User Guide
User Manual: Pdf
Open the PDF directly: View PDF ![]() .
.
Page Count: 35
- 1. What is this?
- 2. Install the ChemoPy package
- 3. Read molecules
- 4. Download molecules from corresponding ID
- 5. Calculating molecular descriptors
- 5.1. Calculating 2-D descriptors
- 5.1.1. Molecular constitutional descriptors
- 5.1.2. Topology descriptors
- 5.1.3. Molecular connectivity indices
- 5.1.4. Kappa shape descriptors
- 5.1.5. Burden descriptors
- 5.1.6. E-state indices
- 5.1.7. Basak information indices
- 5.1.8. Autocorrelation descriptors
- 5.1.9. Molecular properties
- 5.1.10. Charge descriptors
- 5.1.11. MOE-type descriptors
- 5.1.12. Using PyChem2d object
- 5.2. Calculating 3-D descriptors
- 5.3. Molecular fingerprints and chemoinforamtics
- 5.1. Calculating 2-D descriptors
- 6. Calculating all descriptors
- Appendix:

User Guide for ChemoPy 1.0
Dongsheng Cao
Yizeng Liang
©2012 China Computational Biology Drug Design Group

Table of Contents
1. What is this?...........................................................................................................................................3
2. Install the ChemoPy package.................................................................................................................3
3. Read molecules......................................................................................................................................4
4. Download molecules from corresponding ID........................................................................................5
5. Calculating molecular descriptors..........................................................................................................6
5.1. Calculating 2-D descriptors.........................................................................................................7
5.1.1. Molecular constitutional descriptors..................................................................................7
5.1.2. Topology descriptors..........................................................................................................8
5.1.3. Molecular connectivity indices.........................................................................................8
5.1.4. Kappa shape descriptors....................................................................................................9
5.1.5. Burden descriptors.............................................................................................................9
5.1.6. E-state indices....................................................................................................................9
5.1.7. Basak information indices................................................................................................10
5.1.8. Autocorrelation descriptors..............................................................................................10
5.1.9. Molecular properties........................................................................................................10
5.1.10. Charge descriptors..........................................................................................................11
5.1.11. MOE-type descriptors....................................................................................................11
5.1.12. Using PyChem2d object.................................................................................................12
5.2. Calculating 3-D descriptors.......................................................................................................12
5.2.1. Geometric descriptors......................................................................................................12
5.2.2. CPSA descriptors.............................................................................................................13
5.2.3. RDF descriptors...............................................................................................................13
5.2.4. MoRSE descriptors..........................................................................................................13
5.2.5. WHIM descriptors............................................................................................................14
5.2.6. Using PyChem3d object...................................................................................................14
5.3. Molecular fingerprints and chemoinforamtics...........................................................................15
5.3.1. Daylight-type fingerprints................................................................................................15
5.3.2. MACCS keys and FP4 fingerprints.................................................................................15
5.3.3. E-state fingerprints...........................................................................................................16
5.3.4. Atom pairs and topological torsions.................................................................................16
5.3.5. Morgan fingerprints.........................................................................................................16
5.3.6. Using PyChem2d object...................................................................................................17
5.3.7. fingerprint similarity........................................................................................................17
6. Calculating all descriptors....................................................................................................................18
Appendix:.................................................................................................................................................19

1. What is this?
This document is intended to provide an overview of how one can use the ChemoPy functionality from
Python. It’s not comprehensive and it’s not a manual.
If you find mistakes, or have suggestions for improvements, please either fix them yourselves in the
source document (the .py file) or send them to the mailing list: oriental-cds@hotmail.com
2. Install the ChemoPy package
ChemoPy has been successfully tested on Linux and Windows systems. The author could download the
ChemoPy package via: http://code.google.com/p/pychem/downloads/list (.zip and .tar.gz). The install
process of ChemoPy is very easy:
***************************************************************
* You first need to install RDKit, Openbabel, MOPAC and pybel successfully.*
***************************************************************
Openbabel and pybel can be downloaded via: http://openbabel.org/wiki/Main_Page
RDkit can be downloaded via: http://code.google.com/p/rdkit/
MOPAC can be downloaded via: http://openmopac.net/
Note: ChemoPy was tested in MOPAC 7.
On Windows:
(1): download the chemopy package (.zip)
(2): extract or uncompress the .zip file
(3): cd chemopy-1.0
(4): python setup.py install
On Linux:
(1): download the chemopy package (.tar.gz)
(2): tar -zxf chemopy-1.0.tar.gz

(3): cd chemopy-1.0
(4): python setup.py install or sudo python setup.py install
3. Read molecules
The majority of the basic molecular functionality is found in module pychem:
Individual molecule can be constructed using a variety of approaches.
Because we have imported pybel module, all functionalities in pybel can be used to construct a Mol
object. Moreover, the transformation between any two molecular formats are allowed by using pybel.
Note: When computing 2-D descriptors by individual module, we used Mol object from RDKit. When
computing 3-D descriptors by individual module, we used Mol object from pybel. The compatibility
between ChemoPy and other packages ensures that ChemoPy could be conveniently transplanted.
The ChemoPy allow the users to provide different molecular formats when using PyChem2d object or
PyChem3d object.
Using PyChem2d:
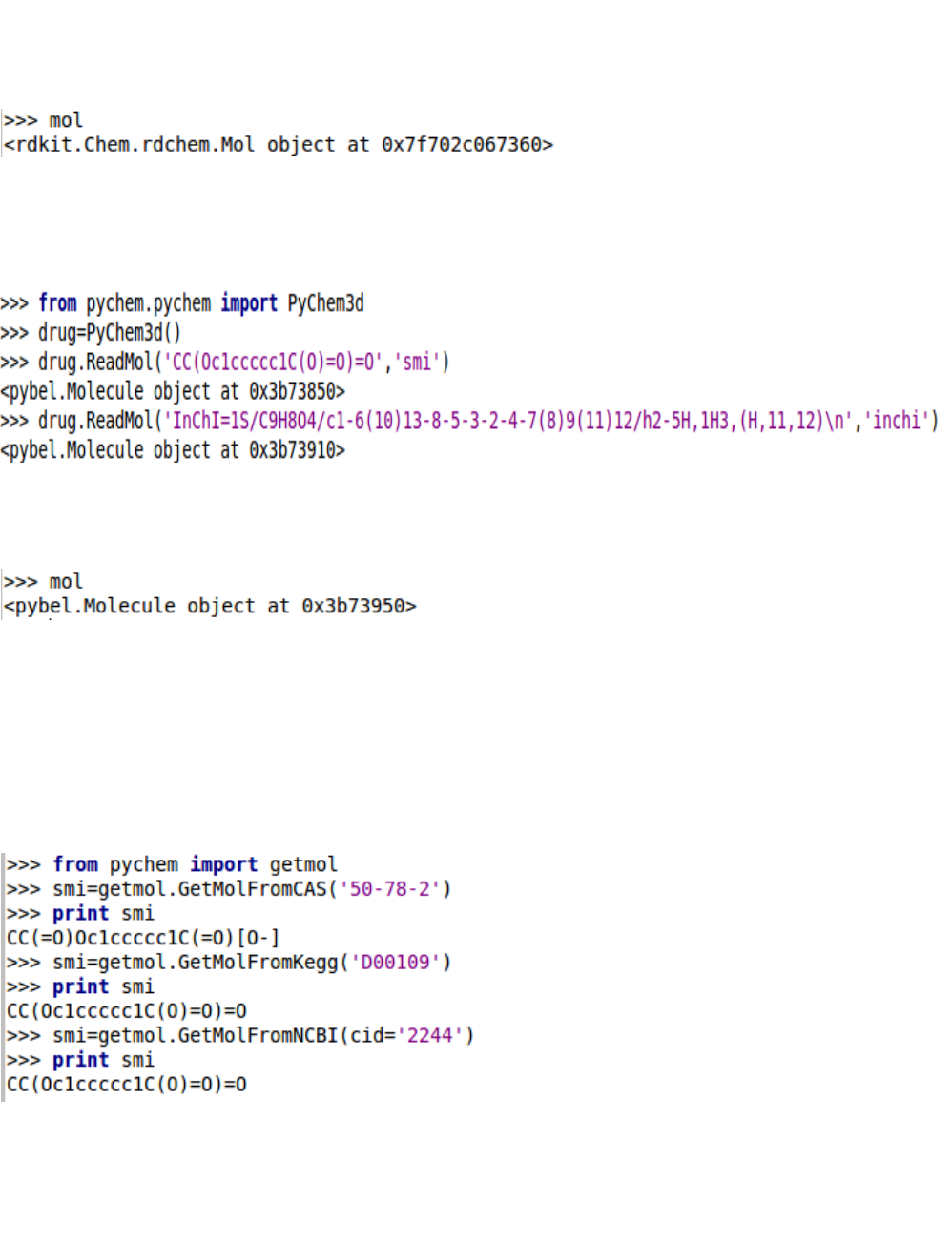
All of these functions return a Mol object on success:
Using PyChem3d:
All of these functions return a Mol object on success:
4. Download molecules from corresponding ID
The PyDPI allows the user to download the molecules by providing their IDs such as CAS, NCBI,
KEGG, EBI and Drugbank.
By providing a aspirin IDs, we could download its SMILES format conveniently.
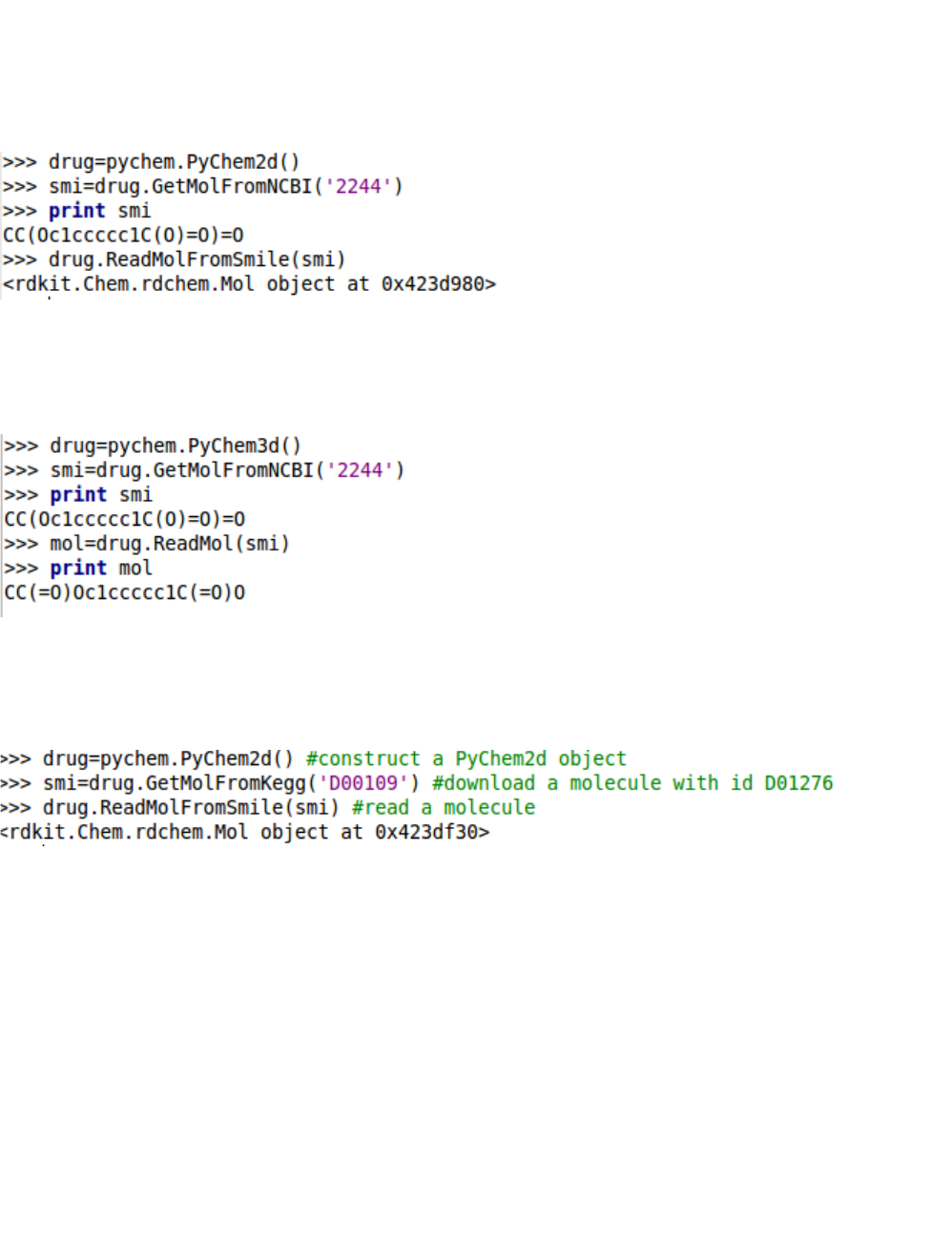
We can also download and read a molecule by constructing a PyChem2d or PyChem3d object, which
contains the majority of the basic drug molecular functionality.
Using PyChem2d object:
Using PyChem3d object:
You could read a molecule by providing a KEGG ID:
5. Calculating molecular descriptors
The ChemoPy package could calculate a large number of molecular descriptors. These descriptors cap-
ture and magnify distinct aspects of chemical structures. Generally speaking, all descriptors could be
divided into two classes: 2-D descriptors and 3-D descriptors. 2-D descriptors only used the property of
molecular topology, including constitutional descriptors, topological descriptors, connectivity indices,
E-state indices, Basak information indices, Burden descriptors, autocorrelation descriptors, charge de-

scriptors, molecular properties, kappa shape indices, MOE-type descriptors, and molecular fingerprints.
3-D descriptors need the optimization of molecular structure. In ChemoPy 1.0, we used a widely used
MOPAC program to optimize molecular structures by the AM1 method. The 3-D descriptors calculated
by ChemoPy include geometric descriptors, CPSA descriptors, RDF descriptors, MoRSE descriptors,
and WHIM descriptors. The ChemoPy package could compute 1135 molecular descriptors.
Once we read a Mol object, we could easily calculate these molecular descriptors:
5.1. Calculating 2-D descriptors
We could import the corresponding module to calculate the molecular descriptors as need. There are 13
modules to compute 2-D descriptors. Moreover, a easier way to compute these descriptors is construct
a PyChem2d object, which encapsulates all methods for the calculation of 2-D descriptors.
5.1.1. Molecular constitutional descriptors
We could calculate any constitution descriptor by calling the corresponding functions. We could also
calculate all 30 descriptors by calling GetConstitution function. The result is given in the form of dic-
tionary.
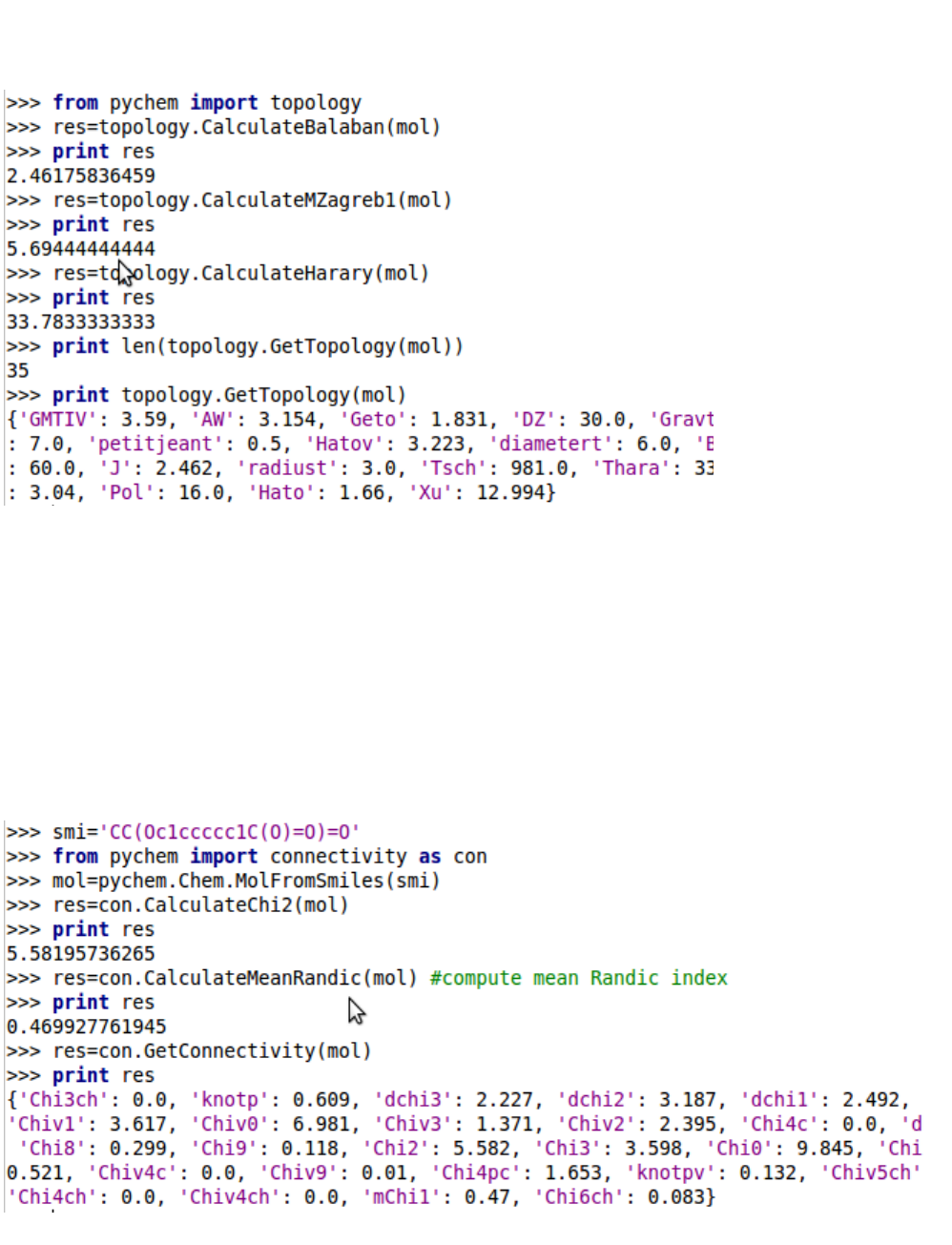
5.1.2. Topology descriptors
35 topology descriptors can be calculated by the ChemoPy package. For detailed information of topol-
ogy descriptors, refer to Table S1 in Appendix and their introductions in Manual.
5.1.3. Molecular connectivity indices
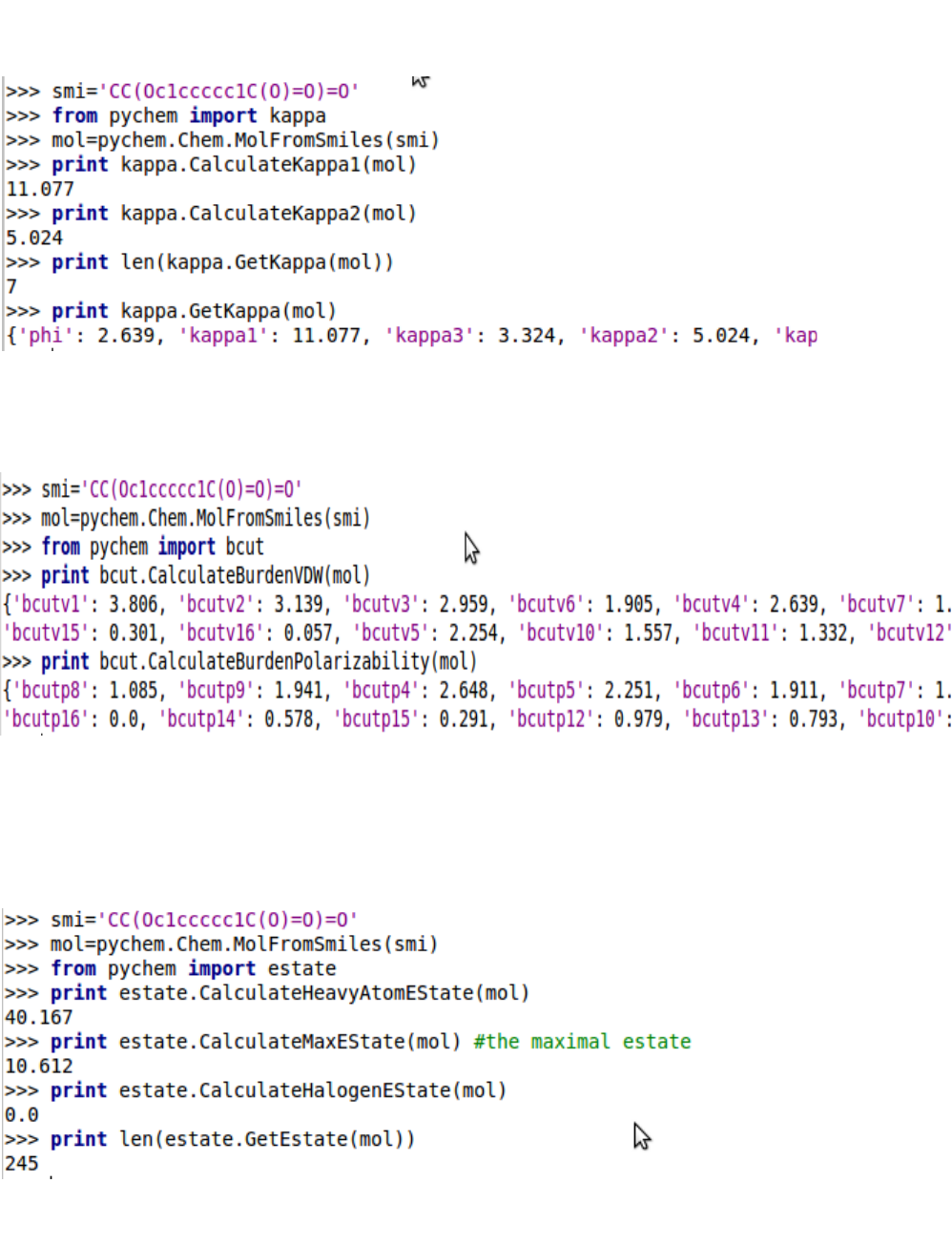
5.1.4. Kappa shape descriptors
5.1.5. Burden descriptors
5.1.6. E-state indices
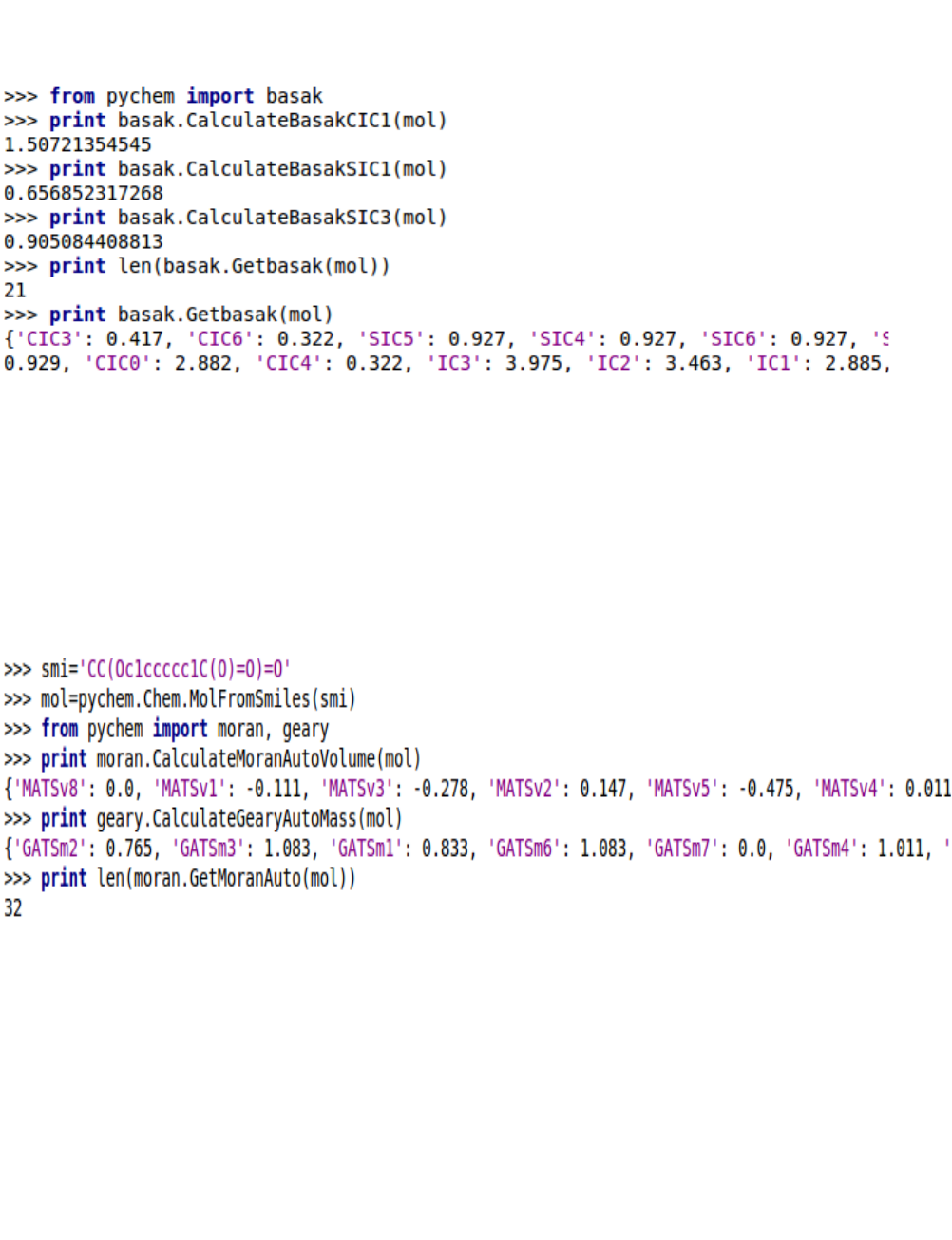
5.1.7. Basak information indices
5.1.8. Autocorrelation descriptors
There are three types of autocorrelation descriptors in the ChemoPy package: Moreau-Broto, Moran,
Geary.
5.1.9. Molecular properties
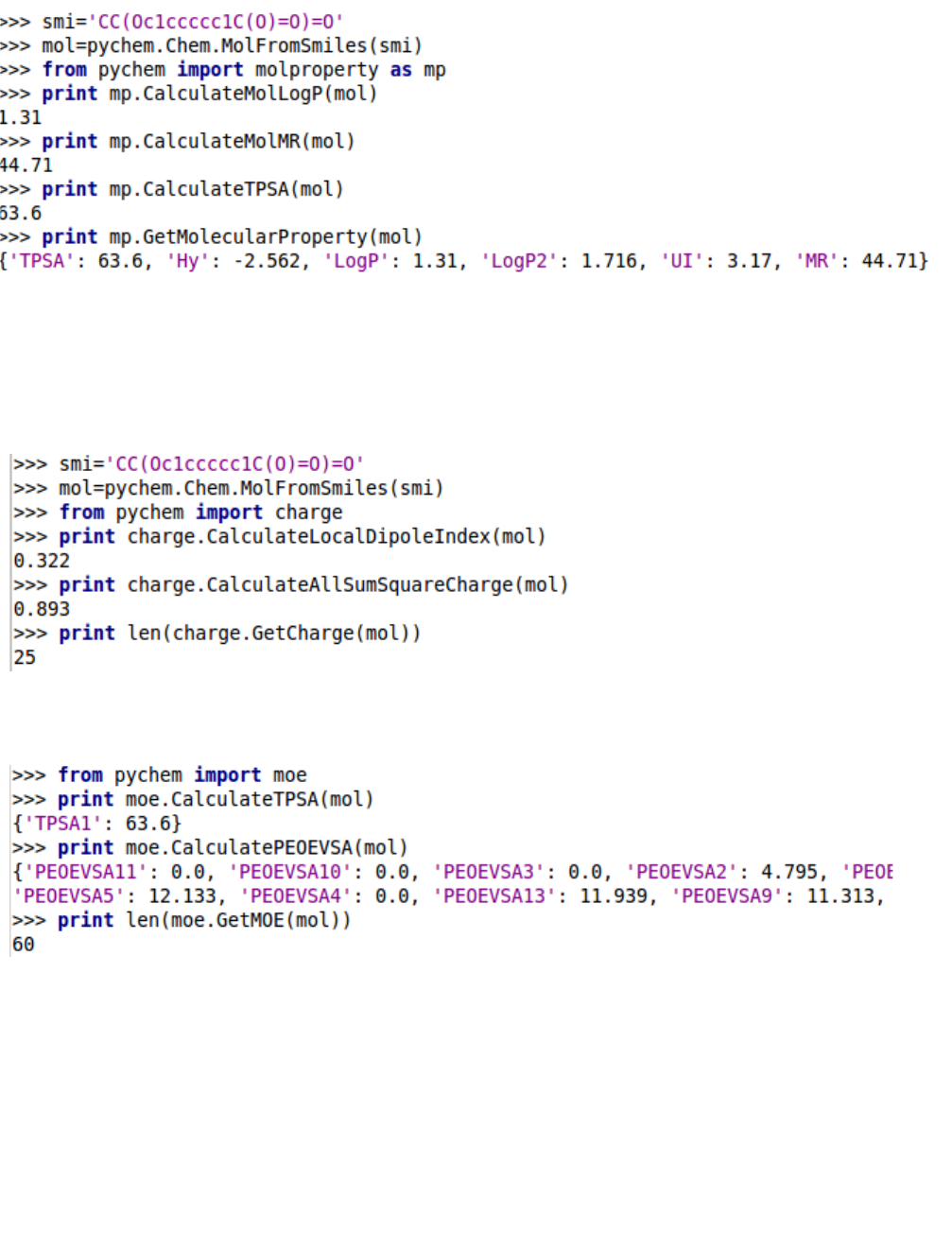
5.1.10. Charge descriptors
5.1.11. MOE-type descriptors
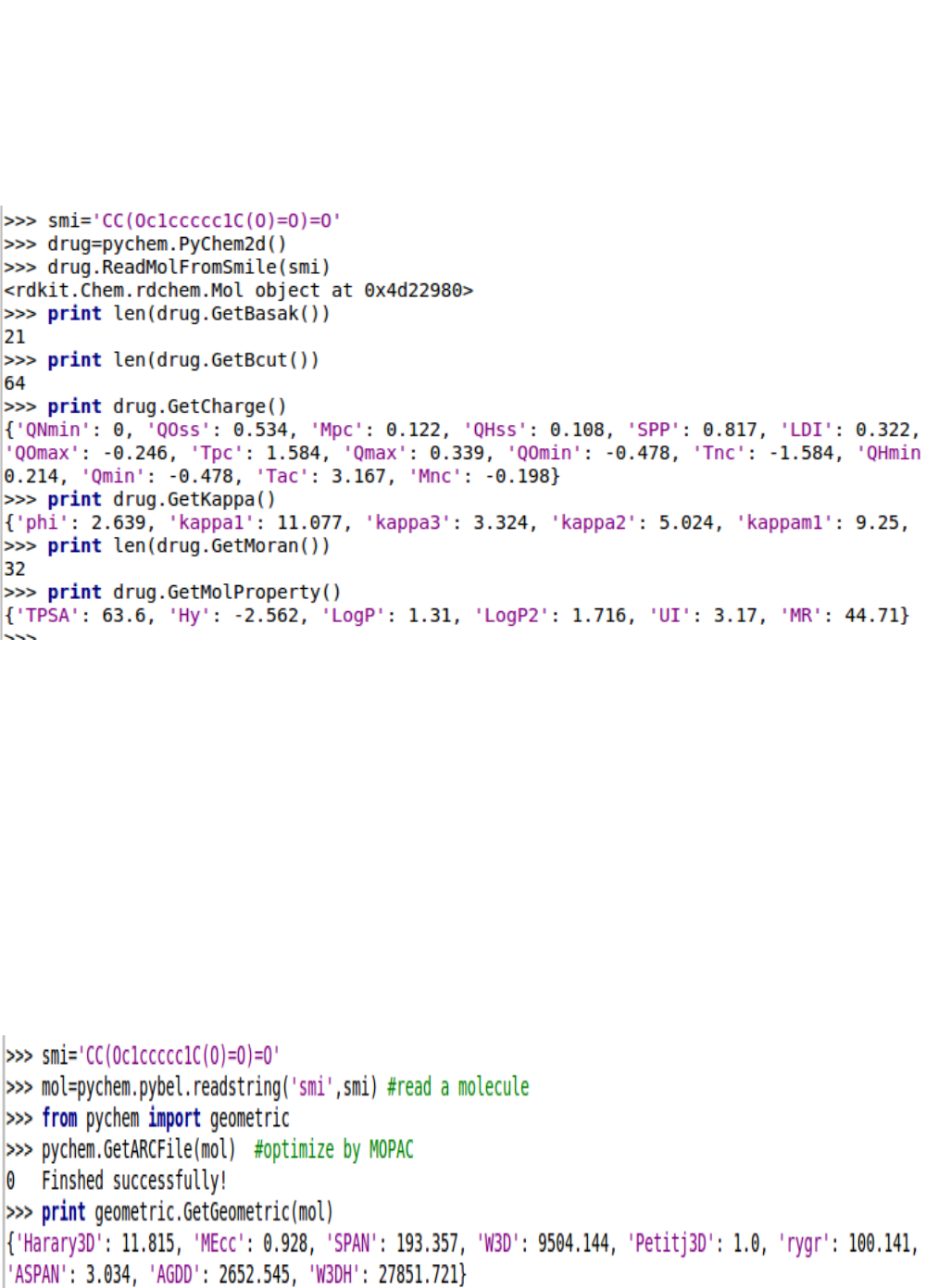
5.1.12. Using PyChem2d object
A easier way to calculate molecular descriptors is to generate a PyChem2d object and then call their
methods. The PyChem2d contains the majority of drug molecule operation functionality.
5.2. Calculating 3-D descriptors
The 3-D molecular descriptors calculated by ChemoPy include geometric descriptors, CPSA descrip-
tors, RDF descriptors, MoRSE descriptors, and WHIM descriptors. We could import the corresponding
module to calculate the molecular descriptors as need. There are 5 modules to compute 3-D descriptors.
Moreover, a easier way to compute these descriptors is construct a PyChem3d object, which encapsu-
lates all methods for the calculation of 3-D descriptors.
5.2.1. Geometric descriptors
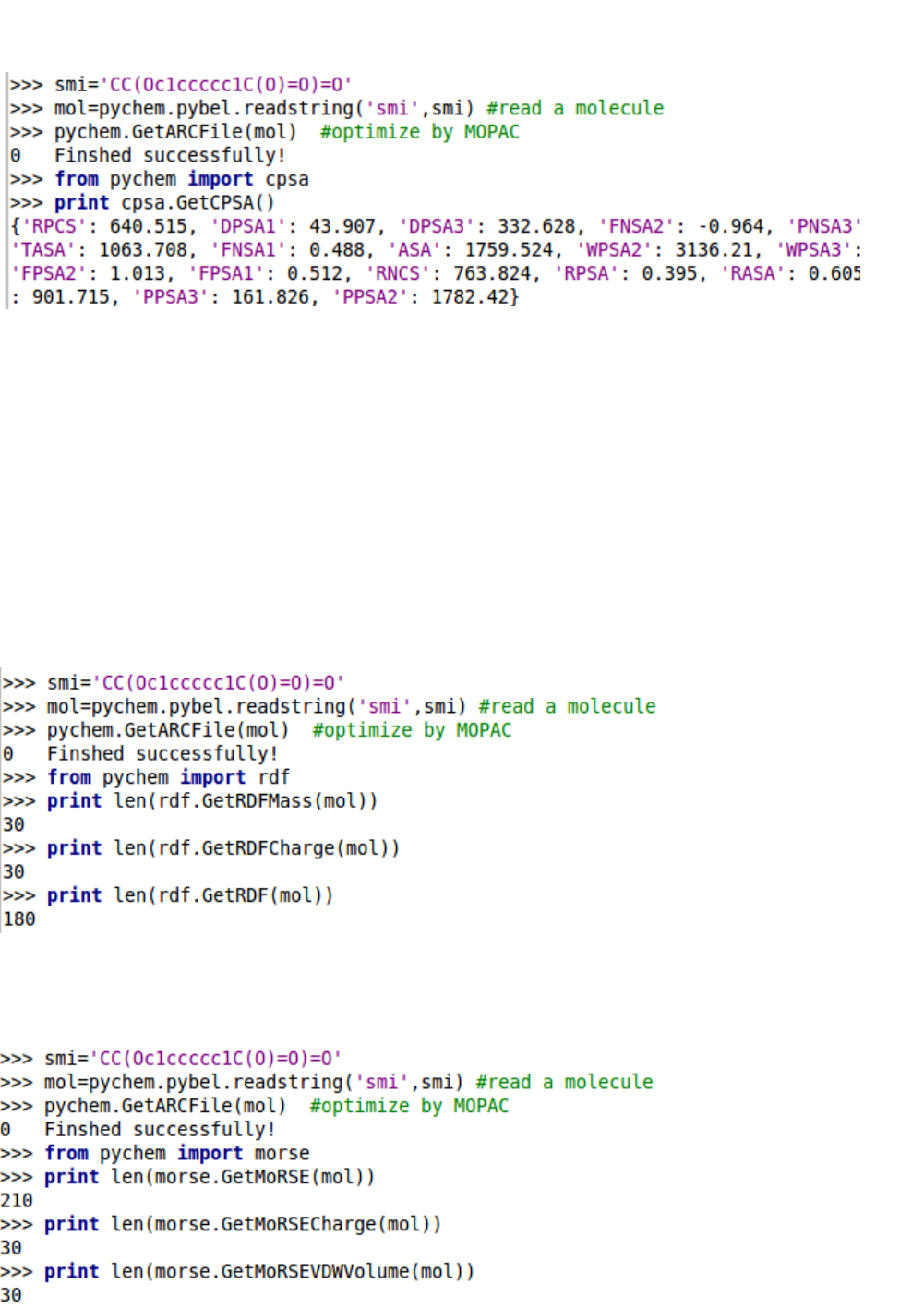
5.2.2. CPSA descriptors
5.2.3. RDF descriptors
5.2.4. MoRSE descriptors
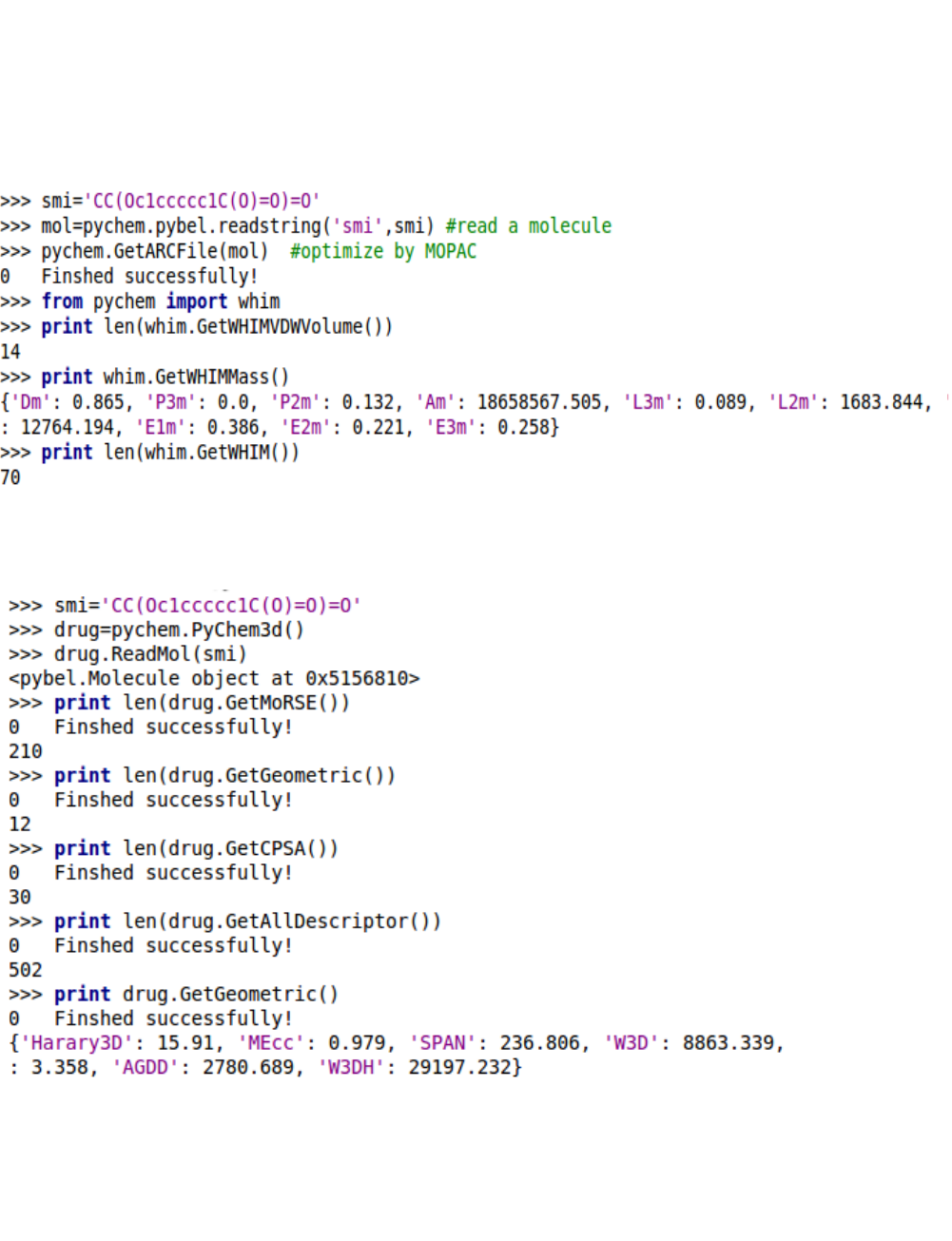
5.2.5. WHIM descriptors
5.2.6. Using PyChem3d object
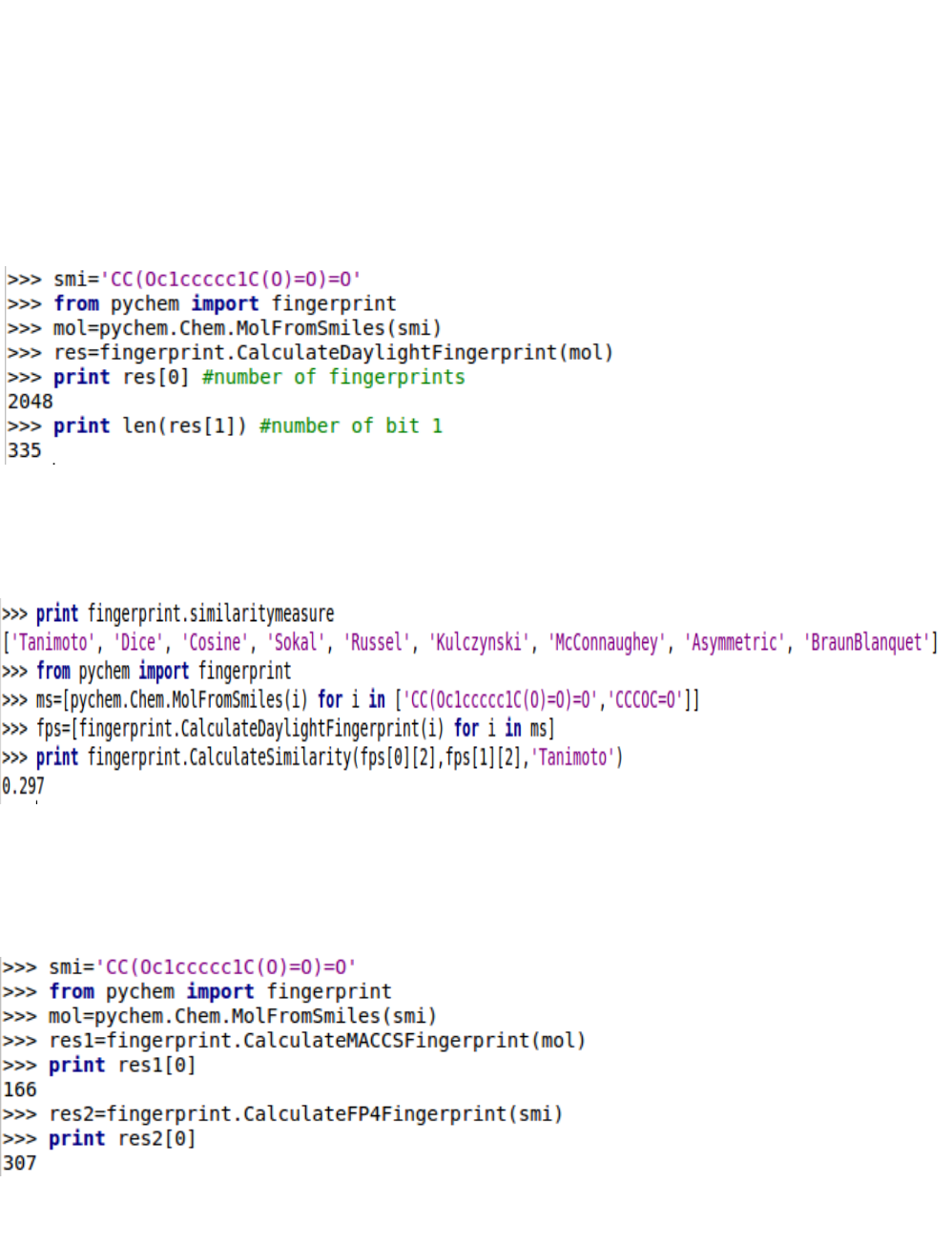
5.3. Molecular fingerprints and chemoinforamtics
In the ChemoPy package, there are seven types of molecular fingerprints which are defined by abstract-
ing and magnifying different aspects of molecular topology.
5.3.1. Daylight-type fingerprints
We can calculate the similarity between two molecules by specifying a type of similarity measure.
There exist to be nine types of similarity measures to calculate the similarity between two molecules.
5.3.2. MACCS keys and FP4 fingerprints

Note that the input of MACCS and FP4 is different.
5.3.3. E-state fingerprints
5.3.4. Atom pairs and topological torsions
5.3.5. Morgan fingerprints
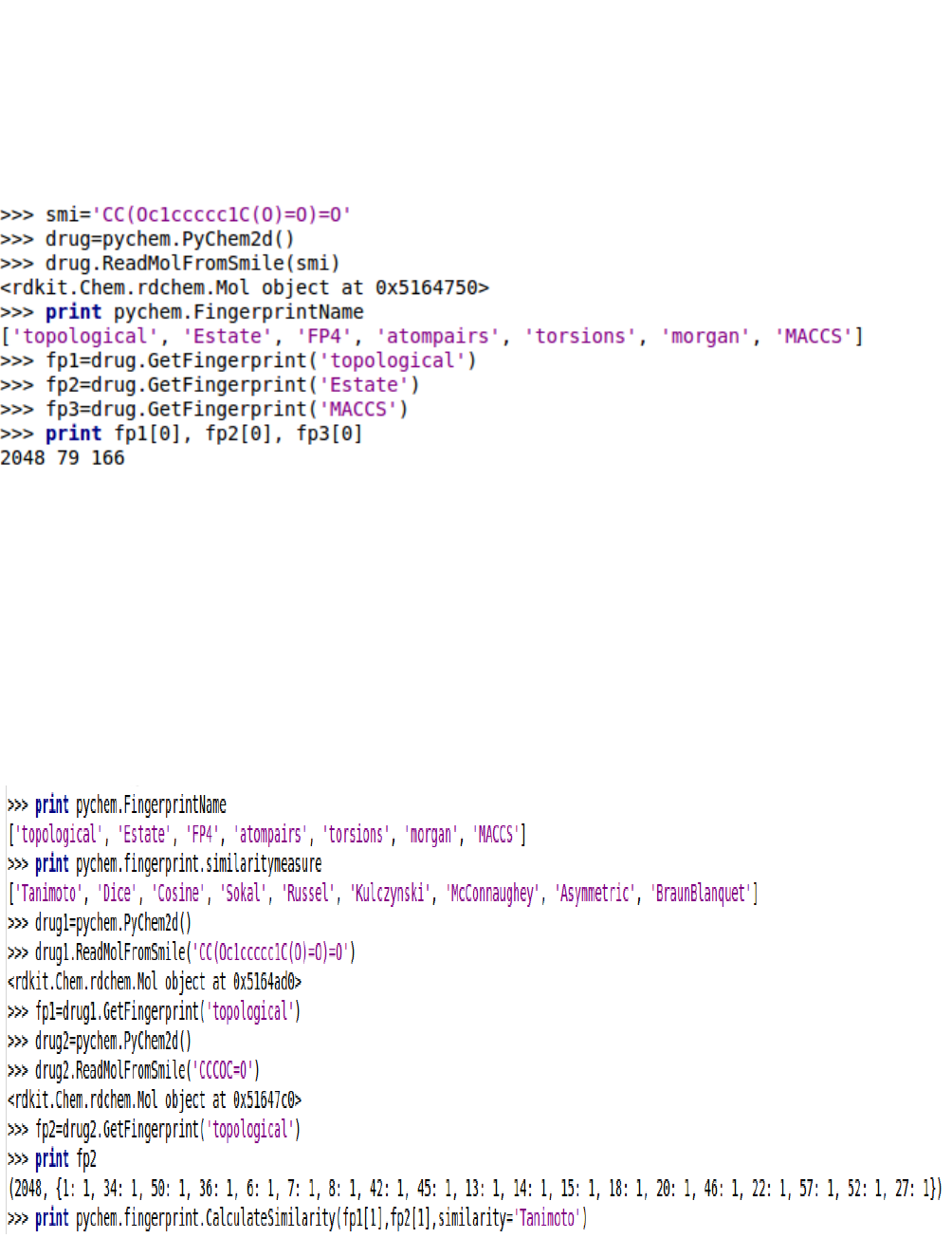
5.3.6. Using PyChem2d object
The convenient way to calculate the fingerprints is to generate a PyDrug object and call GetFingerprint
method.
5.3.7. fingerprint similarity
We could any fingerprint similarity using the nine given similarity measure methods.
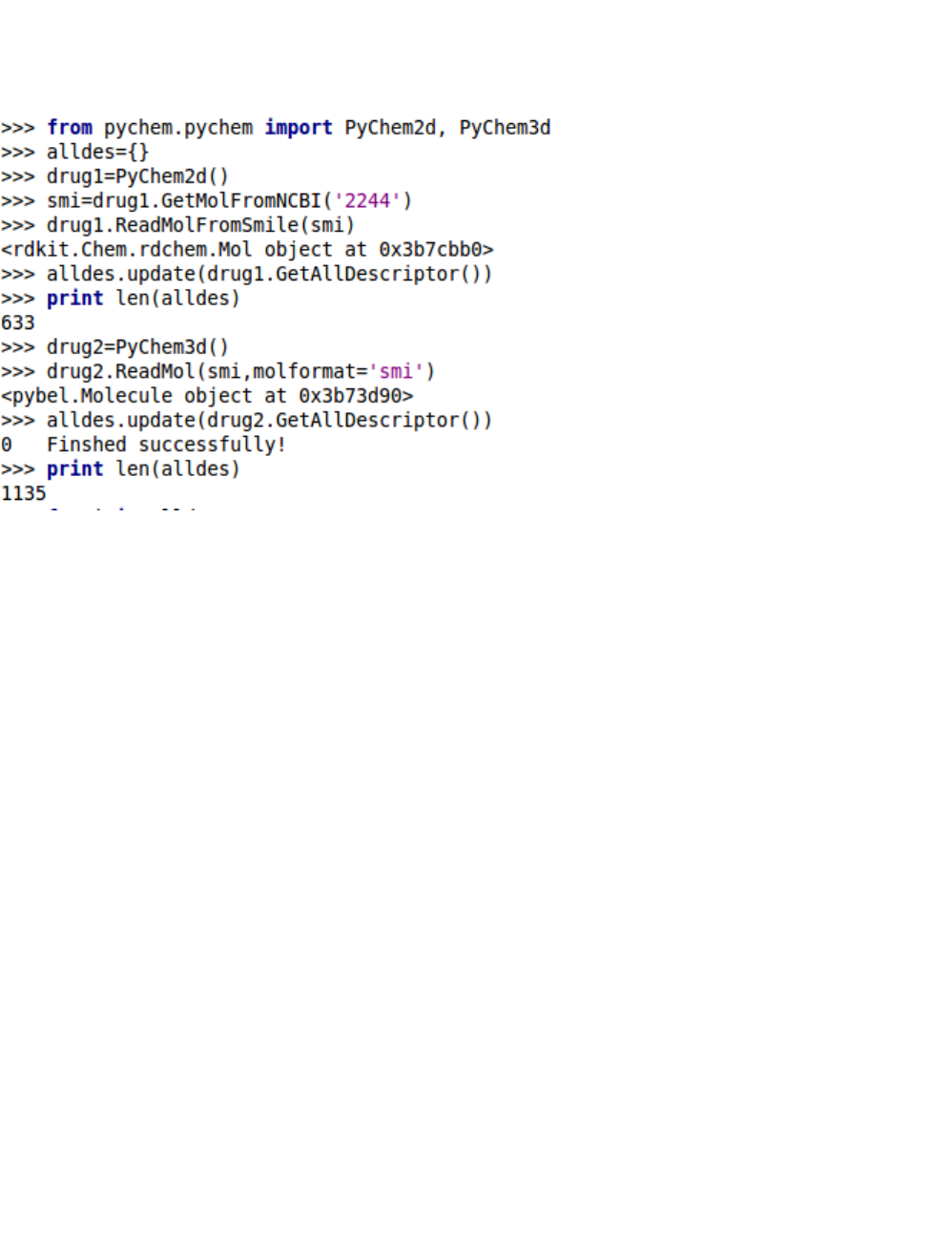
6. Calculating all descriptors
Appendix:
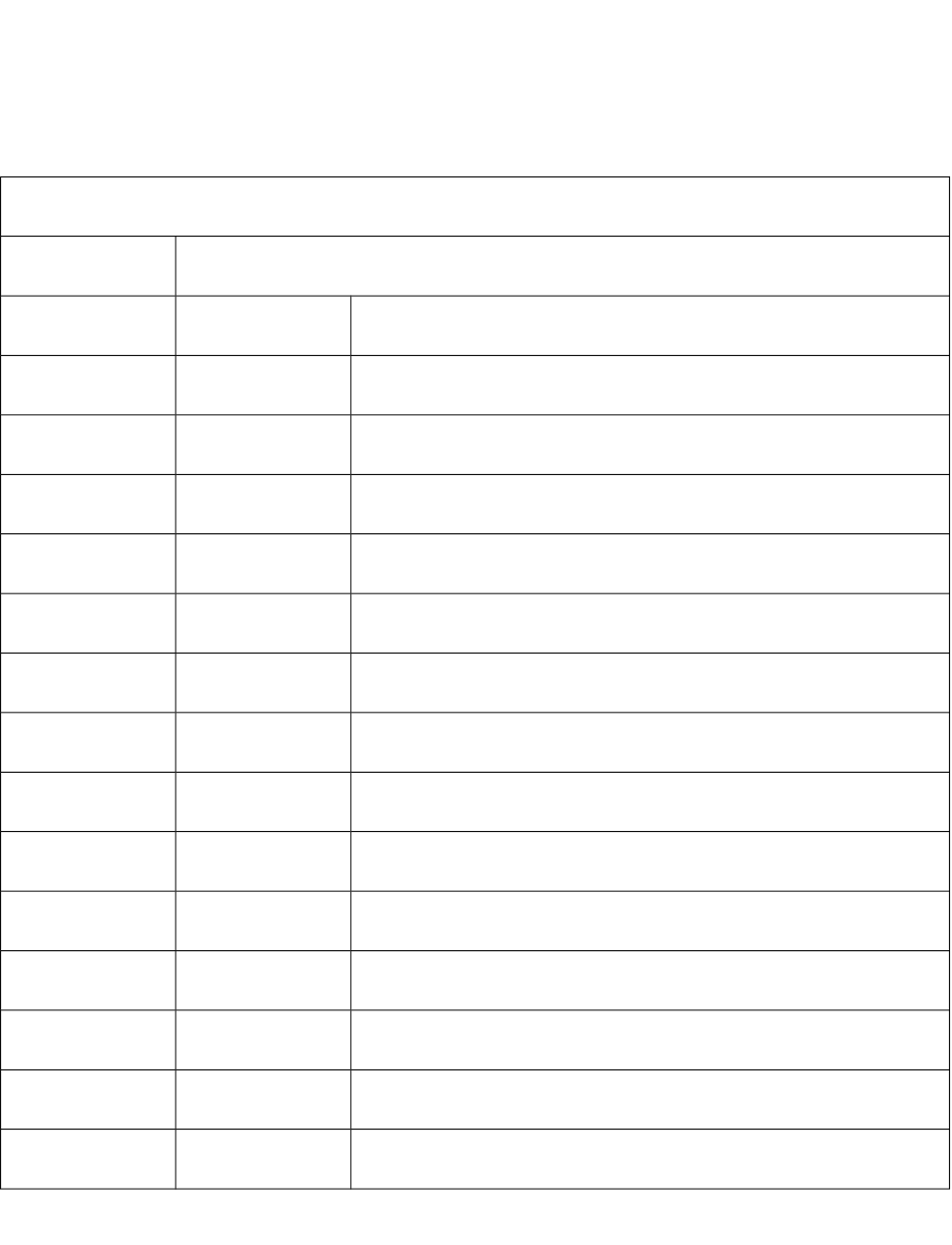
Table S1: List of ChemoPy computed molecular descriptors
Molecular descriptors
Constitutional descriptors (30)
1 Weight Molecular weight
2 nhyd Count of hydrogen atoms
3 nhal Count of halogen atoms
4 nhet Count of hetero atoms
5 nhev Count of heavy atoms
6 ncof Count of F atoms
7 ncocl Count of Cl atoms
8 ncobr Count of Br atoms
9 ncoi Count of I atoms
10 ncarb Count of C atoms
11 nphos Count of P atoms
12 nsulph Count of S atoms
13 noxy Count of O atoms
14 nnitro Count of N atoms
15 nring Number of rings
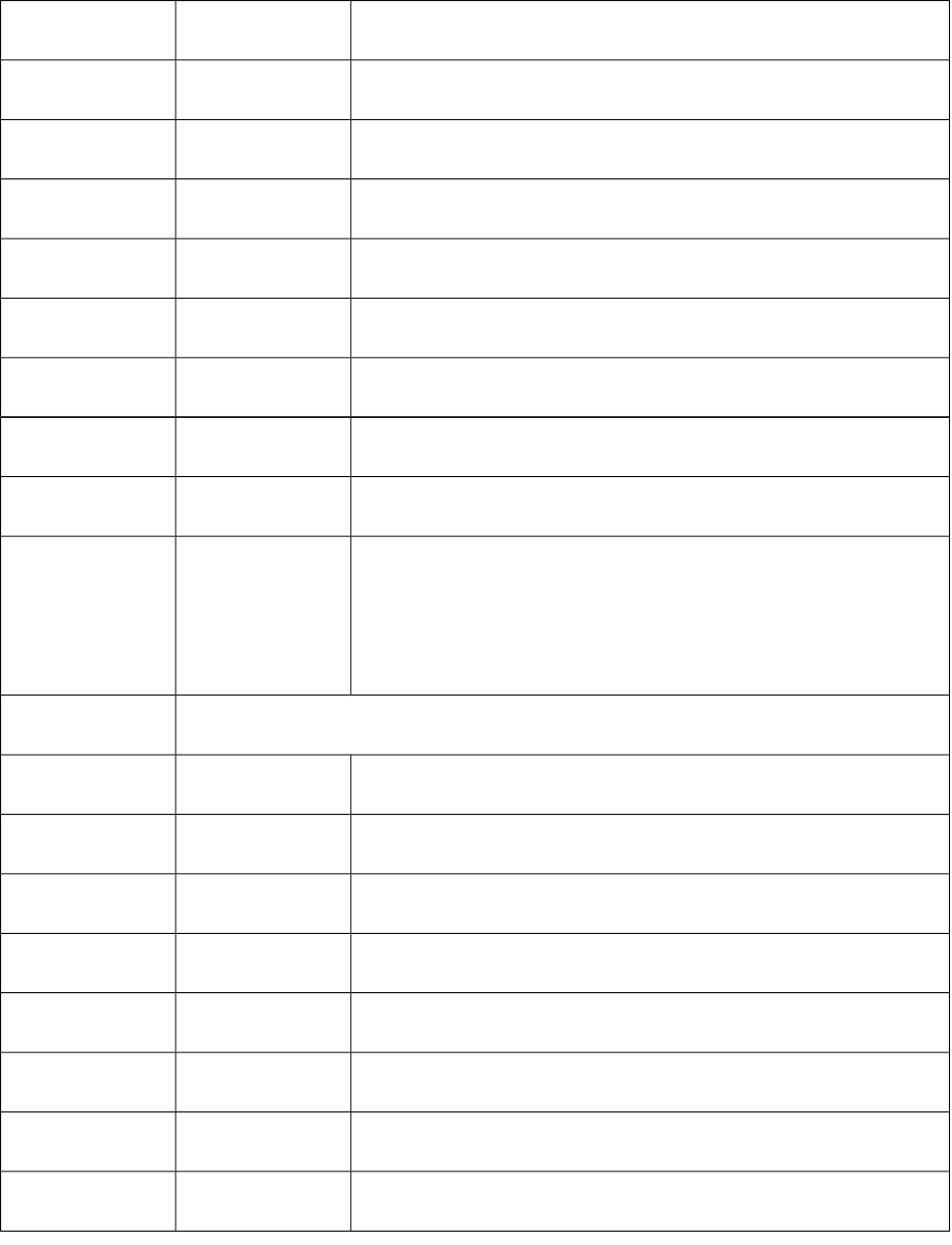
16 nrot Number of rotatable bonds
17 ndonr Number of H-bond donors
18 naccr Number of H-bond acceptors
19 nsb Number of single bonds
20 ndb Number of double bonds
21 ntb Number of triple bonds
22 naro Number of aromatic bonds
23 nta Number of all atoms
24 AWeight Average molecular weight
25-30 PC1
PC2
PC3
PC4
PC5
PC6
Molecular path counts of length 1-6
Topological descriptors (35)
1 W Weiner index
2 AW Average Wiener index
3 J Balaban’s J index
4 Thara Harary number
5 Tsch Schiultz index
6 Tigdi Graph distance index
7 Platt Platt number
8 Xu Xu index
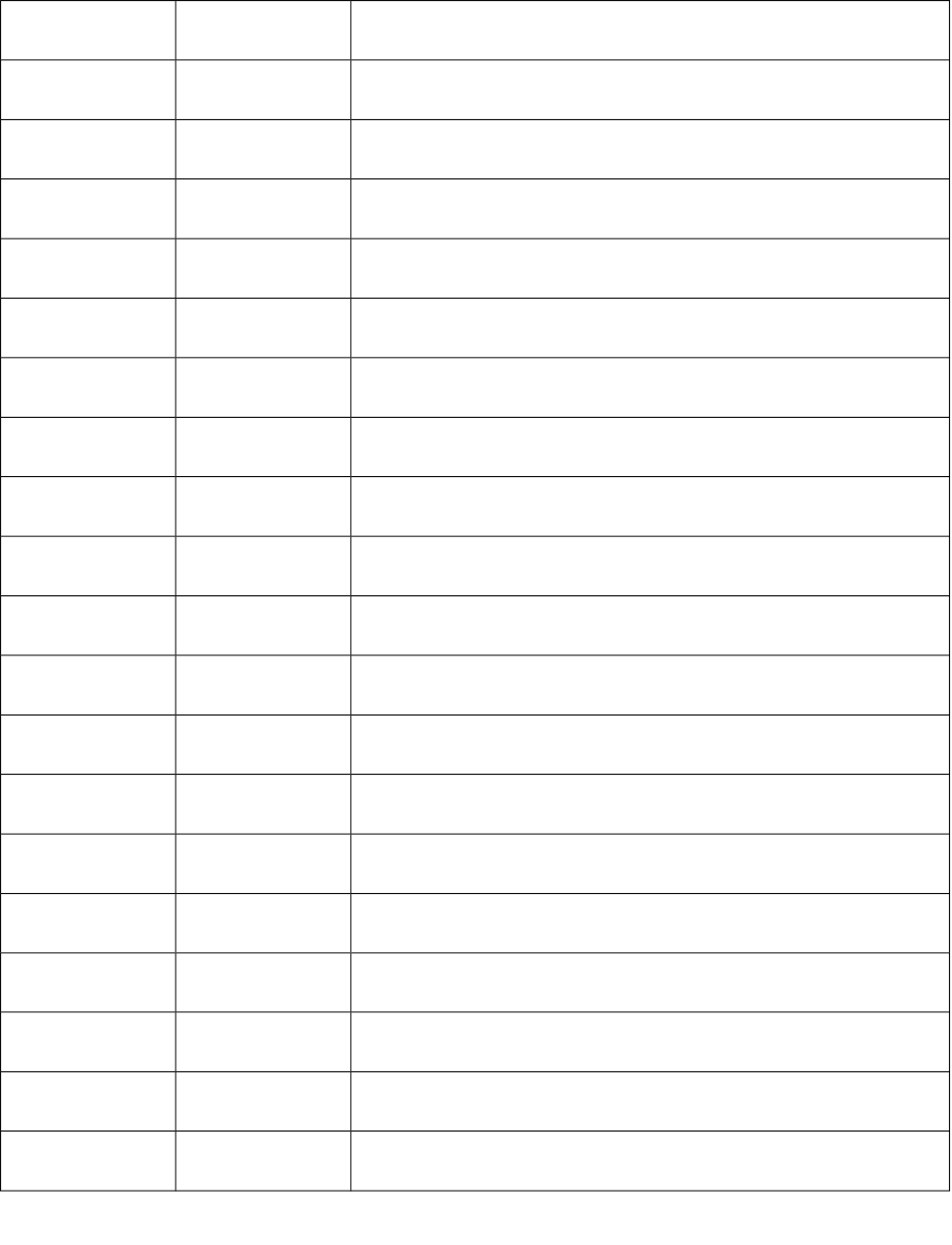
9 Pol Polarity number
10 Dz Pogliani index
11 Ipc Ipc index
12 BertzCT BertzCT
13 GMTI Gutman molecular topological index based on simple vertex
degree
14-15 ZM1
ZM2
Zagreb index with order 1-2
16-17 MZM1
MZM2
Modified Zagreb index with order 1-2
18 Qindex Quadratic index
19 diametert Largest value in the distance matrix
20 radiust radius based on topology
21 petitjeant Petitjean based on topology
22 Sito the logarithm of the simple topological index by Narumi
23 Hato harmonic topological index proposed by Narnumi
24 Geto Geometric topological index by Narumi
25 Arto Arithmetic topological index by Narumi
26 ISIZ Total information index on molecular size
27 TIAC Total information index on atomic composition
28 DET Total information index on distance equality
29 IDE Mean information index on distance equality
30 IVDE Total information index on vertex equality
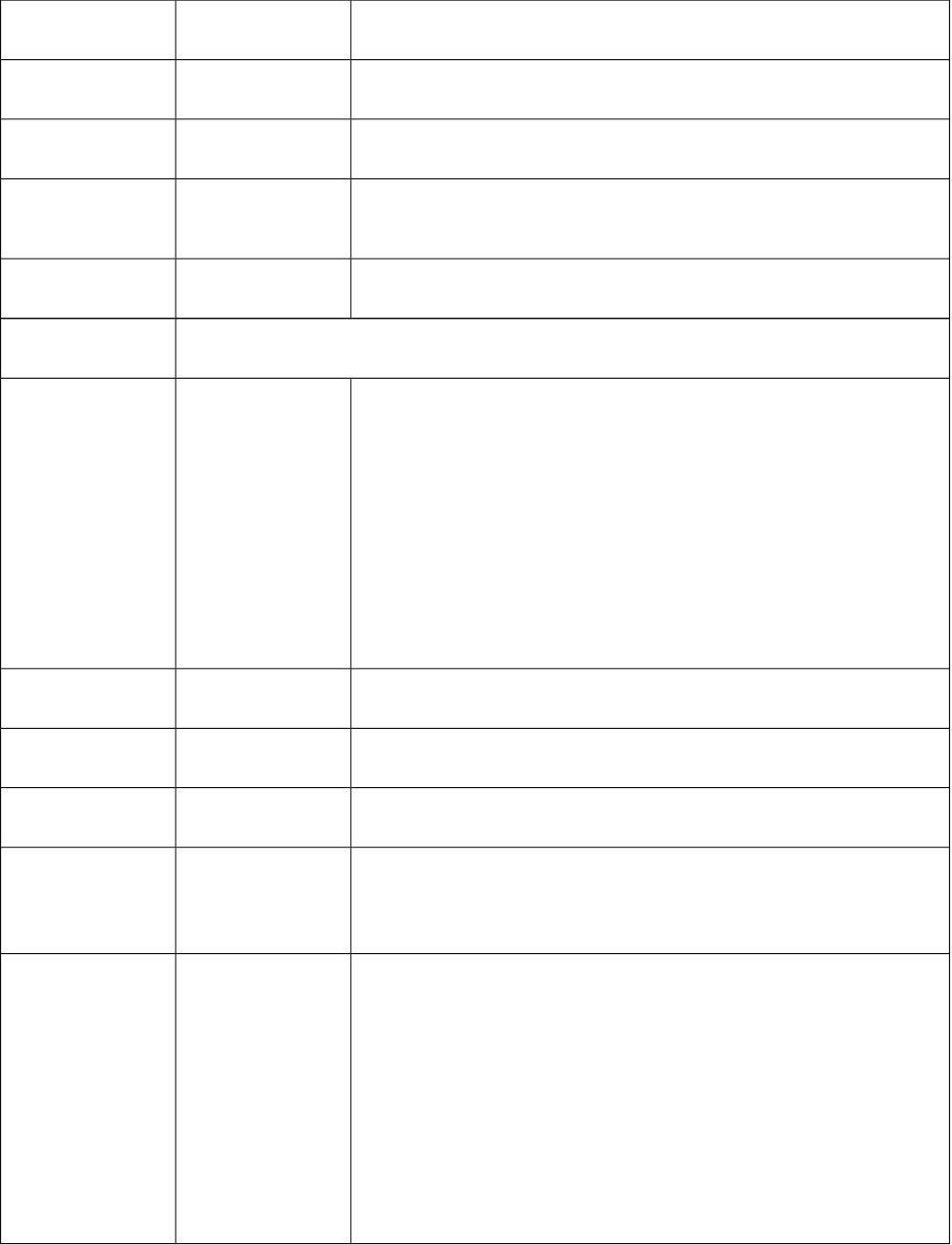
31 Sitov Logarithm of the simple topological index by Narumi
32 Hatov Harmonic topological index proposed by Narnumi
33 Getov Geometric topological index by Narumi
34 Gravto Gravitational topological index based on topological distance
35 GMTIV Gutman molecular topological index based on valence vertex
degree(log10)
Connectivity descriptors (44)
1-11 0χv
1χv
2χv
3χpv
4χpv
5χpv
6χpv
7χpv
8χpv
9χpv
10χpv
Valence molecular connectivity Chi index for path order 0-10
12 3χvcValence molecular connectivity Chi index for three cluster
13 4χvcValence molecular connectivity Chi index for four cluster
14 4χvpc Valence molecular connectivity Chi index for path/cluster
15-18 3χvCH
4χvCH
5χvCH
6χvCH
Valence molecular connectivity Chi index for cycles of 3-6
19-29 0χ
1χ
2χ
3χp
4χp
5χp
6χp
7χp
8χp
9χp
10χp
Simple molecular connectivity Chi indices for path order 0-10
30 3χcSimple molecular connectivity Chi indices for three cluster
31 4χcSimple molecular connectivity Chi indices for four cluster
32 4χpc Simple molecular connectivity Chi indices for path/cluster
33-36 3χCH
4χCH
5χCH
6χCH
Simple molecular connectivity Chi indices for cycles of 3-6
37 mChi1 mean chi1 (Randic) connectivity index
38 knotp the difference between chi3c and chi4pc
39 dchi0 the difference between chi0v and chi0
40 dchi1 the difference between chi1v and chi1
41 dchi2 the difference between chi2v and chi2
42 dchi3 the difference between chi3v and chi3
43 dchi4 the difference between chi4v and chi4
44 knotpv the difference between chiv3c and chiv4pc
Kappa descriptors (7)
11καKappa alpha index for 1 bonded fragment
22καKappa alpha index for 2 bonded fragment
33καKappa alpha index for 3 bonded fragment
4 phi Kier molecular flexibility index
51κ Molecular shape Kappa index for 1 bonded fragment
62κ Molecular shape Kappa index for 2 bonded fragment
73κ Molecular shape Kappa index for 3 bonded fragment
Basak descriptors (21)
1 IC0 Information content with order 0 proposed by Basak
2 IC1 Information content with order 1 proposed by Basak
3 IC2 Information content with order 2 proposed by Basak
4 IC3 Information content with order 3 proposed by Basak
5 IC4 Information content with order 4 proposed by Basak
6 IC5 Information content with order 5 proposed by Basak
7 IC6 Information content with order 6 proposed by Basak
8 SIC0 Complementary information content with order 0
proposed by Basak
9 SIC1 Structural information content with order 1 proposed by Basak
10 SIC2 Structural information content with order 2 proposed by Basak
11 SIC3 Structural information content with order 3 proposed by Basak
12 SIC4 Structural information content with order 4 proposed by Basak
13 SIC5 Structural information content with order 5 proposed by Basak
14 SIC6 Structural information content with order 6 proposed by Basak
15 CIC0 Complementary information content with order 0
proposed by Basak
16 CIC1 Complementary information content with order 1 proposed by
Basak
17 CIC2 Complementary information content with order 2 proposed by
Basak
18 CIC3 Complementary information content with order 3 proposed by
Basak
19 CIC4 Complementary information content with order 4 proposed by
Basak
20 CIC5 Complementary information content with order 5 proposed by
Basak
21 CIC6 Complementary information content with order 6 proposed by
Basak
E-state descriptors (245)
1 S1 Sum of E-State of atom type: sLi
2 S2 Sum of E-State of atom type: ssBe
3 S3 Sum of E-State of atom type: ssssBe
4 S4 Sum of E-State of atom type: ssBH
5 S5 Sum of E-State of atom type: sssB
6 S6 Sum of E-State of atom type: ssssB
7 S7 Sum of E-State of atom type: sCH3
8 S8 Sum of E-State of atom type: dCH2
9 S9 Sum of E-State of atom type: ssCH2
10 S10 Sum of E-State of atom type: tCH
11 S11 Sum of E-State of atom type: dsCH
12 S12 Sum of E-State of atom type: aaCH
13 S13 Sum of E-State of atom type: sssCH
14 S14 Sum of E-State of atom type: ddC
15 S15 Sum of E-State of atom type: tsC
16 S16 Sum of E-State of atom type: dssC
17 S17 Sum of E-State of atom type: aasC
18 S18 Sum of E-State of atom type: aaaC
19 S19 Sum of E-State of atom type: ssssC
20 S20 Sum of E-State of atom type: sNH3
21 S(21) Sum of E-State of atom type: sNH2
22 S22 Sum of E-State of atom type: ssNH2
23 S23 Sum of E-State of atom type: dNH
24 S24 Sum of E-State of atom type: ssNH
25 S25 Sum of E-State of atom type: aaNH
26 S26 Sum of E-State of atom type: tN
27 S27 Sum of E-State of atom type: sssNH
28 S28 Sum of E-State of atom type: dsN
29 S29 Sum of E-State of atom type: aaN
30 S30 Sum of E-State of atom type: sssN
31 S31 Sum of E-State of atom type: ddsN
32 S32 Sum of E-State of atom type: aasN
33 S33 Sum of E-State of atom type: ssssN
34 S34 Sum of E-State of atom type: sOH
35 S35 Sum of E-State of atom type: dO
36 S36 Sum of E-State of atom type: ssO
37 S37 Sum of E-State of atom type: aaO
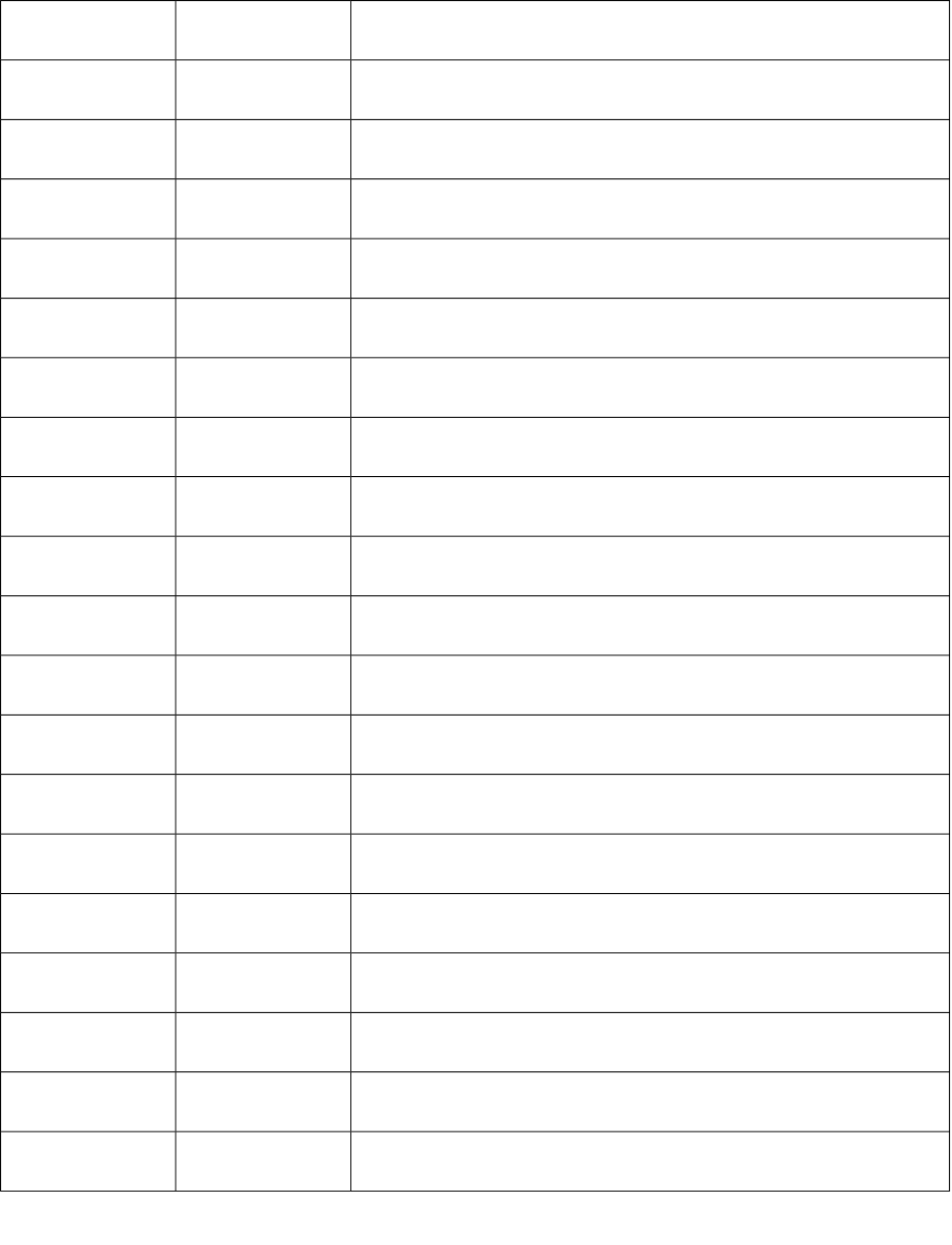
38 S38 Sum of E-State of atom type: sF
39 S39 Sum of E-State of atom type: sSiH3
40 S40 Sum of E-State of atom type: ssSiH2
41 S41 Sum of E-State of atom type: sssSiH
42 S42 Sum of E-State of atom type: ssssSi
43 S43 Sum of E-State of atom type: sPH2
44 S44 Sum of E-State of atom type: ssPH
45 S45 Sum of E-State of atom type: sssP
46 S46 Sum of E-State of atom type: dsssP
47 S47 Sum of E-State of atom type: sssssP
48 S48 Sum of E-State of atom type: sSH
49 S49 Sum of E-State of atom type: dS
50 S50 Sum of E-State of atom type: ssS
51 S51 Sum of E-State of atom type: aaS
52 S52 Sum of E-State of atom type: dssS
53 S53 Sum of E-State of atom type: ddssS
54 S54 Sum of E-State of atom type: sCl
55 S55 Sum of E-State of atom type: sGeH3
56 S56 Sum of E-State of atom type: ssGeH2
57 S57 Sum of E-State of atom type: sssGeH
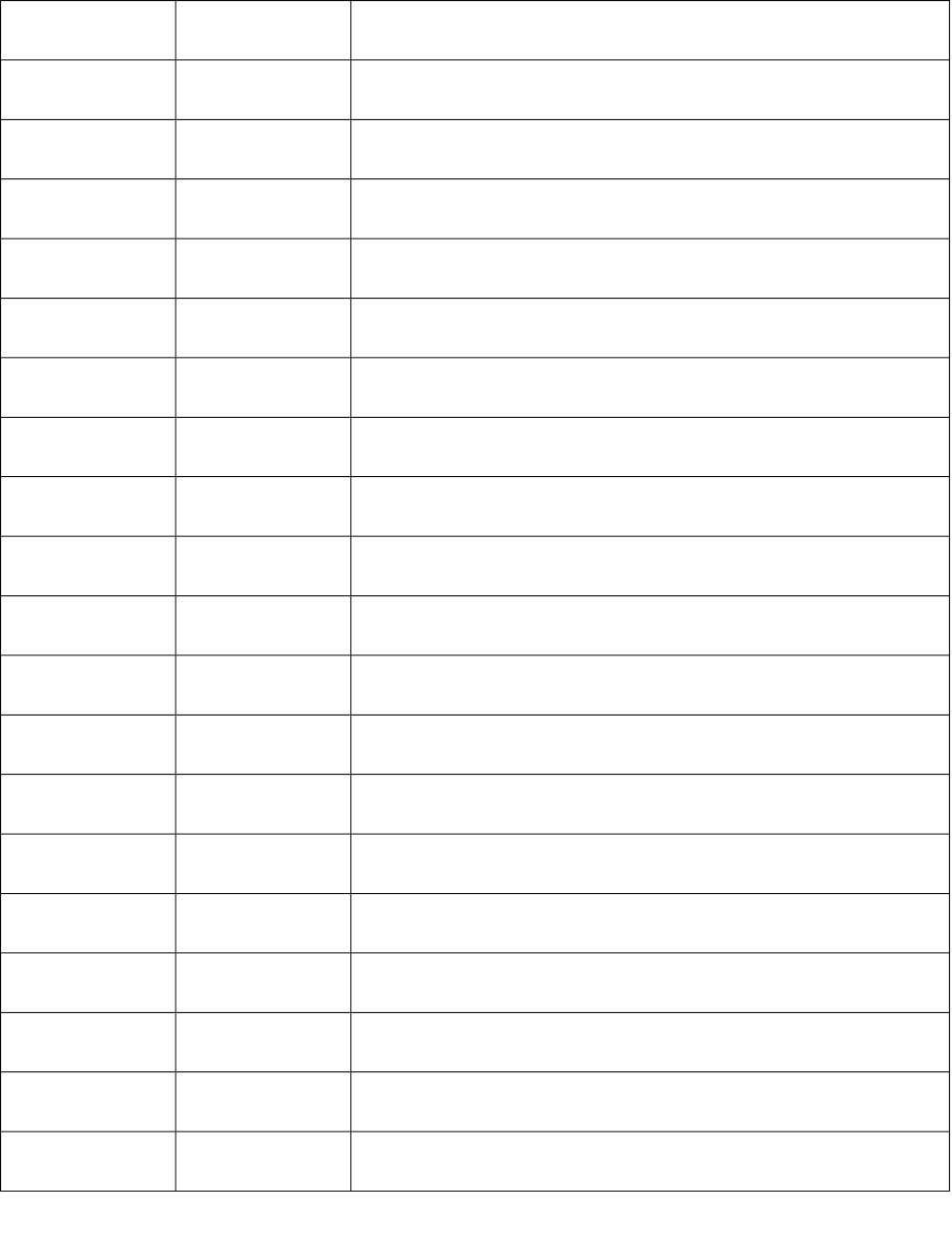
58 S58 Sum of E-State of atom type: ssssGe
59 S59 Sum of E-State of atom type: sAsH2
60 S60 Sum of E-State of atom type: ssAsH
61 S61 Sum of E-State of atom type: sssAs
62 S62 Sum of E-State of atom type: sssdAs
63 S63 Sum of E-State of atom type: sssssAs
64 S64 Sum of E-State of atom type: sSeH
65 S65 Sum of E-State of atom type: dSe
66 S66 Sum of E-State of atom type: ssSe
67 S67 Sum of E-State of atom type: aaSe
68 S68 Sum of E-State of atom type: dssSe
69 S69 Sum of E-State of atom type: ddssSe
70 S70 Sum of E-State of atom type: sBr
71 S71 Sum of E-State of atom type: sSnH3
72 S72 Sum of E-State of atom type: ssSnH2
73 S73 Sum of E-State of atom type: sssSnH
74 S74 Sum of E-State of atom type: ssssSn
75 S75 Sum of E-State of atom type: sI
76 S76 Sum of E-State of atom type: sPbH3
77 S77 Sum of E-State of atom type: ssPbH2
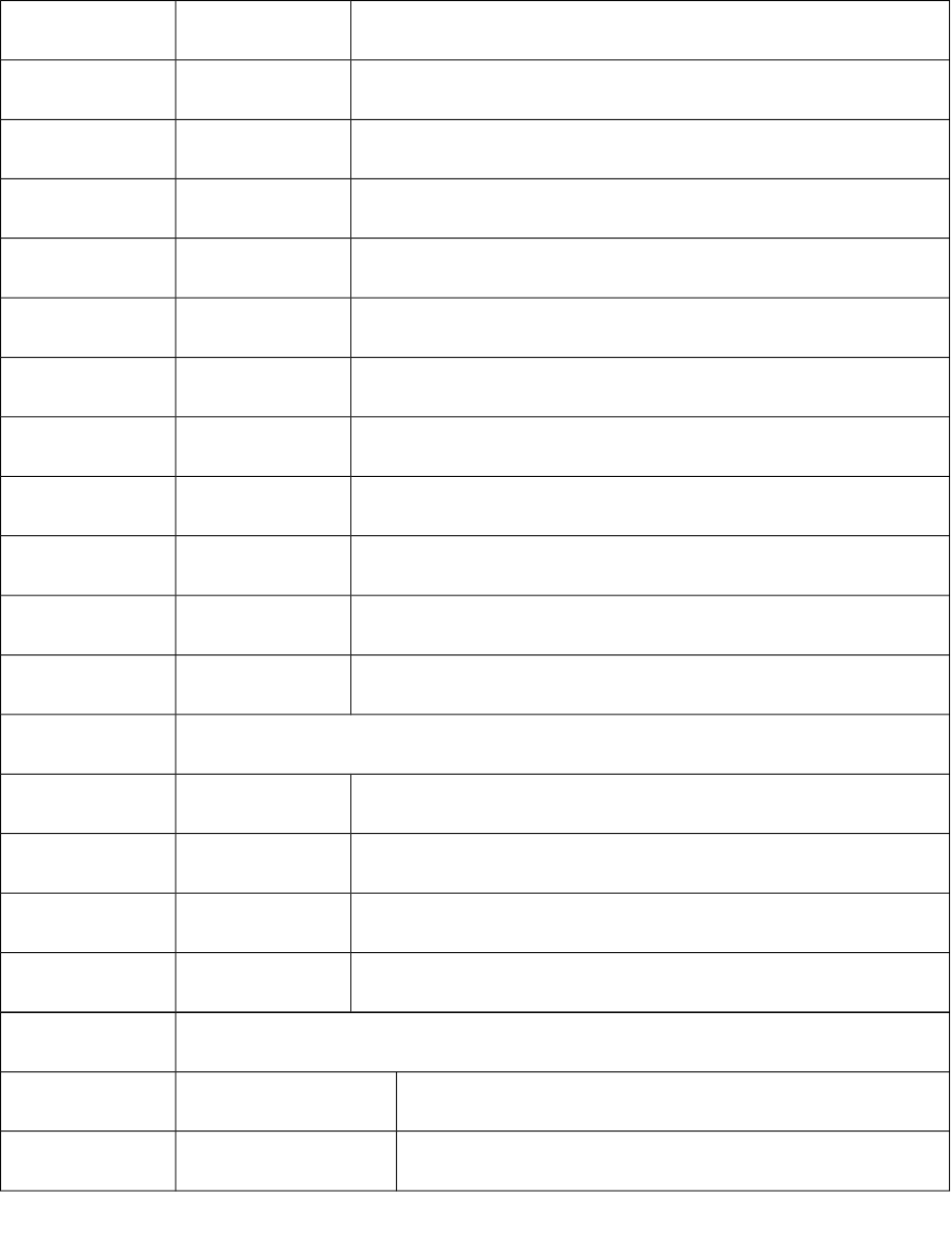
78 S78 Sum of E-State of atom type: sssPbH
79 S79 Sum of E-State of atom type: ssssPb
80-158 Smax1-Smax79 Maxmum of E-State value of specified atom type
159-237 Smin1-Smin79 Minimum of E-State value of specified atom type
238 Shev The sum of the EState indices over all non-hydrogen atoms
239 Scar The sum of the EState indices over all C atoms
240 Shal The sum of the EState indices over all Halogen atoms
241 Shet The sum of the EState indices over all hetero atoms
242 Save The sum of the EState indices over all non-hydrogen atoms
divided by the number of non-hydrogen atoms
243 Smax The maximal Estate value in all atoms
244 Smin The minimal Estate value in all atoms
245 DS The difference between Smax and Smin
Burden descriptors (64)
1-16 bcutm1-16 Burden descriptors based on atomic mass
17-32 bcutv1-16 Burden descriptors based on atomic vloumes
33-48 bcute1-16 Burden descriptors based on atomic electronegativity
49-64 bcutp1-16 Burden descriptors based on polarizability
Autocorrelation descriptors (96)
1-8 ATSm1-ATSm8 Moreau-Broto autocorrelation descriptors based on atom
mass
9-16 ATSv1-ATSv8 Moreau-Broto autocorrelation descriptors based on
atomic van der Waals volume
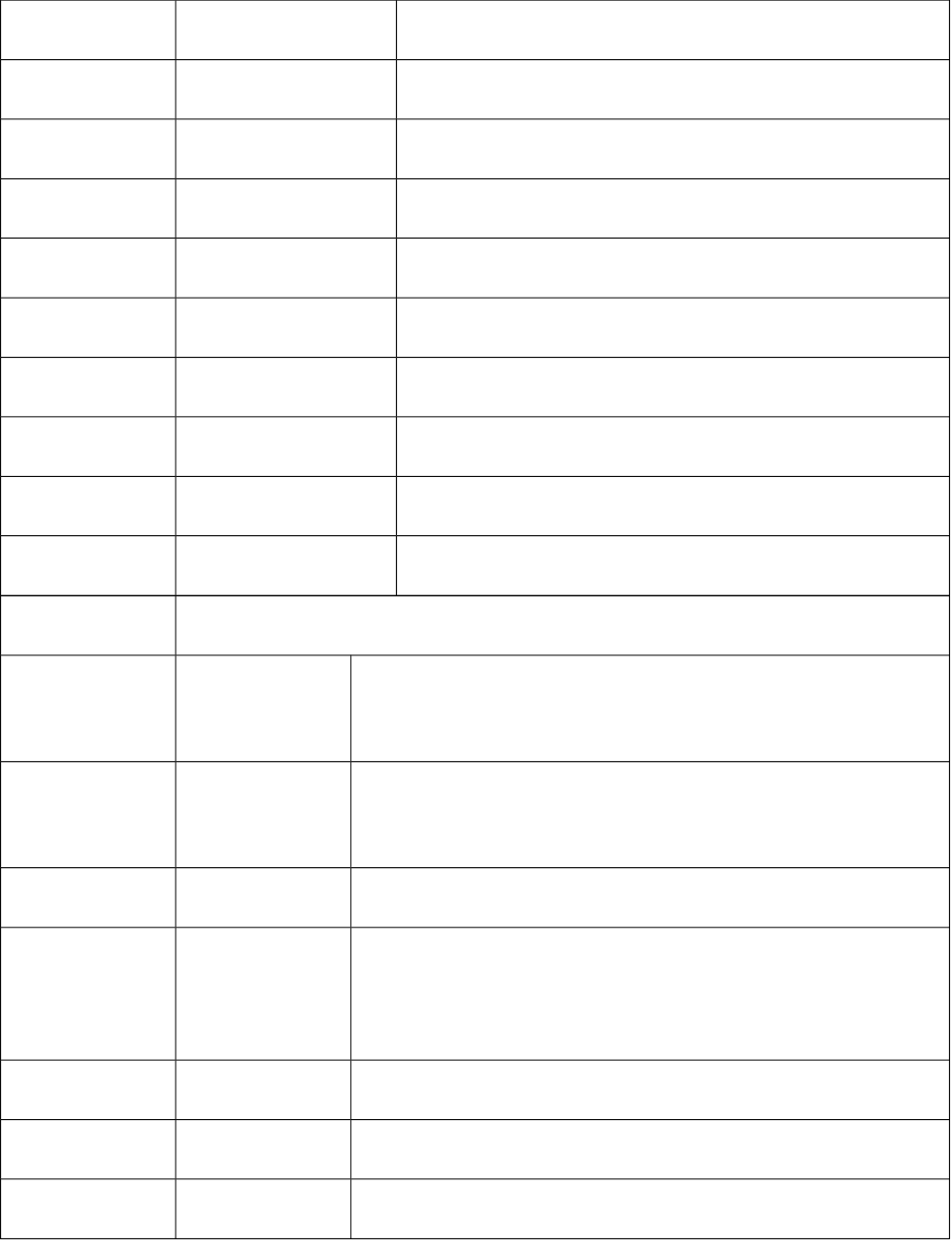
17-24 ATSe1-ATSe8 Moreau-Broto autocorrelation descriptors based on
atomic Sanderson electronegativity
25-32 ATSp1-ATSp8 Moreau-Broto autocorrelation descriptors based on
atomic polarizability
33-40 MATSm1-MATSm8 Moran autocorrelation descriptors based on atom mass
41-48 MATSv1-MATSv8 Moran autocorrelation descriptors based on atomic van
der Waals volume
49-56 MATSe1-MATSe8 Moran autocorrelation descriptors based on atomic
Sanderson electronegativity
57-64 MATSp1-MATSp8 Moran autocorrelation descriptors based on atomic
polarizability
65-72 GATSm1-GATSm8 Geary autocorrelation descriptors based on atom mass
73-80 GATSv1-GATSv8 Geary autocorrelation descriptors based on atomic van
der Waals volume
81-88 GATSe1-GATSe8 Geary autocorrelation descriptors based on atomic
Sanderson electronegativity
89-96 GATSp1-GATSp8 Geary autocorrelation descriptors based on atomic
polarizability
Charge descriptors (25)
1-4 QHmax
QCmax
QNmax
QOmax
Most positive charge on H,C,N,O atoms
5-8 QHmin
QCmin
QNmin
QOmin
Most negative charge on H,C,N,O atoms
9-10 Qmax
Qmin
Most positive and negative charge in a molecule
11-15 QHSS
QCSS
QNSS
QOSS
Qass
Sum of squares of charges on H,C,N,O and all toms
16-17 Mpc
Tpc
Mean and total of positive charges
18-19 Mnc
Tnc
Mean and total of negative charges
20-21 Mac
Tac
Mean and total of absolute charges
22 Rpc Relative positive charge
23 Rnc Relative negative charge
24 SPP Submolecular polarity parameter
25 LDI Local dipole index
Molecular property descriptors (6)
1 MREF Molar refractivity
2 logP LogP value based on the Crippen method
3 logP2 Square of LogP value based on the Crippen method
4 TPSA Topological polarity surface area
5 UI Unsaturation index
6 Hy Hydrophilic index
MOE-type descriptors (60)
1 TPSA Topological polar surface area based on fragments
2 LabuteASA Labute's Approximate Surface Area
3-14 SLOGPVSA MOE-type descriptors using SLogP contributions and surface
area contributions
15-24 SMRVSA MOE-type descriptors using MR contributions and surface
area contributions
25-38 PEOEVSA MOE-type descriptors using partial charges and surface area
contributions
39-49 EstateVSA MOE-type descriptors using Estate indices and surface area
contributions
50-60 VSAEstate MOE-type descriptors using surface area contributions and
Estate indices
Geometric descriptors (12)
1 W3DH 3-D Wiener index based geometrical distance matrix
(including Hs)
2 W3D 3-D Wiener index based geometrical distance matrix (Not
including Hs)
3 Petitj3D Petitjean Index based on molecular geometrical distance
matrix
4 GeDi The longest distance between two atoms (geometrical
diameter)
5 grav1 Gravitational 3D index
6 rygr Radius of gyration
7 Harary3D The 3D-Harary index
8 AGDD The average geometric distance degree
9 SEig The absolute eigenvalue sum on geometry matrix
10 SPAN The span R
11 ASPAN The average span R
12 MEcc The molecular eccentricity
CPSA descriptors (30)
1 ASA Solvent-accessible surface areas
2 MSA Molecular surface areas
3 PNSA1 Partial negative area
4 PPSA1 Partial negative area
5 PNSA2 Total charge weighted negative surface area
6 PPSA2 Total charge weighted negative surface area
7 PNSA3 Atom charge weighted negative surface areas
8 PPSA3 Atom charge weighted positive surface areas
9 DPSA1 Difference in charged partial surface area
10 DPSA2 Difference in total charge weighted partial
surface area
11 DPSA3 Difference in atomic charge weighted surface area
12 FNSA1 Fractional charged partial negative surface areas
13 FNSA2 Fractional charged partial negative surface areas
14 FNSA3 Fractional charged partial negative surface areas
15 FPSA1 Fractional charged partial negative surface areas
16 FPSA2 Fractional charged partial negative surface areas
17 FPSA3 Fractional charged partial negative surface areas
18 WNSA1 Surface weighted charged partial negative surface areas
19 WNSA2 Surface weighted charged partial negative surface areas
20 WNSA3 Surface weighted charged partial negative surface areas
21 WPSA1 Surface weighted charged partial negative surface areas
22 WPSA2 Surface weighted charged partial negative surface areas
23 WPSA3 Surface weighted charged partial negative surface areas
24 TASA Total hydrophobic surface area
25 PSA Total polar surface area
26 FrTATP The fraction between TASA and TPSA
27 RASA Relative hydrophobic surface area
28 RPSA Relative polar surface area
29 RNCS Relative negative charge surface area
30 RPCS Relative positive charge surface area
WHIM descriptors (70)
1-14 --- Unweighted WHIM descriptors
15-28 --- WHIM descriptors based on atomic mass
29-42 --- WHIM descriptors based on Sanderson Electronegativity
43-56 --- WHIM descriptors based on VDW Volume
57-70 --- WHIM descriptors based on Polarizability
MoRSE descriptors (210)
1-30 MoRSEU1-30 Unweighted 3-D MoRse descriptors
31-60 MoRSEC1-30 3-D MoRse descriptors
based on atomic charge
61-90 MoRSEM1-30 3-D MoRse descriptors
based on atomic mass
91-120 MoRSEN1-30 3-D MoRse descriptors
based on atomic number
121-150 MoRSEP1-30 3-D MoRse descriptors
based on atomic polarizablity
151-180 MoRSEE1-30 3-D MoRse descriptors
based on atomic Sanderson electronegativity
181-210 MoRSEV1-30 3-D MoRse descriptors
based on atomic van der Waals volume
RDF descriptors (180)
1-30 RDFU1-30 Unweighted radial distribution function (RDF) descriptors
31-60 RDFC1-30 Radial distribution function (RDF) descriptors based on
atomic charge.
61-90 RDFM1-30 Radial distribution function (RDF) descriptors based on
atomic mass
91-120 RDFP1-30 Radial distribution function (RDF) descriptors based on
atomic polarizability
121-150 RDFE1-30 Radial distribution function (RDF) descriptors based on
atomic electronegativity
151-180 RDFV1-30 Radial distribution function (RDF) descriptors based on
atomic van der Waals volume
Fragment/Fingerprint-based descriptors
1 FP2 (Topological fingerprint) A Daylight-like fingerprint based on
hashing molecular subgraphs
2 MACCS (MACCS keys)Using the 166 public keys implemented as
SMARTS
3 E-state 79 E-state fingerprints or fragments
4 FP4 307 FP4 fingerprints
5 Atom Paris Atom Paris fingerprints
6 Torsions Topological torsion fingerprints
7 Morgan/Circular Fingerprints based on the Morgan algorithm