numbers, or
underscores. Example: SUBJINIT, EYECD.
Req
SDTM 2.2.3,
SDTMIG 4.1.1.9
978H
980HSDTMIG 4.1.2.1
SDTMIG
Appendix C1
SCTEST
Subject Characteristic
Char
*
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the measurement
or finding. The value in SCTEST cannot be longer than 40 characters.
Examples: Subject Initials, Eye Color.
Req
SDTM 2.2.3,
981HSDTMIG 4.1.2.1,
982HSDTMIG 4.1.2.4,
983HSDTMIG 4.1.5.3.1
SCCAT
Category for Subject
Characteristic
Char
*
Grouping
Qualifier
Used to define a category of related records.
Perm
SDTM 2.2.3,
984HSDTMIG 4.1.2.6
SCSCAT
Subcategory for Subject
Characteristic
Char
*
Grouping
Qualifier
A further categorization of the subject characteristic.
Perm
SDTM 2.2.3,
985HSDTMIG 4.1.2.6
SCORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the subject characteristic as originally received or collected.
Exp
SDTM 2.2.3, 986H
987HSDTMIG 4.1.5.1
SCORRESU
Original Units
Char
(988HUNIT)
Variable
Qualifier
Original Unit in which the data were collected. The unit for SCORRES.
Perm
SDTM 2.2.3,
989HSDTMIG 4.1.3.2
990HSDTMIG 4.1.5.1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 151
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled Terms,
Codelist or Format
Role
CDISC Notes
Core
References
SCSTRESC
Character
Result/Finding in Std
Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from
SCORRES in a standard format or standard units. SCSTRESC should
store all results or findings in character format; if results are numeric, they
should also be stored in numeric format in SCSTRESN. For example, if a
test has results ―NONE‖, ―NEG‖, and ―NEGATIVE‖ in SCORRES and
these results effectively have the same meaning, they could be represented
in standard format in SCSTRESC as ―NEGATIVE‖.
Exp
SDTM 2.2.3, 991H
992HSDTMIG 4.1.5.1
SCSTRESN
Numeric Result/Finding
in Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from SCSTRESC. SCSTRESN should store all
numeric test results or findings.
Perm
SDTM 2.2.3,
993HSDTMIG 4.1.5.1
SCSTRESU
Standard Units
Char
(994HUNIT)
Variable
Qualifier
Standardized unit used for SCSTRESC or SCSTRESN.
Perm
SDTM 2.2.3,
995HSDTMIG 4.1.3.2,
996HSDTMIG 4.1.5.1
SCSTAT
Completion Status
Char
(1953HND)
Record
Qualifier
Used to indicate that the measurement was not done. Should be null if a
result exists in SCORRES.
Perm
SDTM 2.2.3,
997HSDTMIG 4.1.5.1,
998HSDTMIG 4.1.5.7,
999HSDTMIG
Appendix C1
SCREASND
Reason Not Performed
Char
Record
Qualifier
Describes why the observation has no result. Example: subject refused.
Used in conjunction with SCSTAT when value is NOT DONE.
Perm
SDTM 2.2.3,
1000HSDTMIG 4.1.5.1,
1001HSDTMIG 4.1.5.7
SCDTC
Date/Time of Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
1002HSDTMIG 4.1.4.1,
SDTMIG 4.1.4.2
1003HSDTMIG 4.1.4.8
SCDY
Study Day of
Examination
Num
Timing
1. Study day of collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm
SDTM 2.2.5,
1004HSDTMIG 4.1.4.4,
1005HSDTMIG 4.1.4.6
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.6.1 ASSUMPTIONS FOR SUBJECT CHARACTERISTICS DOMAIN MODEL
1. SC Definition: Subject Characteristics is for data not collected in other domains that are subject-related. Examples: subject initials, eye color, childbearing
status, etc.
2. The structure for demographic data is fixed and includes date of birth, age, sex, race, ethnicity and country. The structure of subject characteristics is based on
the Findings general observation class and is an extension of the demographics data. Subject Characteristics consists of data that is collected once per
subject (per test). SC contains data that is either not normally expected to change during the trial or whose change is not of interest after the initial collection.
Sponsor should ensure that data considered for submission in SC do not actually belong in another domain.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 152 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
3. The following Qualifiers would not generally be used in SC: --MODIFY, --POS, --BODSYS, --ORNRLO, --ORNRHI, --STNRLO, --STNRHI, --STNRC,
--NRIND, --RESCAT, --XFN, --NAM, --LOINC, --SPEC, --SPCCND, --METHOD, --BLFL, --FAST, --DRVRL, --TOX, --TOXGR, --SEV.
6.3.6.2 EXAMPLE FOR SUBJECT CHARACTISTICS DOMAIN MODEL
The example below shows data that is collected once per subject and does not fit into the Demographics domain. For this example the eye color and initials were
collected.
Row
STUDYID
DOMAIN
USUBJID
SCSEQ
SCTESTCD
SCTEST
SCORRES
SCSTRESC
SCDTC
1
ABC
SC
ABC-001-001
1
EYECD
Eye Color
BROWN
BROWN
1999-06-19
2
ABC
SC
ABC-001-001
2
SUBJINIT
Subject Initials
HLT
HLT
1999-06-19
3
ABC
SC
ABC-001-002
1
EYECD
Eye Color
BLUE
BLUE
1999-03-19
4
ABC
SC
ABC-001-002
2
SUBJINIT
Subject Initials
BAM
BAM
1999-03-19
5
ABC
SC
ABC-001-003
1
EYECD
Eye Color
GREEN
GREEN
1999-05-03
6
ABC
SC
ABC-001-003
2
SUBJINIT
Subject Initials
ALM
ALM
1999-05-03
7
ABC
SC
ABC-002-001
1
EYECD
Eye Color
HAZEL
HAZEL
1999-06-14
8
ABC
SC
ABC-002-001
2
SUBJINIT
Subject Initials
CQH
CQH
1999-06-14
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 153
Final November 12, 2008
6.3.7 VITAL SIGNS — VS
vs.xpt, Vital Signs — Findings, Version 3.1.2. One record per vital sign measurement per time point per visit per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1954HVS
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1006HSDTMIG 4.1.2.2,
1007HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
1008H1009HSDTMIG 4.1.2.3
VSSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
VSGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
1010HSDTMIG 4.1.2.6
VSSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Perm
SDTM 2.2.4,
1011HSDTMIG 4.1.2.6
VSTESTCD
Vital Signs Test Short
Name
Char
(1955HVSTESTCD)
Topic
Short name of the measurement, test, or examination described in
VSTEST. It can be used as a column name when converting a dataset
from a vertical to a horizontal format. The value in VSTESTCD cannot be
longer than 8 characters, nor can it start with a number (e.g.‖1TEST‖).
VSTESTCD cannot contain characters other than letters, numbers, or
underscores. Examples: SYSBP, DIABP, BMI.
Req
SDTM 2.2.3,
1012H1013HSDTMIG 4.1.1.8,
1014HSDTMIG 4.1.2.1,
1015HSDTMIG
Appendix C1
VSTEST
Vital Signs Test Name
Char
(1956HVSTEST)
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the measurement
or finding. The value in VSTEST cannot be longer than 40 characters.
Examples: Systolic Blood Pressure, Diastolic Blood Pressure, Body Mass
Index.
Req
SDTM 2.2.3,
1016HSDTMIG 4.1.2.1,
1017HSDTMIG 4.1.2.4,
1018HSDTMIG 4.1.5.3.1,
1019HSDTMIG
Appendix C1
VSCAT
Category for Vital Signs
Char
*
Grouping
Qualifier
Used to define a category of related records.
Perm
SDTM 2.2.3,
1020HSDTMIG 4.1.2.6
VSSCAT
Subcategory for Vital Signs
Char
*
Grouping
Qualifier
A further categorization of a measurement or examination.
Perm
SDTM 2.2.3,
1021HSDTMIG 4.1.2.6
VSPOS
Vital Signs Position of
Subject
Char
(1022HPOSITION)
Record
Qualifier
Position of the subject during a measurement or examination. Examples:
SUPINE, STANDING, SITTING.
Perm
SDTM 2.2.3
VSORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the vital signs measurement as originally received or collected.
Exp
SDTM 2.2.3, 1023H
1024HSDTMIG 4.1.5.1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 154 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
VSORRESU
Original Units
Char
(1957HVSRESU)
Variable
Qualifier
Original units in which the data were collected. The unit for VSORRES.
Examples: IN, LB, BEATS/MIN.
Exp
SDTM 2.2.3,
1025HSDTMIG 4.1.3.2
1026HSDTMIG 4.1.5.1,
1027HSDTMIG
Appendix C1
VSSTRESC
Character Result/Finding in
Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from
VSORRES in a standard format or standard units. VSSTRESC should
store all results or findings in character format; if results are numeric, they
should also be stored in numeric format in VSSTRESN. For example, if a
test has results ―NONE‖, ―NEG‖, and ―NEGATIVE‖ in VSORRES and
these results effectively have the same meaning, they could be represented
in standard format in VSSTRESC as ―NEGATIVE‖.
Exp
SDTM 2.2.3,
1028H1029HSDTMIG 4.1.5.1
VSSTRESN
Numeric Result/Finding in
Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from VSSTRESC. VSSTRESN should store all
numeric test results or findings.
Exp
SDTM 2.2.3,
1030HSDTMIG 4.1.5.1
VSSTRESU
Standard Units
Char
(1958HVSRESU)
Variable
Qualifier
Standardized unit used for VSSTRESC and VSSTRESN.
Exp
SDTM 2.2.3,
1031HSDTMIG 4.1.3.2,
1032HSDTMIG 4.1.5.1,
1033HSDTMIG
Appendix C1
VSSTAT
Completion Status
Char
(1959HND)
Record
Qualifier
Used to indicate that a vital sign measurement was not done. Should be
null if a result exists in VSORRES.
Perm
SDTM 2.2.3,
1034HSDTMIG 4.1.5.1,
1035HSDTMIG 4.1.5.7,
1036HSDTMIG
Appendix C1
VSREASND
Reason Not Performed
Char
Record
Qualifier
Describes why a measurement or test was not performed. Examples:
BROKEN EQUIPMENT or SUBJECT REFUSED. Used in conjunction
with VSSTAT when value is NOT DONE.
Perm
SDTM 2.2.3,
1037HSDTMIG 4.1.5.1,
1038HSDTMIG 4.1.5.7
VSLOC
Location of Vital Signs
Measurement
Char
(1039HLOC)
Record
Qualifier
Location relevant to the collection of Vital Signs measurement. Example:
LEFT ARM for blood pressure.
Perm
SDTM 2.2.3
VSBLFL
Baseline Flag
Char
(1960HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or
null.
Exp
SDTM 2.2.3,
1040HSDTMIG 4.1.3.7,
1041HSDTMIG
Appendix C1
VSDRVFL
Derived Flag
Char
(1961HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records
which represent the average of other records or which do not come from
the CRF are examples of records that would be derived for the submission
datasets. If VSDRVFL=Y, then VSORRES may be null, with VSSTRESC
and (if numeric) VSSTRESN having the derived value.
Perm
SDTM 2.2.3,
1042HSDTMIG 4.1.3.7,
1043HSDTMIG 4.1.5.1,
1044HSDTMIG
Appendix C1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 155
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
1045HSDTMIG 4.1.4.5,
1046HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
1047HSDTMIG 4.1.4.5,
1048HSDTMIG 7.4
VISITDY
Planned Study Day of Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
1049HSDTMIG 4.1.4.5,
1050HSDTMIG 7.4
VSDTC
Date/Time of
Measurements
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
1051HSDTMIG 4.1.4.1,
1052HSDTMIG 4.1.4.8
VSDY
Study Day of Vital Signs
Num
Timing
1. Study day of vital signs measurements, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm
SDTM 2.2.5,
1053HSDTMIG 4.1.4.4,
1054HSDTMIG 4.1.4.6
VSTPT
Planned Time Point Name
Char
Timing
1. Text Description of time when measurement should be taken.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See VSTPTNUM and VSTPTREF.
Examples: Start, 5 min post.
Perm
SDTM 2.2.5,
1055HSDTMIG 4.1.4.10
VSTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of VSTPT to aid in sorting.
Perm
SDTM 2.2.5,
1056HSDTMIG 4.1.4.10
VSELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Planned Elapsed time (in ISO 8601) relative to a planned fixed reference
(VSTPTREF). This variable is useful where there are repetitive measures.
Not a clock time or a date time variable. Represented as an ISO 8601
Duration. Examples: ―-PT15M‖ to represent the period of 15 minutes
prior to the reference point indicated by VSTPTREF, or ―PT8H‖ to
represent the period of 8 hours after the reference point indicated by
VSTPTREF.
Perm
SDTM 2.2.5,
1057HSDTMIG 4.1.4.3,
1058HSDTMIG 4.1.4.10
VSTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by VSELTM, VSTPTNUM,
and VSTPT. Examples: PREVIOUS DOSE, PREVIOUS MEAL.
Perm
SDTM 2.2.5,
1059HSDTMIG 4.1.4.10
VSRFTDTC
Date/Time of Reference
Time Point
Char
ISO 8601
Timing
Date/time of the reference time point, LBTPTREF.
Perm
SDTM 2.2.5,
1060HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 156 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.7.1 ASSUMPTIONS FOR VITAL SIGNS DOMAIN MODEL
1. VS Definition: CRF data that captures measurements such as blood pressure, height, weight, pulse, and body temperature, or derived data such as body
mass index.
2. In cases where the LOINC dictionary is used for Vital Sign tests, the permissible variable VSLOINC could be used. The sponsor is expected to provide
the dictionary name and version used to map the terms utilizing the define.xml external codelist attributes
3. If a reference range is available for a vital signs test, the variables VSORNRLO,VSORNRHI, VSNRIND from the Findings observation class may be
added to the domain. VSORNRLO and VSORNRHI would represent the reference range , and VSNRIND would be used to indicate where a result falls
in respect to the reference range (examples: HIGH, LOW). Clinical significance would be represented as described in 1061HSection 4.1.5.5 as a record in
SUPPVS with a QNAM of VSCLSIG.
4. The following Qualifiers would not generally be used in VS: --BODSYS, --XFN, --SPEC, --SPCCND, --FAST, --TOX, --TOXGR.
6.3.7.2 EXAMPLE FOR VITAL SIGNS DOMAIN MODEL
The example below shows one subject with two visits, Baseline and Visit 1, including examples of both collected and derived baseline measurements.
Rows 1,2, 4,5, 8, 9: VSTPT and VSTPTNUM are populated since more than one measurement was taken at this visit.
Rows 3, 6: Show an example of a derived value that was not considered to be an original result. In this case the sponsor derived the value in a different
variable in the operational database. VSTPT and VSTPTNUM are not populated for these derived records.
Rows 8, 9: Show two temperatures taken at the baseline visit. Row 9 has a "Y" in the VSBLFL to indicate it was used as the baseline measurement.
Row 14: Shows a value collected in one unit, but converted to selected standard unit.
Row 15: Shows the proper use of the --STAT variable to indicate "NOT DONE" where a reason was collected when a test was not done.
Row
STUDYID
DOMAIN
USUBJID
VSSEQ
VSTESTCD
VSTEST
VSPOS
VSORRES
VSORRESU
VSSTRESC
VSSTRESN
VSSTRESU
1
ABC
VS
ABC-001-001
1
SYSBP
Systolic Blood Pressure
SITTING
154
mmHg
154
154
mmHg
2
ABC
VS
ABC-001-001
2
SYSBP
Systolic Blood Pressure
SITTING
152
mmHg
152
152
mmHg
3
ABC
VS
ABC-001-001
3
SYSBP
Systolic Blood Pressure
SITTING
153
153
mmHg
4
ABC
VS
ABC-001-001
4
DIABP
Diastolic Blood Pressure
SITTING
44
mmHg
44
44
mmHg
5
ABC
VS
ABC-001-001
5
DIABP
Diastolic Blood Pressure
SITTING
48
mmHg
48
48
mmHg
6
ABC
VS
ABC-001-001
6
DIABP
Diastolic Blood Pressure
SITTING
46
46
mmHg
7
ABC
VS
ABC-001-001
7
PULSE
Pulse Rate
SITTING
72
bpm
72
72
bpm
8
ABC
VS
ABC-001-001
8
TEMP
Temperature
34.7
C
34.7
34.7
C
9
ABC
VS
ABC-001-001
9
TEMP
Temperature
36.2
C
36.2
36.2
C
10
ABC
VS
ABC-001-001
10
WEIGHT
Weight
STANDING
90.5
kg
90.5
90.5
kg
11
ABC
VS
ABC-001-001
11
HEIGHT
Height
STANDING
157
cm
157
157
cm
12
ABC
VS
ABC-001-001
12
SYSBP
Systolic Blood Pressure
SITTING
95
mmHg
95
95
mmHg
13
ABC
VS
ABC-001-001
13
DIABP
Diastolic Blood Pressure
SITTING
44
mmHg
44
44
mmHg
14
ABC
VS
ABC-001-001
14
TEMP
Temperature
97.16
F
36.2
36.2
C
15
ABC
VS
ABC-001-001
15
WEIGHT
Weight
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 157
Final November 12, 2008
Row
VSSTAT
VSREASND
VSLOC
VSBLFL
VSDRVFL
VISIT
VISITNUM
VISITDY
VSDTC
VSDY
VSTPT
VSTPTNUM
1 (cont)
LEFT ARM
BASELINE
1
1
1999-06-
19T08:45
1
BASELINE 1
1
2 (cont)
LEFT ARM
BASELINE
1
1
1999-06-
19T09:00
1
BASELINE 2
2
3 (cont)
LEFT ARM
Y
Y
BASELINE
1
1
1999-06-19
1
4 (cont)
LEFT ARM
BASELINE
1
1
1999-06-
19T08:45
1
BASELINE 1
1
5 (cont)
LEFT ARM
BASELINE
1
1
1999-06-
19T09:00
1
BASELINE 2
2
6 (cont)
LEFT ARM
Y
Y
BASELINE
1
1
1999-06-19
1
7 (cont)
LEFT ARM
Y
BASELINE
1
1
1999-06-19
1
8 (cont)
MOUTH
BASELINE
1
1
1999-06-
19T08:45
1
BASELINE 1
1
9 (cont)
MOUTH
Y
BASELINE
1
1
1999-06-
19T09:00
1
BASELINE 2
2
10 (cont)
Y
BASELINE
1
1
1999-06-19
1
11 (cont)
Y
BASELINE
1
1
1999-06-19
1
12 (cont)
LEFT ARM
VISIT 2
2
35
1999-07-21
33
13 (cont)
LEFT ARM
VISIT 2
2
35
1999-07-21
33
14 (cont)
MOUTH
VISIT 2
2
35
1999-07-21
33
15 (cont)
NOT DONE
Subject refused
VISIT 2
2
35
1999-07-21
33
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 158 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.8 DRUG ACCOUNTABILITY — DA
da.xpt, Drug Accountability — Findings, Version 3.1.2. One record per drug accountability finding per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study within the submission.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1962HDA
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1062HSDTMIG 4.1.2.2,
1063HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Unique subject identifier within the submission.
Req
SDTM 2.2.4,
1064H1065HSDTMIG 4.1.2.3
DASEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
DAGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
1066HSDTMIG 4.1.2.6
DAREFID
Reference ID
Char
Identifier
Internal or external identifier such as label number.
Perm
SDTM 2.2.4,
1067HSDTMIG 4.1.2.6
DASPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Examples: Line number on the Drug Accountability page, drug label code.
Perm
SDTM 2.2.4,
1068HSDTMIG 4.1.2.6
DATESTCD
Short Name of
Accountability
Assessment
Char
*
Topic
Short character value for DATEST used as a column name when
converting a dataset from a vertical format to a horizontal format. The
short value can be up to 8 characters and cannot begin with a number or
contain characters other than letters, numbers or underscores. Example:
DISPAMT, RETAMT.
Req
SDTM 2.2.3,
1069H1070HSDTMIG 4.1.1.8
1071HSDTMIG 4.1.2.1
DATEST
Name of Accountability
Assessment
Char
*
Synonym
Qualifier
Verbatim name, corresponding to the topic variable, of the test or
examination used to obtain the drug accountability assessment. The value
in DATEST cannot be longer than 40 characters. Example: Dispensed
Amount, Returned Amount.
Req
SDTM 2.2.3,
1072HSDTMIG 4.1.2.1,
1073HSDTMIG 4.1.2.4,
1074HSDTMIG 4.1.5.3.1
DACAT
Category of Assessment
Char
*
Grouping
Qualifier
Used to define a category of related records. Examples: STUDY
MEDICATION, RESCUE MEDICATION.
Perm
SDTM 2.2.3,
1075HSDTMIG 4.1.2.6
DASCAT
Subcategory of
Assessment
Char
*
Grouping
Qualifier
Used to define a further categorization level for a group of related
records.
Perm
SDTM 2.2.3,
1076HSDTMIG 4.1.2.6
DAORRES
Assessment Result in
Original Units
Char
Result
Qualifier
Result of the Drug Accountability assessment as originally received or
collected.
Exp
SDTM 2.2.3,
1077H1078HSDTMIG 4.1.5.1
DAORRESU
Original Units
Char
(1079HUNIT)
Variable
Qualifier
Unit for DAORRES.
Perm
SDTM 2.2.3,
1080HSDTMIG 4.1.3.2,
1081HSDTMIG 4.1.5.1
SDTMIG
Appendix C1
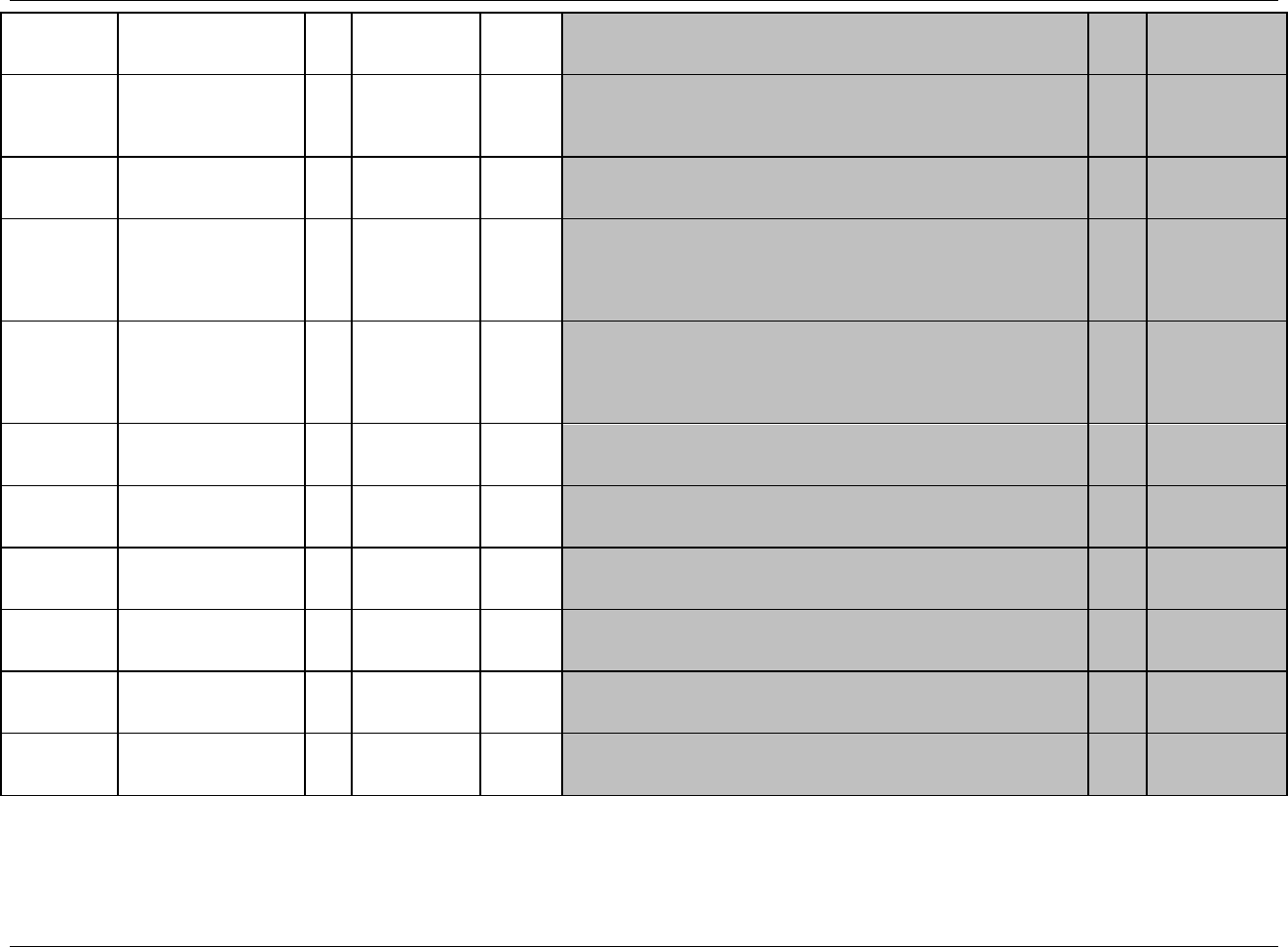
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 159
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
DASTRESC
Assessment Result in Std
Format
Char
Result
Qualifier
Contains the result value for all Drug Accountability assessments, copied or
derived from DAORRES in a standard format or in standard units.
DASTRESC should store all results or findings in character format; if results
are numeric, they should also be stored in numeric format in DASTRESN.
Exp
SDTM 2.2.3,
1082H1083HSDTMIG 4.1.5.1
DASTRESN
Numeric Result/Finding
in Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from DASTRESC. DASTRESN should store
all numeric test results or findings.
Perm
SDTM 2.2.3,
1084HSDTMIG 4.1.5.1
DASTRESU
Assessment Standard
Units
Char
(1085HUNIT)
Variable
Qualifier
Standardized units used for DASTRESC and DASTRESN.
Perm
SDTM 2.2.3,
1086HSDTMIG 4.1.3.2,
1087HSDTMIG 4.1.5.1
SDTMIG
Appendix C1
DASTAT
Completion Status
Char
(1963HND)
Record
Qualifier
Used to indicate that a drug accountability assessment was not done.
Should be null or have a value of NOT DONE.
Perm
SDTM 2.2.3,
1088HSDTMIG 4.1.5.1,
1089HSDTMIG 4.1.5.7,
1090HSDTMIG
Appendix C1
DAREASND
Reason Not Performed
Char
Record
Qualifier
Reason not done. Used in conjunction with DASTAT when value is NOT
DONE.
Perm
SDTM 2.2.3,
1091HSDTMIG 4.1.5.1,
1092HSDTMIG 4.1.5.7 10
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
1094HSDTMIG 4.1.4.5,
1095HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter
2. May be used in addition to VISITNUM and/or VISITDY
Perm
SDTM 2.2.5,
1096HSDTMIG 4.1.4.5,
1097HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
1098HSDTMIG 4.1.4.5,
1099HSDTMIG 7.4
DADTC
Date/Time of
Accountability
Assessment
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
1100HSDTMIG 4.1.4.1,
1101HSDTMIG 4.1.4.8
DADY
Study Day of
Accountability
Assessment
Num
Timing
1. Study day of drug accountability assessment, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm
SDTM 2.2.5,
1102HSDTMIG 4.1.4.4,
1103HSDTMIG 4.1.4.6
*indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.8.1 ASSUMPTIONS FOR DRUG ACCOUNTABILITY DOMAIN MODEL
1. Definition: Drug Accountability is for data regarding the accountability of study drug, such as information on receipt, dispensing, return, and packaging.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 160 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
2. One way a sponsor may choose to distinguish between different types of medications (e.g., study medication, rescue medication, run-in medication) is to use DACAT.
3. DAREFID and DASPID are both available for capturing label information.
4. The following Qualifiers would not generally be used in DA: --MODIFY, --POS, --BODSYS, --ORNRLO, --ORNRHI, --STNRLO, --STNRHI, --STNRC,
--NRIND, --RESCAT, --XFN, --NAM, --LOINC, --SPEC, --SPCCND, --METHOD, --BLFL, --FAST, --DRVRL, --TOX, --TOXGR, --SEV.
6.3.8.2 EXAMPLES FOR DRUG ACCOUNTABILITY DOMAIN MODEL
Example 1
Example 1 below shows drug accounting for a study with two study meds and one rescue med, all of which are measured in tablets. The sponsor has chosen to
add EPOCH from the list of timing variables and to use DASPID and DAREFID for code numbers that appear on the label.
Row
STUDYID
DOMAIN
USUBJID
DASEQ
DAREFID
DASPID
DATESTCD
DATEST
DACAT
DASCAT
1
ABC
DA
ABC/01001
1
XBYCC-E990A
A375827
DISPAMT
Dispensed Amount
Study Medication
Bottle A
2
ABC
DA
ABC/01001
2
XBYCC-E990A
A375827
RETAMT
Returned Amount
Study Medication
Bottle A
3
ABC
DA
ABC/01001
3
XBYCC-E990B
A227588
DISPAMT
Dispensed Amount
Study Medication
Bottle B
4
ABC
DA
ABC/01001
4
XBYCC-E990B
A227588
RETAMT
Returned Amount
Study Medication
Bottle B
5
ABC
DA
ABC/01001
5
DISPAMT
Dispensed Amount
Rescue Medication
6
ABC
DA
ABC/01001
6
RETAMT
Returned Amount
Rescue Medication
Row
DAORRES
DAORRESU
DASTRESC
DASTRESN
DASTRESU
VISITNUM
DADTC
EPOCH
1 (cont)
30
TABLETS
30
30
TABLETS
1
2004-06-15
Study Med Period 1
2 (cont)
5
TABLETS
5
5
TABLETS
2
2004-07-15
Study Med Period 1
3 (cont)
15
TABLETS
15
15
TABLETS
1
2004-06-15
Study Med Period 1
4 (cont)
0
TABLETS
0
0
TABLETS
2
2004-07-15
Study Med Period 1
5 (cont)
10
TABLETS
10
10
TABLETS
1
2004-06-15
Study Med Period 1
6 (cont)
10
TABLETS
10
10
TABLETS
2
2004-07-15
Study Med Period 1
Example 2
Example 2 is for a study where drug containers, rather than their contents, are being accounted for and the sponsor did not track returns. In this case, the purpose
of the accountability tracking is to verify that the containers dispensed were consistent with the randomization. The sponsor has chosen to use DASPID to record
the identifying number of the container dispensed.
Row
STUDYID
DOMAIN
USUBJID
DASEQ
DASPID
DATESTCD
DATEST
DACAT
DASCAT
1
ABC
DA
ABC/01001
1
AB001
DISPAMT
Dispensed Amount
Study Medication
Drug A
2
ABC
DA
ABC/01001
2
AB002
DISPAMT
Dispensed Amount
Study Medication
Drug B
Row
DAORRES
DAORRESU
DASTRESC
DASTRESN
DASTRESU
VISITNUM
DADTC
1 (cont)
1
CONTAINER
1
1
CONTAINER
1
2004-06-15
2 (cont)
1
CONTAINER
1
1
CONTAINER
1
2004-06-15
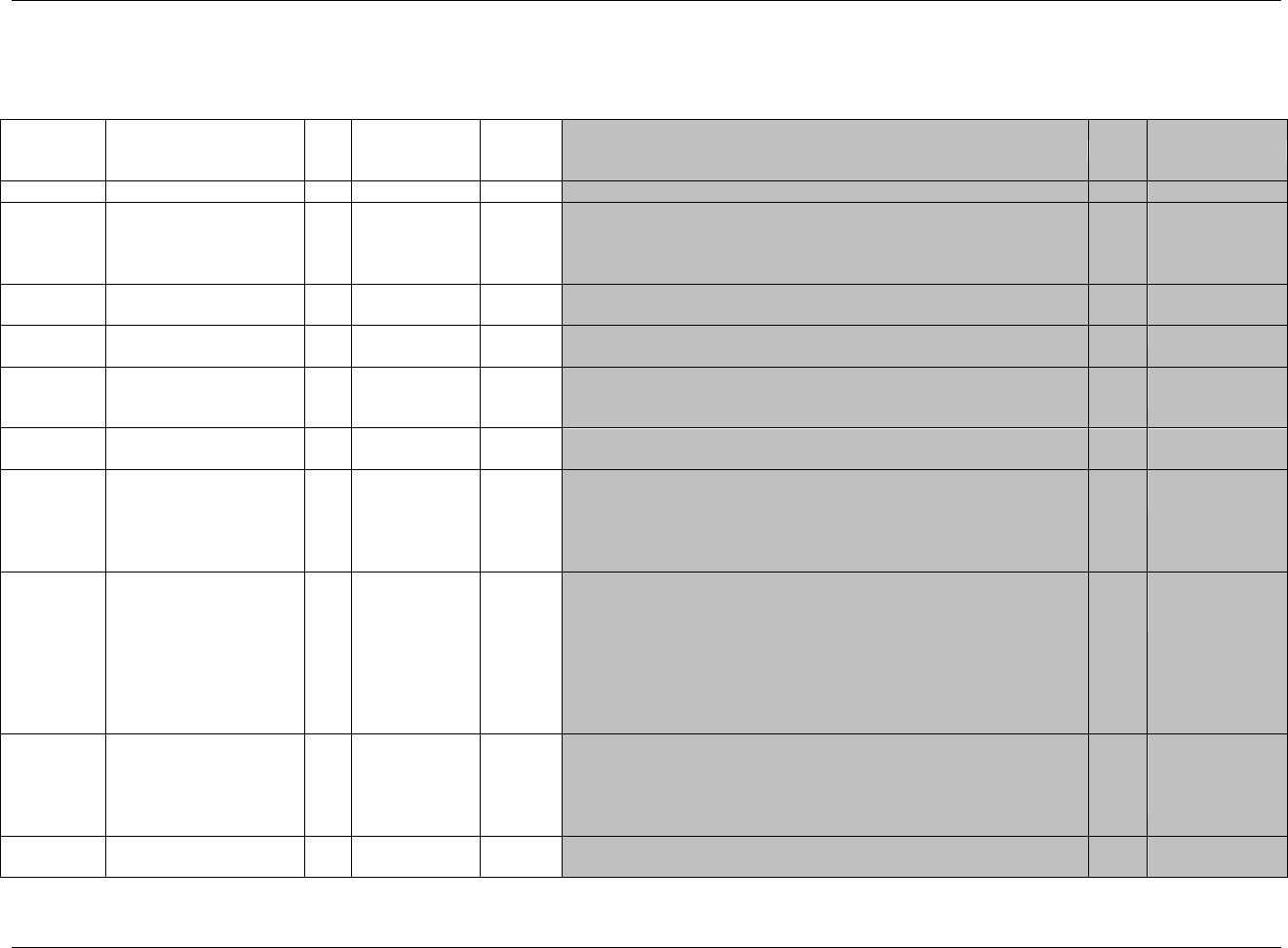
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 161
Final November 12, 2008
6.3.9 MICROBIOLOGY DOMAINS — MB AND 1104HMS
6.3.9.1 MICROBIOLOGY SPECIMEN (MB) DOMAIN MODEL
mb.xpt, Microbiology Specimen — Findings, Version 3.1.2. One record per microbiology specimen finding per time point per visit per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1964HMB
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1105HSDTMIG 4.1.2.2,
1106HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
1107H1108HSDTMIG 4.1.2.3
MBSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
MBGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain to
support relationships within the domain and between domains. In MB,
used to link to findings about organisms which are stored in MS.
Exp
SDTM 2.2.4,
1109HSDTMIG 4.1.2.6
MBREFID
Reference ID
Char
Identifier
Internal or external specimen identifier. Example: Specimen ID
Perm
SDTM 2.2.4,
1110HSDTMIG 4.1.2.6
MBSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor's operational database.
Example: ORGANISM IDENTIFIER. For organism identification, MBSPID
would remain the same each time the same organism is identified in a new
specimen.
Perm
SDTM 2.2.4,
1111HSDTMIG 4.1.2.6
MBTESTCD
Microbiology Test or
Finding Short Name
Char
*
Topic
Short name of the measurement, test, or finding described in MBTEST. It can
be used as a column name when converting a dataset from a vertical to a
horizontal format. The value in MBTESTCD cannot be longer than 8
characters, nor can it start with a number (e.g., ―1TEST‖). MBTESTCD
cannot contain characters other than letters, numbers, or underscores.
Examples for GRAM STAIN findings: GMNROD, GMNCOC,
GMSQEPCE, GMPMNLOW. Examples for CULTURE PLATE findings:
ORGANISM.
Req
SDTM 2.2.3,
1112H1113HSDTMIG 4.1.1.8,
1114HSDTMIG 4.1.2.1
MBTEST
Microbiology Test or
Finding Name
Char
*
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the measurement
or finding. The value in MBTEST cannot be longer than 40 characters.
Examples: GRAM NEGATIVE RODS, GRAM NEGATIVE COCCI,
SQUAMOUS EPITHELIAL CELLS, PMN PER FIELD LOW,
ORGANISM PRESENT
Req
SDTM 2.2.3,
1115HSDTMIG 4.1.2.1,
1116HSDTMIG 4.1.2.4,
1117HSDTMIG 4.1.5.3.1
MBCAT
Category for Microbiology
Finding
Char
*
Grouping
Qualifier
Used to define a category of related records.
Perm
SDTM 2.2.3,
1118HSDTMIG 4.1.2.6
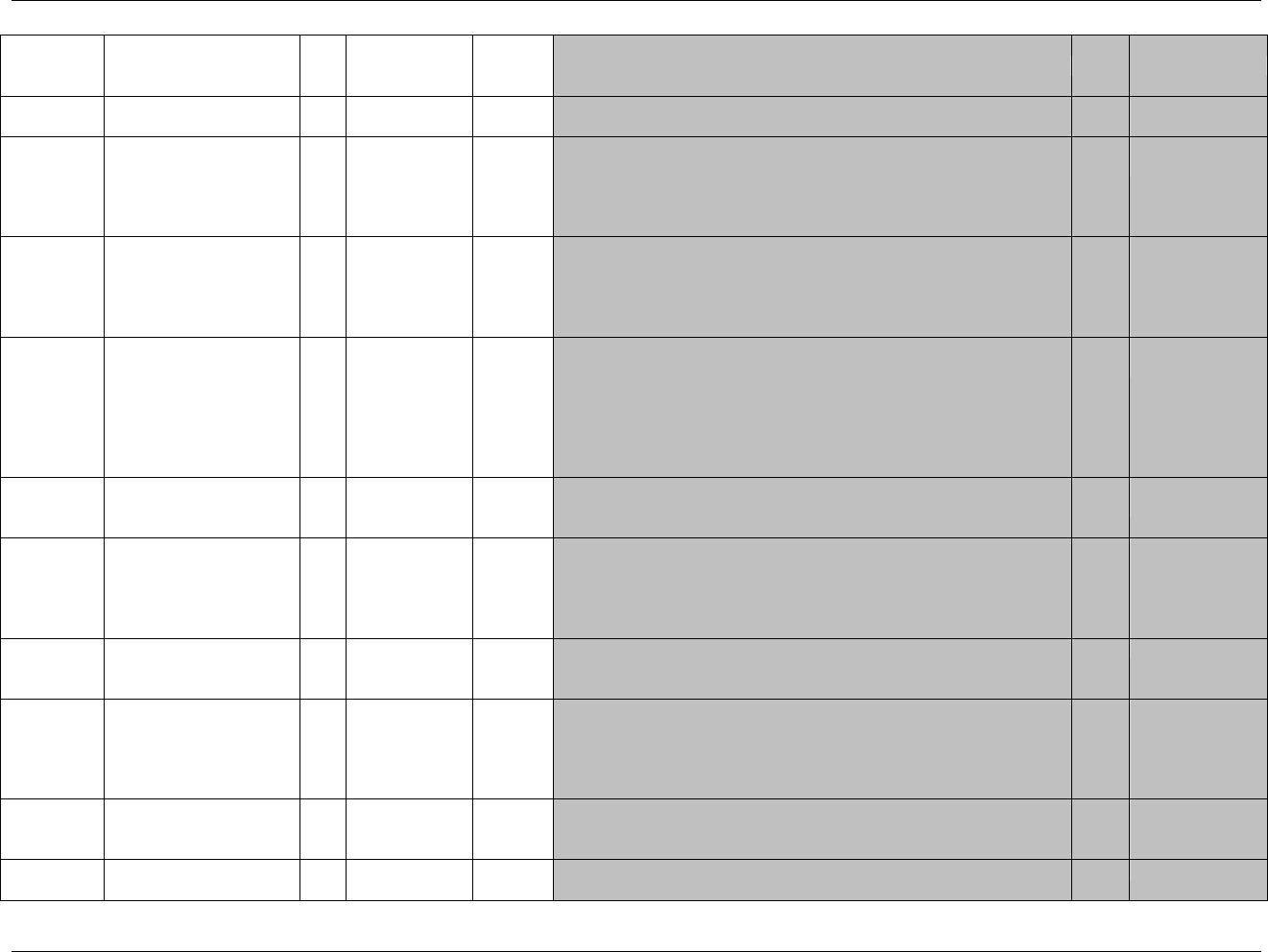
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 162 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MBSCAT
Subcategory for
Microbiology Finding
Char
*
Grouping
Qualifier
Used to define a further categorization of MBCAT.
Perm
SDTM 2.2.3,
1119HSDTMIG 4.1.2.6
MBORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the Microbiology measurement or finding as originally received
or collected. Examples for GRAM STAIN findings: +3 MODERATE, +2
FEW, <10. Examples for CULTURE PLATE (ORGANISM) findings:
KLEBSIELLA PNEUMONIAE, STREPTOCOCCUS PNEUMONIAE
PENICILLIN RESISTANT.
Exp
SDTM 2.2.3,
1120H1121HSDTMIG 4.1.5.1
MBORRESU
Original Units
Char
(1122HUNIT)
Variable
Qualifier
Original unit for MBORRES. Example: mcg/mL
Perm
SDTM 2.2.3,
1123HSDTMIG 4.1.3.2,
1124HSDTMIG 4.1.5.1
SDTMIG
Appendix C1
MBSTRESC
Character Result/Finding in
Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from
MBORRES in a standard format or standard units. MBSTRESC should
store all results or findings in character format; if results are numeric, they
should also be stored in numeric format in MBSTRESN. For example, if a
test has results ―+3 MODERATE‖, ―MOD‖, and ―MODERATE‖ in
MBORRES and these results effectively have the same meaning, they could
be represented in standard format in MBSTRESC as ―MODERATE‖.
Exp
SDTM 2.2.3, 1125H
1126HSDTMIG 4.1.5.1
MBSTRESN
Numeric Result/Finding in
Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from MBSTRESC. MBSTRESN should store
all numeric test results or findings.
Perm
SDTM 2.2.3,
1127HSDTMIG 4.1.5.1
MBSTRESU
Standard Units
Char
(1128HUNIT)
Variable
Qualifier
Standardized unit used for MBSTRESC and MBSTRESN.
Perm
SDTM 2.2.3,
1129HSDTMIG 4.1.3.2,
1130HSDTMIG 4.1.5.1
SDTMIG
Appendix C1
MBRESCAT
Result Category
Char
*
Variable
Qualifier
Used to categorize the result of a finding in a standard format. Example
for ORGANISM finding: INFECTING, COLONIZER,
CONTAMINANT, or NORMAL FLORA.
Exp
SDTM 2.2.3
MBSTAT
Completion Status
Char
(1965HND)
Record
Qualifier
Used to indicate Microbiology was not done, or a test was not done.
Should be null or have a value of NOT DONE.
Perm
SDTM 2.2.3,
1131HSDTMIG 4.1.5.1,
1132HSDTMIG 4.1.5.7,
1133HSDTMIG
Appendix C1
MBREASND
Reason Microbiology Not
Performed
Char
Record
Qualifier
Reason not done. Used in conjunction with MBSTAT when value is NOT
DONE. Examples: BROKEN EQUIPMENT or SUBJECT REFUSED.
Perm
SDTM 2.2.3,
1134HSDTMIG 4.1.5.1,
1135HSDTMIG 4.1.5.7
MBNAM
Vendor Name
Char
Record
Qualifier
Name or identifier of the laboratory or vendor who provides the test
results.
Perm
SDTM 2.2.3
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 163
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MBLOINC
LOINC Code
Char
*
Synonym
Qualifier
1. Dictionary-derived LOINC Code for MBTEST.
2. The sponsor is expected to provide the dictionary name and
version used to map the terms utilizing the define.xml external
codelist attributes
Perm
SDTM 2.2.3,
1136HSDTMIG 4.1.3.2
MBSPEC
Specimen Type
Char
*
Record
Qualifier
Defines the type of specimen used for a measurement. Examples:
SPUTUM, BLOOD, PUS.
Perm
SDTM 2.2.3
MBSPCCND
Specimen Condition
Char
Record
Qualifier
Free or standardized text describing the condition of the specimen.
Example: CONTAMINATED.
Perm
SDTM 2.2.3
MBLOC
Specimen Collection
Location
Char
(1137HLOC)
Record
Qualifier
Location relevant to the collection of the measurement. Examples: LUNG,
VEIN, LEFT KNEE WOUND, ARM ULCER 1, RIGHT THIGH LATERAL
Perm
SDTM 2.2.3,
SDTMIG
Appendix C1
MBMETHOD
Method of Test or
Examination
Char
*
Record
Qualifier
Method of the test or examination. Examples: GRAM STAIN,
CULTURE PLATE, BROTH.
Exp
SDTM 2.2.3
MBBLFL
Baseline Flag
Char
(1966HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or
null.
Perm
SDTM 2.2.3,
1138HSDTMIG 4.1.3.7,
1139HSDTMIG
Appendix C1
MBDRVFL
Derived Flag
Char
(1967HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records
that represent the average of other records or some other derivation, and
those that do not come from the CRF, are examples of records that would
be derived for the submission datasets. If MBDRVFL=Y, then
MBORRES may be null with MBSTRESC and (if numeric) MBSTRESN
having the derived value.
Perm
SDTM 2.2.3,
1140HSDTMIG 4.1.3.7,
1141HSDTMIG 4.1.5.1,
1142HSDTMIG
Appendix C1
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
1143HSDTMIG 4.1.4.5,
1144HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
1145HSDTMIG 4.1.4.5,
1146HSDTMIG 7.4
VISITDY
Planned Study Day of Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
1147HSDTMIG 4.1.4.5,
1148HSDTMIG 7.4
MBDTC
Date/Time of Specimen
Collection
Char
ISO 8601
Timing
Exp
SDTM 2.2.5,
1149HSDTMIG 4.1.4.1,
1150HSDTMIG 4.1.4.8
MBDY
Study Day of MB
Specimen Collection
Num
Timing
1. Study day of the specimen collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5,
1151HSDTMIG 4.1.4.4, 1152H
SDTMIG 4.1.4.6
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 164 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MBTPT
Planned Time Point Name
Char
Timing
1. Text Description of time when specimen should be taken.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See MBTPTNUM and MBTPTREF.
Examples: Start, 5 min post.
Perm
SDTM 2.2.5, 1153H1154H
SDTMIG 4.1.4.10
MBTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of MBTPT to aid in sorting.
Perm
SDTM 2.2.5,
1155HSDTMIG 4.1.4.10
MBELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Planned elapsed time (in ISO 8601) relative to a planned fixed reference
(MBTPTREF). This variable is useful where there are repetitive
measures. Not a clock time or a date time variable. Represented as an ISO
8601 duration. Examples: ―-PT15M‖ to represent the period of 15
minutes prior to the reference point indicated by MBTPTREF, or ―PT8H‖
to represent the period of 8 hours after the reference point indicated by
MBTPTREF.
Perm
SDTM 2.2.5,
1156HSDTMIG 4.1.4.3
1157HSDTMIG 4.1.4.10
MBTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by MBELTM,
MBTPTNUM, and MBTPT. Example: PREVIOUS DOSE.
Perm
SDTM 2.2.5,
1158HSDTMIG 4.1.4.3
1159HSDTMIG 4.1.4.10
MBRFTDTC
Date/Time of Reference
Time Point
Char
ISO 8601
Timing
Date/time of the reference time point, MBTPTREF.
Perm
SDTM 2.2.5,
1160HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.9.2 ASSUMPTIONS FOR MICROBIOLOGY SPECIMEN (MB) DOMAIN MODEL
1. Definition: The MB domain is designed to store microbiology findings that include organisms found, grain stain results and organism growth status.
2. MBSPID is used to uniquely identify an organism. MBSPID would remain the same each time the same organism is identified in a new specimen.
Often the original line number used to record the first occurrence of the organism is used again as an organism identifier when it is found in another
specimen. For example, MBSPID is 01 at visit 10 for organism ―STAPHYLOCOCCUS AUREUS‖. For the same organism at visit 30, MBSPID is
again 01.
3. MBTESTCD value for organisms present in a specimen is "ORGANISM".
4. MBDTC can be used to record the date/time that an organism started to grow in the culture, or the date/time that the culture became positive for the
organism.
5. MBGRPID is used to link to findings related to that organism in the MS domain. For example, if in Specimen 1, organism STREPTOCOCCUS
PNEUMONIAE PENICILLIN RESISTANT is found with MBGRPID=1, then findings such as susceptibility tests, colony count, etc. for that
organism in Specimen 1, would all have the same value of MSGRPID=1 in the MS domain. The use of GRPID to relate MS to MB greatly
simplifies RELREC because only two records are needed in RELREC to describe the relationship of MB to the many related records in MS. With
this method there is no need to create detailed relationships at the subject level.
6. MBRESCAT is expected in all records where a microorganism has been identified to differentiate between colonizing organisms and the one(s) that
are causing the infection. It is not expected when there is ―No growth‖ or when the results are from a gram stain.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 165
Final November 12, 2008
7. The following Qualifiers would not generally be used in MB: --MODIFY, --BODSYS, --FAST, --TOX, --TOXGR --SEV.
6.3.9.3 MICROBIOLOGY SUSCEPTIBILITY (MS) DOMAIN MODEL
ms.xpt, Microbiology Susceptibility Test — Findings, Version 3.1.2. One record per microbiology susceptibility test (or other organism-related finding) per
organism found in MB, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1968HMS
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1161HSDTMIG 4.1.2.2,
1162HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
1163H1164HSDTMIG 4.1.2.3
MSSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
MSGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain to
support relationships within the domain and between domains. In MS,
used to link to organism in MB.
Req
SDTM 2.2.4,
1165HSDTMIG 4.1.2.6
MSREFID
Reference ID
Char
Identifier
Internal or external specimen identifier. Example: Specimen ID.
Perm
SDTM 2.2.4,
1166HSDTMIG 4.1.2.6
MSSPID
Sponsor-Defined Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor's operational database.
Perm
SDTM 2.2.4,
1167HSDTMIG 4.1.2.6
MSTESTCD
Microbiology Organism
Finding Short Name
Char
*
Topic
Short name of the measurement, test, or finding described in MSTEST. It
can be used as a column name when converting a dataset from a vertical
to a horizontal format. The value in MSTESTCD cannot be longer than 8
characters, nor can it start with a number (e.g.‖1TEST). MSTESTCD
cannot contain characters other than letters, numbers, or underscores.
Examples for GROWTH findings: EXTGROW, COLCOUNT. For
SUSCEPTIBILITY findings, the test is the drug the organism was tested
with, i.e. PENICLLN, AMOXCLLN.
Req
SDTM 2.2.3,
1168HSDTMIG 4.1.2.1,
SDTMIG 4.1.1.8
1169H 1170H
MSTEST
Organism Test or Finding
Name
Char
*
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the measurement
or finding. Examples for GROWTH findings: Extent of Growth, Colony
Count. Examples for SUSCEPTIBILITY findings: Amoxicillin
Susceptibility, Penicillin Susceptibility
Req
SDTM 2.2.3,
1171HSDTMIG 4.1.2.1,
1172HSDTMIG 4.1.2.4,
1173HSDTMIG 4.1.5.3.1
MSCAT
Category for Organism
Findings
Char
*
Grouping
Qualifier
Used to define a category of related records. Examples: GROWTH,
SUSCEPTIBILITY.
Req
SDTM 2.2.3,
1174HSDTMIG 4.1.2.6
MSSCAT
Subcategory for Organism
Findings
Char
*
Grouping
Qualifier
A further categorization of a test category. Examples: CULTURE,
ISOLATE
Perm
SDTM 2.2.3,
1175HSDTMIG 4.1.2.6
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 166 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MSORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the Microbiology Organism measurement or finding as
originally received or collected. Examples for GROWTH findings:
GROWTH INTO 3RD QUADRANT. Examples for SUSCEPTIBLITY
findings:.0080,.0023
Exp
SDTM 2.2.3,
1176H1177HSDTMIG 4.1.5.1
MSORRESU
Original Units
Char
(1178HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for MSORRES.
Example: mcg/mL
Exp
SDTM 2.2.3,
1179HSDTMIG 4.1.3.2,
1180HSDTMIG 4.1.5.1
SDTMIG
Appendix C1
MSSTRESC
Character Result/Finding in
Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from
MSORRES in a standard format or standard units. MSSTRESC should
store all results or findings in character format; if results are numeric, they
should also be stored in numeric format in MSSTRESN. For example, if a
test has results ―+3 MODERATE‖, ―MOD‖, and ―MODERATE‖, and in
MSORRES and these results effectively have the same meaning, they
could be represented in standard format in MSSTRESC as
―MODERATE‖.
Exp
SDTM 2.2.3,
1181H1182HSDTMIG 4.1.5.1
MSSTRESN
Numeric Result/Finding in
Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from MSSTRESC. MSSTRESN should store all
numeric test results or findings.
Exp
SDTM 2.2.3,
1183HSDTMIG 4.1.5.1
MSSTRESU
Standard Units
Char
(1184HUNIT)
Variable
Qualifier
Standardized unit used for MSSTRESC and MSSTRESN.
Exp
SDTM 2.2.3,
1185HSDTMIG 4.1.3.2,
1186HSDTMIG 4.1.5.1
SDTMIG
Appendix C1
MSRESCAT
Result Category
Char
*
Variable
Qualifier
Used to categorize the result of a finding in a standard format. Example
for SUSCEPTIBILITY finding: SUSCEPTIBLE, INTERMEDIATE,
RESISTANT, or UNKNOWN.
Exp
SDTM 2.2.3
MSSTAT
Completion Status
Char
(1969HND)
Record
Qualifier
Used to indicate a test on an organism was not done, or a test was not
performed. Should be null if a result exists in MSORRES or have a value
of NOT DONE.
Perm
SDTM 2.2.3,
1187HSDTMIG 4.1.5.1,
1188HSDTMIG 4.1.5.7,
1189HSDTMIG
Appendix C1
MSREASND
Reason Test Not Done
Char
Record
Qualifier
Reason not done. Describes why a measurement or test was not
performed. Used in conjunction with MSSTAT when value is NOT
DONE. Example: SAMPLE LOST
Perm
SDTM 2.2.3,
1190HSDTMIG 4.1.5.1,
1191HSDTMIG 4.1.5.7
MSNAM
Vendor Name
Char
Record
Qualifier
Name or identifier of the laboratory or vendor that provided the test
results.
Perm
SDTM 2.2.3

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 167
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MSLOINC
LOINC Code
Char
*
Synonym
Qualifier
1. Dictionary-derived LOINC Code for MSTEST.
2. The sponsor is expected to provide the dictionary name and
version used to map the terms utilizing the define.xml external
codelist attributes
Perm
SDTM 2.2.3,
1192HSDTMIG 4.1.3.2
MSMETHOD
Method of Test or
Examination
Char
*
Record
Qualifier
Method of the test or examination. Example for SUSCEPTIBILITY:
ETEST, BROTH DILUTION.
Exp
SDTM 2.2.3
MSBLFL
Baseline Flag
Char
(1970HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or
null.
Perm
SDTM 2.2.3,
1193HSDTMIG 4.1.3.7,
1194HSDTMIG
Appendix C1
MSDRVFL
Derived Flag
Char
(1971HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records
that represent the average of other records or some other derivation, and
those that do not come from the CRF, are examples of records that would
be derived for the submission datasets. If MSDRVFL=Y, then MSORRES
may be null, with MSSTRESC and (if numeric) MSSTRESN having the
derived value.
Perm
SDTM 2.2.3,
1195HSDTMIG 4.1.3.7,
1196HSDTMIG 4.1.5.1,
1197HSDTMIG
Appendix C1
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
1198HSDTMIG 4.1.4.5,
1199HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. 1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
1200HSDTMIG 4.1.4.5,
1201HSDTMIG 7.4
VISITDY
Planned Study Day of Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
1202HSDTMIG 4.1.4.5,
1203HSDTMIG 7.4
MSDTC
Date/Time of Test
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
1204HSDTMIG 4.1.4.1,
1205HSDTMIG 4.1.4.8
MSDY
Study Day of Test
Num
Timing
1. Study day of the test, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5,
1206HSDTMIG 4.1.4.4,
1207HSDTMIG 4.1.4.6
MSTPT
Planned Time Point Name
Char
Timing
1. Text Description of time when test should be done.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See MSTPTNUM and MSTPTREF.
Examples: Start, 5 min post.
Perm
SDTM 2.2.5,
1208SDTMIG 4.1.4.10
MSTPTNUM
Planned Time Point
Number
Num
Timing
Numerical version of MSTPT to aid in sorting.
Perm
SDTM 2.2.5,
1209HSDTMIG 4.1.4.10
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 168 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
MSELTM
Planned Elapsed Time
from Time Point Ref
Char
ISO 8601
Timing
Elapsed time (in ISO 8601) relative to a planned fixed reference
(MSTPTREF). This variable is useful where there are repetitive measures.
Not a clock time or a date time variable. Examples: ―-PT15M‖ to
represent the period of 15 minutes prior to the reference point indicated by
MSTPTREF, or ―P8H‖ to represent the period of 8 hours after the
reference point indicated by MSTPTREF.
Perm
SDTM 2.2.5,
1210HSDTMIG 4.1.4.3,
1211HSDTMIG 4.1.4.10
MSTPTREF
Time Point Reference
Char
Timing
Name of the fixed reference point referred to by MSELTM,
MSTPTNUM, and MSTPT. Example: PREVIOUS DOSE.
Perm
SDTM 2.2.5,
SDTMIG 4.1.4.3
1212HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.9.4 ASSUMPTIONS FOR MICROBIOLOGY SUSCEPTIBILITY (MS) DOMAIN MODEL
1. Definition: The MS domain is designed to store any findings related to the organisms found and submitted in MB. This will usually consist of
susceptibility testing results, but can also be other organism-related findings such as extent of growth of an organism. This domain is intended to be
used in conjunction with the MB domain described above.
2. The following Qualifiers would not generally be used in MB: --MODIFY, --BODSYS, --SPEC, --SPCCND, --FAST, --TOX, --TOXGR --SEV.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 169
Final November 12, 2008
6.3.9.5 EXAMPLES FOR MB AND MS DOMAIN MODELS
Example 1: MB specimen findings
Rows 1, 2: Show gram stain results for Specimen 1 (MBREFID=SP01).
Rows 3, 4: Show organisms found in specimen 1 at visit 1. The MBGRPID is used to link these organisms to findings about these organisms in MS,
by MSGRPID.
Row 5: Shows the organism assigned as ORG02 is still present in Specimen 2 at Visit 2.
Row 6: Shows no organisms have grown at Visit 3. Therefore the organism recorded is "NO GROWTH".
Row 1-6: Show MBMETHOD being used for reporting the method of testing the sample, e.g. GRAM STAIN or CULTURE PLATE.
Microbiology Example 1 MB dataset
Row
STUDYID
DOMAIN
USUBJID
MBSEQ
MBGRPID
MBREFID
MBSPID
MBTESTCD
MBTEST
MBORRES
1
ABC
MB
ABC-001-001
1
SP01
GMNCOC
Gram Negative Cocci
2+ FEW
2
ABC
MB
ABC-001-001
2
SP01
GMNROD
Gram Negative Rods
2+ FEW
3
ABC
MB
ABC-001-001
3
1
SP01
ORG01
ORGANISM
Organism Present
STREPTOCOCCUS PNEUMONIAE
PENICILLIN RESISTANT
4
ABC
MB
ABC-001-001
4
2
SP01
ORG02
ORGANISM
Organism Present
KLEBSIELLA PNEUMONIAE
5
ABC
MB
ABC-001-001
5
3
SP02
ORG02
ORGANISM
Organism Present
KLEBSIELLA PNEUMONIAE
6
ABC
MB
ABC-001-001
6
SP03
ORG03
ORGANISM
Organism Present
NO GROWTH
Row
MBSTRESC
MBRESCAT
MBLOC
MBSPEC
MBSPCCND
MBMETHOD
VISITNUM
MBDTC
1 (cont)
FEW
LUNG
SPUTUM
MUCOID
GRAM STAIN
1
2005-06-19T08:00
2 (cont)
FEW
LUNG
SPUTUM
MUCOID
GRAM STAIN
1
2005-06-19T08:00
3 (cont)
STREPTOCOCCUS PNEUMONIAE,
PENICILLIN RESISTANT
INFECTING
LUNG
SPUTUM
MUCOID
CULTURE PLATE
1
2005-06-19T08:00
4 (cont)
KLEBSIELLA PNEUMONIAE
COLONIZER
LUNG
SPUTUM
MUCOID
CULTURE PLATE
1
2005-06-19T08:00
5 (cont)
KLEBSIELLA PNEUMONIAE
COLONIZER
LUNG
SPUTUM
CULTURE PLATE
2
2005-06-26T08:00
6 (cont)
NO GROWTH
LUNG
SPUTUM
CULTURE PLATE
3
2005-07-06T08:00
If the method of the collection of the sputum is reported (e.g., EXPECTORATION or BIOPSY), this information would go into SUPPMB, since MBMETHOD
refers to the method used to obtain the results.
suppmb.xpt
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
ABC
MB
ABC-001-001
MBSEQ
1
COLMETH
Collection Method
EXPECTORATION
CRF
Example 2: MS – Findings about organisms from Example 1, related within USUBJID, by MBGRPID=MSGRPID
Row 1: Shows extent of growth of Organism 1 found at Visit 1 in specimen 1 (MBGRPID=1, Row 3 in MB example above).
Rows 2, 3: Show results of susceptibility testing on Organism 1 found at Visit 1 in specimen 1 (MBGRPID=1, Row 3 in MB example above).
Row 4: Shows extent of growth of Organism 2 found at Visit 1 in specimen 1 (MBGRPID=2, Row 4 in MB example above).
Rows 5, 6: Show results of susceptibility testing on Organism 2 found at Visit 1 in specimen 1 (MBGRPID=2, Row 4 in MB example above).
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 170 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Row 7: Shows results of susceptibility testing on Organism 2 found at Visit 1 in specimen 2 (MBGRPID=3, Row 5 in MB example above).
Row
STUDYID
DOMAIN
USUBJID
MSSEQ
MSGRPID
MSTESTCD
MSTEST
MSCAT
1
ABC
MS
ABC-001-001
1
1
EXTGROW
Extent of Growth
GROWTH
2
ABC
MS
ABC-001-001
2
1
DRUGA
Sponsor Drug
SUSCEPTIBILITY
3
ABC
MS
ABC-001-001
3
1
PENICLLN
Penicillin
SUSCEPTIBILITY
4
ABC
MS
ABC-001-001
4
2
EXTGROW
Extent of Growth
GROWTH
5
ABC
MS
ABC-001-001
5
2
DRUGA
Sponsor Drug
SUSCEPTIBILITY
6
ABC
MS
ABC-001-001
6
2
PENICLLN
Penicillin
SUSCEPTIBILITY
7
ABC
MS
ABC-001-001
7
3
PENICLLN
Penicillin
SUSCEPTIBILITY
Row
MSORRES
MSORRESU
MSSTRESC
MSSTRESN
MSSTRESU
MSRESCAT
MSMETHOD
VISITNUM
1 (cont)
IN 2ND QUADRANT
IN 2ND
QUADRANT
1
2 (cont)
0.004
mcg/mL
0.004
0.004
mcg/mL
SUSCEPTIBLE
E-TEST
1
3 (cont)
0.023
mcg/mL
0.023
0.023
mcg/mL
RESISTANT
E-TEST
1
4 (cont)
>=30 COLONIES IN 2ND
QUADRANT
>=30 COLONIES IN
2ND QUADRANT
1
5 (cont)
0.125
mcg/mL
0.125
0.125
mcg/mL
SUSCEPTIBLE
E-TEST
1
6 (cont)
0.023
mcg/mL
0.023
0.023
mcg/mL
INTERMEDIATE
E-TEST
1
7 (cont’d)
0.026
mcg/mL
0.026
0.026
mcg/mL
INTERMEDIATE
E-TEST
2
Example 3: MB with multiple labs
Row 1, 2: Show the same organism identified by a central and a local lab. Note that MBSPID is different for each lab, and also MBGRPID is different for
each lab. This is because the organism is found and tracked separately for each lab although it came from the same specimen.
Row
STUDYID
DOMAIN
USUBJID
MBSEQ
MBGRPID
MBREFID
MBSPID
MBTESTCD
MBTEST
1
ABC
MB
ABC-001-002
1
1
SPEC01
ORG01
ORGANISM
Organism Present
2
ABC
MB
ABC-001-002
2
2
SPEC01
ORG02
ORGANISM
Organism Present
Row
MBORRES
MBSTRESC
MBRESCAT
MBNAM
MBLOC
MBSPEC
MBMETHOD
VISITNUM
MBDTC
1 (cont)
ENTEROCOCCUS
FAECALIS
ENTEROCOCCUS
FAECALIS
INFECTING
CENTRAL
SKIN SITE 1
FLUID
CULTURE PLATE
1
2005-07-21T08:00
2 (cont)
ENTEROCOCCUS
FAECALIS
ENTEROCOCCUS
FAECALIS
INFECTING
LOCAL
SKIN SITE 1
FLUID
CULTURE PLATE
1
2005-07-21T08:00
Example 4: MS – findings about organisms from Example 3, multiple labs
Rows 1, 2: Show susceptibility test results done by the central lab, for the organism identified by the central lab where MBGRPID=1 in Row 1 of Example 3 above.
Note that the central lab performed only one method of susceptibility testing (the E-TEST) for the two drugs, Sponsor and Amoxicillin.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 171
Final November 12, 2008
Rows 3-8: Show susceptibility test results done by the local lab, for the organism identified by the local lab where MBGRPID=2 in Row 2 of Example 3 above.
Note that the local lab has performed three different methods of susceptibility testing (Broth Dilution, Zone Size, and E-TEST) for two drugs, thus
providing six records for MSGRPID=2.
Row
STUDYID
DOMAIN
USUBJID
MSSEQ
MSGRPID
MSREFID
MSTESTCD
MSTEST
MSCAT
1
ABC
MS
ABC-001-002
1
1
CENTABC
DRUGA
Sponsor Drug
SUSCEPTIBILITY
2
ABC
MS
ABC-001-002
2
1
CENTABC
AMOXCLAV
Amoxicillin / Clavulanate
SUSCEPTIBILITY
3
ABC
MS
ABC-001-002
3
2
LOCXYZ
DRUGA
Sponsor Drug
SUSCEPTIBILITY
4
ABC
MS
ABC-001-002
4
2
LOCXYZ
AMOXCLAV
Amoxicillin / Clavulanate
SUSCEPTIBILITY
5
ABC
MS
ABC-001-002
5
2
LOCXYZ
DRUGA
Sponsor Drug
SUSCEPTIBILITY
6
ABC
MS
ABC-001-002
6
2
LOCXYZ
AMOXCLAV
Amoxicillin / Clavulanate
SUSCEPTIBILITY
7
ABC
MS
ABC-001-002
7
2
LOCXYZ
DRUGA
Sponsor Drug
SUSCEPTIBILITY
8
ABC
MS
ABC-001-002
8
2
LOCXYZ
AMOXCLAV
Amoxicillin / Clavulanate
SUSCEPTIBILITY
Row
MSORRES
MSORRESU
MSSTRESC
MSSTRESN
MSSTRESU
MSRESCAT
MSMETHOD
VISITNUM
1 (cont)
0.25
mcg/mL
0.25
0.25
mcg/mL
SUSCEPTIBLE
E-TEST
1
2 (cont)
1
mcg/mL
1
1
mcg/mL
RESISTANT
E-TEST
1
3 (cont)
0.5
mcg/mL
0.5
0.5
mcg/mL
SUSCEPTIBLE
BROTH DILUTION
1
4 (cont)
0.5
mcg/mL
0.5
0.5
mcg/mL
RESISTANT
BROTH DILUTION
1
5 (cont)
23
mm
23
23
mm
SUSCEPTIBLE
ZONE SIZE
1
6 (cont)
25
mm
25
25
mm
RESISTANT
ZONE SIZE
1
7 (cont)
0.25
mcg/mL
0.25
0.25
mcg/mL
SUSCEPTIBLE
E-TEST
1
8 (cont)
1
mcg/mL
1
1
mcg/mL
RESISTANT
E-TEST
1
Example 5: RELREC to relate MB and MS
Rows 1, 2: Show the one-to-many relationship between MB and MS. For any organism found in a microbiology specimen and recorded in MB, there may be
multiple findings about that organism recorded in MS. The organism in MB can be linked to its findings in MS because the value assigned to
MBGRPID = the value assigned to MSGRPID for any organism within a subject.
Row
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
1
MB
MBGRPID
ONE
A
2
MS
MSGRPID
MANY
A
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 172 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.3.10 PHARMACOKINETICS DOMAINS — PC AND PP
pc.xpt, Pharmacokinetic Concentrations — Findings, Version 3.1.2. One record per sample characteristic or time-point concentration per reference time
point or per analyte per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1972HPC
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1213HSDTMIG 4.1.2.2,
1214HSDTMIG
Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Unique subject identifier within the submission.
Req
SDTM 2.2.4,
1215H1216HSDTMIG 4.1.2.3
PCSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
PCGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain to support
relationships within the domain and between domains.
Perm
SDTM 2.2.4,
1217HSDTMIG 4.1.2.6
PCREFID
Reference ID
Char
Identifier
Internal or external specimen identifier. Example: Specimen ID.
Perm
SDTM 2.2.4,
1218HSDTMIG 4.1.2.6
PCSPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined reference number.
Perm
SDTM 2.2.4,
1219HSDTMIG 4.1.2.6
PCTESTCD
Pharmacokinetic Test
Short Name
Char
Topic
Short name of the analyte or specimen characteristic. It can be used as a
column name when converting a dataset from a vertical to a horizontal format.
The value in PCTESTCD cannot be longer than 8 characters, nor can it start
with a number (e.g., ―1TEST‖). PCTESTCD cannot contain characters other
than letters, numbers, or underscores. Examples: ASA, VOL, SPG.
Req
SDTM 2.2.3,
1220H1221HSDTMIG 4.1.1.8,
1222HSDTMIG 4.1.2.1
PCTEST
Pharmacokinetic Test
Name
Char
Synonym
Qualifier
Name of the analyte or specimen characteristic. Note any test normally
performed by a clinical laboratory is considered a lab test. The value in
PCTEST cannot be longer than 40 characters. Examples: Acetylsalicylic Acid,
Volume, Specific Gravity.
Req
SDTM 2.2.3,
1223HSDTMIG 4.1.2.1,
1224HSDTMIG 4.1.2.4,
1225HSDTMIG 4.1.5.3.1
PCCAT
Test Category
Char
*
Grouping
Qualifier
Used to define a category of related records. Examples: ANALYTE,
SPECIMEN PROPERTY.
Perm
SDTM 2.2.3,
1226HSDTMIG 4.1.2.6
PCSCAT
Test Subcategory
Char
*
Grouping
Qualifier
A further categorization of a test category.
Perm
SDTM 2.2.3,
1227HSDTMIG 4.1.2.6
PCORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the measurement or finding as originally received or collected.
Exp
SDTM 2.2.3,
1228H1229HSDTMIG 4.1.5.1
PCORRESU
Original Units
Char
(1230HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for PCORRES.
Example: mg/L.
Exp
SDTM 2.2.3,
1231HSDTMIG 4.1.3.2,
1232HSDTMIG 4.1.5.1,
SDTMIG
Appendix C1

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 173
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
Reference
PCSTRESC
Character Result/Finding
in Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from PCORRES in
a standard format or standard units. PCSTRESC should store all results or
findings in character format; if results are numeric, they should also be stored
in numeric format in PCSTRESN. For example, if a test has results ―NONE‖,
―NEG‖, and ―NEGATIVE‖ in PCORRES and these results effectively have the
same meaning, they could be represented in standard format in PCSTRESC as
―NEGATIVE‖. For other examples, see general assumptions.
Exp
SDTM 2.2.3, 1233H
1234HSDTMIG 4.1.5.1
PCSTRESN
Numeric Result/Finding
in Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format; copied
in numeric format from PCSTRESC. PCSTRESN should store all numeric test
results or findings.
Exp
SDTM 2.2.3,
1235HSDTMIG 4.1.5.1,
SDTMIG
Appendix C1
PCSTRESU
Standard Units
Char
(1236HUNIT)
Variable
Qualifier
Standardized unit used for PCSTRESC and PCSTRESN.
Exp
SDTM 2.2.3,
1237HSDTMIG 4.1.3.2,
1238HSDTMIG 4.1.5.1
PCSTAT
Completion Status
Char
(1973HND)
Record
Qualifier
Used to indicate a result was not obtained. Should be null if a result exists in
PCORRES.
Perm
SDTM 2.2.3,
1239HSDTMIG 4.1.5.1,
1240HSDTMIG 4.1.5.7,
1241HSDTMIG
Appendix C1
PCREASND
Reason Test Not Done
Char
Record
Qualifier
Describes why a result was not obtained such as SPECIMEN LOST. Used in
conjunction with PCSTAT when value is NOT DONE.
Perm
SDTM 2.2.3,
1242HSDTMIG 4.1.5.1,
1243HSDTMIG 4.1.5.7
PCNAM
Vendor Name
Char
Record
Qualifier
Name or identifier of the laboratory or vendor who provides the test results.
Exp
SDTM 2.2.3
PCSPEC
Specimen Material Type
Char
Record
Qualifier
Defines the type of specimen used for a measurement. Examples: SERUM,
PLASMA, URINE.
Req
SDTM 2.2.3
PCSPCCND
Specimen Condition
Char
Record
Qualifier
Free or standardized text describing the condition of the specimen e.g.
HEMOLYZED, ICTERIC, LIPEMIC etc.
Perm
SDTM 2.2.3
PCMETHOD
Method of Test or
Examination
Char
*
Record
Qualifier
Method of the test or examination. Examples include HPLC/MS, ELISA. This
should contain sufficient information and granularity to allow differentiation of
various methods that might have been used within a study.
Perm
SDTM 2.2.3
PCFAST
Fasting Status
Char
(1974HNY)
Record
Qualifier
Indicator used to identify fasting status.
Perm
SDTM 2.2.3,
1244HSDTMIG
Appendix C1
PCDRVFL
Derived Flag
Char
(1975HNY)
Record
Qualifier
Used to indicate a derived record. The value should be Y or null. Records that
represent the average of other records, which do not come from the CRF, are
examples of records that would be derived for the submission datasets. If
PCDRVFL=Y, then PCORRES may be null with PCSTRESC, and (if numeric)
PCSTRESN having the derived value.
Perm
SDTM 2.2.3,
1245HSDTMIG 4.1.3.7,
1246HSDTMIG 4.1.5.1,
1247HSDTMIG
Appendix C1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 174 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name Variable Label Type Controlled
Terms, Codelist
or Format Role CDISC Notes Core Reference
PCLLOQ Lower Limit of
Quantitation Num Variable
Qualifier Indicates the lower limit of quantitation for an assay. Units should be those
used in PCSTRESU. Exp SDTM 2.2.3
VISITNUM Visit Number Num Timing 1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting. Exp SDTM 2.2.5,
1248HSDTMIG 4.1.4.5,
1249HSDTMIG 7.4
VISIT Visit Name Char Timing 1. Protocol-defined description of clinical encounter
2. May be used in addition to VISITNUM and/or VISITDY Perm SDTM 2.2.5,
1250HSDTMIG 4.1.4.5,
1251HSDTMIG 7.4
VISITDY Planned Study Day of
Visit Num Timing Planned study day of the visit based upon RFSTDTC in Demographics. Perm SDTM 2.2.5,
1252HSDTMIG 4.1.4.5,
1253HSDTMIG 7.4
PCDTC Date/Time of Specimen
Collection Char ISO 8601 Timing Date/time of specimen collection represented in ISO 8601 character format. If
there is no end time, then this will be the collection time. Exp SDTM 2.2.5,
1254HSDTMIG 4.1.4.1,
1255HSDTMIG 4.1.4.8
PCENDTC End Date/Time of
Specimen Collection Char ISO 8601 Timing End date/time of specimen collection represented in ISO 8601 character
format. If there is no end time, the collection time should be stored in PCDTC,
and PCENDTC should be null.
Perm SDTM 2.2.5,
1256HSDTMIG 4.1.4.1,
1257HSDTMIG 4.1.4.8
PCDY Actual Study Day of
Specimen Collection Num Timing 1. Study day of specimen collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics.
Perm SDTM 2.2.5,
1258HSDTMIG 4.1.4.4,
1259HSDTMIG 4.1.4.6
PCTPT Planned Time Point
Name Char Timing 1. Text Description of time when specimen should be taken.
2. This may be represented as an elapsed time relative to a fixed reference
point, such as time of last dose. See PCTPTNUM and PCTPTREF. Examples:
Start, 5 min post.
Perm SDTM 2.2.5,
1260HSDTMIG 4.1.4.10
PCTPTNUM Planned Time Point
Number Num Timing Numerical version of PCTPT to aid in sorting. Perm SDTM 2.2.5,
1261HSDTMIG 4.1.4.10
PCELTM
Planned Elapsed Time
from Time Point Ref
Char ISO 8601 Timing Planned elapsed time (in ISO 8601) relative to a planned fixed reference
(PCTPTREF) such as “PREVIOUS DOSE” or “PREVIOUS MEAL”. This
variable is useful where there are repetitive measures. Not a clock time or a date
time variable.
Perm SDTM 2.2.5,
1262HSDTMIG 4.1.4.3,
1263HSDTMIG 4.1.4.10
PCTPTREF Time Point Reference Char Timing Name of the fixed reference point used as a basis for PCTPT, PCTPTNUM,
and PCELTM. Example: Most Recent Dose. Perm SDTM 2.2.5,
1264HSDTMIG 4.1.4.10
PCRFTDTC Date/Time of Reference
Point Char ISO 8601 Timing Date/time of the reference time point described by PCTPTREF. Perm SDTM 2.2.5,
1265HSDTMIG 4.1.4.10
PCEVLINT Evaluation Interval Char ISO 8601 Timing Evaluation Interval associated with a PCTEST record represented in ISO 8601
character format. Example: "-P2H" to represent an interval of 2 hours prior to a
PCTPT.
Perm SDTM 2.2.5
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 175
Final November 12, 2008
pp.xpt, Pharmacokinetic Parameters — Findings, Version 3.1.2,. One record per PK parameter per time-concentration profile per subject, Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1976HPP
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1266HSDTMIG 4.1.2.2,
1267HSDTMIG Appendix C2
USUBJID
Unique Subject Identifier
Char
Identifier
Unique subject identifier within the submission.
Req
SDTM 2.2.4,
1268H1269HSDTMIG 4.1.2.3
PPSEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
PPGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain to
support relationships within the domain and between domains.
Perm
SDTM 2.2.4,
1270HSDTMIG 4.1.2.6
PPTESTCD
Parameter Short Name
Char
Topic
Short name of the pharmacokinetic parameter. It can be used as a column
name when converting a dataset from a vertical to a horizontal format. The value
in PPTESTCD cannot be longer than 8 characters, nor can it start with a number
(e.g., ―1TEST‖). PPTESTCD cannot contain characters other than letters,
numbers, or underscores. Examples: AUC, TMAX, CMAX.
Req
SDTM 2.2.3,
1271H1272HSDTMIG 4.1.1.8
1273HSDTMIG 4.1.2.1
PPTEST
Parameter Name
Char
Synonym
Qualifier
Name of the pharmacokinetic parameter. The value in PPTEST cannot be
longer than 40 characters. Examples: AUC, Tmax, Cmax.
Req
SDTM 2.2.3,
1274HSDTMIG 4.1.2.1,
1275HSDTMIG 4.1.2.4,
1276HSDTMIG 4.1.5.3.1
PPCAT
Parameter Category
Char
*
Grouping
Qualifier
Used to define a category of related records. For PP, this should be the name
of the analyte in PPTEST whose profile the parameter is associated with.
Exp
SDTM 2.2.3,
1277HSDTMIG 4.1.2.6
PPSCAT
Parameter Subcategory
Char
*
Grouping
Qualifier
Categorization of the model type used to calculate the PK parameters.
Examples include COMPARTMENTAL, NON-COMPARTMENTAL.
Perm
SDTM 2.2.3,
1278HSDTMIG 4.1.2.6
PPORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the measurement or finding as originally received or collected.
Exp
SDTM 2.2.3,
1279H 1280HSDTMIG 4.1.5.1
PPORRESU
Original Units
Char
(1281HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for PPORRES.
Example: ng/L.
Exp
SDTM 2.2.3,
1282HSDTMIG 4.1.3.2,
1283HSDTMIG 4.1.5.1,
SDTMIG Appendix C1
PPSTRESC
Character Result/Finding
in Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from PPORRES in
a standard format or standard units. PPSTRESC should store all results or
findings in character format; if results are numeric, they should also be stored
in numeric format in PPSTRESN.
Exp
SDTM 2.2.3,
1284H 1285HSDTMIG 4.1.5.1
PPSTRESN
Numeric Result/Finding
in Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from PPSTRESC. PPSTRESN should store all
numeric test results or findings.
Exp
SDTM 2.2.3,
1286HSDTMIG 4.1.5.1
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 176 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
PPSTRESU
Standard Units
Char
(1287HUNIT)
Variable
Qualifier
Standardized unit used for PPSTRESC and PPSTRESN.
Exp
SDTM 2.2.3,
1288HSDTMIG 4.1.3.2,
1289HSDTMIG 4.1.5.1,
SDTMIG Appendix C1
PPSTAT
Completion Status
Char
(1977HND)
Record
Qualifier
Used to indicate that a parameter was not calculated. Should be null if a
result exists in PPORRES.
Perm
SDTM 2.2.3,
1290HSDTMIG 4.1.5.1,
1291HSDTMIG 4.1.5.7,
1292HSDTMIG Appendix C1
PPREASND
Reason Parameter Not
Calculated
Char
Record
Qualifier
Describes why a parameter was not calculated, such as INSUFFICIENT
DATA. Used in conjunction with PPSTAT when value is NOT DONE.
Perm
SDTM 2.2.3,
1293HSDTMIG 4.1.5.1,
1294HSDTMIG 4.1.5.7
PPSPEC
Specimen Material Type
Char
*
Record
Qualifier
Defines the type of specimen used for a measurement. If multiple specimen
types are used for a calculation (e.g., serum and urine for renal clearance), then
this field should be left blank. Examples: SERUM, PLASMA, URINE.
Exp
SDTM 2.2.3
PPDTC
Date/Time of Parameter
Calculations
Char
ISO 8601
Timing
Nominal date/time of parameter calculations.
Perm
SDTM 2.2.5,
1295HSDTMIG 4.1.4.1,
1296HSDTMIG 4.1.4.8
PPRFTDTC
Date/Time of Reference
Point
Char
ISO 8601
Timing
Date/time of the reference time point from the PC records used to
calculate a parameter record. The values in PPRFTDTC should be the
same as that in PCRFTDTC for related records.
Exp
SDTM 2.2.5,
1297HSDTMIG 4.1.4.10
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.3.10.1 ASSUMPTIONS FOR PHARMACOKINETIC CONCENTRATIONS (PC) DOMAIN MODEL
1. PC Definition: Data collected about tissue (e.g., serum or plasma) concentrations of analytes (usually study drugs and/or their metabolites) as a function
of time after dosing the study drug.
2. The structure is one record per concentration or sample characteristic per analyte. In addition to one record for each concentration measurement,
specimen properties (e.g., volume and pH) are handled via separate records in this dataset.
3. Due to space limitations, not all expected or permissible Findings variables are included in the example.
4. The following Qualifiers would not generally be used in PC: --BODSYS, --SEV.
6.3.10.2 EXAMPLES FOR PHARMACOKINETIC CONCENTRATIONS (PC) DOMAIN MODEL
Example 1
This example shows concentration data for Drug A and metabolite of Drug A from plasma and from urine (shaded rows) samples collected pre-dose and after
dosing on two different study days, Days 1 and 11.
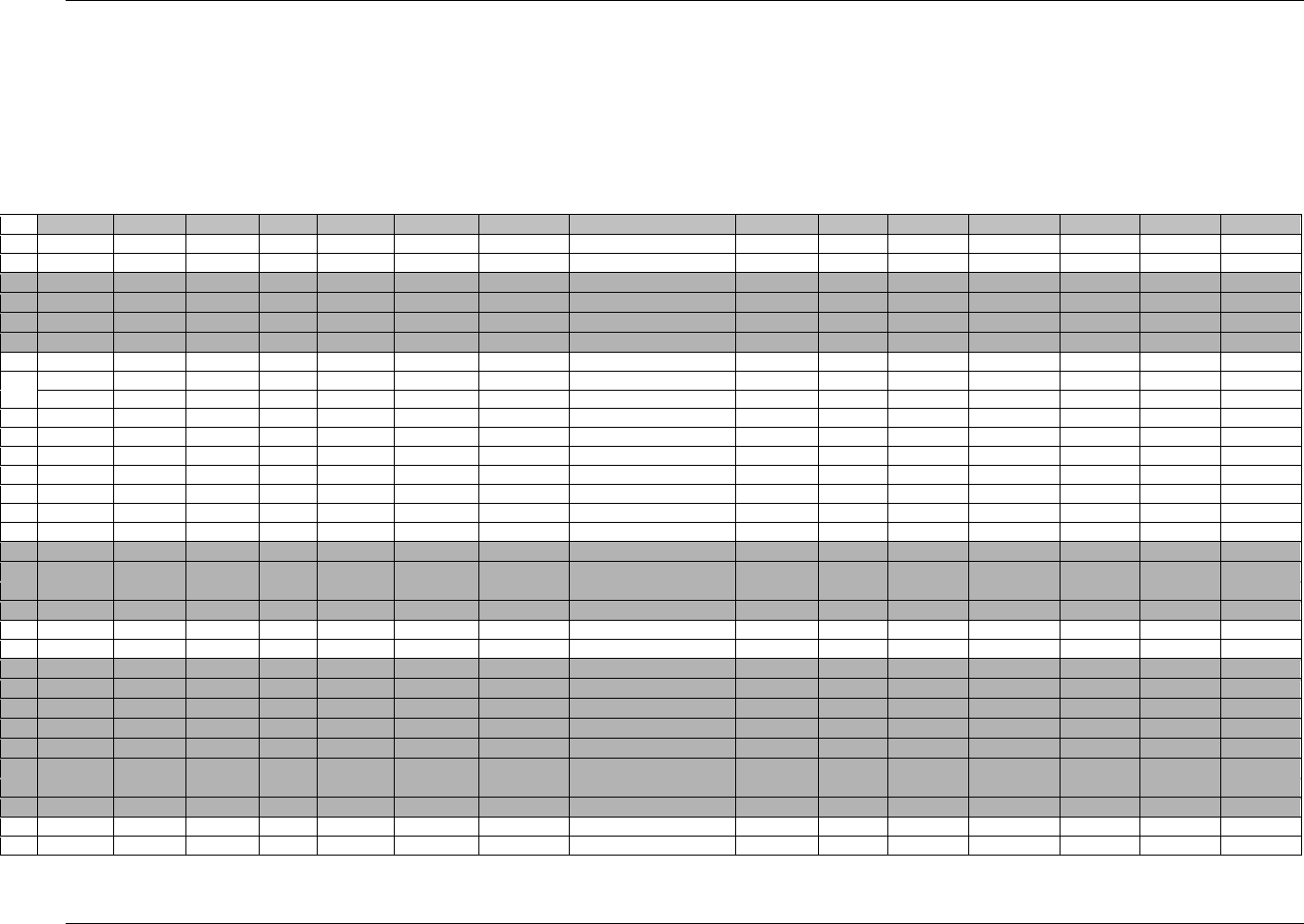
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 177
Final November 12, 2008
All Rows: PCTPTREF is a text value of the description of a “zero” time (e.g. time of dosing). It should be meaningful. If there are
multiple PK profiles being generated, the zero time for each will be different (e.g., a different dose such as "first dose",
"second dose") and, as a result, values for PCTPTREF must be different. In this example it is required to make values of
PCTPTNUM and PCTPT unique (See 1298HSection 4.1.4.10).
Rows 5, 6, 19, 20, 25, 26, 29, and 30: Specimen properties (VOLUME and PH) are submitted as values of PCTESTCD in separate rows.
Rows 3-6, 17-20, 23-30: The elapsed times for urine samples are based upon the elapsed time (from the reference time point, PCTPTREF) for the end of the specimen
collection period. Elapsed time values that are the same for urine and plasma samples have been assigned the same value for PCTPT. For the
urine samples, the value in PCEVLINT describes the planned evaluation (or collection) interval relative to the time point. The
actual evaluation interval can be determined by subtracting PCDTC from PCENDTC.
Row STUDYID DOMAIN USUBJID PCSEQ PCGRPID PCREFID PCTESTCD PCTEST PCCAT PCSPEC PCORRES PCORRESU PCSTRESCPCSTRESNPCSTRESU
1 ABC-123 PC 123-0001 1 Day 1 A554134-10 DRGA_MET Drug A Metabolite ANALYTE PLASMA <0.1 ng/mL <0.1 ng/mL
2 ABC-123 PC 123-0001 2 Day 1 A554134-10 DRGA_PAR Drug A Parent ANALYTE PLASMA <0.1 ng/mL <0.1 ng/mL
3 ABC-123 PC 123-0001 3 Day 1 A554134-11 DRGA_MET Drug A Metabolite ANALYTE URINE <2 ng/mL <2 ng/mL
4 ABC-123 PC 123-0001 4 Day 1 A554134-11 DRGA_PAR Drug A Parent ANALYTE URINE <2 ng/mL <2 ng/mL
5 ABC-123 PC 123-0001 5 Day 1 A554134-11 VOLUME Volume SPECIMEN URINE 3500 mL 100 100 mL
6 ABC-123 PC 123-0001 6 Day 1 A554134-11 PH PH SPECIMEN URINE 5.5 5.5 5.5
7 ABC-123 PC 123-0001 7 Day 1 A554134-12 DRGA_MET Drug A Metabolite ANALYTE PLASMA 5.4 ng/mL 5.4 5.4 ng/mL
8 ABC-123 PC 123-0001 8 Day 1 A554134-12 DRGA_PAR Drug A Parent ANALYTE PLASMA 4.74 ng/mL 4.74 4.74 ng/mL
9 ABC-123 PC 123-0001 9 Day 1 A554134-13 DRGA_MET Drug A Metabolite ANALYTE PLASMA 5.44 ng/mL 5.44 5.44 ng/mL
10 ABC-123 PC 123-0001 10 Day 1 A554134-13 DRGA_PAR Drug A Parent ANALYTE PLASMA 1.09 ng/mL 1.09 1.09 ng/mL
11 ABC-123 PC 123-0001 11 Day 1 A554134-14 DRGA_MET Drug A Metabolite ANALYTE PLASMA
12 ABC-123 PC 123-0001 12 Day 1 A554134-14 DRGA_PAR Drug A Parent ANALYTE PLASMA <0.1 ng/mL <0.1 ng/mL
13 ABC-123 PC 123-0001 13 Day 11 A554134-15 DRGA_MET Drug A Metabolite ANALYTE PLASMA 3.41 ng/mL 3.41 3.41 ng/mL
14 ABC-123 PC 123-0001 14 Day 11 A554134-15 DRGA_PAR Drug A Parent ANALYTE PLASMA <0.1 ng/mL <0.1 ng/mL
15 ABC-123 PC 123-0001 15 Day 11 A554134-16 DRGA_MET Drug A Metabolite ANALYTE PLASMA 8.74 ng/mL 8.74 8.74 ng/mL
16 ABC-123 PC 123-0001 16 Day 11 A554134-16 DRGA_PAR Drug A Parent ANALYTE PLASMA 4.2 ng/mL 4.2 4.2 ng/mL
17 ABC-123 PC 123-0001 17 Day 11 A554134-17 DRGA_MET Drug A Metabolite ANALYTE URINE 245 ng/mL 245 245 ng/mL
18 ABC-123 PC 123-0001 18 Day 11 A554134-17 DRGA_PAR Drug A Parent ANALYTE URINE 13.1 ng/mL 13.1 13.1 ng/mL
19 ABC-123 PC 123-0001 19 Day 11 A554134-17 VOLUME Volume SPECIMEN URINE 574 mL 574 574 mL
20 ABC-123 PC 123-0001 20 Day 11 A554134-17 PH PH SPECIMEN URINE 5.5 5.5 5.5
21 ABC-123 PC 123-0001 21 Day 11 A554134-18 DRGA_MET Drug A Metabolite ANALYTE PLASMA 9.02 ng/mL 9.02 9.02 ng/mL
22 ABC-123 PC 123-0001 22 Day 11 A554134-18 DRGA_PAR Drug A Parent ANALYTE PLASMA 1.18 ng/mL 1.18 1.18 ng/mL
23 ABC-123 PC 123-0001 23 Day 11 A554134-19 DRGA_MET Drug A Metabolite ANALYTE URINE 293 ng/mL 293 293 ng/mL
24 ABC-123 PC 123-0001 24 Day 11 A554134-19 DRGA_PAR Drug A Parent ANALYTE URINE 7.1 ng/mL 7.1 7.1 ng/mL
25 ABC-123 PC 123-0001 25 Day 11 A554134-19 VOLUME Volume SPECIMEN URINE 363 mL 363 363 mL
26 ABC-123 PC 123-0001 26 Day 11 A554134-19 PH PH SPECIMEN URINE 5.5 5.5 5.5
27 ABC-123 PC 123-0001 27 Day 11 A554134-20 DRGA_MET Drug A Metabolite ANALYTE URINE 280 ng/mL 280 280 ng/mL
28 ABC-123 PC 123-0001 28 Day 11 A554134-20 DRGA_PAR Drug A Parent ANALYTE URINE 2.4 ng/mL 2.4 2.4 ng/mL
29 ABC-123 PC 123-0001 29 Day 11 A554134-20 VOLUME Volume SPECIMEN URINE 606 mL 606 606 mL
30 ABC-123 PC 123-0001 30 Day 11 A554134-20 PH PH SPECIMEN URINE 5.5 5.5 5.5
31 ABC-123 PC 123-0001 31 Day 11 A554134-21 DRGA_MET Drug A Metabolite ANALYTE PLASMA 3.73 ng/mL 3.73 3.73 ng/mL
32 ABC-123 PC 123-0001 32 Day 11 A554134-21 DRGA_PAR Drug A Parent ANALYTE PLASMA <0.1 ng/mL <0.1 ng/mL
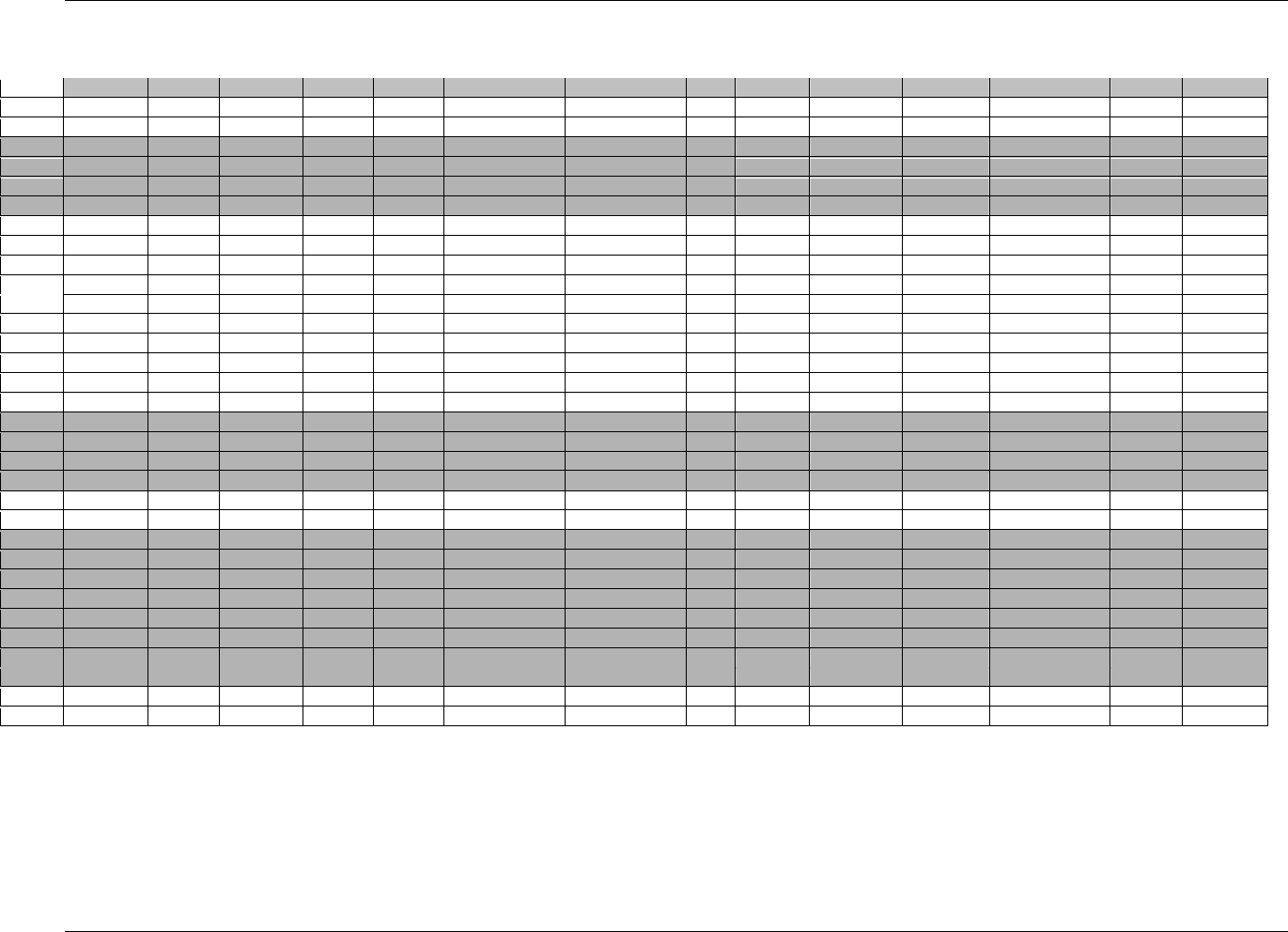
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 178 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
(PC dataset for example 1, continued)
Row
PCSTAT
PCLLOQ
VISITNUM
VISIT
VISITDY
PCDTC
PCENDTC
PCDY
PCTPT
PCTPTNUM
PCTPTREF
PCRFTDTC
PCELTM
PCEVLINT
1 (cont)
0.10
1
DAY 1
1
2001-02-01T07:45
1
PREDOSE
0
Day 1 Dose
2001-02-01T08:00
-PT15M
2 (cont)
0.10
1
DAY 1
1
2001-02-01T07:45
1
PREDOSE
0
Day 1 Dose
2001-02-01T08:00
-PT15M
3 (cont)
2.00
1
DAY 1
1
2001-02-01T07:45
2001-02-01T07:45
1
PREDOSE
0
Day 1 Dose
2001-02-01T08:00
-PT15M
4 (cont)
2.00
1
DAY 1
1
2001-02-01T07:45
2001-02-01T07:45
1
PREDOSE
0
Day 1 Dose
2001-02-01T08:00
-PT15M
5 (cont)
1
DAY 1
1
2001-02-01T07:45
2001-02-01T07:45
1
PREDOSE
0
Day 1 Dose
2001-02-01T08:00
-PT15M
6 (cont)
1
DAY 1
1
2001-02-01T07:45
2001-02-01T07:45
1
PREDOSE
0
Day 1 Dose
2001-02-01T08:00
-PT15M
7 (cont)
0.10
1
DAY 1
1
2001-02-01T09:30
1
1H30MIN
1.5
Day 1 Dose
2001-02-01T08:00
PT1H30M
8 (cont)
0.10
1
DAY 1
1
2001-02-01T09:30
1
1H30MIN
1.5
Day 1 Dose
2001-02-01T08:00
PT1H30M
9 (cont)
0.10
1
DAY 1
1
2001-02-01T14:00
1
6H
6
Day 1 Dose
2001-02-01T08:00
PT6H00M
10 (cont)
0.10
1
DAY 1
1
2001-02-01T14:00
1
6H
6
Day 1 Dose
2001-02-01T08:00
PT6H
11 (cont)
NOT DONE
2
DAY 2
2
2001-02-02T08:00
2
24H
24
Day 1 Dose
2001-02-01T08:00
PT24H
12 (cont)
0.10
2
DAY 2
2
2001-02-02T08:00
2
24H
24
Day 1 Dose
2001-02-01T08:00
PT24H
13 (cont)
0.10
3
DAY 11
11
2001-02-11T07:45
11
PREDOSE
0
Day 11 Dose
2001-02-11T08:00
-PT15M
14 (cont)
0.10
3
DAY 11
11
2001-02-11T07:45
11
PREDOSE
0
Day 11 Dose
2001-02-11T08:00
-PT15M
15 (cont)
0.10
3
DAY 11
11
2001-02-11T09:30
11
1H30MIN
1.5
Day 11 Dose
2001-02-11T08:00
PT1H30M
16 (cont)
0.10
3
DAY 11
11
2001-02-11T09:30
11
1H30MIN
1.5
Day 11 Dose
2001-02-11T08:00
PT1H30M
17 (cont)
2.00
3
DAY 11
11
2001-02-11T08:00
2001-02-11T14:03
11
6H
6
Day 11 Dose
2001-02-11T08:00
PT6H
-PT6H
18 (cont)
2.00
3
DAY 11
11
2001-02-11T08:00
2001-02-11T14:03
11
6H
6
Day 11 Dose
2001-02-11T08:00
PT6H
-PT6H
19 (cont)
3
DAY 11
11
2001-02-11T08:00
2001-02-11T14:03
11
6H
6
Day 11 Dose
2001-02-11T08:00
PT6H
-PT6H
20 (cont)
3
DAY 11
11
2001-02-11T08:00
2001-02-11T14:03
11
6H
6
Day 11 Dose
2001-02-11T08:00
PT6H
-PT6H
21 (cont)
0.10
3
DAY 11
11
2001-02-11T14:00
11
6H
6
Day 11 Dose
2001-02-11T08:00
PT6H
22 (cont)
0.10
3
DAY 11
11
2001-02-11T14:00
11
6H
6
Day 11 Dose
2001-02-11T08:00
PT6H
23 (cont)
2.00
3
DAY 11
11
2001-02-11T14:03
2001-02-11T20:10
11
12H
12
Day 11 Dose
2001-02-11T08:00
PT12H
-PT6H
24 (cont)
2.00
3
DAY 11
11
2001-02-11T14:03
2001-02-11T20:10
11
12H
12
Day 11 Dose
2001-02-11T08:00
PT12H
-PT6H
25 (cont)
3
DAY 11
11
2001-02-11T14:03
2001-02-11T20:10
11
12H
12
Day 11 Dose
2001-02-11T08:00
PT12H
-PT6H
26 (cont)
3
DAY 11
11
2001-02-11T14:03
2001-02-11T20:10
11
12H
12
Day 11 Dose
2001-02-11T08:00
PT12H
-PT6H
27 (cont)
2.00
4
DAY 12
12
2001-02-11T20:03
2001-02-12T08:10
12
24H
24
Day 11 Dose
2001-02-11T08:00
PT24H
-P12H
28 (cont)
2.00
4
DAY 12
12
2001-02-11T20:03
2001-02-12T08:10
12
24H
24
Day 11 Dose
2001-02-11T08:00
PT24H
-P12H
29 (cont)
4
DAY 12
12
2001-02-11T20:03
2001-02-12T08:10
12
24H
24
Day 11 Dose
2001-02-11T08:00
PT24H
-P12H
30 (cont)
4
DAY 12
12
2001-02-11T20:03
2001-02-12T08:10
12
24H
24
Day 11 Dose
2001-02-11T08:00
PT24H
-P12H
31 (cont)
0.10
4
DAY 12
12
2001-02-12T08:00
12
24H
24
Day 11 Dose
2001-02-11T08:00
PT24H
32 (cont)
0.10
4
DAY 12
12
2001-02-12T08:00
12
24H
24
Day 11 Dose
2001-02-11T08:00
PT24H
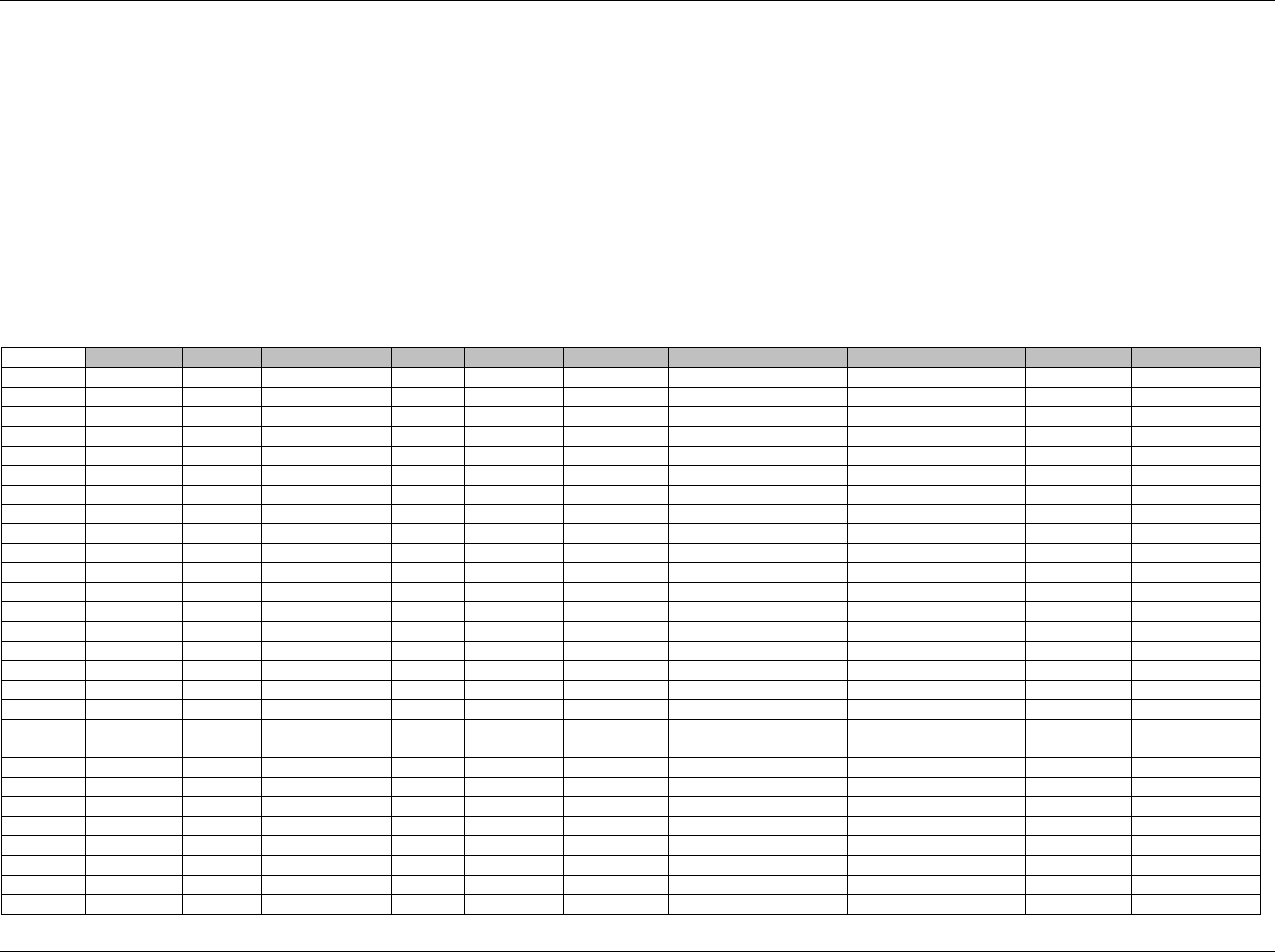
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 179
Final November 12, 2008
6.3.10.3 ASSUMPTIONS FOR PHARMACOKINETIC PARAMETERS (PP) DOMAIN MODEL
1. PP Definition: Data describing the parameters of the time-concentration curve for PC data (e.g., area under the curve, Cmax, Tmax).
2. It is recognized that PP is a derived dataset, and may be produced from an analysis dataset that might have a different structure. As a result, some
sponsors may need to normalize their analysis dataset in order for it to fit into the SDTM-based PP domain.
3. The structure is one record per PK parameter per time-concentration profile per subject
4. Information pertaining to all parameters (e.g., number of exponents, model weighting) should be submitted in the SUPPPP dataset.
5. The following Qualifiers would not generally be used in PP: --BODSYS, --SEV.
6.3.10.4 EXAMPLE FOR PHARMACOKINETIC PARAMETERS (PP) DOMAIN MODEL
Example 1
This example shows PK parameters calculated from time-concentration profiles for parent drug and one metabolite in plasma and urine for one subject on Days 1
(Rows 1-14) and 8 (Rows 15-28). Note that PPRFTDTC is populated in order to link the PP records to the respective PC records. Note that PPSPEC is null for
Clearance records since it is calculated from multiple specimen sources (plasma and urine).
(PP dataset for example 1)
Row
STUDYID
DOMAIN
USUBJID
PPSEQ
PPGRPID
PPTESTCD
PPTEST
PPCAT
PPORRES
PPORRESU
1
ABC-123
PP
ABC-123-0001
1
DAY1_PAR
TMAX
Time to Max Effect
DRUG A PARENT
1.87
h
2
ABC-123
PP
ABC-123-0001
2
DAY1_PAR
CMAX
Max Effect Concentration
DRUG A PARENT
44.5
ug/L
3
ABC-123
PP
ABC-123-0001
3
DAY1_PAR
AUC
Area Under Curve
DRUG A PARENT
294.7
h*mg/L
4
ABC-123
PP
ABC-123-0001
4
DAY1_PAR
THALF_1
Half-life of 1st exp phase
DRUG A PARENT
0.75
h
5
ABC-123
PP
ABC-123-0001
5
DAY1_PAR
THALF_2
Half-life of 2nd exp phase
DRUG A PARENT
4.69
h
6
ABC-123
PP
ABC-123-0001
6
DAY1_PAR
VD
Vol of Distribution
DRUG A PARENT
10.9
L
7
ABC-123
PP
ABC-123-0001
7
DAY1_PAR
CL
Clearance
DRUG A PARENT
1.68
L/h
8
ABC-123
PP
ABC-123-0001
8
DAY1_MET
TMAX
Time to Max Effect
DRUG A METABOLITE
0.94
h
9
ABC-123
PP
ABC-123-0001
9
DAY1_MET
CMAX
Max Effect Concentration
DRUG A METABOLITE
22.27
ug/L
10
ABC-123
PP
ABC-123-0001
10
DAY1_MET
AUC
Area Under Curve
DRUG A METABOLITE
147.35
h*mg/L
11
ABC-123
PP
ABC-123-0001
11
DAY1_MET
THALF_1
Half-life of 1st exp phase
DRUG A METABOLITE
0.38
h
12
ABC-123
PP
ABC-123-0001
12
DAY1_MET
THALF_2
Half-life of 2nd exp phase
DRUG A METABOLITE
2.35
h
13
ABC-123
PP
ABC-123-0001
13
DAY1_MET
VD
Vol of Distribution
DRUG A METABOLITE
5.45
L
14
ABC-123
PP
ABC-123-0001
14
DAY1_MET
CL
Clearance
DRUG A METABOLITE
0.84
L/h
15
ABC-123
PP
ABC-123-0001
15
DAY11_PAR
TMAX
Time to Max Effect
DRUG A PARENT
1.91
h
16
ABC-123
PP
ABC-123-0001
16
DAY11_PAR
CMAX
Max Effect Concentration
DRUG A PARENT
46.0
ug/L
17
ABC-123
PP
ABC-123-0001
17
DAY11_PAR
AUC
Area Under Curve
DRUG A PARENT
289.0
h*mg/L
18
ABC-123
PP
ABC-123-0001
18
DAY11_PAR
THALF_1
Half-life of 1st exp phase
DRUG A PARENT
0.77
h
19
ABC-123
PP
ABC-123-0001
19
DAY11_PAR
THALF_2
Half-life of 2nd exp phase
DRUG A PARENT
4.50
h
20
ABC-123
PP
ABC-123-0001
20
DAY11_PAR
VD
Vol of Distribution
DRUG A PARENT
10.7
L
21
ABC-123
PP
ABC-123-0001
21
DAY11_PAR
CL
Clearance
DRUG A PARENT
1.75
L/h
22
ABC-123
PP
ABC-123-0001
22
DAY11_MET
TMAX
Time to Max Effect
DRUG A METABOLITE
0.96
h
23
ABC-123
PP
ABC-123-0001
23
DAY11_MET
CMAX
Max Effect Concentration
DRUG A METABOLITE
23.00
ug/L
24
ABC-123
PP
ABC-123-0001
24
DAY11_MET
AUC
Area Under Curve
DRUG A METABOLITE
144.50
h*mg/L
25
ABC-123
PP
ABC-123-0001
25
DAY11_MET
THALF_1
Half-life of 1st exp phase
DRUG A METABOLITE
0.39
h
26
ABC-123
PP
ABC-123-0001
26
DAY11_MET
THALF_2
Half-life of 2nd exp phase
DRUG A METABOLITE
2.25
h
27
ABC-123
PP
ABC-123-0001
27
DAY8_MET
VD
Vol of Distribution
DRUG A METABOLITE
5.35
L
28
ABC-123
PP
ABC-123-0001
28
DAY8_MET
CL
Clearance
DRUG A METABOLITE
0.88
L/h

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 180 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
(PP dataset for example 1, continued)
Row
PPSTRESC
PPSTRESN
PPSTRESU
PPSPEC
VISITNUM
VISIT
PPDTC
PPRFTDTC
1 (cont)
1.87
1.87
h
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
2 (cont)
44.5
44.5
ug/L
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
3 (cont)
294.7
294.7
h.mg/L
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
4 (cont)
0.75
0.75
h
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
5 (cont)
4.69
4.69
h
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
6 (cont)
10.9
10.9
L
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
7 (cont)
1.68
1.68
L/h
1
DAY 1
2001-03-01
2001-02-01T08:00
8 (cont)
0.94
0.94
h
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
9 (cont)
22.27
22.27
ug/L
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
10 (cont)
147.35
147.35
h.mg/L
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
11 (cont)
0.38
0.38
h
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
12 (cont)
2.35
2.35
h
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
13 (cont)
5.45
5.45
L
PLASMA
1
DAY 1
2001-03-01
2001-02-01T08:00
14 (cont)
0.84
0.84
L/h
1
DAY 1
2001-03-01
2001-02-01T08:00
15 (cont)
1.91
1.91
h
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
16 (cont)
46.0
46.0
ug/L
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
17 (cont)
289.0
289.0
h.mg/L
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
18 (cont)
0.77
0.77
h
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
19 (cont)
4.50
4.50
h
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
20 (cont)
10.7
10.7
L
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
21 (cont)
1.75
1.75
L/h
2
DAY 11
2001-03-01
2001-02-11T08:00
22 (cont)
0.96
0.96
h
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
23 (cont)
23.00
23.00
ug/L
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
24 (cont)
144.50
144.50
h.mg/L
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
25 (cont)
0.39
0.39
h
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
26 (cont)
2.25
2.25
h
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
27 (cont)
5.35
5.35
L
PLASMA
2
DAY 11
2001-03-01
2001-02-11T08:00
28 (cont)
0.88
0.88
L/h
2
DAY 11
2001-03-01
2001-02-11T08:00
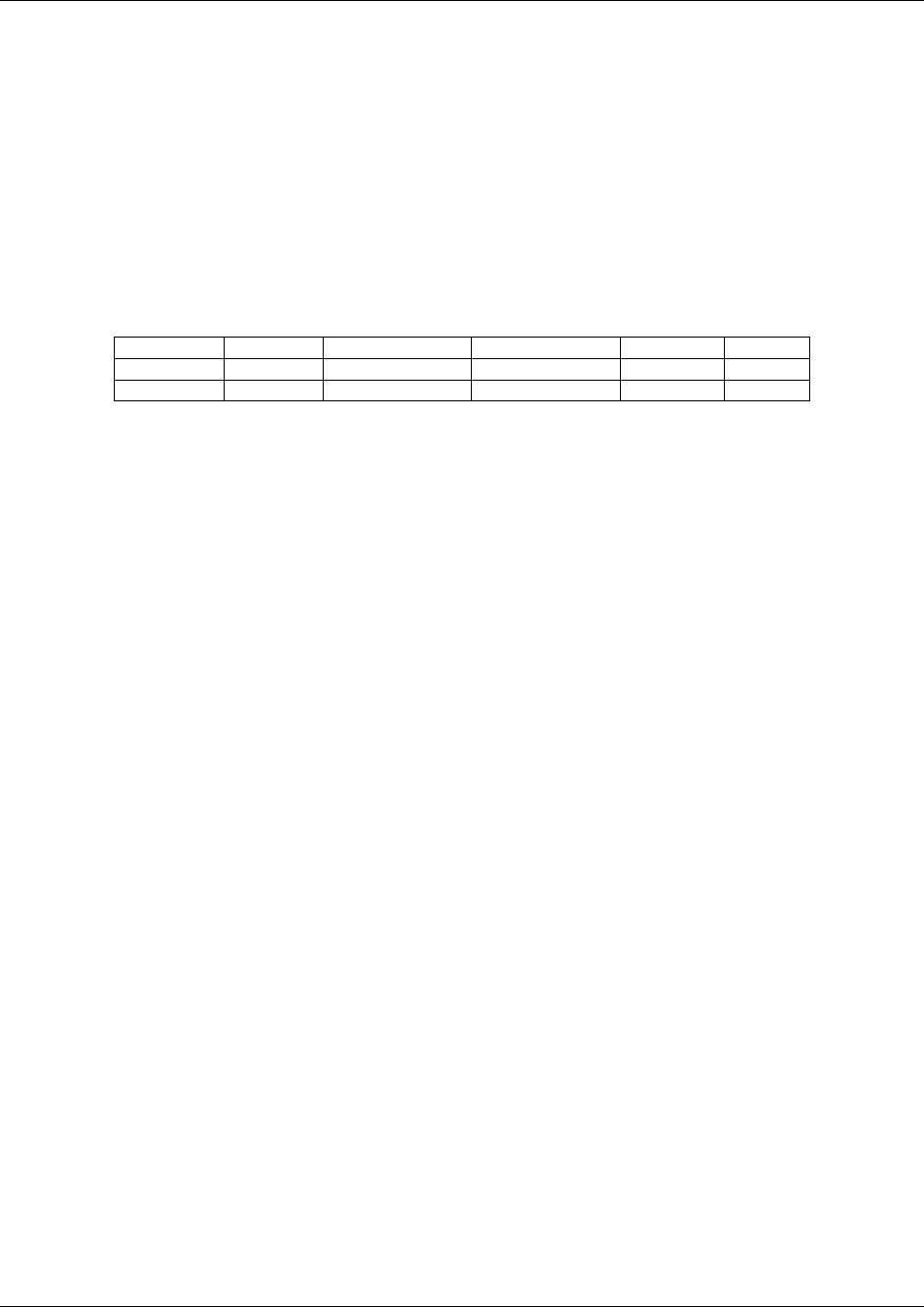
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 181
Final November 12, 2008
6.3.10.5 RELATING PP RECORDS TO PC RECORDS
It is a requirement that sponsors document the concentrations used to calculate each parameter. For many sponsors,
this need is currently met via the analysis metadata. As a result of feedback received from many sponsors on the
draft version of this document, sponsors may continue to document the concentrations used to calculate each
parameter via the analysis datasets.
This section serves as a reference for sponsors who wish to document relationships between PK parameter records in
a Pharmacokinetic Parameter (PP) dataset and specific time-point concentration records in a Pharmacokinetic
Concentration (PC) dataset according to the SDTM using the RELREC table (1299HSection 8.2 and 1300HSection 8.3).
6.3.10.5.1 RELATING DATASETS
If all time-point concentrations in PC are used to calculate all parameters for all subjects, then the relationship
between the two datasets can be documented as shown in the table below.
RDOMAIN USUBJID IDVAR IDVARVAL RELTYPE RELID
PC PCGRPID MANY A
PP PPGRPID MANY A
Note that incorporating the name of the analyte and the day of the collection into the value of --GRPID (or some
equivalent method for assigning different values of --GRPID for all the combinations of analytes and reference time
points) is necessary when there is more than one reference time point (PCRFTDTC and PPRFTDTC, which are the
same for related records), and more than one analyte (PCTESTCD, copied into PPCAT to indicate the analyte with
which the parameters are associated), since these variables are part of the natural key (see 1301HSection 3.2.1.1) for both
datasets. In this case, --GRPID is a surrogate key (1302HSection 3.2.1.1) used for the relationship.
6.3.10.5.2 RELATING RECORDS
Four possible examples of different types of relationships between PC and PP records for one drug (DRUG X in this
case) are described. For all of these, the actual PC and PP data are the same. The only variables whose values change
across the examples are the sponsor-defined PCGRPID and PPGRPID. As in the case for relating datasets above
(1303HSection 6.3.10.5.1), --GRPID values must take into account all the combinations of analytes and reference time
points, since both are part of the natural key (see 1304HSection 3.2.1.1) for both datasets. To conserve space, the PC and
PP domains appear only once, but with four --GRPID columns, one for each of the examples. Note that a submission
dataset would contain only one --GRPID column with a set of values such as those shown in one of the four columns
in the PC and PP datasets, or values defined by the sponsor. Note that --GRPID values in PC and PP do not need to
be the same (e.g., examples show PC with underscores and PP without underscores). The example specifics are as
follows:
Example 1: All PK time-point-concentration values in the PC dataset are used to calculate all the PK parameters in
the PP dataset for both Days 1 and 8 for one subject.
1978HPharmacokinetic Concentrations (PC) Dataset For All Examples
1979HPharmacokinetic Parameters (PP) Dataset For All Examples
1980HRELREC Example 1. All PC records used to calculate all PK parameters
• 1981HMethod A (Many to many, using PCGRPID and PPGRPID)
• 1982HMethod B (One to many, using PCSEQ and PPGRPID)
• 1983HMethod C (Many to one, using PCGRPID and PPSEQ)
• 1984HMethod D (One to one, using PCSEQ and PPSEQ)
Example 2: Two PC values were excluded from the calculation of all PK parameters for the Day 1 data. Day 8
values are related as per Example 1.
1985HRELREC Example 2: Only some records in PC used to calculate all PK parameters
• 1986HMethod A (Many to many, using PCGRPID and PPGRPID)
• 1987HMethod B (One to many, using PCSEQ and PPGRPID)
• 1988HMethod C (Many to one, using PCGRPID and PPSEQ)
• 1989HMethod D (One to one, using PCSEQ and PPSEQ)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 182 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 3: Two PC values were excluded from the calculation of two PK parameters, but used in the others for Day 1.
Day 8 values are related as per Example 1.
1990HRELREC Example 3. Only some records in PC used to calculate some parameters
• 1991HMethod A (Many to many, using PCGRPID and PPGRPID)
• 1992HMethod B (One to many, using PCSEQ and PPGRPID)
• 1993HMethod C (Many to one, using PCGRPID and PPSEQ)
• 1994HMethod D (One to one, using PCSEQ and PPSEQ)
Example 4: Only Some PC records for Day 1 were used to calculate parameters: Time Point 5 was excluded from
Tmax, Time Point 6 from Cmax, and Time Points 11 and 12 were excluded from AUC. Day 8 values are related as
per Example 1.
1995HRELREC Example 4: Only Some records in PC used to calculate parameters
• 1996HMethod A (Many to many, using PCGRPID and PPGRPID)
• Method B (One to many omitted - see note below)
• Method C (Many to one omitted - see note below)
• 1997HMethod D (One to one, using PCSEQ and PPSEQ)
For each example, PCGRPID and PPGRPID were used to group related records within each respective dataset. The
values for these, as well as the values for PCSEQ and PPSEQ, were then used to populate combinations of IDVAR
and IDVARVAL in the RELREC table using four methods (A-D) for Examples 1-3. Only two methods (A and D) are
shown for Example 4, due to its complexity. Since the relationship between PC records and PP records for Day 8
data does not change across the examples, it is shown only for Example 1, and not repeated.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 183
Final November 12, 2008
Pharmacokinetic Concentrations (PC) Dataset For All Examples
Row
STUDYID
DOMAIN
USUBJID
PCSEQ
PCGRPID
PCGRPID
PCGRPID
PCGRPID
PCREFID
PCTESTCD
PCTEST
PCCAT
PCSPEC
PCORRES
PCORRESU
Example 1
Example 2
Example 3
Example 4
1
ABC-123
PC
ABC-123-0001
1
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_A
123-0001-01
DRUG X
Study Drug
ANALYTE
PLASMA
9
ug/mL
2
ABC-123
PC
ABC-123-0001
2
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_A
123-0001-02
DRUG X
Study Drug
ANALYTE
PLASMA
20
ug/mL
3
ABC-123
PC
ABC-123-0001
3
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_A
123-0001-03
DRUG X
Study Drug
ANALYTE
PLASMA
31
ug/mL
4
ABC-123
PC
ABC-123-0001
4
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_A
123-0001-04
DRUG X
Study Drug
ANALYTE
PLASMA
38
ug/mL
5
ABC-123
PC
ABC-123-0001
5
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_B
123-0001-05
DRUG X
Study Drug
ANALYTE
PLASMA
45
ug/mL
6
ABC-123
PC
ABC-123-0001
6
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_C
123-0001-06
DRUG X
Study Drug
ANALYTE
PLASMA
47.5
ug/mL
7
ABC-123
PC
ABC-123-0001
7
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_A
123-0001-07
DRUG X
Study Drug
ANALYTE
PLASMA
41
ug/mL
8
ABC-123
PC
ABC-123-0001
8
DY1_DRGX
EXCLUDE
DY1_DRGX_B
DY1_DRGX_A
123-0001-08
DRUG X
Study Drug
ANALYTE
PLASMA
35
ug/mL
9
ABC-123
PC
ABC-123-0001
9
DY1_DRGX
EXCLUDE
DY1_DRGX_B
DY1_DRGX_A
123-0001-09
DRUG X
Study Drug
ANALYTE
PLASMA
31
ug/mL
10
ABC-123
PC
ABC-123-0001
10
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_A
123-0001-10
DRUG X
Study Drug
ANALYTE
PLASMA
25
ug/mL
11
ABC-123
PC
ABC-123-0001
11
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_D
123-0001-11
DRUG X
Study Drug
ANALYTE
PLASMA
18
ug/mL
12
ABC-123
PC
ABC-123-0001
12
DY1_DRGX
DY1_DRGX
DY1_DRGX_A
DY1_DRGX_D
123-0001-12
DRUG X
Study Drug
ANALYTE
PLASMA
12
ug/mL
13
ABC-123
PC
ABC-123-0001
13
DY11_DRGX
123-0002-13
DRUG X
Study Drug
ANALYTE
PLASMA
10.0
ug/mL
14
ABC-123
PC
ABC-123-0001
14
DY11_DRGX
123-0002-14
DRUG X
Study Drug
ANALYTE
PLASMA
21.0
ug/mL
15
ABC-123
PC
ABC-123-0001
15
DY11_DRGX
123-0002-15
DRUG X
Study Drug
ANALYTE
PLASMA
32.0
ug/mL
16
ABC-123
PC
ABC-123-0001
16
DY11_DRGX
123-0002-16
DRUG X
Study Drug
ANALYTE
PLASMA
39.0
ug/mL
17
ABC-123
PC
ABC-123-0001
17
DY11_DRGX
123-0002-17
DRUG X
Study Drug
ANALYTE
PLASMA
46.0
ug/mL
18
ABC-123
PC
ABC-123-0001
18
DY11_DRGX
123-0002-18
DRUG X
Study Drug
ANALYTE
PLASMA
48.0
ug/mL
19
ABC-123
PC
ABC-123-0001
19
DY11_DRGX
123-0002-19
DRUG X
Study Drug
ANALYTE
PLASMA
40.0
ug/mL
20
ABC-123
PC
ABC-123-0001
20
DY11_DRGX
123-0002-20
DRUG X
Study Drug
ANALYTE
PLASMA
35.0
ug/mL
21
ABC-123
PC
ABC-123-0001
21
DY11_DRGX
123-0002-21
DRUG X
Study Drug
ANALYTE
PLASMA
30.0
ug/mL
22
ABC-123
PC
ABC-123-0001
22
DY11_DRGX
123-0002-22
DRUG X
Study Drug
ANALYTE
PLASMA
24.0
ug/mL
23
ABC-123
PC
ABC-123-0001
23
DY11_DRGX
123-0002-23
DRUG X
Study Drug
ANALYTE
PLASMA
17.0
ug/mL
24
ABC-123
PC
ABC-123-0001
24
DY11_DRGX
123-0002-24
DRUG X
Study Drug
ANALYTE
PLASMA
11.0
ug/mL
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 184 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
(PC dataset for all example, continued)
Row
PCSTRESC
PCSTRESN
PCSTRESU
PCLLOQ
VISITNUM
VISIT
VISITDY
PCDTC
PCDY
PCTPT
PCTPTNUM
PCTPTREF
PCRFTDTC
PCELTM
1 (cont)
9
9
ug/mL
1.00
1
DAY 1
1
2001-02-01T08:35
1
5 min
1
Day 1 Dose
2001-02-01T08:30
PT5M
2 (cont)
20
20
ug/mL
1.00
1
DAY 1
1
2001-02-01T08:55
1
25 min
2
Day 1 Dose
2001-02-01T08:30
PT25M
3 (cont)
31
31
ug/mL
1.00
1
DAY 1
1
2001-02-01T09:20
1
50 min
3
Day 1 Dose
2001-02-01T08:30
PT50M
4 (cont)
38
38
ug/mL
1.00
1
DAY 1
1
2001-02-01T09:45
1
75 min
4
Day 1 Dose
2001-02-01T08:30
PT1H15M
5 (cont)
45
45
ug/mL
1.00
1
DAY 1
1
2001-02-01T10:10
1
100 min
5
Day 1 Dose
2001-02-01T08:30
PT1H40M
6 (cont)
47.5
47.5
ug/mL
1.00
1
DAY 1
1
2001-02-01T10:35
1
125 min
6
Day 1 Dose
2001-02-01T08:30
PT2H5M
7 (cont)
41
41
ug/mL
1.00
1
DAY 1
1
2001-02-01T11:00
1
150 min
7
Day 1 Dose
2001-02-01T08:30
PT2H30M
8 (cont)
35
35
ug/mL
1.00
1
DAY 1
1
2001-02-01T11:50
1
200 min
8
Day 1 Dose
2001-02-01T08:30
PT3H20M
9 (cont)
31
31
ug/mL
1.00
1
DAY 1
1
2001-02-01T12:40
1
250 min
9
Day 1 Dose
2001-02-01T08:30
PT4H10M
10 (cont)
25
25
ug/mL
1.00
1
DAY 1
1
2001-02-01T14:45
1
375 min
10
Day 1 Dose
2001-02-01T08:30
PT6H15M
11 (cont)
18
18
ug/mL
1.00
1
DAY 1
1
2001-02-01T16:50
1
500 min
11
Day 1 Dose
2001-02-01T08:30
PT8H20M
12 (cont)
12
12
ug/mL
1.00
1
DAY 1
1
2001-02-01T18:30
1
600 min
12
Day 1 Dose
2001-02-01T08:30
PT10H
13 (cont)
10.0
10.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T08:35
8
5 min
1
Day 8 Dose
2001-02-08T08:30
PT5M
14 (cont)
21.0
21.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T08:55
8
25 min
2
Day 8 Dose
2001-02-08T08:30
PT25M
15 (cont)
32.0
32.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T09:20
8
50 min
3
Day 8 Dose
2001-02-08T08:30
PT50M
16 (cont)
39.0
39.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T09:45
8
75 min
4
Day 8 Dose
2001-02-08T08:30
PT1H15M
17 (cont)
46.0
46.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T10:10
8
100 min
5
Day 8 Dose
2001-02-08T08:30
PT1H40M
18 (cont)
48.0
48.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T10:35
8
125 min
6
Day 8 Dose
2001-02-08T08:30
PT2H5M
19 (cont)
40.0
40.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T11:00
8
150 min
7
Day 8 Dose
2001-02-08T08:30
PT2H30M
20 (cont)
35.0
35.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T11:50
8
200 min
8
Day 8 Dose
2001-02-08T08:30
PT3H20M
21 (cont)
30.0
30.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T12:40
8
250 min
9
Day 8 Dose
2001-02-08T08:30
PT4H10M
22 (cont)
24.0
24.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T14:45
8
375 min
10
Day 8 Dose
2001-02-08T08:30
PT6H15M
23 (cont)
17.0
17.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T16:50
8
500 min
11
Day 8 Dose
2001-02-08T08:30
PT8H20M
24 (cont)
11.0
11.0
ug/mL
1.00
2
DAY 8
8
2001-02-08T18:30
8
600 min
12
Day 8 Dose
2001-02-08T08:30
PT10H
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 185
Final November 12, 2008
Pharmacokinetic Parameters (PP) Dataset For All Examples
Row
STUDYID
DOMAIN
USUBJID
PPSEQ
PPDTC
PPGRPID
PPGRPID
PPGRPID
PPGRPID
Example 1
Example 2
Example 3
Example 4
1
ABC-123
PP
ABC-123-0001
1
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_A
TMAX
2
ABC-123
PP
ABC-123-0001
2
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_A
CMAX
3
ABC-123
PP
ABC-123-0001
3
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_A
AUC
4
ABC-123
PP
ABC-123-0001
4
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_HALF
OTHER
5
ABC-123
PP
ABC-123-0001
5
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_HALF
OTHER
6
ABC-123
PP
ABC-123-0001
6
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_A
OTHER
7
ABC-123
PP
ABC-123-0001
7
2001-02-01T08:35
DY1DRGX
DY1DRGX
DY1DRGX_A
OTHER
8
ABC-123
PP
ABC-123-0001
8
2001-02-08T08:35
DY11DRGX
9
ABC-123
PP
ABC-123-0001
9
2001-02-08T08:35
DY11DRGX
10
ABC-123
PP
ABC-123-0001
10
2001-02-08T08:35
DY11DRGX
11
ABC-123
PP
ABC-123-0001
11
2001-02-08T08:35
DY11DRGX
12
ABC-123
PP
ABC-123-0001
12
2001-02-08T08:35
DY11DRGX
13
ABC-123
PP
ABC-123-0001
13
2001-02-08T08:35
DY11DRGX
14
ABC-123
PP
ABC-123-0001
14
2001-02-08T08:35
DY11DRGX
Row
PPTESTCD
PPTEST
PPCAT
PPORRES
PPORRESU
PPSTRESC
PPSTRESN
PPSTRESU
1 (cont)
TMAX
Time to Max Effect
DRUG X
1.87
h
1.87
1.87
h
2 (cont)
CMAX
Max Effect Concentration
DRUG X
44.5
ug/L
44.5
44.5
ug/L
3 (cont)
AUC
Area Under Curve
DRUG X
294.7
h*mg/L
294.7
294.7
h*mg/L
4 (cont)
T1/2, 1
Half-life of 1st exp phase
DRUG X
0.75
h
0.75
0.75
h
5 (cont)
T1/2, 2
Half-life of 2nd exp phase
DRUG X
4.69
h
4.69
4.69
h
6 (cont)
VD
Volume of Distribution
DRUG X
10.9
L
10.9
10.9
L
7 (cont)
CL
Clearance
DRUG X
1.68
L/h
1.68
1.68
L/h
8 (cont)
TMAX
Time to Max Effect
DRUG X
1.91
h
1.91
1.91
h
9 (cont)
CMAX
Max Effect Concentration
DRUG X
46.0
ug/L
46.0
46.0
ug/L
10 (cont)
AUC
Area Under Curve
DRUG X
289.0
h*mg/L
289.0
289.0
h*mg/L
11 (cont)
T1/2, 1
Half-life of 1st exp phase
DRUG X
0.77
h
0.77
0.77
h
12 (cont)
T1/2, 2
Half-life of 2nd exp phase
DRUG X
4.50
h
4.50
4.50
h
13 (cont)
VD
Volume of Distribution
DRUG X
10.7
L
10.7
10.7
L
14 (cont)
CL
Clearance
DRUG X
1.75
L/h
1.75
1.75
L/h
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 186 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
RELREC Example 1. All PC records used to calculate all PK parameters.
Method A (Many to many, using PCGRPID and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX
1
2
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX
1
3
ABC-123
PC
ABC-123-0001
PCGRPID
DY11_DRGX
2
4
ABC-123
PP
ABC-123-0001
PPGRPID
DY11DRGX
2
* RELID 1 indicates all PC records with PCGRPID = DY1_DRGX are related to all PP records with PPGRPID = DY1DRGX.
* RELID 2 indicates all PC records with PCGRPID = DY8_DRGX are related to all PP records with PPGRPID = DY8DRGX.
Method B (One to many, using PCSEQ and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
2
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
3
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
4
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
5
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
8
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
9
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
11
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
12
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
13
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX
1
14
ABC-123
PC
ABC-123-0001
PCSEQ
13
2
15
ABC-123
PC
ABC-123-0001
PCSEQ
14
2
16
ABC-123
PC
ABC-123-0001
PCSEQ
15
2
17
ABC-123
PC
ABC-123-0001
PCSEQ
16
2
18
ABC-123
PC
ABC-123-0001
PCSEQ
17
2
19
ABC-123
PC
ABC-123-0001
PCSEQ
18
2
20
ABC-123
PC
ABC-123-0001
PCSEQ
19
2
21
ABC-123
PC
ABC-123-0001
PCSEQ
20
2
22
ABC-123
PC
ABC-123-0001
PCSEQ
21
2
23
ABC-123
PC
ABC-123-0001
PCSEQ
22
2
24
ABC-123
PC
ABC-123-0001
PCSEQ
23
2
25
ABC-123
PC
ABC-123-0001
PCSEQ
24
2
26
ABC-123
PP
ABC-123-0001
PPGRPID
DY8DRGX
2
* RELID 1 indicates records with PCSEQ values of 1-12 are related to records with PPGRPID = DY1DRGX.
* RELID 2 indicates records with PCSEQ values of 13-24 are related to records with PPGRPID = DY8DRGX.
Method C (Many to one, using PCGRPID and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX
1
2
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
3
ABC-123
PP
ABC-123-0001
PPSEQ
2
1
4
ABC-123
PP
ABC-123-0001
PPSEQ
3
1
5
ABC-123
PP
ABC-123-0001
PPSEQ
4
1
6
ABC-123
PP
ABC-123-0001
PPSEQ
5
1
7
ABC-123
PP
ABC-123-0001
PPSEQ
6
1
8
ABC-123
PP
ABC-123-0001
PPSEQ
7
1
9
ABC-123
PC
ABC-123-0001
PCGRPID
DY8_DRGX
2
10
ABC-123
PP
ABC-123-0001
PPSEQ
8
2
11
ABC-123
PP
ABC-123-0001
PPSEQ
9
2
12
ABC-123
PP
ABC-123-0001
PPSEQ
10
2
13
ABC-123
PP
ABC-123-0001
PPSEQ
11
2
14
ABC-123
PP
ABC-123-0001
PPSEQ
12
2
15
ABC-123
PP
ABC-123-0001
PPSEQ
13
2
16
ABC-123
PP
ABC-123-0001
PPSEQ
14
2
* RELID 1 indicates records with a PCGRPID value of DY1_DRGX are related to records with PPSEQ values of 1-7.
* RELID 2 indicates records with a PCGRPID value of DY8_DRGX are related to records with PPSEQ values of 8-14.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 187
Final November 12, 2008
Method D (One to one, using PCSEQ and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
2
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
3
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
4
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
5
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
8
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
9
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
11
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
12
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
13
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
14
ABC-123
PP
ABC-123-0001
PPSEQ
2
1
15
ABC-123
PP
ABC-123-0001
PPSEQ
3
1
16
ABC-123
PP
ABC-123-0001
PPSEQ
4
1
17
ABC-123
PP
ABC-123-0001
PPSEQ
5
1
18
ABC-123
PP
ABC-123-0001
PPSEQ
6
1
19
ABC-123
PP
ABC-123-0001
PPSEQ
7
1
20
ABC-123
PC
ABC-123-0001
PCSEQ
13
2
21
ABC-123
PC
ABC-123-0001
PCSEQ
14
2
22
ABC-123
PC
ABC-123-0001
PCSEQ
15
2
23
ABC-123
PC
ABC-123-0001
PCSEQ
16
2
24
ABC-123
PC
ABC-123-0001
PCSEQ
17
2
25
ABC-123
PC
ABC-123-0001
PCSEQ
18
2
26
ABC-123
PC
ABC-123-0001
PCSEQ
19
2
27
ABC-123
PC
ABC-123-0001
PCSEQ
20
2
28
ABC-123
PC
ABC-123-0001
PCSEQ
21
2
29
ABC-123
PC
ABC-123-0001
PCSEQ
22
2
30
ABC-123
PC
ABC-123-0001
PCSEQ
23
2
31
ABC-123
PC
ABC-123-0001
PCSEQ
24
2
32
ABC-123
PP
ABC-123-0001
PPSEQ
8
2
33
ABC-123
PP
ABC-123-0001
PPSEQ
9
2
34
ABC-123
PP
ABC-123-0001
PPSEQ
10
2
35
ABC-123
PP
ABC-123-0001
PPSEQ
11
2
36
ABC-123
PP
ABC-123-0001
PPSEQ
12
2
37
ABC-123
PP
ABC-123-0001
PPSEQ
13
2
38
ABC-123
PP
ABC-123-0001
PPSEQ
14
2
* RELID 1 indicates records with PCSEQ values of 1-12 are related to records with PPSEQ values of 1-7.
* RELID 2 indicates records with PCSEQ values of 13-24 are related to records with PPSEQ values of 8-14.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 188 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
RELREC Example 2: Only some records in PC used to calculate all PK parameters: Time Points 8 and 9 on
Day 1 not used for any PK parameters.
Method A (Many to many, using PCGRPID and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX
1
2
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX
1
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates only PC records with PCGRPID = DY1_DRGX are related to all PP records with
PPGRPID = DY1DRGX. PC records with PCGRPID = EXCLUDE were not used.
* RELID 2 indicates all PC records with PCGRPID = DY8_DRGX are related to all PP records with PPGRPID =
DY8DRGX.
Method B (One to many, using PCSEQ and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
2
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
3
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
4
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
5
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
11
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX
1
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with PCSEQ values of 1-7 and 10-12 are related to records with PPGRPID = DY1DRGX.
* RELID 2 indicates records with PCSEQ values of 13-24 are related to records with PPGRPID = DY8DRGX.
Method C (Many to one, using PCGRPID and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX
1
2
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
3
ABC-123
PP
ABC-123-0001
PPSEQ
2
1
4
ABC-123
PP
ABC-123-0001
PPSEQ
3
1
5
ABC-123
PP
ABC-123-0001
PPSEQ
4
1
6
ABC-123
PP
ABC-123-0001
PPSEQ
5
1
7
ABC-123
PP
ABC-123-0001
PPSEQ
6
1
8
ABC-123
PP
ABC-123-0001
PPSEQ
7
1
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with a PCGRPID value of DY1_DRGX are related to records with PPSEQ values of 1-7.
Method D (One to one, using PCSEQ and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
2
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
3
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
4
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
5
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
11
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
12
ABC-123
PP
ABC-123-0001
PPSEQ
2
1
13
ABC-123
PP
ABC-123-0001
PPSEQ
3
1
14
ABC-123
PP
ABC-123-0001
PPSEQ
4
1
15
ABC-123
PP
ABC-123-0001
PPSEQ
5
1
16
ABC-123
PP
ABC-123-0001
PPSEQ
6
1
17
ABC-123
PP
ABC-123-0001
PPSEQ
7
1
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with PCSEQ values of 1-7 and 10-12 are related to records with PPSEQ values of 1-7.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 189
Final November 12, 2008
RELREC Example 3. Only some records in PC used to calculate some parameters: Time Points 8 and 9 on
Day 1 not used for half-life calculations, but used for other parameters.
Method A (Many to many, using PCGRPID and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX_A
1
2
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX_B
1
3
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX_A
1
4
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX_A
2
5
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX_HALF
2
The Day 8 relationships are the same as those shown in Example 1.
* RELID of "1" Indicates that all time points on Day 1 (PCGRPID = DY1_DRGX_A and DY1_DRGX_B) were
used to calculate all parameters (PPGRPID = DY1DRGX_A) except half-lives.
* RELID of "2" Indicates only the values for PCGRPID = DY1_DRGX_A were used to calculate the half-lives
(PPGRPID = DY1DRGX_HALF).
Method B (One to many, using PCSEQ and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
2
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
3
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
4
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
5
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
8
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
9
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
11
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
12
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
13
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX_A
1
14
ABC-123
PC
ABC-123-0001
PCSEQ
1
2
15
ABC-123
PC
ABC-123-0001
PCSEQ
2
2
16
ABC-123
PC
ABC-123-0001
PCSEQ
3
2
17
ABC-123
PC
ABC-123-0001
PCSEQ
4
2
18
ABC-123
PC
ABC-123-0001
PCSEQ
5
2
19
ABC-123
PC
ABC-123-0001
PCSEQ
6
2
20
ABC-123
PC
ABC-123-0001
PCSEQ
7
2
21
ABC-123
PC
ABC-123-0001
PCSEQ
10
2
22
ABC-123
PC
ABC-123-0001
PCSEQ
11
2
23
ABC-123
PC
ABC-123-0001
PCSEQ
12
2
24
ABC-123
PP
ABC-123-0001
PPGRPID
DY1DRGX_HALF
2
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with PCSEQ values of 1-12 are related to records with PPGRPID = DY1DRGX_A
* RELID 2 indicates records with PCSEQ values of 1-7 and 10-12 are related to records with PPGRPID = DY1DRGX_HALF.
Method C (Many to one, using PCGRPID and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX_A
1
2
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX_B
1
3
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
4
ABC-123
PP
ABC-123-0001
PPSEQ
2
1
5
ABC-123
PP
ABC-123-0001
PPSEQ
3
1
6
ABC-123
PP
ABC-123-0001
PPSEQ
6
1
7
ABC-123
PP
ABC-123-0001
PPSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCGRPID
DY1_DRGX_A
2
9
ABC-123
PP
ABC-123-0001
PPSEQ
4
2
10
ABC-123
PP
ABC-123-0001
PPSEQ
5
2
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with a PCGRPID value of DY1_DRGX_A and DY1_DRGX_B are related to records
with PPSEQ values of 1-7.
* RELID 2 indicates records with a PCGRPID value of DAYDY1DRGX_A are related to records with
PPSEQ values of 4 and 5.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 190 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Method D (One to one, using PCSEQ and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
2
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
3
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
4
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
5
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
8
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
9
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
11
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
12
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
13
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
14
ABC-123
PP
ABC-123-0001
PPSEQ
2
1
15
ABC-123
PP
ABC-123-0001
PPSEQ
3
1
16
ABC-123
PP
ABC-123-0001
PPSEQ
6
1
17
ABC-123
PP
ABC-123-0001
PPSEQ
7
1
18
ABC-123
PC
ABC-123-0001
PCSEQ
1
2
19
ABC-123
PC
ABC-123-0001
PCSEQ
2
2
20
ABC-123
PC
ABC-123-0001
PCSEQ
3
2
21
ABC-123
PC
ABC-123-0001
PCSEQ
4
2
22
ABC-123
PC
ABC-123-0001
PCSEQ
5
2
23
ABC-123
PC
ABC-123-0001
PCSEQ
6
2
24
ABC-123
PC
ABC-123-0001
PCSEQ
7
2
25
ABC-123
PC
ABC-123-0001
PCSEQ
10
2
26
ABC-123
PC
ABC-123-0001
PCSEQ
11
2
27
ABC-123
PC
ABC-123-0001
PCSEQ
12
2
28
ABC-123
PP
ABC-123-0001
PPSEQ
4
2
29
ABC-123
PP
ABC-123-0001
PPSEQ
5
2
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with PCSEQ values of 1-12 are related to records with PPSEQ values of 1-3 and 6-7.
* RELID 2 indicates records with PCSEQ values of 1-7 and 10-12 are related to records with PPSEQ values of 4 and 5.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 191
Final November 12, 2008
RELREC Example 4: Only Some records in PC used to calculate parameters: Time Point 5 excluded from
Tmax, 6 from Cmax, and Time Points 11 and 12 from AUC
Method A (Many to many, using PCGRPID and PPGRPID)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
1
ABC-123
PP
ABC-123-0001
PPGRPID
TMAX
1
2
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_A
1
3
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_C
1
4
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_D
1
5
ABC-123
PP
ABC-123-0001
PPGRPID
CMAX
2
6
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_A
2
7
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_B
2
8
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_D
2
9
ABC-123
PP
ABC-123-0001
PPGRPID
AUC
3
10
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_A
3
11
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_B
3
12
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_C
3
13
ABC-123
PP
ABC-123-0001
PPGRPID
OTHER
4
14
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_A
4
15
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_B
4
16
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_C
4
17
ABC-123
PC
ABC-123-0001
PCGRPID
DY1DRGX_D
4
The Day 8 relationships are the same as those shown in Example 1.
* Same RELID of "1" Indicates that Tmax used records with PCGRPID values DY1DRGX_A, DY1DRGX_C, and
DY1DRGX_D.
* Same RELID of "2" Indicates that Cmax used records with PCGRPID values DY1DRGX_A, DY1DRGX_B, and
DY1DRGX_D.
* Same RELID of "3" Indicates that AUC used PCGRPID values DY1DRGX_A, DY1DRGX_B, and
DY1DRGX_C.
* Same RELID of "4" Indicates that all other parameters (PPGRPID = OTHER) used all PC time points: PCGRPID
values DY1DRGX_A, DY1DRGX_B, DY1DRGX_C, and DY1DRGX_D.
Note that in the above RELREC table, the single records in rows 1, 3, 5, 7, and 9, represented by their --GRPIDs
(TMAX, DY1DRGX_C, CMAX, DY1DRGX_B, AUC) could have been referenced by their SEQ values, since both
identify the records sufficiently. At least two other hybrid approaches would have been acceptable as well: using
PPSEQ and PCGRPIDs whenever possible, and using PPGRPID and PCSEQ values whenever possible. Method D
below uses only SEQ values.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 192 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Method D (One to one, using PCSEQ and PPSEQ)
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID *
ABC-123
PC
ABC-123-0001
PCSEQ
1
1
ABC-123
PC
ABC-123-0001
PCSEQ
2
1
ABC-123
PC
ABC-123-0001
PCSEQ
3
1
ABC-123
PC
ABC-123-0001
PCSEQ
4
1
5
ABC-123
PC
ABC-123-0001
PCSEQ
6
1
6
ABC-123
PC
ABC-123-0001
PCSEQ
7
1
7
ABC-123
PC
ABC-123-0001
PCSEQ
8
1
8
ABC-123
PC
ABC-123-0001
PCSEQ
9
1
9
ABC-123
PC
ABC-123-0001
PCSEQ
10
1
10
ABC-123
PC
ABC-123-0001
PCSEQ
11
1
11
ABC-123
PC
ABC-123-0001
PCSEQ
12
1
12
ABC-123
PP
ABC-123-0001
PPSEQ
1
1
13
ABC-123
PC
ABC-123-0001
PCSEQ
1
2
14
ABC-123
PC
ABC-123-0001
PCSEQ
2
2
15
ABC-123
PC
ABC-123-0001
PCSEQ
3
2
16
ABC-123
PC
ABC-123-0001
PCSEQ
4
2
17
ABC-123
PC
ABC-123-0001
PCSEQ
5
2
18
ABC-123
PC
ABC-123-0001
PCSEQ
7
2
19
ABC-123
PC
ABC-123-0001
PCSEQ
8
2
20
ABC-123
PC
ABC-123-0001
PCSEQ
9
2
21
ABC-123
PC
ABC-123-0001
PCSEQ
10
2
22
ABC-123
PC
ABC-123-0001
PCSEQ
11
2
23
ABC-123
PC
ABC-123-0001
PCSEQ
12
2
24
ABC-123
PP
ABC-123-0001
PPSEQ
2
2
25
ABC-123
PC
ABC-123-0001
PCSEQ
1
3
26
ABC-123
PC
ABC-123-0001
PCSEQ
2
3
27
ABC-123
PC
ABC-123-0001
PCSEQ
3
3
28
ABC-123
PC
ABC-123-0001
PCSEQ
4
3
29
ABC-123
PC
ABC-123-0001
PCSEQ
5
3
30
ABC-123
PC
ABC-123-0001
PCSEQ
6
3
31
ABC-123
PC
ABC-123-0001
PCSEQ
7
3
32
ABC-123
PC
ABC-123-0001
PCSEQ
8
3
33
ABC-123
PC
ABC-123-0001
PCSEQ
9
3
34
ABC-123
PC
ABC-123-0001
PCSEQ
10
3
35
ABC-123
PP
ABC-123-0001
PPSEQ
3
3
36
ABC-123
PC
ABC-123-0001
PCSEQ
1
4
37
ABC-123
PC
ABC-123-0001
PCSEQ
2
4
38
ABC-123
PC
ABC-123-0001
PCSEQ
3
4
39
ABC-123
PC
ABC-123-0001
PCSEQ
4
4
40
ABC-123
PC
ABC-123-0001
PCSEQ
5
4
41
ABC-123
PC
ABC-123-0001
PCSEQ
6
4
42
ABC-123
PC
ABC-123-0001
PCSEQ
7
4
43
ABC-123
PC
ABC-123-0001
PCSEQ
8
4
44
ABC-123
PC
ABC-123-0001
PCSEQ
9
4
45
ABC-123
PC
ABC-123-0001
PCSEQ
10
4
46
ABC-123
PC
ABC-123-0001
PCSEQ
11
4
47
ABC-123
PC
ABC-123-0001
PCSEQ
12
4
48
ABC-123
PP
ABC-123-0001
PPSEQ
4
4
49
ABC-123
PP
ABC-123-0001
PPSEQ
5
4
50
ABC-123
PP
ABC-123-0001
PPSEQ
6
4
51
ABC-123
PP
ABC-123-0001
PPSEQ
7
4
The Day 8 relationships are the same as those shown in Example 1.
* RELID 1 indicates records with PCSEQ values of 1-4 and 6-12 are related to the record with a PPSEQ value of 1.
* RELID 2 indicates records with PCSEQ values of 1-5 and 7-12 are related to the record with a PPSEQ value of 2.
* RELID 3 indicates records with PCSEQ values of 1-10 are related to the record with a PPSEQ value of 3.
* RELID 4 indicates records with PCSEQ values of 1-12 are related to the records with PPSEQ values of 4-7.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 193
Final November 12, 2008
6.3.10.6 Conclusions
Relating the datasets (1305HSection 6.3.10.5.1, and as described in 1306HSection 8.3) is the simplest method; however, all time-
point concentrations in PC must be used to calculate all parameters for all subjects. If datasets cannot be related,
then individual subject records must be related (1307HSection 8.2). In either case, the values of PCGRPID and PPGRPID
must take into account multiple analytes and multiple reference time points, if they exist.
Method A, is clearly the most efficient in terms of having the least number of RELREC records, but it does require
the assignment of --GRPID values (which are optional) in both the PC and PP datasets. Method D, in contrast, does
not require the assignment of --GRPID values, instead relying on the required --SEQ values in both datasets to relate
the records. Although Method D results in the largest number of RELREC records compared to the other methods, it
may be the easiest to implement consistently across the range of complexities shown in the examples. Two
additional methods, Methods B and C, are also shown for Examples 1-3. They represent hybrid approaches, using
--GRPID values on only one dataset (PP and PC, respectively) and --SEQ values for the other. These methods are
best suited for sponsors who want to minimize the number of RELREC records while not having to assign --GRPID
values in both domains. Methods B and C would not be ideal, however, if one expected complex scenarios such as
that shown in Example 4.
Please note that an attempt has been made to approximate real PK data; however, the example values are not
intended to reflect data used for actual analysis. When certain time-point concentrations have been omitted from PP
calculations in Examples 2-4, the actual parameter values in the PP dataset have not been recalculated from those in
Example 1 to reflect those omissions.
6.3.10.7 Suggestions for Implementing RELREC in the Submission of PK Data
Determine which of the scenarios best reflects how PP data are related to PC data. Questions that should be
considered:
1. Do all parameters for each PK profile use all concentrations for all subjects? If so, create a PPGRPID value
for all PP records and a PCGRPID value for all PC records for each profile for each subject, analyte, and
reference time point. Decide whether to relate datasets (1308HSection 6.3.10.5.1) or records (1309HSection 6.3.10.5.2,
Example 1). If choosing the latter, create records in RELREC for each PCGRPID value and each PPGRPID
value (Method A). Use RELID to show which PCGRPID and PPGRPID records are related. Consider
RELREC Methods B, C, and D as applicable.
2. Do all parameters use the same concentrations, although maybe not all of them (Example 2)? If so, create a
single PPGRPID value for all PP records, and two PCGRPID values for the PC records: a PCGRPID value
for ones that were used and a PCGRPID value for those that were not used. Create records in RELREC for
each PCGRPID value and each PPGRPID value (Method A). Use RELID to show which PCGRPID and
PPGRPID records are related. Consider RELREC Methods B, C, and D as applicable.
3. Do any parameters use the same concentrations, but not as consistently as what is shown in Examples 1 and
2? If so, refer to Example 3. Assign a GRPID value to the PP records that use the same concentrations.
More than one PPGRPID value may be necessary. Assign as many PCGRPID values in the PC domain as
needed to group these records. Create records in RELREC for each PCGRPID value and each PPGRPID
value (Method A). Use RELID to show which PCGRPID and PPGRPID records are related. Consider
RELREC Methods B, C, and D as applicable.
4. If none of the above applies, or the data become difficult to group, then start with Example 4, and decide
which RELREC method would be easiest to implement and represent.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 194 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.4 FINDINGS ABOUT EVENTS OR INTERVENTIONS
Findings About Events or Interventions is a specialization of the Findings General Observation Class. As such, it
shares all qualities and conventions of Findings observations but is specialized by the addition of the --OBJ variable.
6.4.1 WHEN TO USE FINDINGS ABOUT
It is intended, as its name implies, to be used when collected data represent ‖findings about‖ an Event or
Intervention that cannot be represented within an Event or Intervention record or as a Supplemental Qualifier to such
a record. Examples include the following:
Data or observations that have different timing from an associated Event or Intervention as a whole:
For example, if severity of an AE is collected at scheduled time points (e.g., per visit) throughout the
duration of the AE, the severities have timing that are different from that of the AE as a whole. Instead, the
collected severities represent ―snapshots‖ of the AE over time.
Data or observations about an Event or Intervention which have Qualifiers of their own that can be represented
in Findings variables (e.g., units, method):
These Qualifiers can be grouped together in the same record to more accurately describe their context and
meaning (rather than being represented by multiple Supplemental Qualifier records). For example, if the
size of a rash is measured, then the result and measurement unit (e.g., centimeters or inches) can be
represented in the Findings About domain in a single record, while other information regarding the rash
(e.g., start and end times), if collected would appear in an Event record.
Data or observations about an Event or Intervention for which no Event or Intervention record has been
collected or created:
For example, if details about a condition (e.g., primary diagnosis) are collected, but the condition was not
collected as Medical History because it was a prerequisite for study participation, then the data can be
represented as results in the Findings About domain, and the condition as the Object of the Observation
(see 1310HSection 6.4.3).
Data or information about an Event or Intervention that indicate the occurrence of related symptoms or
therapies:
Depending on the Sponsor‘s definitions of reportable events or interventions and regulatory agreements,
representing occurrence observations in either the Findings About domain or the appropriate Event or
Intervention domain(s) is at the Sponsor‘s discretion. For example, in a migraine study, when symptoms
related to a migraine event are queried and their occurrence is not considered either an AE or a record to be
represented in another Events domain, then the symptoms can be represented in the Findings About
domain.
Data or information that indicate the occurrence of pre-specified AEs:
Since there is a requirement that every record in the AE domain represent an event that actually occurred,
AE probing questions that are answered in the negative (e.g., did not occur, unknown, not done) cannot be
stored in the AE domain. Therefore, answers to probing questions about the occurrence of pre-specified
adverse events can be stored in the Findings About domain, and for each positive response (i.e., where
occurrence indicates yes) there should be a record reflected in the AE domain. The Findings About record
and the AE record may be linked via RELREC.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 195
Final November 12, 2008
6.4.2 NAMING FINDINGS ABOUT DOMAINS
The FA domain is defined to store Findings About Events or Interventions. Sponsors may choose to split the domain
into physically separate datasets following guidance described in 1311HSection 4.1.1.7. For example, if Findings About
clinical events and Findings About reproductive events are collected in a study, and are considered separate and
unrelated observations, then they could be split relative to their respective parent domains.
The DOMAIN value would be ―FA‖
Variables that require a prefix would use ―FA‖
Variables of the same name in multiple datasets should have the same SAS Length attribute.
The dataset names would be the domain name plus up to two additional characters indicating the parent
domain (e.g., FACE for the Findings About clinical events and FARE for findings about reproductive events,
where in this example, ―RE‖ is a custom domain to store reproductive events data).
FASEQ must be unique within USUBJID for all records across the split datasets.
Supplemental Qualifier datasets would need to be managed at the split-file level, for example, suppface.xpt
and suppfare.xpt and RDOMAIN would be defined as ―FA‖.
If a dataset-level RELREC is defined (e.g., between the CE and FACE datasets), then RDOMAIN may
contain up to four characters to effectively describe the relationship between the CE parent records with the
FACE child records.
As described above, if domain splitting is implemented then the dataset name will combine the prefix ―FA‖ with the
two-lettered domain code of the parent record. For example, dataset facm.xpt would store Findings About
Concomitant Medications.
6.4.3 VARIABLES UNIQUE TO FINDINGS ABOUT
The variable, --OBJ, is unique to Findings About. In conjunction with FATESTCD, it describes what the topic of the
observation is; therefore both are required to be populated for every record. FATESTCD describes the
measurement/evaluation and FAOBJ describes the Event or Intervention that the measurement/evaluation is about.
When collected data fit a Qualifier variable listed in SDTM Sections 2.2.1 or 2.2.2, and are represented in the
Findings About domain, then the name of the variable should be used as the value of FATESTCD. For example,
FATESTCD
FATEST
OCCUR
Occurrence
SEV
Severity/Intensity
TOXGR
Toxicity Grade
The use of the same names (e.g., SEV, OCCUR) for both Qualifier variables in the observation classes and
FATESTCD is deliberate, but should not lead users to conclude that the collection of such data (e.g.,
severity/intensity, occurrence) must be stored in the Findings About domain. In fact, data should only be stored in
the Findings About domain if they do not fit in the general-observation-class domain. If the data describe the
underlying Event or Intervention as a whole and share it‘s timing, then the data should be stored as a qualifier of the
general-observation-class record.
In general, the value in FAOBJ should match the value in --TERM or --TRT, unless the parent domain is dictionary
coded or subject to controlled terminology, in which case FAOBJ should then match the value in --DECOD.
Representing collected relationships supporting Findings About data are described in 1312HSection 8.6 and are
demonstrated in the examples below (1313HSection 6.4.6).
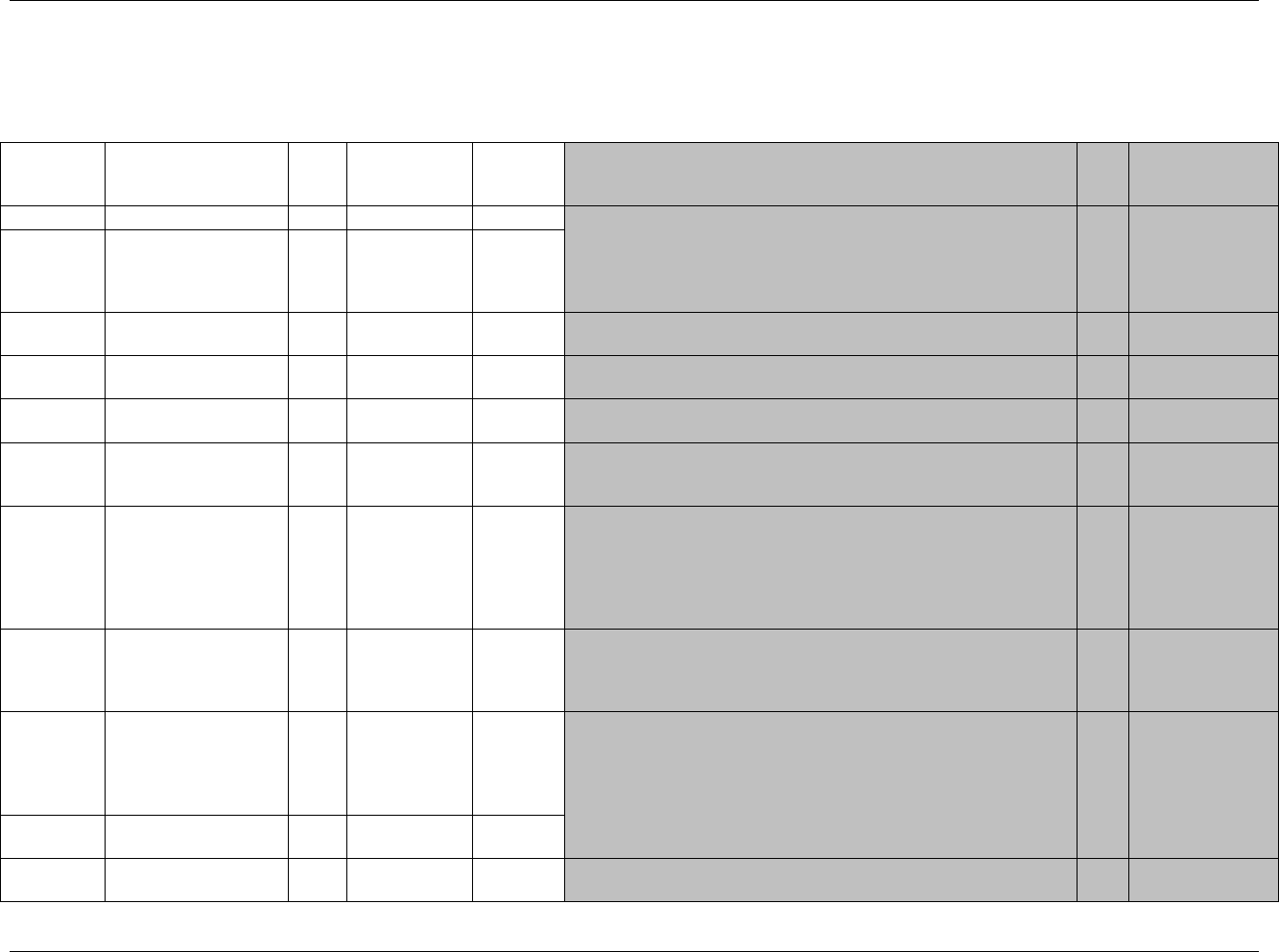
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 196 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
6.4.4 FINDINGS ABOUT (FA) DOMAIN MODEL
fa.xpt, Findings About Events or Interventions — Findings Sub-Class, Version 3.1.2. One record per finding, per object, per time point, per visit per
subject Tabulation
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
SDTM 2.2.4
DOMAIN
Domain Abbreviation
Char
1314H(FA)
Identifier
Two-character abbreviation for the domain.
Req
SDTM 2.2.4,
1315HSDTMIG 4.1.2.2,
1316HSDTMIG Appendix
C2
USUBJID
Unique Subject Identifier
Char
Identifier
Identifier used to uniquely identify a subject across all studies for all
applications or submissions involving the product.
Req
SDTM 2.2.4,
1317HSDTMIG 4.1.2.3
FASEQ
Sequence Number
Num
Identifier
Sequence Number given to ensure uniqueness of subject records within a
domain. May be any valid number.
Req
SDTM 2.2.4
FAGRPID
Group ID
Char
Identifier
Used to tie together a block of related records in a single domain for a
subject.
Perm
SDTM 2.2.4,
1318HSDTMIG 4.1.2.6
FASPID
Sponsor-Defined
Identifier
Char
Identifier
Sponsor-defined reference number. Perhaps pre-printed on the CRF as an
explicit line identifier or defined in the sponsor‘s operational database.
Example: Line number on a CRF.
Perm
SDTM 2.2.4,
1319HSDTMIG 4.1.2.6
FATESTCD
Findings About Test
Short Name
Char
*
Topic
Short name of the measurement, test, or examination described in
FATEST. It can be used as a column name when converting a dataset
from a vertical to a horizontal format. The value in FATESTCD cannot
be longer than 8 characters, nor can it start with a number (e.g.
―1TEST‖). FATESTCD cannot contain characters other than letters,
numbers, or underscores. Example: SEV, OCCUR.
Req
SDTM 2.2.3,
1320H1321HSDTMIG 4.1.1.8,
1322HSDTMIG 4.1.2.1
FATEST
Findings About Test
Name
Char
*
Synonym
Qualifier
Verbatim name of the test or examination used to obtain the
measurement or finding. The value in FATEST cannot be longer than 40
characters. Examples: Severity/Intensity, Occurrence
Req
SDTM 2.2.3,
1323HSDTMIG 4.1.2.1,
1324HSDTMIG 4.1.2.4,
1325H1326HSDTMIG 4.1.5.3.1
FAOBJ
Object of the Observation
Char
Record
Qualifier
Used to describe the object or focal point of the findings observation that
is represented by --TEST. Examples: the term (such as Acne) describing
a clinical sign or symptom that is being measured by a Severity test, or
an event such as VOMIT where the volume of Vomit is being measured
by a VOLUME test.
Req
SDTM 2.2.3.1
FACAT
Category for Findings
About
Char
*
Grouping
Qualifier
Used to define a category of related records. Examples: GERD,
PRE-SPECIFIED AE.
Perm
SDTM 2.2.3,
1327HSDTMIG 4.1.2.6
FASCAT
Subcategory for Findings
About
Char
*
Grouping
Qualifier
A further categorization of FACAT.
Perm
SDTM 2.2.3,
1328HSDTMIG 4.1.2.6
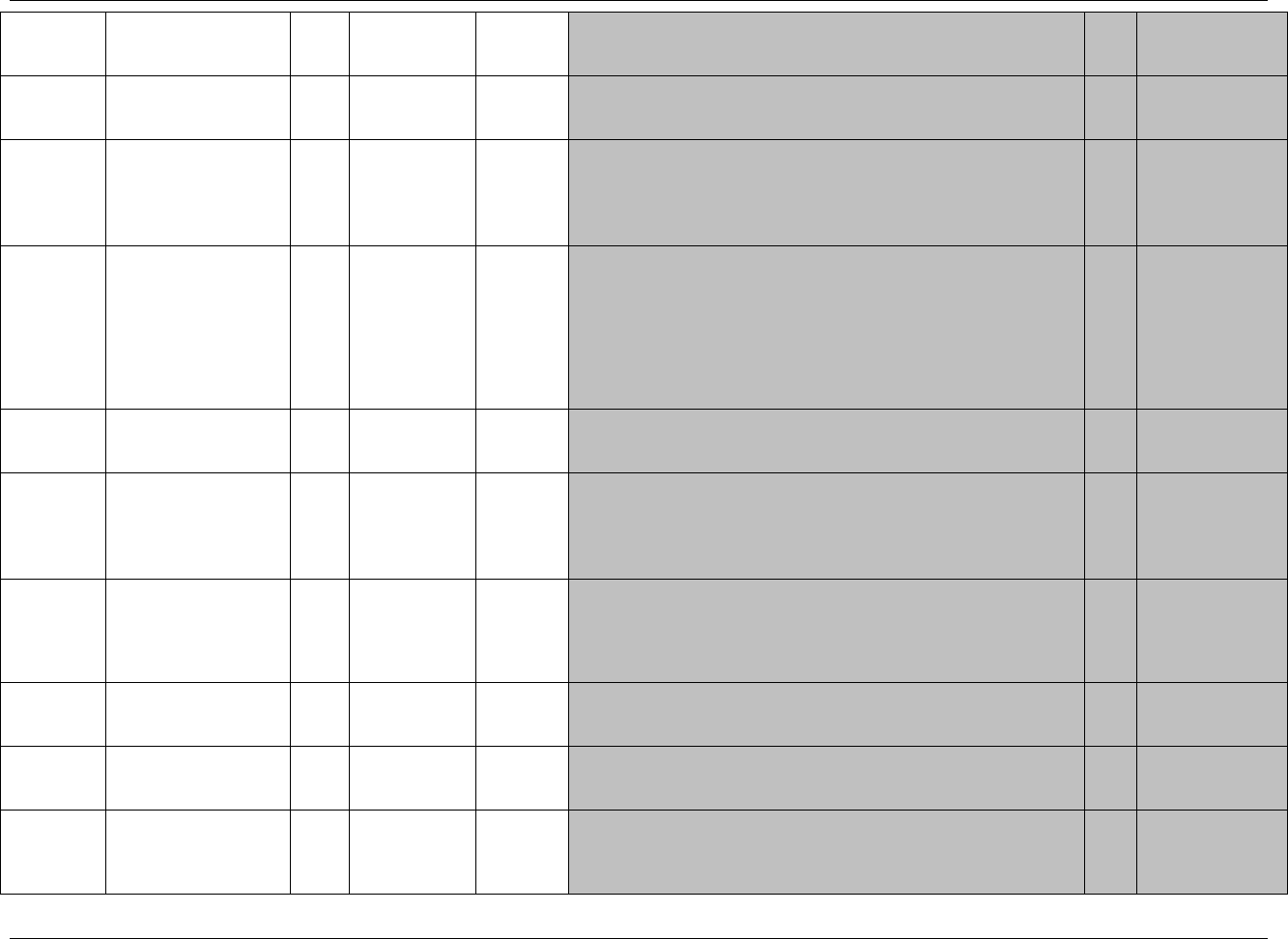
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 197
Final November 12, 2008
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
FAORRES
Result or Finding in
Original Units
Char
Result
Qualifier
Result of the test as originally received or collected.
Exp
SDTM 2.2.3,
1329HSDTMIG 4.1.3.6,
1330HSDTMIG 4.1.5.1
FAORRESU
Original Units
Char
(1331HUNIT)
Variable
Qualifier
Original units in which the data were collected. The unit for FAORRES.
Perm
SDTM 2.2.3,
1332HSDTMIG 4.1.3.2,
1333HSDTMIG 4.1.5.1,
SDTMIG Appendix
C1
FASTRESC
Character Result/Finding
in Std Format
Char
Result
Qualifier
Contains the result value for all findings, copied or derived from
FAORRES in a standard format or standard units. FASTRESC should
store all results or findings in character format; if results are numeric,
they should also be stored in numeric format in FASTRESN. For
example, if a test has results ―NONE‖, ―NEG‖, and ―NEGATIVE‖ in
FAORRES and these results effectively have the same meaning; they
could be represented in standard format in FASTRESC as
―NEGATIVE‖.
Exp
SDTM 2.2.3,
1334HSDTMIG 4.1.3.6,
1335HSDTMIG 4.1.5.1
FASTRESN
Numeric Result/Finding
in Standard Units
Num
Result
Qualifier
Used for continuous or numeric results or findings in standard format;
copied in numeric format from FASTRESC. FASTRESN should store all
numeric test results or findings.
Perm
SDTM 2.2.3,
1336HSDTMIG 4.1.5.1
FASTRESU
Standard Units
Char
(1337HUNIT)
Variable
Qualifier
Standardized unit used for FASTRESC and FASTRESN.
Perm
SDTM 2.2.3,
1338HSDTMIG 4.1.3.2,
1339HSDTMIG 4.1.5.1,
SDTMIG Appendix
C1
FASTAT
Completion Status
Char
(1998HND)
Record
Qualifier
Used to indicate that the measurement was not done. Should be null if a
result exists in FAORRES.
Perm
SDTM 2.2.3,
1340HSDTMIG 4.1.5.1,
1341HSDTMIG 4.1.5.7,
1342HSDTMIG Appendix
C1
FAREASND
Reason Not Performed
Char
Record
Qualifier
Describes why a question was not answered. Example: subject refused.
Used in conjunction with FASTAT when value is NOT DONE.
Perm
SDTM 2.2.3,
1343HSDTMIG 4.1.5.1,
1344HSDTMIG 4.1.5.7
FALOC
Location of the Finding
About
Char
(1345HLOC)
Variable
Qualifier
Used to specify the location of the clinical evaluation. Example: LEFT
ARM
Perm
SDTM 2.2.3,
SDTMIG Appendix
C1
FABLFL
Baseline Flag
Char
(1999HNY)
Record
Qualifier
Indicator used to identify a baseline value. The value should be ―Y‖ or
null.
Perm
SDTM 2.2.3,
1346HSDTMIG 4.1.3.7,
1347HSDTMIG Appendix
C1
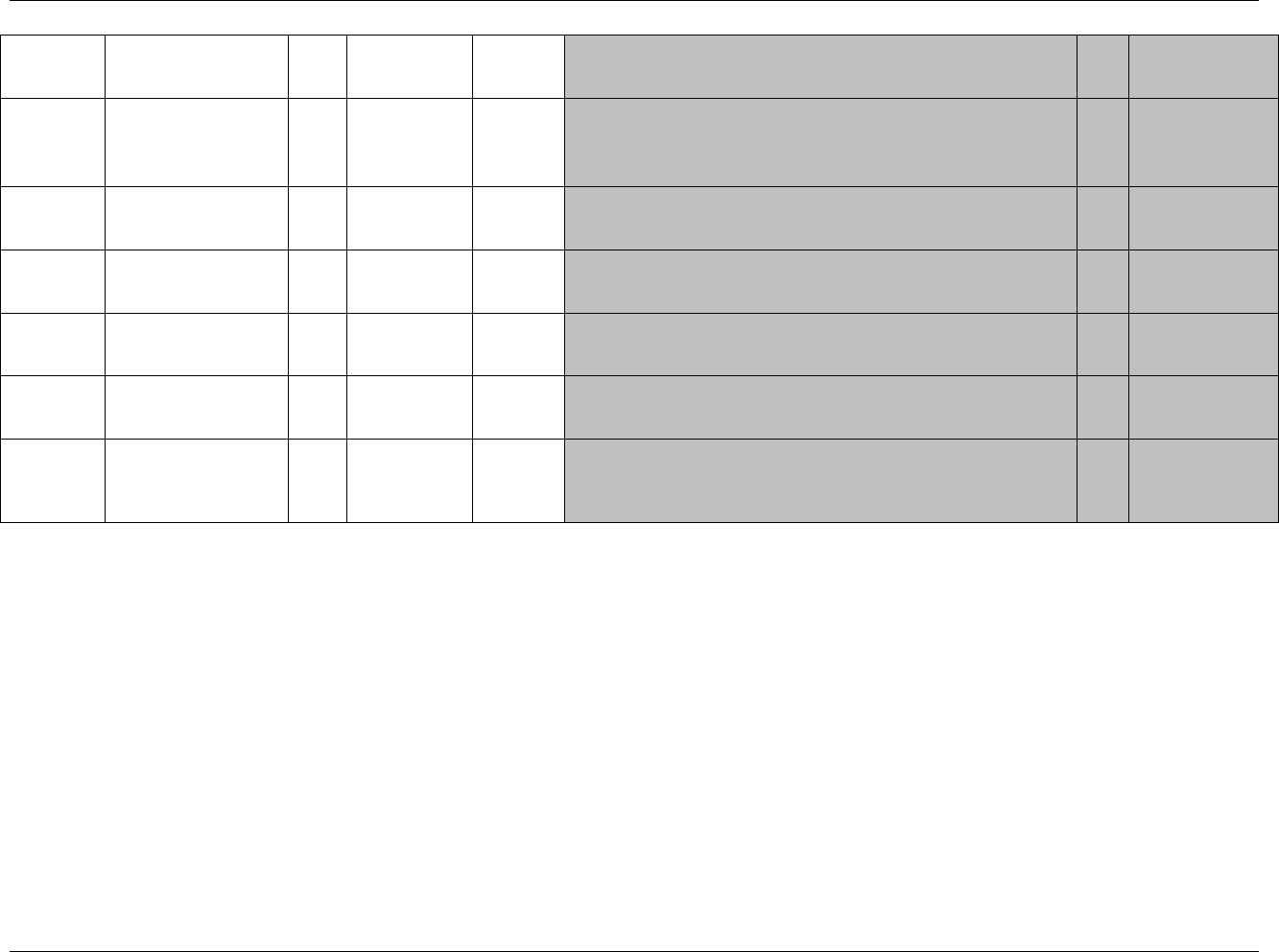
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 198 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Variable
Name
Variable Label
Type
Controlled
Terms, Codelist
or Format
Role
CDISC Notes
Core
References
FAEVAL
Evaluator
Char
*
Record
Qualifier
Role of the person who provided the evaluation. Used only for results
that are subjective (e.g., assigned by a person or a group). Should be null
for records that contain collected or derived data. Examples:
INVESTIGATOR, ADJUDICATION COMMITTEE, VENDOR.
Perm
SDTM 2.2.3,
1348HSDTMIG 4.1.5.4
VISITNUM
Visit Number
Num
Timing
1. Clinical encounter number.
2. Numeric version of VISIT, used for sorting.
Exp
SDTM 2.2.5,
1349HSDTMIG 4.1.4.5,
1350HSDTMIG 7.4
VISIT
Visit Name
Char
Timing
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY.
Perm
SDTM 2.2.5,
1351HSDTMIG 4.1.4.5,
1352HSDTMIG 7.4
VISITDY
Planned Study Day of
Visit
Num
Timing
Planned study day of the visit based upon RFSTDTC in Demographics.
Perm
SDTM 2.2.5,
1353HSDTMIG 4.1.4.5,
1354HSDTMIG 7.4
FADTC
Date/Time of Collection
Char
ISO 8601
Timing
Perm
SDTM 2.2.5,
1355HSDTMIG 4.1.4.1,
1356HSDTMIG 4.1.4.8
FADY
Study Day of Collection
Num
Timing
1. Study day of collection, measured as integer days.
2. Algorithm for calculations must be relative to the sponsor-defined
RFSTDTC variable in Demographics. This formula should be consistent
across the submission.
Perm
SDTM 2.2.5,
1357HSDTMIG 4.1.4.4,
1358HSDTMIG 4.1.4.6
Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
6.4.5 ASSUMPTIONS FOR FINDINGS ABOUT DOMAIN MODEL
1. The following qualifiers should generally not be used in FA: --BODSYS, --MODIFY, --SEV, --TOXGR.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 199
Final November 12, 2008
6.4.6 FINDINGS ABOUT EXAMPLES
Example 1: Migraine Symptoms Diary
The form shown below collects severity and symptoms data at multiple time points about a migraine event.
Migraine Symptoms Diary
Migraine Reference Number
xx
When did the migraine start
DD-MMM-YYYY
HH:MM
Answer the following 5 Minutes BEFORE Dosing
Severity of Migraine
○ Mild ○ Moderate ○ Severe
Associated Symptoms:
Sensitivity to light
Sensitivity to sound
Nausea
Aura
○ No ○ Yes
○ No ○ Yes
○ No ○ Yes
○ No ○ Yes
Answer the following 30 Minutes AFTER Dosing
Severity of Migraine
○ Mild ○ Moderate ○ Severe
Associated Symptoms:
Sensitivity to light
Sensitivity to sound
Nausea
Aura
○ No ○ Yes
○ No ○ Yes
○ No ○ Yes
○ No ○ Yes
Answer the following 90 Minutes AFTER Dosing
Severity of Migraine
○ Mild ○ Moderate ○ Severe
Associated Symptoms:
Sensitivity to light
Sensitivity to sound
Nausea
Aura
○ No ○ Yes
○ No ○ Yes
○ No ○ Yes
○ No ○ Yes
The collected data below the migraine start date on the CRF meet the following Findings About criteria: 1) Data that do not describe an Event or Intervention as a
whole and 2) Data that indicate the occurrence of related symptoms.
In this mock scenario, the Sponsor‘s conventions and/or reporting agreements consider migraine as a clinical event (as opposed to a reportable AE) and consider
the pre-specified symptom responses as findings about the migraine, therefore the data are represented in the Findings About domain with FATESTCD =
‖OCCUR‖ and FAOBJ defined as the symptom description. Therefore, the mock datasets represent (1) The migraine event record in the CE domain, (2) The
severity and symptoms data, per time point, in the Findings About domain, and (3) A dataset-level relationship in RELREC based on the sponsor ID (--SPID)
value which was populated with a system generated identifier unique to each iteration of this form.
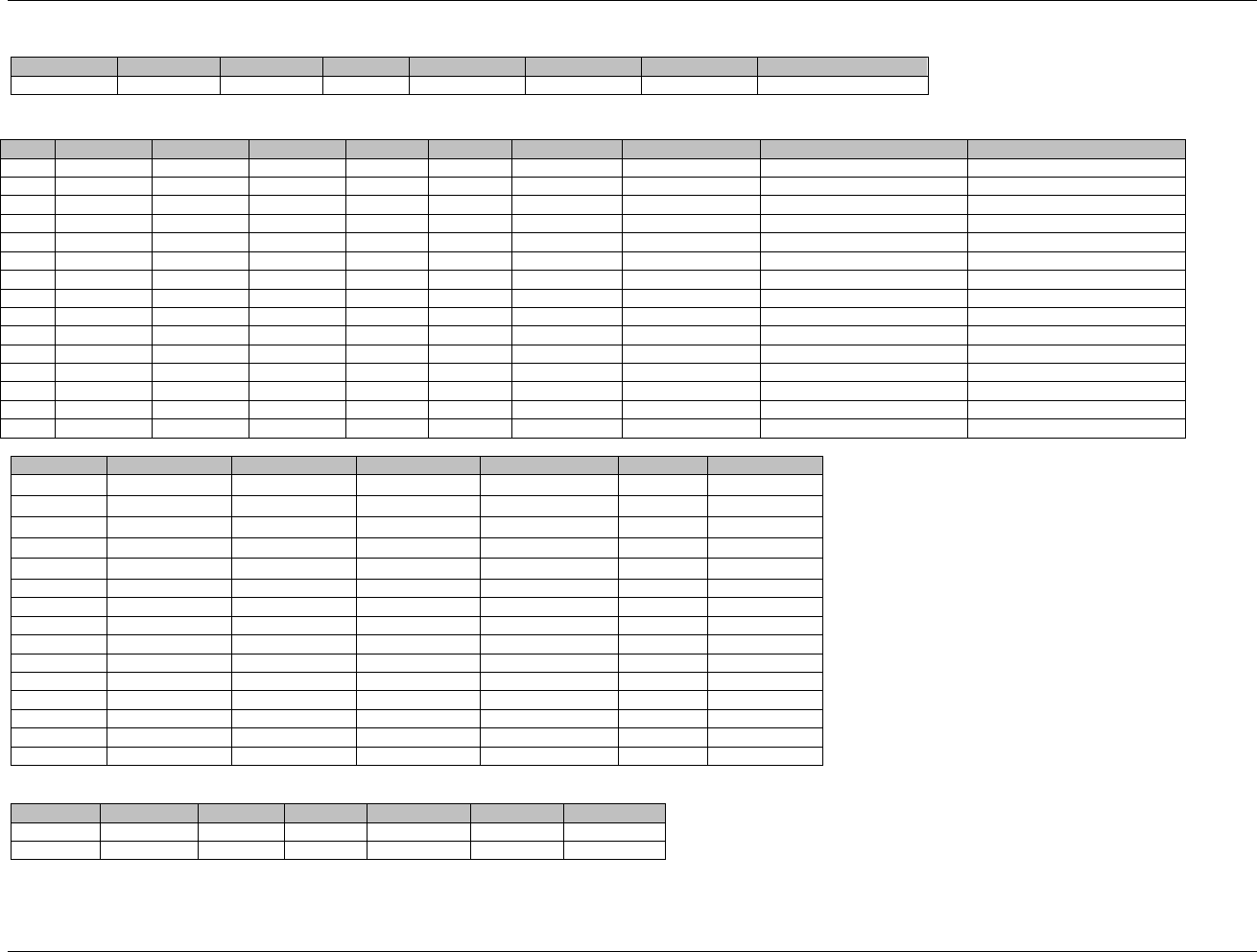
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 200 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
ce.xpt
STUDYID
DOMAIN
USUBJID
CESEQ
CESPID
CETERM
CEDECOD
CESTDTC
ABC
CE
ABC-123
1
90567
Migraine
Migraine
2007-05-16T10:30
fa.xpt
Row
STUDYID
DOMAIN
USUBJID
FASEQ
FASPID
FATESTCD
FATEST
FAOBJ
FACAT
1
ABC
FA
ABC-123
1
90567
SEV
Severity/Intensity
Migraine
MIGRAINE SYMPTOMS
2
ABC
FA
ABC-123
2
90567
OCCUR
Occurrence
Sensitivity To Light
MIGRAINE SYMPTOMS
3
ABC
FA
ABC-123
3
90567
OCCUR
Occurrence
Sensitivity To Sound
MIGRAINE SYMPTOMS
4
ABC
FA
ABC-123
4
90567
OCCUR
Occurrence
Nausea
MIGRAINE SYMPTOMS
5
ABC
FA
ABC-123
6
90567
OCCUR
Occurrence
Aura
MIGRAINE SYMPTOMS
6
ABC
FA
ABC-123
7
90567
SEV
Severity/Intensity
Migraine
MIGRAINE SYMPTOMS
7
ABC
FA
ABC-123
8
90567
OCCUR
Occurrence
Sensitivity To Light
MIGRAINE SYMPTOMS
8
ABC
FA
ABC-123
9
90567
OCCUR
Occurrence
Sensitivity To Sound
MIGRAINE SYMPTOMS
9
ABC
FA
ABC-123
10
90567
OCCUR
Occurrence
Nausea
MIGRAINE SYMPTOMS
10
ABC
FA
ABC-123
12
90567
OCCUR
Occurrence
Aura
MIGRAINE SYMPTOMS
11
ABC
FA
ABC-123
13
90567
SEV
Severity/Intensity
Migraine
MIGRAINE SYMPTOMS
12
ABC
FA
ABC-123
14
90567
OCCUR
Occurrence
Sensitivity To Light
MIGRAINE SYMPTOMS
13
ABC
FA
ABC-123
15
90567
OCCUR
Occurrence
Sensitivity To Sound
MIGRAINE SYMPTOMS
14
ABC
FA
ABC-123
16
90567
OCCUR
Occurrence
Nausea
MIGRAINE SYMPTOMS
15
ABC
FA
ABC-123
18
90567
OCCUR
Occurrence
Aura
MIGRAINE SYMPTOMS
Row
FAORRES
FASTRESC
FADTC
FATPT
FAELTM
FATPTREF
1 (cont’d)
SEVERE
SEVERE
2007-05-16
5M PRE-DOSE
-PT5M
DOSING
2 (cont’d)
Y
Y
2007-05-16
5M PRE-DOSE
-PT5M
DOSING
3 (cont’d)
N
N
2007-05-16
5M PRE-DOSE
-PT5M
DOSING
4 (cont’d)
Y
Y
2007-05-16
5M PRE-DOSE
-PT5M
DOSING
5 (cont’d)
Y
Y
2007-05-16
5M PRE-DOSE
-PT5M
DOSING
6 (cont’d)
MODERATE
MODERATE
2007-05-16
30M POST-DOSE
PT30M
DOSING
7 (cont’d)
Y
Y
2007-05-16
30M POST-DOSE
PT30M
DOSING
8 (cont’d)
N
N
2007-05-16
30M POST-DOSE
PT30M
DOSING
9 (cont’d)
N
N
2007-05-16
30M POST-DOSE
PT30M
DOSING
10 (cont’d)
Y
Y
2007-05-16
30M POST-DOSE
PT30M
DOSING
11 (cont’d)
MILD
MILD
2007-05-16
90M POST-DOSE
PT90M
DOSING
12 (cont’d)
N
N
2007-05-16
90M POST-DOSE
PT90M
DOSING
13 (cont’d)
N
N
2007-05-16
90M POST-DOSE
PT90M
DOSING
14 (cont’d)
N
N
2007-05-16
90M POST-DOSE
PT90M
DOSING
15 (cont’d)
N
N
2007-05-16
90M POST-DOSE
PT90M
DOSING
RELREC
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
ABC
CE
CESPID
ONE
1
ABC
FA
FASPID
MANY
1
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 201
Final November 12, 2008
Example 2: Rash Assessment
This CRF collects details about rash events at each visit, until resolved.
Rash Assessment
Date of Assessment
DD-MMM-YYYY
Associated AE reference number
Rash Size
________ ○ cm ○ in
Lesion Type & Count
Macules
○ 0 ○ 1 to 25 ○ 26 to 100 ○ 101 to 200 ○ 201 to 300 ○ >300
Papules
○ 0 ○ 1 to 25 ○ 26 to 100 ○ 101 to 200 ○ 201 to 300 ○ >300
Vesicles
○ 0 ○ 1 to 25 ○ 26 to 100 ○ 101 to 200 ○ 201 to 300 ○ >300
Pustules
○ 0 ○ 1 to 25 ○ 26 to 100 ○ 101 to 200 ○ 201 to 300 ○ >300
Scabs
○ 0 ○ 1 to 25 ○ 26 to 100 ○ 101 to 200 ○ 201 to 300 ○ >300
Scars
○ 0 ○ 1 to 25 ○ 26 to 100 ○ 101 to 200 ○ 201 to 300 ○ >300
The collected data meet the following Findings About criteria: 1) Data that do not describe an Event or Intervention as a whole and 2) Data (―about‖ an Event or
Intervention) which have Qualifiers of their own that can be represented in Findings variables (e.g., units, method)
In this mock scenario, the rash event is considered a reportable AE; therefore the form design collects a reference number to the AE form where the event is
captured. Data points collected on the Rash Assessment form can be represented in the Findings About domain and related to the AE via RELREC. Note, in the
mock datasets below, the AE started on May 10, 2007 and the Rash assessment was conducted on May 12 and May 19, 2007. Certain Required or Expected
variables have been omitted in consideration of space and clarity.
ae.xpt
STUDYID
DOMAIN
USUBJID
AESEQ
AESPID
AETERM
AEDECOD
AEBODSYS
AELOC
AESEV
AESER
AEACN
AESTDTC
XYZ
AE
XYZ-789
47869
5
Injection
site rash
Injection site
rash
General disorders and
administration site conditions
LEFT
ARM
MILD
N
NOT
APPLICABLE
2007-05-10
fa.xpt
Row
STUDYID
DOMAIN
USUBJID
FASEQ
FASPID
FATESTCD
FATEST
FAOBJ
FAORRES
FAORRESU
VISIT
FADTC
1
XYZ
FA
XYZ-789
123451
5
SIZE
Size
Injection Site Rash
2.5
IN
VISIT 3
2007-05-12
2
XYZ
FA
XYZ-789
123452
5
COUNT
Count
Macules
26 to 100
VISIT 3
2007-05-12
3
XYZ
FA
XYZ-789
123453
5
COUNT
Count
Papules
1 to 25
VISIT 3
2007-05-12
4
XYZ
FA
XYZ-789
123454
5
COUNT
Count
Vesicles
0
VISIT 3
2007-05-12
5
XYZ
FA
XYZ-789
123455
5
COUNT
Count
Pustules
0
VISIT 3
2007-05-12
6
XYZ
FA
XYZ-789
123456
5
COUNT
Count
Scabs
0
VISIT 3
2007-05-12
7
XYZ
FA
XYZ-789
123457
5
COUNT
Count
Scars
0
VISIT 3
2007-05-12
8
XYZ
FA
XYZ-789
123459
5
SIZE
Size
Injection Site Rash
1
IN
VISIT 4
2007-05-19
9
XYZ
FA
XYZ-789
123460
5
COUNT
Count
Macules
1 to 25
VISIT 4
2007-05-19
10
XYZ
FA
XYZ-789
123461
5
COUNT
Count
Papules
1 to 25
VISIT 4
2007-05-19
11
XYZ
FA
XYZ-789
123462
5
COUNT
Count
Vesicles
0
VISIT 4
2007-05-19
12
XYZ
FA
XYZ-789
123463
5
COUNT
Count
Pustules
0
VISIT 4
2007-05-19
13
XYZ
FA
XYZ-789
123464
5
COUNT
Count
Scabs
0
VISIT 4
2007-05-19
14
XYZ
FA
XYZ-789
123465
5
COUNT
Count
Scars
0
VISIT 4
2007-05-19
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 202 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
RELREC
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
XYZ
AE
AESPID
ONE
23
XYZ
FA
FASPID
MANY
23
Example 3: Rheumatoid Arthritis History
The form below collects information about rheumatoid arthritis. In this mock scenario, rheumatoid arthritis is a prerequisite for participation in an osteoporosis
trial and was not collected as a Medical History event.
Rheumatoid Arthritis History
Date of Assessment
DD-MMM-YYYY
During the past 6 months, how would you rate the following:
Joint stiffness
○ MILD ○ MODERATE ○ SEVERE
Inflammation
○ MILD ○ MODERATE ○ SEVERE
Joint swelling
○ MILD ○ MODERATE ○ SEVERE
Joint pain (arthralgia)
○ MILD ○ MODERATE ○ SEVERE
Malaise
○ MILD ○ MODERATE ○ SEVERE
Duration of early morning stiffness (hours and minutes)
_____Hours _____Minutes
The collected data meet the following Findings About criteria: Data (―about‖ an Event or Intervention) for which no Event or Intervention record has been
collected or created
In this mock scenario, the rheumatoid arthritis history was assessed on August 13, 2006.
fa.xpt
Row
STUDYID
DOMAIN
USUBJID
FASEQ
FATESTCD
FATEST
FAOBJ
1
ABC
FA
ABC-123
1
SEV
Severity/Intensity
Joint Stiffness
2
ABC
FA
ABC-123
2
SEV
Severity/Intensity
Inflammation
3
ABC
FA
ABC-123
3
SEV
Severity/Intensity
Joint Swelling
4
ABC
FA
ABC-123
4
SEV
Severity/Intensity
Arthralgia
5
ABC
FA
ABC-123
5
SEV
Severity/Intensity
Malaise
6
ABC
FA
ABC-123
6
DUR
Duration
Early Morning Stiffness
Row
FACAT
FAORRES
FASTRESC
FADTC
FAEVLINT
1 (cont’d)
RHEUMATOID ARTHRITIS HISTORY
SEVERE
SEVERE
2006-08-13
-P6M
2 (cont’d)
RHEUMATOID ARTHRITIS HISTORY
MODERATE
MODERATE
2006-08-13
-P6M
3 (cont’d)
RHEUMATOID ARTHRITIS HISTORY
MODERATE
MODERATE
2006-08-13
-P6M
4 (cont’d)
RHEUMATOID ARTHRITIS HISTORY
MODERATE
MODERATE
2006-08-13
-P6M
5 (cont’d)
RHEUMATOID ARTHRITIS HISTORY
MILD
MILD
2006-08-13
-P6M
6 (cont’d)
RHEUMATOID ARTHRITIS HISTORY
PT1H30M
PT1H30M
2006-08-13
-P6M

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 203
Final November 12, 2008
Example 4: Findings About Fracture Events
In this example, details about bone-fracture events are collected. This form is designed to collect multiple entries of fracture information including an initial entry
for the most recent fracture prior to study participation, as well as entry of information for fractures that occur during the study.
Bone Fracture Assessment
Complete form for most recent fracture prior to study
participation.
Enter Fracture Event Reference Number for all
fractures occurring during study participation:
_____
How did fracture occur
○ Pathologic
○ Fall
○ Other trauma
○ Unknown
What was the outcome
○ Normal Healing
○ Complications
Select all that apply:
□ Complication x
□ Complication y
□ Complication z
Additional therapeutic measures required
○ No
○ Unknown
○ Yes
Select all that apply
□ Therapeutic measure a
□ Therapeutic measure b
□ Therapeutic measure c
The collected data meet the following Findings About criteria: (1) Data (―about‖ an Event or Intervention) that indicate the occurrence of related symptoms or
therapies and (2) Data (―about‖ an event/intervention) for which no Event or Intervention record has been collected or created
Determining when data further describe the parent event record either as Variable Qualifiers or Supplemental Qualifiers may be dependent on data collection
design. In the above form, responses are provided for the most recent fracture but an event record reference number was not collected. But for in-study fracture
events, a reference number is collected which would allow representing the responses as part of the Event record either as Supplemental Qualifiers and/or
variables like --OUT and --CONTRT.
The below domains reflect responses to each Bone Fracture Assessment question. The historical-fracture responses that are without a parent record are
represented in the FA domain, while the current-fracture responses are represented as Event records with Supplemental Qualifiers.
Historical Fractures Having No Event Records

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 204 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
fa.xpt
STUDYID
DOMAIN
USUBJID
FASEQ
FASPID
FATESTCD
FATEST
FAOBJ
FACAT
FAORRES
FADTC
ABC
FA
ABC -US-
701-002
1
798654
REAS
Reason
Bone Fracture
BONE FRACTURE
ASSESSMENT - HISTORY
FALL
2006-04-10
ABC
FA
ABC -US-
701-002
2
798654
OUT
Outcome
Bone Fracture
BONE FRACTURE
ASSESSMENT - HISTORY
COMPLICATIONS
2006-04-10
ABC
FA
ABC -US-
701-002
3
798654
OCCUR
Occurrence
Complications
BONE FRACTURE
ASSESSMENT
Y
2006-04-10
ABC
FA
ABC -US-
701-002
4
798654
OCCUR
Occurrence
Therapeutic Measure
BONE FRACTURE
ASSESSMENT
Y
2006-04-10
suppfa.xpt
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
ABC
FA
ABC -US-701-002
FASEQ
1
FATYP
FA Type
MOST RECENT
CRF
ABC
FA
ABC -US-701-002
FASEQ
2
FATYP
FA Type
MOST RECENT
CRF
ABC
FA
ABC -US-701-002
FASEQ
3
FATYP
FA Type
MOST RECENT
CRF
ABC
FA
ABC -US-701-002
FASEQ
4
FATYP
FA Type
MOST RECENT
CRF
Current Fractures Having Event Records
ce.xpt
STUDYID
DOMAIN
USUBJID
CESEQ
CESPID
CETERM
CELOC
CEOUT
CECONTRT
CESTDTC
ABC
CE
ABC -US-701-002
1
1
Fracture
ARM
NORMAL HEALING
Y
2006-07-03
ABC
CE
ABC -US-701-002
2
2
Fracture
LEG
COMPLICATIONS
N
2006-10-15
suppce.xpt
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
ABC
CE
ABC -US-701-002
CESPID
1
REAS
Reason
FALL
CRF
ABC
CE
ABC -US-701-002
CESPID
2
REAS
Reason
OTHER TRAUMA
CRF
ABC
CE
ABC -US-701-002
CESPID
2
OUT
Outcome
COMPLICATIONS
CRF
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 205
Final November 12, 2008
Example 5: Pre-Specified Adverse Events
In this example, three AEs are pre-specified and are scheduled to be asked at each visit. If the occurrence is yes, then a complete AE record is collected on the AE
form.
Pre-Specified Adverse Events of Clinical Interest
Date of Assessment
DD-MMM-YYYY
Did the following occur? If Yes, then enter a complete record in the AE CRF
Headache
Respiratory infection
Nausea
○ No ○ Yes ○ Not Done
○ No ○ Yes ○ Not Done
○ No ○ Yes ○ Not Done
The collected data meet the following Findings About criteria: Data that indicate the occurrence of pre-specified adverse events.
In this mock scenario, each response to the pre-specified terms is represented in the Findings About domain. For the Y responses, an AE record is represented in
the AE domain with its respective Qualifiers and timing details. In the example below, the AE of ―Headache‖ encompasses multiple pre-specified Y responses
and the AE of ―Nausea‖ asked about on October 10, reported that it occurred and started on October 8 and ended on October 9. Note, in the example below, no
relationship was collected to link the yes responses with the AE entries, therefore no RELREC was created.
fa.xpt
STUDYID
DOMAIN
USUBJID
FASEQ
FATESTCD
FATEST
FAOBJ
FAORRES
FASTRESC
FASTAT
FADTC
VISITNUM
VISIT
QRS
FA
1234
1
OCCUR
Occurrence
Headache
Y
Y
2005-10-01
2
VISIT 2
QRS
FA
1234
2
OCCUR
Occurrence
Respiratory Infection
N
N
2005-10-01
2
VISIT 2
QRS
FA
1234
3
OCCUR
Occurrence
Nausea
NOT DONE
2005-10-01
2
VISIT 2
QRS
FA
1234
4
OCCUR
Occurrence
Headache
Y
Y
2005-10-10
3
VISIT 3
QRS
FA
1234
5
OCCUR
Occurrence
Respiratory Infection
N
N
2005-10-10
3
VISIT 3
QRS
FA
1234
6
OCCUR
Occurrence
Nausea
Y
Y
2005-10-10
3
VISIT 3
ae.xpt
STUDYID
DOMAIN
USUBJID
AESEQ
AETERM
AEDECOD
AEBODSYS
AESEV
AEACN
AEPRESP
AESTDTC
AEENDTC
QRS
AE
1234
1
Headache
Headache
Nervous system disorders
MILD
NONE
Y
2005-09-30
QRS
AE
1234
2
Nausea
Nausea
Gastrointestinal disorders
MODERATE
NONE
Y
2005-10-08
2005-10-09
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 206 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 6: Findings About GERD
In this example, the following CRF is used to capture data about pre-specified symptoms of the disease under study on a daily basis. The date of the
assessment is captured, but start and end timing of the events are not.
SYMPTOMS
INVESTIGATOR GERD SYMPTOM MEASUREMENT
VOLUME (mL)
NUMBER OF
EPISODES
MAXIMUM SEVERITY
None, Mild, Moderate, Severe
Vomiting
Diarrhea
Nausea
The collected data meet the following Findings About criteria: 1) data that do not describe an Event or Intervention as a whole, and 2) data (―about‖ an Event or
Intervention) having Qualifiers that can be represented in Findings variables (e.g., units, method).
The data below represent data from two visits for one subject. Records occur in blocks of three for Vomit, and in blocks of two for Diarrhea and Nausea.
Rows 1 -3: Show the results for the Vomiting tests at Visit 1.
Rows 4 and 5: Show the results for the Diarrhea tests at Visit 1.
Rows 6 and 7: Show the results for the Nausea tests at Visit 1.
Rows 8-10: Show the results for the Vomiting tests at Visit 2. These indicate that Vomiting was absent at Visit 2.
Rows 11 and 12: Show the results for the Diarrhea tests at Visit 2.
Rows 13 and 14: Indicate that Nausea was not assessed at Visit 2.
fa.xpt
Row
STUDYID
DOMAIN
USUBJID
FASEQ
FATESTCD
FATEST
FAOBJ
FACAT
1
XYZ
FA
XYZ-701-002
1
VOL
Volume
Vomit
GERD
2
XYZ
FA
XYZ-701-002
2
NUMEPISD
Number of Episodes
Vomit
GERD
3
XYZ
FA
XYZ-701-002
3
SEV
Severity/Intensity
Vomit
GERD
4
XYZ
FA
XYZ-701-002
4
NUMEPISD
Number of Episodes
Diarrhea
GERD
5
XYZ
FA
XYZ-701-002
5
SEV
Severity/Intensity
Diarrhea
GERD
6
XYZ
FA
XYZ-701-002
6
NUMEPISD
Number of Episodes
Nausea
GERD
7
XYZ
FA
XYZ-701-002
7
SEV
Severity/Intensity
Nausea
GERD
8
XYZ
FA
XYZ-701-002
8
VOL
Volume
Vomit
GERD
9
XYZ
FA
XYZ-701-002
9
NUMEPISD
Number of Episodes
Vomit
GERD
10
XYZ
FA
XYZ-701-002
10
SEV
Severity/Intensity
Vomit
GERD
11
XYZ
FA
XYZ-701-002
11
NUMEPISD
Number of Episodes
Diarrhea
GERD
12
XYZ
FA
XYZ-701-002
12
SEV
Severity/Intensity
Diarrhea
GERD
13
XYZ
FA
XYZ-701-002
13
NUMEPISD
Number of Episodes
Nausea
GERD
14
XYZ
FA
XYZ-701-002
14
SEV
Severity/Intensity
Nausea
GERD
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 207
Final November 12, 2008
Row
FAORRES
FAORRESU
FASTRESC
FASTRESU
VISIT
FASTAT
FADTC
1 (cont’d)
250
mL
250
mL
1
2006-02-02
2 (cont’d)
>10
>10
1
2006-02-02
3 (cont’d)
SEVERE
SEVERE
1
2006-02-02
4 (cont’d)
2
2
1
2006-02-02
5 (cont’d)
SEVERE
SEVERE
1
2006-02-02
6 (cont’d)
1
1
1
2006-02-02
7 (cont’d)
MODERATE
MODERATE
1
2006-02-02
8 (cont’d)
0
mL
0
mL
2
2006-02-03
9 (cont’d)
0
0
2
2006-02-03
10 (cont’d)
NONE
NONE
2
2006-02-03
11 (cont’d)
1
1
2
2006-02-03
12 (cont’d)
SEVERE
SEVERE
2
2006-02-03
13 (cont’d)
2
NOT DONE
2006-02-03
14 (cont’d)
2
NOT DONE
2006-02-03
Example 7: Findings About GERD
This example is similar to the one above except that with the following CRF, which includes a separate column to collect the occurrence of symptoms,
measurements are collected only for symptoms that occurred. There is a record for the occurrence test for each symptom. If Vomiting occurs, there are 3
additional records, and for each occurrence of Diarrhea or Nausea there are two additional records.
Whether there are adverse event records related to these symptoms depends on agreements in place for the study about whether these symptoms are
considered reportable adverse events.
SYMPTOMS
INVESTIGATOR GERD SYMPTOM MEASUREMENT (IF SYMPTOM OCCURRED)
OCCURRED?
Yes/No
VOLUME (mL)
NUMBER OF EPISODES
MAXIMUM SEVERITY
Mild, Moderate, Severe
Vomiting
Diarrhea
Nausea
The collected data meet the following Findings About criteria: 1) data that do not describe an Event or Intervention as a whole, 2) data (―about‖ an
Event or Intervention) having Qualifiers that can be represented in Findings variables (e.g., units, method), and 3) data (―about‖ an Event or
Intervention) that indicate the occurrence of related symptoms or therapies.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 208 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The data below represent data two visits for one subject.
Rows 1-4: Show the results for the Vomiting tests at Visit 1.
Rows 5-7: Show the results for the Diarrhea tests at Visit 1.
Rows 8-10: Show the results for the Nausea tests at Visit 1.
Row 11: Show that Vomiting was absent at Visit 2.
Rows 12-14: Show the results for the Diarrhea tests at Visit 2.
Row 15: Show that Nausea was not assessed at Visit 2.
fa.xpt
Row
STUDYID
DOMAIN
USUBJID
FASEQ
FATESTCD
FATEST
FAOBJ
FACAT
FAORRES
FAORRE
SU
FASTRESC
FASTRESU
VISIT
FASTAT
FADTC
1
XYZ
FA
XYZ-701-002
1
OCCUR
Occurrence
Vomit
GERD
Y
Y
1
2006-02-02
2
XYZ
FA
XYZ-701-002
2
VOL
Volume
Vomit
GERD
250
mL
250
mL
1
2006-02-02
3
XYZ
FA
XYZ-701-002
3
NUMEPISD
Number of Episodes
Vomit
GERD
>10
>10
1
2006-02-02
4
XYZ
FA
XYZ-701-002
4
SEV
Severity/Intensity
Vomit
GERD
SEVERE
SEVERE
1
2006-02-02
5
XYZ
FA
XYZ-701-002
5
OCCUR
Occurrence
Diarrhea
GERD
Y
Y
1
2006-02-02
6
XYZ
FA
XYZ-701-002
6
NUMEPISD
Number of Episodes
Diarrhea
GERD
2
2
1
2006-02-02
7
XYZ
FA
XYZ-701-002
7
SEV
Severity/Intensity
Diarrhea
GERD
SEVERE
SEVERE
1
2006-02-02
8
XYZ
FA
XYZ-701-002
8
OCCUR
Occurrence
Nausea
GERD
Y
Y
1
2006-02-02
9
XYZ
FA
XYZ-701-002
9
NUMEPISD
Number of Episodes
Nausea
GERD
1
1
1
2006-02-02
10
XYZ
FA
XYZ-701-002
10
SEV
Severity/Intensity
Nausea
GERD
MODERATE
MODERATE
1
2006-02-02
11
XYZ
FA
XYZ-701-002
11
OCCUR
Occurrence
Vomit
GERD
N
N
2
2006-02-03
12
XYZ
FA
XYZ-701-002
12
OCCUR
Occurrence
Diarrhea
GERD
Y
Y
2
2006-02-03
13
XYZ
FA
XYZ-701-002
13
NUMEPISD
Number of Episodes
Diarrhea
GERD
1
1
2
2006-02-03
14
XYZ
FA
XYZ-701-002
14
SEV
Severity/Intensity
Diarrhea
GERD
SEVERE
SEVERE
2
2006-02-03
15
XYZ
FA
XYZ-701-002
15
OCCUR
Occurrence
Nausea
GERD
2
NOT
DONE
2006-02-03
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 209
Final November 12, 2008
Example 8: Severity Assessments Per Visit of Adverse Events
The adverse event module collects, instead of a single assessment of severity, assessments of severity at each visit, as follows:
At each visit, record severity of the Adverse Event.
Visit
1
2
3
4
5
6
Severity
The collected data meet the following Findings About criteria: data that do not describe an Event or Intervention as a whole.
AE Domain (For clarity, only selected variables are shown.)
Row 1: Shows the record for a verbatim term of "Morning queasiness", for which the maximum severity over the course of the event was "Moderate."
Row 2: Shows the record for a verbatim term of ―Watery stools‖, for which ―Mild‖ severity was collected at Visits 2 and 3 before the event ended.
ae.xpt
Row
DOMAIN
USUBJID
AESEQ
AETERM
AEDECOD
AESTDTC
AEENDTC
AESEV
1
AE
123
1
Morning queasiness
Nausea
2006-02-01
2006-02-23
MODERATE
2
AE
123
2
Watery stools
Diarrhea
2006-02-01
2006-02-15
MILD
FA domain
Rows 1-4: Show severity data collected at the four visits that occurred between the start and end of the AE, ―Morning queasiness‖. FAOBJ =
NAUSEA, which is the value of AEDECOD in the associated AE record.
Rows 5-6: Show severity data collected at the two visits that occurred between the start and end of the AE, ―Watery stools.‖ FAOBJ = DIARRHEA,
which is the value of AEDECOD in the associated AE record.
fa.xpt
Row
STUDYID
DOMAIN
USUBJID
FASEQ
FATESTCD
FATEST
FAOBJ
FAORRES
VISIT
FADTC
1
XYZ
FA
XYZ-US-701-002
1
SEV
Severity/Intensity
Nausea
MILD
2
2006-02-02
2
XYZ
FA
XYZ-US-701-002
2
SEV
Severity/Intensity
Nausea
MODERATE
3
2006-02-09
3
XYZ
FA
XYZ-US-701-002
3
SEV
Severity/Intensity
Nausea
MODERATE
4
2006-02-16
4
XYZ
FA
XYZ-US-701-002
4
SEV
Severity/Intensity
Nausea
MILD
5
2006-02-23
5
XYZ
FA
XYZ-US-701-002
5
SEV
Severity/Intensity
Diarrhea
MILD
2
2006-02-02
6
XYZ
FA
XYZ-US-701-002
6
SEV
Severity/Intensity
Diarrhea
MILD
3
2006-02-09
RELREC dataset
Depending on how the relationships were collected, in this example, RELREC could be created with either 2 or 6 RELIDs. With 2 RELIDs, the Sponsor
is describing that the severity ratings are related to the AE as well as being related to each other. With 6 RELIDs, the Sponsor is describing that the
severity ratings are related to the AE only (and not to each other).
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 210 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example with two RELIDS
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
ABC
AE
XYZ-US-701-002
AESEQ
1
1
ABC
FA
XYZ-US-701-002
FASEQ
1
1
ABC
FA
XYZ-US-701-002
FASEQ
2
1
ABC
FA
XYZ-US-701-002
FASEQ
3
1
ABC
FA
XYZ-US-701-002
FASEQ
4
1
ABC
AE
XYZ-US-701-002
AESEQ
2
2
ABC
FA
XYZ-US-701-002
FASEQ
5
2
ABC
FA
XYZ-US-701-002
FASEQ
6
2
Example with six RELIDS
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
ABC
AE
XYZ-US-701-002
AESEQ
1
1
ABC
FA
XYZ-US-701-002
FASEQ
1
1
ABC
AE
XYZ-US-701-002
AESEQ
1
2
ABC
FA
XYZ-US-701-002
FASEQ
2
2
ABC
AE
XYZ-US-701-002
AESEQ
1
3
ABC
FA
XYZ-US-701-002
FASEQ
3
3
ABC
AE
XYZ-US-701-002
AESEQ
1
4
ABC
FA
XYZ-US-701-002
FASEQ
4
4
ABC
AE
XYZ-US-701-002
AESEQ
2
5
ABC
FA
XYZ-US-701-002
FASEQ
5
5
ABC
AE
XYZ-US-701-002
AESEQ
2
6
ABC
FA
XYZ-US-701-002
FASEQ
6
6

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 211
Final November 12, 2008
7 Trial Design Datasets
7.1 INTRODUCTION
7.1.1 PURPOSE OF TRIAL DESIGN MODEL
ICH E3, Guidance for Industry, Structure and Content of Clinical Study Reports, Section 9.1, calls for a brief,
clear description of the overall plan and design of the study, and supplies examples of charts and diagrams for this
purpose in Annex IIIa and Annex IIIb. Each Annex corresponds to an example trial, and each shows a diagram
describing the study design and a table showing the schedule of assessments. The Trial Design Model in the
SDTM provides a standardized way to describe those aspects of the planned conduct of a clinical trial shown in
the study design diagrams of these examples. The standard Trial Design Datasets will allow reviewers to:
clearly and quickly grasp the design of a clinical trial
compare the designs of different trials
search a data warehouse for clinical trials with certain features
compare planned and actual treatments and visits for subjects in a clinical trial.
Modeling a clinical trial in this standardized way requires the explicit statement of certain decision rules that may
not be addressed or may be vague or ambiguous in the usual prose protocol document. Prospective modeling of
the design of a clinical trial should lead to a clearer, better protocol. Retrospective modeling of the design of a
clinical trial should ensure a clear description of how the trial protocol was interpreted by the sponsor.
7.1.2 DEFINITIONS OF TRIAL DESIGN CONCEPTS
A clinical trial is a scientific experiment involving human subjects, which is intended to address certain scientific
questions (the objectives of the trial). [See CDISC glossary for more complete definitions of clinical trial and
objective.]
Trial Design: The design of a clinical trial is a plan for what will be done to subjects, and what data will be
collected about them, in the course of the trial, to address the trial's objectives.
Epoch: As part of the design of a trial, the planned period of subjects' participation in the trial is divided into
Epochs. Each Epoch is a period of time that serves a purpose in the trial as a whole. That purpose will be at the
level of the primary objectives of the trial. Typically, the purpose of an Epoch will be to expose subjects to a
treatment, or to prepare for such a treatment period (e.g., determine subject eligibility, wash out previous
treatments) or to gather data on subjects after a treatment has ended. Note that at this high level a ―treatment‖ is a
treatment strategy, which may be simple (e.g., exposure to a single drug at a single dose) or complex. Complex
treatment strategies could involve tapering through several doses, titrating dose according to clinical criteria,
complex regimens involving multiple drugs, or strategies that involve adding or dropping drugs according to
clinical criteria.
Arm: An Arm is a planned path through the trial. This path covers the entire time of the trial. The group of
subjects assigned to a planned path is also often colloquially called an Arm. The group of subjects assigned to an
Arm is also often called a treatment group, and in this sense, an Arm is equivalent to a treatment group.
Study Cell: Since the trial as a whole is divided into Epochs, each planned path through the trial (i.e., each Arm)
is divided into pieces, one for each Epoch. Each of these pieces is called a Study Cell. Thus, there is a study cell
for each combination of Arm and Epoch. Each Study Cell represents an implementation of the purpose of its
associated Epoch. For an Epoch whose purpose is to expose subjects to treatment, each Study Cell associated
with the Epoch has an associated treatment strategy. For example, a three-Arm parallel trial might have a
Treatment Epoch whose purpose is to expose subjects to one of three study treatments: placebo, investigational
product, or active control. There would be three Study Cells associated with the Treatment Epoch, one for each
Arm. Each of these Study Cells exposes the subject to one of the three study treatments. Another example

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 212 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
involving more complex treatment strategies: a trial compares the effects of cycles of chemotherapy drug A given
alone or in combination with drug B, where drug B is given as a pre-treatment to each cycle of drug A.
Element: An Element is a basic building block in the trial design. It involves administering a planned
intervention, which may be treatment or no treatment, during a period of time. Elements for which the planned
intervention is "no treatment" would include Elements for screening, washout, and follow-up.
Study Cells and Elements: Many, perhaps most, clinical trials, involve a single, simple administration of a
planned intervention within a Study Cell, but for some trials, the treatment strategy associated with a Study Cell
may involve a complex series of administrations of treatment. It may be important to track the component steps
in a treatment strategy both operationally and because secondary objectives and safety analyses require that data
be grouped by the treatment step during which it was collected. The steps within a treatment strategy may involve
different doses of drug, different drugs, or different kinds of care, as in pre-operative, operative, and post-
operative periods surrounding surgery. When the treatment strategy for a Study Cell is simple, the Study Cell will
contain a single Element, and for many purposes there is little value in distinguishing between the Study Cell and
the Element. However, when the treatment strategy for a Study Cell consists of a complex series of treatments, a
Study Cell can contain multiple Elements. There may be a fixed sequence of Elements, or a repeating cycle of
Elements, or some other complex pattern. In these cases, the distinction between a Study Cell and an Element is
very useful.
Branch: In a trial with multiple Arms, the protocol plans for each subject to be assigned to one Arm. The time
within the trial at which this assignment takes place is the point at which the Arm paths of the trial diverge, and
so is called a branch point. For many trials, the assignment to an Arm happens all at one time, so the trial has one
branch point. For other trials, there may be two or more branches that collectively assign a subject to an Arm. The
process that makes this assignment may be a randomization, but it need not be.
Treatments: The word "treatment" may be used in connection with Epochs, Study Cells, or Elements, but has
somewhat different meanings in each context:
Since Epochs cut across Arms, an "Epoch treatment" is at a high level that does not specify anything that
differs between Arms. For example, in a three-period crossover study of three doses of Drug X, each
treatment Epoch is associated with Drug X, but not with a specific dose.
A "Study Cell treatment" is specific to a particular Arm. For example, a parallel trial might have Study
Cell treatments Placebo and Drug X, without any additional detail (e.g., dose, frequency, route of
administration) being specified. A Study Cell treatment is at a relatively high level, the level at which
treatments might be planned in an early conceptual draft of the trial, or in the title or objectives of the
trial.
An ―Element treatment‖ may be fairly detailed. For example, for an Element representing a cycle of
chemotherapy, Element treatment might specify 5 daily 100 mg doses of Drug X.
The distinctions between these levels are not rigid, and depend on the objectives of the trial. For example, route is
generally a detail of dosing, but in a bioequivalence trial that compared IV and oral administration of Drug X,
route is clearly part of Study Cell treatment.
Visit: A clinical encounter. The notion of a Visit derives from trials with outpatients, where subjects interact with
the investigator during Visits to the investigator's clinical site. However, the term is used in other trials, where a
trial Visit may not correspond to a physical Visit. For example, in a trial with inpatients, time may be subdivided
into Visits, even though subjects are in hospital throughout the trial. For example, data for a screening Visit may
be collected over the course of more than one physical visit. One of the main purposes of Visits is the
performance of assessments, but not all assessments need take place at clinic Visits; some assessments may be
performed by means of telephone contacts, electronic devices or call-in systems. The protocol should specify
what contacts are considered Visits and how they are defined.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 213
Final November 12, 2008
7.1.3 CURRENT AND FUTURE CONTENTS OF THE TRIAL DESIGN MODEL
The datasets currently included in the Trial Design Model:
Trial Arms: describes the sequences of Elements in each Epoch for each Arm, and thus describes the
complete sequence of Elements in each Arm.
Trial Elements: describes the Elements used in the trial.
Trial Visits: describes the planned schedule of Visits.
Trial Inclusion/Exclusion: describes the inclusion/exclusion criteria used to screen subjects.
Trial Summary: lists key facts (parameters) about the trial that are likely to appear in a registry of clinical
trials.
The Trial Inclusion/Exclusion (TI) is discussed in 1359HSection 7.5. The IE domain (subject specific inclusion/exclusion
criteria not met) described in 1360HSection 6.3.2 contains the actual exceptions to those criteria for enrolled subjects. The
Trial Inclusion/Exclusion dataset was developed before the define.xml standard for metadata. Because the text of all
inclusion/exclusion criteria can now be included in define.xml, this dataset may be deprecated in future versions of
the SDTM.
Future versions of the Trial Design Model are expected to include additional aspects of clinical trials; some of these
additional aspects will be used in submissions, while others are needed for accurate representation of protocols for
the planning stage, but will have limited effects on the SDTM.
Work is underway on representing the schedule of assessments and planned interventions. When this work is
completed, it is expected that the information on planned assessments and interventions will be submitted along with
SDTM datasets containing actual subject data, to allow the comparison of planned and actual assessments and
interventions.
The current Trial Design Model has limitations in representing protocols, which include the following:
plans for indefinite numbers of repeating Elements (e.g., indefinite numbers of chemotherapy cycles)
indefinite numbers of Visits (e.g., periodic follow-up Visits for survival)
indefinite numbers of Epochs
indefinite numbers of Arms.
The last two situations arise in dose-escalation studies where increasing doses are given until stopping criteria are
met. Some dose-escalation studies enroll a new cohort of subjects for each new dose, and so, at the planning stage,
have an indefinite number of Arms. Other dose-escalation studies give new doses to a continuing group of subjects,
and so are planned with an indefinite number of Epochs.
There may also be limitations in representing other patterns of Elements within a Study Cell that are more complex
than a simple sequence. For the purpose of submissions about trials that have already completed, these limitations
are not critical, so it is expected that development of the Trial Design Model to address these limitations will have a
minimal impact on SDTM.
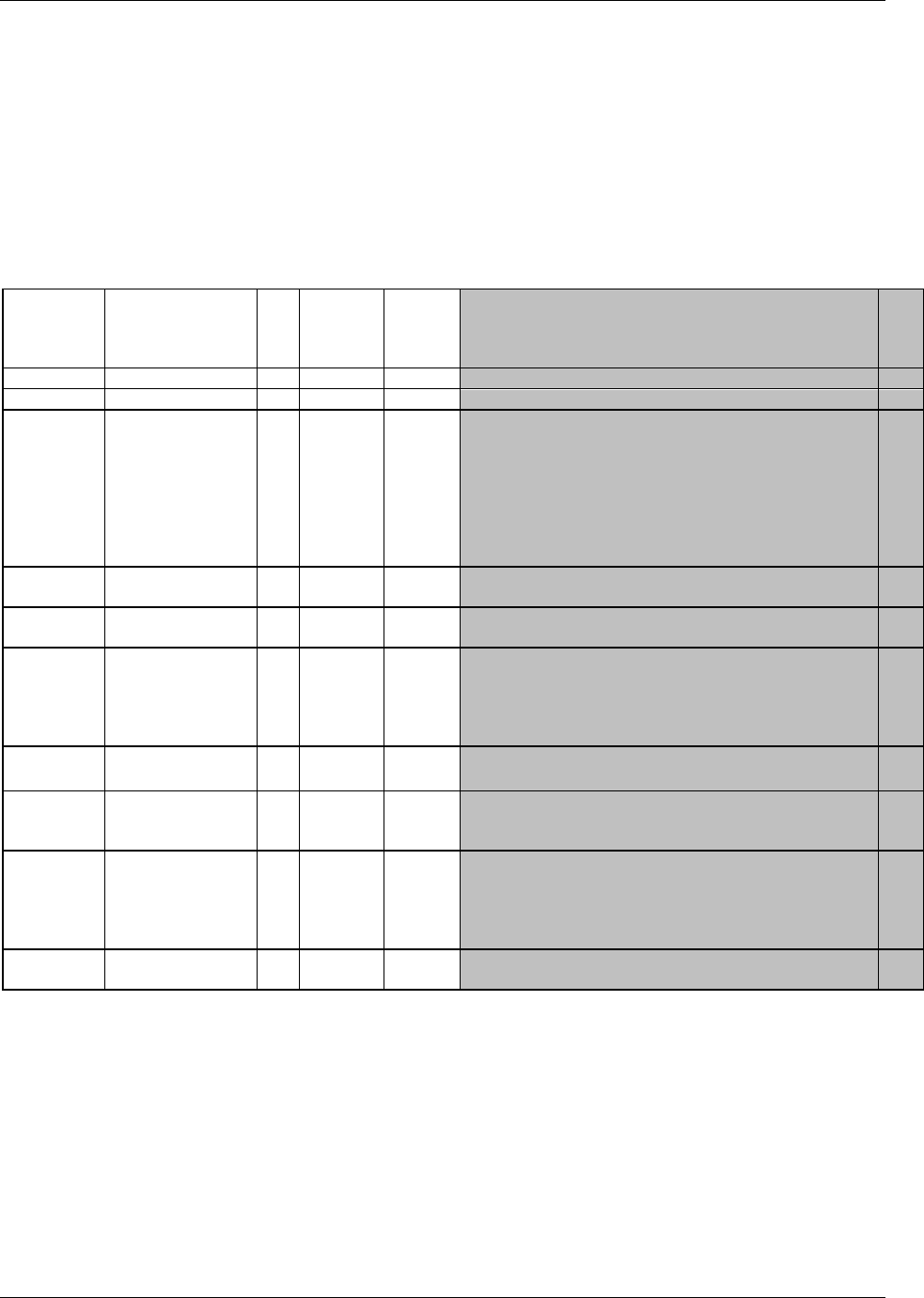
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 214 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.2 TRIAL ARMS
This section contains:
The Trial Arms dataset and assumptions
A series of example trials, which illustrate the development of the Trial Arms dataset
Advice on various issues in the development of the Trial Arms dataset
A recap of the Trial Arms dataset, and the function of its variables.
7.2.1 TRIAL ARMS DATASET — TA
ta.xpt, Trial Arms — Trial Design, Version 3.1.2. One record per planned Element per Arm
Variable
Name
Variable Label
Type
Controlled
Terms,
Codelist or
Format
Role
CDISC Notes
Core
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
DOMAIN
Domain Abbreviation
Char
1361HTA
Identifier
Two-character abbreviation for the domain.
Req
ARMCD
Planned Arm Code
Char
*
Topic
ARMCD is limited to 20 characters and does not have
special character restrictions. The maximum length of
ARMCD is longer than that for other ―short‖ variables to
accommodate the kind of values that are likely to be needed
for crossover trials. For example, if ARMCD values for a
seven-period crossover were constructed using two-character
abbreviations for each treatment and separating hyphens, the
length of ARMCD values would be 20.
Req
ARM
Description of
Planned Arm
Char
*
Synonym
Qualifier
Name given to an Arm or treatment group.
Req
TAETORD
Order of Element
within Arm
Num
Identifier
Number that gives the order of the Element within the Arm.
Req
ETCD
Element Code
Char
*
Record
Qualifier
ETCD (the companion to ELEMENT) is limited to 8
characters and does not have special character restrictions.
These values should be short for ease of use in
programming, but it is not expected that ETCD will need to
serve as a variable name.
Req
ELEMENT
Description of
Element
Char
*
Synonym
Qualifier
The name of the Element. The same Element may occur
more than once within an Arm.
Perm
TABRANCH
Branch
Char
Rule
Condition subject met, at a ―branch‖ in the trial design at the
end of this Element, to be included in this Arm; (e.g.,
randomization to DRUG X).
Exp
TATRANS
Transition Rule
Char
Rule
If the trial design allows a subject to transition to an Element
other than the next Element in sequence, then the conditions
for transitioning to those other Elements, and the alternative
Element sequences, are specified in this rule (e.g.,
Responders go to washout).
Exp
EPOCH
Epoch
Char
*
Timing
Name of the Trial Epoch with which this Element of the
Arm is associated.
Req
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
7.2.2 ASSUMPTIONS FOR TA DATASET
1. TAETORD is an integer. In general the value of TAETORD is 1 for the first Element in each Arm, 2 for the second
Element in each Arm, etc. Occasionally, it may be convenient to skip some values (see 1362HSection 7.2.3.6 for an example).
Although the values of TAETORD need not always be sequential, their order must always be the correct order for the
Elements in the Arm path.
2. Elements in different Arms with the same value of TAETORD may or may not be at the same time, depending on the
design of the trial. The example trials illustrate a variety of possible situations. The same Element may occur more than
once within an Arm.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 215
Final November 12, 2008
3. TABRANCH describes the outcome of a branch decision point in the trial design for subjects in the Arm. A branch
decision point takes place between Epochs, and is associated with the Element that ends at the decision point. For
instance, if subjects are assigned to an Arm where they receive treatment A through a randomization at the end of Element
X, the value of TABRANCH for Element X would be "Randomized to A."
4. Branch decision points may be based on decision processes other than randomizations, such as clinical evaluations of
disease response or subject choice.
5. There is usually some gap in time between the performance of a randomization and the start of randomized treatment.
However, in many trials this gap in time is small and it is highly unlikely that subjects will leave the trial between
randomization and treatment. In these circumstances, the trial does not need to be modeled with this time period between
randomization and start of treatment as a separate Element.
6. Some trials include multiple paths that are closely enough related so that they are all considered to belong to one Arm. In
general, this set of paths will include a "complete" path along with shorter paths that skip some Elements. The sequence of
Elements represented in the Trial Arms should be the complete, longest path. TATRANS describes the decision points that
may lead to a shortened path within the Arm.
7. If an Element does not end with a decision that could lead to a shortened path within the Arm, then TATRANS will be
blank. If there is such a decision, TATRANS will be in a form like, "If condition X is true, then go to Epoch Y" or "If
condition X is true, then go to Element with TAETORD=Z."
8. EPOCH is not strictly necessary for describing the sequence of Elements in an Arm path, but it is the conceptual basis for
comparisons between Arms, and also provides a useful way to talk about what is happening in a blinded trial while it is
blinded. During periods of blinded treatment, blinded participants will not know which Arm and Element a subject is in,
but EPOCH should provide a description of the time period that does not depend on knowing Arm.
9. EPOCH should be assigned in such a way that Elements from different Arms with the same value of EPOCH are
"comparable" in some sense. The degree of similarity across Arms varies considerably in different trials, as illustrated in
the examples.
10. Note that Study Cells are not explicitly defined in the Trial Arms dataset. A set of records with a common value of both
ARMCD and EPOCH constitute the description of a Study Cell. Transition rules within this set of records are also part of
the description of the Study Cell.
11. EPOCH may be used as a timing variable in other datasets, such as EX and DS, and values of EPOCH must be different
for different epochs. For instance, in a crossover trial with three treatment epochs, each must be given a distinct name; all
three cannot be called ―TREATMENT‖.
7.2.3 TRIAL ARMS EXAMPLES
The core of the Trial Design Model is the Trial Arms (TA) dataset. For each Arm of the trial, it contains one record
for each occurrence of an Element in the path of the Arm.
Although the Trial Arms dataset has one record for each trial Element traversed by subjects assigned to the Arm, it is
generally more useful to work out the overall design of the trial at the Study Cell level, then to work out the
Elements within each Study Cell, and finally to develop the definitions of the Elements that are contained in the
Trial Elements table.
It is generally useful to draw diagrams, like those mentioned in ICH E3, when working out the design of a trial. The
protocol may include a diagram that can serve as a starting point. Such a diagram can then be converted into a Trial
Design Matrix, which displays the Study Cells and which can be, in turn, converted into the Trial Arms dataset.
This section uses example trials of increasing complexity, numbered 1 to 7, to illustrate the concepts of trial design. For each
example trial, the process of working out the Trial Arms table is illustrated by means of a series of diagrams and tables,
including the following:
A diagram showing the branching structure of the trial in a ―study schema‖ format such as might appear in
a protocol.
A diagram that shows the ―prospective‖ view of the trial, the view of those participating in the trial. It is
similar to the "study schema" view in that it usually shows a single pool of subjects at the beginning of the
trial, with the pool of subjects being split into separate treatment groups at randomizations and other
branches. They show the epochs of the trial, and, for each group of subjects and each epoch, the sequence
of elements within each epoch for that treatment group. The arms are also indicated on these diagrams.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 216 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
A diagram that shows the ―retrospective‖ view of the trial, the view of the analyst reporting on the trial. The
style of diagram looks more like a matrix; it is also more like the structure of the Trial Arms dataset. It is an
arm-centered view, which shows, for each study cell (epoch/arm combination) the sequence of elements
within that study cell. It can be thought of as showing, for each arm the elements traversed by a subject
who completed that arm as intended.
If the trial is blinded, a diagram that shows the trial as it appears to a blinded participant.
A Trial Design Matrix, an alternative format for representing most of the information in the diagram that
shows Arms and Epochs, and emphasizes the Study Cells.
The Trial Arms dataset.
Readers are advised to read the following section with Example 1 before reading other examples, since Example 1
explains the conventions used for the diagrams and tables.
7.2.3.1 EXAMPLE TRIAL 1, A PARALLEL TRIAL
Diagrams that represent study schemas generally conceive of time as moving from left to right, use horizontal lines
to represent periods of time, and slanting lines to represent branches into separate treatments, convergence into a
common follow-up, or cross-over to a different treatment.
In this document, diagrams are drawn using "blocks" corresponding to trial Elements rather than horizontal lines.
Trial Elements are the various treatment and non-treatment time periods of the trial, and we want to emphasize the
separate trial Elements that might otherwise be "hidden" in a single horizontal line. See 1363HSection 7.3 for more
information about defining trial Elements. In general, the Elements of a trial will be fairly clear. However, in the
process of working out a trial design, alternative definitions of trial Elements may be considered, in which case
diagrams for each alternative may be constructed.
In the study schema diagrams in this document, the only slanting lines used are those that represent branches, the
decision points where subjects are divided into separate treatment groups. One advantage of this style of diagram,
which does not show convergence of separate paths into a single block, is that the number of Arms in the trial can be
determined by counting the number of parallel paths at the right end of the diagram.
Below is the study schema diagram for Example Trial 1, a simple parallel trial. This trial has three Arms, corresponding
to the three possible left-to-right "paths" through the trial. Each path corresponds to one of the three treatment Elements
at the right end of the diagram. Note that the randomization is represented by the three red arrows leading from the
Run-in block.
Example Trial 1: Parallel Design
Study schema
Screen
Placebo
Drug A
Drug B
Run-In
Randomization
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 217
Final November 12, 2008
The next diagram for this trial shows the epochs of the trial, indicates the three Arms, and shows the sequence of
elements for each group of subjects in each epoch. The arrows are at the right hand side of the diagram because it is
at the end of the trial that all the separate paths through the trial can be seen. Note that, in this diagram, the
randomization, which was shown using three red arrows connecting the Run-in block with the three treatment blocks
in the first diagram, is now indicated by a note with an arrow pointing to the line between two epochs.
Treatment
Epoch
Example Trial 1: Parallel Design
Prospective view
Screening
Epoch Run-in
Epoch
Drug A
Drug B
Screen Run-In
Placebo
Randomization
Placebo
Drug A
Drug B
The next diagram can be thought of as the ―retrospective‖ view of a trial, the view back from a point in time when a
subject‘s assignment to an arm is known. In this view, the trial appears as a grid, with an arm represented by a series
of study cells, one for each epoch, and a sequence of elements within each study cell. In this trial, as in many trials,
there is exactly one element in each study cell, but later examples will illustrate that this is not always the case.
Example Trial 1: Parallel Design
Retrospective view
Screening
Epoch Run-in
Epoch
Treatment
Epoch
Drug A
Drug B
Screen
Screen
Run-In Placebo
Drug A
Screen Drug B
Run-In
Run-In
Placebo
Randomization
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 218 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The next diagram shows the trial from the viewpoint of blinded participants. To blinded participants in this trial, all
Arms look alike. They know when a subject is in the Screen Element, or the Run-in Element, but when a subject is
in the Treatment Epoch, they know only that the subject is in an Element which involves receiving study drug, not
which study drug, and therefore not which Element.
Example Trial 1: Parallel Trial
Blinded View
Screening
Epoch Run-in
Epoch
Treatment
Epoch
Blind
Screen Study DrugRun-In
Randomization
A trial design matrix is a table with a row for each Arm in the trial and a column for each Epoch in the trial. It is closely
related to the retrospective view of the trial, and many users may find it easier to construct a table than to draw a
diagram. The cells in the matrix represent the Study Cells, which are populated with trial Elements. In this trial, each
Study Cell contains exactly one Element.
The columns of a Trial Design Matrix are the Epochs of the trial, the rows are the Arms of the trial, and the cells of
the matrix (the Study Cells) contain Elements. Note that the randomization is not represented in the Trial Design
Matrix. All the diagrams above and the trial design matrix below are alternative representations of the trial design.
None of them contains all the information that will be in the finished Trial Arms dataset, but users may find it useful
to draw some or all of them when working out the dataset.
Trial Design Matrix for Example Trial 1
Screen
Run-in
Treatment
Placebo
Screen
Run-in
PLACEBO
A
Screen
Run-in
DRUG A
B
Screen
Run-in
DRUG B
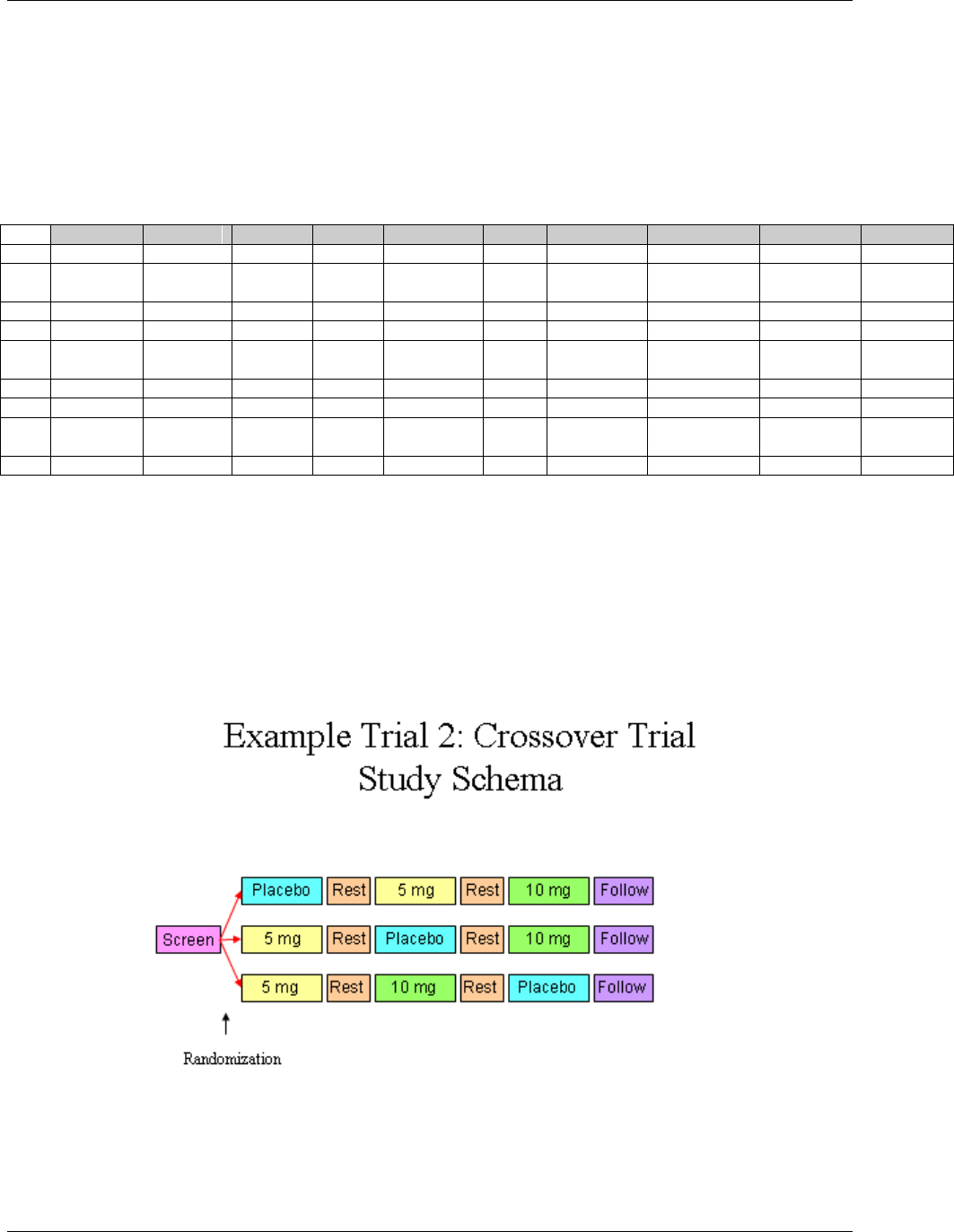
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 219
Final November 12, 2008
For Example Trial 1, the conversion of the Trial Design Matrix into the Trial Arms dataset is straightforward. For
each cell of the matrix, there is a record in the Trial Arms dataset. ARM, EPOCH, and ELEMENT can be populated
directly from the matrix. TAETORD acts as a sequence number for the Elements within an Arm, so it can be
populated by counting across the cells in the matrix. The randomization information, which is not represented in the
Trial Design Matrix, is held in TABRANCH in the Trial Arms dataset. TABRANCH is populated only if there is a
branch at the end of an Element for the Arm. When TABRANCH is populated, it describes how the decision at the
branch point would result in a subject being in this Arm.
Trial Arms Dataset for Example Trial 1
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX1
TA
P
Placebo
1
SCRN
Screen
Screen
2
EX1
TA
P
Placebo
2
RI
Run-In
Randomized
to Placebo
Run-In
3
EX1
TA
P
Placebo
3
P
Placebo
Treatment
4
EX1
TA
A
A
1
SCRN
Screen
Screen
5
EX1
TA
A
A
2
RI
Run-In
Randomized
to Drug A
Run-In
6
EX1
TA
A
A
3
A
Drug A
Treatment
7
EX1
TA
B
B
1
SCRN
Screen
Screen
8
EX1
TA
B
B
2
RI
Run-In
Randomized
to Drug B
Run-In
9
EX1
TA
B
B
3
B
Drug B
Treatment
7.2.3.2 EXAMPLE TRIAL 2, A CROSSOVER TRIAL
The diagram below is for a crossover trial. However, the diagram does not use the crossing slanted lines sometimes
used to represent crossover trials, since the order of the blocks is sufficient to represent the design of the trial.
Slanted lines are used only to represent the branch point at randomization, when a subject is assigned to a sequence
of treatments. As in most crossover trials, the Arms are distinguished by the order of treatments, with the same
treatments present in each Arm. Note that even though all three Arms of this trial end with the same block, the block
for the follow-up Element, the diagram does not show the Arms converging into one block. Also note that the same
block (the "Rest" Element) occurs twice within each Arm. Elements are conceived of as "reusable" and can appear
in more than one Arm, in more than one Epoch, and more than once in an Arm.
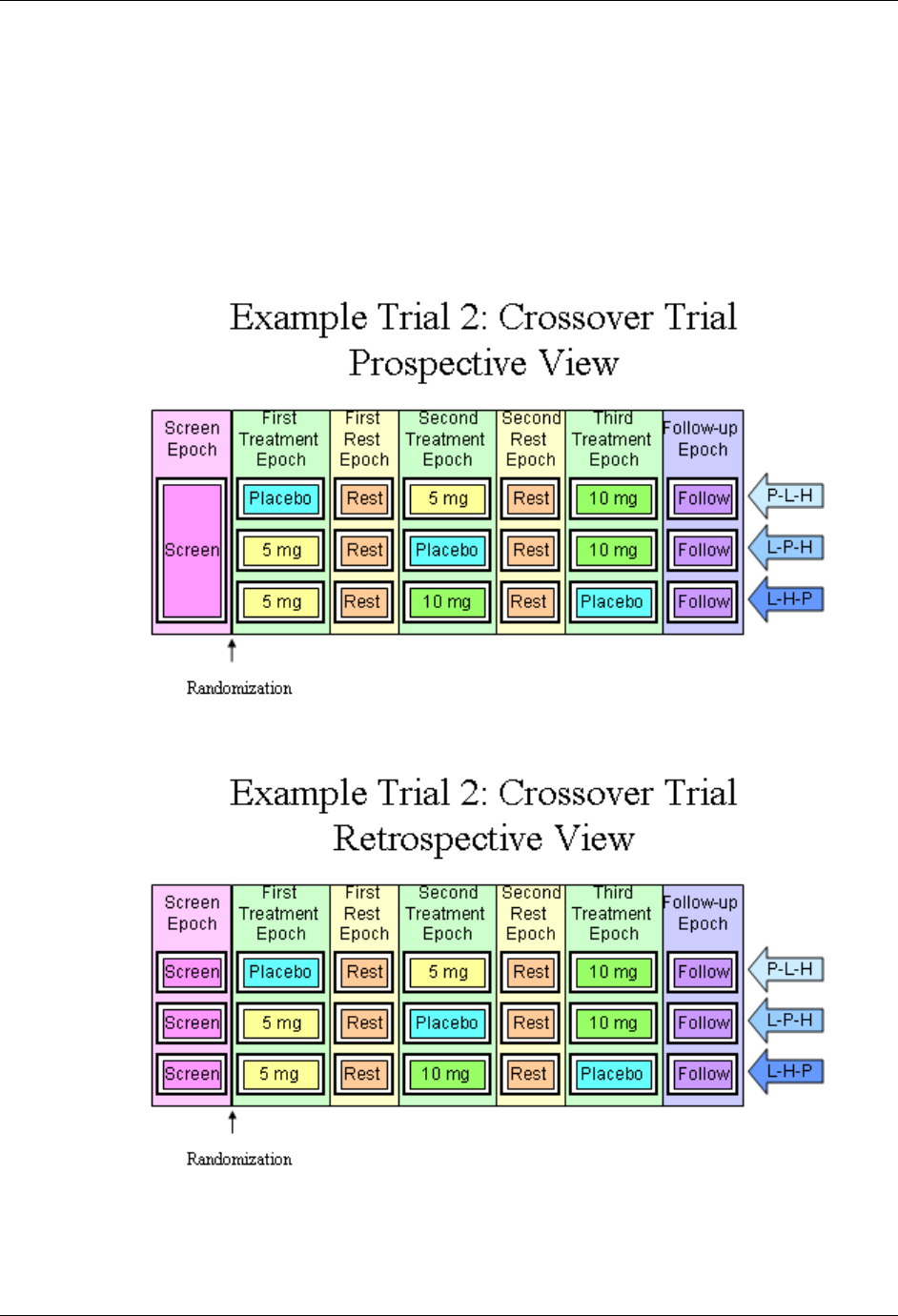
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 220 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The next diagram for this crossover trial shows the prospective view of the trial, identifies the epoch and
arms of the trial, and gives each a name. As for most crossover studies, the objectives of the trial will be
addressed by comparisons between the arms and by within-subject comparisons between treatments. The
design thus depends on differentiating the periods during which the subject receives the three different
treatments and so there are three different treatment epochs. The fact that the rest periods are identified as
separate Epochs suggests that these also play an important part in the design of the trial; they are probably
designed to allow subjects to return to ―baseline‖ with data collected to show that this occurred. Note that
Epochs are not considered "reusable", so each Epoch has a different name, even though all the Treatment
Epochs are similar and both the Rest Epochs are similar. As with the first example trial, there is a one to
one relationship between the Epochs of the trial and the Elements in each Arm.
The next diagram shows the retrospective view of the trial.
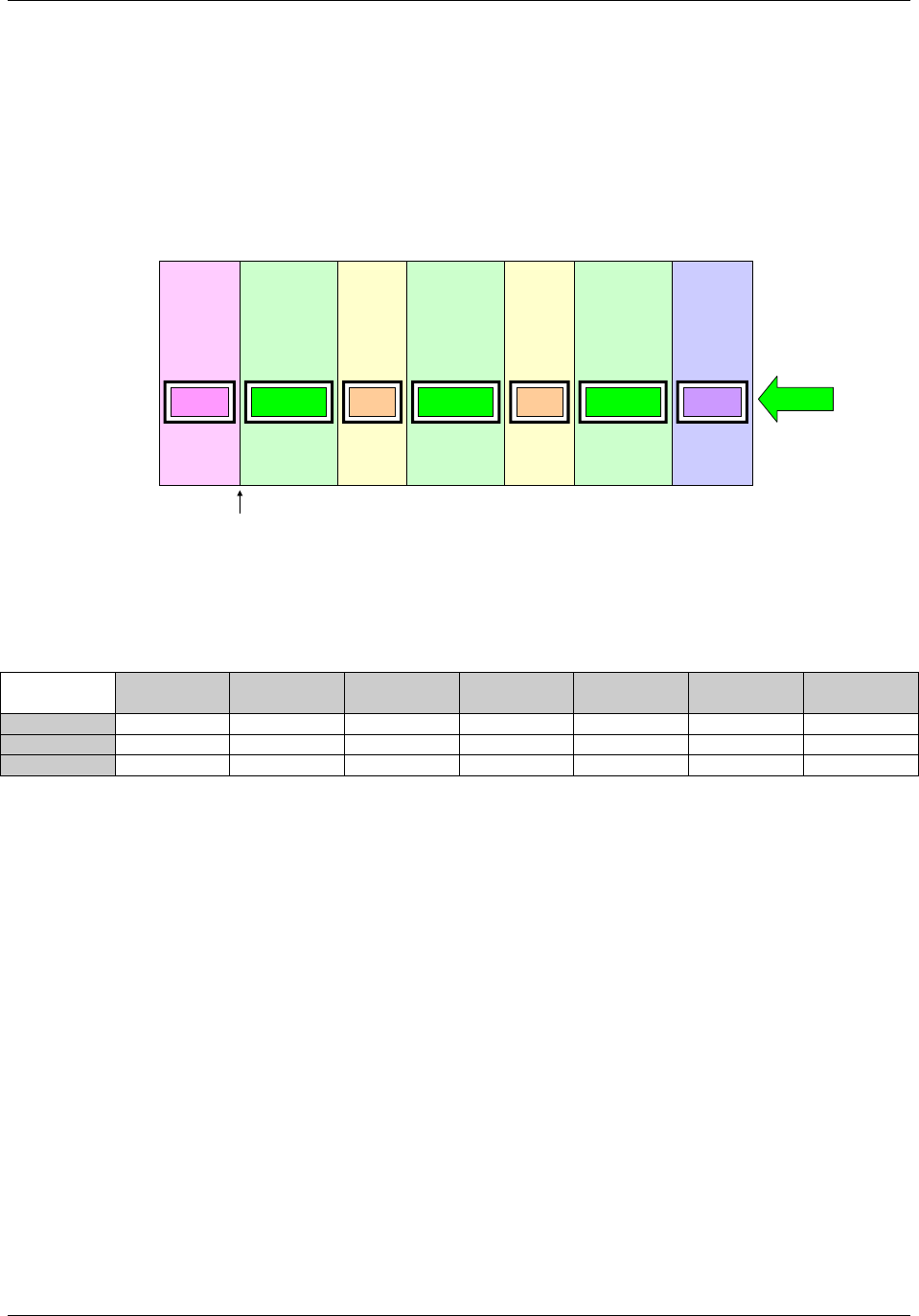
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 221
Final November 12, 2008
The last diagram for this trial shows the trial from the viewpoint of blinded participants. As in the simple
parallel trial above, blinded participants see only one sequence of Elements, since during the treatment Epochs
they do not know which of the treatment Elements a subject is in.
Example Trial 2: Crossover Trial
Blinded View
Follow-up
Epoch
Third
Treatment
Epoch
Second
Rest
Epoch
Second
Treatment
Epoch
First
Rest
Epoch
First
Treatment
Epoch
Screen
Epoch
Drug Rest Drug Rest Drug Follow Blind
Screen
Randomization
The trial design matrix for the crossover example trial is shown below. It corresponds closely to the retrospective
diagram above.
Trial Design Matrix for Example Trial 2
Screen
First
Treatment
First Rest
Second
Treatment
Second Rest
Third
Treatment
Follow-up
P-5-10
Screen
Placebo
Rest
5 mg
Rest
10 mg
Follow-up
5-P-10
Screen
5 mg
Rest
Placebo
Rest
10 mg
Follow-up
5-10-P
Screen
5 mg
Rest
10 mg
Rest
Placebo
Follow-up
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 222 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
It is straightforward to produce the Trial Arms dataset for this crossover trial from the diagram showing Arms and Epochs, or from the Trial Design Matrix. To
avoid confusion between the ―Screen‖ Epoch, and the ―Screen‖ Element, the word ―Epoch‖ has been included in all the Epoch names.
Trial Arms Dataset for Example Trial 2
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX2
TA
P-5-10
Placebo-5mg-10mg
1
SCRN
Screen
Randomized to Placebo - 5 mg - 10 mg
Screen Epoch
2
EX2
TA
P-5-10
Placebo-5mg-10mg
2
P
Placebo
First Treatment Epoch
3
EX2
TA
P-5-10
Placebo-5mg-10mg
3
REST
Rest
First Rest Epoch
4
EX2
TA
P-5-10
Placebo-5mg-10mg
4
5
5 mg
Second Treatment
Epoch
5
EX2
TA
P-5-10
Placebo-5mg-10mg
5
REST
Rest
Second Rest Epoch
6
EX2
TA
P-5-10
Placebo-5mg-10mg
6
10
10 mg
Third Treatment Epoch
7
EX2
TA
P-5-10
Placebo-5mg-10mg
7
FU
Follow-up
Follow-up Epoch
8
EX2
TA
5-P-10
5mg-Placebo-10mg
1
SCRN
Screen
Randomized to 5 mg - Placebo - 10 mg
Screen Epoch
9
EX2
TA
5-P-10
5mg-Placebo-10mg
2
5
5 mg
First Treatment Epoch
10
EX2
TA
5-P-10
5mg-Placebo-10mg
3
REST
Rest
First Rest Epoch
11
EX2
TA
5-P-10
5mg-Placebo-10mg
4
P
Placebo
Second Treatment
Epoch
12
EX2
TA
5-P-10
5mg-Placebo-10mg
5
REST
Rest
Second Rest Epoch
13
EX2
TA
5-P-10
5mg-Placebo-10mg
6
10
10 mg
Third Treatment Epoch
14
EX2
TA
5-P-10
5mg-Placebo-10mg
7
FU
Follow-up
Follow-up Epoch
15
EX2
TA
5-10-P
5mg-10mg-Placebo
1
SCRN
Screen
Randomized to 5 mg - 10 mg – Placebo
Screen Epoch
16
EX2
TA
5-10-P
5mg-10mg-Placebo
2
5
5 mg
First Treatment Epoch
17
EX2
TA
5-10-P
5mg-10mg-Placebo
3
REST
Rest
First Rest Epoch
18
EX2
TA
5-10-P
5mg-10mg-Placebo
4
10
10 mg
Second Treatment
Epoch
19
EX2
TA
5-10-P
5mg-10mg-Placebo
5
REST
Rest
Second Rest Epoch
20
EX2
TA
5-10-P
5mg-10mg-Placebo
6
P
Placebo
Third Treatment Epoch
21
EX2
TA
5-10-P
5mg-10mg-Placebo
7
FU
Follow-up
Follow-up Epoch
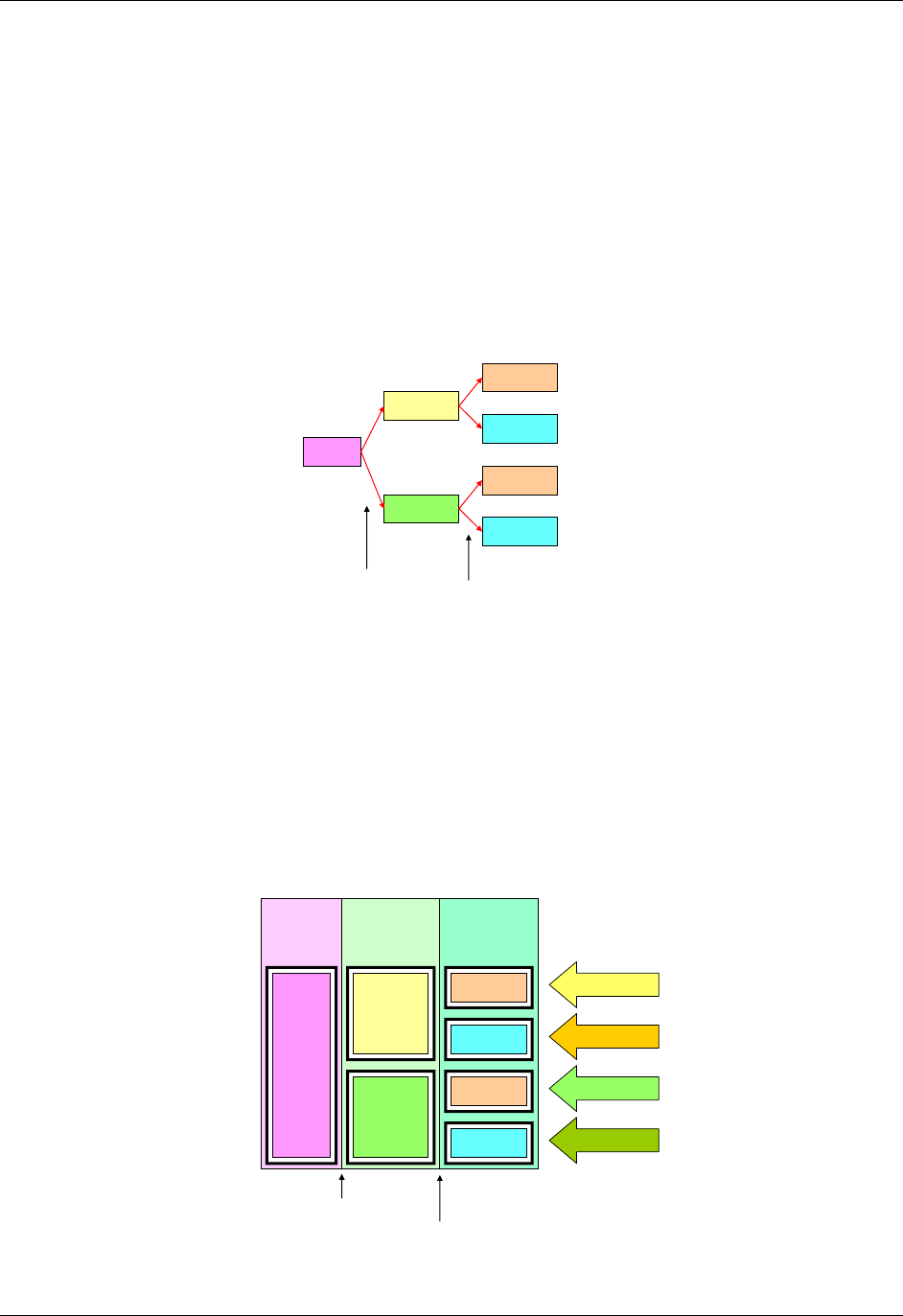
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 223
Final November 12, 2008
7.2.3.3 EXAMPLE TRIAL 3, A TRIAL WITH MULTIPLE BRANCH POINTS
Each of the paths for the trial shown in the diagram below goes through one branch point at randomization, and then
through another branch point when response is evaluated. This results in four Arms, corresponding to the number of
possible paths through the trial, and also to the number of blocks at the right end of the diagram. The fact that there
are only two kinds of block at the right end ("Open DRUG X" and "Rescue") does not affect the fact that there are
four "paths" and thus four Arms.
Example Trial 3: Multiple Branches
Study Schema
Drug A
Open A
Rescue
Open A
Screen
Drug B
Rescue
Randomization
Response Evaluation
The next diagram for this trial is the prospective view. It shows the epochs of the trial and how the initial group of
subjects is split into two treatment groups for the double blind treatment epoch, and how each of those initial treatment
groups is split in two at the response evaluation, resulting in the four Arms of this trial The names of the Arms have
been chosen to represent the outcomes of the successive branches that, together, assign subjects to Arms. These
compound names were chosen to facilitate description of subjects who may drop out of the trial after the first branch
and before the second branch. See 1364HExample 7 in Section 5.1.1.2, which illustrates DM and SE data for such subjects.
Example Trial 3 : Multiple Branches
Prospective View
Open
Treatment
Epoch
Double Blind
Treatment
Epoch
Screen
Epoch
Drug A
Drug B
Open A
Rescue
Open A
A-Rescue
B-Open
A-Open
Rescue
Screen
B-Rescue
Randomization
Response Evaluation
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 224 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The next diagram shows the retrospective view. As with the first two example trials, there is one element in each study cell.
Example Trial 3 : Multiple Branches
Retrospective View
Open
Treatment
Epoch
Double Blind
Treatment
Epoch
Screen
Epoch
Drug A
Drug A
Drug B
Open A
Rescue
Open A
Screen
A-Rescue
B-Open
A-Open
Screen
Screen
Drug B Rescue Screen B-Rescue
Randomization
Response Evaluation
The last diagram for this trial shows the trial from the viewpoint of blinded participants. Since the prospective view is the
view most relevant to study participants, the blinded view shown here is a prospective view. Since blinded participants
can tell which treatment a subject receives in the Open Label Epoch, they see two possible element sequences.
Example Trial 3 : Multiple Branches
Blinded Prospective View
Open
Treatment
Epoch
Double Blind
Treatment
Epoch
Screen
Epoch
Drug
Drug-Open A
Drug-Rescue
Screen
Randomization
Response Evaluation
Open A
Rescue
The trial design matrix for this trial can be constructed easily from the diagram showing Arms and Epochs.
Trial Design Matrix for Example Trial 3
Screen
Double Blind
Open Label
A-Open A
Screen
Treatment A
Open Drug A
A-Rescue
Screen
Treatment A
Rescue
B-Open A
Screen
Treatment B
Open Drug A
B-Rescue
Screen
Treatment B
Rescue
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 225
Final November 12, 2008
Creating the Trial Arms dataset for Example Trial 3 is similarly straightforward. Note that because there are two branch points in this trial, TABRANCH is
populated for two records in each Arm. Note also that the values of ARMCD, like the values of ARM, reflect the two separate processes that result in a subject's
assignment to an Arm.
Trial Arms Dataset for Example Trial 3
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX3
TA
AA
A-Open A
1
SCRN
Screen
Randomized to Treatment A
Screen
2
EX3
TA
AA
A-Open A
2
DBA
Treatment A
Assigned to Open Drug A on basis of
response evaluation
Double Blind
3
EX3
TA
AA
A-Open A
3
OA
Open DRUG
A
Open Label
4
EX3
TA
AR
A-Rescue
1
SCRN
Screen
Randomized to Treatment A
Screen
5
EX3
TA
AR
A-Rescue
2
DBA
Treatment A
Assigned to Rescue on basis of
response evaluation
Double Blind
6
EX3
TA
AR
A-Rescue
3
RSC
Rescue
Open Label
7
EX3
TA
BA
B-Open A
1
SCRN
Screen
Randomized to Treatment B
Screen
8
EX3
TA
BA
B-Open A
2
DBB
Treatment B
Assigned to Open Drug A on basis of
response evaluation
Double Blind
9
EX3
TA
BA
B-Open A
3
OA
Open DRUG
A
Open Label
10
EX3
TA
BR
B-Rescue
1
SCRN
Screen
Randomized to Treatment B
Screen
11
EX3
TA
BR
B-Rescue
2
DBB
Treatment B
Assigned to Rescue on basis of
response evaluation
Double Blind
12
EX3
TA
BR
B-Rescue
3
RSC
Rescue
Open Label
See 1365HSection 7.2.4.1 for additional discussion of when a decision point in a trial design should be considered to give rise to a new Arm.
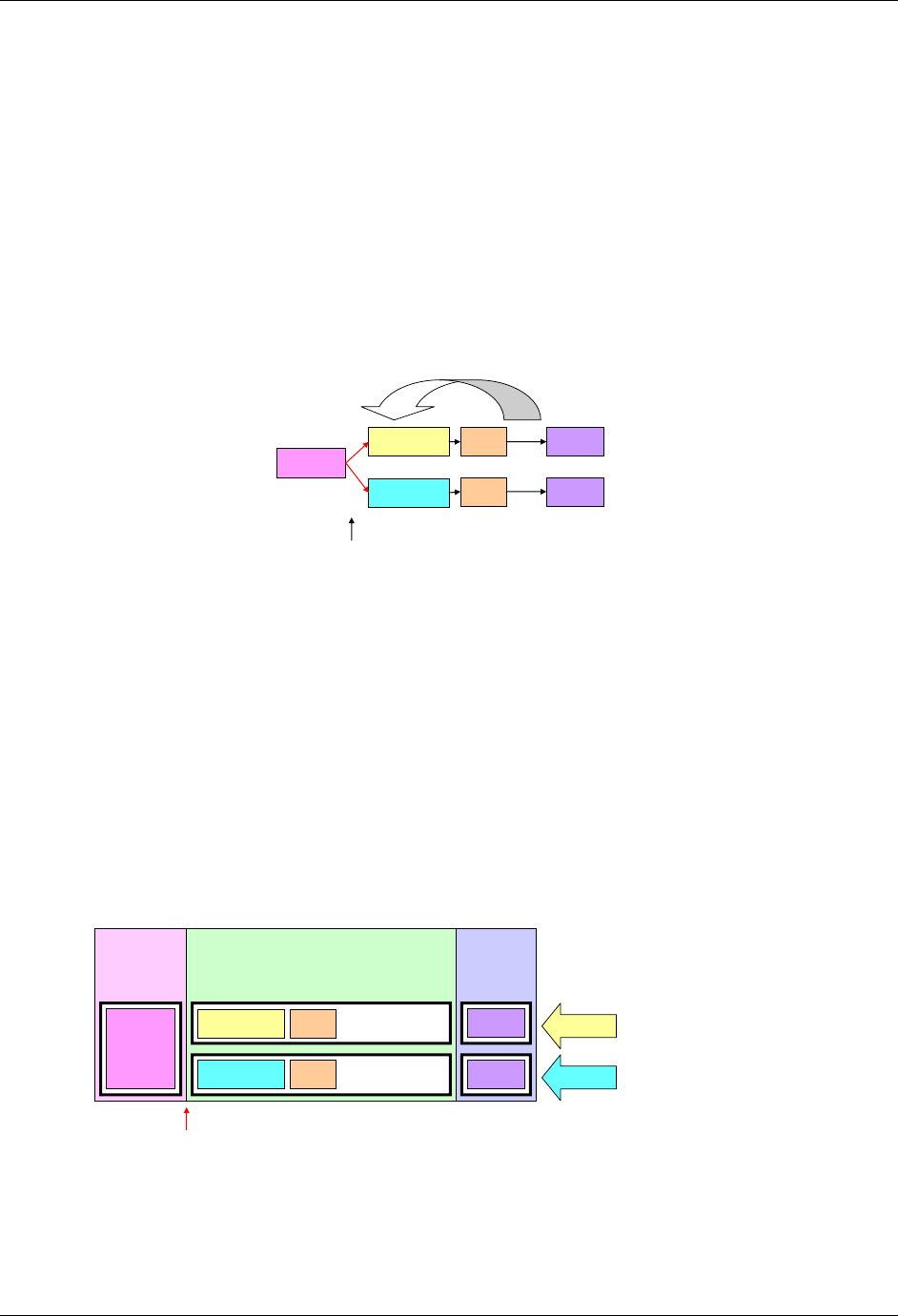
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 226 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.2.3.4 EXAMPLE TRIAL 4, CYCLES OF CHEMOTHERAPY
The diagram below uses a new symbol, a large curved arrow representing the fact that the chemotherapy treatment
(A or B) and the rest period that follows it are to be repeated. In this trial, the chemotherapy "cycles" are to be
repeated until disease progression. Some chemotherapy trials specify a maximum number of cycles, but protocols
that allow an indefinite number of repeats are not uncommon.
Example Trial 4: Cyclical Chemotherapy
Study Schema
Screen Drug A
Drug B
Randomization
Rest
Rest
Repeat until disease progression
Follow
Follow
The next diagram shows the prospective view of this trial. Note that, in spite of the repeating element structure, this is, at its
core, a two-arm parallel study, and thus has two arms. In SDTMIG 3.1.1, there was an implicit assumption that each element
must be in a separate epoch, and trials with cyclical chemotherapy were difficult to handle. The introduction of the concept of
study cells, and the dropping of the assumption that elements and epochs have a one to one relationship resolves these
difficulties. This trial is best treated as having just three epochs, since the main objectives of the trial involve comparisons
between the two treatments, and do not require data to be considered cycle by cycle.
Example Trial 4: Cyclical Chemotherapy
Prospective View
Treatment
Epoch
Screening
Epoch
Drug A
Drug B
Screen
Randomization
Drug A
Drug B
Follow-up
Epoch
Follow
Follow
Rest
Rest
Repeat until
disease progression
Repeat until
disease progression
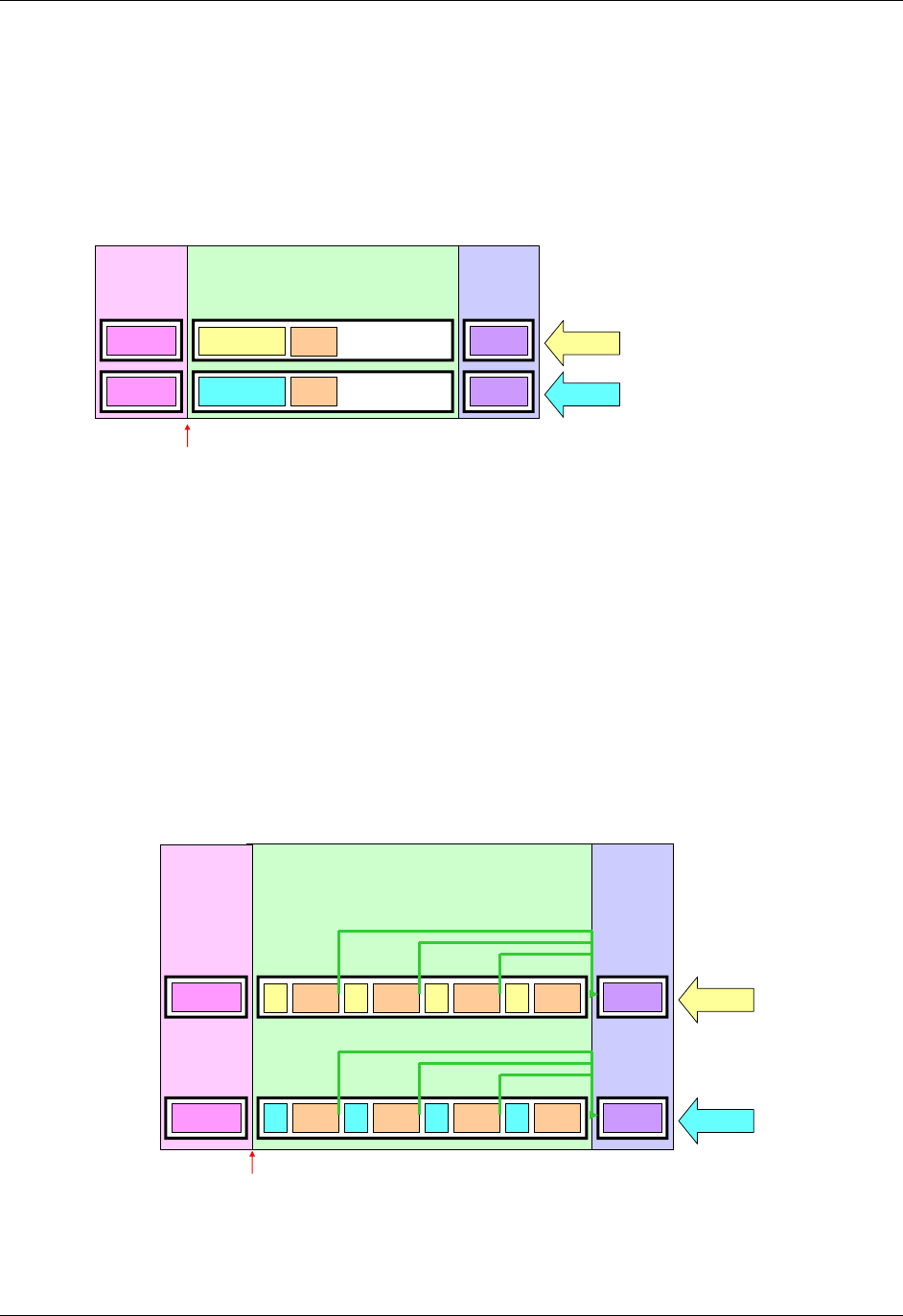
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 227
Final November 12, 2008
The next diagram shows the retrospective view of this trial.
Example Trial 4: Cyclical Chemotherapy
Retrospective View
Treatment
Epoch
Screening
Epoch
Drug A
Drug B
Screen
Randomization
Drug A
Drug B
Follow-up
Epoch
Follow
Follow
Rest
Rest
Repeat until
disease progression
Repeat until
disease progression
Screen
For the purpose of developing a Trial Arms dataset for this oncology trial, the diagram must be redrawn to explicitly represent
multiple treatment and rest elements. If a maximum number of cycles is not given by the protocol, then, for the purposes of
constructing an SDTM Trial Arms dataset for submission, which can only take place after the trial is complete, the number of
repeats included in the Trial Arms dataset should be the maximum number of repeats that occurred in the trial. The next
diagram assumes that the maximum number of cycles that occurred in this trial was four. Some subjects will not have received
all four cycles, because their disease progressed. The rule that directed that they receive no further cycles of chemotherapy is
represented by a set of green arrows, one at the end of each Rest epoch, that shows that a subject ―skips forward‖ if their
disease progresses. In the Trial Arms dataset, each "skip forward" instruction is a transition rule, recorded in the TATRANS
variable; when TATRANS is not populated, the rule is to transition to the next element in sequence.
Example Trial 4: Cyclical Chemotherapy
Retrospective View with Explicit Repeats
Treatment
Epoch
Screening
Epoch
Drug A
Drug B
Screen
Randomization
A
B
Follow-up
Epoch
Follow
Follow
Rest
RestScreen
A
B
Rest
Rest
A
B
Rest
Rest
A
B
Rest
Rest
If disease progression
If disease progression
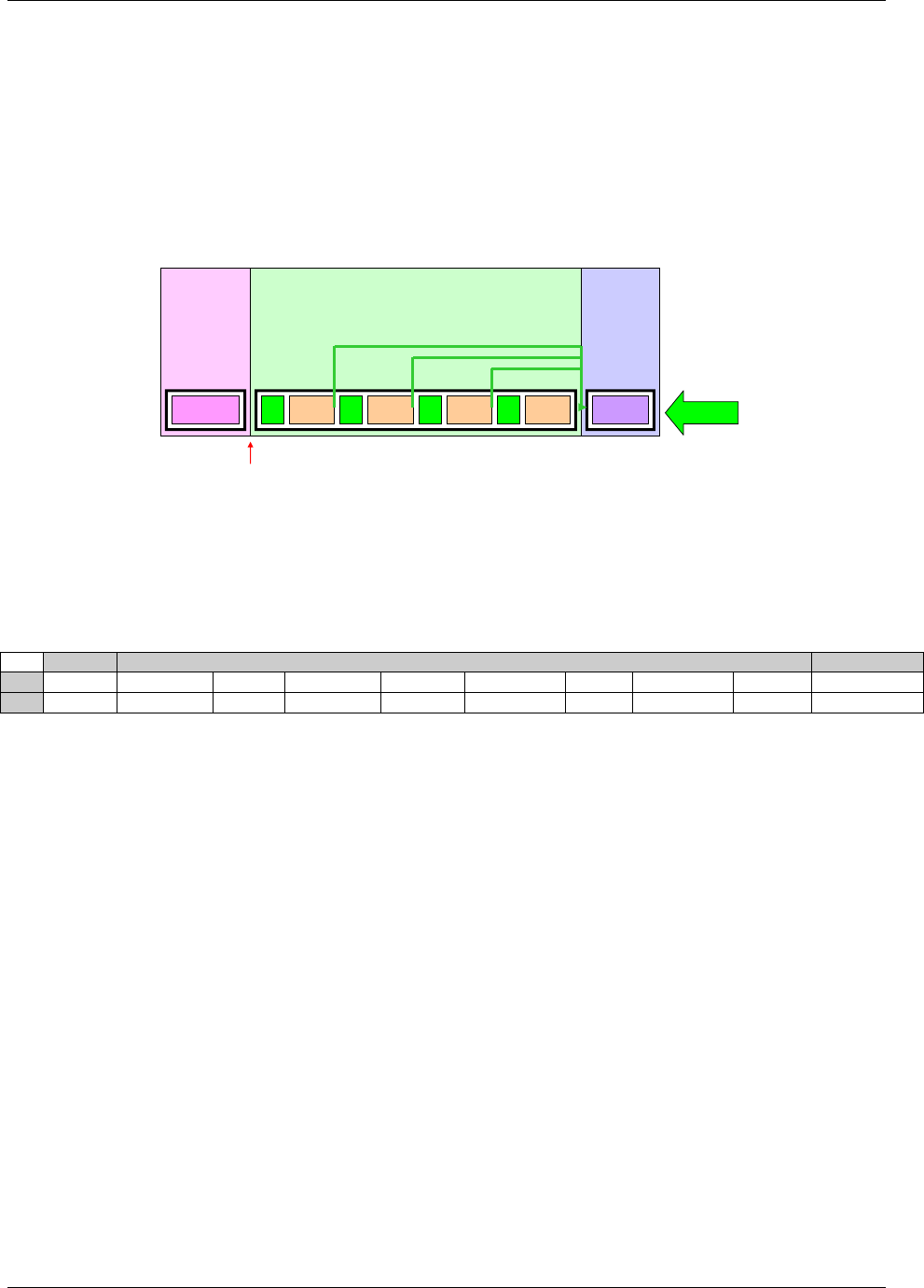
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 228 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The logistics of dosing mean that few oncology trials are blinded, if this trial is blinded, then the next diagram shows
the trial from the viewpoint of blinded participants.
Example Trial 4: Cyclical Chemotherapy
Blinded View
Treatment
Epoch
Screening
Epoch
Blind
Randomization
Rx
Follow-up
Epoch
FollowRestScreen Rx Rest Rx Rest Rx Rest
If disease progression
The Trial Design Matrix for Example Trial 4 corresponds to the diagram showing the retrospective view with
explicit repeats of the treatment and rest elements. As noted above, the Trial Design Matrix does not include
information on when randomization occurs; similarly, information corresponding to the ―skip forward‖ rules is not
represented in the Trial Design Matrix.
Trial Design Matrix for Example Trial 4
Screen
Treatment
Follow-up
A
Screen
Trt A
Rest
Trt A
Rest
Trt A
Rest
Trt A
Rest
Follow-up
B
Screen
Trt B
Rest
Trt B
Rest
Trt B
Rest
Trt B
Rest
Follow-up
The Trial Arms dataset for Example Trial 4 requires the use of the TATRANS variable in the Trial Arms dataset to
represent the "repeat until disease progression" feature. In order to represent this rule in the diagrams that explicitly
represent repeated elements, a green "skip forward" arrow was included at the end of each element where disease
progression is assessed. In the Trial Arms dataset, TATRANS is populated for each element with a green arrow in
the diagram. In other words, if there is a possibility that a subject will, at the end of this Element, "skip forward" to a
later part of the Arm, then TATRANS is populated with the rule describing the conditions under which a subject will
go to a later element. If the subject always goes to the next Element in the Arm (as was the case for the first three
example trials presented here) then TATRANS is null.
The Trial Arms datasets presented below corresponds to the Trial Design Matrix above.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 229
Final November 12, 2008
Trial Arms Dataset for Example Trial 4
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX4
TA
A
A
1
SCRN
Screen
Randomized to A
Screen
2
EX4
TA
A
A
2
A
Trt A
Treatment
3
EX4
TA
A
A
3
REST
Rest
If disease progression, go to Follow-up
Epoch
Treatment
4
EX4
TA
A
A
4
A
Trt A
Treatment
5
EX4
TA
A
A
5
REST
Rest
If disease progression, go to Follow-up
Epoch
Treatment
6
EX4
TA
A
A
6
A
Trt A
Treatment
7
EX4
TA
A
A
7
REST
Rest
If disease progression, go to Follow-up
Epoch
Treatment
8
EX4
TA
A
A
8
A
Trt A
Treatment
9
EX4
TA
A
A
9
REST
Rest
Treatment
10
EX4
TA
A
A
10
FU
Follow-up
Follow-up
11
EX4
TA
B
B
1
SCRN
Screen
Randomized to B
Screen
12
EX4
TA
B
B
2
B
Trt B
Treatment
13
EX4
TA
B
B
3
REST
Rest
If disease progression, go to Follow-up
Epoch
Treatment
14
EX4
TA
B
B
4
B
Trt B
Treatment
15
EX4
TA
B
B
5
REST
Rest
If disease progression, go to Follow-up
Epoch
Treatment
16
EX4
TA
B
B
6
B
Trt B
Treatment
17
EX4
TA
B
B
7
REST
Rest
If disease progression, go to Follow-up
Epoch
Treatment
18
EX4
TA
B
B
8
B
Trt B
Treatment
19
EX4
TA
B
B
9
REST
Rest
Treatment
20
EX4
TA
B
B
10
FU
Follow-up
Follow-up
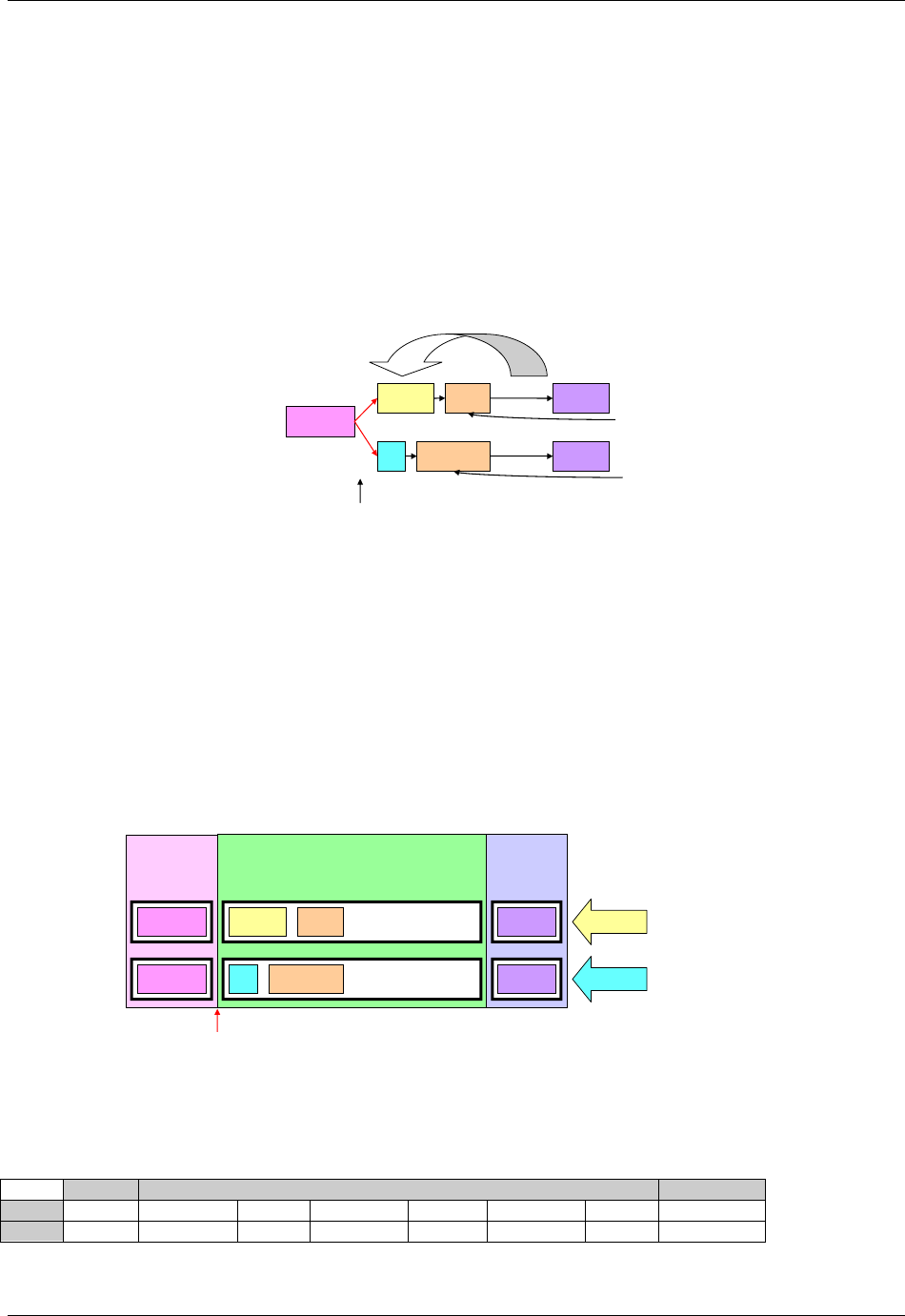
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 230 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.2.3.5 EXAMPLE TRIAL 5, CYCLES WITH DIFFERENT TREATMENT DURATIONS
Example Trial 5 is much like the last oncology trial in that the two treatments being compared are given in cycles,
and the total length of the cycle is the same for both treatments. However, in this trial Treatment A is given over
longer duration than Treatment B. Because of this difference in treatment patterns, this trial cannot be blinded.
Example Trial 5: Different Chemo Durations
Study Schema
Screen
Drug A
B
Randomization
Rest
Rest
Repeat until disease progression
Follow
Follow
Total length Drug A cycle: 3 weeks
Total length Drug B cycle: 3 weeks
In SDTMIG 3.1.1, the assumption of a one to one relationship between elements and epochs made this example
difficult to handle. However, without that assumption, this trial is essentially the same as Trial 4. The next diagram
shows the retrospective view of this trial.
Example Trial 5: Cyclical Chemotherapy
Retrospective View
Treatment
Epoch
Screening
Epoch
Drug A
Drug B
Screen
Randomization
Follow-up
Epoch
Follow
Follow
Repeat until
disease progression
Repeat until
disease progression
Screen
Drug A
B
Rest
Rest
The Trial Design Matrix for this trial is almost the same as for Example Trial 4; the only difference is that the
maximum number of cycles for this trial was assumed to be three.
Trial Design Matrix for Example Trial 5
Screen
Treatment
Follow-up
A
Screen
Trt A
Rest A
Trt A
Rest A
Trt A
Rest A
Follow-up
B
Screen
Trt B
Rest B
Trt B
Rest B
Trt B
Rest B
Follow-up
The Trial Arms dataset for this trial shown below corresponds to the Trial Design Matrix above.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 231
October 2008 Final
Trial Arms Dataset for Example Trial 5, with one Epoch per Cycle
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX5
TA
A
A
1
SCRN
Screen
Randomized to A
Screen
2
EX5
TA
A
A
2
A
Trt A
Treatment
3
EX5
TA
A
A
3
RESTA
Rest A
If disease progression, go to
Follow-up Epoch
Treatment
4
EX5
TA
A
A
4
A
Trt A
Treatment
5
EX5
TA
A
A
5
RESTA
Rest A
If disease progression, go to
Follow-up Epoch
Treatment
6
EX5
TA
A
A
6
A
Trt A
Treatment
7
EX5
TA
A
A
7
RESTA
Rest A
Treatment
8
EX5
TA
A
A
8
FU
Follow-up
Follow-up
9
EX5
TA
B
B
1
SCRN
Screen
Randomized to B
Screen
10
EX5
TA
B
B
2
B
Trt B
Treatment
11
EX5
TA
B
B
3
RESTB
Rest B
If disease progression, go to
Follow-up Epoch
Treatment
12
EX5
TA
B
B
4
B
Trt B
Treatment
13
EX5
TA
B
B
5
RESTB
Rest B
If disease progression, go to
Follow-up Epoch
Treatment
14
EX5
TA
B
B
6
B
Trt B
Treatment
15
EX5
TA
B
B
7
RESTB
Rest B
Treatment
16
EX5
TA
B
B
8
FU
Follow-up
Follow-up
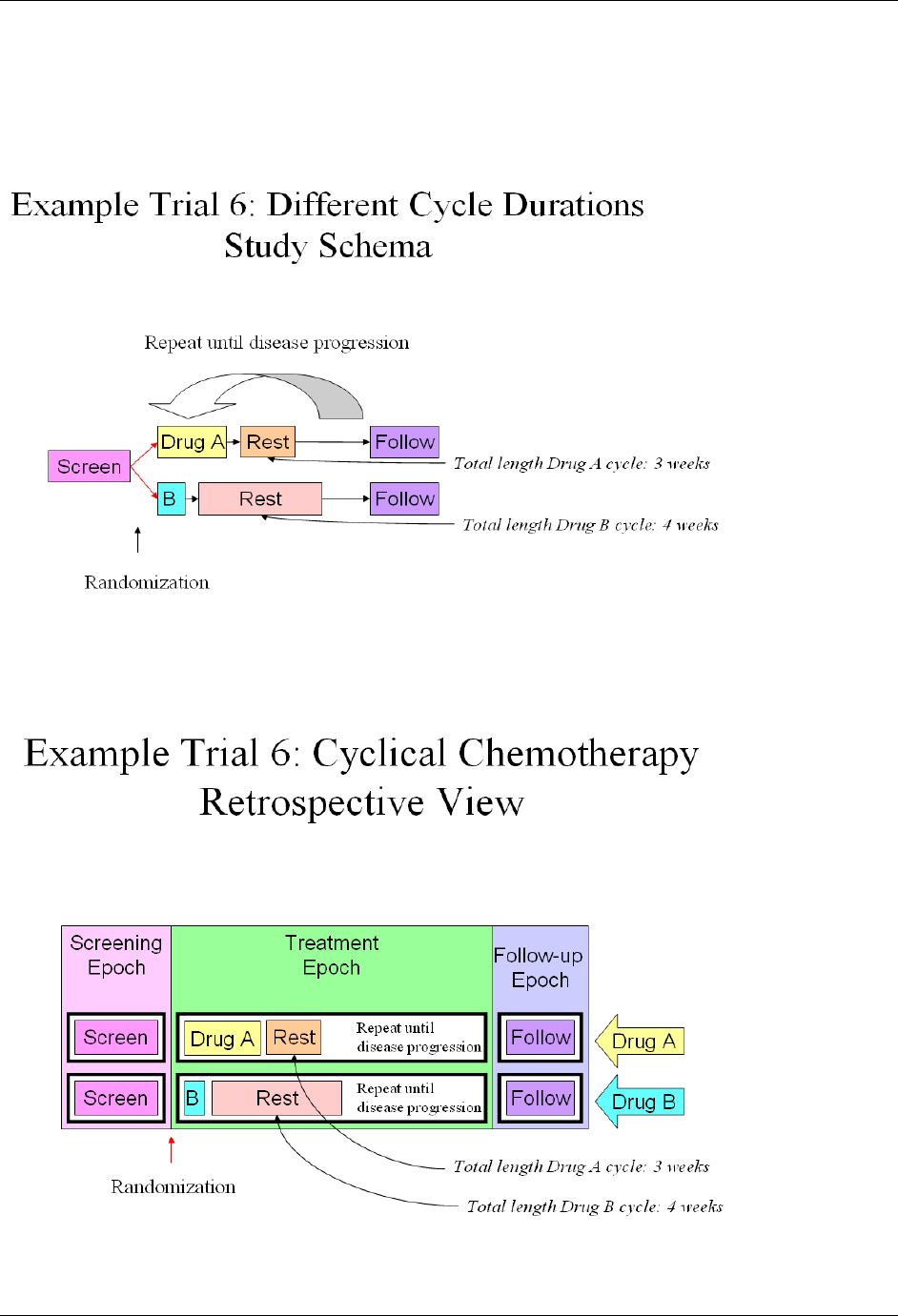
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 232 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.2.3.6 EXAMPLE TRIAL 6, CHEMOTHERAPY TRIAL WITH CYCLES OF DIFFERENT LENGTHS
Example Trial 6 is an oncology trial comparing two types of chemotherapy that are given using cycles of different
lengths with different internal patterns. Treatment A is given in 3-week cycles with a longer duration of treatment
and a short rest, while Treatment B is given in 4-week cycles with a short duration of treatment and a long rest.
The design of this trial is very similar to that for Example Trials 4 and 5. The main difference is that there are two
different rest elements, the short one used with Drug A and the long one used with Drug B. The next diagram shows the
retrospective view of this trial.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 233
Final November 12, 2008
The Trial Design Matrix for this trial assumes that there were a maximum of four cycles of Drug A and a maximum
of three cycles of Drug B.
Trial Design Matrix for Example Trial 6
Screen
Treatment
Follow-up
A
Screen
Trt A
Rest A
Trt A
Rest A
Trt A
Rest A
Trt A
Rest A
Follow-up
B
Screen
Trt
B
Rest B
Trt
B
Rest B
Trt
B
Rest B
Follow-up
In the following Trial Arms dataset, because the Treatment Epoch for Arm A has more Elements than the
Treatment Epoch for Arm B, TAETORD is 10 for the Follow-up Element in Arm A, but 8 for the Follow-up
Element in Arm B. It would also be possible to assign a TAETORD value of 10 to the Follow-up Element in Arm
B. The primary purpose of TAETORD is to order Elements within an Arm; leaving gaps in the series of
TAETORD values does not interfere with this purpose.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 234 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Trial Arms Dataset for Example Trial 6
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX6
TA
A
A
1
SCRN
Screen
Randomized to A
Screen
2
EX6
TA
A
A
2
A
Trt A
Treatment
3
EX6
TA
A
A
3
RESTA
Rest A
If disease progression, go to
Follow-up Epoch
Treatment
4
EX6
TA
A
A
4
A
Trt A
Treatment
5
EX6
TA
A
A
5
RESTA
Rest A
If disease progression, go to
Follow-up Epoch
Treatment
6
EX6
TA
A
A
6
A
Trt A
Treatment
7
EX6
TA
A
A
7
RESTA
Rest A
If disease progression, go to
Follow-up Epoch
Treatment
8
EX6
TA
A
A
8
A
Trt A
Treatment
9
EX6
TA
A
A
9
RESTA
Rest A
Treatment
10
EX6
TA
A
A
10
FU
Follow-up
Follow-up
11
EX6
TA
B
B
1
SCRN
Screen
Randomized to B
Screen
12
EX6
TA
B
B
2
B
Trt B
Treatment
13
EX6
TA
B
B
3
RESTB
Rest B
If disease progression, go to
Follow-up Epoch
Treatment
14
EX6
TA
B
B
4
B
Trt B
Treatment
15
EX6
TA
B
B
5
RESTB
Rest B
If disease progression, go to
Follow-up Epoch
Treatment
16
EX6
TA
B
B
6
B
Trt B
Treatment
17
EX6
TA
B
B
7
RESTB
Rest B
Treatment
18
EX6
TA
B
B
8
FU
Follow-up
Follow-up
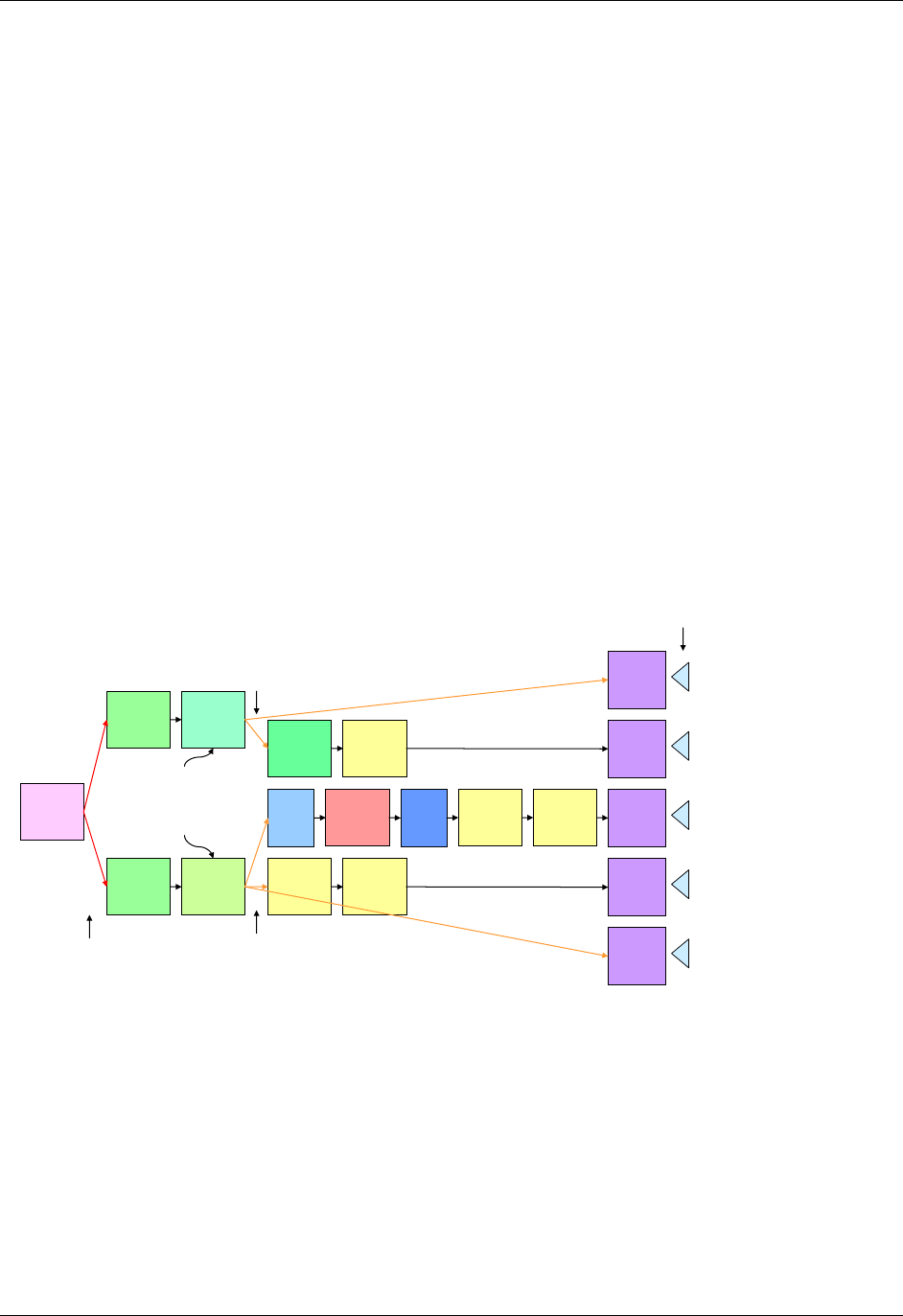
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 235
Final November 12, 2008
7.2.3.7 EXAMPLE TRIAL 7, TRIAL WITH DISPARATE ARMS
In open trials, there is no requirement to maintain a blind, and the Arms of a trial may be quite different from each
other. In such a case, changes in treatment in one Arm may differ in number and timing from changes in treatment in
another Arm, so that there is nothing like a one-to-one match between the Elements in the different Arms. In such a
case, Epochs are likely to be defined as broad intervals of time, spanning several Elements, and be chosen to
correspond to periods of time that will be compared in analyses of the trial.
Example Trial 7, RTOG 93-09, involves treatment of lung cancer with chemotherapy and radiotherapy, with or
without surgery. The protocol (RTOG-93-09), which is available online at the Radiation Oncology Therapy Group
(RTOG) website 1366Hhttp://www.rtog.org/members/numericactive.html , does not include a study schema diagram, but
does include a text-based representation of diverging ―options‖ to which a subject may be assigned. All subjects go
through the branch point at randomization, when subjects are assigned to either Chemotherapy + Radiotherapy (CR)
or Chemotherapy + Radiotherapy + Surgery (CRS). All subjects receive induction chemotherapy and radiation, with
a slight difference between those randomized to the two arms during the second cycle of chemotherapy. Those
randomized to the non-surgery arm are evaluated for disease somewhat earlier, to avoid delays in administering the
radiation boost to those whose disease has not progressed. After induction chemotherapy and radiation, subjects are
evaluated for disease progression, and those whose disease has progressed stop treatment, but enter follow-up. Not
all subjects randomized to receive surgery who do not have disease progression will necessarily receive surgery. If
they are poor candidates for surgery or do not wish to receive surgery, they will not receive surgery, but will receive
further chemotherapy. The diagram below is based on the text ―schema‖ in the protocol, with the five ―options‖ it
names. The diagram in this form might suggest that the trial has five arms.
Example Trial 7: RTOG 93-09
Study schema with 5 ―options‖
Chemo
+ Rad
Follow
3-5 w
Rest
Chemo
+ Rad*
Chemo
+ Rad**
Chemo
Screen
Chemo
+ Rad Chemo
+ Boost
4-6 w
Rest
Follow
Surgery Chemo Chemo Follow
Follow
Follow
Follow
Follow
Chemo Chemo
Randomization Evaluation for
Disease Progression and
Surgical Eligibility
Evaluation for
Disease Progression
Disease evaluation
* earlier
** later
2
5
3
4
1
Options
However, the objectives of the trial make it clear that this trial is designed to compare two treatment strategies,
chemotherapy and radiation with and without surgery, so this study is better modeled as a two-Arm trial, but with
major "skip forward" arrows for some subjects, as illustrated in the following diagram. This diagram also shows
more detail within the blocks labeled ―Induction Chemo + RT‖ and ―Additional Chemo‖ than the diagram above.
Both the ―induction‖ and ―additional‖ chemotherapy are given in two cycles. Also, the second induction cycle is
different for the two arms, since radiation therapy for those assigned to the non-surgery arm includes a ―boost‖
which those assigned to surgery arm do not receive.
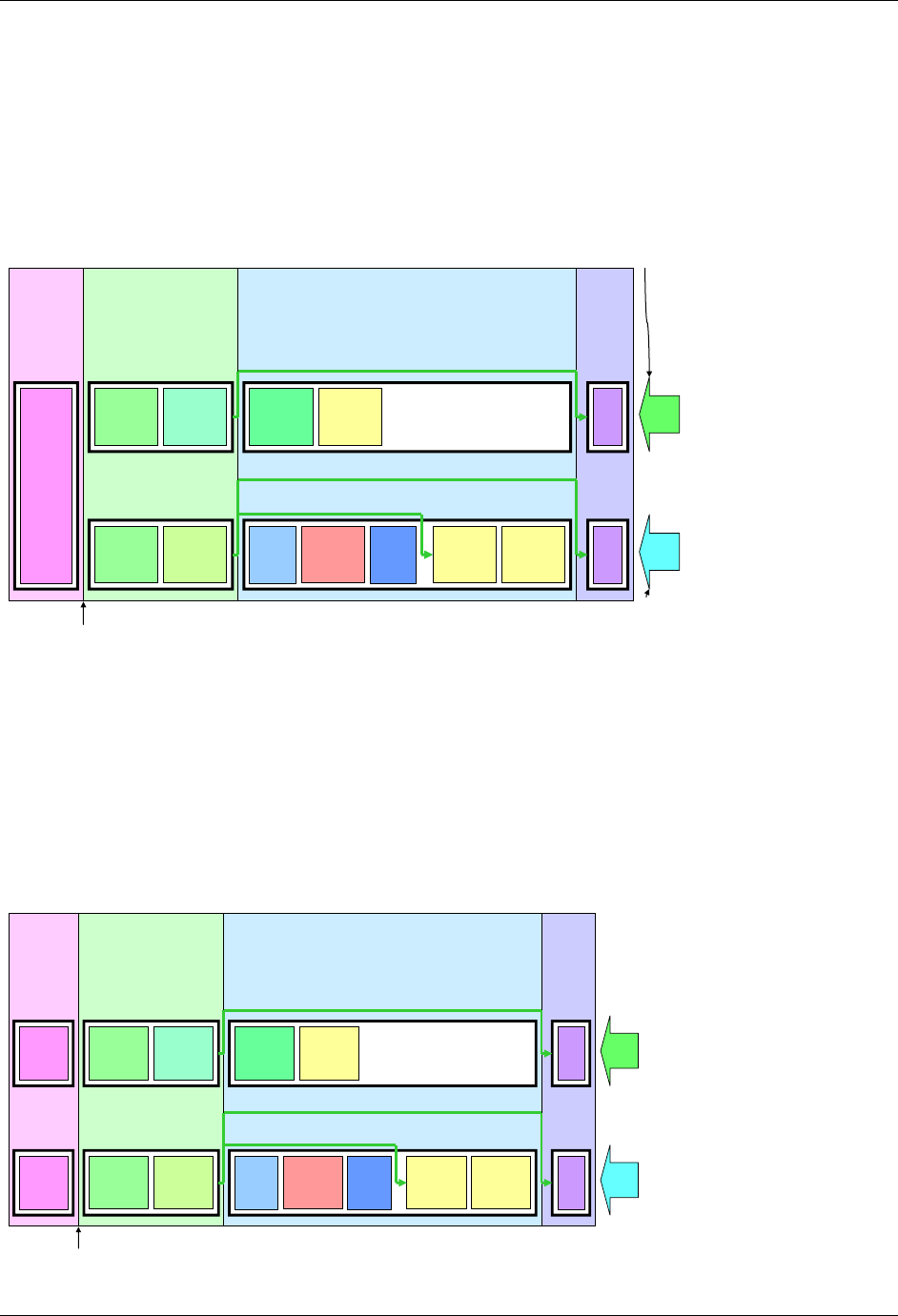
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 236 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The next diagram shows the prospective view of this trial. The protocol conceives of treatment as being divided into
two parts, Induction and Continuation, so these have been treated as two different epochs. This is also an important
point in the trial operationally, the point when subjects are ―registered‖ a second time, and when subjects are
identified who will ―skip forward‖ because of disease progression or ineligibility for surgery.
Follow
Epoch
Continuation
Treatment Epoch
Screen
Epoch Induction
Treatment
Epoch
Example Trial 7: RTOG 93-09
Prospective View
3-5 w
Rest
Chemo
Screen
Chemo
+ Boost
4-6 w
Rest
Surgery Chemo Chemo FU
FU
Randomization
Chemo
+ Rad Chemo
+ Rad**
Chemo
+ Rad*
Chemo
+ Rad
If not eligible for surgery
If disease progression
If disease progression
CR
CRS
Chemotherapy
and Radiation
Arm
Chemotherapy,
Radiation and
Surgery Arm
The next diagram shows the retrospective view of this trial. The fact that the elements in the study cell for the CR arm in the
Continuation Treatment Epoch do not fill the space in the diagram is an artifact of the diagram conventions. Those subjects
who do receive surgery will in fact spend a longer time completing treatment and moving into follow-up. Although it is
tempting to think of the horizontal axis of these diagrams as a timeline, this can sometimes be misleading. The diagrams are
not necessarily ―to scale‖ in the sense that the length of the block representing an element represents its duration, and elements
that line up on the same vertical line in the diagram may not occur at the same relative time within the study.
Follow
Epoch
Continuation
Treatment Epoch
Screen
Epoch Induction
Treatment
Epoch
Example Trial 7: RTOG 93-09
Retrospective View
3-5 w
Rest
Chemo
Screen Chemo
+ Boost
4-6 w
Rest
Surgery Chemo Chemo FU
FU
Randomization
Chemo
+ Rad Chemo
+ Rad**
Chemo
+ Rad*
Chemo
+ Rad
If not eligible for surgery
If disease progression
If disease progression
CR
CRS
Screen
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 237
Final November 12, 2008
The Trial Design Matrix for Example Trial 7, RTOG 93-09, a Two-Arm Trial is shown in the following table.
Screen
Induction
Continuation
Follow-up
CR
Screen
Initial
Chemo + RT
Chemo + RT
(non-Surgery)
Chemo
Chemo
Off Treatment
Follow-up
CRS
Screen
Initial
Chemo + RT
Chemo + RT
(Surgery)
3-5 w
Rest
Surgery
4-6 w
Rest
Chemo
Chemo
Off Treatment Follow-
up
The Trial Arms dataset for the trial is shown below for Example Trial 7, as a two-Arm trial
Row
STUDYID
DOMAIN
ARMCD
ARM
TAETORD
ETCD
ELEMENT
TABRANCH
TATRANS
EPOCH
1
EX7
TA
1
CR
1
SCRN
Screen
Randomized to CR
Screen
2
EX7
TA
1
CR
2
ICR
Initial Chemo + RT
Induction
3
EX7
TA
1
CR
3
CRNS
Chemo+RT (non-Surgery)
If progression, skip to
Follow-up.
Induction
4
EX7
TA
1
CR
4
C
Chemo
Continuation
5
EX7
TA
1
CR
5
C
Chemo
Continuation
6
EX7
TA
1
CR
6
FU
Off Treatment Follow-up
Follow-up
7
EX7
TA
2
CRS
1
SCRN
Screen
Randomized to CRS
Screen
8
EX7
TA
2
CRS
2
ICR
Initial Chemo + RT
Induction
9
EX7
TA
2
CRS
3
CRS
Chemo+RT (Surgery)
If progression, skip to
Follow-up. If no
progression, but
subject is ineligible for
or does not consent to
surgery, skip to Addl
Chemo.
Induction
10
EX7
TA
2
CRS
4
R3
3-5 week rest
Continuation
11
EX7
TA
2
CRS
5
SURG
Surgery
Continuation
12
EX7
TA
2
CRS
6
R4
4-6 week rest
Continuation
13
EX7
TA
2
CRS
7
C
Chemo
Continuation
14
EX7
TA
2
CRS
8
C
Chemo
Continuation
15
EX7
TA
2
CRS
9
FU
Off Treatment Follow-up
Follow-up

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 238 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.2.4 ISSUES IN TRIAL ARMS DATASETS
7.2.4.1 DISTINGUISHING BETWEEN BRANCHES AND TRANSITIONS
Both the Branch and Transition columns contain rules, but the two columns represent two different types of rules.
Branch rules represent forks in the trial flowchart, giving rise to separate Arms. The rule underlying a branch in
the trial design appears in multiple records, once for each "fork" of the branch. Within any one record, there is no
choice (no "if" clause) in the value of the Branch condition. For example, the value of TABRANCH for a record
in Arm A is "Randomized to Arm A" because a subject in Arm A must have been randomized to Arm A.
Transition rules are used for choices within an Arm. The value for TATRANS does contain a choice (an "if"
clause). In Example Trial 4, subjects who receive 1, 2, 3, or 4 cycles of Treatment A are all considered to belong
to Arm A.
In modeling a trial, decisions may have to be made about whether a decision point in the flow chart represents
the separation of paths that represent different Arms, or paths that represent variations within the same Arm, as
illustrated in the discussion of Example Trial 7. This decision will depend on the comparisons of interest in the
trial.
Some trials refer to groups of subjects who follow a particular path through the trial as "cohorts", particularly if
the groups are formed successively over time. The term "cohort" is used with different meanings in different
protocols and does not always correspond to an Arm.
7.2.4.2 SUBJECTS NOT ASSIGNED TO AN ARM
Some trial subjects may drop out of the study before they reach all of the branch points in the trial design. In the
Demographics domain, values of ARM and ARMCD must be supplied for such subjects, but the special values
used for these subjects should not be included in the Trial Arms dataset; only complete Arm paths should be
described in the Trial Arms dataset. Demographics Assumption 4 (1367HSection 5.1.1.1) describes special ARM and
ARMCD values used for subjects who do not reach the first branch point in a trial. When a trial design includes
two or more branches, special values of ARM and ARMCD may be needed for subjects who pass through the
first branch point, but drop out before the final branch point. See 1368HExample 7 in Section 5.1.1.2 for an example of
how to construct values of ARM and ARMCD for such trials.
7.2.4.3 DEFINING EPOCHS
The series of examples in 1369HSection 7.2.3 provides a variety of scenarios and guidance about how to assign Epoch
in those scenarios. In general, assigning Epochs for blinded trials is easier than for unblinded trials. The blinded
view of the trial will generally make the possible choices clear. For unblinded trials, the comparisons that will be
made between Arms can guide the definition of Epochs. For trials that include many variant paths within an Arm,
comparisons of Arms will mean that subjects on a variety of paths will be included in the comparison, and this is
likely to lead to definition of broader Epochs.
7.2.4.4 RULE VARIABLES
The Branch and Transition columns shown in the example tables are variables with a Role of ―Rule.‖ The values
of a Rule variable describe conditions under which something is planned to happen. At the moment, values of
Rule variables are text. At some point in the future, it is expected that these will become executable code. Other
Rule variables are present in the Trial Elements and Trial Visits datasets.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 239
Final November 12, 2008
7.3 TRIAL ELEMENTS
The Trial Elements (TE) dataset contains the definitions of the elements that appear in the Trial Arms (TA) dataset.
An Element may appear multiple times in the Trial Arms table because it appears either 1) in multiple Arms, 2)
multiple times within an Arm, or 3) both. However, an Element will appear only once in the Trial Elements table.
Each row in the TE dataset may be thought of as representing a "unique Element" in the sense of "unique" used
when a case report form template page for a collecting certain type of data is often referred to as "unique page." For
instance, a case report form might be described as containing 87 pages, but only 23 unique pages. By analogy, the
trial design matrix for Example 2000H Trial 1 in 1370HSection 7.2.3.1 has 9 Study Cells, each of which contains one Element,
but the same trial design matrix contains only 5 unique Elements, so the trial Elements dataset for that trial has only
5 records.
An Element is a building block for creating Study Cells and an Arm is composed of Study Cells. Or, from another
point of view, an Arm is composed of Elements, i.e., the trial design assigns subjects to Arms, which are comprised
of a sequence of steps called Elements.
Trial Elements represent an interval of time that serves a purpose in the trial and are associated with certain activities
affecting the subject. ―Week 2 to Week 4‖ is not a valid Element. A valid Element has a name that describes the
purpose of the Element and includes a description of the activity or event that marks the subject's transition into the
Element as well as the conditions for leaving the Element.
7.3.1 TRIAL ELEMENTS DATASET — TE
te.xpt, Trial Elements — Trial Design, Version 3.1.2 One record per planned Element
Variable
Name
Variable Label
Type
Controlled
Terms,
Codelist or
Format
Role
CDISC Notes
Core
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
DOMAIN
Domain Abbreviation
Char
2001HTE
Identifier
Two-character abbreviation for the domain.
Req
ETCD
Element Code
Char
*
Topic
ETCD (the companion to ELEMENT) is limited to 8
characters and does not have special character restrictions.
These values should be short for ease of use in
programming, but it is not expected that ETCD will need
to serve as a variable name.
Req
ELEMENT
Description of Element
Char
*
Synonym
Qualifier
The name of the Element.
Req
TESTRL
Rule for Start of
Element
Char
Rule
Expresses rule for beginning Element.
Req
TEENRL
Rule for End of Element
Char
Rule
Expresses rule for ending Element. Either TEENRL or
TEDUR must be present for each Element.
Perm
TEDUR
Planned Duration of
Element
Char
ISO 8601
Timing
Planned Duration of Element in ISO 8601 format. Used
when the rule for ending the Element is applied after a
fixed duration.
Perm
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 240 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.3.2 ASSUMPTIONS FOR TE DATASET
1. There are no gaps between Elements. The instant one Element ends, the next Element begins. A subject spends
no time ―between‖ Elements.
2. ELEMENT, the Description of the Element, usually indicates the treatment being administered during an Element, or,
if no treatment is being administered, the other activities that are the purpose of this period of time, such as Screening,
Follow-up, Washout. In some cases, this may be quite passive, such as Rest, or Wait (for disease episode).
3. TESTRL, the Rule for Start of Element, identifies the event that marks the transition into this Element. For
Elements that involve treatment, this is the start of treatment.
4. For Elements that do not involve treatment, TESTRL can be more difficult to define. For washout and follow-up
Elements, which always follow treatment Elements, the start of the Element may be defined relative to the end
of a preceding treatment. For example, a washout period might be defined as starting 24 or 48 hours after the
last dose of drug for the preceding treatment Element or Epoch. This definition is not totally independent of the
Trial Arms dataset, since it relies on knowing where in the trial design the Element is used, and that it always
follows a treatment Element. Defining a clear starting point for the start of a non-treatment Element that always
follows another non-treatment Element can be particularly difficult. The transition may be defined by a
decision-making activity such as enrollment or randomization. For example, every Arm of a trial which
involves treating disease episodes might start with a screening Element followed by an Element which consists
of waiting until a disease episode occurs. The activity that marks the beginning of the wait Element might be
randomization.
5. TESTRL for a treatment Element may be thought of as ―active‖ while the start rule for a non-treatment
Element, particularly a follow-up or washout Element, may be ―passive.‖ The start of a treatment Element will
not occur until a dose is given, no matter how long that dose is delayed. Once the last dose is given, the start of
a subsequent non-treatment Element is inevitable, as long as another dose is not given.
6. Note that the date/time of the event described in TESTRL will be used to populate the date/times in the Subject
Elements dataset, so the date/time of the event should be one that will be captured in the CRF.
7. Specifying TESTRL for an Element that serves the first Element of an Arm in the Trial Arms dataset involves
defining the start of the trial. In the examples in this document, obtaining informed consent has been used as
"Trial Entry."
8. TESTRL should be expressed without referring to Arm. If the Element appears in more than one Arm in the
Trial Arms dataset, then the Element description (ELEMENT) must not refer to any Arms.
9. TESTRL should be expressed without referring to Epoch. If the Element appears in more than one Epoch in the
Trial Arms dataset, then the Element description (ELEMENT) must not refer to any Epochs.
10. For a blinded trial, it is useful to describe TESTRL in terms that separate the properties of the event that are
visible to blinded participants from the properties that are visible only to those who are unblinded. For treatment
Elements in blinded trials, wording such as the following is suitable, "First dose of study drug for a treatment
Epoch, where study drug is X."
11. Element end rules are rather different from Element start rules. The actual end of one Element is the beginning
of the next Element. Thus the Element end rule does not give the conditions under which an Element does end,
but the conditions under which it should end or is planned to end.
12. At least one of TEENRL and TEDUR must be populated. Both may be populated.
13. TEENRL describes the circumstances under which a subject should leave this Element. Element end rules may
depend on a variety of conditions. For instance, a typical criterion for ending a rest Element between oncology
chemotherapy-treatment Elements would be, ―15 days after start of Element and after WBC values have
recovered.‖ The Trial Arms dataset, not the Trial Elements dataset, describes where the subject moves next, so
TEENRL must be expressed without referring to Arm.
14. TEDUR serves the same purpose as TEENRL for the special (but very common) case of an Element with a
fixed duration. TEDUR is expressed in ISO 8601. For example, a TEDUR value of P6W is equivalent to a
TEENRL of "6 weeks after the start of the Element."
15. Note that Elements that have different start and end rules are different Elements and must have different values
of ELEMENT and ETCD. For instance, Elements that involve the same treatment but have different durations
are different Elements. The same applies to non-treatment Elements. For instance, a washout with a fixed
duration of 14 days is different from a washout that is to end after 7 days if drug cannot be detected in a blood
sample, or after 14 days otherwise.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 241
Final November 12, 2008
7.3.3 TRIAL ELEMENTS EXAMPLES
Below are Trial Elements datasets for Example Trials 1 and 2 described in 1371HSection 7.2.3.1 and 1372HSection 7.2.3.2. Both
these trials are assumed to have fixed-duration Elements. The wording in TESTRL is intended to separate the
description of the event that starts the Element into the part that would be visible to a blinded participant in the trial
(e.g., "First dose of a treatment Epoch") from the part that is revealed when the study is unblinded (e.g., "where dose
is 5 mg"). Care must be taken in choosing these descriptions to be sure that they are "Arm and Epoch neutral." For
instance, in a crossover trial such as Example 2002HTrial 3 described in Section 7.2.3.3, where an Element may appear in
one of multiple Epochs, the wording must be appropriate for all the possible Epochs. The wording for Example Trial
2 uses the wording "a treatment Epoch." The SDS Team is considering adding a separate variable to the Trial
Elements dataset that would hold information on the treatment that is associated with an Element. This would make
it clearer which Elements are "treatment Elements‖, and therefore, which Epochs contain treatment Elements, and
thus are "treatment Epochs".
Trial Elements Dataset for Example Trial 1
Row
STUDYID
DOMAIN
ETCD
ELEMENT
TESTRL
TEENRL
TEDUR
1
EX1
TE
SCRN
Screen
Informed consent
1 week after start of Element
P7D
2
EX1
TE
RI
Run-In
Eligibility confirmed
2 weeks after start of Element
P14D
3
EX1
TE
P
Placebo
First dose of study drug,
where drug is placebo
2 weeks after start of Element
P14D
4
EX1
TE
A
Drug A
First dose of study drug,
where drug is Drug A
2 weeks after start of Element
P14D
5
EX1
TE
B
Drug B
First dose of study drug,
where drug is Drug B
2 weeks after start of Element
P14D
Trial Elements Dataset for Example Trial 2
Row
STUDYID
DOMAIN
ETCD
ELEME
NT
TESTRL
TEENRL
TEDUR
1
EX2
TE
SCRN
Screen
Informed consent
2 weeks after start of Element
P14D
2
EX2
TE
P
Placebo
First dose of a treatment
Epoch, where dose is
placebo
2 weeks after start of Element
P14D
3
EX2
TE
5
5 mg
First dose of a treatment
Epoch, where dose is 5 mg
drug
2 weeks after start of Element
P14D
4
EX2
TE
10
10 mg
First dose of a treatment
Epoch, where dose is 10
mg drug
2 weeks after start of Element
P14D
5
EX2
TE
REST
Rest
48 hrs after last dose of
preceding treatment Epoch
1 week after start of Element
P7D
6
EX2
TE
FU
Follow-up
48 hrs after last dose of
third treatment Epoch
3 weeks after start of Element
P21D

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 242 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The Trial Elements dataset for Example Trial 4 illustrates Element end rules for Elements that are not of fixed
duration. The Screen Element in this study can be up to 2 weeks long, but may end earlier, so is not of fixed
duration. The Rest Element has a variable length, depending on how quickly WBC recovers. Note that the start rules
for the A and B Elements have been written to be suitable for a blinded study.
Trial Elements Dataset for Example Trial 4
Row
STUDYID
DOMAIN
ETCD
ELEMENT
TESTRL
TEENRL
TEDUR
1
EX4
TA
SCRN
Screen
Informed Consent
Screening assessments are
complete, up to 2 weeks after
start of Element
2
EX4
TA
A
Trt A
First dose of treatment Element,
where drug is Treatment A
5 days after start of Element
P5D
3
EX4
TA
B
Trt B
First dose of treatment Element,
where drug is Treatment B
5 days after start of Element
P5D
4
EX4
TA
REST
Rest
Last dose of previous treatment
cycle + 24 hrs
At least 16 days after start of
Element and WBC recovered
5
EX4
TA
FU
Follow-up
Decision not to treat further
4 weeks
P28D
7.3.4 TRIAL ELEMENTS ISSUES
7.3.4.1 GRANULARITY OF TRIAL ELEMENTS
Deciding how finely to divide trial time when identifying trial Elements is a matter of judgment, as illustrated by the
following examples:
1. Example Trial 2 (described in 1373HSection 7.2.3.2, and with Elements described in 1374HSection 7.3.3) was represented
using three treatment Epochs separated by two washout Epochs and followed by a follow-up Epoch. It might
have been modeled using three treatment Epochs that included both the 2-week treatment period and the 1-week
rest period. Since the first week after the third treatment period would be included in the third treatment Epoch,
the Follow-up Epoch would then have a duration of 2 weeks.
2. In Example Trials 4, 5, and 6 in 1375HSection 7.2.3.4, 1376HSection 7.2.3.5 and 1377HSection 7.2.3.6 separate Treatment and Rest
Elements were identified. However, the combination of treatment and rest could be represented as a single
Element.
3. A trial might include a dose titration, with subjects receiving increasing doses on a weekly basis until certain
conditions are met. The trial design could be modeled in any of the following ways:
using several one-week Elements at specific doses, followed by an Element of variable length at the
chosen dose,
as a titration Element of variable length followed by a constant dosing Element of variable length
one Element with dosing determined by titration
The choice of Elements used to represent this dose titration will depend on the objectives of the trial and how
the data will be analyzed and reported. If it is important to examine side effects or lab values at each individual
dose, the first model is appropriate. If it is important only to identify the time to completion of titration, the
second model might be appropriate. If the titration process is routine and is of little interest, the third model
might be adequate for the purposes of the trial.
7.3.4.2 DISTINGUISHING ELEMENTS, STUDY CELLS, AND EPOCHS
It is easy to confuse Elements, which are reusable trial building blocks, with Study Cells, which contain the
Elements for a particular Epoch and Arm, and with Epochs, which are time periods for the trial as a whole. In part,
this is because many trials have Epochs for which the same Element appears in all Arms. In other words, in the trial
design matrix for many trials, there are columns (Epochs) in which all the Study Cells have the same contents.
Furthermore, it is natural to use the same name (e.g., Screen or Follow-up) for both such an Epoch and the single
Element that appears within it.
Confusion can also arise from the fact that, in the blinded treatment portions of blinded trials, blinded participants do
not know which Element a subject is in, but do know what Epoch the subject is in.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 243
Final November 12, 2008
In describing a trial, one way to avoid confusion between Elements and Epochs is to include "Element" or "Epoch" in
the values of ELEMENT or EPOCH when these values (such as Screening or Follow-up) would otherwise be the same.
It becomes tedious to do this in every case, but can be useful to resolve confusion when it arises or is likely to arise.
The difference between Epoch and Element is perhaps clearest in crossover trials. In Example Trial 2, as for most crossover
trials, the analysis of PK results would include both treatment and period effects in the model. ―Treatment effect‖ derives from
Element (Placebo, 5 mg, or 10 mg), while ―Period effect‖ derives from the Epoch (1st, 2nd, or 3rd Treatment Epoch).
7.3.4.3 TRANSITIONS BETWEEN ELEMENTS
The transition between one Element and the next can be thought of as a three-step process:
Step
Number
Step Question
How step question is answered by information in the Trial Design datasets
1
Should the subject leave
the current Element?
Criteria for ending the current Element are in TEENRL in the TE dataset.
2
Which Element should the
subject enter next?
If there is a branch point at this point in the trial, evaluate criteria described in
TABRANCH (e.g., randomization results) in the TA dataset
otherwise, if TATRANS in the TA dataset is populated in this Arm at this
point, follow those instructions
otherwise, move to the next Element in this Arm as specified by TAETORD in
the TA dataset.
3
What does the subject do
to enter the next Element?
The action or event that marks the start of the next Element is specified in
TESTRL in the TE dataset
Note that the subject is not "in limbo" during this process. The subject remains in the current Element until Step 3, at
which point the subject transitions to the new Element. There are no gaps between Elements.
From this table, it is clear that executing a transition depends on information that is split between the Trial Elements
and the Trial Arms datasets.
It can be useful, in the process of working out the Trial Design datasets, to create a dataset that supplements the Trial
Arms dataset with the TESTRL, TEENRL, and TEDUR variables, so that full information on the transitions is easily
accessible. However, such a working dataset is not an SDTM dataset, and should not be submitted.
The following table shows a fragment of such a table for Example Trial 4. Note that for all records that contain a particular
Element, all the TE variable values are exactly the same. Also, note that when both TABRANCH and TATRANS are
blank, the implicit decision in Step 2 is that the subject moves to the next Element in sequence for the Arm.
ARM
EPOCH
TAETORD
ELEMENT
TESTRL
TEENRL
TEDUR
TABRANCH
TATRANS
A
Screen
1
Screen
Informed Consent
Screening assessments
are complete, up to 2
weeks after start of
Element
Randomized
to A
A
Treatment
2
Trt A
First dose of treatment in
Element, where drug is
Treatment A
5 days after start of
Element
P5D
A
Treatment
3
Rest
Last dose of previous
treatment cycle + 24 hrs
16 days after start of
Element and WBC
recovers
If disease
progression, go to
Follow-up Epoch
A
Treatment
4
Trt A
First dose of treatment in
Element, where drug is
Treatment A
5 days after start of
Element
P5D
Note that both the second and fourth rows of this dataset involve the same Element, Trt A, and so TESTRL is the
same for both. The activity that marks a subject's entry into the fourth Element in Arm A is "First dose of treatment
Element, where drug is Treatment A." This is not the subject's very first dose of Treatment A, but it is their first dose
in this Element.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 244 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.4 TRIAL VISITS
The Trial Visits (TV) dataset describes the planned Visits in a trial. Visits are defined as "clinical encounters" and are
described using the timing variables VISIT, VISITNUM, and VISITDY.
Protocols define Visits in order to describe assessments and procedures that are to be performed at the Visits.
7.4.1 TRIAL VISITS DATASET — TV
tv.xpt, Trial Visits — Trial Design, Version 3.1.2. One record per planned Visit per Arm
Variable
Name
Variable Label
Type
Controlled
Terms,
Codelist or
Format
Role
CDISC Notes
Core
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
DOMAIN
Domain Abbreviation
Char
2003HTV
Identifier
Two-character abbreviation for the domain
Req
VISITNUM
Visit Number
Num
Topic
1. Clinical encounter number
2. Numeric version of VISIT, used for sorting.
Req
VISIT
Visit Name
Char
Synonym
Qualifier
1. Protocol-defined description of clinical encounter.
2. May be used in addition to VISITNUM and/or VISITDY
as a text description of the clinical encounter.
Perm
VISITDY
Planned Study Day of
Visit
Num
Timing
1. Planned study day of VISIT.
2. Due to its sequential nature, used for sorting.
Perm
ARMCD
Planned Arm Code
Char
*
Record
Qualifier
1.ARMCD is limited to 20 characters and does not
have special character restrictions. The maximum
length of ARMCD is longer than for other ―short‖
variables to accommodate the kind of values that are
likely to be needed for crossover trials. For example,
if ARMCD values for a seven-period crossover were
constructed using two-character abbreviations for
each treatment and separating hyphens, the length of
ARMCD values would be 20.
2. If the timing of Visits for a trial does not depend on
which ARM a subject is in, then ARMCD should be null.
Exp
ARM
Description of Planned
Arm
Char
*
Synonym
Qualifier
1. Name given to an Arm or Treatment Group.
2. If the timing of Visits for a trial does not depend on
which Arm a subject is in, then Arm should be left blank.
Perm
TVSTRL
Visit Start Rule
Char
Rule
Rule describing when the Visit starts, in relation to the
sequence of Elements.
Req
TVENRL
Visit End Rule
Char
Rule
Rule describing when the Visit ends, in relation to the
sequence of Elements.
Perm
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
7.4.2 ASSUMPTIONS FOR TV DATASET
1. Although the general structure of the Trial Visits dataset is "One Record per Planned Visit per Arm", for many
clinical trials, particularly blinded clinical trials, the schedule of Visits is the same for all Arms, and the structure of
the Trial Visits dataset will be "One Record per Planned Visit". If the schedule of Visits is the same for all Arms,
ARMCD should be left blank for all records in the TV dataset. For trials with trial Visits that are different for
different Arms, such as Example Trial 7 in 1378HSection 7.2.3.7, ARMCD and ARM should be populated for all records.
If some Visits are the same for all Arms, and some Visits differ by Arm, then ARMCD and ARM should be
populated for all records, to assure clarity, even though this will mean creating near-duplicate records for Visits that
are the same for all Arms.
2. A Visit may start in one Element and end in another. This means that a Visit may start in one Epoch and end in
another. For example, if one of the activities planned for a Visit is the administration of the first dose of study drug,
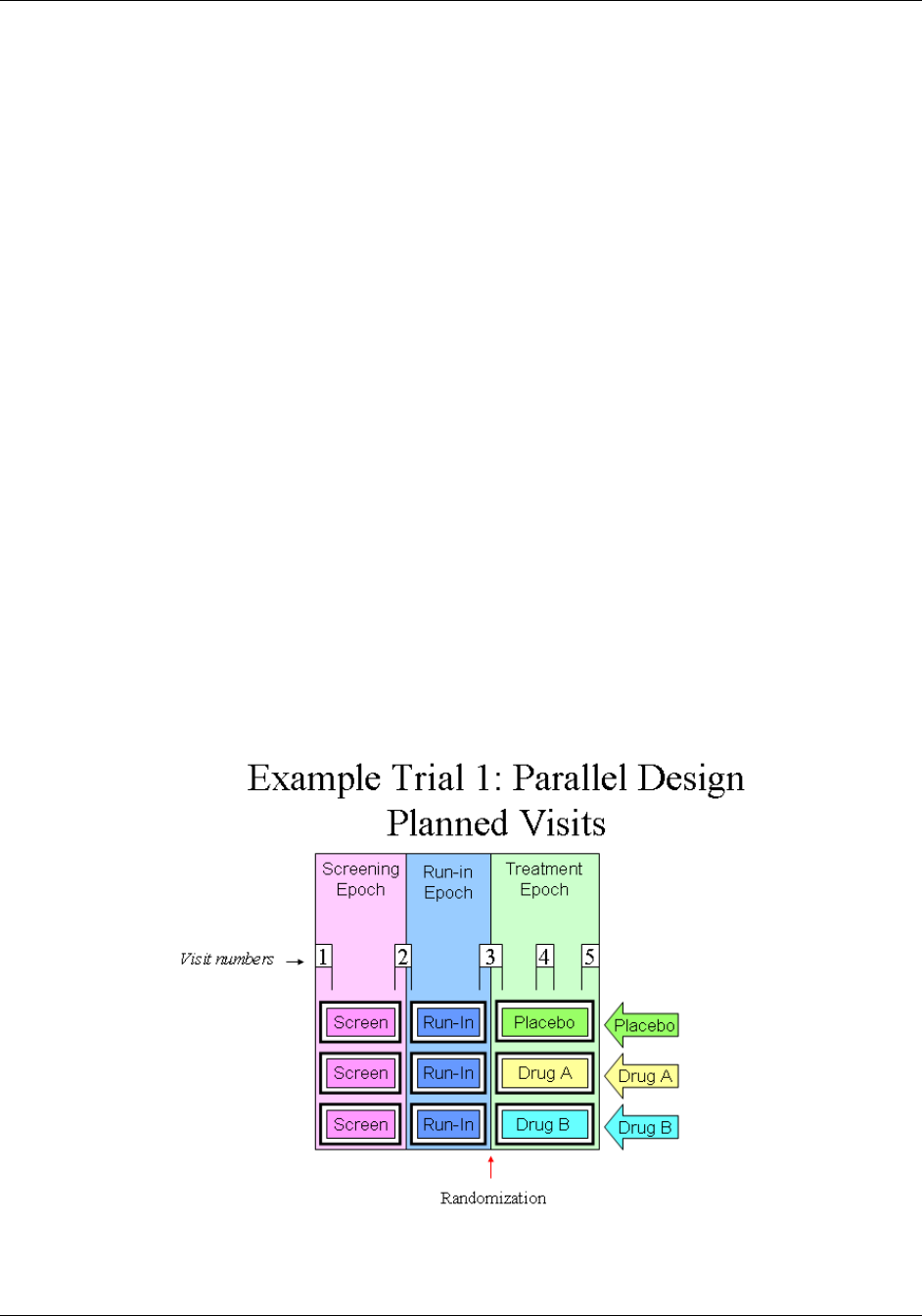
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 245
Final November 12, 2008
the Visit might start in the screen Epoch, in the screen Element, and end in a treatment Epoch, in a treatment
Element.
3. TVSTRL describes the scheduling of the Visit and should reflect the wording in the protocol. In many trials, all
Visits are scheduled relative to the study's Day 1, RFSTDTC. In such trials, it is useful to include VISITDY,
which is, in effect, a special case representation of TVSTRL.
4. Note that there is a subtle difference between the following two examples. In the first case, if Visit 3 were delayed for
some reason, Visit 4 would be unaffected. In the second case, a delay to Visit 3 would result in Visit 4 being delayed
as well.
Case 1: Visit 3 starts 2 weeks after RFSTDTC. Visit 4 starts 4 weeks after RFSTDTC.
Case 2: Visit 3 starts 2 weeks after RFSTDTC. Visit 4 starts 2 weeks after Visit 3.
5. Many protocols do not give any information about Visit ends because Visits are assumed to end on the same day
they start. In such a case, TVENRL may be left blank to indicate that the Visit ends on the same day it starts. Care
should be taken to assure that this is appropriate, since common practice may be to record data collected over more
than one day as occurring within a single Visit. Screening Visits may be particularly prone to collection of data over
multiple days. See 1379H Section 7.4.3 for examples showing how TVENRL could be populated.
6. The values of VISITNUM in the TV dataset are the valid values of VISITNUM for planned Visits. Any values of
VISITNUM that appear in subject-level datasets that are not in the TV dataset are assumed to correspond to
unplanned Visits. This applies, in particular, to the subject-level Subject Visits (SV) dataset; see 1380HSection 5.3.2 on
the SV dataset for additional information about handling unplanned Visits. If a subject-level dataset includes both
VISITNUM and VISIT, then records that include values of VISITNUM that appear in the TV dataset should also
include the corresponding values of VISIT from the TV dataset.
7.4.3 TRIAL VISITS EXAMPLES
The diagram below shows Visits by means of numbered "flags" with Visit Numbers. Each "flag" has two supports,
one at the beginning of the Visit, the other at the end of the Visit. Note that Visits 2 and 3 span Epoch transitions. In
other words, the transition event that marks the beginning of the Run-in Epoch (confirmation of eligibility) occurs
during Visit 2, and the transition event that marks the beginning of the Treatment Epoch (the first dose of study
drug) occurs during Visit 3.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 246 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Two Trial Visits datasets are shown for this trial. The first shows a somewhat idealized situation, where the protocol has given
specific timings for the Visits. The second shows a more usual situation, where the timings have been described only loosely.
Trial Visits Dataset for Example Trial 1 with explicitly scheduled starts and ends of Visits
Row
STUDYID
DOMAIN
VISITNUM
TVSTRL
TVENRL
1
EX1
TV
1
Start of Screen Epoch
1 hour after start of Visit
2
EX1
TV
2
30 minutes before end of Screen Epoch
30 minutes after start of Run-in
Epoch
3
EX1
TV
3
30 minutes before end of Run-in Epoch
1 hour after start of Treatment Epoch
4
EX1
TV
4
1 week after start of Treatment Epoch
1 hour after start of Visit
5
EX1
TV
5
2 weeks after start of Treatment Epoch
1 hour after start of Visit
Trial Visits Dataset for Example Trial 1 with loosely described starts and ends of Visits
Row
STUDYID
DOMAIN
VISITNUM
TVSTRL
TVENRL
1
EX1
TV
1
Start of Screen Epoch
2
EX1
TV
2
On the same day as, but before, the end
of the Screen Epoch
On the same day as, but after, the start
of the Run-in Epoch
3
EX1
TV
3
On the same day as, but before, the end
of the Run-in Epoch
On the same day as, but after, the start
of the Treatment Epoch
4
EX1
TV
4
1 week after start of Treatment Epoch
5
EX1
TV
5
2 weeks after start of Treatment Epoch
At Trial Exit
Although the start and end rules in this example reference the starts and ends of Epochs, the start and end rules of
some Visits for trials with Epochs that span multiple Elements will need to reference Elements rather than Epochs.
When an Arm includes repetitions of the same Element, it may be necessary to use TAETORD as well as an Element
name to specify when a Visit is to occur.
7.4.4 TRIAL VISITS ISSUES
7.4.4.1 IDENTIFYING TRIAL VISITS
In general, a trial's Visits are defined in its protocol. The term ―Visit‖ reflects the fact that data in outpatient studies
is usually collected during a physical Visit by the subject to a clinic. Sometimes a Trial Visit defined by the protocol
may not correspond to a physical Visit. It may span multiple physical Visits, as when screening data may be
collected over several clinic Visits but recorded under one Visit name (VISIT) and number (VISITNUM). A Trial
Visit may also represent only a portion of an extended physical Visit, as when a trial of in-patients collects data
under multiple Trial Visits for a single hospital admission.
Diary data and other data collected outside a clinic may not fit the usual concept of a Trial Visit, but the planned
times of collection of such data may be described as ―Visits‖ in the Trial Visits dataset if desired.
7.4.4.2 TRIAL VISIT RULES
Visit start rules are different from Element start rules because they usually describe when a Visit should occur, while
Element start rules describe the moment at which an Element is considered to start. There are usually gaps between
Visits, periods of time that do not belong to any Visit, so it is usually not necessary to identify the moment when one
Visit stops and another starts. However, some trials of hospitalized subjects may divide time into Visits in a manner
more like that used for Elements, and a transition event may need to be defined in such cases.
Visit start rules are usually expressed relative to the start or end of an Element or Epoch, e.g., ‖1-2 hours before end
of First Wash-out‖ or ―8 weeks after end of 2nd Treatment Epoch.‖ Note that the Visit may or may not occur during
the Element used as the reference for Visit start rule. For example, a trial with Elements based on treatment of
disease episodes might plan a Visit 6 months after the start of the first treatment period, regardless of how many
disease episodes have occurred.
Visit end rules are similar to Element end rules, describing when a Visit should end. They may be expressed relative
to the start or end of an Element or Epoch, or relative to the start of the Visit.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 247
Final November 12, 2008
The timings of Visits relative to Elements may be expressed in terms that cannot be easily quantified. For instance, a
protocol might instruct that at a baseline Visit the subject be randomized, given study drug, and instructed to take the
first dose of study Drug X at bedtime that night. This baseline Visit is thus started and ended before the start of the
treatment Epoch, but we don't know how long before the start of the treatment Epoch the Visit will occur. The trial
start rule might contain the value, "On the day of, but before, the start of the Treatment Epoch."
7.4.4.3 VISIT SCHEDULES EXPRESSED WITH RANGES
Ranges may be used to describe the planned timing of Visits (e.g., 12-16 days after the start of 2nd Element), but
this is different from the ―windows‖ that may be used in selecting data points to be included in an analysis
associated with that Visit. For example, although Visit 2 was planned for 12-16 days after the start of treatment, data
collected 10-18 days after the start of treatment might be included in a ―Visit 1― analysis. The two ranges serve
different purposes.
7.4.4.4 CONTINGENT VISITS
1381HSection 5.3.2, which describes the Subject Visits dataset, describes how records for unplanned Visits are
incorporated. It is sometimes difficult to decide exactly what constitutes an "unplanned Visit" versus a "contingent
Visit, " a Visit that is contingent on a "trigger" event, such as a certain adverse event, a finding above a certain
threshold value, or a decision to discontinue a subject‘s participation in the trial. Contingent Visits may be included
in the Trial Visits table, with start rules that describe the circumstances under which they will take place. Since
values of VISITNUM must be assigned to all records in the Trial Visits dataset, a contingent Visit included in the
Trial Visits dataset must have a VISITNUM, but the VISITNUM value may not be a "chronological" value, due to
the uncertain timing of the Visit.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 248 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.5 TRIAL INCLUSION/EXCLUSION CRITERIA
The Trial Inclusion Exclusion (TI) dataset is not subject oriented. It contains all the inclusion and exclusion criteria
for the trial, and thus provides information that may not be present in the subject-level data on inclusion and
exclusion criteria. The IE domain (described in 1382HSection 6.3.2) contains records only for inclusion and exclusion
criteria that subjects did not meet.
7.5.1 TRIAL INCLUSION/EXCLUSION CRITERIA DATASET — TI
ti.xpt, Trial Inclusion/Exclusion Criteria — Trial Design, Version 3.1.2.One record per I/E criterion
Variable
Name
Variable Label
Type
Controlled
Terms,
Codelist or
Format
Role
CDISC Notes
Core
STUDYID
Study Identifier
Char
Identifier
Unique identifier for a study.
Req
DOMAIN
Domain Abbreviation
Char
2004HTI
Identifier
Two-character abbreviation for the domain.
Req
IETESTCD
Incl/Excl Criterion
Short Name
Char
*
Topic
Short name IETEST. It can be used as a column name
when converting a dataset from a vertical to a horizontal
format. The value in IETESTCD cannot be longer than
8 characters, nor can it start with a number (e.g.,
―1TEST‖). IETESTCD cannot contain characters other
than letters, numbers, or underscores. The prefix ―IE‖ is
used to ensure consistency with the IE domain.
Req
IETEST
Inclusion/Exclusion
Criterion
Char
*
Synonym
Qualifier
Full text of the inclusion or exclusion criterion. The
prefix ―IE‖ is used to ensure consistency with the IE
domain.
Req
IECAT
Inclusion/Exclusion
Category
Char
(2005HIECAT)
Grouping
Qualifier
Used for categorization of the inclusion or exclusion
criteria.
Req
IESCAT
Inclusion/Exclusion
Subcategory
Char
*
Grouping
Qualifier
A further categorization of the exception criterion. Can
be used to distinguish criteria for a sub-study or for to
categorize as a major or minor exceptions. Examples:
MAJOR, MINOR.
Perm
TIRL
Inclusion/Exclusion
Criterion Rule
Char
Rule
Rule that expresses the criterion in computer-executable
form (see assumption 4 below).
Perm
TIVERS
Protocol Criteria
Versions
Char
Record
Qualifier
The number of this version of the Inclusion/Exclusion
criteria. May be omitted if there is only one version.
Perm
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)
7.5.2 ASSUMPTIONS FOR TI DATASET
1. If inclusion/exclusion criteria were amended during the trial, then each complete set of criteria must be included
in the TI domain. TIVERS is used to distinguish between the versions.
2. Protocol version numbers should be used to identify criteria versions, though there may be more versions of the
protocol than versions of the inclusion/exclusion criteria. For example, a protocol might have versions 1, 2, 3, and 4,
but if the inclusion/exclusion criteria in version 1 were unchanged through versions 2 and 3, and only changed in
version 4, then there would be two sets of inclusion/exclusion criteria in TI, one for version 1 and one for version 4.
3. Individual criteria do not have versions. If a criterion changes, it should be treated as a new criterion, with a
new value for IETESTCD. If criteria have been numbered and values of IETESTCD are generally of the form
INCL00n or EXCL00n, and new versions of a criterion have not been given new numbers, separate values of
IETESTCD might be created by appending letters, e.g. INCL003A, INCL003B.
4. IETEST contains the text of the inclusion/exclusion criterion. However, since entry criteria are rules, the
variable TIRL has been included in anticipation of the development of computer executable rules.
5. If a criterion text is <200 characters, it goes in IETEST; if the text is >200 characters, put meaningful text in
IETEST and describe the full text in the study metadata. See 1383HSection 4.1.5.3.1 for further information.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 249
Final November 12, 2008
7.5.3 EXAMPLES FOR TRIAL INCLUSION/EXCLUSION DATASET MODEL
This example shows records for a trial that had two versions of inclusion/exclusion criteria.
Rows 1-3 show the two inclusion criteria and one exclusion criterion for version 1 of the protocol.
Rows 4-6 show the inclusion/exclusion criteria for version 2.2 of the protocol, which changed the minimum age for
entry from 21 to 18.
Row
STUDYID
DOMAIN
IETESTCD
IETEST
IECAT
TIVERS
1
XYZ
TI
INCL01
Has disease under study
INCLUSION
1
2
XYZ
TI
INCL02
Age 21 or greater
INCLUSION
1
3
XYZ
TI
EXCL01
Pregnant or lactating
EXCLUSION
1
4
XYZ
TI
INCL01
Has disease under study
INCLUSION
2.2
5
XYZ
TI
INCL02A
Age 18 or greater
INCLUSION
2.2
6
XYZ
TI
EXCL01
Pregnant or lactating
EXCLUSION
2.2
7.6 TRIAL SUMMARY INFORMATION
The Trial Summary (TS) dataset allows the sponsor to submit a summary of the trial in a structured format. Each
record in the Trial Summary dataset contains the value of a parameter, a characteristic of the trial. For example, Trial
Summary is used to record basic information about the study such as trial phase, protocol title, and trial objectives.
The Trial Summary dataset contains information about the planned trial characteristics; it does not contain subject
level data or data that can be derived from subject data. Thus, for example, it includes the number of subjects
planned for the trial but not the number of subjects enrolled in the trial.
7.6.1 TRIAL SUMMARY DATASET — TS
ts.xpt, Trial Summary — Trial Design, Version 3.1.2.One record per trial summary parameter value
Variable
Name
Variable Label
Type
Controlled
Terms,
Codelist or
Format
Role
CDISC Notes
Core
STUDYID
Study
Identifier
Char
Identifier
Unique identifier for a study.
Req
DOMAIN
Domain
Abbreviation
Char
2006HTS
Identifier
Two-character abbreviation for the domain.
Req
TSSEQ
Sequence
Number
Num
Identifier
Sequence number given to ensure uniqueness within a
dataset. Allows inclusion of multiple records for the same
TSPARMCD, and can be used to join related records.
Req
TSGRPID
Group ID
Char
Identifier
Used to tie together a group of related records
Perm
TSPARMCD
Trial
Summary
Parameter
Short Name
Char
1384HTSPARMCD
Topic
TSPARMCD (the companion to TSPARM) is limited to 8
characters and does not have special character restrictions.
These values should be short for ease of use in programming,
but it is not expected that TSPARMCD will need to serve as
variable names. Examples: AGEMIN, AGEMAX
Req
TSPARM
Trial
Summary
Parameter
Char
1385HTSPARM
Synonym
Qualifier
Term for the Trial Summary Parameter. The value in
TSPARM cannot be longer than 40 characters. Examples
Planned Minimum Age of Subjects, Planned Maximum Age
of Subjects
Req
TSVAL
Parameter
Value
Char
*
Result
Qualifier
Value of TSPARM. Example: ―ASTHMA‖ when TSPARM
value is ―Trial Indication‖. TSVAL cannot be null – a value is
required for the record to be valid. Text over 200 characters
can be added to additional columns TSVAL1-TSVALn.
Req
* Indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code value)

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 250 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.6.2 ASSUMPTIONS FOR TRIAL SUMMARY DATASET MODEL
1. The intent of this dataset is to provide a summary of trial information. This is not subject level data.
2. A list of values for TSPARM and TSPARMCD is included in 1386HAppendix C3. The appendix also includes
assumptions related to particular parameters.
3. TSVAL may have controlled terminology depending on the value of TSPARMCD. See 1387HAppendix C3 for more
information.
4. There is not yet guidance on which Trial Summary parameters are required, but the following minimum
recommended set is based on the WHO International Clinical Trial Registry Platform (ICTRP) Registration
Data Set: TITLE, INDIC, TCNTRL, RANDOM, TRT, COMPTRT (when applicable), AGESPAN, AGEMIN,
AGEMAX, AGEU, SEXPOP, PLANSUB, OBJPRIM, OBJSEC. Parameters to support TRT (e.g., DOSE,
ROUTE, etc.) should be considered. However, for some complex study designs, these simple parameters may
not provide a useful summary of the trial design.
5. Sponsors may include parameters not in 1388HAppendix C3. The meaning of such parameters should be explained
in the metadata for the TS dataset.
6. For some trials, there will be multiple records in the Trial Summary dataset for a single parameter. For
example, a trial that addresses both Safety and Efficacy could have two records with TSPARMCD = TTYPE,
one with the TSVAL = "SAFETY" and the other with TSVAL = "EFFICACY."
7. TSSEQ has a different value for each record for the same parameter. Note that this is different from datasets
that contain subject data, where the --SEQ variable has a different value for each record for the same subject.
8. The method for treating text > 200 characters in Trial Summary is similar to that used for the Comments
special-purpose domain (1389HSection 5.2). If TSVAL is > 200 characters, then it should be split into multiple
variables, TSVAL-TSVALn.
9. Since TS does not contain subject-level data, there is no restriction analogous to the requirement in subject-
level datasets that the blocks bound by TSGRPID are within a subject. TSGRPID can be used to tie together
any block of records in the dataset. GRPID is most likely to be used when the TS dataset includes multiple
records for the same parameter. For example, if a trial compared a dose of 50 mg twice a day with a dose of
100 mg once a day, a record with TSPARMCD = DOSE and TSVAL=50 and a record with TSPARMCD =
DOSFREQ and TSVAL = BID could be assigned one GRPID, while a record with TSPARMCD = DOSE and
TSVAL=100 and a record with TSPARMCD = DOSFREQ and TSVAL = Q24H could be assigned a different
GRPID.
10. The order of parameters in the examples of TD datasets in 1390HSection 7.6.3 should not be taken as a requirement.
There are no requirements or expectations about the order of parameters within the TS dataset.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 251
Final November 12, 2008
7.6.3 EXAMPLES FOR TRIAL SUMMARY DATASET MODEL
Example 1:
Rows 1-5, 10-13,
15 and 16: Use controlled terminology for TSVAL (see 1391HAppendix C3).
Rows 1-6, 8-11,
13 and 14: Contain parameters from the recommended minimal set. The recommended (but not required)
parameters OBJSEC, TCNTRL, and PLANSUB are missing.
Rows 1-2: This trial includes subjects from both the ADULT (18-65) and ELDERLY (>65) age groups, so
there are two records for the AGESPAN parameter.
Row 7: The parameter DESIGN, which is not included in 1392HAppendix C3, was added by the Sponsor.
Rows 15-16: This trial addresses both safety and efficacy, so there are 2 records for the TYPE parameter.
Row
STUDYID
DOMAIN
TSSEQ
TSPARMCD
TSPARM
TSVAL
1
XYZ
TS
1
AGESPAN
Age Span
ADULT (18-65)
2
XYZ
TS
2
AGESPAN
Age Span
ELDERLY (> 65)
3
XYZ
TS
1
AGEMAX
Planned Maximum Age of
Subjects
70
4
XYZ
TS
1
AGEMIN
Planned Minimum Age of
Subjects
18
5
XYZ
TS
1
AGEU
Age Unit
YEARS
6
XYZ
TS
1
COMPTRT
Comparative Treatment
Name
PLACEBO
7
XYZ
TS
1
DESIGN
Description of Trial Design
PARALLEL
8
XYZ
TS
1
INDIC
Trial Indication
Asthma
9
XYZ
TS
1
OBJPRIM
Trial Primary Objective
Reduce the incidence of
exacerbations of asthma
10
XYZ
TS
1
RANDOM
Trial is Randomized
Y
11
XYZ
TS
1
SEXPOP
Sex of Participants
BOTH
12
XYZ
TS
1
TBLIND
Trial Blinding Schema
DOUBLE BLIND
13
XYZ
TS
1
TITLE
Trial Title
A 24 Week Study of Daily
Oral Investigational Drug vs.
Placebo in Subjects with
Asthma
14
XYZ
TS
1
TRT
Reported Name of Test
Product
Investigational New Drug
15
XYZ
TS
1
TTYPE
Trial Type
EFFICACY
16
XYZ
TS
2
TTYPE
Trial Type
SAFETY
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 252 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Example 2
Rows 1-3: AGEMIN, AGEMAX and AGEU are included, but AGESPAN is missing.
Row 5: The parameter DESIGN, which is not included in 1393HAppendix C3, was added by the Sponsor.
Row 7: Note that TSVAL for the LENGTH parameter is expressed in ISO 8601 duration format. Note that
TSVAL for LENGTH is P14W, while the TSVAL for TITLE includes "10-week." The trial involved 10
weeks of treatment, but the planned total duration of a subject's involvement in the study was 14
weeks.
Row 9: The parameter PLANEVAL is not in 1394HAppendix C3, but was added by the sponsor.
Row 15: The title is longer than 200 characters, so it has been separated into two pieces and stored in TSVAL
and TSVAL1.
Row 15: The title includes the information that dosing in the study was flexible. The sponsor felt this flexible
dosing could not be adequately represented using the usual dosing parameters, so none were submitted.
Row
STUDYID
DOMAIN
TSSEQ
TSPARMCD
TSPARM
TSVAL
TSVAL1
1
ABC
TS
1
AGEMIN
Planned Minimum
Age of Subjects
18
2
ABC
TS
1
AGEMAX
Planned
Maximum Age of
Subjects
64
3
ABC
TS
1
AGEU
Age Unit
YEARS
4
ABC
TS
1
COMPTRT
Comparative
Treatment Name
PLACEBO
5
ABC
TS
1
DESIGN
Description of
Trial Design
Parallel
6
ABC
TS
1
INDIC
Trial Indication
Generalized Disease
7
ABC
TS
1
LENGTH
Trial Length
P14W
8
ABC
TS
1
PLANSUB
Planned Number
of Subjects
500
9
ABC
TS
1
PLANEVAL
Planned Number
of Evaluable
Subjects
470
10
ABC
TS
1
SEXPOP
Sex of
Participants
BOTH
11
ABC
TS
1
RANDOM
Trial is
Randomized
Y
12
ABC
TS
1
TBLIND
Trial Blinding
Schema
DOUBLE BLIND
13
ABC
TS
1
TCNTRL
Type of Control
PLACEBO
14
ABC
TS
1
TINDTP
Trial Indication
Type
TREATMENT
15
ABC
TS
1
TITLE
Trial Title
A 10-Week,
Randomized, Double-
Blind, Placebo-
Controlled, Parallel-
Group, Flexible-
Dosage Study to
Evaluate the Efficacy
and Safety of New
Drug (up to
16 mg/day) in the
Treatment of
Adults With
Generalized
Disease
16
ABC
TS
1
TPHASE
Trial Phase
Classification
Phase III Trial
17
ABC
TS
1
TRT
Reported Name of
Test Product
New Drug
18
ABC
TS
1
TTYPE
Trial Type
EFFICACY
19
ABC
TS
2
TTYPE
Trial Type
SAFETY
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 253
Final November 12, 2008
Example 3
Rows 2-6, 11, 13-17,
19, 22, 25, and 26: Contain all the recommended minimum set of parameters except COMPTRT. Since the
TCNTL record indicates that this trial is placebo-controlled, the sponsor decided that
COMPTRT would be redundant, and did not submit it.
Rows 7 and 8: Since this is a trial comparing two doses, there are two DOSE records.
Rows 9, 10 and 17: These rows provide unit, frequency, and route data to support the DOSE records.
Row 12: Note that for the LENGTH parameter, TSVAL is expressed in ISO 8601 duration format.
Standard terminology for TSVAL is in upper case.
Rows 13, 14, and 24: This sponsor has chosen to submit all TSVAL values, including objectives and title, in upper
case.
Row
STUDYID
DOMAIN
TSSEQ
TSPARMCD
TSPARM
TSVAL
1
DEF
TS
1
ADDON
Added on to Existing
Treatments
N
2
DEF
TS
1
AGESPAN
Age Group
ADULT (18-65)
3
DEF
TS
2
AGESPAN
Age Group
ELDERLY (> 65)
4
DEF
TS
1
AGEMAX
Planned Maximum Age of
Subjects
75
5
DEF
TS
1
AGEMIN
Planned Minimum Age of
Subjects
22
6
DEF
TS
1
AGEU
Age Unit
YEARS
7
DEF
TS
1
DOSE
Dose per Administration
100
8
DEF
TS
2
DOSE
Dose per Administration
200
9
DEF
TS
1
DOSFRQ
Frequency
BID
10
DEF
TS
1
DOSEU
Dose Unit
mg
11
DEF
TS
1
INDIC
Trial Indication
TEST INDICATION
12
DEF
TS
1
LENGTH
Trial Length
P30M
13
DEF
TS
1
OBJPRIM
Trial Primary Objective
TO INVESTIGATE THE
SAFETY AND EFFICACY OF
TWO DOSES
14
DEF
TS
1
OBJSEC
Trial Secondary Objective
COMPARE SAFETY
PROFILES OF TWO DOSES
15
DEF
TS
1
PLANSUB
Planned Number of
Subjects
210
16
DEF
TS
1
RANDOM
Trial is Randomized
Y
17
DEF
TS
1
ROUTE
Route of Administration
ORAL
18
DEF
TS
1
SEXPOP
Sex of Participants
BOTH
19
DEF
TS
1
SPONSOR
Sponsoring Organization
SPONSOR NAME
20
DEF
TS
1
TBLIND
Trial Blinding Schema
DOUBLE BLIND
21
DEF
TS
1
TCNTRL
Type of Control
PLACEBO
22
DEF
TS
1
TDIGRP
Diagnosis Group
SUBJECTS DIAGNOSED
WITH DISEASE
23
DEF
TS
1
TINDTP
Trial Indication Type
TREATMENT
24
DEF
TS
1
TITLE
Trial Title
A RANDOMIZED, DOUBLE-
BLIND, PLACEBO-
CONTROLLED, MULTI-
CENTER, PARALLEL GROUP
DOSE RANGING STUDY.
25
DEF
TS
1
TPHASE
Trial Phase Classification
PHASE III TRIAL
26
DEF
TS
1
TRT
Reported Name of Test
Product
STUDY DRUG
27
DEF
TS
1
TTYPE
Trial Type
SAFETY
28
DEF
TS
2
TTYPE
Trial Type
EFFICACY

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 254 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
7.7 HOW TO MODEL THE DESIGN OF A CLINICAL TRIAL
The following steps allow the modeler to move from more-familiar concepts, such as Arms, to less-familiar
concepts, such as Elements and Epochs. The actual process of modeling a trial may depart from these numbered
steps. Some steps will overlap, there may be several iterations, and not all steps are relevant for all studies.
1. Start from the flow chart or schema diagram usually included in the trial protocol. This diagram will show
how many Arms the trial has, and the branch points, or decision points, where the Arms diverge.
2. Write down the decision rule for each branching point in the diagram. Does the assignment of a subject to
an Arm depend on a randomization? On whether the subject responded to treatment? On some other
criterion?
3. If the trial has multiple branching points, check whether all the branches that have been identified really
lead to different Arms. The Arms will relate to the major comparisons the trial is designed to address. For
some trials, there may be a group of somewhat different paths through the trial that are all considered to
belong to a single Arm.
4. For each Arm, identify the major time periods of treatment and non-treatment a subject assigned to that
Arm will go through. These are the Elements, or building blocks, of which the Arm is composed.
5. Define the starting point of each Element. Define the rule for how long the Element should last. Determine
whether the Element is of fixed duration.
6. Re-examine the sequences of Elements that make up the various Arms and consider alternative Element
definitions. Would it be better to ―split‖ some Elements into smaller pieces or ―lump‖ some Elements into
larger pieces? Such decisions will depend on the aims of the trial and plans for analysis.
7. Compare the various Arms. In most clinical trials, especially blinded trials, the pattern of Elements will be
similar for all Arms, and it will make sense to define Trial Epochs. Assign names to these Epochs. During
the conduct of a blinded trial, it will not be known which Arm a subject has been assigned to, or which
treatment Elements they are experiencing, but the Epochs they are passing through will be known.
8. Identify the Visits planned for the trial. Define the planned start timings for each Visit, expressed relative to
the ordered sequences of Elements that make up the Arms. Define the rules for when each Visit should end.
9. Identify the inclusion and exclusion criteria to be able to populate the TI dataset. If inclusion and exclusion
criteria were amended so that subjects entered under different versions, populate TIVERS to represent the
different versions.
10. Populate the TS dataset with summary information.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 255
Final November 12, 2008
8 Representing Relationships
and Data
The defined variables of the SDTM general observation classes could restrict the ability of sponsors to represent all
the data they wish to submit. Collected data that may not entirely fit includes relationships between records within a
domain, records in separate domains, and sponsor-defined ―variables.‖ As a result, the SDTM has methods to
represent five distinct types of relationships, all of which are described in more detail in subsequent sections. These
include the following:
1395HSection 8.1 describes a relationship between a group of records for a given subject within the same dataset.
1396HSection 8.2 describes a relationship between independent records (usually in separate datasets) for a
subject, such as a concomitant medication taken to treat an adverse event.
1397HSection 8.3 describes a relationship between two (or more) datasets where records of one (or more)
dataset(s) are related to record(s) in another dataset (or datasets).
1398HSection 8.4 describes a method for representing the dependent relationship where data that cannot be
represented by a standard variable within a general-observation-class dataset record (or records) can be
related back to that record.
1399HSection 8.5 describes a dependent relationship between a comment in the Comments domain (see also
1400HSection 5.2) and a parent record (or records) in other datasets, such as a comment recorded in association
with an adverse event.
1401HSection 8.6 discusses the concept of related datasets and whether to place additional data in a separate
dataset or a Supplemental Qualifier special-purpose dataset, and the concept of modeling Findings data that
refers to data in another general-observation-class dataset.
All relationships make use of the standard domain identifiers, STUDYID, DOMAIN, and USUBJID. In addition, the
variables IDVAR and IDVARVAL are used for identifying the record-level merge/join keys. These keys are used to
tie information together by linking records. The specific set of identifiers necessary to properly identify each type of
relationship is described in detail in the following sections. Examples of variables that could be used in IDVAR
include the following variables:
The Sequence Number (--SEQ) variable uniquely identifies a record for a given USUBJID within a
domain. The variable --SEQ is required in all domains except DM. For example, if subject 1234-2003 has
25 adverse event records in the adverse event (AE) domain, then 25 unique AESEQ values should be
established for this subject. Conventions for establishing and maintaining --SEQ values are sponsor-
defined. Values may or may not be sequential depending on data processes and sources.
The Reference Identifier (--REFID) variable can be used to capture a sponsor-defined or external identifier,
such as an identifier provided in an electronic data transfer. Some examples are lab-specimen identifiers
and ECG identifiers. --REFID is permissible in all domains, but never required. Values for --REFID are
sponsor -defined and can be any alphanumeric strings the sponsor chooses, consistent with their internal
practices.
The Grouping Identifier (--GRPID) variable, used to link related records for a subject within a dataset, is
explained below in 1402HSection 8.1.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 256 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
8.1 RELATING GROUPS OF RECORDS WITHIN A DOMAIN USING
THE --GRPID VARIABLE
The optional grouping identifier variable --GRPID is permissible in all domains that are based on the general
observation classes. It is used to identify relationships between records within a USUBJID within a single domain.
An example would be Intervention records for a combination therapy where the treatments in the combination varies
from subject to subject. In such a case, the relationship is defined by assigning the same unique character value to
the --GRPID variable. The values used for --GRPID can be any values the sponsor chooses; however, if the sponsor
uses values with some embedded meaning (rather than arbitrary numbers), those values should be consistent across
the submission to avoid confusion. It is important to note that --GRPID has no inherent meaning across subjects or
across domains.
Using --GRPID in the general-observation-class datasets can reduce the number of records in the RELREC, SUPP--,
and CO datasets when those datasets are submitted to describe relationships/associations for records or values to a
―group‖ of general-observation-class records.
8.1.1 --GRPID EXAMPLE
The following table illustrates how to use --GRPID in the Concomitant Medications (CM) domain to identify a
combination therapy. In this example, both subjects 1234 and 5678 have reported two combination therapies, each
consisting of three separate medications. Each component of a combination is given the same value for CMGRPID.
Note that for USUBJID 1234, the medications for CMGRPID = ―COMBO THPY 1‖ (Rows 1-3) are different from the
medications for CMGRPID = ―COMBO THPY 2‖ (Rows 4-6). Likewise, for USUBJID 5678, the medications for
CMGRPID = ―COMBO THPY 1‖ (Rows 7-9) are different from the medications for CMGRPID = ―COMBO THPY 2‖
(Rows 10-12). Additionally, the medications for Subject 1234 CMGRPID = ―COMBO THPY 1‖ and CMGRPID =
―COMBO THPY 2‖ (Rows 1-6) are different from the medications for Subject 5678 CMGRPID = ―COMBO THPY 1‖
and CMGRPID = ―COMBO THPY 2‖ (Rows 7-12). This example illustrates how CMGRPID groups information only
within a subject within a domain.
Row
STUDYID
DOMAIN
USUBJID
CMSEQ
CMGRPID
CMTRT
CMDECOD
CMDOSE
CMDOSU
CMSTDTC
CMENDTC
1
1234
CM
1234
1
COMBO
THPY 1
Verbatim
Med A
Generic Med
A
100
mg
2004-01-17
2004-01-19
2
1234
CM
1234
2
COMBO
THPY 1
Verbatim
Med B
Generic Med
B
50
mg
2004-01-17
2004-01-19
3
1234
CM
1234
3
COMBO
THPY 1
Verbatim
Med C
Generic Med
C
200
mg
2004-01-17
2004-01-19
4
1234
CM
1234
4
COMBO
THPY 2
Verbatim
Med D
Generic Med
D
150
mg
2004-01-21
2004-01-22
5
1234
CM
1234
5
COMBO
THPY 2
Verbatim
Med E
Generic Med
E
100
mg
2004-01-21
2004-01-22
6
1234
CM
1234
6
COMBO
THPY 2
Verbatim
Med F
Generic Med
F
75
mg
2004-01-21
2004-01-22
7
1234
CM
5678
1
COMBO
THPY 1
Verbatim
Med G
Generic Med
G
37.5
mg
2004-03-17
2004-03-25
8
1234
CM
5678
2
COMBO
THPY 1
Verbatim
Med H
Generic Med
H
60
mg
2004-03-17
2004-03-25
9
1234
CM
5678
3
COMBO
THPY 1
Verbatim
Med I
Generic Med I
20
mg
2004-03-17
2004-03-25
10
1234
CM
5678
4
COMBO
THPY 2
Verbatim
Med J
Generic Med
J
100
mg
2004-03-21
2004-03-22
11
1234
CM
5678
5
COMBO
THPY 2
Verbatim
Med K
Generic Med
K
50
mg
2004-03-21
2004-03-22
12
1234
CM
5678
6
COMBO
THPY 2
Verbatim
Med L
Generic Med
L
10
mg
2004-03-21
2004-03-22
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 257
Final November 12, 2008
8.2 RELATING PEER RECORDS
The Related Records (RELREC) special-purpose dataset is used to describe relationships between records for a
subject (as described in this section), and relationships between datasets (as described in 1403HSection 8.3). In both cases,
relationships represented in RELREC are collected relationships, either by explicit references or check boxes on the
CRF, or by design of the CRF, such as vital signs captured during an exercise stress test.
A relationship is defined by adding a record to RELREC for each record to be related and by assigning a unique
character identifier value for the relationship. Each record in the RELREC special-purpose dataset contains keys that
identify a record (or group of records) and the relationship identifier, which is stored in the RELID variable. The value
of RELID is chosen by the sponsor, but must be identical for all related records within USUBJID. It is recommended
that the sponsor use a standard system or naming convention for RELID (e.g., all letters, all numbers, capitalized).
Records expressing a relationship are specified using the key variables STUDYID, RDOMAIN (the two-letter domain
code of the record in the relationship), and USUBJID, along with IDVAR and IDVARVAL. Single records can be
related by using a unique-record-identifier variable such as --SEQ in IDVAR. Groups of records can be related by
using grouping variables such as --GRPID in IDVAR. IDVARVAL would contain the value of the variable described
in IDVAR. Using --GRPID can be a more efficient method of representing relationships in RELREC, such as when
relating an adverse event (or events) to a group of concomitant medications taken to treat the adverse event(s).
The RELREC dataset should be used to represent either:
Explicit relationships, such as concomitant medications taken as a result of an adverse event.
Information of a nature that necessitates using multiple datasets, as described in 1404HSection 8.3.
8.2.1 RELREC DATASET
relrec.xpt, Related Records, Version 3.1.2. One record per related record, group of records or dataset
Variable
Variable Label
Type
Controlled
Terms, Codelist
or Format
CDISC Notes
Core
References
STUDYID
Study Identifier
Char
Unique identifier for a study
Req
RDOMAIN
Related Domain
Abbreviation
Char
1405HDOMAIN
Two-character abbreviation for the domain of the parent
record(s)
Req
1406HSDTMIG
Appendix C2
USUBJID
Unique Subject
Identifier
Char
Identifier used to uniquely identify a subject across all
studies for all applications or submissions involving the
product.
Exp
IDVAR
Identifying
Variable
Char
*
Name of the identifying variable in the general-
observation-class dataset that identifies the related
record(s). Examples include --SEQ and --GRPID.
Req
IDVARVAL
Identifying
Variable Value
Char
Value of identifying variable described in IDVAR. If
--SEQ is the variable being used to describe this record,
then the value of --SEQ would be entered here.
Exp
RELTYPE
Relationship
Type
Char
ONE, MANY
Identifies the hierarchical level of the records in the
relationship. Values should be either ONE or MANY.
Used only when identifying a relationship between
datasets (as described in 1407HSection 8.3).
Exp
RELID
Relationship
Identifier
Char
Unique value within USUBJID that identifies the
relationship. All records for the same USUBJID that have
the same RELID are considered ―related/associated.‖
RELID can be any value the sponsor chooses, and is only
meaningful within the RELREC dataset to identify the
related/associated Domain records.
Req
*indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code
value)
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 258 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
8.2.2 RELREC DATASET EXAMPLES
Example 1:
This example shows how to use the RELREC dataset to relate records stored in separate domains for USUBJID
123456 who had two lab tests performed (Rows 5 and 6) and took two concomitant medications (Rows 2 and 3) as
the result of an adverse event (Rows 1 and 4). This example represents a situation in which the adverse event is
related to both the concomitant medications and the lab tests, but there is no relationship between the lab values and
the concomitant medications
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
1
EFC1234
AE
123456
AESEQ
5
1
2
EFC1234
CM
123456
CMSEQ
11
1
3
EFC1234
CM
123456
CMSEQ
12
1
4
EFC1234
AE
123456
AESEQ
5
2
5
EFC1234
LB
123456
LBSEQ
47
2
6
EFC1234
LB
123456
LBSEQ
48
2
Example 2:
Example 2 is the same scenario as Example 1; however, the relationship between concomitant medications (Rows 2
and 3) and lab values (Rows 4 and 5) and their relationship with the adverse event (Row 1) was collected.
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
1
EFC1234
AE
123456
AESEQ
5
1
2
EFC1234
CM
123456
CMSEQ
11
1
3
EFC1234
CM
123456
CMSEQ
12
1
4
EFC1234
LB
123456
LBSEQ
47
1
5
EFC1234
LB
123456
LBSEQ
48
1
Example 3:
Example 3 is the same scenario as Example 2. However, the two concomitant medications have been grouped by the
sponsor in the CM dataset by assigning a CMGRPID of ―COMBO 1‖, allowing the elimination of a record in the
RELREC dataset.
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
1
EFC1234
AE
123456
AESEQ
5
1
2
EFC1234
CM
123456
CMGRPID
COMBO1
1
3
EFC1234
LB
123456
LBSEQ
47
1
4
EFC1234
LB
123456
LBSEQ
48
1
Additional examples may be found in the domain examples such as the examples for Disposition/Adverse Event
found in 1408HSection 6.2.2.2, Example 4, and all of the Pharmacokinetics examples in 1409HSection 6.3.10.5.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 259
Final November 12, 2008
8.3 RELATING DATASETS
The Related Records (RELREC) special-purpose dataset can also be used to identify relationships between datasets
(e.g., a one-to-many or parent-child relationship). The relationship is defined by including a single record for each
related dataset that identifies the key(s) of the dataset that can be used to relate the respective records.
Relationships between datasets should only be recorded in the RELREC dataset when the sponsor has found it
necessary to split information between datasets that are related, and that may need to be examined together for
analysis or proper interpretation. Note that it is not necessary to use the RELREC dataset to identify associations
from data in the SUPP-- datasets or the CO dataset to their parent general-observation-class dataset records or
special-purpose domain records, as both these datasets include the key variable identifiers of the parent record(s)
that are necessary to make the association.
8.3.1 RELREC DATASET RELATIONSHIP EXAMPLE
This example shows how to use the RELREC dataset to represent related information that is submitted as two
datasets that have a one-to-many relationship. In the example below all the records in one domain are being related
to all of the records in the other, so both USUBJID and IDVARVAL are null.
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
RELTYPE
RELID
1
EFC1234
MB
MBGRPID
ONE
A
2
EFC1234
MS
MSGRPID
MANY
A
In the sponsor's operational database, these datasets may have existed as either separate datasets that were merged
for analysis, or one dataset that may have included observations from more than one general observation class (e.g.,
Events and Findings). The value in IDVAR must be the name of the key used to merge/join the two datasets. In the
above example, the --GRPID variable is used as the key to identify the related observations. The values for the
--GRPID variable in the two datasets are sponsor defined. Although other variables may also serve as a single merge
key when the corresponding values for IDVAR are equal, --GRPID, --SPID, or --REFID are typically used for this
purpose.
The variable RELTYPE identifies the type of relationship between the datasets. The allowable values are ONE and
MANY (controlled terminology is expected). This information defines how a merge/join would be written, and what
would be the result of the merge/join. The possible combinations are the following:
1. ONE and ONE. This combination indicates that there is NO hierarchical relationship between the datasets
and the records in the datasets. Only one record from each dataset will potentially have the same value of
the IDVAR within USUBJID.
2. ONE and MANY. This combination indicates that there IS a hierarchical (parent/child) relationship
between the datasets. One record within USUBJID in the dataset identified by RELTYPE=ONE will
potentially have the same value of the IDVAR with many (one or more) records in the dataset identified
by RELTYPE=MANY.
3. MANY and MANY. This combination is unusual and challenging to manage in a merge/join, and may
represent a relationship that was never intended to convey a usable merge/join (such as in described for
PC and PP in 1410HSection 6.3.10.5).
Since IDVAR identifies the keys that can be used to merge/join records between the datasets, the root values (i.e.,
SPID in the above example) for IDVAR must be the same for both records with the same RELID. --SEQ cannot be
used because --SEQ only has meaning within a subject within a dataset, not across datasets.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 260 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
8.4 RELATING NON-STANDARD VARIABLES VALUES TO A PARENT
DOMAIN
The SDTM does not allow the addition of new variables. Therefore, the Supplemental Qualifiers special purpose
dataset model is used to capture non-standard variables and their association to parent records in general-
observation-class datasets (Events, Findings, Interventions) and Demographics. Supplemental Qualifiers may be
represented as either a single SUPPQUAL dataset per study or via separate SUPP-- datasets for each dataset
containing sponsor-defined variables (see 1411HSection 8.4.2 for more on this topic). Most references in this guide will
use the designation of SUPP-- rather than SUPPQUAL to serve as a reminder of the preferred submission format for
Supplemental Qualifiers.
SUPP-- represents the metadata and data for each non-standard variable/value combination. As the name
"Supplemental Qualifiers" suggests, this dataset is intended to capture additional Qualifiers for an observation. Data
that represent separate observations should be treated as separate observations, either in this domain or another
domain. The Supplemental Qualifiers dataset is structured similarly to the RELREC dataset, in that it uses the same
set of keys to identify parent records. Each SUPP-- record also includes the name of the Qualifier variable being
added (QNAM), the label for the variable (QLABEL), the actual value for each instance or record (QVAL), the
origin (QORIG) of the value (see 1412HSection 4.1.1.8), and the Evaluator (QEVAL) to specify the role of the individual
who assigned the value (such as ADJUDICATION COMMITTEE or SPONSOR). Controlled terminology for
certain expected values for QNAM and QLABEL are included in 2007HAppendix C5.
SUPP-- datasets are also used to capture attributions. An attribution is typically an interpretation or subjective
classification of one or more observations by a specific evaluator, such as a population flag that classifies a subject or their
data according to their evaluability for efficacy analysis, or whether an observation is considered to be clinically
significant. Since it is possible that different attributions may be necessary in some cases, SUPP-- provides a mechanism
for incorporating as many attributions as are necessary. A SUPP-- dataset can contain both objective data (where values
are collected or derived algorithmically) and subjective data (attributions where values are assigned by a person or
committee). For objective data, the value in QEVAL will be null. For subjective data (when QORIG=‖ASSIGNED‖), the
value in QEVAL should reflect the role of the person or institution assigning the value (e.g., SPONSOR or
ADJUDICATION COMMITTEE).
The combined set of values for the first six columns (STUDYID…QNAM) should be unique for every record. That
is, there should not be multiple records in a SUPP-- dataset for the same QNAM value, as it relates to
IDVAR/IDVARVAL for a USUBJID in a domain. For example, if two individuals provide a determination on
whether an Adverse Event is Treatment Emergent (e.g., the investigator and an independent adjudicator) then
separate QNAM values should be used for each set of information, perhaps AETRTEMI and AETRTEMA. This is
necessary to ensure that reviewers can join/merge/transpose the information back with the records in the original
domain without risk of losing information.
When populating a SUPPDM dataset with population flags related to the Demographics domain (subject-level
evaluability), there should be one record for each population flag for each subject. QVAL values for population flags
should be Y or N, with no null values. In the event that evaluability is based upon individual visits or CRF pages,
additional population flags attached to other domains may be included in SUPP-- datasets.
Just as use of the optional grouping identifier variable, --GRPID, can be a more efficient method of representing
relationships in RELREC, it can also be used in a SUPP-- dataset to identify individual qualifier values (SUPP--
records) related to multiple general-observation-class domain records that could be grouped, such as relating an
attribution to a group of ECG measurements.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 261
Final November 12, 2008
8.4.1 SUPPLEMENTAL QUALIFIERS: SUPPQUAL OR SUPP-- DATASETS
supp--.xpt, Supplemental Qualifiers [DOMAIN NAME], Version 3.1.2. One record per IDVAR, IDVARVAL,
and QNAM value per subject
Variable
Variable
Label
Type
Controlled
Terms, Codelist
or Format
CDISC Notes
Core
References
STUDYID
Study
Identifier
Char
Study Identifier of the Parent record(s).
Req
SDTM 2.2.4
RDOMAIN
Related
Domain
Abbreviation
Char
1413HDOMAIN
Two-character abbreviation for the domain of the parent
record(s).
Req
SDTM 2.2.4,
1414HSDTMIG
Appendix C2
USUBJID
Unique
Subject
Identifier
Char
Unique Subject Identifier of the Parent record(s).
Req
SDTM 2.2.4
IDVAR
Identifying
Variable
Char
*
Identifying variable in the dataset that identifies the related
record(s). Examples: --SEQ, --GRPID.
Exp
IDVARVAL
Identifying
Variable Value
Char
Value of identifying variable of the parent record(s).
Exp
QNAM
Qualifier
Variable Name
Char
*
The short name of the Qualifier variable, which is used as
a column name in a domain view with data from the parent
domain. The value in QNAM cannot be longer than 8
characters, nor can it start with a number (e.g., ―1TEST‖).
QNAM cannot contain characters other than letters,
numbers, or underscores. This will often be the column
name in the sponsor‘s operational dataset.
Req
1415HSDTMIG
4.1.2.1,
2008HAppendix C5
QLABEL
Qualifier
Variable Label
Char
This is the long name or label associated with QNAM. The
value in QLABEL cannot be longer than 40 characters.
This will often be the column label in the sponsor‘s
original dataset.
Req
QVAL
Data Value
Char
Result of, response to, or value associated with QNAM. A
value for this column is required; no records can be in
SUPP-- with a null value for QVAL.
Req
QORIG
Origin
Char
Since QVAL can represent a mixture of collected (on a
CRF), derived, or assigned items, QORIG is used to
indicate the origin of this data. Examples include CRF,
ASSIGNED, or DERIVED. See 1416HSection 4.1.1.8.
Req
QEVAL
Evaluator
Char
*
Used only for results that are subjective (e.g., assigned by
a person or a group). Should be null for records that
contain objectively collected or derived data. Some
examples include ADJUDICATION COMMITTEE,
STATISTICIAN, DATABASE ADMINISTRATOR,
CLINICAL COORDINATOR, etc.
Exp
*indicates variable may be subject to controlled terminology, (Parenthesis indicates CDISC/NCI codelist code
value)
A record in a SUPP-- dataset relates back to its parent record(s) via the key identified by the STUDYID, RDOMAIN,
USUBJID and IDVAR/IDVARVAL variables. An exception is SUPP-- dataset records that are related to Demographics
(DM) records, such as the Intent To Treat (ITT) and Safety (SAFETY) subject-level population flags, where both IDVAR
and IDVARVAL will be null because the key variables STUDYID, RDOMAIN, and USUBJID are sufficient to identify
the unique parent record in DM (DM has one record per USUBJID).
All records in the SUPP-- datasets must have a value for QVAL. Transposing source variables with missing/null values
may generate SUPP-- records with null values for QVAL, causing the SUPP-- datasets to be extremely large. When this
happens, the sponsor must delete the records where QVAL is null prior to submission.
See 1417HSection 4.1.5.3 for information on representing information greater than 200 characters in length.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 262 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
See 2009HAppendix C5 for controlled terminology for QNAM and QLABEL for some of the most common Supplemental
Qualifiers. Additional QNAM values may be created as needed, following the guidelines provided in the CDISC
Notes for QVAL.
8.4.2 SUBMITTING SUPPLEMENTAL QUALIFIERS IN SEPARATE DATASETS
In SDTMIG V3.1.1, the preferred approach is to submit Supplemental Qualifiers by domain rather than placing all of the
supplemental information within one dataset. Therefore, it is recommended that sponsors who utilize the single
SUPPQUAL approach begin to transition to individual SUPP-- datasets by domain. The single SUPPQUAL dataset option
will be deprecated (phased out) in the next (post V3.1.2) release.
There is a one-to-one correspondence between a domain dataset and it's Supplemental Qualifier dataset by creating one
SUPPQUAL for each domain dataset. The set of Supplemental Qualifiers for each domain is included in a separate
dataset with the name SUPP-- where ―--― denotes the source domain which the Supplemental Qualifiers relate back to. For
example, population flags and other demographic Qualifiers would be placed in suppdm.xpt. Data may have been
additionally split into multiple datasets (see 1418HSection 4.1.1.7, Splitting Domains).
Sponsors must, however, choose only one approach for each study. Either individual SUPP-- datasets for each domain
where needed should be submitted, or a single SUPPQUAL dataset for the entire study. In other words, separate SUPP--
datasets cannot be used with some domains and SUPPQUAL for the others.
8.4.3 SUPP-- EXAMPLES
The examples below demonstrate how a set of SUPP-- datasets could be used to relate non-standard information to a
parent domain.
Example 1
In the two rows of suppdm.xpt, population flags are defined as supplemental information to a subject‘s demographic
data. IDVAR and IDVARVAL are null because the key variables STUDYID, RDOMAIN, and USUBJID are
sufficient to identify a unique parent record in DM.
suppdm.xpt: Supplemental Qualifiers for DM
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
1996001
DM
99-401
ITT
Intent to Treat
Y
DERIVED
SPONSOR
2
1996001
DM
99-401
PPROT
Per Protocol Set
N
DERIVED
SPONSOR
Example 2
The two rows of suppae.xpt add qualifying information to adverse event data (RDOMAIN=AE). IDVAR defines the
key variable used to link this information to the AE data (AESEQ). IDVARVAL specifies the value of the key
variable within the parent AE record that the SUPPAE record applies to. The remaining columns specify the
supplemental variables‘ names (AESOSP and AETRTEM), labels, values, origin, and who made the evaluation.
suppae.xpt: Supplemental Qualifiers for AE
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
1996001
AE
99-401
AESEQ
1
AESOSP
Other Medically
Important SAE
Spontaneous
Abortion
CRF
2
1996001
AE
99-401
AESEQ
1
AETRTEM
Treatment Emergent
Flag
N
DERIVED
SPONSOR
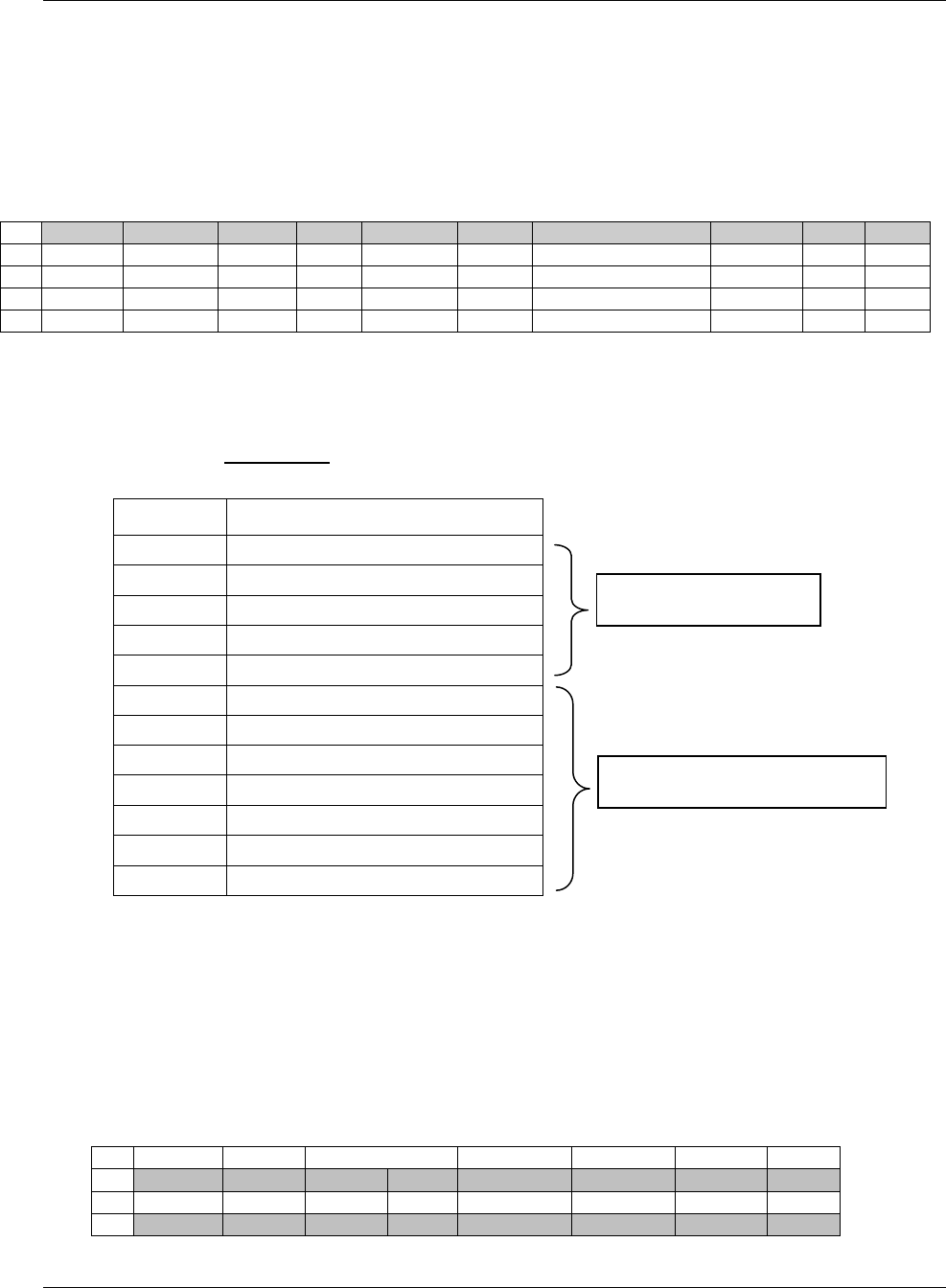
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 263
Final November 12, 2008
Example 3
This is an example of how the language used for a questionnaire might be represented. The parent domain
(RDOMAIN) is QS, and IDVAR is QSCAT. QNAM holds the name of the Supplemental Qualifier variable being
defined (QSLANG). The language recorded in QVAL applies to all of the subject‘s records where IDVAR
(QSCAT) equals the value specified in IDVARVAL. In this case, IDVARVAL has values for two questionnaires
(SF36 and ADAS) for two separate subjects. QVAL identifies the questionnaire language version (French or
German) for each subject.
suppqs.xpt: Supplemental Qualifiers for QS
Row
STUDYID
RDOMAIN
USUBJID
IDVAR
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
1996001
QS
99-401
QSCAT
SF36
QSLANG
Questionnaire Language
FRENCH
CRF
2
1996001
QS
99-401
QSCAT
ADAS
QSLANG
Questionnaire Language
FRENCH
CRF
3
1996001
QS
99-802
QSCAT
SF36
QSLANG
Questionnaire Language
GERMAN
CRF
4
1996001
QS
99-802
QSCAT
ADAS
QSLANG
Questionnaire Language
GERMAN
CRF
Example 4
The following example illustrates how data that may have been represented in an operational database as a single
domain can be expressed using an Events general-observation-class dataset and a Supplemental Qualifiers dataset.
Original Operational (non-SDTM) Dataset:
Variable
Variable Label
HOSEQ
Sequence Number
HOTERM
Term
HOSTDT
Start (Admission) Date/Time
HOENDT
End (Discharge) Date/Time
HODUR
Duration
AEREPF
AE Reported This Episode?
MEDSFL
Meds Prescribed?
PROCFL
Procedures Performed?
PROVNM
Provider Name
SPUNFL
Any Time in Spec. Unit?
SPUNCD
Specialized Unit Type
RLCNDF
Visit Related to Study Med Cond.?
HO Events General Observation Class Custom Dataset with SUPPHO Supplemental Qualifiers dataset:
The shading in the two datasets below is used to differentiate the three hospitalization records for which data are
shown. Note that for Rows 1-7 in the SUPPHO dataset, RDOMAIN= HO, USUBJID = 0001, IDVAR = HOSEQ,
and IDVARVAL = 1. These four values (along with STUDYID) allow these seven SUPPHO records to be linked to
the HO dataset record in Row 1 which has value in HOSEQ = 1 for Subject 0001. Likewise, SUPPHO dataset rows
8-14 are linked to the HO dataset record where HOSEQ = 2 for the same subject, and SUPPHO dataset rows 15-21
are linked to the HO dataset record where HOSEQ =1 for Subject 0002.
ho.xpt: Hospitalization ( )
Row
STUDYID
DOMAIN
USUBJID
HOSEQ
HOTERM
HOSTDTC
HOENDTC
HODUR
1
1999001
HO
0001
1
Hospitalization
2004-01-05
2004-01-12
P1W
2
1999001
HO
0001
2
Hospitalization
2004-01-23
2004-02-07
P15D
3
1999001
HO
0002
1
Hospitalization
2004-01-21
2004-01-22
P1D
SUPPQUAL Variables
Event Variables
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 264 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
suppho.xpt: Supplemental Qualifiers for HO
Row
STUDYID
RDOMAIN
USUBJID
IDVA
R
IDVARVAL
QNAM
QLABEL
QVAL
QORIG
QEVAL
1
1999001
HO
0001
HOSE
Q
1
AEREPF
AE Reported This Episode
Y
CRF
2
1999001
HO
0001
HOSE
Q
1
MEDSFL
Meds Prescribed
Y
CRF
3
1999001
HO
0001
HOSE
Q
1
PROCFL
Procedures Performed
Y
CRF
4
1999001
HO
0001
HOSE
Q
1
PROVNM
Provider Name
General Hosp
CRF
5
1999001
HO
0001
HOSE
Q
1
SPUNCD
Specialized Unit Type
ICU
CRF
6
1999001
HO
0001
HOSE
Q
1
SPUNFL
Any Time in Spec. Unit
Y
CRF
7
1999001
HO
0001
HOSE
Q
1
RLCNDF
Visit Related to Study Med
Cond.
Y
CRF
8
1999001
HO
0001
HOSE
Q
2
AEREPF
AE Reported This Episode
Y
CRF
9
1999001
HO
0001
HOSE
Q
2
MEDSFL
Meds Prescribed
Y
CRF
10
1999001
HO
0001
HOSE
Q
2
PROCFL
Procedures Performed
N
CRF
11
1999001
HO
0001
HOSE
Q
2
PROVNM
Provider Name
Univ Hosp
CRF
12
1999001
HO
0001
HOSE
Q
2
SPUNCD
Specialized Unit Type
CCU
CRF
13
1999001
HO
0001
HOSE
Q
2
SPUNFL
Any Time in Spec. Unit
Y
CRF
14
1999001
HO
0001
HOSE
Q
2
RLCNDF
Visit Related to Study Med
Cond.
Y
CRF
15
1999001
HO
0002
HOSE
Q
1
AEREPF
AE Reported This Episode
Y
CRF
16
1999001
HO
0002
HOSE
Q
1
MEDSFL
Meds Prescribed
N
CRF
17
1999001
HO
0002
HOSE
Q
1
PROCFL
Procedures Performed
Y
CRF
18
1999001
HO
0002
HOSE
Q
1
PROVNM
Provider Name
St. Mary's
CRF
19
1999001
HO
0002
HOSE
Q
1
SPUNCD
Specialized Unit Type
ICU
CRF
20
1999001
HO
0002
HOSE
Q
1
SPUNFL
Any Time in Spec. Unit
N
CRF
21
1999001
HO
0002
HOSE
Q
1
RLCNDF
Visit Related to Study Med
Cond.
Y
CRF
Additional examples may be found in the domain examples such as for Demographics in Examples 3, 4, and 5 under
1419HSection 5.1.1.2, for ECGs in Example 1 under 1420HSection 6.3.1.2, and for Labs in Example 1 under 1421HSection 6.3.3.2
8.4.4 WHEN NOT TO USE SUPPLEMENTAL QUALIFIERS
Examples of data that should not be submitted as Supplemental Qualifiers are the following:
Subject-level objective data that fit in Subject Characteristics (SC). Examples include Subject Initials, Eye
Color.
Findings interpretations that should be added as an additional test code and result. An example of this
would be a record for ECG interpretation where EGTESTCD = ―INTP‖, and the same EGGRPID or
EGREFID value would be assigned for all records associated with that ECG (1422HSection 4.1.5.5).

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 265
Final November 12, 2008
Comments related to a record or records contained within a parent dataset. Although they may have been
collected in the same record by the sponsor, comments should instead be captured in the CO special-
purpose domain.
Data not directly related to records in a parent domain. Such records should instead be captured in either a
separate general observation class or special purpose domain.
8.5 RELATING COMMENTS TO A PARENT DOMAIN
The Comments special-purpose domain, which is also described in 1423HSection 5.2, is used to capture unstructured free text
comments. It allows for the submission of comments related to a particular domain (e.g., adverse events) or those
collected on separate general-comment log-style pages not associated with a domain. Comments may be related to a
Subject, a domain for a Subject, or to specific parent records in any domain. The Comments special-purpose domain is
structured similarly to the Supplemental Qualifiers (SUPP--) dataset, in that it uses the same set of keys (STUDYID,
RDOMAIN, USUBJID, IDVAR, and IDVARVAL) to identify related records.
All comments except those collected on log-style pages not associated with a domain are considered child records of
subject data captured in domains. STUDYID, USUBJID, and DOMAIN (with the value CO) must always be
populated. RDOMAIN, IDVAR, and IDVARVAL should be populated as follows:
1. Comments related only to a subject in general (likely collected on a log-style CRF page/screen) would have
RDOMAIN, IDVAR, IDVARVAL null, as the only key needed to identify the relationship/association to
that subject is USUBJID.
2. Comments related only to a specific domain (and not to any specific record(s)) for a subject would populate
RDOMAIN with the domain code for the domain with which they are associated. IDVAR and IDVARVAL
would be null.
3. Comments related to specific domain record(s) for a subject would populate the RDOMAIN, IDVAR, and
IDVARVAL variables with values that identify the specific parent record(s).
If additional information is collected further describing the comment relationship to a parent record(s), and it cannot
be represented using the relationship variables, RDOMAIN, IDVAR and IDVARVAL, this can be done by two
methods:
1. Values may be placed in COREF, such as the CRF page number or name
2. Timing variables may be added to the CO special-purpose domain, such as VISITNUM and/or VISIT. See
CO special-purpose 1424HSection 5.2.1.1, Assumption 6 for a complete list of Identifier and Timing variables that
can be added to the CO special-purpose domain.
As with Supplemental Qualifiers (SUPP--) and Related Records (RELREC), --GRPID and other grouping variables
can be used as the value in IDVAR to identify comments with relationships to multiple domain records, as a
comment that applies to a group of concomitant medications, perhaps taken as a combination therapy. The limitation
on this is that a single comment may only be related to records in one domain (RDOMAIN can have only one
value). If a single comment relates to records in multiple domains the comment may need to be repeated in the CO
special-purpose domain to facilitate the understanding of the relationships.
Examples for Comments data can be found in 1425HSection 5.2.1.2.
8.6 HOW TO DETERMINE WHERE DATA BELONG IN THE SDTM
8.6.1 GUIDELINES FOR DETERMINING THE GENERAL OBSERVATION CLASS
1426HSection 2.6 discusses when to place data in an existing domain and how to create a new domain. A key part of the
process of creating a new domain is determining whether an observation represents an Event, Intervention, or

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 266 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Finding. Begin by considering the content of the information in the light of the definitions of the three general
observation classes (SDTM Section 2.2) rather than by trying to deduce the class from the information's physical
structure; physical structure can sometimes be misleading. For example, from a structural standpoint, one might
expect Events observations to include a start and stop date. However, Medical History data (data about previous
conditions or events) is Events data regardless of whether dates were collected.
An Intervention is something that is done to a subject (possibly by the subject) that is expected to have a
physiological effect. This concept of an intended effect makes Interventions relatively easy to recognize, although
there are grey areas around some testing procedures. For example, exercise stress tests are designed to produce and
then measure certain physiological effects. The measurements from such a testing procedure are Findings, but some
aspects of the procedure might be modeled as Interventions.
An Event is something that happens to a subject spontaneously. Most, although not all, Events data captured in
clinical trials is about medical events. Since many medical events must, by regulation, be treated as adverse events,
new Events domain will be created only for events that are clearly not adverse events; the existing Medical History
and Clinical Events domain are the appropriate places to store most medical events that are not adverse events.
Many aspects of medical events, including tests performed to evaluate them, interventions that may have caused
them, and interventions given to treat them, may be collected in clinical trials. Where to place data on assessments of
events can be particularly challenging, and is discussed further in 1427HSection 8.6.3.
Findings general-observation-class data are measurements, tests, assessments, or examinations performed on a
subject in the clinical trial. They may be performed on the subject as a whole (e.g., height, heart rate), or on a
"specimen" taken from a subject (e.g., a blood sample, an ECG tracing, a tissue sample). Sometimes the relationship
between a subject and a finding is less direct; a finding may be about an event that happened to the subject or an
intervention they received. Findings about Events and Interventions are discussed further in 1428HSection 8.6.3.
8.6.2 GUIDELINES FOR FORMING NEW DOMAINS
It may not always be clear whether a set of data represents one topic or more than one topic, and thus whether it
should be combined into one dataset (domain) or split into two or more datasets (domains). This implementation
guide shows examples of both.
In some cases, a single data structure works well for a variety of types of data. For example, all questionnaire data is
placed in the QS domain, with particular questionnaires identified by QSCAT (1429HSection 6.3.5). Although some
operational databases may store urinalysis data in a separate dataset, SDTM places all lab data is in the LB domain
(1430HSection 6.3.3) with urinalysis tests identified using LBSPEC.
In other cases, a particular topic may be very broad and/or require more than one data structure (and therefore
require more than one dataset). Two examples in this implementation guide are the topics of microbiology and
pharmacokinetics. Both have been modeled using two domain datasets (see 1431HSection 6.3.9 for Microbiology) and
1432HSection 6.3.10 for Pharmacokinetics). This is because, within these scientific areas, there is more than one topic, and
each topic results in a different data structure. For example, the topic for PC is plasma (or other specimen) drug
concentration as a function of time, and the structure is one record per analyte per time point per reference time
point (e.g., dosing event) subject. PP contains characteristics of the time-concentration curve such as AUC, Cmax,
Tmax, half-life, and elimination rate constant; the structure is one record per parameter per analyte per reference
time point per subject.
8.6.3 GUIDELINES FOR DIFFERENTIATING BETWEEN EVENTS, FINDINGS, AND
FINDINGS ABOUT EVENTS
This section discusses Events, Findings, and Findings about Events. The relationship between Interventions,
Findings, and Findings about Interventions would be handled similarly.
The Findings About domain was specially created to store findings about events. This section discusses Events and
Findings generally, but it is particularly useful for understanding the distinction between the CE and FA domains.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 267
Final November 12, 2008
There may be several sources of confusion about whether a particular piece of data belongs in an Event record or a
Findings record. One generally thinks of an event as something that happens spontaneously, and has a beginning and
end; however, one should consider the following:
Events of interest in a particular trial may be pre-specified, rather than collected as free text.
Some events may be so long lasting in that they are perceived as "conditions" rather than "events", and
their beginning and end dates are not of interest.
Some variables or data items one generally expects to see in an Events record may not be present. For
example, a post-marketing study might collect the occurrence of certain adverse events, but no dates.
Properties of an Event may be measured or assessed, and these are then treated as Findings About Events,
rather than as Events.
Some assessments of events (e.g., severity, relationship to study treatment) have been built into the SDTM
Events model as Qualifiers, rather than being treated as Findings About Events.
Sponsors may choose how they define an Event. For example, adverse event data may be submitted using
one record that summarizes an event from beginning to end, or using one record for each change in
severity.
The structure of the data being considered, although not definitive, will often help determine whether the data
represent an Event or a Finding. The questions below may assist sponsors in deciding where data should be placed
in SDTM.
Question
Interpretation of Answers
Is this a measurement, with
units, etc.?
―Yes‖ answer indicates a Finding.
―No‖ answer is inconclusive.
Is this data collected in a CRF
for each visit, or an overall
CRF log-form?
Collection forms that are independent of visits suggest Event or
Intervention general observation class data
Data collected at visits is usually for items that can be controlled by the
study schedule, namely planned Findings or planned (study) Interventions
or Events.
Data collected at an initial visit may fall into any of the three general
observation classes.
What date/times are
collected?
If the dates collected are start and/or end dates, then data are probably
about an Event or Intervention.
If the dates collected are dates of assessments, then data probably
represents a Finding.
If dates of collection are different from other dates collected, it suggests
that data are historical, or that it is about an Event or Intervention that
happened independently of the study schedule for data collection.
Is verbatim text collected, and
then coded?
―Yes‖ answer suggests that this is Events or Interventions general-
observation-class data. However, Findings general-observation-class data
from an examination that identifies abnormalities may also be coded. Note
that for Events and Interventions general-observation-class data, the topic
variable is coded, while for Findings general-observation-class data, it is
the result that is coded.
A ―No‖ answer is inconclusive. It does not rule out Events or Interventions
general-observation-class data, particularly if Events or Interventions are
pre-specified; it also does not rule out Findings general observation class
data.
If this is data about an event,
does it apply to the event as a
whole?
―Yes‖ answer suggests this is traditional Events general-observation-class
data, and should have a record in an Events domain.
―No‖ answer suggests that there are multiple time-based findings about an
event, and that this data should be treated as Findings About data.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 268 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
The Events general observation class is intended for observations about a clinical event as a whole. Such
observations typically include what the condition was, captured in --TERM (the topic variable), and when it
happened (captured in its start and/or end dates). Other qualifier values collected (severity, seriousness, etc.) apply to
the totality of the event. Note that sponsors may choose how they define the "event as a whole."
Data that does not describe the event as a whole should not be stored in the record for that event or in a --SUPP
record tied to that event. If there are multiple assessments of an event, then each should be stored in a separate FA
record.
When data related to an event does not fit into one of the existing Event general observation class Qualifiers, the
first question to consider is whether the data represents information about the event itself, or whether it represents
data about something (a Finding or Intervention) that is associated with the event.
If the data consist of a finding or intervention that is associated with the event, it is likely that it can be
stored in a relevant Findings or Intervention general observation class dataset, with the connection to the
Event record being captured using RELREC. For example, if a subject had a fever of 102 that was treated
with aspirin, the fever would be stored in an adverse event record, the temperature could be stored in a vital
signs record, and the aspirin could be stored in a concomitant medication record, and RELREC might be
used to link those records.
If the data item contains information about the event, then consider storing it as a Supplemental Qualifier.
However, a number of circumstances may rule out the use of a Supplemental Qualifier:
The data are measurements that need units, normal ranges, etc.
The data are about the non-occurrence or non-evaluation of a pre-specified Adverse Event, data that
may not be stored in the AE domain, since each record in the AE domain must represent a
reportable event that occurred.
If a Supplemental Qualifier is not appropriate, the data may be stored in Findings About. 1433HSection 6.4
provides additional information and examples.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 269
Final November 12, 2008
Appendices
APPENDIX A: CDISC SDS TEAM *
Name
Company
Fred Wood, Team Leader
Octagon Research Solutions, Inc.
Wayne Kubick, Past Team Lead
Lincoln Technologies
Barrie Nelson, SDS Leadership Team
Amgen
Diane Wold, SDS Leadership Team
GlaxoSmithKline
Karen Alexander
Boehringer-Ingelheim
Randall Austin
GlaxoSmithKline
Gary Cunningham
Cephalon
Dan Godoy
Astra Zeneca
Andreas Gromen
Bayer Healthcare
Tom Guinter
Independent
Susan Hamilton
Lilly
Joyce Hernandez
Merck
Jan Hess
Procter & Gamble Pharmaceuticals
Sandy Lei
Johnson and Johnson PRD
Mary Lenzen
Octagon Research Solutions, Inc
Richard Lewis
Octagon Research Solutions, Inc
Tang Li
Cephalon
Musa Nsereko
Shire Pharmaceuticals
Cliff Reinhardt
Schwarz Biosciences, Inc.
Janet Reich
Take Solutions
Gail Stoner
Centocor
Chris Tolk
CDISC
Madhavi Vemuri
Johnson and Johnson PRD
Gary Walker
Quintiles
Carolyn Wilson
Forest Research Institute
Aileen Yam
Sanofi-Aventis
Jay Levine
FDA Liaison
* Individuals having met membership criteria as of publication date.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 270 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
APPENDIX B: GLOSSARY AND ABBREVIATIONS
The following abbreviations and terms are used in this document. Additional definitions can be found in the CDISC
Glossary available at 1434Hhttp://www.cdisc.org/glossary/index.html.
ADaM
CDISC Analysis Dataset Model
ATC code
Anatomic Therapeutic Chemical code from WHO Drug
CDISC
Clinical Data Interchange Standards Consortium
CRF
Case report form (sometimes case record form)
CRT
Case report tabulation
CTCAE
Common Terminology Criteria for Adverse Events
Dataset
A collection of structured data in a single file
Domain
A collection of observations with a topic-specific commonality
eDT
Electronic Data Transfer
FDA
Food and drug Administration
HL7
Health Level 7
ICD9
International Classification of Diseases, 9th revision.
ICH
International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals
for Human Use
ICH E2A
ICH guidelines on Clinical Safety Data Management: Definitions and Standards for Expedited Reporting
ICH E2B
ICH guidelines on Clinical Safety Data Management: Data Elements for Transmission of Individual Cases
Safety Reports
ICH E3
ICH guidelines on Structure and Content of Clinical Study Reports
ICH E9
ICH guidelines on Statistical Principles for Clinical Trials
ISO
International Organization for Standardization
ISO 8601
ISO character representation of dates, date/times, intervals, and durations of time. The SDTM uses the
extended format.
LOINC
Logical Observation, Identifiers, Names, and Codes
MedDRA
Medical Dictionary for Regulatory Activities
NCI
National Cancer Institute (NIH)
SDS
Submission Data Standards. Also the name of the Team that created the SDTM and SDTMIG.
SDTM
Study Data Tabulation Model
SDTMIG
Submission Data Standards Study Data Tabulation Model Implementation Guide: Human Clinical Trials [this
document]
SEND
Standard for Exchange of Non-Clinical Data
SF-36
A multi-purpose, short-form health survey with 36 questions
SNOMED
Systematized Nomenclature of Medicine (a dictionary)
SOC
System Organ Class (from MedDRA)
TDM
Trial Design Model
UUID
Universally Unique Identifier
V3.x
Version 3.1 of the SDTMIG and all subsequent versions of the SDTMIG
WHODRUG
World Health Organization Drug Dictionary
XML
eXtensible Markup Language

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 271
Final November 12, 2008
APPENDIX C: CONTROLLED TERMINOLOGY
The current list of controlled terminology (Appendix C1) is located on the CDISC website at 1435Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC .
Please note that Domain Codes are also listed in 1436HAppendix C2, and Trial Summary Codes are listed in Appendix C3.
APPENDIX C1: CONTROLLED TERMS OR FORMAT FOR SDTM VARIABLES (SEE ALSO 1437H APPENDIX C3: TRIAL
SUMMARY CODES)
Codelist Short
Name
Description
SDTM Variable(s)
Comments
ACN
Action Taken with Study
Treatment
--ACN
Populated using a code value in the list of controlled terms, codelist ACN
(C66767) at
http://www.cancer.gov/cancertopics/terminologyresources/CDISC
AGEU
Age Unit
AGEU
Populated using a code value in the list of controlled terms, codelist
AGEU (C66781) at
http://www.cancer.gov/cancertopics/terminologyresources/CDISC
AESEV
Severity/Intensity Scale
for Adverse Events
AESEV
Populated using a code value in the list of controlled terms, codelist
AESEV (C66769)
athttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
COUNTRY
Country
COUNTRY
Populated using a code value in the list of controlled terms, codelist
COUNTRY (C66786) at
1438Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DSCAT
Category for Disposition
Event
DSCAT
Populated using a code value in the list of controlled terms, codelist
DSCAT (C74558) at
1439Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DOMAIN
Domain Abbreviation
DOMAIN
Please see Appendix C2.
EGMETHOD
ECG Test Method
EGMETHOD
Populated using a code value in the list of controlled terms, codelist
EGMETHOD (C71151) at
1440Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
EGSTRESC
ECG Result
EGSTRESC
Populated using a code value in the list of controlled terms, codelist
EGSTRESC (C71150) at
1441Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
EGTEST
ECG Test Name
EGTEST
Populated using a code value in the list of controlled terms, codelist
EGTEST (C71152) at
1442Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
See also EGTESTCD
EGTESTCD
ECG Test Code
EGTESTCD
Populated using a code value in the list of controlled terms, codelist
EGTESTCD (C71153) at
1443Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
See also EGTEST
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 272 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Codelist Short
Name
Description
SDTM Variable(s)
Comments
ETHNIC
Patient Ethnic Group
ETHNIC
Will be changed to Subject Ethnic Group in the future. Populated using a
code value in the list of controlled terms, codelist ETHNIC (C66790) at
1444Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
FREQ
Frequency
--FRQ
Populated using a code value in the list of controlled terms, codelist
FREQ (C71113) at
1445Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
FRM
Pharmaceutical Dosage
Form
--DOSFRM
Populated using a code value in the list of controlled terms, codelist
FRM (C66726) at
1446Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
IECAT
Category for
Inclusion/Exclusion
IECAT
Populated using a code value in the list of controlled terms, codelist
IECAT (C66797) at
1447Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
LBTEST
Laboratory Test Name
LBTEST
Populated using a code value in the list of controlled terms, codelist
LBTEST (C67154) at
1448Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
See also LBTESTCD
LBTESTCD
Laboratory Test Code
LBTESTCD
Populated using a code value in the list of controlled terms, codelist
LBTESTCD (C65047) at
1449Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
See also LBTEST
LOC
Anatomical Location
--LOC
Populated using a code value in the list of controlled terms, codelist LOC
(C74456) at
1450Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
NCOMPLT
Completion/Reason for
Non-Completion
DSDECOD when DSCAT = ―DISPOSITION
EVENT‖
Populated using a code value in the list of controlled terms, codelist
NCOMPLT (C66727) at
1451Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
ND
Not Done
--STAT
Populated using a code value in the list of controlled terms, codelist ND
(C66789) at
1452Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
NY
No Yes Response
IEORRES, IESTRESC, --OCCUR, --PRESP, --SER
--SCAN, --SCONG, --SDISAB, --SDTH, --SHOSP,
--SLIFE, --SOD, --SMIE, --CONTRT, --BLFL,
--FAST, --DRVFL
Populated using a code value in the list of controlled terms, codelist NY
(C66742) at
1453Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
OUT
Outcome of Event
--OUT
Populated using a code value in the list of controlled terms, codelist OUT
(C66768) at
1454Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
POSITION
Position
--POS
Populated using a code value in the list of controlled terms, codelist
POSITION (C71148) at
1455Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
RACE
RACE
RACE
Populated using a code value in the list of controlled terms, codelist
RACE (C74457) at
1456Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 273
Final November 12, 2008
Codelist Short
Name
Description
SDTM Variable(s)
Comments
ROUTE
Route of Administration
--ROUTE
Populated using a code value in the list of controlled terms, codelist
ROUTE (C66729) at
1457Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SCCD
Subject Characteristic
Code
SCTESTCD
Populated using a code value in the list of controlled terms, codelist
SCCD (C74559) at
1458Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SEX
Sex
SEX
Populated using a code value in the list of controlled terms, codelist SEX
(C66731) at
1459Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SIZE
Size
Controlled Terms for when VSTESTCD =
FRMSIZE (Frame Size)
Populated using a code value in the list of controlled terms, codelist SIZE
(C66733) at
1460Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SOC
CDISC System Organ
Class
Could be used for --BODSYS variables but not
required to be used.
Populated using a code value in the list of controlled terms, codelist SOC
(C66783) at
1461Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
STENRF
Relation to Reference
Period
--STRF, --ENRF
See 1462HSection 4.1.4.7 ―Use of RELATIVE Timing
Variables --STRF, --STTPT, --STRTPT, --ENRF,
--ENTPT, and --ENRTPT‖ for specific regarding
controlled terminology for these variables.
Populated using a code value in the list of controlled terms, codelist
STENRF (C66728) at
1463Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TOXGR
Common Terminology
Criteria for Adverse
Events
Could be used for AETOXGR but not required to be
used.
Populated using a code value in the list of controlled terms, codelist
TOXGR (C66784) at
1464Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
UNIT
Unit
--DOSU, --ORRESU, --STRESU
Populated using a code value in the list of controlled terms, codelist
UNIT (C71620) at
1465Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
VSRESU
Units for Vital Signs
Results
VSORRESU, VSSTRESU
Populated using a code value in the list of controlled terms, codelist
VSRESU (C66770) at
1466Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
VSTEST
Vital Signs Test Name
VSTEST
Populated using a code value in the list of controlled terms, codelist
VSTEST (C67153) at
1467Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
See also VSTESTCD
VSTESTCD
Vital Signs Test Code
VSTESTCD
Populated using a code value in the list of controlled terms, codelist
VSTESTCD (C66741) at
1468Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
See also VSTEST

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 274 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
APPENDIX C2: RESERVED DOMAIN CODES
The following domain codes have been reserved for use with the domain topics listed. CDISC will be preparing additional domain models to describe many of
these over time.
Code
Domain
Class
Description
Status
AD
Analysis Datasets
Not
Applicable
Added as a ―restricted prefix‖ and variable naming prefix - see
2010HAppendix D. Do not use as a Domain Code.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1469Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
AE
Adverse Events
Events
See 1470HSection 6.2.1.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1471Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
CE
Clinical Events
Events
See 1472HSection 6.2.5.1, Assumption 1
Will be added to list of controlled terms on CDISC website.
CM
Concomitant
Medications
Interventions
See 1473HSection 6.1.1.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1474Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
CO
Comments
Special
Purpose
See 1475HSection 5.2.1.1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1476Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DA
Drug
Accountability
Findings
Data regarding the accountability of study drug, such as
information on the receipt, dispensing, return, and packaging. See
1477HSection 6.3.8.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1478Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DM
Demographics
Special
Purpose
Demographics includes a set of essential standard variables that
describe each subject in a clinical study. It is the parent domain
for all other observations for human clinical subjects. See SDTM
2.2.6.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1479Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DS
Disposition
Events
See 1480HSection 6.2.2.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1481Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DV
Protocol
Deviations
Events
See 1482HSection 6.2.4.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1483Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
EG
Electrocardiogram
Test Results
Findings
See 1484HSection 6.3.1.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1485Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
EX
Exposure
Interventions
See 1486HSection 6.1.2.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1487Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
FA
Findings About
Findings
See 1488HSection 6.4.5, Assumption 1.
Will be added to list of controlled terms on CDISC website.
IE
Inclusion/
Exclusion
Criterion not met
Findings
See 1489HSection 6.3.2.1, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1490Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 275
Final November 12, 2008
Code
Domain
Class
Description
Status
LB
Laboratory Data
Findings
See 1491HSection 6.3.3.1, Assumption 1. Does not include
microbiology or PK data, which are stored in separate domains.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1492Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
MB
Microbiology
Specimen
Findings
Microbiology Specimen findings, including gram stain results,
and organisms found. See 1493HSection 6.3.9.2, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1494Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
MH
Medical History
Events
See 1495HSection 6.2.3.1, Assumption 1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1496Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
MS
Microbiology
Susceptibility Test
Findings
Microbiology Susceptibility Test results, plus results of any other
organism-related tests. See 1497HSection 6.3.9.3, Assumption 1.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1498Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
PC
Pharmacokinetic
Concentration
Findings
Concentrations of drugs/metabolites in fluids or tissues as a
function of time.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1499Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
PE
Physical
Examination
Findings
See 1500HSection 6.3.4.1, Assumption 1. Does not include vital signs
measurements, which are stored in the VS domain.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1501Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
PP
Pharmacokinetic
Parameters
Findings
Pharmacokinetic parameters derived from pharmacokinetic
concentration-time (PC) data.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1502Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
QS
Questionnaires
Findings
See 1503HSection 6.3.5.1, Assumption 1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1504Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SC
Subject
Characteristics
Findings
See 1505HSection 6.3.6.1 Assumption 1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1506Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SE
Subject Elements
Special
Purpose
See 1507HSection 5.3.1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1508Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SU
Substance Use
Interventions
See 1509HSection 6.1.3.1, Assumption 1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1510Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SV
Subject Visits
Special
Purpose
See 1511HSection 5.3.2
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1512Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TA
Trial Arms
Trial Design
See 1513HSection 7.2.1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1514Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TE
Trial Elements
Trial Design
See 1515HSection 7.3.1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1516Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 276 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Code
Domain
Class
Description
Status
TI
Trial Inclusion/
Exclusion Criteria
Trial Design
See 1517HSection 7.5.1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1518Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TS
Trial Summary
Trial Design
See 1519HSection 7.6.1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1520Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TV
Trial Visits
Trial Design
See 1521HSection 7.4.1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1522Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
VS
Vital Signs
Findings
See 1523HSection 6.3.7.1, Assumption 1
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1524Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
X-
Sponsor Defined
Sponsor
defined
Reserved for sponsor use; will not be used with SDTM standard
domains. The hyphen may be replaced by any letter or number.
Y-
Sponsor Defined
Sponsor
defined
Z-
Sponsor Defined
Sponsor
defined

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 277
Final November 12, 2008
APPENDIX C2A: RESERVED DOMAIN CODES UNDER DISCUSSION
Code
Domain
Class
Description
Status
BM
Bone Measurements
Findings
Findings resulting from evaluations of bone.
The description of the domain code will be corrected to Bone
Measurements. Bone Measurements is not under development.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1525Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
HO
Hospitalization
Events
Description of Hospitalization events involving research
subjects.
See HU (Healthcare Resource Utilization). Hospitalization events
(HO) will be consolidated with Healthcare Resource Utilization
(HU). The HO domain prefix will be reserved as long as HU is not
developed.
HU
Healthcare Resource
Utilization
Findings
Healthcare resource utilization data such as subject visits to
physicians, hospitalizations, and nursing home stays.
Hospitalization events (HO) will be consolidated with Healthcare
resource utilization (HU). HU is not under development.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1526Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
ML
Meal Data
Interventions
Information regarding the subject's meal consumption, such
as fluid intake, amounts, form (solid or liquid state),
frequency, etc., typically used for PK analysis.
Meal Data (ML) is not under development.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1527Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
NE
Non Subject Events
Events
Used if information is collected on anyone other than the
subject during the trial. It is not limited to just historical data
(before the study). An example of non-subject-event data is
Serious Adverse Events (SAEs) of children born to mothers
participating in the study.
Non Subject Events (NE) is under development.
OM
Organ Measurements
Findings
Findings from organ measurement evaluations.
Organ Measurements is not under development, but under
discussion. Included as a value in the list of controlled terms,
codelist Domain Abbreviation (C66734) at
1528Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
PG
Pharmacogenomics
Findings
Pharmacogenomics findings initially focusing on Genotype
and Gene Expression data.
Pharmacogenomics (PG) is under development.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734)) at
1529Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
PH
Pathology/Histology
Findings
Findings from pathology/histology analysis
Pathology/Histology (PH) is under development.
PF
Pharmacogenomics
Findings
Findings
Findings from genetic testing
Pharmacogenomics Findings (PF) is under development.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 278 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Code
Domain
Class
Description
Status
SG
Surgery
To be
determined
Surgery (SG) might be consolidated with Procedure domain(s).
SG or Procedure domain(s) are not under development, but under
discussion.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1530Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SL
Sleep Data
Findings
Findings from diagnostic sleep tests (e.g., polysomnography).
Sleep Data (SL) is not under development, but under discussion.
Included as a value in the list of controlled terms, codelist Domain
Abbreviation (C66734) at
1531Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TR
Tumor Results
Findings
Results and measurements of tumors.
Tumor Results (TR) is developed and in the public review cycle.
TU
Tumor Identification
Findings
Identification of tumors.
Tumor Identification (TU) is developed and in the public review.

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 279
Final November 12, 2008
APPENDIX C3: TRIAL SUMMARY CODES
Parameters for naming the dictionaries used for a clinical trial (AEDICT, DRUGDICT, MHDICT) were developed, but we now recommend that information on
dictionaries and dictionary versions be included in the SDTM metadata, since the define.xml specification has explicit mechanisms for handling references to
dictionaries and dictionary versions. This recommendation is also based on the fact that Trial Summary is intended to convey information about the planned trial,
while dictionary use, and in particular dictionary version, may not be prospectively defined.
TSPARMCD
TSPARM
TSVAL
Assumptions
Status
ADDON
Added on to
Existing
Treatments
Populated using a code value from the list of
controlled terms, codelist No Yes Response
(C66742) at
1532Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1533Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
AEDICT
Adverse Event
Dictionary
Not applicable
DO NOT USE -
Information on dictionaries
and dictionary versions
should be included in the
SDTM metadata, since the
define.xml specification has
explicit mechanisms for
handling references to
dictionaries and dictionary
versions.
The TSPARMCD code will be removed as a value from the list of
controlled terms, codelist Trial Summary Parameter Test Code
(C66738) at
1534Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
AGEMAX
Planned
Maximum Age
of Subjects
No controlled terminology.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1535Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
AGEMIN
Planned
Minimum Age
of Subjects
No controlled terminology.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1536Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
AGESPAN
Age Group
Populated using a code value from the list of
controlled terms, codelist AGESPAN (C66780)
at
1537Hhttp://www.cancer.gov/cancertopics/termino
logyresources/CDISC
A record for each applicable
category should be included.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1538Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
AGEU
Age Unit
Populated using a code value from the list of
controlled terms, codelist AGEU (C66781) at
1539Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Units are associated with
both AGEMIN and
AGEMAX
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1540Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 280 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
TSPARMCD
TSPARM
TSVAL
Assumptions
Status
COMPTRT
Comparative
Treatment
Name
No controlled terminology.
In the future, may be added to list of controlled terms on
CDISC website.
DOSE
Dose per
Administration
No controlled terminology.
The dose associated with a
test product or comparative
treatment. Records for
dosing parameters may be
grouped using TSGRPID. In
trials with complex dosing,
it may not be useful to
submit dosing parameters, as
the TE and TA datasets are
better suited to describing
such information.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1541Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
DOSFRQ
Dosing
Frequency
Populated using a code value in the list of
controlled terms, codelist FREQ (C71113) at
1542Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Dose frequency associated
with a test product or
comparative treatment.
In the future, may be added to list of controlled terms on
CDISC website
DOSU
Dose Units
Populated using a code value in the list of
controlled terms, codelist UNIT (C71620) at
1543Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Units used with value(s) in
DOSE.
In the future, may be added to list of controlled terms on
CDISC website
DRUGDICT
Drug
Dictionary
Not applicable
DO NOT USE -
Information on dictionaries
and dictionary versions
should be included in the
SDTM metadata, since the
define.xml specification has
explicit mechanisms for
handling references to
dictionaries and dictionary
versions.
The TSPARMCD code will be removed as a value from the list of
controlled terms, codelist Trial Summary Parameter Test Code
(C66738) at
1544Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
INDIC
Trial
Indication
No controlled terminology.
In the future, may be added to list of controlled terms on
CDISC website
LENGTH
Trial Length
No controlled terminology.
Defined as the planned
length of time for a subject's
participation. It should be
recorded using the ISO8601
format for durations, see
1545HSection 4.1.4.3.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1546Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC

CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 281
Final November 12, 2008
TSPARMCD
TSPARM
TSVAL
Assumptions
Status
MHDICT
Medical
History
Dictionary
Not applicable
DO NOT USE -
Information on dictionaries
and dictionary versions
should be included in the
SDTM metadata, since the
define.xml specification has
explicit mechanisms for
handling references to
dictionaries and dictionary
versions.
The TSPARMCD code will be removed as a value from the list of
controlled terms, codelist Trial Summary Parameter Test Code
(C66738) at
1547Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
OBJPRIM
Trial Primary
Objective
No controlled terminology
Should be described in terms
of the desired statement in
labeling.
In the future, may be added to list of controlled terms on
CDISC website
OBJSEC
Trial
Secondary
Objective
No controlled terminology
Should be described in terms
of the desired statement in
labeling.
In the future, may be added to list of controlled terms on
CDISC website
PLANSUB
Planned
Number of
Subjects
No controlled terminology.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1548Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
RANDOM
Trial is
Randomized
Populated using a code value from the list of
controlled terms, codelist NY (C66742) at
1549Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1550Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
ROUTE
Route of
Administration
Populated using a code value from the list of
controlled terms, codelist ROUTE (C66729) at
1551Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
The route associated with a
test product or comparative
treatment.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1552Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SEXPOP
Sex of
Participants
Populated using a code value from the list of
controlled terms, codelist SEXPOP (C66732) at
1553Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1554Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
SPONSOR
Sponsoring
Organization
No controlled terminology.
In the future, may be added to list of controlled terms on
CDISC website
STOPRULE
Study Stop
Rules
If the trial has study stop
rules (STOPRULE is not
equal to "NONE"), contains
a description of the stop
rules.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1555Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TBLIND
Trial Blinding
Schema
Populated using a code value from the list of
controlled terms, codelist TBLIND (C66735) at
1556Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1557Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 282 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
TSPARMCD
TSPARM
TSVAL
Assumptions
Status
TCNTRL
Control Type
Populated using a code value from the list of
controlled terms, codelist TCNTRL (C66785) at
1558Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1559Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TDIGRP
Diagnosis
Group
Populated using a code value from the list of
controlled terms, codelist TDIGRP (C66787) at
1560Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
If trial does not enroll
healthy subjects (TDIGRP is
not equal to "HEALTHY
SUBJECTS"), contains the
diagnosis of subjects to be
enrolled.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1561Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TINDTP
Trial
Indication
Type
Populated using a code value from the list of
controlled terms, codelist TINDTP (C66736) at
1562Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
TINDTP provides a
classification system for the
indication provided as text in
INDIC. MITIGATION is
used narrowly to mean
mitigate the adverse effect of
another treatment.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1563Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TITLE
Trial Title
No controlled terminology.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1564Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TPHASE
Trial Phase
Classification
Populated using a code value from the list of
controlled terms, codelist TPHASE (C66737) at
1565Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
The controlled terminology
for phase includes several
formats as synonyms.
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1566Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
TRT
Reported Name
of Test Product
No controlled terminology.
In the future, may be added to list of controlled terms on
CDISC website
TTYPE
Trial Type
Populated using a code value from the list of
controlled terms, codelist TTYPE (C66739) at
1567Hhttp://www.cancer.gov/cancertopics/terminology
resources/CDISC
Included as a value in the list of controlled terms, codelist Trial
Summary Parameter Test Code (C66738) at
1568Hhttp://www.cancer.gov/cancertopics/terminologyresources/CDISC
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 283
Final November 12, 2008
APPENDIX C4: DRUG ACCOUNTABILITY TEST CODES
The following table contains the test codes suggested by CDISC for use in DRUG Accountability domains.
DATESTCD DATEST
DISPAMT Dispensed Amount
RETAMT Returned Amount
APPENDIX C5: SUPPLEMENTAL QUALIFIERS NAME CODES
The following table contains an initial set of standard name codes for use in the Supplemental Qualifiers (SUPP--)
special-purpose datasets. There are no specific conventions for naming QNAM and some sponsors may choose to
include the 2-character domain in the QNAM variable name. Note that the 2-character domain code is not required
in QNAM since it is present in the variable RDOMAIN in the SUPP-- datasets.
QNAM QLABEL Applicable Domains
AESOSP Other Medically Important SAE AE
AETRTEM Treatment Emergent Flag AE
--CLSIG Clinically Significant Findings
COMPLT Completers Population Flag DM
FULLSET Full Analysis Set Flag DM
ITT Intent to Treat Population Flag DM
PPROT Per Protocol Set Flag DM
SAFETY Safety Population Flag DM
--REAS Reason All general observation classes
--HLGT High Level Group Term AE, MH, PE, and any other domain coded to MedDRA
--HLT High Level Term AE, MH, PE, and any other domain coded to MedDRA
--LLT Lowest Level Term AE, MH, PE, and any other domain coded to MedDRA
--LLTCD Lowest Level Term Code AE, MH, PE, and any other domain coded to MedDRA
--PTCD Preferred Term Code AE, MH, PE, and any other domain coded to MedDRA
--HLTCD High Level Term Code AE, MH, PE, and any other domain coded to MedDRA
--HLGTCD High Level Group Term Code AE, MH, PE, and any other domain coded to MedDRA
--SOCCD System Organ Class Code AE, MH, PE, and any other domain coded to MedDRA
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 284 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
APPENDIX D: CDISC VARIABLE-NAMING FRAGMENTS
The CDISC SDS group has defined a standard list of fragments to use as a guide when naming variables in SUPP--
datasets (as QNAM) or assigning --TESTCD values that could conceivably be treated as variables in a horizontal
listing derived from a V3.x dataset. In some cases, more than one fragment is used for a given keyword. This is
necessary when a shorter fragment must be used for a --TESTCD or QNAM that incorporates several keywords that
must be combined while still meeting the 8-character variable naming limit of SAS transport files. When using
fragments, the general rule is to use the fragment(s) that best conveys the meaning of the variable within the 8-
character limit; thus, the longer fragment should be used when space allows. If the combination of fragments still
exceeds 8 characters, a character should be dropped where most appropriate (while avoiding naming conflicts if
possible) to fit within the 8-character limit.
In other cases the same fragment may be used for more than one meaning, but these would not normally overlap for
the same variable.
Keyword(s)
Fragment
ACTION
ACN
ADJUSTMENT
ADJ
ANALYSIS DATASET
AD
ASSAY
AS
BASELINE
BL
BIRTH
BRTH
BODY
BOD
CANCER
CAN
CATEGORY
CAT
CHARACTER
C
CONDITION
CND
CLASS
CLAS
CLINICAL
CL
CODE
CD
COMMENT
COM
CONCOMITANT
CON
CONGENITAL
CONG
DATE TIME - CHARACTER
DTC
DAY
DY
DEATH
DTH
DECODE
DECOD
DERIVED
DRV
DESCRIPTION
DESC
DISABILITY
DISAB
DOSE, DOSAGE
DOS, DOSE
DURATION
DUR
ELAPSED
EL
ELEMENT
ET
EMERGENT
EM
END
END, EN
ETHNICITY
ETHNIC
EXTERNAL
X
EVALUATOR
EVAL
EVALUATION
EVL
FASTING
FAST
Keyword(s)
Fragment
FILENAME
FN
FLAG
FL
FORMULATION, FORM
FRM
FREQUENCY
FRQ
GRADE
GR
GROUP
GRP
UPPER LIMIT
HI
HOSPITALIZATION
HOSP
IDENTIFIER
ID
INDICATION
INDC
INDICATOR
IND
INTERVAL
INT
INTERPRETATION
INTP
INVESTIGATOR
INV
LIFE-THREATENING
LIFE
LOCATION
LOC
LOINC CODE
LOINC
LOWER LIMIT
LO
MEDICALLY-IMPORTANT EVENT
MIE
NAME
NAM
NON-STUDY THERAPY
NST
NORMAL RANGE
NR
NOT DONE
ND
NUMBER
NUM
NUMERIC
N
OBJECT
OBJ
ONGOING
ONGO
ORDER
ORD
ORIGIN
ORIG
ORIGINAL
OR
OTHER
OTH, O
OUTCOME
OUT
OVERDOSE
OD
PARAMETER
PARM
PATTERN
PATT
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 285
Final November 12, 2008
Keyword(s)
Fragment
POPULATION
POP
POSITION
POS
QUALIFIER
QUAL
REASON
REAS
REFERENCE
REF, RF
REGIMEN
RGM
RELATED
REL, R
RELATIONSHIP
REL
RESULT
RES
RULE
RL
SEQUENCE
SEQ
SERIOUS
S, SER
SEVERITY
SEV
SIGNIFICANT
SIG
SPECIMEN
SPEC, SPC
SPONSOR
SP
STANDARD
ST, STD
START
ST
STATUS
STAT
SUBCATEGORY
SCAT
SUBJECT
SUBJ
SUPPLEMENTAL
SUPP
SYSTEM
SYS
TEXT
TXT
TIME
TM
TIME POINT
TPT
TOTAL
TOT
TOXICITY
TOX
TRANSITION
TRANS
TREATMENT
TRT
UNIT
U
UNIQUE
U
UNPLANNED
UP
VARIABLE
VAR
VALUE
VAL
VEHICLE
V
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 286 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
APPENDIX E: REVISION HISTORY
Changes from CDISC SDTMIG V3.1.1 to V3.1.2
Classification
Type
Section
Description of change
Minor
Addition
2.2
Adds information on how Controlled Terminology (CT) is
represented.
Minor
Deletion
3.2
Reference to an outdated Metadata Model document from November,
2001 is deleted.
Minor
Correction
4.1.1 – General Domain
Assumptions
Section title revised to General Domain Assumptions (from General
Dataset Assumptions).
Minor
Deletion
4.1.1.1 – Review Study Data
Tabulation and Implementation
Guide
Reference to the CDISC Submission Metadata Model was deleted.
Minor
Addition
4.1.1.2 – Relationship to
Analysis Datasets
CDISC ADaM General Considerations referenced.
Minor
Addition
4.1.1.3 – Additional Timing
Variables
General assumption for adding timing variables was expanded to
reference Section 4.1.4.8, domain assumptions and relationship
datasets.
Minor
Addition
4.1.1.4 – Order of the Variables
Additional guidance specified.
Minor
Addition
4.1.1.5 – CDISC Core
Variables
Definitions clarified.
Minor
Addition
4.1.1.6 – Additional Guidance
on Dataset Naming
Guidance for dataset naming described; custom domain codes
beginning with X, Y, or Z will not overlap with future CDISC
reserved codes.
Major
Addition
4.1.1.7 – Splitting Domains
Section and examples added.
Major
Addition
4.1.1.8 – Origin Metadata
Section added.
Major
Addition
4.1.1.9 – Assigning Natural
Keys in the Metadata
Section added.
Minor
Addition
4.1.2.1 – Variable-Naming
Conventions
Conventions for --TESTCDs , QNAMs, and labels clarified.
Minor
Addition
4.1.2.2 – Two-Character
Domain Identifier
Two-character prefixing further explained.
Minor
Addition
4.1.2.3 – Use of ―Subject‖ and
USUBJID
USUBJID expectations further described with an example.
Minor
Addition
4.1.2.5 – Convention for
Missing Values
Missing values for individual data items should be represented by
nulls and convention regarding use of --STAT and --REASND
clarified.
Major
Addition
4.1.2.6 – Grouping Variables
and Categorization
Descriptions of how the following variables group data was clarified:
STUDYID, DOMAIN, --CAT, --SCAT, USUBJID, --GRPID and
--REFID.
Major
Addition
4.1.2.7 – Submitting Free Text
from the CRF
Conventions expanded and examples added.
Major
Addition
4.1.2.8 – Multiple Values for a
Variable
Section added.
Minor
Addition
4.1.3 – Coding and Controlled
Terminology Assumptions
Introductory note added referencing CDISC published controlled
terminology.
Minor
Addition
4.1.3.1 – Types of Controlled
Terminology
Controlled terminology is represented one of three ways: single
asterisk, codelist, external codelist.
Minor
Addition
4.1.3.3 – Controlled
Terminology Values
Convention clarified regarding values to be represented in the
define.xml.
Minor
Addition
4.1.3.4 – Use of Controlled
Terminology and Arbitrary
Number Codes
Description clarified.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 287
Final November 12, 2008
Classification
Type
Section
Description of change
Major
Addition
4.1.3.5 – Storing Controlled
Terminology for Synonym
Qualifier Variables
Convention clarified for values of AEBODSYS, CMCLAS and
expectation to submit dictionary name and version.
Minor
Addition
4.1.3.7 – Use of ―Yes‖ and
―No‖ Values
Note updated to extend values for variables with NY controlled
terminology to include ―NA‖ if collected.
Minor
Addition
4.1.4 – Actual and Relative
Time Assumptions
Introduction to Section 4.1.4 added.
Minor
Deletion
4.1.4.1 – Formats for
Date/Time Variables
References to models prior to SDTMIG v3.1.1 removed.
Minor
Addition
4.1.4.2 – Date/Time Precision
References to models prior to SDTMIG v3.1.1 removed and
description clarified for omitting components for intervals of
uncertainty.
Minor
Addition
4.1.4.3 – Intervals of Time and
Use of Duration for --DUR
Variables
Descriptions and examples expanded.
Major
Change
4.1.4.3 – Removed example of
negative duration, -PT2H
A value containing ―-P‖ cannot be used with a duration, which
signifies a time interval between a start and end of an event. An event
cannot end before it starts, Note that a value containing ―-P‖ can be
used for --ELTM or --EVLINT.
Minor
Addition
4.1.4.5 – Clinical Encounters
and Visits
Conventions for describing clinical encounters clarified.
Major
Addition
4.1.4.6 – Representing
Additional Study Days
Guidance added for representing values like ‗day within element‘ and
‗day within epoch.‘
Major
Addition
4.1.4.7 – Use of Relative
Timing Variables
References to models prior to SDTMIG v3.1.1 removed, conventions
clarified for --STRF and --ENRF and added for --STRTPT, --STTPT,
--ENRTPT and --ENTPT.
Minor
Revised
4.1.4.8 – Date and Time
Reported in a Domain Based on
Findings
Clarified description of interval collections.
Major
Addition
4.1.4.10 – Representing Time
Points
Section added.
Minor
Addition
4.1.5.1 – Original and
Standardized Results of
Findings and Tests Not Done
Descriptions and examples clarified.
Minor
Addition
4.1.5.2 – Linking of Multiple
Observations
Text updated to point to Section 8.
Minor
Addition
4.1.5.3 – Text Strings that
Exceed the Maximum Length
for General-Observation-Class
Domain Variables
Descriptions and examples expanded.
Major
Addition
4.1.5.5 – Clinical Significance
for Findings Observation Class
Data
Section added.
Major
Addition
4.1.5.6 – Supplemental Reason
Variables
Section added.
Major
Addition
4.1.5.7 – Presence or Absence
of Pre-Specified Interventions
and Events
Section added.
Major
Addition
Table 5.1.1 Demographics
The following Timing variables are permissible and may be added as
appropriate: VISITNUM, VISIT, VISITDY. The Record Qualifier
DMXFN (External File Name) is the only additional variable that
may be added, which is adopted from the Findings general
observation class, may also be used to refer to an external file, such
as a subject narrative.
Major
Change
Table 5.1.1 Demographics
Role of RFSTDTC and RFENDTC changed from ―Timing‖ to
―Record Qualifier‖.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 288 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Major
Correction
Table 5.1.1 Demographics
CDISC Notes for SITEID changed from ―Unique identifier for a
study site within a submission.‖ to ―Unique identifier for a site within
a study.‖
Major
Change
Table 5.1.1 Demographics
Role of BRTHDTC changed from ―Result Qualifier‖ to ―Record
Qualifier‖.
Major
Change
Table 5.1.1 Demographics
Role of AGE changed from ―Result Qualifier‖ to ―Record Qualifier‖.
Major
Correction
Table 5.1.1 Demographics
Changed variable label for AGE from ―Age in AGEU at RFSTDTC‖
to ―Age‖ to remove names of other variables in variable labels. AGE
does not have to be derived from RFSTDTC.
Major
Change
Table 5.1.1.1
Demographics
ARMCD is restricted to 20 characters and not 8 characters.
Major
Addition
Table 5.1.1 Demographics -
Assumptions
Added clarifications to Assumption 4 for ARM and ARMCD.
Minor
Deletion
Table 5.1.1.1 Demographics -
Assumptions
Removed Assumption 5. Justification for using SEX vs. GENDER:
Page 71 of 'Providing Regulatory Submissions in Electronic Format -
NDAs' (IT-3, January, 1999), available
at http://www.fda.gov/cder/guidance/2353fnl.pdf specifically lists
SEX as part of demographic data. Similarly, page 60 of 'Guidance for
Industry,
Providing Regulatory Submissions to the Center for Biologics
Evaluation in Electronic Format - Biologics Marketing Applications'
(November, 1999),available at
http://www.fda.gov/cber/guidelines.htm specifically lists SEX as part
of demographic data. SEX is used consistently in both documents
except for one instance where GENDER is used (page 30 for Table 6
which may have been from another writer). 'ICH E3: Structure and
Content of
Clinical Study Reports' (November 30, 1999) only uses SEX (not
GENDER).‖
Major
Addition
Table 5.1.1.1 Demographics -
Assumptions
Added Assumption #6 for submission of multiple races.
Major
Change
Table 5.2.1 Comments
RDOMAIN role changed from ―Identifier‖ to ‖Record Qualifier‖.
Major
Change
Table 5.2.1 Comments
IDVAR role changed from ―Identifier‖ to ‖Record Qualifier‖.
Major
Change
Table 5.2.1 Comments
IDVARVAL role changed from ―Identifier‖ to ‖Record Qualifier‖.
Major
Change
Table 5.2.1 Comments
COVAL role changed from ―Result Qualifier‖ to ‖Topic‖.
Major
Addition
Table 5.2.1 Comments
Added VISITNUM, VISITDY and VISIT
Major
Change
Table 5.2.1 Comments
CODTC is after VISITDY and is now the last variable. Was after
COREF and before COVAL
Major
Addition
Table 5.2.1.1 Comments
Assumptions
Added assumption #6, which no longer restricts the addition of
Identifiers and Timing variables to Comments.
Minor
Change
Table 5.3.1 Subject Elements
Was Table 7.3.1
Major
Addition
Table 5.3.1 Subject Elements
TAETORD and EPOCH added
Minor
Addition
Table 5.3.1 Subject Elements
12 assumptions added
Minor
Addition
Table 5.3.2 Subject Visits
Was Table 7.3.2
Major
Addition
Table 5.3.2 Subject Visits
Added SVSTDY and SVENDY
Minor
Addition
Table 5.3.2 Subject Visits
Added 11 assumptions.
Minor
Change
Table 6.1.1 Concomitant
Medications
Structure of CM domain clarified from 'One record per medication
intervention episode per subject, Tabulation' to 'One record per
recorded intervention occurrence or constant-dosing interval per
subject'
Major
Change
Table 6.1.1 Concomitant
Medications
CMSTAT label changed from 'Concomitant Medication Status' to
'Completion Status' to be compliant with the SDTM
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 289
Final November 12, 2008
Minor
Change
Table 6.1.1 Concomitant
Medications
Assumptions have been modified to more accurately reflect the intent
of the domain
Major
Addition
Table 6.1.1 Concomitant
Medications
Added new variable CMPRESP after CMSCAT and before
CMOCCUR.
Major
Change
Table 6.1.1 Concomitant
Medications
Changed variable label for CMDOSTOT from ―Total Daily Dose
Using DOSU‖ to ―Total Daily Dose‖ to remove names of other
variables in variable labels.
Major
Addition
Table 6.1.1 Concomitant
Medications
Added new variables CMSTRTPT, CMSTTPT, CMENRTPT,
CMENTPT (after CMENRF).
Minor
Addition
Table 6.1.2 Exposure
Added permissible variables EXVAMT and EXVAMTU
Minor
Addition
Table 6.1.2 Exposure
Added permissible variable EPOCH
Major
Change
Table 6.1.2 Exposure
Added assumption that Exposure data is required. Other assumptions
were added and modified.
Minor
Addition
Table 6.1.2 Exposure
Example for submitting placebo data has been added
Major
Change
Table 6.1.2 Exposure
Changed variable label for EXDOSTOT from ―Total Daily Dose
Using DOSU‖ to ―Total Daily Dose‖ to remove names of other
variables in variable labels.
Major
Change
Table 6.1.2 Exposure
Changed variable label for EXELTM from ―Planned Elapsed Time
from Reference Pt‖ to ―Planned Elapsed Time from Time Point Ref‖
to be consistent with SDTM Table 2.2.5 and to more accurately
represent the intent of the variable.
Major
Change
Table 6.1.2 Exposure
EXDOSFRM changed from ―Required‖ to ―Expected‖.
Major
Change
Table 6.1.2 Exposure
EXSTDTC changed from ―Required‖ to ―Expected‖.
Major
Addition
Table 6.1.3 Substance Use
Added new variable SUPRESP after SUSCAT and before
SUOCCUR.
Minor
Change
Table 6.1.3 Substance Use
Structure of SU domain clarified from 'One record per substance type
per visit per subject' to 'One record per substance type per reported
occurrence per subject'
Major
Change
Table 6.1.3 Substance Use
SUSTAT label changed from 'Substance Use Status' to 'Completion
Status' to be more compliant with the SDTM
Major
Change
Table 6.1.3 Substance Use
Changed variable label for SUDOSTOT from ―Total Daily Dose
Using DOSU‖ to ―Total Daily Dose‖ to remove names of other
variables in variable labels.
Minor
Deletion
Table 6.1.3 Substance Use
Removed variables VISIT, VISITNUM and VISITDY but can be
added back in if needed since they are timing variables.
Major
Addition
Table 6.1.3 Substance Use
Added new variables SUSTRTPT, SUSTTPT, SUENRTPT,
SUENTPT (after SUENRF).
Major
Addition
Table 6.2.1 Adverse Events
Added new variable AEPRESP after AESCAT and before
AEBODSYS
Major
Deletion
Table 6.2.1 Adverse Events
Removed variable AEOCCUR. AEOCCUR is not permitted because
the AE domain contains only records for adverse events that actually
occurred.
Major
Change
Table 6.2.1 Adverse Events
Changed variable label for AELOC from ―Location of the Reaction‖
to ―Location of Event‖ to be more generic and not limited to just a
location of a reaction.
Major
Addition
Table 6.2.1 Adverse Events
Added new variables AEENRTPT, AEENTPT (after AEENRF).
Major
Addition
Table 6.2.1 Adverse Events
Added assumption #7 to clarify use of EPOCH and TAETORD.
Major
Change
Table 6.2.1 Adverse Events
Assumption
The adverse event dataset is only for adverse events that happened.
Assumption 4e ―Records should be included in the submission AE
dataset only for adverse events that have actually occurred.‖
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 290 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Major
Addition
Table 6.2.1 Adverse Events
Assumption #8
The following Qualifiers would not be used in AE: --OCCUR,
--STAT, and--REASND. They are the only Qualifiers from the
SDTM Events Class not in the AE domain. They are not permitted
because the AE domain contains only records for adverse events that
actually occurred. See Assumption 4c above for information on how
to deal with negative responses or missing responses to probing
questions for pre-specified adverse events.
Minor
Deletion
Table 6.2.2 Disposition
Removed variables VISIT, VISITNUM and VISITDY but can be
added back in if needed since they are timing variables.
Major
Change
Table 6.2.2 Disposition
EPOCH label changed from ―Trial Epoch‖ to ―Epoch‖. This change
is consistent with SDTM Table 2.2.5, which is the master for the
Timing Variables.
Major
Change
Table 6.2.2 Disposition
DSCAT changed from ―Permissible‖ to ―Expected‖.
Minor
Addition
Table 6.2.2 Disposition
Added assumptions #5 and #6 for ICH E3 guidance.
Major
Change
Table 6.2.3 Medical History
MHSTAT label changed from 'Medical History Status' to
'Completion Status' to be more compliant with the SDTM
Major
Addition
Table 6.2.3 Medical History
Added new variable MHPRESP after MHSCAT and before
MHOCCUR.
Minor
Deletion
Table 6.2.3 Medical History
Removed variables VISIT, VISITNUM and VISITDY but can be
added back in if needed since they are timing variables.
Major
Change
Table 6.2.3 Medical History
Added new variables MHENRTPT, MHENTPT (after MHENRF).
Minor
Addition
Section 6.2.4 Protocol
Deviations
Added new domain model, assumptions and examples.
Major
Addition
Section 6.2.5 Clinical Events
Added new domain model, assumptions and examples.
Minor
Deletion
Table 6.3.1 ECG
EGNRIND removed but can be added back if data is collected or
derived.
Minor
Deletion
Table 6.3.1 ECG
EGLOINC removed but can be added back if data is collected or
derived. EGLOINC was removed from EG because its use is no
longer recommended. Other coding schemes for EGTEST will be
proposed by the CDISC Terminology Team.
Minor
Addition
Table 6.3.1 ECG
Permissible variable EGLOC added but can be dropped if data is not
collected.
Major
Correction
Table 6.3.1 ECG
Order of variables changed to VISITNUM, VISIT from VISIT,
VISITNUM. This change is consistent with SDTM Table 2.2.5,
which is the master for the Timing Variables.
Major
Change
Table 6.3.1 ECG
VISITNUM changed from ―Required‖ to ―Expected‖.
Major
Correction
Table 6.3.1 ECG
Changed variable label for EGELTM from ―Elapsed Time from
Reference Point‖ to ―Planned Elapsed Time from Time Point Ref‖ to
be consistent with SDTM Table 2.2.5 and to more accurately
represent the intent of the variable.
Minor
Addition
Table 6.3.1 ECG
Permissible variable EGRFTDTC added but can be dropped if data is
not collected.
Major\
Change
Table 6.3.1 ECG
EGSTAT label changed to be consistent across domains.
Major
Change
Table 6.3.1 ECG
EGXFN label has the 'F' in 'file' capitalized to be title case.
Major
Change
Table 6.3.1 ECG
EGEVAL changed from Expected to Permissible.
Minor
Change
Table 6.3.1 ECG
EGTPT moved before EGTPTNUM to be consistent with the order in
the SDTM.
Minor
Deletion
Table 6.3.1 ECG
Previous assumption #2 removed because it pertains to EGLOINC,
which has been removed from the model.
Major
Correction
Table 6.3.2 Inclusion/Exclusion
Exceptions
Order of variables changed to VISITNUM, VISIT from VISIT,
VISITNUM. This change is consistent with SDTM Table 2.2.5,
which is the master for the Timing Variables.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 291
Final November 12, 2008
Minor
Change
Table 6.3.2 Inclusion/Exclusion
Structure of IE domain clarified from 'One record per
inclusion/exclusion criteria exception per subject' to 'One record per
inclusion/exclusion criterion not met per subject'
Minor
Addition
Table 6.3.2 Inclusion/Exclusion
3 assumptions added
Minor
Deletion
Table 6.3.2 Inclusion/Exclusion
Previous assumption #2 removed
Major
Change
Table 6.3.3 Lab
LBSTAT label was changed from 'Lab Status' to 'Completion Status'
in order to be more compliant with the SDTM
Major
Change
Table 6.3.3 Lab
VISITNUM changed from Required to Expected
Minor
Addition
Table 6.3.3 Lab
4 assumptions added
Major
Correction
Table 6.3.3
Laboratory Test Results
LBTESTCD variable label changed from ―LAB Test or Examination
Short Name‖ to ‖Lab Test or Examination Short Name‖
Major
Correction
Table 6.3.3
Laboratory Test Results
LBTESTCD variable label changed from ―LAB Test or Examination
Name‖ to ‖Lab Test or Examination Name‖
Major
Correction
Table 6.3.3
Laboratory Test Results
Order of LBSTNRC changed from after LBSTRESC and before
LBSTRESN to after LBSTNRHI and before LBNRIND. This change
is consistent with SDTM Table 2.2.3, which is the master for the
Findings Observation Class.
Major
Correction
Table 6.3.3
Laboratory Test Results
Order of LBDRVFL changed from after LBFAST and before
LBSTRESN to after LBFAST and before LBTOX. This change is
consistent with SDTM Table 2.2.3, which is the master for the
Findings Observation Class.
Major
Correction
Table 6.3.3
Laboratory Test Results
Order of variables changed to VISITNUM, VISIT from VISIT,
VISITNUM. This change is consistent with SDTM Table 2.2.5,
which is the master for the Timing Variables.
Major
Correction
Table 6.3.3
Laboratory Test Results
Changed variable label for LBELTM from ―Elapsed Time from
Reference Point‖ to ―Planned Elapsed Time from Time Point Ref‖ to
be consistent with SDTM Table 2.2.5 and to more accurately
represent the intent of the variable.
Minor
Addition
Table 6.3.3
Laboratory Test Results
Permissible variable LBRFTDTC added but can be dropped if data is
not collected.
Major
Change
Table 6.3.4 Physical
Examination
PESTRESC label had the period after "Std" removed
Major
Deletion
Table 6.3.4 Physical
Examination
Removed expected variable PESTRESN
Major
Deletion
Table 6.3.4 Physical
Examination
Removed expected variable PESTRESU
Major
Change
Table 6.3.4 Physical
Examination
PESTAT had the label changed from 'Examination Status' to
'Completion Status' in order to be more compliant with the SDTM
Minor
Deletion
Table 6.3.4 Physical
Examination
Permissible variable PESEV was dropped from the model, but can be
added back in if collected.
Minor
Addition
Table 6.3.4 Physical
Examination
2 assumptions were added
Major
Correction
Table 6.3.4 Physical
Examination
Order of PELOC changed from after PESCAT and before
PEBODSYS to after PEREASND and before PEMETHOD. This
change is consistent with SDTM Table 2.2.3, which is the master for
the Findings Observation Class.
Minor
Addition
Table 6.3.4 Physical
Examination
Permissible variable PEMETHOD added but can be dropped if data
is not collected.
Major
Correction
Table 6.3.4 Physical
Examination
Removed PEBLFL.
Major
Correction
Table 6.3.4 Physical
Examination
Order of variables changed to VISITNUM, VISIT from VISIT,
VISITNUM. This change is consistent with SDTM Table 2.2.5,
which is the master for the Timing Variables.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 292 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Minor
Change
Table 6.3.5 Questionnaires
Structure of QS domain clarified from 'One record per question per
time point per visit per subject' to 'One record per questionnaire per
question per time point per visit per subject'
Major
Change
Table 6.3.5 Questionnaires
Label of QSSTAT changes from 'Status of Question' to 'Completion
Status' in order to be more compliant with the SDTM
Minor
Change
Table 6.3.5 Questionnaires
The first three assumptions were rearranged for clarity. 3 additional
assumptions were added.
Major
Correction
Table 6.3.5 Questionnaires
Order of variables changed to VISITNUM, VISIT from VISIT,
VISITNUM. This change is consistent with SDTM Table 2.2.5,
which is the master for the Timing Variables.
Major
Correction
Table 6.3.5 Questionnaires
Changed variable label for QSELTM from ―Elapsed Time from
Reference Point‖ to ―Planned Elapsed Time from Time Point Ref‖ to
be consistent with SDTM Table 2.2.5 and to more accurately
represent the intent of the variable.
Minor
Correction
Table 6.3.5 Questionnaires
Permissible variable QSRFTDTC added but can be dropped if data is
not collected.
Major
Change
Table 6.3.6 Subject
Characteristics
SCSTAT label changed from 'Status of SD Measurement' to
'Completion Status' in order to be more compliant with the SDTM
Minor
Addition
Table 6.3.6 Subject
Characteristics
1 assumption added
Major
Deletion
Table 6.3.6 Subject
Characteristics
Example (previously in 9.4.6) had 'Race Other' information removed
Major
Change
Table 6.3.6 Subject
Characteristics
SCDTC changed from ―Expected‖ to ―Permissible‖.
Major
Change
Table 6.3.7 Vital Signs
VISITNUM changed from Required to Expected
Minor
Change
Table 6.3.7 Vital Signs
4 assumptions added
Minor
Addition
Table 6.3.7 Vital Signs
VISITNUM changed from Required to Expected
Minor
Deletion
Table 6.3.7 Vital Signs
VSNRIND removed but can be added back if data is collected or
derived.
Minor
Deletion
Table 6.3.7 Vital Signs
VSLOINC removed. CDISC had defined the controlled terminology
for Vital Signs Tests.
Major
Correction
Table 6.3.7 Vital Signs
Order of variables changed to VISITNUM, VISIT from VISIT,
VISITNUM. This change is consistent with SDTM Table 2.2.5,
which is the master for the Timing Variables.
Major
Correction
Table 6.3.7 Vital Signs
Changed variable label for VSELTM from ―Elapsed Time from
Reference Point‖ to ―Planned Elapsed Time from Time Point Ref‖ to
be consistent with SDTM Table 2.2.5 and to more accurately
represent the intent of the variable.
Minor
Addition
Table 6.3.7 Vital Signs
Permissible variable VSRFTDTC added but can be dropped if data is
not collected.
Major
Addition
Section 6.3.8 Drug
Accountability
Added new domain model, assumptions and examples.
Major
Addition
Section 6.3.9 Microbiology
Added new domain model, assumptions and examples.
Major
Addition
Section 6.3.10 PK
Added new domain model, assumptions and examples.
Major
Addition
Section 6.4 Findings About
Added new domain model, assumptions and examples.
Minor
Change
7.1 Introduction
Expanded introduction and added subsections 7.1.1 Purpose of Trial
Design Model, 7.1.2 Definitions of Trial Design Concepts and 7.1.3
Current and Future Contents of the Trial Design Model.
Minor
Change
Table 7.2.1 Trial Arms
ETCD is restricted to 8 characters. Length was not specified
previously.
Minor
Change
Table 7.2.1 Trial Arms
Was Table 7.2.2
Major
Change
Table 7.2.1 Trial Arms
ARMCD is restricted to 20 characters and not 8 characters.
Major
Change
Table 7.2.1 Trial Arms
ARMCD label changed from ―Arm Code‖ to ―Planned Arm Code‖.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 293
Final November 12, 2008
Major
Change
Table 7.2.1 Trial Arms
ARM label changed from ―Description of Arm‖ to ―Description of
Planned Arm‖.
Major
Change
Table 7.2.1 Trial Arms
EPOCH label changed from ―Trial Epoch‖ to ―Epoch‖. This change
is consistent with SDTM Table 2.2.5, which is the master for the
Timing Variables.
Major
Change
Table 7.2.1 Trial Arms
EPOCH changed from ―Permissible‖ to ―Required‖.
Minor
Change
Table 7.2.1 Trial Arms
11 assumptions added
Minor
Change
Table 7.3.1 Trial Elements
Was Table 7.2.1
Minor
Change
Table 7.3.1 Trial Elements
ETCD is restricted to 8 characters. Length was not specified
previously.
Minor
Change
Table 7.3.1 Trial Elements
Added 15 assumptions
Minor
Change
Table 7.4.1 Trial Visits
Was Table 7.2.3
Major
Change
Table 7.4.1 Trial Visits
ARMCD is restricted to 20 characters and not 8 characters.
Major
Change
Table 7.4.1 Trial Visits
ARMCD label changed from ―Arm Code‖ to ―Planned Arm Code‖.
Major
Change
Table 7.4.1 Trial Visits
ARM label changed from ―Description of Arm‖ to ―Description of
Planned Arm‖.
Major
Change
Table 7.4.1 Trial Visits
TVSTRL changed from ―Permissible‖ to ―Required‖.
Minor
Change
Table 7.4.1 Trial Visits
6 assumptions added
Minor
Change
Table 7.5.1 Trial
Inclusion/Exclusion Criteria
Was Table 7.9
Major
Addition
Table 7.5.1 Trial
Inclusion/Exclusion Criteria
Added new qualifier variable IESCAT to list of qualifiers (after
IECAT and before TIRL).
Major
Addition
Table 7.5.1 Trial
Inclusion/Exclusion Criteria
Added new qualifier variable TIVERS to list of qualifiers (after
TIRL).
Minor
Addition
Table 7.5.1 Trial
Inclusion/Exclusion Criteria
Added 4 assumptions.
Minor
Change
Table 7.6.1 Trial Summary
Was Table 7.10
Major
Addition
Table 7.6.1 Trial Summary
Added new qualifier variable TSGRPID to list of qualifiers (after
TSSEQ and before TSPARMCD).
Minor
Addition
Table 7.6.1 Trial Summary
Added 10 assumptions.
Minor
Change
Section 8 Representing
Relationships and Data
Clarified relationship description. Emphasis was placed on defining
relationships between datasets rather than domains since domains
may occupy multiple datasets.
Minor
Change
Section 8.1 – Relating Groups
of Records within a Domain
using the –GRPID Variable
Simplified wording to clarify concepts especially the use of GRPID
to group records within a subject versus the use of the variable CAT
that can group records across subjects.
Major
Addition
Table 8.2.1 RELREC
Added columns for ―Core‖ and ―References‖.
Minor
Addition
8.3.1 RELREC Dataset
Relationship Example
Added more explanation on the different RELTYPES and the
functionality each provides.
Minor
Change
8.4 Relating Non-Standard
Variables to a Parent Domain
Re-arranged wording to gradually introduce topics by first building
the understanding of foundational concepts such as metadata and
attributions.
Minor
Addition
8.4.1 Supplemental Qualifiers:
SUPPQUAL or SUPP --
Datasets
Added reference to another section for handling data that is greater
than 200 characters. Also added a reference to standard QNAMs for
commonly represented data.
Major
Addition
Table 8.4.1 SUPPQUAL
Added column for ―Core‖.
Minor
Addition
8.4.2 Submitting Supplemental
Qualifiers in Separate Datasets
Added reference to section for additional guidance on splitting
domains.
Minor
Addition
8.4.3 SUPPQUAL Examples
Added an example on how to use SUPPQUAL with a sponsor-
defined domain.
Major
Addition
8.4.4 When not to use
Supplemental Qualifiers
New section with examples that qualify use of SUPPQUAL versus
other domains.
CDISC SDTM Implementation Guide (Version 3.1.2)
Page 294 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Major
Change
8.5 Relating Comments to a
Parent Domain
Add several paragraphs that provide guidance on how to use CO
(Comments) to store information that describes the comment
relationship.
Minor
Change
8.6.1 Guidelines for
Determining the General
Observation Class
Provides more concrete examples for each type of observation class.
Major
Addition
8.6.2 Guidelines for Forming
New Domain
New section that describes how data topics influences whether or not
to create a new domain.
Major
Addition
8.6.3 Guidelines for
Differentiating Between
Events, Findings and Findings
About Events
New section that describes the attributes that may be used to
distinguish between Events and Findings.
Minor
Change
Section 10.1 – CDISC Team
Renamed to Appendix A. Updated to reflect current list of SDS team
members and company affiliation.
Minor
Change
Section 10.2 - Glossary of
Terms
Renamed to Appendix B. Added terms ADaM, ATC code, CRF,
CTCAE, eDT, ICD9, ICH, ICH E2A, ICH E2B, ICH E3, ICH E9,
ISO, ISO 8601, LOINC, MedDRA, NCI, SF-36, SNOMED, SOC,
TDM ,UUID, WHODRUG, XML
Minor
Deletion
Section 10.2 - Glossary of
Terms
Deleted SDSIG (SDS Implementation Guide V3.1, now referred to as
SDTMIG.)
Major
Addition
Appendix C1
New section added: ―Appendix C1: Controlled Terms or Format for
SDTM Variables‖. Replaced values for controlled terminology to
links to CDISC website.
Minor
Change
Section 10.3.1 – Reserved
Domain Codes
Renamed to ―Appendix C2: Reserved Domain Codes‖
Major
Addition
Section C2A
New section added: Appendix C2A: Reserved Domain Codes Under
Discussion
Major
Addition
10.3.1 – Reserved Domain
Codes
Added AD (Analysis Dataset), CE (Clinical Events), FA (Findings
About) and X, Y, Z (used for sponsor defined domains).
Major
Addition
Section C2A
Added HO (Hospitalization), NE (Non Subject Events, PH
(Pathology/Histology), PF (Pharmacogenomics Findings), TR
(Tumor Results), TU (Tumor Identification)
Minor
Change
10.3.1 – Reserved Domain
Codes – EG, PP, and PC
Abbreviations spelled out for ECG (Electrocardiogram), PK
(Pharmacokinetic)
Major
Deletion
10.3.1 – Reserved Domain
Codes – AU (Autopsy)
Information about the autopsy (AU) itself would belong in a
procedures domain. Finding obtained during the autopsy would more
likely belong in the domain based on the topic.
Major
Change
10.3.1 – Reserved Domain
Codes – BM
Bone Mineral Density changed to Bone Measurements to be more
generic since Bone Mineral Density is one type of Bone
Measurement.
Major
Deletion
10.3.1 – Reserved Domain
Codes – BR (Biopsy)
Information about the biopsy (BR) itself would belong in a
procedures domain. Finding obtained during the biopsy would more
likely belong in the domain based on the topic.
Major
Deletion
10.3.1 – Reserved Domain
Codes – DC (Disease
Characteristics)
Disease Characteristics are more likely to be CE (Clinical Events) or
FA (Findings About), which are new models in 3.1.2.
Major
Deletion
10.3.1 – Reserved Domain
Codes – EE
(Electroencephalogram)
Domains are established based on a common topic (i.e., where the
nature of the measurements is the same), rather than by a specific
method of collection such as Electroencephalogram.
Major
Deletion
10.3.1 – Reserved Domain
Codes – IM
Imaging removed as a domain.
Major
Deletion
10.3.1 – Reserved Domain
Codes – SK (Skin Test)
Not under development and concept is too vague for the creation of a
domain model.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 295
Final November 12, 2008
Major
Deletion
10.3.1 – Reserved Domain
Code SL (Sleep
(Polysomnography) Data)
Changed ―SL (Sleep (Polysomnography) Data)‖ to ―Sleep Data‖.
Polysomnography is an example of a finding from diagnostic sleep
tests.
Major
Deletion
10.3.1 – Reserved Domain
Codes – SS (Signs and
Symptoms)
Replaced by Findings About.
Major
Deletion
10.3.1 – Reserved Domain
Codes – ST (Stress (Exercise)
Test Data.
Findings are usually in existing domains such as ECG, laboratory,
vital signs etc.
Major
Deletion
10.3.2: Electrocardiogram Test
Codes and 10.3.3 Vital Signs
Test Codes
Controlled terminology is published on the CDISC website. Section
replaced by ―Appendix C1: Controlled Terms or Format for SDTM
Variables‖.
Major
Deletion
Controlled Terminology - Units
for Vital Signs Results
(VSRESU)
Controlled terminology is published on the CDISC website.
Examples were in CDISC notes for VSORRES. More units were
added and some were modified:
1) INCHES changed to IN
2) FEET is not included
3) POUNDS changed to LB
4) BEATS PER MINUTE changed to BEATS/MINUTE
Major
Deletion
Section 10.3.3 Vital Signs Test
Codes (VSTESTCD, VSTEST)
Controlled terminology is published on the CDISC website and some
changes made from10.3.3 Vital Signs Test Codes Page 167
1) VSTEST ―Frame Size‖ changed to ―Body Frame Size‖
2) VSTEST ―Body Fat‖ changed to ―Adipose Tissue‖
Major
Change
Controlled Terminology –
Action Taken with Study
Treatment
Examples were in CDISC notes for AEACN- DRUG
INTERRUPTED was not included as an example.
Major
Change
Controlled Terminology –SEX
Controlled terms for SEX (Undifferentiated) added to be consistent
with HL7 (Health Level 7).
Major
Change
Controlled Terminology –
Ethnicity
Additions to those listed as controlled terms for ETHNIC. ―NOT
REPORTED‖ and ―UNKNOWN‖ added as terms to match what was
already in NCI caDSR (National Cancer Institute cancer Data
Standards Repository)
Major
Change
Controlled Terminology –
Completion/Reason for Non-
Completion (NCOMPLT)
Examples were in CDISC notes for DSDECOD disposition events.
Added ―RECOVERY‖, Changed ―WITHDRAWAL OF CONSENT‖
to ―WITHDRAWAL BY SUBJECT‖
Major
Change
Controlled Terminology – No
Yes response (NY)
NA (Not Applicable) was added to the list of controlled terms N, Y,
and U. (No, Yes and Unknown).
Major
Change
Controlled Terminology –
Route of Administration
(ROUTE)
Includes many more controlled terms than for those listed in CDISC
Notes for --ROUTE. Includes more specific routes than
INHALATION listed in CDISC notes for SUROUTE.
Major
Change
10.3.5 Trial Summary Codes
Section moved and renamed to ―Appendix C3: Trial Summary
Codes‖.
Major
Change
10.3.5 Trial Summary Codes
All values for TSPARM changed to title case to be consistent with
--TEST.
Major
Change
Controlled Terminology - Trial
Blinding Schema (ADDON)
TSPARMCD value ADDON The TSPARM was changed from
―TEST PRODUCT IS ADDED ON TO EXISTING TREATMENT‖
to ―Added on to Existing Treatments‖.
Major
Change
Controlled Terminology - Trial
Summary Parameter (AEDICT,
DRUGDICT, MHDICT)
AEDICT, DRUGDICT, MHDICT are no longer recommended to be
used as Trial Summary Parameters. Information on dictionaries and
dictionary versions are included in the SDTM metadata, since the
define.xml specification has explicit mechanisms for handling
references to dictionaries and dictionary versions.
Major
Change
Controlled Terminology - Trial
Phase (AGESPAN)
TSPARM for AGESPAN label changed from ―AGE SPAN‖ to ―Age
Group‖ to be more descriptive.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 296 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
Major
Addition
Controlled Terminology - Trial
Phase (AGEU)
AGEU (Age Units) added.
Major
Change
Controlled Terminology - Trial
Blinding Schema (TBLIND)
TSPARMCD was changed from BLIND to TBLIND.
Minor
Change
Controlled Terminology - Trial
Phase (COMPTRT)
TSPARMCD value COMPTRT (Comparative Treatment Name) has
been deferred to a later package based on review comments.
Major
Change
Controlled Terminology - Trial
Control Type (TCNTRL)
TSPARMCD was changed from CONTROL to TCNTRL.
Label changed from ―TYPE OF CONTROL‖ to ―Control Type‖
Major
Change
Controlled Terminology -
Diagnosis Group (TDIGRP)
TSPARMCD was changed from DIAGGRP to TDIGRP
Major
Change
Controlled Terminology -
Diagnosis Group (DOSE)
TSPARMCD value DOSE. The TSPARM was changed from ‖TEST
PRODUCT DOSE PER ADMINISTRATION‖ to ―Dose per
Administration‖.
Major
Change
Controlled Terminology -
Diagnosis Group (DOSFRQ)
TSPARMCD value DOSFRQ. The TSPARM was changed from
‖TEST PRODUCT DOSING FREQUENCY‖ to ―Dosing
Frequency‖.
Minor
Change
Controlled Terminology - Trial
Phase (DOSFRQ)
TSPARMCD value DOSFRQ (Dosing Frequency) has been deferred
to a later package based on review comments.
Major
Change
Controlled Terminology -
Diagnosis Group (DOSU)
TSPARMCD value DOSU. The TSPARM was changed from ‖TEST
PRODUCT DOSE UNITS‖ to ―Dose Units‖.
Minor
Change
Controlled Terminology - Trial
Phase (DOSU)
TSPARMCD value DOSU (TEST PRODUCT DOSE UNITS) has
been deferred to a later package based on review comments.
Major
Correction
Controlled Terminology - Trial
Phase (INDIC)
TSPARMCD value INDIC. The TSPARM was changed from
‖TRIAL INDICATIONS‖ to ―Trial Indication‖. The plural form has
not been used with trial summary parameters so change for
consistency.
Minor
Change
Controlled Terminology - Trial
Phase (INDIC)
TSPARMCD value INDIC (Trial Indication) has been deferred to a
later package since it‘s not apparent what the distinction is between
―Trial Indication‖ and ―Trial Indication Type‖.
Major
Change
Controlled Terminology - Trial
Indication Type (TINDTP)
TSPARMCD was changed from INDICTYP to TINDTP.
Major
Correction
Controlled Terminology - Trial
Phase (LENGTH)
TSPARMCD value LENGTH. The TSPARM was changed from
‖LENGTH OF TRIAL‖ to ―Trial Length‖.
Minor
Change
Controlled Terminology - Trial
Phase (OBJPRIM)
TSPARMCD value OBJPRIM (TRIAL PRIMARY OBJECTIVE)
has been deferred to a later package based on review comments.
Minor
Change
Controlled Terminology - Trial
Phase (OBJSEC)
TSPARMCD value OBJSEC (TRIAL SECONDARY OBJECTIVE)
has been deferred to a later package based on review comments.
Major
Change
Controlled Terminology - Trial
Phase (TPHASE)
TSPARMCD was changed from PHASE to TPHASE. Label changed
from ―TRIAL PHASE‖ to ―Trial Phase Classification‖
Major
Change
Controlled Terminology - Trial
Summary Parameter (ROUTE)
TSPARMCD value ROUTE. The TSPARM was changed from
‖TEST PRODUCT ROUTE OF ADMINISTRATION‖ to ―Route of
Administration‖.
Minor
Change
Controlled Terminology - Trial
Phase (SPONSOR)
TSPARMCD value SPONSOR (SPONSORING ORGANIZATION)
has been deferred to a later package.
Minor
Change
Controlled Terminology - Trial
Phase (TRT)
TSPARMCD value TRT (REPORTED NAME OF TEST
PRODUCT) has been deferred to a later package.
Major
Addition
Appendix C3: Trial Summary
Codes
STOPRULE (Study Stop Rules) added.
CDISC SDTM Implementation Guide (Version 3.1.2)
© 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved Page 297
Final November 12, 2008
Major
Change
Controlled Terminology - Trial
Summary Parameter (TTYPE)
TSPARMCD was changed from TYPE to TTYPE.
Major
Change
Controlled Terminology - Trial
Summary Parameter (TTYPE)
TSPARMCD value TTYPE. The TSPARM was changed from
‖TYPE OF TRIAL‖ to ―Trial Type‖.
Major
Change
Controlled Terminology - Type
of Trial (TTYPE)
TSVAL CONFIRMATORY and EXPLORATORY were
removed based on review comments.
TSVAL PHARMACODYNAMICS changed to
PHARMACODYNAMIC
TSVAL PHARMACOGENOMICS changed to
PHARMACOGENOMIC
TSVAL PHARMACOKINETICS changed to
PHARMACOKINETIC
Minor
Change
Section 10.4 CDISC Variable-
Naming Fragments.
Section renamed to ―Appendix D: CDISC Variable- Naming
Fragments‖. Added ASSAY, CLINICAL, OBJECT, SIGNIFICANT
Minor
Addition
Appendix C4: Drug
Accountability Test Codes
New section added and values: ―Appendix C4: Drug Accountability
Test Codes‖
Minor
Change
Section 10.3.4 Supplemental
Qualifiers Name Codes
Section renamed to ―Appendix C5: Supplemental Qualifiers Name
Codes‖.
Minor
Addition
Section 10.3.4 Supplemental
Qualifiers Name Codes
Added MedDRA specific values (--HLGT = High Level Group
Term, --HLT= High Level Term, --LLT= Lowest Level Term,
--LLTCD = Lowest Level Term Code, --PTCD = Preferred Term
Code, --HLTCD = High Level Term Code, --HLGTCD = High Level
Group Term Code, --SOCCD = System Organ Class Code,)
Minor
Addition
Section 10.3.4 Supplemental
Qualifiers Name Codes
Added --CLSIG = Clinically Significant and --REAS = Reason.
Minor
Deletion
Section 10.5 Lessons Learned
from the Pilot
Section 10.5 Lessons Learned from the Pilot is deleted since it is
historical information and not relevant to this release.

CDISC SDTM Implementation Guide (Version 3.1.2)
Page 298 © 2008 Clinical Data Interchange Standards Consortium, Inc. All rights reserved
November 12, 2008 Final
APPENDIX F: REPRESENTATIONS AND WARRANTIES, LIMITATIONS
OF LIABILITY, AND DISCLAIMERS
CDISC Patent Disclaimers
It is possible that implementation of and compliance with this standard may require use of subject matter covered by
patent rights. By publication of this standard, no position is taken with respect to the existence or validity of any
claim or of any patent rights in connection therewith. CDISC, including the CDISC Board of Directors, shall not be
responsible for identifying patent claims for which a license may be required in order to implement this standard or
for conducting inquiries into the legal validity or scope of those patents or patent claims that are brought to its
attention.
Representations and Warranties
Each Participant in the development of this standard shall be deemed to represent, warrant, and covenant, at the time
of a Contribution by such Participant (or by its Representative), that to the best of its knowledge and ability: (a) it
holds or has the right to grant all relevant licenses to any of its Contributions in all jurisdictions or territories in
which it holds relevant intellectual property rights; (b) there are no limits to the Participant‘s ability to make the
grants, acknowledgments, and agreements herein; and (c) the Contribution does not subject any Contribution, Draft
Standard, Final Standard, or implementations thereof, in whole or in part, to licensing obligations with additional
restrictions or requirements inconsistent with those set forth in this Policy, or that would require any such
Contribution, Final Standard, or implementation, in whole or in part, to be either: (i) disclosed or distributed in
source code form; (ii) licensed for the purpose of making derivative works (other than as set forth in Section 4.2 of
the CDISC Intellectual Property Policy (―the Policy‖)); or (iii) distributed at no charge, except as set forth in
Sections 3, 5.1, and 4.2 of the Policy. If a Participant has knowledge that a Contribution made by any Participant or
any other party may subject any Contribution, Draft Standard, Final Standard, or implementation, in whole or in
part, to one or more of the licensing obligations listed in Section 9.3, such Participant shall give prompt notice of the
same to the CDISC President who shall promptly notify all Participants.
No Other Warranties/Disclaimers. ALL PARTICIPANTS ACKNOWLEDGE THAT, EXCEPT AS PROVIDED
UNDER SECTION 9.3 OF THE CDISC INTELLECTUAL PROPERTY POLICY, ALL DRAFT STANDARDS
AND FINAL STANDARDS, AND ALL CONTRIBUTIONS TO FINAL STANDARDS AND DRAFT
STANDARDS, ARE PROVIDED ―AS IS‖ WITH NO WARRANTIES WHATSOEVER, WHETHER EXPRESS,
IMPLIED, STATUTORY, OR OTHERWISE, AND THE PARTICIPANTS, REPRESENTATIVES, THE CDISC
PRESIDENT, THE CDISC BOARD OF DIRECTORS, AND CDISC EXPRESSLY DISCLAIM ANY
WARRANTY OF MERCHANTABILITY, NONINFRINGEMENT, FITNESS FOR ANY PARTICULAR OR
INTENDED PURPOSE, OR ANY OTHER WARRANTY OTHERWISE ARISING OUT OF ANY PROPOSAL,
FINAL STANDARDS OR DRAFT STANDARDS, OR CONTRIBUTION.
Limitation of Liability
IN NO EVENT WILL CDISC OR ANY OF ITS CONSTITUENT PARTS (INCLUDING, BUT NOT LIMITED TO,
THE CDISC BOARD OF DIRECTORS, THE CDISC PRESIDENT, CDISC STAFF, AND CDISC MEMBERS) BE
LIABLE TO ANY OTHER PERSON OR ENTITY FOR ANY LOSS OF PROFITS, LOSS OF USE, DIRECT,
INDIRECT, INCIDENTAL, CONSEQUENTIAL, OR SPECIAL DAMAGES, WHETHER UNDER CONTRACT,
TORT, WARRANTY, OR OTHERWISE, ARISING IN ANY WAY OUT OF THIS POLICY OR ANY RELATED
AGREEMENT, WHETHER OR NOT SUCH PARTY HAD ADVANCE NOTICE OF THE POSSIBILITY OF
SUCH DAMAGES.
Note: The CDISC Intellectual Property Policy can be found at
http://www.cdisc.org/about/bylaws_pdfs/CDISCIPPolicy-FINAL.pdf.