Adapters Pooling Guide (1000000041074) Illumina 1000000041074 02 20180523
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 26

Index Adapters
Pooling Guide
Introduction 3
Pooling Guidelines 3
Four-Channel Sequencing 3
Two-Channel Sequencing 4
One-Channel Sequencing 4
TruSeq Pooling Guidelines 4
Nextera Pooling Guidelines 12
AmpliSeq for Illumina Pooling Guidelines 19
Next Steps 23
Technical Assistance 24
Revision History 25
Document # 1000000041074 v02
May 2018
ILLUMINA PROPRIETARY
For Research Use Only. Not for use in diagnostic procedures.
This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"), and are intended solely for
the contractual use of its customer in connection with the use of the product(s) described herein and for no other
purpose. This document and its contents shall not be used or distributed for any other purpose and/or otherwise
communicated, disclosed, or reproduced in any way whatsoever without the prior written consent of Illumina. Illumina
does not convey any license under its patent, trademark, copyright, or common-law rights nor similar rights of any third
parties by this document.
The instructions in this document must be strictly and explicitly followed by qualified and properly trained personnel in
order to ensure the proper and safe use of the product(s) described herein. All of the contents of this document must be
fully read and understood prior to using such product(s).
FAILURE TO COMPLETELY READ AND EXPLICITLY FOLLOW ALL OF THE INSTRUCTIONS CONTAINED HEREIN MAY
RESULT IN DAMAGE TO THE PRODUCT(S), INJURY TO PERSONS, INCLUDING TO USERS OR OTHERS, AND DAMAGE
TO OTHER PROPERTY, AND WILL VOID ANY WARRANTY APPLICABLE TO THE PRODUCT(S).
ILLUMINA DOES NOT ASSUME ANY LIABILITY ARISING OUT OF THE IMPROPER USE OF THE PRODUCT(S)
DESCRIBED HEREIN (INCLUDING PARTS THEREOF OR SOFTWARE).
© 2018 Illumina, Inc. All rights reserved.
All trademarks are the property of Illumina, Inc. or their respective owners. For specific trademark information, see
www.illumina.com/company/legal.html.
AmpliSeq is a registered trademark of Thermo Fisher Scientific.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
2
Index Adapters Pooling Guide

Introduction
This document provides Illumina®pooling guidelines for performing library prep for sequencing on Illumina
sequencing systems.
Visit the support page for your library prep kit on the Illumina website at www.illumina.com.
Additional Resources
This document supplements the library prep kit and workflow reference guides. Review the appropriate
reference guide for protocol instructions and product details.
The following documentation is available for download from the Illumina website.
Resource Description
Custom Protocol Selector support.illumina.com/custom-protocol-selector.html
A wizard for generating customized end-to-end documentation that is tailored to the
library prep method, run parameters, and analysis method used for the sequencing run.
Illumina Adapter Sequences
(document # 1000000002694)
Provides the nucleotide sequences that comprise Illumina oligonucleotides used in
Illumina sequencing technologies.
Illumina Experiment Manager
(document # 15031335)
Provides information about creating and editing appropriate sample sheets for Illumina
sequencing systems and analysis software and record parameters for your sample plate.
Nextera DNA Flex Library Prep
Pooling Guide (document #
1000000031471)
Provides pooling guidelines for performing the Nextera™ DNAFlex library prep for
sequencing on Illumina sequencing systems.
Pooling Guidelines
Selecting the correct index combinations avoids Index Read failure due to cluster registration failure and
improves accuracy during data analysis. Illumina sequencing uses a two-channel or four-channel method to
detect individual bases.
It is important to maintain color balance for each base of the Index Read being sequenced, otherwise Index
Read sequencing could fail due to registration failure. This base calling process also ensures accuracy for
data analysis.
Follow these low-plex pooling guidelines, depending on your index adapter component.
Not all color-balanced pools are listed. Check the color balance using Illumina Experiment Manager (IEM) for
the HiSeq worklfow. For more information, see the
Illumina Experiment Manager Guide (document #
15031335)
.
Four-Channel Sequencing
Always use at least two unique and compatible barcodes for each index sequenced.
Four-channel sequencing systems capture four distinct images, which allows cycle-by-cycle observation of
which dye is incorporated into a cluster.
A green laser is used to sequence G and T bases while a red laser sequences A and C bases. To ensure
proper image registration, each cycle reads at least one of two nucleotides per color channel. Therefore, all
four images are required to build the sequence.
The MiSeq system and all HiSeq systems currently use four-channel chemistry.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
3
Index Adapters Pooling Guide
Two-Channel Sequencing
uIndex Reads must begin with at least one base other than G in either of the first two cycles. If an Index
Read begins with two base calls of G, signal intensity is not generated and registration fails. Signal must
be present in either of the first two cycles to ensure demultiplexing performance.
uSelect index sequences that provide signal in at least one channel, preferably both channels, for every
cycle.
uRed channel—A or C
uGreen channel—A or T
Two-channel sequencing simplifies nucleotide detection because only two images are needed to determine
all four base calls. Instead of using a separate dye for each base, two-channel sequencing uses a mix of
dyes. Clusters with intensity in the red channel are C bases, and clusters with intensity in the green channel
are T base. Clusters with intensity in both red and green are A base. Unlabeled clusters are G base.
The NovaSeq, NextSeq, and MiniSeq platforms use two-channel chemistry.
For more information on pooling for two-channel systems, see the
Library pooling guidelines for the NextSeq
and MiniSeq systems
bulletin on the Illumina support site.
One-Channel Sequencing
The first two cycles of an index read cannot start with two G bases, otherwise intensity is not generated. To
ensure demultiplexing performance, intensity must be present in either of the first two cycles.
Make sure that
at least
one index sequence in a library pool does not start with two G bases. Select
balanced index sequences so that signal is present in at least one image (preferably both images) for every
cycle.
The iSeq 100 System uses one-dye sequencing, which requires one dye and two images to encode data for
the four bases.
Intensities extracted from one image and compared to a second image result in four distinct populations,
each corresponding to a nucleotide. Base calling determines which population each cluster belongs to.
TruSeq Pooling Guidelines
This section details pooling strategies for index adapter tubes and index adapter plates for TruSeq based
index adapters (indexed by ligation).
Index Adapter Tubes
When using the Index Adapter Tubes, use the following pooling guidelines for single-indexed sequencing.
TruSeq DNA Single Indexes–Sets A and B
TruSeq DNA Single Index–Sets A and B each contain 12 unique indexes. When using these indexes, use the
following pooling guidelines for single-indexed sequencing.
The following tables detail pooling strategies for two to four samples generated with the index adapters in
each set. For 5–11-plex pools, use the four-plex options with any other available adapters.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
4
Index Adapters Pooling Guide
Plexity Option Set A Only Set B Only
2 1 AD006 and AD012 Not recommended
2 AD005 and AD019
3 1 AD002, AD007, and AD019 AD001, AD010, and AD020
2 AD005, AD006, and AD015 AD003, AD009, and AD025
3 Two-plex options with any other available
adapter
AD008, AD011, and AD022
4 1 AD005, AD006, AD012, and AD019 AD001, AD008, AD010, and AD011
2 AD002, AD004, AD007, and AD016 AD003, AD009, AD022, and AD027
3 Three-plex options with any other available
adapter
Three-plex options with any other available
adapter
Table 1 Single-Index Pooling Strategies for Two-Four Samples (DNA)
Plexity Option Set A Only Set B Only
2 1 AR006 and AR012 Not recommended
2 AR005 and AR019
3 1 AR002, AR007, and AR019 AR001, AR010, and AR020
2 AR005, AR006, and AR015 AR003, AR009, and AR025
3 Two-plex options with any other available
adapter
AR008, AR011, and AR022
4 1 AR005, AR006, AR012, and AR019 AR001, AR008, AR010, and AR011
2 AR002, AR004, AR007, and AR016 AR003, AR009, AR022, and AR027
3 Three-plex options with any other available
adapter
Three-plex options with any other available
adapter
Table 2 Single-Index Pooling Strategies for Two-Four Samples (RNA)
Index Adapter Plate
When using the Index Adapter Plate, use the following pooling guidelines for dual-indexed or single-indexed
sequencing.
Dual-Indexed Sequencing
Dual-indexed libraries help ensure quality data and reliable downstream analyses.
IDT for Illumina-TruSeq Unique Dual (UD) Indexes
The IDT for Illumina-TruSeq UD Indexes are compatible with all Illumina sequencing systems. Use dual-
indexing when pooling > 12 samples in one pool. IDT for Illumina-TruSeq UD Indexes are intended for dual
indexed sequencing. When performing a single indexed sequencing run when using IDT for Illumina-TruSeq
UD Indexes, the same pooling guidelines apply.
The following figure provides example dual-index pooling strategies when using the IDT for Illumina-TruSeq
UD Indexes. Pool libraries of any plexity > 2 down a column (2-plex, 3-plex, 4-plex, 5-plex, etc) as shown in
Figure 1. Do not pool across a row.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
5
Index Adapters Pooling Guide

Figure 1 Example Pooling Strategies
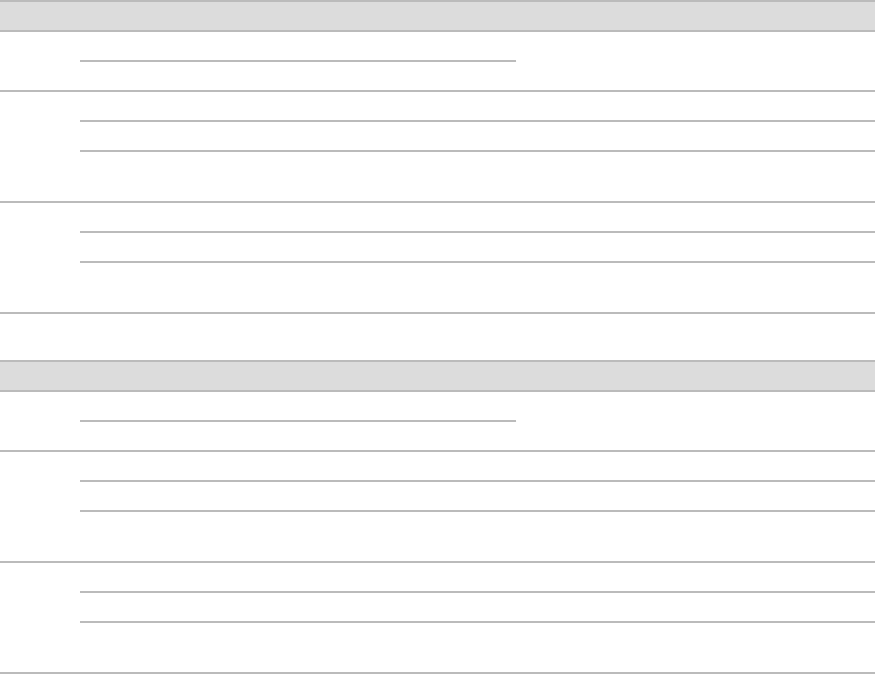
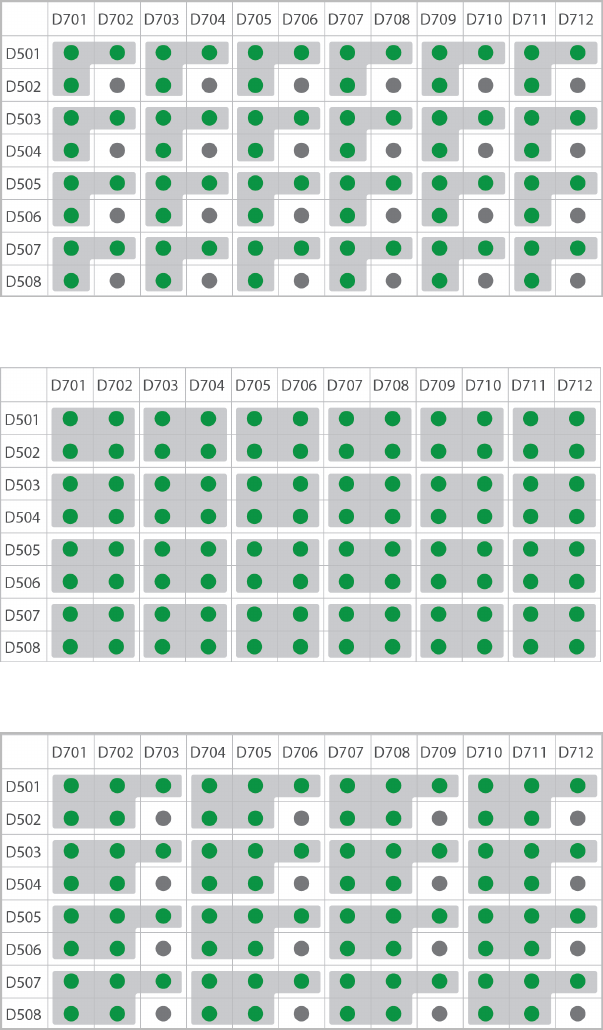
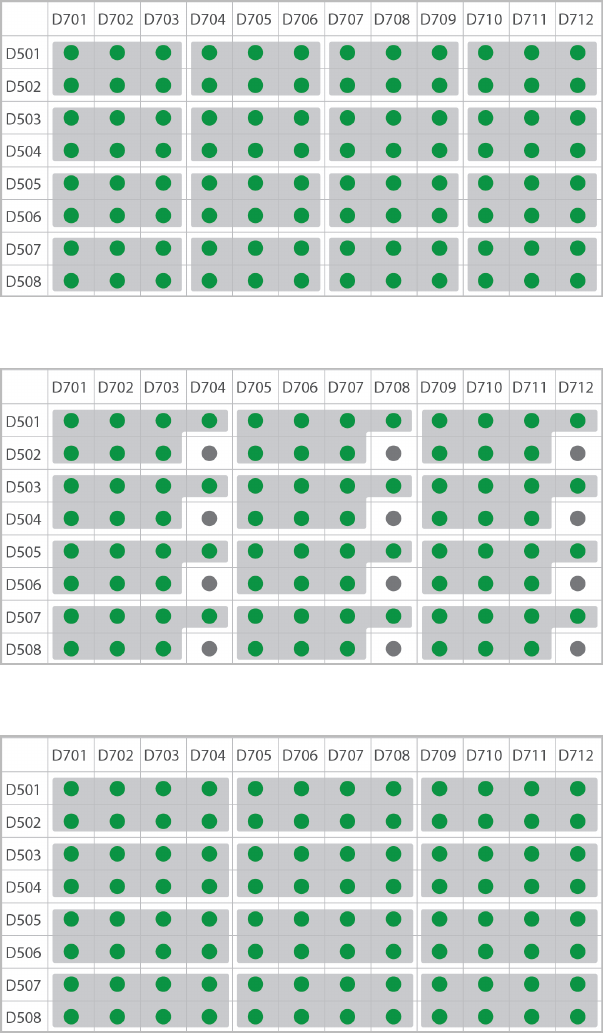
TruSeq DNA and RNA Combinatorial Dual Indexes
The following figures illustrate pooling strategies for 2–16 samples generated with the TruSeq DNA
Combinatorial Dual Indexes (96 indexes, 96 samples) or TruSeq RNA Combinatorial Indexes (96 indexes, 96
samples).
uColor-balanced pools are shaded gray and have green wells.
uTwo-plex pools are diagonal, shaded, and have green wells.
uOdd-numbered pools have dark gray wells. The gray wells are not used for sequencing pooled libraries,
but can be used for sequencing single libraries.
Dual-Indexed, Two-Plex
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
6
Index Adapters Pooling Guide
Dual-Indexed, Three-Plex
Dual-Indexed, Four-Plex
Dual-Indexed, Five-Plex
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
7
Index Adapters Pooling Guide
Dual-Indexed, Six-Plex
Dual-Indexed, Seven-Plex
Dual-Indexed, Eight-Plex (Option 1)
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
8
Index Adapters Pooling Guide
Dual-Indexed, Eight-Plex (Option 2)
Dual-Indexed, 12-Plex
Dual-Indexed, 16-Plex
Single-Indexed Sequencing
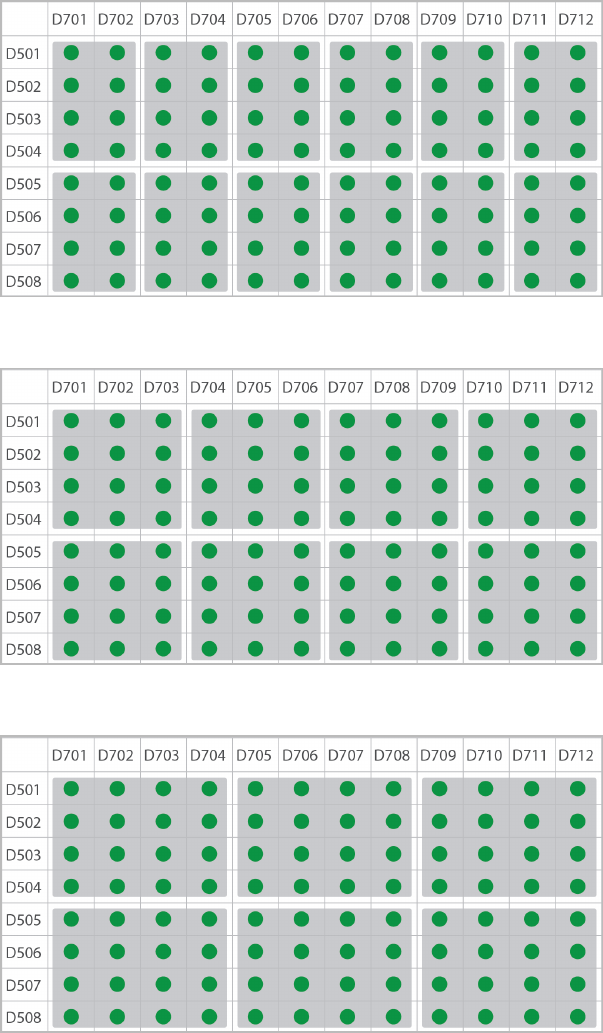
Use single-indexing when pooling ≤ 12 samples in one pool. The following figures illustrate pooling strategies
for 2–12 samples generated with the TruSeq DNA Combinatorial Dual Indexes (96 indexes, 96 samples) or
TruSeq RNA Combinatorial Indexes (96 indexes, 96 samples).
uColor-balanced pools are shaded gray and have green wells.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
9
Index Adapters Pooling Guide

uFive-plex pools have dark gray wells. The gray wells are not used for sequencing pooled libraries, but can
be used for sequencing single libraries.
uFor 7–11-plex pools, combine any of the two to six-plex pools.
Single-Indexed, Two-Plex
Single-Indexed, Three-Plex
Single-Indexed, Four-Plex
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
10
Index Adapters Pooling Guide

Single-Indexed, Five-Plex
Single-Indexed, Six-Plex
Single-Indexed, 12-Plex
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
11
Index Adapters Pooling Guide

Nextera Pooling Guidelines
This section details pooling strategies for index adapter tubes and index adapter plates for Nextera based
index adapters (indexed by ligation).
Nextera DNA 96 CD Indexes (96 indexes, plated)
Kit Contents
Index Name Bases in Adapter Bases for Sample Sheet Type
H701 TCGCCTTA TAAGGCGA i7
H702 CTAGTACG CGTACTAG i7
H703 TTCTGCCT AGGCAGAA i7
H705 AGGAGTCC GGACTCCT i7
H706 CATGCCTA TAGGCATG i7
H707 GTAGAGAG CTCTCTAC i7
H710 CAGCCTCG CGAGGCTG i7
H711 TGCCTCTT AAGAGGCA i7
H712 TCCTCTAC GTAGAGGA i7
H714 TCATGAGC GCTCATGA i7
H720 AGGCTCCG CGGAGCCT i7
H723 GAGCGCTA TAGCGCTC i7
Table 3 Index i7 Adapters
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
12
Index Adapters Pooling Guide
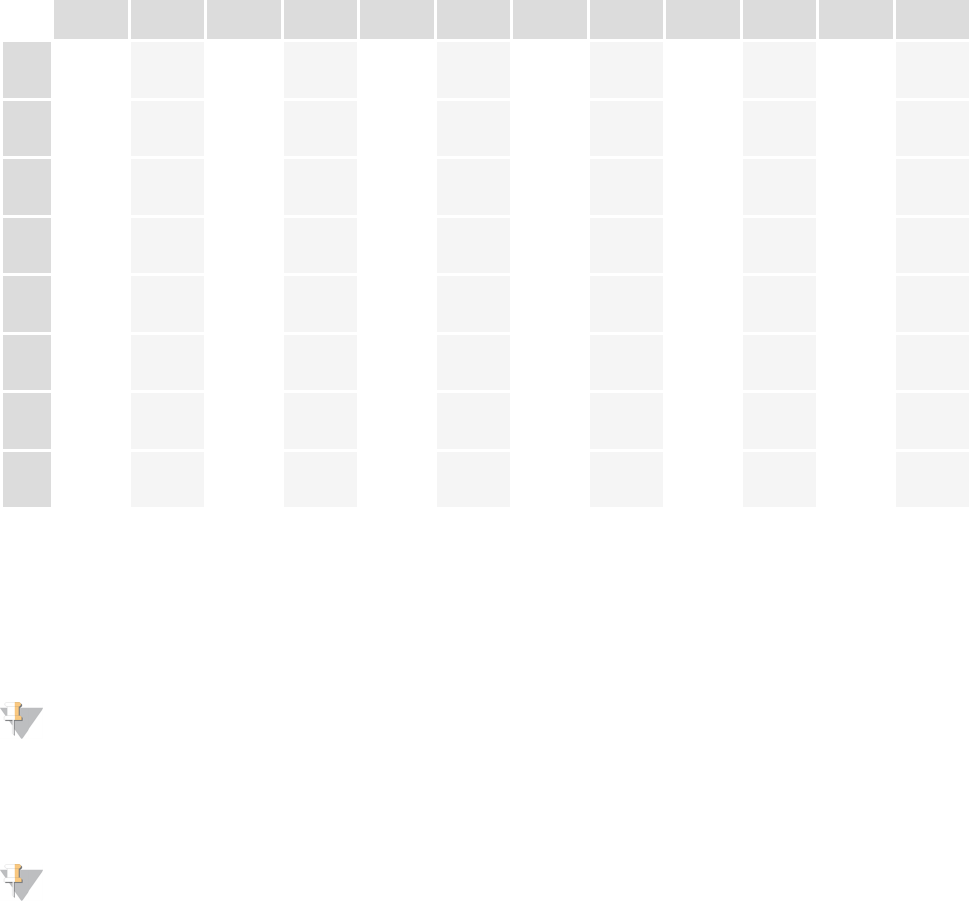
Plate Layout
1 2 3 4 5 6 7 8 9 10 11 12
AH505-
H701
H506-
H702
H517-
H703
H505-
H705
H506-
H707
H517-
H723
H505-
H706
H506-
H712
H517-
H720
H505-
H710
H506-
H711
H517-
H714
BH517-
H702
H505-
H703
H506-
H701
H517-
H707
H505-
H723
H506-
H705
H517-
H712
H505-
H720
H506-
H706
H517-
H711
H505-
H714
H506-
H710
CH506-
H703
H517-
H701
H505-
H702
H506-
H723
H517-
H705
H505-
H707
H506-
H720
H517-
H706
H505-
H712
H506-
H714
H517-
H710
H505-
H711
DH503-
H705
H503-
H707
H503-
H723
H503-
H706
H503-
H712
H503-
H720
H503-
H710
H503-
H711
H503-
H714
H503-
H701
H503-
H702
H503-
H703
EH516-
H706
H516-
H712
H516-
H720
H516-
H710
H516-
H711
H516-
H714
H516-
H701
H516-
H702
H516-
H703
H516-
H705
H516-
H707
H516-
H723
FH522-
H710
H510-
H711
H513-
H714
H522-
H701
H510-
H702
H513-
H703
H522-
H705
H510-
H707
H513-
H723
H522-
H706
H510-
H712
H513-
H720
GH513-
H711
H522-
H714
H510-
H710
H513-
H702
H522-
H703
H510-
H701
H513-
H707
H522-
H723
H510-
H705
H513-
H712
H522-
H720
H510-
H706
HH510-
H714
H513-
H710
H522-
H711
H510-
H703
H513-
H701
H522-
H702
H510-
H723
H513-
H705
H522-
H707
H510-
H720
H513-
H706
H522-
H712
Table 4 96 Plex, Dual Indexed Plate Layout
Low Plexity Guidelines
NovaSeq
The NovaSeq sequencer does not require color balanced indexes; any quantity (including 1-plex), or
combination of indexes, is supported.
NOTE
As an exception, H705 cannot be used as the sole i7 index on a NovaSeq run.
Other Sequencers
All pools listed are color-balanced (2-dye and 4-dye)on any Illumina sequencer.
NOTE
For more information on setting up a run in the MiniSeq Local Run Manager (LRM)software, see
Trim
Adapters for Nextera DNA Flex Kits in MiniSeq LRM Quick Reference Card (document # 1000000040431)
.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
13
Index Adapters Pooling Guide
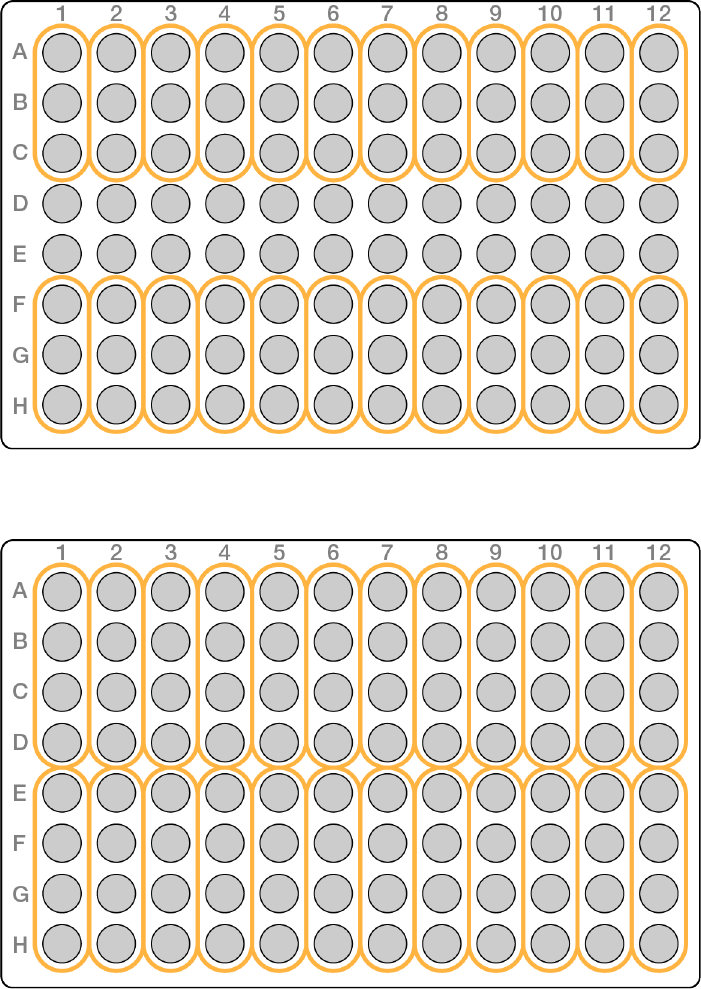
u3-plex—use the first 3, or last 3, wells in a column
uIn 3-plex usage, rows D and E are not used.
Figure 2 3-plex plate layout example
u4-plex—use the first 4, or last 4, wells in a column
Figure 3 4-plex plate layout example
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
14
Index Adapters Pooling Guide
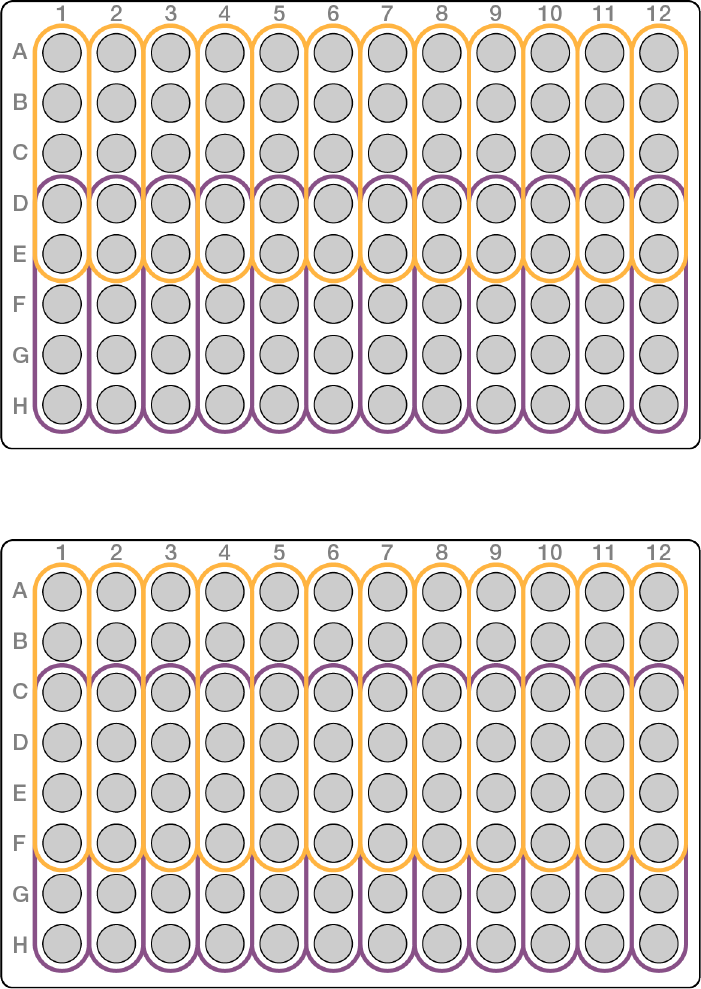
u5-plex—use the first 5 or last 5 wells in a column
Figure 4 5-plex plate layout example
u6-plex—use the first 6 or last 6 wells in a column, or 2 from one column, and 4 from an adjacent column
Figure 5 6-plex plate layout example
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
15
Index Adapters Pooling Guide
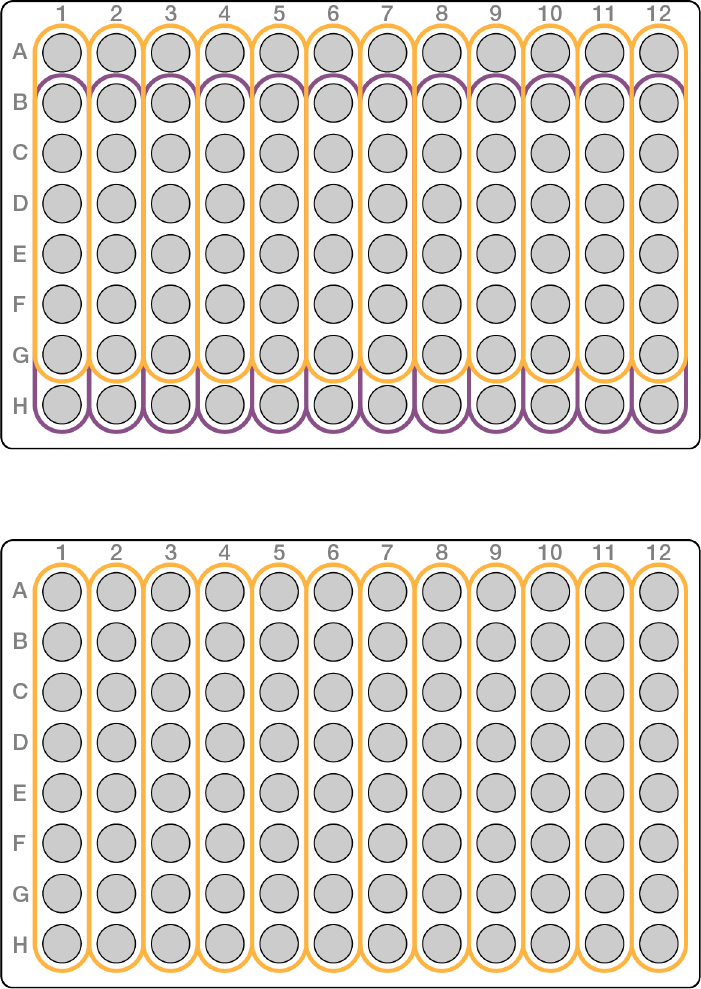
u7-plex—use the first 7, or last 7, wells in a column
Figure 6 7-plex plate layout example
u8-plex—use a full column
Figure 7 8-plex plate layout example
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
16
Index Adapters Pooling Guide
u9-plex+—use any collection of indexes containing color balanced pools (for example, A1-H1 + A2, or A4-
D4 + A5-E5)
Figure 8 Various valid 9-plex plate layout examples
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
17
Index Adapters Pooling Guide
Nextera DNA 24 CD Indexes (24 indexes, tubed)
Kit Contents
Index Name Bases in Adapter Bases for Sample Sheet Type
H705 CGTCTAAT ATTAGACG i7
H706 TCGACTAG CTAGTCGA i7
H707 CCTAGAGT ACTCTAGG i7
H710 GCGTAAGA TCTTACGC i7
H711 TTATGCGA TCGCATAA i7
H714 TCGCCTTA TAAGGCGA i7
Table 5 Index i7 Adapters
Index Name Bases in Adapter
Bases for Sample Sheet
NovaSeq, MiSeq,
HiSeq 2000/2500
Bases for Sample Sheet
MiniSeq, NextSeq,
HiSeq 3000/4000/X
Type
H503 TATCCTCT TATCCTCT AGAGGATA i5
H505 GTAAGGAG GTAAGGAG CTCCTTAC i5
H506 ACTGCATA ACTGCATA TATGCAGT i5
H517 GCGTAAGA GCGTAAGA TCTTACGC i5
Table 6 Index i5 Adapters
Low Throughput Guidelines
NovaSeq
The NovaSeq sequencer does not require color balanced indexes; any quantity (including 1-plex), or
combination of indexes, is supported.
NOTE
As an exception, H705 cannot be used as the sole i7 index on a NovaSeq run.
The pooling guidelines for other Illumina instruments are applicable.
Other Sequencers
All pools listed below are color-balanced (2-dye and 4-dye) on any Illumina sequencer.
u2-plex is not supported
u3-plex
ui5—use H505, H506, and H517
ui7—use either (H705, H706, H707) or (H710, H711, H714)
u4-plex
ui5—use all i5s
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
18
Index Adapters Pooling Guide

ui7—use either 3-plex pool + any i7
uExample:H503-H705, H505-H706, H506-H707, H517-H710
u5-plex
ui5—use the 4-plex pool + any i5
ui7—use either 3-plex pool + any two i7s
uExample: H503-H705, H505-H706, H506-H707, H517-H710, H503-H711
u6-plex
ui5—use the 4-plex pool + any two i5s OR two 3-plex pools
ui7—use all six of the i7s
or
two 3-plex pools
uExample: H503-H705, H505-H706, H506-H707, H517-H710, H503-H711, H505-H714
u7-plex
ui5—use the 4-plex pool + any three i5s OR two 3-plex pools + any i5
ui7—use all six of the i7s + an additional i7
or
two 3-plex pools + any i7
u8-plex
ui5—use all of the i5s (twice each)
ui7—use all six of the i7s + any two additional i7s
u9-plex+
uUse as many valid pools in your set as possible; at a minimum, use at least one valid pool, plus
remaining indexes.
NOTE
For more information on setting up a run in the MiniSeq Local Run Manager (LRM)software, see
Trim
Adapters for Nextera DNA Flex Kits in MiniSeq LRM Quick Reference Card (document # 1000000040431)
.
AmpliSeq for Illumina Pooling Guidelines
This section details pooling strategies for index adapter plates for AmpliSeq for Illumina based index
adapters.
AmpliSeq for Illumina Combinatorial Dual (CD) Indexes
AmpliSeq CD Indexes for Illumina are compatible with all Illumina sequencing systems. Indexes are intended
for dual index sequencing.
NOTE
For pools that contain 8–96 samples, any index combination can be used.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
19
Index Adapters Pooling Guide
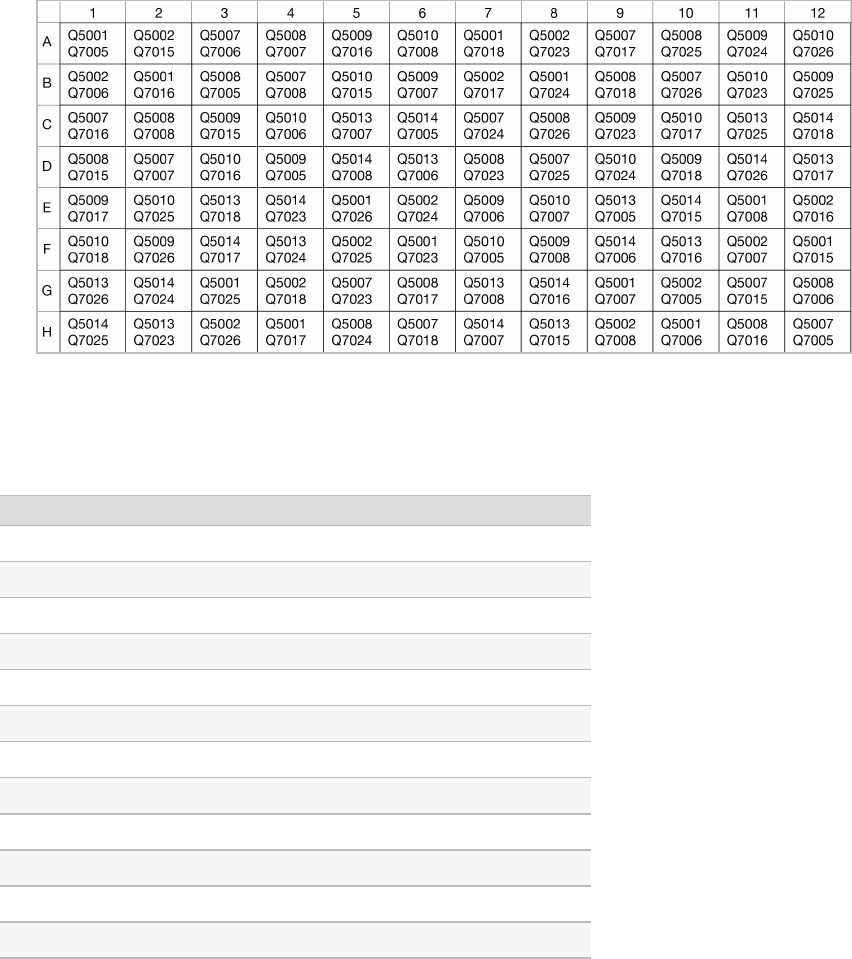
The following table shows the plate layout of AmpliSeq CD indexes for Illumina.
Figure 9 Plate Layout for AmpliSeq CD indexes for Illumina
Index 1 (i7) Adapters
CAAGCAGAAGACGGCATACGAGA[i7]GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG
Index Name Bases for Sample Sheet
Q7005 GTGAATAT
Q7006 ACAGGCGC
Q7007 CATAGAGT
Q7008 TGCGAGAC
Q7015 TCTCTACT
Q7016 CTCTCGTC
Q7017 CCAAGTCT
Q7018 TTGGACTC
Q7023 GCAGAATT
Q7024 ATGAGGCC
Q7025 ACTAAGAT
Q7026 GTCGGAGC
Table 7 Index i7 Adapters
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
20
Index Adapters Pooling Guide
Index 2 (i5) Adapters
AATGATACGGCGACCACCGAGATCTACAC[i5]TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG
Index Name i5 Bases for Sample Sheet MiSeq i5 Bases for Sample Sheet MiniSeq,
NextSeq
Q5001 AGCGCTAG CTAGCGCT
Q5002 GATATCGA TCGATATC
Q5007 ACATAGCG CGCTATGT
Q5008 GTGCGATA TATCGCAC
Q5009 CCAACAGA TCTGTTGG
Q5010 TTGGTGAG CTCACCAA
Q5013 AACCGCGG CCGCGGTT
Q5014 GGTTATAA TTATAACC
Table 8 Index i5 Adapters
Adapter Trimming
The following sequence is needed for adapter trimming.
CTGTCTCTTATACACATCT
Low Plexity Guidelines
To achieve unique dual indexing, pool between two and eight samples according to the following guidelines.
NOTE
These guidelines are for 1–8 samples, there are no specific pooling requirements for pooling more than eight
samples (up to 96).
For columns, the fundamental unit is two, however other combinations are supported. All combinations apply
to any column on the plate.
The example shown in Figure 10 provides dual indexing strategies for columns when using AmpliSeq for
Illumina CD indexes.
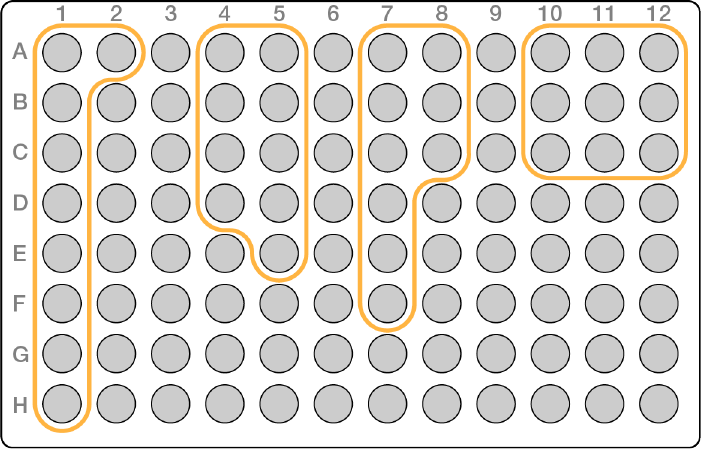
uTwo in a column as shown in orange in the following combinations:
uA-B
uC-D
uE-F
uG-H
uThree in a column, as shown in gray in the following combinations:
uA-C
uD-F
or
uC-E
uF-H
uFour in a column, as shown in purple in the following combinations:
uA-D
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
21
Index Adapters Pooling Guide
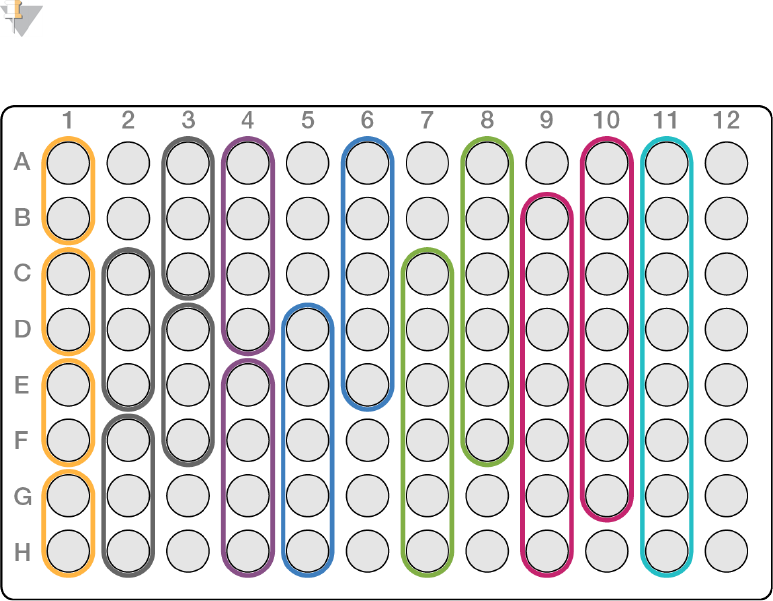
uE-H
uFive in a column, as shown in blue in the following combinations:
uA-E
or
uD-H
uSix in a column, as shown in green in the following combinations:
uA-F
or
uC-H
uSeven in a column, as shown in pink in the following combinations:
uA-G
or
uB-H
uEight in a column, as shown in teal.
NOTE
All eight combinations in any column are unique index combinations.
Figure 10 Example of Column Pooling Guidelines for Low Plexity
For rows, the fundamental unit is three, however any set of pools can be in a row as long as they are
contained between columns 1-6 or columns 7-12. All combinations apply to any row on the plate.
The example shown in Figure 11 provides dual indexing strategies for rows when using AmpliSeq for Illumina
CD indexes.
uThree in a row, as shown in gray in the following combinations:
u1-3
u4-6
u7-9
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
22
Index Adapters Pooling Guide
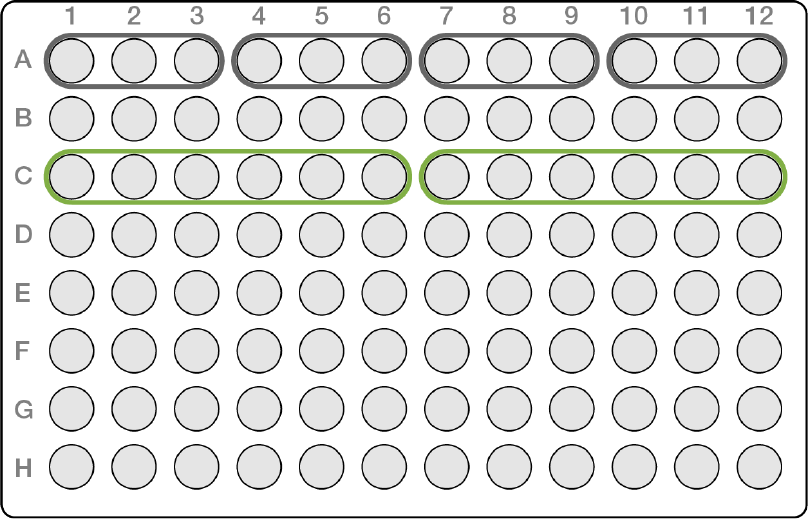
u10-12
uSix in a row, as shown in green in the following combinations:
u1-6
u7-12
Figure 11 Example of Row Pooling Guidelines for Low Plexity
Next Steps
When library prep is complete, you are ready to begin sequencing.
For information on setting up an indexed sequencing run, see the system guide for your sequencing system.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
23
Index Adapters Pooling Guide
Technical Assistance
For technical assistance, contact Illumina Technical Support.
Website: www.illumina.com
Email: techsupport@illumina.com
Illumina Customer Support Telephone Numbers
Region Toll Free Regional
North America +1.800.809.4566
Australia +1.800.775.688
Austria +43 800006249 +43 19286540
Belgium +32 80077160 +32 34002973
China 400.066.5835
Denmark +45 80820183 +45 89871156
Finland +358 800918363 +358 974790110
France +33 805102193 +33 170770446
Germany +49 8001014940 +49 8938035677
Hong Kong 800960230
Ireland +353 1800936608 +353016950506
Italy +39 800985513 +39 236003759
Japan 0800.111.5011
Netherlands +31 8000222493 +31 207132960
New Zealand 0800.451.650
Norway +47 800 16836 +47 21939693
Singapore +1.800.579.2745
Spain +34 911899417 +34 800300143
Sweden +46 850619671 +46 200883979
Switzerland +41 565800000 +41 800200442
Taiwan 00806651752
United Kingdom +44 8000126019 +44 2073057197
Other countries +44.1799.534000
Safety data sheets (SDSs)—Available on the Illumina website at support.illumina.com/sds.html.
Product documentation—Available for download in PDF from the Illumina website. Go to
support.illumina.com, select a product, then select Documentation & Literature.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
24
Index Adapters Pooling Guide

Revision History
Document Date Description of Change
Document
#1000000041074 v02
May
2018
Added One-channel Sequencing section for iSeq.
Document
#1000000041074 v01
January
2018
Added section for AmpliSeq for Illumina pooling guidelines.
Incorporated Nextera DNAFlex pooling guidelines.
Document
#1000000041074 v00
October
2017
Initial release.
Document # 1000000041074 v02
For Research Use Only. Not for use in diagnostic procedures.
25
Index Adapters Pooling Guide

Illumina
5200 Illumina Way
San Diego, California 92122 U.S.A.
+1.800.809.ILMN (4566)
+1.858.202.4566 (outside North America)
techsupport@illumina.com
www.illumina.com
For Research Use Only. Not for use in diagnostic procedures.
© 2018 Illumina, Inc. All rights reserved.
Document # 1000000041074 v02