Phyml Manual
User Manual: Pdf
Open the PDF directly: View PDF ![]() .
.
Page Count: 78
- Availability
- Authors
- Overview
- Bug report
- Installing PhyML
- Program usage.
- Inputs & outputs for command-line and PHYLIP interface
- Inputs & outputs for the XML interface
- Mixture models in PhyML
- Partitions
- Combining mixture and partitions in PhyML: the theory
- The XML format and its use in PhyML
- Setting up mixture and partition models in PhyML: the basics
- XML options
- A simple example: GTR + 4 + I
- A second example: LG4X
- An example with multiple partition elements
- Branch lengths with invariants and partionned data
- Citing PhyML
- Other programs
- Recommendations on program usage
- Frequently asked questions
- Acknowledgements
Contents
1 Availability 5
2 Authors 5
3 Overview 6
4 Bug report 6
5 Installing PhyML 6
5.1 Sources and compilation ..................... 6
5.2 Installing PhyML on UNIX-like systems (including Mac OS) . 7
5.3 Installing PhyML on Microsoft Windows ............ 7
5.4 Installing the parallel version of PhyML ............ 7
5.5 Installing PhyML-BEAGLE ................... 8
6 Program usage. 9
6.1 PHYLIP-like interface ...................... 9
6.1.1 Input Data sub-menu ................... 9
6.1.2 Substitution model sub-menu .............. 10
6.1.3 Tree searching sub-menu ................. 13
6.1.4 Branch support sub-menu ................ 14
6.2 Command-line interface ..................... 15
6.3 XML interface ........................... 21
6.4 Parallel bootstrap ......................... 21
7 Inputs & outputs for command-line and PHYLIP interface 22
7.1 Sequence formats ......................... 22
7.1.1 Gaps and ambiguous characters ............. 24
7.1.2 Specifying outgroup sequences .............. 25
7.2 Tree format ............................ 25
7.3 Multiple alignments and trees .................. 26
7.4 Custom amino-acid rate model .................. 27
7.5 Topological constraint file .................... 27
7.6 Output files ............................ 28
7.7 Treatment of invariable sites with fixed branch lengths . . . . 29
8 Inputs & outputs for the XML interface 30
8.1 Mixture models in PhyML .................... 30
8.2 Partitions ............................. 31
8.3 Combining mixture and partitions in PhyML: the theory . . . 33
2
8.4 The XML format and its use in PhyML ............. 35
8.5 Setting up mixture and partition models in PhyML: the basics 37
8.6 XML options ........................... 40
8.6.1 phyml component ..................... 40
8.6.2 topology component ................... 41
8.6.3 ratematrices component ................ 41
8.6.4 equfreqs component ................... 42
8.6.5 branchlengths component ............... 43
8.6.6 siterates component .................. 43
8.6.7 partitionelem and mixtureelem components ..... 45
8.7 A simple example: GTR + Γ4 + I ............... 46
8.8 A second example: LG4X .................... 48
8.9 An example with multiple partition elements .......... 50
8.10 Branch lengths with invariants and partionned data . . . . . . 52
9 Citing PhyML 53
10 Other programs 53
10.1 PhyTime .............................. 53
10.1.1 Installing PhyTime .................... 54
10.1.2 Running PhyTime .................... 54
10.1.3 PhyTime input ...................... 54
10.1.4 Accounting for calibration uncertainty ......... 57
10.1.5 MCMC settings ...................... 58
10.1.6 PhyTime output ..................... 59
10.1.7 An example of PhyTime input and output files . . . . 60
10.1.8 Citing PhyTime ...................... 62
10.2 PhyloGeo ............................. 64
10.2.1 Installing PhyloGeo .................... 64
10.2.2 Running PhyloGeo .................... 64
10.2.3 Citing PhyloGeo ..................... 65
10.3 PhyREX .............................. 65
10.3.1 Installing PhyREX .................... 66
10.3.2 Running PhyREX ..................... 66
11 Recommendations on program usage 68
11.1 PhyML ............................... 68
11.2 PhyTime .............................. 69
12 Frequently asked questions 70
3
c
Copyright 1999 - 2008 by PhyML Development Team.
The software PhyML is provided “as is” without warranty of any kind. In no
event shall the authors or his employer be held responsible for any damage result-
ing from the use of this software, including but not limited to the frustration that
you may experience in using the package. All parts of the source and documen-
tation except where indicated are distributed under the GNU public licence. See
http://www.opensource.org for details.
1 Availability
•Binaries: http://www.atgc-montpellier.fr/phyml
•Sources: http://stephaneguindon.github.io/phyml-downloads/
•Discussion forum: http://groups.google.com/group/phyml-forum
2 Authors
•St´ephane Guindon and Olivier Gascuel conceived the original PhyML
algorithm.
•St´ephane Guindon conceived the PhyTime method.
•St´ephane Guindon, David Welch and Louis Ranjard conceived the Phy-
loGeo method.
•St´ephane Guindon, Wim Hordjik and Olivier Gascuel conceived the
SPR-based tree search algorithm.
•Maria Anisimova and Olivier Gascuel conceived the aLRT method for
branch support.
•St´ephane Guindon, Franck Lethiec, Jean-Francois Dufayard and Vin-
cent Lefort implemented PhyML.
•Jean-Francois Dufayard created the benchmark and implemented the
tools that are used to check PhyML accuracy and performances.
•Vincent Lefort, St´ephane Guindon, Patrice Duroux and Olivier Gas-
cuel conceived and implemented PhyML web server.
•Imran Fanaswala interfaced PhyML with BEAGLE.
•St´ephane Guindon wrote this document.
5
3 Overview
PhyML [1] is a software package which primary task that is to estimate
maximum likelihood phylogenies from alignments of nucleotide or amino-
acid sequences. It provides a wide range of options that were designed to
facilitate standard phylogenetic analyses. The main strength of PhyML lies
in the large number of substitution models coupled to various options to
search the space of phylogenetic tree topologies, going from very fast and
efficient methods to slower but generally more accurate approaches. It also
implements two methods to evaluate branch supports in a sound statistical
framework (the non-parametric bootstrap and the approximate likelihood
ratio test).
PhyML was designed to process moderate to large data sets. In theory,
alignments with up to 4,000 sequences 2,000,000 character-long can analyzed.
In practice however, the amount of memory required to process a data set
is proportional of the product of the number of sequences by their length.
Hence, a large number of sequences can only be processed provided that
they are short. Also, PhyML can handle long sequences provided that they
are not numerous. With most standard personal computers, the “comfort
zone” for PhyML generally lies around 100-500 sequences less than 10,000
character long. For larger data sets, we recommend using other softwares
such as RAxML [2] or GARLI [3] or Treefinder (http://www.treefinder.de).
4 Bug report
While PhyML is, of course, bug-free (!) (please read the disclaimer care-
fuly...), if you ever come across an issue, please feel free to report it us-
ing the discuss group web site at the following address: https://groups.
google.com/forum/?fromgroups#!forum/phyml-forum. Alternatively, you
can send an email to s.guindon@auckland.ac.nz. Do not forget to mention
the version of PhyML and program options you are using.
5 Installing PhyML
5.1 Sources and compilation
The sources of the program are available free of charge from http:
//stephaneguindon.github.io/phyml-downloads/. The compilation on
UNIX-like systems is fairly standard. It is described in the ‘INSTALL’ file
6

that comes with the sources. In a command-line window, go to the directory
that contains the sources and type:
./configure;
make clean;
make V=0;
By default, PhyML will be compiled with optimization flags turned on. It
is possible to generate a version of PhyML that can run through a debugging
tool (such as ddd) or a profiling tool (such as gprof) using the following
instructions:
./configure --enable-debug;
make clean;
make V=0;
5.2 Installing PhyML on UNIX-like systems (includ-
ing Mac OS)
Copy PhyML binary file in the directory you like. For the operating system
to be able to locate the program, this directory must be specified in the
global variable PATH. In order to achieve this, you will have to add export
PATH="/your path/:$PATH" to the .bashrc or the .bash profile located
in your home directory (your path is the path to the directory that contains
PhyML binary).
5.3 Installing PhyML on Microsoft Windows
Copy the files phyml.exe and phyml.bat in the same directory. To launch
PhyML, click on the icon corresponding to phyml.bat. Clicking on the icon
for phyml.exe works too but the dimensions of the window will not fit PhyML
PHYLIP-like interface.
5.4 Installing the parallel version of PhyML
Bootstrap analysis can run on multiple processors. Each processor analyses
one bootstraped dataset. Therefore, the computing time needed to perform
Rbootstrap replicates is divided by the number of processors available.
This feature of PhyML relies on the MPI (Message Passing In-
terface) library. To use it, your computer must have MPI installed
on it. In case MPI is not installed, you can dowload it from
http://www.mcs.anl.gov/research/projects/mpich2/. Once MPI is installed,
it is necessary to launch the MPI daemon. This can be done by entering
7
the following instruction: mpd &. Note however that in most cases, the MPI
daemon will already be running on your server so that you most likely do not
need to worry about this. You can then just go in the phyml/ directory (the
directory that contains the src/,examples/ and doc/ folders) and enter the
commands below:
./configure --enable-mpi;
make clean;
make;
A binary file named phyml-mpi has now been created in the src/ direc-
tory and is ready to use with MPI. A typical MPI command-line which uses
4 CPUs is given below:
mpirun -n 4 ./phyml-mpi -i myseq -b 100
Please read section 6.4 of this document for more information.
5.5 Installing PhyML-BEAGLE
PhyML can use the BEAGLE [4] library for the likelihood computation.
BEAGLE provides provides significant speed-up: the single core version of
PhyML-BEAGLE can be up to 10 times faster than PhyML on a single core
and up to 150 times on Graphical Processing Units. PhyML-BEAGLE will
eventually have of the features of PhyML, even though at the moment the
boostrap and the invariant site options are not available. Also, please note
that in some cases, the final log-likelihood reported by PhyML and PhyML-
BEALGE may not exactly match, though the differences observed are very
minor (in the 10−4to 10−4range).
In order to install PhyML-BEAGLE, you first need to download and
install the BEAGLE library available from https://code.google.com/p/
beagle-lib/. Then run the following commands:
./configure --enable-beagle;
make clean;
make;
A binary file named phyml-beagle will be created in the src/ directory.
The interface to phyml-beagle (i.e., commandline option of PHYLIP-like
interface) is exactly identical to that of PhyML.
8

6 Program usage.
PhyML has three distinct user-interfaces. The first corresponds to a
PHYLIP-like text interface that makes the choice of the options self-
explanatory. The command-line interface is well-suited for people that are
familiar with PhyML options or for running PhyML in batch mode. The
XML interface is more sophisticated. It allows the user to analyse parti-
tionned data using flexible mixture models of evolution.
6.1 PHYLIP-like interface
The default is to use the PHYLIP-like text interface by simply typing ‘phyml’
in a command-line window or by clicking on the PhyML icon (see Section
5.3). After entering the name of the input sequence file, a list of sub-menus
helps the users set up the analysis. There are currently four distinct sub-
menus:
1. Input Data: specify whether the input file contains amino-acid or nu-
cleotide sequences. What the sequence format is (see Section 7) and
how many data sets should be analysed.
2. Substitution Model: selection of the Markov model of substitution.
3. Tree Searching: selection of the tree topology searching algorithm.
4. Branch Support: selection of the method that is used to measure branch
support.
‘+’ and ‘-’ keys are used to move forward and backward in the sub-menu list.
Once the model parameters have been defined, typing ‘Y’ (or ‘y’) launches the
calculations. The meaning of some options may not be obvious to users that
are not familiar with phylogenetics. In such situation, we strongly recom-
mend to use the default options. As long as the format of the input sequence
file is correctly specified (sub-menu Input data), the safest option for non-
expert users is to use the default settings. The different options provided
within each sub-menu are described in what follows.
6.1.1 Input Data sub-menu
[D] ............................... Data type (DNA/AA)
Type of data in the input file. It can be either DNA or amino-acid sequences
in PHYLIP format (see Section 7). Type Dto change settings.
9

[I] ...... Input sequences interleaved (or sequential)
PHYLIP format comes in two flavours: interleaved or sequential (see Section
7). Type Ito selected among the two formats.
[M] ....................... Analyze multiple data sets
If the input sequence file contains more than one data sets, PhyML can
analyse each of them in a single run of the program. Type Mto change
settings.
[R] ............................................ Run ID
This option allows you to append a string that identifies the current PhyML
run. Say for instance that you want to analyse the same data set with two
models. You can then ‘tag’ the first PhyML run with the name of the first
model while the second run is tagged with the name of the second model.
6.1.2 Substitution model sub-menu
[M] ................. Model of nucleotide substitution
[M] ................ Model of amino-acids substitution
PhyML implements a wide range of substitution models: JC69 [5], K80 [6],
F81 [7], F84 [8], HKY85 [9], TN93 [10] GTR [11,12] and custom for nu-
cleotides; LG [13], WAG [14], Dayhoff [15], JTT [16], Blosum62 [17], mtREV
[18], rtREV [19], cpREV [20], DCMut [21], VT [22] and mtMAM [23] and
custom for amino acids. Cycle through the list of nucleotide or amino-acids
substitution models by typing M. Both nucleotide and amino-acid lists include
a ‘custom’ model. The custom option provides the most flexible way to spec-
ify the nucleotide substitution model. The model is defined by a string made
of six digits. The default string is ‘000000’, which means that the six relative
rates of nucleotide changes: A↔C,A↔G,A↔T,C↔G,C↔Tand
G↔T, are equal. The string ‘010010’ indicates that the rates A↔Gand
C↔Tare equal and distinct from A↔C=A↔T=C↔G=G↔T.
This model corresponds to HKY85 (default) or K80 if the nucleotide fre-
quencies are all set to 0.25. ‘010020’ and ‘012345’ correspond to TN93 and
10

GTR models respectively. The digit string therefore defines groups of rela-
tive substitution rates. The initial rate within each group is set to 1.0, which
corresponds to F81 (JC69 if the base frequencies are equal). Users also have
the opportunity to define their own initial rate values. These rates are then
optimised afterwards (option ‘O’) or fixed to their initial values. The custom
option can be used to implement all substitution models that are special cases
of GTR. Table 1on page 17 gives the correspondence between the ‘standard’
name of the model (see http://mbe.oxfordjournals.org/content/18/6/
897/F2.large.jpg) and the custom model code. The custom model also ex-
ists for protein sequences. It is useful when one wants to use an amino-acid
substitution model that is not hard-coded in PhyML. The symmetric part of
the rate matrix, as well as the equilibrium amino-acid frequencies, are given
in a file which name is given as input of the program. The format of this file
is described in the section 7.4.
[F] ................. Optimise equilibrium frequencies
[E] ......... Equilibrium frequencies (empirical/user)
[F] . Amino acid frequencies (empirical/model defined)
For nucleotide sequences, optimising equilibrium frequencies means that the
values of these parameters are estimated in the maximum likelihood frame-
work. When the custom model option is selected, it is also possible to give
the program a user-defined nucleotide frequency distribution at equilibrium
(option E). For protein sequences, the stationary amino-acid frequencies are
either those defined by the substitution model or those estimated by count-
ing the number of different amino-acids observed in the data. Hence, the
meaning of the Foption depends on the type of the data to be processed.
[T] .................... Ts/tv ratio (fixed/estimated)
Fix or estimate the transition/transversion ratio in the maximum likelihood
framework. This option is only available when DNA sequences are to be
analysed under K80, HKY85 or TN93 models. The definition given to this
parameter by PhyML is the same as PAML’s one. Therefore, the value
of this parameter does not correspond to the ratio between the expected
number of transitions and the expected number of transversions during a
11

unit of time. This last definition is the one used in PHYLIP. PAML’s manual
gives more detail about the distinction between the two definitions (http:
//abacus.gene.ucl.ac.uk/software/paml.html).
[V] . Proportion of invariable sites (fixed/estimated)
The proportion of invariable sites, i.e., the expected frequency of sites that
do not evolve, can be fixed or estimated. The default is to fix this proportion
to 0.0. By doing so, we consider that each site in the sequence may accumu-
late substitutions at some point during its evolution, even if no differences
across sequences are actually observed at that site. Users can also fix this
parameter to any value in the [0.0,1.0] range or estimate it from the data in
the maximum-likelihood framework.
[R] ....... One category of substitution rate (yes/no)
[C] ........... Number of substitution rate categories
[A] ... Gamma distribution parameter (fixed/estimated)
[G] .........‘Middle’ of each rate class (mean/median)
Rates of evolution often vary from site to site. This heterogeneity can be
modelled using a discrete gamma distribution. Type Rto switch this option
on or off. The different categories of this discrete distribution correspond
to different (relative) rates of evolution. The number of categories of this
distribution is set to 4 by default. It is probably not wise to go below this
number. Larger values are generally preferred. However, the computational
burden involved is proportional to the number of categories (i.e., an analysis
with 8 categories will generally take twice the time of the same analysis with
only 4 categories). Note that the likelihood will not necessarily increase as the
number of categories increases. Hence, the number of categories should be
kept below a “reasonable” number, say 20. The default number of categories
can be changed by typing C.
The middle of each discretized substitution rate class can be determined
using the mean or the median. PAML, MrBayes and RAxML use the mean.
However, the median is generally associated with greater likelihoods than the
mean. This conclusion is based on our analysis of several real-world data sets
12
extracted from TreeBase. Despite this, the default option in PhyML is to use
the mean in order to make PhyML likelihoods comparable to those of other
phylogenetic software. One must bare in mind that likelihoods calculated
with the mean approximation are not directly comparable to the likelihoods
calculated using the median approximation.
The shape of the gamma distribution determines the range of rate vari-
ation across sites. Small values, typically in the [0.1,1.0] range, correspond
to large variability. Larger values correspond to moderate to low heterogene-
ity. The gamma shape parameter can be fixed by the user or estimated via
maximum-likelihood. Type Ato select one or the other option.
6.1.3 Tree searching sub-menu
[O] ........................... Optimise tree topology
By default the tree topology is optimised in order to maximise the likelihood.
However, it is also possible to avoid any topological alteration. This option
is useful when one wants to compute the likelihood of a tree given as input
(see below). Type Oto select among these two options.
[S] .................. Tree topology search operations
PhyML proposes three different methods to estimate tree topologies. The
default approach is to use simultaneous NNI. This option corresponds to the
original PhyML algorithm [1]. The second approach relies on subtree pruning
and regrafting (SPR). It generally finds better tree topologies compared to
NNI but is also significantly slower. The third approach, termed BEST,
simply estimates the phylogeny using both methods and returns the best
solution among the two. Type Sto choose among these three choices.
[R] ......................... Use random starting tree
[N] .................. Number of random starting trees
When the SPR or the BEST options are selected, is is possible to use random
trees rather than BioNJ or a user-defined tree, as starting tree. If this option
is turned on (type Rto change), five trees, corresponding to five random
starts, will be estimated. The output tree file will contain the best tree
found among those five. The number of random starts can be modified by
13

typing N. Setting the number of random starting trees to Nmeans that
the analysis will take (slightly more than) Ntimes the time required for a
standard analysis where only one (BioNJ) starting tree is used. However,
the analysis of real data sets shows that the best trees estimated using the
random start option almost systematically have higher likelihoods than those
inferred using a single starting tree.
[U] ........ Starting tree (BioNJ/parsimony/user tree)
When the tree topology optimisation option is turned on, PhyML proceeds
by refining an input tree. By default, this input tree is estimated using
BioNJ [24]. The alternative option is to use a parsimony tree. We found
this option specially useful when analysing large data sets with NNI moves
as it generally leads to greater likelihoods than those obtained when starting
from a BioNJ trees. The user can also to input her/his own tree. This tree
should be in Newick format (see Section 7). This option is useful when one
wants to evaluate the likelihood of a given tree with a fixed topology, using
PhyML. Type Uto choose among these two options.
6.1.4 Branch support sub-menu
[B] ................ Non parametric bootstrap analysis
The support of the data for each internal branch of the phylogeny can be es-
timated using non-parametric bootstrap. By default, this option is switched
off. Typing Bswitches on the bootstrap analysis. The user is then prompted
for a number of bootstrap replicates. The largest this number the more
precise the bootstrap support estimates are. However, for each bootstrap
replicate a phylogeny is estimated. Hence, the time needed to analyse N
bootstrap replicates corresponds to N-times the time spent on the analysis
of the original data set. N= 100 is generally considered as a reasonable
number of replicates.
[A] ................ Approximate likelihood ratio test
When the bootstrap option is switched off (see above), approximate likeli-
hood branch supports are estimated. This approach is considerably faster
than the bootstrap one. However, both methods intend to estimate differ-
ent quantities and conducting a fair comparison between both criteria is not
straightforward. The estimation of approximate likelihood branch support
14
comes in multiple flavours. The default is set to aBayes, corresponding to
the approximate Bayes method described in [25]. The approximate likelihood
ratio test (aLRT) [26], ShimodairaHasegawa aLRT (SH-aLRT) statistics are
the other available options.
6.2 Command-line interface
An alternative to the PHYLIP-like interface is the command-line interface.
Users that do not need to modify the default parameters can launch the
program with the ‘phyml -i seq file name’ command. The list of all com-
mand line arguments and how to use them is given in the ‘Help’ section
which is displayed when entering the ‘phyml --help’ command. The avail-
able command-line options are described in what follows.
•-i (or --input)seq file name
seq file name is the name of the nucleotide or amino-acid sequence
file in PHYLIP format.
•-d (or --datatype)data type
data type is nt for nucleotide (default) and aa for amino-acid se-
quences.
•-q (or --sequential)
Changes interleaved format (default) to sequential format.
•-n (or --multiple)nb data sets
nb data sets is an integer giving the number of data sets to analyse.
•-p (or --pars)
Use a minimum parsimony starting tree. This option is taken into
account when the ‘-u’ option is absent and when tree topology modifi-
cations are to be done.
•-b (or --bootstrap)int
–int >0: int is the number of bootstrap replicates.
–int = 0: neither approximate likelihood ratio test nor bootstrap
values are computed.
–int = -1: approximate likelihood ratio test returning aLRT statis-
tics.
–int = -2: approximate likelihood ratio test returning Chi2-based
parametric branch supports.
15

–int = -4: SH-like branch supports alone.
–int = -5: (default) approximate Bayes branch supports.
•-m (or --model)model name
model name : substitution model name.
–Nucleotide-based models:HKY85 (default) | JC69 | K80 | F81 |
F84 | TN93 | GTR | custom
The custom option can be used to define a new substitution model.
A string of six digits identifies the model. For instance, 000000 cor-
responds to F81 (or JC69 provided the distribution of nucleotide
frequencies is uniform). 012345 corresponds to GTR. This option
can be used for encoding any model that is a nested within GTR.
See Section 6.1.2 and Table 1.NOTE: the substitution parameters
of the custom model will be optimised so as to maximise the like-
lihood. It is possible to specify and fix (i.e., avoid optimisation)
the values of the substitution rates only through the PHYLIP-like
interface.
–Amino-acid based models:LG (default) | WAG | JTT | MtREV
| Dayhoff | DCMut | RtREV | CpREV | VT | Blosum62 |
MtMam | MtArt | HIVw | HIVb | custom
The custom option is useful when one wants to use an amino-acid
substitution model that is not available by default in PhyML.
The symmetric part of the rate matrix, as well as the equilibrium
amino-acid frequencies, are given in a file which name is asked
for by the program. The format of this file is described in section
7.4.
•--aa rate file file name
This option is compulsory when analysing amino-acid sequences under
a ‘custom’ model (see above). file name should provide a rate matrix
and equilibrium amino acid in PAML format (see Section 7.4).
•-f e,m, or “fA,fC,fG,fT”
Nucleotide or amino-acid frequencies.
–e: the character frequencies are determined as follows :
∗Nucleotide sequences: (Empirical) the equilibrium base fre-
quencies are estimated by counting the occurence of the dif-
ferent bases in the alignment.
16

Name Command-line option
JC69 -m 000000 -f 0.25,0.25,0.25,0.25
F81 -m 000000
K80 -m 010010 -f 0.25,0.25,0.25,0.25
HKY85 -m 010010
TrNef -m 010020 -f 0.25,0.25,0.25,0.25
TrN -m 010020
K81 -m 123321 -f 0.25,0.25,0.25,0.25
K81uf -m 123321
TIMef -m 132241 -f 0.25,0.25,0.25,0.25
TIM -m 132241
TVMef -m 102304 -f 0.25,0.25,0.25,0.25
TVM -m 102304
SYM -m 123456 -f 0.25,0.25,0.25,0.25
GTR -m 123456
Table 1. Nucleotide substitution model names (as defined in [27]) and the
corresponding custom model code used in PhyML.
∗Amino-acid sequences: (Empirical) the equilibrium amino-
acid frequencies are estimated by counting the occurence of
the different amino-acids in the alignment.
–m: the character frequencies are determined as follows :
∗Nucleotide sequences: (ML) the equilibrium base frequencies
are estimated using maximum likelihood.
∗Amino-acid sequences: (Model) the equilibrium amino-acid
frequencies are estimated using the frequencies defined by the
substitution model.
–“fA,fC,fG,fT”: only valid for nucleotide-based models. fA,fC,
fG and fT are floating numbers that correspond to the frequencies
of A, C, G and T respectively.
•-t (or --ts/tv)ts/tv ratio
ts/tv ratio: transition/transversion ratio. DNA sequences only. Can
be a fixed positive value (e.g., 4.0) or type eto get the maximum
likelihood estimate.
•-v (or --pinv)prop invar
prop invar: proportion of invariable sites. Can be a fixed value in the
[0,1] range or type eto get the maximum likelihood estimate.
17
•-c (or --nclasses)nb subst cat
nb subst cat: number of relative substitution rate categories. Default:
nb subst cat=4. Must be a positive integer.
•-a (or --alpha)gamma
gamma: value of the gamma shape parameter. Can be a fixed posi-
tive value or e to get the maximum likelihood estimate. The value of
this parameter is estimated in the maximum likelihood framework by
default.
•--use median
The middle of each substitution rate class in the discrete gamma dis-
tribution is taken as the median. The mean is used by default.
•--free rates or --freerates
As an alternative to the discrete gamma model, it is possible to esti-
mate the (relative) rate in each class of the (mixture) model and the
corresponding frequencies directly from the data. This model, called
the FreeRate model, has more parameters than the discrete gamma one
but usually provides a significantly better fit to the data. See [28] for
more information about this model and an illustration of its use.
•--il
il stands here for integrated (branch) length. This model, described
in [29] in the context of molecular dating, provides an efficient way
to implement the covarion model . Under the integrated length (IL)
model, the length of each edge is described by a distribution of val-
ues, instead of a single value corresponding to the expected number of
substitutions per position along the sequence. Let la,s and lb,s be the
number of substitutions at site salong edges aand b, and la,t and lb,t,
the number of substitutions at site t. Standard models have la,s =la,t
and lb,s =lb,t, or la,s =αla,t and lb,s =αlb,t if rates vary across sites.
The IL model has instead la,s =αla,t and lb,s =βlb,t with α6=β, i.e.
substitution rates vary across sites and edges. The IL approach is some-
how an analytical approximation to the covarion model that, unlike the
covarion model, does not incur any computational overhead compared
to the traditional models. A notable difference with the plain vanilla
covarion model and the IL model however is that substitution rates are
not autocorrelated along the phylogeny under the IL model.
•--codpos 1,2 or 3
When analysing an alignment of coding sequences, use this option to
18
consider only the first, second or the third coding position for the esti-
mation.
•-s (or --search)move
Tree topology search operation option. Can be either NNI (default,
fast) or SPR (usually slower than NNI but more accurate) or BEST (best
of NNI and SPR search).
•-u (or --inputtree)user tree file
user tree file: starting tree filename. The tree must be in Newick
format.
•-o params
This option focuses on specific parameter optimisation.
–params=tlr: tree topology (t), branch length (l) and substitution
rate parameters (r) are optimised.
–params=tl: tree topology and branch lengths are optimised.
–params=lr: branch lengths and substitution rate parameters are
optimised.
–params=l: branch lengths are optimised.
–params=r: substitution rate parameters are optimised.
–params=n: no parameter is optimised.
•--rand start
This option sets the initial tree to random. It is only valid if SPR
searches are to be performed.
•--n rand starts num
num is the number of initial random trees to be used. It is only valid if
SPR searches are to be performed.
•--r seed num
num is the seed used to initiate the random number generator. Must
be an integer.
•--print site lnl
Print the likelihood for each site in file * phyml lk.txt. For Γ or Γ+I or
FreeRate models, this option returns the posterior probability of each
relative rate class at each site. Such information can then be used to
identify fast- and slow-evolving regions of the alignment.
19
•--print trace
Print each phylogeny explored during the tree search process in file
* phyml trace.txt. This option can be useful for monitoring the
progress of the analysis for very large data sets and have an approxi-
mate idea of what the final phylogeny will look like.
•--json trace
Print each phylogeny explored during the tree search process in file
* phyml json trace.txt in JSON format (see http://www.json.org/).
This option can be useful for monitoring the progress of the analysis
for very large data sets and have an approximate idea of what the final
phylogeny will look like.
•--run id ID string
Append the string ID string at the end of each PhyML output file. This
option may be useful when running simulations involving PhyML. It
can also be used to ‘tag’ multiple analysis of the same data set with
various program settings.
•--no memory check
By default, when processing a large data set, PhyML will pause and ask
the user to confirm that she/he wants to continue with the execution
of the analysis despite the large amount of memory required. The
--no memory check skips this question. It is especially useful when
running PhyML in batch mode.
•--no colalias
By default, PhyML preprocesses each alignment by putting together
(or aliasing) the columns that are identical. Use this option to skip
this step but be aware that the analysis might then take more time to
complete.
•--constrained lens
When an input tree with branch lengths is provided, this option will
find the branch multiplier that maximises the likelihood (i.e., the rela-
tive branch lengths remain constant)
•--constraint file file name
file name lists the topological constraints under which the tree topol-
ogy search is conducted. This option should be used in conjunction
with -u file name. See Section 7.5 for more information.
20
•--quiet
Runs PhyML in quiet mode. The program will not pause if the memory
required to run the analysis exceeds 256MB and will not output the
progression of the log-likelihood scores on the standard output.
•--ancestral
PhyML calculates the marginal probabilities of each character state at
each internal node and each site of the sequence alignment.
6.3 XML interface
•--xml=xml file name
xml file name is the name of the XML file containing the information
required to run the analysis. More details about this type of file is
given in the section 8.
6.4 Parallel bootstrap
Bootstrapping is a highly parallelizable task. Indeed, bootstrap replicates
are independent from one another. Each bootstrap replicate can then be
analysed separately. Modern computers often have more than one CPU. Each
CPU can therefore be used to process a bootstrap sample. Using this parallel
strategy, performing Rbootstrap replicates on CCPUs ‘costs’ the same
amount of computation time as processing R×Cbootstrap replicates on a
single CPU. In other words, for a given number of replicates, the computation
time is divided by Rcompared to the non-parallel approach.
PhyML sources must be compiled with specific options to turn on the
parallel option (see Section 5.4). Once the binary file (phyml) has been
generated, running a bootstrap analysis with, say 100 replicates on 2 CPUs,
can be done by typing the following command-line:
mpd &;
mpirun -np 2 ./phyml -i seqfile -b 100;
The first command launches the mpi daemon while the second launches the
analysis. Note that launching the daemon needs to be done only once. The
output files are similar to the ones generated using the standard, non-parallel,
analysis (see Section 7). Note that running the program in batch mode, i.e.:
mpirun -np 2 ./phyml -i seqfile -b 100 &
will probably NOT work. I do not know how to run a mpi process in batch
mode yet. Suggestions welcome... Also, at the moment, the number of
21
PHYLIP interleaved
5 80
seq1 CCATCTCACGGTCGGTACGATACACCKGCTTTTGGCAGGAAATGGTCAATATTACAAGGT
seq2 CCATCTCACGGTCAG---GATACACCKGCTTTTGGCGGGAAATGGTCAACATTAAAAGAT
seq3 RCATCTCCCGCTCAG---GATACCCCKGCTGTTG????????????????ATTAAAAGGT
seq4 RCATCTCATGGTCAA---GATACTCCTGCTTTTGGCGGGAAATGGTCAATCTTAAAAGGT
seq5 RCATCTCACGGTCGGTAAGATACACCTGCTTTTGGCGGGAAATGGTCAAT????????GT
ATCKGCTTTTGGCAGGAAAT
ATCKGCTTTTGGCGGGAAAT
AGCKGCTGTTG?????????
ATCTGCTTTTGGCGGGAAAT
ATCTGCTTTTGGCGGGAAAT
PHYLIP sequential
5 40
seq1 CCATCTCANNNNNNNNACGATACACCKGCTTTTGGCAGG
seq2 CCATCTCANNNNNNNNGGGATACACCKGCTTTTGGCGGG
seq3 RCATCTCCCGCTCAGTGAGATACCCCKGCTGTTGXXXXX
seq4 RCATCTCATGGTCAATG-AATACTCCTGCTTTTGXXXXX
seq5 RCATCTCACGGTCGGTAAGATACACCTGCTTTTGxxxxx
Figure 1. PHYLIP interleaved and sequential formats.
bootstrap replicates must be a multiple of the number of CPUs required in
the mpirun command.
7 Inputs & outputs for command-line and
PHYLIP interface
PhyML reads data from standard text files, without the need for any partic-
ular file name extension.
7.1 Sequence formats
Alignments of DNA or protein sequences must be in PHYLIP or NEXUS
[30] sequential or interleaved format (Figures 7.1 and 2). For PHYLIP for-
mated sequence alignments, the first line of the input file contains the number
of species and the number of characters, in free format, separated by blank
characters. One slight difference with PHYLIP format deals with sequence
name lengths. While PHYLIP format limits this length to ten characters,
PhyML can read up to hundred character long sequence names. Blanks
and the symbols “(),:” are not allowed within sequence names because the
Newick tree format makes special use of these symbols. Another slight differ-
ence with PHYLIP format is that actual sequences must be separated from
22

Nexus nucleotides
[ This is a comment ]
#NEXUS
BEGIN DATA;
DIMENSIONS NTAX=10 NCHAR=20;
FORMAT DATATYPE=DNA;
MATRIX
tax1 ?ATGATTTCCTTAGTAGCGG
tax2 CAGGATTTCCTTAGTAGCGG
tax3 ?AGGATTTCCTTAGTAGCGG
tax4 ?????????????GTAGCGG
tax5 CAGGATTTCCTTAGTAGCGG
tax6 CAGGATTTCCTTAGTAGCGG
tax7 ???GATTTCCTTAGTAGCGG
tax8 ????????????????????
tax9 ???GGATTTCTTCGTAGCGG
tax10 ???????????????AGCGG;
END;
Nexus digits
[ This is a comment ]
#NEXUS
BEGIN DATA;
DIMENSIONS NTAX=10 NCHAR=20;
FORMAT DATATYPE=STANDARD SYMBOLS="0 1 2 3";
MATRIX
tax1 ?0320333113302302122
tax2 10220333113302302122
tax3 ?0220333113302302122
tax4 ?????????????2302122
tax5 10220333113302302122
tax6 10220333113302302122
tax7 ???20333113302302122
tax8 ????????????????????
tax9 ???22033313312302122
tax10 ???????????????02122;
END;
Nexus digits
[ This is a comment ]
#NEXUS
BEGIN DATA;
DIMENSIONS NTAX=10 NCHAR=20;
FORMAT DATATYPE=STANDARD SYMBOLS="00 01 02 03";
MATRIX
tax1 ??00030200030303010103030002030002010202
tax2 0100020200030303010103030002030002010202
tax3 ??00020200030303010103030002030002010202
tax4 ??????????????????????????02030002010202
tax5 0100020200030303010103030002030002010202
tax6 0100020200030303010103030002030002010202
tax7 ??????0200030303010103030002030002010202
tax8 ????????????????????????????????????????
tax9 ??????0202000303030103030102030002010202
tax10 ??????????????????????????????0002010202;
END;
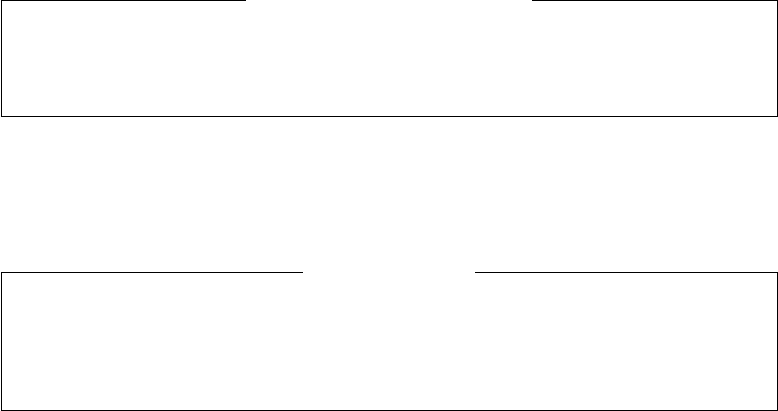
Figure 2. NEXUS formats.
23

their names by at least one blank character.
A PHYLIP input sequence file may also display more than a single data
set. Each of these data sets must be in PHYLIP format and two successive
alignments must be separated by an empty line. Processing multiple data
sets requires to toggle the ‘M’ option in the Input Data sub-menu or use
the ‘-n’ command line option and enter the number of data sets to analyse.
The multiple data set option can be used to process re-sampled data that
were generated using a non-parametric procedure such as cross-validation or
jackknife (a bootstrap option is already included in PhyML). This option
is also useful in multiple gene studies, even if fitting the same substitution
model to all data sets may not be suitable.
PhyML can also process alignments in NEXUS format. Although not
all the options provided by this format are supported by PhyML, a few
specific features are exploited. Of course, this format can handle nucleotide
and protein sequence alignments in sequential or interleaved format. It is also
possible to use custom alphabets, replacing the standard 4-state and 20-state
alphabets for nucleotides and amino-acids respectively. Examples of a 4-state
custom alphabet are given in Figure 2. Each state must here correspond to
one digit or more. The set of states must be a list of consecutive digits
starting from 0. For instance, the list “0, 1, 3, 4” is not a valid alphabet.
Each state in the symbol list must be separated from the next one by a space.
Hence, alphabets with large number of states can be easily defined by using
two-digit number (starting with 00 up to 19 for a 20 state alphabet). Most
importantly, this feature gives the opportunity to analyse data sets made of
presence/absence character states (use the symbols=‘‘0 1’’ option for such
data). Alignments made of custom-defined states will be processed using the
Jukes and Cantor model. Other options of the program (e.g., number of rate
classes, tree topology search algorithm) are freely configurable. Note that, at
the moment, the maximum number of different states is set to 22 in order to
save memory space. It is however possible to lift this threshold by modifiying
the value of the variable T MAX ALPHABET in the file ‘utilities.h’. The
program will then have to be re-compiled.
7.1.1 Gaps and ambiguous characters
Gaps correspond to the ‘-’ symbol. They are systematically treated as un-
known characters “on the grounds that we don’t know what would be there if
something were there” (J. Felsenstein, PHYLIP main documentation). The
likelihood at these sites is summed over all the possible states (i.e., nucleotides
or amino acids) that could actually be observed at these particular positions.
Note however that columns of the alignment that display only gaps or un-
24
Character Nucleotide Character Nucleotide
AAdenosine Y C or T
GGuanosine K G or T
CCytidine B C or Gor T
TThymidine D A or Gor T
UUridine (=T)H A or Cor T
M A or C V A or Cor G
R A or G−or Nor Xor ? unknown
W A or T(=Aor Cor Gor T)
S C or G
Table 2. List of valid characters in DNA sequences and the corre-
sponding nucleotides.
known characters are simply discarded because they do not carry any phylo-
genetic information (they are equally well explained by any model). PhyML
also handles ambiguous characters such as Rfor Aor G(purines) and Yfor
Cor T(pyrimidines). Tables 2and 3give the list of valid characters/symbols
and the corresponding nucleotides or amino acids.
7.1.2 Specifying outgroup sequences
PhyML can return rooted trees provided outgroup taxa are identified from
the sequence file. In order to do so, sequence names that display a ‘*’ char-
acter will be automatically considered as belonging to the outgroup.
The topology of the rooted tree is exactly the same as the unrooted version
of the same tree. In other words, PhyML first ignores the distinction between
ingroup and outgroup sequences, builds a maximum likelihood unrooted tree
and then tries to add the root. If the outgroup has more than one sequence,
the position of the root might be ambiguous. In such situation, PhyML tries
to identify the most relevant position of the root by considering which edge
provides the best separation between ingroup and outgroup taxa (i.e., we are
trying to make the outgroup “as monophyletic as possible”).
7.2 Tree format
PhyML can read one or several phylogenetic trees from an input file. This op-
tion is accessible through the Tree Searching sub menu or the ‘-u’ argument
from the command line. Input trees are generally used as initial maximum
likelihood estimates to be subsequently adjusted by the tree searching al-
25
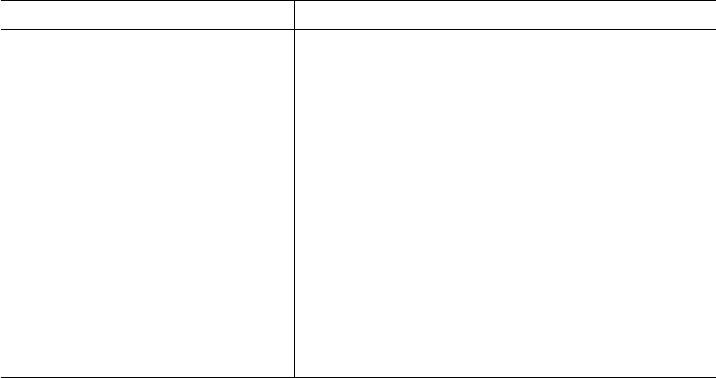
Character Amino-Acid Character Amino-Acid
AAlanine LLeucine
RArginine KLysine
Nor BAsparagine MMethionine
DAspartic acid FPhenylalanine
CCysteine PProline
Qor ZGlutamine SSerine
EGlutamic acid TThreonine
GGlycine WTryptophan
HHistidine YTyrosine
IIsoleucine VValine
LLeucine −or Xor ? unknown
KLysine (can be any amino acid)
Table 3. List of valid characters in protein sequences and the corre-
sponding amino acids.
gorithm. Trees can be either rooted or unrooted and multifurcations are
allowed. Taxa names must, of course, match the corresponding sequence
names.
((seq1:0.03,seq2:0.01):0.04,(seq3:0.01,(seq4:0.2,seq5:0.05):0.2):0.01);
((seq3,seq2),seq1,(seq4,seq5));
Figure 3. Input trees. The first tree (top) is rooted and has branch lengths.
The second tree (bottom) is unrooted and does not have branch lengths.
7.3 Multiple alignments and trees
Single or multiple sequence data sets may be used in combination with single
or multiple input trees. When the number of data sets is one (nD= 1) and
there is only one input tree (nT= 1), then this tree is simply used as input
for the single data set analysis. When nD= 1 and nT>1, each input tree
is used successively for the analysis of the single alignment. PhyML then
outputs the tree with the highest likelihood. If nD>1 and nT= 1, the same
input tree is used for the analysis of each data set. The last combination is
nD>1 and nT>1. In this situation, the i-th tree in the input tree file is
used to analyse the i-th data set. Hence, nDand nTmust be equal here.
26

7.4 Custom amino-acid rate model
The custom amino-acid model of substitutions can be used to implement a
model that is not hard-coded in PhyML. This model must be time-reversible.
Hence, the matrix of substitution rates is symmetrical. The format of the
rate matrix with the associated stationary frequencies is identical to the one
used in PAML. An example is given below:
0.55
0.51 0.64
0.74 0.15 5.43
1.03 0.53 0.27 0.03
0.91 3.04 1.54 0.62 0.10
1.58 0.44 0.95 6.17 0.02 5.47
1.42 0.58 1.13 0.87 0.31 0.33 0.57
0.32 2.14 3.96 0.93 0.25 4.29 0.57 0.25
0.19 0.19 0.55 0.04 0.17 0.11 0.13 0.03 0.14
0.40 0.50 0.13 0.08 0.38 0.87 0.15 0.06 0.50 3.17
0.91 5.35 3.01 0.48 0.07 3.89 2.58 0.37 0.89 0.32 0.26
0.89 0.68 0.20 0.10 0.39 1.55 0.32 0.17 0.40 4.26 4.85 0.93
0.21 0.10 0.10 0.05 0.40 0.10 0.08 0.05 0.68 1.06 2.12 0.09 1.19
1.44 0.68 0.20 0.42 0.11 0.93 0.68 0.24 0.70 0.10 0.42 0.56 0.17 0.16
3.37 1.22 3.97 1.07 1.41 1.03 0.70 1.34 0.74 0.32 0.34 0.97 0.49 0.55 1.61
2.12 0.55 2.03 0.37 0.51 0.86 0.82 0.23 0.47 1.46 0.33 1.39 1.52 0.17 0.80 4.38
0.11 1.16 0.07 0.13 0.72 0.22 0.16 0.34 0.26 0.21 0.67 0.14 0.52 1.53 0.14 0.52 0.11
0.24 0.38 1.09 0.33 0.54 0.23 0.20 0.10 3.87 0.42 0.40 0.13 0.43 6.45 0.22 0.79 0.29 2.49
2.01 0.25 0.20 0.15 1.00 0.30 0.59 0.19 0.12 7.82 1.80 0.31 2.06 0.65 0.31 0.23 1.39 0.37 0.31
8.66 4.40 3.91 5.70 1.93 3.67 5.81 8.33 2.44 4.85 8.62 6.20 1.95 3.84 4.58 6.95 6.10 1.44 3.53 7.09
The entry on the i-th row and j-th column of this matrix corresponds to
the rate of substitutions between amino-acids iand j. The last line in the file
gives the stationary frequencies and must be separated from the rate matrix
by one line. The ordering of the amino-acids is alphabetical, i.e, Ala, Arg,
Asn, Asp, Cys, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Phe, Pro, Ser, Thr,
Trp, Tyr and Val.
7.5 Topological constraint file
PhyML can perform phylogenetic tree estimation under user-specified topo-
logical constraints. In order to do so, one should use the --constraint file
file name command-line option where file name lists the topological con-
straints. Such constraints are straightforward to define. For instance, the
following constraints:
((A,B),C,(D,E,F));
indicate that taxa D, E and F belong to the same clade. A, B and C also
belong to the same clade and the two clades hence defined should not overlap.
Under these two constraints, the tree ((A,B),D,((E,F),C)) is not valid. From
the example above, you will notice that the constraints are defined using a
multifurcating tree in NEWICK format. Note that this tree does not need
27

Sequence file name : ‘seq’
Output file name Content
seq phyml tree.txt ML tree
seq phyml stats.txt ML model parameters
seq phyml boot trees.txt ML trees – bootstrap replicates
seq phyml boot stats.txt ML model parameters – bootstrap replicates
seq phyml rand trees.txt ML trees – multiple random starts
seq phyml ancestral seq.txt ancestral sequences
seq phyml ancestral tree.txt ML tree with node labels as in ancestral sequence file
Table 4. Standard output files
to display the whole list of taxa. For instance, while the only taxa involved
in specifying topological constraints above are A, B, C, D, E & F, the actual
data set could include more than these six taxa only.
PhyML tree topology search algorithms all rely on improving a start-
ing tree. By default, BioNJ is the method of choice for building this tree.
However, there is no guarantee that the phylogeny estimated with PhyML
does comply with the topological constraints. While it is probably possible
to implement BioNJ with topological constraints, we have not done so yet.
Instead, the same multifurcating tree that defines the topological constraints
should also be used as starting tree using the -u (--inputtree) option. Al-
together, the command line should look like the following: -u=file name
--constraint file=file name. It is not possible to use as input tree a
non-binary phylogeny that is distinct from that provided in the constraint
tree file. However, any binary tree compatible with the constraint one can
be used as input tree.
7.6 Output files
Table 4presents the list of files resulting from an analysis. Basically, each
output file name can be divided into three parts. The first part is the se-
quence file name, the second part corresponds to the extension ‘ phyml ’ and
the third part is related to the file content. When launched with the default
options, PhyML only generates two files: the tree file and the model pa-
rameter file. The estimated maximum likelihood tree is in standard Newick
format (see Figure 3). The model parameters file, or statistics file, displays
the maximum likelihood estimates of the substitution model parameters, the
28

likelihood of the maximum likelihood phylogenetic model, and other impor-
tant information concerning the settings of the analysis (e.g., type of data,
name of the substitution model, starting tree, etc.). Two additional out-
put files are created if bootstrap supports were evaluated. These files simply
contain the maximum likelihood trees and the substitution model parameters
estimated from each bootstrap replicate. Such information can be used to
estimate sampling errors around each parameter of the phylogenetic model.
When the random tree option is turned on, the maximum likelihood trees
estimated from each random starting trees are printed in a separate tree file
(see last row of Table 4).
PhyML estimates ancestral sequences by calculating the marginal (as
opposed to the joint) probability of each character state at each inter-
nal node of the phylogeny. These probabilities are given in the file
seq phyml ancestral seq.txt. The bulk of this file is a table where
each row corresponds to a site in the original alignment and a num-
ber corresponding labeling each internal node. It is relatively straight-
forward to identify which number corresponds to which node in the
tree by examining the information provided in the Newick-formatted file
seq phyml ancestral tree.txt.
7.7 Treatment of invariable sites with fixed branch
lengths
PhyML allows users to give an input tree with fixed topology and branch
lengths and find the proportion of invariable sites that maximise the like-
lihood (option -o r). These two options can be considered as conflicting
since branch lengths depend on the proportion of invariants. Hence, chang-
ing the proportion of invariants implies that branch lengths are changing too.
More formally, let ldenote the length of a branch, i.e., the expected number
of substitutions per site, and pbe the proportion of invariants. We have
l= (1 −p)l0, where l0is the expected number of substitutions at variable
sites. When asked to optimize pbut leave lunchanged, PhyML does the
following:
1. Calculate l0=l/(1 −p) and leave l0unchanged throughout the opti-
mization.
2. Find the value of pthat maximises the likelihood. Let p∗denote this
value.
3. Set l∗= (1 −p∗)l0and print out the tree with l∗(instead of l).
29

PhyML therefore assumes that the users wants to fix the branch lengths
measured at variable sites only (i.e., l∗is fixed). This is the reason why
the branch lengths in the input and output trees do differ despite the use
of the the -o r option. While we believe that this approach relies on a
sound rationale, it is not perfect. In particular, the original transformation
of branch lengths (l0=l/(1−p)) relies on a default value for pwith is set to 0.2
in practice. It is difficult to justify the use of this value rather than another
one. One suggestion proposed by Bart Hazes is to avoid fixing the branch
lengths altogether and rather estimate the value of a scaling factor applied
to each branch length in the input tree (option --contrained lens). We
agree that this solution probably matches very well most users expectation,
i.e., “find the best value of pwhile constraining the ratio of branch lengths to
be that given in the input tree”. Please feel free to send us your suggestions
regarding this problem by posting on the forum (http://groups.google.
com/group/phyml-forum).
8 Inputs & outputs for the XML interface
8.1 Mixture models in PhyML
PhyML implements a wide range of mixture models. The discrete gamma
model [31] is arguably the most popular of these models in phylogenetics.
However, in theory, mixture models are not restricted to the description of
the variation of substitution rates across sites. For instance, if there are good
reasons to believe that the relative rates of substitution between nucleotides
vary along the sequence alignments, it makes sense to use a mixture of GTR
models. Consider the case where substitutions between Aand Coccur at high
rate in some regions of the alignment and low rate elsewhere, a mixture with
two classes, each class having its own GTR rate matrix, would be suitable.
The likelihood at any site of the alignment is then obtained by averaging the
likelihoods obtained for each GTR rate matrix, with the same weight given
to each of these matrices.
PhyML implements a generic framework that allows users to define mix-
tures on substitution rates, rate matrices and nucleotide or amino-acid equi-
librium frequencies. Each class of the mixture model is built by assembling
a substitution rate, a rate matrix1and a vector of equilibrium frequencies.
For instance, let {R1, R2, R3}be a set of substitution rates, {M1, M2}a set
of rate matrices and {F1, F2}a set of vectors of equilibrium frequencies. One
1the rate matrix corresponds here the symmetrical matrix giving the so-called “echange-
ability rates”
30
could then define the first class of the mixture model as C1={R1, M1, F1}, a
second class as C2={R2, M1, F1}, and a third class as C3={R3, M2, F2}. If
R1,R2and R3correspond to slow, medium and fast substitution rates, then
this mixture model allows the fast evolving rates to have their own vector
of equilibrium frequencies and rate matrix, distinct from that found at the
medium or slow evolving sites. The likelihood at any given site Dsof the
alignment is then:
Pr(Ds) =
3
X
c=1
Pr(Ds|Cs=c) Pr(Cs=c),
where Pr(Cs=c) is obtained by multiplying the probability (density) of the
three components (i.e., rate, matrix, frequencies). For instance, Pr(C1=
{R1, M1, F1}) = Pr(R1)×Pr(M1)×Pr(F1). We therefore assume here that
substitution rates, rate matrices and equilibrium frequencies are independent
from one another.
Note that, using the same substitution rates, rate matrices and vector
of equilibrium frequencies, it is possible to construct many other mixture
models. For instance, the mixture model with the largest number of classes
can be created by considering all the combinations of these three components.
We would then get a mixture of 3 ×2×2 = 12 classes, corresponding to all
the possible combinations of 3 rates, 2 matrices and 2 vectors of frequencies.
8.2 Partitions
We first introduce some terms of vocabulary that have not been presented
before. A partitionned data set, also referred to as partition, is a set of
partition elements. Typically, a partitionned data set will be made of a set
of distinct gene alignments. A partition element will then correspond to one
(or several) of these gene alignments. Note that the biology litterature often
uses the term partition to refer to an element of a partitionned data. We
thus use here instead the mathematical definition of the terms ‘partition’ and
‘partition element’.
Phylogenetics models usually assume individual columns of an alignment
to evolve independently from one another. Codon-based models (e.g., [32–
35]) are exceptions to this rule since the substitution process applies here to
triplets of consecutive sites of coding sequences. The non-independence of
the substitution process at the three coding positions (due to the specificities
of the genetic code), can therefore be accounted for. Assuming that sites
evolve independently does not mean that a distinct model is fitted to each
site of the alignment. Estimating the parameters of these models would not
31
make much sense in practice due to the very limited amount of phylogenetic
signal conveyed by individual sites. Site independence means instead that
the columns of the observed alignment were sampled randomly from the
same “population of columns”. The stochasticity of the substitution process
running along the tree is deemed responsible to the variability of site patterns.
Some parameters of the phylogenetic model are considered to be common
to all the sites in the alignment. The tree topology is typically one such
parameter. The transition/transversion ratio is also generally assumed to be
the same for all columns. Other parameters can vary from site to site. The
rate at which substitutions accumulate is one of these parameters. Hence,
different sites can have distinct rates. However, such rates are all “drawn”
from the same probabilitic distribution (generally a discrete Gamma density).
Hence, while different sites may have distinct rates of evolution, they all share
the same distribution of rates.
This reasonning also applies on a larger scale. When analysing multiple
genes, one can indeed assume that the same mechanism generated the dif-
ferent site patterns observed for every gene. Here again, we can assume that
all the genes share the same underlying tree topology (commonly refered to
as the “species tree”). Other parameters of the phylogenetic model, such as
branch lengths for instance, might be shared across genes. However, due to
the specificities of the gene evolution processes, some model parameters need
to be adjusted for each gene separately. To sum up, the phylogenetic analysis
of partitionned data requires flexible models with parameters, or distribution
of parameters, shared across several partition elements and other parameters
estimated separately for each element of the partition.
The likelihood of a data set made of the concatenation of nsequence
alignments noted D(1),D(2), . . . , D(n)is then obtained as follows:
Pr(D(1), D(2), . . . , D(n)) =
n
Y
i=1
Pr(D(i))
=
n
Y
i=1
Li
Y
s=1
Pr(D(i)
s),
where Liis the number of site columns in partition element i. Pr(D(i)
s) is
then obtained using Equation 1, i.e., by summing over the different classes of
the mixture model that applies to site sfor partition element i. Hence, the
joint probability of all the partition elements is here broken down into the
product of likelihood of every site for each partition element. As noted just
above, any given component of the mixture model at a given particular site
32
is shared by the other sites that belong to the same partition element and,
for some of them, by sites in other partition elements (e.g., the same tree
topology is shared by all the sites, throughout all the partition elements).
PhyML implements a wide variety of partition models. The only pa-
rameter that is constrained to be shared by all the partition elements is the
tree topology. This constraint makes sense when considering distantly re-
lated taxa, typically inter-species data. For closely related taxa, i.e., when
analysing intra-species or population-level data, not all the genes might have
the same evolutionary history. Recombination events combined to the in-
complete lineage sorting phenomenon can generate discrepancies between the
gene trees and the underlying species tree (see [36] for a review). The phy-
logenetic softwares BEST [37], STEM [38] and *BEAST [39] are dedicated
to the estimation of species tree phylogenies from the analysis of multi-gene
data and allow gene-tree topologies to vary across genes.
Aside from the tree topology that is common to all the sites and all the
partition elements, other parameters of the phylogenetic model can be either
shared across partition elements or estimated separately for each of these.
When analysing three partition elements, A,Band Cfor instance, PhyML
can fit a model where the same set of branch lengths applies to Aand B
while Chas its own estimated lengths. The same goes for the substitution
model: the same GTR model, with identical parameter values, can be fitted
to Aand Cand JC69 for instance can be used for B. The sections below
give more detailed information on the range of models available and how to
set up the corresponding XML configuration files to implement them.
8.3 Combining mixture and partitions in PhyML: the
theory
The rationale behind mixture models as implemented in PhyML lies in (1)
the definition of suitable rate matrices, equilibrium frequency vectors and
relative rates of substitution and (2) the assembly of these components so
as to create the classes of a mixture. The main idea behind partitionned
analysis in PhyML lies in (1) the hypothesis of statistical independance of
the different data partition elements and (2) distinct data partition can share
model components such as rate matrices, equilibrium frequencies or distri-
bution of rates across sites. More formally, the likelihood of a data set made
33

of npartition elements is written as follows:
Pr(D(1), D(2), . . . , D(n)) =
n
Y
i=1
Li
Y
s=1
Pr(D(i)
s)
=
n
Y
i=1
Li
Y
s=1
Ki
X
c=1
Pr(D(i)
s|C =c) Pr(C=c),
where Liis the number of sites in partition element iand Kiis the number
of classes in the mixture model that applies to this same partition element.
Each class of a mixture is made of a rate matrix M, a vector of equilibrium
frequencies Fand a relative rate of substitution R. Branch lengths, Land
tree topology τare also required for the calculation of the likelihood. Hence
we have:
Pr(D(1), D(2), . . . , D(n))
=
n
Y
i=1
Li
Y
s=1
Ki
X
c=1
Pr(D(i)
s|C =c) Pr(C=c)
=
n
Y
i=1
Li
Y
s=1
Mi
X
m
Fi
X
f
Ri
X
r
Pr(D(i)
s|M(i)
m, F (i)
f, R(i)
r, L(i), τ) Pr(M(i)
m, F (i)
f, R(i)
r)I(m, f, r, i)
where Mi,Fiand Riare the number of rate matrices, vector of equilibrium
frequencies and relative rates that apply to partition element irespectively.
I(m, f, r, i) is an indicator function that takes value 1 if the combination Mm,
Ffand Rris acually defined in the model for this particular partition element
i. Its value is 0 otherwise. In the example given in section 8.1 {R1, R2, R3}is
the set of substitution rates, {M1, M2}the set of rate matrices and {F1, F2}
the set of vectors of equilibrium frequencies. We then define the first class of
the mixture model as C1={R1, M1, F1}, a second class as C2={R2, M1, F1}
and the third as C3={R3, M2, F2}. Hence, we have I(1,1,1, i), I(1,1,2, i)
and I(2,2,3, i) equal to one while the nine other values that this indicator
function takes, corresponding to the possible combinations of two vectors of
frequencies, two matrices and three rates, are all zero.
As stated before, our implementation assumes that the different com-
ponents of a mixture are independant. In other words, we have
Pr(M(i)
m, F (i)
f, R(i)
r) = Pr(M(i)
m)×Pr(F(i)
f)×Pr(R(i)
r). In practice, the joint
probability Pr(M(i)
m, F (i)
f, R(i)
r) is obtained as follows:
Pr(M(i)
m, F (i)
f, R(i)
r) = Pr(M(i)
m) Pr(F(i)
f) Pr(R(i)
r)
Pm,f,r Pr(M(i)
m) Pr(F(i)
f) Pr(R(i)
r)I(m, f, r, i)
34

The probabilities Pr(M(i)
m), Pr(F(i)
f) and Pr(R(i)
r), also called ‘weights’, can
be fixed or estimated from the data.
8.4 The XML format and its use in PhyML
The few paragraphs below are largely inspired from the Wikipedia page that
describes the XML format (http://en.wikipedia.org/wiki/XML). XML
(eXtensible Markup Language) is a markup language that defines a set of
rules for encoding documents in a format that is both human-readable and
machine-readable. An XML document is divided into markup and content,
which may be distinguished by the application of simple syntactic rules.
Generally, strings that constitute markup either begin with the character ‘<’
and end with a ‘>’. Strings of characters that are not markup are content:
XML markup and content example
<markup>
content
</markup>
A markup construct that begins with ‘<’ and ends with ‘>’ is called
atag. Tags come in three flavors: (1) start-tags (e.g, <section>), end-
tags (e.g., </section>) and empty-element tags (e.g., <line-break />). A
component either begins with a start-tag and ends with a matching end-
tag or consists only of an empty-element tag. The characters between
the start- and end-tags, if any, are the element’s content, and may con-
tain markup, including other elements, which are called child elements.
In the following example, the element img has two attributes,src and
alt:<img src="madonna.jpg" alt="Foligno Madonna, by Raphael"/>.
Another example would be <step number="3">Connect A to B.</step>
where the name of the attribute is “number” and the value is “3”.
In practice, building a mixture model in a XML file readable by PhyML is
relatively straightforward. The first step is to define the different components
of each class of the mixture. Consider for instance that the fitted model will
have a Gamma distribution with four classes plus a proportion of invariants.
The rate component of the mixture can then be specified using the following
XML code:
35

Γ4+I rates
1
2<siterates id="SiteRates1">
3<weights id="Distrib" family="gamma+inv" alpha=".1" \
4optimise.alpha="yes" pinv="0.4" optimise.pinv="yes">
5</weights>
6<instance id="R1" init.value="1.0"/>
7<instance id="R2" init.value="1.0"/>
8<instance id="R5" init.value="0.0"/>
9<instance id="R3" init.value="1.0"/>
10 <instance id="R4" init.value="1.0"/>
11 </siterates>
12
In the example above, the <siterates> component completely defines a
model of substitution rate variation across sites. This component has a par-
ticular identity, i.e., a name associated to it (“SiteRates1” here), which is
not mandatory. This <siterates> component has six sub-components. The
first is the <weights> component, followed by five <instance> components.
The <weights> component defines the type of distribution that character-
izes the variation of rates across sites. A discrete Gamma plus invariants
is used here. Two parameters specify this distribution: the gamma shape
and the proportion of invariant parameters. Their initial values are set by
using the corresponding attributes and attribute values (alpha="0.1" and
pinv="0.4"). Also, PhyML can optimise these parameters so as to maximise
the likelihood of the whole phylogenetic model (optimise.pinv="yes" and
optimise.alpha="yes"). The following five <instance> components define
the rate classes themselves. The id attribute is here mandatory and must be
unique to each class. Note that one of the initial (relative) rate (init.value
attribute) is set to zero. The corresponding rate class (the third in this
example) will then correspond to the invariant site category.
Having specified the part of the phylogenetic model that describes the
variation of rates across sites, we can now move on to build the rest of the
model. The component below defines two substitution models:
Rate matrices
1
2<ratematrices id="RateMatrices">
3<instance id="M1" model="HKY85" tstv="4.0" optimise.tstv="no"/>
4<instance id="M2" model="GTR" optimise.rr="yes"/>
5</ratematrices>
This <ratematrices> component sets out a list of substitution models
(HKY85 and GTR here). Here again, the different elements in this list
correspond to the <instance> sub-components. Each instance must have
a unique id attribute for a reason that will become obvious shortly. The
remaining attributes and their functions are described in Section 8.6.3.
36
The next “ingredient” in our phylogenetic model are vectors of nucleotide
frequencies. The <equfreqs> component below specifies two of such vectors:
Equilibrium frequencies
1
2<equfreqs id="EquFreq">
3<instance id="F1"/>
4<instance id="F2"/>
5</equfreqs>
6
Now, we need to assemble these three components (rate variation across
sites, rate matrices and vectors of equilibrium frequencies) into a mixture
model. The <partitionelem> component below defines one such model:
Mixture model
1
2<partitionelem id="Part1" file.name="./nucleic.txt" data.type="nt">
3<mixtureelem list="R1, R2, R3, R4, R5"/>
4<mixtureelem list="M1, M1, M1, M2, M2"/>
5<mixtureelem list="F1, F2, F1, F2, F2"/>
6</partitionelem>
7
The <partitionelem> component defines a particular partition element.
In this example, the partition element corresponds to the sequence file
called nucleic.txt, which is an alignment of nucleotide sequences (see the
data.type attribute value). The <mixtureelem> are sub-components of
the <partitionelem> component. Each <mixtureelem> has a list atr-
ribute. Each such list gives the ID of components that have been defined
before. For instance, the first <mixtureelem> refers to the five classes of
the <siterates> component. The ordering of the different term in these
list matters a lot since it is directly related to the elements in each class
of the mixture model. Hence, the first element in the <list> attribute
of the first <mixtureelem> added to the first element in the <list> at-
tribute of the second <mixtureelem> plus the the first element in <list>
attribute of the third <mixtureelem> defines the first class of the mix-
ture model. Therefore, the mixture model defined above has five classes:
C1={R1, M1, F1},C2={R2, M1, F2},C3={R3, M1, F1},C4={R4, M2, F2}
and C5={R5, M2, F2}.
8.5 Setting up mixture and partition models in
PhyML: the basics
Mixture models are particularly relevant to the analysis of partitionned data.
Indeed, some features of evolution are gene-specific (e.g., substitution rates
37

vary across genes). Models that can accomodate for such variation, as mix-
ture models do, are therefore relevant in this context. However, other evolu-
tionary features are shared across loci (e.g., genes located in the same genomic
region usually have similar GC contents). As a consequence, some compo-
nents of mixture models need to be estimated separately for each partition
element while others should be shared by different partition elements.
Below is a simple example with a partitionned data set made of two
elements, corresponding to the sequence alignment files nucleic1.txt and
nucleic2.txt. Importantly, the number and names of sequences in these
two alignments have to match exactly.
Two sets of branch lengths (one per partition element)
1
2<branchlengths id="BranchLens">
3<instance id="L1"/>
4<instance id="L2"/>
5</branchlengths>
6
7<partitionelem id="Part1" file.name="./nucleic1.txt" data.type="nt">
8<mixtureelem list="R1, R2, R3, R4, R5"/>
9<mixtureelem list="L1, L1, L1, L1, L1"/>
10 </partitionelem>
11
12 <partitionelem id="Part2" file.name="./nucleic2.txt" data.type="nt">
13 <mixtureelem list="R1, R2, R3, R4, R5"/>
14 <mixtureelem list="L2, L2, L2, L2, L2"/>
15 </partitionelem>
16
Mixture elements with names R1,. . .,R5 refer to the Γ4+I model defined
previsouly (see Section 8.4). The <branchlengths> XML component defines
a mixture element that had not been introduced before. It defined vectors of
branch lengths that apply to the estimated phylogeny. Two instances of such
vectors are defined: L1 and L2. When examining the two partition elements
(<partitionelem> component), it appears that L1 is associated with Part1
while L2 is associated with Part2. Hence, branch lengths will be estimated
separately for these two partition elements.
Note that a given partition element can only have one branchlengths
instance associated to it. For instance, the example given below is not valid:
Invalid mixture
1
2<partitionelem id="Part1" file.name="./nucleic1.txt" data.type="nt">
3<mixtureelem list="R1, R2, R3, R4, R5"/>
4<mixtureelem list="L1, L1, L1, L2, L2"/>
5</partitionelem>
6
In other words, mixture of branch lengths are forbidden. One reason
for this restriction is that mixture of edge lengths sometimes lead to non-
identifiable models (i.e., models with distinct sets of branch lengths have the
38
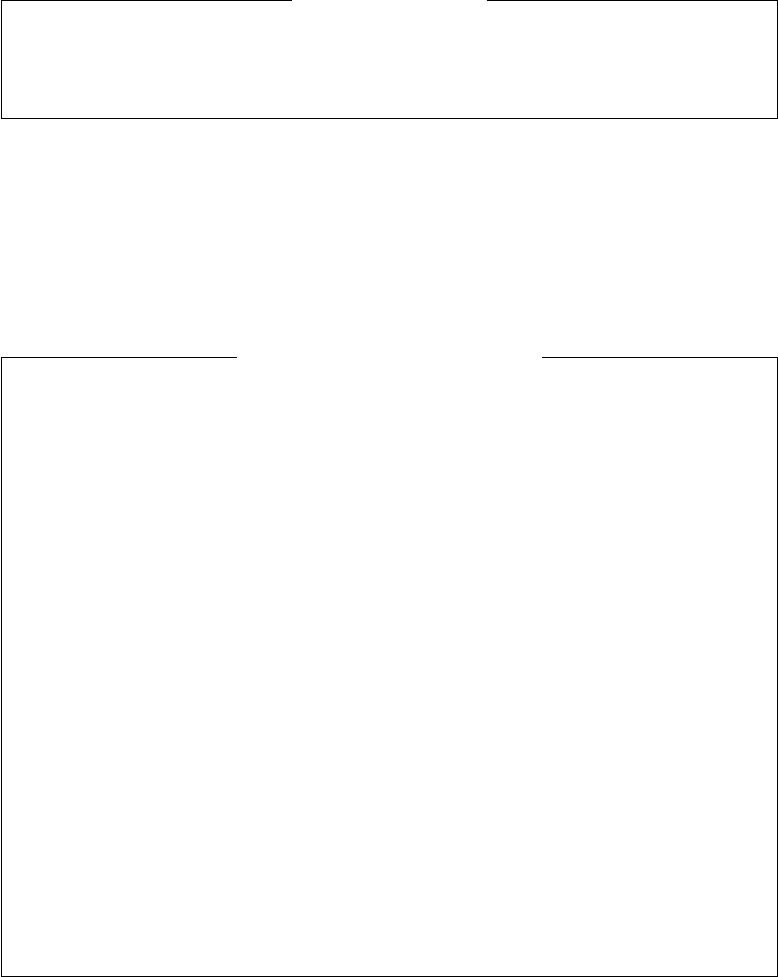
same likelihood) [40]. But mostly, combining mixture of branch lengths with
mixture of rates appears like a deadly combination. Consider for instance
the following model:
Invalid mixture
1
2<partitionelem id="Part1" file.name="./nucleic1.txt" data.type="nt">
3<mixtureelem list="R1, R2, R3"/>
4<mixtureelem list="L1, L2, L3"/>
5</partitionelem>
6
It is here impossible to tell apart branch lengths and substitution rates.
Such model is strongly non-identifiable and therefore not relevant.
In the example given above, the same Γ4+I model (i.e. the same gamma
shape parameter and proportion of invariant ) applies to the two partition
elements. It is possible to use two distinct Γ4+I models instead using the
following XML code:
Two distinct Γ4+I models
1
2<siterates id="SiteRates1">
3<weights id="Distrib1" family="gamma+inv" alpha=".1" \
4optimise.alpha="yes" pinv="0.4" optimise.pinv="yes">
5</weights>
6<instance id="R1" init.value="1.0"/>
7<instance id="R2" init.value="1.0"/>
8<instance id="R5" init.value="0.0"/>
9<instance id="R3" init.value="1.0"/>
10 <instance id="R4" init.value="1.0"/>
11 </siterates>
12
13 <siterates id="SiteRates2">
14 <weights id="Distrib2" family="gamma+inv" alpha=".1" \
15 optimise.alpha="yes" pinv="0.4" optimise.pinv="yes">
16 </weights>
17 <instance id="R6" init.value="1.0"/>
18 <instance id="R7" init.value="1.0"/>
19 <instance id="R8" init.value="0.0"/>
20 <instance id="R9" init.value="1.0"/>
21 <instance id="R10" init.value="1.0"/>
22 </siterates>
23
24 <partitionelem id="Part1" file.name="./nucleic1.txt" data.type="nt">
25 <mixtureelem list="R1, R2, R3, R4, R5"/>
26 <mixtureelem list="L1, L1, L1, L1, L1"/>
27 </partitionelem>
28
29 <partitionelem id="Part2" file.name="./nucleic2.txt" data.type="nt">
30 <mixtureelem list="R6, R7, R8, R9, R10"/>
31 <mixtureelem list="L2, L2, L2, L2, L2"/>
32 </partitionelem>
33
SiteRates1 and SiteRates2 here define two distinct Γ4+I models. Each
of these models apply to one of the two partition elements (nucleic1.txt and
39

nucleic2.txt), allowing them to display different patterns of rate variation
across sites.
8.6 XML options
8.6.1 phyml component
Options:
•output.file="filename". The main output files of PhyML analysis
will be named filename phyml tree and filename phyml stats.
•bootstrap="nreplicates". Run nreplicates replicates for the non-
parametric bootstrap analysis.
•run.id="idstring". PhyML will append the string idstring to each
output file.
•print.json.trace="yes|true|no|false". PhyML will print the es-
timated trees, the corresponding loglikelihoods and various model pa-
rameters at multiple stages of the estimation process. This option is
useful for monitoring the progress of the analysis when processing large
data sets.
•print.trace="yes|true|no|false". PhyML will print the estimated
trees (and the corresponding loglikelihoods) at multiple stages of the es-
timation process. This option is also useful for monitoring the progress
of the analysis when processing large data sets.
•branch.test="aBayes|aLRT|SH|no". Calculate fast branch support
using the aBayes method [25], aLRT [26] or SH [41] tests. These branch
statistics are much faster to estimate than the bootrap proportions and
usually provide good estimates of the probabilities that the correspond-
ing edges are correctly inferred (see Anisimova et al. 2011 for more pre-
cision). By default and if no bootstrap analysis is performed, branch
supports are estimated using the aBayes approach.
•quiet="yes|no". Runs PhyML in quiet mode when quiet=yes. The
program will not pause if the memory required to run the analysis ex-
ceeds 256MB and will not output the progresssion of the log-likelihood
scores on the standard output.
•memory.check="yes|no". By default, when processing a large data
set, PhyML will pause and ask the user to confirm that she/he wants
40

to continue with the execution of the analysis despite the large amount
of memory required. Setting memory.check=no skips this question. It
is especially useful when running PhyML in batch mode.
8.6.2 topology component
Options:
•init.tree="bionj"|"user"|"random". Starting tree. Default is
bionj.
•n.rand.starts="X". Number of random starting trees. Default is 5.
•file.name="name of tree file". In case init.tree="user", this at-
tribute is mandatory. name of tree file is a text file containing a tree
in NEWICK format.
•optimise.tree="yes"|"true"|"no"|"false". The starting tree
topology as defined by init.tree is to be optimised (or not) so as
to maximise the likelihood function.
•search="nni"|"spr"|"none". Tree topology search is conducted us-
ing NNI (fast), SPR (a bit slower but more accurate) or no moves.
Example of ‘topology’ component
1
2<topology>
3<instance id="T1" init.tree="bionj" optimise.tree="true" \
4search="spr"/>
5</topology>
6
8.6.3 ratematrices component
Options:
•model="JC69"|"K80"|"F81"|"F84"|"HKY85"|"TN93"|"GTR"|"custom"
for nucleotide data. The default is "HKY85".
model="LG"|"WAG"|"JTT"|"MtREV"|"Dayhoff"|"DCMut"|"RtREV"|"CpREV"|"VT"
|"Blosum62"|"MtMam"|"MtArt"|"HIVw"|"HIVb"|"customaa" for
amino-acid sequences. The default is "LG".
•model.code="012345". For custom model applied to nucleotide se-
quences: set the string of digits that define a custom substitution
model. See Table 1on page 17 for more information about the model
codes.
41

•ratematrix.code="filename". When used in conjunction with
model="customaa",filename is the name of the file that gives the
rates of substitution between amino-acids as well as their frequences at
equilibrium using PAML rate matrix format. An example of such file
is provided in phyml/examples/X1.mat.
•optimise.rr="yes"|"true"|"no"|"false". For custom and GTR nu-
cleotide models only: optimise the substitution rate model parameters.
•optimise.tstv="yes"|"true"|"no"|"false". For K80,F84,HKY85
and TN93 models only: optimise the transition/transversion rate ratio.
•tstv="value". For K80,HKY85 and TN93 models only: set the transi-
tion/transversion to a given value.
The ratematrices component has the attribute
optimise.weights=yes/no (default is no). If optimise.weights=yes, then
the probabilities (or weights) or each matrix in the set of matrices defined
by this component (see Equation 1), will be estimated from the data.
Example of ‘ratematrices’ component
1
2<ratematrices id="RM1" optimise.weights="yes">
3<instance id="M1" model="custom" model.code="000000"/>
4<instance id="M2" model="GTR" optimise.rr="yes"/>
5<instance id="M3" model="WAG"/>
6</ratematrices>
7
8.6.4 equfreqs component
Options:
•base.freqs="a,b,c,d" where a-d are nucleotide frequencies. Make
sure that these frequencies are separated by comas and no space char-
acter is inserted.
•aa.freqs="empirical|model". Amino-acid frequencies are derived
from counting the number of occurence of each amino-acid in the
alignment (aa.freqs="empirical") or given by the substitution model
(aa.freqs="model").
•optimise.freqs="true|yes|false|no". Nucleotide frequen-
cies can be optimised so as to maximise the likelihood
(optimise.freqs="yes|true").
42

The equfreqs component has the attribute optimise.weights=yes/no
(default is no). If optimise.weights=yes, then the probabilities (or weights)
or each vector of equilibrium frequencies in the set of vectors defined by this
component (see Equation 1), will be estimated from the data.
Example of ‘equfreqs’ component
1
2<equfreqs id="EF1" optimise.weights="yes">
3<instance id="F1" base.freqs="0.25,0.25,0.25,0.25"/>
4<instance id="F2" aa.freqs="empirical"/>
5<instance id="F3" optimise.freqs="yes"/>
6</equfreqs>
7
8.6.5 branchlengths component
Options:
•optimise.lens="yes"|"true"|"no"|"false": branch lengths are op-
timised or not. The default is set to "yes".
Example of ‘branchlengths’ component
1
2<branchlengths id="BL1">
3<instance id="L1" optimise.lens="yes"/>
4<instance id="L2"/>
5<instance id="L3" optimise.lens="false"/>
6</branchlengths>
7
8.6.6 siterates component
Options:
•value="val", where "val" is the relative substitution rate for the cor-
responding class.
Asiterate component generally includes a weights element that specifies
the probabilitic distribution of the relative rates. The available options for
such element are:
•family="gamma|gamma+inv|freerates".gamma indicates that the
distribution of the relative rates is set to be a discrete Gamma den-
sity. gamma+inv indicates that the relative rate model is a mixture of
Gamma and invariant sites (this is the common Γ+I model). FreeRate
is a model that does not use any parametric function to describe the
distribution of the relative rates (see [28]). Under this option, relative
rates and the corresponding frequencies of these classes are directly
43

estimated from the data. While such approach is slightly more com-
putationally demanding than the Γ (or Γ+I) model, it often provides
a significantly better fit to the data.
•alpha="value|optimised", where value is a real positive number.
Use this option to set the gamma shape parameter to the selected
value. optimised: the parameter is estimated from the data (see also
next option).
•optimise.alpha="yes|true|no|false". Optimise the shape of the
Gamma distribution of relative rates (or not).
•pinv="value|optimised", where value is in [0,1]. Use this option to
set the proportion of invariants to the selected value. optimised: the
parameter is estimated from the data (see also next option).
•optimise.pinv="yes|true|no|false". Optimise the proportion of
invariable sites (or not).
•optimise.freerates="yes|true|no|false". Optimise the parame-
ters of the FreeRate model, i.e., the relative rates and the corresponding
frequencies.
Example of ‘siterates’ component (discrete gamma model)
1
2<siterates id="SR1">
3<instance id="R1" init.value="1.0"/>
4<instance id="R2" init.value="1.0"/>
5<instance id="R3" init.value="1.0"/>
6<instance id="R4" init.value="1.0"/>
7<weights id="D1" family="gamma" optimise.alpha="yes" \
8optimise.pinv="no">
9</weights>
10 </siterates>
Setting up an analysis using the FreeRate mixture model of rate variation
across sites [28] requires a bit more work. As in the discrete gamma model,
the FreeRate model relies on definies classes of (relative) rates. In the discrete
gamma model, each class has the same frequency. This is no longer the case
with FreeRate. It is thus necessary to specify the frequency, or weight, of
each class of rate in the XML file. The example below illustrates how these
weights are defined in practice:
44
Example of ‘siterates’ component (FreeRate model)
1<!-- Freerate model of variation of rates across sites -->
2<siterates id="SR1">
3<instance id="R1" init.value="0.1"/>
4<instance id="R2" init.value="0.7"/>
5<instance id="R3" init.value="1.9"/>
6<instance id="R4" init.value="5.1"/>
7<weights id="D1" family="freerates" optimise.freerates="yes">
8<instance appliesto="R1" value="0.4"/>
9<instance appliesto="R2" value="0.3"/>
10 <instance appliesto="R3" value="0.1"/>
11 <instance appliesto="R4" value="0.1"/>
12 </weights>
13 </siterates>
Note that the weights do not have to sum to one. PhyML rescales them
before starting the analysis such that these weights are transformed into
proper probabilities. The relative rates themselves are rescaled too such that
the weighted average (relative) rate is equal to one after rescaling.
8.6.7 partitionelem and mixtureelem components
Options:
•file.name="inputfilename", where inputfilename is the name of
the input sequence file (in PHYLIP format) to be analysed.
•data.type="nt|aa". Specify the type of sequences to be processed
(nucleotide of amino-acid sequences).
•interleaved="yes|true|no|false". Interleaved (yes|true) or se-
quential format (no|false) for the sequence alignment.
•optimise.tree.scale="yes|true|no|false". The sum of edge
length (or tree size) is optimized. This option is relevant when dif-
ferent data partition elements point to the same set of edge lengths so
that setting optimise.tree.scale="yes" will find the optimal ratio
of tree sizes considering the two elements. In other words, the different
trees all share the same (relative) edge lengths but the corresponding
partition elements have different mean rates of substitution.
•tree.scale="val".val is the value of the (relative) substitution rate.
By default, its value is set to 1.0.
•print.site.lk="yes|true|no|false". The likelihood at each
site (and other information) will be written in a file named
inputfilename phyml lk.
45
Each partitionelem element should include exactly four mixtureelem
elements, corresponding to branch lengths, equilibrium frequencies, sub-
stitution rate model and tree topology. The ordering of in which the
mixtureelem elements are given does not matter, though exceptions apply
for the Γ + Imodel (see below). The n-th element in the list attribute
of each mixtureelem defines the n-th class of the mixture model. In the
example given below, the first class of the mixture is made of the following
elements: T1,F1,R1 and L1, the second class is made of T1,F1,R2 and L1,
etc.
Example of ‘partitionelem’ component
1
2<partitionelem id="partition1" file.name="./small_p1.nxs" \
3data.type="nt" interleaved="yes">
4<mixtureelem list="T1, T1, T1, T1"/>
5<mixtureelem list="F1, F1, F1, F1"/>
6<mixtureelem list="R1, R2, R3, R4"/>
7<mixtureelem list="L1, L1, L1, L1"/>
8</partitionelem>
9
In general, the ordering of the mixtureelem elements does not matter.
However, when the model has invariable sites, then the corresponding class
should be first in the list of classes provided by mixtureelem. For instance,
in the example above, if the rates are defined as follows:
Example of ‘siterates’ component
1
2<siterates id="SR1">
3<instance id="R1" init.value="0.0"/>
4<instance id="R2" init.value="1.0"/>
5<instance id="R3" init.value="1.0"/>
6<instance id="R4" init.value="1.0"/>
7<weights id="D1" family="gamma+inv" optimise.alpha="yes" \
8optimise.pinv="no">
9</weights>
10 </siterates>
then R1 corresponds to the invariable rate class (as init.value="0.0").
As R1 is first in the mixtureelem (see line 6 in the example of ‘partionelem’
given above), PhyML will print out an explicit error message and bail out.
One way to avoid this shortcoming is to define mixtureelem as R4, R2, R3,
R1 instead.
8.7 A simple example: GTR + Γ4+I
The example below provides all the required options to fit a Γ4+I model
to a single alignment of nucleotide sequences under the GTR model of sub-
stitution using a SPR search for the best tree topology. The phyml com-
ponent sets the name for the analysis to simple.example, meaning that
46
each output file will display this particular string of characters. Also, the
tree and statistics file names will begin with p1.output. The tree topol-
ogy will be estimated so as to maximise the likelihood and the topology
search algorithm used here is SPR, as indicated by the value of the corre-
sponding attribute (i.e., search="spr"). Only one vector of branch lengths
will be used here since only one partition element will be processed. Hence,
the <branchlengths> component only has one <instance> sub-component.
Also, a single GTR model will apply to all the classes for the mixture model
– the <ratematrices> component has only one <instance> sub-component,
corresponding to this particular substitution model. The next component,
<equfreqs>, indicates that a single vector of equilibrium frequencies will
apply here. Next, the <siterates> component has five <instance> sub-
components. Four of these correspond to the non-zero relative rates of evo-
lution a defined by a discrete Gamma distribution. The last one (<instance
id="R5" value="0.0"/>) defines the class of the mixture corresponding to
invariable sites. The <weight> component indicates that a Γ+I model will
be fitted here. The shape parameter of the Gamma distribution and the pro-
portion of invariants will be estimated from the data. The <partitionelem>
gives information about the sequence alignment (the corresponding file name,
the type of data and the alignment format). The <mixtureelem> components
next define the mixture model. Each class of the fitted model corresponds to
one column, with the first column made of the following elements: T1, M1,
F1, R1 and L1. The second class of the mixture is made of T1, M1, F1,
R2, L1 and so forth.
47

Simple PhyML XML example
1
2<phyml runid="simple.example" output.file="p1.output">
3
4<topology>
5<instance id="T1" init.tree="bionj" optimise.tree="yes" \
6search="spr"/>
7</topology>
8
9<branchlengths id="BL1">
10 <instance id="L1" optimise.lens="yes"/>
11 </branchlengths>
12
13 <ratematrices id="RM1">
14 <instance id="M1" model="GTR"/>
15 </ratematrices>
16
17 <equfreqs id="EF1">
18 <instance id="F1"/>
19 </equfreqs>
20
21 <siterates id="SR1">
22 <instance id="R1" value="1.0"/>
23 <instance id="R2" value="1.0"/>
24 <instance id="R3" value="1.0"/>
25 <instance id="R4" value="1.0"/>
26 <instance id="R5" value="0.0"/>
27 <weights id="D1" family="gamma+inv" optimise.alpha="yes" \
28 optimise.pinv="yes">
29 </weights>
30 </siterates>
31
32 <partitionelem id="partition_elem1" file.name=\
33 "./p1.seq" data.type="nt" interleaved="yes">
34 <mixtureelem list="T1, T1, T1, T1, T1"/>
35 <mixtureelem list="M1, M1, M1, M1, M1"/>
36 <mixtureelem list="F1, F1, F1, F1, F1"/>
37 <mixtureelem list="R1, R2, R3, R4, R5"/>
38 <mixtureelem list="L1, L1, L1, L1, L1"/>
39 </partitionelem>
40
41 </phyml>
42
8.8 A second example: LG4X
The example below shows how to fit the LG4X model [42] to a given align-
ment of amino-acid sequences (file M587.nex.Phy). LG4X is a mixture model
with four classes. Each class has its own rate and corresponding frequencies
(hence the use of the FreeRate model below, see the <siterates> com-
ponent). In the particular example given here, the rate values and fre-
quencies are set by the users. These parameters will then be optimized
by PhyML (optimise.freerates="yes"). Each class also has its own rate
matrix and vector of equilibrium frequencies, which need to be provided
by the user (Note that these matrices can be downloaded from the follow-
ing web address: http://www.atgc-montpellier.fr/download/datasets/
48

models/lg4x/LG4X_4M.txt. They are also provided in the PhyML package
example/lg4x/ directory.)
LG4X
1<phyml run.id="lg4x" output.file="M587.tests" branch.test="no">
2
3<!-- Tree topology: start with BioNJ and then SPRs -->
4<topology>
5<instance id="T1" init.tree="user" file.name="user_tree.txt" \
6search="spr" optimise.tree="no"/>
7</topology>
8
9
10 <!-- Four rate matrices, read from files -->
11 <ratematrices id="RM1">
12 <instance id="M1" model="customaa" ratematrix.file="X1.mat"/>
13 <instance id="M2" model="customaa" ratematrix.file="X2.mat"/>
14 <instance id="M3" model="customaa" ratematrix.file="X3.mat"/>
15 <instance id="M4" model="customaa" ratematrix.file="X4.mat"/>
16 </ratematrices>
17
18 <!-- Freerate model of variation of rates across sites -->
19 <siterates id="SR1">
20 <instance id="R1" init.value="0.197063"/>
21 <instance id="R2" init.value="0.750275"/>
22 <instance id="R3" init.value="1.951569"/>
23 <instance id="R4" init.value="5.161586"/>
24 <weights id="D1" family="freerates" optimise.freerates="yes">
25 <instance appliesto="R1" value="0.422481"/>
26 <instance appliesto="R2" value="0.336848"/>
27 <instance appliesto="R3" value="0.180132"/>
28 <instance appliesto="R4" value="0.060539"/>
29 </weights>
30 </siterates>
31
32 <!-- Amino-acid equilibrium freqs. are given by the models -->
33 <equfreqs id="EF1">
34 <instance id="F1" aa.freqs="model"/>
35 <instance id="F2" aa.freqs="model"/>
36 <instance id="F3" aa.freqs="model"/>
37 <instance id="F4" aa.freqs="model"/>
38 </equfreqs>
39
40
41 <!-- One vector of branch lengths -->
42 <branchlengths id="BL1" >
43 <instance id="L1" optimise.lens="yes"/>
44 </branchlengths>
45
46
47 <!-- Mixture model assemblage -->
48 <partitionelem id="partition1" file.name="M587.nex.Phy" \
49 data.type="aa" interleaved="yes">
50 <mixtureelem list="T1, T1, T1, T1"/>
51 <mixtureelem list="M1, M2, M3, M4"/>
52 <mixtureelem list="F1, F2, F3, F4"/>
53 <mixtureelem list="R1, R2, R3, R4"/>
54 <mixtureelem list="L1, L1, L1, L1"/>
55 </partitionelem>
56
57 </phyml>
In order to fit the LG4X model to the proteic sequence
49

file provided in the examples/ directory, simply type ./phyml
--xml=../examples/lg4x/lg4x.xml (assuming the PhyML binary is in-
stalled in the src/ directory). You can of course slightly tweak the file
../examples/lg4x/lg4x.xml and use it as a template to fit this model to
another data set.
8.9 An example with multiple partition elements
The example below gives the complete XML file to specify the analysis
of three partition elements, corresponding to the nucleotide sequence files
small p1 pos1.seq,small p1 pos2.seq and small p1 pos3.seq in inter-
leaved PHYLIP format. Importantly, the number and names of sequences
in these three alignments match exactly. small p1 pos1.seq is fitted with
the HKY85 model of substitution (with the transition/transversion ratio be-
ing estimated from the data), combined to a Γ4 model of rate variation
across sites (with the gamma shape parameter being estimated from the
data). small p1 pos2.seq is fitted to a custom substitution model with
the constraint A↔G=C↔T. The nucleotide frequencies are set to
1
4here. The model does not allow substitution rates to vary across sites.
small p1 pos3.seq is fitted using a GTR model conbined to a Γ4+I model
of rate variation across sites. Note that the equilibrium nucleotide frequen-
cies for the fourth and fifth class of the mixture are set to be equal to that
estimated from the first partition element (i.e., F1) . The initial phylogeny
is built using BioNJ and the tree topology is to be estimated using a NNI
search algorithm.
50

Example of PhyML XML file
1
2<phyml runid="nnisearch" output.file="small_p1_output">
3
4<topology>
5<instance id="T1" init.tree="bionj" optimise.tree="yes" \
6search="nni"/>
7</topology>
8
9<branchlengths id="BL1">
10 <instance id="L1" optimise.lens="yes"/>
11 <instance id="L2"/>
12 <instance id="L3"/>
13 </branchlengths>
14
15 <ratematrices id="RM1">
16 <instance id="M1" model="HKY85" optimise.tstv="yes"/>
17 <instance id="M2" model="custom" model.code="102304" \
18 optimise.rr="yes"/>
19 <instance id="M3" model="GTR"/>
20 </ratematrices>
21
22 <equfreqs id="EF1">
23 <instance id="F1"/>
24 <instance id="F2" base.freqs="0.25,0.25,0.25,0.25"/>
25 <instance id="F3"/>
26 </equfreqs>
27
28 <siterates id="SR1">
29 <instance id="R1" value="1.0"/>
30 <instance id="R2" value="1.0"/>
31 <instance id="R3" value="1.0"/>
32 <instance id="R4" value="1.0"/>
33 <weights id="D1" family="gamma" optimise.alpha="yes" \
34 optimise.pinv="no">
35 </weights>
36 </siterates>
37
38 <siterates id="SR2">
39 <instance id="R8" value="1.0"/>
40 <weights id="D2" family="gamma" optimise.alpha="yes" \
41 optimise.pinv="yes">
42 </weights>
43 </siterates>
44
45 <siterates id="SR3">
46 <instance id="R10" value="1.0"/>
47 <instance id="R11" value="1.0"/>
48 <instance id="R12" value="1.0"/>
49 <instance id="R13" value="1.0"/>
50 <instance id="R14" value="1.0"/>
51 <weights id="D3" family="gamma" optimise.alpha="yes" \
52 optimise.pinv="yes">
53 </weights>
54 </siterates>
55
51

Example of PhyML XML file (ctnd)
1
2<partitionelem id="partition_elem1" file.name=\
3"./small_p1_pos1.seq" data.type="nt" interleaved="yes">
4<mixtureelem list="T1, T1, T1, T1"/>
5<mixtureelem list="M1, M1, M1, M1"/>
6<mixtureelem list="F1, F1, F1, F1"/>
7<mixtureelem list="R1, R2, R3, R4"/>
8<mixtureelem list="L1, L1, L1, L1"/>
9</partitionelem>
10
11 <partitionelem id="partition_elem2" file.name=\
12 "./small_p1_pos2.seq" data.type="nt" interleaved="yes">
13 <mixtureelem list="T1"/>
14 <mixtureelem list="M2"/>
15 <mixtureelem list="R8"/>
16 <mixtureelem list="F2"/>
17 <mixtureelem list="L2"/>
18 </partitionelem>
19
20 <partitionelem id="partition_elem3" file.name=\
21 "./small_p1_pos3.seq" data.type="nt" interleaved="yes">
22 <mixtureelem list="T1, T1, T1, T1, T1"/>
23 <mixtureelem list="M3, M3, M3, M3, M3"/>
24 <mixtureelem list="R10, R11, R12, R13, R14"/>
25 <mixtureelem list="F3, F3, F3, F1, F1"/>
26 <mixtureelem list="L3, L3, L3, L3, L3"/>
27 </partitionelem>
28
29 </phyml>
30
8.10 Branch lengths with invariants and partionned
data
Accommodating for models with invariable sites applying to some elements of
a partitioned data, with these elements sharing the same set of edge lengths
can lead to inconsistencies. Consider for instance a partitioned data set
with two elements. Assume that these two elements share the same set of
edge lengths. Also, consider that GTR+I applies to the first element and
HKY applies to the second. Now, the expected number of substitutions
per site for the first element of the partition is equal to (1 −p)l, where p
is the estimated proportion of invariants and lis the maximum-likelihood
estimate for the length of that specific edge. For the second element of
the partition, the expected number of substitutions per site is equal to l,
rather than (1 −p)l. While lare common to the two elements, matching
the specification of the input model, the actual edge lengths do differ across
the two partition elements. Please be aware that, due to the programming
structure implemented in PhyML, the program will only return one value
here, which will be equal to (1 −p)l.
52
9 Citing PhyML
The “default citation” for PhyML is:
•“New algorithms and methods to estimate maximum-likelihood phy-
logenies: assessing the performance of PhyML 3.0”. Guindon S., Du-
fayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. 2010,
Systematic Biology, 59(3):307-321
The “historic citation” for PhyML is:
•“A simple, fast and accurate algorithm to estimate large phylogenies by
maximum likelihood” Guindon S., Gascuel O. 2003, Systematic Biology,
52(5):696-704
10 Other programs
PhyML is software package that provides tools to tackle problems other than
estimating maximum likelihood phylogenies. Installing these tools and pro-
cessing data sets is explained is the following sections.
10.1 PhyTime
PhyTime is a program that uses Bayesian sampling techniques to infer phy-
logenies from the analysis of genetic and fossil data. Edge lengths (and node
heights) are expressed in calendar time units as opposed to expected num-
ber of substitutions as in PhyML. PhyTime can thus be used to date past
evolutionary events. The main original features compared to other software
for molecular dating are:
•a model of rate evolution whereby rates can vary along each branch
(other software consider that the rates of evolution vary from one
branch to another but stay constant along any given branch) [29].
•the opportunity to accomodate for uncertainty in calibration infor-
mation. A single time constraint can apply to multiple clades with
corresponding probabilities as determined from prior analysis of fossil
data [43].
53
10.1.1 Installing PhyTime
Compiling PhyTime is straightforward on Unix-like machines (i.e., linux and
MacOS systems). PhyTime is not readily available for Windows machines
but compilation should be easy on this system too. In the ‘phyml’ directory,
where the ‘src/’ and ‘doc/’ directories stand, enter the following commands:
./configure --enable-phytime;
make clean;
make;
This set of commands generates a binary file called phytime which can
be found in the ‘src/’ directory.
10.1.2 Running PhyTime
Passing options and running PhyTime on your data set is quite similar to
running PhyML using an XML parameter input file, i.e., a typical run would
be launched using the following command: ./phytime --xml=./param.xml,
assuming that the phytime binary file is in the same directory as the XML
control file param.xml, which happens to be the current directory here. The
main differences between PhyML and PhyTime are explained below:
•Unlike PhyML, PhyTime requires calibration (along with genetic se-
quence) data as input. The format for defining the relevant time inter-
vals is described below.
•PhyTime does not allow partitionned (i.e., multigene) analysis yet.
10.1.3 PhyTime input
As stated above, PhyTime takes as input an XML file very similar to those
compatible with PhyML. An example is given thereafter. The first part of the
input file (up to the partitionelem block) corresponds to a standard XML
input file for PhyML. Note however that the tag name “phytime” replaces
“phyml”. The second part is PhyTime-specific. Three new tags can be found
here: lineagerates,clade and calibration.
The lineagerates element is used to define the model of rate variation
across lineages. Its only attribute is model which can take the following
values:
•model=strictclock or model=clock implements the strict clock
model, i.e., all lineages evolve at the same instantaneous rate, at any
point in time.
54

•model=lognormal or model=normal. The logarithm of the average rate
along each branch is distributed as a normal distribution. The mean
and variance of this distribution are the same for all branches. More
precisely, let Rbbe the average rate of substitution along edge b. We
have Rb=Xb·µ, where µis the “baseline” (or average over lineages)
rate of substitution and Xbis thus the branch-specific relative rate of
substitution. The lognormal model has log(Xb)∼ N (1, ν), i.e., the
logarithm of the relative rate is normally distributed with mean (mode
and median) set to 1.0 and standard deviation equal to ν.
•model=geometricbrownian or model=brownian or model=geo. The
logarithm of the rate of substitution evolves according to a Brownian
motion process along the tree. More precisely, the logarithm of the rate
of evolution at the end of edge b, noted as Ystop
b= log(Rstop
b), is normally
distributed with variance v:= ν·∆tb, where ∆tbis the (calendar)
time elapsed along edge b, and mean equal to Ystart
b−1
2v, so that
E(Rstop
b) = E(Rstart
b). Under the geometric Brownian motion model,
the average rate of evolution is also random. This variability is taken
into account in the calculation of the transition between nucleotides
or amino-acids along edges. See Guindon, 2012, Syst. Biol. for more
information.
The clade element is used to define a subset of taxa. Each of these taxa
is given in a taxon element. The value that each taxon takes is a string
corresponding to the name of one of the sequences in the alignment file. In
the example that follows, Gymno Cycas,Gymno Ginko and Gymno Juniperus
are three taxa that define a clade called CLADE1. Note that this clade may not
be monophyletic. In fact it is not considered as such during the inference.
The second important element is calibration. It defines time intervals
corresponding to the time of diversification (i.e., the time of the most recent
common ancestor) of the set of taxa the calibration points to. In other words,
a given time interval defines the calibration for the timing of the crown node
of a given clade. Each interval is defined using the upper and lower tags.
The upper (resp. lower) bound for each interval corresponds to the amount
of time elapsed in upper (resp. lower) time units.
55

Example of PhyTime XML file
1<phytime run.id="abc" output.file="def" mcmc.chain.len="1E+7" \
2mcmc.sample.every="100" mcmc.print.every="50" mcmc.burnin="10000">
3
4<!-- Tree topology -->
5<topology>
6<instance id="T1" init.tree="bionj" optimise.tree="yes"/>
7</topology>
8
9
10 <!-- Substitution model -->
11 <ratematrices id="RM1">
12 <instance id="M1" model="HKY85" optimise.tstv="no" tstv="4.0"/>
13 </ratematrices>
14
15
16 <!-- Freerate model of variation of rates across sites -->
17 <siterates id="SR1">
18 <instance id="R3" init.value="0.5"/>
19 <instance id="R2" init.value="1.0"/>
20 <instance id="R1" init.value="2.0"/>
21 <weights id="D1" family="freerates" optimise.freerates="no">
22 <instance appliesto="R3" value="0.33"/>
23 <instance appliesto="R2" value="0.33"/>
24 <instance appliesto="R1" value="0.33"/>
25 </weights>
26 </siterates>
27
28 <!-- Nucleotide frequencies -->
29 <equfreqs id="EF1">
30 <instance id="F1" optimise.freqs="no"/>
31 </equfreqs>
32
33 <!-- Vector of edge lengths -->
34 <branchlengths id="BL1" >
35 <instance id="L1" optimise.lens="no"/>
36 </branchlengths>
37
38 <!-- Model assembly -->
39 <partitionelem id="partition1" file.name="./seq.txt" \
40 data.type="nt" interleaved="no">
41 <mixtureelem list="T1, T1, T1"/>
42 <mixtureelem list="M1, M1, M1"/>
43 <mixtureelem list="F1, F1, F1"/>
44 <mixtureelem list="R1, R2, R3"/>
45 <mixtureelem list="L1, L1, L1"/>
46 </partitionelem>
56

Example of PhyTime XML file (ctnd)
1
2<!-- Model of rate variation across lineages -->
3<lineagerates model="clock"/>
4
5<clade id="CLADE1">
6<taxon value="Gymno_Cycas"/>
7<taxon value="Gymno_Ginkgo"/>
8<taxon value="Gymno_Juniperus"/>
9</clade>
10 <calibration id="CAL1">
11 <lower>290</lower>
12 <upper>452</upper>
13 <appliesto clade.id="CLADE1">
14 </appliesto>
15 </calibration>
16
17 <clade id="CLADE2">
18 <taxon value="Fagales_Juglans"/>
19 <taxon value="Ophioglossum"/>
20 </clade>
21 <calibration id="CAL2">
22 <lower>0</lower>
23 <upper>10000</upper>
24 <appliesto clade.id="CLADE2">
25 </appliesto>
26 </calibration>
27
28 </phytime>
10.1.4 Accounting for calibration uncertainty
It is not always obvious to determine with full precision and accuracy where in
the tree a given fossil branches in a priori. For instance, a first morphological
feature of an ancestral (fossilized) species might be shared by species A but
not B nor C, while a second feature might be displayed by A and B but not
C. Assuming for simplicity that the true tree topology is ((A,B),C), then the
first feature calibrates the most recent common ancestor (MRCA) of A and
B. Indeed, this ancestor cannot be younger than the fossil itself because, if
this was the case, then A and B would display the morphological feature of
interest. In a similar fashion, the second feature suggests that the calibration
should instead apply to the MRCA of A, B and C.
Therefore, assuming that the fossil was discovered in a well-defined ge-
ological layer which age range is known without ambiguity, there is still
uncertainty around the node in the tree this time interval calibrates. In the
particular example given above, it is not clear whether the calibration con-
straints applies to the node corresponding to the MRCA of A and B, or the
root of the tree.
PhyTime accomodates for this uncertainty by giving a probability to the
two scenari: with probability αthe calibration time interval applies to the
MRCA of species A and B and with probability 1−α, this interval calibrates
57

the age of the MRCA of A, B and C.
It is fairly straightforward to set up an analysis that incorporates prob-
abilistic distributions on calibrations in PhyTime. The section of the XML
file below gives a simple example whereby the calibration “CAL1” applies to
“CLADE1” (i.e., the smallest clade in the tree that displays the three taxa
Gymno Juniperus,Gymno Sciadopitys and Gymno Cycas) with probability
0.2 and to “CLADE2” (i.e., the smallest clade that displays Gymno Juniperus
and Gymno Sciadopitys) with probability 0.8. Also, calibration CAL2 applies
to CLADE3 with probability 1.0, which is implicit here and thus does not need
to be specified in the XML file.
Example of PhyTime XML file with calibration uncertainty
1...
2
3<clade id="CLADE1">
4<taxon value="Gymno_Juniperus"/>
5<taxon value="Gymno_Sciadopitys"/>
6<taxon value="Gymno_Cycas"/>
7</clade>
8
9<clade id="CLADE2">
10 <taxon value="Gymno_Juniperus"/>
11 <taxon value="Gymno_Sciadopitys"/>
12 </clade>
13
14 <clade id="CLADE3">
15 <taxon value="Fagales_Juglans"/>
16 <taxon value="Ophioglossum"/>
17 </clade>
18
19
20 <calibration id="CAL1">
21 <lower>50</lower>
22 <upper>400</upper>
23 <appliesto clade.id="CLADE1" probability="0.2"/>
24 <appliesto clade.id="CLADE2" probability="0.8"/>
25 </calibration>
26
27 <calibration id="CAL2">
28 <lower>300</lower>
29 <upper>500</upper>
30 <appliesto clade.id="CLADE3">
31 </appliesto>
32 </calibration>
33
34 </phytime>
10.1.5 MCMC settings
PhyTime estimates the joint posterior distribution of the phylogenetic model
parameters using an MCMC algorithm. This algorithm relies on sampling
values of these parameters iteratively. The default number of iterations is
fixed to 1E+07 and values of parameters are recorded every 1E+03 iter-
ation of the MCMC algorithm. The burn-in period, during which tuning
parameters of the MCMC as adjusted, lasts for 1E+04 iterations. These
58
three parameters can be modified by setting the values of the attributes
mcmc.chain.len,mcmc.sample.every and mcmc.burnin in the XML pa-
rameter file accordingly. For instance, the first line of this file could look as
follows:
MCMC settings
1<phytime
2run.id="example" output.file="small" mcmc.chain.len="1E+7"
3mcmc.sample.every="100" mcmc.print.every="50" mcmc.burnin="10000">
The vast majority of analyses will not warrant 1E+07 iterations to con-
verge to the target distribution (i.e., converge to the solution). I thus rec-
ommend monitoring each analysis by loading on a regular basis the statistics
output file produced by PhyTime and interrupt the analysis once the effective
sample sizes of all parameters have reached 200.
The MCMC algorithm implemented in PhyTime relies on multiple oper-
ators that update different parameters, or the same parameter in different
ways. The weights of these operators, i.e., the frequency with which they are
applied, were adjusted for the analysis of the plant data set described in [43].
If you think these default values are not suitable for your own analysis, please
feel free to have a look at the function MCMC Complete MCMC in the file mcmc.c
to make the appropriate changes to the values of mcmc->move weight[XXX],
where XXX is the name of the operator. Alternatively, you might want to
send me an email (guindon@lirmm.fr) if you are unsure about all this.
It is generally very useful to compare the posterior estimates of model
parameters to that obtained from a sampler that ignores sequence data (i.e.,
the Bayesian sampler only takes as input data the calibration information).
Such analysis helps quantifying how sensitive are the marginal posterior es-
timates to the estimates one would get prior collecting and analysing genetic
sequences. The attribute ignore.sequences used in the phytime tag per-
mits to run such analysis. The first lines of a corresponding XML file would
then look as follows:
Sampling from the prior in PhyTime
1<phytime run.id="prior" output.file="seq" ignore.sequences>
10.1.6 PhyTime output
The program PhyTime generates two output files. Using the XML file given
above, one of the output files is called ‘abc def phytime stats.txt’. It
lists the node times and (relative) substitution rates on edges sampled dur-
ing the estimation process. It also gives the sampled values for other pa-
rameters, such as the autocorrelation of rates (parameter ‘nu’), and the
rate of evolution (parameter ‘clock’) amongst others. This output file
59

can not be analysed with the program Tracer from the BEAST package
(http://beast.bio.ed.ac.uk/Main_Page) or Icylog (http://tgvaughan.
github.io/icylog/) as some of the columns display non-numerical vari-
ables. It is preferable to process this file using R here. Section 10.1.7 of
this document gives more information about the statistics output file. The
second file is called ‘abc def phytime trees.txt’. It is the list of rooted
trees that were collected during the estimation process, i.e., phylogenies
sampled from the posterior density of trees. This file can be processed
using the software TreeAnnotator, also part of the BEAST package (see
http://beast.bio.ed.ac.uk/Main_Page) in order to generate confidence
sets for the node time estimates.
10.1.7 An example of PhyTime input and output files
The directory phyml/examples/phytime/ provides a sequence alignment
(seq.txt) and an XML input file (param.xml) that can be used to run
an analysis (using the following command: phytime --xml=param.xml).
The XML file is very similar to that described above and will
thus not be discussed further. The statistics output file is called
small example phytime stats.txt. The columns of this file are as follows:
•sample: the index of the iteration in the MCMC algorithm. For in-
stance, a sample value of 100 means that 100 “moves” in the MCMC
have been applied so far, one such move can correspond to an attempt
to modify the tree topology, or height of some internal nodes, etc.
•lnL(posterior): the logarithm of the posterior density of the current
model parameters. It is this function that the MCMC attempts to get
samples from.
•lnL(seq): the logarithm of the conditional probability of the sequence
alignment given the current model parameters.
•lnL(times): the logarithm of the probability density of the phylogeny
evaluated under the birth and death branching process.
•lnL(rates): the logarithm of the probability density of the relative
substitution rates along every edge of the current phylogeny.
•birth: sampled values of the birth parameter. Histogram of these
values provide an estimate of the marginal posterior distribution for
this parameter.
•death: death parameter for the birth-death branching process.
60

•clock: mean rate of substitution.
•root: age of the root node, i.e. the MRCA of all taxa in the sample.
•tstv: transition/transversion ratio (the substitution model used here
is HKY, as selected in the param.xml file).
•nu: rate autocorrelation parameter.
•rr0,rr1,rr2,pr0,pr1,pr2: relative rate of substitution and the
corresponding frequencies. The mixture model of rate variation used
in this analysis is the FreeRate model [28] with three rate classes. These
columns give the values of these six parameters (though there are only
four free parameters) sampled from the target (posterior) distribution.
•t(calib:CAL1 clade:CLADE1): sampled age of the MRCA of CLADE1
when calibration CAL1 applies to this particular node.
•t(calib:CAL1 clade:CLADE2): sampled age of the MRCA of CLADE2
when calibration CAL1 applies to this node.
•t(calib:CAL2 clade:CLADE3): sampled age of the MRCA of CLADE3
when calibration CAL2 applies to this node.
•clade(calib:CAL1): the clade id (CLADE1 or CLADE2) calibration CAL1
applies to.
•clade(calib:CAL2): the clade id (CLADE1 or CLADE2) calibration CAL1
applies to.
•br0,. . .,br41: relative rate on all edges in the tree.
•t22,. . .,t42: ages of all internal nodes.
•The remaining columns provides information about the acceptance rate
of various operators in the MCMC and the corresponding tuning pa-
rameters.
The script below can be copied and pasted into R in order to
produce the plots in Figure 4(assuming R was launched from the
phyml/examples/phytime directory). The two plots at the top left and
center give the trace of the node age of the MRCA of CLADE1 when calibra-
tion CAL1 applies to it (left) and that of CLADE2 when the same calibration
CAL1 applies to it (centre). The plot on the top right gives the clade (which
of CLADE1 or CLADE2) the calibration CAL1 applies to. The analysis of these
61

plots shows the impact of applying the calibration constraint to CLADE1 and
CLADE2 alternatively. When CAL1 applies to CLADE2 (i.e., (Gymno Juniperus
and Gymno Sciadopitys) are pushed towards older values compared to the
situation where the constraint applies to CLADE1 (Gymno Juniperus,Gymno
Sciadopitys and Gymno Cycas). This behaviour makes good sense since the
taxa making up CLADE2 define a substet of those making up CLADE1. As a
consequence, when CAL1 applies to CLADE1, the MRCA of CLADE2 is “free”
to wander towards younger ages. When this constraint applies to CLADE2
instead, it pushes the age of that nodes (and that of the MRCA of CLADE1)
towards older values, as expected.
R script to produce traces from the PhyTime statistics file
1file=paste("dating_phytime_stats.txt",sep="");
2d=read.table(file,header=T);
3idx=floor(0.1*length(d$sample)):length(d$sample)
4par(mfrow=c(2,3),mar=c(5,4,2,2));
5plot(d$sample[idx],d$t.calib.CAL1_clade.CLADE1.[idx],type="l",
6col="red",ylab="Time",xlab="Sample",main="Time CLADE1 under CAL1")
7plot(d$sample[idx],d$t.calib.CAL1_clade.CLADE2.[idx],type="l",
8col="red",ylab="Time",xlab="Sample",main="Time CLADE2 under CAL1")
9plot(d$sample[idx],d$clade.calib.CAL1.[idx],type="l",col="black",
10 ylab="Clade",xlab="Sample",main="(1->CLADE1, 2->CLADE2)")
11 plot(d$sample[idx],d$lnL.seq.[idx],type="l",col="orange",
12 ylab="log(Probability)",xlab="Sample",main="Likelihood sequences")
13 plot(d$sample[idx],d$lnL.posterior.[idx],type="l",col="orange",
14 ylab="log(density)",xlab="Sample",main="Joint posterior")
15 plot(d$sample[idx],d$lnL.times.[idx],type="l",col="orange",
16 ylab="log(density)",xlab="Sample",main="Branching process")
10.1.8 Citing PhyTime
The “default citation” is:
•Guindon S. “From trajectories to averages: an improved description
of the heterogeneity of substitution rates along lineages”, Systematic
Biology, 2013. 62(1): 22-34.
An earlier article also described some of the methods implemented in
PhyTime:
•Guindon S. “Bayesian estimation of divergence times from large data
sets”, Molecular Biology and Evolution, 2010, 27(8):1768:81.
62
1e+05 3e+05 5e+05 7e+05
60 80 100 120 140
Time CLADE1 under CAL1
Sample
Time
1e+05 3e+05 5e+05 7e+05
20 30 40 50 60 70 80
Time CLADE2 under CAL1
Sample
Time
1e+05 3e+05 5e+05 7e+05
1.0 1.2 1.4 1.6 1.8 2.0
(1−>CLADE1, 2−>CLADE2)
Sample
Clade
1e+05 3e+05 5e+05 7e+05
−23960 −23950 −23940 −23930 −23920
Likelihood sequences
Sample
log(Probability)
1e+05 3e+05 5e+05 7e+05
−24100 −24080 −24060 −24040 −24020
Joint posterior
Sample
log(density)
1e+05 3e+05 5e+05 7e+05
−70 −65 −60 −55 −50 −45 −40
Branching process
Sample
log(density)
Figure 4. Traces from the statistics output file produced by Phy-
Time.
63

10.2 PhyloGeo
PhyloGeo is a program that implements the competition-dispersal phylogeog-
raphy model described in Ranjard, Welch, Paturel and Guindon “Modelling
competition and dispersal in a statistical phylogeographic framework”. Ac-
cepted for publication in Systematic Biology.
It implements a Markov Chain Monte Carlo approach that samples from
the posterior distribution of the three parameters of interest in this model,
namely the competition intensity λ, the dispersal bias parameter σand the
overal dispersal rate τ. The data consist in a phylogeny with node heights
proportional to their ages and geographical locations for evety taxon in this
tree. An important assumption of the model is that each node in the phy-
logeny corresponds to a speciation and a dispersal event. As a consequence,
this model does not authorize a given taxon to occupy more than one loca-
tions. Note however that the converse is not true: a given location can be
occupied by several different taxa.
10.2.1 Installing PhyloGeo
Compiling PhyloGeo is straightforward on Unix-like machines (i.e., linux and
MacOS systems). PhyloGeo is not readily available for Windows machines
but compilation should be easy on this system too. In the ‘phyml’ directory,
where the ‘src/’ and ‘doc/’ directories stand, enter the following commands:
./configure --enable-geo;
make clean;
make;
This set of commands generates a binary file called phylogeo which can
be found in the ‘src/’ directory.
10.2.2 Running PhyloGeo
PhyloGeo takes as input a rooted tree file in Newick format and a file with
geographical locations for all the tips of the phylogeny. Here is an example
of valid tree and the corresponding spatial locations just below:
Valid PhyloGeo input tree
(((unicaA:1.30202,unicaB:1.30202):1.34596,(((nitidaC:0.94617,(nitidaA:0.31497,
nitidaB:0.31497):0.63120):0.18955,(((mauiensisA:0.00370,mauiensisB:0.00370):0.20068,
(pilimauiensisA:0.05151,pilimauiensisB:0.05151):0.15287):0.78769,(brunneaA:0.10582,
brunneaB:0.10582):0.88625):0.14365):0.80126,(((molokaiensisA:0.03728,
molokaiensisB:0.03728):0.71371,(deplanataA:0.01814,deplanataB:0.01814):0.73285):0.34764,
((parvulaA:0.20487,parvulaB:0.20487):0.40191,(kauaiensisA:0.24276,
kauaiensisB:0.24276):0.36401):0.49186):0.83835):0.71099):1.38043,
(nihoaA:0.05673,nihoaB:0.05673):3.97168);
64

Valid PhyloGeo spatial location file
nihoaA 23.062222 161.926111
nihoaB 23.062222 161.926111
kauaiensisA 22.0644445 159.5455555
kauaiensisB 22.0644445 159.5455555
unicaA 21.436875 158.0524305
unicaB 21.436875 158.0524305
parvulaA 21.436875 158.0524305
parvulaB 21.436875 158.0524305
molokaiensisA 20.90532 156.6499
molokaiensisB 20.90532 156.6499
deplanataA 20.90532 156.6499
deplanataB 20.90532 156.6499
brunneaA 20.90532 156.6499
brunneaB 20.90532 156.6499
mauiensisA 20.90532 156.6499
mauiensisB 20.90532 156.6499
pilimauiensisA 20.90532 156.6499
pilimauiensisB 20.90532 156.6499
nitidaA 19.7362 155.6069
nitidaB 19.7362 155.6069
nitidaC 19.7362 155.6069
In order to run PhyloGeo, enter the following command: ./phylogeo
./tree file ./spatial location file > phylogeo output. PhyloGeo
will then print out the sampled values of the model parameters in the file
phylogeo output. This file can then be used to generate the marginal pos-
terior densities of the model parameters. In particular, evidence for com-
petition corresponds to value of λsmaller than 1.0. Please see the original
article for more information on how to interpret the model parameters.
10.2.3 Citing PhyloGeo
Ranjard, L., Welch D., Paturel M. and Guindon S. “Modelling competition
and dispersal in a statistical phylogeographic framework”. 2014. Systematic
Biology.
10.3 PhyREX
PhyREX is a program that implements the spatial Λ-Fleming-Viot model
[44–49] or ΛV for short. According to this model, the spatial distribution of
individuals in a population is uniform and does not change during the course
of evolution, as opposed to other models such as the very popular isolation
by distance model proposed by Wright and Mal´ecot. The ΛV model does
not suffer from the “pain in the torus” [50] and clumps of individuals with
increasing densities do not arise. Also, estimates of migration parameters
are not sensitive to variation in sampling intensities across regions, unlike
“mugration” or “discrete trait analysis” models [51].
PhyREX implements an original data augmentation technique embedded
in a Bayesian sampler to estimate two important biological parameters: the
neighorhood size (N) and the dispersal intensity (σ2). These two parameters
are closely related to the effective size of the population of interest per unit
65

area since Nis defined as follows: N:= 4πρeσ2, where ρeis the effective
population density.
10.3.1 Installing PhyREX
Compiling PhyREX is straightforward on Unix-like machines (i.e., linux and
MacOS systems). PhyREX is not readily available for Windows machines
but compilation should be easy on this system too. In the ‘phyml’ directory,
where the ‘src/’ and ‘doc/’ directories stand, enter the following commands:
./configure --enable-phyrex;
make clean;
make;
This set of commands generates a binary file called phyrex which can be
found in the ‘src/’ directory.
10.3.2 Running PhyREX
Example input files for PhyREX can be found in the
examples/phyrex input files/ directory. PhyRex takes as input a
sequence alignment, in a PhyML-compatible format as well as a text file
giving the spatial coordinates of each taxon in two dimension. A example of
the coordinates file is given below:
Valid PhyRex spatial location file
# state.name lon lat
|SouthWest| 0 0
|NorthEast| 10 10
levosl 5.082206 4.133893
kmgcwv 5.914294 4.603446
uhfwja 4.990937 4.445124
ndmwkc 5.178017 4.442268
jpadex 3.747484 4.571090
lqcdcw 7.081925 5.133123
wnsbtg 4.164588 4.720346
ptwgnn 5.711159 4.462993
jhqdsm 3.539525 4.537706
vlnoes 4.613251 4.470530
pfrnpk 4.117791 4.489819
elwdvr 5.649958 4.824092
lptxiv 4.563302 4.005124
The first row in this file gives the names of the columns. It starts with the
‘#’ character signaling a comment. The second and third rows define the
limits of the population’s habitat. At the moment, this habitat is assumed
to be a rectangle. The position in space of that rectangle are determined by
the coordinates of the bottom-left corner (|SouthWest| 0 0) and the top-
right one (|NorthEast| 10 10). The ‘|’ characters help identify the terms
66
Figure 5. Statistics file generated by PhyREX and loaded on Icylog
(http://tgvaughan.github.io/icylog/icylog.html). The traces for the
neighborhood size (top) and the dispersal intensity (bottom) are shown here.
in the list of coordinate corresponding indeed to these two particular points
and should thus not be omitted. The remaining row give the coordinates
of each taxon. Every taxon in the sequence file should also be listed in the
coordinate file.
PhyRex uses the same command-line arguments as that used with
PhyML. The only exception being the mandatory argument corresponding
to the coordinate file. A typical command-line will this look as follows:
./phyrex -i sequence file --coord file=./spatial location file
-c 3 --freerates --il. With this particular set of options, the sequence
alignment will be analysed under a FreeRate model [28] with three rate
classes and a covarion-like model of rate variation across lineaged [29].
Please note that the command-line option --coord file is mandatory.
PhyREX generates correlated samples from the posterior distribution of
the ΛV model parameters. The estimated distributions and related summary
statistics can be monitored during the analysis using the MCMC vizualiza-
tion software Icylog. Figure 5shows the traces for two parameters after
about an hour of calculation on the example data set (36 taxa, 50 geographic
locations).
67
11 Recommendations on program usage
11.1 PhyML
The choice of the tree searching algorithm among those provided by PhyML
is generally a tough one. The fastest option relies on local and simultaneous
modifications of the phylogeny using NNI moves. More thorough explorations
of the space of topologies are also available through the SPR options. As these
two classes of tree topology moves involve different computational burdens,
it is important to determine which option is the most suitable for the type of
data set or analysis one wants to perform. Below is a list of recommendations
for typical phylogenetic analyses.
1. Single data set, unlimited computing time. The best option here is
probably to use a SPR search (i.e., straight SPR of best of SPR and
NNI). If the focus is on estimating the relationships between species, it
is a good idea to use more than one starting tree to decrease the chance
of getting stuck in a local maximum of the likelihood function. Using
NNIs is appropriate if the analysis does not mainly focus on estimating
the evolutionary relationships between species (e.g. a tree is needed
to estimate the parameters of codon-based models later on). Branch
supports can be estimated using bootstrap and approximate likelihood
ratios.
2. Single data set, restricted computing time. The three tree searching
options can be used depending on the computing time available and
the size of the data set. For small data sets (i.e., <50 sequences),
NNI will generally perform well provided that the phylogenetic signal
is strong. It is relevant to estimate a first tree using NNI moves and
examine the reconstructed phylogeny in order to have a rough idea of
the strength of the phylogenetic signal (the presence of small internal
branch lengths is generally considered as a sign of a weak phylogenetic
signal, specially when sequences are short). For larger data sets (>50
sequences), a SPR search is recommended if there are good evidence
of a lack of phylogenetic signal. Bootstrap analysis will generally in-
volve large computational burdens. Estimating branch supports using
approximate likelihood ratios therefore provides an interesting alterna-
tive here.
3. Multiple data sets, unlimited computing time. Comparative genomic
analyses sometimes rely on building phylogenies from the analysis of a
large number of gene families. Here again, the NNI option is the most
68
relevant if the focus is not on recovering the most accurate picture
of the evolutionary relationships between species. Slower SPR-based
heuristics should be used when the topology of the tree is an important
parameter of the analysis (e.g., identification of horizontally transferred
genes using phylogenetic tree comparisons). Internal branch support
is generally not a crucial parameter of the multiple data set analyses.
Using approximate likelihood ratio is probably the best choice here.
4. Multiple data sets, limited computing time. The large amount of data to
be processed in a limited time generally requires the use of the fastest
tree searching and branch support estimation methods Hence, NNI
and approximate likelihood ratios rather than SPR and non-parametric
bootstrap are generally the most appropriate here.
Another important point is the choice of the substitution model. While
default options generally provide acceptable results, it is often warranted to
perform a pre-analysis in order to identify the best-fit substitution model.
This pre-analysis can be done using popular software such as Modeltest [52]
or ProtTest [53] for instance. These programs generally recommend the use of
a discrete gamma distribution to model the substitution process as variability
of rates among sites is a common feature of molecular evolution. The choice
of the number of rate classes to use for this distribution is also an important
one. While the default is set to four categories in PhyML, it is recommended
to use larger number of classes if possible in order to best approximate the
patterns of rate variation across sites [54]. Note however that run times
are directly proportional to the number of classes of the discrete gamma
distribution. Here again, a pre-analysis with the simplest model should help
the user to determine the number of rate classes that represents the best
trade-off between computing time and fit of the model to the data.
11.2 PhyTime
Analysing a data set using PhyTime should involve three steps based on the
following questions: (1) do the priors seem to be adequate (2) can I use the
fast approximation of the likelihood and (3) how long shall I run the program
for? I explain below how to provide answers to these questions.
•Are the priors adequate? Bayesian analysis relies on specifiying the joint
prior density of model parameters. In the case of node age estimation,
these priors essentially describe how rates of substitution vary across
lineages and the probabilistic distribution that node ages have when
ignoring the information provided by the genetic sequences. These
69

priors vary from tree to tree. It is therefore essential to check the
adequacy of priors for each user-defined input tree. In order to do so,
PhyTime needs to be run with the --no data option. When this option
is required, the sequence data provided as input will be ignored and
the rest of the analysis will proceed normally. The prior distribution
of model parameters, essentially edge rates and node heights, can then
be checked using the program Tracer as one would do for the standard
‘posterior’ analysis.
•Can I use the fast approximation to the likelihood? The suface of the
log-likelihood function can be approximated using a multivariate nor-
mal density. This technique is saving very substantial amounts of com-
putation time. However, like most approximations, there are situations
where it does not provide a good fit to the actual function. This usu-
ally happens when the phylogeny displays a lot of short branches, i.e.,
the signal conveyed by the sequences is weak. It is therefore important
to first check whether using the approximate likelihood is reasonable.
In order to do so, it is recommended to first run the program without
the approximation, i.e., using the default settings. Once the minimum
value of the ESS of node ages (the last column on the right of the
standard output) has reached 40-50, open the phytime.XXXX output
file with Tracer and examine the correlation between the exact and
approximate likelihood values. If the correlation is deemed to be good
enough, PhyTime can be re-run using the --fast lk option, which
uses the fast normal approximation to the likelihood function.
•How long shall I run the program for? PhyTime should be run long
enough such that the ESS of each parameter is ‘large enough’. The
last column on the right handside of the standard output gives the
minimum ESS across all internal node heights. It is recommended to
run the program so that this number reaches at least 100.
12 Frequently asked questions
1. PhyML crashes before reading the sequences. What’s wrong ?
•The format of your sequence file is not recognized by PhyML. See
Section 7
•The carriage return characters in your sequence files are not rec-
ognized by PhyML. You must make sure that your sequence file
70
is a plain text file, with standard carriage return characters (i.e.,
corresponding to “\n”, or “\r”)
2. The program crashes after reading the sequences. What’s wrong ?
•You analyse protein sequences and did not enter the -d aa option
in the command-line.
•The format of your sequence file is not recognized by PhyML. See
Section 7
3. Does PhyML handle outgroup sequences ?
•Yes, it does. Outgroup taxa are identified by adding the ‘*’ sign at
the end of each corresponding sequence name (see Section 7.1.2)
4. Does PhyML estimate clock-constrained trees ?
•No, the PhyML program does not estimate clock-contrained trees.
One can however use the program PhyTime to perform such anal-
ysis but the tree topology will not be estimated.
5. Can PhyML analyse partitioned data, such as multiple gene sequences ?
•We are currently working on this topic. Future releases of the
program will provide options to estimate trees from phylogenomic
data sets, with the opportunity to use different substitution mod-
els on the different data partitions (e.g., different genes). PhyML
will also include specific algorithms to search the space of tree
topologies for this type of data.
13 Acknowledgements
The development of PhyML since 2000 has been supported by the Centre Na-
tional de la Recherche Scientifique (CNRS) and the Minist`ere de l’´
Education
Nationale.
71
References
[1] Guindon, S. & Gascuel, O. A simple, fast and accurate algorithm to
estimate large phylogenies by maximum likelihood. Systematic Biology
52, 696–704 (2003).
[2] Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phyloge-
netic analyses with thousands of taxa and mixed models. Bioinformatics
22, 2688–2690 (2006).
[3] Zwickl, D. Genetic algorithm approaches for the phylogenetic analysis
of large biological sequence datasets under the maximum likelihood cri-
terion. Ph.D. thesis, The University of Texas at Austin (2006).
[4] Ayres, D. L. et al. Beagle: an application programming interface and
high-performance computing library for statistical phylogenetics. Sys-
tematic Biology 61, 170–173 (2012).
[5] Jukes, T. & Cantor, C. Evolution of protein molecules. In Munro, H.
(ed.) Mammalian Protein Metabolism, vol. III, chap. 24, 21–132 (Aca-
demic Press, New York, 1969).
[6] Kimura, M. A simple method for estimating evolutionary rates of base
substitutions through comparative studies of nucleotide sequences. Jour-
nal of Molecular Evolution 16, 111–120 (1980).
[7] Felsenstein, J. Evolutionary trees from DNA sequences: a maximum
likelihood approach. Journal of Molecular Evolution 17, 368–376 (1981).
[8] Felsenstein, J. PHYLIP (PHYLogeny Inference Package) version 3.6a2
(Distributed by the author, Department of Genetics, University of Wash-
ington, Seattle, 1993).
[9] Hasegawa, M., Kishino, H. & Yano, T. Dating of the Human-Ape split-
ting by a molecular clock of mitochondrial-DNA. Journal of Molecular
Evolution 22, 160–174 (1985).
[10] Tamura, K. & Nei, M. Estimation of the number of nucleotide sub-
stitutions in the control region of mitochondrial DNA in humans and
chimpanzees. Molecular Biology and Evolution 10, 512–526 (1993).
[11] Lanave, C., Preparata, G., Saccone, C. & Serio, G. A new method
for calculating evolutionary substitution rates. Journal of Molecular
Evolution 20, 86–93 (1984).
72
[12] Tavar´e, S. Some probabilistic and statistical problems on the analysis
of DNA sequences. Lectures on Mathematics in the Life Sciences 17,
57–86 (1986).
[13] Le, S. & Gascuel, O. An improved general amino-acid replacement
matrix. Molecular Biology and Evolution 25, 1307–1320 (2008).
[14] Whelan, S. & Goldman, N. A general empirical model of protein evolu-
tion derived from multiple protein families using a maximum-likelihood
approach. Molecular Biology and Evolution 18, 691–699 (2001).
[15] Dayhoff, M., Schwartz, R. & Orcutt, B. A model of evolutionary change
in proteins. In Dayhoff, M. (ed.) Atlas of Protein Sequence and Structure,
vol. 5, 345–352 (National Biomedical Research Foundation, Washington,
D. C., 1978).
[16] Jones, D., Taylor, W. & Thornton, J. The rapid generation of mutation
data matrices from protein sequences. Computer Applications in the
Biosciences (CABIOS) 8, 275–282 (1992).
[17] Henikoff, S. & Henikoff, J. Amino acid substitution matrices from pro-
tein blocks. PNAS 89, 10915–10919 (1992).
[18] Adachi, J. & Hasegawa, M. MOLPHY version 2.3. programs for molec-
ular phylogenetics based on maximum likelihood. In Ishiguro, M. et al.
(eds.) Computer Science Monographs, 28 (The Institute of Statistical
Mathematics, Tokyo, 1996).
[19] Dimmic, M., Rest, J., Mindell, D. & Goldstein, D. rtREV: an amino acid
substitution matrix for inference of retrovirus and reverse transcriptase
phylogeny. Journal of Molecular Evolution 55, 65–73 (2002).
[20] Adachi, J., P., W., Martin, W. & Hasegawa, M. Plastid genome phy-
logeny and a model of amino acid substitution for proteins encoded by
chloroplast DNA. Journal of Molecular Evolution 50, 348–358 (2000).
[21] Kosiol, C. & Goldman, N. Different versions of the Dayhoff rate matrix.
Molecular Biology and Evolution 22, 193–199 (2004).
[22] Muller, T. & Vingron, M. Modeling amino acid replacement. Journal
of Computational Biology 7, 761–776 (2000).
[23] Cao, Y. et al. Conflict among individual mitochondrial proteins in re-
solving the phylogeny of eutherian orders. Journal of Molecular Evolu-
tion 47, 307–322 (1998).
73
[24] Gascuel, O. BioNJ: an improved version of the NJ algorithm based on
a simple model of sequence data. Molecular Biology and Evolution 14,
685–695 (1997).
[25] Anisimova, M., Gil, M., Dufayard, J., Dessimoz, C. & Gascuel, O. Sur-
vey of branch support methods demonstrates accuracy, power, and ro-
bustness of fast likelihood-based approximation schemes. Systematic
Biology 60, 685–699 (2011).
[26] Anisimova, M. & Gascuel, O. Approximate likelihood-ratio test for
branches: a fast, accurate, and powerful alternative. Systematic Biology
55, 539–552 (2006).
[27] Posada, D. & Crandall, K. Selecting models of nucleotide substitution:
an application to human immunodeficiency virus 1 (hiv-1). Molecular
Biology and Evolution 18, 897–906 (2001).
[28] Soubrier, J. et al. The influence of rate heterogeneity among sites on the
time dependence of molecular rates. Molecular Biology and Evolution
(2012).
[29] Guindon, S. From trajectories to averages: an improved description of
the heterogeneity of substitution rates along lineages. Systematic Biology
62, 22–34 (2013).
[30] Maddison, D., Swofford, D. & Maddison, W. NEXUS: an extensible
file format for systematic information. Systematic Biology 46, 590–621
(1997).
[31] Yang, Z. Maximum likelihood phylogenetic estimation from DNA se-
quences with variable rates over sites: approximate methods. Journal
of Molecular Evolution 39, 306–314 (1994).
[32] Yang, Z. Likelihood ratio tests for detecting positive selection and appli-
cation to primate lysozyme evolution. Molecular Biology and Evolution
15, 568–573 (1998).
[33] Yang, Z. & Nielsen, R. Estimating synonymous and nonsynonymous
substitution rates under realistic evolutionary models. Molecular Biology
and Evolution 17, 32–43 (2000).
[34] Yang, Z. & Nielsen, R. Codon-substitution models for detecting molec-
ular adaptation at individual sites along specific lineages. Molecular
Biology and Evolution 19, 908–917 (2002).
74
[35] Guindon, S., Rodrigo, A., Dyer, K. & Huelsenbeck, J. Modeling the
site-specific variation of selection patterns along lineages. PNAS 101,
12957–12962 (2004).
[36] Degnan, J. & Rosenberg, N. Gene tree discordance, phylogenetic in-
ference and the multispecies coalescent. Trends in Ecology & Evolution
24, 332–340 (2009).
[37] Liu, L. BEST: Bayesian estimation of species trees under the coalescent
model. Bioinformatics 24, 2542–2543 (2008).
[38] Kubatko, L., Carstens, B. & Knowles, L. STEM: species tree estimation
using maximum likelihood for gene trees under coalescence. Bioinfor-
matics 25, 971–973 (2009).
[39] Heled, J. & Drummond, A. Bayesian inference of species trees from
multilocus data. Molecular Biology and Evolution 27, 570–580 (2010).
[40] Matsen, F. & Steel, M. Phylogenetic mixtures on a single tree can mimic
a tree of another topology. Systematic Biology 56, 767–775 (2007).
[41] Shimodaira, H. & Hasegawa, M. Multiple comparisons of log-likelihoods
with applications to phylogenetic inference. Molecular Biology and Evo-
lution 16, 1114–1116 (1999).
[42] Le, S. Q., Dang, C. C. & Gascuel, O. Modeling protein evolution with
several amino acid replacement matrices depending on site rates. Molec-
ular Biology and Evolution 29, 2921–2936 (2012).
[43] Guindon, S. Bayesian molecular dating as a “doubly intractable”
problem. BioRxiv http://biorxiv.org/content/early/2017/02/06/106310
(2017).
[44] Etheridge, A. M. Drift, draft and structure: some mathematical models
of evolution. Banach Center Publ. 80, 121–144 (2008).
[45] Berestycki, N., Etheridge, A. & Hutzenthaler, M. Survival, extinction
and ergodicity in a spatially continuous population model. In Markov
Proc. Rel. Fields (2009).
[46] Barton, N., Etheridge, A. & V´eber, A. A new model for evolution in a
spatial continuum. Electronic Journal of Probability 15 (2010).
75
[47] Barton, N. H., Kelleher, J. & Etheridge, A. M. A new model for extinc-
tion and recolonization in two dimensions: quantifying phylogeography.
Evolution 64, 2701–2715 (2010).
[48] V´eber, A. & Wakolbinger, A. The spatial Lambda-Fleming-Viot process:
an event-based construction and a lookdown representation. Annales de
l’Institut Henri Poincar´e in press (2014).
[49] Barton, N., Etheridge, A. & V´eber, A. Modelling evolution in a spatial
continuum. Journal of Statistical Mechanics: Theory and Experiment
2013, P01002 (2013).
[50] Felsenstein, J. A pain in the torus: some difficulties with models of
isolation by distance. American Naturalist 359–368 (1975).
[51] Lemey, P., Rambaut, A., Drummond, A. J. & Suchard, M. A. Bayesian
phylogeography finds its roots. PLoS Comput Biol 5, e1000520 (2009).
[52] Posada, D. & Crandall, K. Modeltest: testing the model of DNA sub-
stitution. Bioinformatics 14, 817–918 (1998).
[53] Abascal, F., Zardoya, R. & Posada, D. Prottest: selection of best-fit
models of protein evolution. Bioinformatics 21, 2104–2105 (2005).
[54] Galtier, N. & Jean-Marie, A. Markov-modulated Markov chains and
the covarion process of molecular evolution. Journal of Computational
Biology 11, 727–733 (2004).
76
Index
κ,11,17
*BEAST, 33
BEAST, 60
BEST, 33
binary characters, 24
BioNJ, 14
bootstrap, 15
parallel, 7,21
bug, 6
command-line options
--constraint file,20
command-line options
--aa rate file,16
--alpha,18
--ancestral,21
--bootstrap,15
--codpos,18
--constrained lens,20
--data type,15
--free rates,18
--il,18
--inputtree,19
--input,15
--json trace,20
--model,16
--multiple,15
--n rand starts,19
--nclasses,18
--no colalias,20
--no memory check,20
--pars,15
--pinv,17
--print site lk,19
--print trace,20
--quiet,21
--r seed,19
--rand start,19
--run id,20
--search,19
--sequential,15
--ts/tv,17
--use median,18
--xml,21
-f,16
-o,19
compilation, 6
covarion, 18
custom models, 17
data partitions, 31,37
FreeRate, 44
frequencies
amino-acid, 16
nucleotide, 16
gamma distribution (discrete)
mean vs. median, 12,18
number of categories, 12,18
shape parameter, 12,18
GARLI, 6
Icylog, 60
input tree, 19
interleaved, 22
invariable sites, 12,17
lg4x, 48
likelihood
print site likelihood, 19
mixture models, 37
mixture models, 30
MPI, 7,21
MRCA, 57
multiple data sets, 15,26
77
Newick format, 26
NEXUS, 22
NNI, 19
optimisation
substitution parameters, 19
topology, 19
PAML, 11,27
partitionned analysis, 31,37
PHYLIP, 12,22
PhyloGeo, 64
PhyRex, 65
PhyTime, 53
calibration, 54
clade, 55
lineagerates, 54
proportion of invariants, 12,17
random number, 19
random tree, 19
RAxML, 6
run ID, 10,20
sequence format
interleaved, 15
sequential, 15
sequential, 22
SPR, 19
stationary frequencies, 16
STEM, 33
substitution models
amino acids, 16
DNA, 16
Tracer, 60
ts/tv ratio, 11,17
user tree, 19
XML, 30
XML options
branchlengths component, 43
equfreqs component, 42
mixtureelem component, 45
partitionelem component, 45
phyml component, 40
ratematrices component, 41
siterates component, 43
topology component, 41
78