Contents
- 1. RF safety Compliance Manual
- 2. MiSeq User Guide
MiSeq User Guide

FOR RESEARCH USE ONLY
ILLUMINA PROPRIETARY
Part # 15027617 Rev. A
August 2011
MiSeq™ System
User Guide
MiSeq System User Guide ii
This document and its contents are proprietary to Illumina, Inc. and its affiliates ("Illumina"), and are
intended solely for the contractual use of its customer in connection with the use of the product(s)
described herein and for no other purpose. This document and its contents shall not be used or distributed
for any other purpose and/or otherwise communicated, disclosed, or reproduced in any way whatsoever
without the prior written consent of Illumina. Illumina does not convey any license under its patent,
trademark, copyright, or common-law rights nor similar rights of any third parties by this document.
The Software is licensed to you under the terms and conditions of the Illumina Sequencing Software
License Agreement in a separate document. If you do not agree to the terms and conditions therein,
Illumina does not license the Software to you, and you should not use or install the Software
The instructions in this document must be strictly and explicitly followed by qualified and properly trained
personnel in order to ensure the proper and safe use of the product(s) described herein. All of the contents
of this document must be fully read and understood prior to using such product(s).
FAILURE TO COMPLETELY READ AND EXPLICITLY FOLLOW ALL OF THE INSTRUCTIONS
CONTAINED HEREIN MAY RESULT IN DAMAGE TO THE PRODUCT(S), INJURY TO PERSONS,
INCLUDING TO USERS OR OTHERS, AND DAMAGE TO OTHER PROPERTY.
ILLUMINA DOES NOT ASSUME ANY LIABILITY ARISING OUT OF THE IMPROPER USE OF THE
PRODUCT(S) DESCRIBED HEREIN (INCLUDING PARTS THEREOF OR SOFTWARE) OR ANY USE
OF SUCH PRODUCT(S) OUTSIDE THE SCOPE OF THE EXPRESS WRITTEN LICENSES OR
PERMISSIONS GRANTED BY ILLUMINA IN CONNECTION WITH CUSTOMER'S ACQUISITION OF
SUCH PRODUCT(S).
FOR RESEARCH USE ONLY
© 2011 Illumina, Inc. All rights reserved.
Illumina,illuminaDx,BeadArray,BeadXpress,cBot,CSPro,DASL,DesignStudio,Eco,GAIIx,
Genetic Energy,Genome Analyzer,GenomeStudio,GoldenGate,HiScan,HiSeq,Infinium,iSelect,
MiSeq,Nextera,Sentrix,Solexa,TruSeq,VeraCode, the pumpkin orange color, and the Genetic Energy
streaming bases design are trademarks or registered trademarks of Illumina, Inc. All other brands and
names contained herein are the property of their respective owners.
This software contains the SeqAn Library, which is licensed to Illumina and distributed under the
following license:
Copyright © 2010, Knut Reinert, FU Berlin, All rights reserved. Redistribution and use in source and
binary forms, with or without modification, are permitted provided that the following conditions are
met:
1 Redistributions of source code must retain the above copyright notice, this list of conditions and
the following disclaimer.
2 Redistributions in binary form must reproduce the above copyright notice, this list of conditions
and the following disclaimer in the documentation and/or other materials provided with the
distribution.
3 Neither the name of the FU Berlin or Knut Reinert nor the names of its contributors may be used
to endorse or promote products derived from this software without specific prior written
permission.
THIS SOFTWARE IS PROVIDED BY THE COPYRIGHT HOLDERS AND CONTRIBUTORS "AS IS"
AND ANY EXPRESS OR IMPLIED WARRANTIES, INCLUDING, BUT NOT LIMITED TO, THE
IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS FOR A PARTICULAR
PURPOSE ARE DISCLAIMED. IN NO EVENT SHALL THE COPYRIGHT HOLDER OR
CONTRIBUTORS BE LIABLE FOR ANY DIRECT, INDIRECT, INCIDENTAL, SPECIAL,
EXEMPLARY, OR CONSEQUENTIAL DAMAGES (INCLUDING, BUT NOT LIMITED TO,
PROCUREMENT OF SUBSTITUTE GOODS OR SERVICES; LOSS OF USE, DATA, OR PROFITS;
OR BUSINESS INTERRUPTION) HOWEVER CAUSED AND ON ANY THEORY OF LIABILITY,
WHETHER IN CONTRACT, STRICT LIABILITY, OR TORT (INCLUDING NEGLIGENCE OR
OTHERWISE) ARISING IN ANY WAY OUT OF THE USE OF THIS SOFTWARE, EVEN IF
ADVISED OF THE POSSIBILITY OF SUCH DAMAGE.

MiSeq System User Guide iii
Revision History
Part # Revision Date Description of Change
15027617 A August 2011 Initial release.
iv Part # 15027617 Rev. A

MiSeq System User Guide v
Table of Contents
Revision History iii
Table of Contents v
Chapter 1 Getting Started 1
Introduction 2
Components 4
Starting the MiSeq System 9
MiSeq Workflow 11
Illumina-Supplied Consumables 13
User-Supplied Consumables 15
Chapter 2 MiSeq Control Software 17
Introduction 18
MiSeq Control Software Interface 19
Stopping or Pausing a Run 29
Sample Sheets 30
Analysis Workflow 35
Run Folders 36
Available Disk Space 37
Chapter 3 Performing a Run 39
Introduction 40
Setting Up the Sample Sheet 41
Preparing the Reagent Cartridge 43
Preparing Your Libraries 44
Clean the Flow Cell 47
Loading the Flow Cell 49
Load Reagents 51
Starting the Run 53
Chapter 4 Post-Run Procedures 55
Introduction 56
Performing a Post-Run Wash 57
Unloading Components 59
Shutting Down the Instrument 61
Chapter 5 On-Instrument Analysis 63
Introduction 64
Primary Analysis Results 65
Secondary Analysis Using MiSeq Reporter 66
Using MiSeq Reporter Off-Instrument 68
Chapter 6 Troubleshooting 69
Getting Started
2Part # 15027617 Rev. A
Introduction
The Illumina MiSeq™ System combines proven sequencing by synthesis (SBS)
technology with a revolutionary workflow that enables you to go from DNA to analyzed
data in as few as eight hours. The MiSeq integrates cluster amplification, sequencing,
and data analysis in a single instrument.
Applications
The MiSeq supports a wide range of applications, including the following:
}De novo sequencing
}Targeted resequencing
}Small genome sequencing
}Highly-multiplexed PCR amplicon sequencing
}Small RNA sequencing
}Library QC
Library Preparation
You can sequence libraries prepared by any of the following Illumina sample
preparation kits:
}TruSeq™ DNA Sample Preparation Kit
}TruSeq™ RNA Sample Preparation Kit
}TruSeq™ Small RNA Sample Preparation Kit
}TruSeq™ Custom Amplicon Kit
}Nextera™ Sample Preparation Kit
For more information, see Applications and Related Consumables on page 13.
Features
}Walkaway Automation—All of the reagents required for cluster generation,
sequencing, and paired-end chemistry are contained in a single reagent cartridge.
After run setup, no additional hands-on time is required.
}Prefilled Reagent Cartridge—A specially designed single-use prefilled reagent
cartridge that provides reagents for cluster generation and sequencing, including
paired-end sequencing reagents. Radio-frequency identification (RFID) tracking
enables positive consumable tracking.
}Interface Controls—MiSeq Control Software (MCS) offers a simple interface to
configure and monitor your run, and perform maintenance procedures on a touch
screen monitor.
}Convenient Flow Cell Loading—A clamping mechanism auto-positions the flow
cell as you load it onto the instrument.
}Innovative Fluidics Architecture—Enables unmatched efficiency in chemistry cycle
time during sequencing.
}Real Time Analysis—Integrated analysis software performs real time on-instrument
data analysis during the sequencing run, which includes image analysis and base
calling, and saves valuable downstream analysis time.
Introduction
MiSeq System User Guide 3
Related Documentation
This guide provides comprehensive information about the MiSeq System. However,
other documentation is available for site preparation and safety guidelines, and as
quick reference guides.
The following list describes related documentation and availability.
}MiSeq System Site Preparation Guide—Provides specifications for laboratory space,
electrical requirements, and environmental considerations.
This guide is available for download from the Illumina website.
}MiSeq System Safety and Compliance Guide—Provides safety-related labeling and
considerations, and information about compliance certifications and labeling.
This guide is provided with the instrument and available for download from the
Illumina website.
}MiSeq System Quick Reference Guide—Provides an overview of the instrument, and
instructions for setting up and performing a run on the MiSeq. Use the quick
reference guide after you are familiar with the contents of the MiSeq System User
Guide.
This guide is provided with the instrument and available for download from the
Illumina website.
}MiSeq Reagent Preparation Guide—Provides a description of kit contents, and
instructions for preparing the reagent cartridge and sample libraries before
beginning your sequencing run.
This guide is provided with the MiSeq Reagent Kit and available for download
from the Illumina website.
}MiSeq Sample Sheet Quick Reference Guide—Provides information about sample
sheets that is included in the MiSeq System User Guide, compiled into a quick
reference guide.
This guide is available for download from the Illumina website.
Illumina Website
Visit the MiSeq support pages on the Illumina website
(http://www.illumina.com/systems/miseq/support.ilmn) for access to documentation,
related downloads, FAQs, and other support information.
Getting Started
4Part # 15027617 Rev. A
Components
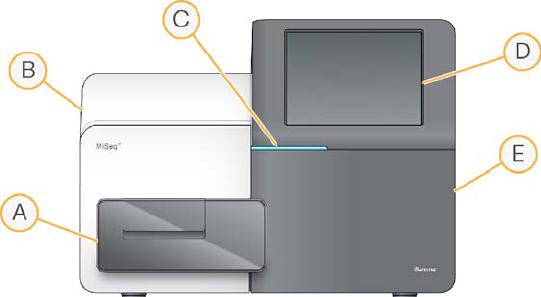
The MiSeq System has the following main components.
Figure 1 Main Compartments
AFlow Cell Compartment—Contains the flow cell stage that houses the flow cell
throughout the run. The flow cell stage is controlled by motors that move the stage
out of the enclosed optical module for flow cell loading, and returns it when the run
begins.
BEnclosed Optics Module—Contains optical components that enable imaging of the
flow cell.
CStatus Bar—Uses three colors to indicate instrument status. Blue indicates the
instrument is running, orange indicates the instrument needs attention, and green
indicates that the instrument is ready to begin the next run.
DTouch Screen Monitor—Enables easy run setup using the software interface and at-
a-glance run monitoring.
EReagent Compartment—Holds reagents used during runs, wash solutions used
during instrument washes, and the waste bottle. The reagent compartment door is
secured by a magnetic latch.
The MiSeq software interface guides you through the run setup steps using the touch
screen monitor. To load run components, you need to access the reagent compartment
and the flow cell compartment.
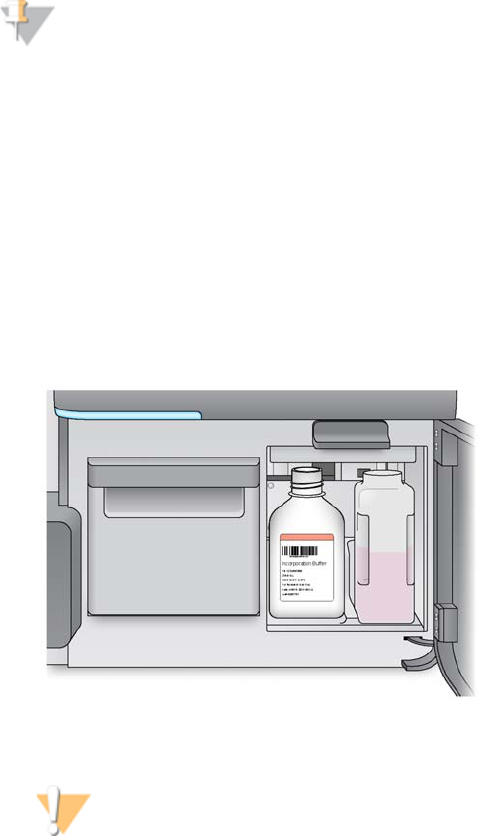
Reagent Compartment
The reagent compartment contains the reagent chiller, and positions for the
incorporation wash buffer (PR2) bottle and the waste bottle.
The reagent chiller holds a single-use reagent cartridge during the run, or a wash
rayduring the instrument wash. The software automatically lowers sippers into the
reagent wells of the cartridge at the appropriate time depending on the process being
performed.
Components
MiSeq System User Guide 5
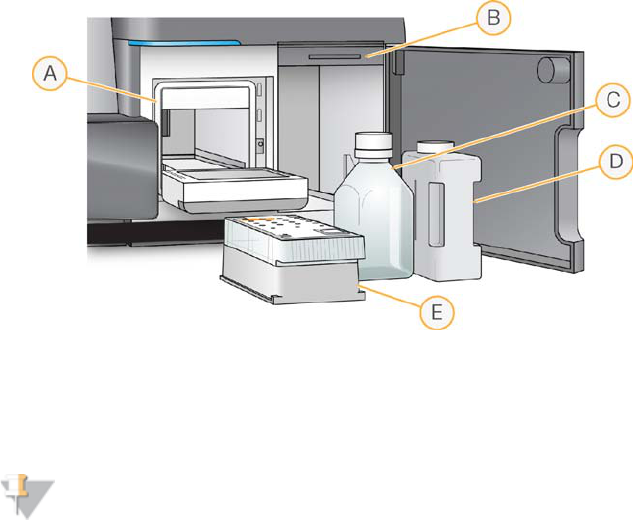
Figure 2 Reagent Compartment
AReagent Chiller
BSipper Handle (shown in raised position)
CReagent (PR2) Bottle
DWaste Bottle
EReagent Cartridge
NOTE
The reagent chiller is designed to maintain reagent temperature at 4°C. The chiller
is not intended to cool down reagents; therefore, reagents must be cool before
they are loaded onto the instrument.
To the right of the reagent chiller are two form-fitted slots, one for the PR2 bottle and
one for the waste bottle. After the bottles are in place, the sipper handle is lowered to
lock the bottles in place and lower the appropriate sipper into each bottle.
Fluidics pumps inside the instrument direct reagents through the sippers, through the
fluidics lines, and then through the flow cell. Reagent waste is delivered to the waste
bottle throughout the process.
Reagent Cartridge
The MiSeq reagent cartridge is a single-use consumable prefilled with cluster generation
and sequencing reagents.

Getting Started
6Part # 15027617 Rev. A
Figure 3 Reagent Cartridge
Each reservoir on the cartridge is numbered. The following table lists the reagent in
each numbered position.
Position Reagent Name Description
1 IMF Incorporation Mix
2 SRE Scan Mix
4 CMF Cleavage Mix
5 AMX1 Amplification Mix
6 AMX2 Read 2 Amplification Mix
7 LPM Linearization Premix
8 LDR Formamide
9 LMX1 Linearization Mix
10 LMX2 Read 2 Linearization Mix
11 RMF Resynthesis Mix
12 HP10 Read 1 Primer Mix
13 HP12 Index Primer Mix
14 HP11 Read 2 Primer Mix
15 PW1 Water
16 PW1 Water
17 Sample Your sample libraries
18 Optional Optional use for custom sequencing primers
19 Optional Optional use for custom sequencing primers
20 Optional Optional use for custom sequencing primers
21 PW1 Water
22 Empty Empty
Before starting a run, your sample libraries must be loaded onto the reagent cartridge in
the designated reservoir. You can sequence a single library or a pool of indexed
libraries.
Components
MiSeq System User Guide 7
Flow Cell Compartment
The flow cell compartment houses the flow cell stage, thermal station, and fluidics
connections to the flow cell.
The flow cell is positioned on the flow cell stage, which moves in and out of the optics
module. The flow cell is auto-positioned on the flow cell holder and held in place by the
flow cell latch. When the flow cell latch closes, two pins near the latch hinge position
the flow cell.
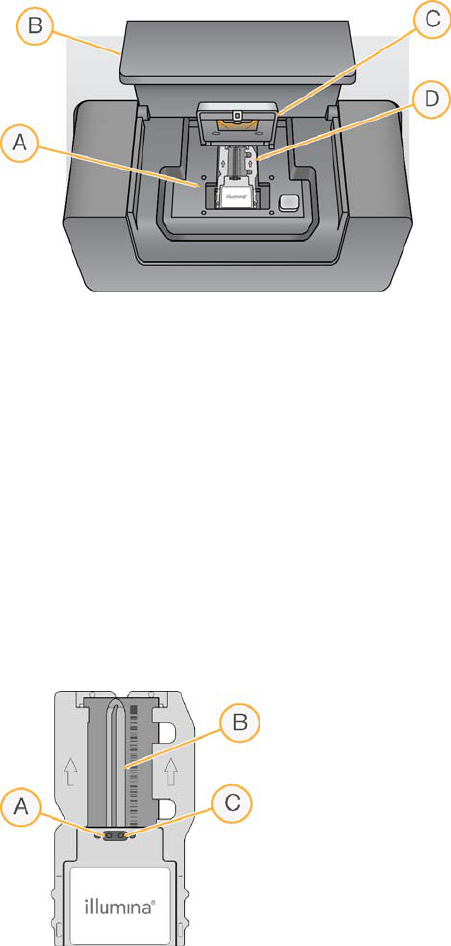
Figure 4 Flow Cell Stage
AFlow Cell Stage
BFlow Cell Compartment Door
CFlow Cell Latch
DFlow Cell
The thermal station, located beneath the flow cell stage, controls changes in flow cell
temperature that are required for cluster generation and sequencing.
Fluidics connections deliver reagents to the flow cell through the inlet port, and return
waste from the flow cell through the outlet port.
Flow Cell
The MiSeq flow cell is a single-lane glass-based substrate in which clusters are
generated and the sequencing reaction is performed. The single lane is divided into
twelve image areas, called tiles, that are imaged by the instrument camera.
Figure 5 Flow Cell
AOutlet Port
BImaging Area
CInlet Port
Getting Started
8Part # 15027617 Rev. A
Samples are loaded onto the reagent cartridge and then automatically transferred to the
flow cell at the beginning of the cluster generation process.
Reagents enter the flow cell through the inlet port, pass through the imaging area, and
then exit the flow cell through the outlet port. Waste exiting the flow cell is delivered to
the waste bottle.
Starting the MiSeq System
MiSeq System User Guide 9
Starting the MiSeq System
Use the following instructions to start the MiSeq and instrument computer.
1Reach around the right side of the instrument to locate the power switch on the
back panel. It is in the lower corner directly above the power cord.
Figure 6 Power Switch Location
2Turn the power switch to the ON position. The integrated instrument computer
starts.
3Log on to the operating system using the default user name. Wait until the
operating system has completely loaded.
•User name: sbsuser
•Password: sbs123
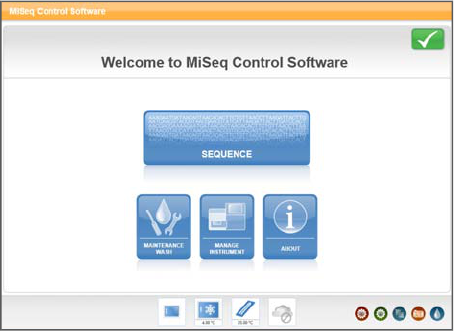
When the system is ready, the MCS (MCS) launches and begins initialization.
Figure 7 MCS Initialization
After initialization is complete, the Welcome screen appears.
Getting Started
10 Part # 15027617 Rev. A
Figure 8 MCS Welcome Screen
4To ensure adequate disk space, archive the data on the instrument computer from
all previous runs to a network location. For more information, see Manage Files on
page 22.
Best Practice
It is best to leave the instrument on at all times. Turn off the instrument only if it will
remain idle for more than ten days. For more information, see Shutting Down the
Instrument on page 61.
If you need to turn off the instrument, wait a minimum of 60seconds before turning the
power switch back to the ON position.

MiSeq Workflow
MiSeq System User Guide 11
MiSeq Workflow
The MiSeq workflow consists of run setup steps, followed by automated cluster
generation, then sequencing and primary analysis, and finally on-instrument secondary
analysis.
The following steps are described in detail in Performing a Run on page 39.
Setting Up Your Run
Create a sample sheet containing parameters
specific to your run.
Prepare the prefilled reagent cartridge for use.
Dilute and denature your libraries, and then load
your libraries onto the reagent cartridge in the
designated reservoir.
From the MCS interface, select Sequence to start the
run set up steps.
Load the flow cell.
Load the PR2 bottle and make sure that the waste
bottle is empty.
Load the reagent cartridge.
Review run parameters and pre-run check results.
Select Start Run.
Monitor your run from the Sequencing screen or on
another computer using Sequencing Analysis
Viewer (SAV).
Cluster Generation
After the run is started, single DNA molecules are bound to the surface of the flow cell,
and then bridge-amplified to form clusters.

Getting Started
12 Part # 15027617 Rev. A
Sequencing
Following cluster generation, clusters are imaged using LED and filter combinations
specific to each of the four fluorescently-labeled dideoxynucleotides. After imaging of
one tile of the flow cell is complete, the flow cell is moved into place to expose the next
tile and the process is repeated. Following image analysis, Real Time Analysis (RTA)
launches to perform basecalling, filtering, and quality scoring.
Analysis
When the run is complete, the MiSeq Reporter analysis software launches automatically
and begins secondary analysis. You can monitor secondary analysis using an internet
connection from another computer.
For more information, see Secondary Analysis Using MiSeq Reporter on page 66.
NOTE
If a new sequencing run is started on the MiSeq before secondary analysis is
complete, the current analysis is stopped. From the MiSeq Reporter interface, you
can requeue secondary analysis for that run after the new sequencing run is
complete. At that point, secondary analysis starts over from the beginning.
Alternatively, MiSeq Reporter can be run off-instrument allowing secondary analysis to
occur during a subsequent sequencing run. For more information, see Using MiSeq
Reporter Off-Instrument on page 68.
Approximate Run Duration
Run duration depends on the number of cycles you perform. You can perform a paired-
end sequencing run up to 2x150 cycles, or as few as 36 cycles for some applications.
Generally, one cycle takes about five minutes. The following table lists the approximate
durations for cluster generation, sequencing, and on-instrument analysis.
Process Read Length Duration
Cluster
Generation
and
Sequencing
36 Cycles (36 bp) Less than 4 hours
50 Cycles (50 bp) Less than 6 hours
Paired-End 150
Cycles (2x150 bp)
Less than 27 hours
On-Instrument
Analysis
and Variant
Calling
Paired-End 150
Cycles (2x150 bp)
Less than 2 hours for all analysis workflows,
except 16s metagenomics.
16s metagenomics completes in less than 24
hours.

Illumina-Supplied Consumables
MiSeq System User Guide 13
Illumina-Supplied Consumables
To perform a run on the MiSeq, you need one MiSeq Reagent Kit.
MiSeq Reagents
The MiSeq Reagent Kit is available in the following sizes:
}300 Cycles—Provides kitted reagents for up to 323 cycles of sequencing on the
MiSeq. This includes sufficient reagents for a 151-cycle paired-end run, plus two
eight-cycle indexed reads.
}50 Cycles—Provides kitted reagents for up to 73 cycles of sequencing on the MiSeq.
This includes sufficient reagents for a 26-cycle paired-end run, or a 51-cycle single-
read run, plus two eight-cycle indexed reads.
MiSeq Reagent Kit Contents
Box1: Store at -15° to -25°C
Quantity Component Storage Description
1Reagent Cartridge -15° to -25°C Single-use pre-filled cartridge
1HT1 -15° to -25°C 5 ml tube, Hybridization Buffer
Box2: Store at 2° to 8°C
Quantity Component Storage Description
1PR2 2° to 8°C 500 ml bottle, Incorporation
Buffer
1Flow Cell 2° to 8°C Single-use flow cell
Applications and Related Consumables
The following table lists applications supported on the MiSeq, the recommended read
length for the run, and the associated Illumina-supplied consumables.
Application Read
Length
MiSeq
Reagent Kit
(# of cycles)
Sample Preparation Kit
TruSeq Custom
Amplicon
2 x 151 300 Cycle Kit TruSeq Custom Amplicon
Library Prep
Nextera Amplicon 1 x 36 50 Cycle Kit Nextera Sample Prep
Standard Amplicon 2 x 151 300 Cycle Kit TruSeq DNA Sample Prep
Small Genome: De novo 2 x 151 300 Cycle Kit TruSeq DNA Sample Prep
Small Genome:
Resequencing
1 x 36 50 Cycle Kit or
300 Cycle Kit
TruSeq DNA Sample Prep or
Nextera Sample Prep
16S Metagenomics 2 x 151 300 Cycle Kit TruSeq DNA Sample Prep or
Nextera Sample Prep

Getting Started
14 Part # 15027617 Rev. A
Application Read
Length
MiSeq
Reagent Kit
(# of cycles)
Sample Preparation Kit
LibraryQC 2 x 25 50 Cycle Kit Open
Small RNA 1 x 36 50 Cycle Kit TruSeq Small RNA Sample Prep
RNA-Seq 1 x 51 50 Cycle Kit TruSeq RNA Sample Prep
TruSeq Custom
Enrichment
2 x 76 300 Cycle Kit TruSeq DNA Sample Prep
User-Supplied Consumables
MiSeq System User Guide 15
User-Supplied Consumables
Make sure that you have all of the user-supplied consumables listed in this section
before you begin your run.
Consumable Supplier Purpose
1 N NaOH General lab supplier Denaturing sample libraries
Diluting PhiX control
Alcohol wipes, 70%
Isopropyl
or
Ethanol, 70%
VWR, catalog#15648-
981
General lab supplier
Cleaning the flow cell holder
Disposable gloves, powder-
free
General lab supplier General use
Lab tissue, low-lint VWR, catalog#21905-
026
Cleaning the flow cell stage
Lens paper, 4x6in. VWR, catalog#52846-
001
Cleaning the flow cell
Tris-Cl 10mM, pH8.5 with
0.1% Tween 20
General lab supplier Diluting PhiX control
Tweezers, square-tip plastic McMaster-Carr,
catalog # 7003A22
Removing flow cell from flow
cell container
Water, laboratory-grade General lab supplier Washing the instrument
Table 1 User-Supplied Consumables
Guidelines for Laboratory-Grade Water
The following list provides acceptable examples of laboratory-grade water. Never use
tap water or deionized water on the instrument.
}Illumina PW1
}18M Ohm water
}Milli-Q water
}Super-Q water
}Molecular biology-grade water
16 Part # 15027617 Rev. A
MiSeq Control Software
18 Part # 15027617 Rev. A
Introduction
The MiSeq Control Software (MCS) interface guides you through the steps to load the
flow cell and reagents prior to starting the run, and then provides an overview of
quality statistics that you can monitor as the run progresses.
During the run, MCS operates the flow cell stage in the optical compartment, gives
commands to dispense reagents, changes temperatures of the flow cell, and captures
images of clusters on the flow cell for image analysis. MCS performs the run based on
parameters specified in the sample sheet. For more information, see Sample Sheets on
page 30.
Real Time Analysis (RTA)
After image analysis is complete, the integrated primary analysis software, Real Time
Analysis (RTA), performs base calling and assigns a quality score to each base for each
cycle. Quality scores can be monitored throughout the run on the MCS Sequencing
screen. For more information, see Sequencing Screen on page 27.
Images are temporarily stored in the run folder for processing by RTA, and then
automatically deleted when RTA analysis is complete.
Sequencing Analysis Viewer (SAV)
To monitor your run in greater detail from another computer, you can use the
Sequencing Analysis Viewer (SAV), an optional software application available for
download from the Illumina website. For more information, see Sequencing Analysis
Viewer on page 28.
MiSeq Control Software Interface
MiSeq System User Guide 19
MiSeq Control Software Interface
The MiSeq Control Software (MCS) includes an intuitive interface that prompts you
through the run setup steps, instrument setup options, and instrument washes. The
interface includes a Sequencing screen that provides information about the run as it
progresses.
Welcome Screen
The interface opens to the Welcome screen when the software launches, and returns to
the Welcome screen when a run is complete.
Figure 9 MCS Welcome Screen
The Welcome screen has the following options:
}Sequence—This option opens a series of run setup screens that guide you through
the setup steps for your run. For more information, see Run Setup Screens on page 24.
}Maintenance Wash—Use this option to perform an instrument wash before a
sequencing run.
}Manage Instrument—Provides links to Setup Options,Manage Runs, and Perform
Diagnostic. For more information, see Manage Instrument on page 20.
}About—Provides information about instrument configuration and software
versions.
}Get Updates—This option appears on the Welcome screen only if a software update
is available. Your MiSeq must be connected to a network to enable this option. For
more information, see Get Updates on page 20.
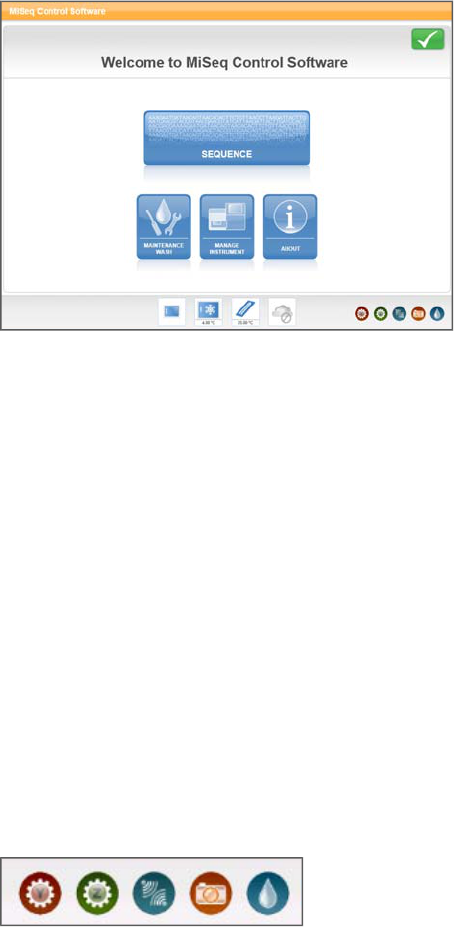
Activity Indicators
In the lower-right corner of the Welcome screen are a series of icons. Each icon is an
activity indicator that tells you which activity the instrument is performing. These
appear throughout the interface and represent the Y-stage motor, Z-stage motor,
electronics functionality, camera, and fluidics system.
Figure 10 Activity Indicators
MiSeq Control Software
20 Part # 15027617 Rev. A
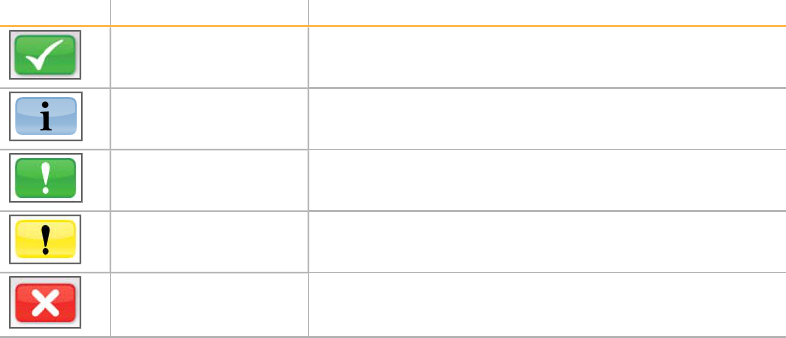
Status Icons
In the top-right corner of the Welcome screen is a status icon that notifies you of any
change in conditions during run setup or during the run.
Status Icon Status Name Description
Status OK No change. System is normal.
Information Information only. No action is required.
Attention Information that might require attention.
Warning Warnings do not stop a run, but might require action
before proceeding.
Error Errors usually stop a run and generally require action
before proceeding with the run.
When a change in condition occurs, the icon changes to the associated image and
blinks to alert you. If this happens, select the icon to open the status window, which
contains a general description of the condition.
}Select any item listed to see a detailed description of the condition and instructions
to resolve the condition, if applicable.
}Select Acknowledge to accept the message and Close to close the dialog box.
You can filter the types of messages that appear in the status window by selecting the
icons along the top margin of the window. Selecting an icon toggles the condition to
show or to hide.
Get Updates
You can update the instrument software from the MCS Welcome screen using the Get
Updates feature. Your system must be connected to a network with internet access to
download updates.
When software updates are available, the Get Updates button appears on the Welcome
screen. Select Get Updates. A dialog box opens asking you to confirm the command,
after which a reboot of the MiSeq is required. Installation of the update begins
automatically upon reboot.
The Get Updates button appears on the Welcome screen only when updates are
available.
Manage Instrument
The Manage Instrument screen contains controls for setting up folder locations and
wash preferences, moving and deleting files, managing recipe files and genome
reference files, and performing an instrument diagnostic.

MiSeq Control Software Interface
MiSeq System User Guide 21
Figure 11 Manage Instrument Screen
}Setup Options—Provides options for changing default locations of data folders. For
more information, see Setup Options on page 21.
}Manage Files—Provides controls for moving and deleting files on the instrument
computer. For more information, see Manage Files on page 22.
}System Settings—Provides the option to change IP Settings, machine name, or
domain. For more information, see System Settings on page 23.
}Perform Diagnostic—Provides instrument diagnostic options for use by an Illumina
Technical Support representative during a Live Help session. Use of this feature is
not required during normal operation or for instrument maintenance.
}Live Help—Connects your instrument to Illumina Technical Support, allowing a
representative to access your instrument computer for troubleshooting. You must
have a network connection to use this feature. For more information, see Live Help
on page 23.
Setup Options
From Setup Options, you can set the default folder locations for recipes, sample sheets,
manifests, and the output folder.
Figure 12 Setup Options
MiSeq Control Software
22 Part # 15027617 Rev. A
Folder Locations
}Recipes—Sets the default location for recipes. Recipes are XML files that the
software uses to perform the sequencing run. A recipe is created at the start of the
run based on parameters that you provide in the sample sheet. After a run-specific
recipe is created, it is copied to the run folder. If you pause the run and resume it
later, this recipe restarts where the run was paused.
}Sample Sheets—Sets the default location for sample sheets. Sample sheets are files
containing run parameters that are copied onto the instrument before you start your
run.
For more information, see Sample Sheets on page 30.
}Manifests—Sets the default location for manifests. Manifests are files used only
with custom amplicon sequencing. They contain the amplicon sequences to which
clusters are aligned during analysis. Manifests are provided when you order your
custom assay (CAT). There is no need to modify the manifest.
}MiSeqOutput—Sets the default location for the output files. Illumina recommends
that you change the default output folder location to a network location.
For more information, see Run Folders on page 36.
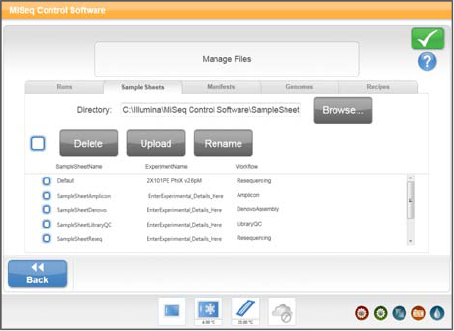
Manage Files
Use the features on the Manage Files screen to move, upload, or delete files on the
instrument computer. The screen is divided into five tabs: Runs, Sample Sheets,
Manifests, Genomes, and Recipes.
Figure 13 Manage Files Screen
}Browse—Select Browse to point to the appropriate output folder or folders other
than the default folders containing sample sheets, manifests, genomes, or recipes.
}Delete—Select the checkbox next to the file or folder listed, and then select Delete.
The Delete feature is available on all tabs.
}Move—Available only for runs. Select the checkbox next to the folder, and then
select Move. A window opens and prompts you to browse to an appropriate
location.
}Select All Files—Select the checkbox to the left of the Delete button, and then select
an action, Delete or Move. The action is applied to all files or folders selected.
MiSeq Control Software Interface
MiSeq System User Guide 23
}Upload Files—Available for sample sheets, manifests, genomes, and recipes. Select
Upload. A window opens that prompts you to browse to a location where the file
resides. The file is uploaded to the folder indicated in the Directory field.
}Rename—Available only for sample sheets. Select the checkbox next to the file, and
then select Rename. Use the online keyboard to rename your sample sheet.
Available Features By Tab
Tab Features
Runs Delete or Move
Sample Sheets Delete, Upload, or Rename
Manifests Delete or Upload
Genomes Delete or Upload
Recipes Delete or Upload
System Settings
System Settings are normally configured when your instrument is initially installed and
started for the first time. If you need to change your settings due to a network or facility
change, use the System Settings feature.
Figure 14 System Settings
System Settings configure the following components:
}IP address and Domain Name Server (DNS) address
}Machine name
}Domain name
}MiSeq Reporter Service credentials
Contact your local IT representative to get information on your network settings.
Live Help
The Live Help feature is an online assistance tool that enables a member of Illumina
Technical Support to view your screen with your permission, and share control of your
instrument. You have overriding control and can end the screen-sharing session at any
time.
To enable a connection, you must obtain a unique connection code from Illumina
Technical Support.

MiSeq Control Software
24 Part # 15027617 Rev. A
Figure 15 Live Help Screen
1Contact Illumina Technical Support to obtain a unique access code.
2Enter your unique access code using the online keyboard.
3Select Next to begin your screen-sharing session.
Run Setup Screens
When you select Sequence on the Welcome screen, a series of four run setup screens
open in the following order: Load Flow Cell, Load Reagents, Review, and Pre-Run
Check.
Set Cloud Option
The first screen in the run setup steps prompts you to log onto the Cloud. Use the Cloud
to store data analysis files. To enable access, you need a network connection and an
iCom account.
Figure 16 Cloud Option Screen
Using the Cloud feature is optional. If you do not want use it, clear the checkbox on the
screen.
MiSeq Control Software Interface
MiSeq System User Guide 25
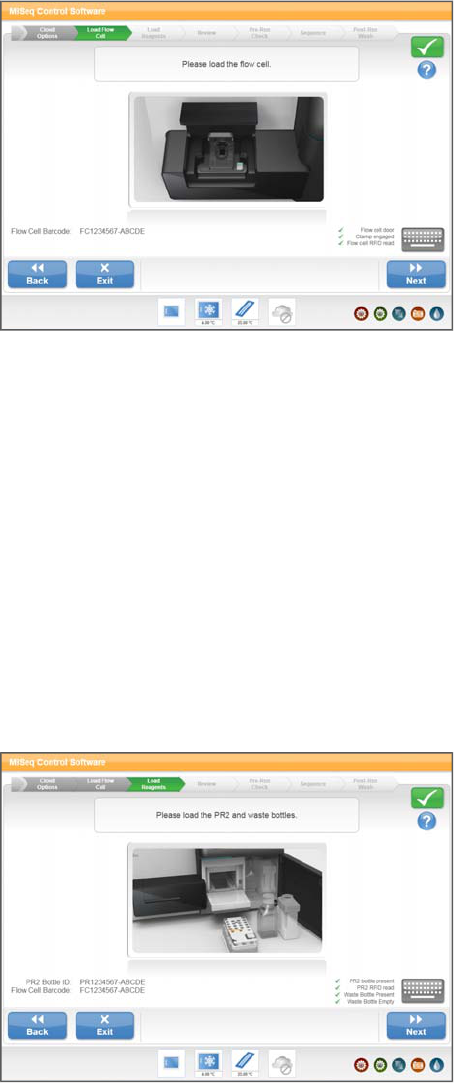
Load Flow Cell
The Load Flow Cell screen prompts you to load the flow cell. When the flow cell is
loaded, the software reads the RFID of the flow cell. A confirmation that the RFID was
successfully read appears in the lower-right corner of the screen.
Figure 17 Load Flow Cell Screen
If for some reason the RFID is not read, select the online keyboard icon and manually
enter the flow cell ID.
After the flow cell is loaded, close the flow cell latch and flow cell compartment door
before proceeding. Both the latch and compartment door must be closed before
beginning the run.
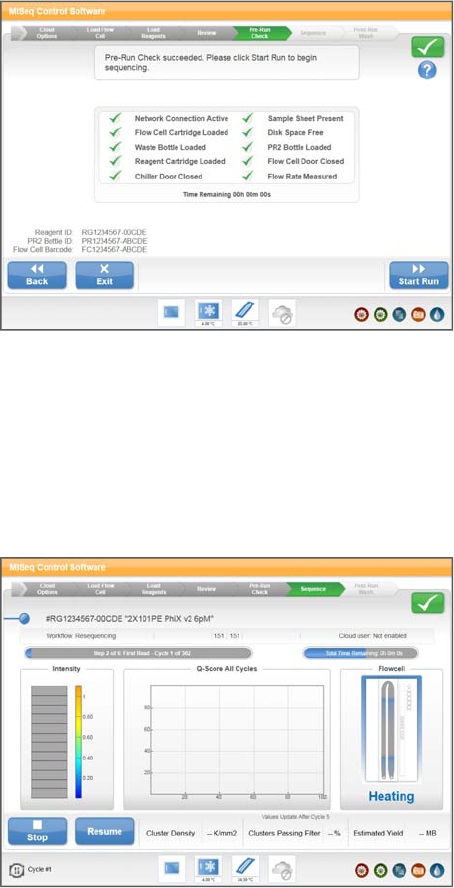
Load Reagents
The Load Reagents screen has two steps: first, you load the PR2 bottle and make sure
that the waste bottle is empty, and then you load the reagent cartridge. When you load
the PR2 bottle and reagent cartridge, the software reads the RFID of the components. A
confirmation that the RFID was successfully read appears in the lower-right corner of
the screen.
If for some reason the RFID is not read, select the online keyboard icon and manually
enter the barcode number located below the barcode on the component label.
Figure 18 Load PR2 Bottle and Waste Bottle

MiSeq Control Software
26 Part # 15027617 Rev. A
Figure 19 Load Reagent Cartridge
Change Sample Sheet
Every run must have a unique sample sheet for the run. If the software cannot find an
appropriately named sample sheet, a message appears that asks you to browse to the
location of your sample sheet.
From the Load Reagents screen, you can use Change Sample Sheet to specify a
different sample sheet for your run. A screen opens that allows you to browse to a
location where you stored the sample sheet.
Review
The Review screen confirms that the software has located the sample sheet for your run.
The Review screen lists the sample sheet name, and the following parameters provided
in the sample sheet:
}Experiment name
}Analysis workflow
}Read length
Figure 20 Review Screen
MiSeq Control Software Interface
MiSeq System User Guide 27
Change Folders
In the lower-left corner of the Review screen, the current folder locations for recipes,
sample sheets, manifests, and output folders are listed. If you want to change folder
locations, select Change Folders, and browse to your preferred locations. Using this
option from the Review screen changes folder locations for the current run only.
Pre-Run Check
The software automatically performs a pre-run check to ensure that required run
components and conditions are satisfied before allowing the run to begin. If any errors
occur during the pre-run check, a message appears on the screen telling you where the
error occurred and what action is needed to correct it. For more information, see Software
Errors on page 71.
Figure 21 Pre-Run Check
When the pre-run check is successful, the Start Run button becomes active. You are
ready to start your run.
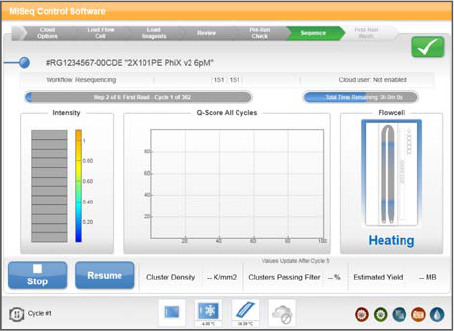
Sequencing Screen
The Sequencing screen provides a visual representation of run progress, intensities, base
call quality scores (Q-scores), and activity on the flow cell.
Figure 22 Sequencing Screen

MiSeq Control Software
28 Part # 15027617 Rev. A
}Run Progress—Shows run progress in a status bar and lists the number of cycles
completed.
}Intensities—Shows the value of cluster intensities of 90% of data for each tile.
}Q-Score All Cycle—Shows the average percentage of bases greater than Q30.
A quality score (Q-score) is a prediction of the probability of an error in base calling.
Q-Score Chance of Wrong Base Call
Q40 1 error in 10,000
Q30 1 error in 1,000
Q20 1 error in 100
Q10 1 error in 10
}Flow Cell—Shows the current temperature of the flow cell indicated by color. Blue
indicates cooler temperatures, while orange and red indicate warmer temperatures.
During imaging, imaged tiles appear dark gray. During chemistry, wavy lines
indicate that reagents are pumping through the flow cell.
NOTE
The MiSeq is sensitive to vibration. Vibrations occurring during a run will
adversely impact sequencing results. Do not touch the instrument or touch screen
monitor during normal run operations unless you plan to stop or pause your run.
Sequencing Analysis Viewer
You can monitor your run in greater detail using the Sequencing Analysis Viewer, a
downloadable software application that is installed onto a computer independent of the
MiSeq, but with access to the same network connected to the instrument. After
launching the software, you can browse to the run folder for your run.
Real Time Analysis (RTA) uses the first four cycles of the sequencing run for template
generation. Template generation is the process by which cluster positions over the entire
flow cell surface are defined along the X and Y axes. After the template of cluster
positions has been generated, the images produced over every subsequent cycle of
imaging are aligned against the template so that individual cluster intensities in all four
nucleotide color channels can be extracted, and basecalls produced from the normalized
cluster intensities.
After cycle 4, the Sequencing Analysis Viewer provides metrics generated by RTA and
organizes the metrics into plots, graphs, and tables. Your MiSeq must be networked to
use Sequencing Analysis Viewer.
NOTE
Sequencing Analysis Viewer is universal to Illumina sequencers, most of which
use an eight-lane flow cell. Some views include drop-down lists showing lanes 1–
8. The MiSeq flow cell is a single-lane flow cell, so your data appears when you
select All or Lane 1.
For more information, see the Sequencing Analysis Viewer User Guide.

Stopping or Pausing a Run
MiSeq System User Guide 29
Stopping or Pausing a Run
The MiSeq is designed to complete a run from beginning to end without user
intervention. However, it is possible to stop a run or pause a run from the Sequencing
screen.
}Stop a run—You might stop a run if the run was set up incorrectly, if the data
quality is bad, or if you experience a hardware error.
}Pause a run—You might pause a run if you suspect that the waste bottle is full or
to check the volume in the PR2 bottle.
Stopping Run
You can stop a run before it has completed using the Stop button on the Sequencing
screen.
Stopping a run is final. You cannot resume a run after it was stopped using the Stop
button. When a run is stopped, the flow cell tray moves to the forward position. You are
ready to unload run components and perform an instrument wash.
Pausing a Run
You can temporarily interrupt a run before it has completed using the Pause button on
the Sequencing screen. When you select Pause, the current command is completed, after
which the run is paused and the flow cell is placed in a safe state.
Best Practice
Empty the waste bottle before each run and make sure that run consumables are
properly loaded before starting the run.
NOTE
On a paused run, the flow cell is stored in safe state for a maximum of three days.
After three days, the instrument flow cell temperature control will turn off.

MiSeq Control Software
30 Part # 15027617 Rev. A
Sample Sheets
The sample sheet is a comma separated values (*.csv) file that stores much of the
information needed to set up, perform, and analyze a sequencing run.
During the run setup steps, the software automatically looks for a sample sheet
associated with reagent cartridge loaded onto the instrument in the previous run setup
step. Therefore, Illumina recommends that you name your sample sheet with the
barcode number of the reagent cartridge that you will use for your run, followed by *.csv
extension. The barcode number is located on the cartridge label directly below the
barcode.
Before starting your run, copy your sample sheet to the instrument computer or to the
network location indicated in Setup Options. When your run begins, the software copies
your sample sheet to the root of your run folder.
At the end of your run, the sample sheet is used for secondary analysis by the MiSeq
Reporter analysis software.
Sample Sheet Template
You can open and edit the sample sheet template in Excel or in Notepad. A sample
sheet template is available for download from the Illumina website at
http://www.illumina.com. Go to the MiSeq support page and click Downloads. An
iCom account is required.
NOTE
Do not use commas within a field in the sample sheet. If a comma is included
within a field, your sample sheet will not be read correctly by the software.
Illumina Experiment Designer
You can use the Illumina Experiment Designer to create your sample sheet, a wizard-
based application for creating the sample sheet. The Experiment Designer guides you
through the steps to create your sample sheet based on the analysis workflow for your
run.
The Experiment Designer provides a feature for recording parameters for your sample
plate, such as sample ID, dual indices, and other parameters applicable to your 96-well
plate for Nextera, TruSeq DNA, TruSeq RNA, Small RNA, and Amplicon library
preparation protocols. Using the Experiment Designer, you can import the sample plate
parameters into your sample sheet. For more information, see the Experiment Designer
Online Help integrated with the application.
The Illumina Experiment Designer can be run on any Windows platform. You can
download the Experiment Designer from the Illumina website at
http://www.illumina.com. Go to the MiSeq support page and click Downloads. An
iCom account is required.
Sample Sheet Parameters
The sample sheet is organized by section text indicated by [ ] brackets, such as [Header],
[Reads], [Manifests], and [Data]. Section text within brackets is case-sensitive. All other
fields are not case-sensitive.
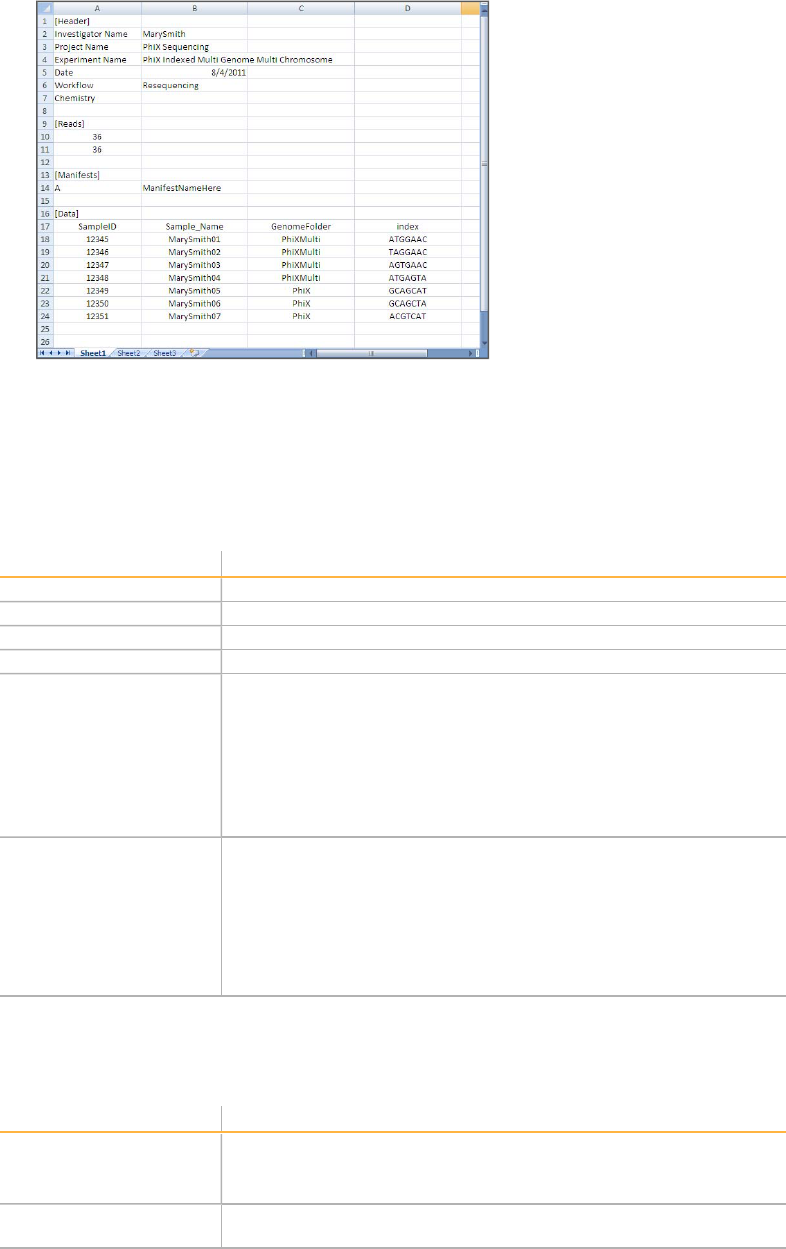
Sample Sheets
MiSeq System User Guide 31
Figure 23 Sample Sheet Example in Excel
Not all sections of the sample sheet are applicable to every run. Applicable sections
depend on the analysis workflow used for your run.
[Header]
The Header section of the sample sheet contains information about your run. Extra
fields are ignored.
Parameter Description
Investigator Name Your name
Project Name Project name of your preference
Experiment Name Experiment name of your preference
Date Date of your experiment
Workflow The analysis workflow for your run.
You must type the analysis workflow name exactly as shown using
one of the following options: Resequencing, Amplicon,
DenovoAssembly, SmallRNA, Metagenomics, or LibraryQC.
If you are using the Experiment Designer, select your workflow
from the drop-down menu.
For more information, see Analysis Workflow on page 35.
Chemistry The folder that contains recipe fragments used to build the recipe. If
this field is blank, the system uses the default recipe fragments.
For TruSeq RNA or TruSeq DNA libraries, this field can be blank
and use the default settings.
For any workflows that use dual indexing, specifically Nextera and
TruSeq Custom Amplicon, the chemistry field is required. Enter
"amplicon" in this field.
[Reads]
Reads indicates the number of sequencing cycles in each read.
Parameter Description
Number of cycles for
Read 1
Read length of a target sequence.
The number of cycles for the index read is indicated by the index
sequence the [Data] section.
Number of cycles for
Read 2
Read length of a target sequence.

MiSeq Control Software
32 Part # 15027617 Rev. A
[Manifests]
This section of the sample sheet is mandatory for the Amplicon analysis workflow. You
do not need to populate this section if you are using other analysis workflows.
Parameter Description
Name of manifest file Name of your manifest file provided by Illumina with your custom
assay (CAT). The manifest file must reside in the same folder
location as the sample sheet.
When you enter your manifest name, omit the file extension (*.txt).
More than one manifest can be specified: one manifest for your
custom CAT, and one manifest for the control CAT.
[SiteReports]
This section of the sample sheet is used by the Resequencing and Amplicon analysis
workflows.
Parameter Description
Name of SiteReport input
file
Each line in the SiteReports section is the name of a SiteReport input
file. This file designates positions on a given chromosome to report
the genotype found at that position.
You can use more than one input file for a run. The SiteReport input
file must reside in the same folder location as the sample sheet.
The SiteReport input file is a comma-separated value (*.csv) format
file that must adhere to the following format:
•The first line are column names. Required columns are Position
and Chromosome.
•Additional user-defined columns are appended to the resulting
analysis report for that position.
When you enter your SiteReport input file name, omit the file
extension (*.csv).
[Data]
In this section, list sample information one sample per line. There must be at least one
sample in this section. Each analysis workflow requires different columns in this
section. Column order is not important.
Parameter Description
SampleID Required. This is usually a barcode but can have any value.
Sample_Name Required. This is used in the reporting web page.
Index Required for multi-sample assays.
Nucleotide sequence in text format. Valid characters are A, C, G, T,
and N, where N matches any base.
Index2 Required for multi-sample assays with dual indexing.
Nucleotide sequence in text format. Valid characters are A, C, G, T,
and N, where N matches any base.
Additional [Data] Columns for Resequencing Workflow
Parameter Description
GenomeFolder Required.
The reference genome folder containing the FASTA files to be used
in the alignment step.

Sample Sheets
MiSeq System User Guide 33
Additional [Data] Columns for Amplicon Workflow
Parameter Description
GenomeFolder Recommended.
It is used to provide variant annotations, if available, and to set the
chromosome sizes in the *.bam file output.
Additional [Data] Columns for DenovoAssembly Workflow
Parameter Description
GenomeFolder
(or Genome)
Optional.
If you populate this field, list the absolute path to the genome folder.
•If the folder listed does not exist, MiSeq Reporter combines the
GenomePath configuration setting with the genome string.
•If the path does not exist, MiSeq Reporter stops processing.
Additional [Data] Columns for SmallRNA Workflow
Parameter Description
GenomeFolder Optional.
A string identifier for the sample. This is usually a barcode but can
have any value. If provided, clusters are aligned against the full
reference genome.
Contaminants String; required.
The path to the folder containing the FASTA files of contaminants.
miRNA Required.
The path to the folder containing FASTA files of mature miRNAs.
RNA Required.
The path to folder containing FASTA files of small RNAs.
Additional [Data] Columns for Library QC Workflow
Parameter Description
GenomeFolder Required.
The reference genome folder containing the FASTA files to be used
in the alignment step.
[Settings]
Settings are optional in the sample sheet. This section describes possible entries in the
Settings section.
Parameter Description
FilterPCRDuplicates Setting are 0 or 1. Default is 1, no filtering.
If set to 0, PCR duplicates are filtered out from subsequent analysis.
Duplicates are defined as paired-end reads where both reads have
the exact same alignment positions.
MinQScore Specifies the Q-score threshold for variant calling.
OnlyGenerateFastq Setting are 0 or 1. Default is 0.
If set to 1, MiSeq Reporter generates FASTQ files and then exits the
workflow. This option is provided as a shortcut if you prefer to use
third-party tools to process FASTQ files.

MiSeq Control Software
34 Part # 15027617 Rev. A
Parameter Description
Adapter It is possible that some clusters will sequence beyond the sample
DNA and read bases from the adapter.
Specify the adapter sequence here to mask the adapter sequence.
Masking the adapter sequence improves accuracy and speed during
secondary analysis.

Analysis Workflow
MiSeq System User Guide 35
Analysis Workflow
The analysis workflow is a procedure performed by MiSeq Reporter, the secondary
analysis software on the MiSeq. Secondary analysis begins after the Real Time Analysis
(RTA) software has completed base call analysis.
For more information, see On-Instrument Analysis on page 63.
The MiSeq supports six analysis workflows. The analysis workflow for your run is
specified in the sample sheet. When you enter the analysis workflow in the sample
sheet, enter the name as shown below in the Workflow column.
Workflow Applications Output
Targeted Resequencing
Enter: Resequencing
Sequencing of an enriched portion of
the human genome, or of a small
genome such as E. coli.
Reads are aligned against the reference
and variants are noted.
Aligned reads in
*.bam format.
Variant calls in *.vcf
format.
Amplicon Sequencing
Enter: Amplicon
Sequencing of PCR amplicons from
probes targeting particular genome
positions (up to approx. 384 loci from
up to approx. 96 samples).
A genomic reference is recommended,
but not required. Including a reference
means that the genome size in the
*.bam file headers is correct, and
enables annotation of variants in dbSNP
or within genes.
Reads are aligned against a manifest file
specified in the sample sheet. Two
manifests can be specified: one for the
control and one for the sample.
Aligned reads in
*.bam format.
Variant calls in *.vcf
format.
De novo Assembly
Enter: DenovoAssembly
Assembly of small (<20 Mb) genomes
from reads without the use of a
genomic reference.
If a genomic reference is included, a dot-
plot is generated with respect to the
reference.
Contigs in FASTA
format.
Small RNA Sequencing
Enter: SmallRNA
Sequencing of miRNA.
Requirements include a genomic
reference, a contaminants database, an
RNA database, and a mature miRNA
database.
Reports on the
relative abundance
of each record.
Metagenomics
Enter: Metagenomics
No genomic reference is required for
metagenomics workflow. Reads are
classified using a database of 16S rRNA
data, which is included with the MiSeq
Reporter software.
Read classifications
by taxonomic group.
Library QC
Enter: LibraryQC
Fast resequencing of a reference
genome to QC the DNA library and
generate per-sample statistics.
Per-sample
summary statistics.
MiSeq Control Software
36 Part # 15027617 Rev. A
Run Folders
Each run on the MiSeq generates a run folder on either the local drive or a network
location. You can specify the location of the run folder in the MiSeqOutput field on the
Setup Options screen. For more information, see Setup Options on page 21.
Run folders contain the data files produced during the run by Real Time Analysis
(RTA), which is integrated with MCS and performs image analysis and base calling.
The data files in the run folder are later used for secondary analysis. For more
information, see On-Instrument Analysis on page 63.
Run Folder Name
By default, the folder is named in the following format:
YYMMDD_<InstrumentNumber>_<Run Number>_<Flow Cell ID>
Example: 110114_SN106_0016_A90095ACXX
The run number increments by one each time you perform a run on a given instrument.
The flow cell ID appends to the end of the run folder name.
Available Disk Space
MiSeq System User Guide 37
Available Disk Space
The software checks available disk space before beginning a run. If there is not enough
disk space for your run, the software pauses the run and a message alerts you. Required
space for your run depends on the number of cycles in your run and the analysis
workflow for your run. The required disk space for your run and the amount of disk
space you must clear, is listed in the message.
If you get this message, you need to make disk space available to continue the run. Go
to Manage Files and click the Runs tab. For more information, see Manage Files on page
22. When sufficient disk space is available, the run automatically resumes.
The integrated instrument computer has a storage capacity of 600GB. A 2x150-cycle
run results in approximately 4.2 GB of stored data.
38 Part # 15027617 Rev. A
Performing a Run
40 Part # 15027617 Rev. A
Introduction
To perform a run on the MiSeq, you first need to prepare for the run using the following
steps described in this chapter:
}Create a sample sheet and copy it to the instrument or an accessible location.
}Prepare the reagent cartridge for use.
}Denature and dilute your sample libraries.
}Load your sample libraries onto the prepared reagent cartridge.
}Set up the run using the MCS interface.
After the run begins, the MiSeq is completely automated and no other user intervention
is required. The sequencing run can be monitored from the Sequencing screen or
monitored remotely using the Sequencing Analysis Viewer, an optional software
application that you can download from the Illumina website.
After your run is complete, perform an instrument wash.

Setting Up the Sample Sheet
MiSeq System User Guide 41
Setting Up the Sample Sheet
To set up a run on the MiSeq, you first need to create the sample sheet for your run. For
more information, see Sample Sheets on page 30.
When your sample sheet is ready, copy the sample sheet to a network location
connected to your instrument. If your instrument is not connected to a network, you can
copy the sample sheet to the instrument using a USB flash drive.
You can create the sample sheet using the sample sheet template or the Illumina
Experiment Designer, a wizard-based software application available for download from
the Illumina website. For more information, see Illumina Experiment Designer on page 30.
NOTE
Illumina recommends that you name your sample sheet with the barcode
number of the reagent cartridge that you will use for your sequencing run. The
barcode number is located on the cartridge label.
1Name your sample sheet with the reagent cartridge barcode number followed by
*.csv extension.
2Create your sample sheet off-instrument using the Illumina Experiment Designer or
the sample sheet template. Enter the following information:
aEnter investigator name, project name, experiment name, and date.
bEnter the appropriate analysis workflow (no spaces): Resequencing, Amplicon,
DenovoAssembly, SmallRNA, Metagenomics, or LibraryQC.
3Enter the number of cycles for each read. To add an Index Read, include an index
sequence for each sample in the [Data] section.
4Complete the remaining sample sheet sections specific to your analysis workflow.
Analysis
Workflow
[Manifests] [SiteReports] [Data]
Resequencing Not
applicable
Optional Required: SampleID, Sample_Name,
GenomeFolder
Optional: Index, Index 2
Amplicon Required Optional Required: SampleID, Sample_Name
Optional: Index, Index 2,
GenomeFolder
DenovoAssembly Not
applicable
Not
applicable
Required: SampleID, Sample_Name
Optional: Index, Index 2,
GenomeFolder
SmallRNA Not
applicable
Not
applicable
Required: SampleID, Sample_Name,
Contaminants, miRNA, RNA
Optional: Index, Index 2,
GenomeFolder

Performing a Run
42 Part # 15027617 Rev. A
Analysis
Workflow
[Manifests] [SiteReports] [Data]
Metagenomics Not
applicable
Not
applicable
Required: SampleID, Sample_Name
Optional: Index, Index 2
LibraryQC Not
applicable
Optional Required: SampleID, Sample_Name,
GenomeFolder
Optional: Index, Index 2
NOTE
For TruSeq Custom Amplicon sample sheets, see the TruSeq Custom Amplicon
Library Preparation Guide for the appropriate index sequences.
For Nextera DNA sample sheets, see the Nextera DNA Sample Preparation Guide
for the appropriate index sequences.
5Copy the sample sheet to the network location specified in Setup Options, or copy it
to the instrument using a USB flash drive and the Manage Files feature accessible
from the Welcome screen.
Preparing the Reagent Cartridge
MiSeq System User Guide 43
Preparing the Reagent Cartridge
1Remove thereagent cartridge from -15° to -25°C storage.
2Place the reagent cartridge in a water bath containing only enough room
temperature deionized water to submerge the base of the reagent cartridge without
reaching the clear cartridge cover.
Figure 24 Maximum Water Line
NOTE
To minimize the risk of cross-contamination, do not allow the water to reach the
clear cartridge cover.
3Allow the reagent tray to thaw in the room temperature water bath for
approximately60minutes or until completely thawed.
4Remove the cartridge from the water bath and gently tap it on the bench to dislodge
water from the base of the cartridge.
5Invert the reagent cartridge ten times to mix the thawed reagents.
6Visually inspect the reagent marked IMF to make sure that it is fully mixed and free
of precipitates.
7Gently tap the cartridge on the bench to dislodge water from the base of the
cartridge and reduce air bubbles in the reagents.
8Place the reagent cartridge on ice until you are ready to load your sample and set
up your run.
WARNING
This set of reagents contains formamide, an aliphatic amide that is a probable
reproductive toxin. Personal injury can occur through inhalation, ingestion, skin
contact, and eye contact.
Dispose of containers and any unused contents in accordance with the
governmental safety standards for your region.
For more information, see the MSDS for this kit, at http://www.illumina.com/msds.

Performing a Run
44 Part # 15027617 Rev. A
Preparing Your Libraries
NOTE
Does Not Apply to TruSeq Custom Amplicon Sequencing—If you prepared your
libraries with the TruSeq Custom Amplicon protocol, do not perform this step.
The TruSeq Custom Amplicon protocol results in a ready-to-use normalized
concentration of pooled libraries for the MiSeq reagent cartridge.
For all types of runs except TruSeq Custom Amplicon sequencing, you must denature
and dilute your sample DNA for cluster generation and sequencing.
Illumina-Supplied Consumables
}HT1 (Hybridization Buffer), pre-chilled
User-Supplied Consumables
}0.1N NaOH
}Tris-Cl 10mM, pH8.5 with 0.1% Tween 20
Denature DNA
NOTE
If your application requires higher than a 20 pM final concentration of your
library, please ensure your concentration of NaOH is not higher than 0.05 N in
the denaturation solution and not more than 0.001 N (1 mM) in the final solution
after diluting with HT1.
Higher concentrations of NaOH in the loaded library will inhibit library
hybridization and decrease cluster density.
Use the following instructions to denature the DNA with 0.1N NaOH to a DNA
concentration of 20pM.
1Combine the following volumes of sample DNA and0.1N NaOH in a
microcentrifuge tube:
•2nM sample DNA (10µl)
•0.1N NaOH (10µl)
2Vortex briefly to mix the sample solution.
3Centrifuge the sample solution to 280xg for one minute.
4Incubate for fiveminutes at room temperature to denature the DNA into single
strands.
5Add the following volume of pre-chilled HT1 to the tube containing denatured
DNA to result in a 20 pM denatured library:
•Denatured DNA (20 µl)
•Pre-chilled HT1 (980µl)
6Place the denatured DNA on ice until you are ready to proceed to final dilution.
Dilute Denatured DNA
Use the following instructions to dilute the denatured DNA with pre-chilled HT1 to a
total volume of 1,000µl.
Preparing Your Libraries
MiSeq System User Guide 45
1Dilute the denatured DNA to the desired concentration using the following
example:
Final Concentration 6pM 8pM 10pM 12pM 15pM
20 pM denatured DNA 300µl 400µl 500µl 600µl 750µl
Pre-chilled HT1 700µl 600µl 500µl 400µl 250µl
2Invert several times to mix the DNA solution.
3Pulse centrifuge the DNA solution.
4Place the denatured and diluted DNA on ice until you are ready to load your
samples onto the MiSeq reagent cartridge.
Denature and Dilute PhiX Control
Use the following instructions to denature and dilute the 10nM PhiX library.
1Combine the following volumes to dilute the PhiX library to 2nM:
•10nM PhiX library (2µl)
•10 mM Tris-Cl, pH 8.5 with 0.1% Tween 20 (8 µl)
2Combine the following volumes of 2nM PhiX library and0.1N NaOH in a
microcentrifuge tube to result in a 1nM PhiX library:
•2nM PhiX library (10µl)
•0.1N NaOH (10µl)
3Vortex briefly to mix the 1nM PhiX library solution.
4Centrifuge the template solution to 280xg for one minute.
5Incubate for fiveminutes at room temperature to denature the PhiX library into
single strands.
6Add the following volume of pre-chilled HT1 to the tube containing denatured PhiX
library to result in a 20pM PhiX library.
•Denatured PhiX library (20µl)
•Pre-chilled HT1 (980µl)
NOTE
The denatured 20pM PhiX library can be stored up to three weeks at -15° to -
25°C. After three weeks, cluster numbers tend to decrease.
7Dilute the denatured 20pM PhiX library to 8 pM as follows:
•20 pM denatured PhiX library (400µl)
•Pre-chilled HT1 (600µl)
Mix Sample Library and PhiX Control
1Combine the following volumes of denatured PhiX control library and your
denatured sample library to result in a 1% volume ratio:
•8 pM PhiX control library (10 µl)
•Denatured sample library (990 µl)
Performing a Run
46 Part # 15027617 Rev. A
2Set the combined sample library and PhiX control aside on ice until you are ready
to load it onto the MiSeq reagent cartridge.
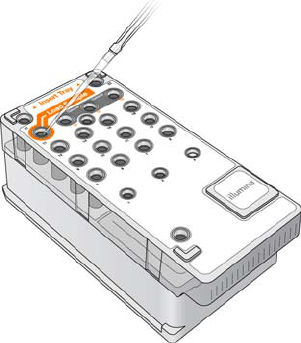
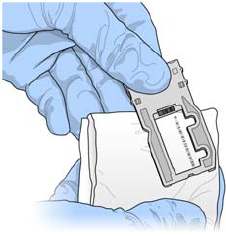
Load Sample Libraries onto Cartridge
When the reagent cartridge is prepared for use on the MiSeq, you are ready to load your
samples. You can load a single library or a pool of indexed libraries.
1Pierce the foil over the reservoir on the reagent cartridge labeled Load Samples.
2Pipette 600 µl of your sample libraries into the Load Samples reservoir.
Figure 25 Load Libraries
3Proceed directly to the run setup steps using the MiSeq Control Software (MCS)
interface.
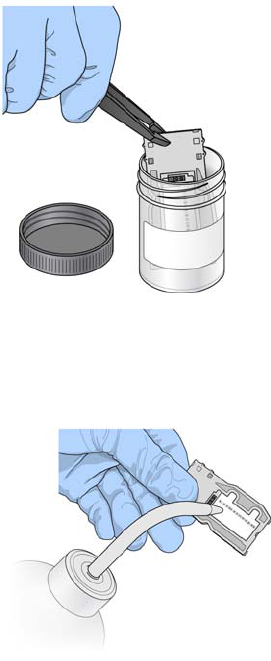
Clean the Flow Cell
MiSeq System User Guide 47
Clean the Flow Cell
The flow cell is provided immersed in storage buffer in a flow cell container. Before
loading the flow cell on the MiSeq, you need to rinse the flow cell and thoroughly dry it,
and then clean the flow cell with an alcohol wipe.
1Using plastic forceps, grip the flow cell by the base of the plastic cartridge and
remove it from the flow cell container.
Figure 26 Remove Flow Cell
2Lightly rinse the flow cell assembly with approximately 2 ml of laboratory-grade
water, making sure that both the glass and plastic cartridge are thoroughly rinsed of
excess salts. Excess salts can affect flow cell seating in the instrument.
Figure 27 Rinse Flow Cell
3Using care around the flow cell port gaskets, thoroughly dry the flow cell and
cartridge using a lint-free lens cleaning tissue. Gently pat dry in the area of the
gaskets and adjacent glass.
4Using an alcohol wipe, clean the flow cell glass, making sure that the glass is free of
streaks and fingerprints. Avoid using the alcohol wipe on the flow cell port gaskets.
Performing a Run
48 Part # 15027617 Rev. A
Figure 28 Dry Flow Cell
5Dry any excess alcohol with a lint-free lens cleaning tissue, and visually inspect to
make sure that no tissue particles are obstructing the flow cell ports and the gaskets
were not at all dislodged from the flow cell ports.
If the gaskets appear to be dislodged at all, gently press them back into place until
they sit squarely around the flow cell ports.

Loading the Flow Cell
MiSeq System User Guide 49
Loading the Flow Cell
The MCS interface will guide you through the run setup steps, starting from the
Welcome screen.
1From the Welcome screen, select Sequence. The Cloud Option screen opens.
2Set your preference for using the Cloud Option and select Next. The Load Flow Cell
screen opens.
3Put on a new pair of powder-free latex gloves.
4Raise the flow cell compartment door.
5Press the silver release button to the right of the flow cell latch. The flow cell latch
raises to open.
Figure 29 Open Flow Cell Latch
6Visually inspect the flow cell stage to make sure it is free of lint.
If lint or other debris is present, clean the flow cell stage using an alcohol wipe or a
lint-free tissue moistened with ethanol or isopropanol. Carefully wipe the surface of
the flow cell stage until it is clean and dry.
7Hold the flow cell by the edges of the flow cell cartridge near the Illumina label.
Make sure the label is facing upward.
8Place the flow cell on the flow cell stage.
Figure 30 Place Flow Cell on Stage
9Gently press down on the flow cell latch to close it over the flow cell. You will hear
a click when the flow cell latch is secure.
NOTE
As you close the flow cell latch, two alignment pins near the hinge of the flow cell

Performing a Run
50 Part # 15027617 Rev. A
latch align and position the flow cell into place.
Figure 31 Close Flow Cell Latch
NOTE
The MiSeq System reads the RFID of the flow cell. If the system is unable to
register the RFID of the flow cell, you can manually enter the barcode ID printed
on the flow cell using the online keyboard. Select the keyboard icon in the lower
right corner of the screen, enter the barcode ID, and select Done to return to the
Load Flow Cell screen.
10 Close the flow cell compartment door.
11 Select Next on the Load Flow Cell screen. The Load Reagents screen opens.
Load Reagents
MiSeq System User Guide 51
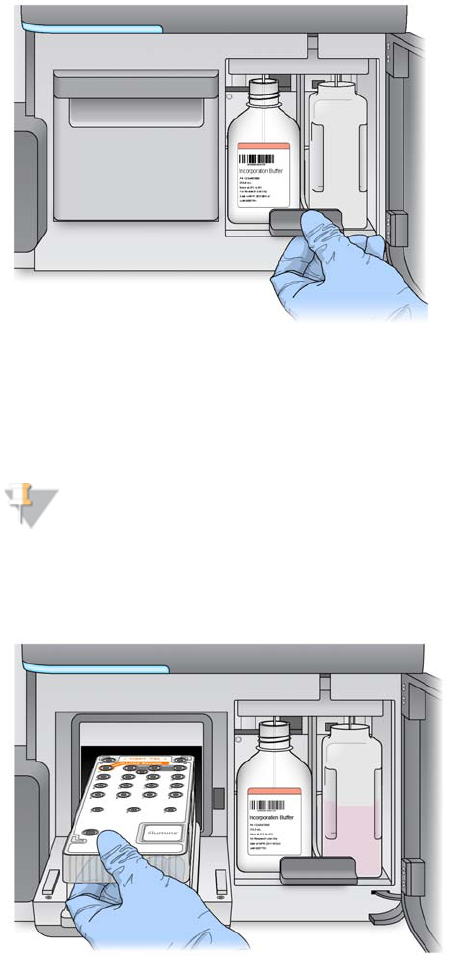
Load Reagents
First, load the PR2 bottle and make sure that the waste bottle is empty, and then load
the reagent cartridge. Check for a message on the lower-right corner of the screen that
confirms the RFID was successfully read for the PR2 bottle and the reagent cartridge.
NOTE
The MiSeq System reads the RFID of the PR2 bottle and reagent cartridge. If the
system is unable to register the RFID, you can manually enter the barcode
number from the label using the online keyboard. Select the keyboard icon in the
lower-right corner of the screen, enter the barcode number, and select Done to
return to the Load Reagents screen.
Load PR2 and Check the Waste Bottle
1Remove the bottle of PR2 from 2° to 8°C storage and remove the lid.
2Open the reagent compartment door.
3Raise the sipper handle until it locks into place.
4Place the PR2 bottle in the indentation to the right of the reagent chiller.
Figure 32 Load the PR2 Bottle
5Confirm that the PR2 RFID was successfully read.
6If necessary, empty the waste bottle into the appropriate waste container.
WARNING
This set of reagents contains formamide, an aliphatic amide that is a probable
reproductive toxin. Personal injury can occur through inhalation, ingestion, skin
contact, and eye contact.
Dispose of containers and any unused contents in accordance with the
governmental safety standards for your region.
For more information, see the MSDS for this kit, at http://www.illumina.com/msds.
7Slowly lower the sipper handle. Make sure that the sippers lower into the PR2 and
waste bottles.
Performing a Run
52 Part # 15027617 Rev. A
Figure 33 Lower Sipper Handle
8Select Next on the Load Reagents screen.
Load the Reagent Cartridge
1Open the reagent chiller door.
NOTE
Do not leave the reagent chiller door open for extended periods of time.
2Hold the reagent cartridge on the end with the Illumina label, and slide the reagent
cartridge into the reagent chiller until the cartridge stops.
Figure 34 Load Reagent Cartridge
3Close the reagent chiller door.
4Confirm that the reagent cartridge RFID was successfully read.
5Close the reagent compartment door.
6Select Next on the Load Reagents screen. The Review screen opens.

Starting the Run
MiSeq System User Guide 53
Starting the Run
After you have loaded the flow cell and reagents, the MCS interface prompts you to
review run parameters and perform a pre-run check before beginning the run.
Review Run Parameters
NOTE
The sample sheet name must match the reagent barcode number. A message
appears if the sample sheet name and reagent barcode number are not identical,
and prompts you to select a sample sheet.
1Review Experiment Name, Analysis Workflow, and Read Length. These parameters
are specified in the sample sheet.
2Review the folder locations in the lower-left corner.
If you need to change any folder locations, select Change Folders. When you have
completed the changes, select Save and then Next.
3Select Next. The Pre-Run Check screen opens.
Review Pre-Run Check
The system performs a check of all run components, disk space, and network
connections before you start the run.
If any items do not pass the pre-run check, a message appears on the screen with
instructions to correct the error. For more information, see Software Errors on page 71.
When all items successfully pass the pre-run check, you are ready to start your run.
Important Note Before Starting the Run
WARNING
The MiSeq is sensitive to vibration. Touching the instrument after starting
your run will adversely impact sequencing results.
After you select Start Run, do not open the flow cell compartment or reagent
compartment doors, or touch the instrument monitor unless you plan to stop
the run.
Wait until the run is complete before touching the monitor or opening
compartment doors.
Monitoring the Run
When the run begins, the RTA window temporarily pops up in front of the Sequencing
screen and then auto-minimizes. This is normal.
Performing a Run
54 Part # 15027617 Rev. A
Figure 35 Sequencing Screen
During your run, use the Sequencing screen to monitor run progress, intensities, quality
scores, fluidics, and imaging.
For more information, see Sequencing Screen on page 27.
To monitor your run in greater detail, use the Sequencing Analysis Viewer. For more
information, see Sequencing Analysis Viewer on page 28 and the Sequencing Analysis User
Guide.
Post-Run Procedures
56 Part # 15027617 Rev. A
Introduction
Perform an instrument wash following the completion of a sequencing run. It is
important to regularly wash the fluidics lines to ensure continued performance.

Performing a Post-Run Wash
MiSeq System User Guide 57
Performing a Post-Run Wash
Always perform a post-run wash after completing a sequencing run. If you did not
perform a wash after the last run, a reminder appears on the MiSeq software interface.
The post-run wash takes approximately 20 minutes.
Illumina-Supplied Accessories
}Wash tray
}Wash bottle, 500 ml
User-Supplied Consumables
}Laboratory-grade water
Procedure
NOTE
Leave the used flow cell on the instrument until the post-run wash is complete.
1When your run is complete, select Start Wash.
2Open the reagent compartment door.
3Open the reagent chiller door and remove the used reagent cartridge from the
chiller. The sippers were automatically raised at the end of the run. The reagent
cartridge should slide easily from the chiller.
4Prepare the wash tray by adding approximately 6ml of laboratory-grade water to
each reservoir.
5Slide the wash tray into the reagent chiller until it stops. Close the reagent chiller
door.
6Raise the sipper handle in front of the PR2 bottle and waste bottle until it locks into
place.
7Prepare the 500ml wash bottle by adding at least 350ml of laboratory-grade water.
8Remove the PR2 bottle from the reagent compartment and replace it with the wash
bottle containing laboratory-grade water.
9Remove the waste bottle and discard the contents appropriately. Return the waste
bottle to the reagent compartment.
WARNING
This set of reagents contains formamide, an aliphatic amide that is a probable
reproductive toxin. Personal injury can occur through inhalation, ingestion, skin
contact, and eye contact.
Dispose of containers and any unused contents in accordance with the
governmental safety standards for your region.
For more information, see the MSDS for this kit, at http://www.illumina.com/msds.
10 Slowly lower the sipper handle, making sure that the sippers lower into the wash
bottle and waste bottle.
11 Close the reagent compartment door.
12 From the Post-Run Wash screen, select Next. The post-run wash begins.
When the post-run wash is complete, a message appears on the screen.
Post-Run Procedures
58 Part # 15027617 Rev. A
13 Select Done. The Welcome screen opens. You are ready to start another sequencing
run.

Unloading Components
MiSeq System User Guide 59
Unloading Components
When the post-run wash is complete you are ready to unload components from the
instrument.
Remove Wash Components
1Open the reagent compartment door and raise the sipper handle to lift the sippers
from the 500ml wash bottle and waste bottle. You should feel the sipper handle
lock into place.
2Remove the wash bottle. Wash the wash bottle with deionized water and
thoroughly dry it so it is ready for a subsequent wash. Store it upside down to
protect from dust.
3Remove the waste bottle and discard the contents appropriately and according to
site standards.
WARNING
This set of reagents contains formamide, an aliphatic amide that is a probable
reproductive toxin. Personal injury can occur through inhalation, ingestion, skin
contact, and eye contact.
Dispose of containers and any unused contents in accordance with the
governmental safety standards for your region.
For more information, see the MSDS for this kit, at http://www.illumina.com/msds.
4Using an alcohol wipe or a lint-free tissue moistened with ethanol or isopropanol,
clean the floor of the reagent compartment of any visible spills or condensation.
5Return the empty waste bottle to the reagent compartment.
6Open the reagent chiller door.
7Remove the wash tray. Wash the wash tray with deionized water and thoroughly
dry it so it is ready for a subsequent wash. Store it upside down to protect from
dust.
8Close the reagent chiller door and the reagent compartment door.
Remove the Flow Cell
1Open the flow cell compartment door.
2Press the release button to the right of the flow cell latch. The flow cell latch raises.
Figure 36 Flow Cell Stage
Post-Run Procedures
60 Part # 15027617 Rev. A
3Hold the used flow cell by the edges of the flow cell cartridge near the Illumina
label, and remove it from the flow cell stage.
4Clean the flow cell stage using an alcohol wipe or a lint-free tissue moistened with
ethanol or isopropanol. Carefully wipe the surface of the flow cell holder until it is
completely clean.
5Gently press down on the flow cell latch to close it. You will hear a click when the
flow cell latch is secure.
6Close the flow cell compartment door.

Shutting Down the Instrument
MiSeq System User Guide 61
Shutting Down the Instrument
Turn off the instrument only if it will not be used for more than ten days. If the
instrument will be used within the next ten days, leave the instrument on.
NOTE
Any time that you turn off the instrument, wait a minimum of 60seconds before
turning the power switch back to the ON position.
Use the following procedure to safely prepare fluidics before shutting down the system.
1Perform a maintenance wash to thoroughly wash the system.
2Leave the used wash tray in the reagent chiller to collect any fluid that may fall
back through the lines.
3Empty the waste bottle and return it to the reagent compartment.
4Place an empty wash bottle in the reagent compartment, lower the sipper handle,
and close the reagent compartment door.
5Remove the flow cell from the flow cell stage.
6Using an alcohol wipe or a lint-free tissue moistened with ethanol or isopropanol,
carefully wipe the surface of the flow cell holder until it is completely clean and dry.
7Press down on the flow cell latch until it locks into place.
8Close the flow cell compartment door.
9Toggle the power switch to turn off the instrument.
10 When you are ready to restart the instrument, load a used flow cell and perform an
instrument wash prior to your next sequencing run.
62 Part # 15027617 Rev. A
On-Instrument Analysis
64 Part # 15027617 Rev. A
Introduction
The MiSeq Control Software (MCS) performs image analysis and base calling using
integrated Real Time Analysis (RTA). Analysis is performed during the sequencing run,
which saves downstream analysis time. RTA runs automatically on the instrument.
The primary analysis output from a sequencing run is a set of quality-scored base call
files (*.bcl files), which are generated from the raw image files and contain base calls per
cycle. By default, images are deleted from the instrument computer after image analysis.
The raw image data are not needed for secondary analysis.
MiSeq Reporter
The MiSeq is the first instrument to provide on-instrument secondary analysis using the
MiSeq Reporter software. MiSeq Reporter is a Windows-based application that
processes base calls generated by RTA. The results include information about
alignment, structural variants, and contig assemblies for each genome requested, and
each sample if your run is a multi-sample run or an indexed run.
Primary Analysis Results
MiSeq System User Guide 65
Primary Analysis Results
The following table describes the folders and files generated by Real Time Analysis
(RTA) during primary analysis. Many of these files are used for secondary analysis by
the MiSeq Reporter software.
Key File Subfolder Description
RTAComplete.txt AnalysisFolder\ A marker file generated when base call
analysis is complete. The presence of this
file triggers the start of secondary analysis.
SampleSheet.csv AnalysisFolder\ This file is read and copied to the run folder
before the run, and later used for secondary
analysis.
RunInfo.xml AnalysisFolder\ Identifies the boundaries of the reads
(including index reads) and the quality table
selected for run.
*.bcl files AnalysisFolder\Data\
Intensities\BaseCalls\
L001\CX.X
Each *.bcl file contains RTA base calling and
base quality scoring results for one cycle,
one tile.
*.stats files AnalysisFolder\Data\
Intensities\BaseCalls\
L001\CX.X
*.stats files contain RTA base calling
statistics for a given cycle/tile.
*.filter files AnalysisFolder\Data\
Intensities\BaseCalls
*.filter files contain filter results per tile.
*.txt AnalysisFolder\Data\RTALogs Log files from primary analysis.
*.cif files AnalysisFolder\Data\
Intensities\L001\CX.X
Each binary *.cif file contains RTA image
analysis results for one cycle, one tile. These
files can be used in the OLB analysis
software. For more information, see the
Off-line Basecaller Software User Guide.
*.locs files AnalysisFolder\Data\
Intensities\BaseCalls\
L001
Reports the cluster coordinates. There is
one *.locs file for each tile..
*.jpg files AnalysisFolder\
Thumbnail_Images\
L001\CX.X
Thumbnail images generated for each cycle
and base, and can be used to troubleshoot a
run.
Table 2 Contents of Analysis Folder
On-Instrument Analysis
66 Part # 15027617 Rev. A
Secondary Analysis Using MiSeq Reporter
MiSeq Reporter runs on the instrument computer and uses the analysis workflow
specified in the sample sheet to perform secondary analysis. For more information, see
Analysis Workflow on page 35.
Even though MiSeq Reporter is running on the instrument, you view the analysis in
progress from any web browser on another computer on the same network as your
MiSeq.
Viewing MiSeq Reporter
MiSeq Reporter launches automatically after RTA completes primary analysis. To view
the MiSeq Reporter interface during analysis, use any web browser on the same network
as your instrument and connect to the MiSeq HTTP service on Port 8042.
}Connect using the instrument IP address:
IP Address HTTP Service Port HTTP Address
10.10.10.10, for example 8042 http://10.10.10.10:8042
}Connect using the your network name:
Network Name HTTP Service Port HTTP Address
YourNetwork, for example 8042 http://yournetwork:8042
}If MiSeq Reporter is installed locally on an off-instrument computer, connect using
"localhost":
Off-Instrument HTTP Service Port HTTP Address
localhost 8042 http://localhost:8042
When you are linked to your MiSeq, the MiSeq Reporter interface opens.
Figure 37 MiSeq Reporter Interface
Secondary Analysis Input Requirements
MiSeq Reporter requires the following primary analysis files to perform secondary
analysis. There is no need to move or copy these files to another location before analysis
begins. The software automatically accesses these files in the run folder.

Secondary Analysis Using MiSeq Reporter
MiSeq System User Guide 67
File Type Description
RTAComplete.txt Marker file that indicates that RTA processing is complete.
SampleSheet.csv Provided at beginning of sequencing run and contains analysis
workflow and important run information.
RunInfo.xml Identifies high-level run information.
*.bcl files Files contain specific run and base calling information.
*.filter files Files contain filter results for each tile.
*.locs files Files contain the locations of all clusters on a tile.
Pre-Installed Databases and Genomes
The MiSeq includes several pre-installed databases and genomes.
Databases
}miRbase for human
}dbSNP for human
}refGene for human
Genomes
}hg19 (human)
}dh10b (E. coli)
}S. aureus
}yeast
}A. thaliana
}mouse
You can upload and use your own reference in FASTA format. Reference FASTA files
should be stored in a single folder and have a *.fa extension.
Use the Manage Files feature in MCS to upload files. For more information, see Manage
Files on page 22.
Sequencing During Analysis
If a new sequencing run is started on the MiSeq before secondary analysis is complete,
the current secondary analysis is stopped. From the MiSeq Reporter interface, you can
requeue secondary analysis for that run after the new sequencing run is complete. At
that point, secondary analysis starts over from the beginning.
For information about using MiSeq Reporter, see MiSeq Reporter Online Help available
from the MiSeq Reporter interface.
On-Instrument Analysis
68 Part # 15027617 Rev. A
Using MiSeq Reporter Off-Instrument
You can install another copy of the MiSeq Reporter software on a separate computer.
Doing so enables you to perform secondary analysis of run data while the MiSeq is
performing a subsequent sequencing run.
Computing Requirements
To run the MiSeq Reporter software, you will need the following computing
requirements:
}64-bit Windows OS (Vista, Windows 7, Windows Server 2008 64-bit)
}>8 GB RAM
}>2.2 GHz
}>1 TB disk space
Installation and Licensing
}You can download a second copy of the MiSeq Reporter software from the Illumina
website.
}After downloading, open the software installer (setup.exe), and accept the end-user
licensing agreement (EULA).
}No license key is required as this additional copy is free of charge with your MiSeq.
Troubleshooting
70 Part # 15027617 Rev. A
Introduction
If an error occurs during the run setup steps, an on-screen message appears with
instructions for correcting the error. Other errors are indicated by the status icon in the
upper-right corner of the screen. For more information, see Status Icons on page 20.
For more information, visit http://www.illumina.com/support.ilmn or contact Illumina
Technical Support.

Software Errors
MiSeq System User Guide 71
Software Errors
This section provides an overview of possible errors using MCS. For most errors, the
action you need to take appears on the screen and the run is not impacted.
Occurs During Error Action
Upon starting the
software.
Initialization failed. The option to re-initialize appears on the
screen. Select Re-initialize.
Pre-Run Check Sample sheet not found. Check the folder location for the sample
sheet in Setup Options on the Welcome
screen.
If the sample sheet is missing, create one
and copy it to the sample sheet locations
specified in Setup Options.
When you finished, select Restart Check.
Pre-Run Check Improperly formatted
sample sheet.
You might have missing information or
syntax errors in your sample sheet. Correct
the errors listed on the screen, and then
select Restart Check.
Pre-Run Check Fluidics check failed. Select Pump. When fluids have been
pumped, select Restart Check.
Pre-Run Check Disk space low. If disk space is low, a message appears
indicating how much disk space is required,
along with the Manage Runs button. Select
Manage Runs and clear the required space
from the instrument computer.
Pre-Run Check Empty waste bottle. Open the reagent compartment door, raise
the sipper handle, and empty the waste
bottle in an appropriate waste container.
Pre-Run Check Network disconnected. Make sure the network cable is plugged into
the instrument.
If the network connection is not resolved,
reboot the instrument by turning off the
power switch. Wait at least 60 seconds, and
then turn the instrument on using the
power switch.
At end of run Data files not copied. Two options appear on the screen: Copy
Data and Delete Data
•Copy Data—Allows you to select an
output folder and copy files.
•Delete Data—Removes temporary files
from the instrument computer.
72 Part # 15027617 Rev. A
Index
MiSeq System User Guide 73
Index
A
activity indicators 19
amplicon sequencing 2, 13
analysis
databases, genomes 67
during sequencing 67
input requirements 66
analysis workflow 30, 35
applications 13
C
Cloud 24
cluster generation 11
components
flow cell 7
flow cell compartment 4, 7
optics module 4
reagent cartridge 5
reagent compartment 4, 51, 59
consumables
Illumina supplied 13
user-supplied 15
consumables, Illumina supplied 13
copying files 20
customer support 75
cycle time 2
D
deleting files 20
diagnostic 20
disk space
checking 37
low 71
documentation 75
E
Experiment Designer 41
F
flow cell
cleaning 47
loading 49
overview 7
single-lane 28
temperature 27
flow cell clamp 7
flow cell compartment 4, 7
fluidics 27
folder locations 21
H
help, technical 75
I
iCloud 24
icons
errors and warnings 20
status alert 20
initialization 71
input requirements 66
instrument setup 20
instrument wash 56
intensities 27
L
Live Help 20
load flow cell 24
load reagents 24
loading reagents 51, 59
M
maintenance wash 56
manifests 21, 41
MiSeq Control Software 19
MiSeq Reporter 66
additional copy of 68
monitoring the run 27, 53
N
network connection 71
O
optics module 4
P
pausing a run 29
post-run wash 57
power switch 9
PR2 51
pre-run check 24
preparing DNA 44
Q
Q-score 27
R
read length 12-13
Index
74 Part # 15027617 Rev. A
reagent cartridge 5, 51
reagent compartment 4, 59
reagents
kitted 13
loading 51, 59
sequencing 51, 59
real time analysis 2, 36, 64
RFID 2, 25
RTA 64
results 65
RTAcomplete.txt 65
run duration 12
run folder 36
run parameters 41
sample sheet 30
run setup screens 24
RunInfo.xml 65
S
sample sheet 30, 41, 65, 71
changing 25
secondary analysis 66
sequencing 11
Sequencing Analysis Viewer 28, 53
sequencing screen 27, 53
setup options 21
sipper handle 4
site reports 41
software
disk space checking 37
initialization 9
instrument setup 20
MiSeq Control 19
run duration 12
sample sheet 30, 41
software updates 20
status alert icon 20
status.xml 65
stopping a run 29
T
technical assistance 75
trurning on the instrument 9
turning off instrument 61
U
user-supplied consumables 15
V
vibration 27
W
washes 21
consumables 57
maintenance wash 56
post-run wash 57
recommendations 56
water wash 56
waste bottle 4
workflow 11
analysis 35
run duration 12

Technical Assistance
MiSeq System User Guide 75
Technical Assistance
For technical assistance, contact Illumina Customer Support.
Illumina Website http://www.illumina.com
Email techsupport@illumina.com
Table 3 Illumina General Contact Information
Region Contact Number Region Contact Number
North America 1.800.809.4566 Italy 800.874909
Austria 0800.296575 Netherlands 0800.0223859
Belgium 0800.81102 Norway 800.16836
Denmark 80882346 Spain 900.812168
Finland 0800.918363 Sweden 020790181
France 0800.911850 Switzerland 0800.563118
Germany 0800.180.8994 United Kingdom 0800.917.0041
Ireland 1.800.812949 Other countries +44.1799.534000
Table 4 Illumina Customer Support Telephone Numbers
MSDSs
Material safety data sheets (MSDSs) are available on the Illumina website at
http://www.illumina.com/msds.
Product Documentation
If you require additional product documentation, you can obtain PDFs from the
Illumina website if PDFs are available. Go to
http://www.illumina.com/support/documentation.ilmn. When you click on a link, you
will be asked to log in to iCom. After you log in, you can view or save the PDF. To
register for an iCom account, please visit https://icom.illumina.com/Account/Register.
Illumina, Inc.
9885 Towne Centre Drive
San Diego, CA 92121-1975
+1.800.809.ILMN (4566)
+1.858.202.4566 (outside North America)
techsupport@illumina.com
www.illumina.com






