Tracefinder User Guide For General Quantitation, Version 4.1 Manual Trace Finder
2017-05-02
User Manual: Pdf Tracefinder User TraceFinder_User TraceFinder Software Manauals User ation freshwebmaterial :
Open the PDF directly: View PDF ![]() .
.
Page Count: 464 [warning: Documents this large are best viewed by clicking the View PDF Link!]
- Contents
- Preface
- Introduction
- Getting Started
- Using the Acquisition Mode
- Using the Analysis Mode for Quantitation Batches
- Using the Analysis Mode for Target Screening Batches
- Using the Analysis Mode for Unknown Screening Batches
- Using the Report Designer
- Using the Audit Viewer
- Using Quick Acquisition
- Using Copy Down and Fill Down
- Moving Data Review Panes
© 2016 Thermo Fisher Scientific Inc. All rights reserved.
TraceFinder, Aria, Q Exactive, FreeStyle, LCquan, ToxID, ExactFinder, Prelude, ISQ, and mzVault are
trademarks, and Exactive, Thermo Scientific, TSQ Quantum, TSQ Endura, TSQ Quantiva, TurboFlow, and
Xcalibur are registered trademarks of Thermo Fisher Scientific Inc. in the United States.
NIST is a registered trademark of the National Institute of Standards and Technology in the United States.
The following are registered trademarks in the United States and other countries: Windows, Active Directory,
Excel, and Microsoft are registered trademarks of Microsoft Corporation. Waters and ACQUITY are registered
trademarks of Waters Corporation.
ChemSpider is a trademark of ChemZoo, Inc.
All other trademarks are the property of Thermo Fisher Scientific Inc. and its subsidiaries.
Thermo Fisher Scientific Inc. provides this document to its customers with a product purchase to use in the
product operation. This document is copyright protected and any reproduction of the whole or any part of this
document is strictly prohibited, except with the written authorization of Thermo Fisher Scientific Inc.
The contents of this document are subject to change without notice. All technical information in this
document is for reference purposes only. System configurations and specifications in this document supersede
all previous information received by the purchaser.
This document is not part of any sales contract between Thermo Fisher Scientific Inc. and a purchaser. This
document shall in no way govern or modify any Terms and Conditions of Sale, which Terms and Conditions of
Sale shall govern all conflicting information between the two documents.
Release history: Revision A, May 2016
Software version: Microsoft Windows 7 Professional SP1; (Thermo) Foundation 3.1 SP3; Xcalibur 4.0;
LC Devices 3.0; GC Devices 3.2
For Research Use Only. Not for use in diagnostic procedures.

Thermo Scientific TraceFinder User Guide iii
C
Preface . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . vii
Accessing Documentation. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . vii
License Activation and Deactivation . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .ix
Special Notices . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . x
Contacting Us . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .xi
Chapter 1 Introduction . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .1
About the TraceFinder Application . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 1
TraceFinder Summary of Features. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 3
TraceFinder Workflow . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 5
Reporting Features . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 6
Chapter 2 Getting Started. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .9
Installing the TraceFinder Application . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 9
Installing the NIST and QED Libraries. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 14
Launching the NIST Library Browser . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 17
Launching a Qualitative Explorer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 18
Thermo Scientific FreeStyle. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 18
Thermo Xcalibur Qual Browser. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 19
Converting Legacy Data . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 20
Converting Methods . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 22
Converting Batches . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 24
Converting Method Templates . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 26
Converting Batch Templates . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 28
Converting Compound Databases. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 30
Choosing a Mode or Console . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 31
Chapter 3 Using the Acquisition Mode . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .35
Working with Batches . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 36
Opening and Navigating the Acquisition Mode . . . . . . . . . . . . . . . . . . . . . . 36
Creating and Submitting Batches . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 38
Contents
Contents
iv TraceFinder User Guide Thermo Scientific
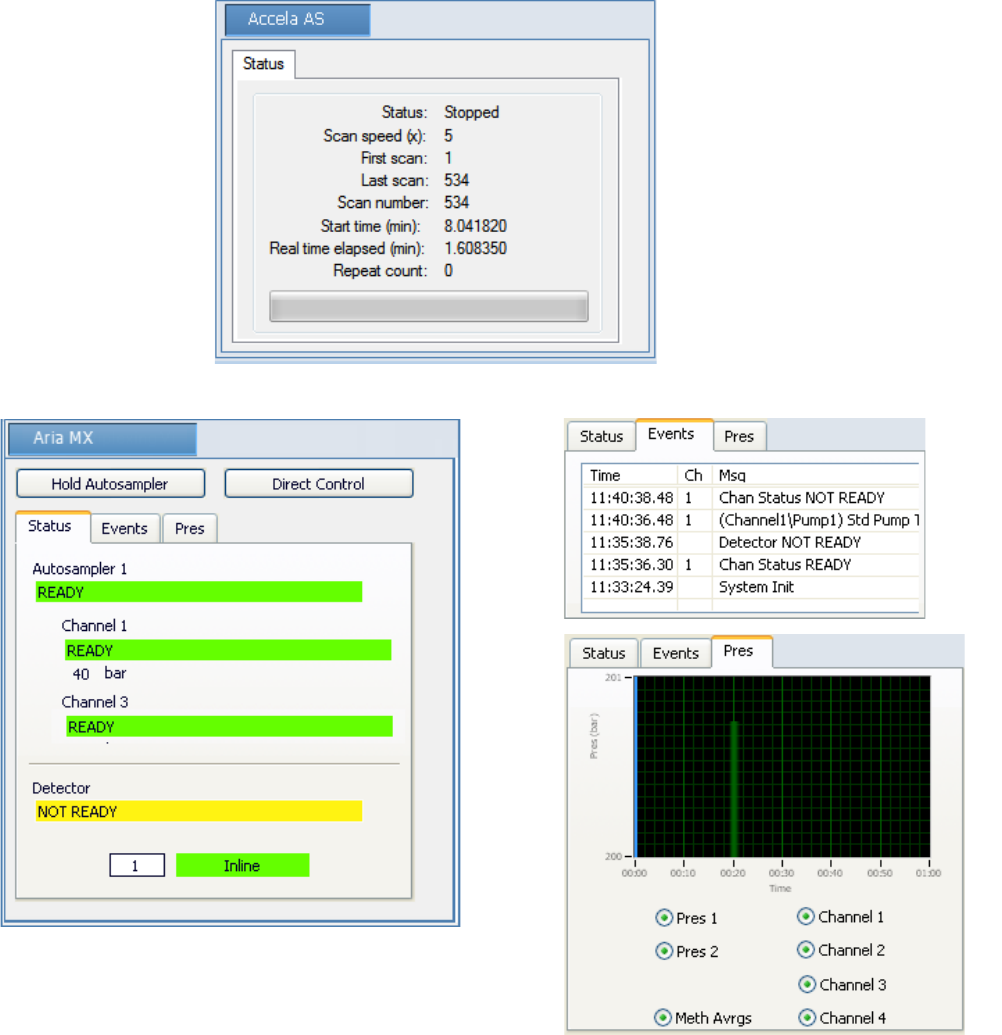
Real Time Status Pane . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 76
Real Time Status – Acquisition Page . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 77
Real Time Status – Instrument Page . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 78
Real Time Status – Queues Page . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 79
Instrument Controls Page . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 84
Real-Time Trace Display. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 89
Sample Types . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 91
Chapter 4 Using the Analysis Mode for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . .93
Working in the Batch View for Quantitation Batches . . . . . . . . . . . . . . . . . . . . 94
Samples Page for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 94
Auto Samples Page for Quantitation Batches. . . . . . . . . . . . . . . . . . . . . . . . 131
Reference Sample Page for Quantitation Batches. . . . . . . . . . . . . . . . . . . . . 133
Threshold Samples Page for Quantitation Batches. . . . . . . . . . . . . . . . . . . . 134
Working in Data Review for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . 135
Sample View for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . . . . . . 136
Compound View for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . . . 151
Compound Details for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . 164
Comparative View for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . . 192
Qualitative View for Quantitation Batches . . . . . . . . . . . . . . . . . . . . . . . . . 200
Column Parameters for Compound Results and Sample Results . . . . . . . . . 214
Working in the Report View for Quantitation Batches . . . . . . . . . . . . . . . . . . 226
Working in the Local Method View for Quantitation Batches . . . . . . . . . . . . 232
Chapter 5 Using the Analysis Mode for Target Screening Batches . . . . . . . . . . . . . . . . . .235
Working in the Batch View for Target Screening Batches . . . . . . . . . . . . . . . . 236
Samples Page Features for Target Screening Batches . . . . . . . . . . . . . . . . . . 237
Creating a New Target Screening Batch . . . . . . . . . . . . . . . . . . . . . . . . . . . 251
Editing a Target Screening Batch. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 258
Submitting a Target Screening Batch. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 261
Working in Data Review for Target Screening Batches . . . . . . . . . . . . . . . . . . 270
Samples Pane. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 272
Compounds Pane . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 273
Chromatogram Pane . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 286
Spectrum Pane . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 286
Working in the Report View for Target Screening Batches . . . . . . . . . . . . . . . 298
Working in the Local Method View for Target Screening Batches. . . . . . . . . . 304
Chapter 6 Using the Analysis Mode for Unknown Screening Batches. . . . . . . . . . . . . . . .307
Working in the Batch View for Unknown Screening Batches . . . . . . . . . . . . . 308
Samples Page Features . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 308
Creating a New Batch . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 320
Editing a Batch . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 328
Submitting a Batch . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 330
Working in Data Review for Unknown Screening Batches . . . . . . . . . . . . . . . 340
Contents
Thermo Scientific TraceFinder User Guide v
Working in the Report View for Unknown Screening Batches . . . . . . . . . . . . 374
Working in the Local Method View for Unknown Screening Batches. . . . . . . 380
Chapter 7 Using the Report Designer. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .383
Editing a Template . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 384
Toolbar Reference . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 396
Home Toolbar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 396
Insert Toolbar – General Features . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 397
Insert Toolbar – Quantitation Reports . . . . . . . . . . . . . . . . . . . . . . . . . . . . 398
Insert Toolbar – Target Screening Reports . . . . . . . . . . . . . . . . . . . . . . . . . 400
Insert Toolbar – Unknown Screening Reports. . . . . . . . . . . . . . . . . . . . . . . 401
Page Layout Toolbar . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 403
Plot Display Properties . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 404
Quantitation and Target Screening Plots. . . . . . . . . . . . . . . . . . . . . . . . . . . 404
Unknown Screening Plots . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 420
Quick Tips . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 432
Chapter 8 Using the Audit Viewer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .433
Audit Trail Log Files . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 433
Audit Log File Functions. . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 434
Audit Viewer . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . 442
Appendix A Using Quick Acquisition . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .445
Appendix B Using Copy Down and Fill Down . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .447
Appendix C Moving Data Review Panes . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .451
Contents
vi TraceFinder User Guide Thermo Scientific

Thermo Scientific TraceFinder User Guide vii
P
Preface
This guide describes how to use the Thermo TraceFinder™ 4.1 application with compatible
Thermo Scientific™ GC/MS and LC/MS analytical instruments.
To suggest changes to the documentation or to the Help
Complete a brief survey about this document by clicking the button below.
Thank you in advance for your help.
Accessing Documentation
The TraceFinder application includes complete documentation. For system requirements,
refer to the Release Notes on the software DVD.
To view the TraceFinder manuals
From the Microsoft™ Windows™ taskbar, choose Start > All Programs > Thermo
TraceFinder > Manuals.
–or–
From the application, choose Help > Manuals.
Contents
•Accessing Documentation
•License Activation and Deactivation
•Special Notices
•Contacting Us
Preface
Accessing Documentation
viii TraceFinder User Guide Thermo Scientific
To view user documentation from the Thermo Fisher Scientific website
1. Go to thermofisher.com.
2. Click the Services & Support tab.
3. On the right, click Manuals & Protocols.
4. In the Refine Your Search box, search by the product name.
5. From the results list, click the title to open the document in your web browser, save it, or
print it.
To return to the document list, click the browser Back button.
To view TraceFinder Help
Open the TraceFinder application and choose Help > TraceFinder Help.
• To find a particular topic, use the Contents, Index, or Search panes.
• To create your own bookmarks, use the Favorites pane.

Preface
License Activation and Deactivation
Thermo Scientific TraceFinder User Guide ix
License Activation and Deactivation
Use the Thermo Scientific Product Licensing wizard to activate or deactivate the license for
the TraceFinder application. To activate the license, you must have an activation code from
Thermo Fisher Scientific. Before you transfer a license to another computer, deactivate the
license.
When you first start the TraceFinder application, a dialog box displays the number of days
remaining in your 120-day free evaluation license. If your evaluation license has expired, the
License Activation wizard opens.
Two types of licenses are available:
• 120-Day Evaluation Version (free of charge)
• Full Version Single License
The evaluation version is full-featured and automatically expires 120 days after activation.
Any attempt to set back the system date automatically terminates this version. You can
purchase and then activate the full version of the application at any time, during or after the
free evaluation, without reinstalling the software.
Each activation key is valid only for a single license. Any additional installation generates a
different license and requires a different activation key.
For software download and licensing questions, contact support at
ThermoMSLicensing@thermo.com.
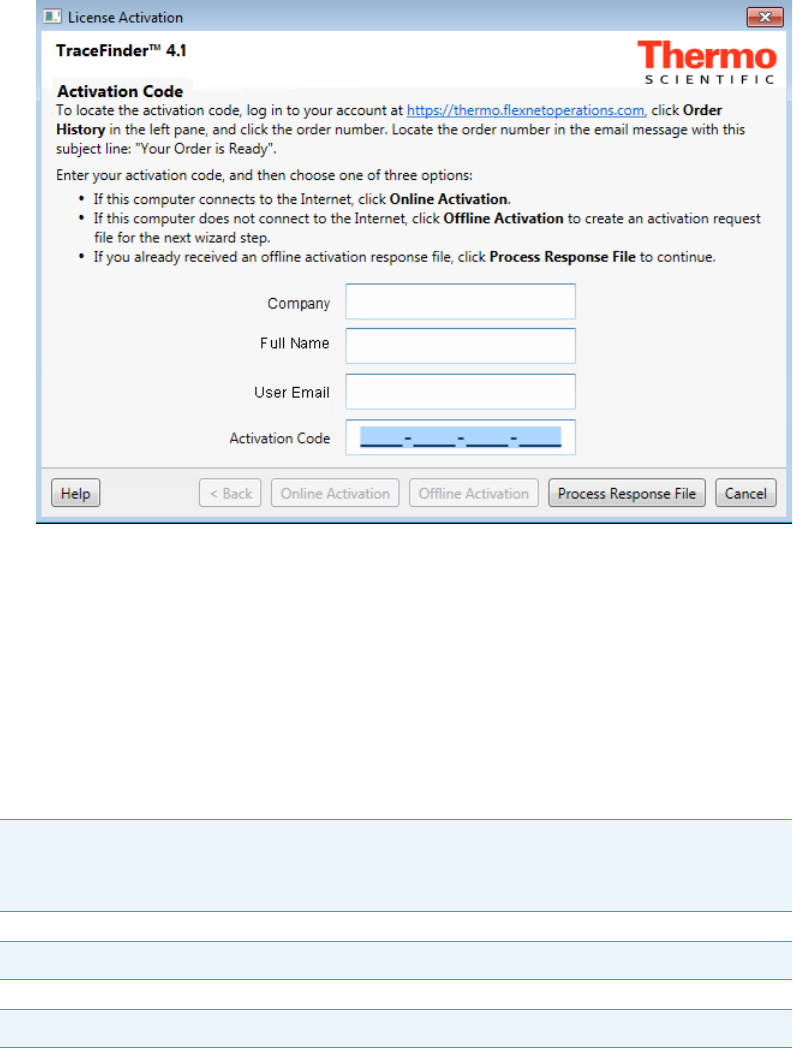
Use the License Activation wizard to activate or deactivate the license for the application. To
activate the license, you must have an activation code from Thermo Fisher Scientific. You
must deactivate the license before you transfer it to another computer.
To start the license activation or deactivation process
1. Open the application.
2. Choose Help > About TraceFinder and Licensing to display the License Activation
wizard.
3. Click Activate (Deactivate) to start the activation or deactivation process, as applicable.
Note You can open the License Activation wizard at any time during your evaluation
period by choosing Help > About TraceFinder and Licensing from the TraceFinder
menu and then clicking Activate. If you already have a permanent license, a message tells
you that your product is fully licensed.
IMPORTANT The 120-day evaluation license includes the basic TraceFinder 4.1 features
and the unknown screening features. When you purchase a permanent license, you have
the option to purchase the unknown screening features. Your permanent license might not
include the unknown screening features.
Preface
Special Notices
xTraceFinder User Guide Thermo Scientific
The License Activation wizard opens.
4. Follow the instructions in the License Activation wizard.
For additional instructions, click Help in the wizard.
Special Notices
Make sure you follow the special notices presented in this guide. Special notices appear in
boxes; those concerning safety or possible system damage also have corresponding caution
symbols.
IMPORTANT Highlights information necessary to prevent damage to software, loss of
data, or invalid test results; or might contain information that is critical for optimal
performance of the system.
Note Highlights information of general interest.
Tip Highlights helpful information that can make a task easier.
Preface
Contacting Us
Thermo Scientific TraceFinder User Guide xi
Contacting Us
There are several ways to contact Thermo Fisher Scientific for the information you need. You
can use your smartphone to scan a QR code, which opens your email application or browser.
Contact us Customer Service and Sales Technical Support
(U.S.) 1 (800) 532-4752 (U.S.) 1 (800) 532-4752
(U.S.) 1 (561) 688-8731 (U.S.) 1 (561) 688-8736
us.customer-support.analyze
@thermofisher.com
us.techsupport.analyze
@thermofisher.com
To find global contact information or customize your request
1. Go to thermofisher.com.
2. Click Contact Us and then select the type of support you need.
3. At the prompt, type the product name.
4. Use the phone number or complete the online form.
To find product support, knowledge bases, and resources
Go to thermofisher.com/us/en/home/technical-resources.
To find product information
Go to thermofisher.com/us/en/home/brands/thermo-scientific.
Note To provide feedback for this document:
• Send an email message to Technical Publications (techpubs-lcms@thermofisher.com).
• Complete a survey at surveymonkey.com/s/PQM6P62.
Preface
Contacting Us
xii TraceFinder User Guide Thermo Scientific
Thermo Scientific TraceFinder User Guide 1
1
Introduction
This chapter describes general features of the TraceFinder application.
About the TraceFinder Application
The TraceFinder application targets the general quantitation market. It supports a focused
quantification workflow for specific nonbioanalytical laboratory use, instrument control, and
method development functionality. TraceFinder is the primary application for the TSQ
Quantum™ XLS triple quadrupole mass spectrometers.
The application can export mass data in the Acquisition List to XML format so that other
systems’ applications, such as for the TSQ 8000, TSQ Quantum™, ISQ, and Q Exactive™,
can import the files into their databases.
The application can import the following file types:
• Sample lists in .csv or .xml format
See Defining a Sample List in Chapter 3, “Using the Acquisition Mode.”
• Compounds from files that use the database (.xml or .cdb) format
Refer to Chapter 2, “Using Compound Databases in the Method Development Mode,”
in the TraceFinder Lab Director User Guide.
• Batches, methods, or templates from the TraceFinder 2.0, 2.1, 3.0, 3.1, 3.2, 3.3, or 4.0
applications.
See Converting Legacy Data.
Contents
•About the TraceFinder Application
•TraceFinder Summary of Features
•TraceFinder Workflow
•Reporting Features
1 Introduction
About the TraceFinder Application
2TraceFinder User Guide Thermo Scientific
• Processing (.pmd) and instrument (.meth) method files from the Xcalibur™ data system
The TraceFinder Lab Director User Guide provides detailed information for creating the
following methods.
The application checks the accuracy and precision of data against systems that have previously
been certified against a standard processing program, such as the Statistical Analysis System
(SAS).
Supported File Types
The application supports the following file types:
• Comma-separated values (.csv): A set of file formats used to store tabular data in which
numbers and text are stored in plain textual form that can be read in a text editor. Lines in
the text file represent rows of a table, and commas in a line separate fields in the tables
row.
• Extensible Markup Language (.xml): A generic framework for storing any amount of text
or any data whose structure can be represented as a tree. The only indispensable
syntactical requirement is that the document has exactly one root element (also called the
document element). This means that the text must be enclosed between a root start-tag
and a corresponding end-tag.
• Instrument method (.meth): A proprietary file format for the Xcalibur data system suite
with specific instructions that enable scientific instruments to perform data acquisition.
• Processing method (.pmd): A proprietary file format for the Xcalibur data system suite
with specific instructions on processing data that was acquired through the instruments
attached to the system.
• Raw data (.raw): The file type for acquired samples on the system.
• Compound database (.cdb): The file type for TraceFinder or ExactFinder™ compound
database data.
• Library (.db): A library used for target screening.
Quantitative
processing methods
Chapter 4, “Using the Method Development Mode for
Quantitation Methods”
Targe t s cre en in g
processing methods
Chapter 5, “Using the Method Development Mode for Target
Screening Methods”
Unknown screening
processing methods
Chapter 6, “Using the Method Development Mode for
Unknown Screening Methods”
Instrument methods Chapter 3, “Using Instrument Methods in the Method
Development Mode”
1 Introduction
TraceFinder Summary of Features
Thermo Scientific TraceFinder User Guide 3
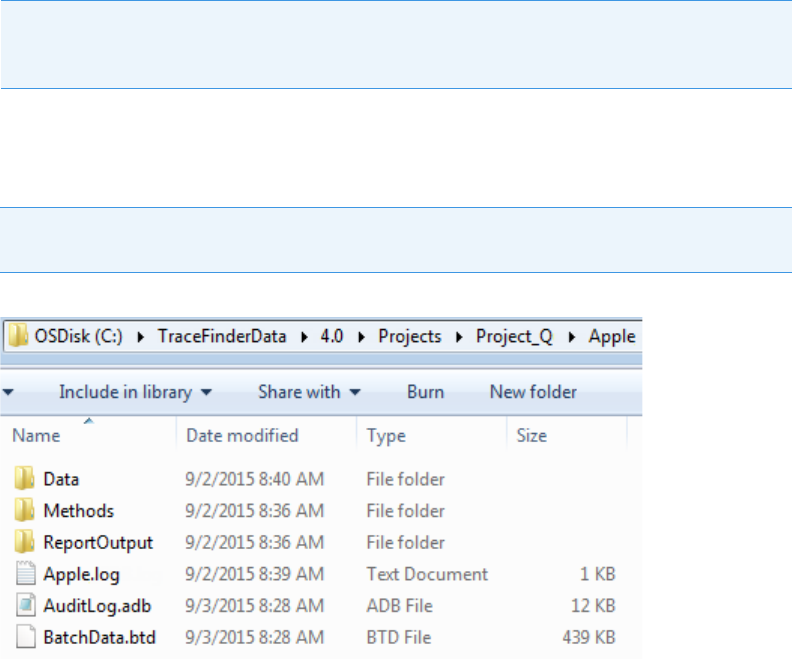
TraceFinder Directory Structure
The application creates folders for batches, methods, and templates in the
…\TraceFinderData directory. Within each batch folder, the application creates folders for
data, methods, and reports.
You can create batches in the TraceFinderData\4.0\Projects folder, or you can create
subfolders within the Projects folder for your batches. You can create as many subfolders as
you want for your batches, but you cannot create a batch within another batch folder.
Figure 1. Example batch directory structure
TraceFinder Summary of Features
The TraceFinder system provides a workflow-oriented approach to high-throughput
quantitation, using tools that automate and speed up the processes of method creation,
loading samples, automatically generating data, manually reviewing and editing results, and
finalizing the data review and reporting process.
The TraceFinder software package includes data acquisition, processing, reviewing, and
reporting capabilities designed to assist analysts in general quantitation applications. The
application has a fully automated acquisition mode and a manual data analysis mode. You can
use the data acquisition system to create and submit batches and monitor real-time review of
results.
IMPORTANT TraceFinder 4.1 uses the same data as TraceFinder 4.0. By default, the application
stores the method, project, template, and compound database data for the 4.1 release in the
TraceFinderData\4.0 folder.
IMPORTANT You cannot rename or move the folders created by the TraceFinder
application.
1 Introduction
TraceFinder Summary of Features
4TraceFinder User Guide Thermo Scientific
The application uses a comprehensive processing method to provide improved handling of
ion ratio calculations, reviewing, and reporting. In addition, it can compare the mass spectra
and integrate the processes of data review and reporting.
Key features include the following:
• Role-based authorization for Security, LabDirector, ITAdmin, Supervisor, Technician,
and QAQC (quality assurance) roles
• Administrator Console for user security, role-based permissions, and data repositories
• Configuration console for report configuration, detection and acquisition defaults,
adduct definitions, screening library selection, and customized columns and flags
• Method Development mode for editing instrument methods, setting processing and error
flag parameters, and setting reporting options
• Acquisition mode that guides you in creating batches and running samples
• Analysis mode with batch views, data review, local method views, and reporting views
• Database-capable method development
• Quantification, target screening, and unknown screening workflows
• Spreadsheet-based report designer
Features of the common workflow core include the following:
• Acquisition and processing
•Peak detection
• Quantification to include calibration
• Error analysis and flag setting
•Reporting
• Data persistence
• Raw data file handling
1 Introduction
TraceFinder Workflow
Thermo Scientific TraceFinder User Guide 5
TraceFinder Workflow
The application is structured with a typical laboratory workflow in mind. You create a batch,
and the system injects samples into the instrument, runs the samples, analyzes the data, and
generates a report. You can set up a master method for specific compound groups or assays
that you expect to run in your laboratory. When you are ready to run a particular type of
sample, select the appropriate method and begin.
When using the application, follow these basic steps:
1. Create and save a master method in the Method Development mode.
A master method combines the instrument method and processing method that define
the following:
• How the raw data is acquired and processed
• How the error checking information evaluates the results
• How the results appear in reports
2. Create and submit a batch using the Acquisition wizard.
A batch lists samples for processing and reporting using a specified method. Each row of a
batch represents a unique sample.
3. Monitor the status of the batch in the Real Time Status view.
The real-time display is visible from all the TraceFinder modes. You can begin another
batch while you watch the real-time display of the currently acquiring batch.
4. Evaluate the data in the Analysis mode.
The Analysis mode includes views where you can review batches, batch data, reports, and
local methods.
5. View and print reports in the Report View of the Analysis mode.
Use the Report View to view or print the reports for the current batch.
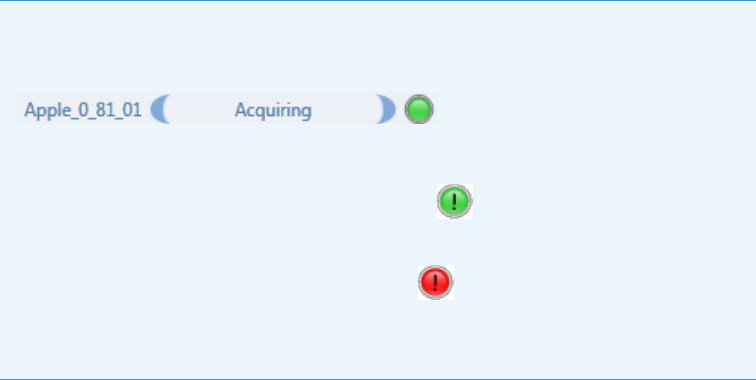
Note At any time, you can quickly view the system status by looking in the upper
right corner of the TraceFinder window. This area displays a green, yellow, or red
status light and a description of any activity in the queues, as in this example:
The status light (in any color) might include an exclamation mark.
• An exclamation mark on a green status light, , might indicate that the
instrument method contains the wrong source (for example, APCI instead of
HESI).
• An exclamation mark on a red status light, , might indicate that a vial is
missing and the batch was forced to stop.
Click the status light to display details of the error.
1 Introduction
Reporting Features
6TraceFinder User Guide Thermo Scientific
Reporting Features
The report engine can generate several different types of reports designed to meet the needs of
the laboratory, the laboratory's customers, and key regulatory agencies that might review the
results. The following types of reports meet the requirements of various methods and global
regulatory agencies, helping to track the performance of LC and GC systems and methods.
Standard Report Types
•Ad Hoc Tune Report
•Batch Report
•Blank Report
• Breakdown Report
•Calibration Report
•Check Standard Report
• Chromatogram Report
• Compound Calibration Report
• Compound Calibration Report - Alternate
•Confirmation Report
•Confirmation Report 2
• High Density Calibration Report
• High Density Internal Standard Report Long
• High Density Sample Report 1 Long
• Intelligent Sequencing
• Internal Standard Summary Report
• Ion Ratio Failure Report
•LCSLCSD Report
• Manual Integration Report
•Method Detection Limit Report
•Method Report
• Method Validation Report
•MSMSD Report
• Quantitation Report
1 Introduction
Reporting Features
Thermo Scientific TraceFinder User Guide 7
• Quantitation Report - 2
• Sample Report
•Solvent Blank Report
• Standard Addition Report
• Surrogate Recovery Report
• Target Screening Height Density Sample Report
• Target Screening Height Density Sample Report 2
• Target Screening Summary Report
•TIC Report
• TIC Summary Report
•Tune Report
• Unknown Screening High Density Report
• Unknown Screening Long Report
• Unknown Screening Summary Report
Use the features in the Report Designer to create custom report types. See Chapter 7, “Using
the Report Designer.”
Thermo Scientific TraceFinder User Guide 9
2
Getting Started
This chapter includes the procedures for getting started with the TraceFinder application.
Installing the TraceFinder Application
To initially install the TraceFinder 4.1 application, follow the instructions in the TraceFin der
Installation Guide. Later, you might need to reinstall the application or other features using
the InstallShield Wizard.
Follow these instructions to reinstall, start, and log in to the application.
To reinstall the TraceFinder application
1. From the Thermo Foundation Instrument Configuration window, remove all
instruments.
2. From the Windows Control Panel, uninstall the application and then uninstall all
Thermo instrument drivers.
3. Insert the TraceFinder DVD, and install both the TraceFinder 4.1 application and the
NIST™ library as follows:
a. Open the TraceFinder launcher and click Next.
Contents
•Installing the TraceFinder Application
•Installing the NIST and QED Libraries
•Launching the NIST Library Browser
•Launching a Qualitative Explorer
•Converting Legacy Data
•Choosing a Mode or Console
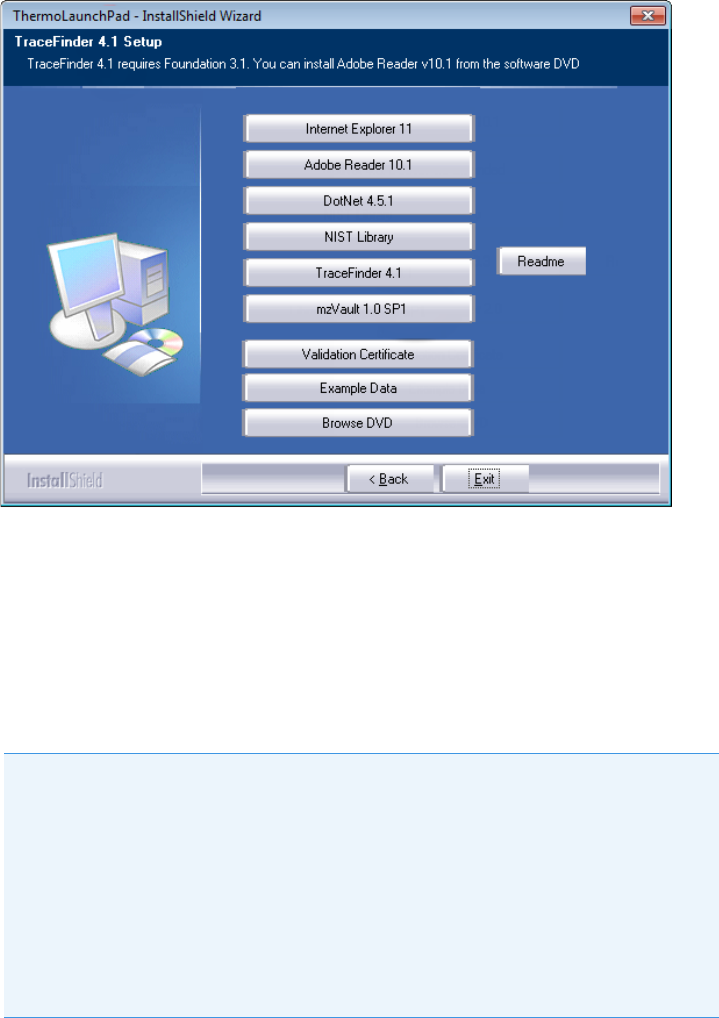
2 Getting Started
Installing the TraceFinder Application
10 TraceFinder User Guide Thermo Scientific
The InstallShield Wizard opens.
b. Click Tr a ce Fi n der 4.1 , and follow the instructions in the InstallShield Wizard.
The installer verifies that you have the appropriate versions of the Thermo
Foundation™ platform and Xcalibur data system and updates them if necessary.
c. At the prompt, click Yes to completely remove any previously installed applications.
d. Open the TraceFinder launcher again and click Next.
e. Click TraceFinde r 4. 1, and follow the instructions in the InstallShield Wizard.
f. When prompted, choose to install either the GC or LC version of the software.
g. When the installation is complete, open the TraceFinder launcher again and click
Next.
h. When prompted to restart your computer, select the Yes option and click Finish.
IMPORTANT For the application to properly install, you might be prompted to
uninstall Foundation platform. Do the following:
1. Click Yes, and then when prompted to restart your computer, click OK.
The wizard continues the TraceFinder installation.
2. When prompted to install Thermo Foundation, click Yes, and then when
prompted to restart your computer, click OK.
The wizard continues the installation.
2 Getting Started
Installing the TraceFinder Application
Thermo Scientific TraceFinder User Guide 11
3. (Optional) After your system restarts, open the TraceFinder launcher again, click Next,
and install any of the following:
a. Click NIST Library and follow the instructions to install it.
b. Click Example Data.
The application installs example compound databases, instrument methods, and
batch data.
c. Click mzVault 1.0 SP1 and follow the instructions to install the Thermo mzVault
application.
4. Install the appropriate device drivers, and configure the instruments in the Thermo
Foundation Instrument Configuration window.
You can now start the TraceFinder application.
To start the TraceFinder application
1. Configure your instruments.
You must close the TraceFinder application before you can configure your instruments.
2. Double-click the TraceFind e r icon on your desktop, or choose Start > All Programs >
Thermo TraceFinder 4.1 > TraceFinder 4.1 General Quan.
By default, user security is not activated and the application does not require a password.
To activate user security, refer to the TraceFinder Administrator Console User Guide.
To log in to the TraceFinder application (when user security is activated)
1. Enter your user name in the TraceFinder login window.
2. Enter your password.
If your user name or password does not match, the system reports this error:
Correct the user name or password, or contact your system administrator.
3. Click Login.
The TraceFinder main window opens.
Note Before you can log in to the application when user security is activated, a system
administrator must set up a user account for you.
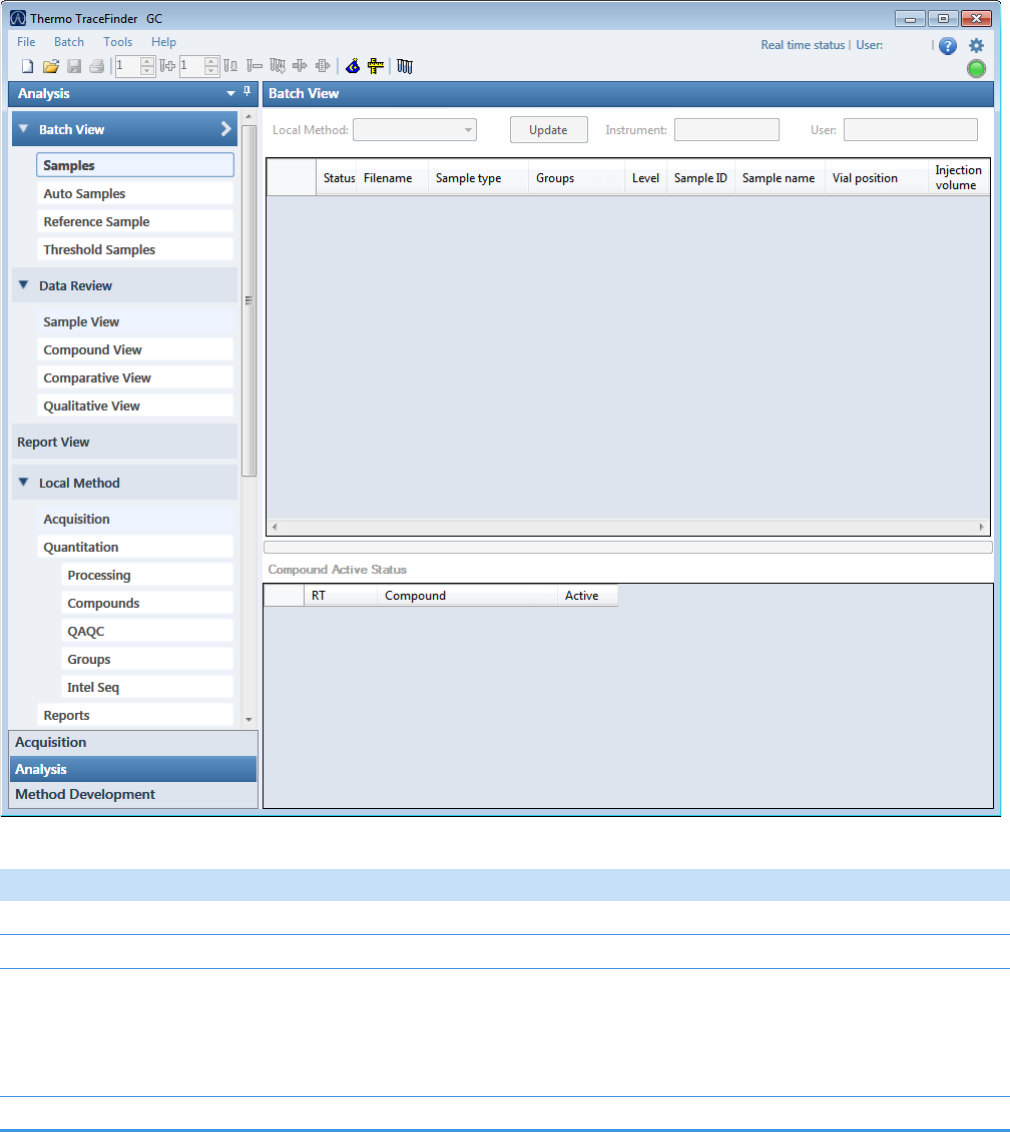
2 Getting Started
Installing the TraceFinder Application
12 TraceFinder User Guide Thermo Scientific
Figure 2. TraceFinder main window, showing the Analysis mode for a quantitation batch
Table 1. TraceFinder main window features (Sheet 1 of 2)
Parameter Description
Modes
Acquisition See Chapter 3, “Using the Acquisition Mode.”
Analysis See Chapter 4, “Using the Analysis Mode for Quantitation Batches.”
See Chapter 5, “Using the Analysis Mode for Target Screening Batches.”
See Chapter 6, “Using the Analysis Mode for Unknown Screening Batches.”
Method Development Refer to the TraceFinder Lab Director User Guide.
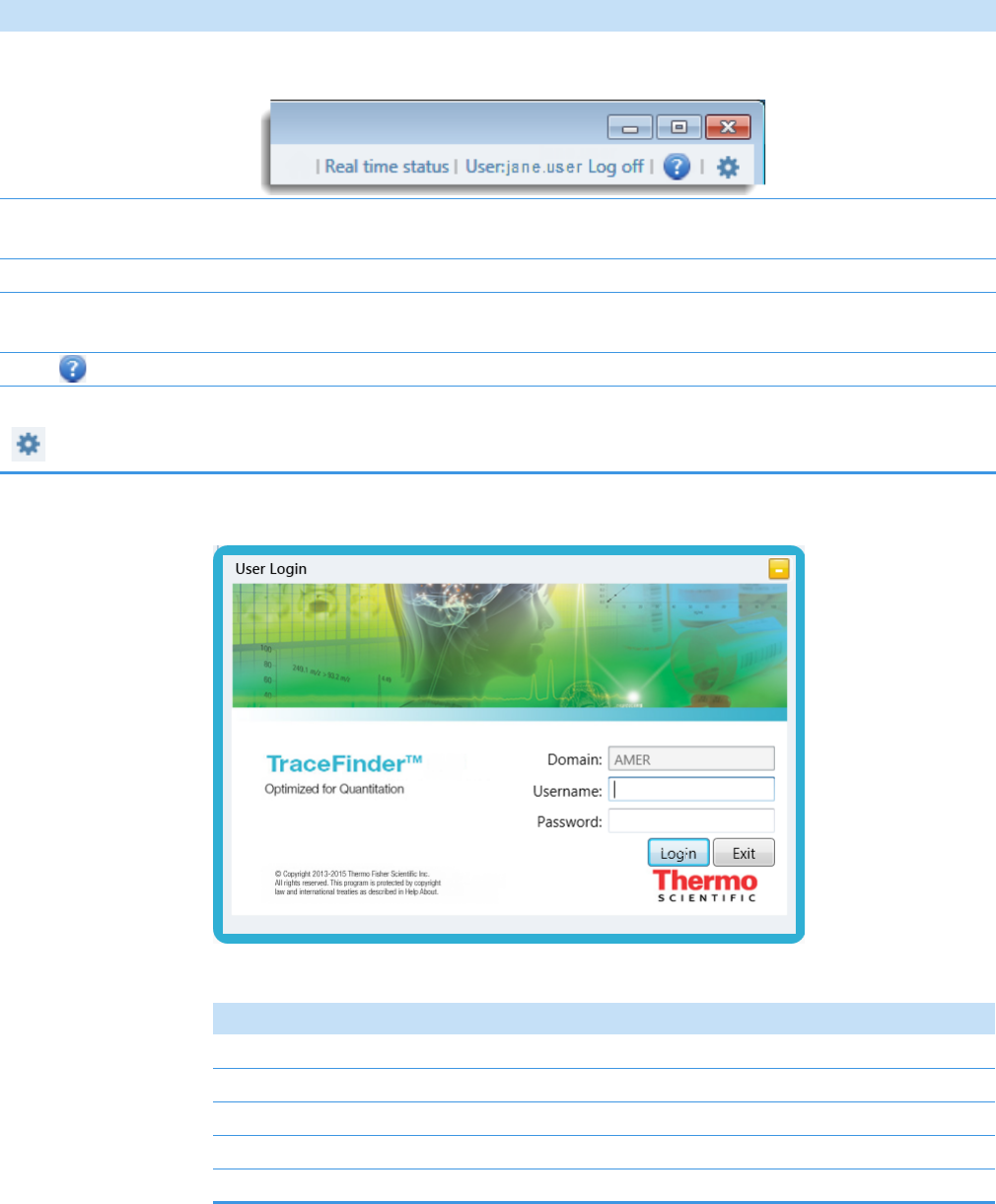
2 Getting Started
Installing the TraceFinder Application
Thermo Scientific TraceFinder User Guide 13
Figure 3. TraceFinder login window
Additional Features
A TraceFinder window with user security for a user in the default LabDirector role has
these functions.
Real Time Status Opens the Real Time Status pane for the current acquisition. The acquisition progress is
displayed within the current mode window.
User Displays the name of the current user.
Log Off Logs off the current user and displays the login screen. This function is available only
when user security is activated.
Help Opens the TraceFinder Help.
Application Configuration Opens the Configuration console where you can configure several options for using the
application. Refer to Chapter 1, “Using the Configuration Console,” in the Trac eFi nd er
Lab Director User Guide.
Table 1. TraceFinder main window features (Sheet 2 of 2)
Parameter Description
Table 2. Login window parameters
Parameter Description
Domain The authentication method.
Username The user’s assigned user name.
Password The assigned password for the user name.
Login Verifies the user name and password, and opens the application.
Exit Closes the TraceFinder login window.
2 Getting Started
Installing the NIST and QED Libraries
14 TraceFinder User Guide Thermo Scientific
Installing the NIST and QED Libraries
When you are using triple quadrupole instruments, such as the TSQ Quantum XLS, follow
these instructions to install the NIST and QED libraries.
To install the NIST library
1. Open the TraceFinder launcher, and click Next.
2. Click NIST Library.
The NIST 08 MS Search and AMDIS Setup wizard opens.
3. Follow the instructions in the setup wizard.
4. When the wizard prompts you to select a destination folder, select
C:\Program Files\NISTMS.
5. Continue to follow the instructions in the wizard until the setup is complete.
To install the QED library
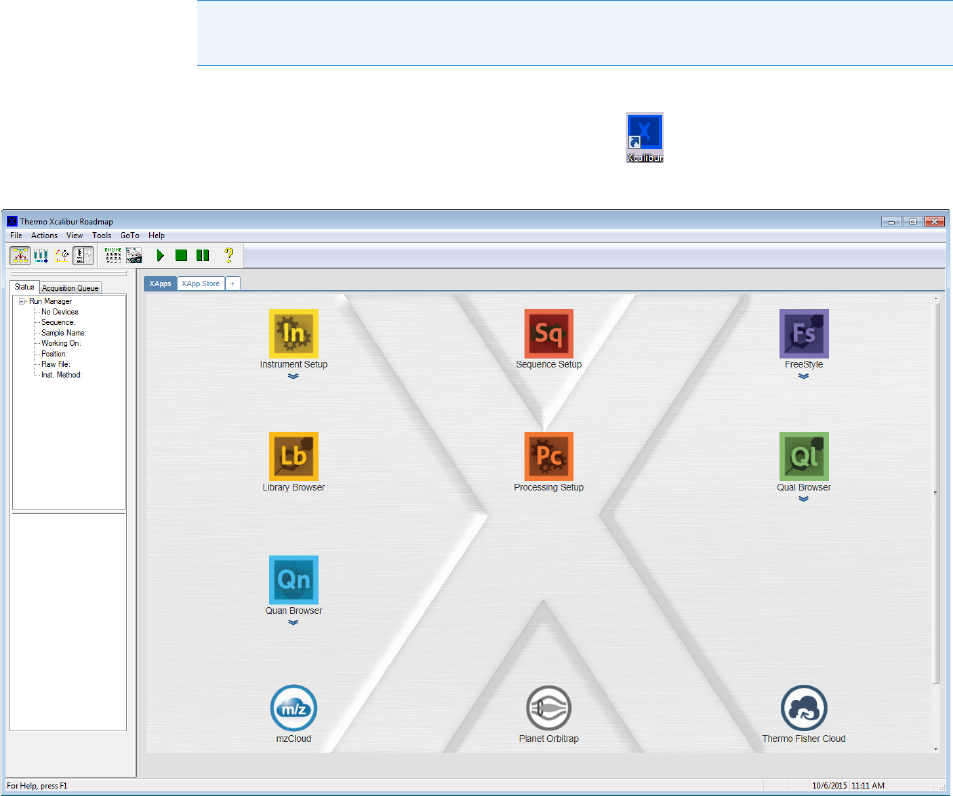
1. On your desktop, double-click the Xcalibur icon, .
The Thermo Xcalibur Roadmap opens.
Note The application installs the NIST 2008 library; however, it also supports the
NIST 2011 and NIST 2014 libraries.
2 Getting Started
Installing the NIST and QED Libraries
Thermo Scientific TraceFinder User Guide 15
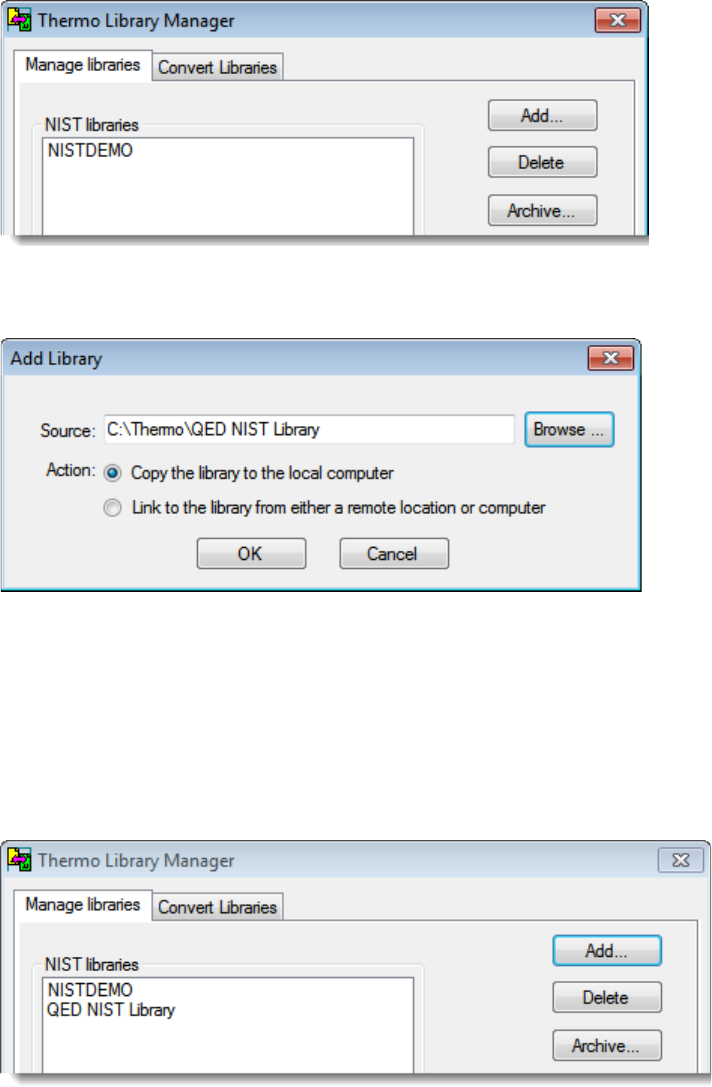
2. Choose To ol s > Li b r a r y M a n a g e r from the main menu.
The Thermo Library Manager dialog box opens, showing the NIST Libraries list.
3. Click Add.
The Add Library dialog box opens.
4. Click Browse, and locate your QED library in the C:\Thermo folder.
5. Click OK.
The Xcalibur data system reports that it has added the library to the NIST application.
6. Click Dismiss to close the message box.
The Xcalibur data system adds the QED library to the NIST Libraries list in the Library
Manager dialog box.
2 Getting Started
Installing the NIST and QED Libraries
16 TraceFinder User Guide Thermo Scientific
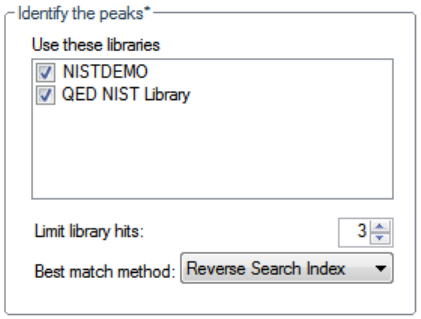
7. Click Exit in the Thermo Library Manager dialog box.
8. To confirm the library installation, do the following:
a. Start the application.
b. Click Method Development in the navigation pane.
c. Click Method View in the Method Development navigation pane.
d. Choose File > New > Method Template from the main menu.
The Method Template Editor displays the QED NIST Library in the Use These
Libraries list.
2 Getting Started
Launching the NIST Library Browser
Thermo Scientific TraceFinder User Guide 17
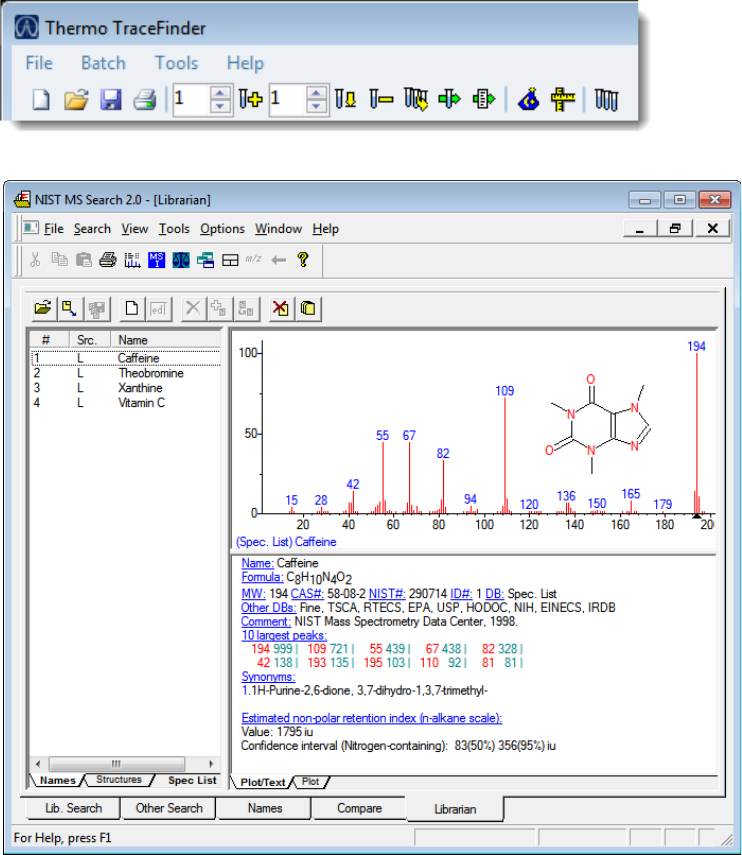
Launching the NIST Library Browser
Use the NIST MS Search tool to search the NIST library.
To open the NIST library browser
Choose Tools > Launch Library Browser from the TraceFinder main menu.
The NIST MS Search window opens.
For detailed instructions about using the library browser, refer to the Help in the NIST MS
Search window.
2 Getting Started
Launching a Qualitative Explorer
18 TraceFinder User Guide Thermo Scientific
Launching a Qualitative Explorer
Use a qualitative explorer application to display chromatograms and spectra, detect
chromatogram peaks, search libraries, simulate spectra, subtract background spectra, apply
filters, add text and graphics, create and save layouts, and view instrument parameters as they
changed during the acquisition.
Your TraceFinder application is configured to use one of the following applications:
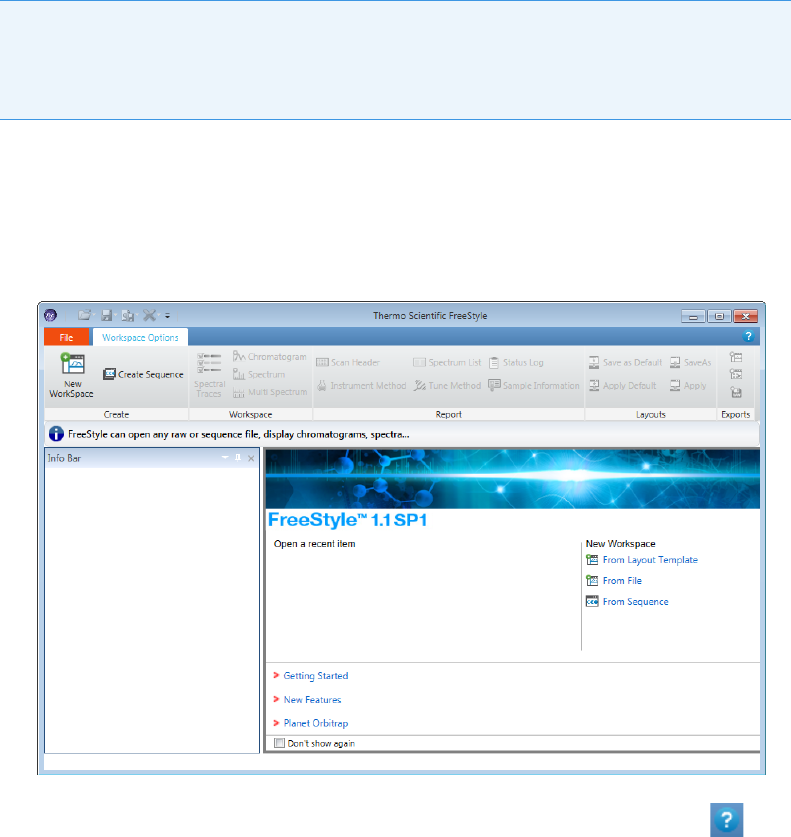
•Thermo Scientific FreeStyle
•Thermo Xcalibur Qual Browser
Thermo Scientific FreeStyle
To open the FreeStyle window
Choose To ol s > La u n ch Q ua l E xp l o rer from the TraceFinder main menu.
The FreeStyle application opens.
Figure 4. FreeStyle main window
For detailed instructions about using the FreeStyle application, click the Help icon, , in
the FreeStyle window.
IMPORTANT The Thermo Scientific FreeStyle™ application is available only when you
configure it as your default qualitative explorer in the Configuration console. Refer to
Chapter 1, “Using the Configuration Console,” in the Trace Fi n der Lab D irecto r Us e r
Guide.
2 Getting Started
Launching a Qualitative Explorer
Thermo Scientific TraceFinder User Guide 19
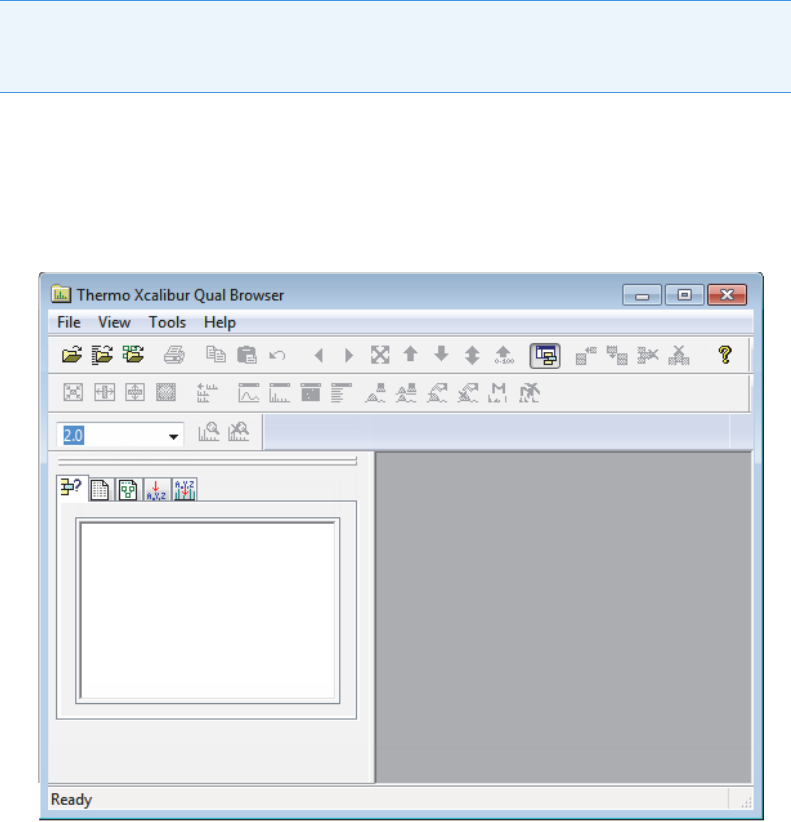
Thermo Xcalibur Qual Browser
To open the Qual Browser window
Choose To ol s > La u n ch Q ua l E xp l o rer from the TraceFinder main menu.
The Thermo Xcalibur Qual Browser application opens.
Figure 5. Qual Browser main window
For detailed instructions about using the Qual Browser application, refer to the Help in the
Qual Browser window.
IMPORTANT The Qual Browser application is available only when you configure it as
your default qualitative explorer in the Configuration console. Refer to Chapter 1, “Using
the Configuration Console,” in the TraceFinder Lab Director User Guide.

2 Getting Started
Converting Legacy Data
20 TraceFinder User Guide Thermo Scientific
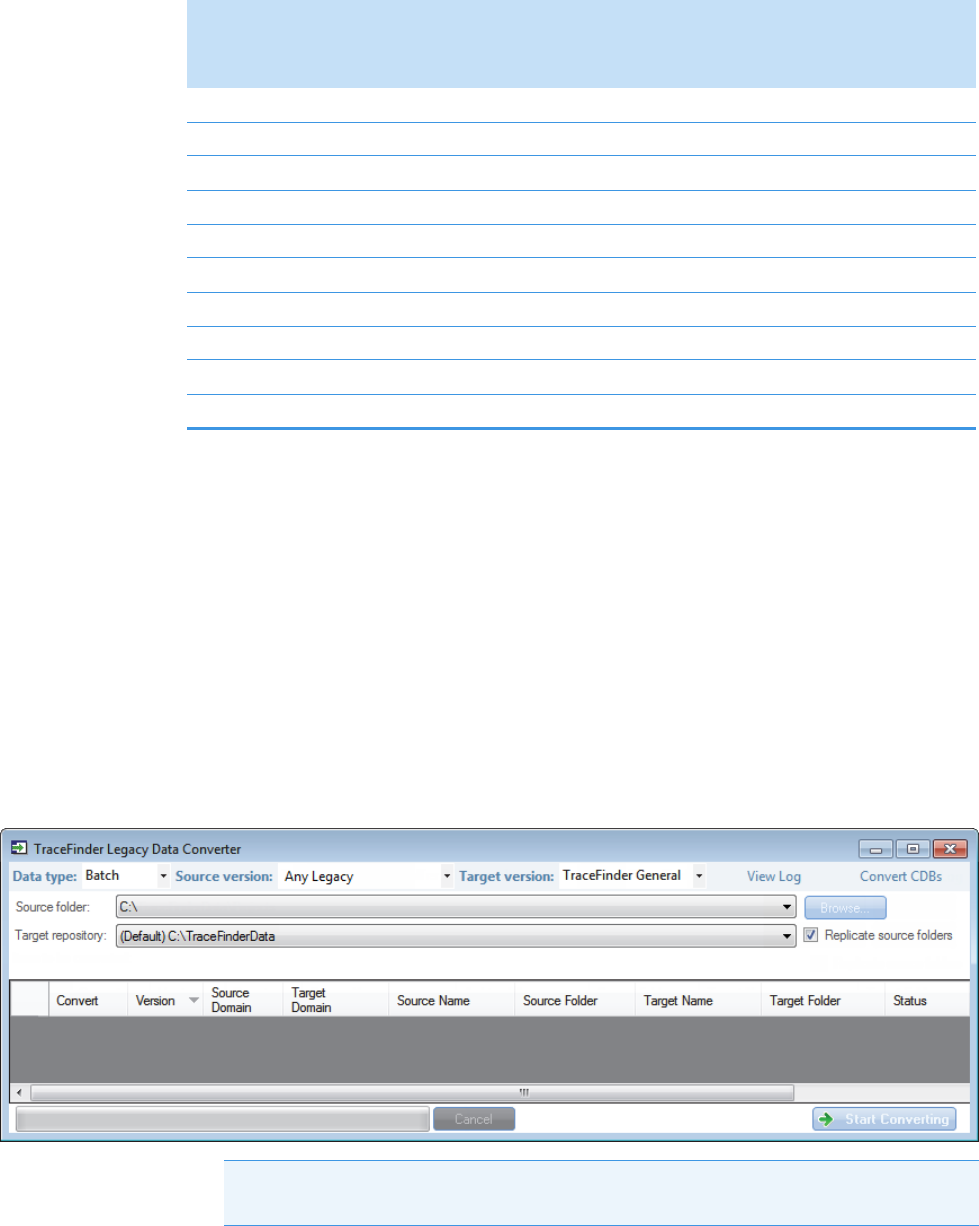
Converting Legacy Data
Use the TraceFinder Legacy Data Converter to convert methods, batches, method templates,
batch templates, or compound databases (CDBs) from the source versions to compatible
TraceFinder 4.0/4.1 target configurations.
• You can convert legacy methods, batches, or method templates from TraceFinder versions
2.0, 2.1, 3.0, 3.1, 3.2, or 3.3.
• You can convert legacy batch templates from TraceFinder versions 3.0, 3.1, 3.2, or 3.3.
• You can convert legacy compound databases from TraceFinder versions 3.0, 3.1, 3.2, or
3.3.
• You can convert data from TraceFinder version 4.0/4.1 for general quantitation to
another installed configuration of TraceFinder 4.0/4.1.
Version Compatibility
This table shows which source versions of methods, batches, method templates, batch
templates, or compound databases are compatible with TraceFinder 4.0/4.1 target
configurations.
IMPORTANT TraceFinder 4.1 uses the same data configuration as TraceFinder 4.0. You
do not need to convert your data from 4.0 to 4.1. Data for the 4.1 release is stored in the
TraceFinderData/4.0 folder.
Table 3. Version compatibility (Sheet 1 of 2)
Source TraceFinder 4.0/4.1 target
General EFSa Clinical
Research
Forensic
Toxicology
TraceFinder 4.0/4.1 General
TraceFinder 3.3 General
Tr a c e Fi n d e r 3 . 3 E F S
TraceFinder 3.3 Clinical Research
TraceFinder 3.3 Forensic Toxicology
TraceFinder 3.2 General
Tr a c e Fi n d e r 3 . 2 E F S
TraceFinder 3.2 Clinical Research
TraceFinder 3.2 Forensic Toxicology
TraceFinder 3.1 General
Tr a c e Fi n d e r 3 . 1 E F S
TraceFinder 3.1 Clinical Research
TraceFinder 3.1 Forensic Toxicology
TraceFinder 3.0 General
2 Getting Started
Converting Legacy Data
Thermo Scientific TraceFinder User Guide 21
See the following topics:
•Converting Methods
•Converting Batches
•Converting Method Templates
•Converting Batch Templates
•Converting Compound Databases
To open the TraceFinder Legacy Data Converter
Choose Tools > Launch Legacy Data Converter from the TraceFinder main menu.
The TraceFinder Legacy Data Converter window opens.
Tr a c e Fi n d e r 3 . 0 E F S
TraceFinder 3.0 Clinical Research
TraceFinder 3.0 Forensic Toxicology
TraceFinder 2.1 General
Tr a c e Fi n d e r 2 . 1 E F S
TraceFinder 2.1 Clinical Research
TraceFinder 2.1 Forensic Toxicology
TraceFinder 2.0 General
Tr a c e Fi n d e r 2 . 0 E F S
TraceFinder 2.0 Clinical Research
aEnvironmental and Food Safety
Table 3. Version compatibility (Sheet 2 of 2)
Source TraceFinder 4.0/4.1 target
General EFSa Clinical
Research
Forensic
Toxicology
Note When you open the application, the system checks for any legacy data and
prompts you to open the Legacy Data Converter.
2 Getting Started
Converting Legacy Data
22 TraceFinder User Guide Thermo Scientific
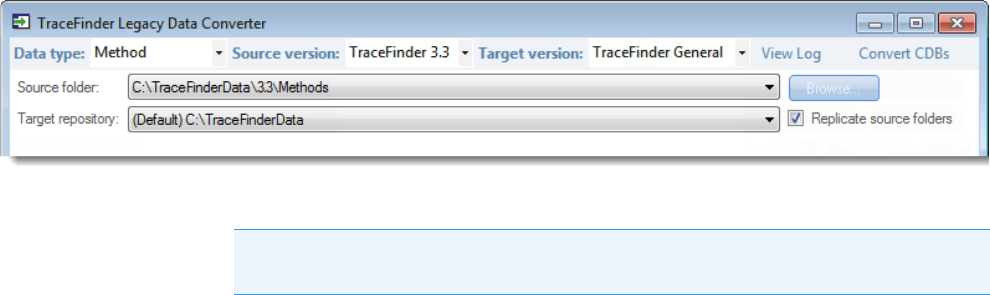
Converting Methods
Use the data converter to convert legacy methods to TraceFinder 4.1 methods.
To convert a method
1. In the Data Type list, select Method.
The TraceFinder Legacy Data Converter displays the interface for converting methods.
The following example shows that you can convert methods from the TraceFinder 3.3
General configuration to the current General configuration. For a complete list of version
compatibilities, see Version Compatibility.
2. In the Source Version list, select the version of the method that you will convert.
The conversion table displays the methods in the Methods folder for the selected source
version. The application verifies that the method file is in the .mmx file format.
3. To convert a method that is not in the default list, do the following:
a. Click Browse and locate a different source method folder.
You can select a specific method folder or a folder that contains multiple methods.
b. Click OK in the Browse for Folder dialog box.
The application displays the selected method folder in the conversion table.
When you select a folder that contains multiple method folders, the application displays
all the methods.
4. In the Target Version list, select the version that you are converting to.
The list displays only TraceFinder configurations with compatible data. See Version
Compatibility.
5. (Optional) In the Target Name column, change the default new name for each method
that you want converted.
Note When you select Any Legacy, the Legacy Data Converter examines all possible
methods in the source folder, regardless of version.
2 Getting Started
Converting Legacy Data
Thermo Scientific TraceFinder User Guide 23
When you populate the conversion table, the application checks each method to see if a
method with this name exists in the target repository.
• If the method name already exists in the target repository, the default new name
appends “_1” to the original name.
• If the method name does not exist in the target repository, the application keeps the
original method name.
6. Select the Convert check box for each method that you will convert, and click
.
The application confirms that all methods to be converted use the .mmx file format.
When the conversion process begins, the application displays a status bar and a Cancel
button. You can cancel pending conversions, but not the method that is currently
converting.
When the Status column reports that a method is successfully converted, the application
writes the converted file to the specified target repository.
7. To view a log of the conversion, click View Log.
The application opens a cumulative log file for the session in a Microsoft Notepad text
editor window.
IMPORTANT The conversion cannot overwrite an existing file name. If the new name
is identical to an existing method file, the conversion will not work. When you
manually enter a new name, you must verify that the name does not already exist.
Note If a method conversion is unsuccessful, the Status column displays “Conversion
failed.” The log file contains details about the failed conversion.
2 Getting Started
Converting Legacy Data
24 TraceFinder User Guide Thermo Scientific
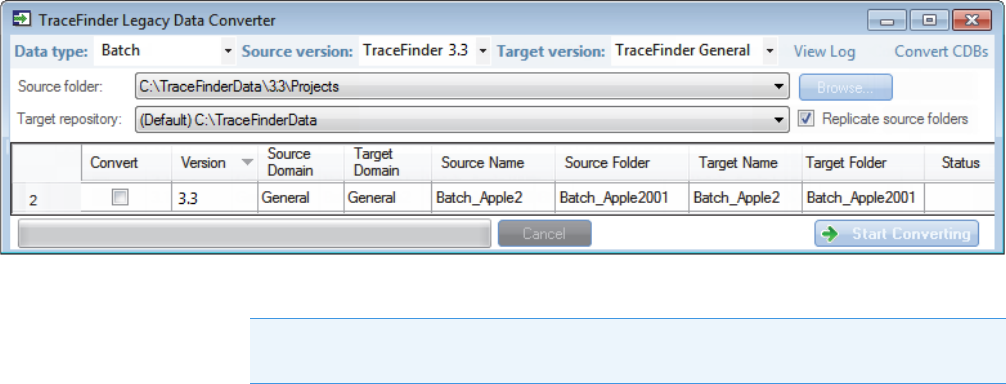
Converting Batches
Use the data converter to convert legacy batches to TraceFinder 4.1 batches.
To convert a batch
1. In the Data Type list, select Batch.
The following example shows that you can convert batches from the TraceFinder 3.3
General configuration to the current General configuration. For a complete list of version
compatibilities, see Version Compatibility.
2. In the Source Version list, select the version of the batch that you will convert.
The conversion table displays all batches in the Projects folder for the selected source
version.
3. In the Target Version list, select the version that you are converting to.
The list displays only TraceFinder configurations with compatible data. See Version
Compatibility.
4. In the Target Default Project and Subproject boxes, type the name of a project and
subproject, or select the Replicate Original Project/Subproject check box.
5. (Optional) In the New Name column, change the default new name for each batch that
you want converted.
When you populate the conversion table, the application checks each batch to see if a
batch with this name exists in the target repository.
• If the batch name already exists in the target repository, the default new name
appends “_1” to the original name.
• If the batch name does not exist in the target repository, the application keeps the
original batch name.
Note When you select Any Legacy, the Legacy Data Converter examines all possible
batches in the source folder, regardless of version.
2 Getting Started
Converting Legacy Data
Thermo Scientific TraceFinder User Guide 25
6. Select the Convert check box for each batch that you will convert, and click
.
The application confirms that all batches to be converted use the .btx file format.
When the conversion process begins, the application displays a status bar and a Cancel
button. You can cancel pending conversions, but not the batch that is currently
converting.
When the Status column reports that a batch is successfully converted, the application
writes the converted batch to the …\TraceFinderData\4.0\Projects folder and uses either
the original project and subproject names or the new names that you entered.
7. To view a log of the conversion, click View Log.
IMPORTANT The conversion cannot overwrite an existing file name. If the new name
is identical to an existing batch folder, the conversion will not work. When you
manually enter a new name, you must verify that the name does not already exist.
Note If a batch conversion is unsuccessful, the Status column displays “Conversion
failed.” The log file contains details about the failed conversion.
2 Getting Started
Converting Legacy Data
26 TraceFinder User Guide Thermo Scientific
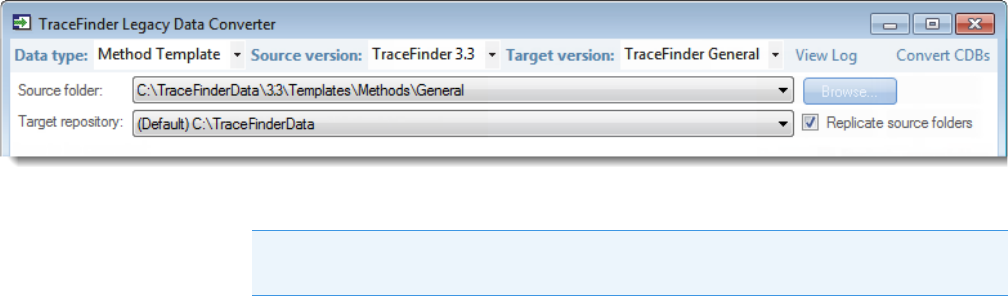
Converting Method Templates
Use the data converter to convert legacy method templates to TraceFinder 4.1 method
templates.
To convert a method template
1. In the Data Type list, select Method Template.
The following example shows that you can convert method templates from the
TraceFinder 3.3 General configuration to the current General configuration. For a
complete list of version compatibilities, see Version Compatibility.
2. In the Source Version list, select the version of the method template that you will convert.
The conversion table displays the method templates in the Templates folder for the
selected source version. The application verifies that the method template file is in
the .pmtx file format.
3. To convert a method template that is not in the default list, do the following:
a. Click Browse and locate a template folder.
You can select a specific template folder or a folder that contains multiple templates.
b. Click OK in the Browse for Folder dialog box.
The application displays the selected folder in the conversion table.
When you select a folder that contains multiple method template folders, the application
displays all the method templates.
4. In the Target Version list, select the version that you are converting to.
The list displays only TraceFinder configurations with compatible data. See Version
Compatibility.
5. (Optional) In the Target Name column, change the default name for each method
template that you want converted.
Note When you select Any Legacy, the Legacy Data Converter examines all possible
method templates in the source folder, regardless of version.
2 Getting Started
Converting Legacy Data
Thermo Scientific TraceFinder User Guide 27
When you populate the conversion table, the application checks each method template to
see if a method template with this name exists in the target repository.
• If the method template name already exists in the target repository, the default new
name appends “_1” to the original name.
• If the method template name does not exist in the target repository, the application
keeps the original method template name.
6. Select the Convert check box for each method template that you will convert, and click
.
The application confirms that all method templates to be converted use the .pmtx file
format.
When the conversion process begins, the application displays a status bar and a Cancel
button. You can cancel pending conversions, but not the template that is currently
converting.
When the Status column reports that the template is successfully converted, the
application writes the converted template to the specified target repository.
7. To view a log of the conversion, click View Log.
The application opens a cumulative log file for the session in a Notepad text editor
window.
IMPORTANT The conversion cannot overwrite an existing file name. If the new name
is identical to an existing method template file, the conversion will fail. When you
manually enter a new name, you must verify that the name does not already exist.
Note If a template conversion fails, the Status column displays “Conversion failed.”
The log file contains details about the failed conversion.
2 Getting Started
Converting Legacy Data
28 TraceFinder User Guide Thermo Scientific
Converting Batch Templates
Use the data converter to convert legacy batch templates to TraceFinder 4.1 batch templates.
To convert a batch template
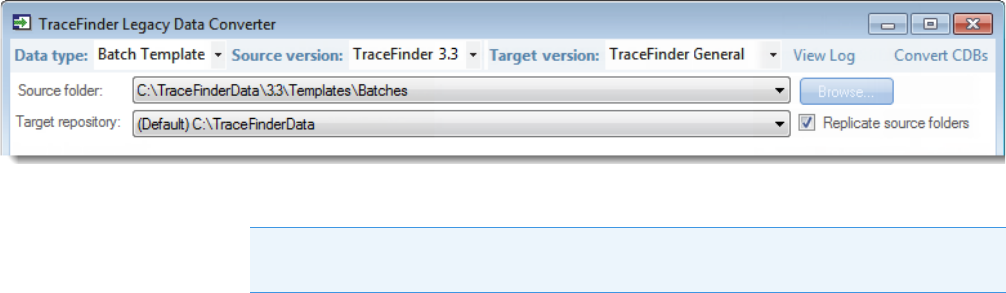
1. In the Data Type list, select Batch Template.
The following example shows that you can convert batch templates from the TraceFinder
3.3 General configuration to the current General configuration. For a complete list of
version compatibilities, see Version Compatibility.
2. In the Source Version list, select the version of the batch template that you will convert.
The conversion table displays the batch templates in the Templates folder for the selected
source version.
3. To convert a batch template that is not in the default list, do the following:
a. Click Browse and locate a template folder.
You can select a specific batch template folder or a folder that contains multiple batch
templates.
b. Click OK in the Browse for Folder dialog box.
The application displays the selected folder in the conversion table.
When you select a folder that contains multiple batch template folders, the application
displays all the batch templates.
4. In the Target Version list, select the version that you are converting to.
The list displays only TraceFinder configurations with compatible data. See Version
Compatibility.
5. (Optional) In the New Name column, change the default new name for each batch
template that you want converted.
When you populate the conversion table, the application checks each batch template to
see if a batch template with this name exists in the target repository.
• If the batch template name already exists in the target repository, the default new
name appends “_1” to the original name.
Note When you select Any Legacy, the Legacy Data Converter examines all possible
batch templates in the source folder, regardless of version.
2 Getting Started
Converting Legacy Data
Thermo Scientific TraceFinder User Guide 29
• If the batch template name does not exist in the target repository, the application
keeps the original batch template name.
6. Select the Convert check box for each batch template that you will convert, and click
.
The application confirms that all batch templates to be converted use the .btx file format.
When the conversion process begins, the application displays a status bar and a Cancel
button. You can cancel pending conversions, but not the template that is currently
converting.
When the Status column reports that the template is successfully converted,
the application writes the converted template folder to the
…\TraceFinderData\4.0\Templates\Batches folder.
7. To view a log of the conversion, click View Log.
The application opens a cumulative log file for the session in a Notepad text editor
window.
IMPORTANT The conversion cannot overwrite an existing file name. If the new name
is identical to an existing batch template file, the conversion will fail. When you
manually enter a new name, you must verify that the name does not already exist.
Note If a template conversion fails, the Status column displays “Conversion failed.”
The log file contains details about the failed conversion.
2 Getting Started
Converting Legacy Data
30 TraceFinder User Guide Thermo Scientific
Converting Compound Databases
Use the data converter to convert legacy compound databases to TraceFinder 4.1 compound
databases.
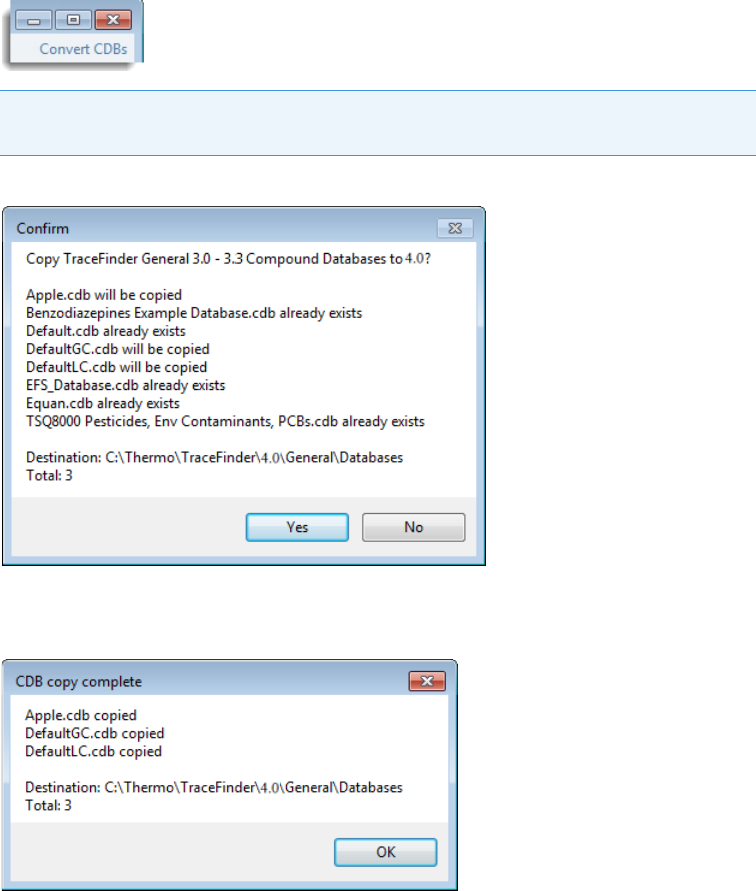
To convert compound databases
1. Click Convert CDBs (in the upper right corner of the window).
The Confirm dialog box lists all databases that will be converted to TraceFinder 4.1.
2. Click Yes.
The Confirm dialog box lists all databases that are converted to TraceFinder 4.1.
3. Click OK.
The converted compound databases are now available in the TraceFinder 4.1 application.
Note If your window is too narrow, you might not see the Convert CDBs button.
Widen the window to expose the button in the upper right corner.
2 Getting Started
Choosing a Mode or Console
Thermo Scientific TraceFinder User Guide 31
Choosing a Mode or Console
When user security is activated, the navigation pane displays the modes and consoles available
to the current user’s assigned roles and permissions. The following table shows the available
modes and consoles for each user role.
Follow these procedures:
•To choose a mode
•To open the Configuration console
•To open the Administrator console
•To display a log of instrument errors
•To monitor instrument status
•To watch acquisition and processing in real time
Table 4. User roles and default access
User role Method
Development Acquisition Analysis Configuration
console
Administrator
Console
Security Security only
LabDirector
ITAdmin
Supervisor
Technician
QAQC
Note When user security is not activated, all modes and consoles are available to all users.
2 Getting Started
Choosing a Mode or Console
32 TraceFinder User Guide Thermo Scientific
To choose a mode
In the navigation pane, click the mode where you want to work.
The navigation pane shows only the modes that you have permission to use.
To open the Configuration console
Click the Application Configuration icon, , in the upper right corner of the
Tr a c e Fi n d e r w i n d o w.
When user security is activated, you must have Configuration permissions to access the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in the
TraceFinder Lab Director User Guide.
To open the Administrator console
Choose Tools > Administrator Console from the TraceFinder main menu.
When user security is activated, you must have Administrator permissions to access the
Administrator Console. Refer to the TraceFinder Administrator Console User Guide.
Mode Description
Acquisition Opens the Acquisition mode where you can create and review batches,
batch data, reports, and local methods.
See Chapter 3, “Using the Acquisition Mode.”
Analysis Opens the Analysis mode where you can review batches, batch data,
reports, and local methods.
See Chapter 4, “Using the Analysis Mode for Quantitation Batches.”
See Chapter 5, “Using the Analysis Mode for Target Screening
Batches.”
See Chapter 6, “Using the Analysis Mode for Unknown Screening
Batches.”
Method
Development
Opens the Method Development mode where you can create a master
method or an instrument method.
Refer to the TraceFinder Lab Director User Guide.
2 Getting Started
Choosing a Mode or Console
Thermo Scientific TraceFinder User Guide 33
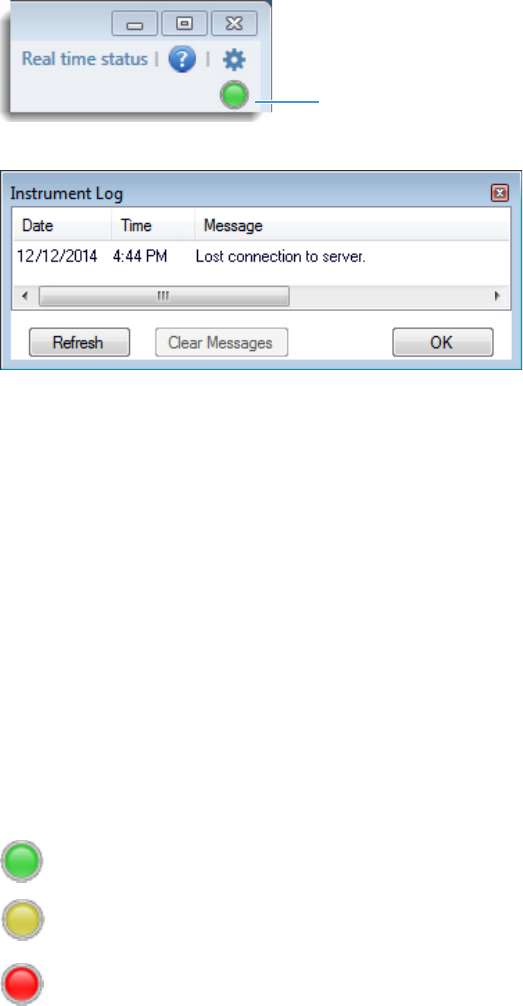
To display a log of instrument errors
1. Click the status light in the upper right corner of the TraceFinder window.
The Instrument Log dialog box opens.
The Instrument Log displays all instrument errors that have occurred since the
application started or since the last time that you cleared the message log.
2. Do any of the following:
• Click Refresh to display errors that occur after you open the Instrument Log dialog
box.
• Click Clear Messages to remove messages from the Instrument Log display.
The application clears messages only from the Instrument Log display. These
messages remain in the following log file:
C:\Thermo\TraceFinder\4.0\General\Logs\TraceFinder.log
• Click OK to dismiss the Instrument Log dialog box.
To monitor instrument status
Look at the status light in the upper right corner of the TraceFinder window.
Green indicates that the instrument is ready.
Yellow indicates that the instrument is in standby mode, waiting for contact
closure, or warming up the lamp.
Red indicates that the instrument is not ready because it is turned off, not
configured or connected, currently initializing, or otherwise busy.
Status light
2 Getting Started
Choosing a Mode or Console
34 TraceFinder User Guide Thermo Scientific
The status light (in any color) might include an exclamation mark.
• An exclamation mark on a green status light, , might indicate that the
instrument method contains the wrong source (for example, APCI instead of HESI).
• An exclamation mark on a red status light, , might indicate that a vial is missing
and the batch was forced to stop.
Click the status light to display details of the error.
To watch acquisition and processing in real time
Click Real Time Status in the upper right corner of the TraceFinder window.
The application displays the Real Time Status pane at the bottom of the window.
For descriptions of all the features of the Real Time Status pane, see Real Time Status
Pane in Chapter 3, “Using the Acquisition Mode.”

Thermo Scientific TraceFinder User Guide 35
3
Using the Acquisition Mode
This chapter describes the tasks associated with the Acquisition mode.
When you plan to work with multiple samples or use similarly designed batches, use the
Acquisition mode to reduce the amount of data you must enter.
Because the nature and types of batches are often similar (in some cases specified by laboratory
standard practices), you can define a batch template that supplies the basic structure of a
batch.
Using a master method, you can create a batch and run the samples. A batch represents one or
more samples that are to be acquired, processed, reviewed, and reported as a set. After you
create a batch of samples, you can submit the batch and review the results in Data Review or
you can go directly to viewing and printing reports.
You can set up a calibration batch with known concentrations of the target compounds and
compare the calibration values against samples in future batches.
You can also use the Quick Acquisition feature to quickly submit samples from any page in
the Acquisition mode. See Appendix A, “Using Quick Acquisition.”
Contents
•Working with Batches
•Real Time Status Pane
•Sample Types
IMPORTANT When user security is activated, you must have Acquisition Wizard –
Template Editing permission to create a batch template.
3 Using the Acquisition Mode
Working with Batches
36 TraceFinder User Guide Thermo Scientific
Working with Batches
See the instructions for the following tasks:
•Opening and Navigating the Acquisition Mode
•Creating and Submitting Batches
Opening and Navigating the Acquisition Mode
To access the Acquisition mode
Click Acquisition in the navigation pane.
The navigation pane for the Acquisition mode opens.
As you progress through the Acquisition mode using any of these methods for creating a
batch, the task pane at the top of the view tracks your progress. As you complete each
stage, you can hold your cursor over the view name in the task pane to display the
parameters that you specified for the batch. See Example task pane when you have
completed the Acquisition mode.
IMPORTANT TraceFinder 4.1 uses the same data as TraceFinder 4.0. By default, the application
stores the batch data for the 4.1 release in the TraceFinderData\4.0\Projects folder.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 37
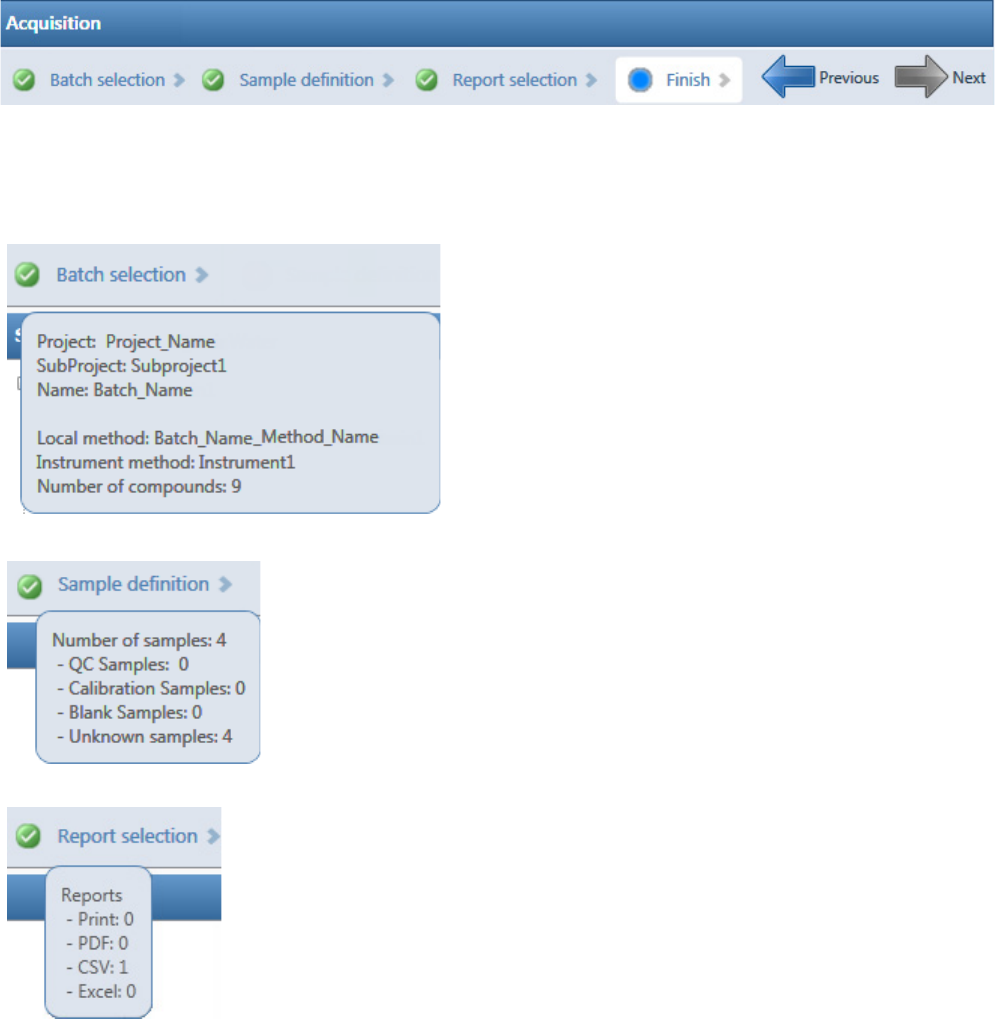
Figure 6. Example task pane when you have completed the Acquisition mode
Categories in the Sample Definition list:
QC Samples: QC Std
Calibration Samples: Cal Std
Blank Samples: Matrix Blank
Unknown Samples: All other samples
Hold your cursor over Batch Selection, Sample Definition,
or Report Selection to view the parameters for your batch.
3 Using the Acquisition Mode
Working with Batches
38 TraceFinder User Guide Thermo Scientific
Creating and Submitting Batches
To create and submit a batch, the Acquisition mode uses a wizard-style interface to guide you
through these major steps:
1. Selecting a Batch
2. Defining a Sample List
3. Selecting and Reviewing Reports
4. Submitting a Batch in the Acquisition Wizard
The Acquisition mode provides multiple techniques for creating either a batch or a batch
template. Each batch creation technique has an associated workflow, as shown in the
following flowcharts. Each workflow uses a different combination of Acquisition mode pages.
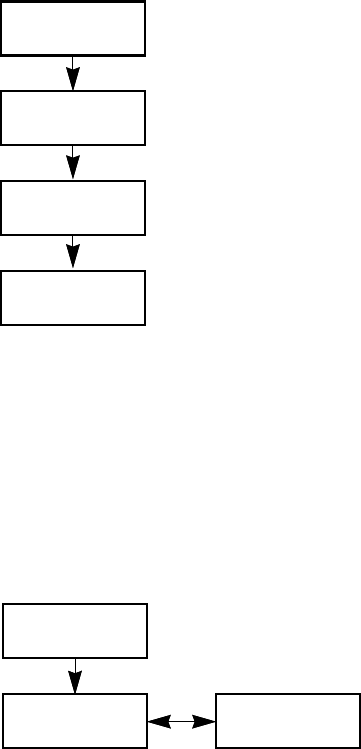
Workflow for Creating an Original Batch
To create an original batch, start with any of these instructions: To start a new quantitation
batch, or To start a new target screening batch, or To start a new unknown screening batch.
To see how to acquire a new batch
1. Choose Help > Animations.
2. From the list of animation topics, click Acquiring a New Batch.
Workflow for Acquiring a Prepared Batch
To acquire a prepared batch, start with the instructions To select a prepared batch.
Batch and
Method
Finish
Reports
Samples
Batch
Finish Samples
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 39
To see how to acquire a prepared batch
1. Choose Help > Animations.
2. From the list of animation topics, click Acquiring a Prepared Batch.
Workflow for Reinjecting a Previously Acquired Batch
To process a previously acquired batch, start with the instructions To reinject samples in a
previously acquired batch.
To see how to reinject samples in a previously acquired batch
1. Choose Help > Animations.
2. From the list of animation topics, click Reinjecting Samples.
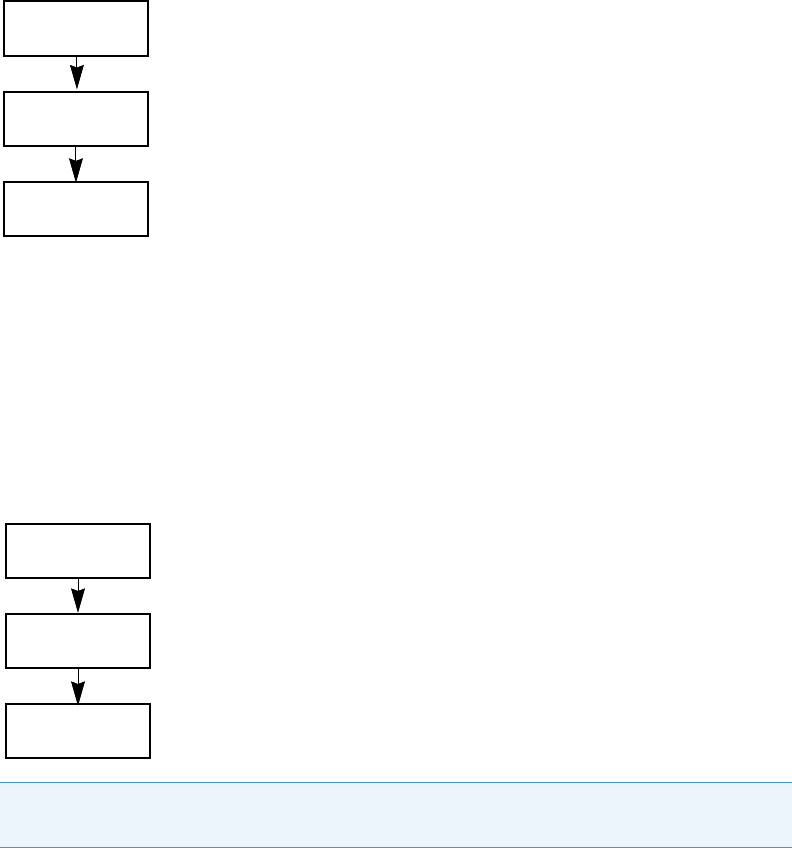
Workflow for Creating or Editing a Batch Template
To create a batch template, start with any of these instructions: To create a quantitation batch
template, To create a target screening batch template, or To c r ea t e an u n k n ow n s c re e n i n g
batch template.
To see how to create a batch template
1. Choose Help > Animations.
2. From the list of animation topics, click Creating a Batch Template.
IMPORTANT When user security is activated, you must have Acquisition Wizard –
Template Editing permission to create a batch template.
Batch
Finish
Samples
Finish
Samples
Template and
Method
3 Using the Acquisition Mode
Working with Batches
40 TraceFinder User Guide Thermo Scientific
Selecting a Batch
On the Batch Selection page of the Acquisition mode, you can create a new quantitative,
target screening, or unknown screening batch in any of your current projects/subprojects. Or,
you can submit a batch that you previously prepared and saved, reinject the samples in a batch
that you previously acquired, or create a batch template to use for future batches.
Follow these procedures:
•To start a new quantitation batch
•To start a new target screening batch
•To start a new unknown screening batch
•To start a new batch from a template
•To select a prepared batch
•To reinject samples in a previously acquired batch
•To create a quantitation batch template
•To create a target screening batch template
•To create an unknown screening batch template
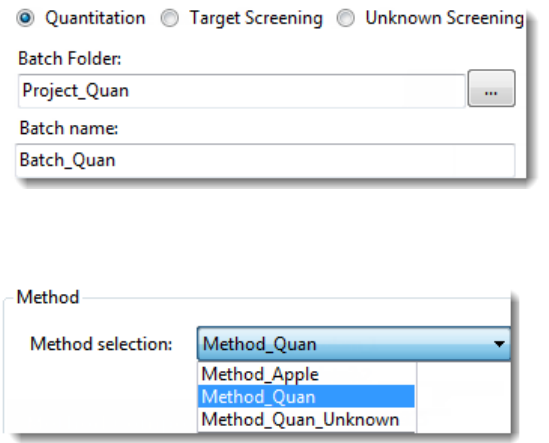
To start a new quantitation batch
1. Click Create a New Batch in the navigation pane.
2. Select the Quantitation option.
3. Select the batch folder where you want to create the new batch.
4. Type a unique name for the new batch in the Batch Name box.
If the name you enter is not unique, a red warning flashes.
5. Select a method from the Method Selection list.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 41
The Method Compound Data pane displays the compounds in the method. You cannot
edit the compounds list from the Acquisition mode.
6. To continue to the next page, click Next.
The Sample Definition page opens. See Defining a Sample List.
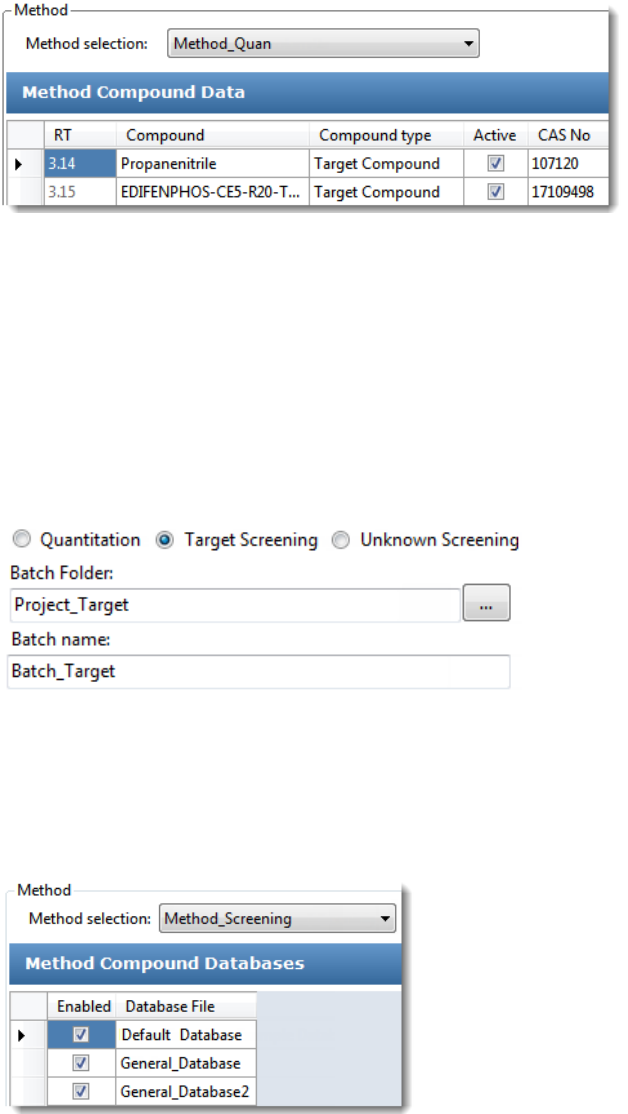
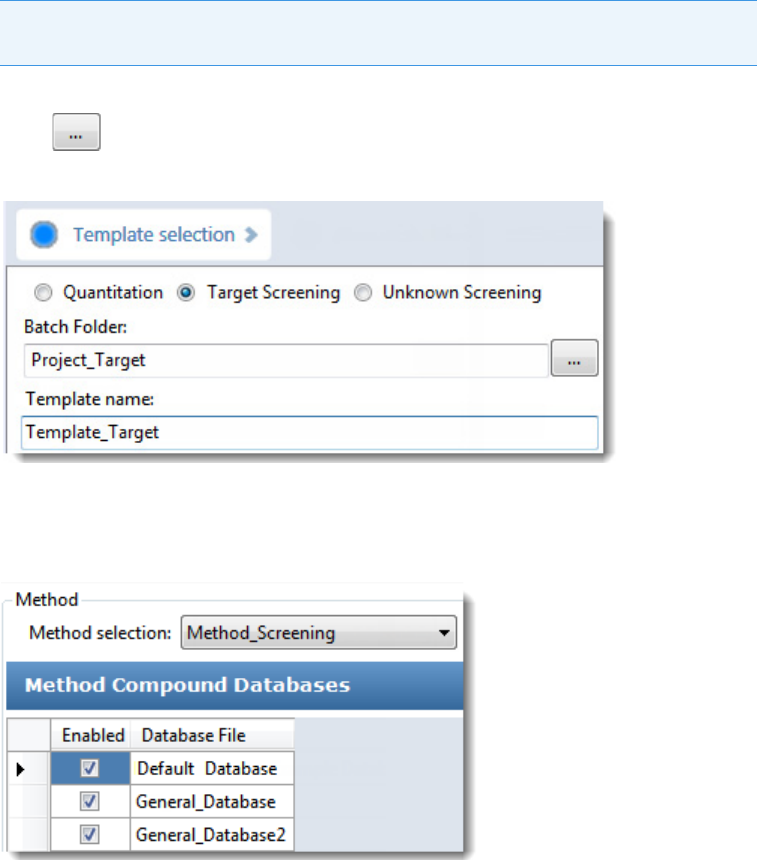
To start a new target screening batch
1. Click Create a New Batch in the navigation pane.
2. Select the Target Screening option.
3. Select the batch folder where you want to create the new batch.
4. Type a name for the new batch in the Batch Name box.
If the name you enter is not unique, a red warning flashes.
5. Select a method from the Method Selection list.
The Method Compound Databases pane displays the compound databases in the
method. The application uses these databases to identify the compounds in the samples.
You cannot edit the compound database list from the Acquisition mode.
6. To continue to the next page, click Next.
The Sample Definition page opens. See Defining a Sample List.
3 Using the Acquisition Mode
Working with Batches
42 TraceFinder User Guide Thermo Scientific
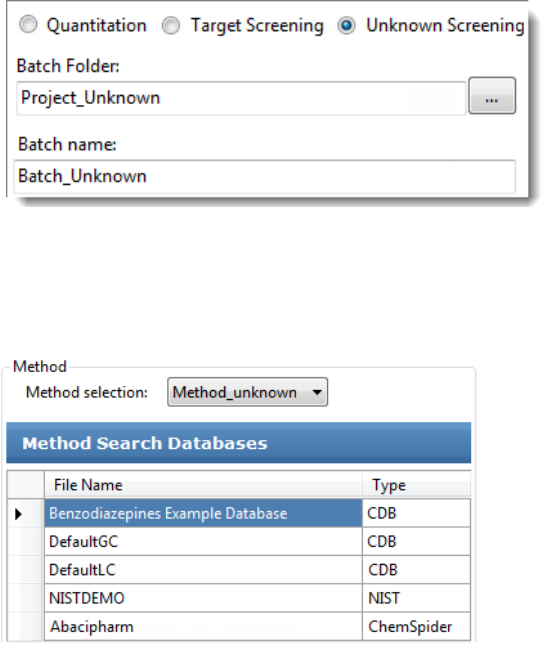
To start a new unknown screening batch
1. Click Create a New Batch in the navigation pane.
2. Select the Unknown Screening option.
3. Select the batch folder where you want to create the new batch.
4. Type a name for the new batch in the Batch Name box.
5. Select a method from the Method Selection list.
The Method Search Databases pane displays the databases that you selected in the
method. The application uses these databases to identify the compounds in the samples.
You cannot edit the database list from the Acquisition mode.
6. To continue to the next page, click Next.
The Sample Definition page opens. See Defining a Sample List.
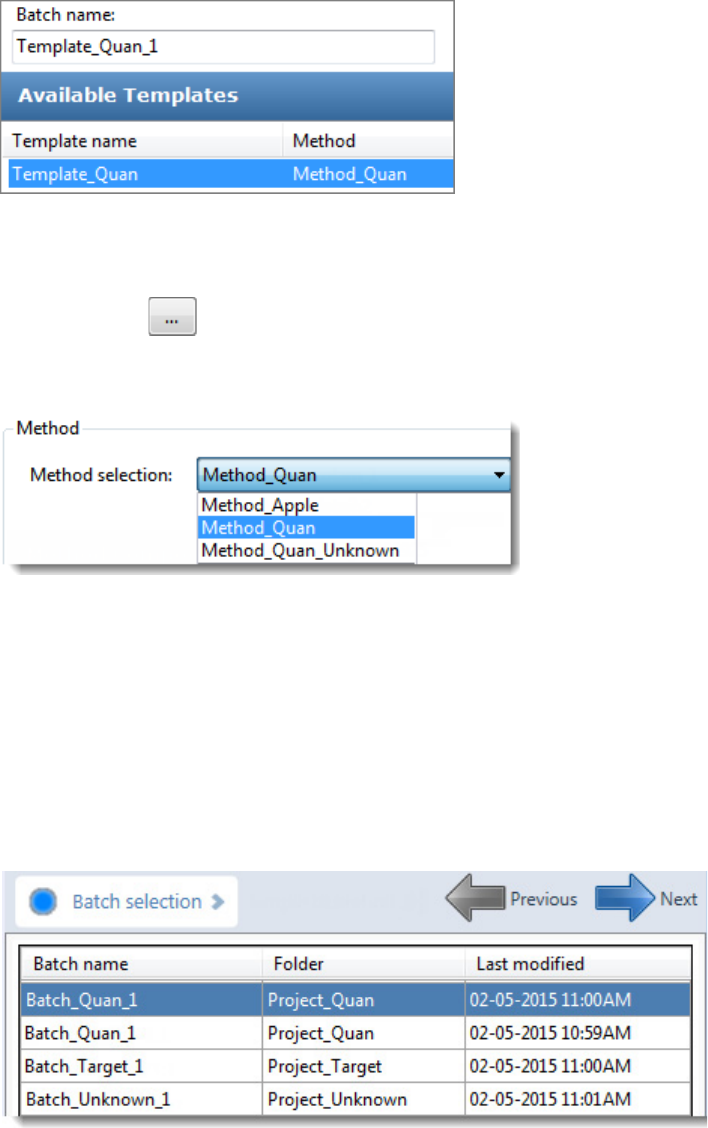
To start a new batch from a template
1. Click Create a New Batch in the navigation pane.
2. Select either the Quantitation, Target Screening, or Unknown Screening option.
The Available Templates pane displays only batch templates for the selected option.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 43
3. In the Available Templates pane, select the template and method combination that you
want to use.
The system creates a batch name with the selected template name and appends the date
and time stamp. You can change the batch name, the default batch folder, or the method
associated with this template.
4. (Optional) Click and select a different batch folder where you want to create the
new batch.
5. (Optional) Select a different method to use for the new batch.
6. To continue to the next page, click Next.
The Sample Definition page of the Acquisition mode opens. See Defining a Sample List.
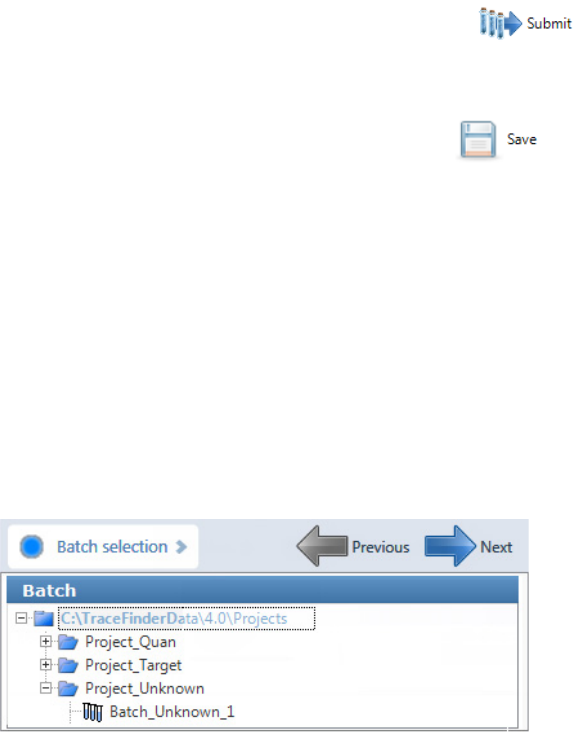
To select a prepared batch
1. Click Submit a Prepared Batch in the navigation pane.
The application displays all your unacquired, saved batches. The application stores all
unacquired batches in the …\TraceFinderData\4.0\Projects\… folder.
2. Select the batch that you want to acquire.
3 Using the Acquisition Mode
Working with Batches
44 TraceFinder User Guide Thermo Scientific
3. To continue to the next page, click Next.
The Finish page of the Acquisition mode opens. From the Finish page, you can save the
batch, submit the batch for acquisition, or go to the Sample Definition page to edit the
sample list for this batch.
• If the batch is unreadable, the application reports that the batch file is not valid and
cannot be opened.
• If a sample in the batch is unreadable, the application cannot open the sample. The
application creates a new sample with the same name and flags the sample. You must
complete the missing information such as Sample Type, Level, and so forth, and then
save the batch before you submit it for acquisition. Or, you can browse in a new raw
data file to replace the corrupt file.
4. Do one of the following:
• To edit the sample list, click Previous.
For detailed instructions, see Defining a Sample List.
–or–
• To prepare the batch for acquisition, click Submit, .
For detailed instructions, see Submitting a Batch in the Acquisition Wizard.
–or–
• To save the batch to be acquired later, click Save, .
The application saves your batch in the following folder:
…\TraceFinderData\4.0\Projects\....
The application closes the Acquisition mode and returns you to the mode you were
last using.
To reinject samples in a previously acquired batch
1. Click Reinject Samples in the navigation pane.
2. On the Batch page, select the batch that you want to reacquire.
The Batch page displays all previously acquired quantitation, target screening, and
unknown screening batches.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 45
3. To continue to the next page, click Next.
The Sample Definition page of the Acquisition mode opens. See Defining a Sample List.
• If the batch is unreadable, the application reports that the batch file is not valid and
cannot be opened.
• If a sample in the batch is unreadable, the application creates a new sample with the
same name and flags the sample. You must complete the missing information, such as
Sample Type, Level, and so forth, and then save the batch before you submit it for
acquisition. Or, you can browse in a new raw data file to replace the corrupt file.
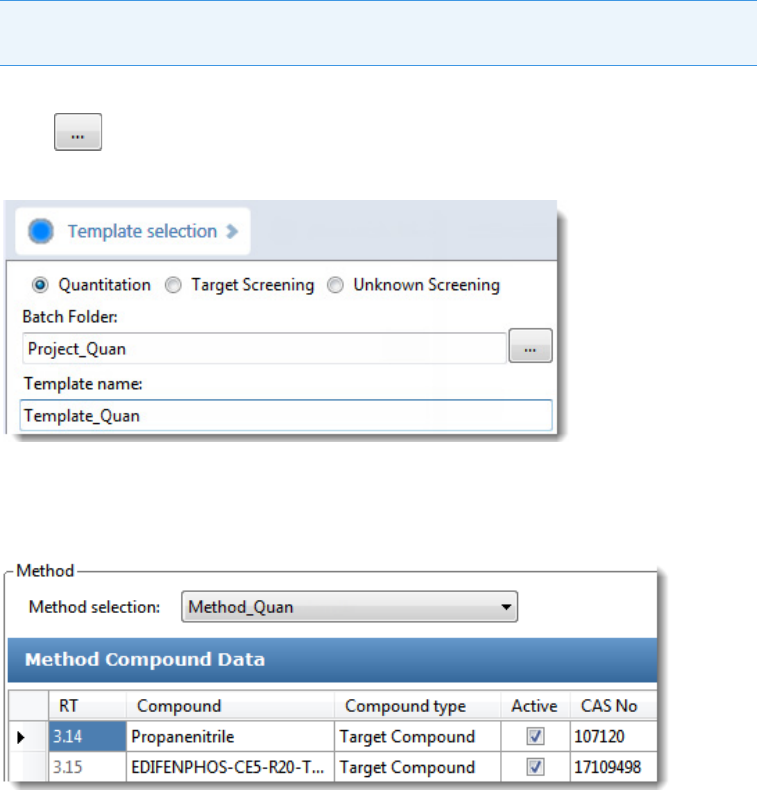
To create a quantitation batch template
1. Click Create or Edit a Template in the navigation pane.
2. Select the Quantitation option.
3. Click and select the folder where you want to create the new batch template.
4. Type a name for the new batch template in the Template Name box.
5. Select a method from the Method Selection list.
The Method Compound Data pane displays the compounds in the method. You cannot
edit the compounds list from the Acquisition mode.
6. To continue to the next page, click Next.
The Sample Definition page of the Acquisition mode opens. See Defining a Sample List.
Note When user security is activated, you must have Acquisition Wizard – Template
Editing permission to create a batch template.
3 Using the Acquisition Mode
Working with Batches
46 TraceFinder User Guide Thermo Scientific
To create a target screening batch template
1. Click Create or Edit a Template in the navigation pane.
2. Select the Target Screening option.
3. Click and select the folder where you want to create the new batch template.
4. Type a name for the new batch template in the Template Name box.
5. Select a method from the Method Selection list.
The Method Compound Databases pane displays the screening databases available for the
selected method.
6. Select the check box for each database that you want to use for screening.
7. To continue to the next page, click Next.
The Sample Definition page of the Acquisition mode opens. See Defining a Sample List.
Note When user security is activated, you must have Acquisition Wizard – Template
Editing permission to create a batch template.
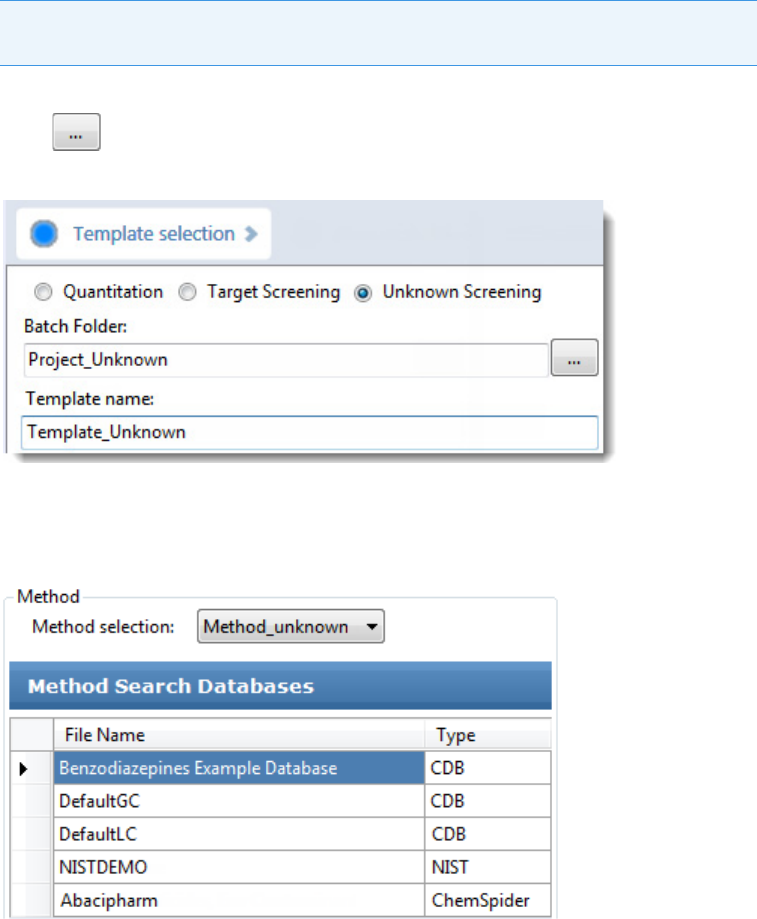
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 47
To create an unknown screening batch template
1. Click Create or Edit a Template in the navigation pane.
2. Select the Unknown Screening option.
3. Click and select the folder where you want to create the new batch template.
4. Type a name for the new batch template in the Template Name box.
5. Select a method from the Method Selection list.
The Method Compound Databases pane displays the unknown screening databases
available for the selected method.
6. Select the check box for each database that you want to use for unknown screening.
7. To continue to the next page, click Next.
The Sample Definition page of the Acquisition mode opens. See Defining a Sample List.
Note When user security is activated, you must have Acquisition Wizard – Template
Editing permission to create a batch template.
3 Using the Acquisition Mode
Working with Batches
48 TraceFinder User Guide Thermo Scientific
Defining a Sample List
Use the Samples page on the Sample Definition page of the Acquisition mode to create a list
of samples for the batch. You can add samples, insert samples, import a sample list, or remove
samples from the list. You can use the Reference Sample page to select a reference sample to
use as a reference peak in the Data Review.
To create the sample list, you can use either of two sets of function buttons (described in the
following graphic) or you can use commands in the shortcut menu (see the Shortcut menu
commands area of the Samples page parameters).
As you enter sample values, you can use the Copy Down and Fill Down commands to quickly
enter column values. For detailed instructions on using Copy Down and Fill Down to enter
column values, see Appendix B, “Using Copy Down and Fill Down.”
Use any of the following procedures to create a sample list. When you finish defining the list
of samples, click Next.
• When you create a batch from scratch and click Next, the Report Selection page opens.
See Selecting and Reviewing Reports.
• When you edit a prepared batch, reinject samples, create a batch from a template, or edit
a batch template and then click Next, the Finish Selection page opens. See Submitting a
Batch in the Acquisition Wizard.
Icons in the toolbar
Buttons at the bottom of the sample definition page
Add samples
Insert samples
Remove samples
Import samples
Add samples Insert samples Import samples
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 49
Follow these procedures:
•To add samples to the list
•To insert samples into the list
•To import samples into the list
•To remove samples from the list
•To reinject a sample from a previously acquired batch
•To select channels for the batch
•To assign a specific channel to a sample
•To select a reference sample
•To add an auto sample type
•To specify different instrument methods for samples
To add samples to the list
1. Select the number of sample rows to add and then click the Add icon, or
.
2. Type a file name in the Filename column for each sample.
Each file name must be unique.
3. Select a sample type from the Sample Type list for each sample.
For a detailed description of each sample type, see Sample Types.
4. For each Cal Std or QC Std sample, select a level from the Level list.
The master method defines the sample levels. If there are no levels to select in the Level
list, ask a user with Method Development permission to edit the method and specify the
levels. Then return to the Acquisition mode, and begin the batch again. The application
does not save a batch when you leave the Acquisition mode.
If you have Method Development permission, do the following:
a. Return to the Method Development mode.
b. Open the method.
c. Click the Compounds tab.
d. Click the Calibration Levels tab.
e. Add the levels.
f. Save the method.
For detailed instructions, refer to Chapter 4, “Using the Method Development Mode for
Quantitation Methods,” in the TraceFinder Lab Director User Guide.
Available sample types
Matrix Blank Solvent QC Std Unknown
Cal Std
3 Using the Acquisition Mode
Working with Batches
50 TraceFinder User Guide Thermo Scientific
5. (Optional) To assign samples to a group, do the following:
a. For each sample, type a group name in the Groups column.
Repeat this for each sample that you want to include in a group.
b. Create as many groups as you want.
• For quantitation experiments, after creating groups, you can choose one of the
samples as a threshold sample for the group and then compare all of the samples in
the Comparative View in Data Review.
For information about specifying a threshold sample for a group of samples, see
Threshold Samples Page for Quantitation Batches. For information about viewing
grouped samples in Data Review, see Comparative View for Quantitation Batches.
For information about Group Averages, see Group Averages.
• For unknown screening experiments, you can create a control group (specify the
group name as Control) to use for group averages in Data Review. For information
about Group Averages, see Group Averages Pane.
6. For each sample, type a vial position in the Vial Position column.
7. For each sample, type a volume in the Injection Volume column.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
8. (Optional) Type or edit the values for the remaining columns.
To automatically copy or fill values in these columns, see Appendix B, “Using Copy
Down and Fill Down.”
Note To assign a sample to multiple groups, separate the groups with a comma.
The application uses only the first group listed for calculating the Maximum Fold
and Group Averages.
For information about Maximum Fold analysis in unknown screening analysis,
see Cross Sample Peak List Pane.
Tip Use the Fill Down command to make entering vial positions easier.
Note When you use the scroll bar at the bottom of the sample list, the following
columns remain fixed: Status, Filename, Sample Type, Groups, Qual Processing
(quantitation) or Blank Subtraction (target screening), Level, Sample ID, and Sample
Name, while the other columns scroll right and left.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 51
To insert samples into the list
1. Select the sample above which you want to insert new Unknown samples.
You cannot use the Insert command to create the first sample row.
2. Select the number of samples to insert and then click the Insert icon, or
.
The application inserts the Unknown samples above the selected sample.
3. For each sample, type a file name in the Filename column.
Each file name must be unique.
4. For each sample, select a sample type from the Sample Type list.
5. For each Cal Std or QC Std sample, click the Level cell and select a level from the list.
The master method defines the sample levels. If there are no levels to select from the Level
list, ask a user with Method Development permission to edit the method and specify the
levels. Then return to the Acquisition mode, and begin the batch again. The application
does not save a batch when you leave the Acquisition mode.
If you have Method Development permission, follow the instructions in step 4 of the
procedure To add samples to the list.
6. (Optional) To assign samples to a group, do the following:
a. For each sample, type the group name in the Groups column.
Repeat this for each sample that you want to include in a group.
b. Create as many groups as you want.
Available sample types
Matrix Blank Solvent QC Std Unknown
Cal Std
Inserted
samples
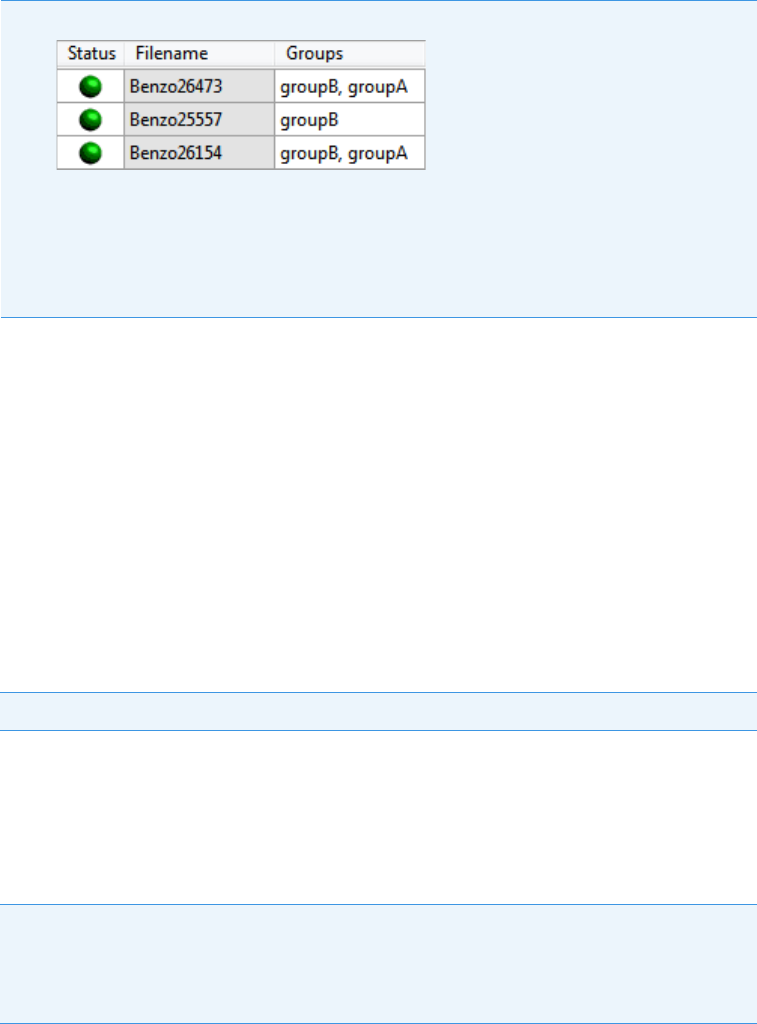
3 Using the Acquisition Mode
Working with Batches
52 TraceFinder User Guide Thermo Scientific
• For quantitation experiments, after creating groups, you can choose one of the
samples as a threshold sample for the group and then compare all of the samples in
the Comparative View in the Analysis mode.
For information about specifying a threshold sample for a group of samples, see
Threshold Samples Page for Quantitation Batches. For information about viewing
grouped samples in Data Review, see Comparative View for Quantitation Batches.
For information about Group Averages, see Group Averages.
• For unknown screening experiments, you can create a control group (specify the
group name as Control) to use for group averages in Data Review. For information
about group averages, see Group Averages Pane.
7. Type a vial position in the Vial Position column for each sample.
8. For each sample, type a volume in the Injection Volume column.
The minimum injection volume allowed is 0.1 μL; the maximum injection volume
allowed is 5000 μL.
9. (Optional) Type or edit the values for the remaining columns.
Note To assign a sample to multiple groups, separate the groups with a comma.
The application uses only the first group listed for calculating the Maximum Fold
and Group Averages.
For information about Maximum Fold analysis in unknown screening analysis,
see Cross Sample Peak List Pane.
Tip Use the Fill Down command to make entering vial positions easier.
Note When you use the scroll bar at the bottom of the sample list, the following
columns remain fixed: Status, Filename, Sample Type, Groups, Qual Processing
(quantitation) or Blank Subtraction (target screening), Level, Sample ID, and Sample
Name, while the other columns scroll right and left.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 53
To import samples into the list
1. Click Import, .
The Sample Import Tool dialog box opens.
Use this dialog box to import a sample list from a CSV, an XML, or an SLD file.
2. Click Browse and select a CSV, an XML, or an SLD file with the sample definitions that
you want to import.
3. From the Imported Samples Will Be list, select either Appended to the End of the List
or Inserted at the Selected Row.
4. Click Import.
The Sample Import Tool dialog box closes, and the application adds the specified samples
to the sample list.
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following column name substitutions.
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following sample type substitutions.
For each imported sample, the application uses the Instrument Method specified in the
local method.
Note The .csv, .xml, or .sld data format must match the TraceFinder data format.
Xcalibur column TraceFinder column
Position Vial position
Inj Vol Injection volume
Dil Factor Conversion Factor
Xcalibur sample type TraceFinder sample type
Blank Matrix Blank
QC QC Std
Std Bracket Cal Std
3 Using the Acquisition Mode
Working with Batches
54 TraceFinder User Guide Thermo Scientific
5. For each Cal Std or QC Std sample, click the Level cell and select a level from the list.
The master method defines the sample levels. If there are no levels to select from the Level
list, ask a user with Method Development permission to edit the method and specify the
levels. Then return to the Acquisition mode, and begin the batch again. The application
does not save a batch when you leave the Acquisition mode.
If you have Method Development permission, follow the instructions in step 4 of the
procedure To add samples to the list.
For detailed instructions about defining calibration levels, refer to Chapter 4, “Using the
Method Development Mode for Quantitation Methods,” in the Trac eFi nd er L a b D ire c t or
User Guide.
6. Type a vial position in the Vial Position column for each sample.
7. Type a volume in the Injection Volume column for each sample.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
8. (Optional) Type or edit the values for the remaining columns.
9. (Optional) When using multiplexing, select a channel for each imported sample.
Imported samples default to Auto.
To remove samples from the list
1. Select the samples that you want to remove.
2. Right-click and choose Remove Selected Samples.
To reinject a sample from a previously acquired batch
1. In the sample list, select the sample to reinject.
2. Right-click and choose Reinject Selected Samples.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the same sample are numbered INJ002, INJ003, and so
forth.
The application copies all parameter values from the original sample.
Tip Use the Fill Down command to make entering vial positions easier.
Note When you use the scroll bar at the bottom of the sample list, the following
columns remain fixed: Status, Filename, Sample Type, Groups, Qual Processing
(quantitation) or Blank Subtraction (target screening), Level, Sample ID, and Sample
Name, while the other columns scroll right and left.
Tip Use the CTRL or SHIFT keys to select multiple samples.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 55
A green status icon indicates previously acquired samples (acquired and processed), and
the sample name is grayed out. A blue status icon indicates samples created for reinjection
(not acquired).
When you submit this batch, the application acquires only the reinjection samples.
To select channels for the batch
To disable a configured channel, clear the check box for the channel in the Multiplexing
Channels area at the bottom of the page.
By default, all configured channels are selected. The configured channels are determined
by the multiplexing settings in the Configuration console. Refer to Chapter 1, “Using the
Configuration Console,” in the TraceFinder Lab Director User Guide.
Clearing a channel in the Multiplexing Channels area does not remove this channel
selection from the Channels list for each sample. When you assign a channel to a sample,
be careful not to assign a channel that is not available.
To assign a specific channel to a sample
1. Scroll to the Channel column.
All samples default to Auto.
2. Select a channel from the Channel list.
Note These features are available only when you have activated multiplexing in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in
the TraceFinder Lab Director User Guide.
Note The Channel column is available only when you have activated multiplexing in
the Configuration console. Refer to Chapter 1, “Using the Configuration Console,”
in the TraceFinder Lab Director User Guide.
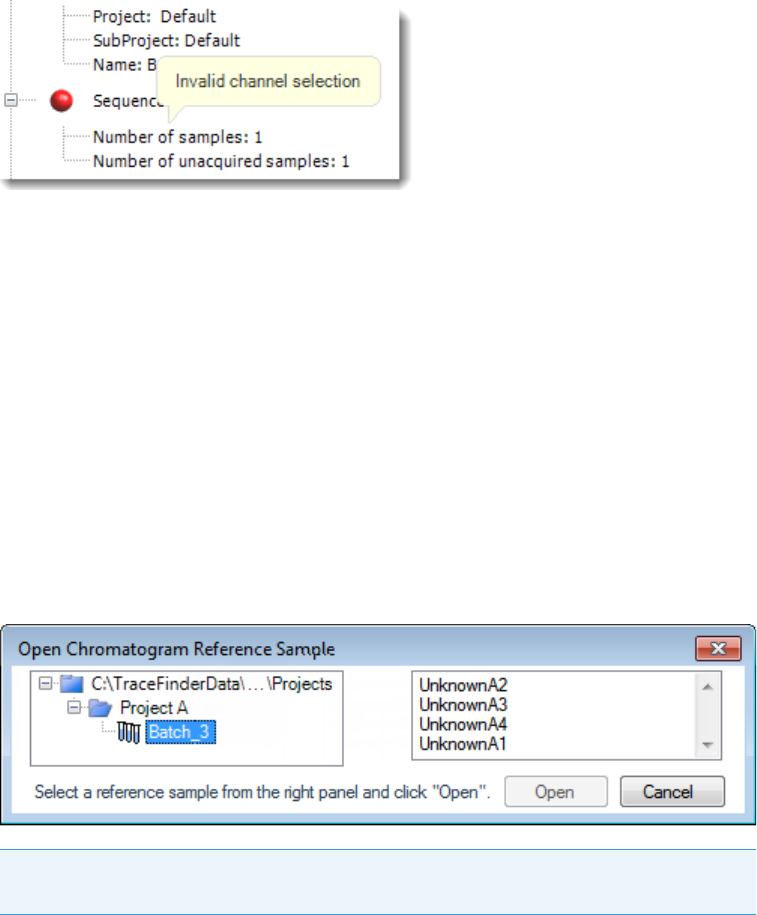
3 Using the Acquisition Mode
Working with Batches
56 TraceFinder User Guide Thermo Scientific
When you submit the batch, samples that are set to Auto run on any of the available
channels and samples that are set to a specific channel run only on that channel.
If you select a channel that is not available for this batch, the application flags the sample
sequence on the Finish page of the Acquisition mode. See the previous procedure, To
select channels for the batch.
3. If you see this error, do the following:
a. Click Previous to return to the Sample Definition page.
The incorrect sample is marked with an error flag.
b. Correct the channel selection.
To select a reference sample
1. Click the Reference Sample tab.
The Reference Sample page on the Sample Definition page opens.
You can select one reference sample to use as a reference peak in the Data Review.
2. Right-click the Reference Sample page and choose Add Reference Sample.
The Open Chromatograph Reference Sample dialog box opens.
3. Select a batch from the list.
The application displays only batches that were created using the current master method.
Note If you are using a new method, you will not see any samples here. You must
create and save a batch using the current method to see available samples in this list.

3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 57
4. Select a sample from the list of processed samples on the right.
The application displays all the processed samples in the selected batch. To use a sample
as a reference sample, the sample must have been processed with the current master
method.
5. Click Open.
The application adds the reference sample to the Reference Sample page.
6. (Optional) Enter values for Sample ID, Sample Name, Comment, and Barcode Actual.
7. (Optional) Change the Vial Position for the sample.
The application uses the peak in this sample as a reference peak in Data Review. See Reference
Peak.
To add an auto sample type
1. Click the Auto Samples tab.
The Auto Samples page opens.
2. Right-click and choose Add Auto Sample, or click the Add New Auto Sample icon,
.
The application adds a Solvent sample to the sample list.
You can add, insert, or remove samples from this list as you would any sample list.
3. To change the sample type to a Matrix Blank, click the Sample Type column and select
Matrix Blank from the list.
4. In the Injection Volume column for the sample, type a volume.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
5. In the Number of Injections column, type the number of injections available in the
designated Solvent or Matrix Blank vial.
After auto sample injections have occurred, you can return to this page to view the
number of Injections Used in each vial.
6. In the Vial Position column, type the vial position for the Solvent or Matrix Blank
sample.
3 Using the Acquisition Mode
Working with Batches
58 TraceFinder User Guide Thermo Scientific
To specify different instrument methods for samples
1. Display the Instrument Method column in the sample list:
a. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
b. In the Available Columns pane, select Instrument Method.
c. Click to move the Instrument Method column to the Displayed Columns
pane.
d. Click OK.
The application displays the Instrument Method column, defaulting to the instrument
method specified in the master method.
2. Click the Instrument Method column and select an instrument method from the list.
This list contains all the available instrument methods. The application prefixes
instrument methods from external sources with “Ext:”.
You can specify a different instrument method for each sample.
Figure 7. Instrument method column
When you submit the batch, the application saves a copy of the selected instrument
methods to the following folders:
External instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method\ExternalMethods
Local instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method
Note By default, the Instrument Method column is not displayed on the Sample
Definition page. See Instrument method column.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 59
Samples
Use the features on the Samples page to create a list of samples for the batch.
Figure 8. Samples page on the Sample Definition page
Table 5. Samples page parameters (Sheet 1 of 2)
Parameter Definition
Previous Returns you to the previous Acquisition page.
Next Takes you to the next Acquisition page.
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Sample Controls
Add Adds the specified number of empty rows to the sample grid.
Insert Inserts the specified number of empty rows above the selected row.
Import Opens the Sample Import Tool to import samples from a CSV, an XML, or an SLD file.
Multiplexing Channels
These features are available only when you have activated multiplexing in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in the
TraceFinder Lab Director User Guide.
All Channels Uses all configured channels to acquire this batch.
Channel 1-nUses only the selected channels to acquire this batch.
Shortcut menu commands
Add Sample Adds a single empty row to the sample grid.
Insert Sample Inserts a single empty row to the sample grid above the selected row.
Insert Copy Sample Copies the currently selected row and inserts a copy above the row.
Reinject Selected
Samples
Creates a copy of the selected sample and appends INJ001 to the file name. Additional
reinjections of the same sample are numbered INJ002, INJ003, and so forth.
3 Using the Acquisition Mode
Working with Batches
60 TraceFinder User Guide Thermo Scientific
Remove Selected
Samples
Removes selected samples from the sample grid.
Import Samples Opens the Sample Import Tool. Follow the instructions To import samples into the list.
Copy Down Copies the value in the selected row to all rows below it. For detailed instructions about
using the Copy Down command, see Appendix B, “Using Copy Down and Fill Down.”
Fill Down Enters sequential values in the column starting with the value in the selected row and
ending with the last row in the column. For detailed instructions about using the Fill
Down command, see Appendix B, “Using Copy Down and Fill Down.”
Modify Columns Opens the Modify Columns dialog box. See Column Display.
Enable/Disable Sample
Weight Calculation
Displays or hides the Sample Volume, Dilution Factor, Sample Weight, Calculation Type,
and Final Units columns.
Copy Copies the data in the selected rows or columns to the Clipboard. Use this command to
copy sample information to a text editor or spreadsheet application. You cannot paste this
data back into the Acquisition mode sample list.
Copy with Headers Copies the data in the selected rows or columns and the associated column headers to the
Clipboard. Use this command to copy sample information to a text editor or spreadsheet
application. You cannot paste this data back into the Acquisition mode sample list.
Paste Pastes a single column of copied data from a text editor or spreadsheet application, into the
selected column.
Undo Last Paste Removes the last pasted item in the Acquisition mode sample list.
Export to CSV File Opens the Save As dialog box where you can save the current sample list to a CSV file.
Edit Instrument Method Opens the Instrument Setup window where you can edit the parameters of the instrument
method.
• When you edit an external method, the application updates the method in the
…\Xcalibur\methods folder.
• When you edit an internal method, the application updates the method in the
…\TraceFinderData\4.0\Projects\project\subproject\batch\Methods\method folder.
For detailed information about editing instrument methods, refer to Chapter 3, “Using
Instrument Methods in the Method Development Mode,” in the TraceFi nde r Lab D irecto r
User Guide.
Table 5. Samples page parameters (Sheet 2 of 2)
Parameter Definition
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 61
Column Display
The Samples page contains many columns of information. You can scroll to see all the
columns, and you can customize which ones to display and their display order.
To scroll the sample list
Use the horizontal scroll bar at the bottom of the sample list to view all the information.
When you use the scroll bar at the bottom of the sample list, the following columns
remain fixed: Status, Filename, Sample Type, Groups, Qual Processing (quantitation) or
Blank Subtraction (target screening) Level, Sample ID, and Sample Name, while the
other columns scroll right and left.
To customize the column display
1. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
2. Use the arrow buttons to move all the columns that you want displayed to the Displayed
Columns pane.
These columns appear after the Status, Filename, Sample Type, Groups, Qual Processing
(quantitation) or Blank Subtraction (target screening), Level, Sample ID, and Sample
Name columns.
3. To arrange the order of the columns, do the following:
a. In the Displayed Columns pane, select a column name.
b. Use Up or Down to move the selected column up or down in the list.
The first column in the list represents the leftmost column in the Samples page
sample list, and the last column in the list represents the rightmost column in the
Samples page sample list.
4. To change the width of a column, do the following:
a. In the Displayed Columns pane, select the column width.
b. Type a new value for the width.
5. Repeat step 4 for all columns whose widths you want to change, and click OK.
The columns in the sample list immediately reflect your changes.
Note The following columns are fixed: Status, Filename, Sample Type, Groups,
Qual Processing (quantitation) or Blank Subtraction (target screening), Level,
Sample ID, and Sample Name.
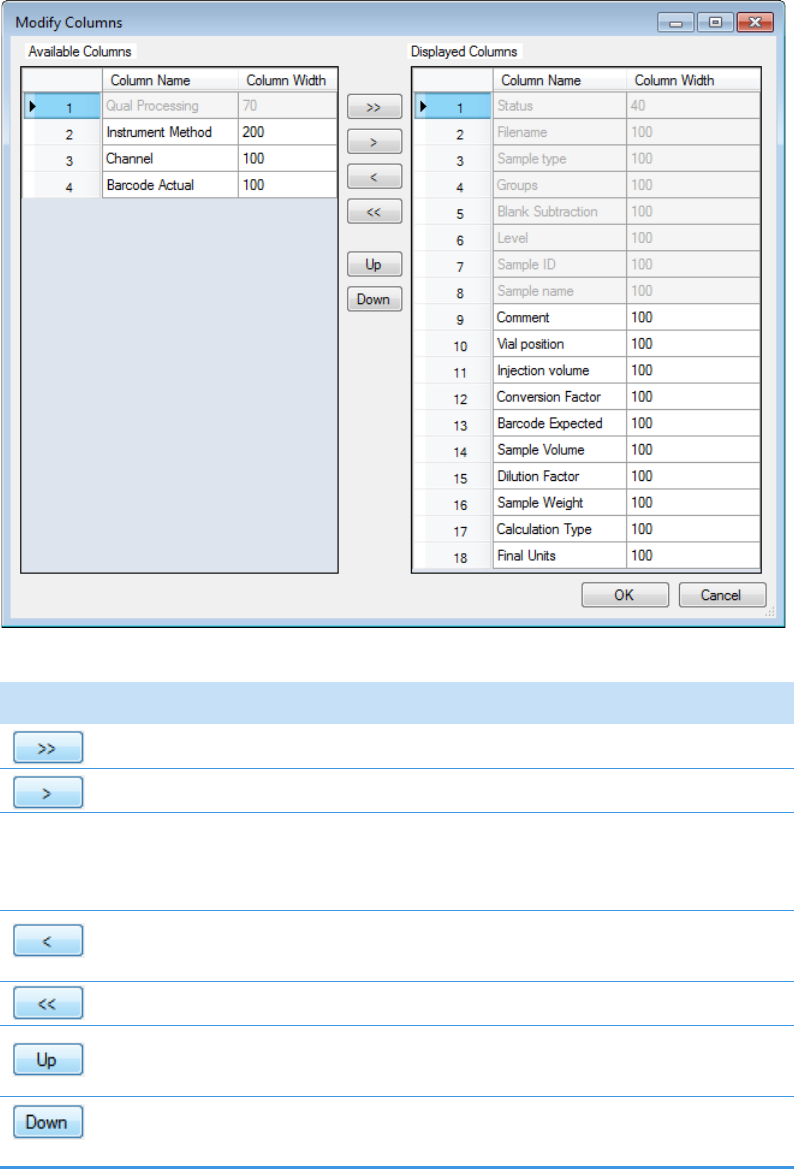
3 Using the Acquisition Mode
Working with Batches
62 TraceFinder User Guide Thermo Scientific
Figure 9. Modify Columns dialog box
Table 6. Button descriptions for the Modify Columns dialog box
Button Description
Moves all columns to the Displayed Columns pane.
Moves the selected column to the Displayed Columns pane.
The following buttons apply to all columns, except for those that are fixed: Status, Filename,
Sample Type, Groups, Qual Processing (quantitation) or Blank Subtraction (target
screening), Level, Sample ID, and Sample Name.
Moves the selected column to the Available Columns pane.
Moves all columns except fixed columns.
Moves the selected column name in the Displayed Columns pane one row
up in the column order.
Moves the selected column name in the Displayed Columns pane one row
down in the column order.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 63
Auto Samples
Use the features on the Auto Samples Sample page to identify the Solvent or Matrix Blank
samples to use for any Auto Sample or Auto Sample and Reinject failure actions as specified
on the Intelligent Sequencing page of the method. Refer to Chapter 4, “Using the Method
Development Mode for Quantitation Methods,” in the TraceFinder Lab Director User Guide.
Each sample type that you specify for a failure action on the Intelligent Sequencing page must
be defined in the samples list on the Auto Samples page.
Figure 10. Auto Samples page on the Sample Definition page
Table 7. Auto Samples page parameters
Column Description
Sample Type The sample type for the auto sample injection as specified on the Intelligent Sequencing page of
the method—either Solvent or Matrix Blank.
Default: Solvent
Injection Volume The injection volume used for the sample acquisition as specified on the Samples page.
Valid range: 0.1 through 5000 μL
Injections Used The number of times a vial has been used. The count is cumulative across all batches.
Number of
Injections
The number of injections available in the designated Solvent or Matrix Blank vial.
Vial Position Vial position for this sample type as specified on the Samples page.
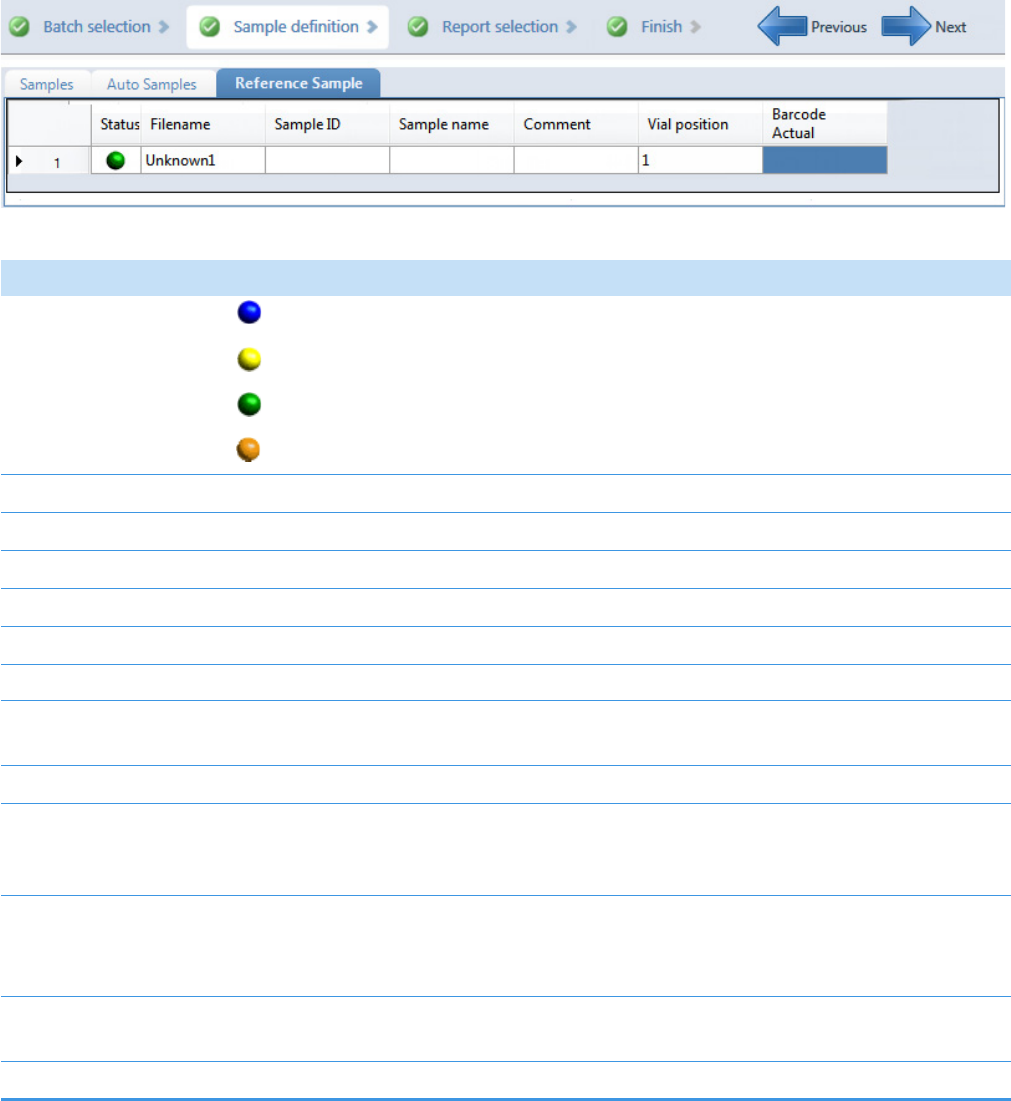
3 Using the Acquisition Mode
Working with Batches
64 TraceFinder User Guide Thermo Scientific
Reference Sample
Use the features on the Reference Sample page to select a sample to use as a reference peak in
Data Review.
Figure 11. Reference Sample page on the Sample Definition page
Table 8. Reference Sample page parameters
Parameter Description
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Filename Name of the raw data file that contains the sample data.
Sample ID A user-defined, alphanumeric string that identifies a sample.
Sample Name A user-defined name that identifies a sample.
Vial Position The tray vial number used for an autosampler acquisition.
Barcode Actual A user-entered barcode for the vial.
Shortcut menu commands
Add Reference Sample Opens the Open Chromatogram Reference Sample dialog box where you can select a
reference sample.
Delete Selected Deletes the reference sample.
Copy Copies the data in the selected rows or columns to the Clipboard. Use this command to
copy sample information to a text editor or spreadsheet application. You cannot paste this
data back into the reference sample list.
Copy with Headers Copies the data in the selected rows or columns and the associated column headers to the
Clipboard. Use this command to copy sample information to a text editor or spreadsheet
application. You cannot paste this data back into the reference sample list.
Paste Pastes a single column of copied data from a text editor or spreadsheet application, into the
selected column.
Export to CSV File Opens the Save As dialog box where you can save the current sample list to a CSV file.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 65
Selecting and Reviewing Reports
On the Report Selection page, you can specify the types of reports that you want to create. See
Report Selection. In addition to the report type, you can specify a report description for each
of your reports.
For each report that you generate, you can create a hard-copy printout, a PDF file, a CSV file,
or an Excel file.
Use any of the following procedures to create a reports list. When you finish specifying your
report options, click Next to go to the Finish page and submit your batch. See Submitting a
Batch in the Acquisition Wizard.
The application writes the resulting output files for your reports to the following folder:
…\TraceFinderData\4.0\Projects\…\batch\Reports
Follow these procedures:
•To edit a report title
•To specify a report in print format or as a PDF, a CSV, or an Excel file
To edit a report title
Select the Report Title column and edit the default title.
The default report title is the same as the report name.
To specify a report in print format or as a PDF, a CSV, or an Excel file
1. For each type of report that you want to create, select the corresponding check box in the
Print, Create PDF, Create CSV, or Create Excel column.
2. To duplicate the output type for all reports, right-click the cell and choose Copy Down.
All check boxes in the column below the selected cell duplicate the selected or cleared
state in the selected cell. This action applies only to report types that make this output
format available.
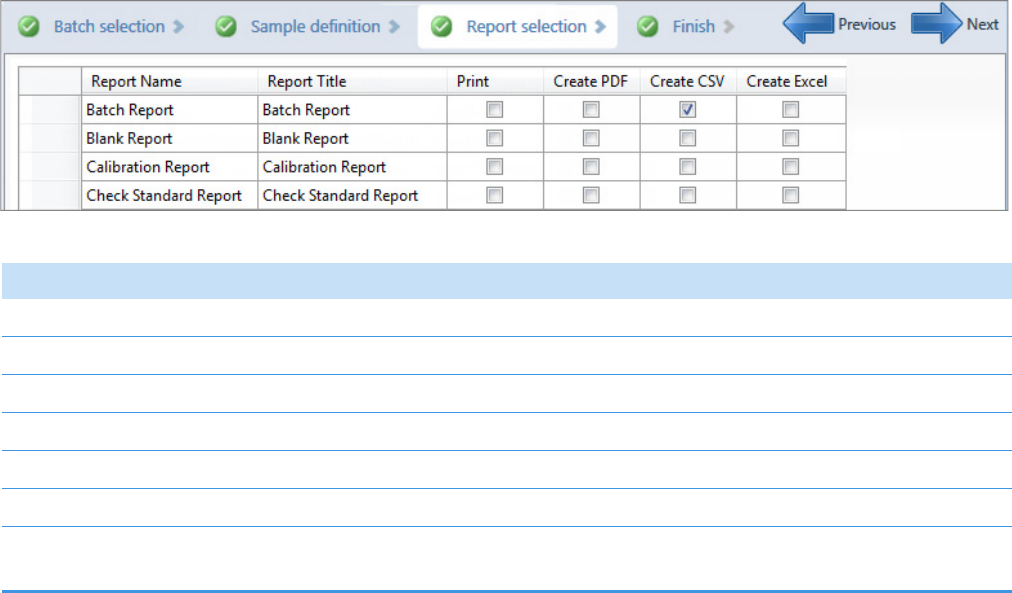
3 Using the Acquisition Mode
Working with Batches
66 TraceFinder User Guide Thermo Scientific
Report Selection
Use the features on the Report Selection page to specify the types of reports that you want to
create.
Figure 12. Report Selection page
Table 9. Report Selection page parameters
Parameter Description
Report Name The name of a report.
Report Title User-editable description to be used on a report.
Print Reports to be sent to the printer.
Create PDF Reports to be saved as PDF files.
Create CSV Reports to be exported as CSV files.
Create Excel Reports to be exported as Excel files.
Shortcut menu:
Copy Down
Copies the selected or cleared state to all subsequent reports in the column.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 67
Submitting a Batch in the Acquisition Wizard
On the Finish page of the Acquisition mode, you can specify a startup method, a shutdown
method, or a calibration batch. You can save the batch to be acquired later, or you can acquire
and process data and optionally create reports.
Follow these procedures:
•To specify startup or shutdown methods
•To automatically update the timed SRM information
•To specify a calibration batch
•To specify device states
•To save a batch for later acquisition or processing
•To start acquisition or processing
•To view the output files
To specify startup or shutdown methods
1. Select a method from the System Startup Method list.
The application runs this method before running the batch. No autosampler injection
takes place. This feature is not available for all instruments.
2. Select a method from the System Shutdown Method list.
The application runs this method after running the batch. This feature is not available for
all instruments.
To automatically update the timed SRM information
Select the Auto TSRM Update check box.
When you submit the batch, the application updates the TSQ method with mass
transitions, collision energy, and other appropriate data for TSRM functionality.
Note If you are working with a batch template, the only available function is Save.
3 Using the Acquisition Mode
Working with Batches
68 TraceFinder User Guide Thermo Scientific
To specify a calibration batch
1. In the Calibration area, select a calibration (.calx) file from the list.
2. To add calibration data from the current batch to the selected calibration file, select the
Extend Calibration option.
To specify device states
In the System Status area, select the name of the device, right-click, and then choose a
device state.
Note You must acquire at least one batch with the current method to create a
calibration (.calx) file.
Table 10. Instrument states (Sheet 1 of 2)
Instrument state Description
Turn Device On Keeps the system in the On state when the current run finishes,
so you can begin another run without waiting. All power and
flows are maintained at operational levels.
Default: On
Turn Device Standby Keeps the system in the Standby state when the current run
finishes, so you can begin another run with only a short delay
between runs.
Some devices do not have a Standby feature. For devices with
this feature, the device enters a power-saving or
consumable-saving mode, and you can switch the device back
on in approximately 15 minutes. Depending on the
instrument, this state turns liquid flows off but maintains
heaters and other subsystems in an On state so that there is no
warm-up time required when you change from Standby to On.
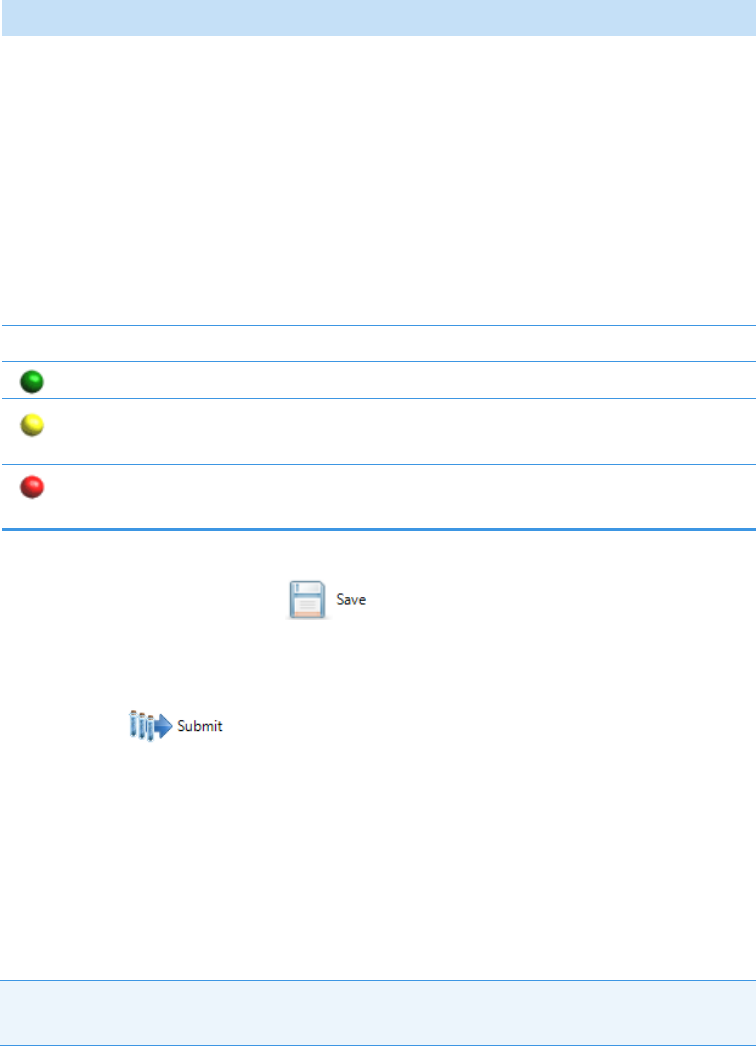
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 69
To save a batch for later acquisition or processing
From the Finish page, click Save, .
The application saves your batch as a prepared file.
To start acquisition or processing
1. Click Submit, .
The Submit Options dialog box opens.
2. To acquire (or reacquire) the submitted samples, select the Acquire Data check box.
• When all submitted samples have been previously acquired, this option is (by default)
not selected.
• When one or more submitted samples have not been acquired, this option is (by
default) selected.
3. To process the submitted samples, select the Process Data check box.
The application displays options for each type of method that the batch uses:
Quantitation, Target Screening, Unknown Screening, or a combination of methods.
Turn Device Off Keeps the system in the Off state when the current run
finishes. The Off state indicates that all power to the
instrument, which the application can control, is turned off.
This includes power to all heaters and subassemblies, but in
some cases not all subassemblies.
Some devices do not have an Off feature. For devices that do
have this feature, the device enters a power-saving or
consumable-saving mode, and you can switch the device back
on. When several runs are queued, the application uses the
system power scheme of the last submitted run.
Instrument status indicators
Green indicates that the device is turned on or is running.
Yellow indicates that the device is in standby mode or is
waiting for contact closure.
Red indicates that the device is turned off or that there is an
error with the device.
Tip You can process the data with or without performing peak detection. For
example, you might want to turn off peak detection when reprocessing samples.
Table 10. Instrument states (Sheet 2 of 2)
Instrument state Description

3 Using the Acquisition Mode
Working with Batches
70 TraceFinder User Guide Thermo Scientific
4. For each method type, select the check box for the options that you want to use.
Peak Detect: Performs peak detection for all method types. You can process the data with
or without performing peak detection. For example, you might want to turn off peak
detection when reprocessing samples.
Quantitate: Performs quantitation for quantitation methods.
Identify: Performs identification for unknown screening methods.
Identify and Confirm: Performs both identification and confirmation for target
screening methods.
With RT Alignment: Performs retention time alignment for unknown screening
methods. This produces the heat map and group averages data in the Unknown Screening
View. When you select this option, the application automatically selects the Peak Detect
option.
5. (Optional) Select the Create Reports check box.
6. (Optional with multiplexing activated) Select the Priority Sequence check box.
The application acquires the priority batch on the next available channel or the assigned
channel.
7. (Optional without multiplexing activated) Select the Priority Sequence check box and
then select one of the following priority options to place the batch in the queue:
•Next Available Batch places the batch immediately after the currently acquiring
batch.
•Next Available Sample places the batch immediately after the currently acquiring
sample.
8. To specify the following optional parameters, click Show Details.
a. Select the Use check box for the device that you want to use for this acquisition.
b. Select the Start Device check box to indicate the device that will initiate
communication with the other instruments.
This is usually the autosampler.
c. Select the Start When Ready check box, which starts all instruments together when
they are all ready.
When you clear this check box, individual instruments can start at different times
and then must wait for the last instrument to be ready.
d. Select the system state after it acquires the last batch: On, Standby, or Off.
Note When you select Full Sequence Submission in the Configuration console, these
options are unavailable because the current batch and the current sample are, in effect,
the same thing.

3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 71
9. To start the selected processes, click OK.
The selected processes begin, and the application shows the real-time display at the
bottom of the current window. You can begin another batch in the Acquisition mode
while you watch the real-time display of the currently acquiring batch.
IMPORTANT When your batch uses unknown screening features, after the first
processing, you might see an error message stating that the total number of results is too
large for a single batch (more than 500 000). Return to the Processing pages for the
unknown screening features (refer to Chapter 6, “Using the Method Development Mode
for Unknown Screening Methods,” in the TraceFinder Lab Director User Guide) and make
one or more of these parameter adjustments:
• Limit the RT range.
• Shorten the signal range.
• Specify a lower value for the Number of Top Matches.
• Specify a Simple Search instead of an Exhaustive Search.
• Specify Top Peaks instead of All Peaks.
• Limit the number of search types.
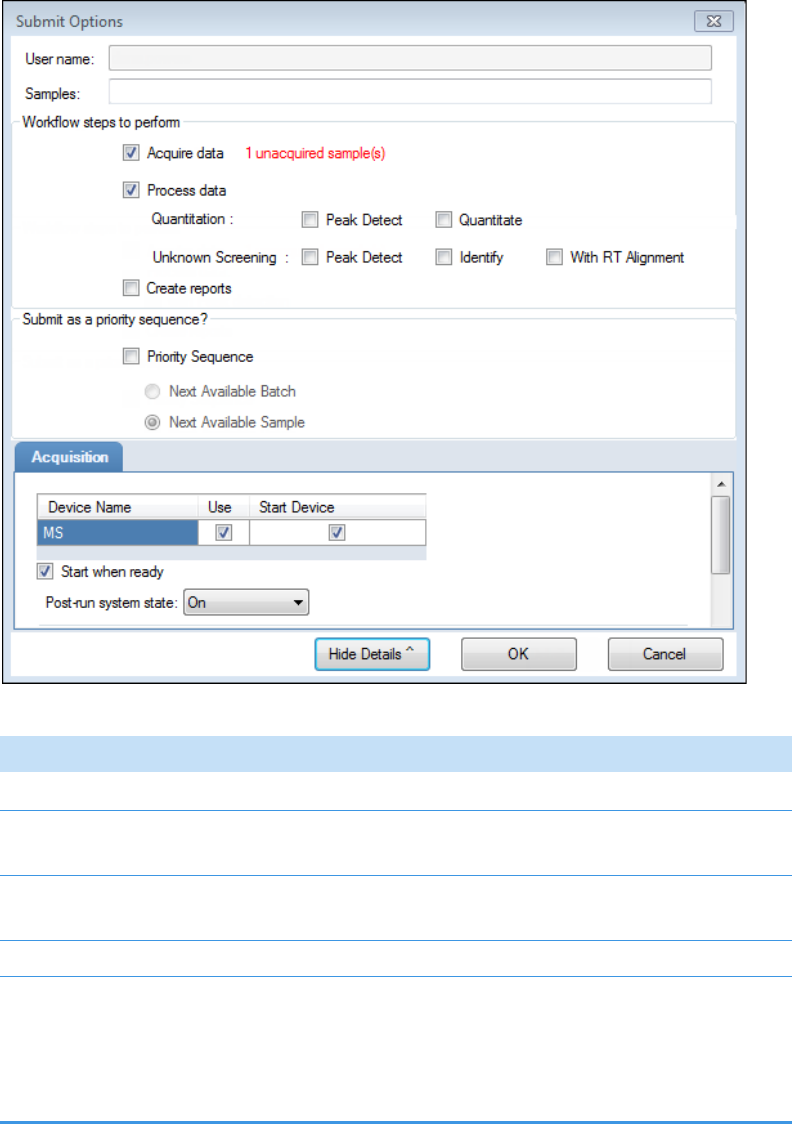
3 Using the Acquisition Mode
Working with Batches
72 TraceFinder User Guide Thermo Scientific
Figure 13. Submit Options dialog box
Table 11. Submit Options dialog box parameters (Sheet 1 of 3)
Parameter Description
User Name Name of the current user.
Samples Number of samples to be submitted for acquisition, upload,
processing, or reporting.
Method Types Lists the types of methods (quantitation, target screening, or unknown
screening) used in the submitted batches.
Workflow Steps to Perform
Acquire Data Submits the current batch to acquisition.
• When all submitted samples have been previously acquired, this
option is (by default) not selected.
• When one or more samples in the batch have not been acquired,
this option is (by default) selected.
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 73
Process Data Processes the data for the current batch using any of the following
options:
Peak Detect: Performs peak detection for all method types. You can
process the data with or without performing peak detection. For
example, you might want to turn off peak detection when reprocessing
samples.
Quantitate: Performs quantitation for quantitation methods.
Identify: Performs identification for unknown screening methods.
Identify and Confirm: Performs both identification and confirmation
for target screening methods.
With RT Alignment: Performs retention time alignment for unknown
screening methods. This produces the heat map and group averages
data in the Unknown Screening View. When you select this option, the
application automatically selects the Peak Detect option.
Create Reports Creates reports for the current batch.
Submit as a Priority Sequence?
Priority Sequence With multiplexing activated, places the batch immediately after the
currently acquiring batch.
Without multiplexing activated, specifies one of the following priority
options to place the batch in the queue:
Next Available Batch: Places the batch immediately after the currently
acquiring batch.
Next Available Sample: Places the batch immediately after the
currently acquiring sample.
Note When you select Full Sequence Submission in the
Configuration console, these options are unavailable because the
current batch and the current sample are, in effect, the same thing.
Acquisition pane
Device Name Lists all configured instruments.
If the instrument that you want to use is not configured, close the
TraceFinder application, configure the instrument, and then reopen
the application. You cannot configure an instrument while the
TraceFinder application is running.
Available only when you select the Acquire Data check box.
Table 11. Submit Options dialog box parameters (Sheet 2 of 3)
Parameter Description
3 Using the Acquisition Mode
Working with Batches
74 TraceFinder User Guide Thermo Scientific
To view the output files
Locate the files to view from the following directories:
The application writes saved batches to the project folder:
…\TraceFinderData\4.0\Projects\…
For each acquired sample, the application writes an RSX file to the batch Data folder:
…\TraceFinderData\4.0\Projects\…\Data
The application saves method information to the batch Methods folder:
…\TraceFinderData\4.0\Projects\…\Methods
The application writes the reports to the batch Reports folder:
…\TraceFinderData\4.0\Projects\…\batch\Reports
Use Specifies the instruments used for this acquisition.
Available only when you select the Acquire Data check box.
Start Device Specifies the instrument that initiates the communication with the
other instruments. This is usually the autosampler.
Available only when you select the Acquire Data check box.
Start When Ready Starts the specified device when all the instruments are ready to acquire
data. When this is cleared, individual instruments can start at different
times and then must wait for the last instrument to be ready.
Post-run System
State
Specifies the system state after it acquires the last batch: On (default),
Standby, or Off.
Buttons
Hide/Show
Details
Collapses or expands the acquisition details of the Submit Options
dialog box.
OK Begins the selected processes.
Cancel Closes the Submit Options dialog box without submitting any tasks.
Table 11. Submit Options dialog box parameters (Sheet 3 of 3)
Parameter Description
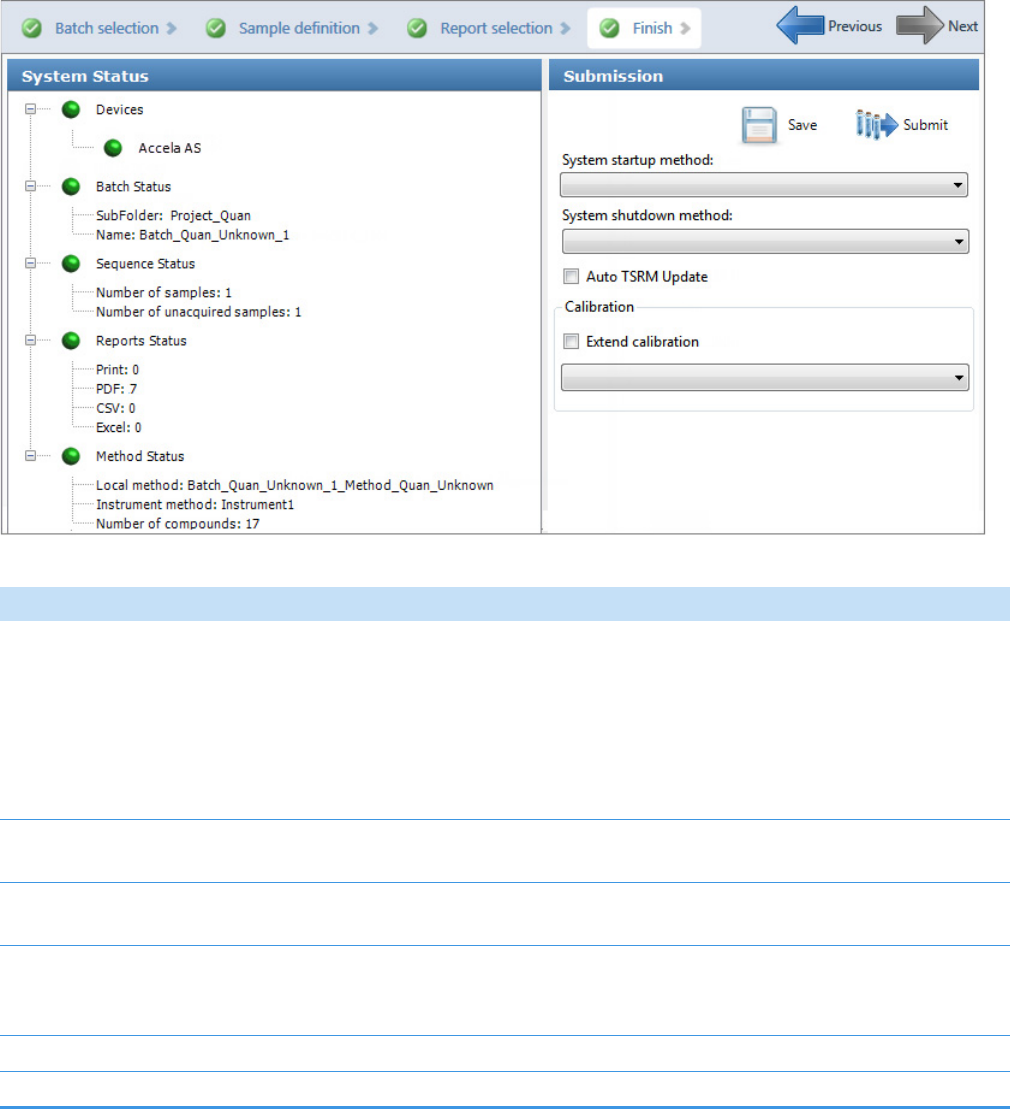
3 Using the Acquisition Mode
Working with Batches
Thermo Scientific TraceFinder User Guide 75
Finish
Use the features on the Finish page to save the batch to be acquired later or acquire and
process data and optionally create reports.
Figure 14. Finish page
Table 12. Finish page parameters
Parameter Description
System Status The System Status pane displays the following:
• Devices used for the acquisition
• Project, subproject, and name of the batch
• Number of acquired and unacquired samples in the batch
• Number of reports to be printed and saved as PDF, CSV, or Excel files
• Local method and instrument method used for the batch
• Number of compounds in the method
System Startup Method
System Shutdown Method
The instrument methods that run before and after the batch. No autosampler injection
takes place. These features are not available for all instruments.
Auto TSRM Update Updates the TSQ method with mass transitions, collision energy, and other appropriate
data for TSRM functionality.
Calibration • Use calibration: Uses the selected calibration file to process the current data.
• Extend calibration: Adds calibration data from the current batch to the selected
calibration file.
Save Saves the current batch as a prepared batch.
Submit Opens the Submit Options dialog box.
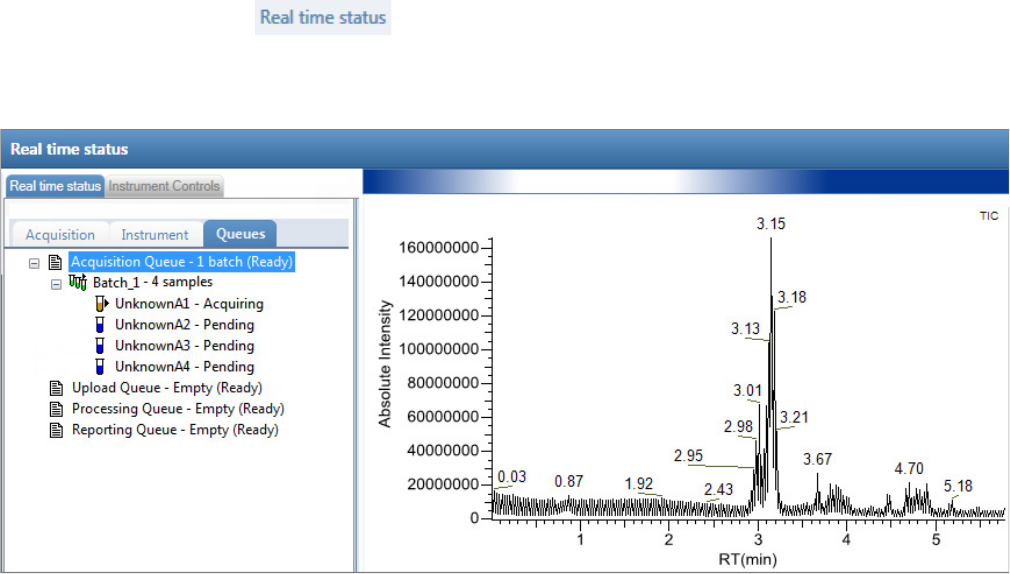
3 Using the Acquisition Mode
Real Time Status Pane
76 TraceFinder User Guide Thermo Scientific
Real Time Status Pane
You can access the Real Time Status pane from any mode in the application.
To access the Real Time Status Pane from any mode
Click Real Time Status in the upper right corner of the TraceFinder window.
The Real Time Status pane opens at the bottom of the current view.
Figure 15. Real Time Status pane
The Real Time Status pane has four pages of information and a real-time trace pane:
•Real Time Status – Acquisition Page
•Real Time Status – Instrument Page
•Real Time Status – Queues Page
•Instrument Controls Page
•Real-Time Trace Display
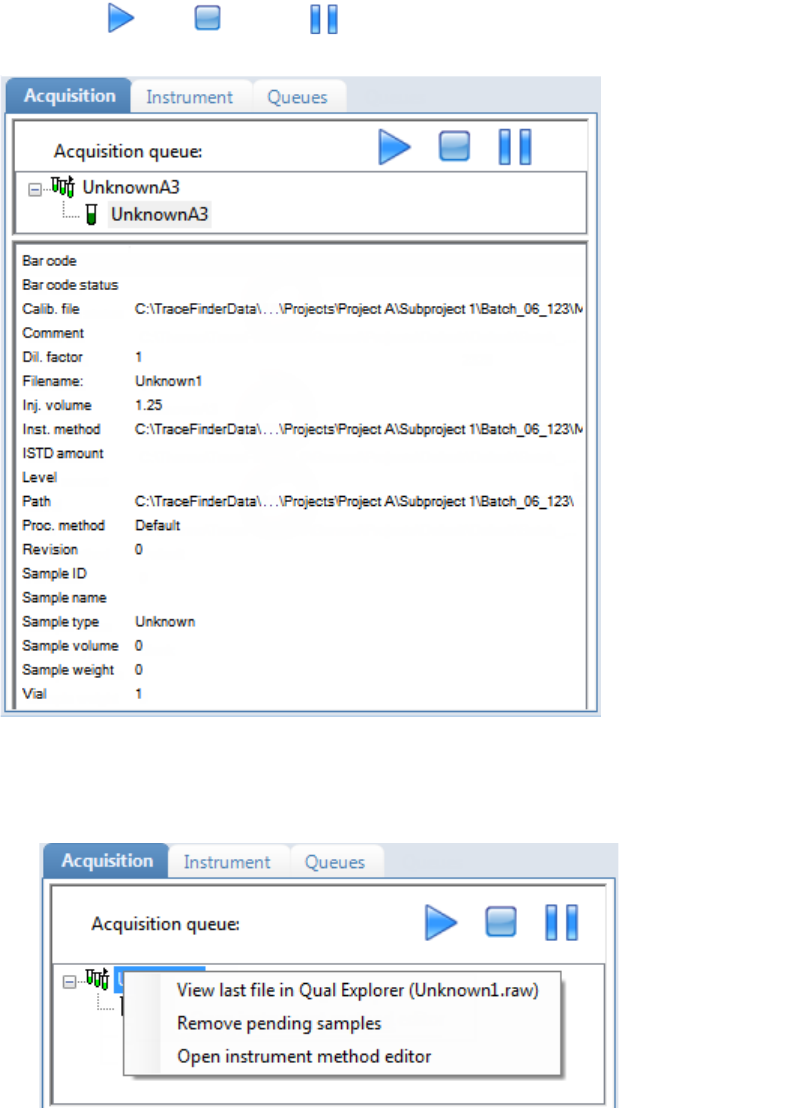
3 Using the Acquisition Mode
Real Time Status Pane
Thermo Scientific TraceFinder User Guide 77
Real Time Status – Acquisition Page
Use the Acquisition page to monitor the progress as the application acquires the samples.
To display the last acquired raw data file in a qualitative browser
On the Acquisition page of the real-time status pane, right-click and choose View Last
File in Qual Explorer.
The last acquired file opens in either the FreeStyle or Qual Browser application.
Use the Start, , Stop, , or Pause, , icons
to control batches in the Acquisition queue.
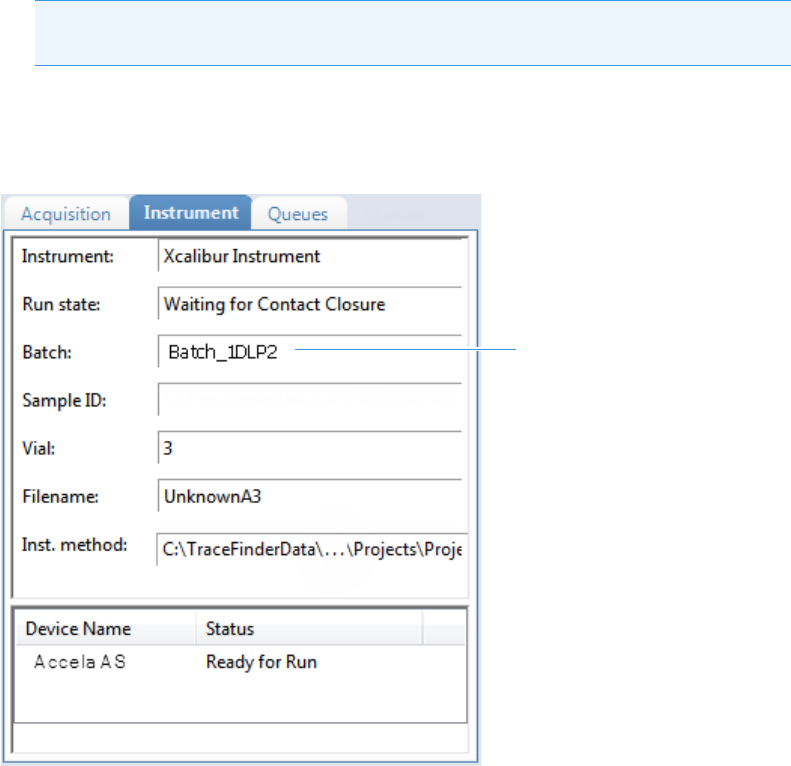
3 Using the Acquisition Mode
Real Time Status Pane
78 TraceFinder User Guide Thermo Scientific
To open the Instrument Setup window
Right-click anywhere in the Acquisition page and choose Open Instrument Method
Editor.
The Thermo Instrument Setup window opens, displaying the currently running
instrument method.
For detailed information about editing instrument methods, refer to Chapter 3, “Using
Instrument Methods in the Method Development Mode,” in the Trace Finde r La b Direct o r
User Guide.
Real Time Status – Instrument Page
Use the Instrument page to monitor the currently acquiring sample.
To view the last acquired file in a qualitative browser
Right-click anywhere in the top pane of the Instrument page and choose View Last File
in Qual Explorer.
The last acquired file opens in either the FreeStyle or Qual Browser application.
Note Changes you make and save to the instrument method do not affect the
currently running batch.
When you run single sample
submission, this displays the
sample name instead of the
batch name.
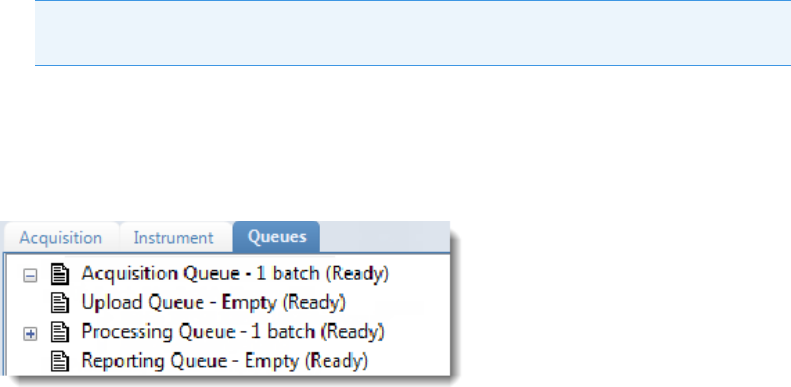
3 Using the Acquisition Mode
Real Time Status Pane
Thermo Scientific TraceFinder User Guide 79
To open the Instrument Setup window
Right-click anywhere in the top pane of the Instrument page and choose Open
Instrument Method Editor.
The Thermo Instrument Setup window opens, displaying the currently running
instrument method.
For detailed information about editing instrument methods, refer to Chapter 3, “Using
Instrument Methods in the Method Development Mode,” in the Trace Finde r La b Direct o r
User Guide.
Real Time Status – Queues Page
Use the Queues page to monitor and control the Acquisition, Upload, Processing, and
Reporting queues.
•Queue-Level Commands: Pause or remove batches in any of the queues.
•Batch-Level Commands: Pause or remove entire batches or samples within batches from
any of the queues.
•Additional Commands: Open the FreeStyle application or the Instrument Setup window.
Queue-Level Commands
Use the queue-level commands to pause or remove batches in any of the queues on the
Queues page. See Queue-Level Shortcut Menu.
Follow these procedures:
•To pause all batches in a queue
•To remove a single batch from a queue
•To remove all batches in a queue
•To re m ove a l l p e n d i n g b a t c h e s
Note Changes you make and save to the instrument method do not affect the
currently running batch.
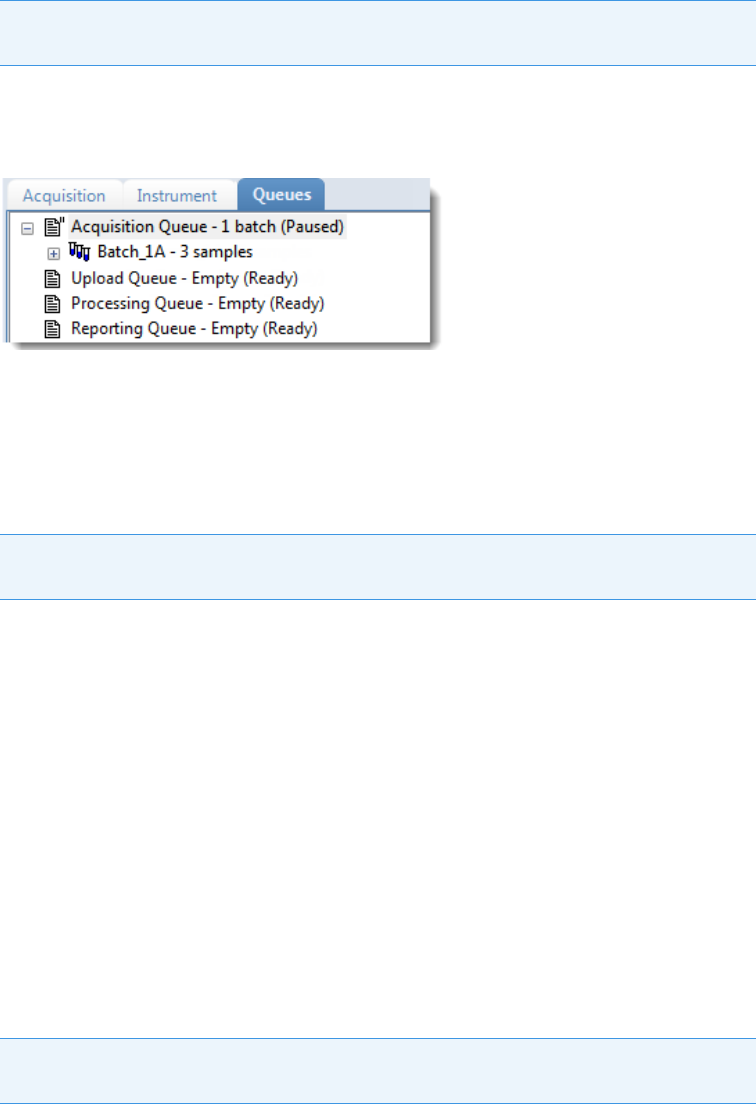
3 Using the Acquisition Mode
Real Time Status Pane
80 TraceFinder User Guide Thermo Scientific
To pause all batches in a queue
1. Select a queue (Acquisition, Upload, Processing, or Reporting).
2. Right-click and choose Pause Queue.
After the current sample is complete, the application pauses all batches and samples in the
specified queue. Only the selected queue is affected.
3. To restart a paused queue, select the queue, right-click, and choose Resume Queue.
To remove a single batch from a queue
1. Select a queue (Acquisition, Upload, Processing, or Reporting).
2. Right-click and choose Stop Active Batch.
The application confirms that you want to remove the active batch from the selected
queue. After the current sample is complete, the application removes the batch and all
pending samples from the queue. Only the selected queue is affected.
To remove all batches in a queue
1. Select a queue (Acquisition, Upload, Processing, or Reporting).
2. Right-click and choose Stop All Batches.
The application removes all batches with pending samples from the selected queue. The
current sample continues to acquire. Only the selected queue is affected.
To remove all pending batches
1. Select a queue (Acquisition, Upload, Processing, or Reporting).
2. Right-click and choose Remove Pending Batches.
The application removes all batches that contain only pending samples. Only the selected
queue is affected.
Note When multiplexing is activated, you can have as many as four samples acquiring
at once. Pausing the Acquisition queue does not affect any acquiring samples.
Note This command is available only when there are active batches in the queue.
Paused batches and batches that contain only pending samples are not “active.”
Note A pending batch is a batch in which all samples are pending. If any sample in
the batch is active, the batch is not affected by this command.
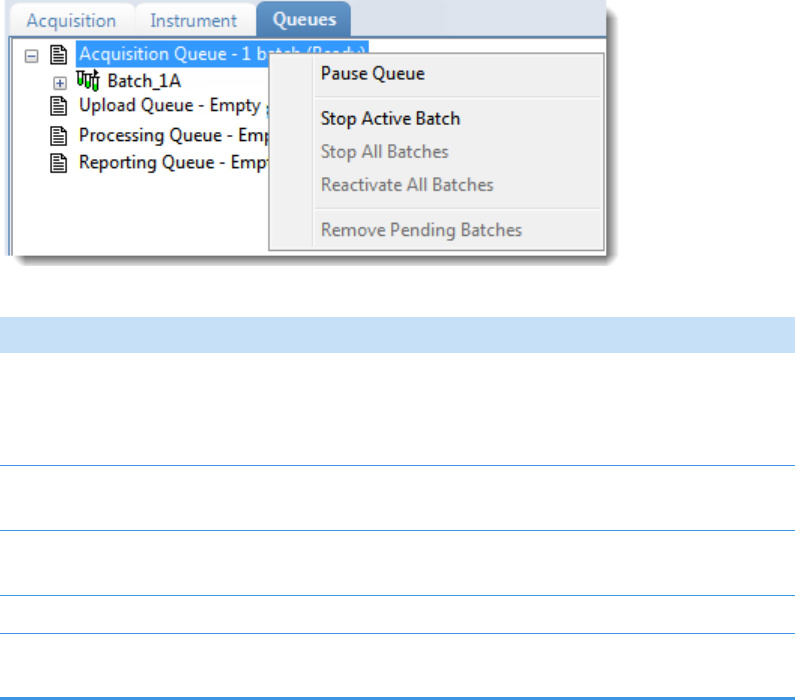
3 Using the Acquisition Mode
Real Time Status Pane
Thermo Scientific TraceFinder User Guide 81
Queue-Level Shortcut Menu
Use the commands in the shortcut menu to pause or remove batches in any of the queues on
the Queues page.
Figure 16. Queue-level shortcut menu
Batch-Level Commands
Use the batch-level commands to pause or remove entire batches or samples within batches
from any of the queues on the Queues page. See Batch-level shortcut menu.
Follow these procedures:
•To remove a single pending sample from a batch
•To remove all pending samples from a batch
•To s t op a b a t ch
•To re m ove a pe n d i n g ba t c h
Table 13. Queue-level shortcut menu commands
Command Description
Pause Queue
Resume Queue
After the current sample is complete, the application pauses the
specified queue. Only the selected queue is affected.
Returns the paused queue to active status.
Stop Active Batch Removes all pending samples from the specified queue. The active
sample is not affected.
Stop All Batches Removes all pending samples and batches from the specified
queue. The active sample is not affected.
Reactivate All Batches Returns all paused batches to active status.
Remove Pending
Batches
Removes all pending batches from the specified queue. The active
batch is not affected.
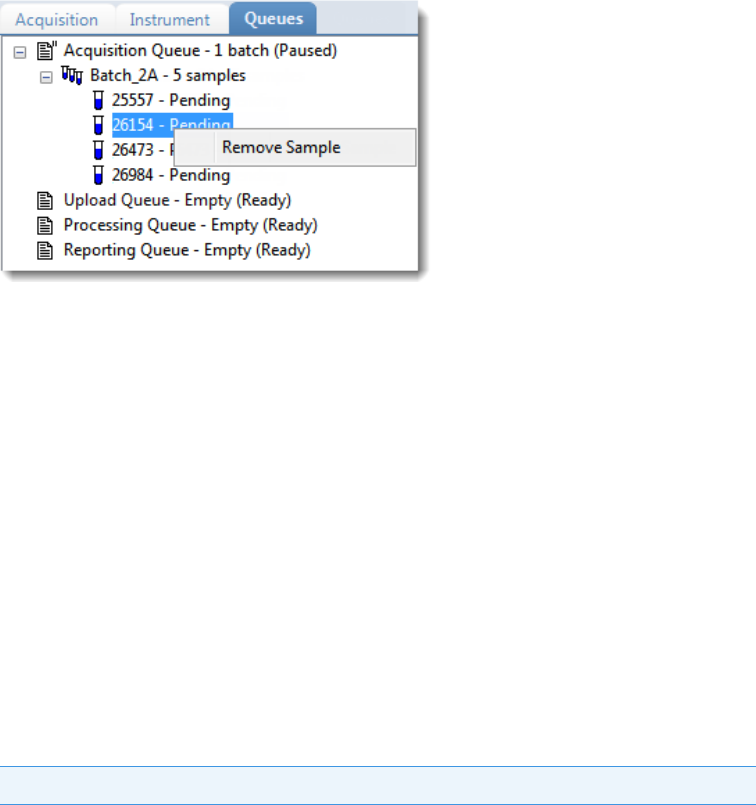
3 Using the Acquisition Mode
Real Time Status Pane
82 TraceFinder User Guide Thermo Scientific
To remove a single pending sample from a batch
1. Select a pending sample.
2. Right-click the sample and choose Remove Sample.
The application confirms that you want to remove the selected sample from the batch and
then removes the sample.
To remove all pending samples from a batch
1. Select a batch in any of the queues (Acquisition, Upload, Processing, or Reporting).
The batch must have at least one pending sample.
2. Right-click and choose Remove Pending Samples.
The application confirms that you want to remove all pending samples from the batch
and then removes the samples. If the batch includes only pending samples, the
application removes the batch from the queue.
To stop a batch
1. Select an active batch in any of the queues (Acquisition, Upload, Processing, or
Reporting).
2. Right-click and choose Stop Batch.
The application confirms that you want to remove the selected batch from the queue.
After the current sample is complete, the application removes the batch and all pending
samples from the queue.
To remove a pending batch
1. Select a pending batch in any of the queues (Acquisition, Upload, Processing, or
Reporting).
Note The batch must have at least one active sample and one pending sample.
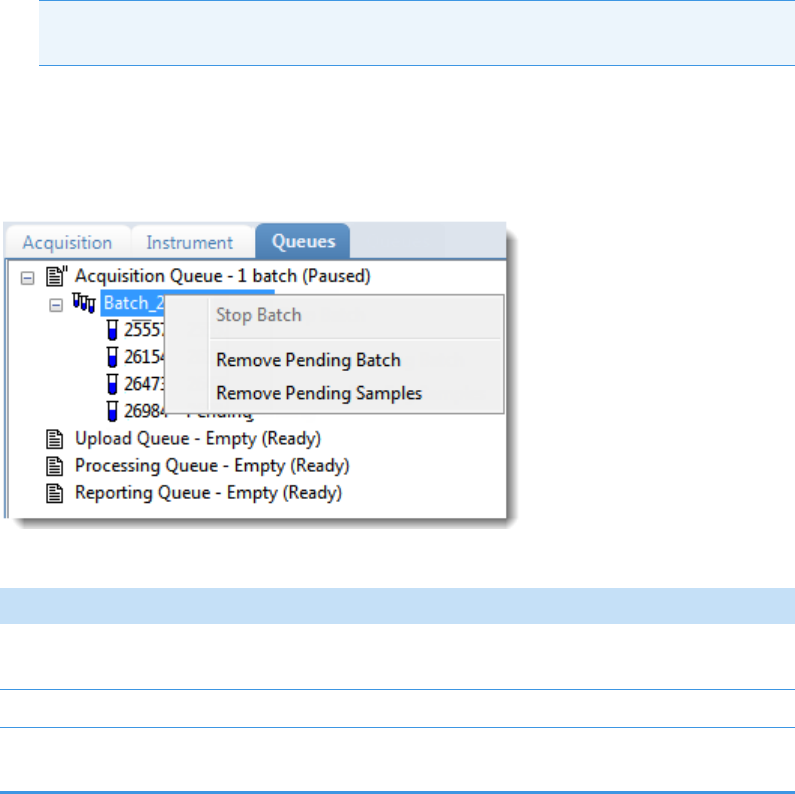
3 Using the Acquisition Mode
Real Time Status Pane
Thermo Scientific TraceFinder User Guide 83
2. Right-click and choose Remove Pending Batch.
The application confirms that you want to remove the selected batch from the queue and
then removes the batch from the queue.
Figure 17. Batch-level shortcut menu
Additional Commands
You can open the FreeStyle application or the Instrument Setup window using additional
commands that you access from the Queues page.
To access the shortcut menu with these commands, you must click in the white space below
the list of queues. If you click on the name of or to the right of the queues, the application
displays queue-, batch-, or sample-level shortcut menus.
Note A pending batch is a batch in which all samples are pending. If any sample in
the batch is active, this command is not available.
Table 14. Batch-level shortcut menu commands
Command Description
Stop Batch After the current sample is complete, the application removes all
samples in the selected batch.
Remove Pending Batch Removes all samples from the selected pending batch.
Remove Pending
Samples
Removes all pending samples from the selected batch.
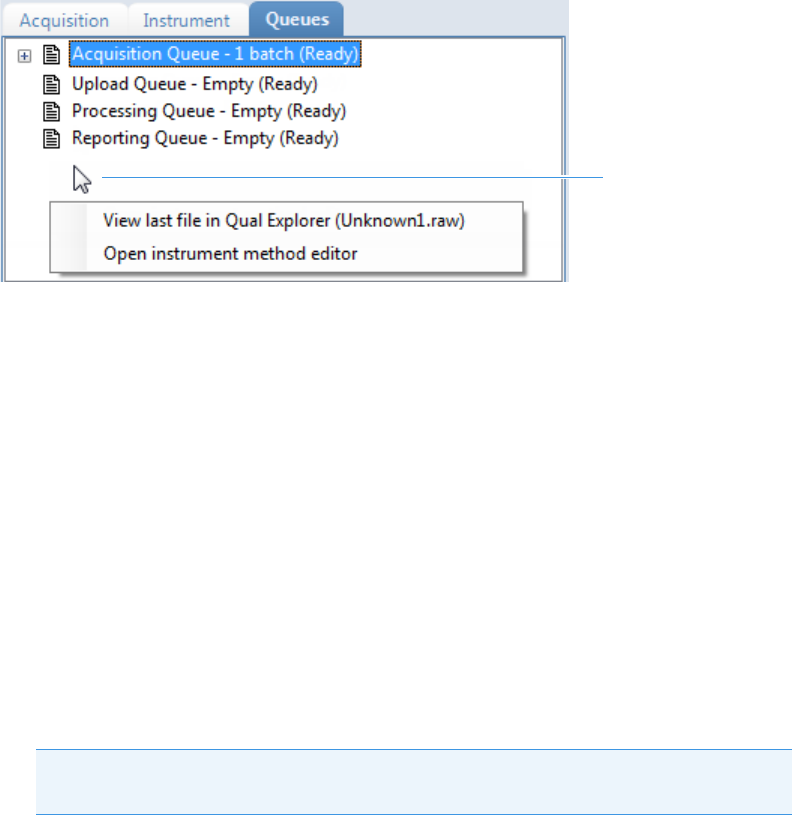
3 Using the Acquisition Mode
Real Time Status Pane
84 TraceFinder User Guide Thermo Scientific
To view the last acquired file in a qualitative browser
Right-click below the queues list on the Queues page and choose View Last File in Qual
Explorer.
The last acquired file opens in either the FreeStyle or Qual Browser application.
To open the Instrument Setup window
Right-click below the queues list on the Queues page and choose Open Instrument
Method Editor.
The Thermo Instrument Setup window opens, displaying the currently running
instrument method.
For detailed information about editing instrument methods, refer to Chapter 3, “Using
Instrument Methods in the Method Development Mode,” in the Trace Finde r La b Direct o r
User Guide.
Instrument Controls Page
Use the Instrument Controls page to monitor the status of the instrument. The feedback you
see on the Instrument Controls page depends on the instrument you are using. The following
examples show an Accela™ autosampler and an Aria™ multiplexing device.
Note Changes you make and save to the instrument method do not affect the
currently running batch.
Right-click below the
list of queues.
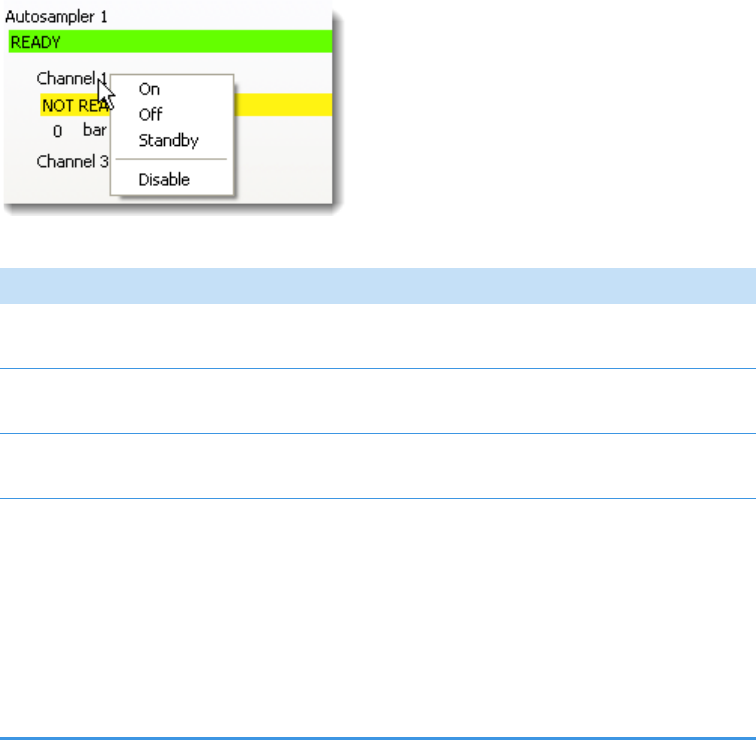
3 Using the Acquisition Mode
Real Time Status Pane
86 TraceFinder User Guide Thermo Scientific
•To view the pressure trace
•To access the Aria multiplexing controls
•To view the last acquired file in a qualitative browser
To pause the autosampler
1. Click Hold Autosampler.
The autosampler finishes the current autosampler step and then pauses. The autosampler
and LC pumps continue to run.
2. To restart the autosampler, click Hold Autosampler again.
To control the channels
Right-click the channel name and choose a command from the shortcut menu.
Table 15. Autosampler shortcut menu commands
Command Description
On Turns on a stopped pump and continues acquiring the sample
list assigned to that channel.
Off After the current sample is complete, the application stops
acquiring and the pump shuts down.
Standby After the current sample is complete, the application stops
acquiring. The pump continues to run.
Disable/Enable Disable: Prevents the channel from receiving samples. When
you choose Disable during a run, the application finishes the
current sample on the channel and then stops.
Enable: Allows the channel to receive samples.
When you disable a channel that is set to On, the channel is
highlighted in green and the status is READY. You can turn
the channel to Off or Standby.
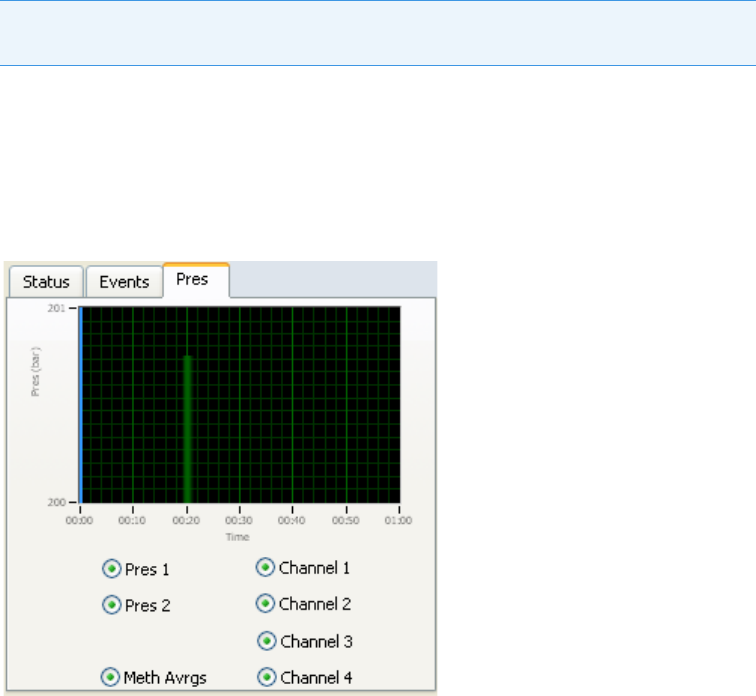
3 Using the Acquisition Mode
Real Time Status Pane
Thermo Scientific TraceFinder User Guide 87
To open the Instrument Setup window
Right-click in the header of the Instrument Controls page and choose Open Instrument
Method Editor.
The Thermo Instrument Setup window opens, displaying the currently running
instrument method.
For detailed information about editing instrument methods, refer to Chapter 3, “Using
Instrument Methods in the Method Development Mode,” in the Trace Finde r La b Direct o r
User Guide.
To view the pressure trace
1. Click the Pres tab.
The Pressure page displays a pump pressure graph for each sample in the batch. A
fluctuation or change in the pump pressure could indicate a change in the
chromatography conditions.
2. To view the pressure for a specific pump, select the Pres 1 or Pres 2 option.
By default, the pressures for all pumps are displayed.
3. To view the pressure for a specific channel, select the corresponding channel number.
By default, the pressures for all channels are displayed.
Note Changes you make and save to the instrument method do not affect the
currently running batch.
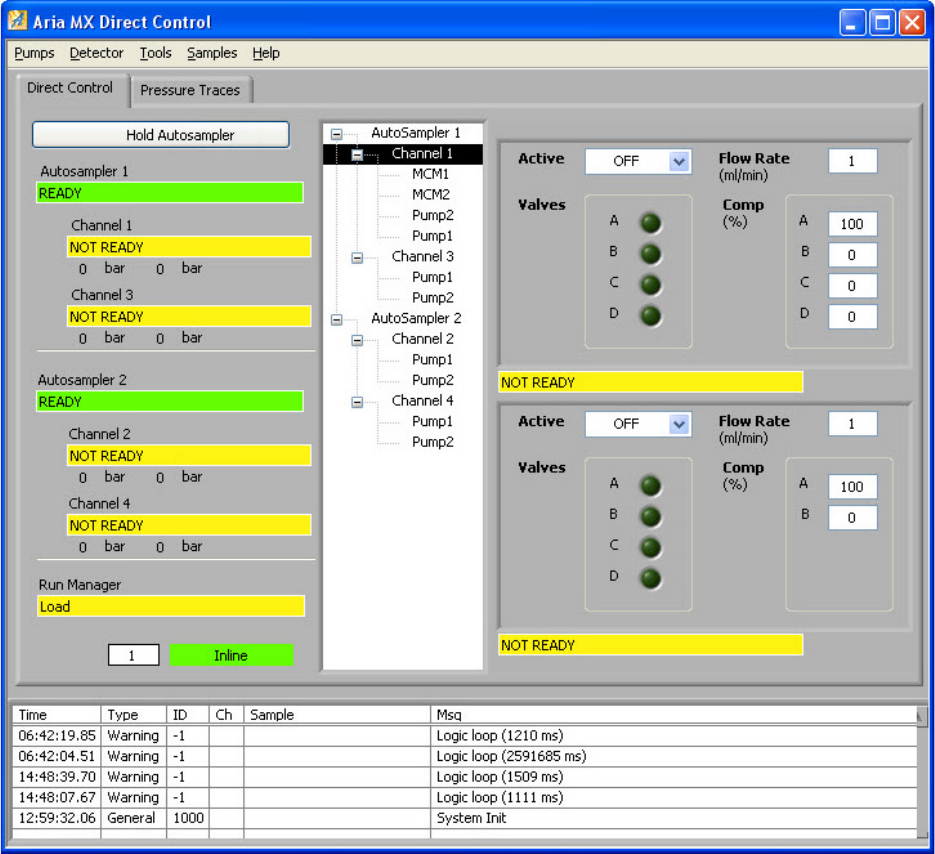
3 Using the Acquisition Mode
Real Time Status Pane
88 TraceFinder User Guide Thermo Scientific
To access the Aria multiplexing controls
Click Direct Control.
The Aria MX Direct Control window opens.
For detailed descriptions of the features in this window, refer to the Transcend Systems with
Xcalibur Software User Guide.
To view the last acquired file in a qualitative browser
Right-click in the header of the Instrument Controls page and choose View Last File in
Qual Explorer.
The last acquired file opens in either the FreeStyle or Qual Browser application.
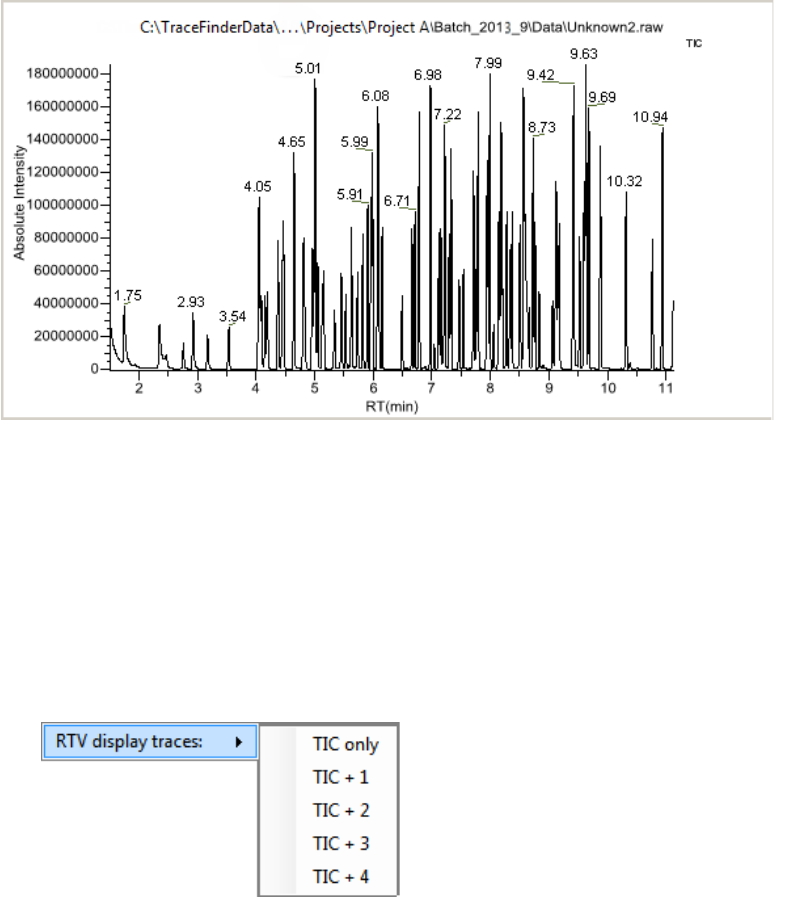
3 Using the Acquisition Mode
Real Time Status Pane
Thermo Scientific TraceFinder User Guide 89
Real-Time Trace Display
As each sample is acquired, the real-time chromatogram pane shows the retention time and
intensity of the TIC trace.
By default, the Real Time Status pane shows only the TIC trace as each sample is acquired. To
observe specific traces, such as the internal standard, use the RTV Display Traces function to
display multiple traces.
When you create your method, you can specify additional traces to display in the real-time
viewer and in which order the traces are displayed. The application always displays the TIC
trace in the top pane. See Real Time Status Pane.
To display multiple traces
Right-click the chromatogram pane and choose the number of traces to display.
The chromatogram pane displays real-time chromatograms for the selected number of
traces.
The TIC is always displayed at the top. When there are more traces than can fit in the
pane, you can scroll through the traces.
For each trace, the application displays the mass or precursor mass. See Real-time trace
display with multiple traces.
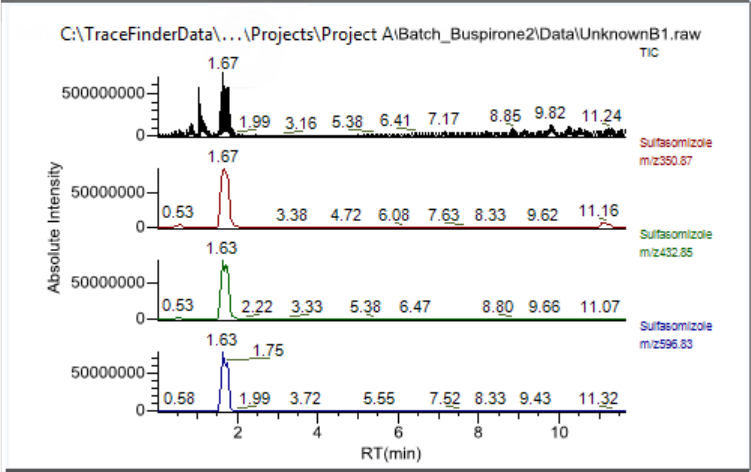
3 Using the Acquisition Mode
Real Time Status Pane
90 TraceFinder User Guide Thermo Scientific
Figure 18. Real-time trace display with multiple traces
3 Using the Acquisition Mode
Sample Types
Thermo Scientific TraceFinder User Guide 91
Sample Types
The TraceFinder application uses the following sample types in all sample definitions and
reports.
Table 16. Sample type definitions
Sample type Definition
Matrix Blank Contains no target compounds but might contain an ISTD when you
use the internal standard quantitative analysis technique. By analyzing a
blank sample, you can confirm that there are no residual compounds in
the solvent system that can cause erroneous results.
Cal Std (Calibration standard) Contains known amounts of all target
compounds. The purpose of a standard is to measure the response of the
instrument to the target compounds so that the processing application
can generate a calibration curve for each compound.
QC Std (Quality Check standard) Contains a known amount of one or more
specific target compounds. The application places check standard
samples in the sequence so that it can test quantitative analysis results
for quality assurance purposes. After the application analyzes the QC
Std sample, it compares the measured quantity with the expected value
and an acceptability range. The quantitative analysis of a QC Std sample
is classified as passed if the difference between the observed and expected
quantities is within the user-defined tolerance. A QC Std sample is
classified as failed if the difference between the observed and expected
quantities is outside the user-defined tolerance.
Solvent Contains only solvent.
Unknown Used for quantitative analysis of samples.
3 Using the Acquisition Mode
Sample Types
92 TraceFinder User Guide Thermo Scientific
Thermo Scientific TraceFinder User Guide 93
4
Using the Analysis Mode for Quantitation Batches
Use the features of the Analysis mode to do the following:
• Create quantitation batches.
• Submit quantitation batches for acquisition, processing, or report generation.
• Review quantitation batches, batch data, reports, and local methods.
To access the Analysis mode
Click Analysis in the navigation pane.
The Analysis navigation pane opens.
IMPORTANT TraceFinder 4.1 uses the same data as TraceFinder 4.0. By default, the application
stores the method, project, template, and compound database data for the 4.1 release in the
TraceFinderData\4.0 folder.
Contents
•Working in the Batch View for Quantitation Batches
•Working in Data Review for Quantitation Batches
•Working in the Report View for Quantitation Batches
•Working in the Local Method View for Quantitation Batches
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
94 TraceFinder User Guide Thermo Scientific
Working in the Batch View for Quantitation Batches
In the Batch View, you can manually create and edit a new quantitation batch or open and
edit a previously saved batch. When you submit a batch, you can acquire and process data and
optionally create reports for the submitted samples.
The Analysis mode includes a toolbar:
Use the Toolbar or the equivalent commands in the Batch View Shortcut Menu to create the
sample list and submit samples for acquisition.
See the following topics:
•Samples Page for Quantitation Batches
•Auto Samples Page for Quantitation Batches
•Reference Sample Page for Quantitation Batches
•Threshold Samples Page for Quantitation Batches
To open the Batch View
1. Click Analysis in the navigation pane of the current mode.
2. Click Batch View.
The Batch View navigation pane opens.
Samples Page for Quantitation Batches
To open the Samples page, click Samples in the Batch View navigation pane.
For details about the Samples page, see the following topics:
•Samples Page Features
•Creating a New Quantitation Batch
•Editing a Quantitation Batch
•Submitting a Quantitation Batch
Available only when you activate Intelligent
Sequencing in the Configuration console
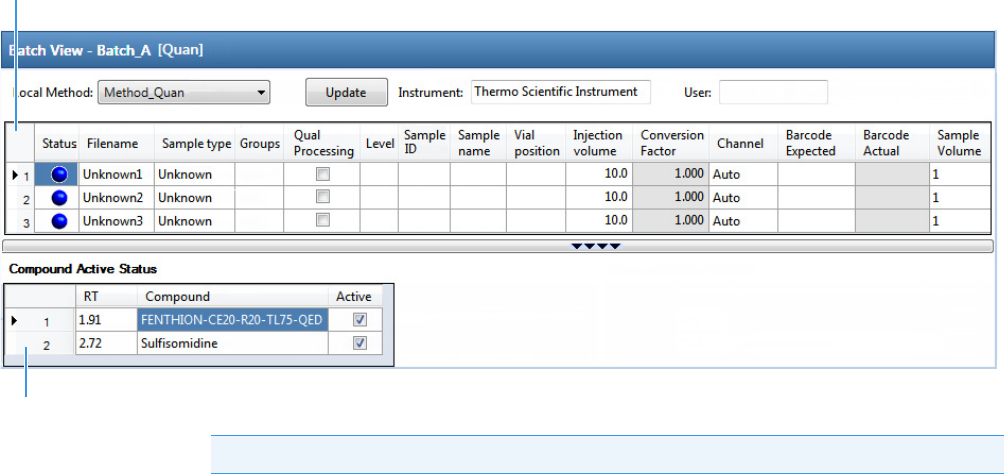
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 95
Samples Page Features
The Samples page is divided into two panes:
• Samples pane
Use the Samples Pane to create a batch.
• Compound Active Status pane
Use the Compound Active Status Pane to make specific compounds active or inactive.
Samples Pane
The samples pane includes the following features:
•Column Display
•Status Indicators
•Groups
•Qual Processing
•Sample Weight Calculation
•Instrument Methods
•Toolbar
•Batch View Sample List
•Batch View Shortcut Menu
Samples pane
Compound Active Status pane
Tip To resize the panes, drag the separator that divides the panes.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
96 TraceFinder User Guide Thermo Scientific
Column Display
The sample list contains many columns of information. You can scroll to see all the columns,
and you can customize which ones to display and their display order.
To scroll the sample list
Use the horizontal scroll bar at the bottom of the sample list to view all the information.
When you use the scroll bar at the bottom of the sample list, the following columns
remain fixed: Status, Filename, Sample Type, Groups, Qual Processing, Level, Sample ID,
and Sample Name, while the other columns scroll right and left.
To customize the column display
1. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
2. Use the arrow buttons to move all the columns that you want displayed to the Displayed
Columns pane.
These columns appear to the right of the sample list’s fixed columns: Status, Filename,
Sample Type, Groups, Qual Processing, Level, Sample ID, and Sample Name.
3. To arrange the order of the columns, do the following:
a. In the Displayed Columns pane, select a column name.
b. Use Up or Down to move the selected column up or down in the list.
The first column in the list represents the leftmost column in the Batch View sample
list, and the last column in the list represents the rightmost column in the Batch View
sample list.
4. To change the width of a column, do the following:
a. In the Displayed Columns pane, select the column width.
b. Type a new value for the width.
5. Repeat step 4 for all columns whose widths you want to change, and click OK.
The columns in the sample list immediately reflect your changes. The application uses
these settings for all sample lists in the Batch View.
Note The following columns are fixed: Status, Filename, Sample Type, Groups,
Qual Processing, Level, Sample ID, and Sample Name.
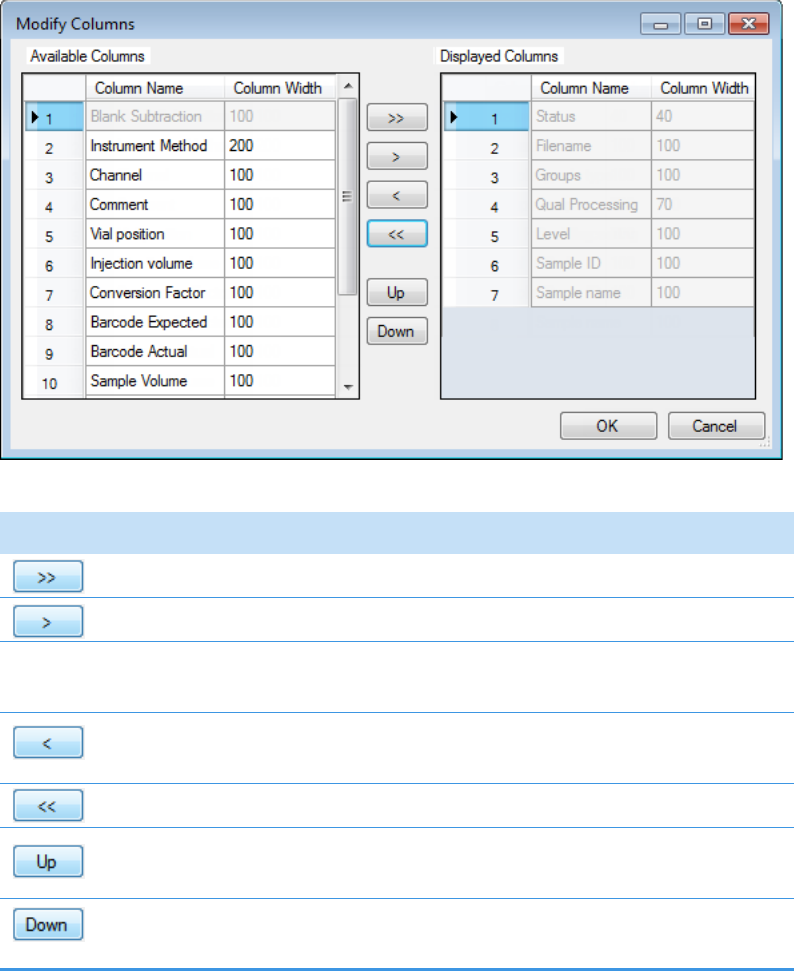
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 97
Figure 19. Modify Columns dialog box
Table 17. Button descriptions for the Modify Columns dialog box
Button Description
Moves all columns to the Displayed Columns pane.
Moves the selected column to the Displayed Columns pane.
The following buttons apply to all columns, except for those that are fixed: Status, Filename,
Sample Type, Groups, Qual Processing, Level, Sample ID, and Sample Name.
Moves the selected column to the Available Columns pane.
Moves all columns except fixed columns.
Moves the selected column name in the Displayed Columns pane one row
up in the column order.
Moves the selected column name in the Displayed Columns pane one row
down in the column order.
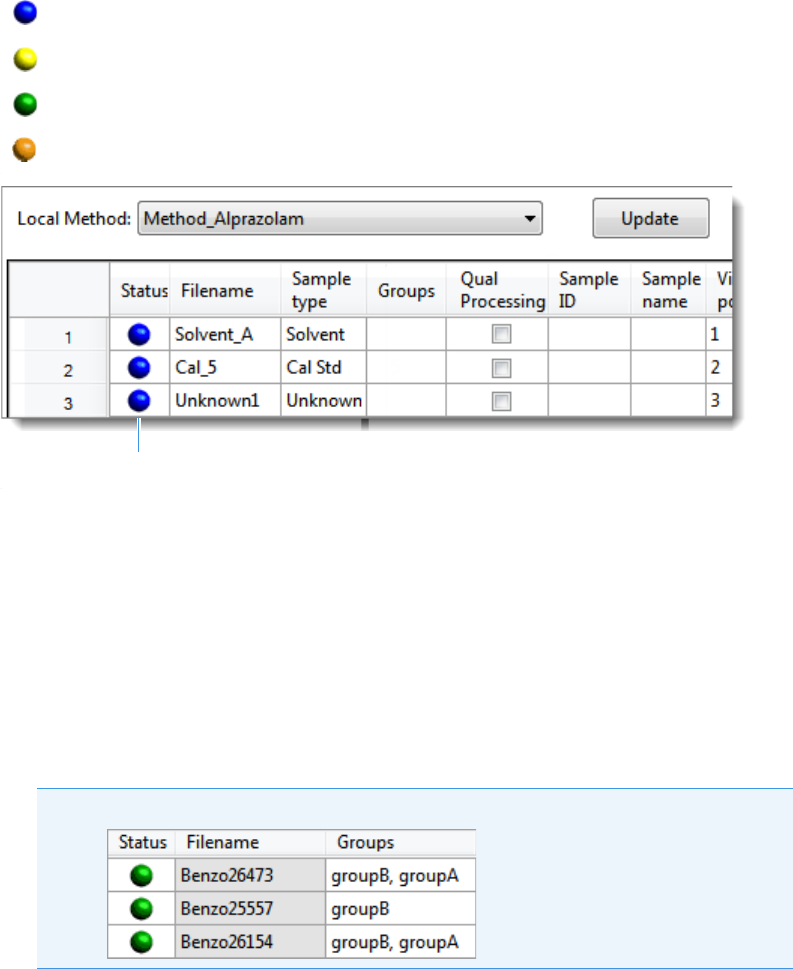
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
98 TraceFinder User Guide Thermo Scientific
Status Indicators
Status indicators show the current status of each sample during the acquisition and
processing.
Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Groups
Use the Groups feature to assign samples to a group.
To create a group
1. For each sample, type the name of a group in the Groups column.
Repeat this for each sample that you want to include in a group.
2. Create as many groups as you want.
Note To assign a sample to multiple groups, separate the groups with a comma.
Status indicators

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 99
Qual Processing
Use the Qual Processing feature to indicate samples to be processed with the qualitative peak
processing criteria specified in the method.
The Qualitative View displays processed data for the selected samples. See Qualitative View
for Quantitation Batches.
Sample Weight Calculation
Use the sample weight features to calculate the conversion factor for a sample. The application
uses different methods to calculate the conversion factor for liquid or solid calculation types.
Liquid: SampleVolume DilutionFactor
Solid: (SampleVolume × DilutionFactor) SampleWeight
Manual: The application does not calculate the Conversion Factor. Instead, you can enter the
Conversion Factor value.
Follow these procedures:
•To display the features for calculating sample weight
•To calculate the conversion factor for a liquid sample
•To calculate the conversion factor for a solid sample
•To manually specify the conversion factor for a sample
To display the features for calculating sample weight
If the Conversion Factor, Sample Volume, Dilution Factor, Sample Weight, Calculation
Type, and Final Units columns are not visible, right-click and choose Enable Sample
Weight Calculation.

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
100 TraceFinder User Guide Thermo Scientific
To calculate the conversion factor for a liquid sample
1. From the Calculation Type list, select Liquid.
For a liquid sample, the Sample Weight value is not editable.
2. In the Sample Volume column, type the volume in ng/mL for your sample.
3. In the Dilution Factor column, type the value for the dilution.
For example, if you have 1000 ng/mL of a substance that is too concentrated for the mass
spectrometer, you can dilute it by 1000. Then your injection volume is 1, your conversion
factor is 1000, and your sample amount is 1000.
4. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
To calculate the conversion factor for a solid sample
1. From the Calculation Type list, select Solid.
2. In the Sample Weight column, type the weight in ng for your sample.
3. In the Sample Volume column, type the volume in ng/ml for your sample.
4. In the Dilution Factor column, type the value for the dilution.
For example, if you have 1000 ng/ml of a substance that is too concentrated for the mass
spectrometer, you can dilute it by 1000. Then your injection volume is 1, your conversion
factor is 1000, and your sample amount is 1000.
5. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
To manually specify the conversion factor for a sample
1. From the Calculation Type list, select Manual.
For a manually calculated sample, the only available columns are the Conversion Factor
and the Final Units.
2. In the Conversion Factor column, type the conversion factor to use for your sample.
3. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
Note The application uses the following formula to calculate the Conversion Factor:
SampleVolume DilutionFactor
Note The application uses the following formula to calculate the Conversion Factor:
(SampleVolume × DilutionFactor) SampleWeight
Note The application uses the specified conversion factor when it calculates the
amount for the sample.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 101
Instrument Methods
Use the Instrument Methods column to specify instrument methods for the samples.
To specify instrument methods for samples
1. Display the Instrument Method column in the sample list:
a. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
b. In the Available Columns pane, select Instrument Method.
c. Click to move the Instrument Method column to the Displayed Columns
pane.
d. Click OK.
The application displays the Instrument Method column, defaulting to the instrument
method specified in the master method.
2. Click the Instrument Method column and select an instrument method from the list.
This list contains all the available instrument methods. Instrument methods from
external sources are prefixed with “Ext:”.
You can specify a different instrument method for each sample.
When you submit the batch for acquisition, the application saves a copy of the selected
instrument methods to the following folders:
External instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method\ExternalMethods
Local instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method
Note By default, the Instrument Method column is not displayed in the Batch View
sample list.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
102 TraceFinder User Guide Thermo Scientific
Toolbar
The Analysis mode includes this toolbar for creating and submitting a batch.
Table 18. Toolbar icons
Icon Description
Adds the specified number of new, empty samples to the end of the
sample list. See the instructions To add samples to the list.
Inserts a new, empty sample or samples above the selected sample. See
the instructions To insert samples into the list.
Removes the selected samples from the sample list. See the
instructions To remove samples from the list.
Adds imported samples from a CSV, an XML, or an SLD file to the
sample list. See the instructions To import samples into the list.
Submits only the selected samples for acquisition, processing, or
report generation. See the instructions To submit samples in the
batch.
Submits the batch for acquisition, processing, or report generation.
See the instructions To submit samples in the batch.
Submits only the selected peaks for processing. See the instructions To
submit selected peaks for processing.
Opens the Acquisition mode where you can use a batch template to
define a standard sequence composed of various sample types to be
assembled into a batch of samples. See Working in Data Review for
Quantitation Batches.
Opens the Acquisition mode where you can create a batch template
that contains the basic settings and sample types for your batches. See
Using the Acquisition Mode.
Opens the Quick Acquisition window where you can quickly submit a
single sample. See Appendix A, “Using Quick Acquisition.”
Opens the Audit Viewer where you can view audit logs. See
Chapter 8, “Using the Audit Viewer.” Available only when you enable
Auditing in the Administrator Console. Refer to the instructions in
the TraceFinder Administrator Console User Guide.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 103
Batch View Sample List
The sample list displays all the quantitative data for the samples of a batch.
Status indicators for each sample indicate if the sample is currently acquiring, not acquired,
acquired, or processed.
The sample list includes the following columns of information:
Figure 20. Batch View sample list

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
104 TraceFinder User Guide Thermo Scientific
Table 19. Batch View sample list columns (Sheet 1 of 2)
Column Description
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Note When you include unknown screening features in the quantitation method and you
choose to process with only the quantitation criteria, the Sample View shows the Status for
the samples as acquired and processed ( ), and the Unknown Screening View shows the
Status for the samples as acquired but not processed ( ).
Note When you include unknown screening features in the quantitation method and you
choose to process with only the unknown screening criteria, both the Sample View and the
Unknown Screening View show the Status for the samples as acquired and processed ( ).
Groups Threshold group to which a sample belongs. Samples can be viewed by group in the
Comparative View of Data Review.
Filename Name of the raw data file that contains the sample data.
Sample Type Defines how the application processes the sample data. Each sample is classified as one of the
following sample types: Matrix Blank, Solvent, Cal Std, QC Std, or Unknown.
Default: Unknown
Qual Processing Indicates samples to be processed with the qualitative peak processing criteria specified in the
method. The Qualitative View displays processed data for the selected samples.
Level The level defined for a calibration sample or quality control sample.
Sample ID A user-defined, alphanumeric string that identifies a sample.
Sample Name A user-defined name that identifies a sample.
Vial Position The tray vial number used for an autosampler acquisition.
Injection Volume The injection volume (in microliters) of the injected sample.
When you are using an autosampler, you can set the default injection volume in the
Autosampler dialog box in the Instrument View. The minimum and maximum injection
volumes that you can use depend on the autosampler you configure. The usable range depends
on the injection mode and might be smaller than the displayed range.
The Injection Volume value set in the master method overwrites the value in the instrument
method.
Valid range: 0.1 through 5000 μL
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 105
Calculation Type Liquid: The application calculates the Conversion Factor as
SampleVolume DilutionFactor
Solid: The application calculates the Conversion Factor as
(SampleVolume × DilutionFactor) SampleWeight
Manual: Sample Volume, Dilution Factor, Sample Weight, and Final Units columns are not
available, and the Conversion Factor value is editable.
Conversion Factor Editable only when Calculation Type is Manual.
Default: 1
Sample Volume Default: 1
Dilution Factor Default: 1
Sample Weight Available only when Calculation Type is Solid.
Default: 1
Final Units Specifies the calculated amount in the Data Review view or in reports.
Default: 1
Instrument Method Specifies the instrument to use for the acquisition. This column is hidden by default. To display
this column, see To customize the column display.
Channel Specifies the channel on which the sample was run. If the sample is not acquired, the value is
Pending. The Channel column is available only when you have activated multiplexing in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in the
TraceFinder Lab Director User Guide.
Barcode Expected A user-entered barcode for the vial.
Barcode Actual An actual barcode for the vial. This value is not editable.
Comment A user-defined comment for the sample.
Table 19. Batch View sample list columns (Sheet 2 of 2)
Column Description

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
106 TraceFinder User Guide Thermo Scientific
Batch View Shortcut Menu
The Batch View includes a shortcut menu for creating a batch.
Table 20. Batch View shortcut menu commands (Sheet 1 of 2)
Command Description
Add Sample Adds a single empty row to the sample grid.
Insert Sample Inserts a single empty row to the sample grid above the selected
row.
Insert Copy Sample Copies the currently selected row and inserts a copy above the
row.
Reinject Selected
Samples
Creates a copy of the selected sample and appends INJ001 to the
file name. Additional reinjections of the same sample are
numbered INJ002, INJ003, and so forth.
Remove Selected
Samples
Removes selected samples from the sample grid.
Import Samples Opens the Sample Import Tool. See To import samples into the
list.
Browse in Raw File
(Move)
Opens a dialog box where you can select a raw data file to use for
the selected sample row. The application removes the raw data
file from the source location.
Browse in Raw File
(Copy)
Opens a dialog box where you can select a raw data file to use for
the selected sample row. The application copies the raw data file
from the source location.
Map Raw Files to
Samples
Opens a dialog box where you can select multiple raw data files
to use for the selected sample rows.
Copy Down Copies the value in the selected row to all rows below it. This
command is available only when you have selected a value that
can be copied down.
Fill Down Enters sequential values in the column starting with the value in
the selected row and ending with the last row in the column.
This command is available only when you have selected a value
that can be filled down.
Modify Columns Opens the Modify Columns Dialog Box.
Hide Sample Weight
Calculation
Displays or hides the Sample Volume, Dilution Factor, Sample
Weight, Calculation Type, and Final Units columns.
Copy Copies the data in the selected rows or columns to the Clipboard.
Use this command to copy sample information into a text editor
or spreadsheet application. You cannot paste this data back into
the Batch View sample list.
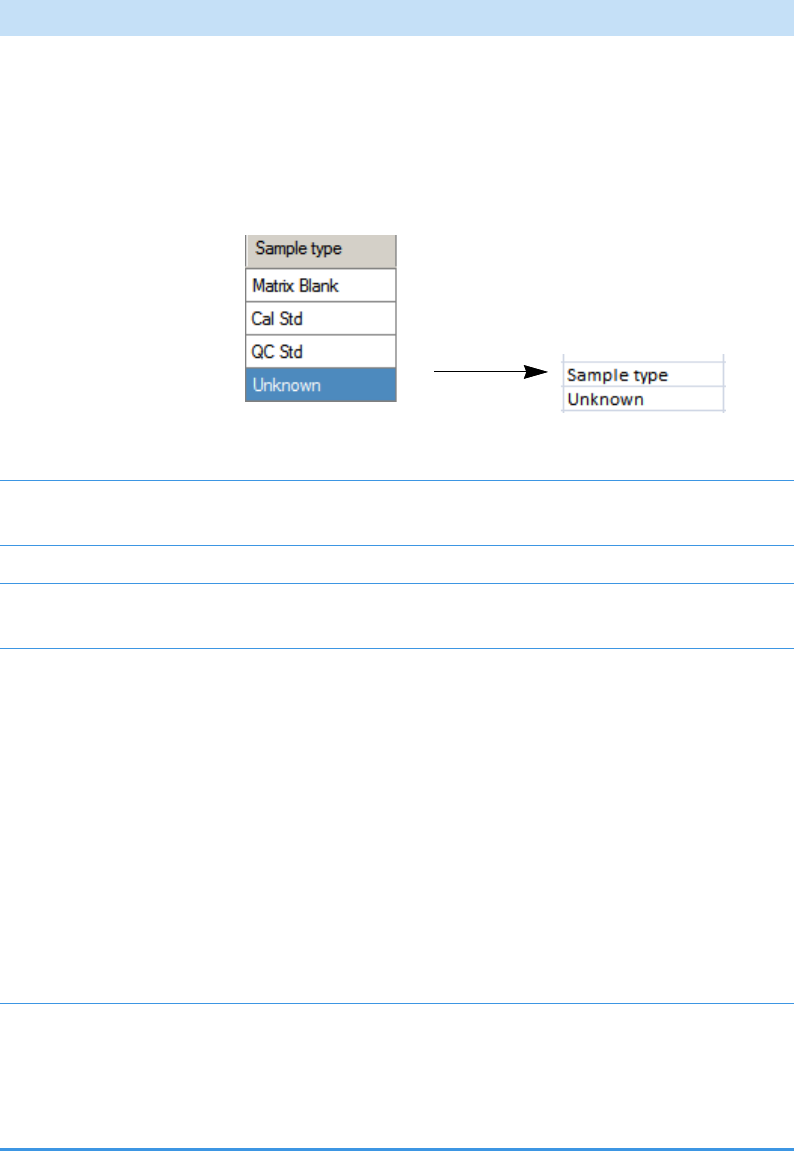
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 107
Copy with Headers Copies the data in the selected rows or columns and the
associated column headers to the Clipboard. Use this command
to copy sample information into another text editor or
spreadsheet application. You cannot paste this data back into the
sample list.
For example
Paste Pastes a single column of copied data from another text editor or
spreadsheet application into the selected column.
Undo Last Paste Removes the last pasted item in the Batch View.
Export to CSV File Opens the Save As dialog box where you can save the current
sample list to a CSV file.
Edit Instrument Method Opens the Instrument Setup window where you can edit the
parameters of the instrument method.
• When you edit an external method, the application updates
the method in the …\Xcalibur\methods folder.
• When you edit an internal method, the application updates
the method in the
…\TraceFinderData\4.0\Projects\…\batch\Methods\method
folder.
For detailed information about editing instrument methods,
refer to Chapter 3, “Using Instrument Methods in the Method
Development Mode,” in the TraceFinder Lab Director User
Guide.
View Sample in Qual
Explorer
Displays the selected sample in the qualitative explorer
application that you configured as your default in the
Configuration console. Refer to Chapter 1, “Using the
Configuration Console,” in the Tra ceFin der L ab D irector Use r
Guide.
Table 20. Batch View shortcut menu commands (Sheet 2 of 2)
Command Description
Copy with Headers
from TraceFinder Paste into an Excel spreadsheet.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
108 TraceFinder User Guide Thermo Scientific
Compound Active Status Pane
In the Compound Active Status pane, you can choose specific compounds to be active or
inactive.
To set a compound as active or inactive
1. In the sample list, select a sample.
All compounds in the selected sample are listed in the Compound Active Status pane.
The default active/inactive status is determined by the identification settings in the local
method. For information about setting the identification parameters, refer to Chapter 4,
“Using the Method Development Mode for Quantitation Methods,” in the Tra ce Fi nder
Lab Director User Guide.
• To display compounds alphabetically, right-click and choose Sort by Compound
Name.
• To display compounds from shorter to longer retention time, right-click and choose
Sort by Retention Time.
2. Select or clear the Active check box for the compound.
To change the active/inactive status in the Data Review view, see Inactive and Excluded
Compounds.
Compound Active Status
You can specify which compounds are active or inactive in the Local Method View or the
Batch View.
Figure 21. Active and inactive compounds in the Local Method View
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 109
For details about setting the status on the Identification page, refer to Chapter 4, “Using the
Method Development Mode for Quantitation Methods,” in the TraceFin d er L ab D irecto r Use r
Guide.
Figure 22. Active and inactive compounds in the Batch View
For details about setting the status in the Batch View, see Compound Active Status Pane.
Creating a New Quantitation Batch
In the Batch View, you can create a new batch.
Follow these procedures:
•To c re a t e a ne w b a t c h
•To add samples to the list
•To insert samples into the list
•To import samples into the list
•To remove samples from the list
•To c o py a sa m pl e
•To reinject a sample
•To edit sample values
•To browse in raw data files
•To customize the column display
To create a new batch
1. Choose File > New > Batch from the main menu.
The Create New Batch Dialog Box opens, displaying all drives that contain projects.
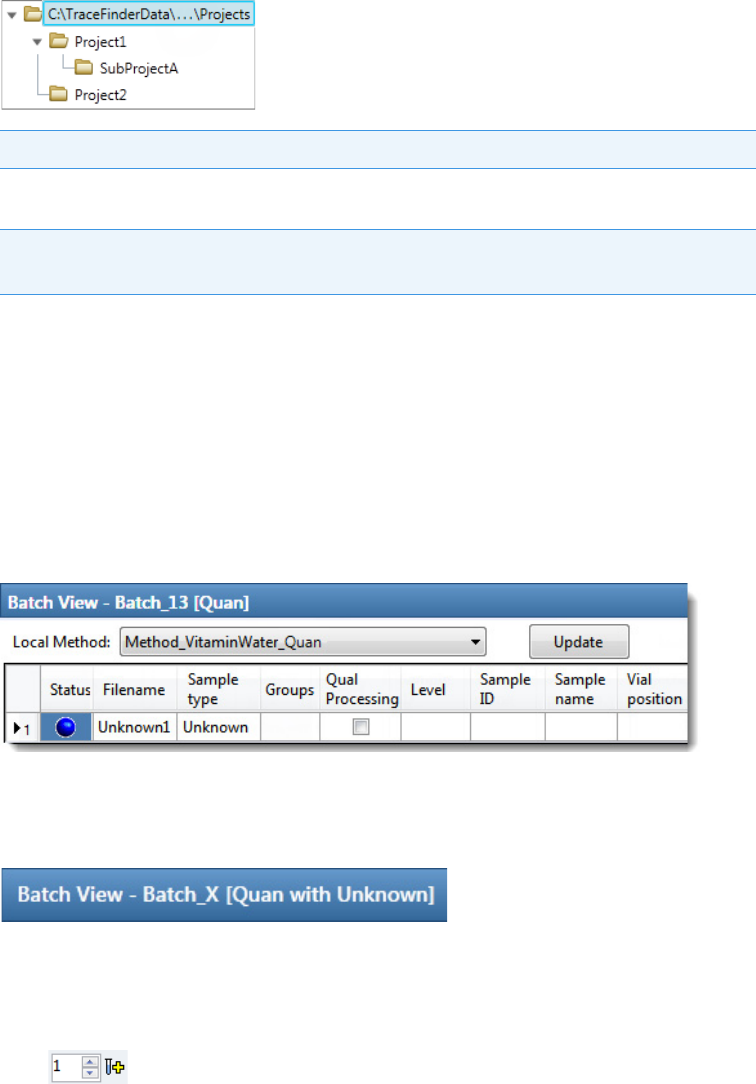
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
110 TraceFinder User Guide Thermo Scientific
2. Select a drive from the list.
3. Select the folder where you want to store your batch.
To create a new folder for the storage location, see Editing Folders for Batches.
4. Select Quan from the Type list.
The batch list displays all batches in the selected folder. The Method list displays all
methods for the quantitation type.
5. Select a master method from the Master Method list.
6. Click Create.
A new batch opens with one Unknown sample.
The batch name in the title bar indicates that you are creating a quantitation batch.
When you create a batch using a quantitation method that includes unknown screening
features, the title bar indicates that the method includes unknown screening.
To add samples to the list
1. To add a single sample row, right-click the sample list and choose Add Sample.
2. To add multiple sample rows, select the number of rows and then click the Add Sample
icon, .
The application adds the specified number of new, empty samples to the end of the
sample list.
Tip The application displays all configured and enabled repositories.
Tip To activate the Create button, you must enter a unique batch name. If the Create
button is not activated, you have entered a batch name that is already used.
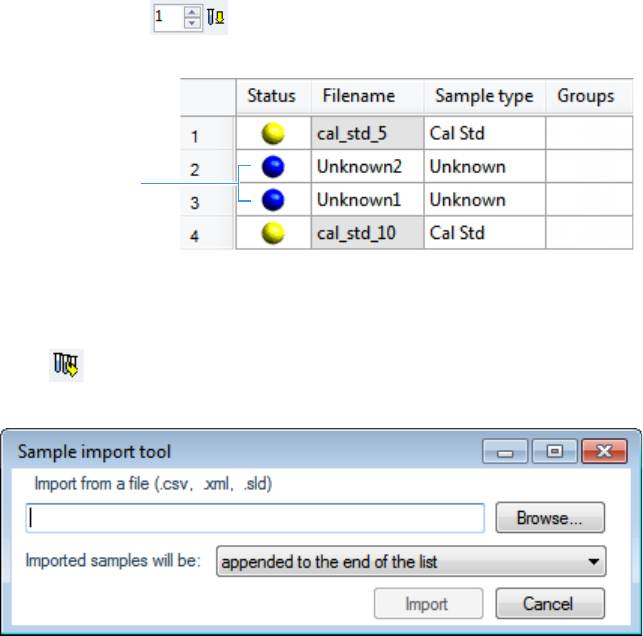
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 111
To insert samples into the list
Select the sample above which you will insert new, Unknown samples, and then do one of
the following:
• To insert a single sample row, right-click and choose Insert Sample.
• To insert multiple sample rows, select the number of rows and then click the Insert
Sample icon .
The application inserts the Unknown samples above the selected sample.
To import samples into the list
1. Choose Batch > Import Samples from the main menu, or click the Import Samples
icon, .
The Sample Import Tool dialog box opens.
From this dialog box, you can import samples from a CSV, an XML, or an SLD file.
2. Click Browse and select a CSV, an XML, or an SLD file that contains the samples to
import.
3. From the Imported Samples Will Be list, select Appended to the End of the List or
Inserted at the Selected Row.
4. Click Import.
The Sample Import Tool dialog box closes, and the application adds the specified samples
to the sample list.
Inserted samples
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
112 TraceFinder User Guide Thermo Scientific
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following column name substitutions.
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following sample type substitutions.
5. (Optional) When using multiplexing, select a channel for each imported sample.
Imported samples default to Auto.
To remove samples from the list
1. Select the samples that you want to remove.
2. Right-click and choose Remove Selected Samples.
To copy a sample
1. Select the sample that you want to copy.
2. Right-click and choose Insert Copy Sample.
The application inserts the copy above the selected sample.
To reinject a sample
1. In the sample list, select the sample that you want to reinject.
2. Right-click and choose Reinject This Sample.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the sample are numbered INJ002, INJ003, and so forth.
The application copies all parameter values from the original sample.
Xcalibur column TraceFinder column
Position Vial Position
Inj Vol Injection Volume
Dil Factor Conversion Factor
Xcalibur sample type TraceFinder sample type
Blank Matrix Blank
QC QC Std
Std Bracket Cal Std
Note The Channel column is available only when you have activated multiplexing in
the Configuration console. Refer to Chapter 1, “Using the Configuration Console,”
in the TraceFinder Lab Director User Guide.
Tip Use the CTRL or SHIFT keys to select multiple samples.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 113
To edit sample values
1. For each sample, do one of the following:
Type a new file name over the current filename.
–or–
Double-click the Filename column and locate a raw data file to use for the sample.
–or–
Right-click and choose Browse in Raw File, and then locate a raw data file to use for the
sample.
By default, the application sets the Sample Type to Unknown.
2. For each sample, click the Sample Type column and select a sample type from the list.
3. For each Cal Std or QC Std sample, select a level from the Level list.
The sample levels are defined in the master method. If there are no levels to select in the
Level list, do the following:
a. Return to the Method Development mode.
b. Open the method.
c. Click Compounds in the navigation pane.
d. Click the Calibration Levels tab.
e. Add the levels.
f. Save the method.
g. Return to the Batch View in the Analysis mode, and then click Update.
The application updates the local method with the new sample levels.
For detailed instructions about specifying calibration levels, refer to Chapter 4, “Using the
Method Development Mode for Quantitation Methods,” in the Trac eFi nd er L a b D ire c t or
User Guide.
4. Type a vial position in the Vial Position column for each sample.
5. Type a volume in the Injection Volume column for each sample.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
Available sample types
Matrix Blank Solvent QC Std Unknown
Cal Std

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
114 TraceFinder User Guide Thermo Scientific
6. (Optional) Type or edit the values for the remaining columns.
To automatically copy or fill values in these columns, see Appendix B, “Using Copy
Down and Fill Down.”
To browse in raw data files
1. Do one of the following:
Double-click the Filename column.
–or–
Right-click and choose Browse in Raw File.
The What Raw File Would You Like to Use dialog box opens.
2. Select a raw data file to use for the sample or use the CTRL key to select multiple files,
and then click Open.
The application overwrites the selected, unacquired sample in the batch with the first
“browsed in” file and adds any additional browsed in files below the selected sample.
For all browsed-in raw data files, the application sets the Status to Acquired, , and sets
the Sample Type to Unknown.
To customize the column display
1. Right-click the Batch View sample list and choose Modify Columns.
The Modify Columns dialog box opens.
2. Use the arrow buttons to move all the columns that you want displayed to the Displayed
Columns pane.
All the columns you select appear after the Status, Filename, Sample Type, Groups, Qual
Processing, Level, Sample ID, and Sample Name columns.
3. To arrange the order of the columns, do the following:
a. In the Displayed Columns pane, select a column name.
b. Use Up or Down to move the selected column up or down in the list.
The first column in the list represents the leftmost column in the Batch View sample
list, and the last column in the list represents the rightmost column in the Batch View
sample list.
Note When you use the scroll bar at the bottom of the sample list, the following
columns remain fixed: Status, Filename, Sample Type, Groups, Qual Processing,
Level, Sample ID, and Sample Name, while the other columns scroll right and left.
Note You cannot overwrite an acquired sample. When you select a sample that is
acquired, the application adds all browsed in files below the selected sample.
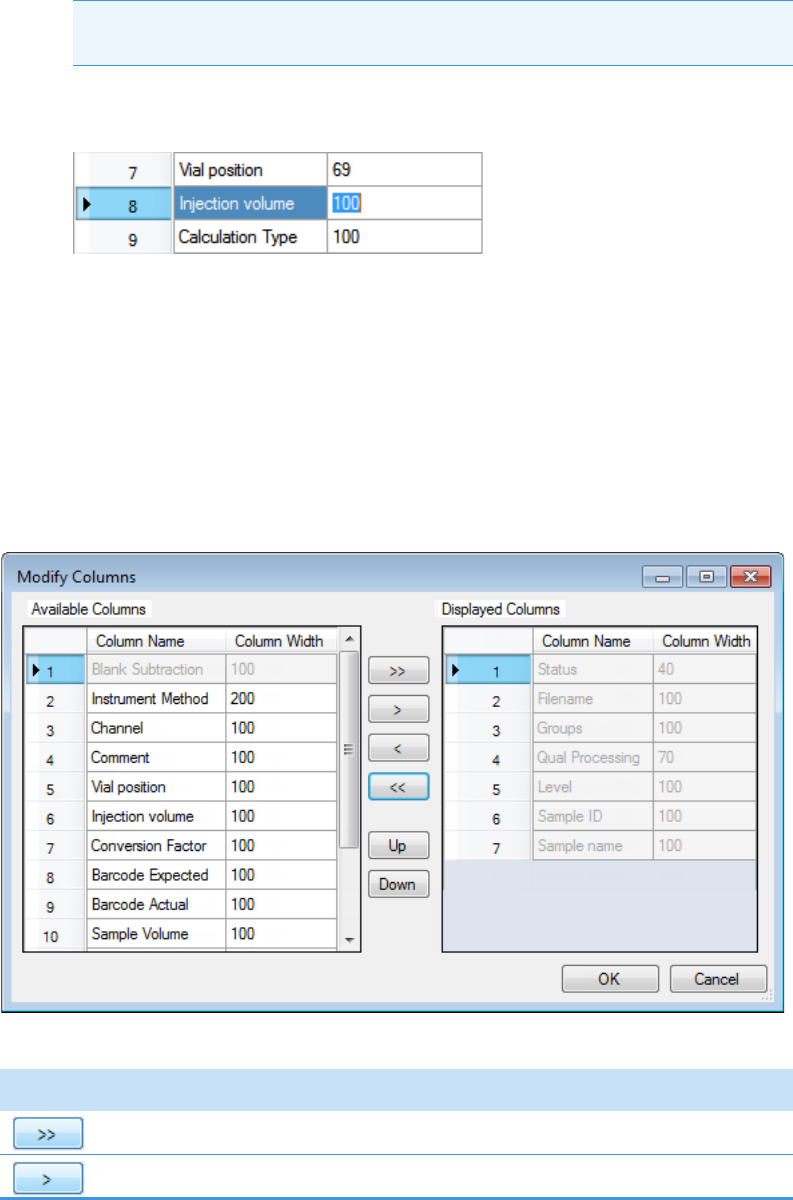
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 115
4. To change the width of a column, do the following:
a. In the Displayed Columns pane, select the column width.
b. Type a new value for the width.
5. When you have completed your changes, click OK.
The columns in the sample list immediately reflect your changes. The application uses
these settings for all sample lists in the Batch View.
Modify Columns Dialog Box
Use the Modify Columns dialog box to select the columns that you want to display.
Figure 23. Modify Columns dialog box
Note The following columns are fixed: Status, Filename, Sample Type, Groups,
Qual Processing, Level, Sample ID, and Sample Name.
Table 21. Button descriptions for the Modify Columns dialog box (Sheet 1 of 2)
Button Description
Moves all columns to the Displayed Columns pane.
Moves the selected column to the Displayed Columns pane.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
116 TraceFinder User Guide Thermo Scientific
Create New Batch Dialog Box
Use the Create New Batch dialog box to select a folder and method for your batch and to
name the new batch.
Figure 24. Create New Batch dialog box
The following buttons apply to all columns, except for those that are fixed: Status, Filename,
Sample Type, Groups, Qual Processing, Level, Sample ID, and Sample Name.
Moves the selected column to the Available Columns pane.
Moves all columns except those that are fixed.
Moves the selected column name in the Displayed Columns pane one row
up in the column order.
Moves the selected column name in the Displayed Columns pane one row
down in the column order.
Table 21. Button descriptions for the Modify Columns dialog box (Sheet 2 of 2)
Button Description
Table 22. Create New Batch dialog box parameters (Sheet 1 of 2)
Parameter Description
Create New Folder
Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Or, you can right-click and choose Create Folder.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 117
Editing Folders for Batches
From the Create New Batch dialog box, you can create new folders for your batches. You can
also delete or rename folders.
Use these procedures:
•To create new project folders
•To delete project folders
•To rename project folders
Delete Folder
With no confirmation prompt, immediately removes the selected folder.
You cannot delete a folder that contains lower-level folders; you must delete the lower-level
folders first.
Or, you can right-click and choose Delete.
Rename Folder
Renames the selected folder.
Or, you can right-click and choose Rename.
Batch table
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date that the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
New batch parameters
New Batch Name of the new batch to create.
Note If the Create button is not activated, you have entered a name that is already used or
you have not selected a method.
Type Type of batch to create: Quan, Screening, or Unknown Only.
Method Method used to create the new batch.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is created.
Buttons
Create Creates the specified batch and opens the Batch View for the new batch.
Cancel Closes the Create New Batch dialog box without creating a batch.
Table 22. Create New Batch dialog box parameters (Sheet 2 of 2)
Parameter Description
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
118 TraceFinder User Guide Thermo Scientific
To create new project folders
1. In the Create New Batch dialog box, select the folder for which you will create a new
lower-level folder.
• You can select the main TraceFinderData\4.0\Projects folder and create a new folder
under it.
• You can select one of the existing folders and create a lower-level folder under it.
2. Click the Create Folder icon, .
The application adds a new lower-level folder to the selected folder.
3. Select the new folder name and type a name for the folder.
Folder names are limited to 30 characters and can contain spaces and special characters,
except for the following special characters: \ / : + ? ” < >
To delete project folders
1. In the Create New Batch dialog box, select the folder to delete.
2. Click the Delete Folder icon, .
With no confirmation prompt, the application immediately removes the selected folder.
To rename project folders
1. In the Create New Batch dialog box, select the folder to rename.
2. Click the Rename Folder icon, .
3. Type a new name for the folder and press ENTER.
The application saves the new folder name.
Editing a Quantitation Batch
In the Batch View, you can open a saved batch and edit the sample list. You can add samples,
edit samples, or remove samples. If the batch has already been acquired, you can select specific
samples for reinjection. If the batch has unacquired samples when you complete your edits,
you can save it as a “ready to acquire” batch.
Note After you add a lower-level folder, you cannot rename the parent folder.
Note You cannot delete folders that contains lower-level folders; you must delete the
lower-level folders first.
Note You cannot rename folders that contain lower-level folders.
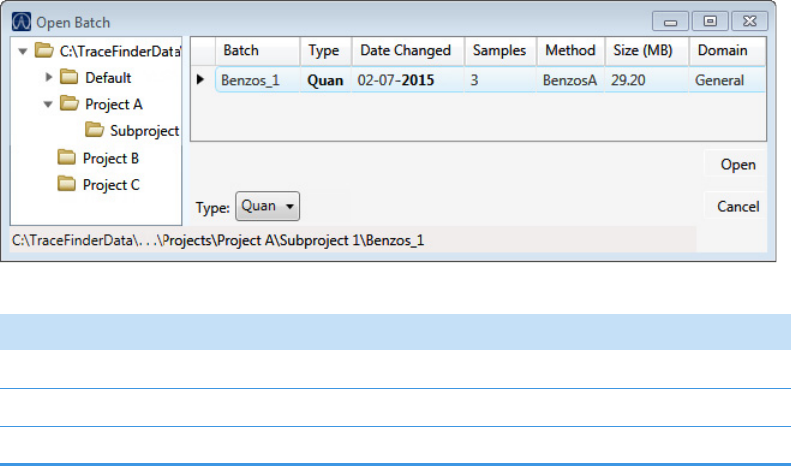
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 119
Follow these procedures:
•To o p e n a sa v e d b at c h
•To open a recent batch
•To edit samples in a batch
•To reinject a sample from a previously acquired batch
To open a saved batch
1. Choose File > Open > Batch from the main menu.
The Open Batch dialog box opens.
2. Select a project and a subproject.
3. Select Quan or Any from the Type list.
The batch list displays all batches created with quantitation methods (or all methods of all
types when you select Any).
4. Select a batch from the list.
5. Click Open.
The selected batch opens in the Batch View.
Open Batch Dialog Box
Use the Open Batch dialog box to select a batch to open.
Figure 25. Open Batch dialog box
Table 23. Open Batch dialog box parameters (Sheet 1 of 2)
Parameter Description
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date the batch was last updated.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
120 TraceFinder User Guide Thermo Scientific
To open a recent batch
Choose File > Recent Files > batch from the main menu.
The selected batch opens in the Batch View.
To edit samples in a batch
Use the commands described in Working in the Batch View for Quantitation Batches.
You can add new samples, edit samples, or delete samples.
To reinject a sample from a previously acquired batch
1. In the sample list, select the sample that you want to reinject.
2. Right-click and choose Reinject This Sample.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the sample are numbered INJ002, INJ003, and so forth.
The application copies all parameter values from the original sample.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
Path Path to the project in the TraceFinderData\4.0\Projects folder
where the batch is stored.
Buttons
Type Type of batch to display in the Batch list: Quan, Screening,
Unknown Only, or Any.
Open Opens the Batch View for the selected batch.
Cancel Closes the Open Batch dialog box without opening a batch.
Table 23. Open Batch dialog box parameters (Sheet 2 of 2)
Parameter Description

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 121
A green status icon indicates previously acquired samples (acquired and processed), and
the sample name is grayed out. A blue status icon indicates samples created for reinjection
(not acquired).
When you submit all samples in this batch, the application acquires all samples (including
previously acquired samples).
3. To save this batch with the new samples for reinjection, choose File > Save > Batch from
the main menu.
The batch is saved as a prepared batch that is ready to submit. You can open this batch
from the Reinject Samples page in the Acquisition mode and submit the batch. The
application acquires only the samples that have not been previously acquired.
Submitting a Quantitation Batch
In the Batch View, you can submit an entire batch or only selected samples in the batch.
When you submit a batch for acquisition and processing, you can choose to create reports for
the submitted samples. See Submit Options dialog box.
For a description of commands in the shortcut menu, see Batch View shortcut menu
commands.
Follow these procedures:
•To submit selected peaks for processing
•To submit samples in the batch
•To view the output files
To submit selected peaks for processing
1. In the Peak List, select the Selected check box for the peaks that you want to process.
2. Click the Submit … for Processing icon, .
The application processes the selected peaks and updates the data in the Data Review
panes.
To submit samples in the batch
1. Do one of the following:
• To submit all samples in the batch, click the Submit Batch icon, .
• To submit specific samples, select the samples and click the Submit Selected
Samples icon, .

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
122 TraceFinder User Guide Thermo Scientific
The Submit Options dialog box opens.
2. To acquire (or reacquire) the submitted samples, select the Acquire Data check box.
• When all submitted samples have been previously acquired, this option is (by default)
not selected.
• When one or more samples in the batch have not been acquired, this option is (by
default) selected.
3. To process the submitted samples, select the Process Data check box.
The application displays processing options for the method.
If the quantitation method includes unknown screening features, the application also
displays unknown screening options. For specifying processing data for batches that also
use unknown screening features, see To submit samples in the batch of Chapter 6, “Using
the Analysis Mode for Unknown Screening Batches.”
4. Select the check box for the quantitation options that you want to use.
Peak Detect: Performs peak detection. You can process the data with or without
performing peak detection. For example, you might want to turn off peak detection when
reprocessing samples.
Quantitate: Performs quantitation.
5. (Optional) Select the Create Reports check box.
6. (Optional with multiplexing activated) Select the Priority Sequence check box.
The application acquires the priority batch on the next available channel or the assigned
channel.
7. (Optional without multiplexing activated) Select the Priority Sequence check box and
then select one of the following priority options to place the batch in the queue:
•Next Available Batch places the batch immediately after the currently acquiring
batch.
•Next Available Sample places the batch immediately after the currently acquiring
sample.
Note You can also submit only selected peaks for processing. See To submit selected
peaks for processing.
Tip You can process the data with or without performing peak detection. For
example, you might want to turn off peak detection when reprocessing samples.
Note When you select Full Sequence Submission in the Configuration console, these
options are unavailable because the current batch and the current sample are, in effect,
the same thing.

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 123
8. To specify the following optional parameters, click Show Details.
a. Select the Use check box for the device that you want to use for this acquisition.
b. Select the Start Device check box to indicate the device that will initiate
communication with the other instruments.
This is usually the autosampler.
c. Select the Start When Ready check box, which starts all instruments together when
they are all ready.
When this is cleared, individual instruments can start at different times and then have
to wait for the last instrument to be ready.
d. Select the system state after it acquires the last batch: On, Standby, or Off.
9. To start the selected processes, click OK.
The selected processes begin, and the application shows the real-time display at the
bottom of the current window. You can begin another batch in the Analysis mode while
you watch the real-time display of the currently acquiring batch.
IMPORTANT When your batch also uses unknown screening features, after the first
processing, you might see an error message stating that the total number of results is too
large for a single batch (more than 500 000). Return to the Processing pages for the
unknown screening features (refer to Chapter 6, “Using the Method Development Mode
for Unknown Screening Methods,” in the TraceFinder Lab Director User Guide) and make
one or more of these parameter adjustments:
• Limit the RT range.
• Shorten the signal range.
• Specify a lower value for the Number of Top Matches.
• Specify a Simple Search instead of an Exhaustive Search.
• Specify Top Peaks instead of All Peaks.
• Limit the number of search types.
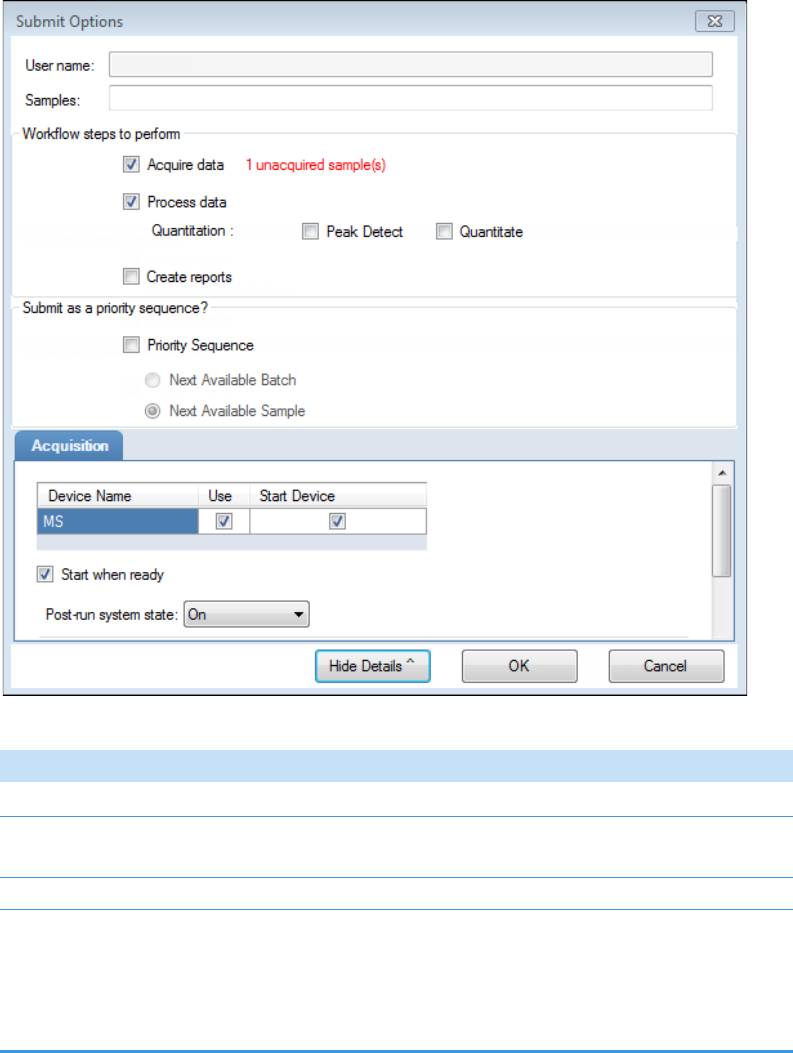
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
124 TraceFinder User Guide Thermo Scientific
Submit Options Dialog Box
Use the Submit Options dialog box to submit a batch for acquisition, processing, or
reporting.
Figure 26. Submit Options dialog box
Table 24. Submit Options dialog box parameters (Sheet 1 of 3)
Parameter Description
User Name Name of the current user.
Samples Number of samples to be submitted for acquisition, processing, or
reporting.
Workflow Steps to Perform
Acquire Data Submits the current batch to acquisition.
• When all submitted samples have been previously acquired, this
option is (by default) not selected.
• When one or more samples in the batch have not been
acquired, this option is (by default) selected.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 125
Process Data Processes the data for the current batch using any of the following
options:
Peak Detect: Performs peak detection. You can process the data
with or without performing peak detection. For example, you
might want to turn off peak detection when reprocessing samples.
Quantitate: Performs quantitation.
Note Quantitation methods can include unknown screening
features. For specifying processing data for batches that also use
unknown screening features, see To submit samples in the batch.
Create Reports Creates reports for the current batch.
Submit as a Priority Sequence?
Priority Sequence With multiplexing activated, places the batch immediately after the
currently acquiring batch.
Without multiplexing activated, specifies one of the following
priority options to place the batch in the queue:
Next Available Batch: Places the batch immediately after the
currently acquiring batch.
Next Available Sample: Places the batch immediately after the
currently acquiring sample.
Note When you select Full Sequence Submission in the
Configuration console, these options are unavailable because the
current batch and the current sample are, in effect, the same
thing.
Acquisition pane
Device Name Lists all configured instruments.
If the instrument that you want to use is not configured, close the
TraceFinder application, configure the instrument, and then reopen
the application. You cannot configure an instrument while the
TraceFinder application is running.
Available only when you select the Acquire Data check box.
Use Specifies the instruments used for this acquisition.
Available only when you select the Acquire Data check box.
Start Device Specifies the instrument that initiates the communication with the
other instruments. This is usually the autosampler.
Available only when you select the Acquire Data check box.
Table 24. Submit Options dialog box parameters (Sheet 2 of 3)
Parameter Description
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
126 TraceFinder User Guide Thermo Scientific
To view the output files
Locate the files to view from the following directories:
The application writes saved batches to the project folder:
…\TraceFinderData\4.0\Projects\…
For each acquired sample, the application writes an RSX file to the batch Data folder:
…\TraceFinderData\4.0\Projects\…\Data
The application saves method information to the batch Methods folder:
…\TraceFinderData\4.0\Projects\…\Methods
The application writes the reports to the batch Reports folder:
…\TraceFinderData\4.0\Projects\…\batch\Reports
Start When Ready Starts the specified device when all the instruments are ready to
acquire data. When this is cleared, individual instruments can start
at different times and then must wait for the last instrument to be
ready.
Post-run System State Specifies the system state after it acquires the last batch: On
(default), Standby, or Off.
Buttons
Hide/Show Details Collapses or expands the acquisition details of the Submit Options
dialog box.
OK Begins the selected processes.
Cancel Closes the Submit Options dialog box without submitting any
tasks.
Table 24. Submit Options dialog box parameters (Sheet 3 of 3)
Parameter Description
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 127
Saving a Batch to a New Location
You can move the current batch to a different project folder, or you can make a copy of the
current batch and save the copy to a different project folder.
Follow these procedures:
•To save a batch to another project folder
•To move a batch to another folder
•To create a new project folder
•To delete project folders
•To rename project folders
To save a batch to another project folder
1. Choose File > Save > Save Batch As from the Batch View main menu in the Analysis
mode.
The Save Batch As Dialog Box opens.
2. Select a storage location.
The default storage location is C:\TraceFinderData\4.0\Projects.
3. Select or create a project folder.
4. Type a name for the new batch.
If you are saving the batch to a different folder, you must give it a unique name. You
cannot overwrite an existing batch in a folder.
5. Click Save.
When you save the batch to a different folder, the reports reflect the original project
folders and the application does not save the calibration history.
To move a batch to another folder
1. Choose File > Save > Move Batch from the Batch View main menu in the Analysis
mode.
The Save Batch As Dialog Box opens.
2. Select a storage location.
The default storage location is C:\TraceFinderData\4.0\Projects.
3. Select or create a project folder.
4. Type a name for the new batch.
You must give the batch a unique name in the new subproject folder. You cannot
overwrite an existing batch.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
128 TraceFinder User Guide Thermo Scientific
5. Click Save.
When you move the batch, the reports reflect the original project and subproject folders
and the application does not save the calibration history.
To create a new project folder
1. In the Save Batch As dialog box, select the folder for which you will create a new
lower-level folder.
• You can select the main TraceFinderData\4.0\Projects folder and create a new folder
under it.
• You can select one of the existing folders and create a lower-level folder under it.
2. Click the Create Folder icon, .
The application adds a new lower-level folder to the selected folder.
3. Select the new folder name and type a name for the folder.
Folder names can contain spaces and special characters, except for the following special
characters: \ / : + ? ” < >
To delete project folders
1. In the Save Batch As dialog box, select the folder to delete.
2. Click the Delete Folder icon, .
With no confirmation prompt, the application immediately removes the selected folder.
To rename project folders
1. In the Save Batch As dialog box, select the folder to rename.
2. Click the Rename Folder icon, .
3. Type a new name for the folder and press ENTER.
The application saves the new folder name.
Note After you add a lower-level folder, you cannot rename the parent folder.
Note This feature is not available for folders that contain lower-level project or batch
folders; you must first delete the lower-level project or batch folders.
Note This feature is not available for folders that contain lower-level project or batch
folders; you must first delete the lower-level project or batch folders.
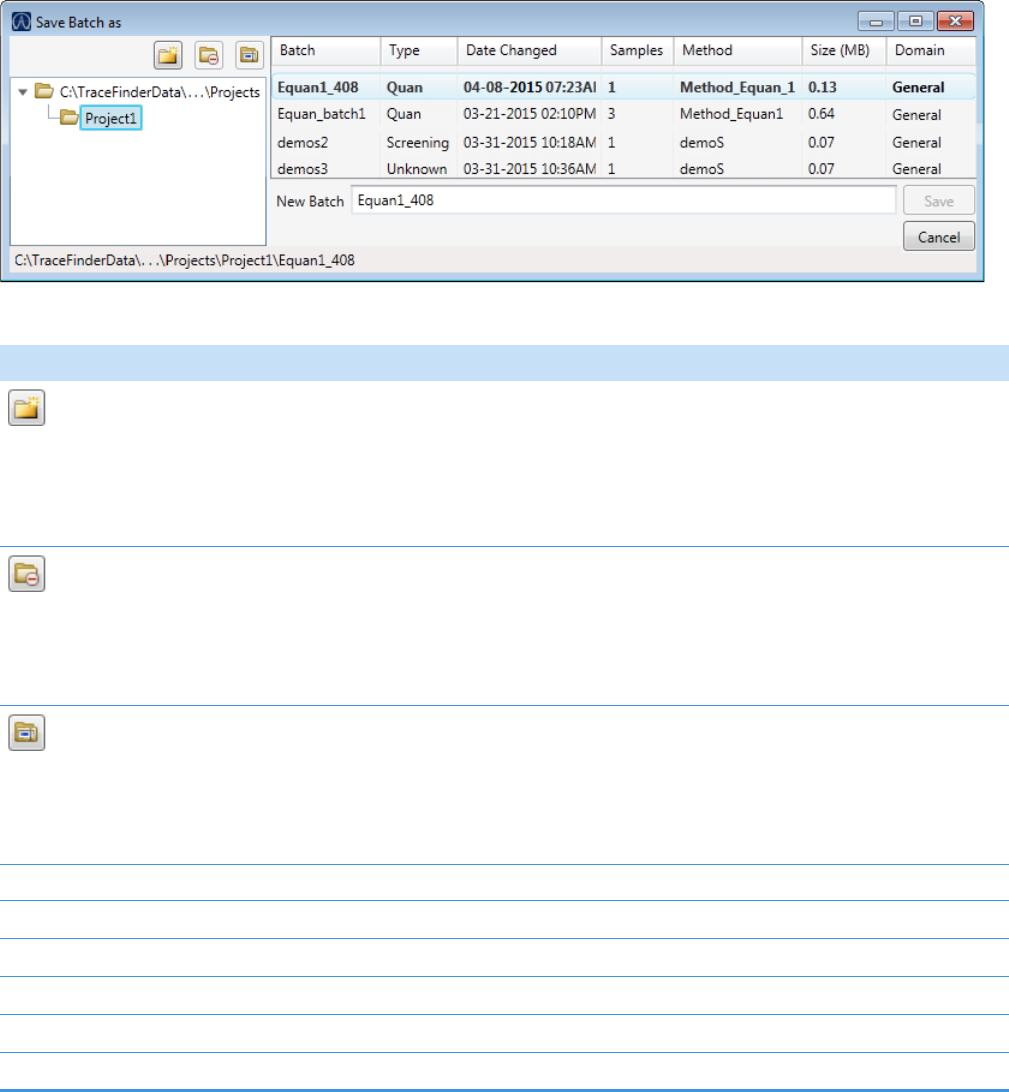
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 129
Save Batch As Dialog Box
Use the features in the Save Batch As dialog box to save a batch to a new name or to move a
batch to a different project folder.
Figure 27. Save Batch As dialog box
Table 25. Save Batch As dialog box parameters (Sheet 1 of 2)
Parameter Description
Create New Folder
Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Or, you can right-click and choose Create Folder.
Delete Folder
With no confirmation prompt, immediately removes the selected folder.
You cannot delete a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Or, you can right-click and choose Delete.
Rename Folder
Renames the selected folder.
You cannot rename a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Or, you can right-click and choose Rename.
Batch table
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date that the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
130 TraceFinder User Guide Thermo Scientific
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
New batch parameters
New Batch Name of the new batch to create.
Note If the Create button is not activated, you have entered a name that is already used or
you have not selected a method.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is created.
Buttons
Save Saves the batch to the specified name and folder and opens the Batch View for the new
batch.
Cancel Closes the Save Batch As dialog box without saving the batch.
Shortcut menu commands
Create Folder Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Delete Folder Immediately removes the selected folder. There is no prompt to confirm that you want to
delete the selected folder.
You cannot delete a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Rename Folder Renames the selected folder.
You cannot rename a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Expand Child Nodes Expands all project and subproject folders in the Project tree.
Collapse Child Nodes Collapses all project and subproject folders in the Project tree.
Table 25. Save Batch As dialog box parameters (Sheet 2 of 2)
Parameter Description

4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 131
Auto Samples Page for Quantitation Batches
The Auto Samples page identifies the Solvent or Matrix Blank samples to use for any Auto
Sample or Auto Sample and Reinject failure actions as specified on the Intelligent Sequencing
page of the method. Refer to Chapter 4, “Using the Method Development Mode for
Quantitation Methods,” in the TraceFinder Lab Director User Guide.
Each sample type that you specify for a failure action on the Intelligent Sequencing page must
be defined in the samples list on the Auto Samples page.
To add an auto sample type
1. Click Auto Samples in the Batch View navigation pane.
The Auto Samples page opens.
2. Right-click and choose Add Auto Sample, or in the toolbar click the Add New Auto
Sample icon, .
The application adds a Solvent sample to the sample list.
You can add, insert, or remove samples from this list as you would any sample list. See
Samples Page for Quantitation Batches.
3. To change the sample type to a Matrix Blank, click the Sample Type column and select
Matrix Blank from the list.
4. In the Injection Volume column for the sample, type a volume.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
5. In the Number of Injections column, type the number of injections available in the
designated Solvent or Matrix Blank vial.
After auto sample injections have occurred, you can return to this page to view the
number of Injections Used in each vial.
6. In the Vial Position column, type the vial position for the Solvent or Matrix Blank
sample.
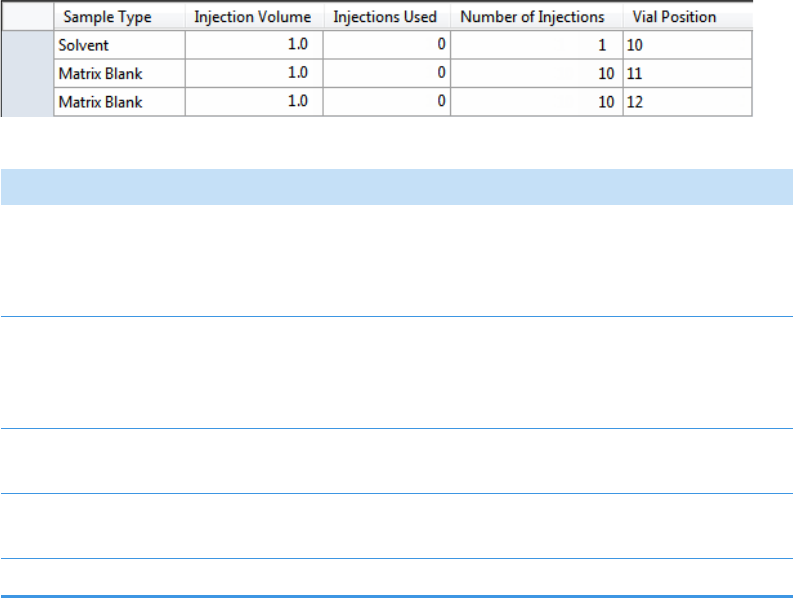
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
132 TraceFinder User Guide Thermo Scientific
Auto Samples Page
Use the Auto Samples page to specify the sample types to use with Intelligent Sequencing.
Figure 28. Auto Samples page
Table 26. Auto Samples page parameters
Column Description
Sample Type The sample type for the auto sample injection as specified on the
Intelligent Sequencing page—either Solvent or Matrix Blank.
Default: Solvent
Injection Volume The injection volume used for the sample acquisition as specified on
the Samples page.
Valid range: 0.1 through 5000 μL
Injections Used The number of times a vial has been used. The count is cumulative
across all batches.
Number of
Injections
The number of injections available in the designated Solvent or Matrix
Blank vial.
Vial Position Vial position for this sample type as specified on the Samples page.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 133
Reference Sample Page for Quantitation Batches
The Reference Sample page displays the reference samples that you selected for this batch.
To specify a chromatogram reference sample
1. In the Batch View, click Reference Sample.
An empty reference sample table opens.
2. Click the Add Reference Sample icon, , in the toolbar, or right-click and choose
Add Reference Sample.
The Open Chromatogram Reference Sample dialog box opens.
3. Select a project from the list of projects.
4. Select a subproject from the list of subprojects.
5. Select a batch from the list of batches.
The application displays only batches that were created using the current master method.
6. Select a sample from the list of processed samples.
The application displays all the processed samples in the selected batch. Before using a
sample as a reference sample, you must have processed the sample with the current master
method.
7. Click Open.
Note If you are using a new method, no reference samples appear here. You must first
process a batch using the current method to see the reference samples in this list.
4 Using the Analysis Mode for Quantitation Batches
Working in the Batch View for Quantitation Batches
134 TraceFinder User Guide Thermo Scientific
Threshold Samples Page for Quantitation Batches
For each group in a batch, you can specify a sample in the group as the threshold sample to
use in the Comparative View.
To specify a threshold sample
1. In the Batch View, click Threshold Samples.
2. Click the Sample list for each group and select a sample in the group to be the threshold
sample.
The Comparative View uses the threshold method and amount you specified in the method,
the group you created on the Samples page, and the threshold sample you selected on this
page to define the threshold guide that it displays in the sample peak plots.
To specify the method for creating a threshold guide, refer to Chapter 4, “Using the Method
Development Mode for Quantitation Methods,” in the TraceFinder Lab Director User Guide.
For information about creating groups, see Groups.
For information about using the threshold guide in the Comparative View, see Comparative
View for Quantitation Batches.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 135
Working in Data Review for Quantitation Batches
In the Data Review view, you can view the data generated by the master method. Use Data
Review to verify the data for a compound before you generate reports.
Follow these procedures:
•To open the Data Review view
•To move, dock, or float Data Review panes
•To restore the default layout
To open the Data Review view
1. Click Analysis in the navigation pane.
The Analysis navigation pane opens.
2. Click Data Review.
The Data Review navigation pane opens.
Choose from a Sample View, Compound View, Comparative View, or Qualitative View to
analyze the data generated by the master method.
In addition to these views, see the following topics:
•Compound Details for Quantitation Batches
•Column Parameters for Compound Results and Sample Results
To move, dock, or float Data Review panes
Follow the instructions in Appendix C, “Moving Data Review Panes.”
To restore the default layout
Choose View > Restore Default Layout.
The Sample View, Compound View, Comparative View, and Qualitative View have their
own defaults for which panes are displayed and in which location.
Displays the Comparative View for Quantitation Batches.
Displays the Compound View for Quantitation Batches.
Displays the Sample View for Quantitation Batches.
Displays the Qualitative View for Quantitation Batches.
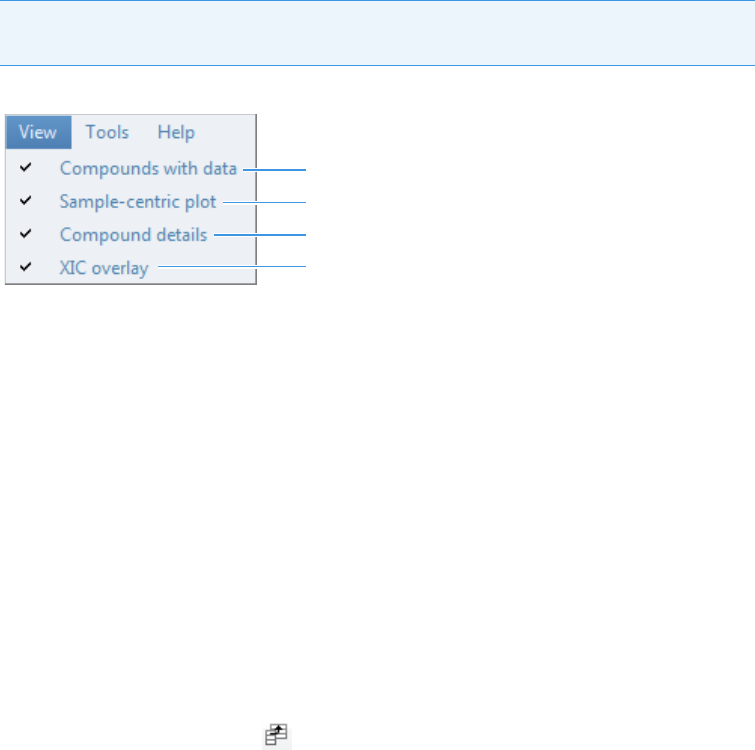
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
136 TraceFinder User Guide Thermo Scientific
Sample View for Quantitation Batches
The Sample View displays a list of all samples in the current batch and the compound results
for all compounds in the method.
To display or hide a pane on the Sample View page
From the View menu, choose to display or hide any of the following panes.
A check mark indicates a displayed pane.
The Sample View display includes the following features:
•Samples Pane
•Compound Results Pane
•Sample-centric Plot Pane
•XIC Overlay
Samples Pane
Use the Samples pane in the Sample View to select a specific sample. The associated
Compound Results Pane displays all compounds in the method and flags any compound with
errors in the selected sample.
To hide or display columns in the Samples pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
Note The Samples pane is required for the Sample View display. You cannot hide the
Samples pane.
Displays the XIC Overlay.
Displays the Sample-centric Plot Pane.
Displays the Compound Results Pane.
Displays the Compound Details for Quantitation Batches.
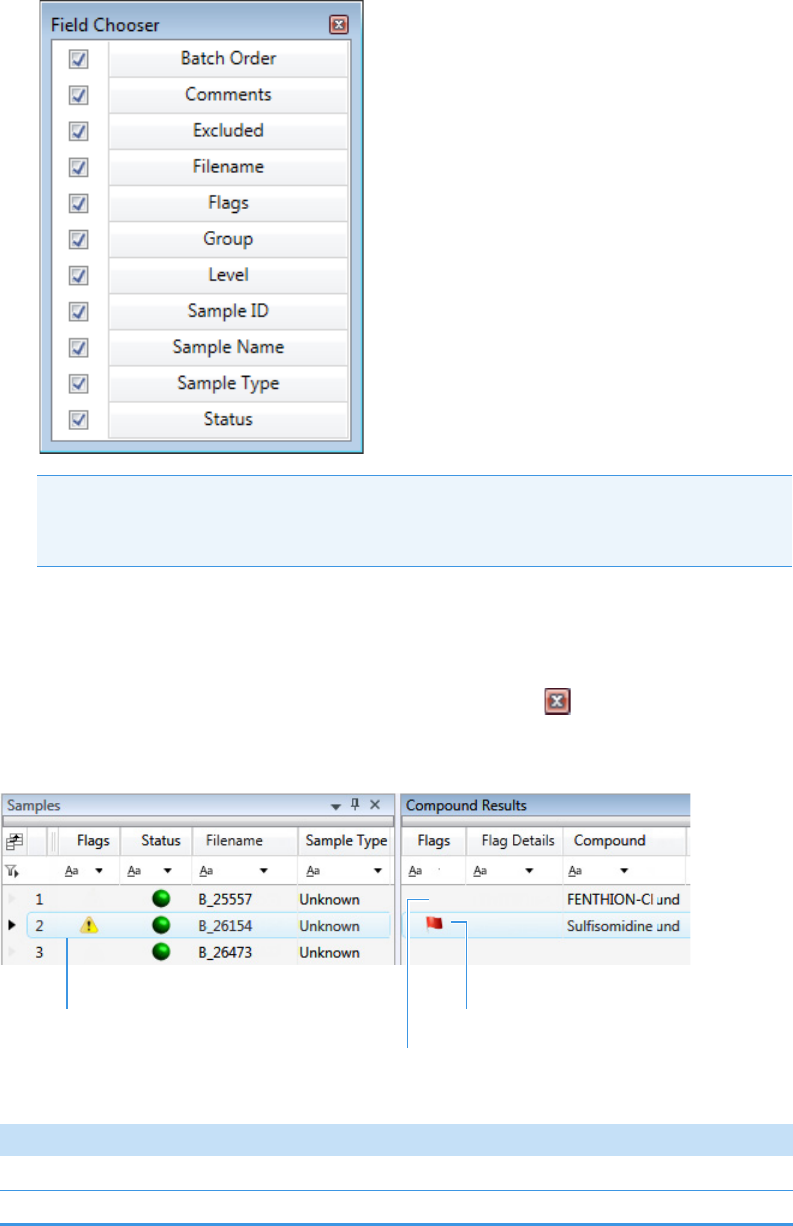
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 137
The Field Chooser displays all available columns of data for the Samples pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Samples pane.
3. When you are finished modifying the column display, click to close the Field
Chooser.
Figure 29. Samples pane
Note The Field Chooser also lists any custom columns that you defined in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in
the TraceFinder Lab Director User Guide.
Table 27. Samples pane columns (Sheet 1 of 2)
Column Description
Batch Order Sequentially numbers the samples.
Comments User-defined comments for the sample.
No compound error in the selected sample
Compound error in the selected sample
Selected sample

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
138 TraceFinder User Guide Thermo Scientific
Excluded Turns a compound on or off in the calibration curve in the Compound
Details pane.
Filename A user-defined name that identifies a sample.
Flags Caution flag displayed when a compound in the sample has an error. See
Caution Flags.
Group Threshold group to which a sample belongs. Samples can be viewed by
group in the Comparative View of Data Review.
Level The level defined for a calibration sample or quality control sample.
Sample ID A user-defined, alphanumeric string that identifies a sample.
Sample Name A user-defined name that identifies a sample.
Sample Type Defines how the application processes the sample data. Each sample is
classified as one of the following sample types: Matrix Blank, Solvent, Cal
Std, QC Std, or Unknown.
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Note When you include unknown screening features in the
quantitation method and you choose to process with only the
quantitation criteria, the Sample View shows the Status for the samples
as acquired and processed ( ), and the Unknown Screening View shows
the Status for the samples as acquired but not processed ( ).
Note When you include unknown screening features in the quantitation
method and you choose to process with only the unknown screening
criteria, both the Sample View and the Unknown Screening View show
the Status for the samples as acquired and processed ( ).
Table 27. Samples pane columns (Sheet 2 of 2)
Column Description
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 139
Compound Results Pane
Use the Compound Results pane in the Sample View to select a specific compound in the
selected sample. The associated Sample-centric Plot Pane highlights the selected compound.
To hide or display columns in the Compound Results pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
The Field Chooser displays all available columns of data for the Compound Results pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application displays or hides the column in the Compound Results pane.
3. When you are finished modifying the column display, click to close the Field
Chooser.
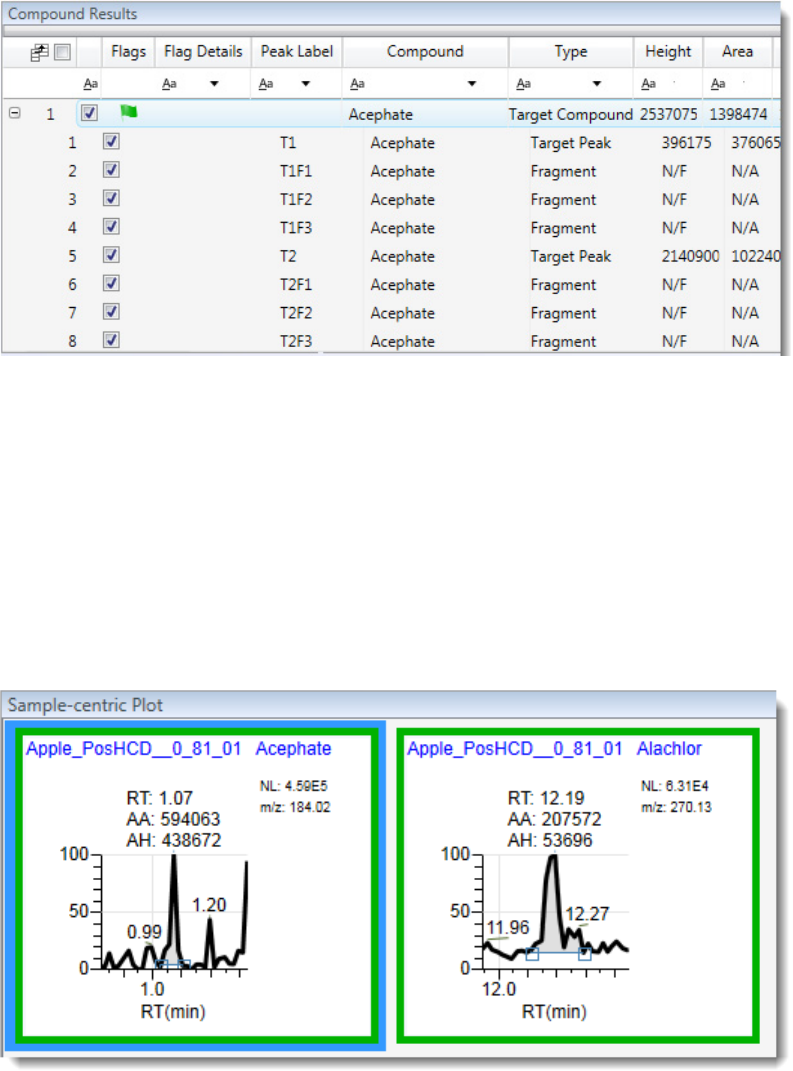
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
140 TraceFinder User Guide Thermo Scientific
Figure 30. Compound Results pane
For descriptions of the column parameters in the Compound Results pane, see Common
Column Parameters.
Sample-centric Plot Pane
The Sample-centric Plot pane in the Sample View displays the chromatogram, retention time,
area, height, and signal-to-noise ratio for each compound in the Compound Results pane.
The application highlights the chromatogram for the compound that is currently selected in
the Compound Results pane.
You can use the Sample-centric Plot pane to view an overlay of the quantitation and
confirming peaks for each sample in the batch.
Follow these procedures:
•To select a sample to view
•To display details for a compound
•To change the number of rows and columns in the display
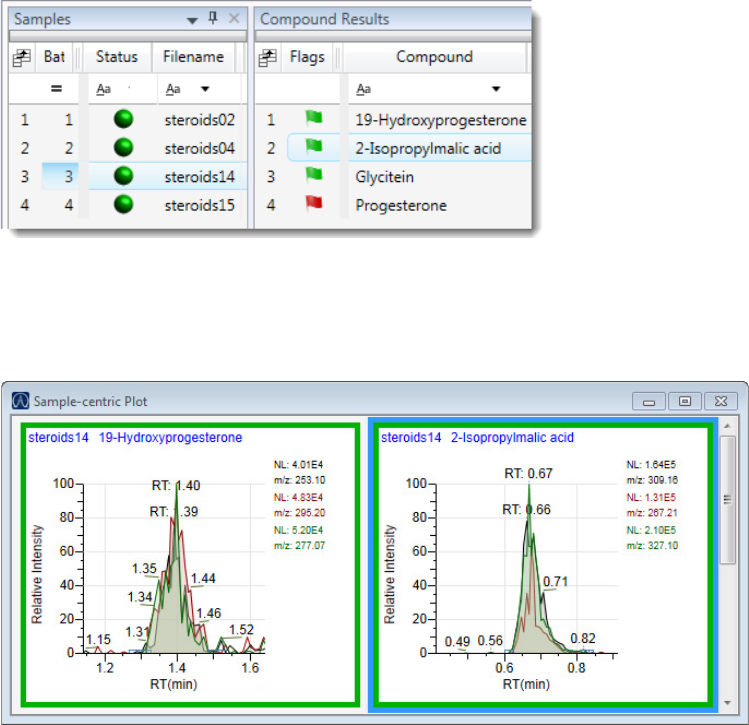
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 141
•To change the target and confirming peaks that are displayed
•To zoom in on a peak
•To copy a plot to another format
To select a sample to view
1. In the Samples pane, select the sample that you want to view.
The Sample-centric Plot pane displays the compound chromatograms for all compounds
in the selected sample. A blue border indicates the compound in the sample that is
currently selected in the Compound Results pane.
Figure 31. Sample-centric Plot pane
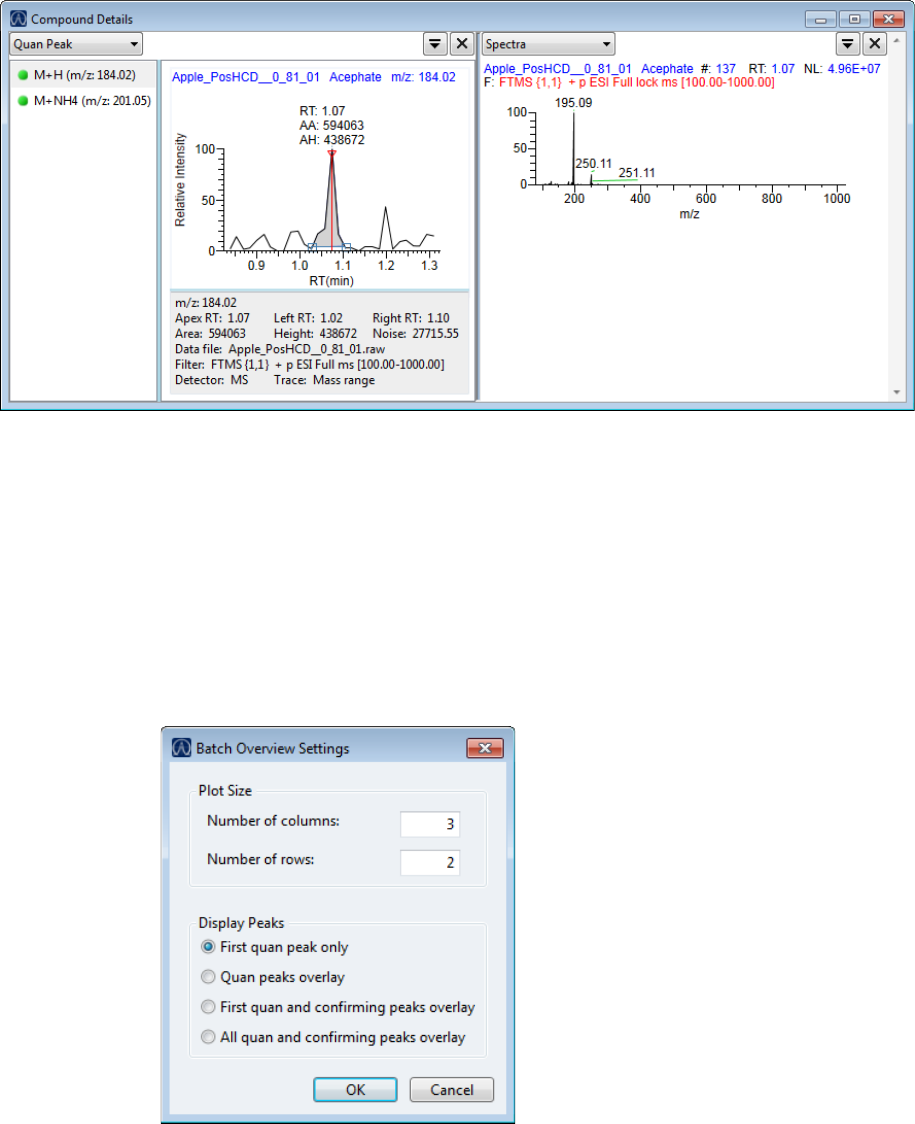
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
142 TraceFinder User Guide Thermo Scientific
To display details for a compound
Double-click the chromatogram in the Sample-centric Plot pane.
The Compound Details pane opens.
The Compound Details pane displays information about the quantitative peak,
calibration curve, confirming ion, internal standard, reference peak, ion overlay, and
spectra for the compound.
For a detailed description of the available information in the Compound Details pane, see
Compound Details for Quantitation Batches.
To change the number of rows and columns in the display
1. Right-click and choose Display Settings.
The Batch Overview Settings dialog box opens.
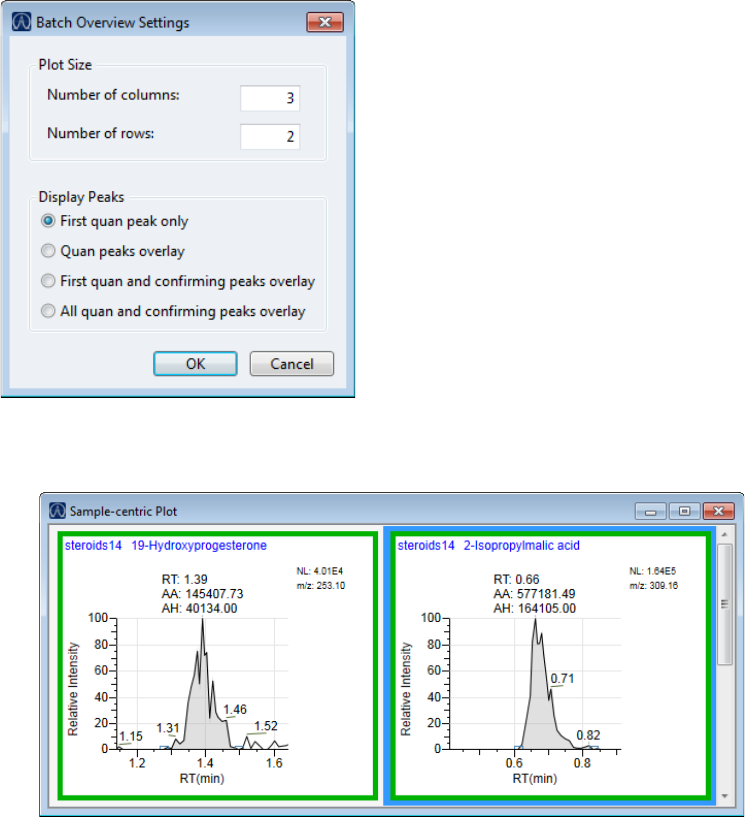
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 143
2. To change the number of rows or columns to fit in the Sample-centric Plot pane, type
new values in the Number of Columns or Number of Rows boxes.
These values do not change the number of rows (compounds) and columns (samples) that
are available in the Sample-centric Plot pane. They determine how many rows and
columns you want to view at one time in the display. The default is two rows and three
columns. Use the scroll bar to view all compound chromatograms.
3. Click OK.
To change the target and confirming peaks that are displayed
1. Right-click and choose Display Settings.
The Batch Overview Settings dialog box opens.
2. Select any of the following display options:
• First Quan Peak Only
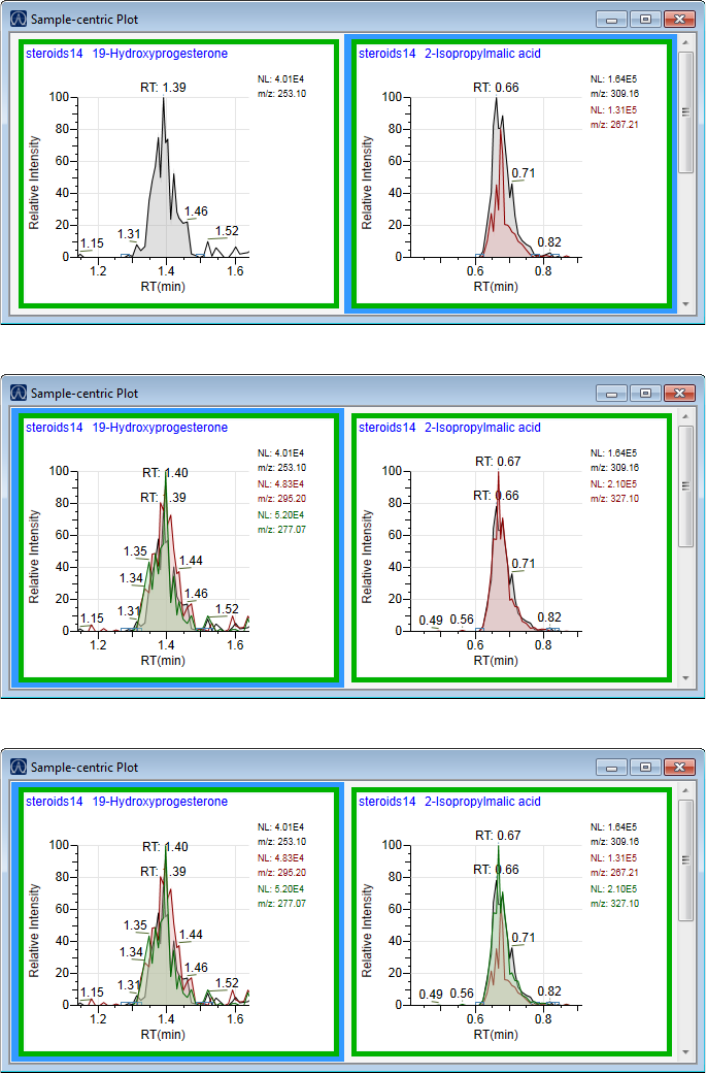
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
144 TraceFinder User Guide Thermo Scientific
•Quan Peaks Overlay
• First Quan and Confirming Peaks Overlay
• All Quan and Confirming Peaks Overlay
3. Click OK.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 145
To zoom in on a peak
1. In any of the chromatogram cells, drag your cursor to delineate a rectangle around the
peak.
The delineated area expands to fill the chromatogram area.
2. To restore the default view, right-click the chromatogram plot and choose Reset Scaling.
To copy a plot to another format
1. Right-click the plot that you want to copy and choose Copy to Clipboard.
The application copies the plot graphic to the Clipboard.
2. In the source format (for example, an Excel spreadsheet or email window), right-click and
choose Paste.
The application pastes the graphic into the new format.
XIC Overlay
Use the XIC Overlay pane to view specific groups of peaks. You can choose to view all peaks,
selected peaks only, or the top 20 most intense peaks (by area). The XIC Overlay plot is a
collection of overlaid, extracted m/z ion plots that use a different color for each peak.
Follow these procedures:
•To display all peaks in a sample
•To display only selected peaks in a sample
•To display the top 20 peaks in a sample
•To focus on a specific peak
To display all peaks in a sample
1. In the Samples pane, select the sample whose peaks you want to view.
2. In the XIC Overlay pane, select the All check box.
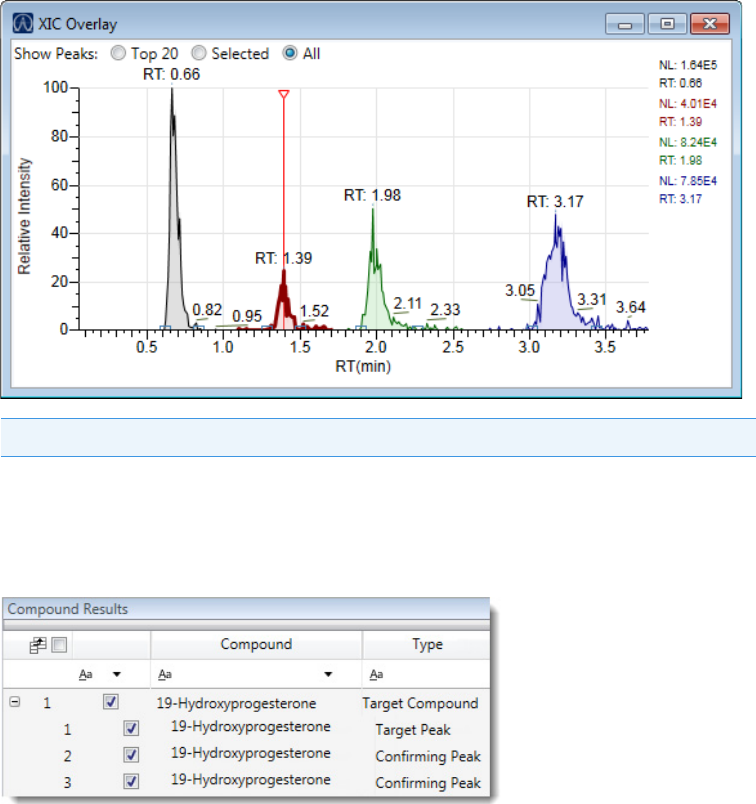
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
146 TraceFinder User Guide Thermo Scientific
The application displays all the peaks in the selected sample.
Figure 32. XIC Overlay pane showing all peaks
To display only selected peaks in a sample
1. In the Samples pane, select the sample whose peaks you want to view.
2. In the Compound Results pane, select the check box for each peak that you want to view.
3. In the XIC Overlay pane, choose the Selected option.
Note If there are more than 1000 peaks in the sample, performance might be slow.
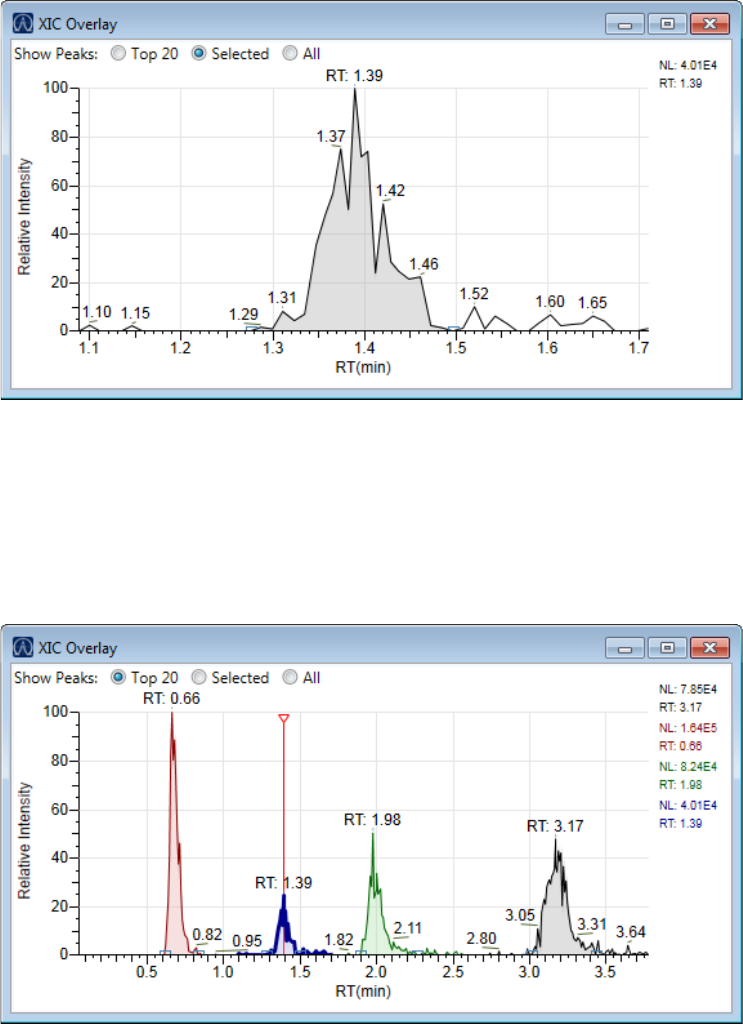
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 147
The application displays only the selected peaks.
Figure 33. XIC Overlay pane showing selected peaks
To display the top 20 peaks in a sample
1. In the Samples pane, select the sample whose peaks you want to view.
2. In the XIC Overlay pane, select the Top 20 option.
The application displays the top 20 peaks in the selected sample.
Figure 34. XIC Overlay pane showing the top 20 peaks
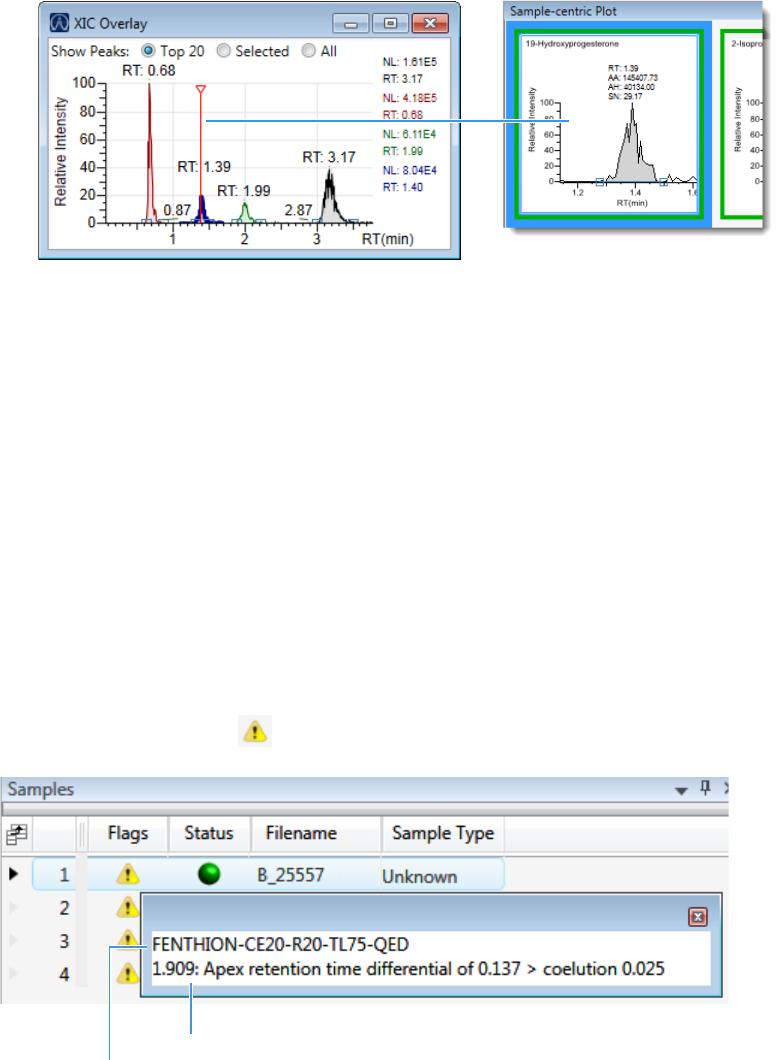
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
148 TraceFinder User Guide Thermo Scientific
To focus on a specific peak
In the XIC Overlay pane, click any displayed peak.
The application indicates the selected peak with a red, vertical marker and updates the
focus in the other Data Review panes with data for that peak.
Caution Flags
In the Sample View, the application displays caution flags in both the Samples pane and the
Compound Results pane.
See the following topics:
•Flags in the Samples Pane
•Flags in the Compound Results Pane
•Error Indicators in the Sample-centric Plot Pane
Flags in the Samples Pane
The Flags column in the Samples pane displays a caution flag if any compound in the sample
is not in compliance with the method criteria.
Click the caution flag icon, , to display the details. Information in the pop-up box shows
the compound that is in error and describes the exact error condition.
Error condition
Compound name

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 149
Flags in the Compound Results Pane
The Flags column in the Compound Results pane displays a flag if the compound in the
selected sample is not in compliance with the method criteria.
Hold your cursor over the flag icon, , to display details for the compound in the selected
sample.
Colored flags in the Compound Results pane have the following meaning:
Red for compounds that have violated (or are activated by) any of the values set in
the method. Refer to Chapter 4, “Using the Method Development Mode for
Quantitation Methods,” in the TraceFinder Lab Director User Guide.
Red for compounds that are outside the specified ion ratio range. See Ion ratio
failure flag.
Orange for compounds that are below the limit of quantitation (LOQ), below the
limit of detection (LOD), or between the LOD and LOQ values specified in the
method. For descriptions of these limits, refer to Chapter 4, “Using the Method
Development Mode for Quantitation Methods,” in the Trace Finde r Lab Direct o r
User Guide.
Green for “found” compounds that are over the limit of reporting (LOR) amount
specified in the method. For a description of the LOR limit, refer to Chapter 4,
“Using the Method Development Mode for Quantitation Methods,” in the
TraceFinder Lab Director User Guide.
Yellow for compounds that are equal to or below the LOR amount specified in the
method.
Yellow for compounds that are not found in Calibrator or QC sample types.
The Compound Results pane does not flag compounds that are not found in
Specimen sample types.
No flag for compounds that have no errors or where no report options are selected.
Note These criteria for flag states do not apply to Matrix Blank sample types when the
compound is an internal standard.
Error condition of the Sulfisomidine
compound in the selected sample
Selected sample
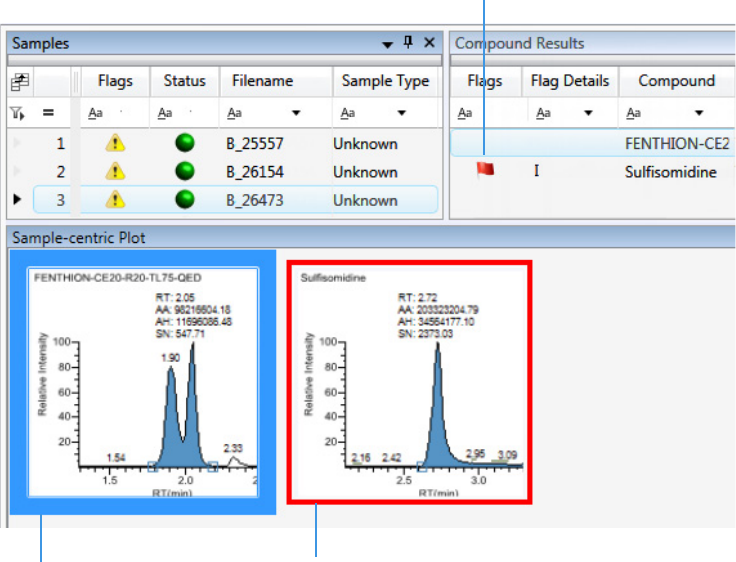
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
150 TraceFinder User Guide Thermo Scientific
Error Indicators in the Sample-centric Plot Pane
In the Sample-centric Plot pane, peak plots are outlined with the color of their associated
error flag. In the following example, the FENTHION peak plot is highlighted in blue to
indicate that FENTHION is the selected compound, and the Sulfisomidine peak plot is
outlined in red to indicate that the Sulfisomidine compound in the selected sample is outside
the specified ion ratio range.
Figure 35. Ion ratio failure flag
Red flag indicating ion ratio failure
Peak with ion ratio failure outlined in red
Selected peak
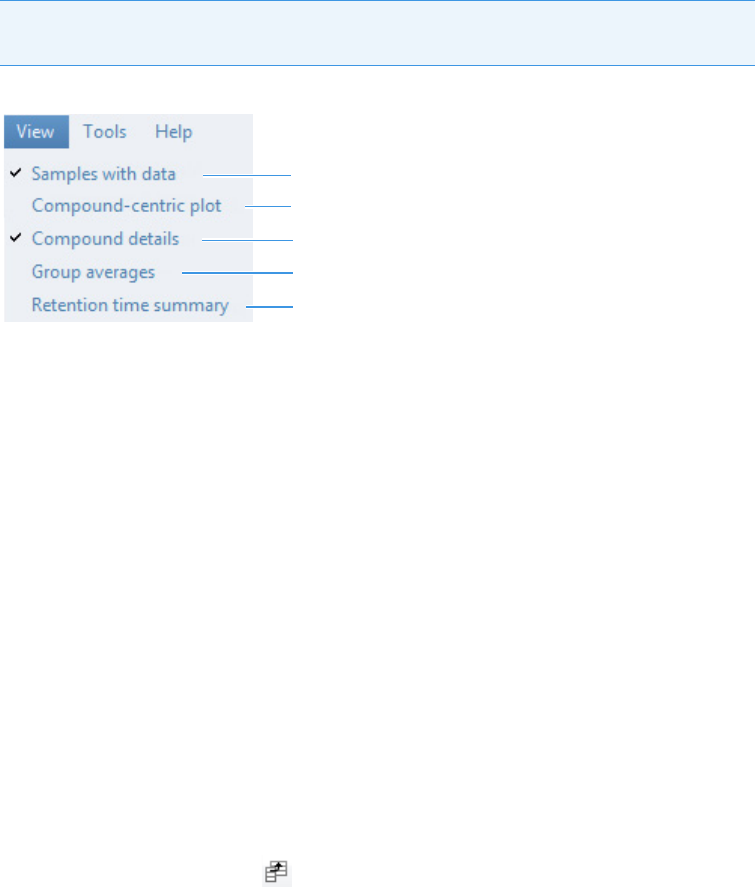
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 151
Compound View for Quantitation Batches
The Compound View displays a list of all compounds available in the method, all samples in
the current batch, and the peak plots for all found compounds.
To display or hide a pane on the Compound View page
From the View menu, choose to display or hide any of the following panes.
A check mark indicates a displayed pane.
The Compound View includes the following features:
•Compounds Pane
•Sample Results Pane
•Compound-centric Plot
•Group Averages
•Retention Time Summary
•Caution Flags
Compounds Pane
Use the Compounds pane in the Compound View to select a specific compound. The Sample
Results Pane displays all samples in the batch and flags any sample that contains errors
associated with the selected compound.
To hide or display columns in the Compounds pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
Note The Compounds pane is required for the Compound View display. You cannot
hide the Compounds pane.
Displays the Compound-centric Plot.
Displays the Sample Results Pane.
Displays the Compound Details for Quantitation Batches.
Displays the Group Averages.
Displays the Retention Time Summary.
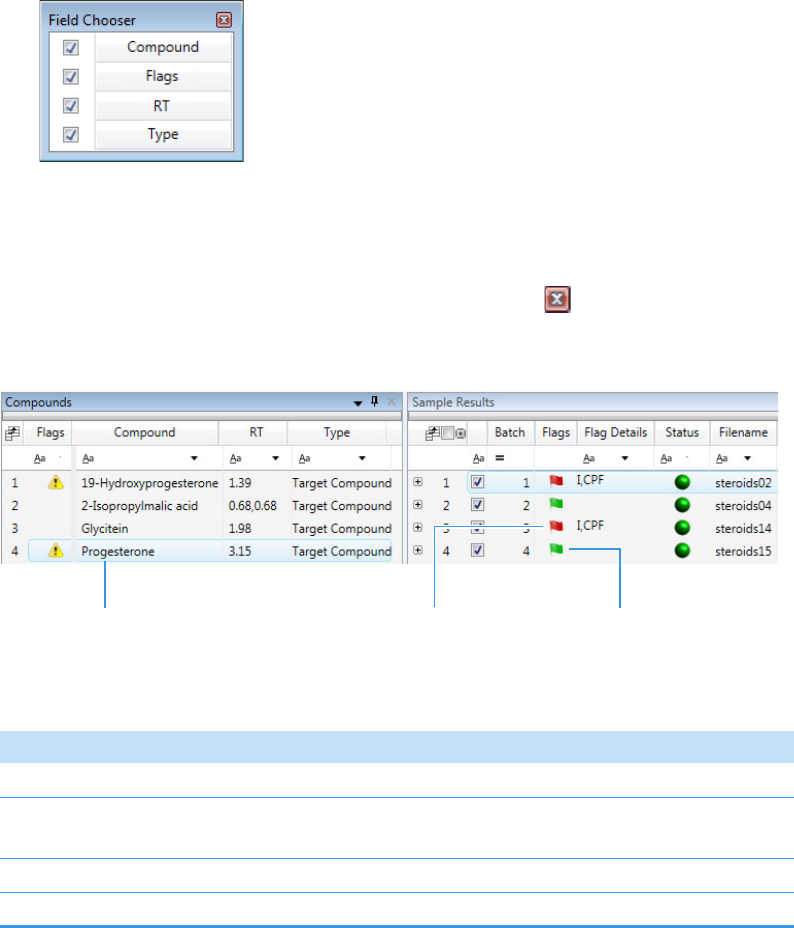
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
152 TraceFinder User Guide Thermo Scientific
The Field Chooser displays all available columns of data for the Compounds pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Compounds pane.
3. When you are finished modifying the column display, click to close the Field
Chooser.
Figure 36. Compounds pane
Table 28. Compounds pane columns
Column Description
Flags Caution flag displayed when a compound has an error in any of the samples.
Compound Compound names as identified in the library. If there is no library selected in
the method template, the compound name is identified as peak@RT.
RT Expected retention time for the compound.
Type Specified compound type: Target Compound or Internal Standard.
Selected compound No error associated with
the selected compound in
this sample
Error associated with
the selected compound
in this sample
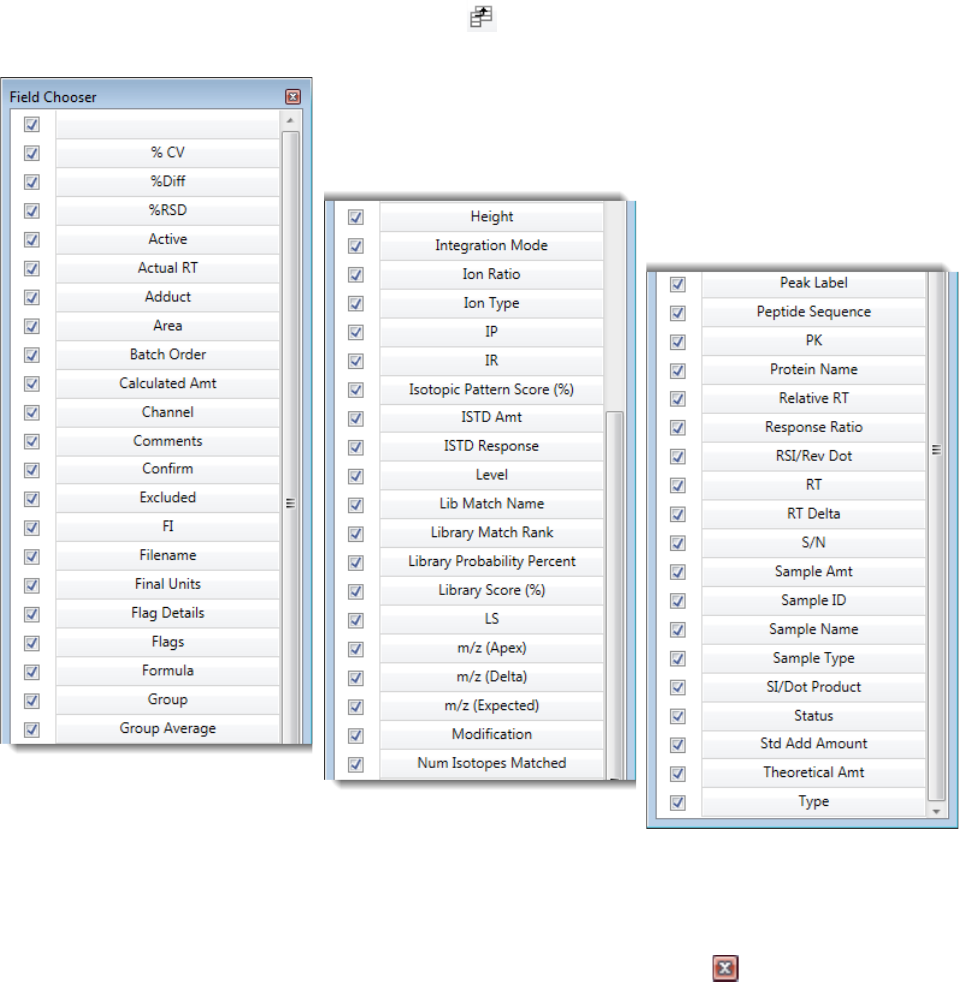
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 153
Sample Results Pane
Use the Sample Results pane in the Compound View to select a specific compound in a
specific sample. The Compound-centric Plot pane highlights the selected sample/compound
and displays the following information about the compound: chromatogram, retention time,
area, height, and signal-to-noise ratio.
To hide or display columns in the Sample Results pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
The Field Chooser displays all default columns of data for the Sample Results pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Sample Results pane.
3. When you are finished modifying the column display, click to close the Field
Chooser.
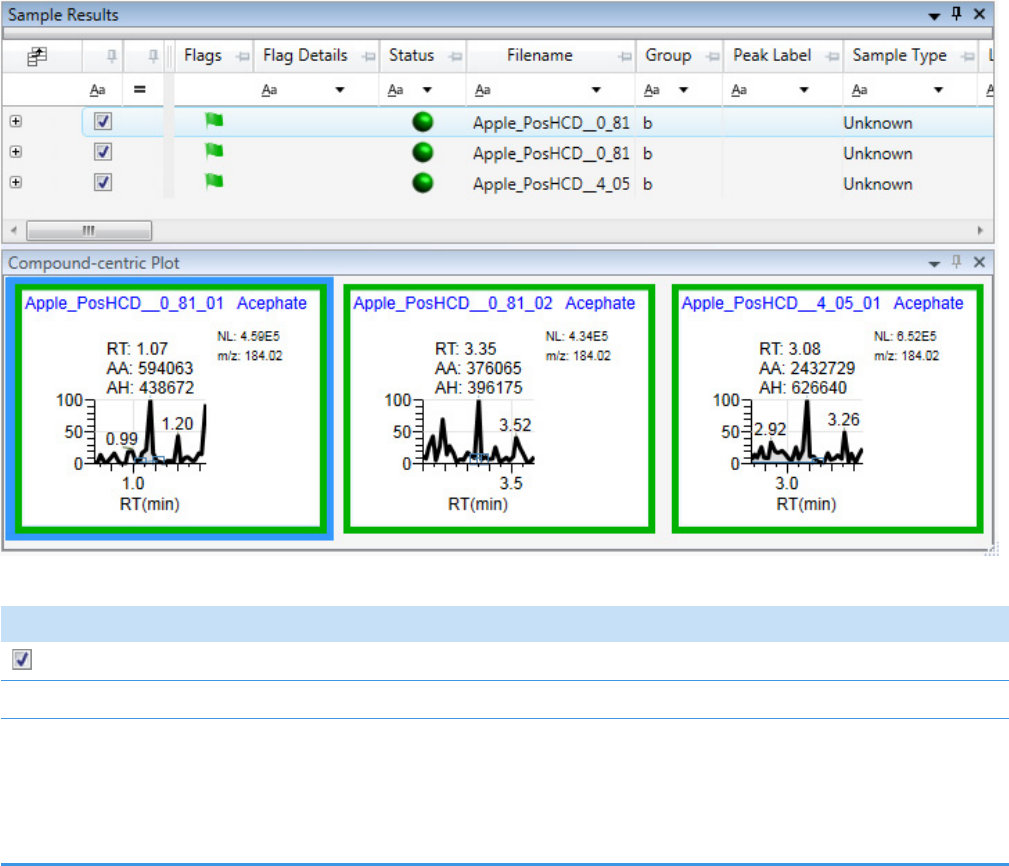
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
154 TraceFinder User Guide Thermo Scientific
Figure 37. Sample Results pane
Table 29. Sample Results pane columns (Sheet 1 of 2)
Column Description
Displays the selected samples in the other Data Review panes.
Flags Displays a caution flag when a compound within the sample has an error.
Flag Details Indicates the type of error:
• I: Confirming ion coelution failure or Ion ratio failure
• A: Amount error
• B: Matrix blank error
• H: Peak not found
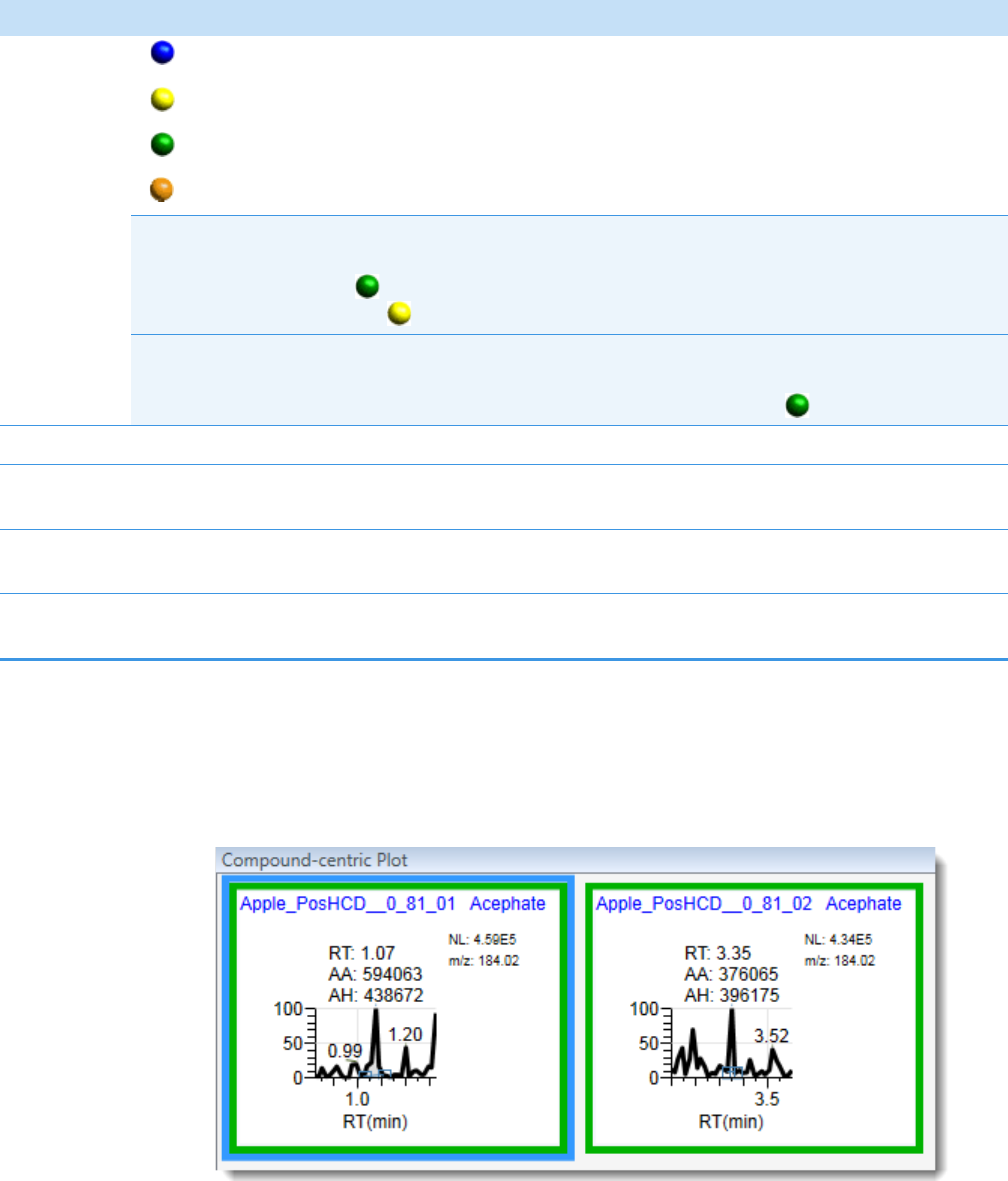
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 155
Compound-centric Plot Pane
The Compound-centric Plot pane in the Compound View displays the compound
chromatogram, retention time, area, height, and signal-to-noise ratio associated with the
selected compound in each of the samples in the batch. The application highlights the
compound chromatogram for the sample that is currently selected in the Sample Results pane.
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Note When you include unknown screening features in the quantitation method and you choose to
process with only the quantitation criteria, the Sample View shows the Status for the samples as
acquired and processed ( ), and the Unknown Screening View shows the Status for the samples as
acquired but not processed ( ).
Note When you include unknown screening features in the quantitation method and you choose to
process with only the unknown screening criteria, both the Sample View and the Unknown
Screening View show the Status for the samples as acquired and processed ( ).
Filename Specifies a user-defined name that identifies a sample.
Group Specifies the threshold group to which a sample belongs. Samples can be viewed by group in the
Comparative View of Data Review.
Sample Type Defines how the application processes the sample data. Each sample is classified as one of the
following sample types: Matrix Blank, Solvent, Cal Std, QC Std, or Unknown.
The remainder of the columns in the Sample Results pane are common to both the Sample View and the Compound
View displays. See Common Column Parameters.
Table 29. Sample Results pane columns (Sheet 2 of 2)
Column Description
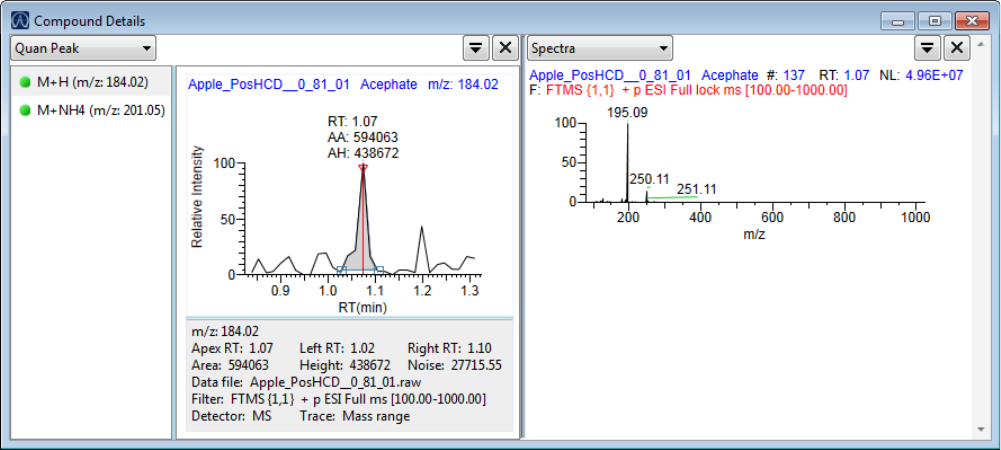
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
156 TraceFinder User Guide Thermo Scientific
To display details for a compound
Double-click the chromatogram in the Compound-centric Plot pane.
The application adds the Compound Details pane to the window.
The Compound Details pane displays information about the quantitative peak, calibration
curve, confirming ion, internal standard, reference peak, ion overlay, and spectra for the
compound.
For details about the available information in the Compound Details pane, see Compound
Details for Quantitation Batches.
Compound-centric Plot
Use the Compound-centric Plot pane to view an overlay of the quantitation and confirming
peaks for each sample in the batch.
Follow these procedures:
•To select a sample to view
•To change the target and confirming peaks that are displayed
•To change the number of rows and columns in the display
•To copy a plot to another format
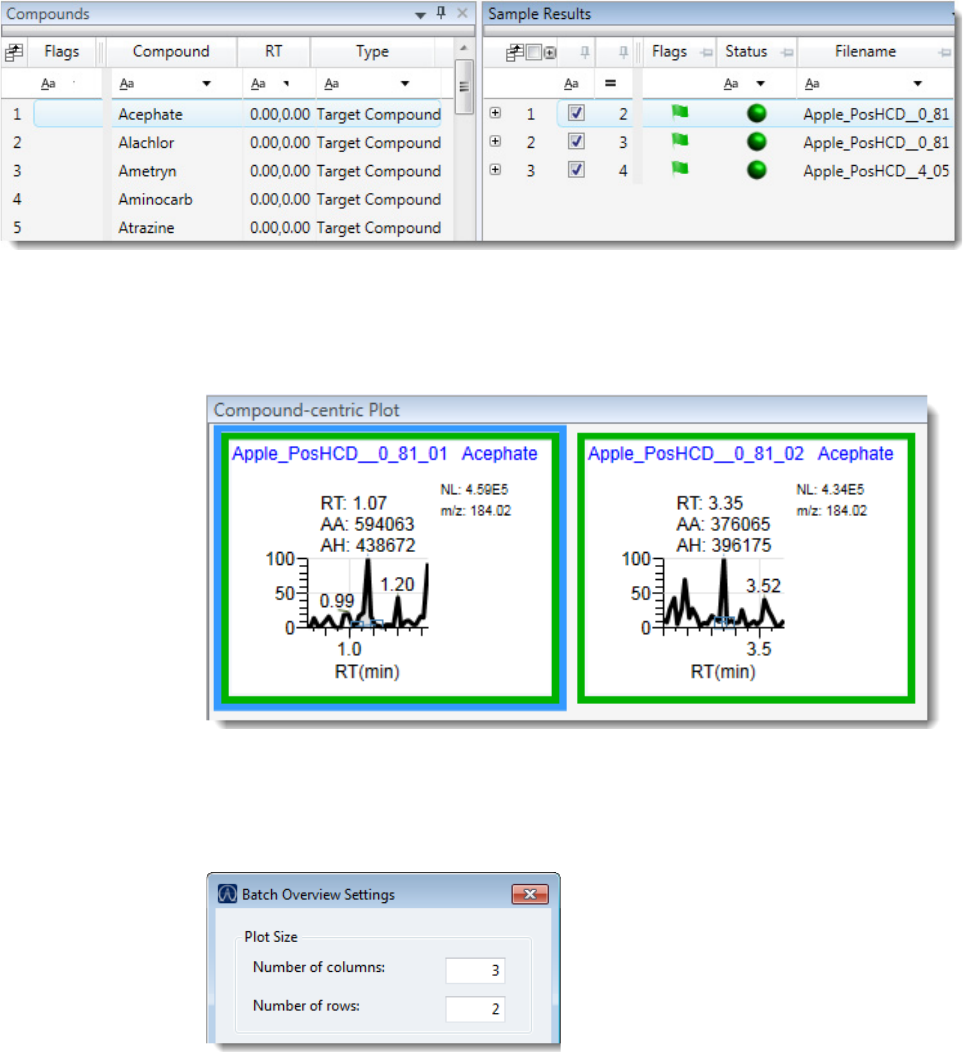
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 157
To select a sample to view
In the Compounds pane, select the compound that you want to view.
The Compound-centric Plot pane displays the compound chromatograms for the
selected compound in each sample in the batch. A blue border indicates the compound in
the sample that is currently selected in the Sample Results pane.
Figure 38. Compound-centric Plot pane
To change the number of rows and columns in the display
1. Right-click and choose Display Settings.
The Batch Overview Settings dialog box opens.
2. To change the number of rows or columns to fit in the Compound-centric Plot pane,
type new values in the Number of Columns or Number of Rows boxes.
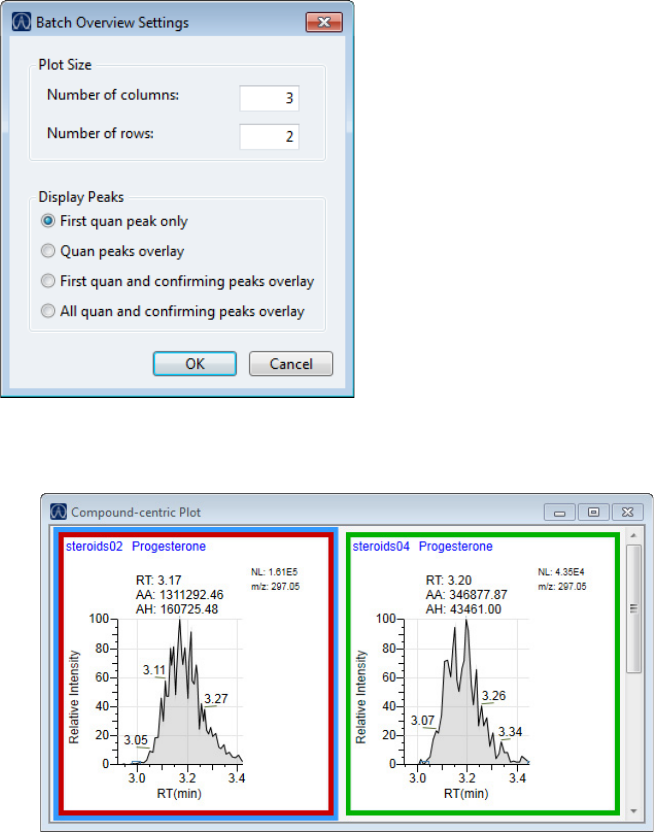
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
158 TraceFinder User Guide Thermo Scientific
These values do not change the number of rows (compounds) and columns (samples) that
are available in the Compound-centric Plot pane. They determine how many rows and
columns you want to view at one time in the display. The default is two rows and three
columns.
3. Click OK.
To change the target and confirming peaks that are displayed
1. Right-click and choose Display Settings.
The Batch Overview Settings dialog box opens.
2. Select any of the following display options:
• First Quan Peak Only
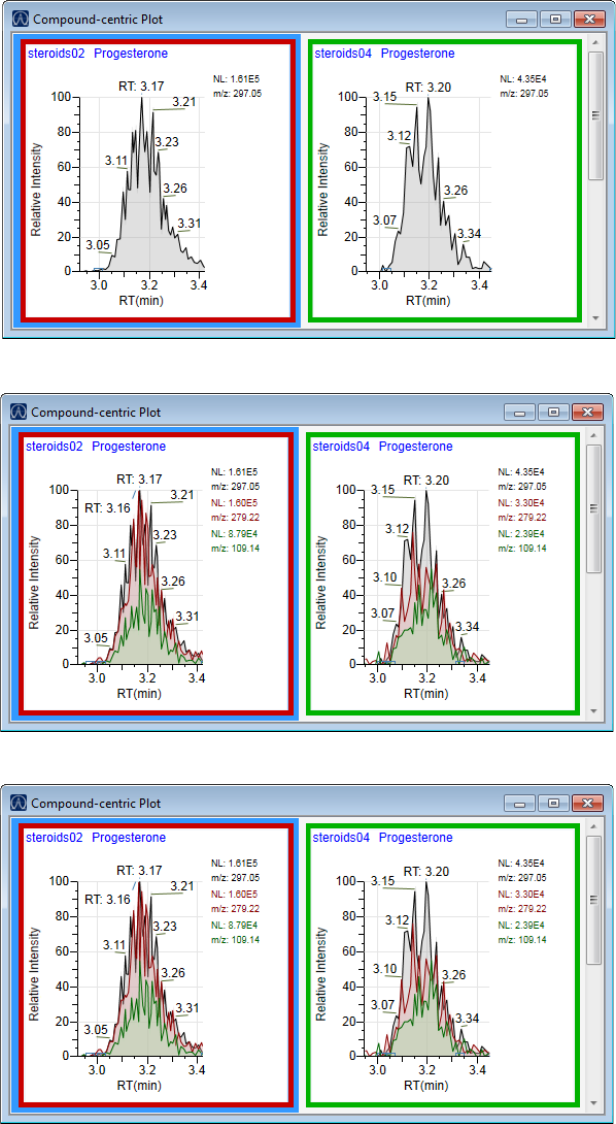
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 159
•Quan Peaks Overlay
• First Quan and Confirming Peaks Overlay
• All Quan and Confirming Peaks Overlay
3. Click OK.
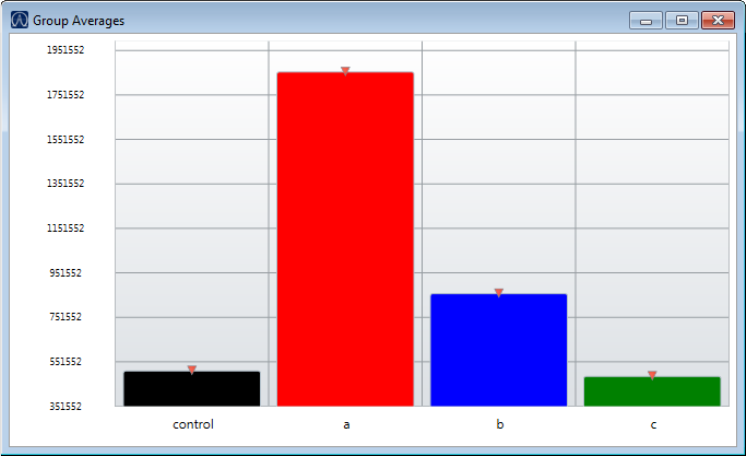
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
160 TraceFinder User Guide Thermo Scientific
To zoom in on a peak
1. In any of the chromatogram cells, drag your cursor to delineate a rectangle around the
peak.
The delineated area expands to fill the chromatogram area.
2. To restore the default view, right-click the chromatogram plot and choose Reset Scaling.
To copy a plot to another format
1. Right-click the plot that you want to copy and choose Copy to Clipboard.
The application copies the plot graphic to the Clipboard.
2. In the source format (for example, an Excel spreadsheet or email window), right-click and
choose Paste.
The application pastes the graphic into the new format.
Group Averages
Use the Group Averages pane to compare the peak areas of different samples to a control
group of samples. You must have at least two groups defined, and one group must be a control
group.
The control group is displayed to the left, and other samples and groups are listed in the order
in which they appear in the Batch View. To specify a group as the control group, see Groups.
Each group displays a colored bar that indicates the average response of the group.
Figure 39. Group Averages pane
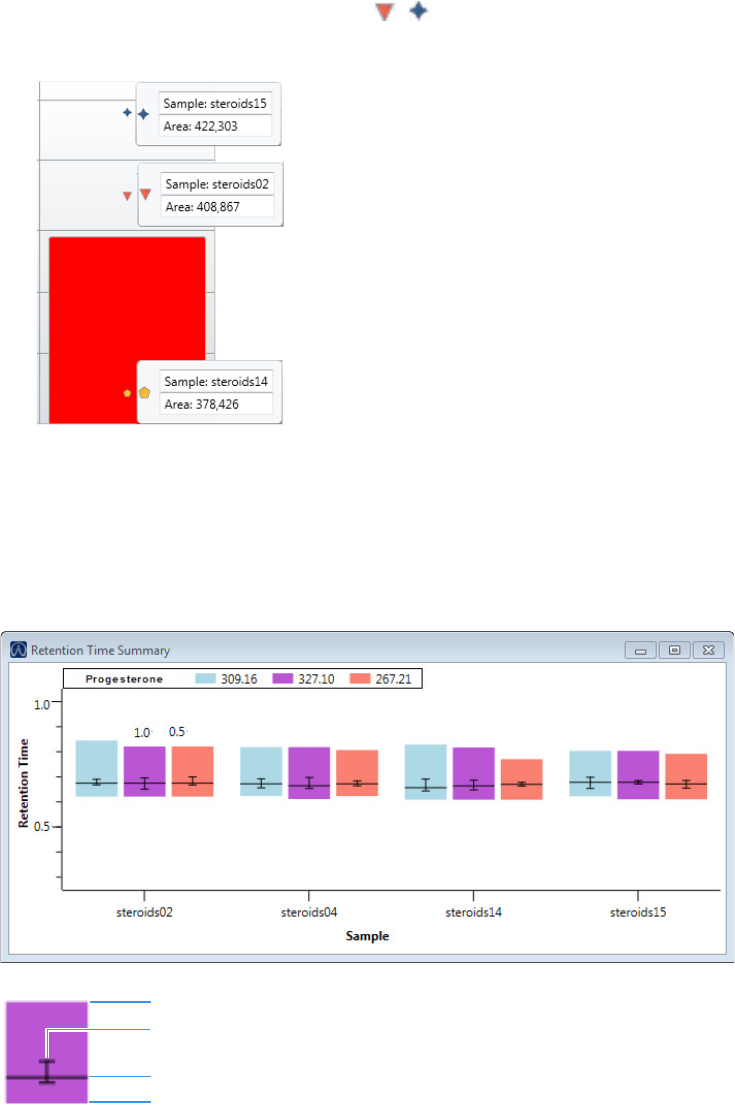
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 161
To display areas for each sample in the batch
Hold the cursor over an indicator icon ( , ).
The application displays the area for the selected compound in each represented sample.
Retention Time Summary
Use the Retention Time Summary pane to view variations of the retention times for a
compound across all samples in a batch. The Retention Time Summary pane displays each
color-coded compound for each sample in the batch.
Figure 40. Retention Time Summary pane
End RT specified in the method
Start RT specified in the method
Deviation
Actual RT
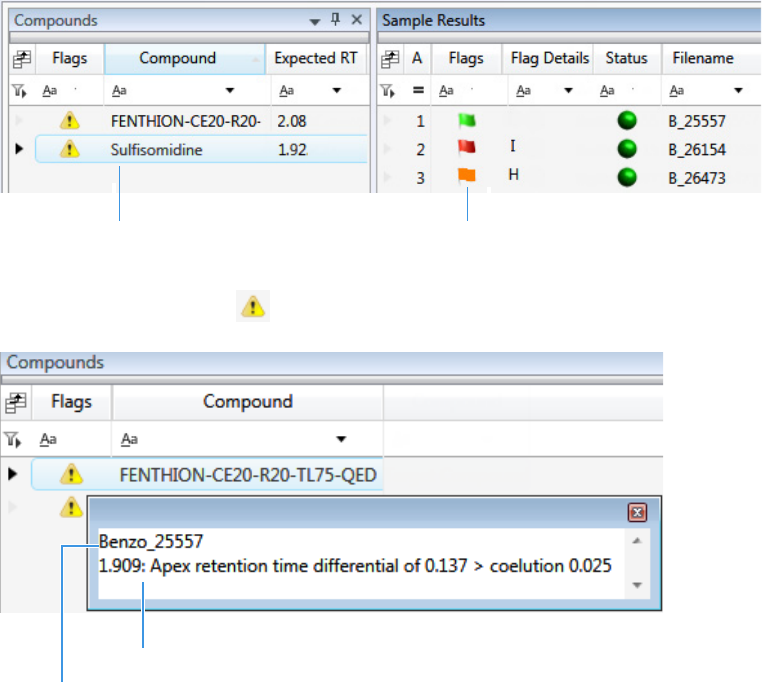
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
162 TraceFinder User Guide Thermo Scientific
Caution Flags
In the Compound View, the application displays caution flags in both the Compounds pane
and in the Sample Results pane.
See the following topics:
•Flags in the Compounds Pane
•Flags in the Sample Results Pane
•Error Indicators in the Compound-centric Plot pane
Flags in the Compounds Pane
The Flags column in the Compounds pane displays a caution flag if the compound in any of
the samples is not in compliance with the method criteria.
Click the caution flag icon, , to display the details. Information in the pop-up box shows
the sample where the compound is in error and describes the exact error condition.
Flags for the Sulfisomidine compound
in each sample
Selected Sulfisomidine compound
Error condition
Sample name
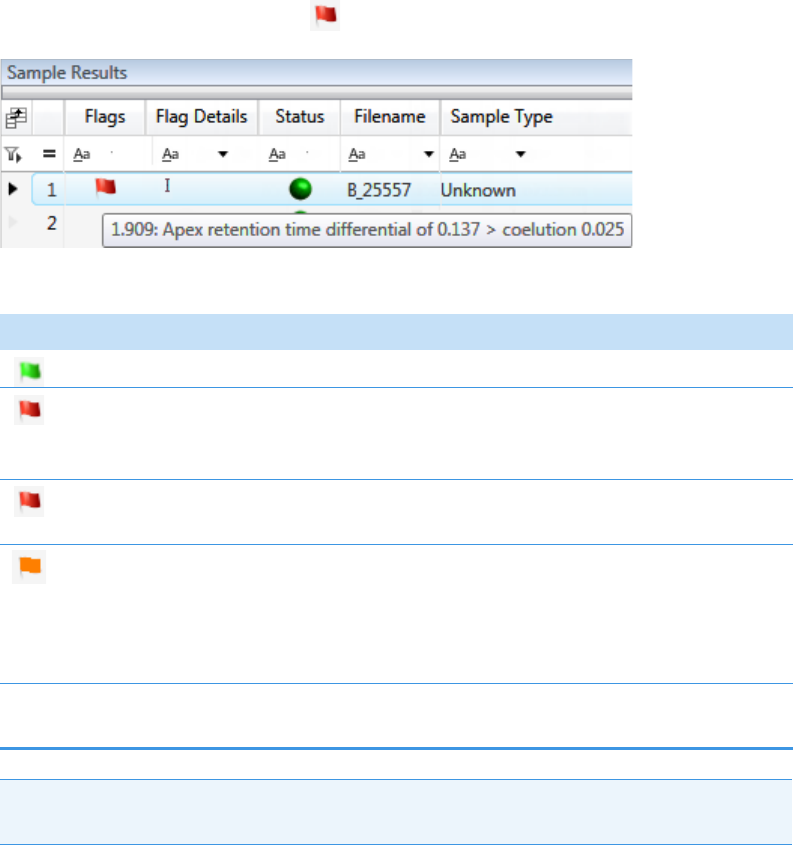
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 163
Flags in the Sample Results Pane
The Flags column in the Sample Results pane displays a flag if the selected compound in the
sample is not in compliance with the method criteria.
Hold your cursor over the flag icon, , to display the details for the selected compound in
the sample.
Colored flags in the Sample Results pane have the following meaning.
Flag Description
Green for compounds that are over the LOR amount specified in the method.
Red for compounds that have violated (or are activated by) any of the values set
in the method. Refer to Chapter 4, “Using the Method Development Mode for
Quantitation Methods,” in the TraceFinder Lab Director User Guide.
Red for compounds that are outside the specified ion ratio range. See Ion ratio
failure flag.
Orange for compounds that are not found in Cal Std or QC Std sample types.
“Not found” compounds are below the LOQ, below the LOD, or between the
LOD and LOQ values specified in the method. The Sample Results pane does
not flag compounds that are not found in Unknown sample types.
No flag for compounds that have no errors or where no report options are
selected.
Note These criteria for flag states do not apply to Matrix Blank sample types when the
compound is an internal standard.
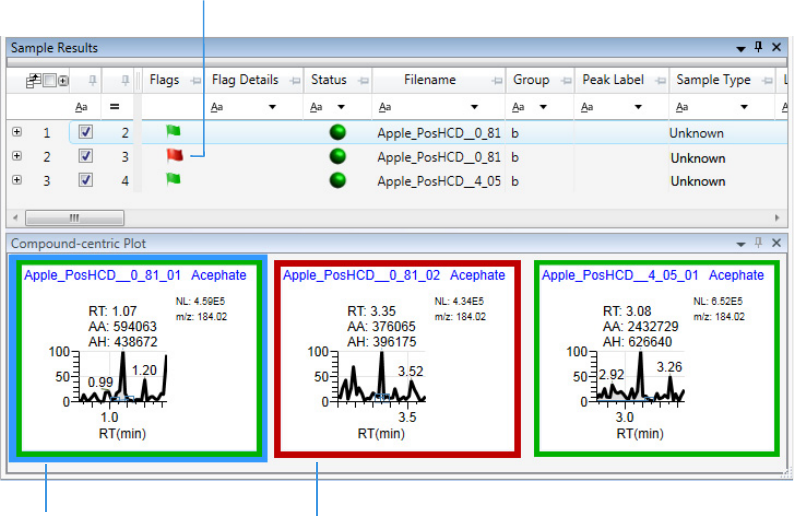
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
164 TraceFinder User Guide Thermo Scientific
Error Indicators in the Compound-centric Plot pane
In the Compound-centric Plot pane, peak plots are outlined with the color of their associated
error flag. In the following example, the peak plot is highlighted in blue to indicate that
Benzo_25557 is the selected sample and outlined in red to indicate that the FENTHION
compound in the selected sample is outside the specified ion ratio range.
Figure 41. Ion ratio failure flag
Compound Details for Quantitation Batches
The Compound Details pane is identical for Sample View and Compound View. Use the
Compounds Details pane to display any of the following types of data:
•Quan Peak
•Confirming Ions
•Calibration Curve
•Ion Overlay
•ISTD
•Reference Peak
•Spectra
•Library Match
Red flag indicating ion ratio failure
Peak with ion ratio failure
Selected peak
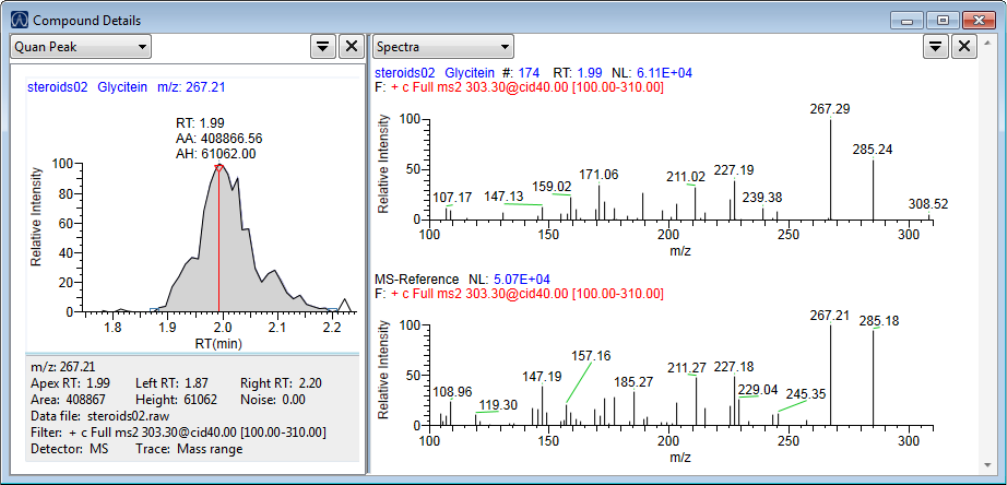
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 165
•Isotope
•Fragments
•Quan Peaks Overlay
•Caution Flags
To open the Compound Details pane
1. Double-click the chromatogram in the Sample-centric plot or the Compound-centric
plot.
The Compound Details pane opens.
By default, the first display pane shows the quantitative peak for the selected compound.
2. In the second pane, select the additional type of data that you want to display.
Follow these procedures to change the display of the peak data in either of the panes:
•To change the peak panes
•To zoom in on a peak
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
166 TraceFinder User Guide Thermo Scientific
To change the peak panes
In any of the peak panes, click to view a list of commands.
To zoom in on a peak
1. In any of the views, drag your cursor to delineate a rectangle around the peak or
spectrum.
The delineated area expands to fill the view.
2. To restore the default view, right-click the chromatogram plot and choose Reset Scaling.
Command Description
Make All Panels
the Same Size
Evenly divides the area to make all panes the same width. This
command does not change the pane height.
Move Panel Left Moves the current pane one space to the left. This command is
not available when the current pane is the leftmost pane.
Move Panel Right Moves the current pane one space to the right. This command is
not available when the current pane is the rightmost pane.
Add a Panel Adds an empty peak pane to the display. You can display a
maximum of four peak panes.
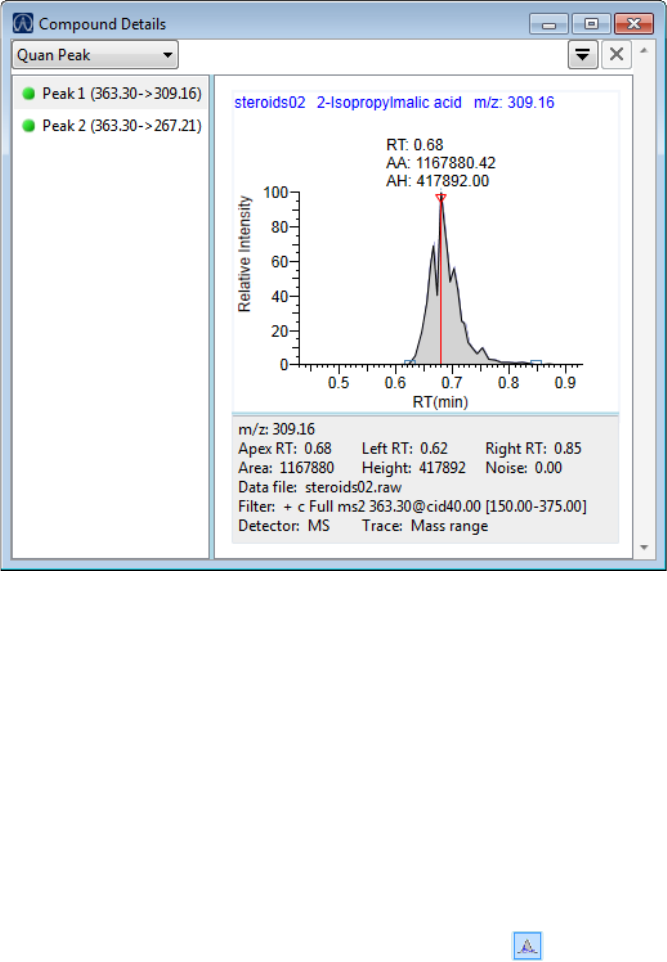
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 167
Quan Peak
A compound can have multiple quantitative peaks. You can switch between quantitative
peaks, but you cannot view multiple quantitative peaks at the same time. The Quan Peak
pane uses a unique shortcut menu. See Quan Peak pane shortcut menu commands.
Figure 42. Quantitative peak pane with multiple quantitative peaks
Follow these procedures to modify the quantitative peak data:
•To manually integrate a peak by swiping the peak
•To manually integrate a peak by moving the peak delimiters
•To remove a manually created peak
•To switch between Method, Manual, and User integration modes
•To customize the labels in the display
•To modify the peak detection settings
To manually integrate a peak by swiping the peak
1. Right-click anywhere in the quantitative peak pane and then choose Swipe Manual
Integration, or click the icon in the upper right corner, .
2. Click the left base of the peak that you want to identify.
3. Drag to the right base and release the mouse.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
168 TraceFinder User Guide Thermo Scientific
The application places the peak delimiter tags at these locations and automatically
updates the peak values (area, height, and so forth) in the result set.
To manually integrate a peak by moving the peak delimiters
1. Hold your cursor over one of the two peak delimiter tags in the peak pane.
When the tag can be selected, the cursor changes to a crosshair-style cursor. You can zoom
in on the baseline to make it easier to select the tag.
2. Drag the peak delimiter tag to another location and automatically update the peak values
(area, height, and so forth) into the result set.
The generated reports for the data identify the manual modifications.
You can store two peak value sets (method and manual integration settings) with each
compound in each file. These settings can result in a different set of stored values. The
application originally calculates the method values based on the processing method
parameters. The manual values are a result of what you edited.
To remove a manually created peak
Right-click the pane and then choose Delete Peak, or click the icon in the upper right
corner, .
The application removes the manual integration from the selected peak.
To switch between Method, Manual, and User integration modes
Right-click the chromatogram pane and choose Integration Settings.
Initially, the method, manual, and user integration settings that are stored for a
compound and file are identical. When you switch modes, the saved result set does not
change. However, when either manual or user data are available, both the chromatogram
plots and the result set update as you switch between modes.
As you switch between modes, each pane in Data Review reflects the changes. The
generated reports for the data identify the manual or user modifications.
Manually added right
and left base points
Crosshair cursor

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 169
To customize the labels in the display
1. Right-click and choose Peak Display Settings.
The Peak Display Settings dialog box opens.
2. To display labels for the peak retention time (RT), area (AA), height (AH), or
signal-to-noise (SN), select the appropriate check box.
3. Click OK in the Peak Display Settings dialog box.
Figure 43. Example of displayed labels
To modify the peak detection settings
1. Right-click the chromatogram view and choose one of the following:
•Peak Detection Settings > Edit Local Method Peak Detection Settings: Makes
changes to the selected compound for all samples in this batch. The application
identifies these settings as “Manual.”
•Peak Detection Settings > Edit User Peak Detection Settings: Makes changes to
the selected compound for only the selected sample. The application saves these
changes with the batch and stops applying the local method detection settings to the
compound for this sample only. The application identifies these settings as “User.”
The Peak Detection Settings dialog box opens where you can adjust detection settings
that were specified in the method. The title bar of the dialog box lists the selected
compound and indicates whether you are making changes to only the selected sample or
to the local method.
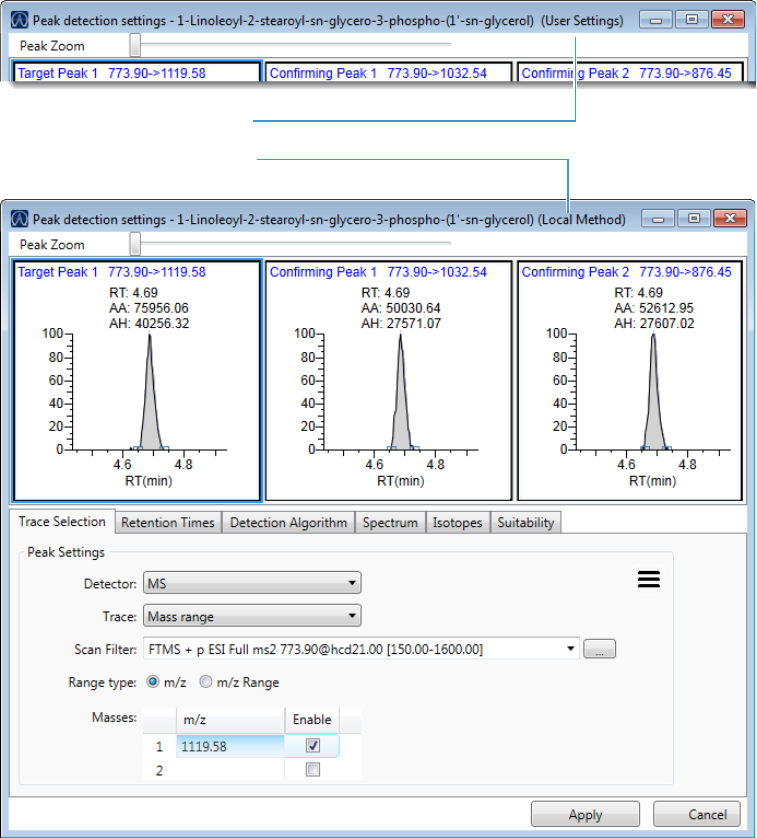
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
170 TraceFinder User Guide Thermo Scientific
Figure 44. Peak Detection Settings dialog box
2. Edit any of the detection settings.
For detailed descriptions of all detection settings, refer to Chapter 4, “Using the Method
Development Mode for Quantitation Methods,” in the TraceFinder Lab Director User
Guide.
3. To save your changes to this compound, click Apply.
• When you are editing a single sample, the application makes changes to the selected
compound for this sample. If the sample is a calibration sample type, this update
changes the calibration curve which, in turn, affects all calculated amounts.
• When you are editing the local method, the application makes changes to the selected
compound for all samples in this batch.
Editing all samples in the batch
Editing only the selected sample
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 171
Table 30. Quan Peak pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Send RT to Method Sets the current retention time as the expected retention time for the compound in the
local method.
Swipe Manual Integration Switches to manual integration mode, where you can manually define the peak.
Also available from the icon in the upper right corner, .
Integration Settings
(Method, Manual, User)
Switches the display between Method, Manual, or User integration modes.
Method (default): The settings defined in the master method.
Manual: Modifications made using the Peak Detection Settings > Edit Local Method
Peak Detection Settings command. The application applies the modified peak
detection settings to all samples in the batch.
User: Modifications made using the Peak Detection Settings > Edit User Peak
Detection Settings command. The application applies the modified peak detection
settings to only the selected sample.
Clear User Peak Detection
Settings
Resets modified peak detection settings to the settings in the master method, and
identifies the integration mode as Method.
Delete Peak Removes the peak and all data for this peak from all Compound View panes.
Also available from the icon in the upper right corner, .
Peak Display Settings Opens the Peak Display Settings dialog box.
Peak Detection Settings Edit Local Method Peak Detection Settings: Edits the peak detection settings for all
samples in the batch. The application identifies these settings as “Manual.”
Edit User Peak Detection Settings: Edits the peak detection settings for only the
selected sample. The application identifies these settings as “User.”
After you apply either of these updates, the application does not retain the modified
integration settings.
The changes that you make on the Signal page in the Peak Detection Settings dialog
box can affect all quantification and ion ratio calculations and are not auto-reversible.
These changes can also create a mismatch with calibration files that are not fully
defined in the batch.
Note The Peak Detection Settings commands are also available in the Confirming
Ions pane.

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
172 TraceFinder User Guide Thermo Scientific
Confirming Ions
The Confirming Ions pane displays a list of peaks, a graphical view of all
qualifying/confirming ions for the selected compound, and calculated ion ratios and ion ratio
acceptance windows. A red border indicates that an ion ratio is outside of its window. See
Quantitative peak with multiple confirming ions. The Confirming Ions pane uses a unique
shortcut menu. See Confirming Ions pane shortcut menu commands.
In the peaks list, the application displays the mass of each confirming ion peak. When a peak
is within the tolerance set, the peak status is green. When a peak is not within the tolerance
set, the peak status is red.
Follow these procedures:
•To remove a peak
•To customize the labels in the display
•To display surrounding apex times
•To manually integrate a peak
•To switch between Method, Manual, and User integration modes
To remove a peak
Right-click the Confirming Ions pane, and choose Delete Peak.
The application removes the peak displayed in the Confirming Ions pane. All data for this
peak are removed from the Compound View panes.
To customize the labels in the display
1. Right-click and choose Peak Display Settings.
The Peak Display Settings Dialog Box opens.
2. To display labels for the peak retention time (RT), mass area (MA), mass height (MH), or
signal-to-noise (SN), select the appropriate check box. See Example of displayed labels.
3. Click OK in the Peak Display Settings dialog box.
Note For compounds with an analog detection type, the application displays “No
Confirming Ions are Enabled” in the Confirming Ions pane.
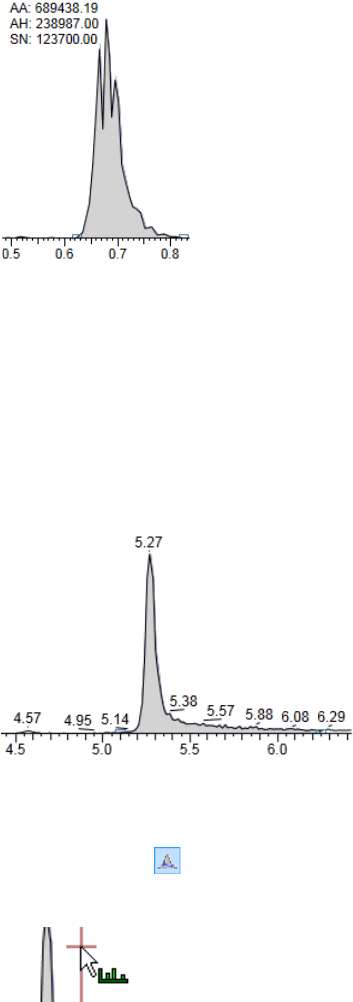
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 173
Figure 45. Example of displayed labels
To display surrounding apex times
1. Right-click and choose Peak Display Settings.
The Peak Display Settings dialog box opens.
2. Select the Show All Apex Times check box.
3. Click OK in the Peak Settings dialog box.
Figure 46. Example of displayed apex times
To manually integrate a peak
1. Right-click and then choose Swipe Manual Integration, or click the Swipe Manual
Integration icon, .
The cursor changes to indicate the manual integration mode.
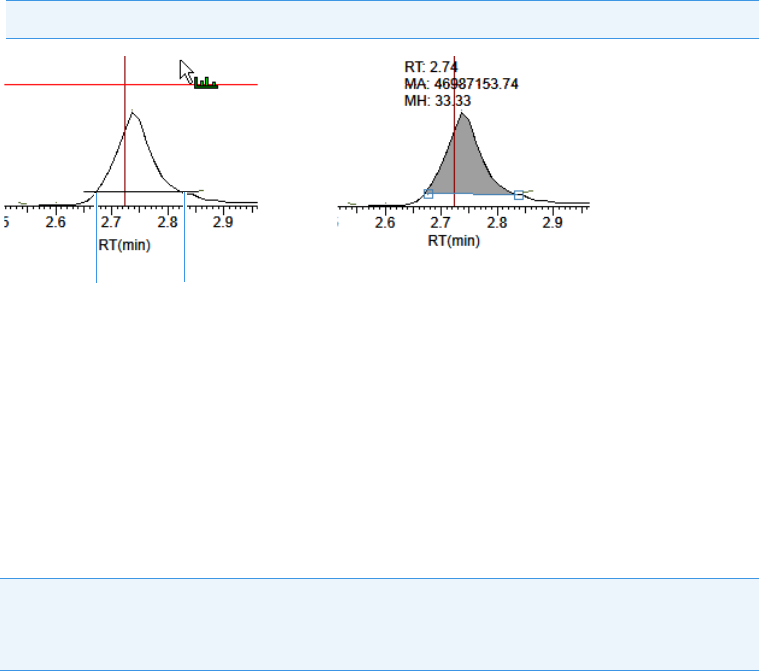
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
174 TraceFinder User Guide Thermo Scientific
2. Drag the cursor to define the beginning and ending base points for the new peak.
The application updates the area of the peak to the baseline that you have just added.
When you regenerate the reports, they display the manually modified data.
You can store three peak value sets (method, manual, and user integration settings) with
each compound in each file. These settings can result in a different set of stored values.
The application originally calculates the method values based on the processing method
parameters. The manual or user values are a result of what you edited.
To switch between Method, Manual, and User integration modes
Right-click the Confirming Ions pane and choose Integration Settings.
Initially, the method, manual, and user integration settings that are stored for a
compound and file are identical. When you switch modes, the saved result set does not
change. However, when either manual or user data are available, both the chromatogram
plots and the result set update as you switch between modes.
As you switch between modes, each pane in Data Review reflects the changes. The
generated reports for the data identify the manual or user modifications.
Note You must drag the cursor inside the x axis and y axis.
Note Because a Blank Report displays only the quantitation mass, when you manually
integrate a confirming ion, the manual integration flag in the report is displayed on
the quantitation mass.
Drag the cursor across
the peak at the base
points for the new peak.
The application identifies the peak
and indicates the manual
integration in the labels.
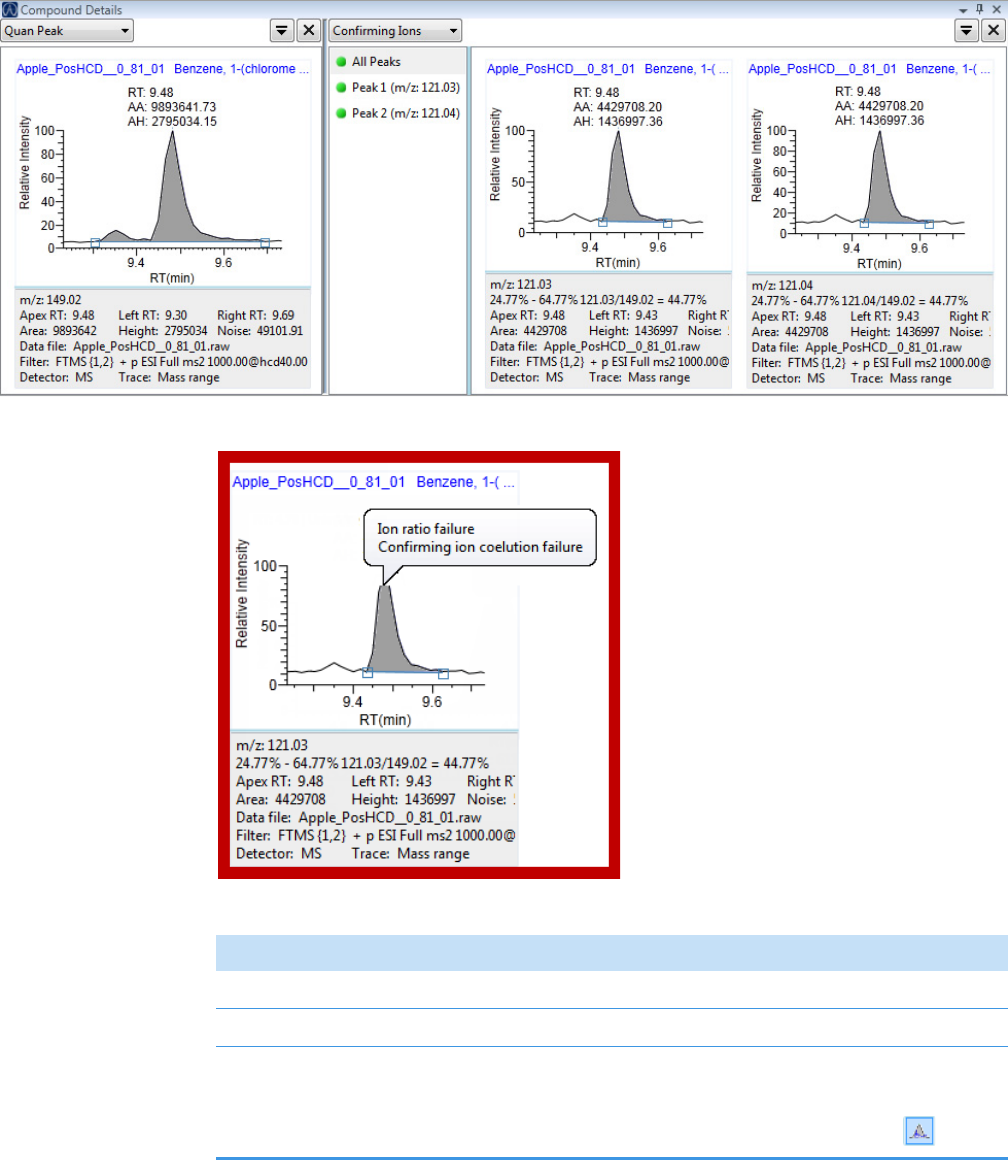
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 175
Figure 47. Quantitative peak with multiple confirming ions
Figure 48. Confirming ion with coelution failure
Table 31. Confirming Ions pane shortcut menu commands (Sheet 1 of 2)
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Swipe Manual
Integration
Switches to manual integration mode, where you can manually
define the peak.
Also available from the icon in the upper right corner, .

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
176 TraceFinder User Guide Thermo Scientific
Integration Settings
(Method, Manual,
User)
Switches the display between Method, Manual, or User
integration modes.
Method (default): The settings defined in the master method.
Manual: Modifications made using the Peak Detection Settings >
Edit Local Method Peak Detection Settings command. The
application applies the modified peak detection settings to all
samples in the batch.
User: Modifications made using the Peak Detection Settings >
Edit User Peak Detection Settings command. The application
applies the modified peak detection settings to only the selected
sample.
Clear User Peak
Detection Settings
Resets modified peak detection settings to the settings in the
master method, and identifies the integration mode as Method.
Delete Peak Removes the peak and all data for this peak from all Compound
View panes.
Also available from the icon in the upper right corner, .
Peak Display Settings Opens the Peak Display Settings dialog box.
Peak Detection
Settings
Edit Local Method Peak Detection Settings: Edits the peak
detection settings for all samples in the batch. The application
identifies these settings as “Manual.”
Edit User Peak Detection Settings: Edits the peak detection
settings for only the selected sample. The application identifies
these settings as “User.”
After you apply either of these updates, the application does not
retain the modified integration settings.
The changes that you make on the Signal page in the Peak
Detection Settings dialog box can affect all quantification and ion
ratio calculations and are not auto-reversible. These changes can
also create a mismatch with calibration files that are not fully
defined in the batch.
Note The Peak Detection Settings commands are also available
in the Confirming Ions pane.
Table 31. Confirming Ions pane shortcut menu commands (Sheet 2 of 2)
Command Description
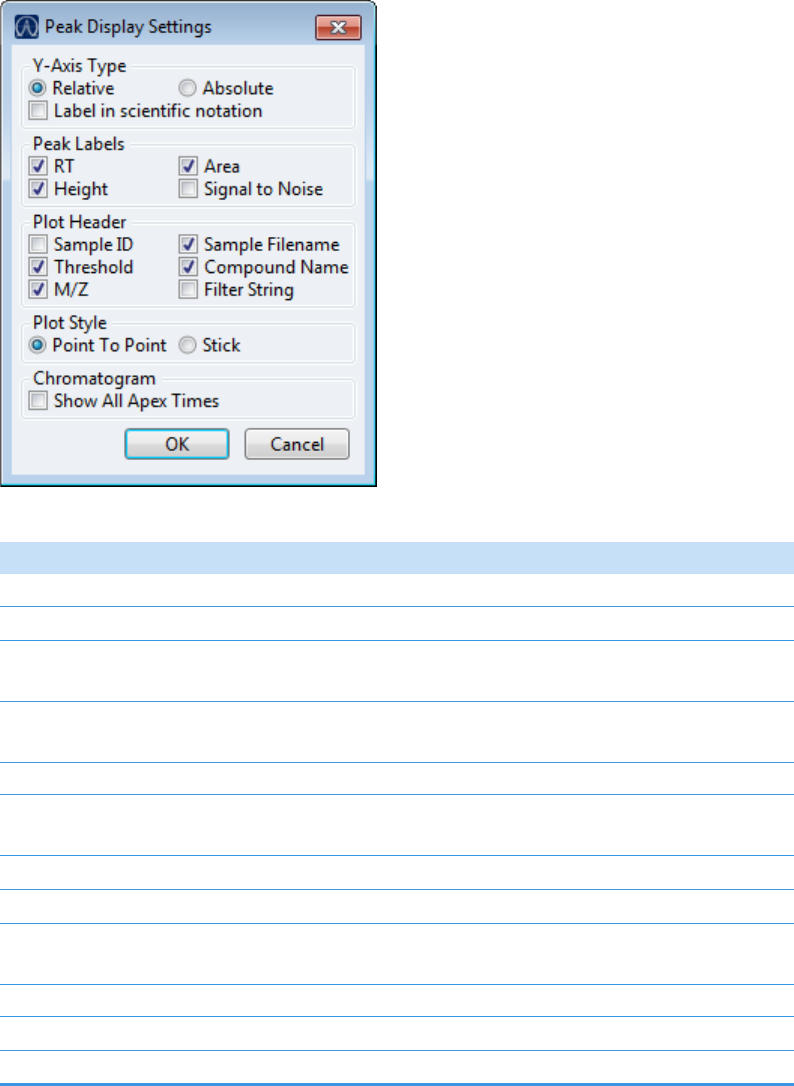
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 177
Peak Display Settings Dialog Box
Use the features in the Peak Display Setting dialog box to change the labels in the Confirming
Ions pane display.
Figure 49. Peak Display Settings dialog box
Table 32. Peak Display Settings dialog box parameters (Sheet 1 of 2)
Command Description
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the most
intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Peak Labels
RT Displays the retention time (RT) label in the peak chromatogram
pane.
Height Displays the peak height (AH) label in the peak chromatogram pane.
Area Displays the peak area (AA) label in the peak chromatogram pane.
Signal to Noise Displays the signal-to-noise (SN) label in the peak chromatogram
pane.
Plot Header
Sample ID Displays a user-defined, alphanumeric string that identifies a sample.
Threshold Displays the peak threshold.
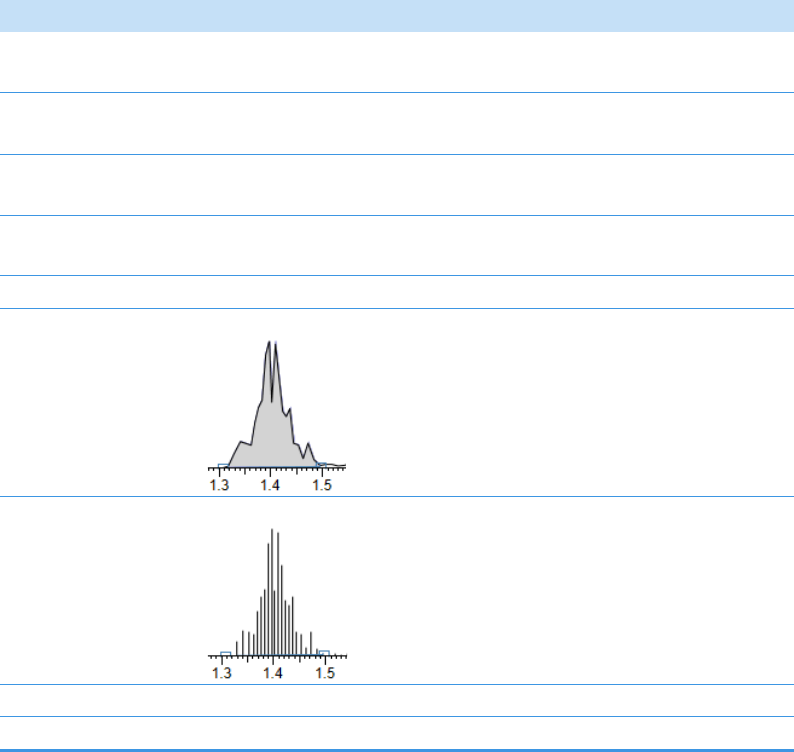
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
178 TraceFinder User Guide Thermo Scientific
m/z Displays the mass-to-charge ratio found in the spectrum for the
peak. Assumes the charge is 1.
Sample Filename Displays the name of the current, selected sample where the peak
was found.
Compound Name Displays the matched compound name found in the library or
compound database.
Filter String Displays the filter used to identify the peak. Specified in the raw data
file or the master method.
Plot Style
Point To Point Displays chromatograms in the following format:
Stick Displays chromatograms in the following format:
Chromatogram
Show All Apex Times Displays all surrounding apex times in the plot.
Table 32. Peak Display Settings dialog box parameters (Sheet 2 of 2)
Command Description
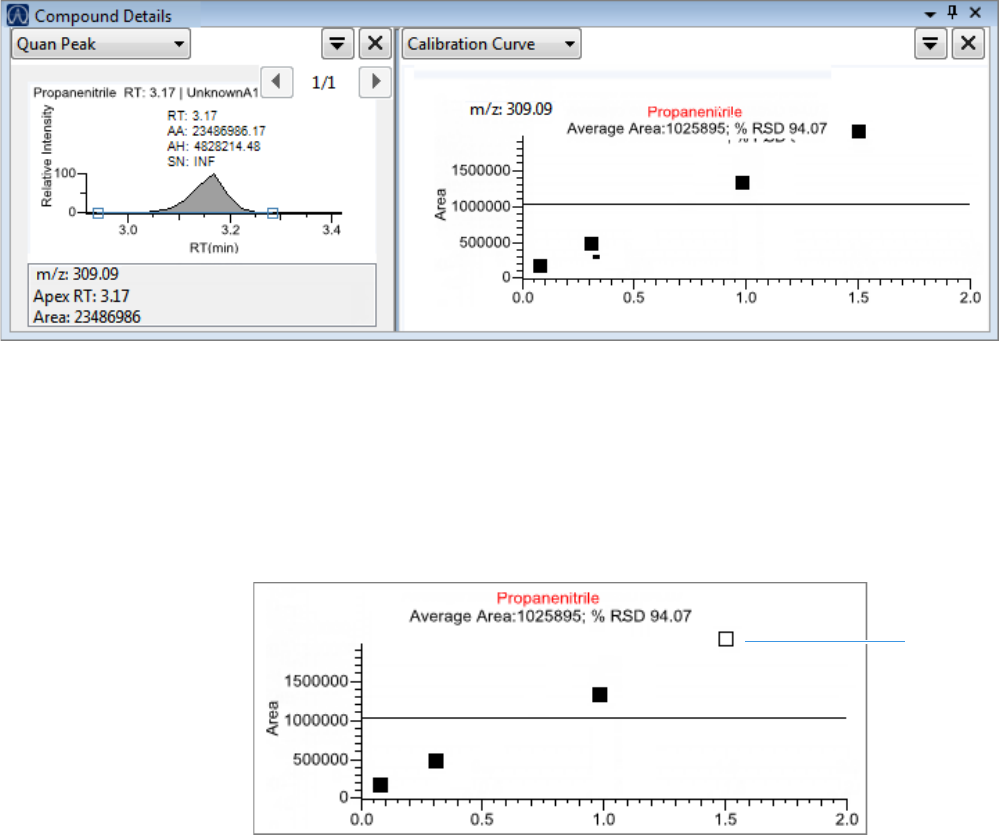
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 179
Calibration Curve
The Calibration Curve pane displays a graphical view of the calibration curve for the selected
compound and key statistical values for evaluating the quality of the calibration. The
Calibration Curve pane uses a unique shortcut menu. See Calibration Curve pane shortcut
menu commands.
Figure 50. Quantitative peak with a calibration curve plot
To manually exclude a calibration point
In the Sample Results pane, select the Excluded check box for the sample.
You might need to use the horizontal scroll bar at the bottom of the table to scroll to the
Excluded column. See Inactive and Excluded Compounds.
When the application no longer uses a calibration point, its value appears as an empty box
in the graphical view of the calibration curve.
Excluded
calibration
point
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
180 TraceFinder User Guide Thermo Scientific
Table 33. Calibration Curve pane shortcut menu commands
Command Description
Standard Type Sets the standard type to External or Internal.
Calibration Curve Type Sets the calibration curve type to one of the following:
• Linear: Uses all settings with this exception: When Origin is
set to Include, all Weighting values are grayed out and
Weighting is set to Equal.
• Quadratic: Uses all settings with this exception: When
Origin is set to Include, all Weighting values are grayed out
and Weighting is set to Equal.
• Average RF: Uses no Weighting or Origin selections. All
Weighting and Origin values are grayed out. Weighting is set
to Equal, and Origin is set to Ignore.
Response Via Sets the response to Area or to Height.
Weighting Sets the weighting to equal, 1/X, 1/X^2, 1/Y, or 1/Y^2.
Origin Sets the origin to Ignore, Force, or Include.
Units Sets the units.
Done with Settings Saves the calibration curve settings.
Reset Scaling Resets the original scale in the calibration curve pane.
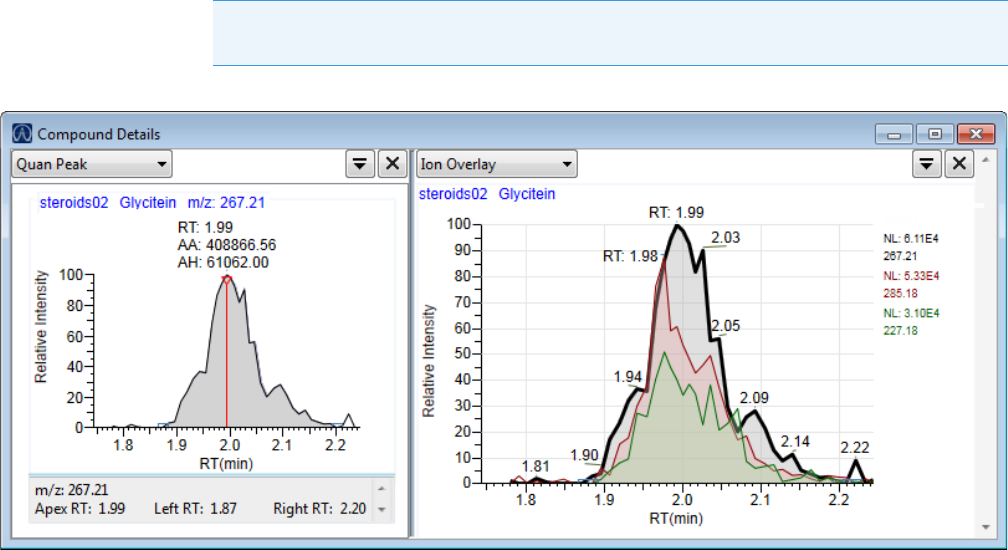
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 181
Ion Overlay
The Ion Overlay pane represents an overlay of the entire ion set—target and confirming—for
the selected compound. Use this pane to graphically review the peak apex alignment and
coeluting peak profiles.
Figure 51. Target peak with a confirming ion overlay
Note For compounds with an analog detection type, the application displays “No Data”
in the Ion Overlay pane.
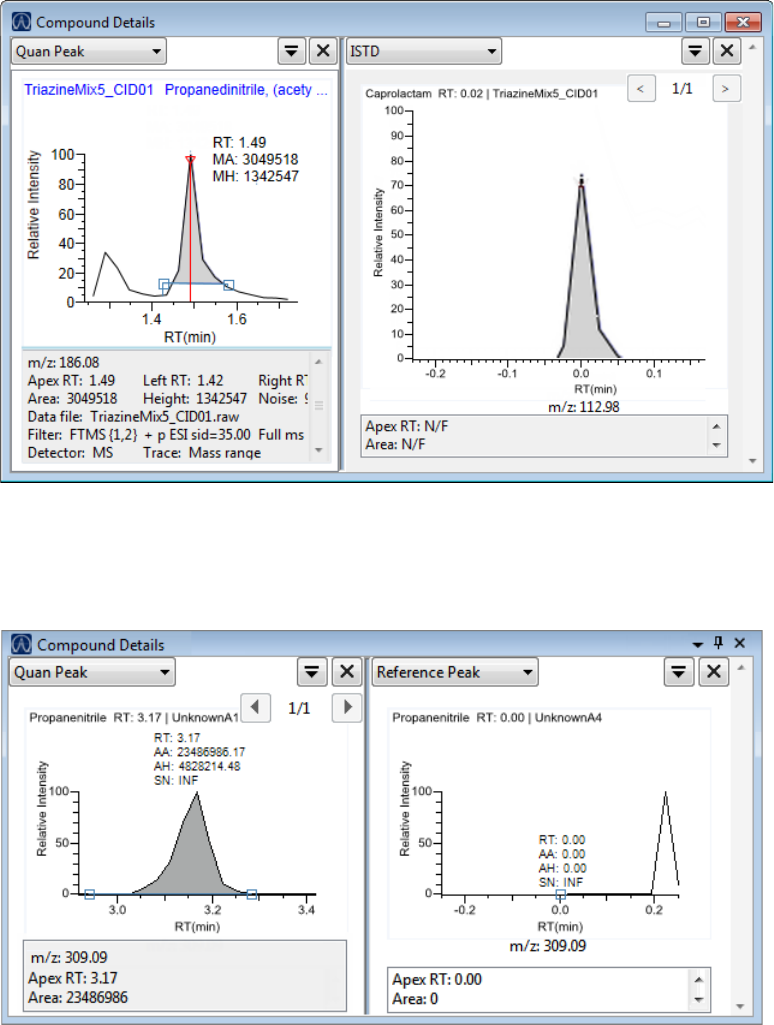
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
182 TraceFinder User Guide Thermo Scientific
ISTD
The ISTD pane displays the internal standard specified for the compound in the method.
Refer to the instructions “To specify an internal standard type for a compound” in Chapter 4,
“Using the Method Development Mode for Quantitation Methods,” in the Tra c eFind er L ab
Director User Guide.
Figure 52. Quantitative peak with an internal standard
Reference Peak
The Reference Peak pane displays the reference peak as specified in the method.
Figure 53. Quantitative peak with a reference peak
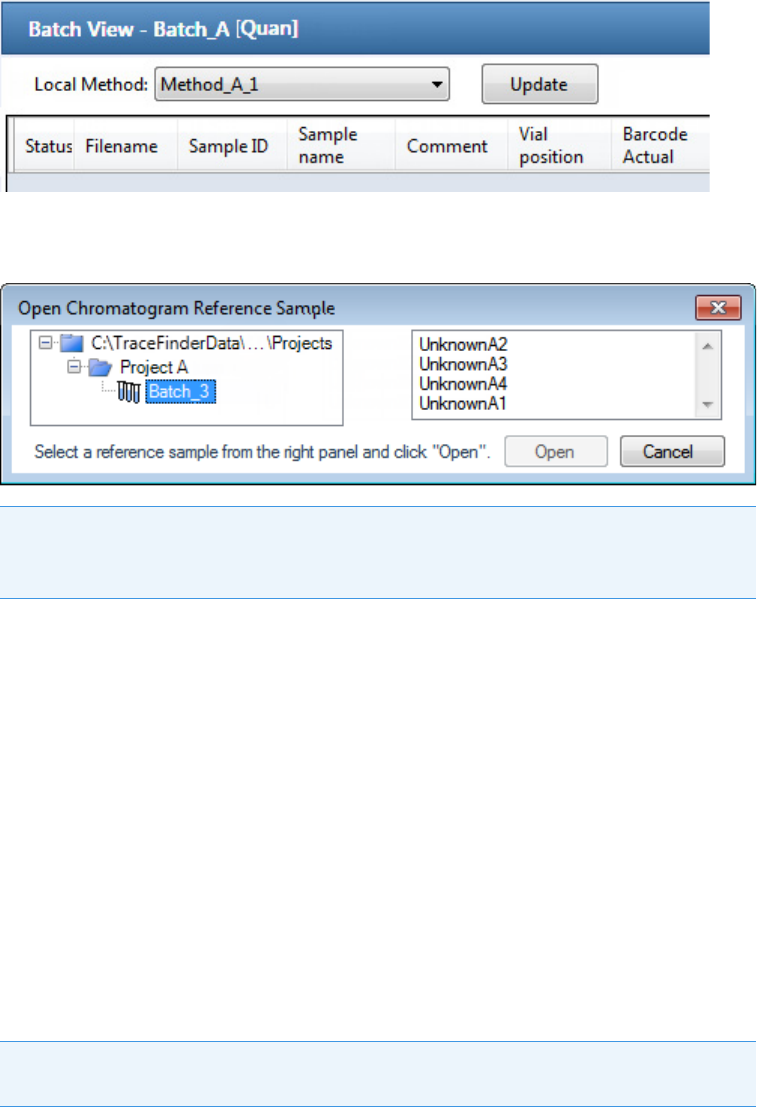
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 183
To specify a chromatogram reference peak
1. In the Batch View task pane, click Reference Sample.
An empty reference sample table opens.
2. Right-click and choose Add Reference Sample.
The Open Chromatogram Reference Sample dialog box opens.
3. Select a project from the list of projects.
4. Select a subproject from the list of subprojects.
5. Select a batch from the list of batches.
The application displays only batches that were created using the current master method.
6. Select a sample from the list of processed samples.
The application displays all the processed samples in the selected batch. To use a sample
as a reference sample, the sample must have been processed with the current master
method.
7. Click Open.
The selected sample is displayed as the chromatogram reference sample in the Method
View in the Method Development mode.
Note If you are creating a new method, you will not see any reference samples here.
You must create and save a batch using the current method to see the reference
samples in this list.
Tip To clear the reference sample from the master method, right-click and choose
Delete Selected.

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
184 TraceFinder User Guide Thermo Scientific
Spectra
The Spectra pane displays a comparison of the spectra found in the data and the method
reference.
Follow these procedures:
•To zoom in on a peak
•To copy a plot to another format
•To display the MS reference
•To customize the labels in a small molecule or peptide spectrum
To zoom in on a peak
1. In the Spectra pane, drag the cursor to delineate a rectangle around the peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the Spectra pane and choose Reset Scaling.
To copy a plot to another format
1. In the data spectrum or the reference spectrum, right-click and choose Copy to
Clipboard.
The application copies the specified plot graphic to the Clipboard.
2. In the source format (for example, an Excel spreadsheet or email window), right-click and
choose Paste.
The application pastes the graphic into the new format.
To display the MS reference
Right-click the Spectra pane and choose Show MS-Reference.
The application displays or hides the reference spectrum in the Spectra pane.
Note For compounds with an analog detection type, the application displays “Not
Available” in the Spectra pane.
Note The application copies only the indicated data spectrum or the reference
spectrum. It does not copy the entire Spectra pane.
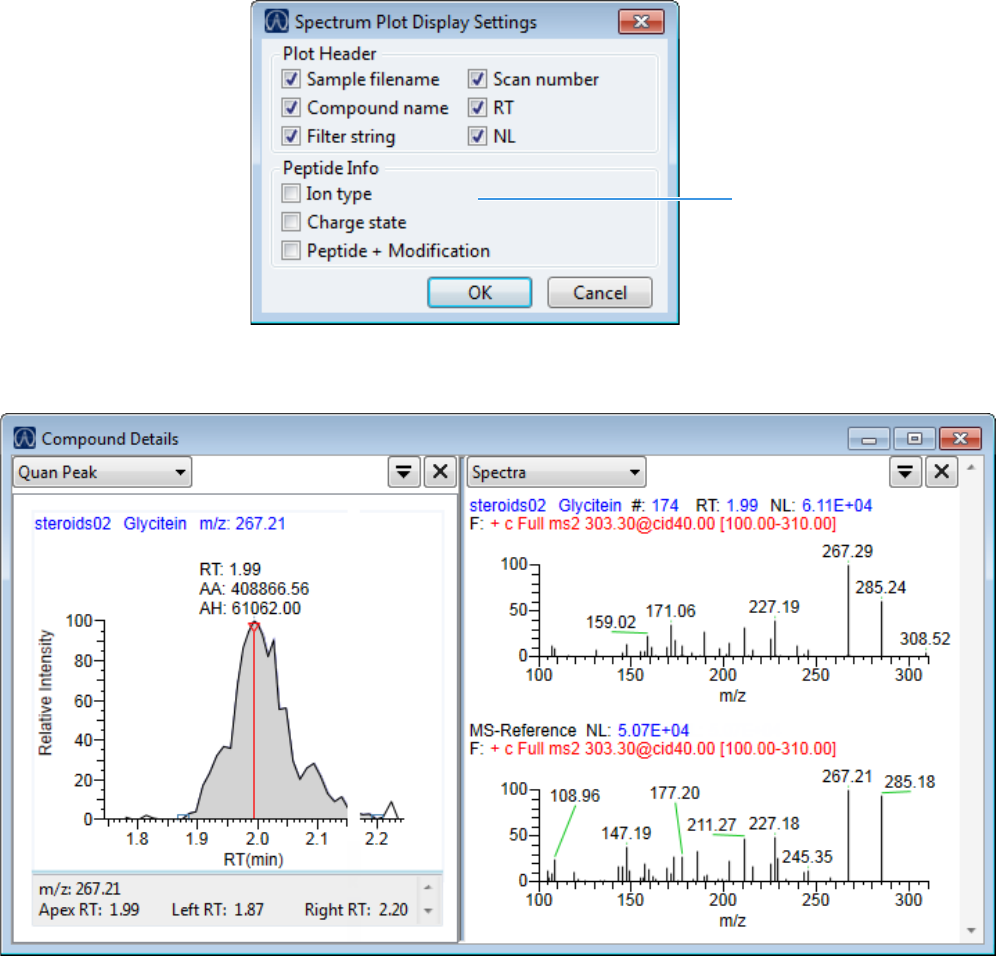
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 185
To customize the labels in a small molecule or peptide spectrum
1. Right-click the Spectra pane and choose Display Settings.
The Spectrum Plot Display Settings dialog box opens.
2. Select the check box for each label that you want to display and click OK.
Figure 54. Quantitative peak with data and reference spectra
Additional options
for a peptide spectrum
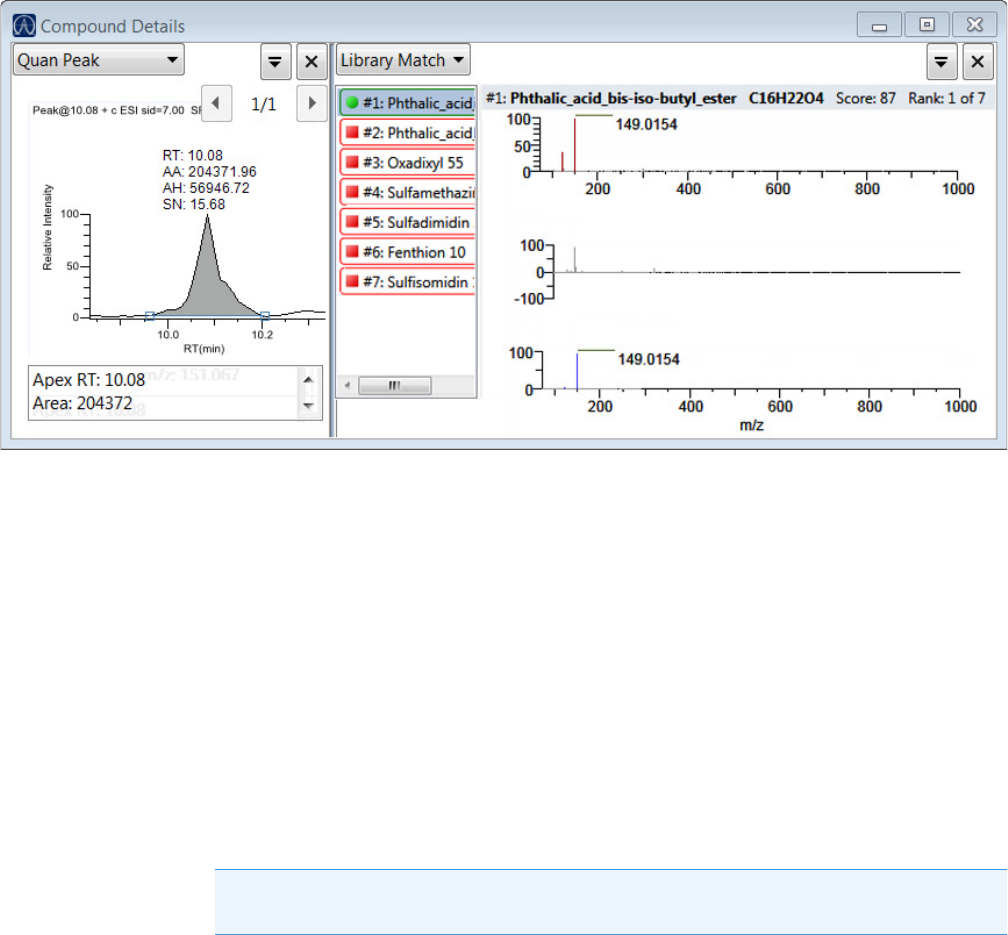
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
186 TraceFinder User Guide Thermo Scientific
Library Match
The Library Match pane displays all library matches for the selected compound.
Figure 55. Library Match for the selected compound
If you have no matches for any of your compounds, make sure you have completed all of the
following:
• Installed a library.
• Selected screening libraries in the Configuration console. Refer to Chapter 1, “Using the
Configuration Console,” in the TraceFinder Lab Director User Guide.
• Enabled Library Matching in the method. Refer to Chapter 4, “Using the Method
Development Mode for Quantitation Methods,” in the TraceFinder Lab Director User
Guide.
Isotope
The Isotope pane displays all isotopes for the selected compound. The Isotope pane includes a
shortcut menu. See Isotope pane shortcut menu commands.
The isotopes pane displays isotopic pattern results for all adducts of a compound according to
the threshold and deviation parameters defined in the method.
Note When you select different samples, the current compound remains selected (as long
as that compound is found in the sample).
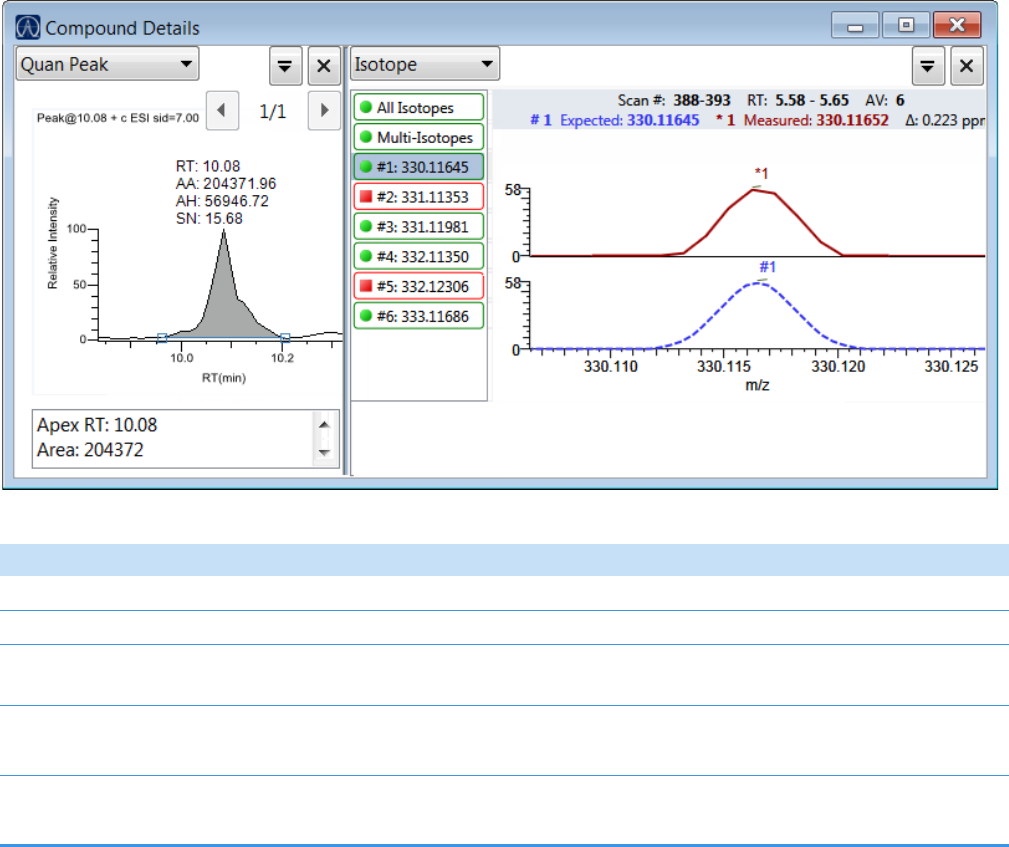
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 187
To identify or confirm the presence of a compound, the resulting score percentage from
isotopic pattern matching must be higher than the specified fit threshold percentage.
• An isotope peak is not found if its intensity, relative to the monoisotopic ion’s intensity, is
more than the specified intensity deviation percentage away from the theoretical relative
intensity of the isotope ion.
• An isotope peak is found if its measured m/z is less than the specified mass deviation
amount away from its expected m/z.
To specify isotopic criteria in a method, refer to Chapter 4, “Using the Method Development
Mode for Quantitation Methods,” in the TraceFinder Lab Director User Guide.
Figure 56. Isotope pane
Table 34. Isotope pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Display Overlay Spectra
Display Stack Spectra
Overlays the two spectrum displays, or stacks the peak apex spectrum over the
simulated spectrum.
Show/Hide Noise Label Adds a noise label to each peak. Expected isotope peaks (displayed in blue) do not
display a noise label.
Show/Hide Resolution Label Adds a resolution label to each peak. Expected isotope peaks (displayed in blue) do not
display a resolution label.
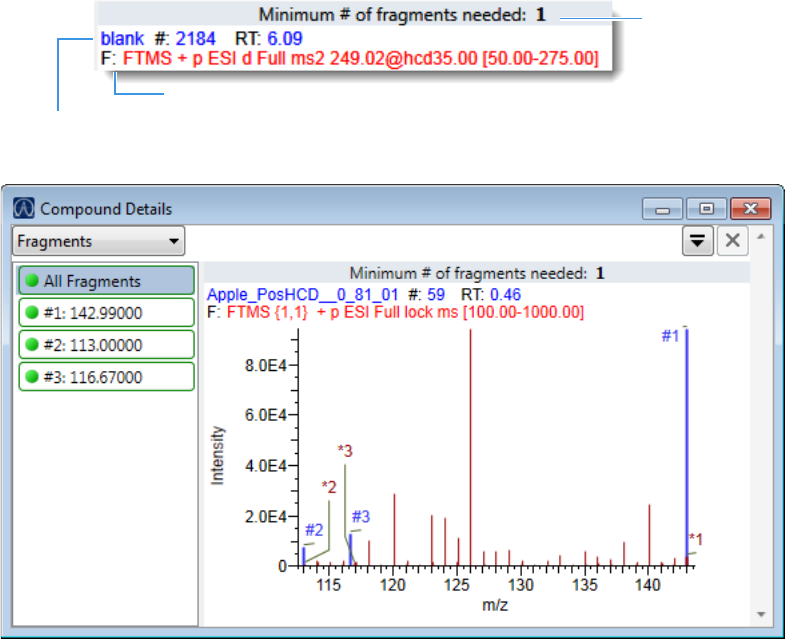
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
188 TraceFinder User Guide Thermo Scientific
Fragments
The Fragments pane displays fragments in one of two ways:
•All Fragments
•Individual Fragments
All Fragments
The All Fragments view displays a composite of all fragments found in the peak. The
application displays the measured peak as a red line; the application displays the expected peak
as a blue line.
The application displays these headers for the All Fragments view:
Figure 57. Fragments pane with overlaid spectra for all fragments
Processing filter
File name
Minimum number
of fragments
specified in the
method
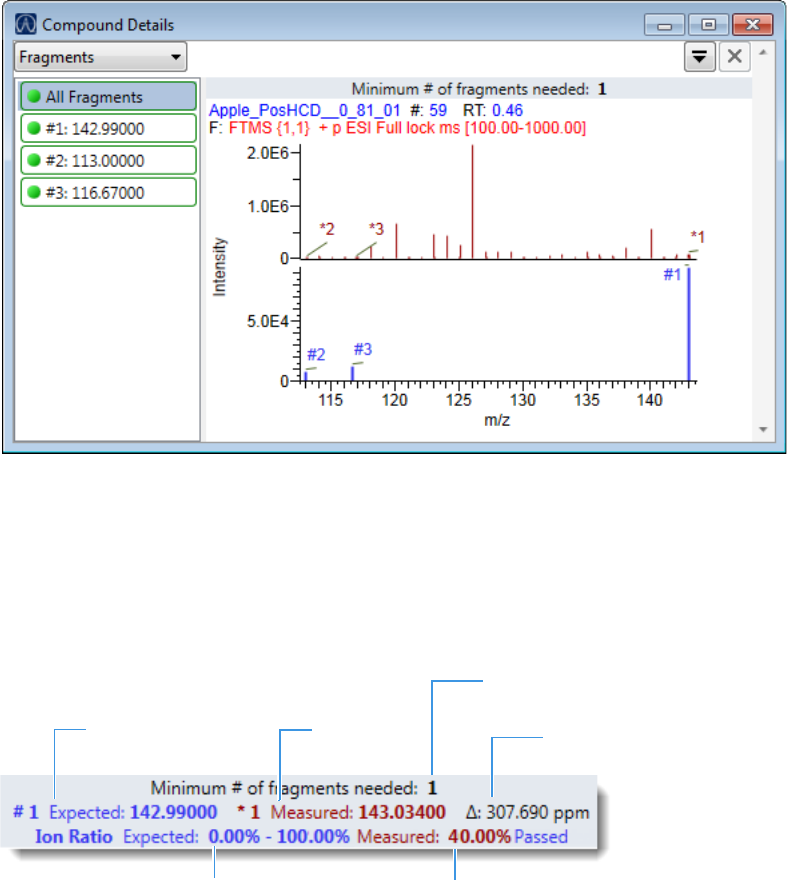
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 189
Figure 58. Fragments pane with stacked spectra for all fragments
Individual Fragments
The individual fragments view displays the expected and measured peaks for a single
fragment.
The application displays these headers for the individual fragments view:
Minimum number of fragments specified
in the method
Expected m/z
for fragment 1
Measured m/z
for fragment 1 Delta between expected
and measured m/z
Expected ion
ratio range
Measured ion ratio
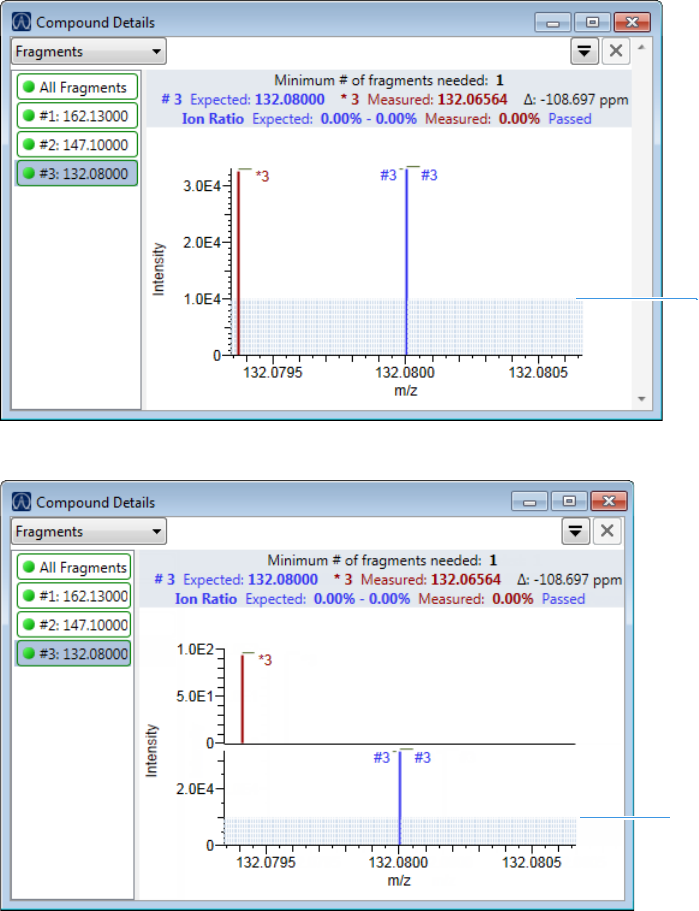
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
190 TraceFinder User Guide Thermo Scientific
Figure 59. Fragments pane with overlaid spectra for a single fragment
Figure 60. Fragments pane with stacked spectra for a single fragment
Intensity
threshold
specified in
the method
Intensity
threshold
specified in the
method
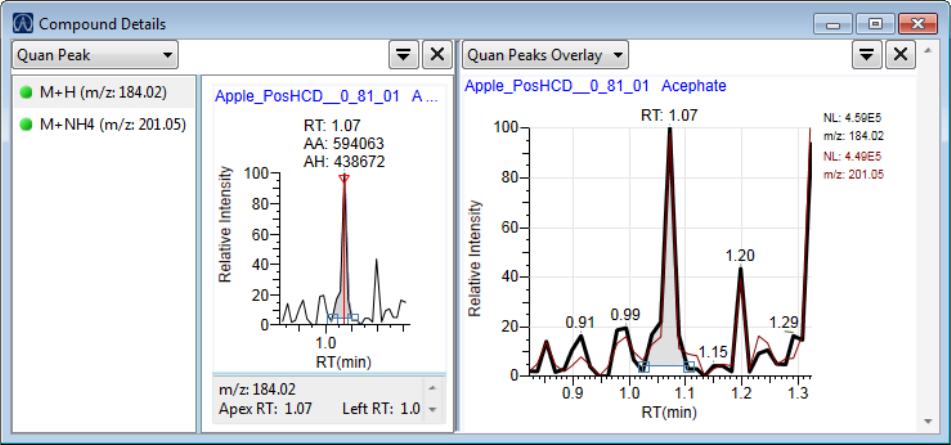
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 191
Quan Peaks Overlay
The Quan Peaks Overlay page displays overlaying quantitation peaks for compounds with
multiple target peaks.
• The first target peak in the peak list appears in black in the overlay pane.
• The second target peak in the peak list appears in red in the overlay pane.
• The selected peak is bolded in the overlay pane.
Figure 61. Quan Peaks Overlay page
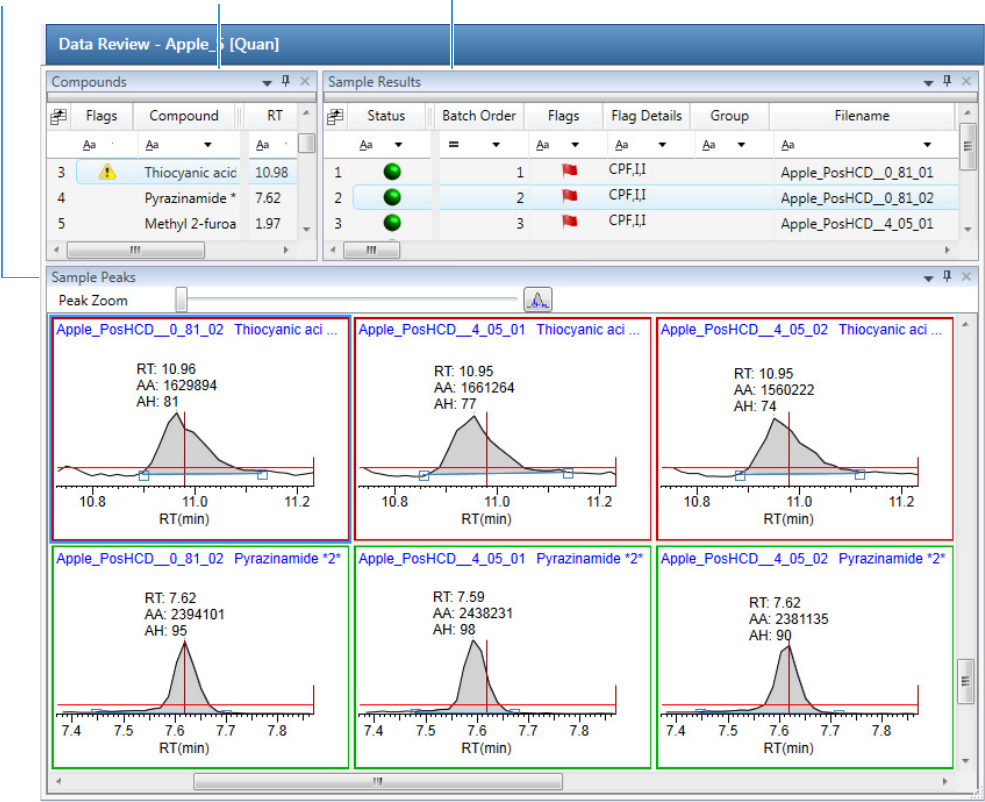
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
192 TraceFinder User Guide Thermo Scientific
Comparative View for Quantitation Batches
The Comparative View uses three panes to display a list of all compounds available in the
method, all samples in the current batch, and the sample peak plots for all compounds found
in the samples with the horizontal threshold guide.
These are the default panes and their locations:
Related Topics
•Using the Threshold Guide
•Configuring Sample Peaks Display Settings
•Manually Integrating Peaks
Compounds pane Sample Results paneSample Peaks pane
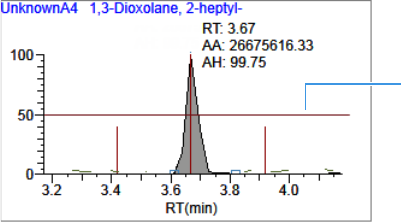
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 193
Using the Threshold Guide
The following factors define the threshold guide that the Comparative View displays in the
sample peak plots:
• The threshold method and amount that you specified in the method
• The group that you created on the Sample page
• The threshold sample that you selected on the Threshold Samples page
For information about creating groups, see Groups.
For information about specifying a threshold sample, see Threshold Samples Page for
Quantitation Batches.
For information about specifying the method to use for creating a threshold guide, refer to
Chapter 4, “Using the Method Development Mode for Quantitation Methods,” in the
TraceFinder Lab Director User Guide.
Configuring Sample Peaks Display Settings
The Sample Peaks pane in the Comparative View displays one row per compound and one
column per sample. The Sample Peaks pane displays all samples in a group when you select
any of the samples belonging to that group.
For information about creating groups, see Groups.
To change the Sample Peaks pane display
1. From the View menu, choose Chromatogram Pane Settings.
The Chromatogram Plot Settings Dialog Box opens.
2. To change the number of rows or columns to fit in the Sample Peaks pane, type new
values in the Number of Rows or Number of Columns boxes.
These values do not change the number of rows (compounds) and columns (samples) that
are available in the Sample Peaks pane. These values determine how many rows and
columns you want to view at one time in the display. The default is three rows and three
columns.
The horizontal guide indicates the threshold
value as specified in the method.
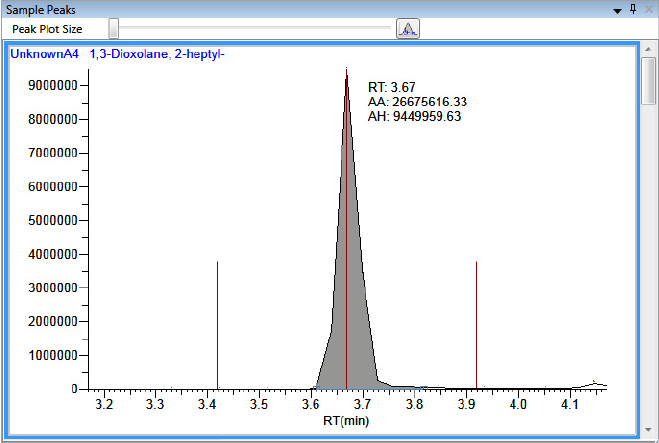
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
194 TraceFinder User Guide Thermo Scientific
In the following examples, the Number of Rows and Number of Columns are set to 1
(Number of Rows equals 1 and Number of Columns equals 1) and the Number of Rows
and Number of Columns are set to 4 (Number of Rows equals 4 and Number of
Columns equals 4).
Figure 62. Number of Rows equals 1 and Number of Columns equals 1
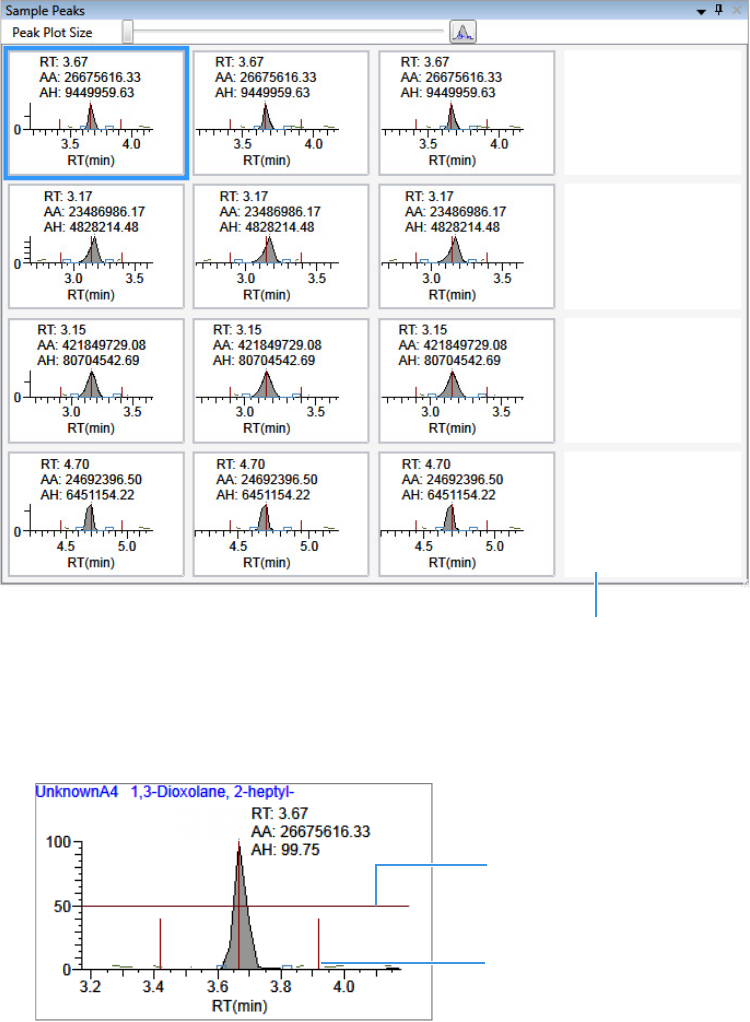
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 195
Figure 63. Number of Rows equals 4 and Number of Columns equals 4
3. To change the display type for the y-axis scale, select one of the following:
•Relative: Displays the y-axis scale from 0 through 100.
Because there are
only three samples,
this column is empty.
Vertical bars indicate the expected
retention time and window, as
specified in the method.
The horizontal bar indicates
the threshold value as
specified in the method.
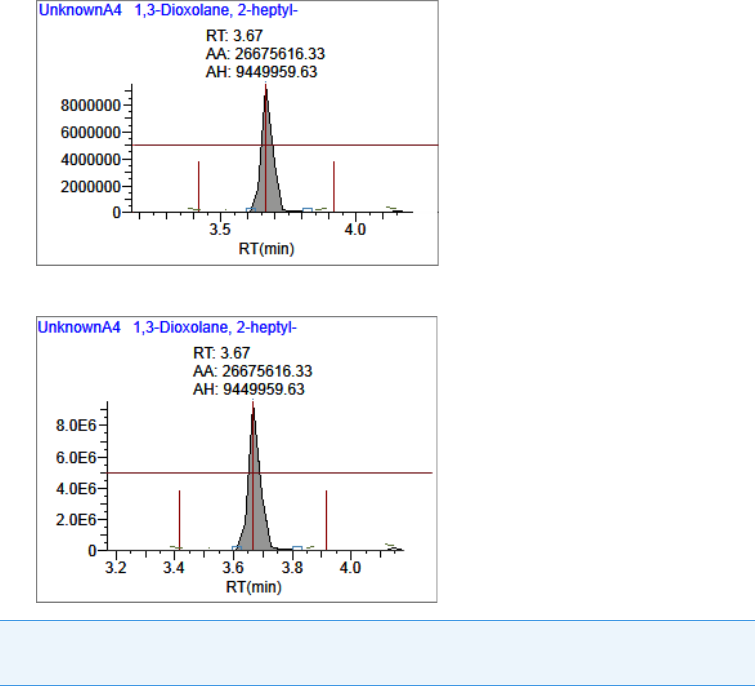
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
196 TraceFinder User Guide Thermo Scientific
•Absolute: Displays the y-axis scale from 0 to the actual value of the most intense peak
in the group.
•Label in Scientific Notation: Displays the y-axis scale in scientific notation.
4. Specify which labels you want to display in the sample peak plots.
For an example of all available peak plot labels, see Peak Plot Labels.
Note The Sample Peaks pane displays a y axis only in the first chromatogram in each
row. The limits of the scale are determined by the most intense peak in the group.
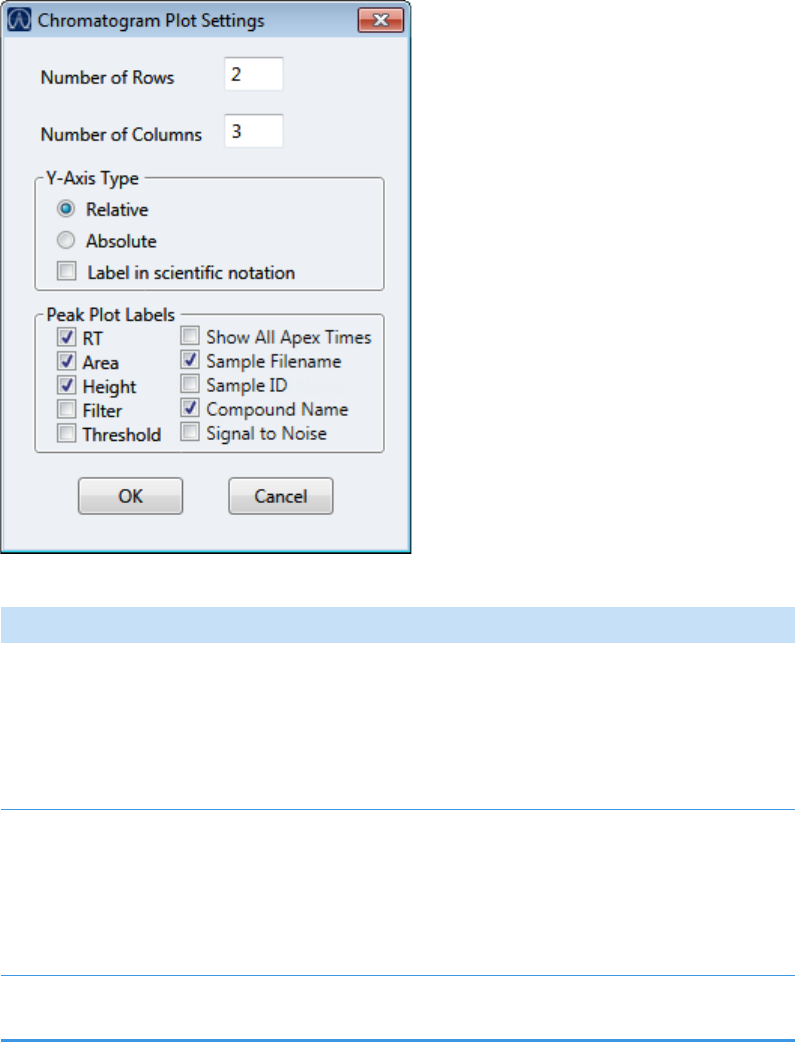
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 197
Chromatogram Plot Settings Dialog Box
Use the Chromatogram Plot Settings dialog box to change the Sample Peaks pane display.
Table 35. Chromatogram Plot Settings dialog box parameters (Sheet 1 of 2)
Parameter Description
Number of Rows Specifies the number of rows visible in the Sample Peaks pane.
When Number of Rows equals 1, the application scales the height
of all chromatograms to fill the Y dimension of the Sample Peaks
pane.
Default: 3
Number of Columns Specifies the number of columns visible in the Sample Peaks pane.
When Number of Columns equals 1, the application scales the
width of all chromatograms to fill the X dimension of the Sample
Peaks pane.
Default: 3
Y-Axis Type Displays the y-axis scale as Relative (to the most intense peak),
Absolute, or in scientific notation.
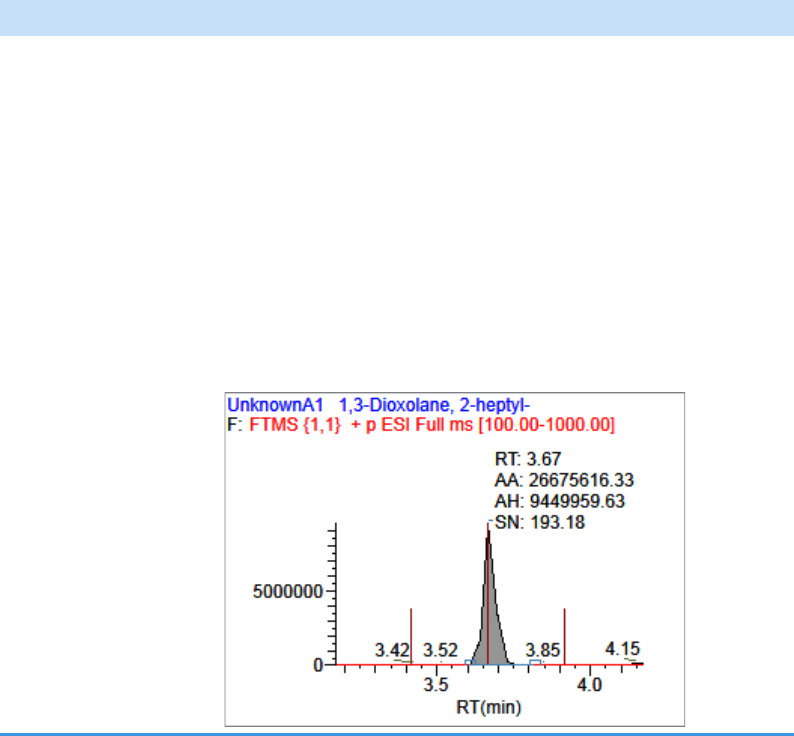
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
198 TraceFinder User Guide Thermo Scientific
Peak Plot Labels Displays or hides the following peak labels:
•RT
•Area
•Height
•Filter
• Threshold
• Show All Apex Times
•Sample Filename
•Sample ID
•Compound Name
• Signal to Noise
Example with all peak labels displayed:
Table 35. Chromatogram Plot Settings dialog box parameters (Sheet 2 of 2)
Parameter Description
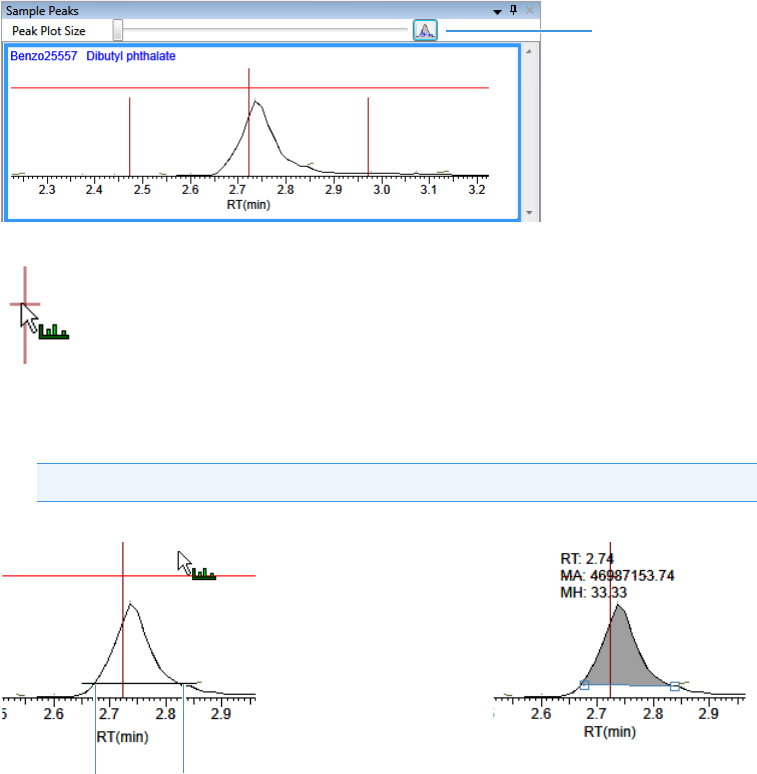
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 199
Manually Integrating Peaks
Use the manual integration feature to manually add a peak. You can manually add a peak in a
chromatogram plot only when the application fails to identify a peak.
To manually integrate a peak
1. In the Sample Peaks plot, click the Manual Integration icon.
The cursor changes to look like this:
2. Integrate a peak as follows:
a. Drag the cursor to define the beginning and ending base points for the new peak.
b. Click outside the plot to refresh the display.
3. To zoom in on an area, do the following:
a. Drag the cursor below the x axis or to the left of the y axis.
The plot zooms to fit the defined X or Y dimension into the entire pane. The
application zooms all compounds in the row to the same scale.
b. To return to the original view, right-click and choose Reset Scaling.
Note You must drag the cursor inside the x axis and y axis.
Manual integration
icon
Click the first base
point, and then drag to
the second base point.
Click outside the plot to
refresh the view.
The application identifies the peak
and indicates the manual
integration in the labels.
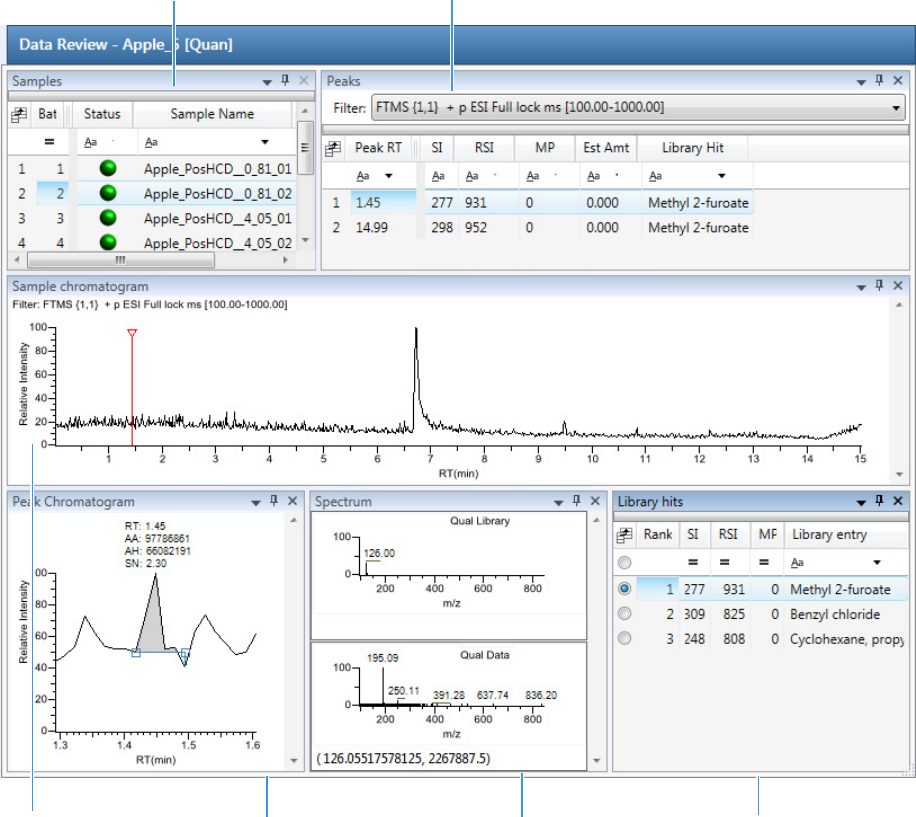
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
200 TraceFinder User Guide Thermo Scientific
Qualitative View for Quantitation Batches
The Qualitative View uses several different panes to display qualitative information for the
selected sample. See Displaying Qualitative View Panes.
If the application finds no detected peaks for the selected sample, you can manually add
peaks.
To see processed data for a sample in the Qualitative View, you must select the Qual
Processing parameter for that sample in the Batch View before you process the batch. See
Batch View Sample List.
These are the default panes and their locations:
Peaks pane
Samples pane
Sample
chromatogram pane
Peak Chromatogram
pane
Spectrum pane Library Hits pane
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 201
The Qualitative View displays data in the following panes:
•Samples Pane
•Peaks Pane
•Sample Chromatogram Pane
•Peak Chromatogram Pane
•Spectrum Pane (Library and Data)
•Library Hits Pane
Displaying Qualitative View Panes
The Qualitative View displays data using multiple panes: Samples, Peaks, Sample
Chromatogram, Peak Chromatogram, Spectrum, and Library Hits. You can display, hide, or
move any of these panes.
To display or hide a Qualitative View pane
From the View menu, choose to display or hide the following:
•Peak List: Displays the Peaks pane
•TIC Chromatogram: Displays the Sample Chromatogram pane
•Peak Chromatogram: Displays the Peak Chromatogram pane
•Spectrum: Displays the Spectrum pane
•Library Search: Displays the Library Hits pane
A check mark indicates a displayed pane.
For procedures about creating docked, floating, or tabbed panes, see Moving Data Review
Panes.
Note The Samples pane is required for the Qualitative View display. You cannot hide
the Samples pane.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
202 TraceFinder User Guide Thermo Scientific
Samples Pane
Use the Samples pane in the Qualitative View to select a specific sample. The associated Peaks
Pane displays all peaks found in the selected sample.
To hide or display columns in the Samples pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
The Field Chooser displays all available columns of data for the Samples pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Samples pane.
3. Click to close the Field Chooser.
Figure 64. Samples pane
Table 36. Samples pane columns (Sheet 1 of 2)
Column Description
Batch
Order
Sequentially numbers the samples.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 203
Peaks Pane
The Peaks Pane in the Qualitative View works with the Samples pane to display graphical
values for a unique sample and peak combination.
Follow these procedures:
•To display peaks for a specific compound
•To hide or display columns in the Peaks pane
To display peaks for a specific compound
1. From the Samples pane, select a sample.
The Peaks pane displays the retention times for peaks identified in the selected sample,
the values for the best match methods for each peak, and the library match.
The method specifies which technique to use for identifying peaks: peaks within a specific
retention time range, as a minimum percentage of the height or area of the largest peak,
or as a minimum percentage of the nearest internal standard peak. You can change the
method for identifying peaks in the Method Template Editor. Refer to Chapter 4, “Using
the Method Development Mode for Quantitation Methods,” in the TraceFi nde r Lab
Director User Guide.
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Note When you include unknown screening features in the quantitation
method and you choose to process with only the quantitation criteria, the
Sample View shows the Status for the samples as acquired and processed
( ), and the Unknown Screening View shows the Status for the samples as
acquired but not processed ( ).
Note When you include unknown screening features in the quantitation
method and you choose to process with only the unknown screening
criteria, both the Sample View and the Unknown Screening View show the
Status for the samples as acquired and processed ( ).
Sample
Name
A user-defined name that identifies a sample.
Sample
Typ e
Defines how the application processes the sample data. Each sample is
classified as one of the following sample types: Matrix Blank, Solvent, Cal Std,
QC Std, or Unknown.
Table 36. Samples pane columns (Sheet 2 of 2)
Column Description
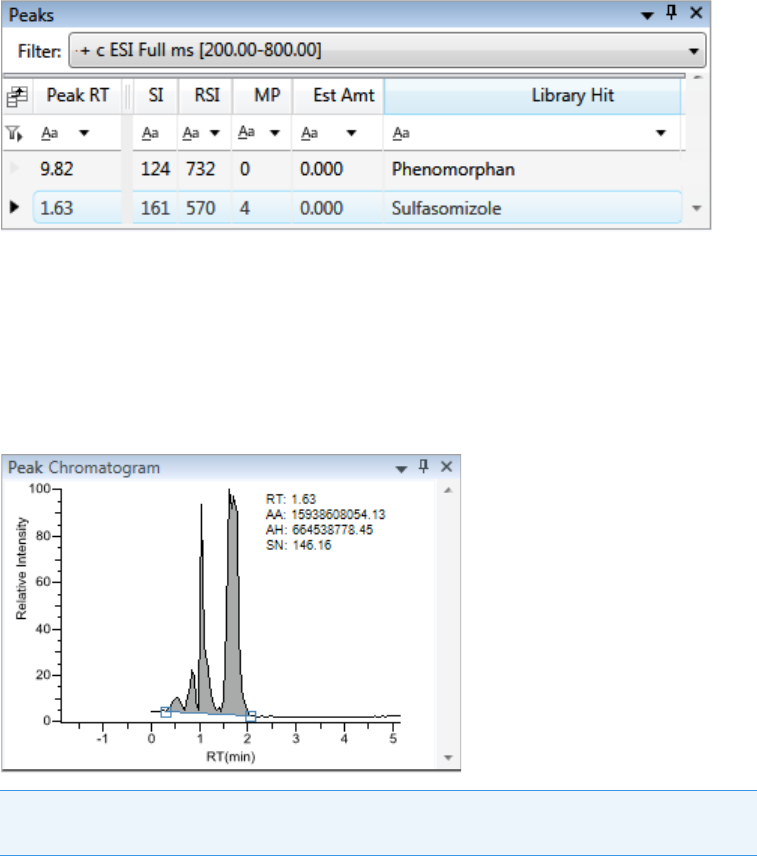
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
204 TraceFinder User Guide Thermo Scientific
2. In the Peaks pane, select a peak in the sample.
The application displays the selected peak in the Peak Chromatogram pane, displays the
Qual Data and Qual Library sections in the Spectrum pane, and locates the selected peak
in the Sample Chromatogram pane.
• The Qual Data section shows spectrum data for the peak in the raw data file.
• The Qual Library section shows actual spectrum for the identified library compound.
Figure 65. Peak Chromatogram pane
Note When you select a data-dependent sample, the peak can be from either a full
scan or a QED spectrum of an SRM-filtered chromatogram.
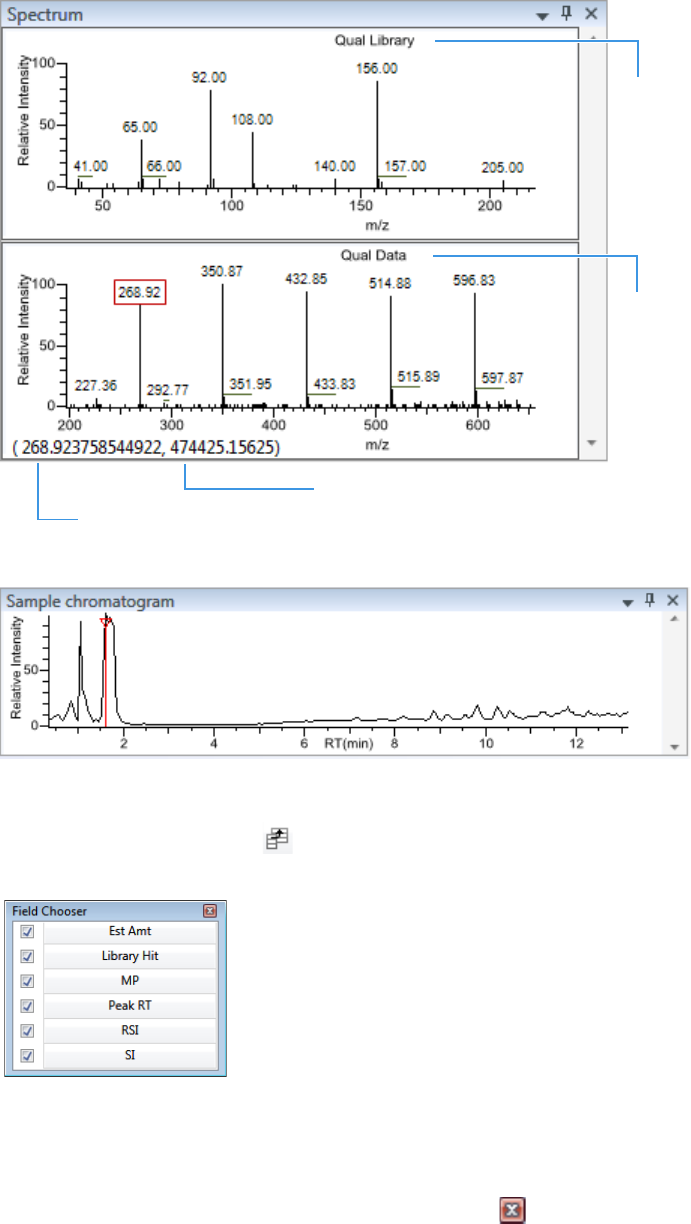
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 205
Figure 66. Spectrum pane
Figure 67. Selected peak in the Sample Chromatogram pane
To hide or display columns in the Peaks pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
The Field Chooser displays all available columns of data for the Peaks pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Peaks pane.
3. When you are finished modifying the column display, click to close the Field Chooser.
Reference
spectrum from
the library
Actual spectrum
data for the
compound
m/z value for the selected spectrum
Absolute intensity for the mass
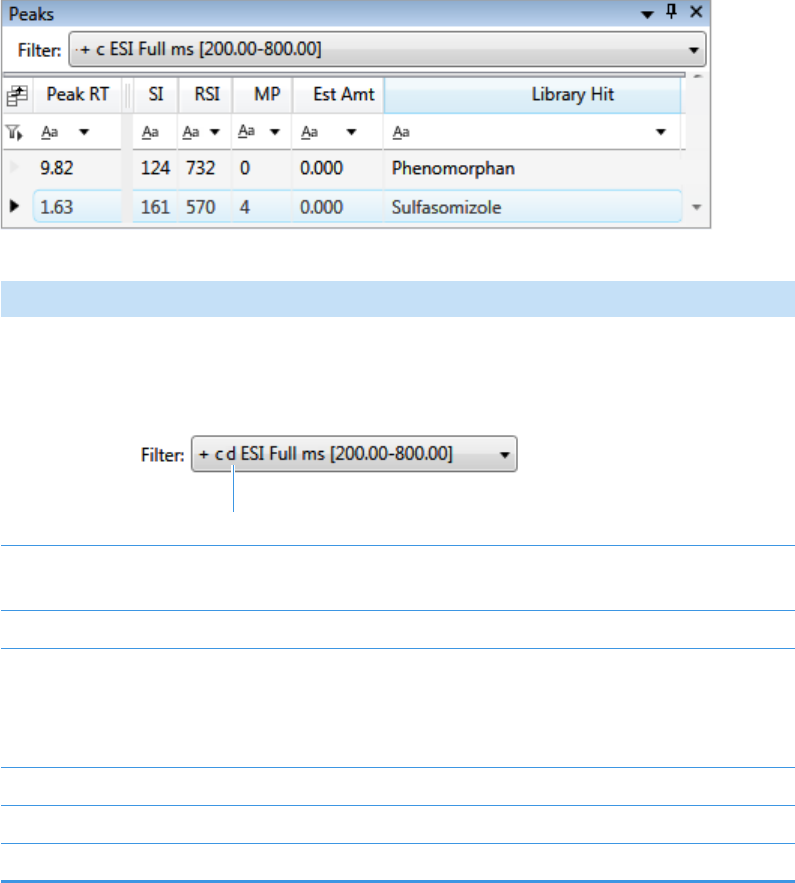
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
206 TraceFinder User Guide Thermo Scientific
Peaks Pane
Use the features in the Peaks pane to display graphical values for each sample and peak
combination.
Figure 68. Peaks pane
Table 37. Peaks pane parameters
Parameter Description
Filter Filter used to identify the peaks. Specified in the raw data file or the master
method.
When your raw data file is data-dependent, the filter indicates this with a “d”.
Peak RT Peak retention time. The time after injection when the compound elutes. The
total time that the compound is retained on the column.
SI Search index method used to search the NIST library.
RSI (Reverse search index) A method used to search the NIST library. A reverse
search compares a library entry to an unknown compound (whereas a forward
search compares the mass spectrum of an unknown compound to a mass
spectral library entry).
MP Match probability.
Est Amt Estimated amount of the compound.
Library Hit Library compound that matches the identified peak.
Data-dependent filter
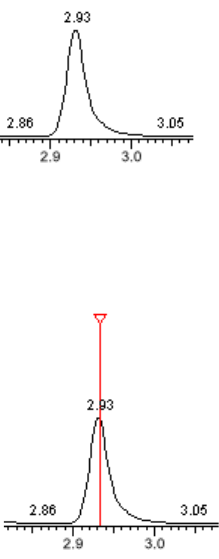
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 207
Sample Chromatogram Pane
The Sample Chromatogram Pane in the Qualitative View displays all peaks in the selected
sample. The peak selected in the Peaks pane displays a red marker.
To zoom in on a peak
1. In the Sample Chromatogram pane, drag the cursor to delineate a rectangle around the
peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the Sample Chromatogram pane and choose Reset
Scaling.
To manually add a peak
1. Zoom in to better identify which peak to add to the results set.
2. Right-click the Sample Chromatogram pane, and choose Add Qual Peak.
3. Click to indicate the left and right base points for the peak.
The application marks the peak in the Sample Chromatogram pane.
The application places the peak delimiter tags at the base point locations and
automatically updates the peak values in the Peaks pane and Peak Chromatogram pane.
See Peak Chromatogram pane with a manually added peak.
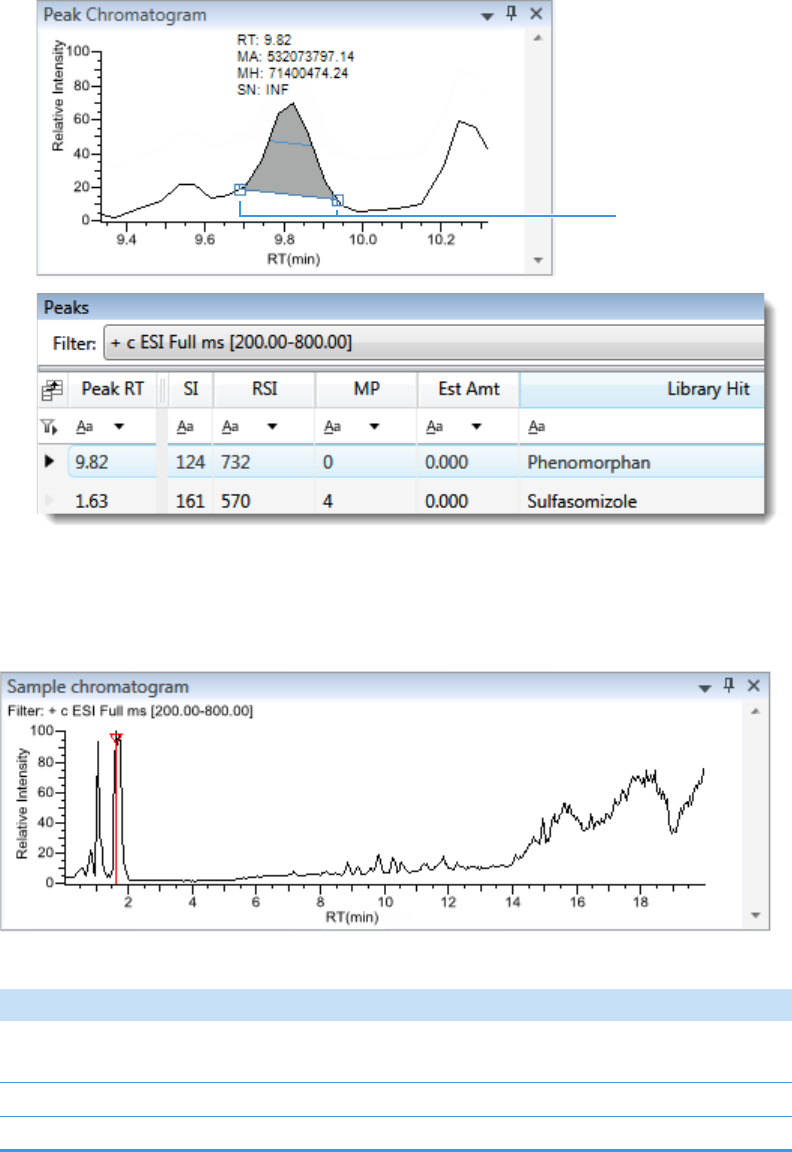
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
208 TraceFinder User Guide Thermo Scientific
Figure 69. Peak Chromatogram pane with a manually added peak
Sample Chromatogram Pane
Use the features in the Sample Chromatogram pane to display peaks in the selected sample.
Figure 70. Sample Chromatogram pane
Table 38. Sample chromatogram pane shortcut menu commands
Command Description
Add Qual Peak Select the beginning and ending base points for a new
qualitative peak. Available only when no peak is detected.
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Manually added
base points
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 209
Peak Chromatogram Pane
The Peak Chromatogram Pane in the Qualitative View displays the selected peak.
Follow these procedures:
•To zoom in on a peak
•To remove a peak
•To switch between method and manual integration modes
•To customize the labels in the display
To zoom in on a peak
1. In the chromatogram plot, drag the cursor to delineate a rectangle around the peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the chromatogram plot and choose Reset Scaling.
To remove a peak
Right-click the Peak Chromatogram pane, and choose Remove Qual Peak.
The application removes the peak displayed in the Peak Chromatogram pane. All data for
this peak are removed from the Qualitative View panes.
To switch between method and manual integration modes
Right-click the Peak Chromatogram pane and choose either Method Integration or
Manual Integration.
Initially, the method integration and manual integration settings that are stored for a
compound and file are identical. When you switch modes, the saved result set does not
change. However, when manual data are available, both the Peak Chromatogram plots
and the result set update as you switch between method and manual modes.
As you switch between modes, each pane reflects the changes. The generated reports for
the data identify the manual modifications.
To customize the labels in the display
1. Right-click and hold the cursor over Peak Labels in the shortcut menu.
2. To display labels for the peak retention time (RT), area (AA), height (AH), or
signal-to-noise (SN), select the appropriate check box.
See Example of displayed labels.
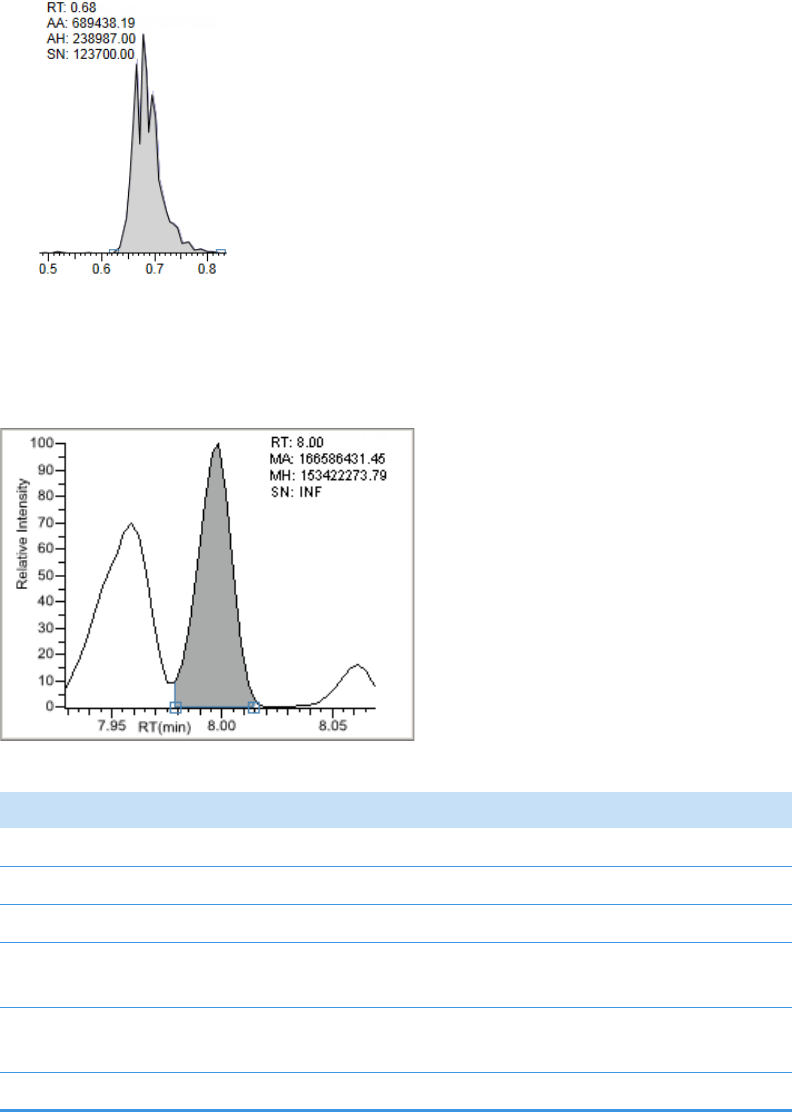
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
210 TraceFinder User Guide Thermo Scientific
Figure 71. Example of displayed labels
Peak Chromatogram Pane
Use the features in the Peak Chromatogram pane to display the selected peak.
Figure 72. Peak Chromatogram pane
Table 39. Peak Chromatogram pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Method Integration Displays method integration settings.
Manual Integration Displays manual integration settings.
Peak Labels Displays or hides the peak labels (Label Area, Label
Retention Time, Label Height, or Label to Noise).
Remove Qual Peak Available only for manually added peaks. Removes the
peak displayed in the Peak Chromatogram pane.
Copy to Clipboard Copies the graphic display to the Clipboard.
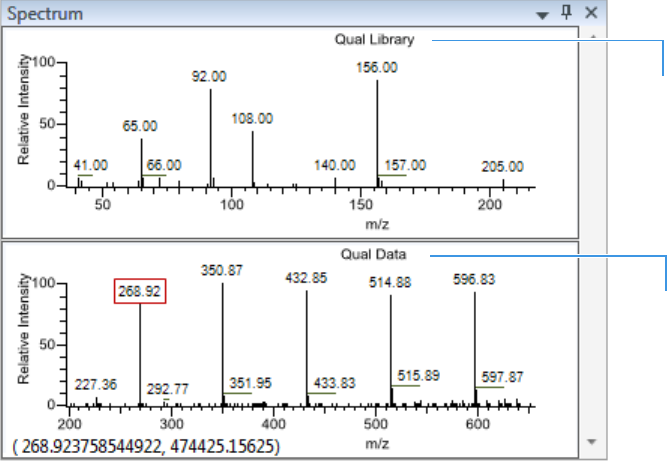
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 211
Spectrum Pane (Library and Data)
The Spectrum pane in the Qualitative View displays the reference spectrum from the library
and the spectrum data for the selected sample. The top pane displays the spectrum for the
identified compound found in the reference library; the bottom pane displays the actual
spectrum data for the selected peak.
Figure 73. Spectrum pane
To zoom in on a peak
1. In either spectrum plot, drag the cursor to delineate a rectangle around the peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the spectrum plot and choose Reset Scaling.
To display detailed m/z values
In the spectrum pane, hold the cursor over the m/z value for a peak.
The application indicates the rounded-off m/z value with a red box and displays the
complete value in the lower left corner of the display.
Reference
spectrum from
the library
Actual spectrum
data for the
compound
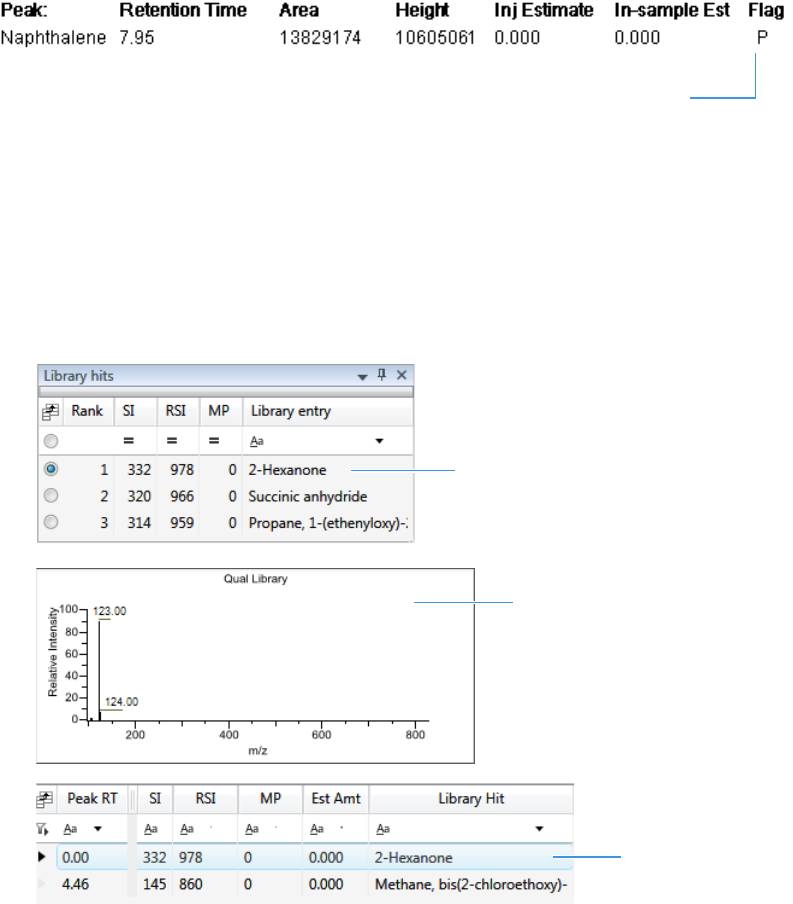
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
212 TraceFinder User Guide Thermo Scientific
Library Hits Pane
The Library Hits pane in the Qualitative View displays the best library matches for the
selected peak. Use this pane to select a different library entry for the peak.
When you select a library entry other than the original entry, the TIC Report and TIC
Summary Report indicate this with a “P” flag:
To change the library entry for a selected peak
In the Library Hits pane, select the check box for the library entry that you want to use to
identify the selected peak.
• In the Spectrum pane, the reference spectra change to show the spectra for the
selected library entry.
• In the Peaks pane, the SI, RSI, MP, and Compound values update to reflect the
selected library entry.
P flag
Selected library entry
in the Library Hits pane
Reference spectra
for Hexanone
Peak list for
Hexanone
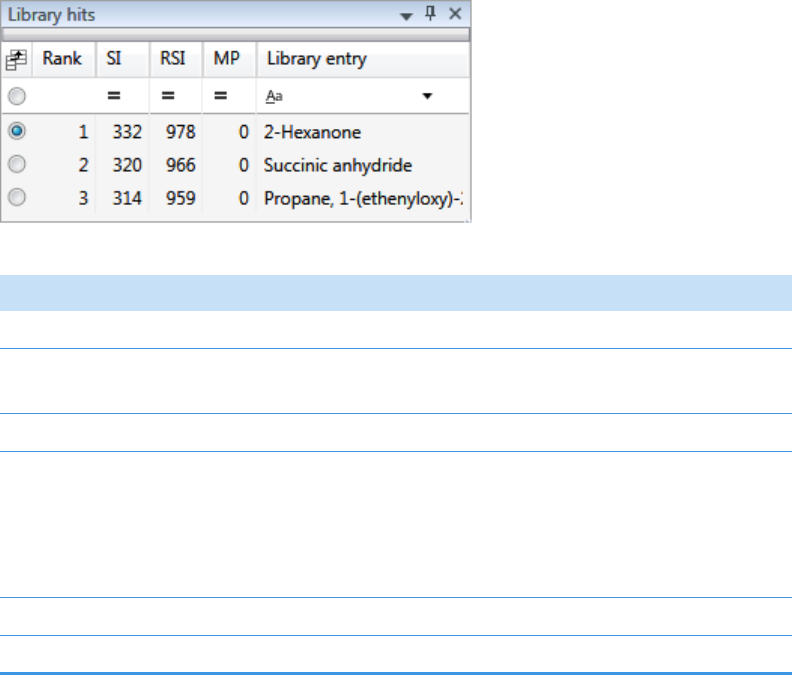
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 213
Figure 74. Library Hits pane
Table 40. Library Hits pane parameters
Parameter Description
<Check box column> Indicates selected library entries for the selected peak.
Rank Indicates the order of best matches between the selected
peak and library entries.
SI (Search index) A method used to search the NIST library.
RSI (Reverse search index) A method used to search the NIST
library. A reverse search compares a library entry to an
unknown compound (whereas a forward search compares
the mass spectrum of an unknown compound to a mass
spectral library entry).
MP Match probability.
Library Entry Library compound that matches the identified peak.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
214 TraceFinder User Guide Thermo Scientific
Column Parameters for Compound Results and Sample Results
The Compound Results and Sample Results data review pages use the following parameters
and features:
•Common Column Parameters
•Inactive and Excluded Compounds
•Exporting Compounds
Common Column Parameters
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 1 of 8)
Column Description
%CV Coefficient of Variance. Standard deviation of the multiple samples of one level, multiplied
by 100, and divided by the average of the multiple samples of that level. This calculation is
based on the areas of the peaks.
%Diff The calculated amount minus the expected amount, divided by the expected amount, and
then multiplied by 100.
%RSD Standard deviation of the multiple samples of one level, multiplied by 100, and divided by
the average of the multiple samples of that level. This calculation is based on the calculated
amounts.
Note This RSD value is not the same as the RSD value used with the Average RF curve
type in the method. Refer to Chapter 4, “Using the Method Development Mode for
Quantitation Methods,” in the TraceFinder Lab Director User Guide.
The application uses this %RSD value in Data Review and in the Compound Calibration
Report when you acquire multiple samples for the same QC Std or Cal Std samples.
Active Displays or hides a compound for a particular sample.
• When a compound is marked inactive, the application does not remove its data and
calculated values from the result set. Instead, the application masks the appearance of
that compound for that particular sample and grays the compound in the compounds
list.
• When a calibration standard is marked inactive, the application no longer uses the data
file’s calibration point for the calibration and removes it from the graphical view of the
calibration curve displayed in the Compound Details pane. It is no longer part of the
result set.
In a Sample View, the Active parameter is in the Compound Results pane.
In a Compound View, the Active parameter is in the Sample Results pane.
Actual RT Actual retention time for the compound. Retention time is the time after injection when a
compound elutes and the total time that the compound is retained on the chromatograph
column.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 215
Adduct The most intense adduct for the retention time for a compound.
Area The area obtained by integrating peak intensities from the start to the end of the peak.
When the Response Ratio is specified as Area, this column displays an asterisk (*Area).
Batch Order
(Sample Results only)
Sequentially numbers the samples in the batch.
Calculated Amt The amount present in the sample, as determined using the calibration curve and the
response ratio.
Channel Specifies the channel on which the sample was run. If the sample is not acquired, the value is
Pending. The Channel column is available only when you have activated multiplexing in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in the
TraceFinder Lab Director User Guide.
Compound
(Compound Results
only)
Name of the identified compound.
Comments
(Sample Results only)
A user-defined comment for the sample.
Confirm Specifies that the compound meets all required processing criteria. Refer to the instructions
“To display only confirmed peaks” in Chapter 4, “Using the Method Development Mode
for Quantitation Methods,” in the TraceFinder Lab Director User Guide.
To display the confirmation criteria, hold your cursor over the indicator.
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 2 of 8)
Column Description
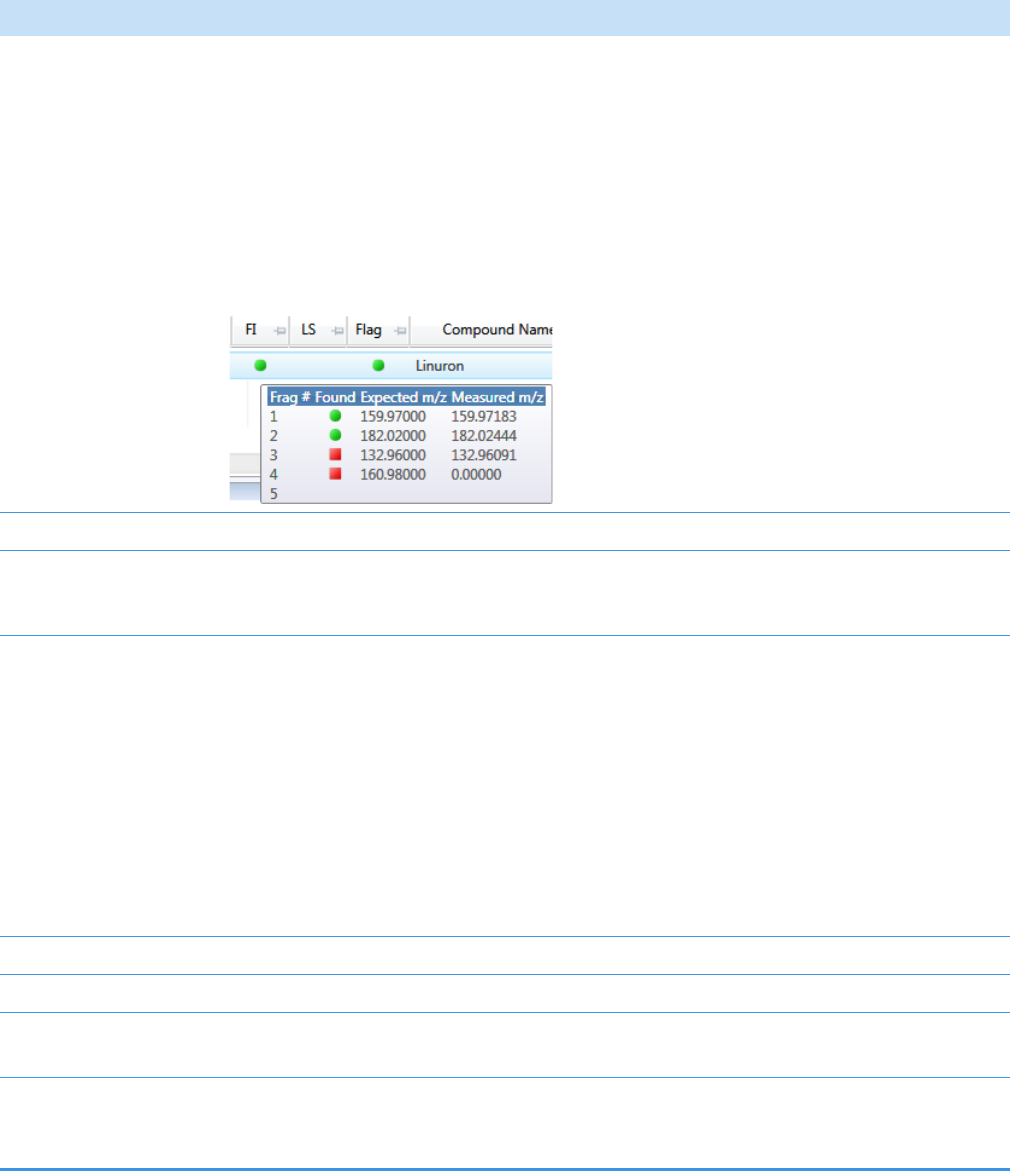
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
216 TraceFinder User Guide Thermo Scientific
FI Fragment Ions flag. The application displays one of these indicators:
• A green circle (pass) when the measured m/z value of any of the fragments is within the
mass tolerance specified in the method. On the Isotopes page in the Spectrum pane, the
All Isotopes and Multi-Isotopes flags are also green.
• A red square (fail) when the measured m/z value of none of the fragments is within the
mass tolerance specified in the method. On the Isotopes page in the Spectrum pane, the
All Isotopes and Multi-Isotopes flags are also red.
• A blank when there are no fragments detected.
To display a list of fragments and their pass/fail status, hold your cursor over the indicator.
Filename A user-defined name that identifies a sample.
Final Units Specifies the calculated amount.
Default: 1
Flag Details Indicates all errors found in the compound.
Typ e o f er ror :
• I: Confirming ion coelution failure
• A: Amount error
• B: Matrix blank error
•PK: Peak not found
• LS: Library matching error
• IP: Isotope error
• FI: Fragment ions error
•IR: Ion ratio error
Flags Caution flag displayed when a compound within the sample has an error.
Formula The formula for the peak as specified in the compound database.
Group
(Sample Results only)
Threshold group to which a sample belongs. You can view samples by group in the
Comparative View.
Group Average
(Compound Results
only)
Average of the samples in a threshold group. You can view averaged samples in the
Compound View.
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 3 of 8)
Column Description
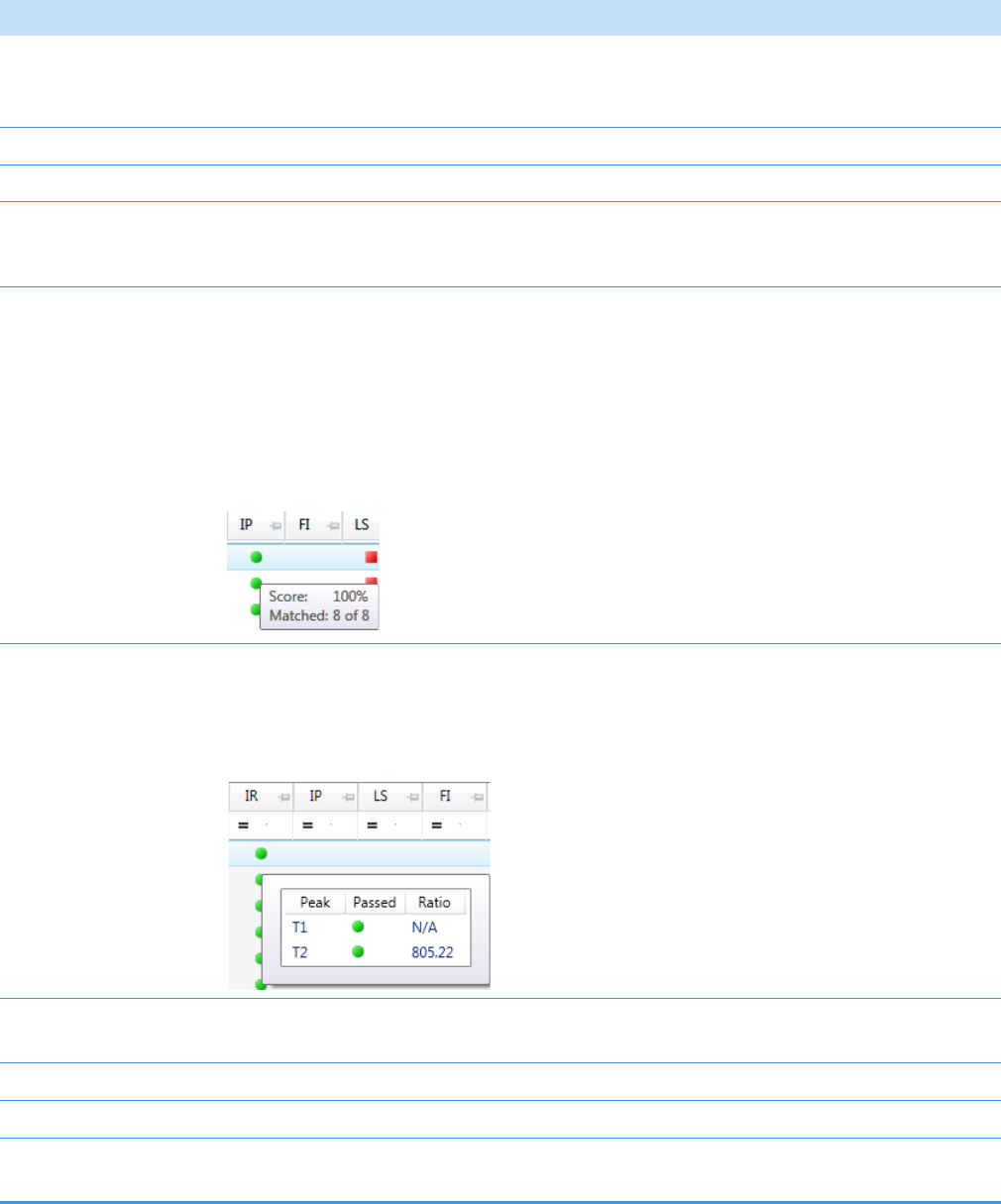
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 217
Height Specifies the distance from the peak maximum to the peak base, measured perpendicular to
the ordinate. When the Response Ratio is specified as Height, this column displays an
asterisk (*Height).
Integration Mode Specifies the integration mode (Method, Manual, or User). See Quan Peak.
Ion Ratio Specifies the ratio of the confirming ion’s response to the target ion’s response.
Ion Type Specified as b or y with a numerical designation.
Available only for peptide databases.
IP Isotopic Pattern flag. The application displays one of these indicators:
• A green circle (pass) when the score percentage is higher than the specified fit threshold
percentage
• A red square (fail) when the score percentage is lower than the specified fit threshold
percentage
• A blank when the parameter is not scored
To display the score of matched isotopes, hold your cursor over the indicator.
IR Ion Ratio flag. The application displays one of these indicators:
• A green circle when the ion ratio is within the acceptable ion ratio range
• A red square when the ion ratio is not within the acceptable ion ratio range
To display the ion ratio results, hold your cursor over the indicator.
Isotopic Pattern Score
(%)
The percentage of the number of total isotopes to the number of matched isotopes.
ISTD Amt Amount of internal standard.
ISTD Response Response of the internal standard.
Level
(Sample Results only)
The level defined for a calibration sample or quality control sample.
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 4 of 8)
Column Description

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
218 TraceFinder User Guide Thermo Scientific
Lib Match Name The name of the best matching compound in the library search. When the application finds
a match in the library, this column displays the matching library entry with the highest
score.
• When the application does not perform a library search, this column displays “N/A” in
black text.
• When the application does not perform an MS/MS scan, this column displays “N/A” in
red text.
Library Match Rank Displays the ranking of the library match. When the application finds a match in the library,
this column displays the library entry’s relative rank, in the format “x of y”, where
• x = the rank of the highest scoring library match.
• y = the total number of library matches from the list of matches for a particular adduct
that contains the highest scoring match.
Results are as follows when the application performs both a library search and an MS/MS
scan and both the library entry and the formula match the target compound:
• A library score that is higher than or equal to the score threshold meets the criteria, and
the values in this column appear in green.
• A library score that is lower than the score threshold fails to meet the criteria, and the
values in this column appear in red.
When the application does not perform a library search, this column displays “N/A” in
black text.
When the application does not perform an MS/MS scan, this column displays “N/A” in red
text.
Library Probability
Percentage
Specifies the probability of the match in the NIST library for forward searches only.
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 5 of 8)
Column Description

4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 219
Library Score (%) Specifies the score from the library fit. When the application finds a match in the high
resolution NIST library or in the mzVault library, this column displays the highest score
associated with the Lib Match Name parameter.
Results are as follows when the application performs both a library search and an MS/MS
scan and both the library entry and the formula match the target compound:
• A library score that is higher than or equal to the score threshold meets the criteria, and
the values in this column appear in green.
• A library score that is lower than the score threshold fails to meet the criteria, and the
values in this column appear in red.
When the application does not perform a library search, this column displays “N/A” in
black text.
When the application does not perform an MS/MS scan, this column displays “N/A” in red
text.
Valid range: 1 through 100%
LS Library Search flag. The application displays one of these flags:
• A green circle when the library search is successful.
• A red square when the library search is not successful.
m/z (Apex) Mass-to-charge ratio found in the spectra for the peak. Assumes that the charge is 1.
When the application successfully integrates the peak, this column displays the charged m/z
value for the compound, which is the highest intensity in the apex scan.
When the application cannot successfully integrate the peak, this column displays “N/F”
(not found).
m/z (Delta) Difference between the m/z (Expected) and m/z (Apex). Assumes that the charge is 1.
When the m/z (Apex) column displays m/z value for the compound, this column displays
the delta m/z corresponding to the highest intensity in the apex scan.
• When the mass tolerance is specified in ppm in the master method, then
m/z (Delta) = 1 000 000 × ([m/z (Apex) – m/z (Expected)] m/z (Expected)).
• When the mass tolerance is specified in mmu in the master method, then
m/z (Delta) = 1000 × (m/z (Apex) – m/z (Expected)).
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 6 of 8)
Column Description
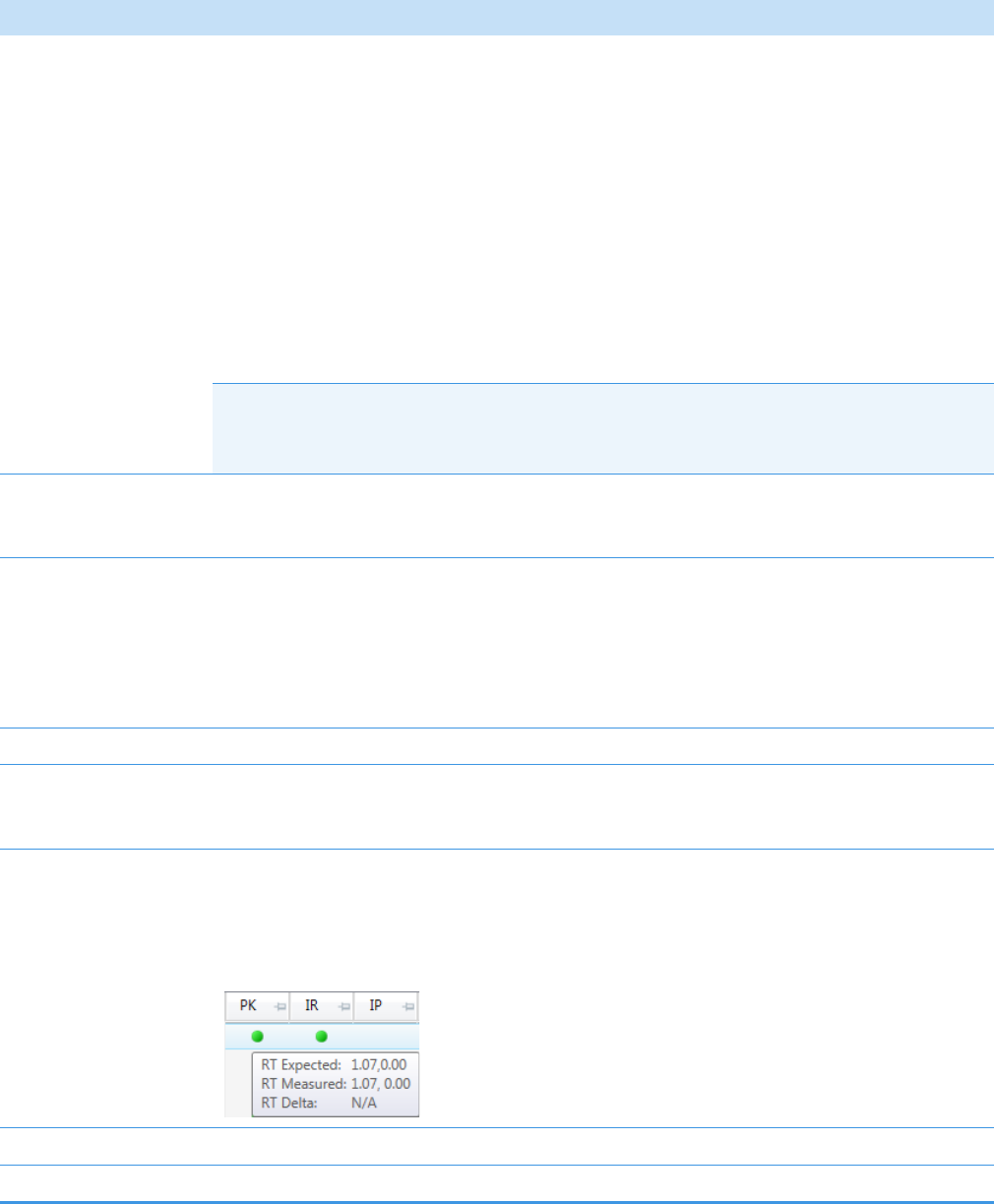
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
220 TraceFinder User Guide Thermo Scientific
m/z (Expected) Mass-to-charge ratio from the compound database. Assumes that the charge is 1.
• When an adduct is found, the application displays the neutral mass value for the
compound (calculated from the neutral formula) ± the mass of the most intense adduct
ion found for the compound.
• When no adduct is found, the application displays the neutral mass value for the
compound ± the mass of the first adduct entered in the compound database.
For details about defining adducts for the compound database, refer to Chapter 1, “Using
the Configuration Console,” in the TraceFinder Lab Director User Guide.
For details about adding adducts to compounds, refer to Chapter 2, “Using Compound
Databases in the Method Development Mode,” in the TraceFinder Lab Director User Guide.
Note When the adduct is a gain, the adduct mass is a positive number. When the adduct
is a loss, the adduct mass is a negative number. The resulting mass value after adding or
subtracting the adduct mass is always a positive number.
Modification Specifies the modifications from imported peptide files or from the Peptide Predictor tool.
Available only for peptide databases.
Num Isotopes Matched The number of isotopes matched in the expected calculated isotope spectra relative to the
total number of isotopes used in the score calculation, in the format “x of y”, where
• x = the number of isotopes matching the elemental composition used for the Isotopic
Pattern Score calculation.
• y = the total number of isotopes considered in the Isotopic Pattern Score calculation.
This is the number of isotope peaks expected to be above the spectral noise.
Peak Label User-specified label.
Peptide Sequence Specifies the peptide sequence that the application uses to calculate the m/z or precursor m/z.
Available only for peptide databases.
PK Specifies if the peak was found. The application displays one of these flags:
• A green circle when the peak is found
• A red square when the peak is not found
To display the peak results, hold your cursor over the indicator.
Protein Name Alphanumeric name assigned to the protein.
Relative RT Specifies the measured retention time relative to the RT Reference values set in the method.
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 7 of 8)
Column Description
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 221
Response Ratio The ratio of the Response value to the IS Response value. If the Response is specified as Area
in the processing method, the units of both Response and IS Response are counts-sec. If the
Response is specified as Height in the processing method, the units of both Response and IS
Response are counts.
RSI/Rev Dot Specifies the results of a reverse search of the NIST library. A reverse search compares a
library entry to an unknown compound.
RT Specifies the retention time for the compound.
RT Delta Specifies the difference between the expected retention time and the measured retention
time.
S/N The signal-to-noise ratio calculated for the found peak.
Sample Amt The injected volume multiplied by the conversion factor. For example, if you have
1000 ng/mL of a substance that is too concentrated for the mass spectrometer, you can
dilute it by 1000. Then your injection volume is 1, your conversion factor is 1000, and your
sample amount is 1000.
SI/Dot Product Specifies the results of a forward search of the NIST library. A forward search compares the
mass spectrum of an unknown compound to a mass spectral library entry.
Sample ID
(Sample Results only)
A user-defined, alphanumeric string that identifies a sample.
Sample Name
(Sample Results only)
A user-defined name that identifies a sample.
Sample Type
(Sample Results only)
Defines how the application processes the sample data. Each sample is classified as one of
the following sample types: Matrix Blank, Solvent, Cal Std, QC Std, or Unknown.
Std Add Amount Specifies the standard amount added to the sample.
Theoretical Amt Theoretical amount of the compound expected in the sample.
Type In the Compound Results pane, specifies Target Compound, Confirming Peak, or
Fragment.
In the Sample Results pane, specifies Sample, Confirming Peak, or Fragment.
Table 41. Common parameters for Compound Results and Sample Results tables (Sheet 8 of 8)
Column Description
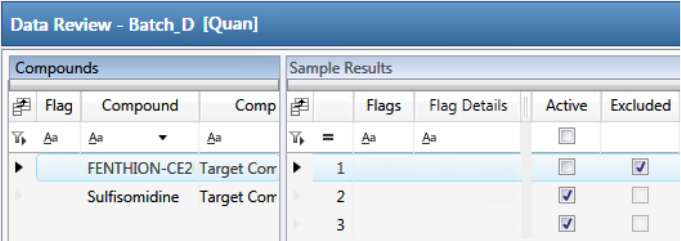
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
222 TraceFinder User Guide Thermo Scientific
Inactive and Excluded Compounds
Use the Active and Excluded columns to control which compounds are used for calculating
the calibration curve and for reporting.
Follow these procedures:
•To make a sample active or inactive
•To exclude a calibration point
To make a sample active or inactive
1. Select the sample in the Sample Results pane.
All compounds in the selected sample appear in the Compounds pane. Inactive
compounds are grayed out.
2. In the Compounds pane, select the compound whose active/inactive status you want to
change.
• When a compound it marked inactive, the application does not remove its data and
calculated values from the result set. Instead, the application masks the appearance of
that compound for that particular sample and grays the compound name in the
compounds list.
• When a calibration standard is marked inactive, the application no longer uses the
data file’s calibration point for the calibration and removes it from the graphical view
of the calibration curve displayed in the Qualification pane. The calibration point is
no longer part of the result set.
3. In the Sample Results pane, select or clear the Active check box.
Use the horizontal scroll bar at the bottom of the table to scroll to the Active column.
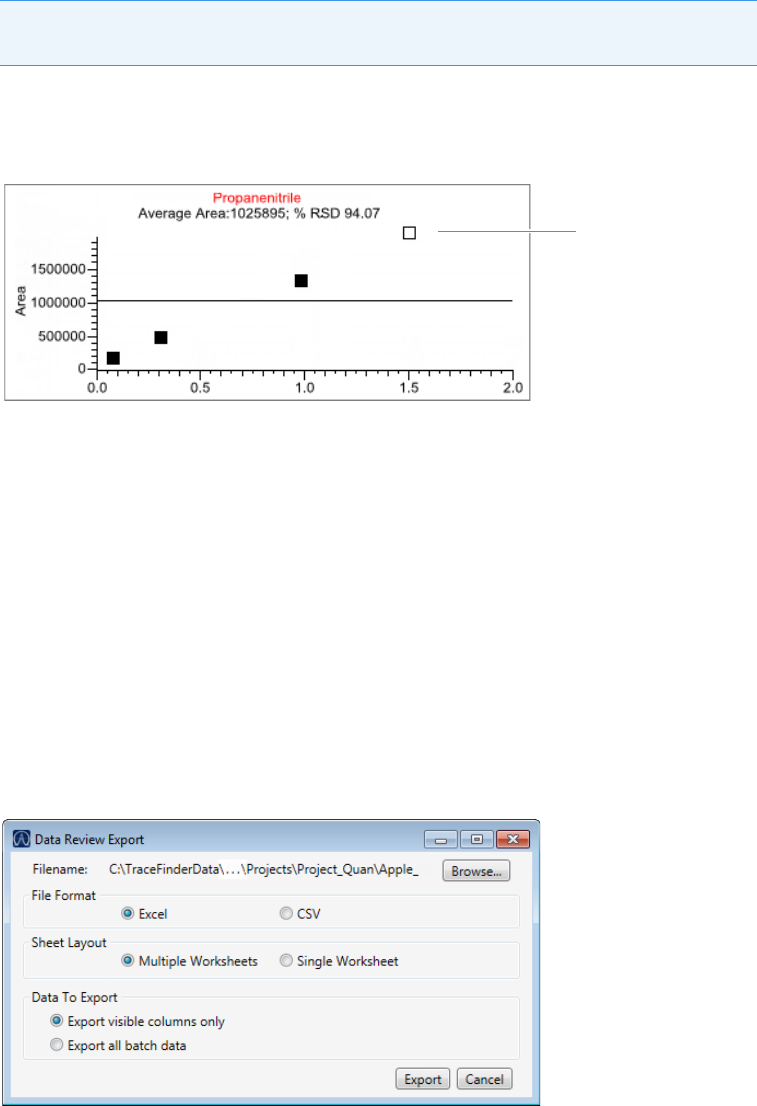
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 223
To exclude a calibration point
In the sample list, select the Excluded check box for the sample.
Use the horizontal scroll bar at the bottom of the table to scroll to the Excluded column.
The application displays a value that is no longer used for calibration as an empty box in
the graphical view of the calibration curve.
Exporting Compounds
You can export compound data to an Excel spreadsheet or to a CSV file. These export
commands are available from the File menu in the Sample View, Compound View, and
Comparative View.
Follow these procedures:
•To export compounds to an Excel spreadsheet
•To export compounds to a CSV file
To export compounds to an Excel spreadsheet
1. Choose File > Export Data To > CSV or Excel from the main menu.
The Data Review Export dialog box opens.
2. Click Browse and, in the Export Data to Excel dialog box, locate the folder where you
want to save the file.
Note Only calibration samples have the Excluded check box available. See Inactive
and Excluded Compounds.
Excluded calibration
point
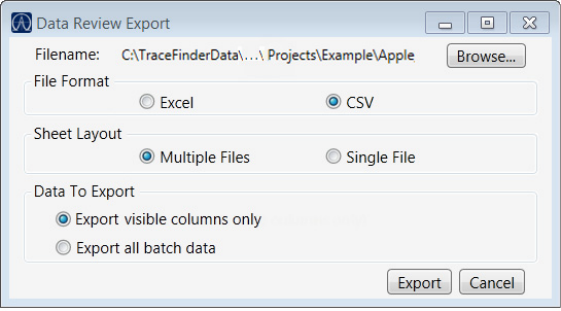
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
224 TraceFinder User Guide Thermo Scientific
3. Type a file name for the XLSX file and click Save.
4. In the File Format area, in the Data Review Export dialog box, select the Excel option.
5. In the Sheet Layout area, select one of the following file formats for the spreadsheet.
•Multiple Worksheets: Writes one sample to each Excel worksheet tab.
•Single Worksheet: Writes all samples to a single Excel worksheet tab.
6. In the Data to Export area, select one of the following sets of data to export.
•Export Visible Columns Only: Writes data from the displayed columns of selected
samples to the specified worksheet format.
•Export All Batch Data: Writes data from all columns (displayed or hidden) of all
samples to the specified worksheet format.
7. Click Export.
The application saves the specified compound data to an Excel spreadsheet and opens the
folder where you saved the file. The application names the file Batch.xlsx.
To export compounds to a CSV file
1. Choose File > Export Data To > CSV or Excel from the main menu.
The Data Review Export dialog box opens.
2. Click Browse and, in the Export Data to Excel dialog box, locate the folder where you
want to save the file.
3. Type a file name for the CSV file and click Save.
4. In the File Format area, in the Data Review Export dialog box, select the CSV option.
5. In the Sheet Layout area, select one of the following file formats for the spreadsheet.
•Multiple Files: Writes one sample to each CSV file.
•Single File: Writes all samples to a single CSV file.
4 Using the Analysis Mode for Quantitation Batches
Working in Data Review for Quantitation Batches
Thermo Scientific TraceFinder User Guide 225
6. In the Data to Export area, select one of the following sets of data to export.
•Export Visible Columns Only: Writes data from the displayed columns of selected
samples to the specified worksheet format.
•Export All Batch Data: Writes all data from all samples in the batch to the specified
worksheet format.
7. Click Export.
The application saves the specified compound data to a CSV spreadsheet.
When you selected to create multiple files, the application opens the folder where you
saved the files. The application names each file Batch_Compound.csv.
When you selected to create a single file, the Excel application opens, displaying the
exported data for all samples. The application names the file Batch.csv.
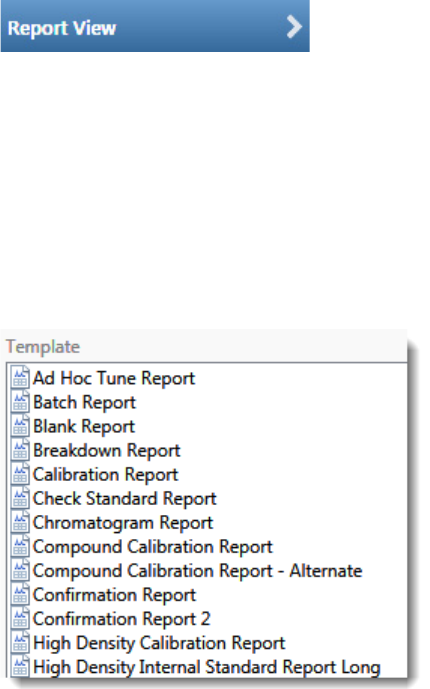
4 Using the Analysis Mode for Quantitation Batches
Working in the Report View for Quantitation Batches
226 TraceFinder User Guide Thermo Scientific
Working in the Report View for Quantitation Batches
The Report View displays example reports for the current batch. You must have an open
batch to use the features in the Report View.
Follow these procedures:
•To open the Report View
•To preview a report
•To generate a report as a PDF, an Excel, or a CSV file
•To print a report
•To display a generated report
•To e d i t a re po r t t e m p l a t e
•To create a new report template
To open the Report View
Click Report View in the navigation pane.
The application opens the Report View.
To preview a report
1. In the Template pane, select a report template.
The template list shows all the quantitation report templates that you configured in the
Configuration console. Refer to “Specifying the Reports” in Chapter 1, “Using the
Configuration Console,” in the TraceFinder Lab Director User Guide.
Figure 75. Example of a report template list
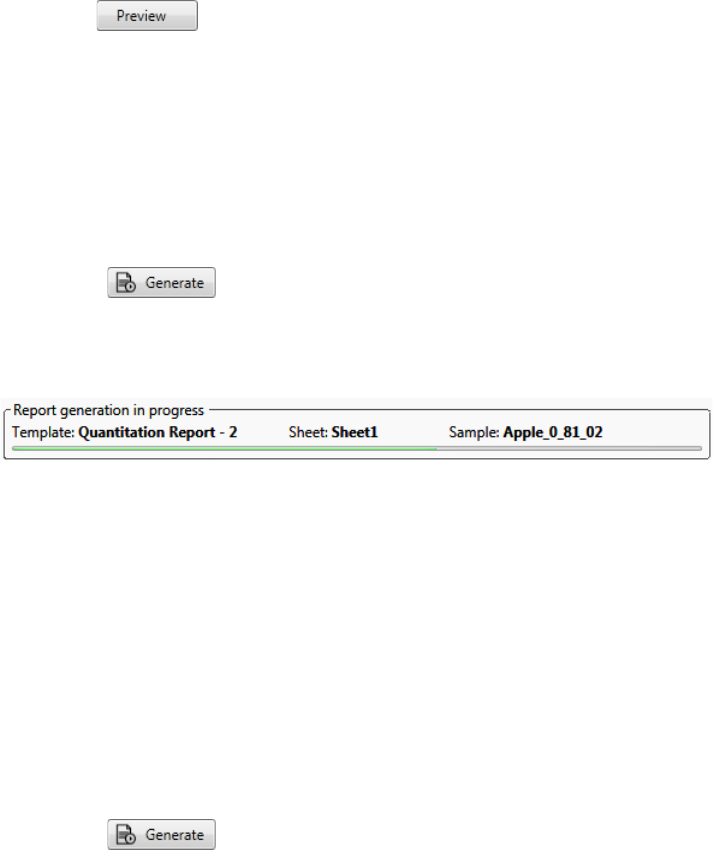
4 Using the Analysis Mode for Quantitation Batches
Working in the Report View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 227
2. Click Preview, .
The application opens a Report Preview, showing the report information for the current
batch in the selected report template format.
To generate a report as a PDF, an Excel, or a CSV file
1. In the Template pane, select a report template.
2. Select the check box for each of the file types that you want to create: PDF, Excel, or
CSV.
3. Click Generate, .
The application does the following:
• Displays a green progress bar as it generates the reports.
• Creates a report for the current batch as a PDF, an Excel, or a CSV file, using the
selected report template format.
• Adds information about the generated report to the Generated Reports pane.
For details about the Generated Reports pane, see Report View.
• Saves the report files to the …\TraceFinderData\4.0\Projects\batch\ReportOutput
folder.
To print a report
1. In the Template pane, select a report template.
2. Select the check box for the Print file format.
3. Click Generate, .
The application does the following:
• Creates a report for the current batch using the selected report template format.
• Prints the report to your default printer.
• Adds information about the generated report to the Generated Reports pane.
For details about the Generated Reports pane, see Report View.
• Saves the report files to the …\TraceFinderData\4.0\Projects\batch\ReportOutput
folder.
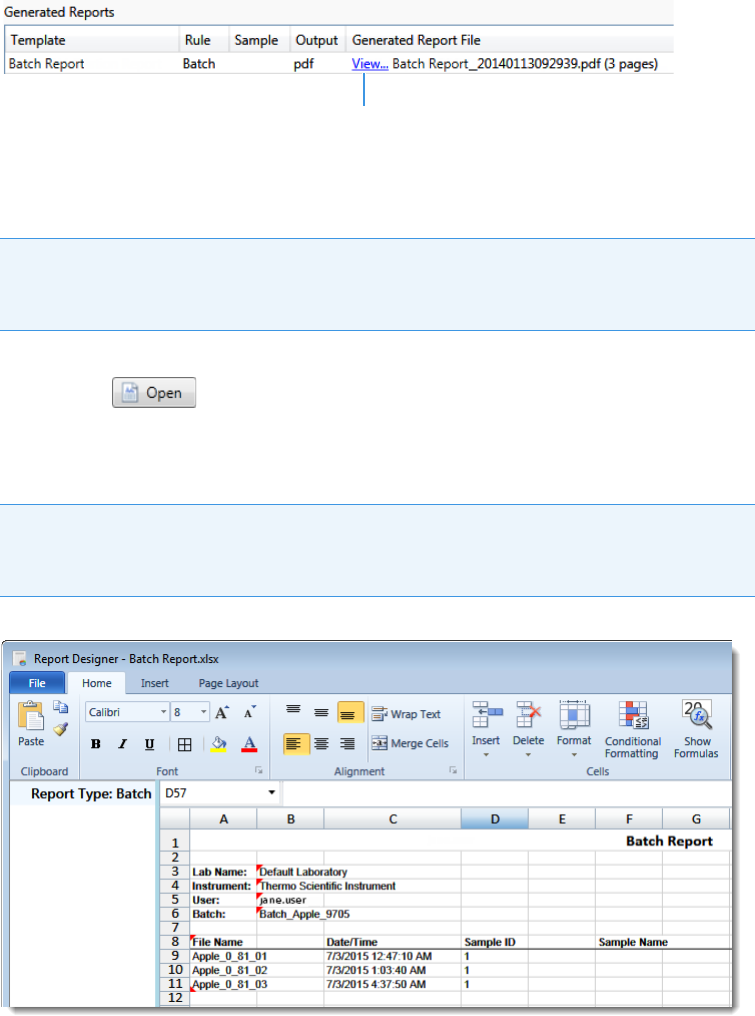
4 Using the Analysis Mode for Quantitation Batches
Working in the Report View for Quantitation Batches
228 TraceFinder User Guide Thermo Scientific
To display a generated report
In the Generated Reports pane, click View for the report that you want to see.
Figure 76. Generated Reports pane showing a PDF report
The application opens the output file.
To edit a report template
1. In the Template pane, select a report template.
2. Click Open, .
The application opens the Report Designer showing the template in an Excel spreadsheet.
See Report Designer showing the template for the selected report.
Figure 77. Report Designer showing the template for the selected report
3. Use the features in the Report Designer to edit the template.
See Chapter 7, “Using the Report Designer.”
4. When you finish your changes, choose File > Save from the Report Designer menu bar.
Note You cannot edit report templates that were provided with the TraceFinder
application; however, you can open a TraceFinder template, make changes, and save it
to a new template name.
Note When user security is activated, you must have Template Editing permission to
edit report templates created by your laboratory. If the Open button is not active, user
security is activated and you do not have Template Editing permission.
Opens the generated file.
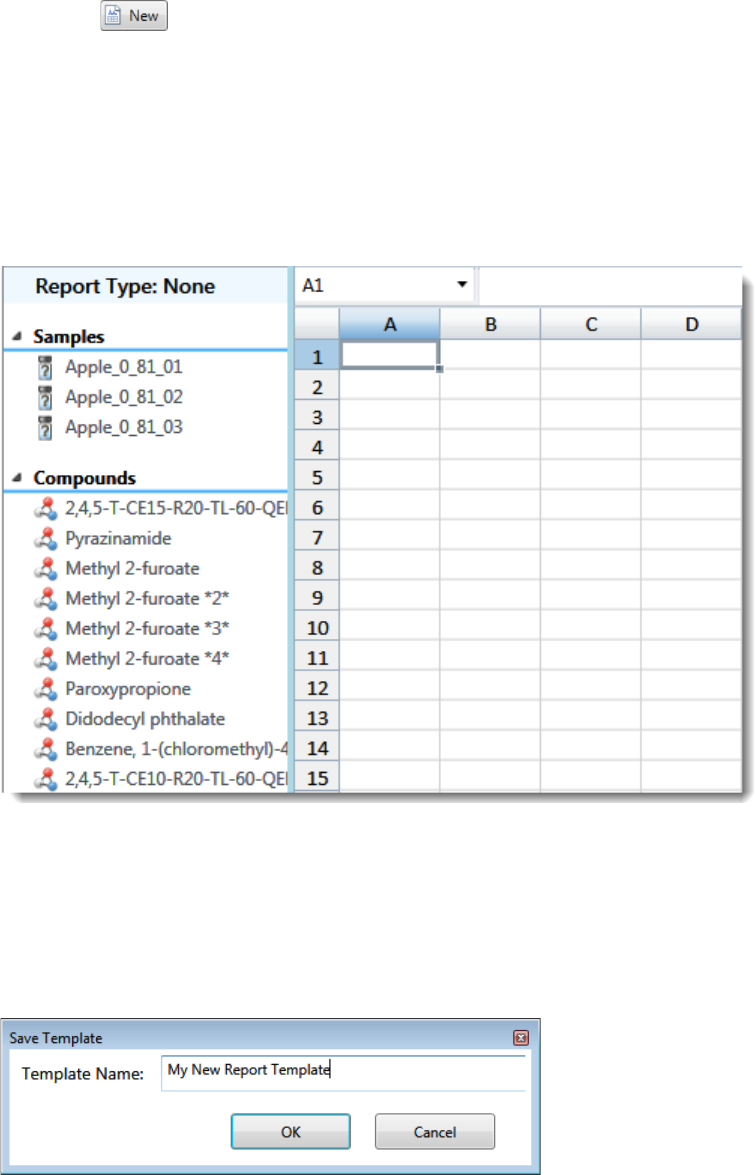
4 Using the Analysis Mode for Quantitation Batches
Working in the Report View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 229
To create a new report template
1. Click New, .
The application opens the Report Designer showing a new, empty template in an Excel
spreadsheet.
The Report Type is None.
In the left pane, the spreadsheet lists all samples in the current batch and all compounds
in the method used for the batch.
Figure 78. Report Designer showing a new, empty template
2. Use the features in the Report Designer to create the report template.
See Chapter 7, “Using the Report Designer.”
3. When you finish your changes, choose File > Save from the Report Designer menu bar.
The Save Template dialog box opens.
Figure 79. Save Template dialog box
4. Type a name for the new report template and click OK.
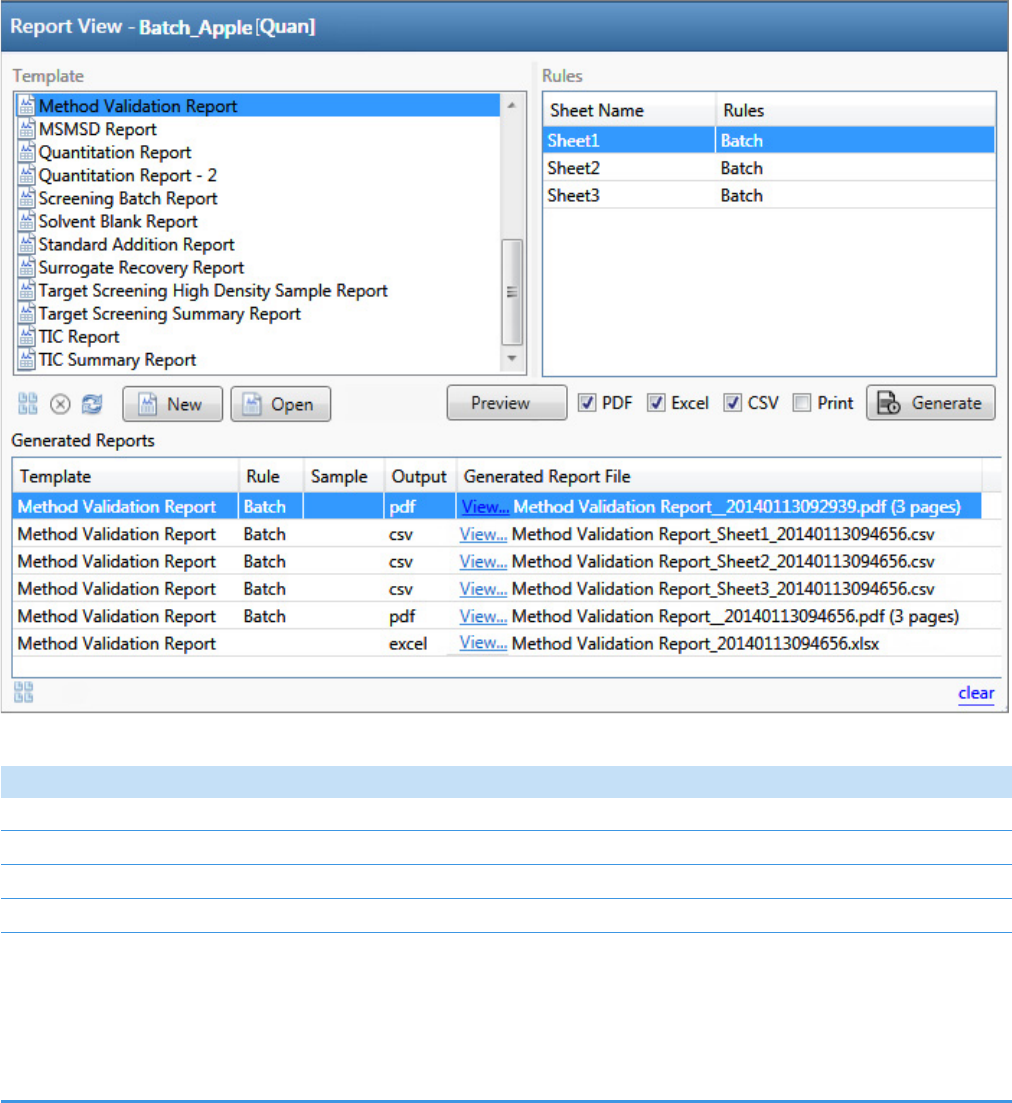
4 Using the Analysis Mode for Quantitation Batches
Working in the Report View for Quantitation Batches
230 TraceFinder User Guide Thermo Scientific
Report View
Use the features in the Report View to display example reports for the current batch.
Figure 80. Report View
Table 42. Report View parameters (Sheet 1 of 2)
Parameter Description
Template
Displays all report templates.
Rules
Sheet Name Specifies each sheet in the report.
Rules Specifies the type of data used in each sheet in the selected report.
•Batch
• EachSample
• SampleType: SampleType
•CompoundType: CompoundType
•SampleCustomFormula:
4 Using the Analysis Mode for Quantitation Batches
Working in the Report View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 231
Buttons
View Report Templates Displays the C:\TraceFinderData\4.0\Templates\ReportTemplates folder that contains
all report templates.
Opens the selected report template in the Report Designer.
Opens a blank report template in the Report Designer.
Opens the Report Designer showing the report information for the current batch in the
selected report template format.
PDF Writes the generated report to a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
Excel Writes the generated report to an Excel file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
CSV Saves the generated report as a CSV file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
When the report contains multiple sheets, the application writes each sheet as a separate
CSV file.
Print Prints the generated report to your default printer.
Generates the selected type of reports for the current batch using the selected report
template.
Generated Reports
Template Report template used for the report. See Example of a report template list.
Rule Type of data used in each sheet of the report. See Rules.
Sample For sample-level reports, the name of each sample in the report.
Output Type of output specified for the report: PDF, Excel, CSV, or Print.
Generated Report File Lists the output file name for each report in the …\TraceFinderData\4.0\Projects folder.
View Displays the generated output file.
View Generated Reports Displays the C:\TraceFinderData\4.0\Projects folder that contains all report outputs.
Clear Removes all reports from the Generated Reports display. This does not delete the reports
from the C:\TraceFinderData\4.0\Projects folder.
Table 42. Report View parameters (Sheet 2 of 2)
Parameter Description
4 Using the Analysis Mode for Quantitation Batches
Working in the Local Method View for Quantitation Batches
232 TraceFinder User Guide Thermo Scientific
Working in the Local Method View for Quantitation Batches
In the Local Method view, you can edit the local method parameters. A local method is a copy
of a master method associated with a batch.
You can edit only the local copy of the method, or you can edit the master method and
overwrite the local copy with the edited master method. Local methods are named
Batch_MasterMethod.
To open the Local Method View
1. Click Analysis in the navigation pane.
2. Click Local Method.
The Local Method view for the currently selected batch opens. See Local Method view.
You can edit many of the method parameters in a local method. Editing the local method
does not affect parameters in the master method.
For detailed descriptions of method parameters, refer to Chapter 4, “Using the Method
Development Mode for Quantitation Methods,” in the TraceFinder Lab Director User
Guide.
3. Enter any local changes to the method.
4. When you have finished editing the local method, choose File > Save.
5. To process the batch or create new reports with the edited local method, return to the
Batch View and submit the batch.
To overwrite the local method with the master method in the Batch View
In the Batch View, click Update.
The application overwrites the local method with the master method of the same name.
You can use this feature to overwrite an edited local method with the original master
method or to overwrite the local method with an updated master method.
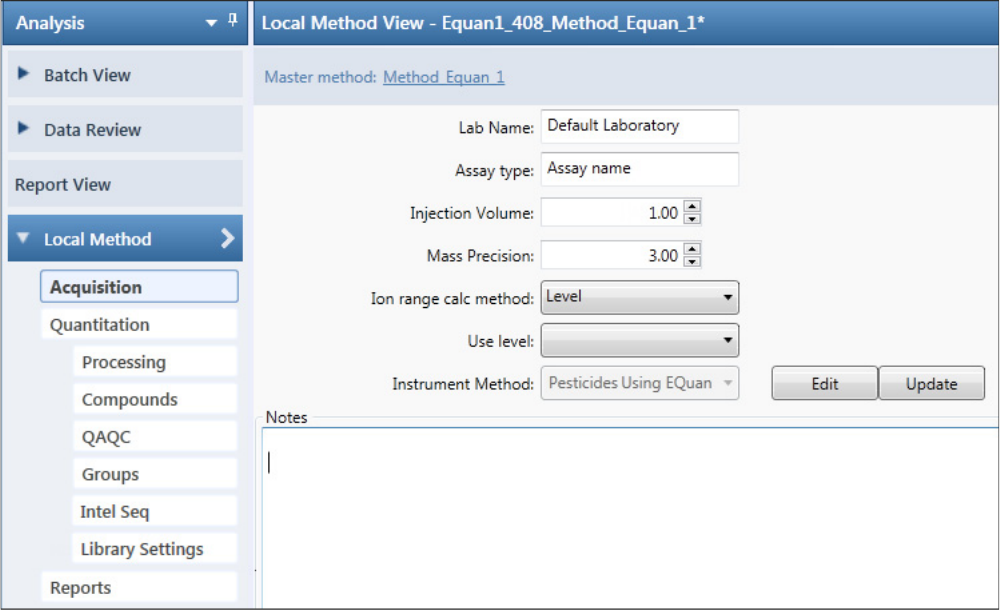
4 Using the Analysis Mode for Quantitation Batches
Working in the Local Method View for Quantitation Batches
Thermo Scientific TraceFinder User Guide 233
Figure 81. Local Method view
4 Using the Analysis Mode for Quantitation Batches
Working in the Local Method View for Quantitation Batches
234 TraceFinder User Guide Thermo Scientific
Thermo Scientific TraceFinder User Guide 235
5
Using the Analysis Mode for Target Screening
Batches
Use the features of the Analysis mode to do the following:
• Create target screening batches.
• Submit target screening batches for acquisition, processing, or report generation.
• Review target screening batches, batch data, reports, and local methods.
To access the Analysis mode
Click Analysis in the navigation pane.
The Analysis navigation pane opens.
IMPORTANT TraceFinder 4.1 uses the same data as TraceFinder 4.0. By default, the application
stores the method, project, template, and compound database data for the 4.1 release in the
TraceFinderData\4.0 folder.
Contents
•Working in the Batch View for Target Screening Batches
•Working in Data Review for Target Screening Batches
•Working in the Report View for Target Screening Batches
•Working in the Local Method View for Target Screening Batches
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
236 TraceFinder User Guide Thermo Scientific
Working in the Batch View for Target Screening Batches
In the Batch View, you can manually create and edit a new target screening batch or open and
edit a previously saved batch. When you submit a batch, you can acquire and process data and
optionally create reports for the submitted samples.
The Analysis mode includes a toolbar:
Use the Toolbar or the equivalent commands in the Batch View Shortcut Menu to create the
sample list and submit samples for acquisition.
To open the Batch View
1. Click Analysis in the navigation pane of the current mode.
2. Click Batch View.
The Batch View navigation pane opens.
The Batch View includes a single page of information, the Samples page.
Related Topics
•Samples Page Features for Target Screening Batches
•Creating a New Target Screening Batch
•Editing a Target Screening Batch
•Submitting a Target Screening Batch
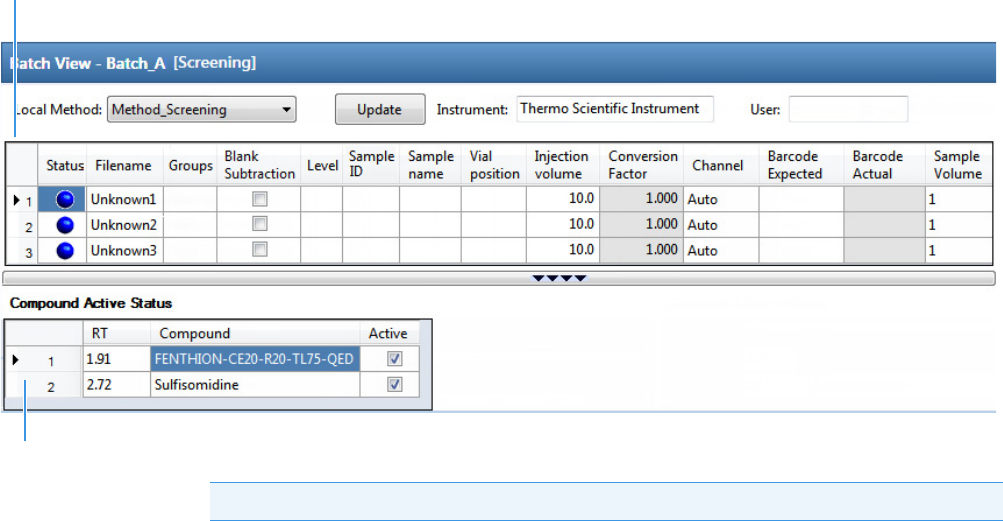
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 237
Samples Page Features for Target Screening Batches
The Samples page is divided into two panes:
• Samples pane
Use the Samples Pane to create a batch.
• Compound Active Status pane
Use the Compound Active Status Pane to make specific compounds active or inactive.
Samples Pane
The samples pane includes the following features:
•Column Display
•Status Indicators
•Groups
•Blank Subtraction
•Sample Weight Calculation
•Instrument Methods
•Toolbar
•Batch View Sample List
•Batch View Shortcut Menu
Samples pane
Compound Active Status pane
Tip To resize the panes, drag the separators that divide the panes.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
238 TraceFinder User Guide Thermo Scientific
Column Display
The sample list contains many columns of information. You can scroll to see all the columns,
and you can customize which ones to display and their display order.
To scroll the sample list
Use the horizontal scroll bar at the bottom of the sample list to view all the information.
When you use the scroll bar at the bottom of the sample list, the following columns
remain fixed: Status, Filename, Groups, Blank Subtraction, Level, Sample ID, and
Sample Name, while the other columns scroll right and left.
To customize the column display
1. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
2. Use the arrow buttons to move all the columns that you want displayed to the Displayed
Columns pane.
These columns appear after the Status, Filename, Groups, Blank Subtraction, Level,
Sample ID, and Sample Name columns.
3. To arrange the order of the columns, do the following:
a. In the Displayed Columns pane, select a column name.
b. Use Up or Down to move the selected column up or down in the list.
The first column in the list represents the leftmost column in the Batch View sample
list, and the last column in the list represents the rightmost column in the Batch View
sample list.
4. To change the width of a column, do the following:
a. In the Displayed Columns pane, select the column width.
b. Type a new value for the width.
5. Repeat step 4 for all columns whose widths you want to change, and click OK.
The columns in the sample list immediately reflect your changes. The application uses
these settings for all sample lists in the Batch View.
Note The following columns are fixed: Status, Filename, Groups, Blank
Subtraction, Level, Sample ID, and Sample Name.
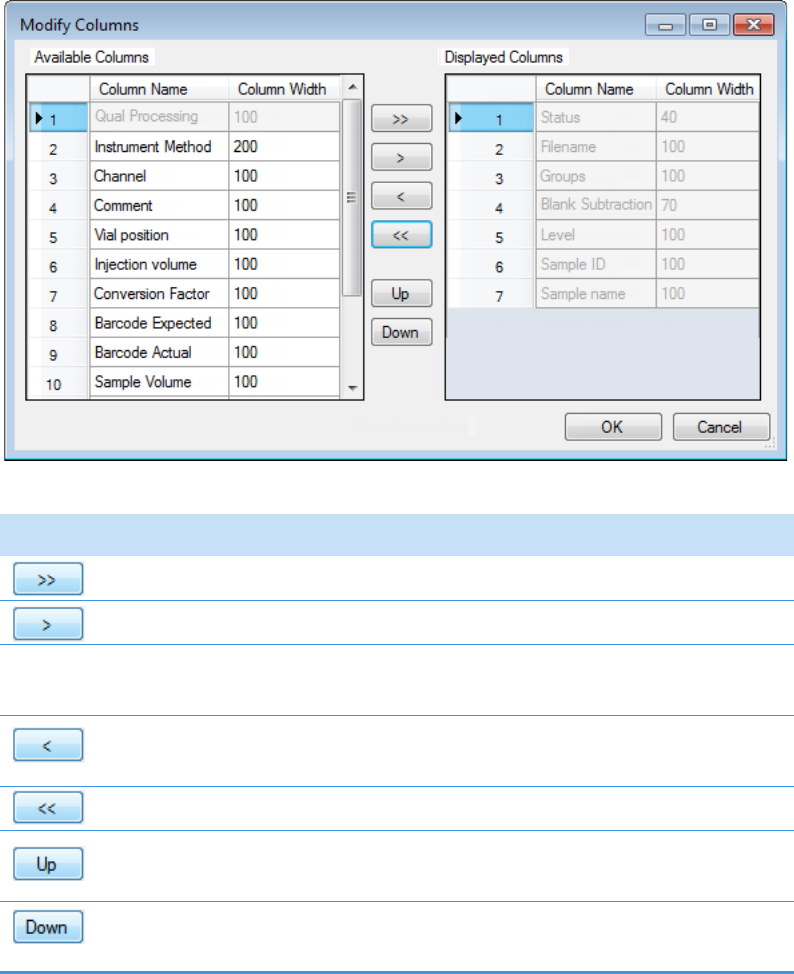
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 239
Figure 82. Modify Columns dialog box
Table 43. Button descriptions for the Modify Columns dialog box
Button Description
Moves all columns to the Displayed Columns pane.
Moves the selected column to the Displayed Columns pane.
The following buttons apply to all columns, except for those that are fixed: Status, Filename,
Sample Type, Groups, Blank Subtraction, Level, Sample ID, and Sample Name.
Moves the selected column to the Available Columns pane.
Moves all columns except fixed columns.
Moves the selected column name in the Displayed Columns pane one row
up in the column order.
Moves the selected column name in the Displayed Columns pane one row
down in the column order.
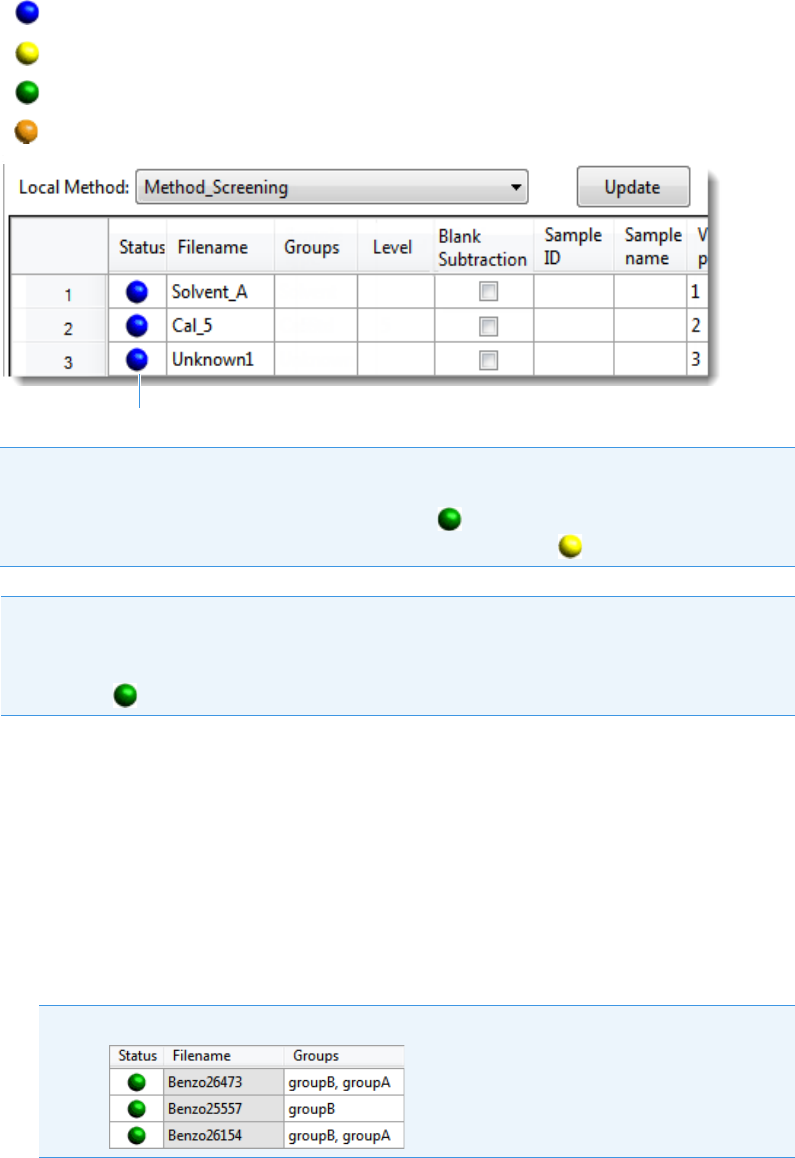
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
240 TraceFinder User Guide Thermo Scientific
Status Indicators
Status indicators show the current status of each sample during the acquisition and
processing.
Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Groups
Use the Groups feature to assign samples to a group.
To create a group
1. For each sample, type the name of a group in the Groups column.
Repeat this for each sample that you want to include in a group.
2. Create as many groups as you want.
Note When you include unknown screening features in the target screening method and
you choose to process with only the target screening criteria, the Samples view shows the
Status for the samples as acquired and processed ( ), and the Unknown Screening View
shows the Status for the samples as acquired but not processed ( ).
Note When you include unknown screening features in the target screening method and
you choose to process with only the unknown screening criteria, both the Samples view
and the Unknown Screening View show the Status for the samples as acquired and
processed ( ).
Status indicators
Note To assign a sample to multiple groups, separate the groups with a comma.

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 241
Blank Subtraction
Use the Blank Subtraction feature to select which matrix blank samples you want to use for
peak subtraction. The application subtracts the areas of the peaks in the selected matrix blank
samples from the matching areas in the unknown samples.
When you process the batch sequence, the application subtracts the peaks in a selected matrix
blank sample from all unknown samples that follow it, until it encounters another matrix
blank sample.
To activate the Blank Subtraction feature, refer to “Editing the Processing Page” in Chapter 5,
“Using the Method Development Mode for Target Screening Methods,” in the Trac eF in der
Lab Director User Guide.
Sample Weight Calculation
Use the sample weight features to calculate the conversion factor for a sample. The application
uses different methods to calculate the conversion factor for liquid or solid calculation types.
Liquid: SampleVolume DilutionFactor
Solid: (SampleVolume × DilutionFactor) SampleWeight
Manual: The application does not calculate the Conversion Factor. Instead, you can enter the
Conversion Factor value.
Follow these procedures:
•To display the features for calculating sample weight
•To calculate the conversion factor for a liquid sample
•To calculate the conversion factor for a solid sample
•To manually specify the conversion factor for a sample
To display the features for calculating sample weight
If the Conversion Factor, Sample Volume, Dilution Factor, Sample Weight, Calculation
Type, and Final Units columns are not visible, right-click and choose Enable Sample
Weight Calculation.

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
242 TraceFinder User Guide Thermo Scientific
To calculate the conversion factor for a liquid sample
1. From the Calculation Type list, select Liquid.
For a liquid sample, the Sample Weight value is not editable.
2. In the Sample Volume column, type the volume in ng/mL for your sample.
3. In the Dilution Factor column, type the value for the dilution.
For example, if you have 1000 ng/mL of a substance that is too concentrated for the mass
spectrometer, you can dilute it by 1000. Then your injection volume is 1, your conversion
factor is 1000, and your sample amount is 1000.
4. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
To calculate the conversion factor for a solid sample
1. From the Calculation Type list, select Solid.
2. In the Sample Weight column, type the weight in ng for your sample.
3. In the Sample Volume column, type the volume in ng/ml for your sample.
4. In the Dilution Factor column, type the value for the dilution.
For example, if you have 1000 ng/ml of a substance that is too concentrated for the mass
spectrometer, you can dilute it by 1000. Then your injection volume is 1, your conversion
factor is 1000, and your sample amount is 1000.
5. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
To manually specify the conversion factor for a sample
1. From the Calculation Type list, select Manual.
For a manually calculated sample, the only available columns are the Conversion Factor
and the Final Units.
2. In the Conversion Factor column, type the conversion factor to use for your sample.
3. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
Note The application uses the following formula to calculate the Conversion Factor:
SampleVolume DilutionFactor
Note The application uses the following formula to calculate the Conversion Factor:
(SampleVolume × DilutionFactor) SampleWeight
Note The application uses the specified conversion factor when it calculates the
amount for the sample.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 243
Instrument Methods
Use the Instrument Methods column to specify instrument methods for the samples.
To specify instrument methods for samples
1. Display the Instrument Method column in the sample list:
a. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
b. In the Available Columns pane, select Instrument Method.
c. Click to move the Instrument Method column to the Displayed Columns
pane.
d. Click OK.
The application displays the Instrument Method column, defaulting to the instrument
method specified in the master method.
2. Click the Instrument Method column and select an instrument method from the list.
This list contains all the available instrument methods. Instrument methods from
external sources are prefixed with “Ext:”.
You can specify a different instrument method for each sample.
When you submit the batch for acquisition, the application saves a copy of the selected
instrument methods to the following folders:
External instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method\ExternalMethods
Local instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method
Note By default, the Instrument Method column is not displayed in the Batch View
sample list.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
244 TraceFinder User Guide Thermo Scientific
Toolbar
The Analysis mode includes this toolbar for creating and submitting a batch.
Table 44. Toolbar icons
Icon Description
Adds the specified number of new, empty samples to the end of the
sample list. See the instructions To add samples to the list.
Inserts a new, empty sample or samples above the selected sample. See
the instructions To insert samples into the list.
Removes the selected samples from the sample list. See the
instructions To remove samples from the list.
Adds imported samples from a CSV, an XML, or an SLD file to the
sample list. See the instructions To import samples into the list.
Submits only the selected samples for acquisition, processing, or
report generation. See the instructions To submit samples in the
batch.
Submits the batch for acquisition, processing, or report generation.
See the instructions To submit samples in the batch.
Submits only the selected samples for processing. See the instructions
To submit selected peaks for processing.
Opens the Acquisition mode where you can use a batch template to
define a standard sequence composed of various sample types to be
assembled into a batch of samples. See Working in Data Review for
Targe t Scree n ing Batc hes .
Opens the Acquisition mode where you can create a batch template
that contains the basic settings and sample types for your batches. See
Using the Acquisition Mode.
Opens the Quick Acquisition window where you can quickly submit a
single sample. See Appendix A, “Using Quick Acquisition.”
Opens the Audit Viewer where you can view audit logs. See
Chapter 8, “Using the Audit Viewer.” Available only when you enable
Auditing in the Administrator Console. Refer to the instructions in
the TraceFinder Administrator Console User Guide.
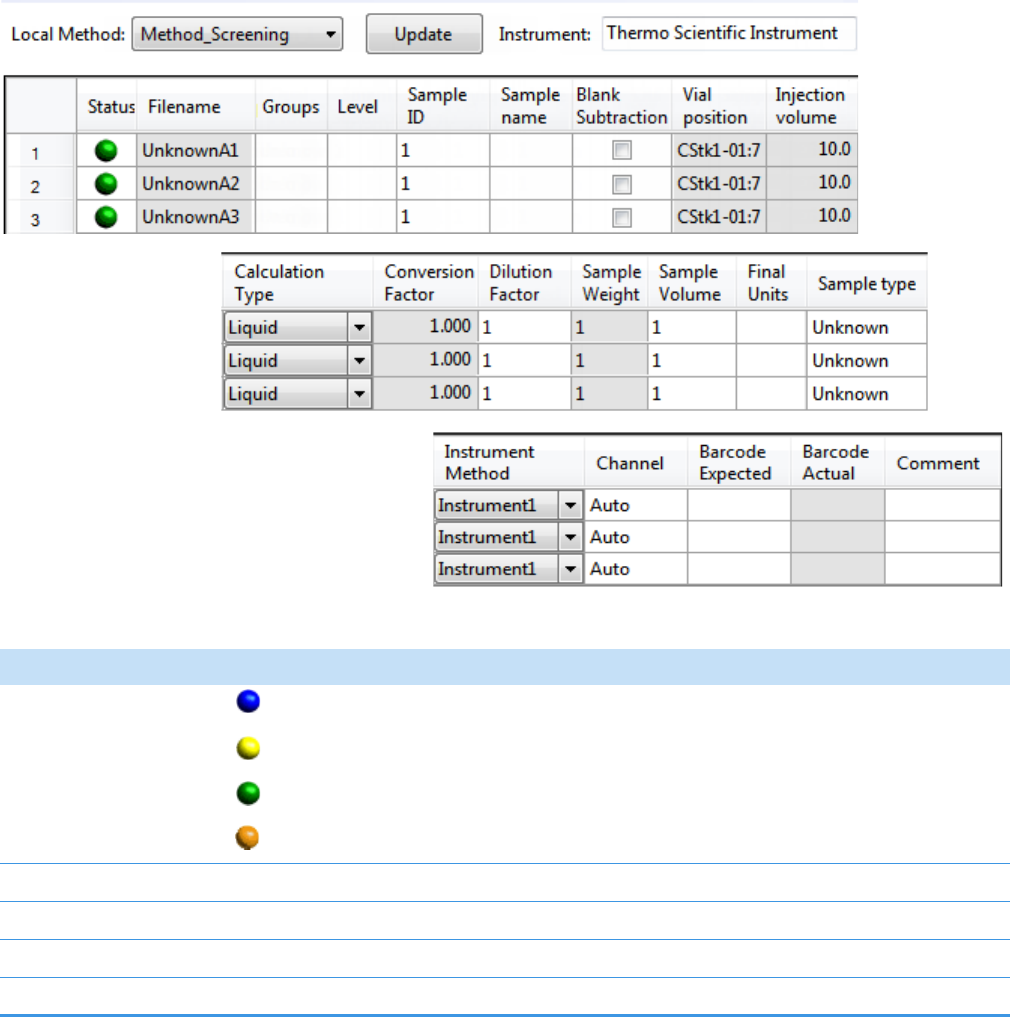
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 245
Batch View Sample List
The sample list displays all the quantitative data for the samples of a batch.
Status indicators for each sample indicate if the sample is currently acquiring, not acquired,
acquired, or processed.
The sample list includes the following columns of information:
Figure 83. Batch View sample list
Table 45. Batch View sample list columns (Sheet 1 of 3)
Column Description
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Filename Name of the raw data file that contains the sample data.
Groups Threshold group to which a sample belongs.
Blank Subtraction Specifies a matrix blank sample to use for blank subtraction.
Level The level defined for a calibration sample or quality control sample.

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
246 TraceFinder User Guide Thermo Scientific
Sample ID A user-defined, alphanumeric string that identifies a sample.
Sample Name A user-defined name that identifies a sample.
Vial Position The tray vial number used for an autosampler acquisition.
Injection Volume The injection volume (in microliters) of the injected sample.
When you are using an autosampler, you can set the default injection volume in the
Autosampler dialog box in the Instrument View. The minimum and maximum injection
volumes that you can use depend on the Autosampler you configure. The usable range
depends on the injection mode and might be smaller than the displayed range.
The Injection Volume value set in the master method overwrites the value in the instrument
method.
Valid range: 0.1 through 5000 μL
Calculation Type Liquid: The application calculates the Conversion Factor as
SampleVolume DilutionFactor
Solid: The application calculates the Conversion Factor as
(SampleVolume × DilutionFactor) SampleWeight
Manual: Sample Volume, Dilution Factor, Sample Weight, and Final Units columns are not
available, and the Conversion Factor value is editable.
Conversion Factor Editable only when Calculation Type is Manual.
Default: 1
Sample Volume Default: 1
Dilution Factor Default: 1
Sample Weight Available only when Calculation Type is Solid.
Default: 1
Final Units Specifies the calculated amount in the Data Review view or in reports.
Default: 1
Instrument Method Specifies the instrument to use for the acquisition. This column is hidden by default. To
display this column, see To customize the column display.
Channel Specifies the channel on which the sample was run. If the sample is not acquired, the value is
Pending. The Channel column is available only when you have activated multiplexing in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in the
TraceFinder Lab Director User Guide.
Table 45. Batch View sample list columns (Sheet 2 of 3)
Column Description

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 247
Batch View Shortcut Menu
The Batch View includes a shortcut menu for creating a batch.
Barcode Expected A user-entered barcode for the vial.
Barcode Actual An actual barcode for the vial. This value is not editable.
Comment A user-defined comment for the sample.
Sample Type Defines how the application processes the sample data. Each sample is classified as one of
the following sample types: Matrix Blank, Solvent, Cal Std, QC Std, or Unknown.
Default: Unknown
Table 45. Batch View sample list columns (Sheet 3 of 3)
Column Description
Table 46. Batch View shortcut menu commands (Sheet 1 of 2)
Command Description
Add Sample Adds a single empty row to the sample grid.
Insert Sample Inserts a single empty row to the sample grid above the selected row.
Insert Copy Sample Copies the currently selected row and inserts a copy above the row.
Reinject Selected
Samples
Creates a copy of the selected sample and appends INJ001 to the file name. Additional
reinjections of the same sample are numbered INJ002, INJ003, and so forth.
Remove Selected
Samples
Removes selected samples from the sample grid.
Import Samples Opens the Sample Import Tool. See To import samples into the list.
Browse in Raw File
(Move)
Opens a dialog box where you can select a raw data file to use for the selected sample row.
The application removes the raw data file from the source location.
Browse in Raw File
(Copy)
Opens a dialog box where you can select a raw data file to use for the selected sample row.
The application copies the raw data file from the source location.
Map Raw Files to
Samples
Opens a dialog box where you can select multiple raw data files to use for the selected
sample rows.
Copy Down Copies the value in the selected row to all rows below it. This command is available only
when you have selected a value that can be copied down.
Fill Down Enters sequential values in the column starting with the value in the selected row and
ending with the last row in the column. This command is available only when you have
selected a value that can be filled down.
Modify Columns Opens the Modify Columns dialog box.
Hide Sample Weight
Calculation
Displays or hides the Sample Volume, Dilution Factor, Sample Weight, Calculation Type,
and Final Units columns.
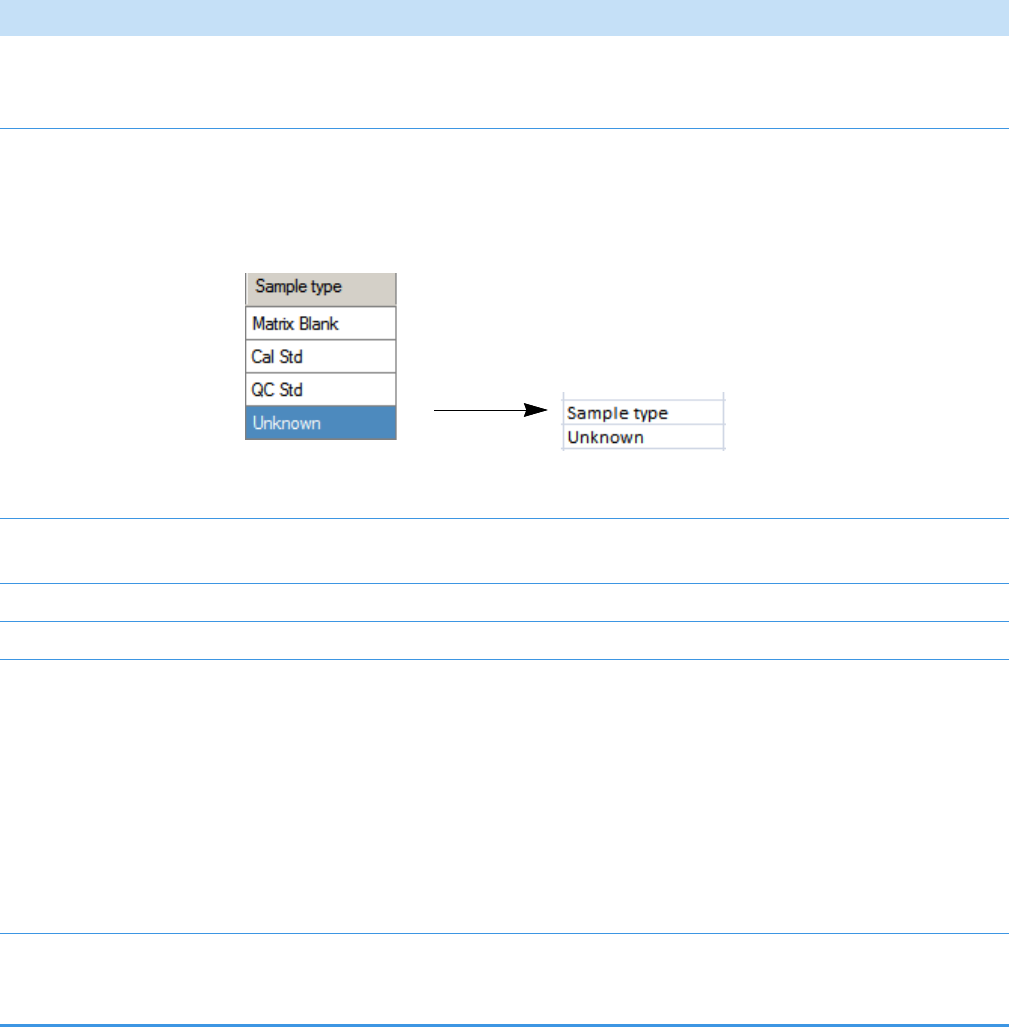
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
248 TraceFinder User Guide Thermo Scientific
Copy Copies the data in the selected rows or columns to the Clipboard. Use this command to
copy sample information into a text editor or spreadsheet application. You cannot paste
this data back into the Batch View sample list.
Copy with Headers Copies the data in the selected rows or columns and the associated column headers to the
Clipboard. Use this command to copy sample information into another text editor or
spreadsheet application. You cannot paste this data back into the sample list.
For example
Paste Pastes a single column of copied data from another text editor or spreadsheet application
into the selected column.
Undo Last Paste Removes the last pasted item in the Batch View.
Export to CSV File Opens the Save As dialog box where you can save the current sample list to a CSV file.
Edit Instrument Method Opens the Instrument Setup window where you can edit the parameters of the instrument
method.
• When you edit an external method, the application updates the method in the
…\Xcalibur\methods folder.
• When you edit an internal method, the application updates the method in the
…\TraceFinderData\4.0\Projects\…\batch\Methods\method folder.
For detailed information about editing instrument methods, refer to Chapter 3, “Using
Instrument Methods in the Method Development Mode,” in the Trace Fi n der La b Di rec t or
User Guide.
View Sample in Qual
Explorer
Displays the selected sample in the qualitative explorer application that you configured as
your default in the Configuration console. Refer to Chapter 1, “Using the Configuration
Console,” in the TraceFinder Lab Director User Guide.
Table 46. Batch View shortcut menu commands (Sheet 2 of 2)
Command Description
Copy with Headers
from TraceFinder Paste into an Excel spreadsheet.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 249
Compound Active Status Pane
In the Compound Active Status pane, you can choose specific compounds to be active or
inactive.
To set a compound as active or inactive
1. In the sample list, select a sample.
All compounds in the selected sample are listed in the Compound Active Status pane.
The default active/inactive status is determined by the identification settings in the local
method. For information about setting the identification parameters, refer to Chapter 4,
“Using the Method Development Mode for Quantitation Methods,” in the Tra ce Fi nder
Lab Director User Guide.
• To display compounds alphabetically, right-click and choose Sort by Compound
Name.
• To display compounds from shorter to longer retention times, right-click and choose
Sort by Retention Time.
2. Select or clear the Active check box for the compound.
Compound Active/Inactive Status
You can specify which compounds are active or inactive in the Local Method View or the
Batch View.
Figure 84. Active and inactive compounds in the Local Method View
For details about setting the status on the Identification page, refer to Chapter 4, “Using the
Method Development Mode for Quantitation Methods,” in the TraceFin d er L ab D irecto r Use r
Guide.
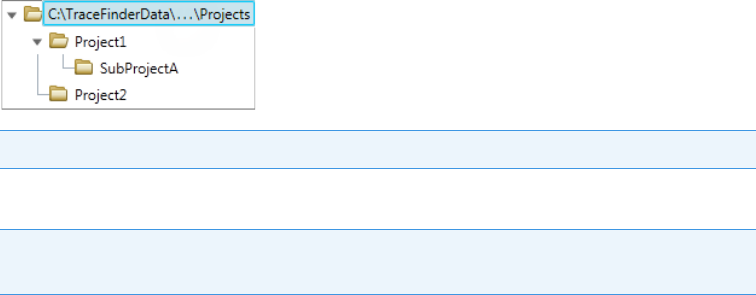
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 251
Creating a New Target Screening Batch
In the Batch View, you can create a new batch.
Follow these procedures:
•To c re a t e a ne w b a t c h
•To add samples to the list
•To insert samples into the list
•To import samples into the list
•To remove samples from the list
•To c o py a sa m pl e
•To reinject a sample
•To edit sample values
•To browse in raw data files
To create a new batch
1. Choose File > New > Batch from the main menu.
The Create New Batch Dialog Box opens and displays all drives that contain projects.
2. Select a drive from the list.
3. Select the folder where you want to store your batch.
To create a new folder for the storage location, see Editing Folders for Batches.
4. Select Screening from the Type list.
The batch list displays all batches in the selected folder. The Method list displays all
methods for the target screening type.
5. Select a master method from the Master Method list.
Tip The application displays all configured and enabled repositories.
Tip To activate the Create button, you must enter a unique batch name. If the Create
button is not activated, you have entered a batch name that is already used.

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
252 TraceFinder User Guide Thermo Scientific
6. Click Create.
A new batch opens with one Unknown sample.
The batch name in the title bar indicates that you are creating a target screening batch.
To add samples to the list
1. To add a single sample row, right-click the sample list and choose Add Sample.
2. To add multiple sample rows, select the number of rows and then click the Add Sample
icon, .
The application adds the specified number of new, empty samples to the end of the
sample list.
To insert samples into the list
Select the sample above which you will insert new, Unknown samples, and then do one of
the following:
• To insert a single sample row, right-click and choose Insert Sample.
• To insert multiple sample rows, select the number of rows and then click the Insert
Sample icon .
The application inserts the Unknown samples above the selected sample.
To import samples into the list
1. Choose Batch > Import Samples from the main menu, or click the Import Samples
icon, .
The Sample Import Tool dialog box opens.
From this dialog box, you can import samples from a CSV, an XML, or an SLD file.
2. Click Browse and select a CSV, an XML, or an SLD file that contains the samples to
import.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 253
3. From the Imported Samples Will Be list, select Appended to the End of the List or
Inserted at the Selected Row.
4. Click Import.
The Sample Import Tool dialog box closes, and the application adds the specified samples
to the sample list.
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following column name substitutions:
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following sample type substitutions:
5. (Optional) When using multiplexing, select a channel for each imported sample.
Imported samples default to Auto.
To remove samples from the list
1. Select the samples that you want to remove.
2. Right-click and choose Remove Selected Samples.
To copy a sample
1. Select the sample that you want to copy.
2. Right-click and choose Insert Copy Sample.
The application inserts the copy above the selected sample.
Xcalibur column TraceFinder column
Position Vial Position
Inj Vol Injection Volume
Dil Factor Conversion Factor
Xcalibur sample type TraceFinder sample type
Blank Matrix Blank
QC QC Std
Std Bracket Cal Std
Note The Channel column is available only when you have activated multiplexing in
the Configuration console. Refer to Chapter 1, “Using the Configuration Console,”
in the TraceFinder Lab Director User Guide.
Tip Use the CTRL or SHIFT keys to select multiple samples.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
254 TraceFinder User Guide Thermo Scientific
To reinject a sample
1. In the sample list, select the sample that you want to reinject.
2. Right-click and choose Reinject This Sample.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the sample are numbered INJ002, INJ003, and so forth.
The application copies all parameter values from the original sample.
To edit sample values
1. For each sample, do one of the following:
Type a new file name over the current filename.
–or–
Double-click the Filename column and locate a raw data file to use for the sample.
–or–
Right-click and choose Browse in Raw File, and then locate a raw data file to use for the
sample.
By default, the application sets the Sample Type to Unknown.
2. For each sample, click the Sample Type column and select a sample type from the list.
3. For each Cal Std or QC Std sample, select a level from the Level list.
The sample levels are defined in the master method. If there are no levels to select in the
Level list, do the following:
a. Return to the Method Development mode.
b. Open the method.
c. Click the Compounds tab.
d. Click the Calibration Levels tab.
e. Add the levels.
f. Save the method.
g. Return to the Batch View in the Analysis mode, and then click Update.
The application updates the local method with the new sample levels.
For detailed instructions about specifying calibration levels, refer to Chapter 4, “Using the
Method Development Mode for Quantitation Methods,” in the Trac eFi nd er L a b D ire c t or
User Guide.
Available sample types
Matrix Blank Solvent QC Std Unknown
Cal Std

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 255
4. Type a vial position in the Vial Position column for each sample.
5. Type a volume in the Injection Volume column for each sample.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
6. (Optional) Type or edit the values for the remaining columns.
To automatically copy or fill values in these columns, see Appendix B, “Using Copy
Down and Fill Down.”
To browse in raw data files
1. Do one of the following:
Double-click the Filename column.
–or–
Right-click and choose Browse in Raw File.
The What Raw File Would You Like to Use dialog box opens.
2. Select a raw data file to use for the sample or use the CTRL key to select multiple files,
and then click Open.
The application overwrites the selected, unacquired sample in the batch with the first
“browsed in” file and adds any additional browsed in files below the selected sample.
For all browsed-in raw data files, the application sets the Status to Acquired, , and sets
the Sample Type to Unknown.
Note When you use the scroll bar at the bottom of the sample list, the following
columns remain fixed: Status, Filename, Sample Type, Groups, Blank Subtraction,
Level, Sample ID, and Sample Name, while the other columns scroll right and left.
Note You cannot overwrite an acquired sample. When you select a sample that is
acquired, the application adds all browsed in files below the selected sample.
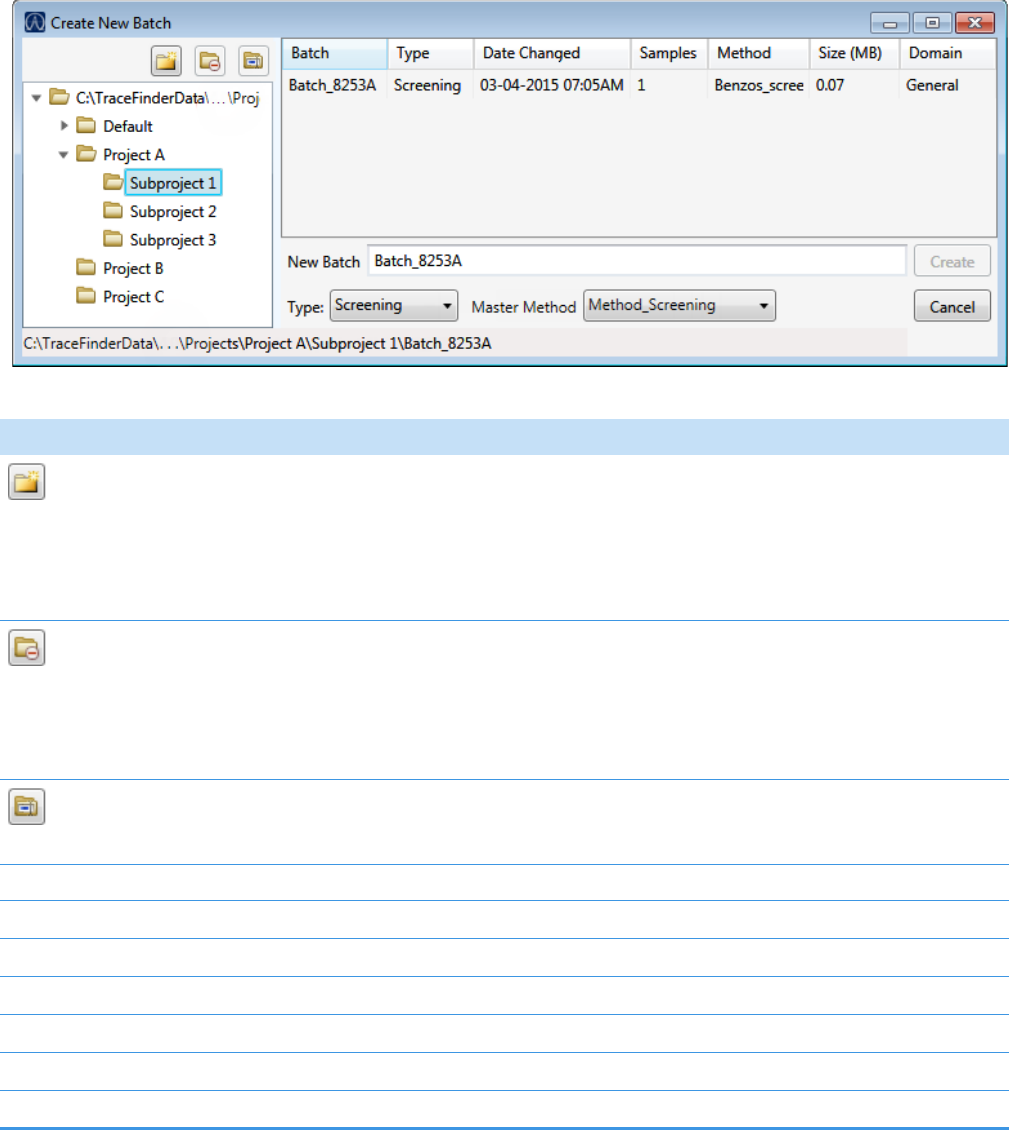
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
256 TraceFinder User Guide Thermo Scientific
Create New Batch Dialog Box
Use the Create New Batch dialog box to select a folder and method for your batch and to
name the new batch.
Table 47. Create New Batch dialog box parameters (Sheet 1 of 2)
Parameter Description
Create New Folder
Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Or, you can right-click and choose Create Folder.
Delete Folder
With no confirmation prompt, immediately removes the selected folder.
You cannot delete a folder that contains lower-level folders; you must delete the lower-level
folders first.
Or, you can right-click and choose Delete.
Rename Folder
Renames the selected folder.
Or, you can right-click and choose Rename.
Batch table
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date that the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 257
Editing Folders for Batches
From the Create New Batch dialog box, you can create new folders for your batches. You can
also delete or rename folders.
Use these procedures:
•To create new project folders
•To delete project folders
•To rename project folders
To create new project folders
1. In the Create New Batch dialog box, select the folder for which you will create a new
lower-level folder.
• You can select the main TraceFinderData\4.0\Projects folder and create a new folder
under it.
• You can select one of the existing folders and create a lower-level folder under it.
2. Click the Create Folder icon, .
The application adds a new lower-level folder to the selected folder.
3. Select the new folder name and type a name for the folder.
Folder names are limited to 30 characters and can contain spaces and special characters,
except for the following special characters: \ / : + ? ” < >
Domain TraceFinder domain in which the batch was created.
New batch parameters
New Batch Name of the new batch to create.
Note If the Create button is not activated, you have entered a name that is already used or
you have not selected a method.
Type Type of batch to create: Quan, Screening, or Unknown Only.
Method Method used to create the new batch.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is created.
Buttons
Create Creates the specified batch and opens the Batch View for the new batch.
Cancel Closes the Create New Batch dialog box without creating a batch.
Table 47. Create New Batch dialog box parameters (Sheet 2 of 2)
Parameter Description
Note After you add a lower-level folder, you cannot rename the parent folder.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
258 TraceFinder User Guide Thermo Scientific
To delete project folders
1. In the Create New Batch dialog box, select the folder to delete.
2. Click the Delete Folder icon, .
With no confirmation prompt, the application immediately removes the selected folder.
To rename project folders
1. In the Create New Batch dialog box, select the folder to rename.
2. Click the Rename Folder icon, .
3. Type a new name for the folder and press ENTER.
The application saves the new folder name.
Editing a Target Screening Batch
In the Batch View, you can open a saved batch and edit the sample list. You can add samples,
edit samples, or remove samples. If the batch has already been acquired, you can select specific
samples for reinjection. If the batch has unacquired samples when you complete your edits,
you can save it as a “ready to acquire” batch.
Follow these procedures:
•To o p e n a sa v e d b at c h
•To open a recent batch
•To edit samples in a batch
•To reinject a sample from a previously acquired batch
To open a saved batch
1. Choose File > Open > Batch from the main menu.
The Open Batch dialog box opens.
2. Select a project and a subproject.
3. Select Screening or Any from the Type list.
The batch list displays all batches created with Target Screening methods (or when you
select Any, all methods of all types).
4. Select a batch from the list.
Note You cannot delete folders that contains lower-level folders; you must delete the
lower-level folders first.
Note You cannot rename folders that contain lower-level folders.
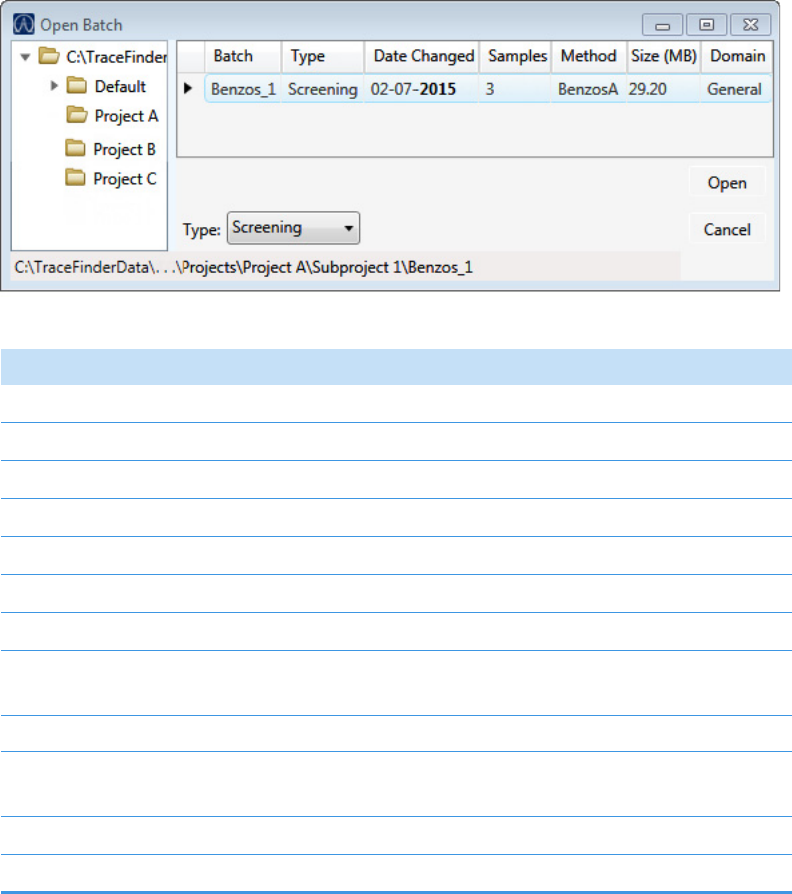
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 259
5. Click Open.
The selected batch opens in the Batch View.
Figure 86. Open Batch dialog box
Table 48. Open Batch dialog box parameters
Parameter Description
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
Path Path to the project in the TraceFinderData\4.0\Projects folder
where the batch is stored.
Buttons
Type Type of batch to display in the Batch list: Quan, Screening,
Unknown Only, or Any.
Open Opens the Batch View for the selected batch.
Cancel Closes the Open Batch dialog box without opening a batch.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
260 TraceFinder User Guide Thermo Scientific
To open a recent batch
Choose File > Recent Files > batch from the main menu.
The selected batch opens in the Batch View.
To edit samples in a batch
Use the commands described in Working in the Batch View for Target Screening Batches.
You can add new samples, edit samples, or delete samples.
To reinject a sample from a previously acquired batch
1. In the sample list, select the sample that you want to reinject.
2. Right-click and choose Reinject This Sample.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the sample are numbered INJ002, INJ003, and so forth.
The application copies all parameter values from the original sample.
A green status icon indicates previously acquired samples (acquired and processed), and
the sample name is grayed out. A blue status icon indicates samples created for reinjection
(not acquired).
When you submit all samples in this batch, the application acquires all samples (including
previously acquired samples).
3. To save this batch with the new samples for reinjection, choose File > Save > Batch from
the main menu.
The batch is saved as a prepared batch that is ready to submit. You can open this batch
from the Reinject Samples page in the Acquisition mode and submit the batch. The
application acquires only the samples that have not been previously acquired.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 261
Submitting a Target Screening Batch
In the Batch View, you can submit an entire batch or only selected samples in the batch.
When you submit a batch for acquisition and processing, you can choose to create reports for
the submitted samples. See Submit Options dialog box.
For a description of commands in the shortcut menu, see Batch View shortcut menu
commands.
Follow these procedures:
•To submit selected peaks for processing
•To submit samples in the batch
•To view the output files
To submit selected peaks for processing
1. In the Peak List, select the Selected check box for the peaks that you want to process.
2. Click the Submit … for Processing icon, .
The application processes the selected peaks and updates the data in the Peak
Identifications pane.
To submit samples in the batch
1. Do one of the following:
• To submit all samples in the batch, click the Submit Batch icon, .
• To submit specific samples, select the samples and click the Submit Selected
Samples icon, .
The Submit Options dialog box opens.
2. To acquire (or reacquire) the submitted samples, select the Acquire Data check box.
• When all submitted samples have been previously acquired, this option is (by default)
not selected.
• When one or more samples in the batch have not been acquired, this option is (by
default) selected.
Note You can also submit only selected peaks for processing. See To submit selected
peaks for processing.
Tip You can process the data with or without performing peak detection. For
example, you might want to turn off peak detection when reprocessing samples.

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
262 TraceFinder User Guide Thermo Scientific
3. To process the submitted samples, select the Process Data check box.
The application displays processing options for the Target Screening method.
If the method includes unknown screening features, the application also displays
unknown screening options. For specifying processing data for batches that also use
unknown screening features, see To submit samples in the batch in Chapter 6, “Using the
Analysis Mode for Unknown Screening Batches.”
4. Select the check box for the options that you want to use.
Peak Detect: Performs peak detection. You can process the data with or without
performing peak detection. For example, you might want to turn off peak detection when
reprocessing samples.
Identify and Confirm: Performs both identification and confirmation.
5. (Optional) Select the Create Reports check box.
6. (Optional with multiplexing activated) Select the Priority Sequence check box.
The application acquires the priority batch on the next available channel or the assigned
channel.
7. (Optional without multiplexing activated) Select the Priority Sequence check box and
then select one of the following priority options to place the batch in the queue:
•Next Available Batch places the batch immediately after the currently acquiring
batch.
•Next Available Sample places the batch immediately after the currently acquiring
sample.
8. To specify the following optional parameters, click Show Details.
a. Select the Use check box for the device that you want to use for this acquisition.
b. Select the Start Device check box to indicate the device that will initiate
communication with the other instruments.
This is usually the autosampler.
c. Select the Start When Ready check box, which starts all instruments together when
they are all ready.
When this is cleared, individual instruments can start at different times and then have
to wait for the last instrument to be ready.
d. Select the system state after it acquires the last batch: On, Standby, or Off.
9. To start the selected processes, click OK.
Note When you select Full Sequence Submission in the Configuration console, these
options are unavailable because the current batch and the current sample are, in effect,
the same thing.
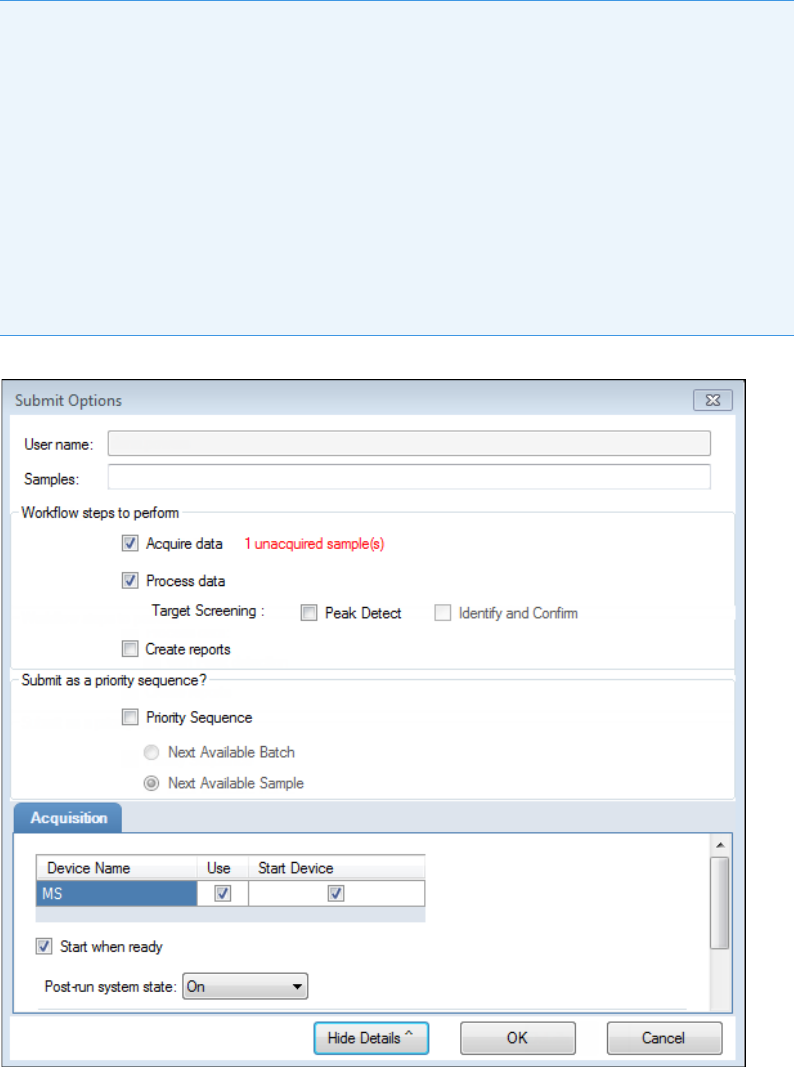
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 263
The selected processes begin, and the application shows the real-time display at the
bottom of the current window. You can begin another batch in the Analysis mode while
you watch the real-time display of the currently acquiring batch.
Figure 87. Submit Options dialog box
IMPORTANT When your batch also uses unknown screening features, after the first
processing, you might see an error message stating that the total number of results is too
large for a single batch (more than 500 000). Return to the Processing pages for the
unknown screening features (refer to Chapter 5, “Using the Method Development Mode
for Unknown Screening Methods,” in the TraceFinder Lab Director User Guide) and make
one or more of these parameter adjustments:
• Limit the RT range.
• Shorten the signal range.
• Specify a lower value for the Number of Top Matches.
• Specify a Simple Search instead of an Exhaustive Search.
• Specify Top Peaks instead of All Peaks.
• Limit the number of search types.

5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
264 TraceFinder User Guide Thermo Scientific
Table 49. Submit Options dialog box parameters (Sheet 1 of 2)
Parameter Description
User Name Name of the current user.
Samples Number of samples to be submitted for acquisition, processing, or
reporting.
Workflow Steps to Perform
Acquire Data Submits the current batch to acquisition.
• When all submitted samples have been previously acquired, this
option is (by default) not selected.
• When one or more samples in the batch have not been
acquired, this option is (by default) selected.
Process Data Processes the data for the current batch using any of the following
options:
Peak Detect: Performs peak detection. You can process the data
with or without performing peak detection. For example, you
might want to turn off peak detection when reprocessing samples.
Identify and Confirm: Performs both identification and
confirmation.
Note Target Screening methods can include unknown screening
features. For specifying processing data for batches that also use
unknown screening features, see To submit samples in the batch.
Create Reports Creates reports for the current batch.
Submit as a Priority Sequence?
Priority Sequence With multiplexing activated, places the batch immediately after the
currently acquiring batch.
Without multiplexing activated, specifies one of the following
priority options to place the batch in the queue:
Next Available Batch: Places the batch immediately after the
currently acquiring batch.
Next Available Sample: Places the batch immediately after the
currently acquiring sample.
Note When you select Full Sequence Submission in the
Configuration console, these options are unavailable because the
current batch and the current sample are, in effect, the same
thing.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 265
To view the output files
Locate the files to view from the following directories:
The application writes saved batches to the project folder:
…\TraceFinderData\4.0\Projects\…
For each acquired sample, the application writes an RSX file to the batch Data folder:
…\TraceFinderData\4.0\Projects\…\Data
The application saves method information to the batch Methods folder:
…\TraceFinderData\4.0\Projects\…\Methods
The application writes the reports to the batch Reports folder:
…\TraceFinderData\4.0\Projects\…\batch\Reports
Acquisition pane
Device Name Lists all configured instruments.
If the instrument that you want to use is not configured, close the
TraceFinder application, configure the instrument, and then reopen
the application. You cannot configure an instrument while the
TraceFinder application is running.
Available only when you select the Acquire Data check box.
Use Specifies the instruments used for this acquisition.
Available only when you select the Acquire Data check box.
Start Device Specifies the instrument that initiates the communication with the
other instruments. This is usually the autosampler.
Available only when you select the Acquire Data check box.
Start When Ready Starts the specified device when all the instruments are ready to
acquire data. When this is cleared, individual instruments can start
at different times and then must wait for the last instrument to be
ready.
Post-run System State Specifies the system state after it acquires the last batch: On
(default), Standby, or Off.
Buttons
Hide/Show Details Collapses or expands the acquisition details of the Submit Options
dialog box.
OK Begins the selected processes.
Cancel Closes the Submit Options dialog box without submitting any
tasks.
Table 49. Submit Options dialog box parameters (Sheet 2 of 2)
Parameter Description
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
266 TraceFinder User Guide Thermo Scientific
Saving a Batch to a New Location
You can move the current batch to a different project folder, or you can make a copy of the
current batch and save the copy to a different project folder.
Follow these procedures:
•To save a batch to another project folder
•To move a batch to another folder
•To create a new project folder
•To delete project folders
•To rename project folders
To save a batch to another project folder
1. Choose File > Save > Save Batch As from the Batch View main menu in the Analysis
mode.
The Save Batch As Dialog Box opens.
2. Select a storage location.
The default storage location is C:\TraceFinderData\4.0\Projects.
3. Select or create a project folder.
4. Type a name for the new batch.
If you are saving the batch to a different folder, you must give it a unique name. You
cannot overwrite an existing batch in a folder.
5. Click Save.
When you save the batch to a different folder, the reports reflect the original project
folders and the application does not save the calibration history.
To move a batch to another folder
1. Choose File > Save > Move Batch from the Batch View main menu in the Analysis
mode.
The Save Batch As Dialog Box opens.
2. Select a storage location.
The default storage location is C:\TraceFinderData\4.0\Projects.
3. Select or create a project folder.
4. Type a name for the new batch.
You must give the batch a unique name in the new subproject folder. You cannot
overwrite an existing batch.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 267
5. Click Save.
When you move the batch, the reports reflect the original project and subproject folders
and the application does not save the calibration history.
To create a new project folder
1. In the Save Batch As dialog box, select the folder for which you will create a new
lower-level folder.
• You can select the main TraceFinderData\4.0\Projects folder and create a new folder
under it.
• You can select one of the existing folders and create a lower-level folder under it.
2. Click the Create Folder icon, .
The application adds a new lower-level folder to the selected folder.
3. Select the new folder name and type a name for the folder.
Folder names can contain spaces and special characters, except for the following special
characters: \ / : + ? ” < >
To delete project folders
1. In the Save Batch As dialog box, select the folder to delete.
2. Click the Delete Folder icon, .
With no confirmation prompt, the application immediately removes the selected folder.
To rename project folders
1. In the Save Batch As dialog box, select the folder to rename.
2. Click the Rename Folder icon, .
3. Type a new name for the folder and press ENTER.
The application saves the new folder name.
Note After you add a lower-level folder, you cannot rename the parent folder.
Note This feature is not available for folders that contain lower-level project or batch
folders; you must first delete the lower-level project or batch folders.
Note This feature is not available for folders that contain lower-level project or batch
folders; you must first delete the lower-level project or batch folders.
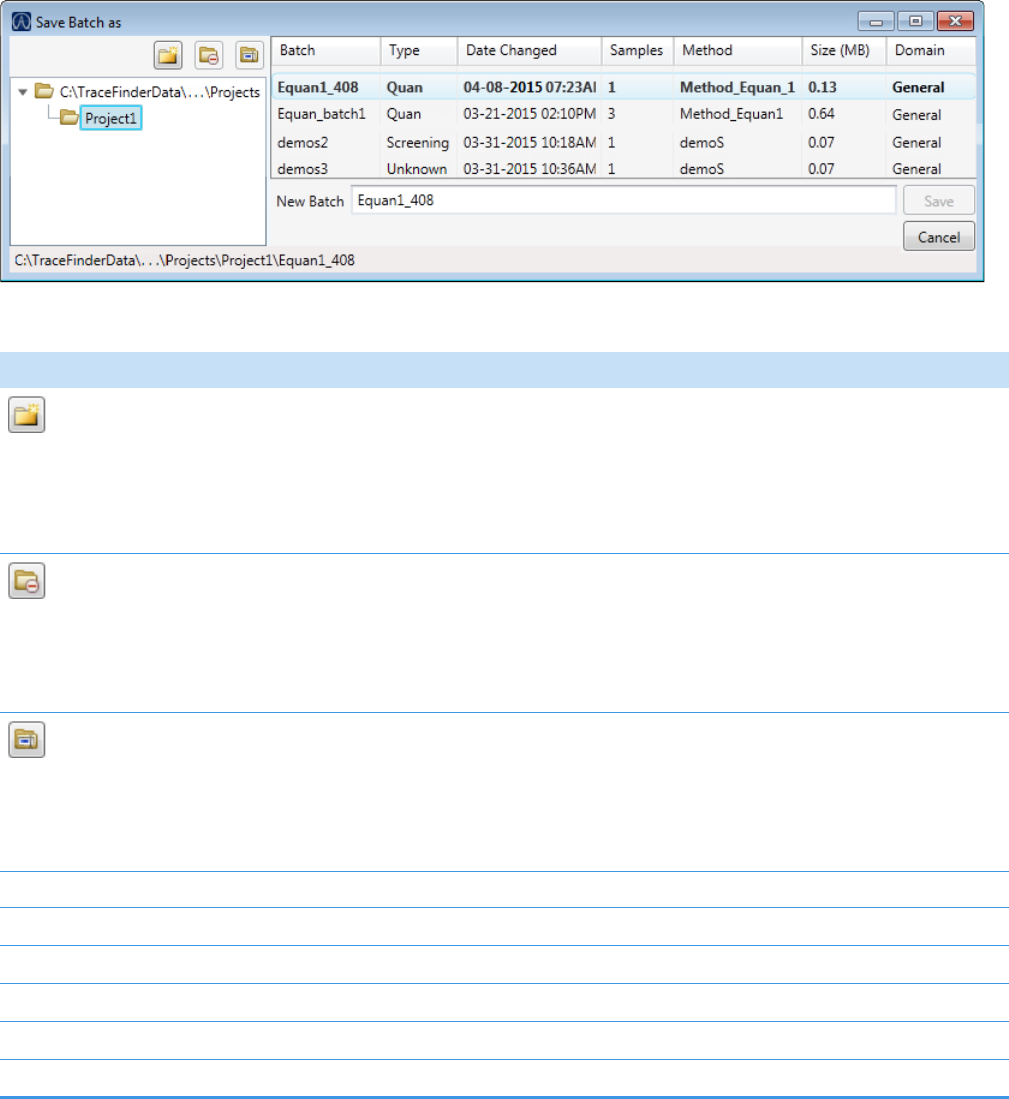
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
268 TraceFinder User Guide Thermo Scientific
Save Batch As Dialog Box
Use the features in the Save Batch As dialog box to save a batch to a new name or to move a
batch to a different project folder.
Figure 88. Save Batch As dialog box
Table 50. Save Batch As dialog box parameters (Sheet 1 of 2)
Parameter Description
Create New Folder
Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Or, you can right-click and choose Create Folder.
Delete Folder
With no confirmation prompt, immediately removes the selected folder.
You cannot delete a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Or, you can right-click and choose Delete.
Rename Folder
Renames the selected folder.
You cannot rename a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Or, you can right-click and choose Rename.
Batch table
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date that the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
5 Using the Analysis Mode for Target Screening Batches
Working in the Batch View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 269
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
New batch parameters
New Batch Name of the new batch to create.
Note If the Create button is not activated, you have entered a name that is already used or
you have not selected a method.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is created.
Buttons
Save Saves the batch to the specified name and folder and opens the Batch View for the new
batch.
Cancel Closes the Save Batch As dialog box without saving the batch.
Shortcut menu commands
Create Folder Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Delete Folder Immediately removes the selected folder. There is no prompt to confirm that you want to
delete the selected folder.
You cannot delete a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Rename Folder Renames the selected folder.
You cannot rename a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Expand Child Nodes Expands all project and subproject folders in the Project tree.
Collapse Child Nodes Collapses all project and subproject folders in the Project tree.
Table 50. Save Batch As dialog box parameters (Sheet 2 of 2)
Parameter Description
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
270 TraceFinder User Guide Thermo Scientific
Working in Data Review for Target Screening Batches
The Data Review view displays the data generated by the master method. Use Data Review to
verify the data for a compound before you generate reports.
Follow these procedures:
•To open the Data Review page
•To display or hide a pane
•To move, dock, or float Data Review panes
•To restore the default layout
To open the Data Review page
1. Click Analysis in the navigation pane.
The Analysis navigation pane opens.
2. Click Data Review.
The Data Review view opens.
On the Data Review page, the panes show the following:
• A list of all samples in the current batch
• The compound results for all compounds in the method
• Chromatogram and spectrum plots for all compounds found in the currently selected
sample
To display or hide a pane
From the View menu, choose to display—indicated with a check mark—or hide the
following panes:
Displays the Samples Pane.
Displays the Compounds Pane.
Displays the Chromatogram Pane.
Displays the Spectrum Pane.
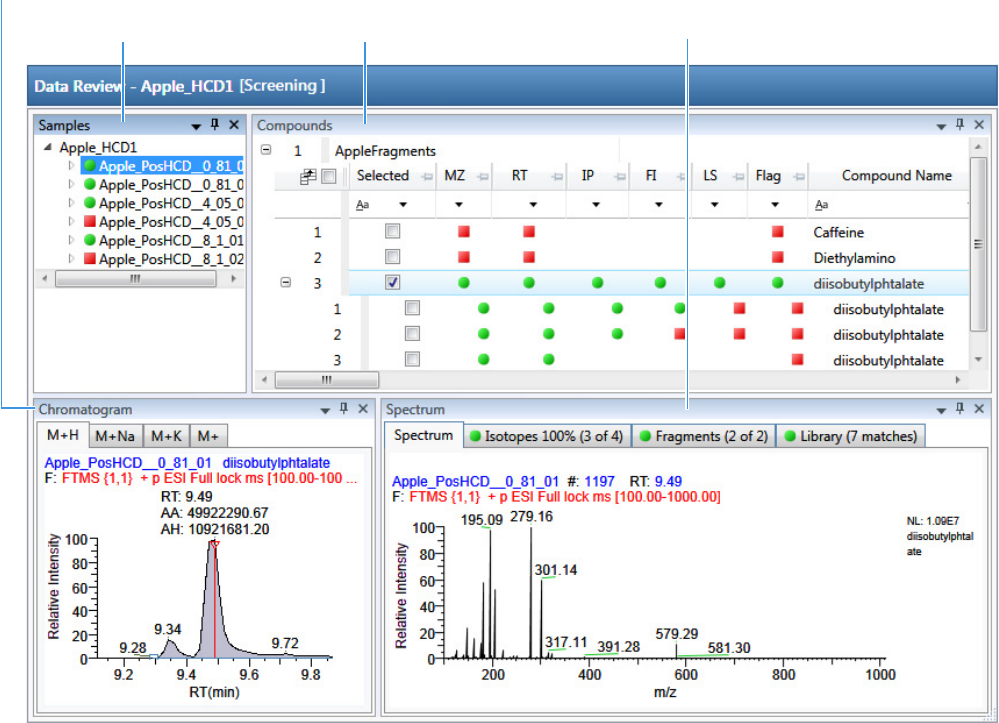
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 271
Figure 89. Data Review panes
When you select a sample in the Samples pane, the associated Compounds pane flags any
compound with errors in the selected sample. The associated Chromatogram pane displays
the chromatogram, retention time, area, height, and signal-to-noise ratio for all compounds in
the selected sample. The Spectrum pane highlights the compound selected in the Compounds
pane. You can display, hide, or move any of these panes.
To move, dock, or float Data Review panes
Follow the instructions in Appendix C, “Moving Data Review Panes.”
To restore the default layout
Choose View > Restore Default Layout.
The Target Screening view displays the default panes in their default locations.
Samples pane Compounds pane
Chromatogram pane
Spectrum pane
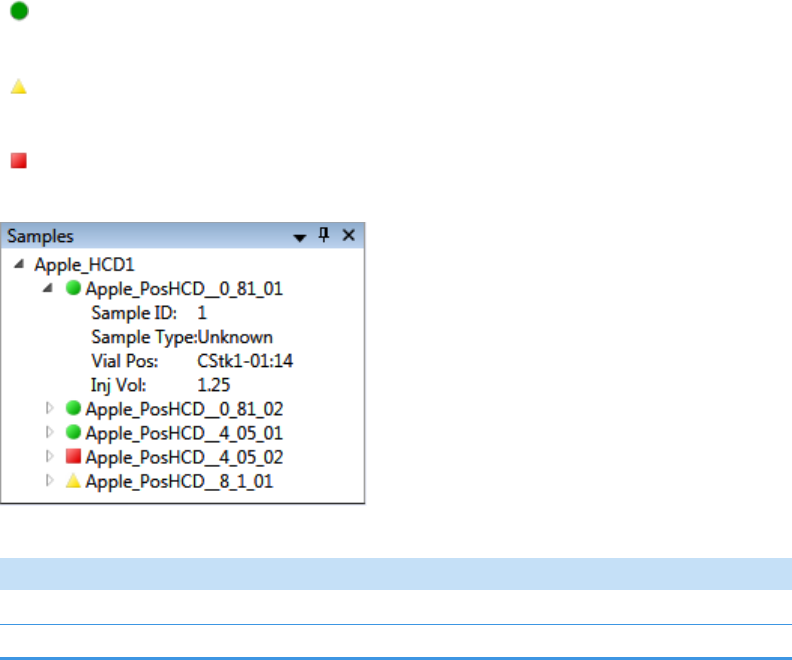
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
272 TraceFinder User Guide Thermo Scientific
The Target Screening view includes the following features:
•Samples Pane
•Compounds Pane
•Chromatogram Pane
•Spectrum Pane
Samples Pane
Use the Samples pane to select a specific sample in the batch. The associated Compounds
Pane displays all compounds in the method and flags any compound with errors in the
selected sample.
Flags in the Samples pane indicate one of the following:
A green circle means that the sample/compound/peak combination is identified and
fully confirmed.
A yellow triangle means that the sample/compound/peak combination is identified but
not fully confirmed.
A red square means that the sample/compound/peak combination is not identified.
Figure 90. Samples pane
Table 51. Samples pane shortcut menu commands
Command Description
Sort by Alphabetical Sorts the samples alphabetically by sample name.
Sort by Import Order Sorts the samples in the order they were processed.
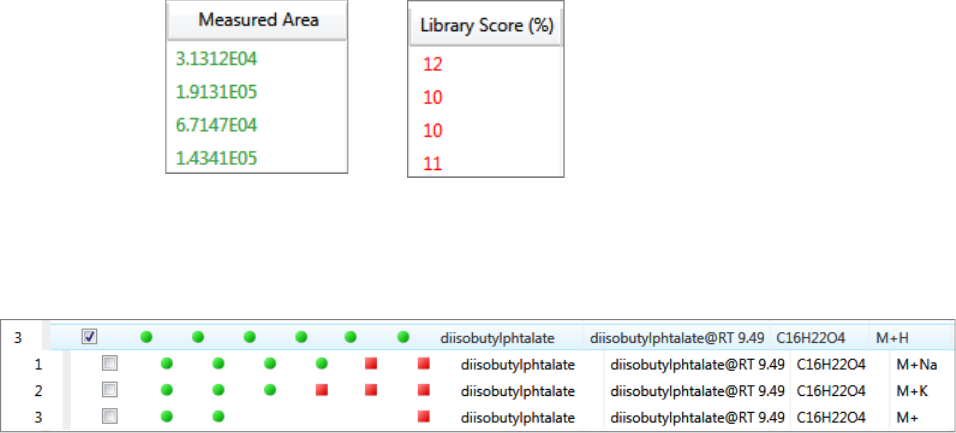
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 273
Compounds Pane
The Compounds pane displays all found peaks in the selected sample and flags any
compound with errors. The compounds table reflects the identified compounds found in the
compound database and the results of the method processing criteria. See Compounds Pane
Columns.
Use the following Compounds pane features:
•Color Coding for Measured or Calculated Values
•Displaying Multiple Adducts
•Exporting Compounds
•Compounds Pane Columns
Color Coding for Measured or Calculated Values
The Compounds pane uses color-coded text to indicate the following:
• Green—Indicates that the measured value of scoring and confirmations pass the criteria
specified in the method.
• Red—Indicates that the measured or calculated value does not pass the criteria specified
in the method.
Displaying Multiple Adducts
When the application finds multiple adducts at the same retention time in a sample, the
Compounds pane displays the adducts on separate rows in the table.
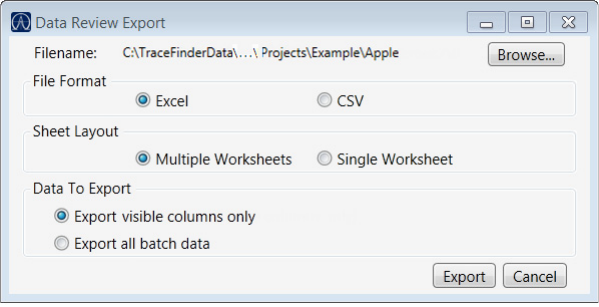
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
274 TraceFinder User Guide Thermo Scientific
Exporting Compounds
Use the commands in the File menu to export data to an Excel spreadsheet, a CSV file, a
compound database, or a new quantitation method.
Follow these procedures:
•To export compounds to an Excel spreadsheet
•To export compounds to a CSV file
•To export compounds to a compound database
•To create a new quantitation method with the selected compounds
•To create a new quantitation method and update the compound database
To export compounds to an Excel spreadsheet
1. For each compound that you want to export to an Excel spreadsheet, select the check box
in the Selected column.
2. Choose File > Export Data To > CSV or Excel from the main menu
The Data Review Export dialog box opens.
3. Click Browse and, in the Export Data to Excel dialog box, locate the folder where you
want to save the file.
4. Type a file name for the XLSX file and click Save.
5. In the File Format area, select the Excel option.
6. In the Sheet Layout area, select one of the following file formats for the spreadsheet.
•Multiple Worksheets: Writes one sample to each Excel worksheet tab.
•Single Worksheet: Writes all samples to a single Excel worksheet tab.
7. In the Data to Export area, select one of the following sets of data to export.
•Export Visible Columns Only: Writes data from the displayed columns of selected
samples to the specified worksheet format.
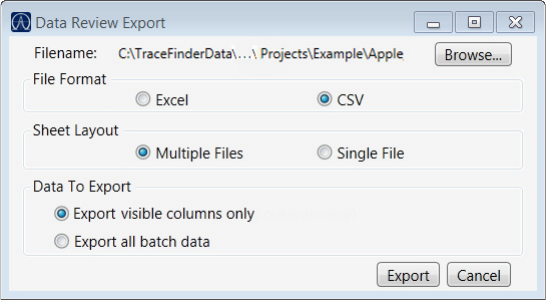
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 275
•Export All Batch Data: Writes data from all columns (displayed or hidden) of all
samples to the specified worksheet format.
8. Click Export.
The application saves the specified compound data to an Excel spreadsheet and opens the
folder where you saved the file.
To export compounds to a CSV file
1. For each compound that you want to export to a CSV file, select the check box in the
Selected column.
2. Choose File > Export Data > To CSV or Excel from the main menu.
The Data Review Export dialog box opens.
3. Click Browse and, in the Export Data to Excel dialog box, locate the folder where you
want to save the file.
4. Type a file name for the CSV file and click Save.
5. In the File Format area, select the CSV option.
6. In the Sheet Layout area, select one of the following file formats for the spreadsheet.
•Multiple Files: Writes one sample to each CSV file.
•Single File: Writes all samples to a single CSV file.
7. In the Data to Export area, select one of the following sets of data to export.
•Export Visible Columns Only: Writes data from the displayed columns of selected
samples to the specified worksheet format.
•Export All Batch Data: Writes all data from all samples in the batch to the specified
worksheet format.
8. Click Export.
The application saves the specified compound data to an CSV spreadsheet and opens the
folder where you saved the file.
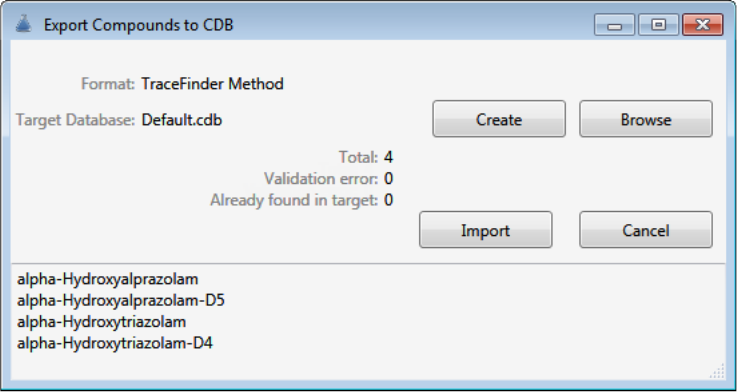
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
276 TraceFinder User Guide Thermo Scientific
To export compounds to a compound database
1. For each compound that you want to export to a compound database, select the check
box in the Selected column, including any adducts that you want to export.
2. Choose File > Export Data To > Compound Database from the main menu.
The Export Compounds to CDB dialog box opens.
3. Do one of the following:
• Accept the default target database.
• Click Create and type the name for a new compound database.
• Click Browse and select from the list of compound databases.
4. Click Import.
• When you export compounds to a database that already contains these compounds,
the application updates the retention times in the database.
• When you add compounds to a database that does not contain these compounds, the
application adds all the compound data to the database.
When you export only one adduct for a compound, the application uses the selected
adduct as the peak in the updated or new compound database. When you export multiple
adducts for export, the application uses the adducts in the order of intensity.
The application does the following:
• Uses the measured retention time value for the compound in the updated or new
compound database.
• Uses the expected m/z value for the compound in the updated or new compound
database.
• Exports all found fragments to the updated or new compound database.
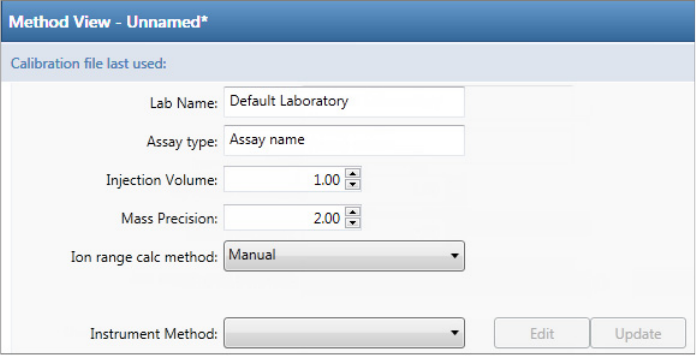
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 277
To create a new quantitation method with the selected compounds
1. For each compound that you want to export to a new quantitation method, select the
check box in the Selected column including any adducts that you want to export.
2. Choose File > Export Data To > New Quantitation Method from the main menu.
The Acquisition page of a new quantitation method opens.
3. From the Instrument Method list, select a method (.meth) file to use for acquiring the
data.
4. Choose File > Save from the main menu.
5. In the Save Master Method dialog box, type a name for the method and click OK.
6. Click Compounds in the Method View navigation pane.
The application exports the selected compounds from the target screening method to the
new quantitation method and uses data from the selected compounds as follows:
• Exports quantitation peaks in the order of intensity.
• Exports measured retention time value for the compounds in the new method.
• Exports expected m/z value for the compounds in the new method.
• Exports all found fragments to the new method.
• Adds a filter for both quantitative peaks and confirming ions.
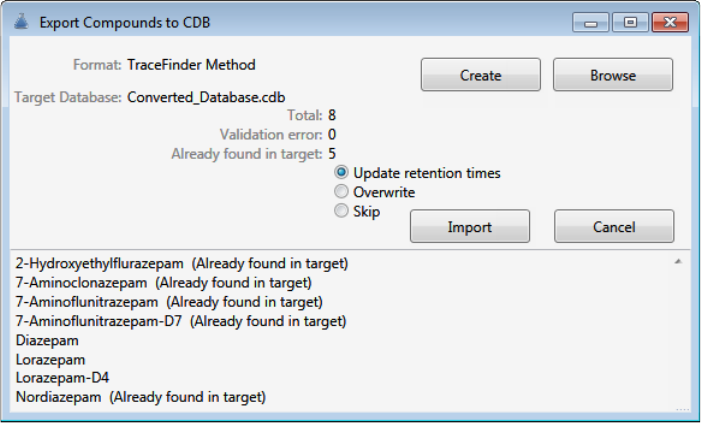
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
278 TraceFinder User Guide Thermo Scientific
To create a new quantitation method and update the compound database
1. For each compound that you want to export, select the check box in the Selected column,
including any adducts that you want to export.
2. Choose File > Export Data > Update Compound Database and Create New
Quantitation Method from the main menu.
The Import Compounds to CDB dialog box opens.
The dialog box lists all compounds selected in the screening batch.
3. Do one of the following:
• Accept the default target database.
• Click Create and type the name for a new target database.
• Click Browse and select from the list of compound databases.
The compounds list indicates which compounds already exist in the target compound
database and which do not.
4. When any of the compounds already exist in the target database, choose one of the
following options:
•Update Retention Times: Updates only the retention times for the duplicate
compounds in the target database.
•Overwrite: Overwrites all compound data for the duplicate compounds in the target
database.
•Skip: Does not write any data from the duplicate compounds to the target database.
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 279
5. Click Import.
The Acquisition page of a new quantitation method opens.
6. From the Instrument Method list, select a method (.meth) file to use for acquiring the
data.
7. Choose File > Save from the main menu.
8. In the Save Master Method dialog box, type a name for the method and click OK.
9. Click Compounds in the Method View navigation pane.
The application exports the selected compounds (with the specified options) from the
target screening method to the new quantitation method.
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
280 TraceFinder User Guide Thermo Scientific
Compounds Pane Columns
The columns of data in the Compounds pane display all parameter values associated with
each compound in the selected sample. See Compounds Pane Parameters.
To hide or display columns in the Compounds pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
The Field Chooser displays all available columns of data for the Compounds pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Compounds pane.
3. When you are finished modifying the column display, click to close the Field
Chooser.
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 281
Compounds Pane Parameters
The Compounds pane displays all parameter values associated with each compound in the
selected sample.
Figure 91. Compounds pane
Table 52. Compounds pane parameters (Sheet 1 of 5)
Column Description
Displays the current compound database. When you have multiple screening databases,
the application lists the results for each database separately.
Adduct The most intense adduct for the retention time for a compound.
Compound Name The compound name match in the compound database.
Confirmed The number of criteria confirmed out of the total number specified in the method.
FI Fragment Ions flag. The application displays one of these indicators:
• A green circle (pass) when the measured m/z value of any of the fragments is within
the mass tolerance specified in the method. On the Isotopes page in the Spectrum
pane, the All Isotopes and Multi-Isotopes flags are also green.
• A red square (fail) when the measured m/z value of none of the fragments is within
the mass tolerance specified in the method. On the Isotopes page in the Spectrum
pane, the All Isotopes and Multi-Isotopes flags are also red.
• A blank when there are no fragments detected.
To display a list of fragments and their pass/fail status, hold your cursor over the
indicator.
Expand the list of found compounds in
the screening database.
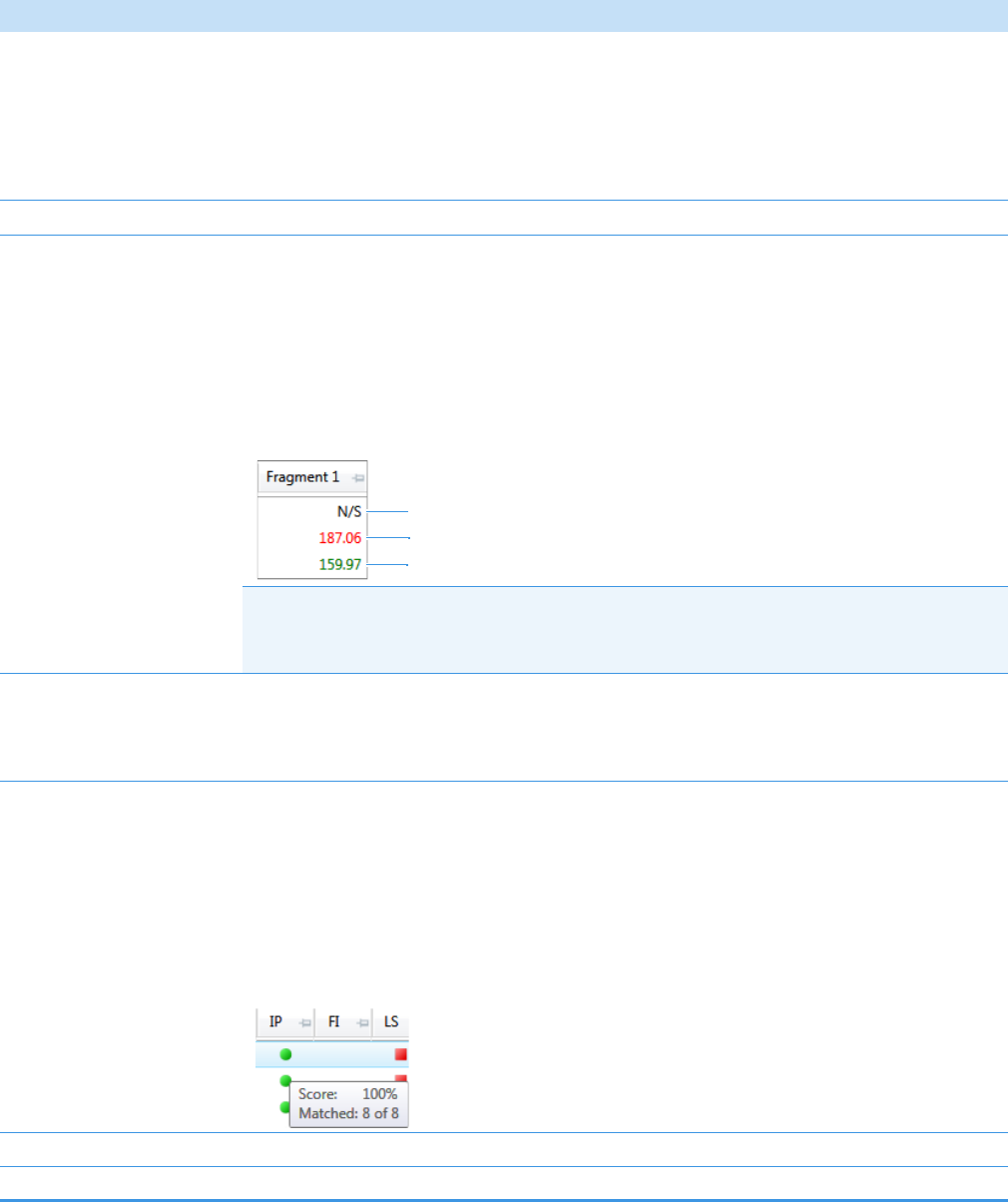
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
282 TraceFinder User Guide Thermo Scientific
Flag Indicates the status of the identification and confirmation criteria.
• A green circle when the sample/compound/peak combination is identified and fully
confirmed.
• A yellow triangle when the sample/compound/peak combination is identified but
not fully confirmed.
• A red square when the sample/compound/peak combination is not identified.
Formula The formula for the peak as specified in the compound database.
Fragment nDisplays the measured m/z for the fragment ion. The application displays a separate
column for each found fragment.
• For each fragment found in the compound database that passes the filter in the
method, the Compounds table displays the m/z value in green text.
• For each fragment found in the compound database that does not pass the filter in
the method, the Compounds table displays the m/z value in red text.
• For each fragment that is not found in the compound database, the Compounds
table displays N/S (none specified).
Note Compounds can have a maximum of five fragments, and the Compounds table
has a maximum of five Fragment columns. When a compound contains fewer than five
fragments, all remaining Fragment columns display N/S.
Fragment n (Delta
(ppm/mmu))
The difference between the expected fragment ion m/z from the compound database and
the measured fragment ion m/z.
The application displays a separate delta column for each identified fragment.
IP Isotopic Pattern flag. The application displays one of these indicators:
• A green circle (pass) when the score percentage is higher than the specified fit
threshold percentage.
• A red square (fail) when the score percentage is lower than the specified fit threshold
percentage.
• A blank when the parameter is not scored.
To display the score of matched isotopes, hold your cursor over the indicator.
Isotopic Pattern Score (%) The percentage of the number of total isotopes to the number of matched isotopes.
Left RT The time point of the left leading edge of the integrated peak.
Table 52. Compounds pane parameters (Sheet 2 of 5)
Column Description
Fragment not found in the compound database
Fragment found but does not meet method parameters
Fragment found and meets method parameters

5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 283
Lib Match Name The name of the best matching compound in the library search. When the application
finds a match in the library, this column displays the matching library entry with the
highest score.
• When the application does not perform a library search, this column displays “N/A”
in black text.
• When the application does not perform an MS/MS scan, this column displays
“N/A” in red text.
Library Match Rank Displays the ranking of the library match. When the application finds a match in the
library, this column displays the library entry’s relative rank, in the format “x of y”, where
• x = the rank of the highest scoring library match.
• y = the total number of library matches from the list of matches for a particular
adduct that contains the highest scoring match.
When the application performs both a library search and an MS/MS scan and both the
library entry and the formula match the target compound:
• A library score that is higher than or equal to the score threshold meets the criteria,
and the values in this column appear in green.
• A library score that is lower than the score threshold fails to meet the criteria, and the
values in this column appear in red.
When the application does not perform a library search, this column displays “N/A” in
black text.
When the application does not perform an MS/MS scan, this column displays “N/A” in
red text.
Library Score (%) The score from the library fit. When the application finds a match in the library, this
column displays the highest score associated with the Lib Match Name parameter.
When the application performs both a library search and an MS/MS scan and both the
library entry and the formula match the target compound:
• A library score that is higher than or equal to the score threshold meets the criteria,
and the values in this column appear in green.
• A library score that is lower than the score threshold fails to meet the criteria, and the
values in this column appear in red.
When the application does not perform a library search, this column displays “N/A” in
black text.
When the application does not perform an MS/MS scan, this column displays “N/A” in
red text.
Valid range: 1 through 100%
LS Library Search flag. The application displays one of these flags:
• A green circle when the library search is successful.
• A red square when the library search is not successful.
Table 52. Compounds pane parameters (Sheet 3 of 5)
Column Description
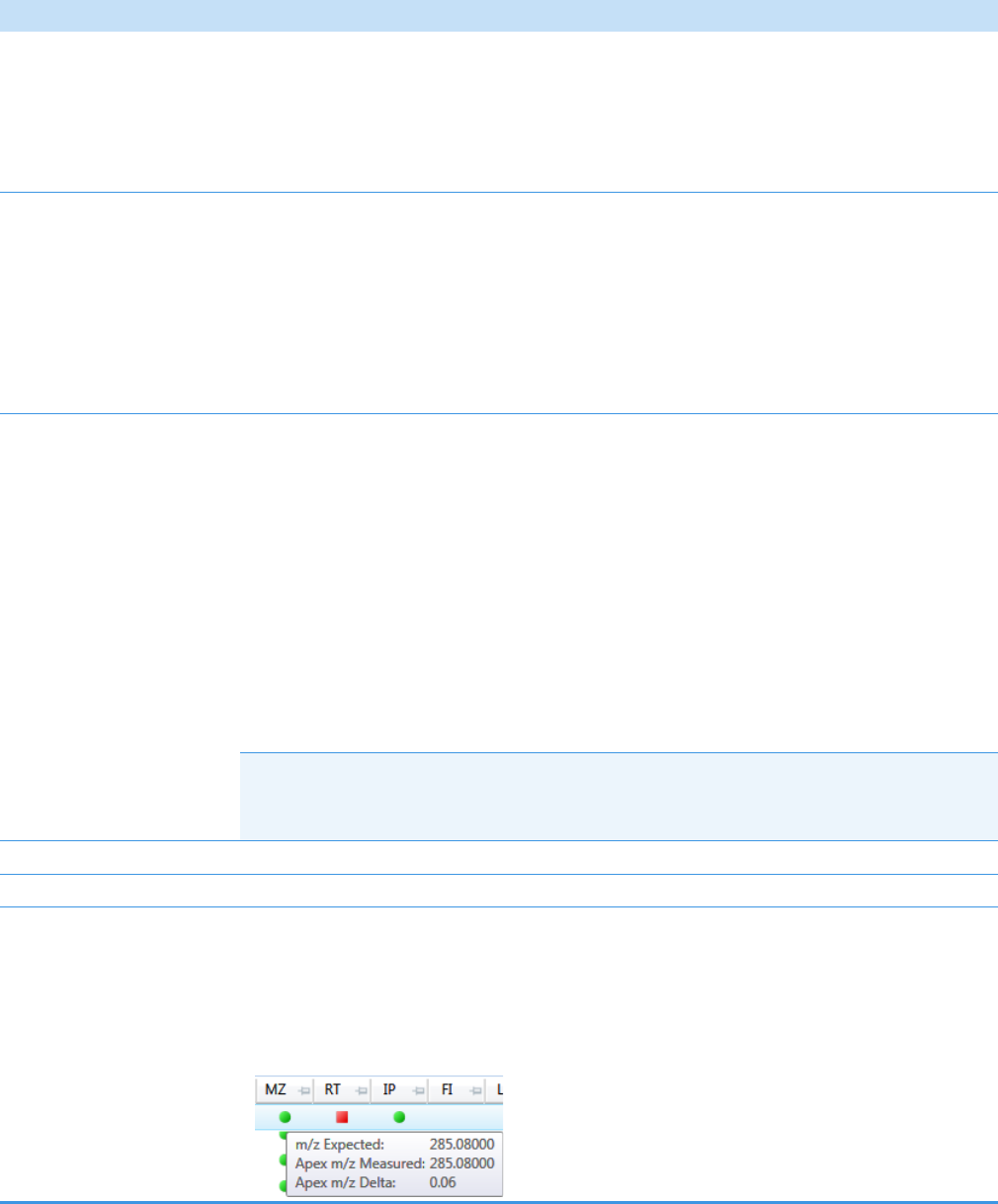
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
284 TraceFinder User Guide Thermo Scientific
m/z (Apex) Mass-to-charge ratio found in the spectra for the peak. Assumes the charge is 1.
When the application successfully integrates the peak, this column displays the charged
m/z value for the compound, which is the highest intensity in the apex scan.
When the application cannot successfully integrate the peak, this column displays “N/F”
(not found).
m/z (Delta) Difference between the m/z (Expected) and m/z (Apex). Assumes the charge is 1.
When the m/z (Apex) column displays m/z value for the compound, this column displays
the delta m/z corresponding to the highest intensity in the apex scan.
• When the mass tolerance is specified in ppm in the master method, then
m/z (Delta) = 1 000 000 × ([m/z (Apex) – m/z (Expected)] m/z (Expected)).
• When the mass tolerance is specified in mmu in the master method, then
m/z (Delta) = 1000 × (m/z (Apex) – m/z (Expected)).
m/z (Expected) Mass-to-charge ratio from the compound database. Assumes the charge is 1.
• When an adduct is found, the application displays the neutral mass value for the
compound (calculated from the neutral formula) ± the mass of the most intense
adduct ion found for the compound.
• When no adduct is found, the application displays the neutral mass value for the
compound ± the mass of the first adduct entered in the compound database.
For details about defining adducts for the compound database, refer to Chapter 1,
“Using the Configuration Console,” in the TraceFinder Lab Director User Guide.
For details about adding adducts to compounds in the database, refer to Chapter 2,
“Using Compound Databases in the Method Development Mode,” in the Tra ce Fi nd er
Lab Director User Guide.
Note When the adduct is a gain, the adduct mass is a positive number. When the
adduct is a loss, the adduct mass is a negative number. The resulting mass value after
adding or subtracting the adduct mass is always a positive number.
Match Result Name The compound name match in the compound database and the retention time.
Measured Area The AA value from the chromatogram pane.
MZ Mass-to-charge ratio flag. The application displays one of these indicators:
• A green circle (pass) when the measured m/z value is within the specified threshold.
• A red square (fail) when the measured m/z value is not within the specified threshold.
• A blank when the mass-to-charge value is unavailable.
To display the expected, measured, and delta m/z, hold your cursor over the indicator.
Table 52. Compounds pane parameters (Sheet 4 of 5)
Column Description
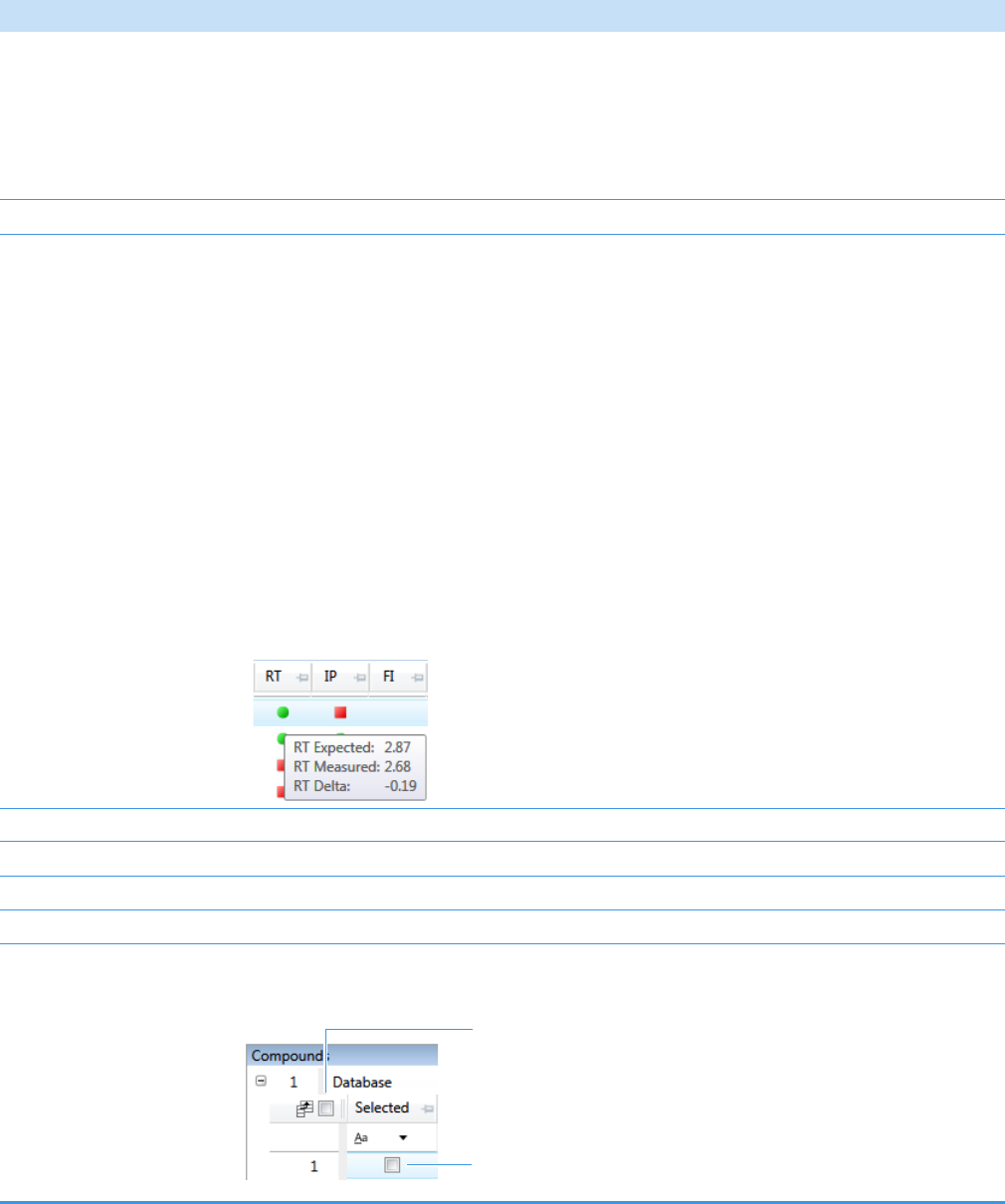
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 285
Num Isotopes Matched The number of isotopes matched in the expected calculated isotope spectra relative to the
total number of isotopes used in the score calculation, in the format “x of y”, where
• x = the number of isotopes matching the elemental composition used for the
Isotopic Pattern Score calculation.
• y = the total number of isotopes considered in the Isotopic Pattern Score calculation.
This is the number of isotope peaks expected to be above the spectral noise.
Right RT The time point of the right trailing edge of the integrated peak.
RT Retention Time flag. The application displays one of these indicators:
• A green circle (pass) when the measured retention time value is within the RT
Window value specified in the compound database.
• A red square (fail) when the measured retention time value is not within the RT
Window value specified in the compound database. In turn, this results in a failure
flag for the m/z value because the application cannot identify an m/z value that meets
the retention time.
• A blank when the retention time value is unavailable. When the retention time is
selected as “confirm” and the m/z is not detected, there is no flag.
Or, you can set a different retention time window on the Processing page in the method.
Refer to Chapter 5, “Using the Method Development Mode for Target Screening
Methods,” in the TraceFinder Lab Director User Guide.
To display the expected, measured, and delta retention times, hold your cursor over the
indicator.
RT (Delta) Difference between the expected and measured retention time for the peak.
RT (Expected) The retention time for the peak as specified in the compound database.
RT (Measured) The found retention time for the peak apex.
S/N The signal-to-noise ratio calculated for the found peak.
Selected Identifies individual compounds for export. To select all compounds for export, select
the check box in the first column.
Table 52. Compounds pane parameters (Sheet 5 of 5)
Column Description
Selects all compounds in the
Compounds pane.
Selects the compound for
export.
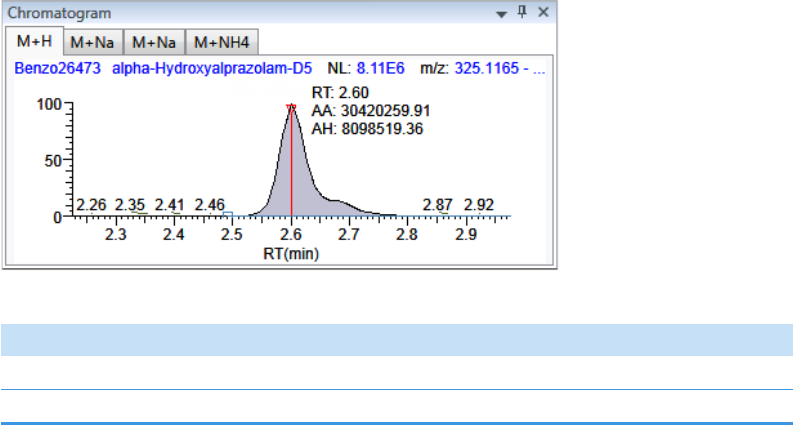
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
286 TraceFinder User Guide Thermo Scientific
Chromatogram Pane
Use the Chromatogram pane to display all extracted chromatograms of all adducts of the
selected compound.
The first tab displays the most intense target adduct for the peak result. Additional (optional)
tabs display extracted ion chromatograms for other adducts for the target compound at the
same retention time in order of intensity. If no signal exists for an adduct, it displays the XIC
of the expected m/z within the specified retention and chromatogram windows. When you do
not specify a retention time or window, the application displays the full time range.
For each adduct, the Spectrum Pane displays the spectrum, isotopes, fragments, and library
matches.
Figure 92. Chromatogram pane
Spectrum Pane
Use the Spectrum pane to display the spectrum, isotopes, fragments, and library search
information for the selected adduct in the Chromatogram pane. The Spectrum pane displays
only the identification and confirmation criteria specified in the method. The confirmations
are based only on the most intense adduct. Refer to “Editing the Processing Page” in
Chapter 5, “Using the Method Development Mode for Target Screening Methods,” in the
TraceFinder Lab Director User Guide.
The Spectrum pane includes the following pages of information (when available) for each
selected sample/compound/peak combination:
•Spectrum
•Isotopes
•Fragments
•Library
Table 53. Chromatogram pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
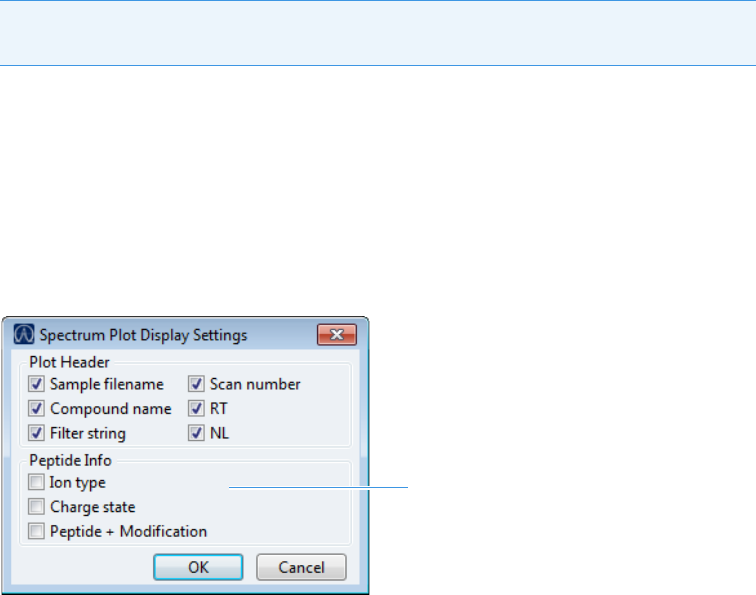
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 287
Spectrum
The application displays the neutral loss (NL) and compound/peak name information on the
right side of the Spectrum page. When data is available, the plot width is the full mass range
in the raw data file. Otherwise, the application scales the width to the scan range.
Follow these procedures:
•To zoom in on a peak
•To copy a plot to another format
•To customize the labels in a small molecule or peptide spectrum
To zoom in on a peak
1. On the Spectrum page, drag the cursor to delineate a rectangle around the peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the Spectrum page and choose Reset Scaling.
To copy a plot to another format
1. In the data spectrum or the reference spectrum, right-click and choose Copy to
Clipboard.
The application copies the specified plot graphic to the Clipboard.
2. In the source format (for example, an Excel spreadsheet or email window), right-click and
choose Paste.
The application pastes the graphic into the new format.
To customize the labels in a small molecule or peptide spectrum
1. Right-click the Spectrum page and choose Display Settings.
The Spectrum Plot Display Settings dialog box opens.
2. Select the check box for each label that you want to display and click OK.
Note The application copies only the indicated data spectrum or the reference
spectrum. It does not copy the entire Spectrum page.
Additional options
for a peptide spectrum
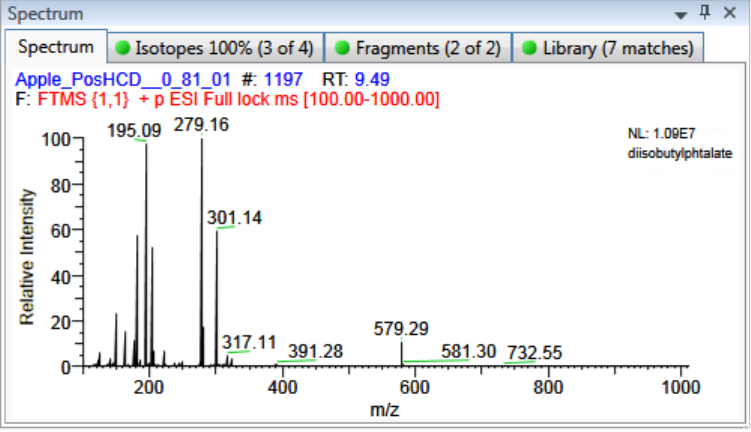
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
288 TraceFinder User Guide Thermo Scientific
Figure 93. Spectrum page
Isotopes
The Isotopes page displays isotopic pattern results for all adducts of a compound according to
the threshold and deviation parameters defined in the screening method.
To identify or confirm the presence of a compound, the resulting score percentage from
isotopic pattern matching must be higher than the specified fit threshold percentage.
• An isotope peak is not found if its intensity, relative to the monoisotopic ion’s intensity, is
more than the specified intensity deviation percentage away from the theoretical relative
intensity of the isotope ion.
• An isotope peak is found if its measured m/z is less than the specified mass deviation
amount away from its expected m/z.
To specify threshold and deviation parameters, refer to “Editing the Processing Page” in
Chapter 5, “Using the Method Development Mode for Target Screening Methods,” in the
TraceFinder Lab Director User Guide.
The Isotopes page displays the isotopes in one of three ways:
•All Isotopes
•Multi-Isotopes
•Individual Isotopes
All isotopes pages use a shortcut menu so that you can specify how to display the data. See
Isotopes Page Shortcut Menu.
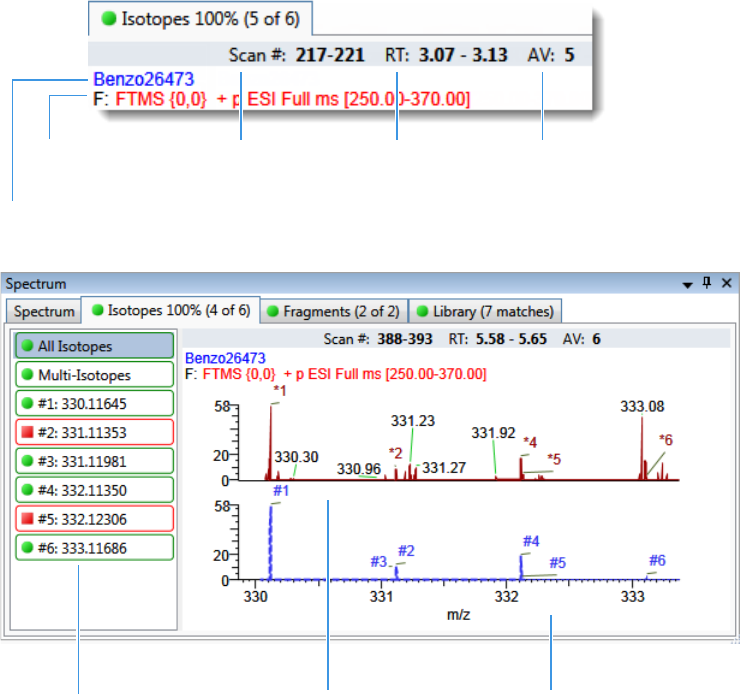
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 289
All Isotopes
The All Isotopes view displays a composite of all isotopes found in the compound. The
application scales the window with respect to the most intense isotope. The most intense
isotope is usually the first isotope unless you are using halogenated compounds. The
application displays the measured peak as a solid red line; the application displays the
expected peak as a dashed blue line.
The application displays these headers for the All Isotopes view:
Figure 94. Isotopes page with stacked spectra for all isotopes
Scan range for
the isotopes
Retention time
range for all
scans
Number of
averaged
scans
Processing
filter
File name
Measured spectrum Expected spectrumExpected m/z of the isotope
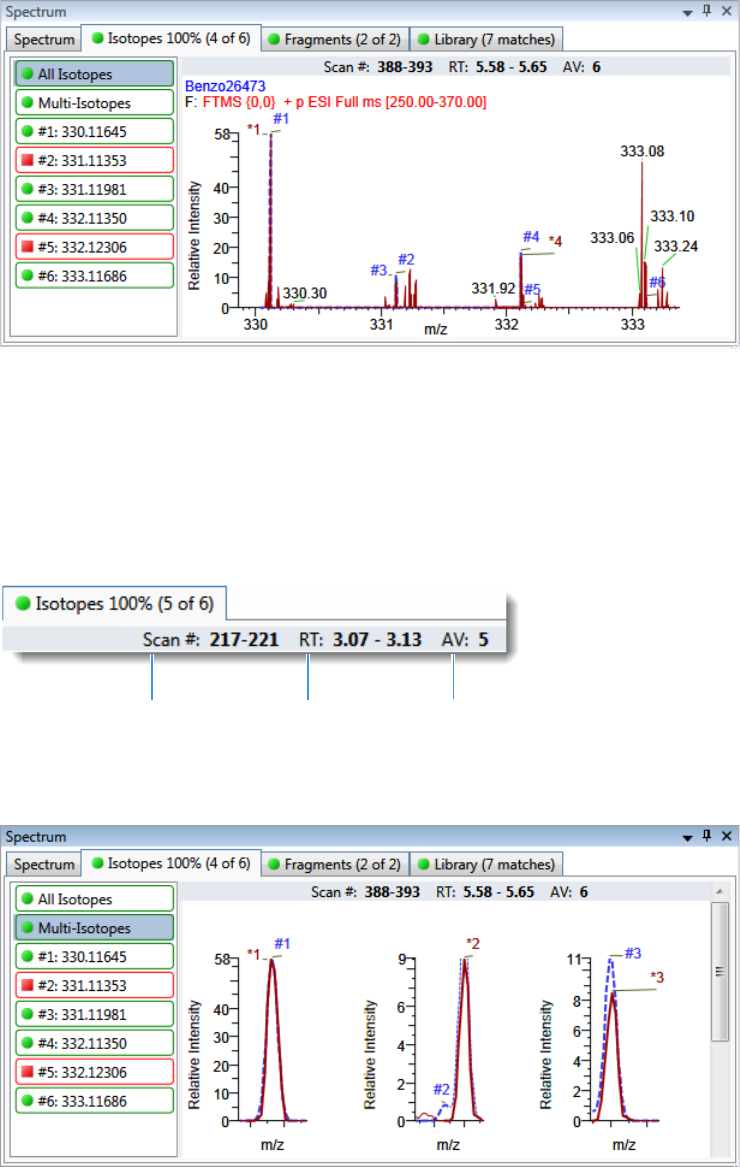
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
290 TraceFinder User Guide Thermo Scientific
Figure 95. Isotopes page with overlaid spectra for all isotopes
Multi-Isotopes
The Multi-Isotopes view displays individual plots for each isotope. You can individually stack
or overlay the plots for each isotope.
The application displays these headers for the Multi-Isotopes view:
Figure 96. Isotopes page with overlaid spectra for multi-isotopes
Measured spectrum in redExpected spectrum in blue
Scan range for
the isotopes
Retention time
range for all
scans
Number of
averaged
scans
Expected spectrum in blue Measured spectrum in red
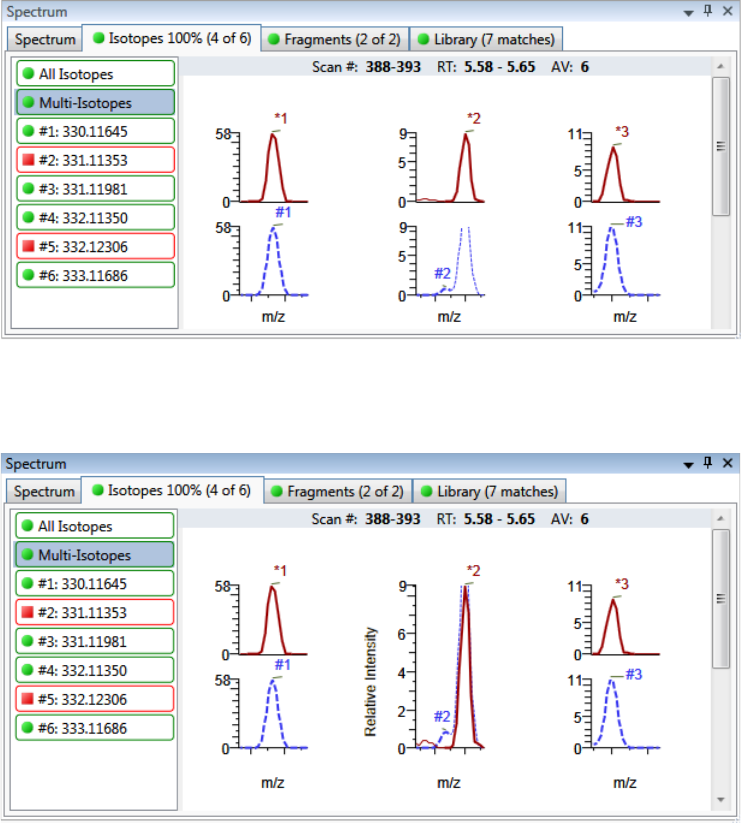
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 291
Figure 97. Isotopes page with stacked spectra for multi-isotopes
Figure 98. Isotopes page with stacked and overlaid spectra for multi-isotopes
Measured spectrum in redExpected spectrum in blue
Measured spectrum in redExpected spectrum in blue
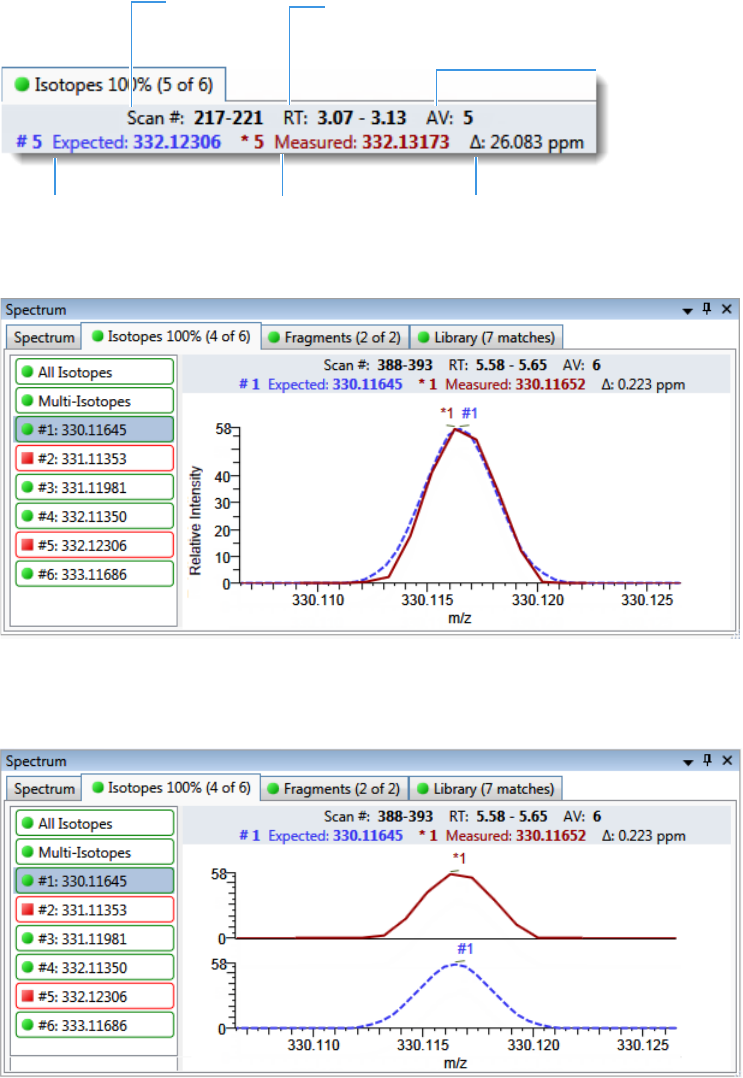
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
292 TraceFinder User Guide Thermo Scientific
Individual Isotopes
The individual isotopes view displays the expected and measured peaks for a single isotope.
The application displays these headers for the individual isotopes view:
Figure 99. Isotopes page with overlaid spectra for a single isotope
Figure 100. Isotopes page with stacked spectra for a single isotope
Scan range for
the isotopes
Retention time range
for all scans
Number of
averaged scans
Expected m/z
for isotope 5
Measured m/z
for isotope 5
Delta between expected and
measured m/z
Measured spectrum in redExpected spectrum in blue
Measured spectrum in redExpected spectrum in blue
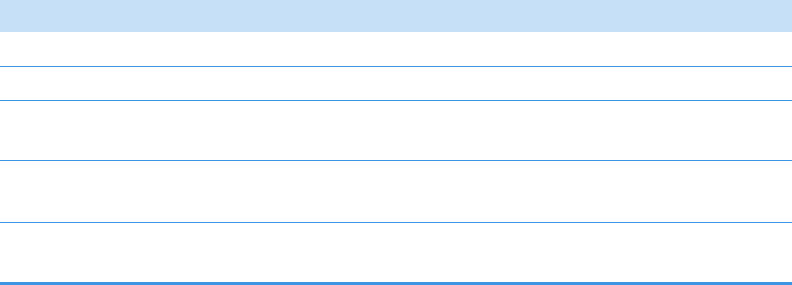
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 293
Isotopes Page Shortcut Menu
Use the commands in the shortcut menu to specify how you want the data displayed.
Fragments
The Fragments page displays the maximum number of fragments as specified in the screening
method. Refer to “Editing the Processing Page” in Chapter 5, “Using the Method
Development Mode for Target Screening Methods,” in the TraceFinder Lab Director User
Guide.
If there are no fragments defined in the screening library for the compound, you can add
fragments to the screening library. Refer to the instructions “To add a fragment to a target
peak” in Chapter 2, “Using Compound Databases in the Method Development Mode,” in the
TraceFinder Lab Director User Guide.
The Fragments page displays the fragments in one of two ways:
•All Fragments
•Individual Fragments
All fragments pages use a shortcut menu so that you can specify how to display the data. See
Fragments Page Shortcut Menu.
Table 54. Isotopes page shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Display Overlay Spectra
Display Stack Spectra
Overlays the two spectrum displays, or stacks the peak apex
spectrum over the simulated spectrum.
Show/Hide Noise Label Adds a noise label to each peak. Expected isotope peaks
(displayed in blue) do not display a noise label.
Show/Hide Resolution
Label
Adds a resolution label to each peak. Expected isotope peaks
(displayed in blue) do not display a resolution label.
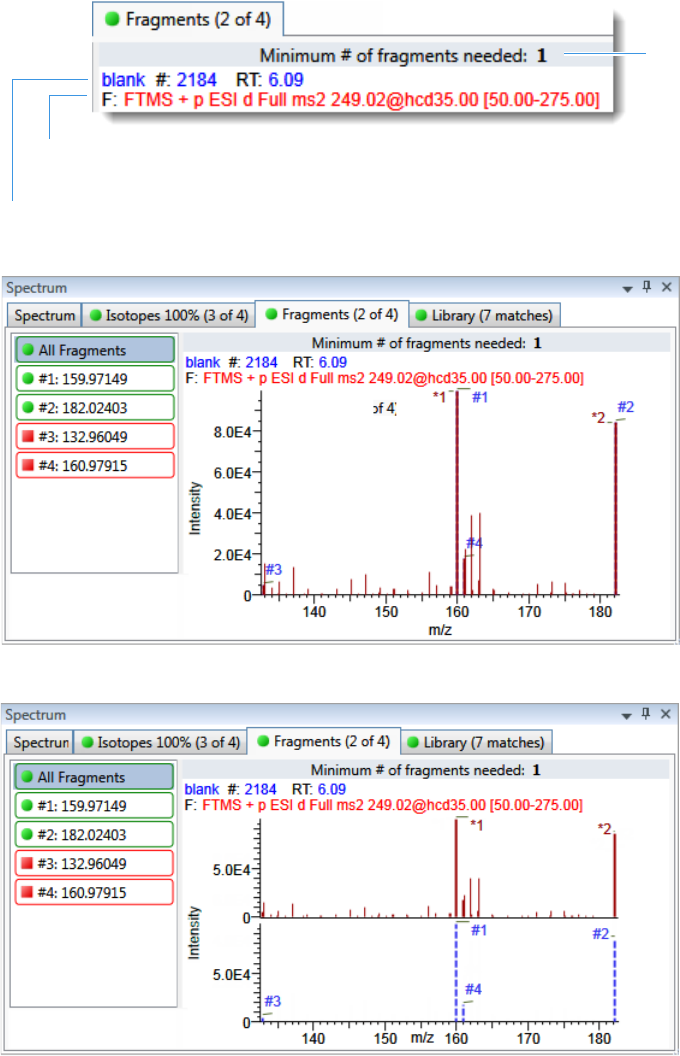
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
294 TraceFinder User Guide Thermo Scientific
All Fragments
The All Fragments view displays a composite of all fragments found in the compound. The
application displays the measured peak as a solid red line; the application displays the
expected peak as a dashed blue line.
The application displays these headers for the All Fragments view:
Figure 101. Fragments page with overlaid spectra for all fragments
Figure 102. Fragments page with stacked spectra for all fragments
Minimum number
of fragments
specified in the
method
Processing
filter
File name
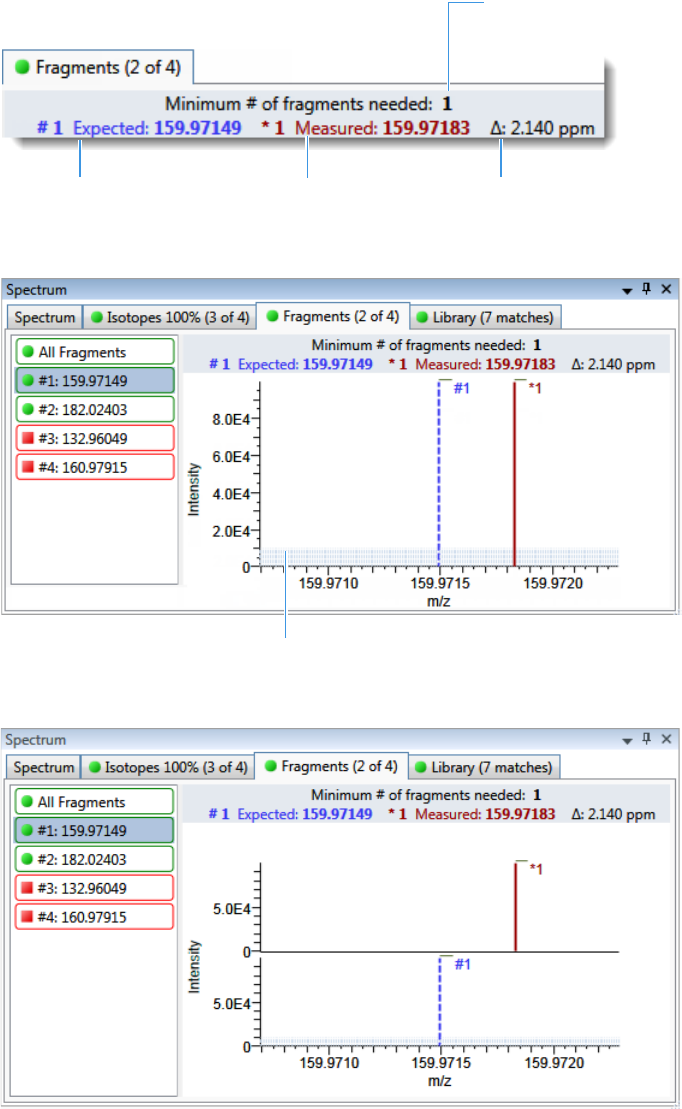
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 295
Individual Fragments
The individual fragments view displays the expected and measured peaks for a single
fragment.
The application displays these headers for the individual fragments view:
Figure 103. Fragments page with overlaid spectra for a single fragment
Figure 104. Fragments page with stacked spectra for a single fragment
Minimum number of fragments specified
in the method
Expected m/z
for fragment 1
Measured m/z
for fragment 1
Delta between expected and
measured m/z
Intensity threshold specified in the method
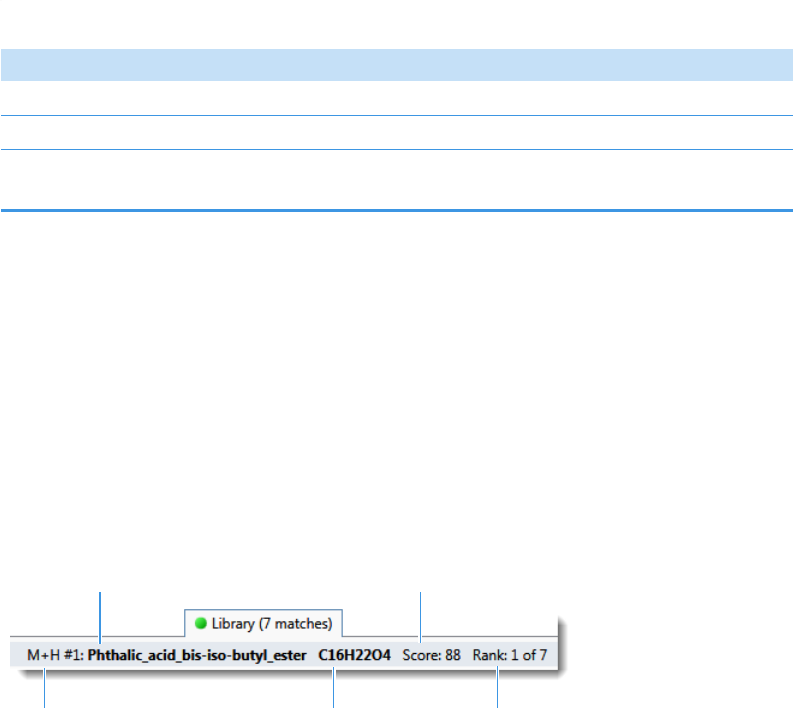
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
296 TraceFinder User Guide Thermo Scientific
Fragments Page Shortcut Menu
Use the commands in the shortcut menu to specify how you want the data displayed.
Library
The Library page displays the matching library spectrum (in blue) and the experimental
spectrum (in black). The resulting score percentage from a library search match must be
higher than your specified threshold value to identify or confirm the presence of a compound.
Refer to “Editing the Processing Page” in Chapter 5, “Using the Method Development Mode
for Target Screening Methods,” in the TraceFinder Lab Director User Guide.
The application scales both the matched library spectrum and the highest peak in the
experimental spectra at 100 percent intensity and displays the resulting neutral loss (NL)
value for the matched library entry name on the right side of the plot.
The application displays these headers for the individual adducts:
Table 55. Fragments page shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Display Overlay Spectra
Display Stack Spectra
Overlays the two spectrum displays, or stacks the simulated
spectrum and the peak apex spectrum.
Library match name
Adduct Formula Library match rank
Library score percentage
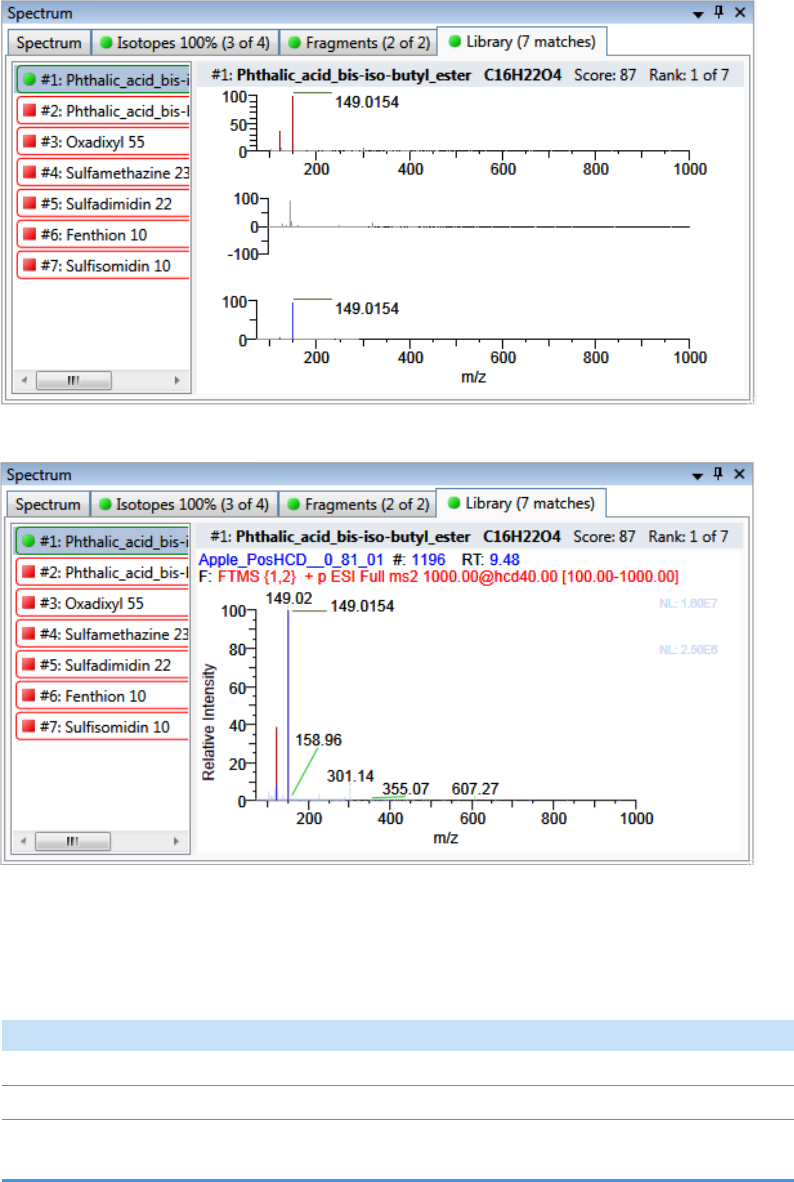
5 Using the Analysis Mode for Target Screening Batches
Working in Data Review for Target Screening Batches
Thermo Scientific TraceFinder User Guide 297
Figure 105. Library page with stacked spectra
Figure 106. Library page with overlaid spectra
Library Page Shortcut Menu
Use the commands in the shortcut menu to specify how you want the data displayed.
Table 56. Library page shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Display Overlay Spectra
Display Stack Spectra
Overlays the two spectrum displays, or stacks the simulated
spectrum and the peak apex spectrum.
5 Using the Analysis Mode for Target Screening Batches
Working in the Report View for Target Screening Batches
298 TraceFinder User Guide Thermo Scientific
Working in the Report View for Target Screening Batches
The Report View displays example reports for the current batch. You must have an open
batch to use the features in the Report View.
Follow these procedures:
•To open the Report View
•To preview a report
•To generate a report as a PDF, an Excel, or a CSV file
•To print a report
•To display a generated report
•To e d i t a re po r t t e m p l a t e
•To create a new report template
To open the Report View
Click Report View in the navigation pane.
The application opens the Report View.
To preview a report
1. In the Template pane, select a report template.
The template list shows all the report templates that you configured in the Configuration
console. Refer to Chapter 1, “Using the Configuration Console,” in the TraceFi n de r La b
Director User Guide.
Figure 107. Example of a report template list
2. Click Preview, .
The application opens the Report Designer, showing the report information for the
current batch in the selected report template format.
For details about using the Report Designer, see Chapter 7, “Using the Report Designer.”
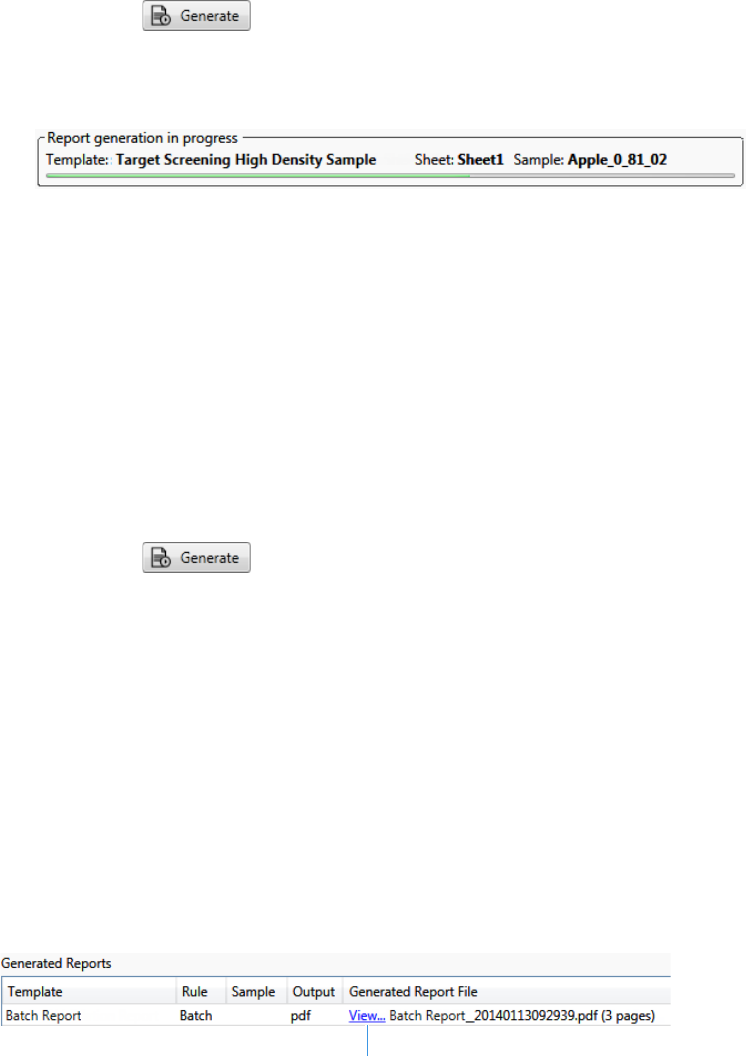
5 Using the Analysis Mode for Target Screening Batches
Working in the Report View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 299
To generate a report as a PDF, an Excel, or a CSV file
1. In the Template pane, select a report template.
2. Select the check box for each of the file types that you want to create: PDF, Excel, or
CSV.
3. Click Generate, .
The application does the following:
• Displays a green progress bar as it generates the reports.
• Creates a report for the current batch as a PDF, an Excel, or a CSV file, using the
selected report template format.
• Adds information about the generated report to the Generated Reports pane.
For details about the Generated Reports pane, see Report View.
• Saves the report files to the …\TraceFinderData\4.0\Projects\batch\ReportOutput
folder.
To print a report
1. In the Template pane, select a report template.
2. Select the check box for the Print file format.
3. Click Generate, .
The application does the following:
• Creates a report for the current batch using the selected report template format.
• Prints the report to your default printer.
• Adds information about the generated report to the Generated Reports pane.
For details about the Generated Reports pane, see Report View.
• Saves the report files to the …\TraceFinderData\4.0\Projects\batch\ReportOutput
folder.
To display a generated report
In the Generated Reports pane, click View for the report that you want to see.
The application opens the output file.
Opens the generated file.
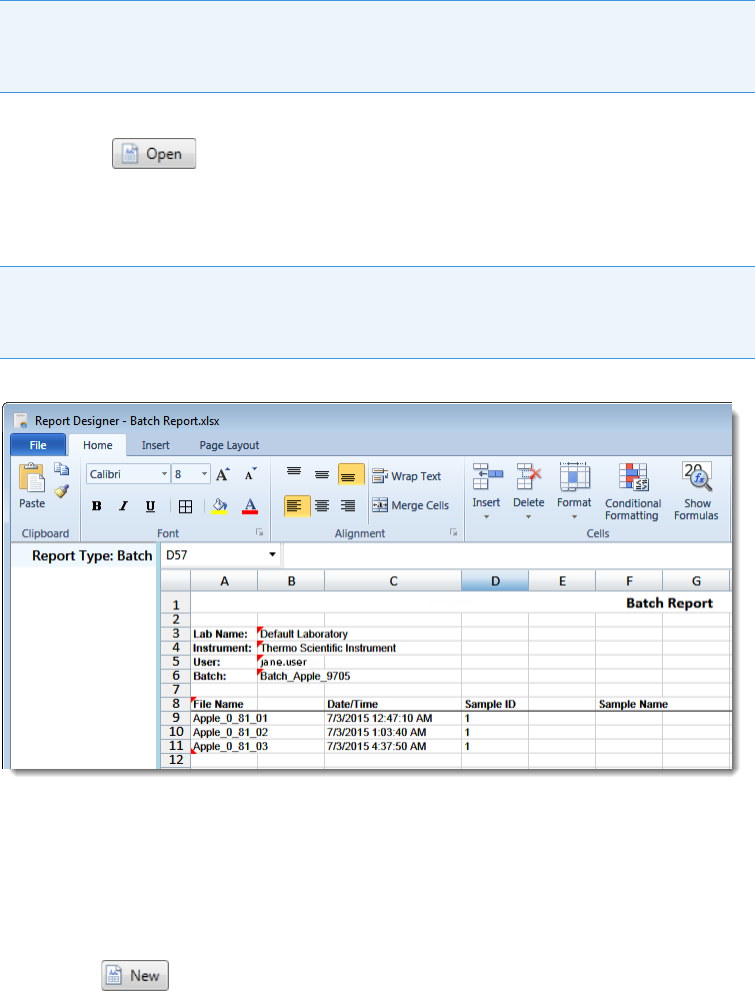
5 Using the Analysis Mode for Target Screening Batches
Working in the Report View for Target Screening Batches
300 TraceFinder User Guide Thermo Scientific
To edit a report template
1. In the Template pane, select a report template.
2. Click Open, .
The application opens the Report Designer showing the template in an Excel spreadsheet.
See Report Designer showing the template for the selected report.
Figure 108. Report Designer showing the template for the selected report
3. Use the features in the Report Designer to edit the template.
For details about using the Report Designer, see Chapter 7, “Using the Report Designer.”
4. When you finish your changes, choose File > Save from the Report Designer menu bar.
To create a new report template
1. Click New, .
The application opens the Report Designer showing an empty template in an Excel
spreadsheet. See Report Designer showing a new, empty template.
The Report Type is None.
In the left pane, the spreadsheet lists all samples in the current batch and all compounds
in the method used for the batch.
Note You cannot edit report templates that were provided with the TraceFinder
application; however, you can open a TraceFinder template, make changes, and save it
to a new template name.
Note When user security is activated, you must have Template Editing permission to
edit report templates created by your laboratory. If the Open button is not active, user
security is activated and you do not have Template Editing permission.
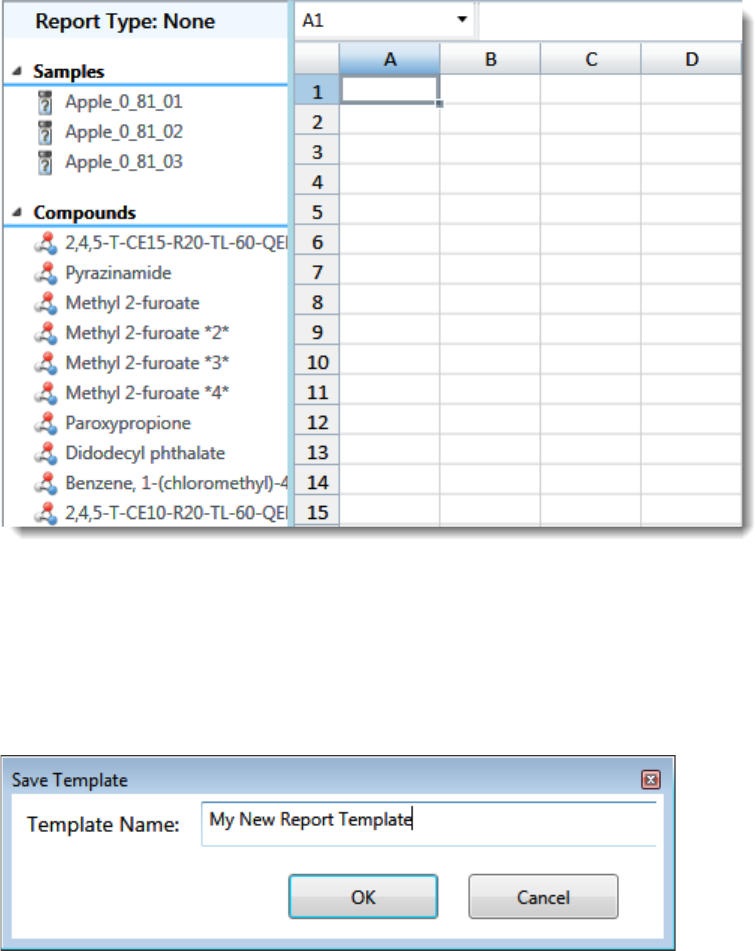
5 Using the Analysis Mode for Target Screening Batches
Working in the Report View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 301
Figure 109. Report Designer showing a new, empty template
2. Use the features in the Report Designer to create the report template.
For details about using the Report Designer, see Chapter 7, “Using the Report Designer.”
3. When you finish your changes, choose File > Save from the Report Designer menu bar.
The Save Template dialog box opens.
Figure 110. Save Template dialog box
4. Type a name for the new report template and click OK.
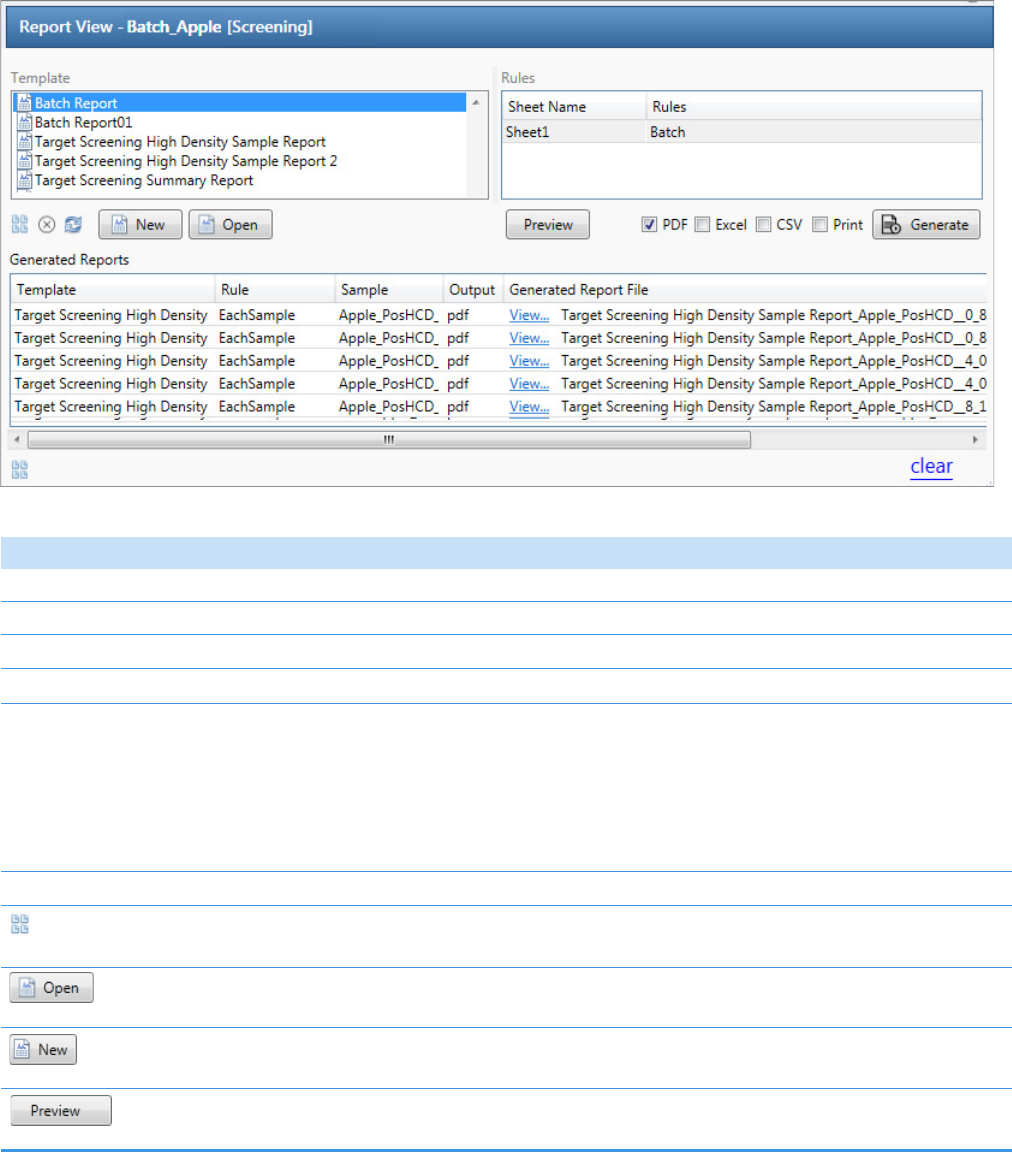
5 Using the Analysis Mode for Target Screening Batches
Working in the Report View for Target Screening Batches
302 TraceFinder User Guide Thermo Scientific
Report View
Use the features in the Report View to display example reports for the current batch.
Figure 111. Report View
Table 57. Report View parameters (Sheet 1 of 2)
Parameter Description
Template
Displays all report templates.
Rules
Sheet Name Specifies each sheet in the report.
Rules Specifies the type of data used in each sheet in the selected report.
•Batch
• EachSample
• SampleType: SampleType
•CompoundType: CompoundType
•SampleCustomFormula:
Buttons
View Report Templates Displays the C:\TraceFinderData\4.0\Templates\ReportTemplates folder that contains
all report templates.
Opens the selected report template in the Report Designer.
Opens a blank report template in the Report Designer.
Opens the Report Designer showing the report information for the current batch in the
selected report template format.
5 Using the Analysis Mode for Target Screening Batches
Working in the Report View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 303
PDF Writes the generated report to a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
Excel Writes the generated report to a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
CSV Saves the generated report as a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
When the report contains multiple sheets, the application writes each sheet as a separate
CSV file.
Print Prints the generated report to your default printer.
Generates the selected type of reports for the current batch using the selected report
template.
Generated Reports
Template Report template used for the report. See Example of a report template list.
Rule Type of data used in each sheet of the report. See Rules.
Sample For sample-level reports, the name of each sample in the report.
Output Type of output specified for the report: PDF, Excel, CSV, or Print.
Generated Report File Lists the output file name for each report in the …\TraceFinderData\4.0\Projects folder.
View Displays the generated output file.
View Generated Reports Displays the C:\TraceFinderData\4.0\Projects folder that contains all report outputs.
Clear Removes all reports from the Generated Reports display. This does not delete the reports
from the C:\TraceFinderData\4.0\Projects folder.
Table 57. Report View parameters (Sheet 2 of 2)
Parameter Description
5 Using the Analysis Mode for Target Screening Batches
Working in the Local Method View for Target Screening Batches
304 TraceFinder User Guide Thermo Scientific
Working in the Local Method View for Target Screening Batches
A local method is a copy of a master method associated with a batch. You can edit only the
local copy of the method, or you can edit the master method and overwrite the local copy
with the edited master method.
In the Local Method view, you can edit the local method parameters. A local method is a copy
of a master method associated with a batch. Local methods are named Batch_MasterMethod.
To open the Local Method View
1. Click Analysis in the navigation pane.
2. Click Local Method.
The Local Method View for the currently selected batch opens.
You can edit many of the method parameters in a local method. Editing the local method
does not affect parameters in the master method.
For detailed descriptions of method parameters, refer to “Editing a Master Method” in
Chapter 5, “Using the Method Development Mode for Target Screening Methods,” in
the TraceFinder Lab Director User Guide.
3. Enter any local changes to the method.
4. When you have finished editing the local method, choose File > Save.
5. To process the batch or create new reports with the edited local method, return to the
Batch View and submit the batch.
To overwrite the local method with the master method in the Batch View
In the Batch View, click Update.
The application overwrites the local method with the master method of the same name.
You can use this feature to overwrite an edited local method with the original master
method or to overwrite the local method with an updated master method.
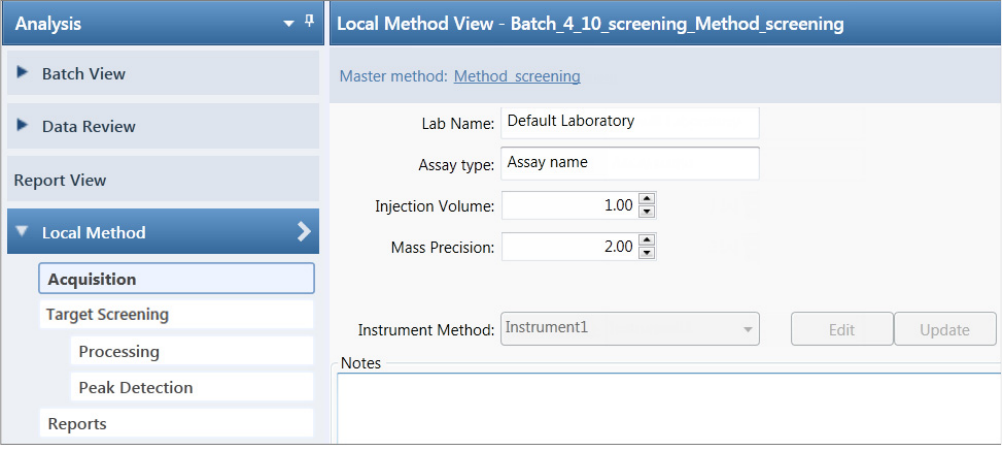
5 Using the Analysis Mode for Target Screening Batches
Working in the Local Method View for Target Screening Batches
Thermo Scientific TraceFinder User Guide 305
Figure 112. Local Method View
5 Using the Analysis Mode for Target Screening Batches
Working in the Local Method View for Target Screening Batches
306 TraceFinder User Guide Thermo Scientific
Thermo Scientific TraceFinder User Guide 307
6
Using the Analysis Mode for Unknown Screening
Batches
Use the features of the Analysis mode to do the following:
• Create unknown screening batches.
• Submit unknown screening batches for acquisition, processing, or report generation.
• Review unknown screening batches, batch data, reports, and local methods.
To access the Analysis mode
Click Analysis in the navigation pane.
The Analysis navigation pane opens.
IMPORTANT TraceFinder 4.1 uses the same data as TraceFinder 4.0. By default, the application
stores the method, project, template, and compound database data for the 4.1 release in the
TraceFinderData\4.0 folder.
Contents
•Working in the Batch View for Unknown Screening Batches
•Working in Data Review for Unknown Screening Batches
•Working in the Report View for Unknown Screening Batches
•Working in the Local Method View for Unknown Screening Batches
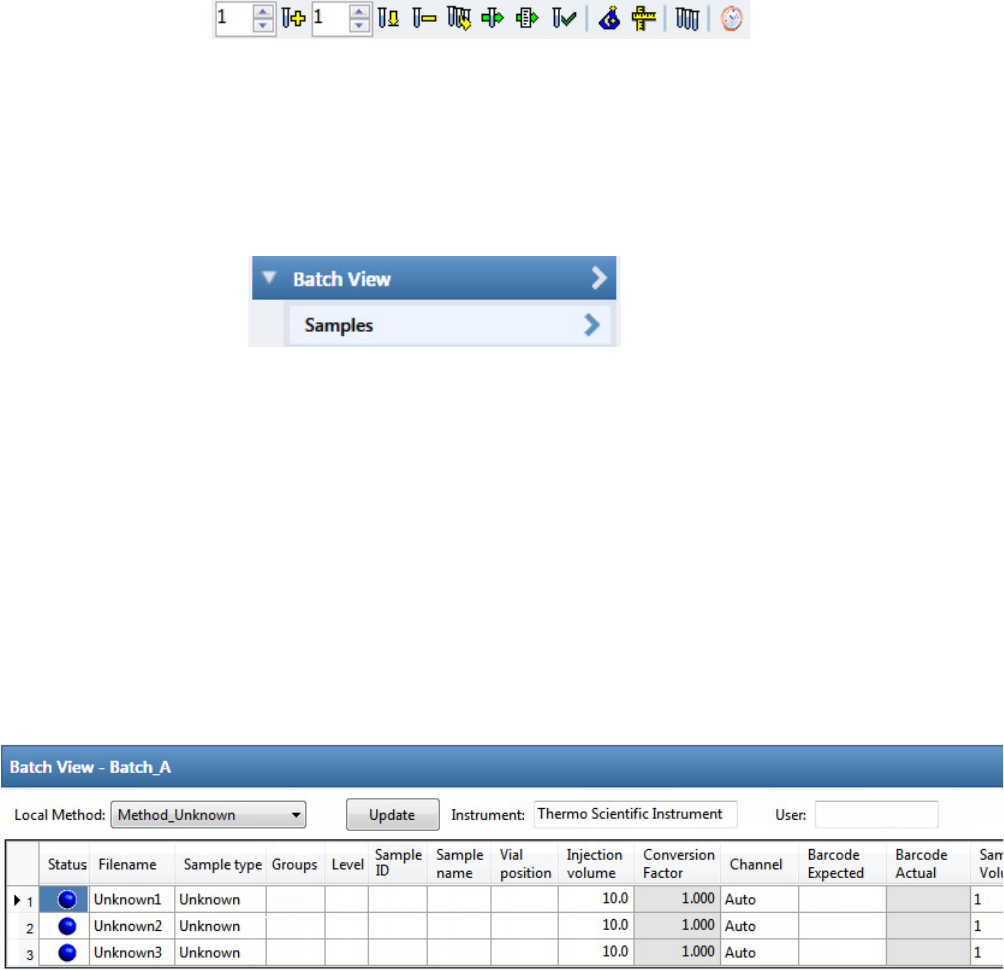
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
308 TraceFinder User Guide Thermo Scientific
Working in the Batch View for Unknown Screening Batches
In the Batch View, you can manually create and edit a new unknown screening batch or open
and edit a previously saved batch. When you submit a batch, you can acquire and process data
and optionally create reports for the submitted samples.
The Analysis mode includes a toolbar:
Use the Toolbar or the equivalent commands in the Batch View Shortcut Menu to create the
sample list and submit samples for acquisition.
To open the Samples page in the Batch View
1. Click Analysis in the navigation pane of the current mode.
2. Click the Batch View down arrow and then click Samples.
The Samples page opens.
Related Topics
•Samples Page Features
•Creating a New Batch
•Editing a Batch
•Submitting a Batch
Samples Page Features
Use the Samples page to create a batch.

6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 309
Samples Page
The Samples page includes the following features:
•Column Display
•Status Indicators
•Groups
•Sample Weight Calculation
•Instrument Methods
•Toolbar
•Batch View Sample List
•Batch View Shortcut Menu
Column Display
The sample list contains many columns of information. You can scroll to see all the columns,
and you can customize which ones to display and their display order.
To scroll the sample list
Use the horizontal scroll bar at the bottom of the sample list to view all the information.
When you use the scroll bar at the bottom of the sample list, the following columns
remain fixed: Status, Filename, Sample Type, Groups, Level, Sample ID, and Sample
Name, while the other columns scroll right and left.
To customize the column display
1. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
2. Use the arrow buttons to move all the columns that you want displayed to the Displayed
Columns pane.
These columns appear after the Status, Filename, Sample Type, Groups, Level, Sample
ID, and Sample Name columns.
3. To arrange the order of the columns, do the following:
a. In the Displayed Columns pane, select a column name.
b. Use Up or Down to move the selected column up or down in the list.
The first column in the list represents the leftmost column in the Batch View sample
list, and the last column in the list represents the rightmost column in the Batch View
sample list.
Note The following columns are fixed: Status, Filename, Sample Type, Groups,
Level, Sample ID, and Sample Name.
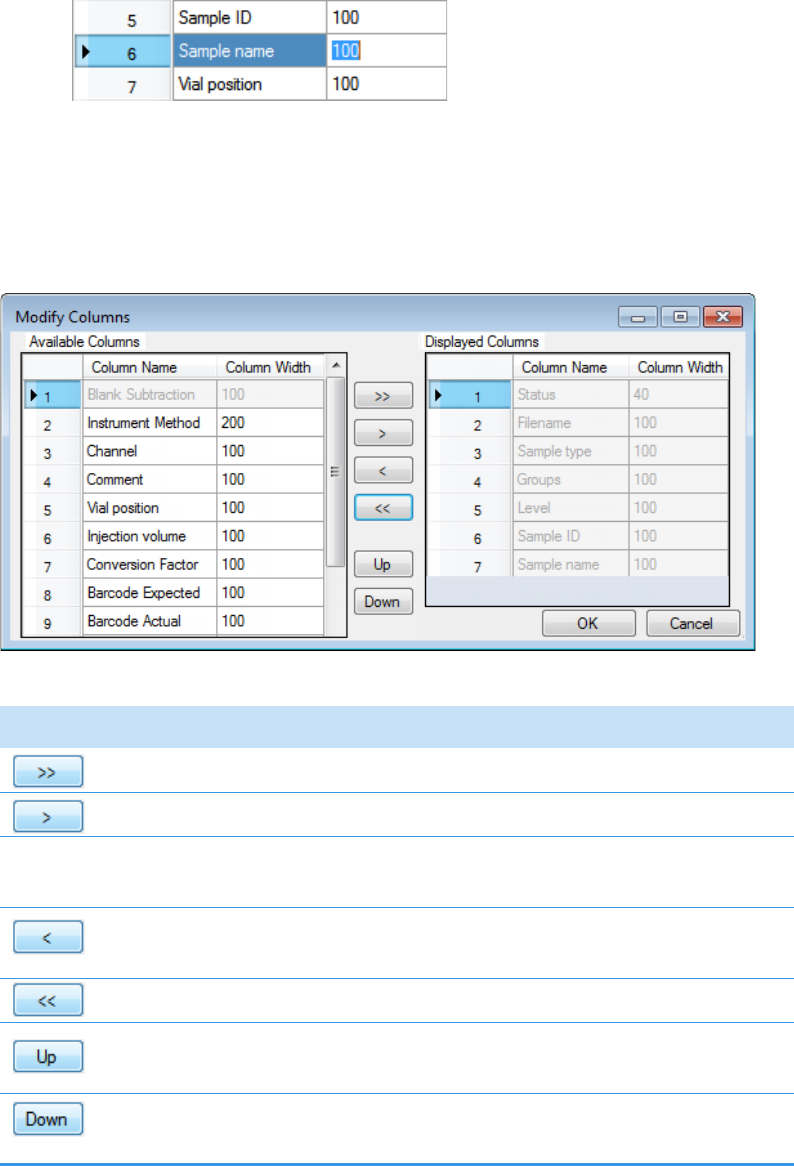
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
310 TraceFinder User Guide Thermo Scientific
4. To change the width of a column, do the following:
a. In the Displayed Columns pane, select the column width.
b. Type a new value for the width.
5. Repeat step 4 for all columns whose widths you want to change, and click OK.
The columns in the sample list immediately reflect your changes. The application uses
these settings for all sample lists in the Batch View.
Figure 113. Modify Columns dialog box
Table 58. Button descriptions for the Modify Columns dialog box
Button Description
Moves all columns to the Displayed Columns pane.
Moves the selected column to the Displayed Columns pane.
The following buttons apply to all columns, except for those that are fixed: Status, Filename,
Sample Type, Groups, Level, Sample ID, and Sample Name.
Moves the selected column to the Available Columns pane.
Moves all columns except fixed columns.
Moves the selected column name in the Displayed Columns pane one row
up in the column order.
Moves the selected column name in the Displayed Columns pane one row
down in the column order.
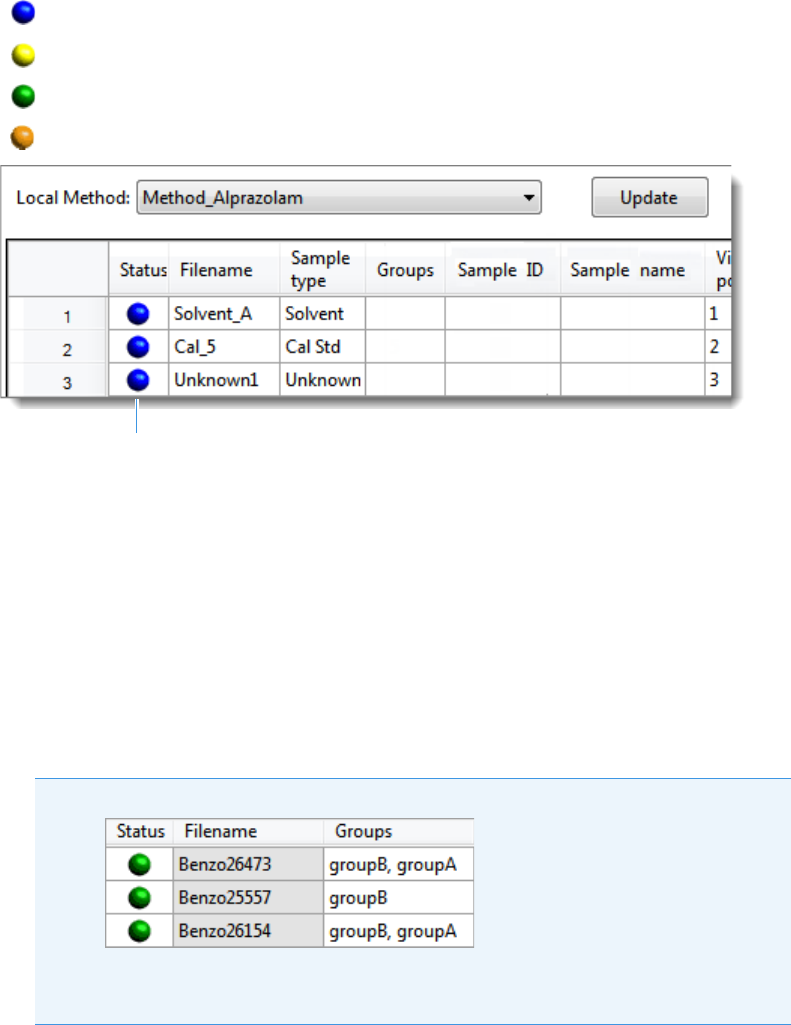
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 311
Status Indicators
Status indicators show the current status of each sample during the acquisition and
processing.
Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Groups
Use the Groups feature to assign samples to a group.
To create a group
1. For each sample, type the name of a group in the Groups column.
Repeat this for each sample that you want to include in a group.
2. Create as many groups as you want.
You can create a control group (specify the group name as Control) to use for group
averages in Data Review.
For information about group averages, see Group Averages Pane. For information about
maximum fold analysis, see Cross Sample Peak List Pane.
Status indicators
Note To assign a sample to multiple groups, separate the groups with a comma.
The application uses the first group listed for the Maximum Fold analysis and in
the Group Averages pane.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
312 TraceFinder User Guide Thermo Scientific
Sample Weight Calculation
Use the sample weight features to calculate the conversion factor for a sample. The application
uses different methods to calculate the conversion factor for liquid or solid calculation types.
Liquid: SampleVolume DilutionFactor
Solid: (SampleVolume × DilutionFactor) SampleWeight
Manual: The application does not calculate the Conversion Factor. Instead, you can enter the
Conversion Factor value.
Follow these procedures:
•To display the features for calculating sample weight
•To calculate the conversion factor for a liquid sample
•To calculate the conversion factor for a solid sample
•To manually specify the conversion factor for a sample
To display the features for calculating sample weight
If the Conversion Factor, Sample Volume, Dilution Factor, Sample Weight, Calculation
Type, and Final Units columns are not visible, right-click and choose Enable Sample
Weight Calculation.
To calculate the conversion factor for a liquid sample
1. From the Calculation Type list, select Liquid.
For a liquid sample, the Sample Weight value is not editable.
2. In the Sample Volume column, type the volume in ng/mL for your sample.
3. In the Dilution Factor column, type the value for the dilution.
For example, if you have 1000 ng/mL of a substance that is too concentrated for the mass
spectrometer, you can dilute it by 1000. Then your injection volume is 1, your conversion
factor is 1000, and your sample amount is 1000.
4. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
Note The application uses the following formula to calculate the Conversion Factor:
SampleVolume DilutionFactor
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 313
To calculate the conversion factor for a solid sample
1. From the Calculation Type list, select Solid.
2. In the Sample Weight column, type the weight in ng for your sample.
3. In the Sample Volume column, type the volume in ng/ml for your sample.
4. In the Dilution Factor column, type the value for the dilution.
For example, if you have 1000 ng/ml of a substance that is too concentrated for the mass
spectrometer, you can dilute it by 1000. Then your injection volume is 1, your conversion
factor is 1000, and your sample amount is 1000.
5. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
To manually specify the conversion factor for a sample
1. From the Calculation Type list, select Manual.
For a manually calculated sample, the only available columns are the Conversion Factor
and the Final Units.
2. In the Conversion Factor column, type the conversion factor to use for your sample.
3. In the Final Units column, type the units that you want to use for the calculated amount
in the Data Review view or in reports.
Instrument Methods
Use the Instrument Methods column to specify instrument methods for the samples.
To specify instrument methods for samples
1. Display the Instrument Method column in the sample list:
a. Right-click the sample list and choose Modify Columns.
The Modify Columns dialog box opens.
b. In the Available Columns pane, select Instrument Method.
c. Click to move the Instrument Method column to the Displayed Columns
pane.
Note The application uses the following formula to calculate the Conversion Factor:
(SampleVolume × DilutionFactor) SampleWeight
Note The application uses the specified conversion factor when it calculates the
amount for the sample.
Note By default, the Instrument Method column is not displayed in the Batch View
sample list.
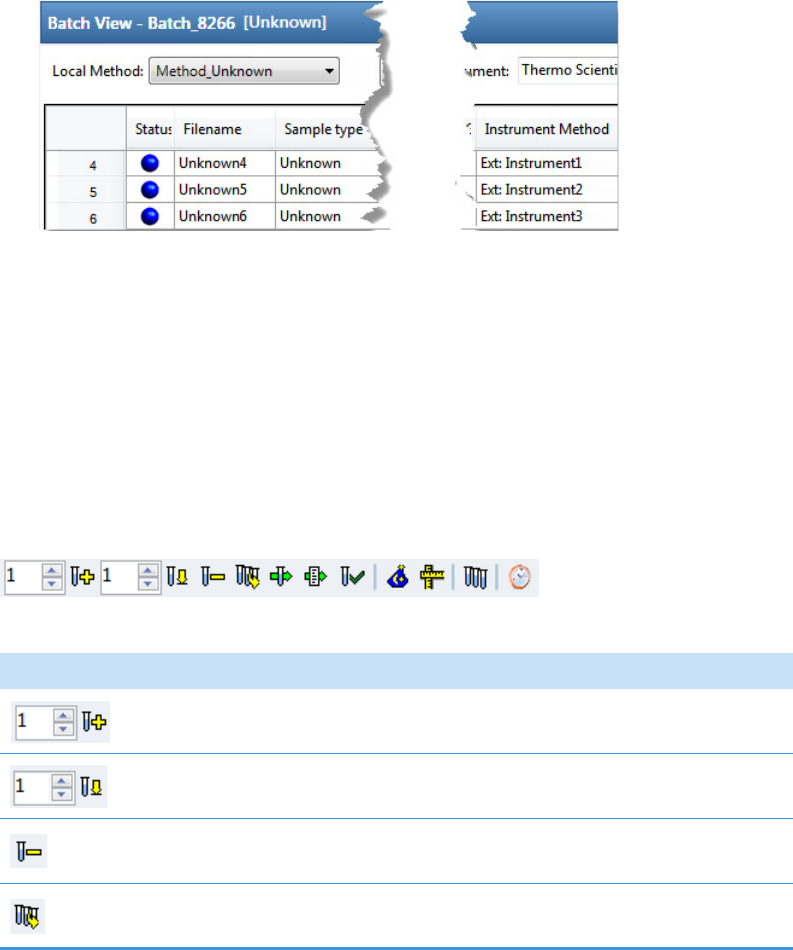
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
314 TraceFinder User Guide Thermo Scientific
d. Click OK.
The application displays the Instrument Method column, defaulting to the instrument
method specified in the master method.
2. Click the Instrument Method column and select an instrument method from the list.
This list contains all the available instrument methods. Instrument methods from
external sources are prefixed with “Ext:”.
You can specify a different instrument method for each sample.
When you submit the batch for acquisition, the application saves a copy of the selected
instrument methods to the following folders:
External instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method\ExternalMethods
Local instrument methods:
…\TraceFinderData\4.0\Projects\…\batch\Methods\method
Toolbar
The Analysis mode includes this toolbar for creating and submitting a batch.
Table 59. Toolbar icons (Sheet 1 of 2)
Icon Description
Adds the specified number of new, empty samples to the end of the
sample list. See the instructions To add samples to the list.
Inserts a new, empty sample or samples above the selected sample. See
the instructions To insert samples into the list.
Removes the selected samples from the sample list. See the
instructions To remove samples from the list.
Adds imported samples from a CSV, an XML, or an SLD file to the
sample list. See the instructions To import samples into the list.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 315
Submits only the selected samples for acquisition, processing, or
report generation. See the instructions To submit samples in the
batch.
Submits the batch for acquisition, processing, or report generation.
See the instructions To submit samples in the batch.
Submits only the selected samples for processing. See the instructions
To submit selected peaks for processing.
Opens the Acquisition mode where you can use a batch template to
define a standard sequence composed of various sample types to be
assembled into a batch of samples. See Working in Data Review for
Unknown Screening Batches.
Opens the Acquisition mode where you can create a batch template
that contains the basic settings and sample types for your batches. See
Using the Acquisition Mode.
Opens the Quick Acquisition window where you can quickly submit a
single sample. See Appendix A, “Using Quick Acquisition.”
Opens the Audit Viewer where you can view audit logs. See
Chapter 8, “Using the Audit Viewer.” Available only when you enable
Auditing in the Administrator Console. Refer to the instructions in
the TraceFinder Administrator Console User Guide.
Table 59. Toolbar icons (Sheet 2 of 2)
Icon Description
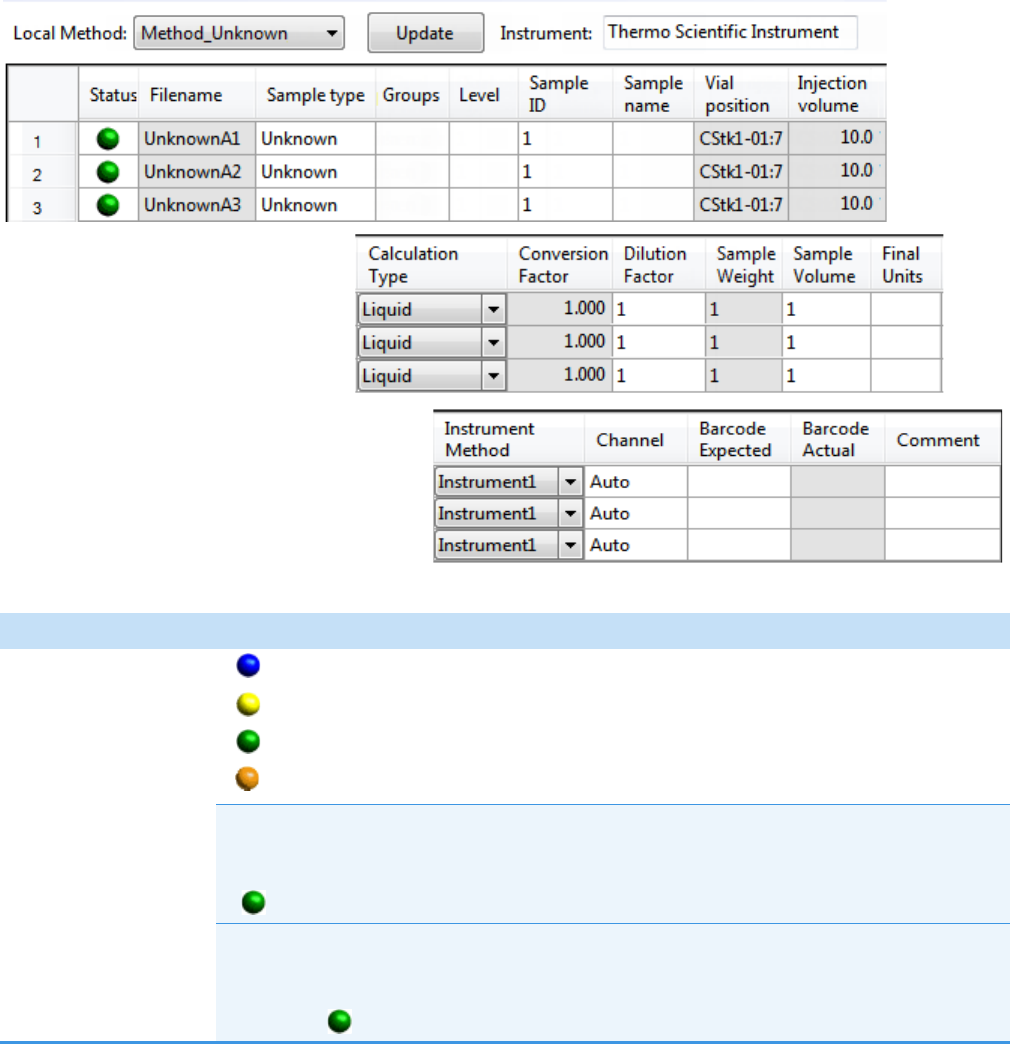
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
316 TraceFinder User Guide Thermo Scientific
Batch View Sample List
The sample list displays all the quantitative data for the samples of a batch. Status indicators
for each sample indicate if the sample is currently acquiring, not acquired, acquired, or
processed.
The sample list includes the following columns of information:
Figure 114. Batch View sample list
Table 60. Batch View sample list columns (Sheet 1 of 3)
Column Description
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
Note When you include unknown screening features in a quantitation method and you
choose to process with only the unknown screening criteria, both the Sample View and
the Unknown Screening View show the Status for the samples as acquired and processed
().
Note When you include unknown screening features in a target screening method and
you choose to process with only the unknown screening criteria, both the Samples view
and the Unknown Screening View show the Status for the samples as acquired and
processed ( ).
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 317
Filename Name of the raw data file that contains the sample data.
Sample Type Defines how the application processes the sample data. Each sample is classified as one of
the following sample types: Matrix Blank, Solvent, QC Std, or Unknown.
Default: Unknown
Note Unknown Screening does not use calibration sample types (Cal Std).
Groups Threshold group to which a sample belongs.
Level Not used in Unknown Screening processing.
Sample ID A user-defined, alphanumeric string that identifies a sample.
Sample Name A user-defined name that identifies a sample.
Vial Position The tray vial number used for an autosampler acquisition.
Injection Volume The injection volume (in microliters) of the injected sample.
When you are using an autosampler, you can set the default injection volume in the
Autosampler dialog box in the Instrument View. The minimum and maximum injection
volumes that you can use depend on the Autosampler you configure. The usable range
depends on the injection mode and might be smaller than the displayed range.
The Injection Volume value set in the master method overwrites the value in the instrument
method.
Valid range: 0.1 through 5000 μL
Calculation Type Liquid: The application calculates the Conversion Factor as
SampleVolume DilutionFactor
Solid: The application calculates the Conversion Factor as
(SampleVolume × DilutionFactor) SampleWeight
Manual: Sample Volume, Dilution Factor, Sample Weight, and Final Units columns are not
available, and the Conversion Factor value is editable.
Conversion Factor Editable only when Calculation Type is Manual.
Default: 1
Sample Volume Default: 1
Dilution Factor Default: 1
Sample Weight Available only when Calculation Type is Solid.
Default: 1
Table 60. Batch View sample list columns (Sheet 2 of 3)
Column Description

6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
318 TraceFinder User Guide Thermo Scientific
Batch View Shortcut Menu
The Batch View includes a shortcut menu for creating a batch.
Final Units Specifies the calculated amount in the Data Review view or in reports.
Default: 1
Instrument Method Specifies the instrument to use for the acquisition. This column is hidden by default. To
display this column, see To customize the column display.
Channel Specifies the channel on which the sample was run. If the sample is not acquired, the value is
Pending. The Channel column is available only when you have activated multiplexing in the
Configuration console. Refer to “Multiplexing” in Chapter 1, “Using the Configuration
Console,” in the TraceFinder Lab Director User Guide.
Barcode Expected A user-entered barcode for the vial.
Barcode Actual An actual barcode for the vial. This value is not editable.
Comment A user-defined comment for the sample.
Table 60. Batch View sample list columns (Sheet 3 of 3)
Column Description
Table 61. Batch View shortcut menu commands (Sheet 1 of 2)
Command Description
Add Sample Adds a single empty row to the sample grid.
Insert Sample Inserts a single empty row to the sample grid above the selected row.
Insert Copy Sample Copies the currently selected row and inserts a copy above the row.
Reinject Selected Samples Creates a copy of the selected sample and appends INJ001 to the file name.
Additional reinjections of the same sample are numbered INJ002, INJ003, and so
forth.
Remove Selected Samples Removes selected samples from the sample grid.
Import Samples Opens the Sample Import Tool. See To import samples into the list.
Browse in Raw File (Move) Opens a dialog box where you can select a raw data file to use for the selected sample
row. The application removes the raw data file from the source location.
Browse in Raw File (Copy) Opens a dialog box where you can select a raw data file to use for the selected sample
row. The application copies the raw data file from the source location.
Map Raw Files to Samples Opens a dialog box where you can select multiple raw data files to use for the selected
sample rows.
Copy Down Copies the value in the selected row to all rows below it. This command is available
only when you have selected a value that can be copied down.
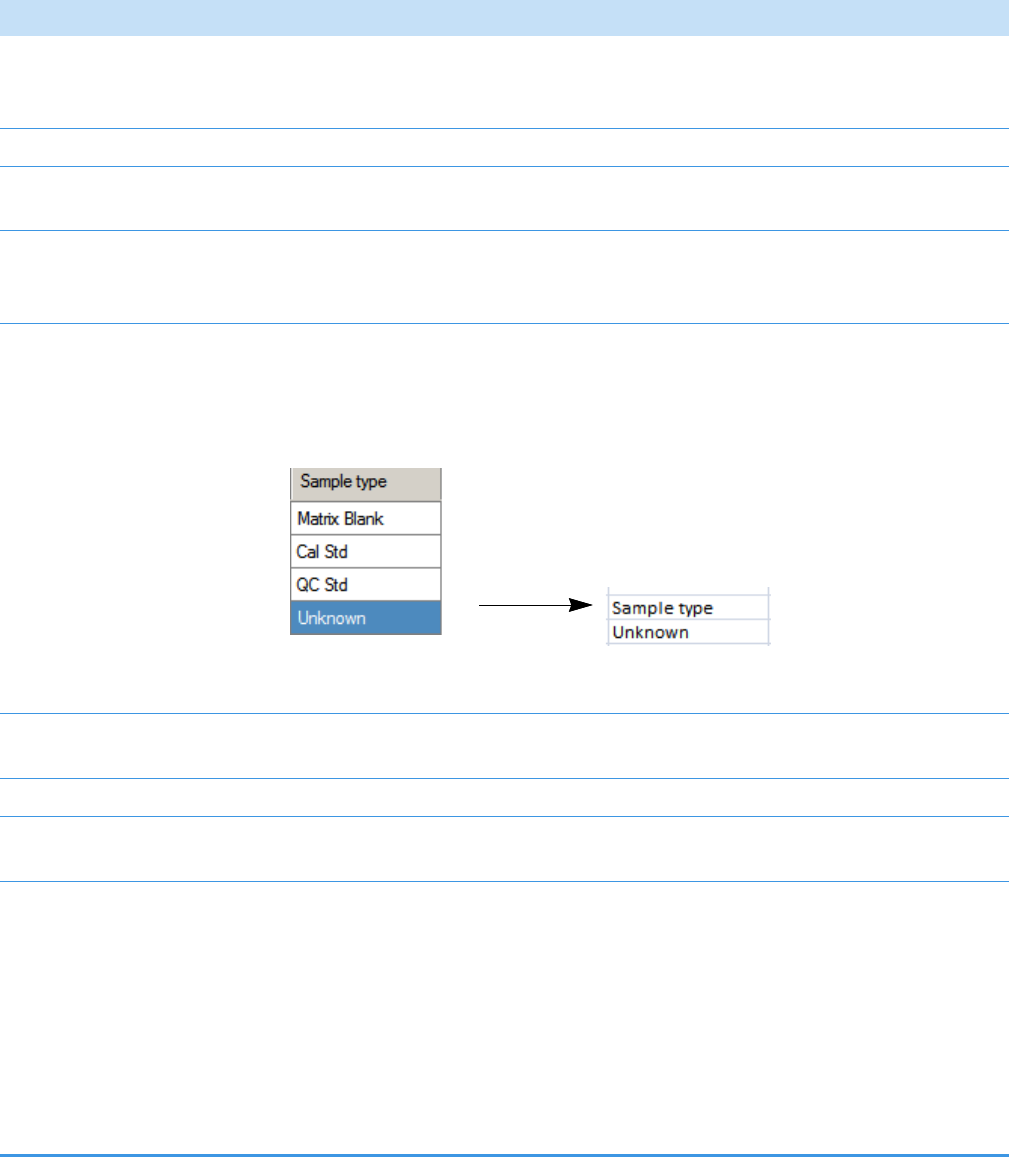
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 319
Fill Down Enters sequential values in the column starting with the value in the selected row and
ending with the last row in the column. This command is available only when you
have selected a value that can be filled down.
Modify Columns Opens the Modify Columns dialog box.
Enable/Disable Sample
Weight Calculation
Displays or hides the Sample Volume, Dilution Factor, Sample Weight, Calculation
Type, and Final Units columns.
Copy Copies the data in the selected rows or columns to the Clipboard. Use this command
to copy sample information into a text editor or spreadsheet application. You cannot
paste this data back into the Batch View sample list.
Copy with Headers Copies the data in the selected rows or columns and the associated column headers to
the Clipboard. Use this command to copy sample information into another text
editor or spreadsheet application. You cannot paste this data back into the sample list.
For example
Paste Pastes a single column of copied data from another text editor or spreadsheet
application into the selected column.
Undo Last Paste Removes the last pasted item in the Batch View.
Export to CSV File Opens the Save As dialog box where you can save the current sample list to a CSV
file.
Edit Instrument Method Opens the Instrument Setup window where you can edit the parameters of the
instrument method.
• When you edit an external method, the application updates the method in the
…\Xcalibur\methods folder.
• When you edit an internal method, the application updates the method in the
…\TraceFinderData\4.0\Projects\…\batch\Methods\method folder.
For detailed information about editing instrument methods, refer to Chapter 3,
“Using Instrument Methods in the Method Development Mode,” in the Trace Fin de r
Lab Director User Guide.
Table 61. Batch View shortcut menu commands (Sheet 2 of 2)
Command Description
Copy with Headers
from TraceFinder Paste into an Excel spreadsheet.
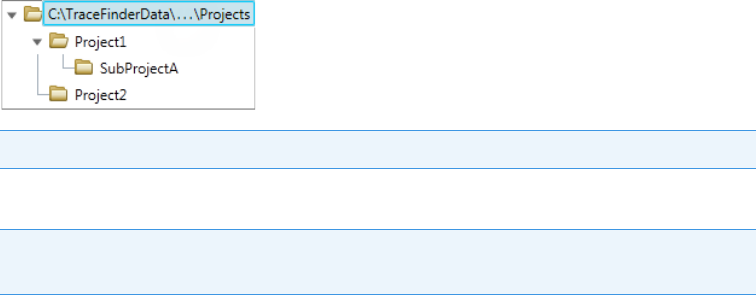
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
320 TraceFinder User Guide Thermo Scientific
Creating a New Batch
In the Batch View, you can create a new batch.
Follow these procedures:
•To c re a t e a ne w b a t c h
•To add samples to the list
•To insert samples into the list
•To import samples into the list
•To remove samples from the list
•To c o py a sa m pl e
•To reinject a sample
•To edit sample values
•To browse in raw data files
•To customize the column display
To create a new batch
1. Choose File > New > Batch from the main menu.
The Create New Batch Dialog Box opens, displaying all drives that contain projects.
2. Select a drive from the list.
3. Select the folder where you want to store your batch.
To create a new folder for the storage location, see Editing Folders for Batches.
4. Select Unknown Only from the Type list.
The batch list displays all batches in the selected folder. The Method list displays all
methods for the unknown screening type.
5. Select a master method from the Master Method list.
Tip The application displays all configured and enabled repositories.
Tip To activate the Create button, you must enter a unique batch name. If the Create
button is not activated, you have entered a batch name that is already used.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 321
6. Click Create.
A new batch opens with one Unknown sample.
The batch name in the title bar indicates that you are creating an unknown screening
batch.
To add samples to the list
1. To add a single sample row, right-click the sample list and choose Add Sample.
2. To add multiple sample rows, select the number of rows and then click the Add Sample
icon, .
The application adds the specified number of new, empty samples to the end of the
sample list.
To insert samples into the list
Select the sample above which you will insert new, Unknown samples, and then do one of
the following:
• To insert a single sample row, right-click and choose Insert Sample.
• To insert multiple sample rows, select the number of rows and then click the Insert
Sample icon .
The application inserts the Unknown samples above the selected sample.
Inserted samples
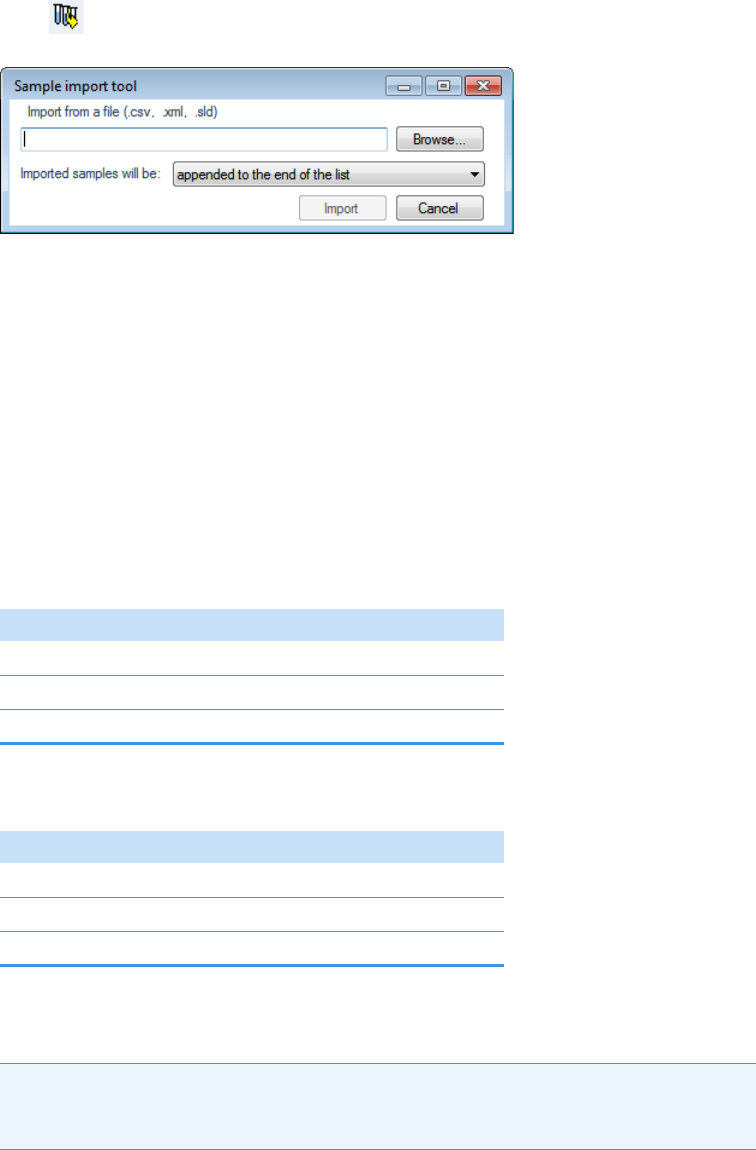
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
322 TraceFinder User Guide Thermo Scientific
To import samples into the list
1. Choose Batch > Import Samples from the main menu, or click the Import Samples
icon, .
The Sample Import Tool dialog box opens.
From this dialog box, you can import samples from a CSV, an XML, or an SLD file.
2. Click Browse and select a CSV, an XML, or an SLD file that contains the samples to
import.
3. From the Imported Samples Will Be list, select Appended to the End of the List or
Inserted at the Selected Row.
4. Click Import.
The Sample Import Tool dialog box closes, and the application adds the specified samples
to the sample list.
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following column name substitutions:
When you import samples from an Xcalibur sequence file (.sld), the application makes
the following sample type substitutions:
5. (Optional) When using multiplexing, select a channel for each imported sample.
Imported samples default to Auto.
Xcalibur column TraceFinder column
Position Vial Position
Inj Vol Injection Volume
Dil Factor Conversion Factor
Xcalibur sample type TraceFinder sample type
Blank Matrix Blank
QC QC Std
Std Bracket Cal Std
Note The Channel column is available only when you have activated multiplexing in
the Configuration console. Refer to “Multiplexing” in Chapter 1, “Using the
Configuration Console,” in the TraceFinder Lab Director User Guide.

6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 323
To remove samples from the list
1. Select the samples that you want to remove.
2. Right-click and choose Remove Selected Samples.
To copy a sample
1. Select the sample that you want to copy.
2. Right-click and choose Insert Copy Sample.
The application inserts the copy above the selected sample.
To reinject a sample
1. In the sample list, select the sample that you want to reinject.
2. Right-click and choose Reinject This Sample.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the sample are numbered INJ002, INJ003, and so forth.
The application copies all parameter values from the original sample.
To edit sample values
1. For each sample, do one of the following:
Type a new file name over the current filename.
–or–
Double-click the Filename column and locate a raw data file to use for the sample.
–or–
Right-click and choose Browse in Raw File, and then locate a raw data file to use for the
sample.
By default, the application sets the Sample Type to Unknown.
2. For each sample, click the Sample Type column and select a sample type from the list.
3. Type a vial position in the Vial Position column for each sample.
4. Type a volume in the Injection Volume column for each sample.
The minimum injection volume value allowed is 0.1 μL; the maximum injection volume
value allowed is 5000 μL.
Tip Use the CTRL or SHIFT keys to select multiple samples.
Available sample types
Matrix Blank Solvent QC Std Unknown
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
324 TraceFinder User Guide Thermo Scientific
5. (Optional) Type or edit the values for the remaining columns.
To automatically copy or fill values in these columns, see Appendix B, “Using Copy
Down and Fill Down.”
To browse in raw data files
1. Do one of the following:
Double-click the Filename column.
–or–
Right-click and choose Browse in Raw File.
The What Raw File Would You Like to Use dialog box opens.
2. Select a raw data file to use for the sample or use the CTRL key to select multiple files,
and then click Open.
The application overwrites the selected, unacquired sample in the batch with the first
“browsed in” file and adds any additional browsed in files below the selected sample.
For all browsed-in raw data files, the application sets the Status to Acquired, , and sets
the Sample Type to Unknown.
To customize the column display
1. Right-click the Batch View sample list and choose Modify Columns.
The Modify Columns dialog box opens.
2. Use the arrow buttons to move all the columns that you want displayed to the Displayed
Columns pane.
All the columns you select appear after the Status, Filename, Sample Type, Groups, Level,
Sample ID, and Sample Name columns.
3. To arrange the order of the columns, do the following:
a. In the Displayed Columns pane, select a column name.
b. Use Up or Down to move the selected column up or down in the list.
The first column in the list represents the leftmost column in the Batch View sample
list, and the last column in the list represents the rightmost column in the Batch View
sample list.
Note When you use the scroll bar at the bottom of the sample list, the following
columns remain fixed: Status, Filename, Sample Type, Groups, Level, Sample ID, and
Sample Name, while the other columns scroll right and left.
Note You cannot overwrite an acquired sample. When you select a sample that is
acquired, the application adds all browsed in files below the selected sample.
Note The following columns are fixed: Status, Filename, Sample Type, Groups,
Level, Sample ID, and Sample Name.
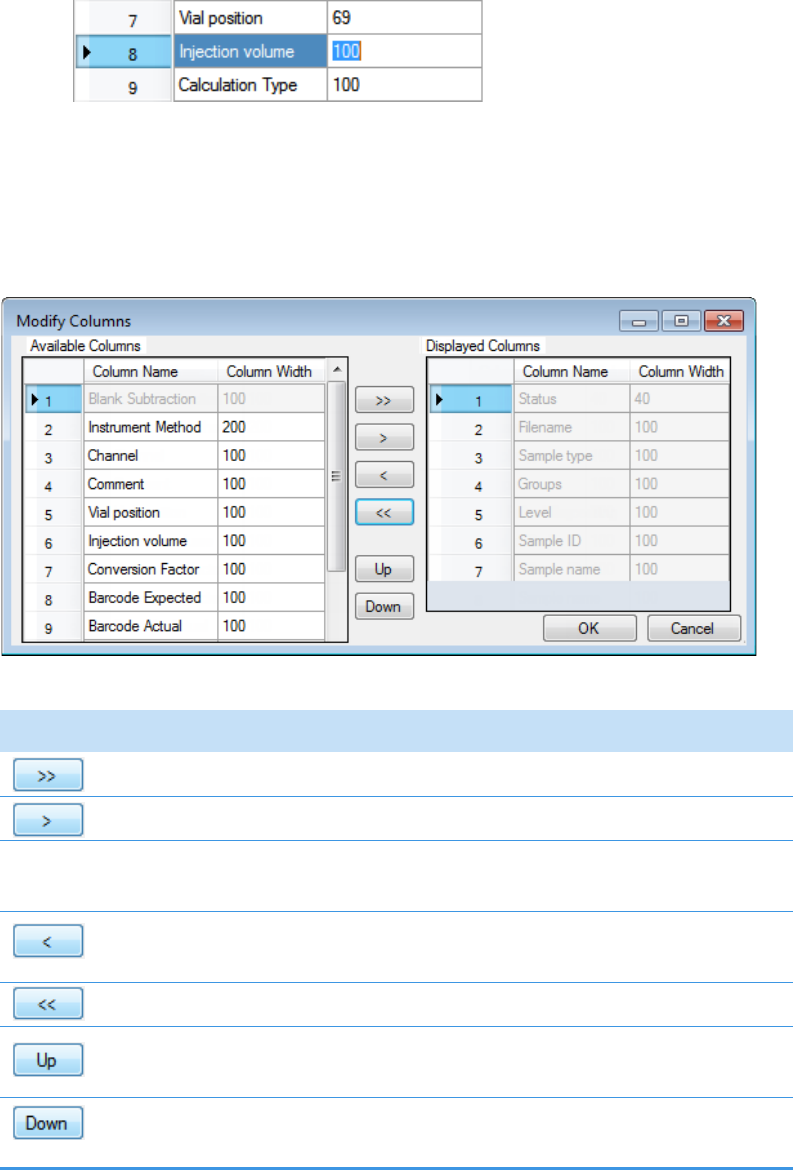
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 325
4. To change the width of a column, do the following:
a. In the Displayed Columns pane, select the column width.
b. Type a new value for the width.
5. When you have completed your changes, click OK.
The columns in the sample list immediately reflect your changes. The application uses
these settings for all sample lists in the Batch View.
Figure 115. Modify Columns dialog box
Table 62. Button descriptions for the Modify Columns dialog box
Button Description
Moves all columns to the Displayed Columns pane.
Moves the selected column to the Displayed Columns pane.
The following buttons apply to all columns, except for those that are fixed: Status, Filename,
Sample Type, Groups, Level, Sample ID, and Sample Name.
Moves the selected column to the Available Columns pane.
Moves all columns except those that are fixed.
Moves the selected column name in the Displayed Columns pane one row up in
the column order.
Moves the selected column name in the Displayed Columns pane one row down
in the column order.
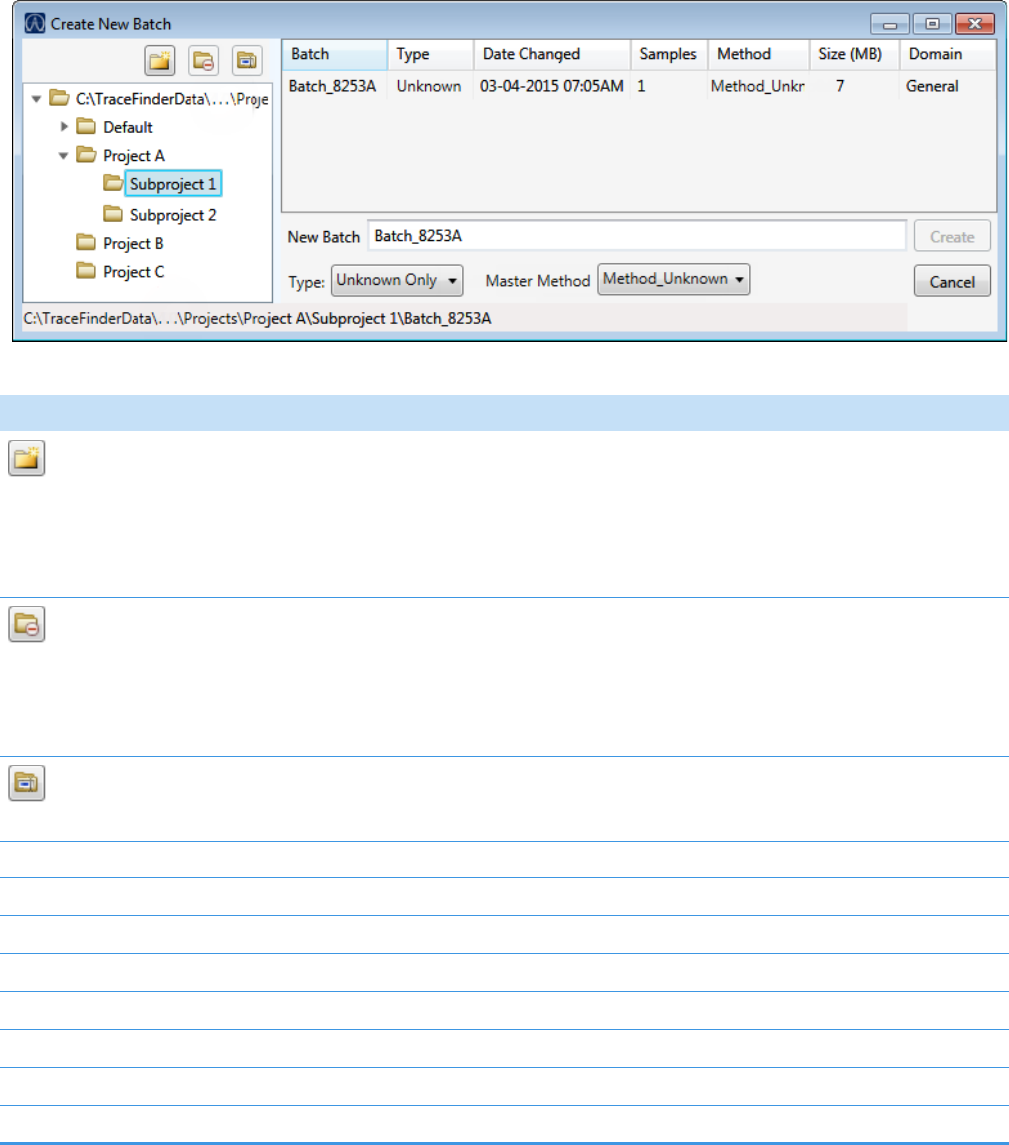
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
326 TraceFinder User Guide Thermo Scientific
Create New Batch Dialog Box
Use the Create New Batch dialog box to select a folder and method for your batch and to
name the new batch.
Table 63. Create New Batch dialog box parameters (Sheet 1 of 2)
Parameter Description
Create New Folder
Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Or, you can right-click and choose Create Folder.
Delete Folder
With no confirmation prompt, immediately removes the selected folder.
You cannot delete a folder that contains lower-level folders; you must delete the lower-level
folders first.
Or, you can right-click and choose Delete.
Rename Folder
Renames the selected folder.
Or, you can right-click and choose Rename.
Batch table
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date that the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 327
Editing Folders for Batches
From the Create New Batch dialog box, you can create new folders for your batches. You can
also delete or rename folders.
Use these procedures:
•To create new project folders
•To delete project folders
•To rename project folders
To create new project folders
1. In the Create New Batch dialog box, select the folder for which you will create a new
lower-level folder.
• You can select the main TraceFinderData\4.0\Projects folder and create a new folder
under it.
• You can select one of the existing folders and create a lower-level folder under it.
2. Click the Create Folder icon, .
The application adds a new lower-level folder to the selected folder.
3. Select the new folder name and type a name for the folder.
Folder names are limited to 30 characters and can contain spaces and special characters,
except for the following special characters: \ / : + ? ” < >
New batch parameters
New Batch Name of the new batch to create.
Note If the Create button is not activated, you have entered a name that is already used or
you have not selected a method.
Type Type of batch to create: Quan, Screening, or Unknown Only.
Method Method used to create the new batch.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is created.
Buttons
Create Creates the specified batch and opens the Batch View for the new batch.
Cancel Closes the Create New Batch dialog box without creating a batch.
Table 63. Create New Batch dialog box parameters (Sheet 2 of 2)
Parameter Description
Note After you add a lower-level folder, you cannot rename the parent folder.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
328 TraceFinder User Guide Thermo Scientific
To delete project folders
1. In the Create New Batch dialog box, select the folder to delete.
2. Click the Delete Folder icon, .
With no confirmation prompt, the application immediately removes the selected folder.
To rename project folders
1. In the Create New Batch dialog box, select the folder to rename.
2. Click the Rename Folder icon, .
3. Type a new name for the folder and press ENTER.
The application saves the new folder name.
Editing a Batch
In the Batch View, you can open a saved batch and edit the sample list. You can add samples,
edit samples, or remove samples. If the batch has already been acquired, you can select specific
samples for reinjection. If the batch has unacquired samples when you complete your edits,
you can save it as a “ready to acquire” batch.
Follow these procedures:
•To o p e n a sa v e d b at c h
•To open a recent batch
•To edit samples in a batch
•To reinject a sample from a previously acquired batch
To open a saved batch
1. Choose File > Open > Batch from the main menu.
The Open Batch dialog box opens.
2. Select a project and a subproject.
3. Select Unknown Only or Any from the Type list.
The batch list displays all batches created with Unknown methods (or all methods of all
types when you select Any).
4. Select a batch from the list.
5. Click Open.
The selected batch opens in the Batch View.
Note You cannot delete folders that contains lower-level folders; you must delete the
lower-level folders first.
Note You cannot rename folders that contain lower-level folders.
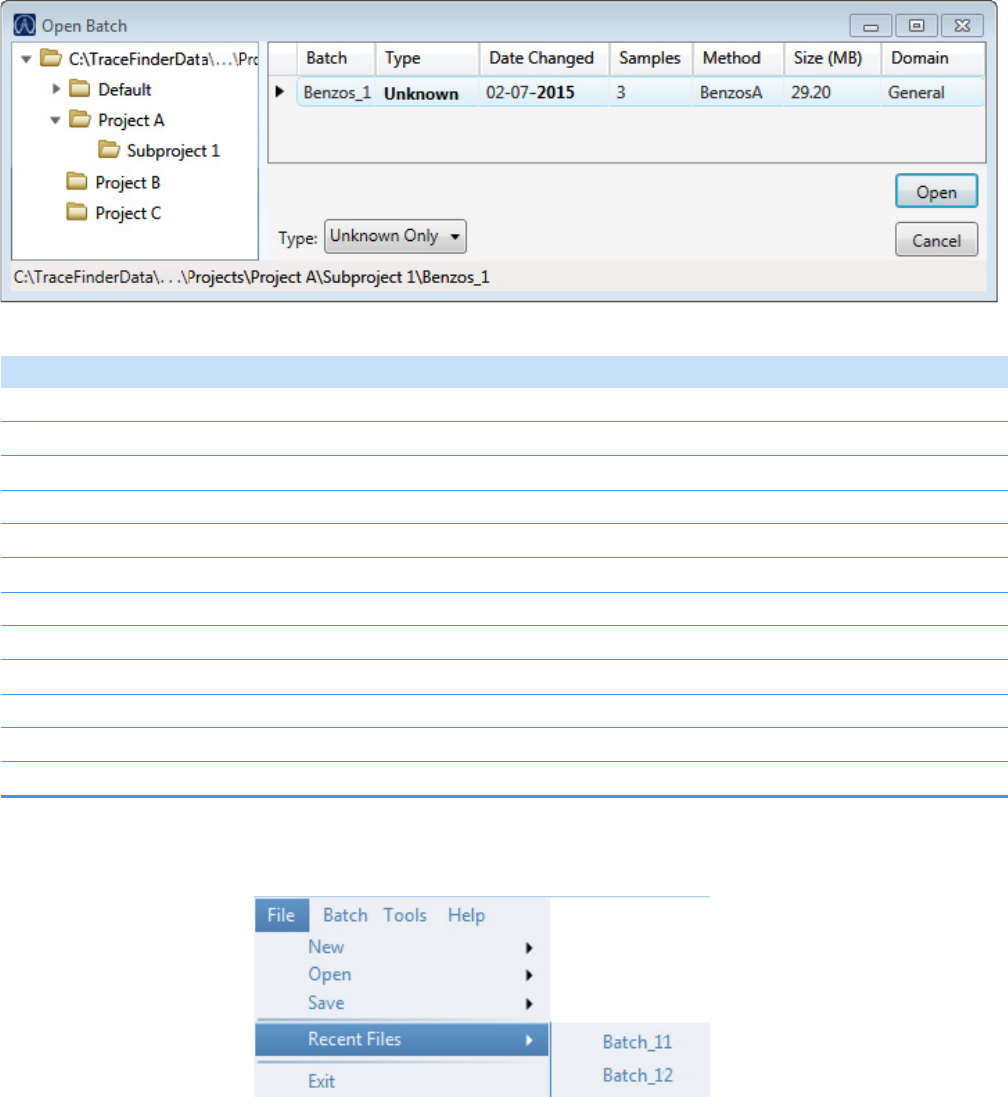
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 329
Figure 116. Open Batch dialog box
To open a recent batch
Choose File > Recent Files > batch from the main menu.
The selected batch opens in the Batch View.
Table 64. Open Batch dialog box parameters
Parameter Description
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is stored.
Buttons
Type Type of batch to display in the Batch list: Quan, Screening, Unknown Only, or Any.
Open Opens the Batch View for the selected batch.
Cancel Closes the Open Batch dialog box without opening a batch.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
330 TraceFinder User Guide Thermo Scientific
To edit samples in a batch
Use the commands described in Working in the Batch View for Unknown Screening
Batches.
You can add new samples, edit samples, or delete samples.
To reinject a sample from a previously acquired batch
1. In the sample list, select the sample that you want to reinject.
2. Right-click and choose Reinject This Sample.
The application creates a copy of the selected sample and appends INJ001 to the file
name. Additional reinjections of the sample are numbered INJ002, INJ003, and so forth.
The application copies all parameter values from the original sample.
A green status icon indicates previously acquired samples (acquired and processed), and
the sample name is grayed out. A blue status icon indicates samples created for reinjection
(not acquired).
When you submit all samples in this batch, the application acquires all samples (including
previously acquired samples).
3. To save this batch with the new samples for reinjection, choose File > Save > Batch from
the main menu.
The batch is saved as a prepared batch that is ready to submit. You can open this batch
from the Reinject Samples page in the Acquisition mode and submit the batch. The
application acquires only the samples that have not been previously acquired.
Submitting a Batch
In the Batch View, you can submit an entire batch or only selected samples in the batch.
When you submit a batch for acquisition and processing, you can choose to create reports for
the submitted samples. See Submit Options dialog box.
For a description of commands in the shortcut menu, see Batch View shortcut menu
commands.
Follow these procedures:
•To submit selected peaks for processing
•To submit samples in the batch
•To view the output files
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 331
To submit selected peaks for processing
1. In the Peak List, select the Selected check box for the peaks that you want to process.
2. Click the Submit … for Processing icon, .
The application processes the selected peaks and updates the data in the Peak
Identifications pane.
To submit samples in the batch
1. Do one of the following:
• To submit all samples in the batch, click the Submit Batch icon, .
• To submit specific samples, select the samples and click the
Submit Selected Samples icon, .
The Submit Options dialog box opens.
2. To acquire (or reacquire) the submitted samples, select the Acquire Data check box.
• When all submitted samples have been previously acquired, this option is (by default)
not selected.
• When one or more samples in the batch have not been acquired, this option is (by
default) selected.
3. To process the submitted samples, select the Process Data check box.
The application displays processing options for the Unknown Screening method.
4. Select the check box for the unknown screening options that you want to use.
Peak Detect: Performs peak detection. You can process the data with or without
performing peak detection. For example, you might want to turn off peak detection when
reprocessing samples.
Identify: Performs identification. This produces the peak identification data in the
Unknown Screening View.
With RT Alignment: Performs retention time alignment. This produces the heat map
and group averages data in the Unknown Screening View. When you select this option,
the application automatically selects the Peak Detect option.
5. (Optional) Select the Create Reports check box.
Note You can also submit only selected peaks for processing. See To submit selected
peaks for processing.
Tip You can process the data with or without performing peak detection. For
example, you might want to turn off peak detection when reprocessing samples.

6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
332 TraceFinder User Guide Thermo Scientific
6. (Optional with multiplexing activated) Select the Priority Sequence check box.
The application acquires the priority batch on the next available channel or the assigned
channel.
7. (Optional without multiplexing activated) Select the Priority Sequence check box and
then select one of the following priority options to place the batch in the queue:
•Next Available Batch places the batch immediately after the currently acquiring
batch.
•Next Available Sample places the batch immediately after the currently acquiring
sample.
8. To specify the following optional parameters, click Show Details.
a. Select the Use check box for the device that you want to use for this acquisition.
b. Select the Start Device check box to indicate the device that will initiate
communication with the other instruments.
This is usually the autosampler.
c. Select the Start When Ready check box, which starts all instruments together when
they are all ready.
When this is cleared, individual instruments can start at different times and then have
to wait for the last instrument to be ready.
d. Select the system state after it acquires the last batch: On, Standby, or Off.
9. To start the selected processes, click OK.
The selected processes begin, and the application shows the real-time display at the
bottom of the current window. You can begin another batch in the Analysis mode while
you watch the real-time display of the currently acquiring batch.
Note When you select Full Sequence Submission in the Configuration console, these
options are unavailable because the current batch and the current sample are, in effect,
the same thing.
IMPORTANT After the first processing, you might see an error message stating that the
total number of results is too large for a single batch (more than 500 000). Return to the
Processing pages in the method (refer to Chapter 6, “Using the Method Development
Mode for Unknown Screening Methods,” in the TraceFinder Lab Director User Guide) and
make one or more of these parameter adjustments:
• Limit the RT range.
• Shorten the signal range.
• Specify a lower value for the Number of Top Matches.
• Specify a Simple Search instead of an Exhaustive Search.
• Specify Top Peaks instead of All Peaks.
• Limit the number of search types.
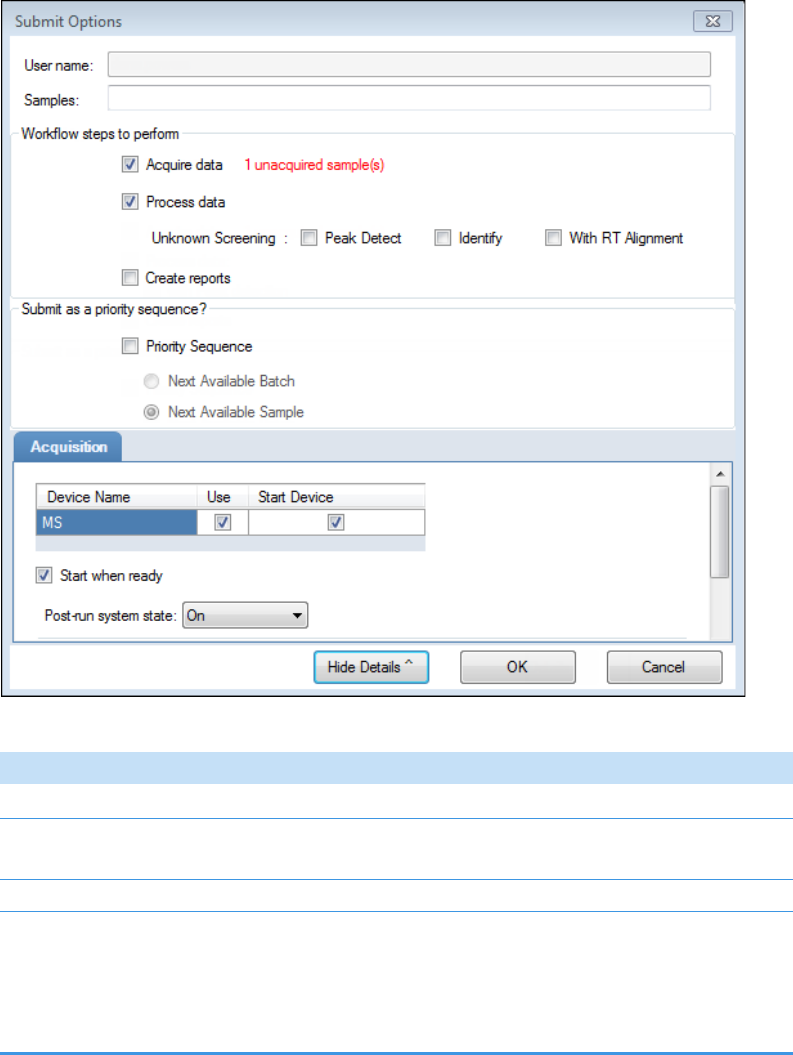
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 333
Figure 117. Submit Options dialog box
Table 65. Submit Options dialog box parameters (Sheet 1 of 3)
Parameter Description
User Name Name of the current user.
Samples Number of samples to be submitted for acquisition, processing, or
reporting.
Workflow Steps to Perform
Acquire Data Submits the current batch to acquisition.
• When all submitted samples have been previously acquired, this
option is (by default) not selected.
• When one or more samples in the batch have not been
acquired, this option is (by default) selected.

6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
334 TraceFinder User Guide Thermo Scientific
Process Data Processes the data for the current batch using any of the following
options:
Peak Detect: Performs peak detection for all method types. You can
process the data with or without performing peak detection. For
example, you might want to turn off peak detection when
reprocessing samples.
Identify: Performs identification for unknown screening methods.
With RT Alignment: Performs retention time alignment for
unknown screening methods. This produces the heat map and
group averages data in the Unknown Screening View. When you
select this option, the application automatically selects the Peak
Detect option.
Note You can use Unknown Screening features in conjunction
with Quantitation or Target Screening methods.
For specifying processing data for batches that also use
Quantitation methods, see To submit samples in the batch.
For specifying processing data for batches that also use Target
Screening methods, see To submit samples in the batch.
Create Reports Creates reports for the current batch.
Submit as a Priority Sequence?
Priority Sequence With multiplexing activated, places the batch immediately after the
currently acquiring batch.
Without multiplexing activated, specifies one of the following
priority options to place the batch in the queue:
Next Available Batch: Places the batch immediately after the
currently acquiring batch.
Next Available Sample: Places the batch immediately after the
currently acquiring sample.
Note When you select Full Sequence Submission in the
Configuration console, these options are unavailable because the
current batch and the current sample are, in effect, the same
thing.
Table 65. Submit Options dialog box parameters (Sheet 2 of 3)
Parameter Description
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 335
To view the output files
Locate the files to view from the following directories:
The application writes saved batches to the project folder:
…\TraceFinderData\4.0\Projects\…
For each acquired sample, the application writes an RSX file to the batch Data folder:
…\TraceFinderData\4.0\Projects\…\Data
The application saves method information to the batch Methods folder:
…\TraceFinderData\4.0\Projects\…\Methods
The application writes the reports to the batch Reports folder:
…\TraceFinderData\4.0\Projects\…\batch\Reports
Acquisition pane
Device Name Lists all configured instruments.
If the instrument that you want to use is not configured, close the
TraceFinder application, configure the instrument, and then reopen
the application. You cannot configure an instrument while the
TraceFinder application is running.
Available only when you select the Acquire Data check box.
Use Specifies the instruments used for this acquisition.
Available only when you select the Acquire Data check box.
Start Device Specifies the instrument that initiates the communication with the
other instruments. This is usually the autosampler.
Available only when you select the Acquire Data check box.
Start When Ready Starts the specified device when all the instruments are ready to
acquire data. When this is cleared, individual instruments can start
at different times and then must wait for the last instrument to be
ready.
Post-run System State Specifies the system state after it acquires the last batch: On
(default), Standby, or Off.
Buttons
Hide/Show Details Collapses or expands the acquisition details of the Submit Options
dialog box.
OK Begins the selected processes.
Cancel Closes the Submit Options dialog box without submitting any
tasks.
Table 65. Submit Options dialog box parameters (Sheet 3 of 3)
Parameter Description
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
336 TraceFinder User Guide Thermo Scientific
Saving a Batch to a New Location
You can move the current batch to a different project folder, or you can make a copy of the
current batch and save the copy to a different project folder.
Follow these procedures:
•To save a batch to another project folder
•To move a batch to another folder
•To create new project folder
•To delete project folders
•To rename project folders
To save a batch to another project folder
1. Choose File > Save > Save Batch As from the Batch View main menu in the Analysis
mode.
The Save Batch As Dialog Box opens.
2. Select a storage location.
The default storage location is C:\TraceFinderData\4.0\Projects.
3. Select or create a project folder.
4. Type a name for the new batch.
If you are saving the batch to a different folder, you must give it a unique name. You
cannot overwrite an existing batch in a folder.
5. Click Save.
When you save the batch to a different folder, the reports reflect the original project
folders and the application does not save the calibration history.
To move a batch to another folder
1. Choose File > Save > Move Batch from the Batch View main menu in the Analysis
mode.
The Save Batch As Dialog Box opens.
2. Select a storage location.
The default storage location is C:\TraceFinderData\4.0\Projects.
3. Select or create a project folder.
4. Type a name for the new batch.
You must give the batch a unique name in the new subproject folder. You cannot
overwrite an existing batch.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 337
5. Click Save.
When you move the batch, the reports reflect the original project and subproject folders
and the application does not save the calibration history.
To create new project folder
1. In the Save Batch As dialog box, select the folder for which you will create a new
lower-level folder.
• You can select the main TraceFinderData\4.0\Projects folder and create a new folder
under it.
• You can select one of the existing folders and create a lower-level folder under it.
2. Click the Create Folder icon, .
The application adds a new lower-level folder to the selected folder.
3. Select the new folder name and type a name for the folder.
Folder names can contain spaces and special characters, except for the following special
characters: \ / : + ? ” < >
To delete project folders
1. In the Save Batch As dialog box, select the folder to delete.
2. Click the Delete Folder icon, .
With no confirmation prompt, the application immediately removes the selected folder.
To rename project folders
1. In the Save Batch As dialog box, select the folder to rename.
2. Click the Rename Folder icon, .
3. Type a new name for the folder and press ENTER.
The application saves the new folder name.
Note After you add a lower-level folder, you cannot rename the parent folder.
Note This feature is not available for folders that contain lower-level project or batch
folders; you must first delete the lower-level project or batch folders.
Note This feature is not available for folders that contain lower-level project or batch
folders; you must first delete the lower-level project or batch folders.
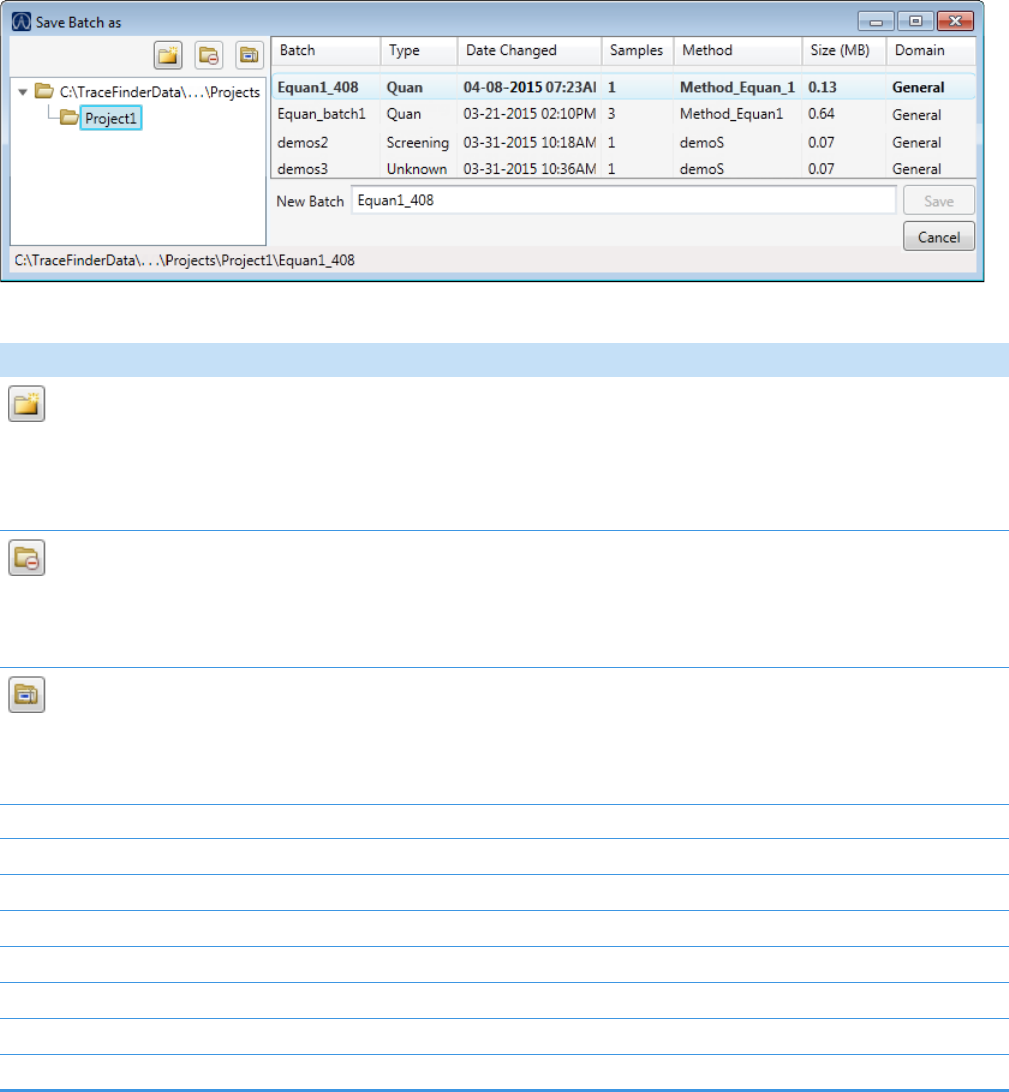
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
338 TraceFinder User Guide Thermo Scientific
Save Batch As Dialog Box
Use the features in the Save Batch As dialog box to save a batch to a new name or to move a
batch to a different project folder.
Figure 118. Save Batch As dialog box
Table 66. Save Batch As dialog box parameters (Sheet 1 of 2)
Parameter Description
Create New Folder
Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Or, you can right-click and choose Create Folder.
Delete Folder
With no confirmation prompt, immediately removes the selected folder.
You cannot delete a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Or, you can right-click and choose Delete.
Rename Folder
Renames the selected folder.
You cannot rename a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Or, you can right-click and choose Rename.
Batch table
Batch Name of batches in the selected project.
Type Type of batch: Quan, Screening, or Unknown Only.
Date Changed Date that the batch was last updated.
Samples Number of samples in the batch.
Method Name of the method used to create the batch.
Size Size of the batch in megabytes.
Domain TraceFinder domain in which the batch was created.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Batch View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 339
New batch parameters
New Batch Name of the new batch to create.
Note If the Create button is not activated, you have entered a name that is already used or
you have not selected a method.
Path Path to the project in the TraceFinderData\4.0\Projects folder where the batch is created.
Buttons
Save Saves the batch to the specified name and folder and opens the Batch View for the new
batch.
Cancel Closes the Save Batch As dialog box without saving the batch.
Shortcut menu commands
Create Folder Adds one of the following:
• When a drive is selected, adds a new project-level folder to the drive.
• When a project folder is selected, adds a subproject-level folder to the selected project.
• When a subproject folder is selected, adds a lower-level folder to the subproject.
Delete Folder Immediately removes the selected folder. There is no prompt to confirm that you want to
delete the selected folder.
You cannot delete a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Rename Folder Renames the selected folder.
You cannot rename a folder that contains lower-level project or batch folders; you must first
delete the lower-level project or batch folders.
Expand Child Nodes Expands all project and subproject folders in the Project tree.
Collapse Child Nodes Collapses all project and subproject folders in the Project tree.
Table 66. Save Batch As dialog box parameters (Sheet 2 of 2)
Parameter Description
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
340 TraceFinder User Guide Thermo Scientific
Working in Data Review for Unknown Screening Batches
The Data Review view displays the data generated by the master method. Use Data Review to
verify the data for identified peaks before you generate reports.
When you run samples in an experiment, you can check for peaks that are not already
identified in either an associated target screening or quantitation method and take appropriate
steps when the application finds these peaks and proposes whether they match one or more
compounds using various identification sources.
Follow these procedures:
•To open the Data Review view
•To display or hide a pane on the Unknown Screening View page
•To move, dock, or float Data Review panes
•To restore the default layout
To open the Data Review view
1. Click Analysis in the navigation pane.
The Analysis navigation pane opens.
2. Click Data Review.
The Data Review navigation pane opens.
IMPORTANT When unknown screening features are associated with a quantitation or
target screening method, the unknown screening results do not include any compounds
(other than internal standards) that were identified by the quantitation or target screening
methods.
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 341
To display or hide a pane on the Unknown Screening View page
From the View menu, choose to display or hide any of the following panes.
A check mark indicates a displayed pane.
To move, dock, or float Data Review panes
Follow the instructions in Appendix C, “Moving Data Review Panes.”
To restore the default layout
Choose View > Restore Default Layout.
The Unknown Screening View displays the default panes in their default locations.
Cross Sample Peak List Pane
Use the Cross Sample Peak List pane to compare peak values across all samples in a batch.
To use the Cross Sample Peak List pane
1. In the Peak List pane, select a peak.
The application scrolls the Cross Sample Peak List pane to the specified peak and selects
the peak.
2. Use the scroll bar at the bottom of the pane to scroll across the values for the selected peak
in all samples and groups in the batch.
Displays the Peak List Pane.
Displays the TIC Chromatogram Pane.
Displays the Spectrum Pane.
Displays the Sample List Pane.
Displays the Fragments Pane.
Displays the Isotopes Pane.
Displays the Cross Sample Peak Overlay Pane.
Displays the Heat Map Pane.
Displays the Peak Identifications Pane.
Displays the Library Search Pane.
Displays the Chemical Structure Pane.
Displays the Group Averages Pane.
Displays the XIC Pane.
Displays the Cross Sample Peak List Pane.
Displays the XIC Overlay Pane.
Displays the Peak Chromatogram Pane.
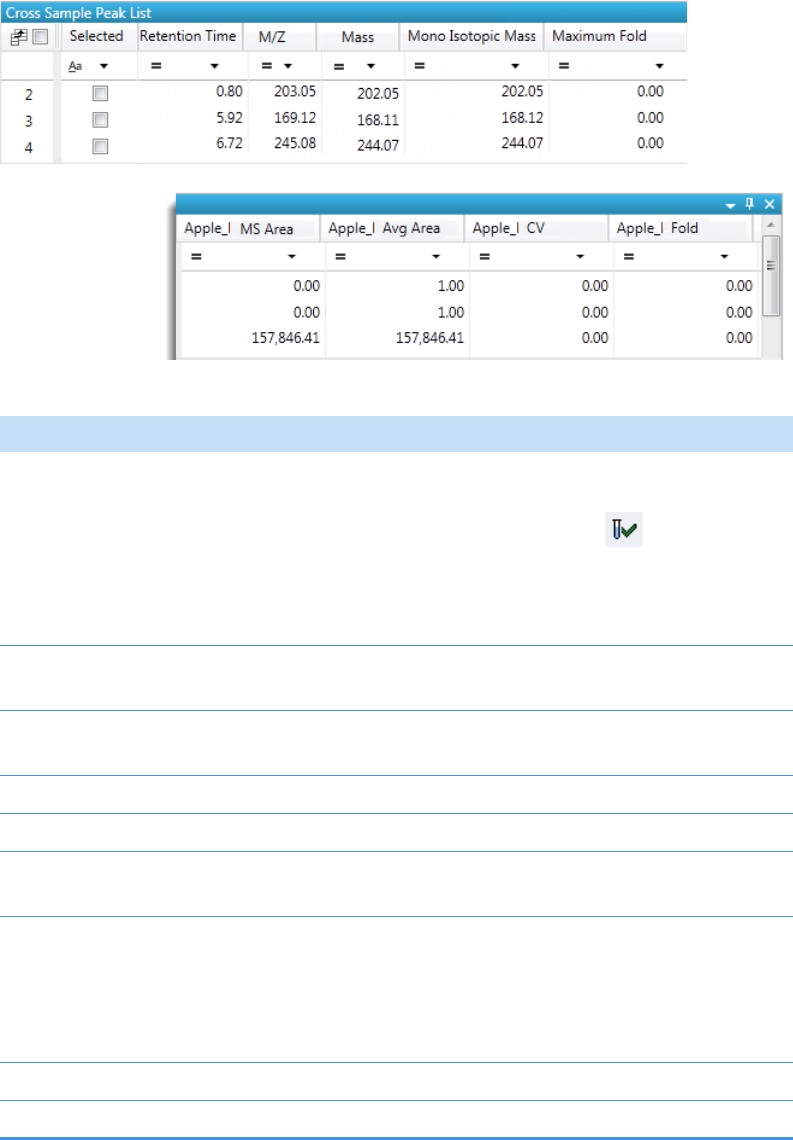
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
342 TraceFinder User Guide Thermo Scientific
Figure 119. Cross Sample Peak List pane
Table 67. Cross Sample Peak List pane columns (Sheet 1 of 2)
Column Description
Selected Identifies peaks to display when you select the Selected option in
the XIC Overlay pane, or identifies which peaks to process when
you click the Submit … for Processing icon, .
Do not confuse a check mark in the Selected column with the
currently selected peak. The row for a currently selected peak is
highlighted in blue.
Retention Time The time after injection when the compound elutes. The total
time that the compound is retained on the column.
M/Z Mass-to-charge ratio found in the spectra for the peak. Assumes
that the charge is 1.
Mass The uncharged mass found in the spectra for the peak.
Monoisotopic Mass The most abundant mass in the spectra for the peak.
Maximum Fold The maximum difference of the group average response from the
control average response.
Compound Name Displays the compound name found in the library or compound
database. This column is displayed only when the peaks are
aligned and you have performed a simple search. Refer to
Chapter 6, “Using the Method Development Mode for Unknown
Screening Methods,” in the TraceFinder Lab Director User Guide.
Sample n MS Area The MS peak area for each sample.
Default Average Area The averaged response of all peak occurrences in the sample.
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 343
Heat Map Pane
Use the Heat Map pane to display the response of each peak occurrence in the batch. The
Heat Map pane displays all MS Area values for all peaks in all samples in the batch that are
above the peak threshold value specified in the method. When you select a peak in the Heat
Map pane, the application displays the associated results for the selected peak in all panes of
the Unknown Screening view.
The Heat Map pane is available only when you select the Alignment and Gap Filling option
in the method (refer to Chapter 6, “Using the Method Development Mode for Unknown
Screening Methods,” in the TraceFinder Lab Director User Guide) or select the RT Alignment
option when you submit the batch (see Submitting a Batch).
Follow these procedures:
•To use the Heat Map pane
•To use the Heat Map pane with the TIC Chromatogram pane
•To use the Heat Map pane with the XIC chromatogram pane
Default % CV The coefficient of variance for each sample.
The coefficient of variance is calculated as the standard deviation
of the multiple samples of one level, multiplied by 100, and
divided by the average of the multiple samples of that level. This
calculation is based on the areas of the peaks.
Default Fold The maximum difference of the sample average response from the
control average response.
Group n Average Area The averaged response of all peak occurrences in the group. The
minimum value is 1, even when the peak never occurs in the
group.
Group n % CV The coefficient of variance for each sample.
The coefficient of variance is calculated as the standard deviation
of the multiple samples of one level, multiplied by 100, and
divided by the average of the multiple samples of that level. This
calculation is based on the areas of all the peaks in the group of
samples.
Group n Fold The maximum difference of the average response of all samples in
a group from the control average response.
Table 67. Cross Sample Peak List pane columns (Sheet 2 of 2)
Column Description
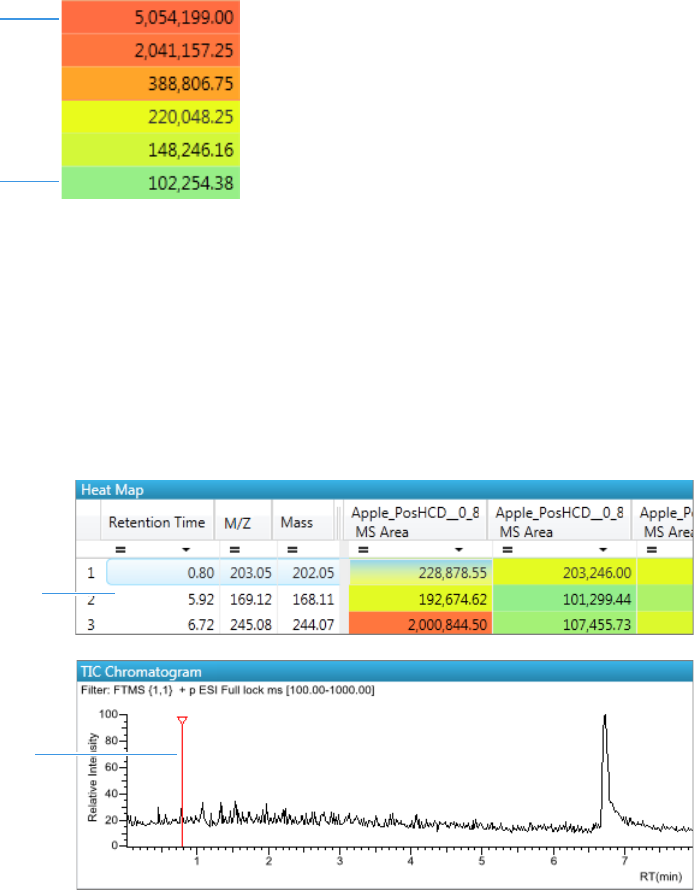
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
344 TraceFinder User Guide Thermo Scientific
To use the Heat Map pane
1. Click the Heat Map tab to display the pane.
For detailed descriptions of all parameters in the Heat Map pane, see Heat Map pane
columns.
2. In the Peak List pane, select a peak.
The application scrolls the Heat Map pane to the specified peak and selects the peak.
For each found peak in the batch, the application assigns a color code indicating the
relative MS area value from highest (orange) to lowest (green).
To use the Heat Map pane with the TIC Chromatogram pane
1. Click the TIC Chromatogram tab to display the pane.
The total ion current (TIC) chromatogram pane displays the relative intensity of all
found peaks in the batch.
2. In the Heat Map pane, select a peak.
The application identifies the selected peak in the TIC Chromatogram pane and indicates
it with a red marker.
Or, you can select a peak in the TIC Chromatogram pane, and the application scrolls the
heat map to the selected peak.
Highest
Lowest
A red marker
indicates the
selected peak in
the TIC
Chromatogram
pane.
Select a peak.
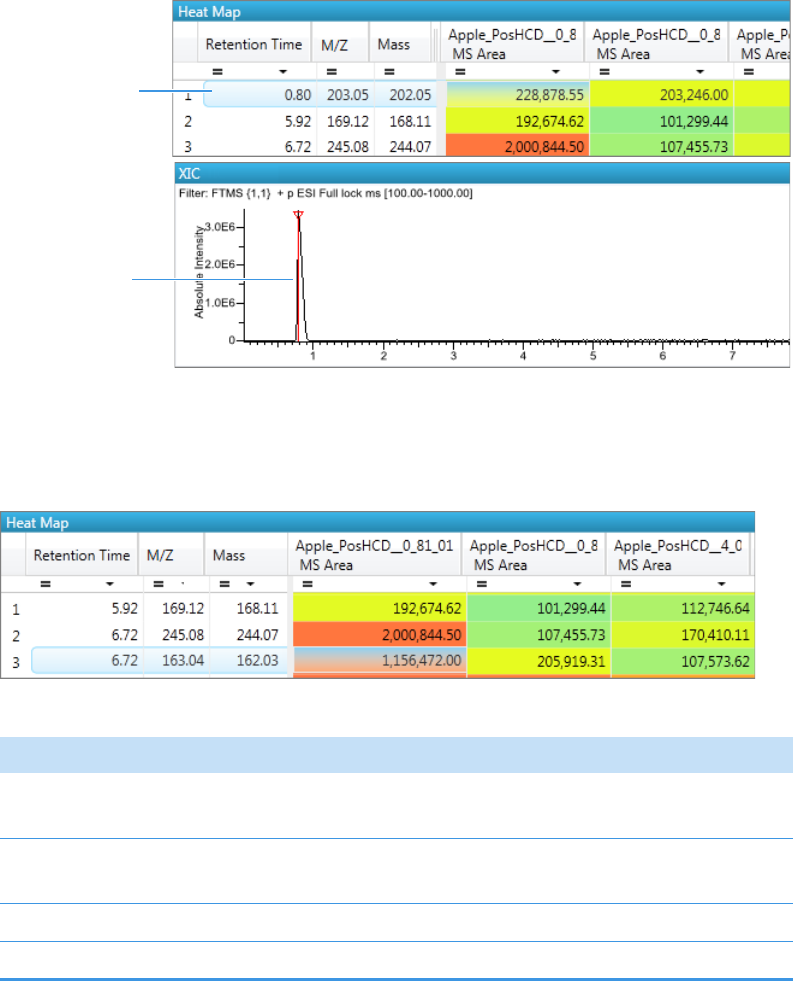
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 345
To use the Heat Map pane with the XIC chromatogram pane
1. Click the XIC tab to display the pane.
The extracted ion chromatogram (XIC) pane displays the absolute intensity of the
selected peak.
2. In the Heat Map pane, select a peak.
The application identifies the selected peak in the XIC, scales the chromatogram’s y axis
to fit the peak, and indicates it with a red marker.
Or, you can select a peak in the XIC pane, and the application scales the chromatogram
to fit the peak, indicates it with a red marker, and scrolls the heat map to the selected
peak.
Figure 120. Heat Map pane
Table 68. Heat Map pane columns
Column Description
Retention Time The time after injection when the compound elutes. The total
time that the compound is retained on the column.
M/Z Mass-to-charge ratio found in the spectra for the peak. Assumes
that the charge is 1.
Mass The uncharged mass found in the spectra for the peak.
Sample n MS Area The MS peak area for each sample.
A red marker
indicates the
selected peak in
the XIC pane.
Select a peak.
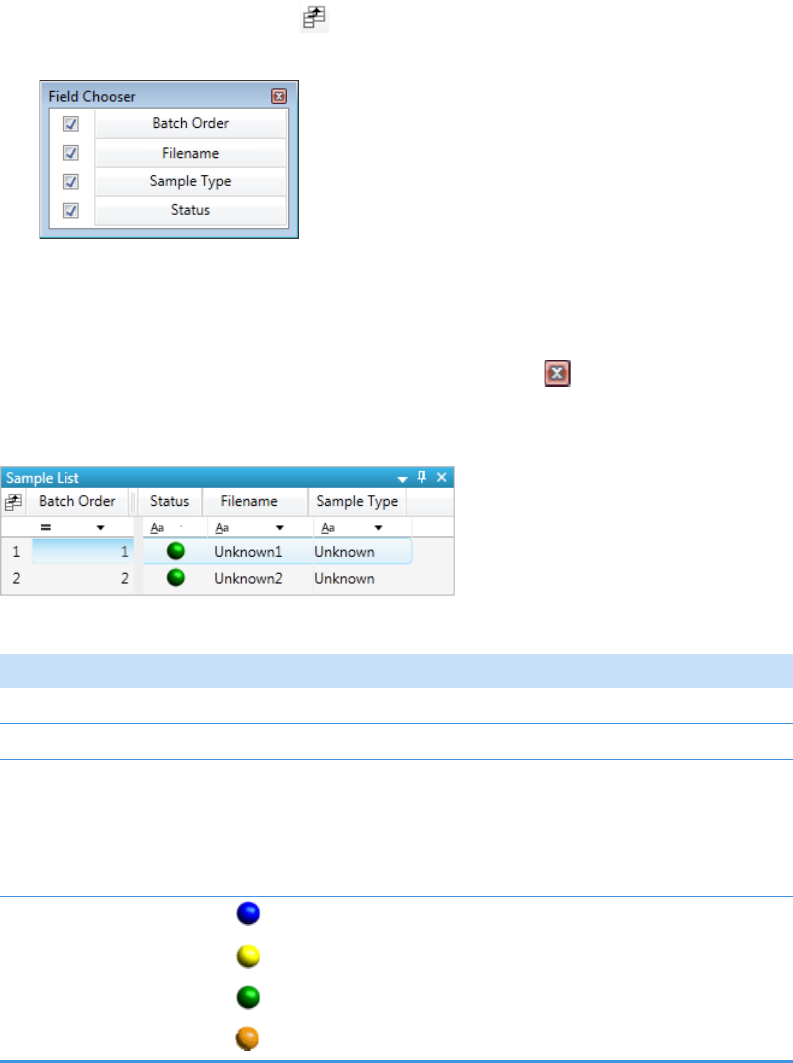
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
346 TraceFinder User Guide Thermo Scientific
Sample List Pane
Use the Sample List pane to select a sample of interest. The application displays the associated
results for the selected sample in all panes of the Unknown Screening view.
To hide or display columns in the Sample List pane
1. Click the Field Chooser icon, , in the upper left corner of the pane.
The Field Chooser displays all available columns of data for the Sample List pane.
2. Select the check box for each column that you want to display, or clear the check box for
each column that you want to hide.
The application immediately displays or hides the column in the Sample List pane.
3. When you are finished modifying the column display, click to close the Field
Chooser.
Figure 121. Sample List pane
Table 69. Sample List pane columns
Column Description
Batch Order The order in which the samples were acquired.
Filename A user-defined name that identifies a sample.
Sample Type Defines how the application processes the sample data. Each
sample is classified as one of the following sample types: Matrix
Blank, Solvent, Cal Std, QC Std, or Unknown.
Default: Unknown
Status Sample is not acquired.
Sample is acquired but not processed.
Sample is acquired and processed.
Sample is currently acquiring.
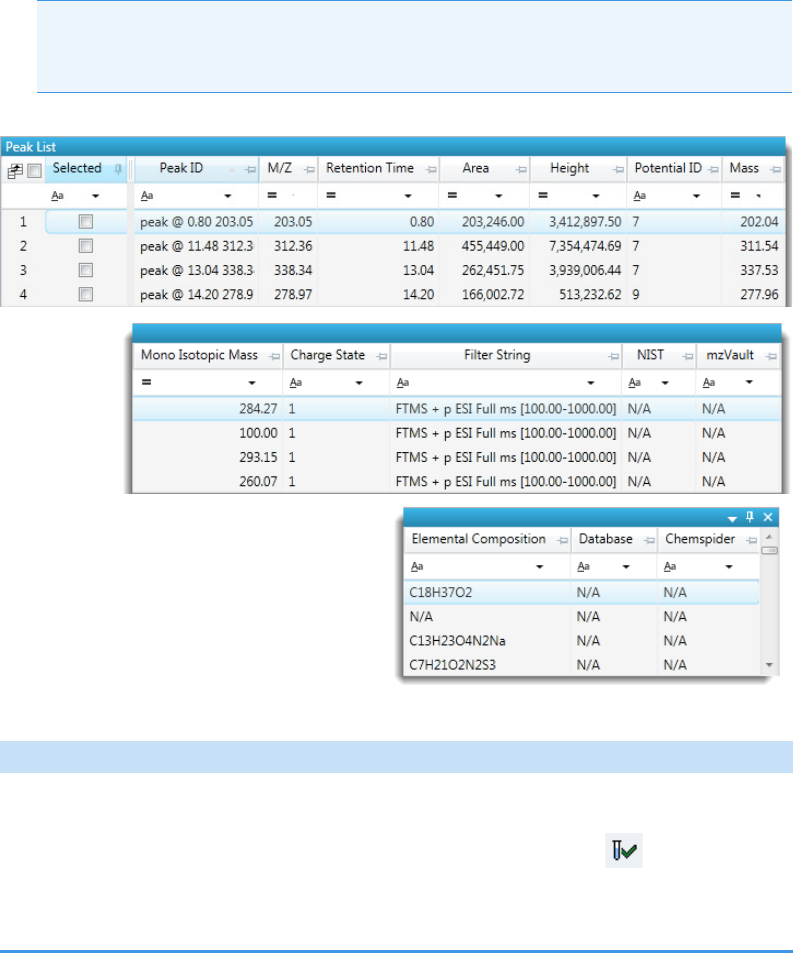
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 347
Peak List Pane
Use the Peak List pane to select a specific peak found in the selected sample. The Peak List
displays each peak that the application identified in the sample.
To display information for a peak
In the Peak List, click anywhere in the peak row.
The application displays information for the selected peak in the other Data Review
panes.
Figure 122. Peak List pane
Note Do not confuse a check mark in the Selected column with the currently selected
peak. The row for the current, selected peak is highlighted in blue. A selected check
box in the Selected column does not indicate the current, selected peak (see Selected).
Table 70. Peak List pane columns (Sheet 1 of 2)
Column Description
Selected Identifies peaks to display when you select the Selected option in
the XIC Overlay pane, or identifies which peaks to process when
you click the Submit … for Processing icon, .
Do not confuse a check mark in the Selected column with the
currently selected peak. The row for the current, selected peak is
highlighted in blue.

6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
348 TraceFinder User Guide Thermo Scientific
Peak ID Identifies the peak as “peak @ retention time m/z.”
Example: peak @ 0.80 203.05
m/z Mass-to-charge ratio found in the spectra for the peak. Assumes
the charge is 1.
Retention Time The time after injection when the compound elutes. The total
time (in minutes) that the compound is retained on the column.
Area The area obtained by integrating peak intensities from the start to
the end of the peak.
Height The distance from the peak maximum to the peak base, measured
perpendicular to the ordinate.
Potential ID The number of returned peak IDs.
Mass The uncharged mass found in the spectra for the peak.
Monoisotopic Mass The sum of the masses of the most abundant isotope.
Charge State The charge state of the ion (the z value in m/z). For example, a
charge state of 2 with a negative polarity means that the
compound has 2 more electrons than protons.
Valid range: 1 through 10
Default: 1
Filter String Filter used to identify the peak. Specified in the raw data file or the
master method.
NIST The NIST match result name for the identified peak. If there is no
match or if the Library search was not specified in the method,
this shows as “N/A.”
mzVault The library match result name for the identified peak. If there is
no match or if the Library Search was not specified in the method,
this shows as “N/A.”
Elemental
Composition
The chemical formula for the identified peak. If there is no match
or the Elemental Composition search was not specified in the
method, this shows as “N/A.”
Database The compound database match result name for the identified
peak. If there is no match or the Database Search was not specified
in the method, this shows as “N/A.”
ChemSpider The ChemSpider database match result name for the identified
peak. If there is no match or the ChemSpider Search was not
specified in the method, this shows as “N/A.”
Table 70. Peak List pane columns (Sheet 2 of 2)
Column Description
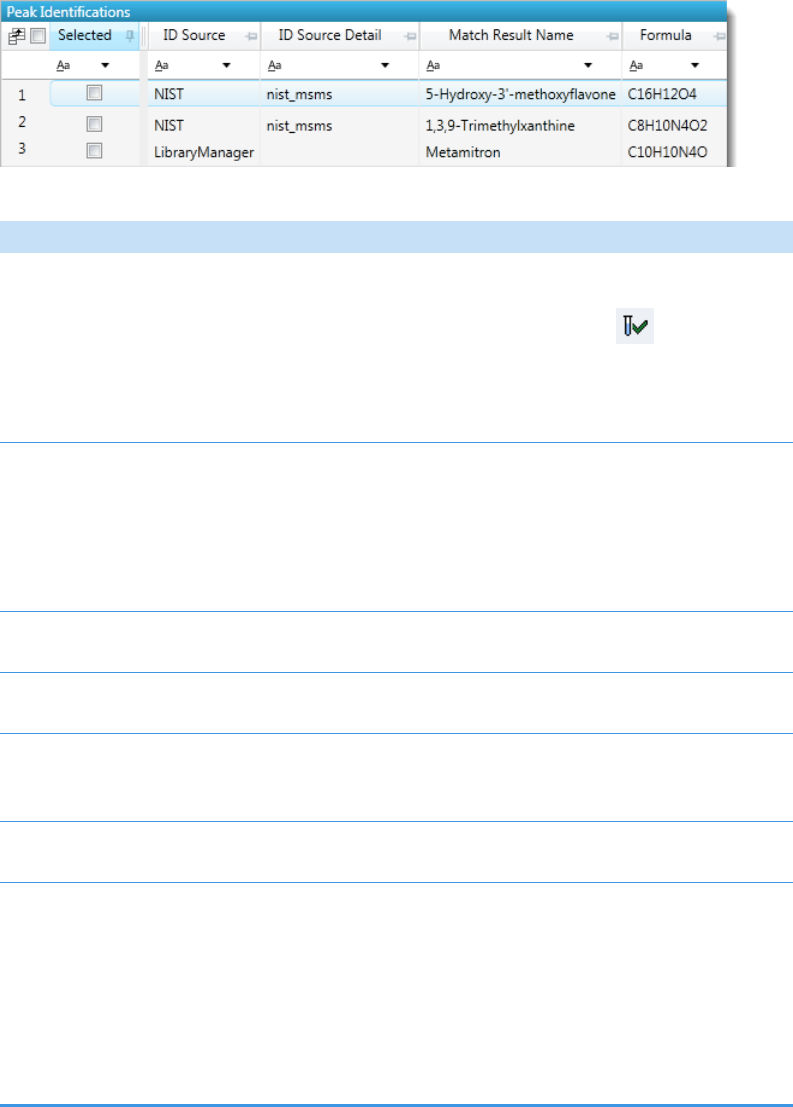
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 349
Peak Identifications Pane
Use the Peak Identifications pane to display the name and formula for identified peaks and
the source of the identification.
Figure 123. Peak Identifications pane
Table 71. Peak Identifications pane columns (Sheet 1 of 2)
Column Description
Selected Identifies peaks to display when you select the Selected option in
the XIC Overlay pane, or identifies which peaks to process when
you click the Submit … for Processing icon, .
Do not confuse a check mark in the Selected column with the
currently selected peak. The row for a currently selected peak is
highlighted in blue.
ID Source Specifies the search type used to identify the peak (Database
Search, Library Search [NIST or mzVault], Elemental
Composition, or ChemSpider Search). Refer to the instructions
“To specify the search options” in Chapter 6, “Using the Method
Development Mode for Unknown Screening Methods,” in the
TraceFinder Lab Director User Guide.
ID Source Detail Specifies the name of the library, the compound database, or the
ChemSpider hyperlink where the peak was identified.
Match Result Name Specifies the compound name that was found in the library, in
the compound database, or in the ChemSpider hyperlink.
Formula Specifies the chemical formula that was found in the library, in
the compound database, in the elemental composition, or in the
ChemSpider hyperlink.
Isotope Pattern Score % Specifies the percentage of matched isotopes in the total number
of isotopes.
Number of Isotopes
Matched
Specifies the number of isotopes matched in the calculated
isotope spectra relative to the total number of isotopes used in
the score calculation, in the format “x of y”, where
• x = the number of isotopes matching the elemental
composition used for the Isotope Pattern Score calculation.
• y = the total number of isotopes considered in the Isotope
Pattern Score calculation. This is the number of isotope
peaks expected to be above the spectral noise.
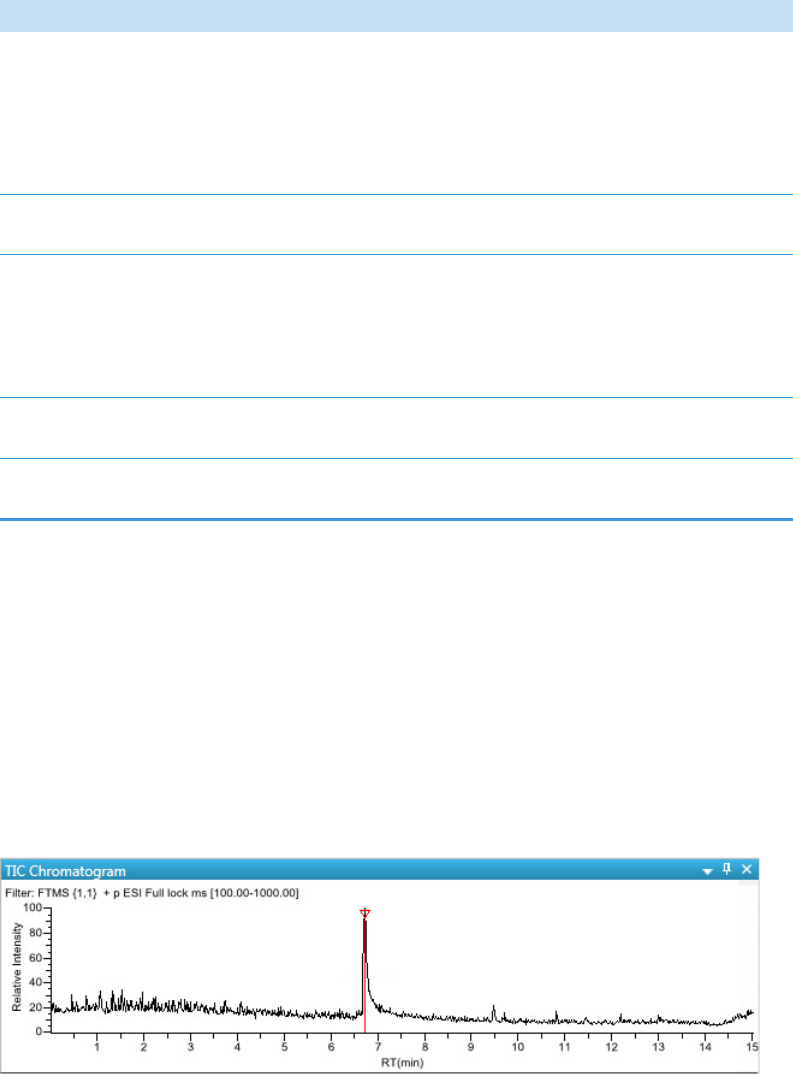
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
350 TraceFinder User Guide Thermo Scientific
TIC Chromatogram Pane
Use the TIC Chromatogram pane to display the relative intensity of a trace along the length
of the sample data retention window.
To focus on a specific peak
In the TIC Chromatogram pane, click any displayed peak.
The application indicates the selected peak with a red, vertical marker and updates the
focus in the other Data Review panes with data for that peak.
Figure 124. TIC Chromatogram pane
Score Specifies the score from the library fit. When the application
finds a match in the library, in the compound database, or
through the ChemSpider hyperlink, this column displays the
highest score associated with the Match Result Name parameter.
Valid range: 1 through 100%
SI/Dot Product Specifies the search index method used to search the NIST
library.
RSI/Rev Product (Reverse search index) Specifies the method used to search the
NIST library. A reverse search compares a library entry to an
unknown compound (whereas a forward search compares the
mass spectrum of an unknown compound to a mass spectral
library entry).
Library Probability
Percent
Specifies the probability ranking from the NIST library. When
you use a reverse search, this value is “N/A.”
Fragments Specifies the number of fragments matched in the calculated
spectra relative to the total number of expected fragments.
Table 71. Peak Identifications pane columns (Sheet 2 of 2)
Column Description
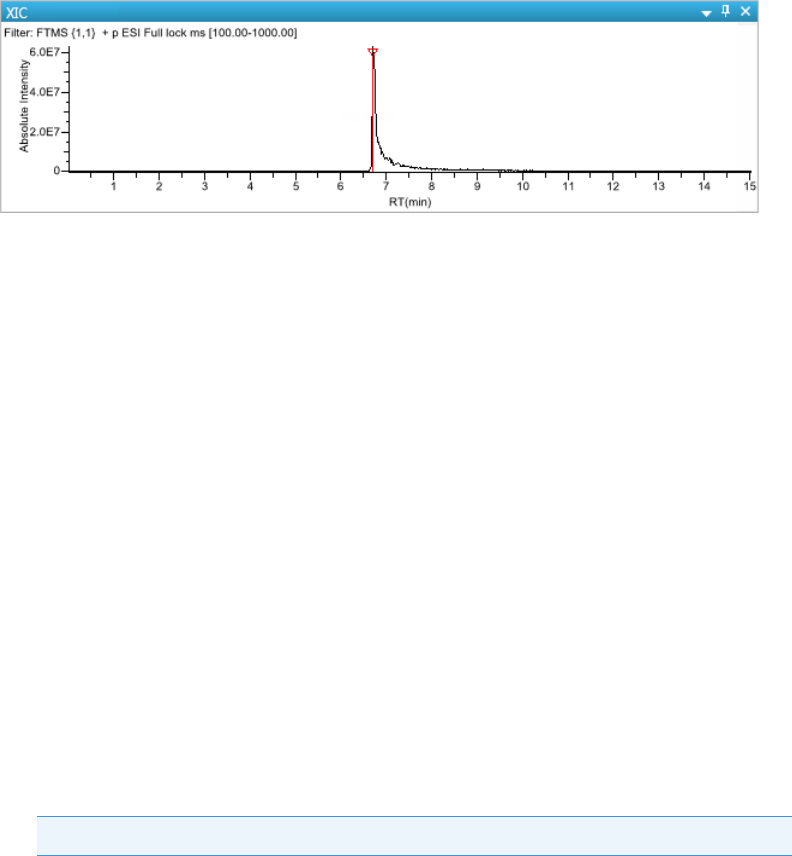
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 351
XIC Pane
Use the XIC pane to display the absolute intensity of an extracted trace along the length of the
sample data retention window.
To focus on a specific peak
In the XIC pane, click any displayed peak.
The application indicates the selected peak with a red, vertical marker and updates the
focus in the other Data Review panes with data for that peak.
Figure 125. XIC pane
XIC Overlay Pane
Use the XIC Overlay pane to view specific groups of peaks. You can choose to view all peaks,
selected peaks only, or the top 20 most intense peaks (by area). The XIC Overlay plot is a
collection of overlaid, extracted m/z ion plots that use a different color for each peak.
Follow these procedures:
•To display all peaks in a sample
•To display only selected peaks in a sample
•To display the top 20 peaks in a sample
•To focus on a specific peak
To display all peaks in a sample
1. In the Sample List pane, select the sample whose peaks you want to view.
2. In the XIC Overlay pane, select the All check box.
The application displays all the peaks in the selected sample. See XIC Overlay plot
showing all peaks.
Note If there are more than 1000 peaks in the sample, performance might be slow.
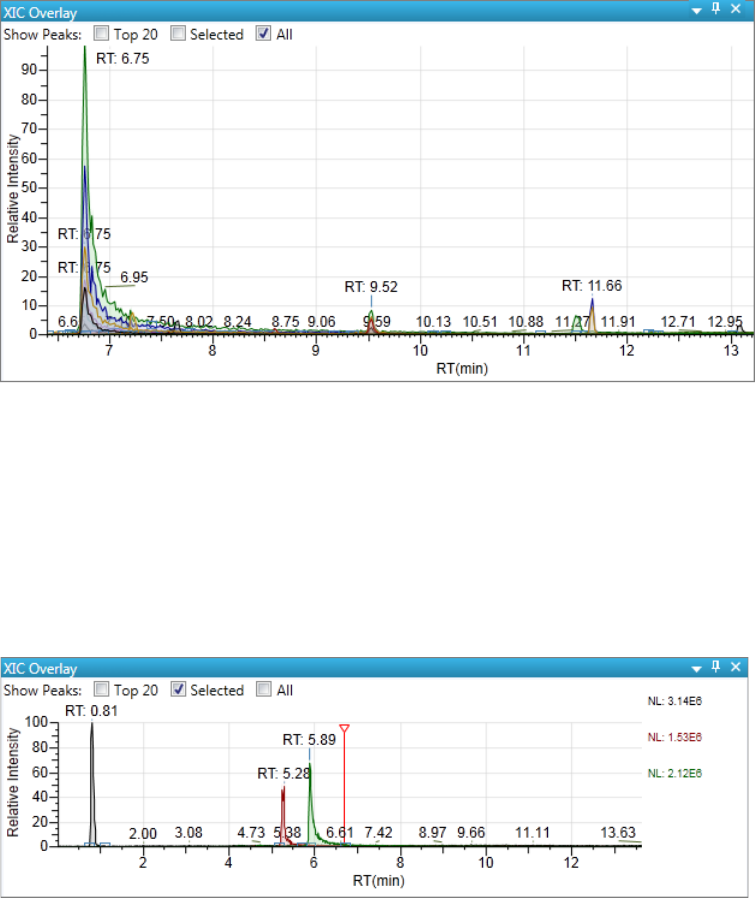
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
352 TraceFinder User Guide Thermo Scientific
Figure 126. XIC Overlay plot showing all peaks
To display only selected peaks in a sample
1. In the Sample List pane, select the sample whose peaks you want to view.
2. In the Peak List pane, select the Selected check box for each peak that you want to view.
3. In the XIC Overlay pane, select the Selected option.
The application displays all selected peaks.
Figure 127. XIC Overlay plot showing selected peaks
To display the top 20 peaks in a sample
1. In the Sample List pane, select the sample whose peaks you want to view.
2. In the XIC Overlay pane, select the Top 20 check box.
The application displays the top 20 peaks in the selected sample.
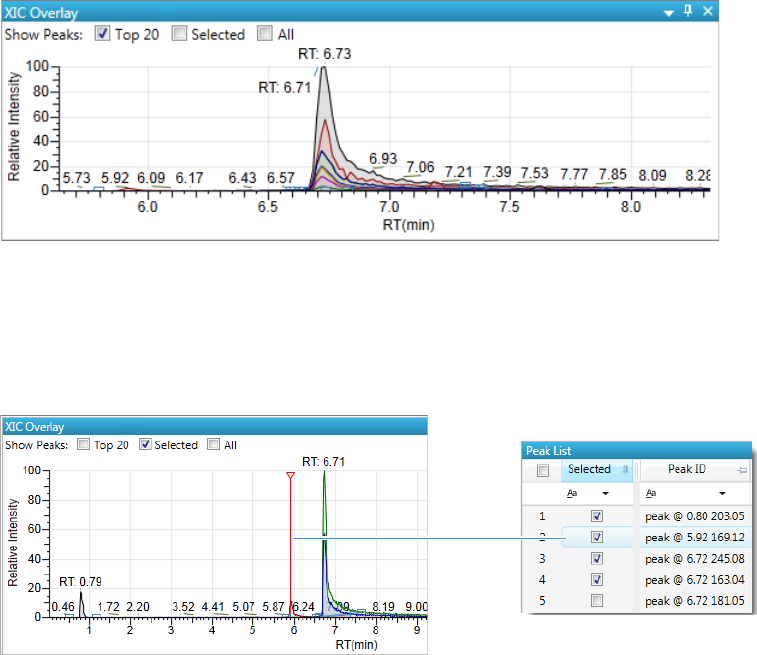
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 353
Figure 128. XIC Overlay plot showing the top 20 peaks
To focus on a specific peak
In the XIC Overlay pane, click any displayed peak.
The application indicates the selected peak with a red, vertical marker and updates the
focus in the other Data Review panes with data for that peak.
Group Averages Pane
Use the Group Averages pane to compare the peak areas of different samples to a control
group of samples.
The Group Averages pane is available only when you select the Alignment and Gap Filling
option in the method (refer to Chapter 6, “Using the Method Development Mode for
Unknown Screening Methods,” in the TraceFinder Lab Director User Guide) or select the RT
Alignment option when you submit the batch (see Submitting a Batch).
The control group is displayed to the left, and other samples and groups are listed in the order
in which they appear in the Batch View. To specify a group as the control group, see Groups.
Each group displays a colored bar that indicates the average response of the group, as
displayed in the Average Area column in the Cross Sample Peak List Pane. The color of the
bar matches the colors assigned to the samples and peaks in the Cross Sample Peak Overlay
Pane.
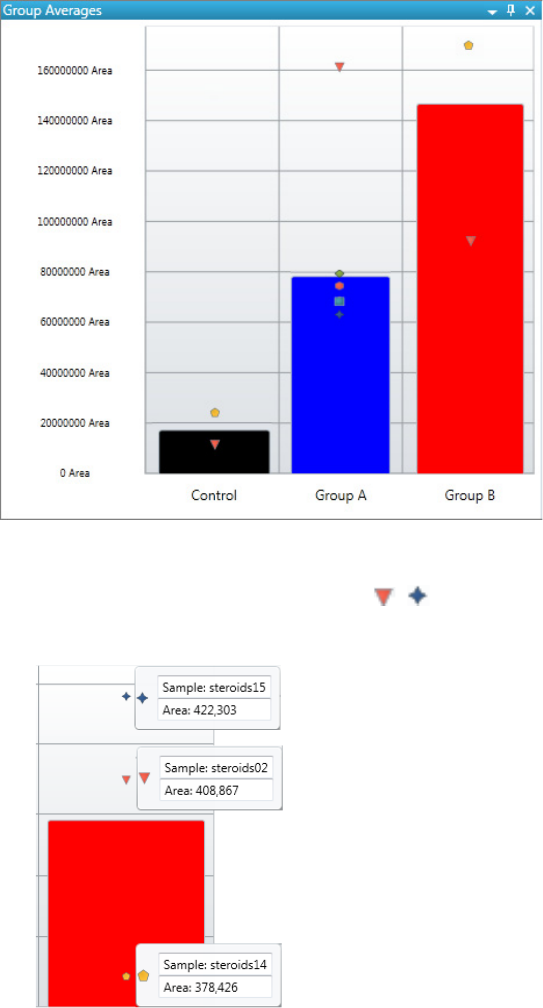
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
354 TraceFinder User Guide Thermo Scientific
Figure 129. Group Averages pane
To display areas for each sample in the batch
Hold the cursor over an indicator icon ( , ).
The application displays the area for the selected compound in each represented sample.
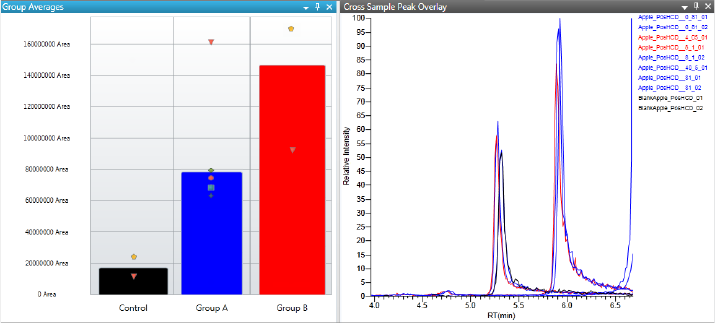
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 355
To use the Group Averages pane with the Cross Sample Peak Overlay pane
Display both the Group Averages pane and the Cross Sample Peak Overlay pane.
The color of the bars in the Group Averages pane matches the colors assigned to the
group samples and peaks in the Cross Sample Peak Overlay pane.
Peak Chromatogram Pane
Use the Peak Chromatogram Pane to display the selected chromatogram peak. The pane
initially displays only the apex scan for the detected peak. You can manually integrate the peak
and use the updated peak to generate reports.
Follow these procedures:
•To zoom in on a peak
•To remove a peak
•To customize the labels in the display
•To display surrounding apex times
•To manually integrate a peak
•To switch between Method and Manual integration modes
To zoom in on a peak
1. In the chromatogram plot, drag the cursor to delineate a rectangle around the peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the chromatogram plot and choose Reset Scaling.
To remove a peak
Right-click the Peak Chromatogram pane, and choose Delete Peak.
The application removes the peak displayed in the Peak Chromatogram pane. All data for
this peak are removed from the Unknown Screening View panes.
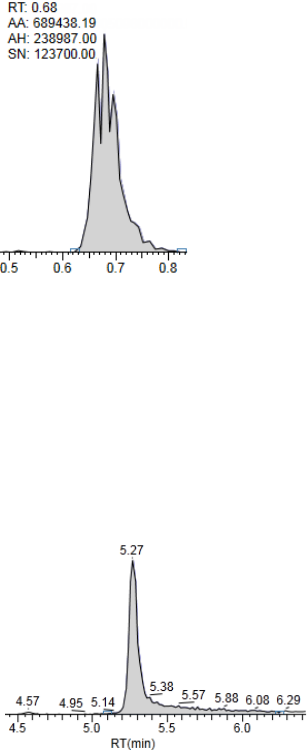
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
356 TraceFinder User Guide Thermo Scientific
To customize the labels in the display
1. Right-click the Peak Chromatogram pane, and choose Peak Display Settings.
The Peak Settings Dialog Box opens.
2. To display labels for the peak retention time (RT), mass height (MH), mass area (MA), or
signal-to-noise (SN), select the appropriate check box.
3. Click OK in the Peak Settings dialog box.
The chromatogram plot displays the selected labels.
Figure 130. Example of displayed labels
To display surrounding apex times
1. Right-click the Peak Chromatogram pane, and choose Peak Display Settings.
The Peak Settings Dialog Box opens.
2. Select the Show All Apex Times check box.
3. Click OK in the Peak Settings dialog box.
Figure 131. Example of displayed apex times
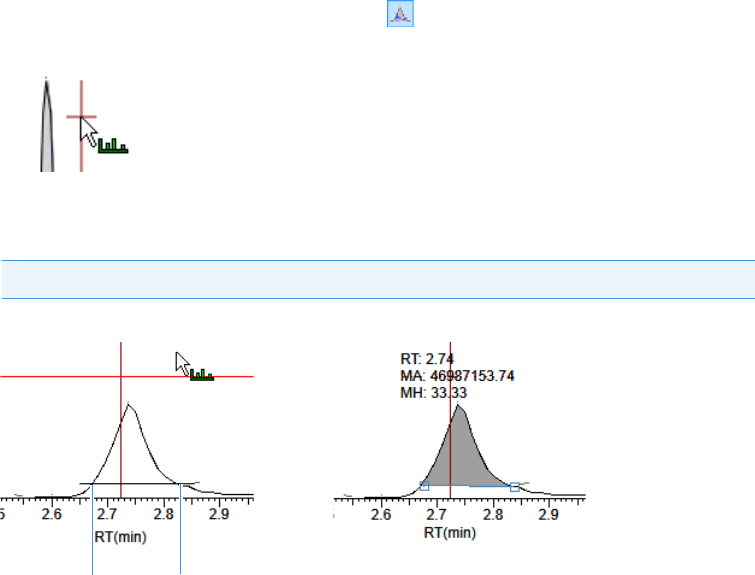
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 357
To manually integrate a peak
1. Right-click the Peak Chromatogram pane and then choose Swipe Manual Integration,
or click the Swipe Manual Integration icon, .
The cursor changes to indicate the manual integration mode.
2. Drag the cursor to define the beginning and ending base points for the new peak. See
Manually integrated peak.
Figure 132. Manually integrated peak
The application updates the area of the peak to the baseline that you just added. When
you regenerate the reports, they display the manually modified data.
3. To return to the original peak as defined by the method, right-click and choose
Integration Settings > Method.
To switch between Method and Manual integration modes
Right-click the Peak Chromatogram pane and choose Integration Settings > Manual or
Integration Settings > Method.
Initially, the method integration and manual integration settings that are stored for a
compound and file are identical. When you switch modes, the saved result set does not
change. However, when manual data are available, both the Peak Chromatogram plots
and the result set update as you switch between method and manual modes.
As you switch between modes, each pane reflects the changes. The generated reports for
the data identify the manual modifications.
Note You must drag the cursor inside the x axis and y axis.
Drag the cursor across the peak at
the base points for the new peak.
The application identifies the peak and
indicates the manual integration in the labels.
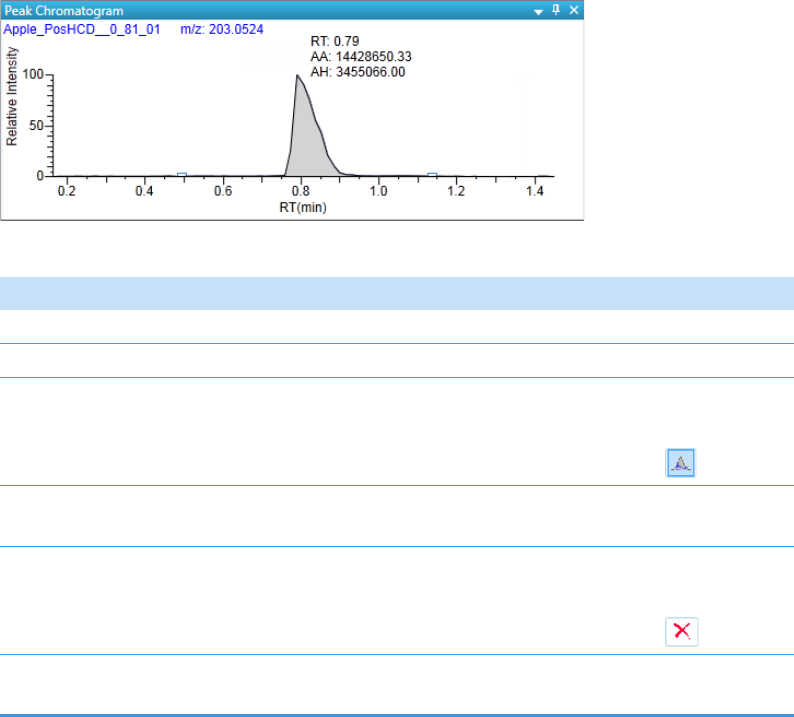
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
358 TraceFinder User Guide Thermo Scientific
Peak Chromatogram Pane
Use the Peak Chromatogram pane to display the selected chromatogram peak.
Figure 133. Peak Chromatogram pane
Table 72. Peak Chromatogram pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Swipe Manual
Integration
Switches to manual integration mode, where you can manually
define the peak.
Also available from the icon in the upper right corner, .
Integration Settings
(Manual or Method)
Switches the display between manual and method integration modes.
Delete Peak Removes the peak and all data for this peak from the Unknown
Screening View panes.
Also available from the icon in the upper right corner, .
Peak Display
Settings
Opens the Peak Settings Dialog Box.
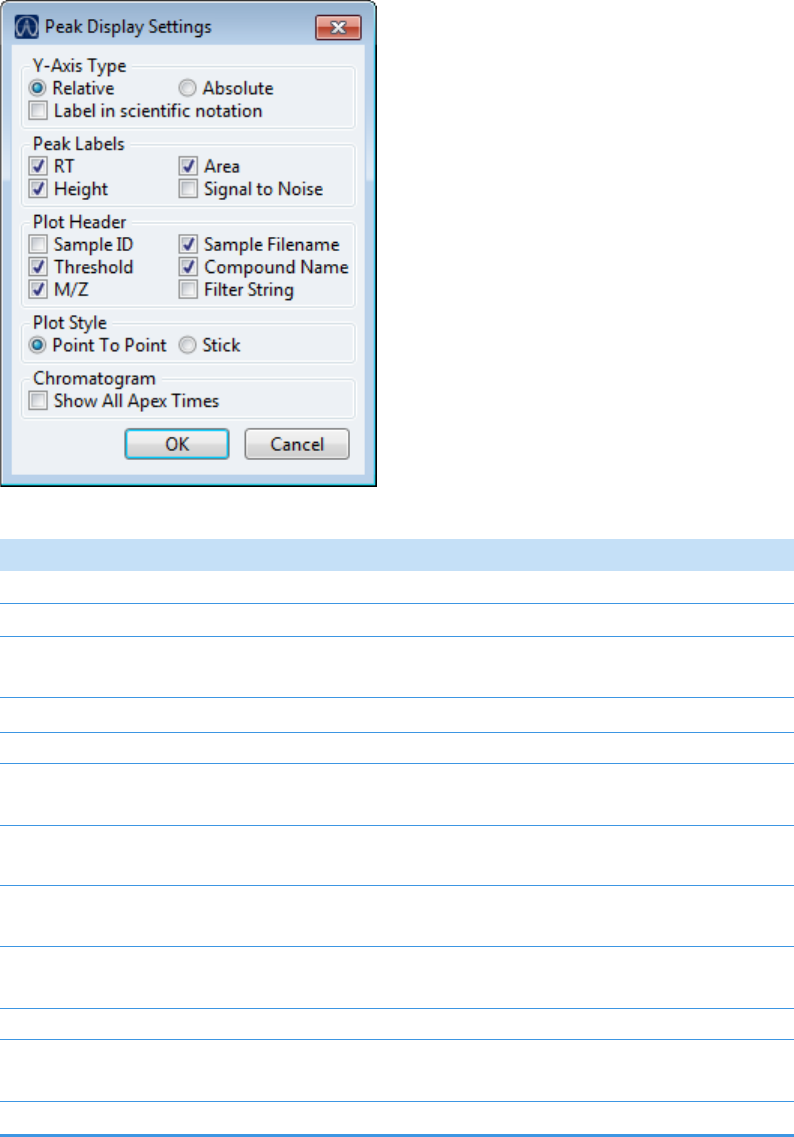
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 359
Peak Settings Dialog Box
Use the Peak Settings dialog box to custom the display settings.
Figure 134. Peak Settings dialog box
Table 73. Peak Settings dialog box parameters (Sheet 1 of 2)
Command Description
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific Notation Displays the y-axis scale in scientific notation.
Peak Labels
RT Displays the retention time (RT) label in the peak
chromatogram pane.
Height Displays the peak height (AH) label in the peak
chromatogram pane.
Area Displays the peak area (AA) label in the peak
chromatogram pane.
Signal to Noise Displays the signal-to-noise (SN) label in the peak
chromatogram pane.
Plot Header
Sample ID Displays a user-defined, alphanumeric string that
identifies a sample.
Threshold Displays the peak threshold.
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
360 TraceFinder User Guide Thermo Scientific
Cross Sample Peak Overlay Pane
Use the Cross Sample Peak Overlay Pane to compare instances of a selected peak across all
samples in the batch. The application overlays all occurrences of the peak in the batch.
The application displays the names of all samples in the batch where the selected peak is
found. Samples that are assigned to groups are color coded, and the peaks found in those
samples are color coded in the plot. To assign samples to groups, see Groups.
The retention time window displayed in the Cross Sample Peak Overlay pane is the RT
window that you specified in the method.
Follow these procedures:
•To select the peak to display in the Cross Sample Peak Overlay pane
•To zoom in on a peak
•To use the Cross Sample Peak Overlay pane with the Group Averages pane
m/z Displays the mass-to-charge ratio found in the spectra for
the peak. Assumes the charge is 1.
Sample Filename Displays the name of the current, selected sample where
the peak was found.
Compound Name Displays the compound name match in the library or
compound database.
Filter String Displays the filter used to identify the peak. Specified in
the raw data file or the master method.
Plot Style
Point To Point Displays chromatograms in the following format:
Stick Displays chromatograms in the following format:
Chromatogram
Show All Apex Times Displays all surrounding apex times in the plot.
Table 73. Peak Settings dialog box parameters (Sheet 2 of 2)
Command Description
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 361
To select the peak to display in the Cross Sample Peak Overlay pane
Select a peak in either the Peak List Pane or the Cross Sample Peak List Pane.
To zoom in on a peak
1. In the chromatogram plot, drag the cursor to delineate a rectangle around the peak.
The delineated area expands to fill the view.
2. To restore the default view, right-click the chromatogram plot and choose Reset Scaling.
To use the Cross Sample Peak Overlay pane with the Group Averages pane
Display both the Cross Sample Peak Overlay pane and the Group Averages pane.
The color of the bars in the Group Averages pane matches the colors assigned to the
group samples and peaks in the Cross Sample Peak Overlay pane.
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
362 TraceFinder User Guide Thermo Scientific
Cross Sample Peak Overlay Pane
Use the Cross Sample Peak Overlay pane to compare instances of a selected peak across all
samples in the batch
Figure 135. Cross Sample Peak Overlay pane
Library Search Pane
The Library Search Pane displays the best library matches for the selected peak, with the
highest score listed first.
Follow these procedures:
•To view the spectrum for a library match
•To display the difference between experimental and library spectra
Table 74. Cross Sample Peak Overlay pane shortcut menu commands
Column Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
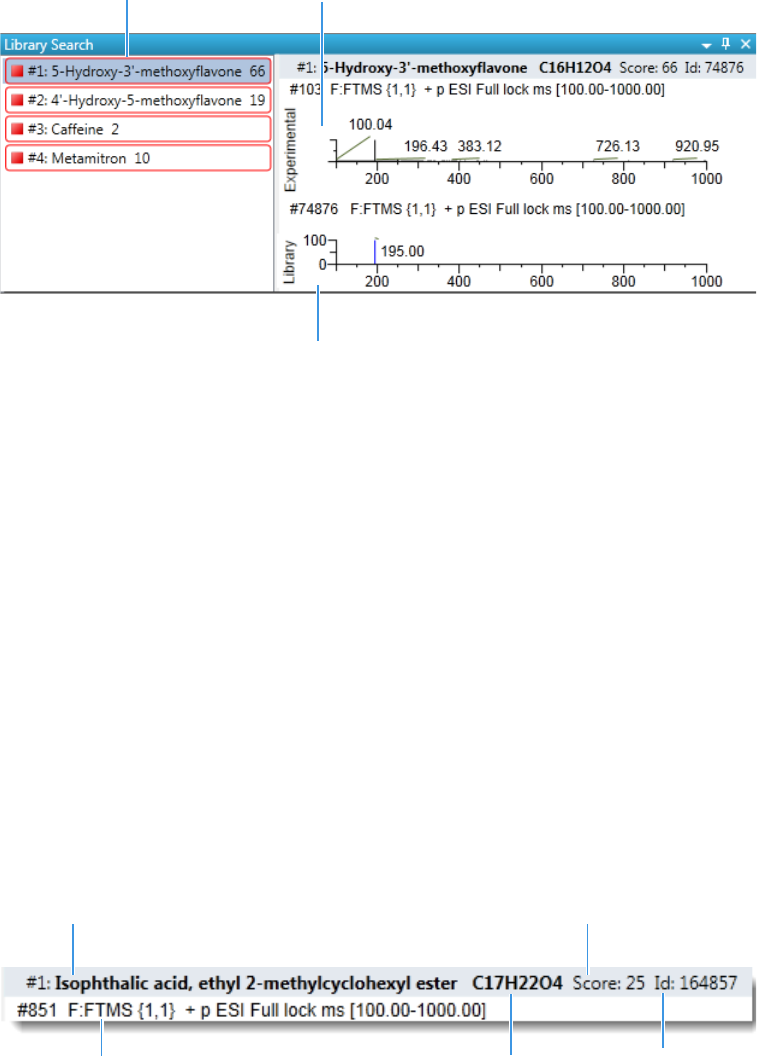
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 363
To view the spectrum for a library match
In the Library Search pane, select a library entry.
The Experimental plot displays the measured spectra for the selected compound. Library
matches are colored red.
The Library plot displays the expected spectra in the library. Library matches are colored
blue.
The spectra header displays the following:
• The library match name
• The chemical formula for the peak
• The score from the library fit when the library type is mzVault or NIST
• SI and RSI when the library type is NIST
• Dot Prod or Rev Dot Prod when the library is HRAM
• Molecular weight when the library type is NIST
• The ID record from the library when the library type is mzVault or NIST
• The scan number and scan filter when either is present in the library
Library matches Experimental spectrum
Library spectrum
Library match name Library score
Score ID Chemical formula Filter
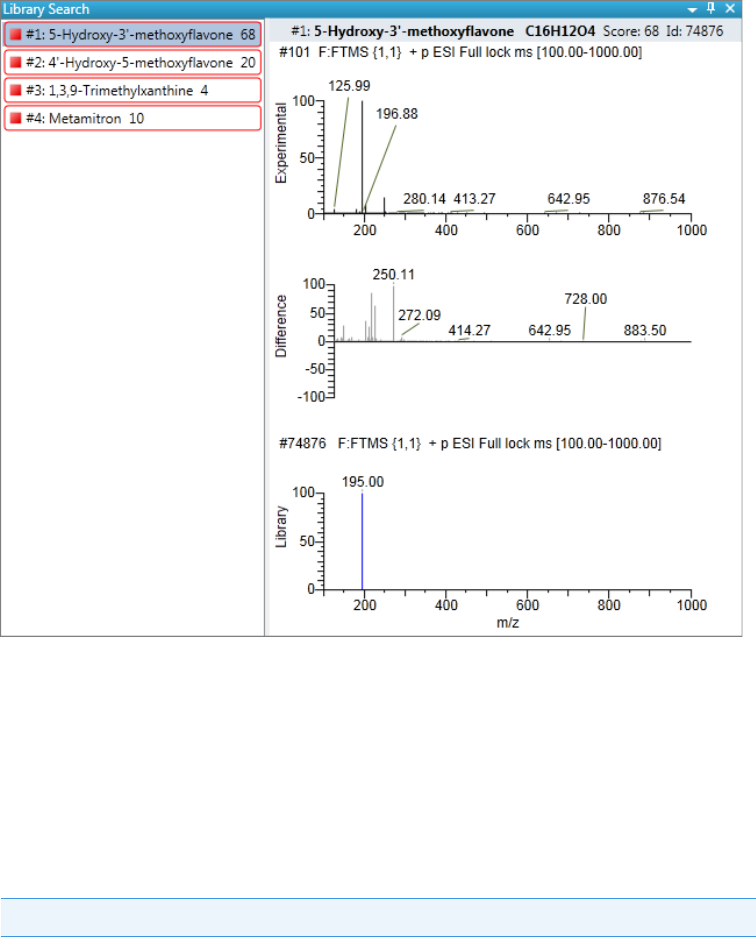
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
364 TraceFinder User Guide Thermo Scientific
To display the difference between experimental and library spectra
1. Right-click the spectrum plot and choose Show Difference Plot.
The application displays the Experimental and Library plots and a Difference plot that
shows the difference between the experimental and library spectra. When you read a
difference plot, the match between the experimental and library spectra is better when
fewer peaks appear.
Figure 136. Library Search pane showing a Difference plot
2. To specify how the difference plot displays the mass tolerance, do the following:
a. Right-click the spectrum plot and choose Set Difference Plot Mass Tolerance.
b. In the dialog box that opens, select either MMU or PPM as the unit of measure.
The default value is the tolerance set in the NIST library or mzVault library.
c. Type a mass tolerance value.
d. Click OK.
Note This command is available only when you display the difference plot.
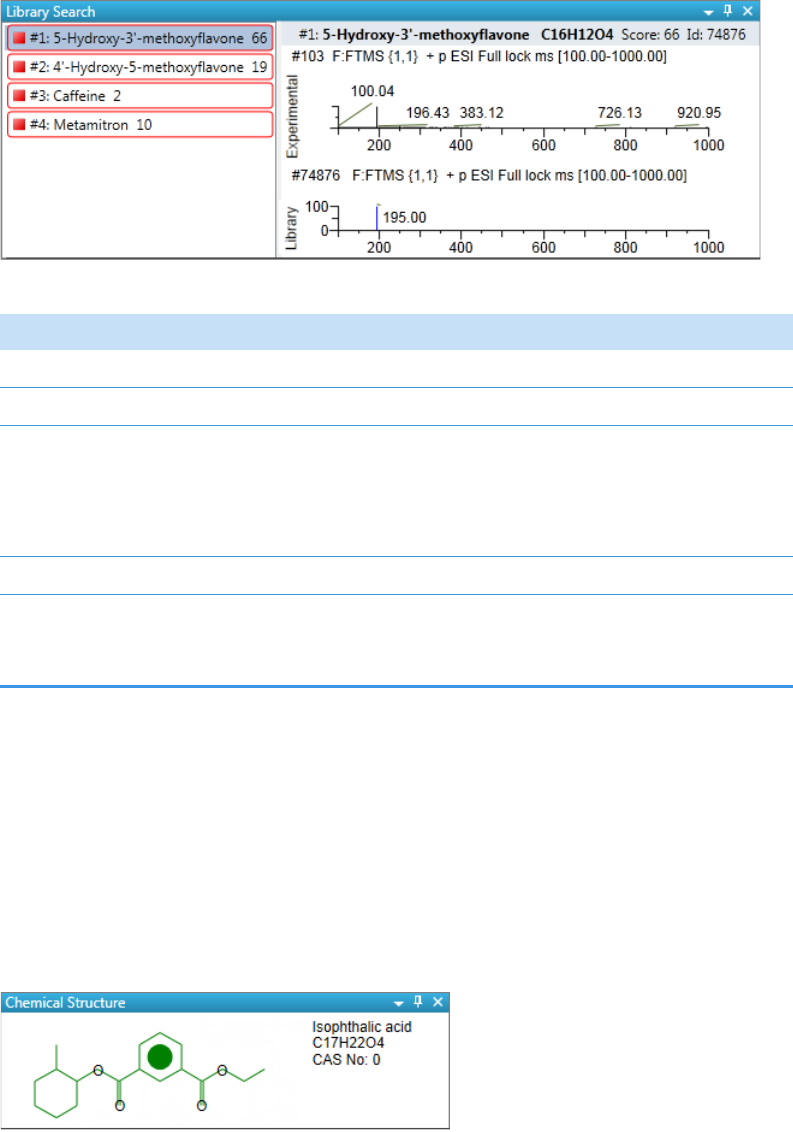
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 365
Library Search Pane
Use the Library Search pane to display the best library matches for the selected peak.
Figure 137. Library Search pane
Chemical Structure Pane
Use the Chemical Structure pane to view the chemical formula and structure for the peak that
is currently selected in the Peak List pane.
The application displays the chemical formula (and CAS number when available) that was
found in the search database that you specified in the method. Refer to Chapter 6, “Using the
Method Development Mode for Unknown Screening Methods,” in the TraceFind er Lab
Director User Guide.
Figure 138. Chemical Structure pane
Table 75. Library Search pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Display Overlay Spectra
Display Stack Spectra
Overlays the two spectrum displays, or stacks the
Experimental and Library spectra.
When you display the Difference plot, this spectrum
also displays the Difference spectrum.
Show/Hide Difference Plot Displays or hides the Difference spectrum plot.
Set Difference Plot Mass Tolerance Opens a dialog box where you specify the mass
tolerance in either MMU or PPM units. Available
only when you display the Difference Plot.
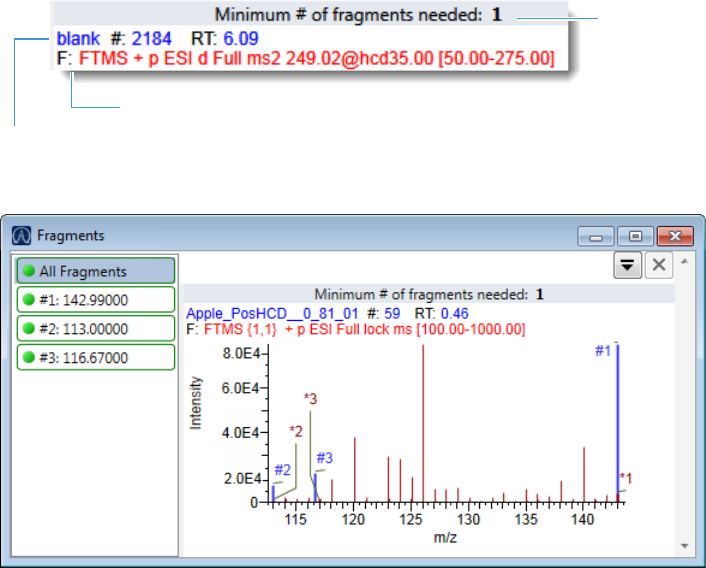
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
366 TraceFinder User Guide Thermo Scientific
Fragments Pane
The Fragments pane displays the maximum number of fragments as specified in the unknown
screening method. Refer to Chapter 6, “Using the Method Development Mode for Unknown
Screening Methods,” in the TraceFinder Lab Director User Guide.
When a compound database search returns a match for a peak that has fragments, the
Fragments pane displays the theoretical fragments.
When the data has an MS/MS element that is identified as belonging to the chromatographic
peak, the Fragments pane displays the fragments found in the MS/MS scan.
The Fragments pane displays the fragments in one of two ways:
•All Fragments
•Individual Fragments
All Fragments
The All Fragments view displays a composite of all fragments found in the peak. The
application displays the measured peak as a red line; the application displays the expected peak
as a blue line. The application displays these headers for the All Fragments view:
Figure 139. Fragments pane with overlaid spectra for all fragments
Processing filter
File name
Minimum number
of fragments
specified in the
method
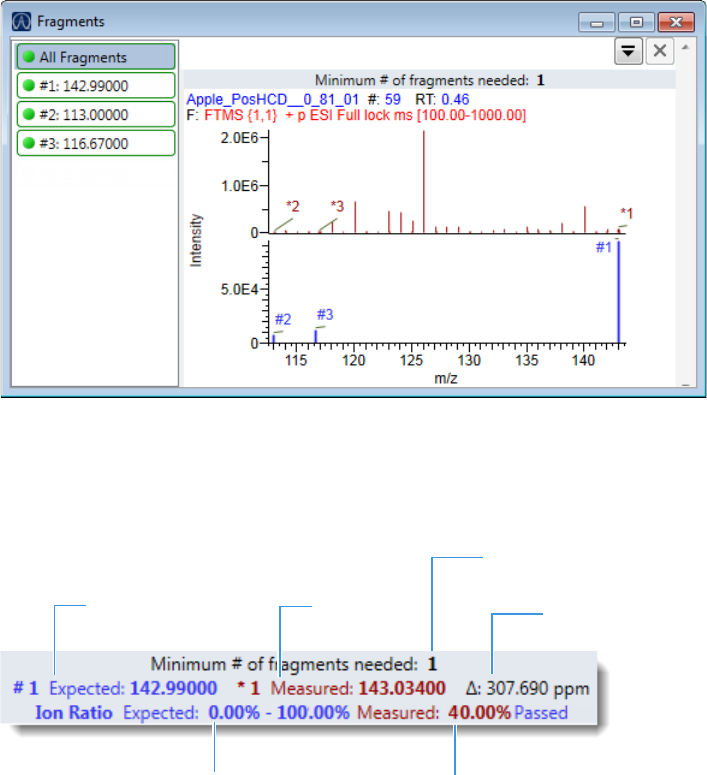
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 367
Figure 140. Fragments pane with stacked spectra for all fragments
Individual Fragments
The individual fragments view displays the expected and measured peaks for a single
fragment. The application displays these headers for the individual fragments view:
Minimum number of fragments specified
in the method
Expected m/z
for fragment 1
Measured m/z
for fragment 1 Delta between expected
and measured m/z
Expected ion ratio range Measured ion ratio
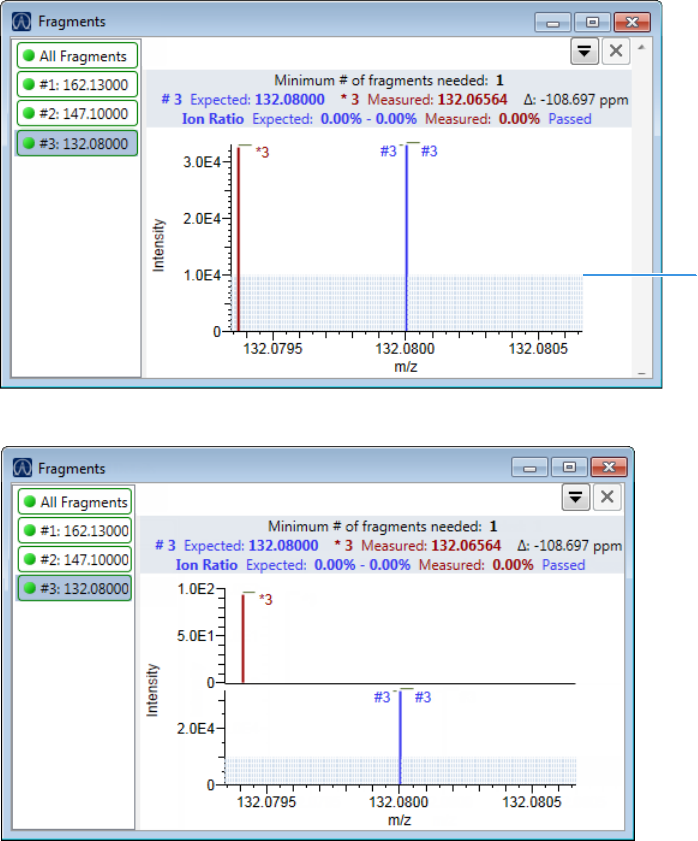
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
368 TraceFinder User Guide Thermo Scientific
Figure 141. Fragments pane with overlaid spectra for a single fragment
Figure 142. Fragments pane with stacked spectra for a single fragment
Isotopes Pane
The isotopes pane displays isotopic pattern results for all adducts of a compound according to
the threshold and deviation parameters defined in the unknown method.
To identify or confirm the presence of a compound, the resulting score percentage from
isotopic pattern matching must be higher than the specified fit threshold percentage.
• An isotope peak is not found if its intensity, relative to the monoisotopic ion’s intensity, is
more than the specified intensity deviation percentage away from the theoretical relative
intensity of the isotope ion.
• An isotope peak is found if its measured m/z is less than the specified mass deviation
amount away from its expected m/z.
Intensity
threshold
specified in
the method
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 369
To specify the peak threshold and deviation parameters, refer to Chapter 6, “Using the
Method Development Mode for Unknown Screening Methods,” in the TraceFind er Lab
Director User Guide.
The Isotopes pane displays the isotopes in one of three ways:
•All Isotopes
•Multi-Isotopes
•Individual Isotopes
All isotopes panes use a shortcut menu so that you can specify how to display the data. See
Isotopes pane shortcut menu commands.
All Isotopes
The All Isotopes view displays a composite of all isotopes found in the compound. The
application scales the window with respect to the most intense isotope. The most intense
isotope is usually the first isotope unless you are using halogenated compounds. The
application displays the measured peak as a solid red line; the application displays the
expected peak as a dashed blue line.
The application displays these headers for the All Isotopes view:
Scan range for
the isotopes
Retention time
range for all scans
Number of averaged scans
Processing filter
Sample file name
Percentage of total isotopes to the
number of matched isotopes
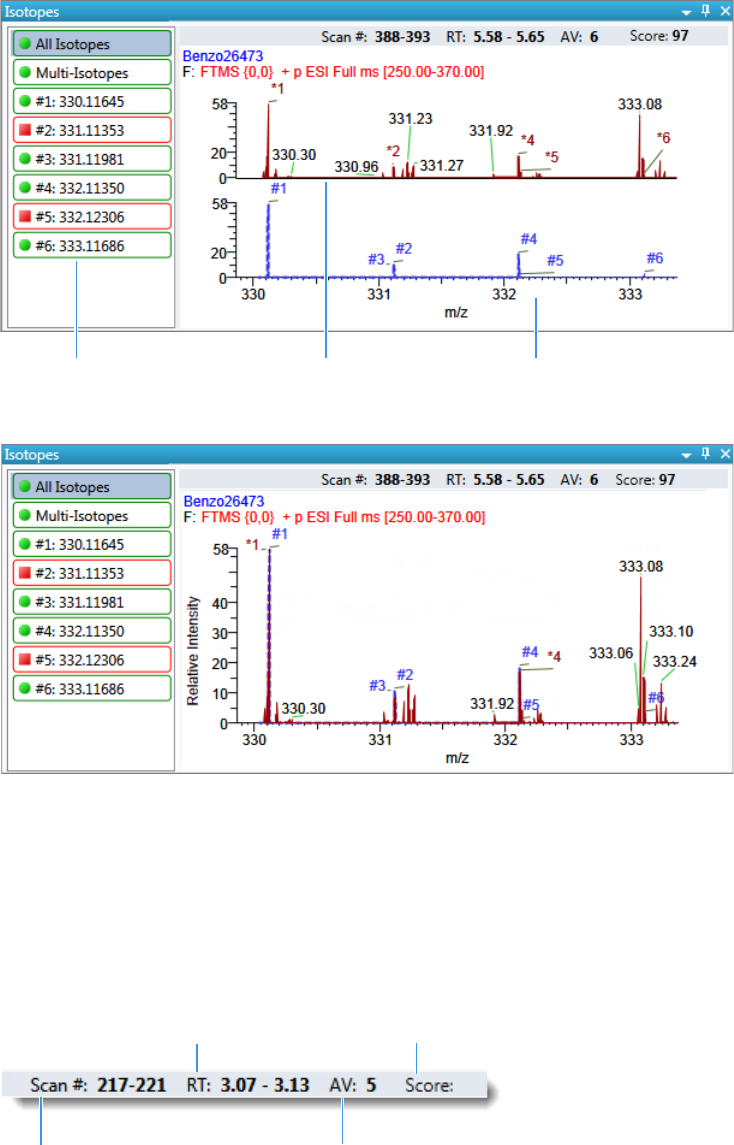
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
370 TraceFinder User Guide Thermo Scientific
Figure 143. Isotopes pane with stacked spectra for all isotopes
Figure 144. Isotopes pane with overlaid spectra for all isotopes
Multi-Isotopes
The Multi-Isotopes view displays individual plots for each isotope. You can individually stack
or overlay the plots for each isotope.
The application displays these headers for the Multi-Isotopes view:
Measured spectrum Expected spectrumExpected m/z of the isotope
Measured spectrum in redExpected spectrum in blue
Scan range for the isotopes
Retention time
range for all scans
Number of averaged scans
Percentage of total isotopes to the
number of matched isotopes
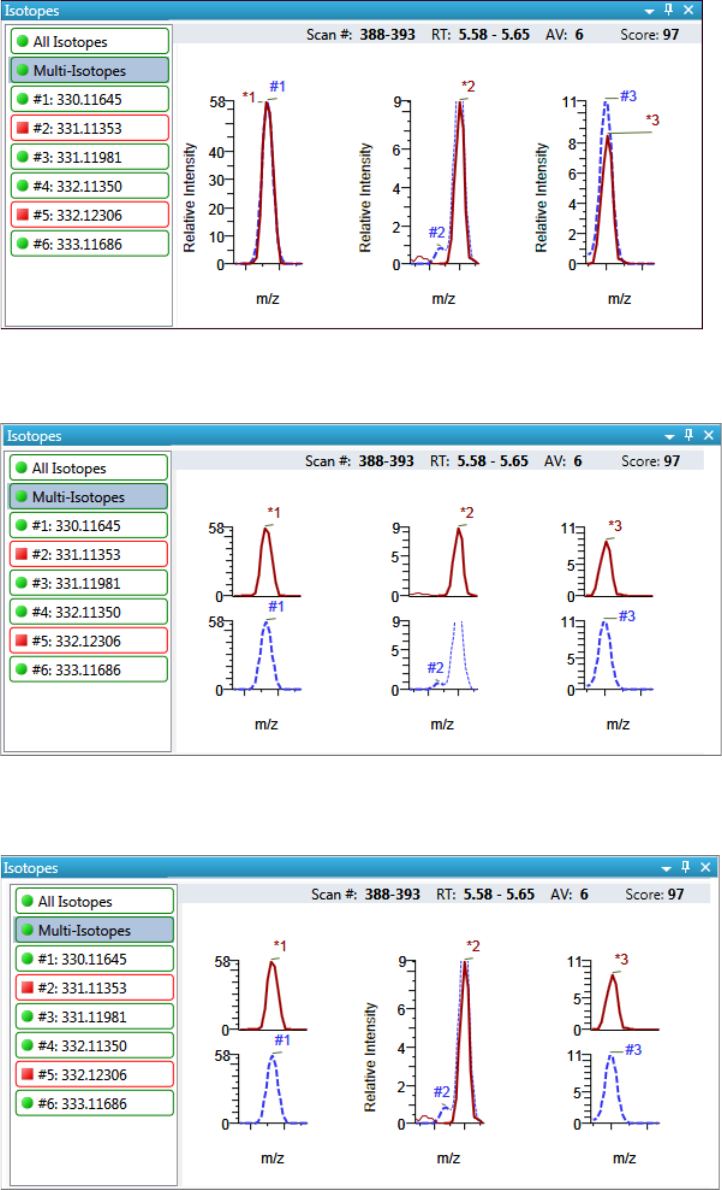
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 371
Figure 145. Isotopes pane with overlaid spectra for multi-isotopes
Figure 146. Isotopes pane with stacked spectra for multi-isotopes
Figure 147. Isotopes pane with stacked and overlaid spectra for multi-isotopes
Expected spectrum in blue Measured spectrum in red
Measured spectrum in redExpected spectrum in blue
Measured spectrum in redExpected spectrum in blue
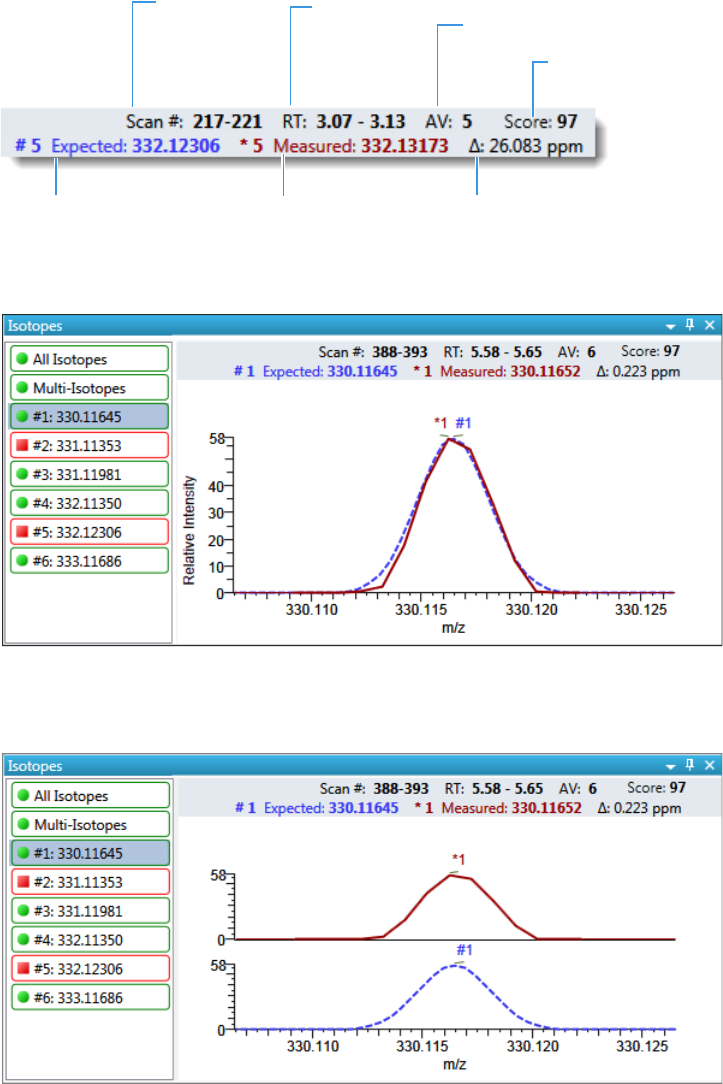
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
372 TraceFinder User Guide Thermo Scientific
Individual Isotopes
The individual isotopes view displays the expected and measured peaks for a single isotope.
The application displays these headers for the individual isotopes view:
Figure 148. Isotopes pane with overlaid spectra for a single isotope
Figure 149. Isotopes pane with stacked spectra for a single isotope
Scan range for
the isotopes
Retention time
range for all
scans
Number of averaged
scans
Expected m/z
for isotope 5
Measured m/z
for isotope 5
Delta between expected
and measured m/z
Percentage of total isotopes to
the number of matched isotopes
Measured spectrum in redExpected spectrum in blue
Measured spectrum in redExpected spectrum in blue
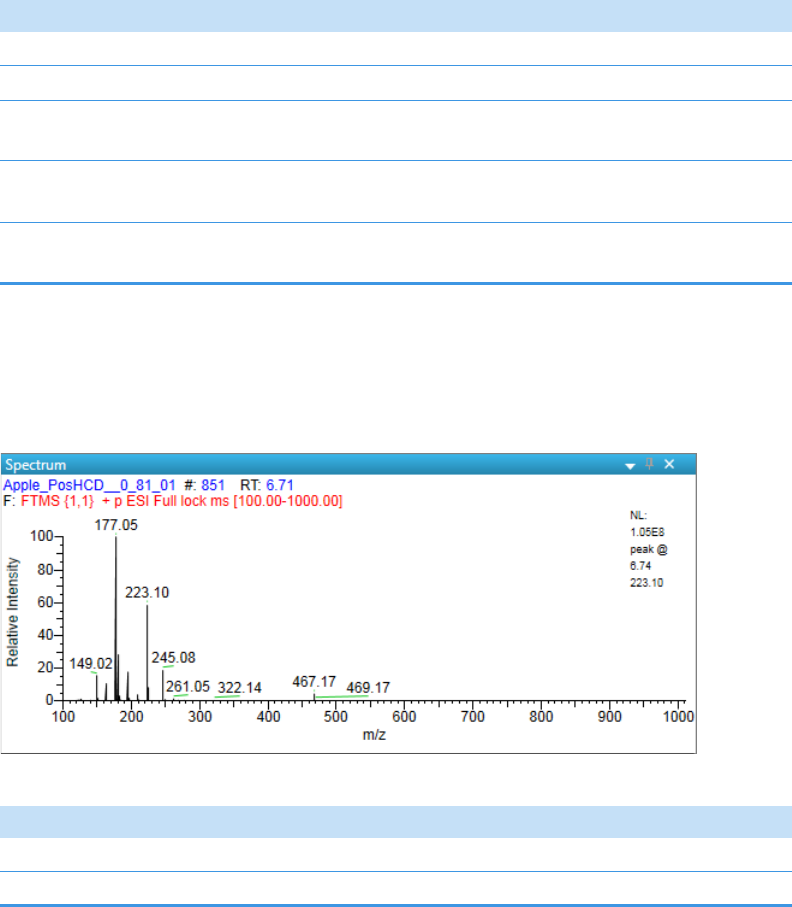
6 Using the Analysis Mode for Unknown Screening Batches
Working in Data Review for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 373
Spectrum Pane
When data is available, the plot width is the full mass range in the raw data file. Otherwise,
the application scales the width to the scan range.
Figure 150. Spectrum pane
Table 76. Isotopes pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
Display Overlay Spectra
Display Stack Spectra
Overlays the two spectrum displays, or stacks the simulated
spectrum and the peak apex spectrum.
Show/Hide Noise Label Adds a noise label to each peak. Expected isotope peaks
(displayed in blue) do not display a noise label.
Show/Hide Resolution
Label
Adds a resolution label to each peak. Expected isotope peaks
(displayed in blue) do not display a resolution label.
Table 77. Spectrum pane shortcut menu commands
Command Description
Reset Scaling Resets the original scaling after a zoom operation.
Copy to Clipboard Copies the graphic display to the Clipboard.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Report View for Unknown Screening Batches
374 TraceFinder User Guide Thermo Scientific
Working in the Report View for Unknown Screening Batches
The Report View displays example reports for the current batch. You must have an open
batch to use the features in the Report View.
Follow these procedures:
•To open the Report View
•To preview a report
•To generate a report as a PDF, an Excel, or a CSV file
•To print a report
•To display a generated report
•To e d i t a re po r t t e m p l a t e
•To create a new report template
To open the Report View
Click Report View in the navigation pane.
The application opens the Report View.
To preview a report
1. In the Template pane, select a report template.
The template list shows all the report templates that you configured in the Configuration
console. Refer to Chapter 1, “Using the Configuration Console,” in the TraceFi nder Lab
Director User Guide.
Figure 151. Example template list for unknown screening batches
2. Click Preview, .
The application opens the Report Designer, showing the report information for the
current batch in the selected report template format.
For details about using the Report Designer, see Chapter 7, “Using the Report Designer.”
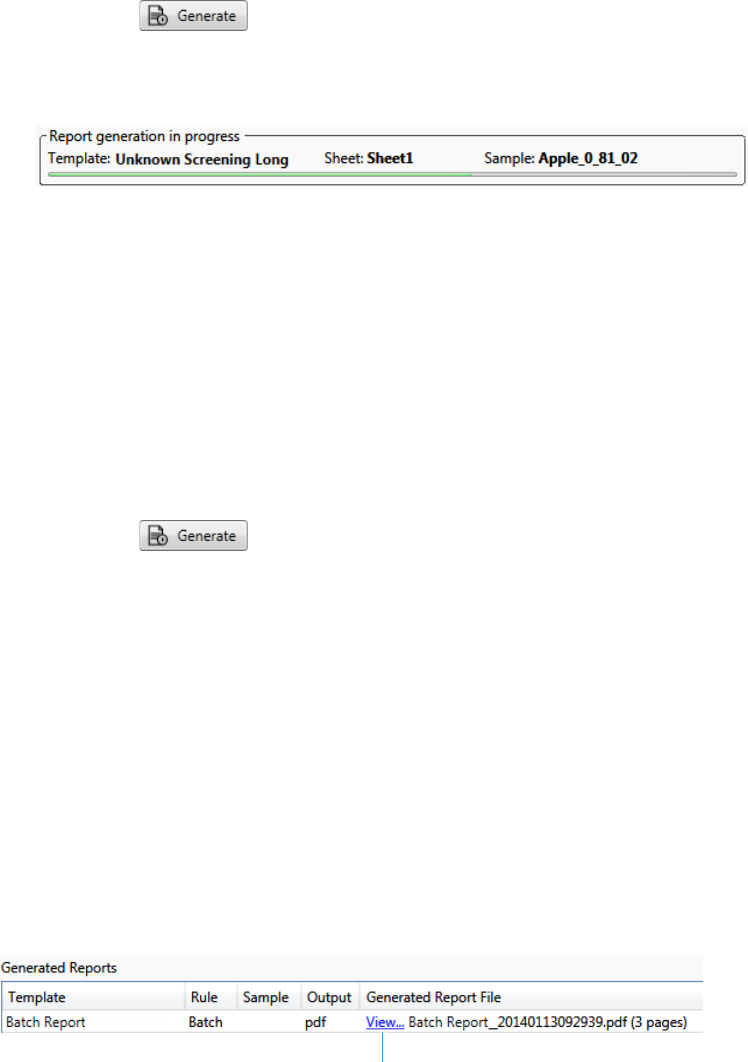
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Report View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 375
To generate a report as a PDF, an Excel, or a CSV file
1. In the Template pane, select a report template.
2. Select the check box for each of the file types that you want to create: PDF, Excel, or
CSV.
3. Click Generate, .
The application does the following:
• Displays a green progress bar as it generates the reports.
• Creates a report for the current batch as a PDF, an Excel, or a CSV file, using the
selected report template format.
• Adds information about the generated report to the Generated Reports pane.
For details about the Generated Reports pane, see Report View.
• Saves the report files to the …\TraceFinderData\4.0\Projects\batch\ReportOutput
folder.
To print a report
1. In the Template pane, select a report template.
2. Select the check box for the Print file format.
3. Click Generate, .
The application does the following:
• Creates a report for the current batch using the selected report template format.
• Prints the report to your default printer.
• Adds information about the generated report to the Generated Reports pane.
For details about the Generated Reports pane, see Report View.
• Saves the report files to the …\TraceFinderData\4.0\Projects\batch\ReportOutput
folder.
To display a generated report
In the Generated Reports pane, click View for the report that you want to see.
Figure 152. Generated Reports pane showing a PDF report
The application opens the output file.
Opens the generated file.
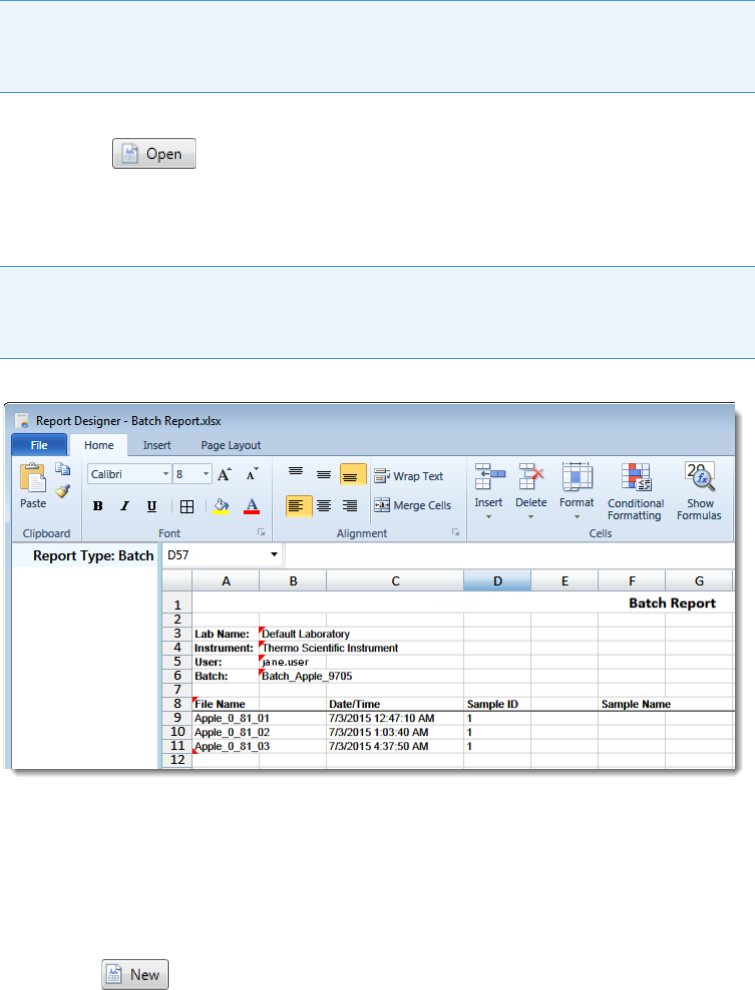
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Report View for Unknown Screening Batches
376 TraceFinder User Guide Thermo Scientific
To edit a report template
1. In the Template pane, select a report template.
2. Click Open, .
The application opens the Report Designer showing the template in an Excel spreadsheet.
See Report Designer showing the template for the selected report.
Figure 153. Report Designer showing the template for the selected report
3. Use the features in the Report Designer to edit the template.
See Chapter 7, “Using the Report Designer.”
4. When you finish your changes, choose File > Save from the Report Designer menu bar.
To create a new report template
1. Click New, .
The application opens the Report Designer showing an empty template in an Excel
spreadsheet. See Report Designer showing a new, empty template.
The Report Type is None.
In the left pane, the spreadsheet lists all samples in the current batch and all compounds
in the method used for the batch.
Note You cannot edit report templates that were provided with the TraceFinder
application; however, you can open a TraceFinder template, make changes, and save it
to a new template name.
Note When user security is activated, you must have Template Editing permission to
edit report templates created by your laboratory. If the Open button is not active, user
security is activated and you do not have Template Editing permission.
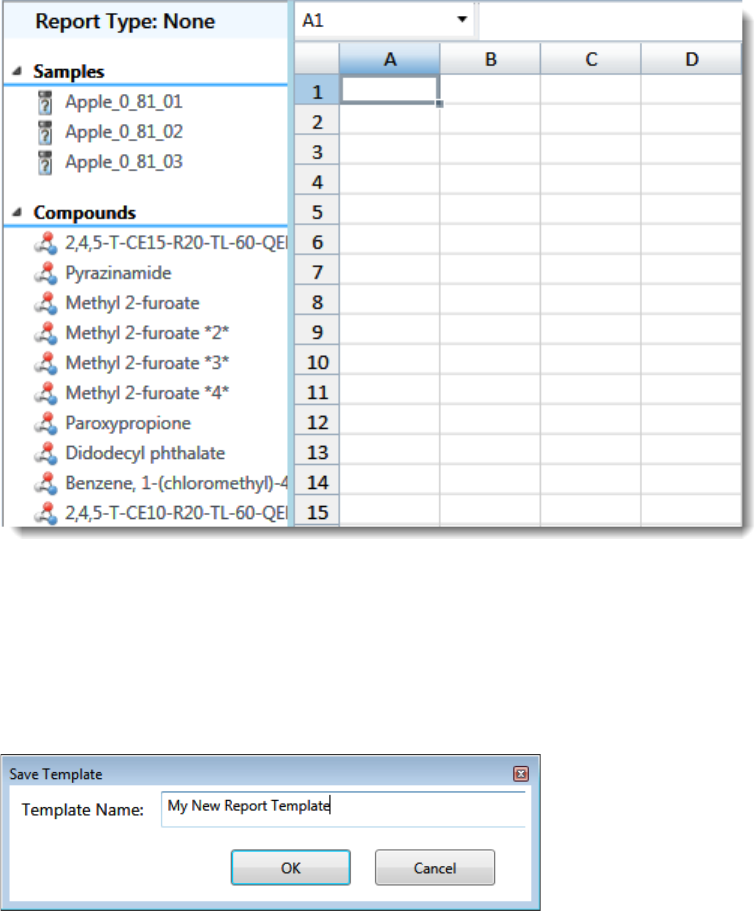
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Report View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 377
Figure 154. Report Designer showing a new, empty template
2. Use the features in the Report Designer to create the report template.
See Chapter 7, “Using the Report Designer.”
3. When you finish your changes, choose File > Save from the Report Designer menu bar.
The Save Template dialog box opens.
Figure 155. Save Template dialog box
4. Type a name for the new report template and click OK.
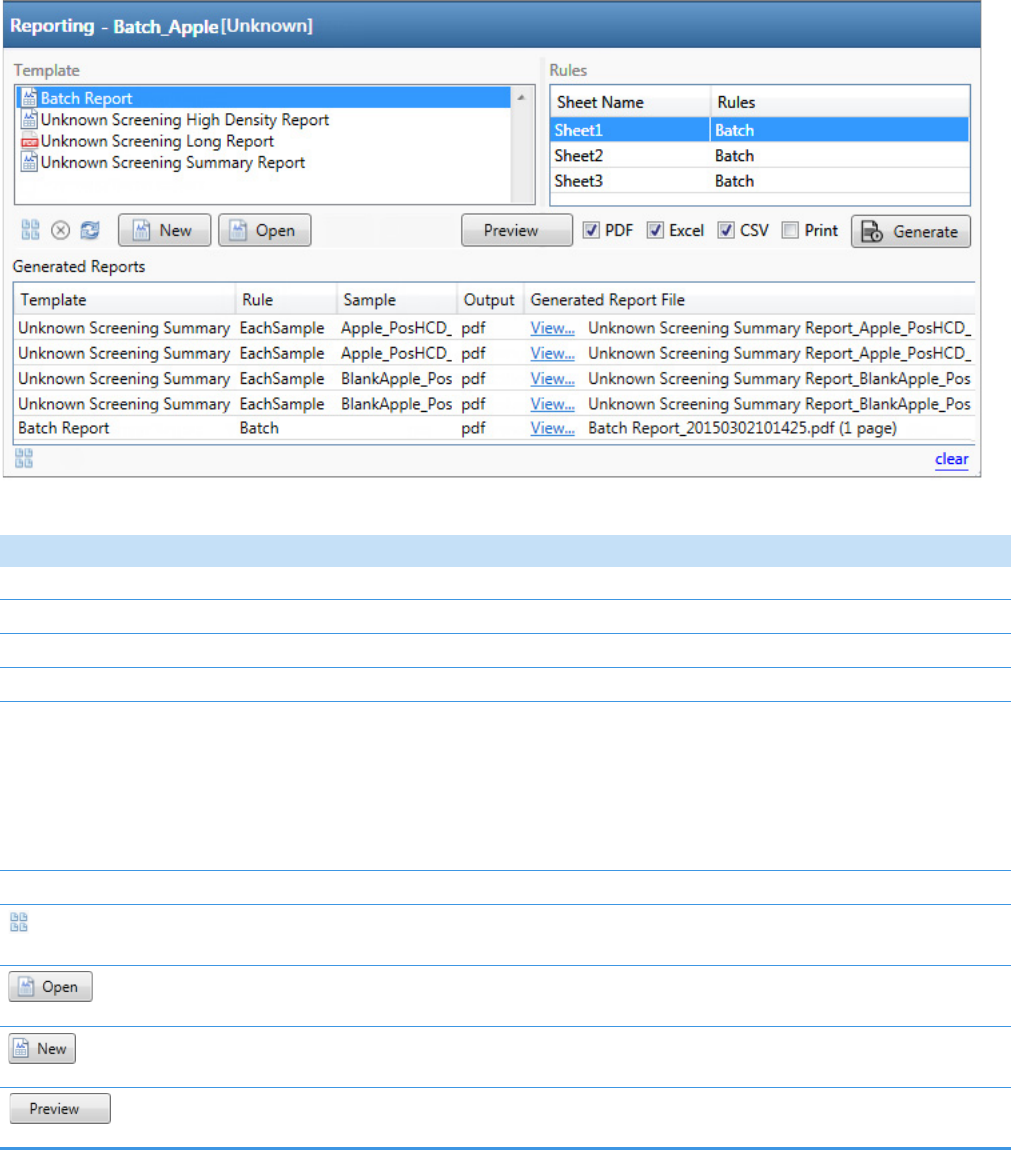
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Report View for Unknown Screening Batches
378 TraceFinder User Guide Thermo Scientific
Report View
Use the features in the Report View to display example reports for the current batch.
Figure 156. Report View
Table 78. Report View parameters (Sheet 1 of 2)
Parameter Description
Template
Displays all available unknown screening report templates.
Rules
Sheet Name Specifies each sheet in the report.
Rules Specifies the type of data used in each sheet in the selected report.
•Batch
• EachSample
• SampleType: SampleType
•CompoundType: CompoundType
•SampleCustomFormula:
Buttons
View Report Templates Displays the C:\TraceFinderData\4.0\Templates\ReportTemplates folder that contains
all report templates.
Opens the selected report template in the Report Designer.
Opens a blank report template in the Report Designer.
Opens the Report Designer showing the report information for the current batch in the
selected report template format.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Report View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 379
PDF Writes the generated report to a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
Excel Writes the generated report to a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
CSV Saves the generated report as a PDF file in the
…\TraceFinderData\4.0\Projects\batch\ReportOutput folder.
When the report contains multiple sheets, the application writes each sheet as a separate
CSV file.
Print Prints the generated report to your default printer.
Generates the selected type of reports for the current batch using the selected report
template.
Generated Reports
Template Report template used for the report. See Example template list for unknown screening
batches.
Rule Type of data used in each sheet of the report. See Rules.
Sample For sample-level reports, the name of each sample in the report.
Output Type of output specified for the report: PDF, Excel, CSV, or Print.
Generated Report File Lists the output file name for each report in the …\TraceFinderData\4.0\Projects folder.
View Displays the generated output file.
View Generated Reports Displays the C:\TraceFinderData\4.0\Projects folder that contains all report outputs.
Clear Removes all reports from the Generated Reports display. This does not delete the reports
from the C:\TraceFinderData\4.0\Projects folder.
Table 78. Report View parameters (Sheet 2 of 2)
Parameter Description
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Local Method View for Unknown Screening Batches
380 TraceFinder User Guide Thermo Scientific
Working in the Local Method View for Unknown Screening Batches
A local method is a copy of a master method associated with a batch. You can edit only the
local copy of the method, or you can edit the master method and overwrite the local copy
with the edited master method.
In the Local Method view, you can edit the local method parameters. A local method is a copy
of a master method associated with a batch. Local methods are named Batch_MasterMethod.
To open the Local Method View
1. Click Analysis in the navigation pane.
2. Click Local Method.
The Local Method View for the currently selected batch opens.
You can edit many of the method parameters in a local method. Editing the local method
does not affect parameters in the master method.
For detailed descriptions of method parameters, refer to Chapter 6, “Using the Method
Development Mode for Unknown Screening Methods,” in the Tra c eFin der L ab D i re ct o r
User Guide.
3. Enter any local changes to the method.
4. When you have finished editing the local method, choose File > Save.
5. To process the batch or create new reports with the edited local method, return to the
Batch View and submit the batch.
To overwrite the local method with the master method in the Batch View
In the Batch View, click Update.
The application overwrites the local method with the master method of the same name.
You can use this feature to overwrite an edited local method with the original master
method or to overwrite the local method with an updated master method.
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Local Method View for Unknown Screening Batches
Thermo Scientific TraceFinder User Guide 381
Figure 157. Local Method View
6 Using the Analysis Mode for Unknown Screening Batches
Working in the Local Method View for Unknown Screening Batches
382 TraceFinder User Guide Thermo Scientific

Thermo Scientific TraceFinder User Guide 383
7
Using the Report Designer
Use the features in the Report Designer to create or edit report templates.
You can create new report templates or edit your laboratory’s report templates. You cannot
edit report templates that were provided with the TraceFinder application; however, you can
open a TraceFinder template, make changes, and save it to a new template name.
When user security is activated, you must have Template Editing permission to edit report
templates created by your laboratory. To use the administration tools to set Reports
permissions, refer to the instructions in the TraceFinder Administrator Console User Guide.
With the Report Designer, you can do any of the following to create or edit a report template:
• Make changes to the template as you would in an Excel application.
• Add or remove graphics.
• Add repeated areas of many types.
• Format fonts and colors.
• Add or edit formulas.
• Edit template text and headers.
In addition to the procedures in this chapter, you can view video instructions at
http://mytracefinder.com/plugins/reporting/.
Contents
•Editing a Template
•Toolbar Reference
•Plot Display Properties
•Quick Tips
Note To edit or create a template, you must have an open batch.
7 Using the Report Designer
Editing a Template
384 TraceFinder User Guide Thermo Scientific
Editing a Template
Use the Report Designer to add or remove graphics, add tables, add repeated areas, and
format headings, data, or cells.
Follow these procedures:
•To insert graphics into a report template
•To resize an inserted graphic
•To move an inserted graphic
•To i n se r t a t a b l e
•To r e m ov e a n i n se r t ed t a bl e
•To make changes to a table
•To format header text in the spreadsheet
•To format data in the report grid
•To format cells in the report grid
•To add repeated areas
•To insert a new sheet in a report
To insert graphics into a report template
1. Click the Insert tab.
The Insert page displays the available objects that you can insert in a quantitation, a target
screening, or an unknown screening report template.
Figure 158. Quan Plots features for quantitation batch report templates
For a description of each graphic element, see Insert Toolbar – General Features.
Figure 159. Screening Plots features for target screening batch report templates
For a description of each graphic element, see Insert Toolbar – Target Screening Reports.
Figure 160. Unknown Plots features for unknown screening batch report templates
7 Using the Report Designer
Editing a Template
Thermo Scientific TraceFinder User Guide 385
For a description of each graphic element, see Insert Toolbar – Unknown Screening
Reports.
2. In the template grid, drag to select cells where you want to place the item.
3. Select a sample from the list of samples in the left pane.
The application uses graphical data from the selected sample when you insert a graphical
plot.
4. Click a graphical plot in the toolbar to insert into the selected cells.
The application inserts the selected plot in the template grid.
Figure 161. Inserted peak chromatogram
To resize an inserted graphic
1. Right-click the graphic to make it editable.
Figure 162. Editable graphic
Note When the method for a quantitation or target screening batch includes
unknown screening, the Insert features for the report templates include Unknown
Plots.
Note You can also select a single row, and the application inserts the upper left corner
of the graphic at the first cell, using a minimum default size.
The border indicates that the
graphic is editable.
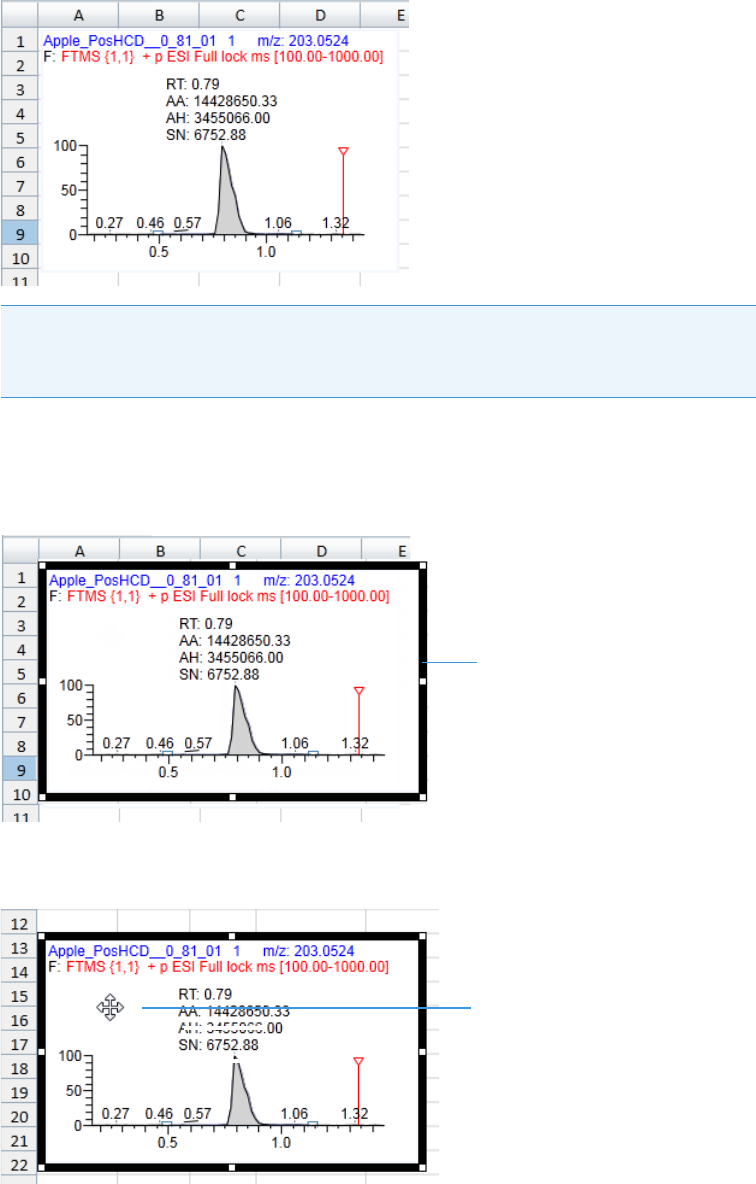
7 Using the Report Designer
Editing a Template
386 TraceFinder User Guide Thermo Scientific
2. Grab a corner of the graphic and stretch it to the new size.
Figure 163. Resized graphic
To move an inserted graphic
1. Right-click the graphic to make it editable.
Figure 164. Editable graphic
2. Click the graphic (notice the cursor change), and drag it to a new location.
Figure 165. Moved graphic
Note In this example, the original graphic was too small to display all the
chromatogram data. Stretching the graphic reveals the sample name, mass, filter, and
labeling.
The border indicates that the
graphic is editable.
Cursor for moving a graphic
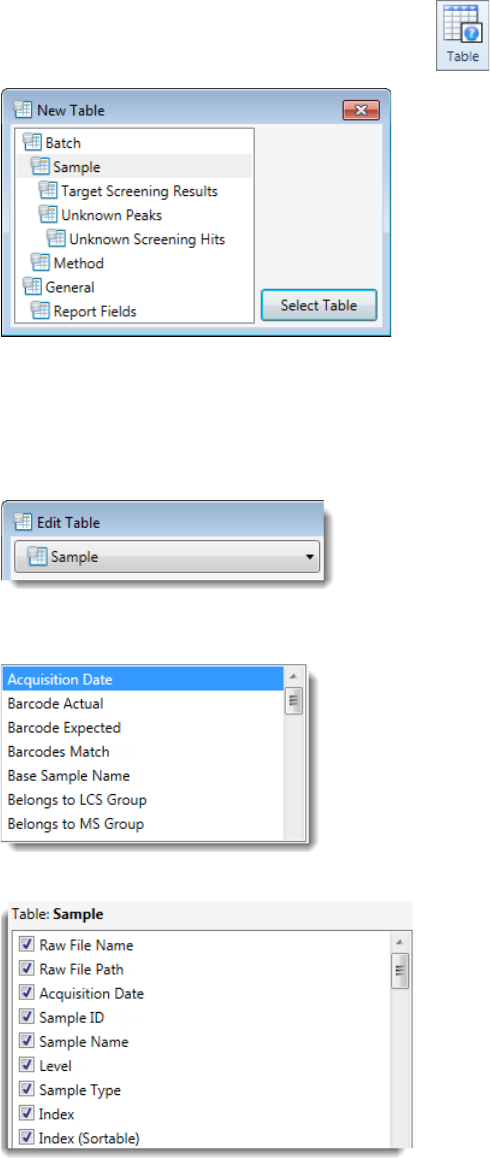
7 Using the Report Designer
Editing a Template
Thermo Scientific TraceFinder User Guide 387
To insert a table
1. Select a row in the report grid where you want to insert the table.
2. Click the Insert tab, and then click the Ta bl e icon, .
The New Table dialog box opens.
3. Select the type of table that you want to insert and click Select Table.
The Edit Table Dialog Box opens.
The dropdown list in the upper left corner displays the data type for the table you
selected.
The left pane displays an alphabetical list of all available fields for the selected data type
that you can include in the table. To display fields for a different data type, see step 4.
The right pane lists the fields that are included in the new table.
7 Using the Report Designer
Editing a Template
388 TraceFinder User Guide Thermo Scientific
You can select as many additional fields as you want. The table you create displays each
selected field as a table column. By default, all available fields for the original data type are
included in the table. The first field in the list is the leftmost column in the table. The last
field in the list is the rightmost column in the table. To change the order of the columns,
see step 6.
4. To add fields to the table from a different data type, do the following:
a. Click the dropdown list and select a different data type.
The left pane displays an alphabetical list of all available fields for the new data type.
b. Select a field in the left pane and then click .
The application moves the selected field to the rightmost pane to indicate that it is
included in the inserted table.
5. To select a field that you do not want in the table, in the rightmost pane, clear the check
box next to the field name.
6. To move a column up or down in the table, select the field name in the rightmost pane
and click the Up arrow, , or Down arrow, .
7. To edit the header for a table column, select the field in the list on the right, and then type
new header text in the Header box in the Field Details area.
The default header name is the same as the field name.
8. To find the formula for a field, select the field.
The application displays the formula in the Formula box.
This video demonstrates how to use the Report Designer to create your own formula:
Working with Formulas.
Field not selected
Field selected (blue background)
Details for the selected field
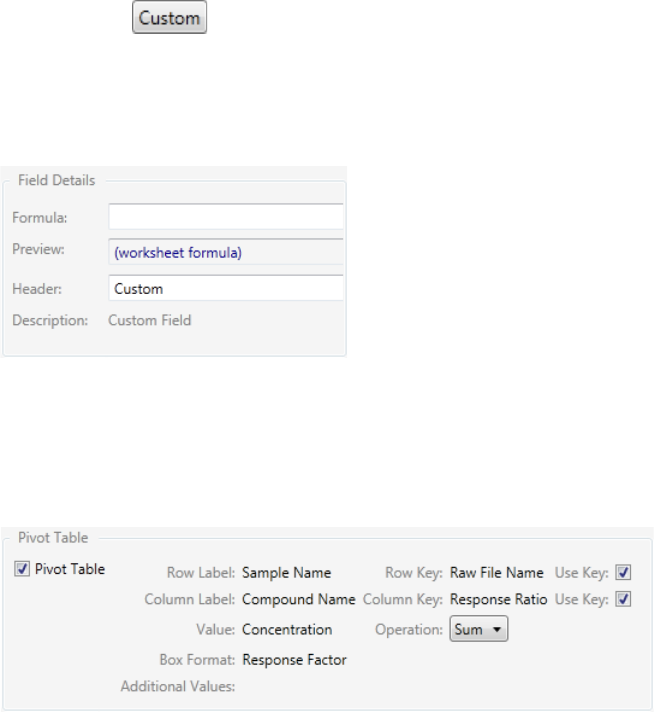
7 Using the Report Designer
Editing a Template
Thermo Scientific TraceFinder User Guide 389
9. To create a custom column, do the following:
a. Click Custom, .
The application adds a Custom field to the fields list.
b. Select the Custom field in the list.
The application display the details for the new, custom column.
c. Enter values for the Formula and Header.
10. To create a pivot table, do the following:
a. Select the Pivot Table check box.
The Pivot Table area expands to display the pivot table options.
b. Select the Use Key check box for the row label.
c. Select the Use Key check box for the column label.
d. Select the operation that you want to use to calculate the aggregate value: Sum,
Count, Average, First, or Last.
11. When you have made all your table selections, click OK.
The application inserts the table into the report template.
To remove an inserted table
1. Select all rows and cells in the table.
2. Right-click and choose Delete – Delete Cells and Shift Cells Left.
The application removes the entire table from the report template.
7 Using the Report Designer
Editing a Template
390 TraceFinder User Guide Thermo Scientific
To make changes to a table
1. In the table that you want to edit, right-click and choose Data Table.
The Edit Table Dialog Box opens, displaying all of the available fields for the selected
template. The Edit Table dialog box is virtually identical to the dialog box that you used
to create a table.
2. Edit the fields for the table and click OK.
To format header text in the spreadsheet
1. In the Report Designer window, select the header in the spreadsheet table.
To change all headers, select the entire header row.
To change a single header, select the specific header cell.
2. Click the Home tab.
3. Use the Font icons to change the font format.
To format data in the report grid
1. Select the data that you want to format.
To format all rows in a column, select the header row.
To format a single cell of data, select only that cell.
2. Click the Home tab and use the toolbar icons to edit the font or cells as appropriate:
• Change the font or font size.
• Make the selected text bold.
• Make the selected text italics.
Tip You can also use the SHIFT key to select sequential cells or the CTRL key to
select nonsequential cells anywhere in the grid.
Tip You can also use the SHIFT key to select sequential cells or the CTRL key to
select nonsequential cells anywhere in the grid.
7 Using the Report Designer
Editing a Template
Thermo Scientific TraceFinder User Guide 391
• Underline the selected text.
• Apply borders to the currently selected cells.
• Increase or decrease font size.
• Apply color to the background for selected cells.
• Change the font color.
To format cells in the report grid
1. Select the cells that you want to modify:
To change all cells in a row, select the entire row.
To change a single cell, select only that cell.
2. Click the Home tab and use the Alignment or Cells toolbar icons to edit the cells.
3. To align the cell text to the top, center, or bottom of the cell, click one of these icons:
.
4. To align the cell text to the left, center, or right of the cell, click one of these icons:
.
5. To make all contents visible within a cell, click Wrap Text.
6. To join selected cells into one cell, click Merge Cells.
7. To insert cells, rows, or columns into the template, click Insert.
8. To delete rows or columns from the template, click Delete.
9. To change the row height or column width, organize sheets, or protect or hide cells, click
Format.
10. To highlight or emphasize useful cells based on specific criteria, click Conditional
Formatting to use data bars, color scales, or icon sets.
11. To show formulas for selected cells instead of the resulting value, click Show Formulas.
Tip You can also use the SHIFT key to select sequential cells or the CTRL key to
select nonsequential cells.
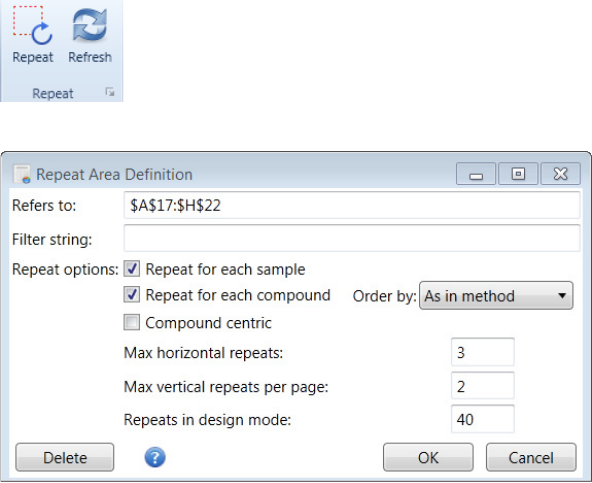
7 Using the Report Designer
Editing a Template
392 TraceFinder User Guide Thermo Scientific
To add repeated areas
1. To define a repeat area, select the cells that you want to repeat.
You can repeat only one area per worksheet, but you can repeat it many times. You cannot
insert data tables beneath a repeated area. See Example of repeated areas.
2. Click the Insert tab and then click Repeat.
The Repeat Area Definition dialog box opens.
3. To filter the repeat area according to the filter criteria, type the string syntax in the Filter
String box.
4. Select to repeat for each sample, for each compound, or for both.
5. Define whether the repeat area repeats first for samples or first for compounds:
• To have the repeat area repeat first for samples and then for compounds, clear the
Compound Centric check box.
• To have the repeat area repeat first for compounds and then for samples, select the
Compound Centric check box.
6. To define the number of times to repeat horizontally before wrapping to the next row,
type a number in the Max Horizontal Repeats box.
7. To define the number of times to repeat vertically before inserting an auto-page break,
type a number in the Max Vertical Repeats Per Page box.
8. To define the maximum suggested number of repeats, type a number in the Repeats in
Design Mode box.
This maximum number of repeats is intended to limit the size of the report while you are
editing the report in the Report Designer. When generating an actual report, the
application does not enforce this limit.
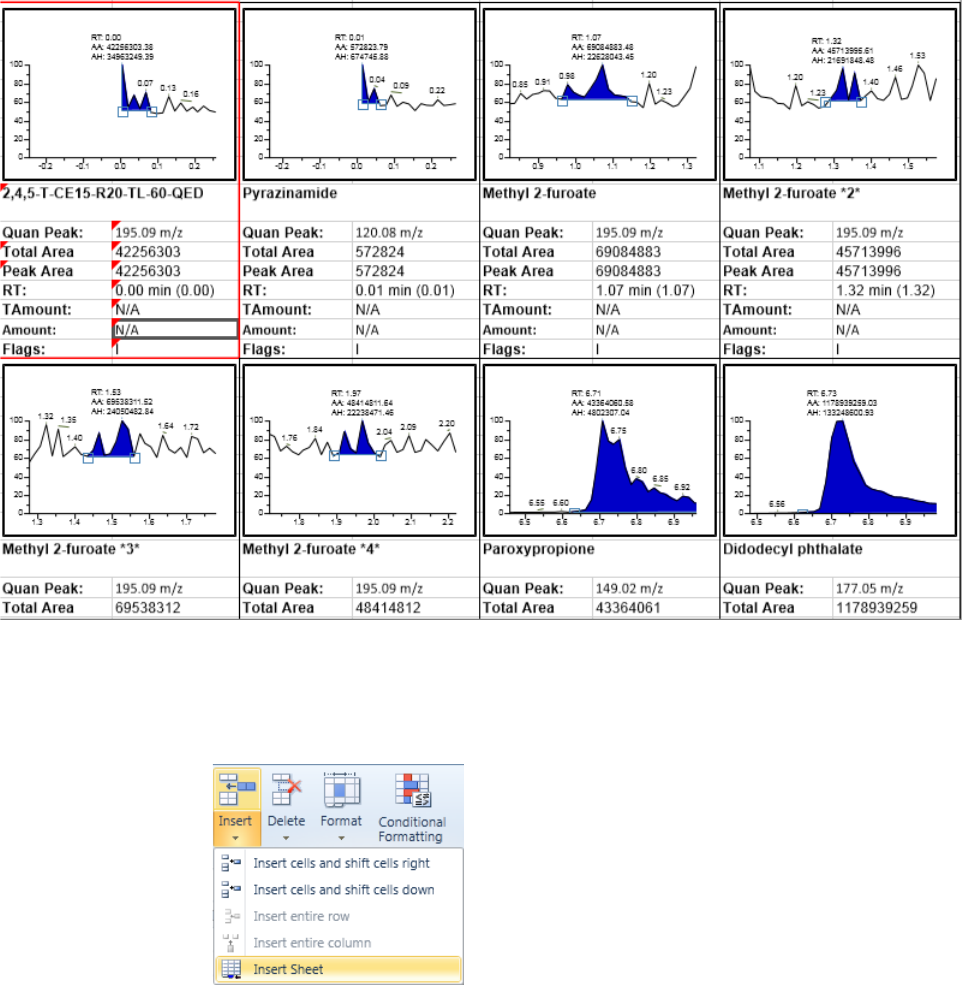
7 Using the Report Designer
Editing a Template
Thermo Scientific TraceFinder User Guide 393
In this example, the repeated area (outlined in red) displays the peak chromatogram, target
peak, total area, peak area, retention time, theoretical amount, actual amount, and reported
flags, if any, for each compound.
Figure 166. Example of repeated areas
To insert a new sheet in a report
1. Click the Home tab.
2. In the Cells area, choose Insert Sheet from the Insert menu.
The application adds a new sheet to the report.
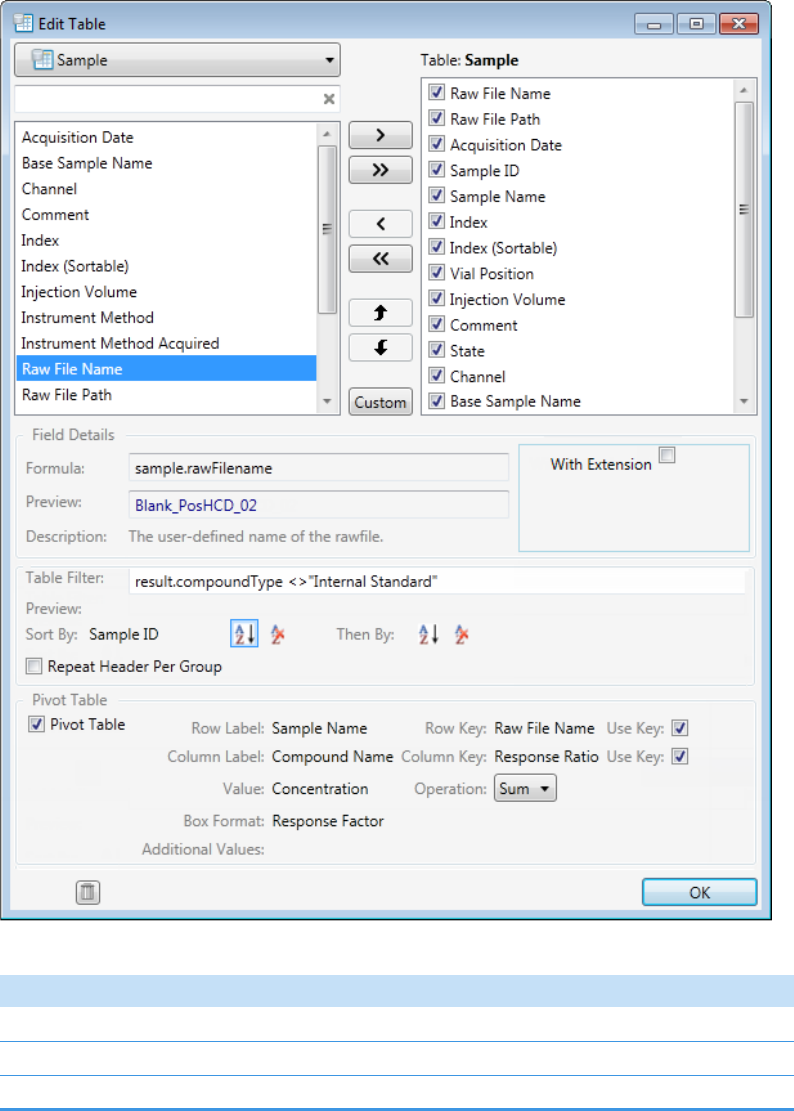
7 Using the Report Designer
Editing a Template
394 TraceFinder User Guide Thermo Scientific
Edit Table Dialog Box
Use the options in the Edit Table dialog box to edit the fields in a table.
Figure 167. Edit Table dialog box
Table 79. Edit Table dialog box parameters (Sheet 1 of 2)
Parameter Description
Dropdown List Specifies the type of table you are editing.
Field List Lists all available table fields.
Table Displays the selected table type and the fields included in the table.
7 Using the Report Designer
Editing a Template
Thermo Scientific TraceFinder User Guide 395
Field Details
Formula Specifies the functions and arguments used to define the field.
Preview Specifies the name of the raw data file used for the report.
Header Specifies the header name for the column in the spreadsheet.
Description Specifies the type of field.
Table Filter
Table Filter Specifies the filter used for the sample compounds.
Preview –
Sort By Sorts the data by ascending or descending values.
Pivot Table
Pivot Table Enables the Pivot Table features.
Row Label –
Row Key –
Column Label –
Column Key –
Value –
Operation Specifies the operation used to calculate the aggregate value.
Valid values: Sum, Count, Average, First, Last
Box Format –
Additional Values –
Delete Deletes the selected table from the template.
Table 79. Edit Table dialog box parameters (Sheet 2 of 2)
Parameter Description
7 Using the Report Designer
Toolbar Reference
396 TraceFinder User Guide Thermo Scientific
Toolbar Reference
The Report Designer includes the following toolbars.
•Home Toolbar
•Insert Toolbar – General Features
•Insert Toolbar – Quantitation Reports
•Insert Toolbar – Target Screening Reports
•Insert Toolbar – Unknown Screening Reports
•Page Layout Toolbar
Home Toolbar
Use the options in the Home toolbar to modify fonts, align cell data, and format cells.
Table 80. Home toolbar options (Sheet 1 of 2)
Option Description
Clipboard
Paste Paste the contents of the Clipboard. You can also paste only formating or only a
formula.
Copy Copy text or graphics to use in another place.
Font
Font and size lists Use the dropdown lists to change the font or font size.
Bold Make the selected text bold.
Italic Make the selected text italics.
Underline Underline the selected text.
Border Apply borders to the currently selected cells.
Font size Increase or decrease the font size.
Fill color Apply color to the background for selected cells.
Font color Change the font color.
Alignment
Align the cell text to the top, center, or bottom of the cell.
Align the cell text to the left, center, or right of the cell.
Wrap text Make all contents visible within a cell.
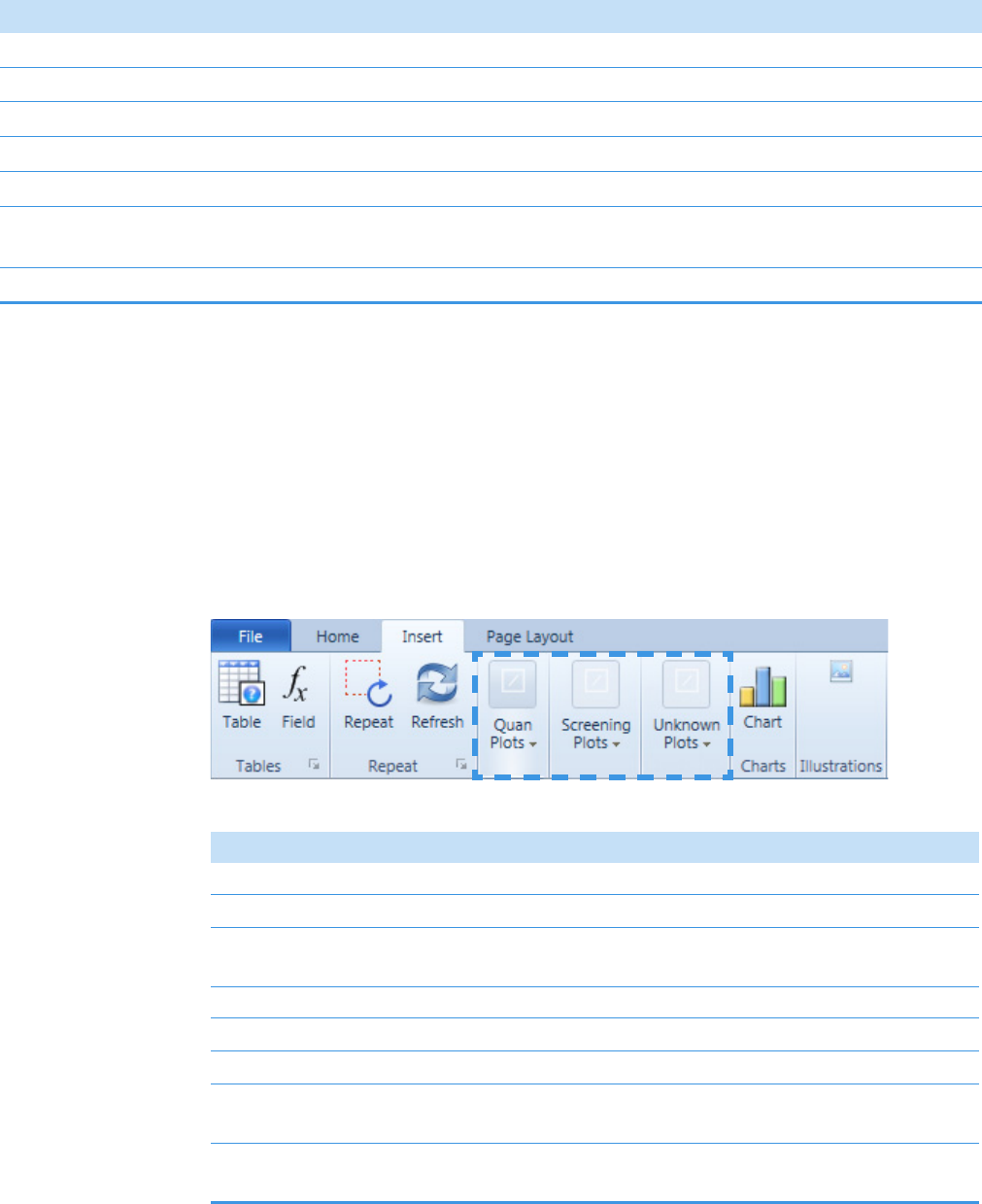
7 Using the Report Designer
Toolbar Reference
Thermo Scientific TraceFinder User Guide 397
Insert Toolbar – General Features
Use the features on the Insert page to create and edit tables, formulas, graphic plots, charts,
and illustrations.
Graphic plots can represent data from quantitative, target screening, or unknown screening
data. The Insert page displays the available data plots for the type of batch: quantitation,
quantitation plus unknown screening, target screening, target screening plus unknown
screening, or unknown screening only.
The following graphic outlines all three plot types for illustrative purposes only.
Merge cells Join selected cells into one cell.
Cells
Insert Insert cells, rows, columns, or sheets into a template.
Delete Delete rows or columns from a template.
Format Change the row height or column width, organize sheets, or protect or hide cells.
Conditional Formatting Based on specific criteria, highlight or emphasize useful cells using data bars, color
scales, and icon sets.
Show Formulas Show formulas for selected cells instead of the resulting value.
Table 80. Home toolbar options (Sheet 2 of 2)
Option Description
Table 81. Insert toolbar options (Sheet 1 of 2)
Option Description
Tables
Table Create a table to manage and analyze data.
Field Edit the formula for a field by choosing functions and editing
arguments.
Repeat
Repeat Repeat text, cell, or graphic elements.
Refresh Update the view to reflect recent changes.
Quan Plots Can be stand-alone or used with Unknown plots. For descriptions of
quantitation plots, see Insert Toolbar – Quantitation Reports.
Screening Plots Can be stand-alone or used with Unknown plots. For descriptions of
target screening plots, see Insert Toolbar – Target Screening Reports.
7 Using the Report Designer
Toolbar Reference
398 TraceFinder User Guide Thermo Scientific
Insert Toolbar – Quantitation Reports
Use the options in the Insert toolbar to add graphics, plots, and other objects to a template or
report. You can also set up repeating objects and define functions.
For each inserted plot, you can customize the display properties. See Quantitation and Target
Screening Plots.
Unknown Plots
Can be stand-alone or used with either Quan plots or Screening plots.
For descriptions of unknown screening plots, see Insert Toolbar –
Unknown Screening Reports.
Charts
Adds a custom chart. To define the chart, double-click the inserted
blank chart.
Illustrations
Insert a picture from a file.
Table 81. Insert toolbar options (Sheet 2 of 2)
Option Description
Table 82. Insert toolbar Quantitation options (Sheet 1 of 2)
Option Description
Adds a sample TIC.
For information about sample TIC data in the Data Review results, see Peaks Pane.
Adds a peak chromatogram.
For information about peak chromatogram data in the Data Review results, see Quan Peak.
Adds an ion overlay.
For information about ion overlay data in the Data Review results, see Ion Overlay.
Adds a quantitation peaks overlay plot.
For information about quantitation peaks overlay data in the Data Review results, see Quan Peaks
Overlay.
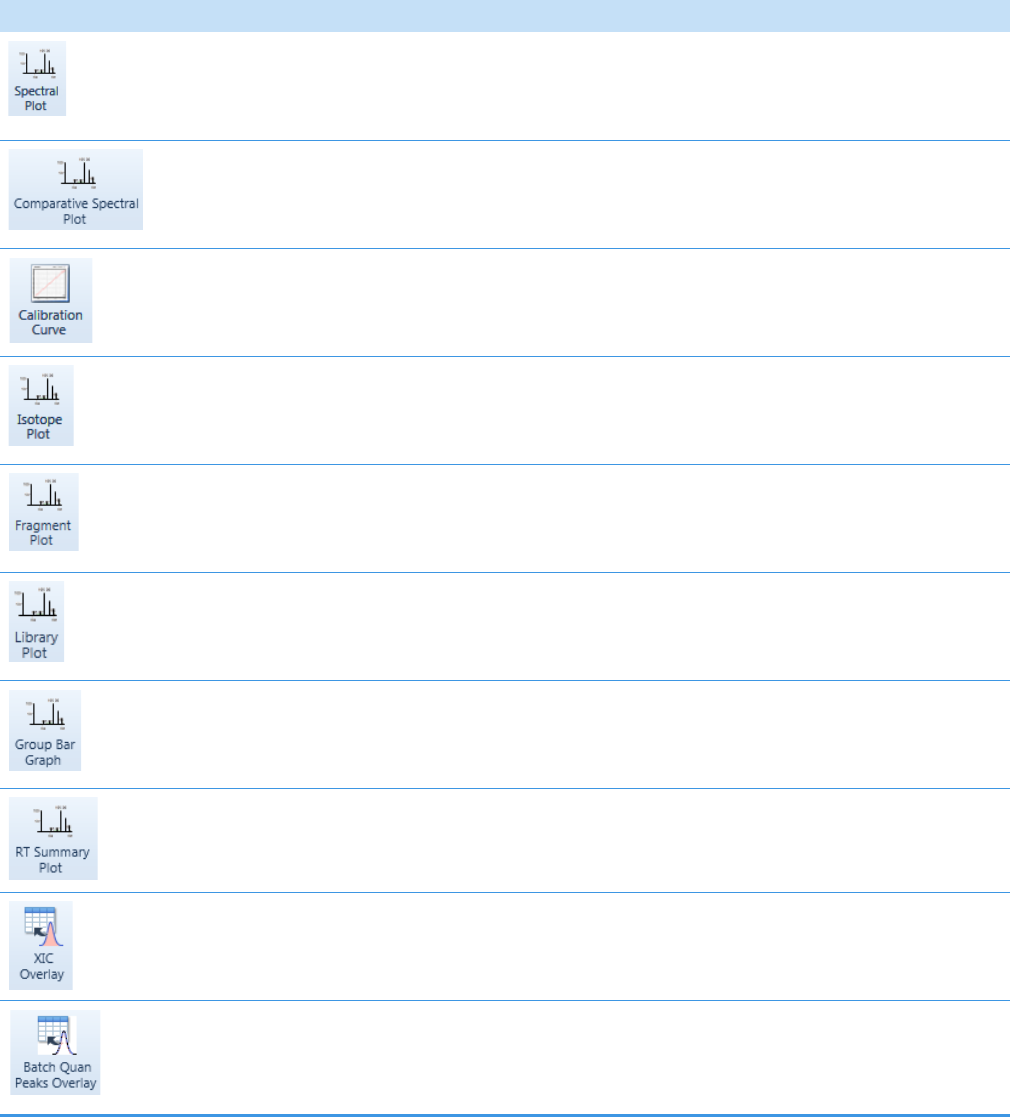
7 Using the Report Designer
Toolbar Reference
Thermo Scientific TraceFinder User Guide 399
Adds a spectral plot.
For information about spectral data in the Data Review results, see Spectra.
Adds a comparative spectral plot.
For information about comparative spectral data in the Data Review results, see Spectra.
Adds a calibration curve plot.
For information about calibration curve data in the Data Review results, see Calibration Curve.
Adds an isotope plot to display the number of isotopes found, the score, a pass/fail flag, and a plot
of the isotopes.
For information about isotope plot data in the Data Review results, see Isotope.
Adds a fragment plot.
For information about fragment plot data in the Data Review results, see Fragments.
Adds a library plot.
For information about library plot data in the Data Review results, see Library Match.
Adds a group averages plot.
For information about group averages data in the Data Review results, see Group Averages.
Adds an RT summary plot.
For information about retention time summary data in the Data Review results, see Retention
Time Summary.
Adds an XIC summary plot.
For information about XIC summary data in the Data Review results, see XIC Overlay.
Adds a Batch Quan Peaks Overlay plot that displays all quantitative peaks in the batch.
Table 82. Insert toolbar Quantitation options (Sheet 2 of 2)
Option Description
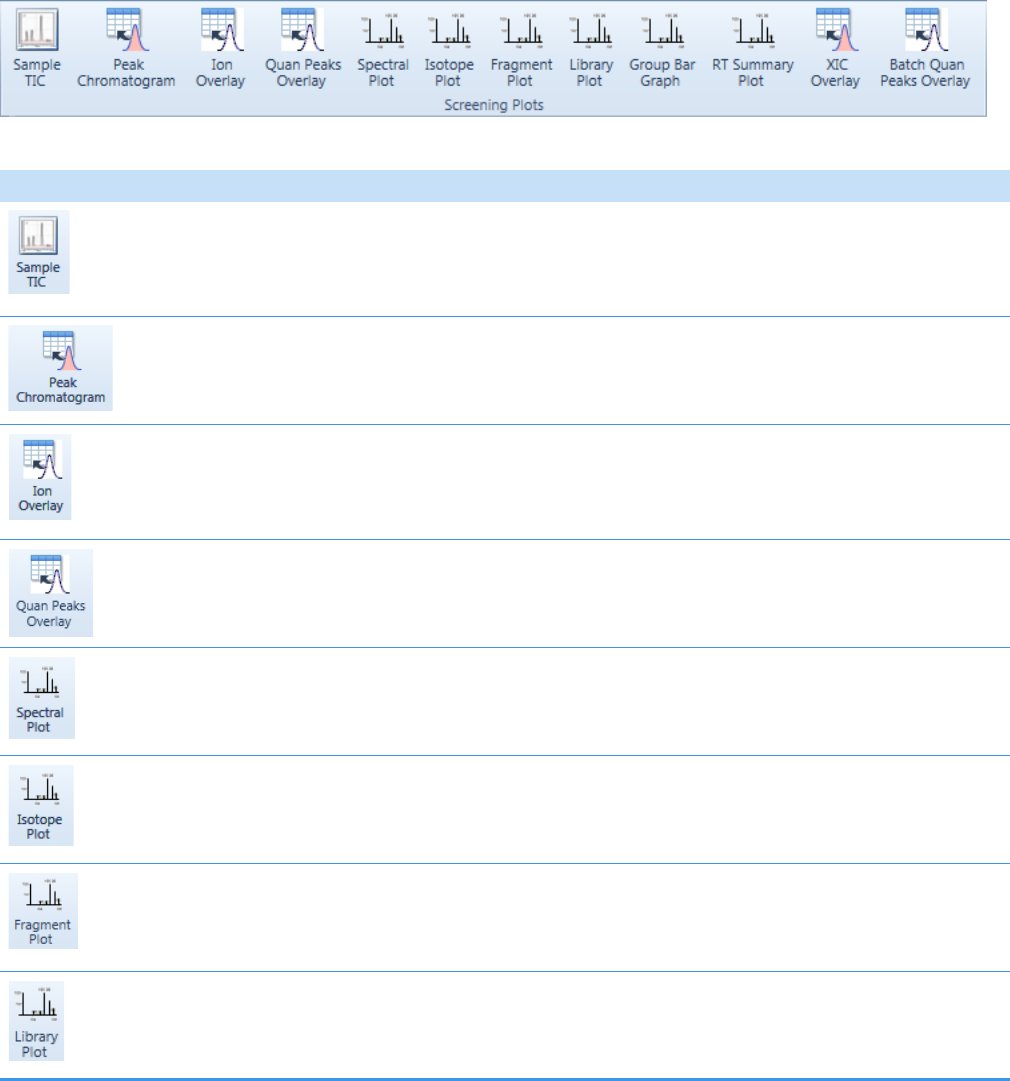
7 Using the Report Designer
Toolbar Reference
400 TraceFinder User Guide Thermo Scientific
Insert Toolbar – Target Screening Reports
Use the options in the Insert toolbar to add graphics, plots, and other objects to a template or
report. You can also set up repeating objects and define functions.
For each inserted plot, you can customize the display properties. See Quantitation and Target
Screening Plots.
Table 83. Insert toolbar Target Screening options (Sheet 1 of 2)
Option Description
Adds a sample TIC plot.
Adds a peak chromatogram plot.
For information about peak chromatogram data in the Data Review results, see
Chromatogram Pane.
Adds an ion overlay plot.
Adds a quantitation peaks overlay plot.
Adds a spectral plot.
For information about spectral plot data in the Data Review results, see Spectrum.
Adds an isotope plot to display the number of isotopes found, the score, a pass/fail flag, and a
plot of the isotopes.
For information about isotope plot data in the Data Review results, see Isotopes.
Adds a fragment plot.
For information about fragment plot data in the Data Review results, see Fragments.
Adds a library plot.
For information about library plot data in the Data Review results, see Library.
7 Using the Report Designer
Toolbar Reference
Thermo Scientific TraceFinder User Guide 401
Insert Toolbar – Unknown Screening Reports
Use the options in the Insert toolbar to add graphics, plots, and other objects to a template or
report. You can also set up repeating objects and define functions.
For each inserted plot, you can customize the display properties. See Unknown Screening
Plots.
Adds a group averages plot.
For information about group averages data in the Data Review results, see Groups.
Adds an RT summary plot.
Adds an XIC summary plot.
Adds a Batch Quan Peaks Overlay plot that displays all quantitative peaks in the batch.
Table 83. Insert toolbar Target Screening options (Sheet 2 of 2)
Option Description
Table 84. Insert toolbar Unknown Screening options (Sheet 1 of 2)
Option Description
Adds a peak chromatogram.
For information about peak chromatogram data in the Data Review results, see Peak
Chromatogram Pane.
Adds a TIC chromatogram.
For information about TIC chromatogram data in the Data Review results, see TIC
Chromatogram Pane.
Adds a spectral plot.
For information about spectral plot data in the Data Review results, see Spectrum Pane.
7 Using the Report Designer
Toolbar Reference
402 TraceFinder User Guide Thermo Scientific
Adds an XIC overlay plot.
For information about XIC overlay data in the Data Review results, see XIC Overlay Pane.
Adds a library plot.
For information about library plot data in the Data Review results, see Library Search Pane.
Adds an XIC plot.
For information about XIC plot data in the Data Review results, see XIC Pane.
Adds a cross-sample peak overlay plot.
For information about cross-sample peak overlay data in the Data Review results, see Cross
Sample Peak Overlay Pane.
Adds a fragments plot.
For information about fragments plot data in the Data Review results, see Fragments Pane.
Adds an isotopes plot.
For information about isotopes plot data in the Data Review results, see Isotopes Pane.
Adds a chemical structure plot.
For information about chemical structure plot data in the Data Review results, see Chemical
Structure Pane.
Adds a group averages plot.
For information about group averages data in the Data Review results, see Group Averages
Pane.
Table 84. Insert toolbar Unknown Screening options (Sheet 2 of 2)
Option Description
7 Using the Report Designer
Toolbar Reference
Thermo Scientific TraceFinder User Guide 403
Page Layout Toolbar
Use the options in the Page Layout toolbar to adjust margins, orientation, paper size, and page
breaks. To see your changes, click Print Preview.
Table 85. Page Layout toolbar options
Option Description
View the report before printing it.
Define page details.
Select page margins for the current view or the entire document.
Switch the pages from portrait to landscape view.
Choose a page size for the current view or the entire document.
Specify where a new page begins in the printed copy.
Template Properties Open the Template Properties dialog box where you can specify
the expected batch type for the report template.
The Template Properties dialog box also displays the Used Range
so that you can identify when there is empty white space in the
report template. Extra white space to the right or left of the
report content can cause unwanted page breaks.

7 Using the Report Designer
Plot Display Properties
404 TraceFinder User Guide Thermo Scientific
Plot Display Properties
For plot types that you insert in the spreadsheet, you can change the display properties.
To change the plot display properties
1. Insert a plot in the spreadsheet.
2. Right-click the plot to select it.
3. Right-click the plot again to display the shortcut menu.
4. Choose Properties.
The Plot Properties dialog box opens, displaying the editable display properties for the
selected plot type.
The following topics show the plots in the report templates and describe their properties.
•Quantitation and Target Screening Plots
•Unknown Screening Plots
Quantitation and Target Screening Plots
To customize the display in the quantitation and target screening reports, use the Plot
Properties dialog box for each plot.
•Sample TIC Plot
•Peak Chromatogram Plot
•Ion Overlay Plot
•Quan Peaks Overlay Plot
•Spectral Plot
•Comparative Spectral Plot (quantitation reports only)
•Calibration Curve Plot (quantitation reports only)
•Isotope Plot, Fragment Plot, or Library Plot
•Group Bar Graph Plot
•RT Summary Plot
•XIC Overlay Plot
•Batch Quan Peaks Overlay Plot
Note This topic shows examples of all plot types; however, the following plots do not have
editable plot properties: Calibration Curve, Group Bar Graph, and RT Summary.
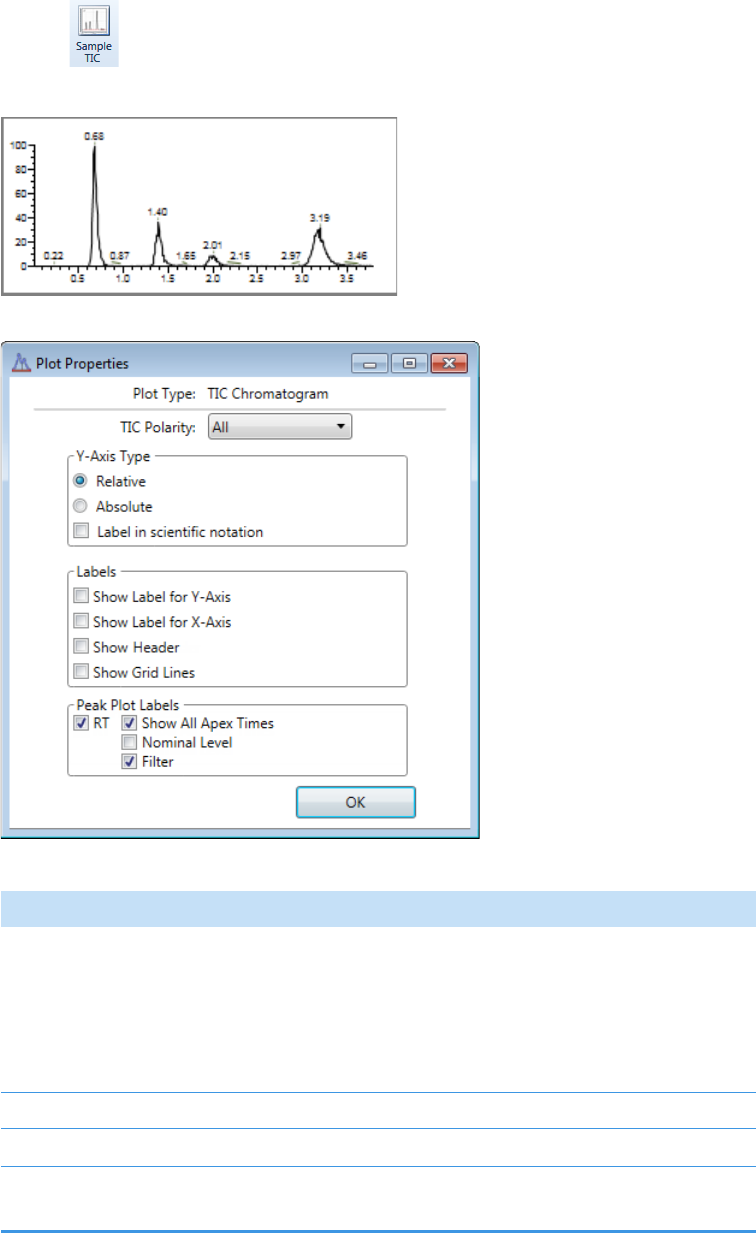
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 405
Sample TIC Plot
Use the icon to insert a sample TIC Chromatogram plot.
Figure 168. TIC Chromatogram plot
Figure 169. Plot Properties dialog box for a TIC Chromatogram plot
Table 86. Plot Properties parameters for a TIC Chromatogram (Sheet 1 of 2)
Command Description
TIC Polarity Specifies the polarity of adducts displayed in the plot.
All: Displays positive, negative, and analog adducts.
Positive: Displays only positive adducts.
Negative: Displays only negative adducts.
Analog: Displays only analog adducts.
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
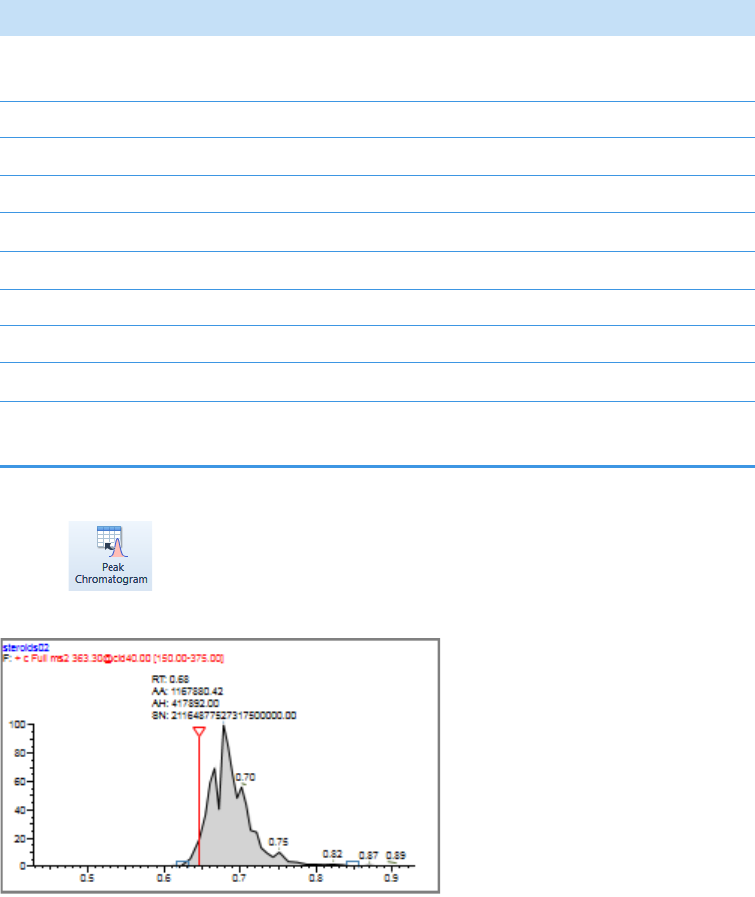
7 Using the Report Designer
Plot Display Properties
406 TraceFinder User Guide Thermo Scientific
Peak Chromatogram Plot
Use the icon to insert a Peak Chromatogram plot.
Figure 170. Peak Chromatogram plot
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the raw data file path at the top of the plot.
Show Grid Lines Displays the grid lines in the plot background.
Peak Plot Labels
RT Displays the retention time (RT) for each peak in the plot.
Show All Apex Times Displays all surrounding apex times in the plot.
Nominal Level Displays the nominal level (NL) values for each peak in the
plot.
Table 86. Plot Properties parameters for a TIC Chromatogram (Sheet 2 of 2)
Command Description
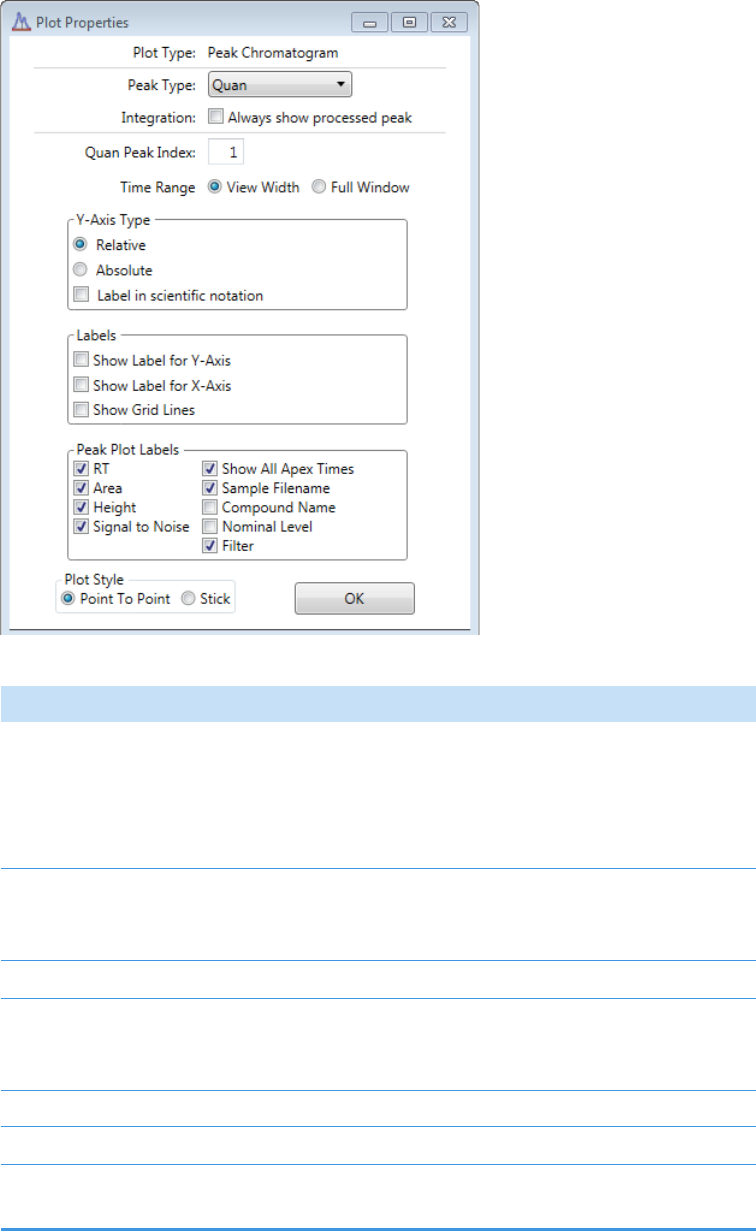
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 407
Figure 171. Plot Properties dialog box for Peak Chromatogram plot
Table 87. Plot Properties parameters for a Peak Chromatogram plot (Sheet 1 of 2)
Command Description
Peak Type Specifies the type of peak to display in the plot:
•Quan
• Confirming
• InternalStandard
• InternalStandardConfirming
Integration:
Always Show Processed
Peak
Specifies that the plot always displays the processed peak.
Quan Peak Index Valid values: 1 through 9
Time Range Specifies that the plot display one of the following:
• View Width: Zooms the display on the peak of interest.
• Full Window: Displays the full width of the scan.
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
7 Using the Report Designer
Plot Display Properties
408 TraceFinder User Guide Thermo Scientific
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Grid Lines Displays the grid lines in the plot background.
Peak Plot Labels
RT Displays the retention time (RT) label in the peak
chromatogram plot.
Area Displays the peak area (AA) label in the peak chromatogram
plot.
Height Displays the peak height (AH) label in the peak
chromatogram plot.
Signal to Noise Displays the signal-to-noise (SN) label in the peak
chromatogram plot.
Show All Apex Times Displays all surrounding apex times in the plot.
Sample Filename Displays the name of the sample file.
Compound Name Displays the compound name.
Nominal Level Displays the nominal level (NL) values for each peak in the
plot.
Filter Displays the filter used for the scan. The plot displays the
filter name below the raw data file name.
Plot Style
Point To Point Displays chromatograms in the following format:
Stick Displays chromatograms in the following format:
Table 87. Plot Properties parameters for a Peak Chromatogram plot (Sheet 2 of 2)
Command Description
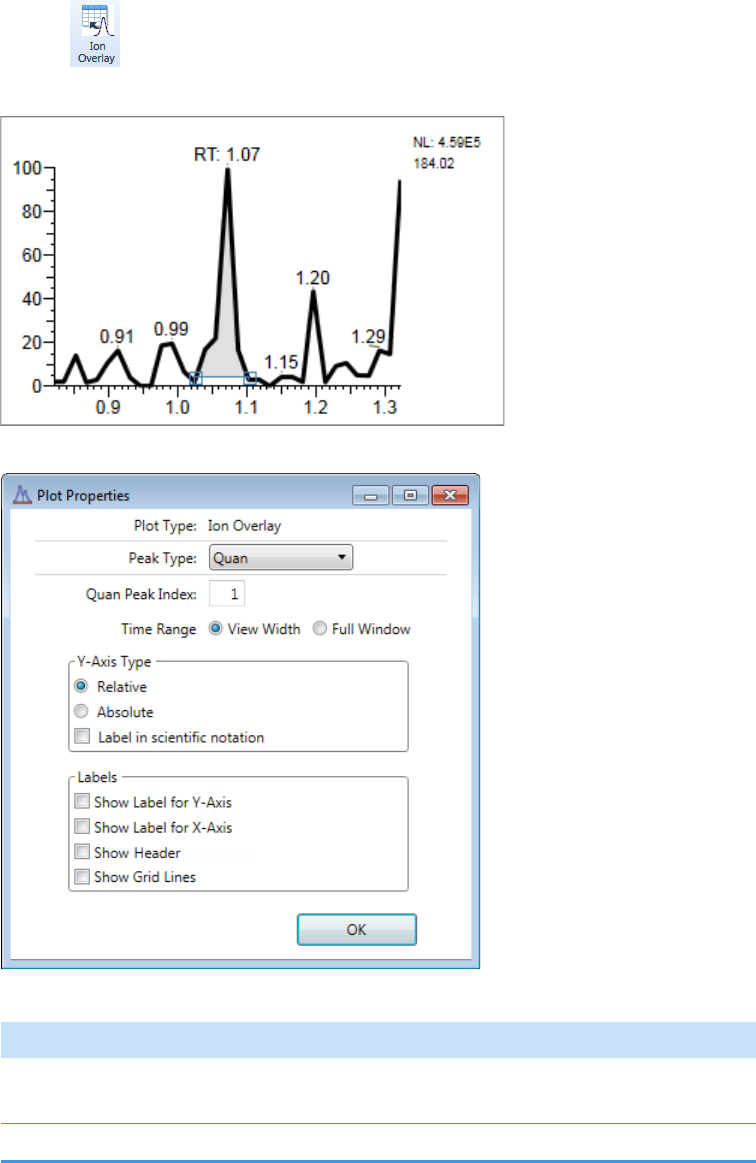
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 409
Ion Overlay Plot
Use the icon to insert an Ion Overlay plot.
Figure 172. Ion Overlay plot
Figure 173. Plot Properties dialog box for Ion Overlay plot
Table 88. Plot Properties parameters for an Ion Overlay plot (Sheet 1 of 2)
Command Description
Peak Type Specifies the type of peak to display in the plot: Quan or
InternalStandard
Quan Peak Index Valid values: 1 through 9
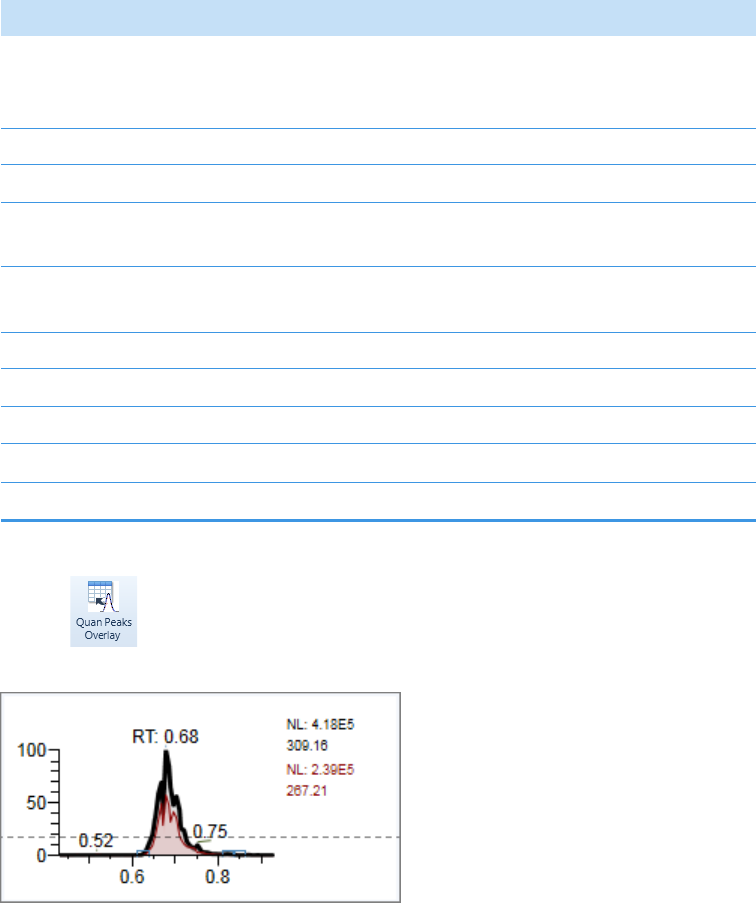
7 Using the Report Designer
Plot Display Properties
410 TraceFinder User Guide Thermo Scientific
Quan Peaks Overlay Plot
Use the icon to insert a quantitation peaks overlay plot.
Figure 174. Quan Peaks Overlay plot
Time Range Specifies that the plot display one of the following:
• View Width: Zooms the display on the peak of interest.
• Full Window: Displays the full width of the scan.
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the raw data file name and compound name.
Show Grid Lines Displays the grid lines in the plot background.
Table 88. Plot Properties parameters for an Ion Overlay plot (Sheet 2 of 2)
Command Description
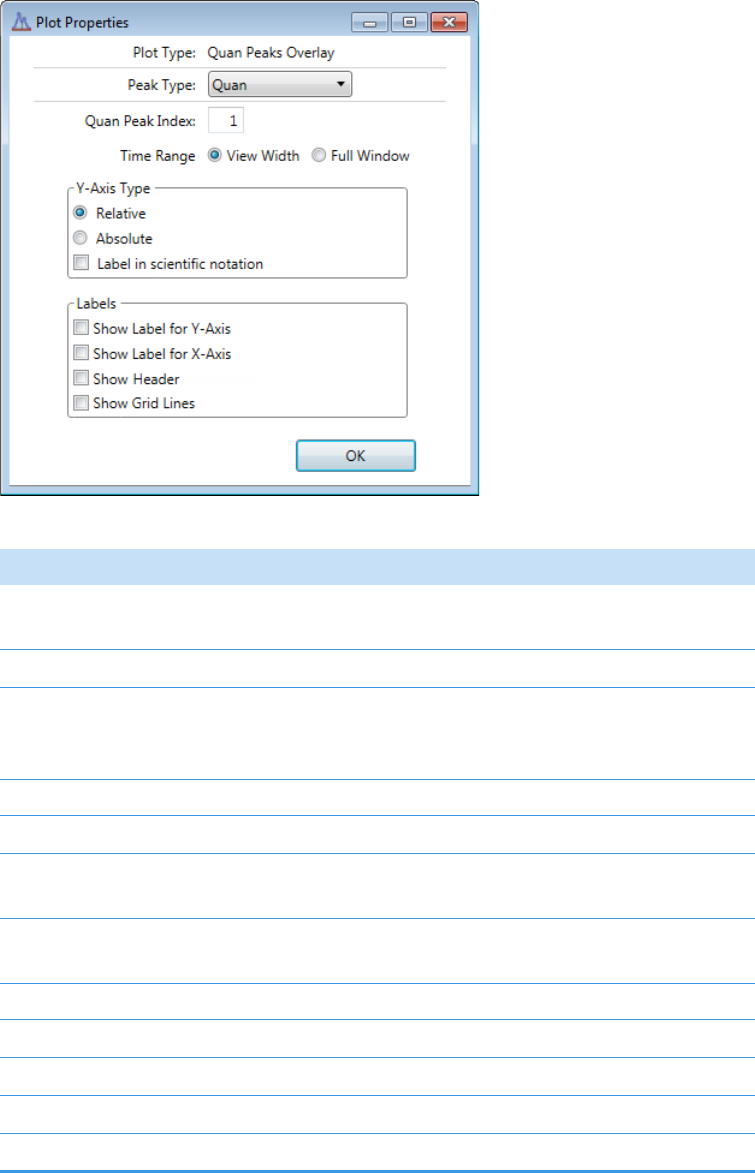
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 411
Figure 175. Plot Properties dialog box for a Quan Peaks Overlay plot
Table 89. Plot Properties parameters for a Quan Peaks Overlay plot
Command Description
Peak Type Specifies the type of peak to display in the plot: Quan or
InternalStandard
Quan Peak Index Valid values: 1 through 9
Time Range Specifies that the plot display one of the following:
• View Width: Zooms the display on the peak of interest.
• Full Window: Displays the full width of the scan.
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the raw data file name and compound name.
Show Grid Lines Displays the grid lines in the plot background.
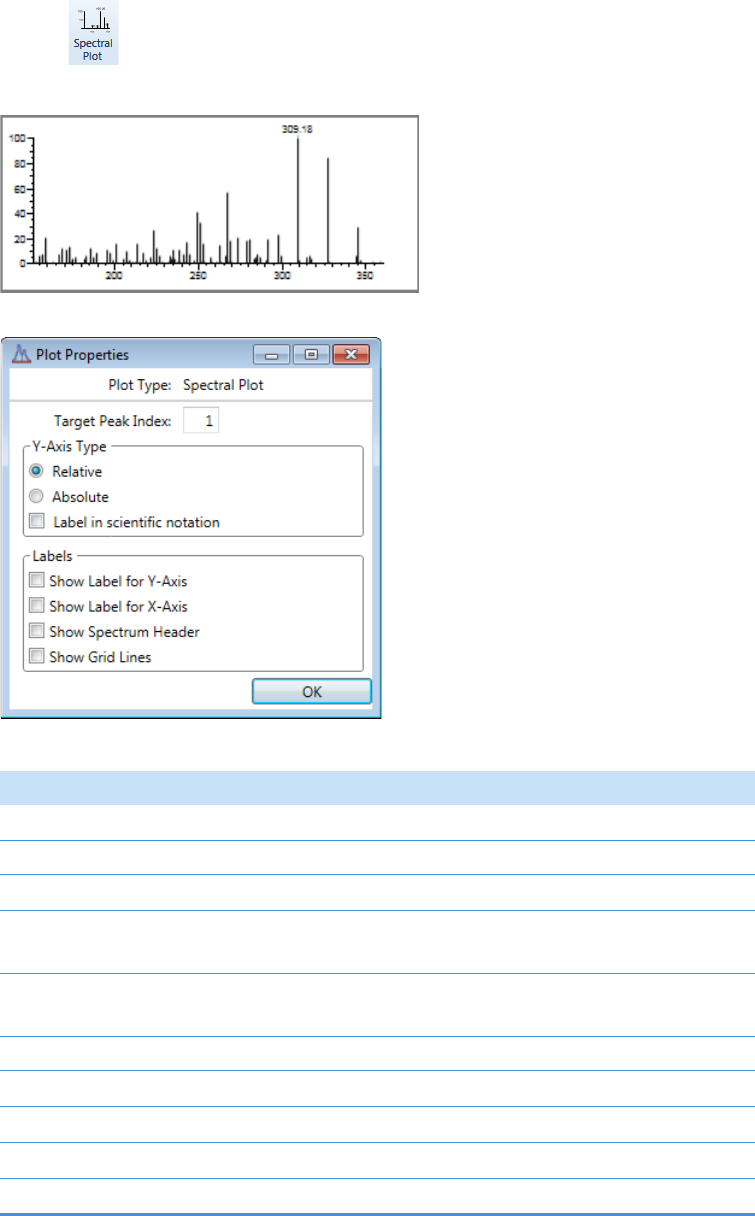
7 Using the Report Designer
Plot Display Properties
412 TraceFinder User Guide Thermo Scientific
Spectral Plot
Use the icon to insert a Spectral plot.
Figure 176. Spectral plot
Figure 177. Plot Properties dialog box for a Spectral plot
Table 90. Plot Properties parameters for a Spectral plot
Command Description
Target Peak Index Valid values: 1 through 9
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the raw data file name and compound name.
Show Grid Lines Displays the grid lines in the plot background.
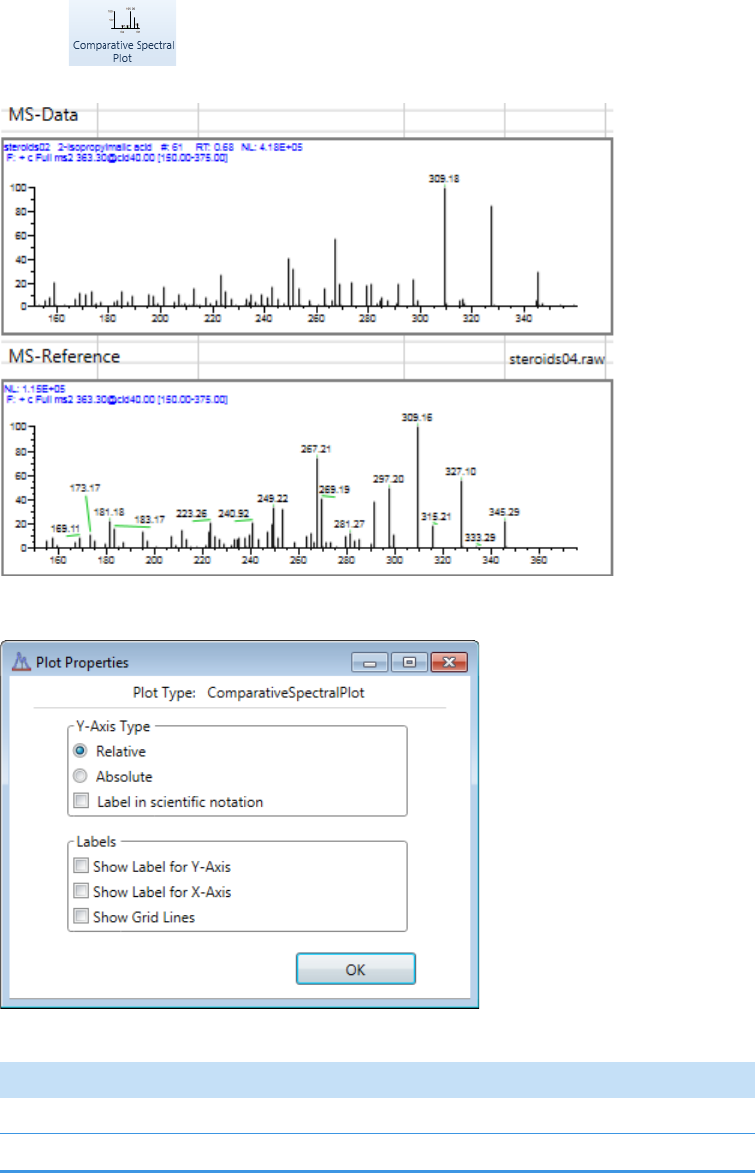
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 413
Comparative Spectral Plot (quantitation reports only)
Use the icon to insert a Comparative Spectral plot.
Figure 178. Comparative Spectral plot
Figure 179. Plot Properties dialog box for a Comparative Spectral plot
Table 91. Plot Properties parameters for a Comparative Spectral plot (Sheet 1 of 2)
Command Description
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
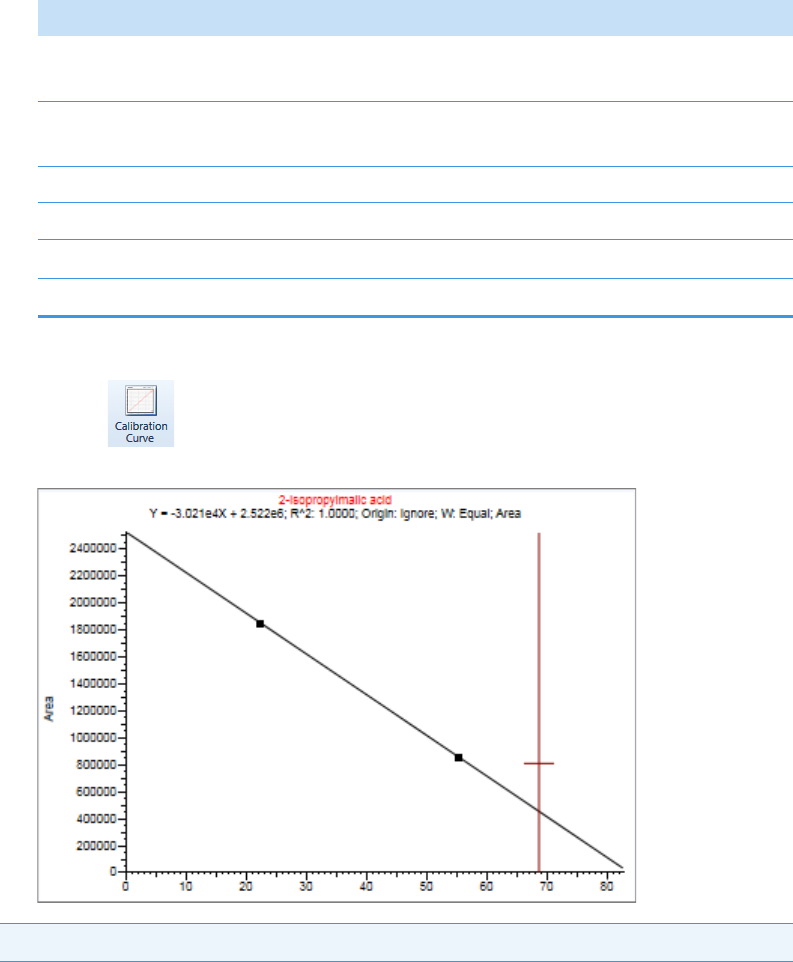
7 Using the Report Designer
Plot Display Properties
414 TraceFinder User Guide Thermo Scientific
Calibration Curve Plot (quantitation reports only)
Use the icon to insert a Calibration Curve plot.
Figure 180. Calibration Curve plot
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Grid Lines Displays the grid lines in the plot background.
Table 91. Plot Properties parameters for a Comparative Spectral plot (Sheet 2 of 2)
Command Description
Note There are no plot properties for a calibration curve plot.
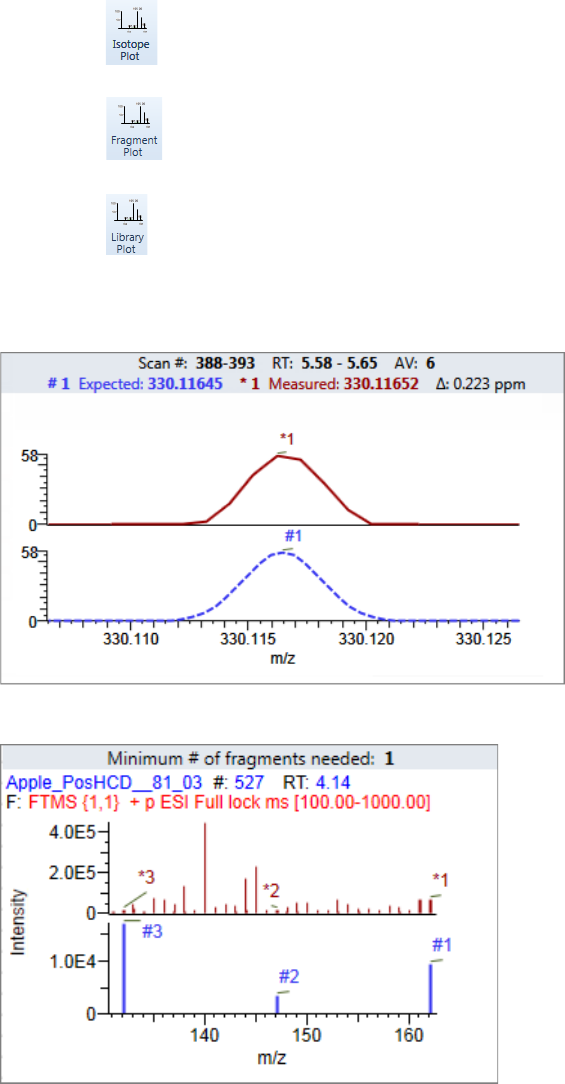
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 415
Isotope Plot, Fragment Plot, or Library Plot
Do one of the following:
• Use the icon to insert an Isotope plot.
• Use the icon to insert a Fragment plot.
• Use the icon to insert a Library plot.
The Isotope, Fragment, and Library plots use identical plot properties.
Figure 181. Isotope plot
Figure 182. Fragment plot
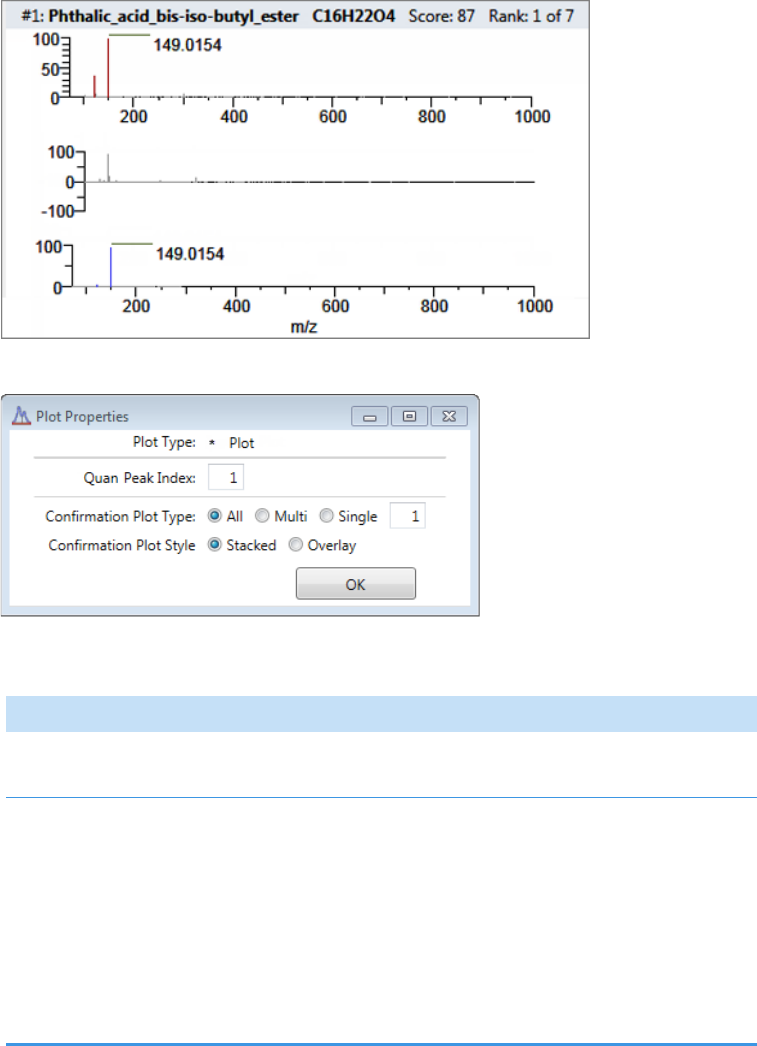
7 Using the Report Designer
Plot Display Properties
416 TraceFinder User Guide Thermo Scientific
Figure 183. Library plot
Figure 184. Plot Properties dialog box for an Isotope plot, a Fragment plot, or a Library plot
Table 92. Plot Properties parameters for an Isotope plot, a Fragment plot, or a Library plot
(Sheet 1 of 2)
Command Description
Target Peak Index Specifies the number of peak plots to display.
Valid values: 1 through 9
Confirmation Plot
Type
Specifies that the plot display one of the following:
• All: Displays isotopes, fragments, or library hits.
• Multi: Displays individual plots for each isotope. You can
individually stack or overlay the plots for each isotope.
• Single: Displays only the selected isotope, fragment, or
library hit.
For examples of these display types, see Isotope, Fragments, or
Library Match.
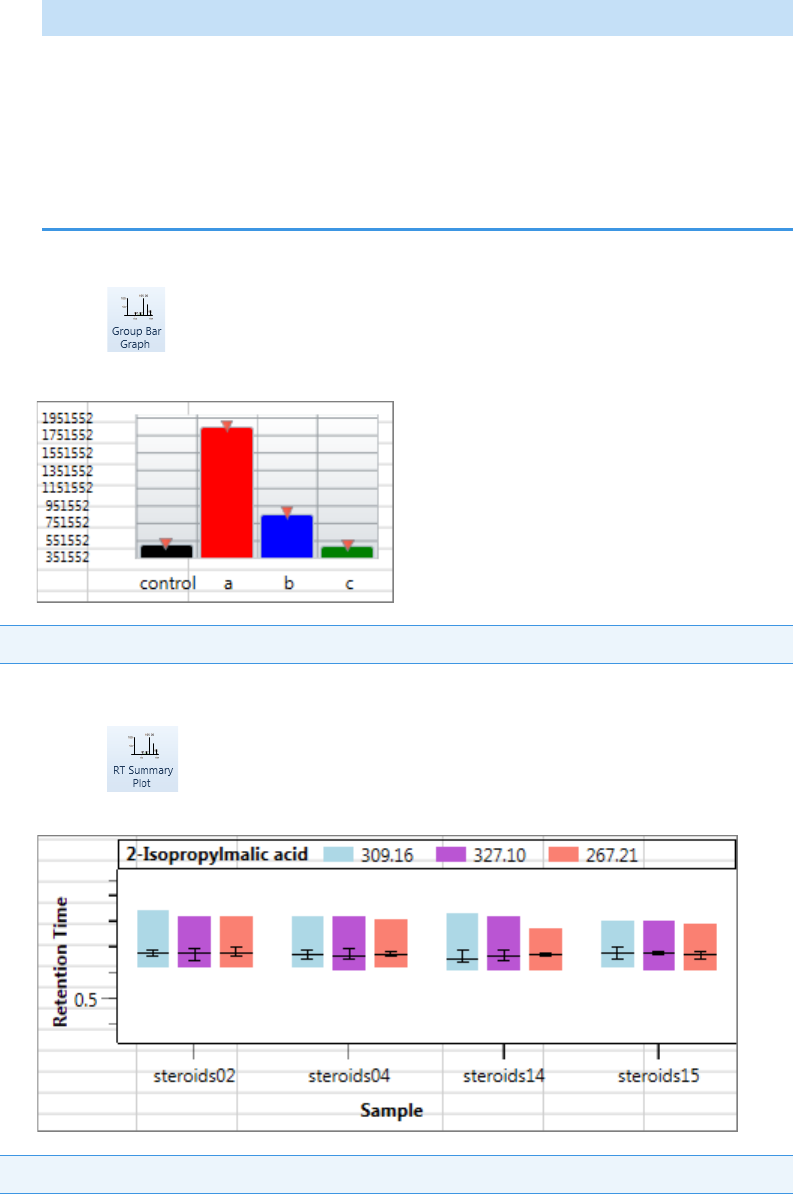
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 417
Group Bar Graph Plot
Use the icon to insert a Group Bar Graph plot.
Figure 185. Group Bar Graph plot
RT Summary Plot
Use the icon to insert an RT Summary plot.
Figure 186. RT Summary plot
Confirmation Plot
Style
Specifies that the plot display one of the following:
• Stacked: Stacks the simulated spectrum and the peak apex
spectrum.
• Overlay: Overlays the two spectrum displays.
For examples of these display types, see Isotope, Fragments, or
Library Match.
Table 92. Plot Properties parameters for an Isotope plot, a Fragment plot, or a Library plot
(Sheet 2 of 2)
Command Description
Note There are no plot properties for a group bar graph plot.
Note There are no plot properties for an RT summary plot.
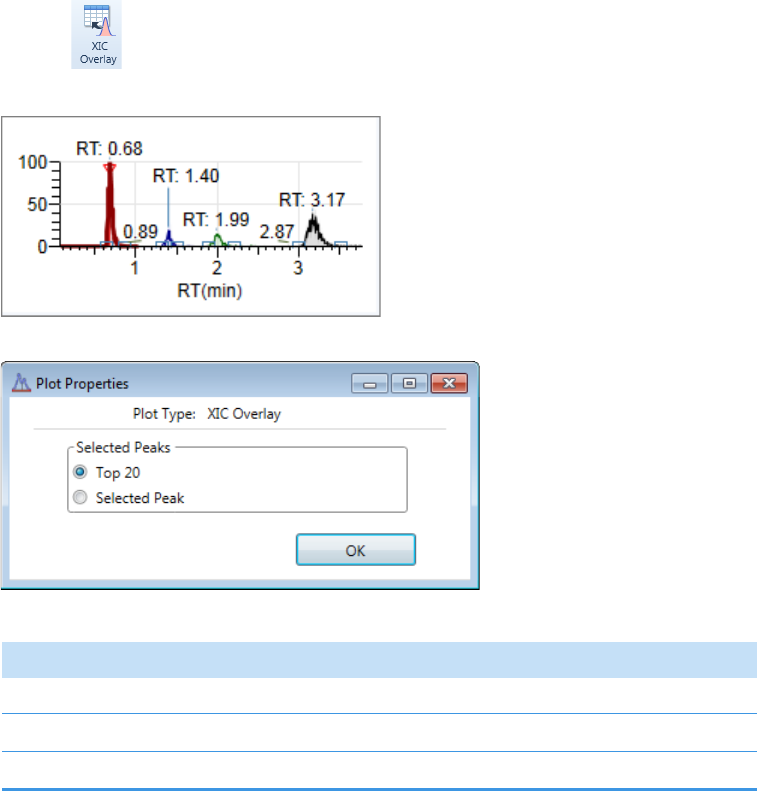
7 Using the Report Designer
Plot Display Properties
418 TraceFinder User Guide Thermo Scientific
XIC Overlay Plot
Use the icon to insert an XIC Overlay plot.
Figure 187. XIC Overlay plot
Figure 188. Plot Properties dialog box for an XIC Overlay plot
Table 93. Plot Properties parameters for an XIC Overlay plot
Command Description
Selected Peaks
Top 20 Displays the top 20 peaks.
Selected Peak Displays only the selected peak.
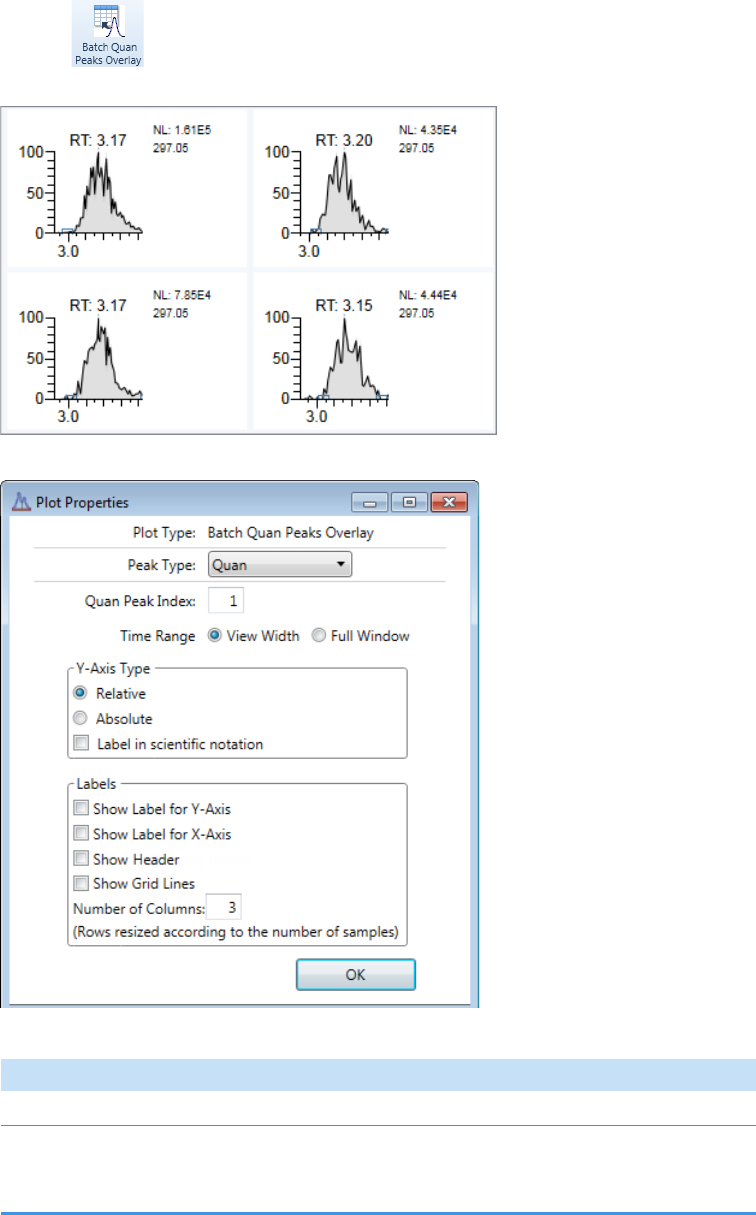
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 419
Batch Quan Peaks Overlay Plot
Use the icon to insert a Batch Quan Peaks Overlay plot.
Figure 189. Batch Quan Peaks Overlay plot
Figure 190. Plot Properties dialog box for a Batch Quan Peaks Overlay plot
Table 94. Plot Properties parameters for a Batch Quan Peaks Overlay plot (Sheet 1 of 2)
Command Description
Quan Peak Index Valid values: 1 through 9
Time Range Specifies that the plot display one of the following:
• View Width: Zooms the display on the peak of interest.
• Full Window: Displays the full width of the scan.

7 Using the Report Designer
Plot Display Properties
420 TraceFinder User Guide Thermo Scientific
Unknown Screening Plots
To customize the display in the unknown screening report, use the Plot Properties dialog box
for each plot.
•Peak Chromatogram Plot
•TIC Chromatogram Plot
•Spectral Plot
•XIC Overlay Plot
•Library Plot, Fragments Plot, or Isotopes Plot
•XIC Plot
•Cross Sample Peak Overlay Plot
•Chemical Structure Plot
•Group Bar Graph Plot
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the raw data file name and compound name.
Show Grid Lines Displays the grid lines in the plot background.
Number of Columns Specifies the number of columns that you want to display in
the inserted graphic. The application resizes the images to fit
the specified number of columns.
Table 94. Plot Properties parameters for a Batch Quan Peaks Overlay plot (Sheet 2 of 2)
Command Description
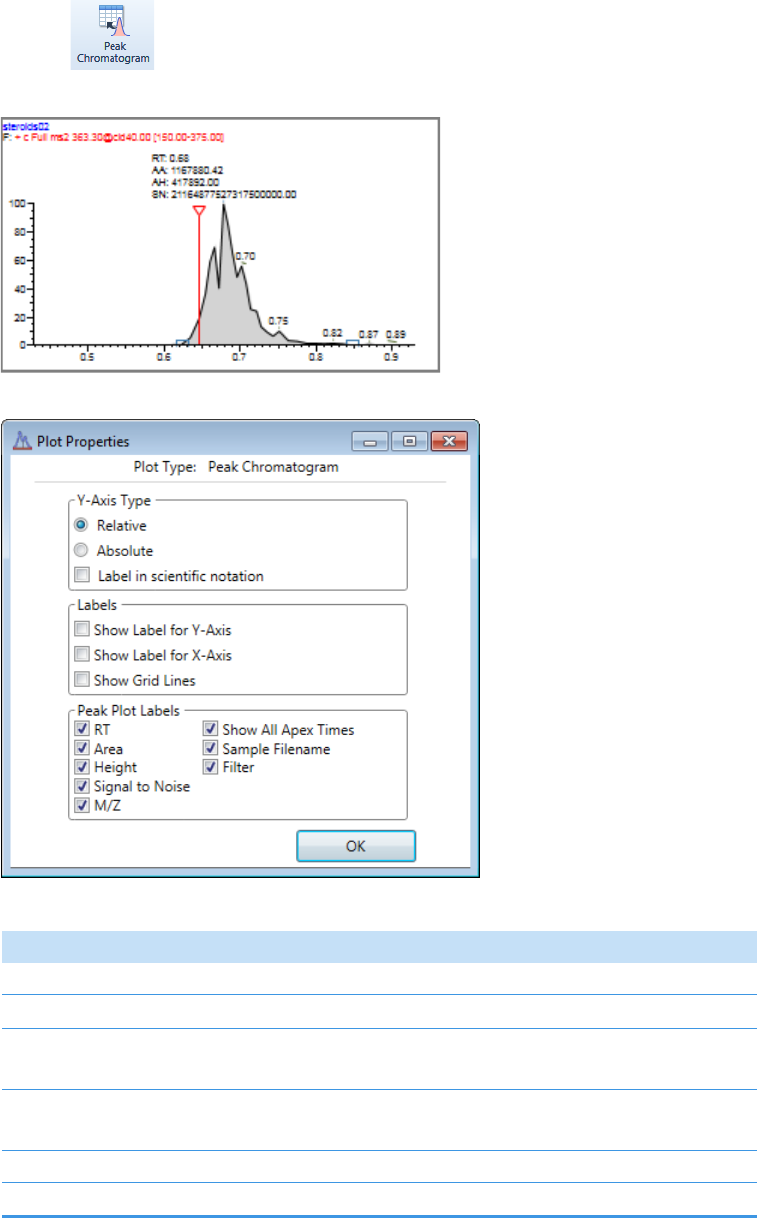
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 421
Peak Chromatogram Plot
Use the icon to insert a Peak Chromatogram plot.
Figure 191. Peak Chromatogram plot
Figure 192. Plot Properties dialog box for Peak Chromatogram plot
Table 95. Plot Properties parameters for a Peak Chromatogram plot (Sheet 1 of 2)
Command Description
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
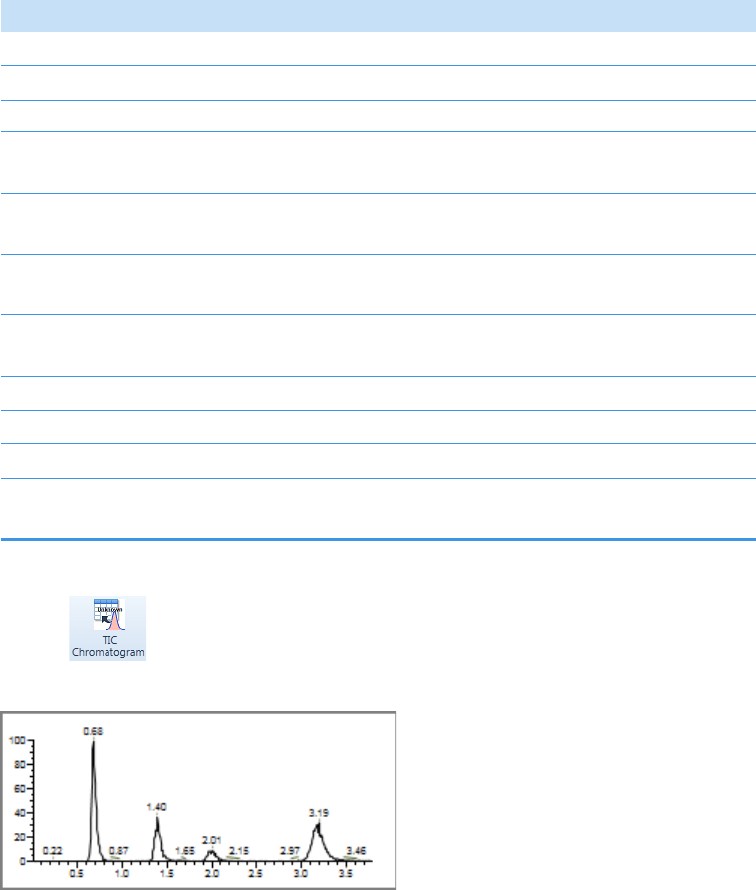
7 Using the Report Designer
Plot Display Properties
422 TraceFinder User Guide Thermo Scientific
TIC Chromatogram Plot
Use the icon to insert a TIC Chromatogram plot.
Figure 193. TIC Chromatogram plot
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Grid Lines Displays the grid lines in the plot background.
Peak Plot Labels
RT Displays the retention time (RT) label in the peak
chromatogram plot.
Area Displays the peak area (AA) label in the peak chromatogram
plot.
Height Displays the peak height (AH) label in the peak
chromatogram plot.
Signal to Noise Displays the signal-to-noise (SN) label in the peak
chromatogram plot.
M/Z Displays the peak mass.
Show All Apex Times Displays all surrounding apex times in the plot.
Sample Filename Displays the name of the sample file.
Filter Displays the filter used for the scan. The plot displays the
filter name below the raw data file name.
Table 95. Plot Properties parameters for a Peak Chromatogram plot (Sheet 2 of 2)
Command Description
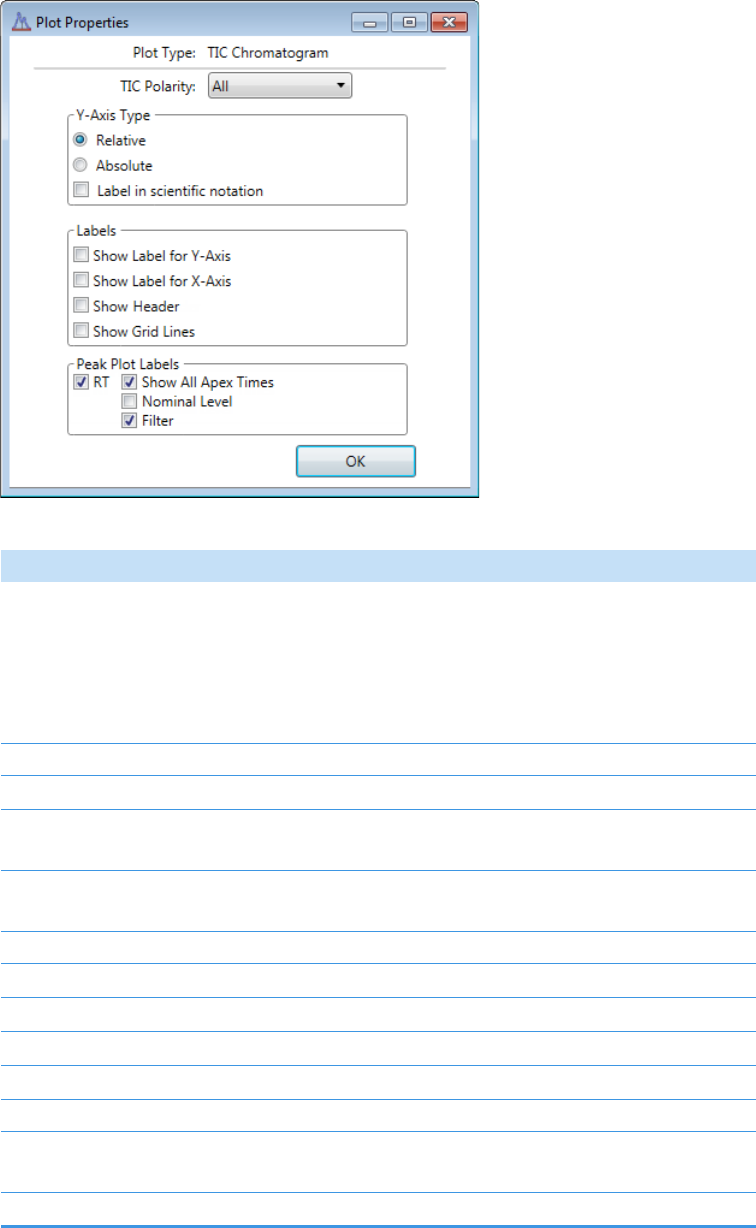
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 423
Figure 194. Plot Properties dialog box for a TIC Chromatogram plot
Table 96. Plot Properties parameters for a TIC Chromatogram plot (Sheet 1 of 2)
Command Description
TIC Polarity Specifies the polarity of adducts displayed in the plot.
All: Displays positive, negative, and analog adducts.
Positive: Displays only positive adducts.
Negative: Displays only negative adducts.
Analog: Displays only analog adducts.
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the raw data file name.
Show Grid Lines Displays the grid lines in the plot background.
Peak Plot Labels
RT Displays the retention time (RT) label in the TIC
chromatogram plot.
Show All Apex Times Displays all surrounding apex times in the plot.
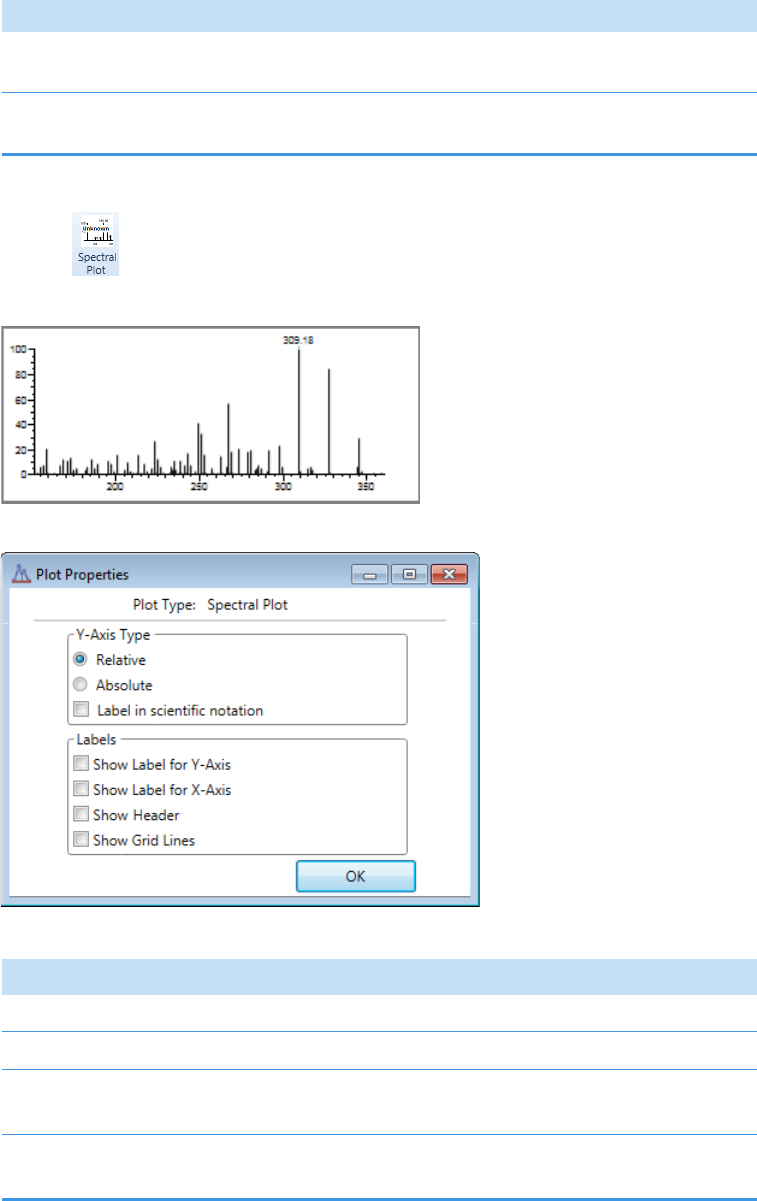
7 Using the Report Designer
Plot Display Properties
424 TraceFinder User Guide Thermo Scientific
Spectral Plot
Use the icon to insert a Spectral plot.
Figure 195. Spectral plot
Figure 196. Plot Properties dialog box for a Spectral plot
Nominal Level Displays the nominal level (NL) values for each peak in the
plot.
Filter Displays the filter used for the scan. The plot displays the
filter name below the raw data file name.
Table 96. Plot Properties parameters for a TIC Chromatogram plot (Sheet 2 of 2)
Command Description
Table 97. Plot Properties parameters for a Spectral plot (Sheet 1 of 2)
Command Description
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
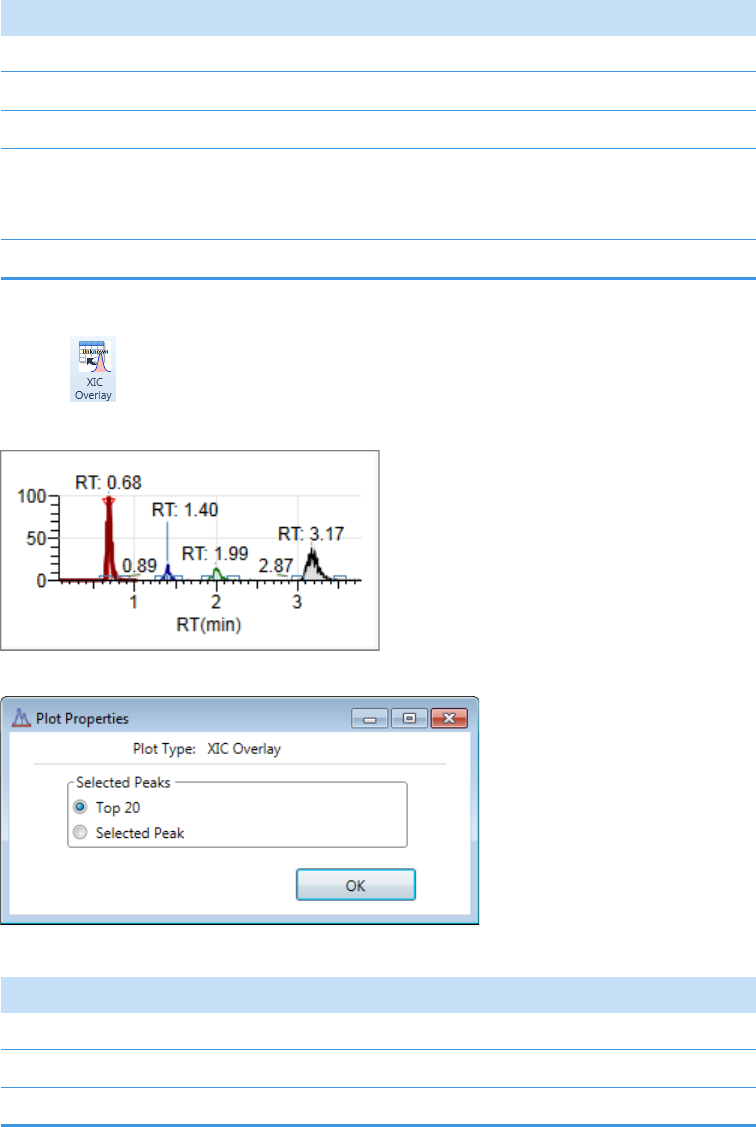
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 425
XIC Overlay Plot
Use the icon to insert an XIC Overlay plot.
Figure 197. XIC Overlay plot
Figure 198. Plot Properties dialog box for an XIC Overlay plot
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header Displays the name of the sample file, the name of the
compound, the retention time, the nominal level, and the
filter used for the scan.
Show Grid Lines Displays the grid lines in the plot background.
Table 97. Plot Properties parameters for a Spectral plot (Sheet 2 of 2)
Command Description
Table 98. Plot Properties parameters for an XIC Overlay plot
Command Description
Selected Peaks
Top 20 Displays the top 20 peaks.
Selected Peak Displays only the selected peak.
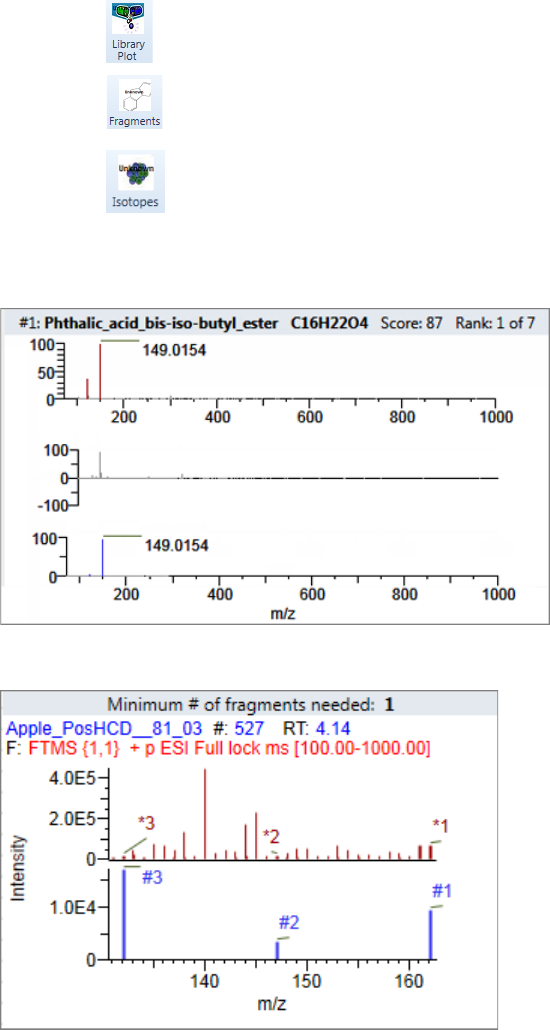
7 Using the Report Designer
Plot Display Properties
426 TraceFinder User Guide Thermo Scientific
Library Plot, Fragments Plot, or Isotopes Plot
Do one of the following:
• Use the icon to insert a Library plot.
• Use the icon to insert a Fragments plot.
Use the icon to insert an Isotope plot.
The Library, Fragments, and Isotopes plots use identical plot properties.
Figure 199. Library plot
Figure 200. Fragments plot
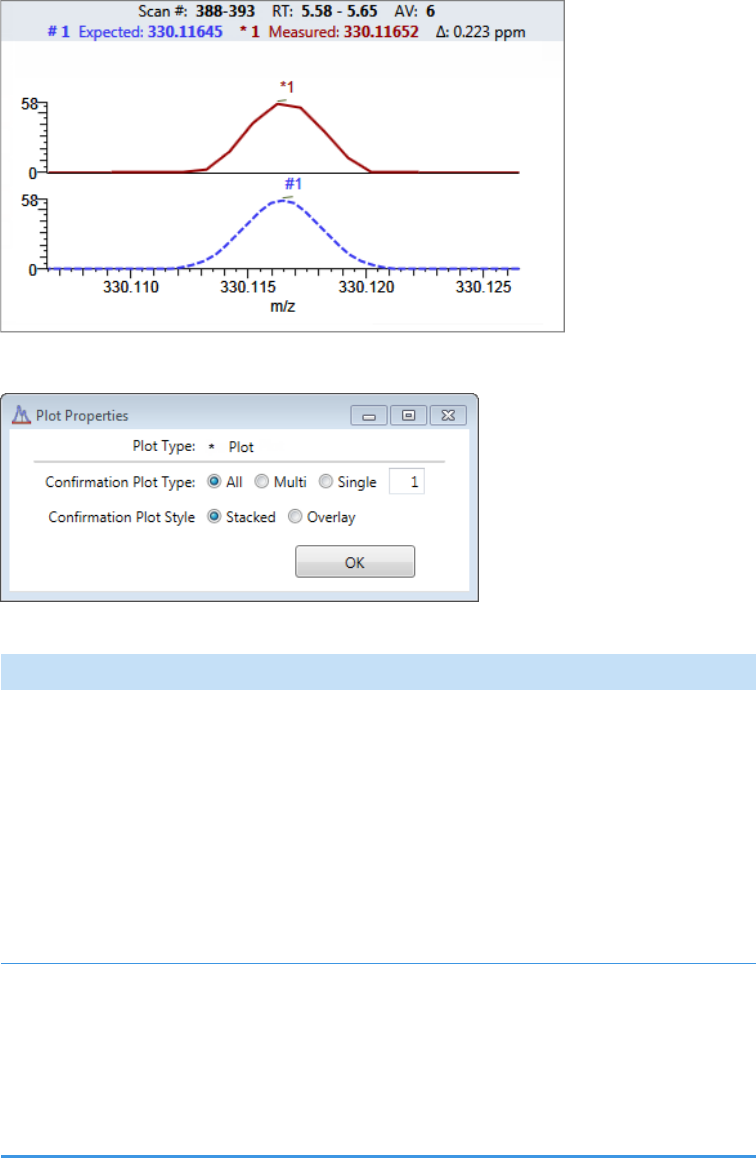
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 427
Figure 201. Isotopes plot
Figure 202. Plot Properties dialog box for a Library plot, a Fragments plot, or an Isotopes plot
Table 99. Plot Properties parameters for a Library plot, a Fragments plot, or an Isotopes plot
Command Description
Confirmation Plot Type Specifies that the plot display one of the following:
• All: Displays isotopes, fragments, or library hits.
• Multi: Displays individual plots for each isotope. You
can individually stack or overlay the plots for each
isotope.
• Single: Displays only the selected isotope, fragment, or
library hit.
For examples of these display types in the Data Review, see
Library Search Pane, Fragments Pane, or Isotopes Pane.
Confirmation Plot Style Specifies that the plot display one of the following:
• Stacked: Stacks the simulated spectrum and the peak
apex spectrum.
• Overlay: Overlays the two spectrum displays.
For examples of these display types in the Data Review, see
Library Search Pane, Fragments Pane, or Isotopes Pane.
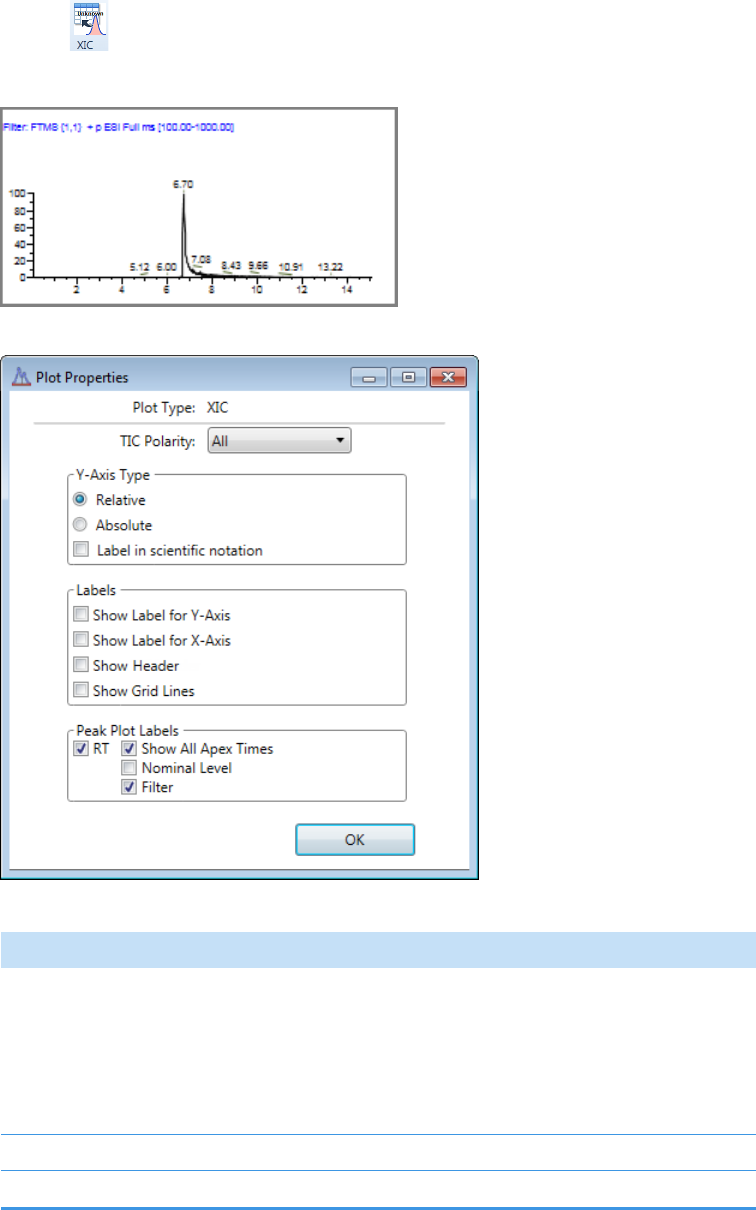
7 Using the Report Designer
Plot Display Properties
428 TraceFinder User Guide Thermo Scientific
XIC Plot
Use the icon to insert an XIC plot.
Figure 203. XIC plot
Figure 204. Plot Properties dialog box for an XIC plot
Table 100. Plot Properties parameters for an XIC plot (Sheet 1 of 2)
Command Description
TIC Polarity Specifies the polarity of adducts displayed in the plot.
All: Displays positive, negative, and analog adducts.
Positive: Displays only positive adducts.
Negative: Displays only negative adducts.
Analog: Displays only analog adducts.
Y-Axis Type
Relative Displays the y-axis scale from 0 through 100.
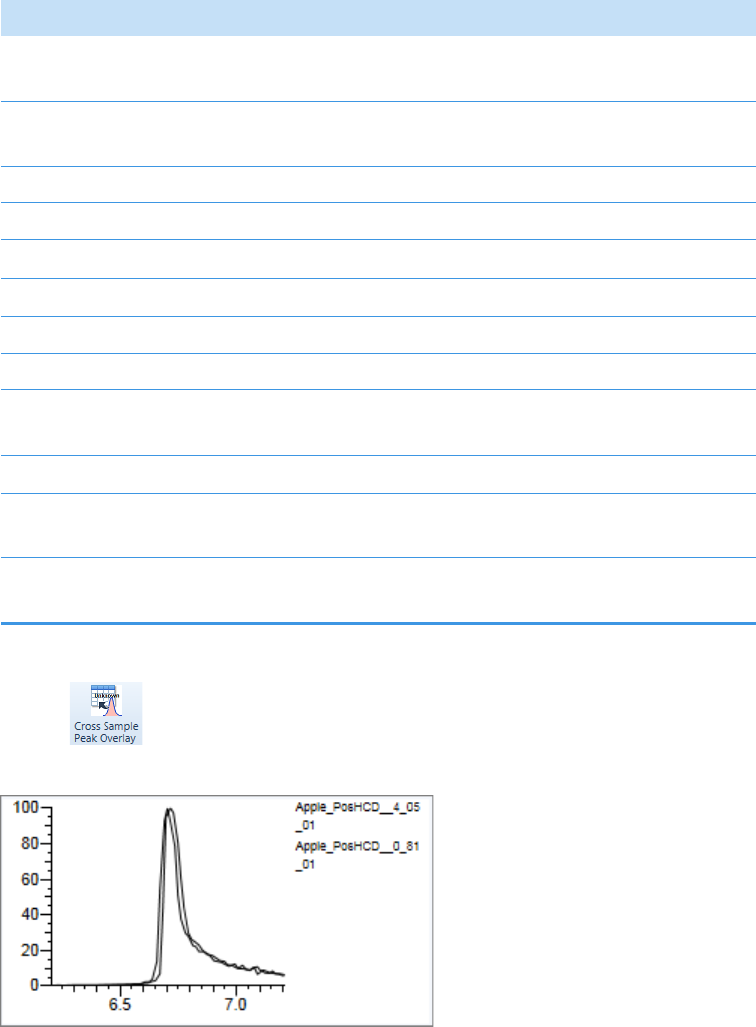
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 429
Cross Sample Peak Overlay Plot
Use the icon to insert a Cross Sample Peak Overlay plot.
Figure 205. Cross Sample Peak Overlay plot
Absolute Displays the y-axis scale from 0 to the actual value of the
most intense peak in the group.
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Labels
Show Label for Y-Axis Displays the relative intensity value on the y axis.
Show Label for X-Axis Displays the retention time (RT) value on the x axis.
Show Header
Show Grid Lines Displays the grid lines in the plot background.
Peak Plot Labels
RT Displays the retention time (RT) values for each peak in the
plot.
Show All Apex Times Displays all surrounding apex times in the plot.
Nominal Level Displays the nominal level (NL) values for each peak in the
plot.
Filter Displays the filter used for the scan. The plot displays the
filter name below the raw data file name.
Table 100. Plot Properties parameters for an XIC plot (Sheet 2 of 2)
Command Description
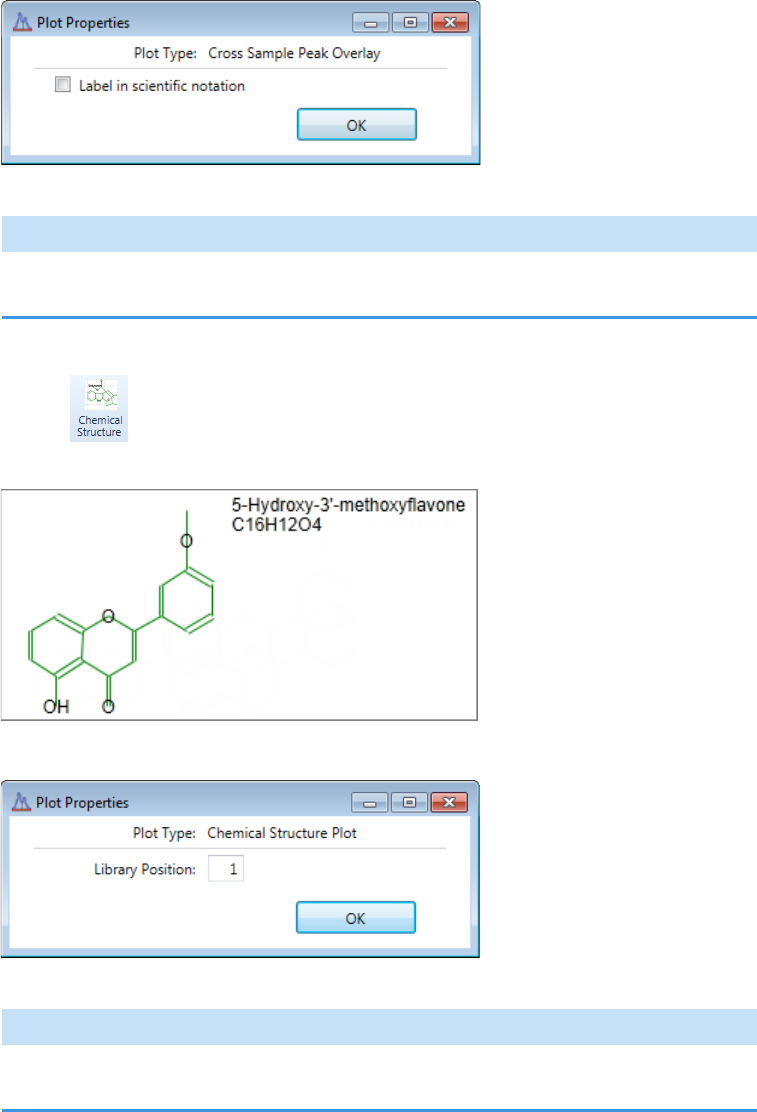
7 Using the Report Designer
Plot Display Properties
430 TraceFinder User Guide Thermo Scientific
Figure 206. Plot Properties dialog box for a Cross Sample Peak Overlay plot
Chemical Structure Plot
Use the icon to insert a Chemical Structure plot.
Figure 207. Chemical Structure plot
Figure 208. Plot Properties dialog box for a Chemical Structure plot
Table 101. Plot Properties parameter for a Cross Sample Peak Overlay plot
Command Description
Label in Scientific
Notation
Displays the y-axis scale in scientific notation.
Table 102. Plot Properties parameter for a chemical structure plot
Command Description
Library Position Specifies the hierarchical position in the library when there
are multiple chemical structures.
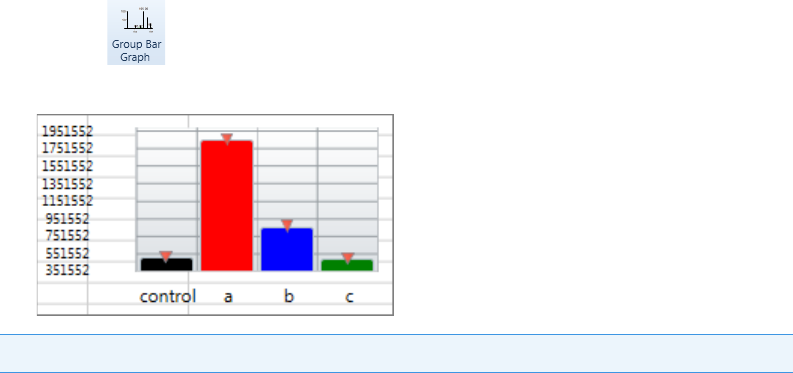
7 Using the Report Designer
Plot Display Properties
Thermo Scientific TraceFinder User Guide 431
Group Bar Graph Plot
Use the icon to insert a Group Bar Graph plot.
Figure 209. Group Bar Graph plot
Note There are no plot properties for a group bar graph plot.
7 Using the Report Designer
Quick Tips
432 TraceFinder User Guide Thermo Scientific
Quick Tips
Follow these tips to make the most of the Report Designer features.
To move a report to the Archive folder
1. With a report open for editing, choose File > Archive from the Report Designer main
menu.
2. At the confirmation prompt, click Yes.
The application deletes the report from the …batch\ReportOutput folder.
To insert items in the Report Designer
• Insert a data table when the selected cell is above or below any other table, but not in the
same row as a field. You cannot insert tables in repeat areas.
• Insert a data field when the selected cell is not in the same row as a table.
• Use these shortcut keys:
To format cells and group headers
Because the first row of a data table retains formatting information, edit the formatting of
the first row.
The application copies the formatting to all other rows.
For group header items, the application copies the formatting of the first group header to
the remaining group headers.
To sort fields
Select a field, and then click A-Z to sort the data in ascending or descending order.
CTRL+T Insert a table.
CTRL+T Edit a table when the selected cell is inside a table.
CTRL+SHIFT+T Insert or edit a field.
CTRL+R Insert or edit a repeating area.

Thermo Scientific TraceFinder User Guide 433
8
Using the Audit Viewer
The TraceFinder application records all user access, including logging in, logging out, data
creation and editing (batches, methods, and templates), and manual integration. You can use
the Audit Viewer to view the resulting log files to track modifications to the data. When an
event requires confirmation (as specified in the Administrator Console), the Audit Viewer
records who confirmed each change to a batch, method, or template. When no confirmation
is required, then the Audit Viewer records the user who was logged in when the change
occurred.
In the Administration Console, a user with Auditing permissions can configure the auditing
service by specifying which events are logged, which events require confirmation, a list of
default reasons for a specific event, and whether a user can submit a custom reason. To use the
auditing administration tools, refer to the instructions in the TraceFinder Administrator
Console User Guide.
Audit Trail Log Files
The application creates the following audit trail log files:
• Application: Records all user access, such as starting and stopping the application, logging
in, logging out, or accessing or saving data in batches and methods. The application saves
the data in the following log file: C:\Thermo\TraceFinder\4.0\Logs\AuditLog.adb.
• Master Method: Records all user interactions with master methods, such as creating,
opening, or editing a master method. The application saves the data in the following log
file: C:\TraceFinderData\4.0\Methods\MasterMethodName\AuditLog.adb.
IMPORTANT TraceFinder 4.1 uses the same data as TraceFinder 4.0. By default, the application
stores the log files for the 4.1 release in the TraceFinderData\4.0 or TraceFinder\4.0 folders.
Contents
•Audit Trail Log Files
•Audit Log File Functions
•Audit Viewer
8 Using the Audit Viewer
Audit Log File Functions
434 TraceFinder User Guide Thermo Scientific
• Batch Template: Records all user interactions with batch templates, such as creating,
opening, or editing a batch template. The application saves the data in the following log
file: C:\TraceFinderData\4.0\Templates\Batches\BatchTemplateName\AuditLog.adb.
• Batch: Records all user interactions with batches, such as creating, opening, editing,
acquiring, processing, or generating reports for a batch. The application saves the data in
the following log file:
C:\TraceFinderData\4.0\Projects\SubFolder\BatchName\AuditLog.adb.
Audit Log File Functions
The Audit Viewer displays all saved audit log files, and you can filter and sort the audit data.
Follow these procedures:
•To access the Audit Viewer
•To select an audit log
•To view only application, method, batch, or batch template events
•To create a filter for audit log events
•To view audit event details
•To create a filter for an audit log history
•To display the history for an event
To access the Audit Viewer
Choose To ol s > Aud i t Tra il from the TraceFinder main menu or click the Audit Viewer
icon, .
The Audit Viewer opens.
Note The Audit Viewer is available only when you have enabled auditing from the
Administration Console. Refer to the TraceFinder Administrator Console User Guide.
Note The Tools > Audit Trail menu command always opens to application log files,
whereas the Audit Viewer icon is context sensitive and opens to the appropriate type
of log files (application, method, batch, or batch template).
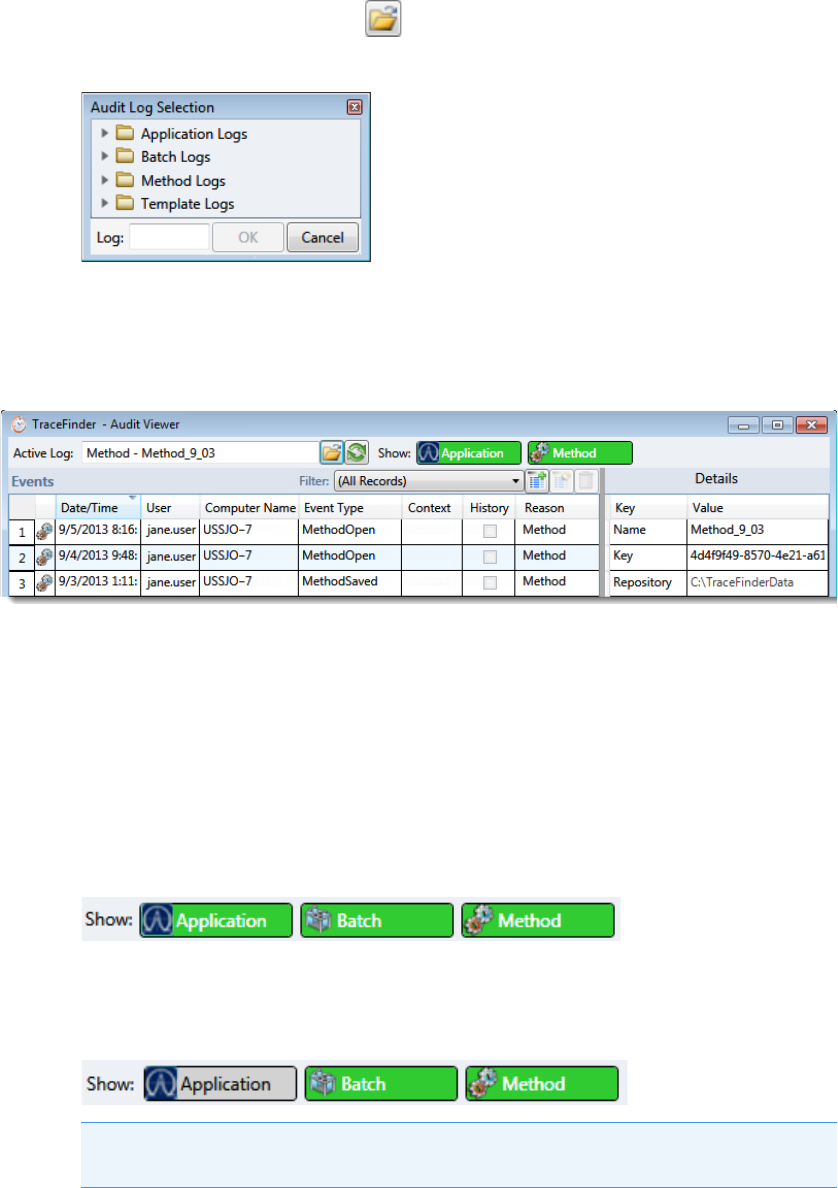
8 Using the Audit Viewer
Audit Log File Functions
Thermo Scientific TraceFinder User Guide 435
To select an audit log
1. Click the Open Audit Log icon, .
The application opens the Audit Log Selection dialog box.
2. Expand a log folder to select an application, batch, method, or batch template audit log
file, and click OK.
The Audit Viewer displays the contents of the selected audit log file, as in this example for
a method.
To view only application, method, batch, or batch template events
1. In the Active Log list, select an Active Log file.
• Application logs include application and security events.
• Method logs include application and method events.
• Batch logs include application, method, and batch events.
• Batch template logs include application and batch template events.
The Audit Viewer displays icons for each of the event types included in the log file.
2. Click an icon to turn the display on or off.
In this example, the Events pane displays Batch and Method events and hides Application
events.
Note There is no icon for security events in an application log file. You cannot hide
the display of security events.
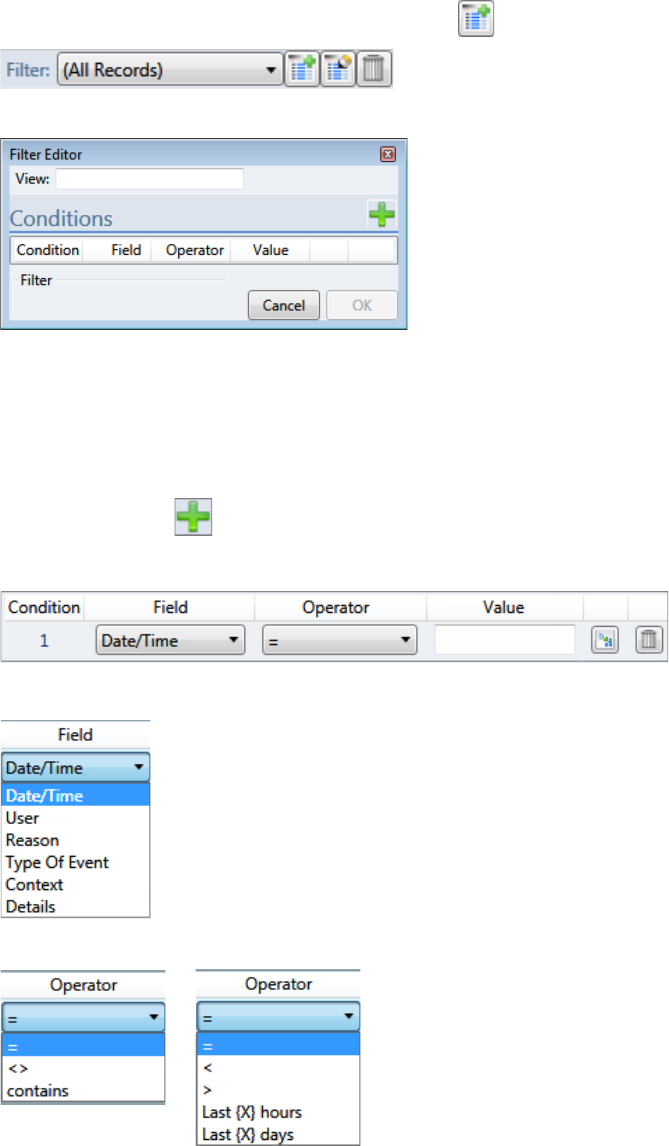
8 Using the Audit Viewer
Audit Log File Functions
436 TraceFinder User Guide Thermo Scientific
To create a filter for audit log events
1. In the Events pane, click the Create New Filter icon, .
The Filter Editor dialog box opens.
2. In the View box, type a name for the new filter.
You can also leave the View box empty when you enter your filter criteria. When you
finish adding conditions and click OK (step 8), the application filters the current events
list based on the filter criteria you specify, but the filter is not saved. The Filter list in the
viewer identifies this filter as (Custom).
3. Click the Add icon, .
The application adds a new, undefined condition to the Condition list.
4. In the Field list, select one of the following field types.
5. In the Operator list, select one of the available operators.
The available operators depend on the field type that you selected.
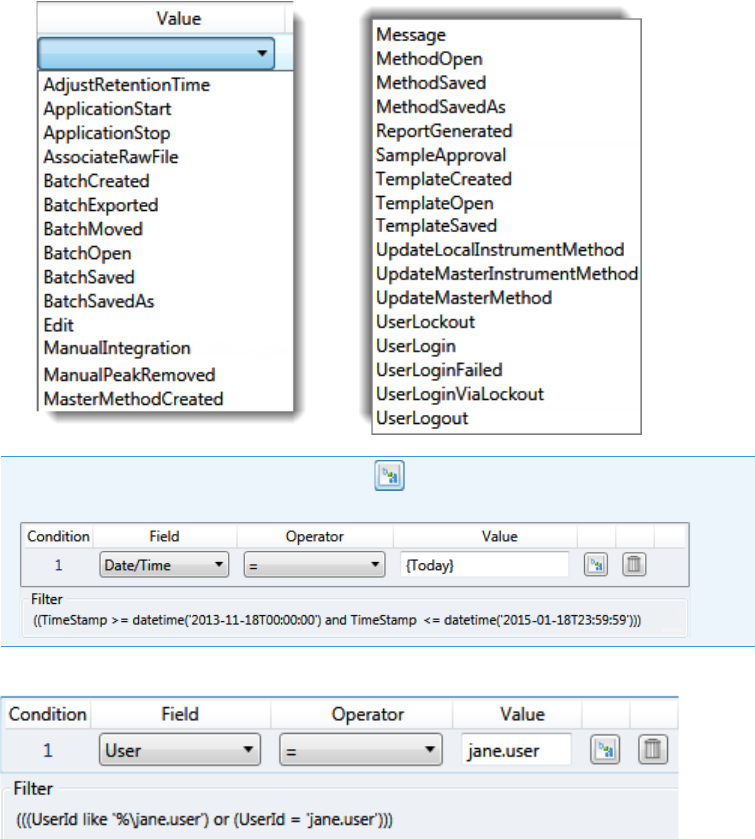
8 Using the Audit Viewer
Audit Log File Functions
Thermo Scientific TraceFinder User Guide 437
6. In the Value box, type a value or select a value from the list.
• For the Date/Time field, type a numerical value in the Value box.
• For the User, Reason, Context, or Details field, type the appropriate text in the Value
box. This value is case sensitive.
• For the Type of Event field, select one of the following values.
The Filter at the bottom of the dialog box displays the complete definition for the filter.
7. Repeat step 3 (on page 436) through step 6 (on this page) for each condition that you
want to include in your filter.
8. When you have added all your conditions, click OK.
The application creates the new filter with the specified conditions.
Note You can also click the Tokens icon, , and select a token for the Value.
Tokens are predefined values, such as dates and sample or compound identifiers.
8 Using the Audit Viewer
Audit Log File Functions
438 TraceFinder User Guide Thermo Scientific
To view audit event details
In the Events pane, select an event.
The Details pane displays key values based on the type of log you select.
To create a filter for an audit log history
1. In the History pane, click the Create New Filter icon, .
The Filter Editor dialog box opens.
2. In the View box, type a name for the new filter.
You can also leave the View box empty when you enter your filter criteria. When you
finish adding conditions and click OK (step 8), the application filters the current history
list based on the criteria you specify, but the filter is not saved. The Filter list in the viewer
identifies this filter as (Custom).
3. Click the Add icon, .
The application adds a new, undefined condition to the Conditions list.
Details for batch log files include the name of the batch.
Details for method log files include the name of the method and
the method type.
Details for batch template log files include the name of the batch
template, the location of the data repository, and the subproject
folder where the template was created.
Details for Application log files include whether the events are for a
batch or method, the location of the data repository, and, for a
batch, the subproject folder where the batch was created.
Details for Security log files include the authentication method
used (Windows Active Directory or local machine) and the
location of the administrator repository.
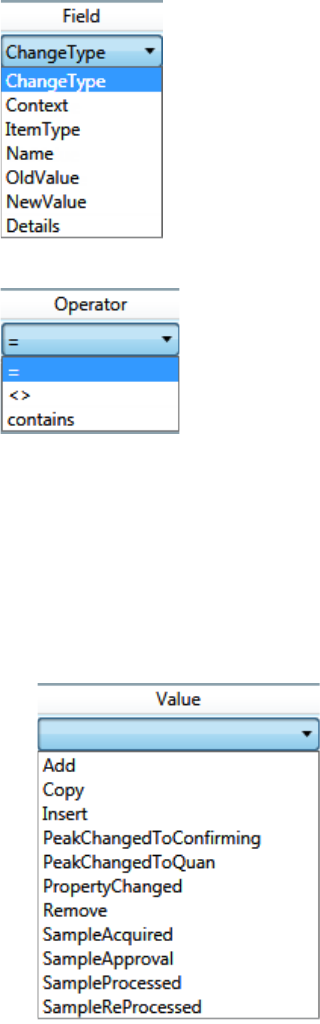
8 Using the Audit Viewer
Audit Log File Functions
Thermo Scientific TraceFinder User Guide 439
4. In the Field list, select one of the following field types.
5. In the Operator list, select from equals, less than/greater than, or contains.
The available operators depend on the field type that you selected.
6. In the Value list, type or select a value from the list.
• For the Name, Context, or Details field, type the appropriate text in the Value box.
This value is case sensitive.
• For the OldValue or NewValue field, type a numerical value in the Value box.
• For the ChangeType field, select one of the following values.
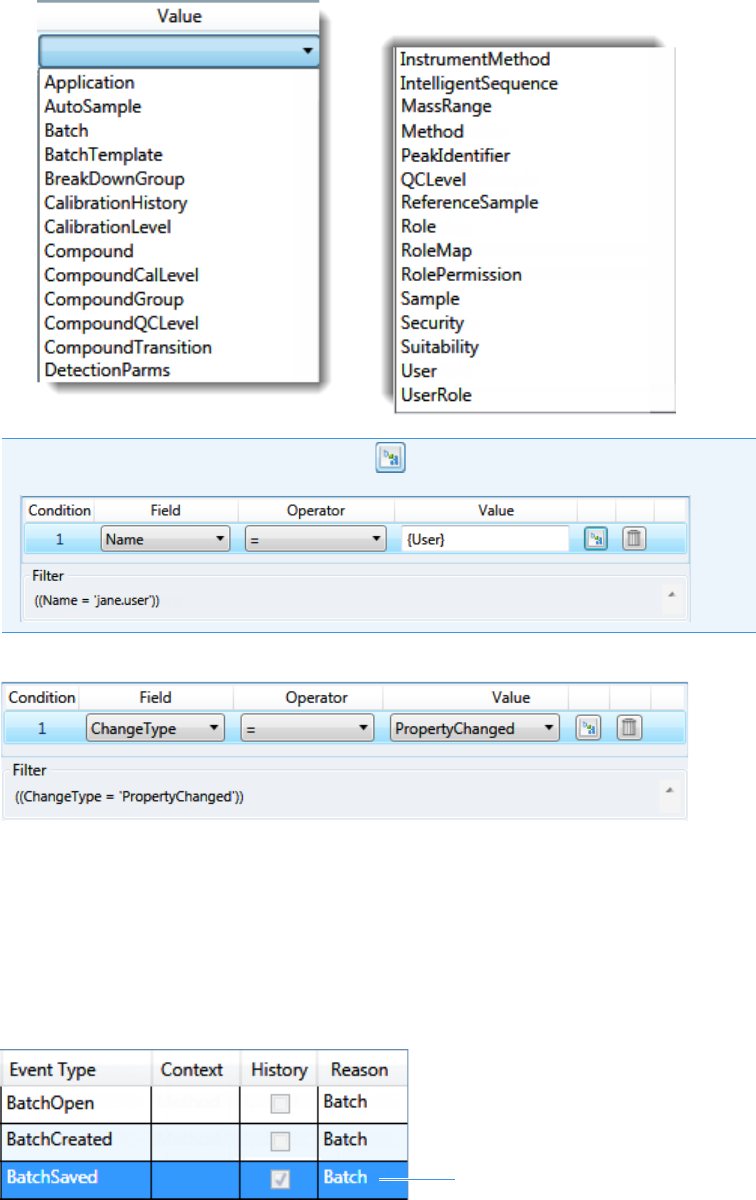
8 Using the Audit Viewer
Audit Log File Functions
440 TraceFinder User Guide Thermo Scientific
• When the selected Field is ItemType, select one of the following values.
The Filter at the bottom of the dialog box displays the complete definition for the filter.
7. Repeat steps 3 through 6 for each condition that you want to include in your filter.
8. When you have added all your conditions, click OK.
The applications created the new filter with the specified conditions.
To display the history for an event
1. In the Events pane, select an event that has a selected check box in the History column.
Note You can also click the Tokens icon, , and select a token for the Value.
Tokens are predefined values, such as dates and sample or compound identifiers.
Select an event that has the
History check box selected.
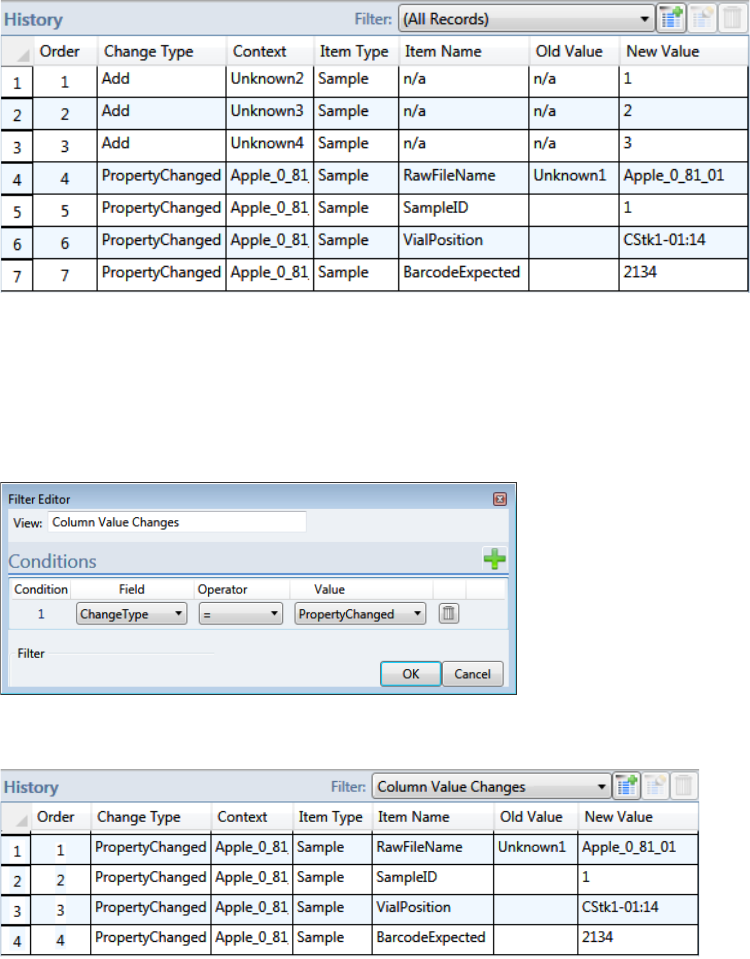
8 Using the Audit Viewer
Audit Log File Functions
Thermo Scientific TraceFinder User Guide 441
The History pane displays all unsaved (queued) actions for the selected event.
2. To limit the actions in the history list, select a filter from the Filter list.
See To create a filter for an audit log history.
Figure 210. Example History filter
When you select this filter,
the History pane displays only these actions for the selected event:
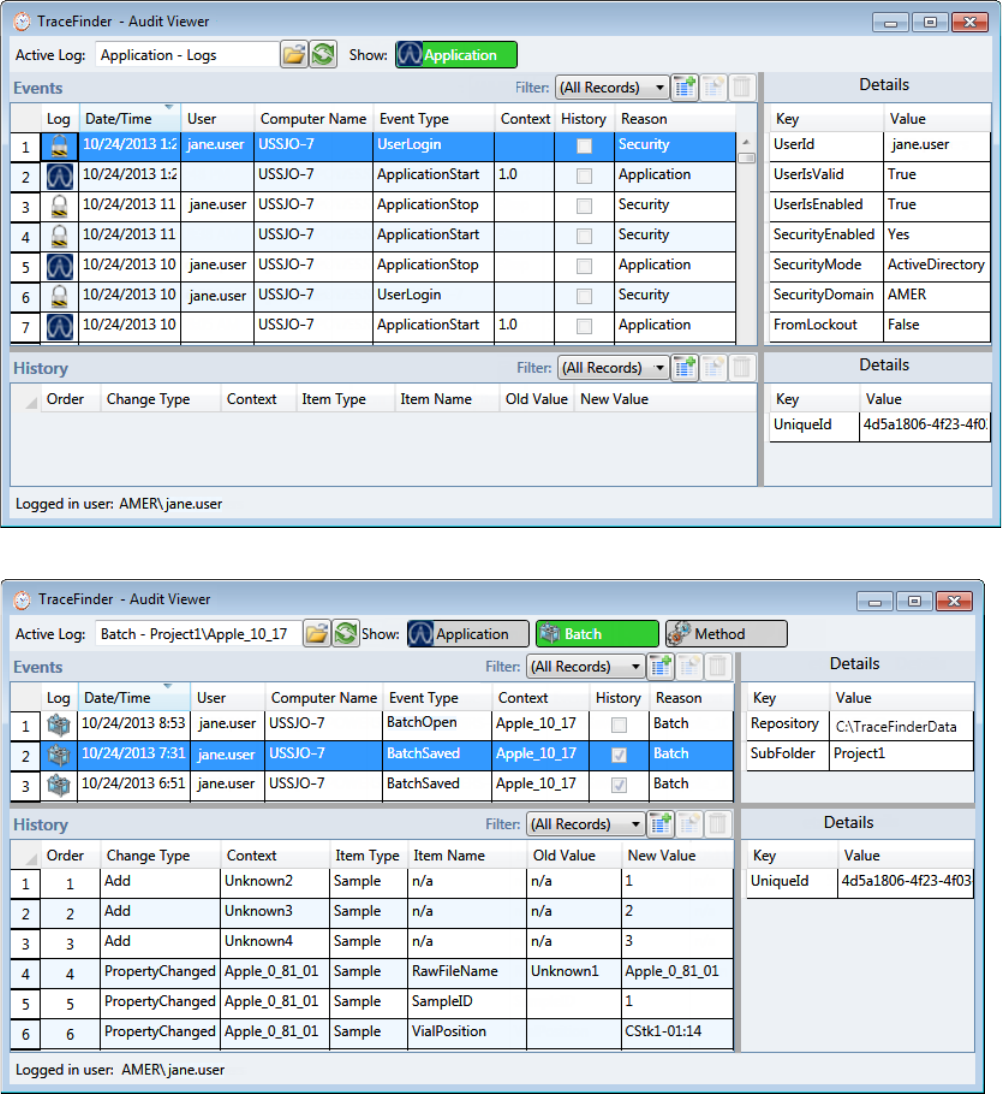
8 Using the Audit Viewer
Audit Viewer
442 TraceFinder User Guide Thermo Scientific
Audit Viewer
Use the Audit Viewer to view the audit log files to track user access and modifications to the
data. See Audit Viewer parameters.
Figure 211. Application log in the Audit Viewer
Figure 212. Batch log in the Audit Viewer
8 Using the Audit Viewer
Audit Viewer
Thermo Scientific TraceFinder User Guide 443
Table 103. Audit Viewer parameters (Sheet 1 of 2)
Parameter Description
Active Log Name of the current audit log file.
Opens the Audit Log Selection dialog box where you can open a different audit log file.
You can select from these audit log files: application, batch, method, or batch template.
Refreshes the current audit log file in the viewer.
Show Select to display only specific types of events in the Events list. The selected log file can
contain batch, method, and application events.
Events
Filter Select a filter view to use for displaying the event log entries.
Opens the Filter Editor dialog box where you can create a filter view.
Opens the Filter Editor dialog box where you can edit the current filter view.
Deletes the current event filter view.
Log
Indicates an event that occurred at the main application level, such as
logging in or opening a batch.
Indicates an event that occurred in the Administration Console.
Indicates an event that occurred in a method template.
Indicates an event that occurred in a method.
Indicates an event that occurred in a batch.
Date/Time Time stamp of the event.
User When an event requires confirmation (as specified in the Administrator Console), User is
the user who confirmed each change to a batch, method, or template.
When no confirmation is required, User is the user who was logged in when the change
occurred.
Computer Name Name of the computer on which the application recorded the event.
Event Type Specific event that triggered the log file entry. For a complete list of event types, refer to
the TraceFinder Administration Console User Guide.
Context Name of the sample, batch, method, or application version where the event occurred.
8 Using the Audit Viewer
Audit Viewer
444 TraceFinder User Guide Thermo Scientific
History Indicates that there is a history log of queued actions for the event.
Reason Default or custom reason that the user entered for the event.
Details
Key/Value
Identifying parameters and their values for the selected auditing event. These key
parameters are different for each type of auditing event.
History
Filter Select a filter view to use for displaying the change history.
Opens the Filter Editor dialog box where you can create a filter view.
Opens the Filter Editor dialog box where you can edit the current filter view.
Deletes the current history filter view.
Order The sequence of actions that occurred.
Change Type One of the predefined ChangeType values.
Context Name of the specific value on which the action occurred.
Item Type One of the predefined ItemType values.
Item Name A user-defined name for the filter.
Old Value/New Value Original parameter value and the changed value.
Details
Key/Value
Identifying parameters and values for the selected history event.
Table 103. Audit Viewer parameters (Sheet 2 of 2)
Parameter Description
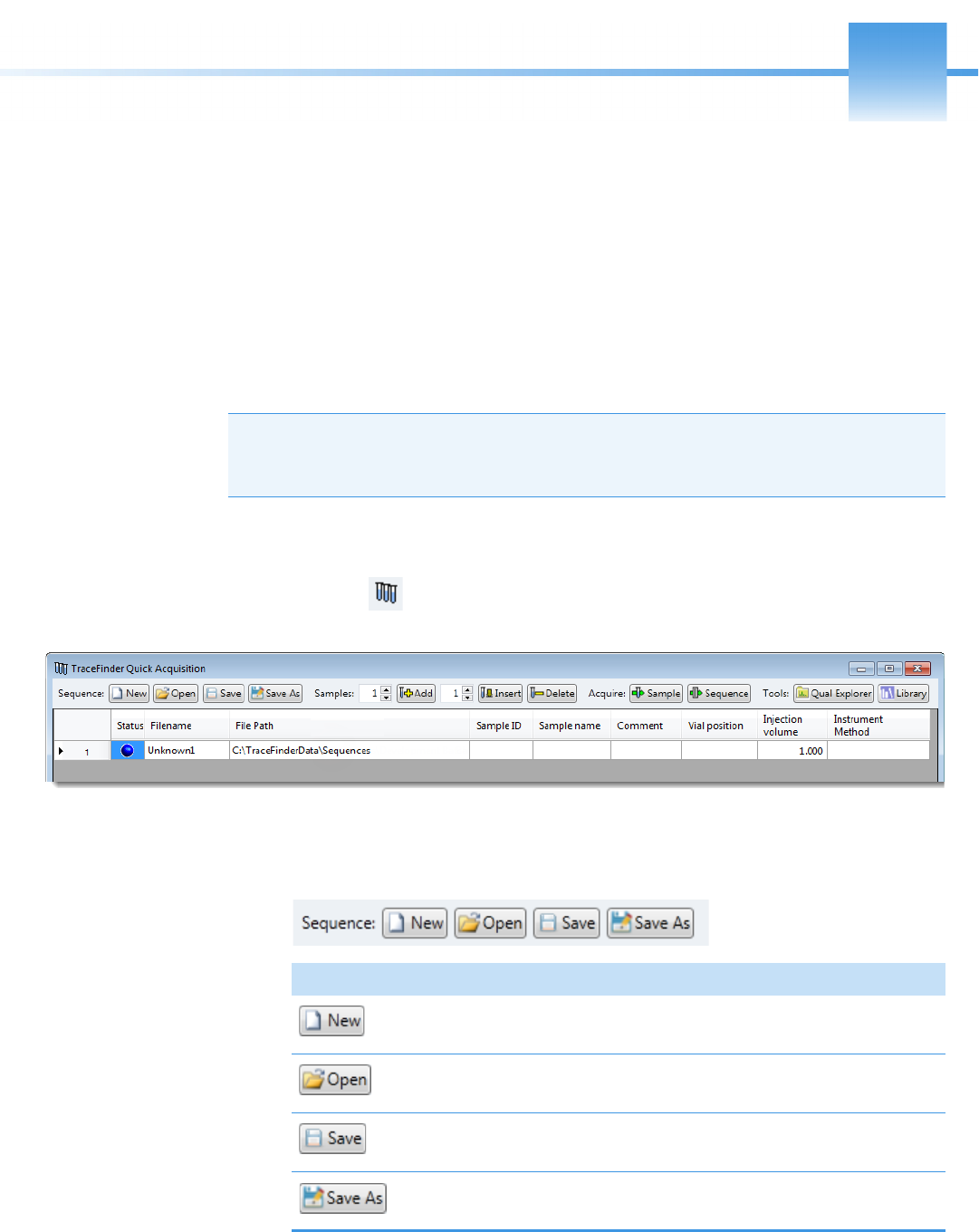
Thermo Scientific TraceFinder User Guide 445
A
Using Quick Acquisition
Use the quick acquisition feature to quickly submit samples from any mode in the
application.
To run a quick acquisition
1. Choose Tools > Quick Acquire Sample from the main menu or click the Quick Acquire
Sample icon, .
The TraceFinder Quick Acquisition window opens.
2. To create the sequence of samples that you want to acquire, do any of the following:
• Use the Sequence buttons in the toolbar to open and save Xcalibur sequence (.sld)
files.
Note The Quick Acquisition feature is available only when you activate it in the
Configuration console. Refer to Chapter 1, “Using the Configuration Console,” in the
TraceFinder Lab Director User Guide.
Button Description
Replaces the current sequence with a new sequence that contains one
Unknown sample.
Opens the Open dialog box where you can open a saved SLD file.
Saves the current sequence as an SLD file in the
C:\TraceFinderData\Sequences folder.
Opens the Save As dialog box where you can save the current sequence
to a new file name or location.
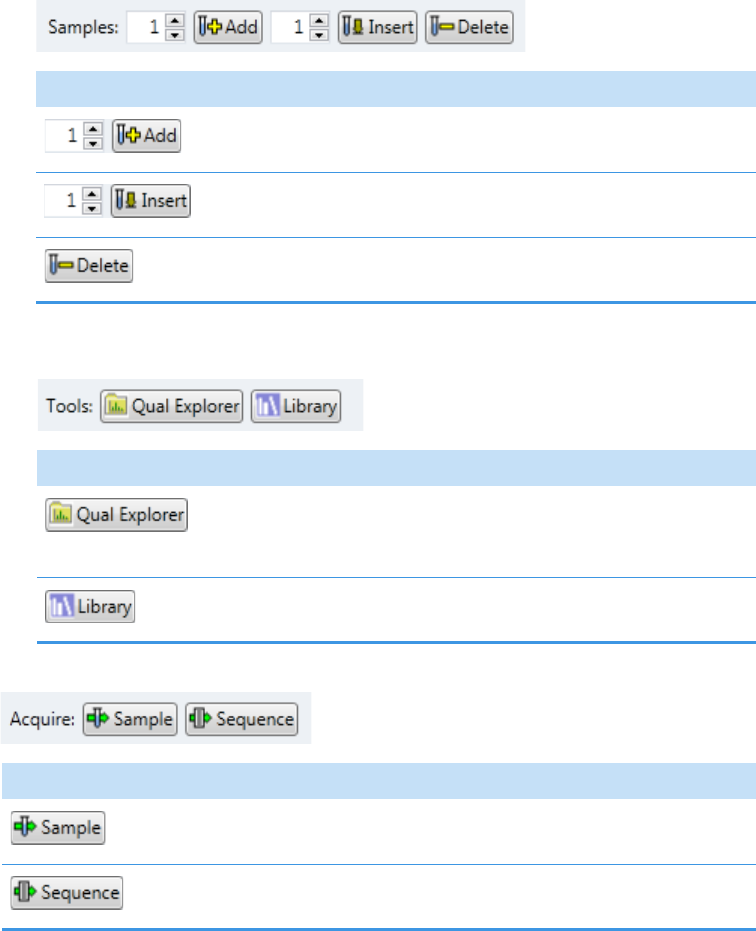
A Using Quick Acquisition
446 TraceFinder User Guide Thermo Scientific
• Use the Samples buttons in the toolbar to create a sequence of samples.
• (Optional) Use the Tools buttons in the toolbar to open a qualitative browser or the
NIST library browser.
3. When you have completed your sequence of samples, click either of the Acquire buttons.
The application submits the samples for acquisition, processing, and report generation.
See Real Time Status – Acquisition Page.
To see how to use the Quick Acquisition wizard
1. Choose Help > Animations.
2. From the list of animation topics, click Quick Acquisition.
Button Description
Adds the specified number of new, empty samples to the end
of the sample list.
Inserts a new, empty sample or samples above the selected
sample.
Removes the selected samples from the sample list.
Button Description
Opens the qualitative explorer that you configured as your
default qualitative explorer in the Configuration console. See
Launching a Qualitative Explorer.
Opens the NIST library browser. See Launching the NIST
Library Browser.
Button Description
Submits only the selected samples for acquisition, processing, or
report generation.
Submits the sequence for acquisition, processing, or report
generation.
Thermo Scientific TraceFinder User Guide 447
B
Using Copy Down and Fill Down
This appendix describes the Copy Down and Fill Down commands that you can use to make
entering column values easier.
• Use the Fill Down command for the Filename, Sample Name, Sample ID, and Vial
Position columns.
• Use the Copy Down command for the Sample Type, Vial Position, Injection Volume,
Conv Factor, Level, Comment, and other columns.
Follow these procedures:
•To automatically copy column values
•To automatically enter sequential column values
•To use Copy Down or Fill Down for a range of samples
To automatically copy column values
1. Select the cell whose value you want to copy to all cells below it.
Observe the difference between a selected and nonselected cell.
2. Right-click and choose Copy Down.
The value is copied to all rows below the selected row.
To automatically enter sequential column values
1. Enter a value for the first row of the fill down sequence.
This does not have to be the first sample row. You can begin the fill down procedure from
any row in the sequence.
Selected
Not selected
B Using Copy Down and Fill Down
448 TraceFinder User Guide Thermo Scientific
2. Select the cell whose value is the first in the fill down sequence.
Observe the difference between a selected and nonselected cell.
3. Right-click and choose Fill Down.
The application enters sequential column values starting with the value in the selected
row and ending with the last row in the column.
You can repeatedly use the Fill Down command to create multiple sequences.
When you use the Fill Down command for the Vial Position column with an autosampler
configured, the application knows the number of vial positions configured in your
autosampler and numbers the positions accordingly.
Selected
Not selected
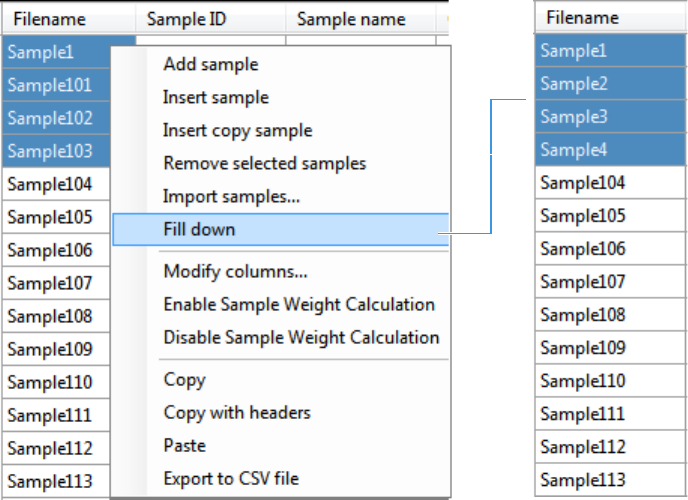
B Using Copy Down and Fill Down
Thermo Scientific TraceFinder User Guide 449
To use Copy Down or Fill Down for a range of samples
1. To select a range of sample values, do one of the following:
Drag your cursor to select a contiguous group of sample values.
–or–
Hold down the SHIFT key to select a contiguous group of sample values.
2. Right-click and choose the appropriate command from the shortcut menu.
The column values are copied or entered sequentially starting with the value in the first
selected row and ending with the last selected row.
Thermo Scientific TraceFinder User Guide 451
C
Moving Data Review Panes
This appendix describes the procedures you can use to move, dock, or float panes in all Data
Review views. The procedures have accompanying animations.
Follow these procedures:
•To move a docked pane
•To make a pane floating or dockable
•To change a pane from a docked pane to a tabbed pane
To move a docked pane
1. Grab the title bar of the pane and begin dragging the pane.
The application displays docking arrows.
2. Drag the pane over one of the arrows.
As you hold the cursor over a docking arrow, the application displays a blue region
indicating where this arrow will place the pane.
3. Drop the pane onto one of the arrows.
This animation shows the various ways that you can use the docking mechanism to move a
pane. To view the animation, click the filmstrip, and then right-click and choose Full Screen
Multimedia. To stop the animation, press ESC.
C Moving Data Review Panes
452 TraceFinder User Guide Thermo Scientific
To make a pane floating or dockable
Do one of the following:
• To make a dockable pane floating, right-click the title bar of the pane and choose
Floating.
While a pane is set as floating, you cannot use the docking arrows to dock it or make
it a tabbed pane.
• To make a floating pane dockable, right-click the title bar of the pane and choose
Dockable.
This animation shows how to switch a pane from docked to floating and back to docked. To
view the animation, click the filmstrip, and then right-click and choose Full Screen
Multimedia. To stop the animation, press ESC.
To change a pane from a docked pane to a tabbed pane
1. Grab the title bar of the pane and begin dragging the pane.
The application displays docking arrows.
2. Hold the cursor over the center of the docking arrows to display a blue region indicating
the location of the tabbed pane.
3. Drop the pane over the center of the docking arrows.
This animation shows how to change a pane from a docked pane to a tabbed pane and back to
a docked pane. To view the animation, click the filmstrip, and then right-click and choose
Full Screen Multimedia. To stop the animation, press ESC.
Note To change a floating pane to a tabbed pane, you must first make the pane a dockable
pane, and then you can make it a tabbed pane.
Drop the pane on the center location to create a tabbed pane.