Ex STra CS V.2.0.2 Users Guide
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 44

User’s Guide
Version 2.0.2 Beta
Ryan J Urbanowicz1, and Jason H Moore2
October 15, 2014
1ryan.j.urbanowicz@dartmouth.edu - ExSTraCS Design and Development
2jason.h.moore@dartmouth.edu - Post-Doctoral Mentor
Contents
1 Introduction 2
1.1 WhatisExSTraCS? ......................... 2
1.2 AlgorithmOverview ......................... 3
1.3 FurtherReading ........................... 4
1.4 Obtaining the Software . . . . . . . . . . . . . . . . . . . . . . . . 5
1.5 Minimum System Requirements . . . . . . . . . . . . . . . . . . . 5
1.6 ExSTraCSVersions.......................... 5
1.7 Contact ................................ 6
2 Using ExSTraCS 7
2.1 RunningExSTraCS.......................... 7
2.2 Configuration File and Run Parameters . . . . . . . . . . . . . . 8
2.2.1 Dataset Parameters . . . . . . . . . . . . . . . . . . . . . 9
2.2.2 General Run Parameters . . . . . . . . . . . . . . . . . . . 13
2.2.3 Supervised Learning Parameters . . . . . . . . . . . . . . 15
2.2.4 Mechanism Parameters . . . . . . . . . . . . . . . . . . . 17
2.3 Overview of ExSTraCS Code . . . . . . . . . . . . . . . . . . . . 24
2.4 OutputFiles ............................. 27
2.4.1 Rule Population . . . . . . . . . . . . . . . . . . . . . . . 27
2.4.2 Population Statistics . . . . . . . . . . . . . . . . . . . . . 32
2.4.3 Co-occurrence......................... 32
2.4.4 Attribute Tracking Scores . . . . . . . . . . . . . . . . . . 33
2.4.5 Predictions .......................... 33
2.4.6 Learning Tracking . . . . . . . . . . . . . . . . . . . . . . 34
2.4.7 Expert Knowledge . . . . . . . . . . . . . . . . . . . . . . 34
2.5 MakingPredictions.......................... 35
3 Future ExSTraCS Expansions 36
3.1 Continuous Attribute Improvements . . . . . . . . . . . . . . . . 36
3.2 Continuous Phenotypes . . . . . . . . . . . . . . . . . . . . . . . 37
3.3 Fitness and Deletion Schemes . . . . . . . . . . . . . . . . . . . . 37
3.4 ExSTraCSGUI............................ 38
4 Version Fixes/Updates 39
4.1 ExSTraCS2.0.2............................ 39
4.2 ExSTraCS2.0.1............................ 39
4.3 ExSTraCS2.0............................. 39
1
Chapter 1
Introduction
1.1 What is ExSTraCS?
Extended Supervised Tracking and Classifying System (ExSTraCS) is a Michigan-
Style learning classifier system (LCS) algorithm developed to specialize in clas-
sification, prediction, data mining, and knowledge discovery tasks. Michigan-
style LCS algorithms constitute a unique class of algorithms that distribute
learned patterns over a collaborative population of individually interpretable
(IF:THEN) rules/classifiers (NOTE: that the terms ‘rule’ and ‘classifier’ are
used interchangeably in this context), allowing them to flexibly and effectively
describe complex and diverse problem spaces. These rule-based algorithms com-
bine the global search of evolutionary computing (i.e. a genetic algorithm)
with the local optimization of supervised machine learning. They apply itera-
tive, rather than batch-wise learning, meaning that classifiers are evaluated and
evolved one data instance at a time. This makes them naturally well-suited
to learning different problem niches found in multi-class, latent-class, or het-
erogeneous problem domains. They are also naturally multi-objective, evolving
classifiers toward maximal accuracy and generality (i.e. classifier simplicity)
to improve predictive performance. ExSTraCS is limited to supervised learn-
ing problems, involving any number of potentially predictive attributes, a finite
number of training instances, and a single discrete class(where class is also re-
ferred to as phenotype, endpoint, or the dependent variable). The size datasets
that ExSTraCS can handle may be limited by memory requirements.
ExSTraCS was primarily developed to address a need in epidemiological
data mining, to identify predictive attribute in noisy datasets, where the rela-
tionship(s) between predictive attributes and disease phenotype is believed to be
complex, multi-locus, and potentially epistatic and/or heterogeneous. These are
all characteristics that make the identification of disease risk factors much more
challenging. ExSTraCS is also designed to be as flexible as possible in handling
the characteristics of different datasets taking into account the following; (1)
discrete or continuous attributes, (2) missing data, (3) scalability in large-scale
2
datasets (large numbers of attributes and/or training instances), (4) balanced
or imbalanced datasets, and (5) binary or many classes. While we do not claim
that ExSTraCS is optimized to all these characteristics, we have extended func-
tionality in each of these areas, and offer ExSTraCS as a platform for further
development/improvements. This current implementation of ExSTraCS is not
set up to handle extremely large-scale analyses. We roughly recommend using
ExSTraCS on datasets of up to 5,000 attributes, and up to 10,000 instances.
ExSTraCS still may function beyond these dataset dimensions, but it has not
yet been explored. Expect both ExSTraCS and the expert knowledge algorithms
to take longer to run as dataset dimensions increase.
ExSTraCS, and LCS algorithms in general, constitute a unique alternative to
other well known machine learning/modeling/classification/data mining strate-
gies that follow the classic paradigm of seeking to identify a ‘best’ model of
disease that can individual be applied to the entire dataset. Examples include
logistic regression, neural networks, decision trees, artificial immune systems,
genetic algorithms, multifactor dimmensionality reduction, and support vector
machines. ExSTraCS relies on a genetic algorithm to evolve it’s classifier pop-
ulation, making it a stochastic algorithm. This means that while ExSTraCS
is not guaranteed to find the optimal solution, it may be reasonably applied
to complex, or larger-scale analyses, to intelligently search the problem space
where deterministic algorithms become intractable options.
ExSTraCS is currently implemented in Python 2.7 and can be run from a
command line or within an editor such as Eclipse with PyDev.
1.2 Algorithm Overview
ExSTraCS is descended from a lineage of Michigan-style LCS algorithms, founded
on the architecture of Wilson’s Extended Classifier System (XCS) [22], the most
successful and best-studied LCS algorithm to date. The Supervised Classifier
System (UCS) [2] replaced XCS’s reinforcement learning scheme with a su-
pervised learning strategy to deal explicitly with single-step problems such as
classification and data mining. Comparing select Michigan and Pittsburgh-
style LCS algorithms, UCS showed particular promise when applied to complex
biomedical data mining problems with patterns of epistasis and heterogeneity
[17, 15]. UCS inspired two algorithmic expansions named Attribute Tracking
and Feedback UCS (AF-UCS) and Expert Knowledge UCS (UCS-EK). AF-
UCS introduced mechanisms that improved learning and uniquely allowed for
the explicit characterization of heterogeneous patterns and the identification
of candidate disease subgroups [18, 21]. UCS-EK incorporated expert knowl-
edge into UCS learning for smart population initialization, directed classifier
discovery, and reduced run time [20]. Additionally, novel rapid rule compaction
strategies were recently developed and evaluated for post-processing classifier
populations to enhance interpretability and improve predictive performance [13].
ExSTraCS merges successful components of this algorithmic lineage with other
valuable LCS research, and a redesigned UCS-like framework with a few novel
3
features. In addition to integrating attribute tracking/feedback, expert knowl-
edge covering, and rapid rule compaction, ExSTraCS (1) adopts a flexible and
efficient classifier representation similar to the one described in [1], to accommo-
date data with both discrete and continuous attributes, (2) outputs attribute
tracking scores and global statistics (in addition to a classifier population) for
significance testing, and visualization-guided knowledge discovery as described
in [19], (3) includes an adaptive data detection scheme to adjust the algorithm
to the characteristics of the dataset, (4) includes a rule specificity limit and
an expert knowledge driven mutation operator to dramatically improve algo-
rithm scalability (5) includes a built-in selection of four attribute weighting
algorithms (ReliefF, SURF, SURF*, and MultiSURF) to discover potentially
useful expert knowledge as a pre-processing step, and (6) includes an expert
knowledge wrapper algorithm that improves the performance of the aforemen-
tioned four attribute weighting algorithms on noisy datasets with a greater
number of attributes. Additionally, ExSTraCS 2.0 includes new features geared
towards algorithm scalability such as a rule specificity limit, and the optional
TuRF expansion to all four attribute weighting algorithms. TuRF improves the
performance of each respective attribute weighting algorithm when the dataset
includes larger number of attributes, and noisy data is an issue.
1.3 Further Reading
•For a complete description and performance evaluation of the ExSTraCS
2.0 algorithm see [16].
•For a complete description and performance evaluation of the ExSTraCS
1.0 algorithm see [14].
•For a detailed explanation of attribute tracking and feedback see [18].
Please note that [14] corrects a subtile miss-statement in [18] about how
the attribute feedback mechanism works.
•For a detailed description of expert knowledge covering see [20].
•For a detailed description of ReliefF (one of the expert knowledge discovery
algorithms), see [9].
•For a detailed description of SURF (one of the expert knowledge discovery
algorithms), see [7].
•For a detailed description of SURF* (one of the expert knowledge discov-
ery algorithms), see [6].
•For a detailed description of MultiSURF (one of the expert knowledge
discovery algorithms), see [5].
•For a detailed description of the 6 rule compaction/filtering strategies that
have been implemented in ExSTraCS, see [13].
4
•For a detailed description of the knowledge representation scheme upon
which ExSTraCS’s scheme is based, see [1].
•For a detailed description of TuRF (an expert knowledge discovery wrap-
per algorithm), see [11].
1.4 Obtaining the Software
ExSTraCS 2.0.2 is available as open-source (GPL) code. It is a cross-platform
program written entirely in Python 2.7. It is freely available for download from
http://sourceforge.net. You may also contact Dr. Ryan Urbanowicz or Dr.
Jason Moore for a copy of the code if you experience difficulties downloading it
from the web site.
1.5 Minimum System Requirements
•Python 2.7 (http://www.python.org).
•1 GHz processor
•256 MB Ram
•800x600 screen resolution
1.6 ExSTraCS Versions
•Version 2.0.2: Made available on 10/14/2014. Fixed functionality of
outputTestPredictions run parameter, and fixed InstanceID’s values
output in the Predictions files.
•Version 2.0.1: Made available on 10/13/2014. Fixed a zero division error
involving balanced accuracy calculation that appeared with very small or
highly class imbalanced datasets. Also added updated balanced accuracy
calculation to rule compaction.
•Version 2.0: Made available on 10/7/2014. This version focused on
improving algorithm scalability, adding a data-driven rule specificity limit
(RSL), an expert knowledge guided mutation operator, and the addition
of the TuRF wrapper for improving expert knowledge generation in noisy
data with a greater number of attributes.
•Version 1.0: Made available on 6/20/2014. This is the initial beta version
of the algorithm/code.
5
1.7 Contact
If you (1) have a question not answered by this user’s guide, (2) would like to
report any potential bugs or issues related to the ExSTraCS 2.0.2 Beta code,
(3) have any suggestions for further improvements or expansions, or (4) have an
interest in collaborating; please contact Dr. Ryan Urbanowicz at the following
email: ryan.j.urbanowicz@dartmouth.edu.
6

Chapter 2
Using ExSTraCS
2.1 Running ExSTraCS
This section describes the bare minimum steps required to get ExSTraCS run-
ning on your dataset of interest. First, make sure that Python 2.7 is installed
on your computer. ExSTraCS is run from the command line, and requires a
properly formatted configuration file. Included with ExSTraCS is an example
configuration file named ExSTraCS Configuration File Minimum.txt. To get
ExSTraCS running on the included training and testing datasets, leave this
configuration file as is. This ‘minimum’ configuration file relies on all avail-
able built-in default ExSTraCS run parameters. Next, from the command line,
navigate to the folder where ExSTraCS has been saved and type the following:
python ./ExSTraCS Main.py ExSTraCS Configuration File Minimum.txt
This is the standard command for running ExSTraCS. Notice that the only
argument required by ExSTraCS is the file path/name for a properly formatted
configuration file.
In order to run ExSTraCS on different datasets using the same default
parameters, users should edit this configuration file to specify respective file
paths/names for the desired training and testing datasets (i.e. edit trainFile
and testFile. Keep in mind that both training and testing datasets should
be tab-delimited .txt format, and columns including attributes, instance identi-
fiers, and class, should be in the same order for both the training and the testing
dataset (if a testing dataset is provided). Additionally ExSTraCS identifies the
class phenotype column using the default specified label ‘Class’, and an optional
column for instance identifiers using the default label ‘InstanceID’. These labels
may be edited in the configuration file to match the labels in the users dataset.
The location of ‘Class’ and ‘instanceID’ columns relative to attribute columns is
not important as ExSTraCS will automatically detect their location using these
unique labels. Note that the only run parameter in ExSTraCS without some
kind of default value is trainFile.
7

The user can also specify a unique file path/name for outFileName, which
gives the location and an optional prefix for the file-name for all standard ExS-
TraCS output text files. Note that ExSTraCS will overwrite any output files with
the same name. By default, (or when the user specifies ’None’ for outFileName),
ExSTraCS will use the name of the training dataset alone as the root of the out-
put file names, and save these files locally in the working directory if no path is
provided in outFileName. Also, whenever expert knowledge is active (as it is by
default), the user can specify a unique file path/name for outEKFileName, which
gives the location and optional prefix for the file-name for the expert knowledge
weights generated by the selected attribute weight algorithm. Similar to with
outFileName, by default,(or when the user specifies ’None’ for outEKFileName),
ExSTraCS will use the name of the training dataset alone as the root of the ex-
pert knowledge output file, and save this file locally in the working directory if
no path is provided in outEKFileName. Since the expert knowledge generation
strategies are deterministic, they need only ever be run once on a given train-
ing dataset, and the weights output to outEKFileName can simply be loaded
in future runs/analyses of the given training dataset. If a outEKFileName al-
ready exists it will not be overwritten, and ExSTraCS will instead automatically
attempt to load the weights from the existing text file.
2.2 Configuration File and Run Parameters
In this section we take a look at two other example configuration files included
with the code and get into all of the editable run parameters available in ExS-
TraCS. First, we refer the reader to...
ExSTraCS Configuration File Complete.txt
This is an example configuration file including all available run paramters
in ExSTraCS. Some may find it convenient to use/edit this configuration file
format so that they have greater control over algorithm parameters, and/or
to have a record of parameters used in an analysis via a copy of a complete
configuration file. Second, we refer the reader to...
ExSTraCS Configuration File Recommended.txt
This is a configuration file example that includes parameters that (1) may
dramatically impact performance given different dataset characteristics (2) are
convenient for data formatting, or (3) give users access to useful optional fea-
tures. We expect that most users would find it convenient to use/edit this
configuration file format, as it leaves out parameters that we have found to
have limited positive impact to shift from the default values included within the
ExSTraCS algorithm.
Now we examine each run parameter in as much detail as possible. The
order in which parameters are discussed is the same order as they are listed
in the ‘complete’ configuration file example. We suggest that readers review
8
parameters as needed. Most default parameter values are somewhat arbitrarily
based on what has been typically used in other LCS algorithms. Default run
parameters do not reflect an optimal set of run parameters for any given dataset.
However, we expect ExSTraCS to function well on most small or modestly sized
datasets using these default parameters.
2.2.1 Dataset Parameters
trainFile
This is the only parameter that must be specified in the configuration file. This
parameter specifies the file path/name for the training dataset. If a path is
not given, it is assumed that the training data is located in the same folder as
the ExSTraCS algorithm (as is the case for the example training dataset). The
training data constitutes the ‘environment’ within which ExSTraCS is seeking
to learn. The training data file should be a tab-delimited .txt file, where the first
row includes column headers (i.e. identifiers for attributes, the class variable,
and optionally instance identifiers). Missing values in the dataset should have
a standard designation (we suggest NA by default).
testFile
This parameter specifies the file path/name for the testing dataset. If a path is
not given, it is assumed that the testing data is located in the same folder as the
ExSTraCS algorithm (as is the case for the example testing dataset). Columns
in the testing data should have all attributes, InstanceID (optional), and Class in
the same order as found in the training dataset. The testing dataset is optional.
If no testing dataset is available (or desired) give the value None instead of
a file path/name. This is the default value for this parameter, therefore if
not specified, ExSTraCS will assume that not testing dataset is available. If a
testing dataset is specified it will be used during complete classifier population
evaluations to determine testing accuracy, a particularly important performance
metric in noisy data where overfitting is a critical concern. Testing accuracy
assesses ExSTraCS’s ability to make predictions on data instances that it has not
yet seen, or in other words, it’s ability to learn patterns that are generalizable
to other instances or situations. Note that ExSTraCS learning is put on hold
whenever evaluations are being completed such that the algorithm does not get
any chance to unfairly learn from testing instances.
outFileName
This parameter specifies the file path/prefix-name for for most standard ExS-
TraCS output files, including (1) the rule population, (2) the population statis-
tics, (3) the co-occurrence values, (4) attribute tracking scores, and (5) learning
tracking. ExSTraCS outputs up to four unique text files every time it completes
a classifier population evaluation, and outputs learning tracking once for the en-
tire learning process. By default, this parameter is set to None. When set to
9
None, ExSTraCS will use the name of the training dataset alone as the root of
the output file names, and save these files locally in the working directory if no
path is provided in outFileName. While the training dataset name will always
be included into the output file names, this parameter allows users to specify
not only a path, but a prefix to the filename. For instance if the training data
was called ‘MyTrain.txt’, and the user set this parameter to C:
Results
Study1 one output file would be C:
Results
Study1 MyTrain ExSTraCS 5000 PopStats.txt. Do not include a file exten-
sion (such as .txt) in this parameter, as this is done automatically. The fol-
lowing text will automatically be added to this parameter for each respective
output file (where [Iteration] indicates the current iteration integer value): (1)
‘ ExSTraCS [Iteration] PopStats.txt’, (2) ‘ ExSTraCS [Iteration] RulePop.txt’,
(3) ‘ ExSTraCS [Iteration] CO.txt’, (4) ‘ ExSTraCS [Iteration] AttTrack.txt’,
(5) ‘ ExSTraCS [Iteration] Predictions.txt’, and (6) ‘ ExSTraCS LearnTrack.txt’.
We recommend that users use this parameter to only specify the desired path
for where output files will be saved, or leave it at None if output files are to be
saved in the working directory. When specifying a file path, end with ‘
’.
offlineData
This parameter specifies whether ExSTraCS will acquire data online or offline.
While ExSTraCS learns iteratively (i.e. one training instance at a time), in-
stances can be made available to ExSTraCS as a complete dataset (i.e. ‘offline’)
or only when they become available, one instance at a time (i.e. ‘online’) By
default, ExSTraCS is specifically designed for ‘offline’ iterative learning, where
a dataset is initially loaded by ExSTraCS in its entirety. However we have in-
cluded the option to learn on data instances that are obtained one instance at a
time each iteration. This option has not yet been fully tested, and is not meant
to be the focus of ExSTraCS. We included this option for development purposes,
in an effort to make ExSTraCS as flexible as possible to the needs of different
users. Currently the only ‘online’ learning data available is accessed using the
included module (Problem Multiplexer.py) that provides a method to randomly
generate and return a single multiplexer training instance for ‘online’ data ac-
quisition. Of further note, when offlineData is set to 0 (i.e. ’onlineData’) the
following mechanisms should not function (and must be deactivatetd); expert
knowledge covering, attribute tracking, attribute feedback, and rule compaction.
Additionally complete classifier population evaluations will not function prop-
erly, since they rely on finite, loaded training and testing datasets. By default,
a value of 1 should be used for this parameter whenever the user is training on
a finite data file. We expect this will be the case for almost all users.
10
internalCrossValidation
This parameter allows the user to perform cross validation analysis internally,
in a serial set of ExSTraCS algorithm runs. A value of 0 or 1 indicates that
no internal cross validation is to be completed, while any other positive integer
value, indicates that internal cross validation is to be completed, and gives the
number of divisions of the data to be included in the cross validation. Specifi-
cally a value of 10 will break the original dataset into 10 random (class balanced)
groups. 10 training and 10 testing datasets will be generated and saved as text
files where in each 9 out of 10 groups make up the training data, and the re-
maining 1/10 makes up the testing dataset, where the 1/10 constituting the
testing data is a different 1/10 for each cross validation set. Activating internal
cross validation will automatically result in these datasets being generated, and
ExSTraCS serially performing a complete run and evaluation on each training
and testing dataset pair. The user can still perform cross validation by generat-
ing their own training and testing datasets outside of ExSTraCS and specifying
these file locations as previously described above. By default we suggest setting
internalCrossValidation to 0, deactivating this option.
randomSeed
This parameter allows the user to set a constant random seed, such that if the
same random seed is used, ExSTraCS will yield the same results in a subsequent
run on the same dataset with the same run parameters. To specify a random
seed value, simply provide any integer value for this parameter. To use a psuedo-
random number that won’t replicate run to run, set this parameter ‘False’ By
default, this parameter is set to ‘False’. However, this parameter should not
meaningfully impact the performance of ExSTraCS, other than any random
chance advantage or disadvantage.
labelInstanceID
This parameter specifies the column header which ExSTraCS uses to identify
the column in dataset files that includes unique identifiers for each attribute
in the respective dataset. This feature serves two purposes. Most simply, if
the dataset includes a column for instance identifiers, the user does not have
to delete or edit this column for formatting reasons. Instead, simply edit this
parameter to match the header label for instance identifiers in the dataset, and
ExSTraCS will know not to treat this column as an attribute upon which to
try and learn. Additionally, the attribute tracking mechanism stores weights
uniquely for every instance in the training dataset. When attribute tracking
scores are output, the instance labels included in the dataset are used to pair
attribute tracking scores to a given instances. If instance identifiers are not
provided in the original dataset, they are generated based on the order of in-
stances in the original dataset, and attribute tracking scores are paired with
these internally generated identifiers in the attribute tracking output file. This
way, users have some way of figuring out which attribute tracking score goes
11
with what instance in the data. This is particularly important, because the
dataset is randomly, internally shuffled at the end of loading/formatting, to try
and remove any potential order bias from the training dataset.
labelPhenotype
This parameter specifies the column header which ExSTraCS uses to identify
the column in the dataset files that includes phenotype values for all instances.
Phenotype is our preferred terminology for ‘class’, ‘endpoint’, or ‘action’ (as it
is often referred to in classic LCS algorithms applied to reinforcement learning
problems). In other words, phenotype refers to the dependent variable. The
phenotype column may be the first, last or any middle column. The value
of this parameter must match the label for phenotype column included in the
dataset. We suggest using ‘Class’ by default.
discreteAttributeLimit
This parameter specifies the number of unique attribute states that may ex-
ist for a given attribute before it is considered to be a continuous attribute
instead. When dealing with datasets that include both discrete and contin-
uous attributes, the user should take some care to ensure that this limit is
set correctly. This parameter has been included as part of the adaptive data
management scheme to automatically detect and discriminate discrete from con-
tinuous attributes. By default, this value is set to 10, which means that if any
attribute has more than 10 possible states, ExSTraCS will treat it as a continu-
ous variable instead. The included example training dataset has 20 attributes,
each with only 3 possible states (0,1, or 2). Based on the default setting of
this parameter, ExSTraCS will treat all 20 attributes as discrete. To ensure
that attributes intended to be considered to be discrete, are treated as such,
we recommend that the user identify (or estimate) the discrete variable in their
dataset with the largest number of states, and set this parameter to that value.
labelMissingData
This parameter specifies the unique designation for a missing data point in a
given training or testing dataset. This allows the user to conveniently change
this parameter instead of changing the identifier for all missing data points
in the dataset. Additionally, this allows ExSTraCS to run on data (without
imputation bias) despite missing data points within instances. Note, the value
specified for this parameter must universally identify missing data points in the
loaded training or testing datsets. We suggest using ‘NA’ by default.
12

2.2.2 General Run Parameters
trackingFrequency
This parameter specifies the frequency with which ExSTraCS performs minor
learning performance tracking estimates (and subsequently outputs these statis-
tics to an output file ending in ‘ ExSTraCS LearnTrack.txt’. This value is set
to zero by default which means that tracking will occur every epoch (i.e. ev-
ery complete cycle through the training dataset). Alternatively, ExSTraCS will
output tracking performance every niterations, where nis the integer value
specified for this parameter. Keep in mind that tracking performance estimates
are based on the last nlearning iterations. More frequent performance evalu-
ations will be less accurate, more variable, and increase overall algorithm run
time. We suggest leaving this parameter at the default, unless the dataset has a
particularly large number of training instances (e.g. greater than 5000), or the
user simply wants more frequent performance estimates.
learningIterations
This parameter is used to specify two things. (1) The last integer value in the
string specifies the maximum number of learning iterations that should be per-
formed using ExSTraCS, and (2) each integer specifies an iteration ‘checkpoint’
at which a complete evaluation of the ExSTraCS classifier population across the
entire training (and testing) datasets is completed and output files are generated.
The user can specify any number of learning checkpoints, however the iteration
numbers should be increasing up to some maximum number of iterations, and
individual values should be separated by a period. Alternatively, the user can
specify a single integer value which indicates that ExSTraCS should run with-
out interim evaluations and file outputs, until that specified iteration is reached.
For larger datasets, or more complex problems, the user may wish to increase
the number of learning iterations. To help estimate the maximum number of
iterations ExSTraCS should be run, consider that XCS [8] can solve the 20-bit
Multiplexer problem in about 50,000 iterations. Some additional examples of
how to properly specify this run parameter include; ’1000.5500’, ’200000’, and
’5000.50000.500000’. Note, do not use commas when specifying learning itera-
tions. The default setting for this parameter is ‘5000.10000.50000.100000’. This
is a largely arbitrary default, and we encourage users to adjust this parameter
to suit their specific needs.
outputSummary
This parameter directs ExSTraCS to output a population statistics summary
file each time a classifier population evaluation is completed (i.e. at each iter-
ation specified in learningIterations). This file includes a number of sum-
mary statistics including training accuracy, testing accuracy, training coverage,
testing coverage, macro-population size, micro-population size, global classifier
generality, three variations of attribute specificity sums (which can be used to
13
identify which attributes ExSTraCS relies on the most to make accurate predic-
tions), and a break down of ExSTraCS run times. To output this file, set this
parameter to 1. To keep it from being generated, set this parameter to 0.
outputPopulation
This parameter directs ExSTraCS to output a text file that specifies the cur-
rent classifier population in it’s entirety. This file allows the user to manually
inspect or visualize the classifier population output by ExSTraCS for knowledge
extraction purposes. This file includes all information required reconstitute the
classifier population from the learning iteration it was output (e.g. condition,
phenotype, fitness, accuracy, numerosity, initial time stamp, etc.). In other
words, ExSTraCS can be ‘reboot’ to continue learning from where a previous
ExSTraCS run left off, by loading this output file (Note that the PopStats.txt file
is also required for a population reboot). To output this file, set this parameter
to 1. To keep it from being generated, set this parameter to 0.
outputAttCoOccur
This parameter directs ExSTraCS to output a text file that ranks top pairs
of attributes that are co-specified in classifiers across the classifier population.
If the loaded training dataset include <= 50 attributes, all attribute pair co-
occurrence scores will be output to this file. If there are more than 50 attributes,
the only top specified 50 attribute will be used to determine the top co-specified
attribute pairs. This is due to the fact that that as the number of attributes
in the dataset increases, the number of attribute pair combinations goes up
exponentially. This co-occurrence metric can be used to better characterize the
relationships between attributes in classifiers across the population. This can be
used, for instance, to help differentiate epistatic interactions from heterogeneous
relationships. To output this file, set this parameter to 1. To keep it from being
generated, set this parameter to 0.
outputTestPredictions
This parameter directs ExSTraCS to output a text file that gives the algorithm’s
predictions for all instances in the loaded testing dataset. For each instance, this
includes the class prediction made, and the respective votes calculated in the
prediction array for each possible class endpoint. These votes may be used as a
fuzzy prediction, Bayesian probabilities, or estimates of prediction confidence.
To output this file, set this parameter to 1. To keep it from being generated,
set this parameter to 0.
14
2.2.3 Supervised Learning Parameters
N
This parameter specifies the maximum population size (i.e. the micro-population
size) that ExSTraCS is allowed to reach before the deletion mechanism turns
on and maintains this maximum number. For reference, the macro-population
size refers to the number of ‘unique’ classifiers in the classifier population, while
micro-population size (N) takes into account the numerosity of unique classifiers.
Numerosity refers to the number of ‘copies’ of a given classifier that currently
exist in the classifier population. Numerosity provides robustness to classifiers,
making it unlikely for for good classifiers to be completely eliminated by chance.
Numerosity also improves performance by not requiring ExSTraCS to maintain
multiple separate copies of the same classifier. The value of the maximum popu-
lation size parameter can have a dramatic influence on ExSTraCS performance.
If N is too small, an LCS algorithm can’t properly explore a given search space,
or maintain ‘good’ classifiers. If N is too large, an LCS algorithm will take
longer to run, and the resulting classifier population will likely be much bigger
than necessary. Properly setting this parameter my require some initial trial
and error or a more formal parameter sweep. By default, ExSTraCS uses a
maximum population size of 1000, which (in our analyses) has yielded reliable
performance on datasets with up to at least 20 attributes. We commonly also
set this parameter to 2000 for more complex, noisy problems, and on datasets
with as many as 2000 attributes. For particularly small datasets, a maximum
population size as low as 500, or even smaller may still function well.
nu
This parameter (ν) specifies the power to which accuracy is raised in calculating
classifier fitness. Tim Kovaks explores this parameter in [10]. In our work,
where we focus on noisy problems/data, it is impossible to achieve a testing
acccuracy of 100%. This these noisy problem domains we have observed that
keeping νlow yields better performance. Essentially, this places less pressure on
the algorithm to evolve classifiers that have a perfect accuracy, which in noisy
problem domains would only occur when a classifier is overfitting. We suggest
a default value of 1, since we assume that we are dealing with noisy problem
domains. For ‘clean’ problem domains such as the multiplexer toy problems, it
might be advantageous to return this parameter value back to the value of 10
used in the original UCS algorithm [2], or the value of 5 used in XCS [4].
chi
This parameter (χ) specifies the probability that the crossover operator will
be applied when the genetic algorithm is activated to discover new offspring
classifiers. This value is typically set within the range of 0.5 to 1.0. We suggest
a default value of 0.8 which has been traditionally used by the XCS and UCS
algorithms [4, 2].
15

upsilon
This parameter (υ) specifies the probability that the mutation operator will
specify or generalize a given attribute state within an offspring classifier any
time the genetic algorithm is activated. This parameter value is typically set
within the range of 0.01 to 0.05. We suggest a default value of 0.04 which has
been traditionally used by the XCS and UCS algorithms [4, 2].
theta GA
This parameter (θGA) is a threshold applied to activating the genetic algorithm.
Specifically, the genetic algorithm is activated when the average number of it-
erations since the genetic algorithm was last applied to classifiers in the correct
set is greater than this threshold. In other words, if the correct set is filled with
very young classifiers, the genetic algorithm is prevented from being applied.
We suggest a default value of 25 which has been traditionally used by the XCS
and UCS algorithms [4, 2].
theta del
This parameter (θdel) is a threshold applied to the deletion mechanism. Specif-
ically, an alternative, more protective deletion vote is calculated for a given
classifier if it’s experience (i.e. the number of instances it has matched so far)
is less than this threshold. We suggest a default value of 20 which has been
traditionally used by the XCS and UCS algorithms [4, 2].
theta sub
This parameter (θsub) is a threshold applied to the subsumption mechanism.
Specifically, a given classifier can only be a potential subsumer if its experience
(i.e. the number of instances it has matched so far) is greater than this threshold.
We suggest a default value of 20 which has been traditionally used by the XCS
and UCS algorithms [4, 2].
acc sub
This parameter is an accuracy threshold applied to the subsumption mechanism.
Specifically, a given classifier can only be a potential subsumer if its accuracy is
greater than this threshold. We suggest a default value of 0.99 which was used
by the UCS algorithm [2]. We suspect that in noisy problem domains it may
be advantageous to lower this threshold, since the expectation that a subsumer
have an accuracy >0.99 is likely poor in noisy problem domains.
beta
This parameter (β) is a learning parameter used to update the average correct
set size value maintained for each classifier. We suggest a default value of 0.2
which has been traditionally used by the XCS and UCS algorithms [4, 2].
16

delta
This parameter (δ) is a deletion parameter applied to the deletion vote cal-
culation for each classifier. We suggest a default value of 0.1 which has been
traditionally used by the XCS and UCS algorithms [4, 2].
init fit
This parameter specifies the initial fitness given to a new classifier. We suggest
a default value of 0.01 which is a fairly typical value for an LCS algorithm. We
expect that any reasonably low-valued setting for this parameter would work.
fitnessReduction
This parameter specifies an initial fitness reduction applied to offspring classi-
fiers generated using the crossover operator in the genetic algorithm. We suggest
a default value of 0.1 which has been traditionally used by the XCS and UCS
algorithms [4, 2].
theta sel
This parameter specifies the fraction of the correct set that will be randomly
included in tournament selection, whenever tournament selection is used to
select parents for the genetic algorithm. We suggest a default value of 0.5 which
is similar to the value of 0.4 utilized in [3].
RSL Override
This parameter is a completely optional override to the rule specificity limit
(RSL). When this parameter is set to 0 (default), ExSTraCS automatically
calculates a reasonable RSL based on the number of instances, and the average
number of attribute states in the training dataset. When set to an integer
greater than 0, but less than or equal to the number of attributes in the dataset,
ExSTraCS will apply the specified integer as the RSL. Setting this value equal to
the number of attributes in the dataset, will allow rules to specify an attribute
state for all attributes in the dataset. Generally, speaking, as the number of
attributes in the dataset becomes large, the use of a large RSL may prevent
ExSTraCS from learning useful generalizations.
2.2.4 Mechanism Parameters
doSubsumption
This parameter specifies whether subsumption (both action/correct set sub-
sumption and genetic algorithm subsumption) is activated or not. A value of 1
activates subsumption and a value of 0 turns it off. Subsumption was introduced
by Wilson [22] as a strategy to promote effective generalization in the classifier
17
population, and get avoid the specification of attributes that confer not accu-
racy advantage. One classifier subsumes (i.e. absorbs) another if the subsuming
classifier is more general and has equal or greater accuracy than the classifier
to be subsumed. The subsuming classifier has it’s numerosity increased by the
current numerosity of the subsumed classifier, while the subsumed classifier is
completely deleted.
selectionMethod
This parameter specifies the selection method that will be used by the ExSTraCS
genetic algorithm to pick parent classifiers. Two methods are currently available
in ExSTraCS including tournament and roulette selection (give a value . We
suggest tournament selection by default based on the results described in [3].
Tournament selection chooses a specified fraction of the classifiers in the correct
set randomly, and deterministically picks the most fit classifier to be a parent.
Roulette wheel selection probabilistically picks a parent classifier based on the
fitness values of all classifiers in the correct set (where a larger fitness equals a
larger selection probability).
doAttributeTracking
This parameter specifies whether the attribute tracking (AT) mechanism is acti-
vated or not. A value of 1 activates AT and a value of 0 turns it off. AT is akin
to long-term memory for supervised, iterative learning. For a finite training
dataset, a vector of accuracy scores is maintained for each instance in the data.
In other words, for every instance in the data we increase attribute weights based
on which attributes are being specified in classifiers found in [C] every iteration.
Post-training, these scores can be applied to characterize patterns of association
in the dataset, in particular heterogeneous patterns which might suggest clinical
patient subgroups that may be targeted for research, treatment, or preventative
measures [18]. Note that using AT alone does not impact learning performance.
Lastly, AT is only useful under the assumption that ExSTraCS will be repeat-
edly exposed to instances in the dataset, so that AT scores become more reliable
through experience. In other words, if the training dataset has a large number
of instances, AT scores may be based on few weight updates, depending on how
many overall learning iterations were completed. An epoch is a complete cycle
through all instances in the training dataset. The more epochs completed by
ExSTraCS, the more useful/reliable we would expect AT scores to be.
doAttributeFeedback
This parameter specifies whether the attribute feedback (AF) mechanism is ac-
tivated or not. A value of 1 activates AF and a value of 0 turns it off. AF
can only be applied if AT is also activated. AF is applied to the GA muta-
tion and crossover operators, probabilistically directing classifier generalization
based on the AT scores from a randomly selected instance in the dataset. The
18

probability that AF will be used in the GA is proportional to the algorithm’s
progress through the specified number of learning iterations (i.e. AF is applied
infrequently early-on, but frequently towards the end). As with AT, we would
expect AF to function better when a greater number of epochs are completed
by ExSTraCS. It is possible that AF may not improve performance in datasets
with a large number of training instances (i.e. where the number of training in-
stances is of similar or greater magnitude than the number of training iterations
to be completed. Note that in developing ExSTraCS we realized that AF-UCS
was not using the AT scores from the current training instance (as mistakenly
described in [18]), but rather the scores from a neighboring instance. This ‘error’
turned out to be essential to recapitulate attribute feedback performance. While
AF using the current training instance is better than no AF at all, AF using a
the AT scores from a random training instance worked even better. AF speeds
up effective learning by gradually guiding the algorithm to more intelligently
explore reliable attribute patterns.
useExpertKnowledge
This parameter specifies whether the expert knowledge (EK) covering mecha-
nism is activated or not. A value of 1 activates EK covering and a value of 0 turns
it off. EK is essentially an external bias introduced to better guide learning, such
that attributes more likely to be important tend to be specified more often when
covering. In other words, classifiers tend to be initialized in parts of the problem
space deemed by the EK to be most useful for predicting class status. Notably,
the utility of EK is only as good as the quality of the information behind the
weights. EK covering is implemented in ExSTraCS as described in [20] including
the calculation of EK probability weights from raw EK scores, and the applica-
tion of these weights within the covering mechanism. In theory, the source of EK
is up to the user (i.e. classifier population initialization can be biased towards
whatever attributes desired). If this parameter is activated there are a number
of associated run parameters described below, that the user may need to specify
accordingly. These include; external EK Generation, outEKFileName, filterAl-
gorithm, turfPercent, reliefNeighbors, reliefSampleFraction, and onlyEKScores.
The most important to consider include; external EK Generation, outEKFile-
Name, filterAlgorithm, and onlyEKScores.
external EK Generation
This parameter specifies whether EK will be generated by one of the four at-
tribute weight/filter algorithms built into ExSTraCS including; ReliefF, SURF,
SURF*, and MultiSURF, or whether it will be loaded from some external source
file. By default ExSTraCS sets this parameter to None, which indicates that
EK will be generated internally (Note: Thus the user may need to consider
the following parameter values; outEKFileName, filterAlgorithm, turfPercent,
reliefNeighbors, reliefSampleFraction, and onlyEKScores). Alternatively this
parameter can be set to the full path/name including the ‘.txt’ file extension
19

pointing to a properly formatted external EK weight file. This tells ExSTraCS
that this file and the included EK weights should be loaded and utilized by the
algorithm. This file should have the same formatting as the EK files output by
ExSTraCS when internally generated. Specifically, there must be three initial
rows in the text file that can be filled with any information, or left blank. The
fourth row should begin the EK scores themselves. Every row from the fourth
down to the last should include the following; (1) the first column should give
an attribute ID as it appears in the training data, and (2) the second column
should include some real-valued (positive or negative) numerical weight for that
respective attribute. Additional columns are optional, and will not be read by
ExSTraCS. For an example of this format, please observe an internally gen-
erated EK file output by ExSTraCS. This parameter only impacts ExSTraCS
if useExpertKnowledge is set to 1. If EK is being internally generated, and
higher dimensional datasets are being considered (with many attributes and/or
many training instances), the python implementation of these attribute weight-
ing algorithms may take a while to run. They have not yet been optimized for
very large datasets. If you notice that EK generation is taking an unreasonable
amount of time for a larger dataset, you might with to refrain from using ExS-
TraCS to generate EK. A faster version of these algorithms that only works on
datasets with discrete, case/control (i.e. two class) datasets is currently available
build into the Multifactor Dimmensionality Reduction (MDR) software [12]. We
plan to make faster versions of these EK generation algorithms available that
can handle discrete and continuous, attributes and endpoints. These currently
implemented versions can indeed handle discrete and continuous, attributes and
endpoints, they have just not yet been optimized for run efficiency.
outEKFileName
This parameter specifies the path/prefix-file root name for the EK score file that
can be internally generated by ExSTraCS. This preserves EK scores for future
reference or use. Similar to with outFileName, by default,(or when the user
specifies ’None’ for outEKFileName), ExSTraCS will use the name of the train-
ing dataset alone as the root of the expert knowledge output file, and save this
file locally in the working directory if no path is provided in outEKFileName.
Also, for this parameter, do not include a file extension such as ‘.txt’. The fol-
lowing text will automatically be added to the EK output filename based on the
weight algorithm [alg] selected for EK generation; ‘ [alg] scores.txt’. [alg] can
be ‘relieff’, ‘surf’, ‘surfstar’, ‘multisurf’, ‘relieff turf’,‘surf turf’,‘surfstar turf’, or
‘multisurf turf’. The value of outEKFileName only matters if useExpertKnowledge
is set to 1 and external EK Generation is set to None. If an expert knowledge
output file of the same name already exists it will not be overwritten, and ExS-
TraCS will instead automatically attempt to load the weights from the existing
text file. We recommend that users use this parameter to only specify the de-
sired path for where output EK files will be saved, or leave it at None if output
EK files are to be saved in the working directory. When specifying a file path,
end with ‘
20

’.
filterAlgorithm
This parameter specifies the weight/filter algorithm that will be used to gen-
erate EK internally within ExSTraCS. The first four valid values for this pa-
rameter include; ‘relieff’, ‘surf’, ‘surfstar’, or ‘multisurf’. These four rapid at-
tribute weighting algorithms were designed to estimate attribute quality, in
terms of predicting class status. For the specifics of a given weighting algo-
rithm please refer to the corresponding reference; ReliefF [9], SURF [7], SURF*
[6], and MultiSURF [5]. We recommend that MultiSURF be used by default,
as it is the newest, and best performing of the four available options, how-
ever MultiSURF can take slightly more time to run compared to the other
three options. New to ExSTraCS 2.0 is the ability to run any of these at-
tribute weighting algorithms in concert with the TuRF wrapper algorithm.
TuRF will run the specified attribute weight algorithm repeatedly, each time
removing a percentage of the lowest scoring attributes. To run an attribute
weight algorithm along with TuRF use one of the following valid values for
this parameter; ‘relieff turf’,‘surf turf’,‘surfstar turf’, or ‘multisurf turf’. The
value of filterAlgorithm only matters if useExpertKnowledge is set to 1 and
external EK Generation is set to None.
turfPercent
This parameter specifies two things. First, it specifies the percent of attributes
that are eliminated from consideration each iteration of the TuRF wrapper
algorithm [11]. Second, this value is inversely proportional to the number of
TuRF iterations that are completed. This way, despite the value chosen for
this parameter, roughly the same number of attributes will remain in the final
attribute weight calculation iteration. While [11] suggests a value of 0.05 for
this parameter, we have adopted a default of 0.2, as requires less time, and
performed well in our preliminary testing.
reliefNeighbors
This parameter is only applicable when using the ReliefF algorithm to gener-
ate EK weights. Specifically this parameter gives the number of ‘neighbors’
used in the calculation of ReliefF scores. Please see [9] for more details on
ReliefF, and the neighbors parameter. We suggest using 10 by default. The
value of reliefNeighbors only matters if useExpertKnowledge is set to 1,
external EK Generation is set to None, and filterAlgorithm is ‘relieff’ or
‘relieff turf’.
reliefSampleFraction
This parameter is only applicable when using either the ReliefF, SURF, or
SURF* algorithm to generate EK weights (i.e. not applicable to MultiSURF).
21

Specifically this parameter gives the number of iterations to be completed by
the respective EK weight algorithm, given as a percent of the training dataset
instances (i.e. between 0 and 1). We suggest using 1 by default, which means
that the respective weight algorithm will iterate over all training instances in the
dataset. The value of reliefNeighbors only matters if useExpertKnowledge,
is set to 1, external EK Generation is set to None, and filterAlgorithm is
either ‘relieff’, ‘surf’, ‘surfstar’, ‘relieff turf’, ‘surf turf’, or ‘surfstar turf’. Please
see ReliefF [9], SURF [7], or SURF* [6] respectively for further discussion.
onlyEKScores
This parameter gives the user the option to use the ExSTraCS platform to only
run one of the accompanied EK generating attribute weight/filter algorithms
including; ReliefF, SURF, SURF*, and MultiSURF. In other words, EK will
be generated, but the rest of the ExSTraCS algorithm will not run. This al-
lows EK generation to be run as a separate pre-processing step. A value of
1 directs ExSTraCS to only perform EK generation, while a value of 0 indi-
cates that ExSTraCS should both perform EK generation and run the core
ExSTraCS LCS algorithm as well. The value of onlyEKScores only matters if
useExpertKnowledge is set to 1.
doRuleCompaction
This parameter specifies whether rule compaction is activated or not. A value
of 1 activates rule compaction and a value of 0 turns it off. ExSTraCS makes
the six rule compaction strategies evaluated in [13] available to post-process the
classifier population. Rule compaction utilizes the whole training dataset to
consolidate the classifier population with the goal of improving interpretation
and knowledge discovery. Comparisons in [13] suggested that simple Quick Rule
Filtering (QRF) was both the fastest, and particularly was well suited to the
theme of global knowledge discovery [19] where it is more important to preserve
or improve performance than to minimize classifier population size(useful for
knowledge discovery by manual rule inspection) [13].
onlyRC
This parameter gives the user the option to use the ExSTraCS platform to only
run one of the accompanied rule compaction strategies on an existing saved
classifier population output file. Essentially this parameter gives the user the
option to try out different rule compaction strategies on any saved classifier
population file output by ExSTraCS. This allows for comparisons between rule
compaction strategies, and the flexibility for a user to return to a previous ExS-
TraCS run in which rule compaction had not been utilized, and quickly compact
this saved classifier population without having to run the entire ExSTraCS al-
gorithm again from scratch. A value of 1 activates this parameter while of value
of 0 indicates that ExSTraCS should be run normally. A value of 1 supersedes
22
most other run parameters, directing ExSTraCS to only run rule compaction
on an existing classifier population, and not running the ExSTraCS core LCS
algorithm at all. When onlyRC is set to 1, doPopulationReboot must also be
set to 1, and popRebootPath must be provided with a valid path/file name for
an existing saved ExSTraCS classifier population file.
ruleCompactionMethod
This parameter specifies the rule compaction or filter algorithm that will be ap-
plied by ExSTraCS after the last learning iteration completes, or when onlyRC
is activated. Valid values for this argument include; ‘QRF’, ‘PDRC’, ‘QRC’,
‘CRA2’, ‘Fu2’ and ‘Fu1’, each which represent an available rule compaction/filter
algorithm implemented in ExSTraCS. We suggest using QRF by default, as it is
extremely fast, simple, and basically just eliminates clearly poor classifiers from
the classifier population.
doPopulationReboot
This parameter gives the user the option to load a previously saved ExSTraCS
classifier population output, and continue to run ExSTraCS from where it left
off. Essentially this prevents the user from having to run the algorithm from
scratch if they wanted to see if additional learning iterations would improve
performance. This parameter must also be activated when onlyRC is set to
1, since rule compaction relies on an existing ExSTraCS classifier population.
A value of 1 activates the population ‘reboot’ (as we refer to it) and a value
of 0 turns it off. If this parameter is set to 1, the user must also specify an
appropriate value for popRebootPath. By default, this option is turned off (i.e.
0)
popRebootPath
This parameter specifies the iteration number for the population file from which
ExSTraCS will ‘reboot’ (i.e. load the saved classifier population, and continue
learning, or run a rule compaction strategy). ExSTraCS will automatically take
from outFileName and trainFile to construct the path/filename for previ-
ously saved output files (including the rule population file and the population
statistics file). Together, these files included everything ExSTraCS needs to
load a previous analyis and rule population and continue learning from where
it left off. No default is available for this parameter, thus it must be specified
if doPopulationReboot is set to 1. Since doPopulationReboot is set to 0 by
default, generally a user does not need to worry about specifying this parameter.
2.3 Overview of ExSTraCS Code
This section gives an overview of the ExSTraCS algorithm code itself, including
the overall organization and function of each file. The ExSTraCS algorithm
23
is open source and coded in Python 2.7. Each following subsection describes
a respective module file within ExSTraCS. We have left out the four EK at-
tribute weight algorithm files (including ReliefF.py, SURF.py, SURFStar.py,
and MultiSURF.py) as well as Problem Multiplexer.py, as they are not strictly
part of the ExSTraCS algorithm code, but rather can optionally be used by the
ExSTraCS algorithm. We have also left out TuRF Me.py below, which serves
as a wrapper algorithm to the aforementioned attribute weighting algorithm.
Problem Multiplexer.py has been included in the software download for users
who may wish to generate multiplexer datasets, or users that might be inter-
ested in trying out the ExSTraCS online learning feature (which has not yet
been fully tested).
Before reviewing individual modules in the code, we begin by pointing out
some of the most notable modules. First, ExSTraCS Main.py is the main run
file for running ExSTraCS from the command line. Second, ExSTraCS Test.py
is a convenient alternative to ExSTraCS Main.py, for running ExSTraCS locally
within a coding environment such as Eclipse with PyDev. Throughout devel-
opment, we used ExSTraCS Test.py to run and debug ExSTraCS in Eclipse
with PyDev. Third, ExSTraCS Algorithm.py initializes the classifier popula-
tion, runs the core learning cycle of the ExSTraCS algorithm, and completes
local and global performance evaluations. Fourth, ExSTraCS ClassifierSet.py
defines and manages all mechanisms operating at the level of classifier sets,
where a classifier set can be the whole population, a match set, or a correct
set. Lastly, ExSTraCS Classifier.py defines and manages individual classifiers
making up the classifier population.
ExSTraCS Main.py
This module is called to run ExSTraCS from the command line. Initialization
of the algorithm and key mechanisms takes place here.
ExSTraCS Test.py
This module is for developing and testing the ExSTraCS algorithm locally. This
module will run ExSTraCS directly within an editor (e.g. Eclipse with PyDev).
Initialization of the algorithm and key mechanisms takes place here.
ExSTraCS Algorithm.py
The major controlling module of ExSTraCS. Includes the major run loop which
controls learning over a specified number of iterations. Also includes periodic
tracking of estimated performance, and checkpoints where complete evaluations
of the ExSTraCS classifier population is performed.
ExSTraCS ClassifierSet.py
This module handles all classifier sets (population, match set, correct set) along
with mechanisms and heuristics that act on these sets.
24
ExSTraCS Classifier.py
This module defines an individual classifier within the classifier population,
along with all respective classifier parameters. Also included are classifier-level
methods, including constructors(covering, copy, reboot) matching, subsump-
tion, crossover, and mutation. Classifier parameter update methods are also
included. Please note that classifier parameters are something different than
‘run parameters’ discussed above in this users guide.
ExSTraCS Constants.py
Stores and makes available all alogrithmic run parameters, and acts as a gateway
for referencing the timer, environment, dataset properties, attribute tracking,
and expert knowledge scores/weights. This is also where the generation expert
knowledge and respective weights is controlled.
ExSTraCS ConfigParser.py
Manages the configuration file by loading, parsing, and passing it’s values to
ExSTraCS Constants. Also includes a method for generating datasets for cross
validation.
ExSTraCS DataManagement.py
Loads the dataset, characterizes and stores critical features of the datasets (in-
cluding discrete vs. continuous attributes and phenotype), determines the rules
specificity limit, handles missing data, and finally formats the data so that it
may be conveniently utilized by ExSTraCS. This is the ‘adaptive data manage-
ment’ component of ExSTraCS.
ExSTraCS Offline Environment.py
In the context of data mining and classification tasks, the ’environment’ for
ExSTraCS is a data set with a limited number of instances with some number
of attributes and a single endpoint (typically a discrete phenotype or class) of
interest. This module manages ExSTraCS’s stepping through learning itera-
tions, and data instances respectively. Special methods are included to jump
from learning to evaluation of a training dataset.
ExSTraCS Online Environment.py
ExSTraCS is best suited to offline iterative learning, however this module has
been implemented as an example of how ExSTraCS may be used to perform
online learning as well. Here, this module has been written to perform online
learning for a n-multiplexer problem, where training instances are generated in
an online fashion. This module has not been fully tested.
25
ExSTraCS Timer.py
This module’s role is largely for development and evaluation purposes. Specifi-
cally it tracks not only global run time for ExSTraCS, but tracks the time uti-
lized by different key mechanisms of the algorithm. This tracking likely wastes a
bit of run time, so for optimal performance check that all ’cons.timer.startXXXX’,
and ’cons.timer.stopXXXX’ commands are commented out within ExSTraCS Main,
ExSTraCS Test, ExSTraCS Algorithm, and ExSTraCS ClassifierSet.
ExSTraCS Prediction.py
Based on a given match set, this module uses a voting scheme to select the
phenotype prediction for ExSTraCS.
ExSTraCS AttributeTracking.py
Handles the storage, update, and application of the attribute tracking and feed-
back heuristics.
ExSTraCS ExpertKnowledge.py
A pre-processing step when activated in ExSTraCS. Converts numerical ex-
pert knowledge scores from any source into probabilities to guide the covering
mechanism in determining which attributes will be specified and which will be
generalized.
ExSTraCS RuleCompaction.py
Includes several rule compaction/rule filter strategies, which can be selected
as a post-processing stage following ExSTraCS classifier population learning.
Fu1, Fu2, and CRA2 were previously proposed/published strategies from other
authors. QRC, PDRC, and QRF were proposed and published by Jie Tan, Jason
Moore, and Ryan Urbanowicz.
ExSTraCS ClassAccuracy.py
Used for global evaluations of the LCS classifier population for problem domains
with a discrete phenotype. Allows for the calculation of balanced accuracy when
a discrete phenotype includes two or more possible classes.
ExSTraCS OutputFileManager.py
This module contains the methods for generating the different output files gen-
erated by ExSTraCS. These files are generated at each learning checkpoint, and
the last iteration. These include...
•writePopStats: Summary of the population statistics
26

•writePop: Outputs a snapshot of the entire classifier population including
classifier conditions, classes, and parameters.
•attCo Occurence: Calculates and outputs co-occurrence scores for each
attribute pair in the dataset.
•writePredictions: Outputs class predictions with corresponding class votes
for each instance in an included testing dataset.
2.4 Output Files
As discussed briefly in the previous section dealing with algorithm run param-
eters, ExSTraCS can output a number of files upon completion. In this section
we review all text files that may be generated as a result of running ExSTraCS,
and provide some direction for interpreting and utilizing these output files as
they were intended. We will begin with files that are generated each time the
classifier population is evaluated globally, and then we will discuss other files
that would be output at most once per run of ExSTraCS.
2.4.1 Rule Population
Likely the most important output file is the rule population file, a.k.a the clas-
sifier population file (the file extension is [Iteration] RulePop.txt). This file
details the classifier population as it exists after [Iteration] learning iterations.
This file can be used to ‘reboot’ ExSTraCS, i.e. to pick up learning from where
it was stopped previously. Additionally this file can be used for knowledge dis-
covery and extraction either by manual rule inspection (where the user can rank
classifiers by numerosity, and examine individual classifiers in search for useful
predictive classifiers), or employ global knowledge discovery strategies to look
for patterns based on what attributes are specified within the classifier popula-
tion as a whole (as described in [19]). Each column in the classifier population
file includes a respective classifier parameter stored for each classifier. Each
row gives the parameter values necessary to remake a respective classifier in
the ExSTraCS classifier population. Below we briefly review each classifier pa-
rameter maintained by ExSTraCS, and saved in the classifier population output
file. Note that these ‘classifier’ parameters, are different than ExSTraCS ‘run’
parameters.
Specified
This parameter gives a list of attribute position identifiers, that indicates which
attributes in the dataset have been specified in a given classifier. For example
the following entry [3,4,10] would indicate that this classifier specifies the fourth,
fifth, and eleventh attributes in the dataset since zero based numbering is used,
where the first attribute in the dataset would be at position zero. This parameter
is never changed for a given classifier.
27
Condition
This parameter gives a list of the state values for each attribute specified in a
given classifier. Using the previous example where the attributes specified for a
classifier included [3,4,10], a condition of [’0’,[0.43,0.78],’red’] indicates the the
fourth attribute must have a state equal to ’0’, the fifth attribute must have a
state value within the range of 0.43 and 0.78, and the eleventh classifier must
have a state equal to ’red’. Note that by storing discrete states as string values
and continuous states as a minimum to maximum value range, ExSTraCS can
easily accommodate continuous attributes, and discrete attributes with numer-
ical or discrete values. This parameter is never changed for a given classifier.
Phenotype
This parameter gives the phenotype (i.e. the endpoint or class) that a respective
classifier predicts given the associated attribute states specified. This parameter
is never changed for a given classifier.
Fitness
This parameter stores the fitness of a respective classifier. Currently, the fitness
of a classifier is equal to classifier accuracy to the power of the run parameter ν.
Since by default this parameter is set to 1, by default fitness is equal to accuracy.
The fitness of a classifier determines (1) it’s likelihood of being selected by the
genetic algorithm to be a parent classifier, (2) it’s likelihood of being selected for
deletion, and (3) it’s vote, when ExSTraCS seeks to apply it’s current classifier
population to make a class prediction. This parameter is updated for a given
classifier any learning iteration that it is included in a match set.
Accuracy
This parameter stores the accuracy of a respective classifier. If you are not
familiar with learning classifier system algorithms, it is important to realize
that the accuracy of a classifier has nothing to do with it’s global accuracy
across a dataset as a whole, rather the accuracy of a classifier is a more local
calculation. Specifically the accuracy of a classifier in ExSTraCS is equal to the
number of times a classifier has been in a correct set, divided by the number of
times it has been in a match set. This parameter is updated for a given classifier
any learning iteration that it is included in a match set.
Numerosity
This parameter stores the numerosity of a respective classifier. The numerosity
of a classifier represents the number of virtual copies of a given classifier are be-
ing maintained within the classifier population. Typically, a high numerosity is
an indicator that a classifier is particularly ‘good’. Traditionally, when seeking
28
to interpret a classifier population, classifiers are ranked by increasing numeros-
ity, and researchers begin by manually inspecting classifiers with the highest
numerosities. However this is not a completely reliable indicator, as numerosity
can sometimes be high by random chance, or simply because a classifier has a
reasonable fitness, but a low specificity (very few attributes specified). In this
situation, the classifier would likely be involved in match sets very frequently,
and thus have a greater opportunity to reproduce through the genetic algorithm,
and thus a greater chance that the same or similar classifiers may be added to
the population. Now we will review some important mechanisms that impact
the numerosity of a classifier. It is useful to note that when a classifier is se-
lected for deletion, it is only completely removed from the population if it has
a numerosity of only one (only one copy of itself). If the classifier’s numerosity
is larger, than deletion will decrement the numerosity of that classifier (i.e. if
it’s numerosity was 5, deletion will reduce it to 4). Numerosity plays a role in
many aspects of the ExSTraCS algorithm including, for example, the prediction
scheme, where classifiers get a vote proportional to their numerosity. When
calculating the micro population size of the classifier population, we sum the
numerosities of all classifiers in the population. Before adding a new classifier
to the population ExSTraCS checks to make sure that a classifier with the same
‘specified’, ‘condition’, and ‘phenotype’ doesn’t already exist. If it does, instead
of adding an entirely new copy of the classifier, the numerosity of the existing
classifier is increased by one. This parameter is updated for a given classifier
whenever it is deleted, whenever a copy of itself is added to the population,
or whenever it subsumes another classifier (in which this classifier’s numerosity
would increase by the numerosity of the classifier it subsumed.
AveMatchSetSize
This parameter stores the estimated average match set size that is maintained
for each classifier. In other words, this parameter is roughly tracking the average
number of classifiers in match sets in which a given classifier is also included.
This parameter is mainly utilized in the calculation of a classifier’s deletion
probability, where a larger AveMatchSetSize yields a larger deletion probability.
Essentially this applies a pressure on the classifier population as a whole to
maintain a diversity of classifiers that apply to different niches of the problem
space. In other words this pressure seeks to balance the number of classifiers
adopting available phenotype/class states, and the number of classifiers that are
specific to different parts of the solution space. This parameter is updated for
a given classifier any learning iteration that it is included in a match set.
TimeStampGA
This parameter stores the last iteration which a given classifier was last in a
correct set upon which the genetic algorithm was activated. This parameter
is in-turn used to determine when the genetic algorithm should be activated.
Generally speaking if the classifiers in a correct set are very young (i.e. the
29
average TimeStampGA of classifiers in the correct set is low, relative to the current
iteration), than the genetic algorithm will not be activated. This parameter is
updated for a given classifier any learning iteration that it is included in a correct
set in which the genetic algorithm was also activated.
InitTimeStamp
This parameter stores the iteration in which a given classifier was first created.
This parameter is never changed for a given classifier. However, note that if a
classifier gets completely deleted, and then the same classifier appears during
some later iteration, it will have a new InitTimeStamp value, since it will be
treated as an entirely new classifier. As currently implemented, this parameter
does not influence learning, but may be used to characterize the classifiers and
the greater classifier population. This parameter is used to determine when
a classifier has been around long enough to have had the opportunity to be
exposed to every instance in the training data (i.e. EpochComplete)
Specificity
This parameter stores the proportion of attributes in the training dataset that
have been specified within a given classifier. This parameter is never changed for
a given classifier. As currently implemented, this parameter does not influence
learning, but may be used to characterize the classifiers and the greater classifier
population.
DeletionProb
This parameter stores the current deletion weight for a given classifier. This pa-
rameter is updated for a given classifier whenever deletion is activated. Other
LCS algorithms calculated this value as needed and do not store it as a parame-
ter. ExSTraCS stores classifier deletion weights to better characterize classifiers
and the greater population after run completion.
CorrectCount
This parameter stores the number of times that a given classifier has been in a
correct set. This parameter along with MatchCount are used to calculate/update
classifier accuracy whenever needed. This parameter is updated any time a given
classifier is in a correct set.
MatchCount
This parameter stores the number of times that a given classifier has been
in a match set. This parameter along with CorrectCount are used to calcu-
late/update classifier accuracy whenever needed. This parameter is updated
any time a given classifier is in a match set.
30

CorrectCover
This parameter stores the number of instances in the training data, for which
this classifier made it into a correct set. This parameter value will be the same
as CorrectCount for a given classifier until an epoch is complete (i.e. until this
classifier has had the opportunity to be exposed to every instance in the training
dataset). At this point CorrectCover becomes a fixed value that can no longer
be updated. This parameter indicates the number of instances in the training
data that are correctly covered by a given classifier. This parameter is currently
used to characterize classifiers in the population and does not impact learning.
MatchCover
This parameter stores the number of instances in the training data, for which
this classifier made it into a match set. This parameter value will be the same
as MatchCount for a given classifier until an epoch is complete (i.e. until this
classifier has had the opportunity to be exposed to every instance in the training
dataset). At this point MatchCover becomes a fixed value that can no longer be
updated. This parameter indicates the number of instances in the training data
that are correctly or incorrectly covered by a given classifier. This parameter is
currently used to characterize classifiers in the population and does not impact
learning.
EpochComplete
This parameter has a boolean value (True/False) and indicates whether a given
classifier has been around long enough to have the opportunity to be exposed
to every instance in the training data. This occurs when the current iteration
number minus InitTimeStamp is larger than the number of training instances
in the dataset. Currently this parameter is used to characterize classifiers in
the population and does not impact learning. However, we expect that in order
to improve learning on datasets with a smaller number of training instances
available, it will be useful to keep track of which classifiers have already seen all
training instances.
2.4.2 Population Statistics
The second file that is output by ExSTraCS each time the classifier popula-
tion is evaluated globally, is the population statistics file (the file extension
is ‘[Iteration] PopStats.txt’). This file is intended to summarize global perfor-
mance of ExSTraCS over the entire training and testing datasets, as well as
characterize global classifier population statistics. Key performance statistics
include, training accuracy, testing accuracy, training coverage, and testing cov-
erage (where coverage refers to the proportion of instances in either the training
or testing datasets that are matched by at least one classifier in the classifier
population). This file also outputs the macro population size, the micro pop-
ulation size, and the average generality of classifiers in the population (where
31

numerosity is taken into account). Next, this file includes three summary statis-
tics introduced in [19] which can be used in knowledge discovery to identify at-
tributes that were of particular importance in making class predictions. These
statistics include the specificity sum, the accuracy sum, and the attribute track-
ing global sum. For each statistic a sum is calculated for every attribute in the
training data. The specificity sum, sums the number of times a respective at-
tribute was specified in classifiers across the population (numerosity taken into
account). The accuracy sum is calculated similarly, but the sum is weighted by
the accuracy of each respective classifier. Lastly, the attribute tracking global
sum is only calculated when attribute tracking has been activated (otherwise
all attribute sums will be zero in this output file). Here, instead of summing
when attributes were specified in the classifier population, attribute tracking
scores for all instances in the dataset are summed for each individual attribute.
Attributes that consistently have the highest sums for these three metrics are
likely to be most important for making accurate predictions.
Next, this file outputs both the global run time (the time in minutes that
ExSTraCS has been running up to the point where this evaluation was con-
ducted), and a breakdown of how much time was required by individual ExS-
TraCS components. The last information included in this output file is the
CorrectTrackerSave. This information has nothing to do with performance
statistics, or a characterization of the classifier population. Rather, this infor-
mation is exclusively used for ‘rebooting’ the classifier population. Specifically
these values capture the prediction successes and failures for the trackingFreq-
uency, which tracks estimated prediction performance of ExSTraCS during
learning. Including this information in this output file allows it to be loaded
back into ExSTraCS’s memory so that the Learning Tracking output file (see
below) can pick up where it left off uninterrupted when the classifier popula-
tion is reboot in ExSTraCS. As a result, this file is required along with the rule
population file in order to successfully reboot a classifier population.
2.4.3 Co-occurrence
The third file that is output by ExSTraCS each time the classifier population is
evaluated globally, is the Co-ocurrence file (the file extension is ‘[Iteration] CO.txt’).
This file ranks top pairs of attributes that are co-specified in classifiers across the
classifier population. If the loaded training dataset include <= 50 attributes,
all attribute pair co-occurrence scores will be output to this file. If there are
more than 50 attributes, the only top specified 50 attribute will be used to de-
termine the top co-specified attribute pairs. This is due to the fact that that as
the number of attributes in the dataset increases, the number of attribute pair
combinations goes up exponentially. This co-occurrence metric can be used to
better characterize the relationships between attributes in classifiers across the
population. This can be used, for instance, to help differentiate epistatic inter-
actions from heterogeneous relationships. Uniquely, this output file includes no
header labels for columns. The first two columns specify a pair of attributes.
The third column gives the co-specification sum for that particular attribute
32

pair (i.e. the number of times both attributes were specified in a classifier to-
gether where numerosity is as usual taken into account). The fourth column is
similar to the third, however the sum is weighted by the respective accuracy of
each classifier in which both attributes were specified. Rows are ranked from
largest co-specification sum to smallest (i.e. based on the values in the third
column).
2.4.4 Attribute Tracking Scores
The fourth file that is output by ExSTraCS each time the classifier population is
evaluated globally (and attribute tracking is activated), is the attribute tracking
score file (the file extension is ‘[Iteration] AttTrack.txt’). This output file should
have the same dimmensionality (i.e. the same number of rows and columns) as
the training data (assuming that the training data includes a column for instance
ID labels). In this output file, the first row gives headers. The first column gives
instance identifiers for all instances in the training dataset. Following columns
give attribute tracking scores for each attribute in the dataset. The last column
is the class/phenotype value for a respective instance in the data. As described
in [18] and [21], hierarchical clustering can be performed on instances, and
attributes within this file to identify groups of instances with similar patterns
of attributes with high attribute tracking scores in order to identify potentially
heterogeneous instance subgroups and better characterize relationships between
attributes predictive of class/phenotype state.
2.4.5 Predictions
The fifth file that is output by ExSTraCS each time the classifier population is
evaluated globally is the Prediction file (the file extension is ‘[Iteration] Predictions.txt’).
This output file will only be able to be generated if a testing dataset has made
available to the algorithm. The first column gives instance identifiers for all
instances in the training dataset. The second column gives the predicted class
made by ExSTraCS for the current instance. The third column gives the true
class value for the current instance. The remaining columns will give respective
votes for each possible class in the dataset. These were the votes that were used
to make the prediction for the given testing instance.
2.4.6 Learning Tracking
Different from the previously described output files, the Learning Tracking file
(the file extension is ‘LearnTrack.txt’) is only output once per run of ExS-
TraCS on a given dataset. The run parameter trackingFrequency, determines
how often a learning tracking estimated performance evaluation is completed
and output to this file. Each evaluation includes the following information:
(1) the current learning iteration, (2) the macro population size, (3) the mi-
cro population size, (4) an accuracy estimate (calculated using the previous
33

trackingFrequency learning iterations, (5) average classifier population gen-
erality, (6) the proportion of experienced classifiers (i.e. classifiers that have
completed at least one epoch), and (7) the amount of run time that has elapsed.
Additionally if an ExSTraCS classifier population is ‘reboot’ to run longer, than
the existing Learning Tracking file will be opened and further learning track-
ing statistics will be added to the original file, picking off from where learning
tracking left off. This file can be used to graph estimated features of learning
progress over time or learning iterations.
2.4.7 Expert Knowledge
The last file that can be output by ExSTraCS is a text file giving the expert
knowledge weights generated internally by ExSTraCS using one of the four in-
cluded attribute filtering/weighting algorithms. This file will only be output if
run parameter useExpertKnowledge is set to 1 and external EK Generation
is set to None. The file extension will be based on the weight algorithm [alg]
selected for EK generation; ‘ [alg] scores.txt’. ExSTraCS will format this ex-
pert knowledge weight file as a tab-delimited .txt file. Specifically, there will
be three initial rows in the text file that serve as a header and give the weight
algorithm run, and the time it took to generate the expert knowledge weights in
seconds. The fourth row will begin the EK scores themselves. Every row from
the fourth down to the last will include the following; (1) the first column will
give an attribute ID as it appears in the training data, (2) the second column
will include the numerical weight for that respective attribute, and (3) the third
column will include the weight rank of an attribute relative to the others. A
user may wish to generate and use these expert knowledge files for a purpose
completely separate from the running the core ExSTraCS algorithm. For this
reason, we included the run parameter onlyEKScores, so that this file could be
produced without running the ExSTraCS algorithm itself.
2.5 Making Predictions
One of the major goals for ExSTraCS is to evolve a prediction model (made up of
a population of classifiers) that can then be applied to the task of class prediction
in data that the algorithm has not yet seen. Currently the simplest way to
examine the predictive ability of the evolved ExSTraCS classifier population is
to include a testing dataset along with the training dataset. While the core
ExSTraCS algorithm is geared towards learning predictive patterns, ExSTraCS
also tests it’s predictive ability on the current training instance each iteration
(to provide a training accuracy estimate for the Learning Tracking output file.
Class predictions are handled by the ExSTraCS Prediction.py module, which
takes all classifiers in the current match set (i.e. all classifiers that match the
current data instance), and sums up a vote score for each possible class state.
The vote score for a given class state sums the fitness values for all classifiers
that specify that given class state. This sum is weighted by classifier numerosity
34
(e.g. a classifier with a numerosity of 3 has it’s fitness value added 3 times to
the vote score sum. New to ExSTraCS 2.0, the votes of individual rules are also
weighted by the class imbalance, such that rules specifying an over-represented
class get a smaller vote than rules from an under-represented class. In balanced
data, this update will not alter algorithm predictive performance. The class
state that is predicted by ExSTraCS is the class state with the largest vote
score. If a tie occurs, than the class state with the largest number of classifiers
(including numerosity) is chosen as the prediction. If a tie still occurs, than the
class state with the youngest average classifiers is chosen as the prediction. If a
tie still exists, than a random choice is made between the possible class states
(this should be very unlikely to occur). This ‘extended’ prediction scheme is
unique to ExSTraCS. Other LCS algorithms tend to use a prediction strictly
based on the fitness vote (as described above), and a tie immediately leads to a
random prediction of state class.
35
Chapter 3
Future ExSTraCS
Expansions
Currently, we have a number of expansions being developed for inclusion in
upcoming versions of ExSTraCS. In the following sections we will briefly discuss
these upcoming additions and the reason for their development.
3.1 Continuous Attribute Improvements
While the knowledge representation scheme adopted by ExSTraCS gives it the
flexibility to handle data with both discrete and continuous attributes, our pre-
liminary analysis has indicated that it does not achieve the same levels of per-
formance when running on discrete vs. continuous attribute datasets with the
same underlying signal strength. This is likely due to the fact that in order to
learn an effective classifier in the context of a continuous attribute, ExSTraCS
has to learn not only ‘which’ attributes to specify, but the optimal range of
continuous values to specify for a given attribute in a classifier. Intuitively this
should be a more difficult, and time consuming task. Additionally, consider
the case where a continuous valued range specified in a classifier covers all or
the majority of the range observed in the training data. In this situation, this
attribute would match for all or most classifiers, providing little or no useful
information pertaining to class prediction. Currently there is no real pressure
to prevent this from happening. Therefore we are exploring an explicit pressure
that will directly encourage narrower continuous valued ranges to be specified
in classifiers, so that such attributes do not become arbitrarily specified within
classifiers, further slowing down matching, and clouding the task of knowledge
discovery.
36
3.2 Continuous Phenotypes
Currently, the adopted ExSTraCS knowledge representation gives this algorithm
the flexibility to handle data with both discrete and continuous attributes. No-
tably, the included expert knowledge attribute weight algorithms have also been
adapted to this end as well. We are interested in giving ExSTraCS the added
flexibility to learn on problems/datasets involving a continuous-valued pheno-
type (where phenotype is also referred to as an endpoint, the dependent variable,
class, or action). While a handful of LCS algorithms have been developed to
handle continuous ‘actions’ (i.e. continuous phenotypes) these expansions have
been primarily geared towards function approximation or behavior modeling
problems. Learning on continuous phenotype data brings about a number of
challenges not found in discrete phenotype learning. For instance, in order for
a classifier to ever generalize to more than one training instance, the phenotype
of a classifier must either be a function of the condition state values, which has
been done previously in computed action research, or classifier phenotypes will
need to be a continuous-valued interval. Additionally predictions will need to be
evaluated based on their error, where error is no longer just a matter of whether
the right or wrong class was predicted, but rather how far the phenotype pre-
diction was from the true phenotype of the data instance. We are currently
evaluating a couple promising implementations that allow for continuous phe-
notype learning.
3.3 Fitness and Deletion Schemes
In the context of modern XCS-based Michigan-style LCS algorithms, it is ex-
tremely common for fitness to be directly based on a power function of classifier
accuracy. The natural multi-objective nature of these algorithms relies on im-
plicit generalization pressures to push classifiers towards a state of maximal
accuracy and maximal generality (i.e. simplicity). However, our experience
with noisy complex data mining problems has indicated some obvious problems
with a fitness scheme based only on accuracy. Upon manual inspection of clas-
sifier populations generated in this way, it is clear that many classifiers that
are actually very poor and over specific, despite the use of the subsumption
mechanism and the implicit generalization pressure derived from the fact that
more general classifiers will tend to appear in match and correct sets more often,
and therefore more often reproduce to potentially yield similar, lower generality
classifiers. Additionally we noted that deletion does not always target classi-
fiers that a user could identify as clearly ‘bad’ (e.g. the classifier only covers
one instance in the data). Presently we are working on a complete revision
both of how fitness is calculated, as well as how the deletion weight of a clas-
sifier is calculated, that will more intelligently improve and speed up learning
mainly geared towards supervised learning in complex, noisy problem domains
(as commonly found in real world problems).
37
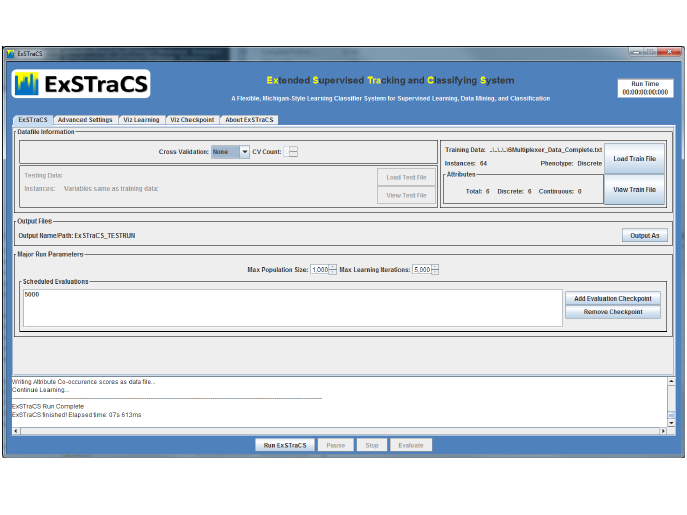
3.4 ExSTraCS GUI
In order to make ExSTraCS even more hands-on, user-friendly, we have de-
veloped a GUI prototype for the software that we hope to finalize and release
sometime early in 2015 (see above). This GUI includes an interface for setting
run parameters, and live visualizations to track algorithm learning progress, and
additional visualizations to guide knowledge discovery.
38
Chapter 4
Version Fixes/Updates
4.1 ExSTraCS 2.0.2
•Fixed the outputTestPredictions run parameter so that users can ac-
tually tell ExSTraCS whether or not to output the Predictions files.
•Fixed the InstanceID values that were being outupt in the Predictions
files. Previously the training data instance ID’s (which had been shuffled
for learning) were being mistakenly output instead of the testing data
instance ID’s.
4.2 ExSTraCS 2.0.1
•Fixed a zero division error involving balanced accuracy calculation that
appeared with very small or highly class imbalanced datasets.
•Added updated balanced accuracy calculation introduced in 2.0 to rule
compaction where it also needed to be fixed.
4.3 ExSTraCS 2.0
•Fixed Mac-related bug: Algorithm crashed tying to load some .txt data
files on the Mac platform. Updated the load file commands to allow for
the different .txt format in dealing with newlines vs return characters.
•Updated ExSTraCS to calculate and utilize a rule specificity limit. This
required alteration to the code dealing with rule covering, mutation, and
crossover, as well as modifications to how expert knowledge and attribute
tracking values were applied in these respective mechanisms. Expert
knowledge module also greatly simplified in how EK scores were trans-
formed and utilized as weights in the algorithm. Eliminated the need
39

for slow, and more complex logistic transformation of weights into scaled
probabilities.
•Updated ExSTraCS to include the TuRF wrapper algorithm as a separate
module. TuRF works in concert with any of the four included attribute
weighting algorithms included in ExSTraCS.
•Updated ExSTraCS Prediction module to more effectively make predic-
tions on imbalanced datasets. Specifically, vote contributions of individual
rules are weighted by class imbalance. In other words, rules that specify
an over-represented class get a proportionally smaller vote, vs. rules that
specify an under-represented class get a proportionally larger vote. This
change has no impact on prediction on balanced datasets.
•Fixed an error in balanced accuracy calculation that would primarily im-
pact accuracy performance evaluations within imbalanced datasets and
datasets with more than two classes. This fix, properly gives the pre-
diction accuracy within each class an equal weight in determining overall
balanced accuracy. This is applied to the calculation of both training and
testing accuracy of the rule population as a whole. Notably, with this
fix, the user should not expect standard accuracy and balanced accuracy
to be equal even when the the datasets are balanced. This is because
the calculation of balanced accuracy independently takes the number of
correctly classified instances for each respective class into consideration.
Thus after an evaluation, 70 out of 100 instances may have been correctly
classified (i.e. 70% standard accuracy), but if 40 of the 70 were from class
A, and 30 of the 70 were from class B, the balanced accuracy will have a
slightly different value. If 35 instances were were correctly classified from
either class, only then would standard accuracy equal balanced accuracy.
•Updated ExSTraCS to simplify run parameters. Added default values for
all parameters except trainFile. Also, to avoid confusion, ExSTraCS
now automatically integrates the name of the training data into the name
of the output files. The output file name parameter can now be left out or
set to None, which will simply save the output files to the working direc-
tory, and name them based on the training dataset name. Additionally,
users can not optionally leave out the ‘.txt’ when specifying the training or
testing dataset names. Lastly, we combined two parameters related to the
use of expert knowledge (i.e. internal EK Generation and EK source)
into the single parameter external EK Generation. If this new parameter
is set to None, ExSTraCS will internally generate EK scores. Otherwise
this parameter should specify the file path/name of the EK source file
giving properly formatted EK scores.
•Fixed small bug involving rule compaction crash on datasets with a smaller
number of attributes than the calculated or manually set rule specificity
limit.
40
Bibliography
[1] Jaume Bacardit and Natalio Krasnogor. A mixed discrete-continuous at-
tribute list representation for large scale classification domains. In Proceed-
ings of the 11th Annual conference on Genetic and evolutionary computa-
tion, pages 1155–1162. ACM, 2009.
[2] E. Bernad´o-Mansilla and J.M. Garrell-Guiu. Accuracy-based learning clas-
sifier system: models, analysis and applications to classification tasks. Evo-
lutionary Computation, 11(3):209–238, 2003.
[3] Martin V Butz, Kumara Sastry, and David E Goldberg. Tournament se-
lection: Stable fitness pressure in xcs. In Genetic and Evolutionary Com-
putationGECCO 2003, pages 1857–1869. Springer, 2003.
[4] Martin V Butz and Stewart W Wilson. An algorithmic description of xcs.
In Advances in Learning Classifier Systems, pages 253–272. Springer, 2001.
[5] Delaney Granizo-Mackenzie and Jason H Moore. Multiple threshold spa-
tially uniform relieff for the genetic analysis of complex human diseases. In
Evolutionary Computation, Machine Learning and Data Mining in Bioin-
formatics, pages 1–10. Springer, 2013.
[6] Casey S Greene, Daniel S Himmelstein, Jeff Kiralis, and Jason H Moore.
The informative extremes: using both nearest and farthest individuals can
improve relief algorithms in the domain of human genetics. In Evolutionary
Computation, Machine Learning and Data Mining in Bioinformatics, pages
182–193. Springer, 2010.
[7] C.S. Greene, N.M. Penrod, J. Kiralis, and J.H. Moore. Spatially uniform
relieff (surf) for computationally-efficient filtering of gene-gene interactions.
BioData mining, 2(1):1–9, 2009.
[8] Muhammad Iqbal, Will N Browne, and Mengjie Zhang. Extending learning
classifier system with cyclic graphs for scalability on complex, large-scale
boolean problems. In Proceeding of the fifteenth annual conference on Ge-
netic and evolutionary computation conference, pages 1045–1052. ACM,
2013.
41
[9] Igor Kononenko. Estimating attributes: analysis and extensions of relief.
In Machine Learning: ECML-94, pages 171–182. Springer, 1994.
[10] Tim Kovacs, Narayanan Edakunni, and Gavin Brown. Accuracy expo-
nentiation in ucs and its effect on voting margins. In Proceedings of the
13th annual conference on Genetic and evolutionary computation, pages
1251–1258. ACM, 2011.
[11] J. Moore and B. White. Tuning relieff for genome-wide genetic analysis.
Evolutionary computation, machine learning and data mining in bioinfor-
matics, pages 166–175, 2007.
[12] Qinxin Pan, Ting Hu, and Jason H Moore. Epistasis, complexity, and
multifactor dimensionality reduction. In Genome-Wide Association Studies
and Genomic Prediction, pages 465–477. Springer, 2013.
[13] Jie Tan, Jason Moore, and Ryan Urbanowicz. Rapid rule compaction
strategies for global knowledge discovery in a supervised learning classifier
system. In Advances in Artificial Life, ECAL, volume 12, pages 110–117,
2013.
[14] R. Urbanowicz, G. Bertasius, and J. Moore. An extended michigan-style
learning classifier system for flexible supervised learning, classification, and
data mining. Parallel Problem Solving from Nature–PPSN XIII, page In
press, 2014.
[15] R. Urbanowicz and J. Moore. The application of pittsburgh-style lcs to
address genetic heterogeneity and epistasis in association studies. Parallel
Problem Solving from Nature–PPSN XI, pages 404–413, 2011.
[16] R. Urbanowicz and J. Moore. Addressing scalability with a rule specificity
limit in a michigan-style supervised learnign classifier system for classifica-
tion, prediction, and knowledge discovery. In Review, 2014.
[17] R.J. Urbanowicz and J.H. Moore. The application of michigan-style learn-
ing classifier systems to address genetic heterogeneity and epistasis in as-
sociation studies. In Proceedings of the 12th annual conference on Genetic
and evolutionary computation, pages 195–202. ACM, 2010.
[18] Ryan Urbanowicz, Ambrose Granizo-Mackenzie, and Jason Moore.
Instance-linked attribute tracking and feedback for michigan-style super-
vised learning classifier systems. In Proceedings of the fourteenth inter-
national conference on Genetic and evolutionary computation conference,
pages 927–934. ACM, 2012.
[19] Ryan J Urbanowicz, Ambrose Granizo-Mackenzie, and Jason H Moore.
An analysis pipeline with statistical and visualization-guided knowledge
discovery for michigan-style learning classifier systems. Computational In-
telligence Magazine, IEEE, 7(4):35–45, 2012.
42
[20] Ryan J Urbanowicz, Delaney Granizo-Mackenzie, and Jason H Moore. Us-
ing expert knowledge to guide covering and mutation in a michigan style
learning classifier system to detect epistasis and heterogeneity. In Parallel
Problem Solving from Nature-PPSN XII, pages 266–275. Springer, 2012.
[21] Ryan John Urbanowicz, Angeline S Andrew, Margaret Rita Karagas, and
Jason H Moore. Role of genetic heterogeneity and epistasis in bladder
cancer susceptibility and outcome: a learning classifier system approach.
Journal of the American Medical Informatics Association, 2013.
[22] S.W. Wilson. Classifier fitness based on accuracy. Evolutionary computa-
tion, 3(2):149–175, 1995.
43