Pvmatlab Manual
Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 19

pvmatlab
MATLAB package for handling ParaVision data
Copyright © 2013
Bruker BioSpin MRI GmbH
D-76275 Ettlingen, Germany
All Rights Reserved

Table of Contents
1 Introduction................................................................................................................3
2 Requirements and Installation...................................................................................3
2.1 System requirements.........................................................................................3
2.2 Installation..........................................................................................................4
3 Quick start..................................................................................................................5
4 Overview....................................................................................................................7
4.1 Data objects........................................................................................................7
4.2 File structure.......................................................................................................7
5 Data objects...............................................................................................................9
5.1 Common properties and methods......................................................................9
5.2 Raw data object................................................................................................10
5.3 Frame data object............................................................................................10
5.4 Cartesian k-space data object..........................................................................11
5.5 Image data object.............................................................................................12
6 Core data handling functions...................................................................................13
6.1 readBrukerParamFile.......................................................................................13
6.2 readBrukerRaw................................................................................................13
6.3 readBruker2dseq..............................................................................................14
6.4 convertRawToFrame........................................................................................15
6.5 convertFrameToCKData...................................................................................15
7 Warnings and Error messages................................................................................16
8 Feedback.................................................................................................................16
9 Appendix..................................................................................................................17
9.1 Some important parameters.............................................................................17
9.2 Required parameters........................................................................................18
10 Legal matters.........................................................................................................19
1 Introduction
The pvmatlab package is a collection of tools for Bruker users who wish to use
MATLAB for processing ParaVision raw data, for performing basic image
reconstruction of Cartesian k-space data and for importing/exporting ParaVision
images.
To get started, it is recommend to read at least section 2.2 (Installation) and to work
through chapter 3 (Quick start), which addresses the most common functions of
pvmatlab using a test data set included in the package.
Chapters 4 to 6 are dedicated to a somewhat more detailed description of the
structure of pvmatlab, its functions and conventions. Some common warnings and
error messages and what to do about them are listed in chapter 7.
A word of caution: pvmatlab is distributed 'as is' without any warranty and without any
claim of being bug-free or complete. The output produced by pvmatlab should always
be checked thoroughly, otherwise the results based on it might be wrong or
meaningless.
Bruker does not provide customer support for pvmatlab like for a regular product and
does not service the package regularly. However, if you notice bugs or malfunctions
you may get a fix and also help other users of pvmatlab by notifying Bruker software
support (see chapter 8).
The pvmatlab package is provided for non-commercial use only, at your Bruker client
site, and may not be modified or redistributed to other institutions. Requests for
pvmatlab can be made via email at: mri-software-support@bruker.de. See also
chapter 10.
2 Requirements and Installation
2.1 System requirements
As pvmatlab is a collection of MATLAB functions, the minimum system requirements
are determined by MATLAB (see http://www.mathworks.de/support/sysreq/). MATLAB
is available for Windows, Linux and Mac platforms, therefore pvmatlab can be run on
all these platforms, however, it has only been tested on Linux.
A basic MATLAB installation, release R2008a or later, is required. Earlier releases do
not support the object oriented code used in pvmatlab. No additional MATLAB
toolboxes are needed. However, the core functions of pvmatlab could be used with
earlier MATLAB releases or with Octave 3.6 or later, see chapter 6.
ParaVision data sets generated by versions 4.0, 5.0, 5.1, 5.1_Icon and 6.0 have been
tested, but data acquired with different versions of ParaVision may work, too.
If large data sets need to be processed, additional memory requirements can be
estimated as follows: For each double precision complex value of raw data, 16 bytes
of memory are needed. For example, a 3D raw data set with a matrix size of
256 x 256 x 256, acquired with a four-channel receive array and four repetitions uses
3

4 GB of memory. Reducing the data precision to “single” (see chapter 5), will bring
the additional memory requirement down to 2 GB. During the processing, copies of
the data need to be made, so the recommended additional RAM size is two times the
required memory for the biggest raw data set, at the very least.
If the memory requirements exceed the available RAM, MATLAB will use swap space
on the file system which will slow down data processing considerably. Exceeding the
swap capacity will cause MATLAB to stop with an “out of memory” error.
2.2 Installation
1. Extract the archive pvmatlab.tar.gz into a folder of your choice. For example
copy pvmatlab.tar.gz to the MATLAB working folder /home/nmrsu/matlab,
and run
[nmrsu@host matlab]$ tar xvzf pvmatlab.tar.gz
By extracting the archive, the subfolder pvtools will be created. This path will
be referred to as “the pvmatlab folder” in the following, for this example it reads
/home/nmrsu/matlab/pvtools.
2. Add the pvmatlab folder including its subfolders to the MATLAB path:
For adding the paths permanently, use the MATLAB command
>> pathtool
Select “Add with Subfolders”, choose the pvmatlab folder and then select
“Save”.
For adding the pvmatlab folder only temporarily, run
>> addBrukerPaths
or include the command inside MATLAB scripts that use pvmatlab functions.
3. For information about the installed version of pvmatlab use
>> bruker_version
This completes the installation of pvmatlab. The file pvmatlab.tar.gz can be deleted or
moved to a different location.
4
3 Quick start
In this chapter, the main functions are presented as one would typically use them.
With the MATLAB script BrukerExample.m provided in the pvmatlab folder, the
steps of this quick start guide can be reproduced in MATLAB. A sample data set of a
kiwi is used in these examples, which is also contained in the pvmatlab folder.
1. Launch MATLAB and change into the pvmatlab folder
2. In the Command Window run
>> edit BrukerExample
MATLAB will open the example script.
3. Initialization of paths
Evaluate the initialization cell with <Ctrl+Enter>. This code sets the paths to
the pvmatlab functions and to the test data set:
addBrukerPaths;
baseDir = fileparts(mfilename('fullpath'));
pathTestData = fullfile(baseDir,'TestData/2/pdata/1');
4. Importing raw data
The next set of commands demonstrates how raw data are imported into a raw
data object (for more details concerning objects see chapter 5):
rawObj = RawDataObject(pathTestData);
numSlices = rawObj.Acqp.NI;
plot(real(rawObj.data{1}(1,:,319)));
The raw data of experiment number 2 can now be accessed (in this example
by plotting echo number 319) as well as the associated method and
acquisition parameters (in this example the number of images, Acqp.NI)
5. Preparing image reconstruction
Before the raw data can be reconstructed, they need to be sorted into k-space
correctly. This is done by creating and initializing a CKDataObject. Also, the
reco parameters need to be read from disk:
kdataObj = CKDataObject(pathTestData);
kdataObj = kdataObj.readReco;
5
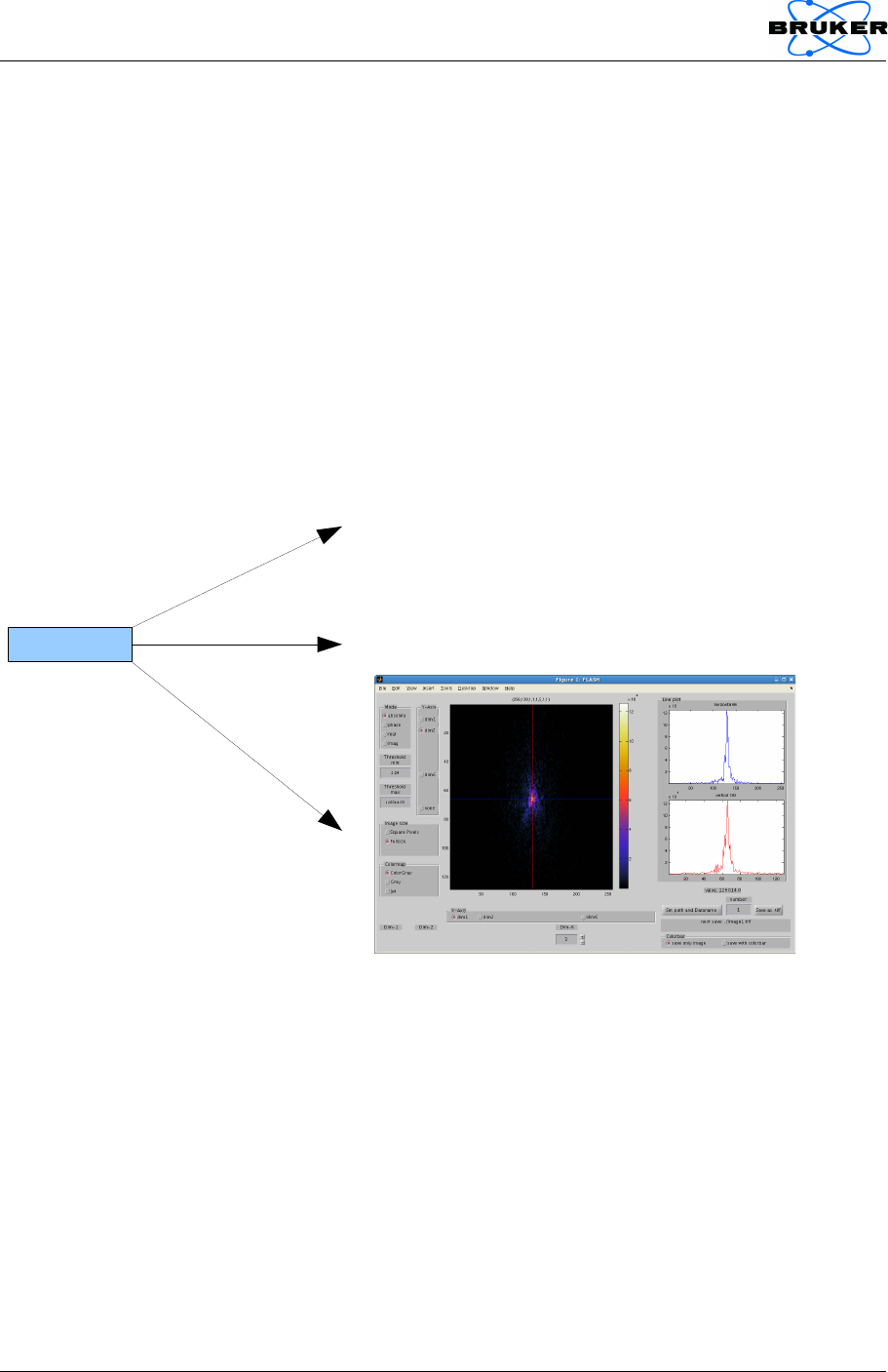
6. Reconstructing images from Cartesian k-space
The k-space object provides a reconstruction method .reco that operates on
the raw data it has been initialized with and creates an image object as output.
Both objects provide a viewer to visualize the k-space data and the image,
respectively:
kdataObj.viewer;
imageObj = kdataObj.reco('all', 'image');
imageObj.viewer;
7. Importing / Exporting ParaVision images
The image object can also be used to import images created by the
ParaVision reconstruction pipeline:
imageObj = ImageDataObject(pathTestData);
imageObj.viewer;
For exporting an image, the path must be specified and visualization
parameters need to be generated. Then the writeImage routine can be called:
imageObj = imageObj.setDataPath('imagewrite', 'export/2/pdata/1');
imageObj = imageObj.genExportVisu('genmode','auto');
imageObj.writeImage;
pvmatlab will then generate a subfolder in the export folder, named
<subjectID>_<studyID> followed by the specified expno/pdata/procno. In this
case the image (2dseq) and visu parameters (visu_pars) are exported to
export/Kiwi_pvmatlab/2/pdata/1/
6
4 Overview
This chapter contains a brief overview of how the pvmatlab package is organized and
how it interacts with the ParaVision data structure. Effectively, pvmatlab translates the
ParaVision data stored in the file system into MATLAB objects and vice versa.
4.1 Data objects
The MATLAB components handling the raw data and image data are implemented as
objects. Apart from the actual data, an object can contain additional information about
the data (acquisition parameters, method parameters, etc.). In addition, the object
contains matlab code (“methods”) working with its data, e. g., an import routine and a
data viewer.
CKDataObj
.data
.viewer
.Acqp.NI 5
There are three types of raw data objects, “RawDataObject”, “FrameDataObject” and
“CKDataObject” and one image object, “ImageObject”.
4.2 File structure
ParaVision stores all data associated with an MR experiment in a file system tree.
Each study (with raw data, image data, parameters) is stored in a separate folder (for
example /opt/PV5.1/data/nmrsu/nmr/TestData) and can be copied to any
other location for processing with MATLAB.
There are binary files containing the actual raw or image data, and parameter files in
text format, containing all necessary parameters to interpret the binary data. For a
detailed description of all parameters please refer to the ParaVision manual. A small
selection of parameters most relevant to pvmatlab is given in section 9.1.
7
size = [256 128 1 1 1 5]
0.0241 + 0.0066i -0.0533 + 0.0210i -0.0258 + 0.0496i
0.0929 + 0.0402i 0.1404 - 0.0049i 0.1222 + 0.0111i
-0.0172 + 0.0620i 0.0002 + 0.0641i -0.0099 + 0.0540i
0.0287 + 0.0527i 0.0809 + 0.0650i 0.1082 + 0.0442i
0.0005 + 0.0112i 0.0370 + 0.0387i 0.0131 + 0.0120i
-0.0094 - 0.0077i 0.0041 + 0.0525i 0.0498 + 0.0789i
-0.0125 + 0.0035i 0.0394 + 0.0220i 0.0536 + 0.0174i
0.0233 - 0.0335i -0.0029 - 0.0074i 0.0022 + 0.0129i
-0.0160 + 0.0175i 0.0302 + 0.0137i 0.0713 – 0.0156i
...
The files that are relevant for pvmatlab can be found in the following locations in the
ParaVision data tree:
TestData/
↳subject
2/
↳acqp
fid
rawdata.job0, .job1, ...
method
pdata/
↳1/
↳2dseq
reco
visu_pars
study folder
subject parameters
folder for each experiment (expno)
acquisition parameters
acquired raw data
acquired raw data
method parameters
folder for processed data
folder for processed data set (procno)
reconstructed image
reconstruction parameters
visualization parameters
text
text
binary
binary
text
text
binary
text
text
The following sketch shows how the data objects and files depend on each other:
job0...nfid
Acquisition Data
RawDataObject FrameDataObject CKDataObject
methodacqp
Acquisition Parameters
Import:
FrameDataObject(RawDataObject)
Import:
CKDataObject(FrameDataObject)
RawDataObject(path, Arguments) FrameDataObject(path, Arguments) CKDataObject(path, Arguments)
Empty:
RawDataObject()
Empty:
FrameDataObject()
Empty:
CKDataObject()
For example, a FrameDataObject can be created from an fid file and an acqp
parameter file, and a CKDataObject can then be created from this FrameDataObject
by calling
frameObj = FrameDataObject(pathTestData);
kdataObj = CKDataObject(frameObj);
8
5 Data objects
Usually, the data objects can be used as described in chapter 3. In that case, they
automatically take care of setting paths, reading the parameter files and then loading
the data. This chapter briefly describes the variables and routines of the data objects
in case you would like to control this process manually.
5.1 Common properties and methods
The data objects “RawDataObject”, “FrameDataObject”, “CKDataObject” and
“ImageObject” have common fields (properties), and matlab code (methods) to
contain and load data and parameters. These are
property method responsible for
Filespath setDataPath Paths to parameter and data files
Acqp readAcqp Acquisition parameters
Method readMethod Method parameters
Visu readVisu Visualization parameters
Reco readReco Reconstruction parameters
Subject readSubject Subject parameters
HeaderInformation - -
- setPrecision Internal representation of data
data specific, see sections 5.2-5.5 Actual data
For example, after creation of an object anyObj, the function
myObj = myObj.setDataPath('auto','/home/nmrsu/TestData/2/pdata/1');
will set the paths to all parameter files automatically. With the command
myObj = myObj.setDataPath('acqp','/home/nmrsu/myacqp');
only the acqp path is set. The property Filespath.path_to_acqp then contains
'/home/nmrsu/myacqp', the other paths are empty. A subsequent call of readAcqp
myObj = myObj.readAcqp;
will result in “myacqp” to be read to myObj.Acqp instead of the acqp file created by
ParaVision in the expno folder. Other parameter structs in myObj remain unchanged.
All objects also provide a function for setting the precision of the data. In case
memory is a limitation, consider setting the precision to 'single' (8 bytes per complex
data point instead of 16 bytes) on initializing. Default is 'double'.
myObj = RawDataObject(pathTestData, 'dataPrecision', 'single' );
9
To reduce the precision of an existing object, use:
myObj = myObj.setPrecision('single');
5.2 Raw data object
The RawDataObject represents the original raw data as an unsorted sequence of
acquired scans of length ScanSize. The data from an fid or job file is stored in a
matrix with dimensions
Raw data sizes
(numReceiveChannels, ScanSize, numScansPerChannel)
These matrices are stored in the data property in a 1D Cell-Array.
The following code creates a raw data object, sets the paths to the parameter files
and to the fid file, reads the acquisition parameters and calls the specific method
“readRaw” to load the raw data:
rawObj = RawDataObject;
rawObj = rawObj.setDataPath('auto','/home/nmrsu/TestData/2');
rawObj = rawObj.readAcqp;
rawObj = rawObj.readRaw;
The fid data matrix is now located in myRawObj.data{1}.
For a minimum set of acqp parameters required for loading raw data see section 9.2.
5.3 Frame data object
The FrameDataObject represents the raw data as a set of frames, i. e., the data are
put into an imaging context. For converting data from a RawDataObject into frames,
more acquisition parameters are required, namely the ACQ_dim, ACQ_phase_factor,
and ACQ_obj_order. Based on these parameters, the scans are sorted into separate
objects (e. g. slices), and reordered in case the acquisition was interleaved. The
resulting data matrix reflects this structure and the number of repetitions:
Frame data sizes
(ScanSize, numScansPerFrame, numReceiveChannels, numObjects, numRepetitions)
A FrameDataObject can be generated from a RawDataObject
frameObj = FrameDataObject(rawObj);
or directly from disk
frameObj = FrameDataObject('/home/nmrsu/TestData/2');
10

5.4 Cartesian k-space data object
The CKDataObject represents the raw data in Cartesian k-space according to the
encoding scheme used in the acquisition method. For converting data from a
FrameDataObject into k-space, method parameters are required, namely
'PVM_Matrix', 'PVM_EncSteps1', and, for 3D, 'PVM_EncSteps2'. Currently, the
methods RARE, FLASH, MSME and FISP are supported. The sizes of the data
matrix are:
Cartesian k-space sizes
(dim1, dim2, dim3, dim4, numReceiveChannels, numObjects, numRepetitions)
A CKDataObject can be created directly from disk as described in chapter 3 or from a
FrameDataObject
kdataObj = CKDataObject(frameObj);
The CKDataObject provides a viewer method (.viewer) to display the k-space data in
a MATLAB figure, and a .reco method to perform basic Fourier reconstruction. The
reconstruction requires a set of parameters in the Reco property.
Usage of .reco method
outObj = kdataObj.reco(recopart,
['image'], ['RECO_Parametername', ParameterValue], ...);
Input (optional in square brackets)
recopart A string or cell array of strings of reconstruction
steps to perform.
'all' = {'quadrature', 'phase_rotate', 'zero_filling',
'FT', 'sumOfSquares', 'transposition'}
'image' If given, outObj is of type ImageObject (see
section 5.5)
'RECO_Parametername',
ParameterValue
Additional reconstruction parameters
Output
outObj A CKDataObject, in which the specified recoparts have
been performed on the data. If the argument 'image' is
provided, outObj is an ImageObject
Example
kdataObj = kdataObj.setDataPath('reco','TestData/2/pdata/1/reco');
kdataObj = kdataObj.readReco;
imageObj = kdataObj.reco('all','image');
11

5.5 Image data object
The ImageObject contains a set of reconstructed images in its “data” property, which
can be created by reconstructing k-space data of a CKDataObject (see section 5.4),
or by loading images reconstructed by ParaVision (see chapter 3). In the latter case,
the data representation as frame groups is read from the visu_pars parameter file.
The sizes of the image matrix in the data property are:
Image sizes
(dim1, dim2, dim3, dim4, FrameGroupDim1, FrameGroupDim2, ...)
The ImageObject provides a viewer method (.viewer) to display the images in a
MATLAB figure, and a .writeImage method to write the image data into the
ParaVision files 2dseq and visu_pars.
For writing images to the disk, a path needs to be set by calling .setDataPath
imageObj = imageObj.setDataPath('imagewrite', 'export/2/pdata/1');
Note: The target folder will not be the exact path specified to setDataPath, but a
folder “<subjectID>_<studyID>” based on the subject and study parameters will be
created above the expno folder. For example, with subjectID “Kiwi” and studyID
“pvmatlab”, the target folder is export/Kiwi_pvmatlab/2/pdata/1
In addition, a valid set of visu parameters in the exportVisu property of the ImageObject
is required, which can be generated via
imageObj = imageObj.genExportVisu('genmode',genmode,
['TemplatePath',path]);
Input (optional in square brackets)
genmode Parameter source for generating the exportVisu struct.
Options:'Visu', 'Template', 'Subject', 'auto', default is 'auto'.
'TemplatePath',
path
Path to a visu template file. Default is expno/visu_pars
The exportVisu parameters are based on the structure of data and on all available
parameter structs (genmode 'auto') or on the parameter source specified in genmode.
Alternatively, the Visu parameters can be set manually using .setExportVisu
imageObj = imageObj.setExportVisu(parameterName,parameterValue);
To clear all or one specific parameter, use
imageObj = imageObj.setExportVisu('all','clear');
imageObj = imageObj.setExportVisu(parameterName,'clear');
With the path and visu parameters set, the image data can be exported by calling
imageObj.writeImage;
12
6 Core data handling functions
This chapter lists the core functions of pvmatlab that are used by the methods of the
data objects. They can also be used on their own and do not require object support,
therefore these functions also work with MATLAB releases earlier than R2008a and
possibly also with Octave.
6.1 readBrukerParamFile
Reads Bruker JCAMP parameter files (subject, acqp, method, reco, visu_pars) into a
struct.
Usage
paramStruct = readBrukerParamFile(filename);
Input
filename Full path and name of the parameter file
Output
paramStruct Structure containing parameter variables. The parameter
names are derived from the JCAMP tags.
Example
Acqp = readBrukerParamFile('TestData/2/acqp');
6.2 readBrukerRaw
Reads Bruker raw data (fid or rawdata.job*) into a matrix or cell array of matrices.
Usage
data = readBrukerRaw(Acqp, path_to_dataFile,
['specified_NRs', NR_array],
['specified_Jobs', job_array],
['precision', precision_str]);
Input arguments (optional in square brackets)
Acqp An acqp struct as generated by the function
readBrukerParamFile('path/acqp')
path_to_dataFile Full path and name (fid/rawdata.job0) of the raw data file
'specified_NRs',
NR_array
A list of repetitions to be read, starting with 1, when
using a standard fid file
'specified_Jobs',
job_array
A list of jobs to read, the first job being job number 0. If
only the fid-file should be read use 'specified_Jobs',[ -1 ]
13

'precision',
precision_string
Precision of the imported data: 'single' or 'double'. Single
precision uses 4 bytes to represent a (real) floating point
number, 'double' uses 8 bytes. Default is 'double'.
Output
data •If path_to_dataFile contains only an fid file, data is a cell with
a 3D matrix of size (ScanSize, numScans, numChannels)
•If path_to_dataFile contains only job files, data is a cell array
{job0, job1, job2, ...}, in which job* are 3D matrices of size
(numChannels, ScanSize, numScans)
•If path_to_dataFile contains an fid and job files, data will be a
cell array {fidFile, job0, ...}
Example
data = readBrukerRaw(Acqp,'TestData/2/fid','precision','single');
6.3 readBruker2dseq
Reads Bruker images (2dseq files) into a matrix
Usage
image = readBruker2dseq(path_to_2dseq, Visu,
['imageType', ForceType],['dim5_n', DimArray]) ;
Input arguments (optional in square brackets)
path_to_2dseq Full path and name of the 2dseq image file
Visu A visu struct as generated by the function
readBrukerParamFile('path/visu_pars')
'imageType',
ForceType
Force image type to be 'complex' or 'real'. Default is 'auto',
which reads the image type from the visu parameters
'dim5_n',
DimArray
Increase output dimensionality. Normally all frames are
indexed in the 5th dimension. dim5_n expands the 5th
dimension, e. g., with 24 visuFrames: 'dim5_n', [3 4 2]
Output
image complex or real image matrix with dimensions (dim1, dim2, dim3,
dim4, dimVisuFrame). With 'dim5_n' option, the dimensions are
(dim1, dim2, dim3, dim4, dim5_n(1), dim5_n(2), dim5_n(3), ...)
14

6.4 convertRawToFrame
Sorts the scans read by readBrukerRaw into frames
Usage
frame = convertRawToFrame(data, Acqp, ['specified_NRs', NRarray]);
Input arguments (optional in square brackets)
data A raw data matrix (fid/rawdata.job*), as read by
the function ReadBrukerRaw. Make sure it is not
a cell or cell array.
Acqp An acqp struct as generated by the function
readBrukerParamFile('path/acqp')
'specified_NRs', NRarray A list of repetitions to be converted.
Output
frame 5D frame matrix with size (ScanSize, numScansPerFrame,
numReceiveChannels, numObjects, numRepetitions)
6.5 convertFrameToCKData
Sorts the frames generated by convertRawToFrame into a k-space matrix. The
methods currently supported are RARE, FLASH, MSME and FISP.
Usage
ckdata = convertFrameToCKData(frame, Acqp, Method,
['specified_NRs', NRarray], ['useMethod', useMethod]);
Input arguments (optional in square brackets)
frame Frame data as generated by convertRawToFrame
Acqp An acqp struct as generated by the function
readBrukerParamFile('path/acqp')
Method A method struct as generated by the function
readBrukerParamFile('path/method') . This input is only
required if useMethod is true
'specified_NRs',
NRarray
A list of repetitions to be converted.
useMethod Indicates if parameters from the method file should be
used. Default is true.
Output
ckdata 7D matrix with Cartesian k-space data, with dimensions
(dim1,dim2,dim3,dim4,numChannels,numObjects,numRepetitions)
15

For more functions see
>> doc Contents
and the MATLAB source code documentation.
7 Warnings and Error messages
•Warning: Template can't be read. Proceeding without template
During image export, the method ImageDataObject.writeImage tries to read a
template visu parameter file in the expno path. If there is none, the remaining
parameter files are used to generate the visu parameters.
•Warning: You are using an unsupported acquisition method. Most probably
your result will be incorrect.
This warning occurs during conversion of frame data k-space data for methods
other than RARE, MSME, FLASH, FISP. Check results carefully!
•Cannot open parameter file. Problem opening file /opt/PV5.1/.../visu_pars
A set of parameters is required for the operation but the parameter file is
missing.
•Warning: It's recommended to read the reco-file first. ??? Reference to non-
existent field 'RECO_transposition'.
For running the CKDataObject.reco function, a set of reco parameters is
needed. Either call kdataObj.readReco to read an existing reco parameter file,
or specify the kdataObj.Reco structure yourself.
8 Feedback
If you notice a problem with a pvmatlab function that is not addressed in this manual,
you should first make sure that the problem persists after reinstalling pvmatlab (see
section 2.2) and is thus not caused by any adaption of the pvmatlab code.
If this is the case, verify that the problem is reproducible and send an email with a
description of the issue to Bruker software support (mri-software-support@bruker.de),
including
•Circumstances in which the malfunction occurs
•Computer platform, ParaVision version, MATLAB version
•Exact contents of error messages (if any)
•Anything else that could help reproduce the problem, e. g. sample data or
saved MATLAB workspace. Attachments should be smaller than 5 MB, send
larger files only when Bruker software support requests them.
16
9 Appendix
9.1 Some important parameters
Parameter name In file Description
ACQ_method acqp Name of the acquisition method e. g. RARE, FLASH
ACQ_dim acqp Acquisition dimension, 1D / 2D / 3D
ACQ_size acqp Dimensions of each object, e. g. [128 128 64] for 3D
NI acqp Number of objects (slices)
NR acqp Number of repetitions
ACQ_obj_order acqp Order in which the objects are measured
ACQ_jobs acqp Size of each job and more
ACQ_jobs_size acqp Number of jobs
PVM_Matrix method Dimension of encoding matrix
PVM_EncSteps1 method Phase encoding steps
PVM_EncSteps2 method Phase encodings in second phase encoding direction (3D)
VisuCoreFrameType visu_pars Type of the image: complex / real
VisuFGOrderDesc visu_pars Contains the FrameGroup dimensions
VisuCoreDim visu_pars Image dimension 1D / 2D / 3D
VisuCoreSize visu_pars Dimensions of each visuFrame e.g. [128 128 64] in 3D
VisuCoreFrameCount visu_pars Number of visuFrames in one data set.
VisuCoreDataSlope visu_pars Scaling factor of image data
VisuCoreDataOffs visu_pars Scaling offset of image data
17

9.2 Required parameters
The following table lists the minimum set of parameters that are required for the
respective functions
Functions Condition Parameter
struct
Required parameters
ReadBrukerRaw
RawDataObject.readRaw
Acqp GO_raw_data_format
BYTORDA
NI
NR
ACQ_size
GO_data_save
GO_block_size
AQ_mod
raw data in job files Acqp ACQ_jobs
ACQ_jobs_size
convertRawToFrame
FrameDataObject.calcFrameData
Acqp NI
NR
ACQ_size
ACQ_phase_factor
ACQ_obj_order
ACQ_dim
convertFrameToCKData
CKDataObject.calcKData
Acqp NI
NR
ACQ_size
ACQ_dim
AQ_mod
useMethod == true Method PVM_Matrix
PVM_EncSteps1
PVM_Matrix is 3D Method PVM_EncSteps2
readBruker2dseq
ImageDataObject.readImage
Visu VisuCoreWordType
VisuCoreByteOrder
VisuCoreSize
VisuCoreFrameCount
VisuCoreDataSlope
VisuCoreDataOffs
VisuCoreDim
VisuCoreDimDesc
ImageType not set
OR
ImageType == 'auto'
Visu VisuCoreFrameType
18

10 Legal matters
In the following, the term “BRUKER” refers to Bruker BioSpin MRI GmbH, Rudolf-
Plank-Str. 23, 76275 Ettlingen, while the term “CLIENT” refers to the person or
institution receiving the MATLAB package for handling ParaVision data, pvmatlab.
The MATLAB package for handling ParaVision (pvmatlab) is provided to CLIENT for
non-commercial use at CLIENT's site. It is not allowed to use pvmatlab or any of its
parts in a product or software package that is distributed or sold to third parties.
The pvmatlab package must not be used for studies on humans.
The pvmatlab package is provided 'AS IS', without warranty of any kind, either
expressed or implied, including but not limited to the implied warranties of
merchantability and fitness for a particular purpose (Some states do not allow the
exclusion of implied warranties, so the exclusion may not apply to you).
Neither BRUKER nor its dealers or distributors assume liability for any alleged or
actual damage, direct, indirect, incidental or consequential, arising in any way out of
the use of or the inability to use pvmatlab.
By providing CLIENT with pvmatlab, it is not implied that BRUKER will also provide
customer support to CLIENT, should problems occur with using the package or as a
consequence thereof. Also BRUKER does not guarantee that pvmatlab is serviced
regularly.
All rights are reserved. In particular, this document, the pvmatlab source code and the
test data are protected by copyright. They may not be reproduced, adapted,
transmitted, or stored in any form by any process (electronic or otherwise) without the
specific written consent of BRUKER. Exempt from this regulation are reproduction,
adaption, transmission and storage that are necessary to use pvmatlab efficiently at
CLIENT's site, and to enable CLIENT's personnel access to pvmatlab functionality.
All product names used in this document are trademarks of their respective owners.
ParaVision is a registered trademark of Bruker BioSpin MRI GmbH in Germany, in
the United States of America and other countries; Bruker is a registered trademark of
BRUKER-PHYSIK AG.
Product names of other manufacturers are used solely to identify their products and
BRUKER is in no way associated or affiliated with these companies. In particular,
MATLAB is a registered trademark of The MathWorks, Inc.; Windows is a registered
trademark of Microsoft Corporation in the United States of America and other
countries; Linux is a registered trademark of Linus Torvalds in the United States of
America and other countries; Mac is a registered trademark of Apple Computer, Inc.
19