User_manual User Manual
user_manual
User Manual: Pdf
Open the PDF directly: View PDF ![]() .
.
Page Count: 7
1
Antimicrobial Resistance Gene Database Integration Toolkit
User manual
Contents
Overview ........................................................................................................................................ 1
Database eligibility ........................................................................................................................ 2
Pre-requisites ................................................................................................................................. 2
Installation ..................................................................................................................................... 2
Important notice............................................................................................................................ 2
Configurations ............................................................................................................................... 3
Sequence header field indexing.................................................................................................... 3
Usage .............................................................................................................................................. 4
Database validation tool .............................................................................................................. 4
Database integration tool ............................................................................................................. 5
Sequence replacement utility ...................................................................................................... 6
Uniprot ID conversion utility ...................................................................................................... 6
Database version diff utility ........................................................................................................ 7
Known issues ................................................................................................................................. 7
Overview
The Antimicrobial Resistance Gene Data Integration Toolkit (ARGDIT) consists of two main tools
and three utilities for users to perform data validation and integration on antimicrobial resistance
gene (ARG) databases. Basically it allows users to validate an ARG database against the coding
sequence/protein information from NCBI databases, and to merge multiple validated databases
into a single ARG database. It also supports re-annotating the output ARG sequences with NCBI
sequence information, as well as predicting their ARG ontology class adopted from an existing
ARG database (called schema database).
Main tools:
ARG database validation tool (check_arg_db.py)
ARG database integration tool (merge_arg_db.py)
Utilities:
Database sequence replacement utility (replace_db_seqs.py)
UniProt identifier to NCBI protein accession number conversion utility
(convert_id_uniprot_to_ncbi.py)
Version diff utility for ARG database (diff_with_old_ver.py)

2
Database eligibility
In order to use the data validation and integration tool, the ARG database (or other bacterial
coding/protein sequence database) must be in FASTA format, and every FASTA sequence header
must contain an NCBI nucleotide/protein accession number. Uniprot ID is an alternative for
protein accession number for protein sequence database (by converting the Uniprot IDs to protein
NCBI accession numbers with the conversion utility provided). ARG ontology class information,
if any, must occupy at least one individual field in the sequence headers, in which all the fields are
separated by the "|" symbol.
Pre-requisites
The followings must be installed for the core ARGDIT operations:
1. Python version 3.5 or higher
2. BioPython version 1.70 or higher
If ARG ontology class prediction or class outlier sequence detection is required, then the
followings must be installed:
1. MUSCLE version 3.8.31 or higher
2. OD-Seq
3. HMMER3 version 3.1b2 or higher
Installation
No installation is required. Make sure all the third-party software in pre-requisites are in the system
path.
In order to access the NCBI databases, users must provide their own contact email addresses along
with their access requests. Fill in your contact email address under the "Entrez" section in the
configuration file (config.ini):
[Entrez]
Email = (your contact email address)
Important notice
All data retrieval of NCBI databases are performed through NCBI Entrez Programming Utilities,
before using ARGDIT it is very important for every user to read its guidelines and requirements
(https://www.ncbi.nlm.nih.gov/books/NBK25497/#chapter2.Usage_Guidelines_and_Requireme
n) and avoid overwhelming the NCBI servers. Based on these requirements, users are required to
provide their contact email addresses (see the Installation section) so that NCBI may attempt
contact before blocking the abusing access. Although this email address is intended for the
software developers, it is more appropriate for the users to fill in their own so that they can be
notified when situation happens.
3
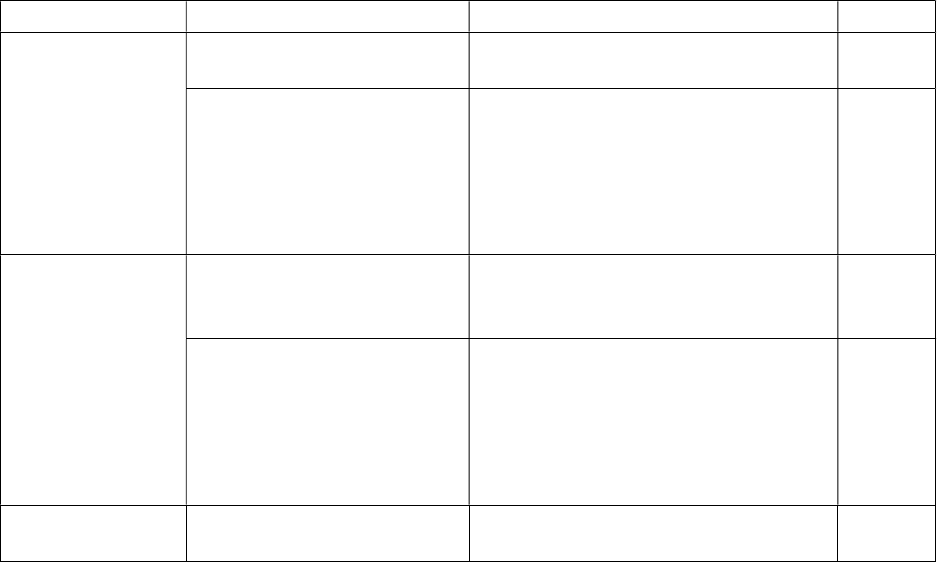
Configurations
The configurable parameters for ARGDIT can be found in the configuration file "config.ini".
These parameters are categorized into different sections listed in the table below:
Section Parameter Description Default
ARGDIT FastaHeaderFieldSeparator
Field separator in the FASTA
sequence header
|
OperationalFieldSeparator FASTA sequence header field
separator to use during program
execution; replaces original field
separator (specified by
FastaHeaderFieldSeparator) during
operation
__
Ontology
annotation check
MinSequenceCount Minimum number of sequences for
an ontology class to be validated or
used for classification
3
BootstrapFactor Determines the number of bootstrap
iterations for sequence outlier
detection according to the formula:
No. of bootstrap iterations = No. of
sequences in an ontology class ×
bootstrap factor
1000
Entrez Email User’s contact email address for the
Entrez utilities
Sequence header field indexing
One-based indexing is applied to index the sequence fields. Assuming the use of the default
FASTA sequence header field separator ("|"), for the sample header at the end of this section, the
third field is "beta-lactamase_CTX-M-134", and the fourth field is empty string "". The fields can
also be indexed from the last field back to the first field, with the last field indexed as -1, the second
last field indexed as -2, and so on. For example, the field with index -4 in the sample header is
"Escherichia_coli". Note that due to input limitation the negative sign "-" is replaced by "~" in the
tool input argument.
Multiple fields can be extracted from the header by slice. For the sample header, by specifying 1-
2 the extracted information is "JX896165|blaCTX-M-134", while "1-876|876|complete" is
extracted with the slice ~1~3.
JX896165|
blaCTX-M-134|
beta-lactamase_CTX-M-134|
|Escherichia_coli|
1-876|
876|
complete
1 2 3 4 5 6 7 8
-8 -7 -6 -5
-4 -3 -2 -1

4
Usage
Database validation tool
Command
./check_arg_db.py [optional arguments] seq_db_path
Mandatory argument
seq_db_path nucleotide/protein database FASTA file path
Optional arguments
-f/--fields FIELD_NUMS specify the ontology label field numbers
FIELD_NUMS to perform
ontology class outlier sequence detection, e.g. -f 4-5, -f ~1-~3
-r/--refine export refined DNA sequences
-e/--exportlog
export validation results and process log
-h/--help show help message and exit
Description
check_arg_db.py performs ARG database validation. The --refine option allows the tool to trim at
most two spurious bases before the start codon or after the stop codon, and export the trimmed
sequences into an individual file specified by the tool. To perform ARG ontology class outlier
sequence detection, specify the ARG ontology class fields after the --fields option. For example,
the hierarchical ontology class can be extracted from MEGARes database by "-f ~1-~3". The
validation results and process log are printed to stdout (i.e. screen) by default, and by specifying
the --exportlog option they will be sent to a .log file in the same directory as the database file.

5
Database integration tool
Command
./merge_arg_db.py [optional arguments] -o OUTPUT_SEQ_DB_PATH seq_db_paths
Mandatory arguments
-o OUTPUT_SEQ_DB_PATH
specify the output database file path
OUTPUT_SEQ_DB_PATH
seq_db_paths
nucleotide/protein database FASTA file paths
Optional arguments
-s/--schema SCHEMA_DB_PATH FIELD_NUMS
specify the schema database
SCHEMA_DB_PATH and ontology label field
numbers FIELD_NUMS to perform sequence
ontology class prediction
-a/--annotate perform automatic re-annotation using NCBI
database information
-p/--protein export protein sequences
-r/--redundant allow redundant sequences
-e/--exportlog export integration results and process log
-h/--help show help message and exit
Description
merge_arg_db.py performs integration of multiple ARG databases. The --annotate option performs
re-annotation of the sequences in the output database. By specifying the schema database file path
and the ARG ontology class fields after the --schema option, the class labels of the output
sequences will be predicted. However, note that the protein sequences of the schema database are
not validated here, so it is advised to validate them using the validation tool. By default only non-
redundant sequences are exported, but this can be overridden with the --redundant option. The tool
provides the --protein option to translate all DNA sequences to protein sequences.

6
Sequence replacement utility
Command
./replace_db_seqs.py [optional argument] seq_db_path replace_seq_file_path
output_seq_db_path
Mandatory arguments
seq_db_path
nucleotide/protein database FASTA file path
replace_seq_file_path
FASTA file path for replacement sequences
output_seq_db_path
output database file path
Optional argument
-h/--help show help message and exit
Description
By matching identical FASTA headers of the sequences, this utility replaces the sequences in the
database FASTA file with those in the replacement sequence file. The database sequences, no
matter replaced or not, are exported to the output database file specified by the user.
Uniprot ID conversion utility
Command
./convert_id_uniprot_to_ncbi.py [optional argument] seq_db_path
output_seq_db_path
Mandatory arguments
seq_db_path
FASTA file path for protein database with Uniprot IDs
output_seq_db_path
output file path for protein database with converted NCBI protein
accession no.
Optional argument
-h/--help show help message and exit
Description
This tool queries the UniProt database for the Uniprot ID to NCBI protein accession number
mappings, and then replaces the UniProt IDs in the sequence headers by the mapped protein
accession numbers. The processed sequences are exported to the output database file specified by
the user.

7
Database version diff utility
Command
./diff_with_old_ver.py [optional argument] seq_db_path old_seq_db_path
Mandatory arguments
seq_db_path
nucleotide/protein database FASTA file path
old_seq_db_path
FASTA file path for previous database release
Optional argument
-h/--help show help message and exit
Description
This utility compares the sequence database with its previous version, and generates a FASTA file
storing all identical sequences, as well as another FASTA file storing those that appear in the
current database only. Two sequences are said to be different when either their nucleotide/protein
sequences or their sequence headers are different.
Known issues
It is sometimes (but not often) possible to have incomplete data retrieval from NCBI databases due
to server-side issues such as heavy workload. This means information for some nucleotide/protein
accession numbers cannot be retrieved at the moment; the outcome is like these accession numbers
are not present in the NCBI databases. When many sequences are spuriously reported as having
their accession numbers not found and/or sequence mismatches, it is advised to try using the
database validation and the integration tools later.