SS33012 User Manual
User Manual:
Open the PDF directly: View PDF ![]() .
.
Page Count: 235 [warning: Documents this large are best viewed by clicking the View PDF Link!]
- Introduction
- New Features Available in Version 3.30
- File Organization
- Starting SS
- Converting Files from 3.24
- Starter File
- Forecast File
- Data File
- Overview of Data File
- Units of Measure
- Time Units
- Terminology
- Model Dimensions
- Fleet Definitions
- Optional Bycatch Fleets
- Catch
- Indices
- Discard
- Mean Body Weight or Length
- Population Length Bins
- Dirichlet Parameter Number and Effective Sample Sizes
- Length Composition Data
- Age Composition Option
- Conditional Age-at-Length
- Mean Length or Body Weight-at-Age
- Environmental Data
- Generalized Size Composition Data
- Tag-Recapture Data
- Stock Composition Data
- Selectivity Empirical Data
- Excluding Data
- Data Super-Periods
- Control File
- Overview of Control File
- Parameter Line Elements
- Terminology
- Beginning of Control File Inputs
- Biology
- Spawner-Recruitment
- Fishing Mortality Method
- Catchability
- Selectivity and Discard
- Tag Recapture Parameters
- Variance Adjustment Factors
- Lambdas (Emphasis Factors)
- Controls for Variance of Derived Quantities
- Using Time-Varying Parameters
- Parameter Priors
- Optional Inputs
- Likelihood components
- Running SS
- Output Files
- Using R To View Model Output (r4ss)
- Special Set-ups
- Change Log
- Appendix A: Recruitment Variability and Bias Correction
- Appendix B: Data Weighting
- Appendix C: Forecast Module
- Appendix D: Code Examples
- Appendix E: In Process and Wish List Items for Future Versions
- Appendix F: Example Model Files

Stock Synthesis User Manual
Version 3.30.12
Richard D. Methot Jr., Chantel R. Wetzel, Ian G. Taylor
NOAA Fisheries
Seattle, WA
August 16, 2018
Contents
Introduction 1
New Features Available in Version 3.30 2
SS v.3.24 Issues Detected ..................................... 5
File Organization 6
Input Files ............................................ 6
Output Files ........................................... 7
Auxiliary Files .......................................... 8
Starting SS 10
Converting Files from 3.24 10
Starter File 11
Jitter ................................................ 19
Forecast File 19
Terminology ........................................... 19
Benchmark Calculations ..................................... 27
Forecast Recruitment Adjustment ................................ 29
Data File 30
Overview of Data File ...................................... 30
Units of Measure ......................................... 31
Time Units ............................................ 31
Seasons ........................................... 32
Subseasons and Timing of events in SS v.3.30 ...................... 32
Terminology ........................................... 33
Model Dimensions ........................................ 33
Season Length and Time Steps .............................. 35
Fleet Definitions ......................................... 35
Optional Bycatch Fleets ..................................... 37
Catch ............................................... 38
Bycatch ........................................... 40
Indices .............................................. 40
Discard .............................................. 44
Mean Body Weight or Length .................................. 45
Population Length Bins ..................................... 47
Dirichlet Parameter Number and Effective Sample Sizes ................... 50
Length Composition Data .................................... 51
Age Composition Option .................................... 52
i

Age Composition Bins ................................... 53
Ageing Error ........................................ 53
Conditional Age-at-Length ................................... 56
Mean Length or Body Weight-at-Age .............................. 56
Environmental Data ....................................... 57
Generalized Size Composition Data ............................... 58
Tag-Recapture Data ....................................... 60
Stock Composition Data ..................................... 62
Selectivity Empirical Data .................................... 63
Excluding Data .......................................... 63
Data Super-Periods ........................................ 63
Control File 65
Overview of Control File .................................... 65
Parameter Line Elements .................................... 67
Terminology ........................................... 68
Beginning of Control File Inputs ................................ 68
Empirical Weight-at-Age ................................. 70
Recruitment Timing and Distribution .......................... 70
Movement ......................................... 73
Time Blocks ........................................ 74
Time-varying Parameter Controls ............................ 75
Biology .............................................. 76
Natural Mortality ..................................... 76
Growth ........................................... 77
Maturity-Fecundity .................................... 80
Hermaphroditism ..................................... 81
Parameter offset method ................................. 81
Catch Multiplier ...................................... 82
Ageing Error Parameters ................................. 82
Sex ratio .......................................... 83
Read Biology Parameters ................................. 83
Time-Varying Biology Parameters ............................ 87
Seasonal Biology Parameters ............................... 88
Spawner-Recruitment ...................................... 89
Spawner-Recruitment Function .............................. 95
Recruitment Eras ..................................... 99
Recruitment Likelihood with Bias Adjustment ...................... 99
Recruitment Autocorrelation ............................... 101
Recruitment Cycle ..................................... 101
Initial Age Composition .................................. 102
Fishing Mortality Method .................................... 102
Initial Fishing Mortality .................................. 104
ii

Catchability ............................................ 104
Selectivity and Discard ...................................... 106
Reading the Selectivity and Retention Parameters .................... 107
Selectivity Patterns ..................................... 108
Selectivity Pattern Details ................................. 111
Retention .......................................... 122
Discard Mortality ..................................... 123
Male Selectivity ...................................... 124
Dirichlet Multinomial Error for Data Weighting ..................... 125
Time-varying Options ................................... 125
Two-Dimensional Auto-Regressive Selectivity ...................... 126
Tag Recapture Parameters .................................... 129
Variance Adjustment Factors .................................. 129
Lambdas (Emphasis Factors) ................................... 131
Controls for Variance of Derived Quantities .......................... 132
Using Time-Varying Parameters ................................. 134
Parameter Priors ......................................... 138
Optional Inputs 142
Empirical Weight-at-Age (wtatage.ss) .............................. 142
runnumbers.ss .......................................... 144
profilevalues.ss .......................................... 145
Likelihood components 145
Running SS 146
Command Line Interface .................................... 146
Example of DOS batch input file ............................. 147
Simple Batch ........................................ 148
Complicated Batch ..................................... 148
Batch Using PROFILEVALUES.SS ............................ 149
Re-Starting a Run ..................................... 149
Debugging Tips .......................................... 150
Keyboard Tips .......................................... 150
Running MCMC ......................................... 151
Output Files 152
Standard ADMB output files ................................... 152
SS Summary ........................................... 153
SIS table .............................................. 153
Derived Quantities ........................................ 153
Virgin Spawning Biomass (B0) vs Unfished Spawning Biomass ............. 153
Metric for Fishing Mortality ............................... 154
iii

Equilibrium SPR ...................................... 154
F std ............................................ 155
F-at-Age .......................................... 155
MSY and other Benchmark Items ............................. 156
Brief cumulative output ..................................... 157
Output for Rebuilder Package .................................. 157
Bootstrap Data Files ....................................... 160
Forecast and Reference Points .................................. 160
Main Output File, report.sso ................................... 165
Using R To View Model Output (r4ss) 171
Special Set-ups 177
Continuous seasonal recruitment ................................ 177
Change Log 178
Appendix A: Recruitment Variability and Bias
Correction 178
Issues with Including Environmental Effects .......................... 184
Initial Age Composition ..................................... 185
Appendix B: Data Weighting 185
Applying the methods ...................................... 186
McAllister-Ianelli ..................................... 186
Francis ........................................... 188
Dirichlet-Multinomial ................................... 190
Appendix C: Forecast Module 193
Introduction ........................................... 193
Multiple Pass Forecast ...................................... 194
Example Effects on Correlations ................................ 196
Future Work ........................................... 197
Appendix D: Code Examples 198
Ageing Error Estimation ..................................... 198
Survival Based SRR Code .................................... 198
Random Walk Selectivity: Pattern 17 .............................. 200
Cubic Spline Selectivity ..................................... 202
Deviation Link .......................................... 203
Appendix E: In Process and Wish List Items for Future Versions 204
Appendix F: Example Model Files 205
iv
Introduction
Fish population (aka “stock”) assessment models determine the impact of past fishing on the
historical and current abundance of the population, evaluate sustainable rates of removals (catch),
and project future levels of catch that will implement risk-averse catch rules. These catch rules are
codified in regional Fishery Management Plans according to requirements of the Sustainable
Fisheries Act. In the U.S., 500 federally managed fish and shellfish populations are under 50 Fishery
Management Plans. About 200 of these populations are assessed according to a prioritized schedule
for their current status each year, but many minor species have never been quantitatively assessed.
Although the pace is slower than weather forecasting, fish stock assessments are operational models
for fisheries management.
Assessment models typically assimilate several decades of annual catch, data on fish abundance from
diverse surveys and fishery sources, and biological information regarding fish body size and the
proportions at age. A suite of models is available depending on the degree of data availability and
unique characteristics of the fish population or its fishery. Where feasible, environmental time series
are used as indicators of changes in population or observation processes, especially to improve the
accuracy of the projections of abundance and sustainable catch into the future. Such linkages are
based principally on correlations given the challenge of conducting field observations on an
appropriate scale. The frontier of model development is in the rapid estimation of parameters to
include random temporal effects, in the simultaneous modeling of a suite of interacting species, and
in the explicit treatment of the spatial distribution of the population.
Assessment models are loosely coupled to other models. For example, an ocean-temperature or
circulation model or benthic-habitat map may be directly included in the pre-processing of the fish
abundance survey. A time series of a derived ocean factor, like the North Atlantic Oscillation, can be
included as an indicator of a change in a population process. Output of a multi-decadal time series of
derived fish abundance can be an input to ecosystem and economic models to better understand
cumulative impacts and benefits.
Stock Synthesis (SS) is an age- and size-structured assessment model in the class of models termed
“integrated analysis models.” SS has a population sub-model that simulates a stock’s growth,
movement, and mortality processes; an observation sub-model estimates expected values for various
types of data; a statistical sub-model characterizes the data’s goodness of fit and obtains best-fitting
parameters with associated variance; and a forecast sub-model projects needed management
quantities. SS outputs the quantities, with confidence intervals, needed to implement risk-averse
fishery control rules. The model is coded in C++ with parameter estimation enabled by automatic
differentiation (www.admb-project.org). Windows, Linux, and iOS versions are available. Output
processing and associated tools are in R, and a graphical interface is in QT. SS is available from
NOAA’s VLAB. The rich feature set in SS allows it to be configured for a wide range of situations and
it has become the basis for a large fraction of U.S. assessments and many other assessments around
the world.
1
This manual provides a guide for using SS. The guide contains a description of the input and output
files and usage instructions. A technical description of the model itself is in Methot and Wetzel
(2013). The model and a graphical user interface are available from the NOAA VLAB at
https://vlab.ncep.noaa.gov/group/stock-synthesis/home. The VLAB site also provides a user forum
for posting Q&A and for accessing various additional materials. An output processor package, r4ss,
in R is available for download from CRAN or GitHub. Additional information about the package can
be located at github.com/r4ss/r4ss.
Additional guidance for new users is available from the NOAA VLAB at
https://vlab.ncep.noaa.gov/group/stock-synthesis/document-library. The "Begin Here - Introduction
to Stock Synthesis" folder located in the Document Library contains step-by-step guidance for
running Stock Synthesis.
New Features Available in Version 3.30
Stock Synthesis version v.3.30 was designed specifically to provided more precise temporal control of
growth, expected values for data, and for recruitment. In additional, a large number of new features
that make substantial changes to the input formats have been introduced. Two executables of SS are
provided. One, ss_trans.exe, will read SS v.3.24 input files and produce SS v.3.30 formatted versions of
those input files. Nearly every feature in v.3.24 can be converted by this program. The other executable,
ss.exe, will then be your primary new assessment tool. Additional information on each new feature
available by clicking on the item.
Category Item Description
General Generic Fleets Fleet specification section of data file is much changed and
now includes fleet type, so fishery fleets, bycatch fleets,
surveys, and someday predators are specified in any order
List-oriented
inputs
Rather than specify the number of items to be read, now SS
can figure it out on its own with lists terminated by -9999 in
first field of the read vector
Internal
sub-seasons
SS v3.24 inherently has 2 subseasons each season (begin and
middle) at which the age-length-key (ALK) is calculated; now
user specifies an even number of sub-seasons to use (2 to
many)
2

Category Item Description
Observation
Timing
Timing of observations now is input as year, month where
month is real; e.g. April 15 is 4.5; age-length-key (ALK) used
for each observation is calculated to the nearest sub-season.
Old "survey_timing" replaced by the month specific inputs.
Season is calculated at runtime from the input month and the
input season durations.
Speed Smarter at when to re-calculate the age-length-key (ALK);
trims tails of size-at-age so calculations avoid many
inconsequential cells of the age-length matrix. ALK tail
compression is specified in the starter file.
Converter Special version of SS, ss_trans.exe, will read files in 3.24
format and write *.ss_new files in 3.30 format. This is the
advised method for converting previous version files, but
always do a side-by-side comparison.
Empirical
Weight-at-Age
Implementing empirical weight-at-age is now specified
separately in the control file rather than under the maturity
options.
Prior Type Change in the prior numbering for parameters. Now, 0
indicates no prior, and 6 indicates a normal distribution prior.
Fishery and
Catch
Catch multiplier Each fishing fleet’s catch can now have a "q" that is a parameter
in the MGparm section.
Catch input Catch input now as list: yr, seas, fleet, amount, se.
Observations Fishery composition observations can be related to season
long catch-at-age, or to a month-specific timing.
Retention Option for dome-shaped retention function and for age-based
retention.
Selectivity Scaling Options A new non-parametric selectivity types that are scaled by the
raw values at particular ages, rather than the max age.
Survey Special Survey
Types
Special selectivity options (type 30 or >) are no longer
specified within the control file. Specifying the use of one
of these selectivity types is now done within the data file by
selecting the survey "units".
3

Category Item Description
Link functions Q_power is now one of several, and growing, set of link
functions.
Catchability setup
reorganization
Major reorganization of catchability (Q) setup, including the
link specification.
Q as a parameter Each survey now must have a Q parameter and its value still
can float (as old option 5).
Recruitment Shepherd SRR A 3-parameter Shepherd stock-recruitment curve is now an
option.
Recruitment
timing
Replace "birthseason" with "settlement event" that has explicit
timing offset from spawning. Month of spawning and
each settlement event must be specified and need not be at
beginning of a season.
Benchmark Global MSY Global MSY based on knife edge age selection; also do
calculation with single age selection. The global MSY value
will automatically be included in the report file.
Mean recruitment
distribution
In multi-area model, can now specify range of years to use
for the average recruitment distribution for forecasting. This
feature is not yet implemented.
Forecast Process error Propagate random walk in MGparms, catchability, and
selectivity into forecast. Specifying the end year for process
error in the forecast period will implement this option. This
option has only been partial implemented at this junction and
will be completed in later versions.
Biology Parameter order MGparms now have maturity, fecundity, sex ratio, and
weight-length by growth pattern.
Sex ratio Change sex ratio at birth from a constant to a morph-specific
MG parameter. This feature was not correctly implemented
in versions of 3.30 earlier than 3.30.12.
Statistical Input variance
adjuster
Added variance adjustment factor for generalized size comp.
Deviation vectors Variance of deviation vectors is now specified with 2
parameters for standard error and auto-correlation (rho), so
can be estimated.
4

Category Item Description
Dirichlet
multinomial
Dirichlet multinomial now a fleet-specific option; takes one
parameter per fleet.
Parameters Parameter order The prior standard deviation column for all parameter
lines has been moved before the prior type column. This
modification improves formatting output between integer
and decimal inputs.
Density
dependence
Beginning of year summary biomass and the recruitment
deviation parameters are mapped to the "environmental"
matrix so that parameters can be density-dependent.
Re-order Pay attention to the new order of the time-varying
adjustments to parameters (block/trend, then environmental,
then deviations).
Time-varying
parameters
Long parameter lines for spawner-recruit relationship
(SRR), catchability (Q), and tag parameters and complete
re-vamp of the way that time-varying parameters are
implemented for SRR and Q. Now shares same internal
code as mortality-growth and selectivity parameters for
time-varying capabilities.
Software
version
control
Version
numbering
The implementation of as new version control has changed
how executable versions will be specified. The executable
releases are now named SS3.3x.xx.xx representing, in order;
major features, minor features, and code fixes.
SS v.3.24 Issues Detected
The process of updating and adding new features within SS v.3.30 expose several issues with the
previous version that have been corrected:
1. Recruitment timing in multi-season models: When spawning occurred in a late season one year
and recruits occurred at beginning of a season the next year, the recruits were starting at age-0,
which was illogical. SS v.3.30 corrects this so that recruits are age-0 only if recruiting at or
between the time of spawning and the end of the year, and recruits after January 1st start at
age-0. A manual option allows users to attempt to replicate the SS v.3.24 protocol.
5
2. Lorenzen Mand time-varying growth interaction: There needs to be a revision to SS v.3.30 so
that growth can be updated each season prior to calculating Lorenzen M.
3. Length at maximum age: SS v.3.24 intended to decay this length over-time at M+Fdecreased
the abundance of fish implicitly older than the maximum age (agemax). However, this decay was
only implemented in years for which time-varying growth was updated. This will go on the the
SS v.3.30 future features wishlist.
4. SS v.3.24 had a lower bound of 1 when adjusting annual sample size (Nsamp) downward for
composition data (length and age). The variance adjustment factors in the specified in the control
file are multiplied across all annual sample size values for each data source (fleet and composition
type). The issue with the lower bound of 1 resulted in sample size adjustment not being constant
across small and large sample size years, possibly resulting in smaller samples have higher impact
than may be desired. SS v3.30 has reduced this lower bound to a value of 0.001 but has retained
user control over this value within the data file ("minsamplesize" column in the Composition
Data Structure matrix at the top of the length and age data sections) to allow comparison with
older model versions.
File Organization
Input Files
1. starter.ss: required file containing filenames of the data file and the control file plus other run
controls (required).
2. datafile: file containing model dimensions and the data (required)
3. control file: file containing set-up for the parameters (required)
4. forecast.ss: file containing specifications for reference points and forecasts (required)
5. ss.par: previously created parameter file that can be read to overwrite the initial parameter values
in the control file (optional)
6. wtatage.ss: file containing empirical input of body weight by fleet and population and empirical
fecundity-at-age (optional)
7. runnumber.ss: file containing a single number used as runnumber in output to CumReport.sso
and in the processing of profilevalues.ss (optional)
8. profilevalues.ss: file contain special conditions for batch file processing (optional)
6
Output Files
1. data.ss_new: contains a user-specified number of datafiles, generated through a parametric
bootstrap procedure, and written sequentially to this file
2. control.ss_new: updated version of the control file with final parameter values replacing the Init
parameter values.
3. starter.ss_new: new version of the starter file with annotations
4. Forecast.ss_new: new version of the forecast file with annotations.
5. warning.sso: this file contains a list of warnings generated during program execution.
6. echoinput.sso: this file is produced while reading the input files and includes an annotated echo
of the input. The sole purpose of this output file is debugging input errors.
7. Report.sso: this file is the primary report file.
8. ss_summary.sso: output file that contains all the likelihood components, parameters, derived
quantities, total biomass, summary biomass, and catch. This file offers an abridged version of
the report file that is useful for quick model evaluation. This file is only available in version
3.30.08.03 and greater.
9. CompReport.sso: observed and expected composition data in a list-based format
10. Forecast-report.sso: output of management quantities and for forecasts
11. CumReport.sso: this file contains a brief version of the run output, output is appended to current
content of file so results of several runs can be collected together. This is useful when a batch of
runs is being processed.
12. Covar.sso: this file replaces the standard ADMB ss.cor with an output of the parameter and
derived quantity correlations in database format
13. ss.par: this file contains all estimated and fixed parameters from the model run.
14. ss.std, ss.rep, ss.cor etc. standard ADMB output files
15. checkup.sso: contains details of selectivity parameters and resulting vectors. This is written
during the first call of the objective function.
16. Gradient.dat: new for SS3.30, this file shows parameter gradients at the end of the run.
17. rebuild.dat: output formatted for direct input to Andre Punt’s rebuilding analysis package.
7
Cumulative output is output to REBUILD.SS (useful when doing MCMC or profiles).
18. SIS_table.sso: output formatted for reading into the NMFS Species Information System.
19. Parmtrace.sso: parameter values at each iteration.
20. posteriors.sso, derived_posteriors.sso, posterior_vectors.sso: files associated with MCMC.
Auxiliary Files
These files are additional files (e.g. excel files) which allow for exploration or understanding of specific
parameterization which can assist in selecting appropriate starting values. These files are available for
download from the VLAB website.
1. SS3-OUTPUT.xls: Excel file with macros to read report.sso and display results.
2. Selex24_dbl_normal.xls:
(a) This excel file is used to show the shape of a double normal selectivity (option number 20
for age-based and 24 for length-based selectivity) given user-selected parameter values.
(b) Instructions are noted in the XLS file but, to summarize
i. Users should only change entries in a yellow box.
ii. Parameter values are changed manually or using sliders, depending on the value of
cell I5.
(c) It is recommend that users select plausible starting values for double-normal selectivity
options, especially when estimating all 6 parameters
(d) Please note that the XLS does NOT show the impact of setting parameters 5 or 6 to ”-999”.
In SS v3.30, this allows the the value of selectivity at the initial and final age or length to be
determined by the shape of the double-normal arising from parameters 1-4, rather than
forcing the selectivity at the intial and final age or length to be estimated separately using
the value of parameters 5 and 6.
3. Selex17_age_randwalk.xls:
(a) This excel file is used to show the shape of age-based selectivity arising from option 17
given user-selected parameter values
(b) Users should only change entries in the yellow box.
8
(c) The red box is the maximum cumulative value, which is subtracted from all cumulative
values. This is then exponentiated to yield the estimated selectivity curve. Positive values
yield increasing selectivity and negative values yield decreasing selectivity.
4. Prior_Tester.xls:
(a) The ’compare’ tab of this spreadsheet shows how the various options for defining
parameter priors work
5. SS_330_Control_Setup.xls:
(a) Shows how to setup an example control file for SS
6. SS_330_Data_Input.xls:
(a) Shows how to setup an example data input for SS
7. SS_330_Starter&Forecast.xls:
(a) Shows how to setup an example data input for SS
8. Growth_Comparison.xls:
(a) Excel file to test parameterization between the growth curve options within SS.
(b) Instructions are noted in the XLS file but, to summarize
i. Users should only change entries in a yellow box.
ii. Entries in a red box are used internally, and can be compared with other
parameterizations, but should not be changed.
(c) The SS-VB is identical to the standard VB, but uses a parameterization where length is
estimated at pre-defined ages, rather than A=0 and A=Inf. The Schnute- Richards is
identical to the Richards-Maunder, but similarly uses the parameterization with length at
pre-defined ages. The Richards coefficient controls curvature, and if the curvature
coefficient = 1, it reverts to the standard VB curve.
9. Movement.xls:
(a) Excel file to explore SS movement parameterization
9
Starting SS
SS is typically run through the command line interface although it can also be called from another
program such as R or the SS-GUI or a script file (such as a DOS batch file). SS is compiled for Windows,
Mac, and Linux operating systems. The memory requirements depend on the complexity of the model
you run, but in general, SS will run much slower on computers with inadequate memory. See the
section for additional notes on methods of running SS.
Communication with the program is through text files. When the program first starts, it reads the file
starter.ss, which typically must be located in the same directory from which SS is being run. The file
starter.ss contains required input information plus references to other required input files, as described
in section . The names of the control and data files must match the names specified in the starter.ss file.
File names, including starter.ss, are case-sensitive on Linux and Mac systems but not on Windows. The
echoinput.sso file outputs how the executable read each input file and can be used for troubleshooting
when trying to get a model setup correctly. Output from SS is as text files containing specific keywords.
Output processing programs, such as the SS GUI, Excel, or R can search for these keywords and parse
the specific information located below that keyword in the text file.
Converting Files from 3.24
Converting files from v.3.24 to v.3.30 can be easily performed by using the program sstrans.exe. The
following file structure and steps are recommended for converting model files:
1. Create "transition" folder. Place four model files from version 3.24 within the transition folder
along with the SS transition executable ("ss_trans.exe"). One tip is to use the control.ss_new from
the 3.24 estimated model rather than the control.ss file which will set all parameter values at the
previous estimated MLE parameters. Run the transition executable with phase = 0 within the
starter file, with the read par file turned off (option 0).
2. Create "converted" folder. Place the ss_new (data.ss_new, control.ss_new, starter.ss_new,
forecast.ss_new)files created by the transition executable contained within the "transition"
folder into this new folder. Rename the ss_new files to the appropriate suffixes and change the
names in the starter.ss file accordingly.
3. Review the control file to determine that all model functions converted correctly. The structural
changes and assumptions for a couple of the advanced model features are too complicated to
convert automatically. See below for some known features that may not convert.
4. Change the max phase to a value greater than the last phase in which the a parameter is set to
estimated within the control file. Run the new 3.30 executable (ss.exe) within the "converted"
10
folder using the renamed ss_new files created from the transition executable.
5. Compare likelihood and model estimates between the 3.24 and 3.30 model versions.
There are some options that have been substantially changed in version 3.30 which impedes the
automatic converting of 3.24 model files. Known examples of 3.24 options that cannot be converted,
but for which better alternatives are available in 3.30 are:
1. The use of Q deviations,
2. Complex birth seasons,
3. Environmental effects on spawner-recruitment parameters,
4. Setup of time-varying quantities for models that used the no-longer-available (e.g logistic bound
constraint).
Starter File
SS begins by reading the file starter.ss. Its format and content is as follows. Note that the term COND
in the Typical Value column means that the existence of input shown there is conditional on a value
specified earlier in the file. Omit or comment out these entries if the appropriate condition has not
been selected.
11

STARTER.SS
Value Options Description
#C this is
a starter
comment
Must begin with #C then rest of the line is
free form
All lines in this file beginning with #C will be retained and written to the top
of several output files
data_file.dat File name of the data file
control_file.ctl File name of the control file
0 Initial Parameter Values: Don’t use this if there have been any changes to the control file that would
alter the number or order of parameters stored in the ss.par file. Values in
ss.par can be edited, carefully. Do not run sstrans.exe from a ss.par from SS
v.3.24.
0 = use values in control file;
1 = use ss.par after reading setup in the
control file
1 Run display detail: With option 2, the display shows value of each -logL component for each
iteration and it displays where crash penalties are created
0 = none other than ADMB outputs;
1 = one brief line of display for each
iteration;
2 = fuller display per iteration
1 Detailed age-structure report Detailed age-structured report in Report.sso
0 = minimal (no Report file);
1 = include all output;
2 = brief output
12

Value Options Description
0 Write 1st iteration details This output is largely unformatted and undocumented and is mostly used by
the developer.
0 = omit
1 = write detailed intermediate calculations
to echoinput.sso during first call
0 Parameter Trace This controls the output to parmtrace.sso. The contents of this output can
be used to determine which values are changing when a model approaches
a crash condition. It also can be used to investigate patterns of parameter
changes as model convergence slowly moves along a ridge.
0 = omit
1 = write good iteration and active
parameters
2 = write good iterations and all parameters
3 = write every iteration and all parameters
4 = write every iteration and active
parameters
1 Cumulative Report Controls reporting to the file Cumreport.sso. This cumulative report is most
useful when accumulating summary information from likelihood profiles or
when simply accumulating a record of all model runs within the current
subdirectory
0 = omit
1 = brief
2 = full
1 Full Priors Turning on this option causes all prior values to be calculated. With this
option off, the total log likelihood, which includes the log likelihood for priors,
would change between model phases as more parameters became active.
0 = only calculate priors for active
parameters
1 = calculate priors for all parameters that
have a defined prior
13

Value Options Description
1 Soft Bounds This option creates a weak symmetric beta penalty for the selectivity
parameters. This becomes important when estimating selectivity functions
in which the values of some parameters cause other parameters to have
negligible gradients, or when bounds have been set too widely such that a
parameter drifts into a region in which it has negligible gradient. The soft
bound creates a weak penalty to move parameters away from the bounds.
0 = omit
1 = use
1 Data File Output All output files are sequentially output to data.ss_new and will need to be
parsed by the user into separate data files. The output of the input data file
makes no changes, so retains the order of the original file. Output files 2-N
contain only observations that have not been excluded through use of the
negative year denotation, and the order of these output observations is as
processed by the model. The N obs values are adjusted accordingly. At this
time, the tag recapture data is not output to DATA.SS_new.
0 = none
1 = output an annotated replicate of the
input data file
2 = add a second data file containing the
model’s expected values with no added
error
3+ = add N-2 parametric bootstrap data
files
8 Turn off estimation The 0 option is useful for (1) quickly reading in a messy set of input files
and producing the annotated control.ss_new and data.ss_new files, or (2)
examining model output based solely on input parameter values. Similarly,
the value option allows examination of model output after completing a
specified phase. Also see usage note for restarting from a specified phase.
-1 = exit after reading input files
0 = exit after one call to the calculation
routines and production of sso and ss_new
files
<positive value> = exit after completing this
phase
10 MCMC burn interval Need to document this and set good default
2 MCMC thin interval Need to document this and set good default
14

Value Options Description
0.0 Jitter The jitter function has been revised with v3.30. Starting values are now
jittered based on a normal distribution based on the pr(PMIN) = 0.1% and the
pr(PMAX) = 99.9%.
A positive value here will add a small
random jitter to the initial parameter
values. When using the jitter option, care
should be given when defining PMIN and
PMAX values and particularly -999 or 999
should not be used to define bounds.
-1 SD Report Start
-1 = begin annual SD report in start year
<year> = begin SD report this year
-1 SD Report End
-1 = end annual SD report in end year
-2 = end annual SD report in last forecast
year
<value> = end SD report in this year
2 Extra SD Report Years In a long time series application, the model variance calculations will be
smaller and faster if not all years are included in the SD reporting. For
example, the annual SD reporting could start in 1960 and the extra option
could select reporting in each decade before then.
0 = none
<value> = number of years to read
COND: If Extra SD report years > 0
1940 1950 Vector of years for additional SD reporting
15

Value Options Description
0.0001 Final convergence This is a reasonable default value for the change in log likelihood denoting
convergence. For applications with much data and thus a large total log
likelihood value, a larger convergence criterion may still provide acceptable
convergence
0 Retrospective year Adjusts the model end year and disregards data after this year. May not handle
time varying parameters completely.
0 = none
-x = retrospective year relative to end year
0 Summary biomass min age Minimum integer age for inclusion in the summary biomass used for
reporting and for calculation of total exploitation rate
1 Depletion basis Selects the basis for the denominator when calculating degree of depletion in
SSB. The calculated values are reported to the SD report.
0 = skip
1 = X*SB0
2 = X*SBMSY
3 = X*SBstyr
4 = X*SBendyr
0.40 Fraction (X) for depletion denominator So would calculate the ratio of SSBy/(0.40*SSB0)
1 SPR report basis SPR is the equilibrium SSB per recruit that would result from the current
year’s pattern and intensity of F’s. The SPR approach to measuring fishing
intensity was implemented because the concept of a single annual F does not
exist in SS. The quantities identified by 1, 2, and 3 here are all calculated in
the benchmarks section. Then the one specified here is used as the selected
denominator in a ratio with the
0 = skip
1 = use 1-SPRtarget
2 = use 1-SPR at MSY
3 = use 1-SPR at Btarget
16

Value Options Description
4 = no denominator, so report actual 1-SPR
values
annual value of (1 – SPR). This ratio (and its variance) is reported to the SD
report output for the years selected above in the SD report year selection.
4 F std report value In addition to SPR, an additional proxy for annual F can be specified here.
As with SPR, the selected quantity will be calculated annually and in the
benchmarks section. The ratio of the annual value to the selected (see F report
basis below) benchmark value is reported to the SD report vector. Options 1
and 2 use total catch for the year and summary abundance at the beginning of
the year, so combines seasons and areas. But if most catch occurs in one area
and there is little movement between areas, this ratio is not informative about
the F in the area where the catch is occurring. Option 3 is a simple sum of
the full F’s by fleet, so may provide non-intuitive results when there are multi
areas or seasons or when the selectivities by fleet do not have good overlap
in age. Option 4 is a real annual F calculated as a numbers weighted F for a
specified range of ages (read below). The F is calculated as Z-M where Z and M
are each calculated an ln(Nt+1/Nt) with and without F active, respectively. The
numbers are summed over all biology morphs and all areas for the beginning
of the year, so subsumes any seasonal pattern.
0 = skip
1 = exploitation rate in biomass
2 = exploitation rate in numbers
3 = sum(full F’s by fleet)
4 = population F for range of ages
5 = unweighted average F for range of ages
COND: If F std reporting > 4 Specify range of ages. Upper age must be less than max age because of
incomplete handling of the accumulator age for this calculation.
13 17 Age range if F std reporting = 4
1 F report basis Selects the denominator to use when reporting the F std report values. Note
that order of these options differs from the biomass report basis options.
0 = not relative, report raw values
1 = use F std value corresponding to
SPRtarget
2 = use F std value corresponding to FMSY
3 = use F std value corresponding to FBtarget
17
Value Options Description
0.01 MCMC output detail Specify format of MCMC output. This input requires the specification of two
items; the output detail and a bump value to be added to the ln(R0) in the first
call to MCMC. A bias adjustment of 1.0 is applied to recruitment deviations
in the MCMC phase, which could result in reduced recruitment estimates
relative to the MLE when a lower bias adjustment value is applied. A small
value, called the "bump", is added to the ln(R0) for the first call to MCMC
in order to prevent the stock from hitting the lower bounds when switching
from MLE to MCMC. If you wanted to select the default output option and
apply a bump value of 0.01 this is specified by 0.01 where the integer value
represents the output detail and the decimal is the bump value.
0 = default
1 = output likelihood components and
associated lambda values
2 = expanded output
3 = make output subdirectory for each
MCMC vector.
0 Age-length-key (ALK) tolerance level, 0 >=
values required
Value of 0 will not apply any compression. Values > 0 (e.g. 0.0001) will apply
compression to the ALK which will increase the speed of calculations. The
size of this value will impact the run time of your model, but one should be
careful to ensure that the value used does not appreciably impact the estimated
quantities relative to no compression of the ALK. The suggested value if
applied is 0.0001.
3.30 3.30: Indicates that the control and data
files are currently in SS v3.30 format.
The transition executable for SS v3.30 will create converted files in the new
format from previous versions (must be 3.24) when 999 is given. All ss_new
files are in the 3.30 format, so starter.ss_new has 3.30 on the last line. Some
Mgparms are in new sequence, so 3.30 cannot read a ss.par file produced by
version 3.24 and earlier, so please ensure that read par file option at the top
of the starter file is set to 0. Please see Converting Files from 3.24 section for
additional information on model features that may impede file conversion.
999: Indicates that the control and data
file are in a previous SS 3.24 version.
The sstrans.exe executable should be
used which will convert the files to the
new format in the control.ss_new and
data.ss_new files.
End of Starter File
18
Jitter
The jitter function has been updated with v.3.30. The following steps are now performed to determine
the jittered starting parameter values:
1. A normal distribution is calculated such that the pr(PMIN) = 0.01% and the pr(PMAX) = 99.9%.
2. A jitter shift value, termed "K", is calculated from the distribution based on the pr(PCURRENT).
3. A random value is drawn, "J", from the range of K-jitter to K+jitter with the constraint that it
cannot be <0.1% or >99.9% of the distribution.
4. Jis a new cumulative normal probability value.
5. Calculate a new parameter value, PJITTERED, such that pr(PJITTERED) = J.
Forecast File
The specification of options for forecasts is contained in the mandatory input file named forecast.ss.
For additional detail on the forecast file see Appendix B.
Terminology
Where the term “COND” appears in the value column of this documentation (it does not actually
appear in the forecast file), it indicates that the following section is omitted except under certain
conditions, or that the factors included in the following section depend upon certain conditions. In
most cases, the description in the definition column is the same as the label output to the
forecast.ss_new file.
19

FORECAST.SS
Value Options Description
1Benchmarks/Reference Points SS checks for consistency of the Forecast specification and the
benchmark specification. It will turn benchmarks on if necessary
and report a warning.
0 = omit
1 = calculate FSPR, FBtarget, and FMSY
2 = calculate FSPR, FBtarget, FMSY, F0.10
1 MSY Method Specifies basis for FMSY.
1=FSPR as proxy
2 = calculate FMSY
3=FBtarget as proxy or F0.10
4=Fend year as proxy
0.45 SPRtarget SS searches for F multiplier that will produce this level of spawning
biomass per recruit (reproductive output) relative to unfished value.
0.40 Relative Biomass Target SS searches for F multiplier that will produce this level of spawning
biomass relative to unfished value. This is not “per recruit” and takes
into account the spawner-recruitment relationship.
0 0 0 0 0 0 0 0 0 0 Benchmark Years Requires 10 values, (1,2) beginning and ending years for biology
(growth, natmort, maturity, fecundity), (3,4) selectivity, (5,6) relative
Fs, (7,8) movement and recruitment distribution; (9,10) SRparms for
averaging years in calculating benchmark quantities
-999: start year
>0: absolute year
<= 0: year relative to end year
20

Value Options Description
1 Benchmark Relative F Basis Does not affect year range for selectivity and biology.
1 = use year range
2 = set range for relF same as forecast below
2 Forecast This input is required but is ignored if benchmarks are turned off. If
FMSY is selected, it uses whatever proxy, e.g. FSPR or FBTGT is selected
in the benchmark section.
0 = none (no forecast years)
1 = use FSPR
2 = use FMSY
3 = use FBtarget or F0.10
4 = set to average F scalar for the forecast
relative F years below
5 = input annual F scalar
10 N forecast years (must be >= 1) At least one forecast year now required which differs from version
3.24 that allowed zero forecast years.
1 F scalar Only used if Forecast option = 5 (input annual F scalar).
0 0 0 0 0 0 Forecast Years Requires 6 values: beginning and ending years for selectivity, relative
Fs, and recruitment distribution that will be used to create averages
to use in forecasts. In future, hope to allow random effects to
propagate into forecast.
NOTE: Relative F for bycatch only fleets is scaled just like other
fleets. More options for this in future.
>0 = absolute year
<= 0 = year relative to end year
21

Value Options Description
0 Forecast Selectivity Option Determines the selectivity used in the forecast years.
0 = forecast selectivity is mean from year
range
1 = forecast selectivity from annual
time-varying parameters
1 Control Rule
1 = catch as function of SSB, buffer on F
2 = F as function of SSB, buffer on F
3 = catch as function of SSB, buffer on catch
4 = F is a function of SSB, buffer on catch
0.40 Control Rule Upper Limit Biomass level (as a fraction of SSB0) above which F is constant at
control rule Ftarget.
0.10 Control Rule Lower Limit Biomass level (as a fraction of SSB0) below which F is set to 0.
0.75 Control Rule Buffer Control rule Ftarget as a fraction of selected FMSY proxy. Note, if using
Pope’s F, then this value will be applied to the catch rather than the F.
Model’s that use either continuous F or the hybrid F will apply this
value directly to the F. Future versions will allow a user to specify
whether the adjustment should be applied to either the catch or F
independent of the fishing mortality method selected.
22

Value Options Description
3 Number of forecast loops (1,2,3) SS sequentially goes through the forecast up to three times.
Maximum number of forecast loops: 1=OFL only, 2=ABC control
rule, 3=set catches equal to control rule or input catch and redo
forecast implementation error.
3First forecast loop with stochastic
recruitment
If this is set to 1 or 2, then OFL and ABC will be as if there was perfect
knowledge about recruitment deviations in the future.
0Forecast recruitment Option 0, ignore input and do forecast recruitment as before SS
v.3.30.10, if 1, then use next value as a multiplier applied after
env/block/regime is applied, if 2, then use value as multiplier times
adjusted virgin recruitment (after time-varying adjustments to R0),
and if 3, then use value as the number of years from end of main
recruitment deviations to average (mean is the recruitments, not the
deviations).
0 = spawner recruit curve
1 = value*(spawner recruit curve)
2 = value*(virgin recruitment)
3 = recent mean from year range above
1 Scaler or N years recent main recruitments
to average
This input depends upon option selected directly above. If option
1 or 2 selected this value should be a scalar value to be applied to
recruitment. If option 3 is selected above this should be input as the
number of years to average recruitment.
0 Forecast loop control #5 Reserved for future model features.
2015 First year for caps and allocations Should be after years with fixed inputs.
23

Value Options Description
0 Implementation Error The standard deviation of the log of the ratio between the realized
catch and the target catch in the forecast. (set value >0.0 to cause
implementation error devs to be an estimated parameter that will
add variance to forecast).
0 Rebuilder Creates a rebuild.dat file to be used for West Coast groundfish
rebuilder program.
0 = omit West Coast rebuilder output
1 = do West Coast rebuilder output
2004 Rebuilder catch (Year Declared)
>0 = year first catch should be set to zero
-1 = set to 1999
2004 Rebuilder start year (Year Initial)
>0 = year for current age structure
-1 = set to end year +1
1 Fleet Relative F
1 = use first-last allocation year
2 = read season(row) x fleet (column) set
below
2 Basis for maximum forecast catch
2 = total catch biomass
3 = retained catch biomass
5 = total catch numbers
6 = retained total numbers
24

Value Options Description
COND 2: Conditional input for fleet relative F
0.1 0.8 0.1 Fleet allocation by relative F fraction The fraction of the forecast F value. For a multiple area model
user must define a fraction for each fleet and each area. The total
fractions must sum to one over all fleets and areas. Starting in
version 3.3 this now also includes surveys which are treated similar
to fleets.
Ex: # Fleet 1 Fleet 2 Survey X (rows are seasons)
1 50 Maximum total catch by fleet Enter fleet number and its max. Last line of the entry must have fleet
number = -9999.
-9999 -1
-9999 -1 Maximum total catch by area Enter area number and its max. Last line of the entry must have area
number = -9999.
-1 = no maximum
1 1 Fleet assignment to allocation group Enter list of fleet number and its allocation group number if it is in
a group. Last line of the entry must have fleet number = -9999.
-9999 -1
COND: if N allocation groups is >0
2002 1 Allocation to each group for each year of
the forecast
Enter a year and the allocation fraction to each group for that year.
SS will fill those values to the end of the forecast, then read another
year from this list. Terminate with -9999 in year field. Annual
values are rescaled to sum to 1.0.
-9999 1
25

Value Options Description
-1 Basis for forecast catch
-1 = Read basis with each observation,
allows for a mixture of dead, retained, or F
basis by different fleets for the fixed catches
below.
2 = Dead catch
3 = Retained catch
99 = Input harvest rate (F)
COND: == -1 Forecasted catches - enter one line per number of fixed forecast year catch
2012 1 1 1200 2 Year & Season & Fleet & Catch or F value & Basis
2013 1 1 1400 3 Year & Season & Fleet & Catch or F value & Basis
-9999 1 1 0000 2 Indicates end of inputted catches to read
COND: > 0 Forecasted catches - enter one line per number of fixed forecast year catch
2012 1 1 1200 Year & Season & Fleet & Catch or F value
2013 1 1 1200 Year & Season & Fleet & Catch or F value
-9999 1 1 0000 Indicates end of inputted catches to read
999 End of Input
End of Forecast File
26
Benchmark Calculations
This feature of SS is designed to calculate an equilibrium fishing rate intended to serve as a proxy for
the fishing rate that would provide maximum sustainable yield (MSY). Then in the forecast module
these fishing rates can be used in the projections.
Four reference points can be calculated by SS:
•FMSY: Search for the F that produces maximum equilibrium (e.g. dead catch), or set FMSY equal
to one of the other three options
•FSPR: Search for the F that produces spawning biomass per recruit this is a specific fraction,
termed SPRtarget, of spawning biomass per recruit under unfished conditions. Note that this is
in relative terms so it does not take into account the spawner-recruit relationship.
•FBtarget: Search for the F that produces an absolute spawning biomass that is a specified fraction,
termed relative biomass target, of the unfished spawning biomass. Note that this is in absolute
terms so takes into account the spawner-recruit relationship.
•F0.10: Search for the F that produces a slope in yield per recruit, dY/dF, that is 10% of the slope at
the origin. Note that this option is mutually exclusive with FBtarget. Only one will be calculated
and the one that is calculated can serve as the proxy for FMSY and forecasting.
Estimation
Each of the potential reference points is calculated by searching across a range of F multiplier levels,
calculating equilibrium biomass and catch at that F, using Newton-Raphson method to calculate a
better F multiplier value, and iterating a fixed number of times to achieve convergence on the desired
level.
Calculations
The calculation of equilibrium biomass and catch uses the same code that is used to calculate the virgin
conditions and the initial equilibrium conditions. This equilibrium calculation code takes into account
all morph, timing, biology, selectivity, and movement conditions as they apply while doing the time
series calculations. You can verify this by running SS to calculate FMSY then hardwire initial F to equal
this value, use the F_method approach 2 so each annual F is equal to FMSY and then set forecast F to be
the same FMSY. Then run SS without estimation and no recruitment deviations. You should see that the
population has an initial equilibrium abundance equal to BMSY and stays at this level during the time
series and forecast.
27
Catch Units
For each fleet, SS always calculates catch in terms of biomass (mt) and numbers (1000s) for encountered
(selected) catch, dead catch, and retained catch. These three categories differ only when some fleets
have discarding or are designated as a bycatch fleet. SS uses total dead catch biomass as the quantity
that is principally reported and the quantity that is optimized when searching for FMSY. The quantity
“dead catch” may occasionally be referred to as “yield”.
Biomass Units
The principle measure of fish abundance, for the purpose of reference point calculation, is female
reproductive output. This is referred to as SSB (spawning stock biomass) and sometimes just “B”
because the typical user settings have one unit of reproductive output (fecundity) per kg of mature
female biomass. So when the output label says BMSY, this is actually the female reproductive output at
the proxy for FMSY.
Fleet Allocation
An important concept for the reference point calculation is the allocation of fishing rate among fleets.
Internally, this is Bmark_relF(f, s) and it is the fraction of the F multiplier assigned to each fleet, fand
season, s. The value, F_multiplier * Bmark_relF(f, s), is the F level for a particular fleet in a particular
season and for the age that has a selectivity of 1.0. Other ages will have different F values according to
their selectivity.
•The Bmark_relF values can be calculated by SS from a range of years specified in the input for
Benchmark Years or it can be set to be the same as the Forecast_RelF, which in turn can be based
on a range of years or can be input as a set of fixed values.
•Note that for Bycatch Fleets, the F’s calculated by application of Bmark_relF for a bycatch fleet
can be overridden by a F value calculated from a range of years or a fixed F value that is input by
the user. If such an override is selected for a bycatch fleet, that F value is not adjusted by changes
to the F multiplier. This allows the user to treat a bycatch fleet as a constant background F while
the optimal F for other fleets is sought. Also for bycatch fleets, there is user control for whether
or not the dead catch from the bycatch fleet is included in the total dead catch that is optimized
when searching for FMSY.
Virgin vs. Unfished
The concept of unfished spawning biomass, SSB_unf, is important to the reference points
calculations. Unfished spawning biomass can be potentially different than virgin spawning biomass,
SSB_virgin.
•Virgin spawning biomass is calculated from the parameter values associated with the start year
of the model configuration and it serves as the basis from which the population model starts and
28
the basis for calculation of stock depletion.
•Unfished spawning biomass can be calculated for any year or range of years, so can change over
time as R0, steepness, or biological parameters change.
•In the reference points calculation, the Benchmark Years input specifies the range of time over
which various quantities are averaged to calculate the reference points. For biology, selectivity,
F’s, and movement the values being averaged are the year-specific derived quantities. But for the
SRparms (R0 and steepness), the parameter values themselves are averaged over time.
•During the time series or forecast, the current year’s unfished spawning output (SSB_unf) is
used as the basis for the spawner-recruitment curve against which deviations from the
spawner-recruitment curve are applied. So if R0 is made time-varying, then the
spawner-recruit curve itself is changed. However, if the regime shift parameter is time-varying,
then this is an offset from the spawner-recruitment curve and not a change in the curve itself.
So changes in R0 will change year-specific reference points and change the expected value for
annual recruitments, but changes in regime shift parameter only change the expected value for
annual recruitments.
•In reporting the time series of depletion level, the denominator can be based on virgin spawning
output (SSB_virgin) or BMSY. Note that BMSY is based on unfished spawning output (SSB_unf)
for the specified range of Benchmark years, not on SSB_virgin.
Forecast Recruitment Adjustment
Recruitment during the forecast years sometimes needs to be set at a level other than that determined
by the spawner-recruitment curve. One way to do this is by an environmental or block effect on the
regime shift parameter. A more straightforward approach is now provided by the special forecast
recruitment feature described here. There are 4 options provided for this feature. These are:
•0 = Do nothing. This is the default and will invoke no special treatment for the forecast
recruitments.
•1 = Multiplier on spawner-recruitment: The expected recruitment from the SRR is multiplied
by this factor.
–This is a multiplier, so null effect comes from a value of 1.0;
–The order of operations is to apply the SRR, then the regime effect, then this special forecast
effect, then bias adjustment, then the deviations;
–In the spawner recruit output of the report.sso there are 4 recruitment values stored.
29
•2 = Multiplier on virgin recruitment: The virgin recruitment is multiplied by this factor.
–This is a multiplier, so null effect comes from a value of 1.0;
–The order of operations is to apply any environmental or block effects to R0, then apply
the special forecast effect, then bias adjustment, then the deviations;
–Note that environmental or block effects on R0 are rare and are different than environment
or block effects on the regime parameter.
•3 = Mean recent recruitment: calculate the mean recruitment and use it.
–Note that bias adjustment is not applied to this mean because the values going into the
mean have already been bias adjusted.
This feature affects the expected recruitment in all years after the last year of the main recruitment
deviations. This means that if the last year of main recruitment deviations is before end year, then
the last few recruitments, termed “late”, are also affected by this forecast option. For example, option
3 would allow you to set the last 2 years of the time series and all forecast years to have recruitment
equal to the mean recruitment for the last 10 years of the main recruitment era.
Data File
Overview of Data File
1. Dimensions (years, ages, N fleets, N surveys, etc.)
2. Fleet and survey names, timing, etc.
3. Catch amount (biomass or numbers)
4. Discard
5. Mean body weight or mean body length
6. Length composition set-up
7. Length composition
8. Age composition set-up
9. Age imprecision definitions
30
10. Age composition
11. Mean length-at-age or mean bodyweight-at-age
12. Generalized size composition (e.g. weight frequency)
13. Tag-recapture
14. Stock composition (e.g. morphs ID’ed by otolith microchemistry)
15. Environmental data
16. Selectivity observations (new placeholder, not yet implemented)
Units of Measure
The normal units of measure are as follows:
•Catch biomass – metric tons
•Body weight – kilograms
•Body length – usually in centimeters. Weight at length parameters must correspond to the units
of body length and body weight.
•Survey abundance – any units if catchability (Q) is freely scaled; metric tons or thousands of fish
if Q has a quantitative interpretation
•Output biomass – metric tons
•Numbers – thousands of fish, because catch is in metric tons and body weight is in kilograms
•Spawning biomass – metric tons of mature females if eggs/kg = 1 for all weights; otherwise has
units that are based on the user-specified fecundity
Time Units
•Spawning is restricted to happening once per year at a specified date (in real months).
•Recruitment happens at specified recruitment events that occur at user-specified dates (in real
months).
31
•There can be 1 to many recruitment events; each producing a platoon as a portion of the total
recruitment.
•A settlement platoon enters the model at age 0 if settlement is between the time of spawning
and the end of the year; it enters at age 1 if settlement is after the first of the year; these ages at
settlement can be overridden in the settlement setup
•All fish advance to the next older integer age on January 1, no matter when they were born during
the year. Consult with your ageing lab to assure consistent interpretation.
•Time-varying parameters are allowed to change annually, not seasonally.
•Rates like growth and mortality are per year.
Seasons
•Seasons are the time step during which constant rates apply
•Catch and discard amounts are per season and F is calculated per season
•The year can have just 1 annual season, or be subdivided into seasons of unequal length.
•Season duration is input in real months and is converted into fractions of an annum. Annual
rate values are multiplied by the per annum season duration.
•If the sum of the input season durations is not close to 12.0, then the input durations is divided
by 12. This allows for a special situation in which the year could be only 0.25 in duration (e.g.
seasons as years) so that spawning and time-varying parameters can occur more frequently.
Subseasons and Timing of events in SS v.3.30
SS v.3.24 and all earlier versions effectively had two subseasons per season because the
age-length-key (ALK) for each observation used the mid-season mean length-at-age and spawning
occurred at the beginning of a specified season. Subseasons in SS v.3.30 provide more precision in
the timing of events.
•Even number (min = 2) of subseasons per season (regardless of season duration):
–2 subseasons will mimic SS v.3.24
–Specifying more sub seasons will give finer temporal resolution, but will slow the model
32

down, the effect of which is mitigated by only calculating growth as needed.
•Survey timing is now cruise-specific and specified in units of months (e.g. April 15 = 4.5).
–sstrans.exe will convert year, season in 3.24 format to year, real month in 3.30 format.
•Survey integer season and spawn integer season assigned at runtime based on real month and
season duration(s).
•The closest subseason is calculated for each observation.
•Growth and the age-length-key (ALK) is only calculated at beginning and mid-season or when
there is an observation in that subseason.
•Fishery body weight uses mid-subseason growth.
•Survey body weight and size composition is calculated using the nearest subseason.
•Reproductive output now has specified spawn timing (in months fraction) and interpolates
growth to that timing.
•Survey numbers calculated at cruise survey timing using e−z.
•Continuous Z for entire season. Same as applied in version 3.24.
Terminology
Where the term “COND” appears in the value column of this documentation (it does not actually
appear in the data file), it indicates that the following section is omitted except under certain
conditions, or that the factors included in the following section depend upon certain conditions. In
most cases, the description in the definition column is the same as the label output to the data.ss_new
file.
Model Dimensions
Value Description
#V3.30.XX.XX Model version number. This is written by SS in the new files and a good
idea to keep updated in the input files.
33

#C data using new
survey
Data file comment. Must start with #C to be retained then written to top
of various output files. These comments can occur anywhere in the data
file, but must have #C in columns 1-2.
1971 Start year
2001 End year
1 Number of seasons per year
12 Vector with N months in each season. These do not need to be integers.
Note: If the sum of this vector is close to 12.0, then it is rescaled to sum
to 1.0 so that season duration is a fraction of a year. But if the sum is not
close to 12.0, then the entered values are simply divided by 12. So with one
season per year and 3 months per season, the calculated season duration
will be 0.25, which allows a quarterly model to be run as if quarters are
years. All rates in SS are calculated by season (growth, mortality, etc.) using
annual rates * season duration.
2The number of subseasons. Entry must be even and the minimum value
is 2. This is for the purpose of finer temporal granularity in calculating
growth and the associated age-length key.
1.5 Spawning month; spawning biomass is calculated at this time of year
(1.5 means January 15) and used as basis for the total recruitment of all
settlement events resulting from this spawning.
2 Number of sexes (1/2)
20 Number of ages. The value here will be the plus-group age. SS starts at age
0.
1 Number of areas
2 Total number of fishing and survey fleets (which now can be in any order).
34
Season Length and Time Steps
Time steps in SS v3.30 can have finer granularity compared to previous versions where season can be
broken into subseason and the age-length key (ALK) can be calculated multiple times over the course
of a year:
ALK ALK* ALK* ALK ALK* ALK
Subseason 1 Subseason 2 Subseason 3 Subseason 4 Subseason 5 Subseason 6
ALK* only re-calculated when there is a survey that subseason
Quantites are calculated at the following times:
•Continuous Z for entire season;
•Even number (min = 2) of subseasons per season (regardless of season duration);
•Fishery body weight uses mid subseason ALK;
•Spawning biomass has specified spawn_timing (in months.fraction); uses closest ALK to that
timing;
•Survey timing is now observation-specific and specified in units of months.fraction (Apr 15 =
4.5);
•Survey season and spawn season assigned at runtime based on month and on season duration(s);
•Survey body weight and length composition uses closest ALK to survey timing;
•Survey numbers calculated at survey timing using e-Z.
Fleet Definitions
The catch data input has been modified to improve the user flexibility to add/subtract fishing and
survey fleets to a model set-up. The fleet setup input is transposed so each fleet is now a row. Previous
versions (3.24 and earlier) required that fishing fleets be listed first followed by survey only fleets. In
version 3.30 all fleets now have the same status within the model structure and each has a specified fleet
type (except for models that use tag recapture data, this will be corrected in future versions). Available
types are: catch fleet, bycatch only, or survey.
35

Inputs that define the fishing and survey fleets:
2 Number of fleets which includes survey in any order
Fleet Type Timing Area Catch Units Catch Mult. Fleet Name
1 -1 1 1 0 FISHERY1
3 1 1 2 0 SURVEY1
Fleet Type
Define the fleet type (e.g., fishery fleet, survey fleet):
•1 = fleet with input catches,
•2 = bycatch fleet (all catch discarded),
•3 = survey: assumes no catch removals even if associated catches are specified below. If you
would like to remove survey catch set fleet type to option = 1 with specific month timing for
removals (defined below in Timing),
•4 = ignored (not yet implemented).
Timing
Timing for data observations has been revised in v3.30:
•Fishery = -1 treat as catch occurred over the whole season or a user can override this
assumption by using the code 10XX (e.g 1007 would indicate that catch was removed mid-year
in July). Fishery fleets can either have a -1 which means that CPUE and composition
observations default to using the total seasonal catch-at-age and midseason length-at-age, or
they can have a timing value of 1 (actually any positive value) in which case the expected value
for CPUE and composition observations will be sampled at the time indicated by the month
value associated with the observation. If the -1 code is entered here, then individual
observations (e.g., compositional data) can override the midseason default by entering the
month as 1000+month. For example, 1004.5 would be entered for a mid-April observation.
•Survey = 1 The fleet timing here for surveys is not used and only the month value with the
observation is relevant (e.g., month specification in the indices of abundance or the month for
composition data).
36

Area
An integer value indicating the area in which a fleet operates.
Catch Units
Ignored for survey fleets, their units are read later:
•1 = biomass (in metric tons),
•2 = numbers (thousands of fish).
See Units of Measure for more information.
Catch Multiplier
Invokes use of a catch multiplier, which is then entered as a parameter in the MG parameter section.
The estimated value or fixed value of the catch multiplier is multiplied by the estimated catch before
being compared to the observed catch:
•0 = No catch multiplier used,
•1 = Apply a catch multiplier which is defined as an estimable parameter in the control file after
the cohort growth deviation in the biology parameter section. The model’s estimated retained
catch will be multiplied by this factor before being compared to the observed retained catch.
Optional Bycatch Fleets
The option to include bycatch fleets was introduced in Stock Synthesis version 3.30.10. This is an
optional input and if no bycatch is to be included in the catches this section can be ignored.
If a fleet above was set as a bycatch fleet (fleet type = 2), the following line is required:
Optional inputs that define bycatch fleet:
Fleet Include F or F or
Index in MSY Fmult First Year Last Year Not used
1 1 1 1982 2010 999
37
Fleet Index
Fleet to include bycatch catch for (fleet number). Fleet number is assigned within the model based on
the order of listed fleets in the Fleet Definition section. If there are multiple bycatch fleets, then a line
for each fleet is required in the bycatch section.
Include in MSY
Options:
•1 = deadfish in MSY, ABC, and other benchmark and forecast output,
•2 = omit from MSY and ABC (but still include the mortality).
Fmult
Options:
•1 = F multiplier scales with other fleets,
•2 = Bycatch F constant at input value,
•3 = Bycatch F from range of years.
F or First Year
F or first year of range.
F or Last Year
F or last year of range.
Not Used
This column is not yet used and is reserved for future features.
Catch
After reading the fleet-specific indicators, a list of catch values by fleet and season are read in by the
model. The format for the catches is year, season that the catch will be attributed to, fleet, a catch value,
and a year specific catch standard error. Only positive catches need to be entered, so there is no need
for records for the survey fleets. To include an equilibrium catch value the year should be noted as
-999 and this is now season specific . There is no longer a need to specify the number of records to be
38
read; instead the list is terminated by entering a record with the value of -9999 in the year field. The
updated list based approach extends throughout the data file (e.g. catch, length- and age-composition
data), the control file (e.g. lambdas), and the forecast file (e.g. total catch by fleet, total catch by area,
allocation groups, forecasted catch).
In addition, it is possible to collapse the number of seasons. So if a season value is greater than the
N seasons for a particular model, that catch is added to the catch for N seasons. This is generally to
collapse a seasonal model into an annual model. In a seasonal model, use of season = 0 will cause
SS to distribute the input value of catch equally among the N seasons. SS assumes that catch occurs
continuously over seasons and hence is not specified as month in the catch data section. However, all
other data types will need to be specified by month.
The new format for version 3.30 for a 2 season model with 2 fisheries looks like the table below. The
example is sorted by fleet, but the sort order does not matter. In data.ss_new, the sort order is fleet,
year, season.
Catches by year, season for every fleet:
Year Season Fleet Catch Catch SE
-999 1 1 56 0.05
-999 2 1 62 0.05
1975 1 1 876 0.05
1975 2 1 343 0.05
... ... ... ... ...
-999 1 2 55 0.05
-999 2 2 22 0.05
1975 1 2 555 0.05
1975 2 2 873 0.05
... ... ... ... ...
-9999 0 0 0 0
•Catch can be in terms of biomass or numbers for each fleet.
•Catch is retained catch. If there is discard also, then it is handled in the discard section below.
This is the recommended setup which results in a model estimated retention curve based upon
the discard data (specifically discard composition data). However, there may be instances where
the data do not support estimation of retention curves. In these instances catches can be specified
as all dead (retained + discard estimates).
•If there is reason to believe that the retained catch values underestimate the true catch, then
it is possible in the retention parameter set up to create the ability for the model to estimate
the degree of unrecorded catch. However, this is better handled with the new catch multiplier
39
option.
Bycatch
Bycatch fleets have an F so impose mortality and catch fish. All this catch is discarded. There must
be a value entered for retained catch so that SS knows to calculate an F for that season, but this catch
amount is ignored in the log likelihood. The amount of discarded catch can be entered as a discard
observation(s). Bycatch fleets have selectivity, which must be specified or estimated if observations of
the size or age composition of the discards is entered.
•Because there is no retained catch amount to match, the F for bycatch only fleets must be by th
continuous F method (F_method = 2).
•MSY and yield per recruit are calculated in terms of dead catch, and they currently include catch
from bycatch fleets. So the search for Fmsy scales the bycatch F along with the F for the fleets
that retain catch.
Indices
Indices are data that are compared to aggregate quantities in the model. Typically the index is a measure
of fish abundance, but this data section also allows for the index to be related to a fishing fleet’s F, or
to another quantity estimated by the model. The first section of the Indices setup contains the fleet ID,
Units, and error distribution for each fleet that has index data.
CPUE and Suvey Abundance Observations:
Fleet/ Error
Survey Units Distribution SD Report
1 1 0 0
2 1 0 0
... ... ... ...
Units
Options:
•0 = numbers,
•1 = biomass,
40
•2 = F.
–Note the F option can only be used for a fishing fleet and not for a survey, even if the
survey selectivity is mirrored to a fishing fleet. The values of these effort data are
interpreted as proportional to the level of the fishery F values. No adjustment is made for
differentiating between continuous F values versus exploitation rate values coming from
Pope’s approximation. A normal error structure is recommended so that the input effort
data are compared directly to the model’s calculated F, rather than to loge(F). The
resultant proportionality constant has units of 1/Q. The options for units are:
•>=30 special survey types. These options bypass the calculation of survey selectivity so no
selectivity parameters should be entered and especially not estimated. The expected values for
these types are:
–30 = spawning biomass (e.g. for an egg and larvae survey),
–31 = exp(recruitment deviation), useful for environmental index affecting recruitment,
–32 = spawning biomass * exp(recruitment deviation), for a pre-recruit survey occurring
before density-dependence,
–33 = recruitment, age-0 recruits,
–34 = depletion (spawning biomass/virgin spawning biomass). Special survey option 34
automatically adjusts phases of parameters. There are options for additional control over
this in the control file Q setup section under the link information column where:
*0 = add 1 to phases of all parameters; only R0 active in new phase 1; mimics the default
option of previous model versions,
*1 = only R0 active in phase 1; then finish with no other parameters becoming active;
useful for data-limited draws of other fixed parameters. Essentially, this option allows
SS to mimic DB-SRA,
*2 = no phase adjustments, can be used when profiling on fixed R0.
–35 = survey of a deviation vector (e(survey(y)) = f(parm_dev(k, y))), can be used for
an environmental time-series with soft linkage to the index. The selected deviation vector
is specified in Q section of the control file. The index of the dev vector to which the index
is related is specified in the 2nd column of the Q setup table (see Catchability).
Error Distribution
Options:
41

•-1 = normal error,
•0 = lognormal error,
•>0 = Student’s t-distribution in log space with degrees of freedom equal to this value. For DF>30,
results will be nearly identical to that for lognormal distribution. A DF value of about 4 gives a
fat-tail to the distribution (see Chen (2003)). The se values entered in the data file must be the
standard error in logespace.
Abundance indices typically have a lognormal error structure with units of standard error of
loge(index). If the variance of the observations is available only as a CV, then the value of se can be
approximated as q(loge(1 + (CV )2)) where CV is the standard error of the observation divided by
the mean value of the observation.
For the normal error structure, the entered values for se are interpreted directly as a se in arithmetic
space and not as a CV. Thus switching from a lognormal to a normal error structure forces the user to
provide different values for the se input in the data file.
If the data exist as a set of normalized Z-scores, you can either: assert a lognormal error structure after
entering the data as exp(Z-score) because it will be logged by SS. Preferably, the Z-scores would be
entered directly and the normal error structure would be used.
Enable SD Report
Indices with SD Report enabled will have the expected values for their historical values appear in the
ss.std and ss.cor files. The default value is for this option is 0.
•0 = SD Report not enabled for this index,
•1 = SD Report enabled for this index.
Data Format
Year Month Fleet/Survey Observation SE
1991 7 3 80000 0.056
1995 7.2 3 65000 0.056
... ... ... ... ...
2000 7.1 3 42000 0.056
-9999 1 1 1 1
42
•For fishing fleets, catch-per-unit-effor (CPUE) is defined in terms of retained catch (biomass or
numbers).
•For fishery independent surveys, retention/discard is not defined so CPUE is implicitly in terms
of total CPUE.
•If a survey has its selectivity mirrored to that of a fishery, only the selectivity is mirrored so the
expected CPUE for this mirrored survey does not use the retention curve (if any) for the fishing
fleet.
•If the fishery or survey has time-varying selectivity, then this changing selectivity will be taken
into account when calculating expected values for the CPUE or survey index.
•Year values that are before start year or after end year are excluded from model, so the easiest
way to include provisional data in a data file is to put a negative sign on its year value.
•Duplicate survey observations are not allowed.
•Observations can be entered in any order, except if the super-year feature is used.
•Observations that are to be included in the model but not included in the negative log likelihood
need to have a negative sign on their fleet ID. Previously the code for not using observations was
to enter the observation itself as a negative value. However, that old approach prevented use of a
Z-score environmental index as a “survey”. This approach is best for single or select years from
an index rather than an approach to remove a whole index. Removing a whole index from the
model should be done through the use of lambdas at the bottom of the control file which will
eliminate the index from model fitting.
•Super-periods are turned on and then turned back off again by putting a negative sign on the
season. Previously, super-periods were started and stopped by entering -9999 and the -9998 in
the SE field. See the "Data Super-Period” section of this manual for more information.
•Research Note: If the statistical analysis used to create the CPUE index of a fishery has been
conducted in such a way that its inherent size/age selectivity differs from the size/age selectivity
estimated from the fishery’s size and age composition, then you may want to enter the CPUE as if
it was a separate survey and with a selectivity that differs from the fishery’s estimated selectivity.
The need for this split arises because the fishery size and age composition should be derived
through a catch-weighted approach (to appropriately represent the removals by the fishery) and
the CPUE should be derived through an area-weighted approach to better serve as a survey of
stock abundance.
43
Discard
If discard is not a feature of the model specification, then just a single input is needed:
0 Number of fleets with discard observations
If discard is being used, the input syntax is:
1 Number of fleets with discard observations
Fleet Units Error Distribution
1 2 -1
Year Month Fleet Observation Standard Error
1980 7 1 0.05 0.25
1991 7 1 0.10 0.25
-9999 1 1 0 0
Discard Units
Options:
•1 = values are amount of discard in either biomass or numbers according to the selection made
for retained catch,
•2 = values are fraction (in biomass or numbers) of total catch discarded; bio/num selection
matches that of retained catch,
•3 = values are in numbers (thousands) of fish discarded, even if retained catch has units of
biomass.
Discard Error Distribution
The four options for discard error are:
•>0 = degrees of freedom for Student’s t-distribution used to scale mean body weight deviations.
Value of error in data file in interpreted as CV of the observation,
•0 = normal distribution, value of error in data file is interpreted as CV of the observation,
•-1 = normal distribution, value of error in data file is interpreted as standard error of the
44
observation,
•-2 = lognormal distribution, value of error in data file is interpreted as standard error of the
observation in log space,
•-3 = truncate normal distribution (new with 3.30, needs further testing), value of error in data
file is interpreted as standard error of the observation. This is a good option for low observed
discard rates.
Discard Notes
•Since discard refers to catch, its time units are in seasons, not months.
•Year values that are before start year or after end year are excluded from model, so the easiest
way to include provisional data in a data file is to put a negative sign on its year value.
•Negative value for fleet causes it to be included in the calculation of expected values, but excluded
from the log likelihood.
•Zero (0.0) is a legitimate discard observation, unless lognormal error structure is used.
•Duplicate discard observations are not allowed.
•Observations can be entered in any order, except if the super-period feature is used.
•Note that in the control file you will enter information for retention such that 1-retention is
the amount discarded. All discard is assumed dead, unless you enter information for discard
mortality. Retention and discard mortality can be either size-based or age-based (new with
v.3.30).
Cautionary Note
The use of CV as the measure of variance can cause a small discard value to appear to be overly precise,
even with the minimum standard error of the discard observation set to 0.001. In the control file, there
is an option to add an extra amount of variance. This amount is added to the standard error, not to the
CV, to help correct this problem of underestimated variance.
Mean Body Weight or Length
This is the overall mean body weight or length across all selected sizes and ages. This may be useful in
situations where individual fish are not measured but mean weight is obtained by counting the number
45

of fish in a specified sample, e.g. a 25 kg basket. Observations can be in terms of mean length by setting
switching the partition code to a negative value (e.g. -0, -1, -2) rather than 0, 1, and 2 typically used
with the mean body weight approach.
Mean Body Weight Data Section:
1 Use mean body size data (0/1)
30 Degrees of freedom for Student’s t-distribution used to evaluate mean
body weight deviation. This is not a conditional input, must be here even if there
are no mean body weight observations.
Year Month Fleet Partition Type Observation Standard Error
1990 7 1 0 1 4.0 0.95
1990 7 1 0 1 1.0 0.95
-9999 0 0 0 0 0 0
Partition
Mean weight data and composition data require specification of what group the sample originated
from (e.g. discard, retained, discard + retained).
•0 = whole catch in units of weight (discard + retained),
•1 = discarded catch in units of weight,
•2 = retained catch in units of weight,
•-0 = whole catch in units of length (discard + retained),
•-1 = discarded catch in units of length,
•-2 = retained catch in units of length.
Type
Specify the type of data:
•1 = mean length,
•2 = mean body weight.
46

Observation - Units
Units must correspond to the units of body weight, normally in kilograms, (or mean length in cm). The
expected value of mean body weight (or mean length) is calculated in a way that incorporates effect of
selectivity and retention.
Error
Error is entered as the CV of the observed mean body weight (or mean length)
Population Length Bins
The first part of the length composition section sets up the bin structure for the population. These
bins define the granularity of the age-length key and the coarseness of the length selectivity. Fine bins
create smoother distributions, but a larger and slower running model. First read a single value to select
one of three population length bin methods, then any conditional input for options 2 and 3:
1 Use data bins to be read later. No additional input here.
2 generate from bin width min max, read next:
2 Bin width
10 Lower size of first bin
82 Lower size of largest bin
The number of bins is then calculated from: (max Lread - min Lread)/(bin width) + 1
3 Read 1 value for number of bins, and then read vector of bin boundaries
25 Number of population length bins to be read
26 28 30 ... Vector containing lower edge of each population
size bin
Notes
For option 2, bin width should be a factor of min size and max size. For options 2 and 3, the population
length bins must not be wider than the length data bins, but the boundaries of the bins do not have to
align. The transition matrix between population and data length bins is output to echoinput.sso.
The mean size at settlement (virtual recruitment age) is set equal to the min size of the first population
length bin.
When using more, finer population length bins, SS will create smoother length selectivity curves and
smoother length distributions in the age-length key, but run slower (more calculations to do).
47

The mean weight-at-length, maturity-at-length and size-selectivity are based on the mid-length of the
population bins. So these quantities will be rougher approximations if broad bins are defined.
Provide a wide enough range of population size bins so that the mean body weight-at-age will be
calculated correctly for the youngest and oldest fish. If the growth curve extends beyond the largest
size bin, then these fish will be assigned a length equal to the mid-bin size for the purpose of
calculating their body weight.
While exploring the performance of models with finer bin structure, a potentially pathological
situation has been identified. When the bin structure is coarse (note that some applications have used
10 cm bin widths for the largest fish), it is possible for a selectivity slope parameter or a retention
parameter to become so steep that all of the action occurs within the range of a single size bin. In this
case, the model will see zero gradient of the log likelihood with respect to that parameter and
convergence will be hampered.
ALK Tolerance: A value read near the end of the starter.ss file defines the degree of tail compression
used for age-length key. If this is set to 0.0, then no compression is used and all cells of the age-length
key are processed, even though they may contain trivial (e.g. 1 e-13) fraction of the fish at a given age.
With tail compression of, say 0.0001, SS will at the beginning of each phase calculate the min and max
length bin to process for each age of each morphs ALK and compress accordingly. Depending on how
many extra bins are outside this range, you may see speed increases near 10-20%. Large values of ALK
tolerance, say 0.1, will create a sharp end to each distribution and likely will impede convergence. Try
out ALK tolerance.
Length Compostion Data Structure
Enter a code to indicate whether or not length composition data will be used:
1 Use length composition data (0/1)
If the value 0 is entered, then skip all length related inputs below and skip to the age data setup
section. Otherwise continue:
Specify bin compression and error structure for length composition data for each fleet:
Min. Constant Combine Comp. Dirichlet Min.
Tail added males & Compress. Error Param. Sample
Compress. to prop. females Bins Dist. Select Size
0 0.0001 0 0 0 0 1
0 0.0001 0 0 0 0 1
48
Minimum Tail Compression
Compress tails of composition until observed proportion is greater than this value; negative value
causes no compression; Advise using no compression if data are very sparse, and especially if the set-up
is using agecomp within length bins because of the sparseness of these data.
Added Constant to Proportions
Constant added to observed and expected proportions at length and age to make logL calculations
more robust. Tail compression occurs before adding this constant. Proportions are renormalized to
sum to 1.0 after constant is added.
Combine Males % Females
Combine males into females at or below this bin number. This is useful if the sex determination of
very small fish is doubtful so allows the small fish to be treated as combined sex. If Combine Males
& Females > 0, then add males into females for bins 1 thru this number, zero out the males, set male
data to start at the first bin above this bin. Note that Combine Males & Females > 0 is entered as a
bin index, not as the size associated with that bin. Comparable option is available for age composition
data.
Compress Bins
This options allows for the compression of length or age bins beyond a specific length or age by each
data source. As an example, a value of 5 in the compress bins column would condense the final five
length bins for the specified data source.
Composition Error Distribution
Options:
•0 = Multinomial Error,
•1 = Dirichlet Multinomial Error.
–The Dirichlet Multinomial Error distribution requires the addition of a parameter lines
for the natural log of the effective sample size multiplier (θ) at the end of the selectivity
parameter section in the control file. Click here to see the parameter setup in the control file.
–The Parameter Select option needs be used to specified which data sources should be
weighted together or separate.
Dirichlet Parameter Select
Value that indicates the groups of composition data for estimation of the Dirichlet parameter for
49

weighting composition data.
•0 = Default,
•1-N = Only used for the Dirichlet option. Set to a sequence of numbers from 1 to N where N
is the total number of combinations of fleet and age/length. That is, if you have 3 fleets with
length data, but only 2 also have age data, you would have values 1 to 3 in the length comp setup
and 4 to 5 in the age comp setup. You can also have a data weight that is shared across fleets by
repeating values in Parameter Select.
Minimum Sample Size
The minimum value (floor) for all sample sizes. This value must be at least 0.001. Conditional
age-at-length data may have observations with sample sizes less than 1. Stock Synthesis 3.24 has an
implicit minimum sample size value of 1.
Dirichlet Parameter Number and Effective Sample Sizes
If the Dirichlet multinomial error distribution is selected, indicate here which of a list of Dirichlet
multinomial parameters will be used for this fleet. So each fleet could use a unique Dirichlet
multinomial parameter, or all could share the same, or any combination of unique and shared. The
requested number of Dirichlet multinomial parameters will be read from the control file. Please note
that age-compositions Dirichlet multinomial parameters are continued after length-compositions, so
a model with one fleet and both data types would presumably require two new Dirichlet multinomial
parameters.
The Dirichlet estimates the effective sample size as Neff =1
1+θ+Nθ
1+θwhere θis the estimated
parameter and Nis the input sample size. Stock Synthesis estimates the log of the Dirichlet
multinomial parameter such that ˆ
θfishery =e−0.6072 = 0.54 where assuming N= 100 for the fishery
would result in an effective sample size equal to 35.7.
This formula for effective sample size implies that, as the Stock Synthesis parameter log_Theta goes
to large values (i.e., 20), then Neff will converge to the input sample size (Ninput). In this case, small
changes in the value of the log_Theta parameter has no action, and the derivative of the negative
log-likelihood is zero with respect to the parameter, which means the Hessian will be singular and
cannot be inverted. To avoid this non-invertible Hessian when the log_Theta parameter becomes
large, turn it off while fixing it at the high value. In summary, we recommend setting the upper bound
for the Dirichlet multinomial parameter log_Theta to a high value (i.e., 20-25), and then if any fleet
has an estimate of log_Theta >15 then turn that Dirichlet multinomial parameter off while starting
it at the estimated high value. This is equivalent to turning off down-weighting of fleets where
evidence suggests that Neff =Ninput.
50
For additional information about the Dirichlet multinomial please see Thorson et al. 2017.
Model-based estimates of effective sample size in stock assessment models using the Dirichlet
multinomial distribution. Fisheries Research, 192: 84-93.
Length Composition Data
Specify the length compostion data:
30 Number of length bins for data
26 28 30 ... 88 90 Vector of length bins associated with the length data
Example of a single length composition observation:
Year Month Fleet Sex Partition Nsamp data vector
1986 1 1 3 0 20 <female then male data>
... ... ... ... ... ... ...
-9999 0 0 0 0 0 0
Sex
If model has only one sex defined in the set-up, all observations must have sex set equal to 0 or 1. In a
2 sex model, the data vector always has female data followed by male data, even if only one of the two
sexes has data that will be used.
•Sex = 0 means combined male and female (must already be combined and information placed in
the female portion of the data vector) (male entries must exist for correct data reading, then will
be ignored).
•Sex = 1 means female only (male entries must exist for correct data reading, then will be ignored).
•Sex = 2 means male only (female entries must exist and will be ignored after being read).
•Sex = 3 means data from both sexes will be used and they are scaled so that they together sum
to 1.0; i.e. sex ratio is preserved.
Partition
Partition indicates samples from either discards,retained, or combined.
51

•0 = combined,
•1 = discard,
•2 = retained.
Excluding Data
•If the value of year is negative, then that observation is not transferred into the working array.
This feature is the easiest way to include observations in a data file but not to use them in a
particular model scenario.
•If the value of fleet is negative, then the observation is processed and its expected value and log
likelihood is calculated, but this log likelihood is not included in the total log likelihood. This
feature allows the user to see the fit to a provisional observation without having that observation
affect the model.
Note
Version 3.30 no longer requires that the number of length composition data lines be specified. Entering
-9999 at the end of the data matrix will indicate to the model the end of length composition lines to be
read.
Each observation can be stored as one row for ease of data management in a spreadsheet and for sorting
of the observations. However, the 6 header values, the female vector and the male vector could each be
on a separate line because ADMB reads values consecutively from the input file and will move to the
next line as necessary to read additional values.
The composition observations can be in any order and replicate observations are allowed (unlike
survey and discard data). However, if the super-period approach is used, then each super-periods’
observations must be contiguous in the data file.
Age Composition Option
The age composition section begins by reading the number of age bins. If the value 0 is entered for the
number of age bins, then SS skips reading the bin structure and all reading of other age composition
data inputs.
17 Number of age’ bins; can be equal to 0 if age data not used; do not include a vector
of agebins if Nage’ bins is set equal to 0.
52

Age Composition Bins
If a positive number of age bins is read, then SS reads the bin definition next.
1 2 3 ... 20 25 Vector of ages
The bins are in terms of observed age (here age’) and entered as the lower edge of each bin. Each ageing
imprecision definition is used to create a matrix that translates true age structure into age’ structure.
The first and last age’ bins work as accumulators. So in the example any age 0 fish that are caught
would be assigned to the age’ = 1 bin.
Ageing Error
Here, the capability to create a distribution of age’ (e.g. age with possible bias and imprecision) from
true age is created. One or many age error definitions can be created. For each, there is input of a
vector of mean age’ and stddev of age’. For one definition, the input vectors can be replaced by vectors
created from estimable parameters. In the future, capability to read a full age’ – age matrix could be
created. The dimension of the ageing error matrix requires the column length match the population
maximum age specified at the top of the data file. However, the maximum age for binning of age data
may be lower that the population maximum age.
2 Number of ageing error matrices to generate
Age-0 Age-1 Age-2 ... Max Age
-1 -1 -1 ... -1 #Mean Age
0.001 0.001 0.001 ... 0.001 #SD
0.5 1.5 2.3 ... Max Age + 0.5 #Mean Age
0.5 0.65 0.67 ... 4.3 #SD Age
The above table shows the values for the first 3 ages for each of two age transition definitions: the first
defines a matrix with no bias and negligible imprecision and the second shows a small negative bias
beginning at age 2.
Note
•In principle, one could have year or laboratory specific matrices.
53

•For each matrix, enter a vector with mean age’ for each true age; if there is no ageing bias, then
set age’ equal to true age + 0.5. Alternatively, -1 value for mean age’ means to set it equal to true
age plus 0.5. The addition of +0.5 is needed so that fish will get assigned to the intended interger
age’.
•The length of the input vector is Nage+1, with the first entry being for age 0 fish and the last for
fish of age Nage. The following line is a a vector with the standard deviation (stddev) of age’ for
each true age.
•SS is able to create one ageing error matrix from parameters, rather than from an input vector.
The range of conditions in which this new feature will perform well has not been evaluated, so
it should be considered as a preliminary implementation and subject to modification.
–To invoke this option, for the selected ageing error vector, set the stddev of ageing error to a
negative value for age 0. This will cause creation of an ageing error matrix from parameters
and any age or size-at-age data that specify use of this age error pattern will use this matrix.
Then in the control file, add 7 parameters below the cohort growth dev parameter. These
parameters are described in the control file section of this manual.
Specify bin compression and error structure for age composition data for each fleet:
Min. Constant Combine Comp. Dirichlet Min.
Tail added males & Compress. Error Param. Sample
Compress. to prop. females Bins Dist. Select Size
0 0.0001 1 0 0 0 1
0 0.0001 1 0 0 0 1
Specify method by which length bin range for age obs will be interpreted:
1 Bin method for age data
1 = value refers to population bin index
2 = value refers to data bin index
3 = value is actual length (which must correspond to population length bin
boundary)
54
An example age composition observation:
Year Month Fleet Sex Partition Age
Err
Lbin
lo
Lbin
hi
Nsamp Data Vector
1987 1 1 3 0 2 -1 -1 79 <enter data
values>
-9999 0 0 0 0 0 0 0 0 0
Note
•Syntax for Sex, Partition, and data vector are same as for length.
•Ageerr identifies which ageing error matrix to use to generate expected value for this
observation.
•The data vector has female values then male values, just as for the length composition data.
•As with the length comp data, a negative value for year causes the observation to not be read
into the working matrix, a negative value for fleet causes the observation to be included in
expected values calculation, but not in contribution to total logL, a negative value for month
causes start-stop of super-period.
•Lbin lo, and Lbin hi are the range of length bins that this age composition observation refers to.
Normally these are entered with a value of -1 and -1 to select the full size range. Whether these
are entered as population bin number, length data bin number, or actual length is controlled by
the value of the length bin range method above.
–Entering value of 0 or -1 for Lbin lo converts Lbin lo to 1;
–Entering value of 0 or -1 for Lbin hi converts Lbin hi to Maxbin;
–It is strongly advised to use the “-1” codes to select the full size range. If you use explicit
values, then the model could unintentionally exclude information from some size range if
the population bin structure is changed.
–In reporting to the comp_report.sso, the reported Lbin_lo and Lbin_hi values are always
converted to actual length.
55
Conditional Age-at-Length
Use of conditional age’-at-length will greatly increase the total number of age’ composition
observations and associated model run time, but it is a superior approach for several reasons. First, it
avoids double use of fish for both age’ and size information because the age’ information is
considered conditional on the length information. Second, it contains more detailed information
about the relationship between size and age so provides stronger ability to estimate growth
parameters, especially the variance of size-at-age. Lastly, where age data are collected in a
length-stratified program, the conditional age’-at-length approach can directly match the protocols
of the sampling program.
In a two sex model, it is best to enter these conditional age’-at-length data as single sex observations
(sex =1 for females and = 2 for males), rather than as joint sex observations (sex = 3). Inputting joint
sex observations comes with a more rigid assumption about sex ratios within each length bin. Using
separate vectors with sex = 1 and 2 allows 100% of the expected comp to be fit to 100% observations
within each sex, whereas with sex=3, you would have a bad fit if the sex ratio were out of balance with
the model expectation, even if the observed proportion at age within each sex exactly matched the
model expectation for that age. Additionally, inputting the conditional age-at-length data as single sex
observations isolates the age composition data from any sex selectivity as well.
When Lbin_lo and Lbin_hi are used to select a subset of the total size range, the expected value for
these age’ data is calculated within that specified size range, so is age’ conditional on length.
Mean Length or Body Weight-at-Age
SS also accepts input of mean length-at-age’ or mean bodywt-at-age’. This is done in terms of age’, not
true age, to take into account the effects of ageing imprecision on expected mean size-at-age’. If the
value of “AgeErr” is positive, then the observation is interpreted as mean length-at-age’. If the value of
“AgeErr” is negative, then the observation is interpreted as mean bodywt-at-age’ and the abs(AgeErr)
is used as AgeErr.
1 Use mean size-at-age obsevation (0 = none, 1 = read data matrix)
An example observation:
Age Data Vector Sample Size
Yr Month Fleet Sex Part. Err. Ignore (Female - Male) (Female - Male)
1989 7 1 3 0 1 999 <Mean Size values> <Sample Sizes>
...
-9999 7 1 3 0 1 999
56

Note
•Negatively valued mean size entries with be ignored in fitting.
•Nfish value of 0 will cause mean size value to be ignored in fitting.
•Negative value for year causes observation to not be included in the working matrix.
•Each sexes’ data vector and N fish vector has length equal to the number of age’ bins.
•The "Ignore" column is not used but still needs to have default values in that column.
•Where age data are being entered as conditional age’-at-length and growth parameters are being
estimated, it may be useful to include a mean length-at-age vector with nil emphasis to provide
another view on the model’s estimates.
•An experiment that may be of interest might be to take the body weight-at-age data an enter it to
SS as empirical body wt-at-true age in the wtatage.ss file, and to contrast results to entering the
same body weight-at-age data here and to attempt to estimate growth parameters, potentially
time-varying, that match these body weight data.
Environmental Data
SS accepts input of time series of environmental data. Parameters can be made to be time-varying by
making them a function of one of these environmental time series.
Parameter values can be a function of an environmental data series:
1 Number of environmental variables
COND > 0 Example of 2 environmental observations:
Year Variable Value
1990 1 0.10
1991 1 0.15
-9999 0 0
Note
•Any years for which environmental data are not read are assigned a value of 0.0.
57
•It is permissible to include a year that is one year before the start year in order to assign
environmental conditions for the initial equilibrium year. But this works only for recruitment
parameters, not biology or selectivity parameters.
•Environmental data can be read for up to 100 years after the end year of the model. Then, if the
recruitment-environment link has been activated, the future recruitments will be influenced
by any future environmental data. This could be used to create a future “regime shift” by
setting historical values of the relevant environmental variable equal to zero and future values
equal to 1, in which case the magnitude of the regime shift would be dictated by the value of
the environmental linkage parameter. Note that only future recruitment and growth can be
modified by the environmental inputs; there are no options to allow environmentally-linked
selectivity in the forecast years.
•Note that some model derived quantities like summary biomass and recruitment deviation are
assigned to some negative valued environmental variables. This is a stepping stone towards
creating ability for parameters to be density-dependent.
Generalized Size Composition Data
A flexible feature with SS is a generalized approach to size composition information. It was designed
initially to provide a means to include weight frequency data, but was implemented to provide a
generalized capability. The user can define as many size frequency methods as necessary.
•Each method has a specified number of bins.
•Each method has "units" so the frequencies can be in units of biomass or numbers.
•Each method has “scale” so the bins can be in terms of weight or length (including ability to
convert bin definitions in pounds or inches to kg or cm).
•The composition data is input as females then males, just like all other composition data in SS.
So, in a two-sex model, the new composition data can be combined sex, single sex, or both sex.
•If a retention function has been defined, then the new composition data can be from the
combined discard + retained, discard only or retained only.
58
Example entry:
0 N of size frequency methods
COND > 0
25 15 #Nbins per method
2 3 #Units per each method (1 = biomass, 2 = numbers)
3 3 #Scale per each method (1 = kg, 2 = lbs, 3 = cm, 4 = inches)
1e-9 1e-9 #Min compression to add to each observation (entry for each method)
2 2 #N observations per weight frequency method
Then enter the lower edge of the bins for each method. The two row vectors shown
below contain the bin definitions for methods 1 and 2 respectively:
-26 28 30 32 34 36 38 40 42 ... 60 62 64 68 72 76 80 90
-26 28 30 32 34 36 38 40 42 44 46 48 50 52 54
Note
•There is no tail compression for generalized size frequency data.
•Super-period capability is enabled in same way as for length and age composition data.
•There are two options for treating fish that in population size bins that are smaller than the
smallest size frequency bin.
–Option 1: By default, these fish are excluded (unlike length composition data where the
small fish are automatically accumulated up into the first bin.
–Option 2: If the first size bin is given a negative value, then: accumulation is turned on and
the negative of the entered value is used as the lower edge of the first size bin;
•By choosing units=2 and scale=3, the size comp method can be nearly identical to the length
comp method if the bins are set identically;
•Bin boundaries can be real numbers so obviously do not have to align with population length
bin boundaries, SS interpolates as necessary;
•Size bins cannot be defined to be narrower than the population binwidth; an untrapped error
will occur;
•Because the transition matrix can depend upon weight-at-length, it is calculated internally for
59
each sex and for each season because weight-at-length can differ between sexes and can vary
seasonally.
An example observation is below. Note that its format is identical to the length composition data,
including sex and partition options, except for the addition of the first column to indicate the size
frequency method.
Sample <composition
Method Year Month Fleet Sex Part Size females then males>
1 1975 1 1 3 0 43 <data>
1 1977 1 1 3 0 43 <data>
1 1979 1 1 3 0 43 <data>
1 1980 1 1 3 0 43 <data>
Tag-Recapture Data
An ability to analyze tag-recapture data is available with SS. Each released tag group is characterized by
an area, time, sex and age at release. Each recapture event is characterized by a time and fleet. Because
SS fleet’s each operate in only one area, it is not necessary to record the area of recapture. Inside the
model, the tagged cohort is apportioned across all growth patterns in that area at that time (with options
to apportion to only one sex or to both). The tag cohort x growth pattern then behaves according to the
movement and mortality of that growth pattern. The number of tagged fish is modeled as a negligible
fraction of the total population. This means that a tagging event does not move fish from an untagged
group to a tagged group. Instead it acts as if the tags are seeded into the population with no impact at all
on the total population abundance or mortality. The choice to require assignment of a predominant age
at release for each tag group is a pragmatic coding and model efficiency choice. By assigning a tag group
to a single age, rather than distributing it across all possible ages according to the size composition of
the release group, it can be tracked as a single diagonal cohort through the age x time matrix with
minimal overhead to the rest of the model. Tags are considered to be released at the beginning of a
season (period) and recaptures follow the timing of the fleet that made the recapture.
60

Example set-up for tagging data:
1 #Do tags - if this value is 0, then omit all entries below
COND = 1 All subsequent tag-recapture entries must be omitted if "Do Tags" = 0
3 #Number of tag groups
12 #Number of recapture events
2 #Mixing latency period: N periods to delay before comparing
observed to expected recoveries (0 = release period)
10 #Max periods (months) to track recoveries, after which tags enter
accumulator
#Release Data
#TG Area Year Month <tfill> Sex Age N Release
1 1 1980 1 999 0 24 2000
2 1 1995 1 999 1 24 1000
3 1 1985 1 999 2 24 10
#Recapture Data
#TG Year Month Fleet Number
1 1982 1 1 7
1 1982 1 2 5
1 1985 1 2 0
2 1997 1 1 6
2 1997 2 1 4
3 1986 1 1 7
3 1986 2 1 5
Note
•The release data must be enter in TG order.
•<tfill> values are place holders and are replaced by program generated values for model time.
•Analysis of the tag-recapture data has one -logL component for the distribution of recaptures
across areas and another -logL component for the decay of tag recaptures from a group over
61
time, hence informative about mortality. More on this in the control file.
Stock Composition Data
It is sometimes possible to observe the fraction of a sample that is composed of fish from different
stocks. These data could come from genetics, otolith microchemistry, tags or other means. The growth
pattern feature in SS allows definition of cohorts of fish that have different biological characteristics
and which are independently tracked as they move among areas. SS now incorporates the capability
to calculate the expected proportion of a sample of fish that come from different growth patterns.
In the inaugural application of this feature, there was a 3 area model with one stock spawning and
recruiting in area 1, the other stock in area 3, then seasonally the stocks would move into area 2 where
stock composition observations were collected, then they moved back to their natal area later in the
year.
Stock composition data can be entered in SS as follows:
1 #Do morphcomp (if zero, then do not enter any further input below)
COND = 1
3 #Number of observations
2 #Number of stocks
0.0001 #Minimum Compression
#Year Month Fleet Part Nsamp Data Vector
1980 1 1 0 36 0.4 0.6 ...
1981 1 1 0 40 0.44 0.62 ...
1982 1 1 0 50 0.49 0.50 ...
Note
•The N stocks entered with these data must match the N growth patterns in the control file.
•The expected value is combined across sexes.
•The “partition” flag is included here in the data, but cannot be used because the expected value
is calculated before the catch is partitioned into discard and retained components.
62
•Note that there is a specific value of mincomp to add to all values of observed and expected.
Selectivity Empirical Data
It is sometimes possible to conduct field experiments or other studies to provide direct information
about the selectivity of a particular length or age relative to the length or age that has peak selectivity, or
to have a prior for selectivity that is more easily stated than a prior on a highly transformed selectivity
parameter. This section provides a way to input data that would be compared to the specified derived
value for selectivity. This is a placeholder at this time and will be fully implemented soon.
Selectivity data can be entered in SS as follows:
0 #Do data read for selectivity (if zero, then do not enter any further input below)
#Year Month Fleet Age/Size Bin # Datum DatumSE
End of Data File
999 #End of data file marker
Excluding Data
Data that are before the model start year or greater than the retrospective year are not moved into the
internal working arrays at all. So if you have any alternative observations that are used in some model
runs and not in others, you can simply give them a negative year value rather than having to
comment them out. The first output to data.ss_new has the unaltered and complete input data.
Subsequent reports to data.ss_new produce expected values or bootstraps only for the data that are
being used.
Data that are to be included in the calculations of expected values, but excluded from the calculation
of negative log likelihood, are flagged by use of a negative value for fleet ID.
Data Super-Periods
The super-period capability allows the user to introduce data that represent a blend across a set of time
steps and to cause the model to create an expected value for this observation that uses the same set of
time steps. The option is available for all types of data and a similar syntax is used. Super-periods are
63
started with a negative value for month, and then stopped with a negative value for month, placeholder
observations within the super-period are designated with a negative fleet field. The standard error or
Nsamp field is now used for weighting of the expected values. An error message is generated if the
super-period does not contain exactly one observation with a positive fleet field.
All super-period observations must be contiguous in the data file. All but one of the observations in the
sequence will have a negative value for fleet ID so the data associated with these dummy observations
will be ignored. The observed values must be combined outside of the model and then inserted into the
data file for the one observation with a positive fleet ID. An expected value for the observation will be
computed for each selected time period within in the super-period. The expected values are weighted
according to the values entered in the standar error (or Nsamp) field for all observations expect the
single observation holding the combined data. The expected value for that year gets a relative weight of
1.0. So in the example below, the relative weights are: 1982, 1.0 (fixed); 1983, 0.85; 1985, 0.4; 1986, 0.4.
These weights are summed and rescaled to sum to 1.0, and are output in the echoinput.sso file.
Not all time steps within the extent of a super-period need be included. For example, in a 3 season
model a super-period could be set up to combine information from season 2 across 3 years, e.g. skip
over the season 1 and season 2 for the purposes of calculating the expected value for the super-period.
The key is to create a dummy observation (negative fleet value) for all time steps, except 1, that will be
included in the super-period and to include one real observation (positive fleet value; which contains
the real combined data from all the specified time steps).
Super-period example:
#Year Month Fleet Obs SE Comment
1982 -2 3 34.2 0.3 Start super-period. This observation has positive fleet
value, so is expected to contain combined data from all
identified periods of the super-period. The se entered
here is use as the se of the combined observation. The
expected value for the survey in 1982 will have a relative
weight of 1.0 (default) in calculating the combined
expected value.
1983 2 -3 55 0.3 In super-period; entered observation is ignored. The
expected value for the survey in 1983 will have a relative
weight equal to the value in the standard error field
(0.85) in calculating the combined expected value.
1985 2 -3 88 0.40 Note that 1984 is not included in the supe-rperiod.
Relative weight for 1985 is 0.4
1986 -2 -3 88 0.40 End super-period
64
A time step that is within the time extent of the super-period can still have its own separate observation.
In the above example, the survey observation in 1984 could be entered as a separate observation, but
it must not be entered inside of the contiguous block of super-period observations. For composition
data (which allow for replicate observations), a particular time steps’ observations could be entered as
a member of a super-period and as a separate observation.
The super-period concept can also be used to combine seasons within a year with multiple seasons.
This usage could be preferred if fish are growing rapidly within the year so their effective age selectivity
is changing within year as they grow; fish are growing within the year so fishery data collected year
round have a broader size-at-age modes than a mid-year model approximation can produce; and it
could be useful in situations with very high fishing mortality.
Control File
Overview of Control File
These listed model features are denoted in the control file in the following order:
1. Number of growth patterns and platoons
2. Design matrix for assignment of recruitment to area/settle_event/growth pattern
3. Design matrix for movement between areas
4. Definition of time blocks that can be used for time-varying parameters
5. Controls far all time-varying parameters
6. Specification for growth and fecundity
7. Natural mortality and growth parameters for each sex x growth pattern
8. Maturity, fecundity and weight-length for each sex
9. Recruitment distribution parameters for each area, settle_event, growth pattern
10. Cohort growth deviation
11. Movement between areas
12. Catch Multiplier
65
13. Fraction female
14. Setup for any MG parameters are time-varying
15. Seasonal effects on biology parameters
16. Spawner-recruitment parameters
17. Setup for any SR parameters are time-varying
18. Recruitment deviations
19. F ballpark value in specified year
20. Method for calculating fishing mortality (F)
21. Initial equilibrium F for each fleet
22. Catchability (Q) setup for each fleet and survey
23. Catchability parameters
24. Setup for any Q parameters are time-varying
25. Length selectivity, retention, discard mortality setup for each fleet and survey
26. Age selectivity setup for each fleet and survey
27. Parameters for length selectivity, retention, discard mortality for each fleet and survey
28. Parameters for age selectivity, retention, discard mortality for each fleet and survey
29. Setup for any selectivity parameters are time-varying
30. Tag-recapture parameters
31. Variance adjustments
32. Lambdas for likelihood components
66

The order in which they appear in the control file has grown over time rather opportunistically, so it
may not appear particularly logical at this time, especially various aspects of recruitment distribution
and growth. When the same information is entered via the GUI, it is organized more logically and then
written in this form to the text control file.
Parameter Line Elements
The primary role of the SS control file is to define the parameters to be used by the model. The general
syntax of the 14 elements of a long parameter line is described here. The first seven elements of a
parameter line are used for time-varying parameters that cannot themselves be time-varying and will
be referred to as a short parameter line. Three types of time-varying properties can be applied to a
base parameter: blocks or trend, environmental linkage, and random deviation. Each parameter line
contains:
Column Element Description
1 LO Minimum value for the parameter
2 HI Maximum value for the parameter
3 INIT Initial value for the parameter. If the ss.par file is read, it
overwrites these INIT values.
4 PRIOR Expected value for the parameter. This value is ignored if the
prior type is 0 (no prior) or 1 (symmetric beta).
5 PRIOR STDEV Standard deviation for the prior, used to calculate likelihood
of the current parameter value. This value is ignored if prior
type is 0.
6PRIOR TYPE 0 = none,
1 = symmetric beta,
2 = full beta,
3 = lognormal without bias adjustment,
4 = lognormal with bias adjustment,
5 = gamma,
6 = normal.
7 PHASE Phase in which parameter begins to be estimated. A negative
value causes the parameter to retain its INIT value (or value
read from the ss.par file).
Note that relative to SS v.3.24, the order of Prior StDev and Prior Type have been switched and the
67

Prior Type options have been renumbered.
Short parameter lines have only the above 7 elements. The full parameter line syntax for the
mortality-growth, spawn-recruitment, selectivity, and Q sections provides additional controls to give
the parameter time-varying properties. These are listed briefly below and described in more detail in
the section time varying parameter options found in the Time-Varying Parameters section.
Column Element Description
8 Env Var & Link Create a linkage to an input environmental time-series
9 Dev Link Invokes use of the deviation vector in the linkage function
10 Dev min yr Beginning year for the deviation vector
11 Dev max yr Ending year for the deviation vector
12 Dev Phase Phase for estimation for elements in the deviation vector
13 Block Specify time block or trend to be applied
14 Block Fxn Function form for the block offset
Terminology
Where the term “COND” appears in the value column of this documentation (it does not actually
appear in the control file), it indicates that the following section is omitted except under certain
conditions, or that the factors included in the following section depend upon certain conditions. In
most cases, the description in the Definition column is the same as the label output to the
control.ss_new file.
Beginning of Control File Inputs
Typical Value Description and Options
#C comment Comments beginning with #C at the top of the file will be retained and
included in output.
0 0 = do not read the wtatage.ss file,
1 = read the wtatage.ss file, also read and use the growth parameters,
2 = Future option to read the wtatage.ss file, then omit reading and using
growth parameters and all length-based data.
68

Typical Value Description and Options
2 N growth patterns (GP)
These are collections of fish with unique biological characteristics
(growth, M, wt-len, reproduction.) The GP x Sex x Settlement Events
constitute unique morphs that are tracked in SS. They are assigned these
characteristics at birth and retain them throughout their lifetime. At
recruitment, morph members are distributed across areas (if any) and they
retain their biological characteristics even if they move to another area
in which a different cohort with different biological characteristics might
predominate. For example, one could assign a fast-growing morph to
recruit predominately in a southern areas and a slow-growing morph to a
northern area. The natural mortality and growth parameters are specified
for each growth pattern in the MG parameters section in the order of
females growth pattern 1 to growth pattern N followed by males growth
pattern 1 to growth pattern N.
3 Number of platoons within a morph.
This allows exploration of size-dependent survivorship. A value of 1
will not create additional platoons. Odd-numbered values of 3 - 5 will
break the overall morph into that number of platoons. More platoons
slows model execution, so values above 5 not advised. The fraction of
each morph assigned to each platoon is custom-input or designated to
be a normal approximation. When multiple platoons are designated,
an additional input is the ratio of between platoon to within platoon
variability in size-at-age. This is used to partition the total growth
variability. For the platoons, their size-at-age is calculated as a factor
(determined from the between-within variability calculation) times the
size-at-age of the central morph which is determined from the growth
parameters for that Growth Pattern x Sex.
COND > 1 Following 2 lines are conditional on N platoons > 1
0.7 Platoon between/within stdev ratio. Ratio of the amount of variability in
length-at-age between platoons to within platoons.
0.2 0.6 0.2 Distribution among platoons. Enter custom vector or enter -1 to first value
of vector to get a normal approximation: (0.15, 0.70, 0.15) for 3 platoons,
(0.031, 0.237, 0.464, 0.237, 0.031) for 5 platoons.
69
Empirical Weight-at-Age
With version 3.04, SS added the capability to read empirical body weight at age for the population and
each fleet, in lieu of generating these weights internally from the growth parameters, weight-at-length,
and size-selectivity. The values are read from a separate file named, wtatage.ss. This file is only required
to exist if this option is selected. See the section on weight-at-age for additional information on file
formatting for empirical weight-at-age.
Recruitment Timing and Distribution
In older versions of SS one value of spawning biomass was calculated annually at the beginning of
one specified spawning season and this spawning biomass produces one annual total recruitment
value and this annual recruitment was distributed among seasons, areas, and growth types according
to other model parameters.
In SS v.3.30, more control of the seasonal timing is provided and there now is an explicit time delta
between spawning and recruitment. Spawning still occurs just once per year, but its timing can be
at any specified time, not just the beginning of a season. Recruitment of the progeny from an annual
spawning enter the population in one or more settlement events.
Example set-up where there are multiple settlement events:
3 Number of recruitment settlement events
0 Unused option
Growth Pattern Month Area Age (for each settlement assignment)
1 11.0 1 0
1 12.0 1 0
1 1.0 1 1
Recruitment Timing and Settlement
Details regarding recruitment timing and settlement:
•Recruitment happens in specified settlement events (growth pattern, Month, Area);
•Number of unique settlement timings calculated at runtime;
•Now there is explicit elapsed time between spawning and recruitment;
70

•Growth and natural mortality of the platoon begins at time of settlement, which is its real age
0.0 for growth; but pre-settlement fish exist from the beginning of the season of settlement, so
can be caught if selected;
•Age at recruitment now user-controlled (should be 0 if in year of spawning)
•All fish become integer age 1 (for age determination) on their first January 1st;
•Recruitment can occur >12 months after spawning
The distribution of recruitment among these settlement events is controlled by recruitment
apportionment parameters. There must be a parameter line for each GP, then for each area, then for
each settlement. All of these are required, but only those GP x area x settlements designated to
receive recruits in the recruitment design matrix will have the parameter used in the recruitment
distribution calculation. For the recruitment apportionment, the parameter values are the
ln(apportionment weight). The sum of all apportionment weights is calculated for each pattern x area
x settlements that have been designated to receive recruits in the recruitment design matrix. Then the
apportionment weights are scaled to sum to 1.0 so that the total recruitment from the spawning event
is distributed among the cells designated to receive recruitment. These distribution parameters can
be time-varying, so the fraction of the recruits that occur in a particular GP, area, or settlement can
change from year to year.
Recruitment Distribution and Parameters
•SS processes the parameter values according to the following equation:
apportionmenti=epi
PN
j=1 epi(1)
•Set the value for one of these parameters to 0.0 and not estimate it so that other parameters will
be estimated relative to its fixed value.
•Give the estimated parameters a min-max of something like -5 and 5, so they have a good range
relative to the base parameter.
•In order to get a different distribution of recruitments in different years, you will need to make
at least one of the recruitment distribution parameters time-varying.
•In a seasonal model, all cohorts graduate to the age of 1 when they first reach January 1, even if
the seasonal structure of the model has them being born in the late fall. In general, this means
that SS operates under the assumption that all age data have been adjusted so that fish are age 0
at the time of spawning and all fish graduate to the next age on Jan 1. This can be problematic
71

if the ageing structures deposit a ring at another time of year. Consequently, you may need to
add or subtract a year to some of your age data to make it conform to the SS structure, or you
may need to define the SS calendar year to start at the beginning of the season at which ring
deposition occurs. Talk with your ageing lab about their criteria for seasonal ring deposition!
•Seasonal recruitment is coded to work smoothly with growth. If the recruitment occurring in
each season is assigned the same growth pattern, then each seasonal cohort’s growth trajectory
is simply shifted along the age/time axis. At the end of the year, the early born cohorts will be
larger, but all are growing with the same growth parameters so all will converge in size as they
approach their common Lmax.
•At the time of settlement, fish are assigned a size equal to the lower edge of the first population
size bin and they grow linearly until they reach the age A1. SS generates a warning if the first
population length bin is greater than 10 cm as this seems an unreasonably large value for a larval
fish. A1 is in terms of real age elapsed since birth. All fish advance to the next integer age on
Jan 1, regardless of birth season. For example, consider a 2 season model with some recruitment
in each season and with each season’s recruits coming from the same GP. At the end of the first
year, the early born fish will be larger but both of the seasonal cohorts will advance to an integer
age of 1 on Jan 1 of the next year. The full growth curve is still calculated below A1, but the
size-at-age used by SS is the linear replacement. Because the linear growth trajectory can never
go negative, there is no need for the additive constant to the standard deviation (necessary for
the growth model used in SS2 V1.x), but the option to add a constant has been retained in the
model.
Typical Value Description and Options
1 Recruitment distribution method. This section controls which
combinations of Growth Pattern x Area x Settlement will get a portion of
the total recruitment coming from each spawning. Options:
1 = use the 3.24 or earlier setup,
2 = main effects for GP, settle timing, and area,
3 = each settle entity,
4=no parameters (only if Growth Pattern x Settlement x Area = 1).
1 Spawner-Recruitment, options:
1 = global,
2 = by area (by area is not yet implemented; there is a conceptual challenge
to doing the equilibrium calculation when there is fishing).
1 Number of recruitment settlement assignments. Must be at least 1 even if
only 1 settlement and 1 area because the timing of that settlement must be
specified.
72

Typical Value Description and Options
0 Year x Area x Settlement Event Interaction Requested (only for recruitment
distribution method = 1).
1 5.5 1 0 Recruitment assignment to grwoth pattern, month, area, and age (for each
settlement event). Here settlement is set to mid-May (month 5.5). Note:
Normally the calendar age at settlement is 0 if settlement happens between
the time of spawning and the end of that year, and at age 1 if settlement is in
the year after spawning. In 3.24, settlement always happened at age 0 even
if in following year. That is illogical, but this age option allows replication
of 3.24 for testing purposes.
Movement
Here we define movement among the areas. This is a box transfer with no explicit adjacency of areas, so
fish can move from any area to any other area in each time step. Future Need: augment the capability
further to allow sex-specific movement, and also to allow some sort of mirroring so that sexes and
growth patterns can share the same movement parameters if desired.
Typical Value Description and Options
COND: only if areas > 1
4 Enter Number of movement definitions.
1.0 First age that moves. This value is a real number, not an integer, to allow
for an in-year start to movement in a multi-season model. It is the real age
at the beginning of a season, even though movement does not occur until
the end of the season. For example, in a setup with two 6-month seasons a
value of 0.5 will cause the age 0 fish to not move when they complete their
first 6 month season of life, and then to move at the end of their second
season because they start movement capability when they reach the age of
0.5 years (6 months).
1112410 The four requested movement definitions appear here. Each definition
specifies: season, morph, source area, destination age1, age2. The rate
of movement will be controlled by the movement parameters later. Here
the age1 and age2 controls specify the range over which the movement
parameters are interpolated with movement costant below age1 and above
age2.
1121410
1212410
1221410
73

Two parameters will be entered later for each growth pattern, area pair and season.
•movement is constant at P1 below the specified minage for movement change, constant at P2
above maxage for movement change, and linearly interpolated for intermediate ages;
•For each source area the implicit movement parameter value is 0.0, but this default value is
replaced if the stay movement is selected to have an explicit pair of parameter (e.g. specify
movement rate for area 1 to area 1).
•the parameter is exponentiated so that a movement parameter value of 0 becomes 1.0;
•for each source area, all movement rates are then summed and divided by this sum so that 100%
of the fish are accounted for in the movement calculations;
ratei=epi
PN
j=1 epi(2)
•at least one movement parameter must be fixed so that all other movement parameters are
estimated relative to it. This is achieved naturally by not specifying the stay rate parameter so it
has a fixed value of 0.0;
•the resultant movement rates are multiplied by season duration in a seasonal model;
Time Blocks
Typical Value Description and Options
3Number of block patterns. These patterns can be referred to in the
parameter sections to create a separate parameter value for each block.
COND > 0: Following inputs are omitted if N Block patterns equals 0.
321 Blocks per pattern
1975 1985
1986 1990
1995 2001
Beginning and ending years for blocks in design 1; years not assigned to a
block period retain the baseline value for a parameter that uses this
pattern.
1987 1990
1995 2001
Beginning and ending years for blocks in design 2.
74

1999 2002 Beginning and ending years for blocks in design 3.
When using time blocks, it is important to consider which parameters will affect which years of the
time series. There are three main situations:
1. Offset approach: One or more time blocks are created and cover all or a subset of the years. Each
block gets a parameter that is used as an offset from the base parameter. In this situation you
typically will allow SS to estimate the base parameter and each of the offset parameters. In years
not covered by blocks, the base parameter alone is used. However, if blocks cover all the years,
then the value of the block parameter is completely correlated with the mean of the block offsets,
so model convergence and variance estimation could be affected. The recommended approach
when using offsets is to not have all the years be covered by blocks when doing offsets, or to fix
the base parameter value at a reasonable level when doing offsets for all years.
2. Replacement approach-Option A: Here time blocks are created which cover a subset of the
years. The base parameter is used in the non-block years and the value of that base parameter
is replaced by the block parameter in each respective block. In this situation, you typically
allow SS to estimate the base parameter and each of the block parameters.
3. Replacement-Option B: Here replacement time blocks are created for all the years. In this case
the base parameter is simply a placeholder that is always replaced by a block parameter. In this
situation, do not allow SS to estimate the base parameter and only estimate the corresponding
block replacement parameters, otherwise, the search algorithm will be attempting to estimate
parameters that do not contribute to the log-likelihood, so model convergence and variance
estimation could be effected. Note however, that the minimum and maximum for the base
parameter are used as checks on the minimum and maximum of the blocks.
Regardless of the block set-up approach, special consideration should be given regarding which
parameter values are should be applied during forecast years. The model will default to use all base
parameter values during the forecast period. However, there are controls in the forecast file which
allow the user to specify specific parameter years to be applied during the forecast period for
selectivity, relative F, and recruitment.
Time-varying Parameter Controls
In SS v.3.30, several changes are introduced to the implementation of time-varying parameters:
•Time-varying parameters for biology, spawner-recruitment, catchability, and selectivity are
implemented using the same approach and share code.
•The block feature that allowed input of one block parameter line and replication of that line
75

by SS as often as needed has been replaced. Now there is a complete time-varying parameter
auto-generation capability.
•The logistic bound constraint is no longer implemented due to the challenges it created to
interpreting parameter values. Instead, the auto-generate feature now creates bounds on
time-varying parameters for blocks such that the combination of a bounded value of the
time-varying parameter and the base parameter will not violate the base parameter bounds.
•For more information on the implementation of time-varying parameters, see the Using
Time-varying Parameters section
Typical Value Description and Options
1 Environmental/Block/Deviation adjust method for all time-vary
parameters.
1 = warning relative to baseparameter bounds,
3 = no bound check). Note: logistic bound check form previous SS versions
(e.g., 3.24) is no longer an option.
0 0 0 0 0 Five values control auto-generation for: 1-biology, 2-spawnrecr,
3-catchability, 4-tag (future), 5-selectivity.
The accepted values are:
0 = auto-generate all time-varying parameters,
1 = read each time-varying parameter line,
2 = read each line and auto-generation if read value for parameter min =
-12345.
Biology
Natural Mortality
Natural mortality (M) has some options that are referenced to integer age, and some to real age since
settlement. So, if M varies by age, M will change by season and cohorts born early in the year will have
different M than late born cohorts.
Lorenzen natural mortality is based on the concept that natural mortality varies over the life cycle of
a fish, which is driven by physiological and ecological processes. So, natural mortality is scaled by the
length of the fish.
76

Typical Value Description and Options
1 Natural Mortality Options:
0 = A single parameter,
1 = N breakpoints,
2 = Lorenzen,
3 = Read age specific M and do not do seasonal interpolation.
4 = Read age specific and do seasonal interpolation, if appropriate.
COND = 0 No additional natural mortality controls.
COND = 1
4 Number of breakpoints. Then read a vector of ages for these breakpoints. Later,
per sex x GP, read N parameters for the natural mortality at each breakpoint.
2.5 4.5 9.0 15.0 Vector of age breakpoints.
COND = 2
4 Reference age for Lorenzen Natural Mortality: read one additional integer
value that is the reference age. Later read one parameter for each sex x growth
pattern that will be the M at the reference age. Other ages will have an M scaled
to its body size-at-age. However, if platoons are used, all will have the same M
as their growth pattern. Lorenzen M calculation will be updated if the starting
year growth parameters are active, but if growth parameters vary during the
time-series, the M is not further updated. So be careful in using Lorenzen when
there is time-varying growth.
COND = 3 or 4 Do not read any natural mortality parameters. With option 2, these M values
are held fixed for the integer age (no seasonality or birth season considerations).
With option 4, there is seasonal interpolation based on real age, just as in
options 1 and 2.
0.20 0.25 ...
0.20 0.23 ...
Age-specific M values: row 1 is female growth pattern 1, row 2 is female 2
growth pattern 2, row 3 is male growth pattern1, etc.
Growth
Timing
When fish recruit at the real age of 0.0 at settlement, they have body size equal to the lower edge of
the first population size bin. The fish then grow linearly until they reach a real age equal to the input
value “growth_age_for_L1” and have a size equal to the parameter value for L1. As they age further,
77
they grow according the selected growth equation. The growth curve is calibrated to go through the
size L2 when they reach the age “Growth_age_for_L2”.
Linf
If “Growth_age_for_L2” is set equal to 999, then the size L2 is used as Linf. If MGparm_def option
= 1 (direct estimate, not offsets), then setting a male growth or natural mortality parameter value to
0.0 and not estimating it will cause SS to use the corresponding female parameter value for the males.
This check is done on a parameter, by parameter basis and is probably most useful for setting male L1
equal to female L1, then letting males and females have separate K and Linf parameters.
Schnute growth function
The Schnute implementation of a 3-parameter growth function is invoked by entering 2 in the grow
type field. Then a fourth parameter is read after reading the von Bertalanffy K parameter. When this
fourth parameter has a value of 1.0, it is equivalent to the standard von Bertalanffy growth curve. When
this function was first introduced in SS, it required that A0 be set to 0.0.
Mean sizea-at-maximum age
The mean size of fish in the max age age bin depends upon how close the growth curve is to Linf by
the time it reaches max age AND the mortality rate of fish after they reach max age. SS provides an
option for the mortality rate to use in this calculation during the initial equilibrium year. This must
be specified by the user and should be reasonably close to M plus initial F. In SS v.3.30, this uses the
von Bertalanffy growth out to 3*nages and decays the numbers at age by exp(-value set here). For
subsequent years of the time series, SS should update the size-at-maxage according to the weighted
average mean size of fish already at max age and the size of fish just graduating into max age.
Unfortunately, this updating is only happening in years with time-varying growth. Hope to fix that in
the future.
Age-specific K
This option creates age-specific K multipliers for each age of a user-specified age range, with
independent multiplicative factors for each age in the range and for each growth pattern / sex. The
null value is 1.0 and each age’s K is set to the next earlier age’s K times the value of the current aged’s
multiplier. Each of these multipliers is entered as a full parameter line, so inherits all time-varying
capabilities of full parameters. The lower end of this age range cannot extend younger than the
specified age for which the first growth parameter applies. This is a beta model feature, so examine
output closely to assure you are getting the size-at-age pattern you expect. Beware of using this
option in a model with seasons within year because the K deviations are indexed solely by integer age
according to birth year. There is no offset for birth season timing effects, nor is there any seasonal
interpolation of the age-varying K.
78

Typical Value Description and Options
1 Growth Model:
1 = von Bertalanffy (2 parameters),
2 = Schnute’s generalized growth curve (aka Richards curve) with 3
parameters. Third parameter has null value of 1.0,
3 = von Bertalanffy with age-specific kmultipliers for specified range of
ages,
4 = age specific K. Set base kas kfor age = nages and working backwards
and the age-specific k=kfor the next older age * multiplier,
5 = age specific k. Set base kas kfor nages and work backwards and the
age-specific k= base k* multiplier.
1.66 Growth Amin (A1): Reference age for first size-at-age parameter.
25 Growth Amax (A2): Reference age for second size-at-age parameter (999
to use as L infinity).
0.20 Exponential decay for growth above maximum age (plus group: fixed at
0.20 in 3.24; should approximate initial Z).
Alternative Options:
-998 = Disable growth above maximum age (plus group) similar to earlier
versions of SS (prior to SS v3.24),
-999 = Replicate the simpler calculation done in SS v.3.24.
0 Placeholder for future growth feature.
COND >= 3 Growth option age-specific k
2 Number of (k) multipliers to read
5 Minimum age for age-specific k
7 Maximum age for age-specific k
0 Standard deviation added to length-at-age: Enter 0.10 to mimic SS2 V1.xx.
Recommend using a value of 0.0. Click here for more information.
1 CV Pattern
0: CV=f(LAA), so the 2 parameters are in terms of CV of the distribution
of length-at-age and the interpolation between these 2 parameters is a
function of mean length-at-age,
1: CV=f(A), so interpolation is a function of age,
2: SD=f(LAA), so parameters define the standard deviations of
length-at-age and interpolation is a function of mean length-at-age,
79

Typical Value Description and Options
3: SD=f(A),
4: Lognormal distribution of size-at-age. Input parameters will specify
the standard deviation of loge size at age. E.g. entered values will typically
be between 0.05 and 0.15. A bias adjustment is applied so the lognormal
distribution of size-at-age will have the same mean size as when a normal
distribution is used.
Maturity-Fecundity
Typical Value Description and Options
2 Maturity Option:
1 = length logistic,
2 = age logistic,
3 = read age-maturity for each female growth pattern,
4 = read an empirical age-maturity vector for all ages,
Note: need to read 2 parameters even if option 3 or 4 is selected.
COND = 3 or 4 Maturity Option
0 0.05 0.10 ... Vector of age-specific maturity or fecundity. One row of length Nages + 1 for
each female growth pattern.
1 First Mature Age: Overridden if maturity option is 3 or 4 or if empirical
wtatage.ss is used, but still must exist here. therwise, all ages below the first
mature age will have maturity set to zero.
1 Fecundity Option (irrelevant if maturity option is 4 or wtatage.ss is used):
1 = to interpret the 2 egg parameters as linear eggs/kg on body weight (current
SS default), so fecundity = wt ∗(a+b∗wt), so value of a=1, b=0 causes eggs to
be equivalent to spawning biomass,
2 = to set fecundity= a∗Lb,
3 = to set fecundity= a∗Wb, so values of a=1, b=1 causes fecundity to be equiv
to spawning biomass,
4 = fecundity = a+b∗L,
5 = Eggs = a+b∗wt.
80

Hermaphroditism
Typical Value Description and Options
0 Hermaphroditism Option:
0 = not used,
1 = invoke female to male age-specific function,
-1 = invoke male to female age-specific function,
Note: this creates the annual, age-specific fraction that change sex, it is not the
fraction that is each sex.
COND = 1 Read 2 lines below if hermaphroditism. is selected; also read 3 parameters after
reading the male weight-length parameter.
-1 Hermaphroditism Season:
-1 to do transition at the end of each season (after mortality and before
movement),
<positive integer> to select just one season,
1 Include males in spawning biomass,
0 = no males in spawning biomass,
1 = simple addition of males to females,
xx = more options to come later.
Parameter offset method
Typical Value Description and Options
2 Parameter Offset Method:
1 = direct assignment,
2 = for each growth pattern x sex, parameter defines offset from sex 1, offsets are
in exponential terms, so for example: Mold male =Mold female ∗exp(Mold male),
3 = for each growth pattern x sex, parameter defines offset from growth pattern
1 sex 1. For females, given that “natM option” is breakpoint and there are
two breakpoints, parameter defines offset from early age (e.g., Mold female =
Myoung female ∗exp(Mold female). For males, given that “natM option” is breakpoint
and there are two breakpoints, parameter is defined as offset from females AND
from early age (e.g., Mold male =Myoung female ∗exp(Myoung male)∗exp(Mold male)).
81
Catch Multiplier
These parameter lines are only included in the control file if the catch multiplier field in the data file
is set to 1 for a fleet:
Cadj =Cexp ∗cmult (3)
where Cexp is the expected catch from the fishing mortality and cmult is the catch multiplier. It has
year-specific, not season-specific, time-varying capabilities. In the catch likelihood calculation,
expected catch is multiplied by the catch multiplier by year and fishery to get Cadj before being
compared to the observed retained catch, so Cadj value of 1.1 means that the observed catch is 10%
greater than modeled catch.
Ageing Error Parameters
These parameters are only included in the control file if one of the ageing error definitions in the data
file has requested this feature (by putting a negative value for the ageing error of the age zero fish of
one ageing error definition). As of version 3.30.12, these parameters now have time-varying capability.
Seven additional full parameter lines are required. The parameter lines specify:
•age at which the estimated pattern begins (just linear below this age). This is “start age”
•bias at start age (as additive offset from unbiased age’)
•bias at maxage (as additive offset from unbiased age’)
•power function coefficient for interpolating between those 2 values (value of 0.0 produces linear
interpolation in the bias)
•standard deviation at age
•standard deviation at max age
•power function coefficient for interpolating between those 2 values
Code for implementing vectors of mean age’ and standard deviation of age’ within SS can be located
in Appendix C.
82

Sex ratio
The last line in the mortality-growth parameter section allows the user to fix or estimate the sex ratio
between female and male fish. The parameter is specified in the fraction of female fish. The default
option is a sex ratio of 0.50 with this parameter not being estimated. Estimation of the sex ratio is a
new feature within SS and should be done with care with the user checking that the answer is reflective
of the data.
As of v.3.30.12, this parameter now has time-varying capability similar to other parameters in the
mortality-growth section.
Read Biology Parameters
Next, SS reads the mortality-gowth (MG) parameters in generally the following order (may vary based
on selected options):
Parameter Description
Females Female natural mortality and growth parameters in the
following order by growth pattern.
natM Natural mortality for female growth pattern 1, where the
number of natural mortality parameters depends on the
option selected.
Lmin Length at Amin (units in cm) for female, growth pattern 1.
Lmax Length at Amax (units in cm) for female, growth pattern 1.
VBK Von Bertanlaffy growth coefficient (units are per year) for
females, growt pattern 1.
COND if growth type =2
Richards
Coefficient
Only include this parameter if Richards growth function is
used. If included, a parameter value of 1.0 will have a null
effect and produce a growth curve identical to Bertalanffy.
COND if growth type >=3 Age-Specific k
kdeviations for first age in range
kdeviations for next age in range
...
kdeviations for last age in range
83

Parameter Description
CV young Variability for size at age <= AFIX (units are fraction) for
females, growth pattern 1. Note that CV cannot vary over
time, so do not set up env-link or a deviation vector. Also,
units are either as CV or as standard deviation, depending on
assigned value of CV pattern.
CV old Variability for size at age >= AFIX (units are fraction) for
females, growth pattern 1. For intermediate ages, do a linear
interpolation of CV on means size-at-age. Note that the
units for CV will depend on the CV pattern and the value of
MGparm as offset. The CV value cannot vary over time.
WtLen scale Coefficient to convert length in cm to weight in kg for females.
WtLen exp Exponent in to convert length to weight for females.
Mat-50 Maturity logistic inflection (in cm or years). Where female
maturity-at-length (or age) is a logistic function: Ml= 1/(1+
exp(α∗(la−β))) where αis the slope, lais the size-at-age,
and βis the inflection of the maturity curve.
Mat-slope Logistic slope (must have negative value).
Eggs-alpha Two fecundity parameters; usage depends on the selected
fecundity option. Must be included here eve if vector is read
in the control section above.
Eggs-beta
COND: GP > 1 Repeat female parameters in the above order for growth
pattern 2.
Males Male natural mortality and growth parameters in the
following order by growth pattern.
natM Natural mortality for male GP1, where the number of natural
mortality parameters depends on the option selected.
Lmin Length at Amin (units in cm) for male, GP1
Lmax Length at Amax (units in cm) for male, GP1
VBK Von Bertanlaffy growth coefficient (units are per year) for
males, growth pattern1.
COND if growth type = 2
Richards
Coefficient
Only include this parameter if Richards growth function is
used. If included, a parameter value of 1.0 will have a null
effect and produce a growth curve identical to Bertalanffy.
84

Parameter Description
COND if growth type = 3 Age-Specific K
K deviations for first age in range
K deviations for next age in range
...
K deviations for last age in range
CV young Variability for size at age <= AFIX (units are fraction) for
males, GP1. Note that CV cannot vary over time, so do not
set up env-link or a dev vector. Also, units are either as CV
or as standard deviation, depending on assigned value of CV
pattern.
CV old Variability for size at age >= AFIX (units are fraction) for
males, GP1. For intermediate ages, do a linear interpolation
of CV on means size-at-age. Note that the units for CV will
depend on the CV pattern and the value of MGparm as offset.
COND: GP > 1 Repeat male parameters in the above order for growth pattern
2.
WtLen scale Coefficient to convert length in cm to weight in kg for males.
WtLen exp Exponent in to convert length to weight for males.
COND: GP > 1 Repeat male parameters in the above order for growth pattern
2.
COND: Hermaphrodism 3 parameters define a normal distribution for the transition
rate of females to males (or vice versa).
Inflect Age Hermaphrodite inflection age.
StDev Hermaphrodite standard deviation (in age) .
Asmp Rate Hermaphrodite asymptotic rate.
Recr Dist GP Recruitment apportionment by growth pattern, if multiple
growth patterns, multiple entries required.
Recr Dist Area Recruitment apportionment by area, if multiple areas,
multiple entries required
Recr Dist Seas Recruitment apportionment by season, if multiple seasons,
multiple entries required
COND: If recruitment distribution interaction = 1 (on).
85
Parameter Description
N patterns x
N areas x N
seasons
Note that the order of recruitment distribution parameters
has areas then seasons for main effect, and seasons then areas
for interactions.
Cohort growth deviation Set equal to 1.0 and do not estimate; it is deviations from this
base that matter.
2 x N selected movement pairs Movement parameters
COND: The following lines are only required when the associated features are turned on.
Ageing Error Turned on in the data file.
Catch
Multiplier
For each fleet selected for this option in the data file.
Fraction female Fraction female by growth pattern, if multiple growth
patterns, multiple entries required.
Example format for MG parameter section with 2 sexes, 2 areas. Parameters marked with COND are
conditional on selecting that feature:
Prior <other Block
LO HI INIT Value entries> Fxn Parameter Label
0 0.50 0.15 0.1 ... 0 #NatM_p_1_Fem_GP_1
0 45 21 36 ... 0 #L_at_Amin_Fem_GP_1
40 90 70 70 ... 0 #L_at_Amax_Fem_GP_1
0 0.25 0.15 0.10 ... 0 #VonBert_K_Fem_GP_1
0.10 0.25 0.15 0.20 ... 0 #CV_young_Fem_GP_1
0.10 0.25 0.15 0.20 ... 0 #CV_old_Fem_GP_1
-3 3 2e-6 0 ... 0 #Wtlen_1_Fem
-3 4 3 3 ... 0 #Wtlen_2_Fem
50 60 55 55 ... 0 #Mat50%_Fem
-3 3 -0.2 -0.2 ... 0 #Mat_slope_Fem
-5 5 0 0 ... 0 #Eggs/kg_inter_Fem
-50 5 0 0 ... 0 #Eggs/kg_slope_wt_Fem
0 0.50 0.15 0.1 ... 0 #NatM_p_1_Mal_GP_1
0 45 21 36 ... 0 #L_at_Amin_Mal_GP_1
40 90 70 70 ... 0 #L_at_Amax_Mal_GP_1
0 0.25 0.15 0.10 ... 0 #VonBert_K_Mal_GP_1
86

Prior <other Block
LO HI INIT Value entries> Type Parameter Label
0.10 0.25 0.15 0.20 ... 0 #CV_young_Mal_GP_1
0.10 0.25 0.15 0.20 ... 0 #CV_old_Mal_GP_1
-3 3 2e-6 0 ... 0 #Wtlen_1_Mal
-3 4 3 3 ... 0 #Wtlen_2_Mal
0 0 0 0 ... 0 #RecrDist_GP_1
0 0 0 0 ... 0 #RecrDist_Area_1
0 0 0 0 ... 0 #RecrDist_Area_2
0 0 0 0 ... 0 #RecrDist_Settlement_1
0.2 5 1 1 ... 0 #CohortGrowDev
-5 5 -4 1 ... 0 #Move_A_seas1_GP1_from_1to2
(COND)
-5 5 -4 1 ... 0 #Move_B_seas1_GP1_from_1to2
(COND)
-99 99 1 0 ... 0 #AgeKeyParm1 (COND)
-99 99 0.288 0 ... 0 #AgeKeyParms 2 to 5 (COND)
-99 99 0.715 0 ... 0 #AgeKeyParm6 (COND)
0.2 3.0 1.0 0 ... 0 #Catch_mult_fleet1 (COND)
0.001 0.999 0.5 0.5 ... 0 #FracFemale_GP_1
Time-Varying Biology Parameters
Any of the parameters defined above can be made time-varying through linkage to an environmental
data series, through time blocks or trend, or by setting up annual deviations. The options for making
biology, spawner-recruitment, catchability and selectivity parameters change over time is detailed in
the section labeled Time-Varying Parameters. After reading the biology parameters above, which will
include possible instructions to create environmental link, blocks, or deviation vectors, then read the
following section. Note that all inputs in this section are conditional (COND) on entries in the biology
parameter section. So if no biology parameters invoke any time-varying properties, this section is left
blank (or completely commented out with #).
When time-varying growth is used, there are some additional considerations to be aware of:
•Growth deviations propagate into the forecast. The user can select which growth parameters
get used during the forecast by setting the end year of the last block. If the last block ends in
the model’s end year, then the growth parameters in effect during the forecast will revert to the
“no-block” baseline level. By setting the end year of the last block to end year (endyr) + 1, the
87
model will continue the last block’s growth parameter levels throughout the forecast.
•The equilibrium benchmark quantities (MSY, F40%, etc.) previously used end year (endyr) body
size-at-age, which is a disequilibrium vector. There is a capability to specify a range of years
over which to average the size-at-age used in the benchmark calculations.
•An addition issue occurred in versions prior to 3.20. Its description is retained here, but it was
resolved with the growth code modification for version 3.20.
–Issue for versions prior to 3.20: When the growth reference ages have A1>0 and A2<999,
the effect of time-varying K has a non-intuitive aspect. This occurs because the virtual
size at age 0.0 and the actual Linf are calculated annually from the current L1, L2 and K
parameters. Because these calculated quantities are outside the age range A1, A2, a
reduction in K will cause an increase in the calculated size-at-age 0.0 that year. So there is
a ripple effect as the block’s growth parameters affect the young cohorts in existence at
the time of the change. The workaround for this is to set A1=0 and A2=999. However,
this may create another incompatibility because the size-at-age 0.0 cannot be allowed to
be negative and should not be allowed to be less than the size of the first population
length bin. Therefore, previous use of A1=2 might have implied a virtual size at age 0.0
that was negative (which is ok), but setting A1=0 does not allow the size at age=A1 to be
negative.
Time-varying parameter specification:
Prior Prior Prior
LO HI INIT Value SD Type Phase Parameter Label
COND: Only if MG parameters are time-varying
-99 99 1 0 0.01 0 -1 #Wtlen_1_Fem_ENV_add
-99 99 1 0 0.01 0 -1 #Wtlen_2_Fem_ENV_add
Seasonal Biology Parameters
Seasonal effects are available for weight-length parameters and for the growth K. Seasonality is not
needed for the maturity and fecundity parameters because spawning is only defined to occur in one
season. Seasonal L1 may be implemented at a later date. The seasonal parameter values adjust the base
parameter value for that season.
P0=P∗exp(seas_value)(4)
88

Control file continued:
Value Description
0 0 0 0 0 0 0 0 0 0 Seasonality for selected biology parameters (not a conditional
input). Read 10 integers to specify which biology parameters
have seasonality: fem-wtlen1, fem-wtlen2, mat1, mat2, fec1,
fec2, male-wtlen1, male-wtlen2, L1, K. Reading a positive
value selects that factor for seasonality.
COND: If any factors have seasonality, then read N seasons parameters that define the
seasonal offsets from the base parameter value.
<short parameter line(s)> Read N seasons short parameter lines for each factor selected
for seasonality. The parameter values define an exponential
offset from the base parameter value.
Spawner-Recruitment
The spawner-recruitment section starts by specification of the functional relationship that will be
used.
Control file continued:
Value Label Description
3 Spawner- The options are:
Recruitment 1: null,
Relationship 2: Ricker (2 parameters: log(R0) and steepness),
3: standard Beverton-Holt (2 parameters: log(R0) and steepness),
4: ignore steepness and no bias adjustment. Use this in
conjunction with very low emphasis on recruitment deviations
to get CAGEAN-like unconstrained recruitment estimates (2
parameters, but only uses the first one),
5: Hockey stick (3 parameters: log(R0), steepness, and Rmin) for
ln(R0), fraction of virgin SSB at which inflection occurs, and the R
level at SSB=0.0.
6: Beverton-Holt with flat-top beyond Bzero (2 parameters: log(R0)
and steepness),
7: Survivorship function (3 parameters: log(R0), zf rac, and β).
Suitable for sharks and low fecundity stocks to assure recruits are
<= population production.
89
Value Label Description
8: Shepherd (3 parameters: log(R0), steepness, and shape parameter,
c).
9(beta): Shepherd re-parameterization (3 parameters: log(R0),
steepness, and shape parameter, c).
10(beta): Ricker re-parameterization (3 parameters: log(R0),
steepness, and Ricker power, γ). This SRR is was added to version
3.30.11 and is in beta mode.
1 Equilibrium
recruitment
Use steepness in initial equilibrium recruitment calculation
0 = none
1 = use steepness
0 Future Feature Reserved for the future option to make realized sigmaR a function
of the stock-recruitment curve.
The number of parameters needed by each relationship is stored internally. In SS v.3.24 and before,
only a short parameter line was used for the spawner-recruitment section. SS v3.30 now requires
long parameter lines in the spawner-recruitment section because it now uses the same time-varying
parameter approach as the biology and selectivity parameters. This generic time-varying approach
replaces the SR envlink concept in SS v.3.24. Also the R1 offset was effectively a block to implement a
regime shift for the initial equilibrium year. Now in v.3.30, the R1 offset parameter is replaced by a
parameter termed "regime shift". The SR regime parameter is intended to have a base value of 0.0 and
not be estimated. Similar to the cohort-growth deviation, it serves simply as a base for adding
time-varying adjustments. This regime shift parameter can have blocks, environmental links or
random deviations.
If a SS v.3.24 model implement the R1 offset approach this can be mimicked in v.3.30 by the SR regime
parameter. For any given year, including startyr - 1, the R0 is replaced by R0*exp(SR_regime[y]). So
to mimic the R1_offset, you need to put a block on SR_regime for y = startyr - 1. But if SR_regime
has some change during your main time series, that change will be filtered through the stock-recruit
relationship into an impact on the numbers at age for whatever years are impacted. This can also
apply to the forecast. A block on SR_regime is also easier than some of the old dummy environmental
variables that were created in the past to adjust recruitment up and down for long periods.
If the R0 or steepness parameters are time-varying, then SS will use the current year’s parameters to
calculate recruits as a function of the spawning biomass. If the SR regime parameter is time-varying,
then SS applies this offset after calculating recruits as function of spawning biomass. The expected
deviations can no longer be linked to "env": instead the environmental effect will be on the
"regime".
90
Read the required number of long parameter set-up lines (e.g. LO, HI, INIT, PRIOR, PRIOR TYPE, SD,
PHASE, ..., and BLOCK TYPE). These parameters are:
Value Label Description
8.5 log(R0) Log of virgin recruitment level.
0.60 Steepness Steepness of S-R, bound by 0.2 and 1.0 for the Beverton-Holt.
COND: If SRR = 5, 7, or 8
3rd Parameter Optional depending on which SRR function is used.
0.60 sigma-R Standard deviation of log recruitment. This parameter has two
related roles. It penalizes deviations from the spawner-recruitment
curve, and it defines the offset between the arithmetic mean
spawner-recruitment curve (as calculated from log(R0) and
steepness) and the expected geometric mean (which is the basis
from which the deviations are calculated. Thus the value of sigmaR
must be selected to approximate the true average recruitment
deviation.
0 Regime Parameter This replaces the R1 offset parameter. It can have a block for
the initial equilibrium year, so can fully replicate the functionality
of the previous R1 offset approach. The SR regime parameter
is intended to have a base value of 0.0 and not be estimated.
Similar to cohort-growth deviation, it serves simply as a based for
adding time-varying adjustments. This concept is similar to the old
environment effect on deviates feature in v.3.24 and earlier.
0 Autocorrelation Autocorrelation in recruitment.
Example set-up of the spawner-recruitment section:
LO HI INIT PRIOR <other entries> Block Fxn Parameter Label
3 31 8.81 10.3 ... 0 #SR_LN(R0)
0.2 1 0.61 0.70 ... 0 #SR_BH_steep
0 2 0.60 0.80 ... 0 #SR_sigmaR
-5 5 0 0 ... 0 #SR_regime
-99 99 0 0 ... 0 #SR_autocorr
91
•The R0, steepness, and regime shift parameters can be time-varying by blocks, trends,
environmental linkages, or random deviation. However, not all of these options make sense for
all parameters. Before discussing these, another important change must be noted.
•The sigmaR and autocorrelation parameters can not be time-varying.
•In SS v.3.24, the R0 and steepness parameter was used for both the virgin calculation and for the
MSY (benchmark) calculations. In SS v.3.30, these usages are more explicitly defined. The value
of R0 and steepness in the initial year is used for virgin and for calculation the denominator in
depletion estimates. The average value of R0 and steepness in the range of years specified as the
benchmark years 9 and 10 (see forecast.ss) is used for MSY type calculations. So, for example, a
long-term climate effect could cause R0 to change over time and BMSY could now be calculated
for some future range of years.
•Since R0 can be time-varying, what is regime shift for? Regime shift is for multi-year or
environmentally driven deviations from R0 without changing R0 itself. Then the recruitment
deviations are annual deviations from the current regime. And these recruitment deviations
can have autocorrelation.
–Warning: Use these options judiciously because the same algebraic effect on the calculated
recruitment can be achieved by different combinations of these options.
•Preliminary recommendation: Use block, trend or environmental effects on R0 only for very
long-term and highly persistent effects; use time-vary effects on regime shift for transitory, but
multi-year deviations from R0.
•The time-vary parameter lines are short lines because they themselves cannot be time-varying.
•The order of time-vary parameters are; R0, steepness, then regime shift. The number of
time-varying parameters from each of these can range from zero to many.
•Note that setting a block for just the initial equilibrium year is equivalent to R1_offset. If R1
offset is being used in 3.24, then sstrans.exe will automatically add a new block for the initial
equilibrium year so that you can easily re-implement a R1 offset effect.
Control file continued:
Value Label Description
1 Do Recruitment
Deviations
This selects the way in which recruitment deviations are coded:
0: None (so all recruitments come from S-R curve),
92

Value Label Description
1: Deviation vector (previously the only option). Here the deviations
are encoded as a deviation vector, so ADMB enforces a sum-to-zero
constraint.
2: Simple deviations. Here the deviations do not have an explicit
constraint to sum to zero, although they still should end up having
close to a zero sum. The difference in model performance between
options (1) and (2) has not been fully explored to date.
1971 Main recruitment
deviations begin
year
If begin year is less than the model start year, then the early
deviations are used to modify the initial age composition. However,
if set to be more than Nages before start year, it is changed to equal
Nages before start year.
1999 Main recruitment
deviations end
year
If recruitment deviations end year is later than retro year, it is reset
to equal retro year.
3 Main recruitment
deviations phase
1 Advanced 0: Use default values for advanced options
Options 1: Read values for the 11 advanced options.
COND = 1 Beginning of advanced options
1950 Early Recruitment Deviation Start Year:
0: skip (default),
+year: absolute year (must be less than begin year of main
recruitment deviations),
-integer: set relative to main recruitment deviations start year.
Note: because this is a deviation vector, it should be long enough
so that recruitment deviations for individual years are not unduly
constrained.
6 Early Recruitment Deviation Phase:
Negative value: default value to not estimate early deviations.
Users may want to set to a late phase if there is not much early data.
0 Forecast Recruitment Phase:
0 = Default value.
93

Value Label Description
Forecast recruitment deviations always begin in the first year
after the end of the main recruitment deviations. Recruitment
in the forecast period is deterministic derived from the specified
stock-recruitment relationship. Setting their phase to 0 causes their
phase to be set to max lambda phase +1 (so that they become active
after rest of parameters have converged.). However, it is possible
here to set an earlier phase for their estimation, or to set a negative
phase to keep the forecast recruitment deviations at a constant level.
1 Forecast Recruitment Deviations Lambda:
1 = Default value.
This lambda is for the log-likelihood of the forecast recruitment
deviations that occur before endyr + 1. Use a larger value here if
solitary, noisy data at end of time series cause unruly recruitment
deviation estimation.
1956 Last year wth no bias adjustment.
1970 First year wth full bias adjustment.
2001 Last year wth full bias adjustment.
2002 First recent year with no bias adjustment.
These four entries control how the bias adjustment is phased in and
then phased back out when the model is searching for the maximum
log-likelihood. Bias adjustment is automatically turned off when in
MCMC mode. For intervening years between the first and second
years in this list, the amount of bias adjustment that will be applied
is linearly phased in. The first year with full bias adjustment should
be a few years into the data-rich period so that SS will apply the
full bias-correction only to those recruitment deviations that have
enough data to inform the model about the full range of recruitment
variability. Defaults for the four year values: start year – 1000, start
year – Nages, main recruitment deviation final year, end year + 1.
Click here for more information..
0.85 Max Bias Adjustment:
Value for the maximum bias adjustment during the MLE mode.
A value of -1 will set the bias adjustment to 1.0 for all years
with estimated recruitment deviations. Likewise, all estimated
recruitment deviations, even those within a ramped era, switch to
maxbias=1.0 during MCMC.
0 Period For Recruitment Cycles:
94

Value Label Description
Use this when SS is configured to model seasons as years and there
is a need to impose a periodicity to the expected recruitment level.
If value is >0, then read that number of full parameter lines below
define the recruitment cycle
-5 Minimum Recruitment Deviation: Min value for recruitment
deviation.
Negative phase = Default value.
5 Maximum Recruitment Deviation: Max value for recruitment
deviation.
Late Phase = Default value (e.g., 5)
2 Number of Explicit Recruitment Deviations to Read:
0: Default, do not read any recruitment deviations; Integer: read this
number of recruitment deviations.
End of advanced options
COND = If N recruitment cycle is > 0, enter N full parameter lines below.
<parameter line> Full parameter line for each of the N periods of recruitment cycle.
COND = If N explicit recruitment deviations is > 0, then enter N lines below.
1977 3.0 Enter year and deviation.
1984 3.0 Two example recruitment deviations being read. Note: SS will
rescale the entire vector of recrdevs after reading these deviations,
so by reading two positive values, all other recrdevs will be scaled
to a small negative value to achieve a sum to zero condition before
starting model estimation.
Spawner-Recruitment Function
The number of age-0 fish is related to spawning biomass according to a stock-recruitment relationship.
SS has the option of the Beverton-Holt, Ricker, Hockey-Stick, and a survival-based stock recruitment
relationship.
95
Beverton-Holt
The Beverton-Holt Stock Recruitment curve is calculated as:
Ry=4hR0SBy
SB0(1 −h) + SBy(5h−1)e−0.5byσ2
R+˜
Ry˜
Ry∼N(0; σ2
R)(5)
where R0is the unfished equilibrium recruitment, SB0is the unfished equilibrium spawning
biomass (corresponding to R0), SByis the spawning biomass at the start of the spawning season
during year y, h is the steepness parameter, by is the bias adjustment fraction applied during year y, is
the standard deviation among recruitment deviations in log space, and is the lognormal recruitment
deviation for year y. The bias-adjustment factor (Methot and Taylor 2011) ensures unbiased
estimation of mean recruitment even during data-poor eras in which the maximum likelihood
estimate of the is near 0.0.
Ricker
The Ricker Stock Recruitment curve is calculated as:
Ry=R0SBy
SB0
eh(1−SBy/SB0)e−0.5byσ2
R+˜
Ry˜
Ry∼N(0; σ2
R)(6)
Hockey-Stick
The hockey-stick recruitment curve is calculated as:
Ry=RminR0+SBy
hSB0
(R0−Rmin(join) + R0(1 −join)(7)
where Rmin is the minimum recruitment level predicted at a spawning size of zero and is set by the
user in the control file, his defined as the fraction of SB0below which recruitment declines linearly,
and join is defined as:
join ="1 + e1000∗(SB0−hSB0)
SB0#−1
(8)
Survivorship
The survivorship stock recruitment relationship based on Taylor et al. 2013 is a stock-recruitment
model that enables explicit modeling of survival between embryos and age 0 recruits, and allows the
description of a wide rage of pre-recruit survival curves based. The model is especially useful for low
fecundity species that produce relatively few offspring per litter and exhibit a more direct connection
between spawning output and recruitment than species generating millions of eggs.
Survival-based recruitment is constrained so that the recruitment rate cannot exceed fecundity. The
relationship between survival and spawning output is based on parameters which are on a log scale.
These are:
z0=−log(S0)(9)
96

which is the negative of the log of the equilibrium survival S0, and can be thought of as a pre-recruit
instantaneous mortality rate at equilibrium, and
zmin =−log(Smax) = z0(1 −zfrac)(10)
which is the negative of the log of the maximum pre-recruit survival rate (Smax, the limit as spawning
output approaches 0), and is parameterized as a function of zfrac (which represents the reduction in
mortality as a fraction of z0) so the expression is well defined over the parameter range 0<zfrac<1.
Recruitment at age 0 for each year in the time series is calculated as:
Ry=SBye−z0+(z0−zmin)1−(SBy/SB0)βe˜
Ry˜
Ry∼N(0; σ2
R)(11)
where SByis the spawning output in year y, βis a parameter controlling the shape of
density-dependent relationship between relative spawning depletion SBy/SB0and pre-recruit
survival (with limit β< 1), ˜
Ryis the recruitment in year y, and σRis the standard deviation of
recruitment in log space.
As implemented in Stock Synthesis, the parameters needed to apply the stock-recruitment
relationship based on the pre-recruit survival are log(R0), zfrac, and β. The value of log(R0) defines
the equilibrium quantities of SB0,S0, and z0for a given set of biological inputs (either estimated of
fixed), regardless of the values of zfrac and β.
The interpretation of the quantity z0=−log(S0)as pre-recruit instantaneous mortality rate at
unfished equilibrium is imperfect because the recruitment in a given year is calculated as a product of
the survival fraction Syand the spawning output SByfor that same time period so that there is not a
1-year lag between quantification of eggs or pups and recruitment at age 0, which is when recruits
are calculated within Stock Synthesis. However, if age 0 or some set of youngest ages is not selected
by any fishery of survey, then density dependent survival may be assumed to occur anywhere before
the first appearance of any cohort in the data or model expectations. In such cases, the upper limit on
survival up to age ais given by Smaxe−aM .
Nevertheless, interpreting z0as an instantaneous mortality helps with the understanding of zfrac. This
parameter controls the magnitude of the density-dependent increase in survival associated with a
reduction in spawning output. It represents the fraction by which this mortality-like rate is reduced
as spawning output is reduced from SB0to 0. This is approximately equal to the increase in survival
as a fraction of the maximum possible increase in survival. That is:
zfrac =log(Smax)−log(S0)
−log(S0)≈Smax −S0
1−S0
(12)
97
For example, if S0= 0.4,zfrac = 0.8, then the resulting fraction increase in survival is
(Smax −S0)/(1 −S0)=0.72.
The parameter βcontrols the point where survival changes fastest as a function of spawning
depletion. A value of β= 1 corresponds to a linear change in log survival and an approximately linear
relationship between survival and spawning depletion. Values of β<1 have survival increasing fastest
at low spawning output (concave decreasing survival) whereas β>1 has the increase in survival
occurring fastest closer to the unfished equilibrium (convex decreasing survival).
The steepness (h) of the spawner-recruit curve (defined as recruitment relative to R0 at a spawning
depletion level of 0.2) based on pre-recruit survival can be derived from the parameters discussed above
according to the relationship and associated inequality:
h= 0.2ez0zfrac(1−0.2β)<0.2ez0=1
5S0
=SB0
5R0
(13)
Unlike the Beverton-Holt stock-recruitment relationship, recruitment can increase above R0for
stocks that are below SB0and thus the steepness is not fundamentally constrained below 1.
However, in many cases, steepness will be limited well below 1 by the inequality above, which implies
an inverse relationship between the maximum steepness and equilibrium survival. Specifically, the
inequality above bounds steepness below 1 for all cases where S0>0.2, which are those with the
lowest fecundity, an intuitively reasonable result. For example, with S0=0.4, the steepness is limited
below 0.5, regardless of the choice of zfrac or β. This natural limit on steepness may be one of the
most valuable aspects of this stock-recruitment relationship.
Code for the survival based recruitment can be found in Appendix C.
Shepherd
The Shepherd stock recruit curve is calculated as:
Ry= SBy
SB0!5hadj R0(1 −0.2c)
(1 −5hadj 0.2c) + (5hadj −1)(SBy
SB0)ce−0.5byσ2
R+˜
Ry˜
Ry∼N(0; σ2
R)(14)
where c is the shape parameter for the stock recruitment curve, and hadj is the transformed steepness
parameter defined as:
hadj = 0.2 + h−0.2
0.8! 1
5∗0.2c−0.2!(15)
Shepherd Re-parameterization
The Shepherd stock recruit curve re-parameterized version (More details can be found in Punt and
98

Cope, in press)
Ry=R0SBy
SB0 5h(1 −0.2c)
1−5h0.2c+ (5h−1)(SBy/SB0)c!(16)
where cis the shape parameter for the stock recruitment curve.
Ricker Re-parameterization
The Ricker stock recruit curve re-parameterized version (More details can be found in Punt and Cope,
in press)
Ry=R0∗(1 −temp)∗eln(5h)(1−SBy/SB0)γ/0.8γ(17)
where γis the Ricker shape parameter and temp is defined as:
temp =
1−SBy/SB0if 1−SBy/SB0>0
0.001 if 1−SBy/SB0≤0(18)
where temp stabilizes recruitment at R0if SBy> SB0.
Recruitment Eras
Conceptually, SS treats the early, data-poor period, the main data-rich period, and the recent/forecast
time period as three eras along a continuum. The user has control of the break year between eras.
Each era has its own vector. The early era is defined as a vector (prior to V3.10 this was a deviation
vector) so it can have zeros during the earliest years not informed by data and then a few years with
non-zero values without imposing a zero-centering on this collection of deviations. The main era can
be a vector of simple deviations, or a deviation vector but it is normally implemented as a deviation
vector so that the spawner-recruitment function is its central tendency. The last era does not force a
zero-centered deviation vector so it can have zeros during the actual forecast and non-zero values in
last few years of the time series. The early and last eras are optional, but their use can help prevent
SS from balancing a preponderance of negative deviations in early years against a preponderance of
positive deviations in later years. When the 3 eras are used, it would be typically to turn on the main
era during an early model phase, turn on the early era during a later phase, then have the last era turn
on in the final phase.
Recruitment Likelihood with Bias Adjustment
For each year in the total recruitment deviation time series (early, mid, late/forecast) the contribution of
that year to the log-likelihood is equal to: dev2/(2.0∗sigmaR2)+offset∗log(sigmaR); where offset
is the recruitment bias adjustment between the arithmetic and geometricmean of expected recruitment
for that year. With this approach, years with a zero or small offset value do not contribute to the
99

second component. SigmaR may be estimable when there is good data to establish the time series of
recruitment deviations, but see recent work (Thorson et al) on use of a superior approach.
The recruitment bias adjustment implemented in SS is based upon the work documented in Methot
and Taylor (2011) and following the work of Maunder and Deriso (2003). The concept is based upon
the following logic. SigmaR represents the true variability of recruitment in the population. It
provides the constraining penalty for the estimates of recruitment deviations and it is not affected by
data. Where data that are informative about recruitment deviations are available, the total variability
in recruitment, sigmaR, is partitioned into a signal (the variability among the recruitment estimates)
and the residual, the variance of each recruitment estimate (see equation below). Where there are no
data, no signal can be estimated and the individual recruitment deviations collapse towards 0.0 and
the variance of each recruitment deviation approaches sigmaR. Conversely, where there highly
informative data about the recruitment deviations, then the variability among the estimated
recruitment deviations will approach sigmaR and the variance of each recruitment deviation will
approach zero. Perfect data will estimate the recruitment time series signal perfectly. Of course, we
never have perfect data so we should always expect the estimated signal (variability among the
recruitment deviations) to be less than the true population recruitment variability.
SE(ˆry)2+SD(ˆr)2=
1
σ2
d
+1
σ2
R!−1/2
2
+
σ2
R
(σ2
R+σ2
d)1/2
2
=σ2
R(19)
The correct offset (bias adjustment) to apply to the expected value for recruitment is based on the
concept that a time series of estimated recruitments should be mean unbiased, not median unbiased,
because the biomass of a stock depends upon the cumulative number of recruits, which is dominated
by the large recruitments. The degree of offset depends upon the degree of recruitment signal that can
be estimated. Where no recruitment signal can be estimated, the median recruitment is the same as
the mean recruitment, so no offset is applied. Where lognormal recruitment signal can be estimated,
the mean recruitment will be greater than the median recruitment. The value
by=
ESD(ˆry)2
σ2
R
= 1 −SE(ˆry)2
σ2
R
(20)
of the offset then depends upon the partitioning of sigmaR into between and within recruitment
variability. The most appropriate degree of bias adjustment can be approximated from the
relationship among sigmaR, recruitment variability (the signal), and recruitment residual error.
100
Because the quantity and quality of data varies during a time series, SS allows the user to control the
rate at which the offset is ramped in during the early, data-poor years, and then ramped back to zero
for the forecast years. On output to report.sso, SS calculates the mean bias adjustment during the
early and main eras and compares it to the RMSE of estimated recruitment deviations. A warning is
generated if the RMSE is small and the bias adjustment is larger than 2.0 times the ratio of rmse2to
sigmaR2.
In MCMC mode, the model still draws recruitment deviations from the lognormal distribution, so the
full offset is used such that the expected mean recruitment from this lognormal distribution will stay
equal to the mean from the spawner-recruitment curve. When SS reaches the MCMC and MCEVAL
phases, all bias adjustment values are set to 1.0 for all active recruitment deviations because the model
is now re-sampling from the full lognormal distribution of each recruitment.
Recruitment Autocorrelation
The autocorrelation parameter is implemented. It is not performance tested and it has no effect on the
calculation of the offsets described in the section above.
Recruitment Cycle
When SS is configured such that seasons are modeled as years, the concept of season within year
disappears. However, there may be reason to still want to model a repeating pattern in expected
recruitment to track an actual seasonal cycle in recruitment. If the recruitment cycle factor is set to a
101
positive integer, this value is interpreted as the number of time units in the cycle and this number of
full parameter lines will be read. The cyclic effect is modeled as an exp(p) factor times R0, so a
parameter value of 0.0 has nil effect. In order to maintain the same number of total recruits over the
duration of the cycle, a penalty is introduced so that the cumulative effect of the cycle produces the
same number of recruits as Ncycles * R0. Because the cyclic factor operates as an exponential, this
penalty is different than a penalty that would cause the sum of the cyclic factors to be 0.0. This is
done by adding a penalty to the parameter likelihood, where:
X=X(ep)
Y=Ncycle
P enalty = 100000 ∗(X−Y)2
(21)
Initial Age Composition
A non-equilibrium initial age composition is achieved by setting the first year of the recruitment
deviations before the model start year. These pre-start year recruitment deviations will be applied to
the initial equilibrium age composition to adjust this composition before starting the time series. The
model first applies the initial F level to an equilibrium age composition to get a preliminary N-at-age
vector and the catch that comes from applying the F’s to that vector, then it applies the recruitment
deviations for the specified number of younger ages in this vector. If the number of estimated ages in
the initial age composition is less than Nages, then the older ages will retain their equilibrium levels.
Because the older ages in the initial age composition will have progressively less information from
which to estimate their true deviation, the start of the bias adjustment should be set
accordingly.
Fishing Mortality Method
There are three methods available for calculation of fishing mortality. These are: Pope’s approximation,
continuous F with each F as a model parameter, and a hybrid method that does a Pope’s approximation
to provide initial values for iterative adjustment of the continuous F values to closely approximate the
observed catch. With the hybrid method, the final values are in terms of continuous F, but do not need
to be specified as full parameters. In a 2 fishery, low F case, the hybrid method is just as fast as the Pope
approx. and produces identical result. When F is very high, the problem becomes quite stiff for Pope’s
and the hybrid method so convergence may slow. It may be better to use F option 2 (continuous F as full
parameters) in these high F cases. F as parameter is also preferred for situations where catch is known
imprecisely and you are willing to accept a solution in which the final F values do not reproduce the
input catch levels exactly. For the F as parameter approach, there is an option to do early phases using
hybrid, then switch to F as parameter in later phases and transfer the hybrid F values to the parameter
initial values.
Option 1 (Pope’s approx) still exists, but it is recommended to switch to option 3.
102
Control file continued:
Typical Value Description and Options
0.2 F ballpark
This value is compared to the sum of the F’s for the specified
year. The sum is over all seasons and areas. The lambda for the
comparison goes down by a factor of 10 each phase and goes to 0.0
in the final phase.
-1990 F ballpark year
Negative value disable F ballpark.
3 F Method
1 = Pope’s (discrete),
2 = Baranov (continuous) F as a parameter,
3 = Hybrid F (recommended).
2.9 Maximum F
This maximum is applied within each season and area. A value
of 0.9 is recommended for F method 1, and a value of about 4 is
recommended for F method 2 and 3.
COND: F method = 1, no additional input for Pope’s approximation.
COND: F method = 2:
0.10 1 1 Inital F value, Phase, N F detail setup lines to read. Starting value for
each F. Initializing value for each F parameter.
For phases prior to the phase of the F value becoming active, SS
will use the hybrid option and the F values so calculated become the
starting values for the F parameters when this phase is reached.
If F method = 2 and N for F detail is > 0
1 1980 1 0.20 0.05 4 fleet, year, season, F, SE, phase - these values override the catch se
values in the data file and the overall starting F value and phase read
just above.
COND: Fmethod = 3
4 Number of tuning iterations in hybrid method. A value of 2 or 3 is
sufficient with a single fleet and low Fs. A value of 5 or so may be
needed to match the catch near exactly when there are many fleets
and high F.
103
Initial Fishing Mortality
Read a short parameter setup line for each fishery. The parameters are the fishing mortalities for the
initial equilibrium. Do not try to estimate parameters for fisheries with zero initial equilibrium catch.
If there is catch, then give a starting value greater than zero and it generally is best to estimate the
parameter in phase 1.
In SS3.30, the initial equilibrium year has explicit seasons, so the needed initial F values will also be by
season. Initial F values are only needed for fleet/seasons that have catch. So if no fleet/season combo
has catch, then no parameters are needed.
It is possible to use the initial F method to achieve an estimate of the initial equilibrium Z in cases
where the initial equilibrium catch is unknown. To do this:
•Include a positive value for the initial equilibrium catch;
•Set the lambda for the logL for initial equilibrium catch to a nil value (hence causing SS to ignore
the lack of fit to the input catch level;
•Allow the initial F parameter to be estimated. It will be influenced by the early age and size
comps which should have some information about the early levels of Z.
Catchability
Catchability is the scaling factor that relates a model quantity to the expected value for some type
of data (index). Typically this is used to converted selected numbers or biomass for a fleet into the
expected value for a survey or CPUE by that fleet. In SS, the concept has been extended so that, for
example, a time series of an environmental factor could be treated as a survey of the time series of
deviations for some parameter. This flexibility means that a family of link functions beyond simple
proportionality is needed.
For each fishery and survey with an index, enter a row with the entries as described below:
1. Fleet Number
2. Link type or index of dev vector: An assumed functional form between Q, the expected value,
and the survey observation.
(a) 1 = simple Q, proportional assumption about Q: y=q∗x.
(b) 2 = mirror simple Q, 1 mirrored parameter.
104
(c) 3 = Q with power, 2 parameters establish a parameter for non-linearity in
survey-abundance linkage. Assumes proportional with offset and power function:
y=qxcwhere q=exp(lnQbase)) thus the cis not related to expected biomass but
vulnerable biomass to Q. Therefore, c < 0 leads to hyper-stability and c > 0leads to
hyper-depletion.
(d) If the parameter is for an index of a dev vector (index units = 35), use this column to enter
the index of the dev vector to which the index is related
3. Extra input for link (i.e. mirror fleet)
(a) >0 = mirror the Q from another (lower numbered survey designated by abs(value))
4. Do extra SD
(a) 0 = skip (typical)
(b) 1 = estimate a parameter that will contain an additive constant to be added to the input
standard deviation of the survey variability. This extra SD approach accomplishes the same
thing in principle as the older code, but may not give exactly the same answer as the older
code. The newer code for extra SD estimation is recommended.
5. Bias adjustment
(a) 0 = no bias adjustment applied
(b) 1 = apply bias adjustment
6. Q float
(a) 0 = no float (parameter is estimated)
(b) 1 = float (analytical solution is used, but parameter line still required)
For a setup with a single survey, the Q setup matrix could be:
Fleet Link Link Extra Bias
Num. Type Info SD Adjust Float Label
3 1 0 1 1 0 #Survey
-9999 0 0 0 0 0 #End Read
105

A long parameter line is expected for each link parameter (i.e. Q) and for the
LO HI INIT PRIOR <other entries> Block Fxn Parameter Label
-5 5 -0.12 0 ... 0 #Survey1 LnQ base
0 0.5 0.1 0 ... 0 #Survey1 Extra SD
If the Q base parameter specifies that it is time-varying by the annual dev method, short parameter
lines to specify the specifications of the deviation vector come after all the base Q parameters.
Note: In 3.24 it was common to use the deviation approach and this deviation approach in SS v.3.24
was implemented as if it was survey specific blocks. In some cases, only one year’s deviation was made
active in order to implement, in effect, a block for Q. sstrans.exe cannot convert this, but an analogous
approach is available in SS3.30 because true blocks can now be used, as well as env links and annual
deviations. Also note that deviations in 3.24 were survey specific (so no parameter for years with no
survey). But in SS3.30, deviations are always year-specific, so you might have a deviation created for a
year with no survey.
Selectivity and Discard
For each fleet and survey, read a definition line for size selectivity and retention. The four values read
from each line are:
Pattern
Valid length selectivity pattern code.
•Discard (0/1/2/3/4 or -index) If value is 1, then program will read 4 retention parameters after
reading the specified number of selectivity parameters and all discarded fish are assumed dead.
If the value is 2, then the program will read 4 retention parameters and 4 discard mortality
parameters. If the value is 3, then no additional parameters are read and all fish are assumed
discarded and dead. If the value is 4, then the program will read 7 retention parameters (for
dome-shaped retention) and 4 discard mortality parameters. If the value is a negative number,
then it will mirror the retention and discard mortality pattern of the lower numbered fleet.
•Male (0/1/2/3/4) If value is 1, then program will read 4 additional parameters to define the
male selectivity relative to the female selectivity. Anytime the male selectivity is caused to be
greater than 1.0; the entire male/female matrix of selectivity values is scaled by the max so that
the realized max is 1.0. Hopefully this does not cause gradient problems. If the value is 2, then
the main selectivity parameters define male selectivity and female selectivity is estimated as an
offset from male selectivity. This alternative is preferable if female selectivity is less than male
106

selectivity. The option 3 is only available if the selectivity pattern is 1, 20, or 24 and it causes
the male selectivity parameters to be offset from the female parameters, rather than the male
selectivity being an offset from the female selectivity.
•Special (0/value). This value is used in different ways depending on the context. If the selectivity
type is to mirror another selectivity type, then put the index of that source fleet or survey here.
It must refer to a lower numbered fleet/survey. If the selectivity type is 6 (linear segment), then
put the number of segments here. If the selectivity type is 7, then put a 1 here to keep selectivity
constant above the mean average size for old fish of morph 1.
For each fleet and survey, read a definition line for age selectivity. The 4 values to be read are the same
as for the size-selectivity. However, the retention value must be set to 0.
Example Setup for Size Selectivity:
Pattern Discard Male Special Label
1 2 0 0 #Fishery1
1 0 0 0 #Survey1
0 0 0 0 #Survey2
Age Selectivity Types:
Pattern Discard Male Special Label
11 0 0 0 #Fishery1
11 0 0 0 #Survey1
11 0 0 0 #Survey2
Reading the Selectivity and Retention Parameters
Read the required number of parameter setup lines as specified by the definition lines above. The
complete order of the parameter setup lines is:
1. Size selectivity for fishery 1
2. Retention for fishery 1 (if discard specified)
3. Discard Mortality for fishery 1 (if discard specified)
4. Male offsets for size selectivity for fishery 1 (if offsets used)
107

5. <repeat for additional fleets and surveys>
6. Age selectivity for fishery 1
7. Male offsets for age selectivity for fishery 1 (if offsets used)
8. <repeat for additional fleets and surveys>.
The list of parameters to be read from the above setup would be:
LO HI INIT PRIOR <other entries> Block Fxn Parameter Label
19 80 53.5 50 ... 0 #SizeSel p1 fishery 1
0.01 60 18.9 15 ... 0 #SizeSel p2 fishery 1
20 70 38.6 40 ... 0 #Retain p1 fishery 1
0.1 10 6.5 1 ... 0 #Retain p2 fishery 1
0.001 1 0.98 1 ... 0 #Retain p3 fishery 1
-10 10 1 0 ... 0 #Retain p4 fishery 1
0.1 1 0.6 0.6 ... 0 #DiscMort p1 fishery 1
-2 2 0 0 ... 0 #DiscMort p2 fishery 1
20 70 40 40 ... 0 #DiscMort p3 fishery 1
0.1 10 1 1 ... 0 #DiscMort p4 fishery 1
19 80 53.5 50 ... 0 #SizeSel p1 survey 1
0.01 60 18.9 15 ... 0 #SizeSel p2 survey 1
0 40 0 5 ... 0 #AgeSel p1 fishery 1
0 40 40 5 ... 0 #AgeSel p2 fishery 1
0 40 0 5 ... 0 #AgeSel p1 survey 1
0 40 40 5 ... 0 #AgeSel p2 survey 1
0 40 0 5 ... 0 #AgeSel p1 survey 2
0 40 0 5 ... 0 #AgeSel p2 survey 2
Selectivity Patterns
The currently defined selectivity patterns, and corresponding required number of parameters,
are:
SIZE BASED SELECTIVITY
Pattern N Parameters Description
0 0 Selectivity = 1.0 for all sizes.
108

Pattern N Parameters Description
1 2 Logistic selectivity.
2 8 Discontinued: Double logistic with defined peak (uses IF
joiners). Use pattern 8 instead.
3 6 Discontinued
4 0 Discontinued: Set size selectivity equal to female fecundity.
Use special survey pattern 30 located in the index section
within the data file instead.
5 2 Mirror another selectivity. The two parameters select bin
range.
6 2 + special value Non-parametric
7 8 Discontinued: Double logistic with defined peak, uses smooth
joiners; special = 1 causes constant selectivity above Linf for
morph 1. Use pattern #8.
8 8 Double logistic, with defined peak, uses smooth joiners;
special=1 causes constant selectivity above Linf for morph 1.
9 6 Simple double logistic with no defined peak.
15 0 Mirror another selectivity (same as for age selectivity).
21 1 + special value Non-parametric size selectivity
22 4 Double normal; similar to CASAL.
23 6 Same as the double normal pattern 24 except the final
selectivity is now directly interpreted as the terminal
selectivity value. Cannot be used with Pope’s F method
because maximum selectivity may be greater than 1.
24 6 Double normal with defined initial and final selectivity level –
Recommended option. Test using SELEX-24.xls.
25 3 Exponential-logistic.
27 3 + 2*N nodes Cubic spline with N nodes.
42 5 + 2*x Selectivity pattern 27 but with 2 additional scaling
parameters.
43 4 + x Selectivity pattern 6 but with 2 additional scaling parameters.
109

AGE BASED SELECTIVITY
Pattern N Parameters Description
0 0 Selectivity = 1.0 for ages 0+.
10 0 Selectivity = 1.0 for all ages beginning at age 1. If it is
desired that age-0 fish be selected, then use pattern #11 and
set minimum age to 0.0.
11 2 Selectivity = 1.0 for a specified age range.
12 2 Logistic selectivity.
13 8 Double logistic, IF joiners. Use discouraged. Use pattern 18
instead.
14 nages + 1 Separate parameter for each age (empirical), value at age is
1
1+exp(−x).
15 0 Mirror another age-specific selectivity pattern.
16 2 Coleraine single Gaussian
17 nages + 1 Empirical as a random walk from previous age. For all ages
in the population beginning with Amin = 1 for the fishery
and 2 for the survey, there is a corresponding set of selectivity
parameters for each fleet, pa.
18 8 Double logistic, with defined peak, uses smooth joiners.
19 6 Simple double logistic with no defined peak.
20 6 Double normal with defined initial and final level.
Recommended option. Test using SELEX-24.xls.
26 3 Exponential logistic.
27 3 + 2*N nodes Cubic spline in age based on N nodes.
41 2 + nages + 1 Selectivity pattern 17 but with 2 additional scaling
parameters.
42 5 + 2*N nodes Selectivity pattern 27 but with 2 additional scaling
parameters.
44 4 + nages Similar to age selectivity pattern 17 but with separate
parameters for males and with revised controls.
45 4 + nages Similar to age selectivity pattern 14 but with separate
parameters for males and with revised controls.
110

Special Selectivity Options
Special selectivity options (type 30 in size based selectivity) are no longer specified within the control
file. Specifying the use of one of these selectivity types is now done within the data file by selecting the
survey "units" (see the section on Index units).
Selectivity Pattern Details
Pattern 1 (size) and 12 (age) - Simple Logistic
Within SS logistic selectivity for the primary sex (if selectivity varies by sex) is formulated as:
Sl=1.0
1 + exp(−ln(19)(Ll−p1)/p2) (22)
where Llis the length bin. If age based selectivity is selected then the length bin is replaced by the
age vector. If sex specific selectivity is specified the non-primary sex the p1 and p2 parameters are
estimated as offsets. Note that with a large p2 parameter, selectivity may not reach 1.0 at the largest
size bin. The parameters are:
•p1 - size/age at inflection
•p2 - width for 95% selection; a negative width causes a descending curve.
Pattern 5 (size) - Mirror Selectivity
Two parameters select the min and max bin number (not min max size) of the source pattern. If first
parameter has value <=0, then interpreted as a value of 1 (e.g. first bin). If second parameter has value
<=0, then interpreted as nlength (e.g. last bin). The source pattern must have a lower type
number
Pattern 6 (size) - Non-parametric Selectivity
Non-parametric size selectivity uses a set of linear segments. The first waypoint is at Length = p1 and
the last waypoint is at Length = p2. The total number of waypoints is specified by the value of the
Special factor in the selectivity set-up, so the N intervals is one less than the number of waypoints.
Intermediate waypoints are located at equidistant intervals between p1 and p2. Parameters 3 to N are
the selectivity values at the waypoints, entered as logistic, e.g. 1/(1 + exp(−x)). Ramps from –10 to
p3 if L<p1. Constant at pN if L>p2. Note that prior to version 3.03 the waypoints were specified in
terms of bin number, rather than length.
Pattern 8 (size) and 18 (age) - Double Logistic
111

•p1 – PEAK: size (age) for peak. Should be an integer and should be at bin boundary and not
estimated. But options 7 and 18 may allow estimation.
•p2 – INIT: selectivity at lengthbin=1 (minL) or age=0.
•p3 – INIT: selectivity at lengthbin=1 (minL) or age=0. A logit transform (1/(1 + exp(−x))
is used so that the transformed value will be between 0 and 1. So a p1 value of –1.1 will be
transformed to 0.25 and used to set the selectivity equal to 0.5 at a size (age) equal to 0.25 of the
way between minL and PEAK.
•p4 – SLOPE1: log(slope) of left side (ascending) selectivity.
•p5 – FINAL: logit transform for selectivity at maxL (or maxage).
•p6 – INFL2: logit transform for size(age) at right side selectivity equal to half way between
PEAK+PEAKWIDTH and maxL (or max age).
•p7 – SLOPE2: log(slope) of right side (descending) selectivity
•p8 – PEAKWIDTH: in width of flattop.
Pattern 14 (age) - Revise Age
Age-selectivity pattern 14 to allow selectivity-at-age to be the same as selectivity at the next younger
age. When using this option, the range on each parameter should be approximately -5 to 9 to prevent
the parameters from drifting into extreme values with nil gradient. SS calculates the age-based
selectivity as where a= 1 to a=Amax + 1:
temp = 9 −max(p(a))
Sa=1
1 + exp(−(p(a+ 1) + temp))
(23)
Pattern 17 (age) - Random Walk
This selectivity pattern provides for a random walk in ln(selectivity). For each age a≥Amin, where
Amin is the minimum age for which selectivity is allowed to be non-zero, there is a selectivity parameter,
pa, controlling the changein selectivity from age a−1to age a.
The selectivity at age ais computed as
Sa= exp(S0
a−S0
max),(24)
where
S0
a=
a
X
i=Amin
pi(25)
112
and
S0
max =max{S0
a}.(26)
Selectivity is fixed at Sa= 0 for a<Amin.
This formulation has the properties that the maximum selectivity equals 1, positive values of paare
associated with increasing selectivity between ages a−1and a, and negative values are associated
with decreasing selectivity between those ages and pa= 0 gives constant selectivity.
The condition that maximum selectivity equals 1 results in one fewer degree of freedom than the
number of estimated pa. Therefore, at least one parameter should be fixed at an arbitrary value,
typically pAmin = 0.
In typical usage:
•First parameter (for age 0) could have a value of -1000 so that the age 0 fish would get a selectivity
of 0.0;
•Second parameter (for age 1) could have a value of 0.0 and not be estimated, so age 1 is the
reference age against which subsequent changes occur;
•Next parameters get estimated values. To assure that selectivity increases for the younger ages,
the parameter min for these parameters could be set to 0.0 or a slightly negative value.
•If dome-shaped selectivity is expected, then the parameters for older ages could have a range
with the max set to 0.0 so they cannot increase further.
•To keep selectivity at a particular age the same as selectivity at the next younger age, set its
parameter value to 0.0 and not estimated. This allows for all older ages to have the same
selectivity.
•To keep a constant rate of change in selectivity across a range of ages, use the -999 flag to keep
the same rate of change in ln(selectivity) as for the previous age.
•Code for implementing random walk selectivity within SS can be found in Appendix C.
Pattern 9 (size) and 19 (age) - Simple Double Logistic with no defined peak
•p1 - INFL1: ascending inflection size (in cm)
•p2 – SLOPE1: ascending slope
•p3 – INFL2: descending inflection size (in cm)
113
•p4 – SLOPE2: descending slope
•p5 – first BIN: bin number for the first bin with non-zero selectivity (must be an integer bin
number, not a size)
•p6 – offset: enter 0 if P3 is independent of P1; enter 1 if P3 is an offset from P1
Pattern 22 (size) - Double Normal with Plateau
•p1 – PEAK1: beginning size for the plateau (in cm)
•p2 – PEAK2: ending size for the plateau. Calculated as a fraction of the distance between PEAK1
and 99% of the lower edge of the last size bin in the model. Transformed as (1/(1+exp(-p2)). So
a value of 0 results in PEAK2 being halfway between PEAK1 and 99% of the last bin
•p3 – upslope: ln(variance) on ascending side
•p4 – downslope: ln(variance) on descending side
Pattern 23 (size) and 24 (size) - Double Normal Selectivity
•p1 – PEAK: beginning size for the plateau (in cm)
•p2 – TOP: width of plateau, as logistic between PEAK and MAXLEN
•p3 – ASC-WIDTH: parameter value is ln(width)
•p4 – DESC-WIDTH: parameter value is ln(width)
•p5 – INIT: selectivity at first bin, as logistic between 0 and 1.
•p6 – FINAL: selectivity at last bin, as logistic between 0 and 1. (for pattern 24) or
•p6 – FINAL: selectivity at last bin, as absolute value, so can be >1.0. (for pattern 23). Warning: Do
not allow this value to go above 1.0 if the F_method uses Pope’s approximation. OK to go above
1.0 when F is in exponential form. When this parameter is above 1.0, the overall selectivity
pattern will have an intermediate plateau at 1.0 (according to peak and top), then will ascend
further to the final value.
Notes for Double Normal Selectivity:
114
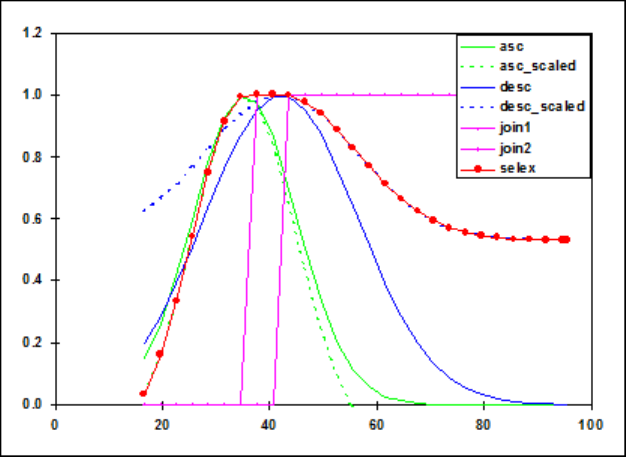
•See spreadsheet SELEX-24.xls for parameterization example.
•For the initial selectivity parameter (parameter 5)
–-999 or –1000: ignore the initial selectivity algorithm and simply decay the small fish
selectivity according to P3,
–< -1000: ignore the initial selectivity algorithm as above and then set selectivity equal to
1.0e-06 for size bins 1 through bin = -1001 –value. So a value of –1003 would set selectivity
to a nil level for bins 1 through 2 and begin using the modeled selectivity in bin 3.
•For the final selectivity parameter (parameter 6)
–-999 or –1000: ignore the final selectivity algorithm and simply decay the large fish
selectivity according to parameter 4,
–<-1000: set selectivity constant for bins greater than bin number = -1000 – value.
Selectivity pattern 24, double normal, showing sub-functions and steep logistic joiners:
Pattern 15 (age) - Mirror
No parameters. Whole age range is mirrored from a user-specified fleet.
Pattern 16 - Gaussian (similar to Coleraine)
115

•p1 – age below which selectivity declines
•p2 – scaling factor for decline
Pattern 9 (size) and 19 (age) - Simple Double Logistic
•p1 – ascending inflection age/size
•p2 – ascending slope
•p3 – descending inflection age/size
•p4 – descending slope
•p5 – age or size at first selection; this is a specification parameter, so must not be estimated.
Enter integer that is age for pattern 19 and is bin number for pattern 9
•p6 – (0/1) where a value of 0 causes the descending inflection to be a standalone parameter, and
a value of 1 causes the descending inflection to be interpreted as an offset from the ascending
inflection. This is a specification parameter, so must not be estimated.
A value of 1.0e-6 is added to the selectivity for all ages, even those below the minage.
Pattern 25 (size) and 26 (age) - Exponential logistic
•p1 – ascending rate, min: 0.02, max: 1.0, reasonable start value: 0.1
•p2 – peak, as fraction of way between min size and max size. Parameter min value: 0.01; max:
0.99; reasonable start value: 0.5
•p2 – minsize + p2*(maxsize-minsize)
•p3 – descending rate, min: 0.001, max: 0.5, reasonable start value: 0.01. A value of 0.001 provides
a nearly asymptotic curve. Values above 0.2 provide strongly dome-shaped function in which
the p3 and p1 parameters interact strongly.
ep3∗p1(p20−size)
1−p3(1 −ep1(p20−size))(27)
Example: Exponential logistic selectivity with p1 = 0.30, p2 = 0.50, and p3 = 0.02:
116
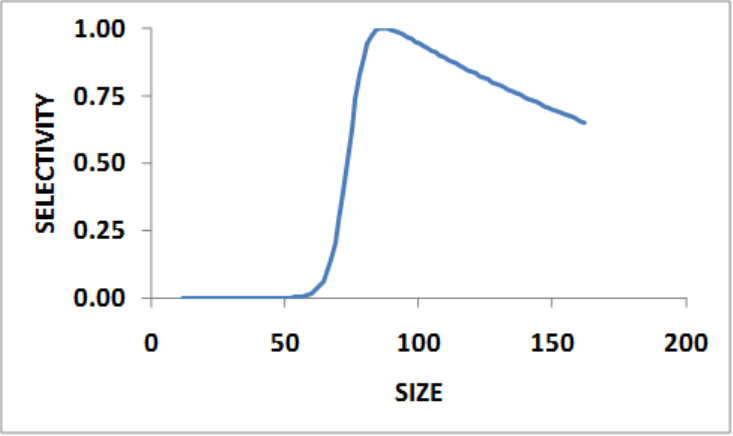
Pattern 27 (size and age)- Cubic Spline
This selectivity pattern uses the ADMB implementation of the cubic spline function. This function
requires input of the number of nodes, the positions of those nodes, the parameter values at those
nodes, and the slope of the function at the first and last node. In SS, the number of nodes is specified
in the “special” column of the selectivity set-up. The pattern number 27 is used to invoke cubic spline
for size selectivity and for age selectivity; the input syntax is identical.
For a 3 node setup, the SS input parameters would be:
•p1 – code for initial set-up (0, 1 or 2) as explained below
•p2 – gradient at the first node (should be a small positive value)
•p3 – gradient at the last node (should be zero or a small negative value)
•p4-p6 – the nodes in units of cm; must be in rank order and inside of the range of the population
length bins. These must be held constant (not estimated, e.g. negative phase value) during a
model run.
•p7-p9 – the values at the nodes. Units are ln(selectivity).
Notes:
•There must be at least 3 nodes.
•One of these selectivity parameter values should be held constant so others are estimated relative
to it.
117
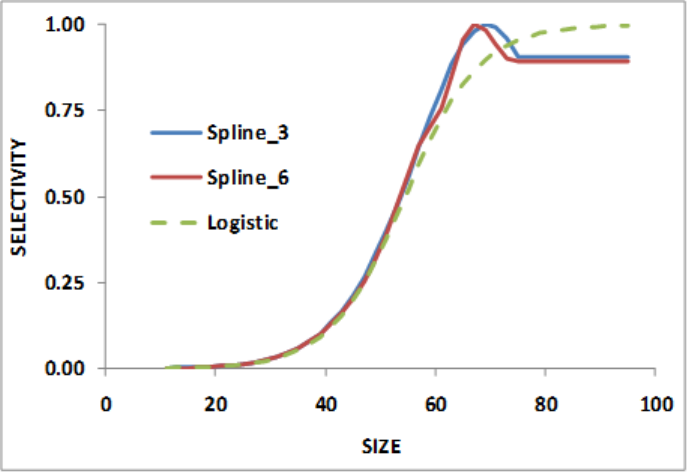
•Selectivity is forced to be constant for sizes greater than the size at the last node
•The overall selectivity curve is scaled to have a peak equal to 1.0.
•Terminal nodes cannot be at the min or max population length bins.
Code for implementing cubic spline selectivity within SS can be found in Appendix C.
The figure below compares a 3 node and a 6 node cubic spline with a 2 parameter logistic function.
In fitting these functions, the 2 cubic spline approaches fit slightly better than the logistic, presumably
because the data were slightly indicative of a small dome in selectivity.
Auto-Generation of Cubic Spline Control File Set-Up: A New SS feature pioneered with the cubic
spline function is a capability to produce more specific parameter labels and to auto-generate
selectivity parameter setup. The auto-generation feature is controlled by the first selectivity
parameter value for each fleet that is specified to use the cubic spline. There are 3 possible values for
this setup parameter:
•0: no auto-generation, process parameter setup as read.
•1: auto-generate the node locations based on the specified number of nodes and on the
cumulative size distribution of the data for this fleet/survey.
•2: auto-generate the nodes and also the min, max, prior, init, and phase for each parameter.
With either the auto-generate option #1 or #2, it still is necessary to include in the parameter file
placeholder rows of values so that the init_matrix command can input the current number of values
118

because all selectivity parameter lines are read as a single matrix dimensioned as N parameters x 14
columns. The read values of min, max, init, prior, prior type, prior stddev, and phase will be
overwritten.
Cumulative size and age distribution is calculated for each fleet, summing across all samples and both
sexes. These distributions are output in echoinput.sso and in a new OVERALL_COMPS section of
report.sso.
When the nodes are auto-generated, the first node is placed at the size corresponding to the 2.5%
percentile of the cumulative size distribution, the last is placed at the 97.5% percentile of the size
distribution, and the remainder are placed at equally spaced percentiles along the cumulative size
distribution. These calculated node values are output into control.ss_new. So, the user could extract
these nodes from control.ss_new, edit them to desired values, then, insert them into the input control
file. Remember to turn off auto-generation in the revised control file.
When the complete auto-generation is selected, the control.ss_new would look like the table
below:
LO HI INIT <other entries> Block Fxn Parameter Label
0 2 2.0 ... 0 #SizeSpline Code
-0.001 1 0.13 ... 0 #SizeSpline GradLo
-1 0.001 -0.03 ... 0 #SizeSpline GradHi
11 95 38 ... 0 #SizeSpline Knot1
11 95 59 ... 0 #SizeSpline Knot2
11 95 74 ... 0 #SizeSpline Knot3
-9 7 -3 ... 0 #SizeSpline Value1
-9 7 -1 ... 0 #SizeSpline Value2
-9 7 -0.78 ... 0 #SizeSpline Value3
Pattern 41 (age) - Random Walk with User-Defined Scaling
Selectivity pattern 17 with two additional parameters. The two additional parameters are the bin
numbers to define the range of bins for scaling. All of the selectivity values will be scaled (divided) by
the mean value over this range. The low and high bin numbers are defined before the other selectivity
parameters.
LO HI INIT <other entries> Block Fxn Parameter Label
0 20 10 ... 0 #AgeSel_ScaleAgeLo
0 20 20 ... 0 #AgeSel_ScaleAgeHi
119

Pattern 42 (size and age) - Cubic Spline with User-Defined Scaling
Selectivity pattern 27 with two additional parameters. The two additional parameters are the bin
numbers to define the range of bins for scaling. All of the selectivity values will be scaled (divided) by
the mean value over this range. The low and high bin numbers are defined before the other selectivity
parameters.
LO HI INIT <other entries> Block Fxn Parameter Label
0 20 10 ... 0 #AgeSpline_ScaleAgeLo
0 20 20 ... 0 #AgeSpline_ScaleAgeHi
Pattern 43 (size) - Non-parametric with User-Defined Scaling
Selectivity pattern 6 with two additional parameters. The two additional parameters are the bin
numbers to define the range of bins for scaling. All of the selectivity values will be scaled (divided) by
the mean value over this range. The low and high bin numbers are defined before the other selectivity
parameters.
LO HI INIT <other entries> Block Fxn Parameter Label
1 80 50 ... 0 #SizeSel_ScaleBinLo
1 80 70 ... 0 #SizeSel_ScaleBinHi
Pattern 44 (age)
Similar to pattern 17 but with separate parameters for males and females. This selectivity pattern
provides for a random walk in ln(selectivity). In typical usage:
•p1 - First parameter (for age 0) could have a value of -1000 so that the age 0 fish would get a
selectivity of 0.0.
•p2 - The first age for which mean selectivity = 1.
•p3 - The last age for which mean selectivity = 1.
•p4 - Male mean selectivity relative to the female selectivity mean entered as ln(ratio) for the male
relative female selectivity.
•p5-pn- Additional parameter lines for the log of the selectivity change between ages
corresponding to the user specified number of changes in the "special" column for the
selectivity specification for each sex with females entered first then males.
120

•-999 input indicates to the model to keep the change unchanged from the previous age (keeps
same rate of change).
•-1000 input indicates used only for male selectivity indicates to the model to set the change in
male selectivity equal to the female change in selectivity.
An example specification and setup for this selectivity option where selectivity is dome-shaped,
peaking at age 2 with female and male selectivity are equal with 4 change points per sex:
#Pattern Discard Male Special
44 0 0 4
LO HI INIT <other entries> Block Fxn Parameter Label
0 20 0 ... 0 #first selex age
0 20 2 ... 0 #first age peak selex (mean)
0 20 2 ... 0 #last age peak selex (mean)
-1 2 -0.001 ... 0 #male ln(ratio)
-10 10 3.01 ... 0 #female ln(selex) change 1
-10 10 1.56 ... 0 #female ln(selex) change 2
-10 10 -0.15 ... 0 #female ln(selex) change 3
-10 10 -0.15 ... 0 #female ln(selex) change 4
-1000 10 -1000 ... 0 #male ln(selex) change 1
-1000 10 -1000 ... 0 #male ln(selex) change 2
-1000 10 -1000 ... 0 #male ln(selex) change 3
-1000 10 -1000 ... 0 #male ln(selex) change 4
Pattern 45 (age) - Revise Age
Similar to pattern 14 but with separate parameters for males and females. Age-selectivity pattern 45
to allow selectivity-at-age to be the same as selectivity at the next younger age.
•p1 - is the first age with non-zero selectivity.
•p2 - The first age in mean for peak selectivity
•p3 - The last age in mean for peak selectivity
121

•p4 - The male mean selectivity relative to the female mean, entered as ln(ratio) for the male
relative female selectivity
•-999 input indicates to the model to keep the change unchanged from the previous age (keeps
same rate of change).
•-1000 input indicates used only for male selectivity indicates to the model to set the change in
male selectivity equal to the female change in selectivity.
An example specification and setup for this selectivity option where selectivity is asymptotic, with
female and male selectivity are equal with 4 change points per sex:
#Pattern Discard Male Special
45 0 0 3
LO HI INIT <other entries> Block Fxn Parameter Label
0 20 2 ... 0 #first selex age
0 20 5 ... 0 #first age peak selex (mean)
0 20 5 ... 0 #last age peak selex (mean)
-1 2 -0.001 ... 0 #male ln(ratio)
-10 10 -8.1 ... 0 #female ln(selex) change 1
-10 10 -4.1 ... 0 #female ln(selex) change 2
-10 10 -1.8 ... 0 #female ln(selex) change 3
-1000 10 -1000 ... 0 #male ln(selex) change 1
-1000 10 -1000 ... 0 #male ln(selex) change 2
-1000 10 -1000 ... 0 #male ln(selex) change 3
Retention
Retention is defined as a logistic function of size. It does not apply to surveys. Four parameters (for
asymptotic retention) or seven parameters (for dome-shaped retention) are used:
•p1 – ascending inflection
•p2 – ascending slope
122

•p3 – maximum retention (often a time-varying quantity to match the observed amount of
discard)
•p4 – male offset to ascending inflection (arithmetic, not multiplicative)
•p5 – descending inflection
•p6 – descending slope
•p7 – male offset to descending inflection (arithmetic, not multiplicative)
Retention = P3
1 + e−(L−(P1+P4∗male))
P2!∗ 1−1
1 + e−(L−(P5+P7∗male))
P6!(28)
Discard Mortality
Discard mortality is defined as a logistic function of size such that mortality declines from 1.0 to an
asymptotic level as fish get larger. It does not apply to surveys and it does not affect the calculation of
expected values for discard data. It is applied so that the total mortality rate is:
deadfish = selex * (retain + (1.0-retain)*discmort)
If discmort is 1.0, all selected fish are dead; if discmort is 0.0, only the retained fish are dead.
Four parameters are used:
•p1 – descending inflection
•p2 – descending slope
•p3 – maximum discard mortality
•p4 – male offset to descending inflection (arithmetic, not multiplicative)
Discard mortality is calculated as:
Mortality = 1−1−P3
1 + e−(L−(P1+P4∗male))
P2!(29)
123
Male Selectivity
There are two approaches to specifying sex specific selectivity. One approach allows male selectivity
to be specified as a fraction of female selectivity (or vice versa). This first approach can be used for
any selectivity pattern. The other option allows for separate selectivity parameters for each sex plus
an additional parameter to define the scaling of one sex’s peak selectivity relative to the other sex’s
peak. This second approach has only been implemented for a few selectivity patterns.
Approach #1:
If the “domale” flag is set to 1, then the selectivity parameters define female selectivity and the offset
defined below sets male selectivity relative to female selectivity. The two sexes switch roles if the
“domale” flag is set to 2. Generally it is best to select the option so that the dependent sex has lower
selectivity, thus obviating the need to rescale for selectivities that are greater than 1.0. Sex specific
selectivity is done the same way for all size and age selectivity options.
•P1 – size (age) at which a dogleg occurs (set to an integer at a bin boundary and do not estimate)
•P2 – log(relative selectivity) at minL or age=0. Typically this will be set to a value of 0.0 (for no
offset) and not estimated. It would be a rare circumstance in which the youngest/smallest fish
had sex-specific selectivity.
•P3 – log(relative selectivity) at the dogleg
•P4 – log(relative selectivity) at maxL or max age.
For intermediate ages, the log values are linearly interpolated on size (age).
If selectivity for the dependent sex is greater than the selectivity for the first sex (which always peaks
at 1.0), then the male-female selectivity matrix is rescaled to have a maximum of 1.0.
Approach #2:
A new sex selectivity option (3 or 4) has been implemented for size selectivity patterns 1 (logistic) and
23 and 24 (double normal) or age selectivity pattern 20 (double normal age). Rather than calculate
male selectivity as an offset from female selectivity, here the male selectivity is calculated by making
the male parameters an offset from the female parameters (option 3), or females are offset from males
with option 4. The description below applies to option 3. If the size selectivity pattern is 1 (logistic),
then read 3 parameters:
•male parameter 1 is added to the first selectivity parameter (inflection)
•male parameter 2 is added to the second selectivity parameter (width of curve)
•male parameter 3 is the asymptotic selectivity
124

If the size selectivity pattern is 20, 23 or 24 (double normal), then:
•male parameter 1 is added to the first selectivity parameter (peak)
•male parameter 2 is added to the third selectivity parameter (width of ascending side); then
exp(this sum) per previous transform
•male parameter 3 is added to the fourth selectivity parameter (width of descending side); then
exp(sum) per previous transform
•male parameter 4 is added to the sixth selectivity parameter (selectivity at final size bin); then
1/(1+exp(-sum)) per previous transform
•male parameter 5 is the apical selectivity for males
Note that the male selectivity offsets currently cannot be time-varying (need to check on this).
Because they are offsets from female selectivity, they inherit the time-varying characteristics of the
female selectivity.
Dirichlet Multinomial Error for Data Weighting
If the Dirichlet multinomial error distribution was selected in the data file for either length or age data
weighting an additional parameter line is required immediately following the selectivity parameter
block.
The list of parameters to be read from the above setup would be:
LO HI INIT <other entries> Block Fxn Parameter Label
1 25 1 ... 0 #ln(EffN mult) Age or Length 1
1 25 1 ... 0 #ln(EffN mult) Age or Length 2
Time-varying Options
The time-varying options for selectivity parameters are identical to the time-varying options for
biology parameters. These options are described below in the Using Time-Varying Parameter Options.
After reading the selectivity parameters, which will include possible instructions to create
environmental link, blocks, or deviation vectors, then read the following section. Note that all inputs
in this section are conditional (COND) on entries in the selectivity parameter section. So if no
selectivity parameters invoke any time-varying properties, this section is left blank (or completely
commented out with #).
125

Example short parameter lines for selectivity time-varying parameters:
PR PRIOR
LO HI INIT PRIOR SD TYPE PHASE Label
0.01 2.0 0.58 0.58 0.5 4 -5 #AgeSel_P4_Fishery_dev_se
1 80 70 70 0.5 4 -5 #AgeSel_P4_Fishery_dev_autocorr
Two-Dimensional Auto-Regressive Selectivity
A new experimental feature added within SS v. 3.30.03.02. Earlier versions do not have this feature
and hence this input is not expected. This features allows for auto-correlation by age and/or time. The
age based random walk selectivity, pattern 17, should be selected to implement the semiparametric
auto-regressive selectivity.
Typical Value Description and Options
1 Two-dimensional auto-regressive selectivity:
0 = Not used,
1 = Use 2D AR.
COND = 1 Read the following long parameter lines:
Sigma Use Len(1)/ Before After
Fleet Ymin Ymax Amin Amax Amax Rho Age(2) Phase Range Range
1 1979 2015 2 10 1 1 2 5 1980 2007
Continued:
PRIOR PRIOR
LO HI INIT PRIOR SD TYPE PHASE LABEL
0 4 1 1 0.1 6 -4 #Sigma selex
-1 1 0 0 0.1 6 -4 #Rho year
-1 1 0 0 0.1 6 -4 #Rho age
#Terminator line of 11 in length indicates the end of parameter input lines
-9999 1 1 1 1 1 1 1 1 1 1
Parameter Definitions:
126
•Fleet: Fleet number to which semi-parametric deviations should be added
•Ymin: First year with deviations
•Ymax: Last year with deviations
•Amin: First integer age (or population length bin index) with deviations
•Amax: Last integer age (or population length bin index) with deviations
•Sigma Amax: Not currently implemented. It is recommended to set the Sigma Amax equal to
the Amin value. In future, Sigma Amax will be set to last age (or population length bin index) for
which a separate sigma should be read
•Use Rho: Use autocorrelation parameters
•Len(1)/Age(2): 1 or 2 to specify whether the deviations should be applied to length- or age-based
selectivity
•Phase: Phase to begin estimation of the deviation parameters
•Before Range: How should selectivity be modeled in the years prior to Ymin? Available options
are (0) apply no deviations, (1) use deviations from the first year with deviations (Ymin), and (3)
use average across all years with deviations (Ymin to Ymax)
•After Range: Similar to Before Range but defines how selectivity should be modeled after Ymax
Parameterizing the Two-Dimensional Autoregressive Selectivity
When the two-dimensional autoregressive selectivity feature in turned on for a fleet, the selectivity
of which is calculated as a product of the assumed selectivity pattern and a non-parametric deviation
term away from this assumed pattern:
ˆ
Sa,t =Saexpa,t (30)
where Sais specified in the corresponding age/length selectivity types section and it can be either
parametric (recommended) or non-parametric (including any of the existing selectivity options in
Stock Synthesis); a,t is simulated as a two-dimensional first-order autoregressive (2D AR1) process:
vec()∼MV N(0, σ2
sRtotal)(31)
where is the two-dimensional deviation matrix and σ2
sRtotal is the covariance matrix for the 2D AR1
process. More specifically, σ2
squantifies the variance in selectivity deviations and Rtotal is equal to
the knonecker product (⊗) of the two correlation matrices for the among-age and among-year AR1
processes:
Rtotal =R⊗˜
R(32)
127
Ra,˜a=ρ|a−˜a|
a(33)
˜
Rt,˜
t=ρ|t−˜
t|
t(34)
where ρaand ρtare the among-age year AR1 coefficients, respectively. When both of them are zero,
Rand ˜
Rare two identity matrices and their Kronecker product, Rtotal, is also an identity matrix. In
this case selectivity deviations are essentially identical and mutually independent:
a,t ∼N(0, σ2
s)(35)
Using the Two-Dimensional Autoregressive Selectivity
First, fix the two AR1 coefficients (ρaand ρt) at 0 and tune σsiteratively to match the relationship:
σ2
s=SD()2+1
(amax −amin + 1)(tmax −tmin + 1)
amax
X
a=amin
tmax
X
t=tmin
SE(a,t)2(36)
The minimal and maximal ages/lengths and years for the 2D AR1 process can be freely specified by
users in the control file. However, we recommend specifying the minimal and maximal ages and years
to cover the relatively "data-rich" age/length and year ranges only. Particularly we introduce:
b= 1 −
1
(amax−amin+1)(tmax−tmin+1) Pamax
a=amin Ptmax
t=tmin SE(a,t)2
σ2
s
(37)
as a measure of how rich the composition data is regarding estimating selectivity deviations. We also
recommend using the Dirichlet-Multinomial method to "weight" the corresponding composition data
while σsis interactively tuned in this step.
Second, fix σsat the value iteratively tuned in the previous step and estimate a,t. Plot both Pearson
residuals and a,t out on the age-year surface to check their 2D dimensions. If their distributions seems
to be not random but rather be autocorrelated (deviation estimates have the same sign several ages
and/or years in a row), users should consider estimating and then including the autocorrelations in
a,t.
Third, extract the estimated selectivity deviation samples from the previous step for estimating ρaand
ρtexternally by fitting the samples to a stand-alone model written in Template-Model Builder (TMB).
In this model, both ρaand ρtare bounded between 0 and 1 via applying a logic transformation. If
at least one of the two AR1 coefficients are notably different from 0, Stock Synthesis should be run
one more time by fixing the two AR1 coefficients at their values externally estimated from deviation
samples. The Pearson residuals and a,t from this run are expected to distribute more randomly as the
autocorrelations in selectivity deviations can be at least partially included in the 2D AR1 process.
128

Tag Recapture Parameters
Specify if tagging data are being used:
Typical Value Description and Options
1 Tagging Data Present:
0 = No tagging data,
1 = Read following lines of tagging data.
COND = 1 Read the following long parameter lines:
LO HI INIT PRIOR <other entries> Block
Fxn
Parameter Label
-10 10 9 9 ... 0 #TG loss init 1
-10 10 9 9 ... 0 #TG loss init 2
-10 10 9 9 ... 0 #TG loss init 3
-10 10 9 9 ... 0 #TG loss chronic1
-10 10 9 9 ... 0 #TG loss chronic2
-10 10 9 9 ... 0 #TG loss chronic3
1 10 2 2 ... 0 #TG loss overdisperion1
1 10 2 2 ... 0 #TG loss overdisperion2
1 10 2 2 ... 0 #TG loss overdisperion3
-10 10 9 9 ... 0 #TG report fleet1
-10 10 9 9 ... 0 #TG report fleet2
-4 0 0 0 ... 0 #TG report decay1
-4 0 0 0 ... 0 #TG report decay2
The tagging reporting rate parameter is transformed within SS during estimation to maintain a positive
value and is reported according to the transformation:
Tagging Reporting Rate =einput parameter
1 + einput parameter (38)
Variance Adjustment Factors
When doing iterative reweighting of the input variance factors, it is convenient to do this in the control
file, rather than the data file. This section creates that capability.
129

Read variance adjustment factors to be applied:
Factor Fleet Value Description
1 2 0.5 # Survey CV for survey/fleet 2
4 1 0.25 # Length data for fleet 1
4 2 0.75 # Length data for fleet 2
-9999 0 0 # End read
Additive Survey CV - Factor 1
The survey input variance (labeled survey CV) is actually the standard deviation of the ln(survey). The
variance adjustment is added directly to this standard deviation. Set to 0.0 for no effect. Negative
values are OK, but will crash if adjusted value becomes negative.
Additive Discard - Factor 2
The input variance is the CV of the observation. Because this will cause observations of near zero
discard to appear overly precise, the variance adjustment is added to the discard standard deviation,
not to the CV. Set to 0.0 for no effect.
Additive Mean Body Weight - Factor 3
The input variance is in terms of the CV of the observation. Because such data are typically not very
noisy, the variance adjustment is added to the CV and then multiplied by the observation to get the
adjusted standard deviation of the observation.
Multiplicative Length Composition - Factor 4
The input variance is in terms of an effective sample size. The variance adjustment is multiplied times
this sample size. Set variance adjustment to 1.0 for no effect.
Multiplicative Age Composition - Factor 5
Age composition is treated the same way as length composition.
Multiplicative Size-at-Age - Factor 6
Size-at-age input variance is the sample size for the N observations at each age. The variance
adjustment is multiplied by these N values. Set to 1.0 for no effect.
130

Multiplicative Generalized Size Composition - Factor 7
Generalized size composition input variance is the sample size for each observation. The variance
adjustment for each fleet is multiplied by these sample sizes. Set to 1.0 for no effect.
Variance Adjustment Usage Notes
The report.sso output file contains information useful for determining if an adjustment of these input
values is warranted to better match the scale of the average residual to the input variance scale.
Because the actual input variance factors are modified, it is these modified variance factors that are
used when creating parametric bootstrap data files. So, the control files used to analyze bootstrap
generated data files should have the variance adjustment factors reset to null levels.
Lambdas (Emphasis Factors)
These values are multiplied by the corresponding likelihood component to calculate the overall
negative log likelihood to be minimized.
Typical Value Description and Options
4 Max lambda phase: read this number of lambda values for each element below.
The last lambda value is used for all higher numbered phases.
1 SD offset:
0 = The log(like) to omit the +log(s) term,
1 = The log(like) to include the log(s) term for CPUE, discard, growth CV,
mean body weight, recruitment deviations. If you are estimating any variance
parameters, SD offset must be set to 1.
Lambda Usage Notes
If the CV for size-at-age is being estimated and the model contains mean size-at-age data, then the
flag for inclusion of the +log(stddev) term in the likelihood must be included. Otherwise, the model
will always get a better fit to the mean size-at-age data by increasing the parameter for CV of
size-at-age.
The reading of the lambda values has been substantially altered with SS v3.30. Instead of reading a
matrix containing all the needed lambda values, SS now just reads those elements that will be given
a value other than 1.0. After reading the datafile, SS sets lambda equal to 0.0 if there are no data for
a particular fleet/data type, and a value of 1.0 if data exist. So beware if your data files had data but
you had set the lambda to 0.0 in a previous version of SS. First read an integer for the number of
131
changes.
Read the lambda adjustments by fleet and data type:
Likelihood Lambda SizeFreq
Component Fleet Phase Value Method
1 2 2 1.5 1
4 2 2 10 1
4 2 3 0.2 1
-9999 1 1 1 1
The codes for component are:
1 = survey 10 = recruitment deviations
2 = discard 11 = parameter priors
3 = mean weight 12 = parameter deviations
4 = length 13 = crash penalty
5 = age 14 = morph composition
6 = size frequency 15 = tag composition
7 = size-at-age 16 = tag negative binomial
8 = catch 17 = F ballpark
9 = initial equilibrium catch 18 = regime shift
Controls for Variance of Derived Quantities
Additional standard deviation reported may be selected:
Typical Value Description and Options
1 0 = No additional std dev reporting,
1 = read values below.
COND > 0 : Read the 4 following lines:
Selex
Type
Len/Age
/Both
Year Nselex
Bins
Growth
Pattern
N
Growth
Ages
NatAge
Area
NatAge
year
N
Natage
132
1 1 -1 5 1 5 1 -1 5
Vector (length of 5) with selex std bin picks (-1 in first bin to self-generate).
5 15 25 35 43
Vector (length of 5) with growth std bin picks (-1 in first bin to self-generate).
1 2 14 26 40
Vector (length of 5) with NatAge std bin picks (-1 in first bin to self-generate).
1 2 14 26 40
999 #End of the control file input
Where:
•Selex fleet: The index of the fleet to be output.
–0 = No selectivity variance output,
–1 = Selectivity variance output.
•Len/Age/Both:
–1 = Select length selectivity,
–2 = Select age selectivity,
–3 = Both length and age selectivity.
•Year:
–year = Enter a value for the selected year,
–-1 = To get the selectivity in the end year.
•N Selex bins: enter the number of bins for which selectivity will be output. This number controls
the number of items to be read below, even if the Selex fleet is set to zero. In other words, the
read occurs even if the effect of the read is disabled.
•Growth pattern: growth pattern is the number of the growth pattern to be output. Note that
133
in a multiple season model, SS will output the size-at-age for the last birth season that gets any
recruits within the year. Also, if growth parameters are not estimated, then standard deviation
output of mean size-at-age is disabled.
–postitive value = Growth pattern,
–0 = No variance output for size-at-age.
•N growth bins: Number of ages for which size-at-age variance is requested. This number
controls the number of items to be read below, even if the growth pattern selection is set to
zero. In other words, the read occurs even if the effect of the read is disabled.
•Area for Natage: specifies the area for which output of numbers at age is requested.
–positive value = area to output
–0 = Disables this output,
–-1 = Requests that numbers-at-age be summed across all areas. In all cases, numbers-at-age
is summed across all growth patterns and platoons and output for each sex.
•NatAge Year: specifies the year for which numbers-at-age are output.
–year = The year to output,
–-1 = Requests output for year equal to end year + 1, hence the year that starts the forecast
period.
•N Natage bins: as with the N growth bins.
Using Time-Varying Parameters
The approach to allowing parameters to have time-varying values has been completely overhauled
in the transition from SS v.3.24 to SS v.3.30. Fortunately, the sstrans.exe will do the conversion for
you, but you should review the new control file closely before simply running with it, especially for
time-varying catchability parameters.
Time-Varying Parameter Change from Earlier SS Versions
In SS v.3.24, the group of biology parameters (termed MGparm) and the selectivity parameters used
the same long parameter line approach, but it was implemented with entirely different code, and
hence was inefficient. The spawner-recruitment parameters used short parameter lines and a
different approach for linkage to an environmental variable and the R1 offset provided a limited type
134
of block. The catchability parameters also used short parameter lines and had its own approach to
doing environmental linkage and random deviations, but not blocks. Then finally, the tagging
parameters had long parameter lines, but there was no code to interpret any time-varying info in
those lines. The situation was begging for a more modular approach.
Code Flow Version 3.30
In SS v.3.30, mortality-growth, selectivity, stock recruitment relationship, catchability, and tag (soon
but not as of v3.30.12) base parameters all will use long parameter lines and will invoke blocks,
trends, environmental linkages, and random deviations using identical syntax. As SS v.3.30 executes
the SS_readcontrol code, it calls a function in SS_global called “create_timevary” whenever a base
parameter has any one of the 4 types of time-varying options. In fact, block/trend, env and devs all
can be applied to the same base parameter. Only blocks and trends are mutually exclusive, but any
combined effect could be used together judiciously. “Create_timevary” creates all needed information
to describe and index a list of time varying parameter specifications. in fact, if the auto-generation
switch has been set to zero, then you will omit all of the needed parameters for implementing the
time-varying effect and SS will autogenerate and use the needed parameters and write them out in
the control.ss_new file. Then as SS gets into iterative parameter updating it starts by calling a
function in SS_timevaryparm that processes each time-varying parameter specification (each of
which can contain any combination of block/trend, env and dev specs) and creates a time-series of
the parameter value that are used as SS subsequently loops through the years.
Parameter Order
The order of parameters has changed and the re-ordering is handled by the sstrans.exe. Previously,
for each of mortality-growth and selectivity parameters all environmental link parameters were
listed first, then block/trend parameters and then deviation parameters. In SS v.3.30, these
parameters are re-organized such that all parameters that affect a base parameter are clustered
together with block/trend first, then environmental, then deviation. So, if mortality-growth (MG)
base parameters 3 and 7 had time varying changes, the order would look like:
MG base parameter 3 Block parameter 3-1
Block parameter 3-2
Environmental link parameter 3-1
Deviation se parameter 3
Deviation rho parameter 3
MG base parameter 7 Block parameter 7-1
Deviation se parameter 7
Deviation rho parameter 7
135
Link Functions
The functional form by which a time-varying parameter, Q, changes a base parameter, P, is a link
function: P0
y=f(P, Q). Typically, this is additive or multiplicative function, but the parameter
mirroring feature is essentially a link that takes no parameter. Another type of link in SS is between a
model state variable, such as available biomass, and the expected value for a survey. Typically, this is a
simple proportional link taking one parameter, q, but the q power feature is essentially a 2 parameter
link function. So, a parameter link function can change q over time, and a survey link function then
uses the annual value of q to link the annual value of a state variable to the expected value for a
survey. In SS v.3.24, various usages of positive and negative codes and other conventions were used
to invoke additive vs multiplication links and other options. But as v.3.30 builds capability to allow an
environment index to be a “survey” of a parameter deviation, we need a larger family of link
functions such as logistic and even dome-shaped.
The link specifications in SS v.3.30 has been updated from v.3.24. Take special note of the
environmental linkage specification where two bits of information are coded into one number. The
new specification has the environmental link function denoted by the first environmental index to
use specified by two additional. (e.g. environmental link specification of 204 is parsed by SS to use
link type 2 using environmental variable 4).
The new available options for time-varying parameters in SS v3.30 are described below:
•Environmental Link and Variance - Element 8 in parameter setup
–env_data is a dvar_matrix populated with the read environmental data for columns
1-N_envvariables and derived quantities mapped to columns -1 to -4 to
density-dependence:
*-1; for log(relative spawning biomass);
*-2; for recdev;
*-3; for log(relative summary biomass) (e.g. smrybio/smrybio in initial equilibrium);
*-4; for log(relative summary numbers).
–So, environmental input 103 would use link type 1 and apply it to environmental data
column 3 and environmental input -103 would use link type 1 and apply it to the "-3"
column which is log(relative summary biomass).
–These four derived quantities are all calculated at the beginning of each year within the
model, so they are available inside SS to use as the basis for time-varying parameter links
136
without violating any oder of operations rule.
•Deviation Link - Element 9 in parameter setup
–1 = multiplicative (P(y)∗=exp(dev(y)∗devse),
–2 = additive (P(y)+ = env(y)∗devse),
–3 = random walk options are now implemented by using rho in the objective function. SS
now expects the estimated deviations to be normal in distribution and the deviation values
are multiplied by the standard error parameter as they are used,
–4 = zero reverting random walk with rho. The deviation parameter is now multiplied
by the standard error parameter, rather than deviations being penalized according to a
specified standard error (the approach in v.3.24).
–Click here to see the code behind each of these options.
•Deviation Minimum Year - Element 10 in parameter setup
–Year for deviations to start for parameter
•Deviation Maximum Year - Element 11 in parameter setup
–Year for deviations to end for parameter
•Deviation Phase - Element 12 in parameter setup
–integer, this available element in the long parameter line is now a deviation vector specific
phase control
•Blocks - Element 13 in parameter setup. Currently, there are four options for applying blocks:
–>0: block index for parameter
–-1: trend with final as offset from base parameter and offset values is in log space, also
inflection year is in log space and the offset from log(0.5). No additional parameter lines
are required. Three parameters will be estimated; end trend parameter value logistic offset,
inflection year logistic offset, and slope.
–-2: trend with final as standalone value. No additional parameter lines are required. Three
parameters will be estimated; end trend parameter value, inflection year, and slope.
–-3 end value is a fraction of base parameter maximum - minimum; inflection year is
fraction of end year - start year. No additional parameter lines are required. Three
137
parameters will be estimated; end trend parameter value as a fraction, inflection year as a
fraction, and slope.
•Block Functional Form: Element 14 in parameter setup
–0: multiplicative (Pblock =Pbase ∗exp(tvpara)),
–1: additive (Pblock =Pbase +tvpara),
–2: replace (Pblock =tvpara),
–3: random walk across blocks (Pblock =Pblock,−1+tvpara),
–4: mean reverting random walk
Block Trends
Additional information regarding the options for applying blocks (element 13):
•-1: Trend bounded by base parm min-max and parms in transformed units (use with caution),
–Logistic approach to trend as offset from base parameter
–Transform the base parameter
temp =−0.5∗log
MGparm1(j, 2) −MGparm1(j, 1) + 0.0000002
MGparm(j)−MGparm1(j, 1) + 0.0000001 −1
(39)
–Add the offset. Note, that offset values in in the transform space.
temp+ = MGparm(k+ 1) (40)
–Back transform
temp1 = MGparm1(j, 1) + MGparm1(j, 2) −MGparm1(j, 1)
1 + e−2∗temp (41)
Parameter Priors
Priors on parameters fulfill two roles in SS. First, for parameters provided with an informative prior,
SS is receiving additional information about the true value of the parameter. This information works
with the information in the data through the overall log-likelihood function to arrive at the final
138
parameter estimate. Second, diffuse priors provide only weak information about the value of a prior
and serve to manage model performance during execution. For example, some selectivity parameters
may become unimportant depending upon the values of other parameters of that selectivity function.
In the double normal selectivity function, the parameters controlling the width of the peak and the
slope of the descending side become redundant if the parameter controlling the final selectivity
moves to a value indicating asymptotic selectivity. The width and slope parameters would no longer
have any effect on the log-likelihood, so they would have no gradient in the log-likelihood and would
drift aimlessly. A diffuse prior would then steer them towards a central value and avoid them
crashing into the bounds. Another benefit of diffuse priors is the control of parameters that are given
unnaturally wide bounds. When a parameter is given too broad of a bound, then early in a model run
it could drift into this tail and potentially get into a situation where the gradient with respect that
parameter approaches zero even though it is not at its global best value. Here the diffuse prior helps
move the parameter back towards the middle of its range where it presumably will be more
influential and estimable.
The options for parameter priors are as follows., described as a function of P val, the value of the
parameter for which a prior is being calculated, as well as the parameter bounds (P max and P min),
and the input values for P rior and P r_SD, which in some cases are the mean and standard deviation,
but interpretation depends on the prior type. Finally, P const is a small constant, currently hardwired
at 0.0001. The Prior Likelihoods below represent the negative log-likelihood in all cases.
Prior Types
Note that the numbering in SS version 3.30 is different from that used in 3.24 (where confusingly -1
indicated no prior and 0 indicated a normal prior). The calculation of the negative log-likelihood is
provided below for each prior types, as a function of the following inputs:
pThe value of the parameter for which a prior is being calculated
Pmin The lower bound of the parameter (1st column in control file)
Pmax The upper bound of the parameter (2nd column in control file)
P rior The input value for the PRIOR input (4th column in control file)
P r_SD The input value for the PR_SD input (5th column in control file)
•Prior Type = 0 = No prior applied
In a Bayesian context this is equivalent to a uniform prior between the parameter bounds.
•Prior Type = 1 = Symmetric beta prior
The symmetric beta is scaled between parameter bounds, imposing a larger penalty near the
bounds. Prior standard deviation of 0.05 is very diffuse and a value of 5.0 provides a smooth
139
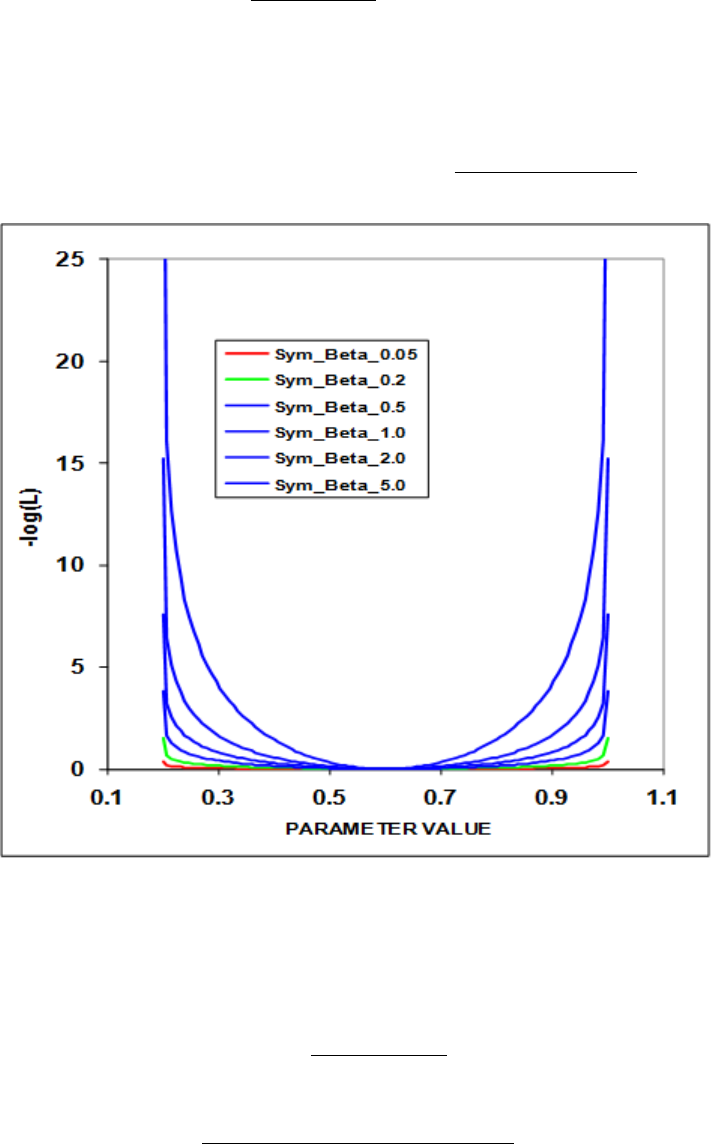
U-shaped prior. The PRIOR input is ignored for this prior type.
µ=−P r_SD ·log Pmax +Pmin
2−Pmin−P r_SD ·log(0.5) (42)
Prior Likelihood =−µ−P r_SD ·log (p−Pmin + 0.0001)
−P r_SD ·log 1−p−Pmin −0.0001
Pmax −Pmin (43)
Prior distributions for the symmetric beta distribution.
•Prior Type = 2 = Beta prior
The definition of µis consistent with CASAL’s formulation with the Bprior and Aprior
corresponding to the mand nparameters.
µ=P rior −Pmin
Pmax −Pmin
(44)
τ=(P rior −Pmin)(Pmax −P rior)
P r_SD2−1(45)
140
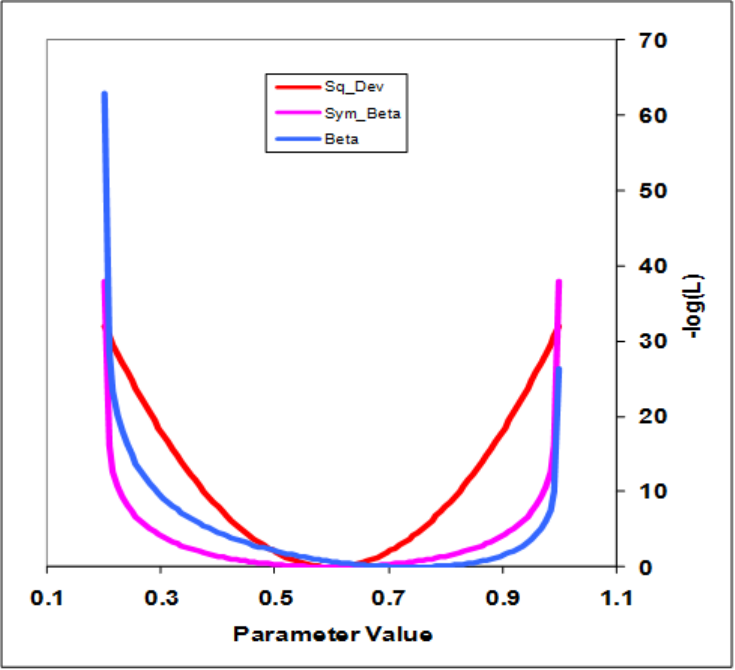
Bprior =τ·µ(46)
Aprior =τ(1 −µ)(47)
Prior Likelihood =(1 −Bprior)·log(0.0001 + p−Pmin)
+ (1 −Aprior)·log(0.0001 + Pmax −p)
−(1 −Bprior)·log(0.0001 + P rior −Pmin)
−(1 −Aprior)·log(0.0001 + Pmax −P rior)
(48)
Comparison of the symmetric beta and the beta prior functions
•Prior Type 3 = Lognormal prior
Note that this is undefined for p <= 0 so the lower bound on the parameter must be > 0.
The prior value is input into the parameter line in log space while the initial parameter value is
141
defined in normal space (e.g. INIT = 0.20, PRIOR = -1.609438).
Prior Likelihood =1
2 log(p)−P rior
P r_SD !2
(49)
•Prior Type 4 = Lognormal prior with bias correction
This option is thanks to Larry Jacobson. Note that this is undefined for p <= 0 so the lower
bound on the parameter must be > 0.
Prior Likelihood =1
2 log(p)−P rior +1
2P r_SD2
P r_SD !2
(50)
•Prior Type 5 = Gamma prior
This option is thanks to Larry Jacobson. The lower bound should be 0 or greater.
scale =P r_SD2
P rior (51)
shape =P rior
scale (52)
Prior Likelihood =−shape·log(scale)−logΓ(shape)+(shape−1)·log(p)−p
scale (53)
•Prior Type 6 = Normal prior
Note that this function is independent of the parameter bounds.
Prior Likelihood =1
2p−P rior
P r_SD 2
(54)
Optional Inputs
Empirical Weight-at-Age (wtatage.ss)
SS has the capability to read empirical body weight at age for the population and each fleet, in lieu of
generating these weights internally from the growth parameters, weight-at-length, and size-selectivity.
Selection of this option is done by setting an explicit switch near the top of the control file. The values
are read from a separate file named, wtatage.ss. This file is only required to exist if this option is
selected.
142
The first value read is a single integer for the maxage used in reading this file. So if the maximum age
is 40, there will be 41 columns of weight-at-age entries to read, with the first column being for age
0.
If N ages in this table is greater than maximum age in the model, the extra weight-at-age values are
ignored.
If N ages in this table is less than maximum age in the model, the weight-at-age for N ages in the file is
filled in for all unread ages out to maximum age.
The format of this input file is:
Growth Birth
Year Season Gender Pattern Season Fleet Age-0 Age-1 ...
1971 1 1 1 1 -2 0 0 0.1003
1971 1 1 1 1 -1 0.0169 0.0864 0.2495
1971 1 1 1 1 0 ... ... ...
1971 1 1 1 1 1 ... ... ...
1971 1 1 1 1 2 ... ... ...
-9999 1 1 1 1 0 ... ... ...
where:
•Fleet = -2 is age-specific fecundity*maturity, so time-varying fecundity is possible to implement.
•Fleet = -1 is population wt-at-age at middle of the season.
•Fleet = 0 is population wt-at-age at the beginning of the season.
•There must be an entry for fecundity*maturity, population wt-at-age at the middle of the season,
population wt-at-age at the beginning of the season, and wt-at-age for each fleet (even if these
vectors are identical in some cases)
•Fleets that do not use biomass do not need to have wt-at-age assigned.
•GP and birthseas probably will never be used, but are included for completeness.
•A negative value for year will fill the table from that year through the ending year of the forecast,
overwriting anything that has already been read for those years.
•Judicious use of negative years in the right order will allow user to enter blocks without having
to enter a row of info for each year
143
•There is no internal error checking to verify that weight-at-age has been read for every fleet and
every year.
•In the future, there could be an option to use another value of the control file switch to turn off
all aspects of growth parameters and size selectivity.
•The values entered for endyr+1 will be used for the benchmark calculations and for the forecast;
this aspect needs a bit more checking.
Caveats
•SS will still calculate growth curves from the input parameters and can still calculate
size-selectivity and can still examine size composition data.
•However, there is no calculation of wt-at-age from the growth input, so no way to compare the
input wt-at-age from the wt-at-age derived from the growth parameters.
•If wt-at-age is read and size-selectivity is used, a warning is generated.
•If wt-at-age is read and discard/retention is invoked, then a BEWARE warning is generated
because of untested consequences for the body wt of discarded fish.
•Warning: age 0 fish seem to need to have weight=0 for spawning biomass calculation (code -2).
Testing
•A model was setup with age-maturity (option 2) and only age selectivity.
•The output calculation of wt-at-age and fecundity-at-age was taken from report.sso and put into
wtatage.ss (as shown above).
•Re-running SS with this input wt-at-age (Maturity_Option 5) produced identical results to the
run that had generated the weight-at-age from the growth parameters.
runnumbers.ss
This file contains a single integer value. It is read when the program starts, incremented by 1, used
when processing the profile value inputs (see below), used as an identifier in the batch output, then
saved with the incremented value. Note that this incrementation may not occur if a run crashes.
144

profilevalues.ss
This file contains information for changing the value of selected parameters for each run in a batch.
In the ctl file, each parameter that will be subject to modification by profilevalues.ss is designated by
setting its phase to -9999 .
The first value in profilevalues.ss is the number of parameters to be batched. This value MUST match
the number of parameters with phase set equal to -9999 in the ctl file. The program performs no
checks for this equality. If the value is zero in the first field, then nothing else will be read. Otherwise,
the model will read runnumber * Nparameters values and use the last Nparameters of these to replace
the initial values of parameters designated with phase = –9999 in the ctl file.
Usage Note: If one of the batch runs crashes before saving the updated value of runnumber.ss, then
the processing of the profilevalue.ss will not proceed as expected. Check the output carefully until a
more robust procedure is developed. Usage Note: This options was created before use of R became
widespread. You probably can create a more flexible approach using R today.
Likelihood components
The objective function Lis the weighted sum of the individual components indexed by kind of data i,
and fishery/survey fas appropriate:
L=
I
X
i=1
Af
X
f=1
ωi,f Li,f +ωRLR+X
θ
ωθLθ+X
P
ωPLP(55)
where Lis the total objective function, iis the index of the objective function component, Li,f is
the objective function for data kind ifor the fishery/survey f, and ωi,f is a weighting factor for each
objective function component.
The components of the objective function based on the model set-up and data are:
Index Source Kind Error structure
ifishery/survey fCPUE or Abundance index user choice
ifishery fDiscard biomass user choice
ifishery/survey fMean body weight normal
ifishery/survey fLength composition multinomial or log-gamma
145

ifishery/survey fAge composition multinomial or log-gamma
ifishery/survey fMean size-at-age normal
ifishery/survey fTag-recapture 1 multinomial
ifishery/survey fTag-recapture 2 negative binomial
ifishery fInitial equilibrium catch normal
RRecruitment deviations lognormal
PRandom parameter devs normal
θParameter priors user choice
FBF ballpark penalty
CPCrash penalty
Full description of likelihood distributions by source will be added in the future.
Running SS
Command Line Interface
The name of the SS executable files often contains the phrase “safe” or “opt” (for optimized). The safe
version includes checking for out of bounds values and should always be used whenever there is a
change to the data file. The optimized version runs slightly faster but can result in data not being
included in the model as intended if the safe version hasn’t been run first. A file named “ss.exe” is
typically the safe version unless the result of renaming by the user. In some situations, users may wish
to rename the file they are using to ss.exe, but the longer file name can be used.
On Mac and Linux computers, the executable does not include an extension (like .exe on Windows).
Running the executable on from the DOS command line in Windows simply require typing the
executable name (without the .exe extension):
> ss
On Mac and Linux computers, the executable name must be preceded by a period and slash (unless it’s
location has been added to the user’s PATH):
> ./ss
146
Additional ADMB commands can follow the executable name, such as “ –nohess” to avoid calculating
the Hessian matrix. To see a full list of options, add “ -?” after the executable name (with a space in
between).
On all operating systems, a copy of the SS executable can either be located in the same directory as
the model input files or in a central location and referenced either by adding it to the PATH or by a
script files. Further discussion on script files for Windows is below. Editing the PATH is not covered
here.
Example of DOS batch input file
One file management approach is to put ss.exe in its own folder (example: C:\SS_model) and to put
your input files in separate folder (example: C:\My Documents \SS_runs). Then a DOS batch file in
the SS_runs folder can be run at the command line to start ss.exe. All output will appear in SS_runs
folder.
A DOS batch file (e.g. SS.bat) might contain some explicit ADMB commands, some implicit commands,
and some DOS commands:
c:\SS_model\ss.exe -cbs 5000000000 -gbs 50000000000 \%1 \%2 \%3 \%4
del ss.r0*
del ss.p0*
del ss.b0*
In this batch file, the –cbs and –gbs arguments allocate a large amount of memory for SS to use (you
may need to edit these for your computer and SS configuration), and the %1, %2 etc. allows passing of
command line arguments such as –nox or –nohess. You add more items to the list of % arguments as
needed.
An easy way to start a command line in your current directory (SS_runs) is to create a shortcut to the
DOS command line prompt. The shortcut’s target would be:
> %SystemRoot%\system32\cmd.exe
And it would start in:
> %CURRDIR%
147
Simple Batch
This first example relies upon having a set of prototype files that can be renamed to starter.ss and
then used to direct the running of SS. The example also copies one of the output files to save it from
being overwritten. This sequence is repeated 3 times here and can be repeated an unlimited number
of times. The batch file can have any name with the .bat extension, and there is no particular limit to
the DOS commands invoked. Note that brief output from each run will be appended to cumreport.sso
(see below).
del ss.cor
del ss.std
copy starter.r01 starter.ss
c:\admodel\ss\ss.exe -sdonly
copy ss.std ss-std01.txt
copy starter.r01 starter.ss
c:\admodel\ss\ss.exe -sdonly
copy ss.std ss-std02.txt
Complicated Batch
This second example processes 25 dat files from a different directory, each time using the same ctl and
nam file. The loop index is used in the file names, and the output is searched for particular keywords
to accumulate a few key results into the file SUMMARY.TXT. Comparable batch processing can be
accomplished by using R or other script processing programs.
del summary.txt
del ss-report.txt
copy /Y runnumber.zero runnumber.ss
FOR /L \%\%i IN (1,1,25) DO (
copy /Y ..\MakeData\A1-D1-%%i.dat Asel.dat
del ss.std
del ss.cor
del ss.par
c:\admodel\ss\ss.exe
copy /Y ss.par A1-D1-A1-%%i.par
copy /Y ss.std A1-D1-A1-%%i.std
find "Number" A1-D1-A1-%%i.par >> Summary.txt
find "hessian" ss.cor >> Summary.txt)
148
Batch Using PROFILEVALUES.SS
This example will run a profile on natural mortality and spawner-recruitment steepness, of course.
Edit the control file so that the natural mortality parameter and steepness parameter lines have the
phase set to –9999. Edit STARTER.SS to refer to this control file and the appropriate data file.
Create a PROFILEVALUES.SS file
2 # number of parameters using profile feature
0.16 # value for first selected parameter when runnumber equals 1
0.35 # value for second selected parameter when runnumber equals 1
0.16 # value for first selected parameter when runnumber equals 2
0.40 # value for second selected parameter when runnumber equals 2
0.18 # value for first selected parameter when runnumber equals 3
0.40 # value for second selected parameter when runnumber equals 3
etc.; make it as long as you like.
Create a batch file that looks something like this. Or make it more complicated as in the example
above.
del cumreport.sso
copy /Y runnumber.zero runnumber.ss % so you will start with runnumber=0
C:\SS330\ss.exe
C:\SS330\ss.exe
C:\SS330\ss.exe
Repeat as many times as you have set up conditions in the PROFILEVALUES.SS file. The summary
results will all be collected in the cumreport.sso file. Each step of the profile will have an unique
Runnumber and its output will include the values of the natmort and steepness parameters for that
run.
Re-Starting a Run
SS model runs can be restarted from a previously estimated set of parameter values. In the starter.ss
file, enter a value of 1 on the first numeric input line. This will cause SS to read the file ss.par and use
these parameter values in place of the initial values in the control file. This option only works if the
number of parameters to be estimated in the new run is the same as the number of parameters in the
previous run because only actively estimated parameters are saved to the file ss.par. The file ss.par can
be edited with a text editor, so values can be changed and rows can be added or deleted. However, if the
resulting number of elements does not match the setup in the control file, then unpredictable results
will occur. Because ss.par is a text file, the values stored in it will not give exactly the same initial results
149
as the run just completed. To achieve greater numerical accuracy, SS can also restart from ss.bar which
is the binary version of ss.par. In order to do this, the user must make the change described above to
the starter.ss file and must also enter –binp ss.bar as one of the command line options.
Debugging Tips
When SS input files are causing the program to crash or fail to produce sensible results, there are
a few steps that can be taken to diagnose the problem. Before trying the steps below, examine the
ECHOINPUT.SSO file. It is highly annotated, so you should be able to see if SS is interpreting your
input files as you intended.
1. Set the turn_off_phase switch to 0 in the STARTER.SS file. This will cause the mode to not
attempt to adjust any parameters and simply converges a dummy parameter. It will still produce
a REPORT.SSO file, which can be examined to see what has been calculated from the initial
parameter values.
2. Turn the verbosity level to 2 in the STARTER.SS file. This will cause the program to display the
value of each likelihood component to the screen on each iteration. So it the program is
creating an illegal computation (e.g. divide by zero), it may show you which likelihood
component contains the problematic calculation. If the program is producing a REPORT.SSO
file, you may then see which observation is causing the illegal calculation.
3. Run the program with the command ss »SSpipe.txt. This will cause all screen display to go to
the specified text file (note, delete this file before running because it will be appended to).
Examination of this file will show detailed statements produced during the reading and
preprocessing of input files.
4. CHECKUP.SSO: This file can be written during the first iteration of the program. It contains
details of selectivity and other calculations as an aid to debugging model problems.
5. If SS fails to achieve a proper Hessian it exits without writing the detailed outputs in the
FINAL_SECTION. If this happens, you can do a run with the –nohess option so you can view
the report.sso to attempt to diagnose the problem.
Keyboard Tips
Typing “N” during a run will cause ADMB to immediately advance to the next phase of
estimation.
Typing “Q” during a run will cause ADMB to immediately go to the final phase. This bypasses
150
estimation of the Hessian and will produce all of the SS outputs, which are coded in the
FINAL_SECTION.
Running MCMC
Run SS v3.30
•This gives MPD estimates, report file, Hessian matrix and the .cor file
•(Recommended) Look for parameters stuck on bounds which will degrade efficiency of MCMC
implementation
•(Recommended) Look for very high correlations that may degrade the efficiency of MCMC
implementation
Run SS v3.30 with arguments -mcmc xxxx -mcsave yyyy
•Where: xxxx is the number of iterations for the chain, and yyyy is the thinning interval (1000 is
a good place to start)
•MCMC chain starts at the MPD values
•(Recommended) Remove existing .psv files in run directory to generate a new chain.
•(Recommended) Set DOS run detail switch in starter file to 0; reporting to screen will
dramatically slow MCMC progress
•(Optional) Add -nohess to use the existing Hessian file without re-estimating
•(Optional) To start the MCMC chain from specific values change the par file; run the model with
estimation, adjust the par file to the values that the chain should start from, change within the
starter file for the model to begin from the par file, and call the MCMC function using ss –mcmc
xxxx – mcsave yyyy -nohess –noest.
•(Optional) Add -noest -nohess and modify starter file so that run will now start from the
converged (or modified) parameter estimates in "ss.par"
Run SS v3.30 with argument -mceval
•This generates the posterior output files.
•(Optional) Modify starter file entries to add a burn-in and thinning interval above and beyond
the ADMB thinning interval applied at run time.
151
•(Recommended) MCMC always begins with the MPD values and so a burn-in >0 should always
be used.
•This step can be repeated for alternate forecast options (e.g. catch levels) without repeating step
2.
(Optional) Run SS v3.30 with arguments -mcr -mcmc xxxx -mcsave yyyy ...
•This restarts and extends an uninterrupted chain previously completed (note that any
intermediate runs without the -mcr command in the same directory will break this option).
NOTES:
When SS switches to MCMC or MCEVAL mode, it sets all the bias adjustment factors to 1.0 for any
years with recruitment deviations defined. SS does not create a report file after completing MCMC
because it would show values based on the last MCMC step.
Output Files
Standard ADMB output files
Standard ADMB files are created by SS. These are:
SS.PAR – This file has the final parameter values. They are listed in the order they are declared in SS.
This file can be read back into SS to restart a run with these values (see running SS).
SS.STD – This file has the parameter values and their estimated standard deviation for those
parameters that were active during the model run. It also contains the derived quantities declared as
sdreport variables. All of this information is also report in the covar.sso. Also, the parameter section
of report.sso lists all the parameters with their SS generated names, denotes which were active in the
reported run, displays the parameter standard deviations, then displays the derived quantities with
their standard deviations.
SS.REP – This report file is created between phases so, unlike report.sso, will be created even if the
Hessian fails. It does not contain as much output as shown in report.sso.
SS.COR – This is the standard ADMB report for parameter and sdreport correlations. It is in matrix
form and challenging to interpret. This same information is reported in covar.sso.
152
SS Summary
The ss_summary.sso file (available for versions 3.30.08.03 and later) is designed to put key model
outputs all in one concise place. It is organized as a list. At the top of the file are some descriptors,
followed by the likelihoods for each component, then the parameters and their standard errors, then
the derived quantities and their standard errors. The total biomass, summary biomass, and catch
were added to the quantities reported in this file in version 3.30.11 and later. This output was created
to make it easy to compare the results between different versions of the executable, however, this file
could be useful for numerous other uses.
SIS table
The SIS_table.sso file contains model output formatted for reading into the NMFS Species
Information System (SIS). This file includes an assessment summary for categories of information
(abundance, recruitment, spawners, catch estimates) that are input into the SIS database. A
time-series of estimated quantities which aggregates estimates across multiple areas and seasons are
provided to summarize model results. Access to the SIS database is granted to all NOAA employees
and can be accessed as: https://www.st.nmfs.noaa.gov/sis/.
Derived Quantities
Before listing the derived quantities reported to the sdreport, there are a couple of topics that deserve
further explanation.
Virgin Spawning Biomass (B0) vs Unfished Spawning
Biomass
Unfished is the condition for which reference points (benchmark) are calculated. Virgin Spawning
Biomass (B0) is the initial condition on which the start of the time-series depends. If biology or SR
parameters are time-varying, then the benchmark year input in the forecast file tells SS which years
to average in order to calculate "unfished". In this case, virgin recruitment and/or the virgin spawning
biomass will differ from their unfished counterparts. Virgin recruitment and spawning biomass are
reported in the mgmt_quant portion of the sd_report and are now labeled as "unfished" for clarity.
Note that if ln(R0) is time-varying, then this will cause unfished to differ from virgin. However, if
regime shift parameter is time-varying, then unfished will remain the same as virgin because the regime
shift is treated as a temporary offset from virgin. Virgin spawning biomass is denoted as SPB_virgin
and spawning biomass unfished is denoted as SPB_unf in the report file.
153
Virgin Spawning Biomass (B0) is used in four ways within SS:
1. Anchor for the spawner-recruitment relationship as virgin spawning biomass.
2. Basis for the initial equilibrium abundance.
3. Basis against which annual depletion is calculated.
4. Benchmark calculations.
However, if there is time-varying biology, then the 4th usage can have a different B0 calculation
compared to the other usages.
Metric for Fishing Mortality
A generic single metric of annual fishing mortality is difficult to define in a generalized model that
admits multiple areas, multiple biological cohorts, dome-shaped selectivity in size and age for each of
many fleets. Several separate indices are provided and others could be calculated by a user from the
detailed information in report.sso.
Equilibrium SPR
This index focuses on the effect of fishing on the spawning potential of the stock. It is calculated as
the ratio of the equilibrium reproductive output per recruit that would occur with the current year’s F
intensities and biology, to the equilibrium reproductive output per recruit that would occur with the
current year’s biology and no fishing. Thus it internalizes all seasonality, movement, weird selectivity
patterns, and other factors. Because this index moves in the opposite direction than F intensity itself,
it is usually reported as 1-SPR. A benefit of this index is that it is a direct measure of common proxies
used for FMSY, such as F40%. A shortcoming of this index is that it does not directly demonstrate the
fraction of the stock that is caught each year. The SPR value is also calculated in the benchmarks (see
below). The derived quantities report shows an annual SPR statistic. The options, as specified in the
starter.ss file, are:
•0 = skip
•1 = (1-SPR)/(1-SPRTGT)
•2 = (1-SPR)/(1-SPRMSY)
•3 = (1-SPR)/(1-SPRBtarget)
154
•4 = raw SPR
F std
This index provides a direct measure of fishing mortality. The options are:
•0 = skip
•1 = exploitation(Bio)
•2 = exploitation(Num)
•3 = sum(Frates)
The exploitation rates are calculated as the ratio of the total annual catch (in either biomass or numbers
as specified) to the summary biomass or summary numbers on Jan 1. The sum of the F rates is simply
the sum of all the apical Fs. This makes sense if the F method is in terms of instantaneous F (not Pope’s
approximation) and if there are not fleets with widely different size/age at peak selectivity, and if there
is no seasonality, and especially if there is only one area. In the derived quantities, there is an annual
statistic that is the ratio of the can be annual F_std value to the corresponding benchmark statistic.
The available options for the denominator are:
•0 = raw
•1 = F/FSPR
•2 = F/FMSY
•3 = F/FBtarget
F-at-Age
Because the annual F is so difficult to interpret as a sum of individual F components, an indirect
calculation of F-at-age is reported at the end of the report.sso file. This section of the report
calculates Z-at-age simply as ln(Na+1,t+1/Na,t). This is done on an annual basis and summed over all
areas. It is done once using the fishing intensities as estimated (to get Z), and once with the F
intensities set to 0.0 to get M-at-age. This latter sequence also provides a measure of dynamic Bzero.
The user can then subtract the table of M-at-age/year from the table of Z-at-age/year to get a table of
F-at-age/year. From this apical F, average F over a range of ages, or other user-desired statistics could
be calculated. Further work within SS with this table of values is anticipated.
155

MSY and other Benchmark Items
The following quantities are included in the sdreport vector mgmt_quantities, so obtain estimates of
variance. Some additional quantities can be found in the benchmarks section of the
forecast_report.sso.
Benchmark Item Description
SSB_Unfished Unfished reproductive potential (SSB is commonly female mature
spawning biomass).
TotBio_Unfished Total age 0+ biomass on January 1.
SmryBio_Unfished Biomass for ages at or above the summary age on January 1.
Recr_Unfished Unfished recruitment.
SSB_Btgt SSB at user specified SSB target.
SPR_Btgt Spawner potential ratio (SPR) at F intensity that produces user
specified SSB target.
Fstd_Btgt F statistic at F intensity that produces user specified SSB target.
TotYield_Btgt Total yield at F intensity that produces user specified SSB target.
SSB_SPRtgt SSB at user specified SPR target (but taking into account the
spawner-recruitment relationship).
Fstd_SPRtgt F intensity that produces user specified SPR target.
TotYield_SPRtgt Total yield at F intensity that produces user specified SPR target.
SSB_MSY SSB at F intensity that is associated with MSY; this F intensity may
be directly calculated to produce MSY, or can be mapped to F_SPR
or F_Btgt.
SPR_MSY Spawner potential ratio (SPR) at F intensity associated with MSY.
Fstd_MSY F statistic at F intensity associated with MSY.
TotYield_MSY Total yield (biomass) at MSY.
RetYield_MSY Retained yield (biomass) at MSY.
156
Brief cumulative output
Cum_Report.sso: contains a brief version of the run output, which is appended to current content of
file so results of several runs can be collected together. This is especially useful when a batch of runs is
being processed. Unless this file is deleted, it will contain a cumulative record of all runs done in that
subdirectory. The first column contains the run number.
Output for Rebuilder Package
Output filename is REBUILD.DAT
#Title # various run summary outputs
SS#_default_rebuild.dat
# Number of sexes
2
# Age range to consider (minimum age; maximum age)
0 40
# Number of fleets
3
# First year of projection (Yinit)
2002
# First Year of rebuilding period (Ydecl)
1999
# Number of simulations
1000
# Maximum number of years
500
# Conduct proejctions with multiple starting values (0 = No, 1 = Yes)
0
# Number of parameter vectors
1000
# Is the maximum age a plus-group (1 = Yes; 2 = No)
1
#Generate future recruitments using historical recruitments (1) historical recruits/spawner (2) or a
stock-recruitment (3)
3
# Constant fishing mortality (1) or constant Catch (2) projections
1
# Fishing mortality based on SPR (1) or actual rate (2)
1v # Pre-specify the year of recovery (or -1) to ignore
157
-1
# Fecundity-at-age
# 0 1 2 3 4 5 6 7 8 9 10 <deleted values>
0 0.000450117 0.00436298 0.0271371 <deleted values>
# Age specific selectivity and weight adjusted for discard and discard mortality
#wt and selex for gender, fleet: 1 1
0.146708 0.320119 0.555587 0.830467 <deleted values>
0.0122887 0.0351722 0.0838682 0.165479 <deleted values>
#wt and selex for gender ,fleet: 2 1
0.150944 0.33768 0.588317 0.874376 <deleted values>
0.0127241 0.0380999 0.0922667 <deleted values>
# M and current age-structure in year Yinit: 2002
# gender = 1
0.1 0.1 0.1 0.1 0.1 <deleted values>
1425.96 797.624 1234.77 428.207 <deleted values>
# gender = 2
0.1 0.1 0.1 0.1 0.1 <deleted values>
1425.96 797.531 1233.66 <deleted values>
# Age-structure at Ydeclare= 1999
598.671 652.739 2925.76 2227.69 <deleted values>
598.671 652.666 2923.27 2221.05 <deleted values>
# Year for Tmin Age-structure (set to Ydecl by SS) 1999
1999
# recruitment and biomass
# Number of historical assessment years
33
# Historical data
# year recruitment spawner in B0 in R project in R/S project
1970 1971 1972 1973 1974 1975 1976 <deleted values> 2001 2002
#years (with first value representing R0)
8853.43 8658.22 8651.96 8645.41 8638.43 8630.75 <deleted values> 1594.53 2075.34 #recruits; first
value is R0 (virgin)
63679.5 63679.5 63679.3 63678.3 63673.9 63661.6 <deleted values> 8614.18 7313.2 #spbio; first
value is S0 (virgin)
10000000000000000000000<deletedvalues>00#inBzero
0 1 1 1 1 1 1 <deleted values> 1 1 0 0 0 # in R project
0 1 1 1 1 1 1 <deleted values> 1 1 0 0 0 # in R/S project
# Number of years with pre-specified catches
0
# catches for years with pre-specified catches go next
# Number of future recruitments to override
3
158
# Process for overriding (-1 for average otherwise index in data list)
2000 1 2000
2001 1 2001
2002 1 2002
# Which probability to product detailed results for (1=0.5; 2=0.6; etc.)
3
# Steepness sigma-R Auto-correlation
0.610789 0.6 0
# Target SPR rate (FMSY Proxy); manually change to SPR_MSY if not using SPR_target
0.5
# Target SPR information: Use (1=Yes) and power
0 20
# Discount rate (for cumulative catch)
0.1
# Truncate the series when 0.4B0 is reached (1=Yes)
0
# Set F to FMSY once 0.4B0 is reached (1=Yes)
0
# Maximum possible F for projection (-1 to set to FMSY)
-1
# Defintion of recovery (1=now only; 2=now or before)
2
# Projection type
4
# Definition of the 40-10 rule
10 40
# Produce the risk-reward plots (1=Yes)
0
# Calculate coefficients of variation (1=Yes)
0
# Number of replicates to use
10
# Random number seed
-99004
# File with multiple parameter vectors
rebuild.SS0
# User-specific projection (1=Yes); Output replaced (1->9)
0 5
# Catches and Fs (Year; 1/2/3 (F or C or SPR); value); Final row is -1v 2002 1 1
-1 -1 -1
# Split of Fs
2002 1
159
-1111
# Yrs to define TTARGET for projection type 4 (aka 5 pre-specified inputs)
2011 2012 2013 2014 2015 2016 2017 2018
# Time varying weight-at-age (1=Yes;0=No)
0
# File with time series of weight-at-age data
none
# Use bisection (0) or linear interpolation (1)
1
# Target Depletion
0.4
# CV of implementation error
0
Bootstrap Data Files
Data.ss_new: contains a user-specified number of data files, generated through a parametric bootstrap
procedure, and written sequentially to this file. These can be parsed into individual data files and re-run
with the model. The first output provides the unaltered input data file (with annotations added). The
second provides the expected values for only the data elements used in the model run. The third and
subsequent outputs provide parametric bootstraps around the expected values.
Forecast and Reference Points
FORECAST-REPORT.sso: This file contains output of fishery reference points and forecasts. It is
designed to meet the needs of the Pacific Fishery Management Council’s Groundfish Fishery
Management Plan, but it should be quite feasible to develop other regionally specific variants of this
output.
The vector of forecast recruitment deviations is estimated during an additional model estimation
phase. This vector includes any years after the end of the recrdev time series and before or at the end
year. When this vector starts before the ending year of the time series, then the estimates of these
recruitments will be influenced by the data in these final years. This is problematic, because the
original reason for not estimating these recruitments at the end of the time series was the poor
signal/noise ratio in the available data. It is not that these data are worse than data from earlier in the
time series, but the low amount of data accumulated for each cohort allows an individual datum to
dominate the model’s fit. Thus, an additional control is provided so that forecast recruitment
deviations during these years can receive an extra weighting in order to counter-balance the
160
influence of noisy data at the end of the time series.
An additional control is provided for the fraction of the log-bias adjustment to apply to the forecast
recruitments. Recall that R is the expected mean level of recruitment for a particular year as specified
by the spawner-recruitment curve and R’ is the geometric mean recruitment level calculated by
discounting R with the log-bias correction factor e-0.5sˆ
2. Thus a lognormal distribution of
recruitment deviations centered on R’ will produce a mean level of recruitment equal to R. During
the modeled time series, the virgin recruitment level and any recruitments prior to the first year of
recruitment deviations are set at the level of R, and the lognormal recruitment deviations are
centered on the R’ level. For the forecast recruitments, the fraction control can be set to 1.0 so that
100% of the log-bias correction is applied and the forecast recruitment deviations will be based on
the R’ level. This is certainly the configuration to use when the model is in MCMC mode. Setting the
fraction to 0.0 during maximum likelihood forecasts would center the recruitment deviations, which
all have a value of 0.0 in ML mode, on R. Thus would provide a mean forecast that would be more
comparable to the mean of the ensemble of forecasts produced in MCMC mode. Further work on
this topic is underway.
Note:
•Cohorts continue growing according to their specific growth parameters in the forecast period
rather than staying static at the endyr values.
•Environmental data entered for future years can be used to adjust expected recruitment levels.
However, environmental data will not affect growth or selectivity parameters in the forecast.
The top of this file shows the search for FSPR and the search for FMSY so the user can verify convergence.
Note: if the STD file shows aberrant results, such as all the standard deviations being the same value
for all recruitments, then check the FMSY search for convergence.
The FMSY can be calculated, or set equal to one of the other F reference points per the selection made
in STARTER.SS.
The reference point output is shown int he table below:
161
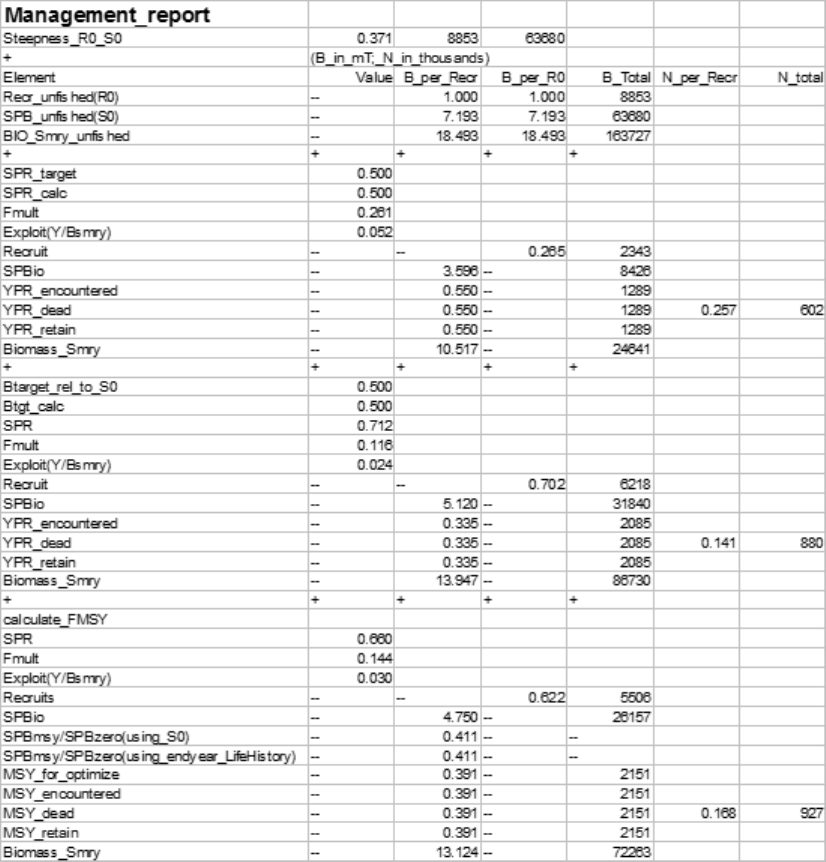
162
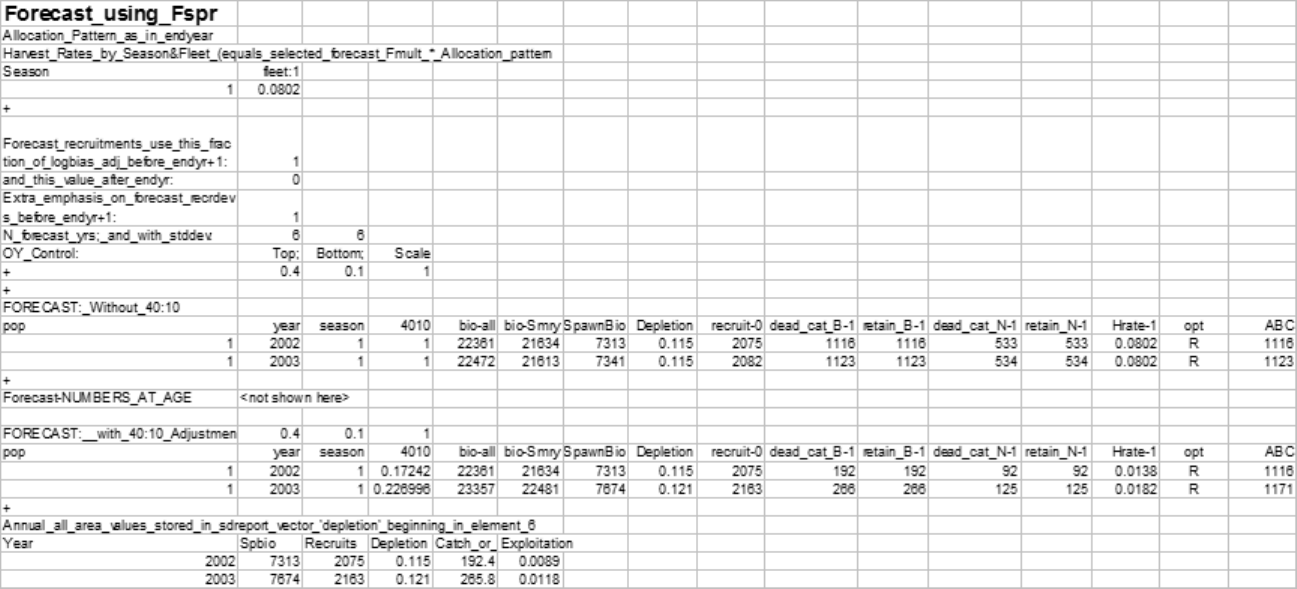
The forecast is done once using the Target SPR and once using the adjustments specified in the 40:10 section of forecast.ss input.
Each section contains a time series of seasonal biomass and catch, followed by a time series of population numbers-at-age for each
platoon.
163
where:
•40:10 is the magnitude of the adjustment of harvest multiplier to implement the OY policy
•bio-all is the biomass of all ages
•bio-smry is the biomass for ages at or above the summary age
•Spawnbio - is the female spawning output
•Depletion is the spawnbio divided by the unfished spawnbio
•Recruit-0 is the recruitment of age-o fish in this year
•Dead_cat_B-1 is the total dead (retained plus dead discard) catch in MT for fleet 1
•Retain_B-1 is fleet 1’s retained catch in MT
•Equivalent catch in numbers is then reported.
•Hrate-1 is the harvest rate, as adjusted by the 40:10 policy. The units will depend on the F method
selected (Pope’s method giving mid-year harvest rate or the continuous F.
•Opt=C means that the rate was calculated from an input catch level (and crashed means that this
caused an excessive harvest rate.
•Opt=R means that the catch was calculated from the target harvest rate.
•ABC is equal to the Total-Catch when the 40:10 option is not used (upper portion of table). When
the 40:10 is on (lower table), the ABC is the catch level corresponding to no 40:10 adjustment
after accounting for catch in previous year’s from the 40:10.
The time series output described above is detailed by season, area, platoon and fishery. It is usually
more convenient to have annual values summed across areas, platoons and fisheries. This is done for
the 40:10 output and a subset of these values are replicated in the depletion vector in the sd_report
so that variance estimates can be obtained. The elements of the depletion vector in the sd_report
are:
•depletion level in end year
•depletion level in end year+1
•MSY (if calculated, else spbio in endyr-1)
•BMSY (if calculated, else spbio in endyr)
164
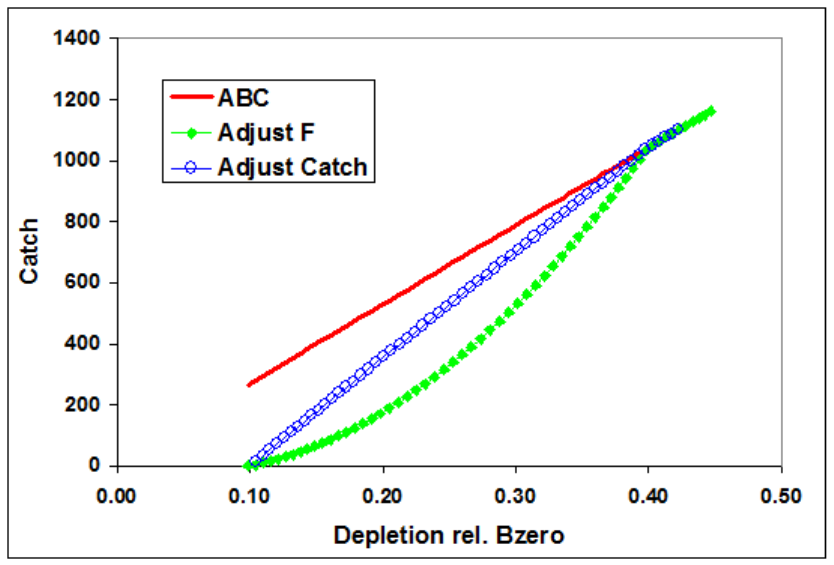
•SPRMSY (if calculated, else spbio in endyr+1) then the time series of:
–Spawning biomass
–Recruitment
–Depletion level
–Total catch (if forecast calculated catch from rates) or sum of fishery-specific harvest rates
(if forecast is based on fixed input catch level in this year)
–Total exploitation rate (total dead catch divided by the summary biomass at the beginning
of the year).
Two examples of harvest forecast adjustment: one adjusts catch and the other adjusts F.
Main Output File, report.sso
This is the primary output file. Its major sections are listed below.
The sections of the output file are:
165
•SS version number with date compiled. Time and date of model run. This info appears at the
top of all output files.
•Comments
–Input file lines starting with #C are echoed here
•Keywords
–List of keywords used in searching for output sections.
•Fleet Names
–List of fishing fleet and survey names assigned in the data file
•Likelihood
–Final values of the negative log(likelihood) are presented.
•Input Variance Adjustments
–The matrix of input variance adjustments is output here because these values affect the
logL calculations
•Parameters
–The parameters are listed here. For the estimated parameters, the display shows: Num
(count of parameters), Label (as internally generated by SS), Value, Active_Cnt, Phase,
Min, Max, Init, Prior, Prior_type, Prior_SD, Prior_Like, Parm_StD (standard deviation
of parameter as calculated from inverse Hessian), Status (e.g. near bound), Pr_atMin
(value of prior penalty if parameter was near bound), and Pr_atMin. The Active_Cnt
entry is a count of the parameters in the same order they appear in the ss.cor file.
•Derived Quantities
–This section starts by showing the options selected from the starter.ss and forecast.ss input
files:
*SPR ratio basis
*F report basis
*B ratio denominator
Then the time series of output, with standard deviation of estimates, are produced with internally
generated labels. Note that these time series extend through the forecast era. The order of the output
166
is: spawning biomass, recruitment, SPRratio, Fratio, Bratio, management quantities, forecast catch (as a
target level), forecast catch as a limit level (OFL), Selex_std, Grow_std, NatAge_std. For the three “ratio”
quantities, there is an additional column of output showing a Z-score calculation of the probability
that the ratio differs from 1.0. The “management quantities” section is designed to meet the terms of
reference for west coast groundfish assessments; other formats could be made available upon request.
The std quantities at the end are set up according to specifications at the end of the control input file.
In some cases, a user may specify that no derived quantity output of a certain type be produced. In
those cases, SS substitutes a repeat output of the virgin spawning biomass so that vectors of null length
are not created.
ADMB NOTE: while vectors of null length are very useful for controlling optional model inputs, they
cannot be used with current version of ADMB for sdreport quantities.
•MGparm by year after adjustments
–This block shows the time series of Mgparms by year after adjustment by environmental
links, blocks and deviations.
•SELparm (size) by year after adjustments
–This block shows the size selectivity parameters, after adjustment, for each year in which
a change occurs.
•SELparm (age) by year after adjustments
–This block shows the age selectivity parameters, after adjustment, for each year in which a
change occurs.
•Recruitment Distribution
–This block shows the distribution of recruitment across growth patterns, genders,
birthseasons, and areas in the endyr of the model.
•Platoon Indexing
–This block shows the internal index values for various quantities. It can be a useful
reference for complex model setups. The vocabulary is: Bio_Pattern refers to a collection
of cohorts with the same defined growth and natural mortality parameters; sex is the
next main index. If recruitment occurs in multiple seasons, then Birthseas is the index for
that factor. The index labeled “Platoon” is used as a continuous index across all the other
factor-specific indices. If sub-platoons are used, they are nested within the Bio_Pattern x
Sex x Birthseas platoon. However, some of the output tables use the column label
“platoon” as a continuous index across platoons and sub-platoons. Note that there is no
index here for area. Each of the cohorts is distributed across areas and they retain their
167
biological characteristics as they move among areas.
•Size Freq Translation
–If the generalize size frequency approach is used, this block shows the translation
probabilities between population length bins and the units of the defined size frequency
method. If the method uses body weight as the accumulator, then output is in
corresponding units.
•Movement
–This block shows movement rate between areas in a multi-area model.
•Exploitation
–This block shows the time series of the selected F_std unit and the F multiplier for each
fleet in terms of harvest rate (if Pope’s approximation is used) or fully selected F.
•Index 2
–This section reports the observed and expected values for each index. All are reported in
one list with index number included as a selection field. At the bottom of this section, the
root mean squared error of the fit to each index is compared to the mean input error level
to assist the user in gaging the goodness-of-fit and potentially adjusting the input level of
imprecision.
•Index 3
–This section shows the parameter number assigned to each parameter used in this section.
•Discard
–This is the list of observed and expected values for the amount (or fraction) discard.
•Mean Body Wt
–This is the list of observed and expected values for the mean body weight.
•Fit Len Comps
–This is the list of the goodness of fit to the length compositions. The input and output
levels of effective sample size are shown as a guide to adjusting the input levels to better
match the model’s ability to replicate these observations.
•Fit Age Comps
168
–This has the same format as the length composition section.
•Fit Size Comps
–This has the same format as the length composition section and is used for the generalized
size composition summary.
•Len Selex
–Here is the length selectivity and other length specific quantities for each fishery and
survey.
•Age Selex
–Here is reported the time series of age selectivity and other age-related quantities for each
fishery and survey. Some are directly computed in terms of age, and others are derived
from the combination of a length-based factor and the distribution of size-at-age.
•Environmental Data
–The input values of environmental data are echoed here. In the future, the summary
biomass in the previous year will be mirrored into environmental column –2 and that the
age zero recruitment deviation into environmental column –1. Once so mirrored, they
may enable density-dependent effects on model parameters.
•Numbers at Age
–The output (in thousands of fish) is shown for each cohort tracked in the model.
•Numbers at Length
–The output is shown for each cohort tracked in the model.
•Catch at Age
–The output is shown for each fleet. It is not necessary to show by area because each fleet
operates in only one area.
•Biology
–The first biology section shows the length-specific quantities in the ending year of the time
series only. The derived quantity spawn is the product of female body weight, maturity and
fecundity per weight. The second section shows natural mortality.
•Growth Parameters
169
–This section shows the growth parameters, and associated derived quantities, for each year
in which a change is estimated.
•Biology at Age
–This section shows derived size-at-age and other quantities. It is the basis for the Bio report
page of the Excel output processor.
•Mean Body Wt (begin)
–This section reports the time series of mean body weight for each platoon. Values shown
are for the beginning of each season of each year.
•Mean Size Timeseries
–This section shows the time series of mean length-at-age for each platoon. At the bottom
is the average mean size as the weighted average across all platoons for each gender.
•Age Length Key
–This is reported for the midpoint of each season in the ending year.
•Age Age Key
–This is the calculated distribution of observed ages for each true age for each of the defined
ageing keys.
•Selectivity Database
–This section contains the selectivities organized as a database, rather than as a set of
vectors.
•Spawning Biomass Report 2, etc.
–The section shows annual total spawning biomass, then numbers-at-age at the beginning
of each year for each Bio_Pattern and Sex as summed over sub-platoons and areas. Then
Z-at-age is reported simply as ln(Nt+1,a+1Nt,a). Then the Report_1 section loops back
through the time series with all F values set to zero so that a dynamic Bzero, N-at-age, and
M-at-age can be reported. The difference between Report_1 and Report_2 can be used to
create an aggregate F-at-age.
•Composition Database
–This section is reported to a separate file, compreport.sso, and contains the length
composition, age composition, and mean size-at-age observed and expected values. It is
170
arranged in a database format, rather than an array of vectors. Software to filter the
output allows display of subsets of the database.
Using R To View Model Output (r4ss)
A collection of functions developed as a package, r4ss, for the statistical software R has been created
to explore SS model output. The functions include tools for summarizing and plotting results,
manipulating files, visualizing model parameterizations, and various other tasks. Currently,
information on the code, including installation instructions, can be found at
https://github.com/r4ss/r4ss. The software package is under constant development to maintain
compatibility with new versions of SS and to improve functionality.
Two of the most commonly used functions for model diagnostics are SS_output and SS_plots. After
running a model using SS, the report can be read into R by the SS_output function which stores
quantities in a list with named objects. This list can then be passed to the SS_plots function which
creates a series of over 100 plots that are useful to visualize output such as model fit to the data and
time series of quantities of interest.
The latest r4ss version on CRAN can be installed using a command like:
> install.packages("r4ss")
However, more frequent enhancements and bug fixes are posted to the GitHub project. The latest
version of r4ss can be installed directly from GitHub at any time via the devtools package in R with
the following commands:
> install.packages("devtools")
> devtools::install_github("r4ss/r4ss")
Note: devtools will give this message: "WARNING: Rtools is required to build R packages, but is not
currently installed." However, Rtools is NOT required for installing r4ss via devtools, so ignore
this warning.
Once you have installed the r4ss package, it can be loaded in the regular manner:
> library(r4ss)
The results from a model run can be read in and plots created using the following commands:
> setwd("C:\directory where model was run")
> base.model = SS_output(getwd())
> SS_plots(base.model)
171
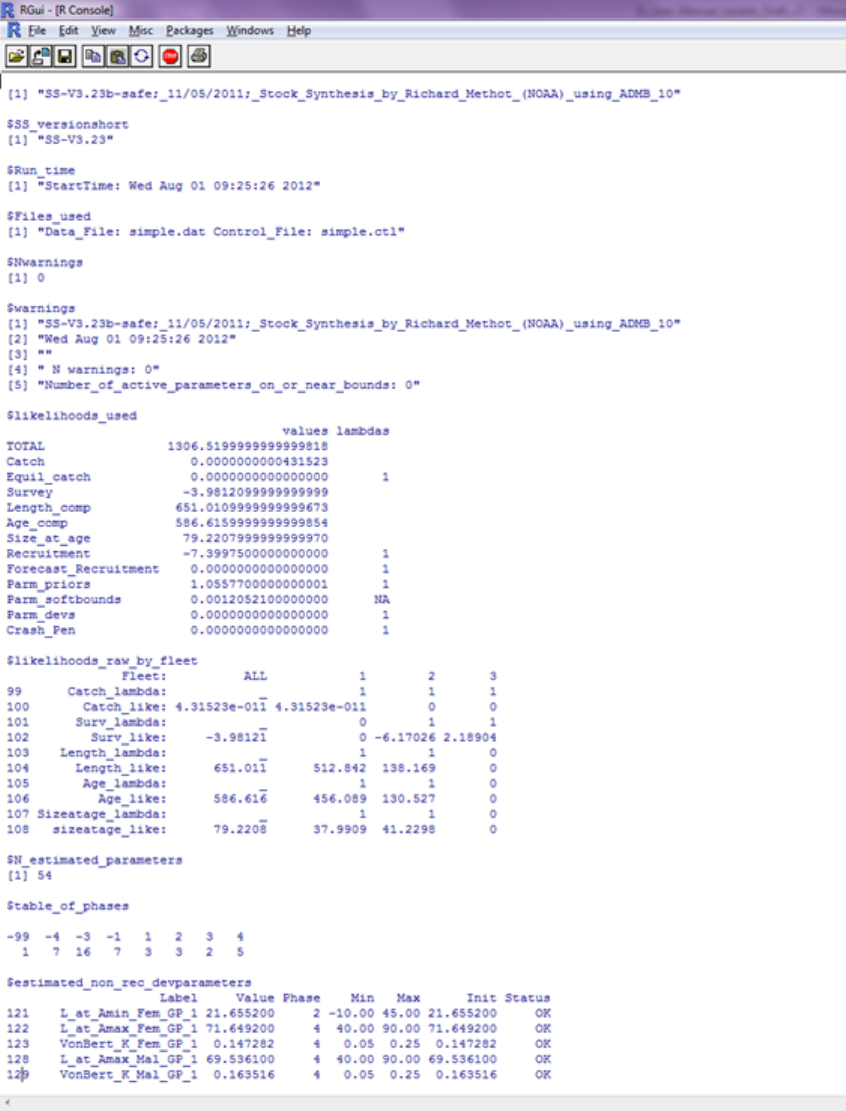
Example of the data displayed used by the SS_output function:
172
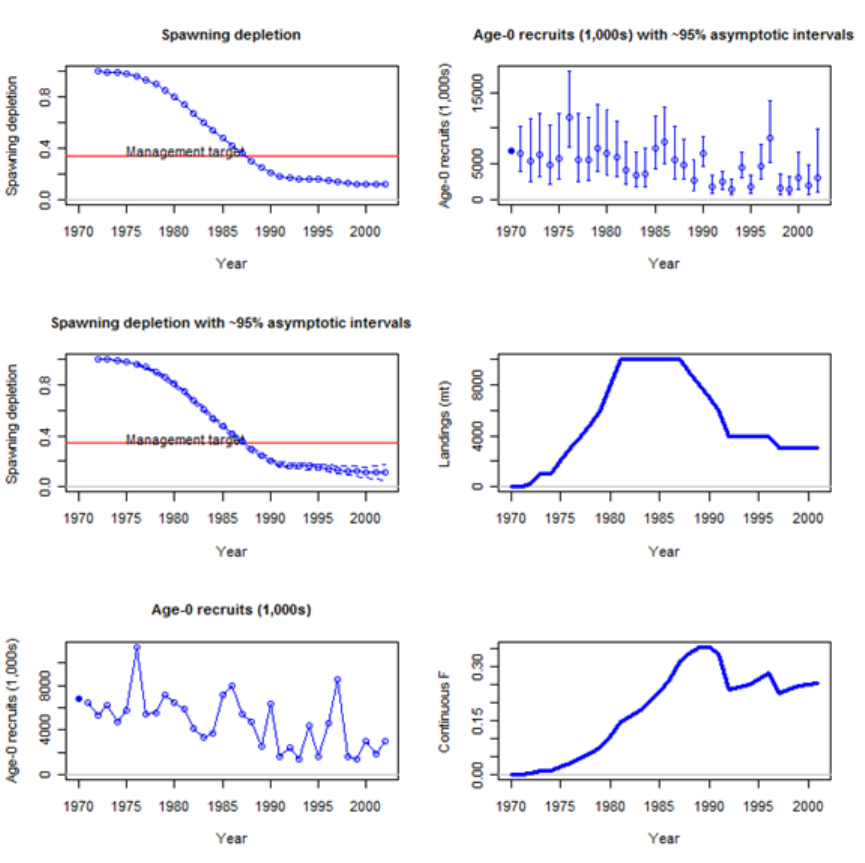
Example of the plots created using the SS_plots function:
173
The functions included in r4ss ranging from general use to functions developed for specific model
applications:
Core Functions
SS_output A function to create a list object for the output from Stock
Synthesis
SS_plots Plot many quantities related to output from Stock Synthesis
Plot functions called by SS_plots:
SSplotBiology Plot biology related quantities from Stock Synthesis model
output, including mean weight, maturity, fecundity, and
spawning output.
SSplotCatch Plot catch related quantities
SSplotCohorts Plot cumulative catch by cohort
SSplotComps Plot composition data and fits
SSplotData Timeline of presence/absence data by type, year, and fleet
SSplotDiscard Plot fit to discard fraction
SSplotIndices Plot indices of abundance and associated quantities
SSplotMnwt Plot mean weight data and fits
SSplotMovementMap Show movement rates on a map
SSplotMovementRates Show movement rates on a map
SSplotNumbers Plot numbers-at-age related data and fits
SSplotRecdevs Plot recruitment deviations
SSplotRecdist Plot of recruitment distribution among areas and seasons
SSplotSelex Plot selectivity
SSplotSexRatio Plot sex ratios
SSplotSummaryF Plot time series summary of F (or harvest rate)
SSplotSpawnrecruit Plot spawner-recruit curve
SSplotSPR Plot SPR quantities
SSplotTags Plot tagging data and fits
SSplotTimeseries Plot time series data
174

SSplotYield Plot yield and surplus production
SS_html Create HTML files to view figures in browser
SS_fitbiasramp Estimate bias adjustment for recruitment deviates
Model Comparisons and other diagnostics:
SSplotPars Plot distributions of priors, posteriors, and estimates
SSplotProfile Plot likelihood profile results
PinerPlot Plot fleet-specific contributions to likelihood profile
SSplotRetroRecruits Make retrospective pattern of recruitment estimates (a.k.a. squid
plot) as seen in Pacific hake assessments
Functions related to MCMC diagnostics:
mcmc_nuisance Summarize nuisance MCMC output
mcmc_out Summarize, analyze, and plot key MCMC output
SSgetMCMC Read MCMC output
SSplotMCMC_ExtraSelex Plot uncertainty around chosen selectivity ogive from MCMC
Interactive tools for exploring functional forms:
movepars Explore movement parameterization
selfit A function to visualize parameterization of double normal and
double logistic selectivity in SS
selfit_spline Visualize parameterization of cubic spline selectivity in SS
sel_line A function for drawing selectivity curves
File manipulation for inputs:
SS_readdat Read data file
SS_readforecast Read forecast file
SS_readstarter Read starter file
SS_writedat Write data file
SS_writeforecast Write forecast file
SS_writestarter Write starter file
175

SS_makedatlist Make a list for SS data
SS_parlines Get parameter lines from SS control file
SS_changepars Change parameters in the control file
SSmakeMmatrix Create inputs for entering a matrix of natural mortality by age
and year
SS_profile Run a likelihood profile in SS (incomplete)
NegLogInt_Fn Calculated variances of time-varying parameters using SS
implementation of the Laplace Approximation
File manipulations for outputs:
SS_recdevs Insert a vector of recruitment deviations into the control file
SS_splitdat Split apart bootstrap data to make input file
Minor plotting functions:
bubble3 Create a bubble plot
make_multifig Create multi-figure plots
Make_multifig_sexratio Create multi-figure plots of sex ratios
plotCI Plot points with confidence intervals
rich_colors_short Make a vector of colors
stackpoly Plot stacked polygons
mountains Make shaded polygons with a mountain-like appearance
Really specialized functions:
DoProjectPlots Make plots from Rebuilder program
IOTCmove Make a map of movement for a 5-area Indian Ocean model
SSFishGraph A function for converting SS output to the format used by
FishGraph
TSCplot Create a plot for the TSC report
176
Special Set-ups
Continuous seasonal recruitment
It is awkward in SS to set up a seasonal model such that recruitment can occur with similar and
independent probability in any season of any year. Consequently, some users have attempted to setup
SS so that each quarter appears as a year. They have set up all the data and parameters to treat
quarters as if they were years (i.e. each still has a duration of 1.0 time step). This can work, but
requires that all rate parameters be re-scaled to be correct for the quarters being treated as
years.
Another option is available. If there is one season per year and the season duration is set to 3 (rather
than the normal 12), then the season duration is calculated to be 3/12 or 0.25. This means that the
rate parameters can stay in their normal per year scaling and this shorter season duration makes the
necessary adjustments internally. Some other adjustments to make when doing quarters as years
include:
•re-index all "year seas" inputs to be in terms of quarter-year because all are now season 1; increase
endyr value in sync with this
•increase max age because age is now in quarters
•in the age error definitions, increase the number of entries in accord with new max age
•in the age error definitions, recode so that each quarter-age gets assigned to the correct agebin;
This is because the age data are still in terms of agebins; i.e. the first 4 entries for quarter-ages 1
through 4 will all be assigned to agebin 1.5; the next four to agebin 2.5; you cannot accomplish
the same result by editing the age bin values because the stddev of ageing error is in terms of
agebin
•in the control file, multiple the natM age breakpoints and growth AFIX values by 1/seasdur
•decrease the R0 parameter starting value because it is now the average number of recruitments
per qtryear
•edit the rec_dev start and endyrs to be in terms of qtryear
•edit any age selectivity parameters that refer to age to now refer to qtrage
•if there needs to be some degree of seasonality to recruitment or some parameter, then you
could create a cyclic pattern in the environmental input and make recruitment or some other
parameter a function of this cyclic pattern
177
A good test showing comparability of the 3 approaches to setting up a quarterly model should be
done.
Change Log
This section has been removed from the user manual. Information on changes to SS is now recorded in
the spreadsheet database, SS_Changes.xlsx. Fields include date, version number, category (e.g. growth,
selectivity), type (e.g. new, clarify, fix). Occasional model tips will be added with the type=”Tip”.
Appendix A: Recruitment Variability and Bias
Correction
Recruitments in SS are defined as lognormal deviates around a log-bias adjusted
spawner-recruitment curve. The magnitude of the log-bias adjustment is calculated from the level of
σR, which is the standard deviation of the recruitment deviations (in log-space). There are 5
segments of the time series in which to consider the effect of the log-bias adjustment: virgin; initial
equilibrium; early data-poor period; data-rich period; very-recent/forecast. The choice of break
points between these segments need not correspond directly with the settings for the bias
adjustment, although some alignment might be desired. Methot and Taylor (2011) provide more
detailed discussion of the bias adjustment than what is provided below but do not address the
separation of time periods into separate segments. The approach is illustrated with figures associated
with a recent assessment for darkblotched rockfish (Gertseva and Thorson, 2013).
178
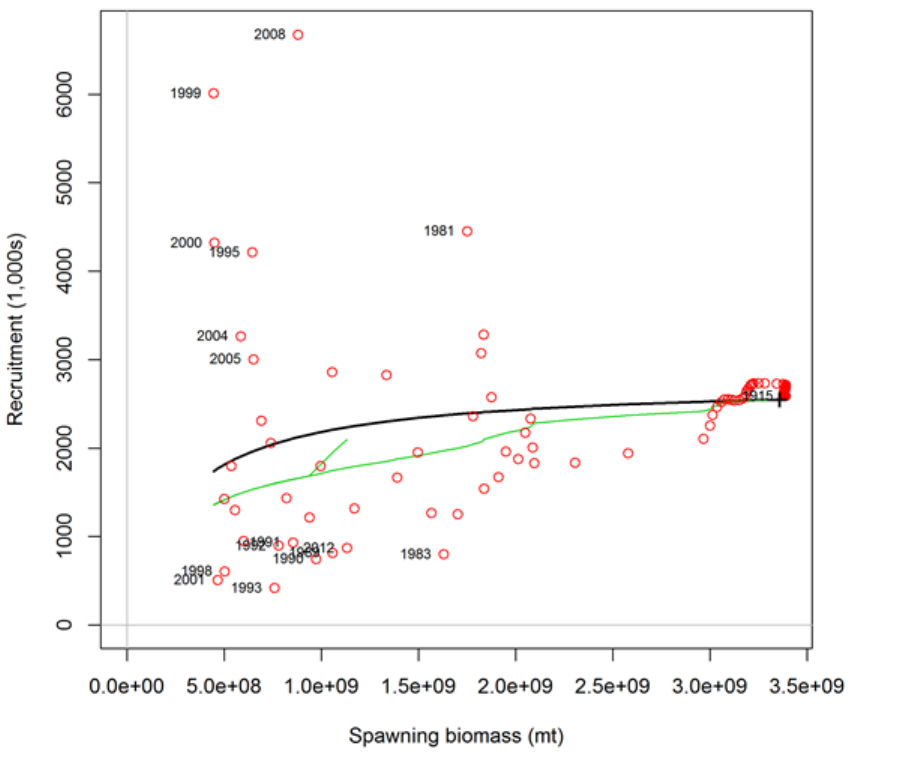
Figure A.1. Spawner-recruitment relationship for darkblotched rockfish (Gertseva and Thorson,
2013). Red points represent estimated recruitments, the solid black line is the stock-recruit
relationship and the green line represents the adjustment to this relationship after adjustment to
account for the lognormal distribution associated with each year. The “+” symbol labeled 1915 near
the right side represents both the virgin and initial equilibrium of the model. The numerous red
points close to the initial conditions correspond to the early years of the model with low harvest
rates.
179
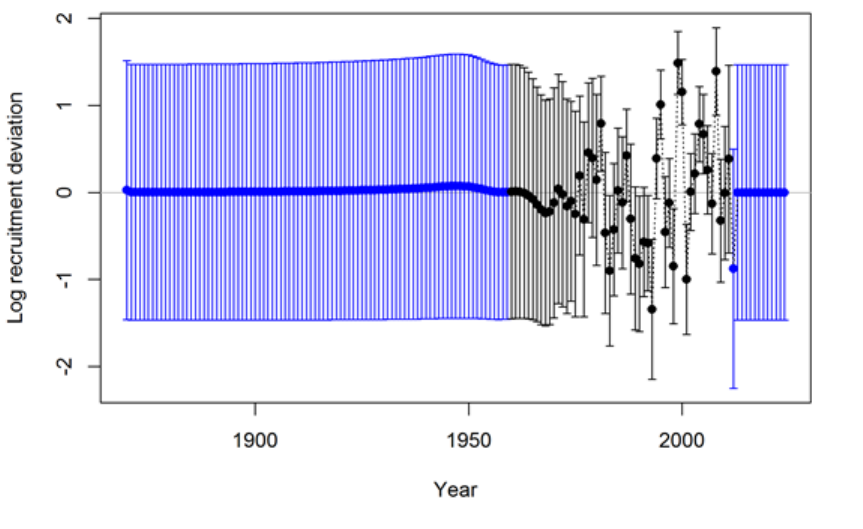
Figure A.2. Timeseries of log recruitment deviations for darkblotched rockfish with 95% uncertainty
intervals. The start year of the model is 1915, but recruitment deviations are estimates starting in 1870.
The 45 deviation estimates for 1870–1914 inform the age structure used in the start year. The black
color for the years 1960–2011 indicates the “main” recruitment deviation vector, while the blue color
for the years 1870–1959 and 2012–2024 indicates the “early” and “late/forecast” recruitment deviation
vectors, respectively.
Virgin Biomass
The R0 level of recruitment is a parameter of the spawner-recruitment curve. This recruitment and the
corresponding spawning biomass, S0, are expected to represent the long-term arithmetic mean.
Initial Equilibrium
The level of recruitment is typically maintained at the R0 level even though the initial equilibrium catch
will reduce the spawning biomass below the virgin level. If steepness is moderately low or the initial F
is high, then the lack of response in recruitment level may appear paradoxical. The logic is that building
in the spawner-recruitment response to initial F would significantly complicate the calculations and
would imply that the initial equilibrium catch level had been going on for multiple generations. If the
lack of response is considered to be problematic in a particular application, then start the model at an
earlier year and with a lower initial equilibrium catch so that the dynamics of the spawner-recruitment
response get captured in the early period, rather than getting lost in the initial equilibrium.
180
Early Data-Poor Period
This is the early part of the time series where the only data typically are landed catch. There are no data
to inform the model about the specific year-to-year fluctuations in recruitment, although the ending
years of this period will begin to be influenced by the data. The “early time period” is not a formal
concept. It is up to the user to decide whether to start estimating recruitment deviations beginning
with the first year of the model, or to delay such estimation until the data become more informative.
Modeling recruitment deviations in this period may lead to a more realistic portrayal of the uncertainty
in depletion, but can also lead to spurious patterns in estimated recruitments that may be driven by
the fit to index data or other sources that would not be expected to have accurate information on
recruitment.
•Option A: Do not estimate recruitment deviations during this early period. During years prior
to the first year of recruitment deviations, the model will set the recruitment equal to the level
of the spawner-recruitment curve. Thus, it is a mean-based level of recruitment. Because these
annual parameters are fixed to the level of the spawner-recruitment curve, they have no
additional uncertainty and make no contribution to the variance of the model. This approach
may produce relatively large, or small, magnitude deviations at the very beginning of the
subsequent period, as the model “catches up” to any slight signal that could not be captured
through estimated deviations in the early data-poor period. There may be some effect on the
estimate of R0 as a result of lack of model flexibility in balancing early period removals with
signal in the early portion of the data-rich period.
•Option B: Estimate recruitment deviations for all the early years. Each of these recruitment
deviations is now a dev parameter so will have a variance that contributes to the total model
variance. The estimated standard deviation of each of these dev parameters should be similar to
σRbecause σRis the only constraint on these parameters (however, the last few in the sequence
will begin to feel the effect of the data so may have lower standard deviations).
Data-Rich Period
Here the data inform the model on the year-to-year level of recruitment. These fluctuations in
recruitment are assumed to have a lognormal distribution around the log-bias adjusted
spawner-recruitment curve. The level of σRinput to the model should match this level of fluctuation
to a reasonable degree. Because the recruitments are lognormal, they produce a mean biomass level
that is comparable to the virgin biomass and thus the depletion level can be calculated without bias.
However, if the early period has recruitment deviations estimated by MPD, then the depletion levels
during the early part of the data-rich period may have some lingering effect of negative bias during
the early time period. The level of σRshould be at least as large as the level of variability in these
estimated recruitments. If too high a level of σRis used, then a bias can occur in the estimate of
spawner-recruitment steepness, which determines the trend in recruitment. This occurs when the
early recruitments are taken directly from the spawner- recruitment curve, so are mean unbiased,
then the later recruitments are estimated as deviations from the log-bias adjusted curve. If σRis too
large, then the bias-adjustment is too large, and the model may compensate by increasing steepness to
181
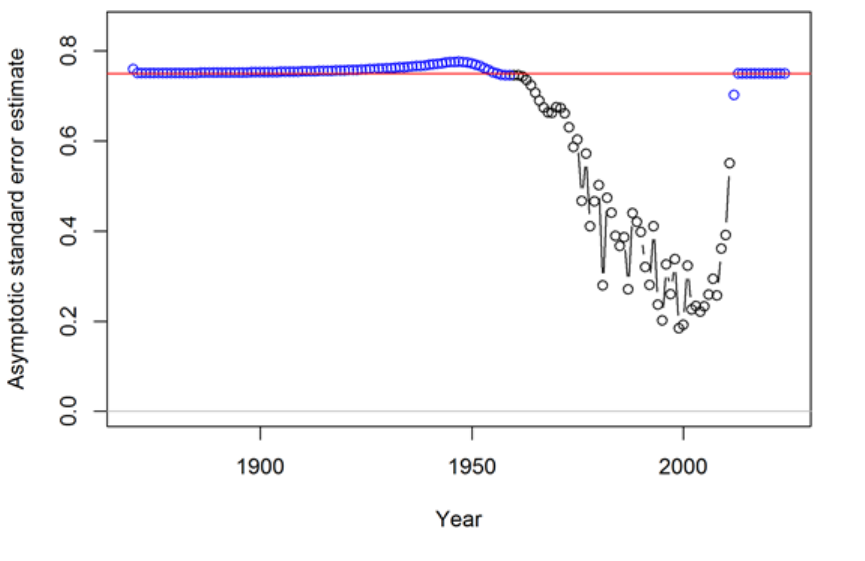
keep the mean level of recent recruitments at the correct level.
Recent Years/Forecast
Here the situation is very similar to the early time period in that there are no data to inform the model
about the year-to-year pattern in recruitment fluctuations so all devs will be pulled to a zero level
in the MPD. The structure of SS creates no sharp dividing line between the estimation period and
the forecast period. In many cases one or more recruitments at the end of the time series will lack
appreciable signal in the data and should therefore be treated as forecast recruit deviations. To the
degree that some variability is observed in these recruitments, partial or full bias correction may be
desirable for these devs separate from the purely forecast devs, there is therefore an additional control
for the level of bias correction applied to forecast deviations occurring prior to endyear+1.
Figure A.3. Timeseries of standard error estimates for the log recruitment deviations for
darkblotched rockfish with 95% uncertainty intervals. As in Figure A.2, the black color indicates the
main recruitment period. This period with lower standard error is associated with higher variability
among deviations (Figure A.2). The red line at 0.75 indicates the σRvalue in this model.
182
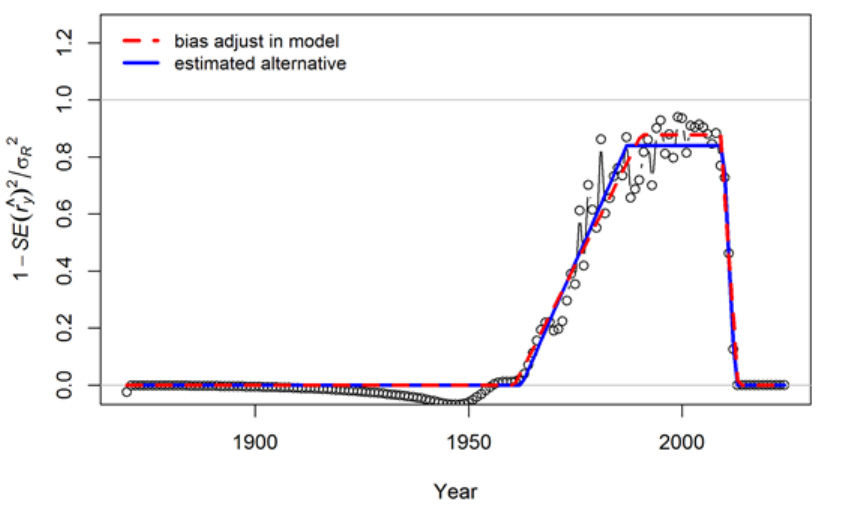
Figure A.4. Transformation of the standard error estimates (shown in Figure A.3) for darkblotched
rockfish following the approach suggested by Methot and Taylor (2011). These values were used to
set the 5 values controlling the degree of bias adjustment (as a fraction of σR/2) to account for
differences in the mean and median of the lognormal distribution from which the recruitment
deviations are drawn. The red line indicates a bias adjustment of 0 up to the 1960.75, ramping up to a
maximum adjustment level of 0.877 for the period 1990.4–2008.98,and reducing back to 0 starting in
2013.08. Note that these values controlling the bias adjustment need not be integer year values. Also
the break points in the bias adjustment function need not match the break points between early,
main, and late/forecast recruitment deviation vectors (indicated by blue and black colors in Figures
A.2 and A.3). The blue line indicates a functional form that minimizes the sum of squared differences
between the bias adjustment function and the transformed standard error values. The subtle
differences between red and blue lines are unlikely to have any appreciable effect on the model
results.
183
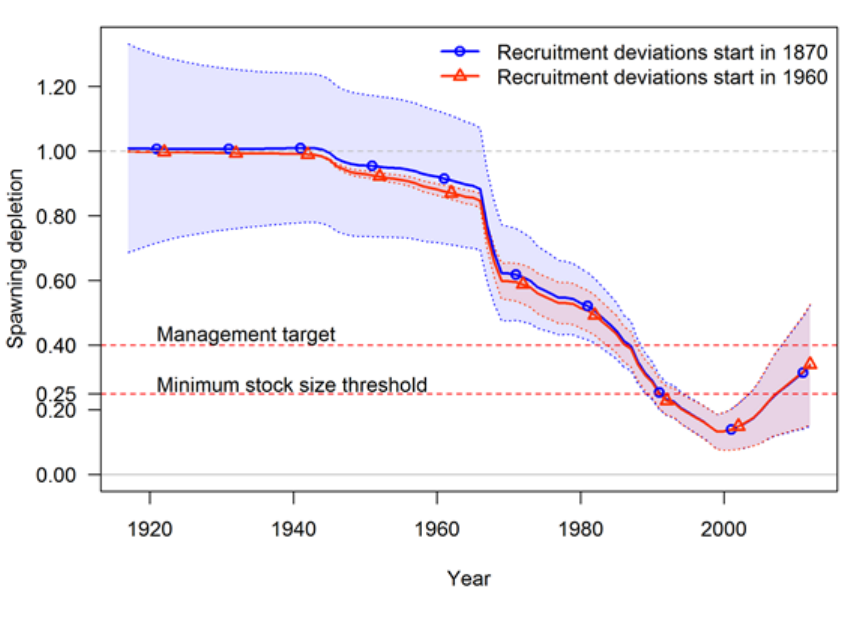
Figure A.5. Comparison of timeseries of spawning depletion for darkblotched rockfish models with
early recruitment deviations (starting in 1870) and without early deviations (only main recruitment
deviations starting in 1960). The point estimates are similar, but the 95% uncertainty intervals are
substantially different. With no recruitment deviations for the early period, the estimates of spawning
depletion in the early years are very precise and uncertainty increases as the stock moves into the data
rich period. In contrast, the addition of the early recruitment deviations results in a large uncertainty
in spawning depletion for the early years and an increase in precision as the stock moves into the data
rich period. In this application, the uncertainty associated with the recent years is independent of the
assumptions about early recruitments.
Issues with Including Environmental Effects
The expected level of recruitment is a function of spawning biomass, an environmental time series,
and a log-bias adjustment.
E(Recruitment) = f(SpBio)∗exp(β∗envdata)∗exp(−0.5∗σ2
R)(56)
σRis the variability of the deviations, so it is in addition to the variance “created” by the environmental
effect. So, as more of the total recruitment variability is explained by the environmental effect, the
residual σRshould be decreased. The model does not do this automatically.
184
The environmental effect is inherently lognormal. So when an environmental effect is included in the
model, the arithmetic mean recruitment level will be increased above the level predicted by f(SpBio)
alone. The consequences of this have not yet been thoroughly investigated, but there probably should
be another bias correction based on the variability of the environmental data as scaled by the estimated
linkage parameter, β. It is also problematic that the environmental effect time series used as input is
assumed to be measured without error.
The preferred approach to including environmental effects on recruitment is not to use the
environmental effect in the direct calculation of the expected level of recruitment. Instead, the
environmental data would be used as if it was a survey observation of the recruitment deviation. This
approach is similar to using the environmental index as if it was a survey of age 0 recruitment
abundance because by focusing on the fit to the deviations it removes the effect of SpBio on
recruitment. In this alternative, the σRwould not be changed by the environmental data; instead the
environmental data would be used to explain some of the total variability represented by σR. This
approach may also allow greater uncertainty in forecasts, as the variability in projected recruitments
would reflect both the uncertainty in the environmental observations themselves and the model fit to
these observations.
Initial Age Composition
If the first year with recruitment deviations is set less than the start year of the model, then these
early deviations will modify the initial age composition. The amount of information on historical
recruitment variability certainly will degrade as the model attempts to estimate deviations for older
age groups in the initial equilibrium. So the degree of bias correction is reduced linearly in proportion
to age so that the correction disappears when maximum age is reached. The initial age composition
approach normally produces a result that is indistinguishable from a configuration that starts earlier
in the time series and estimates a longer time series of recruitments. However, because the initial
equilibrium is calculated from a recruitment level unaffected by spawner-recruitment steepness and
initial age composition adjustments are applied after the initial equilibrium is calculated, it is possible
that the initial age composition approach will produce a slightly different result than if the time series
was started earlier and the deviations were being applied to the recruitment levels predicted from the
spawner-recruitment curve.
Appendix B: Data Weighting
In 2015 there was a CAPAM workshop dedicated to data-weighting. Description of the workshop
can be found at http://www.capamresearch.org/data-weighting/workshop. The presentations from
the workshop are available through that website and many of them were included in a special issue of
Fisheries Research a https://www.sciencedirect.com/journal/fisheries-research/vol/192.
185
Currently, there are three main methods for weighting length and data applied for U.S. West Coast
assessments using Stock Synthesis.
1. McAllister - Ianelli: Effective sample size is calculated from fit of observed to expected length
or age compositions. Tuning algorithm is intended to make the arithmetic mean of the input
sample size equal to the harmonic mean of the effective sample size. Reference: McAllister,
M.K. and J.N. Ianelli 1997. Bayesian stock assessment using catch-age data and the sampling -
importance resampling algorithm. Can. J. Fish. Aquat. Sci, 1997, 54(2): 284-300.
2. Francis: Based on variability in the observed mean length or age by year, where the sample sizes
are adjusted such that the fit of the expected mean length or age should fit within the uncertainty
intervals at a rate which is consistent with variability expected based on the adjusted sample sizes
(Method "TA1.8"). Reference: Francis, R.I.C.C. (2011). Data weighting in statistical fisheries
stock assessment models. Can. J. Fish. Aquat. Sci. 68: 1124-1138.
3. Dirichlet-Multinomial: A new likelihood (as opposed to the standard multinomial) which
includes an estimable parameter (theta) which scales the input sample size. In this case, the
term “Effective sample size” has a different interpretation than in the McAllister-Ianelli
approach. References: Thorson, J.T., Johnson, K.F., Methot, R.D. and Taylor, I.G. 2017.
Model-based estimates of effective sample size in stock assessment models using the
Dirichlet-multinomial distribution. Fish. Res. 192: 84-93. Thorson, J.T. 2018. Perspective:
Let’s simplify stock assessment by replacing tuning algorithms with statistics. Fish. Res.
Applying the methods
McAllister-Ianelli
The “Length_Comp_Fit_Summary” and “Age_Comp_Fit_Summary” sections in the Report file
include information on the harmonic mean of the effective sample size and arithmetic mean of the
input sample size used in this tuning method. In the r4ss package, these tables are returned by the
SS_output function as $Length_comp_Eff_N_tuning_check and
$Age_comp_Eff_N_tuning_check.
A convenient way to process these values into the format required by the control file is to use the
function:
SS_tune_comps(replist, option = "MI")
where the input “replist” is the object created by SS_output. This function will return a table and also
write a matching file called “suggested_tuning.ss” to the directory where the model was run.
186
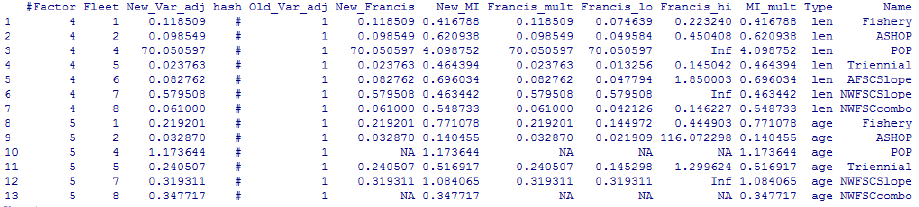
For models using SS version 3.30, the table created by SS_tune_comps can be pasted into the bottom
of the control file in the section labeled “Input variance adjustments”, followed by the terminator line
which indicates the end of the section. Here’s an example of the first few rows of the table followed by
the terminator line (not added by the function):
Also see the help page for the r4ss function SS_varadjust which can be used to automatically write
a new control file if you want to streamline the process of applying multiple iterations of this tuning
method.
If the tuning has been implemented, the green lines in the figure below would intersect at a point which
is on the black 1-to-1 diagonal line in this figure created by the r4ss function SS_plots.
187
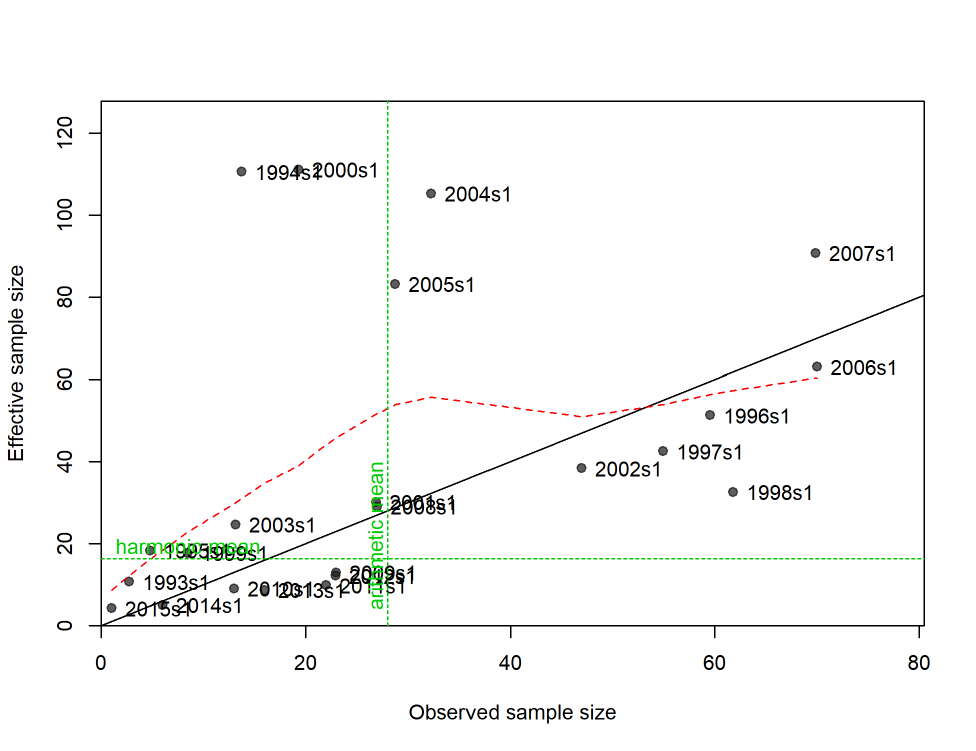
Figure B.1.N vs. EffN comparison, Length composition data as plotted by r4ss.
There are a couple of challenges posed by the McAllister-Ianelli data-weighting approach:
1. Subjective choice of how many iterations to take to achieve adequate convergence. Often just
one iteration is applied.
2. Takes time to implement so tuning is rarely repeated during retrospective or sensitivity analyses.
Francis
Implementation: recommended adjustments are calculated by the r4ss functions SSMethod.TA1.8 and
SSMethod.Cond.TA1.8. These functions are rarely used alone but are called by the SS_plots function
when making plots like the one below. For SS 3.30 models, the simplest way to get the adjustments in
the format required by the control file is to use the SS_tune_comps function (described above under
188
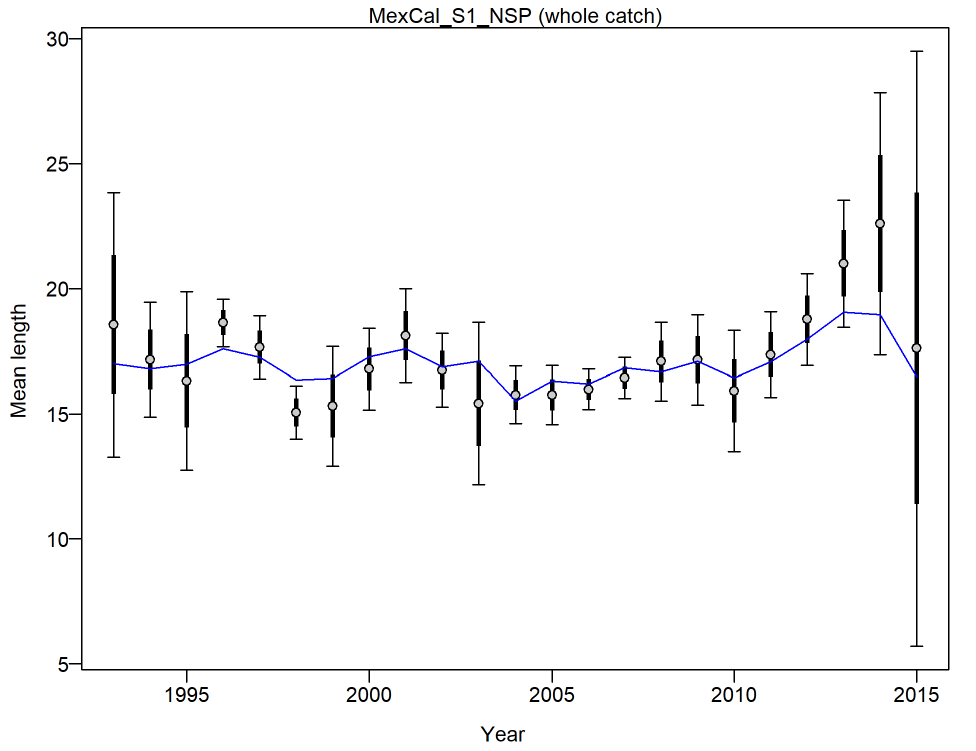
the McAllister-Ianelli method), but with a different option specified:
SS_tune_comps(replist, option = "Francis")
The figure below shows the estimated 95% intervals around the observed mean length by year based on
the input sample size (thick lines) and the increase in that uncertainty which would occur if the sample
sizes were adjusted according to the proposed multiplier.
Figure B.2.Mean length for Fleet1 with 95% confidence intervals based on current samples sizes.
Francis data weighting method TA1.8: thinner intervals (with capped ends) show result of further
adjusting sample sizes based on suggested multiplier (with 95% interval) for len data from Fleet1:
0.2739 (0.1661-0.6305)
There are a several of challenges posed by the Francis data-weighting approach:
189

1. Subjective choice of how many iterations to take to achieve adequate convergence. Often just
one iteration is applied.
2. Takes time to implement so tuning is rarely repeated during retrospective or sensitivity analyses.
3. Recommended adjustment can be sensitive to outliers (remove a few years of anomalous
composition data can lead to large change in recommended adjustment).
Dirichlet-Multinomial
Change the choice of likelihood and set parameter choices in the data file:
•In the specification of the length and/or age data, change "CompError" column in age and length
comp specifications from 0 to 1 and "ParmSelect" from 0 to a sequence of numbers from 1 to N
where N is the total number of combinations of fleet and age/length.
•Resulting input should look similar to:
•The ParmSelect column can also have repeated values so that multiple fleets share the same
log(theta) parameter.
•If you have both length and age data, the ParmSelect should have separate numbers for each, e.g.
1 and 2 for the length comps and 3 and 4 for the age comps for the same two fleets.
Add parameter lines to the control file:
•Add as many parameter lines as the maximum numbers in the ParmSelect column. The new
parameter lines go after the main selectivity parameters but before any time-varying selectivity
parameters
•Jim Thorson initially recommended bounds of -5 to 20, with a starting value of 0 (which
corresponds to a weight of about 50% of the input sample size). However, parameter estimates
above 5 are associated with 99-100% weight with little information in the likelihood about the
parameter value. Therefore, an upper bound of 5 may help identify cases that otherwise would
have convergence issues.
•Example parameter lines are below:
190
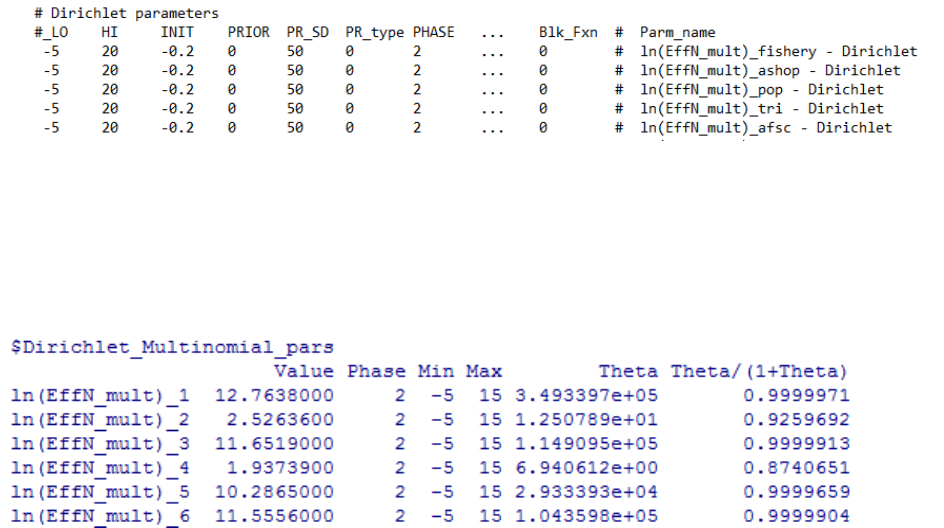
•Reset any existing variance adjustments factors that might have been used for the
McAllister-Ianelli or Francis tuning methods. In 3.24 this means setting the values to 1, in SS
version 3.30, you can simply delete or comment-out the rows with the adjustments.
The SS_output function in r4ss returns table like the following:
The ratio shown in the final column is the estimated multiplier which can be compared to the sample
size adjustment estimated in the other tuning methods above (the New_Var_adj column in the table
produced by the SS_tune_comps function in r4ss).
If the reported θ/(1 + θ)ratio is close to 1.0, that indicates that the model is trying to tune the sample
size as high as possible. In this case, the ln(θ)parameters should be fixed at a high value, like the upper
bound of 20, which will result in 100% weight being applied to the input sample sizes. An alternative
would be to manually change the input sample sizes to a higher value so that the estimated weighting
will be less than 100
There is also information about this result produced in the plots created by the SS_plots
function:
191
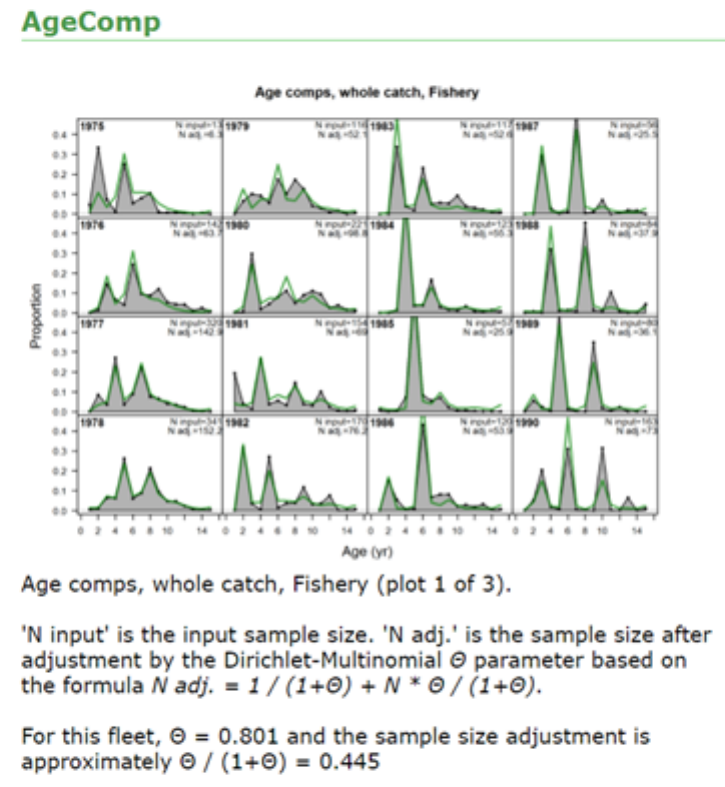
There are a several of challenges posed by the Dirichlet-Multinomial data-weighting approach:
1. Not yet widely used so little guidance is available.
2. Does not allow weights above 100% (by design) so it is not yet clear how best to deal with the
situation when the estimated weight is close to 100%.
3. Parameterization has potential to cause convergence issues or inefficient MCMC sampling
when weights are close to 100% (Jim Thorson has proposed a prior distribution that may help
with this, but has not yet been tested).
192
Appendix C: Forecast Module
Introduction
Version 3.20 of Stock Synthesis (SS) introduced substantial upgrades to the benchmark and forecast
module. The general intent was to make the forecast outputs more consistent with the requirement
to set catch limits that have a known probability of exceeding the overfishing limit. In addition, this
upgrade addressed several inadequacies with the previous module, including:
•The average selectivity and relative F was the same for the benchmark and the forecast
calculations;
•The biology-at-age in endyr+1 was used as the biology for the benchmark, but biology–at-age
propagated forward in the forecast if there was time-varying growth;
•The forecast module had a kluge approach to calculation of OFL conditioned on previously
catching ABC;
•The forecast module implementation of catch caps was incomplete and applied some caps on a
seasonally, rather than the more logical annual basis;
•The Fmult scalar for fishing intensity presented a confusing concept for many users;
•No provision for specification of catch allocation among fleets;
•The forecast allowed for a blend of fixed input catches and catches calculated from target F; this
is not optimal for calculation of the variance of F conditioned on a catch policy that sets ACLs.
The V3.20 module addressed these issues by:
•Providing for unique specification of a range of years from which to calculate average
selectivity for benchmark, average selectivity for forecast, relative F for benchmark, and
relative F for forecast;
•Create a new specification for the range of years over which to average size-at-age and
fecundity-at-age for the benchmark calculation. In a setup with time-varying growth, it may
make sense to do this over the entire range of years in the time series. Note that some
additional quantities still use their endyr values, notably the migration rates and the allocation
of recruitments among areas. This will be addressed shortly;
•Create a multiple pass approach that rectifies the OFL calculation;
•Improve the specification of catch caps and implement specification of catch allocations so that
193
there can be an annual cap for each fleet, an annual cap for each area, and an annual allocation
among groups of fleets (e.g. all recreational fleets vs. all commercial fleets);
•Introduce capability to have implementation error in the forecast catch (single value applied to
all fleets in all seasons of the year).
Multiple Pass Forecast
The most complicated aspect of the changes is with regard to the multiple pass aspect of the forecast.
This multiple pass approach is needed to calculate both OFL and ABC in a single model run. More
importantly, the multiple passes are needed in order to mimic the actual sequence of
assessment-management action – catch over a multi-year period. The first pass calculates OFL based
on catching OFL each year, so presents the absolute maximum upper limit to catches. The second
pass forecasts a catch based on a harvest policy, then applies catch caps and allocations, then updates
the F’s to match these catches. In the third pass, stochastic recruitment and catch implementation
error are implemented and SS calculates the F that would be needed in order to catch the adjusted
catch amount previously calculated in the second pass. With this approach, SS is able to produce
improved estimates of the probability that F would exceed the overfishing F. In effect it is the
complement of the P* approach. Rather than the P* approach that calculates the stream of annual
catches that would have an annual probability of F>Flimit, SS calculates the expected time series of
P* that would result from a specified harvest policy implemented as a buffer between Ftarget and
Flimit.
The sequence of multiple forecast passes is as follows:
1. Pass 1 (a.k.a. Fcast_Loop1)
(a) Loop Years
i. SubLoop (a.k.a. ABC_Loop) = 1
A. R=f(SSB) with no deviations
B. F=Flimit
C. Fixed input catch amounts ignored
D. No catch adjustments (caps and allocations)
E. No implementation error
F. Result: OFL conditioned on catching OFL each year
194
2. Pass 2
(a) Loop Years
i. SubLoop = 1
A. R=f(SSB) with no deviations
B. F=Flimit
C. Fixed input catch amounts ignored
D. No catch adjustments (caps and allocations)
E. No implementation error
F. Result: OFL conditioned on catching ABC previous year. Stored in std_vector.
ii. SubLoop = 2
A. R=f(SSB) with no deviations
B. F=Ftarget to get catc for each fleet in each season
C. Fixed input catch amounts replace catch from step 2
D. Catch adjustments (caps and allocations) applied on annual basis (after looping
through seasons and and areas within this year). These adjustments utilize the
logistic joiner approach common in SS so the overall results remain completely
differentiable.
E. No implementation error
F. Result: ABC as adjusted for caps and allocations
iii. SubLoop = 3
A. R=f(SSB) with no deviations
B. Catches from Pass 2 multiplied by the random term for implementation error
C. F=adjusted to match the catch*error while taking into account the random
recruitments. This is most easily visualized in a MCMC context where the
recruitment deviation and the implementation error deviations take on
non-zero values in each instance. In MLE, because the forecast recruitments
and implementation error are estimated parameters with variance, their
195

variance still propagates to the derived quantities in the forecast.
D. Result: Values for F, SSB, Recruitment, Catch are stored in std-vectors
•In addition, the ratios F/Flimit and SSB/SSBlimit or SSB/SSBtarget are also
stored in std_vectors.
•Estimated variance in these ratios allows calculation of annual probability that
F>Flimit or B<Blimit. This is essentially the realized P* conditioned on the
specified harvest policy.
Example Effects on Correlations
An example that illustrates the above process was conducted. The situation was a low M,
late-maturing species, so changes are not dramatic. The example conducted a 10 year forecast and
examined correlations with derived quantities in the last year of the forecast. This was done once
with the full set of 3 passes as described above, and again with only 2 passes and stochastic
recruitment occurring in pass 2, rather than 3. This alternative setup is more similar to forecasts
done using previous model versions.
2 Forecast Passes with F from 2 Forecast Passes with catch from
ABC and random recruitment target F and equilibrium recruitment
Factor X Factor Y Corr Factor X Factor Y Corr
A1 F 2011 RecrDev 2002 -0.126 A2 F 2011 RecrDev
2002
0.090
B1 F 2011 Recr 2002 0.312 B2 F 2011 Recr 2002 0.518
C1 ForeCatch 2011 RecrDev 2002 0.000 C2 ForeCatch 2011 RecrDev
2002
0.129
D1 ForeCatch 2011 Recr 2002 0.455 D2 Forecatch 2011 Recr 2002 0.555
Correlation A2 shows a small positive correlation between the recruitment deviation in 2002 and the
F in 2011. This is probably due to the fact that a positive deviation in recruitment in 2002 will reduce
the chances that the biomass in 2011 will be below the inflection point in the control rule. This
occurs because in calculating catch from F, the model effectively “knows” the future recruitments. I
predict that this B1 correlation would be near zero if there was no inflection in the control rule.
Correlation A1 shows this turning into a negative correlation. This is because the future catches are
first calculated from equilibrium recruitment, then when random recruitments are implemented, a
positive recruitment deviation will cause a negative deviation in the F needed to catch that now
196
“fixed” amount of future catch.
Correlations B1 and B2 are in terms of absolute recruitment, not recruitment deviation. Now overall
model conditions that cause a higher absolute recruitment level will also result in a higher forecast
level. No surprise there, and the correlation is stronger when variance is based on catch is calculated
from F (B2).
Correlation C2 shows a positive correlation between recruitment deviation in 2002 and forecast catch
in 2011. However, correlation C1 is 0.0 because the forecast catch in 2011 is set based on equilibrium
recruitment and is not influenced by the recruitment deviations.
Future Work
•More testing with high M, rapid turnover conditions
•Testing without inflection in control rule
•Consider separating implementation error into a pass #4 so results will more clearly show effect
of assessment uncertainty separate from implementation uncertainty
•Consider adding a random “assessment” error which essentially is a random variable that scales
population abundance before passing into the forecast stage. Complication is figuring out how
to link it to the correlated error in the benchmark quantities
•Because all of these calculations occur only in the sdphase or the mceval phase, it would be
feasible for mceval calls to add an additional pass that is implemented many times and in which
random forecast recruitment draws are made.
•Factors like selectivity and fleet relative F levels are calculated as an average of these values
during the time series. This is internally consistent if these factors do not vary during the time
series (although clearly this is a stiff model that will underestimate process variance). However,
if these factors do vary over time, then the average used for the forecast will under-represent
the variance. A better approach would be to set up the parameters of selectivity as a random
process that extends throughout the forecast period, and to update estimated selectivity in each
year of the forecast based upon the random realization of these parameters.
197
Appendix D: Code Examples
Ageing Error Estimation
/* SS_Label_FUNCTION 45 get_age_age */
FUNCTION void get_age_age(const int Keynum, const int AgeKey_StartAge, const int AgeKey_Linear1, const int AgeKey_Linear2)
{
// FUTURE: calculate adjustment to oldest age based on continued ageing of old fish
age_age(Keynum).initialize();
dvariable age;
dvar_vector age_err_parm(1,7);
dvariable temp;
if(Keynum==Use_AgeKeyZero)
{
// SS_Label_45.1 set age_err_parm to mgp_adj, so can be time-varying according to MGparm options
for (a=1;a<=7;a++)
{age_err_parm(a)=mgp_adj(AgeKeyParm-1+a);}
age_err(Use_AgeKeyZero,1)(0,AgeKey_StartAge)=r_ages(0,AgeKey_StartAge)+0.5;
age_err(Use_AgeKeyZero,2)(0,AgeKey_StartAge)=age_err_parm(5)*(r_ages(0,AgeKey_StartAge)+0.5)/
(age_err_parm(1)+0.5);
// SS_Label_45.3 calc ageing bias
if(AgeKey_Linear1==0)
{
age_err(Use_AgeKeyZero,1)(AgeKey_StartAge,nages)=0.5 + r_ages(AgeKey_StartAge,nages) +
age_err_parm(2)+(age_err_parm(3)-age_err_parm(2))*(1.0-mfexp(-age_err_parm(4)*
(r_ages(AgeKey_StartAge,nages)-age_err_parm(1)))) / (1.0-mfexp(-age_err_parm(4)*
(r_ages(nages)-age_err_parm(1))));
}
else
{
age_err(Use_AgeKeyZero,1)(AgeKey_StartAge,nages)=0.5 + r_ages(AgeKey_StartAge,nages) +
age_err_parm(2)+(age_err_parm(3)-age_err_parm(2))*
(r_ages(AgeKey_StartAge,nages)-age_err_parm(1))/(r_ages(nages)-age_err_parm(1));
}
// SS_Label_45.4 calc ageing variance
if(AgeKey_Linear2==0)
{
age_err(Use_AgeKeyZero,2)(AgeKey_StartAge,nages)=age_err_parm(5)+(age_err_parm(6)-age_err_parm(5))*
(1.0-mfexp(-age_err_parm(7)*(r_ages(AgeKey_StartAge,nages)-age_err_parm(1)))) /
(1.0-mfexp(-age_err_parm(7)*(r_ages(nages)-age_err_parm(1))));
}
else
{
age_err(Use_AgeKeyZero,2)(AgeKey_StartAge,nages)=age_err_parm(5)+(age_err_parm(6)-age_err_parm(5))*
(r_ages(AgeKey_StartAge,nages)-age_err_parm(1))/(r_ages(nages)-age_err_parm(1));
}
}
Survival Based SRR Code
Code for the survival based recruitment is shown below:
// SS_Label_43.3.7 survival based
case 7: // survival based, so constrained such that recruits cannot exceed fecundity
{
198
SRZ_0=log(1.0/(SSB_virgin_adj/Recr_virgin_adj));
SRZ_max=SRZ_0+SR_parm_work(2)*(0.0-SRZ_0);
SRZ_surv=mfexp((1.-pow((SSB_curr_adj/SSB_virgin_adj),SR_parm_work(3)) )*(SRZ_max-SRZ_0)+SRZ_0); // survival
NewRecruits=SSB_curr_adj*SRZ_surv;
exp_rec(y,1)=NewRecruits; // expected arithmetic mean recruitment
// SS_Label_43.3.7.1 Do variation in recruitment by adjusting survival
if(recdev_cycle>0)
{
gg=y - (styr+(int((y-styr)/recdev_cycle))*recdev_cycle)+1;
SRZ_surv*=mfexp(recdev_cycle_parm(gg));
}
exp_rec(y,2)=SSB_curr_adj*SRZ_surv;
SRZ_surv*=mfexp(-biasadj(y)*half_sigmaRsq); // bias adjustment
exp_rec(y,3)=SSB_curr_adj*SRZ_surv;
if(y <=recdev_end)
{
if(recdev_doit(y)>0) SRZ_surv*=mfexp(recdev(y)); // recruitment deviation
}
else if(Do_Forecast>0)
{
SRZ_surv *= mfexp(Fcast_recruitments(y));
}
join=1./(1.+mfexp(100*(SRZ_surv-1.)));
SRZ_surv=SRZ_surv*join + (1.-join)*1.0;
NewRecruits=SSB_curr_adj*SRZ_surv;
exp_rec(y,4) = NewRecruits;
break;
}
199
Random Walk Selectivity: Pattern 17
Code for selectivity pattern 17, random walk shown below:
// SS_Label_Info_22.7.17 #age selectivity: each age has parameter as random walk
// #41 each age has parameter as random walk scaled by average of values at low age through high age
// transformation as selex=exp(parm); some special codes */
case 41:
scaling_offset = 2;
case 17: //
{
lastsel=0.0; // value is the change in log(selex); this is the reference value for age 0
tempvec_a=-999.;
tempvec_a(0)=0.0; // so do not try to estimate the first value
int lastage;
if(seltype(f,4)==0)
{lastage=nages;}
else
{lastage=abs(seltype(f,4));}
for (a=1;a<=lastage;a++)
{
// with use of -999, lastsel stays constant until changed, so could create a linear change in ln(selex)
// use of (a+1) is because the first element, sp(1), is for age zero
if(sp(a+1+scaling_offset)>-999.) {lastsel=sp(a+1+scaling_offset);}
tempvec_a(a)=tempvec_a(a-1)+lastsel; // cumulative log(selex)
}
if (scaling_offset == 0)
{
temp=max(tempvec_a); // find max so at least one age will have selex=1.
}
else
{
int low_bin = int(value(sp(1)));
int high_bin = int(value(sp(2)));
if (low_bin < 0)
{
low_bin = 0;
N_warn++; warning<<" selex pattern 41; value for low bin is less than 0, so set to 0 "<<endl;
}
if (high_bin > nages)
{
high_bin = nages;
N_warn++; warning<<" selex pattern 41; value for high bin is greater than "<<nages<<", so set to "<<nages<<" "<<endl;
}
if (high_bin < low_bin) high_bin = low_bin;
if (low_bin > high_bin) low_bin = high_bin;
sp(1) = low_bin;
sp(2) = high_bin;
temp=mean(tempvec_a(low_bin,high_bin));
}
sel_a(y,fs,1)=mfexp(tempvec_a-temp);
a=0;
while(sp(a+1+scaling_offset)==-1000) // reset range of young ages to selex=0.0
{
sel_a(y,fs,1,a)=0.0;
a++;
}
scaling_offset = 0; // reset scaling offset
if(lastage<nages)
{
for (a=lastage+1;a<=nages;a++)
200
{
if(seltype(f,4)>0)
{sel_a(y,fs,1,a)=sel_a(y,fs,1,a-1);}
else
{sel_a(y,fs,1,a)=0.0;}
}
}
break;
}
201
Cubic Spline Selectivity
Code for cubic spline selectivity, option 42, shown below:
// SS_Label_Info_22.7.27 #age selectivity: cubic spline
// #42 cubic spline scaled by average of values at low age through high age
case 42:
scaling_offset = 2;
case 27:
{
k=seltype(f,4); // n points to include in cubic spline
for (i=1;i<=k;i++)
{
splineX(i)=value(sp(i+3+scaling_offset)); // "value" required to avoid error, but values should be always fixed anyway
splineY(i)=sp(i+3+k+scaling_offset);
}
z=nages;
while(r_ages(z)>splineX(k)) {z--;}
j2=z+1; // first age beyond last node
vcubic_spline_function splinefn=vcubic_spline_function(splineX(1,k),splineY(1,k),sp(2+scaling_offset),sp(3+scaling_offset));
tempvec_a= splinefn(r_ages); // interpolate selectivity at each age
if (scaling_offset == 0)
{
temp=max(tempvec_a(0,j2));
}
else
{
int low_bin = int(value(sp(1)));
int high_bin = int(value(sp(2)));
if (low_bin < 0)
{
low_bin = 0;
N_warn++; warning<<" selex pattern 42; value for low bin is less than 0, so set to 0 "<<endl;
}
if (high_bin > nages)
{
high_bin = nages;
N_warn++; warning<<" selex pattern 42; value for high bin is greater than "<<nages<<", so set to "<<nages<<" "<<endl;
}
if (high_bin < low_bin) high_bin = low_bin;
if (low_bin > high_bin) low_bin = high_bin;
sp(1) = low_bin;
sp(2) = high_bin;
temp=mean(tempvec_a(low_bin,high_bin));
scaling_offset = 0; // reset scaling offset
}
tempvec_a-=temp; // rescale to get max of 0.0
tempvec_a(j2+1,nages) = tempvec_a(j2); // set constant above last node
sel_a(y,fs,1)=mfexp(tempvec_a);
break;
}
202
Deviation Link
Code for alternative deviation links shown below:
case 1 (multiplicative):
{
for (j=timevary_setup(10);j<=timevary_setup(11);j++)
{
parm_timevary(tvary,j)*=mfexp(parm_dev(k,j)*parm_dev_stddev(k));
}
break;
}
case 2 (additive):
{
for (j=timevary_setup(10);j<=timevary_setup(11);j++)
{
parm_timevary(tvary,j)+=parm_dev(k,j)*parm_dev_stddev(k);
}
break;
}
case 3 (random walk):
{
parm_dev_rwalk(k,timevary_setup(10))=parm_dev(k,timevary_setup(10))*parm_dev_stddev(k);
parm_timevary(tvary,timevary_setup(10))+=parm_dev_rwalk(k,timevary_setup(10));
for (j=timevary_setup(10)+1;j<=timevary_setup(11);j++)
{
parm_dev_rwalk(k,j)=parm_dev_rwalk(k,j-1)+parm_dev(k,j)*parm_dev_stddev(k);
parm_timevary(tvary,j)+=parm_dev_rwalk(k,j);
}
break;
}
case 4 (mean reverting random walk)
{
parm_dev_rwalk(k,timevary_setup(10))=parm_dev(k,timevary_setup(10))*parm_dev_stddev(k);
parm_timevary(tvary,timevary_setup(10))+=parm_dev_rwalk(k,timevary_setup(10));
for (j=timevary_setup(10)+1;j<=timevary_setup(11);j++)
{
// =(1-rho)*mean + rho*prevval + dev // where mean = 0.0
parm_dev_rwalk(k,j)=parm_dev_rho(k)*parm_dev_rwalk(k,j-1)+parm_dev(k,j)*parm_dev_stddev(k);
parm_timevary(tvary,j)+=parm_dev_rwalk(k,j);
}
break;
}
203

Appendix E: In Process and Wish List Items for
Future Versions
Category Item Description
In process Environmental survey of a
deviation vector
Environmental survey data can be related to f(a deviation vector), but needs more Q_link
functions.
Retrospective Make blocks work when doing retrospective analyses.
Wishlist Tag-recapture major revamp
Area specific spawner-recruitment
Add automatic set-up for size selectivity option 6 and age selectivity option 17 given the data
(comparable to the current capacity for cubic spline selectivity).
More error checking on read of empirical weight-at-age and composition data.
New version of age-specific K needed because current version is inefficient.
Mean size in plus group uses a fixed erosion factor of 0.2; should be context specific.
Consider Tim Miller’s state space model approach.
Add other options to CV growth patter for log(SD).
Add measure of auto-correlation in composition and in survey residuals.
Add SD report for survey expected values.
Add calculation of Francis composition weighting method.
Year-specific MSY.
204
Appendix F: Example Model Files
starter.ss
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
#_user_support_available_at:NMFS.Stock.Synthesis@noaa.gov
#_user_info_available_at:https://vlab.ncep.noaa.gov/group/stock-synthesis
#C starter comment here
simple.dat
simple.ctl
0 # 0=use init values in control file; 1=use ss.par
1 # run display detail (0,1,2)
1 # detailed age-structured reports in REPORT.SSO (0=low,1=high,2=low for data-limited)
0 # write detailed checkup.sso file (0,1)
4 # write parm values to ParmTrace.sso (0=no,1=good,active; 2=good,all; 3=every_iter,all_parms; 4=every,active)
1 # write to cumreport.sso (0=no,1=like×eries; 2=add survey fits)
1 # Include prior_like for non-estimated parameters (0,1)
1 # Use Soft Boundaries to aid convergence (0,1) (recommended)
3 # Number of datafiles to produce: 1st is input, 2nd is estimates, 3rd and higher are bootstrap
10 # Turn off estimation for parameters entering after this phase
0 # MCeval burn interval
1 # MCeval thin interval
0 # jitter initial parm value by this fraction
1969 # min yr for sdreport outputs (-1 for styr)
2011 # max yr for sdreport outputs (-1 for endyr; -2 for endyr+Nforecastyrs
0 # N individual STD years
#vector of year values
0.0001 # final convergence criteria (e.g. 1.0e-04)
0 # retrospective year relative to end year (e.g. -4)
1 # min age for calc of summary biomass
1 # Depletion basis: denom is: 0=skip; 1=rel X*B0; 2=rel X*Bmsy; 3=rel X*B_styr
0.4 # Fraction (X) for Depletion denominator (e.g. 0.4)
1 # SPR_report_basis: 0=skip; 1=(1-SPR)/(1-SPR_tgt); 2=(1-SPR)/(1-SPR_MSY); 3=(1-SPR)/(1-SPR_Btarget); 4=rawSPR
4 # F_report_units: 0=skip; 1=exploitation(Bio); 2=exploitation(Num); 3=sum(Frates); 4=true F for range of ages
20 23 #_min and max age over which average F will be calculated
1 # F_report_basis: 0=raw_F_report; 1=F/Fspr; 2=F/Fmsy ; 3=F/Fbtgt
0 # MCMC output detail (0=default; 1=obj func components; 2=expanded; 3=make output subdir for each MCMC vector)
0 # ALK tolerance (example 0.0001)
3.30 # check value for end of file and for version control
205
forecast.ss
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
#C generic forecast file
# for all year entries except rebuilder; enter either: actual year, -999 for styr, 0 for endyr, neg number for rel. endyr
1 # Benchmarks: 0=skip; 1=calc F_spr,F_btgt,F_msy; 2=calc F_spr,F0.1,F_msy
2 # MSY: 1= set to F(SPR); 2=calc F(MSY); 3=set to F(Btgt) or F0.1; 4=set to F(endyr)
0.4 # SPR target (e.g. 0.40)
0.342 # Biomass target (e.g. 0.40)
#_Bmark_years: beg_bio, end_bio, beg_selex, end_selex, beg_relF, end_relF, beg_recr_dist, end_recr_dist, beg_SRparm, end_SRparm (enter actual year, or values of 0 or -integer to be rel. endyr)
2001 2001 2001 2001 2001 2001 1971 2001 1971 2001
1 #Bmark_relF_Basis: 1 = use year range; 2 = set relF same as forecast below
#
1 # Forecast: 0=none; 1=F(SPR); 2=F(MSY) 3=F(Btgt) or F0.1; 4=Ave F (uses first-last relF yrs); 5=input annual F scalar
10 # N forecast years
0.2 # F scalar (only used for Do_Forecast==5)
#_Fcast_years: beg_selex, end_selex, beg_relF, end_relF, beg_recruits, end_recruits (enter actual year, or values of 0 or -integer to be rel. endyr)
0 0 -10 0 -999 0
0 # Forecast selectivity (0=fcast selex is mean from year range; 1=fcast selectivity from annual time-vary parms)
1 # Control rule method (1=catch=f(SSB) west coast; 2=F=f(SSB) )
0.4 # Control rule Biomass level for constant F (as frac of Bzero, e.g. 0.40); (Must be > the no F level below)
0.1 # Control rule Biomass level for no F (as frac of Bzero, e.g. 0.10)
0.75 # Control rule target as fraction of Flimit (e.g. 0.75)
3 #_N forecast loops (1=OFL only; 2=ABC; 3=get F from forecast ABC catch with allocations applied)
3 #_First forecast loop with stochastic recruitment
0 #_Forecast recruitment: 0= spawn_recr; 1=value*spawn_recr_fxn; 2=value*VirginRecr; 3=recent mean)
1 # value is ignored
0 #_Forecast loop control #5 (reserved for future bells&whistles)
2010 #FirstYear for caps and allocations (should be after years with fixed inputs)
0 # stddev of log(realized catch/target catch) in forecast (set value>0.0 to cause active impl_error)
0 # Do West Coast gfish rebuilder output (0/1)
1999 # Rebuilder: first year catch could have been set to zero (Ydecl)(-1 to set to 1999)
2002 # Rebuilder: year for current age structure (Yinit) (-1 to set to endyear+1)
1 # fleet relative F: 1=use first-last alloc year; 2=read seas, fleet, alloc list below
# Note that fleet allocation is used directly as average F if Do_Forecast=4
2 # basis for fcast catch tuning and for fcast catch caps and allocation (2=deadbio; 3=retainbio; 5=deadnum; 6=retainnum)
# Conditional input if relative F choice = 2
# enter list of: season, fleet, relF; if used, terminate with season=-9999
# 1 1 1
# enter list of: fleet number, max annual catch for fleets with a max; terminate with fleet=-9999
-9999 -1
# enter list of area ID and max annual catch; terminate with area=-9999
-9999 -1
# enter list of fleet number and allocation group assignment, if any; terminate with fleet=-9999
-9999 -1
#_if N allocation groups >0, list year, allocation fraction for each group
# list sequentially because read values fill to end of N forecast
# terminate with -9999 in year field
206
# no allocation groups
2 # basis for input Fcast catch: -1=read basis with each obs; 2=dead catch; 3=retained catch; 99=input Hrate(F)
#enter list of Fcast catches; terminate with line having year=-9999
#_Yr Seas Fleet Catch(or_F)
-9999 1 1 0
#
999 # verify end of input
data.ss
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
#_user_support_available_at:NMFS.Stock.Synthesis@noaa.gov
#_user_info_available_at:https://vlab.ncep.noaa.gov/group/stock-synthesis
#_Start_time: Fri Feb 2 10:54:21 2018
#_Number_of_datafiles: 3
#C data file for simple example
#_observed data:
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
1971 #_StartYr
2001 #_EndYr
1 #_Nseas
12 #_months/season
2 #_Nsubseasons (even number, minimum is 2)
1 #_spawn_month
2 #_Ngenders
40 #_Nages=accumulator age
1 #_Nareas
3 #_Nfleets (including surveys)
#_fleet_type: 1=catch fleet; 2=bycatch only fleet; 3=survey; 4=ignore
#_survey_timing: -1=for use of catch-at-age to override the month value associated with a datum
#_fleet_area: area the fleet/survey operates in
#_units of catch: 1=bio; 2=num (ignored for surveys; their units read later)
#_catch_mult: 0=no; 1=yes
#_rows are fleets
#_fleet_type timing area units need_catch_mult fleetname
1 0.5 1 1 0 FISHERY1 # 1
3 0.5 1 2 0 SURVEY1 # 2
3 0.5 1 2 0 SURVEY2 # 3
#Bycatch_fleet_input_goes_next
#a: fleet index
#b: 1=include dead bycatch in total dead catch for F0.1 and MSY optimizations and forecast ABC; 2=omit from total catch for these purposes (but still include the mortality)
#c: 1=Fmult scales with other fleets; 2=bycatch F constant at input value; 3=bycatch F from range of years
#d: F or first year of range
#e: last year of range
#f: not used
207
#abcdef
#_Catch data: yr, seas, fleet, catch, catch_se
#_catch_se: standard error of log(catch)
#_NOTE: catch data is ignored for survey fleets
-999 1 1 0 0.01
1971 1 1 0 0.01
1972 1 1 200 0.01
1973 1 1 1000 0.01
1974 1 1 1000 0.01
1975 1 1 2000 0.01
1976 1 1 3000 0.01
1977 1 1 4000 0.01
1978 1 1 5000 0.01
1979 1 1 6000 0.01
1980 1 1 8000 0.01
1981 1 1 10000 0.01
1982 1 1 10000 0.01
1983 1 1 10000 0.01
1984 1 1 10000 0.01
1985 1 1 10000 0.01
1986 1 1 10000 0.01
1987 1 1 10000 0.01
1988 1 1 9000 0.01
1989 1 1 8000 0.01
1990 1 1 7000 0.01
1991 1 1 6000 0.01
1992 1 1 4000 0.01
1993 1 1 4000 0.01
1994 1 1 4000 0.01
1995 1 1 4000 0.01
1996 1 1 4000 0.01
1997 1 1 3000 0.01
1998 1 1 3000 0.01
1999 1 1 3000 0.01
2000 1 1 3000 0.01
2001 1 1 3000 0.01
-9999 0 0 0 0
#
#_CPUE_and_surveyabundance_observations
#_Units: 0=numbers; 1=biomass; 2=F; >=30 for special types
#_Errtype: -1=normal; 0=lognormal; >0=T
#_SD_Report: 0=no sdreport; 1=enable sdreport
#_Fleet Units Errtype SD_Report
1 1 0 0 # FISHERY1
2 1 0 0 # SURVEY1
3 0 0 0 # SURVEY2
#_yr month fleet obs stderr
1977 7 2 339689 0.3 #_ SURVEY1
1980 7 2 193353 0.3 #_ SURVEY1
208
1983 7 2 151984 0.3 #_ SURVEY1
1986 7 2 55221.8 0.3 #_ SURVEY1
1989 7 2 59232.3 0.3 #_ SURVEY1
1992 7 2 31137.5 0.3 #_ SURVEY1
1995 7 2 35845.4 0.3 #_ SURVEY1
1998 7 2 27492.6 0.3 #_ SURVEY1
2001 7 2 37338.3 0.3 #_ SURVEY1
1990 7 3 5.19333 0.7 #_ SURVEY2
1991 7 3 1.1784 0.7 #_ SURVEY2
1992 7 3 5.94383 0.7 #_ SURVEY2
1993 7 3 0.770106 0.7 #_ SURVEY2
1994 7 3 16.318 0.7 #_ SURVEY2
1995 7 3 1.36339 0.7 #_ SURVEY2
1996 7 3 4.76482 0.7 #_ SURVEY2
1997 7 3 51.0707 0.7 #_ SURVEY2
1998 7 3 1.36095 0.7 #_ SURVEY2
1999 7 3 0.862531 0.7 #_ SURVEY2
2000 7 3 5.97125 0.7 #_ SURVEY2
2001 7 3 1.69379 0.7 #_ SURVEY2
-9999 1 1 1 1 # terminator for survey observations
#
0 #_N_fleets_with_discard
#_discard_units (1=same_as_catchunits(bio/num); 2=fraction; 3=numbers)
#_discard_errtype: >0 for DF of T-dist(read CV below); 0 for normal with CV; -1 for normal with se; -2 for lognormal; -3 for trunc normal with CV
# note, only have units and errtype for fleets with discard
#_Fleet units errtype
# -9999 0 0 0.0 0.0 # terminator for discard data
#
0 #_use meanbodysize_data (0/1)
#_COND_0 #_DF_for_meanbodysize_T-distribution_like
# note: use positive partition value for mean body wt, negative partition for mean body length
#_yr month fleet part obs stderr
# -9999 0 0 0 0 0 # terminator for mean body size data
#
# set up population length bin structure (note - irrelevant if not using size data and using empirical wtatage
2 # length bin method: 1=use databins; 2=generate from binwidth,min,max below; 3=read vector
2 # binwidth for population size comp
10 # minimum size in the population (lower edge of first bin and size at age 0.00)
94 # maximum size in the population (lower edge of last bin)
1 # use length composition data (0/1)
#_mintailcomp: upper and lower distribution for females and males separately are accumulated until exceeding this level.
#_addtocomp: after accumulation of tails; this value added to all bins
#_males and females treated as combined gender below this bin number
#_compressbins: accumulate upper tail by this number of bins; acts simultaneous with mintailcomp; set=0 for no forced accumulation
#_Comp_Error: 0=multinomial, 1=dirichlet
#_Comp_Error2: parm number for dirichlet
#_minsamplesize: minimum sample size; set to 1 to match 3.24, minimum value is 0.001
#_mintailcomp addtocomp combM+F CompressBins CompError ParmSelect minsamplesize
0 1e-07 0 0 0 0 0.001 #_fleet:1_FISHERY1
209
0 1e-07 0 0 0 0 0.001 #_fleet:2_SURVEY1
0 1e-07 0 0 0 0 0.001 #_fleet:3_SURVEY2
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
25 #_N_LengthBins; then enter lower edge of each length bin
26 28 30 32 34 36 38 40 42 44 46 48 50 52 54 56 58 60 62 64 68 72 76 80 90
#_yr month fleet sex part Nsamp datavector(female-male)
197171301250000000004112415623118450000000000101303424591783800
19727130125000000000301211627410104530000000001324131447381141000
1973713012500000000000073456310126109000000000000030130726785530
1974713012500000000022011145388104700000000012040015664615115030
19757130125000000021211302562359101000000000000422123514513116400
1976713012500000002102203233371814422000000010000012466571264300
19777130125000000010202240267511785400000021301332014537795300
19787130125000000511101318446598365000000002112112241411396400
19797130125000000000035215055274755000000000221327244581086410
198071301250000000400102432323161112420000000001411235263110114200
198171301250000001000312245273139840000000211122331612175106700
1982713012500000000521323825446101100000000001030215618551052500
19837130125000000000071154226281386000000000004103304954786600
198471301250000001004303125247119680000000003311333224481145200
198571301250000000011225033511489324000000001012038343841374100
19867130125000310120420042835115661000000022012134234655646500
19877130125000011111021642763511954000000000210524344424765200
1988713012500000201421122174569921000000021131363304535998000
19897130125000001021332144342395112000000003621304332577933400
1990713012500000002222222944668441000000011223828663234651200
19917130125000000030335543301610440000000111134653566664733000
19927130125000022011133276442563600000000005313535834631341000
199371301250000001222224510573212760000000000311326484642431100
19947130125000000000414344946785320000000002021144105863561300
199571301250001001111225841155487000000001001133126344831243000
199671301250001021024332366334116600000000012033105467451034100
19977130125000200220031646294591200000000031053241164166564000
199871301250000312222313620745123120000004110220114625461374100
19997130125000010113012283473565700000000073423252113515742000
200071301250000010012431644334511000000000244336341835141115500
20017130125000021011027694256476400000000201023253833521063000
19777230125000030022312505653384100000000006332252338116583200
19807230125000011132213612513383341000001123444441115351475200
19837230125000023352452325565331800000002212242623524416100000
1986723012500002114623111555337732000001213215025673523744000
1989723012500000583351241224323320000002235258873243631800000
1992723012500000566532566551313400000000024365366254313123000
1995723012500002004755562565603410000000230121534953342543000
19987230125000311234646531211152200000001054237214453231862000
2001723012500000235759295441122800000000214656434451321320000
-99990000000000000000000000000000000000000000000000000000000
#
210
17 #_N_age_bins
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 20 25
2 #_N_ageerror_definitions
0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5 16.5 17.5 18.5 19.5 20.5 21.5 22.5 23.5 24.5 25.5 26.5 27.5 28.5 29.5 30.5 31.5 32.5 33.5 34.5 35.5 36.5 37.5 38.5 39.5 40.5
0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001
0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5 16.5 17.5 18.5 19.5 20.5 21.5 22.5 23.5 24.5 25.5 26.5 27.5 28.5 29.5 30.5 31.5 32.5 33.5 34.5 35.5 36.5 37.5 38.5 39.5 40.5
0.5 0.65 0.67 0.7 0.73 0.76 0.8 0.84 0.88 0.92 0.97 1.03 1.09 1.16 1.23 1.32 1.41 1.51 1.62 1.75 1.89 2.05 2.23 2.45 2.71 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
#_mintailcomp: upper and lower distribution for females and males separately are accumulated until exceeding this level.
#_addtocomp: after accumulation of tails; this value added to all bins
#_males and females treated as combined gender below this bin number
#_compressbins: accumulate upper tail by this number of bins; acts simultaneous with mintailcomp; set=0 for no forced accumulation
#_Comp_Error: 0=multinomial, 1=dirichlet
#_Comp_Error2: parm number for dirichlet
#_minsamplesize: minimum sample size; set to 1 to match 3.24, minimum value is 0.001
#_mintailcomp addtocomp combM+F CompressBins CompError ParmSelect minsamplesize
0 1e-07 1 0 0 0 0.001 #_fleet:1_FISHERY1
0 1e-07 1 0 0 0 0.001 #_fleet:2_SURVEY1
0 1e-07 1 0 0 0 0.001 #_fleet:3_SURVEY2
1 #_Lbin_method_for_Age_Data: 1=poplenbins; 2=datalenbins; 3=lengths
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
#_yr month fleet sex part ageerr Lbin_lo Lbin_hi Nsamp datavector(female-male)
1971713021-17500003114210122132300421121221212658
1972713021-1752111031225312298300123130513021232
1973713021-1750010112331152274300041351231320536
1974713021-1750020142224111266600412212001211657
1975713021-175001231112122231034000010123210000936
1976713021-17500102221312311713000074321244008100
1977713021-1750000710024223172300214233422201834
1978713021-1750032110202431094600225102324204433
1979713021-1752015212333221037000201023251312691
1980713021-17501020112232110780000321112242221138
1981713021-1750403722221122144600322113220122533
1982713021-1750211332112210263900003501411121890
1983713021-1750006122211450062700313510113033534
1984713021-1750003403631402072300315423512120125
1985713021-1750005124502432334500012324202311722
1986713021-1750221374322222042200004441234001570
1987713021-1750313123423322132000715142432310214
1988713021-1751050233343310335001332214321240530
1989713021-1750311437151141017000534115315210220
1990713021-1750073730130111134001084332451510120
1991713021-1750041742321011333003425441330420410
1992713021-17500745104303102021100513833120130110
1993713021-1750074375721010400000334370042111500
1994713021-1750036444945100000300090722340320000
1995713021-1753120852625021400000025323561011311
1996713021-1750011543723233151002505412342301200
1997713021-1750535024345113220000031655234123000
211
1998713021-1755314123432020150000464272116300210
1999713021-1752233633383330110001333540424010100
2000713021-17502194422431010500008113122112102300
2001713021-1750116811052220340000534633143112300
1977723021-17521210433211011470022710101241227100
1980723021-1753346520230322221402353121121110314
1983723021-1753432300700311056002241234320112712
1986723021-1753025355313211130200236613311112230
1989723021-1757373210321211500004861235112040000
1992723021-17525340505200010300045510862120010100
1995723021-17505232354211200300023511265121200200
1998723021-1759443111133121700006535133232010000
2001723021-175404115342200000200024711520222000100
-9999 000000000000000000000000000000000000000000
#
1 #_Use_MeanSize-at-Age_obs (0/1)
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
# ageerr codes: positive means mean length-at-age; negative means mean bodywt_at_age
#_yr month fleet sex part ageerr ignore datavector(female-male)
# samplesize(female-male)
1971 7 1 3 0 1 2 29.8931 40.6872 44.7411 50.027 52.5794 56.1489 57.1033 61.1728 61.7417 63.368 64.4088 65.6889 67.616 68.5972 69.9177 71.0443 72.3609 32.8188 39.5964 43.988 50.1693 53.1729 54.9822 55.3463 60.3509 60.7439 62.3432 64.3224 65.1032 64.1965 66.7452 67.5154 70.8749 71.2768 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1995 7 1 3 0 1 2 32.8974 38.2709 43.8878 49.2745 53.5343 55.1978 57.4389 62.0368 62.1445 62.9579 65.0857 65.6433 66.082 65.6117 67.0784 69.3493 72.2966 32.6552 40.5546 44.6292 50.4063 52.0796 56.1529 56.9004 60.218 61.5894 63.6613 64.0222 63.4926 65.8115 69.5357 68.2448 66.881 71.5122 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1971 7 2 3 0 1 2 34.1574 38.8017 43.122 47.2042 49.0502 51.6446 56.3201 56.3038 60.5509 60.2537 59.8042 62.9309 66.842 67.8089 71.1612 70.7693 74.5593 35.3811 40.7375 44.5192 47.6261 52.5298 53.5552 54.9851 58.9231 58.9932 61.8625 64.0366 62.7507 63.9754 64.5102 66.9779 67.7361 69.1298 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1995 7 2 3 0 1 2 34.6022 38.3176 42.9052 48.2752 50.6189 53.476 56.7806 59.4127 60.5964 60.5537 65.3608 64.7263 67.4315 67.1405 68.9908 71.9886 74.1594 35.169 40.2404 43.8878 47.3519 49.9906 52.2207 54.9035 58.6058 60.0957 62.4046 62.2298 62.1437 66.2116 65.7657 69.9544 70.6518 71.4371 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
-9999 00000000000000000000000000000000000000000000000000000000000000000000000000
#
0 #_N_environ_variables
#Yr Variable Value
#
0 # N sizefreq methods to read
#
0 # do tags (0/1)
#
0 # morphcomp data(0/1)
# Nobs, Nmorphs, mincomp
# yr, seas, type, partition, Nsamp, datavector_by_Nmorphs
#
0 # Do dataread for selectivity priors(0/1)
# Yr, Seas, Fleet, Age/Size, Bin, selex_prior, prior_sd
# feature not yet implemented
#
999
#_expected values with no error added
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
1971 #_StartYr
2001 #_EndYr
1 #_Nseas
212
12 #_months/season
2 #_Nsubseasons (even number, minimum is 2)
1 #_spawn_month
2 #_Ngenders
40 #_Nages=accumulator age
1 #_Nareas
3 #_Nfleets (including surveys)
#_fleet_type: 1=catch fleet; 2=bycatch only fleet; 3=survey; 4=ignore
#_survey_timing: -1=for use of catch-at-age to override the month value associated with a datum
#_fleet_area: area the fleet/survey operates in
#_units of catch: 1=bio; 2=num (ignored for surveys; their units read later)
#_catch_mult: 0=no; 1=yes
#_rows are fleets
#_fleet_type timing area units need_catch_mult fleetname
1 0.5 1 1 0 FISHERY1 # 1
3 0.5 1 2 0 SURVEY1 # 2
3 0.5 1 2 0 SURVEY2 # 3
#Bycatch_fleet_input_goes_next
#a: fleet index
#b: 1=include dead bycatch in total dead catch for F0.1 and MSY optimizations and forecast ABC; 2=omit from total catch for these purposes (but still include the mortality)
#c: 1=Fmult scales with other fleets; 2=bycatch F constant at input value; 3=bycatch F from range of years
#d: F or first year of range
#e: last year of range
#f: not used
#abcdef
#_catch:_columns_are_year,season,fleet,catch,catch_se
#_Catch data: yr, seas, fleet, catch, catch_se
-999 1 1 0 0.01
1971 1 1 0 0.01
1972 1 1 200 0.01
1973 1 1 1000 0.01
1974 1 1 1000 0.01
1975 1 1 2000 0.01
1976 1 1 3000 0.01
1977 1 1 4000 0.01
1978 1 1 5000 0.01
1979 1 1 6000 0.01
1980 1 1 8000 0.01
1981 1 1 10000 0.01
1982 1 1 10000 0.01
1983 1 1 10000 0.01
1984 1 1 10000 0.01
1985 1 1 10000 0.01
1986 1 1 10000 0.01
1987 1 1 10000 0.01
1988 1 1 9000 0.01
1989 1 1 8000 0.01
1990 1 1 7000 0.01
1991 1 1 6000 0.01
213
1992 1 1 4000 0.01
1993 1 1 4000 0.01
1994 1 1 4000 0.01
1995 1 1 4000 0.01
1996 1 1 4000 0.01
1997 1 1 3000 0.01
1998 1 1 3000 0.01
1999 1 1 3000 0.01
2000 1 1 3000 0.01
2001 1 1 3000 0.01
-9999 0 0 0 0
#
#
#_CPUE_and_surveyabundance_observations
#_Units: 0=numbers; 1=biomass; 2=F; >=30 for special types
#_Errtype: -1=normal; 0=lognormal; >0=T
#_SD_Report: 0=no sdreport; 1=enable sdreport
#_Fleet Units Errtype SD_Report
1 1 0 0 # FISHERY1
2 1 0 0 # SURVEY1
3 0 0 0 # SURVEY2
#_year month index obs err
1977 7 2 185035 0.3 #_orig_obs: 339689 SURVEY1
1980 7 2 162288 0.3 #_orig_obs: 193353 SURVEY1
1983 7 2 124799 0.3 #_orig_obs: 151984 SURVEY1
1986 7 2 86178.3 0.3 #_orig_obs: 55221.8 SURVEY1
1989 7 2 58379.5 0.3 #_orig_obs: 59232.3 SURVEY1
1992 7 2 45851.3 0.3 #_orig_obs: 31137.5 SURVEY1
1995 7 2 38866.3 0.3 #_orig_obs: 35845.4 SURVEY1
1998 7 2 33173.2 0.3 #_orig_obs: 27492.6 SURVEY1
2001 7 2 33563.3 0.3 #_orig_obs: 37338.3 SURVEY1
1990 7 3 7.9626 0.7 #_orig_obs: 5.19333 SURVEY2
1991 7 3 2.07335 0.7 #_orig_obs: 1.1784 SURVEY2
1992 7 3 2.97781 0.7 #_orig_obs: 5.94383 SURVEY2
1993 7 3 1.6979 0.7 #_orig_obs: 0.770106 SURVEY2
1994 7 3 5.55711 0.7 #_orig_obs: 16.318 SURVEY2
1995 7 3 2.07302 0.7 #_orig_obs: 1.36339 SURVEY2
1996 7 3 5.81354 0.7 #_orig_obs: 4.76482 SURVEY2
1997 7 3 10.7034 0.7 #_orig_obs: 51.0707 SURVEY2
1998 7 3 1.98023 0.7 #_orig_obs: 1.36095 SURVEY2
1999 7 3 1.71246 0.7 #_orig_obs: 0.862531 SURVEY2
2000 7 3 3.80169 0.7 #_orig_obs: 5.97125 SURVEY2
2001 7 3 2.3342 0.7 #_orig_obs: 1.69379 SURVEY2
-9999 1 1 1 1 # terminator for survey observations
#
0 #_N_fleets_with_discard
#_discard_units (1=same_as_catchunits(bio/num); 2=fraction; 3=numbers)
#_discard_errtype: >0 for DF of T-dist(read CV below); 0 for normal with CV; -1 for normal with se; -2 for lognormal; -3 for trunc normal with CV
# note, only have units and errtype for fleets with discard
214
#_Fleet units errtype
# -9999 0 0 0.0 0.0 # terminator for discard data
#
0 #_use meanbodysize_data (0/1)
#_COND_0 #_DF_for_meanbodysize_T-distribution_like
# note: use positive partition value for mean body wt, negative partition for mean body length
#_yr month fleet part obs stderr
# -9999 0 0 0 0 0 # terminator for mean body size data
#
# set up population length bin structure (note - irrelevant if not using size data and using empirical wtatage
2 # length bin method: 1=use databins; 2=generate from binwidth,min,max below; 3=read vector
2 # binwidth for population size comp
10 # minimum size in the population (lower edge of first bin and size at age 0.00)
94 # maximum size in the population (lower edge of last bin)
1 # use length composition data (0/1)
#_mintailcomp: upper and lower distribution for females and males separately are accumulated until exceeding this level.
#_addtocomp: after accumulation of tails; this value added to all bins
#_males and females treated as combined gender below this bin number
#_compressbins: accumulate upper tail by this number of bins; acts simultaneous with mintailcomp; set=0 for no forced accumulation
#_Comp_Error: 0=multinomial, 1=dirichlet
#_Comp_Error2: parm number for dirichlet
#_minsamplesize: minimum sample size; set to 1 to match 3.24, minimum value is 0.001
#_mintailcomp addtocomp combM+F CompressBins CompError ParmSelect minsamplesize
0 1e-07 0 0 0 0 0.001 #_fleet:1_FISHERY1
0 1e-07 0 0 0 0 0.001 #_fleet:2_SURVEY1
0 1e-07 0 0 0 0 0.001 #_fleet:3_SURVEY2
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
25 #_N_LengthBins
26 28 30 32 34 36 38 40 42 44 46 48 50 52 54 56 58 60 62 64 68 72 76 80 90
#_yr month fleet sex part Nsamp datavector(female-male)
1971 7 1 3 0 125 0 0 0 0 0 0 0 0 0 3.34814 1.15397 1.48309 1.87166 2.31601 2.80561 3.32197 3.83722 4.31242 4.69843 9.9337 9.25965 7.05824 7.32944 0 0 0 0 0 0 0 0 0 0 2.43891 0.896971 1.1836 1.54052 1.97211 2.4785 3.0501 3.66233 4.27121 4.81209 5.2061 10.6473 9.13652 6.13614 4.83803 0 0
1972 7 1 3 0 125 0 0 0 0 0 0 0 0 0 3.30584 1.15533 1.48473 1.8736 2.31826 2.80815 3.32479 3.84026 4.31564 4.70176 9.94029 9.26543 7.06248 4.28859 3.04513 0 0 0 0 0 0 0 0 0 2.39332 0.89806 1.18498 1.54221 1.97415 2.48089 3.05284 3.6654 4.27458 4.81569 5.20981 10.6545 9.14238 6.13996 4.84096 0 0
1973 7 1 3 0 125 0 0 0 0 0 0 0 0 0 0 0 0 7.76387 2.32478 2.81491 3.33151 3.84669 4.32157 4.70703 9.94858 9.27077 7.06544 7.33584 0 0 0 0 0 0 0 0 0 0 0 0 0 5.94964 1.98029 2.4877 3.06 3.67263 4.28161 4.82226 5.21575 10.664 9.14859 6.14334 3.16205 1.6811 0
1974 7 1 3 0 125 0 0 0 0 0 0 0 0 0 3.10018 1.15174 1.49144 1.88767 2.33691 2.8291 3.3464 3.86141 4.33553 4.71984 9.96983 9.28568 7.07455 7.34341 0 0 0 0 0 0 0 0 0 1.56019 0.63785 0.875966 1.1772 1.54635 1.98709 2.49957 3.07464 3.68841 4.29747 4.83751 5.22987 10.6875 9.16472 6.15255 3.16628 1.68314 0
1975 7 1 3 0 125 0 0 0 0 0 0 0 1.56742 0.610316 0.825134 1.10897 1.46133 1.875 2.33992 2.84306 3.36642 3.88371 4.35769 4.74048 10.0041 9.30955 7.08899 7.35528 0 0 0 0 0 0 0 0 0 0 0 2.98359 1.13045 1.5099 1.96849 2.49864 3.08688 3.70844 4.3209 4.86137 5.25239 10.7251 9.1903 6.16702 4.85918 0 0
1976 7 1 3 0 125 0 0 0 0 0 0 0 1.69087 0.601649 0.813398 1.08189 1.41842 1.82867 2.30516 2.82748 3.36837 3.89651 4.37436 4.75612 10.0241 9.31455 7.08609 4.29781 3.04883 0 0 0 0 0 0 0 1.23532 0.430718 0.601139 0.822434 1.10705 1.46947 1.92064 2.45902 3.06574 3.70627 4.33162 4.87758 5.26853 10.7473 9.19845 6.16769 4.85674 0 0
1977 7 1 3 0 125 0 0 0 0 0 0 0 1.98149 0.585955 0.787468 1.05544 1.39145 1.79511 2.26387 2.78647 3.33816 3.88186 4.37236 4.76002 10.0278 9.3026 7.06711 4.28241 3.03574 0 0 0 0 0 0 1.18691 0.351071 0.445308 0.590067 0.796228 1.07723 1.4398 1.8868 2.4183 3.02381 3.67314 4.31323 4.87242 5.27053 10.751 9.19064 6.15559 4.8426 0 0
1978 7 1 3 0 125 0 0 0 0 0 0 1.67363 0.625767 0.724506 0.860404 1.07132 1.37077 1.75642 2.21746 2.73584 3.28603 3.83293 4.33074 4.72602 9.96921 9.24076 7.009 4.24187 3.00385 0 0 0 0 0 0 0 0 2.27401 0.75635 0.89549 1.11074 1.42695 1.84878 2.36932 2.97023 3.61827 4.26111 4.82684 5.23241 10.688 9.13376 6.11042 4.80083 0 0
1979 7 1 3 0 125 0 0 0 0 0 0 0 0 0 0 5.53701 1.54817 1.835 2.21776 2.68938 3.21688 3.75454 4.24989 4.64631 9.81826 9.10294 6.89572 4.16726 2.94685 0 0 0 0 0 0 0 0 0 3.01978 1.15802 1.39298 1.64248 1.96044 2.38884 2.93 3.54731 4.17731 4.73946 5.14631 10.5293 9.0026 6.01792 3.08592 1.63533 0
1980 7 1 3 0 125 0 0 0 0 0 0 0 2.10473 0.838413 1.15359 1.50311 1.84208 2.15266 2.46113 2.81291 3.23115 3.69577 4.15204 4.5311 9.57695 8.88052 6.72058 4.05536 2.86261 0 0 0 0 0 0 0 0 0 2.89064 1.15143 1.52823 1.91438 2.2853 2.66025 3.08399 3.58115 4.12457 4.63826 5.02393 10.2784 8.79077 5.87315 4.60079 0 0
1981 7 1 3 0 125 0 0 0 0 0 0 1.6538 0.615708 0.814006 1.09088 1.45902 1.89383 2.34301 2.75687 3.12002 3.45553 3.79324 4.13243 4.43102 9.27274 8.56856 6.47471 6.64806 0 0 0 0 0 0 0 1.15675 0.463222 0.617275 0.818947 1.09767 1.47509 1.93732 2.43842 2.92808 3.38287 3.81302 4.23331 4.62454 4.92281 9.96849 8.4953 5.66948 4.43394 0 0
1982 7 1 3 0 125 0 0 0 0 0 0 0 0 3.25409 1.16001 1.48825 1.89301 2.36413 2.85971 3.32102 3.70688 4.01237 4.25318 4.43342 9.02176 8.20934 12.498 0 0 0 0 0 0 0 0 0 0 2.30815 0.904803 1.17896 1.52285 1.95032 2.45742 3.01037 3.55292 4.0349 4.43155 4.73277 4.91753 9.7124 8.15757 5.42165 4.23065 0 0
1983 7 1 3 0 125 0 0 0 0 0 0 0 0 0 0 6.08421 2.00466 2.45079 2.93052 3.41006 3.84025 4.17781 4.40274 4.51499 8.91198 7.88161 5.86043 5.96657 0 0 0 0 0 0 0 0 0 0 0 4.42928 1.64033 2.07202 2.55996 3.09322 3.64048 4.1503 4.56742 4.84967 4.97074 9.57436 7.84775 5.15994 4.00791 0 0
1984 7 1 3 0 125 0 0 0 0 0 0 1.4006 0.672662 0.949526 1.29195 1.69257 2.13665 2.6074 3.08567 3.54737 3.96176 4.29326 4.50999 4.59325 8.88769 7.62634 5.55881 5.59803 0 0 0 0 0 0 0 0 0 2.00382 0.938454 1.29339 1.71746 2.19748 2.71662 3.25514 3.7865 4.27422 4.67234 4.93288 5.01728 9.49893 7.5952 4.90974 3.77705 0 0
1985 7 1 3 0 125 0 0 0 0 0 0 0 0 3.00292 1.22483 1.67116 2.18163 2.72072 3.24881 3.72936 4.13261 4.43432 4.61407 4.65663 8.86849 7.41585 5.28036 3.08758 2.13519 0 0 0 0 0 0 0 0 2.08878 0.849989 1.21497 1.67913 2.22378 2.81413 3.4083 3.96427 4.44315 4.8081 5.02329 5.05777 9.42796 7.37378 4.67544 3.54262 0 0
1986 7 1 3 0 125 0 0 0 1.04524 0.338462 0.374853 0.45157 0.592879 0.811061 1.12772 1.55757 2.09054 2.68904 3.29466 3.84323 4.28252 4.58126 4.72686 4.71821 8.82608 7.19806 5.0043 4.83351 0 0 0 0 0 0 0 1.75517 0.45517 0.591322 0.807103 1.12389 1.56235 2.11959 2.76217 3.43147 4.05729 4.57546 4.93887 5.11802 5.09732 9.33833 7.14143 4.43857 3.29884 0 0
1987 7 1 3 0 125 0 0 0 0 1.73527 0.637043 0.753544 0.87393 1.00849 1.21104 1.53204 1.98589 2.55145 3.17506 3.77823 4.27928 4.61784 4.7668 4.7284 8.69845 6.91024 4.68513 4.40013 0 0 0 0 0 0 0 0 0 4.0085 1.04804 1.24892 1.56757 2.03135 2.6258 3.29923 3.96953 4.54389 4.94478 5.12635 5.07578 9.15663 6.84008 4.16123 3.02407 0 0
1988 7 1 3 0 125 0 0 0 0 0 2.47684 1.06644 1.32582 1.5464 1.74113 1.94601 2.20918 2.57545 3.04939 3.57731 4.06837 4.43224 4.60887 4.57968 8.35213 6.48518 4.28426 3.89865 0 0 0 0 0 0 0 2.38741 1.05897 1.34675 1.60015 1.82181 2.04731 2.32562 2.70614 3.20415 3.77276 4.31572 4.72715 4.92804 4.88432 8.74114 6.40076 6.50845 0 0 0
215
1989 7 1 3 0 125 0 0 0 0 0 2.17903 1.06477 1.47913 1.93178 2.3598 2.70534 2.95215 3.14212 3.34191 3.59197 3.8764 4.12956 4.27198 4.24867 7.73692 5.92407 3.82426 3.36562 0 0 0 0 0 0 0 0 3.1228 1.4549 1.93436 2.40714 2.80981 3.11199 3.3437 3.57168 3.84373 4.14909 4.41918 4.56356 4.51437 8.05497 5.8253 3.40042 2.34754 0 0
1990 7 1 3 0 125 0 0 0 0 0 0 0 4.39296 1.88743 2.4911 3.10798 3.62645 3.96201 4.10583 4.12011 4.08631 4.0479 3.99144 3.87086 6.95294 5.26655 3.34192 2.8483 0 0 0 0 0 0 0 1.98931 0.919147 1.33824 1.86327 2.48558 3.1448 3.73013 4.1425 4.35412 4.41435 4.40172 4.36236 4.28412 4.1189 7.22229 5.16043 2.96875 1.99991 0 0
1991 7 1 3 0 125 0 0 0 0 0 0 0 4.09296 1.64299 2.29507 3.03157 3.76509 4.37775 4.75688 4.85257 4.7066 4.41883 4.08131 3.73599 6.36937 4.67843 5.34272 0 0 0 0 0 0 0 1.57251 0.593367 0.757213 1.09034 1.60054 2.26364 3.02957 3.81101 4.48825 4.9417 5.10659 5.00768 4.73428 4.37673 3.98083 6.62084 4.57618 4.30059 0 0 0
1992 7 1 3 0 125 0 0 0 0 1.14817 0.714632 0.985086 1.23854 1.53592 1.99136 2.65426 3.45904 4.26297 4.90477 5.25657 5.26781 4.98156 4.5057 3.95521 6.2929 4.33271 4.74929 0 0 0 0 0 0 0 0 0 0 3.99585 1.56349 2.00186 2.65192 3.47212 4.32171 5.02694 5.44403 5.50747 5.2482 4.76673 4.17729 6.52145 4.23454 3.8299 0 0 0
1993 7 1 3 0 125 0 0 0 0 0 0 2.04406 1.13504 1.62108 2.12832 2.65352 3.25053 3.94618 4.66017 5.2264 5.49062 5.39221 4.97951 4.36912 6.70701 4.30723 4.41978 0 0 0 0 0 0 0 0 0 0 3.03125 1.59563 2.14632 2.71432 3.33539 4.04254 4.77541 5.37553 5.67713 5.6019 5.18653 4.54485 6.8763 4.19419 2.19774 1.37418 0 0
1994 7 1 3 0 125 0 0 0 0 0 0 0 0 0 5.89568 2.54279 3.27569 3.96913 4.59323 5.11492 5.4558 5.5195 5.26134 4.72559 7.29227 4.50824 2.45522 1.81618 0 0 0 0 0 0 0 0 0 2.70496 1.22716 1.80442 2.53058 3.32009 4.08106 4.7581 5.30752 5.65531 5.71096 5.42979 4.85414 7.37608 4.35271 2.16859 1.293 0 0
1995 7 1 3 0 125 0 0 0 0.961078 0.389094 0.406131 0.49252 0.694918 1.02484 1.49848 2.13254 2.9096 3.7518 4.53684 5.15066 5.52286 5.61943 5.42856 4.96962 7.84754 9.16501 0 0 0 0 0 0 0 0 1.33251 0.422612 0.492032 0.678865 1.00156 1.47487 2.11687 2.91726 3.80581 4.65641 5.33423 5.74328 5.83645 5.60586 5.084 7.86174 4.63441 3.49972 0 0 0
1996 7 1 3 0 125 0 0 0 1.04596 0.427028 0.62106 0.806664 0.949113 1.09574 1.35729 1.81475 2.47428 3.28097 4.13316 4.89479 5.43082 5.65278 5.54037 5.13151 8.24457 5.17611 4.45139 0 0 0 0 0 0 0 0 0 2.83787 0.984372 1.14101 1.38891 1.83091 2.49407 3.32847 4.22708 5.04277 5.62313 5.86006 5.72165 5.24965 8.23062 4.90294 2.34016 1.26797 0 0
1997 7 1 3 0 125 0 0 0 2.03415 0.511234 0.582804 0.756954 1.04813 1.38788 1.70904 2.01607 2.39164 2.92223 3.60821 4.34449 4.97249 5.34895 5.39415 5.1078 8.39217 9.95858 0 0 0 0 0 0 0 0 0 3.12949 0.744185 1.02847 1.39057 1.75462 2.10238 2.49917 3.03669 3.73442 4.4959 5.15162 5.53831 5.561 5.21689 8.35954 5.06466 3.70511 0 0 0
1998 7 1 3 0 125 0 0 0 0 3.02856 1.0242 1.10074 1.24198 1.45902 1.78319 2.19937 2.63924 3.06239 3.49381 3.96613 4.44458 4.82046 4.9725 4.83339 8.20873 5.42056 2.86441 1.80793 0 0 0 0 0 0 2.9636 1.07213 1.14496 1.28632 1.50089 1.81874 2.24684 2.72886 3.20817 3.68538 4.18228 4.6657 5.02779 5.14252 4.93944 8.16368 5.0899 2.46695 1.29465 0 0
1999 7 1 3 0 125 0 0 0 0 1.72612 1.29625 1.76775 2.07943 2.23115 2.3464 2.53382 2.82342 3.18493 3.56005 3.89911 4.17946 4.38159 4.46089 4.35973 7.5943 5.19991 4.59897 0 0 0 0 0 0 0 0 0 4.57648 2.15669 2.3674 2.49776 2.6776 2.96275 3.33678 3.74416 4.12333 4.42953 4.62563 4.66204 4.48681 7.56893 4.87277 3.68807 0 0 0
2000 7 1 3 0 125 0 0 0 0 0 1.79151 1.1645 1.88241 2.6607 3.29 3.64346 3.76672 3.80757 3.87654 3.98777 4.09501 4.14508 4.09994 3.93398 6.83379 9.16204 0 0 0 0 0 0 0 0 0 1.68287 1.07457 1.78751 2.61863 3.35623 3.82835 4.02801 4.08981 4.15054 4.25273 4.35623 4.395 4.31457 4.08243 6.85492 4.48806 2.2765 1.22207 0 0
2001 7 1 3 0 125 0 0 0 0 1.25614 0.444031 0.652456 1.0826 1.78799 2.73227 3.73653 4.53623 4.94475 4.97007 4.76953 4.50621 4.25343 4.00181 3.71259 6.28507 4.38296 4.12477 0 0 0 0 0 0 0 0 1.67609 0.624044 1.01745 1.69079 2.63393 3.69795 4.61856 5.16606 5.2923 5.12801 4.84807 4.55038 4.23881 3.8756 6.3447 4.1299 3.28794 0 0 0
1977 7 2 3 0 125 0 0 0 0 2.0441 1.40472 1.7148 1.96091 2.14696 2.31272 2.47604 2.63516 2.79381 2.95915 3.13169 3.30296 3.45834 3.57953 3.64503 7.14723 6.2374 9.20265 0 0 0 0 0 0 0 0 0 5.23899 1.97425 2.16203 2.33845 2.52717 2.72673 2.93651 3.16101 3.39844 3.6344 3.84264 3.98893 4.03596 7.66495 6.16419 3.9879 3.06424 0 0
1980 7 2 3 0 125 0 0 0 0 1.63467 1.36466 1.9349 2.46741 2.89668 3.19466 3.32507 3.2895 3.15911 3.03342 2.98101 3.01463 3.10466 3.20519 3.27173 6.43629 5.61465 4.10455 2.42964 1.6933 0 0 0 0 0 1.60989 1.35451 1.89855 2.4191 2.85694 3.18868 3.38062 3.41862 3.35376 3.27884 3.26829 3.34118 3.46487 3.58053 3.62759 6.90976 5.55957 3.58782 2.74513 0 0
1983 7 2 3 0 125 0 0 0 0 1.48096 1.53513 2.28053 2.83829 3.15536 3.31546 3.38779 3.41598 3.43201 3.44664 3.44844 3.41893 3.34897 3.24317 3.11089 5.7173 4.7554 6.80872 0 0 0 0 0 0 0 1.41799 1.48406 2.22345 2.80566 3.15648 3.35125 3.46254 3.53077 3.58489 3.638 3.68145 3.69496 3.66129 3.57239 3.4249 6.14378 10.0262 0 0 0 0
1986 7 2 3 0 125 0 0 0 0 2.44255 1.61281 2.00577 2.35511 2.66266 2.96752 3.27399 3.5473 3.74978 3.85858 3.8701 3.79661 3.6569 3.46724 3.23721 5.64057 4.32564 2.90446 2.7376 0 0 0 0 0 1.25091 1.20453 1.65837 2.02176 2.34893 2.64966 2.95743 3.28402 3.5966 3.85176 4.0188 4.08566 4.05631 3.94235 3.75417 3.49732 5.96919 4.2927 4.44715 0 0 0
1989 7 2 3 0 125 0 0 0 0 0 5.07382 3.78492 4.70213 5.07531 4.96947 4.55082 4.00885 3.50648 3.13222 2.89467 2.7502 2.63798 2.50773 2.33284 3.95796 2.84961 3.30236 0 0 0 0 0 0 1.03471 1.39403 2.46395 3.67087 4.62511 5.08207 5.06915 4.72656 4.22589 3.73143 3.34756 3.09755 2.94367 2.82299 2.6789 2.47874 9.56949 0 0 0 0 0
1992 7 2 3 0 125 0 0 0 0 0 4.51119 3.50728 3.94361 4.04173 4.20028 4.47207 4.7047 4.76493 4.60438 4.24291 3.74336 3.18735 2.64917 2.1752 3.22675 4.27431 0 0 0 0 0 0 0 0 0 4.26267 3.48252 4.01084 4.11428 4.22244 4.46812 4.72249 4.83058 4.71907 4.39422 3.91366 3.35794 2.80265 2.29733 3.3446 2.04106 1.76633 0 0 0
1995 7 2 3 0 125 0 0 0 0 2.48215 1.57748 1.97494 2.49204 3.03733 3.55975 4.04668 4.45705 4.72304 4.79672 4.68234 4.42011 4.04943 3.59476 3.07815 4.53333 2.63449 2.23882 0 0 0 0 0 0 0 2.50918 1.6415 1.97298 2.43447 2.96834 3.50366 4.01695 4.46879 4.79103 4.92313 4.84921 4.59652 4.20582 3.71217 3.149 4.54232 2.51675 1.81957 0 0 0
1998 7 2 3 0 125 0 0 0 2.57479 2.69615 3.50824 3.89239 3.92768 3.81327 3.73563 3.68043 3.56526 3.3997 3.25752 3.17954 3.13688 3.06329 2.90374 2.64008 4.17992 2.59515 2.14045 0 0 0 0 0 0 0 5.28151 3.67241 4.04877 4.06789 3.9227 3.8101 3.75986 3.68633 3.56154 3.43614 3.35282 3.29294 3.19504 3.00303 2.698 4.15791 2.43734 1.72558 0 0 0
2001 7 2 3 0 125 0 0 0 0 0 3.44211 2.26956 3.3678 4.59686 5.63053 6.15076 6.02792 5.39989 4.55839 3.76126 3.12852 2.65888 2.29879 1.99481 3.14773 3.92195 0 0 0 0 0 0 0 0 1.94163 1.4934 2.17073 3.16514 4.34695 5.42789 6.08727 6.13733 5.64156 4.85392 4.04396 3.36586 2.84451 2.43493 2.0824 3.17847 3.42833 0 0 0 0
-99990000000000000000000000000000000000000000000000000000000
#
17 #_N_age_bins
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 20 25
2 #_N_ageerror_definitions
0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5 16.5 17.5 18.5 19.5 20.5 21.5 22.5 23.5 24.5 25.5 26.5 27.5 28.5 29.5 30.5 31.5 32.5 33.5 34.5 35.5 36.5 37.5 38.5 39.5 40.5
0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001
0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5 16.5 17.5 18.5 19.5 20.5 21.5 22.5 23.5 24.5 25.5 26.5 27.5 28.5 29.5 30.5 31.5 32.5 33.5 34.5 35.5 36.5 37.5 38.5 39.5 40.5
0.5 0.65 0.67 0.7 0.73 0.76 0.8 0.84 0.88 0.92 0.97 1.03 1.09 1.16 1.23 1.32 1.41 1.51 1.62 1.75 1.89 2.05 2.23 2.45 2.71 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
#_mintailcomp: upper and lower distribution for females and males separately are accumulated until exceeding this level.
#_addtocomp: after accumulation of tails; this value added to all bins
#_males and females treated as combined gender below this bin number
#_compressbins: accumulate upper tail by this number of bins; acts simultaneous with mintailcomp; set=0 for no forced accumulation
#_Comp_Error: 0=multinomial, 1=dirichlet
#_Comp_Error2: parm number for dirichlet
#_minsamplesize: minimum sample size; set to 1 to match 3.24, minimum value is 0.001
#_mintailcomp addtocomp combM+F CompressBins CompError ParmSelect minsamplesize
0 1e-07 1 0 0 0 0.001 #_fleet:1_FISHERY1
0 1e-07 1 0 0 0 0.001 #_fleet:2_SURVEY1
0 1e-07 1 0 0 0 0.001 #_fleet:3_SURVEY2
1 #_Lbin_method_for_Age_Data: 1=poplenbins; 2=datalenbins; 3=lengths
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
#_yr month fleet sex part ageerr Lbin_lo Lbin_hi Nsamp datavector(female-male)
1971 7 1 3 0 2 1 -1 75 0 0 0 0 5.35843 1.94253 2.08004 2.12736 2.10638 2.02926 1.9194 1.79896 1.67404 1.54816 6.02373 3.61648 5.84912 0 0 1.58776 1.40238 1.75166 1.97907 2.09454 2.12335 2.08885 2.00299 1.88818 1.76539 1.63985 1.5145 5.87928 3.52222 5.6861
1972 7 1 3 0 2 1 -1 75 0.795707 0.553478 0.929128 1.33609 1.69628 1.94449 2.08195 2.12915 2.10803 2.03076 1.92074 1.80017 1.67513 1.54914 6.02735 3.61855 5.85237 0 0 1.58696 1.40406 1.75358 1.98105 2.09646 2.12515 2.0905 2.00449 1.88953 1.7666 1.64094 1.51549 5.88294 3.52433 5.68942
1973 7 1 3 0 2 1 -1 75 0 0 2.2116 1.34153 1.70253 1.95043 2.08714 2.13345 2.11147 2.03344 1.92282 1.80178 1.67638 1.55011 6.02989 3.61942 5.85318 0 0 0 2.97363 1.7599 1.98699 2.10162 2.12944 2.09397 2.00726 1.89172 1.76834 1.64234 1.51661 5.8862 3.5257 5.69113
216
1974 7 1 3 0 2 1 -1 75 0 0 2.11019 1.34178 1.7148 1.9632 2.09862 2.14318 2.11945 2.03984 1.92791 1.80584 1.67964 1.55276 6.03764 3.62277 5.85742 0 0 1.47936 1.4095 1.77233 1.99976 2.11299 2.13907 2.1019 2.01368 1.89691 1.77256 1.64579 1.51947 5.89503 3.52982 5.69679
1975 7 1 3 0 2 1 -1 75 0 0 2.02583 1.29155 1.71719 1.98234 2.11725 2.1591 2.13252 2.0503 1.93619 1.81241 1.68489 1.55699 6.04989 3.62796 5.86392 0 0 0 0 4.53713 2.01885 2.13145 2.15478 2.11485 2.02414 1.9053 1.77933 1.65131 1.52401 5.90887 3.5362 5.70546
1976 7 1 3 0 2 1 -1 75 0 0 2.21108 1.20612 1.65157 1.98516 2.13969 2.17889 2.14744 2.06087 1.94328 1.81691 1.6875 1.55827 6.04721 3.62248 5.85171 0 0 0 0 4.36188 2.02064 2.15348 2.1742 2.12956 2.03474 1.91264 1.78425 1.65447 1.5259 5.9094 9.23063 0
1977 7 1 3 0 2 1 -1 75 0 0 0 0 5.25183 1.90846 2.14136 2.20214 2.16649 2.07345 1.95062 1.82037 1.68822 1.55712 6.03064 3.60648 5.8207 0 0 1.42577 1.27892 1.60897 1.94074 2.15391 2.1968 2.14824 2.04728 1.92028 1.78829 1.65597 1.52565 5.89762 3.52063 5.67305
1978 7 1 3 0 2 1 -1 75 0 0 2.765 1.17991 1.5686 1.81277 2.05133 2.19339 2.18086 2.08308 1.95331 1.8179 1.68225 1.54892 5.98095 3.56788 5.751 0 0 1.86173 1.23781 1.6185 1.84347 2.0616 2.18688 2.16201 2.05679 1.92332 1.78659 1.65105 1.5187 5.85469 3.48709 5.61261
1979 7 1 3 0 2 1 -1 75 0.908257 0.732806 1.37032 1.43928 1.53175 1.80957 1.95332 2.09031 2.15586 2.08242 1.94868 1.80696 1.66695 1.53103 5.88653 9.12915 0 0 0 2.25121 1.5156 1.58031 1.83886 1.96279 2.0826 2.13627 2.05592 1.91909 1.77657 1.63708 1.50241 5.76914 3.42502 5.50394
1980 7 1 3 0 2 1 -1 75 0 1.61676 1.23679 2.02299 1.92391 1.79026 1.93565 1.99222 2.0447 2.04179 1.93282 1.78834 1.64294 1.50363 5.74535 8.8506 0 0 0 0 4.08221 1.99144 1.81944 1.94334 1.9843 2.02511 2.01541 1.90381 1.75915 1.61479 1.47709 5.63964 3.33247 5.34307
1981 7 1 3 0 2 1 -1 75 0 1.78475 1.05936 1.80412 2.61666 2.29988 1.943 1.96155 1.94639 1.92558 1.87688 1.75658 1.60994 1.46648 5.55322 3.25952 5.21193 0 0 1.8746 1.88349 2.69669 2.342 1.95081 1.95212 1.92722 1.90043 1.84916 1.72908 1.58405 1.44266 5.46272 3.20588 5.12328
1982 7 1 3 0 2 1 -1 75 0 1.81054 1.24262 1.6111 2.31959 3.0094 2.52465 1.99406 1.90241 1.82712 1.75896 1.68836 1.5655 1.423 5.3261 3.09426 4.92326 0 0 0 0 6.2186 3.05011 2.53646 1.98425 1.88235 1.80295 1.73333 1.663 1.54204 1.40204 5.25221 7.91174 0
1983 7 1 3 0 2 1 -1 75 0 0 0 4.92087 2.13505 2.66731 3.17673 2.59009 1.95391 1.77266 1.66328 1.57448 1.49104 1.37166 5.08561 2.9158 4.61012 0 0 2.27222 1.97671 2.19499 2.69372 3.17831 2.57645 1.93272 1.74827 1.63896 1.55153 1.47008 1.35336 5.02761 2.88778 4.56869
1984 7 1 3 0 2 1 -1 75 0 0 0 4.90433 2.49274 2.51636 2.82998 3.15121 2.52332 1.83994 1.60336 1.48256 1.38477 1.29552 4.83436 2.72994 4.28156 0 0 2.15621 2.15517 2.56185 2.54535 2.82408 3.12421 2.49353 1.81375 1.57941 1.46124 1.36638 1.27994 4.79141 2.71431 4.26323
1985 7 1 3 0 2 1 -1 75 0 0 0 4.8915 2.70801 2.91581 2.71922 2.8256 2.97718 2.34887 1.67732 1.42007 1.29537 1.1957 4.54784 2.53598 3.93641 0 0 0 4.01847 2.77636 2.94641 2.71523 2.79623 2.93495 2.31265 1.65138 1.39964 1.27889 1.18277 4.51938 2.53231 3.94045
1986 7 1 3 0 2 1 -1 75 0 2.4258 1.06945 1.80133 2.65304 3.14247 3.11228 2.7443 2.67809 2.68028 2.10105 1.49008 1.22905 1.10542 4.20147 2.32758 3.56513 0 0 0 0 6.45935 3.16783 3.10327 2.71519 2.6368 2.63504 2.0663 1.46801 1.21393 1.09467 4.18702 5.92578 0
1987 7 1 3 0 2 1 -1 75 0 3.43121 1.30377 1.64253 2.34107 3.03681 3.29908 3.06986 2.59654 2.3996 2.29818 1.80436 1.28006 1.03075 3.78768 5.23761 0 0 0 2.65186 1.71068 2.38934 3.05347 3.282 3.03249 2.55466 2.35757 2.25905 1.77652 1.26422 1.02182 3.78645 2.10854 3.19223
1988 7 1 3 0 2 1 -1 75 2.22044 1.67316 2.20793 2.06151 2.1412 2.6263 3.09343 3.14888 2.79418 2.28202 2.02203 1.87345 1.47273 1.04776 3.32321 4.49088 0 0 1.79682 2.35148 2.15651 2.18716 2.63508 3.06973 3.10413 2.74483 2.23984 1.98733 1.84524 1.4544 1.03895 3.33231 4.57709 0
1989 7 1 3 0 2 1 -1 75 0 3.50542 2.96223 3.37401 2.72236 2.38659 2.6021 2.84669 2.75683 2.35158 1.87594 1.61315 1.44996 1.14239 2.93934 3.71232 0 0 0 4.895 3.52817 2.79251 2.3963 2.57716 2.80003 2.70338 2.30472 1.84165 1.58911 1.43328 1.13318 2.95353 3.81107 0
1990 7 1 3 0 2 1 -1 75 0 0 6.01594 4.25252 4.26614 3.05059 2.35091 2.33648 2.41087 2.23796 1.85646 1.46341 1.22895 1.07577 2.69653 2.97293 0 0 1.48717 2.92904 4.42487 4.36881 3.0723 2.32914 2.29414 2.35964 2.19006 1.81993 1.43978 1.21513 1.06845 2.7122 3.07386 0
1991 7 1 3 0 2 1 -1 75 0 0 5.49326 3.87979 5.14446 4.63047 3.05353 2.14359 1.97476 1.93663 1.74408 1.42601 1.11538 0.916779 2.49994 2.36724 0 0 1.24159 2.45681 4.02428 5.24249 4.65324 3.03009 2.10423 1.92947 1.89192 1.7073 1.40081 1.10116 0.910568 2.51638 2.46373 0
1992 7 1 3 0 2 1 -1 75 0 0 4.98534 3.20138 4.61525 5.40504 4.53089 2.84257 1.87087 1.61823 1.52818 1.35255 1.09592 0.853092 2.3324 0.818778 1.11626 0 0 3.61164 3.32124 4.69449 5.41165 4.4874 2.79116 1.82579 1.57733 1.49262 1.32577 1.07923 0.845094 2.34666 2.02317 0
1993 7 1 3 0 2 1 -1 75 0 0 4.59845 2.78357 3.80734 4.86652 5.22348 4.17673 2.54588 1.60063 1.3196 1.21459 1.06112 4.75829 0 0 0 0 0 3.74952 2.9016 3.87535 4.86934 5.16156 4.09521 2.48289 1.55701 1.2854 1.18737 1.04208 0.844815 3.99166 0 0
1994 7 1 3 0 2 1 -1 75 0 0 3.86802 3.36864 3.48569 4.0618 4.75692 4.7854 3.69901 2.2112 1.34757 1.07146 0.963143 0.832101 2.20375 0.669769 0.793073 0 0 0 6.24891 3.5685 4.06755 4.69997 4.6857 3.6035 2.14823 1.3094 1.04467 5.50603 0 0 0 0
1995 7 1 3 0 2 1 -1 75 1.67878 0.75097 1.30344 2.52186 4.13693 3.91958 4.04003 4.42554 4.23777 3.18446 1.8883 1.13363 0.871994 4.27314 0 0 0 0 0 0 4.79846 4.2284 3.94602 3.99604 4.33467 4.12628 3.09165 1.8319 1.10221 0.852069 0.752419 2.16992 0.675134 0.728414
1996 7 1 3 0 2 1 -1 75 0 0 4.52033 1.89583 3.07003 4.47156 4.03 3.79493 3.93045 3.60551 2.6585 1.58396 0.943903 0.702695 2.09572 1.22865 0 0 1.29502 1.42735 1.97239 3.12137 4.48944 4.00255 3.72214 3.82985 3.50135 2.57858 1.53749 0.919621 0.688902 3.38188 0 0
1997 7 1 3 0 2 1 -1 75 0 4.38331 2.07909 1.99907 2.33057 3.26667 4.3321 3.79694 3.35711 3.30635 2.93716 2.15052 1.29434 0.768088 3.1056 0 0 0 0 0 5.63026 2.37884 3.26973 4.29168 3.73447 3.27561 3.21344 2.85029 2.08701 1.25837 3.90337 0 0 0
1998 7 1 3 0 2 1 -1 75 3.87916 1.72279 2.04903 2.91731 2.51123 2.47973 3.13601 3.8696 3.34012 2.81875 2.66932 2.32399 1.69431 1.0325 2.98234 0 0 0 0 4.02341 3.05125 2.58088 2.49048 3.10248 3.79941 3.2649 2.74295 2.59255 2.25669 1.64676 1.00605 1.92446 1.09152 0
1999 7 1 3 0 2 1 -1 75 2.48817 2.72909 2.9563 2.83206 3.43092 2.71678 2.36485 2.76277 3.21911 2.74605 2.24345 2.06077 1.76392 1.28442 3.02286 0 0 0 2.93749 3.15626 2.95518 3.5163 2.74478 2.34578 2.71138 3.14378 2.67549 2.18177 2.00344 1.71615 1.25161 3.03907 0 0
2000 7 1 3 0 2 1 -1 75 0 3.22365 4.33057 4.21197 3.36249 3.47941 2.62525 2.09167 2.28238 2.52437 2.1484 1.72542 1.53869 1.29772 3.16886 0 0 0 0 6.5466 4.42566 3.44791 3.50353 2.61518 2.05701 2.22922 2.45904 2.09123 1.67962 1.49917 1.26624 3.16872 0 0
2001 7 1 3 0 2 1 -1 75 0 2.38814 2.71103 5.7528 5.15529 3.56656 3.23803 2.3685 1.77842 1.82494 1.93187 1.65256 1.31657 1.14095 3.28171 0 0 0 0 3.7471 6.00213 5.31227 3.59882 3.21679 2.33528 1.7396 1.77853 1.88112 1.60986 1.28435 1.11506 3.27174 0 0
1977 7 2 3 0 2 1 -1 75 3.24228 1.8733 2.27587 2.56585 2.48241 2.41674 2.32132 2.12807 1.92246 1.72835 1.55236 1.39933 1.264 1.14234 4.27212 6.41306 0 0 1.99619 2.33891 2.58973 2.48857 2.41648 2.31975 2.12686 1.92205 1.72872 1.55334 1.40072 1.26562 1.14407 4.28036 6.42879 0
1980 7 2 3 0 2 1 -1 75 2.59348 1.96897 3.15167 4.08931 3.09903 2.18911 1.9788 1.81393 1.70426 1.60254 1.45068 1.29676 1.16023 1.04036 3.83836 2.20207 3.47541 0 2.08325 3.2221 4.1198 3.1047 2.18528 1.97374 1.81012 1.70234 1.60251 1.45233 1.29969 1.16401 1.04463 3.86006 2.21753 3.50295
1983 7 2 3 0 2 1 -1 75 2.42912 2.43525 3.52387 3.67488 3.11797 2.99719 3.10825 2.31823 1.5789 1.33016 1.19103 1.088 1.00412 0.90575 3.24346 4.60723 0 0 2.56667 3.6087 3.69883 3.11163 2.97971 3.08816 2.30496 1.57313 1.32873 1.1928 1.09244 1.01055 0.913422 3.28485 1.83378 2.85822
1986 7 2 3 0 2 1 -1 75 3.82413 2.15877 2.70555 3.29115 3.72473 3.60891 3.07345 2.40323 2.11963 2.00496 1.51721 1.03492 0.825918 0.726875 2.66793 1.43376 2.15924 0 0 5.06345 3.29933 3.70204 3.57438 3.04086 2.37991 2.10467 1.99628 1.51504 1.03811 0.832512 0.735678 2.72361 3.71376 0
1989 7 2 3 0 2 1 -1 75 4.07818 4.10452 6.19585 5.52908 3.39519 2.23352 2.01207 1.96476 1.76115 1.41734 1.07782 0.890167 0.780278 3.91551 0 0 0 0 4.31016 6.32505 5.55001 3.37123 2.20259 1.9807 1.93641 1.74044 1.40599 1.07473 0.89326 4.85398 0 0 0 0
1992 7 2 3 0 2 1 -1 75 3.65422 3.89317 4.05137 4.69301 5.20119 5.00778 3.6873 2.06819 1.21549 0.969712 0.873742 0.749373 0.59246 0.451552 2.13994 0 0 0 4.12476 4.16045 4.68947 5.14332 4.92428 3.61664 2.02734 1.1941 0.957196 0.867228 0.748114 0.595233 0.457003 2.24639 0 0
1995 7 2 3 0 2 1 -1 75 0 5.8697 2.96959 3.96372 5.28192 4.20195 3.52575 3.44573 3.0616 2.1897 1.24439 0.714116 0.52963 0.454938 1.98912 0 0 0 2.17603 3.03227 3.96461 5.23791 4.15016 3.46733 3.38251 3.00423 2.15024 1.2248 0.706656 0.528061 0.456873 2.07646 0 0
1998 7 2 3 0 2 1 -1 75 8.52241 4.44497 4.15715 4.54022 3.10089 2.26106 2.3673 2.64921 2.14571 1.67935 1.51559 1.28095 0.914811 2.04397 0 0 0 0 4.76022 4.2718 4.55423 3.08608 2.23338 2.33148 2.60713 2.1123 1.65513 1.4961 1.26688 3.00169 0 0 0 0
2001 7 2 3 0 2 1 -1 75 2.95482 1.91293 4.65293 8.4292 6.39117 3.3858 2.51127 1.66752 1.11572 1.05885 1.07659 0.897032 0.694079 0.58832 1.63368 0 0 0 2.01759 4.70452 8.43215 6.3611 3.34957 2.4753 1.64253 1.09978 1.04588 1.06563 0.889927 0.690928 0.587606 1.66755 0 0
-9999 000000000000000000000000000000000000000000
#
1 #_Use_MeanSize-at-Age_obs (0/1)
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
# ageerr codes: positive means mean length-at-age; negative means mean bodywt_at_age
#_yr month fleet sex part ageerr ignore datavector(female-male)
# samplesize(female-male)
1971 7 1 3 0 1 2 31.4528 39.4399 44.5453 48.7796 52.2939 55.2412 57.7416 59.8818 61.7242 63.3154 64.692 65.8838 66.9158 67.8093 69.6533 71.6518 73.0013 31.8953 40.019 45.1253 49.2868 52.6886 55.5009 57.8521 59.8337 61.5124 62.9384 64.1514 65.184 66.063 66.8113 68.3029 69.8373 70.7638
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1995 7 1 3 0 1 2 32.4644 39.4399 44.5453 48.7796 52.2939 55.2412 57.7416 59.8818 61.7242 63.3154 64.692 65.8838 66.9158 67.8093 69.5291 71.6122 72.9698 32.9214 40.019 45.1253 49.2868 52.6886 55.5009 57.8521 59.8337 61.5124 62.9384 64.1514 65.184 66.063 66.8113 68.2111 69.8107 70.7458
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
217
1971 7 2 3 0 1 2 34.4005 39.3378 43.2293 46.9111 50.3042 53.3329 55.9881 58.2964 60.2957 62.0243 63.5177 64.8072 65.9205 66.8816 68.8439 70.9782 72.4046 34.7723 39.7586 43.7062 47.3832 50.7015 53.6057 56.1066 58.2444 60.0656 61.6148 62.9316 64.0504 65.0008 65.808 67.4026 69.0506 70.0375
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1995 7 2 3 0 1 2 34.6678 39.3378 43.2293 46.9111 50.3042 53.3329 55.9881 58.2964 60.2957 62.0243 63.5177 64.8072 65.9205 66.8816 68.7102 70.9362 72.3713 35.0356 39.7586 43.7062 47.3832 50.7015 53.6057 56.1066 58.2444 60.0656 61.6148 62.9316 64.0504 65.0008 65.808 67.3033 69.0222 70.0183
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
-9999 0000000000000000000000000000000000000000
0000000000000000000000000000000000
#
0 #_N_environ_variables
#Yr Variable Value
#
0 # N sizefreq methods to read
#
0 # do tags (0/1)
#
0 # morphcomp data(0/1)
# Nobs, Nmorphs, mincomp
# yr, seas, type, partition, Nsamp, datavector_by_Nmorphs
#
0 # Do dataread for selectivity priors(0/1)
# Yr, Seas, Fleet, Age/Size, Bin, selex_prior, prior_sd
# feature not yet implemented
#
999
#_bootstrap file: 1
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
1971 #_StartYr
2001 #_EndYr
1 #_Nseas
12 #_months/season
2 #_Nsubseasons (even number, minimum is 2)
1 #_spawn_month
2 #_Ngenders
40 #_Nages=accumulator age
1 #_Nareas
3 #_Nfleets (including surveys)
#_fleet_type: 1=catch fleet; 2=bycatch only fleet; 3=survey; 4=ignore
#_survey_timing: -1=for use of catch-at-age to override the month value associated with a datum
#_fleet_area: area the fleet/survey operates in
#_units of catch: 1=bio; 2=num (ignored for surveys; their units read later)
#_catch_mult: 0=no; 1=yes
#_rows are fleets
#_fleet_type timing area units need_catch_mult fleetname
1 0.5 1 1 0 FISHERY1 # 1
3 0.5 1 2 0 SURVEY1 # 2
3 0.5 1 2 0 SURVEY2 # 3
#Bycatch_fleet_input_goes_next
#a: fleet index
#b: 1=include dead bycatch in total dead catch for F0.1 and MSY optimizations and forecast ABC; 2=omit from total catch for these purposes (but still include the mortality)
218
#c: 1=Fmult scales with other fleets; 2=bycatch F constant at input value; 3=bycatch F from range of years
#d: F or first year of range
#e: last year of range
#f: not used
#abcdef
#_catch_biomass(mtons):_columns_are_fisheries,year,season
#_catch:_columns_are_year,season,fleet,catch,catch_se
#_Catch data: yr, seas, fleet, catch, catch_se
-999 1 1 0 0.01
1971 1 1 0 0.01
1972 1 1 202.538 0.01
1973 1 1 986.866 0.01
1974 1 1 1002.43 0.01
1975 1 1 2018.8 0.01
1976 1 1 3065.7 0.01
1977 1 1 4001.68 0.01
1978 1 1 5009.88 0.01
1979 1 1 6009.19 0.01
1980 1 1 8114.01 0.01
1981 1 1 9933.76 0.01
1982 1 1 9966.44 0.01
1983 1 1 10047.9 0.01
1984 1 1 10060.2 0.01
1985 1 1 9845.76 0.01
1986 1 1 9870.06 0.01
1987 1 1 9877.96 0.01
1988 1 1 8992.91 0.01
1989 1 1 7803.49 0.01
1990 1 1 6993.29 0.01
1991 1 1 6021.81 0.01
1992 1 1 3999.49 0.01
1993 1 1 3988.58 0.01
1994 1 1 3987.35 0.01
1995 1 1 3989.31 0.01
1996 1 1 4005.95 0.01
1997 1 1 2957.3 0.01
1998 1 1 3005.3 0.01
1999 1 1 3023.5 0.01
2000 1 1 2976.62 0.01
2001 1 1 3057.63 0.01
-9999 0 0 0 0
#
#_CPUE_and_surveyabundance_observations
#_Units: 0=numbers; 1=biomass; 2=F; >=30 for special types
#_Errtype: -1=normal; 0=lognormal; >0=T
#_SD_Report: 0=no sdreport; 1=enable sdreport
#_Fleet Units Errtype SD_Report
1 1 0 0 # FISHERY1
2 1 0 0 # SURVEY1
219
3 0 0 0 # SURVEY2
#_year month index obs err
1977 7 2 206844 0.3 #_orig_obs: 339689 SURVEY1
1980 7 2 155990 0.3 #_orig_obs: 193353 SURVEY1
1983 7 2 79533.6 0.3 #_orig_obs: 151984 SURVEY1
1986 7 2 105534 0.3 #_orig_obs: 55221.8 SURVEY1
1989 7 2 66258.4 0.3 #_orig_obs: 59232.3 SURVEY1
1992 7 2 38114.7 0.3 #_orig_obs: 31137.5 SURVEY1
1995 7 2 52082.6 0.3 #_orig_obs: 35845.4 SURVEY1
1998 7 2 58874.6 0.3 #_orig_obs: 27492.6 SURVEY1
2001 7 2 36889.2 0.3 #_orig_obs: 37338.3 SURVEY1
1990 7 3 7.07716 0.7 #_orig_obs: 5.19333 SURVEY2
1991 7 3 1.5901 0.7 #_orig_obs: 1.1784 SURVEY2
1992 7 3 0.759259 0.7 #_orig_obs: 5.94383 SURVEY2
1993 7 3 1.76131 0.7 #_orig_obs: 0.770106 SURVEY2
1994 7 3 6.10527 0.7 #_orig_obs: 16.318 SURVEY2
1995 7 3 0.93583 0.7 #_orig_obs: 1.36339 SURVEY2
1996 7 3 11.0008 0.7 #_orig_obs: 4.76482 SURVEY2
1997 7 3 12.8587 0.7 #_orig_obs: 51.0707 SURVEY2
1998 7 3 0.965074 0.7 #_orig_obs: 1.36095 SURVEY2
1999 7 3 3.70735 0.7 #_orig_obs: 0.862531 SURVEY2
2000 7 3 0.704704 0.7 #_orig_obs: 5.97125 SURVEY2
2001 7 3 2.22424 0.7 #_orig_obs: 1.69379 SURVEY2
-9999 1 1 1 1 # terminator for survey observations
#
0 #_N_fleets_with_discard
#_discard_units (1=same_as_catchunits(bio/num); 2=fraction; 3=numbers)
#_discard_errtype: >0 for DF of T-dist(read CV below); 0 for normal with CV; -1 for normal with se; -2 for lognormal; -3 for trunc normal with CV
# note, only have units and errtype for fleets with discard
#_Fleet units errtype
# -9999 0 0 0.0 0.0 # terminator for discard data
#
0 #_use meanbodysize_data (0/1)
#_COND_0 #_DF_for_meanbodysize_T-distribution_like
# note: use positive partition value for mean body wt, negative partition for mean body length
#_yr month fleet part obs stderr
# -9999 0 0 0 0 0 # terminator for mean body size data
#
# set up population length bin structure (note - irrelevant if not using size data and using empirical wtatage
2 # length bin method: 1=use databins; 2=generate from binwidth,min,max below; 3=read vector
2 # binwidth for population size comp
10 # minimum size in the population (lower edge of first bin and size at age 0.00)
94 # maximum size in the population (lower edge of last bin)
1 # use length composition data (0/1)
#_mintailcomp: upper and lower distribution for females and males separately are accumulated until exceeding this level.
#_addtocomp: after accumulation of tails; this value added to all bins
#_males and females treated as combined sex below this bin number
#_compressbins: accumulate upper tail by this number of bins; acts simultaneous with mintailcomp; set=0 for no forced accumulation
#_Comp_Error: 0=multinomial, 1=dirichlet
220
#_Comp_Error2: parm number for dirichlet
#_minsamplesize: minimum sample size; set to 1 to match 3.24, minimum value is 0.001
#_mintailcomp addtocomp combM+F CompressBins CompError ParmSelect minsamplesize
0 1e-07 0 0 0 0 0.001 #_fleet:1_FISHERY1
0 1e-07 0 0 0 0 0.001 #_fleet:2_SURVEY1
0 1e-07 0 0 0 0 0.001 #_fleet:3_SURVEY2
25 #_N_LengthBins
26 28 30 32 34 36 38 40 42 44 46 48 50 52 54 56 58 60 62 64 68 72 76 80 90
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
#_yr month fleet sex part Nsamp datavector(female-male)
19717130125 00000000062201422461289900000000002004122364211125400
19727130125 000000000411232124611910200000000003102423343214139400
19737130125 00000000000074206231271010000000000000061327484797220
19747130125 00000000060323215455191120000000002210202624537107310
19757130125 000000001011304626109149600000000000301130435112811100
19767130125 0000000211012652375612111500000000011020129637783500
19777130125 0000000010311421355841151000000211111103618481097600
19787130125 0000001103202314556768310000000020002010546514146800
19797130125 0000000000432202444913743000000000021131655241089520
19807130125 000000032334131224393692000000000402013253978128100
19817130125 0000002013112224165131261200000000020000503014210116800
19827130125 0000000031121345536881200000000003013212363641373600
19837130125 0000000000511352264109790000000000041325326449104400
19847130125 000000111201323335101112420000000002022003437461376200
19857130125 0000000011131135287109524000000003004453213241087600
19867130125 0001000011114343236159103000000010010211243845955700
19877130125 000000310011338652614833000000000710130348252685300
19887130125 000001111022311366681159000000010151114443775644000
19897130125 00000430124246334532663000000004044182235462954100
19907130125 00000006055223414627657000000010135442417564652000
19917130125 0000000432424661014110020000000411212422565626548000
19927130125 00001102342026335269660000000000506226757432942000
19937130125 00000022102473755447350000000000221743635346755100
19947130125 00000000054812663219534000000000223433814777447000
19957130125 0002000001233243113898000000001101214444644161076000
19967130125 000201121133554712373520000000003100135264884634000
19977130125 000102320234302695271300000000020311115874369640000
19987130125 000021103304546555892210000001001102165745103913000
19997130125 000020353444053438210520000000007415421235253545000
20007130125 00000212035726636218600000000010128476432173744100
20017130125 00001011241449644158540000000021121448532445843000
19777230125 000011232352213134567150000000004133324464522843100
19807230125 000012134022324731210684200000101412333155145691300
19837230125 000012483242432331464100000000313535266224323270000
19867230125 000050112117245723110543000000204213183515616544000
19897230125 0000064563321312100554000000143934311113532205500000
19927230125 00000246432856056224200000000076323994434242002000
19957230125 00002003051447595435340000000411632525624613343000
221
19987230125 00016644421535442204500000000337277145131340660000
20017230125 00000332247361446226900000000003263559625311550000
-99990000000000000000000000000000000000000000000000000000000
#
17 #_N_age_bins
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 20 25
2 #_N_ageerror_definitions
0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5 16.5 17.5 18.5 19.5 20.5 21.5 22.5 23.5 24.5 25.5 26.5 27.5 28.5 29.5 30.5 31.5 32.5 33.5 34.5 35.5 36.5 37.5 38.5 39.5 40.5
0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001 0.001
0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5 9.5 10.5 11.5 12.5 13.5 14.5 15.5 16.5 17.5 18.5 19.5 20.5 21.5 22.5 23.5 24.5 25.5 26.5 27.5 28.5 29.5 30.5 31.5 32.5 33.5 34.5 35.5 36.5 37.5 38.5 39.5 40.5
0.5 0.65 0.67 0.7 0.73 0.76 0.8 0.84 0.88 0.92 0.97 1.03 1.09 1.16 1.23 1.32 1.41 1.51 1.62 1.75 1.89 2.05 2.23 2.45 2.71 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3
#_mintailcomp: upper and lower distribution for females and males separately are accumulated until exceeding this level.
#_addtocomp: after accumulation of tails; this value added to all bins
#_males and females treated as combined sex below this bin number
#_compressbins: accumulate upper tail by this number of bins; acts simultaneous with mintailcomp; set=0 for no forced accumulation
#_Comp_Error: 0=multinomial, 1=dirichlet
#_Comp_Error2: parm number for dirichlet
#_minsamplesize: minimum sample size; set to 1 to match 3.24, minimum value is 0.001
#_mintailcomp addtocomp combM+F CompressBins CompError ParmSelect minsamplesize
0 1e-07 1 0 0 0 0.001 #_fleet:1_FISHERY1
0 1e-07 1 0 0 0 0.001 #_fleet:2_SURVEY1
0 1e-07 1 0 0 0 0.001 #_fleet:3_SURVEY2
1 #_Lbin_method_for_Age_Data: 1=poplenbins; 2=datalenbins; 3=lengths
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
#_yr month fleet sex part ageerr Lbin_lo Lbin_hi Nsamp datavector(female-male)
1971 71 3021-175 0000921111200234800222421133111639
1972 71 3021-175 10103223211110611000322231134110458
1973 71 3021-175 00210313310221103700045131224003533
1974 71 3021-175 0011061143111334400025222211010869
1975 71 3021-175 00221131322421724000041101131009611
1976 71 3021-175 00402014303412122500005240515010270
1977 71 3021-175 0000513543311121600112315450220454
1978 71 3021-175 0002213104210178500403350401241344
1979 71 3021-175 3002021462131468000412011600331523
1980 71 3021-175 0315121140133067000054132111221554
1981 71 3021-175 0210240531110152700232130402113657
1982 71 3021-175 0301232032303185200002343322211950
1983 71 3021-175 0003124211110066800001432510314339
1984 71 3021-175 0005252201131253600007350132142414
1985 71 3021-175 0004505040210451300024320884021304
1986 71 3021-175 0304011734203162300006044351102360
1987 71 3021-175 0300233440234155000235362123110313
1988 71 3021-175 2520321132221156000206133413102290
1989 71 3021-175 0423231341532121000571233321011720
1990 71 3021-175 0062714221210224001143421354213320
1991 71 3021-175 0042463312000442002178552310110220
1992 71 3021-175 00344537131000220003381234110200210
1993 71 3021-175 00443641220021400000627362011120200
222
1994 71 3021-175 00207233402110110000714748430100000
1995 71 3021-175 2211331781411200000044526841100300
1996 71 3021-175 0022435355232133000003233580200600
1997 71 3021-175 0600234762430020000041464414215000
1998 71 3021-175 4036122382440070000421051442121110
1999 71 3021-175 3142235525142240003332014112240400
2000 71 3021-175 0364552335113120000622421242212100
2001 71 3021-175 0206763103221210000274532314301400
1977 72 3021-175 6122421020121228000326512232111640
1980 72 3021-175 2304323111100372401240611421210535
1983 72 3021-175 6501311215200143002364452111100541
1986 72 3021-175 4115511127002163100544730510010130
1989 72 3021-175 5235410321110600008865101212250000
1992 72 3021-175 2537482300110000001587434211010200
1995 72 3021-175 0467223452001110001738624121110000
1998 72 3021-175 13522323455110200005332231410120000
2001 72 3021-175 105511425012101100036117201020120100
-9999 000000000000000000000000000000000000000000
#
1 #_Use_MeanSize-at-Age_obs (0/1)
# sex codes: 0=combined; 1=use female only; 2=use male only; 3=use both as joint sexxlength distribution
# partition codes: (0=combined; 1=discard; 2=retained
# ageerr codes: positive means mean length-at-age; negative means mean bodywt_at_age
#_yr month fleet sex part ageerr ignore datavector(female-male)
# samplesize(female-male)
1971 7 1 3 0 1 2 31.4162 38.6229 43.6231 48.0368 51.0399 54.4278 58.476 56.9059 63.0114 65.1603 63.9478 65.0109 66.4932 67.1987 69.9286 68.6615 71.6184 32.1109 39.5534 44.3028 50.4847 51.9323 54.4694 56.2276 60.83 60.4257 64.2749 63.3618 64.6688 66.3104 67.5702 67.9094 68.6782 70.722
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1995 7 1 3 0 1 2 32.0027 38.7135 44.5384 49.3326 51.0093 55.1973 57.6228 59.3377 60.3642 62.1497 66.4974 62.3795 67.867 66.0373 69.7154 69.3185 72.2666 32.7867 41.3142 43.3295 48.046 50.7067 56.4385 57.7696 62.1555 62.3235 62.8589 62.6859 66.7405 66.9157 65.5271 68.9712 69.9304 66.7984
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1971 7 2 3 0 1 2 33.3397 39.5351 43.8641 47.2807 51.3397 53.109 56.8232 60.592 58.6174 63.1289 62.9685 66.7526 66.6988 66.7211 69.473 70.579 72.5308 34.1885 40.2613 43.2173 49.2213 51.8578 52.4676 56.0092 60.0047 61.6688 61.8141 64.1234 63.6133 65.1565 67.0794 65.309 67.5909 69.4096
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
1995 7 2 3 0 1 2 34.3832 38.5101 43.913 46.921 49.815 53.992 57.6028 61.0538 59.5837 62.3403 63.837 66.6111 66.8381 68.6201 69.7881 71.8883 74.1751 34.7775 39.3016 43.5807 47.3892 52.9023 56.0222 56.1842 57.0555 61.8584 60.2383 62.0913 65.1373 63.0871 64.3621 67.1485 69.945 73.3388
20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20 20
-9999 0000000000000000000000000000000000000000
0000000000000000000000000000000000
#
0 #_N_environ_variables
#Yr Variable Value
#
0 # N sizefreq methods to read
#
0 # do tags (0/1)
#
0 # morphcomp data(0/1)
# Nobs, Nmorphs, mincomp
# yr, seas, type, partition, Nsamp, datavector_by_Nmorphs
#
0 # Do dataread for selectivity priors(0/1)
# Yr, Seas, Fleet, Age/Size, Bin, selex_prior, prior_sd
223
# feature not yet implemented
#
999
control.ss
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
#C growth parameters are estimated
#C spawner-recruitment bias adjustment Not tuned For optimality
#_data_and_control_files: simple.dat // simple.ctl
#V3.30.10.00-safe;_2018_01_09;_Stock_Synthesis_by_Richard_Methot_(NOAA)_using_ADMB_11.6
#_user_support_available_at:NMFS.Stock.Synthesis@noaa.gov
#_user_info_available_at:https://vlab.ncep.noaa.gov/group/stock-synthesis
0 # 0 means do not read wtatage.ss; 1 means read and use wtatage.ss and also read and use growth parameters
1 #_N_Growth_Patterns
1 #_N_platoons_Within_GrowthPattern
#_Cond 1 #_Morph_between/within_stdev_ratio (no read if N_morphs=1)
#_Cond 1 #vector_Morphdist_(-1_in_first_val_gives_normal_approx)
#
2 # recr_dist_method for parameters: 2=main effects for GP, Area, Settle timing; 3=each Settle entity
1 # not yet implemented; Future usage: Spawner-Recruitment: 1=global; 2=by area
1 # number of recruitment settlement assignments
0 # unused option
#GPattern month area age (for each settlement assignment)
1 1 1 0
#
#_Cond 0 # N_movement_definitions goes here if Nareas > 1
#_Cond 1.0 # first age that moves (real age at begin of season, not integer) also cond on do_migration>0
#_Cond 1 1 1 2 4 10 # example move definition for seas=1, morph=1, source=1 dest=2, age1=4, age2=10
#
1 #_Nblock_Patterns
1 #_blocks_per_pattern
# begin and end years of blocks
1970 1970
#
# controls for all timevary parameters
1 #_env/block/dev_adjust_method for all time-vary parms (1=warn relative to base parm bounds; 3=no bound check)
# autogen
0 0 0 0 0 # autogen: 1st element for biology, 2nd for SR, 3rd for Q, 4th reserved, 5th for selex
# where: 0 = autogen all time-varying parms; 1 = read each time-varying parm line; 2 = read then autogen if parm min==-12345
#
#
# setup for M, growth, maturity, fecundity, recruitment distibution, movement
#
0 #_natM_type:_0=1Parm; 1=N_breakpoints;_2=Lorenzen;_3=agespecific;_4=agespec_withseasinterpolate
224
#_no additional input for selected M option; read 1P per morph
1 # GrowthModel: 1=vonBert with L1&L2; 2=Richards with L1&L2; 3=age_specific_K; 4=not implemented
0 #_Age(post-settlement)_for_L1;linear growth below this
25 #_Growth_Age_for_L2 (999 to use as Linf)
-999 #_exponential decay for growth above maxage (fixed at 0.2 in 3.24; value should approx initial Z; -999 replicates 3.24)
0 #_placeholder for future growth feature
0 #_SD_add_to_LAA (set to 0.1 for SS2 V1.x compatibility)
0 #_CV_Growth_Pattern: 0 CV=f(LAA); 1 CV=F(A); 2 SD=F(LAA); 3 SD=F(A); 4 logSD=F(A)
1 #_maturity_option: 1=length logistic; 2=age logistic; 3=read age-maturity matrix by growth_pattern; 4=read age-fecundity; 5=disabled; 6=read length-maturity
1 #_First_Mature_Age
1 #_fecundity option:(1)eggs=Wt*(a+b*Wt);(2)eggs=a*L^b;(3)eggs=a*Wt^b; (4)eggs=a+b*L; (5)eggs=a+b*W
0 #_hermaphroditism option: 0=none; 1=female-to-male age-specific fxn; -1=male-to-female age-specific fxn
1 #_parameter_offset_approach (1=none, 2= M, G, CV_G as offset from female-GP1, 3=like SS2 V1.x)
#
#_growth_parms
#_ LO HI INIT PRIOR PR_SD PR_type PHASE env_var&link dev_link dev_minyr dev_maxyr dev_PH Block Block_Fxn
0.05 0.15 0.1 0.1 0.8 0 -3 0 0 0 0 0 0 0 # NatM_p_1_Fem_GP_1
-10 45 21.6552 36 10 6 2 0 0 0 0 0 0 0 # L_at_Amin_Fem_GP_1
40 90 71.6492 70 10 6 4 0 0 0 0 0 0 0 # L_at_Amax_Fem_GP_1
0.05 0.25 0.147282 0.15 0.8 6 4 0 0 0 0 0 0 0 # VonBert_K_Fem_GP_1
0.05 0.25 0.1 0.1 0.8 0 -3 0 0 0 0 0 0 0 # CV_young_Fem_GP_1
0.05 0.25 0.1 0.1 0.8 0 -3 0 0 0 0 0 0 0 # CV_old_Fem_GP_1
-3 3 2.44e-06 2.44e-06 0.8 0 -3 0 0 0 0 0 0 0 # Wtlen_1_Fem_GP_1
-3 4 3.34694 3.34694 0.8 0 -3 0 0 0 0 0 0 0 # Wtlen_2_Fem_GP_1
50 60 55 55 0.8 0 -3 0 0 0 0 0 0 0 # Mat50%_Fem_GP_1
-3 3 -0.25 -0.25 0.8 0 -3 0 0 0 0 0 0 0 # Mat_slope_Fem_GP_1
-3 3 1 1 0.8 0 -3 0 0 0 0 0 0 0 # Eggs/kg_inter_Fem_GP_1
-3 3 0 0 0.8 0 -3 0 0 0 0 0 0 0 # Eggs/kg_slope_wt_Fem_GP_1
0.05 0.15 0.1 0.1 0.8 0 -3 0 0 0 0 0 0 0 # NatM_p_1_Mal_GP_1
1 45 0 36 10 0 -3 0 0 0 0 0 0 0 # L_at_Amin_Mal_GP_1
40 90 69.5361 70 10 6 4 0 0 0 0 0 0 0 # L_at_Amax_Mal_GP_1
0.05 0.25 0.163516 0.15 0.8 6 4 0 0 0 0 0 0 0 # VonBert_K_Mal_GP_1
0.05 0.25 0.1 0.1 0.8 0 -3 0 0 0 0 0 0 0 # CV_young_Mal_GP_1
0.05 0.25 0.1 0.1 0.8 0 -3 0 0 0 0 0 0 0 # CV_old_Mal_GP_1
-3 3 2.44e-06 2.44e-06 0.8 0 -3 0 0 0 0 0 0 0 # Wtlen_1_Mal_GP_1
-3 4 3.34694 3.34694 0.8 0 -3 0 0 0 0 0 0 0 # Wtlen_2_Mal_GP_1
0 0 0 0 0 0 -4 0 0 0 0 0 0 0 # RecrDist_GP_1
0 0 0 0 0 0 -4 0 0 0 0 0 0 0 # RecrDist_Area_1
0 0 0 0 0 0 -4 0 0 0 0 0 0 0 # RecrDist_month_1
1 1 1 1 1 0 -1 0 0 0 0 0 0 0 # CohortGrowDev
1e-06 0.999999 0.5 0.5 0.5 0 -99 0 0 0 0 0 0 0 # FracFemale_GP_1
#
#_no timevary MG parameters
#
#_seasonal_effects_on_biology_parms
0 0 0 0 0 0 0 0 0 0 #_femwtlen1,femwtlen2,mat1,mat2,fec1,fec2,Malewtlen1,malewtlen2,L1,K
#_ LO HI INIT PRIOR PR_SD PR_type PHASE
#_Cond -2 2 0 0 -1 99 -2 #_placeholder when no seasonal MG parameters
#
225
#_Spawner-Recruitment
3 #_SR_function: 2=Ricker; 3=std_B-H; 4=SCAA; 5=Hockey; 6=B-H_flattop; 7=survival_3Parm; 8=Shepard_3Parm
0 # 0/1 to use steepness in initial equ recruitment calculation
0 # future feature: 0/1 to make realized sigmaR a function of SR curvature
#_ LO HI INIT PRIOR PR_SD PR_type PHASE env-var use_dev dev_mnyr dev_mxyr dev_PH Block Blk_Fxn # parm_name
3 31 8.81544 10.3 10 0 1 0 0 0 0 0 0 0 # SR_LN(R0)
0.2 1 0.613717 0.7 0.05 1 4 0 0 0 0 0 0 0 # SR_BH_steep
0 2 0.6 0.8 0.8 0 -4 0 0 0 0 0 0 0 # SR_sigmaR
-5 5 0 0 1 0 -4 0 0 0 0 0 0 0 # SR_regime
0 0 0 0 0 0 -99 0 0 0 0 0 0 0 # SR_autocorr
1 #do_recdev: 0=none; 1=devvector; 2=simple deviations
1971 # first year of main recr_devs; early devs can preceed this era
2001 # last year of main recr_devs; forecast devs start in following year
2 #_recdev phase
1 # (0/1) to read 13 advanced options
0 #_recdev_early_start (0=none; neg value makes relative to recdev_start)
-4 #_recdev_early_phase
0 #_forecast_recruitment phase (incl. late recr) (0 value resets to maxphase+1)
1 #_lambda for Fcast_recr_like occurring before endyr+1
1900 #_last_early_yr_nobias_adj_in_MPD
1900 #_first_yr_fullbias_adj_in_MPD
2001 #_last_yr_fullbias_adj_in_MPD
2002 #_first_recent_yr_nobias_adj_in_MPD
1 #_max_bias_adj_in_MPD (-1 to override ramp and set biasadj=1.0 for all estimated recdevs)
0 #_period of cycles in recruitment (N parms read below)
-5 #min rec_dev
5 #max rec_dev
0 #_read_recdevs
#_end of advanced SR options
#
#_placeholder for full parameter lines for recruitment cycles
# read specified recr devs
#_Yr Input_value
#
# all recruitment deviations
# 1971R 1972R 1973R 1974R 1975R 1976R 1977R 1978R 1979R 1980R 1981R 1982R 1983R 1984R 1985R 1986R 1987R 1988R 1989R 1990R 1991R 1992R 1993R 1994R 1995R 1996R 1997R 1998R 1999R 2000R 2001R 2002F 2003F 2004F 2005F 2006F 2007F 2008F 2009F 2010F 2011F
# 0.127812 -0.0629072 0.0999946 -0.173914 0.0306907 0.714754 -0.0229372 0.00347081 0.260891 0.173281 0.0891999 -0.227374 -0.440643 -0.312905 0.391936 0.551136 0.218287 0.15476 -0.384699 0.596713 -0.68218 -0.273103 -0.829262 0.365425 -0.604348 0.455566 1.11144 -0.545922 -0.65609 0.172111 -0.301188 0 0 0 0 0 0 0 0 0 0
# implementation error by year in forecast: 0 0 0 0 0 0 0 0 0 0
#
#Fishing Mortality info
0.3 # F ballpark
-2001 # F ballpark year (neg value to disable)
3 # F_Method: 1=Pope; 2=instan. F; 3=hybrid (hybrid is recommended)
2.9 # max F or harvest rate, depends on F_Method
# no additional F input needed for Fmethod 1
# if Fmethod=2; read overall start F value; overall phase; N detailed inputs to read
# if Fmethod=3; read N iterations for tuning for Fmethod 3
4 # N iterations for tuning F in hybrid method (recommend 3 to 7)
#
226
#_initial_F_parms; count = 0
#_ LO HI INIT PRIOR PR_SD PR_type PHASE
#2011 2022
# F rates by fleet
# Yr: 1971 1972 1973 1974 1975 1976 1977 1978 1979 1980 1981 1982 1983 1984 1985 1986 1987 1988 1989 1990 1991 1992 1993 1994 1995 1996 1997 1998 1999 2000 2001 2002 2003 2004 2005 2006 2007 2008 2009 2010 2011
#seas: 11111111111111111111111111111111111111111
# FISHERY1 0 0.00211063 0.010608 0.0107027 0.0217039 0.0333291 0.0459439 0.0599346 0.0757011 0.107712 0.146835 0.162479 0.180802 0.202809 0.230256 0.26605 0.314454 0.337993 0.354248 0.355798 0.338692 0.23792 0.242784 0.250578 0.263425 0.283222 0.227015 0.238035 0.247381 0.252164 0.252998 0.0130853 0.0280047 0.0380878 0.0447957 0.0493817 0.0527536 0.0555053 0.0579621 0.0602612 0.0624351
#
#_Q_setup for fleets with cpue or survey data
#_1: link type: (1=simple q, 1 parm; 2=mirror simple q, 1 mirrored parm; 3=q and power, 2 parm)
#_2: extra input for link, i.e. mirror fleet
#_3: 0/1 to select extra sd parameter
#_4: 0/1 for biasadj or not
#_5: 0/1 to float
#_ fleet link link_info extra_se biasadj float # fleetname
2 1 0 1 0 0 # SURVEY1
3 1 0 0 0 0 # SURVEY2
-9999 0 0 0 0 0
#
#_Q_parms(if_any);Qunits_are_ln(q)
#_ LO HI INIT PRIOR PR_SD PR_type PHASE env-var use_dev dev_mnyr dev_mxyr dev_PH Block Blk_Fxn # parm_name
-7 5 0.515263 0 1 0 1 0 0 0 0 0 0 0 # LnQ_base_SURVEY1(2)
0 0.5 0 0.05 1 0 -4 0 0 0 0 0 0 0 # Q_extraSD_SURVEY1(2)
-7 5 -6.62828 0 1 0 1 0 0 0 0 0 0 0 # LnQ_base_SURVEY2(3)
#_no timevary Q parameters
#
#_size_selex_patterns
#Pattern:_0; parm=0; selex=1.0 for all sizes
#Pattern:_1; parm=2; logistic; with 95% width specification
#Pattern:_5; parm=2; mirror another size selex; PARMS pick the min-max bin to mirror
#Pattern:_15; parm=0; mirror another age or length selex
#Pattern:_6; parm=2+special; non-parm len selex
#Pattern:_43; parm=2+special+2; like 6, with 2 additional param for scaling (average over bin range)
#Pattern:_8; parm=8; New doublelogistic with smooth transitions and constant above Linf option
#Pattern:_9; parm=6; simple 4-parm double logistic with starting length; parm 5 is first length; parm 6=1 does desc as offset
#Pattern:_21; parm=2+special; non-parm len selex, read as pairs of size, then selex
#Pattern:_22; parm=4; double_normal as in CASAL
#Pattern:_23; parm=6; double_normal where final value is directly equal to sp(6) so can be >1.0
#Pattern:_24; parm=6; double_normal with sel(minL) and sel(maxL), using joiners
#Pattern:_25; parm=3; exponential-logistic in size
#Pattern:_27; parm=3+special; cubic spline
#Pattern:_42; parm=2+special+3; // like 27, with 2 additional param for scaling (average over bin range)
#_discard_options:_0=none;_1=define_retention;_2=retention&mortality;_3=all_discarded_dead;_4=define_dome-shaped_retention
#_Pattern Discard Male Special
1 0 0 0 # 1 FISHERY1
1 0 0 0 # 2 SURVEY1
0 0 0 0 # 3 SURVEY2
#
#_age_selex_types
227
#Pattern:_0; parm=0; selex=1.0 for ages 0 to maxage
#Pattern:_10; parm=0; selex=1.0 for ages 1 to maxage
#Pattern:_11; parm=2; selex=1.0 for specified min-max age
#Pattern:_12; parm=2; age logistic
#Pattern:_13; parm=8; age double logistic
#Pattern:_14; parm=nages+1; age empirical
#Pattern:_15; parm=0; mirror another age or length selex
#Pattern:_16; parm=2; Coleraine - Gaussian
#Pattern:_17; parm=nages+1; empirical as random walk N parameters to read can be overridden by setting special to non-zero
#Pattern:_41; parm=2+nages+1; // like 17, with 2 additional param for scaling (average over bin range)
#Pattern:_18; parm=8; double logistic - smooth transition
#Pattern:_19; parm=6; simple 4-parm double logistic with starting age
#Pattern:_20; parm=6; double_normal,using joiners
#Pattern:_26; parm=3; exponential-logistic in age
#Pattern:_27; parm=3+special; cubic spline in age
#Pattern:_42; parm=2+nages+1; // cubic spline; with 2 additional param for scaling (average over bin range)
#_Pattern Discard Male Special
11 0 0 0 # 1 FISHERY1
11 0 0 0 # 2 SURVEY1
11 0 0 0 # 3 SURVEY2
#
#_ LO HI INIT PRIOR PR_SD PR_type PHASE env-var use_dev dev_mnyr dev_mxyr dev_PH Block Blk_Fxn # parm_name
19 80 53.6526 50 0.01 1 2 0 0 0 0 0 0 0 # SizeSel_P1_FISHERY1(1)
0.01 60 18.9204 15 0.01 1 3 0 0 0 0 0 0 0 # SizeSel_P2_FISHERY1(1)
19 70 36.6499 30 0.01 1 2 0 0 0 0 0 0 0 # SizeSel_P1_SURVEY1(2)
0.01 60 6.58921 10 0.01 1 3 0 0 0 0 0 0 0 # SizeSel_P2_SURVEY1(2)
0 40 0 5 99 0 -1 0 0 0 0 0 0 0 # AgeSel_P1_FISHERY1(1)
0 40 40 6 99 0 -1 0 0 0 0 0 0 0 # AgeSel_P2_FISHERY1(1)
0 40 0 5 99 0 -1 0 0 0 0 0 0 0 # AgeSel_P1_SURVEY1(2)
0 40 40 6 99 0 -1 0 0 0 0 0 0 0 # AgeSel_P2_SURVEY1(2)
0 40 0 5 99 0 -1 0 0 0 0 0 0 0 # AgeSel_P1_SURVEY2(3)
0 40 0 6 99 0 -1 0 0 0 0 0 0 0 # AgeSel_P2_SURVEY2(3)
#_no timevary selex parameters
#
0 # use 2D_AR1 selectivity(0/1): experimental feature
#_no 2D_AR1 selex offset used
#
# Tag loss and Tag reporting parameters go next
0 # TG_custom: 0=no read; 1=read if tags exist
#_Cond -6 6 1 1 2 0.01 -4 0 0 0 0 0 0 0 #_placeholder if no parameters
#
# no timevary parameters
#
#
# Input variance adjustments factors:
#_1=add_to_survey_CV
#_2=add_to_discard_stddev
#_3=add_to_bodywt_CV
#_4=mult_by_lencomp_N
228
#_5=mult_by_agecomp_N
#_6=mult_by_size-at-age_N
#_7=mult_by_generalized_sizecomp
#_Factor Fleet Value
-9999 1 0 # terminator
#
4 #_maxlambdaphase
1 #_sd_offset; must be 1 if any growthCV, sigmaR, or survey extraSD is an estimated parameter
# read 3 changes to default Lambdas (default value is 1.0)
# Like_comp codes: 1=surv; 2=disc; 3=mnwt; 4=length; 5=age; 6=SizeFreq; 7=sizeage; 8=catch; 9=init_equ_catch;
# 10=recrdev; 11=parm_prior; 12=parm_dev; 13=CrashPen; 14=Morphcomp; 15=Tag-comp; 16=Tag-negbin; 17=F_ballpark
#like_comp fleet phase value sizefreq_method
12211
42211
42311
-9999 1 1 1 1 # terminator
#
# lambdas (for info only; columns are phases)
# 0 0 0 0 #_CPUE/survey:_1
# 1 1 1 1 #_CPUE/survey:_2
# 1 1 1 1 #_CPUE/survey:_3
# 1 1 1 1 #_lencomp:_1
# 1 1 1 1 #_lencomp:_2
# 0 0 0 0 #_lencomp:_3
# 1 1 1 1 #_agecomp:_1
# 1 1 1 1 #_agecomp:_2
# 0 0 0 0 #_agecomp:_3
# 1 1 1 1 #_size-age:_1
# 1 1 1 1 #_size-age:_2
# 0 0 0 0 #_size-age:_3
# 1 1 1 1 #_init_equ_catch
# 1 1 1 1 #_recruitments
# 1 1 1 1 #_parameter-priors
# 1 1 1 1 #_parameter-dev-vectors
# 1 1 1 1 #_crashPenLambda
# 0 0 0 0 # F_ballpark_lambda
1 # (0/1) read specs for more stddev reporting
1 1 -1 5 1 5 1 -1 5 # selex type, len/age, year, N selex bins, Growth pattern, N growth ages, NatAge_area(-1 for all), NatAge_yr, N Natages
5 15 25 35 43 # vector with selex std bin picks (-1 in first bin to self-generate)
1 2 14 26 40 # vector with growth std bin picks (-1 in first bin to self-generate)
1 2 14 26 40 # vector with NatAge std bin picks (-1 in first bin to self-generate)
999
229